- 1Guangdong Technology Research Center for Marine Algal Bioengineering, Guangdong Provincial Key Laboratory for Plant Epigenetics, Shenzhen Engineering Laboratory for Marine Algal Biotechnology, Longhua Innovation Institute for Biotechnology, College of Life Sciences and Oceanography, Shenzhen University, Shenzhen, China
- 2College of Physics and Optoelectronic Engineering, Shenzhen University, Shenzhen, China
Hazardous impacts caused by harmful algal blooms (HABs) have facilitated the development of novel effective approaches for controlling the frequent HABs. Recently, bacteria with algicidal effects, as a promising alternative tool for eliminating the HABs-forming species have received wide concerns. In the present study, a marine bacterial strain 6A1 with potent algicidal activity was successfully isolated from a bloom of Akashiwo sanguinea, and the characteristics related to its algicidal action were systematically investigated. Sequencing results of 16S rDNA suggested that this bacterium belongs to the genus Arenibacter in the family Flavobacteriaceae, while the phylogenetic analysis revealed that Arenibacter sp. strain 6A1 was the only algicidal bacterium in Arenibacter to date, according to the best of our knowledge. Arenibacter sp. strain 6A1 had an extraordinarily high algicidal performance when it was added to A. sanguinea culture with a volume fraction of 1.5% for 2 h. The algicidal performance of strain 6A1 was concentration-dependent and time-dependent. The best algicidal efficacy of strain 6A1 was seen during the stationary phase, while the resistance of A. sanguinea to strain 6A1 declined as the growth cycle progressed. Subsequently, the indirect algicidal mode of Arenibacter sp. strain 6A1 was verified and the algicidal compounds produced by strain 6A1 were proved to be heat-resistant. Further research showed that strain 6A1 caused a severe damage in photosynthesis, oxidative stress, and also damage in three HABs-forming algae to varying degrees. The greatest algicidal performance was found on unarmored dinoflagellate A. sanguinea, followed by raphidophyte Chattonella antiqua and armored dinoflagellate Alexandrium tamarense. The Arenibacter sp. strain 6A1 exhibited selective algicidal activity on a wide range of algal species, primarily targeting Pyrrophyta and Ochrophyta. The proposed bacteria (strain 6A1) can be a powerful and eco-friendly tool for emergency treatment of HABs caused by certain algal species in coastal water bodies.
1 Introduction
Frequently occurring harmful algal bloom (HABs) has profound and deleterious effects on fisheries, tourism, ecosystem, and even public health (Van Dolah, 2000; Lewitus et al., 2012; Berdalet et al., 2015; Brown et al., 2019). The over-proliferation of HABs-forming algae, especially those toxic species, may cause massive fish kills, destroy or poison shellfish beds, contribute to wildlife mortality, human illness even death (Van Dolah, 2000). With the predictable climate changes in the next decades that will favor the HABs outbreak in coastal regions, as well as an expected increase in human activities in coastal areas, HABs will be a growing concern in a global context (Harvell et al., 1999; Hallegraeff, 2010; Griffith and Gobler, 2020). It is increasingly urgent to explore effective strategies to control HABs and reduce their negative impact (Anderson et al., 2012).
Over the past decades, various physical and chemical approaches for controlling HABs have been implemented to mitigate the resultant damage of HABs (Gallardo-Rodríguez et al., 2019). Physical treatments, such as mixing, filtering, flocculation, sediment burial, dredging, and using ultrasound, have been adopted to eliminate HABs (Kim, 2006; Park et al., 2017; Yu et al., 2018; Jiang et al., 2021). Most chemical methods for HABs control have emphasized the use of biosurfactants, biocides or allelochemicals, synthetic chemicals, isolated algicidal compounds, and metallic compounds such as copper sulfate (Wang et al., 2005; Anderson, 2009; Zhu et al., 2021b). These methods can remove HABs efficiently and are used operationally as a last resort, but they can be costly, lack specificity to HABs, and are generally less effective in coastal situations (Sellner and Rensel, 2018). Therefore, more effective and eco-friendly methodologies are urgently needed to counteract HABs in coastal water bodies.
Contrary to the above-mentioned methods, biological measures include the application of microbe (viral, bacterial, fungal, and/or protistan) that are capable of infecting/lysing cells or inhibiting the growth of HABs-forming species (Sun et al., 2018). Recent evidence suggests that biological methods have been recognized as one of the most attractive and eco-friendly tools in mitigating HABs, as they can be potentially more specific for individual HABs-forming species, minimizing the impact on other non-target species (Pal et al., 2020). In addition, the algicidal microbe can be native species isolated from environments with frequent outbreaks of HABs, and therefore, the balance of previous coastal ecosystems is easier to restore after biological treatment (Sun et al., 2016).
Recent studies showed that many genera of marine bacteria demonstrated algicidal performance and play important roles in the eradication of HAB in natural marine environments. For instance, some bacteria are known to have been involved in the elimination of algal blooms caused by Prorocentrum minimum (Kim et al., 2009a), Prorocentrum donghaiense (Shi et al., 2018; Zhang et al., 2018; Zhang et al., 2020), Akashiwo sanguinea (Wang et al., 2020b), Alexandrium catenella (Kim et al., 2015), Alexandrium tamarense (Wang et al., 2012; Li et al., 2015), Karenia brevis (Roth et al., 2008), Karenia mikimotoi (Lu et al., 2016), Skeletonema costatum (Shi et al., 2013), Chattonella marina (Kim et al., 2009b). Most known algicidal bacteria within the Cytophaga/Flavobacterium/Bacteroides (CFB) group or the γ-Proteobacteria group are known to, directly and indirectly, attack harmful algae in a variety of ways (Mayali and Azam, 2004; Wang et al., 2020a). Based on the literature, about 50% of the algicidal strains belong to the CFB group, while about 45% are members of γ-Proteobacteria (Wang et al., 2020a). The remaining strains are from Saprospire sp., Vibrio, and Bacillus sp. (Wang et al., 2020a). These findings have raised the possibility of utilizing bacteria for regulating HABs in the natural environment. However, it is remarkable that some of the reported algicidal bacteria belong to pathogenic bacteria and could be potential threats to human health. Previous studies found that γ-Proteobacteria is related to endogenous alcohol production which could result in liver damage (Ren et al., 2007; Rizzatti et al., 2017). What’s more, most strains of Vibrio are associated with gastroenteritis (West, 1989; Froelich and Noble, 2016). Hence, much effort needs to be made to investigate more eco-friendly and safe bacterial strains from diverse aquatic environments with more efficient algicidal capability.
To date, few studies have investigated possible control of HABs using Arenibacter. Members of Arenibacter have frequently been reported to be useful for degradation of polycyclic aromatic hydrocarbons (PAH) and crab chitin (Gutierrez et al., 2014; Gutierrez et al., 2016; Gurav et al., 2019). In the present study, a novel effective algicidal marine bacterial strain 6A1 within the genus Arenibacter was isolated in the course of screening for algicidal bacteria in an A. sanguinea bloom event. Akashiwo sanguinea is a notorious HABs-forming marine dinoflagellate widely distributed in coastal areas which definitively results in the mortality of several marine species (Du et al., 2011; White et al., 2014; Jones et al., 2017). The main aims of this study were: (1) identify the taxonomic status of strain 6A1 based on 16S rDNA results and the difference between strain 6A1 and its related bacteria; (2) investigate the algicidal performance of strain 6A1 on A. sanguinea; (3) demonstrate the feasibility of strain 6A1 for HABs-control in natural water by testing and comparing the effect of strain 6A1 on a wide range of aquatic algal species, including HABs-forming species and beneficial species. To the best of our knowledge, this is the first investigation of the algicidal activity of the genus Arenibacter. Overall, the results of this study provide an alternative way of preventing HABs.
2 Materials and methods
2.1 Algal cultures
Three HABs-forming species (Akashiwo sanguinea, Chattonella antiqua, and Alexandrium tamarense) used in the present study were provided by the Algal Culture Collection, Institute of Hydrobiology, Jinan University, Guangzhou, China. The culture of each strain was grown in sterile f/2 seawater medium at 20 ± 1°C under a 12 h:12 h light-dark cycle with a light intensity of 35 μmol photons m-2 s-1. Aged seawater filtered through 0.22 μm mixed cellulose esters (MCE) filters were used for medium preparation.
2.2 Screening for algicidal bacteria from A. sanguinea bloom
Offshore seawater samples from 0 to 0.5 m were collected for algicidal bacteria isolation on Feb 24, 2016, during an A. sanguinea bloom (approx. 1.1×107 cell L-1) in the eastern coastal area of Shenzhen, China (N 22°28’25”, E 114°32’21”).
For screening algicidal bacteria, 1 L seawater was filtered through 0.22 μm MCE filters, the filter was then put into 100 mL 2216E medium and incubated under 25°C for 24 h. After incubation, 0.1 mL aliquots were serially diluted and streaked on 2216E agar (1% agar in 2216E medium) to isolate single colonies. The isolated culturable bacterial strains were then transferred into liquid 2216E medium for incubation over 24 h. Then, bacterial cultures were added into exponentially growing A. sanguinea culture with a ratio of 10% (v: v) and cultured in the light incubator overnight. For each bacterial strain, three replicate co-cultures were made, and algal cultures added with 10% (v: v) 2216E medium were used as the negative control. After incubation, 1 mL of each co-culture mixture was added into 12 well microtiter plates and checked under an optical microscope (Nikon, Tokyo, Japan). The bacterial strains that showed significant algicidal symptoms (cell shrinking, stopping swimming, or cell lysing) compared with control treatments were defined as algicidal and labeled for further investigation. Over ten isolates were observed to have algicidal activity on A. sanguinea in the preliminary screening. Among these isolates, a strain named 6A1 exhibited the strongest algicidal activity (Supplementary Table 1). Thus, strain 6A1 was used for all following tests.
