- 1Centro de Biotecnología y Genómica de Plantas, Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA-CSIC), Universidad Politécnica de Madrid (UPM), Madrid, Spain
- 2Departamento de Biotecnología-Biología Vegetal, Escuela Técnica Superior de Ingeniería Agronómica, Alimentaria y de Biosistemas, Universidad Politécnica de Madrid (UPM), Madrid, Spain
Introduction
Coastal lagoons represent transitional ecosystems between terrestrial and marine environments and play a central role in nutrient cycling, carbon flux, and biodiversity conservation (Kjerfve, 1994; Schubert and Telesh, 2017). These characteristics and their location on low-lying coasts are responsible for coastal lagoons being among the most productive and valuable ecosystems in the world, providing and supporting a wide range of ecological services and social benefits (Pérez-Ruzafa et al., 2020). Coastal lagoons are classified as one of the most sensitive ecosystems to changes in their environment (Velasco et al., 2018) especially to anthropogenic eutrophication and pollutant accumulation (Barbier et al., 2011), which explains why most coastal lagoons around the world show signs of ecological imbalance and degradation.
Marine coastal sediments are complex ecosystems that are influenced by the interaction of various geological, hydrological, physical, chemical and biological factors (Zhang et al., 1999). Coastal sediments formed by continental transport and sedimentation of biological products provide abundant nutrients for microorganisms (Parkes et al., 2014). In coastal marine ecosystems, microorganisms play a central role in shaping nutrient dynamics and biogeochemical cycles by transforming and metabolizing nutrients and pollutants (Nogales et al., 2011). Microorganisms can degrade sediment organic matter, promote sulfate reduction, sulfide/sulfur oxidation, iron reduction, nitrification and pollutant degradation, improve sediment structure and increase the stability of ecosystems (Behera et al., 2017). However, microorganisms in coastal sediments are influenced by the physico-chemical properties of the sediment, including salinity, pH, nutrients and are especially vulnerable to pollutants (Rodríguez et al., 2018; Liang et al., 2023). Changes in these properties can have a cascading effect on the microbial growth, metabolism, and activity, thereby affecting the structure of the sediment-associated microbial communities (Jackson and Vallaire, 2009; Liang et al., 2023).
The Mar Menor, located in the southeast of Spain, is the largest hypersaline coastal lagoon in Europe, with a surface area of 135 km2 (Conesa and Jiménez-Cárceles, 2007). The lagoon and its adjacent areas are protected at the national and international levels. Mar Menor is included in the Ramsar wetland sites of international importance and the Specially Protected Areas of Mediterranean Importance [SPAMI; (Álvarez-Rogel et al., 2020)]. In recent years, increasing human activities, especially mining, agriculture and tourism, have caused significant negative changes in the Mar Menor, leading to a considerable input of sediments, nutrients (nitrates and phosphates) and pollutants (pesticides and metals) into the lagoon (Velasco et al., 2006; Pérez-Ruzafa et al., 2019). These activities have caused a decline in water quality, leading to the proliferation of harmful algal blooms, fish mortality events, and degradation of seagrass beds (Pérez Ruzafa et al., 2004; Pérez-Ruzafa et al., 2006; Zamora-López et al., 2022). In 2015, the ecological condition of the lagoon deteriorated significantly due to a massive phytoplankton bloom, resulting in the loss of macrophytes and benthic fauna (Ruiz-Fernández et al., 2019). This disturbance destabilized the natural ecological balance of the lagoon leading to several hypoxic episodes in recent years (Álvarez-Rogel et al., 2020). Many of these negative impacts could also affect the sediment-associated microbiota, which is not only an important contributor to biogeochemical cycles but could also help to counteract these impacts by accumulating and managing nutrients and contaminants (Dash et al., 2013; Nzila, 2013; Rodríguez et al., 2018). However, our knowledge of the microbial communities in the Mar Menor remains quite limited (Aldeguer-Riquelme et al., 2022).
This study aims to preliminarily characterize the microbial communities in different zones of the Mar Menor impacted by tourism, mining, or agricultural activities. Its objective is to identify microbial biomarkers that serve as indicators of the lagoon’s environmental conditions. We anticipate that these findings will provide valuable insight into the microbial biosphere of the region and make a significant contribution to the establishment of effective conservation and management practices for the sustainability of this important ecosystem.
