- 1Institute of Oceanography, College of Geography and Oceanography, Minjiang University, Fuzhou, China
- 2Agricultural and Rural Development Service Centre, Pingtan Comprehensive Pilot Zone, Fuzhou, China
- 3College of Biological Science and Engineering, Fuzhou University, Fujian, China
Introduction: Harmful algal blooms (HABs) have been increasing in frequency and expanding their ranges on coastlines worldwide in recent decades. Algicidal bacteria play a pivotal role in eliminating HABs, yet the characteristics of bacterial communities and their algicidal activity during a Karenia longicanalis bloom remain poorly understood.
Methods: In this study, we investigated bacterial communities using 16S rRNA sequencing during a K. longicanalis bloom to identify bacteria with high algicidal activity that could be isolated. Five sampling sites in Tongxin Bay, located in Lianjiang County, China, including TX1 to TX5, were selected based on the concentration of K. longicanalis cells.
Results: Our 16S rRNA sequencing results revealed that the TX4 site was enriched with genera known to contain algicidal bacteria, such as Pseudoalteromonas and Alteromonas, which are members of the Gammaproteobacteria class, while Sulfitobacter, a member of the Alphaproteobacteria class, was enriched in the TX5 site. Among the 100 cultivable bacteria isolated from the 5 sampling sites, 6 exhibited an algicidal rate of over 80%, with FDHY-MQ5, isolated from the TX4 site, exhibiting an algicidal rate of approximately 100% against Karenia mikimotoi after 48 hours of challenge with 2% (v/v) bacterial volume (OD600=4.5) concentration. Our 16S rRNA sequencing result showed FDHY-MQ5 was a member of the Pseudoalteromonas genus, and this bacterium also demonstrated high algicidal activity against Heterosigma akashiwo and Alexandrium tamarense.
Discussion: Our findings shed light on the changes in bacterial community structure and the algicidal behavior of bacteria towards algae during a K. longicanalis bloom, providing a research basis for a better understanding of HAB management.
1 Introduction
In recent decades, harmful algal blooms (HABs) have shown increased frequency and range expansion in coastlines worldwide, primarily due to eutrophication, climatic change, and human activities, among other factors (Chen et al., 2019). These HABs have had a severe impact on fishery resources and marine ecosystems. In particular, HAB events caused by toxic algae have resulted in massive losses for the aquaculture industry and public health (Shi et al., 2022). Dinoflagellates are major causative agents of HABs, and many dinoflagellates can produce a variety of toxins, such as fish-killing poisoning, paralytic shellfish poisoning, diarrhetic shellfish poisoning, yessotoxins and azaspiracids (Heredia-Tapia et al., 2002; Krock et al., 2009; Richlen et al., 2010; Pitcher et al., 2019; Lee et al., 2020). These toxins can either directly kill fish and shellfish, causing significant losses to aquaculture, or accumulate in shellfish that is then consumed, which can cause illness or death in humans (Wang et al., 2011). Thus, marine dinoflagellate blooms have become a major concern over the past decades.
Karenia species are notorious for causing HABs in many parts of the world (Bechard and Lang, 2023), including China (Gu et al., 2022). Of particular concern are blooms of Karenia brevis, which have negatively impacted the waters of the Gulf of Mexico off the coast of southwest Florida for a long time (Hu et al., 2016). Severe K. brevis blooms have caused large-scale mortality of fish, sea birds and even some larger marine animals (Hoagland et al., 2020). The ‘mikimotoi-complex’, a subset of HAB species that includes Karenia mikimotoi, Karenia digitata and Karenia longicanalis, is classified under the broader Karenia genus (Yang et al., 2001). These species share significant morphological similarities and have been reported to concurrently form HABs in several instances (Yang et al., 2001; Orlova et al., 2022). K. mikimotoi is responsible for large-scale fish and abalone kills in China, making it possibly the most notorious species in this region (Gu et al., 2022). The inaugural bloom of K. mikimotoi was first documented in Hong Kong during March and April of 1998. This algal outbreak led to devastating fish mortality in the afflicted areas, resulting in an estimated economic loss of HK$315,000,000 (Lu and Hodgkiss, 2004). Notably, the species K. longicanalis was also present during this event (Yang et al., 2001). Since 2005, recurrent blooms of K. mikimotoi have frequently occurred in the East China Sea and in the coastal waters of Zhejiang and Fujian Provinces in particular (Gu et al., 2022). Notably, K. mikimotoi has caused high mortality of cultured abalones, resulting in a total loss of 2 billion RMB in Fujian Province (including the Tongxin Bay) in 2012 (Guo et al., 2014). Karenia longicanalis, a related species of K. mikomotoi, was originally observed in 1998 in the Hong Kong harbor (Yang et al., 2001) and subsequently found in 2016 in Hong Kong harbor, the East China Sea and other Chinese coasts (Chan et al., 2004; Wang et al., 2018). However, compared with K. mikimotoi, the blooms of K. longicanalis have been rarely reported in Fujian Province.
The formation and demise of HABs are influenced by a variety of abiotic and biotic factors (Zhou et al., 2020). Among the biotic factors, microorganisms play a pivotal role in HAB formation and termination (Zhou et al., 2020). Marine microalgae, including dinoflagellates, coexist with a complex microbial community that forms an important component of phytoplankton physiology and ecology (Cole, 1982). The interactions between the microbial community and microalgae are diverse and include highly specific interactions, such as symbiosis, mutualism, or parasitic/algicidal behaviors, as well as less-specific interactions, such as nutrient competition/modification (Van Mooy et al., 2009; Bolch et al., 2017). These interactions can alter the behavior and physiology of the organisms involved. For example, microalgae can grow rapidly in the presence of certain types of bacteria by absorbing essential nutrients and vitamins produced by the bacteria (Croft et al., 2005). Certain bacterial growth factors also impact phytoplankton growth, such as indole-3-acetic acid (Amin et al., 2015). In contrast, during blooms, microalgae can be lysed by algicidal bacteria (Shi et al., 2022). Bacteria can also accelerate their growth by absorbing the organic matter (Lau et al., 2007), or be inhibited by antibiotics produced by microalgae (Senhorinho et al., 2015).
Algicidal bacteria, designated as critical for the termination of HABs, have been the focus of studies aimed at developing biological agents to inhibit HABs (Meyer et al., 2017). These bacteria have been isolated from diverse regions of marine systems, including seagrass (Inaba et al., 2017), sediments (Shi et al., 2013a), surface seawater (Jung et al., 2008) and algal bloom samples (Shi et al., 2018). The majority of isolates of algicidal bacteria are Gram-negative bacteria, with over half of these bacteria belonging to the Cytophaga-Flavobacteria-Bacteroidetes (CFB) group, and the remainder belonging to the Proteobacteria (Coyne et al., 2022). A tiny percentage of documented algicidal bacteria are Gram-positive bacteria from the phylum Firmicutes and the Terrabacteria group of the phylum Actinobacteria (Meyer et al., 2017). Despite many studies focusing on the bacterial community during HABs, the characteristics of the bacteria community and algicidal activity during K. longicanalis bloom remain unclear. Tongxin Bay, located in Lianjiang County, China, is a typical abalone culture area and one of the largest bays in China for abalone farming. This bay has experienced dinoflagellate blooms almost every year in recent decades, with K. mikimotoi blooms causing economic losses (Guo et al., 2014). Therefore, this study investigated the bacterial communities that were in dinoflagellate-bloom samples from Tongxin Bay. Considering the genetic similarity between K. longicanalis and K. mikimotoi, their phycosphere bacterial communities during outbreak events in the region could possibly exhibit similar characteristics. Concurrently, K. longicanalis incidences within this area are still somewhat infrequent. However, its initial emergence may foreshadow recurring, and potentially multiple, future outbreaks of this alga, which sheds light on the anticipatory importance of such studies. Algicidal bacteria were further isolated from surface seawater in bloom samples. Additionally, to gain a better understanding of the underlying mechanisms of the activity of the new algicidal bacterial strain, the basic characteristics and algicidal action mode of this strain were investigated.
2 Materials and methods
2.1 Study area, sampling, and environmental parameters
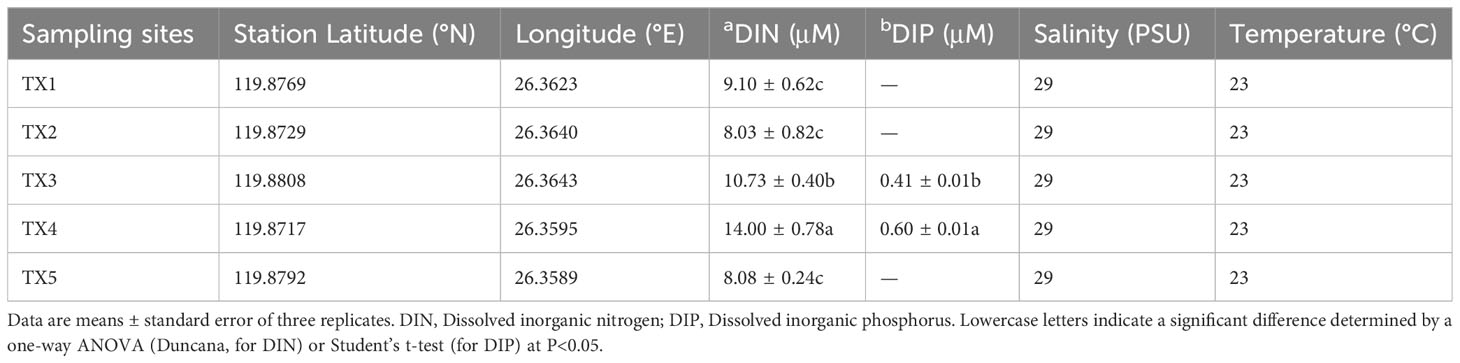
The field experiments were conducted on June 13, 2018 in Tongxin Bay of Lianjiang sea area of China (Figure 1A). The density of dinoflagellate cells was determined as previously described (Shi et al., 2022). For each sample, 2 L of surface seawater was pre-filtered using a 200 µm sieve to remove the large zooplankton, and then filtered onto 47 mm in diameter isopore filters with a 0.22 µm pore size under low vacuum pressure (< 10 PSI) using a vacuum pump. The filtrate was transferred to 2 mL tubes which contained 0.4 mL DNA lysis buffer (100 mM Tris-HCl and 50 mM EDTA, pH 8.0) and stored at −20°C until their DNA extraction. The filtrate was stored at -20°C for nutrient analysis. The concentrations of phosphate and inorganic nitrogen were determined as previously described (Shi et al., 2017; Shi et al., 2021). The temperature and salinity of each sample site were measured using a CTD (SBE 17plus V2 Searam Recording & AutoFire Module; Sea-Bird Scientific, Bellevue, MD, USA). Statistical analyses were performed using SPSS (version 19.0, IBM) and statistically different groups were determined by a one–way ANOVA or Student’s t-test at p< 0.05.
