- 1Division of Environmental Science & Ecological Engineering, College of Life Science & Biotechnology, Korea University, Seoul, Republic of Korea
- 2Division of Life Sciences, Korea Polar Research Institute, Incheon, Republic of Korea
- 3School of Earth and Environmental Sciences & Research Institute of Oceanography, Seoul National University, Seoul, Republic of Korea
Solar salterns were placed along the coast and were frequently left unattended after use. While many studies have isolated and identified microorganisms from hypersaline environments, their role and adaptation mechanisms are still unclear. Herein, we elucidated the role of halophiles in salt-polluted areas through the recently reported Halomonas getboli YJPS3-2 from the abandoned saltern. We analyzed the expression levels of genes in the YJPS3-2 strain to identify its adaptation mechanisms to high salinity environments, by representing the process from tidal flats to abandoned salterns with varying salinity gradients. The YJPS3-2 strain primarily overexpresses genes associated with ABC transport to adapt to hypersaline environments. Interestingly, the cheA gene, which recognizes changes in the surrounding, was the most upregulated, and it was also associated with the overexpression of the MS ring and T3SS mechanisms relating to the flagellar activity. The YJPS3-2 recognized the high salt concentration in its surroundings and attempted to accumulate compatible solutes that could withstand high osmotic pressure inside the cell to adapt to the high salinity environment. Furthermore, during this process, the YJPS3-2 strain removed surrounding pollutants and secreted secondary metabolites that could be utilized by neighboring organisms. Our results suggested that this halophilic bacterium has the potential to serve as a pioneering species for thriving the surrounding while adapting to saline environments.
1 Introduction
Coastal tidal flats are a unique ecosystem that is alternately covered and exposed by the tides, and it includes various habitats such as tidal flats and salt marshes (Chen et al., 2016). The intertidal zones, where seawater and freshwater meet, create unique environments due to the presence of salinity, which limits biodiversity. When salt concentrations increases, the biological diversity decreases, however, halotolerant microorganisms often require high-salt environments and can even be found at the highest salt concentrations (Gerday and Glansdorff, 2007). In particularly, coastal solar salterns are noteworthy as they utilize these environmental characteristics of tidal flat for salt production. They are man-made, thalassohaline hypersaline environments (Oren, 2007) created by evaporating seawater and are located along the coastline (Paul et al., 2020). These artificial solar salterns create an extremely hypersaline environment that results in biodiversity loss, providing evidence that the region has undergone “environmental degradation” (Rodriguez-Valera et al., 1985).
Hypersaline environments, such as solar salterns and salt lakes, are characterized by high salt levels (> 3.5%, higher than seawater), and are inhabited by halophiles (Ventosa, 2006; Javor, 2012), a group of microorganisms adapted to these extreme conditions. Halophilic bacteria can be classified based on their salt requirement and growth patterns. Slight halophiles grow optimally at 2 – 5% NaCl, moderate halophiles at 5 – 20% NaCl, and extreme halophiles at 20 – 30% NaCl (Irshad et al., 2014). To cope with high extracellular salinity, halophilic microorganisms have adopted various strategies. Bacteria and archaea accumulate K+ and Cl– ions while maintaining a low Na+ concentration in response to external osmotic pressure. Most bacteria and eukaryotes store or synthesize organic solutes, such as glycine betaine, ectoine, and other amino acids and sugar derivatives, to deal with high salinity (Oren, 2002; Roberts, 2005). Halophiles have developed basic biochemical adaptations in their proteins, osmoregulation systems, nucleic acids, and lipids to thrive and reproduce in such high-salt, low-water activity conditions (Javor, 2012).
Getbol, tidal flats in Korean, are located along the west coast of the Republic of Korea and include several manmade salterns, such as Taepyeong salt farms in Sinan-gun, Jeung-do (Lee et al., 2018), and Yubu-do solar saltern in Taean-gun. The coastal tidal flats have been extensively studied, and numerous microorganisms have been reported and isolated from this ecosystem (Yeon et al., 2005). However, it is worth noting that microorganisms are also being isolated, particularly from abandoned salterns. Among them, the genus Halomonas is frequently isolated from high-salt environments such as abandoned salterns (Lim et al., 2004; Lee et al., 2005; Lee et al., 2016). The genus Halomonas belongs to the phylum Gammaproteobacteria, class Oceanospirillales, family Halomonadaceae, and is one of the largest halophilic genus so far with over 100 species and diverse salinity growth range.
In a previous study, the comparative evaluation of bacterial diversity and community structure was conducted on three abandoned salterns with different natural restoration periods (Lee et al., 2020). As the salterns were abandoned for longer periods of time, the microbial community structure became increasingly similar to that of the adjacent natural tidal flats. In a recent study, a novel halophilic bacterial species named “Halomonas getboli YJPS3-2” was isolated and reported from the sediment of an abandoned saltern in Yongyu-do, where the difference in microbial community structure between the saltern and the adjacent natural tidal flat was the greatest in previous studies (Lee et al., 2020; Yoo et al., 2022). Therefore, this study was conducted as a follow-up to the previous research to evaluate the ecological role of halophilic bacteria in abandoned saltern, using newly identified halophilic bacteria “YJPS3-2” as a research subject. The specific objectives of this study were: 1) to reveal adaptation strategies to the hypersaline environment by exposing the strain to various salt conditions and 2) to report halotolerant-related genes through the newly identified species.
2 Materials and methods
2.1 Isolation of bacterial strains and culture conditions
Our previous studies reported the study site (Figure 1) and detailed procedures for the isolation of bacterial strains (Lee et al., 2020; Yoo et al., 2022). Most of the culturable bacteria isolated from Yongyu-do grew well on a basal medium ISP2 (Islam and Hernández, 1966) with seawater (4 g L-1 yeast extract (Difco), 10 g L-1 malt extract (Difco), 4 g L-1 dextrose (Difco), and 10 g L-1 artificial sea salt (Merck), 10 ppt, pH 7.0), as well as marine agar (MA 2216, Difco).
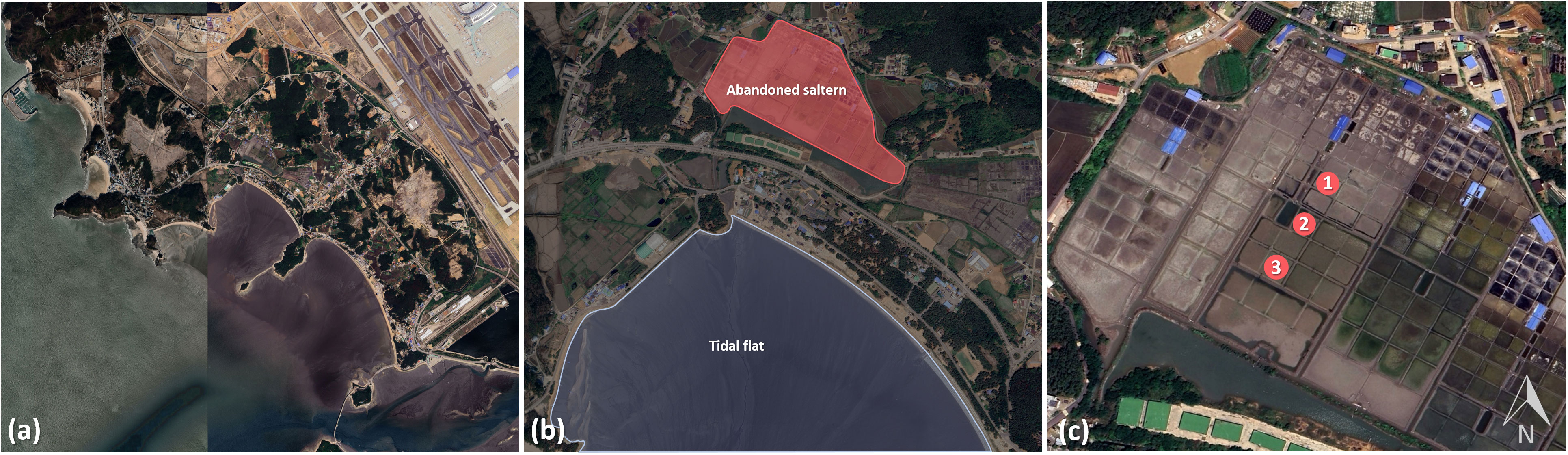
Figure 1 Map showing the sampling location of Yongyudo in the Yellow Sea, Korea. (A) Yongyudo coastline, (B) the sampling sites of tidal flats and abandoned saltern, (C) the three sampling sites of abandoned saltern.
