
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 24 November 2023
Sec. Marine Biology
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1266330
 Jianpeng Zhang1,2
Jianpeng Zhang1,2 Yi Wang1,2
Yi Wang1,2 Zekun Huang1,2
Zekun Huang1,2 Mingyi Cai3
Mingyi Cai3 Weiwei You1,2
Weiwei You1,2 Ying Lu1,2
Ying Lu1,2 Xuan Luo1,2*
Xuan Luo1,2* Caihuan Ke1,2*
Caihuan Ke1,2*Pacific abalone (Haliotis discus hannai) is an economically important marine shellfish for aquaculture and is distributed throughout eastern Asia. Although a lot of genetic breeding work has been carried out, chromosome identification in abalone is still a challenging task. Here, we developed a set of BACs to be chromosome-specific probes in Pacific abalone, and to study chromosome evolution in the related species. Through BAC paired-end sequencing and sequence alignment, we were able to in silico anchor 168 BACs onto 18 pseudochromosomes of Pacific abalone genome. After selecting 42 BACs that contained DNA inserts with minimal repetitive sequences, we validated them through PCR and Fluorescence in situ hybridization (FISH) test. As a result, We obtained specific FISH signals for 26 clones on the chromosomes of Pacific abalone with at least one BAC mapped per chromosome. We also applied the chromosome-specific BAC-FISH probes to a close relative of Pacific abalone, Xishi abalone (H. gigantea), which revealed that chromosome 13 and 15 between the two species underwent a chromosomes rearrangement event. This study provides the first set of chromosome-specific probes for the family Haliotidae, which can serve as an important tool for future cytogenetics and genomics research.
The family Haliotidae, which contains only one genus (Haliotis), includes more than 70 abalone species (Nguyen et al., 2022). Some of which are significant commercial fishery resources and aquaculture species (Morash and Alter, 2015). Abalone karyotypes exhibit distinct geographic characteristics, with cytogenetic studies classifying the species into three groups based on their chromosome numbers and karyotypes: European species (2n = 28), Indo-Pacific species (2n = 32) and Northeast and Northwest Pacific species (2n = 36) (Franchini et al., 2010). However, although there are clear differences in karyotypes among abalones in different geographic locations, there is still a lack of cytogenetic tools that enable further chromosome studies. The chromosomes of the family Haliotidae are small, have similar sizes and arm ratios, and are different to identify through karyotypic measurement (Palma-Rojas and von Brand, 2008; Amar-Basulto et al., 2011). As such, it is evident that traditional cytogenetic tools are insufficient for effective abalone chromosome identification.
Fluorescence in situ hybridization (FISH) is an essential technique in identifying abalone chromosomes. FISH with chromosome-specific probes offers a rich and dependable source of markers for cytogenetic research including cytogenetic map, chromosome identification, chromosome rearrangement, and chromosome evolution across specie (Pinkel et al., 1986; Yang et al., 2019; Bielski et al., 2020; do Vale Martins et al., 2021). Chromosome-specific FISH probes were developed using various approaches, such as DOP-PCR following microdissection or flow sorting (Telenius et al., 1992; Vandewoestyne et al., 2009), as well as screening BACs from a constructed library (Dong et al., 2000), and synthesizing bulky oligonucleotides based on the assembled genomes (Han et al., 2015). With the improvement of genome assembly level in recent years, the efficiency of screening and obtaining chromosome-specific probes has been greatly enhanced, both for BAC screening and bulk oligonucleotide synthesis (Han et al., 2015; Dong et al., 2018).
The Pacific abalone, Haliotis discus hannai is one of the most commercially important marine shellfish for aquaculture which is distributed throughout eastern Asia (Wang et al., 2015). Given its high market value and nutrient content, the research on this species has received significant attention, not only in aquaculture technology, but also in nutrition, ecology, molecular and cell biology (Sekino and Hara, 2007; Kobayashi and Kijima, 2010; An et al., 2011; Nie et al., 2012). What’s more, the genetic map and chromosome-level reference genomes of Pacific Abalone are available and had been applied in genetic breeding research (Liu et al., 2006; Nam et al., 2017). In the 1980s, Arai et al. (Arai et al., 1982) firstly reported the karyotype of Pacific Abalone, and the results showed that Pacific Abalone has 36 chromosomes, and the karyotype formula was 2n= 20m+16sm. However, the karyotype of Pacific Abalone was revealed with variation in different research (Arai et al., 1982; Okumura et al., 1995; Cai et al., 2013). Banding and FISH were carried out to provided markers for identifying individual chromosome of Pacific Abalone (Cai et al., 2010; Wang et al., 2015). However, there is no method to identify the whole set chromosomes in Pacific Abalone yet. The Xishi abalone, H.gigantea is a closely related species of Pacific Abalone, which is a valuable commercial species alone the coast of Japan and was introduced in to China in 2003 (Luo et al., 2010). In chromosome morphology, Xishi abalone and Pacific abalone are basically the same (Miyaki et al., 1997). However, interspecific variation in chromosome structure is common in this genus (Franchini et al., 2010), so whether the internal structure of chromosomes between the two species is similar still needs further study. In this study, we used chromosome-specific BAC clones as probes to construct a cytogenetic map and identify chromosome in H. discus hannai through multiple FISH. We also tested the cross-species universality of the probes in its cross relative, H.gigantea in order to explore the differences in chromosome structure between the two abalone species.
Samples of abalones for chromosome preparation used in this study were all collected from the Fuda Abalone Aquaculture Co. Ltd., Fujian Province, China. Chromosome preparations were obtained according to the conventional method with slight modifications (Cai et al., 2010). The trochophores were collected with filter mesh cultured in 0.03% colchicine at 20~25°C for 1 hour. Then the trochophores were exposed to 0.075 mol/L KCl solution for 1 hour, fixed three times for 30 min each with ethanol/glacial acetic acid (3:1), and stored at -20°C until use. Fixed larvae were dissociated into fine pieces in 50% acetic acid solution, the resulting cell suspensions were dropped onto preheated slides and air dried. Then heating 1-3 hours in a 60°C hot plate. The chromosome preparations were preserved at -20°C for Fluorescence in situ hybridization.