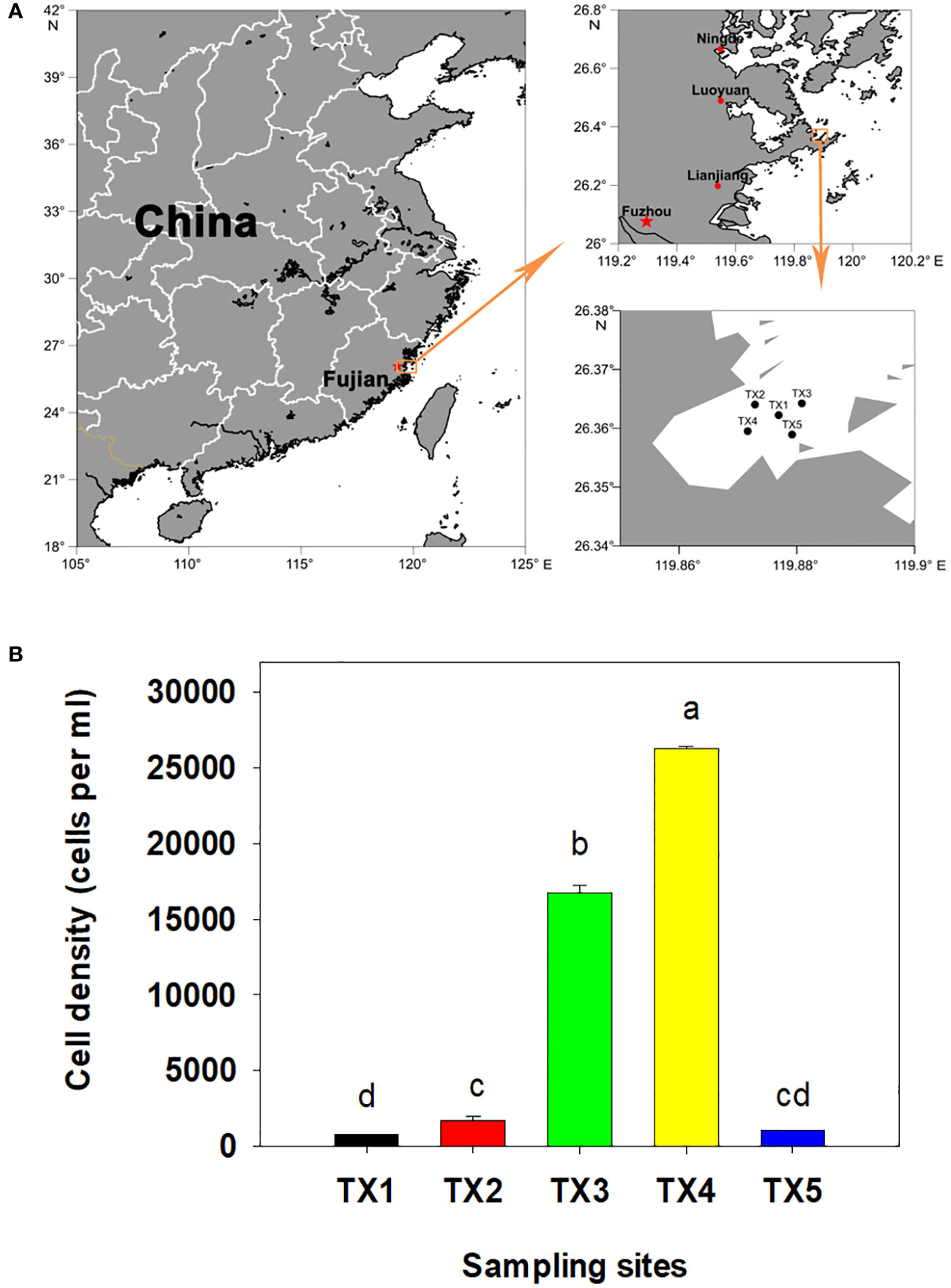
Figure 1 Location of the sample sites. (A) Bloom and study sites selected in Tongxin Bay in Fujian province. (B) Cell density of Karenia longicanalis in the five sample sites. Bars with different letters were significantly different at p<0.05.
2.2 Identification of the dinoflagellate species and high-throughput DNA sequencing of bacteria
The DNA was extracted from the dinoflagellates using a CTAB protocol coupled with Zymo DNA Clean & Concentrator columns (Zymo Research Corp., Irvine, CA, USA) as previously described (Shi et al., 2013b). The concentration and quality of DNA was determined using a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The ITS rRNA gene sequencing was used to identify the dinoflagellate species as previously described (Wang et al., 2014). The dominant species was identified by direct sequencing of PCR product. Then the PCR product was cloned into T vector. The clones were random picked up and sequenced to validate the result of PCR product direct sequencing.
Amplification of the prokaryotic 16S rRNA was performed using primers 515F (5′-GTGYCAGCMGCCGCGGTAA -3′) and 926R (5′-CCGYCAATTYMTTTRAGTTT-3′) (Parada et al., 2016). These primers were demonstrated to generate the V4–V5 hypervariable region of all archaea and bacteria and more accurately estimate the community abundances (Needham and Fuhrman, 2016). PCR was performed as follows: 95°C 2 min; 95°C 45 s, 50°C 45 s, 68°C 90 s for 25 cycles; and a final step of 68°C for 5 min. The samples were amplified in triplicate. Triplicate PCR products were pooled, barcoded, and sequenced with TruSeq DNA PCR-Free Sample Preparation Kit (250 paired-end reads) on an Illumina NovaSeq 6000 platform (San Diego, CA, USA) according to the standard protocols by Novogene Biotech Co., Ltd. (Beijing, China).
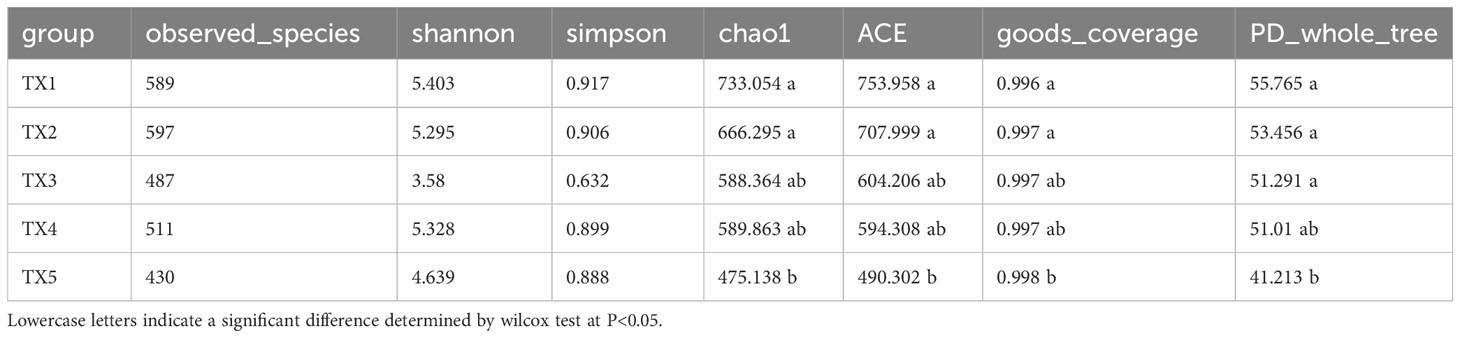
Amplification primers and barcode sequences were removed, and the reads were merged using FLASH (V1.2.7; Magoc and Salzberg, 2011) and quality-filtered as previously described (Bokulich et al., 2013). These clean tags underwent quality control using QIIME (v. 1.9.1; Caporaso et al., 2010). The tags were clustered into operational taxonomic units (OTUs) based on 97% identity using scripts from the software Uparse (v7.0.1001; Haas et al., 2011). The samples were rarefied to the minimum sequencing depth to conduct a rarefaction analysis on the OTUs. The α-diversity, including observed OTUs, Chao1, Shannon, Simpson, Ace, Goods-coverage, and PD whole tree, was determined using QIIME (V1.9.1) and the Wilcox test (P<0.05). Nonmetric multidimensional scaling (NMDS) ordinations were based on Bray-Curtis matrices, which were analyzed using the R package vegan and the Wilcox test (P<0.05). Taxonomic composition was analyzed using the small subunit (SSU) rRNA database of SILVA132 and the Mothur method for bacteria (Edgar, 2013). The results were summarized in profiling tables and histograms at the phylum, order, class, family, and genus levels. The correlations between the bacterial community and environmental factors, such as nitrogen, phosphorus, and the interaction between nitrogen and phosphorus, were analyzed using the Mantel test (P<0.05). The linear discriminant analysis (LDA) effect size (LEfSe) was applied to the taxonomic levels ranging from phylum to genus (with an LDA score of 4; www.bioincloud.tech). The Canonical Correspondence Analysis (CCA) was carried out using the ‘cca’ function from the ‘vegan’ package for sorting analysis. The ‘envfit’ function was utilized to compute the R-squared and P-values, which estimate each environmental factor’s influence on species distribution.
