- 1Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 2Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao, China
- 3Abteilung Marine Zoologie, Forschungsinstitut und Naturmuseum Senckenberg, Frankfurt, Germany
- 4Independent Researcher, Munich, Germany
- 5Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 6Graduate University, University of Chinese Academy of Sciences, Beijing, China
A new species, belonging to the rossellid subfamily Lanuginellinae Gray, 1872, is described based on two specimens collected from two different seamounts in the western Pacific Ocean. Species characterization was approached by analyzing the morphological and skeletal features as well as the complete mitochondrial genome. Caulophacus (Caulodiscus) iocasicus sp. nov. is distinguishable from its congeners by its branched external shape and a unique combination of microscleres. It only has hemionychohexasters while other C. (Caulodiscus) species additionally have either onychohexasters or discohexasters. Furthermore, the new species has microhexactins, which have not been reported from the other six species of C. (Caulodiscus). Regarding the mitochondrial genome, the occurrence of the atp8 gene, the absence of tRNA E, the translocation of tRNA D between cob and nad6, and the rearrangement of nad6-nad4 distinguish the new species from other rossellids and even other hexactinellids for which mitogenomic information is available. The herein revised morphological and molecular information of the genus Caulophacus also suggests that the monospecific subgenus C. (Caulophacella) should be removed from Caulophacus and reinstated as a separate genus in the subfamily Lanuginellinae.
Introduction
Caulophacus Schulze, 1886 is a pedunculate genus within Lanuginellinae Gray, 1872, with 32 species currently recognized (de Voogd et al., 2022). Caulophacus is cosmopolitan (Figure 1), with species recorded from the Pacific, Atlantic, and Indian oceans (Supplementary Excel S1). It is essentially a deep-sea genus. Caulophacus (Caulodiscus) leonieae Buskowiak & Janussen, 2021 is reported to live at a depth of 296 m, which is the shallowest recorded depth for the genus. Generally, only a few species are reported to occur in the mesobenthic zone (200–1000 m), while most of the species occur in bathyal (1000–4000 m) and abyssal (4000–6000 m) waters. The deepest record for Caulophacus has been, until now, 6770 m (hadal), held by Caulophacus (Caulophacus) hadalis Lévi, 1964. The highest abundance of Caulophacus tends to be reported at depths of 3000–5000 m. Some species have a large depth range, such as C. (Caulophacus) latus Schulze, 1886, from 2926 m (Schulze, 1887) to 6710 m (Koltun, 1970).
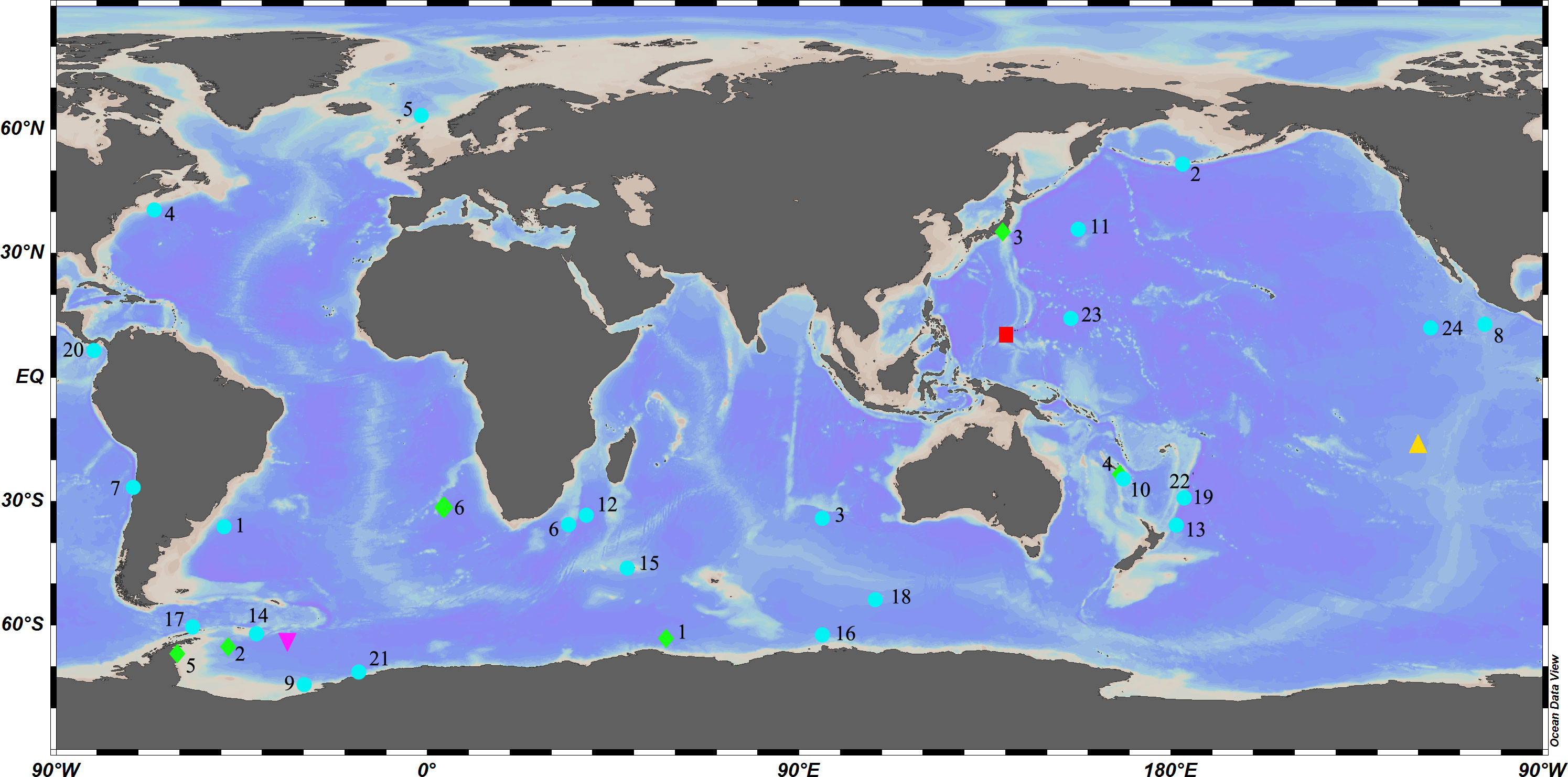
Figure 1 Global diversity of Caulophacus:  Caulophacus (Caulophacella) tenuis.
Caulophacus (Caulophacella) tenuis.  Caulophacus (Oxydiscus) weddelli.
Caulophacus (Oxydiscus) weddelli.  Caulophacus (Caulodiscus): 1, C. valdiviae; 2, C. brandti; 3, C. lotifolium; 4, C. onychohexactinus; 5, C. leonieae; 6, C. polyspiculus.
Caulophacus (Caulodiscus): 1, C. valdiviae; 2, C. brandti; 3, C. lotifolium; 4, C. onychohexactinus; 5, C. leonieae; 6, C. polyspiculus.  Caulophacus (Caulophacus): 1, C. abyssalis; 2, C. adakensis; 3, C. antarcticus; 4, C. agassizi; 5, C. arcticus; 6, C. basispinosus; 7, C. chilense; 8, C. cyanae; 9, C. discohexactinus; 10, C. discohexaster; 11, C. elegans; 12, C. galatheae; 13, C. hadalis; 14, C. instabilis; 15, C. latus; 16, C. oviformis; 17, C. palmeri; 18, C. pipetta; 19, C. ramosus; 20, C. schulzei; 21, C. scotiae; 22, C. serpens; 23, C. variens; 24, C. wilsoni.
