- 1Ocean Georesources Research Department, Korea Institute of Ocean Science and Technology, Busan, Republic of Korea
- 2BK21 Center for Precision Medicine & Smart Engineering, Inha University, Incheon, Republic of Korea
- 3Program in Biomedical Science and Engineering, Inha University, Incheon, Republic of Korea
- 4Department of Ocean Sciences, Inha University, Incheon, Republic of Korea
- 5Marine Animal Research and Conservation, Jeju, Republic of Korea
- 6Graduate School of Informatics, Kyoto University, Kyoto, Japan
- 7Golden Honu Services of Oceania, Honolulu, HI, United States
- 8Department of Life Sciences, Yeungnam University, Gyeongsan, Republic of Korea
- 9College of Ocean Sciences, Jeju National University, Jeju, Republic of Korea
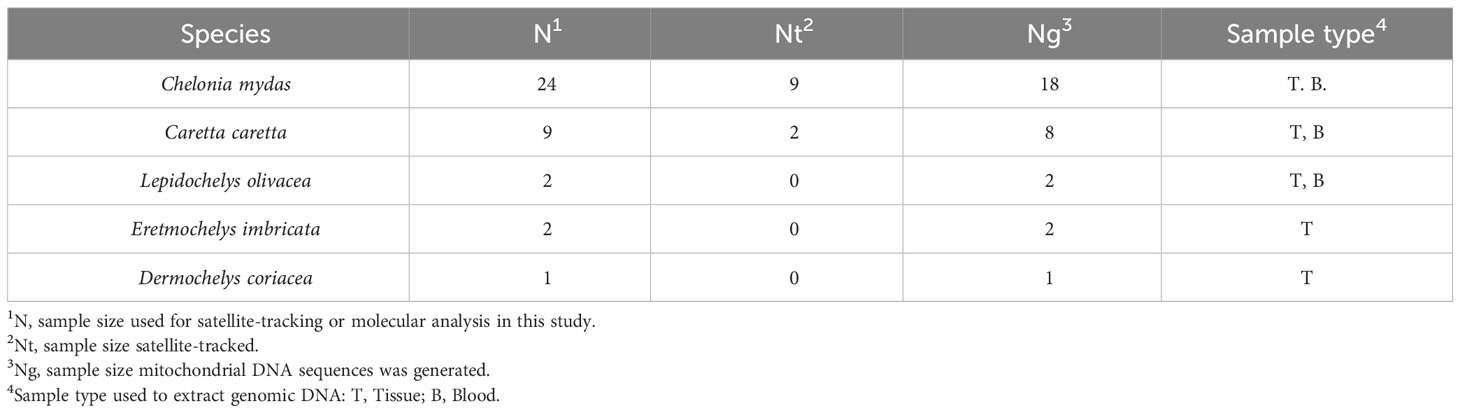
The northwestern Pacific region is an important habitat for sea turtles, hosting five species out of seven. There is still limited information available about the sea turtle aggregations around the Korean Peninsula, which is the northern boundary for many sea turtle species in the western Pacific area. The present study aims to investigate the migratory route of sea turtles visiting Jeju Island. Five species of sea turtles were identified from by-catch and stranding data between 2013 and 2022 on Jeju Island in Korea: green (Chelonia mydas; 24 individuals), loggerhead (Caretta caretta; 9), hawksbill (Eretmochelys imbricata; 2), olive ridley (Lepidochelys olivacea; 2), and leatherback (Dermochelys coriacea; 1). Mixed stock analysis using mitochondrial DNA haplotypes revealed that Jeju green turtles primarily originate from the rookeries of the Japanese Archipelago. This connectivity between two regions was also supported by the similar genetic composition of loggerhead turtles. Similarly, satellite tracking data showed that several green turtles originating from Jeju Island migrated to waters near the Ryukyu Archipelago in Japan. Nevertheless, about 60% of the tracked green turtles stayed near Jeju Island, with most overwintering there, indicating the long residency in Jeju Island. This study also provides the genetic sequences of other three species including new orphan haplotypes of hawksbill and olive ridley turtles. Our findings suggest that Jeju Island serves as a stable foraging habitat and provide insight into understanding the habitat range of sea turtles in the western Pacific.
1 Introduction
Sea turtle populations experienced a major decline worldwide in the past due to anthropogenic activities, such as bycatch (Lewison et al., 2004), coastal development (Fuentes et al., 2020), and habitat change resulting from global warming (Reece et al., 2013). Currently, most species are classified as vulnerable, endangered, or critically endangered (IUCN, 2017). Globally, fundamental investigations encompassing the habitat ranges of sea turtles, spatial and seasonal migratory patterns, and changes in population size at habitat sites have been ongoing to design effective conservation strategies. As a result of this effort, recent research confirmed increasing trends in population abundance for specific sea turtle species at nesting sites in various global management regions (Mazaris et al., 2017). Furthermore, Mazaris et al. (2017) demonstrated the importance of long-term monitoring for accurately detecting abundance trends. Nonetheless, most of these investigations to date have focused on specific regions, leaving some areas poorly studied. To advance conservation strategies, it is a priority to fill knowledge gaps in underrepresented sites and develop strategies at both local and regional scales.
Genetic data, particularly genetic variation and population structure information derived from mitochondrial DNA sequences, have been an important foundation for the establishment of conservation units for a range of sea turtle management approaches including Evolutionary Significant Units (ESUs), Distinct Population Segments (DPSs), Regional Management Units (RMUs), and Management Units (MUs) (Komoroske et al., 2017). All of these conservation units are premised on shared genetic variation within a population segment, though their designations differ spatially and temporally (USFWS and NMFS, 1996; Wallace et al., 2010). To define conservation units more precisely, the sampling of turtle foraging grounds is required, which would allow the connectivity between foraging aggregates and nesting populations to be understood. The Bayesian approach, Mixed Stock Analysis (MSA), has been employed as the primary method to identify the major contributing rookery populations to the foraging aggregate, based on relative genotype frequency (Pella and Masuda, 2001).
Jeju Island on the Korean Peninsula is the northern boundary for most sea turtle species inhabiting the western Pacific region and has been recognized as a foraging ground for green (Chelonia mydas) and loggerhead turtles (Caretta caretta) (Jung et al., 2012b; Jang et al., 2018). To date, all five species of sea turtles reported in the northwestern Pacific have also been observed in the waters around the Korean Peninsula: green, loggerhead, hawksbill (Eretmochelys imbricata), olive ridley (Lepidochelys olivacea), and leatherback turtles (Dermochelys coriacea) (Kim et al., 2017; Kim et al., 2022). Satellite tracking data of green turtles suggest that the aggregation off Jeju originates from neighboring rookeries in Japan, Taiwan, and China (Moon et al., 2011; Jang et al., 2018). On the other hand, Jeju island also has suitable physical conditions for nesting and hatchling survival, and nesting behaviors of sea turtles were recorded in the past despite occasional events (Jung et al., 2012a). There are substantial gaps in the understanding of the ecology and habitat range of sea turtles around Jeju Island. To fill the gaps, comprehensive investigations are needed into their genetic features and migration patterns. As of now, mitochondrial sequences for sea turtles on the Korean Peninsula have been officially reported for only a few individuals of hawksbill, olive ridley, and leatherback turtles (Cho et al., 2018). In the northwestern Pacific region, there have been few studies on these species despite regular observations (Hamann et al., 2006; Chan et al., 2007; Nishizawa et al., 2012; Fukuoka et al., 2019; Ng and Matsuzawa, 2021).
In this study, we analyzed the genetic composition of five sea turtle species collected around Jeju Island and explored the population connectivity to the neighboring rookeries to identify their natal origin. Additionally, we traced migratory routes for several green and loggerhead turtles using satellite tracking. This comprehensive analysis based on multidisciplinary data enhances the understanding of migration patterns in the northwestern Pacific region; in addition, the DNA sequences obtained for hawksbill, olive ridley, and leatherback turtles contribute to filling the sampling gap in the northwestern Pacific allowing more informed assumptions to be made about their movements.
2 Materials and methods
2.1 Sampling and sequencing of genomic DNAs
A total of 38 stranded dead or bycaught sea turtles were collected on and around Jeju Island between 2013 and 2022 (Figure 1, Table 1, Supplementary Table 1). Of these, 15 turtles (4 loggerheads, 10 greens, and 1 olive ridley) were captured alive, while the rest were found dead. For most of dead turtles, the carcass condition was considered as fresh or slightly decomposed (fresh dead or early moderately decomposed state described in Reneker et al. (2018); decomposition code 1 or 2, respectively, described in Santos et al. (2018)). Based on the time carcass decomposition takes to proceed (1–8 days estimated in Santos et al. (2018), we considered these turtles to be from the same stock as the bycaught turtles rather than having been transported from other stocks.
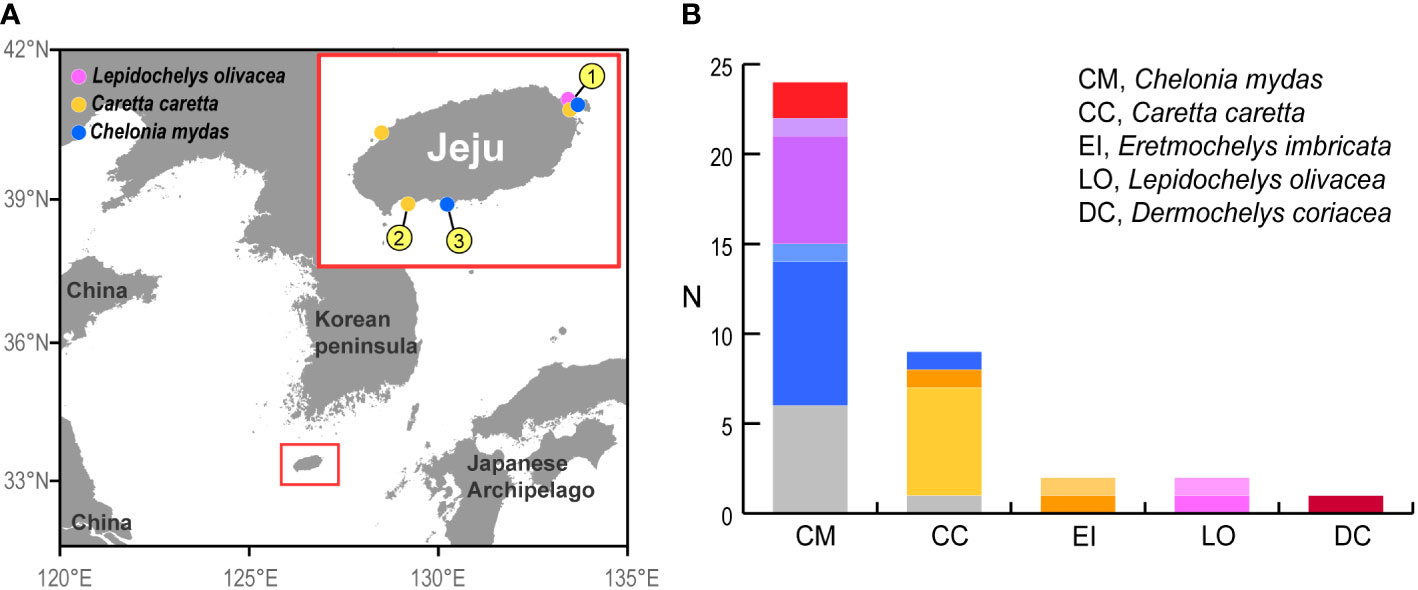
Figure 1 Location of Jeju Island and the samples employed in the present study. (A) Map of the Korean Peninsula and the surrounding seas with an enlarged map of Jeju Island. The colored circles represent the locations where bycaught turtles were captured, and the numbers represent the capture locations of turtles tracked by satellite, as detailed in Table 2. (B) Sample size for each turtle species analyzed in this study. Individuals with distinct haplotypes are differentiated by color.
