
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mar. Sci. , 20 September 2023
Sec. Marine Ecosystem Ecology
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1227560
This article is part of the Research Topic Restoration of Coastal Marine Ecosystems View all 22 articles
 Cinzia Corinaldesi1,2*
Cinzia Corinaldesi1,2* Silvia Bianchelli2,3
Silvia Bianchelli2,3 Marco Candela2,4
Marco Candela2,4 Antonio Dell’Anno2,3
Antonio Dell’Anno2,3 Cristina Gambi2,3
Cristina Gambi2,3 Eugenio Rastelli5
Eugenio Rastelli5 Stefano Varrella2,3
Stefano Varrella2,3 Roberto Danovaro2,3
Roberto Danovaro2,3Microorganisms interact with all biological components in a variety of ways. They contribute to increase the efficiency of marine food webs and facilitate the adaptation of multicellular organisms to climate change and other human-induced impacts. Increasing evidence suggests that microbiomes are essential for the health of marine species, for maintaining productive marine ecosystems, and thus for the sustainable functioning of the global biosphere. Marine microbiomes are typically species- or habitat-specific and are susceptible to environmental and human-driven changes. The microbiota of seagrasses, macroalgae, mangroves or tropical corals benefits their hosts by increasing their fitness, contributing to the removal of toxic compounds, conferring protection against pathogens, and/or supporting nutrient requirements. Alterations of the microbiomes might have negative consequences on species’ health, survival, and overall ecosystem functioning. Despite the key ecological role of microbiomes in all ecosystems, their potential for the restoration of degraded habitats is still largely unexplored. Here we present a literature survey of the existing information on the microbiota associated with habitat-forming species and suggest that the resilience/recovery of damaged marine habitats can depend largely on the changes in the microbiota. Nature-based solutions relying on microbiome analyses (also through omics approaches) enable health monitoring of transplanted organisms/metacommunities and potential identification/production of probiotics/bio-promoters to stabilize unhealthy conditions of transplants. In the context of international strategies concerning ecological restoration, the use of the scientific knowledge acquired on the marine microbiome deserves to be exploited to assist both traditional and innovative restoration approaches. The success of habitat restoration may depend on our ability to maintain, along with the restored species and habitats, a functional microbiota.
Microorganisms support the existence and resilience of all life forms (Gilbert and Neufeld, 2014). They are crucial for the maintenance of healthy and productive marine ecosystems and for the sustainable functioning of the global biosphere (Trevelline et al., 2019). Microorganisms are intimately associated with animals, plants, and algae, and contribute to their nutrition, defence, immunity, and development (Pita et al., 2018; Stévenne et al., 2021). The prevalence of these associations implies that multicellular organisms can no longer be considered as individual entities, but rather as “holobionts” (consisting of the host, the microbiota, and the interactions among them). Consequently, the impact of climate changes and human pressures on multicellular organisms as well as their resilience will also depend on the responses of their associated microbiota, which will be essential to achieve an environmentally sustainable future (Doney et al., 2012; McFall-Ngai et al., 2013; Petersen and Osvatic, 2018).
It is recognized that microorganisms have the potential to adapt to climate change scenarios (e.g., global warming, ocean acidification, oxygen depletion) and human-induced impacts (e.g., pollution due to chemical contaminants, eutrophication) (Danovaro et al., 2011; Torda et al., 2017; Cavicchioli et al., 2019). Micro-organisms, indeed, thrive in the most extreme environments on Earth, from the coldest to warmest habitats, from the acid and hypersaline environments to anoxic systems, even at the highest pressures of the deep sea (Boetius et al., 2015; Dick, 2019; Manea et al., 2019; Hiraoka et al., 2020). It has been also extensively documented that marine microbiota can allow multicellular organisms to thrive in extreme marine habitats by conferring them the ability to cope with conditions incompatible with life (Danovaro et al., 2017; Sogin et al., 2020) by regulating the maintenance of their fitness, conferring them resistance and resilience (Bang et al., 2018). Microorganisms can also be sentinels of the impact of climate change and/or pollution on the holobionts (Conte et al., 2021; Corinaldesi et al., 2022) revealing early their health conditions (Peixoto et al., 2022).
Despite the key ecological role of microbiota in all ecosystems, their potential for restoration actions of degraded habitats and pollution control is still largely unexplored. Due to the rapid decline of marine ecosystems and their associated biodiversity (Danovaro et al., 2020; Orfanidis et al., 2021), both in coastal and deep-sea habitats, ecological restoration actions appear to be the most promising strategy to rebuild them (Cebrian et al., 2021; Fraschetti et al., 2021). This practice refers to the process able to assist the recovery of degraded, damaged, and destroyed ecosystems (Gann et al., 2019), thus retrieving lost biodiversity and ecosystem services (i.e., nutrient cycling, primary and secondary production, maintenance of genetic diversity, habitat provisioning, pollution control, carbon storage, erosion prevention). To revert the trajectory of degradation of most ecosystems on Earth and to accelerate the recovery of damaged ecosystems, the United Nations has declared the Decade on “Ecosystem Restoration” 2021-2030.
The strategies and techniques for ecological restoration of different marine habitats/ecosystems (e.g., seagrass beds, macroalgae forests, reef-forming corals) are mostly already available, but these do not consider the role of the microbial component. The International Coral Reef Society (ICRS) has recently published a scientific policy document that underlines the importance of “driving innovation by developing new approaches, where current solutions are not sufficient to address the coral reef emergency” (Knowlton, 2021). Accordingly, the declaration of the Decade on Ecosystem Restoration by the United Nations has sparked interest in the methods needed to implement best practices for maximum gain.
In the present literature survey, we provide an overview of the diversity and role of the microbiome (Berg et al., 2020) in marine habitats/ecosystems to shed light on the importance of integrating this component into future restoration plans. To the extent possible, we will draw parallels with microbiome-based restoration in the terrestrial environment, which is a much more advanced scientific field than in the marine environment, to identify key aspects and pathways in the recovery of degraded marine ecosystems. In this regard, the use of microbial inoculants and microbiome engineering will be addressed, emphasizing the challenges and pitfalls of these practices in the marine environment.
Ecological restoration in the marine environment is certainly a much younger field of scientific application than the terrestrial one (Saunders et al., 2020). In terrestrial ecosystems, it has been reported that the success of ecological restoration largely depends on soil microbial assemblages that degrade plant-associated organic substances, thus improving the physicochemical characteristics of the soil (Trivedi et al., 2013; Calderón et al., 2017). In addition, plant-associated microbes (e.g., the rhizosphere and endophytic microbiomes) provide nutrients, minerals, and vitamins and protect plants against biotic and abiotic stress (Vandenkoornhuyse et al., 2015; Dubey et al., 2019).
Especially, with the advent of the -omics approaches it has been possible to identify key microbiomes and their metabolites in different environments at unprecedented resolution. In terrestrial ecosystems, there is evidence that the core microbiota is essential for the maintenance of the functional stability of soil microbiomes, nutrient cycling, and plant establishment in reforested ecosystems and that it should be integrated into the policy and management strategies of ecological restoration plans (Jiao et al., 2022). In this regard, soil inoculation techniques and the use of carriers for inoculants have been assessed to promote plant growth. In particular, Rhizobium inoculants have been commercially produced worldwide (Catroux et al., 2001; Deaker et al., 2004) as well as Azospirillum brasilense and Bacillus pumilus (Cassán et al., 2020). Manipulative experiments highlighted that the soil community is an important driver of plant-community development and that manipulation of the soil community is a key step for successful restoration (Wubs et al., 2016).
A large-scale, six-year-old field experiment on terrestrial habitats has shown that the application of soil inoculants is a powerful tool to promote ecosystem restoration and steer plant community development depending on the origin of the soil inoculum (Wubs et al., 2016). Other studies have reported that inoculation of soils with mycorrhizal fungi can also increase the growth performance of plants and that local mycorrhizal fungi strains outperform commercial strains. Co-inoculation of different native mycorrhizal fungi species can improve plant growth with positive implications for restoration ecology (Crossay et al., 2019). A key role is indeed represented by the species richness of the inoculum (Hu et al., 2021). Previous investigations indicated that the use of consortia that contained more Pseudomonas strains produced beneficial effects on plant growth. Consistently, it has been also documented that the transfer of whole soil communities, enriched in plant grow-promoting consortia, is more effective than the addition of individual species or strains in restoration actions (Emam, 2016; Bulot et al., 2017). Another interesting finding obtained in terrestrial ecology is that soil transplantation experiments at large-spatial scales are more successful than at smaller-spatial scales (Kardol et al., 2009; Pywell et al., 2011; Jaunatre et al., 2014). Several investigations have also indicated that environmental and biological factors such as nutrient loads and interactions among soil biological communities can influence the success of transplantation (De Deyn et al., 2004; Van Elsas et al., 2012). However, several studies concluded that it is currently unclear if the inoculant or soil transplantation effects will be transient or whether they will last through plant generations, and it is necessary to better deepen scientific research on plant-microbe interactions for sustainable agriculture (Hu et al., 2021).
In terrestrial ecology, it has been also reported that plants and their roots have evolved “a cry for help” response to resist environmental and anthropogenic stress, by developing adaptive strategies including the production of metabolites with direct defensive effects and molecules that attract beneficial (micro)organisms for protection (Rizaludin et al., 2021). In a recent study, the “cry for help” concept was also supported by the results of a field experiment, in which durum wheat (Triticum turgidum L. var. durum) naturally infected with the crown rot pathogen Fusarium graminearum enriched for Stenotrophomonas rhizophila (SR80) in the rhizosphere and root endosphere (Liu et al., 2021). After reintroduction, the SR80 strain induced resistance against canopy rot and improved grain growth. The rhizosphere microbiome is thus a reservoir of efficient helpers that plants can specifically exploit to cope with one or more stressors. Deciphering their communication can provide fundamental knowledge for the future development of plants resistant to multiple stressors also in marine ecosystems.
If the integration of the microbiome in the design and management of terrestrial restoration actions is under experimentation, in the marine environment this is still far from being taken into consideration. This wide gap is due to the greater complexity of the marine submersed environment compared to the terrestrial one, its high variability due to hydrodynamic forcings, and the difficulty of applying and managing bacterial inoculants. Furthermore, while on land, vegetation is the main (practically the only) biological component that structures the landscape, in the marine environment even animal components such as corals can form habitats. Therefore, scientific research on the role and application of the microbiome in restoration ecology has a more complex job to do. However, knowledge accumulated in terrestrial microbiome-based restoration may inspire marine ecosystem restoration.
As it becomes more evident that microbiomes power ecosystem-level processes and resilience to anthropogenic and climate-driven impacts, scientific interest in microbiome engineering is increasing. Microbiome selection (which is part of the wider field of microbiome engineering but excludes any form of genetic manipulation) aims at improving host fitness with designed microbial communities (Mueller and Sachs, 2015). Microbiome selection thus foresees the desired modulation of the tailored microbiome to enhance a selected microbiome-dependent host phenotype. To this aim chemical-based (prebiotics and antibiotics), cellular-based (probiotics and microbiota transplants), phage-based (bacteriophages), or host-mediated microbiome-modulation approaches can be implemented (Li et al., 2022).
Traditionally, the modulation of the host-associated microbiome to tailor a desired phenotype was mainly performed empirically, thus completely blind concerning the compositional and functional features of the target microbiome, just relying on the use of allochthonous probiotics and prebiotics (i.e., compounds that foster growth or activity of beneficial microorganisms such as bacteria and fungi) as possible providers of the desired functions (Peixoto et al., 2022; Figure 1). The current multi-omics era is changing our capacity to study and understand the microbiome in their native ecosystems. From an operational point of view, the identification of the microbiome features being responsible for a desired function foresees 4 phases: (i) microbiome sampling, biobanking and multi-omics assessment; (ii) microbiome modelling and networks construction; (iii) assembly of de novo Microbiome-Assembled Genomes (MAGs) and (iv) construction of genome-scale metabolic models. The identification of the microbiome modules and their metabolic interactions allows the knowledge-based implementation of microbiome-based approaches - in terms of selected bio-promoters and bio-inocula – piloting these modules, their components, and their web of interactions.
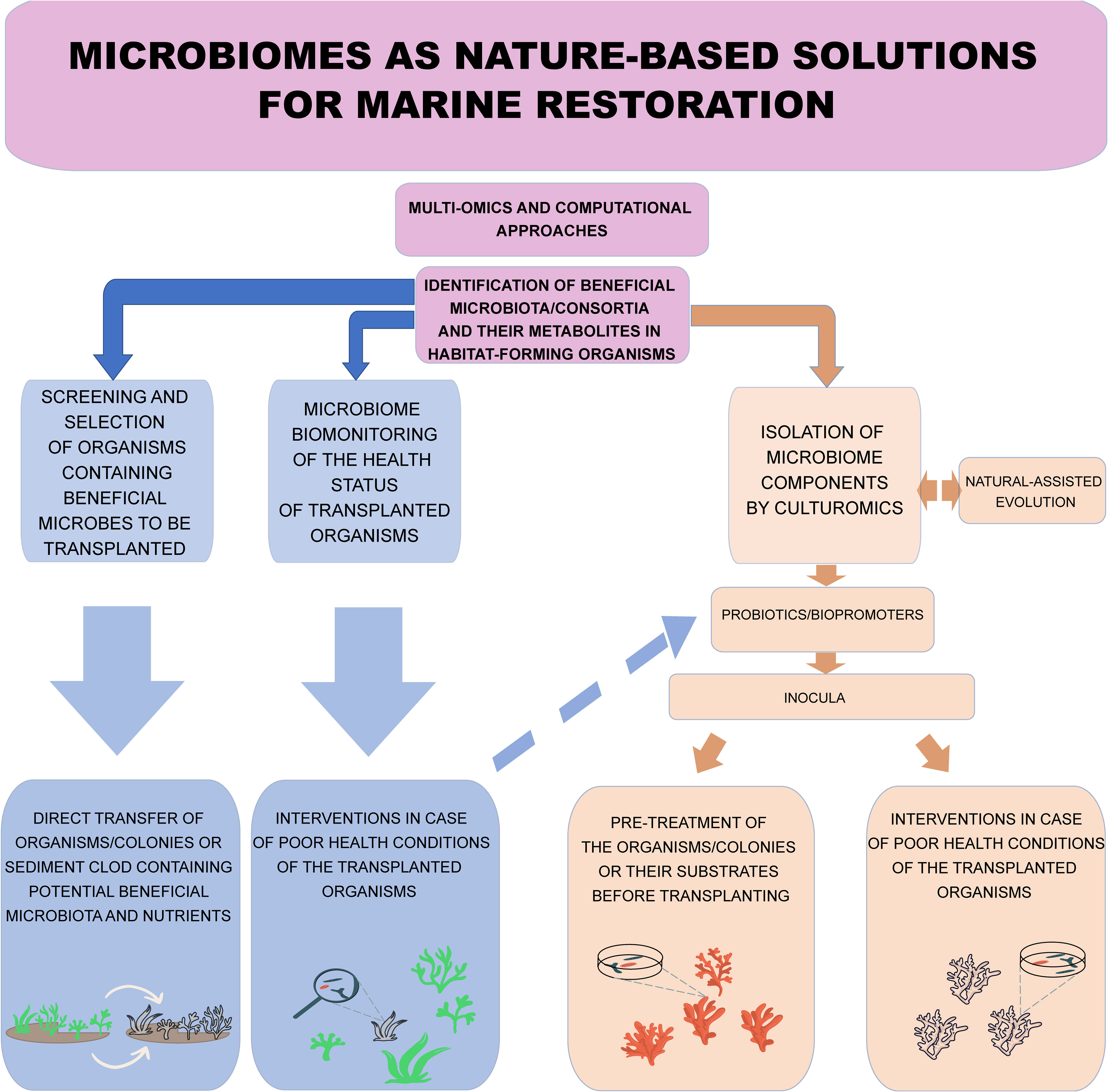
Figure 1 Conceptual model of the potential nature-based solutions relying on the microbiome to be applied to ecological restoration of habitat-forming organisms. Nature-based solutions can be represented by a) the screening and selection, through the microbiome analyses, of habitat-forming organisms or metacommunities containing beneficial microbes for transplanting and b) the biomonitoring of the health status of transplanted organisms to make decisions (including the adoption of probiotics/bio-promoters based on the microbiome-culture approach, see the light blue dashed arrow) in case of poor health conditions of the transplanted organisms. Microbiome manipulation allows the identification of keystone, beneficial and/or resistant microbiota, and their functions through -omics technologies and their selection through culturomics-based approaches, which should provide usable probiotics and or bio-promoters to be inoculated to transplanted organisms.
The recovery and selection of beneficial and resistant microorganisms can be obtained through specific culture media (Peixoto et al., 2017), whose selection and implementation can be also carried out based on the genome-scale metabolic models of the tailored beneficial microorganisms, allowing the in-depth understanding of their nutritional requirements (Liu et al., 2010). However, only a minor fraction of marine microbes are actually cultivable even using the best available colturomics-based approaches (Rodrigues and de Carvalho, 2022). In addition, if we consider that only a few marine isolates are endowed with the technological properties necessary for their implementation in concrete biotechnological applications, such as marine probiotics (e.g., the ability to grow as axenic culture in pilot bioreactors) it is clear that at present only a tiny fraction of the diversity of marine microbiomes can be exploited to produce probiotics (Wang et al., 2021a). These selected microorganisms can be inoculated at different life stages of the host using similar approaches to those already used in agriculture for probiotics (Backer et al., 2018). The inoculation of beneficial microorganisms can be performed using microencapsulation and nanoparticles as well as saline suspension and substrates for the microbial immobilization adopted in terrestrial ecosystems (Vassilev et al., 2020; Balla et al., 2022) and aquaculture systems (Prado et al., 2020). However, the delivery of microorganisms (microbial inoculum) in marine environments is a major challenge due to dilution in the water column and dispersion by currents and the difficulty of controlling interactions with a wide range of micro- and macro-organisms naturally present in these systems. Therefore, the immobilization of the microbial cells could be one of the approaches that might be adopted to overcome this problem (Peixoto et al., 2021).
