
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 21 September 2023
Sec. Aquatic Physiology
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1270776
This article is part of the Research Topic Endocrine Regulation and Physiological Adaptation of Stress Response in Aquatic Organisms View all 18 articles
 Wan-Hong Zeng1,2†
Wan-Hong Zeng1,2† Xiu-Ying Wei2†
Xiu-Ying Wei2† Wei Qin2
Wei Qin2 Chuan-Jie Qin2
Chuan-Jie Qin2 Qiong Shi2,3,4
Qiong Shi2,3,4 Sheng-Tao Guo5
Sheng-Tao Guo5 Panita Prathomya6
Panita Prathomya6 Shi-Yong Zhang7
Shi-Yong Zhang7 Peng Fu8
Peng Fu8 Wei Hu1*
Wei Hu1* Han-Wen Yuan1*
Han-Wen Yuan1* Zheng-Yong Wen1,2,3*
Zheng-Yong Wen1,2,3*Elongase of very long-chain fatty acid 8 (Elovl8) is a new member identified in the Elovl family that is involved in the synthesis of highly unsaturated fatty acids (HUFAs). However, the evolutionary and physiological roles of this enzyme are still largely unknown. In the present study, the elovl8 gene was identified and characterized from yellow catfish Pelteobagrus fulvidraco, and then its evolutionary and molecular characteristics as well as transcriptional changes in response to various nutritional status were determined. Results showed that the open reading frame (ORF) of elovl8 was 795 bp in length, encoding a protein of 264 amino acids. Multiple sequences alignment showed that the yellow catfish Elovl8 was highly conserved with other homologs in teleosts, sharing similar structural characteristics (including six conserved transmembrane α-helical domains, four conserved elongase motifs, and three highly conserved cysteine residues). Meanwhile, comparisons of genetic synteny confirmed that the elovl8 gene identified from the yellow catfish was the homolog of elovl8b in other teleosts, and thus, the elovl8a gene was lost in the genome of the yellow catfish. Gene structure analysis revealed that the elovl8b gene contained eight exons and seven introns, which was highly conserved in teleosts, implying the functional conservation among various fish species. Tissue distribution analysis detected by real-time quantitative PCR (RT-qPCR) showed that the elovl8 gene was extensively expressed in all detected tissues except eyes, with high expression levels in the intestine and liver. Temporal expression analysis revealed that the expression level of elovl8 was stably expressed in the early 12 h after fertilization, and then dramatically decreased at 24, 48, 72, and 96 h after fertilization, implying that elovl8 is required for HUFA biosynthesis in the early development stages. Functional experiments showed that the expression of the elovl8 gene was stimulated after feeding with egg yolk but was not obviously affected after feeding with halogenated worms, indicating that diets full of HUFAs can inhibit the expression of elovl8 in yellow catfish. Our findings will help us to better understand the evolutionary and functional characteristics of elovl8 in teleosts, and lay a solid basis for investigating the regulation mechanism of HUFA biosynthesis.
Highly unsaturated fatty acids (HUFAs) are a series of straight-chain fatty acids with three or more double bonds and 20 or more carbon atoms, which play important roles in maintaining cell membrane fluidity, regulating fat metabolism, enhancing immunity, and reducing inflammation (Zhang et al., 2019; Ri et al., 2022). Notably, eicosapentaenoic acid (EPA, 20:5n-3) and docosahexaenoic acid (DHA, 22:6n-3) are the most prominent HUFAs that play critical roles involved in promoting growth and development, and preventing cardiovascular and neurodevelopmental disorders (De Giuseppe et al., 2014; Zárate et al., 2017).
Usually, vertebrates possess the ability to biosynthesize HUFAs by using essential polyunsaturated fatty acids as precursors with two critical rate-limiting enzymes including elongases of very long chain fatty acids (Elovls) and fatty acid desaturases (Fads) (Oboh et al., 2017). The former can elongate the carbon chain of polyunsaturated fatty acids while the latter can introduce a double bond at a specific position on the carbon chain to produce desaturation (Nakamura and Nara, 2004; Guillou et al., 2010). In teleosts, it is commonly accepted that freshwater and migratory fishes have obvious capacity to biosynthesize HUFAs while seawater fishes usually have lower or no capacity to achieve this goal because they are surrounded by an environment with abundant HUFAs in diets (Li et al., 2010; Jaya-Ram et al., 2011). Meanwhile, it seems that food habitats also have effects on the HUFA biosynthesis in teleosts. For example, herbivorous or omnivorous freshwater fishes such as grass carp (Du et al., 2006) and zebrafish (Ishak et al., 2008) are proved to have the ability to synthesize arachidonic acid (AA), EPA, and DHA with linoleic acid (LA) and linolenic acid (LNA) as precursors. Differently, carnivorous fishes such as groupers (Li et al., 2016) and gilthead seabream (Ganga et al., 2005) lack this kind of capacity or have a weak capacity, and thus higher levels of dietary HUFAs are required to meet their requirements for growth, development, and reproduction. However, more studies are still required to investigate the exact mechanisms involved in HUFA biosynthesis in teleosts.
Thus far, seven members named ELOVL1–ELOVL7 in the ELOVL family were identified in mammals, and these members showed different substrate specificities for various fatty acid substrates (Jakobsson et al., 2006; Ohno et al., 2010). Overall, ELOVL1, ELOVL3, ELOVL6, and ELOVL7 primarily extend saturated fatty acids as well as monounsaturated fatty acids; ELOVL2, ELOVL4, and ELOVL5 mainly catalyze the elongation reactions of polyunsaturated fatty acids, while Elovl4 prolongs both very-long-chain saturated fatty acids and very-long-chain polyunsaturated fatty acids (Agbaga et al., 2008; Guillou et al., 2010). Recently, a novel teleost-specific member of the Elovl family, named Elovl8, had been identified in a marine rabbitfish (Siganus canaliculatus) (Li et al., 2020). Genetic synteny and gene phylogeny revealed that this new member commonly contains two isoforms, and it seemed that the elovl8a gene has been lost in some fish genomes (Li et al., 2020). Further functional experiments indicated that Elovl8b but not Elovl8a possesses the capacity to biosynthesize HUFAs (Li et al., 2020). Consistently, a single elovl8b gene was identified in hybrid grouper (Epinephelus fuscoguttatus ♀ × Epinephelus lanceolatus ♂), and functional experiments revealed that diets full of HUFAs can inhibit elovl8b transcription in liver, suggesting that a negative feedback regulation of the HUFA synthetic pathway existed (Wu et al., 2022). However, the evolutionary and functional characteristics of this new member are still largely unclear, and much more studies are required to illustrate these issues.
Yellow catfish (Pelteobagrus fulvidraco) belongs to family Bagridae, order Siluriformes, and is an omnivorous freshwater fish that is widely cultured in China due to its rapid growth, good taste, and valuable nutrition values (Guo et al., 2023; Wei et al., 2023). Thus far, two key genes involved in HUFA biosynthesis including fad2 and elovl5 have been identified in yellow catfish (Song et al., 2015; Qin et al., 2017), but their exact roles have not been well investigated. In the present study, we identified a teleost-specific elovl8 gene from yellow catfish, and then its evolutionary and molecular characteristics, spatiotemporal expression patterns, and functional traits were determined for the first time. Our findings will provide not only a novel insight into mechanisms involved in HUFA biosynthesis in yellow catfish, but also valuable data for better understanding the evolutionary and functional traits of elovl8 in teleosts.