2.3 Identification of algicidal bacterial strains 6A1
The isolated bacterial strain 6A1 was inoculated in a flask containing 100 mL 2216E medium. The culture was maintained at 30°C with shaking at 200 rpm until it reached a cell density of 9×108 cell mL-1. The bacterial cells were collected by centrifugation at 12000 rpm for 3 min and the pellet was resuspended using DNA lysis buffer. The sample was incubated for 1 h at 55°C after adding 25 μL of proteinase K. DNA extraction was then carried out using a Bacterial DNA Kit (Omega Inc., U.S.A.). The 16S rRNA gene was amplified by PCR using primers 27 F and 1492 R with 10 ng of the isolated genomic DNA as a template. The amplified fragments were sent to TaKaRa Company (Shanghai, China) for sequencing. The sequence was analyzed by BLAST using the GenBank database. A phylogenetic tree was constructed to determine the affinity of strain 6A1 by the maximum likelihood method with the representative bacteria in MEGA (ver.10).
2.4 Algicidal activity of strain 6A1 against A. sanguine
The growth phase of A. sanguinea was monitored for several days. Bacterial cultures at the stationary stage were inoculated into 50 mL algal cultures at distinct growth stages (lag stage, logarithmic phase, and stationary stage) with a volume fraction of 1.5%. For each growth stage, three replicate co-cultures were made, and algal cultures added with 1.5% (v: v) 2216E medium were used as control. The cell density of A. sanguinea was counted under optical microscope at the beginning, 15 min, and 120 min. Then the algicidal ratio was estimated according to the following formula and presented as mean ± standard deviation (SD).
Nc is the number of algal cells in the control group; Nt is the number of algal cells in the treatment group.
2.5 Algicidal characteristics of strain 6A1
To investigate the algicidal mode of strain 6A1, the bacterial culture was divided into four parts (whole culture, culture medium, supernatant, pellet). The supernatant was acquired by filtering the bacterial culture through 0.22 mm MCE filters. To obtain the pellet of bacterium, the same amount of bacterial culture was centrifugated and the bacterial pellet was washed twice using sterile f/2 medium and resuspended in equivalent sterile f/2. Following that, each component was resuspended or added into 25 mL stationary growing A. sanguinea culture with a final volume rate of 1.5%. The cell density of A. sanguinea was counted under optical microscope at the beginning, 15 min, and 120 min to evaluate the algicidal ratio.
To evaluate the algicidal performance of strain 6A1 cultures at different growth stages, the growth curve of strain 6A1 was inspected, and then the bacterial cultures at distinct growth phases (lag phase, logarithmic phase, and stationary phase) were inoculated into 50 mL exponentially growing A. sanguinea cultures with a volume fraction of 1.5%. For each growth phase, three replicate co-cultures were made, and algal cultures added with 1.5% (v: v) 2216E medium were used as control. The cell density of A. sanguinea was counted under the optical microscope at the beginning, 15 min, and 120 min, respectively to evaluate the algicidal ratio.
To test the temperature stability of strain 6A1, the 6A1 culture was held at 100°C for 30 min, after which the temperature of the cultures was allowed to resume at room temperature. The bacterial cultures were inoculated into stationary growing A. sanguinea cultures to determine the algicidal activity. The algal culture added with 6A1 culture held at room temperature was used as the control.
2.6 Algicidal performance of strain 6A1 on three typical HABs-forming species
The algicidal activity of strain 6A1 was also investigated in typical algal species including A. sanguinea, C. antiqua, and A. tamarense. Strain 6A1 was added to algae culture during their stationary growth stage. According to our previous study, the optimal algicidal dosage of strain 6A1 was 1.0% (v: v) and 1.5% (v: v) for A. sanguinea, 1.5% (v: v) and 2.5% (v: v) for C. antiqua, and 5.0% (v: v) and 10.0% (v: v) for A. tamarense. The algicidal process of each species was observed under an optical microscope (Nikon, Tokyo, Japan) and scanning electron microscope (SEM) (Thermo Scientific, Netherlands) at the beginning, 15 min, 1 h, and 2 h, respectively. Additionally, the algal cell density and maximum photosynthesis quantum yields (Fv/Fm) of algal cells were detected at the beginning, 1 h, 2 h, 4 h, 6 h, and 8 h respectively to evaluate the level of photosynthetic activity in algae. Fv/Fm was measured using a chlorophyll fluorometer (FL3500, Czech Republic). All samples were dark-adapted for 15 min before measurement. The superoxide dismutase (SOD) activity, catalase (CAT) activity, malondialdehyde (MDA) content, and reactive oxygen species (ROS) levels in algal cells were detected at the beginning, 2 h, 4 h, and 6 h respectively to evaluate the level of oxidative stress in algae. SOD activity, CAT activity, MDA content, and ROS production were measured using a total SOD activity detection kit, catalase assay kit, MDA assay kit, and Reactive oxygen species Assay Kit (Nanjing jiancheng bioengineering institute), respectively according to the manufacturer’s manual. Each treatment was repeated three times independently.
2.7 Assay of the algicidal spectrum of strain 6A1
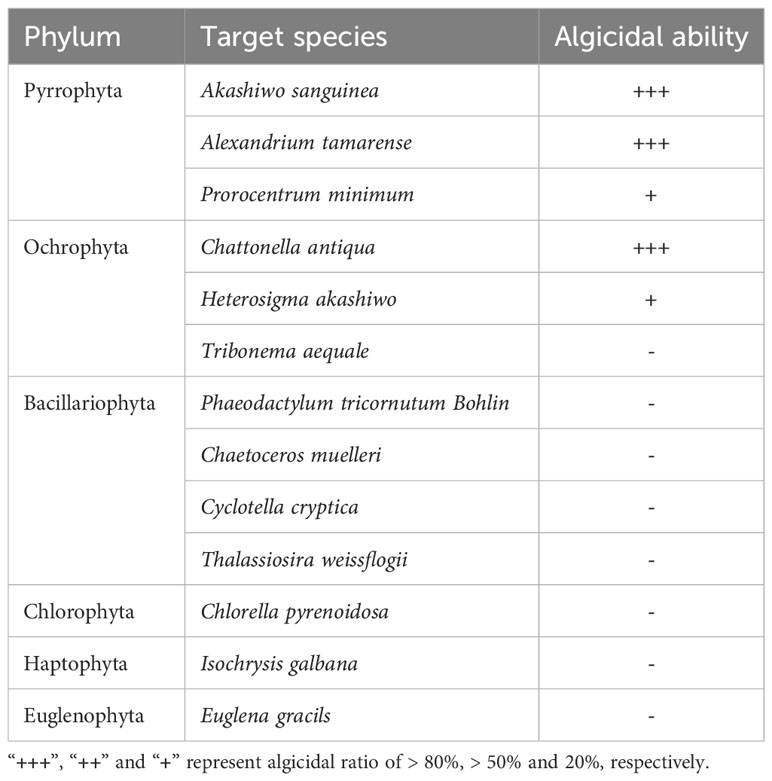
To check whether strain 6A1 exhibits effect on other algae in aquatic environment, we tested the algicidal performance on 13 different algae, which included typical HABs-forming species and beneficial algae: Akashiwo sanguinea, Alexandrium tamarense, Prorocentrum minimum, Chattonella antiqua, Heterosigma akashiwo, Tribonema aequale, Phaeodactylum tricornutum Bohlin, Chaetoceros muelleri, Cyclotella cryptica, Thalassiosira weissflogii, Chlorella pyrenoidosa, Isochrysis galbana, Euglena gracils (provided by Algal Culture Collection, Institute of Hydrobiology, Jinan University, Guangzhou, China), adding strain 6A1 into algal culture with the volume fraction of 10%.
2.8 Statistics
In this study, each experiment was carried out at least in triplicate and presented as mean ± standard deviation (SD) of three replicates. All data were evaluated using one-way analysis of variance (ANOVA) and paired t-test (SPSS 13.0 for Windows).
3 Results and discussion
3.1 Isolation and identification of strain 6A1
Strain 6A1 was a gram-negative, non-motile, rod-shaped bacterium with an average size of 1.70× 0.55 μm (Figure 1A). Strain 6A1 colonies on the agar plate were yellow, wet, and transparent with a flat edge (Figure 1B). Sequence analysis of the 16S rDNA gene showed that strain 6A1 was Arenibacter sp. (The sequence data were submitted to NCBI under accession number MG396982). In addition, the draft genome sequence of Arenibacter sp. strain 6A1 was reported in our previous study (Chen et al., 2020). Phylogenetic analysis based on the 16S rDNA sequences of strain 6A1 and other reported strains further confirmed that strain 6A1 belongs to the genus Arenibacter (Figure 1C).