Genomic DNA was extracted from the tissue or blood of 32 turtles, with a DNA sample from one satellite-tagged green turtle tracked (Tracking ID: 53759) in a previous study (Supplementary Table 1) (Jang et al., 2018). Six bycaught sea turtles (Tracking IDs: 718336, 710313, 710314, 710315, 724924, and 724925) were excluded from the sequencing analysis due to absence of samples. For the bycaught specimens, tissue samples were taken from the soft part behind the hind flipper when a tag was placed and blood samples were obtained from the bilateral cervical sinus, following the protocol recommended by NMFS-SEFSC (2008). Stranded dead specimens were stored at –80°C until fixation. All tissue samples were preserved in 99% ethanol for transportation to the laboratory and subsequently stored at –20°C until DNA extraction. Genomic DNA was extracted using a QIAamp Fast DNA Tissue Kit (Qiagen, Inc., Hilden, Germany) and a DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA) for tissue and blood samples, respectively. The mitochondrial control region was amplified using the primers LCM15382 and H950 (Abreu-Grobois et al., 2006). The PCR mixture consisted of IP-Taq PCR Master mix (Cosmogenetech, Seoul, Korea) with 1 μl of genomic DNA, 10 pmoles of each forward and reverse primer, and distilled water adjusted to a final volume of 20 μl. Thermocycling was conducted under the following conditions: 5 min at 94°C for initial denaturation; 35 cycles including 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C; and 7 min at 72°C for the final extension. Raw sequences were generated using a Big Dye Terminator V3.1 Cycle Sequencing kit on an Applied Biosystems 3730xl DNA Analyzer (Applied Biosystems Inc, South Korea) and modified manually using Geneious Prime v. 2022.2. The sequence of a tracked green turtle (ID 724927) was omitted from the subsequent analysis due to the presence of an ambiguous site.
2.2 Molecular analysis
The mitochondrial DNA sequences varied in length depending on the species but were at least 700 bp. The nomenclature of the mitochondrial sequences was initially identified by comparing with databases of NCBI GenBank using BLAST searches, and the results were subsequently compared with published data specific to each species. The sequence lengths for subsequent molecular analysis were adjusted to be either long or short based on the haplotype datasets available for each species and the specific aims of the analysis. For both long and short fragments, the lengths were adjusted to produce overlapping fragments after alignment using MUSCLE v. 2.8.425 (Edgar, 2004) in Geneious Prime. Haplotype networks were constructed based on the median-joining algorithm using PopART v. 1.7 (Leigh and Bryant, 2015). Indel sites were not considered in this analysis. The estimation of genetic diversity and the exact test of population differentiation in the following analyses were conducted using Arlequin v. 3.5 (Excoffier and Lischer, 2010).
For green turtles, mitochondrial haplotype data based on short sequences (<400 bp) were compared with the data from nesting and foraging aggregations in the northwestern Pacific RMU (Wallace et al., 2010) including the Japanese Archipelago, Taiwan and South China region (Nishizawa et al., 2011; Nishizawa et al., 2013; Hamabata et al., 2014; Ng et al., 2014; Nishizawa et al., 2014b; Hamabata et al., 2015; Ng et al., 2017; Hamabata et al., 2018) (Supplementary Table 2). The haplotypes were clustered in accordance with the phylogenetic clade defined in Jensen et al. (2019) and the haplotype network reconstructed in this study (Supplementary Figure 1).
Mixed-stock analysis (MSA) was conducted to determine the natal origin of the green turtles around Jeju Island using the MIXSTOCK package in R via Bayesian approach (Bolker et al., 2003). This MSA was conducted using haplotype data from short fragments in accordance with the available reference data. We included potential source rookeries in the north and central western Pacific following the method outlined by Hamabata et al. (2018). Haplotype data for rookeries were derived from Dethmers et al. (2006); Cheng et al. (2008); Nishizawa et al. (2011); Nishizawa et al. (2013); Dutton et al. (2014a; 2014b); Hamabata et al. (2014); Ng et al. (2014); Jensen et al. (2016); Gaillard et al. (2020); Song et al. (2022), and Li et al. (2023). According to the MIXSTOCK protocol, unique haplotypes in mixed populations were excluded, and haplotypes found only in a single population were lumped (Pella and Masuda, 2001). Each run consisted of the 100,000 MCMC iterations with a burn-in period of 10,000 steps. MCMC convergence was diagnosed using a Gelman-Rubin criterion (Gelman et al., 1996). Haplotype frequency of green turtles around Jeju Island was compared with nearby Japanese rookeries by the exact test. Taking into account the genetic subdivision patterns reported by Hamabata et al. (2014) and Nishizawa et al. (2011) and the geographical distance, we divided the Japanese rookeries into four separate populations: Central Ryukyus, Ishigaki, southwestern Iriomote in Yaeyama, and Ogasawara.
The sequences from longer fragments (>700 bp) were utilized to construct a haplotype network for Jeju green turtles and to estimate the genetic diversity, which was compared with neighboring foraging aggregations to improve the accuracy of the estimation based on the greater amount of genetic variation (Supplementary Table 3).
For loggerhead turtles, we did not use MSA due to the small sample size. Long sequences of mitochondrial fragments (>800 bp) were used to construct a haplotype network for Jeju loggerhead turtles and to analyze the exact test between Jeju Island and nearby nesting aggregations in the Japanese Archipelago, which are the only reported nesting places in the same RMU, north Pacific (Matsuzawa et al., 2016) (Supplementary Table 4). Genetic diversity was estimated based on both short sequences (Bowen et al., 1995; Nishizawa et al., 2014a) and long fragments because comparable foraging aggregations were limited to a single location (Sanriku in the Japanese Archipelago) when using long fragments only.
For the remaining three species, mitochondrial sequences were compared with previously reported haplotype data from the Pacific region, given the lack of defined RMUs. For hawksbill turtles, the long sequences (>700 bp) were compared to haplotypes from the western Pacific (Nishizawa et al., 2010; Nishizawa et al., 2012; Nishizawa et al., 2016; Vargas et al., 2016; Kim et al., 2020) and the eastern Pacific (Zuñiga-Marroquin and Espinosa de los Monteros, 2017; Gaos et al., 2018; Gaos et al., 2020) to identify the nomenclature of newly obtained sequences and compare the regional haplotype frequency (Supplementary Table 5). Haplotypes were distinguished according to the phylogenetic clades in Arantes et al. (2020). Shorter sequences (~500 bp) were used to compare the haplotypes with the prefix ‘EIJ’ reported in the northwestern Pacific.
For olive ridley turtles, the genetic sequences from Jeju were identified based on comprehensively rearranged haplotype data (> 600 bp) from Vilaça et al. (2022). Haplotype compositions for specific habitats were derived from studies conducted in the western Pacific (Jensen et al., 2013; Kim et al., 2019; Adnyana et al., 2020; Vilaça et al., 2022) and the eastern Pacific (Duchene et al., 2012; Pinou et al., 2018; Campista León et al., 2019; Silver-Gorges et al., 2020; Vilaça et al., 2022; Martín-del-Campo et al., 2023) (Supplementary Table 6). Clustering of these haplotypes followed the divergence patterns identified in Vilaça et al. (2022).
For leatherback turtles, the genetic sequences from Jeju were identified based on both short sequences of 496 bp (Dutton et al., 1999) and long sequences of 763 bp (Yoshikawa et al., 2016; Cho et al., 2018). Haplotype compositions for specific habitats based on a sequence length of 496 bp were derived from studies conducted in the western Pacific (Dutton et al., 2007; Yoshikawa et al., 2016) and the eastern Pacific (Dutton et al., 1999) (Supplementary Table 7). Haplotype data from long fragments (>700 bp) were not used for the comparison because several haplotype sequences available in GenBank remain unpublished.