Methods
Study area and field sampling
The Mar Menor is a hypersaline coastal lagoon that occupies 135 km2 of surface, its volume is 653 Hm3, and its maximum depth is 7 m. It is located in Campo de Cartagena, in the Region of Murcia, southeast of Spain. The lagoon is one of the largest in the Mediterranean basin and the largest on the Spanish Mediterranean coast. It is separated from the open sea by a 22-km-long sand bar (La Manga), with three channels through which it exchanges water with the sea.
Five zones, characterized by different levels of anthropogenic impact, were selected for sediment sampling along the Mar Menor during May 2022 (Supplementary Figure S1). Zone 1 (Z1; Playa de la Mota, 4 samples) and Zone 2 (Z2; Playa de las Salinas, 4 samples) are located in the northern part of the lagoon and are the subject of extensive urban and maritime activities. Zone 3 (Z3; Albujón estuary, 4 samples) is located in the western area of the lagoon and is directly influenced by the discharge of the Albujón watercourse. Inputs to the lagoon from the Albujón wadi include organic particles, dissolved inorganic nitrogen (mainly as nitrate), and phosphorus derived from agriculture (Velasco et al., 2006). Zone 4 (Z4; Beal dry riverbed, 3 samples), the Beal River is one of the main collectors of the Cartagena-La Unión mine and is polluted with heavy metals. Zone 5 (Z5; Playa del Vivero, 3 samples) is located in the southern part of the lagoon. This area is surrounded by hotels and restaurants and is influenced mainly by tourism. At each sampling zone, surface sediments (5-10 cm) were collected from 3-4 different locations with the help of a sediment core sampler (3 cm diameter). All the samples were promptly transferred into sterile plastic bags and stored in boxes containing dry ice, maintaining temperatures below -20°C for further analysis. After being transported to the laboratory, the sediments were homogenized completely and stored at -80°C.
DNA extraction and 16S rRNA Illumina Novaseqalanala sequencing
The total DNA of the 18 sediment samples (250 mg) was extracted with the DNeasy PowerSoil Pro Kit (QIAGEN), according to the manufacturer’s instructions. DNA concentration was calculated with a Qubit™ fluorometer using the dsDNA High Sensitivity (HS; ThermoFisher) kit. The V3–V4 region of the microbial 16S rRNA gene was amplified using primers 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′). The PCR products with the proper size were selected by 2% agarose gel electrophoresis. The same amount of PCR products from each sample were pooled, end-repaired, A-tailed, and further ligated with Illumina adapters. Libraries were sequenced using Novaseq 6000 Illumina platform to generate 250bp paired-end raw reads. The DNA sequencing and library preparation were carried out by the Novogene (Cambridge, UK) Sequencing Centre. The sequencing process resulted in more than 2,280,000 raw reads of V3-V4 amplicons from 18 samples (Supplementary Table S1). After filtering out low-quality reads with Q values < 10 and N > 10% and chimera filtering, a total of 1,353,236 high-quality sequences with an average read length of around 418bp were obtained (Supplementary Table S1).
Bioinformatic and statistical analysis
Paired-end reads were assigned to samples based on their unique barcodes and truncated by cutting off the barcode and primer sequences. FLASH v1.2.1 (Magoč and Salzberg, 2011) was used to merge the reads to get raw tags. High quality clean tags were obtained using FASTP v0.20.0 (Chen et al., 2018). To obtain effective tags, chimera tags were removed with Vsearch v2.15.0 (Rognes et al., 2016). The DADA2 module (Callahan et al., 2016) in QIIME 2 (v2020.6) was used for denoising effective tags, and the sequences with a frequency below 5 were filtered out to obtain the final Amplicon Sequence Variables (ASV). Subsequently, the species assignment of each ASV was performed with the q2-feature-classifier plugin (Bokulich et al., 2018) QIIME 2’s (v2020.6). Core microbiome analysis was performed using the Microbiome (Lahti and Shetty, 2019) package (v1.18.0) in R software (v4.2.1) using Rstudio (v2022.02.2). Alpha and beta diversity indices were calculated with the Phyloseq package v1.40.0 (McMurdie and Holmes, 2013) and PERMANOVA analysis was performed using the Vegan package v2.6.4 (Oksanen et al., 2020) in R software (v4.2.1) using Rstudio (v2022.02.2). To normalize the data, the absolute frequency of ASVs in all samples was standardized based on the sequence count of the sample with the lowest number of sequences. LEfSe analysis was performed by LEfSe software v1.0 (Segata et al., 2011).