Yellow catfish (weight = 32.6 ± 3.1 g) used in this study were purchased from a fishery farm in Sichuan province of China, and fishes were transported to the laboratory and temporarily reared in 1 m × 1 m × 1 m net boxes set up in a 16-m2 microfluidic pond. Fishes were reared for 2 weeks in a natural light condition (12 L/12 D), and water temperature was maintained at 24–26°C. Fishes were fed with commercial feed (approximately 3%–4% of body weight) at 19:00 every day as described in our previous studies (Da et al., 2022). Experimental fishes showed normal feeding and activity during the accumulation period. After that, five fishes were randomly selected and used for tissue distribution studies. Fishes were anesthetized with MS-222 before decapitated, and then tissues including adipose, brain, barbel, eye, gill, gonad, heart, intestine, kidney, liver, muscle, spleen, and stomach were collected. Tissue samples were immediately frozen in liquid nitrogen, and subsequently stored at −80°C until further utilization.
Meanwhile, fertilized eggs of yellow catfish used in this study were obtained by artificial breeding in our laboratory. During incubation, the development status of yellow catfish embryos was observed with a microscope, and five embryos and larvae were respectively sampled at each time point including 0 h, 3 h, 6 h, 12 h, 24 h, 36 h, 48 h, 72 h, and 96 h after fertilization in the early developmental stages. Similarly, samples were frozen rapidly with liquid nitrogen and then stored at −80°C for further utilization.
Diets with different content of HUFAs, including Artemia nauplii and egg yolk, were separately used as larval stage feeds to investigate the potential effects of various diets on transcription of elovl8 in yellow catfish. Meanwhile, approximately 200 yellow catfish larvae were randomly selected and fed with different diets on day 4 after hatching, and three parallel experiments were designed for each group. During the experiment, 25% water changes were carried out daily, the oxygenation pump was kept continuously oxygenated, and the dissolved oxygen in the water was maintained above 6 mg/L. Fishes were fed at 08:00, 12:00, 16:00 and 20:00 to ensure sufficient diet supplement in the experimental tanks. The experiment lasted for 3 days, and five fishes from each tank were collected at 24, 48, and 72 h after feeding. Finally, samples were immediately frozen in liquid nitrogen and stored at −80 °C for further utilization.
Protein similarity BLAST was conducted to identify the genomic and transcriptomic sequences from yellow catfish genome data in NCBI database and transcriptome database using several valid fish Elovl8 protein sequences as queries. Subsequently, potential sequences were verified and confirmed by NCBI-BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Meanwhile, specific primers used for cloning experiment were designed with the Primer Premier 5.0 software (www.PremierBiosoft.com) (Supplementary Table 1).
PCR was performed to amplify the aimed elovl8 gene fragment with liver cDNA as template. Then, PCR products were tested by agarose gel electrophoresis and isolated and purified using the TaKaRa gel recovery kit. Finally, PCR products were cloned into the pMD-19T vector (TaKaRa, Dalian, China) and then sequenced at Sangon Co. Ltd. (Guangzhou, China) to ensure the accuracy of the potential elovl8 gene in yellow catfish.
The nucleotide sequence of elovl8 was obtained from yellow catfish using BLAST in the NCBI database (http://www.ncbi.nlm.nih.gov/BLAST). After validation, the nucleotide sequences were translated into protein sequences using the Primer Premier 5.0 software. Multiple Elovl8 protein sequence alignment was conducted by using Clustal X software as described in our previous studies (Li et al., 2020). Meanwhile, comparative analyses of genetic synteny and gene structure were conducted to determine the evolutionary pattern of elovl8 genes in teleosts. Additionally, a phylogeny was constructed on the basis of the protein sequences of various teleosts to investigate the evolutionary history of the elovl8 gene family. Elovl8 protein sequences used for phylogeny were downloaded from the NCBI or Ensembl databases. After calculation, JTT + G was selected as the best model, and the neighbor-joining (NJ) method was chosen to construct the phylogenetic tree. Finally, the robustness of the tree topology was assessed by a nonparametric bootstrap analysis with 1,000 resampling replicates. Details of the selected Elovls family proteins are given in Supplementary Table 2.
Total RNA was extracted with Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol, and then the concentration and quality of total RNA were confirmed by a spectrophotometer (Nano Drop 2000, Thermo Scientific, USA) as described in our previous studies (Wen et al., 2020; Wen et al., 2021). Subsequently, 1 μg of total RNA was reverse transcribed to cDNA using the QuantiTect® Reverse Transcription kit (Takara Biotech, Dalian, China). Quantitative real-time PCR (qPCR) was conducted to measure the mRNA level of rabbitfish elovl8 on a Light Cycler Real-Time system with a final volume of 20 μL. Meanwhile, the relative expression level of mRNA was normalized with β-actin after assessing the stability of five reference genes. Finally, the relative transcription level of elovl8 was calculated using the Pfaffl method (Yang et al., 2018; Wen et al., 2019). Sequences of the specific primers used for qPCRs are provided in Supplementary Table 1.
SPSS 22.0 (IBM, Armonk, NY, USA) and GraphPad Prism 7.0 (GraphPad, Prism Software Inc., San Diego, California) were used for statistical analysis. All data were shown as mean normalized values ± standard error of the mean (SEM). Significant differences were determined using one-way analysis of variance (ANOVA) followed by Tukey’s test, and differences were significant when p < 0.05.
In the present study, we identified the elovl8 gene from yellow catfish for the first time. NCBI-Blast revealed that the identified elovl8 gene is the homolog of elovl8b gene in teleost. The open reading frame (ORF) of yellow catfish elovl8 was 795 bp in length and encoded a protein with 264 amino acids. Multiple Elovl8b protein sequence alignment showed that Elovl8b is highly conserved (protein similarity > 84.4%) in teleosts and that they shared similar structural features, containing six conserved transmembrane α-helix structural domains, four conserved elongase motifs, and three highly conserved cysteine residues (Figure 1).
