
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 22 January 2024
Sec. Marine Evolutionary Biology, Biogeography and Species Diversity
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1323156
This article is part of the Research TopicEVOLMAR 2023: r-Evolutionary routes in the SeaView all articles
 Elisa Nocella1,2*
Elisa Nocella1,2* Sofya Sergeevna Zvonareva3
Sofya Sergeevna Zvonareva3 Giulia Fassio1
Giulia Fassio1 Daniela Pica4
Daniela Pica4 Barbara Buge5
Barbara Buge5 Raimondo Villa6
Raimondo Villa6 Nicolas Puillandre5
Nicolas Puillandre5 Maria Vittoria Modica2†
Maria Vittoria Modica2† Marco Oliverio1†
Marco Oliverio1†Introduction: Host-parasite associations provide very useful models to study adaptive processes. We investigated the interaction between carnivorous marine gastropods, the Ovulidae or egg-cowries, and their cnidarian food targets. Ovulidae (Fleming, 1828), is a family of specialized carnivorous caenogastropods that feed by browsing on octocorals (Anthozoa: Octocorallia: Malacalcyonacea and Scleralcyonacea) or, to a much lesser degree, on antipatharians (Anthozoa: Hexacorallia: Antipatharia) and Stylasteridae (Hydrozoa: Hydroidolina: Anthoathecata). Very scanty information is available on the phylogenetic relationships and the degree of specificity of the relationship with the cnidarians of this corallivorous lineage, especially for deep-water taxa.
Methods: To assess taxonomic identifications and investigate cnidarian/ovulid relationships in the context of their evolution, we generated an extensive molecular dataset comprising two mitochondrial (cox1 and 16S rDNA) and one nuclear gene (28S rDNA) from 524 specimens collected worldwide. The coral hosts of the ovulid species have been identified by integrating literature data with new records, employing morphological and/or molecular (the mitochondrial 16S rDNA and mtMSH, and the nuclear ITS2) markers.
Results: We obtained a molecular phylogenetic framework for the Ovulidae, time-calibrated with nine reliable fossil records. An ancestral state reconstruction allowed to identify Hexacorallia or Hydroidolina as the most likely ancestral cnidarian host for the Ovulidae.
Discussion: Our phylogenetic hypothesis revealed the existence of groups that do not completely correspond to the currently employed subfamilial arrangement. Concerning trophic ecology, while only pediculariines (Pedicularia and allied) are associated with hydrozoans (Stylasteridae), our results suggest that some ovulid lineages shifted independently between octocorals and hexacorals.
The egg-cowries comprise a group of specialized parasitic caenogastropods that feed by browsing on octocorals (Anthozoa: Octocorallia: Malacalcyonacea and Scleralcyonacea) or, to a much lesser extent, on hexacorals (Anthozoa: Hexacorallia: Antipatharia and Scleractinia) (Tazioli et al., 2007) or stylasterid corals (Hydrozoa: Hydroidolina: Anthoathecata: Stylasteridae) (Lorenz and Fehse, 2009). Egg-cowries are predominantly tropical, with only few species inhabiting temperate areas (Zvonareva et al., 2020). Most species live in shallow waters, between 10 and 50 meters, but some are collected as deep as 1100 m (Lorenz and Fehse, 2009). Their shells can be pyriform, ovate, cylindrical or lanceolate with a slit-shaped aperture (Schiaparelli et al., 2005); in living specimens of most species the shell is almost entirely covered by the mantle, that camouflages the gastropod on its cnidarian host, and a few species have aposematic colorations (Rosenberg, 1992) (Figure 1). Egg-cowries are mostly classified in the family Ovulidae Fleming, 1828; however, the species of the genus Pedicularia differ from the remaining egg-cowries by not covering their shell with the mantle (Lorenz and Fehse, 2009) and in their radular morphology (Simone, 2004). Due to these features, Pedicularia and a few additional taxa are currently classified in a separate family Pediculariidae Gray, 1853 (Fehse, 2007; Lorenz and Fehse, 2009).
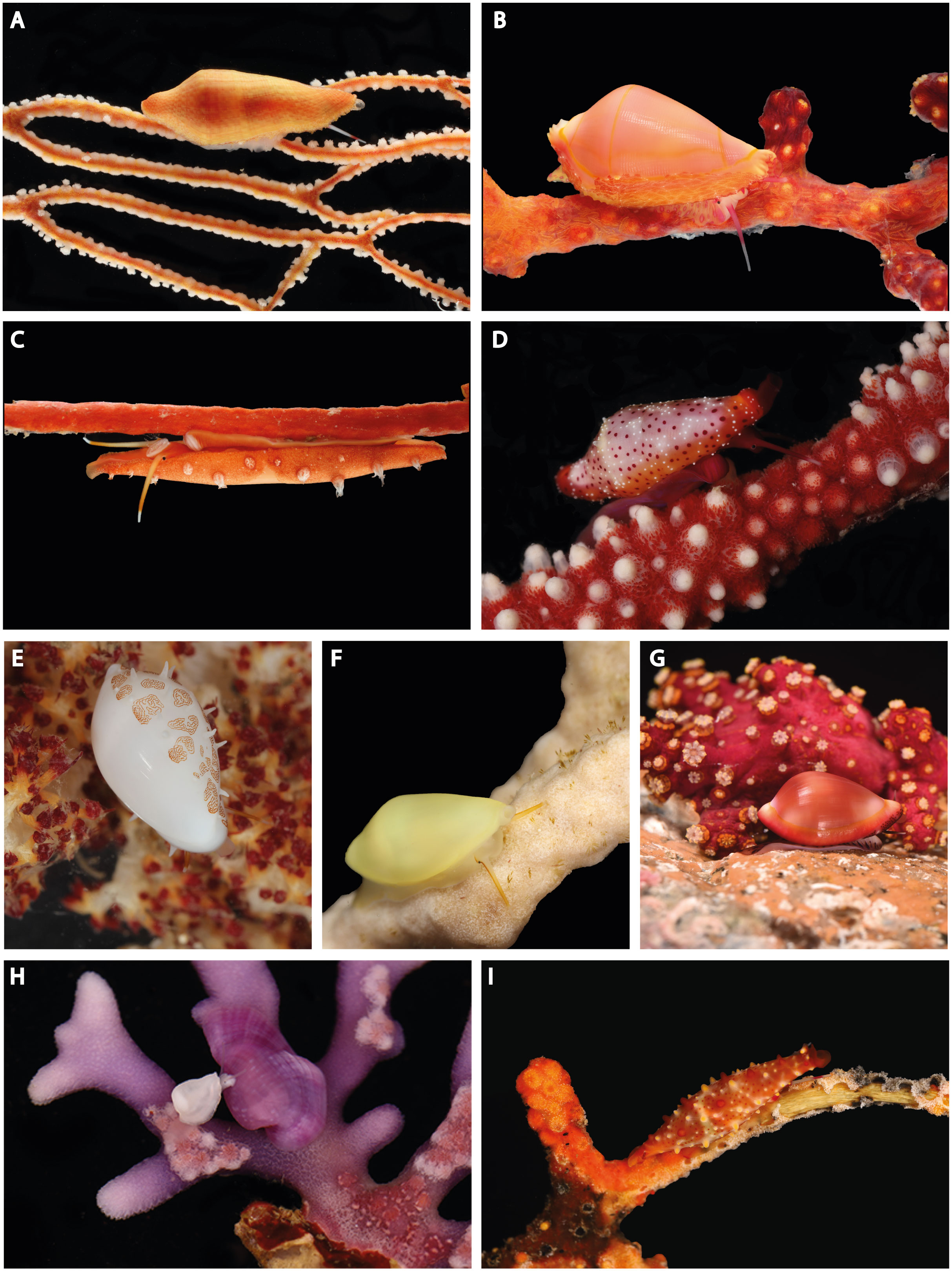
Figure 1 Living specimens on host analyzed in this work. (A) Prosimnia semperi (MNHN-IM-2013-4178) on Melithaeidae, Papua New Guinea. (B) Dentiovula dorsuosa (MNHN-IM-2013-4461) on Siphonogorgiidae, Papua New Guinea. (C) Aclyvolva coarcata (MNHN-IM-2013-11745) on Ellisellidae, Papua New Guinea. (D) Crenavolva traillii (Ov-QL-1361) on Paramuriceidae, Vietnam. (E) Habuprionovolva sp. (MNHN-IM-2013-17977) on Neptheidae, Papua New Guinea. (F) Naviculavolva kurziana (MNHN-IM-2019-8310) on Isididae, New Caledonia. (G) Pseudosimnia carnea (MNHN-IM-2019-18135) on Alcyoniidae, Corse. (H) Pedicularia pacifica (male and female, Ov-NT-1736 and Ov-NT-1737) on Stylasteridae, Vietnam. (I) Phenacovolva rosea (Ov-NT-1733) on Acanthogorgiidae, Vietnam. Photograph credits: (A–C, E, F), Laurent Charles (MNHN); (D, H, I), Elena Mekhova (IEE); (G), Gilles Devauchelle (MNHN).
Ovulids and pediculariids reside permanently on their cnidarian host or in its close vicinity, feeding on polyps and their secretions. Certain species seem to favor a single cnidarian host while others have a more varied diet including hosts of different species, genera, and even families of Cnidaria (Lorenz and Fehse, 2009). However, little information is available on ovulid-cnidarian interactions (e.g. Reijnen et al., 2010; Zvonareva et al., 2020). We use the parasite-host categorization for all relationships between the ovulids and their cnidarian food targets. However, ovulids that feed on colonial cnidarians may be regarded as parasites of the colony, and as predators of the single polyps, still behaving as carnivorous grazers (see e.g. Schiaparelli et al., 2011).
The taxonomy of Ovulidae and Pediculariidae has been restructured several times (e.g. Schilder, 1932; Fehse, 2001; Lorenz and Fehse, 2009). The families as currently conceived encompass 292 accepted species (274 Ovulidae and 18 Pediculariidae: MolluscaBase, 2023), although Schiaparelli et al. (2005) suggested that the actual number of living species could be reduced to a more reasonable estimation of c. 100. According to Rosenberg (1992) the reason for this profusion of nominal taxa is the remarkable variation in their mantle color and shell shape.
We have here generated the largest molecular dataset so far for egg-cowries, including one nuclear (28S rDNA) and two mitochondrial (cox1 and 16S rDNA) molecular markers, from 524 specimens collected worldwide, to provide a robust molecular phylogenetic framework for taxonomic and evolutionary studies of ovulids and pediculariids. We aim to i) gather all available knowledge on trophic associations between egg-cowries and cnidarians and integrate it with new data including cnidarian identification through DNA-barcoding; ii) reconstruct the evolution of trophic ecology for egg-cowries within a robust phylogenetic framework; iii) analyze the relationship between trophic specialization and phyletic diversification.
The dataset consisted of 524 specimens, morphologically ascribed to species belonging to 36 of the 39 accepted extant genera of Ovulidae and Pediculariidae (MolluscaBase, 2023); specimens from tropical, subtropical and temperate areas were included (Figure 2, created using QGIS Geographic Information System. http://qgis.org). A total of 60 specimens were collected by the authors (MO, SSZ, DP) or kindly provided by Paolo Mariottini of the University of Roma Tre. A significant proportion of the samples (197 specimens) were collected during several scientific expeditions by the Muséum national d’Histoire Naturelle (MNHN — www.expeditions.mnhn.fr). Samples were collected from 0 m to approximately 800 m depth and specifically fixed in the field for molecular analyses. Specimens collected after 2012 were processed with a microwave oven to facilitate removal of soft tissue from the shell (Galindo et al., 2014). Whole specimens were almost invariably preserved in 95-98% ethanol. The majority of shells were kept intact for identification and deposited as vouchers in the MNHN, Australian Museum (AM) and Biologia Ambientale e dell’Uomo (BAU) collections. In some cases, ovulid samples have been collected with their host and stored together. In the lack of preserved host, when the ovulid whole body was available, the stomach and foregut were dissected and processed for the amplification of cnidarian DNA directly from the snail’s digestive system (Oliverio et al., 2009a) using cnidarian primer sets (see below).