The construction of the H. discus hannai BAC library followed previously published methods (Luo et al., 2006; Zhang et al., 2022). Firstly, High molecular genomic DNA was extracted from the adductor muscle of H. discus hannai and digested with HindIII restriction enzyme into 110 - 220 kb. Then, the size-selected DNA fragments were ligated with HindIII-digested pHZAUBAC1 vector (Eight Star Bio-tech Co., Ltd., Wuhan, China). After the ligation products were transformed into Escherichia coli strain DH10B T1 Phage-Resistant (Invitrogen), the transformed cells were plated on LB agar plates supplemented with appropriate chloramphenicol, X-gal, and IPTG. At last, the clones were randomly picked and arrayed into 384-well plates containing LB media and then frozen at -80°C. Thirty random clones were selected to evaluate the quality of the BAC library. The extracted plasmids were digested with I-Sce I enzyme (Fermentas), as previously described (Shi et al., 2011). And then the pulsed-field gel electrophoresis was used to examine the size of the inserted fragments and the nulling rate.
The methodology was based on previous studies (Zhang et al., 2022). A total of 1044 BAC clones were selected randomly from the BAC library of H. discus hannai. The paired-end sequences of the BAC clones were obtained by Sanger sequencing with Applied Biosystems 3730 DNA Analyzer. The Phred (http://www.phrap.org/) and Seqclean (https://sourceforge.net/projects/seqclean) were used for quality control of BAC end sequences (BESs). Then, the end sequences were aligned to the reference genome of H. discus hannai (not published data) using Blast (Altschul et al., 1997), with an E-value threshold of 1 × 10−5. In subsequent analysis, the sequences of insert of the mapped BACs were deduced according to the genomic sequences between the paired BESs. And then, the size and the content of repetitive sequences was estimated. The repetitive sequences were found by using Repeatmasker (http://www.repeatmasker.org/RepeatMasker/). The BACs with insert size around the estimated average were selected as candidate chromosome specific cytogenetic markers, and their position of BACs on the chromosome was visualized with Mapchart v2.30.
Refer to previous methods for validating candidate clones (Zhang et al., 2022). PCR method was used to verify the correspondence between BAC clones and pseudochromosomes. The primers were designed using Primer Premier 5 software based on the deduced sequence of the BAC insert, and were synthesized by BGI Genomics Inc. PCR was carried out using a commercially available kit following the manufacturer’s instructions, with amplification conditions including pre-denaturation at 94°C for 3 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at Tm for 30 s, and extension at 72°C for 30 s. The results of the PCR amplification were evaluated by analyzing the number and size of the products using 1% agarose gel electrophoresis.
FISH experiment was performed following the methods published previously with minor modifications (Cai et al., 2010). In this study, BACs were labeled with either Digoxigenin or biotin. The resulting labeled probe was added to a hybridization solution consisting of 50% deionized formamide and 2 × SSC, following denaturation at 72°C for 8 min and cooling on ice for over 10 min. Slides of chromosome preparations were performed by treating them with 75% formamide/2×SSC at 74°C for 2.5 min, followed by dehydration in a gradient ethanol solution. After hybridizing the probe solution with denatured chromosomes on the slides overnight at 37°C, the slides were washed with 10% formamide/2×SSC, and twice with 4×SSC before adding 10 μg/mL anti-Digoxigenin-fluorescein rhodamine 100 μL or 10 μg/mL streptavidin 100 μL onto the slide. Subsequently, the slides were incubated in a wet box filled with 2 × SSC for 30 min. Once the coverslip was removed, the slides were washed again with 4×SSC-0.1% Triton for 5 min, and then twice with 4×SSC for 5 min each time. Following a slight cleaning with purified water, the slides were air-dried at room temperature and treated with DAPI (4’,6-diamidino-2-phenylindole) in 20 μL. The exposed slide was visualized using an Olympus epifluorescence microscope equipped with a CCD camera, capturing gray-scale images for each color channel through a digital image capture system (Olympus DP 80). Background subtraction and image feature intensification were carried out following the previous studies (Zhang et al., 2022). And the chromosome pairing and alignment were performed in Adobe photoshop CC 2017.
H. gigantea is a closely related species to the Pacific Abalone, sharing similar karyotypic characteristics with H. discus hannai. In order to examine the variations in genome sequence structure between these two species and assess the cross-species adaptability of the developed H. discus hannai BAC-FISH probes, the BESs of H. discus hannai were aligned with the reference genome of H. gigantea (unpublished data). Based on the results of this sequence alignment, clones demonstrating significant differences in expected positions between the two species selected for further FISH validation.
A BAC library of H. discus hannai was construed using DNA extracted from the muscle of a male. The library contains 16,128 BAC clones, and its quality was evaluated by examining thirty randomly selected clones with pulsed field gel electrophoresis (Supplementary Figure 1). No empty clone was found while some clones were polyclonal. The size of the insert fragments ranged from 110 kb to 170 kb, with an average of 143.3 kb (Supplementary Figure 2). Extrapolating from the above results, the library was estimated to cover 1.2 times genome of H. discus hannai.
A total of 1044 BAC clones were randomly selected from the BAC library for paired end sequencing and subjected to paired-end sequencing (The number of clones to be sequenced is determined by the genome coverage of the BAC library and the average insertion fragment size of the BAC clone). Subsequently, 1908 high-quality BAC end sequences (BESs) were obtained (Table 1), with an average length of 949 bp and a range of 100 bp to 1200 bp. Using BLAST analysis, 245 BAC sequences were identified that corresponded to the 18 chromosomes of H. discus hannai. These sequences were demonstrated good collinearity and sequence specificity.
To minimize the effect of repeat sequences on FISH analysis, the BAC sequences were screened using RepeatMasker against the high-repeat H. discus hannai DNA database to filter repeat sequences (Supplementary Table 1). Following this step, a total of 168 BAC clones were identified and distributed across 18 chromosomes of H. discus hannai for use in chromosome identification. The genomic locations of these BACs were documented in detail in Supplementary Table 1 and visualized in Figure 1. According to the putative content of repeat sequences and chromosome position, 3 clones for each chromosome were selected for further PCR and FISH verification (Figure 1).
