- 1State Key Laboratory of Mycology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China
- 2College of Life Sciences, University of Chinese Academy of Sciences, Beijing, China
- 3Department of Biological Sciences & Biotechnology, Faculty of Science & Technology, Universiti Kebangsaan Malaysia, Selangor, Malaysia
- 4Engineering Laboratory of Microbial Breeding and Preservation of Hebei Province, School of Life Sciences, Institute of Life Sciences and Green Development, Hebei University, Baoding, China
China has the second greatest extent of intertidal zones in the world. The intertidal zone is the most dynamic environments in the biosphere and potentially supports high biodiversity. Marine yeasts show excellent performance in various industrial, environmental and medical applications, however, the marine yeast diversity has rarely been studied in China. In this study, we collected 1241 samples including marine sediments, marine water, plants, and benthos at 161 GPS sites in different types of intertidal zones along the Chinese coastline from north to south. A total of 4436 strains were isolated from these samples using different methods and 286 species including 39 potential novel species were identified from these strains based on the internal transcribed spacer (ITS) region or the D1/D2 domain of the large subunit rRNA gene sequence analysis. The majority of the yeast species in different geographical locations belong to the five orders Serinales, Saccharomycetales, Tremellales, Sporidiobolales, and Pichiales. The yeast species diversity varied depending on sample types, depth of marine sediments, intertidal zone types and geographical locations. Mean annual temperature (MAT), salinity and pH had the greatest effect on the community structures of the yeasts isolated from the intertidal zones. This study represents one of the most comprehensive surveys of marine yeasts in China to date and provides a better understanding of marine yeast diversity and distribution.
1 Introduction
Tidal flat, or intertidal zone refers to the area above the low and below the high tide line, which provides a broad range of habitats such as mangrove, mud flat, sandy beach, rocky shore, coral reef and aquaculture area. China has the second largest intertidal zone in the world, extending from the tropical to the temperate climate zones (Liu, 2013; Murray et al., 2019). The Chinese intertidal zones potentially harbour high biodiversity of microorganisms. Marine yeasts are ubiquitous in marine environments and some of them show better growth in seawater than in fresh water (Chi et al., 2010; Kaewkrajay et al., 2020). Due to their high pressure tolerance ability, marine yeasts show outstanding performance in various industrial and medical applications, and remediation of marine environments. In industrial applications, marine yeasts not only play a significant role in the production of various enzymes (Chi et al., 2009; Chi et al., 2010), but also possess enormous potential for bioethanol and biodiesel production (Zaky et al., 2014; Wang et al., 2017). In addition, marine yeasts also exhibit high potentials in the production of single cell protein, polysaccharides, vitamins, killer toxins, pigments etc. In medical applications, marine yeasts can be used to produce pharmaceutical products including astaxanthin, siderophore and riboflavin (Wang et al., 2008; Wang et al., 2009; Nath Ushakumari and Ramanujan, 2013; Zaky et al., 2014). A previous study showed that Yarrowia lipolytica isolated from marine environments can produce nanoparticles (Chi et al., 2010). Many marine yeasts can remove organic pollutants and heavy metals, so they can be applied to the remediation of marine environments (Chi et al., 2010).
However, yeast diversity in marine environments has been much less studied compared to that in terrestrial environments in China. Yeast diversity in terrestrial natural habitats has been extensively studied in China, for examples, on the surface and gut of insects (Lou et al., 2014), sediments or soil related to glacier, forest, orchard, and desert (Luo et al., 2019; Li et al., 2020; Zhu et al., 2021; Wei et al., 2022), on the surface of the grapes and plant leaves (Li et al., 2010; Li et al., 2020; Wei et al., 2022), oral cavity (Wang et al., 2007), samples associated with Chinese Baijiu fermentation environments (Wu et al., 2012; Lei et al., 2022) and rotting wood (Gao et al., 2017; Lu et al., 2017; Zheng et al., 2017; Gao et al., 2018; Huang et al., 2018; Huang et al., 2019; Ke et al., 2019; Xi et al., 2019; Chai et al., 2020; Jia et al., 2020; Gao et al., 2021; Shi et al., 2021). Compared to the studies on terrestrial environments, only limited studies focused on yeasts in marine environments in China. Yang et al. (2011); Chi et al. (2012); Zhu et al. (2023a), and Zhu et al. (2023b) described some known and novel marine yeast species in diverse marine environments of China. Wang et al. (2017) found one marine yeast strain which possesses the capability for biodiesel production from renewable feedstocks.
In the course of microbial resource investigation in intertidal zones of China performed in recent years, we collected more than 1200 diversified marine samples along the Chinese coastline from north to south. Based on cultivation-dependent method, we revealed species diversity and distribution of yeasts in Chinese intertidal zones. We also identified key environmental factors affecting the yeast diversity in these intertidal zones.
2 Materials and methods
2.1 Samples collection
A total of 1241 marine samples including marine sediments, marine water, plants, and benthos were collected from different types of intertidal zones, including sand beach, rocky beach, mud flat, grass lands with different dominant plants, and mangrove (Figure 1) along the coastline of China during July 2019 to July 2023. A total of 161 GPS sites covering 11 coastal provinces of China, including Fujian, Guangdong, Guangxi, Hainan, Hebei, Jiangsu, Liaoning, Shandong, Shanghai, Tianjin, and Zhejiang were sampled (Figure 2). The samples were collected using sterile plastic bags and transferred in ice boxes to the laboratory for yeast isolation and physicochemical property measurement (Table S1).

Figure 1 Different types of intertidal zones in China sampled in this study. (A) sand beach; (B) rocky beach; (C) mud flat; (D) grass land with plant Phragmites australis; (E) grass land with plant Suaeda salsa; (F) mangrove.
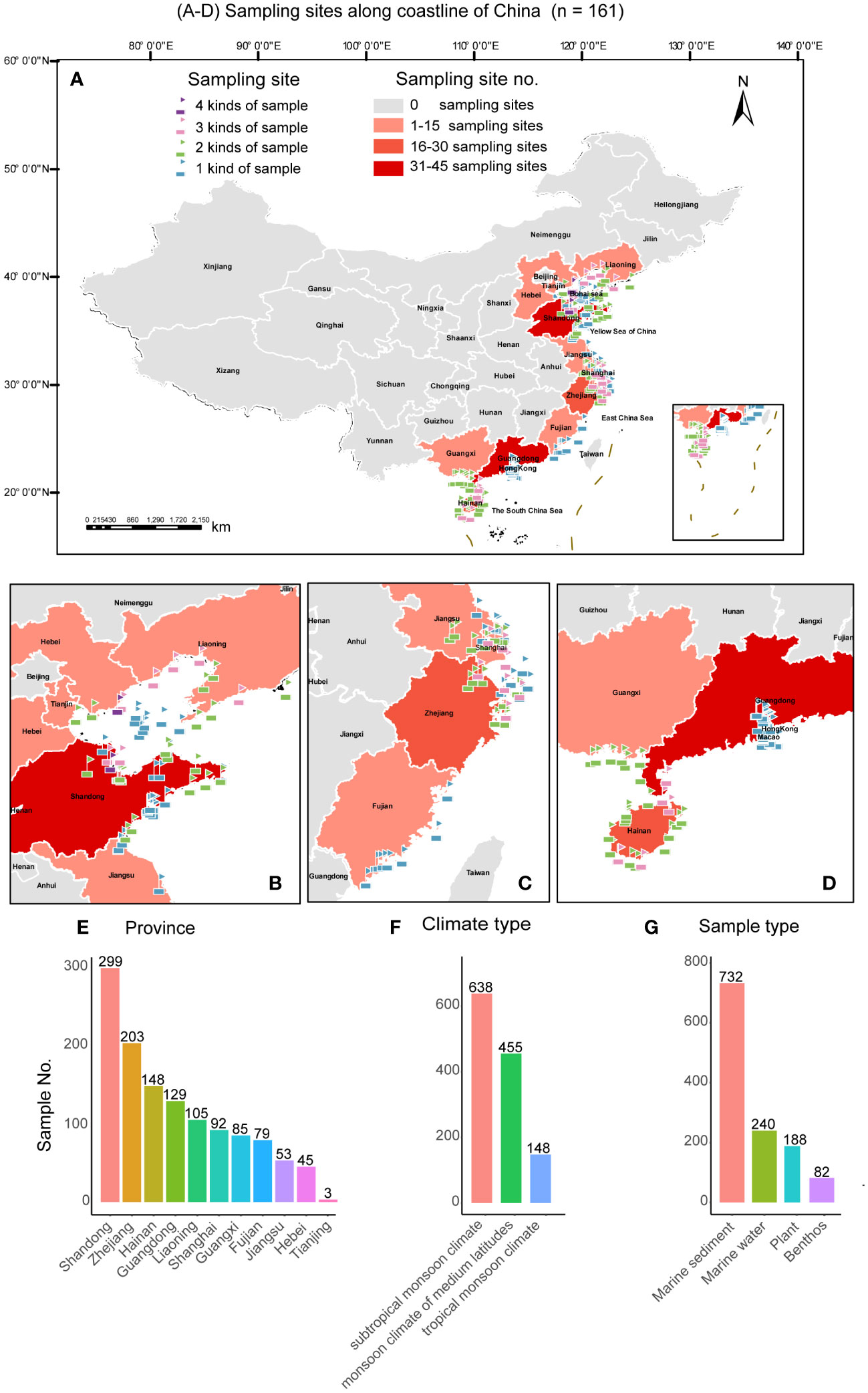
Figure 2 The geographic locations of the sampling sites in this study. (A) Map showing the distribution of all the sampling sites in 11 provinces in China; (B) Map showing the distribution of the samples collected from the Bohai Wan (BHW) Bay; (C) Map showing the distribution of the samples collected from the Hangzhou Wan (HZW) Bay; (D) Map showing the distribution of the samples collected from the Beibu Wan (BBW) Bay; (E−G) Characteristics of collected marine samples. In (A−D), Maps were generated by the ArcGIS v. 10.8 software (Esri, Redlands, CA, USA).
