- 1Key Laboratory of Marine Genetic Resources, Third Institute of Oceanography, Ministry of Natural Resources of China, Xiamen, China
- 2State Key Laboratory for Marine Environmental Science, Institute of Marine Microbes and Ecospheres, College of Ocean and Earth Sciences, Xiamen University, Xiamen, China
- 3State Key Laboratory Breeding Base of Marine Genetic Resources, Third Institute of Oceanography, Ministry of Natural Resources, Xiamen, China
- 4Key Laboratory of Marine Genetic Resources of Fujian Province, Third Institute of Oceanography, Ministry of Natural Resources, Xiamen, China
- 5School of Marine Biology, Xiamen Ocean Vocational College, Xiamen, China
Bacteria of the genus Sulfurovum within the class Campylobacteria are widespread in global oceans and are predominant in sulfide-rich environments. However, little is known about their adaptation to such harsh environments owing to their resistance to cultivation. In this study, we obtained three pure cultures of this genus from marine coastal environments and compared them with those obtained from the deep sea. Phylogenetic analysis of 16S rRNA gene sequences indicated that they represent two novel species of the genus, sharing 95.9%–96.1% sequence similarities to Sulfurovum aggregans Monchim33T. Based on the polyphasic classification results, the type strains XTW-4T and zt1-1T were proposed to represent two new species: Sulfurovum xiamenensis sp. nov. and Sulfurovum zhangzhouensis sp. nov., respectively. These coastal isolates were also obligate chemoautotrophs featuring molecular hydrogen as an electron donor and molecular oxygen, thiosulfate, or elemental sulfur as the sole electron acceptor. Comparative genomic analyses based on 11 Sulfurovum species further revealed a clear differentiation between hydrothermal vent and non-vent origins. The non-vent Sulfurovum can use thiosulfate as an electron acceptor but lacks denitrification pathways, whereas the vent bacteria can respire nitrate through complete denitrification pathways. Moreover, the non-vent Sulfurovum contained a nitrogen fixation pathway, implying their adaptation to nitrogen source-deficit niches. In addition, non-vent Sulfurovum species adapted to a higher oxygen concentration via multiple antioxidative defense mechanisms. These phenotypic and genotypic features help us to understand the ecological role of Sulfurovum bacteria in marine ecosystems.
Introduction
The genus Sulfurovum is an important taxon of sulfur-oxidizing bacteria belonging to the family Sulfurovaceae of the phylum Campylobacterota (Waite et al., 2018) and can be found in various habitats, including mangroves (Lin et al., 2019), nearshore sediments (Marziah et al., 2016), shallow hydrothermal zones (Wang et al., 2017), glaciers (Wright et al., 2013), seagrass beds (Sun Y. Y. et al., 2020), and deep-sea hydrothermal vents (Nakagawa et al., 2006; Dahle et al., 2013; Meier et al., 2017). Sulfurovum bacteria are especially abundant in deep-sea hydrothermal vent environments and are widely distributed in various hydrothermal habitats, including chimneys, sediments, plumes, diffuse-flow vent fluids, and symbionts with animals (Akerman et al., 2013; Sheik et al., 2015; Motoki et al., 2020; Moulana et al., 2020; Lee et al., 2021; Chiu et al., 2022). Sulfurovum species are strict chemolithoautotrophs that have diverse energy metabolic pathways (Meier et al., 2017; Wang et al., 2023). They usually use hydrogen and reduced sulfur compounds as electron donors and oxygen, nitrate, and elemental sulfur as electron acceptors to fix CO2, thus contributing to primary productivity in chemosynthetic ecosystems (Wright et al., 2013; Sheik et al., 2015). Recently, Sulfurovum was found to have potential applications in environmental remediation by removing hydrogen sulfide and nitrite from industrial wastewater (Li W. et al., 2022; Wang K. Q. et al., 2022; Zheng et al., 2022).
Currently, the genus Sulfurovum comprises only five species with validly published names and one provisional species. Five species, Sulfurovum lithotrophicum 42BKTT, Sulfurovum aggregans Monchim33T, Sulfurovum riftiae 1812ET, Sulfurovum denitrificans eps51T, and Sulfurovum indicum ST-419T, were isolated from deep-sea hydrothermal vent environments, including vent sediments, plumes, chimneys, and outer biofilms of the polychaete Riftia pachyptila (Inagaki et al., 2004; Mino et al., 2014; Giovannelli et al., 2016; Mori et al., 2018; Xie et al., 2021). One provisional species, “Candidatus Sulfurovum sediminum”, strain AR, was isolated from a non-vent environment. It was an enrichment culture from 78-m deep marine sediment collected near Svalbard Island in the Arctic Circle (Park et al., 2012). Recently, two strains Sulfurovum sp. TSL1 and TSL 6 were isolated from a tsunami-launched marine sediment based only on genomic analysis (Guo et al., 2022). Thus, in view of their widespread distribution and potentially high diversity, more bacteria of this genus are being isolated from different environments.
Although sequences affiliated with genus Sulfurovum have been detected from worldwide marine systems (Teske et al., 2002; Takai et al., 2004; Sylvan et al., 2012; Takai et al., 2015), physiological diversity and the ecological role of this genus have not yet been fully elucidated due to the lack of cultivated strains. Compared to vent Sulfurovum species, which has been studied extensively, there has been less exploration of non-vent Sulfurovum bacteria owing to fewer pure cultures being available from non-vent marine environments. In this study, three strains of the genus Sulfurovum were isolated from intertidal sediments, mangrove sediments, and mariculture pond sediments in subtropical estuary areas along the west bank of the Taiwan Strait. To elucidate their ecological roles and environmental adaptations in coastal marine ecosystems, we characterized strains belonging to two novel species using a polyphasic taxonomic approach. In addition, we performed comparative genomic analyses of all members of the genus Sulfurovum, including those with vent and non-vent origins, to gain insights into the environmental adaptation mechanisms underlying their widespread distribution.
Materials and methods
Bacterial enrichment and isolation
The marine sediments were sampled from intertidal zone of Xiamen island on April 12, 2021 (11812′25′′E 2429′52′′N), the mangrove garden of Xia-Tan-Wei at the estuary of Xixi River of Xiamen on June 27, 2022 (11812′4′′E 2439′14′′N), and an abandoned pond nearby the mangrove conservation area at the estuary of Zhangjiang River, Zhangzhou on September 14, 2022 (11725′55′′E 2357′18′′N) (Supplementary Figure S1). These samples are black and smell like rotten eggs, implying these sediments are enriched in sulfide. The samples were kept cool and returned to the laboratory. Each of 2 g (wet mass) was suspended with artificial seawater, and then 1 ml was transferred into 50 ml anaerobic bottles respectively containing 10 ml MMJHS medium supplied with 76% H2/20% CO2/4% O2 (200kPa) with hydrogen and thiosulfate as energy sources, or 10 ml of MMJS medium, which contains 76% N2/20% CO2/4% O2 (200 kPa) with thiosulfate as the sole energy source, or 10 ml MMJH medium which contains 76% H2/20% CO2/4% O2 (200 kPa) with hydrogen as the energy source (See supplementary material for medium composition). All of them were incubated at 32°C according to a method described previously (Jiang et al., 2017). After 3-round enrichment, the cell growth in MMJH medium was observed. Subsequently, cells were purified three times with the dilution-to-extinction technique. The culture in the serum bottle showing growth at the highest dilution was designated as strains XGS-02, XTW-4, and zt1-1. The purity of these cultures was confirmed by microscopic examination and 16S rRNA gene sequencing.
Phenotypic and chemotaxonomic characterization
Cultures were grown in MMJH medium at 30°C for 2 days, cell morphology was observed under transmission electron microscopy (HT7800, Hitachi, Japan). The physiological characterization of three strains was performed in MMJHS medium and growth was measured by direct cell counting using a phase contrast microscope (Eclipse 80i, Nikon) according to the method previously described (Jiang et al., 2017). The temperature range for growth was determined at 4, 10, 15, 20, 25, 28, 30,32, 35, 37, 40, 45, 50 and 60°C. The salinity range for growth was examined by adjusting the concentrations of NaCl between 0 and 9.0% (w/v) at 0.5 (w/v) intervals. Oxygen sensitivity was examined using MMJHS medium with different O2 concentrations (0%, 1%, 2%, 4%, 6%, 8%, 10%, 15% and 20%) in the headspace gas. In the case of oxygen absence, 10 mM nitrate was added as a potential electron acceptor. The pH range was investigated from 3.5 to 9.0 with a 0.5 pH unit interval by altering pH with several buffers, including 30 mM acetate/acetic acid buffer (pH 3.0–5.0), MES (pH 5.5–6.0), PIPES (pH 6.5–7.0), HEPES (pH 7.5–8.0), Tris and CAPSO (pH 8.0 and above).
To determine the electron donors and acceptors, MMJ synthetic seawater containing 0.1% (w/v) NaHCO3 was used as a basal medium, and H2 was tested as the electron donor for the growth of three strains, nitrate (0.1%, w/v), nitrite (0.1 and 0.01%, w/v), thiosulfate (0.1%, w/v), sulfite (0.01%, w/v), elemental sulfur (1%, w/v), or molecular oxygen (1%), were tested as the only electron acceptor. The sulfur oxidation was tested with MMJS medium, using sodium thiosulfate (0.1%, w/v) or elemental sulfur (S) (1%, w/v) as sole electron donor, nitrate (0.1%, w/v), nitrite (0.1 and 0.01%, w/v), or molecular oxygen (0.1 and 1%, v/v) were tested as the only electron acceptor. Heterotrophic growth was tested in MMJ medium in the absence of NaHCO3 and in a 94% N2/6% O2 (200 kPa) gas phase. The following potential organic carbon compounds were tested: 0.02% (w/v) glucose, galactose and fructose, 0.1% (w/v) peptone, starch and casein, 5 mM formate, acetate, citrate, fumarate and pyruvate.
For fatty acids analysis, cells grown on MMJHS medium at 30C for 24h were saponified, methylated, and extracted following the standard MIDI protocol (Sherlock Microbial Identification System, version 6.0B). The fatty acids were analyzed by gas chromatography (Agilent Technologies 6850), and then the result was identified using the TSBA6.0 database of the Microbial Identification System.
