
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 15 February 2024
Sec. Marine Biology
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1332011
The taxon Prolecithophora, which is closely related to the well-known clade Tricladida, is rarely studied, particularly in terms of molecular phylogeny and behavioral characteristics. In this study, we employed an integrative analysis of molecular phylogeny, histology, and ethology to describe a new marine species, Plagiostomum robusta A.Wang, sp. nov. of the order Prolecithophora. Additionally, we obtained its nearly complete mitogenome sequence with annotations of 12 protein-coding genes, two rRNA genes and 22 tRNA genes. P. robusta is characterized by several features, including a pair of short tentacles, a terminal mouth and a terminal gonopore, a large variable pharynx, two extended front ends of the intestine, paired testes located behind the ovaries and outside the vitellaria, a muscular and movable distal sac, and a long tubular penis surrounded by a sheath. The phylogenetic analysis, based on 18S rDNA and 28S rDNA, revealed that the new species formed a clade with six Plagiostomum flatworms, indicating a close kinship with Plagiostomum. Despite contradictions between the traditional morphological classification system and the results of molecular phylogenetics, we chose to assign the new species to Plagiostomum based on comprehensive considerations. Moreover, our study has provided more insights into the behavioral features of Plagiostomidae species, as we found that P. robusta fed on other flatworms and engaged in cannibalism, and unexpectedly it mated via hypodermic inpregnation. This study represents the first comprehensive description of the reproductive behavior of Plagiostomidae species, contributing to the more in-depth understanding of the biological characteristics of this group.
Prolecithophora Karling, 1940, belonging to Adiaphanida, forms a clade together with the parasitic Fecampiida and the well-studied order Tricladida. Despite the increasing use of integrative taxonomy in studying some widely known clades such as Tricladida, the order Prolecithophora has received little attention beyond morphological analysis. Flatworms of the Prolecithophora are generally small species that are mostly distributed in marine habitats, living in soft sediments or on algae. Their body is spindle-shaped or elongated, cylindrical in the middle, and lacks cement glands in the anterior part. Most species have two pairs of eyes and a ciliated groove in the anterior body, with an encapsulated brain (Jondelius et al., 2001; Caira and Littlewood, 2013). The pharynx can be simple, plicate, or variable, and they possess a simple, elongated sac-like intestine. The oral and genital openings may be located together or separately in the anterior and posterior body portions (Karling, 1940; Grosbusch et al., 2021). Their testes and ovaries can be compact or diffuse, single or paired. Most species have diffuse vitellaria and the eggs are ectolecithal (Caira and Littlewood, 2013). Currently, Prolecithophora consists of 5 families, primarily Plagiostomidae, which has the largest number of recorded species within the order (World Register of Marine Species, 2023). Plagiostomidae species have an oblong body, often lacking a brain capsule and a ciliated groove, and typically have one or two pairs of eyes. The pharynx is located in the anterior body, with the genital opening near the posterior end (Grosbusch et al., 2022). Plagiostomidae species are further classified into nine genera, with Plagiostomum Schmidt, 1852, and Vorticeros Schmidt, 1852 being studied for longer. These two genera share many morphological similarities, with the presence or absence of tentacles being the main distinguishing feature (Kozloff and Westervelt, 2001). However, Jondelius et al. (2001) demonstrated that such a classification system renders Plagiostomum non-monophyletic.
Most research on prolecithophorans was conducted in the 19th and 20th centuries, leaving a lack of recent advances in taxonomy. This lack of data poses a challenge for phylogenetic analysis. In molecular phylogenetic studies, mitochondrial genes and mitogenomes are commonly used as molecular markers due to their conserved gene composition, high base substitution rate, maternal inheritance, and low recombination rate (Brand et al., 2022). However, mitochondrial data of prolecithophorans are largely unknown. Additionally, while the morphology of the reproductive organs serves as an important classification basis for flatworm groups, reproductive behavior is significant for supporting the categorization, but little attention has been given to this aspect. In this study, we report the discovery of a flatworm species in the coastal region of South China, identified as a member of the Plagiostomidae based on its external form and internal structure. We also conducted a molecular phylogenetic analysis using 18S rDNA and 28S rDNA sequences, supporting its assignment to Plagiostomum. Moreover, in order to expand phylogenetic resources for the order, we generated the first, nearly complete mitochondrial genome data for Prolecithophora. Furthermore, we observed and described in detail the reproductive behavior, ontogenesis, and predation behavior of the new species. By synthesizing these findings, our aim is to discuss the evolutionary relationship within Plagiostomidae and the significance of hypodermic inpregnation in the mating behavior of the new species Plagiostomum robusta A. Wang, sp. nov.
Specimens of Plagiostomum robusta were collected from an estuary in Xichong, Shenzhen, China (22°28’12”N, 114°31’23”E) on December 7th, 2021. The water temperature and the salinity at the time of collection were 21°C and 25‰, respectively. To collect flatworms, the algae from the sampling site were rinsed with habitat water. The water samples were then transported to the laboratory of Shenzhen University and examined under a stereomicroscope (Nikon SMZ445, Japan). All of the obtained flatworms were cultured separately in glass bowls at a temperature of 23 ± 0.5°C and with artificial seawater of 25‰ salinity, which was prepared using Red Sea Salt (Red Sea, USA). Living flatworms were fed daily with the marine planarian Paucumara falcata Wang & Li, 2019.
Three specimens were starved for three days before genomic DNA extraction. The E.Z.N.A™ Mollusk DNA Isolation Kit (Omega, Norcross, GA, USA) was used for DNA extraction. Nested PCR amplification of 18S rDNA was performed using two pairs of primers - TimA/18S1100R and TimB/18S600F, following the method described by Noren and Jondelius (2002). The PCR cycles started with an initial denaturation at 95°C for 5min, followed by 35 circles of 30s at 94°C, 30s at 55°C, 50s at 72°C, and a final extension at 72°C for 5 minutes. For amplification of 28S rDNA, primers LSU5 and LSU6-3 (Littlewood et al., 2000) were used. The cycling conditions were as follows: 95°C for 5 minutes, followed by 35 cycles of 94°C for 60 seconds, 50°C for 60 seconds, and 72°C for 50 seconds. The final step was at 72°C for 5 minutes. PCR reactions were performed using KOD One™ PCR Master Mix polymerase (TOYOBO, Japan). The amplified DNA fragments were Sanger sequenced by Beijing Genomics Institute (BGI, Shenzhen, China).
In order to reconstruct phylogenetic trees for the estimation of the phylogenetic position of Plagiostomum robusta within Prolecithophora, the 18S rDNA and 28S rDNA sequences of 32 Prolecithophora species were downloaded from Genbank database (Supplementary Table 1). Two Tricladida species were used as outgroups. Multiple sequence alignment of nucleotide sequences was performed using MAFFT version 7 (Katoh and Standley, 2013) with the E-INS-i iterative refinement method, while conservative region selection was done using GBLOCKS v.0.91b (Castresana, 2000) with the “Allowed All Gap Positions” parameter. The concatenated dataset was obtained using Sequence Matrix v.1.7.8 (Vaidya et al., 2011). PartitionFinder2 (Lanfear et al., 2017) was utilized to select best-fit partition models for Bayesian inference (BI) and Maximum likelihood (ML) analyses, respectively. BI phylogeny was inferred by MrBayes v.3.2.6 (Ronquist et al., 2012), with two simultaneous runs of 2,000,000 generations. Trees were sampled every 2,000 generations with three heated chains and one cold chain to encourage swapping among the Markov Chain Monte Carlo (MCMC) chains, using a 25% burn-in. After 2,000,000 generations, the standard deviation of split frequencies was below 0.01, and the potential scale reduction factor (PSRF) was close to 1.0 for all parameters. The credibility of the result was assessed using TRACER v1.7.1 (Rambaut et al., 2018) to check if all the Estimated Sample Size values (ESS) were above 200. ML analysis with 10,000 bootstrap replicates was performed using IQ-TREE v2.1.2 (Nguyen et al., 2015). The phylogenetic trees were visualized in iTOL v6.8 (Letunic and Bork, 2021).
REPLI-g Mini Kit (QIAGEN, Germany) was used for multiple displacement amplification of the three above-mentioned genomic DNA samples. After quality testing, the samples were sent to BGI (Shenzhen, China) for library construction and whole genome sequencing on the DNBSEQ platform. Approximately 40 million clean PE150 reads were obtained for each sample. MitoFlex v0.2.9 (Li et al., 2021) was utilized to conduct de novo assembly of the mitochondrial genome, referencing the Platyhelminthes dataset. The mitogenome sequence was determined by examining the interactive mapping results in IGV 2.16.2 (Thorvaldsdóttir et al., 2013) and aligning three assembly sequences by using MEGA 11 (Tamura et al., 2021).
Gene annotation and tRNA secondary structure prediction were performed using MITOS2 (Donath et al., 2019) with the “RefSeq 89 Metazoa” option and the echinoderm and flatworm mitochondrial code. The atp8 gene was manually annotated firstly using ORFfinder, to search for open reading frames (ORFs) with a length ranging from 100 to 300 bp. Then, the hydrophobicity maps of each ORF were analyzed on ExPasy (Duvaud et al., 2021) based on the principle of “Kyte and Doolittle”, while SMART (Letunic et al., 2021) was utilized to predict the structural characteristics of the corresponding protein. Candidate ORFs were selected based on the above analysis. Annotated protein-coding genes and rRNA genes were verified through NCBI BLAST by comparing them with the published sequences. The mitogenome was visualized by Proksee v1.0.0a6 (Grant et al., 2023). Tandem Repeats Finder (Benson, 1999) was used to identify tandem repeats in the large non-coding region with default parameters. PhyloSuite v1.2.3 (Zhang et al., 2020) was employed to calculate the relative synonymous codon usage (RSCU) for protein-coding genes, excluding incomplete stop codons, and to generate a stacked bar plot. The composition skewness of each component of the genome was calculated according to the following formula: AT-skew = (A-T)/(A+T); GC-skew = (G-C)/(G+C).
The continuous tissue sections were prepared referring to the method of Yang et al. (2020). The flatworms were starved for three days and placed on glass slides. Fully stretched worms were killed by soaking them in modified Bouin’s fluid, which is a mixture of saturated aqueous picric acid, formaldehyde, and acetic acid (70/25/5, v/v/v). They were then transferred to a weighing bottle and fixed with Bouin’s fluid for 4 hours. Specimens were dehydrated with ethanol, cleared with xylene and embedded in paraffin. The Leica RM2235 manual rotary microtome (Leica Biosystems, Germany) was used to obtain histological sections with a thickness of 6 μm. After being stained with modified Cason’s Mallory-Heidenhain stain, the specimens were sealed in neutral balsam (Maclin, Shanghai, China).