Results and discussion
Summary 16S rRNA data and microbial diversity
A total of 15,117 ASVs were identified in the 18 samples analyzed (Supplementary Table S2). The rarefaction curves showed that the number of ASVs reached the saturation plateau at ~20K sequences, suggesting adequate sampling effort at the five sampling stations (Supplementary Figure S2). The abundance of ASVs in the sediment samples in Z5 (965-1540) was significantly lower than in the other sampling zones (1573 - 2854; p < 0.05) (Supplementary Table S1). Comparison of alpha diversity values showed that there were differences in Chao1 and Shannon indices between Z5 and the other zones (Figures 1A, B). Both alpha diversity indices of Z1-Z4 were significantly (p < 0.05) higher than those of Z5. In addition, Z4 had the highest Shannon diversity index, indicating a greater diversity of microbial communities in this zone compared to the other regions (Figure 1B).
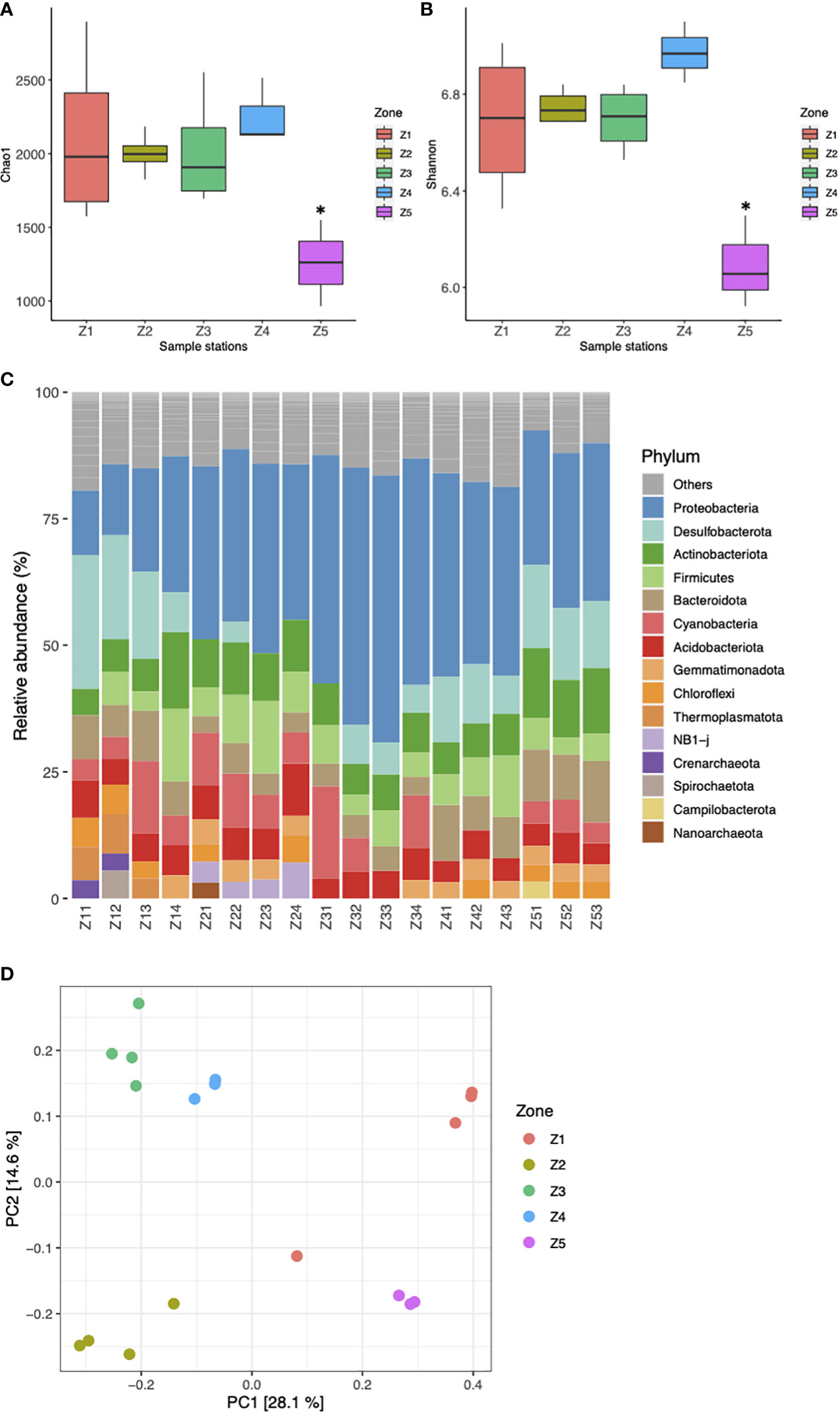
Figure 1 Comparison of sediment bacterial composition between sampling stations in the Mar Menor. Boxplots with alpha diversity metrics for the five sampling stations (Z1–Z5) calculated with Chao1 (A) and Shannon index (B), bars are colored by sampling site (*Kruskal-Wallis test, p-value 0.05). (C) Relative abundance (%) of bacterial and archaeal top phyla. The category ‘Others’ accounted for phyla that were represented by < 2.5% abundance. (D) PCoA of Bray-Curtis distances between all samples, colored by sampling station (Z1-Z5).
