
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 19 December 2023
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1270968
 Christopher J. Payne1
Christopher J. Payne1 Kathryn Grace1
Kathryn Grace1 Vo Hong Phuong2
Vo Hong Phuong2 Nguyen Ngoc Phuoc3
Nguyen Ngoc Phuoc3 Tu Thanh Dung4
Tu Thanh Dung4 Le Hong Phuoc2
Le Hong Phuoc2 Margaret Crumlish1*
Margaret Crumlish1*Since first being identified in 1999, disease outbreaks from Edwardsiella ictaluri remain a significant health challenge for the farmed catfish sector in Vietnam. To better understand the population structure of E. ictaluri circulating in Vietnamese striped catfish (Pangasianodon hypophthalmus) farms, this study aimed to investigate the genetic diversity of 83 E. ictaluri isolates recovered from natural disease outbreaks occurring in the Mekong Delta region between 2001 and 2021. Pulsed-field gel electrophoresis resolved isolates into 15 pulsotypes following restriction digest with speI, with a Simpson’s diversity index of 0.8548. The genetic fingerprints of isolates recovered from recent outbreaks across different provinces were highly shared (100% similarity), suggesting clonal expansion within the production systems situated in the Mekong Delta region. Findings from this study also showed that E. ictaluri populations circulating this region have changed over time, as isolates recovered between 2001 and 2011 were genetically distinct from those recovered after 2017. Furthermore, isolates recovered after 2004 and 2010 lacked the virulence gene traD and a 5.8 Kb plasmid DNA fragment, respectively. Findings from this study illustrate the need for continued epidemiological monitoring of E. ictaluri to ensure prevention and treatment strategies in the Vietnamese striped catfish sector remain robust and effective.
Edwardsiella ictaluri is described as an intracellular, facultative, Gram-negative rod-shaped bacterium and member of the Hafniaceae family (Adeolu et al., 2016). Historically, E. ictaluri was described as biochemically homogenous, testing positive for lysine decarboxylase activity and glucose metabolism, however host-dependent differences have expanded this description to include isolates positive for ornithine decarboxylase and cytochrome oxidase activities as well as citrate utilisation and H2S production (Rogge et al., 2013; Machimbirike et al., 2022). Recently, published genomes of E. ictaluri have revealed a genome size ranging from 3.6 – 4.0 Mb, with G+C (%) of 57.0 to 57.6 and the presence of a diverse collection of antimicrobial resistance and virulence genes (Abdelhamed et al., 2018; Machimbirike et al., 2021). The bacterium E. ictaluri was initially reported to have a narrow host range (Hawke et al., 1981), however has since been found to infect 44 susceptible hosts, either naturally or experimentally, including striped catfish (Pangasianodon hypophthalmus) (Crumlish et al., 2002; Ngoc Phuoc et al., 2020; Machimbirike et al., 2022).
Vietnamese production of striped catfish has intensified in recent years to support the growing demand for aquatic animal protein, increasing from 40,000 tonnes in 1997 to 1.5 million tonnes in 2020, worth $1.7 billion USD (FAO, 2021). However, this intensification has been accompanied by increased incidence of disease outbreaks across the production sector (Hoa et al., 2021). The bacterium E. ictaluri was first identified as the aetiological agent of bacillary necrosis of Pangasius (BNP) in striped catfish from Vietnam over two decades ago (Crumlish et al., 2002). The clinical signs of BNP have remained consistent over time, where diseased fish typically show erratic swimming as well as swollen abdomens, and internally present multifocal white lesions on the liver, kidney and spleen (Ngoc Phuoc et al., 2020). Despite its historical presence within the country, recent surveillance of the production sector suggests outbreaks of BNP are on the rise (Hoa et al., 2021). This is particularly concerning given that the disease causes significant economic losses on the farm, affecting all stages of development with up to 90% mortality rates reported (Vu et al., 2019), thereby requiring effective control strategies to combat infection.
Epidemiological investigation of disease outbreaks is vital to inform health management policies, as it provides criteria on which to base surveillance and vaccination programmes. Whilst alternative high-resolution methods are available for bacterial identification (e.g., whole genome sequencing), pulsed-field gel electrophoresis (PFGE) is considered the gold-standard of genotyping methods for molecular characterisation and to determine the genetic relatedness of bacterial isolates during disease outbreaks, due to its low cost, high discriminatory power, reproducibility and ease of interpretation (Duman et al., 2022; Ramadan, 2022; Sciuto et al., 2022). Indeed, this method has been used previously to type several aquatic bacterial pathogens including Aeromonas salmonicida subsp. salmonicida, Edwardsiella tarda, Flavobacterium psychrophilum, Vibrio parahaemolyticus and Yersinia ruckeri (García et al., 2000; Yang et al., 2013; Narayanan et al., 2020; Calvez et al., 2021; Feng et al., 2022). Furthermore, PFGE has been previously applied to understand the genetic diversity of E. ictaluri isolates recovered from diseased striped catfish in Vietnam between 2001 and 2004 (Bartie et al., 2012), which found that Vietnamese isolates were genetically distinct from those recovered from channel catfish in United States (Bartie et al., 2012). Geographical or host-dependant genotypes in E. ictaluri have also been confirmed through other typing methods, whereby isolates from Southeast Asia can differ in their plasmid DNA and virulence gene profiles compared with American isolates (Rogge et al., 2013; Dong et al., 2019). Since the early studies by Bartie et al. (2012) and Rogge et al. (2013), and subsequent whole genome sequencing of an isolate from Northern Vietnam (Machimbirike et al., 2021) were completed, no further genotyping studies on Vietnamese E. ictaluri have been performed to the best of our knowledge. There is therefore a lack of understanding on the genetic diversity that exists in E. ictaluri populations currently circulating the striped catfish sector in Vietnam. This information is vital to understand the epidemiological spread of E. ictaluri populations in the Vietnamese striped catfish sector and better inform effective control strategies.
Considering the epidemic nature of E. ictaluri in Vietnam and scarcity in knowledge surrounding the epidemiology of this species, the aim of this study was to characterise the genetic diversity of E. ictaluri recovered from clinical disease outbreaks in Vietnamese farmed striped catfish over a twenty-year period (2001 and 2021). The use of PFGE as well as profiling plasmid DNA and virulence genes was applied to better understand the structure of E. ictaluri populations currently circulating striped catfish farms in Vietnam and how these populations have changed over time.
The 83 E. ictaluri isolates used in this study (Table S1) were recovered from naturally infected striped catfish between 2001 and 2021 across six provinces in Vietnam (Figure 1). All E. ictaluri isolates were recovered from the kidney or liver of fish showing clinical signs of BNP including white spots on the kidney, liver or spleen, during an active disease outbreak. The isolates E. ictaluri NCIMB 13272, E. ictaluri ATCC 93-146 and Edwardsiella piscicida NCIMB 14824, served as an American representative and controls in different assays, respectively (Table S2). All bacterial isolates were recovered from storage on Protect Beads (SWA1147; SLS, UK) at -80°C onto tryptone soya agar (TSA) (CM0131, Oxoid, UK), where they were incubated at 28°C for 72 hours.
