- 1Department of Environmental Science and Engineering, Kyung Hee University, Yongin, Republic of Korea
- 2Department of Ocean Integrated Science, Chonnam National University, Yeosu, Republic of Korea
- 3Institute for Environment and Energy, Pusan National University, Busan, Republic of Korea
In aquatic ecosystem, the food web structure based on composition and abundance of prey species influences the feeding characteristics of predators, resulting in changes to the structure and function of the entire food web. Zooplankton mainly feed on phytoplankton, but the method of analyzing the feed source through visual and microscopic identification of gut contents has limitations in applicable species and the low resolution of the analysis results. In this study, potential vs. eaten food sources of Sinocalanus tenellus, a small and medium-sized copepod species dominant in brackish waters, were analyzed based on phytoplankton operational taxonomic units detected from the habitat’s raw water and treated whole bodies of S. tenellus. The alpha diversity and dominant/subdominant species of each potential and eaten food source were identified, and they were compared across seasons. At the same time, the feeding selectivity index (Ei) was calculated by identifying overlapping species in the potential vs. eaten food source pool. As a result, it was confirmed that not all surrounding potential food sources are fed on by S. tenellus, and that its diet can vary depending on the presence/absence of preferred prey. Metabarcoding techniques is considered to be an effective way to identify fluctuations in major food sources of zooplankton at the genus or species level. If uncertainties such as incomplete species identification as identified from the results of this study are improved through improving of analysis methods such as application of species-specific primers in the future, it will be useful for securing information on the feeding characteristics of small to medium-sized zooplankton.
1 Introduction
Zooplankton, as primary consumers, feed on lower-level trophic organisms such as phytoplankton, while facilitating material and energy transfers in aquatic ecosystems through consumption by higher-level trophic organisms, such as fish (Turner, 2004). Nutrient and energy transfers through the zooplankton community in the aquatic ecosystem food web are strongly influenced by species-specific feeding characteristics and preferences according to prey species composition and abundance (Dam and Baumann, 2017). Species diversity of phytoplankton, the main food source of zooplankton, causes changes in the structure and function of aquatic food webs, specifically of the grazing food web (Polis et al., 1997; Thébault and Loreau, 2005; Narwani and Mazumder, 2012). In other words, identifying the feeding characteristics of zooplankton species present in a water body and their interactions with phytoplankton—the predator-prey relationship—can be fundamental in understanding and predicting the response of primary producers in the food web to environmental changes. For example, in brackish environments, an increase in water temperature limits zooplankton abundance, which can affect increases in phytoplankton biomass (Oda et al., 2018). In this way, the changes in the responses of phytoplankton and zooplankton communities, along with their consequent interactions’ changes, in response to the environmental conditions of the water body are considered important information for understanding the relationships within food web network (Kruk et al., 2021).
The traditional method to analyze the feeding characteristics of organisms is to identify the gut contents via the naked eye or with a microscope (Chang et al., 2011; Furgała-Selezniow et al., 2014). However, these methods have limited results because they can be derived from the decomposition of feeding sources remaining in the gut. Therefore, by applying DNA technology to gut content analysis, it is possible to identify species at a high resolution with only short regions (approximately 150~700 bp) (Pompanon et al., 2012). Because the meta-barcoding technique has the advantage of being able to detect many species simultaneously, it helps identify species that have been eaten based on DNA present in the gut (Vacher et al., 2016). Therefore, recently, through meta-barcoding analysis of gut contents, studies on the feeding characteristics of target organisms, such as fish and benthic macroinvertebrates, have been conducted (Su et al., 2018; Jo et al., 2020; Kuo et al., 2021; Vasquez et al., 2021). Nevertheless, this technology is rarely applied to zooplankton, whose gut contents are difficult to extract and handle due to their small body size and the problem of extracellular DNA contamination.
Copepods play an intermediate role because they are phytoplankton consumers and comprise the largest component of the zooplankton biomass in brackish ecosystems (Atkinson, 1995; Bollens and Penry, 2003; Olson et al., 2006; Chen et al., 2018). Copepods selectively feed according to the quality, ease of digestion, and toxicity of food sources (Lee et al., 2020). As a result, studies have been conducted on the feeding characteristics of copepod species because their food preferences vary depending on prey diversity within the habitat, affecting the entire aquatic ecosystem food web (Boltizár et al., 2017; Cleary et al., 2017; Ho et al., 2017; Yeh et al., 2020).
Copepod feeding characteristics are primarily obtained from seawater species, such as the genus Calanus, whose gut contents are relatively easy to sort and extract because of their large size (Gabrielsen et al., 2012). Conversely, in the case of small- and medium-sized copepods dominant in brackish water, it is difficult to analyse food sources in the same way as for large copepods, resulting in limited information on their feeding characteristics (Mäkinen et al., 2017; Soulié et al., 2022). In addition, when gut contents are extracted, there is a risk of contamination from external unnecessary genes or loss of genes in the gut contents. To solve this, a method of analysing food sources through gene extraction from the whole copepod body by removing only extracellular DNA without dissection has been proposed (Oh et al., 2020; Chae et al., 2021). In this case, an additional pre-treatment process is required to remove unnecessary genes, such as DNA remaining in the raw water surrounding or attached to the individual copepod. Various chemicals, such as commercial bleaches containing sodium hypochlorite, can be used to remove contaminants during genetic analyses (Greenstone et al., 2012; Oh et al., 2020; Chae et al., 2021).
Here, the feeding characteristics of (Sinocalanus tenellus, a representative species of brackish reservoirs in Korea (Kim et al., 2000), were identified using meta-barcoding analysis and an appropriate pre-treatment method. S. tenellus is a dominant brackish water species worldwide (Uye et al., 2000; Kang and Kim, 2003; Asami, 2004; Chen et al., 2018; Oda et al., 2018); therefore, they function as the main food source for fry and play an important role as food sources in maintaining fisheries. We (1) compared potential food sources in the raw water of habitats and the eaten food sources of S. tenellus to identify feeding selectivity and (2) analysed the relationship between potential and eaten food sources and seasonal environmental factors.
2 Materials and methods
2.1 Study site and sampling period
The Saemangeum Reservoir is a semi-closed coastal ecosystem that controls seawater inflow using two sluice gates on the southern part of the dykes. The simultaneous inflows of seawater from the sluice gates and freshwater from the Mangyeong and Dongjin Rivers adjacent to the Saemangeum Reservoir create a brackish zone. Here, we selected one site within the Saemangeum Reservoir: the point (35°49′17.03″ N, 126°37′38.60″ E) of freshwater inflow from the Dongjin River to the reservoir (Figure 1). Seasonal water quality and plankton community analyses were conducted through field surveys in the winter (February), spring (May), summer (August), and fall (November) of 2020. Simultaneously, raw water and zooplankton samples were collected to analyse potential and consumed food sources (individual gut contents) of the targeted copepod species S. tenellus. Ethical review and approval were not required for the study on animals in accordance with the local legislation and institutional requirements.