Microbial community composition
A total of 74 bacterial phyla were identified at the 18 sampling sites. The predominant bacterial phyla were Proteobacteria (36.1%), Desulfobacterota (13.1%), Actinobacteriota (12.7%), Firmicutes (6.5%), Bacteroidota (4.7%), and Cyanobacteria (4.2%), which accounted for 77.9% of the total sequences in all sediment samples (Figure 1C). In contrast to the bacterial kingdom, only 2.27% of the sequences (11 phyla) were identified as archaea, dominated by the phyla Thermoplasmatota (1.2%) and Nanoarchaeota (0.7%; Figure 1C). The relative abundance of the 3 main phyla, Proteobacteria, Desulfobacterota and Actinobacteria was significantly different among the 18 sampling sites. Z3 (48.9%) and Z2 (17.5%) had the highest abundance of phyla Proteobacteria and Actinobacteria, respectively, while the relative abundance of Desulfobacterota was highest in both Z5 (21.7%) and Z1 (19.8%; Figure 1C). Previous studies of coastal and estuarine sediments affected by contaminants from anthropogenic activities such as agriculture, industry and urban wastewater have consistently identified Proteobacteria, Desulfobacterota, Actinobacteriota and Bacteroidota as the most abundant phyla (Mahmoudi et al., 2015; Quero et al., 2015; Guo et al., 2018). Consistent with these observations, a recent study identified a similar pattern in the sediment of Mar Menor, with Proteobacteria, Bacteroidota, and Desulfobacterota being the most abundant prokaryotic phyla (Aldeguer-Riquelme et al., 2022). The prevalence of these phyla in contaminated sediments seems to be related to their capacity to tolerate and metabolize various contaminants. Proteobacteria play an important role in degrading organic materia and hydrocarbon compounds (Liang et al., 2023) while members of Desulfobacterota are critically involved in the anaerobic degradation of organic pollutants and the cycling of sulfur compounds (Gillan et al., 2005; Quero et al., 2015). Actinobacteriota and Bacteridoidota are known to metabolize contaminants like hidrocarbons and PCBs and to tolerate heavy metals. (Zhang et al., 2008; Nogales et al., 2011; Zanaroli et al., 2012; Quero et al., 2015). Overall, these results indicate a considerable influence of pollutants on the microbial community in the sediment of the Mar Menor.
Analysis of the core microbiome showed that 7 classes and 4 families co-occurred in the 18 samples at the minimum detection threshold of 0.01% (Supplementary Figure S3). Accordingly, the core microbiome of the Mar Menor sediment was formed by the classes Gammaproteobacteria, Acidimicrobiia, Alphaproteobacteria, Bacteroidia, Clostridia, Polyangia, and Thermoanaerobaculia, and the families Woeseiaceae, Rhodobacteraceae, Ilumatobacteraceae, as well as Thermoanaerobaculaceae. Previous studies in contaminated coastal areas in Italy and Australia have also found the presence of Gammaproteobacteria, Acidimicrobiia, Alphaproteobacteria, Bacteroidia, and Clostridia in the core microbiome (Sun et al., 2013; Quero et al., 2015). Interestingly, to our knowledge, this is the first time that Polyangia and Thermoanaerobaculia clades have been found as part of a core microbiome in coastal sediments.
Prokaryotic structure of the sediment microbial communities and taxonomic biomarkers
The Principal Coordination Analysis (PCoA) based on Bray–Curtis distance (Figure 1D) showed the dissimilarity of bacterial communities among sampling sites, and it was significant as revealed by a PERMANOVA test (R2 = 0.426, p < 0.001; Supplementary Table S3). The sediment samples from Z4 and Z5 were highly clustered, while the samples from Z1 showed the greatest dissimilarity in composition. PCoA clearly distinguished Z1, Z2 and Z5 from the sampling sites near the estuaries, Z3 and Z4.