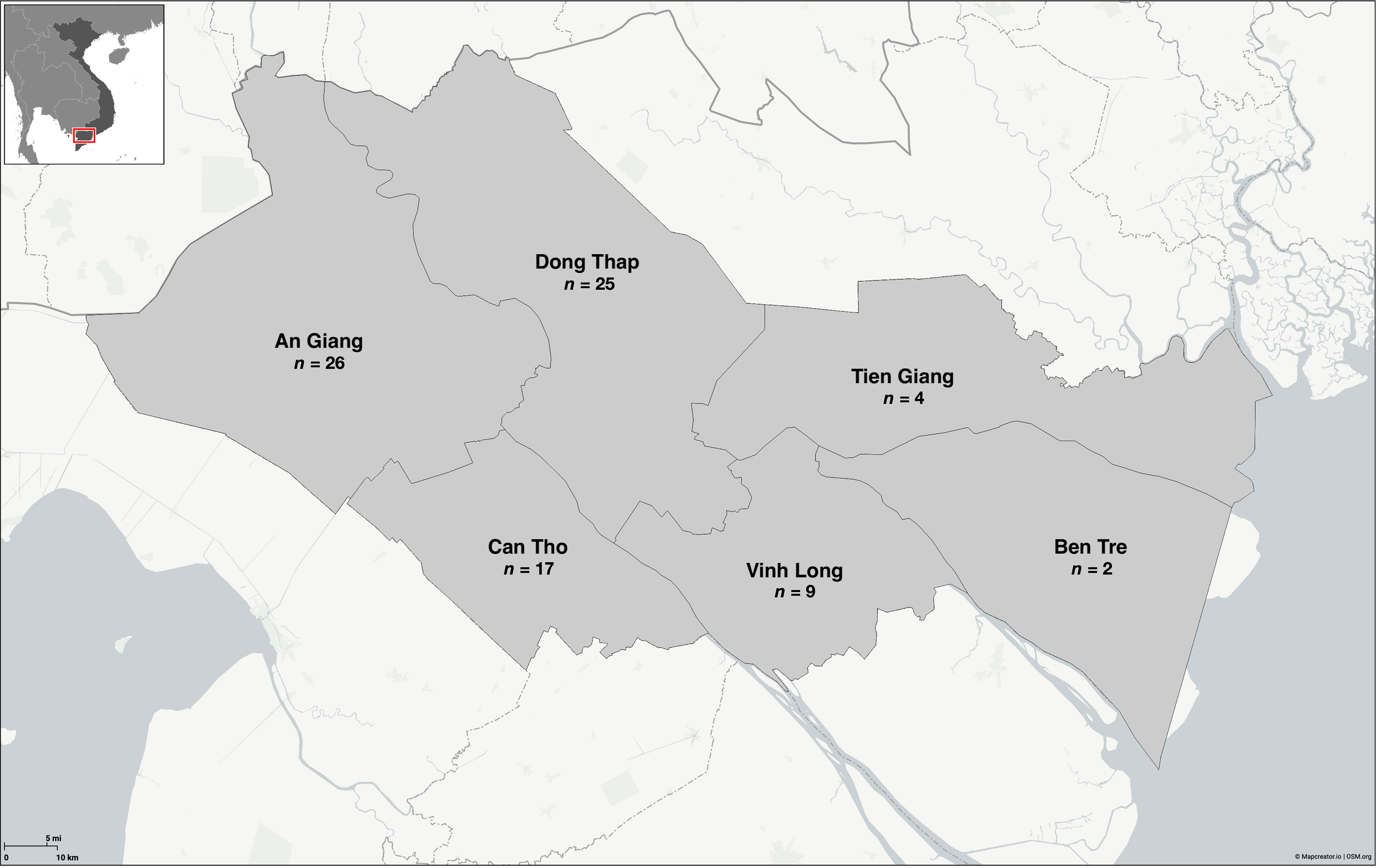
Figure 1 Distribution of Edwardsiella ictaluri isolates recovered from disease outbreaks in striped catfish (Pangasianodon hypophthalmus) farms in the Mekong Delta region of Vietnam. Map created using Mapcreator (https://mapcreator.io).
Genomic DNA was extracted from bacteria using the method described by Queipo-Ortuńo et al. (2008), except final DNA was eluted in 500 μL TE (10 mM tris pH 8.0, 1 mM EDTA pH 8.0). The quality and concentration of extracted DNA samples was measured using the Nanodrop 2000c spectrophotometer (Fisher Scientific, UK) and 10 μL aliquots stored at -20°C, until required.
Primary identification was performed as described by Frerichs and Millar (1993) and biochemical profiles obtained using the commercial API®20E biochemical strips (20100, BioMerieux Ltd, UK), following the manufacturer’s instructions, except incubating strips at 28°C for 48 hours before recording results.
All bacterial isolates were identified as E. ictaluri by species-specific PCR using the primer set EDi-F (5’- CAGATGAGCGGATTTCACAG –’3) and EDi-R (5’- CGCGCAATTAACATAGAGCC -’3) (Sakai et al., 2009). Briefly, a 10 μL PCR reaction was prepared for each bacterial isolate, which comprised 5 μL 2 X HS mytaQ mastermix (BIO-25045, Bioline, UK), 0.5 μL (5 pM) of each forward and reverse primer, 3 μL nuclease-free water and 10 ng DNA. Genomic DNA from E. ictaluri NCIMB 13272 and E. piscicida NCIMB 14824 was also included in the analysis, where they served as positive and negative controls, respectively. In addition, a no template control (NTC) was also included to confirm PCR reagents and subsequent reactions were free from microbial DNA contamination. The E. ictaluri specific fimbrial gene was then amplified using the PCR conditions; 95°C for two minutes, then 30 x cycles at 95°C for 15 seconds, 65°C for 20 seconds, 72 °C for 30 seconds, with a final extension at 72°C for two minutes. Following PCR, PCR products were visualised on a 1.5% agarose gel containing ethidium bromide (0.06 μg/mL), following electrophoresis at 11 V/cm for 20 minutes. Isolates were identified as E. ictaluri based on the presence of a single PCR product with a molecular weight of 470 bp.
Macrorestriction analysis of bacterial isolates was performed using PFGE, following the protocol described by Bartie et al. (2012) with minor modifications. Briefly, 24-hour bacterial cultures were washed in 1 mL STE buffer (10 mM tris pH 8.0, 1 mM EDTA pH 8.0, 100 mM NaCl) before being resuspended in STE buffer to an OD600nm of 0.8, as measured by a cell density meter (Biowave CO 8000; Biochrom, UK). One percent agar plugs were then prepared using equal volumes (100 μL) of bacterial suspension and a 2% PFGE agarose (A2929, Sigma, UK) solution. In addition, bacteria cells were lysed in 3 mL lysis buffer (10 mM tris pH 8.0, 1 mM EDTA pH 8.0, 1% sarcosine, 1 mg/mL proteinase K) at 55°C and 175 rpm for 18 hours. Plug washing, restriction digest of DNA with speI-HF (R3133, NEB, UK) and electrophoresis was performed following Bartie et al. (2012). Following PFGE, gels were stained in an ethidium bromide solution (1 μg/mL) for 30 minutes, followed by two consecutive washes in distilled water for 20 minutes and visualised under UV illumination.
Plasmid DNA was extracted from 17-hour cultures using the QIAprep® Spin Miniprep Kit (27104, Qiagen, UK), following the manufacturer’s instructions with minor modifications. Briefly, the QIAprep® columns were washed in 500 μL buffer PB. In addition, plasmid DNA was eluted in 100 μL pre-heated buffer EB. Following extraction, 250 ng plasmid DNA was digested with EcoRI-HF (R3101, NEB, UK) (Dong et al., 2019) and visualised on a 0.8% agarose gel in 1 X TAE (40 mM tris, 20 mM acetic acid, 1 mM EDTA) and ethidium bromide (0.25 μg/mL), following electrophoresis at 5 V/cm for 90 minutes. Plasmid DNA was visualised alongside a 1 Kb DNA ladder (Fisher Scientific, UK) under UV illumination.