For the marine sediment samples aiming to detailed and extensive yeast diversity and ecological analyses, four to six replicates with the same distance (>50 m) between adjacent samples were collected and mixed together for each site. To reveal the vertical profile of culturable marine yeast communities, at each sampling site, a marine sediment column with 10 cm diameter and 50 cm length was collected using a plastic sampler. The column was separated into three samples with different depth layers (oxic zone, 0 to 5 cm; anoxic zones, 5 to 15cm and 15 to 25 cm). For each marine water sample, 300 ml was collected in a sterile bag.
2.2 Yeast isolation
For the benthos, marine sediment and water samples, yeast strains were isolated using the enrichment method described previously by Zaky et al. (2016), but additional media were used in this study. For each marine sediment or benthos sample, two grams were suspended in 20 ml sterile water and shaken at 200 rpm for 30 minutes at 25°C. Then each suspension was diluted to 1×10-1 and 1×10-2 and 200 μl of each dilution was respectively plated on 1/5 malt extract agar (1/5MEA, 0.4% glucose, 0.4% malt extract, 0.02% peptone, 2% agar), corn meal agar (CMA, 2.5% corn starch, 2% agar), potato dextrose agar (PDA, 20% potato infusion, 2% glucose, 2% agar), Rhodotorula isolation agar (RM, 1% glucose, 0.1% yeast extract, 0.2% peptone, 2% agar, 1 mg/L sodium glutamate), Yeast malt agar (YM, 1% glucose, 0.3% yeast extract, 0.3% malt extract, 0.5% peptone, 2% agar), and yeast extract peptone dextrose (YPD, 2% glucose, 1% yeast extract, 2% peptone, 2% agar) plates containing 1.5% sea salt, 500 mg/L penicillin and 500 mg/L streptomycin sulphate. For each medium, two plates were inoculated and incubated at 17°C and 25°C, respectively, for 3–5 days. For each marine water sample, 200 ml was filtered through a filter membrane with 100 mm diameter and then one-fourth of the filter was put in 20 ml sterile water and then treated in the same way as the soil suspension. For the plant leaves and stems, yeast strains were isolated by an enrichment method according to Bai et al. (2002) and Wang et al. (2012) with minor modifications. Specifically, two grams of each sample were placed into 10 ml YM broth supplemented with 7% ethanol and 200 mg/L chloramphenicol in a 15 ml sterile centrifuge tube and then incubated at 25 °C for one week. Then 100 μl enrichment culture and appropriate decimal dilutions were spread on YM agar plates supplemented with 200 mg/L chloramphenicol and then incubated at 25 °C for 3–4 days. In the meantime, for the plant leaves, yeast strains were also isolated using ballistoconidia-fall method described by Nakase and Takashima (1993). Yeast and yeast like colonies that appeared on the plates were picked, purified, and preserved in 25% glycerol at –80 °C.
2.3 Measurement of environmental factors and physiochemical properties
The pH, salinity, dissolved oxygen (DO) and oxidation reduction potential (ORP) for marine water were determined using the HQ40d Portable Meter (Hach Company, Loveland, CO, USA). The mean annual temperature (MAT) and mean annual precipitation (MAP) of the sampling areas were obtained from the WorldClim database (http://www.worldclim.org).
2.4 DNA extraction, sequencing and yeast identification
Nuclear DNA of the yeast strains was extracted using the method described previously (Wang and Bai, 2008). The D1/D2 domain of the 26S ribosomal RNA gene and the internal transcribed spacer (ITS) region was amplified and sequenced using the method described by Bai et al. (2002). Yeasts were identified by analysis of the sequence similarity of the D1/D2 domain using the BLASTn search program (https://blast.ncbi.nlm.nih.gov). For the identification of yeasts, the strains with 0–3 nucleotide substitutions in the D1/D2 domain were designated as conspecific, while the strains showing greater than 1% nucleotide substitutions (six or more nucleotides) were considered as different species (Kurtzman and Robnett, 1998; Fell et al., 2000; Scorzetti et al., 2002; Vu et al., 2016). When a strain showed more than three nucleotide substitutions from the type strain of the most closely related known species, the ITS sequence was compared further. If more than 1% mismatch was found, the strain was treated as a “potential novel species” in this study.
2.5 Phylogenetic analyses
Phylogenetic analysis based on the D1/D2 domain or ITS region was performed to verify the identification using sequence similarity analysis. The sequences of representative strains were aligned by MAFFT v. 7 (Katoh and Standley, 2013) and manually improved where necessary using MEGA v.7 (Kumar et al., 2016). A phylogenetic tree was constructed from Kimura’s two parameter model (Kimura, 1980) using the neighbour-joining algorithm executed in MEGA v.7 (Kumar et al., 2016; Lachance, 2022). Confidence levels of the clades were estimated from bootstrap analysis (1000 replicates) (Felsenstein, 1985). The phylogenetic trees were visualized and annotated using the online open-source tool Interactive Tree of Life (iTOL) (Letunic and Bork, 2019).
2.6 Statistical analysis
Alpha diversity analysis was calculated in R using the package vegan (Oksanen et al., 2022). Beta diversity analysis was conducted based on Sørensen dissimilarity. This analysis was performed among different collection sites based on the yeast species using the R packages vegan and ape (Paradis and Schliep, 2019). The statistical significance was calculated using the ANOVA or Kruskal–Wallis test executed by the aov function and kruskal.test function of R package multcomp and FSA, respectively. Non-metric multidimensional scaling (NMDS) plot was created using a Bray–Curtis dissimilarity matrix of samples in the R package phyloseq version 1.25.2 (McMurdie and Holmes, 2013). Redundancy analysis (RDA) was used to explore effect of environmental factors on the yeast community. Correlation between yeast species and environmental factors as well as physiochemical properties profiles was analyzed using Spearman’s ρ with Padj < 0.05 (Best and Roberts, 1975). Differences in environmental factors, physiochemical property profiles, and yeast species across different sites were assessed using a two-way ANOVA, with the Bonferroni post hoc test used for repeated measurements. All significant yeast species and environmental factors, along with the physicochemical properties, were visualized by heatmap graphs. All statistical analyses were performed using R, version v.4.3.1 (R Core Team, 2023).
3 Results
3.1 The overall yeast diversity from all the samples collected in intertidal zones of China
The 1241 marine samples collected in this study yielded a total of 4436 yeast strains. Altogether, 286 yeast species including 141 ascomycetous and 145 basidiomycetous species were identified from these strains (Figures 3 and 4; Table S2). Among them, 39 are potential novel species (Table S3). Among the species discovered, 41 yeast species were isolated from more than thirty samples and 33 species were isolated from more than five provinces in this study (Figures 3 and 4; Table S2). A total of 21 yeast species were the most frequently isolated yeasts, including 15 ascomycetous species, namely Candida parapsilosis, Candida pseudolambica, Candida tropicalis, Diutina catenulate, Geotrichum candidum, Kluyveromyces siamensis, Kodamaea ohmeri, Meyerozyma caribbica, Meyerozyma guilliermondii, Nakaseomyces glabratus, Pichia kudriavzevii, Saccharomyces cerevisiae, Saccharomycopsis fibuligera, Scheffersomyces spartinae, and Wickerhamomyces anomalus; and six basidiomycetous species, namely, Cystobasidium minutum, Cystobasidium slooffiae, Moesziomyces aphidis, Papiliotrema laurentii, Saitozyma podzolica, Trichosporon asahii (Figures 3 and 4; Table S2). Furthermore, three ascomycetous yeast species, Candida parapsilosis, Candida tropicalis, and Scheffersomyces spartinae were isolated from all the provinces sampled except Tianjin. Only three samples were collected from Tianjin and one yeast strain identified as Tilletiopsis washingtonensis was isolated from the Tianjin samples (Figure 2E; Table S2).

Figure 3 Phylogenies and geographic distributions of the ascomycetous yeast species discovered in this study. The trees were constructed based on the D1/D2 sequences. The green squares represent the 11 sampling provinces in this study. Solid and hollow squares indicate the presence and absence of the species in the locations, respectively. Yellow solid circles that each of indicate the species was isolated more than 30 samples. The blue bar charts indicate the number of strains isolated.