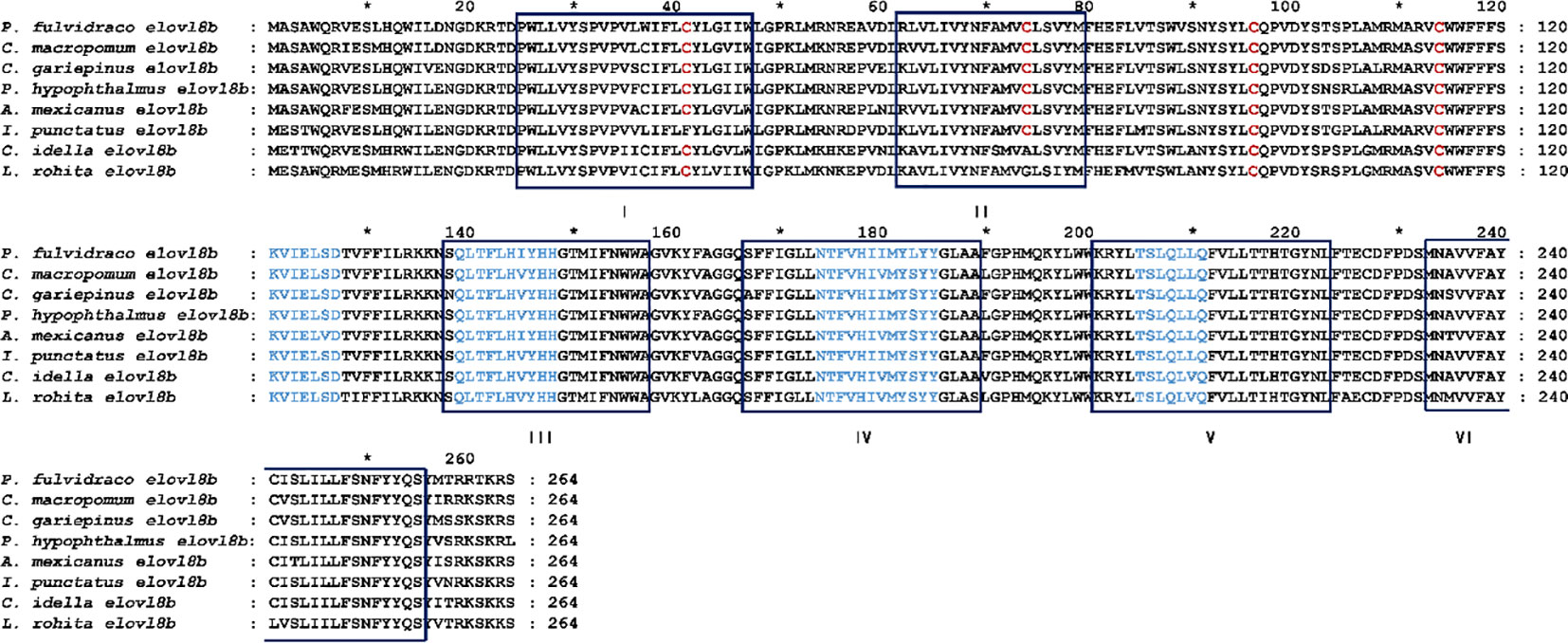
Figure 1 Multiple protein sequence alignment of Elovl8b between yellow catfish and other representative teleosts. Six conserved transmembrane α-helix structural domains are labeled with I–VI. The four conserved elongase motifs are labeled in blue font. Cysteines are shown in red. C-terminal ER retrieval signals are marked with dashed boxes. Asterisks (*) indicate the amino acids conserved among all members of the proteins.
To better understand the evolutionary relationships of the elovl8 gene family in teleosts, a phylogenetic tree was constructed using the NJ method with MEGA-X software. Results showed that the elovl8 family was divided into two clades, namely, elovl8a and elovl8b subfamilies, and the yellow catfish elovl8 was clustered into the elovl8b clade (Figure 2). Notably, red-bellied piranha (P. nattereri), chum salmon (O. keta), zebrafish (D. rerio), Roho labeo (L. rohita), fathead minnows (P. promelas), grass carp (C. idella), cachama (C. macropomum), and delta smelt (H. transpacificus) contained two elovl8 paralogs. Differently, north African catfish (C. gariepinus), channel catfish (I. punctatus), yellow catfish (P. fulvidraco), freshwater shark (P. hypophthalmus), southern catfish (S. meridionalis), Mexican tetra (A. mexicanus), the sharp-headed submarginal fish (X. texanus), and the common carp (C. carpio) only contained the elovl8b subtype (Figure 2). Moreover, yellow catfish elovl8b shared a close relationship with channel catfish and African catfish elovl8b (Figure 2).
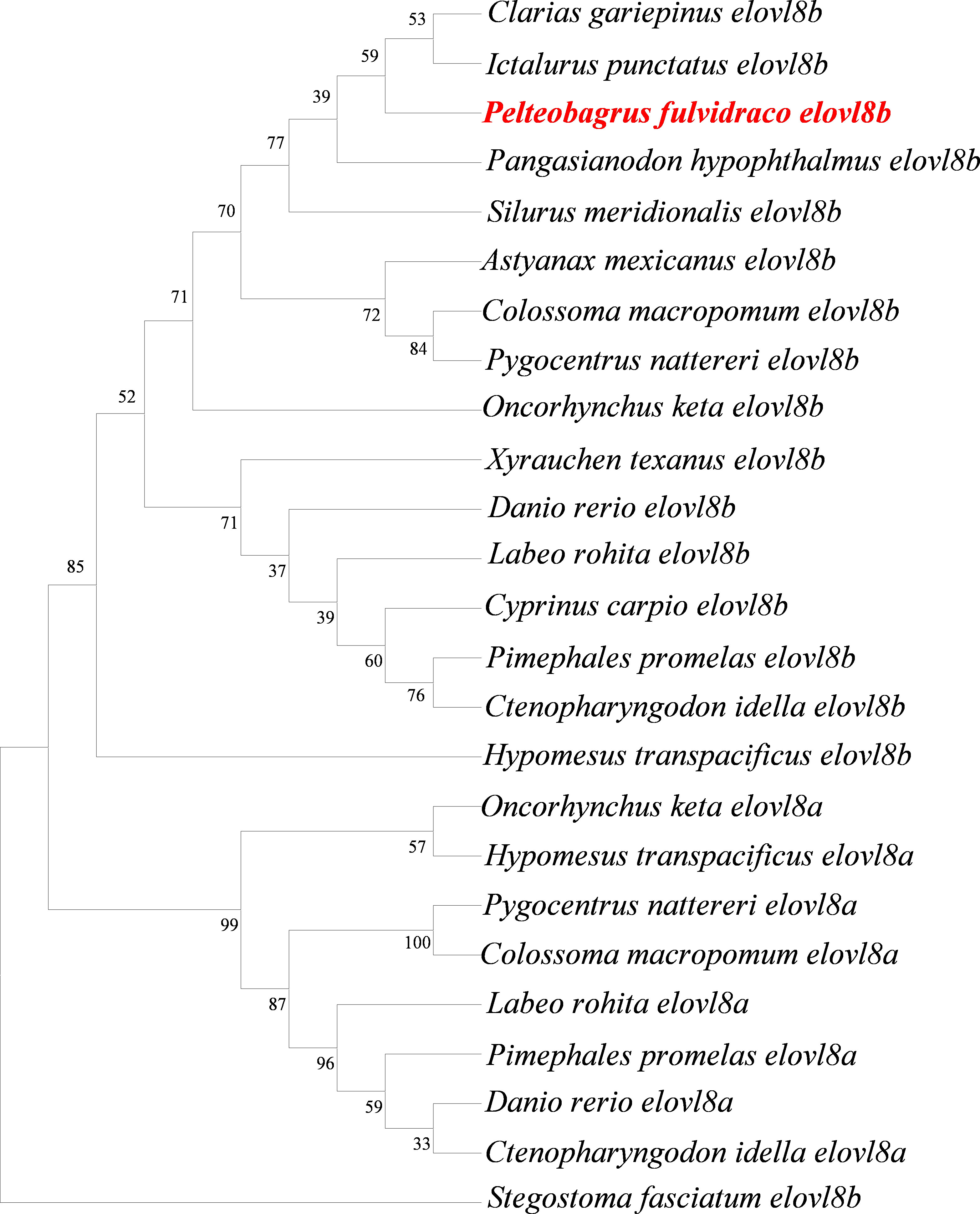
Figure 2 Phylogeny reveals the evolutionary history of the elovl8 gene family in teleosts. Protein sequences were used to construct a phylogenetic tree with the NJ method using MEGA-X software. Numbers up the clades represent the bootstrap percentages from 1,000 replicates and the yellow catfish elovl8 is labeled in red color. The leopard shark (Stegostoma fasciatum) was used as the outgroup.