Caulophacus (Caulophacus): 1, C. abyssalis; 2, C. adakensis; 3, C. antarcticus; 4, C. agassizi; 5, C. arcticus; 6, C. basispinosus; 7, C. chilense; 8, C. cyanae; 9, C. discohexactinus; 10, C. discohexaster; 11, C. elegans; 12, C. galatheae; 13, C. hadalis; 14, C. instabilis; 15, C. latus; 16, C. oviformis; 17, C. palmeri; 18, C. pipetta; 19, C. ramosus; 20, C. schulzei; 21, C. scotiae; 22, C. serpens; 23, C. variens; 24, C. wilsoni.  Caulophacus (Caulodiscus) iocasicus sp. nov.
Caulophacus (Caulodiscus) iocasicus sp. nov.
Caulophacus once contained two subgenera (C. (Caulophacus) Schulze, 1886 and C. (Caulodiscus) Ijima, 1927) (Tabachnick, 2002). Janussen et al. (2004) established a new subgenus C. (Oxydiscus) based on specimens from the Weddell Sea, which is characterized by microscleres having oxyoidal and discoidal endings. They suggested the rossellid genus Caulophacella Lendenfeld, 1915 should be included in Caulophacus as another subgenus with microscleres having exclusively oxyoidal endings, which was formally implemented by Boury-Esnault et al. (2015). Therefore, there are currently 4 subgenera of Caulophacus, which are differentiated by the terminations of microscleres: C. (Caulodiscus) having discoidal and onychoidal terminations, C. (Caulophacella) having exclusively oxyoidal terminations, C. (Caulophacus) mainly having discoidal terminations, and C. (Oxydiscus) having oxyoidal and discoidal terminations (Tabachnick, 2002; Janussen et al., 2004). However, molecular phylogenetic analyses indicate that inclusion of Caulophacella in Caulophacus is not justified and that C. (Caulodiscus) and C. (Caulophacus) are not monophyletic groups (Dohrmann et al., 2008; Dohrmann et al., 2017; Kersken et al., 2018; Dohrmann, 2019). Thus, the intrageneric classification scheme of Caulophacus requires revision based on morphological and molecular data.
Mitochondria play important roles in providing energy for eukaryotic cells. Genetic-level changes in mitochondria can reflect physiological changes and evolutionary pressures of the species (Plese et al., 2021). Moreover, mitochondria have their own genetic system, which contains useful information for investigating phylogenetic relationships. The mitogenome of most metazoans is a closed circular molecule of 15–20 kb in length, usually containing 37 genes: 13 protein-coding genes (PCGs), 2 ribosomal RNA (rRNA) genes, and 22 transfer RNA (tRNA) genes (Boore, 1999). However, the mitogenome characteristics of sponges differ between classes. For example, Demospongiae and Homoscleromorpha have 14 PCGs (Gazave et al., 2010; Ereskovsky et al., 2017; Plese et al., 2021), Hexactinellida have 13 PCGs (atp8 missing) (Haen et al., 2014), and Calcarea have 11 PCGs (atp8, nad6, and nad4L missing) (Lavrov et al., 2016). To date, only a few mitochondrial genomes of hexactinellid sponges have been sequenced. Most of the mitogenome sequences are incomplete and only those of four species are complete (Aphrocallistes vastus, Vazella pourtalesii, Tabachnickia sp., and Lophophysema eversa). We analyzed the incomplete and complete mitogenomes of hexactinellid sponges available in GenBank, which range from 13,588 bp to 20,591 bp in length and contain 1–2 rRNA genes, 10–14 PCGs and 11–22 tRNA genes. Many important taxa of hexactinellid sponges still lack mitochondrial genomes, and deeper phylogenetic relationships could be investigated with more comprehensive taxon sampling.
In 2017 and 2019, we conducted cruises to two different seamounts in the northwestern Pacific Ocean to survey seamount biodiversity with the research vessel Ke Xue. Two remarkable bush-like sponge specimens were collected at depths of 884 m and 1055.5 m, respectively. Based on the morphological and molecular analyses, they are described and illustrated herein as new to science. We also report the complete mitogenome of the new species and we hope to broaden our view on the diversity of mt genomes through the first mt genome of Caulophacus.
Materials and methods
Sample collection
The specimens were collected from two seamounts by the submersible ROV Fa Xian with the R/V Ke Xue during its cruises in 2017 and 2019 in the western Pacific Ocean, and preserved in 75% ethanol. The samples are deposited in the Marine Biological Museum (MBM) of the Chinese Academy of Sciences, Qingdao, China.
Spicule analysis
Concentrated nitric acid was used to digest a small piece of sponge tissue and clean spicules were isolated by rinsing several times with distilled water and dehydrated with 95% ethanol. A Hitachi S-3400N scanning electron microscope (SEM) was used to observe the spicules. Additional spicule measurements were performed using an Olympus DSX500 Optodigital light microscope (LM) (Table 1).
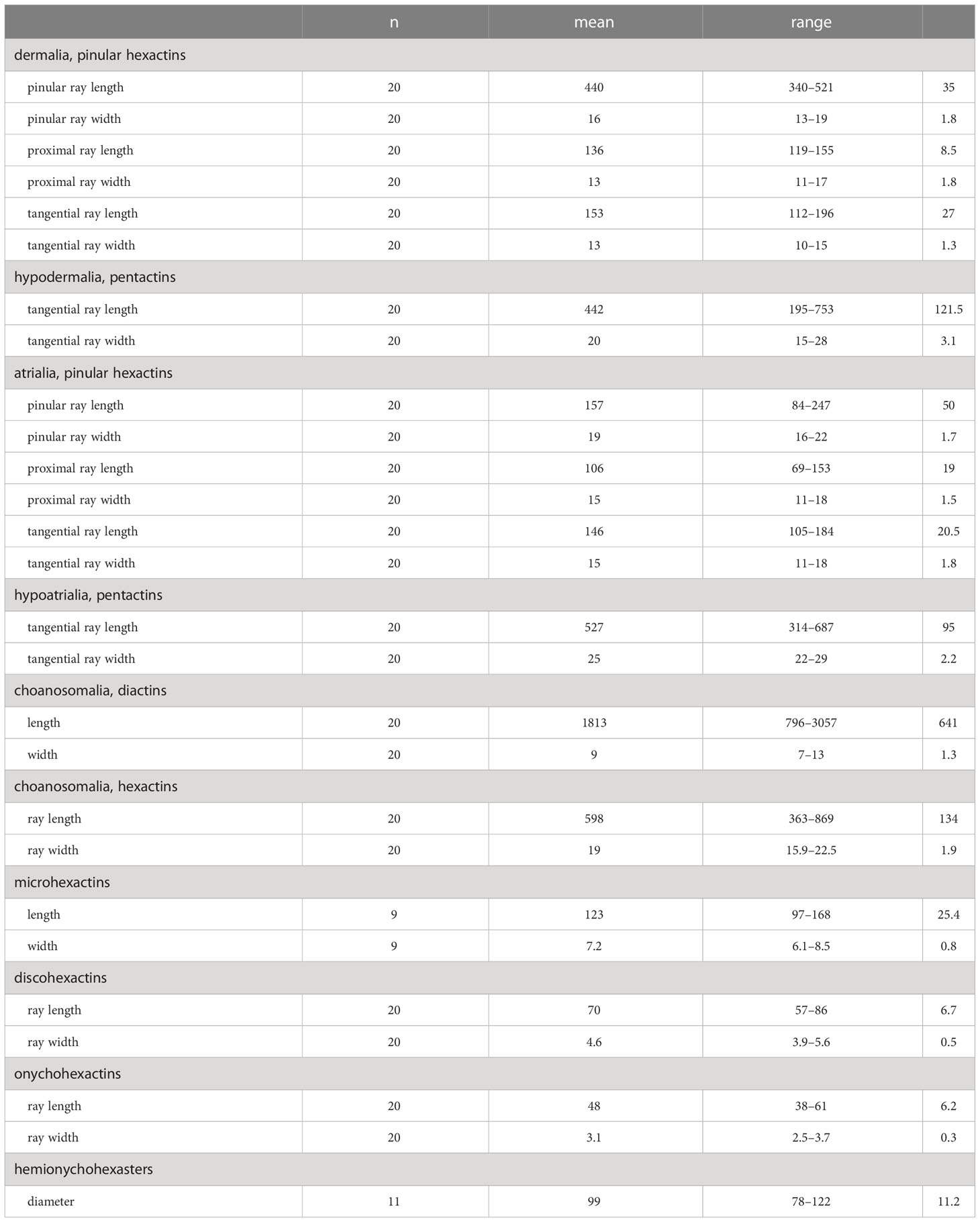
Table 1 Measurements of the spicules of Caulophacus (Caulodiscus) iocasicus sp. nov. (in µm); “n”, number of spicules measured; “s.d.”, standard deviation; “range”, range from the minimum to the maximum.