Microbiome manipulation to improve host health could be based on other approaches such as microbiome-assisted evolution, used to accelerate adaptation on natural taxa to select those that show the highest resilience against stress and thus increase the fitness of the transplanted species (Van Oppen et al., 2015). In this case, experimental evolution studies involve the maintenance of organisms under controlled conditions and the monitoring of their response over several generations (Figure 1). Organisms are expected to adapt to these conditions through selection due to random natural genetic mutations, trans-generationally stable epigenetic modifications and/or through the acquirement of adaptive changes in the associated microbiome (Henry et al., 2021; Maire and van Oppen, 2022; Mueller and Linksvayer, 2022). Even though the selected pressures cannot dictate what adaptive traits will appear, these factors may select beneficial changes at the microbiome level (Maire and van Oppen, 2022). These microbiome-dependent adaptive changes can be characterized by multi-omic approaches, allowing the complete understanding of the correspondent functional traits, and opening the opportunity for their engineering.
The experimental evolution of coral-associated bacteria is at the stage of hypothesis, and it has been not implemented yet (Rosado et al., 2019; Maire and van Oppen, 2022). Although scientific literature is available on the possibilities of exploiting microbiome selection and probiotic supply to improve the resistance and resilience of natural ecosystems (especially concerning coral reefs), the implementation of these technologies in restoration plans is still far from being reached. Several gaps in knowledge and practical issues (e.g., in the approaches of target microorganisms and the industrial production of pro-biotics and bio-promoters) need to be filled (Peixoto et al., 2022) to make the most of the potential of the microbiome in the restoration ecology.
Many of the marine restoration efforts have been focused on coral reefs of shallow-coastal ecosystems contrary to what has been carried out for cold-water corals, and mesophotic bioconstructions (Fraschetti et al., 2021). Tropical coral transplantation is also one of the pioneering approaches to marine ecological restoration (Boström-Einarsson et al., 2020). This is due to several reasons, including the high ecological and economic importance of these habitats, their easy accessibility, and the rapid decline they are facing due to climate change and other anthropogenic impacts (Rivera et al., 2020; Ferse et al., 2021). At the same time, wide information is available on microbial assemblages associated with coral reefs, also justified by the desire to identify microbial diseases that contribute to their decline in the world’s oceans (Li et al., 2022). Despite this, the microbiome has not yet been factored into effective plans to restore coral reef ecosystems. However, it could contribute to the success of their restoration, and to that of other bioconstructions and even deep habitat-forming species (Figure 2). The general habitat characteristics of the microbiomes and their potential in the restoration of different bioconstructions is described here below.
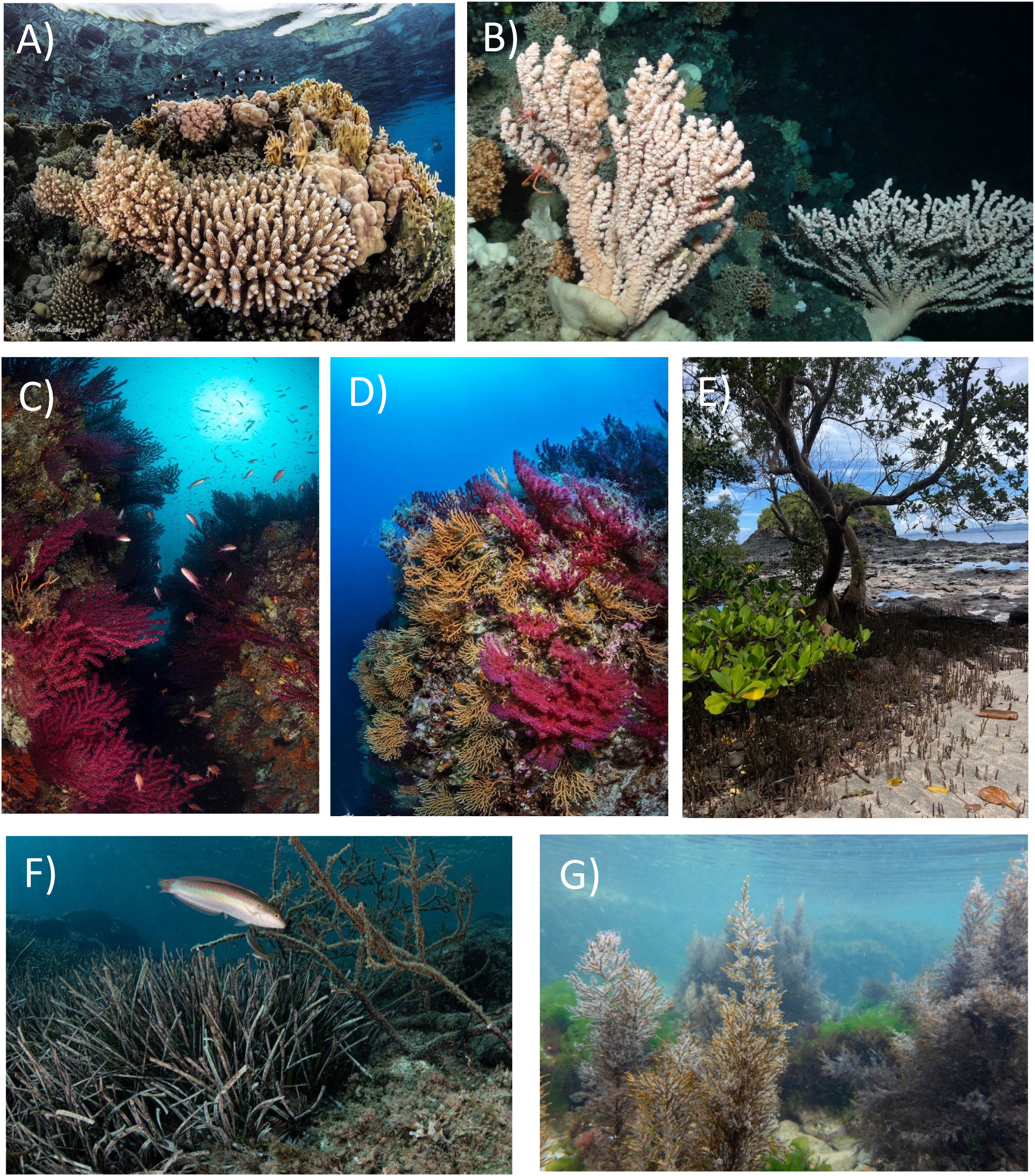
Figure 2 Habitat-forming species, which can be involved in the microbiome-based-restoration. (A) Coral reef (image credits by Gabriella Luongo); (B) cold-water corals; (C, D) coral gardens (credits by Gabriella Luongo); (E) mangroves; (F) seagrass meadows (credits by Gabriella Luongo), and (G) macroalgal forests (credits by AFRIMED project).
Reef-building corals (Figure 2) - Awareness of the rapid decline of coral reefs worldwide due to anthropogenic impacts and climate change has spurred restoration efforts, which are now widely applied using several approaches (Pandolfi et al., 2003; Bellwood et al., 2004; Vaughan, 2021) including coral gardening, transplantation of coral fragments, micro-fragmentation, and the use of artificial substrates (Boström-Einarsson et al., 2020). In the last decade studies of the coral microbiome have allowed expanding knowledge of the diversity and functions of microorganisms associated with reef-building corals (Van Oppen and Blackall, 2019). There is evidence that coral-associated microorganisms play a key role in the cycling and/or supply of nutrients (e.g. carbon, nitrogen, and sulfur) to corals or their symbiotic algae (e.g., Symbiodiniaceae), in the protection of corals (possibly through the production of antimicrobial compounds), in improving larval settlement (Van Oppen and Blackall, 2019) and resilience to environmental stressors, which can lead to coral diseases (Bourne et al., 2016; Vanwonterghem and Webster, 2020). Available studies indicate that the diversity of coral-associated bacteria far outnumbers that of archaea (two archaeal phyla versus at least 39 bacterial phyla described and candidate; Gardner et al., 2023). However, few studies have specifically focused on archaea, mainly because of technical limitations e.g., primer specificity (Gantner et al., 2011; Apprill et al., 2015; Parada et al., 2016), thus it is possible that their diversity is greater than what is known to date.
The most common coral-associated bacterial taxa belong to Gammaproteobacteria, Alphaproteobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, and Tenericutes, while the coral-associated archaea mostly affiliate with Euryarchaeota and Nitrososphaerota (syn. Thaumarchaeota) (Table 1; Huggett and Apprill, 2019). Available data for coral-associated fungi are currently limited and only in recent research, some taxa such as Aspergillus, Clonostachys, Mortierella, Cladosporium, Wicherhamomyces, Simplicillium, Cutaneotrichosporon, and Penicillium have been identified (Cheng et al., 2023). The microbiome of different coral species can change over time and across different geographic regions, and microbiome shifts can depend also upon environmental stressors as well as coral health (Hernandez-Agreda et al., 2018; Morrow et al., 2022). It has been also reported that coral adults have a less diverse and more stable microbiome than larvae and early recruits, suggesting a selection process during coral maturation (Epstein et al., 2019) and that microbiome is fine-tuned to meet the needs of a particular host in a certain environment (Van Oppen and Blackall, 2019). Given the complexity of studying the whole coral microbiome, scientific research is increasingly focusing on core microbial taxa that persist within the host across different spatial and temporal scales (Hernandez-Agreda et al., 2017; Dunphy et al., 2019). The core microbiome is indeed composed of common taxa, host and habitat-specific (Sweet and Bulling, 2017), which form stable and persistent symbiotic interactions with corals to provide key functions (Shade and Handelsman, 2012; Shafquat et al., 2014; Ainsworth et al., 2015; Chu and Vollmer, 2016). A less stable core microbiome could indicate a higher susceptibility of the holobiont to environmental stress (Ziegler et al., 2016). Therefore, the analysis of the core microbiome is essential to inform holobiont health status, which in turn is also fundamental for outlining strategies for coral restoration (Hernandez-Agreda et al., 2016; Sweet and Bulling, 2017).
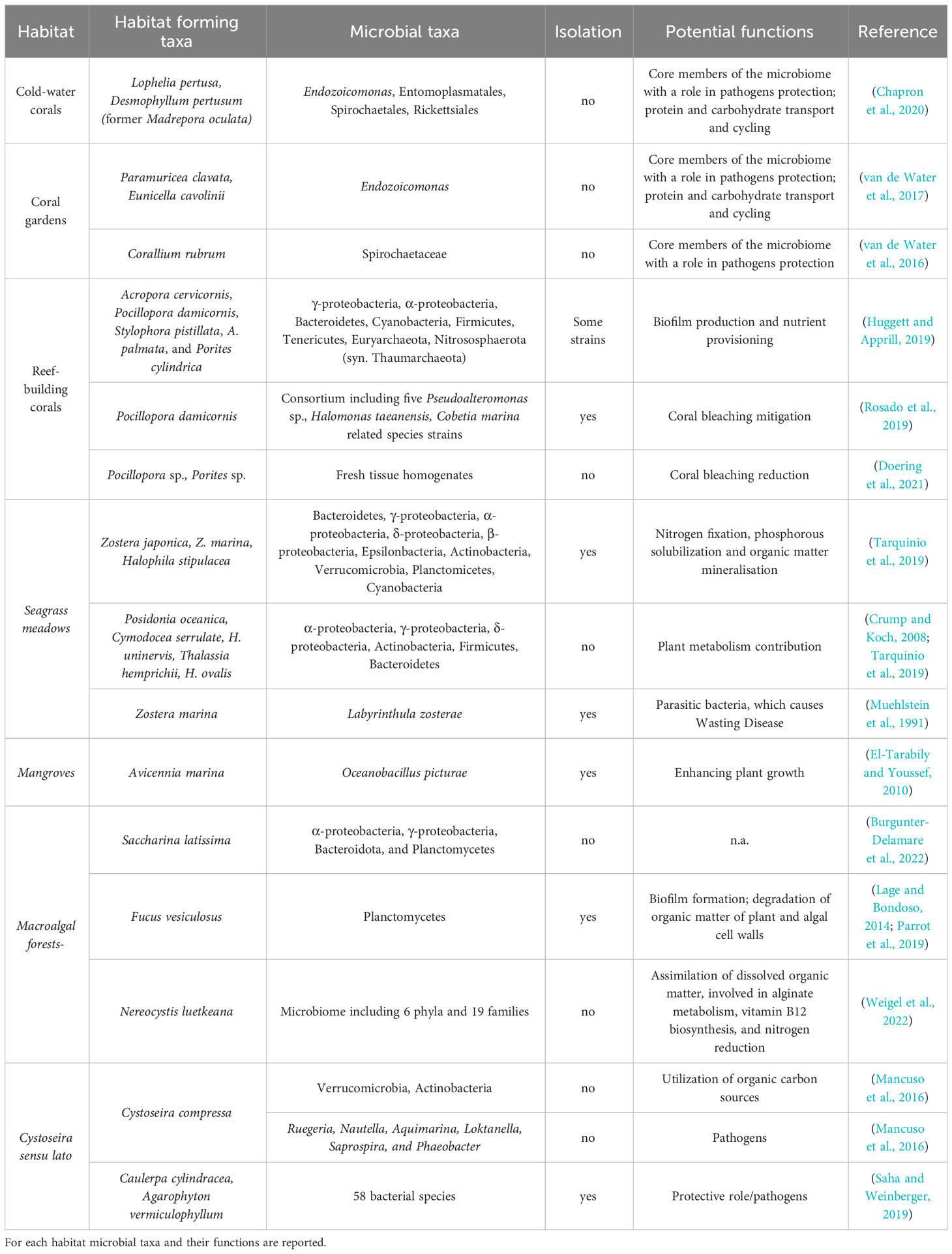
Table 1 Examples of microbial taxa of habitat-forming species which could have a key role in restoration activities.
The top five reef-building coral species used in restoration projects are Acropora cervicornis, Pocillopora damicornis, Stylophora pistillata, A. palmata, and Porites cylindrica (Boström-Einarsson et al., 2020). In general, the dominant taxa of bacterial assemblages associated with corals are assumed to be their healthy symbiotic state (Hernandez-Agreda et al., 2017 and references within). Bacterial members belonging to the genus Endozoicomonas are dominant in corals although their contribution to the entire microbial assemblage is highly variable within individuals of the same species and among different species (Hernandez-Agreda et al., 2017; Robbins et al., 2019; Rosales et al., 2019; Vanwonterghem and Webster, 2020). There is evidence that Endozoicomonas members are beneficial symbionts of the coral hosts with a role in biofilm production (that promotes surface colonization of other bacteria), sulphur metabolism, the cycling of carbohydrates and the provision of proteins to the host (Jessen et al., 2013; Ainsworth et al., 2015; Pootakham et al., 2019; Krishnaswamy et al., 2023). Endozoicomonas can have a mutualistic relationship with the endosymbiotic dinoflagellate partner Symbiodinium. This relationship has been suggested to be critical to the success of coral restoration (Bernasconi et al., 2019). However, a broad spectrum of other microorganisms (e.g., bacteria, archaea, viruses and eukaryotic microbes) may play a key role in maintaining coral health (Van Oppen et al., 2009; Ainsworth et al., 2017) and beneficial microorganisms should be identified specifically for each coral species, regional location, and stage of development (Peixoto et al., 2017).
Previous studies suggested that coral transplantation can affect coral microbial assemblages thus increasing the susceptibility of the corals to diseases (Casey et al., 2015). This finding reinforces the need to investigate the microbiome of coral reefs to increase the success of the restoration actions (Garren and Azam, 2012; Moriarty et al., 2020). In this regard, probiotics can also be adopted for disease mitigation (Peixoto et al., 2019). For example, marine bacteria isolated from Acropora palmata and other corals were found to produce anti-bacterial molecules against a broad spectrum of pathogens, including Serratia marcescens which is a pathogen known to lead to the disease ‘white pox’ (Ritchie, 2006; Alagely et al., 2011). This method was also tested on cnidarians affected by whitepox disease revealing beneficial effects.
Transplant-induced coral diseases could combine with the adverse effects of climate change, thus hampering successful restoration. Despite it has been reported that the coral microbiome can acclimatize to climate-driven changing conditions (including pH acidification, and heatwaves) enriching its functions related to nitrogen metabolism, host nutrition, and increased resistance (Biagi et al., 2020; Palladino et al., 2022; Prada et al., 2023), monitoring microbiome changes in restoration projects could be useful for predicting the effects of multiple stressors.
Recent studies have also suggested that microbiome manipulation could enhance heat tolerance and help corals survive the effects of ocean warming (Doering et al., 2021). From this perspective, microbiome engineering could represent a powerful tool to mitigate disease-associated threats, increase corals’ stress tolerance to perturbations, and improve coral resilience in the face of ongoing climate change (Li et al., 2022).