2.3 Satellite tracking
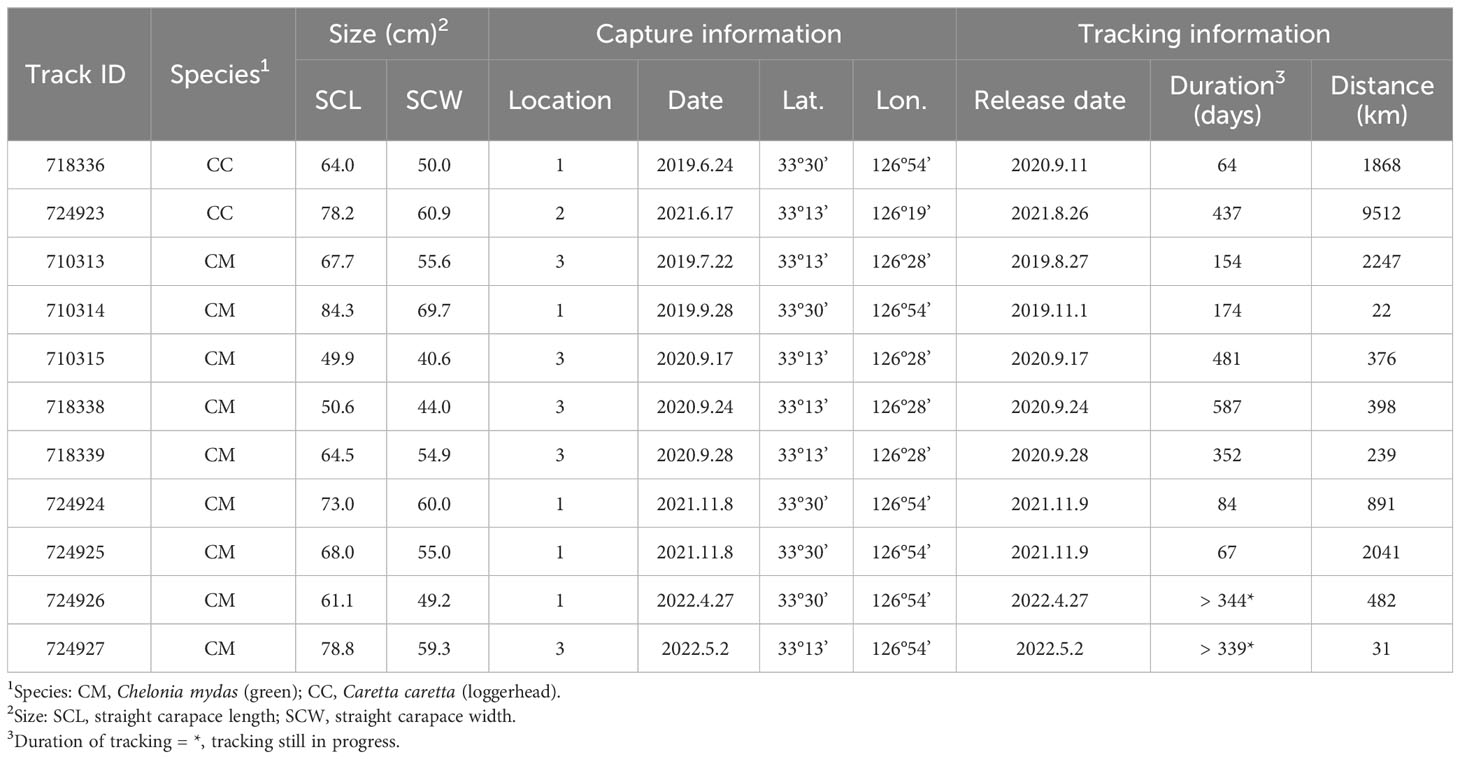
Ten green and two loggerhead turtles were tracked by satellite after being bycaught in pound nets (Table 2), which allowed the turtles to breathe by providing open access to the sea surface (Jang et al., 2018). Following standard legal requirements, the turtles were reported to the marine police of Jeju and visually inspected for potential injury or diseases. Based on the opinion of a veterinarian and the animal rescue institute involved, satellite tracking was only conducted on individuals without health problems. The straight carapace length (SCL) and straight carapace width (SCW) were measured to the nearest 0.1 cm. An Inconel tag provided by the Marine Biodiversity Institute of Korea was attached to the flipper for future identification. An Iridium satellite transmitter (SeaTrkr-4370 or 4375, Telonics, Mesa, AZ, United States) was deployed using polyester resin and glass fiber. The weight of the satellite transmitter (190–210 g) was estimated to not exceed 5% of the body weight of the turtles. Although the actual weights of the turtles were unknown, this estimate was based on a SCL of at least 45 cm and good body condition, indicating sufficient body weight.
Once the resin had hardened, the turtles were released at a location close to their point of capture (Figure 1; Table 2). The iridium satellite transmitter used Quick Fix Pseudoranging (QFP) to transmit GPS positions with a 25 m error range at 1 to 4 hour intervals to the orbiting IRIDIUM satellite system (Jim et al., 2022). The transmitters employed a two-way system to confirm whether the data had been successfully received, with the data automatically retransmitted in order to resolve problems with the original transmission. Both successful and resolved QFP data were plotted on a Google map using the “geoscatter” function in the Matlab program. Data that were not correctly transmitted (labeled “unresolved” and “resolved (uncertain)”) and any two consecutive points where the speed required to cover the distance between them exceeded 5 km/h were excluded from the analysis (Luschi et al., 1998). Satellite tracking was terminated when the transmitter did not send signals for more than three months.
3 Results
A total of 38 turtles, including green, loggerhead, hawksbill, olive ridley and leatherback, were obtained around Jeju Island on the Korean Peninsula. Of the 38 turtles, green turtles were the most frequently encountered (N=24, 61.5%), followed by loggerhead turtles (N = 9, 23.1%) (Figure 1, Table 2). The satellite-tracked green and loggerhead turtles were captured around southwestern or eastern side of Jeju Island (Figure 1, Tables 1, 2). Other bycaught sea turtles were captured on the northwestern side (loggerhead) and on the northeastern side (olive ridley) (Figure 1). Mitochondrial sequences longer than 700 bp were obtained from 31 sea turtles (Table 1).
3.1 Genetic composition of the Jeju sea turtles
3.1.1 Green turtle
We acquired mitochondrial DNA sequences with lengths of 706–864 bp from 18 individuals. After alignment, the 706 bp sequences corresponded to five distinct haplotypes previously identified in the western Pacific and were separated into three clades (Figure 2A, 3A, Supplementary Table 1). Clade VII with haplotypes CmP39.1 (8 individuals) and CmP213.1 (1 individual), was the most common, followed by Clade VIII with CmP50.1 (6 individuals) and CmP57.2 (1 individual) and Clade III with haplotype CmP54.1 (2 individuals) (Figure 2A). The satellite-tracked individuals with ID 718338, 718339, and 724926 were identified as haplotypes CmP50.1, CmP213.1, and CmP39.1, respectively. Both the haplotype (h) and nucleotide (π) diversity in Jeju (h = 0.7124, π = 0.0217) was higher than that of the foraging aggregations around the main islands of Japan but lower than that of the foraging aggregations around the Ryukyu Archipelago and around the South China region (Supplementary Table 8). MSA revealed that the rookeries in Japan were the primary contributors to the foraging aggregation around Jeju, accounting for 89.57% [95% confidence interval (CI): 72.02–99.01%]. In contrast, other rookeries, including the populations of Hong Kong and Taiwan, which are in close vicinity, exhibited a low contribution level of less than 1% (Figure 4, Supplementary Table 9). The exact test revealed significant population differentiation between the Jeju aggregate and rookeries in the Yaeyama Islands situated in the southernmost part of Ryukyu Archipelago (Table 3).
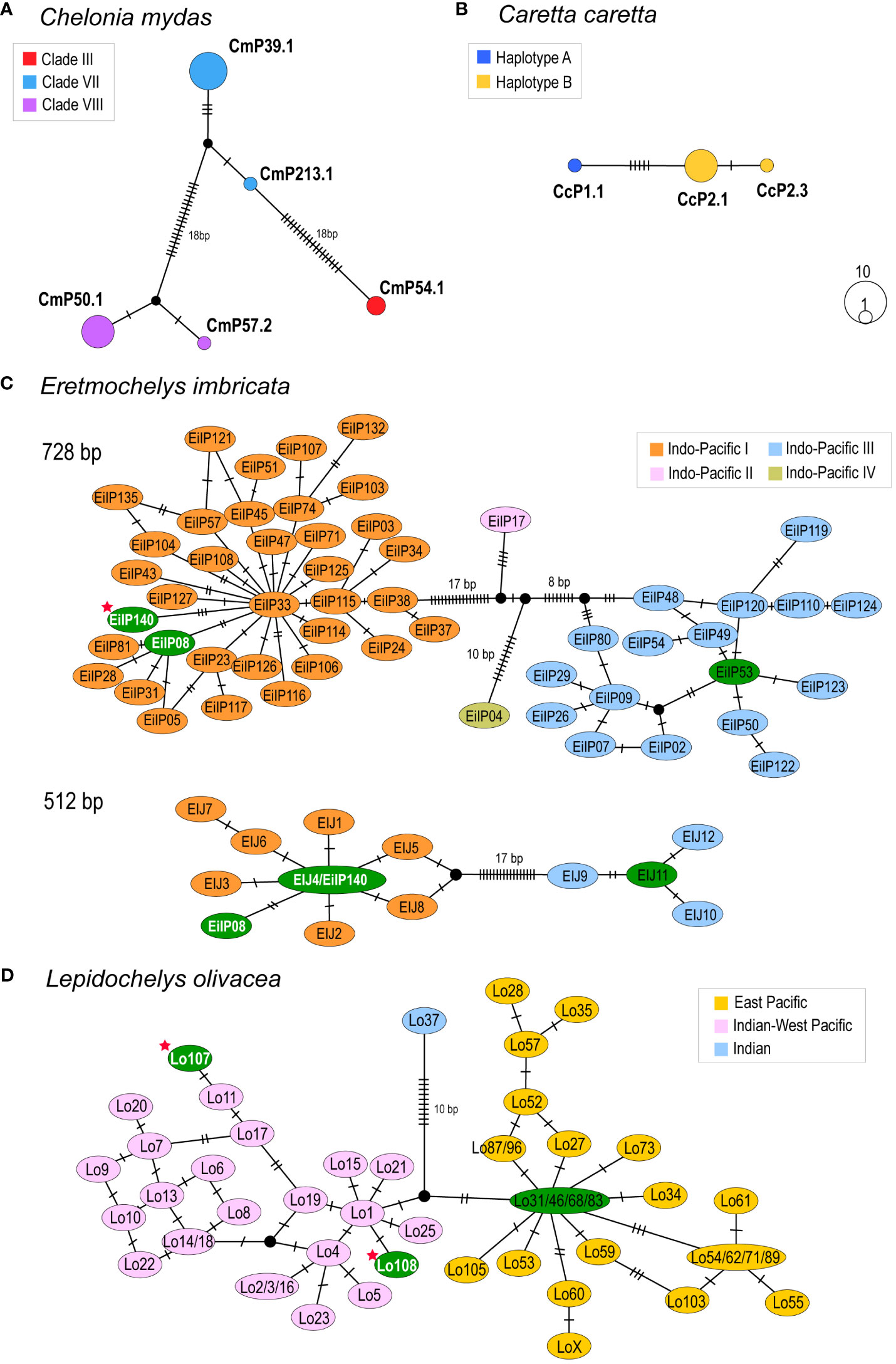
Figure 2 Median-joining networks of Jeju turtles. Haplotypes are colored by clade or clustering, following the approaches of Jensen et al. (2019) for green turtles (A), Bowen et al. (1995) for loggerhead turtles (B), Arantes et al. (2020) for hawksbill turtles (C), and Vilaça et al. (2022) for olive ridley turtles. (D) The black dots represent a hypothetical haplotype. The hatch marks represent a single nucleotide mutation, and mutations of more than 5 bp are represented as a number. (A, B) represents the haplotype network among sequences obtained in this study. (A) Green turtles (706 bp). (B) Loggerhead turtles (808 bp). (C, D) represents haplotype networks among previously reported sequences from Pacific region and newly obtained sequences from this study for hawksbill and olive ridley turtles. Haplotype colored green denotes that have been found on the Korean Peninsula to date, white font denotes that was identified in the our study, and the red star denote novel haplotypes. (C) Hawksbill turtles (728 bp and 512 bp). Two different sequence lengths were used in accordance with comparable data. (D) Olive ridley turtles (642 bp).