2.3 Cultures of harmful algal species
The K. mikimotoi culture (CCM-083 strain) was provided by the Center for Collections of Marine Algae at Xiamen University, China. The diatoms Phaeodactylum tricomutum and Skeletonema costatum; the dinoflagellates Prorocentrum shikokuense, Alexandrium tamarense and Alexandrium carterae; and the Raphidophyceae Heterosigma akashiwo were maintained in Fuzhou University. All these cultures were prepared in glass bottles with L1 medium (Guillard and Hargraves, 1993) prepared with filtered (0.22 mm), autoclaved natural seawater (28 PSU) at 20°C under a 14 h:10 h light: dark photoperiod at a light intensity of approximately 100 μmol photons. m−2. s−1 provided by fluorescent lamps. Before test, all the cultures were treated with a cocktail of antibiotics, including ampicillin (200 mg/L), kanamycin (100 mg/L), and streptomycin (100 mg/L) as previously described (Lin et al., 2015) to suppress the proliferation of potential existing bacteria.
2.4 Screening and identification of algicidal bacteria
At each sampling site, 100 mL of seawater was drawn and subsequently filtered through a sieve with a 200 µm mesh size. The samples were then stored at a temperature of 4°C to isolate algicidal bacteria. The pre-filtered seawater was subjected to 10-fold serial dilution, and 0.1 mL of each dilution was spread on a sterile Zobell 2216E agar plate, followed by incubation at 25°C for 7 days. From each sampling site, 20 individual colonies with distinct morphologies were selected and transferred to a flask containing sterile 2216E broth. The flasks were agitated at 150 rpm and 25°C for 24 h. An aliquot of each bacterial culture was then inoculated into triplicate samples of exponentially growing K. mikimotoi cultures. The final volume ratio of bacterial to algae was 2% (v/v). Negative controls were also established by adding the same volume of sterile 2216E broth without algicidal bacteria to the algal cultures. The algicidal rate of each bacterial concentration was calculated using the following formula: NT represented the algal cell density in the treated algal culture, while NC was the algal cell density in the negative control (Shi et al., 2018). A bacterial strain that killed over 80% of the algae within 48 h was defined as algicidal.
The strain FDHY-MQ5 exhibited the highest algicidal rate and was cultured in a flask with shaking, as previously described. Genomic DNA was extracted from the strain using a CTAB protocol coupled with Zymo DNA Clean & Concentrator by Shi et al. (2013b). The 16S rRNA gene was amplified by PCR using primers 27F and 1492R (DeLong, 1992), with the isolated genomic DNA serving as template. The resulting amplicons were purified and cloned into vector pMD18-T (TaKaRa, Dalian, China) for sequencing. Phylogenetic trees were inferred using MEGA version 7.0 with neighbor-joining (NJ) (Saitou and Nei, 1987) and maximum-likelihood (ML) algorithms, and reference sequences of 16S rDNA were retrieved from the GenBank database and combined with the sequences generated from this study to form a dataset.
2.5 Algicidal activity and characteristics of strain FDHY-MQ5 against harmful algal species
Approximately 1mL of exponential-phased FDHY-MQ5 culture was collected by centrifugation at 4000 rpm for 5 min at 20°C. The harvested cells were suspended in 0.1 M PBS (pH 7.4) containing 2.5% glutaraldehyde (v/v) and incubated for 2 h. The cells were then gently rinsed twice with PBS and post-fixed in 1.0% OsO4 in the same buffer for 2 h. After two additional rinses with PBS, the samples were dehydrated in a graded ethanol series (30, 50, 70, 90, 95, and 100%) and stored in pure tertiary butanol at 4°C overnight. The dehydrated samples were then critical-point-dried and mounted on stubs for imaging with a scanning electron microscope (Nova NanoSEM 230, FEI), enabling visualization of the algicidal cells.
To test the algicidal effect of strain FDHY-MQ5 against general harmful algal species, the diatoms P. tricomutum and S. costatum; the dinoflagellates P. shikokuense, A. tamarense and A. carterae; and the Raphidophyceae H. akashiwo were selected for this assay and all algal cultures were in exponential phase. FDHY-MQ5 was applied to algal cultures at a final ratio of 1.0% (v/v). The algal cell density was measured after 24 h incubation with strain FDHY-MQ5 or sterile 2216E broth, without the algicidal bacteria (negative controls). The algicidal rate was calculated as previously described.
To investigate the algicidal activity of strain FDHY-MQ5 against K. mikimotoi, triplicate sets of 25 mL exponentially growing cultures of K. mikimotoi were treated with different concentrations (0.5%, 1% and 2% [v/v]) of FDHY-MQ5. The OD600 value of the original bacteria culture was 4.5. A negative control consisting of the same amount of 2216E broth was added to the algal cultures. The algal cell concentration and rate were measured as previously described (Shi et al., 2022).
To determine the algicidal attack process of strain FDHY-MQ5 against K. mikimotoi, treated K. mikimotoi was examined using a fluorescence microscope (Nikon ECLIPSE 80i, Tokyo, Japan). Approximately 100 mL of exponentially growing K. mikimotoi cultures were prepared. One milliliter of the supernatant of strain FDHY-MQ5 bacterial cells (OD600 = 4.5) was added to the algal cultures, and at several time points (0, 6, 12, 18 and 24 h), the incubated algal cells were observed under the microscope. Additionally, 10 mL of incubated algal culture was collected at each time point using centrifugation at 4000 rpm at 20°C for 5 min. The cell pellet was resuspended in 0.1 M phosphate-buffered saline (PBS) (8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4, 0.24 g KH2PO4, and 1 L distilled water, at pH 7.4) with 4% (v/v) paraformaldehyde for 10 min. The cells were then washed twice with PBS and stained with 5 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) for 5 min in the dark at room temperature. Bright-field and fluorescence of the samples were simultaneously evaluated by fluorescence microscopy.
3 Results
3.1 General characteristics of the dinoflagellate bloom
In this bloom event, K. longicanalis was identified as the dominant species through morphological and direct sequencing analysis of its ITS PCR product. The percent identity is 99.55%. As depicted in Figure 1B, the algal density of dominant species in five sampling sites were 780 (TX1), 1,700 (TX2), 16,750 (TX3), 26,250 (TX4) and 1,000 (TX5) cells per mL, respectively. While the water in TX1 was clean, the foam produced by algae in TX5 was evident. Based on the progression of the bloom and the cell concentration of dinoflagellate cells, the five samples were inferred as pre-bloom (TX1), onset phase (TX2), exponential phase (TX3), decline phase (TX4) and termination phase (TX5). The dissolved inorganic nitrogen (DIN) concentration ranged from 8.03 to 14.0 µM, with the highest concentrations observed in TX4 and TX3 (Table 1). Dissolved inorganic phosphorus (DIP) was only detected in TX4 and TX3, and below the limit of detection in the other sites (Table 1).
3.2 Microbiome diversity of different samples
Table S1 presents the quality control profile of 16S rRNA sequencing. The majority of prokaryotic OTUs were found in all the samples (Figure 2A). In total, 1,468 OTUs were identified for prokaryotes with 97% sequence similarity. There were significant differences in bacterial richness between the samples for the Chao1, ACE, good coverage, and PD whole tree. However, there were no significant differences in microbial diversity (P > 0.05) among all the samples for the observed species and the Shannon and Simpson indices (Table 2). The Non-Metric Multi-Dimensional Scaling (NMDS) of the Bray-Curtis distance matrix indicated that samples from the different sites exhibited clear separation in their prokaryotic communities (Figure 2B). Significant correlations were observed between bacterial community and nitrogen, phosphorus, and the interaction between nitrogen and phosphorus at the levels of OTU, genus, family, class, and order (Table 3).
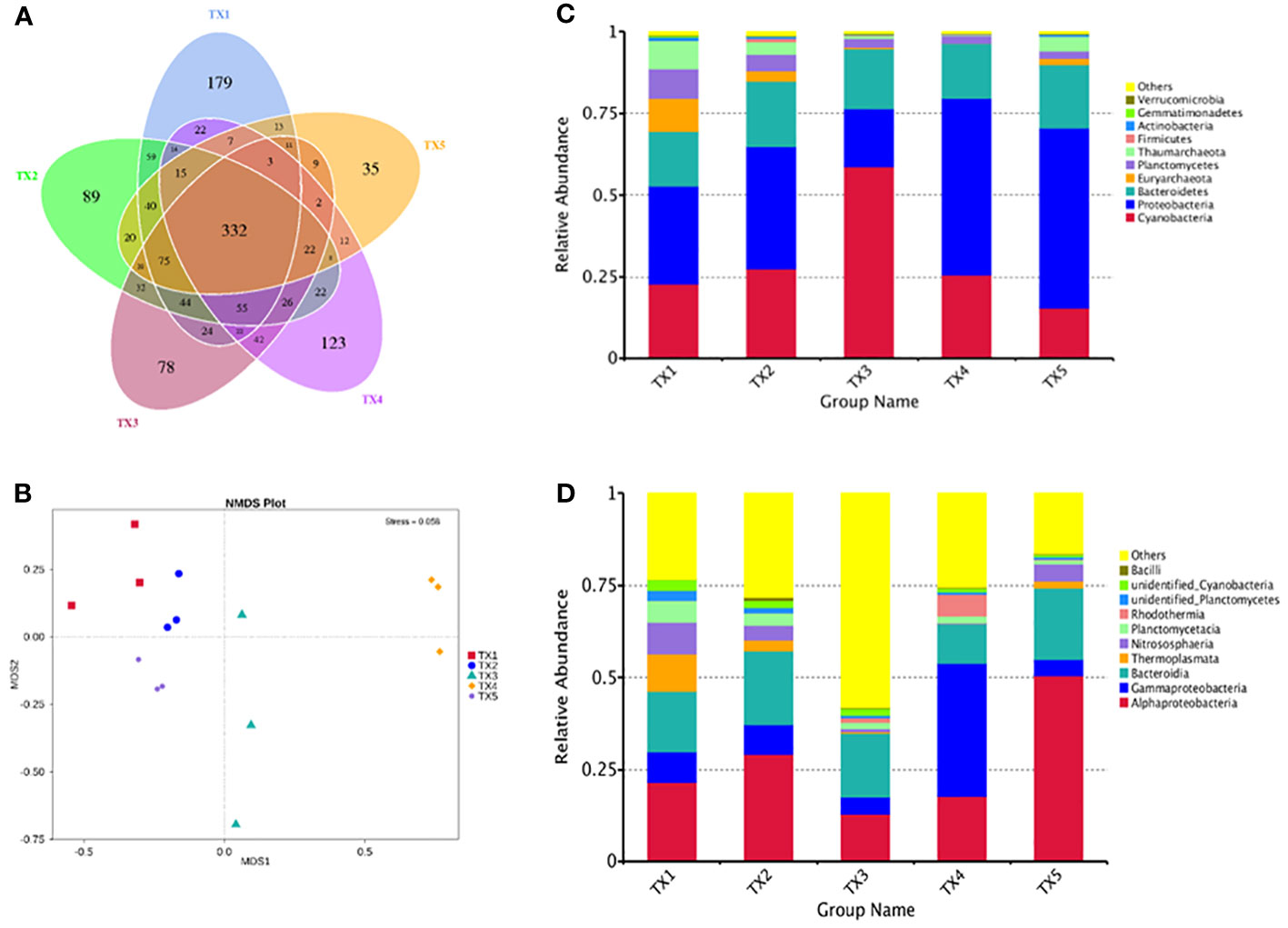
Figure 2 Comparison of bacterial OTUs and relative abundance of the bacterial communities in samples from different sample sites. (A) Venn diagrams of shared OTUs of samples. (B) NMDS for samples. (C) Phylum level and (D) class level relative abundance of the top 10 bacterial communities in samples.
