- 1College of Fisheies, Guangdong Qcean University, Zhanjiang, China
- 2Agricultural Service Center, Agricultural and Rural Bureau of Sanjiao Town, Zhongshan, China
NHE and NKA are important regulators of ion transport in fish and play a pivotal role in maintaining osmotic balance and adapting to salinity changes. However, no systematic identification and functional analysis has been conducted for NHEs and NKAs in the cobia (Rachycentron canadum), a commercially important worldwide flatfish. Herein,12 NHE genes were found to be distributed on 10 chromosomes and 12 NKA genes were found to be distributed on 9 chromosomes were identified in the R. canadum at the genome-wide level. Histopathological examination of the gills demonstrated the response of gill lamellae and chloride cells to salinity, while the microstructure of the intestine and kidney exhibited changes associated with salinity. The findings show that members of the NHE and NKA gene families are widely distributed in gill, brain, and heart tissues. Specifically, NHE genes exhibited high expression levels in the gill, somatic kidney, and brain, whereas NKA genes displayed prominent expression in the gill, brain, and heart. Moreover, salinity adaptation experiments were conducted to examine the response of NHE and NKA genes. In the intestine, NHE1 expression was significantly upregulated following both high and low salt stimulation, while the somatic kidney exhibited a proportional response to changes in salinity. Notably, a significant downward trend in NHE2c expression was observed in the gill, intestine, and somatic kidney with increasing salinity. Following low-salt acclimation, NKAα1b and NKAβ3a were significantly down-regulated in the gill, whereas NKAα3a and NKAβ3a displayed significant up-regulation and down-regulation in the intestine, respectively. In the somatic kidney, NKAα1b, NKAα3a, and NKAβ3a were significantly up-regulated. During high-salt acclimation, the expression patterns of NKAα1b and NKAβ3a in the gill were consistent with those observed during low-salt acclimation, while NKAα3a and NKAβ1b exhibited significant upregulation. Our findings underscore the high conservation of NHE and NKA gene family members in R. canadum and highlight tissue-specific expression patterns and their responses to salinity changes. These results provide valuable insights into the molecular mechanisms governing ion transport and osmoregulation in R. canadum, contributing to the development of novel strategies for enhancing aquaculture practices of this species.
1 Introduction
The cobia, Rachycentron canadum, is a euryhaline teleost known for its ability to tolerate a wide range of salinity levels, ranging from 22.5 to 44.5 ‰, and it exhibits excellent growth performance (Shaffer and Nakamura, 1989; Smith, 1995). In addition to its fast growth rate, R. canadum is a carnivorous fish species highly valued for its delicious meat, nutritional content, and robust resistance. As a result, R. canadum has become an important mariculture species in the southeast coast of China (Zhou et al., 2006; Chen et al., 2009). However, the culture of R. canadum in seawater nets is vulnerable to extreme weather conditions such as typhoons, heavy rains, and cold fronts. The churning of upwelling and surface seawater can lead to significant fluctuations in seawater salinity and temperature, which have a significant impact on the growth and survival of teleost (Benetti et al., 2021). Salinity is one of the key environmental factors that affect the growth and reproduction of fish (Zhang et al., 2017). Teleost can be classified into euryhaline and non-euryhaline species based on their salinity tolerance. Euryhaline teleost often have well-developed mechanisms of osmoregulation that activate osmoregulatory cells, ion channels, enzymes, and hormones in response to salinity stress, allowing them to survive and thrive in a wide range of salinity levels (Fiol and Kültz, 2007; Holmes et al., 2022). Therefore, studying the salinity adaptation mechanisms of euryhaline teleost can provide a theoretical basis for understanding how they maintain organismal homeostasis in different salinity environments.
Euryhaline teleosts are able to adapt to various salinity levels by regulating ion transport, neuroendocrine function, and energy metabolism through osmoregulatory organs such as the gills, intestines, and kidneys (Yamaguchi et al., 2018). At the molecular level, specific genes have been identified as playing a role in regulating salinity adaptation. These genes include ion transporters such as the sodium potassium pump (NKA), Na+/K+/2Cl- cotransporter 1 (NKCC1), Na+/H+ exchanger 3 (NHE3), Na+/Cl- cotransporters (NCC), and cystic fibrosis transmembrane conductance regulator (CFTR) (Marshall, 2011; Hwang et al., 2018). In addition, endocrine hormone genes such as growth hormone (GH), hydrocortisone (COR), prolactin (PRL), and insulin-like growth factor-1 (IGF-1) have also been implicated in the regulation of osmolarity (Jia and Lu, 2016).
NHE proteins are widely distributed ion transporter protein, that play a crucial role in regulating Na+ and H+ concentration gradients, which is important for physiological processes (Orlowski and Grinstein, 2004). Members of the NHE gene family are involved in a range of cellular processes, including intracellular acid-base homeostasis, cell volume regulation, and Na+ reabsorption in the kidney and gastrointestine (Counillon and Pouysségur, 2000). In studies on osmolarity in fish, NHE1, NHE2, and NHE3 are commonly considered as main candidate genes of the NHE family, with research primarily focused on exploring their relationship with dynamic pH balance, osmolarity homeostasis, and ammonia excretion activity (Edwards et al., 2005).
During salinity acclimation, euryhaline teleosts maintain intracellular homeostasis through the action of ion transport proteins and channels mediated by Na+/K+-ATPase (NKA) in the gills (Upling, 2020). NKA is a transmembrane protein composed of α and β subunits that are widely distributed in the gill filaments and body kidneys of fish (Han et al., 2022). It mainly relies on the energy generated by ATP hydrolysis to regulate ion concentration homeostasis in the body, achieve active transmembrane transport of Na+ and K+, maintain cellular ion homeostasis, and can be used as an important indicator of osmotic pressure regulation in fish(Jiang et al., 2022). In addition, a study on the salinity experiments of migratory Arctic charr(Salvelinus alpinus) suggested that inefficient regulation of osmolality may be due to the failure of NKAα1b expression (Bystriansky et al., 2007). NKA activity is closely related to environmental salinity (Yang et al., 2022). Generally, in teleost, NKA activity is positively correlated with increasing salinity. For instance, juvenile Turbot(Scophthalmus maximus) showed that gill filament NKA activity and plasma osmolality were highest at a salinity of 33.5 ‰ and lowest at 15 ‰ (Imsland et al., 2003). In Gadus morhua, NKAα expression in gill filaments and body kidneys significantly decreased in hypoosmotic acclimation experiments, while showing an increasing trend in hypertonic water bodies (Larsen et al., 2012). Furthermore, (Shi et al., 2017) investigated the effect of salinity gradient on Epinephelus moara and found that NKA activity initially increased and then decreased in all treatment groups (except the 9‰ group) with a sudden decrease in salinity, while the 9‰ group always showed a decrease in NKA enzyme activity, suggesting that very low salinity leads to a decrease in NKA activity, impaired ion transport efficiency in the gills, and prevents excessive ion loss. The pattern of changes in NKA enzyme activity in the gill filaments and liver of juvenile Amphiprion clarkii was consistent, with a constant increase within 24 h of low salt stress and a return to normal or slightly below normal enzyme activity at 48 h and 96 h (Hu et al., 2016). In a seawater desalination experiment with Lateolabrax japonicus, gill tissue NKA enzyme activity initially decreased and then increased, following a “U” shape. During the desalination adaptation phase, the activity gradually recovered and stabilized but remained lower than the control group (salinity 30), with a significant difference between the two groups (Zhang et al., 2018). However, the change pattern of NKA activity in teleost was inconsistent, affected by the intensity of salinity adaptation and could be divided into two contradictory types: positive and negative correlation of salinity change. The former, such as A. clarkii, Oreochromis mossambicus, and E. moara, and the latter, such as Cleisthenes herzensteini and Sparus macrocephlus, may be related to the strength of osmotic stress tolerance of the species (Lin et al., 2006). Additionally, it has been suggested that NKA activity reaches a minimum when the salinity of the water column reaches the isotonic point in fish (Wang et al., 2011). Differences in osmoregulatory capacity and regulation in different fish species, changes in NKA activity involving individual development (adults vs. juveniles) and salinity adaptation patterns (acute vs. chronic, long-term vs. short-term), are closely related to species evolution.
In this study, we conducted a comprehensive analysis of the NHE and NKA gene families in R. canadum to investigate their roles in osmoregulation. Our objectives included identifying and characterizing the members of these gene families using genomic data. We examined the conserved structures of the genes, established an evolutionary tree for the species, and performed transcriptome sequencing to explore the expression patterns of the NHE and NKA gene families in R. canadum under various salinity conditions. This research provides valuable insights into the involvement of these gene families in osmoregulation and contributes to our understanding of how R. canadum adapts to different salinity environments.
2 Materials and methods
2.1 Experimental fish and sampling
The fish used for the experiment were the juvenile fish artificially hatched and cultured by our group. The fish were temporarily kept in a bucket of 1.5 cubic meters of water in a salinity of 28-30‰, a water temperature of 26-28 °C, and a DO of not less than 6 mg·L-1. 180 fish of uniform size, healthy and vigorous with no damage on the body surface were selected for the salinity adaptation experiment after one week of temporary rearing, and the initial weight of the fish was 9.74 ± 0.85 g. The fish were divided into 10‰ salinity group, 30‰ salinity group and 35‰ salinity group with three biological replicates in each group. The fish were cultured in 9 buckets of 500 L size for 4 weeks, with 20 fish randomly placed in each bucket. The culture water salinity was adjusted downward by 4‰/d using fully aerated dechlorinated fresh water or upward by 4‰/d using sea crystals until the salinity of the experimental group reached the preset salinity and then the experiment was officially started. During the culture period, the water was fed twice daily with 6% body weight of commercial compound feed (46% crude protein and 8% crude lipid) without interruption of aeration and the water exchange rate was 30%.
Two sampling were conducted. In the first sampling, five R. canadum were randomly selected after seven days of culture, and eight tissues, including gill, intestine, body kidney, brain, stomach, muscle, spleen, and heart, were collected after anesthesia with eugenol (200 mg/L) for tissue distribution assay. In the second sampling, after four weeks of culture, five fish were randomly selected from each barrel, with a total of 15 fish. Among them,6 fish were anesthetized and three tissues of gill, intestine, and body kidney were taken for phenotypic analysis and qPCR detection, and 9 fish were mixed for transcriptome sequencing. All molecular samples were snap frozen in liquid nitrogen and stored at -80 °C after collection.
2.2 HE staining
The fresh tissue was fixed using paraformaldehyde (4%) for 24 h. Afterwards, the tissue was orderly dehydrated using gradient alcohol, and the wax-soaked tissue was embedded in the embedding machine. And further the tissue was cut into slices with its thickness 4 μm, and the paraffin sections were dewaxed and further washed by distilled water. Lastly, the nucleus and cytoplasm were stained by hematoxylin and eosin, respectively.
2.3 Identification of NHE and NKA gene family members in R. canadum
For the complete identification of NHE and NKA gene family members in R. canadum, this study was based on the whole genome data of R. canadum (PRJNA634421) obtained in our laboratory and the NCBI public database, blast identification of NHE and NKA gene family members, recorded as the first round of screening results. The NHE and NKA gene family features were obtained from the Pfam database (http://pfam.xfam.org/) as PF00999 (Sodium/hydrogen exchanger family), PF00287 (Sodium/potassium ATPase beta chain), and PF00690 (Cation transporter/ATPase, N-terminus), respectively. The Hidden Markov Models (HMM) were used to obtain the features of the gene family. The HMMER 3.0 software was used to retrieve the whole-genome data, and the results of the second round of screening were tallied. Integrate the results, delete the mutilated or duplicate sequences and upload to SMART website and NCBI database for duplicate checks. The naming of NHE and NKA gene family members was based on reference comparisons and NCBI search results.
2.4 Structural analysis and genomic localization of the NHE and NKA gene families in R. canadum
In order to further investigate the NHE and NKA gene family members, we conducted several analyses. Firstly, we determined the intron and exon length information, as well as the genomic localization, based on the genome annotation file gff. Additionally, we predicted the molecular weight (MW) and isoelectric point (PI) of the family members using the ExPasy website (http://web.expasy.org/). Moreover, we employed the MEME (Bailey et al., 2015) website (http://meme-suite.org/) to predict amino acid conserved motifs. Furthermore, we predicted the protein structural domains of NHE and NKA gene family members using the SMATR website. Finally, we utilized the TBtools software to map the NHE and NKA gene family structures and genomic localization.