DNA extraction and PCR amplification
Total genomic DNA was extracted using a Tissue DNA Kit (OMEGA Bio-Tek) following the manufacturer’s instructions. Polymerase chain reaction (PCR) reaction mix (25 μL) comprised 12.5 μL of Premix Taq™ (Takara, Otsu, Shiga, Japan), 1 μL of each primer, 2 μL of template DNA, and 8.5 μL DNase-free ddH2O. The 16S rDNA was amplified with the primers 16S1fw/16SH_mod (Dohrmann et al., 2008). The amplification was performed using the following procedure: 5 min/94°C; 30 cycles (30 s/94°C, 30 s/48°C, 60 s/72°C); 5 min/72°C.
Illumina sequencing, mitochondrial genome assembly and annotation
Short-insert libraries (insert size of 430 bp) were constructed according to the manufacturer’s instructions of the TruSeq™ Nano DNA Sample Prep Kit (Illumina) and sequenced on an Illumina HiSeq 4000 instrument (San Diego, USA). Quality control of the raw data was performed using Trimmomatic (Bolger et al., 2014) by removing adapters, duplicated sequences, reads with a quality score below 20 (Q < 20), and reads containing a percentage of uncalled bases (“N” characters) equal to or greater than 10%. Clean data were reconstructed using a combination of de novo and reference-guided assemblies. De novo assembly with GetOrganelle v1.6.4 (Jin et al., 2020) was performed using the mitochondrial genome of Vazella pourtalesii (NC_028054.1) as the reference. Potential mitochondrial reads were extracted from the Illumina reads using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The filtered reads were assembled into contigs using SPAdes-3.13.1 (Bankevich et al., 2012). Finally, the assembled sequence was reordered and oriented according to the reference mitochondrial genome, thus generating the final assembled sequence. The mitochondrial genes were annotated using the MITOS2 webserver (Bernt et al., 2013). Locations and sizes of the protein coding genes (PCGs) were identified using Open Reading Frame Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) with the invertebrate mitochondrial code. Transfer RNA (tRNA) genes were predicted by ARWEN (Laslett & Canback, 2008) and DOGMA (Wyman et al., 2004). Ribosomal RNA (rRNA) genes were identified by the rRNAmmer 1.2 webserver (Lagesen et al., 2007). The mitochondrial genome map was generated by CGView (Stothard & Wishart, 2005). Nucleotide composition and codon usage were calculated using DnaSP 5.1 (Librado & Rozas, 2009). The values of AT and GC skews were calculated according to the following formulas: AT skew = (A − T)/(A + T) and GC skew = (G − C)/(G + C) (Perna & Kocher, 1995). The relative synonymous codon usage (RSCU) data were obtained with MEGA 7 (Kumar et al., 2016).
Phylogenetic analysis
A phylogenetic tree was reconstructed from partial 16S rDNA and COI gene sequences of Lanuginellinae. We obtained 11 16S rDNA sequences and 7 COI sequences of Lanuginellinae from GenBank, including all four subgenera of Caulophacus. Rossella levis was used as an outgroup (Table 2).
We used the workflow desktop platform of PhyloSuite (Zhang et al., 2020) to build the phylogenetic tree. The sequences were aligned using MAFFT (Katoh & Standley, 2013) with the default parameters. The concatenated dataset consisted of 1052 bp (16S/COI = 493/559bp). The best-fitting nucleotide substitution model, which was determined by ModelFinder 2 (Kalyaanamoorthy et al., 2017) with the BIC (Bayesian Information Criterion), was GTR+F+G4. Bayesian inference (BI) analysis was carried out in MrBayes 3.2 (Ronquist et al., 2012) with Markov Chains run for 2 million generations, sampling every 1000 generations. The first 25% of trees were discarded as burn-in. The average standard deviation of split frequencies reached 0.0045. Phylogenetic trees were annotated in iTOL (Letunic & Bork, 2021; https://itol.embl.de/).
We also built a phylogeny based on the 14 PCGs of mitochondrial sequences, also using PhyloSuite. We downloaded 13 relatively complete or complete mitogenomes of hexactinellid sponges belonging to two subclasses, three orders, and seven families (Table 2). Alignments were built in MAFFT. GBlocks v0.91b (Castresana, 2000) was used to eliminate ambiguously aligned regions and gaps of each gene. Then the trimmed alignments were concatenated into a single dataset using the Concatenate Sequence function in PhyloSuite. For BI and Maximum Likelihood (ML) analyses, the best-fit substitution models and partition schemes were inferred by ModelFinder (Kalyaanamoorthy et al., 2017) (Supplementary Table S1). Maximum likelihood (ML) analysis was performed in IQ-TREE 1.6.10 (Nguyen et al., 2015). Topological robustness for the ML analysis was evaluated with 1000 bootstrap replicates. Bayesian inference was conducted in MrBayes 3.2, using four chains run for 10 million generations, sampling every 1000 generations and discarding the first 25% of samples as burn-in.
Results
Taxonomy
Family Rossellidae Schulze, 1885
Subfamily Lanuginellinae Gray, 1872
Genus Caulophacus Schulze, 1886
Subgenus Caulophacus (Caulodiscus) Ijima, 1927
Caulophacus (Caulodiscus) iocasicus Gong & Li, sp. nov.
ZooBank registration LSID: urn:lsid:zoobank.org:act:0A0CB816-1E8B-485A-91C3-AEDA19A0B762
Material examined
Holotype
MBM287354, Caroline seamount located south of the Mariana Trench and north of the Yap Trench (10°30’58.559’’N, 140°10’23.238’’E), Pacific Ocean, 25 August 2017, 1055.5 m depth.
Paratype
MBM287355, Mariana M5 seamount near Mariana Trench (10°04’40.001’’N, 140°11’57.426’’E), Pacific Ocean, 28 May 2019, 884 m depth.
Description
The new species has 8–10 branches like a luxuriant tree growing on the bottom of the sea. There is a mushroom-shaped body at the end of each branch. The holotype has 8 mushroom-shaped spheres (Figures 2A, C). The overall dimension of each sphere ranges from 38 mm to 170 mm in diameter. The sponge is approximately 110 cm in height with a main stalk having a diameter of 17 mm and many stems having a thinnest diameter of 6 mm. The lateral stalks can be as strong as the main stalk and the length of each lateral stalk varies from 19 mm to 65 mm. The paratype has 10 spheres (Figures 2B, D) with diameters ranging from 40 mm to 160 mm. The sponge is approximately 120 cm in height, with a prominent main stalk having a diameter of 16 mm. The stalks of the sponge are hollow and there are no visible spicules on the surface. Color of the body in life is white and pale brown when preserved in ethanol.