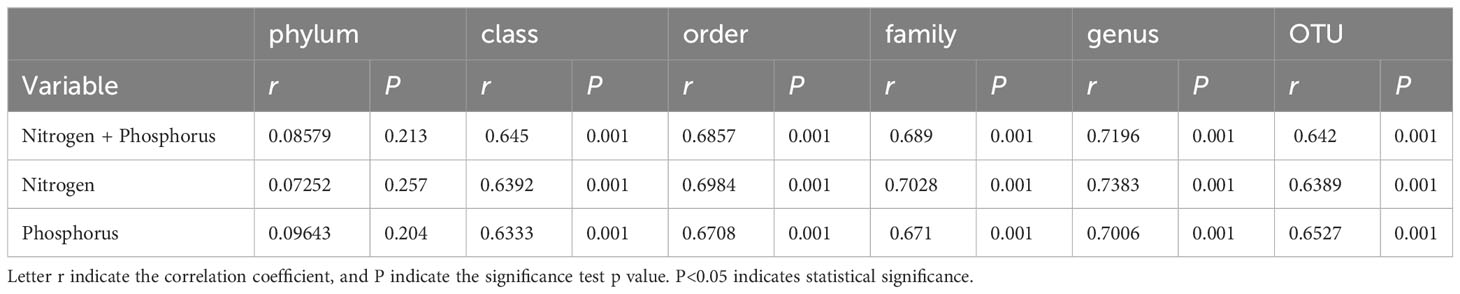
Table 3 Mantel test showed the correlation of nitrogen, phosphorus, interaction between nitrogen and phosphorus with bacterial community.
3.3 Composition of the prokaryotic community
The composition of prokaryotic community was investigated using 16S rRNA sequencing (Figures 2C, D). A total of 34 distinct prokaryotic phyla were detected across all the samples, although only six of these (Cyanobacteria, Proteobacteria, Bacteroidetes, Euryarchaeota, Planctomycetes and Thaumarchaeota) comprised > 97% of each sample (Figure 2C). Inside these phyla, Cyanobacteria, Proteobacteria, Bacteroidetes accounted 69% (lowest in TX1) to 96% (highest in TX5) of the five sites. The relative abundance of Proteobacteria was significantly higher in the TX4 and TX5 samples than that in the TX1 and TX3 samples, while the relative abundance of Cyanobacteria and Bacteroidetes did not differ significantly among all the samples. Further analysis identified a differential abundance of Euryarchaeota and Thaumarchaeota in sample TX1, while Proteobacteria showed a significant abundance in TX5 (Figure 3). At the genus level, TX4 revealed a differential abundance of Alteromonas, Pseudoalteromonas, Rubrivirga, Vibrio, Idiomarinaceae, and Halomonas, and their presence positively correlated with nitrogen and phosphorous levels. Conversely, in TX5, Sulfitobacter and Formosa were differentially abundant and showed a negative correlation with nitrogen and phosphorous levels (Figure 3). These findings suggest that the TX4 and TX5 sites host a unique assortment of bacterial genera compared to other sites.
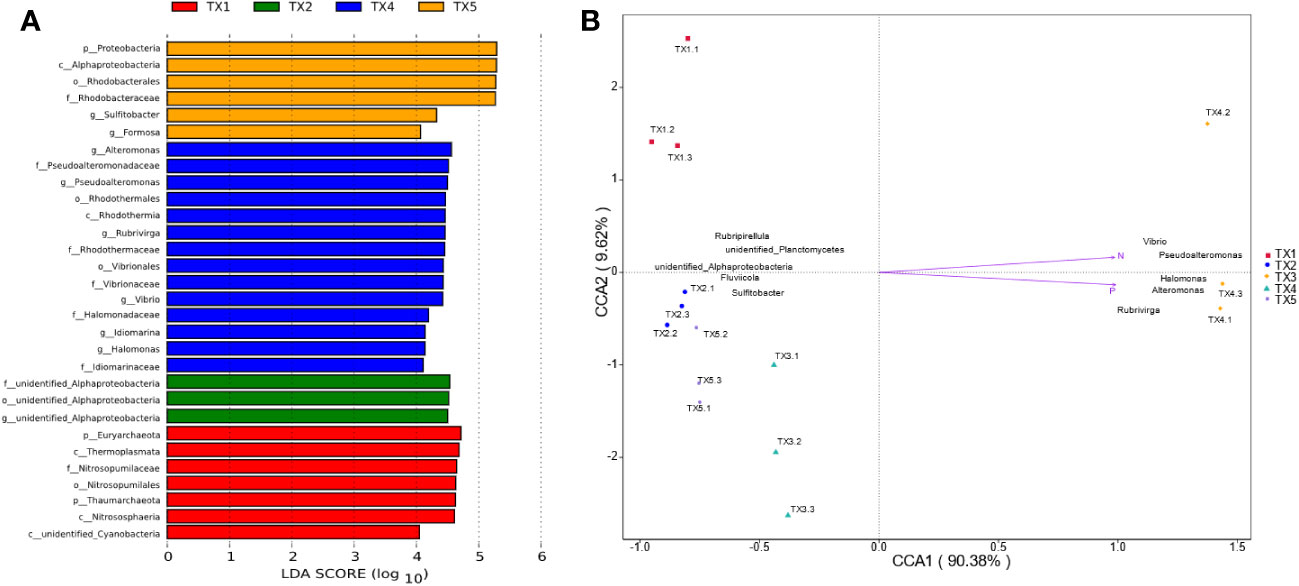
Figure 3 Identification of distinct taxa across various sampling sites and their correlations with environmental factors. (A) LDA across all sampling sites - note that TX3 exhibited no significantly varied taxa. The different letters in the bar plot denote bacterial taxonomic levels: phylum (p_), class (c_), order (o_), family (f_), genus (g_); lineages with LDA > 4 are displayed. (B) The CCA demonstrates relationships between the top 10 bacterial communities, environmental factors, and all sampling sites.
3.4 Isolation of bacteria and the assessment of algicidal activity
In order to assess the algicidal activity of bacteria, a total of 100 individual colonies were collected from five different sampling sites. Out of these colonies, 6 exhibited an algicidal rate over 80%, as indicated in Table S2. Interestingly, no algicidal bacteria were isolated from the TX1 sampling site. However, on the TX2, TX3, TX4, and TX5 plates, one, one, three, and two colonies, respectively, exhibited algicidal activity. The strain FDHY-MQ5 was found to exhibit 100% algicidal activity against K. mikimotoi after 48 h of challenge with 2% (v/v) of bacterial volume (OD600 = 4.5) concentration, as shown in Table S2. As a result, this strain was selected for further analysis, which will include identification, assessment of algicidal activity against various harmful algal species, investigation of algicidal characteristics, and examination of its algicidal process to lyse K. mikimotoi.
The bacterial strain FDHY-MQ5 was first identified using a scanning electron microscope, and then through analysis of its 16S rDNA sequence. The strain was about 0.5 to 1.0 μm in diameter and 1.0 to 3.5 μm in length, with flagella (Figure 4A). The sequence exhibited the greatest similarity to that of Pseudoalteromonas flavipulchra strain CSMA-N1 (GenBank accession no. MK482011) which was isolated from the Bohai Bay area of China, with 99% identity (Figure 4B). Based on a phylogenetic analysis, the shortest genetic distance among the Pseudoalteromonas species and strain FDHY-MQ5 was calculated. Thus, this bacterial strain was classified as P. flavipulchra FDHY-MQ5. Therefore, this bacterial strain was placed in the same subcluster of Gammaproteobacteria class as other species of Pseudoalteromonas and Alteromonas (Figure 4B).