Phylogenetic analysis and genome sequencing
The total DNA of the three strains was extracted and the 16S rRNA gene was amplified as described previously (Jiang et al., 2009). The similarity of 16S rRNA gene sequences was determined using the EzBioCloud server (http://www.ezbiocloud.net/) and NCBI database, and sequences of close strains were downloaded for the phylogenetic analysis. Sequence comparison was performed using CLUSTAL_W (Larkin et al., 2007) and phylogenetic trees were constructed using MEGA X (Kumar et al., 2018) according to the Neighbor-Joining (Tamura et al., 2004), Maximum likelihood and Maximum Parsimony methods (Felsenstein, 1981). The online website TVBOT was used to embellish the phylogenetic tree (Xie et al., 2023). Genetic distance was calculated using Kimura two-parameter model. Bootstrap analysis was calculated based on 1000 replications. The complete genome of strain XGS-02 was sequenced by Tianjin Biochip Corporation (Tianjin, PR China) using the single molecule real-time (SMRT) technology on the Pacific Biosciences (PacBio) sequencing platform. De novo assembly was performed using the Hierarchical Genome Assembly Process (Version 4) workflow, and the complete circular genome was derived. Draft genome sequences of strains XTW-4 and zt1-1 were sequenced using Illumina Hiseq 2000 platform (Major Bio-Pharm Technology Co., Ltd, China). Raw reads were clipped and trimmed using Trimmomatic version 0.32 (Bolger et al., 2014). Trimmed reads were assembled using SOAPdenovo (Luo et al., 2015). The G+C content of the genomic DNA was determined from the whole genome sequence. Gene prediction and annotation were performed using NCBI Prokaryotic Genomes Annotation Pipeline (PGAP) (Tatusova et al., 2016) and the Rapid Annotation using Subsystem Technology (RAST) pipeline (Overbeek et al., 2014)
Genomic properties and comparative genomic analysis
We downloaded all culturable Sulfurovum genomes available from NCBI database, including four strains Sulfurovum indicum ST-419T, Sulfurovum riftiae 1812ET, Sulfurovum lithotrophicum ATCC BAA-797T, and Sulfurovum sp. NBC37-1 from deep-sea hydrothermal vent environments, four strains, Sulfurovum sp. TSL1, Sulfurovum sp. TSL6, Candidatus Sulfurovum sp. AR, and Sulfurovum sp. HSL1-3 from coastal sediments. A total of eleven genomic sequences, including three genomes from our newly isolated strains XGS-02, XTW-4, and zt1-1, were checked for completeness and contamination using CheckM (Parks et al., 2015). The phylogenomic tree was constructed based on an up-to-date 92 bacterial core gene sets by UBCG version 3.0 (Na et al., 2018), the ANI was compared using the pyani tool (Pritchard et al., 2016), Pan-genomic analysis was performed with BPGA software (Chaudhari et al., 2016). Protein sequences were identified using PowerBLAST (Zhang and Madden, 1997). Sequence comparison was performed using MEGA X (Kumar et al., 2018), and phylogenetic trees based on amino acid sequences were constructed using the Maximum Likelihood method with bootstrap values based on 1000 replicates.
Data availability
The GenBank accession numbers for 16S rRNA gene sequence of strains XGS-02, XTW-4 and zt1-1 are OM960633, OP810811 and OP810810, respectively. The genome sequences have been deposited at GenBank under the accession numbers of CP093312, JAQIBC000000000 and JAQIBD000000000, respectively. The three Sulfurovum strains were deposited in the Marine Culture Collection of China (MCCC) under collection numbers MCCC 1A18820, MCCC 1A19406, MCCC 1A19490.
Results
Phylogenetic analysis based on 16S rRNA gene and core genome
With enrichments with H2 as the sole energy source, three strains of Sulfurovum were isolated from coastal sediments and designated as strains XGS-02, XTW-4, and zt1-1. In other enriched media, the isolates belonged to genus Thiomicrorhabdus and Sulfurimonas, which were not the focus of this study. The full-length 16S rRNA gene sequences of strains XGS-02 and zt1-1 retrieved from genomes were 1,503 bp and 1,513 bp, respectively. The 16S rRNA gene sequences of XTW-4 was 1,419 bp. BLAST analysis of the 16S rRNA gene sequences indicated that strains XGS-02, XTW-4, and zt1-1 shared the highest sequence similarity with S. aggregans Monchim33T, which was isolated from a deep-sea hydrothermal vent chimney with similarities of 96.4%, 96.1%, and 95.9%, respectively. They were also closely related to S. indicum ST-419T from the deep-sea hydrothermal plume with 95.6%, 94.6%, and 95.2% similarity, respectively. Furthermore, strains XGS-02 and XTW-4 shared 99.6% sequence similarity with each other and 95.9% and 96.0% similarity with strain zt1-1, respectively. The results indicate that strains XTW-4 and zt1-1 represent two novel species of the genus Sulfurovum.
Based on 16S rRNA gene sequences, the phylogenetic tree indicated that strains XGS-02 and XTW-4 clustered with Sulfurovum species from non-vent marine habitats, whereas those from vent environments formed separate clusters. Notably, strains zt1-1 and HSL1-3 represented a new branch located differently from other Sulfurovum species (Figure 1, Supplementary Figures S2, S3). The phylogeny was confirmed by whole-genome phylogenetic analysis based on 92 core gene sequences (Figure 2), which was consistent with the phylogenetic tree based on the 16S rRNA gene sequences. Combining the phylogenetic tree based on 16S rRNA gene sequences and whole genomes, three different evolutionary branches of the genus Sulfurovum were revealed. Correspondingly, three groups were named: Group I, bacteria from intertidal sediments; Group II, bacteria from hydrothermal vent areas; and Group III, bacteria from intertidal sediments (Figure 2).
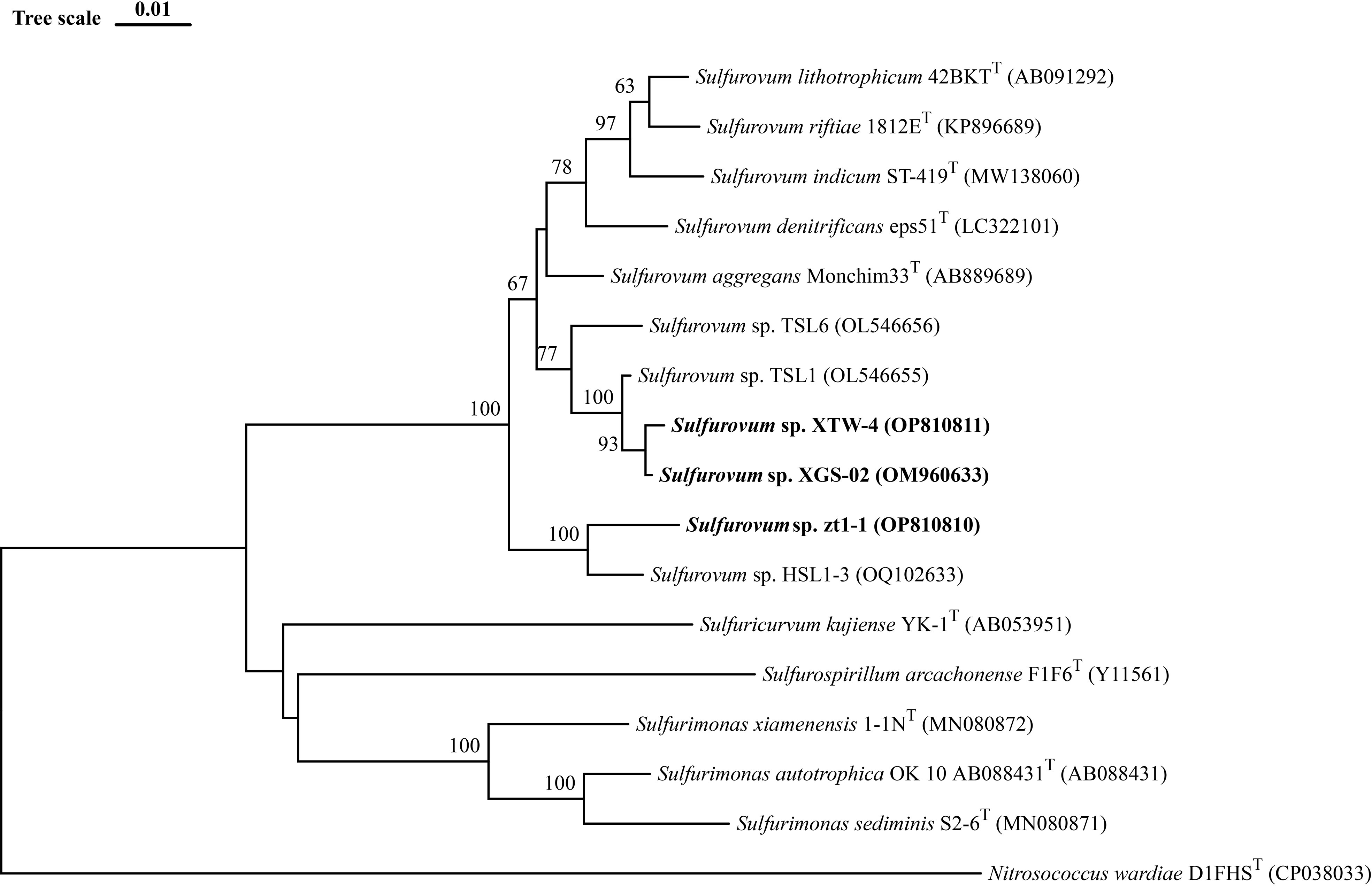
Figure 1 Neighbor-Joining phylogenetic tree based on 16S rRNA gene sequences showing the phylogenetic positions of strains XGS-02, XTW-4, zt1-1 and other members of the genus Sulfurovum. The tree was rooted using Nitrosococcus wardiae as an outgroup. Bootstrap values based on 1000 replicates are shown at branch node. Higher than 50 % bootstrap values are shown. Bar, 0.01 substitutions per nucleotide position.
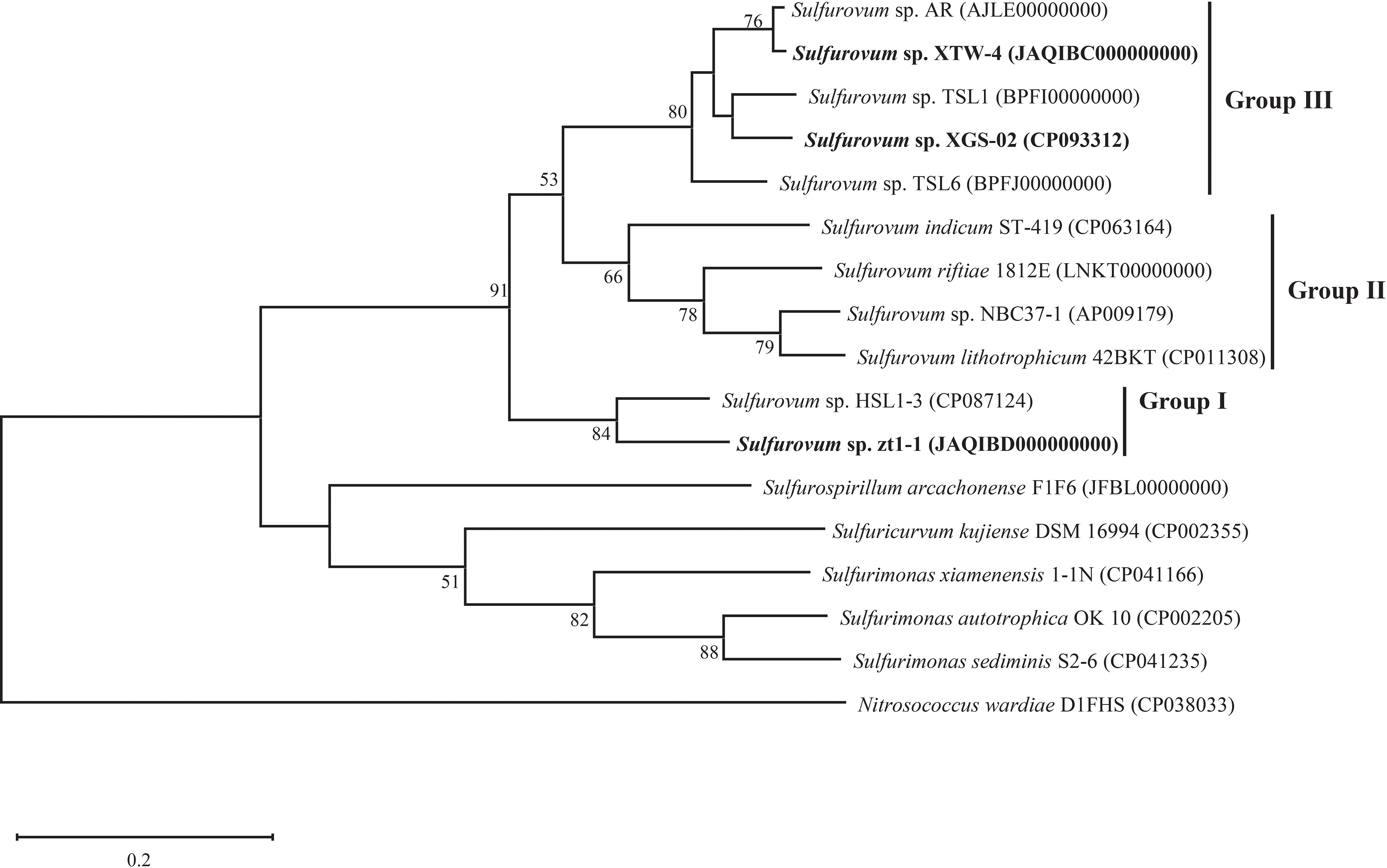
Figure 2 Whole genome phylogenetic tree based on 92 core gene sequences showing the position of XGS-02, XTW-4, zt1-1 and closely related taxa within the genus Sulfurovum using the maximum-likelihood algorithm. The node is labeled with Gene Support Index (GSI) values. Branch node values below 50% are not shown. The accession numbers of the genomes are shown in parentheses. Bar, 0.2 substitutions per position.