2.5 Evolutionary analysis of the NHE and NKA gene families in R. canadum
The NHE and NKA family members from Homo sapiens, Mus musculus, zebrafish (Danio rerio), S. maximus, Rainbow Trout (Oncorhynchus mykiss), and L. japonicus were retrieved from the NCBI database. These sequences served as references for multiple amino acid sequence comparisons and homology analyses, which were performed using ClustalX1.83. The resulting phylogenetic tree was constructed by applying the neighbor-joining method (NJ) through MEGA-X (Kumar et al., 2016) software.
2.6 RNA-seq of NHE and NKA gene families in R. canadum
To investigate the impact of different salinity acclimation conditions on the expression patterns of NHE and NKA, RNA was extracted from the gills, intestine, and body kidney of R. canadum following 4 weeks of culture in salinities of 10‰, 30‰, and 35‰. RNA from nine fish in each salinity group was pooled to obtain one sample, and Illumina Hiseq™2000 was used to sequence the transcriptome. The raw mRNA sequencing data has been deposited in the NCBI Sequence Read Archive (SRA) under the accession number SRP202920 (published by our research groups) (Cao et al., 2020).
The raw mRNA sequencing data was processed using fastp (Chen et al., 2018) to remove low-quality data, and the remaining clean reads were mapped to the R. canadum genome (PRJNA634421) using HISAT2 software (Kim et al., 2015). StringTie software (Pertea et al., 2015) was then used to assemble the mapped reads. The expression of all genes in each sample (FPKM and reads count) was then calculated using RSEM (Li and Dewey, 2011), and the read count was normalized and analyzed for differentially expressed genes using edgeR (Robinson et al., 2010) (P<0.05 for significantly differentially expressed genes, FDR<0.05 and |log2FC|> 1 for highly significant differentially expressed genes).The resulting expression data (log2FPKM) were utilized to generate a gene expression heat map using TBtools, and correlation analysis was performed.
2.7 Analysis of qPCR expression of NHE and NKA genes in R. canadum
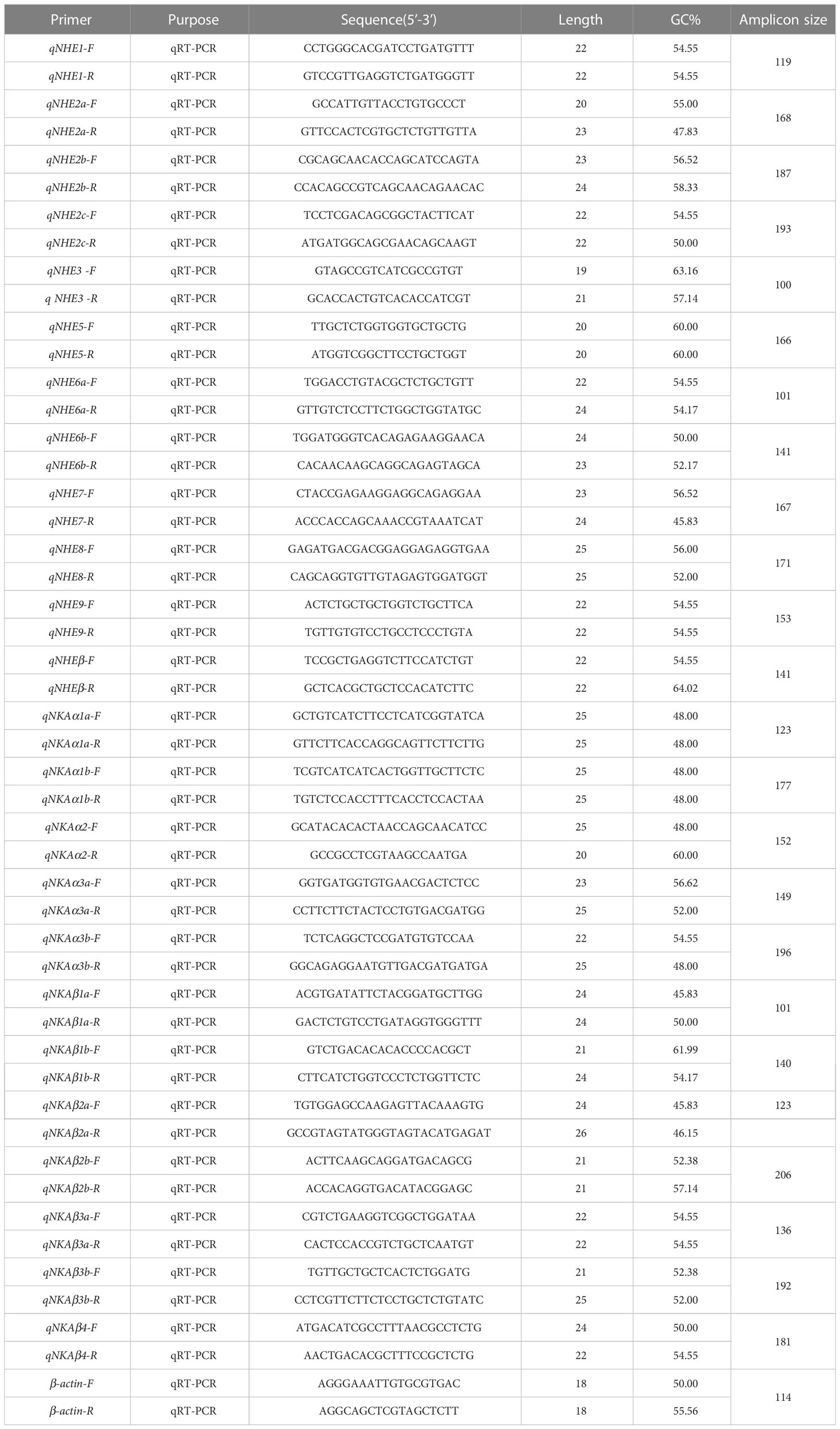
Gene-specific primers were designed based on the cDNA sequences for NHE and NKA gene family members, resulting in the amplification of fragments ranging from 100-230 bp (Table 1). β-actin was chosen as the reference gene. qRT-PCR was performed using a Roche Light Cycler™ 96 real-time PCR machine and SYBR®Select Master Mix. The expression levels of the three genes in 9 tissues and the expression levels of the genes in osmoregulatory organs such as gills, intestines, and kidneys after salinity adaptation were determined.
The amplification program consisted of an initial denaturation step at 95°C for 10 min followed by 40 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 20 s, and extension at 72°C for 20 s. To minimize errors, three different R. canadum individuals were sampled for each salt treatment, and qPCR was repeated three times for each individual. The expression levels of NHE1, NHE2a, NHE2c, NHE5, NKAα1b, NKAα3a, NKAβ1b, and NKAβ3a genes were analyzed using the 2-ΔΔCt method, and one-way ANOVA (LSD, Duncan) was performed using SPSS22.0 software.
3 Results
3.1 Analysis of organizational structure of R. canadum after salinity adaptation
After 30 days of domestication in low salinity water (10 ppt), the length (width) of gill filaments and gill lamellae of R. canadum increased significantly. The spacing between gill lamellae decreased, and the cells of gill lamellae were rounded and full. The number of chloride-secreting cells on gill filaments and gill lamellae decreased significantly. In the high salinity group (35 ppt), the number of chloride-secreting cells on gill filaments and gill lamellae increased slightly but not significantly. The width of gill filaments, gill lamellae, and cartilage tissues decreased significantly, and the spacing of gill lamellae increased (Figure 1A).
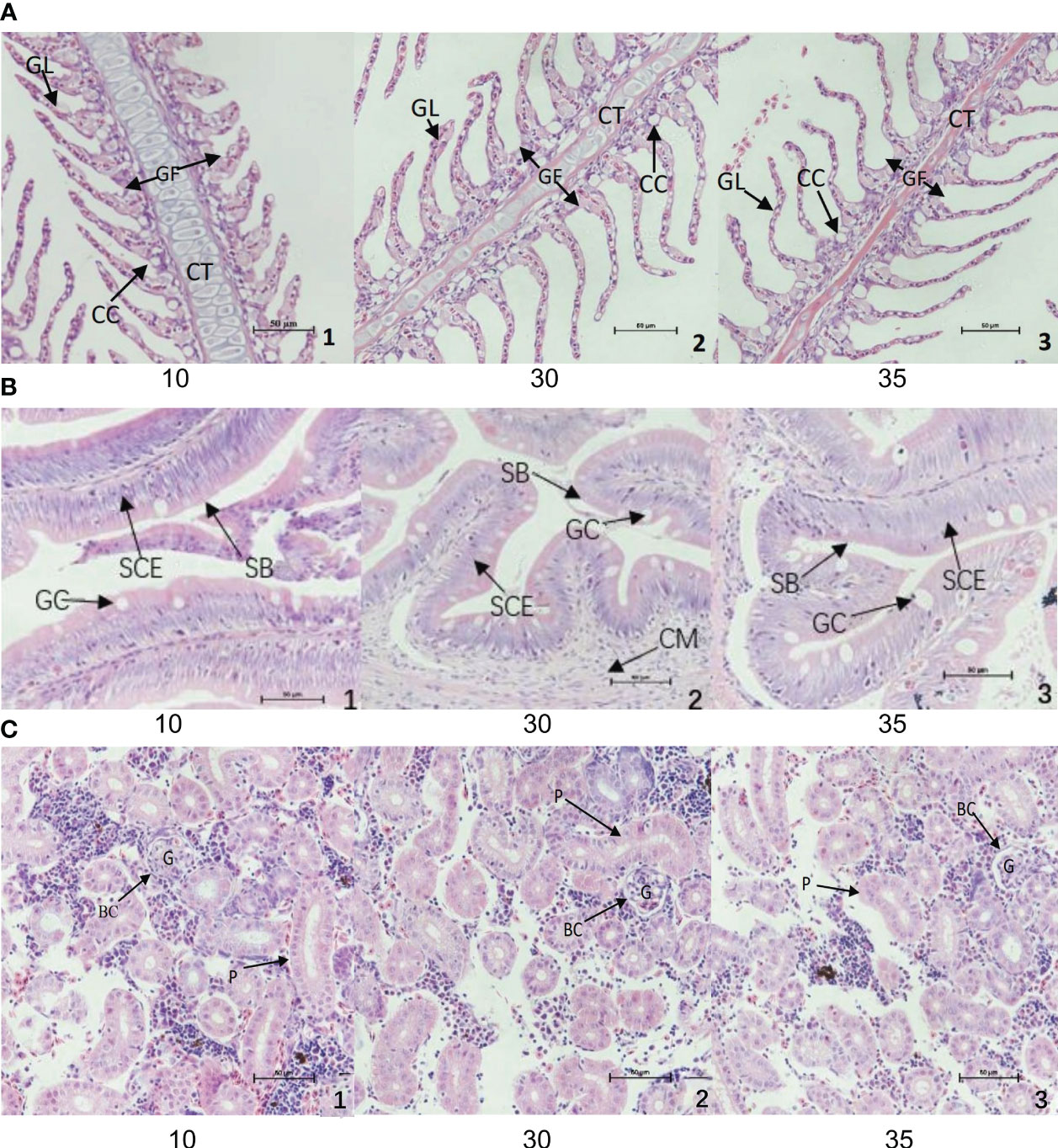
Figure 1 Structural changes of gill (A), intestine (B) and body kidney (C) of juvenile of R. canadum. 10, salinity 10 ‰; 30, salinity 30 ‰; 35, salinity 35 ‰. GF, gill filaments; GL, gill small pieces; CC, chlorinated cells; CT, cartilage tissue; SCE, monolayer columnar epithelium; SB, striatum; GC, goblet cells; CM, ring muscle; G, glomerulus; BC, renal capsule; P, renal tubule.
The microstructure of the intestine of juvenile R. canadum in the control group (30 ppt) showed that the single layer of columnar epithelium on the intestinal villi of juvenile R. canadum in the low salinity group became thicker, and the number of cupped cells decreased significantly. The size of the cupped cells did not change significantly. In the high-salinity group, the cytosol of the cup-shaped cells was enlarged, and the thickness of the unilamellar columnar epithelium and the number of cup-shaped cells on the intestinal villi did not change significantly (Figure 1B).