For whole mount samples, the worm body was flattened with a cover glass and then transferred to a weighing bottle and soaked in Bouin’s fluid for 30 minutes. Then the specimen was washed using phosphoric acid buffer. Afterwards it was stained in Hematoxylin for 30 minutes, followed by rinsing with 1% hydrochloric acid. Subsequently, gradient ethanol dehydration and xylene clearing were performed. The specimens were then transferred to glass slides and sealed with neutral balsam.
All specimens were observed using a differential interference microscope (Olympus BX51, PA, USA). Photography and measurements were performed using a digital camera (Olympus DP72, PA, USA) and DP-2-BSW professional software (Olympus, version 2.2, PA, USA). Adobe Photoshop 2021 (Adobe, USA) was used to edit photos, adjust contrast and brightness, and draw reconstructions of specimens. Type materials were deposited in the Institute of Zoology, Chinese Academy of Sciences (IZCAS).
Sexually mature flatworms were randomly selected and placed together in a concave slide with an aperture of 2.5 cm and a depth of 0.5 cm. Mating was considered to be complete when one individual injected a sperm mass into the body of its partner, after which the copulatants separated from each other. After that, the flatworms were immediately isolated. Two pairs of flatworms were sectioned in order to explore the location and means of sperm transfer. Several fertilized flatworms were cultured separately to observe egg deposition and ontogenesis. Every day, new eggs were numbered and picked out, scraped off from the wall of the glass bowl using an insect needle, and then cultured in 12-well plates. One hundred juveniles were cultured individually on 12-well plates, while the remaining juveniles were placed in a glass bowl (90 mL) for co-culture. The mating behavior and developmental process of the flatworms were observed using a stereomicroscope (Leica EZ4, Leica Microsystems, Germany). Images were captured using Olympus DP-2-BSW and Image-Pro Plus v.6.0 software. Brightness and contrast were adjusted using Photoshop.
br. brain; cg. cement glands; ds. distal sac; e. eye; go. gonopore; in. intestine; mo. mouth; ov. ovary; p. penis; pg. prostatic glands; ph. pharynx; ps. penis sheath; sg. shell glands; sv. seminal vesicle; t. tentacle; te. testis; va. vagina; vd. vitelline duct; vi. vitellaria.
In the present study, we used 18S rDNA and 28S rDNA sequences of 32 Prolecithophora species to construct the phylogenetic tree. Dugesia japonica Ichikawa & Kawakatsu, 1964 and Dugesia umbonata Song & Wang, 2020 were used as outgroups. Some Prolecithophora species in the GenBank database do not have data for either 18S rDNA or even both genes, so that they were excluded from the phylogenetic analysis. After processing the sequences with GBLOCKS, the 18S rDNA dataset consisted of 39 sequences with a length of 1,764 bp, while the 28S rDNA dataset contained 27 sequences with a length of 1,022 bp. According to PartitionFinder2, the optimal models for both partitions were GIR+I+G, which applied to both BI and ML analyses. Since the BI and ML trees had similar topologies, we merged them into a single one.
The phylogenetic tree (Figure 1) revealed two main clades within Prolecithophora with strong support values. The first clade consisted of Plagiostomidae, which formed a highly supported monophyletic group (1.00 posterior probability - pp, 100% bootstrap - bs). This clade was separated from another highly supported monophylum that included species from Pseudostomidae, Protomonotresidae, and Sclerocaulophoridae. Within the Plagiostomidae, Acmostomum dioicum was positioned at the base, with its sister group comprising two clades that shared a high support sister-group relationship (1.00 pp, 100% bs). In Clade 1, three individuals of Plagiostomum robusta clustered together with 1.00 pp and 100% bs. They formed a relationship with six Plagiostomum species as ((P. robusta + P. striatum) + (P. cuticulata + (P. whitmani + (P. lemani + P. album)))) + Plagiostomum sp. MW-2019. The branch of P. lemani + P. album and the branch of Plagiostomum sp. MW-2019 with six other species received strong support (pp > 0.90, bs > 80%). The branch of (P. cuticulata + (P. whitmani + (P. lemani + P. album)) and the branch of (P. whitmani + (P. lemani + P. album)) had high posterior probabilities but low bootstraps. The support values for other branches were relatively low (pp < 0.90, bs < 80%). Clade 2 consisted of (a) the Vorticeros species V. auriculatum and V. ijimai, (b) the Plagiostomum species P. vittatum, P. ochroleucum, P. stellatum, Plagiostomum sp. SL-2020, and (c) the Torgea Jondelius, 1997 species T. phukettensis. All branches within this clade had high support values. Notably, Vorticeros (two species) and T. phukettensis were located within the Plagiostomum branch, which is consistent with the result of Noren (2004), suggesting that Plagiostomum is not a monophyletic group. The phylogenetic tree supports that P. robusta is a new species that belongs to the Plagiostomidae.
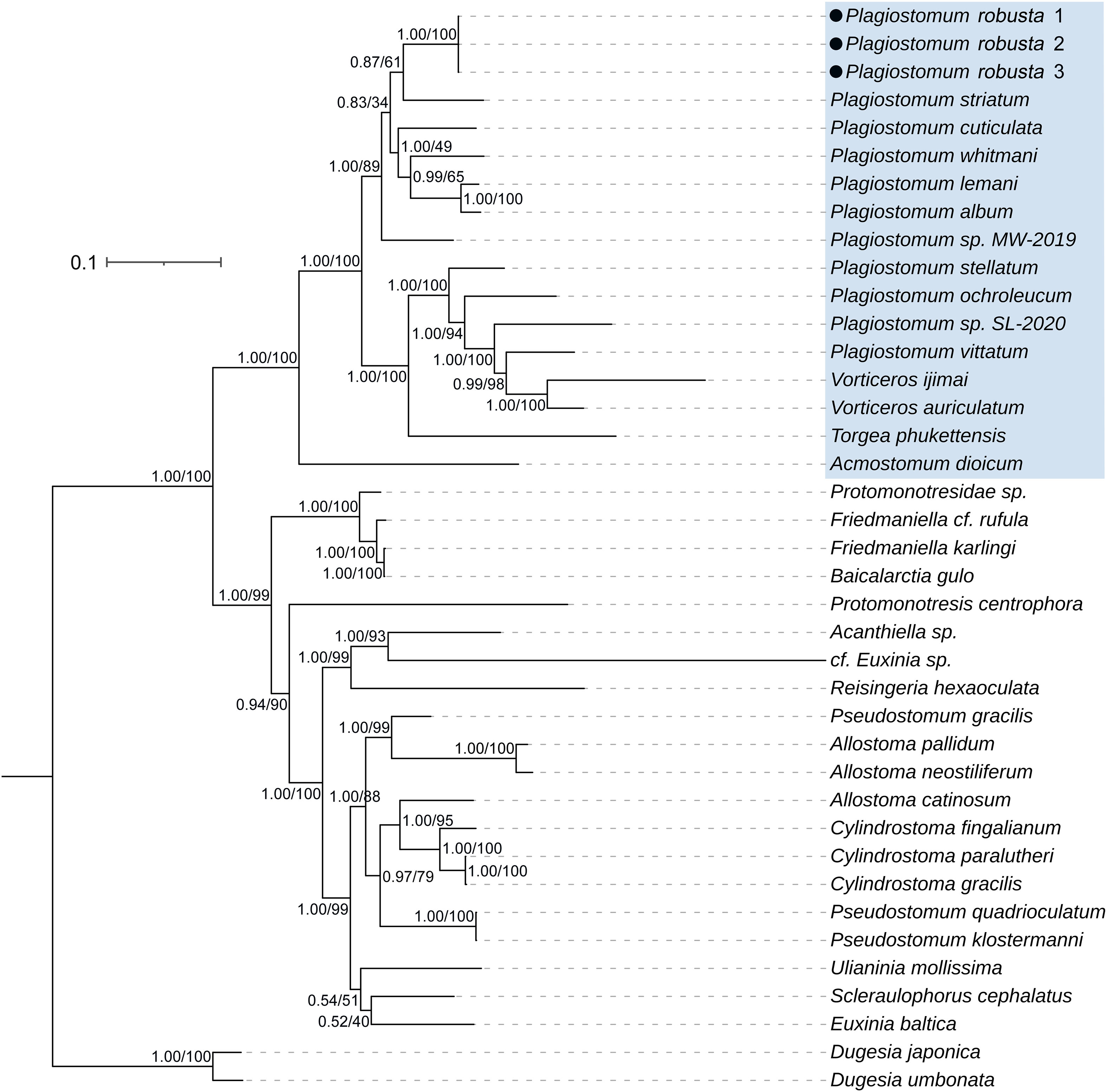
Figure 1 Phylogenetic tree inferred from a concatenated matrix of 18S and 28S rDNA (2786bp) using BI and ML methods (pp/bs), depicting the position of Plagiostomum robusta within Prolecithophora. Plagiostomidae is covered in blue.
Family Plagiostomidae Graff, 1882
Genus Plagiostomum Schmidt, 1852
Plagiostomum robusta A. Wang, sp. nov.
Material examined. Holotype, PLA-Pl001, collected by Yujia Wang on December 7th, 2021 from an estuary in Xichong, Shenzhen, China (22°28’12”N, 114°31’23”E), sagittal sections on 11 slides. Paratypes: (collection information same as the holotype), PLA-Pl002, sagittal sections on 20 slides; PLA-Pl003, sagittal sections on 8 slides; PLA-Pl004, transverse sections on 31 slides; PLA-Pl005, transverse sections on 12 slides; PLA-Pl006, unsliced whole mount on one slide; PLA-Pl007, whole mount on one slide.
Etymology. The species name is derived from the Latin adjective robustus, “strong”, thus alluding to the fact that the pharynx and mating organ of the new species are well developed and characterized by their large size.
Diagnosis. A species of Plagiostomum characterized by a pair of short tentacles on the anterior dorsal side of the body. It has a body length averaging 7.9 ± 1.45 mm, with a light yellow body surface. The species possesses a pair of eyes with three retinal cells per eyecup. The mouth opening is situated at the anterior end of the body. The pharynx is large and variable, occupying about one third of the body length. The intestine is a simple sac-like structure with two projecting front ends. The reproductive system includes a pair of ovaries located ventro-laterally, paired testes situated behind the ovaries and external to the vitellaria, a muscular and movable distal sac, a long tubular penis surrounded by a muscular sheath, and a gonopore located at the posterior end of the body. The sperm possess a short contact region and a long tail region, and the body exhibit a coiled spiral enclosed by fusiform membrane.