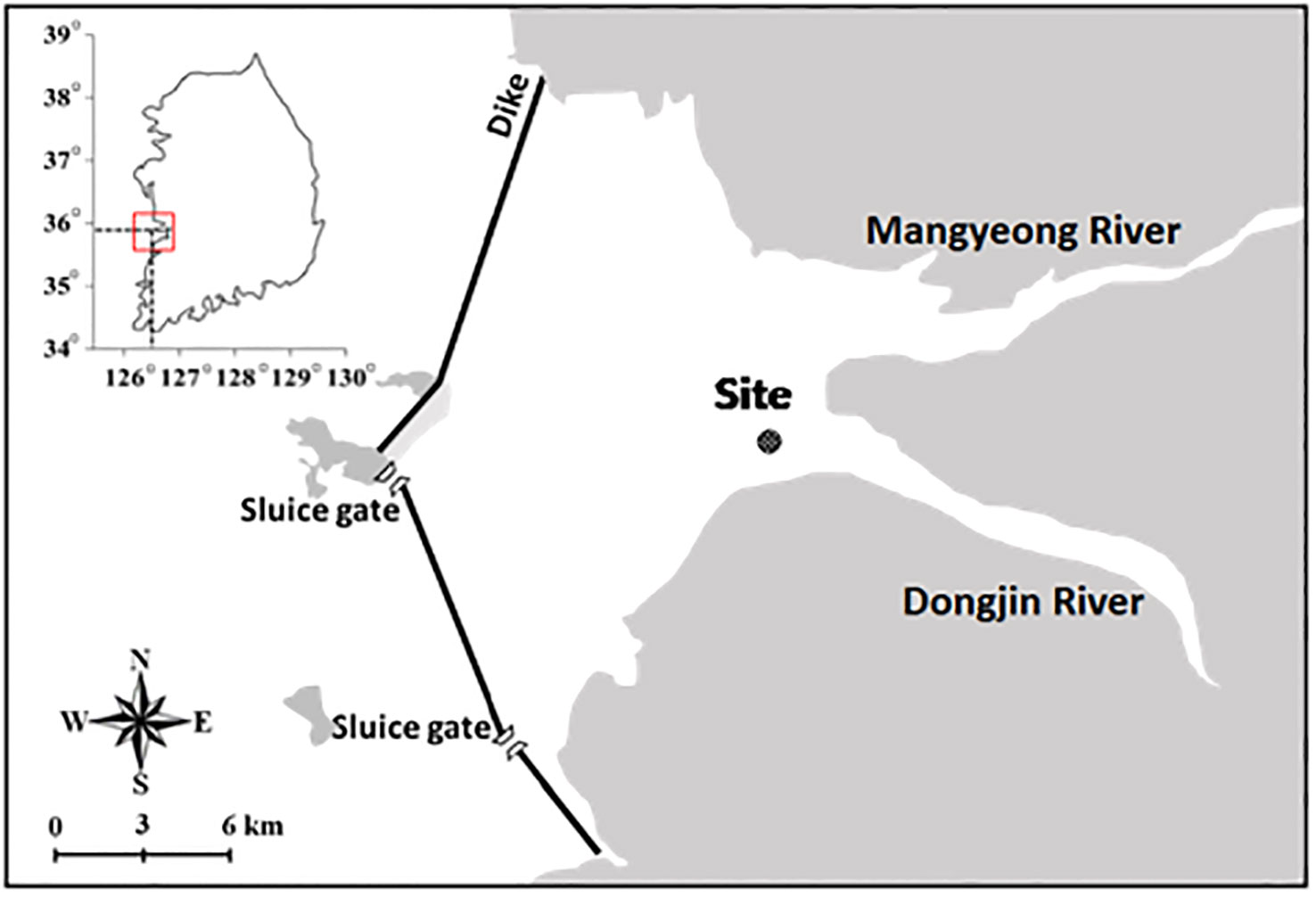
Figure 1 Study site in the Saemangeum Reservoir. (modified from Chae et al., 2021).
2.2 Water quality analysis
The water quality parameters were measured within the surface layer (0–0.5 m). Temperature (°C), pH, electrical conductivity (S/m), and salinity (‰) data were collected using a portable multi-item water quality meter (U-20; Horiba, Kyoto, Japan). Dissolved oxygen (mg/L, DO), chemical oxygen demand (mg/L, COD), chlorophyll-a (µg/L, chl-a), total nitrogen (mg/L, TN), and total phosphorus (mg/L, TP) were analysed in the laboratory following the Korean standards for water pollution process tests (ME, 2018) and marine environmental process tests (MOF, 2018).
2.3 Sample collection, pre-treatment, and dna extraction
Zooplankton samples were collected using a 100-μm mesh-sized Kitahara zooplankton net with a diameter of 0.3 m from the bottom to the surface until a sufficient number of individuals (approximately 3 to 5 times of vertical towing) were obtained in each season; February, May, August, and November in 2020. These samples were kept frozen at -20°C until further processing. Among the collected copepod species, S. tenellus, observed throughout the year at the study site, was selected as the target species for the gut content analysis. S. tenellus individuals were identified and sorted from zooplankton samples under a microscope (Leica S8 APO, Leica, Wetzlar, Germany), 10 individuals were randomly selected from sorted population of each season, except for summer (n=8) when the number of S. tenellus individuals was low (in total 38 individuals). Each specimen was stored separately in glass vials filled with 70% ethanol in a refrigerator to prevent cross-contamination. To remove unnecessary DNA surrounding individuals, S. tenellus samples were pre-treated with commercial bleach diluted to a final concentration of 2.5% for 2 minutes and washed twice with distilled water (Chae et al., 2021). Pre-treated samples were homogenised using a Bead Ruptor 12 (Omni International, Kennesaw, GA, USA), and genomic DNA of each individual was extracted using the DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany).
Simultaneously, raw water samples were collected on-site and used to analyze potential food sources for S. tenellus in the water and to validate the commercial bleach pre-treatment method aimed at removing unnecessary DNA that might be attached to the exoskeleton surface when analyzing the eaten food sources by S. tenellus. The collected raw water samples were filtered through a 20-μm mesh to remove suspended cells, aiming to extract only dissolved eDNA (Chae et al., 2021). Raw water samples for the analysis of potential food sources for S. tenellus were filtered through GF/F filter paper (0.45-μm pore size; Whatman, Maidstone, UK), with filtration volumes ranging from 500 mL to 1 L, depending on the concentration of particles in the water and consequent degree of pore clogging. For the validation of the pre-treatment method, 50 mL of the same raw water were treated with commercial bleach (final concentration of 2.5%) (we refer to this as the treated water), filtered through GF/F filter paper after 2 minutes, and then washed twice with distilled water to prevent the effect of continuous chemical treatment. Nuclease-free water (QIAGEN, Hilden, Germany) was also filtered through GF/F filter paper and used as a negative control. Genomic DNA was extracted from these samples using a DNeasy PowerWater Kit (QIAGEN, Hilden, Germany).
The DNA extraction processes for S. tenellus and the water samples were performed according to the manufacturer’s instructions, with only the reagent dose in the yield stage adjusted to 50 μL to increase the extracted gene concentration. Finally, DNA was obtained from seasonal raw water samples (n=4), seasonal treated water samples (n=4), and pre-treated S. tenellus individuals (total n=38 from four seasons) collected at the study site, as well as a negative control sample (n=4). The extracted DNA samples were frozen at -20°C. In the pre-treatment and DNA extraction processing of the samples, we used disposable gloves, instruments sterilised with commercial bleach, and ethanol to minimise contamination from the surrounding environment.
2.4 23S rRNA amplicon library generation
The genomic DNA from 10 individuals of S. tenellus was pooled from each season, except for summer (n=8). The extracted DNA for sequencing was prepared according to the Illumina 23S metagenomic sequencing library protocols (Illumina, San Diego, CA, USA). DNA quantity, quality, and integrity were measured using PicoGreen (Thermo Fisher Scientific, Waltham, MA, USA) and a VICTOR Nivo Multimode Microplate Reader (PerkinElmer, Waltham, MA, USA). Amplification was performed using AccuPower Hot Start PCR PreMix (Bioneer, Daejeon, Korea) with genomic DNA and primers in a final volume of 20 μL (Table 1). For DNA amplification, the 23S rRNA universal algal primer, capable of detecting various taxonomic groups of phytoplankton, such as diatoms, cyanobacteria, and green algae, was used (Yang et al., 2018). A gradient PCR was performed using a thermal cycler (T-100; Bio-Rad, Hercules, CA, USA).
First, amplification products were separated on 1.5% agarose gel by electrophoresis. Second, to produce an indexing PCR, the first PCR products were amplified under the same conditions. The final products were normalised and pooled using PicoGreen, and the size of libraries was verified using the LabChip GX HT DNA High Sensitivity Kit (PerkinElmer, Waltham, MA, USA). NGS analysis, including index PCR, was completed by Macrogen Inc. (Seoul, Korea). Sequencing libraries were prepared by random fragmentation of the DNA sample, followed by 5’ and 3’ adapter ligation. Alternatively, ‘tagmentation’ combined the fragmentation and ligation reactions into a single step that greatly increased the efficiency of the library preparation process. The adapter-ligated fragments were then PCR-amplified and gel-purified. PCR products were sequenced using the MiSeq™ platform (Illumina, San Diego, CA, USA) from a commercial service (Macrogen Inc.).
2.5 Data analysis
The detected organisms, excluding phytoplankton but including Cyanobacteria, were eliminated. Species with a total operational taxonomic unit (OTU) of less than 100 for the entire sample were eliminated due to the high probability of false positives. Raw reads were trimmed using CD-HIT-OUT software, and chimeras were identified and removed using rDNATools. Fast length adjustment of short reads (FLASH v.1.2.11) was used for paired-end merging, and the reads were processed and clustered into OTUs using the bioinformatics algorithm UCLUST at a 97% OTU cut-off value. Taxonomy was assigned to the representative sequences obtained using BLAST and UCLUST. We classified each OTU according to the identity percentage (%): species level ≥ 97%, genus level ≥ 90%, and family level; ≥ 84% reads with less than 84% identity were excluded. OTUs detected in commercial bleach treated water samples were treated following the experimental protocols of Oh et al. (2020) and Chae et al. (2021).