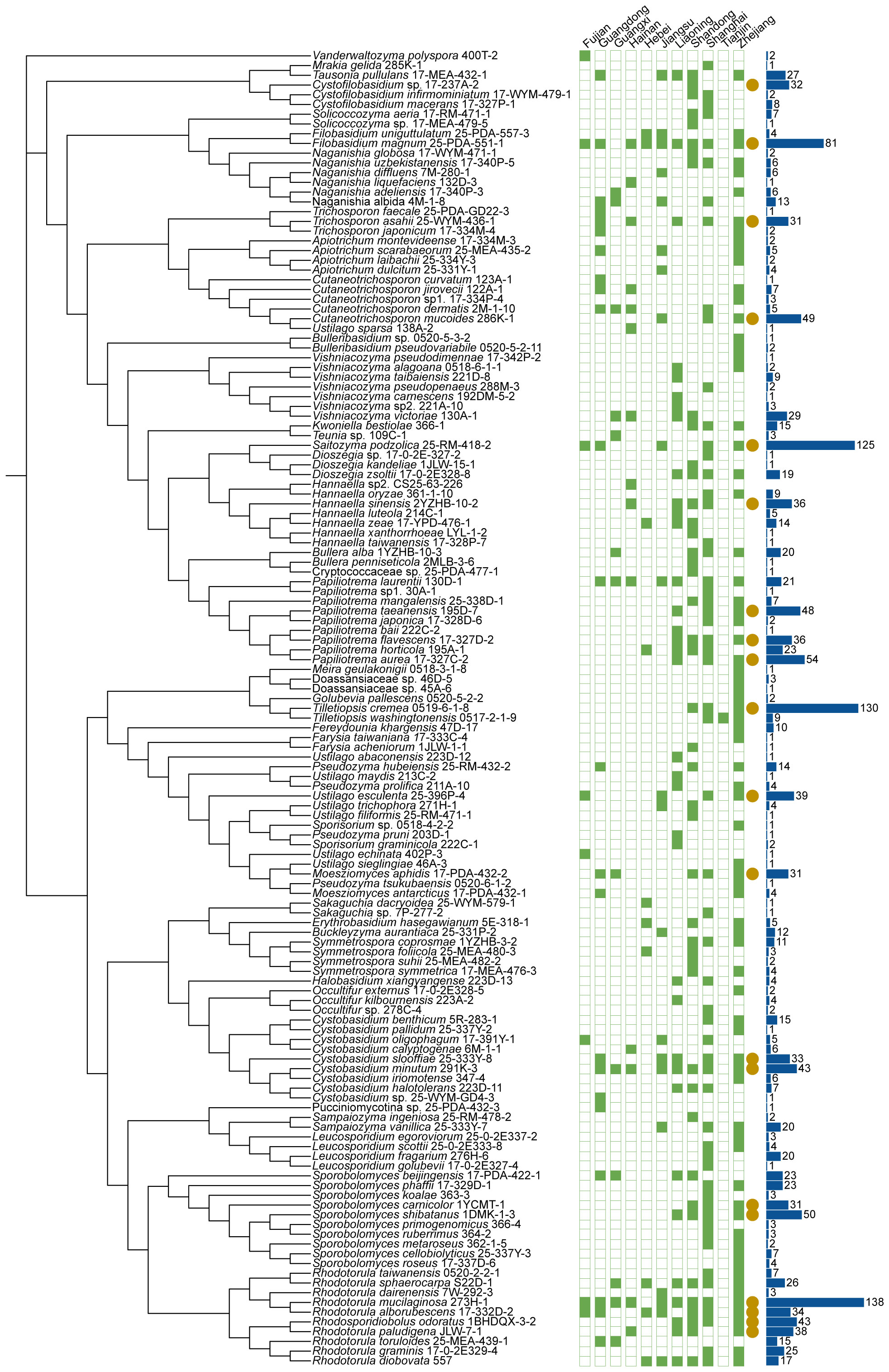
Figure 4 Phylogenies and geographic distributions of the basidiomycetous yeast species discovered in this study. The trees were constructed based on the D1/D2 sequences. The green squares represent 11 sampling provinces in this study. Solid and hollow squares indicate the presence and absence of the species in the locations, respectively. Yellow solid circles indicate the species were isolated more than 30 samples. The blue bar charts indicate the number of strains isolated.
3.2 Yeast diversity in representative GPS sites from intertidal zones of China
In order to perform detailed biodiversity analyses of yeasts in depth, we selected 41 representative GPS sites from intertidal zones of China. The samples from these sites were collected using the same sampling method and strategy and were subjected to yeast isolation using the same media and the same processing procedure. Three marine samples including one surface sediment (0−5cm), one subsurface sediment (5−25cm), and one water samples were isolated from per representative GPS site. We divided all the samples from these 41 sites into three groups according to their geographical locations. The Beibu Wan (BBW) Bay group included 17 GPS sites located in the Beibu Gulf, South China (Figure 2D; Table S4); the Hangzhou Wan (HZW) Bay group included 15 GPS sites located in the Hangzhou Bay, East China (Figure 2C; Table S4); and the Bohai Wan (BHW) Bay group included 9 GPS sites located in the Bohai Bay, Northeast China (Figure 2B; Table S4). These samples were subjected to yeast isolation using the same media PDA, RM and YM at 25°C. A total of 776 yeast strains belonging to 115 species and 63 genera were isolated from these samples. Specifically, 117 strains of 49 species were isolated from HZW; 324 strains of 42 species from BBW; and 335 strains of 58 species from BHW (Figure 5A).
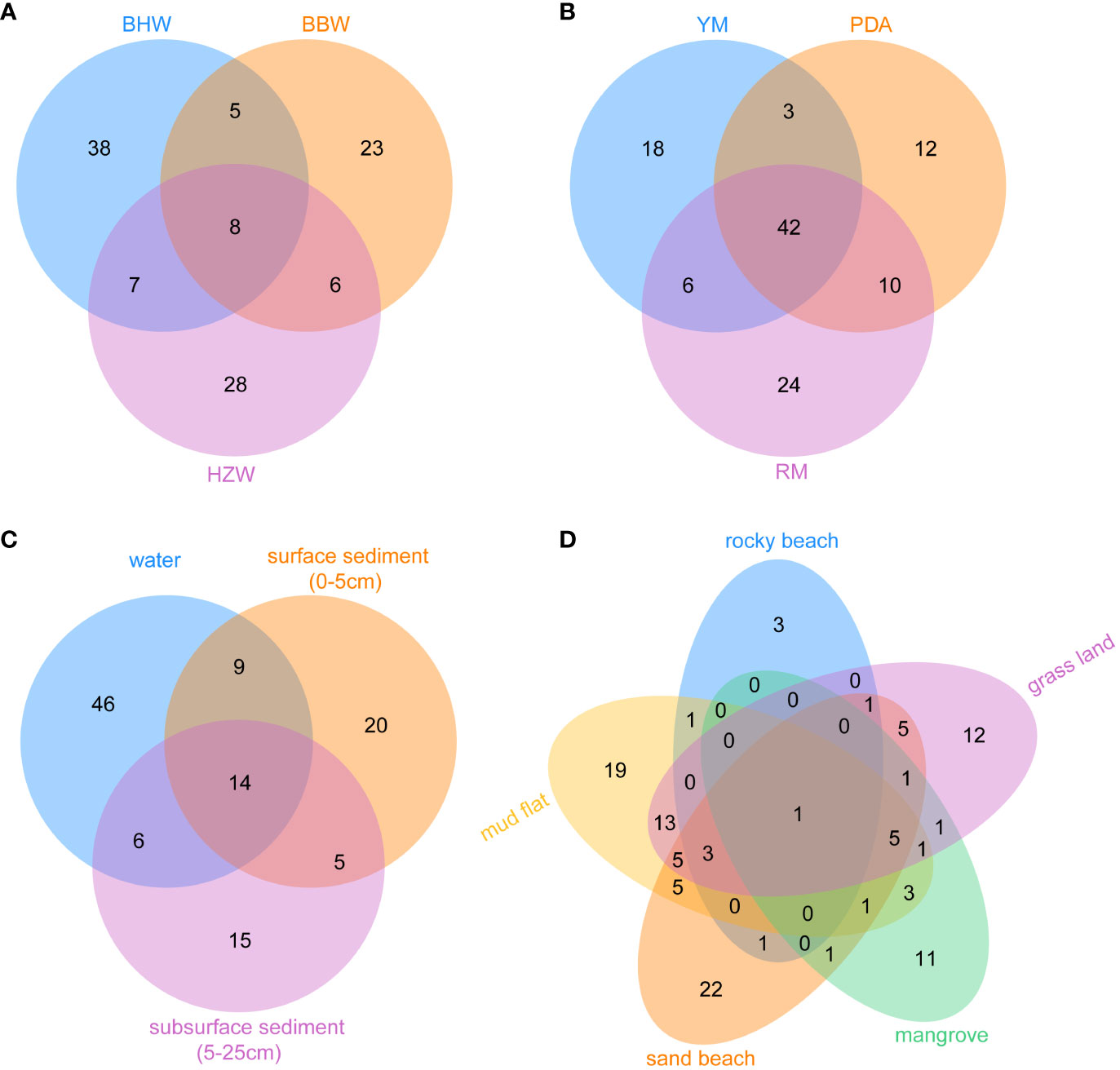
Figure 5 The number of unique and shared yeast species from 41 representative GPS sites of intertidal zones in different locations (BBW, the Beibu Wan Bay; BHW, the Bohai Wan Bay; and HZW, the Hangzhou Wan Bay) (A), different media (PDA, potato dextrose agar; RM, Rhodotorula isolation magar; YM, yeast malt agar) (B), different substrates (C) and different intertidal zone types (D).
