- 1Department of Life Science, Division of EcoScience, Ewha Womans University, Seoul, Republic of Korea
- 2Laboratory of Ecology and Environmental Management, Science and Technology Advanced Institute, Van Lang University, Ho Chi Minh City, Vietnam
- 3Department of International Program, US Vietnam Talent International School, Ho Chi Minh City, Vietnam
The introduced Asian mud-tidal gastropod Batillaria attramentaria has been reported to quickly dominate its new habitat, Elkhorn Slough, USA, which has a different osmotic condition from its native habitat. This species has also been reported to have a substantial impact on the ecosystem of the new habitat. In this study, we compared the gene expression profiles in response to temporary salinity variation in native (Asian) and introduced (North American) snails and elucidated the genetic mechanism underlying such rapid adaptation of the introduced species. We examined the transcriptomes of four B. attramentaria populations, including three from the native habitats and one from the introduced habitat, in response to salinity variation. We found that 2,353, 2,505, 10,362, and 16,381 genes were differentially expressed due to their lineages (Kuroshio vs. Tsushima), origins (native vs. introduced), locations (Korea, Japan, or the USA), and salinity variations (stressful: 13, 23, and 43 PSU vs. optimal: 33 PSU), respectively. We found that GO-enriched differentially expressed genes involved in the detection of various abiotic and biotic stimuli through sensory perception and genes involved in response to abiotic stimulus and stress were upregulated when exposed to lower-salinity conditions in all locations. The results indicated that B. attramentaria has adapted well to varied salinity conditions and evolved after around 100 years of invasion in Elkhorn Slough. This study provides valuable transcriptomic data on salinity stress response genes in the tidal gastropod and contributes to the research on the adaptive evolution of coastal gastropods.
1 Introduction
The invasions of biological species, particularly those involving rapid expansion from native ranges into new ecological habitats, often cause evolutionary problems for native species and the ecology of the encroached habitat. In biological invasions, invasiveness is characterized by the selection process of nonindigenous species, which is influenced by various factors (Lee, 2002). Abiotic (Spalding et al., 2007) and biotic factors include advantageous traits, such as dispersal ability or reproductive assurance, female-biased sex ratio, gamete quality, and fecundity (Reznick and Ghalambor, 2001; Le Cam et al., 2009; Sussarellu et al., 2015); these traits have attracted scientific attention. Moreover, the absence of parasites or predator species specific to invading hosts in the new habitats enables their successful inhabitation and expansion (Colautti et al., 2004; Dunn, 2009; Strayer, 2012). Several studies conducted over the last few decades have shown that the successful invasion of coastal habitats depends not only on physiology and plasticity but also on adaptive responses (Baker, 1965; Strayer, 1999; Wolff, 2000; Stockwell et al., 2003; Sax et al., 2007). As insufficient genomic information is available on invading marine organisms, research on their genetic mechanisms of invasions requires more attention (Prentis et al., 2008; Bock et al., 2016). Transcriptomic studies on Mytilus bivalves (Lockwood et al., 2010; Lockwood and Somero, 2011) and Pacific oysters (Sussarellu et al., 2015) will provide further insights in this direction.
Among various introduced organisms, the intertidal snail Batillaria attramentaria is considered a model for studying biological invasion because it exhibits direct development and limited dispersal ability (Kojima et al., 2004) and forms a relatively closed local population after translocation through various human-mediated activities. B. attramentaria is the most common Batillaria species in the Northwest Pacific Ocean. Its range extends from southern Taiwan (Golikov, 1967) and Hong Kong (Golikov and Gulbin, 1978) to southern Primorye, Sakhalin, and northern Kuriles (Golikov, 1967; Golikov and Gulbin, 1978; Golikov and Scarlato, 1985), encompassing the seacoasts of China, Korea, and Japan between the latitudinal limits. Originally from northeast Asia, B. attramentaria was introduced into the bays and estuaries of the Pacific coast of North America by human-mediated transport along with the importation of the Pacific oyster Crassostrea gigas from Japan in the early nineteenth century (Fisheries and Galtsoff, 1932). It appeared 30 years later in Monterey Bay, Elkhorn Slough (Bonnot, 1935). B. attramentaria then reproduced rapidly, and their small population increased. As a result, these introduced snails have now become one of the most common residents in these areas in the present time. The introduced B. attramentaria have severely impacted other snail populations on California mud flats. The native snail Cerithidea californica and the introduced snail B. attramentaria feed on epibenthic diatoms. As revealed in previous manipulative field experiments and modeling projections (Byers, 2000a), because of exploitative competition for food resources, B. attramentaria has completely displaced the native snail C. californica in Elkhorn Slough (Monterey County, California). It has also impacted other native snails in the bay and estuary regions along several northern California marshes (Byers, 2000a; Byers, 2000b). On the other hand, B. attramentaria species have positively impacted native species in several areas in Washington, such as Padilla Bay, by providing their dead shells as new rigid substrates for native hermit crabs (Pagurus hirsutiusculus and P. granosimanus), along with the introduced Atlantic slipper snail Crepidula convexa and the Asian sea anemone Diadumene lineata (Wonham et al., 2005). While the ecological aspects of the invasion of this species have been reported, its adaptive physiological and genetic responses during the invasion remain unclear because of the lack of genomic resources. However, our previous study provided behavioral and transcriptomic data of native B. attramentaria from Korea in response to salinity variation; these findings can serve as the basis for studying the adaptive evolution of snails (Ho et al., 2019; Ho et al., 2021).
In the intertidal zone, ecosystems are highly complex because of the continuous fluctuation between dry and submerged conditions, temperature changes, and salinity variation caused by river effluxes and tides. Among these factors, salinity variation affects the ability of marine organisms to adapt to this habitat (Somero, 2012; Shui et al., 2022). It is therefore convincing that the intertidal snail B. attramentaria is well adapted to salinity fluctuations. The salinity fluctuation occurring throughout the year varied among the sampling sites and could be categorized as follows: low (29–33 PSU) in Japan (Fukuda et al., 2016; Okumura et al., 2021), moderate (18–32 PSU) in Korea (Park et al., 2020), and high (0–30 PSU) in the USA (Pennington and Chavez, 2000). In this study, B. attramentaria snail populations belonging to two lineages, Tushima and Kuroshio, were used based on our previous findings (Ho et al., 2015; Ho et al., 2021). These two lineages are classified according to the northward Pacific Sea current passing by the north and south of the Japanese archipelago (Kojima et al., 2004), respectively. In this study, we assessed the differences in gene expression patterns between the native and introduced B. attramentaria snails and between the Tsushima and Kuroshio lineage snails on continuous exposure to a range of salinity conditions (13–43 PSU).
We also identified genes associated with salinity stress response by comparing gene expression patterns between native and introduced snails. The results may provide insights into how B. attramentaria snails cope with life under varied salinity conditions. They may also increase our understanding of the response patterns of mud-tidal snails and highlight potentially relevant genes for future research. To understand the genetic regulation mechanism to withstand salinity fluctuation, we studied the transcriptomic responses of native and introduced populations of B. attramentaria to different salinity conditions (43, 33, 23, and 13 PSU). The findings will also help unravel the genetic mechanism that has evolved between two divergent genetic lineages and between native and introduced B. attramentaria populations. This study extends our previous evolutionary, physiological, behavioral, and transcriptional analyses of the salinity tolerance of B. attramentaria from the Korean coast (Ho et al., 2015; Ho et al., 2019; Ho et al., 2021). The findings will also deepen our understanding of the biological invasion and adaptive evolution of B. attramentaria and thus provide insights into the adaptive molecular evolution of other marine invertebrates during their invasion of new habitats.
2 Materials and methods
2.1 Sample collection
Native B. attramentaria (G. B. Sowerby I, 1855) snails were collected from Hacheon-ri, Cheonlabuk-do, Republic of Korea (ROK, 35°32′N, 126°33′E); from Nemuro city, Hokkaido Prefecture, Japan (43°15′N, 145°28′E); and from Matsushima Bay, Miyagi Prefecture, Japan (38°22′N, 141°4′E). The introduced snails were collected from Monterey Bay, Elkhorn Slough, CA, USA (36°49′N, 121°45′W) (Figure 1). Three native populations, including one Korean and two Japanese snail populations (80 individuals per population), were collected from sites with a moderate surface salinity of 29–33 PSU. On the other hand, the intrusive population consisting of 40 individuals was collected from a site with a low surface salinity of 4 PSU. Surface salinities were measured at the sampling time. Only adult individuals with equivalent shell lengths (approximately 2.5 cm for native populations and approximately 4 cm for the introduced population) were used for salinity acclimation, behavioral observations, and transcriptomics experiments. All specimens were grown in a plastic aquarium (25°C room temperature and 12-h light:12-h dark cycle) for 2 days before the long-term acclimation experiments to reduce the effects of transport, as described by Ho et al. (2019).