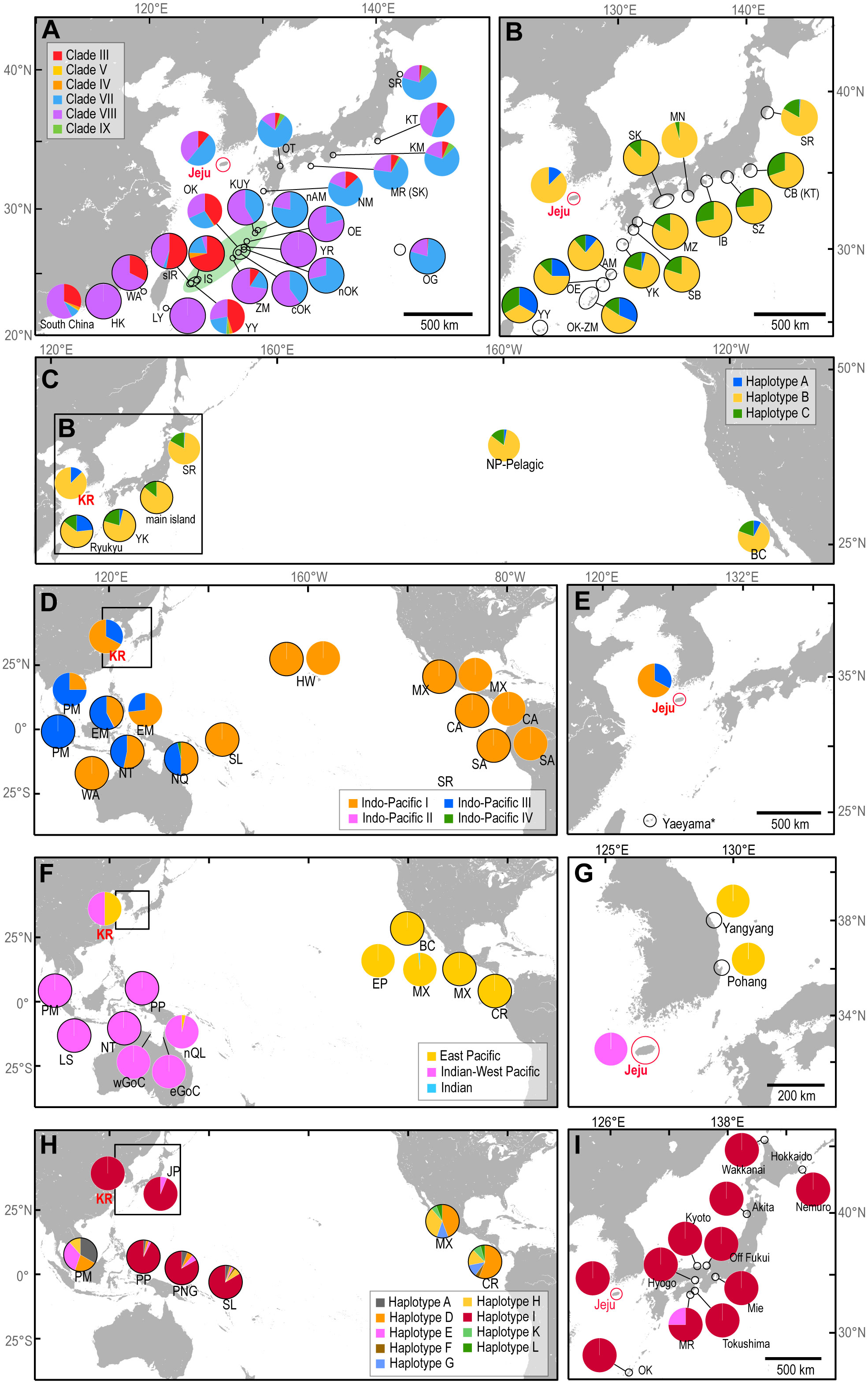
Figure 3 Haplotype frequency for foraging aggregations and rookeries for five sea turtle species in RMUs. Pie diagrams outlined with black lines represent rookery populations. Color of pie in (A–G) corresponds to the scheme used in Figure 2. (A) Green turtles in the northwestern Pacific. The region colored green denotes the Ryukyu Island of Japan. (B, C) Loggerhead turtles in the north Pacific. (D, E) Hawksbill aggregates in the Pacific. (F, G) Olive ridley turtles in the Pacific. (H, I) Leatherback turtles in Pacific. Designation of haplotypes follows the approach by Dutton et al. (1999). BC, Baja California; CA, Central America; CB, Chiba; cOK, central western Okinawajima; CR, Costa Rica; eGoC, eastern Gulf of Carpentaria; EM, East Malaysia; GN, Ginoza; IB, Ise Bay; IS, Ishigakijima; JP, Japan; KM, Kumano-nada; KR, Korea; KT, Kanto; KUY, Kakeromajima-Ukejima-Yorojima; LY, Lanyu; NM, Nomaike; MN, Minabe; MR, Muroto; MX, Mexico; MZ, Miyazaki; nAM, northwestern Amami Oshima; nOK, northern Okinawajima; NP, North Pacific; NQ, North Queensland; NT, Northern Territory; OE, Okinoerabujima; OG, Ogasawara; OT, Oita; PM, Peninsula Malaysia; PNG, Papua New Guinea; PP, Papua; SA, South America; sIR, southwestern Iriomote; SK, Shikoku; SL, Solomon Islands; SR, Sanriku; SR, Sanriku; SZ, Shizuoka; WA, Wan-an; WA, Western Australia; wGoC, western GoC; YK, Yakushima; YR, Yoronjima; YY, Yaeyama; ZM, Zamamijima.
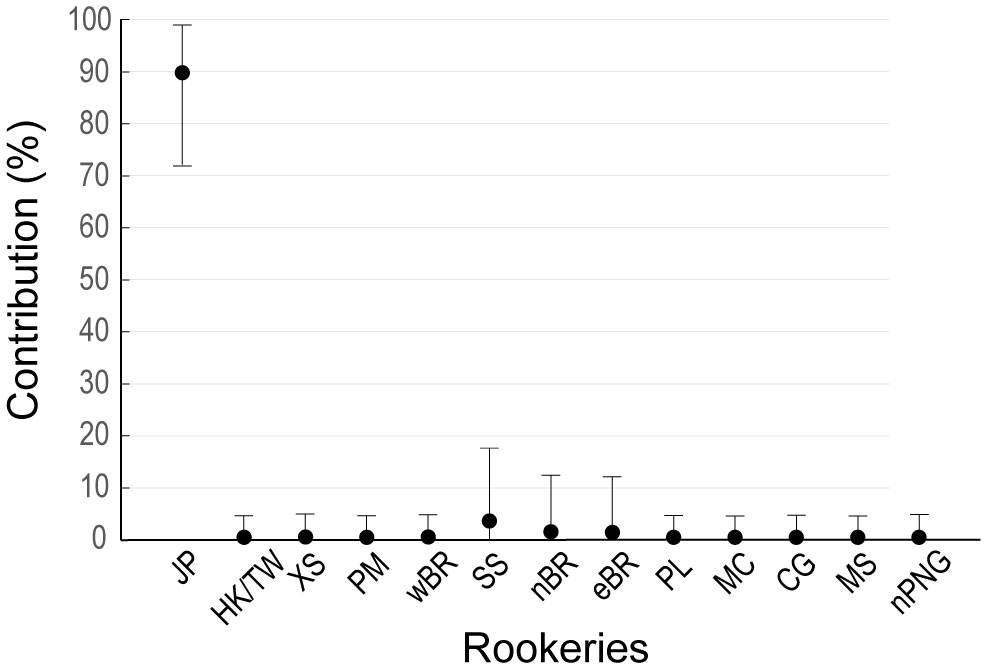
Figure 4 Mixed stock analysis (MSA) of green turtles from western Pacific rookeries contributing to the Jeju foraging ground. CG, CNMI/Guam; eBR, eastern BR; HK, Hong Kong; JP, Japan; MC, Micronesia; MS, Marshall; nBR, northeast Borneo; nPNG, northern Papua New Guinea; PL, Palau; SS, Sulu Sea; TW, Taiwan; wBR, western BR; XS, Xisha Island in China.
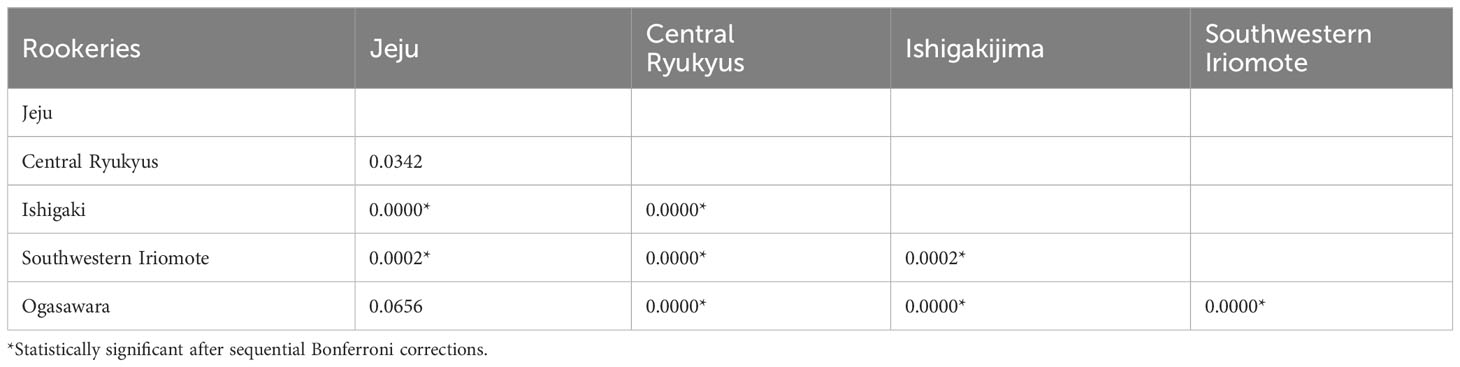
Table 3 Exact test (p values) of population differentiation between green turtles (C. mydas) in Jeju foraging ground and Japanese rookeries.
3.1.2 Loggerhead turtle
We obtained mitochondrial DNA sequences with lengths of 894–896 bp from eight individuals. After alignment, the 808 bp sequences corresponded to three distinct haplotypes identical to those reported for the northwestern Pacific (Figure 2B, Supplementary Table 1): CcP2.1 (75%), CcP2.3 (12.5%), and CcP1.1 (12.5%). One satellite-tracked specimen (ID. 724923) had the haplotype CcP2.1. Given the shorter fragment (350 bp) previously employed by Bowen et al. (1995), the genetic composition was simplified to include only two haplotypes, Haplotype A and Haplotype B (Figures 3B, C, Supplementary Table 4). The genetic diversity in Jeju was lower than that of the foraging aggregations in the north Pacific region (Supplementary Table 10). The exact test found statistically non-significant population differentiation between the Jeju aggregates and rookeries in the Japanese Archipelago (Supplementary Table 11).
3.1.3 Hawksbill turtle
From the two hawksbill turtle specimens, we identified two haplotypes with a length of 892 bp with five of polymorphic sites, which clustered into clade Indo-Pacific I (Figure 2C). Based on long sequence alignment (718–719 bp), one sequence was identical to the haplotype EiIP08 within the Indo-Pacific I clade (Figures 2C, 3D, E, Supplementary Table 1). The other was identified as a previously unreported orphan haplotype that diverged from haplotype EiIP33 by three transitions. The novel haplotype was assigned the name ‘EiIP140’ according to the standardized nomenclature from previous studies and was deposited in GenBank (accession ID: OR400415). A comparison based on a shorter sequence alignment (512–513 bp) revealed a consistent pattern of sequence divergence (Figure 2C).