Knowledge of corals’ ability to acquire new microbiome components to mitigate the negative effects of environmental stress led to the development of the “Coral Probiotic Hypothesis” based on the identification of strains of dinoflagellates and microbes able to promote the growth and persistence of the host under shifting environmental conditions (Epstein et al., 2019). Theoretically, a coral treated with an inoculum of these microorganisms taken from a stress-adapted conspecific should adapt or acclimatize to that stress more rapidly (Epstein et al., 2019). The “Coral Beneficial Microorganisms” (CBM) concept provides procedures to further apply the “Coral Probiotic Hypothesis” by identifying potential selected advantageous mechanisms provided by the microbiome, isolating these beneficial microbes, and experimentally testing them for their role in coral resilience to environmental perturbations, both in situ and in controlled conditions (e.g., aquaria, Peixoto et al., 2017). This concept and its applications might be also translated to other habitat-forming species, including vegetated habitats.
Pioneering studies have shown that the microbiome can be shaped through inoculation with cultured bacterial isolates (Welsh et al., 2017; Damjanovic et al., 2019), while other investigations have tested the probiotic potential of these inoculations to assess improvements in coral health and resistance under different types of stress (Fragoso Ados Santos et al., 2015; Jacquemot et al., 2018; Rosado et al., 2019). For example, a consortium of native (isolated from Pocillopora damicornis and surrounding seawater) putatively beneficial microorganisms for corals, including Pseudoalteromonas sp., Halomonas taeanensis and Cobetia marina strains was added to coral colonies to increase resistance to bleaching (Table 1; Rosado et al., 2019). In a recent transplantation study of two cosmopolitan reef-building corals, Pocillopora sp. and Porites sp., from the Andaman Sea in Thailand, heat-sensitive corals were transplanted with donor microbiomes using fresh tissue homogenates produced from heat-tolerant conspecific donor corals, thus bypassing time-consuming culturing and screening for beneficial bacteria from healthy donors and allowing the transmission of the “unculturable” microbiome fraction (Table 1; Doering et al., 2021). The inoculation reduced coral bleaching under heat stress suggesting its beneficial effect and the potential of a probiotic intervention to support success in coral restoration.
Overall, based on the available information, although the microbiome offers important opportunities for coral restoration and re-establishing self-sustaining and functional habitats, its use has been made on a limited spatial scale and is still at an experimental level.
Gorgonian forests (Figure 2) - Gorgonian forests include corals belonging to the Anthozoan subclass of Octocorallia, which comprise soft corals (e.g., sea fans and sea whips - order Alcyonacea-, sea pens -order Pennatulacea-), and blue corals (order Helioporacea). They represent corals of conservationist interest as they act as ecosystem engineers, supporting high biodiversity levels (Ballesteros, 2006; Poulos et al., 2013; Sánchez, 2017; Angiolillo and Canese, 2018; van De Water et al., 2018a). Over the last 30 years, gorgonian gardens have been reported to be highly vulnerable to episodic climate-driven events as well as to anthropogenic activities and diseases (Smith and Weil, 2004; Sánchez et al., 2014; Girard and Fisher, 2018; Garrabou et al., 2019). Some coral species in the Mediterranean Sea including Corallium rubrum, Paramuricea clavata, and Eunicella cavolini, are regressing in different coastal areas (Barnes, 2010; Angiolillo and Canese, 2018; Verdura et al., 2019; Gómez-Gras et al., 2021; Topçu et al., 2023). Similarly, the soft coral Dendronephthya australis has been listed as endangered in Australia (Larkin et al., 2023). For this reason, these species have been a target of restoration experiments in the field and in aquaria (i.e., to assist in situ recovery) (Ballesteros, 2006; Ponti et al., 2014; Ingrosso et al., 2018; Montero-Serra et al., 2018; Verdura et al., 2019; Steinberg et al., 2020; Casoli et al., 2022; Topçu et al., 2023). Also, gorgonians collected as by-catch can be a valuable resource for the restoration of impacted ecosystems (Montseny et al., 2019; Casoli et al., 2020; Montseny et al., 2020).
Only a few studies have been carried out to test the efficiency of transplanting to rehabilitate gorgonian populations in impacted areas reporting contrasting results in terms of survival rate (30%- 90% in 2.5 years; Linares et al., 2008; Fava et al., 2010; Montseny et al., 2019; Casoli et al., 2022). The design of any restoration procedure must take into account the likelihood of mortality of coral colonies before transplanting since this can affect the initial attachment effort necessary to increase the success of the restoration process. To this aim, microbiome analyses including the detection of pathogens can help to anticipate these actions by identifying stressed colonies that visually do not show any signs of alteration (Corinaldesi et al., 2022).
Octocoral-microbe interactions compared to those of Scleractinia have attracted a lower interest, with only a limited number of studies focused on their associated microbiota (van De Water et al., 2018a), thus limiting understanding of the role of the microbiome in gorgonian health. Available information indicates that in natural conditions, the core microbiome of gorgonians is stable (van de Water et al., 2016; van de Water et al., 2018b) over space and seasons as in the case of E. cavolini, P. clavata and different Caribbean Octocorals, which are generally dominated by Endozoicomonadaceae (Table 1; Bayer et al., 2013; Vezzulli et al., 2013; van de Water et al., 2017; McCauley et al., 2020; van de Water et al., 2020) or Spirochaetaceae mostly associated with C. rubrum (van de Water et al., 2016; van de Water et al., 2018b; Corinaldesi et al., 2022). Thus, a shift in the composition of the core microbiome of these species after transplantation actions can represent an early warning indicator of stressful conditions that can decrease the success rate of the restoration actions. Considering that newly established coral populations need 30-40 years to show an adequate colony size distribution (Montero-Serra et al., 2018), it is quite difficult to assess the restoration success after only a few years (Linares et al., 2008; Duarte et al., 2020). This constraint is mainly due to the short time scale (3-4 years) of funded projects aimed at promoting some restoration actions and experiments. Consequently, long-term monitoring programs, including analyses of the microbiome associated with transplanted organisms, should be planned over a longer time scale to provide more comprehensive information about the factors influencing microbiome dysbiosis, thus allowing the implementation of possible intervention actions to keep microbiome eubiosis in future restoration activities.
Cold-water corals (Figure 2) - Cold-water corals are key habitat-forming species, producing complex three-dimensional structures over wide areas, which foster biodiversity hotspots including species of high commercial interest (Roberts et al., 2009; Angiolillo and Canese, 2018). These corals are generally found in the deep sea (Roberts et al., 2009), particularly on continental margins, seamounts and canyons (Montseny et al., 2021). These ecosystems are threatened by multiple human activities such as commercial bottom fisheries, hydrocarbon exploration and extraction (Ramirez-Llodra et al., 2011; Ragnarsson et al., 2017; Montseny et al., 2021) and ongoing climate changes (Danovaro et al., 2017; Portilho-Ramos et al., 2022). Most restoration actions in marine ecosystems have been conducted in shallow waters (Bakker et al., 2002; Hughes and Paramor, 2004; Fraschetti et al., 2021), including tropical coral reefs (Rinkevich, 2005; Precht and Robbart, 2006; Young et al., 2012; Deignan and McDougald, 2022) and gorgonian forests (Linares et al., 2008; Fava et al., 2010; Weinberg, 2019; Basconi et al., 2020; Duarte et al., 2020; Montseny et al., 2021). The restoration of deep-sea habitats, including cold-water corals has been delayed due to the technological challenges associated with their remoteness, water depth (below 200 m), and high costs of intervention (Van Dover et al., 2014; Da Ros et al., 2019). It has been estimated that the cost of deep-sea ecosystem restoration might be up to 3–4 orders of magnitude higher than that of shallow-water ecosystems (Barbier et al., 2014). The most used techniques for active cold-water coral restoration are transplantation techniques from a healthy donor reef to a degraded reef, and the use of artificial structures (Brooke et al., 2006; Dahl et al., 2012; Jonsson et al., 2015; Montseny et al., 2021). Additionally, it may be necessary to harvest and maintain cold-water corals ex situ (i.e., in aquaria before returning them to their natural habitat. However, the slow growth rates of cold-water corals, the stress experienced by corals during harvesting, such as thermal changes, and the complexity of replicating their natural environment in the laboratory are some of the most important barriers to successful active restoration (Orejas et al., 2019). To overcome all these problems, improving underwater technologies and long-term monitoring strategies, as well as expanding knowledge about species interactions, are key steps in deep-sea restoration. To increase the success rate of transplantations, we would also benefit from a better understanding of the role and composition of coral-associated microbiomes and particularly of the beneficial taxa, which can be markers useful to select healthy coral colonies suitable for transplantation actions and to monitor over time the holobiont health status.
Information on the microbiomes of the cold-water corals in the frame of deep-sea restoration is still very limited. Spirochaetes or Endozoicomonas have been suggested to be bacterial taxa supporting the physiology of Desmophyllum pertusum (former Lophelia pertusa) and Madrepora oculata (Kellogg et al., 2009; Meistertzheim et al., 2016; Neave et al., 2016). However, the Desmophyllum pertusum microbiome appears to be more variable in terms of composition than the microbiome of M. oculata (Meistertzheim et al., 2016; Galand et al., 2018). An investigation of the microbiome during an in situ transplanting of Desmophyllum pertusum and M. oculata revealed shifts of some bacterial taxa (such as Entomoplasmatales, Spirochaetales, Rickettsiales and Endozoicomonas; Table 1; Chapron et al., 2020). In addition, the microbiome-based analyses were useful in investigating the habitat preference for the two cold-water species (i.e., D. pertusum did not show a strict preference for habitat due to its higher capacity to acquire bacteria from the surrounding waters, whereas M. oculata did) (Chapron et al., 2020). This could be crucial information in deep-water coral restoration as well as the potential presence of pathogens in gorgonians, which could hinder their recovery. Deep-sea ecosystem restoration appears to be the biggest challenge among marine habitat restoration actions; the microbiome could help address this challenge.
Seagrass meadows, mangroves and macroalgal forests support high biodiversity levels by providing food, habitat, and shelter to a variety of marine organisms. They also underpin many important ecosystem functions and services such as primary and secondary productivity, C sequestration (acting as a “blue carbon” storage system; Duarte, 2017), the provision of nursery areas for endangered and commercially important species, protection from coastal erosion (Duffy et al., 2019). However, these meadows/forests are facing dramatic decline worldwide, so effective restoration efforts are urgently needed to recover their associated key ecosystem functions and services (Paling et al., 2009; Tan et al., 2020; Cebrian et al., 2021; Dahdouh-Guebas and Cannicci, 2021; Dunic et al., 2021). We detail below the general characteristics of the microbiomes of these vegetated ecosystems and perspectives for their potential application to the restoration of such habitats.
Seagrass meadows (Figure 2) - Classical seagrass restoration practices have obtained contrasting results in different coastal habitats, either testing bare-root transplanting, alternative methods such as deploying multiple plants in larger cores, planting seagrass seeds or using biodegradable containers (Zhou et al., 2014; van Katwijk et al., 2016; Da Ros et al., 2021; Mokumo et al., 2023). There is broad consensus that harnessing positive biological interactions, including those between seagrass and microbiota, can increase restoration success (Halpern et al., 2007; Silliman et al., 2015; Tan et al., 2020; Valdez et al., 2020). Different members of the seagrass microbiota can indeed benefit their hosts (recently reviewed by (Ugarelli et al., 2017; Tarquinio et al., 2019; Conte et al., 2021). Epiphytic diazotrophic bacteria can increase nutrient bioavailability through nitrogen fixation and phosphorous solubilization while sulphate-reducing bacteria play a similar role in the rhizosphere and roots through anaerobic nitrogen fixation and organic matter mineralisation (Table 1; Welsh, 2000; Hamisi et al., 2009; Sharma et al., 2013; Tarquinio et al., 2019; Mohr et al., 2021). Several microbes involved in growth-promotion activities in plants or macroalgae (like Kocuria sp., Vibrio sp., Methylophilus sp., Alteromonas macleodii, and Marinomonas sp.) have been described also as epi- or endophytic microbes of seagrasses, hence hypothesized to play a similar role (Tarquinio et al., 2019). As in terrestrial plants, seagrass roots and rhizosphere are also enriched with beneficial sulphide-oxidising bacteria able to remove the otherwise toxic H2S produced by the sulphate-reducing bacteria during organic matter degradation (Conte et al., 2021) while sulphate-reducing bacteria have been suggested to detoxify from ethanol (another phytotoxin produced by the seagrass fermentative metabolism in the dark) (Ugarelli et al., 2017). Similarly, methanol (a waste by-product of seagrass metabolism known to inhibit plant growth and seed germination; Abanda-Nkpwatt et al., 2006) has been suggested to be consumed by methylotrophic bacteria (Methylophilaceae and Methylophagaceae), which are typically associated with seagrasses (Adamczyk et al., 2022). An additional protective mechanism of seagrasses can be attributed to microbes capable of producing antioxidant enzymes (e.g., Marinomonas mediterranea MMB-1), which can alleviate their oxidative stress by scavenging extracellular toxic radicals (Sanchez-Amat et al., 2010; Tarquinio et al., 2019). Additional plant-protection features, including the production of antiviral, antiparasitic, antibacterial and anti-biofouling compounds have been described or inferred for several bacteria (e.g., Actinobacteria) directly isolated from seagrasses or identified by metabarcoding (Marhaeni et al., 2010; Tarquinio et al., 2019; Boontanom and Chantarasiri, 2020). Such microbial products are supposed to benefit the seagrass by promoting healthy microbe-host interactions and contrasting potentially harmful outbreaks.
Bacteria have been by far the main target of seagrass microbiome studies, even though it can be expected that future research will provide evidence also on the relevance of archaea, as anticipated from terrestrial plants (Jung et al., 2020; Ayangbenro and Babalola, 2021). Similarly, the importance of fungi in fostering plant fitness is well-known in terrestrial ecosystems (Averill et al., 2022; Busby et al., 2022). As far as seagrasses are concerned, studies on fungi and other eukaryotic microbiota associated with them have mainly focused on pathogens and parasites like labyrinthulids, oomycetes and Phytomyxea (Sullivan et al., 2018; Ettinger and Eisen, 2019; Ettinger and Eisen, 2020), therefore the potentially beneficial functions and implications of microbial eukaryotes for the seagrass holobiont are still largely unknown (Wainwright et al., 2019; Ettinger et al., 2021; Liu et al., 2022).
Despite the growing awareness of the key role of the microbiome in seagrass health, little is still known about the possibility of using and/or modulating specific components of the seagrass microbiota to enhance transplantation success at large spatial scale (Trevathan-Tackett et al., 2019; Unsworth et al., 2019; Tan et al., 2020; Unsworth et al., 2022). Recent investigations on seagrass transplantation have demonstrated the importance of implementing a more holistic approach based on the “holobiont” concept, which argues that ‘macrobial’ hosts and their associated microbiota form a coherent biological entity and we need to consider them together to understand the ecology of hosts and their interactions (McFall-Ngai et al., 2013; Wang et al., 2021b). Milbrandt et al. (2008) showed that seagrass transplanting performed using portions of the original bulk sediments can increase transplant success, compared to the use of sterile sediment. Similarly, preliminary studies reported that seagrass seeds germinate and grow more quickly after the addition of their original sediments than in fully sterile sand (Boyer and Wyllie-Echeverria, 2010). These results suggested that seagrass transplanting may be more successful if using the plant’s autochthonous sediments, possibly also thanks to the beneficial microbes living in it (Tarquinio et al., 2019). Indeed, as for other terrestrial organisms (Jiang et al., 2022; Sorbara and Pamer, 2022; Morales Moreira et al., 2023), also the seagrass microbiome (including bacteria, archaea, fungi and other microbial eukaryotes associated with the leaves, roots and surrounding sediments) has been proposed to potentially increase the seagrass host fitness, resistance to environmental stressors and hazards and resilience (Fahimipour et al., 2017; Crump et al., 2018; Tarquinio et al., 2019; Trevathan-Tackett et al., 2019; Unsworth et al., 2019; Wainwright et al., 2019; Chen et al., 2022; Unsworth et al., 2022). For example, Fuggle et al. (2023), investigated the influence of root and sediment microbiomes on Zostera muelleri growth under stressful conditions (i.e., excess nutrient load). They found that the disruption of the root microbiome caused reduced seagrass growth under stressful conditions, thus inducing an uncoupling between bacteria involved in sulphate reduction and sulphide oxidation. These findings suggest a “cry for help” mechanism also for seagrass as observed in terrestrial plants, which under stress conditions attract beneficial (micro)organisms for protection (Meena et al., 2017; Khan et al., 2021). This mechanism indicates that the seagrass-microbiota interactions can influence host resilience. A recent study on in-situ seagrass (Zostera marina) transplanting investigated the changes in microbial assemblages after transplanting (Adamczyk et al., 2022), showing that the microbiota rapidly changed after transplanting and was strongly influenced by environmental conditions. In addition, this investigation revealed a stable microbiome core of the seagrass leaves represented by methanol-utilizing bacteria suggesting their relevant role for seagrass health, in view of transplanting and ecosystem restoration purposes.
Further investigations pointed out the need to re-think the holobiont concept in a “meta-holobiont” perspective (i.e., a network of holobionts that can exchange biomolecules and microbiota across generations, thus impacting the fitness of both biological scales: holobionts and meta-holobionts; Vannier et al., 2019). This new concept could have direct implications for seagrass restoration, as also exemplified by the in-situ evidence that mutualism between seagrasses and lucinid bivalves, which host sulphide-oxidizing gill symbionts, can reduce the sulphide-induced stress (Valdez et al., 2020; van der Geest et al., 2020). Other examples include the investigation conducted by Schenck et al. (2023), which performed a transplanting experiment of Zostera marina ex-situ, with its pathogenic parasite Labyrinthula zosterae (Table 1). The authors reported that changes in the host microbiome may influence the outcome of the host–parasite interactions, which would have direct implications for seagrass restoration and conservation management. Similarly, another study carried out by O’Connor et al. (2022) conducted an in situ reciprocal transplanting experiment with Zostera marina with and without its epiphytic macroalgal symbiont Smithora naiadum, showing that the presence of Smithora changes the Zostera leaf microbiota, thus suggesting a possible role of the microbiota in the plant-macroalgae symbiosis.