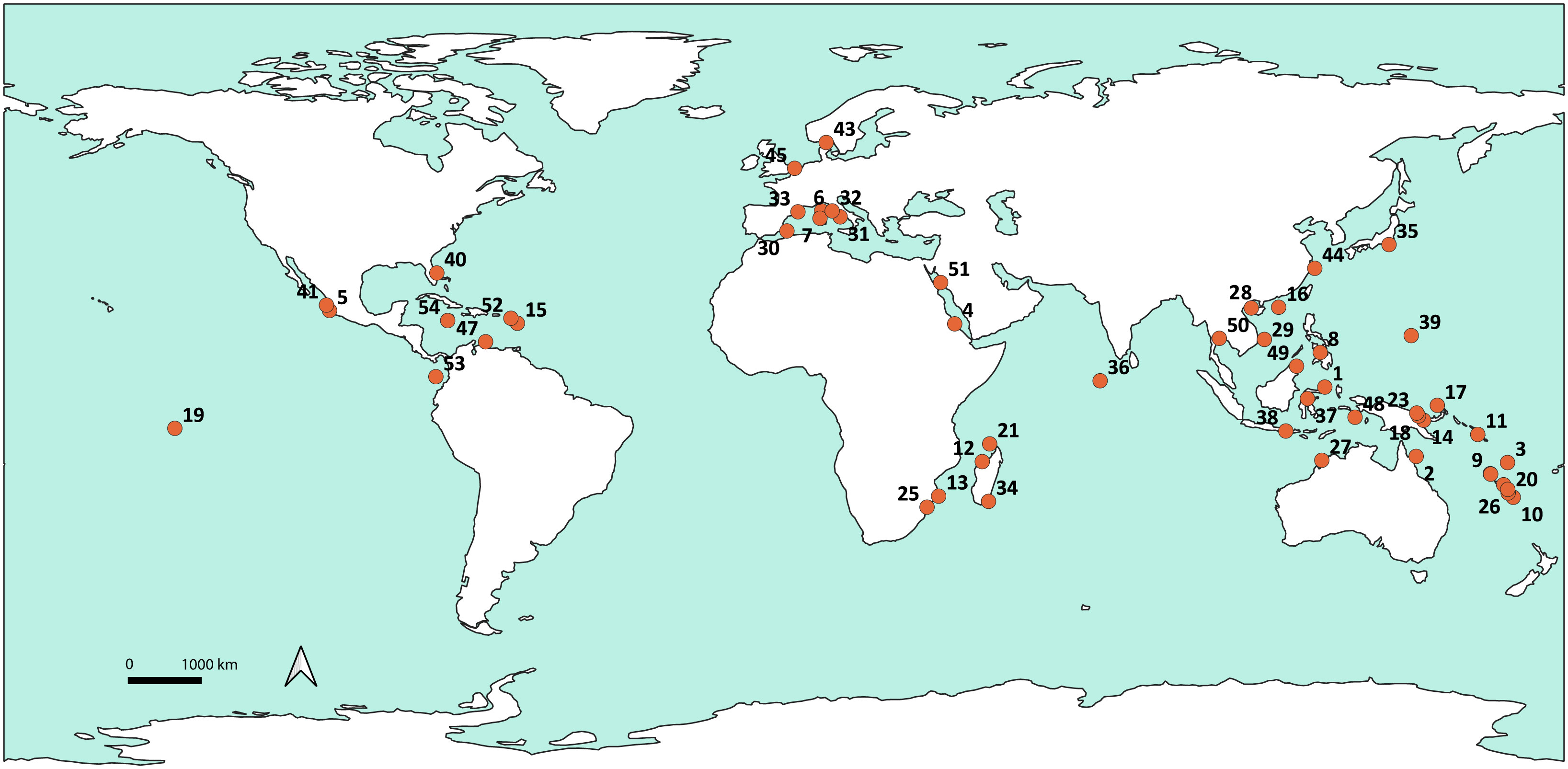
Figure 2 Global map featuring sampling locations of the ovulids dataset created using QGIS. See Table S1 for corresponding locality identifiers.
Sequences (total: 558 of gastropods, 57 of cnidarians) were produced at the Muséum national d’Histoire Naturelle (20 of gastropods), the A.N. Severtsov Institute of Ecology and Evolution, Moscow, Russia (19 of gastropods, 2 of cnidarians), and the remaining at the Molecular Systematics Laboratory of the Department of Biology and Biotechnologies ‘Charles Darwin’, Sapienza University of Rome, Italy (517 of gastropods, 55 of cnidarians). Sequences of 273 additional specimens were obtained from GenBank, including the sequences from the six species used as outgroup [Surrepifungium costulatum (Kiener, 1838), Epitoniidae; Lamellaria latens (O. F. Müller, 1776), Velutinidae; Trivia arctica (Pulteney, 1799), Triviidae; Alvania angioyi Van Aartsen, 1982, Rissoidae; Littorina littorea (Linnaeus, 1758), Littorinidae; and Tonna galea (Linnaeus, 1758), Tonnidae]. For details on samples and GenBank accession numbers see the Supporting Materials (Tables S1).
Whole gastropod genomic DNA was extracted from tissue samples with the Macherey-Nagel NucleoSpin 96 Tissue Kit following the manufacturer’s protocol (at the Service de Systématique Moléculaire of the MNHN), or with a ‘salting out’ protocol (modified from Aljanabi and Martinez, 1997, following Fassio et al., 2023) (at Sapienza University of Rome), or finally following Zvonareva et al. (2020) (at the A.N. Severtsov Institute of Ecology and Evolution). For cnidarians, the ‘salting out’ protocol was used, but the calcareous skeleton, when present, was broken with a sterile pestle, and tissues were incubated overnight on a thermo-block shaker at 54°C with 430 μL of PK buffer and 20 μL of proteinase K (20 mg/mL).
Two mitochondrial and one nuclear marker were amplified for gastropods. A 658 bp region of the mitochondrial cytochrome oxidase subunit 1, cox1, was amplified using several different primer pairs: LCO1490 and HCO2198 (Folmer et al., 1994) and CoxAF and CoxAR (Colgan et al., 2003). When amplification of this region failed, possibly due to DNA degradation, a shorter “minibarcode” region of ~350 bp was amplified, replacing the forward primer with the internal primer mlCOIintF (Leray et al., 2013). A ~700 bp region of the mitochondrial 16S rDNA gene was amplified with the primers 16SA (Palumbi et al., 1991), and CGLeuR (Hayashi, 2003) or 16SH (Espiritu et al., 2001). A ~800 bp region of the nuclear 28S rDNA gene was amplified with the primers C1 and D2 (Jovelin and Justine, 2001). For cnidarians and gut contents, two mitochondrial and one nuclear marker were amplified A ~700 bp region of the mitochondrial 16S rDNA gene was amplified with the primers 16SHA and 16SHB (Cunningham and Buss, 1993) and a ~800 bp region of the mitochondrial mtMSH gene was amplified with the primers ND42599F (France and Hoover, 2002) and Mut3458R (Sánchez et al., 2003). A ~350 bp region of the nuclear second internal transcribed spacer (ITS2) was amplified with the primers 5.8S–436 and 28S-663 (Grajales et al., 2007).
Polymerase chain reactions were performed following Russini et al. (2023). The PCR conditions were as follows: initial denaturation (5 min at 94°C); 35 cycles of denaturation (30 s at 94°C), annealing (40 s at 48–50°C for cox1, 40 s 50-52 for gastropod 16S; 40 s at 56–60°C for 28S; 1.5 min 58°C for mtMSH and 40 sec 50-54 for ITS2°C) and extension (50 sec-1 min at 72°C); final extension (7 min at 72°C). For amplification of cnidarian 16S, protocol described by Cunningham and Buss (1993) was used; for amplification of the minibarcodes, the ‘touchdown’ profile described by Leray et al. (2013) was used following Chiappa et al. (2022).
The PCR products were purified using ExoSAP-IT (79 µL H20, 20 µL rAPid Alkaline Phosphatase, 1 µL Exonuclease I) or Exo/SAP Go PCR Purification Kit (Grisp, Portugal) and both strands were sequenced at Macrogen Europe, Inc or at the laboratory of molecular genetics, IEE RAS or at Eurofins Scientific.
The cox1, 16S and 28S sequences were aligned following Fassio et al. (2022), while the ITS2 sequences were aligned with MAFFT v.7 online (Katoh and Standley, 2013; Katoh et al., 2019) using the E-INS-i algorithm.
To assess species diversity in our dataset of ovulid samples, we adopted an integrative taxonomy approach for large datasets (Puillandre et al., 2012). Primary species hypotheses (PSH) were proposed based on the examination of shell characters, coloration of the body, and morphology of the mantle, based on taxonomic literature, with special reference to Lorenz and Fehse (2009). Shell features were examined for each specimen and integrated with published data; when available, the coloration of the living animal was noted. Alcohol-preserved specimens were observed and dissected under a Wild M6 microscope. Shell photographs were taken at the MNHN molluscs molecular collection using the BK imaging system with a Canon 5DSr.
These morphological species hypotheses were validated by the analyses of molecular data in the framework of an integrative taxonomy approach (Barberousse and Samadi, 2010).
First a genetic distance approach was used through the program ASAP (Assemble Species by Automatic Partitioning) that implements an ascendent hierarchical clustering algorithm (Puillandre et al., 2021). ASAP analyses were conducted on the cox1 ingroup dataset after removal of 8 short minibarcode sequences (Kimura two-parameter substitution model, with other parameters as default). A first ASAP analysis was conducted to detect potential contaminations or individuals clearly misplaced based on morphological characteristics. Following the removal of contaminants and correction of any morphological discrepancies, a second ASAP analysis was carried out. The morphology-defined PSH were then compared to the partitions obtained with ASAP and discussed in the context of a phylogenetic tree derived from the combined genetic dataset (details provided below in the phylogenetic reconstruction section) and considering additional factors such as host associations, depth, and geographic range. This comprehensive assessment led to the proposal of Secondary Species Hypotheses (SSH).
Cnidarians (for a few of which field observation and/or photographs were also available) were observed under a Wild M6 dissection microscope for the examination of polyps arrangement and distributions, and calcareous skeleton when present, and identified following Fabricius and Alderslade (2001) and Grasshoff (1999).
All 16S, MSH and ITS2 cnidarian nucleotide sequences were matched with sequences available in Genbank with the NCBI BLAST web interface (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Molecular taxonomic classifications were determined using the BLAST taxonomy lineage report, taking in consideration all IDs with percent of identity ≥ 87% and e-value ≤ 6e-43. Preliminary taxonomic identifications were checked for consistency with morphology (when the host sample was available), geographical range and cnidarian host information available in the. Cnidarian identification was conducted to the lowest taxonomic level possible (ideally the species), at least to the family level. For details on samples and GenBank accession numbers see Supporting Material (Table S2).
Due to the high phenotypic plasticity of ovulids and cnidarians, literature records of ovulid-coral associations were considered reliable only when supported by morphological (on actual specimens or field photographs) or genetic identification pertaining to both gastropods and cnidarians. For ovulid species for which no such data were available, field observations by the authors or communicated by colleagues were taken into consideration.
Phylogenetic analyses were performed on the three single-gene datasets (cox1 partitioned by codon, 16S and 28S) and on the combined dataset (cox1 partitioned by codon + 16S + 28S) with both maximum likelihood (ML) and Bayesian inference (BI). Some samples for which only 16S and/or 28S sequences were present in the GenBank database (no cox1 available), but the identification was not consistent with the phylogenetic results, were discarded from all analyses. All the specimens with the cox1 data were kept in the combined dataset; when the cox1 was missing, only the specimens with both the 28S and the 16S were retained in the combined dataset. The cox1 alignment was partitioned by codon position using PAUP V4.0 (Wilgenbusch and Swofford, 2003). The best fitting nucleotide substitution model for each partition was chosen with jModelTest 2.1.6 (Darriba et al., 2012) on the CIPRES portal (www.phylo.org, Miller et al., 2010), using the Bayesian information criterion (BIC; cox1, 1st codon position of the open reading frame=SYM+I+G; cox1, 2nd codon position=HKY+G; cox1, 3rd codon position=GTR+G; 16S=HKY+I+G and 28S=GTR+I+G). Maximum likelihood analyses were done using IQ-TREE (Trifinopoulos et al., 2016), with 10,000 ultrafast bootstrap replicates for node support. Bayesian analyses were performed using MrBayes v. 3.2.3 [(Ronquist and Huelsenbeck, 2003) for 2 x 108 generations, trees sampled every 1,000 generations, 25% burn-in] on the CIPRES Science Gateway (Miller et al., 2010). Convergence of MCMC was analyzed with Tracer 1.7.2 (Rambaut et al., 2018) and was assumed to have occurred when the effective sample size was >200. Only nodes with ultrafast bootstrap values (UfB) ≥95 or posterior probabilities (PP) values ≥0.95 were considered to be supported. Phylogenetic trees were visualized using FigTree v.1.4.4 (Rambaut et al., 2018).
We identified 14 reliable fossil first occurence of egg-cowries in the literature. Among them, we selected nine records that were suitable for calibration of relevant nodes in our time-calibrate phylogenetic reconstruction (Table 1). The ovulids are a relatively young gastropod group from a geological point of view. The first representatives are known from the Early Eocene (47.8–56 mya) of Western Europe, but their diversity exploded in the Plio–Pleistocene. Since shell morphology of the first representatives of the family hardly differs from some of their extant descendants, the first appearance record of this caenogastropod lineage is particularly reliable (Fehse, 2001). Concerning the genus Pseudocypraea F. A. Schilder, 1927, the oldest known record is from the Middle Eocene (37.8–41.2 mya) with Pseudocypraea dolini, (Pacaud, 2003). The first certain appearance of Pedicularia Swainson, 1840 was during the Late Eocene (33.9–37.8 mya) (Ladd, 1977). The first appearance of the genus Pellasimnia Schilder, 1939 was in the Late Eocene (33.9–37.8 mya) with Pellasimnia maxwelli (Beu and Marshall, 2011). The fossil record of Cyphoma Röding, 1798 dates back to the early Miocene (15.97–23.44 mya) with Cyphoma intermedium (G.B. Sowerby I, 1828) (Woodring, 1973). Fossils belonging to the genus Primovula Thiele, 1925 are common in the Pliocene (2.58–5.3 mya) with the earliest known species Primovula beckeri G.B. Sowerby III, 1900 (Ladd, 1977). The first certain appearance of Volva Röding, 1798, is Late Pliocene (2.58–3.6 mya) fossils of Volva javana Martin, 1899 (Schilder, 1932). The Pleistocene records of Diminovula margarita G. B. Sowerby I, 1828 and Simnia spelta Linnaeus, 1758 provided the upper bound at 2.5 mya (Schilder, 1932) for those species.