3.1.4 Olive ridley turtle
In the two olive ridley turtles, we identified two distinct haplotypes (874 bp) with eight polymorphic sites, which clustered with haplotypes from Indian-West Pacific (Figures 2D, 3F, G). Based on sequence alignment with reference data (642–650 bp), two sequences were identified as previously unreported orphan haplotypes that diverged from Lo1 and Lo11 within the Indian-West Pacific clade, respectively (Figure 2D). These new sequences were assigned names ‘Lo107’ and ‘Lo108’ in line with NOAA’s nomenclature and were deposited in GenBank (accession ID: OR400416 and OR400417).
3.1.5 Leatherback turtle
The mitochondrial sequence of the one specimen was classified as haplotype Dc9.1 for the long fragment (890 bp), and Haplotype I for the shorter fragment (496 bp) (Figures 3H, I, Supplementary Table 1).
3.2 Tracking migratory pathways
Of the 12 satellite tagged sea turtles, 11 were tracked successfully, including nine green turtles and two loggerhead turtles (Table 2). The green turtles had an SCL of 66.43 ± 11.59 cm (mean ± SD) with a range of 49.9–84.3 cm, while the loggerhead turtles had an SCL of 64.0 cm and 78.2 cm. The average number of days over which the turtles were tracked was 288.09 ± 182.06.
Of the nine green turtles, three moved away from the release site on Jeju Island (Figure 5A, Table 2). The turtle with ID 710313 followed a dispersal route slightly distinct from the other two for 154 days. It stayed around Jeju Island initially post-release, then moved north along the western coast to the south of the Korean Peninsula, setting there during October and November 2019. In December, it headed southward, passing the eastern coast of Jeju Island, and arrived northeast of Fukue Island in the Japanese Archipelago. The other two turtles, ID 724924 and 724925, moved south for a month after released in November 2021. However, the final detected locations were distinct. The turtle with ID 724924 reached the Amami Oshima in Japanese Archipelago, while the turtle with ID 724925 moved further southward along the western side of Amami Oshima and Okinawa Island in the Japanese Archipelago. It then turned northwest towards China from at Kume Island, and its signal was eventually lost.
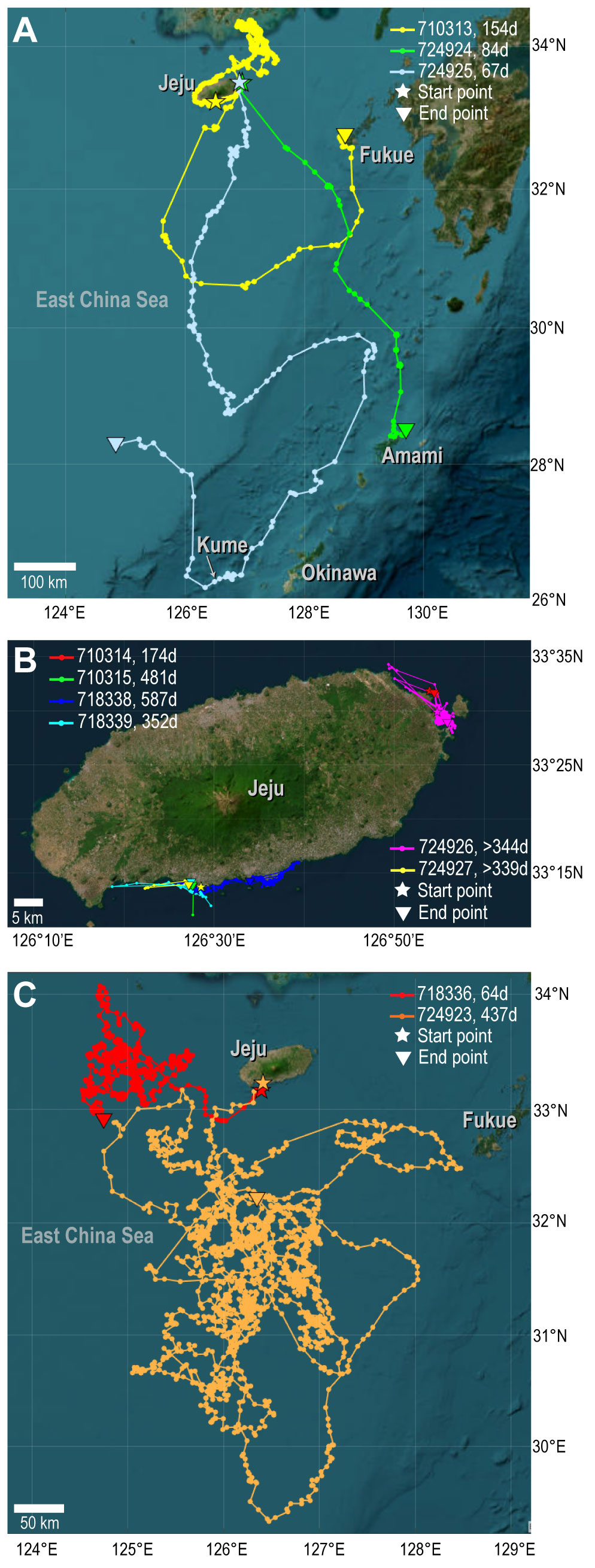
Figure 5 Migration pathways tracked by satellite. Each line represents an individual with a corresponding Track ID. The duration of tracking (in days) is represented alongside each ID. (A) Green turtles moving away from Jeju Island. (B) Green turtles remaining near Jeju Island. (C) Loggerhead turtles moving away from Jeju Island. Geobasemap from Earthstar Geographics Copyright: © 2022b MathWorks, Inc. Licensed for use.
Six green turtles remained in the vicinity of the site where they were released, and these were tracked for longer (174 to 587 days, or still in progress) than the three turtles that migrated away (Figure 5B, Table 2). The turtle with ID 718338, which was tracked for the longest period, 587 days (excluding individuals currently being tracked), spent at least two winters around Jeju Island.
As shown in Figure 5C and Table 2, the loggerhead turtle with ID 718336 moved to the west of Jeju Island post-release and roamed for 64 days. The other loggerhead with ID 724923 (identified as CcP2.1) roamed within a broader area in southern region of the island. This turtle journeyed the longest distance of the tagged turtles, heading southward until October 2021. In November, it turned eastward to the southern region of Fukue Island, then shifted back westward. Subsequently, this individual proceeded southward and then remained within the geographical area spanning 29.32°N to 32.32°N and 125.38°E to 127.14°E until the signal was finally lost in September 2022.
4 Discussion
Here, we discussed comprehensively the genetic connectivity between the Jeju foraging aggregates of five sea turtle species in Jeju Island and adjacent rookeries. Green and loggerhead turtles exhibited a high connectivity with rookeries in Japan. The satellite tracking data revealed migratory patterns southward for a few green turtles. On the other hand, about 60% of the green turtles (6 out of 9) remained in the vicinity of Jeju Island, with five of them staying for an extended period more than 300 days. Furthermore, the relatively high level of genetic diversity of Jeju green turtles indicates that Jeju Island serves as a stable foraging habitat.
4.1 Connectivity with neighboring rookeries
The MSA for green turtles revealed that the main natal population of the Jeju aggregate originated from the Japanese populations among the rookeries in the western Pacific. The presence of widely observed haplotypes (CmP39, CmP50, and CmP54) from rookeries in the Japanese Archipelago (Nishizawa et al., 2011; Nishizawa et al., 2013; Hamabata et al., 2014) supports the genetic connectivity between the Jeju aggregation and the Japanese rookeries. In contrast with the initial expectation, contribution from populations in China and Taiwan to the Jeju aggregation were not supported. The exact test indicated a stronger connection with the rookeries in Central Ryukyus and Ogasawara among Japanese rookeries. This connectivity was supported by satellite tracking data of several turtles. Three of nine turtles exhibited dispersal to the southern islands of Japanese Archipelago. Considering their genetic stock and life stage as subadults (<80 cm in accordance with Phillips et al., 2021), the migratory pathways are likely associated with the behavior returning to their natal origins. It is also interpreted as the traveling of subadults to adult habitats for foraging or breeding (Pelletier et al., 2003; Chambault et al., 2018; Doherty et al., 2020).
The connectivity with Japanese rookeries, particularly with Ryukyu Archipelago, was also implied from the genetic composition of loggerhead turtles. Nesting sites in the north Pacific are predominantly located in the Japanese main islands and Ryukyu Archipelago (Iwamoto et al., 1985; Hatase et al., 2002a; Watanabe et al., 2011; Matsuzawa et al., 2016), with additional site in China’s Xisha Archipelago (Chan et al., 2007). The detection of Haplotype A in the Jeju aggregate, a haplotype previously identified exclusively in Ryukyu Archipelago, suggests that the Ryukyu rookeries are candidates for its natal origins. However, the satellite tracking data was inconclusive regarding the genetic connectivity between Jeju Island and nesting sites in the Japanese Archipelago. The migration to the neritic zone of the East China Sea has previously been observed from Taiwan (Kobayashi et al., 2011) and the Japanese Archipelago (Hatase et al., 2002b; Hatase et al., 2007; Saito et al., 2015). Therefore, we provide evidence that the loggerhead turtles visiting Jeju Island are also utilizing the East China Sea as feeding habitats, but further discussion is limited due to the small sample size and the loss of tracking signals before the turtles reached land.
Interestingly, the genetic connectivity between Jeju foraging aggregates and central area of the western Pacific was also detected. The haplotype CmP57.2 found in the Jeju green turtles has not been reported in Japanese rookeries previously but was observed in the Sulu Sea and in northeastern and eastern Borneo rookeries (Jensen et al., 2016). The genetic sequences from hawksbill, olive ridley, and leatherback turtles of Jeju aggregates supported the connectivity. The haplotype EiIP08 has been observed exclusively in the western Pacific, particularly from rookeries in northern Australia (Vargas et al., 2016). This connection was further substantiated by the novel orphan haplotype EiIP140. It diverged from the haplotype EiIP33, which is typically observed in rookeries around northern Australia and the Solomon Islands (Vargas et al., 2016; Gaos et al., 2018; Gaos et al., 2020). Additionally, haplotype EiIP53 (EIJ11) detected previously in a turtle stranded on Jeju Island (Kim et al., 2020) has been identified in rookeries in Malaysia (Nishizawa et al., 2016; Vargas et al., 2016). For olive ridley turtles, haplotype Lo1, from which Lo108 diverged, has been observed in rookeries in Malaysia, Indonesia, and northern Australia (Jensen et al., 2013; Vilaça et al., 2022). For leatherback turtles, Haplotype I is one of haplotypes observed only in the western Pacific region (Dutton et al., 1999) and this sequence is also widely identified in stranded turtles from Korean Peninsula (Cho et al., 2018) and Japanese Archipelago (Yoshikawa et al., 2016). In the western Pacific, rookeries of leatherback turtles distributed only in the central and southern areas (Wallace et al., 2010), leading to conclusion that the leatherback turtles visited Jeju Island likely migrated from these areas. Nesting leatherback turtles exhibit diverse seasonal migratory pathways (Benson et al., 2011). These genetic compositions of Jeju foraging turtles suggests the migration from Southeast Asia or northern Australia. Nevertheless, we cannot entirely exclude the possibility of migration from other nesting sites due to the small sample size in the present study and the lack of sequence data for rookeries in the northwestern Pacific.