3.3 Yeast species distributions vary depending on geographic regions, substrates and intertidal zone types
Different regions, media, substrates, and tidal flats harbored their own yeast species (Figure 5). Although only nine GPS sites in BHW were selected (Table S4), 68 yeast species were isolated from this region, while 42 species from 17 GPS sites in BBW and 49 species from 15 GPS sites in HZW were discovered. Only eight species were shared by the three regions. BHW possesses the highest yeast species diversity among these three regions (Figure 5A). Although 42 species were commonly found in three different media, each medium yielded its own unique species. Specifically, 18, 12, and 24 unique species were obtained using YM, PDA, and RM, respectively (Figure 5B). A total of 75 species were isolated from marine water samples, while 48 and 40 species were isolated from surface and subsurface sediment samples, respectively (Figure 5C). Different types of intertidal zones also harbour different yeast diversities. A total of 57 species were isolated from mud flat samples, 51 species from sand beach, 48 species from grass land, 25 species from mangrove, and only 10 from rocky beach samples. The yeast species shared by different types of intertidal zones were limited (Figure 5D).
3.4 Yeast community composition in different intertidal zones of China
At the order level, the five orders Serinales, Saccharomycetales, Tremellales, Sporidiobolales, and Pichiales were the dominant orders in all the three groups, collectively accounting for 84.11% in BBW group, 78.55% in BHW group, and 52.84% in HZW group (Figure 6A; Table S5). The order Saccharomycetales was much more abundant in the BBW group (25.23%) than in the BHW group (8.00%) and HZW group (18.18%); the order Serinales was much more abundant in the BHW group (26.18%) than in the BBW group (14.64%) and HZW group (9.66%) (Figure 6A; Table S5). At the family level, Sporidiobolaceae was the dominant family in both BBW (20.66%) and HZW groups (13.56%), meanwhile, Bulleribasidiaceae is the dominant family in BHW (15.64%) (Figure 6B; Table S5). At the genus level, the dominant genera were also different among the three groups (Figure 6C; Table S5). Both Candida and Rhodotorula accounted for over 20% each in the BBW group, whereas in the BHW group, only Metschnikowia accounted for over 20%. Rhodotorula was the dominant genus in HZW group but with a less ratio (15%) (Figure 6C; Table S5).
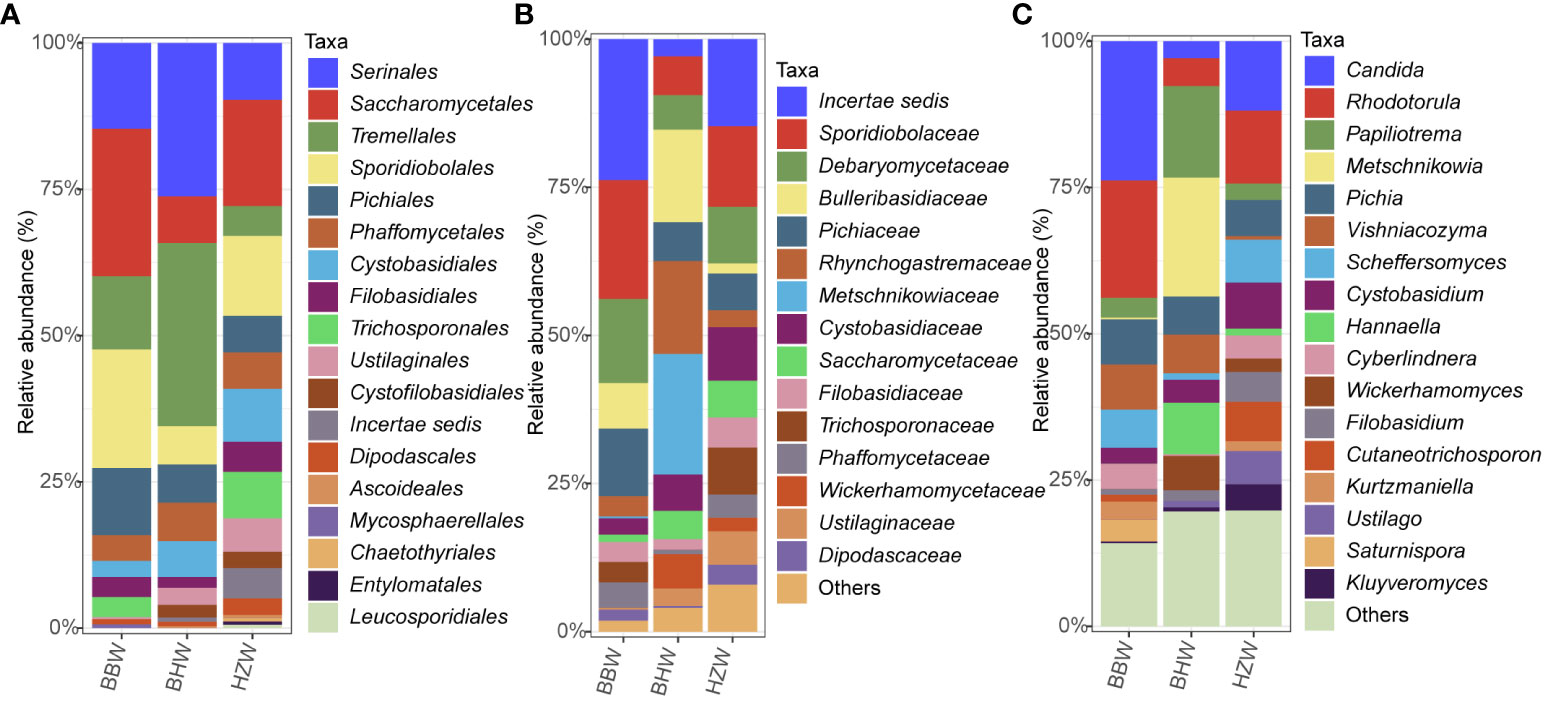
Figure 6 Relative abundances of yeast species in different locations at the order (A), family (B) and genus (C) levels. BBW, the Beibu Wan Bay; BHW, the Bohai Wan Bay; HZW, the Hangzhou Wan Bay.
3.5 Alpha and beta diversities of yeasts in different intertidal zones of China
The alpha diversity analysis showed that the Shannon index of yeasts was higher in BHW group than those in BBW and HZW groups, but the difference was not statistically significant (Figure 7A). The estimated richness (ACE index) of yeasts in BHW group was significantly higher than those in BBW and HZW groups (P < 0.05) (Figure 7B), suggesting that the number of yeast species found in BHW group was significantly higher than those in the latter regions.
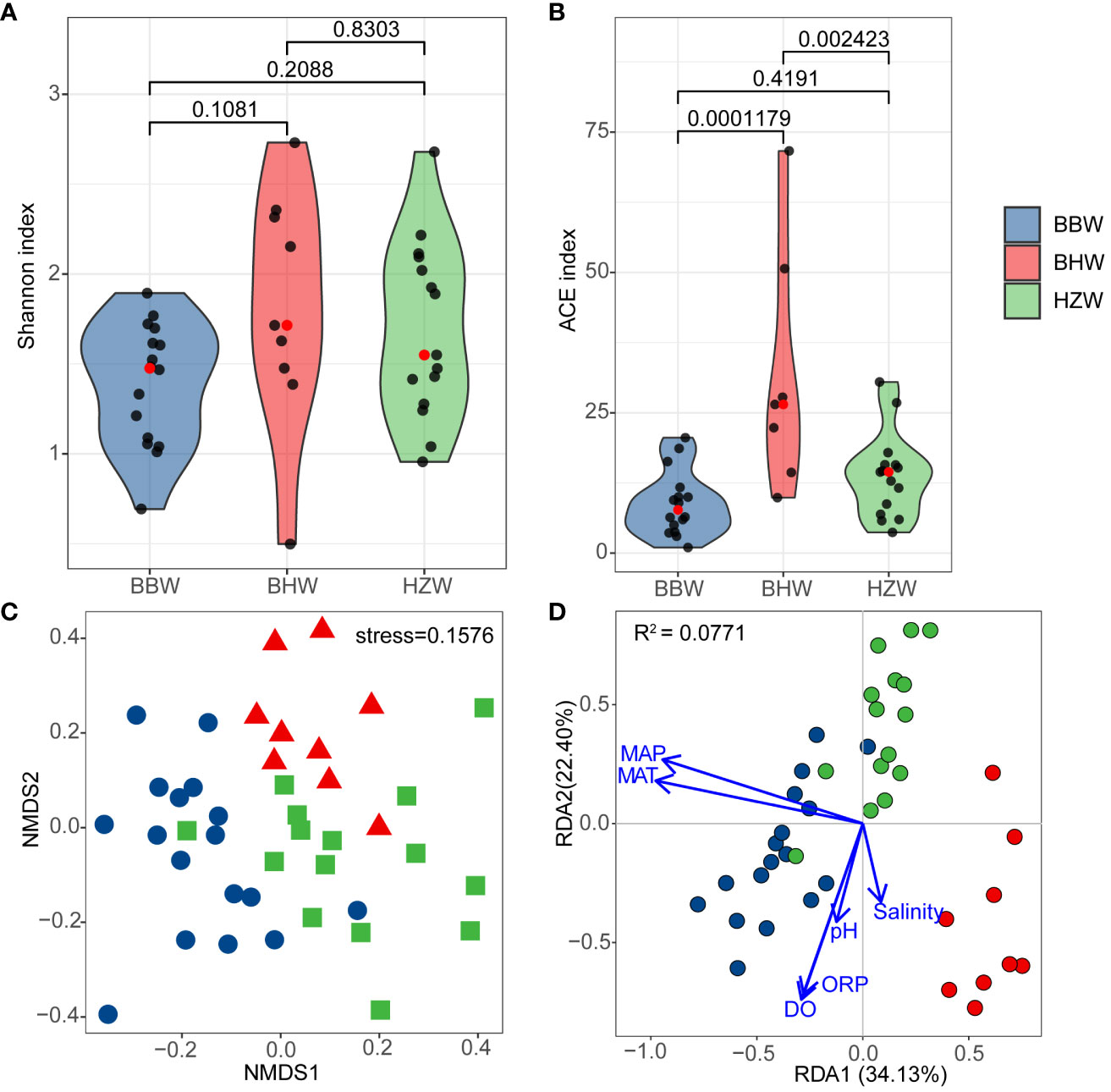
Figure 7 Alpha and Beta diversity of marine yeasts from 41 representative GPS sites of intertidal zones of China. (A) Shannon index; (B) ACE index; (C) Non-metric multidimensional scaling; (D) Redundancy analysis based on four physiochemical properties, pH, salinity, dissolved oxygen (DO) and oxidation reduction potential (ORP) and two environmental factors, the mean annual temperature (MAT) and mean annual precipitation (MAP). BBW, the Beibu Wan Bay; BHW, the Bohai Wan Bay; HZW, the Hangzhou Wan Bay.