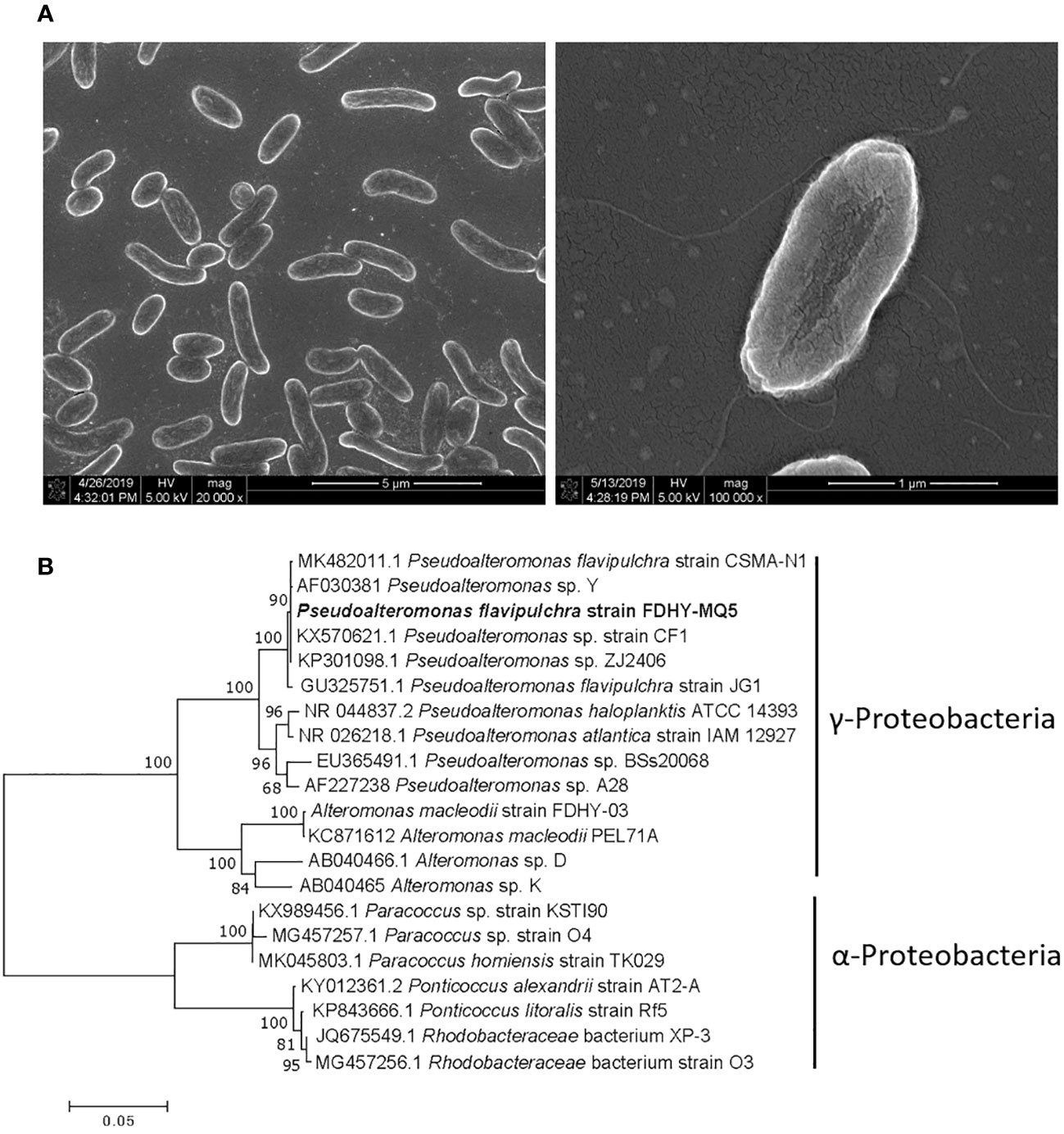
Figure 4 One cultivable bacterium Pseudoalteromonas flavipulchra FDHY-MQ5 was isolated from the TX4 site. (A) The morphology of FDHY-MQ5 under scanning electron microscopy. (B) Phylogenetic tree of known algicidal bacteria, including strain FDHY-MQ5. The tree topology shown was obtained from the neighbor-joining analysis. Support of node >50% is shown. The codes in front of the names are the GenBank accession numbers.
After the addition of 1% (v/v) of bacterial strain to the final volume rate, FDHY-MQ5 exhibited exceptional algicidal activity against K. mikimotoi (97% ± 2) and H. akashiwo (93% ± 0.1) after 24h of treatment (Figure 5). Subsequently, it displayed moderate activity against A. tamarense (50% ± 4) and S. costatum (29% ± 6), algicidal activity against and, while showing less than 10% algicidal activity against A. carterae, P. tricornutum and P. shikokuense. Further analysis of the algicidal rate of different bacterial concentrations against K. mikimotoi revealed that a bacterial concentration of 2% (OD600 = 4.5), resulted in a 100% algicidal rate after 6 hours of treatment. However, a 48-hour treatment was required to achieve the same rate when the bacterial concentration was reduced to 1%. A concentration of 0.5% resulted in only a 76% algicidal rate after 72 hours (Figure 6).
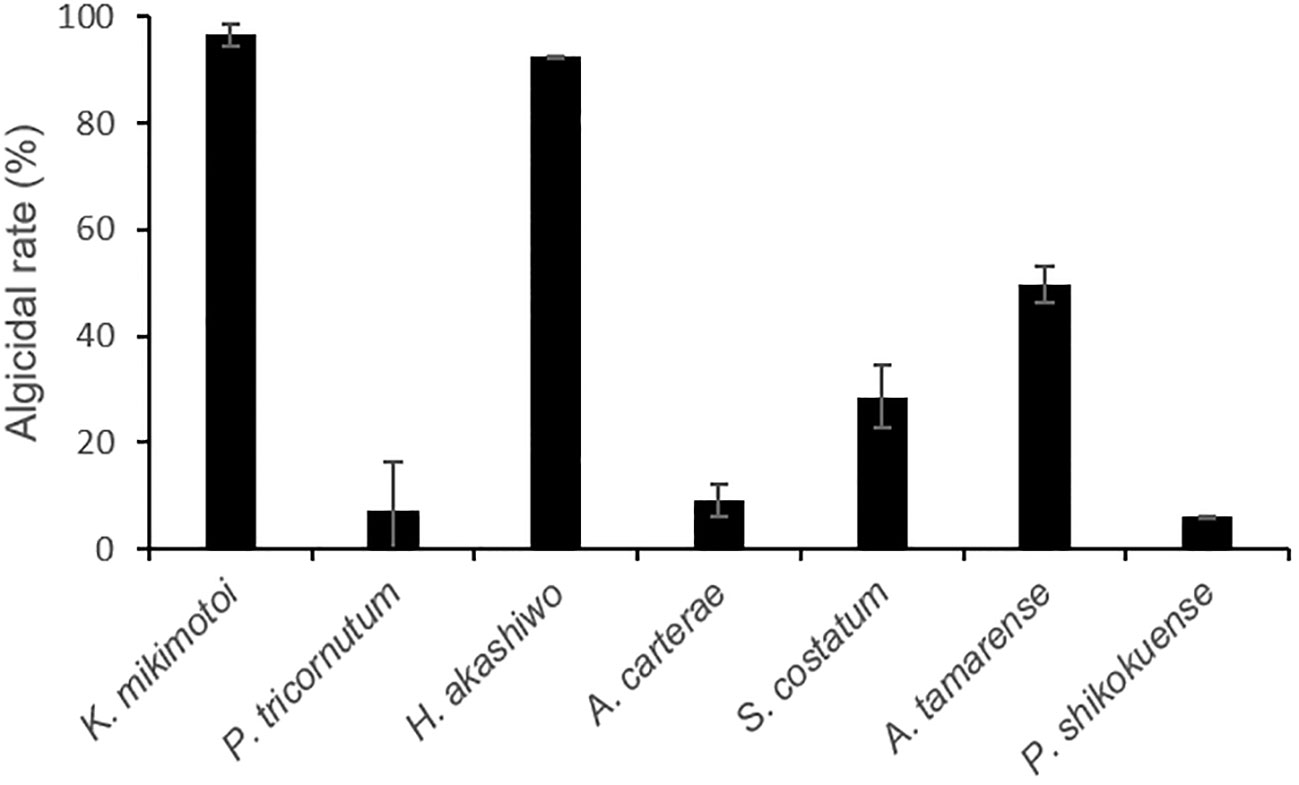
Figure 5 Algicidal activity of FDHY-MQ5 against several types of phytoplankton (Karenia mikimotoi, Phaeodactylum tricomutum, Heterosigma akashiwo, Alexandrium carterae, Skeletonema costatum, Alexandrium tamarense, Prorocentrum shikokuense) after 24 h of lysing. Bars represent triplicate means of % activity. Error bars indicate ± SD of biological triplicates.
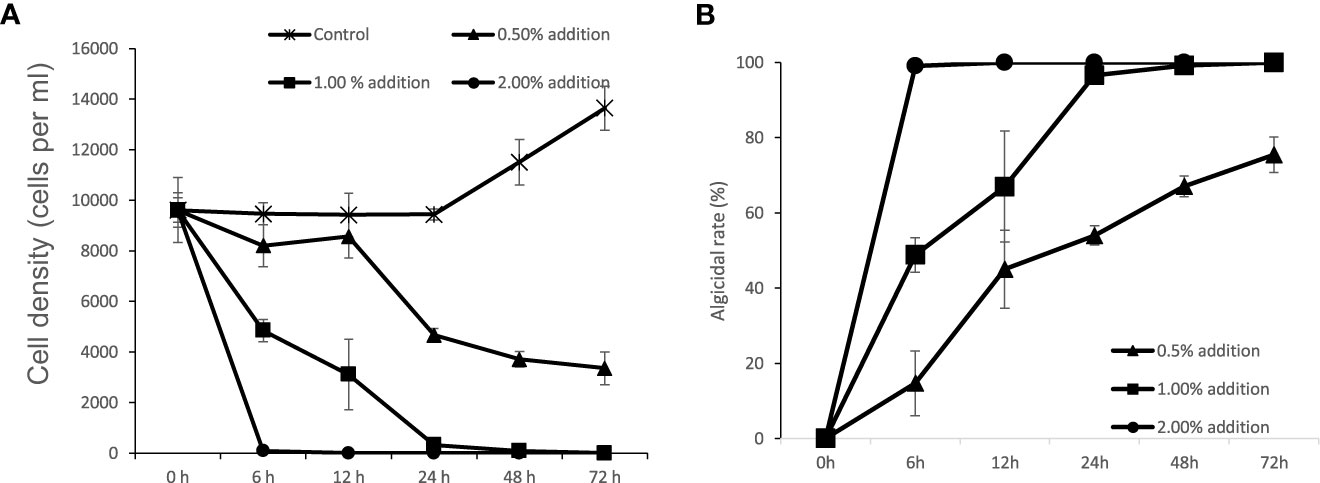
Figure 6 Algicidal modes of strain FDHY-MQ5 against Karenia mikimotoi. Growth dynamics (A) and algicidal rate (B) of algal cultures with different fractions of bacterial culture. Error bars indicate ± SD of biological triplicates.