Overall, recent studies support the expectation that since the seagrass microbiome has a crucial role in the seagrass meta-holobiont (Vannier et al., 2019; Valdez et al., 2020) by enhancing nutrient uptake, immunity, and tolerance to environmental and anthropogenic stressors, its examination can represent a useful tool to increase restoration success. In this regard, the first milestone would be to define best practices in meta-holobiont transplanting, by emulating terrestrial microbiome studies (Emam, 2016; Bulot et al., 2017).
Mangroves (Figure 2) - Mangroves are highly productive and valuable habitat-forming species in tropical marine ecosystems, that are facing a rapid human-induced decline worldwide and need large-scale restoration interventions (Romañach et al., 2018; Birnbaum and Trevathan-Tackett, 2022). Mangrove restoration is technically feasible in appropriate locations (Lewis, 2005), but projects often have failed because of the unsuitability of the transplanting sites (Lee et al., 2019), largely associated with the presence of plant growth-promoting microbiota living in the sediment (Holguin et al., 2001).
The failures obtained so far in mangrove restoration, suggest the need to explore in more detail the mangrove-microbe interactions and develop microbial-based interventions (Lee et al., 2019; Allard et al., 2020; Birnbaum and Trevathan-Tackett, 2022).
Classical microbiological research (mainly, non-omics approaches) has provided plentiful examples of mangrove-associated microbes (i.e., bacteria and fungi) with a role in plant health such as the production of growth-promoting phytohormones (Xu et al., 2018), metabolites that protect the plant against stress (Lata et al., 2018), the provision of nutrients through nitrogen fixation and the phosphate solubilization (Jha et al., 2011; Fukami et al., 2018), as well as the degradation of contaminants (Sipahutar and Vangnai, 2017), which can alleviate the toxic effects of soil pollutants on the plant (Siraj et al., 2023 and references within). However, mangrove-microbiota interactions have been scarcely explored for their potential to enhance mangrove restoration success (Holguin et al., 2001; Birnbaum and Trevathan-Tackett, 2022). Pioneering laboratory work inspired by terrestrial research on plant-growth-promoting bacteria (PGPB) found that the inoculation of mangroves with diazotrophic cyanobacteria isolated by the mangroves themselves resulted in rapid colonization of the roots and the transfer of N from the N2-fixing bacteria to the plant (Holguin et al., 2001 and references therein). Successively, El-Tarabily and Youssef (2010), reported a phosphate-solubilizing bacterium (Oceanobacillus picturae) isolated from the mangrove Avicennia marina rhizosphere, able to strongly enhance plant growth, and suggested its possible use in reforestation programs (Table 1). The same authors also studied endophytic mangrove actinobacteria, describing one of the most promising strains (Streptomyces mutabilis) able to promote mangrove growth under laboratory conditions (El-Tarabily et al., 2021). Janarthine and Eganathan (2012), isolated one endophytic bacterial strain (Sporosarcina aquimarina) from pneumatophores of Avicennia marina, which enhanced the growth of different species of mangroves. Similarly, Soldan et al. (2019) isolated bacterial endophytes from mangrove propagules, describing one Gordonia terrae strain able to strongly promote the mangrove propagule germination and root colonization ability.
Overall, the available evidence supports the expectation that the mangrove microbiota can have great potential to help increase mangrove restoration success. However, additional research is needed to advance both theoretical and applied knowledge, by integrating classical microbiological studies with meta-omics, also performing microbiome-assisted mangrove transplant experiments in the field (Allard et al., 2020; Birnbaum and Trevathan-Tackett, 2022).
Macroalgal forests (Figure 2) - Among macroalgal forests, kelps (brown algae in the order Laminariales) play essential roles as ecosystem engineers in temperate coastal marine environments. Besides the ecosystem services provided, they are also important in many industries to produce alginates, human food supplements or food for abalone aquaculture (Burgunter-Delamare et al., 2022 and citations therein). Climate change and anthropogenic impacts are contributing to the global decline of the macroalgal forests making it increasingly urgent to restore these habitats (Mills et al., 2017). The history of kelp forest restoration, based on transplanting, seeding, grazer control, and artificial reefs (Eger et al., 2022), revealed that interactions between kelps and its associated microbiome can be crucial for obtaining successful restoration (Eger et al., 2022). The microbial partners, indeed, regulate and support their metabolism, health, fitness, resistance to pathogens, and adaptation to environmental changes (Wiese et al., 2009; Goecke et al., 2010; Dittami et al., 2016; Burgunter-Delamare et al., 2020).
Most available studies on the kelp microbiome are descriptive (i.e., focused on microbiome composition and distribution, and the interactions with environmental conditions; Lachnit et al., 2011; King et al., 2023). Knowing the environment and host characteristics shaping the kelp microbiota is important, as this may have implications on how we design restoration and/or future-proofing programs (Wood et al., 2019). Previous investigations documented that kelp and brown algae-associated microbiota is typically dominated by Alphaproteobacteria, Gammaproteobacteria, and Bacteroidota (Burgunter-Delamare et al., 2022 and citations therein; Table 1). In perennial species, such as Saccharina latissima, differences in the assemblage composition have been observed between different parts of the individuals, for example between apex and meristem. This is due to the algal growth mainly occurring in the meristem region, and younger meristem tissues are typically less colonised by bacteria (Burgunter-Delamare et al., 2022). Planctomycetes, which were observed predominantly in the S. latissima apex, have been observed also in other species, such as the brown algae Fucus vesiculosus, and are typical components of algal biofilms (Table 1; Lage and Bondoso, 2014; Parrot et al., 2019). The N. luetkeana blades’ bacterial genomes spanned 6 phyla and 19 families and included common alga-associated microbial symbionts such as those belonging to the genus Granulosicoccus. Key functions encoded in kelp-associated bacterial genomes included dissolved organic matter assimilation, alginate metabolism, vitamin B12 biosynthesis, and nitrogen reduction from nitrate and urea to ammonium, potentially providing the host kelp with vitamins and reduced nitrogen compounds (Weigel et al., 2022). Field studies conducted on the bull kelp (Nereocystis luetkeana) microbiota, compared with other different substrates and the surrounding water column, revealed that new kelp blade tissues were colonized by markedly distinct microbial taxa (Table 1; Weigel and Pfister, 2021; Ramírez-Puebla et al., 2022). This suggests that microbial establishment on algae surfaces is more than just an attraction to a polysaccharide-rich surface and that host-specific factors may deter some surface-associated marine microbial taxa (Ramírez-Puebla et al., 2022; Weigel et al., 2022). Microbes can also be useful to sustain food webs in kelp forests as indicated by an experimental study based on the manipulation of bacterial abundances in kelp biofilm (Singh et al., 2021).
Despite the increasing interest in the diversity and functions of the algal microbiome, only a few studies have gone beyond the structural and functional description of the microbiomes and evaluated the responses to experimental inoculation of target microorganisms (Vairappan et al., 2001; Peng and Li, 2013) or assessed the role of microbiome in restoration actions (Eger et al., 2020). These approaches may be useful to identify beneficial or pathogenic microbes and to help in enhancing kelp performance and/or confer resistance or resilience to future environmental conditions. Although some studies tested the responses of the microbiomes of different kelp species (Macrocystis pyrifera and Ecklonia radiata; Minich et al., 2018; Qiu et al., 2019) to climate changes (e.g., increased temperature and partial pressure of carbon dioxide (pCO2), how these changes will module the holobiont responses in restoration interventions is still practically unknown. This will be another challenge for restoration practices.
The canopy-forming macroalgae Cystoseira sensu lato (s.l.) (Figure 2G; including the Cystoseira, Gongolaria and Ericaria genera; Smith et al., 2023), are among the most important brown macroalgal forests, which are object of restoration projects for their key ecological role in coastal habitats (Cebrian et al., 2021) and provision of ecosystem services (including potential pharmaceuticals; Zbakh et al., 2020). Although available studies on the microbiome of Cystoseira s.l. forests are very limited, an investigation on Ericaria amantacea showed a core of associated bacterial families (including Rhodobacteraceae, Flavobacteriaceae, Alteromonadaceae, Vibrionaceae, and Halomonadaceae) also present in other brown algae suggesting a great plasticity of the microbiome of these macroalgae (Malfatti et al., 2023). Other studies carried out on C. compressa showed a distinct microbiota from the surrounding water column, with only a few taxa in common (Table 1; Mancuso et al., 2016). At the same time, a clear successional pattern in the epiphytic bacteria of C. compressa was observed, with the gradual appearance of specific bacterial taxa (e.g., Verrucomicrobia, Actinobacteria) over the growing season (Mancuso et al., 2016). An increase of bacteria affiliated with Rhodobacteraceae, which comprise six potential pathogenic genera, Ruegeria, Nautella, Aquimarina, Loktanella, Saprospira, and Phaeobacter, was also observed in aged thalli of C. compressa. These bacteria could influence the health and ecology of the algae, suggesting a possible role of the microbiome in contributing to the extensive ongoing declines of populations of Cystoseira s.l. in the Mediterranean Sea. In this regard, information on red macroalgae revealed that their decline is caused by bacterial diseases, such as the case of Delisea pulchra affected by bleaching also in combination with temperature rise events (Case et al., 2011; Campbell et al., 2014). However, the microbiome can also prevent the establishment of pathogenic microbes (Abdul Malik et al., 2020). An example is the study conducted on the seaweed Phyllospora comosa (Fucales, Seirococcaceae), where a “core” of microbial taxa (i.e., present on all individuals sampled) was considered responsible for multiple functions, including biological defence (Wood et al., 2022). The capacity of aquatic macrophytes to “garden” protective microorganisms to the benefit of strengthened disease resistance has been demonstrated in the model invasive seaweed holobiont Agarophyton vermiculophyllum (Saha and Weinberger, 2019). This species is characterized by beneficial microbiota on its surface that protects from bacterial pathogens. Metabolites from the holobiont surface reduce the settlement of opportunistic pathogens and promote the settlement of other beneficial bacteria (Saha and Weinberger, 2019). Other functions can be attributed to the interactions between seaweeds and microbiomes; for example, investigations conducted on non-indigenous and invasive algae, such as Caulerpa cylindracea and Agarophyton vermiculophyllum (Table 1; Saha and Weinberger, 2019; Morrissey et al., 2021), reported a key role of microbiomes in their success in quickly colonizing new areas.
The tight relationship between macroalgae and microbiota suggests that they interact as a unified functional entity (i.e., holobiont), therefore, future restoration interventions cannot ignore the microbiome component (Morrissey et al., 2021). This holds true especially in the light of future climate change scenarios, which can determine shifts in macroalgal microbiomes and may lead to host disease, with potential cascading impacts on associated ecosystems (Qiu et al., 2019). In this regard, it remains largely unexplored how the interactions between macroalgae and microbes are affected by environmental stressors (van der Loos et al., 2019). Macroalgae–microbe interactions have been recognized as drivers of acclimatization to environmental changes as in the case of the brown alga Ectocarpus, whose ability to acclimate to salinity gradients has been attributed to their microbial associations (Dittami et al., 2016). In addition, it has been reported that the responses of macroalgae to various environmental perturbations, such as heat stress and nutrient enrichment, may depend on the niche-specific microbiota of macroalgae (i.e., endo-microbiota, epi-microbiota and rhizo-microbiota) (Morrissey et al., 2021). For example, in C. cylindracea, endomicrobiota has been reported to have the highest resistance, but the lowest resilience to environmental stress.
Given the rate at which macroalgal forests and other marine habitats are changing and given the predicted increases in the frequency and magnitude of multiple stressors, the need for subtidal marine macrophyte restoration efforts is evident (Wood et al., 2022). Future macroalgal forest restoration may rely on resistance and resilience of the associated microbiomes, therefore exploiting the provided probiotic functions to facilitate the holobiont adaptation to environmental stress, will be a step forward in the macroalgae forest restoration (Ghaderiardakani et al., 2020). Investigations into the selection and use of inoculants and probiotics in macroalgae restoration are very limited. A recent study based on metabarcoding analysis suggested that some bacterial taxa (Postechiella, Winogradskyella, Roseovarius and Arenibacter) are the main responsible for algal growth and potential candidates for probiotic consortia promoting the growth of Ericaria amentacea (Phaeophyceae) seedlings (Malfatti et al., 2023). Molecular analysis of the microbiome of macroalgal forests based on metagenomic approaches can be a valuable tool for biomonitoring and predicting their health status, thus guiding restoration actions. However, we urgently need to increase the number of in situ investigations of the macroalgal forest microbiome in different habitats and environmental conditions and at different spatial scales to understand more about how to use the microbiome in restoration actions.
Nature-based solutions (i.e., actions that are inspired by, supported by, or copied from nature), also those relying on microbes, are gaining momentum globally as a concept to address ecological and societal challenges. Here, we advocate for combining these two concepts to incorporate microbiome analysis into nature-based solutions for the restoration actions of different habitat-forming marine species (Figure 1).
Microbiome characterization and identification of beneficial microbial taxa or their potential metabolites can be used for screening and selection of the “meta-community and meta-ecosystem” (e.g., plants and their sediment containing microbiota and nutrients) to be transplanted. Traditionally, this screening and selection was done empirically (e.g., visually) but, nowadays we can leverage multi-omics and computational approaches (e.g., metabarcoding, metagenomics and meta-transcriptomics) to identify organisms, colonies or sediment clods to be translocated to the restoration site or to identify the most suitable substrate for larval/propagule settlements (Jorissen et al., 2021). Changes in the composition of microbiome are, indeed, considered an early warning of holobiont health (Glasl et al., 2019; Corinaldesi et al., 2022), therefore microbiome analysis might be particularly useful in deep-sea ecosystems (e.g., cold-water corals) for the selection of healthy organisms and their monitoring once translocated/transplanted. The biomonitoring of the microbiome of habitat-forming species is now possible thanks to well-consolidated methodologies and ready-to-use technologies. However, it does not represent a way to remediate failed restoration interventions, but rather a strategy for the early detection of potential pathogens and for making quick decisions, for example, to stop the spread of disease (Pollock et al., 2011; Traylor-Knowles et al., 2022). Probiotic and bio-promoter supplements could, once their efficacy has been demonstrated, not only increase the resistance of the holobiont (e.g., to coral bleaching and other stress factors) before transplanting, but also stabilize it or improve its health conditions after transplanting. Other microbiome-based strategies for addressing restoration intervention include isolating microbiome components through culturomics-based approaches, which allow the selection of microbes producing usable probiotics and or bio-promoters to be inoculated to transplanted organisms. Other methods based on culturomics include the natural microbiome-assisted evolution, used to select microbial taxa that show the greatest resilience to stress and thereby increase the fitness of transplanted species. However, these approaches are characterized by drawbacks (e.g., difficulties in culturing marine bacteria and in producing probiotics), which make their application difficult, especially for large-scale interventions. Given the urgency of implementing marine ecosystem restoration, culturomics-based approaches need to be optimized.
Microbiota can respond and adapt more rapidly than their associated pluricellular organisms to climate change and anthropogenic stressors, potentially increasing holobiont resilience to novel environmental conditions (Duarte, 2017; Martin et al., 2019). Due to their great potential, microbiome-based approaches can be very useful for the recovery of degraded ecosystems, contributing to their resilience and the achievement of the UN objectives of sustainability and habitat restoration. In the context of international strategies concerning ecological restoration, the use of the scientific knowledge acquired on the marine microbiome deserves to be exploited to assist both traditional and innovative restoration approaches. Taking a cue from the positive results obtained in terrestrial ecosystems, especially from reforestation experiments, there is ample space to exploit the potential of the microbiome in the restoration of different habitat-forming species. Although the use of microbiomes is still in its infancy in marine ecosystem restoration, it is being successfully applied in the restoration of coral reefs and experimentally in seagrass beds and macroalgal forests. The delay in the use of microbiomes in marine restoration is more relevant for deep-sea ecosystems due to technological and logistical difficulties. Therefore, upgrading the portfolio of technology tools for supporting such approaches, especially in deep-sea ecosystems (e.g., collecting samples in the long-term monitoring of deep-sea communities) is, as well, needed to effectively make microbiome-based restoration feasible in the future. Marine microbial nature-based restoration is an emerging and promising sector to accelerate and expand the spatial scale of blue restoration plans and to increase success of restoration initiatives.
CC conceived the review and its structure. RD, MC, AD’A and CG contributed to the manuscript design. CC, SB, MC, ER, SV contributed to the first draft and the different sections of the manuscript. All authors contributed to the article and approved the submitted version.
This study was conducted in the framework of the National Recovery and Resilience Plan (NRRP), Mission 4 Component 2 Investment 1.4 - Call for tender No. 3138 of 16 December 2021, rectified by Decree n.3175 of 18 December 2021 of Italian Ministry of University and Research funded by the European Union – NextGenerationEU; Award Number: Project code CN_00000033, Concession Decree No. 1034 of 17 June 2022 adopted by the Italian Ministry of University and Research, Project title “National Biodiversity Future Center - NBFC”. The review was also produced in the framework of the EASME–EMFF (Sustainable Blue Economy) Project AFRIMED (http://afrimed-project.eu/, grant agreement N. 789059), supported by the European Community, and the project BioDiversa + 2021-134 (https://www.biodiversa.eu/2023/04/19/forescue/).