Phenotypic and chemotaxonomic characterization
Morphological observation by TEM showed that strains XTW-4 and zt1-1 had no flagella. The result was confirmed by a genomic analysis, without genes encoding flagella in their genomes. The cells of strain XTW-4 was short rods, 0.2–0.4 µm wide and 0.4–1.2 µm long (Supplementary Figure S4). The morphology of strain zt1-1 was short rod-shaped, 0.2–0.5 µm wide, and 0.4–1.2 µm long, which was similar to strain XTW-4 (Supplementary Figure S4).
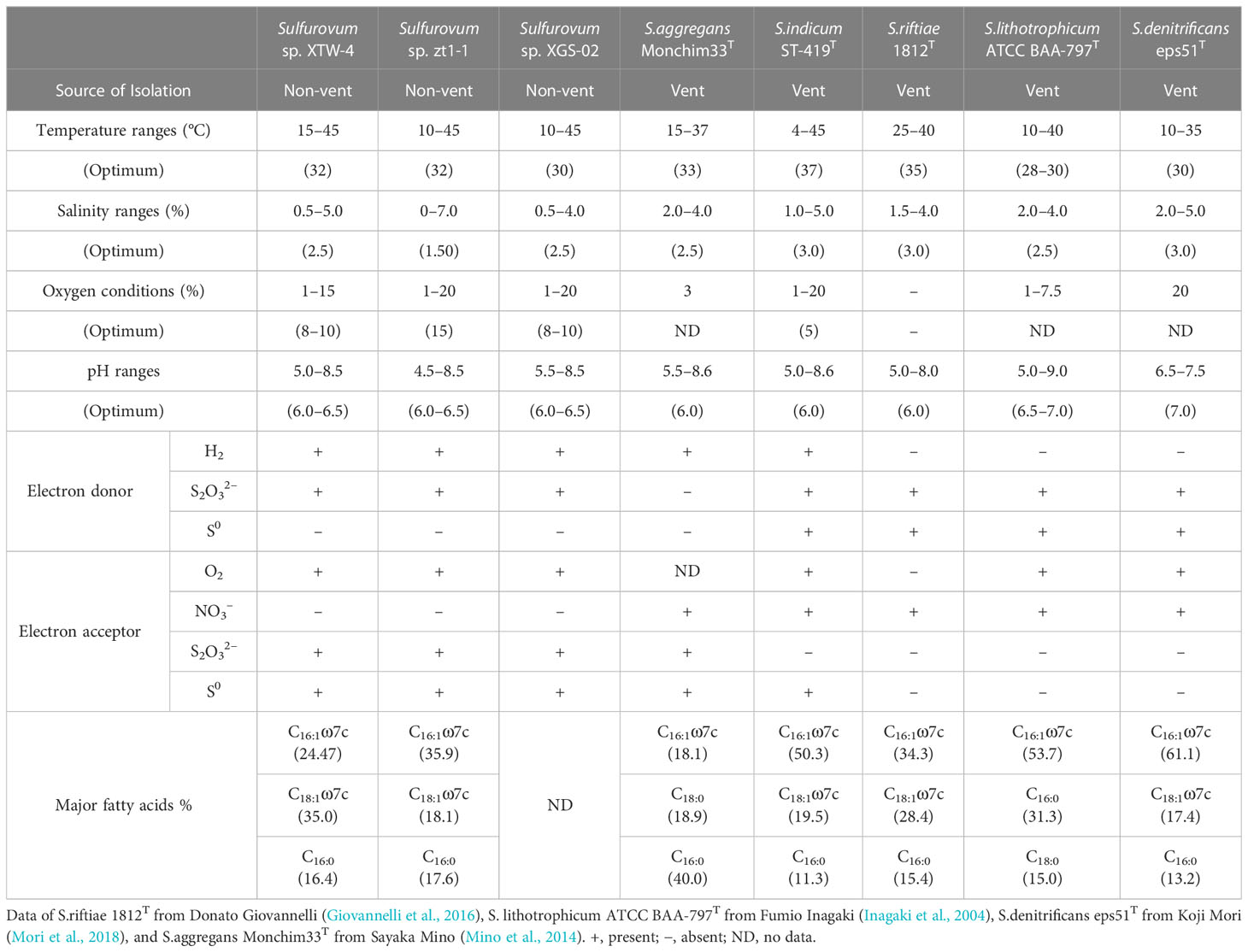
The growth experiment showed that strain XGS-02 could grow at a temperature range of 10–45°C, salinity range of 0.5–4.0% (w/v) NaCl, pH range of 5.5–8.5, and oxygen concentration range of 1–20%. Strain XTW-4 could grow at temperature (15–45°C), salinity (0.5–5.0% (w/v) NaCl), pH (5.0–8.5), and oxygen (1–15%) ranges. Strain zt1-1 could grow at 10–45°C, 0%–7.0% (w/v) NaCl, 4.5–8.5 pH, and 1%–20% O2. The optimal temperature for the three newly isolated strains was approximately 30°C and the optimum pH was 6.0–6.5. The optimal salinity was 2.5% (w/v) NaCl for strains XGS-02 and XTW-4, and 1.5% (w/v) NaCl for strain zt1-1. The optimum oxygen concentrations were 8% for XGS-02 and XTW-4 and 15% for zt1-1. Chemoautotrophic growth showed that the three strains could utilize hydrogen and thiosulfate as electron donors but fail to oxidize elemental sulfur. They had the highest biomass with hydrogen as the electron donor. Thus, hydrogen may be the preferred energy source for these bacteria. All oxygen, thiosulfate and elemental sulfur can be used as electron acceptors. It is worth mentioning that the strains grew weakly when thiosulfate acted as the electron donor and oxygen acted as electron acceptor. All strains could not use nitrate and nitrite as electron acceptors.
We further compared the physiological characteristics of the three new non-vent strains with those of the vent-type strains, S. indicum ST-419T (Xie et al., 2021), S. riftiae 1812ET (Giovannelli et al., 2016), S. lithotrophicum ATCC BAA-797T (Inagaki et al., 2004), S. denitrificans eps51T (Mori et al., 2018), and S. aggregans Monchim33T (Mino et al., 2014) (Table 1). The results revealed an evident divergence between the vent and non-vent strains. The non-vent strains had an optimal oxygen range of 8–15%, showing higher oxygen adaptation than the vent strains. Moreover, the three non-vent strains could not use nitrate as an electron acceptor, whereas all the vent strains could. All the non-vent Sulfurovum-type strains could reduce thiosulfate, but all those of the vent species could not, except for S. aggregans. In the case of electron donor utilization, all non-vent strains can utilize hydrogen as an electron donor, while most (3/5) vent strains cannot. In contrast, none of the non-vent strains can grow with elemental sulfur as the sole energy source, whereas most (4/5) vent strains can grow.
The fatty acid profiles of strains XTW-4 and zt1-1 and related type strains are listed in Table 1. Strain XTW-4 contained the predominant cellular fatty acids C18:1ω7c (35.0%), C16:1ω7c (24.5%), and C16:0 (16.4%), and strain zt1-1 mainly contained C16:1ω7c (35.9%), C18:1ω7c (18.1%), and C16:0 (17.6%). These profiles are similar to those of S. indicum ST-419T, S. riftiae 1812T, and S. denitrificans eps51T.
Genomic features of the two novel species of Sulfurovum
The genome size of strain XTW-4 was 2.20 Mb with a GC content of 39.1%, and the genome size of zt1-1 was 2.18 Mb with a GC content of 39.3%. A total of 2,268 genes were predicted in the genome of strain XTW-4, including 2,203 protein-coding and 65 RNA genes. The genome of strain zt1-1 contains 2,267 genes, including 2,209 protein-coding genes and 58 RNA genes. The genomic similarity of strains XTW-4 and zt1-1 with other species of the genus Sulfurovum was determined using ANI values. The paired ANI values between strain XTW-4 and its close relatives, S. indicum and S. riftiae, were 86.0% and 85.7%, respectively. The paired ANI values between strain zt1-1 and its close relatives, S. indicum and S. riftiae, were 84.0% and 84.4%, respectively. These findings support the classification of strains XTW-4 and zt1-1 as two distinct species based on the cut-off threshold of ANI (95–96%) for the delineation of prokaryotic species. Strains XTW-4, zt1-1, and others of Sulfurovum species shared less than 40.0 % DDH values (Table S2). The low DDH values (< 70%) confirmed that strains XTW-4 and zt1-1 represented two novel species within the genus Sulfurovum (Stackebrandt and Goebel, 1994; Auch et al., 2010; Meier-Kolthoff et al., 2014).
Comparative genomic analysis within the genus
A total of 11 Sulfurovum genomes from hydrothermal vents and intertidal environments were used for comparative analysis in this study. The genome size range of the non-vent Sulfurovum species was 2.1–2.6 Mb, and the G+C content ranged from 38.4 to 43.6%, while the genome size range of the vent strain was 2.2–2.6 Mb and the range of G+C content was 42.5–45.7%. The results showed that Sulfurovum species from deep-sea hydrothermal vent environments had relatively higher G+C values (Table 2). In addition, comparative genomic analysis revealed 1051 core genes in all Sulfurovum genomes (Supplementary Figure S5). The percentage of core genes per genome ranged from 42.8% to 50.9%, and the percentage of unique genes per genome ranged from 5.4% to 19.6%. Sulfurovum genomes belonging to Group I had 12.2–19.3% of unique genes, and those genomes from Group II had 11.9–17.9%. Group III from non-vent genomes had the lowest percentage of unique genes (< 9%) (Table S1).