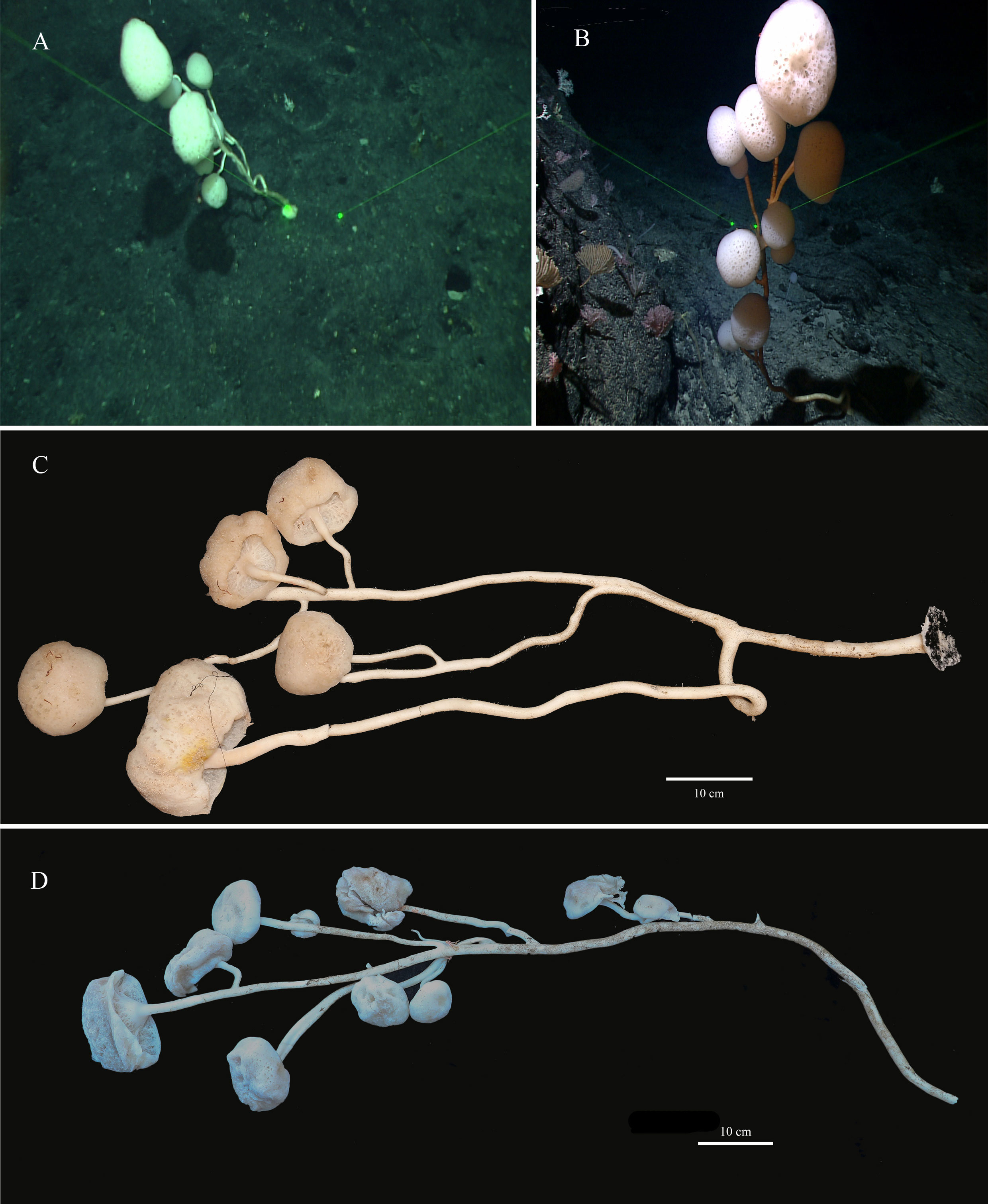
Figure 2 Caulophacus (Caulodiscus) iocasicus sp. nov. (A), photograph showing the holotype specimen in its natural habitat. (B), photograph showing the paratype specimen in its natural habitat. (C), external morphology of the holotype after removal from seawater. (D), external morphology of the paratype after removal from seawater.
Dermalia are pinular hexactins (Figure 3D), with pinular ray gradually tapering towards the end (340–(440)–521 µm in length), slightly spiny tangential rays 112–(153)–196 µm in length, and proximal ray 119–(136)–155 µm in length. Atrialia are pinular hexactins (Figure 3C) with spindle-like pinular ray (84–(157)–247 µm in length). The tangential and proximal rays, covered with many small spines, are 105–(146)–184 µm and 69–(106)–153 µm in length, respectively. Hypodermalia (Figure 3A) and hypoatrialia (Figure 3B) are pentactins with smooth rays. Tangential rays of hypodermalia and hypoatrialia are 195–(442)–753 µm and 314–(527)–687 µm in length, respectively. The length of the proximal ray is difficult to measure under a light microscope. Choanosomal spicules are diactins and hexactins. Diactins (796–(1813)–3057 µm in length; 7–(9)–13 µm in width) are smooth with sparsely spined bluntly conical terminations (Figure 3E). Hexactins (363–(598)–869 µm in length; 15.9–(19.4)–22.5 µm in width) are smooth with conical terminations.