Species detected in the environmental DNA analysis of water samples were considered to be potential food sources of S. tenellus, and the gut contents of copepod samples were determined to be eaten by S. tenellus. We analysed the alpha diversity indices (Species richness, Chao-1, Shannon’s diversity, Simpson’s evenness, and Simpson’s dominance) of the potential and eaten food sources and confirmed the core taxa using a Venn diagram for each season (with 0.001 detection probability of at least 90% of samples and 0.75 prevalence). Redundancy analysis (RDA) was performed to identify changes in the phytoplankton community and potential food sources for S. tenellus according to environmental factors. These analyses were performed using the R package “Microbiome” in R studio (v.4.1.1) using the remaining OTUs after undergoing the OTU data processing steps (Lahti and Shetty, 2018). The feeding selectivity index (Ei) was calculated for species that were equally detected as potential food sources and eaten in the same season (Ivlev, 1961).
Feeding selectivity
where ri is the relative proportion of organism i in consumed food sources; pi is the relative proportion of i in potential food sources; Ei represents a value between -1 and 1. When the value is 0, it corresponds to random feeding; a value closer to 1 indicates a higher preference for feeding. If the ratio of reads present in the gut of S. tenellus is higher than that in potential food sources, it can be determined that the food is selectively eaten. The feeding preferences of S. tenellus were determined by identifying phytoplankton species that were simultaneously detected as both potential food sources and eaten food sources by S. tenellus within the same season.
3 Result
3.1 Water quality and food environment of study sites
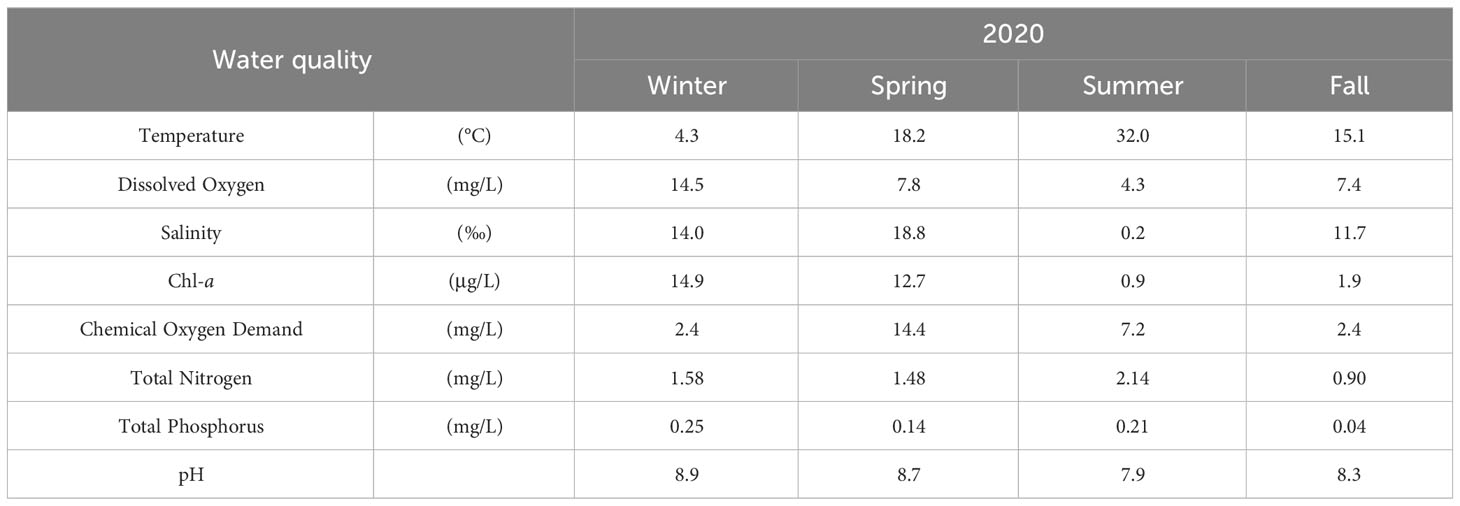
The factors showed different patterns according to the season. In summer, the water temperature increased to 32°C, and DO showed the lowest value. Salinity ranged from 11 to 19 ‰, indicating brackish water; however, in summer it had a very low value of 0.2 ‰, seemingly strongly affected by the inflow of freshwater during the rainy season. Chl-a increased from winter to spring compared to that in summer and fall, whereas nutrients, including COD, TN, and TP, increased in the summer and decreased in the fall (Table 2).
3.2 Comparison of phytoplankton communities in potential and eaten food sources of Sinocalanus tenellus
Species detected in raw water samples were considered to be potential food sources for S. tenellus. Amplicon sequencing of these species produced an average of 58,931 ± 11,064 reads per sample. A total of 16 high-abundance OTUs were identified after the removal of non-phytoplankton or less than unique 100 OTUs of the detected organisms. The final number of reads obtained was 21,965 ± 18,571 (Table S1). As the seasons change, the habitat and the diversity of potential food sources of S. tenellus show a different trend. In winter and spring, the number and abundance were the same; the diversity was higher in spring. The number of observed species increased in summer, indicating the highest diversity among the four seasons. Fewer species were detected in fall than in winter, but the diversity was higher (Table 3A).
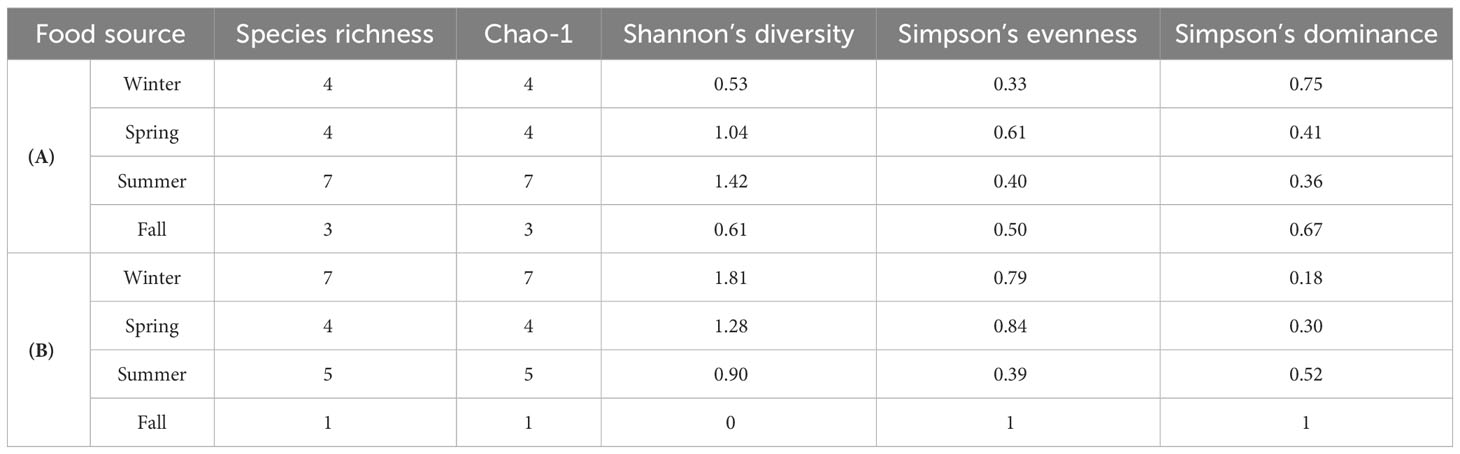
Table 3 Alpha-diversity of the phytoplankton communities of potential food sources (A) and eaten food sources (B) of S. tenellus at each season.
The gut contents of S. tenellus individuals were identified as the eaten food source, and amplicon sequencing produced an average of 75,253 ± 27,913 reads per sample. Eighteen high-abundance OTUs were identified after the removal of non-phytoplankton or less than unique 100 OTUs of the detected organisms, and the final number of reads obtained was 7,904 ± 9,435 (Table S2). For consumed food sources, the alpha diversity index was markedly higher in winter than in other seasons. In spring and summer, the number of species was similar, but spring showed a higher level of diversity, while diversity decreased from winter to fall and only one species was detected. In the fall, the diversity was calculated to be 0, and evenness and dominance were 1 (Table 3B).