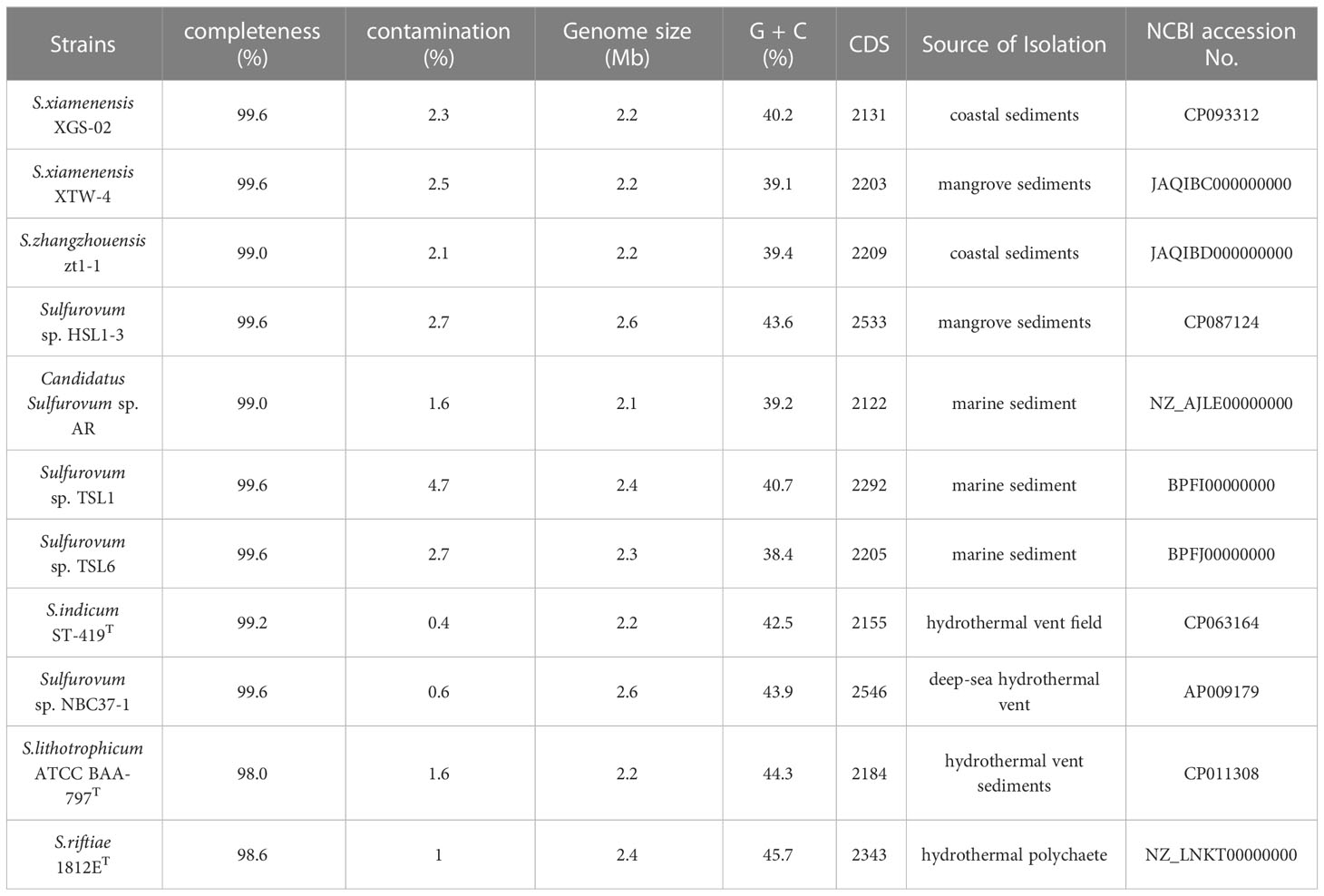
Table 2 Genomic information of all Sulfurovum strains available in NCBI database and obtained in this study.
Central metabolisms of Sulfurovum
Nitrogen metabolism and its differentiation among vent and non-vent origins
Comparative genomic analysis showed significant differences in nitrogen metabolism between the vent and non-vent Sulfurovum species (Table 3). Sulfurovum species from hydrothermal vent habitats contain genes required for the complete denitrification pathway, including the napAB gene encoding outer membrane nitrate reductase, the nirS gene encoding nitrite reductase, the norBC gene encoding nitric oxide reductase, and the nosZ gene encoding nitric oxide reduction enzyme. In addition, the genes nrfA and napA, performing a heterotrimeric nitrate reduction pathway, were present in all vent Sulfurovum genomes. Intriguingly, the denitrification and dissimilatory nitrate reduction pathways were completely absent in all non-vent Sulfurovum genomes, except for strain TSL1, which only has a denitrification-related nirk gene. Interestingly, key genes encoding nitrogen-fixing enzymes were observed for the first time in Sulfurovum genomes, including nifH, nifD, and nifK, but only in the non-vent strains zt1-1 and HSL1-3. In the phylogenetic tree, we found that the nifH of Sulfurovum represents a different branch from that of Betaproteobacteria and Alphaproteobacteria (Supplementary Figure S6).
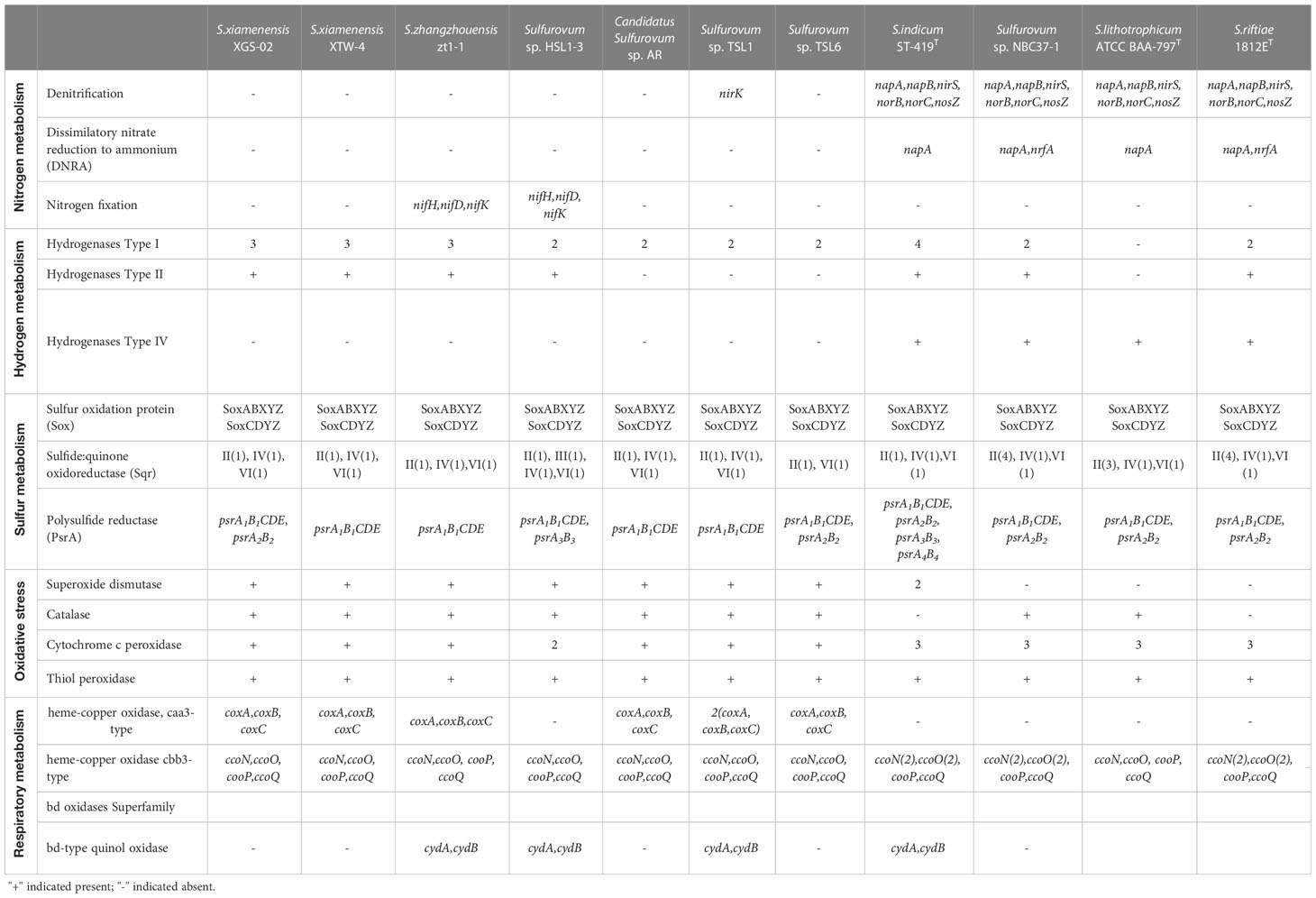
Table 3 Comparison of key enzymes for nitrogen, hydrogen, and sulfur metabolisms in Sulfurovum species based on RAST annotations in this study.
Hydrogen metabolism and the differentiation within the genus
Sulfurovum species contain three types of hydrogenases (Table 3): Type I, Type II, and Type IV. However, S. lithotrophicum only contains Type IV hydrogenase. Type I hydrogenases are the most widely distributed among members of the genus Sulfurovum. They usually have multiple copies of Type I hydrogenases, and S. indicum contains four copies of Type I hydrogenases (Figure 3). Type II was not conserved in Sulfurovum species, as in Type I, which was absent in some non-vent isolates, such as Sulfurovum sp. AR, TSL1, TSL6, and the vent-type S. lithotrophicum strains. Intriguingly, Type IV was only detected in the vent isolates, which belong to the hydrogenase Hyc type, except for S. indicum with a Coo type. In addition, phylogenetic analysis based on the large subunit of hydrogenase showed that hydrogenases from vent genomes and non-vent genomes of the genus Sulfurovum clustered together.