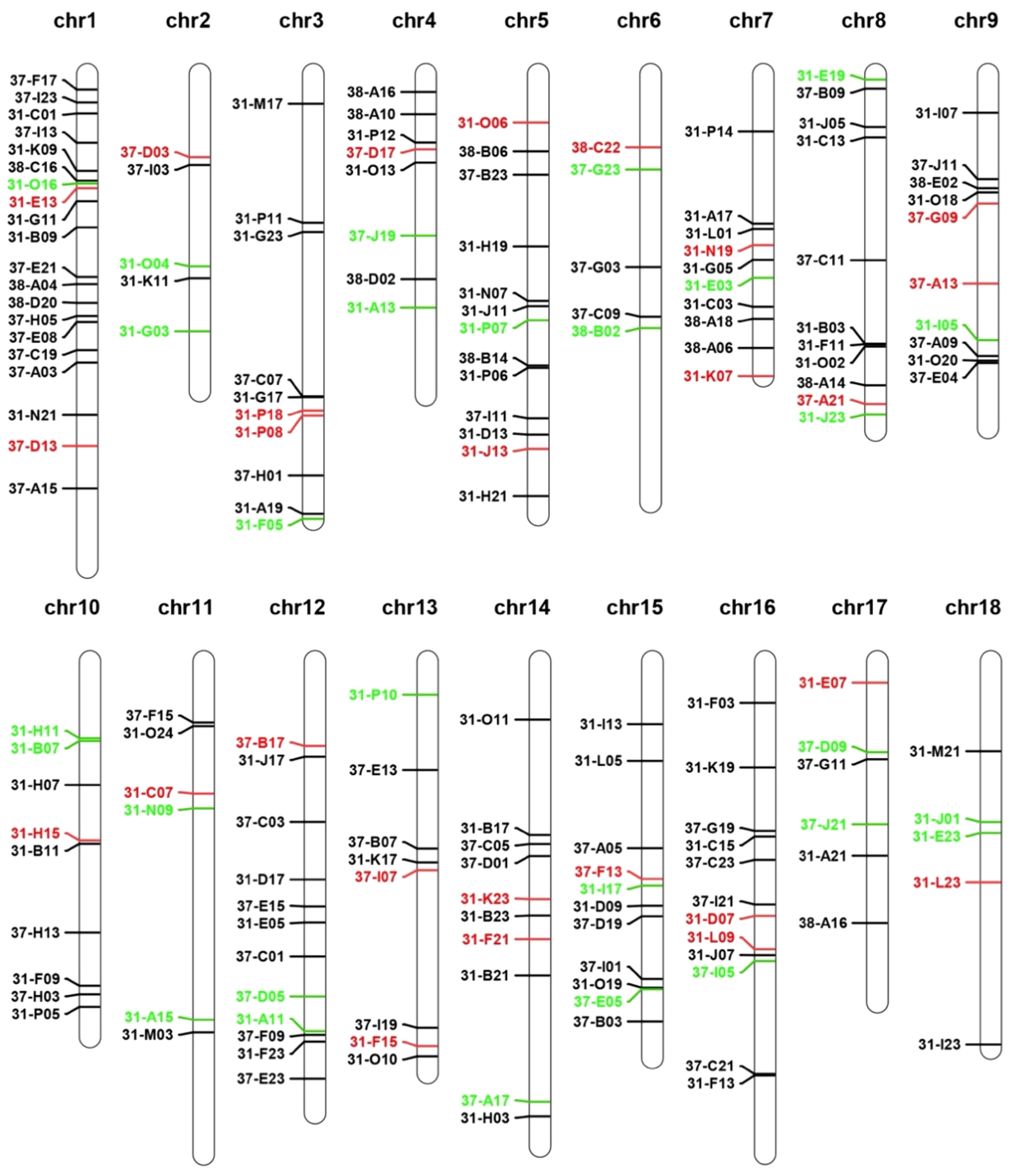
Figure 1 Genomic localization of BAC clones on the reference genome of H. discus hannai. The cloned clone representation shown in the figure will be used for further FISH verification. Red and green indicating passed and unpassed, respectively.
To verify the in-silico location of the BACs, a pair of primers were designed for each clone based on the putative insertion sequence (Supplementary Table 2). PCR analysis of the BAC clones using the primers resulted in the amplification of a single product of the expected length. The representative results of PCR verification were shown in Supplementary Figure 3. There finding confirm the accuracy of the predicted locations of the BACs.
The detection of selected BACs clones using FISH was carried out based on two criteria: (1) production of a single pair FISH signals and (2) collinearity with the putative adjacent BACs. A total of 26 BACs was verified, with at least one verified clone for each chromosome of H. discus hannai. The signals pattern of these probes on the chromosome of H. discus hannai is shown in Supplementary Figure 4, while the position of 26 chromosome-specific BAC clones on metaphase chromosome was showed in Figure 2. The unpassed BAC clones were rechecked with independent FISH test with single BAC.