A comparison of the relative abundance, based on the OTUs of classes of the detected species in each sample, revealed different trends, not only in seasons but also between potential food sources and food eaten by S. tenellus. In winter, Bacillariophyceae dominated the potential food sources, accounting for a relatively small percentage of eaten food. In the case of spring, Cryptophyceae and Prymnesiophyceae, which were identified as potential food sources, were not detected in the eaten food, while Trebouxiophyceae represented 31.6% of eaten food. In addition, Cyanobacteria, which are sub-dominant as potential food sources, accounted for a low percentage of eaten food, while Bacillariophyceae accounted for the second largest proportion. In summer, Cyanobacteria was dominant in both samples, while Bacillariophyceae accounted for a small percentage of eaten food compared to potential food sources. Cryptophyceae, Eustigmatophyceae, and Trebouxiophyceae were detected only in potential food sources. In the fall, Cyanobacteria dominated in both samples, while in eaten food, only one Cyanobacteria species was detected. Trebouxiophyceae, which were identified in potential food sources, were not detected in the eaten food (Figure 2).
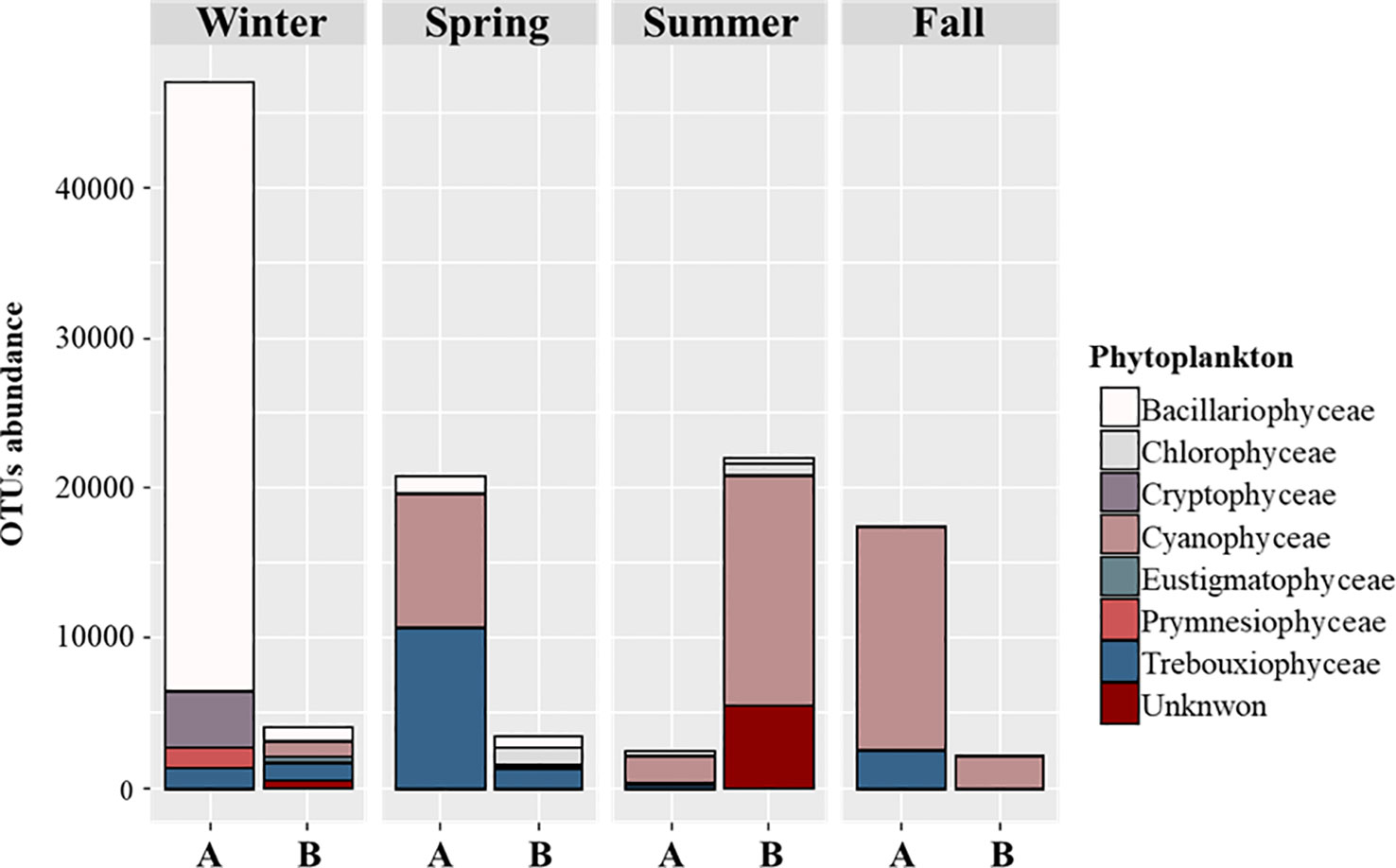
Figure 2 OTU abundance of phytoplankton detected in potential food sources (A) and eaten food sources (B) of S. tenellus in each season.
Chlorophyta and Cyanobacteria (Geminella minor and Synechococcus spp.) are generally dominant as potential food sources, except in winter, during which, with little precipitation, Bacillariophyta (Actinocyclus subtilis) accounted for more than 85% of the total. G. minor was detected in all the seasons, while A. subtilis and Synechococcus spp. were detected continuously, except in the fall and winter, respectively (Table S1, Figure 3A). In addition, Cyanobium sp., Synechococcus rubescens, Cryptophyta sp., Chroomonas sp., Microchloropsis salina, Isochrysis galbana, genus Chroococcidiopsis, and order Pleurocapsales were analysed as potential food sources.
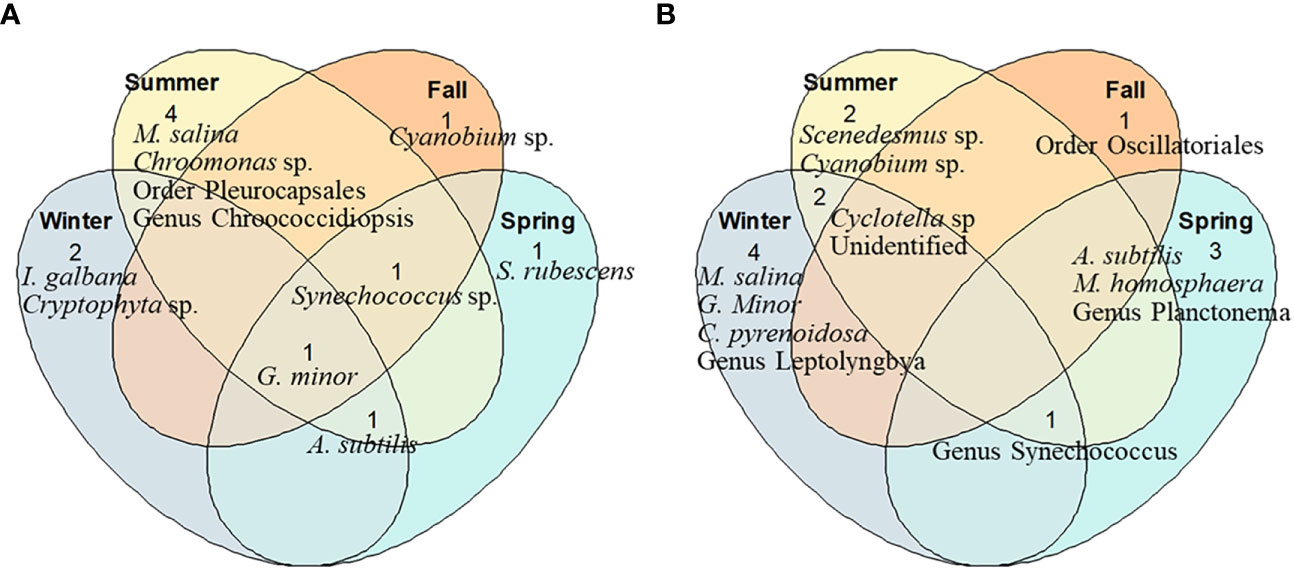
Figure 3 Venn diagrams comparing the phytoplankton species composition detected in potential food sources (A) and eaten food sources (B) of S. tenellus at each season (C, Chlorophyta: Geminella minor, Mychonastes homosphaera, Chlorella pyrenoidosa, Scenedesmus sp., Genus Planctonema; Cy, Cyanobacteria: Synechococcus rubescens, Cyanobium sp., Synechococcus spp., Genus Leptolyngbya, Genus Chroococcidiopsis, Order Oscillatoriales, Order Pleurocapsales; Cr, Crytophyta: Cryptophyta sp., Chroomonas sp.; O, Ochrophyta: Microchloropsis salina; B, Bacillariophyta: Actinocyclus subtilis, Cyclotella sp.; H, Haptista: Isochrysis galbana).