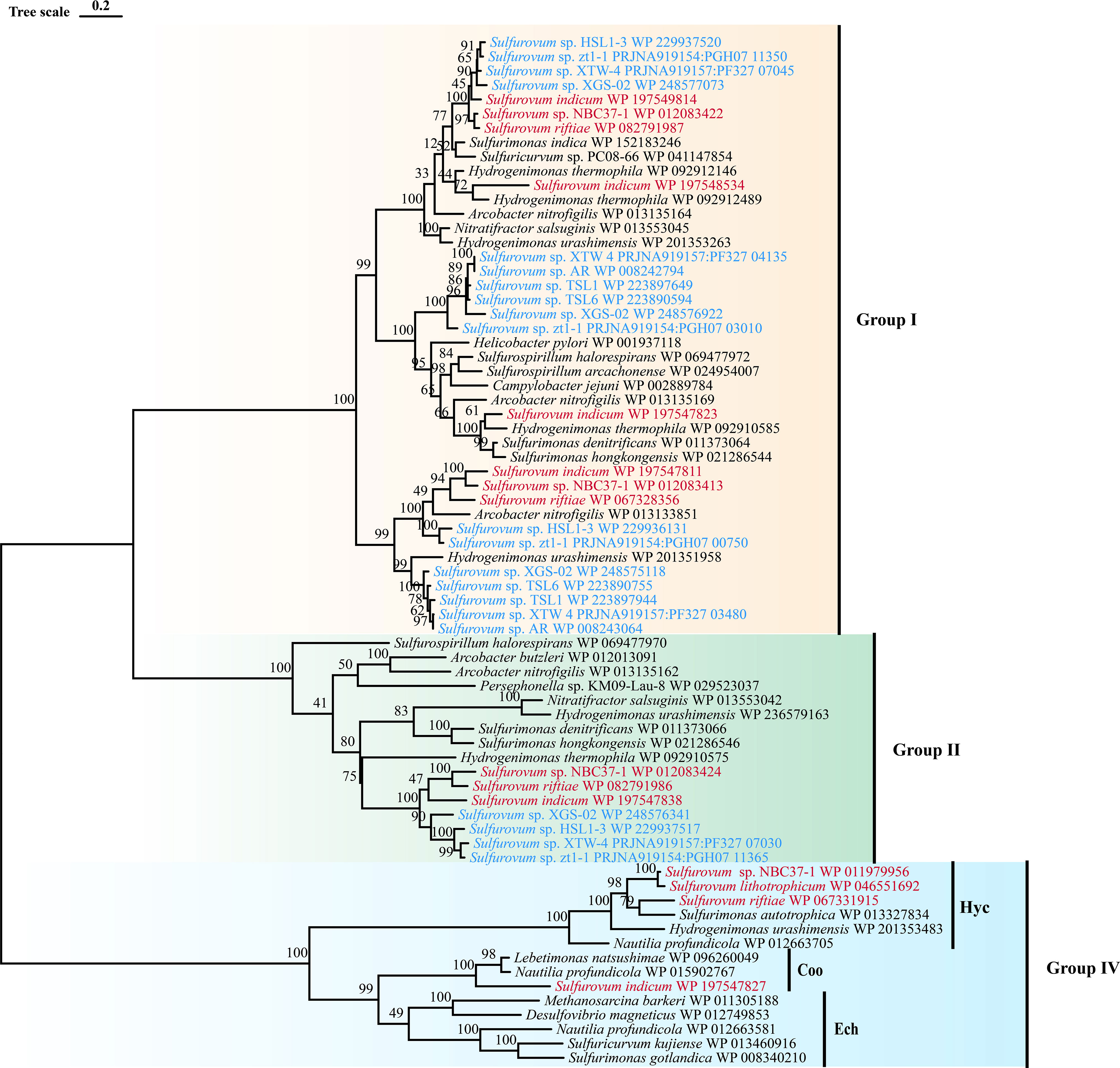
Figure 3 Maximum-likelihood tree based on large subunit sequences of hydrogenase from Sulfurovum. Bootstrap values shown on each node are based on a total of 1000 bootstrap replicates. Branch node values below 50% are not shown. The isolation source of the Sulfurovum species were indicated by different font colors: red, deep-sea hydrothermal vent environments; blue, marine non-vent system.
Differentiation in sulfur metabolism among vent and non-vent origins
For sulfide oxidation, Sulfurovum species contain four sulfoquinone oxidoreductase (Sqr) types, including Type II, Type III, Type IV, and Type VI (Supplementary Figure S7). Type II Sqr was present in all strains (Table 3), implying that it is essential for sulfide oxidation in Sulfurovum species. Notably, the vent strains contained more copies of Type II Sqr, up to four copies in strain NBC37-1 and S. riftiae, in contrast to the single copy in the non-vent strains. Type III Sqr was present only in the non-vent strain HSL1-3, whereas Type VI Sqr was present in all strains in a single copy. Phylogenetic analysis showed that the vent and non-vent branches of Sqr were located in different evolutionary clusters, and Sqr from different groups of Sulfurovum clustered together.
In addition, all Sulfurovum species encode the complete sox gene cluster (soxABXYZ and soxCDYZ) for thiosulfate oxidation. The phylogenetic tree based on the SoxB protein showed a topology similar to that of the phylogenomic analysis of Sulfurovum, suggesting that the SoxB gene in Sulfurovum species is acquired by vertical inheritance within the lineage (Supplementary Figure S8). For elemental sulfur respiration, all Sulfurovum strains have polysulfide reductase (Psr) of the three groups, which can reduce elemental sulfur to sulfide (Table 3). Phylogenetic analysis showed that polysulfide reductase Group I was detected in all Sulfurovum species from non-vent environments, encoded by five genes, psrA1B1CDE. Groups II and III were also present in some non-vent strains, such as TSL6, XGS-02, and HSL1-3 (Supplementary Figure S9).
Differentiation in antioxidation and terminal oxidases
Genes encoding superoxide dismutase and catalase, which are involved in oxidative stress, were found in non-vent strains (Table 3), while superoxide dismutase was absent in vent strains NBC37-1, S. lithotrophicum and S. riftiae, and catalase was absent in S. indicum and S. riftiae. In addition, all Sulfurovum strains contain genes encoding cytochrome c peroxidase and thiol peroxidase, which scavenge peroxides and provide defense against oxidative damage (Missall et al., 2004). Vent Sulfurovum genomes usually have more copies of cytochrome c peroxidase than those from non-vent environments. Multiple terminal oxidases are present in the Sulfurovum genomes, all containing cbb3-type cytochrome c oxidase (Table 3). Phylogenetic analysis showed that cbb3 type cytochrome c oxidase clustered into two groups: CcoN I and CcoN II (Supplementary Figure S10). All strains containing CcoN I and CcoN II were of vent origin. The coxA-encoded caa3 type terminal oxidase was present in non-vent strains and absent in vent strains (Table 3). In addition, some Sulfurovum strains, such as zt1-1, HSL1-3, TSL1, and S. indicum, contained genes encoding cytochrome bd ubiquinone oxidase.
Discussion
Sulfurovum spp. in the Campylobacteria class are important chemolithoautotrophic bacteria with wide distribution in sulfidic habitats in the ocean, and they play an essential role in marine carbon, nitrogen, and sulfur biogeochemical cycling (Patwardhan et al., 2018; Sun Q. L. et al., 2020; Huang L. B. et al., 2021; Wang Y. et al., 2022). However, knowledge about this important bacterial taxon has been limited to deep-sea hydrothermal vent environments, and little is known about its diversity and ecological role in non-vent environments. In the present study, we isolated these species from coastal sediments. Further comparative analyses revealed the presence of unique metabolic features. Evident diversification was confirmed by phylogenetic analysis of vent and non-vent origins.
Two novel species of non-vent origin of the genus
To our knowledge, this is the first report of a novel species of non-vent origin in Sulfurovum. The combined phenotypic, chemotaxonomic, and phylogenetic characteristics demonstrated that strains XTW-4 and zt1-1 represent two new species in the genus Sulfurovum, for which the names Sulfurovum xiamenensis sp. nov. and Sulfurovum zhangzhouensis sp. nov. are proposed, both of which showed the closest 16S rRNA gene sequence similarity to S. aggregans Monchim33T (95.8% and 95.4%, respectively). They differed from other type strains in phenotypical features, such as optimum salinity and oxygen concentrations and the utilization of electron acceptors. Intriguingly, the two coastal species both respired for thiosulfate, while the vent strains could not; however, while the vent strains could respire nitrate, the two coastal strains could not (Table 1).
Environmental adaptations of Sulfurovum revealed by comparative genomic analyses
To understand their environmental adaptation, we performed comparative genomic analyses between the vent and non-vent Sulfurovum strains. Significant differences were observed in key metabolic mechanisms and stress responses between the vent and non-vent strains, implying that their adaptive evolution and species diversification were related to inhabitance.
Correspondingly, we found that all members of the genus Sulfurovum from deep-sea hydrothermal vents contained a complete denitrification pathway in addition to some genes involved in dissimilatory nitrate reduction to ammonia (DNRA) in part vent strains. Denitrification and DNRA processes are currently considered to be two competing pathways for nitrate reduction (Bu et al., 2017; Jia et al., 2020; Pandey et al., 2020; Li S. et al., 2022), and few studies have reported their coexistence except for Pseudomonas and Shewanella (Kuypers et al., 2018; Huang X. J. et al., 2021; Liu et al., 2021). The nitrate reduction pathway (NAP) is considered conserved and widespread in the phylum Campylobacterota based on bacteria from deep-sea hydrothermal vent habitats (Vetriani et al., 2014). However, these two processes were absent in our coastal strains. Sulfur oxidation coupled with nitrate reduction is considered the primary source of energy in vent ecosystems (Wang et al., 2009), whereas this process does not occur in coastal Sulfurovum species. The genome revealed that the denitrification and DNRA pathways were completely absent, and experimental validation showed that they could not use nitrate as an electron acceptor. To compensate, our two Sulfurovum species adapted to higher oxygen concentrations and preferred to use oxygen as an electron acceptor, which is in agreement with the loss of denitrification and DNRA-related genes, as well as other non-vent Sulfurovum strains (unpublished data).
Intriguingly, members of the genus Sulfurovum from coastal sediments possess genes for nitrogen fixation, which are absent in deep-sea vent Sulfurovum. Coastal mangrove sediments are recognized to be nitrogen-limited due to the unbalance of carbon and nitrogen caused by burying of carbon-rich plant litter (Lin et al., 2019; Luo et al., 2021). Nitrogen-fixing bacteria are considered to play a key role in nitrogen amendment (Alfaro-Espinoza and Ullrich, 2015; Tang et al., 2020; Luo et al., 2021). Therefore, Sulfurovum containing nitrogenase genes are of competitive advantage in coastal environments. Recently, the expression of Nif genes associated with Sulfurovum spp., as well as the presence of the nifH gene in a Sulfurovum metagenome-assembled genome (MAG) was demonstrated at a coastal gas vent site (Patwardhan et al., 2021). Certainly, not all coastal Sulfurovum have nitrogenase genes. This is probably due to highly variable environmental parameters in coastal sediments. In contrast, ammonia is relatively abundant in the vent fluid, which can reach micromolar level in situ (Xu et al., 2014), and can be oxidized to nitrate by ammonia-oxidizing archaea (Crepeau et al., 2011; Zhang et al., 2016; Ding et al., 2017). Thus, genes for nitrogen fixation in deep-sea vent may be redundant and lost in all vent Sulfurovum. The differentiation of Sulfurovum in accordance with unique habitats expands our understanding of the genus in terms of environmental adaptation (Figure 4).