In the low salinity group, the tubular diameter of all levels of renal tubules of R. canadum increased, and the glomerulus was enlarged, full, and filled. The lumen of its capsule was small. In the high salinity group, the glomerulus atrophied, the lumen of the glomerular capsule increased, and the tubular diameter of all levels of renal tubules decreased slightly (Figure 1C).
3.2 Identification of NHE and NKA gene family members in R. canadum
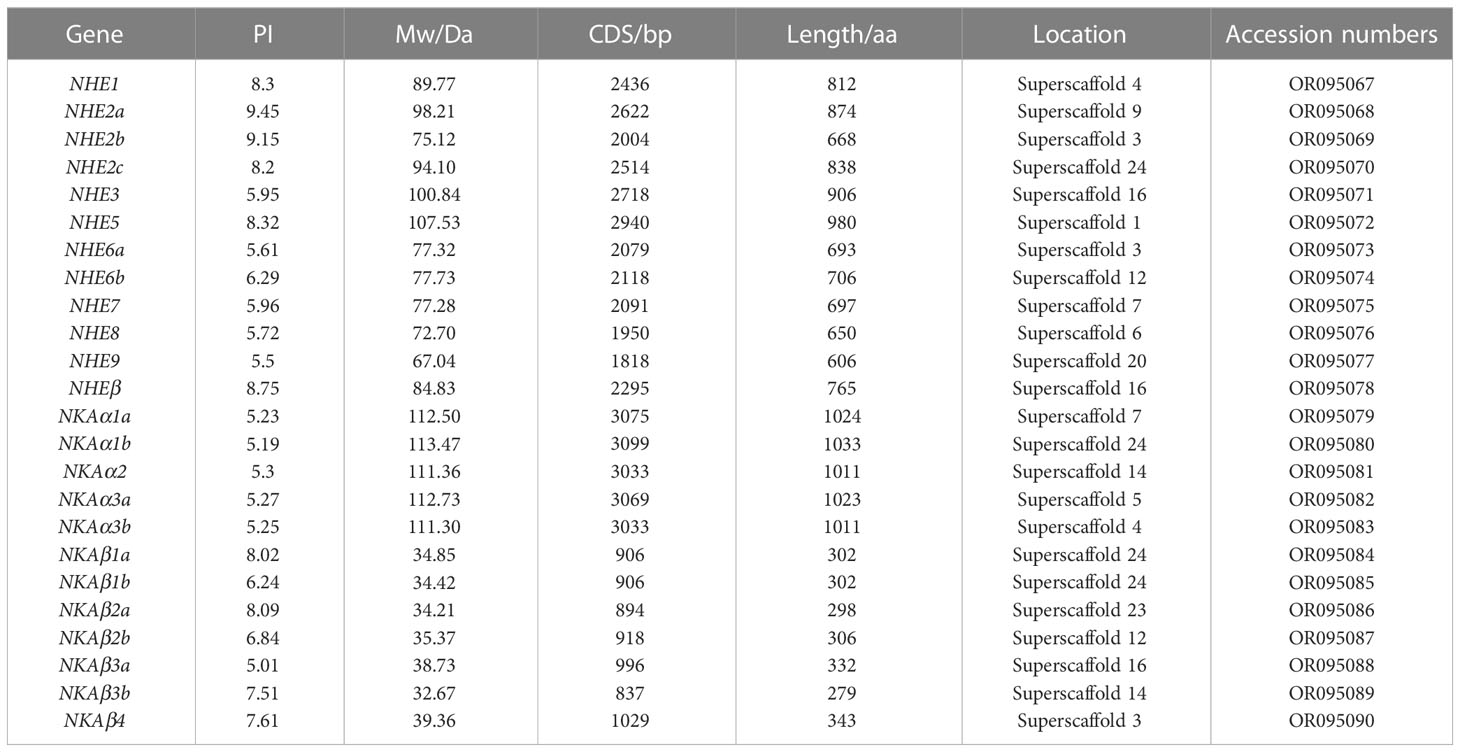
In this study, the NHE and NKA families were characterized using genome−wide data and an HMM model. The analysis identified a total of 12 NHE family members and 12 NKA family members, including both single-copy and multi-copy genes. Specifically, NHE2 and NHE6 were found to be multi-copy genes, while NKA1, NKA3, and NKAβ1~3 were also identified as multi-copy genes. The coding sequence (CDS) of NHE genes ranged from 1818 to 2940 bp in length, with amino acid sizes ranging from 606 to 980 aa. The PI ranged from 5.5 to 9.45, and the Mw ranged from 67.04 kD to 107.53 kD. Similarly, the CDS of NKA genes ranged from 837 to 3099 bp in length, with amino acid sizes ranging from 279 to 1033 aa. The PI ranged from 5.01 to 8.09, and the Mw ranged from 32.67 kD to 113.47 kD (Table 2).
3.3 Structural analysis and genomic localization of the NHE and NKA gene families in R. canadum
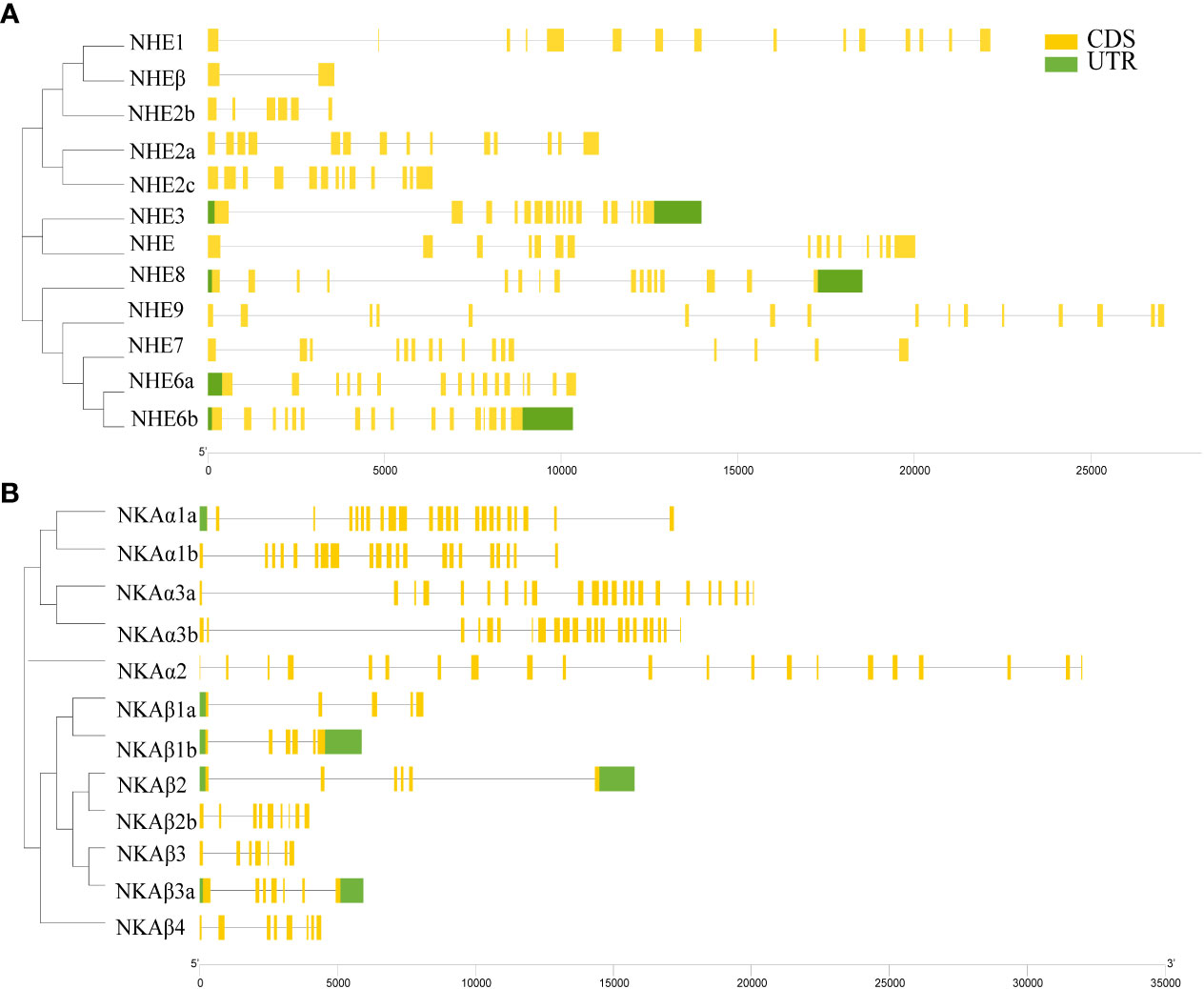
The NHE family members of R. canadum had 2 to 16 Coding DNA Sequence (CDS), with NHE3, NHE7, NHE8, and NHE9 having 16 CDS, and NHEβ having the least number of CDS with only 2, which might be attributed to genome assembly issues. In contrast, NHE1 had the highest number of CDS with 15 (Figure 2A). The number of CDS of NKA family members ranged from 5 to 23, with NKAα members and NKAβ members showing polarized CDS numbers, where none of the former had less than 21 and all of the latter had less than 10 CDS, implying a correlation between CDS numbers and subtype classification (Figure 2B).
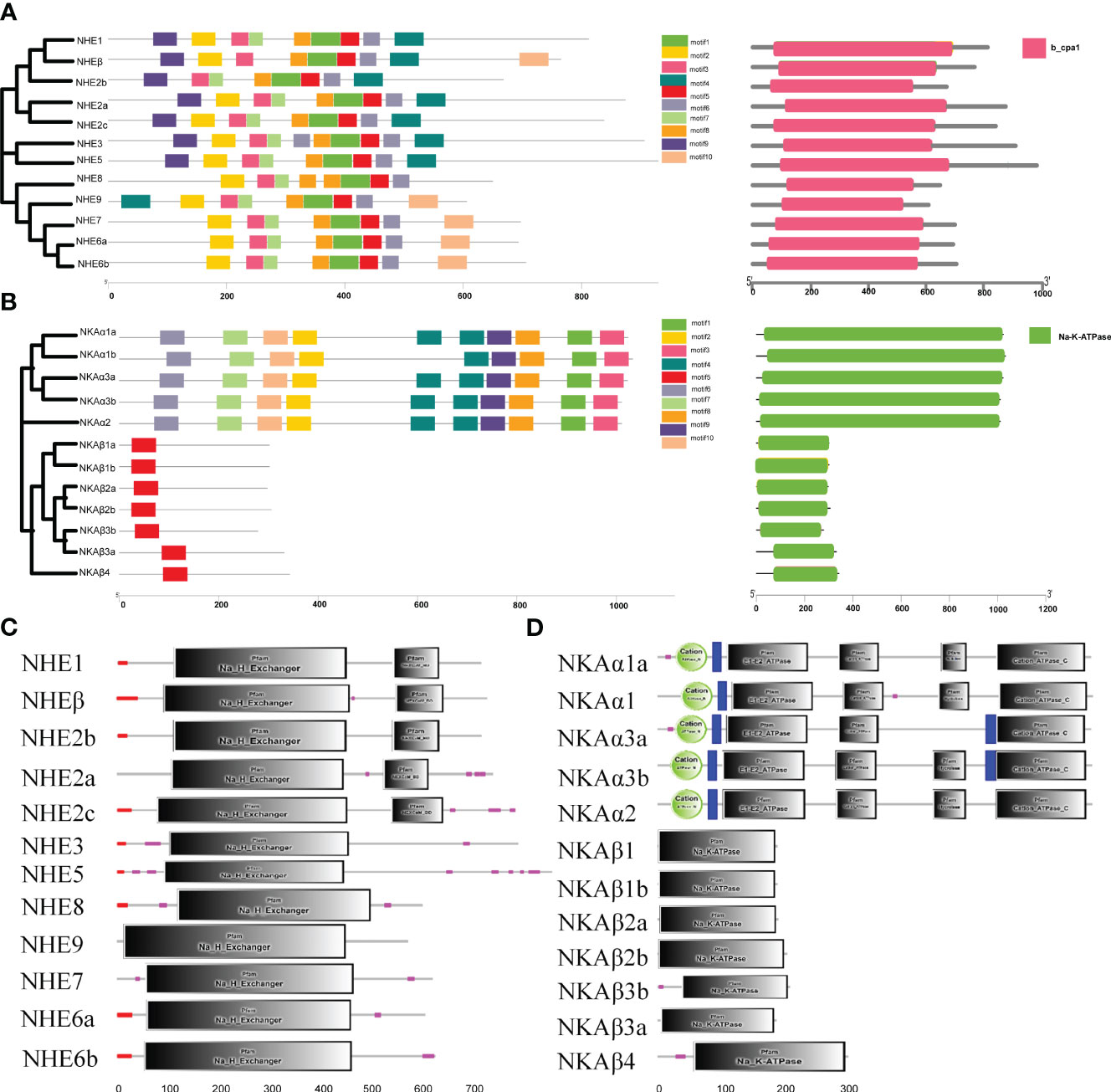
The present study aimed to analyze the motif composition of NHE and NKA genes in R. canadum using the MEME website. The results showed that both gene families contained 10 motifs arranged in an organized and regular manner. Most motifs of NHE genes were associated with Na+/H+ exchanger structural domains, except for motif4, while all motifs identified in NKA were associated with Cation transporter/ATPase and Hydrolase structural domains. Further analysis revealed that NHE motif1 and motif4 were mainly identified in NHE1~5, and motif 8 appeared only twice in NHE8 and once in all other members. In contrast, motif4 appeared in the anterior segment of the NHE9 sequence. Furthermore, NHEβ lacked motif7 and had more motif10 compared to NHE1(Figure 3A). Concerning the NKA gene family, motif10 was found only in the NKAβ isoform, while the remaining nine motifs were ordered in the NKAα isoform (Figure 3B). The only difference was that motif4 was missing in NKAα1b.