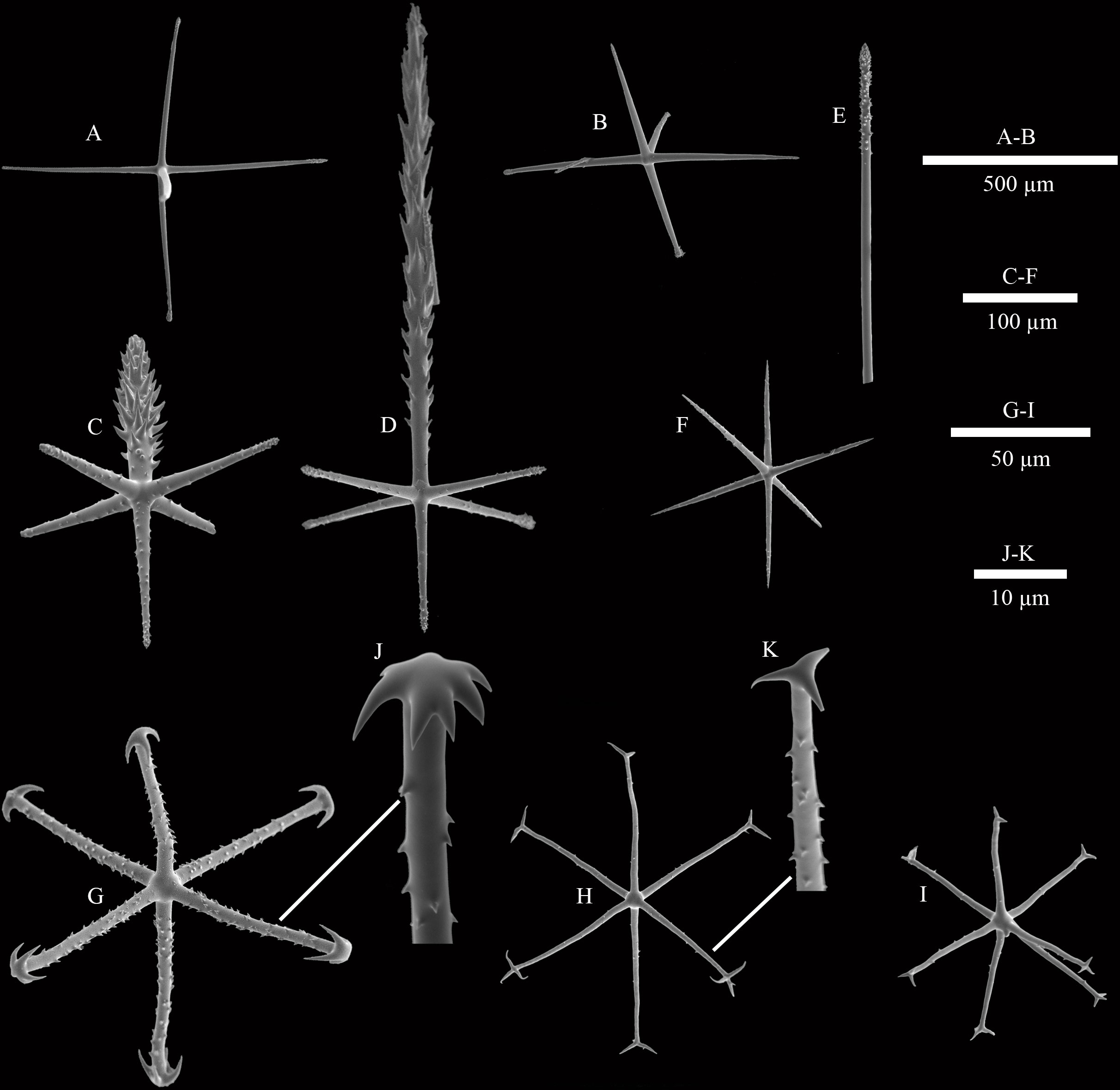
Figure 3 SEM images of spicules of Caulophacus (Caulodiscus) iocasicus sp. nov. (A), hypodermal pentactin; (B), hypoatrial pentactin; (C), atrial pinular hexactin; (D), dermal pinular hexactin; (E), details of the termination of choanosomal diactin; (F), microhexactin; (G), discohexactin; (H), onychohexactin; (I), hemionychohexaster; (J), detail of the toothed disc of discohexactin; (K), detail of the termination of onychohexactin.
Microscleres are discohexactins, onychohexactins, hemionychohexasters and microhexactins. Discohexactins (Figure 3G) have six slightly curved rays (57–(70)–86 µm in length; 3.9–(4.6)–5.6 µm in width) covered with numerous spines (Figure 3J). Most of the onychoidal microscleres are onychohexactins (Figure 3H) with rays covered with small spines (Figure 3K). The ray length of onychohexactins is 38–(48)–61 µm and the width is 2.5–(3.1)–3.7 µm. Hemionychohexasters (Figure 3I) are 78–(99)–122 µm in diameter. Microhexactins (Figure 3F) with smooth rays (97–(123)–168 µm in length and 6.1–(7.2)–8.5 µm in width) occur infrequently.
Etymology
iocasicus is named after iocas, celebrating the 70th anniversary of the Institute of Oceanology, Chinese Academy of Sciences (IOCAS) from its establishment in 1950.
Type locality
Seamounts near Mariana Trench and Yap Trench (western Pacific) with hard bottom, 884–1055.5 m.
Mitochondrial genome organization and characterization
The complete mitogenome of C. (Caulodiscus) iocasicus sp. nov. is 19,930 bp in length. The genome encodes 37 genes, including 14 PCGs, 2 rRNA genes, and 21 tRNA genes (duplication of tRNALeu and tRNASer) (Figure 4). All of the genes are encoded on the heavy (H) strand. There were 5 overlaps between adjacent genes with a size range of 3 to 83 bp (Table 3). The combined length of 14 PCGs was 12,041 bp, accounting for 60.42% of the complete mitochondrial genome. Two of the PCGs started with ATA as initiation codons, while the others started with ATG. All of the PCGs ended with TAA as the stop codon (Table 3). The 14 PCGs encode a total of 4,033 amino acids, Met (12.77%) and Cys (0.92%) are the most and the least frequently used amino acids, respectively (Figure 5). The relative synonymous codon usage (RSCU) values show that the six most commonly used codons are AGA (Ser), GUA (Val), CGA (Arg), GGA (Gly), UUA (Leu), and CUA (Leu) (Figure 5), all of which have A in their third codon position. These features are similar to many metazoans, where codon usage is biased toward A and T at the third codon position (Zhang et al., 2017; Yang et al., 2019). The lengths of the 21 tRNA genes range from 63 (tRNAMet) to 73 bp (tRNASer (AGA)), and all tRNAs can be folded into classic clover leaf structures (Supplementary Figure S1). The rRNA genes of rrnS and rrnL of the new species are 940 bp and 1493 bp in length, respectively (Table 3).
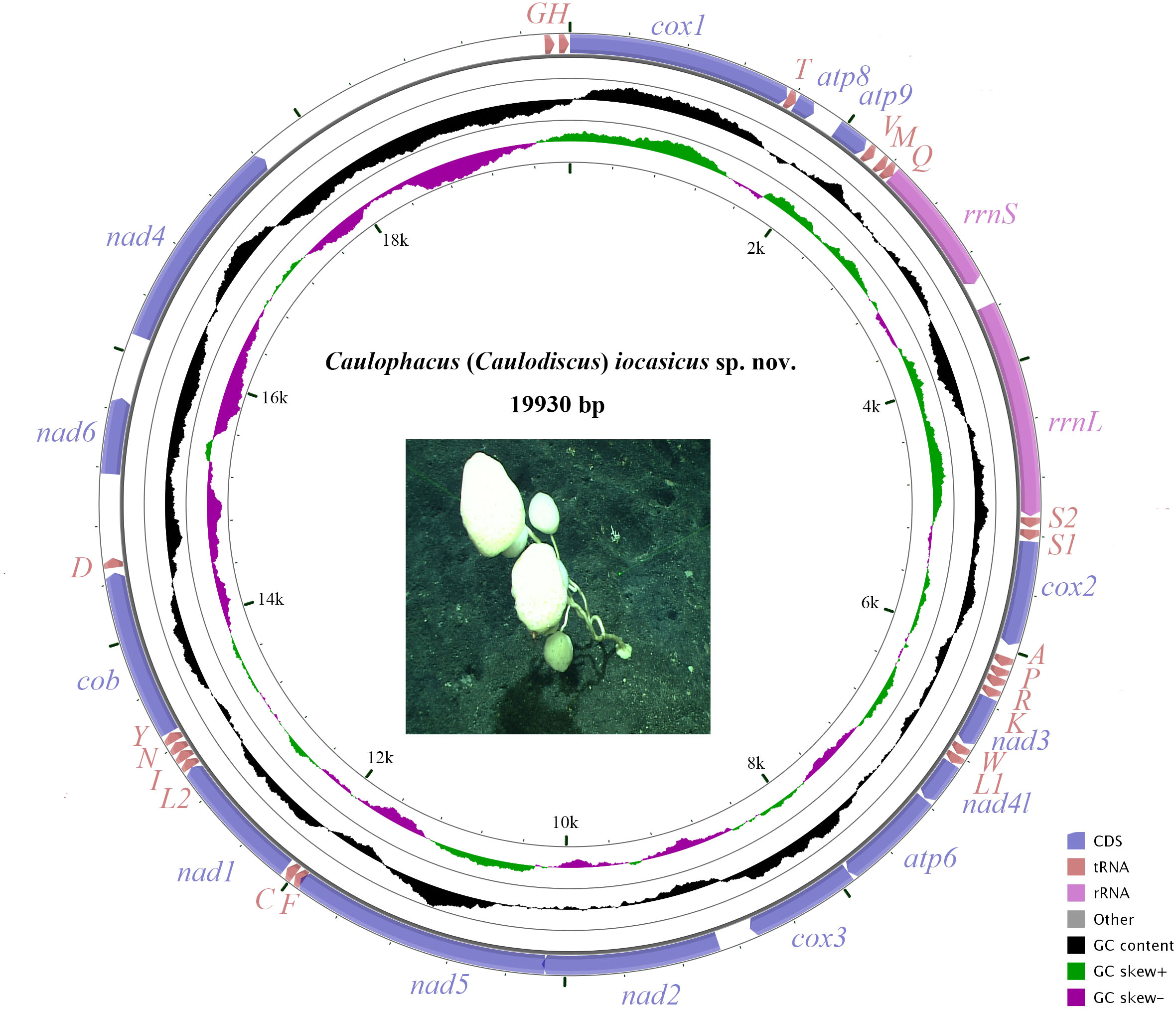
Figure 4 The organization of the mitochondrial genome of Caulophacus (Caulodiscus) iocasicus sp. nov. Genes for protein coding genes are shown with standard abbreviations. 12S and 16S ribosomal RNA genes are are indicated as rrnS and rrnL, respectively. Genes for tRNAs are labeled by one-letter code for their corresponding amino acid.