The composition of phytoplankton species detected differed according to the season. Chlorophyta occupied the highest proportion in winter (Chlorella pyrenoidosa, Mychonastes homosphaera) and spring (genus Planctonema). In contrast, Cyanobacteria were predominant in summer (Synechococcus spp.) and fall (order Oscillatoriales). Synechococcus spp., which were dominant in summer, were detected in all seasons except for fall. Cyclotella sp. and unidentified eukaryotic phytoplankton were detected in both summer and winter (Table S2, Figure 3B).
3.3 Feeding selectivity index (Ei) of Sinocalanus tenellus
One species was identified in winter and summer and two were identified in spring. As a result of calculating the feeding selectivity index (Ei) using these proportions, G. minor and A. subtilis showed a value >0, indicating that they were selectively eaten. Conversely, the index of Synechococcus spp. was <0 in spring but ≈0 in summer. A value close to -1 indicates intentional avoidance of feeding and a value ≈0 indicates random consumption. The seasonal variation in Ei is due to the dominance of potential food sources in summer (Table 4).
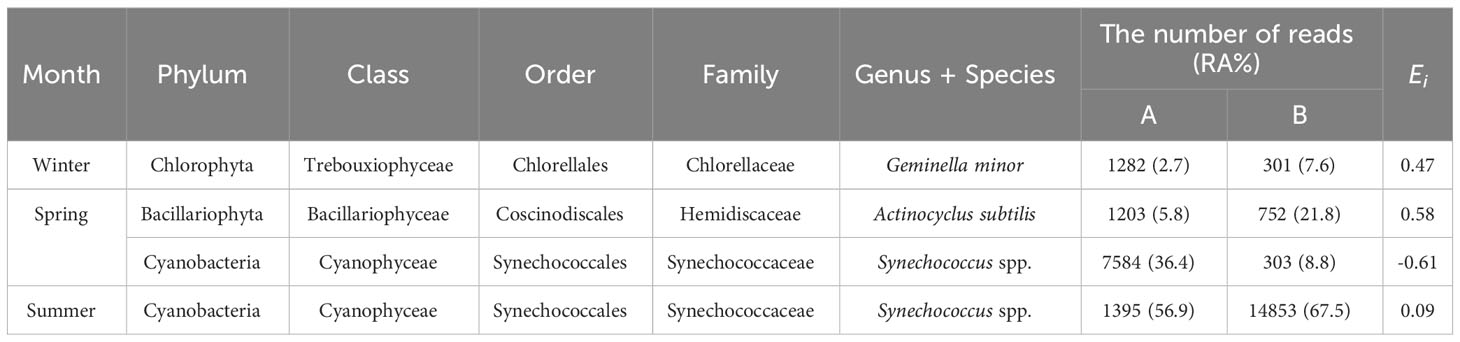
Table 4 Results of the feeding selectivity index (Ei) calculations of Sinocalanus tenellus for phytoplankton species detected as both potential food sources (A) and eaten food sources (B) for S. tenellus in each season: the number of reads and their relative abundance (RA%) within the potential and eaten food source pools are indicated.
3.4 Correlation between detected phytoplankton communities and environmental factors
For the potential food sources of S. tenellus in the study site, the RDA 1 axis explained 85.22% and the RDA 2 axis explained 11.48% of the total variation. Based on the RDA 1 axis, Bacillariophyceae and Cyanobacteria tended to increase and decrease, respectively, as TP increased. In terms of RDA 2 axis, there was tendency for Trebouxiophyceae to increase with an increase in chl-a concentration (Figure 4A; Table 5A).
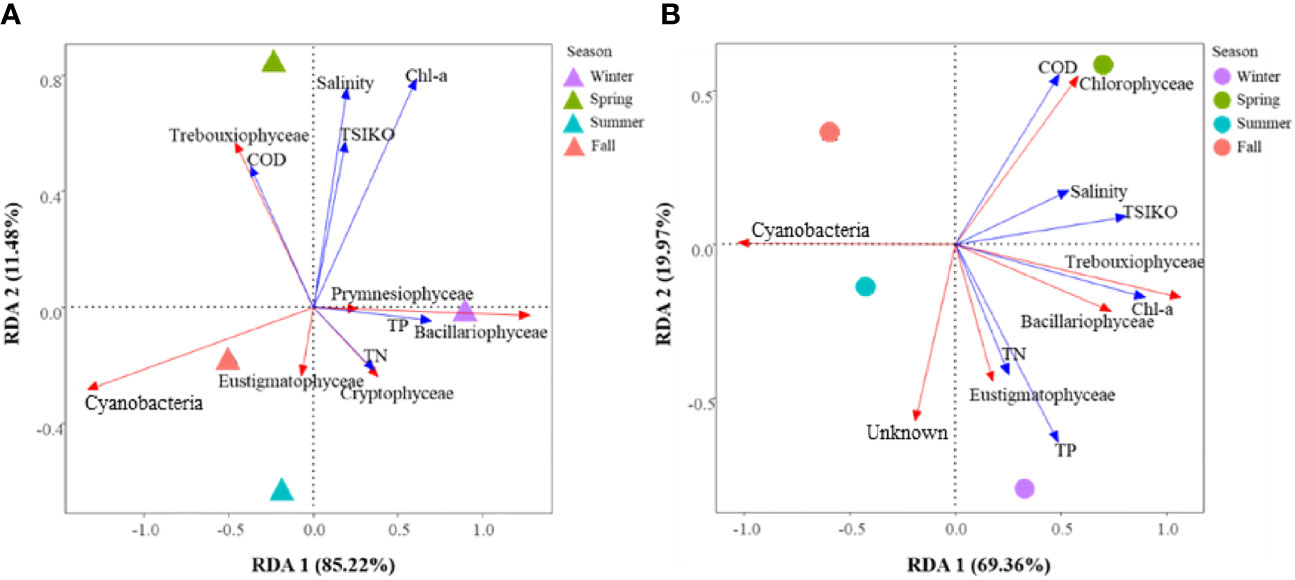
Figure 4 RDA of water quality and phytoplankton communities in potential food sources (A) and eaten food sources (B) of S. tenellus at the study site.
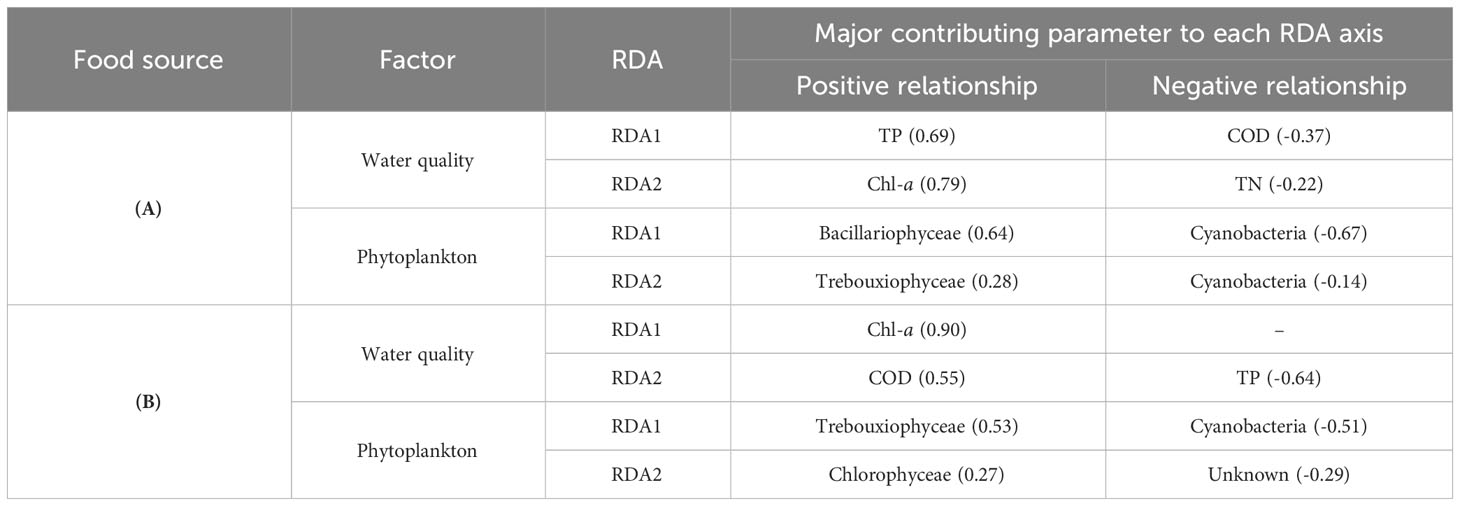
Table 5 Summary of the RDA of water quality and phytoplankton communities in potential food sources (A) and eaten food sources (B) of S. tenellus at the study site focusing on the major contributing variables to each RDA axis.