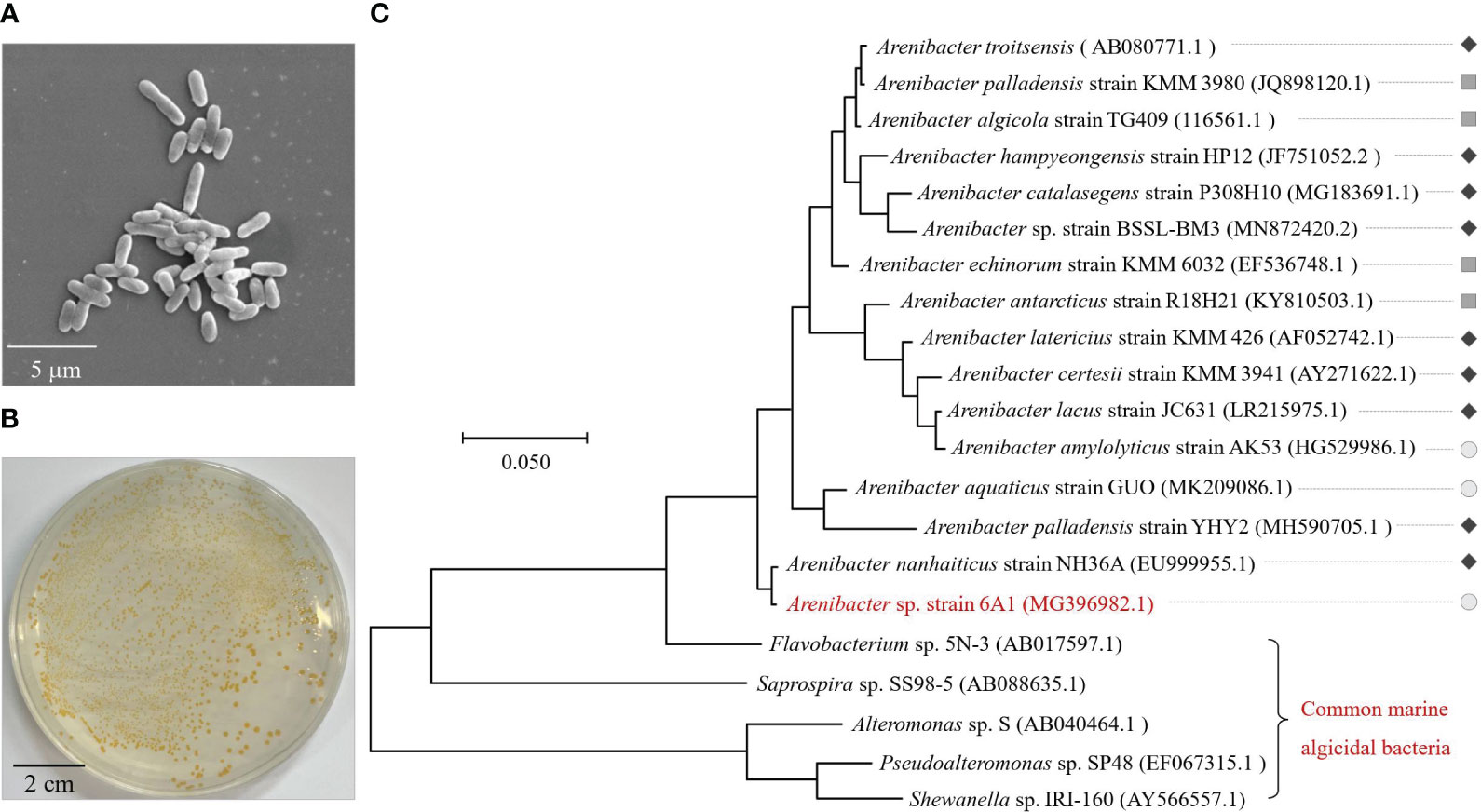
Figure 1 Morphology (A), colonial morphology (B), and phylogenetic identification (C) of strain 6A1. The phylogenetic tree is constructed based on bacterial 16S rDNA gene sequences of strain 6A1, other reported strains in genus Arenibacter, and common marine algicidal bacteria. The color bar is used to distinguish the habitat of strains in genus Arenibacter (“ ”, “
”, “ ” and “
” and “ ” represent strains isolated from sediment, marine organisms, and surface seawater, respectively). The diameter of the flat plate is 9 cm (B).
” represent strains isolated from sediment, marine organisms, and surface seawater, respectively). The diameter of the flat plate is 9 cm (B).
The genus Arenibacter is a highly taxonomic diverse group of microorganisms, which can participate in the degradation of natural polymers in marine environments depending on their niche and habitat adaptations (Bakunina et al., 2013). So far, most of the strains belonging to Arenibacter have been isolated from sediment (Figure 1C) (Nedashkovskaya et al., 2003; Sun et al., 2010; Ito et al., 2016; Li et al., 2018; Kumar et al., 2020; Park et al., 2021). Among these, Arenibacter TG409 has been proven to be able to degrade PAH (Gutierrez et al., 2014; Gutierrez et al., 2016). The capability of crab chitin hydrolyzation was also verified in Arenibacter palladensis YHY2 (Gurav et al., 2019). In addition, the antimicrobial activity of Arenibacter nanhaiticus sp. nov. NH36A was also investigated (Chen et al., 2013). However, no reports have documented that a bacterium belonging to Arenibacter could lyse algal cells. Most known algicidal bacteria phylogenetically fall into either the CFB group or the γ-Proteobacteria group and to the genera Alteromonas, Bacillus, Cellulophaga, Cytophaga, Flavobacterium, Micrococcus, Planomicrobium, Pseudoalteromonas, Pseudomonas, Saprospira, Vibrio, and Zobellia (Wang et al., 2020a). It is worth noting that this is the first evidence of an algicidal bacterium within the genus Arenibacter with algicidal ability to A. sanguinea. Remarkably, the results of comparative genomics indicated that the number of genes related to other glycan degradation in strain 6A1 was notably higher than in Arenibacter algicola, and Arenibacter latericius (Supplementary Figure 1), which were isolated from a laboratory culture of the marine diatom Skeletonema costatum and bottom sediments, respectively. Algae synthesize structurally complex glycans to build a protective barrier, also called the extracellular matrix (Bligh et al., 2022). A large amount of glycan degradation gene may also contribute to the strong algicidal capacity of strain 6A1. The algicidal activity of strain 6A1 might be related to its niche and habitat adaptations. This finding has enhanced the understanding of the potential function of genus Arenibacter, broadened the diversity of algicidal bacteria, and revealed the wide distribution of the algicidal microorganism in coastal environments.
3.2 Algicidal activity of Arenibacter sp. strain 6A1 on A. sanguinea
The concentration and the duration of bacterium treatment are generally seen as factors strongly related to the algicidal performance. The higher concentrations of bacterial culture added or the longer time treated, the greater algicidal performance was achieved (Guan et al., 2014). To evaluate the algicidal activity, an initial test was carried out to explore the optimal condition of 6A1 for lysing A. sanguinea cells. The results suggested that optimal algicidal conditions for 6A1 should be a volume ratio of 1.5% (bacterium/algae ratio) with a duration of 2 h (data not shown). Compared with Pseudoalteromonas S1, an algicidal bacterium reported in our previous study that can lyse A. sanguinea with a bacteria/algae ratio of 50:1 in 4 h (Sun et al., 2016), strain 6A1 showed a much stronger algicidal ability to remove A. sanguinea cells with lower dosage. In addition, strain 6A1 showed the strongest algicidal performance in a very short period of time in contrast to other algicidal bacteria listed in Supplementary Table 2, most of which lysis target species within several days. Therefore, strain 6A1 could be expected to become a powerful tool for emergency treatment of A. sanguinea bloom.
The growth curve of A. sanguinea was also inspected every 2 days to assess the algicidal ability of strain 6A1 against A. sanguinea from different growth stages as shown in Figure 2A. The algicidal efficiency of strain 6A1 to A. sanguinea from different growth stages (day 5, day 12, and day 20) was determined and compared. The algicidal ratio of strain 6A1 was 5.26% in 15 min and 82.53% in 120 min on day 5 (Figure 2B). Until day 12, the algicidal ratio was up to 23.89% in 15 min and 100% in 120 min, respectively. On day 20, the algicidal ratio was notably increased to 94% in 15 min (p < 0.01) (Figure 2B). The algicidal efficiency of 6A1 was significantly increased as the growth cycle of A. sanguinea progressed (p < 0.05). The resistance of A. sanguinea cell in the stationary stage was the weakest against strain 6A1 which suggested the secondary metabolites of the algal cell may participate in the algal-bacterial interaction through quorum sensing (Amin et al., 2015; Zhou et al., 2016; Chen et al., 2021), in turn enhancing the algicidal efficiency of strain 6A1. These results implied that strain 6A1 is suitable for eliminating A. sanguinea bloom, especially during the outbreak stage. Besides, the algicidal efficiency was largely related to treatment time so the algicidal ratio in 120 min was always higher than 15 min. Algal cells treated with a volume fraction of 1% strain 6A1 exhibited rounding in 15 min, then cell colors faded in 1 h, and finally cells were disrupted and cellular substances were decomposed and released in 2 h (Figure 2C). Marine phytoplankton species are able to survive when environmental variables exceed the tolerance range for vegetative cells by producing resting cysts and resuming vegetative growth in favorable conditions (Tang and Gobler, 2015; Rubino et al., 2017). A previous study has reported that bacterial strains promoted the formation of cyst-like structures in the K. brevis cultures (Roth et al., 2008). However, there are no resting cysts of A. sanguinea appeared during the algicidal process, which indicated that there was no potential threat for the resuming of A. sanguinea cells after algicidal treatment by strain 6A1.