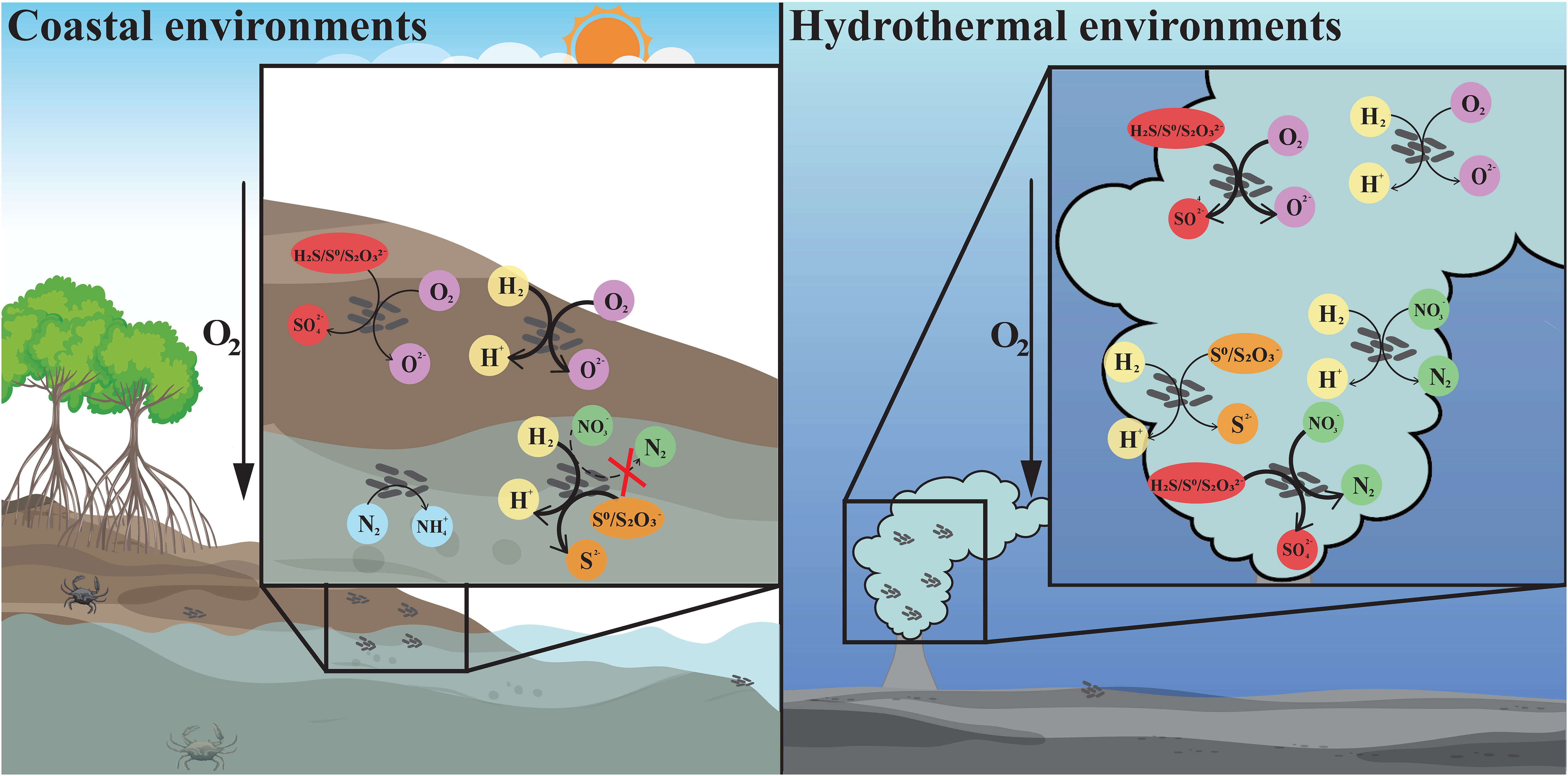
Figure 4 The differentiation and adaptation of Sulfurovum bacteria in coastal sediment and deep-sea hydrothermal vent environments. Metabolic features of Sulfurovum with different origins, including hydrogen oxidation (yellow), sulfur oxidation (red), oxygen reduction (purple), sulfur reduction (orange), denitrification (green), and nitrogen fixation (blue). The arrows with different styles indicate the presence or absence of the related metabolic pathway, and whether it is the major energy-acquiring pathway. Thick solid line: the process is present and probably the main process; thin solid line: the relevant gene is present; thin dashed line: the relevant gene is absent. Left: coastal environments represented by a mangrove ecosystem; right: deep-sea hydrothermal vent environment.
Reduced sulfur compounds, such as sulfides, are important energy sources for chemolithoautotrophs in deep-sea vents. Unexpectedly, we found that coastal isolates of Sulfurovum grew best with hydrogen as an electron donor and weakly with thiosulfate and sulfide. Moreover, all members of this genus contain an extensive suite of hydrogenases, including [NiFe]-hydrogenase groups 1b, 2d, and 4 (Hyc and Coo). The most frequently occurring hydrogenases belonged to Group I. Almost all Sulfurovum members contained multiple copies of this group, probably in response to different hydrogen concentrations, suggesting that hydrogenase in Group I plays an important role in energy draining to support bacterial growth (Grote et al., 2012; Berney et al., 2014).
Regarding sulfide oxidation, Sulfurovum species contain genes encoding different types of Sqr, classified as types II, III, IV, and VI. The diversification of these Sqr genes is thought to play important roles in sulfide oxidation, sulfide assimilation, energy production, heavy metal tolerance, detoxification, and sulfide signaling (Marcia et al., 2010). Additionally, Sulfurovum bacteria have one or more copies of Type II Sqr (Table 3), which may compensate for other Sqrs under specific environmental conditions (Wang et al., 2021). Thiosulfate (Figure 4) can commonly be oxidized by Sulfurovum of any origin, but as an electron acceptor, it can be used by non-vent Sulfurovum, as confirmed by our coastal pure cultures (Table 1).
Sulfurovum usually grows in a mixed zone with unsaturated oxygen concentration (Meier et al., 2017). Antioxidation systems may be essential for traveling across redox gradients to oxic areas. Reactive oxygen species (ROS) are formed by the intermediate products of oxygen reduction and can cause damage to cellular macromolecules (Cabiscol et al., 2000). Superoxide dismutase and catalase are the most well-characterized ROS defense mechanisms (Johnson and Hug, 2019). Interestingly, we found that the genes encoding both superoxide dismutase and catalase were present in the non-vent strains but were partially or completely absent in the vent strains (Table 3). In addition, some terminal oxidases function in the defense against ROS (Kaminski et al., 1996; Hassani et al., 2010; Borisov and Siletsky, 2019), allowing certain strictly anaerobic bacteria to grow in microaerobic environments, such as cbb3-type hemocopper oxidase and cytochrome bd ubiquinone oxidase. All pure culture strains of Sulfurovum contain the CcoNOQP operon, which encodes a cbb3-type cytochrome c oxidase that supports bacterial growth under anaerobic and microaerobic conditions owing to its high affinity for oxygen. Some strains also encode cytochrome bd ubiquinone oxidase, which is an alternative oxidase found in many bacteria that oxidizes ubiquinone and reduces oxygen as part of the electron transport chain. In addition, unlike vent strains, non-vent strains have a caa3-type hemocopper oxidase that catalyzes the reduction of oxygen to water and may play an important role in oxygen adaptation. Hence, it is possible that the ROS defense mechanisms enable coastal Sulfurovum bacteria to deal with oxidative stress caused by increased O2 concentrations in coastal environments. This is consistent with their wider growth ranges with regard to oxygen concentrations (1–15%) and higher optimum oxygen concentration compared the vent Sulfurovum bacteria (Table 1).
Conclusions
Two novel Sulfurovum species were characterized from coastal marine habitats for the first time, Sulfurovum xiamenensis and Sulfurovum zhangzhouensis, contributing to the understanding of the ecological role of Sulfurovum in marine environments. As summarized in Figure 4, the coastal isolates significantly differed from the vent species of this genus in energy metabolism and environmental adaptation. Non-vent Sulfurovum bacteria cannot respire nitrate because of the lack of nitrate reduction pathways but can reduce thiosulfate coupling hydrogen oxidation in addition to high oxygen concentrations. Moreover, non-vent members can perform nitrogen fixation, which might facilitate their survival in niches with limited N sources. These results reveal the diversification among members inhabiting deep-sea vent and non-vent ecosystems and highlight the unique roles of Sulfurovum in coastal marine environments.
Description of Sulfurovum xiamenensis sp. nov.
Sulfurovum xiamenensis (xia.men.en’sis. N.L. masc. adj. xiamenensis of Xiamen, a city in Fujian, China, where the type strain was isolated).
Cells are Gram-negative short rods (0.4-1.2 × 0.2-0.4 μm) without flagella. Optimum oxygen concentration 8%-10%. Growth occurs at 15-45°C (optimum 32°C), pH 5.0-8.5 (optimum pH 6.0-6.5), and 0.5-5.0% (w/v) NaCl (optimum 2.5%). Obligate chemolithoautotrophic growth occurs with H2 as electron donor, and oxygen, thiosulfate, and S0 can be utilized as electron acceptors. It also growth occurs with thiosulfate as an electron donor and oxygen as an electron acceptor. Organic substrates are not utilized as carbon sources and energy sources. Major cellular fatty acids are C18:1ω7c, C16:1ω7c and C16:0.
The type strain, XTW-4T (=MCCC 1A19406), was isolated from mangrove sediments in, Xiamen, Fujian Province, PR China. The genomic G+C content of the type strain is 39.1 mol%.
Description of Sulfurovum zhangzhouensis sp. nov.
Sulfurovum zhangzhouensis (zhang.zhou.en′sis. N.L. masc. adj. zhangzhouensis of Zhangzhou, a city in Fujian, China, where the type strain was isolated).
Cells are Gram-negative short rods (0.4-1.2 × 0.2-0.5 μm) without flagella. Optimum oxygen concentration 15%. Growth occurs at 10-45°C (optimum 32°C), pH 4.5-8.5 (optimum pH 6.0-6.5), and 0.5-7.0% (w/v) NaCl (optimum 1.5%). Obligate chemolithoautotrophic growth occurs with H2 as electron donor, and oxygen, thiosulfate, and S0 can be utilized as electron acceptors. It also growth occurs with thiosulfate as an electron donor and oxygen as an electron acceptor. Organic substrates are not utilized as carbon sources and energy sources. Major cellular fatty acids are C16:1ω7c, C18:1ω7c and C16:0.
The type strain, zt1-1T (=MCCC 1A19490), was isolated from the sediment of retirement ponds in Yunxiao County, Zhangzhou City, Fujian Province, PR China. The genomic G+C content of the type strain is 39.4 mol%.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
ZS and LJ conceived the study. JW, LJ, QZ, and ZS designed the experiments. JW, YZ, and QY participated in the sample collection. JW performed the experiments and analyzed the data. JW and JZ drafted the manuscript. SW, QZ, LJ, and ZS revised the manuscript, which was finally read and corrected by ZS. All authors read and approved the final manuscript. All authors contributed to the article.
Funding
Financial support was provided by the National Natural Science Foundation of China (No. 42176134 to LJ; No.42030412 to ZS), the China Ocean Mineral Resources R&D Association (COMRA) program (No. DY135-B2-01 to ZS), National Key Research and Development Program of China (No. 2018YFC0310701 to ZS), the Scientific Research Foundation of Third Institute of Oceanography, MNR (No. 2019021 to ZS).