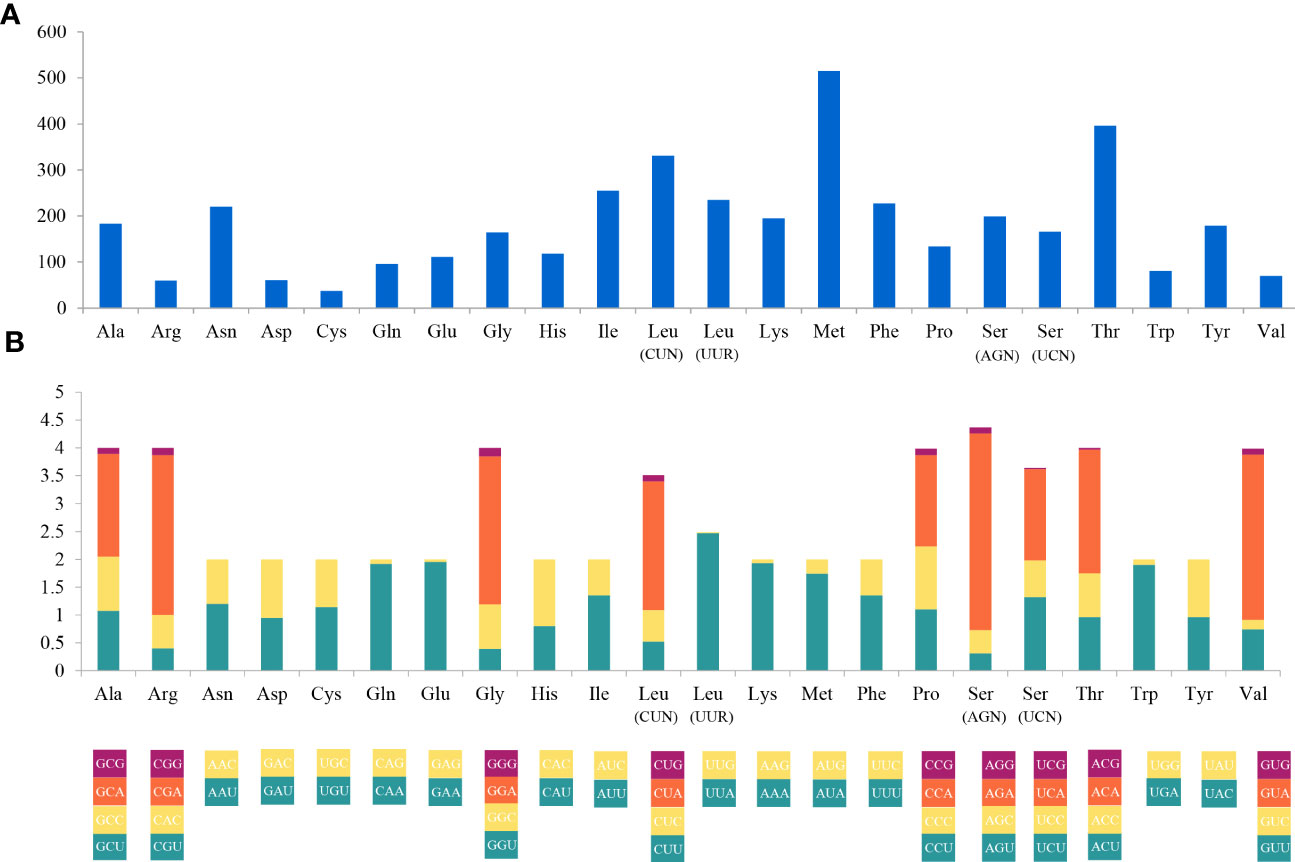
Figure 5 Codon usage (A) and the relative synonymous codon usage (RSCU) (B) of the Caulophacus (Caulodiscus) iocasicus sp. nov. mitogenome. Numbers to the left refer to the total number of codons (A) and the RSCU values (B).
Phylogenetic analysis
16S rDNA and COI gene phylogeny
In the BI tree (Figure 6) based on the 16S rDNA and COI sequences, C. (Caulodiscus) iocasicus sp. nov. is nested within the genus Caulophacus, confirming the validity of our genus assignment. The new species is not closely related to C. (Oxydiscus), and relationships to C. (Caulodiscus) or C. (Caulophacus) are not resolved. Caulophacella groups outside of Caulophacus, as sister to all other Lanuginellinae included here. This is different from previous studies that resolved a closer relationship of Caulophacella to Doconesthes and Lophocalyx (Dohrmann et al., 2017; Kersken et al., 2018; Dohrmann, 2019).

Figure 6 Phylogenetic tree obtained by Bayesian inference analysis based on 16S rDNA and COI gene sequences. The numbers at each node are Bayesian posterior probabilities (PP).
Mitochondrial protein-coding gene phylogeny
The mitogenomic phylogenetic tree based on the nucleotide sequences of 14 PCGs (Supplementary Figure S2) is consistent with previous works (e.g., Haen et al., 2014; Dohrmann, 2019). The new species is nested within the family Rossellidae as sister to the other included lanuginelline, Sympagella nux.
Mitochondrial gene order and rearrangements
The mitogenome of C. (Caulodiscus) iocasicus sp. nov. contains atp9, which has only been found in sponges among Metazoa (Haen et al., 2007). The new species is only the second species with atp8, which is lacking in most of the hexactinellid sponges investigated so far (except Oopsacas minuta (Jourda et al., 2015)). Additionally, it misses tRNA E, unlike other rossellids. The gene orders vary considerably among hexactinellid sponges, while the more closely related taxonomic groups are generally stable (Figure 7). Within Rossellidae, the orders of protein coding genes are generally conserved except for the absence of nad6 in most species. Transposition of nad4 and nad6 occurred in the two completed sequences of V. pourtalesii and the new species. In rossellid sponges, two conserved gene clusters are shared: M–Q–rrnS–rrnL–S2 and K–nad3–W–L1–nad4–atp6–cox3–nad2–nad5–F–C–nad1–L2–I–N. Although most of the gene sequences are incomplete, we can see a gene rearrangement of tRNAD in the new species.