Description. The animals are elongated, measuring approximately 7.9 ± 1.45 mm in length (n=10), being about 10 times longer than their width. They are cylindrical throughout most of the body and taper at the posterior end. Live specimens are light yellow and translucent (Figure 2A). The color of the intestine varies, depending on the contents of the food, ranging from dark brown to yellow or gray. In larger individuals, the length can be approximately 1 cm, while the pharynx appears orange in color (Figure 3D, 60d). The epidermis is covered by short cilia. On the anterior dorsal side of the body, there are two tentacles that point anteriorly, measuring approximately 0.13 ± 0.01 mm in length (n=10) and being spaced about 0.3 ± 0.02 mm apart at the base (n=10). The tentacles have a blunt tip and are adorned with several tactile hairs on their tips (Figure 2B).
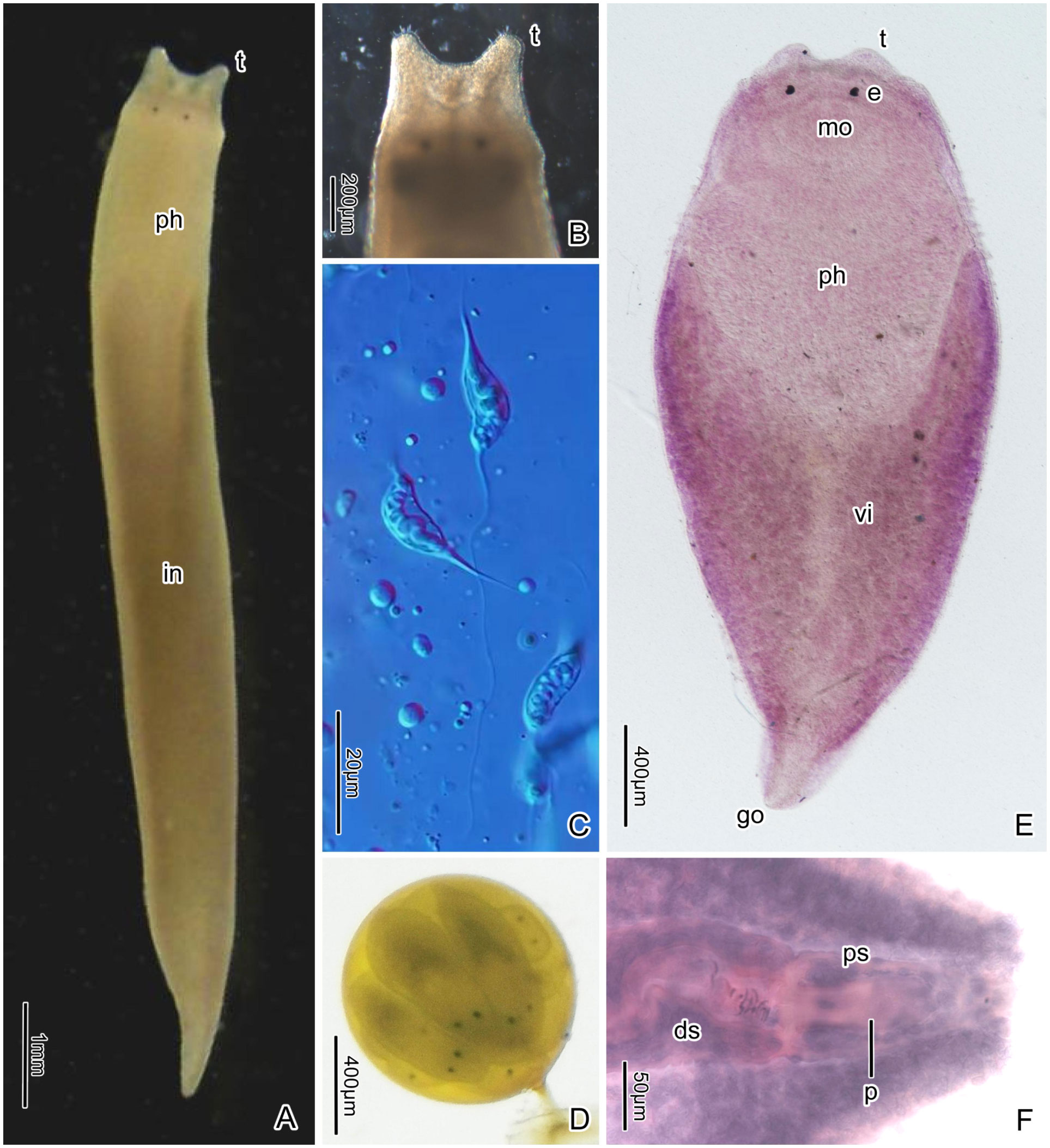
Figure 2 Plagiostomum robusta. (A) Living specimen, dorsal view. (B) Anterior end of a living specimen, showing the tentacles and tactile hairs. (C) Spermatozoa. (D) Egg containing multiple embryos. (E) PLA-Pl006, whole mounted specimen, ventral view. (F) Posterior end of the whole mounted specimen PLA-Pl007, showing the joint of the distal sac with the penis and the penis sheath.
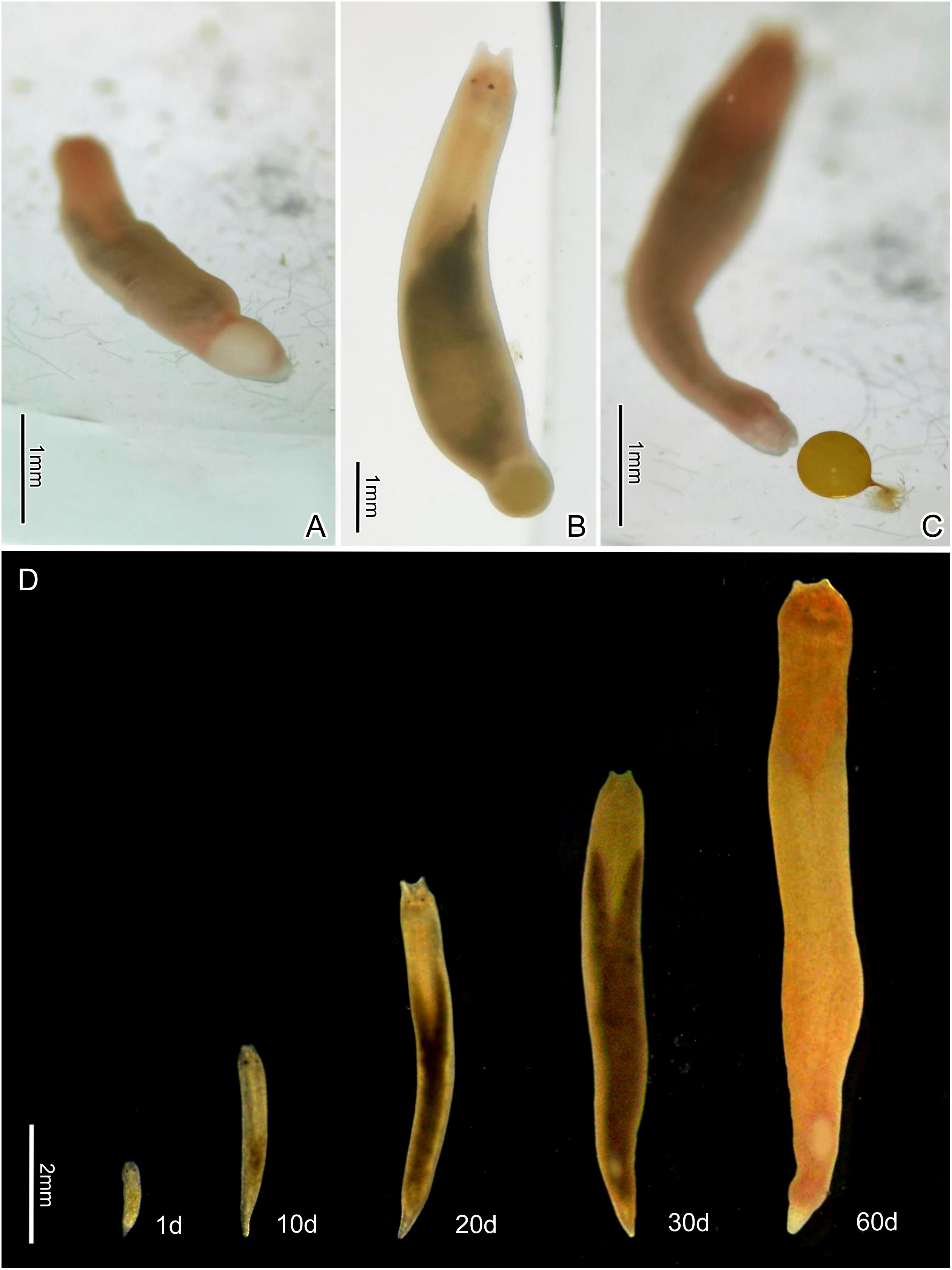
Figure 3 The egg laying process of Plagiostomum robusta, showing the egg changing color from white to orange-yellow (A, B) and being discharged through the terminal gonopore (C). (D) Living specimens at different developmental stages.
The brain is situated dorsally and anterior to the pharynx (Figures 4A, 5A, G). In living specimens, two nerve cords extending forward to the tentacles are evident. Two eye spots are located at the front of the brain. The two black, round eyes have an average distance of 0.2 ± 0.02 mm (n=10) between them; each eyecup contains three retinal cells (Figures 2A, E, 4B).