We selected a single exemplar (indicated by an asterisk in Table S1) for each SSH based on ASAP and combined tree results to create a new combined dataset comprising 73 samples (including the outgroup). This dataset was used to produce a time-calibrated tree to estimate the node ages of each clade, using the software BEAST 1.10.0 (Suchard et al., 2018). The heterogeneity of the mutation rate across lineages was set under uncorrelated lognormal distributed relaxed clocks for the six partitions mentioned above (except for SYM that is not implemented in BEAST), and the Yule process (Gernhard, 2008) was chosen. All other priors were set with default values. The nine calibration points were set under exponential prior (Ho and Phillips, 2009) following Fassio et al. (2021), with the major distributions within the boundaries of the relative stage age of identification. We performed two runs of 28 generations, sampled every 10,000 steps. Results were analyzed with Tracer 1.7.2 (Rambaut et al., 2018) and all runs were pooled and re-sampled using LogCombiner 1.10.4 (Rambaut et al., 2018), after 25% samples were discarded as burn–in. The maximum clade credibility tree was estimated with TreeAnnotator 1.10.4.
Macroevolutionary dynamics of diversification were modeled across the phylogeny using the software Bayesian Analysis of Macroevolutionary Mixtures (BAMM) v.2.5.0 (Rabosky, 2013; Rabosky, 2014) on the Maximum Clade Credibility tree obtained in BEAST, after outgroup removal using TreeGraph2 (Stöver and Müller, 2010). Ten million generations of reversible jump Markov Chain Monte Carlo were run, drawing samples from the posterior every 10,000 generations following Modica et al. (2020). Priors were chosen using the setBAMMpriors command in the R package BAMMtools (Rabosky, 2014), except for the prior probability of rate shift, which has been shown to affect BAMM results (Moore et al., 2016). For this prior, we tested values ranging from 0.1 to 10 and chose the value leading to the highest Effective Sample Size (ESS) values for LogLikelihood and NumberOfShifts. After removing a 10% burn–in, we processed the output data using BAMMtools statistics and plotted diversification rate through time.
Ancestral state reconstruction was conducted using the Bayesian Binary MCMC (BBM; Ali et al., 2012) on RASP (Reconstruct Ancestral State in Phylogenies, version 4.2) (Yu et al., 2015). This software was chosen since it allows reconstructing ancestral states of multiple discrete characters: either the families or the subclasses (hexacorals, octocorals, or hydroidolinas) of cnidarian hosts exploited by each ovulid species were used as characters in two distinct analyses. The calibrated combined tree obtained by BEAST (after outgroup removal) was used as a species phylogeny. Analyses were based on 50,000 trees, sampling every 100 steps, of which 125 were discarded (number of discrete areas=3, Among– Site rate variation= Gamma +G; all other parameters set with default values). The families and the subclasses of the host cnidarians were chosen as prior for the states following Ploch et al. (2022). Since the host data for the genus Pseudocypraea, which are currently missing, may prove crucial to the ancestral state reconstruction, we conducted two ancestral state analyses using subclasses as priors: one with missing data for Pseudocypraea; the other with Hexacorallia as the cnidarian subclass associated with Pseudocypraea exquisita and P. adamsonii, assuming that the frequent observation of living specimens crawling amidst sponges (Pacaud, 2003; F. Lorenz, pers. comm; M.O., unpublished observation), may be suggestive of an association with sponge-associated Zoantharia (Gray, 1832).
The molecular dataset was composed of 1158 sequences (454 cox1, 450 16S and 254 28S) for a total of 524 ovulids and pediculariids specimens. Out of them, three specimens were excluded from defining the PSH due to their juvenile shell, which made it challenging to identify morphologically. The dataset of the combined tree included sequences from 468 samples (the others were excluded because only 16S or 28S sequences were available). All the 477-658 bp long cox1 sequences (440) were included in the ASAP analysis.
The 521 specimens for which morphological characters were analyzed, were ascribed to 76 PSH. The ten best partitions found by ASAP divided the dataset into 57-73 hypothetical species. The best partition according to the ASAP score split our dataset into 66 hypothetical species (threshold distance=4.6%) and the second best into 67 (threshold distance=3.7%; Supporting Information, Figure S1). The other ASAP partitions were not taken into account due to their high asap-score. The difference between first- and second-best partitions concerned the sample MNHN-IM-2013-63254 Quasisimnia sp. (considered as a separate species in the second, included in the same subset of Quasisimnia hirasei by the first).
All the hypothetical species identified by ASAP, in both the first- and the second-best partitions (except Pellasimnia brunneiterma, Ufb=95, PP=0.95), corresponded to monophyletic groups highly supported in the ML and BI phylogenetic reconstructions (Ufb=100, PP=0.98-1). The specimens that were not included in the ASAP analysis did not represent new independent lineages in the phylogenetic analyses, but ended up in hypothetical species already identified by ASAP, except MNHN-IM-2013-63255 and MNHN-IM-2019-15204. These two samples formed two distinct lineages separated from the other clades in the combined tree and may represent two additional hypothetical species. However, they are not included in the count of the SSH since we were unable to confirm it through robust analyses, either morphological or by integrative taxonomy.
In six cases, two or more PSH were considered by both ASAP and phylogenetic analysis as one hypothetical species: (1) Simnialena rufa and Simnia avena (Ufb=1, PP=1); (2) Cyphoma gibbosum, C. signatum, C. intermedium and C. mcgintyi (Ufb=100, PP=1); (3) Pellasimnia annabelae and P. brunneiterma (Ufb=0.95, PP=95); (4) Phenacovolva rosea, P. rosea B and P. nectarea (Ufb=1, PP=1); (5) Pseudosimnia carnea and P. adriatica (Ufb=100 PP=1); (6) Crenavolva aureola and C. chiapponii (Ufb =100, PP=1). Given the well-documented morphological plasticity within Ovulidae, characterized by significant variations in mantle color and shell shape, for these six dubious cases we considered more reliable the hypothetical species identified by ASAP and corresponding to well supported monophyletic clades.
Regarding MNHN-IM-2013-63254 Quasismnia sp., this sample was associated with a single PSH. The best ASAP partition grouped it within the same subset as MNHN-IM-2007-38930 Quasisimnia hirasei, while the second partition separated them. However, in the combined tree, MNHN-IM-2013-63254 Quasismnia sp. represented a distinct lineage from MNHN-IM-2007-38930 Quasisimnia hirasei (Ufb=100, PP=1). Thus, it is more reliable to consider the hypothesis that they represent two distinct species.
In the integrative taxonomic process, we identified a total of 67 SSH as depicted in the collapsed tree (Figure 3), for which the oldest species name has been used.
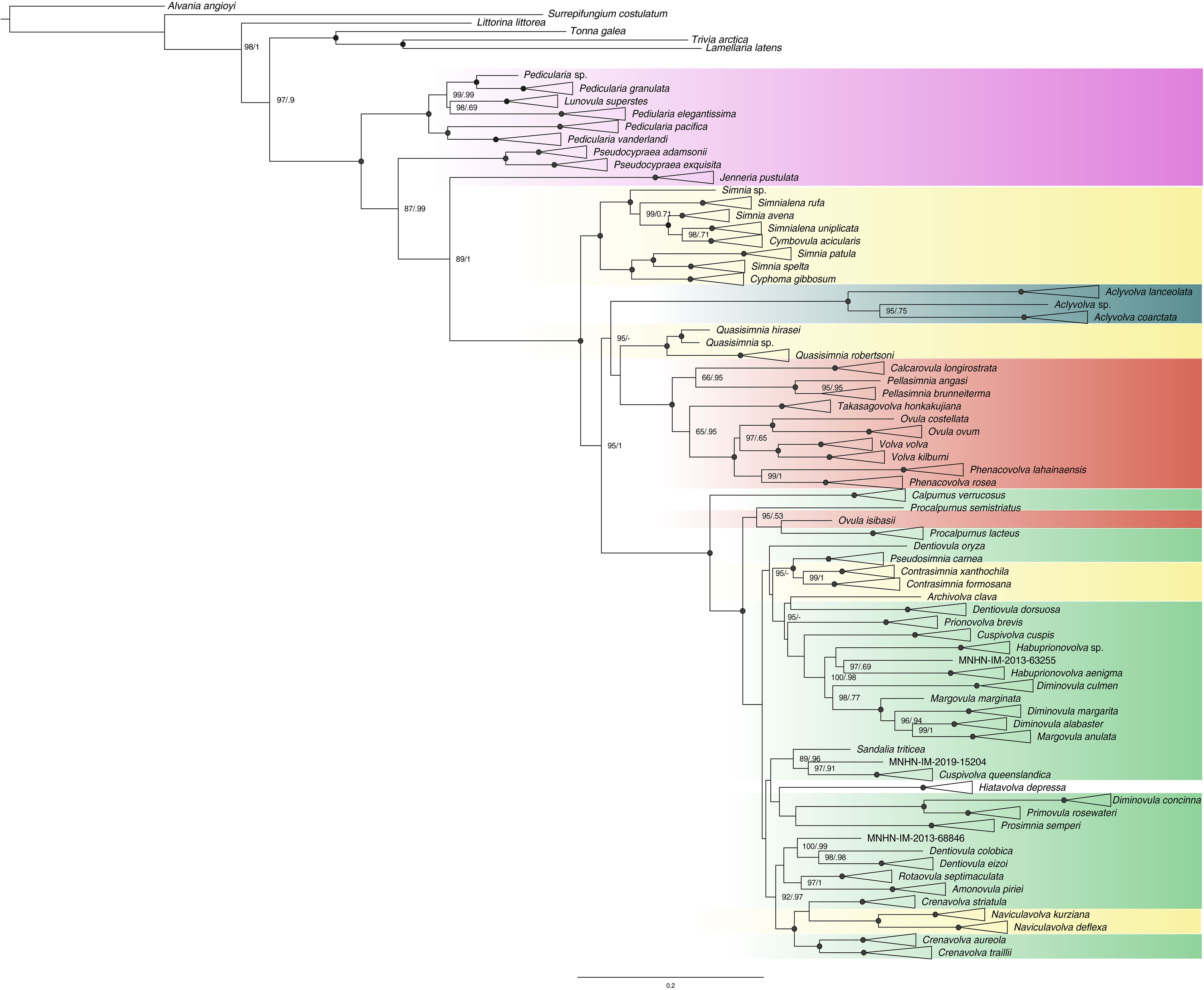
Figure 3 Phylogenetic relationships of the family Ovulidae (Maximum Likelihood tree based on the combined dataset), with clades collapsed by species. Numbers at nodes indicate branch support values [ultrafast bootstrap (Ufb) values and posterior probability (PP), respectively]; support values are shown only when at least one of them is ≥ 95%; black circles at nodes indicate maximum support (PP=1, Ufb=100). Colors at collapsed clades and taxa indicate the traditional subfamilial classification of the taxa represented: pink=Pediculariinae; yellow=Simniinae; dark green=Aclyvolvinae; red=Ovulinae; green=Prionovolvinae.
Comparison of the major nodes among all trees, both for single genes and combined datasets, revealed no inconsistencies, except that the 28S trees split Aclyvolva coarctata in two strongly supported clades, Ufb=99, PP=1, while in all the other trees it represented a strongly supported single clade. Within the combined trees, the monophyly of a group including ovulids and pediculariids was strongly supported (Ufb=100, PP=1), while some genera as traditionally conceived did not prove monophyletic. The genus Lunovula was nested inside a major Pedicularia clade, making the latter polyphyletic. The genus Margovula was split into one clade of species Margovula anulata, sister to Diminovula alabaster (Ufb =99, PP=1), and another lineage represented by Margovula marginata. The genus Ovula was split into one clade of two species including the type species Ovula ovum (Ufb=100, PP=1), and a lineage of Ovula isibasii, sister to Procalpurnus lacteus (not supported). The genus Dentiovula was retrieved as polyphyletic and split into three lineages: one represented by Dentiovula oryza (Ufb=95, PP=/), another represented by Dentiovula dorsuosa, sister to Archivolva clava (not supported), and a third one including the type species Dentiovula colobica and Dentiovula eizoi (Ufb=100, PP=0.99). The genus Diminovula was split into one lineage of Diminovula concinna, sisters to Primovula rosewateri (Ufb=100, PP=1), one of Diminovula culmen, and a clade including the other two species of Diminovula and Margovula anulata (Ufb=98, PP=0.77). The genus Procalpurnus was split into two lineages: one including the type species Procalpurnus lacteus, sister to Ovula isibasii (not supported), and one represented by Procalpurnus semistriatus Ufb=95, PP=0.53). The genus Crenavolva was split into two lineages: one including the type species Crenavolva striatula, sister to the genus Naviculavovla (not supported), and one including Crenavolva aureola and Crenavolva traillii (Ufb =100, PP=1). The genus Simnia was split into three lineages: one clade including the type species Simnia spelta (Ufb=100, PP=1) and Simnia patula, one lineage represented by Simnia avena (Ufb=100, PP=1) and a third lineage represented by Simnia sp. (voucher MNHN-IM-2013-60781, Ufb=100, PP=1).
Pediculariids as currently conceived did not prove monophyletic (Ufb=100, PP=1): the genera Pseudocypraea and Jenneria resulted in two distinct lineages more closely related to the rest of ovulids than to Pedicularia (Ufb=87, PP=0.99).