To find more detailed differences in the taxonomic composition of the 5 microbial communities associated with the 5 sampling zones, a Linear discriminant analysis Effect Size (LEfSe) was performed using phylum-to-species level data. This analysis allowed us to identify discriminating bacterial and archaeal taxa from marine sediments that could serve as ecological biomarkers. The LEfSe of all species found in the five sediment sampling zones (Z1, Z2, Z3, Z4 and Z5) were analyzed together, resulting in 26 taxa with significant differences (LDA Score > 10,000; Figure 2). Members of Desulfobacterota (class Desulfobacteria, order Desulfobacterales, family Desulfosarcinaceae) and Chloroflexi phylum, Thiotrichales order (family Thiotrichaceae) and the uncultivated family CG2-30-50-142 (phylum Actinobacteriota) were identified as bacterial biomarkers of the Z1. Interestingly, Z1 was the only microbial community showing an Archaea biomarker, as it was enriched in the Thermoplasmata class. Z2 was enriched with members of the Acidobacteriota and the non-cultivated phyla NB1-J, the classes Acidimicrobiia and Alphaproteobacteria as well as the order Rhizobiales. In the case of Z3, most bacterial taxa identified as bacterial biomarkers belong to the phylum Proteobacteria, including the class Gammaproteobacteria and the order Steroidobacterales (family Woeseiaceae), while in Z4 only two bacterial taxa were consistently enriched, the phylum Bacteroidia and the order Rhodobacterales (family Rhodobacteraceae). Finally, the LEfSe analysis in Z5 shows a significant enrichment in members of the phylum Desulfobacterota (class Desulfobulbia, order Desulfobulbales, class Desulfocapsacea), the order Chromatiales (family Chromatiaceae), the class Bacteroida and the genus Desulfosarcina (order Desulfobacterales).
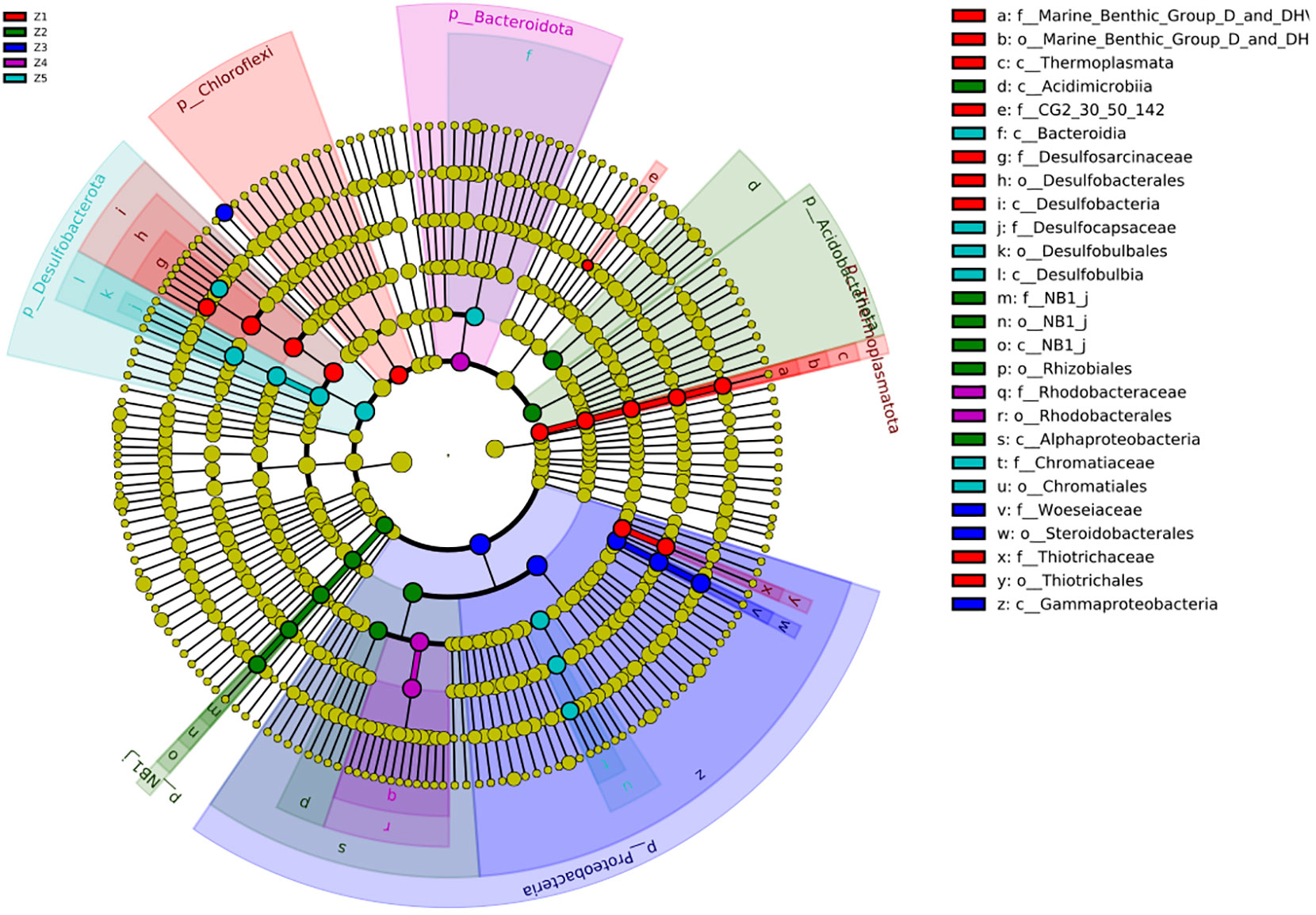
Figure 2 Circular LEfSe cladogram showing the phylogenetic distribution of microbial lineage in the five sampling regions. Linages with LDA values of 3.0 or higher are indicated. The dot in the center represents phylum-level ASVs, while the outer circle of dots denotes genus-level ASVs. The order, family, and class that differ significantly between two groups are indicated in the upper right corner with corresponding color codes.
Conclusions
In this study, we investigated microbial diversity and community structure by 16S metagenomic sequencing of the sediment-associated microbiota in the Mar Menor coastal lagoon. We identified 74 bacterial phyla, with Proteobacteria, Desulfobacterota, Actinobacteriota, and Bacteroidota being the most represented. The occurrence of lineages belonging to the Archaea domain was quite limited, accounting for only 2.25% of the sequences. The southern region of the coastal lagoon (Z5), which is most strongly influenced by local tourism, showed the lowest ecological diversity values. The Mar Menor sediment’s core microbiome comprised seven classes and four families. The LefSE analysis revealed a strong correlation between the phyla Acidobacteria, NB1_J, Chloroflexi, and Thermoplasmatota with the north, while the phylum Bacteroidota emerging as an ecological indicator for the southern region. The findings from this investigation will prove useful for broadening our understanding of microbial communities in lagoon sediments and for future efforts aimed at promoting the sustainability of this significant ecosystem.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA961079, SRA NCBI.