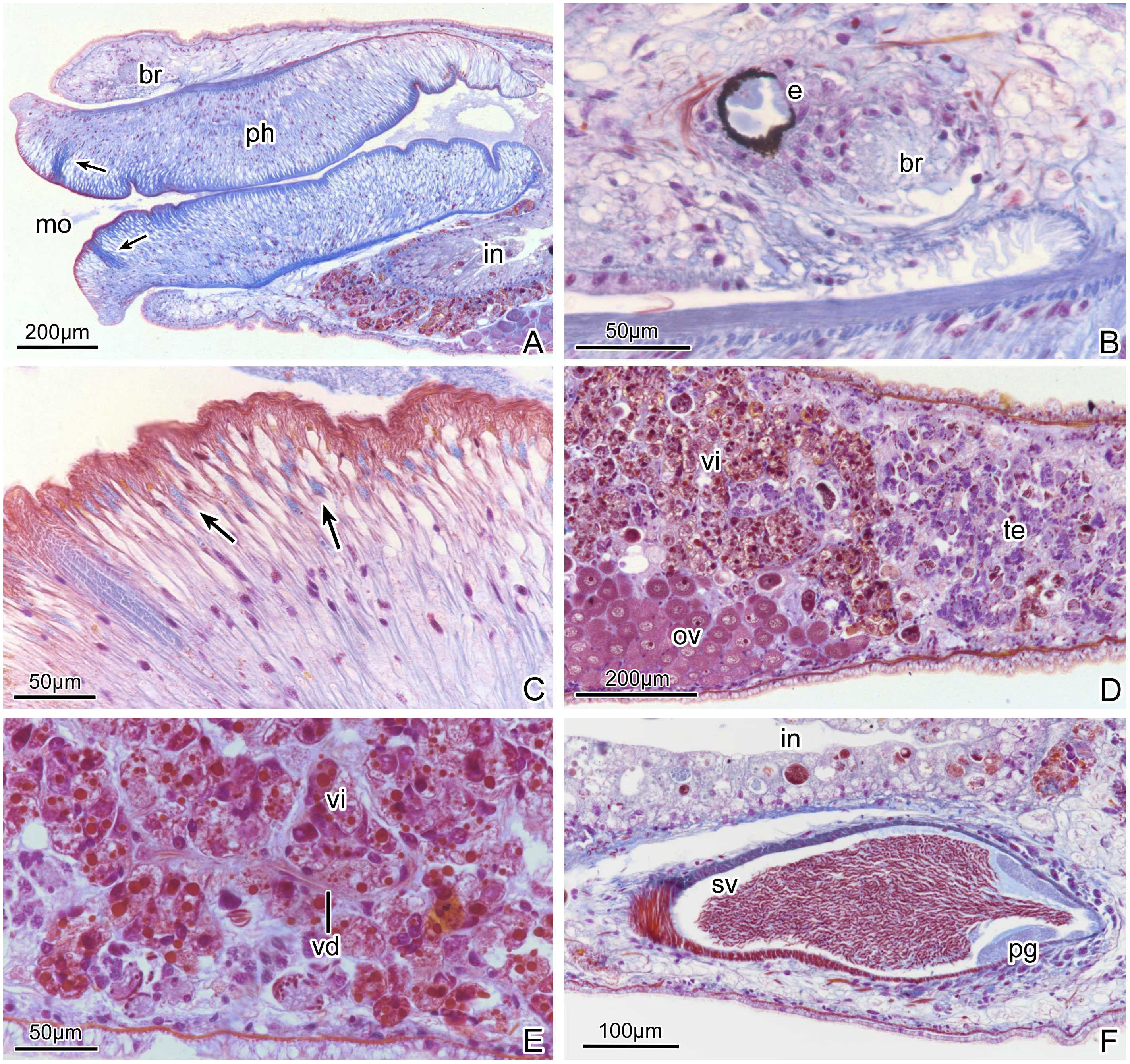
Figure 4 Plagiostomum robusta. Photomicrographs of sagittal histological sections: (A) Anterior portion, showing the terminal mouth and large muscular pharynx, arrows mark the stuck of circular muscles. (B) Brain and eyes. (C) Details of the pharyngeal tissue, arrows mark granular pharyngeal glands. (D) Holotype PLA-Pl001, showing the position relationship between ovaries and testes. (E) Vitelline duct between the vitellaria. (F) Seminal vesicle and prostatic glands.
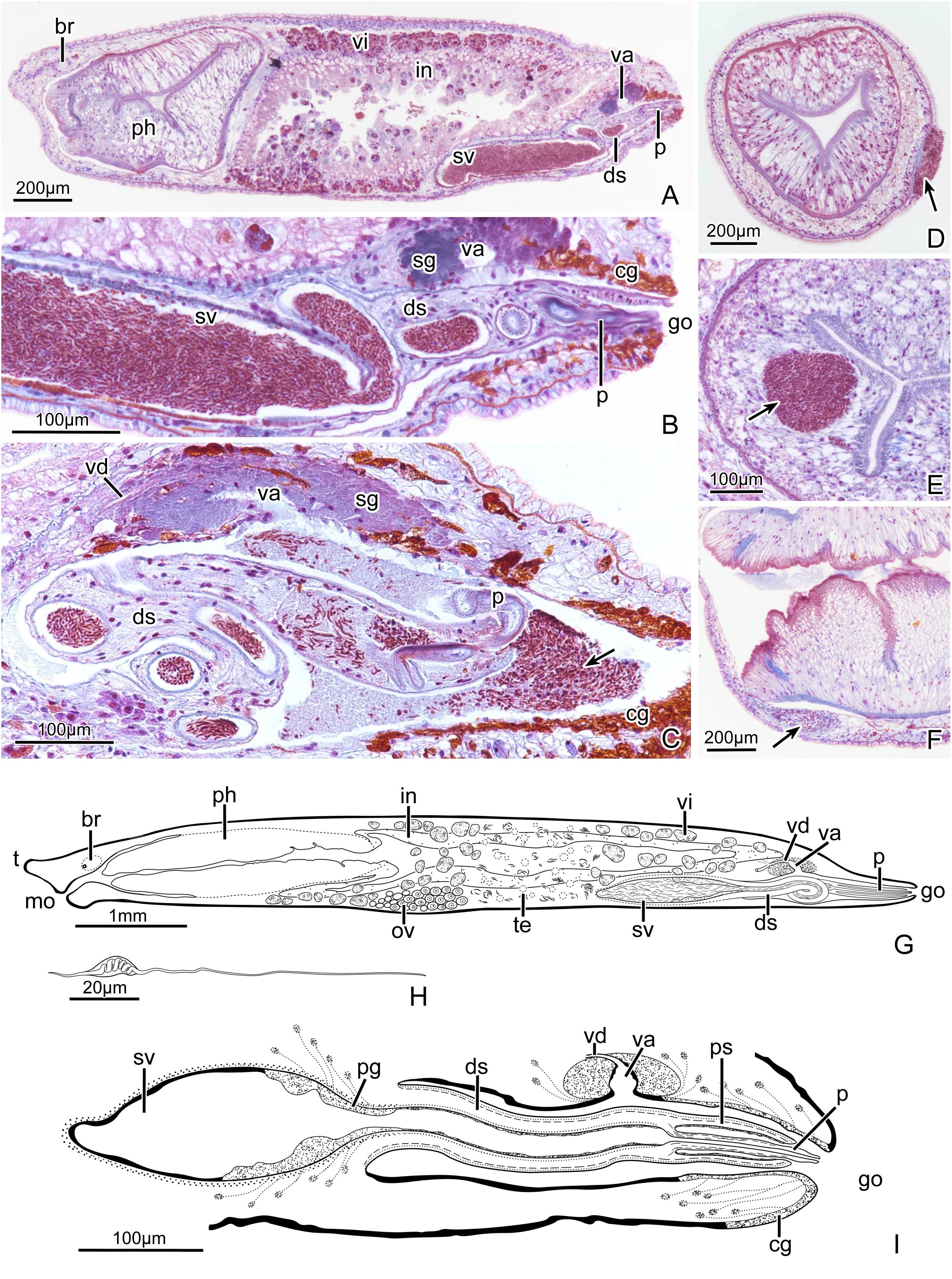
Figure 5 Plagiostomum robusta. (A) PLA-Pl003, sagittal section through entire specimen. (B) PLA-Pl003, sagittal section of posterior end. (C) PLA-Pl002, sagittal section of the copulatory organ, arrow marks semen that spills from the penis into the common atrium. (D-F) Sections of individuals impregnated at body wall (D, F) or pharynx (E), arrows mark sperm mass. (G) Sagittal view of the specimen as a whole. (H) Reconstruction of the spermatozoa. (I) Reconstruction of the copulatory apparatus.
The mouth is located below the tentacles, at the anterior end of the body (Figures 2E, 4A, 5G). The pharynx is highly developed and makes up approximately one third of the body length (Figures 2A, 5A, G, 6D). It measures around 2.82 ± 0.4 mm in length (n=10) and 0.68 ± 0.04 mm in width (n=10). The pharyngeal cavity wall is covered with cilia. Both the outer and inner surfaces of the pharynx have several longitudinal muscle layers, followed by a layer of circular muscles. Numerous radial fibers are distributed between the internal and external muscle layers (Figures 4A, 7A). The upper, lower left, and lower right regions of the pharyngeal tube are the thickest parts. In horizontal sections, the pharynx lumen appears as a compressed triangle (Figures 7A, 5D). Near the first 1/8 of the pharynx, there are multiple layers of circular muscles surrounding the pharyngeal cavity (Figures 4A, C, 5A, F). Granular pharyngeal glands are distributed in strips near the inner surface of the pharynx (Figure 4C).