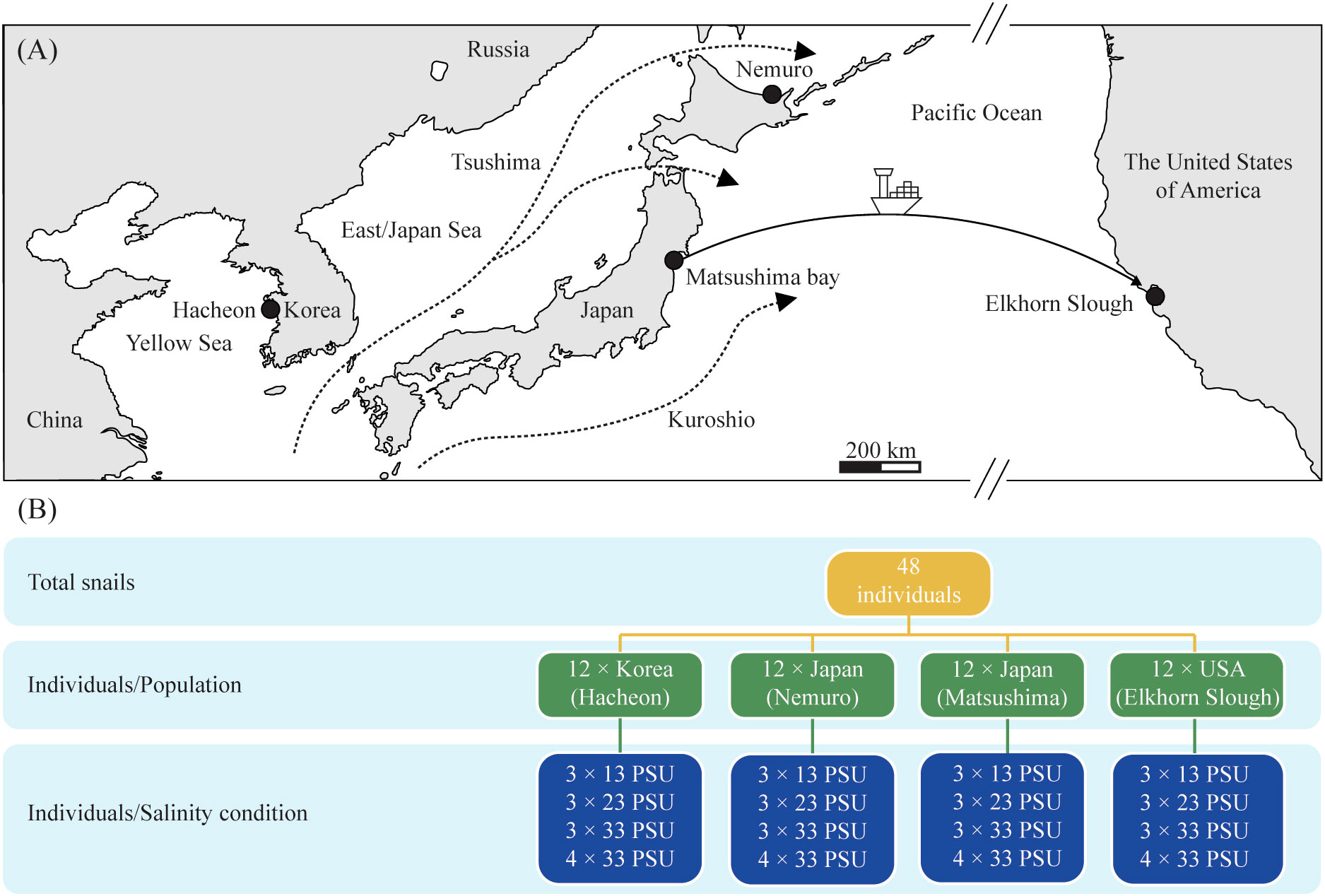
Figure 1 (A) Map showing the sampling sites of the Batillaria attramentaria snails used in the present study. Gray color represents continental areas. Black dots are sampling sites. (B) Schematic view of the experimental design displaying snails treated with each salinity conditions used for transcriptomic analysis.
2.2 Salinity acclimation experiments
After performing species identification for a representative snail, the snails from each sampling sites were randomly divided into four groups for exposure to different salinity conditions (13, 23, 33, and 43 PSU) equally (20 snails per treatment for the Japanese population and 10 snails per treatment for the USA population). These four groups of each snail population were reared in separate plastic aquaria (40 × 23 × 21 cm3) with an inclined layer of sea sand on the bottom and fresh aerated artificial sea at salinities of 13, 23, 33, and 43 PSU for 30 days. Each snail in each group was marked using nail polish (Eco Nail color, Innisfree, ROK) for their identification. All animals were fed to satiation with excised fresh seaweed (Ottogi, ROK) every 2 days throughout the acclimation experiments.
2.3 Transcriptomic sequencing and assembly
Only gill tissues of snails that were thoroughly acclimated to 13, 23, 33, and 43 PSU salinity regimes were used for de novo transcriptomic analysis with three biological replicates per salinity condition. Sample preparation and RNA isolation were performed using the RNeasy Mini kit (Qiagen, Venlo, Netherlands). RNA integrity was determined using a NanoDrop 1000 Spectrometer (Thermo Scientific, DE, USA) and an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA). Next, library construction was performed using the TruSeq RNA Library Prep Kit (Illumina, CA, USA). Transcriptomic sequencing was performed on an Illumina HiSeq 4000 Genome Analyzer platform (Illumina, CA, USA). All sequences were trimmed from adapter fragments using Trimmomatic v.0.36 (Bolger et al., 2014) before further processing. Trinity v.r20140717 (the Broad Institute and the Hebrew University of Jerusalem) with default parameters was used for the de novo assembly of short reads with at least the N50 values into longer fragments or contigs without N gaps and to create the reference transcriptome (Grabherr et al., 2011). A total of 5 billion reads containing 528 billion bases were produced with an average GC content of 44.52% and an average Q30 value of 95.60%. In the following step, all the trimmed reads of each sample were aligned to the de novo reference genome using Bowtie v.1.1.2 (Langmead et al., 2009) to generate a total of 626,765 genes and obtain an estimated count of reads that would be used for creating a read count matrix using HTSeq v.0.6.0 (Anders and Huber, 2010). Moreover, all the identified contigs were clustered into a non-redundant transcript, called Unigenes using the CD-HIT-EST v.4.6 program, which was further used for annotation, open reading frame prediction, and expression analysis (Fu et al., 2012).
2.4 Multidimensional scaling
Multidimensional scaling (MDS) was performed to visualize the level of similarity of the experimental variables of salinity levels and animal populations of the variations in gene expression patterns (Borg and Groenen, 2005). The results of MDS were assessed using loading plots; the spatial clustering of experimental samples on loading plots reflects the degree of similarity between transcriptomes. The abundance values of all genes were normalized to log2 (FPKM + 1) before MDS analysis. This allowed the identification of any outlier samples or similar expression patterns between sample groups. The results of MDS were computed using a correlation matrix. The analysis was performed using the R Statistics software (R Development Core Team, 2011).
2.5 Ecological factors
In addition to exposure salinity, four ecological factors that affect the behavioral and transcriptional differences among groups were assessed: (1) genetic clades of each divergent lineage, i.e., lineage, (2) sample origin, (3) location, and (4) population. Lineage was created by matching mitochondrial COI gene haplotypes of each individual and categorized into two groups: Tsushima (including Hacheon and Nemuro populations) and Kuroshio (including Matsushima and Elkhorn Slough populations). Origin was categorized into two groups: native (including populations collected from the coastline of Korea and Japan) and introduced (including a population collected from Elkhorn Slough, CA, USA) groups. Location was categorized by the nation from which the samples were collected: Korea, Japan, and the USA. Population referred to the snails collected from the sampling sites (Hacheon in Korea, Nemuro and Matsushima in Japan, and Elkhorn Slough in the USA).