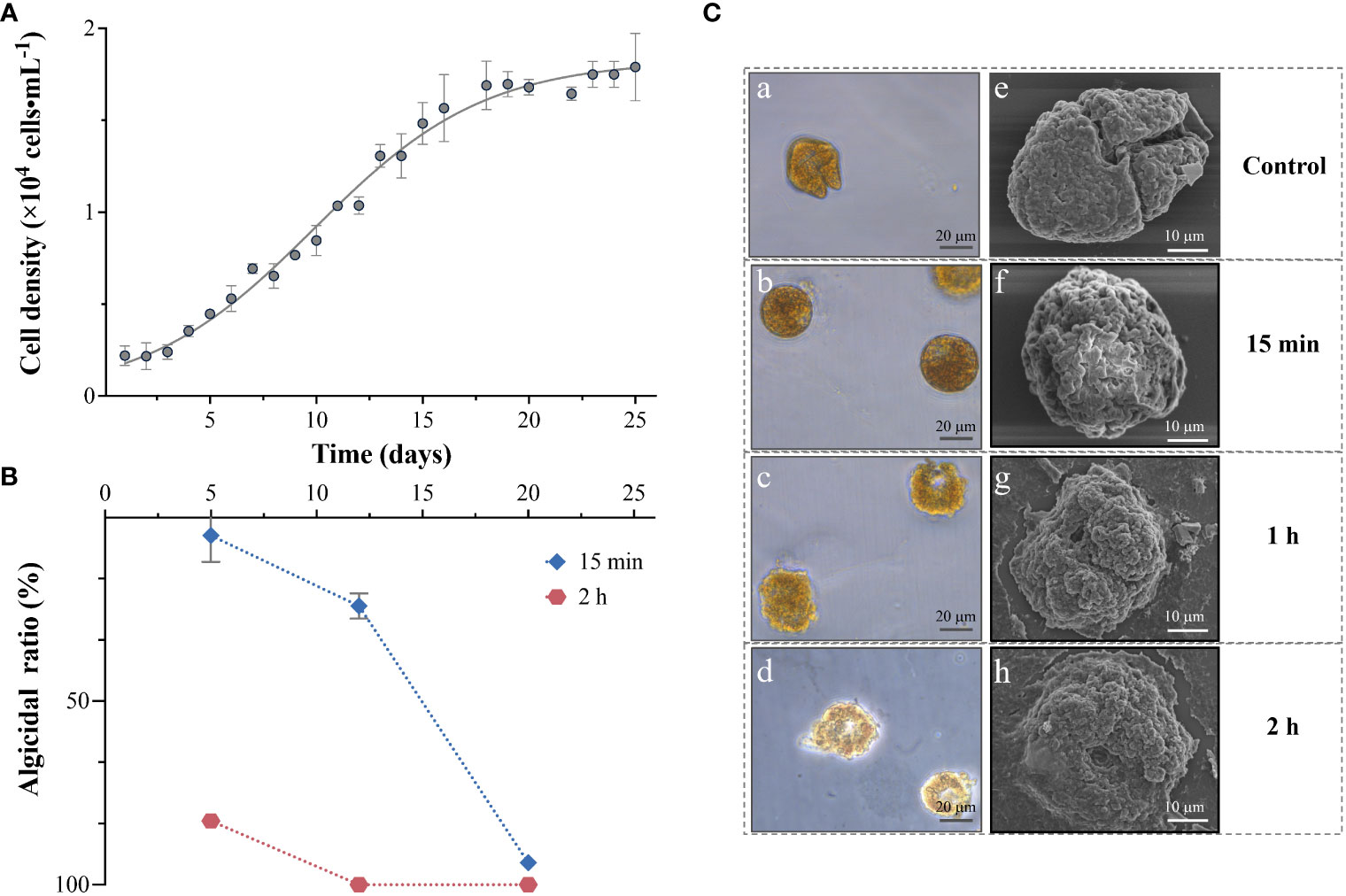
Figure 2 The growth kinetics (A), algicidal ratio (B), and cell lysis process (C) of Akashiwo sanguinea when exposed to strain 6A1. Results are displayed as mean ± SD, n = 3.
3.3 Algicidal characteristics of strain 6A1
According to the previous reports, two algicidal modes of bacteria (direct and indirect) are broadly proposed based on whether the algicidal action requires direct contact with the target algae (Wang et al., 2020a). So far, approximately 70% of strains exercised indirect algicidal properties, while the remaining 30% exercised direct algicidal properties (Demuez et al., 2015). In this study, the bacterial cultures were divided into four parts (whole culture, culture medium, supernatant, and pellet) for inoculation into the algal cultures from the logarithmic stage. The algicidal mode of Arenibacter sp. strain 6A1 was explored by evaluating the algicidal performance of each separated component. As it was indicated in Figure 3A, the whole culture and supernatant of strain 6A1 showed a similar algicidal performance with more than 95% algicidal ratio after being treated for 15 min, which revealed that strain 6A1 lysed A. sanguinea cells by releasing an active compound as an indirect attacker into the supernatant. However, the pellet and culture medium 2216E had no remarkable algicidal activity (Figure 3A), which means the contact between algal cells and bacterial cells was not necessary. Therefore, it was obvious that strain 6A1 is considered using primarily the algicidal mechanism of indirect attack. The negligible algicidal activity observed in bacterial cells might have originated from the compounds produced by the bacterial cells after they were inoculated into the algal cultures (Guan et al., 2014). Compared to bacterial cells, the algicidal compounds produced by bacteria should be more feasible in eliminating HABs in that the direct bacterial cell addition will increase the ecological burden (Ouyang et al., 2021). So, it is a prospective method to implement the immobilized strain 6A1 to control A. sanguinea bloom.
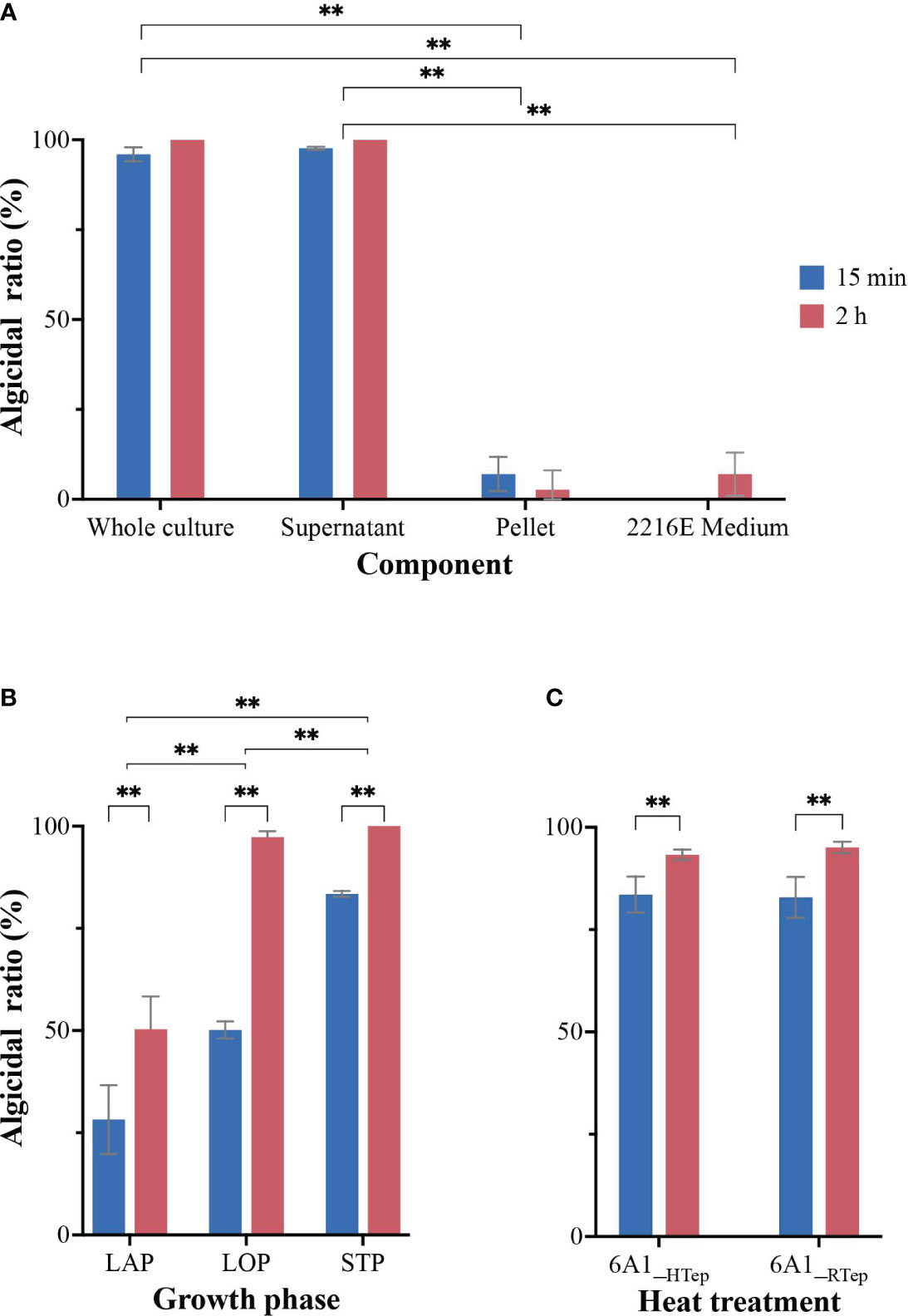
Figure 3 The algicidal performance of four components of 6A1 culture (A), the algicidal activity of 6A1 supernatant from different growth phases (B), and the algicidal ratio of 6A1 supernatant after high-temperature processing (C). LAP, lag phase; LOP, logarithmic stage; STP, stationary stage. 6A1_RTep represented 6A1 in room temperature; 6A1_HTep represented 6A1 after being treated under high temperature (100°C) for 2 h. Results are displayed as mean ± SD, n = 3. ** p < 0.05.
The growth curve of strain 6A1 culture was inspected every 2 h and the bacterial culture at distinct growth stages was harvested and added into the logarithmic growing algal culture with a volume fraction of 1.5% for 2 h, respectively. The results indicated that the algicidal activity of 6A1 culture from different growth stages varied significantly (p < 0.05) (Figure 3B). In the lag phase (10 h), the algicidal ratio of strain 6A1 was 28% in 15 min and 47% in 120 min. In the logarithmic phase (24 h), the algicidal ratio was dramatically increased to 50% in 15 min and 97% in 120 min. In the stationary phase (30 h), the algicidal activity was more effective and the algicidal ratio was 83% in 15 min and 100% in 120 min (Figure 3B). Thus, the greatest algicidal activity was achieved while strain 6A1 was in the stationary phase. Therefore, the algicidal performance of 6A1 was increased with the culture time, suggesting that the accumulation of certain secondary metabolites might benefit the algicidal activity.