Comparative yeast diversity analysis was performed after the strain numbers were normalized. NMDS analysis based on Bray-Curtis dissimilarity distances revealed that the yeast communities in BBW, BHW and HZW groups were separated with limited intersection (Figure 7C), indicating that the yeast species compositions in different geographic locations are different.
The RDA analysis based on yeast species and environmental factors and physico-chemical properties of the samples from different regions showed that the first and second RDA components explained 56.53% of the total variation (Figure 7D). MAT, salinity and pH were significantly associated with the yeast community (P < 0.05) (Figure 7D; Table S6). The results suggest a significant correlation between the yeast community and physicochemical property and environmental climate, particularly MAT in the sampling regions.
3.6 Influence of physiochemical and environmental factors on the occurrence of yeast species in intertidal zones of China
To identify the preliminary factors affecting the occurrence of yeast species in intertidal zones of China, the top 30 most frequently isolated yeast species were selected and the Spearman correlation coefficients between the isolation frequencies of these yeast species and four physicochemical properties and two environmental factors were calculated (Table S4). The result showed that nine yeast species were significantly influenced by MAT and MAP. Among these species, Rhodotorula toruloides and Papiliotrema laurentii showed a significantly positive correlation with MAP and MAT (P < 0.05); while Wickerhamomyces anomalus, Geotrichum candidum, Cyberlindnera jadinii, Kluyveromyces lactis, Rhodotorula mucilaginosa, Tausonia pullulans, and Pichia pseudolambica exhibited a negative correlation with MAP and MAT (P < 0.05) (Figure 8). pH positively affected the occurrence of Candida tropicalis but negatively affected the occurrence of Hannaella zeae, Papiliotrema aurea, Papiliotrema flavescens, and Saccharomyces cerevisiae (P < 0.05). Limited species were significantly influenced by DO, ORP and salinity (Figure 8). Among the 30 yeast species analyzed, nine species, including Cutaneotrichosporon mucoides, Filobasidium magnum, Saturnispora suwanaritii, Ustilago esculenta, Vishniacozyma taibaiensis, Hannaella sinensis, Pichia kudriavzevii, and Candida parapsilosis, did not show any significant correlation with the physiochemical and environmental factors analyzed (Figure 8).
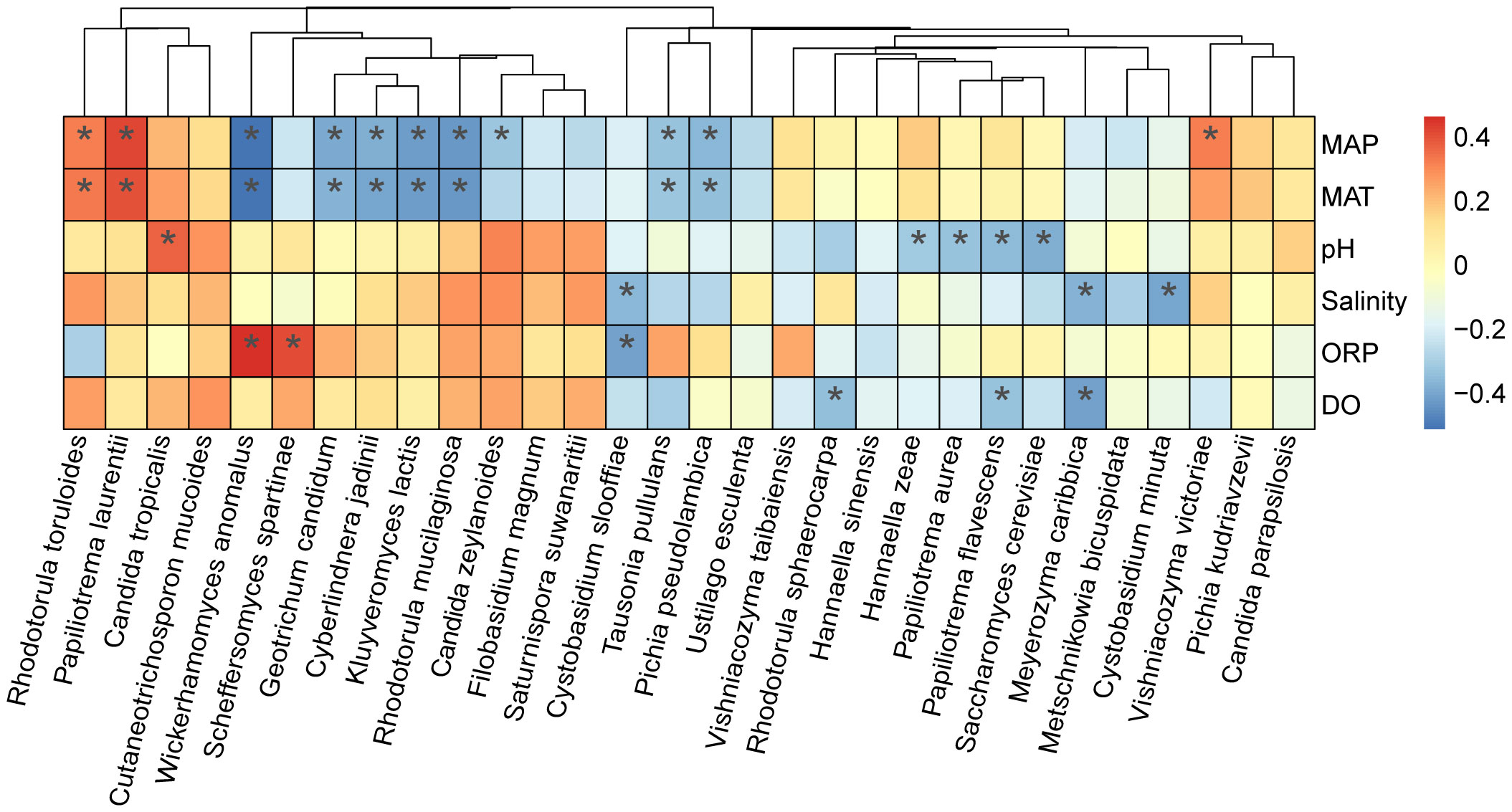
Figure 8 Correlation of the occurrence of marine yeast species with physiochemical properties pH, salinity, dissolved oxygen (DO) and oxidation reduction potential (ORP) and environmental factors mean annual temperature (MAT) and mean annual precipitation (MAP). Heatmaps were created based on Spearman correlation coefficients with P < 0.05 between the isolation frequency of the yeast species accounting for > 1% of the yeast strains isolated and physiochemical properties and environmental factors. Heat map colors reflect different degrees of negative (blue) and positive (red) correlations according to the scale on the right. * 0.01 < P < 0.05.
4 Discussion
To our knowledge, this the most extensive study on yeast diversity in marine environment, especially in intertidal regions, in terms of the sample number collected and the yeast strains isolated and the geographic regions covered. In this study, we obtained 286 yeast species, including 140 ascomycetous and 146 basidiomycetous species associated with intertidal zones of China (Table S2). Boekhout et al. (2022) summarized that at least 782 yeast species have been recorded from China recorded until 2020 and 49 additional yeast species were reported from China in the past three years (Gao et al., 2021; Shi et al., 2021; Chai et al., 2022a; Chai et al., 2022b; Chai et al., 2022c; Chu et al., 2022; Li et al., 2022; Wei et al., 2022; Chai et al., 2023; Liu et al., 2023; Qiao et al., 2023; Yu et al., 2023; Zhu et al., 2023a; Zhu et al., 2023b). The number of yeast species discovered in this study accounts for 34.7% in the total yeast species that have been found from China, indicating that there are abundant resources of culturable yeasts in the intertidal zones of China. Jones et al. (2015) summarized that there are 213 marine yeasts, including 138 ascomycetous species, 75 basidiomycetous species, which were reported from marine habitats and even if they are facultative. Our study provides a wider perspective to know the marine yeast species.