The presence of seven putative virulence genes (eseJ, traD, hcp, fldA, wzx, fhuC and ureE) was detected in this study using primer pairs listed in Table 1. Briefly, a 10 μL PCR reaction was prepared for each bacterial isolate, which comprised 5 μL 2 X HS mytaQ mastermix, 0.5 μL (5 pM) of each forward and reverse primer, 3 μL nuclease-free water and 10 ng DNA. Primer sets were designed against the genome of E. ictaluri ATCC 93-146 (Accession: NC_012779.2) using the NCBI primer design software (Ye et al., 2012). Genomic DNA from E. ictaluri ATCC 93-146 and E. piscicida NCIMB 14824 was also included in the analysis, where they served as positive and negative controls, respectively. In addition, a NTC was also included to confirm PCR reagents and subsequent reactions were free from microbial DNA contamination. Virulence genes were amplified in DNA samples using the following PCR conditions: 95°C for two minutes, then 30 x cycles at 95°C for 15 seconds, 62-64°C for 15 seconds and 72°C for 30 seconds, followed by a final elongation stage at 72°C for two minutes. Following PCR, PCR products were visualised on a 1.5% agarose gel containing ethidium bromide (0.06 μg/mL), following electrophoresis at 11 V/cm for 20 minutes. The annealing temperature and product size of each virulence gene is given in Table 1.
Genetic fingerprints were analysed by GelJ (Version 2) (Heras et al., 2015). Fingerprints for each bacterial isolate were clustered based on the Dice Coefficient with tolerance set to 1%. A dendrogram was then generated to visualise clusters using the unweighted pair group method with arithmetic mean or “UPGMA” algorithm. Isolates that had DNA fingerprints with similarities of more than 86% were grouped into the same pulsotype and considered closely or possibly related (Feng et al., 2022). Isolates which had identical DNA fingerprints were termed clonal isolates. To determine the genetic diversity of the bacterial isolate collection from Vietnam, Simpson’s Diversity Index (SDI) was calculated based on pulsotypes detected and with a confidence interval (CI) of 95% (Carriço et al., 2006). A binary matrix of presence/absence data was generated for plasmid DNA band and virulence gene datasets to determine similarities between isolates based on Euclidean distance and complete linkage (UPGMA) using Rstudio 1.4.1717. Dendograms and heatmaps for plasmid DNA and virulence gene data were generated using Rstudio 1.4.1717 and “factoextra” and “ggplot2” packages. The distribution of plasmid DNA bands and virulence genes were compared between PFGE groups, province and year using the Chi-square (X2) or Fisher’s exact tests on JMP Pro software (Version 17.0.0), with significance determined when p < 0.05.
All isolates were confirmed to be E. ictaluri and possessed the fimbrial gene as detected by species-specific PCR (Figure S1). All isolates were described as being Gram-negative rods, non-motile, negative for cytochrome oxidase and fermentative by the OF test (Table S1). Eighty-one (98%) isolates were positive for lysine decarboxylase and glucose metabolism on the API20E system, giving the numerical profile 4004000 (Table S1). Two (2%) isolates, isolated in 2003 and 2017, were found to also be positive for citrate utilisation in addition to lysine decarboxylase and glucose metabolism, giving a numerical profile of 4204000 (Table S1).
Eighty-four E. ictaluri isolates were successfully typed by PFGE using speI, which yielded at least 21 well-resolved DNA fragments ranging in size from 20 – 540 Kb (Figure 2). Isolates were distributed into three distinct clusters based on their DNA fingerprint (≤ 75% similarity). Within these clusters, 13 pulsotypes (≥ 86% similarity) and three singletons were identified (Figure 2). Cluster I was represented by the American type isolate (Pulsotype A), which was distinct from the Vietnamese isolates that clustered into pulsotypes B – D and E – P, within Clusters II and III, respectively (Figure 2). Cluster II was represented by 10% of isolates (n = 8) recovered from multiple provinces in Vietnam between 2001 and 2003 (Figure 2). The remaining Vietnamese isolates (89%; n = 75) representing disease outbreaks across all surveyed provinces between 2001 and 2021 were distributed into Cluster III (Figure 2). Within Cluster III, the DNA fingerprints of isolates recovered between 2001 and 2011 (pulsotypes H - L) were found to group separately from those of isolates recovered after 2017 (pulsotypes E – G and M - P) (Figure 2). The SDI of Vietnamese isolates in this study was 0.8548 (95% CI 0.8545 – 0.8551). Furthermore, the SDI was found to reduce over time as E. ictaluri isolates recovered between 2001 and 2011 had an SDI of 0.7503 (95% CI of 0.7497 – 0.7509), whereas those recovered after 2017 had an SDI of 0.6598 (95% CI of 0.6595 – 0.6600). Clonal isolates were detected in several pulsotypes including M & P, which comprised clonal isolates recovered from An Giang and Dong Thap in 2019 and 2020 (Figure 2). Genetically distinct isolates (< 86% similarity) recovered from the same farm in the same year were also detected on several occasions, including An Giang in 2002, Can Tho in 2002 and 2020, Dong Thap in 2021, Tien Giang in 2020 and Vihn Long in 2003 and 2020 (Figure 2).

Figure 2 Cluster analysis of speI macrorestriction profiles of 84 Edwardsiella ictaluri isolates. Dendogram was generated using DICE and UPGMA algorithms with band tolerance set to 1%. Pulsotypes were defined by a cut-off value of ≥ 86% similarity (dashed line). For each isolate, PFGE cluster, pulsotype, year of recovery, province and farm are shown. Provinces include An Giang (AG), Ben Tre (BT), Can Tho (CT), Dong Thap (DT), Tien Giang (TG), United States of America (USA) and Vinh Long (VL). UF, unknown farm.
Pulsotype P was the most prevalent of all types, accounting for 27% (n = 23) of all isolates, closely followed by K (23%; n = 19) (Tables 2, 3). An Giang, Dong Thap and Can Tho were the most represented provinces surveyed, contributing 31% (n = 26), 30% (n = 25) and 20% (n = 17) of Vietnamese isolates investigated, respectively (Table 2). Within these provinces, pulsotype K was found to dominate An Giang and Can Tho, accounting for 29% (n = 10) and 41% (n = 7) of isolates within this pulsotype, respectively, whereas pulsotype P was found to dominate Dong Thap (44%; n = 11) (Table 2). On further investigation, pulsotype K was detected between 2001 and 2006, whereas pulsotype P was only detected after 2017 (Table 3).

Table 2 Distribution of pulsotypes of Edwardsiella ictaluri isolates recovered from disease outbreaks in striped catfish (Pangasianodon hypophthalmus) farms in different provinces of Vietnam between 2001 and 2021.