The monophyly of the subfamily Ovulinae was strongly supported (Ufb=100, PP=1), with the exception of Ovula isibasii that ended up inside a distinct prionovolvine clade (Ufb=100, PP=1). The monophyly was also supported for the subfamily Aclyvolvinae (Ufb=100, PP=1). The other traditional subfamilies were polyphyletic. The Simniinae were split into five different lineages: (i) Simniinae sensu stricto, that included Simnia spelta (type species of the type genus Simnia), Simnia patula, Simnialena rufa, Simnialena uniplicata, Simnia sp., Cymbovula acicularis and Cyphoma gibbosum; (ii) Quasisimnia (sister to Ovulinae, not supported); (iii) Contrasimnia; (iv) Archivolva; and (v) Naviculavolva. A large clade of core Prionovolvinae was defined (Ufb=100, PP=1), including the type genus Prionovolva (type species Prionovolva brevis), but also other species traditionally ascribed to different subfamilies. Hiatavolva depressa (subfamily unclear according to MolluscaBase, 2023) also ended up in the prionovolvine clade, even if their relationships with other nearby clades were not resolved.
For both ML and BI trees, see Supporting Information, Figures S2–S9).
From this point onward, we shall use Ovulidae to indicate the clade including all lineages studied herein (thus comprising ovulids and pediculariids as previously conceived); Pediculariinae for the clade comprising the genera Pedicularia and Lunovula; Ovulinae, for the clade comprising all species traditionally included in this subfamily except Ovula isibasii, along with the genus Quasisimnia; Simniinae, for the clade comprising the genera Simnia, Simnialena, Cyphoma and Cymbovula; Prionovolvinae, for the clade comprising all species traditionally included in this subfamily plus Ovula isibasii and the genera Contrasimnia, Naviculavolva, Archivolva.
The time-calibrated phylogeny (Figure 4; see Table 1 for 95% HPD) estimated the origin of the family Ovulidae at 54 mya (95% HPD 47.8–63.16) during the Ypresian (Early Eocene). The origin of the Pediculariinae was dated at 35 mya (95% HPD 33.9–36.54) during the Priabonian (Upper Eocene). The subfamily Ovulinae was estimated as having originated 42 mya (95% HPD 38.33–44.68) during the Bartonian (Middle–Eocene). The origin of the core Prionovolvinae was dated at 30 mya (95% HPD 20.95–39.76) during the Rupelian (Early Oligocene). The Simniinae were estimated as having originate during the Burdigalian (Early Miocene), with the node dated at 17 mya (95% HPD 15.07–21.63). The Aclyvolvinae resulted to be the most recent subfamily, with the origin estimated around 15 mya (95% HPD 5.94– 31.55) in the Langhian (Middle–Miocene). Except for Cyphoma, which exhibited a discrepancy of approximately 15 million years compared to its fossil record, all of our calibration nodes were dated congruently with the utilized fossil data.
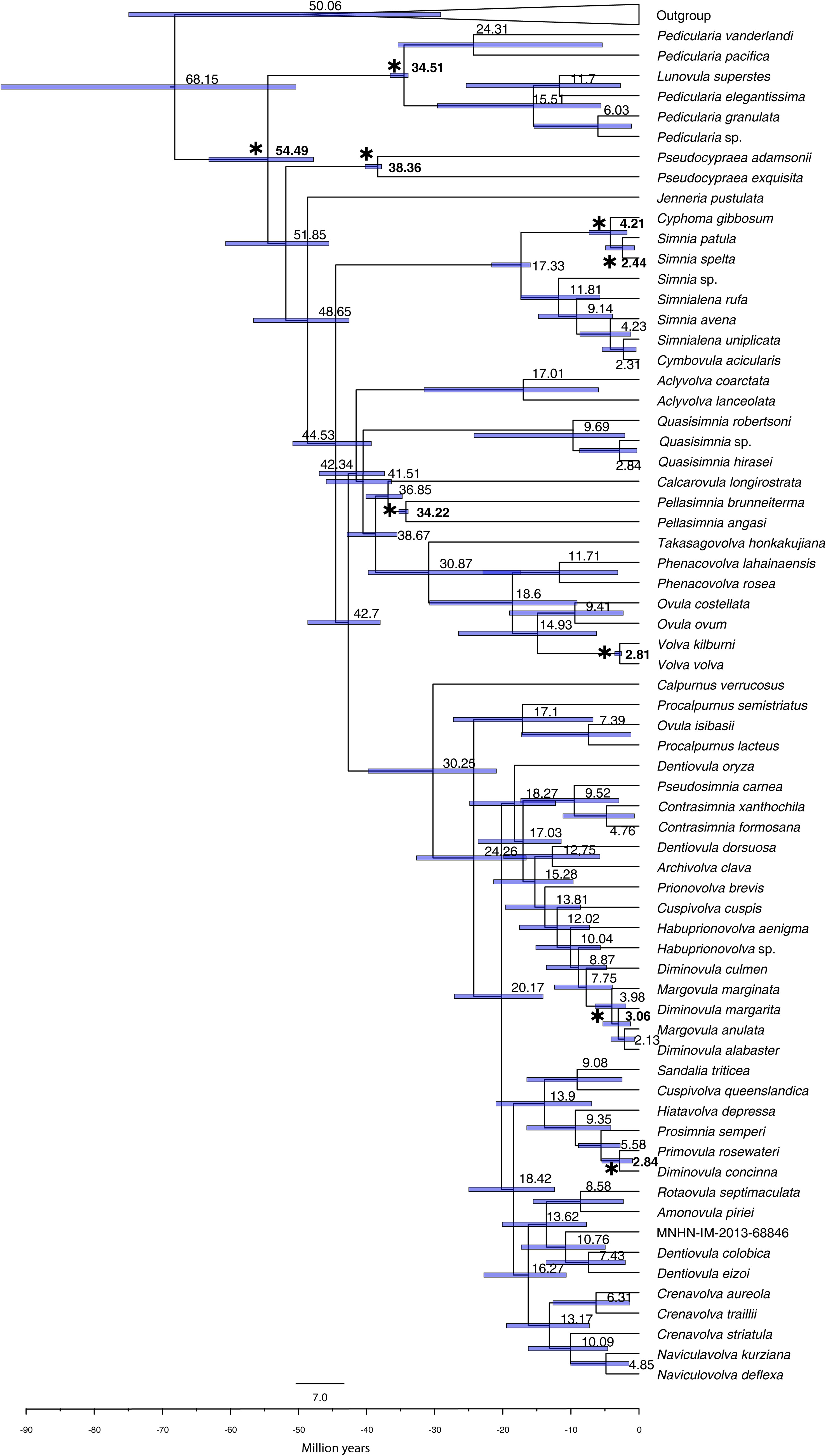
Figure 4 Bayesian Inference, time calibrated, phylogenetic reconstruction of the family Ovulidae using BEAST from the combined dataset, comprising one exemplar for each SSH. Bars at nodes indicate 95% confidence intervals of ages expressed in mya. Asterisks (and numbers in bold) indicate the nodes (and the relevant fossil based dates) used to calibrate the tree.
We modeled ovulid diversification rates as a function of time. The best model supported by BAMM analysis indicated a steady rate of diversification over time, with a 0.98 posterior probability. The analyses inferred a diversification model with the highest probability of 0.1 major shift in the rate of speciation in the Ovulidae. Both posterior probabilities and Bayes factors were remarkably lower for alternative models with more rate shifts. The credible shifts plot depicted a non-core shift for the Ovulidae (Figure 5A) with a 98% posterior probability. The rate-through-time BAMM plot supported a scenario of an initial higher rate of diversification (speciation rate 0.13) that decreased gradually over time (to 0.054) across all lineages of Ovulidae (Figure 5B).
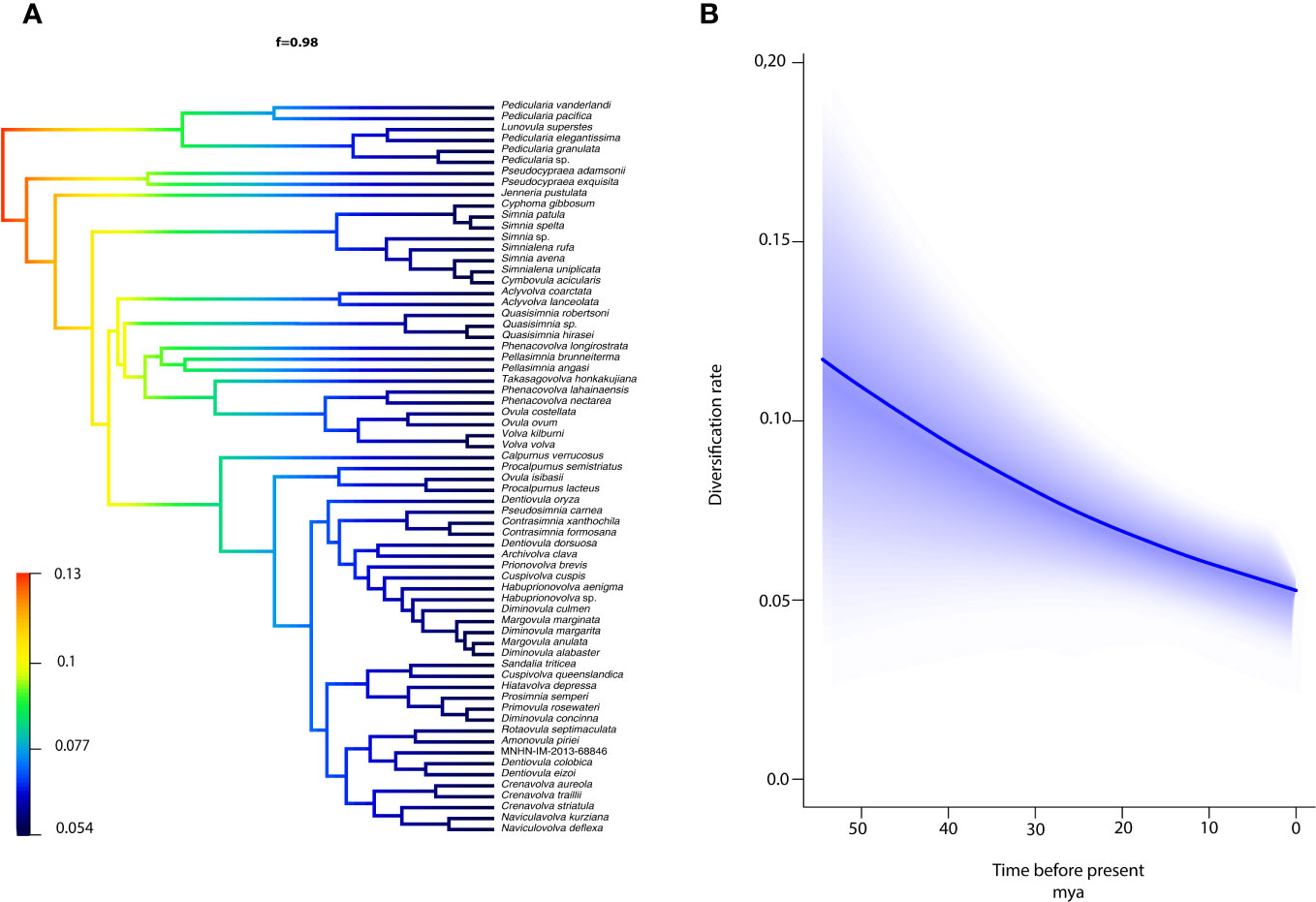
Figure 5 The ovulid diversification rates vary across clades and time. (A) The single BAMM credible shifts plot representing the rate-shift configuration and a posterior probability shift configuration corresponding to 0.98. (B) BAMM plot depicting the net diversification rates through time.
The ovulid-cnidarian associations identified in this work, integrated with literature data, are summarized in Table 2. We have identified the cnidarian hosts (26 families) of 82% (55) of the assayed species. Among these, the host of 27 species was sourced from literature, whereas for the remaining 28 species, the host identification was based on our molecular and/or morphological analyses. These data included 57 cnidarian samples identified by DNA-barcoding of the collected sample, while 69 were exclusively identified by morphology due to the unavailability of coral tissue or unsuccessful DNA amplification. None of the 39 DNA samples extracted from the foregut and the stomach tissue displayed successful sequencing with specific cnidarian primers. When multiple records were available, usually, each ovulid species has been recorded as feeding on the same cnidarian family; nevertheless, in some cases (11 species) more cnidarian families belonging to the same subclass have been recorded for a single ovulid species.