To determine the optimal growth conditions of the strain YJPS3-2 used in this study, it was grown in a basal medium (ISP2, without artificial sea salt) with various concentrations of NaCl (DUKSAN, grade=99.5%), including 0%, 1%, 5%, 10%, 15%, and 20% (w/v), and incubated at 30 °C with shaking at 180 rpm. The concentration of strain YJPS3-2 (OD600) was monitored every 6 h until 96 h using a spectrophotometer (SPARK 10M), with three replicates.
2.2 Extraction of genomic DNA and total RNA of strain YJPS3-2
Strain YJPS3-2 was cultured in a basal medium with 1% NaCl at 30°C for 18 h, and genomic DNA was extracted using the method described in Yoo et al. (2021). Whole-genome sequencing was performed by use of the MiSeq (2 x 300) sequencing platform (Illumina). The sequencing reads were obtained from the paired-end sequencing of a genomic library, with an average insert size of 500 bp. The low-quality reads were trimmed with a quality threshold of Q30. The filtered reads were de novo assembled using the SPAdes genome assembler with default parameters (Bankevich et al., 2012).
For total RNAs, strain YJPS3-2 cells (OD600 = 2.0) were grown in a medium containing 1%, 5%, and 15% NaCl, and RNA was extracted using TruSeq Total RNA with Ribo-Zero (Illumina) according to the manufacturer’s instructions. Whole-transcriptome sequencing was performed on the HiseqXten, NextSeq500 sequencing platform (Illumina). Total RNA from one control group (1% NaCl) and three replicates of strains in 5% and 15% NaCl each was prepared for transcriptomic sequencing.
2.3 Genome and transcriptome data processing and differential gene expression analysis
The resulting genomic sequence was submitted to the RAST server (Aziz et al., 2008), which predicted the open reading frames (ORFs), tRNAs, and rRNAs. The predicted ORFs were annotated by querying the SEED (http://pubseed.theseed.org/) (Disz et al., 2010) and Kyoto Encyclopedia of Genes and Genomes (KEEG, KO numbers) databases. The PAI Finder (Yoon et al., 2007), a web-based search tool of the pathogenicity island database (PAIDB), was used to identify potential pathogenic islands (PAIs). Biosynthetic gene clusters for secondary metabolites were predicted using the antiSMASH tool (Weber et al., 2015).
For gene expression analysis, transcriptomic reads from the 5% and 15% NaCl samples were aligned to the whole transcriptome sequencing of the 1% NaCl strain of YJPS3-2. De novo assembly of the merged data was carried out using Trinity with the default parameters (Grabherr et al., 2011). For the assembled genes, the longest contigs of the assembled contigs were filtered and clustered into non-redundant transcripts using the CD-HIT-EST program (Fu et al., 2012). We defined these transcripts as “unigenes”, which were used for predicting ORFs, annotating against several known sequence databases, and analyzing differentially expressed genes (DEGs). Fragments Per Kilobase Million (FPKM) metrics were used to quantify the total transcription levels of the genes, and genes with expression level P-values of < 0.05 were identified as DEGs (Anders and Huber, 2012). The transcription expression levels were represented using FPKM values evaluated with eXpress. Genes were considered to be differentially expressed if the absolute value 2 of the log2FC (Fold change) in FPKM exceeded.
For functional annotation of the unigenes, the Gene Ontology (GO) database, and Kyoto Encyclopedia of Genes and Genomes (KEGG) database were applied to classify the annotated unigenes using BLASTX of DIAMOND with an E-value cut-off of 1.0E5. Classification of the GO terms were subsequently performed using an in-house script. Bi-directional best hit (BBH), which is a widely used method to infer orthology, was used to search against the KEGG database to obtain the KO (reference pathway) number of the KEGG annotation. The KO number of the transcriptome was also obtained according to the KEGG annotation.
3 Results
3.1 Bacterial isolation and selection of the experimental strain
In our recent study, we examined the sediment microbiome from Yongyudo and isolated a total of 104 culturable bacteria (Lee et al., 2020) (Table S1). Of these isolates, 73 strains (23 genera and 61 species) were obtained from tidal flats, while 31 strains (16 genera and 27 species) were obtained from abandoned salterns. This result indicated lower biodiversity in salterns, which is consistent with our previous research (Lee et al., 2020). We conducted a salt-tolerant test and found that nine bacterial species were able to grow in a slightly halophilic environment (5% NaCl), while four of them were able to grow in a moderately halophilic environment (15% NaCl) (Table 1). The NaCl concentration gradient used for the salt-tolerant test was selected based on the standards for classifying halophilic bacteria, as well as the salinity of tidal flats, oceans (around 32 – 33‰) (Choi et al., 2021), and solar salterns (Song et al., 2022). Among them, strain YJPS3-2 was newly reported, which was belonging to the genus Halomonas with a 16S rRNA gene similarity of 98.2% (Yoo et al., 2022). The genus Halomonas is classified within the family Halomonadaceae in the class Gammaproteobacteria and is also well-known as halophilic bacteria, in which the cells are Gram-negative and chemoorganotrophic (Ventosa et al., 2015). Therefore, this strain was selected as the experimental strain in the present study, as it was considered to be appropriate for revealing the mechanism of adaption to halophilic environments.
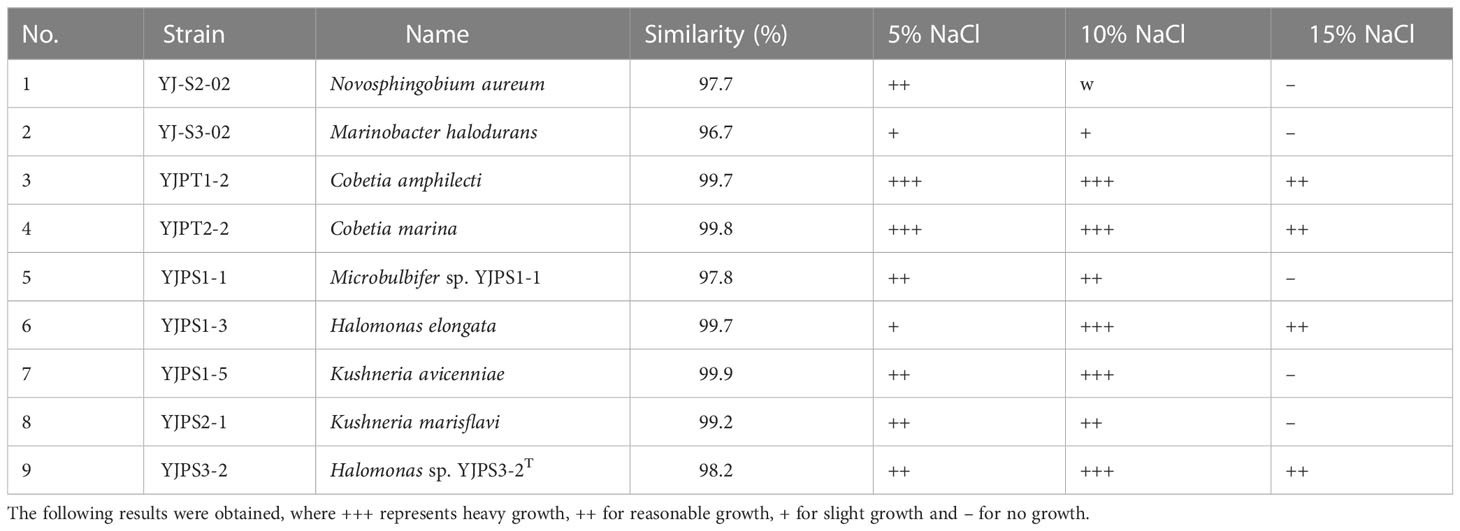
Table 1 List of salt-tolerant tests for halophilic and halotolerant bacteria isolated from the study area.
3.2 Growth properties of strain YJPS3-2 under various salt conditions
Strain YJPS3-2 showed a minimum requirement of 1% NaCl for survival, with no bacterial growth observed in either 0% or 20% NaCl conditions (Figure 2). Growth curves revealed a higher growth rate in 1% NaCl compared to 5%, 10%, or 15% NaCl between 3 h and 24 h. These results indicate that NaCl is necessary for growth and that the strain prefers a halophilic environment, classifying it as a halophilic bacterium.