Meanwhile, two haplotypes of olive ridley turtles from Jeju Island were distinguishable from the haplotypes Lo46/Lo68 reported in the East Sea of Korea (also known as the Sea of Japan), which are associated with rookeries in the eastern Pacific (Kim et al., 2019) (Figure 2). To date, the olive ridley rookeries have not been reported in the northwestern Pacific. Only individuals stranded or found in bycatch have been reported (Chan et al., 2007; Fukuoka et al., 2019; Kim et al., 2021) and the northwestern Pacific has been assumed to be a seasonal foraging area (Fukuoka et al., 2019). A genetic composition consisting of distinct clades not only provokes curiosity about the migratory pathways for olive ridley turtles in the Pacific but also highlights the need for more expansion of genetic data from more locations in the northwestern Pacific.
4.2 Role of the Jeju Island as a habitat for sea turtles
High fidelity (up to 500 days) to the foraging grounds was evidenced based on the satellite tracks of six green turtles. This result is consistent with previous studies (Moon et al., 2011; Jang et al., 2018), and stomach contents analysis confirm that green turtles feed in the local waters (Kim et al., 2021). Interestingly, three of the resident turtles had the same genetic sequences (CmP39.1 CmP213.1, and CmP50.1) with those found in Japanese rookeries. These combined satellite tracking and molecular data suggest that green turtles originating from Japanese rookeries might utilize Jeju Island as a long-term habitat. This habitat range for sea turtles underscores the importance of international cooperation in the efforts to protect the endangered species.
Our satellite tracking results also raised another intriguing possibility for green turtles. In the Okinawa foraging aggregations, green turtles within 50–70 cm SCL are less observed, suggesting the existence of alternative developmental habitats in that stage (Hamabata et al., 2015; Hayashi and Nishizawa, 2015; Hamabata et al., 2018). In fact, ontogenetic shift of foraging grounds in green turtles has been previously evidenced through isotope ratio (Shimada et al., 2014). The prevalence of Jeju green turtles within 50–70 cm SCL range (6 out of the 9 tracked individuals) indicates movement between foraging grounds and that Jeju Island is utilized as a developmental feeding ground by green turtles within this size range. Despite the relatively small sample size of our study, size distribution for green turtles (<70 cm for 7 out of 8 individuals) reported by Jang et al. (2018) further supports this hypothesis.
5 Conclusion
In this study, we revealed the genetic composition of five sea turtle species found in Jeju Island, with a particular focus on the genetic stock of the green turtles. The combination of satellite tracking data and genetic data allowed to infer the nesting population and migratory routes of green and loggerhead turtles. The long-term presence of multiple individuals off the coast of Jeju Island emphasized the importance of Jeju Island as a developmental habitat for sea turtle conservation. The mitochondrial DNA sequences for hawksbill, olive ridley, and leatherback turtles analyzed in the present study suggests a connection between the Jeju aggregates and more distant rookeries in the Southeast Asia and northern Australia. However, it remains difficult to definitively identify source populations due to the small sample size and the lack of reference data for neighboring rookeries in the northwestern Pacific. Relatively little is understood about the migratory patterns of hawksbill, olive ridley, and leatherback turtles, especially in the northwestern Pacific. The genetic sequences presented in this study provide an essential basis for delineating conversation units at a more refined scale and bridging existing sampling gaps. The information gathered in this study also highlights the need for further research to fully understand the ecology of sea turtles.
Data availability statement
The datasets presented in this study can be found in online repositories. Mitochondrial DNA sequences were deposited in GenBank (accession IDs: OR400415-OR400417), and metadata of satellite trackingcan be available on BiP (https://www.bip-earth.com/). The genetic data utilized in this study are available at Figshare (10.6084/m9.figshare.23897895).
Ethics statement
The animal study was approved by Jeju Special Self-Governing Province. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
S-JJ: Data curation, Formal analysis, Investigation, Validation, Visualization, Writing – original draft, Writing – review & editing. JK: Data curation, Formal analysis, Investigation, Validation, Visualization, Writing – original draft, Writing – review & editing. SJ: Data curation, Resources, Writing – review & editing. HN: Writing – review & editing. MK: Data curation, Resources, Writing – review & editing. GB: Writing – review & editing. JI: Data curation, Investigation, Writing – review & editing. HYS: Data curation, Writing – review & editing. B-YK: Conceptualization, Data curation, Resources, Writing – review & editing. TK: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the PICES (North Pacific Marine Science Organization) Special Research Project and by the Korea Institute of Marine Science & Technology (KIMST) funded by the Ministry of Oceans and Fisheries (20210642 and RS-2023-00256330, Development of risk managing technology tackling ocean and fisheries crisis around Korean Peninsula by Kuroshio Current).
Acknowledgments
We appreciate research team of Hanwha Aqua Planet Jeju for proving samples and related information. We are also grateful to local fishermen, Jeju Regional Headquarters Korea Coast Guard, and others who reported the stranding and bycatch of the sea turtles and assisted with the sampling.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1281897/full#supplementary-material
References
Abreu-Grobois F. A., Harrocks J., Formia A., R L., Beggs J. (2006). “New mtDNA dloop primers which work for a variety of marine turtle species may increase the resolution of mixed stock analyses,” in Twenty Sixth Annual Symposium on Sea Turtle Biology and Conservation. Eds. Frick M. P. A., Rees A. F., Williams K. (Athens, Greece: International Sea Turtle Society), 179.
Adnyana W., Jayaratha M., Purwanasari H., Wandia I., Dethmers K., Limpus C. (2020). Post-nesting migration and mitochondrial DNA structures of olive ridley turtles (Lepidochelys olivacea) nested on beaches of the bird’s head of papua and the lesser Sunda regions, Indonesia. Int. J. Sci. Res. Publ. 10 (7), 809–817. doi: 10.29322/IJSRP.10.07.2020.p10390
Arantes L. S., Vargas S. M., Santos F. R. (2020). Global phylogeography of the critically endangered hawksbill turtle (Eretmochelys imbricata). Genet. Mol. Biol. 43(2):e20190264. doi: 10.1590/1678-4685-GMB-2019-0264
Benson S. R., Eguchi T., Foley D. G., Forney K. A., Bailey H., Hitipeuw C., et al. (2011). Large-scale movements and high-use areas of western Pacific leatherback turtles, Dermochelys coriacea. Ecosphere 2 (7), 1–27. doi: 10.1890/ES11-00053.1
Bolker B., Okuyama T., Bjorndal K., Bolten A. (2003). Sea turtle stock estimation using genetic markers: accounting for sampling error of rare genotypes. Ecol. Appl. 13 (3), 763–775. doi: 10.1890/1051-0761(2003)013[0763:STSEUG]2.0.CO;2
Bowen B., Abreu-Grobois F., Balazs G., Kamezaki N., Limpus C., Ferl R. (1995). Trans-Pacific migrations of the loggerhead turtle (Caretta caretta) demonstrated with mitochondrial DNA markers. Proc. Natl. Acad. Sci. 92 (9), 3731–3734. doi: 10.1073/pnas.92.9.3731
Campista León S. C., Beltrán Espinoza J. A., Sosa Cornejo I., Castillo Ureta H., Martín del Campo Flores J. R., Sánchez Zazueta J. G., et al. (2019). Haplotypic characterization of the olive ridley turtle (Lepidochelys olivacea) in northwest Mexico: the northernmost limit of its distribution. Anim. Biodiv. Conserv. 42 (1), 113–126. doi: 10.32800/abc.2019.42.0113
Chambault P., de Thoisy B., Huguin M., Martin J., Bonola M., Etienne D., et al. (2018). Connecting paths between juvenile and adult habitats in the Atlantic green turtle using genetics and satellite tracking. Ecol. Evol. 8 (24), 12790–12802. doi: 10.1002/ece3.4708
Chan S. K.-F., Cheng I.-J., Zhou T., Wang H.-J., Gu H.-X., Song X.-J. (2007). A comprehensive overview of the population and conservation status of sea turtles in China. Chelonian. Conserv. Biol. 6 (2), 185–198. doi: 10.2744/1071-8443(2007)6[185:ACOOTP]2.0.CO;2
Cheng I. J., Dutton P., Chen C. L., Chen H. C., Chen Y. H., Shea J. W. (2008). Comparison of the genetics and nesting ecology of two green turtle rookeries. J. Zool. 276 (4), 375–384. doi: 10.1111/j.1469-7998.2008.00501.x
Cho Y., Kim H. K., Lee K., Kim H. W., Park K. J., Sohn J., et al. (2018). Determination of the haplotype and complete mitochondrial genome of the leatherback turtle Dermochelys coriacea (Testudines: Dermochelyidae) found in the vicinity of Korea. Conservation Genetics Resources 10, 701–704. doi: 10.1007/s12686-017-0905-z
Dethmers K. E., Broderick D., Moritz C., Fitzsimmons N. N., Limpus C. J., Lavery S., et al. (2006). The genetic structure of Australasian green turtles (Chelonia mydas): exploring the geographical scale of genetic exchange. Mol. Ecol. 15 (13), 3931–3946. doi: 10.1111/j.1365-294X.2006.03070.x
Doherty P. D., Broderick A. C., Godley B. J., Hart K., Phillips Q., Sanghera A., et al. (2020). Spatial ecology of sub-adult green turtles in coastal waters of the Turks and Caicos Islands: implications for conservation management. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00690
Duchene S., Frey A., Alfaro-Núñez A., Dutton P. H., Gilbert M. T. P., Morin P. A. (2012). Marine turtle mitogenome phylogenetics and evolution. Mol. Phylogenet. Evol. 65 (1), 241–250. doi: 10.1016/j.ympev.2012.06.010
Dutton P. H., Bowen B. W., Owens D. W., Barragan A., Davis S. K. (1999). Global phylogeography of the leatherback turtle (Dermochelys coriacea). J. Zool. 248 (3), 397–409. doi: 10.1017/S0952836999007116
Dutton P. H., Hitipeuw C., Zein M., Benson S. R., Petro G., Pita J., et al. (2007). Status and genetic structure of nesting populations of leatherback turtles (Dermochelys coriacea) in the western Pacific. Chelonian. Conserv. Biol. 6 (1), 47–53. doi: 10.2744/1071-8443(2007)6[47:SAGSON]2.0.CO;2
Dutton P. H., Jensen M. P., Frey A., LaCasella E., Balazs G. H., Zárate P., et al. (2014a). Population structure and phylogeography reveal pathways of colonization by a migratory marine reptile (Chelonia mydas) in the central and eastern Pacific. Ecol. Evol. 4 (22), 4317–4331. doi: 10.1007/s10592-013-0456-0
Dutton P. H., Jensen M. P., Frutchey K., Frey A., LaCasella E., Balazs G. H., et al. (2014b). Genetic stock structure of green turtle (Chelonia mydas) nesting populations across the Pacific islands. Pac. Sci. 68 (4), 451–464. doi: 10.2984/68.4.1
Edgar R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5 (1), 1–19. doi: 10.1186/1471-2105-5-113
Excoffier L., Lischer H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10 (3), 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fuentes M. M., Allstadt A. J., Ceriani S. A., Godfrey M. H., Gredzens C., Helmers D., et al. (2020). Potential adaptability of marine turtles to climate change may be hindered by coastal development in the. Reg. Environ. Change. 20, 1–14. doi: 10.1007/s10113-020-01689-4
Fukuoka T., Kinoshita C., Sato K. (2019). Northernmost bycatch record of an olive ridley turtle (Lepidochelys olivacea) in the Pacific Coast of Japan. Mar. Turt. Newsl. 159, 17–22.