2.6 Determination of significant changes in gene expression
The DESeq2 package (Bioconductor) was used for the analysis of differentially expressed genes (DEGs) (Anders and Huber, 2010; Ching et al., 2014; Costa-Silva et al., 2017). DEGs were analyzed to identify significant changes in gene expression in response to salinity variation (13, 23, 33, and 43 PSU) between individuals from different origins (native and introduced), lineages (Tsushima and Kuroshio), locations (Korea, Japan, and the USA), and populations (Hacheon, Nemuro, Matsushima, and Elkhorn Slough). Two-way analysis of variance (two-way ANOVA) was conducted to assess raw reads of pooled transcript data of all snails from the four aforementioned locations (N = 48). In the two-way ANOVA, exposure salinity, origin, location, population, and lineage were modeled as fixed effects. We focused on genes that were significant for the interactions of exposure salinity × origin, exposure salinity × location, exposure salinity × population, and exposure salinity × lineage. Subsequently, we identified origin-, location-, and lineage-specific genes that may have contributed to the adaptive evolutionary process of the species after approximately 100 years of introduction into the new habitat and determined the genetic divergence among the two lineages, respectively. These statistical analyses were performed using the same count matrix and measuring fold change (fc) measurement with |fc| ≥ 2. All p values were adjusted with a false discovery rate (FDR) correction for multiple testing using the Benjamini–Hochberg method (Benjamini and Hochberg, 1995). These analyses were conducted using R Statistics (R Development Core Team, 2011).
2.7 Gene annotation and functional analysis
For the annotation of the transcriptome, all transcripts were aligned against the protein reference databases Gene Ontology Consortium (GO v.20150407, http://www.geneontology.org/) and UniProtKB (reviewed as Swiss-Prot, http://www.uniprot.org/downloads, released on 2012/04/20) using DIAMOND-FAST V.0.4.7 (Buchfink et al., 2015). GO enrichment analysis of DEGs was performed on GOATOOLS (Klopfenstein et al., 2018) using Fisher’s exact test, with the p-value corrected using the Benjamini–Hochberg FDR statistical model. GO enrichment bubble plots were drawn using the ggplot2 package in R (Wickham, 2011).
2.8 Short-read submission
The short-read (Illumina HiSeq 4000) data of the Korean population (SRA accession number: SUB2798442), Nemuro and USA populations (SRA accession number: SUB3156821), and Matsushima population (SRA accession number: PRJNA980419) can be found online at the NCBI website.
3 Results
3.1 Transcriptomic responses of the snails to salinity variation
Trinity assembly generated 626,765 contigs from 48 samples (Table 1). After normalization, there were 41,194 contigs that remained for DEG analyses. DEGs across samples collected under control (33 PSU) and stress (13, 23, and 43 PSU) conditions were counted with fold change |fc| ≥2 and FDR-corrected p-value <0.05. Both upregulated and downregulated DEGs were detected based on ecological factors (Figure 2). An MDS plot, based on the pooled wide transcriptomes of all 48 samples from Korea, Japan, and the USA, was used to visualize the relationship of the DEGs with exposure salinity, lineage, origin, location, and population (Figure 3). Among all parameters, principal component 1 (PC1) explained 18% of the variance in gene expression, whereas principal component 2 (PC2) explained 11.9% of the variance in gene expression.
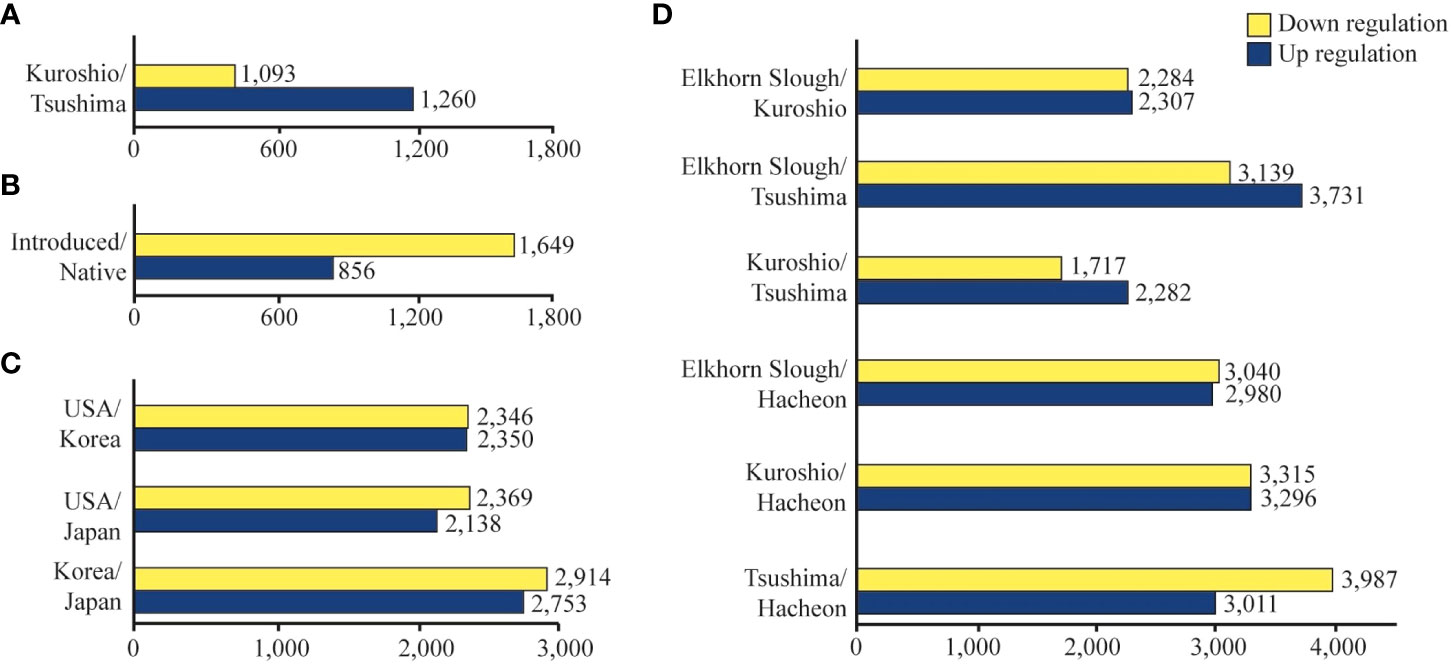
Figure 2 Bar plot showing the number of upregulated and downregulated DEGs in snail populations under salinity variations. DEGs according to (A) lineage, (B) origin, (C) location, and (D) population.
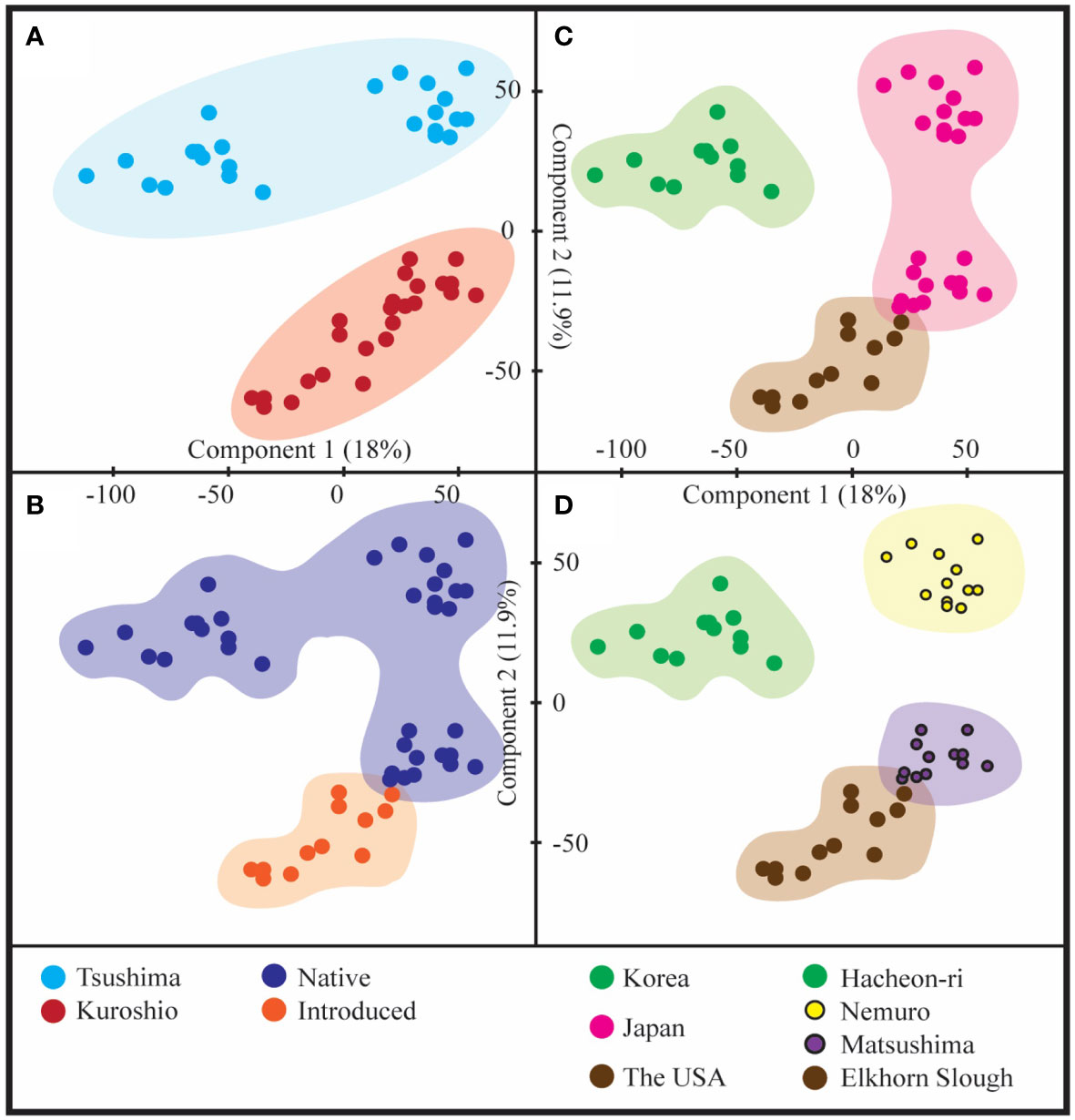
Figure 3 Multidimensional scaling (MDS) loading plot of principal component 1 (PC1) vs. principal component 2 (PC2) based on the transcriptomes of all the samples. (A) Lineage, (B) origin, (C) location, and (D) population based MDS.