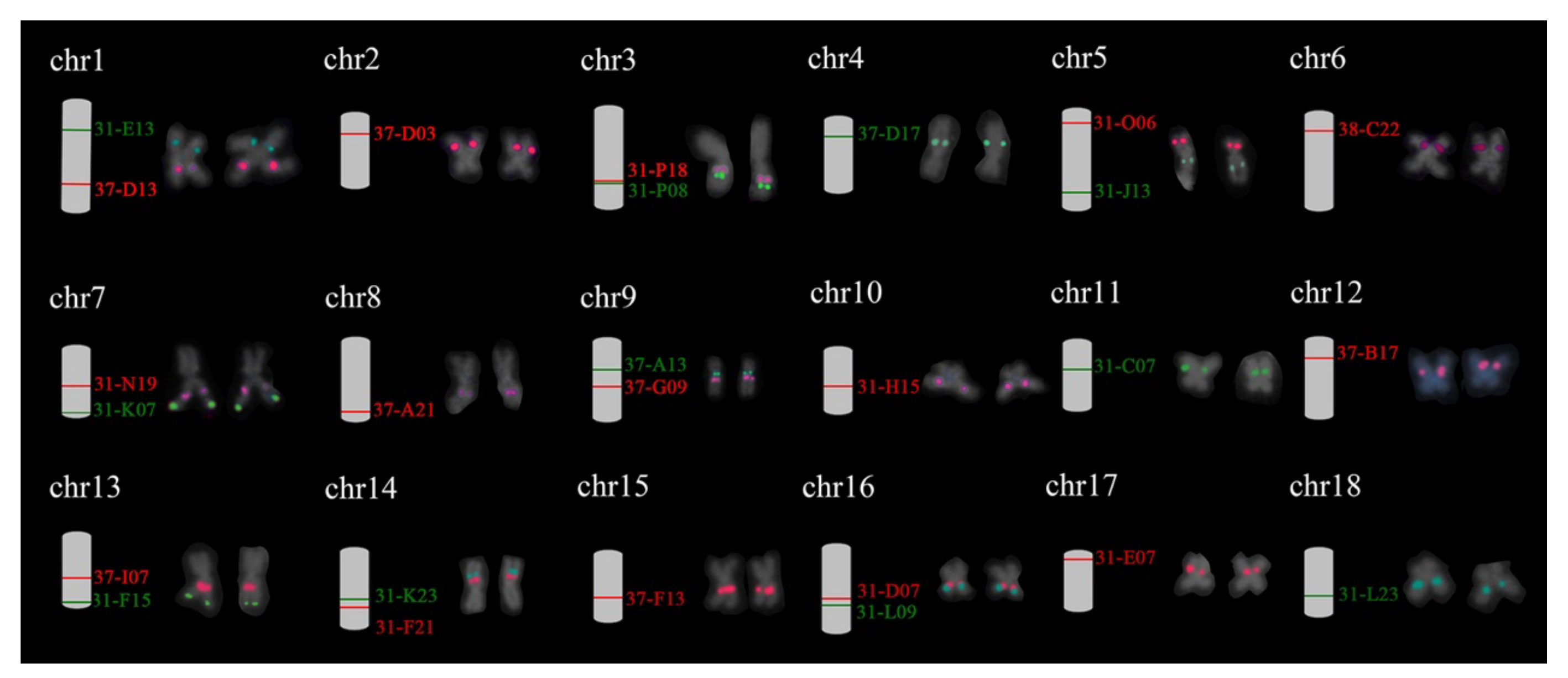
Figure 2 Eighteen individual chromosomes of Pacific abalone with dual-color FISH signals derived from BAC clones. The expected chromosome location of BAC clones is shown on the right side. The corresponding metaphases seen in Supplementary Figure 4. Red and green correspond to digoxigenin- and biotin-labeled probes.
The BESs of H. discus hannai were mapped to the reference genome of H. gigantea. The results showed that there were 200 pairs of end sequences aligned to the genome of H. gigantea (Supplementary Figure 5). Based on the localization results of Abalone, we selected the clones whose BAC clones had different chromosomal positions between the two species for cytogenetic verification. Two BACs derived from the Chr13, and two BACs derived from the Chr15 were further examined through FISH.
The results showed that 37-I07 and 31-F15 were co-located on the on the chromosome 13 of H. gigantea (Figure 3), which implied that the chromosome-specific FISH probe of H. discus hannai might be partially applied to H. gigantea. In H. discus hannai, the clones 31-F15 and 37-I07 were found to be co-located on the short arm of a chromosome, with clone 31-F15 being located near the telomere. However, in H. gigantea, clone 31-F15 and 37-I07 were located on different short arms of chromosomes, and clone 31-F15 was positioned near the centromere. A similar observation was made on chromosome 15. In H. discus hannai, the clones 37-B03 and 31-I13 were found to be located on different short arms of the chromosome. However, in H. gigantea, they were both positioned on the same short arm of the chromosome. The location of chromosome specific clones of chromosome 13 and 15 revealed that chromosome rearrangement might have occurred between the H. discus hannai and the H. gigantea.
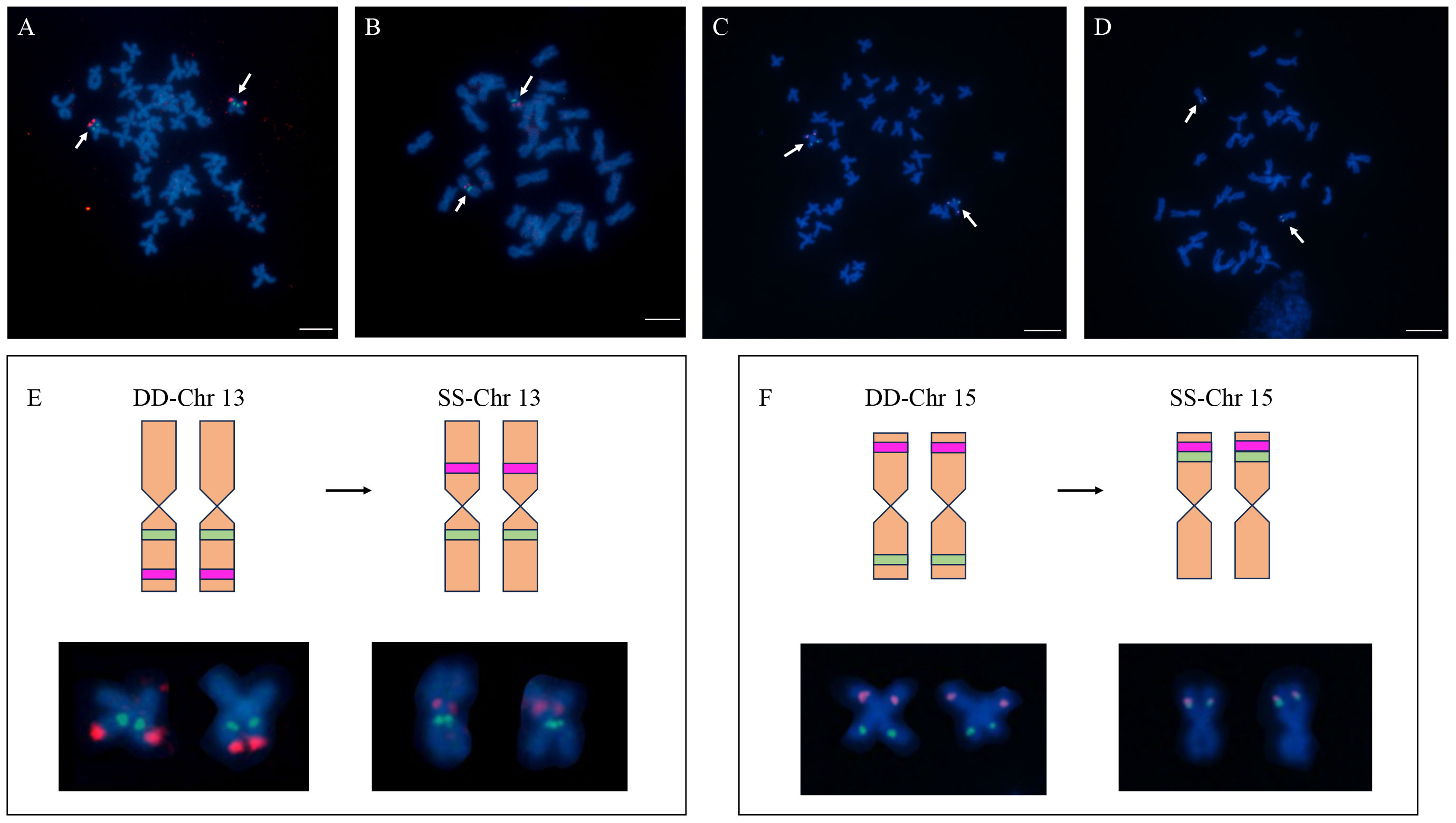
Figure 3 The location of Chromosome-specific probes on chromosome 13 and 15 in the H. discus hannai and H. gigantea. (A) Localization of clones (37-I07 and 31-F15) on the metaphase of H. discus hannai; (B) Localization of clones (37-I07 and 31-F15) on the metaphase of H. gigantea; (C) Localization of clones (37-B03 and 31-I13) on the metaphase of H. discus hannai; (D) Localization of clones (37-B03 and 31-I13) on the metaphase of H. gigantea; (E, F) Chromosome cloning refers to the distribution of chromosomes in different species. The diagram presented here shows a comparison between a schematic representation and a normal chromosome, highlighting the occurrence of chromosomal rearrangements among different species. DD represents the H. discus hannai; SS represents the H. gigantea; The green and red blocks on the diagram represent the locations where signals occur. Bar represents 5 μm for metaphase chromosome.
As far as we know, of all the species of Haliotidae described, only 18 species have been karyotyped (Franchini et al., 2010). Summarizing these studies, it can be found that the number of abalone chromosomes is geographically regular, with the original H. tuberculate having the fewest number of chromosomes (28) (Arai and Wilkins, 1986; Colombera and Tagliaferri, 2014), followed by species living in tropical areas (32) and epigenetic species (36) (Nakamura, 1986; Arai et al., 1988; Ebied et al., 2000; Van der Merwe and Roodt-Wilding, 2008; Cai et al., 2010). The distribution of chromosomes also verifies the evolution theory of abalone. Although lots of studies