Author contributions
JG-L: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Project administration, Writing – original draft, Writing – review & editing. JH-C: Funding acquisition, Resources, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the PADI Foundation [project 75263 to JG-L] and the National Programme for Fostering Excellence in Scientific and Technical Research [PID2021-127210NB-I00 MCIU/AEI/FEDER, UE to JH-C and JG-L].
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1319961/full#supplementary-material
References
Aldeguer-Riquelme B., Rubio-Portillo E., Álvarez-Rogel J., Giménez-Casalduero F., Otero X. L., Belando M.-D., et al. (2022). Factors structuring microbial communities in highly impacted coastal marine sediments (Mar Menor lagoon, SE Spain). Front. Microbiol. 13. doi: 10.3389/fmicb.2022.937683
Álvarez-Rogel J., Barberá G. G., Maxwell B., Guerrero-Brotons M., Díaz-García C., Martínez-Sánchez J. J., et al. (2020). The case of Mar Menor eutrophication: State of the art and description of tested Nature-Based Solutions. Ecol. Eng. 158, 106086. doi: 10.1016/j.ecoleng.2020.106086
Barbier E. B., Hacker S. D., Kennedy C., Koch E. W., Stier A. C., Silliman B. R. (2011). The value of estuarine and coastal ecosystem services. Ecol. Monogr. 81, 169–193. doi: 10.1890/10-1510.1
Behera P., Mahapatra S., Mohapatra M., Kim J. Y., Adhya T. K., Raina V., et al. (2017). Salinity and macrophyte drive the biogeography of the sedimentary bacterial communities in a brackish water tropical coastal lagoon. Sci. Total Environ. 595, 472–485. doi: 10.1016/j.scitotenv.2017.03.271
Bokulich N. A., Kaehler B. D., Rideout J. R., Dillon M., Bolyen E., Knight R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 90. doi: 10.1186/s40168-018-0470-z
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chen S., Zhou Y., Chen Y., Gu J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Conesa H. M., Jiménez-Cárceles F. J. (2007). The Mar Menor lagoon (SE Spain): A singular natural ecosystem threatened by human activities. Mar. pollut. Bull. 54, 839–849. doi: 10.1016/j.marpolbul.2007.05.007
Dash H. R., Mangwani N., Chakraborty J., Kumari S., Das S. (2013). Marine bacteria: potential candidates for enhanced bioremediation. Appl. Microbiol. Biotechnol. 97, 561–571. doi: 10.1007/s00253-012-4584-0
Gillan D. C., Danis B., Pernet P., Joly G., Dubois P. (2005). Structure of sediment-associated microbial communities along a heavy-metal contamination gradient in the marine environment. Appl. Environ. Microbiol. 71, 679–690. doi: 10.1128/AEM.71.2.679-690.2005