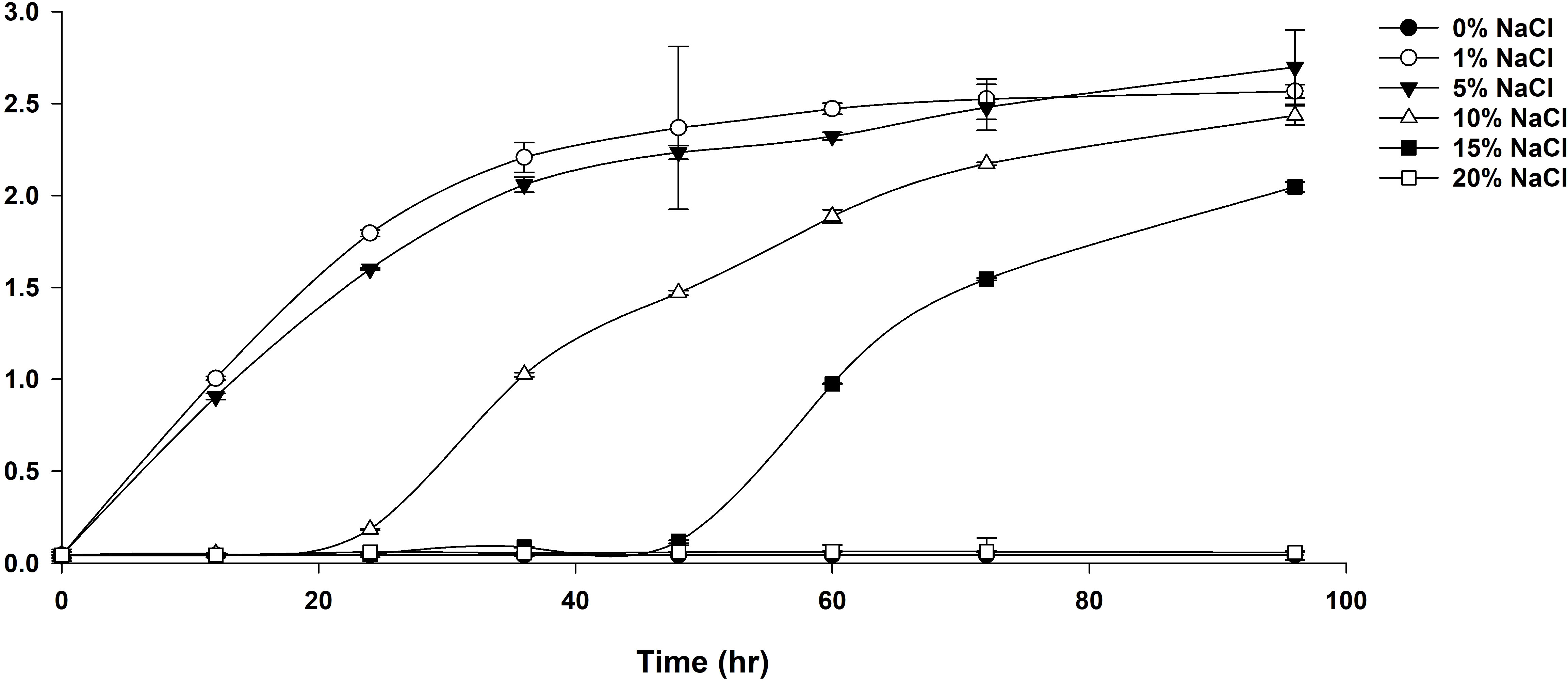
Figure 2 Growth curve of strain YJPS3-2 in media containing various concentrations of NaCl, including 0%, 1%, 5%, 10%, 15%, and 20% (w/v) NaCl.
3.3 Genomic features and transcriptomic profiles of strain YJPS3-2
The whole-genome sequencing of strain YJPS3-2 resulted in the annotation of a total of 1,348 genes (Figure 3A), which were categorized as follows (>10%): amino acids and derivatives (19.40%); protein metabolism (12.73%); cofactors, vitamins, prosthetic groups, and pigments (10.59%); and carbohydrates (10.59%) (Figure 3B).
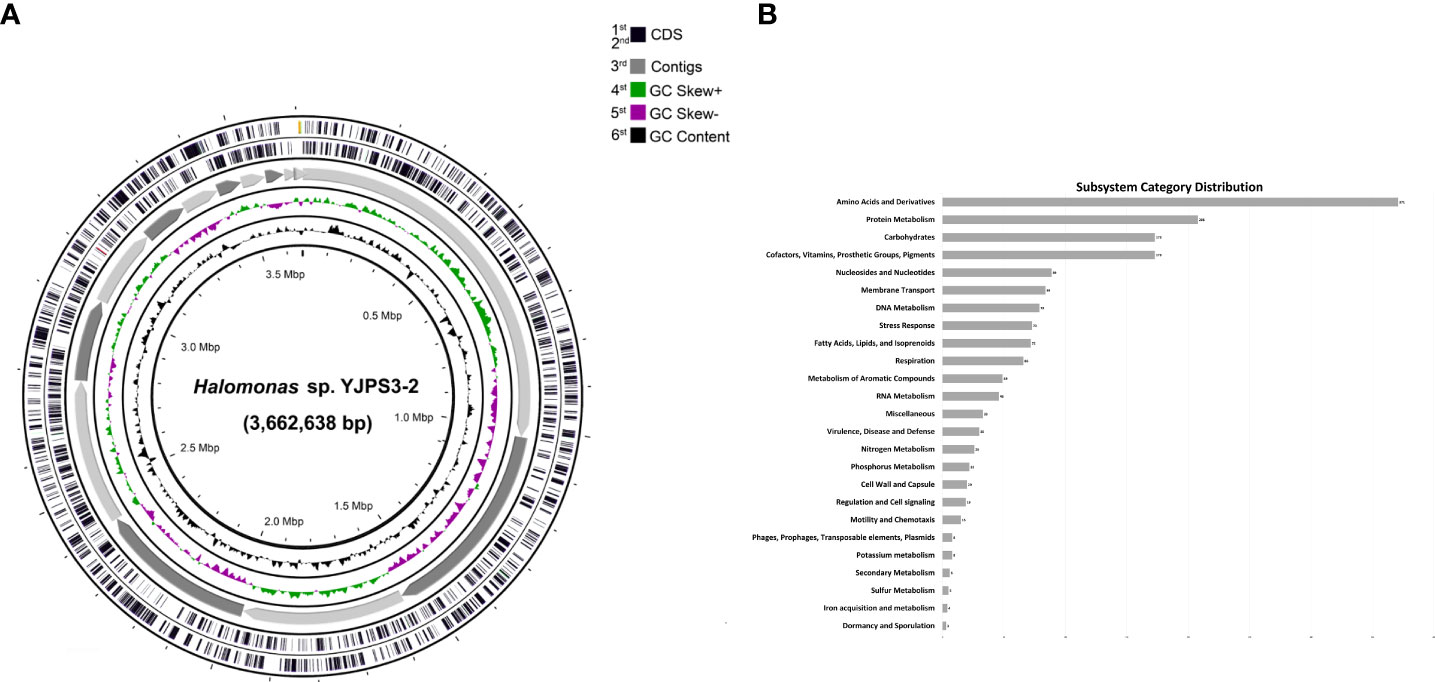
Figure 3 Genome map of Halomonas sp. YJPS3-2. (A) Rings from the outside are as follows: 1st and 2nd circles, predicted genes on the coding sequence (CDS); 3rd circle, mapping count contigs; 4th circle, GC skew + strand; 5th circle, GC skew – strand; and 6th circle, CG content. (B) Diagram of subsystem category distribution results from whole genome sequencing analysis of strain YJPS3-2.
Genomic analysis of strain YJPS3-2 revealed that it is incapable of carbon fixation and instead utilizes glycoside hydrolase, peptidase, and gluconeogenesis to obtain carbon sources. The Embden-Meyerhof pathway and Entner-Doudoroff pathway are engaged during glycolysis, with the TCA cycle being utilized. YJPS3-2 is classified as a mixotroph, capable of using nitrogen via dissimilatory nitrate reduction when oxygen is not available as an electron acceptor. Furthermore, the genomic data indicated that YJPS3-2 can degrade pollutants by utilizing the aroD gene for quinate degradation, and benK, benD, benB, and pobA for benzonate degradation. Additionally, it can decompose aromatic compounds via hmgA, fahA, hppD, maiA, hppD, and phhA.