Comparative genomics were conducted to further explore the exact evolutionary status of elovl8 genes in eight representative fishes, namely, L. rohita, C. macropomum, C. idella, A. mexicanus, P. hypophthalmus, I. punctatus, C. gariepinus, and P. fulvidraco (Figure 3). Results showed that three species, namely, L. rohita, C. macropomum, and C. idella, contained the elovl8a gene, and a conserved gene cluster tesk2-toe1-selenop2-elovl8a-zswim5-urod was identified in these fishes (Figure 3A). Notably, the elovl8a gene had been lost in the other five fish species, namely, P. fulvidraco, I. punctatus, C. gariepinus, P. hypophthalmus, and A. mexicanus (Figure 3A). Moreover, a highly conserved gene cluster dmap1-guk1b-armh1-mutyh-elovl8b was identified in all the representative fish genomes (Figure 3B).

Figure 3 Genetic synteny of elovl8a (A) and elovl8b (B) in representative fish genomes. The colorful blocks represent different genes. The solid lines represent intergenic regions. The target species in the present study are marked in red front.
Gene structure comparison was conducted to reveal the structural and functional differentiation of the elovl8b gene in eight representative fish species. Results showed that the elovl8b gene of seven selected fish species, namely, P. fulvidraco, A. mexicanus, C. gariepinus, C. idella, C. macropomum, L. rohita, and P. hypophthalmus, had similar gene structures, containing eight exons and seven introns (Figure 4). Differently, the gene structure of I. punctatus elovl8b contained nine exons and eight introns, but showed a conserved coding sequence (CDS) region with the seven other fish elovl8b genes (Figure 4).
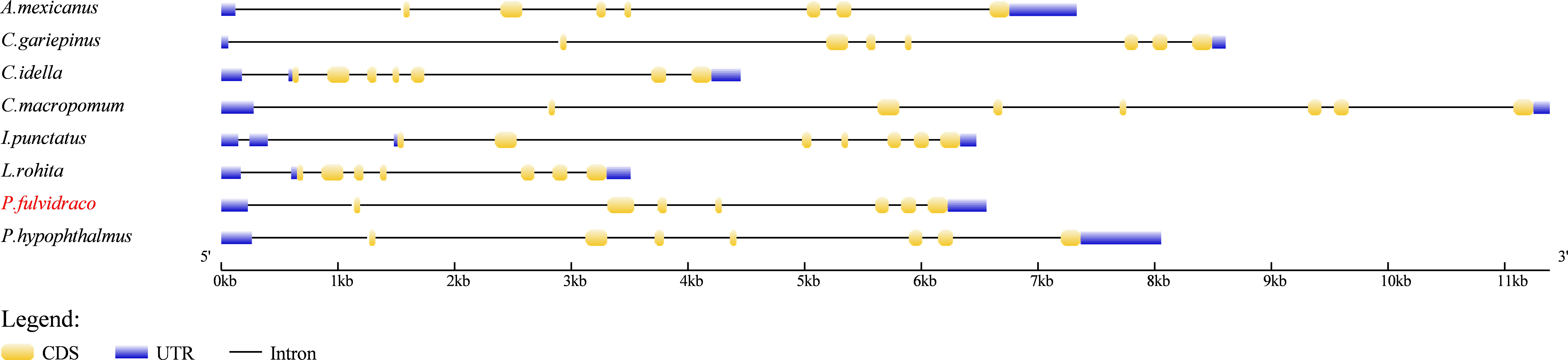
Figure 4 Comparison of the gene structure of elovl8b in eight representative teleosts. The colorful blocks and solid lines represent the exons and introns, respectively. UTR and CDS are marked with blue and yellow blocks, respectively.
The distribution pattern of elovl8 in yellow catfish was determined by real-time quantitative PCR. Results showed that the elovl8 gene of yellow catfish was widely expressed in most selected tissues including adipose, barbel, brain, gill, gonad, heart, intestine, kidney, liver, muscle, spleen, and stomach, and relative high expression levels were observed in intestine and liver tissues (Figure 5). Differently, no expression was detected in the eyes (Figure 5).
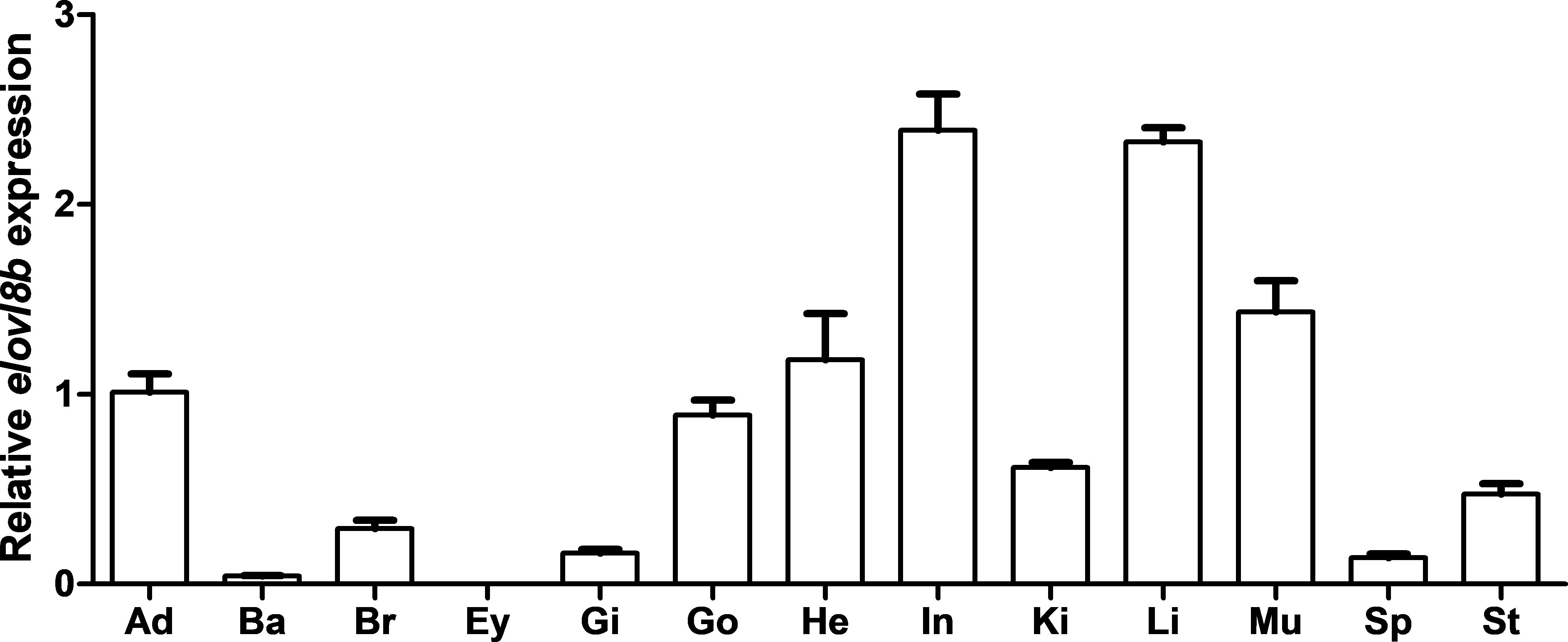
Figure 5 Tissue distribution of the elovl8 gene in yellow catfish. Adipose (Ad); Barbel (Ba); Brain (Br); Eye (Ey); Gill (Gi); Gonad (Go); Heart (He); Intestine (In); Kidney (Ki); Liver (Li); Muscle (Mu); Spleen (Sp); Stomach (St). β-actin was selected as the internal reference gene. Data are expressed as mean ± standard error (n = 5).