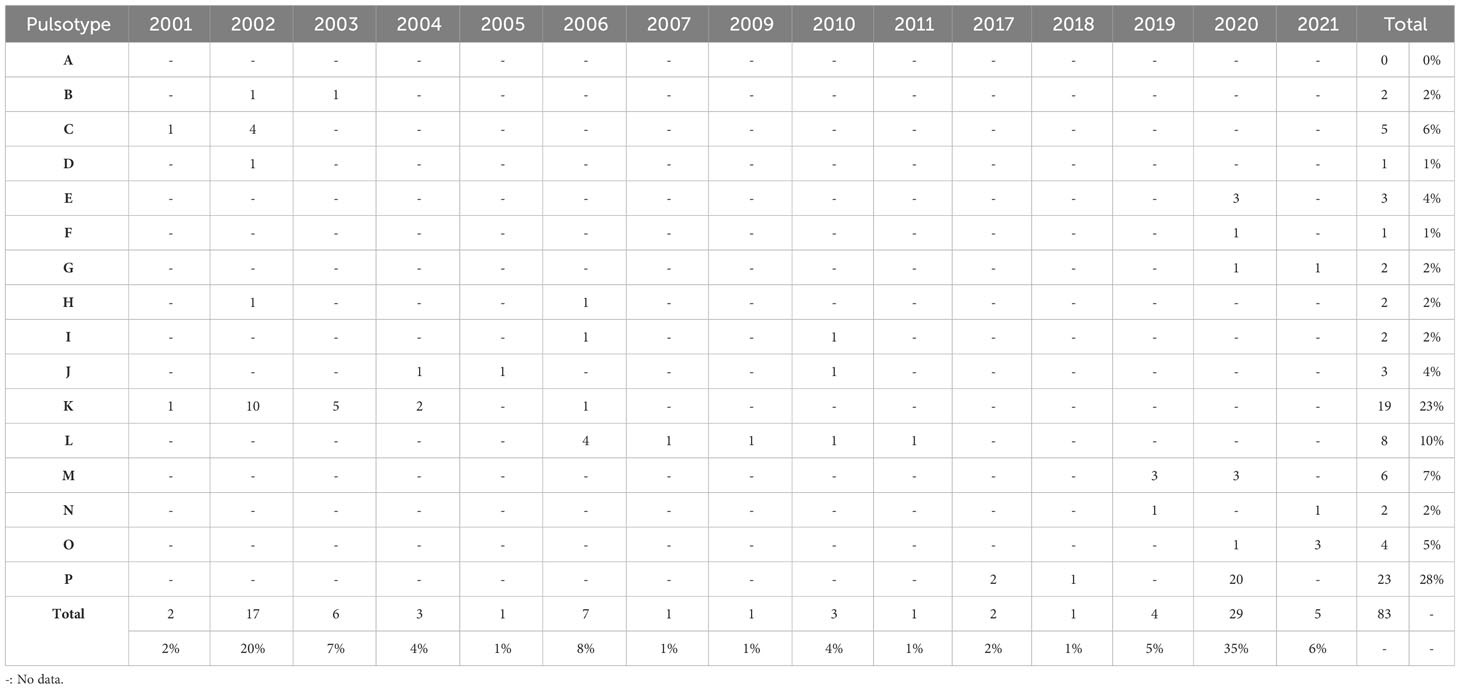
Table 3 Distribution of pulsotypes of Edwardsiella ictaluri isolates recovered from disease outbreaks in striped catfish (Pangasianodon hypophthalmus) farms in Vietnam between 2001 and 2021.
The EcoRI digestion of plasmid DNA was successfully performed on all E. ictaluri isolates screened and revealed 30 distinct plasmid DNA patterns across the collection (Figure 3). All isolates had at least one plasmid DNA band, however variability was noted between isolates in both the number (range: 1 – 8 bands) and size of plasmid DNA bands (range: 2.3 – more than 20 Kb) detected (Figure 3). No conserved plasmid DNA band was detected across the isolate collection (Figure 3). Within the Vietnamese isolates, a plasmid DNA band at 3.8 Kb was the most frequently detected, found in 98% (n = 81) of isolates screened, followed by a 4.8 Kb plasmid DNA band (52%; n = 43) (Figure 3). Forty (48%) Vietnamese isolates were found to harbour both 3.8 Kb and 4.8 Kb plasmid DNA bands (Figure 3). The American type isolate was found to have a distinct plasmid restriction profile, with two plasmid DNA bands observed at 4.8 Kb and 5.8 Kb (Figure 3). A similar sized plasmid DNA band at 5.8 Kb was also detected in 39% (n = 32) of Vietnamese isolates, although both year of recovery (p < 0.0001) and PFGE cluster (p = 0.0002) was found to influence the detection of this band across the isolate collection. In fact, the 5.8 Kb plasmid DNA band was only detected in those isolates recovered between 2001 and 2009 and was more commonly associated with isolates within PFGE Cluster II compared with Cluster III (Figure 3).
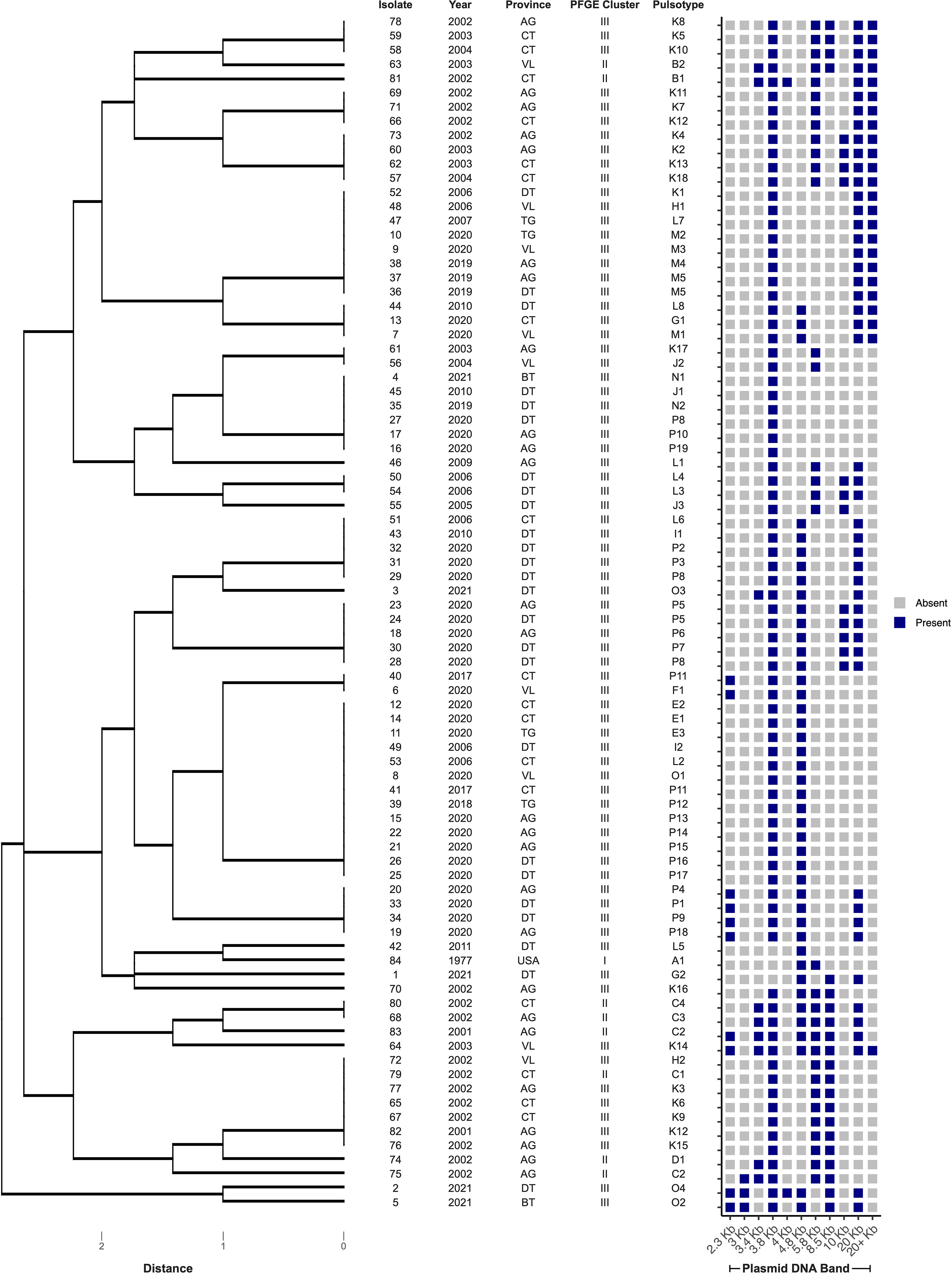
Figure 3 Cluster analysis of plasmid restriction DNA profiles of 84 Edwardsiella ictaluri isolates following digestion by EcoRI. Dendograms were generated using Euclidean distance and UPGMA. Provinces include An Giang (AG), Ben Tre (BT), Can Tho (CT), Dong Thap (DT), Tien Giang (TG), United States of America (USA) and Vinh Long (VL).