have given new insight into the phylogenetic status of Haliotidae, the mechanism involve in chromosomal variability are still unknown. The cytogenetic research of Haliotidae mainly focused on the traditional karyotype analysis, chromosome banding and chromosome localization of rDNA (Okumura et al., 1995; Hernández-Ibarra et al., 2007; Wang et al., 2015). But the chromosome evolution in Haliotidae may be more dynamic beyond the previous knowledge. For instance, the karyotype reported previously in the literature for H. discus hannai showed a small difference in the chromosomal structure. Arai et al. found a chromosome composition of 10M +8SM, Okumura et al. found 11M + 7SM, while Wang et al. found a rare type of 10M + 7SM +1ST (Arai et al., 1982; Okumura et al., 1995; Wang et al., 2015). What’s more, the number of NORs and rDNA clusters always varied in many species of Haliotidae revealed by chromosome banding technique and FISH (Gallardo-Escárate et al., 2005; Wang et al., 2015). Therefore, more cytogenetic research should be carried out to build a set of tools for chromosome identification in abalone.
Chromosome-specific BACs have proven to be excellent bridges between DNA and chromosomes (Dong et al., 2000; Schubert et al., 2001). Traditional methods of screening these BACs from constructed library using 3D-PCR were very inefficient and tedious (Taboada et al., 2014), as the power of the chromosome painting probe was often compromised by a high content of repetitive sequence or the innate incorrect location (Trifonov et al., 2009). However, in this study, a bioinformation-based method was used to efficiently screen chromosome specific BACs for chromosome identification in Haliotidae. The method resulted in a higher throughput and more informative approach, with 172 BACs being in silico anchored to the chromosomes. Increasing the number of BACs for ending sequencing could improve the results further, while the negative impact of repetitive sequence could be minimized by evaluating them with bioinformation methods. With the recent assembly of chromosome-level genomes in 135 mollusks species (https://www.ncbi.nlm.nih.gov/) and the BAC libraries in many shellfish species to support the research on genome and genes (Zhang et al., 2008; Jiang et al., 2016; Ma et al., 2019), the information-based method would be a promising way to screen chromosome-specific BACs for chromosome identification.
The versatility of BAC as chromosome- specific probes has been demonstrated across various species, starting with mammalian species and expanding to include many others (Rens et al., 2006). For example, cross-species BAC-FISH painting has been used to study tomato and potato chromosome 6, revealing previously unknown chromosomal rearrangements (Szinay et al., 2012). In strawberry, chromosome-specific oligo probes have shown that the karyotype is highly conserved across 16 different species (Qu et al., 2021). FISH mapping has also been used in fish, with the use of SATA and telomeric probes to identify three fusions in O. karonge that are not present in the typical karyotype (Mota-Velasco et al., 2010). In this study, four chromosome-specific BACs derived from H. discus hannai were hybridized with chromosomes of H. gigantea, which revealed chromosomal specific probes can also be used interchangeably between abalone species, providing a research foundation for subsequent studies on abalone chromosomal evolution.