3.2 Lineage-specific DEGs in response to salinity variation
The analysis of lineage-specific DEGs revealed a total of 2,353 significant DEGs in response to salinity variation between Kuroshio and Tsushima snails (1,093 up- and 1,260 downregulated genes (Figure 2A). The MDS analysis revealed that the USA and Japanese (Matsushima) snail populations originated from the Kuroshio. On the other hand, the Korean and Japanese (Nemuro) snail populations were derived from the Tsushima lineage (Figure 3A). Hierarchical clustering analysis revealed several gene expression pattern changes among the two lineages, which can be seen as two distinguished blocks in Figure 4A. GO enrichment analysis suggested that among the many genes, vesicle and regulation of metabolic process and sulfotransferase activity were upregulated whereas organic cyclic compound binding and carbohydrate binding genes were downregulated. By contrast, transferase and catalytic activity genes were both upregulated and downregulated (Figure 5). We also analyzed the gene expression patterns of Kuroshio and Tsushima snail populations in response to different salinity variations (Supplementary Figure 1). At a salinity of 13 PSU, both Kuroshio and Tsushima lineage snails exhibited a higher number of overexpressed genes. Tsushima lineage snails had 1,799 genes and Kuroshio lineage snails had 1,1779 that were highly expressed under the 13 PSU salinity condition (Supplementary Figure 1). GO enrichment analysis of DEGs under the same condition suggested that genes related to sulfur compound metabolic process, peptidyl-amino acid modification, organic acid metabolic process, molybdoprotein cofactor metabolic process, methionine and aspartate family amino acid metabolic process, response to stimulus and stress, and cellular metabolic process were overexpressed in Tsushima lineage snails (Supplementary Figure 2). On the other hand, genes related to transferase activity; response to virus and other organisms; defense response to virus and other organisms; transmembrane transporter activity; organic ion binding; metal ion binding; and cation, calcium ion, cadherin, and bile acid binding were overexpressed in Kuroshio lineage snails (Supplementary Figure 2).
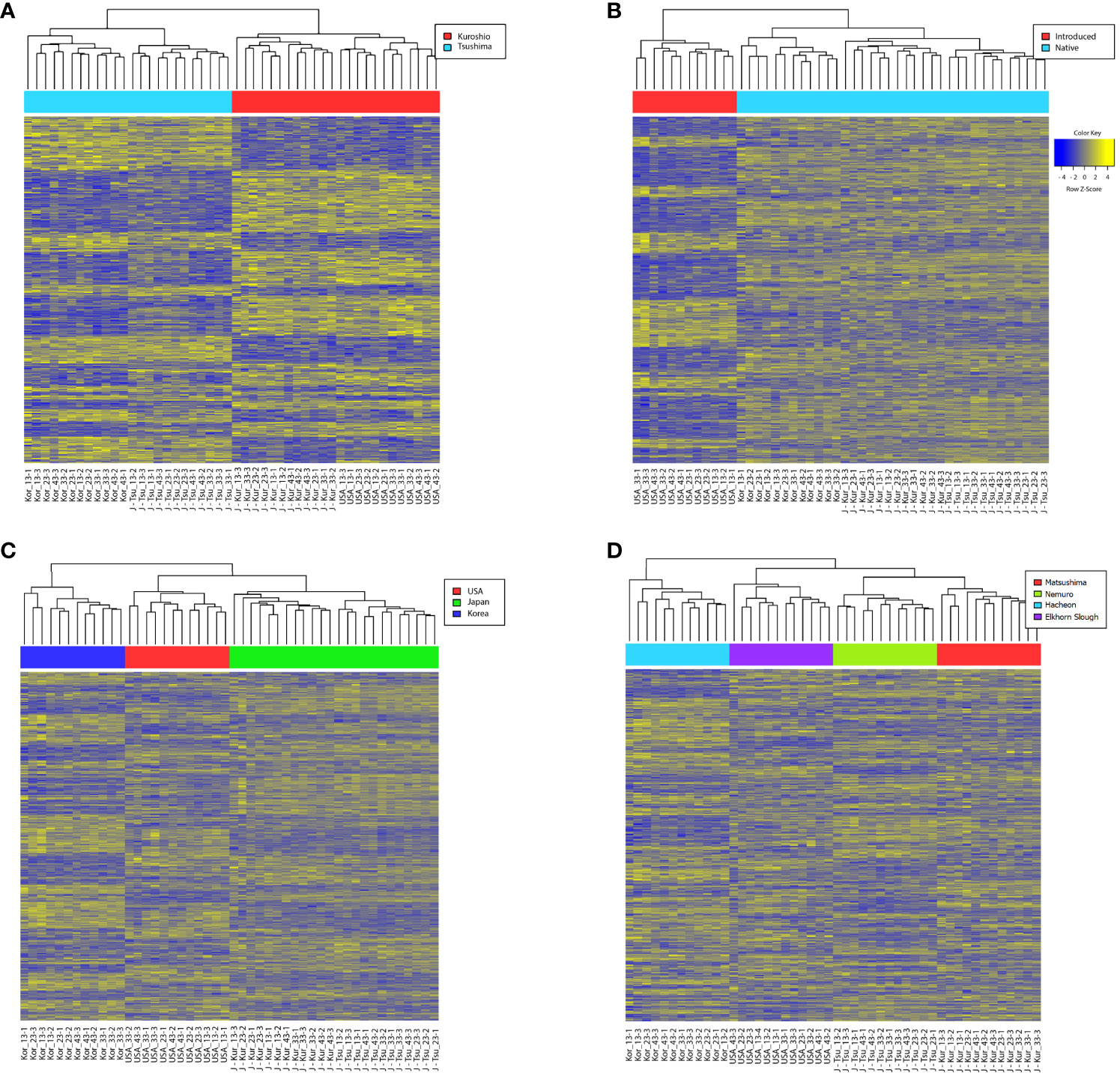
Figure 4 Heatmap of DEGs (|fc| ≥2, FDR-corrected p < 0.05) in snail populations. Genes are ordered in rows. DEG analyses of groups of snails based on (A) lineage (B) origin (C) location and (D) population.