Several studies worked on the investigation of algicidal compounds produced by marine bacteria, which are involved in the lysis of algal cells. There are a few already known algicidal compounds that have been proven to be effective in inhibiting the growth of marine harmful algae including prodigiosin (Pokrzywinski et al., 2012; Zhang et al., 2020), pyrrole-2-carboxylic acid (Ko et al., 2023), Nω-acetylhistamine (N-AcH) (Zhu et al., 2021a), hexahydro pyrrolo [1,2-a] pyrazine-1,4-dione (Wang et al., 2020b), questiomycins (Umetsu et al., 2019), and (2-iso butoxy phenyl) amine (An et al., 2015). However, there are still a number of substances produced by algicidal bacteria that have yet to be isolated and verified. In order to understand the characteristics of algicidal compounds generated by strain 6A1, we examined the thermal stability of the algicidal activity of strain 6A1. The strain 6A1 culture after being treated at 100°C for 30 min presented outstanding algicidal activity similar to the normal 6A1 culture (Figure 3C). These results demonstrated that the algicidal compounds secreted by Arenibacter sp. strain 6A1 are tolerant to heat, which implies that the algicidal compounds were not proteinaceous. The active substance produced by strain 6A1 with its strong algicidal activity is still under research. Our preliminary study found that pyrrole derivative was the main algicidal substance in the algicidal process of bacteria 6A1. However, the algicidal effect of the pyrrole derivative is not as good as that of algae-lysing bacteria 6A1 itself (data not shown), which suggests there may be other substances involved in the algicidal process of 6A1. The exact algicidal compounds need to be further purified and verified.
3.4 Comparison of algicidal performance of 6A1 to three typical HABs-forming species
In order to further explore the algicidal characteristics of 6A1, the resistance of three harmful algae (including A. sanguinea, C. antiqua, and A. tamarense) to 6A1 were investigated and compared.
3.4.1 Morphological and ultrastructure changes of tested algae
Morphological alterations of three algae cells during the algicidal process were observed by both optical microscope (Supplementary Figure 2) and SEM (Figure 4). Throughout the experiment, three tested algae exposed to f/2 medium showed normal morphological characteristics in the photograph. The morphological change varied among these three species after being treated with 6A1 culture. For A. sanguinea, cells treated with a volume fraction of 1% strain 6A1 exhibited rounding first, then cell colors faded, and finally cells were disrupted and cellular substances were decomposed and released in 1 h. For C. antiqua, cells treated with a volume fraction of 5% strain 6A1 drastically changed to ovoid after 15min, and the cellular components were aggregated afterward in 1 h. Finally, the cell colors faded and the cell component was released in 2 h. While A. tamarense cells treated with a volume fraction of 10% strain 6A1 exposed loss of armor in 15 min. The cells were collapsed and cellular substances were released in 2 h. These results illustrated that strain 6A1 apparently damaged the morphology of the three above-mentioned HABs-forming species and the optimal algicidal dosage for each species is subtly different. The armored A. tamarense cells showed the strongest tolerance to 6A1. Notably, there are no resting cysts of any species that appeared during the algicidal process.
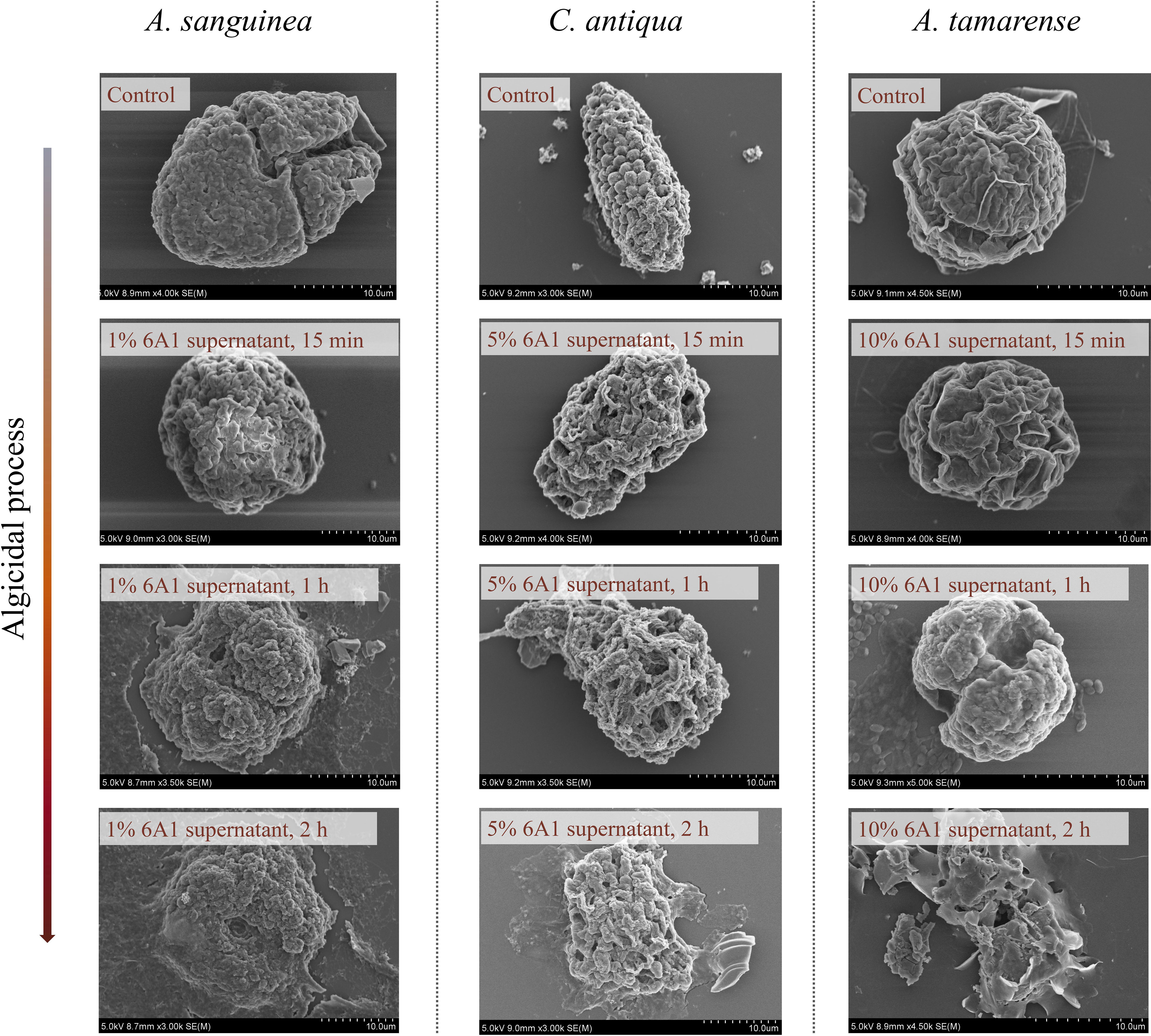
Figure 4 Changes in the subcellular ultrastructure of A. sanguinea, C. antiqua, and A. tamarense after exposure to 6A1, observed by scanning electron microscope (SEM). The algal culture without 6A1 addition was used as the control. (Bar = 10 μm).
3.4.2 The photosynthetic damage and oxidative stress of tested algae induced by 6A1
In the present study, different concentration of strains 6A1 supernatant was added to three tested algae to evaluate the algicidal performance. The results indicated that 1% (v: v) supernatant could inhibit more than 50% of the growth of A. sanguinea cells after 2-h exposure, and 1.5% (v: v) supernatant had approximately 90% algicidal ratio. While the C. antiqua cells density decreased dramatically in the first 2 h by adding 2.5% (v: v) supernatant and more than 95% of C. antiqua cells were eliminated after 4 h. A. tamarense also obviously declined after being treated with 10% (v: v) supernatant and 95% A. tamarense cells have been lysed for 2 h (Supplementary Figures 3A–C). As described above, A. sanguinea was the most sensitive to strain 6A1, followed by C. antiqua and finally A. tamarense. Therefore, strain 6A1 showed selective algicidal capability and exhibited significant dosage-dependent and species-specific algicidal performance. Photosynthesis is vital to the photoautotrophic algal lifestyle. Marine bacteria with indirect algicidal activity played an important role in the changes during the photosynthetic process in algal cells (Yang et al., 2014; Zhang et al., 2014; Hou et al., 2016; Tan et al., 2016). The Fv/Fm values of three tested algae cells decreased significantly with time after exposure to strain 6A1 supernatant (Supplementary Figures 3D–F). The rate of decline in Fv/Fm value varied among the tested algae. The Fv/Fm values of A. sanguinea dropped to zero within 2 h when treated with 1.5% (v: v) 6A1 supernatant. However, The Fv/Fm values of C. antiqua were down to zero after being treated with 2.5% (v: v) 6A1 supernatant for 4 h. Significantly, A. tamarense was the least sensitive against 6A1 so that the Fv/Fm values fall to zero until 8 h even treated by 10% (v: v) 6A1 supernatant. The different response in Fv/Fm values of algae cells to 6A1 supernatant suggested that strain 6A1 showed selectivity in terms of the algicidal performance on three HABs-forming species.
To further confirm the oxidative stress induced by 6A1, we also investigated SOD activity, MDA content, CAT activity, and ROS production of three tested algae after exposure to 6A1. Both A. sanguinea and A. tamarense cells may suffer from oxidative damage, reflected by a significant increase in CAT activity (Supplementary Figures 4A, C). However, the C. antiqua cells may still suffer from oxidative damage, reflected by a significant increase in MDA content (Supplementary Figure 4B). In addition, a significant increase of ROS was detected in A. sanguinea and A. tamarense after being treated with strain 6A1 in 2 h, while the ROS accumulated in C. antiqua cells was slightly inconspicuous (Figure 5). Besides, the ROS in A. sanguinea cells was dramatically increased with the raising of 6A1 concentration (p < 0.05), and similar results were not found in C. antiqua and A. tamarense (Figure 5), which displayed the A. sanguinea cells was the most sensitive target against strain 6A1 among the tested algae. MDA is a product of lipid peroxidation, which represents the degree of damage to the cell membrane under environmental stress. The role of CAT is to promote the decomposition of hydrogen peroxide into molecular oxygen and water, thus protecting cells from H2O2 poisoning. Intracellular ROS accumulation induced protoplast shrinkage and DNA fragmentation prior to cell death (Vardi et al., 1999; Sheyn et al., 2016). The ROS levels are important parameters that can indicate the oxidative damage of the cellular components (Zhang et al., 2014). The CAT, MDA, and ROS changes, along with photosynthetic damage, indicate significant oxidative stress and cell damage, which is likely the mechanism underlying the lysis of 6A1 to algae cells. Besides, the algicidal activity of strain 6A1 on HABs-forming algae has strong selectivity.