Microscopy observations of the algal lysis of strain FDHY-MQ5 against K. mikimotoi were conducted (Figure 7). The bacterial supernatant was filtered, and upon the addition of this filtrate, the K. mikimotoi cell chains partially broke up and fell apart after 6 hours of incubation. After 12 hours of treatment, the entire cell wall shattered into small pieces (Figure 7D1). Subsequently, the cell walls were steadily breached, and after 18 hours of incubation, more than half of cell walls were lysed (Figure 7E1). Based on the results of DAPI staining, only the cell wall and/or cell membranes were lysed at this moment, while the nuclei remained intact, and the lipids and proteins were packaged in the cell (Figure 7E2). After 24 h of treatment, the algal cells were completely degraded (Figures 7F1, F2). These results indicate that FDHY-MQ5 can destroy the cell wall and membrane of K. mikimotoi, leading to its cytolysis within 24 h.
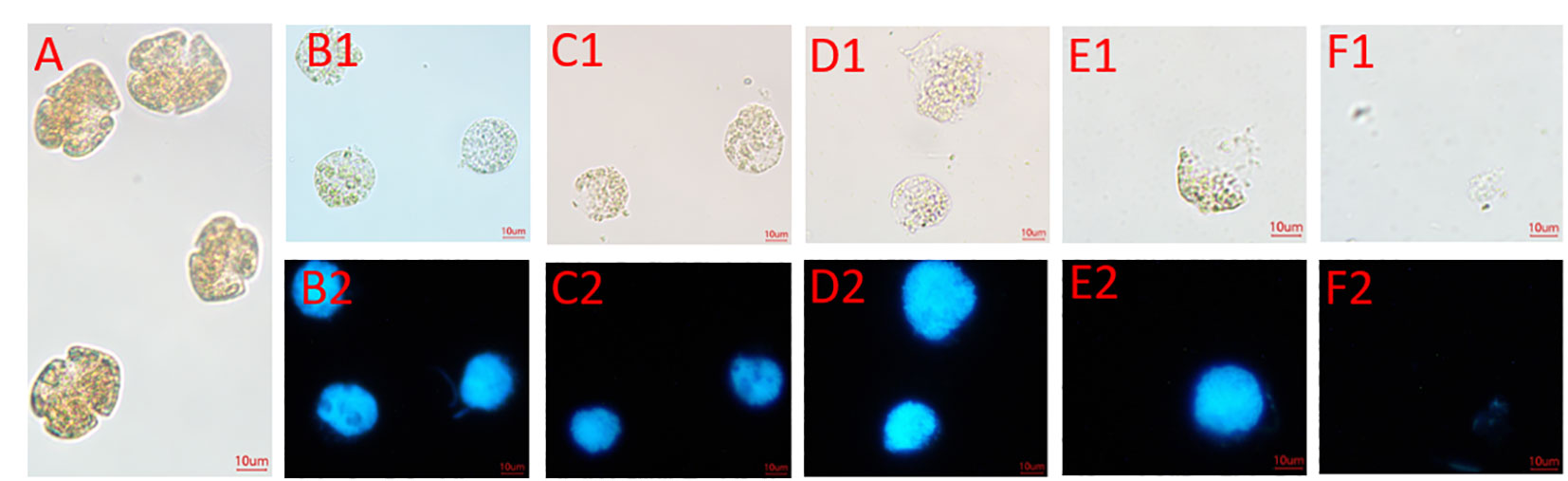
Figure 7 Changes in the morphology and nuclei of Karenia mikimotoi cells lysed with FDHY-MQ5 bacteria. (A), Bright-field illumination of K. mikimotoi cells; (B1-F1) indicated that algal cells treated with FDHY-MQ5 for 0 h, 6 h, 12 h, 18 h and 24 h under bright-field, respectively. B2-F2 represented that UV illumination for DAPI (5 g/ml) stained nuclei for the cell (B1-F1), respectively. DAPI, 4′,6-diamidino-2-phenylindole.
4 Discussion
Harmful algal blooms (HABs) have become increasingly frequent and intense in coastal zones worldwide in recent decades (Bechard and Lang, 2023). The interactions between bacteria and algae, including competition, symbiosis, and predation, have garnered growing attention due to their association with HABs (Kazamia et al., 2012; Kim et al., 2023). The Tongxin bay is subject to a variety of environmental pressures induced by aquaculture, such as eutrophication. In this study, K. longicanalis dominated the bloom in Tongxin Bay, with TX3 and TX4 exhibiting the highest DIN and DIP, respectively, as well as the highest algal cell density (Figure 1; Table 1). The Mantel test results indicated that nitrogen, phosphorus, and the interaction between nitrogen and phosphorus significantly impacted the bacterial community at the OTU, genus, family, class, and order level (Table 3). Nitrogen and phosphorus have been extensively studied as crucial factors with a tremendous impact on bacterial diversity in both terrestrial and the marine environments (Meyer-Reil and Köster, 2000; Zhang et al., 2019a; Zhang et al., 2019b). Eukaryotic phytoplankton require nitrogen and phosphorus as essential macronutrients (Zhang et al., 2015). Initially, eukaryotic phytoplankton can increase by taking up DIN and DIP from surrounding waters and releasing organic phosphorus into the water (Kang et al., 2021; Eigemann et al., 2022). When phosphate is scarce, eukaryotic phytoplankton and cyanobacteria can adapt to low phosphorus levels by replacing phospholipids with non-phosphorus membrane lipids, while marine heterotrophic bacteria only contain phospholipids and no substitute lipids (Van Mooy et al., 2009). The bacterial community is capable of exclusively meeting its nitrogen and phosphorus demands with organic forms provided by phytoplankton exudates (Eigemann et al., 2022). Thus, dinoflagellate blooms in the Tongxin bay, as well as the interaction between microalgae and bacteria, are significantly influenced by nitrogen and phosphorus.
A previous study has shown that phytoplankton contributes nearly half of the global primary production through photosynthetic activity (Behrenfeld et al., 2006). In the microbial loop, heterotrophic bacteria recycle approximately 90% of the organic materials generated by phytoplankton (Azam et al., 1983). Clues from previous studies suggest that the numbers of algicidal bacteria increase during later bloom phases or in nutrient hotspots dominated by specific phytoplankton (Meyer et al., 2017). Certain microorganisms can lyse algae cells during the progression of HABs, and are thus critical for their termination (Coyne et al., 2022). Algicidal bacteria are of substantial interest for application in biotechnology and environmental engineering (Meyer et al., 2017). In this study, bacterial communities were significantly different in five sites. At the phylum level, Proteobacteria had the highest relative abundance in the TX4 and TX5 sites (Figure 2C). At the class level, Gammaproteobacteria, including Pseudoalteromonas and Alteromonas, and Alphaproteobacteria (primarily Sulfitobacter) were enriched in the TX4 and TX5 sites, respectively (Figure 2D and 3). Many bacteria belonging to Pseudoalteromonas, Alteromonas, and Sulfitobacter were found to be algicidal against dinoflagellates (Skerratt et al., 2002; Cai et al., 2011; Zhang et al., 2020; Hu et al., 2021; Wang et al., 2022). Furthermore, some Pseudoalteromonas isolated from the digestive tracts of blue mussels contaminated with paralytic shellfish toxins (PSTs) may degrade PSTs (Donovan et al., 2009). These results suggest that bacteria in the Pseudoalteromonas, Alteromonas and Sulfitobacter genera could play an important role in the late phases of blooms due to their dominance in the decline and termination phases of the process.
Several strains from the Pseudoalteromonas genus have been found to exhibit algicidal activity against harmful algae (Meyer et al., 2017). However, their algicidal activity manifested differently toward target algal cells. For instance, Pseudoalteromonas sp48 showed high algicidal activity against the dinoflagellate A. tamarense (Su et al., 2007; Lyu et al., 2019), while the strain Pseudoalteromonas FDHY-MQ5 was only 50% algicidal active against A. tamarense in 24 h (Figure 5). In our previous study, we isolated a Pseudoalteromonas strain FDHY-MZ2 (Shi et al., 2022), which showed significantly lower algicidal activity against H. akashiwo compared to FDHY-MQ5. These differences could be associated with the significant differences in the genome specific species of Pseudoalteromonas (Alker et al., 2021). Simultaneously, it is plausible that microbial competition, coupled with its impacts on the co-culture of algicidal bacteria and phytoplankton, might contribute to the observed differential species representation. These bactria may generate differental algicidal metabolites, which may have a defined target of cell membranes component and therefore lead specific host cell death (Meyer et al., 2017). Based on the result of our study, the strain FDHY-MQ5 exhibited high algicidal activity against naked dinoflagellates and harmful red tide algae that lack cell walls (Figures 5 and 6). Compared with K. mikimotoi targeted algicidal bacteria reported, the algicidal rate of the strain FDHY-MQ5 is relatively high (Zheng et al., 2018; Du et al., 2023). Bacteria with the ability to dissolve cell walls typically posses a comprehensive system for chitin degradation. This is often observed in pigmented strains belonging to the genus Pseudoalteromonas. While non-pigmented strains are usually capable of degrading carbohydrates derived from algae (Paulsen et al., 2019). The strain FDHY-MQ5 showed no pigmentation during the culturing process, thus we believe it falls in the latter category. Thus, This strain may have potential applications in the control of algal blooms caused by these type of algae. Our study also found that the strain FDHY-MQ5 could rapidly and effectively lyse the cell membrane of the algal cell, as determined by the process of algae lysis and morphology changes of K. mikimotoi (Figure 7). Compared to previously reported algicidal bacteria on K. mikimotoi (Shi et al., 2022; Du et al., 2023), the strain FDHY-MQ5 was among the highest group with algicidal activity and thus has the potential to be a valuable source of bacterial agents for controlling K. mikimotoi blooms. K. longicanalis closely resembles morphologically K. mikimotoi. Both of them belong to the ‘mikimotoi-complex’ (Yang et al., 2001). In HABs events, they coexist in the water body, thus we hypothesize that they may have a similar econiche in the progression of HABs events (Yang et al., 2001; Orlova et al., 2022). There was no culture for K. longicanalis available for this study; therefore, whether the strain FDHY-MQ5 also has high algicidal rate on this species needs further study.