The presence of seven genes, representing suspected virulence factors in E. ictaluri, were screened by PCR with bands at the expected molecular size considered positive results (Figure S2). Four virulence gene profiles were identified across the isolate collection in this study, where profile III was most frequently detected (61%; n = 51) (Table 4). Further, virulence gene profiles I and III were more associated with isolates in PFGE Cluster III, whereas profiles II and IV were more associated with PFGE Cluster II (p < 0.0001) (Figure 4). All isolates possessed the genes hcp, fldA, wzx, fhuC and ureE but there was variation in whether isolates harboured the eseJ and traD genes (Figure 4). A total of 56 (67%) isolates, were positive for carrying eseJ (Figure 4). Whilst the proportion of isolates harbouring eseJ was higher in An Giang, Ben Tre, Dong Thap and Vinh Long (p = 0.0306), the presence of eseJ did not relate with specific PFGE clusters or year of recovery (p > 0.05) (Figure 4). The gene traD was only detected in 12% of isolates (n = 10) and was more associated with isolates recovered between 2001 and 2003 (p = 0.0188) and PFGE cluster II (p < 0.0001) (Figure 4). Six (7%) isolates including the American type isolate were found to harbour all seven virulence genes screened (Figure 4) and for Vietnamese isolates were more likely to be associated with PFGE Cluster II (p < 0.0001).
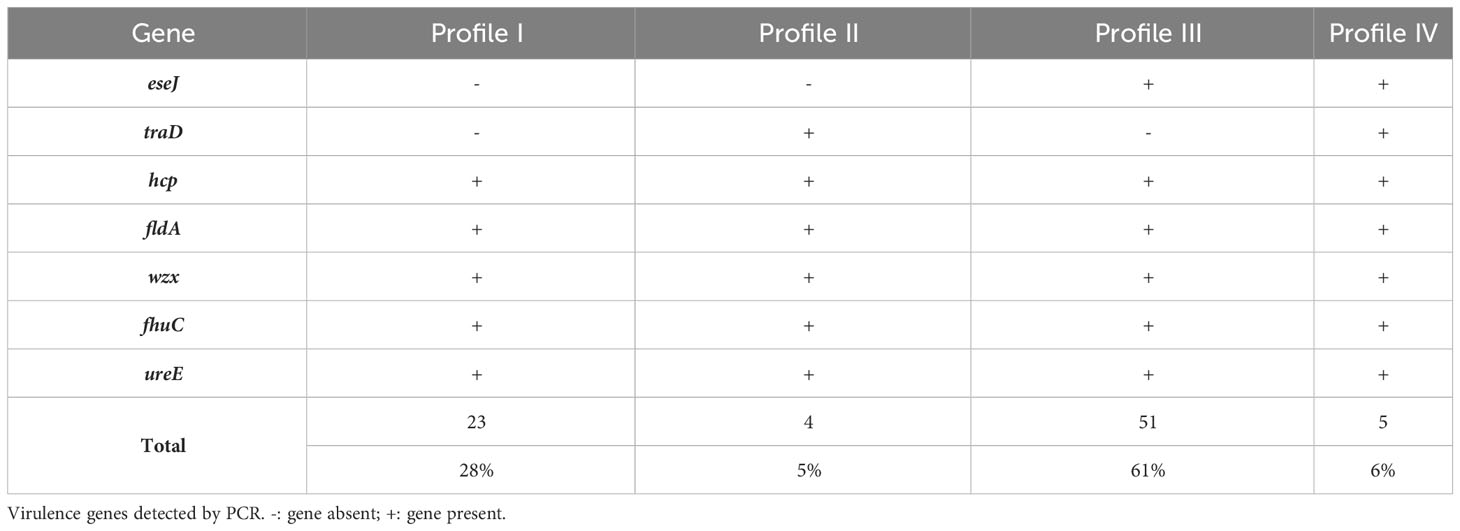
Table 4 Virulence gene profiles of Edwardsiella ictaluri isolates recovered from disease outbreaks in striped catfish (Pangasianodon hypophthalmus) farms in Vietnam between 2001 and 2021.
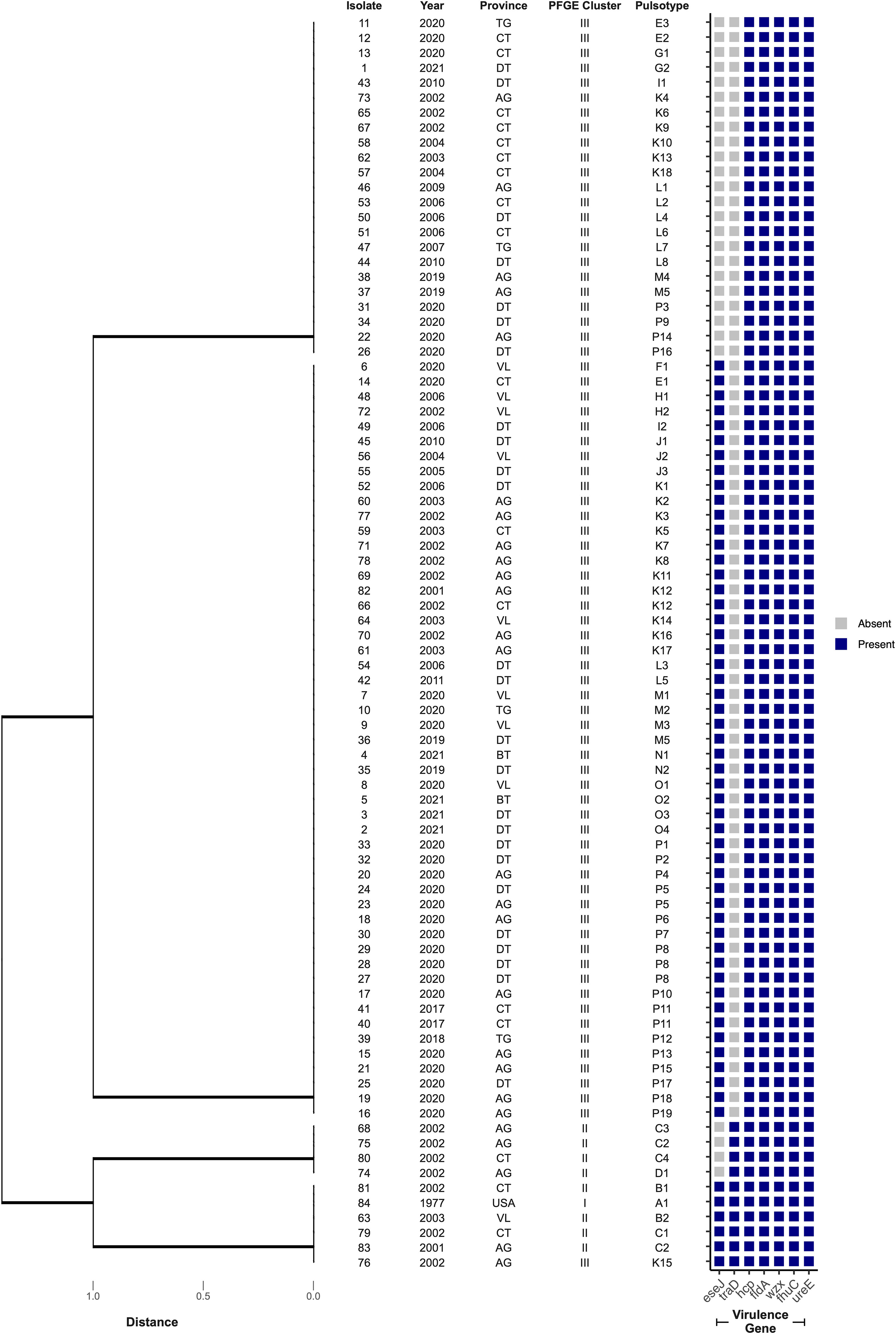
Figure 4 Cluster analysis of virulence gene profiles of 84 Edwardsiella ictaluri isolates. Dendograms were generated using Euclidean distance and UPGMA. Provinces include An Giang (AG), Ben Tre (BT), Can Tho (CT), Dong Thap (DT), Tien Giang (TG), United States of America (USA) and Vinh Long (VL).