Quantitative PCR was conducted to detect the expression level of the elovl8 gene in the early developmental stages of yellow catfish. Results showed that the expression level was not significantly changed at 0, 3, 6, and 12 h, but dramatically decreased at 24, 36, 48, and 72 h, and reached the lowest expression level at 96 h after fertilization (Figure 6).
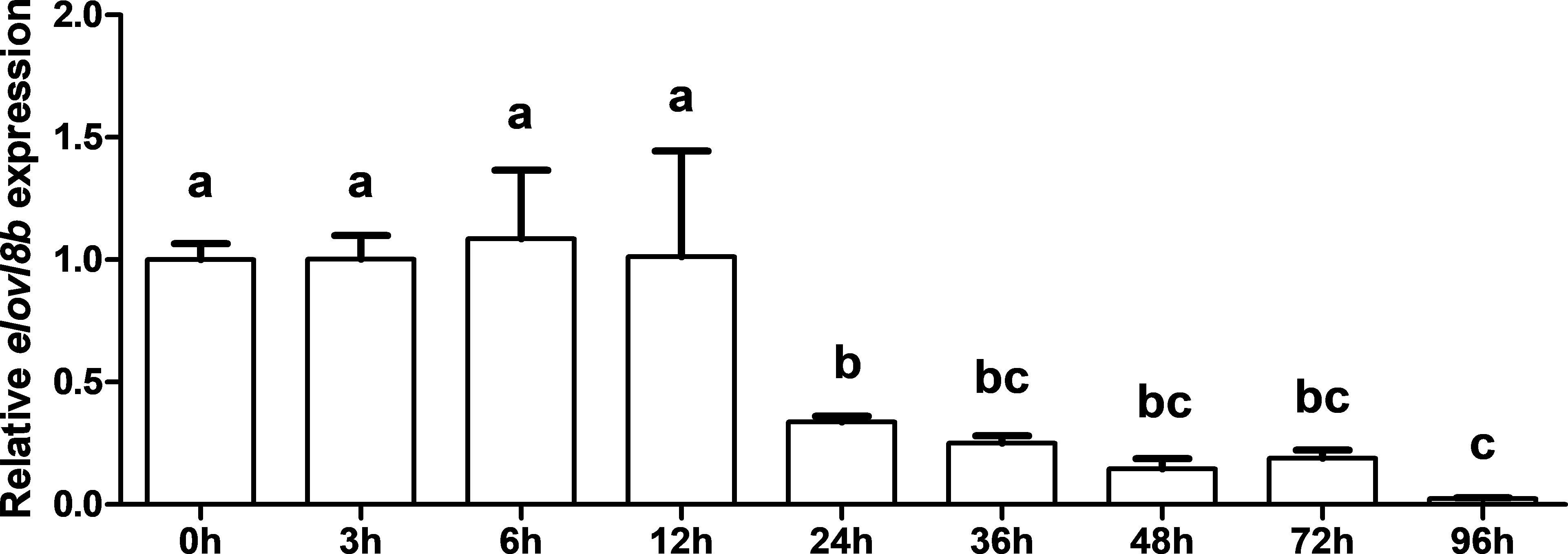
Figure 6 Gene expression pattern of the elovl8b gene in the early developmental stage of yellow catfish. β-actin was selected as the internal reference gene. Data are expressed as mean ± standard error (n = 5). Groups that differ significantly are indicated by different lowercase letters above bars.
Transcriptional change patterns of the yellow catfish elovl8b gene in response to larval stage diets with different HUFA content were determined (Figure 7). Results showed that the expression level of the elovl8b gene was slightly increased but not significantly changed at 1, 2, and 3 days after feeding with A. nauplii as in yellow catfish (Figure 7A). Differently, the expression level of the elovl8b gene was dramatically increased at 2 days and then significantly decreased at 3 days after feeding with egg yolk in yellow catfish (Figure 7B).
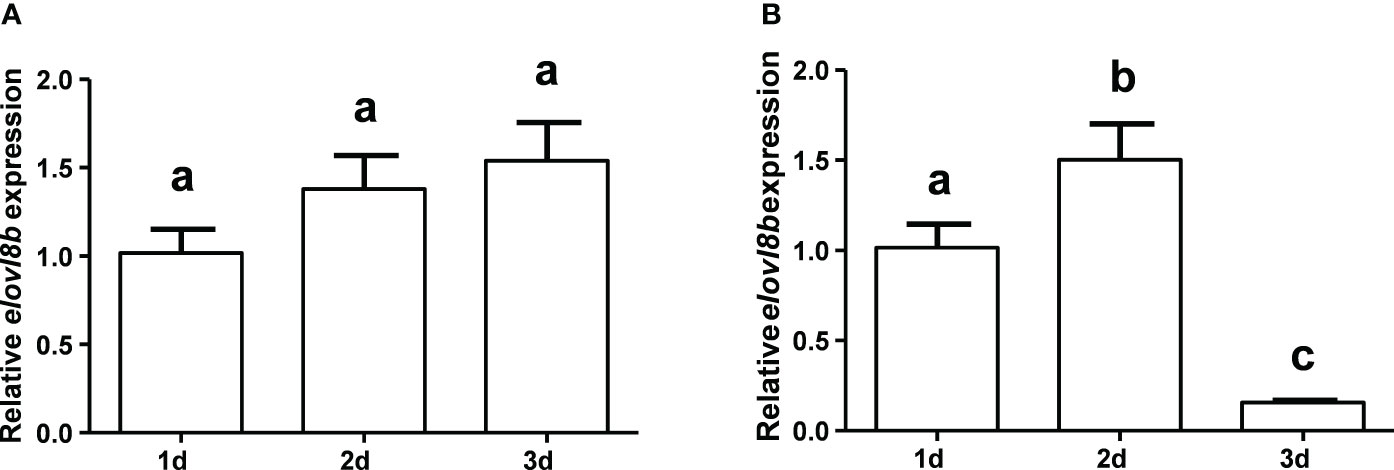
Figure 7 Effect of dietary A nauplii (A) and egg yolk (B) on elovl8 gene expression in yellow catfish larvae. β-actin was selected as the internal reference gene. Data are expressed as mean ± standard error (n = 5). Groups that differ significantly are indicated by different lowercase letters above bars.