5 Conclusions
Overall, our results suggest that the bacterial communities differ significantly in five sites and algicidal bacteria mainly in later phase of dinoflagellate bloom. Our findings shed light on the potential of algicidal bacteria for controlling HABs and their potential applications in biotechnology and environmental engineering.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov, BioProject: PRJNA987320.
Author contributions
YZ, XZ and XS contributed to conception and design of the study. YZ, XZ, and XS performed the experiments. WT and KZ performed the statistical analysis. YZ, XS, and ZW wrote the first draft of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the National Natural Science Foundation of China (41976130); the Special Fund Project for Marine and Fishery Protection and Development in Fujian Province, China (FZJZ-2022-1); the Special Foundation for Young Scientists of Minjiang University (MJY20008); the Fuzhou Forestry Science and Technology Research Project (2023RLKY03); and the Project of Fujian Provincial Department of Education (JAT190627).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1242319/full#supplementary-material
References
Alker A. T., Gode B. S., Aspiras A. E., Jones J. E., Michael S. R., Aguilar D., et al. (2021). Draft genome sequences of 10 bacteria from the marine pseudoalteromonas group. Microbiol. Resource Announcements 10 (32), e00404-21:1-3. doi: 10.1128/mra.00404-21
Amin S. A., Hmelo L. R., Van Tol H. M., Durham B. P., Carlson L. T., Heal K. R., et al. (2015). Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522 (7554), 98–101. doi: 10.1038/nature14488
Azam F., Fenchel T., Field J. G., Gray J. S., Meyer-Reil L.-A., Thingstad F. (1983). The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser. 10, 251–263. doi: 10.7208/chicago/9780226125534-024
Bechard A., Lang C. (2023). Seafood consumption during harmful algal blooms: The impact of information regarding safety and health. Harmful Algae 123, 102387. doi: 10.1016/j.hal.2023.102387
Behrenfeld M. J., O’malley R. T., Siegel D. A., Mcclain C. R., Sarmiento J. L., Feldman G. C., et al. (2006). Climate-driven trends in contemporary ocean productivity. Nature 444 (7120), 752–755. doi: 10.1038/nature05317
Bokulich N. A., Subramanian S., Faith J. J., Gevers D., Gordon J. I., Knight R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10 (1), 57–59. doi: 10.1038/nmeth.2276
Bolch C. J. S., Bejoy T. A., Green D. H. (2017). Bacterial associates modify growth dynamics of the dinoflagellate gymnodinium catenatum. Front. Microbiol. 8, 670. doi: 10.3389/fmicb.2017.00670
Cai W., Wang H., Tian Y., Chen F., Zheng T. (2011). Influence of a bacteriophage on the population dynamics of toxic dinoflagellates by lysis of algicidal bacteria. Appl. Environ. Microbiol. 77 (21), 7837–7840. doi: 10.1128/AEM.05783-11
Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 (5), 335–336. doi: 10.1038/nmeth.f.303
Chan L. L., Hodgkiss I. J., Lu S., Lo S. C.-L. (2004). Use of two-dimensional gel electrophoresis proteome reference maps of dinoflagellates for species recognition of causative agents of harmful algal blooms. Proteomics 4 (1), 180–192. doi: 10.1002/pmic.200300548
Chen T., Xiao J., Liu Y., Song S., Li C. (2019). Distribution and genetic diversity of the parasitic dinoflagellate Amoebophrya in coastal waters of China. Harmful Algae 89, 101633. doi: 10.1016/j.hal.2019.101633
Cole J. J. (1982). Interactions between bacteria and algae in aquatic ecosystems. Annu. Rev. Ecol. Systematics 13, 291–314. doi: 10.1146/annurev.es.13.110182.001451
Coyne K. J., Wang Y., Johnson G. (2022). Algicidal bacteria: A review of current knowledge and applications to control harmful algal blooms. Front. Microbiol. 13, 871177–871177. doi: 10.3389/fmicb.2022.871177
Croft M. T., Lawrence A. D., Raux-Deery E., Warren M. J., Smith A. G. (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438 (7064), 90–93. doi: 10.1038/nature04056
Delong E. F. (1992). Archaea in coastal marine environments. Proc Natl Acad Sci USA 89 (12), 5685–5689. doi: 10.1073/pnas.89.12.5685
Donovan C. J., Garduño R. A., Kalmokoff M., Ku J. C., Quilliam M. A., Gill T. A. (2009). Pseudoalteromonas bacteria are capable of degrading paralytic shellfish toxins. Appl. Environ. Microbiol. 75 (21), 6919–6923. doi: 10.1128/AEM.01384-09
Du W., Ding N., Wang R., Gao P., Zhang W., Lin F., et al. (2023). Characterization of cell death in harmful Karenia mikimotoi under algicidal activity of Marinobacter sp. O-7. J. Sea Res. 191, 102326. doi: 10.1016/j.seares.2022.102326
Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10 (10), 996–998. doi: 10.1038/nmeth.2604
Eigemann F., Rahav E., Grossart H.-P., Aharonovich D., Sher D., Vogts A., et al. (2022). Phytoplankton exudates provide full nutrition to a subset of accompanying heterotrophic bacteria via carbon, nitrogen and phosphorus allocation. Environ. Microbiol. 24 (5), 2467–2483. doi: 10.1111/1462-2920.15933
Gu H., Wu Y., Lu S., Lu D., Tang Y. Z., Qi Y. (2022). Emerging harmful algal bloom species over the last four decades in China. Harmful Algae 111, 102059. doi: 10.1016/j.hal.2021.102059
Guillard R. R. L., Hargraves P. E. (1993). Stichochrysis immobilis is a diatom, not a chrysophyte. Phycologia 32 (3), 234–236. doi: 10.2216/i0031-8884-32-3-234.1
Guo X. L. H., Ding D., Guan C., Yi X. (2014). The economic cost of red tides in China from 2008-2012. PICES Sci. Rep. 47 (27), 27–34.
Haas B. J., Gevers D., Earl A. M., Feldgarden M., Ward D. V., Giannoukos G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21 (3), 494–504. doi: 10.1101/gr.112730.110
Heredia-Tapia A., Arredondo-Vega B. O., Nuñez-Vázquez E. J., Yasumoto T., Yasuda M., Ochoa J. L. (2002). Isolation of Prorocentrum lima (Syn. Exuviaella lima) and diarrhetic shellfish poisoning (DSP) risk assessment in the Gulf of California, Mexico. Toxicon 40 (8), 1121–1127. doi: 10.1016/S0041-0101(02)00111-3
Hoagland P., Kirkpatrick B., Jin D., Kirkpatrick G., Fleming L. E., Ullmann S. G., et al. (2020). Lessening the hazards of Florida red tides: a common sense approach. Front. Mar. Sci. 7, 538. doi: 10.3389/fmars.2020.00538
Hu C., Barnes B. B., Qi L., Lembke C., English D. (2016). Vertical migration of Karenia brevis in the northeastern Gulf of Mexico observed from glider measurements. Harmful Algae 58, 59–65. doi: 10.1016/j.hal.2016.07.005
Hu T., Wang S., Shan Y., Zhang Y., Zhu Y., Zheng L. (2021). Complete genome of marine microalgae associated algicidal bacterium sulfitobacter pseudonitzschiae H46 with quorum sensing system. Curr. Microbiol. 78 (10), 3741–3750. doi: 10.1007/s00284-021-02632-4
Inaba N., Trainer V. L., Onishi Y., Ishii K.-I., Wyllie-Echeverria S., Imai I. (2017). Algicidal and growth-inhibiting bacteria associated with seagrass and macroalgae beds in Puget Sound, WA, USA. Harmful Algae 62, 136–147. doi: 10.1016/j.hal.2016.04.004
Jung S., Kim B. H., Katano T., Kong D. S., Han M. S. (2008). Pseudomonas fluorescens HYK0210-SK09 offers species-specific biological control of winter algal blooms caused by freshwater diatom Stephanodiscus hantzschii. J. Appl. Microbiol. 105 (1), 186–195. doi: 10.1111/j.1365-2672.2008.03733.x
Kang J., Park J. S., Jung S. W., Kim H.-J., Joo H. M., Kang D., et al. (2021). Zooming on dynamics of marine microbial communities in the phycosphere of Akashiwo sanGuinea (Dinophyta) blooms. Mol. Ecol. 30 (1), 207–221. doi: 10.1111/mec.15714
Kazamia E., Czesnick H., Nguyen T. T. V., Croft M. T., Sherwood E., Sasso S., et al. (2012). Mutualistic interactions between vitamin B12-dependent algae and heterotrophic bacteria exhibit regulation. Environ. Microbiol. 14 (6), 1466–1476. doi: 10.1111/j.1462-2920.2012.02733.x
Kim H.-J., Jeoung G., Kim K. E., Park J. S., Kang D., Baek S. H., et al. (2023). Co-variance between free-living bacteria and Cochlodinium polykrikoides (Dinophyta) harmful algal blooms, South Korea. Harmful Algae 122, 102371. doi: 10.1016/j.hal.2022.102371
Krock B., Tillmann U., John U., Cembella A. D. (2009). Characterization of azaspiracids in plankton size-fractions and isolation of an azaspiracid-producing dinoflagellate from the North Sea. Harmful Algae 8 (2), 254–263. doi: 10.1016/j.hal.2008.06.003
Lau W. W. Y., Keil R. G., Armbrust E. V. (2007). Succession and diel transcriptional response of the glycolate-utilizing component of the bacterial community during a spring phytoplankton bloom. Appl. Environ. Microbiol. 73 (8), 2440–2450. doi: 10.1128/AEM.01965-06