Many of the taxa isolated in this study were previously known as opportunistic pathogens of human, including Candida albicans, Candida parapsilosis, Candida tropicalis, Pichia kudriavzeveii, Nakaseomyces glabrata which are recorded in the WHO fungal priority pathogens list (World Health Organization, 2022). Among these five yeast species, Candida parapsilosis and Candida tropicalis were the most prevalent which were distributed in 10 of the 11 provinces samples in this study (Figure 3; Table S2). Only the samples from Tianjin showed negative isolation of these two species most probably because of the very limited samples collected from this region. Pichia kudriavzeveii distributed in eight provinces was the second prevalent species (Figure 3; Table S2). Candida albicans and Nakaseomyces glabrata were discovered in five and two provinces, respectively (Figure 3; Table S2). All five species were discovered in more than one province, suggesting their existence in intertidal zone is not a one-off event. Besides the above-mentioned five yeast species, there are many species which are opportunistic pathogenic yeast in previous publications, for example, Aureobasidium melanogenum (Chen et al., 2016), A. pullulans (Pikazis et al., 2009), Candida allociferrii (Soki et al., 2015), Geotrichum candidum (Keene et al., 2019), Wickerhamomyces anomalus (Aboutalebian et al., 2023), Yarrowia lipolytica (Desnos-Ollivier et al., 2020) and so on. Every coin has two sides. For instance, although Pichia kudriavzeveii is a globally distributed opportunistic pathogenic yeast (Pfaller et al., 2008), whose infections frequently acquired from the environment was verified by Douglass et al. (2018), it plays a significant role in bioethanol production (Hoppert et al., 2022), cocoa fermentation (Pereira et al., 2017), single−cell protein production (Hashem et al., 2022). Meanwhile, high resistance to fluconazole is common in environmental and clinical isolates without distinction in Pichia kudriavzeveii (Douglass et al., 2018), which make the treatment for fungal infections more difficult undoubtedly. There is a question that whether the fungal resistance is common in other opportunistic pathogenic yeasts. In search of the solution, our study maybe could provide vast environmental isolates.
Apart from the dominant species Pichia kudriavzeveii, many other dominant species demonstrate remarkable performance in numerous industrial and medical applications. For instance, Candida tropicalis, Debaryomyces hansenii, and Saccharomyces cerevisiae were frequently utilized in the production of bio-ethanol (Zaky et al., 2014). The two species Candida tropicalis and Debaryomyces hansenii also play a significant role in the biomaterial industry in producing silver nanoparticles. The Rhodotorula mucilaginosa can be utilized not only in biodiesel industries for the production of microbial oil, but also in food colorings to generate carotene and feed industries for the production of protease (Zaky et al., 2014). Despite their small size, yeast cells possess a boundless potential. Our study could provide the support for the marine yeast application.
As China is one of the largest coastal countries in the world, our sampling still exists the room for extending sampling, which is requiring us to conduct research continuously in the intertidal zones in the future. Although our representative sampling sites from south (BBW) to north (BHW) are gradually decreasing (from 17 to nine) (Table S2), the number of yeasts isolated is gradually increasing (Figure 5A). With the rise of the latitude, the MAT is gradually increasing (Table S4). RDA analysis results indicates that MAT, salinity and pH had the significant effect on the community structure of the yeasts isolated from the intertidal zones, especially the MAT, which is consistent with Zhou et al. (2016) (Figure 7D). Climate warming is increasingly leading to marked changes in biodiversity, including plant, animal, and microorganism (Zhou et al., 2016). Meanwhile, with the climate warming, the change of carbon dioxide in marine water will dramatically change, which could lead to Ocean acidification (Anand et al. 2021). It will affect the growth of microorganisms. The above-mentioned results are reminding us that it is urgent to rescue the lost microbial resources, including the yeasts.
Yeast species distributions vary depending on geographic regions, isolation media, substrates and intertidal zone types. During the initial medium screening stage, we utilized not only YM, PDA, and RM, but also other three media, namely, YPD, CMA, and 1/5MEA (data not shown). Different media yielded their own unique yeast species, among these media tested, and the three media YM, PDA and RM had the best isolation results. Therefore, these three media were used throughout our entire research. The isolation results from different substrates, including marine water and marine sediment with different depth, show that the marine water possesses more yeast species. Our study provides valuable suggestions and clues for sampling and yeast isolation from marine environments in the future.
Data availability statement
The sequence data of the study are deposited in the NCBI database with accession numbers which can be found in the article/Supplementary Material.
Author contributions
H-YZ: Investigation, Data curation, Visualization, Writing original draft. D-YH: Investigation, Data curation, Visualization. L-CG: Investigation, Data curation, Visualization. J-NL: Investigation. X-YW: Investigation. R-PZ: Investigation. Q-MW: Investigation, Data curation. Y-JS: Investigation. L-JL: Investigation. Y-HW: Data curation, Visualization. X-ZL: Investigation. F-YB: Funding acquisition, Investigation, Supervision, Writing review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Science & Technology Fundamental Resources Investigation Program of China (grant No. 2019FY100700).
Acknowledgments
We thank Fang Liu and Chang Liu from the Institute of Microbiology, Chinese Academy of Sciences, Wen-Jun Li from Sun Yat-sen University, Cong Sun and Jun-Jie Ying from Zhejiang Sci-Tech University, Zhi-Cheng Wu from Zhejiang University and Du-Qiang Luo from Hebei University for providing some marine samples.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1286511/full#supplementary-material
Abbreviations
1/5MEA, 1/5 malt extract agar; CMA, corn meal agar; DO, dissolved oxygen; ITS, the internal transcribed spacer region; NMDS, Non-metric multidimensional scaling; ORP, oxidation reduction potential; PDA, potato dextrose agar; RDA, Redundancy analysis; RM, Rhodotorula isolation medium; YM, Yeast malt medium; YPD, yeast extract peptone dextrose.
References
Aboutalebian S., Mirhendi H., Eshaghi H., Nikmanesh B., Charsizadeh A. (2023). The first case of Wickerhamomyces anomalus fungemia in Iran in an immunedeficient child, a review on the literature. J. Med. Mycology 33, 101351. doi: 10.1016/j.mycmed.2022.101351
Anand M., Rangesh K., Maruthupandy M., Jayanthi G., Rajeswari B., Priya R. J. (2021). Effect of CO2 driven ocean acidification on calcification, physiology and ovarian cells of tropical sea urchin Salmacis virgulata – A microcosm approach. Heliyon 7, e05970. doi: 10.1016/j.heliyon.2021.e05970
Bai F. Y., Zhao J. H., Takashima M., Jia J. H., Boekhout T., Nakase T. (2002). Reclassification of the Sporobolomyces roseus and Sporidiobolus pararoseus complexes, with the description of Sporobolomyces phaffii sp. nov. Int. J. Syst. Evol. Microbiol. 52, 2309–2314. doi: 10.1099/00207713-52-6-2309
Best D. J., Roberts D. E. (1975). Algorithm AS 89: the upper tail probabilities of spearman’s Rho. Appl. Stat. 24, 377. doi: 10.2307/2347111
Boekhout T., Amend A. S., El Baidouri F., Gabaldón T., Geml J., Mittelbach M., et al. (2022). Trends in yeast diversity discovery. Fungal Divers 114, 491–537. doi: 10.1007/s13225-021-00494-6
Chai C. Y., Gao W. L., Li Y., Yan Z. L., Hui F. L. (2022a). Kodamaea hongheensis f.a., sp. nov., Kodamaea ovata f.a., sp. nov. and Kodamaea yamadae f.a., sp. nov., three new yeast species of Kodamaea (Saccharomycetales, Debaryomycetacae) from China. MycoKeys 89, 121–137. doi: 10.3897/mycokeys.89.81119
Chai C. Y., Gao W. L., Yan Z. L., Hui F. L. (2022c). Four new species of Trichomonascaceae (Saccharomycetales, Saccharomycetes) from Central China. MycoKeys 90, 1–18. doi: 10.5962/bhl.part.10003
Chai C. Y., Jia R. R., Chen C. Y., Hui F. L. (2020). Blastobotrys baotianmanensis sp. nov. and Blastobotrys xishuangbannaensis f.a., sp. nov., two novel yeast species associated with insects and rotting wood. Int. J. Syst. Evol. Microbiol. 70, 4217–4223. doi: 10.1099/ijsem.0.004275