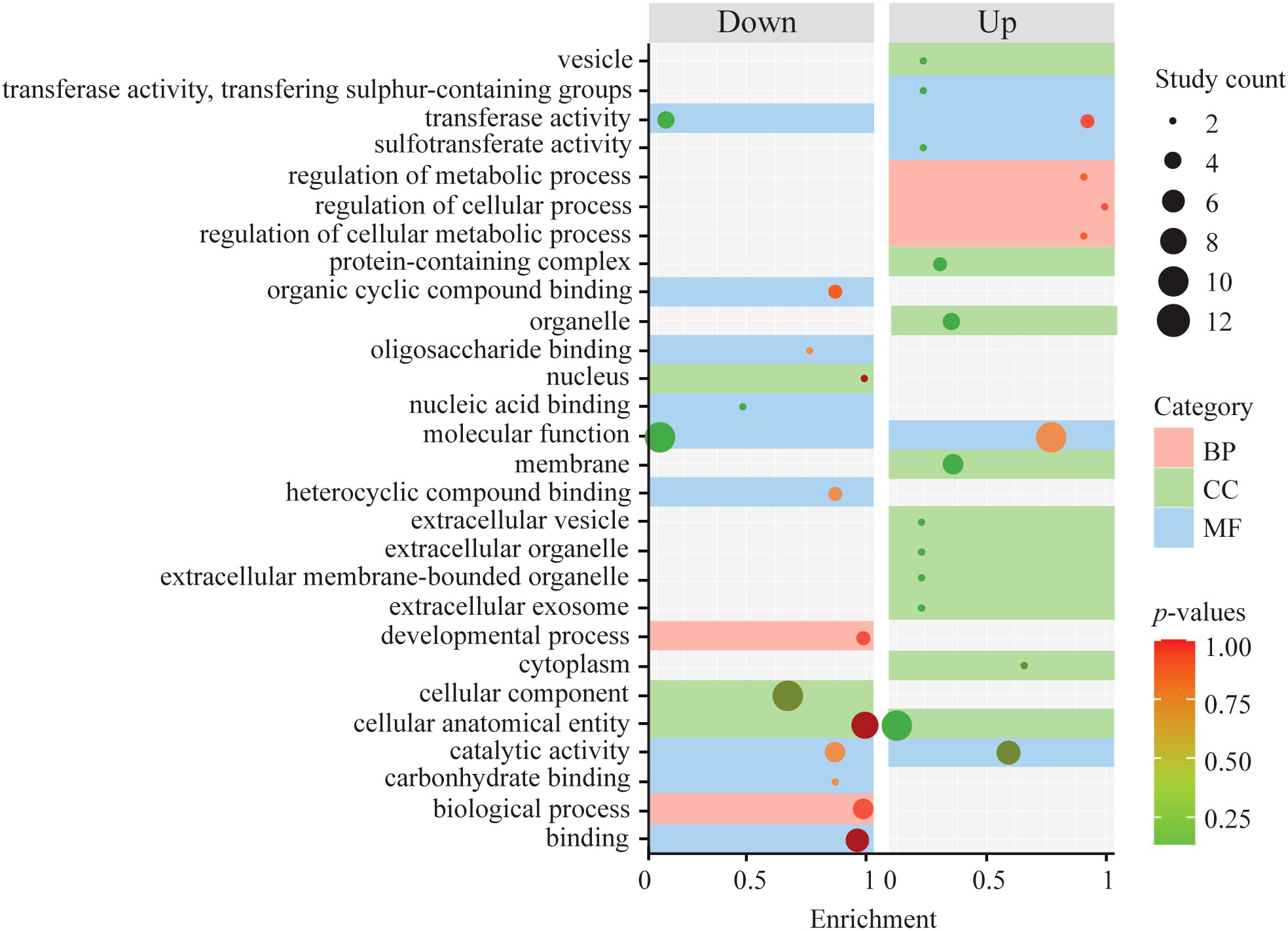
Figure 5 Plot showing GO enrichment of upregulated and downregulated DEGs based on lineages (Kuroshio vs. Tsushima).
3.3 Geographic origin-specific responses to salinity variation
The analysis of geographical origin-specific DEGs revealed a response of a total of 2,505 significant DEGs to salinity variation between native and introduced snails (856 upregulated and 1,649 downregulated genes) (Figure 2B). Two-way ANOVA considering the effect of salinity × geographical origin (introduced vs. native populations) × haplogroup interaction on the transcriptomic response of the snail B. attramentaria revealed clustering of the Matsushima population belonging to the native lineage with the introduced population in the USA (Figure 3B). Hierarchical clustering analysis revealed several gene expression pattern changes among the geographical groups of native and introduced species, which can be seen as two distinguished blocks in Figure 4B. GO enrichment analysis suggested that transport, transferase activity, and ion-binding and cation-binding genes were upregulated, whereas cellular component organization genes were downregulated (Figure 6). We also analyzed the gene expression patterns of introduced and native snail populations in response to different salinity variations (Supplementary Figure 3). In the introduced snail population, a higher number of genes (990 genes) were expressed under the 13 PSU salinity condition. Of these, 631 genes were specific to the 13 PSU salinity condition (Supplementary Figure 3A). GO enrichment analysis revealed that the Biological Process (BP) category consisted of genes related to the regulation of monoatomic ion and cation transport, metal ion transport, and calcium ion transport (Supplementary Figure 4A). The Molecular Function (MF) category consisted of genes related to serine and serine-type peptidase activity, potassium:chloride symporter activity, transmembrane transporter activity of several ions (such as monoatomic anions and cations, metal ions, and inorganic ions), and cadherin-binding activity. All these genes were highly expressed in the introduced snail population (Supplementary Figure 4C). In the native snail populations, the highest number of genes (1,669 genes) were expressed under the 13 PSU salinity condition. Of these, 810 genes are specific to the 13 PSU salinity condition (Supplementary Figure 1B). In the BP category, genes are related to cell division and cell cycles were highly expressed in the native snail populations (Supplementary Figure 4B). In the MF category, genes related to transferase activity, catalytic activity, and carbohydrate-binding activity were downregulated (Supplementary Figure 4D).
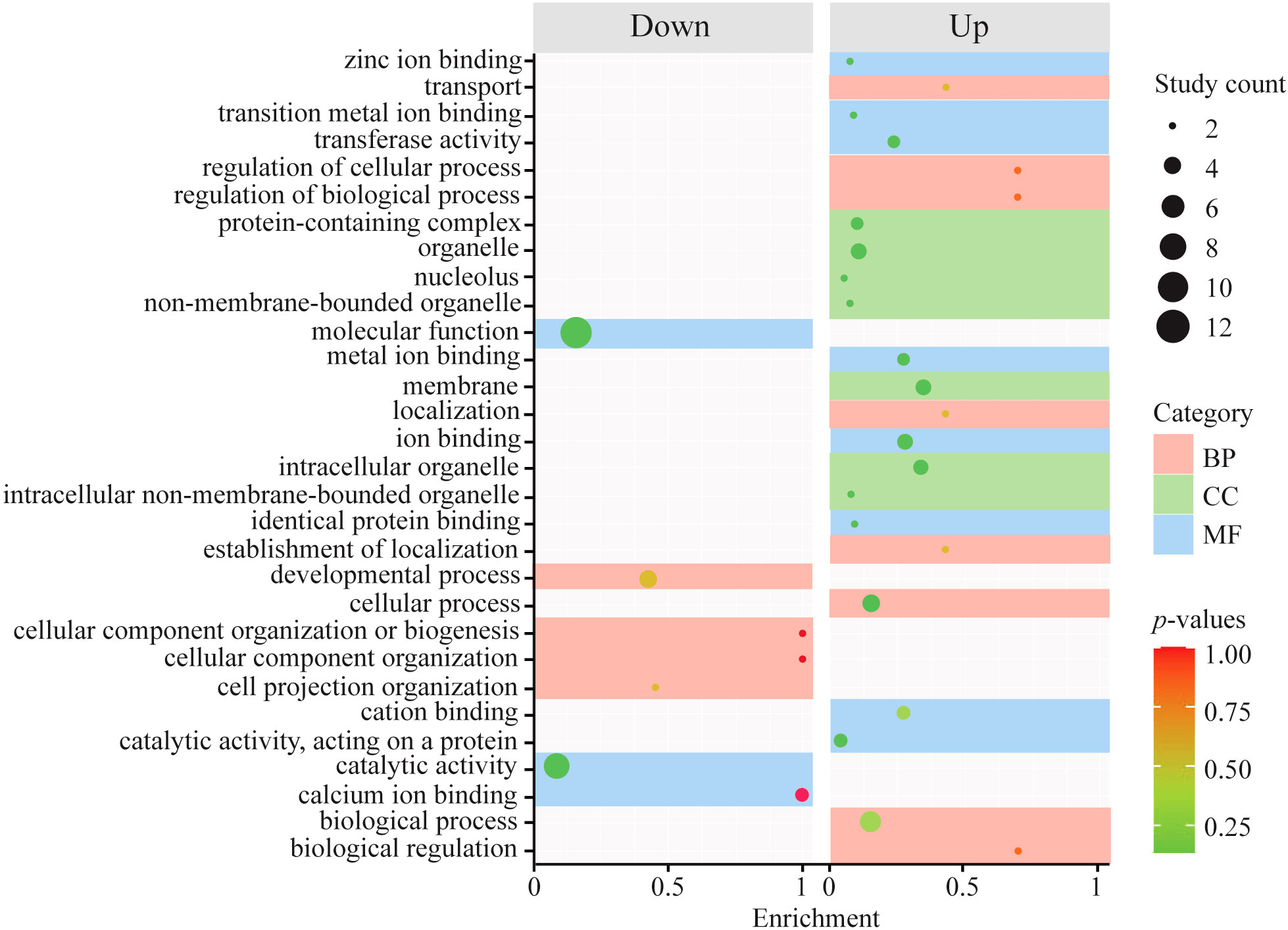
Figure 6 Plot showing GO enrichment of upregulated and downregulated DEGs based on origin (introduced vs. native).