For the eaten food sources of S. tenellus, the RDA 1 axis explained 69.36% and the RDA 2 axis explained 19.97% of the total variation. Based on the RDA 1 axis, Trebouxiophyceae increased with increasing chl-a, while Cyanobacteria decreased. COD and TP were extracted as relatively high contributing parameters to the RDA 2 axis, but did not show a clear phytoplankton community response (Figure 4B; Table 5B).
When viewed in terms of RDA 1 axes, which had a high explanatory power for the total variation in both potential and eaten food sources of S. tenellus, there was a relatively high similarity in phytoplankton composition between summer and fall compared to other seasons. Cyanobacteria appeared to be a major contributor to this similarity, while no distinct common patterns were observed in other phytoplankton classification and water quality variables. Considering both RDA axes, during the high TN and TP winter season, Bacillariophyceae were predominant as potential food sources, whereas Eustigmatophyceae showed a significant contribution in eaten food sources. In the case of the high COD spring season, Trebouxiophyceae were found to be dominant among potential food sources, while Chlorophyceae played a significant role among eaten food sources (Figure 4; Table 5).
4 Discussion
In this study, we applied meta-barcoding technique to the raw water of S. tenellus’ habitat and their individuals to identify potential and eaten food sources focusing on phytoplankton community at a high resolution. In the case of potential food sources, G. minor, which dominated in spring, was a common freshwater species, except in summer (Satpati et al., 2016). In addition, Synechococcus spp., which dominate in summer, inhabit brackish water or seawater and prefer a nutrient-rich, warm environment and euphotic zones where sufficient light is transmitted for photosynthesis (Partensky et al., 1999; Six et al., 2021). This is because the study site is where freshwater flows into the reservoir from the Dongjin River and mixes with seawater. A. subtilis, which dominates in winter, is a Bacillariophyta commonly found in marine ecosystems and generally has the highest biomass in late winter and early spring (Barone and Naselli-Flores, 2003). This consistency increased the reliability of the meta-barcoding analysis results.
Consequently, we found that in formation on the potential and eaten sources of S. tenellus obtained through DNA analysis was useful for analysing their feeding selectivity. To date, studies on the feeding characteristics of S. tenellus have been conducted under limited food and environmental conditions (Hada and Uye, 1991); therefore, it is difficult to represent a real environment where several factors act simultaneously. Here, we can understand the actual ecological interactions in the ecosystem through meta-barcoding of the gut contents of individuals that respond directly to changes in habitat and feeding environments. Furthermore, this research approach can be utilized as valuable foundational data amidst the ongoing need for studying the qualitative and quantitative characteristics of zooplankton within aquatic ecosystems (Goździejewska et al., 2010; Majeed et al., 2022).
When a high proportion of OTU reads occur in potential food sources but are lacking in a low proportion as eaten food sources, it can be considered that S. tenellus has a low preference for that food. On the other hand, the inverse situation—low reads of potential food sources and a high proportion of these in the diet—would imply a high preference for this food source. S. tenellus showed a low food preference for Bacillariophyceae and Trebouxiophyceae, except in spring and winter, respectively, while Cryptophyceae and Prymnesiophyceae were not consumed by them. In winter, Chlorophyceae and Cyanobacteria were detected only as eaten food sources of S. tenellus; therefore, they were considered to have been selectively consumed. However, S. tenellus’ preference for Cyanobacteria changed according to season, showing low preference in spring and randomly consumption in summer and fall, as analysed. Cyanobacteria, which dominate colonies in freshwater, have been reported as inefficient food sources for copepods because they produce toxic substances and have low nutrient value (Carmichael, 1992; Müller-Navarra et al., 2000; Müller-Navarra, 2008). Nevertheless, our results suggest that when Cyanobacteria constitute a high proportion of the potential food source, S. tenellus consume low-quality food sources, albeit involuntarily. In previous studies, the genus Synechococcus, a marine unicellular cyanobacteria, was detected in the gut contents of copepods in seawater (Ho et al., 2017; Chen et al., 2018; Yeh et al., 2020). However, it is difficult to ascertain whether the copepods selectively fed on them, as they are known to produce toxic substances (Jakubowska and Szeląg-Wasielewska, 2015) and are very small (6~1.6 μm). In the case of Bacillariophyceae, despite they being commonly known as a typical food source for copepods (Kruse et al., 2009; Russo et al., 2019), the results of this study did not show a high consumption preference for them by S. tenellus. According to the literature, some species of Bacillariophyceae can have negative effects on copepod egg formation and hatching due to their toxicity, and there are research findings indicating insufficient nutrients like fatty acids (Miralto et al., 1999; Carotenuto et al., 2002; Ianora et al., 2004). Accordingly, we confirmed that S. tenellus does not feed on all surrounding food sources but can change its food source depending on the presence or absence of a preferred food source.
In this study, there were some phytoplankton species that were not detected in the eDNA analysis of the potential food pool from raw water but were detected as eaten food by S. tenellus (Table S1; S2). These results suggest that these species were present at very low concentrations in the water and were not detected as potential food sources due to insufficient raw water filtration volume for eDNA extraction. S. tenellus, on the other hand, continuously filters the surrounding water during its feeding activity, and during this process, it accumulates substances present in the surrounding water. For species whose proportion in the potential food pool was not specified, it is difficult to analyse whether S. tenellus feeding was intentional or the result of random feeding using the feeding selectivity index. Therefore, further studies should be conducted to analyse the feeding selectivity of S. tenellus for phytoplankton species detected only in the eaten food pool, using technical enhancements such as maximizing the amount of raw water filtration or applying primers with high specificity for phytoplankton taxa.
Since copepods have diverse species-specific feeding characteristics, long-term analysis of more diverse samples, using specific primers for the target taxa, is required to understand the complex interactions in the entire food web. In this study, we analysed phytoplankton as food sources for S. tenellus using 23S rRNA universal algal primer, which, among primers targeting various regions (23S rRNA, 18S rRNA, PC-IGS) of phytoplankton, allowed the detection of several taxa including Cyanobacteria through preliminary testing. Nevertheless, this primer has a low detection resolution for dinoflagellates (Howe et al., 2008; Hancock et al., 2021), therefore we were unable to analyse the feeding selectivity of S. tenellus for dinoflagellates in this study. Using a primer that can detect more universal or supplementary information will help clarify the food sources of S. tenellus because dinoflagellates appear in many brackish environments and can be a food source for copepod species. In addition, as 23S rRNA universal alga primer has a wide detection range as long as allows for the detection of Cyanobacteria, several bacterial species were detected in addition to Cyanobacteria and had to be excluded during our OTU data processing. The bacterial species detected in the gut contents of S. tenellus cannot be considered intentionally ingested due to their feeding habits, which is mainly to feed on phytoplankton, and can be assumed to be unintentionally introduced from the surrounding water or gut microbiota (Chae et al., 2021). Based on the results of this study, predation experiments in limited environments would provide more reliable results on zooplankton feeding selectivity based on meta-barcoding. In addition, although this study pooled DNA from S. tenellus individuals by season and focused on seasonal changes in their feeding selectivity, an individual-specific approach would provide more statistically significant results and allow for discussion of differences between individuals within the same species.
It is concluded that meta-barcoding of the gut contents of small- and medium-sized copepod species, such as S. tenellus, is an effective method for identifying their food sources and understanding the biological interactions in the food web. It can be used in methodological studies of the food sources and feeding characteristics of zooplankton species of which little is known.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical review and approval was not required for the study on animals in accordance with the local legislation and institutional requirements.