Chai C. Y., Lei T., Chu X. Y., Hui F. L. (2023). Multi-gene phylogeny and taxonomy of the genus Bannoa with the addition of three new species from central China. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1143156
Chai C. Y., Li Y., Yan Z. L., Hui F. L. (2022b). Phylogenetic and genomic analyses of two new species of Clavispora (Metschnikowiaceae, Saccharomycetales) from Central China. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1019599
Chen W. T., Tu M. E., Sun P. L. (2016). Superficial phaeohyphomycosis caused by Aureobasidium melanogenum mimicking tinea nigra in an immunocompetent patient and review of published reports. Mycopathologia 181, 555–560. doi: 10.1007/s11046-016-9989-3
Chi Z., Chi Z., Zhang T., Liu G., Li J., Wang X. (2009). Production, characterization and gene cloning of the extracellular enzymes from the marine-derived yeasts and their potential applications. Biotechnol. Adv. 27, 236–255. doi: 10.1016/j.bioteChadv.2009.01.002
Chi Z. M., Liu T. T., Chi Z., Liu G. L., Wang Z. P. (2012). Occurrence and diversity of yeasts in the mangrove ecosystems in Fujian, Guangdong and Hainan Provinces of China. Indian J. Microbiol. 52, 346–353. doi: 10.1007/s12088-012-0251-5
Chi Z. M., Liu G., Zhao S., Li J., Peng Y. (2010). Marine yeasts as biocontrol agents and producers of bio-products. Appl. Microbiol. Biotechnol. 86, 1227–1241. doi: 10.1007/s00253-010-2483-9
Chu S. B., Hu W. T., Hui F. L. (2022). Torulaspora jiuxiensis sp. nov., a novel yeast species isolated from rotting wood. Int. J. Syst. Evol. Microbiol. 72, 005629. doi: 10.1099/ijsem.0.005629
Desnos-Ollivier M., Letscher-Bru V., Neuvéglise C., Dromer F. (2020). Yarrowia lipolytica causes sporadic cases and local outbreaks of infections and colonisation. Mycoses 63, 737–745. doi: 10.1111/myc.13095
Douglass A. P., Offei B., Braun-Galleani S., Coughlan A. Y., Martos A. A. R., Ortiz-Merino R. A., et al. (2018). Population genomics shows no distinction between pathogenic Candida krusei and environmental Pichia kudriavzevii: one species, four names. PloS Pathog. 14, 1–27. doi: 10.1371/journal.ppat.1007138
Fell J. W., Boekhout T., Fonseca A., Scorzetti G., Statzell-Tallman A. (2000). Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 50, 1351–1371. doi: 10.1099/00207713-50-3-1351
Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Gao W. L., Li Y., Chai C. Y., Yan Z. L., Hui F. L. (2021). New species of Yamadazyma from rotting wood in China. MycoKeys 83, 69. doi: 10.3897/MYCOKEYS.83.71156
Gao W. L., Liu K. F., Yao L. G., Hui F. L. (2018). Pichia nanzhaoensis sp. nov. and Pichia paraexigua f.a., sp. nov., two yeast species isolated from rotting wood. Int. J. Syst. Evol. Microbiol. 68, 3311–3315. doi: 10.1099/ijsem.0.002989
Gao W. L., Liu T. T., Zheng J., Hui F. L. (2017). Kodamaea neixiangensis f.a., sp. nov. and Kodamaea jinghongensis f.a., sp. nov., two yeast species isolated from rotting wood. Int. J. Syst. Evol. Microbiol. 67, 3358–3362. doi: 10.1099/ijsem.0.002117
Hashem M., Al-Qahtani M. S., Alamri S. A., Moustafa Y. S., Lyberatos G., Ntaikou I. (2022). Valorizing food wastes: assessment of novel yeast strains for enhanced production of single-cell protein from wasted date molasses. Biomass Convers Biorefin 12, 4491–4502. doi: 10.1007/s13399-022-02415-2
Hoppert L., Kölling R., Einfalt D. (2022). Investigation of stress tolerance of Pichia kudriavzevii for high gravity bioethanol production from steam–exploded wheat straw hydrolysate. Bioresour. Technol. 364, 128079. doi: 10.1016/j.biortech.2022.128079
Huang L. N., Xi Z. W., Li Y., Hui F. L. (2018). Sugiyamaella xiaguanensis f.a., sp. nov., a yeast species isolated from rotting wood. Int. J. Syst. Evol. Microbiol. 68, 3307–3310. doi: 10.1099/ijsem.0.002988
Huang L. N., Xi Z. W., Zhai Y. C., Chai C. Y., Hui F. L. (2019). Saturnispora galanensis sp. nov., a yeast species isolated from rotting wood. Int. J. Syst. Evol. Microbiol. 69, 2658–2661. doi: 10.1099/ijsem.0.003501
Jia R., Lv S., Chai C., Hui F. L. (2020). Three new Scheffersomyces species associated with insects and rotting wood in China. MycoKeys 71, 87–99. doi: 10.3897/mycokeys.87.56168
Jones E. B. G., Suetrong S., Sakayaroj J., Bahkali A. H., Abdel-Wahab M. A., Boekhout T., et al. (2015). Classification of marine ascomycota, basidiomycota, blastocladiomycota and chytridiomycota. Fungal Divers. 73, 1–72. doi: 10.1007/s13225-015-0339-4
Kaewkrajay C., Chanmethakul T., Limtong S. (2020). Assessment of diversity of culturable marine yeasts associated with corals and zoanthids in the Gulf of Thailand, south China sea. Microorganisms 8, 474. doi: 10.3390/microorganisms8040474
Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Ke T., Zhai Y. C., Yan Z. L., Hui F. L. (2019). Kazachstania jinghongensis sp. nov. and Kazachstania menglunensis f.a., sp. nov., two yeast species isolated from rotting wood. Int. J. Syst. Evol. Microbiol. 69, 3623–3628. doi: 10.1099/ijsem.0.003670
Keene S., Sarao M. S., McDonald P. J., Veltman J. (2019). Cutaneous geotrichosis due to Geotrichum candidum in a burn patient. Access Microbiol. 1, 1–6. doi: 10.1099/acmi.0.000001
Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Nordic Hydrology 34, 281–294. doi: 10.2166/nh.2003.0008
Kurtzman C. P., Robnett C. J. (1998). Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 98, 331–371. doi: 10.1023/A:1001761008817
Lachance M. A. (2022). Phylogenies in yeast species descriptions: In defense of neighbor-joining. Yeast 39, 513–520. doi: 10.1002/yea.3812
Lei X., Zheng J., Zhao D., Qiao Z., An M., Zhang X. (2022). Moniliella aeria sp. nov., a novel yeast isolated from the air of a Wuliangye baijiu-making workshop. Int. J. Syst. Evol. Microbiol. 72, 005464. doi: 10.1099/ijsem.0.005464
Letunic I., Bork P. (2019). Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, 256–259. doi: 10.1093/nar/gkz239
Li S. S., Cheng C., Li Z., Chen J. Y., Yan B., Han B. Z., et al. (2010). Yeast species associated with wine grapes in China. Int. J. Food Microbiol. 138, 85–90. doi: 10.1016/j.ijfoodmicro.2010.01.009
Li Y. Y., Wang M. M., Groenewald M., Li A. H., Guo Y. T., Wu F., et al. (2022). Proposal of two new combinations, twenty new species, four new genera, one new family, and one new order for the anamorphic basidiomycetous yeast species in Ustilaginomycotina. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.777338
Li A. H., Yuan F. X., Groenewald M., Bensch K., Yurkov A. M., Li K., et al. (2020). Diversity and phylogeny of basidiomycetous yeasts from plant leaves and soil: proposal of two new orders, three new families, eight new genera and one hundred and seven new species. Stud. Mycol 96, 17–140. doi: 10.1016/j.simyco.2020.01.002
Liu J. Y. (2013). Status of marine biodiversity of the China seas. PloS One 8, 1–24. doi: 10.1371/journal.pone.0050719
Liu S., Guo Q. C., An Z. R., Hui F. L. (2023). Danielozyma pruni sp. nov., an asexual yeast species isolated from insect frass. Int. J. Syst. Evol. Microbiol. 73, 006124. doi: 10.1099/ijsem.0.006124
Lou Q. Z., Lu M., Sun J. H. (2014). Yeast diversity associated with invasive dendroctonus valens killing Pinus tabuliformis in China using culturing and molecular methods. Microb. Ecol. 68, 397–415. doi: 10.1007/s00248-014-0413-6
Lu Y. F., Wang M., Zheng J., Hui F. L. (2017). Ogataea neixiangensis sp. nov. and Ogataea paraovalis f.a., sp. nov., two methanol-assimilating yeast species isolated from rotting wood. Int. J. Syst. Evol. Microbiol. 67, 3038–3042. doi: 10.1099/ijsem.0.002075
Luo B., Sun H., Zhang Y., Gu Y., Yan W., Zhang R., et al. (2019). Habitat-specificity and diversity of culturable cold-adapted yeasts of a cold-based glacier in the Tianshan Mountains, northwestern China. Appl. Microbiol. Biotechnol. 103, 2311–2327. doi: 10.1007/s00253-018-9512-5
McMurdie P. J., Holmes S. (2013). Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8, e61217. doi: 10.1371/journal.pone.0061217
Murray N. J., Phinn S. R., DeWitt M., Ferrari R., Johnston R., Lyons M. B., et al. (2019). The global distribution and trajectory of tidal flats. Nature 565, 222–225. doi: 10.1038/s41586-018-0805-8
Nakase T., Takashima M. (1993). A simple procedure for the high frequency isolation of new taxa of ballistosporous yeasts living on the surfaces of plants. Riken Rev. 3, 33–34.
Nath U. U., Ravi R. (2013). Isolation of astaxanthin from marine yeast and study of its pharmacological activity. Int. curr. pharm. j. 2, 67–69. doi: 10.3329/icpj.v2i3.13584
Oksanen r J., Simpson G. L., Blanchet F. G., Solymos P., Stevens M. H. H., Szoecs E., et al. (2022). vegan: community ecology package. Available at: https://github.com/vegandevs/vegan.