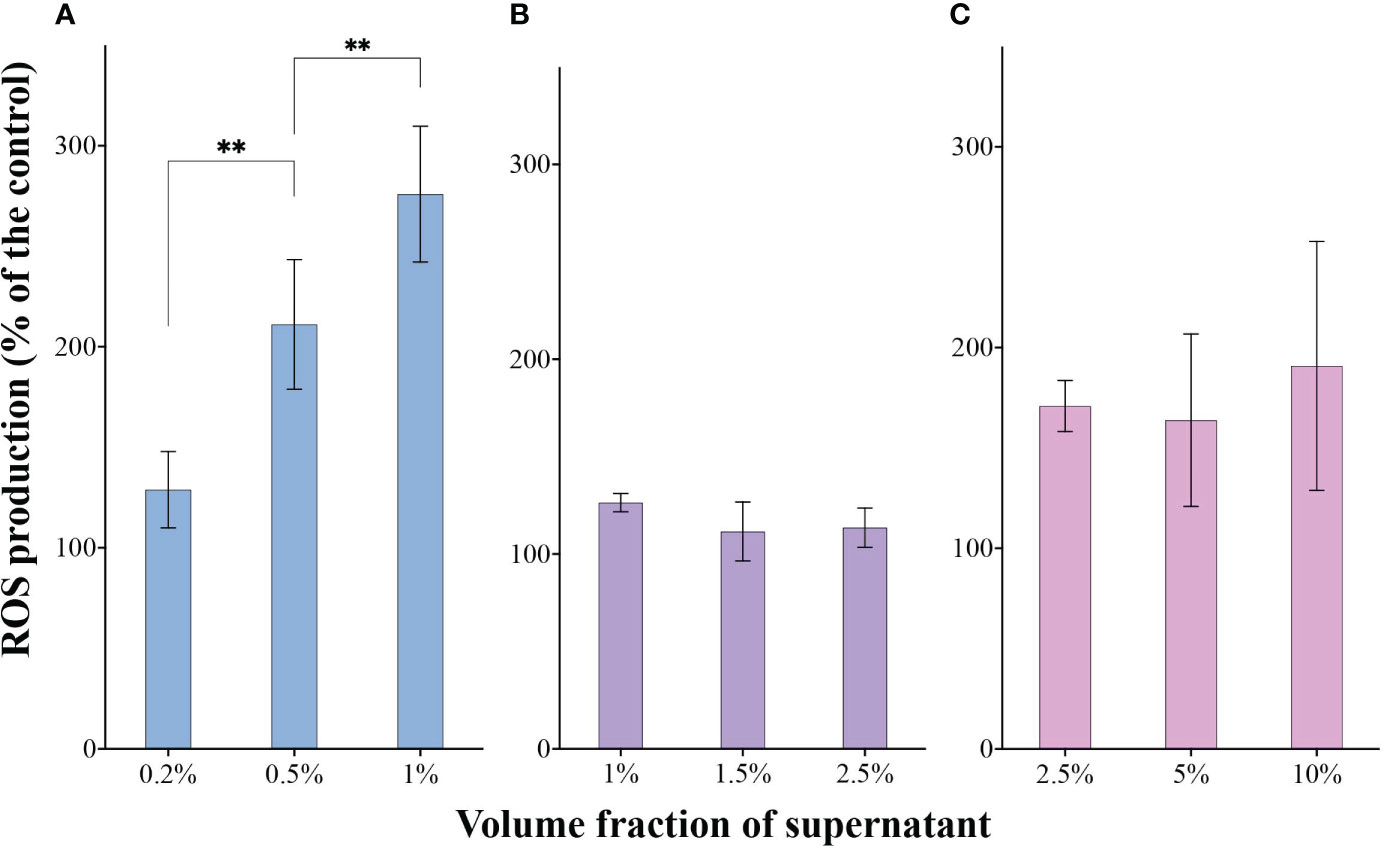
Figure 5 The ROS production in A. sanguinea (A), C. antiqua (B), and A. tamarense (C) cells after being exposed to different volume fractions of strain 6A1 supernatant for 2 h. The ROS level of tested algae with the addition of 2216E medium was used as a control. Results are displayed as mean ± SD, n = 3. ** p < 0.05.
3.5 Algicidal spectrum of Arenibacter sp. strain 6A1
Algicidal bacteria are highly species-specific in controlling harmful algae blooms in many cases, however, certain species have a wide target (Wang et al., 2020a). Whether strain 6A1 was an excellent algicide against a wide range of algae or not is still unknown. In the present study, strain 6A1 showed algicidal activity against 5 algae cells among the 13 tested algae, A. sanguinea, A. tamarense, P. minimum, C. antiqua and H. akashiwo (Table 1). The results strongly suggested that strain 6A1 primarily targets Pyrrophyta and Ochrophyta and had no lytic activity toward Bacillariophyta, Chlorophyta, Haptophyta, and Euglenophyta. This result indicated that the application of strain 6A1 to HABs control should be feasible because it would not influence the growth of other microalgae.
4 Conclusion
In this study, a novel potent algicidal bacterium Arenibacter sp. strain 6A1 has been isolated and identified. The algicidal performance of Arenibacter sp. strain 6A1 relates to the growth stage of both A. sanguinea and Arenibacter sp. strain 6A1. The indirect algicidal mode of Arenibacter sp. strain 6A1 is verified and the algicidal compounds have excellent thermal stability. Arenibacter sp. strain 6A1 has been proven to be useful for removing other HABs-forming algae. The most sensitive species against Arenibacter sp. strain 6A1 is A. sanguinea, followed by C. antiqua and A. tamarense in order. In addition, 6A1 exhibited selective algicidal activity on a wide range of algal species and had no lytic activity toward beneficial microalgae. This study provides an important insight into the further utilization of algicidal bacteria in controlling HABs. Nevertheless, further studies, regarding the algicidal mechanism of 6A1 on different HABs-forming algae, are needed.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, MG396982.
Author contributions
SW: formal analysis, writing-original draft. JT: investigation, methodology, and formal analysis. JC: investigation and methodology. MC: investigation and methodology. LW: conceptualization. SL: conceptualization. ZH: conceptualization and supervision. HC: writing-review & editing, conceptualization, project administration, funding acquisition, and supervision. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Funding for this study was provided by Sustainable Development Special Project of Shenzhen Science and Technology Innovation Committee (KCXFZ20230731093402005) to Dr. HC, the National Natural Science Foundation of China (32273118), The Science and Technology Program of Guangdong Province of China (2020B121202014), Shenzhen Special Fund for Sustainable Development (KCXFZ20211020164013021) and Shenzhen University 2035 Program for Excellent Research (2022B010) to Dr. ZH, Shenzhen Science and Technology Program (KCXST20221021111206015 and KCXFZ20201221173404012) to Dr. SL, China Postdoctoral Science Foundation (2021M692195) to Dr. SW.