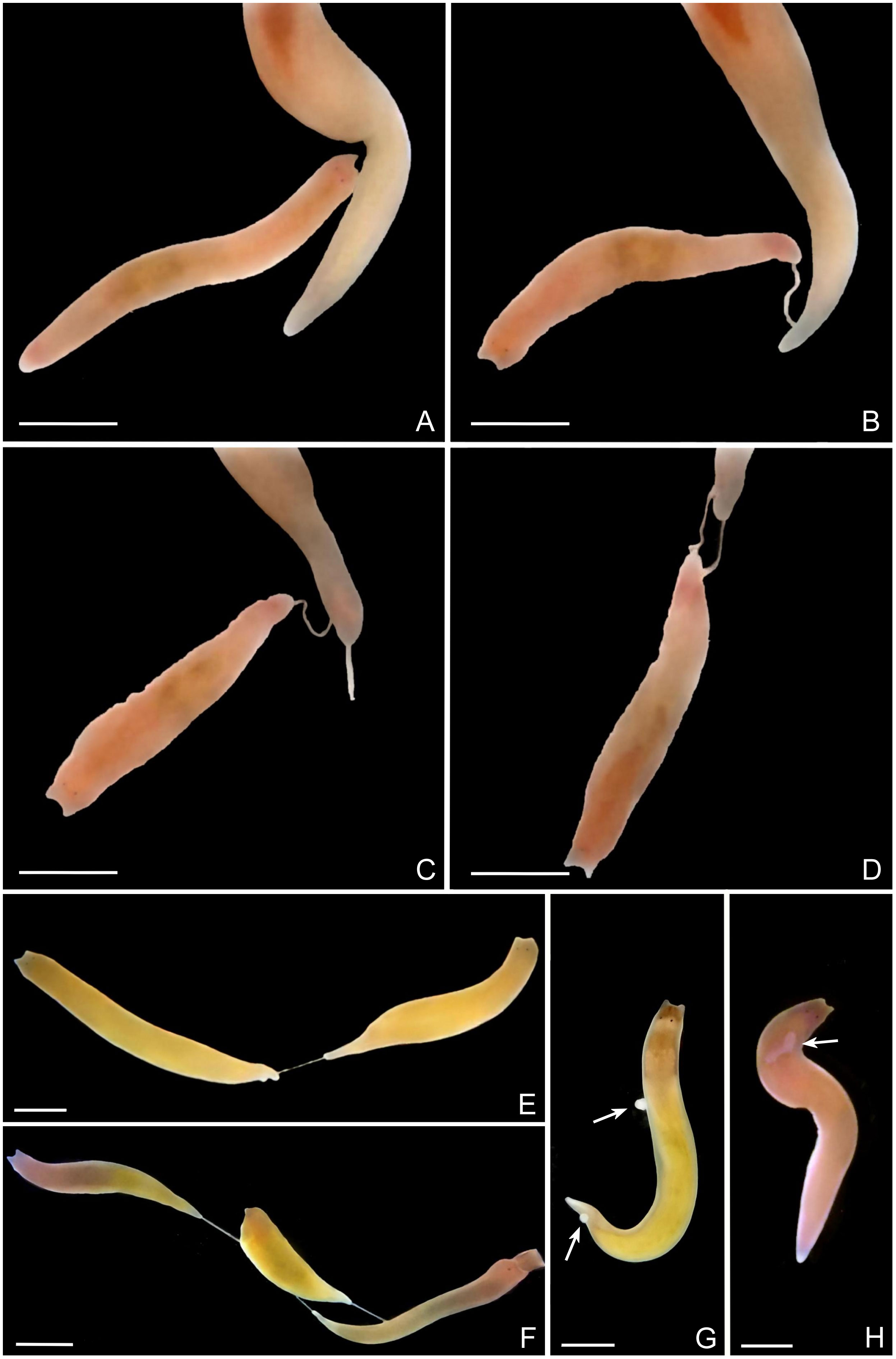
Figure 6 Hypodermic inpregnation of Plagiostomum robusta. (A-D) Process of the beginning of reciprocal inpregnation (a scenario). (E) Unilateral inpregnation. (F) Multidirectional impregnation. (G, H) Living sperm receptors, arrows mark sperm mass on the body wall (G) or in the subcutaneous tissue (H). All the scale bars: 2mm.
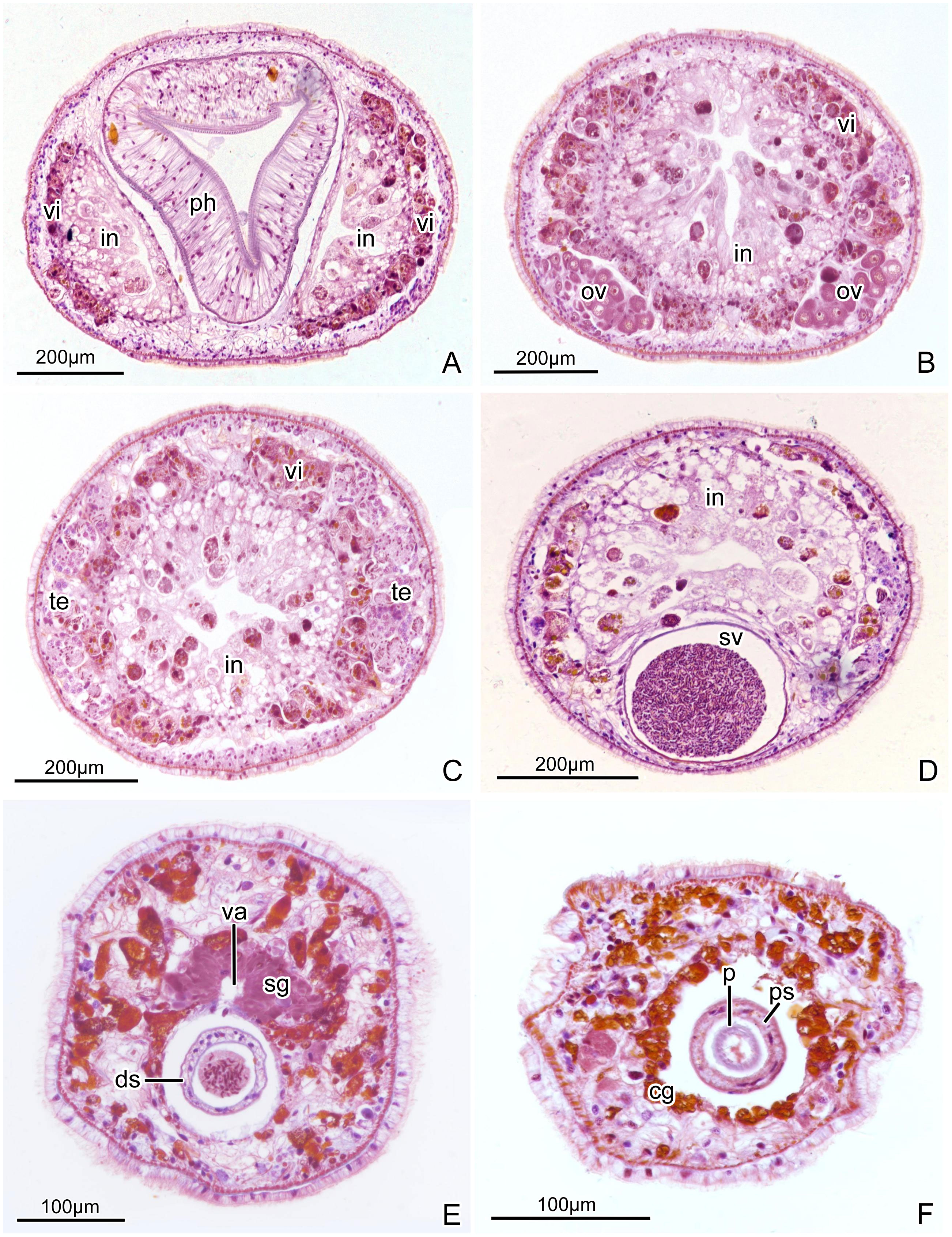
Figure 7 Plagiostomum robusta. Photomicrographs of transverse histological sections: (A) Section through pharynx and intestine, showing the extended front end of the intestine on both left and right side. (B) Section through ovaries. (C) Section through testes. (D) Section through seminal vesicle. (E) Section through anterior portion of common atrium and distal sac, showing the entrance of vagina into common atrium. (F) Section through penis and penis sheath.
The intestine is long and sac-like, extending to around 1/12-1/10 of the posterior part of the body (Figures 5A, G). At the junction with the pharynx, the intestine projects forward on the left and right sides, giving it a “Y” shape (Figures 2A, E, 3D, 7A). The intestine is lined with vacuolated, columnar epithelial cells.
One common atrium and one gonopore are shared by the male and female reproductive systems. The gonopore is located at the tail end (Figures 2E, F, 5A, B, G, I). A pair of ovaries, consisting of closely arranged cells, are positioned ventro-laterally to the middle of the projecting portions of the intestine (Figures 4D, 7B). In histological sections it was common to find sperm being present in mesenchymatic tissues, clearly being on their way towards the oocytes. There are no apparent oviducts extending from the ovaries. Surrounding the intestine, numerous vitellaria extend from the front to the end of the intestine where is close to the seminal vesicle (Figures 4A, 7A–D). Traces of winding vitelline ducts can be found between the vitellaria (Figure 4E) on the left and right sides of the body. These ducts have a small diameter of only 2-4 μm in sagittal sections, which then conjoin to form a common duct near the end of the intestine and lead to the vagina. The vagina, located at the dorsal middle of the common atrium, is very short (Figures 5A–C, G, I, 7E). Shell glands are distributed around the vagina and tightly cluster. Cement glands are abundant around the gonopore and the posterior segment of the common atrium (Figures 5A–C, I, 7F).
The testes, located behind the ovaries and laterally to the vitellaria, are situated close to the left and right body walls and extend to the seminal vesicle (Figures 4D, 7C). The body of mature spermatozoa is enclosed by a long, fusiform membrane, measuring 27-35 μm in length (n=3) and 4-6 μm in width (n=3). The structure within this membrane coils in a spring-like spiral and has a short, tapered filament, known as the contact region, at one end, and a slender, linear flagella at the other end, approximately four times the length of the sperm body, referred to as the tail region (Figures 2C, 5H).
The seminal vesicle has a rugby ball or long oval shape when filled with sperm and contains circular muscles in the outer wall (Figures 4F, 5A, B, G, I, 7D). The prostatic layer, which has a bumpy surface, is attached to the inner wall of the posterior end of the seminal vesicle, extending to the narrow “neck”, where the seminal vesicle joins the distal sac (Figures 4F, 5G, I). There is no separate prostate sac. No obvious vas deferens was observed. The distal sac is long tubular, coiled and constricted in the common atrium, often filled with sperm from the seminal vesicle (Figures 2F, 5A–C, G, 7E). Its inner and outer surfaces are covered by circular muscles, with longitudinal muscles below. The distal sac connects the penis and its sheath (Figures 2F, 5A, B, G, I). The penis sheath has a muscle layer distribution similar to that of the penis sac. The penis is slender, tubular (Figures 5A–C, G, I), and consists mainly of longitudinal muscles. In specimen PLA-Pl002, the penis is extended outside the penis sheath into the common atrium, while in the specimen PLA-Pl003, it is contained within the sheath (Figures 5A-C). The common atrium is densely lined by short cilia.