The authors wish to thank the two reviewers for their helpful comments and suggestions on this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abanda-Nkpwatt D., Krimm U., Coiner H. A., Schreiber L., Schwab W. (2006). Plant volatiles can minimize the growth suppression of epiphytic bacteria by the phytopathogenic fungus Botrytis cinerea in co-culture experiments. Environ. Exp. Bot. 56, 108–119. doi: 10.1016/j.envexpbot.2005.01.010
Abdul Malik S. A., Bedoux G., Garcia Maldonado J. Q., Freile-Pelegrín Y., Robledo D., Bourgougnon N. (2020). Defence on surface: macroalgae and their surface-associated microbiome. Adv. Bot. Res. 95, 327–368. doi: 10.1016/bs.abr.2019.11.009
Adamczyk E. M., O’Connor M. I., Parfrey L. W. (2022). Seagrass (Zostera marina) transplant experiment reveals core microbiota and resistance to environmental change. Mol. Ecol. 31, 5107–5123. doi: 10.1111/mec.16641
Ainsworth T. D., Fordyce A. J., Camp E. F. (2017). The other microeukaryotes of the coral reef microbiome. Trends Microbiol. 25, 980–991. doi: 10.1016/j.tim.2017.06.007
Ainsworth T. D., Krause L., Bridge T., Torda G., Raina J. B., Zakrzewski M., et al. (2015). The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME. J. 9, 2261–2274. doi: 10.1038/ismej.2015.39
Alagely A., Krediet C. J., Ritchie K. B., Teplitski M. (2011). Signaling-mediated cross-talk modulates swarming and biofilm formation in a coral pathogen Serratia marcescens. ISME. J. 5, 1609–1620. doi: 10.1038/ismej.2011.45
Allard S. M., Costa M. T., Bulseco A. N., Helfer V., Wilkins L. G. E., Hassenrück C., et al. (2020). Introducing the mangrove microbiome initiative: identifying microbial research priorities and approaches to better understand, protect, and rehabilitate mangrove ecosystems. mSystems 5, 1–10. doi: 10.1128/mSystems.00658-20
Angiolillo M., Canese S. (2018). “Deep gorgonians and corals of the mediterranean sea,” in Corals in a changing world. Eds. Duque C., Camacho E. T. (London, United Kingdom: InTech), 29–49. doi: 10.5772/intechopen.69686
Apprill A., Mcnally S., Parsons R., Weber L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microbial. Ecol. 75, 129–137. doi: 10.3354/ame01753
Averill C., Anthony M. A., Baldrian P., Finkbeiner F., van den Hoogen J., Kiers T., et al. (2022). Defending Earth’s terrestrial microbiome. Nat. Microbiol. 7, 1717–1725. doi: 10.1038/s41564-022-01228-3
Ayangbenro A. S., Babalola O. O. (2021). Reclamation of arid and semi-arid soils: The role of plant growth-promoting archaea and bacteria. Curr. Plant Biol. 25, 100173. doi: 10.1016/j.cpb.2020.100173
Backer R., Rokem J. S., Ilangumaran G., Lamont J., Praslickova D., Ricci E., et al. (2018). Plant growth-promoting rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 871. doi: 10.3389/fpls.2018.01473
Bakker J. P., Esselink P., Dijkema K. S., Van Duin W. E., De Jong D. J. (2002). Restoration of salt marshes in the Netherlands. Hydrobiologia 478, 29–51. doi: 10.1023/A:1021066311728
Balla A., Silini A., Cherif-Silini H., Chenari Bouket A., Alenezi F. N., Belbahri L. (2022). Recent advances in encapsulation techniques of plant growth-promoting microorganisms and their prospects in the sustainable agriculture. Appl. Sci. 12, 1–18. doi: 10.3390/app12189020
Ballesteros E. (2006). Mediterranean coralligenous assemblages: a synthesis of present knowledge. Oceanogr. Mar. Biol. 44, 123–195. doi: 10.1016/j.jenvman.2012.07.024
Bang C., Dagan T., Deines P., Dubilier N., Duschl W. J., Fraune S., et al. (2018). Metaorganisms in extreme environments: do microbes play a role in organismal adaptation? Zoology 127, 1–19. doi: 10.1016/j.zool.2018.02.004
Barbier E. B., Moreno-Mateos D., Rogers A. D., Aronson J., Pendleton L., Danovaro R., et al. (2014). Protect the deep sea. Nature 505, 475–477. doi: 10.1038/505475a
Barnes H. (2010). Oceanography and marine biology. Eds. Gibson R. N., Atkinson R. J. A., Gordon J. D. M. (New York, USA: Chapman and Hall/CRC). doi: 10.1201/EBK1439821169
Basconi L., Cadier C., Guerrero-Limón G. (2020). “Challenges in marine restoration ecology: how techniques, assessment metrics, and ecosystem valuation can lead to improved restoration success,” in YOUMARES 9-the oceans: our research, our future: proceedings of the 2018 conference for YOUng MArine RESearcher in oldenburg, Germany (London, United Kingdom: Springer International Publishing), 83–99.
Bayer T., Arif C., Ferrier-Pagès C., Zoccola D., Aranda M., Voolstra C. R. (2013). Bacteria of the genus Endozoicomonas dominate the microbiome of the Mediterranean gorgonian coral Eunicella cavolini. Mar. Ecol. Prog. Ser. 479, 75–84. doi: 10.3354/meps10197
Bellwood D. R., Hughes T. P., Folke C., Nyström M. (2004). Confronting the coral reef crisis. Nature 429, 827–833. doi: 10.1038/nature02691
Berg G., Rybakova D., Fischer D., Cernava T., Vergès M. C. C., Charles T., et al. (2020). Microbiome definition re-visited: old concepts and new challenges. Microbiome 8, 1–22. doi: 10.1186/S40168-020-00875-0
Bernasconi R., Stat M., Koenders A., Huggett M. J. (2019). Global networks of symbiodinium-bacteria within the coral holobiont. Microb. Ecol. 77, 794–807. doi: 10.1007/s00248-018-1255-4
Biagi E., Caroselli E., Barone M., Pezzimenti M., Teixido N., Soverini M., et al. (2020). Patterns in microbiome composition differ with ocean acidification in anatomic compartments of the Mediterranean coral Astroides calycularis living at CO2 vents. Sci. Total. Environ. 724, 138048. doi: 10.1016/j.scitotenv.2020.138048
Birnbaum C., Trevathan-Tackett S. M. (2022). Aiding coastal wetland restoration via the belowground soil microbiome: an overview. Restor. Ecol. 31, e13824. doi: 10.1111/REC.13824
Boetius A., Anesio A. M., Deming J. W., Mikucki J. A., Rapp J. Z. (2015). Microbial ecology of the cryosphere: Sea ice and glacial habitats. Nat. Rev. Microbiol. 13, 677–690. doi: 10.1038/nrmicro3522
Boontanom P., Chantarasiri A. (2020). Short Communication: Diversity of culturable epiphytic bacteria isolated from seagrass (Halodule uninervis) in Thailand and their preliminary antibacterial activity. Biodiversitas 21, 2907–2913. doi: 10.13057/biodiv/d210706
Boström-Einarsson L., Babcock R. C., Bayraktarov E., Ceccarelli D., Cook N., Ferse S. C. A., et al. (2020). Coral restoration – A systematic review of current methods, successes, failures and future directions. PloS One 15, e0226631. doi: 10.1371/journal.pone.0226631
Bourne D. G., Morrow K. M., Webster N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Boyer K. E., Wyllie-Echeverria S. (2010) Eelgrass conservation and restoration in San Francisco Bay: Opportunities and constraints. Available at: http://www.sfbaysubtidal.org/PDFS/Ap8-1Eelgrass.pdf.
Brooke S., Koenig C. C., Shepard A. N. (2006). Oculina banks restoration project : description and preliminary assessment. Proc. 57th. Gulf. Caribbean. Fisheries. Inst. 607–620.
Bulot A., Potard K., Bureau F., Bérard A., Dutoit T. (2017). Ecological restoration by soil transfer: impacts on restored soil profiles and topsoil functions. Restor. Ecol. 25, 354–366. doi: 10.1111/rec.12424
Burgunter-Delamare B., KleinJan H., Frioux C., Fremy E., Wagner M., Corre E., et al. (2020). Metabolic complementarity between a brown alga and associated cultivable bacteria provide indications of beneficial interactions. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00085
Burgunter-Delamare B., Tanguy G., Legeay E., Boyen C., Dittami S. M. (2022). Effects of sampling and storage procedures on 16S rDNA amplicon sequencing results of kelp microbiomes. Mar. Genomics 63, 100944. doi: 10.1016/j.margen.2022.100944
Busby P. E., Newcombe G., Neat A. S., Averill C. (2022). Facilitating reforestation through the plant microbiome: perspectives from the phyllosphere. Annu. Rev. Phytopathol. 60, 337–356. doi: 10.1146/annurev-phyto-021320-010717
Calderón K., Spor A., Breuil M. C., Bru D., Bizouard F., Violle C., et al. (2017). Effectiveness of ecological rescue for altered soil microbial communities and functions. ISME. J. 11, 272–283. doi: 10.1038/ismej.2016.86
Campbell A. H., VergéS A., Steinberg P. D. (2014). Demographic consequences of disease in a habitat-forming seaweed and impacts on interactions between natural enemies. Ecology 95, 142–152. doi: 10.1890/13-0213.1
Case R. J., Longford S. R., Campbell A. H., Low A., Tujula N., Steinberg P. D., et al. (2011). Temperature induced bacterial virulence and bleaching disease in a chemically defended marine macroalga. Environ. Microbiol. 13, 529–537. doi: 10.1111/j.1462-2920.2010.02356.x
Casey J. M., Connolly S. R., Ainsworth T. D. (2015). Coral transplantation triggers shift in microbiome and promotion of coral disease associated potential pathogens. Sci. Rep. 5, 11903. doi: 10.1038/srep11903
Casoli E., Mancini G., Ventura D., Pace D. S., Belluscio A., Ardizzone G. D. (2020). Reteporella spp. success in the re-colonization of bare coralligenous reefs impacted by Costa Concordia shipwreck: The pioneer species you did not expect. Mar. pollut. Bull. 161, 111808. doi: 10.1016/j.marpolbul.2020.111808
Casoli E., Ventura D., Mancini G., Cardone S., Farina F., Donnini L., et al. (2022). Rehabilitation of Mediterranean animal forests using gorgonians from fisheries by-catch. Restor. Ecol. 30, e13465. doi: 10.1111/rec.13465
Cassán F., Coniglio A., López G., Molina R., Nievas S., de Carlan C. L. N., et al. (2020). Everything you must know about Azospirillum and its impact on agriculture and beyond. Biol. Fertil. Soils. 56, 461–479. doi: 10.1007/s00374-020-01463-y
Catroux G., Hartmann A., Revellin C. (2001). Trends in rhizobial inoculant production and use. Plant Soil 230, 21–30. doi: 10.1023/A:1004777115628
Cavicchioli R., Ripple W. J., Timmis K. N., Azam F., Bakken L. R., Baylis M., et al. (2019). Scientists’ warning to humanity: microorganisms and climate change. Nat. Rev. Microbiol. 17, 569–586. doi: 10.1038/s41579-019-0222-5
Cebrian E., Tamburello L., Verdura J., Guarnieri G., Medrano A., Linares C., et al. (2021). A roadmap for the restoration of mediterranean macroalgal forests. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.709219
Chapron L., Lartaud F., Le Bris N., Peru E., Galand P. E. (2020). Local variability in microbiome composition and growth suggests habitat preferences for two reef-building cold-water coral species. Front. Microbiol. 11. doi: 10.3389/FMICB.2020.00275
Chen J., Zang Y., Yang Z., Qu T., Sun T., Liang S., et al. (2022). Composition and functional diversity of epiphytic bacterial and fungal communities on marine macrophytes in an intertidal zone. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.839465
Cheng K., Tong M., Cai Z., Jong M. C., Zhou J., Xiao B. (2023). Prokaryotic and eukaryotic microbial communities associated with coral species have high host specificity in the South China Sea. Sci. Total. Environ. 867, 161185. doi: 10.1016/j.scitotenv.2022.161185
Chu N. D., Vollmer S. V. (2016). Caribbean corals house shared and host-specific microbial symbionts over time and space. Environ. Microbiol. Rep. 8, 493–500. doi: 10.1111/1758-2229.12412
Conte C., Rotini A., Manfra L., D’Andrea M. M., Winters G., Migliore L. (2021). The seagrass holobiont: what we know and what we still need to disclose for its possible use as an ecological indicator. Water 13, 406. doi: 10.3390/W13040406
Corinaldesi C., Varrella S., Tangherlini M., Dell’Anno A., Canensi S., Cerrano C., et al. (2022). Changes in coral forest microbiomes predict the impact of marine heatwaves on habitat-forming species down to mesophotic depths. Sci. Total. Environ. 823, 153701. doi: 10.1016/j.scitotenv.2022.153701
Crossay T., Majorel C., Redecker D., Gensous S., Medevielle V., Durrieu G., et al. (2019). Is a mixture of arbuscular mycorrhizal fungi better for plant growth than single-species inoculants? Mycorrhiza 29, 325–339. doi: 10.1007/s00572-019-00898-y
Crump B. C., Koch E. W. (2008). Attached bacterial populations shared by four species of aquatic angiosperms. Appl. Environ. Microbiol. 74, 5948–5957. doi: 10.1128/AEM.00952-08
Crump B. C., Wojahn J. M., Tomas F., Mueller R. S. (2018). Metatranscriptomics and amplicon sequencing reveal mutualisms in seagrass microbiomes. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00388
Dahdouh-Guebas F., Cannicci S. (2021). Mangrove restoration under shifted baselines and future uncertainty. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.799543
Dahl M. P., Pereyra R. T., Lundälv T., André C. (2012). Fine-scale spatial genetic structure and clonal distribution of the cold-water coral Lophelia pertusa. Coral. Reefs. 31, 1135–1148. doi: 10.1007/s00338-012-0937-5
Damjanovic K., Van Oppen M. J. H., Menéndez P., Blackall L. L. (2019). Experimental inoculation of coral recruits with marine bacteria indicates scope for microbiome manipulation in Acropora tenuis and Platygyra daedalea. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01702
Danovaro R., Corinaldesi C., Dell’Anno A., Fuhrman J. A., Middelburg J. J., Noble R. T., et al. (2011). Marine viruses and global climate change. FEMS Microbiol. Rev. 35, 993–1034. doi: 10.1111/J.1574-6976.2010.00258.X
Danovaro R., Corinaldesi C., Dell’Anno A., Snelgrove P. V. R. (2017). The deep-sea under global change. Curr. Biol. 27, R461–R465. doi: 10.1016/j.cub.2017.02.046
Danovaro R., Nepote E., Lo Martire M., Carugati L., Da Ros Z., Torsani F., et al. (2020). Multiple declines and recoveries of Adriatic seagrass meadows over forty years of investigation. Mar. pollut. Bull. 161, 111804. doi: 10.1016/j.marpolbul.2020.111804
Da Ros Z., Corinaldesi C., Dell’Anno A., Gambi C., Torsani F., Danovaro R. (2021). Restoration of Cymodocea nodosa seagrass meadows: efficiency and ecological implications. Restor. Ecol. 29, e13313. doi: 10.1111/rec.13313
Da Ros Z., Dell’Anno A., Morato T., Sweetman A. K., Carreiro-Silva M., Smith C. J., et al. (2019). The deep sea: The new frontier for ecological restoration. Mar. Policy 108, 103642. doi: 10.1016/j.marpol.2019.103642
Deaker R., Roughley R. J., Kennedy I. R. (2004). Legume seed inoculation technology - A review. Soil Biol. Biochem. 36, 1275–1288. doi: 10.1016/j.soilbio.2004.04.009
De Deyn G. B., Raaijmakers C. E., van der Putten W. H. (2004). Plant community development is affected by nutrients and soil biota. J. Ecol. 92, 824–834. doi: 10.1111/j.0022-0477.2004.00924.x
Deignan L. K., McDougald D. (2022). Differential response of the microbiome of pocillopora acuta to reciprocal transplantation within Singapore. Microb. Ecol. 83, 608–618. doi: 10.1007/s00248-021-01793-w
Dick G. J. (2019). The microbiomes of deep-sea hydrothermal vents: distributed globally, shaped locally. Nat. Rev. Microbiol. 17, 271–283. doi: 10.1038/s41579-019-0160-2
Dittami S. M., Duboscq-Bidot L., Perennou M., Gobet A., Corre E., Boyen C., et al. (2016). Host–microbe interactions as a driver of acclimation to salinity gradients in brown algal cultures. ISME. J. 10, 51–63. doi: 10.1038/ismej.2015.104
Doering T., Wall M., Putchim L., Rattanawongwan T., Schroeder R., Hentschel U., et al. (2021). Towards enhancing coral heat tolerance: a “microbiome transplantation” treatment using inoculations of homogenized coral tissues. Microbiome 9, 102. doi: 10.1186/s40168-021-01053-6
Doney S. C., Ruckelshaus M., Emmett Duffy J., Barry J. P., Chan F., English C. A., et al. (2012). Climate change impacts on marine ecosystems. Ann. Rev. Mar. Sci. 4, 11–37. doi: 10.1146/annurev-marine-041911-111611
Duarte C. M. (2017). Reviews and syntheses: Hidden forests, the role of vegetated coastal habitats in the ocean carbon budget. Biogeosciences 14, 301–310. doi: 10.5194/bg-14-301-2017
Duarte C. M., Agusti S., Barbier E., Britten G. L., Castilla J. C., Gattuso J.-P., et al. (2020). Rebuilding marine life. Nature 580, 39–51. doi: 10.1038/s41586-020-2146-7
Dubey A., Malla M. A., Khan F., Chowdhary K., Yadav S., Kumar A., et al. (2019). Soil microbiome: a key player for conservation of soil health under changing climate. Biodivers. Conserv. 28, 2405–2429. doi: 10.1007/s10531-019-01760-5
Duffy J. E., Benedetti-Cecchi L., Trinanes J., Muller-Karger F. E., Ambo-Rappe R., Boström C., et al. (2019). Toward a coordinated global observing system for seagrasses and marine macroalgae. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00317