Guo X.-P., Lu D.-P., Niu Z.-S., Feng J.-N., Chen Y.-R., Tou F.-Y., et al. (2018). Bacterial community structure in response to environmental impacts in the intertidal sediments along the Yangtze Estuary, China. Mar. pollut. Bull. 126, 141–149. doi: 10.1016/j.marpolbul.2017.11.003
Jackson C. R., Vallaire S. C. (2009). Effects of salinity and nutrients on microbial assemblages in Louisiana wetland sediments. Wetlands 29, 277–287. doi: 10.1672/08-86.1
Kjerfve B. (1994). “Chapter 1 Coastal Lagoons,” in Elsevier Oceanography Series. Ed. Kjerfve B. (Elsevier), 1–8. doi: 10.1016/S0422-9894(08)70006-0
Lahti L., Shetty S. (2019) Tools for Microbiome Analysis in R. Available at: http://microbiome.github.com/microbiome.
Liang S., Li H., Wu H., Yan B., Song A. (2023). Microorganisms in coastal wetland sediments: a review on microbial community structure, functional gene, and environmental potential. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1163896
Magoč T., Salzberg S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mahmoudi N., Robeson M. S., Castro H. F., Fortney J. L., Techtmann S. M., Joyner D. C., et al. (2015). Microbial community composition and diversity in Caspian Sea sediments. FEMS Microbiol. Ecol. 91, 1–11. doi: 10.1093/femsec/fiu013
McMurdie P. J., Holmes S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8, e61217. doi: 10.1371/journal.pone.0061217
Nogales B., Lanfranconi M. P., Piña-Villalonga J. M., Bosch R. (2011). Anthropogenic perturbations in marine microbial communities. FEMS Microbiol. Rev. 35, 275–298. doi: 10.1111/j.1574-6976.2010.00248.x
Nzila A. (2013). Update on the cometabolism of organic pollutants by bacteria. Environ. pollut. 178, 474–482. doi: 10.1016/j.envpol.2013.03.042
Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D. (2020) Vegan Community Ecology Package. Available at: https://cran.r-project.orghttps://github.com/vegandevs/vegan.
Parkes R. J., Cragg B., Roussel E., Webster G., Weightman A., Sass H. (2014). A review of prokaryotic populations and processes in sub-seafloor sediments, including biosphere:geosphere interactions. Mar. Geol. 352, 409–425. doi: 10.1016/j.margeo.2014.02.009
Pérez-Ruzafa A., Campillo S., Fernández-Palacios J. M., García-Lacunza A., García-Oliva M., Ibañez H., et al. (2019). Long-term dynamic in nutrients, chlorophyll a, and water quality parameters in a coastal lagoon during a process of eutrophication for decades, a sudden break and a relatively rapid recovery. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00026
Pérez-Ruzafa A., García-Charton J. A., Barcala E., Marcos C. (2006). Changes in benthic fish assemblages as a consequence of coastal works in a coastal lagoon: The Mar Menor (Spain, Western Mediterranean). Mar. pollut. Bull. 53, 107–120. doi: 10.1016/j.marpolbul.2005.09.014
Pérez Ruzafa Á., Marcos Diego C., Gilabert Cervera F. J. (2004) Coastal lagoons: Ecosystem processes and modeling for sustainable use and development. (FL: CRC Press). Available at: https://repositorio.upct.es/handle/10317/10309 (Accessed March 7, 2023).