H. discus hannai and H. gigantea both originated from North America (Lee and Vacquier, 1995). The two species are capable mating and produce offspring, indicating a close genetic relationship between them (Luo et al., 2010). Previous research has shown that both the H. discus hannai and the H. gigantea have the same number of chromosomes and the same chromosome karyotype (Miyaki et al., 1997; Cai et al., 2010). However, there is a difference in the number of rDNA loci between H. discus hannai and H. gigantea, with the former having fewer loci (Wang et al., 2015). This suggests that during the evolutionary process, recombination of repetitive sequences may have occurred in these two abalone species. In fact, karyotype rearrangements are very common in this genus. For example, the karyotype and genome size analysis of Haliotis maidae and three other abalone suggested that there were some preferential karyotype rearrangements during the evolution of the genus (Franchini et al., 2010). Cytogenetic data about three California abalone species showed that significant chromosomal rearrangements occurred during the evolution of Haliotidae on the California coast (Gallardo-Escárate and Del Río-Portilla, 2007). In this study, we found that chromosome 13 and 15 between the two species underwent a chromosome rearrangement event. The results confirmed the conjecture obtained by previous karyotype analysis, and the application potential of chromosome-specific probes developed in this study will be further explored in the future, so as to contribute more data to solving the problem of abalone chromosome evolution.
This study aims to develop a comprehensive set of chromosome-specific BAC-FISH probes that cover all chromosomes in H. discus hannai. The probes were not only useful for improving chromosome identification in metaphase spreads, but also for linking metaphase chromosomes to pseudochromosome of the assembled genome. Additionally, partial cross-species applicability of the developed chromosome-specific BAC-FISH probes in H.gigantea was demonstrated, and cross-species FISH, revealed that there have been chromosome rearrangements in certain chromosomes between the two species. This study marks the first successful development of a chromosome-specific BAC-FISH probe set for Haliotidae, which will enhance the ability to perform cytological studies on abalone species and provide new insight into genome structure and evolution in Haliotidae.
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/ BioProject : PRJNA1041788.
The animal study was approved by Laboratory Animal Management and Ethics Committee of Xiamen University. The study was conducted in accordance with the local legislation and institutional requirements.
JZ: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft, Writing – review and editing, Project administration, Software. YW: Supervision, Validation, Writing – review and editing. ZH: Resources, Data curation, Supervision, Validation, Writing – review and editing. MC: Software, Supervision, Validation, Writing – review and editing. YL: Data curation, Formal Analysis, Supervision, Writing – review and editing. WY: Funding acquisition, Supervision, Validation, Visualization, Writing – review and editing, Resources. XL: Funding acquisition, Resources, Supervision, Validation, Visualization, Writing – review and editing. CK: Funding acquisition, Resources, Supervision, Validation, Visualization, Writing – review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from National Natural Science Foundation of China (No. 32172961), Seed Industry Innovation and Industrialization in Fujian Province (No.2021FJSCZY02), Earmarked Fund for CARS (No. CARS-49), Fundamental Research Funds for the Central Universities (2072022). We’d like to express our gratitude for the assistance provided by the Germplasm Resource-sharing Platform of Aquatic Species in Fujian Province and the XMU-MRB Abalone Research Center.
We would like to thank Songyu Luo for their assistance in BAC library construction of Pacific abalone. We are also grateful to the reviewers for review our manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1266330/full#supplementary-material
Altschul S. F., Madden T. L., Schaffer A. A., Zhang J. H., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25 (17), 3389–3402. doi: 10.1093/nar/25.17.3389
Amar-Basulto G., FL-dl C., Iturra-Constant P., Gallardo-Escárate C. (2011). Karyotype analysis of interspecific hybrids between Haliotis rufescens and Haliotis discus hannai. Aquacult. Res. 42, 1460–1466. doi: 10.1111/j.1365-2109.2010.02738.x
An H. S., Lee J. W., Kim H. C., Myeong J. I. (2011). Genetic characterization of five hatchery populations of the Pacific Abalone (Haliotis discus hannai) using microsatellite markers. Int. J. Mol. Sci. 12 (8), 4836–4849. doi: 10.3390/ijms12084836
Arai K., Fujino K., Kudo M. (1988). Karyotype and Zymogram Differences among Three Species of the Abalones Haliotis planata, H. varia, and H. diversicolor diversicolor. Nippon. Suisan. Gakkaishi. 54 (12), 2055–2064. doi: 10.2331/suisan.54.2055
Arai K., Tsubaki H., Ishitani Y., Fujino K. (1982). Chromosomes of Haliotis discus hannai and Haliotis discus. Bull. Japanese. Soc. Sci. Fisheries. 48 (12), 1689–1692. doi: 10.2331/suisan.48.1689
Arai K., Wilkins N. P. (1986). Chromosomes of haliotis tuberculata L. Aquaculture 58 (3), 305–308. doi: 10.1016/0044-8486(86)90096-7
Bielski W., Książkiewicz M., Šimoníková D., Hřibová E., Susek K., Naganowska B. (2020). The puzzling fate of a lupin chromosome revealed by reciprocal oligo-FISH and BAC-FISH mapping. Genes 11 (12), 1489. doi: 10.3390/genes11121489
Cai M. Y., Ke C. H., Luo X., Wang G., Wang Z., Wang Y. (2010). Karyological Studies of the Hybrid Larvae of Haliotis disversicolor Supertexta Female and Haliotis discus discus Male. J. Shellfish. Res. 29 (3), 735–740. doi: 10.2983/035.029.0327
Cai M. Y., Liu X. D., Chen Z. Y., Cai B. B., Ke C. H. (2013). Characterization of Pacific abalone (Haliotis discus hannai) karyotype by C-banding and fluorescence in situ hybridization with rDNA. J. Fisheries. China 37, 1002–1008. doi: 10.3724/SP.J.1231.2013.38481
Colombera D., Tagliaferri F. (2014). Chromosomes from Male Gonads of Haliotis tubercolata and Haliotis lamellosa (Haliotidae, Archeogasteropoda, Mollusca). Caryologia 36, 231–234. doi: 10.1080/00087114.1983.10797663
Dong F., Song J., Naess S. K., Helgeson J. P., Gebhardt C., Jiang J. (2000). Development and applications of a set of chromosome-specific cytogenetic DNA markers in potato. Theor. Appl. Genet. 101 (7), 1001–1007. doi: 10.1007/s001220051573
Dong G., Shen J., Zhang Q., Wang J., Yu Q., Ming R., et al. (2018). Development and applications of chromosome-specific cytogenetic BAC-FISH probes in S. spontaneum. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00218
do Vale Martins L., de Oliveira Bustamante F., da Silva Oliveira A. R., da Costa A. F., de Lima Feitoza L., Liang Q., et al. (2021). BAC- and oligo-FISH mapping reveals chromosome evolution among Vigna angularis, V. unguiculata, and Phaseolus vulgaris. Chromosoma 130 (2-3), 133–147. doi: 10.1007/s00412-021-00758-9
Ebied A. M., Hassan H. A., Abu Almaaty A., Yaseen A. E. (2000). Cytogenetic studies on metaphase chromosomes of eight gastropod species of orders Mesogastropoda and Neogastropoda (Prosobranchiata – Mollusca). JEgyptGerSocZool 33, 317–339.