3.4 Location-specific responses to salinity variation
Two-way ANOVA of the pooled transcriptomes of all snails based on location-specific DEGs revealed 10,362 significant DEGs considering the effect of salinity × location, as illustrated in a heatmap in Figure 4C. Three distinct blocks could be seen: I (Japan), II (Korea), and III (USA). The gene expression patterns were similar with evident shifts among the blocks (Figure 4C), suggesting the effect of location. Moreover, the gene expression varied between columns within the block, possibly because of salinity variation and plasticity among individuals.
Two-way ANOVA of the pairwise integrated DEGs of snails from different locations (the USA vs. Korea, the USA vs. Japan, and Korea vs. Japan) was conducted to investigate the gene expression patterns with a significant salinity effect based on geographical locations. Assessment of pooled DEGs of these populations indicated that populations within the native group (Korea vs. Japan) had a higher number of expressed genes (2,753 upregulated and 2,914 downregulated genes) than those among the other groups (the USA vs. Japan and the USA vs. Korea: 2,138 and 2,350 up-regulated and 2,369 and 2,346 down-regulated genes, respectively; Figure 2C). GO enrichment analysis of the Japanese vs. Korean snails suggested that genes involved in response to temperature stimulus and abiotic stimulus through the regulation of metabolic process, macromolecule metabolic process-related genes, and immune response-related genes were up-regulated (Supplementary Figure 5A). Similarly, in the USA vs. Japanese and Korean snail populations, genes related to the detection of stimulus due to temperature/abiotic factors/pain, genes related to response to stress/other organisms/organonitrogen compounds/external biotic stimulus, and immune response-related genes were downregulated (Supplementary Figure 5B).
A two-way ANOVA on population-specific genes revealed 17,358 significant DEGs found in the analysis based on the effect of salinity × population (Figure 4D). In addition, we investigated whether some of the genes were shared between samples collected under control and stress conditions from populations of Korea, Japan, and the USA. The Venn diagrams revealed that the number of shared DEGs was significantly altered in the stressed condition samples compared with the control samples (13 vs. 43, 23 vs. 33, and 43 vs. 33) in all populations (Figure 7). Compared with other salinity conditions, more genes were expressed (both upregulated and downregulated) in all populations under low salinity (13 PSU) (Figure 7; Supplementary Figure 6). In particular, the USA snail population expressed more genes (2,398 upregulated and 1,740 downregulated) under low salinity than the other populations (Figure 7). The USA snail population had 1,533 highly expressed and 1,473 less expressed unique genes, whereas it shared the maximum number of genes (368 genes upregulated and 144 genes downregulated genes) with the Korean snail population. In total, 49 highly expressed and 9 less expressed genes were shared among all snail populations (Figure 7). Under the salinity 13 vs. 33 PSU salinity condition, DEGs were mainly enriched in the BP category of GO terms. For instance, genes related to response to temperature stimulus, response to abiotic stimulus, catalytic activity, and transferase activity were upregulated in both Japanese B. attramentaria populations, compared with genes related to transport, calcium ion binding, ATP binding, zinc ion binding, phosphatidylinositol binding, phospholipid binding, and transition metal ion binding in Korean and the USA populations (Supplementary Figures 7A–D). Genes related to defense response to viruses, symbionts, and other organisms were highly expressed specifically in the Korean snail population (Supplementary Figure 7C). GO enrichment analysis revealed that the highly expressed genes uniquely found in the USA snail population were mostly related to cellular component organization and transmembrane transport in the BP category and to organic cyclic compound, carbohydrate, anion, cation, metal and calcium ion binding in the MF category.
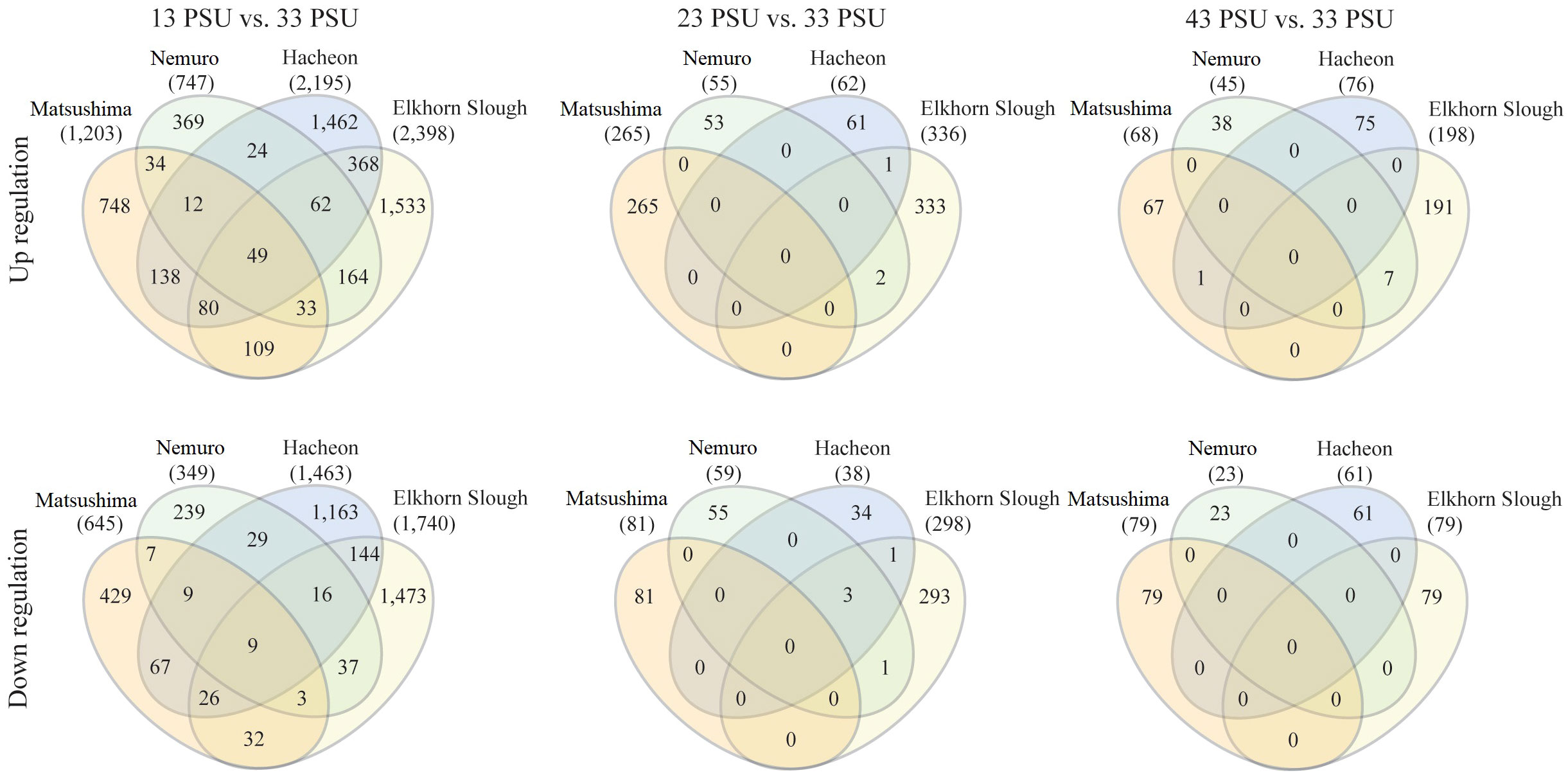
Figure 7 Venn diagram showing the number of DEGs of the snail population from different geographical locations exposed to different salinities (13, 23, and 43 PSU) in comparison with the control (33 PSU). The expressed results are with (|fc| ≥ 2 and p < 0.05). The Kuroshio and Tsushima snail populations are from Japan, the Hacheon population is from Korea, and the Elkhorn Slough population is from the USA.