Exploring strain diversity of bacterial pathogens over time is critical to the development and maintenance of efficacious prevention strategies. In this study, the PFGE fingerprints of isolates from Vietnam clustered away from the type isolate recovered from United States, supporting previous work which suggest that American and Asian E. ictaluri isolates are genetically distinct (Bartie et al., 2012). Furthermore, results from the present study demonstrated low genetic diversity in E. ictaluri populations recovered from recent outbreaks of BNP in the Mekong Delta region of Vietnam, when compared with populations from historical outbreaks. These findings also support previous work by Bartie et al. (2012), who successfully typed 47 E. ictaluri isolates from disease outbreaks in the same region between 2001 and 2004 using the same restriction enzyme. In their study, isolates resolved into 16 pulsotypes, corresponding to an SDI of 0.8936 (95% CI of 0.8874 – 0.8999), when typed using speI and applying a similar clustering threshold of ≥ 86% (data not shown). Findings from the current study identified a low genetic diversity of E. ictaluri overall, irrespective of outbreak period, as the SDI of the complete Vietnamese collection in this study was lower than the SDIs of other aquatic bacterial pathogen populations including E. tarda (SDI = 0.897) and Y. ruckeri (SDI = 0.93), when typed using speI or other restriction enzymes, respectively (Yang et al., 2013; Calvez et al., 2015). These findings are promising from the perspective of developing effective control strategies, as high pathogen diversity can be a major challenge for the development of cross-protective vaccines (Kennedy et al., 2020; Munang’andu et al., 2020).
The genomic profiles presented in this study identified genetic changes in the E. ictaluri populations circulating within the Vietnamese catfish farms. The mechanisms behind the genetic shifts identified within this bacterial population have not been elucidated in this study, but the virulence profile data may provide insight into changes in the pathogenesis from the varied pulsotypes. The gene traD, encoded within the T4SS, was only detected in isolates recovered before 2004 and similar omissions were reported by Rogge et al. (2013), who were unable to detect virD4, a homologue of traD, in E. ictaluri isolates recovered from Vietnam between 2006 and 2011. The T4SS is an important virulence system for bacterial pathogens, as the genes encoded mediate the translocation of macromolecules such as DNA and proteins across the cell envelope into eukaryotic or prokaryotic host cells (Mary et al., 2018). Indeed, the T4SS has been demonstrated to play a role in the virulence of other aquatic bacterial pathogens including E. piscicida and Y. ruckeri, through the transfer of plasmid DNA as well as secretion of proteins that promote immune evasion, respectively (Liu et al., 2017; Liu et al., 2022). Although its function in E. ictaluri is not presently known, traD/virD4 has been identified as an ATP-ase “coupling protein” functioning in substrate recruitment in other bacterial species (Grohmann et al., 2018). In addition, this gene has also been implicated in F pilus assembly in E. tarda (Verjan-Garcia et al., 2015), thus similar functions may exist in E. ictaluri. Nonetheless, findings from this study suggest a shift in T4SS-driven virulence pathways for E. ictaluri isolates recently recovered from diseased striped catfish in Vietnam. These shifts may be driven by host-dependent evolution of E. ictaluri genotypes as reported by Machimbirike et al. (2021). This would certainly be supported by findings from previous studies, which found host-dependent variability in virulence factors including traD/virD4 in channel catfish, Nile tilapia (Oreochromis niloticus), striped catfish and zebrafish (Danio rerio) (Griffin et al., 2016; Nhinh et al., 2022).
The PFGE of E. ictaluri isolates in this study found that a number of outbreaks from different provinces in 2019 and 2020 originated from a single population or a common source of contamination, as these isolates shared identical DNA fingerprints following restriction digest with speI. Furthermore, clonal isolates were more commonly shared between neighbouring provinces. Together, these findings would indicate either a single source of infection or a cross-contamination between farms and suggest that similar clones are circulating this region. These findings reflect the structure of the striped catfish sector within Vietnam, as clonal isolates were frequently identified in An Giang and Dong Thap provinces, where hatchery and nursery systems are concentrated (Hasan and Shipton, 2021), therefore the movement of fish stocks may be encouraging the spread of pathogenic populations including E. ictaluri. Likewise, production within open systems whereby water sources are shared, for example on the Mekong River, as well as the emergence of large scale vertically integrated companies across the Mekong Delta (Phu et al., 2016; Hasan and Shipton, 2021), may also account for the detection of clonal isolates from distant provinces. If anthropogenic factors are driving the spread of E. ictaluri populations across the sector, then biosecurity measures may help to mitigate the ongoing BNP epidemic in Vietnam. However, more work is required to investigate the spatiotemporal transmission of E. ictaluri in the region, especially under different active and passive transmission (e.g., through water currents) mechanisms.
In this study, subpopulations were also found to be circulating several farms across the Mekong Delta in the same year, as isolates recovered from these sites were clustered into different pulsotypes with varying genomic fingerprints. The presence of genetic subpopulations circulating the same fish farm has also been reported for F. psychrophilum in rainbow trout production systems in France (Calvez et al., 2021). In the present study, all isolates were recovered from diseased fish, therefore it is unlikely that the subpopulations detected were associated to the health status (e.g., carrier vs diseased) of fish. However, if fish originated from different production systems (e.g., ponds), the occurrence of different pulsotypes within these farms may therefore represent local adaptations of the subpopulations to the environmental niche within the individual production system. Local adaptation of bacterial pathogens is highly likely if these production systems experienced low level disease outbreaks which had been tolerated or poorly controlled. In fact, local adaptation in aquatic bacterial populations has previously been reported in other aquatic pathogenic species (Du et al., 2019; Fu et al., 2021), thus the same may also be true for E. ictaluri. These findings present a challenge for ongoing vaccination programs and current work is exploring the influence of genotypic shifts in relation to circulating antigenic profiles.
Considerable variation was noted in the plasmid profiles of Vietnamese E. ictaluri isolates in this study. No Vietnamese isolate was found to share the same plasmid profile as the American type isolate, which was found to harbour two plasmid bands at approximately 4.8 Kb and 5.8 Kb in size and likely the pEI1 (ca. 4.8 Kb) and pEI2 (ca. 5.6 Kb) plasmids detected previously in E. ictaluri isolates from channel catfish (Hawke et al., 2013). Similar sized plasmid bands were only found in 52% and 39% of the Vietnamese isolates, respectively. These findings follow that by Rogge et al. (2013), in which geographical differences were noted in plasmid DNA profiles between isolates from Vietnam and USA. Furthermore, in this study, the 5.8 Kb plasmid band was only detected in Vietnamese isolates recovered before 2010 suggesting a change in circulating plasmid populations within the region overtime, although sequencing of detected plasmid bands would be required to confirm this. Most isolates in this study were found to harbour two plasmid bands at 3.8 Kb and 4.8 Kb, with the former being detected in 98% of Vietnamese isolates. A similar sized plasmid has also been reported in E. ictaluri from striped catfish in Vietnam (4.0 Kb) as well as zebrafish from United States (3.9 Kb) (Rogge et al., 2013; Griffin et al., 2016). Furthermore, in the work by Griffin et al. (2016), a 3.9 Kb plasmid detected from zebrafish isolates was found to be a derivative of the channel catfish pEI2 plasmid, therefore, the plasmids at 3.8 and 4.8 Kb detected in Vietnamese E. ictaluri isolates in this study could be homologous to the pEI2 and pEI1 plasmids, respectively. Considerably larger plasmids (> 8 Kb) were also detected in several isolates in this study. Previous work by Bartie et al. (2012) also detected large plasmids (> 7 Kb) in E. ictaluri isolates recovered from Vietnam. However, it is difficult to compare these findings with our own, as the aforementioned study did not use restriction digestion to linearise extracted plasmids, therefore profiles may contain other forms (e.g., nicked or supercoiled) of plasmid DNA.