The PAI Finder was used to compare the PAI regions of Halomonas elongata and YJPS3-2. Consequently, nPAIs (PAI-like regions not overlapping genomic islands) but not PAI-like regions were present in YJPS3-2. The detected nPAI region, HELO_2920 gene was present in the genome of YJPS3-2. However, there was no significant expression in the transcriptome under 5% and 15% NaCl conditions. In addition, an ectoine synthase (ectC) gene cluster encoding enzymes involved in the ectoine transport system was identified by antiSMASH (node 827,601 – 827,993 bps) and showed 75% similarity with the ectoine biosynthetic gene cluster from Methylomicrobium kenyense. The siderophore biosynthesis protein (lucA/lucC) gene cluster was identified by antiSMASH (node 79,902 – 81,779 bps) and showed 66% similarity with the desferrioxamine cluste from Streptomyces argillaceus. Also, RiPP-like (Ribosomally synthesized and post-translationally modified peptides) enzyme gene cluster was identified by antiSMASH (node 421,288 – 433,486 bps) and showed 17% similarity with fengycin biosynthetic gene cluster from Bacillus velezensis FZB42 (Figure 4).
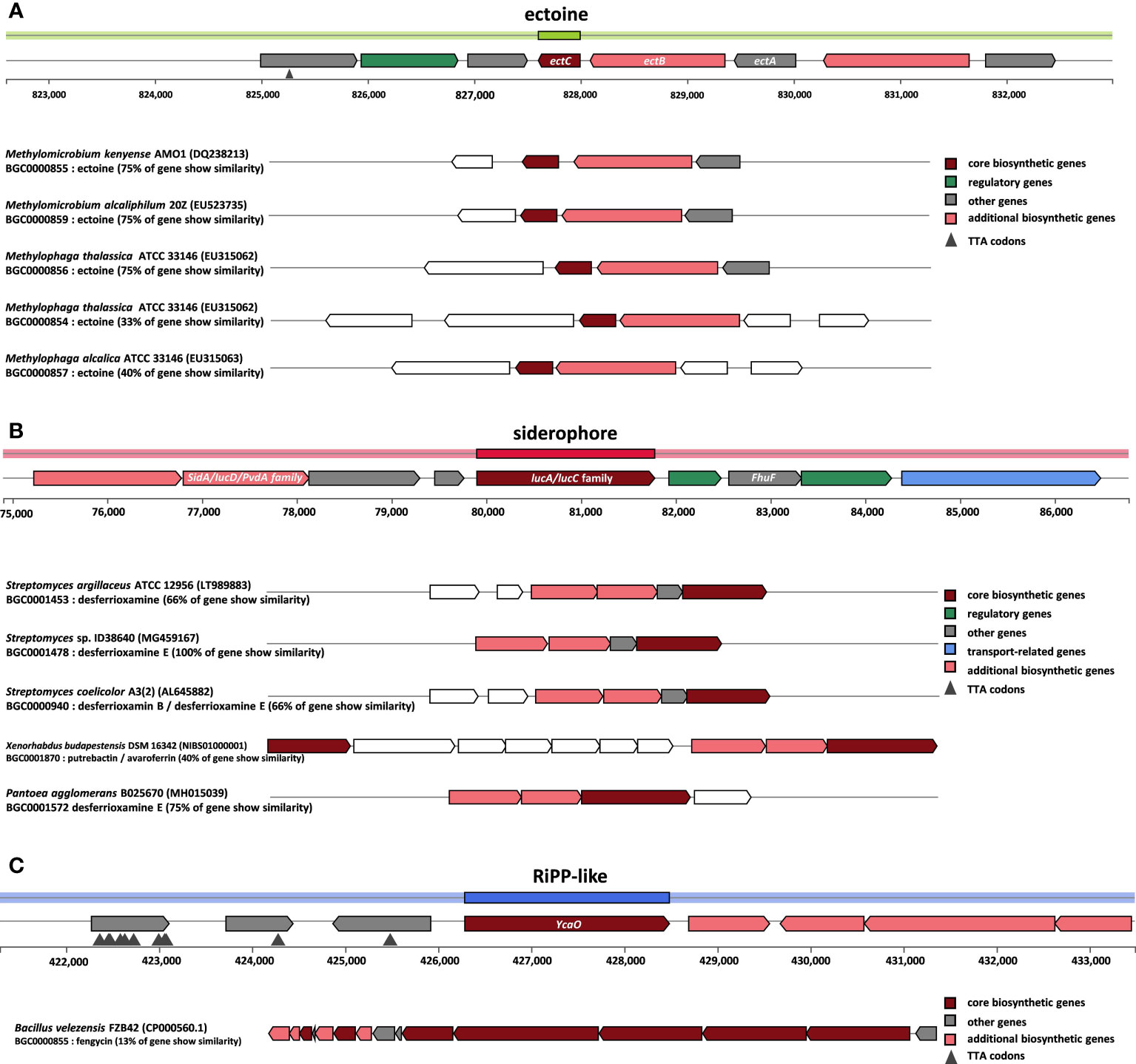
Figure 4 Schematic overview of the genomic region containing the strain YJPS3-2 secondary metabolites gene cluster. biosynthetic gene clusters (BGC) comparison of the (A) ectoine synthesis and (B) siderophore synthesis (C) RiPP-like gene clusters of the isolate and related strains as predicted by antiSMASH. Based on sequence similarity and gene function prediction.
Transcriptomic analysis of strain YJPS3-2 under varying NaCl conditions was performed by de novo sequencing. A total of seven samples were analyzed, including one control strain (1% NaCl) and three replicates each in 5% and 15% NaCl conditions, resulting in six designed comparison analyses. The raw reads were generated, on average, for 1% (22 Mbps), 5% (24 Mbps), and 15% NaCl (24 Mbps) conditions, respectively (Table S2). To avoid biased analysis, only 570 contigs were used for statistical analysis after excluding 1,838 out of 2,408 contigs that had more than one read count value of 0 (Figure S1).
3.4 Halotolerance-related genes from differential expression analysis
The different expression levels of genes are presented as a heat map (Figure 5) and notable metabolic pathways in the saline conditions were predicted based on the DEG analysis data and KO number annotations (Figure 6; Tables S4–S6). Compared to the condition with 1% NaCl, both 5% and 15% NaCl conditions had a significant impact on membrane transporter-related genes. The majority of genes were related to ATP-binding cassette (ABC) transporters, two-component systems, and the Na+ symporter, as shown in Figures 5, 6. Among the affected genes, afuA (iron complex), phoR and phoD (phosphate), glnL, glnG, and glnD (glutamate), modB (molybdate), aapJ, aapQ, and aapP (general L-amino acid), and livM (branched-amino acid) are involved in ion transport, and malK (maltose), thuF, thuG, and thuK (trehalose), smoK (mannitol), and araG (L-arabinose) are genes involved in solutes compatible with the ABC transporter. In the ABC transporter smoK, thuF, thuG, thuK, malK, modB, araG, and phoD were upregulated, and glnL, glnG, and phoR were downregulated under 5% NaCl conditions.