4 Discussions
Invertebrates, particularly mollusks, are equipped with genetic and behavioral mechanisms to tackle the constantly varying biotic and abiotic factors in the coastal regions (Vermeij, 1973; Lockwood et al., 2010; Lockwood and Somero, 2011; Gao et al., 2017; Leung et al., 2019). The intertidal snail B. attramentaria lives in the coastal region and is daily exposed to the various of environmental conditions (Ho et al., 2019; Ho et al., 2021). To understand the adaptive process of the intertidal gastropod B. attramentaria, we examined its transcriptional response to one of the most significant abiotic factors influencing adaptivity: salinity (13, 23, and 43 PSU). We compared the transcriptomic response to this stressed salinity to that of its optimal salinity (33 PSU) in experimental conditions. Our study emphasizes the change in molecular mechanisms toward the salinity variation behind the successful invasion of the B. attramentaria to its new habitats. B. attramentaria species are more susceptible to additional salinity changes because tidal fluctuation often exposes them to extreme salinity. They respond instantly when exposed to lower salinity (3 to 13 PSU) by hermitization (closing of the mantle cavity) to impede water–salt exchange with the low-salinity seawater (Ho et al., 2019; Ho et al., 2021). Regardless of the population in different geographical locations, B. attramentaria significantly expressed more genes when exposed to low salinity (13 PSU) in this study (Figure 7). Most of these highly expressed genes are related to stress detection and response to salinity stress, e.g., genes related to the detection of response to abiotic stress and genes related to the transportation of ions for regulating osmoregulation. Similar findings have been reported in the invasive marine mussel M. galloprovincialis. In this mussel, the gene related to the efflux of inorganic ions and amino acids is upregulated when exposed to lower salinity conditions (Lockwood and Somero, 2011). During the low-salinity salinity phase in the intertidal region resulting from freshwater efflux from the terrestrial region or rain, the osmolality of seawater decreases, causing water to flood into the cellular cytoplasm. In response to this stress, cells simultaneously reduce the concentration of solutes in their cytoplasm to prevent uncontrolled swelling by allowing ions to move freely through the transmembrane ion transporter across the cell membrane and equalizing the ion concentration on either side of the cell membrane (Silva and Wright, 1992; Lockwood and Somero, 2011). However, under salinity levels of 23 and 43 PSU, no significantly expressed genes related to stress response were detected.
B. attramentaria has two divergent haplogroups, Tsushima and Kuroshio, based on the northward Pacific Sea current passing by the north and south of the Japanese archipelago (Kojima et al., 2004). MDS analysis confirmed that the Kuroshio lineage was associated with Japan (Matsushima) and the USA because of the shipment of the Matsushima lineage of B. attramentaria shipment to the American continent approximately 100 years ago (Bonnot, 1935). On comparing the gene expression of the introduced snail population with that of the native population, we found that the introduced snail population (the USA) had a higher number of significantly expressed genes.
Our findings suggest that the introduced snail population inhabits environments with salinities ranging from 0 to 30 PSU, compared with the native populations which inhabit ecosystems with salinities that are always higher than 13 PSU (Pennington and Chavez, 2000; Ho et al., 2015; Fukuda et al., 2016; Park et al., 2020; Ho et al., 2021; Okumura et al., 2021). The genes primarily expressed in the introduced population under salinity stress are related to cellular component organization and other GO categories. By contrast, the native populations expressed genes related to cation/ion/metal ion/zinc ion/calcium-binding activity, transferase activity, and cellular and biological process regulation. Thus, under low salinity (13 PSU), the introduced B. attramentaria species can activate normal biological functions, such as growth, which may explain the high expression of genes related to the organization of cellular components. These findings are consistent with those reported in Mytilus (Silva and Wright, 1992; Lockwood and Somero, 2011), suggesting that cell-cycle process regulation is an important factor affecting the salinity stress response of B. attramentaria. By contrast, native populations of B. attramentaria, which are not accustomed to such low salinity in their natural habitat, activate osmotic regulatory mechanisms to cope with unusual changes in salinity, which may explain the higher expression of genes related to ion exchange noted in these populations.
Under salinity stress, we found that the genes of the two snail populations lineages were expressed differently; however, little variation existed within each population regardless of its geographical location and origin.
5 Conclusion
This study supports the idea that the historical distribution of B. attramentaria, the intraspecific variation in its genetic makeup, the interaction, and the past exhibition of different salinity fluctuation profiles have a specific effect on the behavioral and transcriptomic responses to salinity variation. Consequently, these location- and geographical origin-specific gene-expression responses can help elucidate differences in salinity tolerance capabilities. The present findings serve as an important step toward understanding the genetic mechanisms underlying the salinity tolerance and partial adaptation of the introduced gastropod in its new habitat. This study sheds light on fundamental processes that typically occur over prolonged periods but in a contemporary timeframe by comparing the whole transcriptome between populations and understanding the mechanism underlying the response to salinity stress and the differences in the capacities of B. attramentaria lineages to respond to the salinity stress. Understanding how acclimation ability varies between populations and species in nature and knowing the relevant genes and mechanisms will be important for characterizing and predicting the ecological and evolutionary consequences of human-accelerated environmental change.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical review and approval was not required for the study of the mud-tidal snail B. attramentaria in accordance with the local legislation and institutional requirements. This invertebrate species is neither an endangered species in Korea nor elsewhere.
Author contributions
Y-JW conceptualized the project. P-TH and Y-JW collected the sample. P-TH purified and sequenced RNA. AP and P-TH analyzed the data. All authors wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by a project titled “Understanding the deep-sea biosphere on seafloor hydrothermal vents in the Indian Ridge (No. 20170411)” funded by the Ministry of Oceans and Fisheries, Korea, to Y-JW; by the National Research Foundation of Korea (NRF) of Korea and the Korean government, Ministry of Science, Information and Communication Technology, and Future Planning of Korea (MSIP) (NFR-2015R1A4A1041997) to Y-JW; and by the Basic Science Research Program through the NRF funded by the Ministry of Education (2019R1I1A2A02057134) to Y-JW.
Acknowledgments
We are grateful to our lab members, Sook-Jin Jang and Bit-Na Lee, and our long-time colleague, Bob Vrijenhoek, for their help during our trips for sample collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1251815/full#supplementary-material
References
Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. doi: 10.1186/gb-2010-11-10-r106
Baker H. G. (1965). Characteristics and modes of origin of weeds. Characteristics modes origin weeds, 147–172.
Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. society Ser. B (Methodological) 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bock D. G., Caseys C., Cousens R. D., Hahn M. A., Heredia S. M., Hübner S., et al. (2016). What we still don’t know about invasion genetics. Invasion genetics: baker stebbins legacy, 346–370. doi: 10.1002/9781119072799.ch20
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bonnot P. (1935). A recent introduction of exotic species of molluscs into California waters from Japan. Nautilus 49, 1–2.
Borg I., Groenen P. J. (2005). Modern multidimensional scaling: Theory and applications (The USA: Springer Science & Business Media).
Buchfink B., Xie C., Huson D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Byers J. E. (2000a). Competition between two estuarine snails: implications for invasions of exotic species. Ecology 81, 1225–1239. doi: 10.1890/0012-9658(2000)081[1225:CBTESI]2.0.CO;2
Byers J. E. (2000b). Effects of body size and resource availability on dispersal in a native and a non-native estuarine snail. J. Exp. Mar. Biol. Ecol. 248, 133–150. doi: 10.1016/S0022-0981(00)00163-5
Ching T., Huang S., Garmire L. X. (2014). Power analysis and sample size estimation for RNA-Seq differential expression. RNA 20, 1684–1696. doi: 10.1261/rna.046011.114
Colautti R. I., Ricciardi A., Grigorovich I. A., Macisaac H. J. (2004). Is invasion success explained by the enemy release hypothesis? Ecol. Lett. 7, 721–733. doi: 10.1111/j.1461-0248.2004.00616.x
Costa-Silva J., Domingues D., Lopes F. M. (2017). RNA-Seq differential expression analysis: An extended review and a software tool. PloS One 12, e0190152. doi: 10.1371/journal.pone.0190152
Dunn A. M. (2009). Parasites and biological invasions. Adv. Parasitol. 68, 161–184. doi: 10.1016/S0065-308X(08)00607-6
Fisheries U. S. B. O., Galtsoff P. S. (1932). Introduction of Japanese oysters into the United States. (The USA: US Government Printing Office).