Author contributions
Conceptualization, H-JO and K-HC; methodology, H-JO, K-HC and HJ; validation, K-HC and HJ; formal analysis, Y-JC, H-JO and HJ; investigation, Y-JC, H-JO and K-HC; resources, Y-JC, H-JO and K-HC; data curation, Y-JC, H-JO and HJ; writing original draft preparation, Y-JC, H-JO and K-HC; writing—review and editing, Y-JC, H-JO, K-HC, and HJ; visualization, Y-JC and H-JO; supervision, K-HC, I-SK and HJ; project administration, I-SK; funding acquisition, HJ. All authors contributed to the article and approved the submitted version.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Research Foundation of Korea (grant number NRF- 2020R1C1C1009066).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1234754/full#supplementary-material
References
Asami H. (2004). Early life ecology of Japanese smelt (Hypomesus nipponensis) in Lake Abashiri, a brackish water, eastern Hokkaido, Japan. Sci. Rep. Hokkaido Fish. Exp. Stn. (Japan) 67. Availabe at: https://www.hro.or.jp/list/fisheries/marine/o7u1kr0000000xuh.html
Atkinson A. (1995). Omnivory and feeding selectivity in five copepod species during spring in the Bellingshausen Sea, Antarctica. ICES J. Mar. Sci. 52 (3-4), 385–396. doi: 10.1016/1054-3139(95)80054-9
Barone R., Naselli-Flores L. (2003). Distribution and seasonal dynamics of Cryptomonads in Sicilian water bodies. Hydrobiologia 502 (1), 325–329. doi: 10.1023/B:HYDR.0000004290.22289.c2
Bollens G. C. R., Penry D. L. (2003). Feeding dynamics of Acartia spp. copepods in a large, temperate estuary (San Francisco Bay, CA). Mar. Ecol. Prog. Ser. 257, 139–158. doi: 10.3354/meps257139
Boltizár O., Müller T., Urbányi B., Csenki Z., Bakos K., Staszny Á., et al. (2017). Predatory effect of Copepods on the larvae of some freshwater fish. Fundam. Appl. Limnol. 190 (4), 349–356. doi: 10.1127/fal/2017/0682
Carmichael W. W. (1992). Cyanobacteria secondary metabolites-the cyanotoxins. J. Appl. Bacteriol. 72 (6), 445–459. doi: 10.1111/j.1365-2672.1992.tb01858.x
Carotenuto Y., Ianora A., Buttino I., ROmano G., Miralto A. (2002). Is postembryonic development in the copepod Temora stylifera negatively affected by diatom diets? J. Exp. Mar. Biol. Ecol. 276 (1-2), 49–66. doi: 10.1016/S0022-0981(02)00237-X
Chae Y. J., Oh H. J., Chang K. H., Kwak I. S., Jo H. (2021). Application of Next-Generation Sequencing for the Determination of the Bacterial Community in the Gut Contents of Brackish Copepod Species (Acartia hudsonica, Sinocalanus tenellus, and Pseudodiaptomus inopinus). Animals 11 (2), 542. doi: 10.3390/ani11020542
Chang K. H., Sakamoto M., Ha J. Y., Miyabara Y., Nakano S. I., Doi H., et al. (2011). Response of the plankton community to herbicide application (triazine herbicide, simetryn) in a eutrophicated system: short-term exposure experiment using microcosms. Limnology 12 (1), 11–16. doi: 10.1007/s10201-010-0317-1
Chen M., Kim D., Liu H., Kang C. K. (2018). Variability in copepod trophic levels and feeding selectivity based on stable isotope analysis in Gwangyang Bay of the southern coast of the Korean Peninsula. Biogeosciences 15 (7), 2055–2073. doi: 10.5194/bg-15-2055-2018
Cleary A. C., Søreide J. E., Freese D., Niehoff B., Gabrielsen T. M. (2017). Feeding by Calanus glacialis in a high arctic fjord: potential seasonal importance of alternative prey. ICES J. Mar. Sci. 74 (7), 1937–1946. doi: 10.1093/icesjms/fsx106
Dam H. G., Baumann H. (2017). Climate change, zooplankton and fisheries. Clim. Change Impacts Fish. Aquac. 2, 851–874. doi: 10.1002/9781119154051.ch25
Furgała-Selezniow G., Skrzypczak A., Kucharczyk D., Kujawa R., Mamcarz A., Żarski D., et al. (2014). Food selection of burbot (Lota lota L.) larvae reared in illuminated net cages in mesotrophic Lake Maróz (north-eastern Poland). Aquac. Int. 22, 41–52. doi: 10.1007/s10499-013-9707-9
Gabrielsen T. M., Merkel B., Søreide J. E., Johansson-Karlsson E., Bailey A., Vogedes D., et al. (2012). Potential misidentifications of two climate indicator species of the marine arctic ecosystem: Calanus glacialis and C. finmarchicus. Polar. Biol. 35 (11), 1621–1628. doi: 10.1007/s00300-012-1202-7
Goździejewska A., Skrzypczak A., Furgała-Selezniow G., Koszałka J., Mamcarz A. (2010). Zooplankton in the Nida River (the upper Wkra river) subjected to revitalization treatments. Pol. J. Natur. Sc 25 (4), 387–400. doi: 10.2478/v10020-010-0036-4
Greenstone M. H., Weber D. C., Coudron T. A., Payton M. E., Hu J. S. (2012). Removing external DNA contamination from arthropod predators destined for molecular gut-content analysis. Mol. Ecol. Resour. 12 (3), 464–469. doi: 10.1111/j.1755-0998.2012.03112.x
Hada A., Uye S. I. (1991). Cannibalistic feeding behavior of the brackish-water copepod Sinocalanus tenellus. J. Plankton Res. 13 (1), 155–166. doi: 10.1093/plankt/13.1.155
Hancock T. L., Blonder S. L., Bury A. A., Smolinski R. A., Parsons M. L., Robertson A., et al. (2021). Succession pattern and phylotype analysis of microphytobenthic communities in a simulated oil spill seagrass mesocosm experiment. Sci. Total. Environ. 784, 147053. doi: 10.1016/j.scitotenv.2021.147053
Ho T. W., Hwang J. S., Cheung M. K., Kwan H. S., Wong C. K. (2017). DNA-based study of the diet of the marine calanoid copepod Calanus sinicus. J. Exp. Mar. Biol. Ecol. 494, 1–9. doi: 10.1016/j.jembe.2017.04.004
Howe C. J., Nisbet R. E. R., Barbrook A. C. (2008). The remarkable chloroplast genome of dinoflagellates. J. Exp. Bot. 59 (5), 1035–1045. doi: 10.1093/jxb/erm292
Ianora A., Miralto A., Poulet S. A., Carotenuto Y., Buttino I., ROmano G., et al. (2004). Aldehyde suppression of copepod recruitment in blooms of a ubiquitous planktonic diatom. Nature 429 (6990), 403–407. doi: 10.1038/nature02526
Ivlev V. S. (1961). Experimental ecology of the feeding of fishes (New Haven: Yale University Press), 41–115.
Jakubowska N., Szeląg-Wasielewska E. (2015). Toxic picoplanktonic cyanobacteria. Mar. Drugs 13 (3), 1497–1518. doi: 10.3390/md13031497
Jo H., Choi B., Park K., Kim W. S., Kwak I. S. (2020). First gut content analysis of 4th instar midge larvae (Diptera: Chronomidae) in large-scale weirs using a DNA meta-barcoding approach. Int. J. Environ. Res. Public Health 17 (8), 2856. doi: 10.3390/ijerph17082856
Kang Y. S., Kim S. W. (2003). Brackish lakes in Shinpo District, North Korea. I. Zooplankton. J. Ecol. Environ. 36 (2), 215–220.
Kim K. S., Lee J. B., Lee K. S., Kang J. W., Yoo H. B. (2000). Ecological study of copepoda community in the lower Seomjin River system, Korea. J. Ecol. Environ. 33 (2), 176–186.
Kruk M., Paturej E., Obolewski K. (2021). Zooplankton predator–prey network relationships indicates the saline gradient of coastal lakes. Machine learning and meta-network approach. Ecol. Indic. 125, 107550. doi: 10.1016/j.ecolind.2021.107550
Kruse S., Jansen S., Krägefsky S., Bathmann U. (2009). Gut content analyses of three dominant Antarctic copepod species during an induced phytoplankton bloom EIFEX (European iron fertilization experiment). Mar. Ecol. 30 (3), 301–312. doi: 10.1111/j.1439-0485.2009.00284.x
Kuo J., Chen C. Y., Han C. C., Ju Y. M., Tew K. S. (2021). Analyses of diet preference of larval orange-spotted grouper (Epinephelus coioides) grown under inorganic fertilization method using next-generation sequencing. Aquaculture 531, 735916. doi: 10.1016/j.aquaculture.2020.735916
Lahti L., Shetty S. (2018) Introduction to the microbiome R package. Available at: http://microbiome.github.com/microbiome.