Figure 7 Mitochondrial gene organization and gene rearrangement in Hexactinellida. The length of mitochondrial genomes is presented in brackets. Conserved gene blocks within Rossellidae are shown in pink line; gene arrangements of tRNAD within Rossellidae are shown as green triangles; =atp8 genes are shown as blue triangles.
Discussion
Morphological differences
The new species is attributed to the genus Caulophacus by having long peduncles, dermalia and atrialia being pentactins and hexactins, and microscleres having discoidal and onychoidal terminations. The most outstanding character of the new species is its external shape with many branches. Other pedunculate branched Caulophacus only have small branches, such as C. (Caulophacus) ramosus Reiswig, Dohrmann & Kelly, 2021. However, their spicules are very different. Caulophacus (Caulophacus) ramosus has thin-rayed stellate discohexasters and lacks onychohexasters (Reiswig et al., 2021).
The new species is assigned to C. (Caulodiscus) by having microscleres with discoidal and onychoidal terminations. Two of the six described species of C. (Caulodiscus) have onychohexasters (C. (Caulodiscus) onychohexactinus Tabachnick & Lévi, 2004 and C. (Caulodiscus) lotifolium Ijima, 1903), and the other four species have discohexasters, while the new species only has hemionychohexasters. It also has microhexactins, which were not found in other C. (Caulodiscus) species. Therefore, the unique combination of microscleres justifies the proposal that the species is new to science. Another distinctive characteristic of the new species is the multiramose body shape, unknown from other C. (Caulodiscus) species, which only have a single unbranched peduncle.
Molecular data
Molecular data show that the new species does not appear to be closely related to any of the included C. (Caulophacus) or C. (Caulodiscus) species. However, both of these subgenera are polyphyletic groups in the molecular phylogeny. Increased taxon sampling of Caulophacus for molecular phylogenetics combined with thorough morphological revision will be necessary to resolve its internal relationships and propose a natural subgeneric classification of this large genus.
Establishment of the genus Caulophacella
In molecular phylogenies, C. (Caulophacella) does not nest within the clade of Caulophacus (Dohrmann et al., 2017; Kersken et al., 2018; Dohrmann, 2019; this study: Figure 6). Also, all Caulophacus species have discoidal microscleres, except for C. (Caulophacella) tenuis. Therefore, we remove Caulophacella from Caulophacus and reinstate it as a separate genus in the subfamily Lanuginellinae, reversing the move of Boury-Esnault et al. (2015).
Conclusion
Caulophacus species are mostly deep-sea dwellers, and their richest abundance is at depths of 3000–5000 m. Caulophacus (Caulodiscus) iocasicus sp. nov. is reported from depths of 884–1055.5 m, somewhat shallower than most species (Supplementary Excel S1). The new species has a highly branched stalk with many mushroom-shaped bodies while most of its congeners only have a single unbranched stalk. Also, its spicule composition is unique among C. (Caulodiscus).
The complete mitogenome of the new species has 14 PCGs, 2 rRNA genes, and 21 tRNA genes. It is the second hexactinellid genome that contains atp8, but it lacks tRNA E, which is reported from all other rossellids investigated so far. A higher taxon coverage of mitogenomes will be needed to explore the mitochondrial characteristics of Rossellidae and to obtain a more comprehensive understanding of Caulophacus.
Caulophacus (Caulophacella) does not group within Caulophacus in molecular phylogenies and lacks discoidal microscleres, which are always present in the other subgenera. Therefore, we reinstate it as a separate genus in the subfamily Lanuginellinae.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
LG drafted the manuscript. MY conducted the analysis of the mitochondrial genome. DJ and MD supervised the identification of the new species from morphology and molecular data, respectively. MD revised and edited the manuscript. XL designed the study and revised the manuscript. We confirm that all the listed authors have participated actively in the study, and have seen and approved the submitted manuscript. The authors declare that they have no conflict of interest. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Senior User Project of RV KEXUE (KEXUE2020GZ01) and the Biological Resources Program, Chinese Academy of Sciences (No. KFJ-BRP-017-37).
Acknowledgments
Special thanks to Dr. Kuidong Xu (Institute of Oceanology, Chinese Academy of Sciences, Qingdao) for providing the deep-sea sponge specimens and the in situ pictures. We are grateful for the crews of the R/V ‘Kexue’ for their support in collecting the deep-sea sponge specimens during the cruises. We sincerely express our gratitude to the editor, Prof. Manuel Maldonado, and the reviewers for investing their time to help to improve this paper.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.979912/full#supplementary-material
References
Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bernt M., Donath A., Jühling F., Externbrink F., Florentz C., Fritzsch G., et al. (2013). MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69, 313–319. doi: 10.1016/j.ympev.2012.08.023
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boore J. L. (1999). Animal mitochondrial genomes. Nucleic Acids Res. 27, 1767–1780. doi: 10.1093/nar/27.8.1767
Boury-Esnault N., Vacelet J., Reiswig H. M., Fourt M., Aguilar R., Chevaldonné P. (2015). Mediterranean hexactinellid sponges, with the description of a new Sympagella species (Porifera, Hexactinellida). J. Mar. Biol. Ass. U.K. 95, 1353–1364. doi: 10.1017/S0025315414001891
Buskowiak D., Janussen D. (2021). An exceptional occurrence of deep-sea sponges in the region of former Larsen ice shelves, Antarctic peninsula, with the description of two new species. Mar. Biodivers. 51, 8. doi: 10.1007/s12526-020-01134-z
Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
de Voogd N. J., Alvarez B., Boury-Esnault N., Carballo J. L., Cárdenas P., Díaz M.-C., et al. (2022) World Porifera database. Available at: https://www.marinespecies.org/porifera (Accessed April 8, 2022).
Dohrmann M. (2019). Progress in glass sponge phylogenetics: a comment on Kersken et al. (2018). Hydrobiologia 843, 51–59. doi: 10.1007/s10750-018-3708-7
Dohrmann M., Janussen D., Reitner J., Collins A. G., Wörheide G. (2008). Phylogeny and evolution of glass sponges (Porifera, Hexactinellida). Syst. Biol. 57 (3), 388–405. doi: 10.1080/10635150802161088
Dohrmann M., Kelley C., Kelly M., Pisera A., Hooper J. N. A., Reiswig H. M. (2017). An integrative systematic framework helps to reconstruct skeletal evolution of glass sponges (Porifera, Hexactinellida). Front. Zool. 14, 18. doi: 10.1186/s12983-017-0191-3
Ereskovsky A. V., Richter D. J., Lavrov D. V., Schippers K. J., Nichols S. A. (2017). Transcriptome sequencing and delimitation of sympatric Oscarella species (O. carmela and o. pearsei sp. nov) from California, USA. PloS One 12 (9), e0183002. doi: 10.1371/journal.pone.0183002
Gazave E., Lapébie P., Renard E., Vacelet J., Rocher C., Ereskovsky A. V., et al. (2010). Molecular phylogeny restores the supra-generic subdivision of homoscleromorph sponges (Porifera, Homoscleromorpha). PloS One 5 (12), e14290. doi: 10.1371/journal.pone.0014290
Gray J. E. (1872). Notes on the classification of the sponges. Ann. Mag. Nat. Hist. 9 (54), 442–461. doi: 10.1080/00222937208696616
Haen K. M., Lang B. F., Pomponi S. A., Lavrov D. V. (2007). Glass sponges and bilaterian animals share derived mitochondrial genomic features: a common ancestry or parallel evolution? Mol. Biol. Evol. 24 (7), 1518–1527. doi: 10.1093/molbev/msm070
Haen K. M., Pett W., Lavrov D. V. (2014). Eight new mtDNA sequences of glass sponges reveal an extensive usage of +1 frameshifting in mitochondrial translation. Gene 535 (2), 336–344. doi: 10.1016/j.gene.2013.10.041
Ijima I. (1903). Studies on the Hexactinellida. Contribution III. (Placosoma, a new euplectellid; Leucopsacidae and Caulophacidae). J. Coll. Sci. Imp. Univ. Tokyo 18 (1), 1–124.