Elovls are key rate-limiting enzymes to extend the carbon chain of polyunsaturated fatty acids by catalyzing condensation reaction, which play important roles involved in the biosynthesis of HUFAs in animals (Nugteren, 1965). Thus far, elovl1–elovl7 are widely studied both in vertebrates and in invertebrates, but less is known about elovl8, the newly identified member of the elovl family (Ohno et al., 2010; Naganuma et al., 2011; Gregory and James, 2014; Yan et al., 2018). In this study, the elovl8 gene of the yellow catfish was identified and characterized; it contained a 795- bp-long ORF that encodes a protein of 264 amino acids. Similar results were also observed in rabbitfish (Siganus canaliculatus) (Li et al., 2020) and hybrid grouper (Epinephelus fuscoguttatus♀ × Epinephelus lanceolatus♂) (Wu et al., 2022), suggesting that the elovl8 genes are conserved among different fishes.
Multiple protein sequence comparison showed that yellow catfish Elovl8 possessed all the characteristics of ELOVL protein family members, including four conserved regions (KXXEXXDT, QXXFLHXYHH, NXXXHXXMYXYY, and TXXQXXQ), endoplasmic reticulum (ER)-resident signals, six transmembrane regions, and a histidine cluster (HXXHH) that is involved in fatty acid elongation processes (Jakobsson et al., 2006). The secondary characteristics are similar to that of Elovl8b in the rabbitfish (Li et al., 2020) and hybrid grouper (Wu et al., 2022), implying the evolutionary and functional conservation of the Elovl8b among various teleosts.
Phylogenetic analysis showed that the elovl8 gene family was clustered into two branches of elovl8a and elovl8b subfamilies, which was consistent with the findings in previous studies, suggesting that teleost-specific elovl8 family may commonly contain two paralogs in teleost genomes, and this phenomenon may be caused by an additional teleost-specific genome duplication (TSGD) event (Kuraku and Meyer, 2009; Sun et al., 2021). Genetic synteny further supported teleost lineage, which usually contains elovl8a and elovl8b isoforms, such as fishes that belong to the orders Cyprinodontiformes, Salmoniformes, and Osmeriformes. However, it seemed that the elovl8a gene had been lost in Siluriformes teleosts, including C. gariepinus, P. hypophthalmus, I. punctatus, and P. fulvidraco, suggesting that elovl8b should be more conserved than elovl8a, and thus, the former but not the latter may play important roles in HUFA biosynthesis (Li et al., 2020). To our knowledge, the elovl8a gene lost in some teleost genomes may be caused by functional redundancy between elovl8a and elovl8b isoforms in the early period of evolution. In addition, the structure of the elovl8b gene commonly contained eight exons and seven introns in teleosts, suggesting that this gene was highly conserved and might play similar functions among various teleosts.
Tissue distribution experiments showed that elovl8b was widely expressed in various tissues with high expression levels in liver and intestine, which was different with rabbitfish and hybrid grouper (Li et al., 2020; Wu et al., 2022), indicating that the tissue distribution pattern of elovl8b is species-specific and it may play diverse roles in various teleosts. Indeed, this phenomenon may be caused by the different ability to synthesize HUFAs in different fishes (Gong et al., 2014). Notably, the elovl8b was highly expressed in liver in most teleosts, suggesting that this gene may also play similar roles in HUFA biosynthesis because liver is the major organ that is involved in HUFA production.
The gene expression pattern of the elovl8b gene during the early development stage of yellow catfish was also determined, and results showed that elovl8b was stably expressed within 12 h after fertilization, which was slightly different with other HUFA biosynthesis-related genes in rabbitfish (You et al., 2017), suggesting that HUFA biosynthesis is very important for the early development of embryo despite the fact that the pattern may be different among various species. However, the expression level of yellow catfish elovl8 was significantly decreased from 24 h to 96 h. It has been shown that during the early stage of embryonic development in yellow catfish, the PUFAs in yolk are involved in metabolism and catabolism to provide energy preparation for embryonic development, with C18:2n-6, C20:4n-3, and DHA dominating (Yao et al., 2009). Interestingly, the elovl8b gene was shown to have the ability to prolong C18 (18:2n-6, 18:3n-3, and 18:4n-3) and C20 (20:4n-6 and 20:5n-3) HUFAs to long-chain polyunsaturated fatty acids (LC-HUFAs) in rabbitfish (Li et al., 2020). This expression pattern suggests that yellow catfish elovl8 is also involved in the synthesis of long-chain polyunsaturated fatty acids and plays an important role in early embryonic development.
Previous studies have shown that diets full of HUFAs could inhibit the transcriptional expression in rabbitfish and hybrid grouper (Li et al., 2020; Wu et al., 2022). In the present study, functional experiments showed that the transcription of elovl8 was not significantly affected by feeding with A. salina, whereas it was inhibited after feeding with egg yolk in yellow catfish. It is shown that the lack of HUFAs in A. salina might explain why dietary A. salina slightly increased elovl8 gene expression (Tocher et al., 2003; Garcia et al., 2008). Meanwhile, dietary egg yolk induced but then inhibited elovl8 expression, which may be caused by the high content of phospholipid in egg yolk. Our findings further supported the idea that a compensatory mechanism also existed in yellow catfish, which was similar to previous reports and could demonstrate a key role for elovl8b in LC-PUFA biosynthesis (Cho et al., 1999; Seiliez et al., 2001).
In summary, an elovl8 gene was identified and characterized from yellow catfish for the first time. Multiple protein sequence alignment, genetic synteny, gene structure comparisons, and phylogeny revealed that elovl8a had been lost in Siluriformes, and elovl8b was highly conserved among various teleosts. Moreover, spatiotemporal expression analyses suggested that elovl8 was widely distributed, and it could play important roles in the early development stage in yellow catfish. Finally, functional experiments showed that a diet full of HUFAs may inhibit the transcription of elovl8 in yellow catfish. Our findings will help us to better understand the evolutionary and functional characteristics of elovl8 in teleosts, and lay a solid basis for investigating the regulation mechanism of HUFA biosynthesis.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was approved by Neijiang Normal University Animal Care and Use Committee. The study was conducted in accordance with the local legislation and institutional requirements.