Dunic J. C., Brown C. J., Connolly R. M., Turschwell M. P., Côté I. M. (2021). Long-term declines and recovery of meadow area across the world’s seagrass bioregions. Glob. Chang. Biol. 27, 4096–4109. doi: 10.1111/gcb.15684
Dunphy C. M., Gouhier T. C., Chu N. D., Vollmer S. V. (2019). Structure and stability of the coral microbiome in space and time. Sci. Rep. 9, 1–13. doi: 10.1038/s41598-019-43268-6
Eger A. M., Marzinelli E. M., Christie H., Fagerli C. W., Fujita D., Gonzalez A. P., et al. (2022). Global kelp forest restoration: past lessons, present status, and future directions. Biol. Rev. 97, 1449–1475. doi: 10.1111/brv.12850
Eger A. M., Marzinelli E., Gribben P., Johnson C. R., Layton C., Steinberg P. D., et al. (2020). Playing to the positives: using synergies to enhance kelp forest restoration. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00544
El-Tarabily K. A., Ramadan G. A., Elbadawi A. A., Hassan A. H., Tariq S., Ghazal E. W., et al. (2021). The Marine Endophytic Polyamine-Producing Streptomyces mutabilis UAE1 Isolated From Extreme Niches in the Arabian Gulf Promotes the Performance of Mangrove (Avicennia marina) Seedlings Under Greenhouse Conditions. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.710200
El-Tarabily K. A., Youssef T. (2010). Enhancement of morphological, anatomical and physiological characteristics of seedlings of the mangrove Avicennia marina inoculated with a native phosphate-solubilizing isolate of Oceanobacillus picturae under greenhouse conditions. Plant Soil 332, 147–162. doi: 10.1007/s11104-010-0280-y
Emam T. (2016). Local soil, but not commercial AMF inoculum, increases native and non-native grass growth at a mine restoration site. Restor. Ecol. 24, 35–44. doi: 10.1111/rec.12287
Epstein H. E., Smith H. A., Torda G., van Oppen M. J. H. (2019). Microbiome engineering: enhancing climate resilience in corals. Front. Ecol. Environ. 17, 100–108. doi: 10.1002/fee.2001
Ettinger C. L., Eisen J. A. (2019). Characterization of the mycobiome of the seagrass, zostera marina, reveals putative associations with marine chytrids. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02476
Ettinger C. L., Eisen J. A. (2020). Fungi, bacteria and oomycota opportunistically isolated from the seagrass, Zostera marina. PloS One 15, e0236135. doi: 10.1371/journal.pone.0236135
Ettinger C. L., Vann L. E., Eisen J. A. (2021). Global diversity and biogeography of the zostera marina mycobiome. Appl. Environ. Microbiol. 87, 1–19. doi: 10.1128/AEM.02795-20
Fahimipour A. K., Kardish M. R., Lang J. M., Green J. L., Eisen J. A., Stachowicz J. J. (2017). Globalscale structure of the eelgrass microbiome. Appl. Environ. Microbiol. 83, e03391–e03316. doi: 10.1128/AEM.03391-16
Fava F., Bavestrello G., Valisano L., Cerrano C. (2010). Survival, growth and regeneration in explants of four temperate gorgonian species in the Mediterranean Sea. Ital. J. Zool. 77, 44–52. doi: 10.1080/11250000902769680
Ferse S. C. A., Hein M. Y., Rölfer L. (2021). A survey of current trends and suggested future directions in coral transplantation for reef restoration. PloS One 16, e0249966. doi: 10.1371/journal.pone.0249966
Fragoso Ados Santos H., Duarte G. A. S., Rachid C. T. D. C., Chaloub R. M., Calderon E. N., Marangoni L. F. D. B., et al. (2015). Impact of oil spills on coral reefs can be reduced by bioremediation using probiotic microbiota. Sci. Rep. 5, 1–11. doi: 10.1038/srep18268
Fraschetti S., McOwen C., Papa L., Papadopoulou N., Bilan M., Boström C., et al. (2021). Where is more important than how in coastal and marine ecosystems restoration. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.626843
Fuggle R. E., Gribben P. E., Marzinelli E. M. (2023). Experimental evidence root-associated microbes mediate seagrass response to environmental stress. J. Ecol. 111, 1079–1093. doi: 10.1111/1365-2745.14081
Fukami J., Cerezini P., Hungria M. (2018). Azospirillum: benefits that go far beyond biological nitrogen fixation. AMB. Express. 8, 1–12. doi: 10.1186/S13568-018-0608-1
Galand P. E., Chapron L., Meistertzheim A. L., Peru E., Lartaud F. (2018). The effect of captivity on the dynamics of active bacterial communities differs between two deep-sea coral species. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02565
Gann G. D., McDonald T., Walder B., Aronson J., Nelson C. R., Jonson J., et al. (2019). International principles and standards for the practice of ecological restoration. Second edition. Restor. Ecol. 27, S1–S46. doi: 10.1111/rec.13035
Gantner S., Andersson A. F., Alonso-Sáez L., Bertilsson S. (2011). Novel primers for 16S rRNA-based archaeal community analyses in environmental samples. J. Microbiol. Methods 84, 12–18. doi: 10.1016/j.mimet.2010.10.001
Gardner S. G., Leggat W., Ainsworth T. D. (2023). The microbiome of the endosymbiotic Symbiodiniaceae in corals exposed to thermal stress. Hydrobiologia 1–20, 3685–3704. doi: 10.1007/s10750-023-05221-7
Garrabou J., Gómez-Gras D., Ledoux J. B., Linares C., Bensoussan N., López-Sendino P., et al. (2019). Collaborative database to track mass mortality events in the mediterranean sea. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00707
Garren M., Azam F. (2012). New directions in coral reef microbial ecology. Environ. Microbiol. 14, 833–844. doi: 10.1111/j.1462-2920.2011.02597.x
Ghaderiardakani F., Quartino M. L., Wichard T. (2020). Microbiome-dependent adaptation of seaweeds under environmental stresses: A perspective. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.575228
Gilbert J. A., Neufeld J. D. (2014). Life in a world without microbes. PloS Biol. 12, 1–3. doi: 10.1371/journal.pbio.1002020
Girard F., Fisher C. R. (2018). Long-term impact of the Deepwater Horizon oil spill on deep-sea corals detected after seven years of monitoring. Biol. Conserv. 225, 117–127. doi: 10.1016/J.BIOCON.2018.06.028
Glasl B., Bourne D. G., Frade P. R., Thomas T., Schaffelke B., Webster N. S. (2019). Microbial indicators of environmental perturbations in coral reef ecosystems. Microbiome 7, 1–13. doi: 10.1186/s40168-019-0705-7
Goecke F., Labes A., Wiese J., Imhoff J. F. (2010). Review chemical interactions between Marine macroalgae and bacteria. Mar. Ecol. Prog. Ser. 409, 267–300. doi: 10.3354/meps08607
Gómez-Gras D., Linares C., Dornelas M., Madin J. S., Brambilla V., Ledoux J. B., et al. (2021). Climate change transforms the functional identity of Mediterranean coralligenous assemblages. Ecol. Lett. 24, 1038–1051. doi: 10.1111/ele.13718
Halpern B. S., Silliman B. R., Olden J. D., Bruno J. P., Bertness M. D. (2007). Incorporating positive interactions in aquatic restoration and conservation. Front. Ecol. Environ. 5, 153–160. doi: 10.1890/1540-9295
Hamisi M. I., Lyimo T. J., Muruke M. H. S., Bergman B. (2009). Nitrogen fixation by epiphytic and epibenthic diazotrophs associated with seagrass meadows along the Tanzanian coast, Western Indian Ocean. Aquat. Microb. Ecol. 57, 33–42. doi: 10.3354/ame01323
Henry L. P., Bruijning M., Forsberg S. K. G., Ayroles J. F. (2021). The microbiome extends host evolutionary potential. Nat. Commun. 12, 1–13. doi: 10.1038/s41467-021-25315-x
Hernandez-Agreda A., Gates R. D., Ainsworth T. D. (2017). Defining the core microbiome in corals’ Microbial soup. Trends Microbiol. 25, 125–140. doi: 10.1016/j.tim.2016.11.003
Hernandez-Agreda A., Leggat W., Bongaerts P., Ainsworth T. D. (2016). The microbial signature provides insight into the mechanistic basis of coral success across reef habitats. mBio 7, e00560–e00516. doi: 10.1128/mBio.00560-16
Hernandez-Agreda A., Leggat W., Bongaerts P., Herrera C., Ainsworth T. D. (2018). Rethinking the coral microbiome: Simplicity exists within a diverse microbial biosphere. mBio 9. doi: 10.1128/mBio.00812-18
Hiraoka S., Hirai M., Matsui Y., Makabe A., Minegishi H., Tsuda M., et al. (2020). Microbial community and geochemical analyses of trans-trench sediments for understanding the roles of hadal environments. ISME. J. 14, 740–756. doi: 10.1038/s41396-019-0564-z
Holguin G., Vazquez P., Bashan Y. (2001). The role of sediment microorganisms in the productivity, conservation, and rehabilitation of mangrove ecosystems: An overview. Biol. Fertil. Soils. 33, 265–278. doi: 10.1007/s003740000319
Hu J., Yang T., Friman V. P., Kowalchuk G. A., Hautier Y., Li M., et al. (2021). Introduction of probiotic bacterial consortia promotes plant growth via impacts on the resident rhizosphere microbiome. Proc. R. Soc B.: Biol. Sci. 288, 20211396. doi: 10.1098/rspb.2021.1396
Huggett M. J., Apprill A. (2019). Coral microbiome database: Integration of sequences reveals high diversity and relatedness of coral-associated microbes. Environ. Microbiol. Rep. 11, 372. doi: 10.1111/1758-2229.12686
Hughes R. G., Paramor O. A. L. (2004). On the loss of saltmarshes in south-east England and methods for their restoration. J. Appl. Ecol. 41, 440–448. doi: 10.1111/J.0021-8901.2004.00915.X
Ingrosso G., Abbiati M., Badalamenti F., Bavestrello G., Belmonte G., Cannas R., et al. (2018). Mediterranean bioconstructions along the italian coast. Adv. Mar. Biol. 79, 61–136. doi: 10.1016/bs.amb.2018.05.001
Jacquemot L., Bettarel Y., Monjol J., Corre E., Halary S., Desnues C., et al. (2018). Therapeutic potential of a new jumbo phage that infects Vibrio coralliilyticus, a widespread coral pathogen. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02501
Janarthine S. R. S., Eganathan P. (2012). Plant growth promoting of endophytic Sporosarcina aquimarina SjAM16103 isolated from the pneumatophores of Avicennia marina L. Int. J. Microbiol. 2012, 1–10. doi: 10.1155/2012/532060
Jaunatre R., Buisson E., Dutoit T. (2014). Can ecological engineering restore Mediterranean rangeland after intensive cultivation? A large-scale experiment in southern France. Ecol. Eng. 64, 202–212. doi: 10.1016/j.ecoleng.2013.12.022
Jessen C., Villa Lizcano J. F., Bayer T., Roder C., Aranda M., Wild C., et al. (2013). In-situ effects of eutrophication and overfishing on physiology and bacterial diversity of the red sea coral acropora hemprichii. PloS One 8, e62091. doi: 10.1371/journal.pone.0062091
Jha C. K., Aeron A., Patel B. V., Maheshwari D. K., Saraf M. (2011). “Enterobacter: role in plant growth promotion,” in Bacteria in agrobiology: plant growth responses (Cham, Switzerland: Springer), 159–182. doi: 10.1007/978-3-642-20332-9_8
Jiang G., Zhang Y., Gan G., Li W., Wan W., Jiang Y., et al. (2022). Exploring rhizo-microbiome transplants as a tool for protective plant-microbiome manipulation. ISME. Commun. 2, 10. doi: 10.1038/s43705-022-00094-8
Jiao S., Chen W., Wei G. (2022). Core microbiota drive functional stability of soil microbiome in reforestation ecosystems. Glob. Chang. Biol. 28, 1038–1047. doi: 10.1111/gcb.16024
Jonsson L., Dahl M., Strömberg S., Lindegarth M., André C., Lundälv T. (2015). In situ measurement of survival and growth of genotyped transplanted fragments of the cold-water coral Lophelia pertusa (Sweden: University of Gothenburg).
Jorissen H., Galand P. E., Bonnard I., Meiling S., Raviglione D., Meistertzheim A. L., et al. (2021). Coral larval settlement preferences linked to crustose coralline algae with distinct chemical and microbial signatures. Sci. Rep. 11, 1–11. doi: 10.1038/s41598-021-94096-6
Jung J., Kim J.-S., Taffner J., Berg G., Ryu C.-M. (2020). Archaea, tiny helpers of land plants. Comput. Struct. Biotechnol. J. 18, 2494–2500. doi: 10.1016/j.csbj.2020.09.005
Kardol P., Bezemer T. M., van der Putten W. H. (2009). Soil organism and plant introductions in restoration of species-rich grassland communities. Restor. Ecol. 17, 258–269. doi: 10.1111/j.1526-100X.2007.00351.x
Kellogg C. A., Lisle J. T., Galkiewicz J. P. (2009). Culture-independent characterization of bacterial communities associated with the cold-water coral lophelia pertusa in the northeastern gulf of Mexico. Appl. Environ. Microbiol. 75, 2294–2303. doi: 10.1128/AEM.02357-08
Khan N., Ali S., Shahid M. A., Mustafa A., Sayyed R. Z., Curá J. A. (2021). Insights into the interactions among roots, rhizosphere, and rhizobacteria for improving plant growth and tolerance to abiotic stresses: A review. Cells 10, 1551. doi: 10.3390/cells10061551
King N. G., Moore P. J., Thorpe J. M., Smale D. A. (2023). Consistency and variation in the kelp microbiota: patterns of bacterial community structure across spatial scales. Microb. Ecol. 85, 1265–1275. doi: 10.1007/S00248-022-02038-0
Knowlton N. (2021). Local management matters for coral reefs. Sci. (1979) 372, 908–909. doi: 10.1126/science.abi7286
Krishnaswamy V. G., Mani K., Senthil Kumar P., Rangasamy G., Sridharan R., Rethnaraj C., et al. (2023). Prevalence of differential microbiome in healthy, diseased and nipped colonies of corals, Porites lutea in the Gulf of Kachchh, north-west coast of India. Environ. Res. 216, 114622. doi: 10.1016/j.envres.2022.114622
Lachnit T., Meske D., Wahl M., Harder T., Schmitz R. (2011). Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environ. Microbiol. 13, 655–665. doi: 10.1111/j.1462-2920.2010.02371.x
Lage O. M., Bondoso J. (2014). Planctomycetes and macroalgae, a striking association. Front. Microbiol. 5. doi: 10.3389/FMICB.2014.00267
Larkin M. F., Davis T. R., Harasti D., Benkendorff K., Smith S. D. A. (2023). Substantial advancement in aquaria rearing methods to assist recovery of an Endangered soft coral. Aquat. Conserv. 33, 1–14. doi: 10.1002/AQC.3895
Lata R., Chowdhury S., Gond S. K., White J. F. (2018). Induction of abiotic stress tolerance in plants by endophytic microbes. Lett. Appl. Microbiol. 66, 268–276. doi: 10.1111/LAM.12855
Lee S. Y., Hamilton S., Barbier E. B., Primavera J., Lewis R. R. (2019). Better restoration policies are needed to conserve mangrove ecosystems. Nat. Ecol. Evol. 3, 870–872. doi: 10.1038/S41559-019-0861-Y
Lewis R. R. (2005). Ecological engineering for successful management and restoration of mangrove forests. Ecol. Eng. 24, 403–418. doi: 10.1016/J.ECOLENG.2004.10.003
Li J., Yang Q., Dong J., Sweet M., Zhang Y., Liu C., et al. (2022). Microbiome engineering: A promising approach to improve coral health. Engineering. doi: 10.1016/j.eng.2022.07.010
Linares C., Coma R., Zabala M. (2008). Restoration of threatened red gorgonian populations: An experimental and modelling approach. Biol. Conserv. 141, 427–437. doi: 10.1016/j.biocon.2007.10.012
Liu L., Agren R., Bordel S., Nielsen J. (2010). Use of genome-scale metabolic models for understanding microbial physiology. FEBS Lett. 584, 2556–2564. doi: 10.1016/j.febslet.2010.04.052
Liu H., Li J., Carvalhais L. C., Percy C. D., Prakash Verma J., Schenk P. M., et al. (2021). Evidence for the plant recruitment of beneficial microbes to suppress soil-borne pathogens. New Phytol. 229, 2873–2885. doi: 10.1111/nph.17057
Liu P, Zhang H., Sun Y., Wang C., Hu X. (2022). Molecular diversity and biogeography of benthic microeukaryotes in temperate seagrass (Zostera japonica) systems of northern China. Acta Oceanol. Sin. 41, 115–125. doi: 10.1007/s13131-021-1960-6
Maire J., van Oppen M. J. H. (2022). A role for bacterial experimental evolution in coral bleaching mitigation? Trends Microbiol. 30, 217–228. doi: 10.1016/j.tim.2021.07.006
Malfatti F., Kaleb S., Saidi A., Pallavicini A., Agostini L., Gionechetti F., et al. (2023). Microbe-assisted seedling crop improvement by a seaweed extract to address fucalean forest restoration. Front. Mar. Sci. 10. doi: 10.3389/FMARS.2023.1181685
Mancuso F. P., D’Hondt S., Willems A., Airoldi L., De Clerck O. (2016). Diversity and temporal dynamics of the epiphytic bacterial communities associated with the canopy-forming seaweed Cystoseira compressa (Esper) Gerloff and Nizamuddin. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00476
Manea E., Dell’anno A., Rastelli E., Tangherlini M., Nunoura T., Nomaki H., et al. (2019). Viral infections boost prokaryotic biomass production and organic C cycling in hadal trench sediments. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01952
Marhaeni B., Radjasa O. K., Bengen D. G., Kaswadji R. F. (2010). Screening of bacterial symbionts of seagrass Enhalus sp. against biofilm-forming bacteria. J. Coast. Dev. 13, 126–132.