Franchini P., Slabbert R., van der Merwe M., Roux A., Roodt-Wilding R. (2010). Karyotype and genome size estimation of Haliotis midae: estimators to assist future studies on the evolutionary history of Haliotidae. J. Shellfish. Res. 29 (4), 945–950. doi: 10.2983/035.029.0428
Gallardo-Escárate C., Álvarez-Borrego J., Del Río-Portilla M. A., Cross I., Merlo A., Rebordinos L. (2005). Fluorescence in situ hybridization of rDNA, telomeric (TTAGGG)n and (GATA)n repeats in the red abalone Haliotis rufescens (Archaeogastropoda: Haliotidae). Hereditas 142, 73–79. doi: 10.1111/j.1601-5223.2005.01909.x
Gallardo-Escárate C., Del Río-Portilla M. A. (2007). Kartotype composition in three california abalones and their relationship with genome size. J. Shellfish. Res. 26, 825–832. doi: 10.2983/0730-8000(2007)26[825:KCITCA]2.0.CO;2
Han Y. H., Zhang T., Thammapichai P., Weng Y. Q., Jiang J. M. (2015). Chromosome-specific painting in cucumis species using bulked oligonucleotides. Genetics 200 (3), 771–779. doi: 10.1002/gcc.2870040311
Hernández-Ibarra N., Ibarra A. M., Cruz P., Leitch A. (2007). FISH mapping of 5S rRNA genes in chromosomes of North American abalone species, Haliotis rufescens and H. fulgens. Aquaculture 272, S268. doi: 10.1016/j.aquaculture.2007.07.086
Jiang L., You W., Zhang X., Xu J., Jiang Y., Wang K., et al. (2016). Construction of the BAC library of small abalone (Haliotis diversicolor) for gene screening and genome characterization. Mar. Biotechnol. 18 (1), 49–56. doi: 10.1007/s10126-015-9666-4
Kobayashi T., Kijima A. (2010). Effects of Inbreeding Depression in Pacific Abalone Haliotis discus hannai. J. Shellfish. Res. 29 (3), 643–9, 7. doi: 10.2983/035.029.0313
Lee Y. H., Vacquier V. (1995). Evolution and systematics in Haliotidae (Mollusca: Gastropoda): inferences from DNA sequences of sperm lysin. Mar. Biol. 124, 267–278. doi: 10.1007/BF00347131
Liu X., Liu X., Guo X., Gao Q., Zhao H., Zhang G. (2006). A preliminary genetic linkage map of the Pacific abalone Haliotis discus hannai Ino. Mar. Biotechnol. 8, 386–397. doi: 10.1007/s10126-005-6133-7
Luo M., Kim H., Kudrna D., Sisneros N. B., Lee S. J., Mueller C., et al. (2006). Construction of a nurse shark (Ginglymostoma cirratum) bacterial artificial chromosome (BAC) library and a preliminary genome survey. BMC Genomics 7, 106. doi: 10.1186/1471-2164-7-106
Luo X., Ke C., You W., Wang D. (2010). Factors affecting the fertilization success in laboratory hybridization between Haliotis discus hannai and Haliotis gigantea. J. Shellfish. Res. 29, 621–625. doi: 10.2983/035.029.0310
Ma K. Y., Yu S. H., Du Y. X., Feng S. Q., Qiu L. J., Ke D. Y., et al. (2019). Construction of a genomic bacterial artificial chromosome (BAC) library for the prawn Macrobrachium rosenbergii and initial analysis of ZW chromosome-derived BAC inserts. Mar. Biotechnol. 21 (2), 206–216. doi: 10.1007/s00343-008-0215-0
Miyaki K., Tabeta O., Kayano H. (1997). Karyotypes of the Two Species of Abalones Nordotis discus and N. gigantea. Fisheries. Sci. 63, 179–180. doi: 10.2331/fishsci.63.179
Morash A., Alter K. (2015). Effects of environmental and farm stress on abalone physiology: Perspectives for abalone aquaculture in the face of global climate change. Rev. Aquacult. 8 (4), 342–368. doi: 10.1111/raq.12097
Mota-Velasco J. C., Ferreira I. A., Cioffi M. B., Ocalewicz K., Campos-Ramos R., Shirak A., et al. (2010). Characterisation of the chromosome fusions in Oreochromis karongae. Chromosome Res. 18 (5), 575–586. doi: 10.1007/s10577-010-9141-z
Nakamura H. K. (1986). Chromosomes of archaeogastropoda (Mollusca: prosobranchia), with some remarks on their cytotaxonomy and phylogeny. Publications. Seto. Mar. Biol. Laboratory. 31, 191–267. doi: 10.5134/176126
Nam B., Kwak W., Kim Y., Kim D., Kong H., Kim W., et al. (2017). Genome sequence of Pacific Abalone (Haliotis discus hannai): the first draft genome in family Haliotidae. Gigascience 6, gix014. doi: 10.1093/gigascience/gix014
Nguyen T. V., Alfaro A. C., Mundy C., Petersen J., Ragg N. L. C. (2022). Omics research on abalone (Haliotis spp.): Current state and perspectives. Aquaculture 547, 737438. doi: 10.1016/j.aquaculture.2021.737438
Nie H., Li Q., Kong L. (2012). Centromere mapping in the Pacific abalone (Haliotis discus hannai) through half-tetrad analysis in gynogenetic diploid families. Anim. Genet. 43 (3), 290–297. doi: 10.1111/j.1365-2052.2011.02254.x
Okumura S., Yamada S., Sugie T., Sekiyama D., Toda A., Hajima H., et al. (1995). C-banding study of chromosomes in Pacific abalone, Haliotis discus hannai (Archaeogastropoda: Haliotidae). Chromosome Inf. Service.: CIS. 59, 7–9.