Fu L., Niu B., Zhu Z., Wu S., Li W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinf. (Oxford England) 28, 3150–3152. doi: 10.1093/bioinformatics/bts565
Fukuda H., Katayama R., Yang Y., Takasu H., Nishibe Y., Tsuda A., et al. (2016). Nutrient status of Otsuchi Bay (northeastern Japan) following the 2011 off the Pacific coast of Tohoku Earthquake. J. oceanogr. 72, 39–52. doi: 10.1007/s10872-015-0296-2
Gao X., Li Y., Li X., Wu F., Song C., Liu Y. (2017). The response and osmotic pressure regulation mechanism of Haliotis discus hannai (Mollusca, Gastropoda) to sudden salinity changes. Hydrobiologia 795, 181–198. doi: 10.1007/s10750-017-3129-z
Golikov A. (1967). Molluscs of Possiet Bay (the Sea of Japan) and their ecology. Trans. Zoological Institute 42, 5–154.
Golikov A., Gulbin V. (1978). “Prosobranch gastropod molluscs (Gastropoda, Prosobranchiata) of the shelf of the Kurile Islands. I. Orders Docoglossa—Entomostoma,” in Animal and plant kingdoms of shelf zones of the Kurile Islands. Ed. Kussakin O. G. (Moscow: Nauka).
Golikov A., Scarlato O. (1985). Shell-bearing gastropod and bivalve molluscs of the shelf of southern Sakhalin and their ecology. Biocenoses fauna shelf South Sakhalin Issledovaniya Fauny Morei 30, 360–487.
Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Ho P. T., Kwan Y. S., Kim B., Won Y. J. (2015). Postglacial range shift and demographic expansion of the marine intertidal snail Batillaria attramentaria. Ecol. Evol. 5, 419–435. doi: 10.1002/ece3.1374
Ho P. T., Nguyen H. Q., Kern E. M., Won Y. J. (2021). Locomotor responses to salt stress in native and invasive mud-tidal gastropod populations (Batillaria). Ecol. Evol. 11, 458–470. doi: 10.1002/ece3.7065
Ho P.-T., Rhee H., Kim J., Seo C., Park J. K., Young C. R., et al. (2019). Impacts of salt stress on locomotor and transcriptomic responses in the intertidal gastropod Batillaria attramentaria. Biol. Bull. 236, 224–241. doi: 10.1086/703186
Klopfenstein D., Zhang L., Pedersen B. S., Ramírez F., Warwick Vesztrocy A., Naldi A., et al. (2018). GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 8, 1–17. doi: 10.1038/s41598-018-28948-z
Kojima S., Hayashi I., Kim D., Iijima A., Furota T. (2004). Phylogeography of an intertidal direct-developing gastropod Batillaria cumingi around the Japanese Islands. Mar. Ecol. Prog. Ser. 276, 161–172. doi: 10.3354/meps276161
Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, 25. doi: 10.1186/gb-2009-10-3-r25
Le Cam S., Pechenik J. A., Cagnon M., Viard F. (2009). Fast versus slow larval growth in an invasive marine mollusc: does paternity matter? J. Heredity 100, 455–464. doi: 10.1093/jhered/esp007
Lee C. E. (2002). Evolutionary genetics of invasive species. Trends Ecol. Evol. 17, 386–391. doi: 10.1016/S0169-5347(02)02554-5
Leung J. Y., Russell B. D., Connell S. D. (2019). Adaptive responses of marine gastropods to heatwaves. One Earth 1, 374–381. doi: 10.1016/j.oneear.2019.10.025
Lockwood B. L., Sanders J. G., Somero G. N. (2010). Transcriptomic responses to heat stress in invasive and native blue mussels (genus Mytilus): molecular correlates of invasive success. J. Exp. Biol. 213, 3548–3558. doi: 10.1242/jeb.046094
Lockwood B. L., Somero G. N. (2011). Transcriptomic responses to salinity stress in invasive and native blue mussels (genus Mytilus). Mol. Ecol. 20, 517–529. doi: 10.1111/j.1365-294X.2010.04973.x
Okumura Y., Masuda Y., Suzuki N., Kakehi S., Hara M. (2021). Temporal changes in the nutrient status of Matsushima Bay after a wastewater plant was destroyed by a tsunami on 11 March 2011. Fisheries Sci. 87, 845–859. doi: 10.1007/s12562-021-01553-2
Park S., Choi M., Jang D., Joe D., Park K., Lee H., et al. (2020). Spatiotemporal distribution of dissolved heavy metals in Gyeonggi Bay, Korea. Ocean Sci. J. 55, 69–84. doi: 10.1007/s12601-020-0002-1
Pennington J. T., Chavez F. P. (2000). Seasonal fluctuations of temperature, salinity, nitrate, chlorophyll and primary production at station H3/M1 over 1989–1996 in Monterey Bay, California. Deep Sea Res. Part II: Topical Stud. Oceanogr. 47, 947–973. doi: 10.1016/S0967-0645(99)00132-0
Prentis P. J., Wilson J. R., Dormontt E. E., Richardson D. M., Lowe A. J. (2008). Adaptive evolution in invasive species. Trends Plant Sci. 13, 288–294. doi: 10.1016/j.tplants.2008.03.004
R Development Core Team (2011). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical computing).
Reznick D. N., Ghalambor C. K. (2001). The population ecology of contemporary adaptations: what empirical studies reveal about the conditions that promote adaptive evolution. Microevolution rate pattern process, 183–198. doi: 10.1007/978-94-010-0585-2_12
Sax D. F., Stachowicz J. J., Brown J. H., Bruno J. F., Dawson M. N., Gaines S. D., et al. (2007). Ecological and evolutionary insights from species invasions. Trends Ecol. Evol. 22, 465–471. doi: 10.1016/j.tree.2007.06.009
Shui B., Wang Y., Lou F., Han Z. (2022). Salinity fluctuation on the genetic regulatory mechanisms of the crustacean, Charybdis japonica. Front. Mar. Sci. 9, 870891. doi: 10.3389/fmars.2022.870891
Silva A. L., Wright S. H. (1992). Integumental taurine transport in Mytilus gill: short-term adaptation to reduced salinity. J. Exp. Biol. 162, 265–279. doi: 10.1242/jeb.162.1.265
Somero G. N. (2012). The physiology of global change: linking patterns to mechanisms. Annu. Rev. Mar. Sci. 4, 39–61. doi: 10.1146/annurev-marine-120710-100935
Spalding M. D., Fox H. E., Allen G. R., Davidson N., Ferdaña Z. A., Finlayson M., et al. (2007). Marine ecoregions of the world: a bioregionalization of coastal and shelf areas. BioScience 57, 573–583. doi: 10.1641/B570707
Stockwell C. A., Hendry A. P., Kinnison M. T. (2003). Contemporary evolution meets conservation biology. Trends Ecol. Evol. 18, 94–101. doi: 10.1016/S0169-5347(02)00044-7
Strayer D. (1999). Invasion of fresh waters by saltwater animals. Trends Ecol. Evol. 14, 448–449. doi: 10.1016/S0169-5347(99)01712-7
Strayer D. L. (2012). Eight questions about invasions and ecosystem functioning. Ecol. Lett. 15, 1199–1210. doi: 10.1111/j.1461-0248.2012.01817.x
Sussarellu R., Huvet A., Lapègue S., Quillen V., Lelong C., Cornette F., et al. (2015). Additive transcriptomic variation associated with reproductive traits suggest local adaptation in a recently settled population of the Pacific oyster, Crassostrea gigas. BMC Genomics 16, 1–12. doi: 10.1186/s12864-015-1972-8
Vermeij G. J. (1973). Morphological patterns in high-intertidal gastropods: adaptive strategies and their limitations. Mar. Biol. 20, 319–346. doi: 10.1007/BF00354275
Wickham H. (2011). ggplot2. Wiley Interdiscip. reviews: Comput. Stat 3, 180–185. doi: 10.1002/wics.147
Wolff W. J. (2000). Recent human-induced invasions of fresh waters by saltwater animals? Aquat. Ecol. 34, 319–321.
Keywords: Batillaria attramentaria, biological invasion, adaptive evolution, mud-tidal gastropod, salinity variation
Citation: Patra AK, Ho P-T and Won Y-J (2023) Transcriptomic response to salinity variation in native and introduced mud-tidal gastropod Batillaria attramentaria. Front. Mar. Sci. 10:1251815. doi: 10.3389/fmars.2023.1251815
Received: 02 July 2023; Accepted: 13 November 2023;
Published: 29 November 2023.
Edited by:
Xiao Liang, Shanghai Ocean University, ChinaReviewed by:
Mbaye Tine, Gaston Berger University, SenegalShikai Liu, Ocean University of China, China
Copyright © 2023 Patra, Ho and Won. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong-Jin Won, d29uQGV3aGEuYWMua3I=
†These authors have contributed equally to this work
 Ajit Kumar Patra
Ajit Kumar Patra Phuong-Thao Ho
Phuong-Thao Ho Yong-Jin Won
Yong-Jin Won