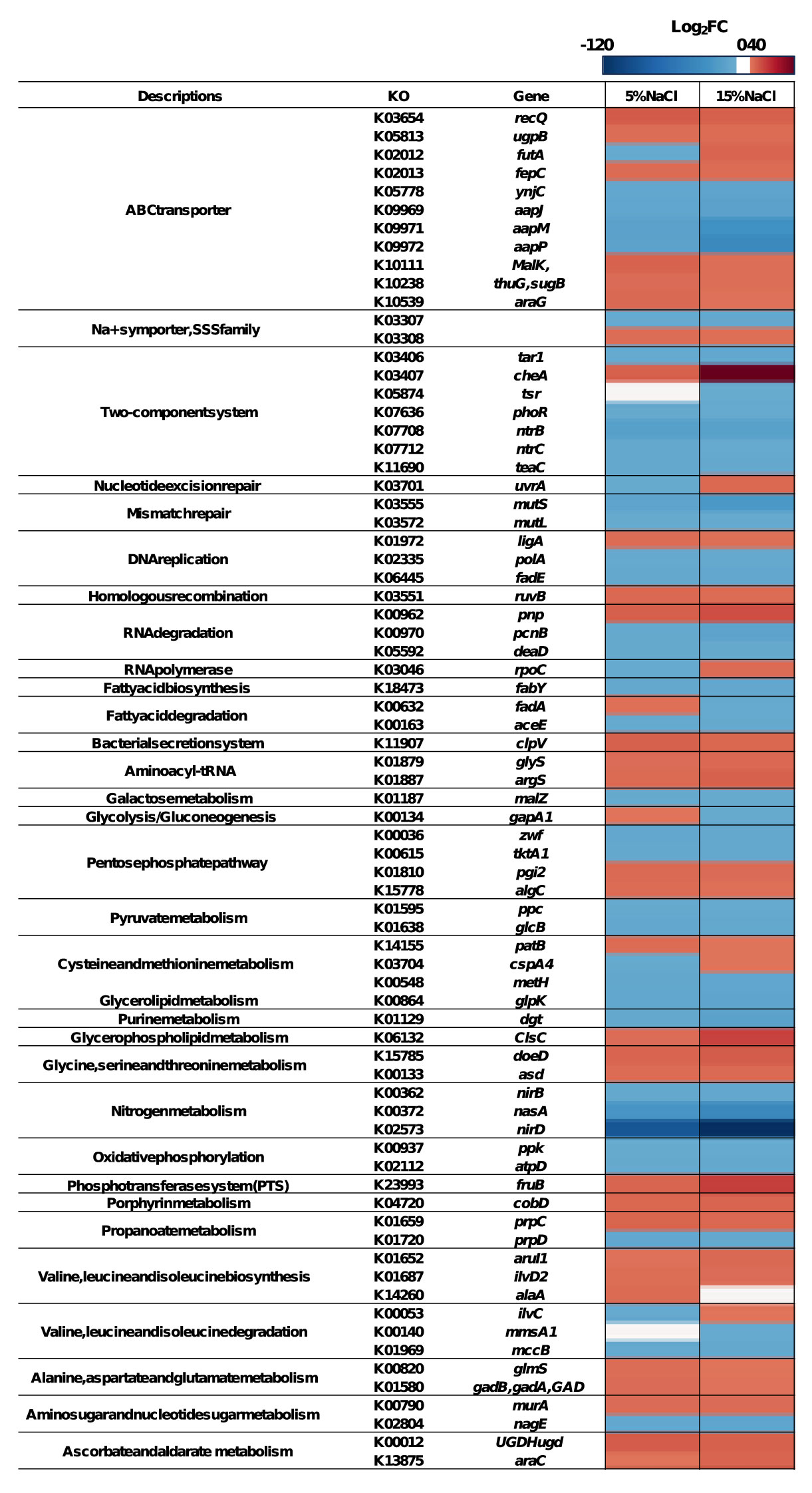
Figure 5 The differentially expressed genes (DEGs) of strain YJPS3-2 between 5% NaCl and 20% NaCl conditions. The red columns represent upregulated DEGs, for which the log2FC of FPKM was higher than 2; the blue columns represent downregulated DEGs, for which log2FC in FPKM was lower than -2. The genes without significant expression change were colored white.
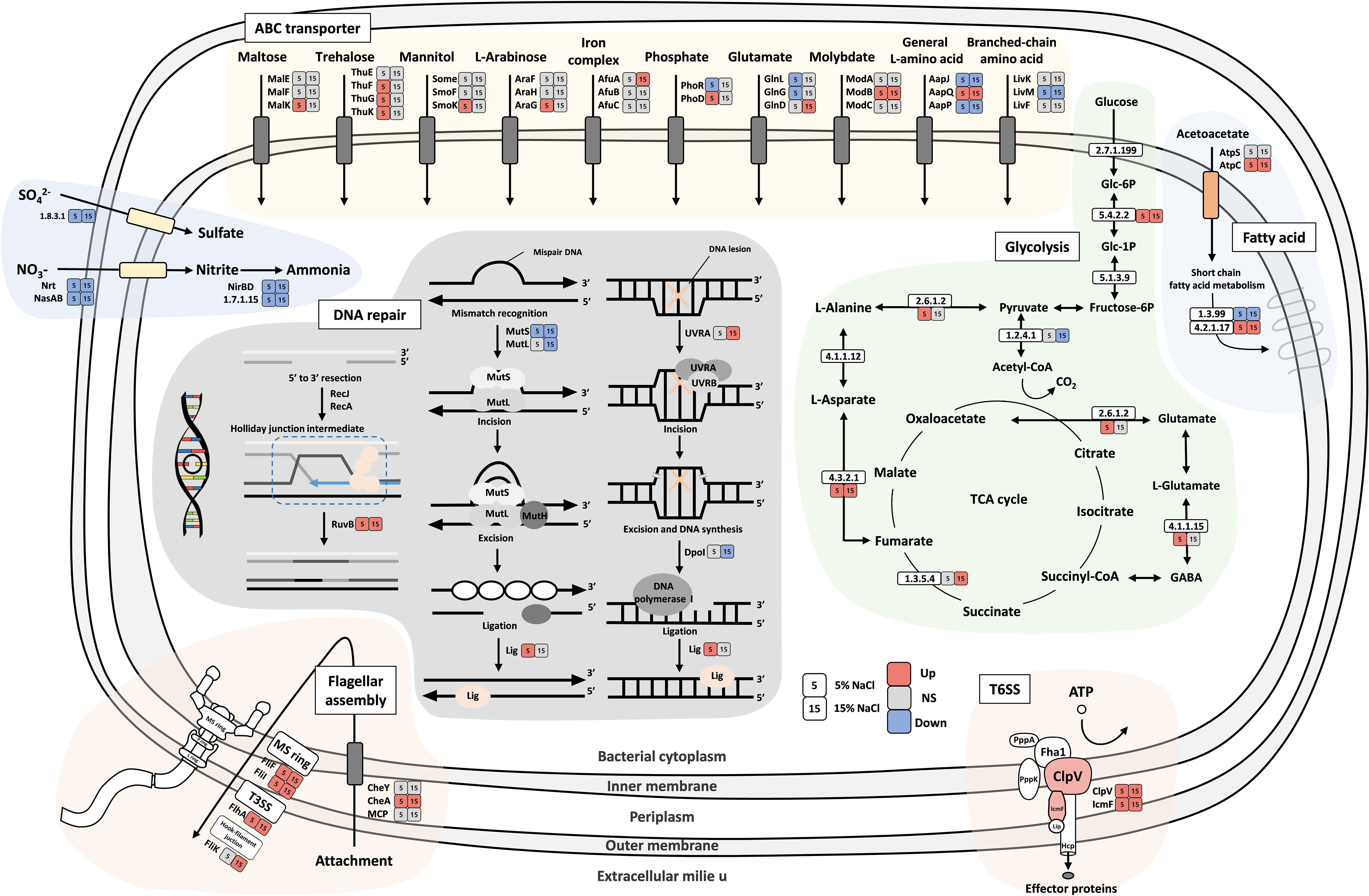
Figure 6 Schematic diagram of carbohydrate metabolism and membrane transport in strain YJPS3-2. Colored boxes indicate differentially expressed profiles (upregulated, red; downregulated, blue; no significance (NS), gray) in 5% and 15% NaCl conditions compared to optimum NaCl conditions (1% NaCl).
Figure 6 shows the differential expression levels of genes involved in the glycolysis pathway under high salinity concentrations. This pathway involves complex genes, such as those related to the pentose phosphate pathway; alanine, aspartate, and glutamate metabolism; valine, leucine, and isoleucine biosynthesis; galactose metabolism, and citrate cycle (TCA cycle) functions. Furthermore, the flagellar assembly and pathway-related genes (cheA, flaG, filF, fliI, and flhA) from DEG analysis were upregulated in both 5% and 15% NaCl conditions except for filK. Notably, the DNA repair pathways, including nucleotide excision repair, mismatch repair, DNA replication, and homologous recombinations, were significantly affected by saline conditions. Among these pathways, mutS and mutL were downregulated, while ruvB, uvrA, and ligA were upregulated in saline conditions.