Palma-Rojas C., von Brand E. (2008). Preliminary results of C-banding pattern in the red abalone Haliotis rufescens. J. Shellfish. Res. 27 (4), 1037–1038.
Pinkel D., Straume T., Gray J. W. (1986). Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. P Natl. Acad. Sci. U.S.A. 83 (9), 2934–2938. doi: 10.1073/pnas.83.9.2934
Qu M., Zhang L., Li K., Sun J., Li Z., Han Y. (2021). Karyotypic stability of Fragaria (strawberry) species revealed by cross-species chromosome painting. Chromosome Res. 29 (3-4), 285–300. doi: 10.1007/s10577-021-09666-1
Rens W., Moderegger K., Skelton H., Clarke O., Trifonov V., Ferguson-Smith M. (2006). A procedure for image enhancement in chromosome painting. Chromosome Res. 14, 497–503. doi: 10.1007/s10577-006-1056-3
Schubert I., Fransz P. F., Fuchs J., Jong J. (2001). Chromosome painting in plants. Methods Cell Sci. 23, 57–69. doi: 10.1023/A:1013137415093
Sekino M., Hara M. (2007). Linkage maps for the Pacific abalone (genus Haliotis) based on microsatellite DNA markers. Genetics 175 (2), 945–958. doi: 10.1534/genetics.106.065839
Shi X., Zeng H., Xue Y., Luo M. (2011). A pair of new BAC and BIBAC vectors that facilitate BAC/BIBAC library construction and intact large genomic DNA insert exchange. Plant Methods 7 (1), 33. doi: 10.1186/1746-4811-7-33
Szinay D., Wijnker E., Berg R., Visser R., Jong H., Bai Y. (2012). Chromosome evolution in Solanum traced by cross-species BAC-FISH. New Phytol. 195, 688–698. doi: 10.1111/j.1469-8137.2012.04195.x
Taboada X., Pansonato-Alves J. C., Foresti F., Martinez P., Vinas A., Pardo B. G., et al. (2014). Consolidation of the genetic and cytogenetic maps of turbot (Scophthalmus maximus) using FISH with BAC clones. Chromosoma 123 (3), 281–291. doi: 10.1007/s00412-014-0452-2
Telenius H., Pelmear A. H., Tunnacliffe A., Carter N. P., Behmel A., Fergusonsmith M. A., et al. (1992). Cytogenetic analysis by chromosome painting using DOP-PCR amplified flow-sorted chromosomes. Gene Chromosome Canc. 4 (3), 257–263. doi: 10.1002/gcc.2870040311
Trifonov V. A., Vorobieva N. N., Rens W. (2009). “FISH With and Without COT1 DNA,” in Fluorescence In Situ Hybridization (FISH) — Application Guide. Ed. Liehr T. (Berlin, Heidelberg: Springer Berlin Heidelberg), 99–109. doi: 10.1007/978-3-540-70581-9_9
Van der Merwe M., Roodt-Wilding R. (2008). Chromosome number of the South African abalone Haliotis midae. Afr. J. Mar. Sci. 30, 195–198. doi: 10.2989/AJMS.2008.30.1.21.471
Vandewoestyne M., Van Hoofstat D., Van Nieuwerburgh F., Deforce D. (2009). Suspension fluorescence in situ hybridization (S-FISH) combined with automatic detection and laser microdissection for STR profiling of male cells in male/female mixtures. Int. J. Legal. Med. 123 (5), 441–447. doi: 10.1007/s00414-009-0341-z
Wang H. S., Luo X., You W. W., Dong Y. W., Ke C. H. (2015). Cytogenetic analysis and chromosomal characteristics of the polymorphic 18S rDNA of Haliotis discus hannai from Fujian, China. PloS One 10 (2), e0113816. doi: 10.1371/journal.pone.0113816
Yang Z., Li X., Liao H., Hu L., Peng C., Wang S., et al. (2019). A molecular cytogenetic map of scallop (Patinopecten yessoensis). Mar. Biotechnol. 21 (6), 731–742. doi: 10.1007/s10126-019-09918-6
Zhang J., Xie J., Liu B., Wang Z., Wang Y., Jiang Y., et al. (2022). Development and applications of chromosome-specific cytogenetic BAC-FISH probes in Larimichthys crocea. Front. Mar. Sci. 9, 1078110. doi: 10.3389/fmars.2022.1078110
Keywords: Haliotis discus hannai, chromosome identification, fluorescence in situ hybridization (FISH), bacterial artificial chromosome (BAC), chromosome evolution Haliotis discus hannai
Citation: Zhang J, Wang Y, Huang Z, Cai M, You W, Lu Y, Luo X and Ke C (2023) Development and applications of chromosome-specific BAC-FISH probes in Pacific abalone (Haliotis discus hannai). Front. Mar. Sci. 10:1266330. doi: 10.3389/fmars.2023.1266330
Received: 24 July 2023; Accepted: 02 November 2023;
Published: 24 November 2023.
Edited by:
Yunyan Deng, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Liang Guo, Hunan Normal University, ChinaCopyright © 2023 Zhang, Wang, Huang, Cai, You, Lu, Luo and Ke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caihuan Ke, Y2hrZUB4bXUuZWR1LmNu; Xuan Luo, eGx1b0B4bXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.