Comparative discussion. According to the traditional classification system, the tentacled Plagiostomidae species have generally been classified into the genus Vorticeros. However, Plagiostomum robusta, unlike all other recorded species of Vorticeros, has its gonopore situated at the tail tip. Among Vorticeros, V. luteum Hallez, 1879 is described as chromium-yellow in color (Kozloff and Westervelt, 2001), similar to P. robusta. However, V. luteum is plumper and more spindle-shaped, with tentacles that are about 1/7-1/6 of the body length, which is apparently larger than the tentacle length ratio of P. robusta. Vorticeros lobatum Tozawa, 1918, found in Japan, has short tentacles at the termination of its head (Tozawa, 1918), similar to P. robusta. But V. lobatum has auricular grooves on both sides of its head, a milky white body, and a dark purple reticular band on the mid-dorsal surface, all of which are clearly distinct from P. robusta. On the other hand, P. robusta can be easily distinguished from other Plagiostomum species due to its short, blunt tentacles and unique copulatory organ structure. The characteristic combination of having the oral opening at the front end and the genital opening at the tail end is also rare within this genus, further establishing its status as a new species. Although P. album Hyman, 1938 is considered to be most similar to P. robusta, as they share at least four features, including a large muscular pharynx, two extended front ends of the intestine, a long tube-like penis, and a genital opening at the tail end (Hyman, 1944), P. robusta possesses an apparent difference in that its distal sac is connected to the wall of the common atrium only at the front end, with the penis sheath and penis connected to the distal sac end. In contrast, the distal sac of P. album is a tube surrounded by parenchymal tissue, with the penis sheath and penis connected to the anterior end of the common atrium. Plagiostomum album also differs from P. robusta in terms of body color (white), brown stripes on the back, two pairs of eyes, and the location of the mouth and ovaries behind the testes. While the structural pattern of copulatory organs in most Plagiostomum flatworms is similar to that of P. album, P. robusta stands out with its unique movable distal sac, which is not found in any of the Plagiostomum species described so far. Interestingly, the characteristic combination of a movable distal sac and an elongated penis is present in Vorticeros cyrtum Marcus, 1947 (Marcus, 1947). However, V. cyrtum differs from the new species in its smaller and plumper body, larger tentacle-to-body length ratio, and pigment distribution extending down to the middle of its two tentacles, making it easily distinguishable from P. robusta.
Mating behavior. The mating activity of 20 pairs of flatworms was observed, and it was found that Plagiostomum robusta mated through hypodermic inpregnation. This process involved the elongation of the long muscular penis, with the part protruding out of the gonopore reaching 1/6-1/5 of the body length. The penis papilla has the ability to move rapidly and flexibly. Once the papilla of the penis touched the mating partner, it became tightly anchored to the epidermis (Figures 6B–D). During mating, the flatworm wiggled and squeezed to ejaculate sperm bundles into the tissues of the mating partner, after which it retracted the penis to complete the mating process. When sexually mature individuals recognized their partners using tentacles (Figure 6A), they displayed three types of reactions. (a) The flatworm directly turned its body to extend the penis, indicating its intention to mate (Figure 6B). (b) It evaded and moved away, showing no mating behavior. (c) It stretched out the pharynx to attack the other flatworm. If the other individual is smaller, it may prey on the other one. If the other individual is similar in size and cannot be swallowed, it will either extend the penis to mate or move away. During mating, if both flatworms inject sperm bundles into each other’s tissue, reciprocal inpregnation was observed (Figure 6D). Six pairs were observed in this regard. However, if only one flatworm successfully inserts the penis into the other, and the other partner fails to inject sperm due to orientation or other reasons, or does not show mating intentions, then unilateral inpregnation was observed (Figure 6E). In total, unilateral inpregnation of six pairs were recorded. When only one partner showed willingness to mate, inpregnation often failed because the penis could not reach the other flatworm, which moved away (n=4 pairs). In the presence of multiple flatworms, multidirectional inpregnation could be observed (Figure 6F). An individual could be inseminated by multiple individuals, indicating the randomness of mating partner. The sperm recipient displayed strong resistance, regardless of the direction of impregnation. This resistance included constant swimming, in order to try to get rid of the inserted penis and attacking the sperm donor using the pharynx. In some cases, the sperm donor retracted its penis due to injury or stimulation, thus ending the mating process (n=4 pairs). The sperm bundles in the tissue of the sperm donor appeared white (Figures 6G, H) and were partially or completely buried under the epidermis. This could cause varying degrees of tissue damage. The sperm bundles were absorbed within a few hours. The site of inpregnation was random, depending on where the sperm donor’s penis touched the body of the sperm recipient. As a result, it often occurred that sperm was injected into the “wrong” place, such as the pharynx or intestine (n=5) (Figures 5D–F, 6F–H).
Ontogenesis and Behavior. Plagiostomum robusta had no fixed egg deposition area. During the process of egg laying, the flatworm attached its tail end to the wall of the container and underwent a wriggling motion from front to back, leading to the discharge of the eggs through the gonopore. The mucus secreted by the gonopore solidified with sea water, resulting in the formation of an egg stalk. The eggs were spherical or drop-shaped and initially appeared white (Figures 2D, 3C), but they soon underwent a color change to orange-yellow (Figures 3A-C). The translucent egg shell enclosed a dense fertilized egg. A total of 17.4 ± 3.14 eggs were laid in each spawning cycle (n=10), with a cycle duration of 18.4 ± 2.46 days (n=10). The incubation cycle lasted for 13.4 ± 1.72 days (n=30), and the hatching rate was 71% (n=10), with an average of 24.1 ± 7.76 juveniles incubated per egg (n=30). Within 24 hours of spawning, the fertilized egg divided into multiple spherical early embryos. During days 2 to 7 of embryonic development, no apparent changes were observed. On day 8, eye spots became visible and the embryo assumed an elliptical shape. Upon exposure to cold light, peristaltic movements could be observed in the embryo. Between day 8 and day 12, the embryo underwent elongation, and the tail transformed into a cone shape. At this stage, the body surface lacked pigmentation, and the intestine appeared opaque (Figure 2D). Notably, avoidance and withdrawal behaviors were exhibited when the heads of two embryos came into contact with each other. On the 13th day, a round crack with rough edges appeared on the egg shell, enabling the juveniles to hatch. In some cases, a portion of the fully developed embryo failed to break through the shell, leading to eventual inactivity and disintegration.
Parental care has not been observed in Plagiostomum robusta. The newly hatched juveniles had a transparent body, long sac-like intestines, indistinct tentacles, and a slender tail (Figure 3D, 1d). The juveniles turned yellowish at about ten days of age, and their intestines became yellowish brown or gray after ingestion of food (Figure 3D, 10d). Seminal vesicle development occurred around day 20, accompanied by an enlarged tail and elongated tentacles (Figure 3D, 20d). Mating behavior between individuals began when male mating organs matured at around day 30. As the flatworms continued to grow, they became opaque, with further enlargement of the seminal vesicle and penis (Figure 3D, 30d). After approximately 60 days, development slowed down. On average, it took 30.1 ± 2.26 days (n=30) from hatching to reach sexual maturity. Under the culture conditions of this study, the flatworms typically resided on the side walls or bottom of container in lucifugal areas. When in motion, they slid along the container wall, swinging their heads and continually probing with the tentacles. Upon contact with a foreign object, they immediately retracted their tentacles and changed direction. Furthermore, the worms exhibited obvious light avoidance behavior. When multiple flatworms coexisted, they tended to disperse instead of forming clusters. Plagiostomum robusta fed on other flatworms and was observed to engage in cannibalism. Using their tentacles, they sensed their prey and rapidly opened their mouths to extend their pharynx into a trumpet-like shape. When they caught relatively small prey by their pharynx, they quickly engulfed it into the intestinal sac. When encountering prey larger than themselves, the worms would catapult the pharynx to attack or tightly grasp the prey, tearing off its tissues before swallowing it.
The lengths of the three assembled mitochondrial genome sequences were 14,491 bp, 14,466 bp, and 14,426 bp, respectively. Each of the three sequences was annotated with 36 genes, including 12 protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes, and 22 transfer RNA (tRNA) genes. These genes are transcribed by the same strand (Supplementary Table 2). In these assembled sequences, there is a large non-coding region that contains a gap. However, due to interference from tandem repeats, accurate sequencing and assembly of all three sequences at this position could not be achieved. Finally, the mitochondrial genome with a length of 14,491 bp (GenBank accession number: OR260862) was chosen for further analysis (Figure 8A; Supplementary Table 3).
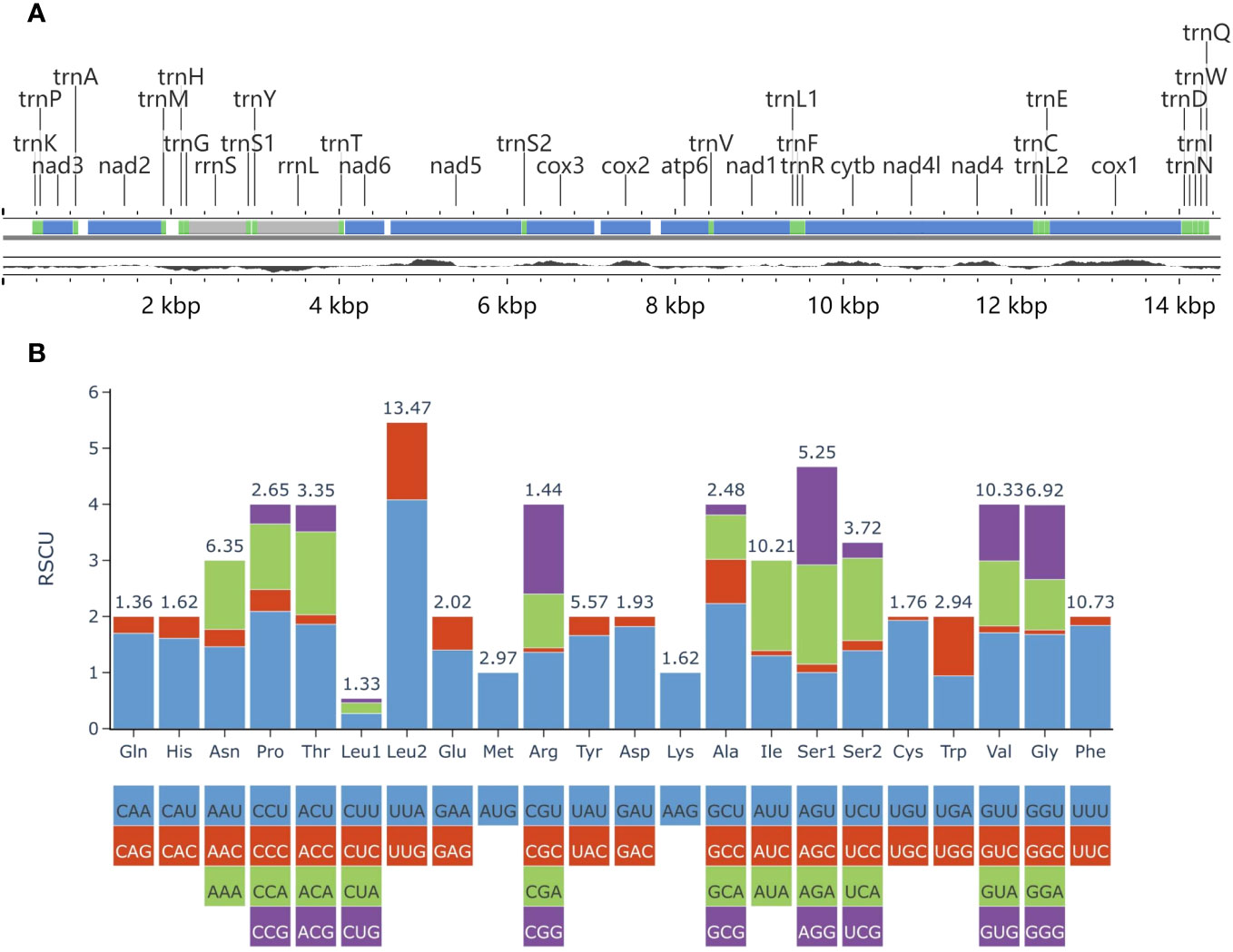
Figure 8 (A) Partial mitochondrial genome map of Plagiostomum robusta. The top line shows the gene annotation, with protein-coding genes, ribosomal RNA genes and transfer RNA genes represented by blue, gray and green, respectively. The bottom black line displays the GC content. (B) The RSCU of 12 protein-coding genes in the mitogenome of P. robusta. Codon families are provided on the X-axis.