Through DEG analysis, the upregulated genes were sorted under both 5% and 15% NaCl conditions (Table S4). Among the upregulated genes, cheA, which encodes taxis sensor histidine kinase for a two-component system, exhibited the highest expression followed by cslC, fruB, filF, and ABC transporter-related genes. On the other hand, the genes that showed downregulated expression under both 5% and 15% NaCl conditions included nirD, aapP, aapM, nasA, mutS, dgt, pcnB, aapJ, and ynjC, as well as the majority of the transporter-related genes and DNA repair-related genes indicated in Figure 6. The genes that were upregulated only under 5% NaCl conditions were lepA, ligA, algC, patB, glmS, thuG, sugarB, gadB, gadA, AllaA, malZ, araG, and ugpB. In contrast, the ntrC, phoR, uvrA, ilvC, and metG genes were downregulated only in 5% NaCl conditions (Table S4). Additionally, ispH, tar1, tsr, flgK, futA, and araC were upregulated, and fdhA, yheS, tar1, pnp, htpG, phbC, and cspA4 were downregulated only under 15% NaCl conditions (Table S5).
4 Discussion
This study focused on halophilic bacteria isolated from abandoned solar salterns, aiming to reveal adaptation strategies under hypersaline environments and designating this halophilic bacterium as a pioneering species in the field. The study utilized the halophilic bacterial strain YJPS3-2, which was found to primarily adapt to the halophilic environment by modulating the ABC transporter to maintain osmotic balance, regulating flagellar assembly, and enhancing the DNA repair pathway (Figure 6). Additionally, the strain also acted as a pioneering species, allowing other organisms to thrive by settling first in hypersaline environments and producing a variety of secondary metabolites (Figure 4).
ABC transporters, which should be initially discussed when studying the halophilic adaptive mechanism of strain YJPS3-2, use the energy of ATP hydrolysis for the uptake and efflux of solutes across the cell membrane. These transporters are crucial for halophiles, which produce or acquire compatible solutes (such as ectoine, trehalose, glycine betaine, and choline) and anions into the cytoplasm to balance the osmotic pressure (van der Heide and Poolman, 2000; Lewis et al., 2012; Das et al., 2015). Genes related to the ABC transporter for maltose (malK), trehalose (thuF, thuG, and thuK), mannitol (smoK), and L-arabinose (araG) were upregulated in a 5% NaCl environment. However, genes associated with negatively charged ions such as phosphate (PO43-), glutamate, and amino acids were downregulated, indicating that the bacterium avoids importing them to adapt to osmotic pressure.
Motility is a common strategy used by various types of bacteria to respond to adversity (Li et al., 1993), and it has been found that it is higher in 25% NaCl environments compared to 0% NaCl (Remonsellez et al., 2018). The bacterial flagellum functions as a motor, protein exporter, and assembly apparatus (Macnab, 2003), and is composed of an MS ring formed by the inner-membrane protein FliF (Kubori et al., 1992), T3SS (type III secretion system), and a hook-filament junction. Chemotaxis is a mechanism by which bacteria efficiently and rapidly respond to changes in chemical composition using methyl-accepting chemotaxis proteins (MCPs) and cheA and cheY genes (Bren and Eisenbach, 2000). In response to changes, bacteria use flagella to attach to and invade targets (Bange et al., 2010), promote plant growth (Glick, 1995), and remove contaminants (Omotayo et al., 2013). In the study, genes related to flagellar assembly, including MS ring formation (filF and fliI), T3SS (flhA and filK), and attachment (cheA), were overexpressed, with the most highly overexpressed gene being cheA, which regulates gene expression in response to changing environmental conditions (Stock et al., 1988). The cheA was expressed more in 15% NaCl than in the 5% NaCl conditions, which might be attributed to its role in the bacterial chemotactic system.
The overexpression of ligA and ruvB, which plays an important role in DNA processes (Alomari, 2018) and recombination, respectively, was upregulated in both conditions. Nucleotide repair-related genes in bacteria have previously been reported to be important for bacterial survival under salt stress (Mirete et al., 2015), and their over and under-expression is expected to be common in extreme environments (Puig et al., 2021).
In contrast to T3SS, T6SS (type VI secretion system) is a versatile mechanism that can transfer toxins to both eukaryotic and bacterial cells (Douzi et al., 2016), increasing the survival rate of bacteria by removing risk factors from their surrounding environment. T6SS component genes, namely clpV and lcmF were found to be upregulated in 5% and 15% NaCl conditions, respectively. The genus Halomonas has been studied for its biochemical potential, but some studies have reported its pathogenicity (Rojas et al., 2009; Tsuji et al., 2022) and potential harm to humans (Stevens et al., 2009; Kim et al., 2012). HELO_2920 gene encodes the T6SS cluster protein VasA, which is a macromolecular machine that plays an important role in the pathogenicity of Gram-negative bacteria (Wang et al., 2021) by transferring and secreting effectors (Mariano et al., 2019) and injecting a toxin into eukaryotic cells to hydrolyze the cell walls. The existence of such a potential halophilic human pathogen in abandoned salterns near tidal flats that are easily accessible to people may pose a danger.
During the process of adapting to high salinity conditions, it is crucial to investigate not only the overexpressed genes but also to genes that are downregulated or underexpressed.
In this study, we observed a substantial underexpression of genes associated with nitrogen metabolism, including nirB, nirD, and nasA Notably, nirD exhibited the most pronounced underexpression among all the downregulated genes. This finding is consistent with previous studies reporting a decrease in nitrogen metabolism in bacteria under high salinity environments (Li et al., 2021). The observed decrease in nitrogen metabolism suggests a strategic energy conservation strategy employed by bacteria to optimize their physiological responses to high salinity stress and allocate resources primarily to essential metabolic pathways required for their adaptation and survival.
The intriguing similarity in gene expression patterns across major pathways is noteworthy, wherein genes that were overexpressed in 5% NaCl conditions were under-expressed in 15% NaCl conditions, and vice versa. Nevertheless, some genes displayed higher expression in 15% NaCl conditions compared to the 5% NaCl conditions (Figure 5). This suggested that the genes that enable adaption to the halophilic environment may also contribute to survival in a subsequent hypersaline environment.
Halophilic microorganisms, while adapting to hypersaline environments, produce various secondary metabolites that aid the survival of other organisms (Chung et al., 2020). The genomic analysis of strain YJPS3-2 by antiSMASH predicted the production of ectoine and siderophore, which was consistent with the transcriptome data. Ectoine helps the organism tolerate osmotic pressure (Hahn et al., 2016), while the compatible solutes produced by the bacteria are absorbed by algae and plants in a halophilic environment, enabling them to withstand osmotic stress (Rontein et al., 2002; Moghaieb et al., 2006). Moreover, siderophores, which are peptidic chelates excreted by microorganisms to uptake iron (Scavino and Pedraza, 2013), contribute to plant growth and phytosanitary protection (Compant et al., 2005; Maheshwari, 2011). Thus, halophilic strains living in an extremely halophilic environment produce various secondary metabolites that assist other organisms in surviving in saline environments, thereby promoting the recovery these areas through the bloom of algae and plant growth and the increase in biodiversity (Scavino and Pedraza, 2013). This highlights the significance of the “halophiles settlement” phenomenon.
The genus Halomonas is known for its ability to degrade aromatic compounds (García et al., 2005; Oie et al., 2007; Mnif et al., 2009), as well as its high tolerance to hypersaline environments. Strain YJPS3-2 also contains genes involved in the degradation of quinate, biphenyl, benzoate, p-hydroxybenzene, and generate. However, these genes were not expressed in either 5% or 15% NaCl conditions, suggesting that aromatic compounds were not supplied to the medium during transcriptomic analysis. This finding implies that this strain has the potential to act as a pioneer species in both salt and oil contamination.
In the present study, we confirmed low microbial diversity in hypersaline environments and investigated the adaptive genes of a halophilic bacterium, with a focus on its potential to serve as a pioneering species in harsh environments. Genomic and transcriptomic analysis of strain YJPS3-2 revealed that this strain adapts to halophilic environments by secreting substances that can be used by surrounding organisms, aiding in the thriving of algae and plants in hypersaline environments.