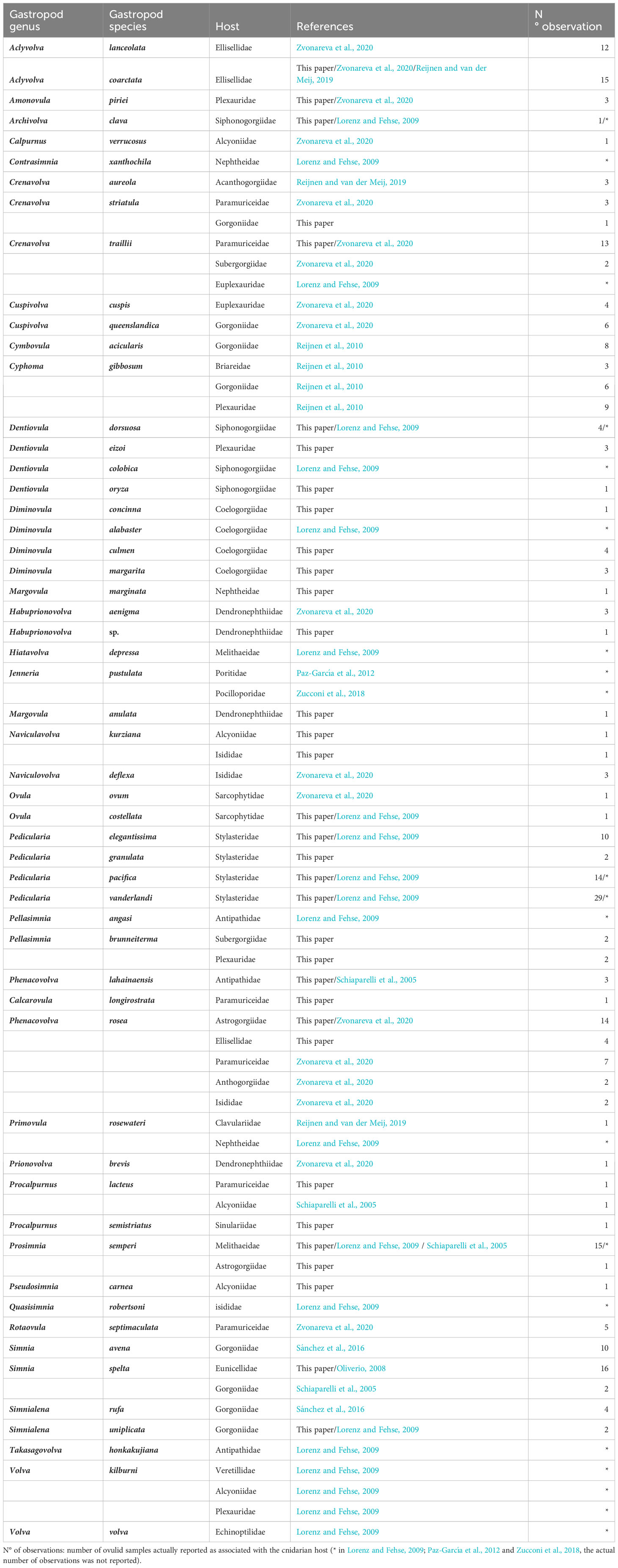
Table 2 List of 55 Ovulidae species associated with their respective cnidarian family host as documented in both literature and this paper.
The results of the ancestral state reconstruction for BBM using cnidarian families as prior for the states are shown in Figure 6 (nodes are numbered to ease following the description). Node 133 represented the ancestral state for the Ovulidae; the hydroid family Stylasteridae was estimated the most likely ancestral host for this node (79.4% marginal probability), followed by an uncertain other cnidarian family (20.6%). At node 72, Stylasteridae were estimated as the ancestral host of the Pediculariinae (98.43%). The ancestral reconstruction at node 93 indicated that the hexacoral Antipathidae was the most likely ancestral host of the Ovulinae (64.12%). The Prionovolvinae (node 128) showed Alcyoniidae as the most likely ancestral host (85.02%). The ancestor of the subfamily Simniinae (node 80) had Gorgoniidae as the most likely host (90.38%). The ancestor of Aclyvolvinae (node 81) had most likely the octocoral family Ellisellidae as host (78.63%), followed by a multiple association with Ellisellidae and Subergorgiidae (18.99%).
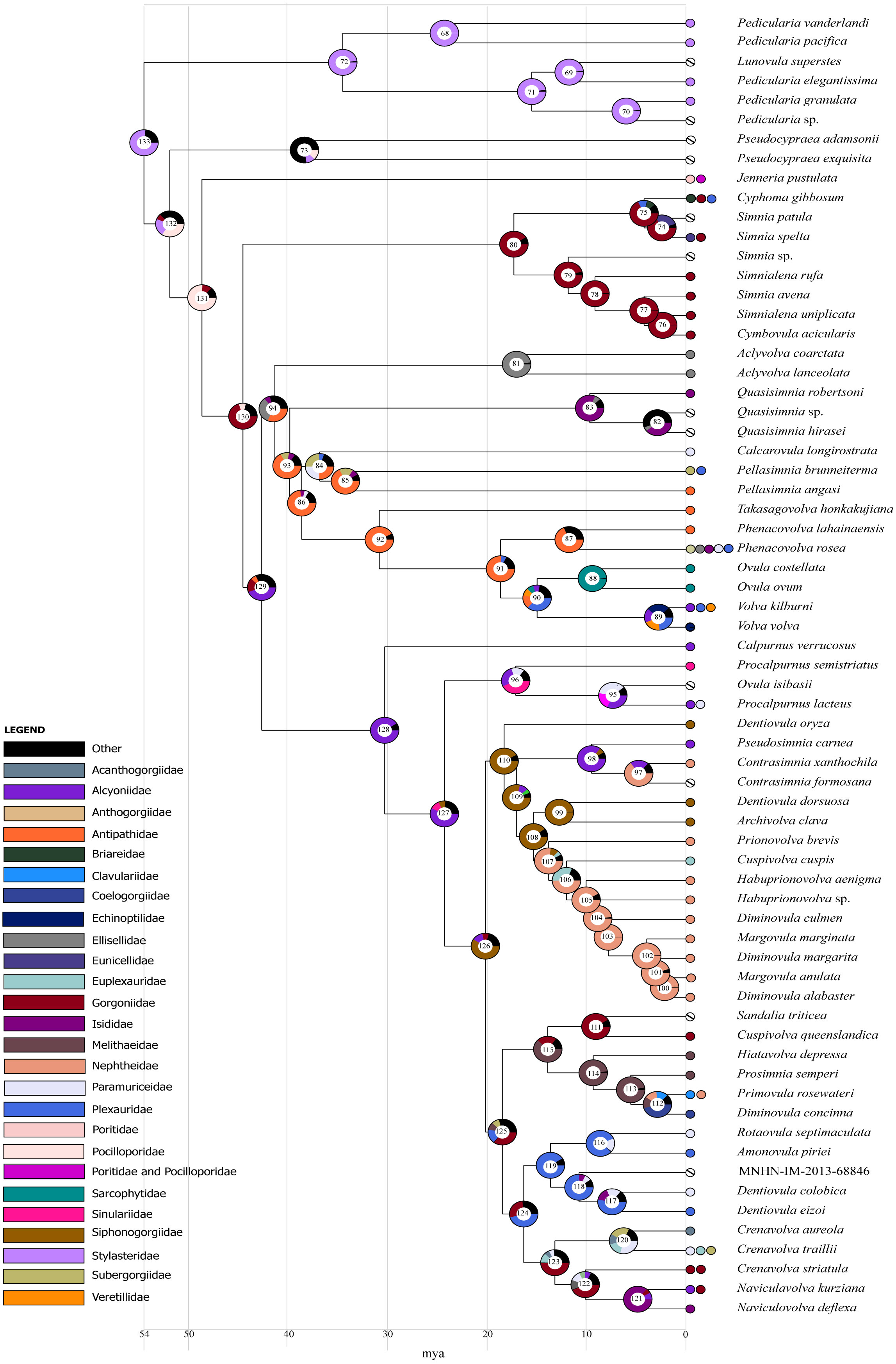
Figure 6 Graphical representation of ancestral state reconstruction at each node of the phylogeny of the family Ovulidae obtained by BBM analysis (exported from RASP) using cnidarian families as a prior. Pie charts at each node (from 68 to 133) show the probabilities of alternative ancestral states; numbers inside the pie charts identify each node. The legend shows the color key to the hosts; black represents other unknown ancestral states. White barred circles represent no host information. X-axis represents time in millions of years.
The results of the ancestral state reconstruction using cnidarian subclasses as prior for the states are shown in Figure 7. The host of the ancestral Ovulidae (node 133) was estimated to be a Hydroidolina (92.81% marginal probability) followed by Hexacorallia (5.26%). For the Pediculariinae (node 72), the reconstruction indicated Hydroidolina (99.78%) as the most likely ancestral host; the Octocorallia were estimated to be the most likely ancestral state of Ovulinae (node 93, 95.32%), Prionovolvinae (node 128, 99.86%), Simniinae (node 80, 99.63%), and Aclyvolvinae (node 81, 99.97%).
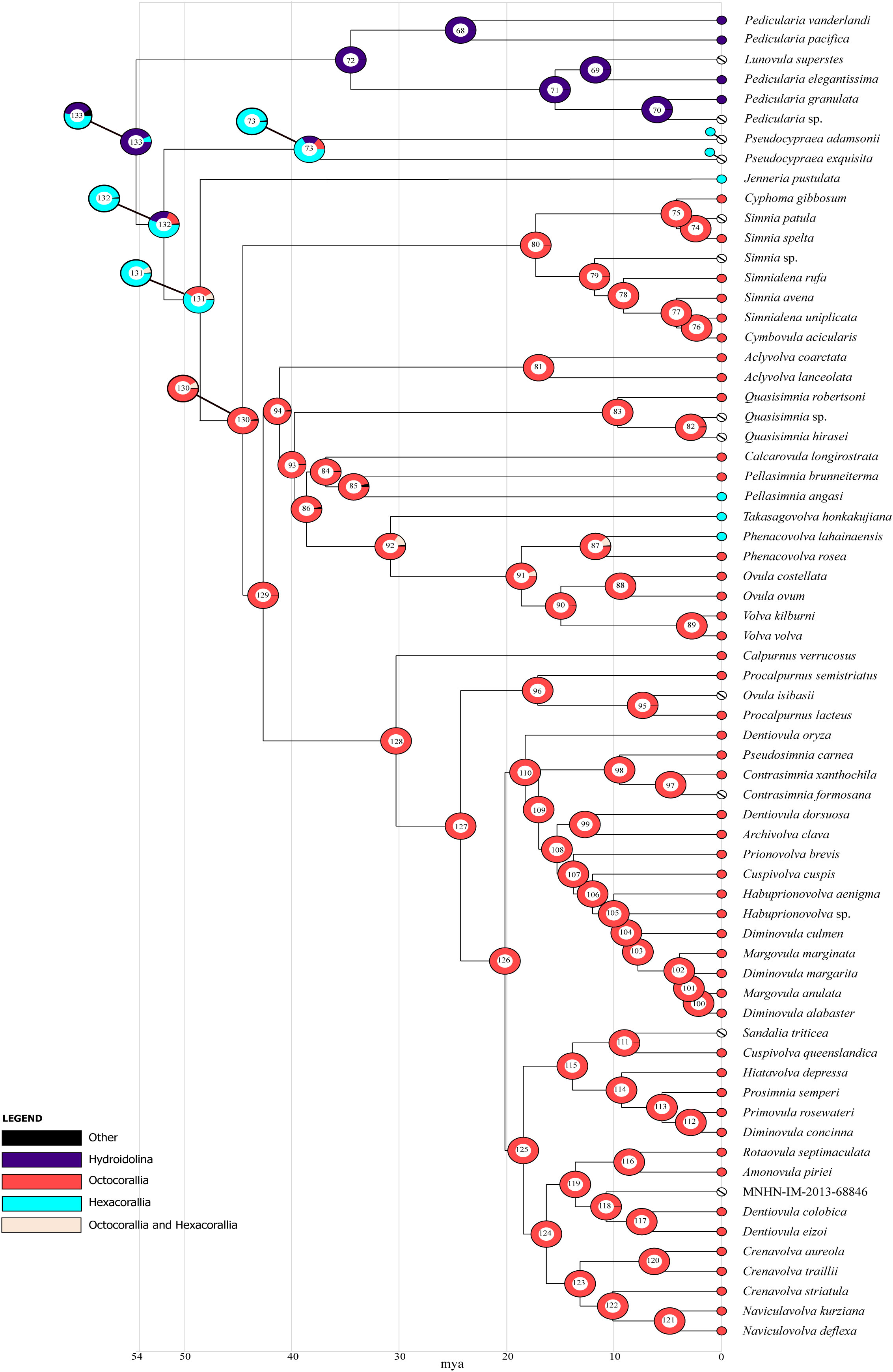
Figure 7 Graphical representation of ancestral state reconstruction at each node of the phylogeny of the family Ovulidae obtained by BBM analysis (exported from RASP) using cnidarian subclasses as a prior. Pie charts at each node (from 68 to 133) show the probabilities of alternative ancestral states; numbers inside the pie charts identify each node. The legend shows the color key to the ancestral hosts; black represents other unknown ancestral states. White barred circles represent no host information. Nodes 73, 130, 131, 132, 133 are also represented with the corresponding ancestral state reconstruction from the analysis with the Pseudocypraea-hexacorals association. X-axis represents time in millions of years.
The results of the ancestral state reconstruction at relevant nodes, using hexacorals as a prior of Pseudocypraea, are presented in Figure 6, juxtaposed with the corresponding node outcomes from the prior analysis. The main differences from the previous analysis concerned the ancestral host of the family (node 133); Hexacorallia was estimated as the most likely ancestral host for this node (54.09% marginal probability), followed by Hydroidolina (38.79%) and by an uncertain other cnidarian subclass (7.12%).
The molecular data presented here span a significant increase in the taxonomic coverage of egg-cowries compared with previous works on the group (Schiaparelli et al., 2005; Reijnen et al., 2010; Zvonareva et al., 2020).
According to our integrative taxonomy approach, the 78 morphospecies initially identified, encompass 67 distinct species hypotheses, with some remarkable cases discussed below:
(1) Simnialena rufa/Simnia avena – We confirm the results by Sánchez et al. (2016) who found that specimens morphologically identified as Simnia avena fall in two distinct clades: one conspecific with specimens morphologically identified as Simnialena rufa, and another group of individuals, which we retrieved as sister to the clade Simnialena uniplicata + Cymbovula acicularis. An integrative taxonomy study based on a wider sampling, and including topotypes of the involved nominal taxa, is urged.
(2) Cyphoma – Reijnen and van der Meij (2017) showed that all their Cyphoma morphospecies (C. gibbosum, C. mcgintyi, and C. signatum, identified mostly based on their mantle patterns and coloration) were distributed randomly in a clade representing one genetically homogeneous species. In our work, an additional morphospecies, C. intermedium, clustered within the same clade as the rest of the Cyphoma specimens.