Lee H.-G., Kim H. M., Min J., Park C., Jeong H. J., Lee K., et al. (2020). Quantification of the paralytic shellfish poisoning dinoflagellate Alexandrium species using a digital PCR. Harmful Algae 92, 101726. doi: 10.1016/j.hal.2019.101726
Lin S., Cheng S., Song B., Zhong X., Lin X., Li W., et al. (2015). The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 350 (6261), 691–694. doi: 10.1126/science.aad0408
Lu S., Hodgkiss I. J. (2004). Harmful algal bloom causative collected from Hong Kong waters. Hydrobiologia 512 (1), 231–238. doi: 10.1023/B:HYDR.0000020331.75003.18
Lyu Y. H., Zhou Y. X., Li Y., Zhou J., Xu Y. X. (2019). Optimized culturing conditions for an algicidal bacterium Pseudoalteromonas sp. SP 48 on harmful algal blooms caused by Alexandrium tamarense. MicrobiologyOpen 8 (8). doi: 10.1002/mbo3.803
Magoc T., Salzberg S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27 (21), 2957–2963. doi: 10.1093/bioinformatics/btr507
Meyer N., Bigalke A., Kaulfuß A., Pohnert G. (2017). Strategies and ecological roles of algicidal bacteria. FEMS Microbiol. Rev. 41 (6), 880–899. doi: 10.1093/femsre/fux029
Meyer-Reil L.-A., Köster M. (2000). Eutrophication of marine waters: effects on benthic microbial communities. Mar. pollut. Bull. 41 (1), 255–263. doi: 10.1016/S0025-326X(00)00114-4
Needham D. M., Fuhrman J. A. (2016). Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat. Microbiol. 1 (4), 16005. doi: 10.1038/nmicrobiol.2016.5
Orlova T. Y., Aleksanin A. I., Lepskaya E. V., Efimova K. V., Selina M. S., Morozova T. V., et al. (2022). A massive bloom of Karenia species (Dinophyceae) off the Kamchatka coast, Russia, in the fall of 2020. Harmful Algae 120, 102337. doi: 10.1016/j.hal.2022.102337
Parada A. E., Needham D. M., Fuhrman J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18 (5), 1403–1414. doi: 10.1111/1462-2920.13023
Paulsen S. S., Strube M. L., Bech P. K., Gram L., Sonnenschein E. C. (2019). Marine chitinolytic Pseudoalteromonas represents an untapped reservoir of bioactive potential. Msystems e00060-19, 1–12. doi: 10.1128/msystems.00060-19
Pitcher G. C., Foord C. J., Macey B. M., Mansfield L., Mouton A., Smith M. E., et al. (2019). Devastating farmed abalone mortalities attributed to yessotoxin-producing dinoflagellates. Harmful Algae 81, 30–41. doi: 10.1016/j.hal.2018.11.006
Richlen M. L., Morton S. L., Jamali E. A., Rajan A., Anderson D. M. (2010). The catastrophic 2008–2009 red tide in the Arabian gulf region, with observations on the identification and phylogeny of the fish-killing dinoflagellate Cochlodinium polykrikoides. Harmful Algae 9 (2), 163–172. doi: 10.1016/j.hal.2009.08.013
Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 (4), 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Senhorinho G. N. A., Ross G. M., Scott J. A. (2015). Cyanobacteria and eukaryotic microalgae as potential sources of antibiotics. Phycologia 54 (3), 271–282. doi: 10.2216/14-092.1
Shi R., Huang H., Qi Z., Hu W., Tian Z., Dai M. (2013a). Algicidal activity against Skeletonema costatum by marine bacteria isolated from a high frequency harmful algal blooms area in southern Chinese coast. World J. Microbiol. Biotechnol. 29 (1), 153–162. doi: 10.1007/s11274-012-1168-1
Shi X., Lin X., Li L., Li M., Palenik B., Lin S. (2017). Transcriptomic and microRNAomic profiling reveals multi-faceted mechanisms to cope with phosphate stress in a dinoflagellate. ISME J. 11, 2209–2218. doi: 10.1038/ismej.2017.81
Shi X., Liu L., Li Y., Xiao Y., Ding G., Lin S., et al. (2018). Isolation of an algicidal bacterium and its effects against the harmful-algal-bloom dinoflagellate Prorocentrum donghaiense (Dinophyceae). Harmful Algae 80, 72–79. doi: 10.1016/j.hal.2018.09.003
Shi X., Xiao Y., Liu L., Xie Y., Ma R., Chen J. (2021). Transcriptome responses of the dinoflagellate Karenia mikimotoi driven by nitrogen deficiency. Harmful Algae 103, 101977. doi: 10.1016/j.hal.2021.101977
Shi X., Zhang H., Lin S. (2013b). Tandem repeats, high copy number and remarkable diel expression rhythm of form II ruBisCO in prorocentrum donghaiense (Dinophyceae). PloS One 8 (8), e71232. doi: 10.1371/journal.pone.0071232
Shi X., Zou Y., Zheng W., Liu L., Xie Y., Ma R., et al. (2022). A Novel Algicidal Bacterium and Its Effects against the Toxic Dinoflagellate Karenia mikimotoi (Dinophyceae). Microbiol. Spectr. 10 (3), e00429–e00422. doi: 10.1128/spectrum.00429-22
Skerratt J. H., Bowman J. P., Hallegraeff G., James S., Nichols P. D. (2002). Algicidal bacteria associated with blooms of a toxic dinoflagellate in a temperate Australian estuary. Mar. Ecol. Prog. Ser. 244, 1–15. doi: 10.3354/meps244001
Su J. Q., Yang X. R., Zheng T. L., Tian Y., Jiao N. Z., Cai L. Z., et al. (2007). Isolation and characterization of a marine algicidal bacterium against the toxic dinoflagellate Alexandrium tamarense. Harmful Algae 6 (6), 799–810. doi: 10.1016/j.hal.2007.04.004
Van Mooy B., Fredricks H. F., Pedler B. E., Dyhrman S. T., Karl D. M., Koblížek M., et al. (2009). Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature 458 (7234), 69–72. doi: 10.1038/nature07659
Wang J., Cen J., Li S., Lü S., Moestrup Ø., Chan K.-K., et al. (2018). A re-investigation of the bloom-forming unarmored dinoflagellate Karenia longicanalis (syn. Karenia umbella) from Chinese coastal waters. J. Oceanology Limnology 36 (6), 2202–2215. doi: 10.1007/s00343-019-7191-4
Wang Z.-H., Nie X.-P., Jiang S.-J., Zhao J.-G., Cao Y., Zhang Y.-J., et al. (2011). Source and profile of paralytic shellfish poisoning toxins in shellfish in Daya Bay, South China Sea. Mar. Environ. Res. 72 (1), 53–59. doi: 10.1016/j.marenvres.2011.04.007
Wang Q., Shi X., Guo Y., Lv P., Zhong Y., Xie H., et al. (2022). Optimization of Algicidal Activity for Alteromonas sp. FDHY-03 against Harmful Dinoflagellate. J. Mar. Sci. Eng. 10 (9), 1274. doi: 10.3390/jmse10091274
Wang L., Zhuang Y., Zhang H., Lin X., Lin S. (2014). DNA barcoding species in Alexandrium tamarense complex using ITS and proposing designation of five species. Harmful Algae 31, 100–113. doi: 10.1016/j.hal.2013.10.013
Yang Z. B., Hodgkiss I. J., Hansen G. (2001). Karenia longicanalis sp nov. (Dinophyceae): a new bloom-forming species isolated from Hong Kong, May 1998. Botanica Marina 44 (1), 67–74. doi: 10.1515/BOT.2001.009
Zhang F., Fan Y., Zhang D., Chen S., Bai X., Ma X., et al. (2020). Effect and mechanism of the algicidal bacterium Sulfitobacter porphyrae ZFX1 on the mitigation of harmful algal blooms caused by Prorocentrum donghaiense. Environ. pollut. 263, 114475. doi: 10.1016/j.envpol.2020.114475
Zhang J., Liu Y. X., Zhang N., Hu B., Jin T., Xu H., et al. (2019a). NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 37 (6), 676–684. doi: 10.1038/s41587-019-0104-4
Zhang Y., Wang X., Xu F., Song T., Du H., Gui Y., et al. (2019b). Combining irrigation scheme and phosphorous application levels for grain yield and their impacts on rhizosphere microbial communities of two rice varieties in a field trial. J. Agric. Food Chem. 67 (38), 10577–10586. doi: 10.1021/acs.jafc.9b03124
Zhang Y.-J., Zhang S.-F., He Z.-P., Lin L., Wang D.-Z. (2015). Proteomic analysis provides new insights into the adaptive response of a dinoflagellate Prorocentrum donghaiense to changing ambient nitrogen. Plant Cell Environ. 38 (10), 2128–2142. doi: 10.1111/pce.12538
Zheng N., Ding N., Gao P., Han M., Liu X., Wang J., et al. (2018). Diverse algicidal bacteria associated with harmful bloom-forming Karenia mikimotoi in estuarine soil and seawater. Sci. Total Environ. 631-632, 1415–1420. doi: 10.1016/j.scitotenv.2018.03.035
Keywords: algicidal bacterium, bacterial communities, harmful algal bloom, Karenia longicanalis, Karenia mikimotoi
Citation: Zhang Y, Zhou X, Tang W, Zhou K, Wang Z and Shi X (2023) Decoding the influence of bacterial community structure and algicidal bacteria in a Karenia longicanalis bloom event. Front. Mar. Sci. 10:1242319. doi: 10.3389/fmars.2023.1242319
Received: 20 June 2023; Accepted: 16 October 2023;
Published: 30 October 2023.
Edited by:
Judith Marie O’Neil, University of Maryland, College Park, United StatesReviewed by:
Cong Fei, New York University Abu Dhabi, United Arab EmiratesHao Zhang, Xiamen University, China
Copyright © 2023 Zhang, Zhou, Tang, Zhou, Wang and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinguo Shi, eHNoaUBmenUuZWR1LmNu
 Yingjiao Zhang
Yingjiao Zhang Xiaowen Zhou2
Xiaowen Zhou2 Weiqi Tang
Weiqi Tang Konglin Zhou
Konglin Zhou Zonghua Wang
Zonghua Wang Xinguo Shi
Xinguo Shi