The domain information of NHE and NKA genes in R. canadum was predicted using the SMART website, which revealed that NHE genes contained CPA1 and NHE structural domains (Figure 3C), and NKA genes contained NKA and CPA-N/C structural domains (Figure 3D).
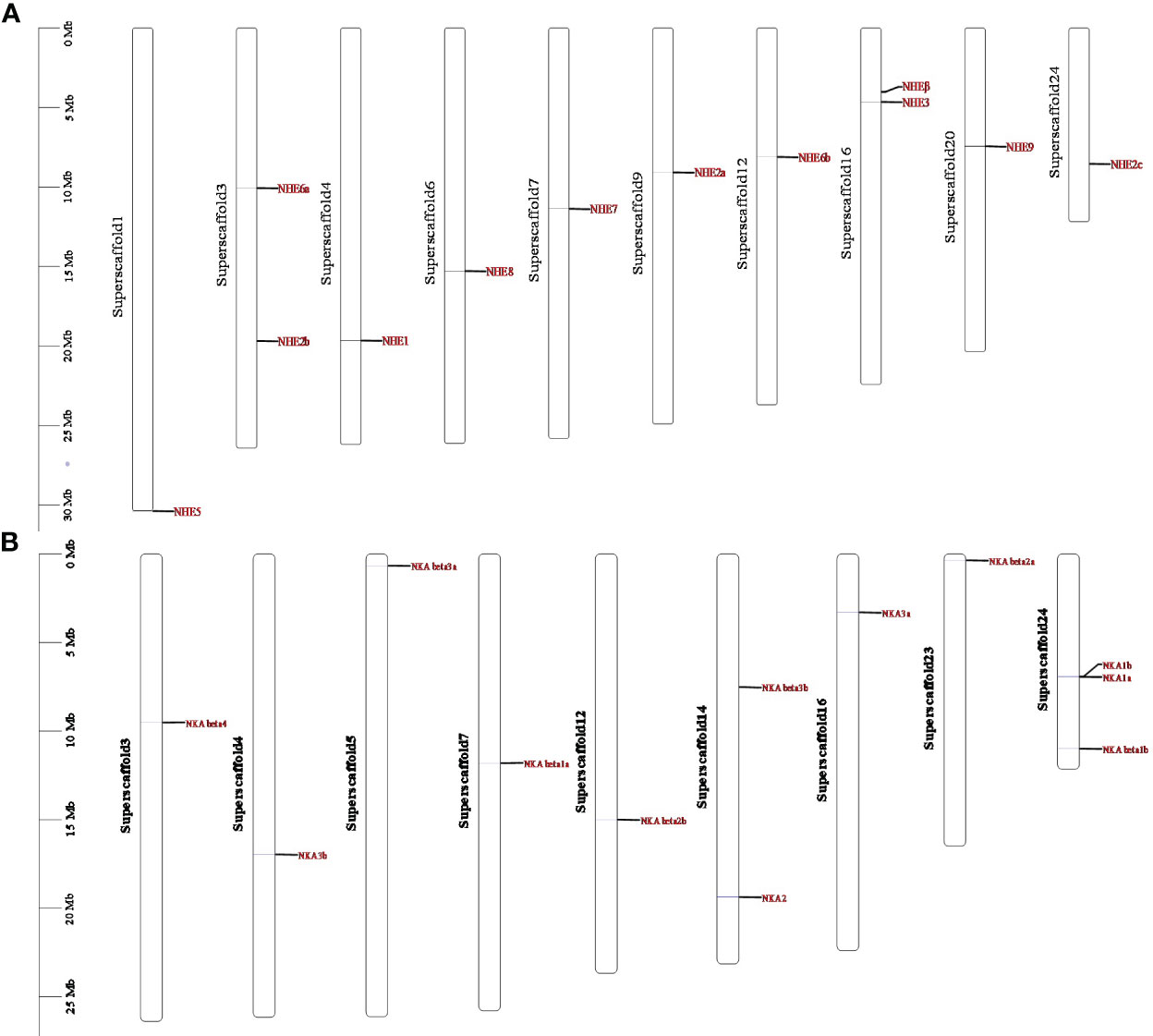
Genomic localization shows that members of the NHE and NKA gene families of R. canadum localize to 10 and 9 superscaffolds, respectively (Figure 4). Specifically, NHE2b and NHE6a were present simultaneously on superscaffold16 and superscaffold3, respectively, while the remaining eight members of NHE family were distributed randomly on a single superscaffold (Figure 4A). Similarly, 2-3 members of NKA family (NKAβ1b, NKAα1b, and NKAα1a; NKAα2 and NKAβ3) were simultaneously present on superscaffold24 and superscaffold14, respectively. In contrast, the remaining eight members of NKA family were randomly distributed on a single superscaffold (Figure 4B). Moreover, the multi-copy members of NHE family, NHE2a-c, were localized to the 9th, 3rd, and 24th superscaffold, respectively, whereas NHE6a and NHE6b were localized to the 3rd and 12th superscaffold, respectively (Figure 4A) multi-copy. Additionally, the multi-copy members of NKA family, NKAβ1-3 and NKAα3a and NKAα3b, were identified in the 7th, 24th, 23rd, 12th, 5th, and 14th superscaffold, respectively, and NKAα1a and NKAα1b were localized in both superscaffold 24 (Figure 4B).
3.4 Phylogenetic analysis of the NHE and NKA gene families in R. canadum
The NHE and NKA gene family members of six species, including Homo sapiens, Mus musculus, D. rerio, S. maximus, O. mykiss, and Lateolabrax maculatus, were used as references to construct a phylogenetic evolutionary tree, which further verified the accuracy of the annotation of the NHE and NKA genes of R. canadum and revealed the variation of these genes during species evolution (Figure 5). The analysis indicated that NHE1~9 of R. canadum were most closely related to teleost and furthest from mammals. NHE1 and NHEβ were clustered into a single clade with teleost such as D. rerio and S. maximus, respectively, before merging into one clade; NHE3 and NHE5 of R. canadum were merged into one clade, while H. sapiens and M. musculus were separate clades. The multi-copy genes NHE2a~c were clustered with other species and re-clustered with mammalian NHE4, respectively; NHE6a and NHE6b were alone, and their closest relatives were L. japonicus, S. maximus, D. rerio and O. mykiss (Figure 5A). Additionally, the NKAα subtype gene and NKAβ subtype gene of R. canadum were separately divided into the same branch with other species and were most closely located with teleost. Among them, NKAα4 of H. sapiens and M. musculus were separately merged with NKAβ1a of R. canadum. Moreover, NKAα1a~b and NKAα2 of R. canadum were closest together and merged into one branch, and NKAα1 and NKAα2 of H. sapiens and M. musculus were independently into one branch (Figure 5B).
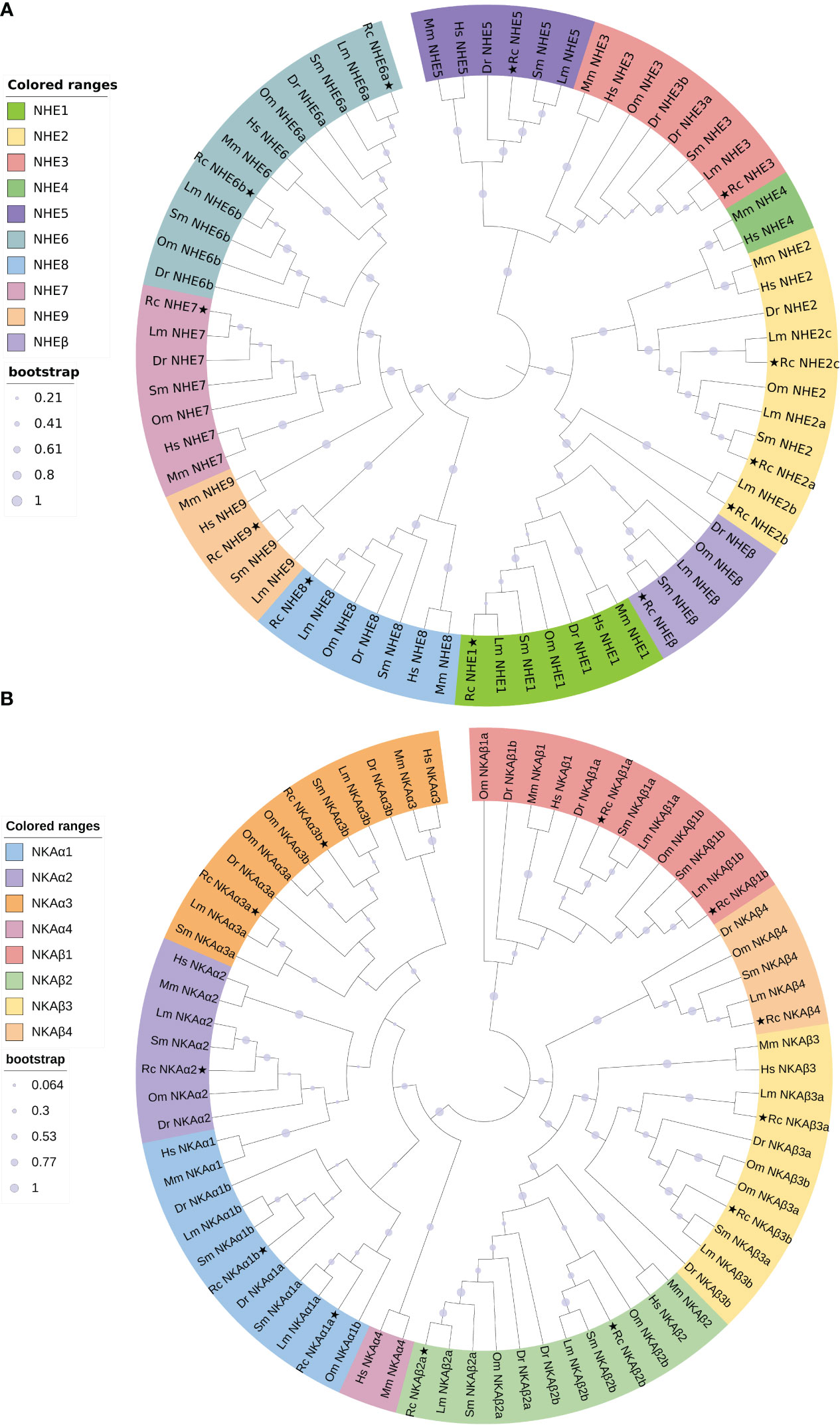
Figure 5 Phylogenetic analysis trees of R. canadum NHE (A) and NKA (B) gene families. Rc, R. canadum; Dr, D. rerio; Sm, S. maximus; Lm, L. maculatus; Hs, H. sapiens; Mm, M. musculus.