Pérez-Ruzafa A., Pérez-Marcos M., Marcos C. (2020). Coastal lagoons in focus: Their environmental and socioeconomic importance. J. Nat. Conserv. 57, 125886. doi: 10.1016/j.jnc.2020.125886
Quero G. M., Cassin D., Botter M., Perini L., Luna G. M. (2015). Patterns of benthic bacterial diversity in coastal areas contaminated by heavy metals, polycyclic aromatic hydrocarbons (PAHs) and polychlorinated biphenyls (PCBs). Front. Microbiol. 6. doi: 10.3389/fmicb.2015.01053
Rodríguez J., Gallampois C. M. J., Timonen S., Andersson A., Sinkko H., Haglund P., et al. (2018). Effects of organic pollutants on bacterial communities under future climate change scenarios. Front. Microbiol. 9, 2926. doi: 10.3389/fmicb.2018.02926
Rognes T., Flouri T., Nichols B., Quince C., Mahé F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584. doi: 10.7717/peerj.2584
Ruiz-Fernández J., León V. M., Marín-Guirao L. (2019). Synthesis report of the current state of Mar Menor lagoon and its causes in relation to the nutrient contents. Technical report. doi: 10.13140/RG.2.2.20978.04808
Schubert H., Telesh I. (2017). “Estuaries and coastal lagoons,” in Biological Oceanography of the Baltic Sea. Eds. Snoeijs-Leijonmalm P., Schubert H., Radziejewska T. (Dordrecht: Springer Netherlands), 483–509. doi: 10.1007/978-94-007-0668-2_13
Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60. doi: 10.1186/gb-2011-12-6-r60
Sun M. Y., Dafforn K. A., Johnston E. L., Brown M. V. (2013). Core sediment bacteria drive community response to anthropogenic contamination over multiple environmental gradients. Environ. Microbiol. 15, 2517–2531. doi: 10.1111/1462-2920.12133
Velasco J., Lloret J., Millan A., Marin A., Barahona J., Abellan P., et al. (2006). Nutrient and particulate inputs into the mar Menor lagoon (Se Spain) from an intensive agricultural watershed. Water Air Soil pollut. 176, 37–56. doi: 10.1007/s11270-006-2859-8
Velasco A. M., Pérez-Ruzafa A., Martínez-Paz J. M., Marcos C. (2018). Ecosystem services and main environmental risks in a coastal lagoon (Mar Menor, Murcia, SE Spain): The public perception. J. Nat. Conserv. 43, 180–189. doi: 10.1016/j.jnc.2017.11.002
Zamora-López A., Guerrero-Gómez A., Torralva M., Zamora-Marín J. M., Guillén-Beltrán A., López-Martínez de la Plaza P., et al. (2022). Fish assemblage response to eutrophic-mediated environmental stress events in the mar menor coastal lagoon (SE of Spain). Biol. Life Sci. Forum 13, 5. doi: 10.3390/blsf2022013005
Zanaroli G., Negroni A., Vignola M., Nuzzo A., Shu H.-Y., Fava F. (2012). Enhancement of microbial reductive dechlorination of polychlorinated biphenyls (PCBs) in a marine sediment by nanoscale zerovalent iron (NZVI) particles. J. Chem. Technol. Biotechnol. 87, 1246–1253. doi: 10.1002/jctb.3835
Zhang W., Ki J.-S., Qian P.-Y. (2008). Microbial diversity in polluted harbor sediments I: Bacterial community assessment based on four clone libraries of 16S rDNA. Estuar. Coast. Shelf Sci. 76, 668–681. doi: 10.1016/j.ecss.2007.07.040
Keywords: Mar Menor (SE Spain), coastal lagoon, coastal sediments, microbial communities, ecological biomarkers
Citation: Giner-Lamia J and Huerta-Cepas J (2024) Exploring the sediment-associated microbiota of the Mar Menor coastal lagoon. Front. Mar. Sci. 11:1319961. doi: 10.3389/fmars.2024.1319961
Received: 11 October 2023; Accepted: 02 January 2024;
Published: 23 January 2024.
Edited by:
Katherine Dafforn, Macquarie University, AustraliaReviewed by:
Giulia Filippini, Macquarie University, AustraliaJurica Zucko, University of Zagreb, Croatia
Copyright © 2024 Giner-Lamia and Huerta-Cepas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joaquín Giner-Lamia, amdpbmVyMUB1cy5lcw==; Jaime Huerta-Cepas, ai5odWVydGFAY3NpYy5lcw==
†Present address: Joaquín Giner-Lamia, Departamento de Bioquímica Vegetal y Biología Molecular, Facultad de Biología, Universidad de Sevilla, Sevilla, Spain
Instituto de Bioquímica Vegetal y Fotosíntesis (IBVF), Universidad de Sevilla-CSIC, Sevilla, Spain
 Joaquín Giner-Lamia
Joaquín Giner-Lamia Jaime Huerta-Cepas1*
Jaime Huerta-Cepas1*