W-HZ: Formal Analysis, Investigation, Writing – original draft. X-YW: Writing – original draft, Formal Analysis, Investigation, Software. WQ: Writing – original draft, Investigation. C-JQ: Writing – review & editing. QS: Writing – review & editing. SG: Writing – review & editing, Formal Analysis, Software. PP: Software, Writing – review & editing. SZ: Writing – review & editing, Funding acquisition, Investigation. PF: Writing – review & editing, Formal Analysis, Investigation. WH: Methodology, Supervision, Writing – review & editing. H-WY: Supervision, Writing – review & editing. Z-YW: Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing.
This work was financially supported by the Research Fund from Key Laboratory of Sichuan Province for Fishes Conservation and Utilization in the Upper Reaches of the Yangtze River (No. NJTCSC23-3), the Natural Science Fund of Sichuan Province of China (No. 2023NSFSC1221), the Project of Sichuan Provincial Department of Science and Technology (No. 2021YFYZ0015), the Cooperation Fund from Sichuan University (No. 2022H013), and the Important New Varieties Selection Project of Jiangsu Province (No. PZCZ201742).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1270776/full#supplementary-material
Agbaga M. P., Brush R. S., Mandal M. N., Henry K., Elliott M. H., Anderson R. E. (2008). Role of Stargardt-3 macular dystrophy protein (ELOVL4) in the biosynthesis of very long chain fatty acids. Proc. Natl. Acad. Sci. U.S.A. 105, 12843–12848. doi: 10.1073/pnas.0802607105
Cho H. P., Nakamura M., Clarke S. D. (1999). Cloning, expression, and fatty acid regulation of the human delta-5 desaturase. J. Biol. Chem. 274, 37335–37339. doi: 10.1074/jbc.274.52.37335
Da F., Wen Z., Wang X.-D., Luo Y. (2022). Molecular identification, tissue distribution, and effects of fasting and refeeding on the transcription of uncoupling protein 2 in yellow catfish, Pelteobagrus vachelli. Pak J. Zool. 54, 1539–1547. doi: 10.17582/journal.pjz/20200316080346
De Giuseppe R., Roggi C., Cena H. (2014). n-3 LC-PUFA supplementation: effects on infant and maternal outcomes. Eur. J. Nutr. 53, 1147–1154. doi: 10.1007/s00394-014-0660-9
Du Z. Y., Clouet P., Zheng W. H., Degrace P., Tian L. X., Liu Y. J. (2006). Biochemical hepatic alterations and body lipid composition in the herbivorous grass carp (Ctenopharyngodon idella) fed high-fat diets. Br. J. Nutr. 95, 905–915. doi: 10.1079/bjn20061733
Ganga R., Bell J. G., Montero D., Robaina L., Caballero M. J., Izquierdo M. S. (2005). Effect of dietary lipids on plasma fatty acid profiles and prostaglandin and leptin production in gilthead seabream (Sparus aurata). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 142, 410–418. doi: 10.1016/j.cbpb.2005.09.010
Garcia A. S., Parrish C. C., Brown J. A. (2008). Growth and lipid composition of Atlantic cod (Gadus morhua) larvae in response to differently enriched Artemia franciscana. Fish. Physiol. Biochem. 34, 77–94. doi: 10.1007/s10695-007-9149-2
Gong Y., Wan X., Jiang M., Hu C., Hu H., Huang F. (2014). Metabolic engineering of microorganisms to produce omega-3 very long-chain polyunsaturated fatty acids. Prog. Lipid Res. 56, 19–35. doi: 10.1016/j.plipres.2014.07.001
Gregory M. K., James M. J. (2014). Rainbow trout (Oncorhynchus mykiss) Elov15 and Elov12 differ in selectivity for elongation of omega-3 docosapentaenoic acid. Biochim. Biophys. Acta 1841, 1656–1660. doi: 10.1016/j.bbalip.2014.10.001
Guillou H., Zadravec D., Martin P. G., Jacobsson A. (2010). The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res. 49, 186–199. doi: 10.1016/j.plipres.2009.12.002
Guo S., Zeng M., Gao W., Li F., Wei X., Shi Q., et al. (2023). Toll-like receptor 3 in the hybrid yellow catfish (Pelteobagrus fulvidraco ♀ × P. vachelli ♂): Protein structure, evolution and immune response to exogenous Aeromonas hydrophila and Poly (I:C) stimuli. Animals 13 (2), 288. doi: 10.3390/ani13020288
Ishak S. D., Tan S. H., Khong H. K., Jaya-Ram A., Enyu Y. L., Kuah M. K., et al. (2008). Upregulated mRNA expression of desaturase and elongase, two enzymes involved in highly unsaturated fatty acids biosynthesis pathways during follicle maturation in zebrafish. Reprod. Biol. Endocrinol. 6, 56. doi: 10.1186/1477-7827-6-56
Jakobsson A., Westerberg R., Jacobsson A. (2006). Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog. Lipid Res. 45, 237–249. doi: 10.1016/j.plipres.2006.01.004
Jaya-Ram A., Ishak S. D., Enyu Y. L., Kuah M. K., Wong K. L., Shu-Chien A. C. (2011). Molecular cloning and ontogenic mRNA expression of fatty acid desaturase in the carnivorous striped snakehead fish (Channa striata). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 158, 415–422. doi: 10.1016/j.cbpa.2010.11.018
Kuraku S., Meyer A. (2009). The evolution and maintenance of Hox gene clusters in vertebrates and the teleost-specific genome duplication. Int. J. Dev. Biol. 53 (5-6), 765–773. doi: 10.1387/ijdb.072533km
Li Y., Monroig O., Zhang L., Wang S., Zheng X., Dick J. R., et al. (2010). Vertebrate fatty acyl desaturase with Δ4 activity. Proc. Natl. Acad. Sci. U.S.A. 107, 16840–16845. doi: 10.1073/pnas.1008429107
Li Y., Wen Z., You C., Xie Z., Tocher D. R., Zhang Y., et al. (2020). Genome wide identification and functional characterization of two LC-PUFA biosynthesis elongase (elovl8) genes in rabbitfish (Siganus canaliculatus). Aquaculture 522, 735127. doi: 10.1016/j.aquaculture.2020.735127
Li S., Yuan Y., Wang T., Xu W., Li M., Mai K., et al. (2016). Molecular cloning, functional characterization and nutritional regulation of the putative elongase elovl5 in the orange-spotted grouper (Epinephelus coioides). PloS One 11, e0150544. doi: 10.1371/journal.pone.0150544
Naganuma T., Sato Y., Sassa T., Ohno Y., Kihara A. (2011). Biochemical characterization of the very long-chain fatty acid elongase ELOVL7. FEBS Lett. 585, 3337–3341. doi: 10.1016/j.febslet.2011.09.024
Nakamura M. T., Nara T. Y. (2004). Structure, function, and dietary regulation of delta6, delta5, and delta9 desaturases. Annu. Rev. Nutr. 24, 345–376. doi: 10.1146/annurev.nutr.24.121803.063211
Nugteren D. H. (1965). The enzymic chain elongation of fatty acids by rat-liver microsomes. Biochim. Biophys. Acta 106, 280–290. doi: 10.1016/0005-2760(65)90036-6
Oboh A., Navarro J. C., Tocher D. R., Monroig O. (2017). Elongation of very Long-Chain (>C(24)) Fatty Acids in Clarias gariepinus: Cloning, Functional Characterization and Tissue Expression of elovl4 Elongases. Lipids 52, 837–848. doi: 10.1007/s11745-017-4289-3
Ohno Y., Suto S., Yamanaka M., Mizutani Y., Mitsutake S., Igarashi Y., et al. (2010). ELOVL1 production of C24 acyl-CoAs is linked to C24 sphingolipid synthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 18439–18444. doi: 10.1073/pnas.1005572107
Qin C.-J., Wen Z.-Y., Yuan D.-Y., Shao T., Gong Q. (2017). Molecular cloning and expression of fatty acid desaturase and elongase genes in darkbarbel (Pelteobagrus vachellii). J. Oceanol. Limnol. 48, 884–893.