3.5 Analysis of the expression patterns of NHE and NKA gene families in R. canadum
To investigate the expression patterns of the NHE and NKA gene families in various tissues of R. canadum under normal seawater salinity, qRT-PCR was used to determine the gene expression abundance in nine different tissues. The results demonstrated that the NHE and NKA family members of R. canadum were widely expressed in all tissues, including the gill, brain, heart, intestine, kidney, liver, spleen, stomach and muscle(Figure 6). Specifically, the tissues with high expression of NHE were the gill, somatic kidney, and brain, while NKA was highly expressed in the gill, brain, and heart.
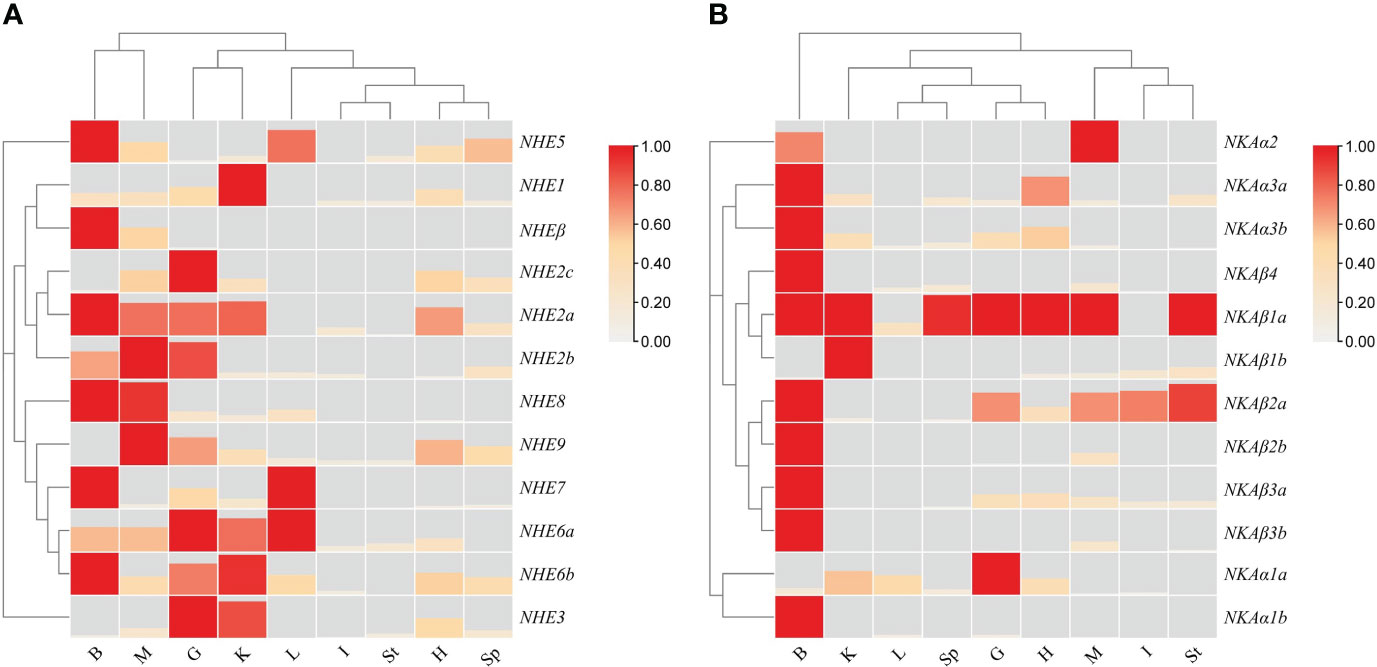
Figure 6 Heat map of tissue expression of NHEs (A) and NKAs (B) in R. canadum. G, gill; I:intestine; K,Kidney; L, Liver; H, Heart; B, Brain; Sp, Spleen; St, Stomach; M, Muscle.
Of the single-copy genes, NHE1 and NHE3 showed similar expression patterns and were mainly concentrated in the gill and somatic kidney. On the other hand, NHE5 and NHE7 were highly expressed in the liver and brain, respectively. The highest expression signals of multi-copy genes NHE2a and NHE2b were detected in the brain and muscle, respectively, while the highest expression tissues were the gill and liver for NHE6a, and the somatic kidney and brain for NHE6b multi-copy. Furthermore, NHEβ and NHE1 expression patterns were inconsistent, with high expression detected only in the brain and trace expression in other tissues (Figure 6A).
The NKA family members NKAα1a, NKAβ1b, NKAβ2a, and NKAβ3a were highly expressed in osmolarity-regulating organs, such as the gill, intestine, and somatic kidney, respectively. Among them, NKAα1a was the most highly expressed in the gill. NKAα2 was expressed only in the brain and muscle, while NKAβ4 was highly expressed in the brain, with lower expression levels in other tissues. Most of the multi-copy genes (NKAα1b, NKAα3a, NKAα3b, NKAβ1a, NKAβ2a, NKAβ2b, NKAβ3a, and NKAβ3b) were highly expressed in brain tissues, with NKAα1b only detected as a fluorescent signal in brain tissues, not consistent with NKAα1a. NKAα3a and NKAα3b showed similar expression patterns, with high expression in the brain, heart, somatic kidney, and gill in descending order. NKAβ1a was highly expressed in several tissues, mainly in the brain, stomach, and muscle, while NKAβ1b was highly expressed mainly in the somatic kidney. NKAβ2a was highly expressed in the gill, intestine, brain, stomach, and muscle, while NKAβ2b was hardly expressed except in the brain and muscle. The expression pattern difference of NKAβ3a-NKAβ3b was similar to that of NKAβ2a-NKAβ2b. Additionally, fluorescent signals of other members were less frequently detected in the liver, spleen, and stomach, except for NKAα1a, NKAβ1a, and NKAβ2a, which showed expression (Figure 6B).
3.6 Transcriptome analysis of the NHE and NKA gene families in R. canadum
The effects of salinity acclimation on the expression patterns of NHE and NKA gene families in different tissues of R. canadum was investigated using a reference transcriptome sequencing approach and validated by qRT-PCR. The results demonstrated that the expression patterns of NHE and NKA genes were influenced by salinity and exhibited tissue-specific characteristics (Figure 7). In gill tissues, the expression of NHE3 was significantly down-regulated under salinity 10 and 35 acclimation conditions compared to the control group acclimated at salinity 30. Additionally, NHE6b and NHE8 were significantly down-regulated under salinity 35 acclimation conditions, while NHE2a was significantly down-regulated under salinity 10 acclimation conditions. However, the experimental group exhibited significant upregulation in NHE2c expression, and NHE7 showed significant upregulation under salinity 35 acclimation conditions. In intestinal tissues, the expression levels of NHE genes were significantly increased under experimental conditions (salinity 10 and 35) compared to the salinity 30 control. Specifically, NHE7 was significantly upregulated under both low (10) and high (35) salinity acclimation conditions, while NHE1 and NHE6a-b were only significantly upregulated under salinity 35 acclimation conditions. NHE9 exhibited significant upregulation under salinity 10 acclimation conditions. In kidney tissues, NHE3 expression increased with higher salinity and showed highly significant upregulation at salinity 35, along with significant downregulation at salinity 10 compared to the salinity 30 control. NHE6a exhibited significant upregulation at salinity 35, whereas NHE2a showed significant downregulation at salinity 10. NHE5 and NHEβ expression levels remained relatively stable across all three salinity acclimation conditions in all tissues, while NHE2b and NHE5 expression levels were comparatively low under all conditions (Figure 7A).
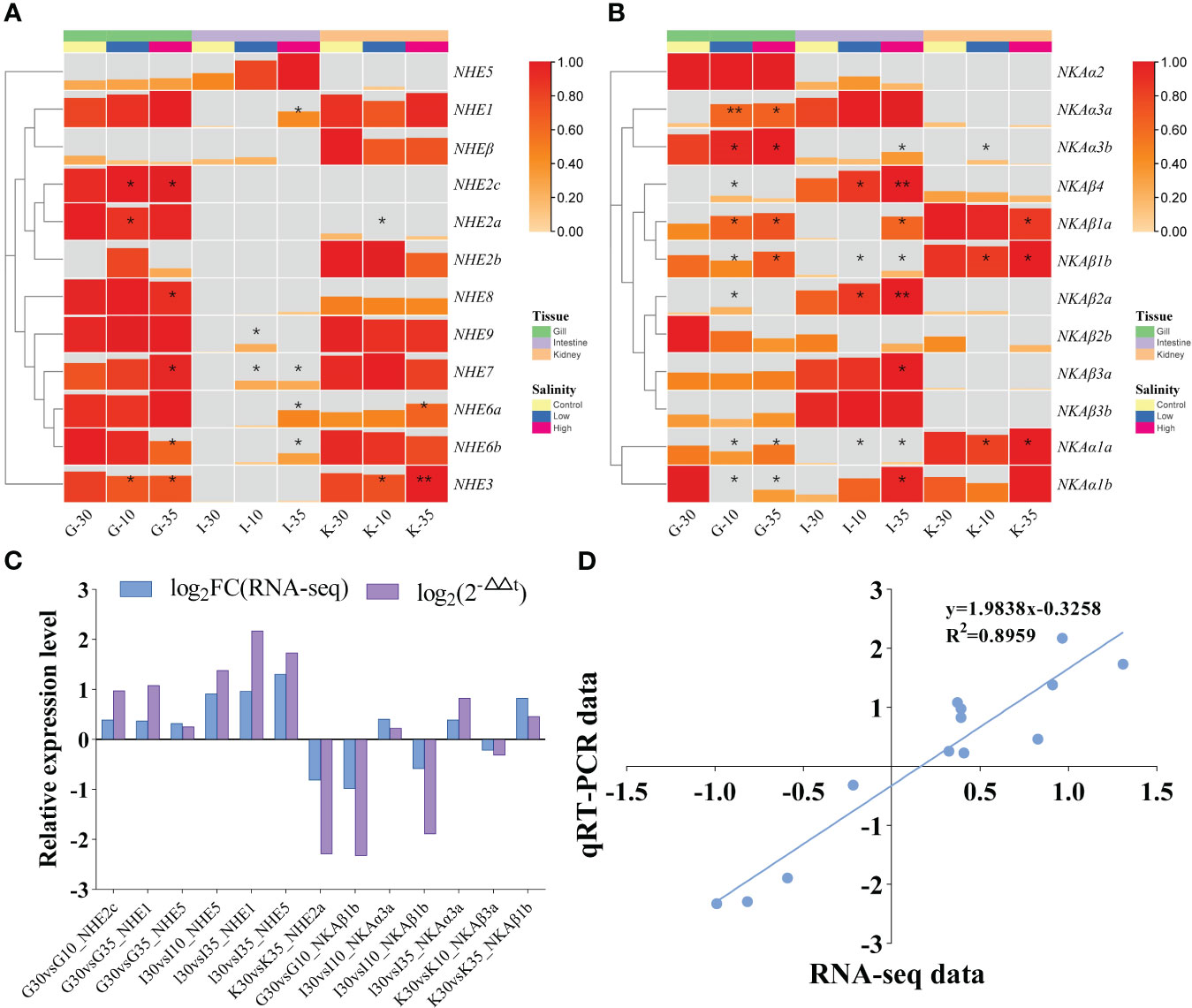
Figure 7 Transcriptome expression patterns of NHEs (A) and NKAs (B) of R. canadum under different salinity acclimation. Relative expression level (C) and linear regression (D) of RNA-Seq and qRT-PCR data that are expressed as a log2 fold change. 10, salinity 10 ‰; 30, salinity 30 ‰; 35, salinity 35 ‰; G, gill; I, intestine; K, kidney.
In gill tissues, NKAα3a, NKAα3b, and NKAβ1a expression levels were significantly upregulated under salinity acclimation conditions of 10 and 35, with NKAα3a displaying particularly highly significant upregulation at salinity 10 compared to the control group at salinity 30. Conversely, NKAα1b expression was significantly downregulated at both salinities 10 and 35. In gill, intestinal, and kidney tissues, the expression levels of NKAα1a and NKAβ1b showed significant increases with increasing salinity. In intestinal tissues, NKAβ2a and NKAβ4 expression levels were significantly upregulated at salinity acclimation condition of 10, and exhibited highly significant upregulation at salinity acclimation condition of 35. Additionally, NKAα1b, NKAα3b, NKAβ1a, and NKAβ3a were significantly upregulated at salinity acclimation condition of 35 compared to the control. In kidney tissues, NKAβ1a expression was significantly downregulated at salinity 35, while NKAα3b showed significant upregulation at salinity 10 compared to the control group. However, the expression levels of NKAα2, NKAα3a, and NKAβ2b were relatively low under all conditions (Figure 7B). The qRT-PCR validation and RNA-seq were in general agreement in terms of the fold change in differential expression (Figure 7C). In the linear regression analysis of trend changes, R2 = 0.8959 (Figure 7D). The results indicate that the gene expression analysis based on RNA-Seq data is reliable.