In conclusion, findings from this study demonstrate clonal expansion of E. ictaluri populations across several provinces within the Mekong Delta region of Vietnam, likely associated with anthropogenic factors. Hence, improving biosecurity strategies may offer an effective approach to mitigating ongoing BNP challenges in the sector. Furthermore, our data also shows that the E. ictaluri populations circulating the striped catfish sector have undergone genetic changes overtime, as evidenced in the observed changes in pulsotype, plasmid DNA and virulence gene profiles of isolates recovered from disease outbreaks between 2001 and 2021. These findings highlight the current and continued needs for surveillance of E. ictaluri in Vietnam, as any genetic changes may confer changes in antigenic proteins, thereby compromising the effectiveness of available vaccines to promote protection. Effective vaccination programs will be vital to the striped catfish sector to protect against future BNP outbreaks and curb any development of antibiotic resistance in E. ictaluri within the Mekong Delta region.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
CJP: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. KG: Methodology, Writing – review & editing. VHP: Methodology, Writing – review & editing. NNP: Methodology, Writing – review & editing. TTD: Methodology, Writing – review & editing. LHP: Conceptualization, Funding acquisition, Writing – review & editing. MC: Conceptualization, Funding acquisition, Project administration, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded through an InnoVet-AMR project (ID: 109057) with financial support from the UK Government Department of Health and Social Care (DHSC), the Global AMR Innovation Fund (GAMRIF), and the International Development Research Centre (IDRC). The APC was funded by the University of Stirling.
The authors would like to thank Mr Truong Hong Viet and Professor James Turnbull for their help with bacterial recovery/identification and data analysis, respectively.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1270968/full#supplementary-material
Abdelhamed H., Tekedar H. C., Ozdemir O., Hsu C.-Y., Arick M. A., Karsi A., et al. (2018). Complete genome sequence of multidrug-resistant edwardsiella ictaluri strain MS-17-156. Genome Announc. 6, e00477-18. doi: 10.1128/genomeA.00477-18
Adeolu M., Alnajar S., Naushad S., Gupta R. (2016). Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 66, 5575–5599. doi: 10.1099/ijsem.0.001485
Bartie K. L., Austin F. W., Diab A., Dickson C., Dung T. T., Giacomini M., et al. (2012). Intraspecific diversity of Edwardsiella ictaluri isolates from diseased freshwater catfish, Pangasianodon hypophthalmus (Sauvage), cultured in the Mekong Delta, Vietnam. J. Fish Dis. 35, 671–682. doi: 10.1111/j.1365-2761.2012.01376.x
Calvez S., Fournel C., Douet D.-G., Daniel P. (2015). Pulsed-field gel electrophoresis and multi locus sequence typing for characterizing genotype variability of Yersinia ruckeri isolated from farmed fish in France. Vet. Res. 46, 73. doi: 10.1186/s13567-015-0200-5
Calvez S., Navarro-Gonzalez N., Siekoula-Nguedia C., Fournel C., Duchaud E. (2021). High Genetic Diversity in Flavobacterium psychrophilum Isolates from Healthy Rainbow Trout (Oncorhynchus mykiss) Farmed in the Same Watershed, Revealed by Two Typing Methods. Appl. Environ. Microbiol. 87, e01398-20. doi: 10.1128/AEM.01398-20
Carriço J. A., Silva-Costa C., Melo-Cristino J., Pinto F. R., de Lencastre H., Almeida J. S., et al. (2006). Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant streptococcus pyogenes. J. Clin. Microbiol. 44, 2524–2532. doi: 10.1128/JCM.02536-05
Chen L., Yang J., Yu J., Yao Z., Sun L., Shen Y., et al. (2005). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328. doi: 10.1093/nar/gki008
Crumlish M., Dung T. T., Turnbull J. F., Ngoc N. T. N., Ferguson H. W. (2002). Identification of Edwardsiella ictaluri from diseased freshwater catfish, Pangasius hypophthalmus (Sauvage), cultured in the Mekong Delta, Vietnam. J. Fish Dis. 25, 733–736. doi: 10.1046/j.1365-2761.2002.00412.x
Dong H. T., Senapin S., Jeamkunakorn C., Nguyen V. V., Nguyen N. T., Rodkhum C., et al. (2019). Natural occurrence of edwardsiellosis caused by Edwardsiella ictaluri in farmed hybrid red tilapia (Oreochromis sp.) in Southeast Asia. Aquaculture 499, 17–23. doi: 10.1016/j.aquaculture.2018.09.007
Du X., Bayliss S. C., Feil E. J., Liu Y., Wang C., Zhang G., et al. (2019). Real time monitoring of Aeromonas salmonicida evolution in response to successive antibiotic therapies in a commercial fish farm. Environ. Microbiol. 21, 1113–1123. doi: 10.1111/1462-2920.14531
Duman M., Altun S., Saticioglu I. B. (2022). General assessment of approaches to the identification of aquatic bacterial pathogens: A methodological review. N. Am. J. Aquac. 84, 405–426. doi: 10.1002/naaq.10260
FAO (2021). Fisheries and aquaculture software (Rome, Italy: FishStat Plus - Universal software for fishery statistical time series. Bibliographic citation).
Feng Y., Cao S., Qin Z., Ouyang P., Chen D., Guo H., et al. (2022). Comparative analysis of sturgeon- and catfish-derived Yersinia ruckeri reveals the genetic variation and the risk of heavy antibiotic resistance. Aquac. Rep. 25, 101231. doi: 10.1016/j.aqrep.2022.101231
Frerichs G. N., Millar S. D. (1993). Manual for the isolation and identification of fish bacterial pathogens. (Stirling: Pisces Press).