Acknowledgments
We thank Instrumental Analysis Center of Shenzhen University for the assistance with the High Resolution Scanning Electron Microscope (Thermo APREO S(A5-112)).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1287998/full#supplementary-material
References
Amin S. A., Hmelo L. R., van Tol H. M., Durham B. P., Carlson L. T., Heal K. R., et al. (2015). Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522 (7554), 98–101. doi: 10.1038/nature14488
An X., Zhang B., Zhang H., Li Y., Zheng W., Yu Z., et al. (2015). Discovery of an algicidal compound from Brevibacterium sp. BS01 and its effect on a harmful algal bloom-causing species, Alexandrium tamarense. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.01235
Anderson D. M. (2009). Approaches to monitoring, control and management of harmful algal blooms (HABs). Ocean Coast. Manage. 52 (7), 342–347. doi: 10.1016/j.ocecoaman.2009.04.006
Anderson D. M., Cembella A. D., Hallegraeff G. M. (2012). Progress in understanding harmful algal blooms: paradigm shifts and new technologies for research, monitoring, and management. Ann. Rev. Mar. Sci. 4, 143–176. doi: 10.1146/annurev-marine-120308-081121
Bakunina I., Nedashkovskaya O., Balabanova L., Zvyagintseva T., Rasskasov V., Mikhailov V. (2013). Comparative analysis of glycoside hydrolases activities from phylogenetically diverse marine bacteria of the genus Arenibacter. Mar. Drugs 11 (6), 1977–1998. doi: 10.3390/md11061977
Berdalet E., Fleming L. E., Gowen R., Davidson K., Hess P., Backer L. C., et al. (2015). Marine harmful algal blooms, human health and wellbeing: challenges and opportunities in the 21st century. J. Mar. Biol. Assoc. 96 (1), 61–91. doi: 10.1017/S0025315415001733
Bligh M., Nguyen N., Buck-Wiese H., Vidal-Melgosa S., Hehemann J.-H. (2022). Structures and functions of algal glycans shape their capacity to sequester carbon in the ocean. Curr. Opin. Chem. Biol. 71, 102204. doi: 10.1016/j.cbpa.2022.102204
Brown A. R., Lilley M., Shutler J., Lowe C., Artioli Y., Torres R., et al. (2019). Assessing risks and mitigating impacts of harmful algal blooms on mariculture and marine fisheries. Rev. Aquaculture. 12, 1663–1688. doi: 10.1111/raq.12403
Chen H., Jiang F., Li S., Hu Z. (2021). Temporal variability of free-living microbial culturability and community composition after an Akashiwo sanguinea bloom in Shenzhen, China. Ecotoxicology 30 (5), 975–985. doi: 10.1007/s10646-021-02407-4
Chen J., Tong J., Li S., Hu Z., Wang L., Chen H. (2020). Draft Genome Sequence of an Algicidal Bacterium, Arenibacter sp. strain 6A1, Isolated from Seawater during an Akashiwo sanguinea Bloom in Shenzhen, China. Microbiol. Resour. Announc 9 (50). doi: 10.1128/MRA.00964-20
Chen Y., Tang J., Tang X., Wang C., Lian Y., Shao Z., et al. (2013). New phenethylamine derivatives from Arenibacter nanhaiticus sp. nov. NH36A(T) and their antimicrobial activity. J. Antibiot. (Tokyo) 66 (11), 655–661. doi: 10.1038/ja.2013.65
Demuez M., Gonzalez-Fernandez C., Ballesteros M. (2015). Algicidal microorganisms and secreted algicides: New tools to induce microalgal cell disruption. Biotechnol. Adv. 33 (8), 1615–1625. doi: 10.1016/j.bioteChadv.2015.08.003
Du X., Peterson W., McCulloch A., Liu G. (2011). An unusual bloom of the dinoflagellate Akashiwo sanguinea off the central Oregon, USA, coast in autumn 2009. Harmful Algae 10 (6), 784–793. doi: 10.1016/j.hal.2011.06.011
Froelich B. A., Noble R. T. (2016). Vibrio bacteria in raw oysters: managing risks to human health. Philos. Trans. R. Soc Lond. B. Biol. Sci. 371 (1689), 20150209. doi: 10.1098/rstb.2015.0209
Gallardo-Rodríguez J. J., Astuya-Villalón A., Llanos-Rivera A., Avello-Fontalba V., Ulloa-Jofré V. (2019). A critical review on control methods for harmful algal blooms. Rev. Aquaculture 11 (3), 661–684. doi: 10.1111/raq.12251
Griffith A. W., Gobler C. J. (2020). Harmful algal blooms: A climate change co-stressor in marine and freshwater ecosystems. Harmful Algae 91, 101590. doi: 10.1016/j.hal.2019.03.008
Guan C., Guo X., Cai G., Zhang H., Li Y., Zheng W., et al. (2014). Novel algicidal evidence of a bacterium Bacillus sp. LP-10 killing Phaeocystis globosa, a harmful algal bloom causing species. Biol. Control 76, 79–86. doi: 10.1016/j.biocontrol.2014.05.007
Gurav R., Bhatia S. K., Moon Y.-M., Choi T.-R., Jung H.-R., Yang S.-Y., et al. (2019). One-pot exploitation of chitin biomass for simultaneous production of electricity, n-acetylglucosamine and polyhydroxyalkanoates in microbial fuel cell using novel marine bacterium Arenibacter palladensis YHY2. J. Cleaner Production 209, 324–332. doi: 10.1016/j.jclepro.2018.10.252
Gutierrez T., Rhodes G., Mishamandani S., Berry D., Whitman W. B., Nichols P. D., et al. (2014). Polycyclic aromatic hydrocarbon degradation of phytoplankton-associated Arenibacter spp. and description of Arenibacter algicola sp. nov., an aromatic hydrocarbon-degrading bacterium. Appl. Environ. Microbiol. 80 (2), 618–628. doi: 10.1128/AEM.03104-13
Gutierrez T., Whitman W. B., Huntemann M., Copeland A., Chen A., Kyrpides N., et al. (2016). Genome sequence of Arenibacter algicola strain TG409, a hydrocarbon-degrading bacterium associated with marine eukaryotic phytoplankton. Genome Announc 4 (4). doi: 10.1128/genomeA.00765-16
Hallegraeff G. M. (2010). Ocean climate change, phytoplankton community responses, and harmful algal blooms: a formidable predictive challenge. J. Phycology 46 (2), 220–235. doi: 10.1111/j.1529-8817.2010.00815.x
Harvell C. K., Burkholder J. M., Colwell R. R., Epstein P. R., Grimes D. J., Hofmann E. E., et al. (1999). Review: marine ecology - emerging marine diseases - climate links and anthropogenic factors. Science 285 (5433), 1505–1510. doi: 10.1126/science.285.5433.1505
Hou S., Shu W., Tan S., Zhao L., Yin P. (2016). Exploration of the antioxidant system and photosynthetic system of a marine algicidal Bacillus and its effect on four harmful algal bloom species. Can. J. Microbiol. 62 (1), 49–59. doi: 10.1139/cjm-2015-0425
Ito K., Nakajima N., Yamamura S., Tomita M., Suzuki H., Amachi S. (2016). Draft genome sequence of Arenibacter sp. strain C-21, an iodine-accumulating bacterium isolated from surface marine sediment. Genome Announc 4 (5). doi: 10.1128/genomeA.01155-16
Jiang W., Yu Z., Cao X., Jiang K., Yuan Y., Anderson D. M., et al. (2021). Effects of soluble organics on the settling rate of modified clay and development of improved clay formulations for harmful algal bloom control. Environ. pollut. 289, 117964. doi: 10.1016/j.envpol.2021.117964
Jones T., Parrish J. K., Punt A. E., Trainer V. L., Kudela R., Lang J., et al. (2017). Mass mortality of marine birds in the Northeast Pacific caused by Akashiwo sanguinea. Mar. Ecol. Prog. Ser. 579, 111–127. doi: 10.3354/meps12253
Kim H. (2006). Mitigation and controls of HABs. In: Edna G., Turner J. T. (Eds.), Ecology of Harmful Algae (Berlin: Springer-Verlag), 327–338.
Kim J. D., Kim J. Y., Park J. K., Lee C. G. (2009a). Selective control of the Prorocentrum minimum harmful algal blooms by a novel algal-lytic bacterium Pseudoalteromonas haloplanktis AFMB-008041. Mar. Biotechnol. (NY) 11 (4), 463–472. doi: 10.1007/s10126-008-9167-9
Kim Y. S., Lee D.-S., Jeong S.-Y., Lee W. J. L., M.-S. (2009b). Isolation and characterization of a marine algicidal bacterium against the harmful raphidophyceae Chattonella marina. J. Microbiol. 47 (1), 9–18. doi: 10.1007/s12275-008-0141-z
Kim Y. S., Son H. J., Jeong S. Y. (2015). Isolation of an algicide from a marine bacterium and its effects against the toxic dinoflagellate Alexandrium catenella and other harmful algal bloom species. J. Microbiol. 53 (8), 511–517. doi: 10.1007/s12275-015-5303-1
Ko S.-R., Le V. V., Srivastava A., Kang M., Oh H.-M., Ahn C.-Y. (2023). Algicidal activity of a novel bacterium, Qipengyuania sp. 3-20A1M, against harmful Margalefidinium polykrikoides: effects of its active compound. Mar. pollut. Bull. 186, 114397. doi: 10.1016/j.marpolbul.2022.114397
Kumar D., Smita N., Kumar G., Suresh G., Jagadeeshwari U., Sasikala C., et al. (2020). Arenibacter lacus sp. nov., Isolated from Chilika Lagoon, India. Curr. Microbiol. 77 (12), 4152–4159. doi: 10.1007/s00284-020-02205-x
Lewitus A. J., Horner R. A., Caron D. A., Garcia-Mendoza E., Hickey B. M., Hunter M., et al. (2012). Harmful algal blooms along the north American west coast region: history, trends, causes, and impacts. Harmful Algae 19, 133–159. doi: 10.1016/j.hal.2012.06.009
Li A. Z., Lin L. Z., Zhang M. X., Lv Y., Zhu H. H. (2018). Arenibacter catalasegens sp. nov., isolated from marine surface sediment, and emended description of the genus Arenibacter. Int. J. Syst. Evol. Microbiol. 68 (3), 758–763. doi: 10.1099/ijsem.0.002576
Li Y., Zhu H., Lei X., Zhang H., Guan C., Chen Z., et al. (2015). The first evidence of deinoxanthin from Deinococcus sp. Y35 with strong algicidal performance on the toxic dinoflagellate Alexandrium tamarense. J. Hazard. Mater. 290, 87–95. doi: 10.1016/j.jhazmat.2015.02.070
Lu X., Zhou B., Xu L., Liu L., Wang G., Liu X., et al. (2016). A marine algicidal Thalassospira and its active substance against the harmful algal bloom species Karenia mikimotoi. Appl. Microbiol. Biotechnol. 100 (11), 5131–5139. doi: 10.1007/s00253-016-7352-8