Gaillard D., Yeh F. C., Lin L., Chen H.-Q., Zhang T., Luo S.-J., et al. (2020). Lost at sea: determining geographic origins of illegally traded green sea turtles (Chelonia mydas) rescued on Hainan Island, China. Wildlife Res. 48 (1), 55–63. doi: 10.1071/WR19127
Gaos A. R., LaCasella E. L., Kurpita L., Balazs G., Hargrove S., King C., et al. (2020). Hawaiian hawksbills: a distinct and isolated nesting colony in the Central North Pacific Ocean revealed by mitochondrial DNA. Conserv. Genet. 21, 771–783. doi: 10.1007/s10592-020-01287-1
Gaos A. R., Lewison R. L., Jensen M. P., Liles M. J., Henriquez A., Chavarria S., et al. (2018). Rookery contributions, movements and conservation needs of hawksbill turtles at foraging grounds in the eastern Pacific Ocean. Mar. Ecol. Prog. Ser. 586, 203–216. doi: 10.3354/meps12391
Gelman A., Meng X.-L., Stern H. (1996). Posterior predictive assessment of model fitness via realized discrepancies. Stat. Sin. 6 (4), 733–760.
Hamabata T., Hikida T., Okamoto K., Watanabe S., Kamezaki N. (2015). Ontogenetic habitat shifts of green turtles (Chelonia mydas) suggested by the size modality in foraging aggregations along the coasts of the western Japanese main islands. J. Exp. Mar. Biol. Ecol. 463, 181–188. doi: 10.1016/j.jembe.2014.12.007
Hamabata T., Kamezaki N., Hikida T. (2014). Genetic structure of green turtle (Chelonia mydas) peripheral populations nesting in the northwestern Pacific rookeries: evidence for northern refugia and postglacial colonization. Mar. Biol. 161, 495–507. doi: 10.1007/s00227-013-2352-z
Hamabata T., Nishizawa H., Kawazu I., Kameda K., Kamezaki N., Hikida T. (2018). Stock composition of green turtles Chelonia mydas foraging in the Ryukyu Archipelago differs with size class. Mar. Ecol. Prog. Ser. 600, 151–163. doi: 10.3354/meps12657
Hamann M., Limpus C., Kamezaki N. (2006). “Status of leatherback turtles in Japan,” in Assessment of the Conservation Status of the Leatherback Turtle in the Indian ocean and South East Asia, IOSEA species assessment volume I. Eds. Hamann M., Limpus C., Hughes G., Mortimer J., Pilcher N. (Bangkok: IOSEA Marine Turtle Memorandum of Understanding).
Hatase H., Kinoshita M., Bando T., Kamezaki N., Sato K., Matsuzawa Y., et al. (2002a). Population structure of loggerhead turtles, Caretta caretta, nesting in Japan: bottlenecks on the Pacific population. Mar. Biol. 141, 299–305. doi: 10.1007/s00227-002-0819-4
Hatase H., Omuta K., Tsukamoto K. (2007). Bottom or midwater: alternative foraging behaviors in adult female loggerhead sea turtles. J. Zool. 273 (1), 46–55. doi: 10.1111/j.1469-7998.2007.00298.x
Hatase H., Takai N., Matsuzawa Y., Sakamoto W., Omuta K., Goto K., et al. (2002b). Size-related differences in feeding habitat use of adult female loggerhead turtles Caretta caretta around Japan determined by stable isotope analyses and satellite telemetry. Mar. Ecol. Prog. Ser. 233, 273–281. doi: 10.3354/meps233273
Hayashi R., Nishizawa H. (2015). Body size distribution demonstrates flexible habitat shift of green turtle (Chelonia mydas). Glob. Ecol. Conserv. 3, 115–120. doi: 10.1016/j.gecco.2014.11.008
Iwamoto T., Ishii M., Nakashima Y., Takeshita H., Itoh A. (1985). Nesting cycles and migrations of the loggerhead sea turtle in Miyazaki, Japan. Jpn. J. Ecol. 35 (4), 505–511. doi: 10.18960/seitai.35.4_505
Jang S., Balazs G. H., Parker D. M., Kim B.-Y., Kim M. Y., Ng C. K. Y., et al. (2018). Movements of green turtles (Chelonia mydas) rescued from pound nets near Jeju Island, Republic of Korea. Chelonian. Conserv. Biol. 17 (2), 236–244. doi: 10.2744/CCB-1279.1
Jensen M., Bell I., Limpus C., Hamann M., Ambar S., Whap T., et al. (2016). Spatial and temporal genetic variation among size classes of green turtles (Chelonia mydas) provides information on oceanic dispersal and population dynamics. Mar. Ecol. Prog. Ser. 543, 241–256. doi: 10.3354/meps11521
Jensen M. P., FitzSimmons N. N., Bourjea J., Hamabata T., Reece J., Dutton P. H. (2019). The evolutionary history and global phylogeography of the green turtle (Chelonia mydas). J. Biogeogr. 46 (5), 860–870. doi: 10.1111/jbi.13483
Jensen M. P., Limpus C. J., Whiting S. D., Guinea M., Prince R. I., Dethmers K. E., et al. (2013). Defining olive ridley turtle Lepidochelys olivacea management units in Australia and assessing the potential impact of mortality in ghost nets. Endanger. Species Res. 21 (3), 241–253. doi: 10.3354/esr00521
Jim L. M. R., Rice M. R., Hickey F. R., Balazs G. H. (2022). Destination revealed: post-nesting migrations of hawksbill turtles (Eretmochelys imbricata) from Moso Island, Republic of Vanuatu. Micronesica 1, 1–16.
Jung M.-M., Moon D.-Y., Kim S.-H., Kim H.-S., Kim J.-W. (2012a). Environmental conditions as accidental nesting place of sea turtle located in Jeju Island of Korea. J. Fish. Mar. Sci. Edu. 24 (4), 507–515. doi: 10.13000/JFMSE.2012.24.4.507
Jung M. M., Moon D. Y., Kim S. H., Kim J. W. (2012b). Observation and record of sea turtles in bycatch and stranding from Jeju Island of Korea. J. Fish. Mar. Sci. Edu. 24 (5), 662–669.
Kim H. K., Chan B. K., Yi C., Kim I.-H., Choi Y. N. (2022). Barnacle epibiosis on sea turtles in korea: A West Pacific region with low occurrence and intensity of Chelonibia testudinaria (Cirripedia: chelonibiidae). Front. Ecol. Evol. 10. doi: 10.3389/fevo.2022.785692
Kim I.-H., Moon D. Y., Cho I. Y., Kim M. S., An Y. R., Han D, et al. (2017). Occurrence of sea turtles in the Korean waters and the morphological characteristics of two major species. Korean J. Fish. Aquat. Sci. 50(3), 311–318.
Kim J., Kim I.-H., Kim M.-S., Lee H. R., Kim Y. J., Park S., et al. (2021). Occurrence and diet analysis of sea turtles in Korean shore. J. Ecol. Environ. 45 (1), 1–15. doi: 10.1186/s41610-021-00206-w
Kim I.-H., Yi C.-H., Han D.-J., Park D., Park J., Cho I.-Y., et al. (2020). First record of the hawksbill turtle (Eretmochelys imbricata, Reptilia: Testudines: Cheloniidae) from South Korea. J. Asia-Pac. Biodivers. 13 (2), 151–155. doi: 10.1016/j.japb.2020.02.006
Kim I.-H., Yi C.-H., Lee J.-H., Park D., Cho I.-Y., Han D.-J., et al. (2019). First record of the olive ridley sea turtle Lepidochelys olivacea (Reptilia: Testudines: Cheloniidae) from South Korea. Curr. Herpetol. 38 (2), 153–159. doi: 10.5358/hsj.38.153
Kobayashi D. R., Cheng I. J., Parker D. M., Polovina J. J., Kamezaki N., Balazs G. H. (2011). Loggerhead turtle (Caretta caretta) movement off the coast of Taiwan: characterization of a hotspot in the East China Sea and investigation of mesoscale eddies. ICES J. Mar. Sci. 68 (4), 707–718. doi: 10.1093/icesjms/fsq185
Komoroske L. M., Jensen M. P., Stewart K. R., Shamblin B. M., Dutton P. H. (2017). Advances in the application of genetics in marine turtle biology and conservation. Front. Mar. Sci. 4. doi: 10.3389/fmars.2017.00156
Leigh J. W., Bryant D. (2015). POPART: full-feature software for haplotype network construction. Methods Ecol. Evol. 6 (9), 1110–1116. doi: 10.1111/2041-210X.12410
Lewison R. L., Crowder L. B., Read A. J., Freeman S. A. (2004). Understanding impacts of fisheries bycatch on marine megafauna. Trends Ecol. Evol. 19 (11), 598–604. doi: 10.1016/j.tree.2004.09.004
Li M., Zhang T., Liu Y., Li Y., Fong J. J., Yu Y., et al. (2023). Revisiting the genetic diversity and population structure of the endangered Green Sea Turtle (Chelonia mydas) breeding populations in the Xisha (Paracel) Islands, South China Sea. PeerJ 11, e15115. doi: 10.7717/peerj.15115
Luschi P., Hays G., Del Seppia C., Marsh R., Papi F. (1998). The navigational feats of green sea turtles migrating from Ascension Island investigated by satellite telemetry. Proc. R. Soc B:Biol. Sci. 265 (1412), 2279–2284. doi: 10.1098/rspb.1998.0571
Matsuzawa Y., Kamezaki N., Ishihara T., Omuta K., Takeshita H., Goto K., et al. (2016). Fine-scale genetic population structure of loggerhead turtles in the Northwest Pacific. Endanger. Species Res. 30, 83–93. doi: 10.3354/esr00724
Martín-del-Campo R., Ortega-Ortiz C. D., Abreu-Grobois A., Enríquez-Paredes L. M., Petatán-Ramírez D., Quijano-Scheggia S. I. (2023). Genetic Evidence for Indo-Western Pacific Olive Ridley Sea Turtles in Mexican Waters. Diversity 15 (3), 430. doi: 10.3390/d15030430
Mazaris A. D., Schofield G., Gkazinou C., Almpanidou V., Hays G. C. (2017). Global sea turtle conservation successes. Sci. Adv. 3 (9), e1600730. doi: 10.1126/sciadv.1600730
Moon D. Y., An Y. R., Jung M. M., Kim S. Y., Choi S. K., Lee H. Y., et al. (2011). Satellite tracking of green sea turtles Chelonia mydas in Korean waters. Korean. J. Fish. Aquat. Sci. 44 (6), 709–716. doi: 10.5657/KFAS.2011.0709
National Marine Fisheries Service (NMFS) Southeast Fisheries Science Center(SEFSC). (2008). Sea turtle research techniques manual. (NOAA Technical Memorandum NMFS-SEFSC-579), 92pp.