In the mitochondrial genome, the 12 PCGs have a total length of 10,435 bp. All PCGs, except for nad5, start with the codon ATG. However, nad5 starts with GTG. cox1 was found to contain an incomplete termination codon T. Four PCGs (nad5, nad1, cytb, nad4l) use TAG as the stop codon, while the rest use TAA. The A + T content of the 12 PCGs is 71.14%. Calculated values for relative synonymous codon usage (RSCU) were provided (Figure 8B; Supplementary Table 3). The total number of codons in the 12 PCGs is 3,467, excluding the stop codons. Among these codons, leucine (Leu2) (UUR), phenylalanine (Phe), valine (Val), and isoleucine (Ile) are the most frequently used. On the other hand, codons encoding leucine (Leu1) (CUN) and glutamine (Gln) are rare. The AT skew in PCGs is negative (-0.212), while the GC skew is positive (0.408).
Between trnM and trnH, we found a putative atp8 ORF at positions 1,941-2,093 bp that is 153 bp long. The ORF starts with ATG as the start codon and ends with TAA as the stop codon. It overlaps with trnH by 5 bp. The amino acid sequence translated from the ORF starts with MAQV, which has a comparability with the widely conserved MPQL. The hydrophobicity profile showed a trend of increasing and then decreasing, with positive hydrophobicity scores at the N-terminus and mostly negative scores at the C-terminus (Supplementary Figure 1). A transmembrane domain was predicted to be present within the 13-32 amino acids. The above two features are similar to those of ATP8 in other animals. However, no signal peptide was predicted. We performed NCBI BLASTp, but the amino acid sequence did not show significant homology with any of the published sequences. Therefore, we merely refer to this ORF as putative atp8 until further support from the transcriptome data regarding the presence of atp8 in Plagiostomum robusta and the actual location of atp8 is acquired.
There are 22 tRNA genes in the genome, with the longest being 66 bp (trnK, trnP, trnS1, trnY, trnL2) and the shortest being 59 bp (trnH). Apart from trnS2, which lacks dihydrouracil (DHU) arms, all tRNA genes possess a typical cloverleaf structure (Supplementary Figure 2). The total length of the tRNA genes is 1,396 bp, and they have an A + T content of 77.72%. The two rRNA genes have a combined length of 1,597 bp and an A + T content of 77.35%. The rrnS gene is 730 bp long and is situated between trnG and trnS1, overlapping by 52 bp and 7 bp, respectively. The rrnL gene is 867 bp long and is located between trnY and trnT.
The large non-coding region of the genome, located between trnQ and trnK, contains many tandem repeats. The tandem repeat region near trnQ consists of a 25 bp unit and is repeated 5.7 times in the assembly sequence. While the tandem repeat region near trnK consists of a 191 bp unit and is repeated 2.1 times in the assembly sequence. In addition to these tandem repeats, 20 short non-coding regions were identified in the mitochondrial genome with a length varying from 1 bp to 114 bp (excluding the assumed location of atp8).
According to the results of our comprehensive analysis, the taxonomic status of Plagiostomum robusta at the family level can be easily determined. As for the genus to which it belongs, we believe it is more reasonable to classify it into Plagiostomum. In this study, phylogenetic trees were built using the 18S rDNA and 28S rDNA concatenated dataset, which revealed that P. robusta did not cluster with two species of Vorticeros, but instead formed a clade with six species of Plagiostomum. Based on Laumer and Giribet (2017) phylogenetic trees constructed using 18S rDNA and 28S rDNA, P. robusta and two undetermined species tentatively classified into Plagiostomum were added in this work. Compared with the phylogenetic trees in previous studies (Jondelius et al., 2001; Noren and Jondelius, 2002; Noren, 2004; Laumer and Giribet, 2017), it was observed that the branching topology of the clade that contains P. striatum, P. cuticulata, P. whitmani, P. lemani and P. cinctum was inconsistent between different trees. Noren and Jondelius (2002) constructed two trees based on two datasets with different numbers of species, and the above problem was also reflected in their results. The inconsistency could be mainly attributed to the differences in species sampling, the number of genes or sites, and the tree-building methods used. In our study, despite selecting a different evolutionary model than Laumer and Giribet (2017), the phylogenetic tree topology is consistent with their results (excluding the newly added species). Our resulting BI and ML trees have a similar structure and mostly high support values. At present, there is only a very limited amount of sequence data available for Plagiostomidae species on GenBank. Under these circumstances, we believe that, nevertheless, a rather credible result was achieved. Protein-coding genes in mitochondrial genomes evolve at different rates, providing richer differentiation information than the conserved 18S rDNA and 28S rDNA, which is of great significance for the analysis of evolutionary relationships between species. To supplement the data available for phylogenetic analysis, we provide an almost complete mitochondrial genome sequence of P. robusta, consisting of 12 PCGs, an atp8 putative ORF, two rRNA genes, 22 tRNA genes, and a large non-coding region. However, the large non-coding region sequence could not be analyzed due to a gap containing tandem repeats at both ends. Our attempts to close the gap through second-generation sequencing and Sanger sequencing were unsuccessful, similar to the situation reported for Dugesia ryukuensis (Sakai and Sakaizumi, 2012). In the future, using long-read sequencing may be an appropriate solution to this problem. Currently, there are multiple controversies regarding the classification system of Prolecithophora due to inconsistencies between morphological characteristics and molecular phylogenetic analyses. It is necessary to reconfigure the taxa under Prolecithophora by supplementing molecular data for a larger number of species and taking into account various aspects more comprehensively.
Morphologically, Plagiostomum robusta resembles the species in Clade 1 identified in our phylogenetic analysis in terms of body size, pharynx-to-body ratio and penis morphology. In Clade 1, P. album, P. lemani, P. striatum, P. whitmani, Plagiostomum sp. MW-2019 have a relatively large and variable pharynx (>1/4 body length) and a long tubular penis, similar to P. robusta. This may suggest that these traits are synapomorphies of Clade 1 (detailed data on P. culticulata is not available to us). On the other hand, Clade 2 consists of seven species with a relatively small pharynx and a relatively short penis. Besides, none of the nine documented species of the genus Vorticeros has a relatively large pharynx. Although the copulatory organ of V. cyrtum shows a similar structural pattern to P. robusta, the elongated proximal penis of V. cyrtum is folded into the distal sac. In contrast, the long tubular penis of P. robusta usually points towards the common atrium. Our behavioral observations also suggested that it will be extended out of the gonopore during mating. Since the mating behavior of Plagiostomidae worms have not been studied before, it is still unknown whether the corresponding parts of species with long tubular penis perform the same function. In general, the group relationships shown in our phylogenetic tree are in agreement with the morphological observations, indicating a close relationship between P. robusta and Plagiostomum species in Clade 1, suggesting that the tentacles of P. robusta may be the result of convergent evolution.
Due to the lack of molecular data, the phylogenetic position of Vorticeros species other than V. auriculatum and V. ijimai cannot be determined. According to a recent phylogenetic tree, Jondelius et al. (2001) suggested to place both Vorticeros and Torgea within Plagiosomum. However, as T. phukettensis exhibits unique pharyngeal structures (Noren, 2004), the independence of Torgea cannot be denied at this stage. Therefore, even if Vorticeros would be merged with Plagiostomum, the non-monophyletic problem cannot be resolved yet. To address the conflict between different taxonomic systems, it is considered more reasonable to re-split the Plagiostomidae. This work requires detailed morphological characteristics of each species to determine more appropriate taxonomic characteristics, with molecular phylogenetic analysis as a powerful reference. In brief, based on results of our integrative analysis, since P. robusta appears to be more related to species in Clade 1 we choose to classify the new species under the genus Plagiostomum.
There are two main mating patterns in flatworms. One of them is reciprocal copulation, where two individuals join their gonopores and transfer sperm to each other via the female atrium. The other method is hypodermic impregnation, which involves a scleroid penis that is shaped like a hook or a syringe needle (Brand et al., 2022). These flatworms inject sperm into the mesenchymatic tissues of their mating partners. Interestingly, our results showed that although Plagiostomum robusta exhibits hypodermic impregnation mating behavior, its copulatory organ lacks needles or spiny structures and, instead, consists of muscular penis papilla. Nonetheless, the terminal location of the gonopore, the strong and flexible penis, and the highly agile movement of the penial papilla indicate its suitability for hypodermic impregnation.
The mating mode of Plagiostomum robusta is simple and primitive, as indicated by two characteristics. First, it lacks a sperm receiving organ and instead has a random impregnation site on the body surface. After inpregnation, allogeneic sperm swim disorderly through the tissue and migrate towards the egg cells. Second, there is no courtship behavior and the partner is chosen randomly. In the presence of multiple flatworms, an individual even can be inseminated by multiple partners simultaneously. Mating through hypodermic injection is widespread among animals that are simultaneous hermaphrodites (Schärer et al., 2015). However, this method is costly and risky for the sperm receptor, as it must undergo tissue restoration, clearance of foreign matter, and assume responsibility for laying the fertilized egg. For the sperm donor, this approach may increase frequency of insemination and potentially enable sperm to reach the egg cell directly, thus enhancing sperm competition. We also observed the occurrence of multidirectional inpregnation in this study. Since P. robusta does not exhibit gregarious behavior, this phenomenon is likely rare under natural conditions. However, similar situations, wherein an individual is inseminated by different partners within a relatively short time interval, may have implications for inseminating more eggs and improving the success rate of sperm delivery.