(3) Pellasimnia annabelae/P. brunneiterma – As already suggested by Zvonareva et al. (2020), specimens morphologically identified as Pellasimnia annabelae, P. cf. annabelae and P. brunneiterma represented a single species.
(4) Phenacovolva rosea/P. nectarea – As previously proposed by Zvonareva et al. (2020), all the specimens morphologically assigned to Phenacovolva rosea and P. nectarea belonged to a single species. In our analyses, in contrast to their results, we could not find any robust support to the distinction of the two sub-lineages, that Zvonareva et al. (2020) called P. rosea A and P. rosea B.
(5) Pseudosimnia – The specimens ascribed to the two iconic and morphologically readily identified Mediterranean nominal species Pseudosimnia carnea and P. adriatica were shown to represent a single lineage based on molecular data.
(6) Crenavolva aureola/C. chiapponii – As shown by Reijnen (2016), specimens ascribed to Crenavola chiapponii and C. aureola were not distinguishable based on genetic data.
It is remarkable that taxonomic changes suggested by molecular data in ovulids almost entirely consist of lumping species, an uncommon situation in molluscs where cryptic species complexes are particularly abundant. We confirm the hypothesis proposed by Schiaparelli et al. (2005) and Zvonareva et al. (2020) that the morphological plasticity has an important, and probably largely neglected relevance for taxonomy of ovulids: there are possibly many more names available than actually existing species.
Out of the 36 nominal extant genera represented in our dataset (92% of total existent), a total of eight (Pedicularia, Ovula, Dentiovula, Diminovula, Margovula, Procalpurnus, Crenavolva, and Simnia) were not monophyletic as traditionally conceived. Notably, among these, the genera Pedicularia, Ovula, and Simnia are the type genera of the subfamilies Pediculariinae, Ovulinae, and Simniinae, respectively, but their polyphyly does not affect the subfamily definition. In particular, Ovula isibasii must be classified in a distinct genus (whose relationships with Procalpurnus remain to be defined); the systematics of pediculariines should be reassessed, probably with more lineages worthy of genus recognition; the close relationship among the lineages morphologically ascribed to Simnia and Cyphoma, besides the need to restrict the use of the genus Simnia, is remarkable for what concerns the evolution of the peculiar Cyphoma-like morphology (see below).
Our study has also highlighted the need for a revision of the current suprageneric classification of the egg-cowries. We have found no major evidence in support of or against the proposal by Lorenz and Fehse (2009) to classify Pedicularia and allied in a distinct family Pediculariidae. However, given the rather strong evidence in our analyses that the genera Jenneria and Pseudocypraea are not in the same clade as Pedicularia and Lunovula, but rather represent two independent lineages, we would tend towards maintaining the whole set of egg-cowries, including Pedicularia and allies, within a single family Ovulidae. The following classification scheme is a working hypothesis to be tested by further studies (Figure 8):
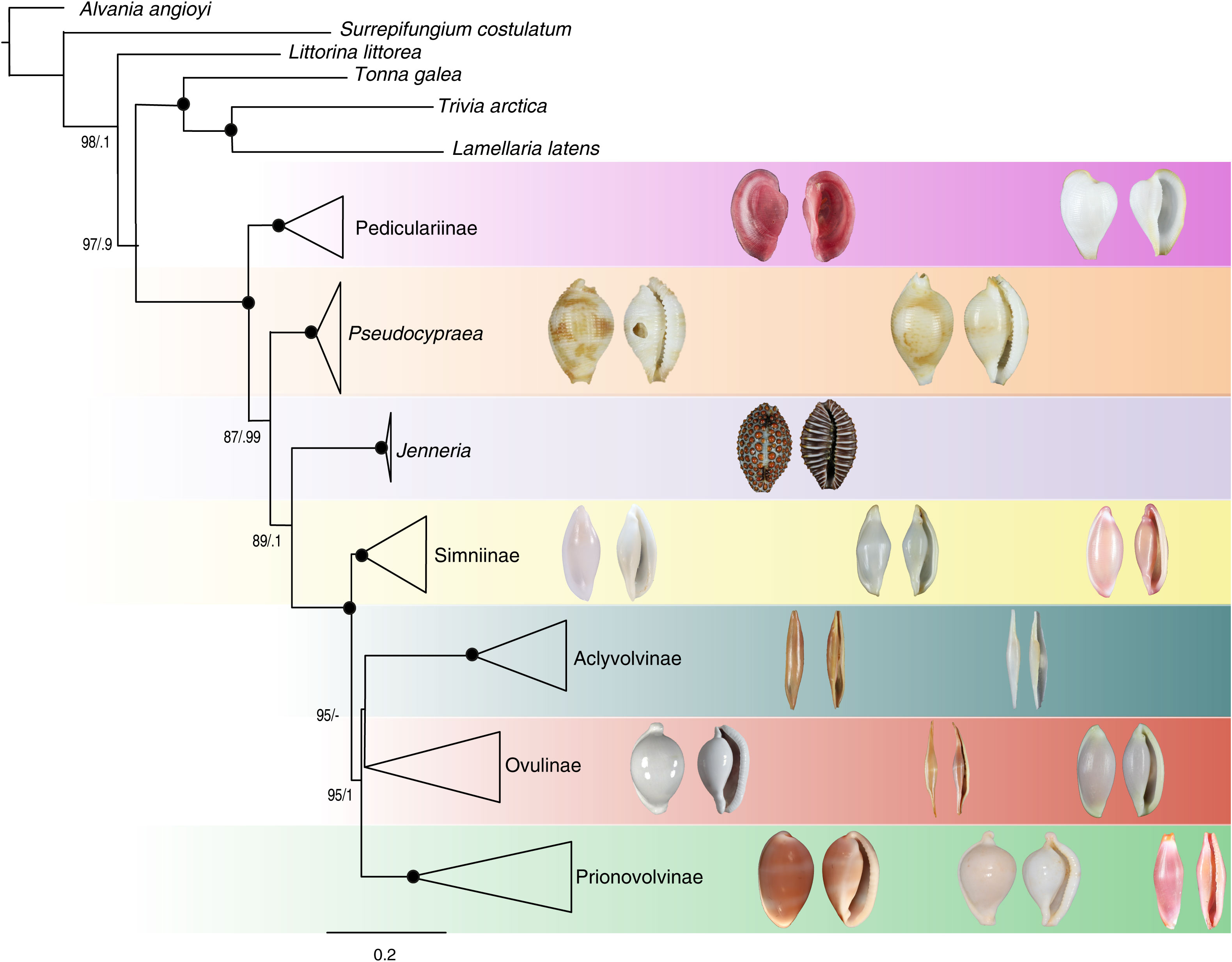
Figure 8 Phylogenetic relationships of the family Ovulidae (Maximum Likelihood tree based on the combined dataset), with clades collapsed by subfamily-level lineages. Numbers at nodes indicate branch support values [ultrafast bootstrap (Ufb) and posterior probability (PP) values, respectively]; support values are shown only when at least one of them is ≥ 95%; black circles at nodes indicate maximum support (PP=1, Ufb=100). Shells of representative taxa, not to scale (from top-left): MNHN-IM-2013-49448 Pedicularia pacifica; MNHN-IM-2019-18297 Lunovula superstes; MNHN-IM-2013-3598 Pseudocypraea adamsonii; MNHN-IM-2013-63257 Pseudocypraea exquisita; BAU-4297 Jenneria pustulata; MNHN-IM-2019-6537 Simnia spelta; MNHN-IM-2013-8520 Cyphoma gibbosum; MNHN-IM-2013-56966 Simnialena uniplicata; MNHN-IM-2013-11745 Aclyvolva coarctata; MNHN- IM-2013-69669 Aclyvolva sp.; MNHN-IM-2007-32155 Ovula ovum; MNHN-IM-2013-53966 Phenacovolva rosea; MNHN-IM-2013-44511 Quasisimnia robertsoni; Ov-NT-982 Prionovolva brevis; MNHN-IM-2013-68826 Ovula isibasii; MNHN-IM-2013-85207 Crenavolva traillii. Photograph credits: Elisa Nocella, Elena Mekhova.
Family Ovulidae J. Fleming, 1822
Subfamily Pediculariinae Gray, 1853
Genus Jenneria Jousseaume, 1884 (lineage probably worthy of a subfamily on its own).
Genus Pseudocypraea F.A. Schilder, 1927 (lineage probably worthy of a subfamily on its own).
Subfamily Simniinae F.A. Schilder, 1927 (the scope should be restricted to encompass only the genera Simnia, Simnialena, Cymbovula and Cyphoma).
Subfamily Ovulinae J. Fleming, 1822 (the actual position of the genus Quasisimnia is obscured by unsupported nodes, although it seems likely a relationship with the ovulines).
Subfamily Aclyvolvinae Fehse, 2007
Subfamily Prionovolvinae Fehse, 2007 [here redefined to comprise, besides the traditional genera, also the genera Contrasimnia, Archivolva, and Naviculavolva (traditionally attributed to Simniinae), Ovula isibasii (formerly in Ovulinae) and Hiatavolva depressa (currently considered as incertae sedis, according to MolluscaBase, 2023)].
Robust molecular evidence indicates that the origin and early diversification of the Ovulidae could have occurred between 47 and 63 mya. The early diversification of major lineages corresponding to subfamilies has been estimated as having occurred between 17 and 41 mya, which largely agrees with the available knowledge from the fossil records, including the oldest genus-level lineage, Pseudocypraea, which was dated between 37 and 40 mya (congruent with the age of the fossil Pseudocypraea dolini: Pacaud, 2003). The observed disparity between the younger dating of the Cyphoma node (1-7 mya) in our analysis compared to its reliable fossil records (15-23 mya), could be explained considering that species currently ascribed to Cyphoma and Simnia actually represent a single lineage. This is evidently characterized by a high degree of morphological plasticity, and we suspect that the Cyphoma-like morphology is plesiomorphic in this clade, as witnessed by the old Cyphoma-like fossils.
The results obtained by the BAMM analysis of ovulid diversification rates across time did not support any core-shift in diversification rates across the phylogeny of the family (Figure 5A). The shape of the rate-through-time plot suggests that the diversification rate started decreasing at the beginning of the ovulid history and continued to decrease steadily until the present day (Figure 5B). We suggest that during the initial stages of the evolution of the Ovulidae, a higher diversification rate (0.1) existed only until the split between Pediculariinae and the rest of ovulids; subsequently, this rate gradually declined in both lineages. In marine gastropods, the lack of distinct shifts in diversification rates within specific clades, despite variations in species richness, has been reported in Terebridae, where it was associated with a pattern of slow, steady increase in diversification across the family due to ecological release corresponding to the colonization of deep waters (Modica et al., 2020). The opposite trend of a steady decrease in the overall diversification rate observed in Ovulidae (Figure 5B) could instead be explained as a diversity-dependent diversification pattern. Diversity-dependent diversification slowdowns are commonly observed and might be explained by various niche-related factors (Moen and Morlon, 2014). In particular, after an initial burst of diversification, the rate of speciation can decelerate due to the occupation of all available niches. In some cases, niche saturation can be also favored by the presence of ecological constraints on clade diversity (Moen and Morlon, 2014). Diversity-dependent diversification slowdowns can be prevented by the repeated evolution of key innovations, each of which could allow subclades to diverge from the trajectory of their parent clade. This pattern may be explained considering the evolution of trophic ecology in ovulids. The initial higher diversification rate was probably related to the onset, in the ancestral ovulids, of the ability to exploit cnidarians as a new trophic resource; it may have been briefly supported by a host-shift between stylasterid corals (Pediculariinae) and anthozoans (non-pediculariines). The initial key innovation, their association with corals, yielded relationships robust enough to limit diversification rates for more than 50 million years.
However, many biases in the methods used to estimate phylogeny and diversification rates have been shown to lead to apparent slowdowns in diversification rates, the main issue being insufficient taxonomic sampling (Moen and Morlon, 2014). Despite our analyses including only 23% of the currently recognized ovulid species, the results also highlighted that the inflation of nominal species in the systematics of ovulids can be ascribed to the extreme morphological plasticity of most of the species, suggesting that our taxon sampling represents a higher fraction of the diversity of egg-cowries, possibly not weakening the validity of the estimation of diversification rates of the family.
As suggested by the available literature, we confirmed in the present work (including 20 new ovulid-cnidarian associations) that the vast majority of ovulid species is rather specialized in its parasitic behavior. Although, admittedly, our dataset (Table 2) was based on a majority of single records per ovulid species (which may bias the perception toward a “one ovulid-one cnidarian” pattern), many of these specialized associations are confirmed by anecdotal reports or photographical material on scuba divers’ websites (e.g.: http://www.underwaterkwaj.com) which however were not included in Table 2. Even taking the potential bias into consideration, in the majority of cases, individuals of the same ovulid species feed on cnidarians belonging to the same family, with a few documented exceptions where a single ovulid species feeds on corals of two or three different families. An exceptional case is represented by Phenacovolva rosea, which displays the broadest known diet, feeding on corals of five different families.
Using coral families as a prior in our efforts to reconstruct ancestral states has been instrumental in identifying the ancestral host for each ovulid subfamily-level lineage. However, especially in the deep part of the tree, this yielded nodes with 20-77% probability of “unknown” host. This uncertainty in the reconstruction of the ancestral hosts might be due to the high number of states with respect to the number of taxa (25 cnidarian families for a dataset of 67 ovulid species). In contrast, the analyses in which the subclasses were used as a prior for the host (with only three states), yielded more robust inferences (1-7% probability of “unknown” host at the base of the tree).