Fu S., Yang Q., Wang Q., Pang B., Lan R., Wei D., et al. (2021). Continuous genomic surveillance monitored the in vivo evolutionary trajectories of vibrio parahaemolyticus and identified a new virulent genotype. mSystems 6, e01254-20. doi: 10.1128/mSystems.01254-20
García J. A., Larsen J. L., Dalsgaard I., Pedersen K. (2000). Pulsed-field gel electrophoresis analysis of Aeromonas salmonicida ssp. salmonicida. FEMS Microbiol. Lett. 190, 163–166. doi: 10.1111/j.1574-6968.2000.tb09280.x
Griffin M. J., Reichley S. R., Greenway T. E., Quiniou S. M., Ware C., Gao D. X., et al. (2016). Comparison of Edwardsiella ictaluri isolates from different hosts and geographic origins. J. Fish Dis. 39, 947–969. doi: 10.1111/jfd.12431
Grohmann E., Christie P. J., Waksman G., Backert S. (2018). Type IV secretion in Gram-negative and Gram-positive bacteria. Mol. Microbiol. 107, 455–471. doi: 10.1111/mmi.13896
Hasan M. R., Shipton T. A. (2021). Aquafeed value chain analysis of striped catfish in Vietnam. Aquaculture 541, 736798. doi: 10.1016/j.aquaculture.2021.736798
Hawke J. P., Kent M., Rogge M., Baumgartner W., Wiles J., Shelley J., et al. (2013). Edwardsiellosis Caused by Edwardsiella ictaluri in Laboratory Populations of Zebrafish Danio rerio. J. Aquat. Anim. Health 25, 171–183. doi: 10.1080/08997659.2013.782226
Hawke J. P., McWhorter A. C., Steigerwalt A. G., Brenner D. J (1981). Edwardsiella ictaluri sp. nov., the Causative Agent of Enteric Septicemia of Catfish. Int. J. Syst. Bacteriol. 31, 396–400. doi: 10.1099/00207713-31-4-396
Heras J., Domínguez C., Mata E., Pascual V., Lozano C., Torres C., et al. (2015). GelJ – a tool for analysing DNA fingerprint gel images. BMC Bioinf. 16, 270. doi: 10.1186/s12859-015-0703-0
Hoa T. T. T., Boerlage A. S., Duyen T. T. M., Thy D. T. M., Hang N. T. T., Humphry R. W., et al. (2021). Nursing stages of striped catfish (Pangasianodon hypophthalmus) in Vietnam: Pathogens, diseases and husbandry practices. Aquaculture 533, 736114. doi: 10.1016/j.aquaculture.2020.736114
Kennedy R. B., Ovsyannikova I. G., Palese P., Poland G. A. (2020). Current challenges in vaccinology. Front. Immunol. 11. doi: 10.3389/fimmu.2020.01181
Liu T., Zhou Z.-Y., Lu C., Bai S.-J., Wang E.-L., Wang J., et al. (2022). Yersinia ruckeri strain SC09 restrains innate immunity to promote infection process in rainbow trout (Oncorhynchus mykiss) via the type IV secretion system (T4SS). Aquaculture 560, 738564. doi: 10.1016/j.aquaculture.2022.738564
Liu Y., Gao Y., Liu X., Liu Q., Zhang Y., Wang Q., et al. (2017). Transposon insertion sequencing reveals T4SS as the major genetic trait for conjugation transfer of multi-drug resistance pEIB202 from Edwardsiella. BMC Microbiol. 17, 112. doi: 10.1186/s12866-017-1013-7
Machimbirike V. I., Crumlish M., Dong H. T., Santander J., Khunrae P., Rattanarojpong T. (2022). Edwardsiella ictaluri: A systemic review and future perspectives on disease management. Rev. Aquac. 14, 1613–1636. doi: 10.1111/raq.12665
Machimbirike V. I., Uthaipaisanwong P., Khunrae P., Dong H. T., Senapin S., Rattanarojpong T., et al. (2021). Comparative genomics of Edwardsiella ictaluri revealed four distinct host-specific genotypes and thirteen potential vaccine candidates. Genomics 113, 1976–1987. doi: 10.1016/j.ygeno.2021.04.016
Mary C., Fouillen A., Bessette B., Nanci A., Baron C. (2018). Interaction via the N terminus of the type IV secretion system (T4SS) protein VirB6 with VirB10 is required for VirB2 and VirB5 incorporation into T-pili and for T4SS function. J. Biol. Chem. 293, 13415–13426. doi: 10.1074/jbc.RA118.002751
Munang’andu H. M., Salinas I., Tafalla C., Dalmo R. A. (2020). Vaccines and immunostimulants for finfish. Front. Immunol. 11. doi: 10.3389/fimmu.2020.573771
Narayanan S. V., Joseph T. C., Peeralil S., Koombankallil R., Vaiyapuri M., Mothadaka M. P., et al. (2020). Tropical shrimp aquaculture farms harbour pathogenic Vibrio parahaemolyticus with high genetic diversity and Carbapenam resistance. Mar. pollut. Bull. 160, 111551. doi: 10.1016/j.marpolbul.2020.111551
Ngoc Phuoc N., Richards R., Crumlish M. (2020). Establishing bacterial infectivity models in striped Catfish Pangasianodon hypophthalmus (Sauvage) with Edwardsiella ictaluri. J. Fish Dis. 43, 371–378. doi: 10.1111/jfd.13135
Nhinh D. T., Giang N. T. H., van Van K., Dang L. T., Dong H. T., Hoai T. D. (2022). Widespread presence of a highly virulent Edwardsiella ictaluri strain in farmed tilapia, Oreochromis spp. Transbound Emerg. Dis. 69, e2276-e2290. doi: 10.1111/tbed.14568
Phu T. M., Phuong N. T., Dung T. T., Hai D. M., Son V. N., Rico A., et al. (2016). An evaluation of fish health-management practices and occupational health hazards associated with Pangasius catfish (Pangasianodon hypophthalmus ) aquaculture in the Mekong Delta, Vietnam. Aquac. Res. 47, 2778–2794. doi: 10.1111/are.12728
Queipo-Ortuńo M. I., Colmenero J. D. D., Macias M., Bravo M. J., Morata P. (2008). Preparation of bacterial DNA template by boiling and effect of immunoglobulin G as an inhibitor in real-time PCR for serum samples from patients with brucellosis. Clin. Vaccine Immunol. 15, 293–296. doi: 10.1128/CVI.00270-07
Ramadan A. A. (2022). Bacterial typing methods from past to present: A comprehensive overview. Gene Rep. 29, 101675. doi: 10.1016/j.genrep.2022.101675
Rogge M., Dubytska L., Jung T., Wiles J., Elkamel A., Rennhoff A., et al. (2013). Comparison of Vietnamese and US isolates of Edwardsiella ictaluri. Dis. Aquat Organ 106, 17–29. doi: 10.3354/dao02620
Sakai T., Yuasa K., Sano M., Iida T. (2009). Identification of Edwardsiella ictaluri and E. tarda by species-specific polymerase chain reaction targeted to the upstream region of the fimbrial gene. J. Aquat. Anim. Health 21, 124–132. doi: 10.1577/H08-061.1
Sciuto S., Colli L., Fabris A., Pastorino P., Stoppani N., Esposito G., et al. (2022). What can genetics do for the control of infectious diseases in aquaculture? Animals 12, 2176. doi: 10.3390/ani12172176
Verjan-Garcia N., Iregui C. A., Hirono I. (2015). Novel type 4 secretion system (T4SS)-related genes of Edwardsiella tarda. Orinoquia 19, 186–194. doi: 10.22579/20112629.318
Vu N. T., Sang N., Trong T. Q., Duy N. H., Dang N. T., Nguyen N. H. (2019). Breeding for improved resistance to Edwardsiella ictaluri in striped catfish (Pangasianodon hypophthalmus ): Quantitative genetic parameters. J. Fish Dis. 42, 1409–1417. doi: 10.1111/jfd.13067
Yang M., Shao S., Xiao J., Wang Q., Zhang Y. (2013). Phylogenetic investigation of Edwardsiella tarda with multilocus sequence typing (MLST) and pulsed field gel electrophoresis (PFGE) typing methods. Aquaculture 410–411, 79–85. doi: 10.1016/j.aquaculture.2013.05.024
Keywords: Edwardsiella ictaluri, Bacillary Necrosis of Pangasius, Pangasianodon hypophthalmus, Vietnam, genotyping, plasmids, virulence
Citation: Payne CJ, Grace K, Phuong VH, Phuoc NN, Dung TT, Phuoc LH and Crumlish M (2023) Exploring the genetic diversity of Edwardsiella ictaluri in Vietnamese striped catfish (Pangasianodon hypophthalmus) farms over a 20-year period. Front. Mar. Sci. 10:1270968. doi: 10.3389/fmars.2023.1270968
Received: 03 August 2023; Accepted: 01 December 2023;
Published: 19 December 2023.
Edited by:
Ji Hyung Kim, Gachon University, Republic of KoreaReviewed by:
Muhammed Duman, Bursa Uludağ University, TürkiyeCopyright © 2023 Payne, Grace, Phuong, Phuoc, Dung, Phuoc and Crumlish. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Margaret Crumlish, bWFyZ2FyZXQuY3J1bWxpc2hAc3Rpci5hYy51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.