Acknowledgments
We thank Dr. Ding Li and M.S. Xuying Zhu from the Third Institute of Oceanography for their help in the collection of XGS-02 samples.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1222526/full#supplementary-material
References
Akerman N. H., Butterfield D. A., Huber J. A. (2013). Phylogenetic diversity and functional gene patterns of sulfur-oxidizing subseafloor Epsilonproteobacteria in diffuse hydrothermal vent fluids. Front. Microbiol. 4. doi: 10.3389/fmicb.2013.00185
Alfaro-Espinoza G., Ullrich M. S. (2015). Bacterial N-2-fixation in mangrove ecosystems: insights from a diazotroph-mangrove interaction. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00445
Auch A. F., von Jan M., Klenk H.-P., Goeker M. (2010). Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genomic Sci. 2 (1), 117–134. doi: 10.4056/sigs.531120
Berney M., Greening C., Hards K., Collins D., Cook G. M. (2014). Three different NiFe hydrogenases confer metabolic flexibility in the obligate aerobe Mycobacterium smegmatis. Environ. Microbiol. 16 (1), 318–330. doi: 10.1111/1462-2920.12320
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Borisov V. B., Siletsky S. A. (2019). Features of organization and mechanism of catalysis of two families of terminal oxidases: Heme-copper and bd-type. Biochem. (Mosc) 84 (11), 1390–1402. doi: 10.1134/s0006297919110130
Bu C., Wang Y., Ge C., Ahmad H. A., Gao B., Ni S.-Q. (2017). Dissimilatory nitrate reduction to ammonium in the yellow river estuary: Rates, abundance, and community diversity. Sci. Rep. 7 (1), 6830. doi: 10.1038/s41598-017-06404-8
Cabiscol E., Tamarit J., Ros J. (2000). Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 3 (1), 3–8.
Chaudhari N. M., Gupta V. K., Dutta C. (2016). ). BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6, 24373. doi: 10.1038/srep24373
Chiu L., Wang M.-C., Tseng K.-Y., Wei C.-L., Lin H.-T., Yang S.-H., et al. (2022). Shallow-water hydrothermal vent system as an extreme proxy for discovery of microbiome significance in a crustacean holobiont. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.976255
Crepeau V., Bonavita M. A. C., Lesongeur F., Randrianalivelo H., Sarradin P. M., Sarrazin J., et al. (2011). Diversity and function in microbial mats from the Lucky Strike hydrothermal vent field. FEMS Microbiol. Ecol. 76 (3), 524–540. doi: 10.1111/j.1574-6941.2011.01070.x
Dahle H., Roalkvam I., Thorseth I. H., Pedersen R. B., Steen I. H. (2013). The versatile in situ gene expression of an Epsilonproteobacteria-dominated biofilm from a hydrothermal chimney. Environ. Microbiol. Rep. 5 (2), 282–290. doi: 10.1111/1758-2229.12016
Ding J., Zhang Y., Wang H., Jian H. H., Leng H., Xiao X. (2017). Microbial community structure of deep-sea hydrothermal vents on the ultraslow spreading southwest Indian ridge. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01012
Felsenstein J. (1981). Evolutionary trees from dna-sequences - A maximum-likelihood approach. J. Mol. Evol. 17 (6), 368–376. doi: 10.1007/bf01734359
Giovannelli D., Chung M., Staley J., Starovoytov V., Le Bris N., Vetriani C. (2016). Sulfurovum riftiae sp nov., a mesophilic, thiosulfate-oxidizing, nitrate-reducing chemolithoautotrophic epsilonproteobacterium isolated from the tube of the deep-sea hydrothermal vent polychaete Riftia pachyptila. Int. J. Syst. Evol. Microbiol. 66, 2697–2701. doi: 10.1099/ijsem.0.001106
Grote J., Schott T., Bruckner C. G., Gloockner F. O., Jost G., Teeling H., et al. (2012). Genome and physiology of a model Epsilonproteobacterium responsible for sulfide detoxification in marine oxygen depletion zones. PNAS 109 (2), 506–510. doi: 10.1073/pnas.1111262109
Guo Y., Ihara H., Aoyagi T., Hori T., Katayama Y. (2022). ). Draft genome sequences of sulfurovum spp. TSL1 and TSL6, two sulfur-oxidizing bacteria isolated from marine sediment. Microbiol. Resour. Announce. 11 (1), e0092221. doi: 10.1128/mra.00922-21
Hassani B. K., Steunou A. S., Liotenberg S., Reiss-Husson F., Astier C., Ouchane S. (2010). Adaptation to oxygen: role of terminal oxidases in photosynthesis initiation in the purple photosynthetic bacterium, Rubrivivax gelatinosus. J. Biol. Chem. 285 (26), 19891–19899. doi: 10.1074/jbc.M109.086066
Huang L. B., Bae H. S., Young C., Pain A. J., Martin J. B., Ogram A. (2021). Campylobacterota dominate the microbial communities in a tropical karst subterranean estuary, with implications for cycling and export of nitrogen to coastal waters. Environ. Microbiol. 23 (11), 6749–6763. doi: 10.1111/1462-2920.15746
Huang X. J., Tie W. Z., Xie D. T., Li Z. L. (2021). Low C/N Ratios Promote Dissimilatory Nitrite Reduction to Ammonium in Pseudomonas putida Y-9 under Aerobic Conditions. Microorganisms 9 (7), 1524. doi: 10.3390/microorganisms9071524
Inagaki F., Takai K., Nealson K. H., Horikoshi K. (2004). Sulfurovum lithotrophicum gen. nov., sp nov., a novel sulfur-oxidizing chemolithoautotroph within the epsilon-Proteobacteria isolated from Okinawa Trough hydrothermal sediments. Int. J. Syst. Evol. Microbiol. 54, 1477–1482. doi: 10.1099/ijs.0.03042-0
Jia M. S., Winkler M. K. H., Volcke E. I. P. (2020). Elucidating the competition between heterotrophic denitrification and DNRA using the resource-ratio theory. Environ. Sci. Technol. 54 (21), 13953–13962. doi: 10.1021/acs.est.0c01776
Jiang L. J., Lyu J., Shao Z. Z. (2017). Sulfur metabolism of hydrogenovibrio thermophilus strain S5 and its adaptations to deep-sea hydrothermal vent environment. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02513
Jiang L., Zheng Y., Peng X., Zhou H., Zhang C., Xiao X., et al. (2009). Vertical distribution and diversity of sulfate-reducing prokaryotes in the Pearl River estuarine sediments, Southern China. FEMS Microbiol. Ecol. 70 (2), 93–106. doi: 10.1111/j.1574-6941.2009.00758.x
Johnson L. A., Hug L. A. (2019). Distribution of reactive oxygen species defense mechanisms across domain bacteria. Free Radic. Biol. Med. 140, 93–102. doi: 10.1016/j.freeradbiomed.2019.03.032
Kaminski P. A., Kitts C. L., Zimmerman Z., Ludwig R. A. (1996). Azorhizobium caulinodans uses both cytochrome bd (quinol) and cytochrome cbb3 (cytochrome c) terminal oxidases for symbiotic N2 fixation. J. Bacteriol 178 (20), 5989–5994. doi: 10.1128/jb.178.20.5989-5994.1996
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi: 10.1093/molbev/msy096
Kuypers M. M. M., Marchant H. K., Kartal B. (2018). The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 16 (5), 263–276. doi: 10.1038/nrmicro.2018.9
Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23 (21), 2947–2948. doi: 10.1093/bioinformatics/btm404
Lee W. K., Juniper S. K., Perez M., Ju S. J., Kim S. J. (2021). Diversity and characterization of bacterial communities of five co-occurring species at a hydrothermal vent on the Tonga Arc. Ecol. Evol. 11 (9), 4481–4493. doi: 10.1002/ece3.7343
Li S., Jiang Z., Ji G. (2022). Effect of sulfur sources on the competition between denitrification and DNRA. Environ. Pollut. 305, 119322. doi: 10.1016/j.envpol.2022.119322
Li W., Zhen Y. M., Li N., Wang H. Q., Lin M. H., Sui X. T., et al. (2022). Sulfur transformation and bacterial community dynamics in both desulfurization-denitrification biofilm and suspended activated sludge. Bioresour. Technol. 343, 126108. doi: 10.1016/j.biortech.2021.126108
Lin X., Hetharua B., Lin L., Xu H., Zheng T., He Z., et al. (2019). Mangrove sediment microbiome: Adaptive microbial assemblages and their routed biogeochemical processes in yunxiao mangrove national nature reserve, China. Microb. Ecol. 78 (1), 57–69. doi: 10.1007/s00248-018-1261-6
Liu S. Y., Dai J. C., Wei H. H., Li S. Y., Wang P., Zhu T. B., et al. (2021). Dissimilatory nitrate reduction to ammonium (DNRA) and denitrification pathways are leveraged by cyclic AMP receptor protein (CRP) paralogues based on electron donor/acceptor limitation in shewanella loihica PV-4. Appl. Environ. Microbiol. 87 (2), e01964–20. doi: 10.1128/aem.01964-20
Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., et al. (2015). Erratum: SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 4, 30. doi: 10.1186/s13742-015-0069-2
Luo Z. W., Zhong Q. P., Han X. G., Hu R. W., Liu X. Y., Xu W. J., et al. (2021). Depth-dependent variability of biological nitrogen fixation and diazotrophic communities in mangrove sediments. Microbiome 9 (1), 212. doi: 10.1186/s40168-021-01164-0
Marcia M., Ermler U., Peng G. H., Michel H. (2010). A new structure-based classification of sulfide:quinone oxidoreductases. Proteins-Structure Funct. Bioinf. 78 (5), 1073–1083. doi: 10.1002/prot.22665
Marziah Z., Mahdzir A., Musa M. N., Jaafar A. B., Azhim A., Hara H. (2016). Abundance of sulfur-degrading bacteria in a benthic bacterial community of shallow sea sediment in the off-Terengganu coast of the South China Sea. Microbiologyopen 5 (6), 967–978. doi: 10.1002/mbo3.380
Meier D. V., Pjevac P., Bach W., Hourdez S., Girguis P. R., Vidoudez C., et al. (2017). Niche partitioning of diverse sulfur-oxidizing bacteria at hydrothermal vents. ISME J. 11 (7), 1545–1558. doi: 10.1038/ismej.2017.37
Meier-Kolthoff J. P., Klenk H.-P., Goeker M. (2014). Taxonomic use of DNA G plus C content and DNA-DNA hybridization in the genomic age. Int. J. Syst. Evol. Microbiol. 64, 352–356. doi: 10.1099/ijs.0.056994-0
Mino S., Kudo H., Arai T., Sawabe T., Takai K., Nakagawa S. (2014). Sulfurovum aggregans sp nov., a hydrogenoxidizing, thiosulfate-reducing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent chimney, and an emended description of the genus Sulfurovum. Int. J. Syst. Evol. Microbiol. 64, 3195–3201. doi: 10.1099/ijs.0.065094-0
Missall T. A., Pusateri M. E., Lodge J. K. (2004). Thiol peroxidase is critical for virulence and resistance to nitric oxide and peroxide in the fungal pathogen, Cryptococcus neoformans. Mol. Microbiol. 51 (5), 1447–1458. doi: 10.1111/j.1365-2958.2004.03921.x
Mori K., Yamaguchi K., Hanada S. (2018). Sulfurovum denitrificans sp nov., an obligately chemolithoautotrophic sulfur-oxidizing epsilonproteobacterium isolated from a hydrothermal field. Int. J. Syst. Evol. Microbiol. 68 (7), 2183–2187. doi: 10.1099/ijsem.0.002803