Ijima I. (1927). “The Hexactinellida of the Siboga expedition,” in Siboga-expeditie. uitkomsten op zoologisch, botanisch, oceanographisch en geologisch gebied verzameld in nederlandsch oost-lndië 1899- 1900 aan boord H.M. Ed. Weber M. (Leiden: E.J. Brill), 1–383.
Janussen D., Tabachnick K. R., Tendal O. S. (2004). Deep-sea Hexactinellida (Porifera) of the Weddell Sea. Deep-Sea Res. Part II 51, 1857–1882. doi: 10.1016/j.dsr2.2004.07.018
Jin J. J., Yu W. B., Yang J. B., Song Y., de Pamphilis C. W., Ting-Shuang Yi T. S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21, 241. doi: 10.1186/s13059-020-02154-5
Jourda C., Santini S., Rocher C., Le Bivic A., Claverie J. M. (2015). Mitochondrial genome sequence of the glass sponge Oopsacas minuta. Genome Announc. 3 (4), e00823–15. doi: 10.1128/genomeA.00823-15
Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14 (6), 587–589. doi: 10.1038/nmeth.4285
Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30 (4), 772–780. doi: 10.1093/molbev/mst010
Kersken D., Kocot K., Janussen D., Schell T., Pfenninger M., Arbizu. P. M. (2018). First insights into the phylogeny of deep-sea glass sponges (Hexactinellida) from polymetallic nodule fields in the Clarion-Clipperton Fracture Zone (CCFZ), northeastern Pacific. Hydrobiologia 811 (1), 283–293. doi: 10.1007/s10750-017-3498-3
Koltun V. M. (1970). “Sponge fauna of the northwestern Pacific from the shallows to the hadal depths,” in Fauna of the Kurile-Kamchatka trench and its environment. Institute of oceanology of the academy of sciences of the U.S.S.R. Ed. Bogorov V. G. (Moskwa: Akademiya Nauk SSSR), 165–221.
Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lagesen K., Hallin P., Rodland E. A., Staerfeldt H. H., Rognes T., Ussery D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic. Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Laslett D., Canback B. (2008). ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 24, 172–175. doi: 10.1093/bioinformatics/btm573
Lavrov D. V., Adamski M., Chevaldonné P., Adamska M. (2016). Extensive mitochondrial mRNA editing and unusual mitochondrial genome organization in calcaronean sponges. Curr. Biol. 26, 86–92. doi: 10.1016/j.cub.2015.11.043
Lendenfeld R. (1915). The sponges. 3. Hexactinellida. Memoirs Mus. Comp. Zoology Harv. Coll. 42 (2), 1–396.
Letunic I., Bork P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucl. Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Lévi C. (1964). Spongiaires des zones bathyale, abyssale et hadale. Galathea Rep. Sci. Result. Dan. Deep-Sea Exped. Round World 1950-52 7, 63–112.
Librado P., Rozas J. (2009). DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Perna N. T., Kocher T. D. (1995). Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 41, 353–358. doi: 10.1007/BF01215182
Plese B., Kenny N. J., Rossi M. E., Cardenas P., Schuster A., Taboada S., et al. (2021). Mitochondrial evolution in the Demospongiae (Porifera): Phylogeny, divergence time, and genome biology. Mol. Phylogenet. Evol. 155, 107011. doi: 10.1016/j.ympev.2020.107011
Reiswig H. M., Dohrmann M., Kelly M., Mills S., Schupp P. J., Wörheide G. (2021). Rossellid glass sponges (Porifera, Hexactinellida) from New Zealand waters, with description of one new genus and six new species. ZooKeys 1060, 33–84. doi: 10.3897/zookeys.1060.63307
Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61 (3), 539–542. doi: 10.1093/sysbio/sys029
Schulze F. E. (1885). “The Hexactinellida,” in Report on the scientific results of the voyage of H.M.S. ‘Challenger’, 1873-1876. Eds. Tizard T. H., Moseley H. M., Buchanan J. Y., Murray J. (Edinburgh: Her Majesty Stationery office), 437–451.
Schulze F. E. (1886). Über den Bau und das System der Hexactinelliden Phys. Abh. königl. preuß. Akad. Wiss. Berlin (Phys.-Math. Cl.) 1, 1–97.
Schulze F. E. (1887). Report on the Hexactinellida collected by H.M.S. ‘Challenger’ during the years 1873–1876. Rep. Sci. Challenger Zoology 21, 1–514.
Stothard P., Wishart D. S. (2005). Circular genome visualization and exploration using CGView. Bioinformatics 21, 537–539. doi: 10.1093/bioinformatics/bti054
Tabachnick K. R. (2002). “Family Rossellidae Schulze, 1885,” in Systema Porifera - a guide to the classification of sponges. Eds. Hooper J. N. A., Soest R. W. M. V. (New York, Boston, Dordrecht, London, Moscow: Kluwer Academic/Plenum Publishers), 1441–1505.
Tabachnick K. R., Lévi C. (2004). “Lyssacinosida du pacifique sud-ouest (Porifera: Hexactinellida),” in Tropical deep-Sea benthos 23. Eds. Marshall B. A., de Forg R. B. Mém. Mus. Natl. Hist. Nat. 191, 11–71.
Wyman S. K., Jansen R. K., Boore J. L. (2004). Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20, 3252–3255. doi: 10.1093/bioinformatics/bth352
Yang M., Gong L., Sui J., Li X. (2019). The complete mitochondrial genome of Calyptogena marissinica (Heterodonta: Veneroida: Vesicomyidae): Insight into the deep-sea adaptive evolution of vesicomyids. PloS One 14 (9), e0217952. doi: 10.1371/journal.pone.0217952
Zhang D., Gao F., Jakovli I., Zou H., Wang G. T. (2020). Phylosuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20 (1), 348–355. doi: 10.1111/1755-0998.13096
Keywords: Porifera, Caulophacus (Caulodiscus) iocasicus sp. nov., Caulophacella, seamount, mitochondrial genome
Citation: Gong L, Yang M, Janussen D, Dohrmann M and Li X (2023) A new species of Caulophacus (Hexactinellida: Lyssacinosida: Rossellidae) from the western Pacific Ocean, with new insights into the mitochondrial genome characteristics of hexactinellid sponges. Front. Mar. Sci. 10:979912. doi: 10.3389/fmars.2023.979912
Received: 04 July 2022; Accepted: 25 January 2023;
Published: 09 February 2023.
Edited by:
Manuel Maldonado, Center for Advanced Studies of Blanes (CSIC), SpainReviewed by:
Swee Cheng Lim, National University of Singapore, SingaporePilar Ríos, Spanish Institute of Oceanography, Spain
Laia Leria, Centro de Estudios Avanzados de Blanes (CEAB-CSIC), Spain
Copyright © 2023 Gong, Yang, Janussen, Dohrmann and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinzheng Li, bGl4emhAcWRpby5hYy5jbg==
†These authors have contributed equally to this work
 Lin Gong
Lin Gong Mei Yang1,2†
Mei Yang1,2† Dorte Janussen
Dorte Janussen Martin Dohrmann
Martin Dohrmann Xinzheng Li
Xinzheng Li