Ri K., Lee-Okada H. C., Yokomizo T. (2022). Omega-6 highly unsaturated fatty acids in Leydig cells facilitate male sex hormone production. Commun. Biol. 5, 1001. doi: 10.1038/s42003-022-03972-y
Seiliez I., Panserat S., Kaushik S., Bergot P. (2001). Cloning, tissue distribution and nutritional regulation of a Delta6-desaturase-like enzyme in rainbow trout. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 130, 83–93. doi: 10.1016/s1096-4959(01)00410-9
Song Y. F., Luo Z., Pan Y. X., Zhang L. H., Chen Q. L., Zheng J. L. (2015). Three unsaturated fatty acid biosynthesis-related genes in yellow catfish Pelteobagrus fulvidraco: Molecular characterization, tissue expression and transcriptional regulation by leptin. Gene 563 (1), 1–9. doi: 10.1016/j.gene.2014.12.014
Sun S., Wang Y., Goh P. T., Lopes-Marques M., Castro L. F. C., Monroig Ó., et al. (2021). Evolution and functional characteristics of the novel elovl8 that play pivotal roles in fatty acid biosynthesis. Genes 12 (8), 271. doi: 10.3390/genes12081287
Tocher D. R., Bell J. G., Dick J. R., Crampton V. O. (2003). Effects of dietary vegetable oil on Atlantic salmon hepatocyte fatty acid desaturation and liver fatty acid compositions. Lipids 38 (7), 723–732. doi: 10.1007/s11745-003-1120-y
Wei X. Y., Wang J., Guo S. T., Lv Y. Y., Li Y. P., Qin C. J., et al. (2023). Molecular characterization of a teleost-specific toll-like receptor 22 (tlr22) gene from yellow catfish (Pelteobagrus fulvidraco) and its transcriptional change in response to poly I:C and Aeromonas hydrophila stimuli. Fish. Shellfish Immunol. 134, 108579. doi: 10.1016/j.fsi.2023.108579
Wen Z. Y., Liu T., Qin C. J., Zou Y. C., Wang J., Li R., et al. (2021). MRAP2 interaction with melanocortin-4 receptor in snakehead (Channa argus). Biomolecules 11 (3), 481. doi: 10.3390/biom11030481
Wen Z. Y., Qin C. J., Wang J., He Y., Li H. T., Li R., et al. (2020). Molecular characterization of two leptin genes and their transcriptional changes in response to fasting and refeeding in Northern snakehead (Channa argus). Gene 736, 144420. doi: 10.1016/j.gene.2020.144420
Wen Z. Y., Wang J., Bian C., Zhang X., Li J., Peng Y., et al. (2019). Molecular cloning of two kcnk3 genes from the Northern snakehead (Channa argus) for quantification of their transcriptions in response to fasting and refeeding. Gen. Comp. Endocrinol. 281, 49–57. doi: 10.1016/j.ygcen.2019.05.016
Wu Q., Zheng Z., Wang C., Wang Y., Sun Y., Gao Y. (2022). Molecular characterization, tissue distribution and differential nutritional regulation of three n-3 LC-PUFA biosynthesis-related genes in hybrid grouper (Epinephelus fuscoguttatus ♀ × Epinephelus lanceolatus ♂). Animals 12 (3), 234. doi: 10.3390/ani12030234
Yan J., Liang X., Cui Y., Cao X., Gao J. (2018). Elovl4 can effectively elongate C18 polyunsaturated fatty acids in loach Misgurnus anguillicaudatus. Biochem. Biophys. Res. Commun. 495, 2637–2642. doi: 10.1016/j.bbrc.2017.12.123
Yang S., Wen Z. Y., Zou Y. C., Qin C. J., Wang J., Yuan D. Y., et al. (2018). Molecular cloning, tissue distribution, and effect of fasting and refeeding on the expression of neuropeptide Y in Channa argus. Gen. Comp. Endocrinol. 259, 147–153. doi: 10.1016/j.ygcen.2017.11.017
Yao J., Zhao H. Y., Li C., He D., Hu C. (2009). Changes in Fatty Acid Composition During Early Embryonic Development of Yellow Catfish Pelteobagrus f iulvidraco. Fish. Sci. 28, 644–647.
You C., Miao S., Lin S., Wang S., Waiho K., Li Y. (2017). Expression of long-chain polyunsaturated fatty acids (LC-PUFA) biosynthesis genes and utilization of fatty acids during early development in rabbitfish Siganus canaliculatus. Aquaculture 479, 774–779. doi: 10.1016/j.aquaculture.2017.07.028
Zárate R., El Jaber-Vazdekis N., Tejera N., Pérez J. A., Rodríguez C. (2017). Significance of long chain polyunsaturated fatty acids in human health. Clin. Transl. Med. 6, 25. doi: 10.1186/s40169-017-0153-6
Keywords: yellow catfish, elovl8, gene cloning, gene expression, HUFA biosynthesis
Citation: Zeng W-H, Wei X-Y, Qin W, Qin C-J, Shi Q, Guo S-T, Prathomya P, Zhang S-Y, Fu P, Hu W, Yuan H-W and Wen Z-Y (2023) Molecular characterization, spatiotemporal expression patterns of fatty acid elongase (elovl8) gene, and its transcription changes in response to different diet stimuli in yellow catfish (Pelteobagrus fulvidraco). Front. Mar. Sci. 10:1270776. doi: 10.3389/fmars.2023.1270776
Received: 01 August 2023; Accepted: 29 August 2023;
Published: 21 September 2023.
Edited by:
Yiming Li, Fishery Machinery and Instrument Research Institute, ChinaReviewed by:
Zhendong Qin, Zhongkai University of Agriculture and Engineering, ChinaCopyright © 2023 Zeng, Wei, Qin, Qin, Shi, Guo, Prathomya, Zhang, Fu, Hu, Yuan and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Hu, aHV3ZWkxOTg3MjAwNkAxNjMuY29t; Han-Wen Yuan, aGFud2VuX3l1YW5AMTI2LmNvbQ==; Zheng-Yong Wen, emhlbmd5b25nX3dlbkAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.