3.7 Salinity-adapted qPCR expression analysis of the NHE and NKA gene families of R. canadum
To investigate the differences in the expression patterns of some members of the NHE and NKA gene families of R. canadum under salinity acclimation, qPCR was performed on R. canadum acclimated to salinities of 10‰, 30‰ and 35‰ for 4 weeks in this study. The results showed that the relative expression of NHE1 was significantly up-regulated in the intestine and down-regulated in the kidney after increasing or decreasing salinity, while the expression was not significantly increased in the gills after decreasing salinity (Figure 8A). In addition, NHE2a expression in the gill did not change significantly in low-salt acclimation, while it was significantly down-regulated in high-salt acclimation (Figure 8B). NHE2c was significantly down-regulated in gill, intestine and somatic kidney with increasing salinity (Figure 8C), and NHE5 was also significantly down-regulated in somatic kidney. In addition, the expression pattern of NHE5 in both gill and intestine was significantly different in a “U” pattern (Figure 8D).
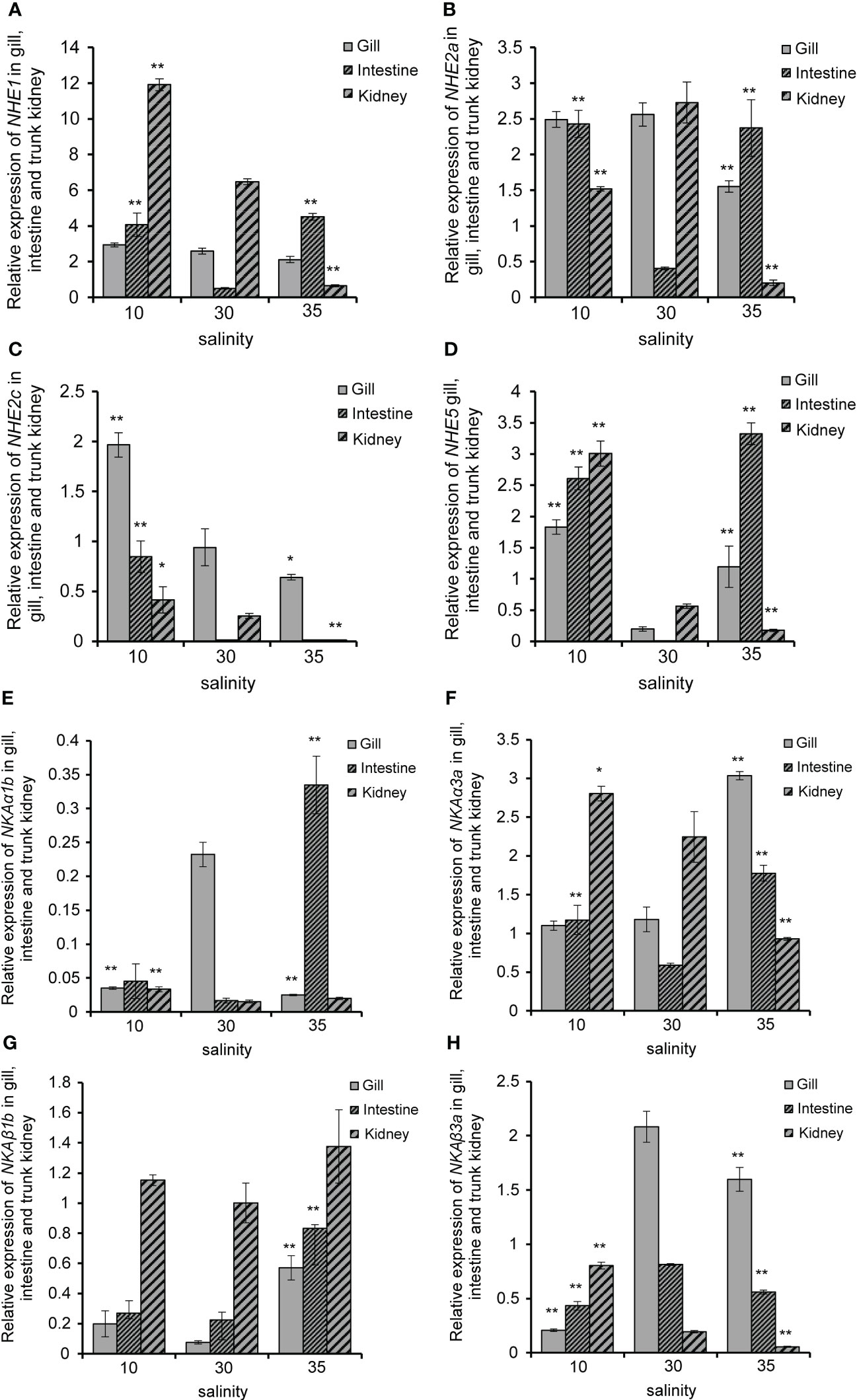
Figure 8 Relative expression of R. canadum NHE1 (A), NHE2a (B), NHE2 (C), NHE5 (D), NKAα1b(E), NKAα3a(F), NKAβ1b (G) and NKAβ3a (H) in different tissue after salinity adaption. 10, salinity 10 ‰; 30, salinity 30 ‰; 35, salinity 35 ‰. * and* * indicated that the expression of each gene was significantly different from that of the control group, and the significance levels were P < 0.05,0.01, respectively.
Following low-salt acclimation, significant down-regulation of NKAα1b and NKAβ3a was observed in the gills of R. canadum (Figures 8E, H), while NKAα3a and NKAβ1b did not exhibit significant changes (Figures 8F, G). In the intestine, NKAα1b and NKAβ1b did not exhibit significant changes (Figures 8E, G), whereas NKAα3a and NKAβ3a were significantly up-regulated and down-regulated, respectively (Figures 8F, H). In the somatic kidney, NKAα1b, NKAα3a, and NKAβ3a showed significant up-regulation (Figures 8E, F, H), while NKAβ1b did not change significantly (Figure 8G). Upon high-salt acclimation, the expression of NKAα1b and NKAβ3a in the gills remained consistent with low-salt acclimation (Figures 8E, H), while NKAα3a and NKAβ1b exhibited significant up-regulation (Figures 8F, G). In the intestine, the expression levels of NKAα1b, NKAα3a, and NKAβ1b were significantly increased (Figures 8E, F, G), while NKAβ3a expression levels were significantly decreased (Figure 8H). The expression pattern of NKAα3a and NKAβ3a in the somatic kidney was consistent with low-salt adaptation (Figures 8F, H), while NKAα1b and NKAβ1b expression were unaffected by salinity (Figures 8E, G).
4 Discussions
The process of salinity adaptation in fish can be divided into two stages: passive adaptation to the external environment and active osmoregulation (Li et al., 2022). The key to salinity adaptation in fish is ion transport, and this regulation is primarily performed by osmoregulatory organs such as the gills, kidneys, and intestines (Whittamore, 2012; Dawood et al., 2021; Ali et al., 2022). The gills are the major organ of osmoregulation in fish and play an important role in maintaining the balance between the internal and external environment of the fish, which is closely related to the ion transport gene sodium/potassium pump (NKA) on the cell membrane (Dawood et al., 2021). In this study, the gill filaments and gill lamellae of juvenile R. canadum in the low salinity group were more round and full, and wider than those in the control group. On L. japonicus, Acipenser schrenckii and Gymnocypris przewalskii, gill tissues underwent similar adaptive changes in order to maintain osmoregulatory homeostasis (Hou et al., 2006; Wang and Hu, 2009; Huang et al., 2022). This is because the gill filaments and gill lamellae of juvenile fish living in a desalinated environment tend to change to wider and longer to facilitate sufficient contact with the water column and thus take up inorganic ions in the water to adapt to the hypotonic environment (Yu et al., 2012; Li et al., 2014). Conversely, gill filament cells shrank, and gill lamellae spacing increased at high salinity. These changes increased water and oxygen exchange between the fish and the external environment, which promoted better survival of fish under high salinity conditions (Yang et al., 2014). In this experiment, the number of chloride-secreting cells in R. canadum decreased at low salinity and increased at high salinity. Chloride-secreting cells are important regulatory cells that adapt to different salinities in euryhaline fish, and they have abundant Na+/K+-ATPase on the microtubule system in their cytoplasm (Sakamoto et al., 2001). During salinity changes, chloride-secreting cells secrete Cl- in hypotonic regulation and absorb Na+ and Cl- in hypertonic regulation to regulate osmotic pressure homeostasis (Foskett et al., 1983; Marshall, 2011). This shows that the gills of R. canadum have certain adaptability when the external salinity changes.
The intestine of fish plays a smaller role in osmoregulation, but it has slao been shown that the intestinal epithelial cell membrane can be involved in active transmembrane transport of Na+, K+ and Cl- inside and outside the cell through ion channel transporter proteins, combined with passive transport of water to maintain osmotic pressure homeostasis in the body (Li et al., 2011). In our study, we observed changes in the intestinal villi structure of juvenile R. canadum in the low salinity group, where the monolayer columnar epithelium of intestinal villi became thicker. These results are consistent with the speculation of Sun et al. (2016). who hypothesized that low salinity has a detrimental effect on the intestine of Takifugu rubripes. We also observed changes in cupped cells, which secrete mucus to lubricate the epithelial surface and remove waste (Kibenge and Strange, 2021). The number of cupped cells decreased at low salinity, while the cytosol of cupped cells became larger at high salinity. Further research is necessary to determine whether these changes in cupped cells are related to salinity adaptation in juvenile R. canadum. The kidney is another important organ in the regulation of osmotic pressure in fish, as the glomerulus filters blood cells and macromolecular proteins from the blood, and the renal tubules reabsorb water, glucose and amino acids (Smith, 1932; Gonzalez, 2012). In this study, the tubular diameter of all levels of renal tubules in the low-salinity group became larger, the glomeruli were expanded, full and filled, and the gap in the inner wall of the renal capsule was reduced; in the high-salinity group, the glomeruli were atrophied and the gap in the inner wall of the renal capsule was increased. This structural change was beneficial to the filtration and reabsorption of nutrients in the blood of juvenile R. canadum.
Changes in water salinity have a significant impact on the osmotic pressure of fish organisms, and fish have developed various mechanisms to counteract the negative effects of salinity by activating activities such as their own water-salt regulation (Gonzalez, 2012). Sodium hydrogen exchanger (NHE) is a class of ion channel proteins present in most species, involved in the intracellular and extracellular transport of Na+ and H+ ions, and plays a critical role in cellular acid-base homeostasis, cell volume regulation, and osmotic pressure regulatory networks in fish, such as Na+ reabsorption in the kidney, stomach, and intestine (Edwards et al., 2005). NHE3, one of the NHE family members, also forms an ion regulatory network in teleost with NKA, NKCC1, NCC, and CFTR, which together maintain salt secretion and ion transport of Na+, K+, and Cl- in and out of the membrane (Marshall, 2011). Current studies in fish osmoregulation have focused on NHE2 and NHE3. For example, Yan et al. (2007) reported that D. rerio treated with soft water had increased expression of DrNHE2 and DrNHE3 genes in the gills and were enriched in mitochondrion-rich cells(MRCs). The gill of LjNHE3 expression level of L. japonicus was upregulated after low salt stress (Inokuchi et al., 2017). The number of NHE family members varies somewhat among species; for instance, NHEβ4 is identified in mammals such as H. sapiens and M. musculus, but not in teleost such as D. rerio, Oryzias latipes, and Dicentrarchus labrax (Tine et al., 2014). Sodium/potassium-transporting ATPase (NKA), the main active pump in the gill, is responsible for Na+ ion transport as well as NHE, except that it empowers the ion transport system of multiple osmolarity-regulated epithelial cells by hydrolyzing ATP (Lin et al., 2003). It is now known that in teleosts, NKA exists in both α and β subunits and is classified as such (Zhang et al., 2019). Among them, the former is responsible for adenosine triphosphatase catalysis and Na+ and K+ transport activities, while the latter is mainly responsible for auxiliary α-subunit folding (Sundh et al., 2014). A variety of NKAα isoforms are expressed in fish osmoregulatory organs, and their expression levels are positively or negatively correlated with salinity changes, indicating that different NKA isoforms are involved in osmoregulatory processes and have different roles (Yang et al., 2016; Hu et al., 2017).Further studies on the NHE and NKA gene families may provide insights into the potential functions of the remaining members and enhance our understanding of osmoregulation in fish.