Paradis E., Schliep K. (2019). Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528. doi: 10.1093/bioinformatics/bty633
Pereira G. V. M., Alvarez J. P., Neto D. P., de C., Soccol V. T., Tanobe V. O. A., et al. (2017). Great intraspecies diversity of Pichia kudriavzevii in cocoa fermentation highlights the importance of yeast strain selection for flavor modulation of cocoa beans. Lwt 84, 290–297. doi: 10.1016/j.lwt.2017.05.073
Pfaller M. A., Diekema D. J., Gibbs D. L., Newell V. A., Nagy E., Dobiasova S., et al. (2008). Candida krusei, a multidrug-resistant opportunistic fungal pathogen: geographic and temporal trends from the ARTEMIS DISK Antifungal Surveillance Progra to 2005. J. Clin. Microbiol. 46, 515–521. doi: 10.1128/JCM.01915-07
Pikazis D., Xynos I. D., Xila V., Velegraki A., Aroni K. (2009). Extended fungal skin infection due to Aureobasidium pullulans. Clin. Exp. Dermatol. 34, 892–894. doi: 10.1111/j.1365-2230.2009.03663.x
Qiao Y. Z., Chen X., Hui F. L. (2023). Barnettozyma menglunensis f.a., sp. nov., a novel yeast species isolated from rotting wood. Int. J. Syst. Evol. Microbiol. 73, 1–5. doi: 10.1099/ijsem.0.005711
R Core Team (2023) R: A language and environment for statistical computing (RVienna, Austria: Foundation for Statistical Computing). Available at: https://www.R-project.org (Accessed 16 June 2023).
Scorzetti G., Fell J. W., Fonseca A., Statzell-Tallman A. (2002). Systematics of basidiomycetous yeasts: A comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2, 495–517. doi: 10.1016/S1567-1356(02)00128-9
Shi C. F., Zhang K. H., Chai C. Y., Yan Z. L., Hui F. L. (2021). Diversity of the genus Sugiyamaella and description of two new species from rotting wood in China. MycoKeys 77, 27–39. doi: 10.3897/MYCOKEYS.77.60077
Soki H., Abo K., Yamazaki K., Kojima T., Oda T., Uzawa Y., et al. (2015). First report of intraorbital abscess caused by Candida allociferrii and specific PCR for differentiating Stephanoascus ciferrii complex species. Med. Mycol J. 56, E9–E14. doi: 10.3314/mmj.56.E9
Vu D., Groenewald M., Szöke S., Cardinali G., Eberhardt U., Stielow B., et al. (2016). DNA barcoding analysis of more than 9 000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud. Mycol. 85, 91–105. doi: 10.1016/j.simyco.2016.11.007
Wang Q. M., Bai F. Y. (2008). Molecular phylogeny of basidiomycetous yeasts in the Cryptococcus luteolus lineage (Tremellales) based on nuclear rRNA and mitochondrial cytochrome b gene sequence analyses: Proposal of Derxomyces gen. nov. and Hannaella gen. nov., and description of eight novel Derxomyces species. FEMS Yeast Res. 8, 799–814. doi: 10.1111/j.1567-1364.2008.00403.x
Wang W. L., Chi Z. M., Chi Z., Li J., Wang X. H. (2009). Siderophore production by the marine-derived Aureobasidium pullulans and its antimicrobial activity. Bioresour. Technol. 100, 2639–2641. doi: 10.1016/j.biortech.2008.12.010
Wang L., Chi Z., Wang X., Ju L., Chi Z., Guo N. (2008). Isolation and characterization of Candida membranifaciens subsp. flavinogenie W14-3, a novel riboflavin-producing marine yeast. Microbiol. Res. 163, 255–266. doi: 10.1016/j.micres.2007.12.001
Wang Q., Cui Y., Sen B., Ma W., Zheng R. L., Liu X., et al. (2017). Characterization and robust nature of newly isolated oleaginous marine yeast Rhodosporidium spp. from coastal water of Northern China. AMB Expr. 7, 30. doi: 10.1186/s13568-017-0329-x
Wang H., Wang Y., Chen J., Zhan Z., Li Y., Xu J. (2007). Oral yeast flora and its ITS sequence diversity among a large cohort of medical students in Hainan, China. Mycopathologia 164, 65–72. doi: 10.1007/s11046-007-9028-5
Wang Q. M., Liu W. Q., Liti G., Wang S. A., Bai F. Y., et al. (2012). Surprisingly diverged populations of Saccharomyces cerevisiae in natural environments remote from human activity. Mol. Ecol. 21, 54045417. doi: 10.1111/j.1365-294X.2012.05732.x
Wei X. Y., Zhu H. Y., Song L., Zhang R. P., Li A. H., Niu Q. H., et al. (2022). Yeast diversity in the Qaidam basin desert in China with the description of five new yeast species. J. Fungi 8, 858. doi: 10.3390/jof8080858
World Health Organization (2022) WHO fungal priority pathogens list to guide research, development and public health action. Available at: https://www.who.int/publications/i/item/9789240060241 (Accessed October 25, 2022).
Wu Q., Xu Y., Chen L. (2012). Diversity of yeast species during fermentative process contributing to chinese maotai-flavour liquor making. Lett. Appl. Microbiol. 55, 301–307. doi: 10.1111/j.1472-765X.2012.03294.x
Xi Z. W., Huang L. N., Li Y., Hui F. L. (2019). Vanrija jinghongensis sp. nov., an asexual basidiomycetous yeast from rotting wood. Int. J. Syst. Evol. Microbiol. 69, 105–108. doi: 10.1099/ijsem.0.003108
Yang S. P., Wu Z. H., Jian J. C. (2011). Distribution of marine red yeasts in shrimps and the environments of shrimp culture. Curr. Microbiol. 62, 1638–1642. doi: 10.1007/s00284-011-9910-8
Yu H. T., Shang Y. J., Zhu H. Y., Han P. J., Wang Q. M., Santos A. R. O., et al (2023). Yueomyces silvicola sp. nov., a novel ascomycetous yeast species unable to utilize ammonium, glutamate, and glutamine as sole nitrogen sources. Yeast 110. doi: 10.1002/yea.3901
Zaky A. S., Greetham D., Louis E. J., Tucker G. A., Du C. (2016). A new isolation and evaluation method for marine-derived yeast spp. with potential applications in industrial biotechnology. J. Microbiol. Biotechnol. 26, 1891–1907. doi: 10.4014/jmb.1605.05074
Zaky A. S., Tucker G. A., Daw Z. Y., Du C. (2014). Marine yeast isolation and industrial application. FEMS Yeast Res. 14, 813–825. doi: 10.1111/1567-1364.12158
Zheng J., Lu Y. F., Liu X. J., Hui F. L. (2017). Cyberlindnera xishuangbannaensis f.a., sp. nov., a yeast isolated from rotting wood. Int. J. Syst. Evol. Microbiol. 67, 5051–5055. doi: 10.1099/ijsem.0.002411
Zhou J., Deng Y., Shen L., Wen C., Yan Q., Ning D., et al. (2016). Temperature mediates continental-scale diversity of microbes in forest soils. Nat. Commun. 7, 12083. doi: 10.1038/ncomms12083
Zhu S. S., Lei Y. H., Wang C., Wei Y. M., Wang C. C., Sun Y. F. (2021). Patterns of yeast diversity distribution and its drivers in rhizosphere soil of Hami melon orchards in different regions of Xinjiang. BMC Microbiol. 21, 1–18. doi: 10.1186/s12866-021-02222-1
Zhu H. Y., Wei Y. H., Guo L. C., Wei X. Y., Li J. N., Zhang R. P., et al. (2023a). Vishniacozyma pseudocarnescens sp. nov., a new anamorphic tremellomycetous. Int. J. Syst. Evol. Microbiol. 73, 6076. doi: 10.1099/ijsem.0.006076
Keywords: marine yeasts, species diversity, ecology, intertidal zones, China
Citation: Zhu H-Y, Han D-Y, Guo L-C, Li J-N, Wei X-Y, Zhang R-P, Wang Q-M, Shang Y-J, Luo L-J, Wei Y-H, Liu X-Z and Bai F-Y (2023) Diversity and distribution of yeasts in intertidal zones of China. Front. Mar. Sci. 10:1286511. doi: 10.3389/fmars.2023.1286511
Received: 31 August 2023; Accepted: 13 November 2023;
Published: 07 December 2023.
Edited by:
Chenyu Du, University of Huddersfield, United KingdomReviewed by:
Darren Greetham, University of Leeds, United KingdomBao-zhu Fang, Chinese Academy of Sciences (CAS), China
Copyright © 2023 Zhu, Han, Guo, Li, Wei, Zhang, Wang, Shang, Luo, Wei, Liu and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng-Yan Bai, YmFpZnlAaW0uYWMuY24=
†These authors have contributed equally to this work
 Hai-Yan Zhu
Hai-Yan Zhu Da-Yong Han
Da-Yong Han Liang-Chen Guo1,2
Liang-Chen Guo1,2 Jun-Ning Li
Jun-Ning Li Ri-Peng Zhang
Ri-Peng Zhang Qi-Ming Wang
Qi-Ming Wang Lu-Jun Luo
Lu-Jun Luo Xin-Zhan Liu
Xin-Zhan Liu Feng-Yan Bai
Feng-Yan Bai