Sexual conflicts in hermaphrodite animals are mostly due to conflicts in individual sexual interests (Michiels and Newman, 1998), which is reflected in the imbalance between the individual’s willingness to donate and receive sperm. For animals that undergo traumatic mating through injection, the conflict between the benefits of insemination and the high cost of being inseminated is strong. However, unlike Polycladida species that perform compulsive unilateral insemination through “penis fencing” to compete for the right to inseminate (Ramm, 2017), Plagiostomum robusta does not actively avoid the partner’s penis during insemination, but rather shows resistance after being stabbed. Our observations revealed that for sexually mature mating pairs, the likelihood of inseminating each other is about the same, and resistance does not usually interrupt the ongoing injection. It may, however, affect the amount of ejaculated sperm. This suggests that the male and female roles assumed by the flatworms are basically in balance, and sexual conflicts are resolved in some way. We propose that P. robusta and its mating partner somehow select each other before mating. One possible route is the pharyngeal tussle described in the present study. The strength of the pharynx is an important reflection of survivability, and this predation-like behavior may help to detect the size and strength of the other individual so as to identify strong and viable sperm recipients and/or donors, with the weaker individuals serving as nourishment. However, some individuals did not experience this process. They were observed to mate after one flatworm touched the other’s body through its tentacles or after both tentacles touch each other, suggesting that before the tussle of the pharynx, some “signal” of the tentacles may function as a means of communication, although for now the mechanism involved is unclear. Furthermore, an individual’s sex role preference and behavior are related to resource conditions and sex allocation (Ramm, 2017). The level of sperm competition in P. robusta may be so intense that it causes a decrease in male function allocation. Further experimental evidence is needed to test this hypothesis. In summary, we suppose that the result of selection is such that in P. robusta mating partners decide to perform the male and female roles simultaneously. This is consistent with its ability to anchor its penis for a long duration, instead of a split-second, during hypodermic impregnation, thus allowing for reciprocal mating rather than just unilateral mating.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was approved by Animal Ethical and Welfare Committee of Shenzhen University. The study was conducted in accordance with the local legislation and institutional requirements.
YW: Data curation, Funding acquisition, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. JH: Data curation, Methodology, Writing – review & editing. YZ: Funding acquisition, Supervision, Writing – review & editing. AW: Conceptualization, Project administration, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by Special Funds for the Cultivation of Guangdong College Students’ Scientific and Technological Innovation (“Climbing Plan” Special Funds key project approval; grant no. pdjh2023a0440), China Undergraduate Training Program for Innovation and Entrepreneurship (grant no. 202210590035), and the Shenzhen University Innovation Development Fund (grant no. 2022305), as well as grants from Scientific and Technical Innovation Council of Shenzhen Government (grant nos. jcyj20210324093412035 and kcxfz20201221173404012) and Innovation Team Project of Universities in Guangdong Province (no. 2023KCXTD028).
We sincerely thank Dahao Lin and Jixiang Li for culturing Paucumara falcata and helping with photo-documenting, Chuyi Huang for examining the specimens, and Prof. Ronald Sluys (Naturalis Biodiversity Center, P. O. Box 9517, 2300 RA Leiden, The Netherlands) for providing valuable comments on this work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1332011/full#supplementary-material
Benson G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27 (2), 573–580. doi: 10.1093/nar/27.2.573
Brand J. N., Viktorin G., Wiberg R. A. W., Beisel C., Schärer L. (2022). Large-scale phylogenomics of the genus Macrostomum (Platyhelminthes) reveals cryptic diversity and novel sexual traits. Mol. Phylogenet Evol. 166, 107296. doi: 10.1016/j.ympev.2021.107296
Caira J. N., Littlewood D. T. J. (2013). ““Worms, platyhelminthes,”,” in Encyclopedia of biodiversity, 2nd. Ed. Scheiner S. M. (New York, NY: Academic Press), 437–469.
Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Donath A., Jühling F., Al-Arab M., Bernhart S. H., Reinhardt F., Stadler P. F., et al. (2019). Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47 (20), 10543–10552. doi: 10.1093/nar/gkz833
Duvaud S., Gabella C., Lisacek F., Stockinger H., Ioannidis V., Durinx C. (2021). Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 49 (W1), W216–W227. doi: 10.1093/nar/gkab225
Grant J. R., Enns E., Marinier E., Mandal A., Herman E. K., Chen C. Y., et al. (2023). Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 51 (W1), W484–W492. doi: 10.1093/nar/gkad326
Grosbusch A. L., Bertemes P., Egger B. (2021). The serotonergic nervous system of prolecithophorans shows a closer similarity to fecampiids than to triclads (Platyhelminthes). J. Morphol. 282 (4), 574–587. doi: 10.3390/biology11111588
Grosbusch A. L., Bertemes P., Kauffmann B., Gotsis C., Egger B. (2022). Do not lose your head over the unequal regeneration capacity in prolecithophoran flatworms. Biology. 11 (11), 1588. doi: 10.3390/biology11111588
Hyman L. H. (1944). Marine turbellaria from the atlantic coast of north america. Am. Mus Novit. 1266, 1–15.
Jondelius U., Norén M., Hendelberg J. (2001). ““The prolecithophora,”,” in Interrelationships of the platyhelminthes. Eds. Littlewood D. T. J., Bray R. A. (London: CRC Press), 74–80.
Karling T. G. (1940). Zur Morphologie und Systematik der Alloeocoela cumulate und Rhabdocoela lecithophora (Turbellaria). Acta Zool Fenn. 26, 1–260.
Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 (4), 772–780. doi: 10.1093/molbev/mst010
Kozloff E. N., Westervelt C. A. (2001). Vorticeros praedatorium sp. nov. (Platyhelminthes: Proleccithophora: Plagiostomidae) from the Pacific coast of North America. Cah Biol. Mar. 42 (4), 303–314.
Lanfear R., Frandsen P. B., Wright A. M., Senfeld T., Calcott B. (2017). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34 (3), 772–773. doi: 10.1093/molbev/msw260
Laumer C. E., Giribet G. (2017). Phylogenetic relationships within Adiaphanida (Phylum Platyhelminthes) and the status of the crustacean-parasitic genus Genostoma. Invertebr Biol. 136 (2), 184–198. doi: 10.1111/ivb.12169
Letunic I., Bork P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49 (W1), W293–W296. doi: 10.1093/nar/gkab301
Letunic I., Khedkar S., Bork P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49 (D1), D458–D460. doi: 10.1093/nar/gkaa937
Li J. Y., Li W. X., Wang A. T., Zhang Y. (2021). MitoFlex: an efficient, high-performance toolkit for animal mitogenome assembly, annotation and visualization. Bioinformatics 37 (18), 3001–3003. doi: 10.1093/bioinformatics/btab111
Littlewood D. T. J., Curini-Galletti M., Herniou E. A. (2000). The interrelationships of Proseriata (Platyhelminthes: Seriata) tested with molecules and morphology. Mol. Phylogenet Evol. 16 (3), 449–466. doi: 10.1006/mpev.2000.0802
Marcus E. (1947). Turbellarios marinhos do brasil. Bolm Fac Fil Cienc SPaulo Zool. 12, 99–215, 1-12.
Michiels N. K., Newman L. J. (1998). Sex and violence in hermaphrodites. Nature 391 (6668), 647. doi: 10.1038/35527
Nguyen L. T., Schmidt H. A., Von Haeseler A., Minh B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32 (1), 268–274. doi: 10.1093/molbev/msu300
Noren M. (2004). Four new plagiostomidae (Prolecithophora: platyhelminthes) from phuket, Thailand, with a re-evaluation of torgeidae jondelius 1997, and paramultipeniata kulinich 1974. Res. Bull. 65, 9–22.
Noren M., Jondelius U. (2002). The phylogenetic position of the Prolecithophora (Rhabditophora,’Platyhelminthes’). Zool. Scr. 31 (4), 403–414. doi: 10.1046/j.1463-6409.2002.00082.x
Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A. (2018). Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67 (5), 901–904. doi: 10.1093/sysbio/syy032
Ramm S. A. (2017). Exploring the sexual diversity of flatworms: Ecology, evolution, and the molecular biology of reproduction. Mol. Reprod. Dev. 84 (2), 120–131. doi: 10.1002/mrd.22669
Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61 (3), 539–542. doi: 10.1093/sysbio/sys029
Sakai M., Sakaizumi M. (2012). The complete mitochondrial genome of Dugesia japonica (Platyhelminthes; order Tricladida). Zoolog Science. 29 (10), 672–680. doi: 10.2108/zsj.29.672
Schärer L., Janicke T., Ramm S. A. (2015). Sexual conflict in hermaphrodites. Cold Spring Harb. Perspect. Biol. 7 (1), a017673. doi: 10.1101/cshperspect.a017673
Tamura K., Stecher G., Kumar S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi: 10.1093/molbev/msab120
Thorvaldsdóttir H., Robinson J. T., Mesirov J. P. (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14 (2), 178–192. doi: 10.1093/bib/bbs017
Tozawa T. (1918). Vorticeros ijimai and Vorticeros lobatum spp. n. from Misaki. Zool. Mag. Tokyo. 30, 77–80; 111-115; 196-199.
Vaidya G., Lohman D. J., Meier R. (2011). SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27 (2), 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
World Register of Marine Species (2023) Prolecithophora. Available at: https://marinespecies.org/aphia.php?p=taxdetails&id=2854 (Accessed September 30, 2023).
Yang Y., Li J. Y., Sluys R., Li W. X., Li S. F., Wang A. T. (2020). Unique mating behavior, and reproductive biology of a simultaneous hermaphroditic marine flatworm (Platyhelminthes, Tricladida, Maricola). Invertebr. Biol. 139 (1), e12282. doi: 10.1111/ivb.12282
Keywords: marine flatworm, Plagiostomidae, integrative taxonomy, phylogeny, mitogenome, mating behavior
Citation: Wang Y, Huang J, Zhang Y and Wang A (2024) Molecular phylogeny and ethology of the Family Plagiostomidae (Platyhelminthes, Prolecithophora), with integrative description of a new species, Plagiostomum robusta A. Wang, sp. nov.. Front. Mar. Sci. 11:1332011. doi: 10.3389/fmars.2024.1332011
Received: 03 November 2023; Accepted: 26 January 2024;
Published: 15 February 2024.
Edited by:
Qiang Li, Chengdu University, ChinaReviewed by:
Xidong Mu, Chinese Academy of Fishery Sciences, ChinaCopyright © 2024 Wang, Huang, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Zhang, YmlvenlAc3p1LmVkdS5jbg==; Antai Wang, d2FuZzExOEBzenUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.