A total of 12 NHE genes and 12 NKA genes were identified in R. canadum. These genes were found to be distributed randomly on 10 and 9 superscaffolds, respectively, which is consistent with the findings reported by Zhang et al. (2019). The gene sequences of NHE and NKA gene family members were compared with those of O. mykiss, L. japonicus and S. maximus to name them and analyze the conserved motifs and structural domain characteristics (Berthelot et al., 2014; Figueras et al., 2016; Gao et al., 2022). The results revealed that the motifs and domains of the NHE genes were concentrated in the Na+/H+ exchanger domains, which the identification of the NKA also showed that they closely association with Cation transporter/ATPase and Hydrolase domains, and the motifs and domains of different members were arranged in different patterns. This indicates that the sequence characteristics of NHE and NKA members are closely related to their taxonomy and functions. In other teleost fishes, NHE4 is commonly missing and NHEβ is only found in teleost fishes (Kasahara et al., 2007; Howe et al., 2013; Tine et al., 2014). In this study, NHE4 was lost but NHEβ was identified by phylogenetic analysis. The results shown that NHE and NKA genes were the closest to teleosts in taxonomic status and the farthest from mammals, indicating that the protein sequences of the two family members were highly conserved in species evolution. The results of qRT-PCR showed that NHE and NKA family members were widely distributed in nine tissues such as heart, liver and spleen, and the expression patterns of NHE multi-copy gene members were different, such as NHE2a and NHE2b high expression tissues. However, the expression patterns of NKA multi-copy gene members are partially similar, all of which are highly expressed in brain tissue, but NKAβ1a and NKAβ1b are highly expressed in stomach, muscle and body kidney, respectively. There are significant differences between the two, indicating that multi-copy genes are functionally different.
The expression patterns of NHE and NKA gene families in R. canadum differed significantly in different salinities. Significant up-regulation of gill NHE2c and NHE5 expression in R. canadum occurred after low-salt acclimation, while no significant changes were found in NHE2a, and similar results were found in L. maculatus, suggesting that NHE2c and NHE5 may play an important role in the gill tissue of R. canadum in low-salt acclimation (Zhang et al., 2019). Studies have reported that NHE1 expression trends in S. maximus gill, intestine and somatic kidney were negatively correlated with salinity changes and showed low salt adaptation (Zhang et al., 2020). In the present study, significant upregulation of NHE1 in the intestine and somatic kidney of R. canadum was similarly identified during hyposalinity acclimation, suggesting that NHE1 can be involved in hyposalinity acclimation in R. canadum through the intestine and somatic kidney. Meanwhile, the expression of NHE1 and NHE2a increased in gill with decreasing salinity, but the degree of difference was not significant, and a significant down-regulation was observed in high-salt acclimation. It is speculated that the reduced expression of NHE1 and NHE2a in high salt is to reduce the ion transport efficiency of gill epithelial cells and prevent the decrease of osmotic pressure in vivo.
Few studies related to NKAα3a have been reported, and it has been suggested that its role in osmolarity regulation is relatively weak compared to NKAα1a (Nilsen et al., 2007). In the present study, the expression of NKAα3a and NKAβ1b in the gills of R. canadum decreased significantly with increasing salinity. Similar results could be found in O. mossambicus (Feng et al., 2002). As multi-copy genes, NKAα1a and NKAα1b are often compared together. In the present study, NKAα1b was found to be significantly downregulated in both high and low salt suits, in contrast to NKAα1a expression pattern. NKAα1b was similarly found to be strongly affected by salinity in O. mossambicus and Galaxias rostratus, similar to NKAα1a, further suggesting that NKAα1 isoforms appear to differ in function (Tipsmark et al., 2011; Urbina et al., 2013). In addition, two NKAα isoforms (NKAα1a and NKAα3b) were highly expressed in the gills after high salt domestication in L. maculatus, indicating the importance of NKAα isoform genes in the salt stress response of fish (Zhang et al., 2019).
5 Conclusion
In the present study,12 NHE genes and 12 NKA genes were systematically identified from R. canadum genome. These genes were found to be distributed across 10 and 9 superscaffolds. NHE and NKA members of R. canadum are closest in taxonomic position to teleosts and furthest from mammals, indicating that the protein sequences of both family members are highly conserved in species evolution. The histology of the gills, intestine and kidneys exhibited changes associated with salinity adaptation. Different expression patterns of R. canadum NHE genes and NKA genes were displayed in multiple tissues. At the same time, transcriptome sequencing and qPCR results showed that there were differences in the expression patterns of NHE and NKA gene families under different salinities, which provided research data for the osmotic pressure regulation mechanism of R. canadum.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRP202920 https://www.ncbi.nlm.nih.gov/, PRJNA634421.
Ethics statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee (IACUC), Fisheries College, Guangdong Ocean University.
Author contributions
ZW, ZC, and BH contributed to conception and design of the study. ZW organized the database. BH performed the statistical analysis. ZC wrote the first draft of the manuscript. ZY, MZ, MJ, and AZ wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
Guangdong University Innovation Team Project (2021KCXTD026, 2022KCXTD013).
Acknowledgments
We are grateful to Guangzhou Genedenovo Biotechnology Co., Ltd for assisting in sequencing and bioinformatics analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1228933/full#supplementary-material
References
Ali A., Azom M. G., Sarker B. S., Rani H., Alam M. S., Islam M. S. (2022). Repercussion of salinity on hematological parameters and tissue morphology of gill and kidney at early life of tilapia. Aquaculture Fisheries. doi: 10.1016/j.aaf.2022.04.006
Bailey T. L., Johnson J., Grant C. E., Noble W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Benetti D. D., Suarez J., Camperio J., Hoenig R. H., Tudela C. E., Daugherty Z., et al. (2021). A review on cobia, Rachycentron canadum, aquaculture. J. World Aquaculture Soc. 52, 691–709. doi: 10.1111/jwas.12810
Berthelot C., Brunet F., Chalopin D., Juanchich A., Bernard M., Noël B., et al. (2014). The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat. Commun. 5, 3657. doi: 10.1038/ncomms4657
Bystriansky J. S., Frick N. T., Richards J. G., Schulte P. M., Ballantyne J. S. (2007). Failure to up-regulate gill Na+, K+-ATPase α-subunit isoform α1b may limit seawater tolerance of land-locked Arctic char (Salvelinus alpinus). Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 148, 332–338. doi: 10.1016/j.cbpa.2007.05.007
Cao D. Y., Li J. F., Huang B. S., Zhang J. D., Pan C. H., Huang J. S., et al. (2020). RNA-seq analysis reveals divergent adaptive response to hyper- and hypo-salinity in cobia, Rachycentron canadum. Fish Physiol. Biochem. 46, 1713–1727. doi: 10.1007/s10695-020-00823-7
Chen G., Wang Z. L., Wu Z. H., Gu B. H. (2009). Effects of salinity on growth and energy budget of juvenile cobia, rachycentron canadum. J. World Aquaculture Soc. 40, 374–382. doi: 10.1111/j.1749-7345.2009.00257.x
Chen S., Zhou Y., Chen Y., Gu J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Counillon L., Pouysségur J. (2000). The expanding family of eucaryotic Na+/H+ exchangers. J. Biol. Chem. 275, 1–4. doi: 10.1074/jbc.275.1.1
Dawood M. A.O., Noreldin A. E., Sewilam H. (2021). Long term salinity disrupts the hepatic function, intestinal health, and gills antioxidative status in Nile tilapia stressed with hypoxia. Ecotox Environ. Safe 220, 112412. doi: 10.1016/j.ecoenv.2021.112412
Edwards S. L., Wall B. P., Morrison-Shetlar A., Sligh S., Weakley J. C., Claiborne J. B. (2005). The effect of environmental hypercapnia and salinity on the expression of NHE-like isoforms in the gills of a euryhaline fish (Fundulus heteroclitus). J. Exp. Zoology Part A: Comp. Exp. Biol. 303, 464–475. doi: 10.1002/jez.a.175
Feng S. H., Leu J. H., Yang C. H., Fang M. J., Huang C. J., Hwang P. P. (2002). Gene expression of na+-k+-atpase a1 and a3 subunits in gills of the teleost Oreochromis mossambicus, adapted to different environmental salinities. Mar. Biotechnol. 4, 379–391. doi: 10.1007/s10126-002-0006-0
Figueras A., Robledo D., Corvelo A., Hermida M., Pereiro P., Rubiolo J. A., et al. (2016). Whole genome sequencing of turbot (Scophthalmus maximus; Pleuronectiformes): a fish adapted to demersal life. DNA Res. 23, 181–192. doi: 10.1093/dnares/dsw007
Fiol D. F., Kültz D. (2007). Osmotic stress sensing and signaling in fishes. FEBS J. 274, 5790–5798. doi: 10.1111/j.1742-4658.2007.06099.x
Foskett J. K., Bern H. A., Machen T. E., Conner M. (1983). Chloride cells and the hormonal control of teleost fish osmoregulation. J. Exp. Biol. 106, 255–281. doi: 10.1242/jeb.106.1.255
Gao J., Nie Z. J., Xu G. C., Xu P. (2022). Genome-wide identification of the NHE gene family in Coilia nasus and its response to salinity challenge and ammonia stress. BMC Genomics 23, 1–14. doi: 10.1186/s12864-022-08761-9
Gonzalez R. J. (2012). The physiology of hyper-salinity tolerance in teleost fish: a review. J. Comp. Physiol. B 182, 321–329. doi: 10.1007/s00360-011-0624-9
Han K., Zhou L., Zeng X., Zhang Z., Zou P., Huang W., et al. (2022). Effects of low-salinity acclimation on the Na+/K+ ATPase activity and expression of osmoregulatory-related genes in large yellow croaker (Larimichthys crocea). Aquaculture Rep. 26, 101326. doi: 10.1016/j.aqrep.2022.101326
Holmes B. J., Williams S. M., Barnett A., Awruch C. A., Currey-Randall L. M., Ferreira L. C., et al. (2022). “Research methods for marine and estuarine fishes,” in Wildlife Research in Australia: Practical and Applied Methods (Australia: Csiro Publishing), 257–286.
Hou J. L., Chen L. Q., Zhuang P., Zhang L. Z., Tian H. J., Wang W., et al. (2006). Structural changes of chloride cells in gills epithelia of juvenile Acipenser schrenckii acclimated to various salinities. J. Fisheries China 30, 316–322. doi: 10.1360/aps050066
Howe K., Clark M. D., Torroja C. F., Torrance J., Berthelot C., Muffato M., et al. (2013). The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503. doi: 10.1038/nature12111
Hu Y. C., Chu K. F., Yang W. K., Lee T. H. (2017). Na+, K+-ATPase β1 subunit associates with α1 subunit modulating a “higher-NKA-in-hyposmotic media” response in gills of euryhaline milkfish, Chanos chanos. J. Comp. Physiol. B 187, 995–1007. doi: 10.1007/s00360-017-1066-9
Hu J., Ye L., Wu K. C., Wang Y. (2016). Effect of acute salinity stress on serum cortisol and activity of Na+-K+-ATPase of juvinile Amphiprion clarkii. South. Aquat. Sci. China 12, 116–120. doi: 10.3969/j.issn.2095-0780.2016.02.017
Huang S., Li C. Z., Li Z. X., Duanzhi D., Liu Y. H., Ran F. X., et al. (2022). Short-term exposure to 5‰ and 15‰ salinity causes the dynamic changes of the NKA gene, enzyme activities and morphological characteristics in fish tissues of Gymnocypris przewalskii. Aquac Res. 53, 6389–6398. doi: 10.1111/are.16112
Hwang J., Kim S., Seo Y., Lee K., Park C., Choi Y., et al. (2018). Mechanisms of salinity control in sea bass. Biotechnol. Bioproc E 23, 271–277. doi: 10.1007/s12257-018-0049-3
Imsland A. K., Gunnarsson S., Foss A., Stefansson S. O. (2003). Gill Na+, K+-ATPase activity, plasma chloride and osmolality in juvenile turbot (Scophthalmus maximus) reared at different temperatures and salinities. Aquaculture 218, 671–683. doi: 10.1016/S0044-8486(02)00423-4
Inokuchi M., Nakamura M., Miyanishi H., Hiroi J., Kaneko T. (2017). Functional classification of gill ionocytes and spatiotemporal changes in their distribution after transfer from seawater to fresh water in Japanese seabass. J. Exp. Biol. 220, 4720–4732. doi: 10.1242/jeb.167320
Jia Q., Lu W. (2016). Effects of low salinity stress on plasma osmolality, Cortisol, prolactin and growth hormone of Japanese flounder, Paralichthys olivaceus. J. Shanghai Ocean Univ. 25, 71–77.