5 Conclusions
We investigated the adaptative strategies of halophilic bacteria in hypersaline environments, using a newly identified strain YJPS3-2 as a model approach. The transmembrane protein in the cell membrane seemed to play a key role in withstanding osmotic pressure and this bacterium could aid other living organisms to survive in harsh environment by adapting to a salt-stress environment. Although whole-genome sequencing analysis and transcriptomic analysis determined the gene expression levels under various environmental conditions, only known and annotated genes were interpreted. Further research would be necessary to evaluate the newly identified salt-stress-related gene and, particularly identification of the secondary metabolites. Our findings suggest YJPS3-2 would be considered as a pioneer in improving the surrounding by first settling in hypersaline areas near the coast, and elsewhere.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
YY: Conceptualization, formal analysis, statistical analyses, visualization, writing – original draft, writing - review & editing. HL: Conceptualization, formal analysis, statistical analyses, visualization, writing – original draft, writing - review & editing. JL: Visualization, writing - review & editing. JK: Conceptualization, data curation, writing - review & editing, project administration. J-JK: Conceptualization, writing – review & editing, project administration, funding acquisition, supervision. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the project entitled “Development of Living Shoreline Technology Based on Blue Carbon Science Toward Climate Change Adaptation (20220526)” in Marine Environments Program of Korea Institute of Marine Science & Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries of Korea.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor YH declared a past collaboration with the authors YY, J-JK, and HL.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1229444/full#supplementary-material
References
Alomari A. (2018). Biophysical and kinetic analysis of escherichia coli DNA ligase activity and inhibition (University of Portsmouth).
Anders S., Huber W. (2012). Differential expression of RNA-seq data at the gene level–the DESeq package Vol. 10 (Heidelberg, Germany: European Molecular Biology Laboratory (EMBL), f1000research.
Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9, 1–15. doi: 10.1186/1471-2164-9-75
Bange G., Kümmerer N., Engel C., Bozkurt G., Wild K., Sinning I. (2010). FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc. Natl. Acad. Sci. 107, 11295–11300. doi: 10.1073/pnas.1001383107
Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bren A., Eisenbach M. (2000). How signals are heard during bacterial chemotaxis: protein-protein interactions in sensory signal propagation. J. bacteriology 182, 6865–6873. doi: 10.1128/JB.182.24.6865-6873.2000
Chen Y., Dong J., Xiao X., Zhang M., Tian B., Zhou Y., et al. (2016). Land claim and loss of tidal flats in the Yangtze estuary. Sci. Rep. 6, 1–10. doi: 10.1038/srep24018
Choi Y., Park Y., Choi M., Jung K. T., Kim K. O. (2021). A fine grid tide-Wave-Ocean circulation coupled model for the yellow Sea: comparison of turbulence closure schemes in reproducing temperature distributions. J. Mar. Sci. Eng. 9, 1460. doi: 10.3390/jmse9121460
Chung D., Kim H., Park H.-S., Kim H. S., Yang D., Kim J. G. (2020). Extracellular enzyme activities in the solar saltern sediments. Microbiological Soc. Korea 56, 133–139. doi: 10.7845/kjm.2020.0003
Compant S., Duffy B., Nowak J., Clément C., Barka E. A. (2005). Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71, 4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005
Das P., Behera B. K., Meena D. K., Azmi S. A., Chatterjee S., Meena K., et al. (2015). Salt stress tolerant genes in halophilic and halotolerant bacteria: paradigm for salt stress adaptation and osmoprotection. Int. J. Curr. Microbiol. Appl. Sci. 4, 642–658.
Disz T., Akhter S., Cuevas D., Olson R., Overbeek R., Vonstein V., et al. (2010). Accessing the SEED genome databases via web services API: tools for programmers. BMC Bioinf. 11, 1–11. doi: 10.1186/1471-2105-11-319
Douzi B., Brunet Y. R., Spinelli S., Lensi V., Legrand P., Blangy S., et al. (2016). Structure and specificity of the type VI secretion system ClpV-TssC interaction in enteroaggregative escherichia coli. Sci. Rep. 6, 1–13. doi: 10.1038/srep34405
Fu L., Niu B., Zhu Z., Wu S., Li W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. doi: 10.1093/bioinformatics/bts565
García M. T., Ventosa A., Mellado E. (2005). Catabolic versatility of aromatic compound-degrading halophilic bacteria. FEMS Microbiol. Ecol. 54, 97–109. doi: 10.1016/j.femsec.2005.03.009
Glick B. R. (1995). The enhancement of plant growth by free-living bacteria. Can. J. Microbiol. 41, 109–117. doi: 10.1139/m95-015
Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., et al. (2011). Trinity: reconstructing a full-length transcriptome without a genome from RNA-seq data. Nat. Biotechnol. 29, 644. doi: 10.1038/nbt.1883
Hahn M. B., Uhlig F., Solomun T., Smiatek J., Sturm H. (2016). Combined influence of ectoine and salt: spectroscopic and numerical evidence for compensating effects on aqueous solutions. Phys. Chem. Chem. Phys. 18, 28398–28402. doi: 10.1039/C6CP05417J
Irshad A., Ahmad I., Kim S. B. (2014). Culturable diversity of halophilic bacteria in foreshore soils. Braz. J. Microbiol. 45, 563–572. doi: 10.1590/S1517-83822014005000050
Islam A., Hernández F. L. (1966). Methods for characterization of streptomyces species. METHODS 16, 313–340. doi: 10.1099/00207713-16-3-313
Javor B. J. (2012). Hypersaline environments: microbiology and biogeochemistry (Springer Science & Business Media).
Kim K. K., Lee K. C., Jeong H., Stevens D. A., Lee J.-S. (2012). Draft genome sequence of the human pathogen halomonas stevensii S18214T. Am. Soc. Microbiol. doi: 10.1128/JB.01071-12
Kubori T., Shimamoto N., Yamaguchi S., Namba K., Aizawa S.-I. (1992). Morphological pathway of flagellar assembly in salmonella typhimurium. J. Mol. Biol. 226, 433–446. doi: 10.1016/0022-2836(92)90958-M
Lee H., Heo Y. M., Kwon S. L., Yoo Y., Lee A. H., Kwon B.-O., et al. (2020). Recovery of the benthic bacterial community in coastal abandoned saltern requires over 35 years: a comparative case study in the yellow Sea. Environ. Int. 135, 105412. doi: 10.1016/j.envint.2019.105412
Lee J.-C., Jeon C. O., Lim J.-M., Lee S.-M., Lee J.-M., Song S.-M., et al. (2005). Halomonas taeanensis sp. nov., a novel moderately halophilic bacterium isolated from a solar saltern in Korea. Int. J. systematic evolutionary Microbiol. 55, 2027–2032. doi: 10.1099/ijs.0.63616-0
Lee J.-C., Kim S.-J., Whang K.-S. (2016). Halomonas sediminicola sp. nov., a moderately halophilic bacterium isolated from a solar saltern sediment. Int. J. Systematic Evolutionary Microbiol. 66, 3865–3872. doi: 10.1099/ijsem.0.001278
Lee K. W., Shim J. M., Kim D. W., Yao Z., Kim J. A., Kim H.-J., et al. (2018). Effects of different types of salts on the growth of lactic acid bacteria and yeasts during kimchi fermentation. Food Sci. Biotechnol. 27, 489–498. doi: 10.1007/s10068-017-0251-7
Lewis V. G., Ween M. P., McDevitt C. A. (2012). The role of ATP-binding cassette transporters in bacterial pathogenicity. Protoplasma 249, 919–942. doi: 10.1007/s00709-011-0360-8
Li C., Louise C., Shi W., Adler J. (1993). Adverse conditions which cause lack of flagella in escherichia coli. J. bacteriology 175, 2229–2235. doi: 10.1128/jb.175.8.2229-2235.1993
Li X., Wan W., Luo X., Zheng L., He G., Huang D., et al. (2021). High salinity inhibits soil bacterial community mediating nitrogen cycling. Appl. Environ. Microbiol. 87, e01366–e01321. doi: 10.1128/AEM.01366-21
Lim J.-M., Yoon J.-H., Lee J.-C., Jeon C. O., Park D.-J., Sung C., et al. (2004). Halomonas koreensis sp. nov., a novel moderately halophilic bacterium isolated from a solar saltern in Korea. Int. J. systematic evolutionary Microbiol. 54, 2037–2042. doi: 10.1099/ijs.0.63194-0
Macnab R. M. (2003). How bacteria assemble flagella. Annu. Rev. Microbiol. 57, 77. doi: 10.1146/annurev.micro.57.030502.090832
Maheshwari D. K. (2011). Bacteria in agrobiology: plant growth responses (Springer Science & Business Media). doi: 10.1007/978-3-642-20332-9
Mariano G., Trunk K., Williams D. J., Monlezun L., Strahl H., Pitt S. J., et al. (2019). A family of type VI secretion system effector proteins that form ion-selective pores. Nat. Commun. 10, 1–15. doi: 10.1038/s41467-019-13439-0
Mirete S., Mora-Ruiz M. R., Lamprecht-Grandío M., De Figueras C. G., Rosselló-Móra R., González-Pastor J. E. (2015). Salt resistance genes revealed by functional metagenomics from brines and moderate-salinity rhizosphere within a hypersaline environment. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.01121
Mnif S., Chamkha M., Sayadi S. (2009). Isolation and characterization of halomonas sp. strain C2SS100, a hydrocarbon-degrading bacterium under hypersaline conditions. J. Appl. Microbiol. 107, 785–794. doi: 10.1111/j.1365-2672.2009.04251.x
Moghaieb R., Tanaka N., Saneoka H., Murooka Y., Ono H., Morikawa H., et al. (2006). Characterization of salt tolerance in ectoine-transformed tobacco plants (Nicotiana tabaccum): photosynthesis, osmotic adjustment, and nitrogen partitioning. Plant Cell Environ. 29, 173–182. doi: 10.1111/j.1365-3040.2005.01410.x
Oie C. S., Albaugh C. E., Peyton B. M. (2007). Benzoate and salicylate degradation by halomonas campisalis, an alkaliphilic and moderately halophilic microorganism. Water Res. 41, 1235–1242. doi: 10.1016/j.watres.2006.12.029
Omotayo A., Egbomeade L., Taiwo O., Oyebamiji O., Ilori M. (2013). Hydrocarbon degradation by free-living nitrogen-fixing bacteria 2013. 14, 75–84.