Mayali X., Azam F. (2004). Algicidal bacteria in the sea and their impact on algal blooms. J. Eukaryotic Microbiol. 51 (2), 139–144. doi: 10.1111/j.1550-7408.2004.tb00538.x
Nedashkovskaya O. I., Suzuki M., Vysotskii M. V., Mikhailov V. V. (2003). Arenibacter troitsensis sp. nov., isolated from marine bottom sediment. Int. J. Syst. Evol. Microbiol. 53 (Pt 5), 1287–1290. doi: 10.1099/ijs.0.02384-0
Ouyang L., Liu Y., Chen H., Zaynab M., Yang X., Wang S., et al. (2021). Encapsulation and algicidal properties of fermentation products from Vibrio brasiliensis H115. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.676913
Pal M., Yesankar P. J., Dwivedi A., Qureshi A. (2020). Biotic control of harmful algal blooms (HABs): A brief review. J. Environ. Manage. 268, 110687. doi: 10.1016/j.jenvman.2020.110687
Park J., Church J., Son Y., Kim K.-T., Lee W. H. (2017). Recent advances in ultrasonic treatment: challenges and field applications for controlling harmful algal blooms (HABs). Ultrasonics sonochemistry 38, 326–334. doi: 10.1016/j.ultsonch.2017.03.003
Park S., Lee S. Y., Lee J.-S., Kim W., Yoon J.-H. (2021). Arenibacter arenosicollis sp. nov., isolated from a sand dune. Int. J. Systematic Evolutionary Microbiol. 71 (3), 004709. doi: 10.1099/ijsem.0.004709
Pokrzywinski K. L., Place A. R., Warner M. E., Coyne K. J. (2012). Investigation of the algicidal exudate produced by Shewanella sp. IRI-160 and its effect on dinoflagellates. Harmful Algae 19, 23–29. doi: 10.1016/j.hal.2012.05.002
Ren N., Xing D., Rittmann B. E., Zhao L., Xie T., Zhao X. (2007). Microbial community structure of ethanol type fermentation in bio-hydrogen production. Environ. Microbiol. 9 (5), 1112–1125. doi: 10.1111/j.1462-2920.2006.01234.x
Rizzatti G., Lopetuso L. R., Gibiino G., Binda C., Gasbarrini A. (2017). Proteobacteria: A common factor in human diseases. BioMed. Res. Int. 2017, 9351507. doi: 10.1155/2017/9351507
Roth P. B., Twiner M. J., Mikulski C. M., Barnhorst A. B., Doucette G. J. (2008). Comparative analysis of two algicidal bacteria active against the red tide dinoflagellate Karenia brevis. Harmful Algae 7 (5), 682–691. doi: 10.1016/j.hal.2008.02.002
Rubino F., Belmonte M., Galil B. S. (2017). Plankton resting stages in recent sediments of Haifa port, Israel (Eastern Mediterranean) - Distribution, viability and potential environmental consequences. Mar. pollut. Bull. 116 (1-2), 258–269. doi: 10.1016/j.marpolbul.2016.12.078
Sellner K. G., Rensel J. (2018). Prevention, control, and mitigation of harmful algal bloom impacts on fish, shellfish, and human consumers. In Harmful Algal Blooms (eds Shumway S. E., Burkholder J. M., Morton S. L.) 435–492. doi: 10.1002/9781118994672.ch12
Sheyn U., Rosenwasser S., Ben-Dor S., Porat Z., Vardi A. (2016). Modulation of host ROS metabolism is essential for viral infection of a bloom-forming coccolithophore in the ocean. ISME J. 10 (7), 1742–1754. doi: 10.1038/ismej.2015.228
Shi R., Huang H., Qi Z., Hu W., Tian Z., Dai M. (2013). Algicidal activity against Skeletonema costatum by marine bacteria isolated from a high frequency harmful algal blooms area in southern Chinese coast. World J. Microbiol. Biotechnol. 29 (1), 153–162. doi: 10.1007/s11274-012-1168-1
Shi X., Liu L., Li Y., Xiao Y., Ding G., Lin S., et al. (2018). Isolation of an algicidal bacterium and its effects against the harmful-algal- bloom dinoflagellate Prorocentrum donghaiense (Dinophyceae). Harmful Algae 80, 72–79. doi: 10.1016/j.hal.2018.09.003
Sun F., Wang B., Du Y., Liu X., Lai Q., Li G., et al. (2010). Arenibacter nanhaiticus sp. nov., isolated from marine sediment of the South China Sea. Int. J. Syst. Evol. Microbiol. 60 (Pt 1), 78–83. doi: 10.1099/ijs.0.008573-0
Sun H.-Y., Zhang Y., Chen H.-R., Hu C.-X., Li H., Hu Z.-L. (2016). Isolation and characterization of the marine algicidal bacterium Pseudoalteromonas S1 against the harmful alga Akashiwo sanguinea. Mar. Biol. 163 (3). doi: 10.1007/s00227-016-2836-8
Sun R., Sun P., Zhang J., Esquivel-Elizondo S., Wu Y. (2018). Microorganisms-based methods for harmful algal blooms control: a review. Bioresour. Technol 248, 12–20. doi: 10.1016/j.biortech.2017.07.175
Tan S., Hu X., Yin P., Zhao L. (2016). Photosynthetic inhibition and oxidative stress to the toxic Phaeocystis globosa caused by a diketopiperazine isolated from products of algicidal bacterium metabolism. J. Microbiol. 54 (5), 364–375. doi: 10.1007/s12275-016-6012-0
Tang Y. Z., Gobler C. J. (2015). Sexual resting cyst production by the dinoflagellate Akashiwo sanguinea: a potential mechanism contributing to the ubiquitous distribution of a harmful alga. J. Phycol. 51 (2), 298–309. doi: 10.1111/jpy.12274
Umetsu S., Kanda M., Imai I., Sakai R., Fujita M. J. (2019). Questiomycins, algicidal compounds produced by the marine bacterium Alteromonas sp. D and their production cue. Molecules 24 (24). doi: 10.3390/molecules24244522
Van Dolah F. M. (2000). Marine algal toxins origins, health effects, and their increased occurrence. Environ. Health Perspect. 108, 133–141. doi: 10.1289/ehp.00108s1133
Vardi A., Berman-Frank I., Rozenberg T., Hadas O., Kaplan A., Levine A. (1999). Programmed cell death of the dinoflagellate Peridinium gatunense is mediated by CO2 limitation and oxidative stress. Curr. Biol. 9, 1061–1064. doi: 10.1016/S0960-9822(99)80459-X
Wang M., Chen S., Zhou W., Yuan W., Wang D. (2020a). Algal cell lysis by bacteria: A review and comparison to conventional methods. Algal Res. 46. doi: 10.1016/j.algal.2020.101794
Wang X., Gong L., Liang S., Han X., Zhu C., Li Y. (2005). Algicidal activity of rhamnolipid biosurfactants produced by Pseudomonas aeruginosa. Harmful algae 4 (2), 433–443. doi: 10.1016/j.hal.2004.06.001
Wang Y., Li S., Liu G., Li X., Yang Q., Xu Y., et al. (2020b). Continuous production of algicidal compounds against Akashiwo sanguinea via a Vibrio sp. co-culture. Bioresour. Technol. 295, 122246. doi: 10.1016/j.biortech.2019.122246
Wang B., Yang X., Lu J., Zhou Y., Su J., Tian Y., et al. (2012). A marine bacterium producing protein with algicidal activity against Alexandrium tamarense. Harmful Algae 13, 83–88. doi: 10.1016/j.hal.2011.10.006
West P. A. (1989). The human pathogenic vibrios–a public health update with environmental perspectives. Epidemiol. Infect. 103 (1), 1–34. doi: 10.1017/s0950268800030326
White A. E., Watkins-Brandt K. S., McKibben S. M., Wood A. M., Hunter M., Forster Z., et al. (2014). Large-scale bloom of Akashiwo sanguinea in the northern California current system in 2009. Harmful Algae 37, 38–46. doi: 10.1016/j.hal.2014.05.004
Yang X., Li X., Zhou Y., Zheng W., Yu C., Zheng T. (2014). Novel insights into the algicidal bacterium DH77-1 killing the toxic dinoflagellate Alexandrium tamarense. Sci. Total Environ. 482-483, 116–124. doi: 10.1016/j.scitotenv.2014.02.125
Yu Z., Song X., Cao X., Liu Y. (2018). “Mitigation and control of harmful algal blooms,” in Global Ecology and Oceanography of Harmful Algal Blooms. Eds. Glibert P. M., Berdalet E., Burford M. A., Pitcher G. C., Zhou M. (Cham: Springer International Publishing), 403–423. doi: 10.1007/978-3-319-70069-4_21
Zhang F., Fan Y., Zhang D., Chen S., Bai X., Ma X., et al. (2020). Effect and mechanism of the algicidal bacterium Sulfitobacter porphyrae ZFX1 on the mitigation of harmful algal blooms caused by Prorocentrum donghaiense. Environ. pollut. 263 (Pt A), 114475. doi: 10.1016/j.envpol.2020.114475
Zhang H., Lv J., Peng Y., Zhang S., An X., Xu H., et al. (2014). Cell death in a harmful algal bloom causing species Alexandrium tamarense upon an algicidal bacterium induction. Appl. Microbiol. Biotechnol. 98 (18), 7949–7958. doi: 10.1007/s00253-014-5886-1
Zhang F., Ye Q., Chen Q., Yang K., Zhang D., Chen Z., et al. (2018). Algicidal activity of novel marine bacterium Paracoccus sp. Strain Y42 against a Harmful Algal-Bloom-Causing Dinoflagellate, Prorocentrum donghaiense. Appl. Environ. Microbiol. 84 (19). doi: 10.1128/AEM.01015-18
Zhou J., Lyu Y., Richlen M., Anderson D. M., Cai Z. (2016). Quorum sensing is a language of chemical signals and plays an ecological role in algal-bacterial interactions. CRC Crit. Rev. Plant Sci. 35 (2), 81–105. doi: 10.1080/07352689.2016.1172461
Zhu X., Dao G., Tao Y., Zhan X., Hu H. (2021b). A review on control of harmful algal blooms by plant-derived allelochemicals. J. Hazard. Mater. 401, 123403. doi: 10.1016/j.jhazmat.2020.123403
Keywords: Arenibacter sp. strain 6A1, Akashiwo sanguinea, algicidal bacteria, algicidal activity, harmful algal blooms (HABs)
Citation: Wu S, Tong J, Chen J, Chen M, Wang L, Li S, Hu Z and Chen H (2024) Characterization of a novel algicidal bacteria Arenibacter sp. strain 6A1 and its application to eliminate harmful algal blooms. Front. Mar. Sci. 10:1287998. doi: 10.3389/fmars.2023.1287998
Received: 03 September 2023; Accepted: 28 December 2023;
Published: 31 January 2024.
Edited by:
Jinhyun Kim, Korea Polar Research Institute, Republic of KoreaReviewed by:
Wanchun Guan, Wenzhou Medical University, ChinaChuner Cai, Shanghai Ocean University, China
Jin Zhou, Tsinghua University, China
Copyright © 2024 Wu, Tong, Chen, Chen, Wang, Li, Hu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huirong Chen, Y2hlbmhyQHN6dS5lZHUuY24=
 Sha Wu
Sha Wu Jing Tong1
Jing Tong1 Liyan Wang
Liyan Wang Zhangli Hu
Zhangli Hu Huirong Chen
Huirong Chen