Jiang Y. H., Yuan C., Qi M., Liu Q. G., Hu Z. J. (2022). The effect of salinity stress on enzyme activities, histology, and transcriptome of silver carp (Hypophthalmichthys molitrix). Biology 11, 1580. doi: 10.3390/biology11111580
Kasahara M., Naruse K., Sasaki S., Nakatani Y., Qu W., Ahsan B., et al. (2007). The medaka draft genome and insights into vertebrate genome evolution. Nature 447, 714–719. doi: 10.1038/nature05846
Kibenge F. S., Strange R. J. (2021). “Introduction to the anatomy and physiology of the major aquatic animal species in aquaculture,” in Aquaculture Pharmacology (Netherlands: Elsevier), 1–111.
Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Larsen P. F., Nielsen E. E., Meier K., Olsvik P. A., Hansen M. M., Loeschcke V. (2012). Differences in salinity tolerance and gene expression between two populations of Atlantic cod (Gadus morhua) in response to salinity stress. Biochem. Genet. 50, 454–466. doi: 10.1007/s10528-011-9490-0
Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12, 1–16. doi: 10.1186/1471-2105-12-323
Li L., Jiang M., Wang Y., Wu Q., Niu J., Shen X. (2014). Effects of low salinity stress on Na+-K+-ATPase activities, expression of Na+-K+-ATPase β-subunit mRNA and microscopical structure in gill filaments of juvenile Mugil cephalus. J. Zhejiang Univ. (Agriculture Life Sciences) 40, 223–230. doi: 10.3785/j.issn.1008-9209.2013.09.031
Li X. J., Shen Y. D., Bao Y. G., Wu Z. X., Yang B. Q., Jiao L. F., et al. (2022). Physiological responses and adaptive strategies to acute low-salinity environmental stress of the euryhaline marine fish black seabream (Acanthopagrus schlegelii). Aquat. Toxicol. 554, 738117. doi: 10.1016/j.aquaculture.2022.738117
Li H. Y., Zhu J. Q., Chen F., Ding L. F. (2011). Themorphology of the digestive tract of Centropristis striata. J. Biol. China 28, 31–34+46. doi: 10.3969/j.issn.2095-1736.2011.04.031
Lin Y. M., Chen C. N., Lee T. H. (2003). The expression of gill Na, K-ATPase in milkfish, Chanos chanos, acclimated to seawater, brackish water and fresh water. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 135, 489–497. doi: 10.1016/S1095-6433(03)00136-3
Lin Y. M., Chen C. N., Yoshinaga T., Tsai S. C., Shen I. D., Lee T. H. (2006). Short-term effects of hyposmotic shock on Na+/K+-ATPase expression in gills of the euryhaline milkfish, Chanos chanos. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 143, 406–415. doi: 10.1016/j.cbpa.2005.12.031
Marshall W. S. (2011). Mechanosensitive signalling in fish gill and other ion transporting epithelia. Acta Physiologica 202, 487–499. doi: 10.1111/j.1748-1716.2010.02189.x
Nilsen T. O., Ebbesson L. O., Madsen S. S., McCormick S. D., Andersson E., Björnsson B. T., et al. (2007). Differential expression of gill Na+, K+-ATPaseα-and β-subunits, Na+, K+, 2Cl-cotransporter and CFTR anion channel in juvenile anadromous and landlocked Atlantic salmon Salmo salar. J. Exp. Biol. 210, 2885–2896. doi: 10.1242/jeb.002873
Orlowski J., Grinstein S. (2004). Diversity of the mammalian sodium/proton exchanger SLC9 gene family. Pflügers Archiv 447, 549–565. doi: 10.1007/s00424-003-1110-3
Pertea M., Pertea G. M., Antonescu C. M., Chang T.-C., Mendell J. T., Salzberg S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Sakamoto T., Uchida K., Yokota S. (2001). Regulation of the ion-transporting mitochondrion-rich cell during adaptation of teleost fishes to different salinities. Zoological Sci. 18, 1163–1174. doi: 10.2108/zsj.18.1163
Shaffer R. V., Nakamura E. L. (1989). Synopsis of biological data on the cobia Rachycentron canadum (Pisces: Rachycentridae). USA: NOAA/National Marine Fisheries Service. Available at: http://hdl.handle.net/1834/20527.
Shi Z. H., Liao Y. L., Wang X. S., Zhang C. J., Peng S. M., Gao Q. X. (2017). Impact of the abrupt salinity decrease on ion-regulation enzyme activity in the gill and serum osmolality from Epinehelus moara. J. Saf. Environ. China 17, 1210–1214. doi: 10.13637/j.issn.1009-6094.2017.03.074
Smith H. W. (1932). Water regulation and its evolution in the fishes. Q. Rev. Biol. 7, 1–26. doi: 10.1086/394393
Smith J. W. (1995). Life history of cobia, Rachycentron canadum (Osteichthyes: Rachycentridae), in North Carolina waters. Brimleyana 23, 1–23.
Sun M. L., Jiang J. L., Wang L. P., Chen F., Han Y. Z., Jiang Z. Q., et al. (2016). Structural Changes in Gill, Kidney and Intestine of Juvenile Takifugu rubripes under Low Salinity Treatment. J. Guangdong Ocean Univ. China 36, 38–43. doi: 10.3969/j.issn.1673-9159.2016.06.007
Sundh H., Nilsen T. O., Lindström J., Hasselberg-Frank L., Stefansson S. O., McCormick S. D., et al. (2014). Development of intestinal ion-transporting mechanisms during smoltification and seawater acclimation in Atlantic salmon Salmo salar. J. Fish Biol. 85, 1227–1252. doi: 10.1111/jfb.12531
Tine M., Kuhl H., Gagnaire P.-A., Louro B., Desmarais E., Martins R. S., et al. (2014). European sea bass genome and its variation provide insights into adaptation to euryhalinity and speciation. Nat. Commun. 5, 5770. doi: 10.1038/ncomms6770
Tipsmark C. K., Breves J. P., Seale A. P., Lerner D. T., Hirano T., Grau E. G. (2011). Switching of Na+, K+-ATPase isoforms by salinity and prolactin in the gill of a cichlid fish. J. Endocrinol. 209, 237. doi: 10.1530/JOE-10-0495
Upling J. Y. (2020). A review on the activity of Na+/K+-ATPase in branchial ionocytes and its role in salinity adaptation among diadromous species. World J. Advanced Res. Rev. 6, 201–211. doi: 10.30574/wjarr.2020.6.2.0158
Urbina M. A., Schulte P. M., Bystriansky J. S., Glover C. N. (2013). Differential expression of Na+, K+-ATPase α-1 isoforms during seawater acclimation in the amphidromous galaxiid fish Galaxias maculatus. J. Comp. Physiol. B 183, 345–357. doi: 10.1007/s00360-012-0719-y
Wang Y., Hu X. C. (2009). Microscopical observation on the gill structure of juvenile Lateolabrax japonicus under different salinities. Mar. Sci. 33, 138–142. doi: 10.1016/j.elecom.2008.10.019
Wang S. J., Zhang H. F., Zhao J., Yang Y. Q., Yang S. S. (2011). Effects of different salinities on growth and physiology of orange-spotted grouper, epinephelus coioides. J. Guangdong Ocean Univ. China 31, 39–44. doi: 10.3969/j.issn.1673-9159.2011.06.006
Whittamore J. M. (2012). Osmoregulation and epithelial water transport: lessons from the intestine of marine teleost fish. J. Comp. Physiol. B 182, 1–39. doi: 10.1007/s00360-011-0601-3
Yamaguchi Y., Breves J. P., Haws M. C., Lerner D. T., Grau E. G., Seale A. P. (2018). Acute salinity tolerance and the control of two prolactins and their receptors in the Nile tilapia (Oreochromis niloticus) and Mozambique tilapia (O. mossambicus): a comparative study. Gen. Comp. Endocrinol. 257, 168–176. doi: 10.1016/j.ygcen.2017.06.018
Yan J. J., Chou M. Y., Kaneko T., Hwang P. P. (2007). Gene expression of Na+/H+ exchanger in zebrafish H+-ATPase-rich cells during acclimation to low-Na+ and acidic environments. Am. J. Physiology-Cell Physiol. 293, C1814–C1823. doi: 10.1152/ajpcell.00358.2007
Yang W. K., Chung C. H., Cheng H. C., Tang C. H., Lee T. H. (2016). Different expression patterns of renal Na+/K+-ATPase α-isoform-like proteins between tilapia and milkfish following salinity challenges. Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 202, 23–30. doi: 10.1016/j.cbpb.2016.07.008
Yang J., Xu W., Geng L. W., Guan H. H., Dang Y. F., Jiang H. F. (2014). Effects of salinity on survival,gill and kidney tissue in juveniles of 5 species. Freshw. Fisheries China 44, 7–12. doi: 10.13721/j.cnki.dsyy.2014.04.002
Yang J. R., Yang J. L., Chen M. Q., Fu Z., Sun J., Yu G., et al. (2022). Physical responses of Pinctada fucata to salinity stress. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.792179
Yu N., Li J., Ou Y. J., Wang Y. C., Su H. (2012). Structural changes in gill and kidney of juvenile grey mullet under different salinity. Ecol. Sci. China 31, 424–428.
Zhang J. S., Liu Z. F., Ma A. J., Cui W. X., Qu J. B. (2020). Response of aquaporin (AQP1, AQP3) and ion channel protein (CFTR, NHE1) of turbot (Scophthalmus maximus) to low-salinity stress. Prog. IN FISHERY Sci. OF China 41, 41–49. doi: 10.19663/j.issn2095-9869.20190410003
Zhang X. Y., Wen H. S., Qi X., Zhang K. Q., Liu Y., Fan H. Y., et al. (2019). Na+-K+-ATPase and nka genes in spotted sea bass (Lateolabrax maculatus) and their involvement in salinity adaptation. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 235, 69–81. doi: 10.1016/j.cbpa.2019.05.017
Zhang X. Y., Wen H. S., Wang H. L., Ren Y. Y., Zhao J., Li Y. (2017). RNA-Seq analysis of salinity stress–responsive transcriptome in the liver of spotted sea bass (Lateolabrax maculatus). PloS One 12, e0173238. doi: 10.1371/journal.pone.0173238
Zhang X. Y., Wen H. S., Zhang K. Q., Liu Y., Fang X., Li Y. (2018). Analysis of the isotonic point and effects of seawater desalination on the Na+/K+ /Cl concentration, Na+-K+ -ATPase activity and relative gene expressions in Lateolabrax maculatus. J. Fisheries China 42, 1199–1208. doi: 10.11964/jfc.20170410780
Keywords: Rachycentron canadum, salinity adaptation, NHE, NKA, histopathology, RNA-seq, qRT-PCR
Citation: Chen Z, Huang B, Yan Z, Hong Y, Zhao M, Jin M, Zheng A and Wang Z (2023) Genome−wide expression profile analysis of the NHE and NKA gene family in Rachycentron canadum (Linnaeus, 1766) and its response to salinity adaptation. Front. Mar. Sci. 10:1228933. doi: 10.3389/fmars.2023.1228933
Received: 25 May 2023; Accepted: 05 July 2023;
Published: 21 July 2023.
Edited by:
Yi-Feng Li, Shanghai Ocean University, ChinaReviewed by:
Sang Yoon Lee, Cellqua., Inc, Republic of KoreaChunyan Zhao, Qingdao Agricultural University, China
Copyright © 2023 Chen, Huang, Yan, Hong, Zhao, Jin, Zheng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongliang Wang, emhvbmdsaWFuZ3dhbmdAdmlwLjE2My5jb20=
 Zongfa Chen
Zongfa Chen Baosong Huang2
Baosong Huang2 Zhongliang Wang
Zhongliang Wang