Oren A. (2002). Diversity of halophilic microorganisms: environments, phylogeny, physiology, and applications. J. Ind. Microbiol. Biotechnol. 28, 56–63. doi: 10.1038/sj/jim/7000176
Oren A. (2007). Biodiversity in highly saline environments. Physiol. Biochem. Extremophiles. doi: 10.1128/9781555815813.ch17:221-231
Paul V., Banerjee Y., Ghosh P., Busi S. B. (2020). Depthwise microbiome and isotopic profiling of a moderately saline microbial mat in a solar saltern. Sci. Rep. 10, 1–16. doi: 10.1038/s41598-020-77622-w
Puig J., Knödlseder N., Quera J., Algara M., Güell M. (2021). DNA Damage protection for enhanced bacterial survival under simulated low earth orbit environmental conditions in escherichia coli. Front. Microbiol. 2021. doi: 10.3389/fmicb.2021.789668:3898
Remonsellez F., Castro-Severyn J., Pardo-Esté C., Aguilar P., Fortt J., Salinas C., et al. (2018). Characterization and salt response in recurrent halotolerant exiguobacterium sp. SH31 isolated from sediments of salar de huasco, Chilean altiplano. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02228
Roberts M. F. (2005). Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst. 1, 1–30. doi: 10.1186/1746-1448-1-5
Rodriguez-Valera F., Ventosa A., Juez G., Imhoff J. F. (1985). Variation of environmental features and microbial populations with salt concentrations in a multi-pond saltern. Microbial Ecol. 11, 107–115. doi: 10.1007/BF02010483
Rojas R., Miranda C. D., Amaro A. M. (2009). Pathogenicity of a highly exopolysaccharide-producing halomonas strain causing epizootics in larval cultures of the Chilean scallop argopecten purpuratus (Lamarck 1819). Microbial Ecol. 57, 129–139. doi: 10.1007/s00248-008-9401-z
Rontein D., Basset G., Hanson A. D. (2002). Metabolic engineering of osmoprotectant accumulation in plants. Metab. Eng. 4, 49–56. doi: 10.1006/mben.2001.0208
Scavino A. F., Pedraza R. O. (2013). “The role of siderophores in plant growth-promoting bacteria,” in Bacteria in agrobiology: crop productivity (Springer). doi: 10.1007/978-3-642-37241-4_11
Song T., Liang Q., Du Z., Wang X., Chen G., Du Z., et al. (2022). Salinity gradient controls microbial community structure and assembly in coastal solar salterns. Genes 13, 385. doi: 10.3390/genes13020385
Stevens D. A., Hamilton J. R., Johnson N., Kim K. K., Lee J.-S. (2009). Halomonas, a newly recognized human pathogen causing infections and contamination in a dialysis center: three new species. Medicine 88, 244–249. doi: 10.1097/MD.0b013e3181aede29
Stock A., Chen T., Welsh D., Stock J. (1988). CheA protein, a central regulator of bacterial chemotaxis, belongs to a family of proteins that control gene expression in response to changing environmental conditions. Proc. Natl. Acad. Sci. 85, 1403–1407. doi: 10.1073/pnas.85.5.1403
Tsuji A., Takei Y., Nishimura T., Azuma Y. (2022). Identification of new halomonas strains from food-related environments. Microbes environments 37, ME21052. doi: 10.1264/jsme2.ME21052
van der Heide T., Poolman B. (2000). Osmoregulated ABC-transport system of lactococcus lactis senses water stress via changes in the physical state of the membrane. Proc. Natl. Acad. Sci. 97, 7102–7106. doi: 10.1073/pnas.97.13.7102
Ventosa A. (2006). “Unusual micro-organisms from unusual habitats: hypersaline environments,” in Symposia-society for general microbiology, vol. 1999. (Cambridge: Cambridge University Press). doi: 10.1017/CBO9780511754913.015
Ventosa A., de la Haba R. R., Arahal D. R., Sánchez-Porro C. (2015). “Halomonas,” in Bergey's manual of systematics of archaea and bacteria, vol. 2015. doi: 10.1002/9781118960608.gbm01190:1-111
Wang N., Han N., Tian R., Chen J., Gao X., Wu Z., et al. (2021). Role of the type VI secretion system in the pathogenicity of pseudomonas syringae pv. actinidiae, the causative agent of kiwifruit bacterial canker. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.627785
Weber T., Blin K., Duddela S., Krug D., Kim H. U., Bruccoleri R., et al. (2015). antiSMASH 3.0–a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–W243. doi: 10.1093/nar/gkv437
Yeon S.-H., Jeong W.-J., Park J.-S. (2005). The diversity of culturable organotrophic bacteria from local solar salterns. J. Microbiol. 43, 1–10.
Yoo Y., Kim D., Lee H., Khim J. S., Kim B., Yang D., et al. (2021). Novosphingobium aureum sp. nov., a marine bacterium isolated from salt flat sediment. Int. J. Syst. Evol. Microbiol. 71, 004930.
Yoo Y., Lee H., Khim J. S., Xu X., Kim B., Choi I.-G., et al. (2022). Halomonas getboli sp. nov., a halotolerant bacteria isolated from a salt flat. Int. J. Systematic Evolutionary Microbiol. 72, 5634. doi: 10.1099/ijsem.0.005634
Keywords: halophiles, halomonas, abandoned solar salterns, hypersaline, whole genome sequence, transcriptome
Citation: Yoo Y, Lee H, Lee J, Khim JS and Kim J-J (2023) Insights into saline adaptation strategies through a novel halophilic bacterium isolated from solar saltern of Yellow sea. Front. Mar. Sci. 10:1229444. doi: 10.3389/fmars.2023.1229444
Received: 26 May 2023; Accepted: 20 June 2023;
Published: 19 July 2023.
Edited by:
Young Mok Heo, COSMAX BTI, Republic of KoreaReviewed by:
Min Seo Jeon, Korea Atomic Energy Research Institute (KAERI), Republic of KoreaSeyoung Mun, Dankook University, Republic of Korea
Copyright © 2023 Yoo, Lee, Lee, Khim and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jong Seong Khim, anNrb2NlYW5Ac251LmFjLmty; Jae-Jin Kim, amFlLWppbmtpbUBrb3JlYS5hYy5rcg==
†These authors have contributed equally to this work and share first authorship
 Yeonjae Yoo
Yeonjae Yoo Hanbyul Lee
Hanbyul Lee Junghyun Lee
Junghyun Lee Jong Seong Khim3*
Jong Seong Khim3* Jae-Jin Kim
Jae-Jin Kim