Martin B. C., Bougoure J., Ryan M. H., Bennett W. W., Colmer T. D., Joyce N. K., et al. (2019). Oxygen loss from seagrass roots coincides with colonisation of sulphide-oxidising cable bacteria and reduces sulphide stress. ISME. J. 13, 707–719. doi: 10.1038/s41396-018-0308-5
McCauley M., Jackson C. R., Goulet T. L. (2020). Microbiomes of caribbean octocorals vary over time but are resistant to environmental change. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01272
McFall-Ngai M., Hadfield M. G., Bosch T. C. G., Carey H. V., Domazet-Lošo T., Douglas A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236. doi: 10.1073/pnas.1218525110
Meena K. K., Sorty A. M., Bitla U. M., Choudhary K., Gupta P., Pareek A., et al. (2017). Abiotic stress responses and microbe-mediated mitigation in plants: The omics strategies. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00172
Meistertzheim A. L., Lartaud F., Arnaud-Haond S., Kalenitchenko D., Bessalam M., Le Bris N., et al. (2016). Patterns of bacteria-host associations suggest different ecological strategies between two reef building cold-water coral species. Deep. Sea. Res. Part I. Oceanogr. Res. Pap. 114, 12–22. doi: 10.1016/J.DSR.2016.04.013
Milbrandt E. C., Greenawalt-Boswell J., Sokoloff P. D. (2008). Short-term indicators of seagrass transplant stress in response to sediment bacterial community disruption. botm 51, 103–111. doi: 10.1515/BOT.2008.020
Mills J. G., Weinstein P., Gellie N. J. C., Weyrich L. S., Lowe A. J., Breed M. F. (2017). Urban habitat restoration provides a human health benefit through microbiome rewilding: the Microbiome Rewilding Hypothesis. Restor. Ecol. 25, 866–872. doi: 10.1111/REC.12610
Minich J. J., Morris M. M., Brown M., Doane M., Edwards M. S., Michael T. P., et al. (2018). Elevated temperature drives kelp microbiome dysbiosis, while elevated carbon dioxide induces water microbiome disruption. PloS One 13, e0192772. doi: 10.1371/journal.pone.0192772
Mohr W., Lehnen N., Ahmerkamp S., Marchant H. K., Graf J. S., Tschitschko B., et al. (2021). Terrestrial-type nitrogen-fixing symbiosis between seagrass and a marine bacterium. Nature 600, 105–109. doi: 10.1038/s41586-021-04063-4
Mokumo M. F., Adams J. B., von der Heyden S. (2023). Investigating transplantation as a mechanism for seagrass restoration in South Africa. Restor. Ecol. 31, e13941. doi: 10.1111/rec.13941
Montero-Serra I., Garrabou J., Doak D. F., Figuerola L., Hereu B., Ledoux J. B., et al. (2018). Accounting for life-history strategies and timescales in marine restoration. Conserv. Lett. 11, e12341. doi: 10.1111/CONL.12341
Montseny M., Linares C., Carreiro-Silva M., Henry L.-A., Billett D., Cordes E. E., et al. (2021). Active ecological restoration of cold-water corals: techniques, challenges, costs and future directions. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.621151
Montseny M., Linares C., Viladrich N., Capdevila P., Ambroso S., Díaz D., et al. (2020). A new large-scale and cost-effective restoration method for cold-water coral gardens. Aquat. Conserv. 30, 977–987. doi: 10.1002/aqc.3303
Montseny M., Linares C., Viladrich N., Olariaga A., Carreras M., Palomeras N., et al. (2019). First attempts towards the restoration of gorgonian populations on the Mediterranean continental shelf. Aquat. Conserv. 29, 1278–1284. doi: 10.1002/aqc.3118
Morales Moreira Z. P., Chen M. Y., Yanez Ortuno D. L., Haney C. H. (2023). Engineering plant microbiomes by integrating eco-evolutionary principles into current strategies. Curr. Opin. Plant Biol. 71, 102316. doi: 10.1016/j.pbi.2022.102316
Moriarty T., Leggat W., Huggett M. J., Ainsworth T. D. (2020). Coral disease causes, consequences, and risk within coral restoration. Trends Microbiol. 28, 793–807. doi: 10.1016/J.TIM.2020.06.002
Morrissey K. L., Iveša L., Delva S., D’Hondt S., Willems A., De Clerck O. (2021). Impacts of environmental stress on resistance and resilience of algal-associated bacterial communities. Ecol. Evol. 11, 15004–15019. doi: 10.1002/ece3.8184
Morrow K. M., Pankey M. S., Lesser M. P. (2022). Community structure of coral microbiomes is dependent on host morphology. Microbiome 10, 1–19. doi: 10.1186/s40168-022-01308-w
Muehlstein L. K., Porter D., Short F. T. (1991). Labyrinthula zosterae sp. nov., the causative agent of wasting disease of eelgrass, Zostera marina. Mycologia 83, 180–191. doi: 10.2307/3759933
Mueller U. G., Linksvayer T. A. (2022). Microbiome breeding: conceptual and practical issues. Trends Microbiol. 30, 997–1011. doi: 10.1016/j.tim.2022.04.003
Mueller U. G., Sachs J. L. (2015). Engineering microbiomes to improve plant and animal health. Trends Microbiol. 23, 606–617. doi: 10.1016/j.tim.2015.07.009
Neave M. J., Apprill A., Ferrier-Pagès C., Voolstra C. R. (2016). Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Appl. Microbiol. Biotechnol. 100, 8315–8324. doi: 10.1007/s00253-016-7777-0
O’Connor M. I., Griffiths G., Sanders-Smith R., Hessing-Lewis M., Davis K. M., Forbes C., et al. (2022). A reciprocal transplant experiment sheds new light on a classic marine seagrass-algal symbiosis and suggests influence of epiphytic symbiont on seagrass microbiota. Aquat. Bot. 179, 103511. doi: 10.1016/j.aquabot.2022.103511
Orejas C., Taviani M., Ambroso S., Andreou V., Bilan M., Bo M., et al. (2019). “38 cold-water coral in aquaria: advances and challenges. A focus on the mediterranean,” in Mediterranean cold-water corals: past, present and future: understanding the deep-sea realms of coral. (Cham, Switzerland: Springer Nature) 435–471. doi: 10.1007/978-3-319-91608-8_38
Orfanidis S., Rindi F., Cebrian E., Fraschetti S., Nasto I., Taskin E., et al. (2021). Effects of natural and anthropogenic stressors on fucalean brown seaweeds across different spatial scales in the mediterranean sea. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.658417
Paling E. I., Fonseca M., van Katwijk M. M., van Keulen M. (2009). “Seagrass restoration,” in Coastal Wetlands - An integrated ecosystem approach (Amsterdam, The Netherlands: Elsevier Amsterdam), 687–713.
Palladino G., Caroselli E., Tavella T., D’Amico F., Prada F., Mancuso A., et al. (2022). Metagenomic shifts in mucus, tissue and skeleton of the coral Balanophyllia europaea living along a natural CO2 gradient. ISME. Comm. 2, 65. doi: 10.1038/s43705-022-00165-w
Pandolfi J. M., Bradbury R. H., Sala E., Hughes T. P., Bjorndal K. A., Cooke R. G., et al. (2003). Global trajectories of the long-term decline of coral reef ecosystems. Sci. (1979) 301, 955–958. doi: 10.1126/science.1085706
Parada A. E., Needham D. M., Fuhrman J. A. (2016). Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Parrot D., Blümel M., Utermann C., Chianese G., Krause S., Kovalev A., et al. (2019). Mapping the surface microbiome and metabolome of brown seaweed fucus vesiculosus by amplicon sequencing, integrated metabolomics and imaging techniques. Sci. Rep. 9, 1–17. doi: 10.1038/s41598-018-37914-8
Peixoto R. S., Rosado P. M., Leite D. C., de A., Rosado A. S., Bourne D. G. (2017). Beneficial microorganisms for corals (BMC): Proposed mechanisms for coral health and resilience. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00341
Peixoto R. S., Sweet M., Bourne D. G. (2019). Customized medicine for corals. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00686
Peixoto R. S., Sweet M., Villela H. D. M., Cardoso P., Thomas T., Voolstra C. R., et al. (2021). Coral probiotics: premise, promise, prospects. Annu. Rev. Anim. Biosci. 9, 265–288. doi: 10.1146/annurev-animal-090120-115444
Peixoto R. S., Voolstra C. R., Sweet M., Duarte C. M., Carvalho S., Villela H., et al. (2022). Harnessing the microbiome to prevent global biodiversity loss. Nat. Microbiol. 7, 1726–1735. doi: 10.1038/s41564-022-01173-1
Peng Y., Li W. (2013). A bacterial pathogen infecting gametophytes of Saccharina japonica (Laminariales, Phaeophyceae). Chin. J. Oceanol. Limnol. 31, 366–373. doi: 10.1007/s00343-013-2136-9
Petersen J. M., Osvatic J. (2018). Microbiomes in natura : importance of invertebrates in understanding the natural variety of animal-microbe interactions. mSystems 3, e00179–e00117. doi: 10.1128/msystems.00179-17
Pita L., Rix L., Slaby B. M., Franke A., Hentschel U. (2018). The sponge holobiont in a changing ocean: from microbes to ecosystems. Microbiome 6, 1–18. doi: 10.1186/S40168-018-0428-1
Pollock F. J., Morris P. J., Willis B. L., Bourne D. G. (2011). The urgent need for robust coral disease diagnostics. PloS Pathog. 7, e1002183. doi: 10.1371/journal.ppat.1002183
Ponti M., Perlini R. A., Ventra V., Grech D., Abbiati M., Cerrano C. (2014). Ecological shifts in mediterranean coralligenous assemblages related to gorgonian forest loss. PloS One 9, e102782. doi: 10.1371/journal.pone.0102782
Pootakham W., Mhuantong W., Yoocha T., Putchim L., Jomchai N., Sonthirod C., et al. (2019). Heat-induced shift in coral microbiome reveals several members of the Rhodobacteraceae family as indicator species for thermal stress in Porites lutea. Microbiologyopen 8, 1–20. doi: 10.1002/mbo3.935
Portilho-Ramos R., da C., Titschack J., Wienberg C., Siccha Rojas M. G., Yokoyama Y., et al. (2022). Major environmental drivers determining life and death of cold-water corals through time. PloS Biol. 20, e3001628. doi: 10.1371/journal.pbio.3001628
Poulos D. E., Harasti D., Gallen C., Booth D. J. (2013). Biodiversity value of a geographically restricted soft coral species within a temperate estuary. Aquat. Conserv. 23, 838–849. doi: 10.1002/AQC.2362
Prada F., Franzellitti S., Caroselli E., Cohen I., Marini M., Campanelli A., et al. (2023). Acclimatization of a coral-dinoflagellate mutualism at a CO2 vent. Commun. Biol. 6, 66. doi: 10.1038/s42003-022-04327-3
Prado S., Barja J. L., Luzardo A., Dubert J., Blanco J. (2020). Encapsulation of live marine bacteria for use in aquaculture facilities and process evaluation using response surface methodology. Appl. Microbiol. Biotechnol. 104, 1993–2006. doi: 10.1007/s00253-019-10332-0
Precht W. F., Robbart M. (2006). “Coral reef restoration: the rehabilitation of an ecosystem under siege,” in Coral reef restoration handbook. (London: CRC Press) 1–24.
Pywell R. F., Meek W. R., Webb N. R., Putwain P. D., Bullock J. M. (2011). Long-term heathland restoration on former grassland: The results of a 17-year experiment. Biol. Conserv. 144, 1602–1609. doi: 10.1016/j.biocon.2011.02.010
Qiu Z., Coleman M. A., Provost E., Campbell A. H., Kelaher B. P., Dalton S. J., et al. (2019). Future climate change is predicted to affect the microbiome and condition of habitat-forming kelp. Proc. R. Soc B.: Biol. Sci. 286, 20181887. doi: 10.1128/AEM.02326-20
Ragnarsson S. Á., Burgos J. M., Kutti T., van den Beld I., Egilsdóttir H., Arnaud-Haond S., et al. (2017). “The impact of anthropogenic activity on cold-water corals,” in Marine animal forests: the ecology of benthic biodiversity hotspots. (Cham, Switzerland: Springer Nature), 989–1023. doi: 10.1007/978-3-319-21012-4_27
Ramirez-Llodra E., Tyler P. A., Baker M. C., Bergstad O. A., Clark M. R., Escobar E., et al. (2011). Man and the last great wilderness: human impact on the deep sea. PloS One 6, e22588. doi: 10.1371/journal.pone.0022588
Ramírez-Puebla S. T., Weigel B. L., Jack L., Schlundt C., Pfister C. A., Mark Welch J. L. (2022). Spatial organization of the kelp microbiome at micron scales. Microbiome 10, 1–20. doi: 10.1186/s40168-022-01235-w
Rinkevich B. (2005). Conservation of coral reefs through active restoration measures: recent approaches and last decade progress. Environ. Sci. Technol. 39, 4333–4342. doi: 10.1021/ES0482583
Ritchie K. B. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14. doi: 10.3354/meps322001
Rivera H., Chan A., Luu V. (2020). Coral reefs are critical for our food supply, tourism, and ocean health. We can protect them from climate change. MIT. Sci. Policy Rev. 1, 18–33. doi: 10.38105/spr.7vn798jnsk
Rizaludin M. S., Stopnisek N., Raaijmakers J. M., Garbeva P. (2021). The chemistry of stress: Understanding the ‘cry for help’ of plant roots. Metabolites 11, 357. doi: 10.3390/metabo11060357
Robbins S. J., Singleton C. M., Chan C. X., Messer L. F., Geers A. U., Ying H., et al. (2019). A genomic view of the reef-building coral Porites lutea and its microbial symbionts. Nat. Microbiol. 4, 2090–2100. doi: 10.1038/s41564-019-0532-4
Roberts J. M., Wheeler A., Freiwald A., Cairns S. (2009). Cold-water corals: the biology and geology of deep-sea coral habitats (Cambridge, United Kingdom: Cambridge University Press).