Our analysis results in a diversified framework at the subfamily level: while certain subfamilies display a clear pattern of specialization in some cases feeding on a single family of coral, others exhibit a higher degree of dietary variance. Notably, the subfamilies Pediculariinae and Simniinae represent extreme cases of specialization, including species that predominantly feed on Stylasteridae and Gorgoniidae, respectively. We have been unable to detect any major adaptations corresponding to the major shifts; apomorphic features of pediculariines (shell, mantle and radular characters) may have adaptive values in their exclusive relationship with stylasterids, but this must still be defined. The origin of Ovulidae is estimated to have occurred after the Mesozoic marine revolution (Vermeij, 1977), and we have detected no evident relations of the shifts with major geological events in the oceans.
We can reasonably infer that the ancestor of the Pediculariinae, evolved in association with Stylasteridae approximately 38 to 54 million years ago, and maintained this dietary preference up to the present day. Our working hypothesis is that also Lunovula and Pedicularia sp. predominantly (or perhaps exclusively) feed on Stylasteridae, especially in deep-sea environments (supported by the abundant stylasterids collected at the same sampling stations).
The ancestor of Simniinae similarly evolved a strong association with Gorgoniidae sea-fans 18 to 45 mya, apparently retained as the unique food source. Despite the limited representation of Aclyvolvinae (only two out of the five recognised species represented in our dataset), we can reasonably estimate that the shift to Ellisellidae sea-fans could have occurred between 18 to 42 mya.
Pseudocypraea and Jenneria represent crucial lineages for understanding the evolution of the egg-cowries. Whereas for Jenneria the association with hexacorals is well known, for Pseudocypraea it can only be hypothesized (based on ancestral state reconstruction analysis, see below).
Prionovolvinae is the subfamily with the highest diversification of cnidarian hosts in the present day. Prionovolvinae are estimated to have first evolved a strong association with Alcyoniidae (30-45 mya), subsequently followed by a broad host diversification (11 cnidarian families in our dataset) with a remarkable increase in species diversity of the family, which, however, did not represent a rate shift in our BAMM analysis.
A remarkable host shift identified in this study involves the subfamily Ovulinae. Somewhere along their evolutionary history they shifted from octocorals to the hexacoral family Anthipatidae. This is estimated to have occurred at the origin of the subfamily (between 38 and 43 mya) in the analysis based on cnidarian families. However, the high number of cnidarian families associated with Ovulinae (12 in total) compared to the relatively limited number of assayed ovuline species (13), may have biased the results. In fact, the shift was more plausibly estimated as a series of (possibly independent) very recent events in the analysis based on cnidarian subclasses.
The reconstruction of the ancestral state using coral subclasses as prior of the analyses allowed us to provide two different hypotheses about the ancestral ovulids host:
1) Hydroidolina (Stylasteridae) resulted as the ancestral host from the first BBM analysis carried out without defining the host of Pseudocypraea. In this hypothesis the ancestral ovulids diversified while primarily feeding on Stylasteridae over a considerable time frame spanning from 47 to 56 mya. This dietary adaptation would have persisted up to the present day only in the subfamily Pediculariinae. In contrast, non-pediculariine ovulids underwent a distinct dietary shift to hexacorals from 52 to 54 mya, an adaptation that has been maintained in Jenneria (and possibly also in Pseudocypraea). From approximately 32 to 48 mya, the remaining ovulids transitioned to octocorals as their primary hosts, a trophic preference that persisted until today. Interestingly, a subsequent dietary return to hexacorals is estimated to have occurred (independently) in three ovuline species: Pellasimnia angasi, Takasagovolva honkakuijana, and Phenacovolva lahainensis.
The Hydroidolina hypothesis raises a problem with the depth range of the ancestral egg-cowries. In fact, it is currently accepted (Lindner et al., 2008) that stylasterids first appeared in deep-water environments during the lower Palaeocene (c. 65 mya), and only later (32-38 mya) invaded shallow waters. This would imply that, while the majority of Ovulidae currently inhabit shallow waters, their ancestors might have been deep-water dwellers (associated with deep-water stylasterids), with a subsequent colonization of shallow waters. However, a Distichopora fossil from the Lutetian (41-47 mya: Lindner et al., 2008) opens the possibility that these corals might have existed in shallow waters earlier than supposed (Distichopora is one of the most common stylasterids on which Pedicularia spp. currently feed in shallow waters), allowing for an origin of ovulids in shallow water, associated with stylasterid corals, and offering some support to this hypothesis.
2) Hexacorallia resulted as the ancestral host from the BBM analysis carried out with hexacorals as a diet prior for Pseudocypraea. In this hypothesis, over a period of time ranging from 47 to 56 mya, the ancestral Ovulidae would have evolved in association with Hexacorallia. This adaptation was then maintained in the ancestor of non-pediculariines and up to the present in Jenneria and Pseudocypraea. The shift to Octocorallia occurred about 32 to 48 mya in the other members of the family; this adaptation has persisted in these lineages as their primary dietary preference up to the present day. A subsequent return to hexacorals occurred (possibly independently) in three ovulines: Pellasimnia angasi, Takasagovolva honkakuijana, and Phenacovolva lahainensis. The shift to Hydroidolina is estimated to have taken place 35 to 54 mya in ancestral members of the Pedicularinae.
According to this hypothesis, the ancestral Ovulidae should have evolved in shallow waters feeding on Hexacorallia, probably belonging to the order Scleractinia (such as done by Jenneria). The origin of scleractinians is dated from the Carboniferous to Silurian (324–447 mya) (McFadden et al., 2021). Scleractinia rapidly expanded and diversified across shallow marine environments about 240 mya during the Ladinian (Frankowiak et al., 2023), thus providing an increasingly broad trophic niche available to corallivorous species. In addition, according to this hypothesis, the association between the subfamily Pediculariinae and the stylasterid corals would have started around 35 mya, broadly the same time when Stylasteridae invaded shallow waters, as suggested by Lindner et al. (2008).
Both hypotheses (Hydroidolina or Hexacorallia) are biologically plausible, although we would favor an ancestral exploitation of shallow water hexacorals by the first egg-cowries that could have subsequently colonized deeper habitats following an onshore-offshore trend widely documented in marine fauna in agreement with previous hypotheses (Jacobs and Lindberg, 1998; Oliverio et al., 2009b).
In any case, it seems evident that the diversification of Ovulidae has not been strictly coupled with cnidarian evolution (no pervasive co-evolutionary pattern). All families within Octocorallia, Hexacorallia, and Hydroidolina originated significantly earlier than Ovulidae, with the most recent emergence occurring approximately 100 mya (Lindner et al., 2008; McFadden et al., 2021). Overall, our analyses, as also anticipated by Reijnen (2016), challenge the notion of coevolution and cospeciation as governing factors in the evolutionary history of Ovulidae and Cnidaria. Instead, the evolutionary trajectories of egg-cowries are better defined as a pattern of sequential evolution. Ovulids should have selected their hosts based on contingent ecological factors (e.g.: host abundance, lack of competitors, etc.) rather than being constrained by a phylogenetic coevolutionary process; associations have remained rather stable across time, with shifts mostly consisting of changes of families within the same cnidarian subclass.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The manuscript presents research on animals that do not require ethical approval for their study.
EN: Writing – original draft, Data curation, Formal analysis, Investigation, Methodology, Software, Visualization. SZ: Formal analysis, Resources, Writing – review & editing. GF: Methodology, Writing – review & editing. DP: Formal analysis, Resources, Writing – review & editing. BB: Data curation, Writing – review & editing. RV: Data curation, Writing – review & editing. NP: Writing – review & editing. MM: Conceptualization, Funding acquisition, Project administration, Writing – review & editing, Validation. MO: Conceptualization, Funding acquisition, Project administration, Writing – review & editing, Validation.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The research was partially funded by the Doctorate School in Evolutionary and Environmental Biology of Sapienza University of Rome, and by the 2023 Unitas Malacologica Student Research Awards (to EN); by the Sapienza 2020 INV-EVO/2 grant (RM12017276B846BB) and Sapienza 2021 SEED-3840974 grant (to MO); and by the Russian Science Foundation (Grant number [22-74-00113], to SZ).
Many of the specimens sequenced herein were collected under the auspices of the Our Planet Reviewed Tropical Deep Sea Benthos programs at the MNHN (expeditions ATIMO VATAE, DOI:10.17600/10110040; AURORA 2007; BIOMAGLO, DOI: 10.17600/17004000; CONCALIS, DOI: 10.17600/8100010; CORSICABENTHOS 1; CORSICABENTHOS 2; CORSICABENTHOS 3; EBISCO, DOI: 10.17600/5100080; EXBODI, DOI: 10.17600/11100080; ILES DU SALUT; KANACONO, DOI: 10.17600/16003900; KANADEEP, DOI: 10.17600/17003800; KARUBENTHOS 2, DOI: 10.17600/15005400; KAVIENG 2014, DOI: 10.17600/14004400; KOUMAC 2.3; MADEEP, DOI: 10.17600/14004000; MAINBAZA; MIRIKI; NanHai 2014; MADIBENTHOS; NORFOLK 2, DOI: 10.17600/3100030; PAKAIHI I TE MOANA; PANGLAO 2004; PAPUA NIUGINI, DOI:10.17600/18000841; SALOMONBOA 3, DOI: 10.17600/7100070; SANTO 2006; SPANBIOS, DOI: 10.17600/18000701; ZhongSha 2015). These expeditions operated under the regulations then in force in the countries in question and satisfy the conditions set by the Nagoya Protocol for access to genetic resources. We extend our acknowledgements to Paolo Mariottini (University of Roma Tre), Justine Siegwald (University of Bergen) and Francesco Colloca (Stazione Zoologica Anton Dohrn) for providing molecular grade materials. We are grateful to Philippe Maestrati (MNHN), Laurent Charles (Muséum de Bordeaux), Gill Devauchelle, Elena Mekhova (A.N. Severtsov Institute of Ecology and Evolution) for providing precious photographic material. We thank Dario Zuccon (MNHN) for his help with MNHN molecular sequences. Lucrezia Leotta (Sapienza University of Rome) and Giacomo Chiappa (Sapienza University of Rome) are acknowledged for their help with the laboratory work, and Claudio Pica (University of Southern Denmark) for help with bioinformatic analysis. Luigi Romani is thanked for his help with bibliographic research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1323156/full#supplementary-material
Ali S. S., Yu Y., Pfosser M., Wetschnig W. (2012). Inferences of biogeographical histories within subfamily Hyacinthoideae using S-DIVA and Bayesian binary MCMC analysis implemented in RASP (Reconstruct Ancestral State in Phylogenies). Ann. Bot. 109, 95–107. doi: 10.1093/aob/mcr274
Aljanabi S. M., Martinez I. (1997). Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 25, 4692–4693. doi: 10.1093/nar/25.22.4692
Barberousse A., Samadi S. (2010). Species from Darwin onward. Integr. Zool. 5, 187–197. doi: 10.1111/j.1749-4877.2010.00204.x
Beu A. G., Marshall B. A. (2011). New Cenozoic records of genera and families from New Zealand (Mollusca, Gastropoda): Highlights from Phillip Maxwell’s collection. New Z. J. Geol. Geophys. 54, 13–34. doi: 10.1080/00288306.2011.536517
Chiappa G., Fassio G., Corso A., Crocetta F., Modica M. V., Oliverio M. (2022). How many abalone species live in the mediterranean sea? Diversity (Basel) 14 (12), 1107. doi: 10.3390/d14121107
Colgan D. J., Ponder W. F., Beacham E., Macaranas J. M. (2003). Gastropod phylogeny based on six segments from four genes representing coding or non-coding and mitochondrial or nuclear DNA. Molluscan Res. 23, 123–148. doi: 10.1071/MR03002
Cunningham C. W., Buss L. W. (1993). Molecular evidence for multiple episodes of paedomorphosis in the family Hydractiniidae. Biochem. Syst. Ecol. 21, 57–69. doi: 10.1016/0305-1978(93)90009-G
Darriba D., Taboada G. L., Doallo R., Posada D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772. doi: 10.1038/nmeth.2109
Espiritu D. J. D., Watkins M., Dia-Monje V., Cartier G. E., Cruz L. J., Olivera B. M. (2001). Venomous cone snails: molecular phylogeny and the generation of toxin diversity. Toxicon 39, 1899–1916. doi: 10.1016/S0041-0101(01)00175-1
Fabricius K., Alderslade P. (2001). Soft corals and sea fans: a comprehensive guide to the tropical shallow water genera of the central-west Pacific, the Indian Ocean and the Red Sea (Townsville: Australian Institute of Marine Science).
Fassio G., Bouchet P., Lozouet P., Modica M. V., Russini V., Schiaparelli S., et al. (2021). Becoming a limpet: An ‘intermittent limpetization’ process driven by host features in the kleptoparasitic gastropod family Capulidae. Mol. Phylogenet. Evol. 155, 107014. doi: 10.1016/j.ympev.2020.107014
Fassio G., Russo P., Bonomolo G., Fedosov A. E., Modica M. V., Nocella E., et al. (2022). A molecular framework for the systematics of the Mediterranean spindle-shells (Gastropoda, Neogastropoda, Fasciolariidae, Fusininae). Mediterr. Mar. Sci. 23, 623–636. doi: 10.12681/mms.29935
Fassio G., Stefani M., Russini V., Buge B., Bouchet P., Treneman N., et al. (2023). Neither slugs nor snails: a molecular reappraisal of the gastropod family Velutinidae. Zool. J. Linn Soc. 197, 924–964. doi: 10.1093/zoolinnean/zlac091
Fehse (2001). Beiträge zur Kenntnis der Ovulidae (Mollusca: Cypraeoidea) VIII. Einleitung sur Familie sowie Katalog, Taxonomie und Bibliographie und Bemerkungen zu verwandten Gruppen. Acta Conchyliorum (Ludwigsburg) 5, 3–47.