Lee M. C., Choi H., Park J. C., Yoon D. S., Lee Y., Hagiwara A., et al. (2020). A comparative study of food selectivity of the benthic copepod Tigriopus japonicus and the pelagic copepod Paracyclopina nana: a genome-wide identification of fatty acid conversion genes and nitrogen isotope investigation. Aquaculture 521, 734930. doi: 10.1016/j.aquaculture.2020.734930
Majeed O. S., Nashaat M. R., Al-Azawi A. J. (2022). “Physicochemical parameters of river water and their relation to zooplankton: A review,” in IOP Conference Series: Earth and Environmental Science. (England: IOP Publishing), Vol. 1120. 012040.
Mäkinen K., Vuorinen I., Hänninen J. (2017). Climate-induced hydrography change favours small-bodied zooplankton in a coastal ecosystem. Hydrobiologia 792, 83–96. doi: 10.1007/s10750-016-3046-6
ME, Ministry of Environment (2018). Water pollution process standard (Sejong, Korea: Ministry of Environment).
Miralto A., Barone G., ROmano G., Poulet S. A., Ianora A., Russo G. L., et al. (1999). The insidious effect of diatoms on copepod reproduction. Nature 402 (6758), 173–176. doi: 10.1038/46023
MOF, Ministry of Oceans and Fisheries (2018). Marine environmental process standards (Sejong, Korea: Ministry of Oceans and Fisheries), 1–258.
Müller-Navarra D. C. (2008). Food web paradigms: the biochemical view on trophic interactions. Int. Rev. Hydrobiol. 93 (4-5), 489–505. doi: 10.1002/iroh.200711046
Müller-Navarra D. C., Brett M. T., Liston A. M., Goldman C. R. (2000). A highly unsaturated fatty acid predicts carbon transfer between primary producers and consumers. Nature 403, 74–77. doi: 10.1038/47469
Narwani A., Mazumder A. (2012). Bottom-up effects of species diversity on the functioning and stability of food webs. J. Anim. Ecol. 81 (3), 701–713. doi: 10.1111/j.1365-2656.2011.01949.x
Oda Y., Nakano S., Suh J. M., Oh H. J., Jin M. Y., Kim Y. J., et al. (2018). Spatiotemporal variability in a copepod community associated with fluctuations in salinity and trophic state in an artificial brackish reservoir at Saemangeum, South Korea. PloS One 13 (12), e0209403. doi: 10.1371/journal.pone.0209403
Oh H. J., Krogh P. H., Jeong H. G., Joo G. J., Kwak I. S., Hwang S. J., et al. (2020). Pretreatment method for DNA barcoding to analyze gut contents of rotifers. Appl. Sci. 10 (3), 1064. doi: 10.3390/app10031064
Olson M. B., Lessard E. J., Wong C. H. J., Bernhardt M. J. (2006). Copepod feeding selectivity on microplankton, including the toxigenic diatoms Pseudo-nitzschia spp., in the coastal Pacific Northwest. Mar. Ecol. Prog. Ser. 326, 207–220. doi: 10.3354/meps326207
Partensky F., Blanchot J., Vaulot D. (1999). Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: a review. Bull. Inst. Oceanogr. 19, 457–475.
Polis G. A., Anderson W. B., Holt R. D. (1997). Toward an integration of landscape and food web ecology: the dynamics of spatially subsidized food webs. AnnuRev. Ecol. Syst. 28 (1), 289–316. doi: 10.1146/annurev.ecolsys.28.1.289
Pompanon F., Deagle B. E., Symondson W. O., Brown D. S., Jarman S. N., Taberlet P. (2012). Who is eating what: diet assessment using next generation sequencing. Mol. Ecol. 21 (8), 1931–1950. doi: 10.1111/j.1365-294X.2011.05403.x
Russo E., Ianora A., Carotenuto Y. (2019). Re-shaping marine plankton communities: Effects of diatom oxylipins on copepods and beyond. Mar. Biol. 166, 9. doi: 10.1007/s00227-018-3456-2
Satpati G. G., Pal R., Pal R. (2016). New and rare records of filamentous green algae from Indian Sundarbans Biosphere Reserve. J. Algal Biomass Util. 7 (2), 159–175.
Six C., Ratin M., Marie D., Corre E. (2021). Marine Synechococcus picocyanobacteria: Light utilization across latitudes. Proc. Natl. Acad. Sci. 118 (38), e2111300118. doi: 10.1073/pnas.2111300118
Soulié T., Engström-Öst J., Glippa O. (2022). Copepod oxygen consumption along a salinity gradient. Mar. Freshw. Behav. Physiol. 55 (5–6), 107–119. doi: 10.1080/10236244.2022.2104720
Su M., Liu H., Liang X., Gui L., Zhang J. (2018). Dietary analysis of marine fish species: enhancing the detection of prey-specific DNA sequences via high-throughput sequencing using blocking primers. Estuaries Coasts 41 (2), 560–571. doi: 10.1007/s12237-017-0279-1
Thébault E., Loreau M. (2005). Trophic interactions and the relationship between species diversity and ecosystem stability. Am. Nat. 166 (4), E95–E114. doi: 10.1086/444403
Turner J. T. (2004). The importance of small planktonic copepods and their roles in pelagic marine food webs. Zool. Stud. 43 (2), 255–266.
Uye S., Shimazu T., Yamamuro M., Ishitobi Y., Kamiya H. (2000). Geographical and seasonal variations in mesozooplankton abundance and biomass in relation to environmental parameters in Lake Shinji–Ohashi River–Lake Nakaumi brackish-water system, Japan. J. Mar. Syst. 26 (2), 193–207. doi: 10.1016/S0924-7963(00)00054-3
Vacher C., Tamaddoni-Nezhad A., Kamenova S., Peyrard N., Moalic Y., Sabbadin R., et al. (2016). Learning ecological networks from next-generation sequencing data. Adv. Ecol. Res. 54, 1–39. doi: 10.1016/bs.aecr.2015.10.004
Vasquez A. A., Mohiddin O., Li Z., Bonnici B. L., Gurdziel K., Ram J. L. (2021). Molecular diet studies of water mites reveal prey biodiversity. PloS One 16 (7), e0254598. doi: 10.1371/journal.pone.0254598
Yang J., Jiang H., Liu W., Wang B. (2018). Benthic algal community structures and their response to geographic distance and environmental variables in the Qinghai-Tibetan lakes with different salinity. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00578
Keywords: brackish reservoir, gut contents DNA, next-generation sequencing, feeding selectivity index, feeding characteristics
Citation: Chae Y-J, Oh H-J, Kwak I-S, Chang K-H and Jo H (2023) A study of the feeding characteristics of a small and medium-sized copepod species (Sinocalanus tenellus) using genetic analysis techniques: seasonal comparison of potential/eaten food sources focused on phytoplankton. Front. Mar. Sci. 10:1234754. doi: 10.3389/fmars.2023.1234754
Received: 13 June 2023; Accepted: 22 August 2023;
Published: 12 September 2023.
Edited by:
Valerio Matozzo, University of Padua, ItalyReviewed by:
Dariusz Kucharczyk, University of Warmia and Mazury in Olsztyn, PolandGesche Winkler, Université du Québec à Rimouski, QC, Canada
Copyright © 2023 Chae, Oh, Kwak, Chang and Jo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hye-Ji Oh, b2hnMjA5MEBuYXZlci5jb20=; Kwang-Hyeon Chang, Y2hhbmczOEBraHUuYWMua3I=; Hyunbin Jo, cHJvemV2YUBwdXNhbi5hYy5rcg==
†ORCID: Yeon-Ji Chae, orcid.org/0000-0002-1185-7945
Hye-Ji Oh, orcid.org/0000-0003-2098-8485
Ihn-Sil Kwak, orcid.org/0000-0002-1010-3965
Kwang-Hyeon Chang, orcid.org/0000-0002-7952-4047
Hyunbin Jo, orcid.org/0000-0001-8064-7880
 Yeon-Ji Chae1†
Yeon-Ji Chae1† Hye-Ji Oh
Hye-Ji Oh Hyunbin Jo
Hyunbin Jo