Rodrigues C. J. C., de Carvalho C. C. C. R. (2022). Cultivating marine bacteria under laboratory conditions: Overcoming the “unculturable” dogma. Front. Bioeng. Biotechnol. 10. doi: 10.3389/FBIOE.2022.964589
Romañach S. S., DeAngelis D. L., Koh H. L., Li Y., Teh S. Y., Raja Barizan R. S., et al. (2018). Conservation and restoration of mangroves: Global status, perspectives, and prognosis. Ocean. Coast. Manag. 154, 72–82. doi: 10.1016/J.OCECOAMAN.2018.01.009
Rosado P. M., Leite D. C. A., Duarte G. A. S., Chaloub R. M., Jospin G., Nunes da Rocha U., et al. (2019). Marine probiotics: increasing coral resistance to bleaching through microbiome manipulation. ISME. J. 13, 921–936. doi: 10.1038/s41396-018-0323-6
Rosales S. M., Miller M. W., Williams D. E., Traylor-Knowles N., Young B., Serrano X. M. (2019). Microbiome differences in disease-resistant vs. susceptible Acropora corals subjected to disease challenge assays. Sci. Rep. 9, 18279. doi: 10.1038/s41598-019-54855-y
Saha M., Weinberger F. (2019). Microbial “gardening” by a seaweed holobiont: Surface metabolites attract protective and deter pathogenic epibacterial settlement. J. Ecol. 107, 2255–2265. doi: 10.1111/1365-2745.13193
Sánchez J. A. (2017). “Diversity and evolution of octocoral animal forests at both sides of tropical america,” in Marine animal forests (Cham: Springer International Publishing), 111–143. doi: 10.1007/978-3-319-21012-4_39
Sánchez J. A., Ardila N. E., Andrade J., Dueñas L. F., Navas R., Ballesteros D. (2014). Octocoral densities and mortalities in Gorgona Island, Colombia, Tropical Eastern Pacific. Rev. Biol. Trop. 62, 209. doi: 10.15517/rbt.v62i0.16277
Sanchez-Amat A., Solano F., Lucas-Elío P. (2010). Finding new enzymes from bacterial physiology: A successful approach illustrated by the detection of novel oxidases in Marinomonas mediterranea. Mar. Drugs 8, 519–541. doi: 10.3390/md8030519
Saunders M. I., Doropoulos C., Bayraktarov E., Babcock R. C., Gorman D., Eger A. M., et al. (2020). Bright spots in coastal marine ecosystem restoration. Curr. Biol. 30, R1500–R1510. doi: 10.1016/j.cub.2020.10.056
Schenck F. R., DuBois K., Kardish M. R., Stachowicz J. J., Hughes A. R. (2023). The effect of warming on seagrass wasting disease depends on host genotypic identity and diversity. Ecology 104, e3959. doi: 10.1002/ecy.3959
Shade A., Handelsman J. (2012). Beyond the Venn diagram: The hunt for a core microbiome. Environ. Microbiol. 14, 4–12. doi: 10.1111/j.1462-2920.2011.02585.x
Shafquat A., Joice R., Simmons S. L., Huttenhower C. (2014). Functional and phylogenetic assembly of microbial communities in the human microbiome. Trends Microbiol. 22, 261–266. doi: 10.1016/j.tim.2014.01.011
Sharma S. B., Sayyed R. Z., Trivedi M. H., Gobi T. A. (2013). Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus 2, 587. doi: 10.1186/2193-1801-2-587
Silliman B. R., Schrack E., He Q., Cope R., Santoni A., van der Heide T., et al. (2015). Facilitation shifts paradigms and can amplify coastal Restoration efforts. Proc. Natl. Acad. Sci. U.S.A. 112, 14295–14300. doi: 10.1073/PNAS.1515297112
Singh C. L., Huggett M. J., Lavery P. S., Säwström C., Hyndes G. A. (2021). Kelp-associated microbes facilitate spatial subsidy in a detrital-based food web in a shoreline ecosystem. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.678222
Sipahutar M. K., Vangnai A. S. (2017). Role of plant growth-promoting Ochrobactrum sp. MC22 on triclocarban degradation and toxicity mitigation to legume plants. J. Hazard. Mater. 329, 38–48. doi: 10.1016/j.jhazmat.2017.01.020
Siraj M. A., Medha M. M., Nahar A. U., Islam M. A., Seidel V. (2023). “Mangrove endophytes and their natural metabolites: role in promoting plant health,” in Microbial endophytes and plant growth (Cambridge, United States: Elsevier), 99–116. doi: 10.1016/b978-0-323-90620-3.00015-5
Smith C. J., Verdura J., Papadopoulou N., Fraschetti S., Cebrian E., Fabbrizzi E., et al. (2023). A decision-support framework for the restoration of Cystoseira sensu lato forests. Front. Mar. Sci. 10. doi: 10.3389/fmars.2023.1159262
Smith G. W., Weil E. (2004). “Aspergillosis of gorgonians,” in Coral health and disease (Cham, Switzerland: Springer), 279–287.
Sogin E. M., Leisch N., Dubilier N. (2020). Chemosynthetic symbioses. Curr. Biol. 30, R1137–R1142. doi: 10.1016/J.CUB.2020.07.050
Soldan R., Mapelli F., Crotti E., Schnell S., Daffonchio D., Marasco R., et al. (2019). Bacterial endophytes of mangrove propagules elicit early establishment of the natural host and promote growth of cereal crops under salt stress. Microbiol. Res. 223–225, 33–43. doi: 10.1016/j.micres.2019.03.008
Sorbara M. T., Pamer E. G. (2022). Microbiome-based therapeutics. Nat. Rev. Microbiol. 20, 365–380. doi: 10.1038/s41579-021-00667-9
Steinberg R. K., Dafforn K. A., Ainsworth T., Johnston E. L. (2020). Know thy anemone: A review of threats to octocorals and anemones and opportunities for their restoration. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00590
Stévenne C., Micha M., Plumier J. C., Roberty S. (2021). Corals and sponges under the light of the holobiont concept: how microbiomes underpin our understanding of marine ecosystems. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.698853
Sullivan B. K., Trevathan-Tackett S. M., Neuhauser S., Govers L. L. (2018). Review: Host-pathogen dynamics of seagrass diseases under future global change. Mar. pollut. Bull. 134, 75–88. doi: 10.1016/j.marpolbul.2017.09.030
Sweet M. J., Bulling M. T. (2017). On the importance of the microbiome and pathobiome in coral health and disease. Front. Mar. Sci. 4. doi: 10.3389/fmars.2017.00009
Tan Y. M., Dalby O., Kendrick G. A., Statton J., Sinclair E. A., Fraser M. W., et al. (2020). Seagrass restoration is possible: insights and lessons from Australia and New Zealand. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00617
Tarquinio F., Hyndes G. A., Laverock B., Koenders A., Säwström C. (2019). The seagrass holobiont: understanding seagrass-bacteria interactions and their role in seagrass ecosystem functioning. FEMS Microbiol. Lett. 366, fnz057. doi: 10.1093/femsle/fnz057
Topçu N. E., Yılmaz I. N., Saraçoğlu C., Barraud T., Öztürk B. (2023). Size matters when it comes to the survival of transplanted yellow gorgonian fragments. J. Nat. Conserv. 71, 126326. doi: 10.1016/j.jnc.2022.126326
Torda G., Donelson J. M., Aranda M., Barshis D. J., Bay L., Berumen M. L., et al. (2017). Rapid adaptive responses to climate change in corals. Nat. Clim. Chang. 7, 627–636. doi: 10.1038/nclimate3374
Traylor-Knowles N., Baker A. C., Beavers K. M., Garg N., Guyon J. R., Hawthorn A., et al. (2022). Advances in coral immunity ‘omics in response to disease outbreaks. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.952199
Trevathan-Tackett S. M., Sherman C. D. H., Huggett M. J., Campbell A. H., Laverock B., Hurtado-McCormick V., et al. (2019). A horizon scan of priorities for coastal marine microbiome research. Nat. Ecol. Evol. 3, 1509–1520. doi: 10.1038/s41559-019-0999-7
Trevelline B. K., Fontaine S. S., Hartup B. K., Kohl K. D. (2019). Conservation biology needs a microbial renaissance: A call for the consideration of host-associated microbiota in wildlife management practices. Proc. R. Soc. B.: Biol. Sci. 286, 1–9. doi: 10.1098/rspb.2018.2448
Trivedi P., Anderson I. C., Singh B. K. (2013). Microbial modulators of soil carbon storage: Integrating genomic and metabolic knowledge for global prediction. Trends Microbiol. 21, 641–651. doi: 10.1016/j.tim.2013.09.005
Ugarelli K., Chakrabarti S., Laas P., Stingl U. (2017). The seagrass holobiont and its microbiome. Microorganisms 5, 81. doi: 10.3390/microorganisms5040081
Unsworth R. K. F., Bertelli C. M., Cullen-Unsworth L. C., Esteban N., Jones B. L., Lilley R., et al. (2019). Sowing the seeds of seagrass recovery using hessian bags. Front. Ecol. Evol. 7. doi: 10.3389/fevo.2019.00311
Unsworth R. K. F., Rees S. C., Bertelli C. M., Esteban N. E., Furness E. J., Walter B. (2022). Nutrient additions to seagrass seed planting improve seedling emergence and growth. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1013222
Vairappan C. S., Suzuki M., Motomura T., Ichimura T. (2001). Pathogenic bacteria associated with lesions and thallus bleaching symptoms in the Japanese kelp Laminaria religiosa Miyabe (Laminariales, Phaeophyceae). Hydrobiologia 445, 183–191. doi: 10.1023/A:1017517832302
Valdez S. R., Zhang Y. S., van der Heide T., Vanderklift M. A., Tarquinio F., Orth R. J., et al. (2020). Positive ecological interactions and the success of seagrass restoration. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00091
Vandenkoornhuyse P., Quaiser A., Duhamel M., Le Van A., Dufresne A. (2015). The importance of the microbiome of the plant holobiont. New Phytol. 206, 1196–1206. doi: 10.1111/nph.13312
van der Geest M., van der Heide T., Holmer M., de Wit R. (2020). First field-based evidence that the seagrass-lucinid mutualism can mitigate sulfide stress in seagrasses. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00011
van der Loos L. M., Eriksson B. K., Falcão Salles J. (2019). The macroalgal holobiont in a changing sea. Trends Microbiol. 27, 635–650. doi: 10.1016/j.tim.2019.03.002
van De Water J. A. J. M., Allemand D., Ferrier-Pagès C. (2018a). Host-microbe interactions in octocoral holobionts - recent advances and perspectives. Microbiome 6, 64. doi: 10.1186/s40168-018-0431-6
van de Water J. A. J. M., Coppari M., Enrichetti F., Ferrier-Pagès C., Bo M. (2020). Local conditions influence the prokaryotic communities associated with the mesophotic black coral antipathella subpinnata. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.537813
van de Water J. A. J. M., Melkonian R., Junca H., Voolstra C. R., Reynaud S., Allemand D., et al. (2016). Spirochaetes dominate the microbial community associated with the red coral Corallium rubrum on a broad geographic scale. Sci. Rep. 6, 27277. doi: 10.1038/srep27277
van de Water J. A. J. M., Melkonian R., Voolstra C. R., Junca H., Beraud E., Allemand D., et al. (2017). Comparative assessment of mediterranean gorgonian-associated microbial communities reveals conserved core and locally variant bacteria. Microb. Ecol. 73, 466–478. doi: 10.1007/s00248-016-0858-x
van de Water J. A. J. M., Voolstra C. R., Rottier C., Cocito S., Peirano A., Allemand D., et al. (2018b). Seasonal stability in the microbiomes of temperate gorgonians and the red coral corallium rubrum across the mediterranean sea. Microb. Ecol. 75, 274–288. doi: 10.1007/s00248-017-1006-y
Van Dover C. L., Aronson J., Pendleton L., Smith S., Arnaud-Haond S., Moreno-Mateos D., et al. (2014). Ecological restoration in the deep sea: Desiderata. Mar. Policy 44, 98–106. doi: 10.1016/j.marpol.2013.07.006
Van Elsas J. D., Chiurazzi M., Mallon C. A., Elhottova D., Krištůfek V., Salles J. F. (2012). Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. U.S.A. 109, 1159–1164. doi: 10.1073/pnas.1109326109
van Katwijk M. M., Thorhaug A., Marbà N., Orth R. J., Duarte C. M., Kendrick G. A., et al. (2016). Global analysis of seagrass restoration: The importance of large-scale planting. J. Appl. Ecol. 53, 567–578. doi: 10.1111/1365-2664.12562
Vannier N., Mony C., Bittebiere A.-K., Theis K. R., Rosenberg E., Vandenkoornhuyse P. (2019). Clonal plants as meta-holobionts. mSystems 4, e00213–e00218. doi: 10.1128/msystems.00213-18
Van Oppen M. J. H., Blackall L. L. (2019). Coral microbiome dynamics, functions and design in a changing world. Nat. Rev. Microbiol. 17, 557–567. doi: 10.1038/s41579-019-0223-4
Van Oppen M. J. H., Leong J. A., Gates R. D. (2009). Coral-virus interactions: A double-edged sword? Symbiosis 47, 1–8. doi: 10.1007/BF03179964
Van Oppen M. J. H., Oliver J. K., Putnam H. M., Gates R. D. (2015). Building coral reef resilience through assisted evolution. Proc. Natl. Acad. Sci. U.S.A. 112, 2307–2313. doi: 10.1073/pnas.1422301112
Vanwonterghem I., Webster N. S. (2020). Coral reef microorganisms in a changing climate. iScience 23, 100972. doi: 10.1016/j.isci.2020.100972
Vassilev N., Vassileva M., Martos V., Garcia del Moral L. F., Kowalska J., Tylkowski B., et al. (2020). Formulation of microbial inoculants by encapsulation in natural polysaccharides: focus on beneficial properties of carrier additives and derivatives. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00270
Vaughan D. E. (2021). Active coral restoration: techniques for a changing planet (Fort Lauderdale, FL, United States: J. Ross Publishing).
Verdura J., Linares C., Ballesteros E., Coma R., Uriz M. J., Bensoussan N., et al. (2019). Biodiversity loss in a Mediterranean ecosystem due to an extreme warming event unveils the role of an engineering gorgonian species. Sci. Rep. 9, 1–11. doi: 10.1038/s41598-019-41929-0
Vezzulli L., Pezzati E., Huete-Stauffer C., Pruzzo C., Cerrano C. (2013). 16SrDNA Pyrosequencing of the Mediterranean Gorgonian Paramuricea clavata Reveals a Link among Alterations in Bacterial Holobiont Members, Anthropogenic Influence and Disease Outbreaks. PloS One 8, e67745. doi: 10.1371/journal.pone.0067745
Wainwright B. J., Zahn G. L., Zushi J., Lee N. L. Y., Ooi J. L. S., Lee J. N., et al. (2019). Seagrass-associated fungal communities show distance decay of similarity that has implications for seagrass management and restoration. Ecol. Evol. 9, 11288–11297. doi: 10.1002/ece3.5631
Wang L., English M. K., Tomas F., Muellera R. S. (2021b). Recovery and community succession of the zostera marina rhizobiome after transplantation. Appl. Environ. Microbiol. 87, 1–16. doi: 10.1128/AEM.02326-20
Wang F., Li M., Huang L., Zhang X.-H. (2021a). Cultivation of uncultured marine microorganisms. Mar. Life Sci. Technol. 3, 117–120. doi: 10.1007/s42995-021-00093-z
Weigel B. L., Miranda K. K., Fogarty E. C., Watson A. R., Pfister C. A. (2022). Functional insights into the kelp microbiome from metagenome-assembled genomes. mSystems 7, e01422–e01421. doi: 10.1128/msystems.01422-21
Weigel B. L., Pfister C. A. (2021). Oxygen metabolism shapes microbial settlement on photosynthetic kelp blades compared to artificial kelp substrates. Environ. Microbiol. Rep. 13, 176–184. doi: 10.1111/1758-2229.12923
Weinberg S. (2019). Transplantation experiments with mediterranean gorgonians. Bijdragen. tot. Dierkunde. 49, 31–41. doi: 10.1163/26660644-04901003
Welsh D. T. (2000). Nitrogen fixation in seagrass meadows: Regulation, plant-bacteria interactions and significance to primary productivity. Ecol. Lett. 3, 58–71. doi: 10.1046/j.1461-0248.2000.00111.x
Welsh R. M., Rosales S. M., Zaneveld J. R., Payet J. P., McMinds R., Hubbs S. L., et al. (2017). Alien vs. predator: bacterial challenge alters coral microbiomes unless controlled by Halobacteriovorax predators. PeerJ 5, e3315. doi: 10.7717/peerj.3315
Wiese J., Thiel V., Nagel K., Staufenberger T., Imhoff J. F. (2009). Diversity of antibiotic-active bacteria associated with the brown alga Laminaria saccharina from the baltic sea. Mar. Biotechnol. 11, 287–300. doi: 10.1007/s10126-008-9143-4
Wood G., Marzinelli E. M., Coleman M. A., Campbell A. H., Santini N. S., Kajlich L., et al. (2019). Restoring subtidal marine macrophytes in the Anthropocene: Trajectories and future-proofing. Mar. Freshw. Res. 70, 936–951. doi: 10.1071/MF18226
Wood G., Steinberg P. D., Campbell A. H., Vergés A., Coleman M. A., Marzinelli E. M. (2022). Host genetics, phenotype and geography structure the microbiome of a foundational seaweed. Mol. Ecol. 31, 2189–2206. doi: 10.1111/mec.16378
Wubs E. R. J., van der Putten W. H., Bosch M., Bezemer T. M. (2016). Soil inoculation steers restoration of terrestrial Ecosystems. Nat. Plants 2, 1–5. doi: 10.1038/NPLANTS.2016.107
Xu G., Yang S., Meng L., Wang B. G. (2018). The plant hormone abscisic acid regulates the growth and metabolism of endophytic fungus Aspergillus nidulans. Sci. Rep. 8, 1–9. doi: 10.1038/s41598-018-24770-9
Young C. N., Schopmeyer S. A., Lirman D. (2012). A review of reef restoration and Coral propagation using the threatened genus Acropora in the Caribbean and western Atlantic. Bull. Mar. Sci. 88, 1075–1098. doi: 10.5343/bms.2011.1143
Zbakh H., Zubía E., de los Reyes C., Calderón-Montaño J. M., López-Lázaro M., Motilva V. (2020). Meroterpenoids from the brown alga Cystoseira usneoides as potential anti-inflammatory and lung anticancer agents. Mar. Drugs 18, 207. doi: 10.3390/md18040207
Zhou Y., Liu P., Liu B., Liu X., Zhang X., Wang F., et al. (2014). Restoring eelgrass (Zostera marina L.) habitats using a simple and effective transplanting technique. PloS One 9, e92982. doi: 10.1371/journal.pone.0092982
Keywords: marine ecosystem restoration, microbiome, corals, seagrasses, macroalgae, habitat-forming species
Citation: Corinaldesi C, Bianchelli S, Candela M, Dell’Anno A, Gambi C, Rastelli E, Varrella S and Danovaro R (2023) Microbiome-assisted restoration of degraded marine habitats: a new nature-based solution? Front. Mar. Sci. 10:1227560. doi: 10.3389/fmars.2023.1227560
Received: 23 May 2023; Accepted: 04 September 2023;
Published: 20 September 2023.
Edited by:
Katherine Dafforn, Macquarie University, AustraliaReviewed by:
Paul Gribben, University of New South Wales, AustraliaCopyright © 2023 Corinaldesi, Bianchelli, Candela, Dell’Anno, Gambi, Rastelli, Varrella and Danovaro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cinzia Corinaldesi, Yy5jb3JpbmFsZGVzaUB1bml2cG0uaXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.