Fehse D. (2007). Contributions to the knowledge of the ovulidae. XVI. the higher systematics. Spixiana 30, 121–125.
Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3 (5), 294–299.
France S. C., Hoover L. L. (2002). DNA sequences of the mitochondrial COI gene have low levels of divergence among deep-sea octocorals (Cnidaria: Anthozoa). Hydrobiologia 471, 149–155. doi: 10.1023/A:1016517724749
Frankowiak K., Wang X. T., Sigman D. M., Gothmann A. M., Kitahara M. V., Mazur M., et al. (2023). Photosymbiosis and the expansion of shallow-water corals. Sci. Adv. 2, e1601122. doi: 10.1126/sciadv.1601122
Galindo L. A., Puillandre N., Strong E. E., Bouchet P. (2014). Using microwaves to prepare gastropods for DNA barcoding. Mol. Ecol. Resour. 14, 700–705. doi: 10.1111/1755-0998.12231
Gernhard T. (2008). The conditioned reconstructed process. J. Theor. Biol. 253, 769–778. doi: 10.1016/j.jtbi.2008.04.005
Grajales A., Aguilar C., Sánchez J. A. (2007). Phylogenetic reconstruction using secondary structures of Internal Transcribed Spacer 2 (ITS2, rDNA): finding the molecular and morphological gap in Caribbean gorgonian corals. BMC Evol. Biol. 7, 90. doi: 10.1186/1471-2148-7-90
Grasshoff M. (1999). The shallow water gorgonians of New Caledonia and adjacent islands. Senckenb Biol. 78 (1/2), 1–121.
Hayashi S. (2003). The molecular phylogeny of the buccinidae (Caenogastropoda: Neogastropoda) as inferred from the complete mitochondrial 16S rRNA gene sequences of selected representatives. Molluscan Res. 25, 85–98.
Ho S. Y. W., Phillips M. J. (2009). Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst. Biol. 58, 367–380. doi: 10.1093/sysbio/syp035
Jacobs D. K., Lindberg D. R. (1998). Oxygen and evolutionary patterns in the sea: Onshoreoffshore trends and recent recruitment of deep-sea faunas. Proc. Natnl Acad. Sci. 95 (16), 9396–9401. doi: 10.1073/pnas.95.16.9396
Jovelin R., Justine J. L. (2001). Phylogenetic relationships within the polyopisthocotylean monogeneans (Platyhelminthes) inferred from partial 28S rDNA sequences. Int. J. Parasitol. 31, 393–401. doi: 10.1016/S0020-7519(01)00114-X
Katoh K., Rozewicki J., Yamada K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Leray M., Yang J. Y., Meyer C. P., Mills S. C., Agudelo N., Ranwez V., et al. (2013). A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Front. Zool. 10, 34. doi: 10.1186/1742-9994-10-34
Lindner A., Cairns S. D., Cunningham C. W. (2008). From offshore to onshore: Multiple origins of shallow-water corals from deep-dea ancestors. PloS One 3 (6), e2429. doi: 10.1371/journal.pone.0002429
Lorenz F., Fehse D. (2009). The living Ovulidae: a manual of the families of allied cowries: Ovulidae, Pediculariidae and Eocypraeidae (Hackenheim: ConchBook).
McFadden C. S., Quattrini A. M., Brugler M. R., Cowman P. F., Dueñas L. F., Kitahara M. V., et al. (2021). Phylogenomics, origin, and diversification of anthozoans (Phylum cnidaria). Syst. Biol. 70, 635–647. doi: 10.1093/sysbio/syaa103
Miller M. A., Pfeiffer W., Schwartz T. (2010). "Creating the CIPRES Science Gateway for inference of large phylogenetic trees," in. 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA., pp. 1–8. doi: 10.1109/GCE.2010.5676129
Modica M. V., Gorson J., Fedosov A. E., Malcolm G., Terryn Y., Puillandre N., et al. (2020). Macroevolutionary analyses suggest that environmental factors, not venom apparatus, play key role in terebridae marine snail diversification. Syst. Biol. 69, 413–430. doi: 10.1093/sysbio/syz059
Moen D., Morlon H. (2014). Why does diversification slow down? Trends Ecol. Evol. 29, 190–197. doi: 10.1016/j.tree.2014.01.010
MolluscaBase (2023). Cypraeoidea. Available at: https://www.molluscabase.org/aphia.php?p=taxdetails&id=14774.
Moore B. R., Höhna S., May M. R., Rannala B., Huelsenbeck J. P. (2016). Critically evaluating the theory and performance of Bayesian analysis of macroevolutionary mixtures. Proc. Natnl Acad. Sci. 113, 9569–9574. doi: 10.1073/pnas.1518659113
Oliverio M. (2008). “Coralliophilinae (Neogastropoda: Muricidae) from the southwest pacific,” in Tropical deep-sea benthos 25, vol. 196. Eds. Héros V., Cowie R., Bouchet P. (Mémoires du Muséum National d'Histoire Naturelle), 451–555.
Oliverio M., Barco A., Modica M. V., Richter A., Mariottini P. (2009a). Ecological barcoding of corallivory by second internal transcribed spacer sequences: hosts of coralliophiline gastropods detected by the cnidarian DNA in their stomach. Mol. Ecol. Resour. 9, 94–103. doi: 10.1111/j.1755-0998.2008.02388.x
Oliverio M., Barco A., Richter A., Modica M. V. (2009b). The coralliophiline (Gastropoda: Muricidae) radiation: repeated colonizations of the deep sea? Nautilus 123 (3), 113–120.
Pacaud J. M. (2003). First fossil records of the Recent Ovulid genus Pseudocypraea Schilder 1927 (Mollusca, Gastropoda) with description of a new species. Geodiversitas 25 (3), 451–462.
Palumbi S. R., Martin A. P., Romano S. L., McMillan W. O., Stice L., Grabowski G. (1991). The simple fool’s guide to PCR (Honolulu: Dept. of Zoology, University of Hawaii).
Paz-García D. A., Aldana-Moreno A., Cabral-Tena R. A., Balart E. F. (2012). High predation by the corallivore sea snail Jenneria pustulata in a high-latitude reef in the Gulf of California. Mar. Biodivers. Rec. 5, e94. doi: 10.1017/S1755267212000826
Ploch S., Kruse J., Choi Y.-J., Thiel H., Thines M. (2022). Ancestral state reconstruction in Peronospora provides further evidence for host jumping as a key element in the diversification of obligate parasites. Mol. Phylogenet. Evol. 166, 107321. doi: 10.1016/j.ympev.2021.107321
Puillandre N., Brouillet S., Achaz G. (2021). ASAP: assemble species by automatic partitioning. Mol. Ecol. Resour. 21, 609–620. doi: 10.1111/1755-0998.13281
Puillandre N., Modica M. V., Zhang Y., Sirovich L., Boisselier M. C., Cruaud C., et al. (2012). Large-scale species delimitation method for hyperdiverse groups. Mol. Ecol. 21, 2671–2691. doi: 10.1111/j.1365-294X.2012.05559.x
Rabosky D. L. (2013). Diversity-dependence, ecological speciation, and the role of competition in macroevolution. Annu. Rev. Ecol. Evol. Syst. 44, 481–502. doi: 10.1146/annurev-ecolsys-110512-135800
Rabosky D. L. (2014). Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PloS One 9, e89543. doi: 10.1371/journal.pone.0089543
Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A. (2018). Posterior summarization in bayesian phylogenetics using tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Reijnen B. T. (2016). Phylogenetic ecology of octocoral-gastropod associations. [PhD’s thesis] (Leiden: Naturalis Biodiversity Center, Science, Leiden University). Available at: http://hdl.handle.net/1887/43471.
Reijnen B. T., Hoeksema B. W., Gittenberger E., Arntzen J. W. (2010). Host specificity and phylogenetic relationships among Atlantic Ovulidae (Mollusca: Gastropoda). Contrib. to Zool. 79, 69–78. doi: 10.1163/18759866-07902002
Reijnen B. T., van der Meij S. E. T. (2017). Coat of many colours—DNA reveals polymorphism of mantle patterns and colouration in Caribbean Cyphoma Röding 1798 (Gastropoda, Ovulidae). PeerJ 5, e3018. doi: 10.7717/peerj.3018
Reijnen B. T., van der Meij S. E. T. (2019). Systematics of the subfamily aclyvolvinae (Caenogastropoda: Ovulidae) based on molecular and morphometric analyses. J. Molluscan Stud. 85 (3), 336–347.
Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Rosenberg G. (1992). An introduction to the Ovulidae (Gastropoda: Cypraeacea). Amer Conchol. 20, 4–7.
Russini V., Fassio G., Nocella E., Houart R., Barco A., Puillandre N., et al. (2023). Whelks, rock-snails, and allied: a new phylogenetic framework for the family Muricidae (Mollusca: Gastropoda). Eur. Zool. J. 90, 856–868. doi: 10.1080/24750263.2023.2283517
Sánchez J. A., Fuentes-Pardo A. P., Ní Almhain Í., Ardila-Espitia N. E., Cantera-Kintz J., Forero-Shelton M. (2016). The masquerade game: marine mimicry adaptation between egg-cowries and octocorals. PeerJ 4, e2051. doi: 10.7717/peerj.2051
Sánchez J. A., McFadden C. S., France S. C., Lasker H. R. (2003). Molecular phylogenetic analyses of shallow-water Caribbean octocorals. Mar. Biol. 142, 975–987. doi: 10.1007/s00227-003-1018-7
Schiaparelli S., Barucca M., Olmo E., Boyer M., Canapa A. (2005). Phylogenetic relationships within Ovulidae (Gastropoda: Cypraeoidea) based on molecular data from the 16S rRNA gene. Mar. Biol. 147, 411–420. doi: 10.1007/s00227-005-1566-0
Schiaparelli S., Fransen C., Oliverio M. (2011). "Marine partnerships in Santo’s reef environments: parasites, commensals, and other organisms that live in close association," in The Natural History of Santo. MNHN, Paris; IRD, Marseille; PNI, Paris. Eds P. Bouchet, H. Le Guyader and O. Pascal (Patrimoines naturels; 70), pp. 449–457.
Simone L. R. L. (2004). Morphology and phylogeny of the Cypraeoidea (Mollusca, Caenogastropoda) (Rio de Janeiro: Papel Virtual).
Stöver B. C., Müller K. F. (2010). TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinf. 11, 7. doi: 10.1186/1471-2105-11-7
Suchard M. A., Lemey P., Baele G., Ayres D. L., Drummond A. J., Rambaut A. (2018). Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4 (1), vey016. doi: 10.1093/ve/vey016
Tazioli S., Bo M., Boyer M., Rotinsulu H., Bavestrello G. (2007). Ecological observations of some common antipatharian corals in the marine park of Bunaken (North Sulawesi, Indonesia). Zool. Stud. 46 (2), 227–241.
Trifinopoulos J., Nguyen L.-T., von Haeseler A., Minh B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Vermeij G. J. (1977). The Mesozoic marine revolution: evidence from snails, predators and grazers. Paleobiology 3, 245–258. doi: 10.1017/S0094837300005352
Wilgenbusch J. C., Swofford D. (2003). Inferring evolutionary trees with PAUP. Curr. Protoc. Bioinf. 1), 6–4.
Woodring W. P. (1973). Geology and Paleontology of Canal Zone and Adjoining Parts of Panama Description of Tertiary Mollusks (Additions to gastropods, scaphopods, pelecypods: Nuculidae to Malleidae). Geol. Survey Prof. Pap. 306-E, 453–539. doi: 10.3133/pp306E
Yu Y., Harris A. J., Blair C., He X. (2015). RASP (Reconstruct Ancestral State in Phylogenies): A tool for historical biogeography. Mol. Phylogenet. Evol. 87, 46–49. doi: 10.1016/j.ympev.2015.03.008
Zucconi M. G., Obonaga L. D., Londoño-Cruz E. (2018). Corallivory of the gastropod Jenneria pustulata (Ovulidae: Pediculariinae) in two coral reefs at Gorgona Island NNP. Boletin Investigaciones Marinas y Costeras 47, 11–23. doi: 10.25268/bimc.invemar.2018.47.2.744
Keywords: Ovulidae, Cnidaria, trophic ecology, molecular phylogeny, macroevolution, host-parasite association, adaptation, corallivory
Citation: Nocella E, Zvonareva SS, Fassio G, Pica D, Buge B, Villa R, Puillandre N, Modica MV and Oliverio M (2024) Spicy food for the egg-cowries: the evolution of corallivory in the Ovulidae (Gastropoda: Cypraeoidea). Front. Mar. Sci. 10:1323156. doi: 10.3389/fmars.2023.1323156
Received: 17 October 2023; Accepted: 27 December 2023;
Published: 22 January 2024.
Edited by:
Juan Armando Sanchez, University of Los Andes, ColombiaReviewed by:
Joachim Langeneck, Consorzio Nazionale Interuniversitario per le Scienze del Mare (CoNISMa), ItalyCopyright © 2024 Nocella, Zvonareva, Fassio, Pica, Buge, Villa, Puillandre, Modica and Oliverio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elisa Nocella, ZWxpc2Eubm9jZWxsYUB1bmlyb21hMS5pdA==
†These authors have contributed equally to this work and share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.