National Marine Fisheries Service (NMFS) and U.S. Fish and Wildlife Service (USFWS) (1996). Policy regarding the recognition of distinct vertebrate population segments under the endangered species act. Fed. Regist. 61, 4722–4725.
Ng C. K. Y., Dutton P. H., Chan S. K. F., Cheung K.-S., Qiu J.-W., Sun Y.-N. (2014). Characterization and conservation concerns of green turtles (Chelonia mydas) nesting in Hong Kong, China. Pac. Sci. 68 (2), 231–243. doi: 10.2984/68.2.5
Ng C. K. Y., Dutton P. H., Gu H. X., Li T. H., Ye M. B., Xia Z. R., et al. (2017). Regional conservation implications of green turtle (Chelonia mydas) genetic stock composition in China. Chelonian. Conserv. Biol. 16 (2), 139–150. doi: 10.2744/CCB-1253.1
Ng C. K. Y., Matsuzawa Y. (2021). Sea Turtles in the East Asia Region. Marine Turtle Specialist Group (MTSG) Annual Regional Report 2021. Draft report to the IUCN-SSC Marine Turtle Specialist Group. 129pp.
Nishizawa H., Abe O., Okuyama J., Kobayashi M., Arai N. (2011). Population genetic structure and implications for natal philopatry of nesting green turtles Chelonia mydas in the Yaeyama Islands, Japan. Endanger. Species Res. 14 (2), 141–148. doi: 10.3354/esr00355
Nishizawa H., Joseph J., Chong Y. K. (2016). Spatio-temporal patterns of mitochondrial DNA variation in hawksbill turtles (Eretmochelys imbricata) in Southeast Asia. J. Exp. Mar. Biol. Ecol. 474, 164–170. doi: 10.1016/j.jembe.2015.10.015
Nishizawa H., Naito Y., Suganuma H., Abe O., Okuyama J., Hirate K., et al. (2013). Composition of green turtle feeding aggregations along the Japanese archipelago: implications for changes in composition with current flow. Mar. Biol. 160 (10), 2671–2685. doi: 10.1007/s00227-013-2261-1
Nishizawa H., Narazaki T., Fukuoka T., Sato K., Hamabata T., Kinoshita M., et al. (2014a). Genetic composition of loggerhead turtle feeding aggregations: migration patterns in the North Pacific. Endanger. Species Res. 24 (1), 85–93. doi: 10.3354/esr00588
Nishizawa H., Narazaki T., Fukuoka T., Sato K., Hamabata T., Kinoshita M., et al. (2014b). Juvenile green turtles on the northern edge of their range: mtDNA evidence of long-distance westward dispersals in the northern Pacific Ocean. Endanger. Species Res. 24 (2), 171–179. doi: 10.3354/esr00592
Nishizawa H., Okuyama J., Abe O., Kobayashi M., Arai N. (2012). Mitochondrial DNA variation in hawksbill turtles (Eretmochelys imbricata) nesting on Ishigaki Island. Japan. Mar. Turt. Newsl. 132 (1).
Nishizawa H., Okuyama J., Kobayashi M., Abe O., Arai N. (2010). Comparative phylogeny and historical perspectives on population genetics of the Pacific hawksbill (Eretmochelys imbricata) and green turtles (Chelonia mydas), inferred from feeding populations in the Yaeyama Islands, Japan. Zool. Sci. 27 (1), 14–18. doi: 10.2108/zsj.27.14
Pella J., Masuda M. (2001). Bayesian methods for analysis of stock mixtures from genetic characters. Fish. Bull. 99 (1), 151–167.
Pelletier D., Roos D., Ciccione S. (2003). Oceanic survival and movements of wild and captive-reared immature green turtles (Chelonia mydas) in the Indian Ocean. Aquat. Living Resour. 16 (1), 35–41. doi: 10.1016/S0990-7440(03)00005-6
Phillips K. F., Stahelin G. D., Chabot R. M., Mansfield K. L. (2021). Long-term trends in marine turtle size at maturity at an important Atlantic rookery. Ecosphere 12 (7), e03631. doi: 10.1002/ecs2.3631
Pinou T., Prunier R., Bresson M., Padilla I. E., Javier Perez J. F., Robles J. A. T., et al. (2018). Repeated sampling adds to the genetic diversity of Lepidochelys olivacea (Eschscholtz 1829) olive ridley sea turtle. J. Nat. Hist. 52 (45-46), 2899–2917. doi: 10.1080/00222933.2018.1557755
Reece J. S., Passeri D., Ehrhart L., Hagen S. C., Hays A., Long C., et al. (2013). Sea level rise, land use, and climate change influence the distribution of loggerhead turtle nests at the largest USA rookery (Melbourne Beach, Florida). Mar. Ecol. Prog. Ser. 493, 259–274. doi: 10.3354/meps10531
Reneker J. L., Cook M., Nero R. W. (2018). Preparation of fresh dead sea turtle carcasses for at-sea drift experiments (Mississippi, USA: NOAA Technical Memorandum NMFS-SEFSC-731), 14pp. doi: 10.25923/9hgx-fn38
Saito T., Kurita M., Okamoto H., Uchida I., Parker D., Balazs G. (2015). Tracking male loggerhead turtle migrations around southwestern Japan using satellite telemetry. Chelonian. Conserv. Biol. 14 (1), 82–87. doi: 10.2744/ccab-14-01-82-87.1
Santos B. S., Kaplan D. M., Friedrichs M. A., Barco S. G., Mansfield K. L., Manning J. P. (2018). Consequences of drift and carcass decomposition for estimating sea turtle mortality hotspots. Ecol. Indic. 84, 319–336. doi: 10.1016/j.ecolind.2017.08.064
Shimada T., Aoki S., Kameda K., Hazel J., Reich K., Kamezaki N. (2014). Site fidelity, ontogenetic shift and diet composition of green turtles Chelonia mydas in Japan inferred from stable isotope analysis. Endangered Species Res. 25 (2), 151–164. doi: 10.3354/esr00616
Silver-Gorges I., Koval J., Rodriguez-Zarate C. J., Paladino F. V., Jordan M. (2020). Large-scale connectivity, cryptic population structure, and relatedness in Eastern Pacific Olive ridley sea turtles (Lepidochelys olivacea). Ecol. Evol. 10 (16), 8688–8704. doi: 10.1002/ece3.6564
Song J.-H., Lin B.-A., Jia Y.-Y., Dutton P. H., Kang B., Balazs G. H., et al. (2022). New management unit for conservation of the Endangered green turtle Chelonia mydas at the Xisha (Paracel) Islands, South China Sea. Endanger. Species Res. 47, 145–154. doi: 10.3354/ESR01172
Stokes L., Epperly S. P., Avens L. I., Belskis L. C., Benson S. R., Braun-McNeill J., et al. (2008). Sea turtle research techniques manual.
Vargas S. M., Jensen M. P., Ho S. Y., Mobaraki A., Broderick D., Mortimer J. A., et al. (2016). Phylogeography, genetic diversity, and management units of hawksbill turtles in the Indo-Pacific. J. Hered. 107 (3), 199–213. doi: 10.1093/jhered/esv091
Vilaça S. T., Hahn A. T., Naro-Maciel E., Abreu-Grobois F. A., Bowen B. W., Castilhos J. C., et al. (2022). Global phylogeography of ridley sea turtles (Lepidochelys spp.): evolution, demography, connectivity, and conservation. Conserv. Genet. 23, 995–1010. doi: 10.1007/s10592-022-01465-3
Wallace B. P., DiMatteo A. D., Hurley B. J., Finkbeiner E. M., Bolten A. B., Chaloupka M. Y., et al. (2010). Regional management units for marine turtles: a novel framework for prioritizing conservation and research across multiple scales. PloS One 5 (12), e15465. doi: 10.1371/journal.pone.0015465
Watanabe K. K., Hatase H., Kinoshita M., Omuta K., Bando T., Kamezaki N., et al. (2011). Population structure of the loggerhead turtle Caretta caretta, a large marine carnivore that exhibits alternative foraging behaviors. Mar. Ecol. Prog. Ser. 424, 273–283. doi: 10.3354/meps08989
Yoshikawa N., Kamezaki N., Kawazu I., Hirai S., Taguchi S. (2016). Stock origin of the leatherback turtles (Dermochelys coriacea) found in the vicinity of Japan revealed by mtDNA Haplotypes. Curr. Herpetol. 35 (2), 115–121. doi: 10.5358/hsj.35.115
Keywords: sea turtles, genetic connectivity, satellite tracking, natal origin, foraging ground
Citation: Jang S-J, Jo K, Jang S, Nishizawa H, Kim M, Balazs G, Im J, Suk HY, Kim B-Y and Kim T (2024) Connectivity between sea turtles off Jeju Island on the Korean Peninsula, and other populations in the western Pacific. Front. Mar. Sci. 11:1281897. doi: 10.3389/fmars.2024.1281897
Received: 23 August 2023; Accepted: 25 January 2024;
Published: 16 February 2024.
Edited by:
Laura Prosdocimi, Museo Argentino de Ciencias Naturales Bernardino Rivadavia, ArgentinaReviewed by:
Natalie Elizabeth Wildermann, King Abdullah University of Science and Technology, Saudi ArabiaNicolas James Pilcher, Marine Research Foundation, Malaysia
Copyright © 2024 Jang, Jo, Jang, Nishizawa, Kim, Balazs, Im, Suk, Kim and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Byung-Yeob Kim, a2ltYnlAamVqdW51LmFjLmty; Taewon Kim, a3R3b25AaW5oYS5hYy5rcg==
 Sook-Jin Jang
Sook-Jin Jang Kyungsik Jo
Kyungsik Jo Soojin Jang
Soojin Jang Hideaki Nishizawa
Hideaki Nishizawa Miyeon Kim5
Miyeon Kim5 George Balazs
George Balazs Ho Young Suk
Ho Young Suk Taewon Kim
Taewon Kim