Motoki K., Watsuji T. O., Takaki Y., Takai K., Iwasaki W. (2020). Metatranscriptomics by in situ RNA stabilization directly and comprehensively revealed episymbiotic microbial communities of deep-sea squat lobsters. Msystems 5 (5), e00551–20. doi: 10.1128/mSystems.00551-20
Moulana A., Anderson R. E., Fortunato C. S., Huber J. A. (2020). Selection is a significant driver of gene gain and loss in the pangenome of the bacterial genus sulfurovum in geographically distinct deep-sea hydrothermal vents. Msystems 5 (2), e00673–19. doi: 10.1128/mSystems.00673-19
Na S.-I., Kim Y. O., Yoon S.-H., Ha S. M., Baek I., Chun J. (2018). UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 56 (4), 280–285. doi: 10.1007/s12275-018-8014-6
Nakagawa T., Takai K., Suzuki Y., Hirayama H., Konno U., Tsunogai U., et al. (2006). Geomicrobiological exploration and characterization of a novel deep-sea hydrothermal system at the TOTO caldera in the Mariana Volcanic Arc. Environ. Microbiol. 8 (1), 37–49. doi: 10.1111/j.1462-2920.2005.00884.x
Overbeek R., Olson R., Pusch G. D., Olsen G. J., Davis J. J., Disz T., et al. (2014). The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42 (D1), D206–D214. doi: 10.1093/nar/gkt1226
Pandey C. B., Kumar U., Kaviraj M., Minick K. J., Mishra A. K., Singh J. S. (2020). DNRA: A short-circuit in biological N-cycling to conserve nitrogen in terrestrial. Sci. Total Environ. 738, 139710. doi: 10.1016/j.scitotenv.2020.139710
Park S.-J., Ghai R., Martin-Cuadrado A.-B., Rodriguez-Valera F., Jung M.-Y., Kim J.-G., et al. (2012). Draft genome sequence of the sulfur-oxidizing bacterium "Candidatus sulfurovum sediminum" AR, which belongs to the epsilonproteobacteria. J. Bacteriol. 194 (15), 4128–4129. doi: 10.1128/jb.00741-12
Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25 (7), 1043–1055. doi: 10.1101/gr.186072.114
Patwardhan S., Foustoukos D. I., Giovannelli D., Yucel M., Vetriani C. (2018). Ecological succession of sulfur-oxidizing epsilon- and gammaproteobacteria during colonization of a shallow-water gas vent. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02970
Patwardhan S., Smedile F., Giovannelli D., Vetriani C. (2021). Metaproteogenomic profiling of chemosynthetic microbial biofilms reveals metabolic flexibility during colonization of a shallow-water gas vent. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.638300
Pritchard L., Glover R. H., Humphris S., Elphinstone J. G., Toth I. K. (2016). Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal. Methods 8 (1), 12–24. doi: 10.1039/c5ay02550h
Sheik C. S., Anantharaman K., Breier J. A., Sylvan J. B., Edwards K. J., Dick G. J. (2015). Spatially resolved sampling reveals dynamic microbial communities in rising hydrothermal plumes across a back-arc basin. ISME J. 9 (6), 1434–1445. doi: 10.1038/ismej.2014.228
Stackebrandt E., Goebel B. M. (1994). A place for dna-dna reassociation and 16s ribosomal-rna sequence-analysis in the present species definition in bacteriology. Int. J. Systematic Bacteriology 44 (4), 846–849. doi: 10.1099/00207713-44-4-846
Sun Y. Y., Song Z. L., Zhang H. K., Liu P. Y., Hu X. K. (2020). Seagrass vegetation affect the vertical organization of microbial communities in sediment. Mar. Environ. Res. 162, 105174. doi: 10.1016/j.marenvres.2020.105174
Sun Q. L., Zhang J., Wang M. X., Cao L., Du Z. F., Sun Y. Y., et al. (2020). High-throughput sequencing reveals a potentially novel sulfurovum species dominating the microbial communities of the seawater-sediment interface of a deep-sea cold seep in South China sea. Microorganisms 8 (5), 687. doi: 10.3390/microorganisms8050687
Sylvan J. B., Pyenson B. C., Rouxel O., German C. R., Edwards K. J. (2012). Time-series analysis of two hydrothermal plumes at 950′N East Pacific Rise reveals distinct, heterogeneous bacterial populations. Geobiology 10 (2), 178–192. doi: 10.1111/j.1472-4669.2011.00315.x
Takai K., Gamo T., Tsunogai U., Nakayama N., Hirayama H., Nealson K. H., et al. (2004). Geochemical and microbiological evidence for a hydrogen-based, hyperthermophilic subsurface lithoautotrophic microbial ecosystem (HyperSLiME) beneath an active deep-sea hydrothermal field. Extremophiles 8 (4), 269–282. doi: 10.1007/s00792-004-0386-3
Takai K., Nakagawa S., Nunoura T. (2015). “Comparative Investigation of Microbial Communities Associated with Hydrothermal Activities in the Okinawa Trough,” in Subseafloor Biosphere Linked to Hydrothermal Systems: TAIGA Concept. Eds. Ishibashi J. I., Okino K., Sunamura M. (Tokyo: Springer Japan), 421–435.
Tamura K., Nei M., Kumar S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. PNAS 101 (30), 11030–11035. doi: 10.1073/pnas.0404206101
Tang W. Y., Cerdan-Garcia E., Berthelot H., Polyviou D., Wang S. V., Baylay A., et al. (2020). New insights into the distributions of nitrogen fixation and diazotrophs revealed by high-resolution sensing and sampling methods. ISME J. 14 (10), 2514–2526. doi: 10.1038/s41396-020-0703-6
Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Nawrocki E. P., Zaslavsky L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44 (14), 6614–6624. doi: 10.1093/nar/gkw569
Teske A., Hinrichs K.-U., Edgcomb V., de Vera Gomez A., Kysela D., Sylva S. P., et al. (2002). Microbial diversity of hydrothermal sediments in the guaymas basin: Evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68 (4), 1994–2007. doi: 10.1128/aem.68.4.1994-2007.2002
Vetriani C., Voordeckers J. W., Crespo-Medina M., O'Brien C. E., Giovannelli D., Lutz R. A. (2014). Deep-sea hydrothermal vent Epsilonproteobacteria encode a conserved and widespread nitrate reduction pathway (Nap). Isme J. 8 (7), 1510–1521. doi: 10.1038/ismej.2013.246
Waite D. W., Vanwonterghem I., Rinke C., Parks D. H., Zhang Y., Takai K., et al. (2018). Comparative genomic analysis of the class epsilonproteobacteria and proposed reclassification to epsilonbacteraeota (phyl. nov.). Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00772
Wang Y., Bi H. Y., Chen H. G., Zheng P. F., Zhou Y. L., Li J. T. (2022). Metagenomics reveals dominant unusual sulfur oxidizers inhabiting active hydrothermal chimneys from the Southwest Indian ridge. Front. Microbiol. 13, 861795. doi: 10.3389/fmicb.2022.861795
Wang L., Cheung M. K., Liu R., Wong C. K., Kwan H. S., Hwang J.-S. (2017). Diversity of total bacterial communities and chemoautotrophic populations in sulfur-rich sediments of shallow-water hydrothermal vents off kueishan island, Taiwan. Microb. Ecol. 73 (3), 571–582. doi: 10.1007/s00248-016-0898-2
Wang S., Jiang L., Hu Q., Cui L., Zhu B., Fu X., et al. (2021). Characterization of Sulfurimonas hydrogeniphila sp. nov., a Novel Bacterium Predominant in Deep-Sea Hydrothermal Vents and Comparative Genomic Analyses of the Genus Sulfurimonas. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.626705
Wang S. S., Jiang L. J., Xie S. B., Alain K., Wang Z. D., Wang J., et al. (2023). Disproportionation of inorganic sulfur compounds by mesophilic chemolithoautotrophic campylobacterota. Msystems 8 (1), e0095422. doi: 10.1128/msystems.00954-22
Wang K. Q., Qaisar M., Chen B. L., Cai J. (2022). Response difference of simultaneous sulfide and nitrite removal process to different cooling modes. Bioresour. Technol. 346. doi: 10.1016/j.biortech.2021.126601
Wang F., Zhou H., Meng J., Peng X., Jiang L., Sun P., et al. (2009). GeoChip-based analysis of metabolic diversity of microbial communities at the Juan de Fuca Ridge hydrothermal vent. PNAS 106 (12), 4840–4845. doi: 10.1073/pnas.0810418106
Wright K. E., Williamson C., Grasby S. E., Spear J. R., Templeton A. S. (2013). Metagenomic evidence for sulfur lithotrophy by Epsilonproteobacteria as the major energy source for primary productivity in a sub-aerial arctic glacial deposit Borup Fiord Pass. Front. Microbiol. 4. doi: 10.3389/fmicb.2013.00063
Xie J., Chen Y., Cai G., Cai R., Hu Z., Wang H. (2023). Tree Visualization By One Table (tvBOT): a web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 51 (W1), W587–W592. doi: 10.1093/nar/gkad359
Xie S., Wang S., Li D., Shao Z., Lai Q., Wang Y., et al. (2021). Sulfurovum indicum sp. nov., a novel hydrogen- and sulfur-oxidizing chemolithoautotroph isolated from a deep-sea hydrothermal plume in the Northwestern Indian Ocean. Int. J. Syst. Evol. Microbiol. 71 (3). doi: 10.1099/ijsem.0.004748
Xu W., Li M., Ding J. F., Gu J. D., Luo Z. H. (2014). Bacteria dominate the ammonia-oxidizing community in a hydrothermal vent site at the Mid-Atlantic Ridge of the South Atlantic Ocean. Appl. Microbiol. Biotechnol. 98 (18), 7993–8004. doi: 10.1007/s00253-014-5833-1
Zhang L. K., Kang M. Y., Xu J. J., Xu J., Shuai Y. J., Zhou X. J., et al (2016). Bacterial and archaeal communities in the deep-sea sediments of inactive hydrothermal vents in the Southwest India Ridge. Sci. Rep. 6, 25982. doi: 10.1038/srep25982
Zhang J., Madden T. L. (1997). PowerBLAST: a new network BLAST application for interactive or automated sequence analysis and annotation. Genome Res. 7 (6), 649–656. doi: 10.1101/gr.7.6.649
Keywords: Sulfurovum xiamenensis, Sulfurovum zhangzhouensis, nitrogen fixation, coastal environment, hydrothermal vent, environmental adaptation
Citation: Wang J, Zheng Q, Wang S, Zeng J, Yuan Q, Zhong Y, Jiang L and Shao Z (2023) Characterization of two novel chemolithoautotrophic bacteria of Sulfurovum from marine coastal environments and further comparative genomic analyses revealed species differentiation among deep-sea hydrothermal vent and non-vent origins. Front. Mar. Sci. 10:1222526. doi: 10.3389/fmars.2023.1222526
Received: 14 May 2023; Accepted: 24 July 2023;
Published: 11 August 2023.
Edited by:
Xue-Wei Xu, Ministry of Natural Resources, ChinaReviewed by:
Min Yu, Ocean University of China, ChinaCostantino Vetriani, The State University of New Jersey, United States
Michael Hügler, Technologiezentrum Wasser, Germany
Copyright © 2023 Wang, Zheng, Wang, Zeng, Yuan, Zhong, Jiang and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lijing Jiang, amlhbmdsaWppbmdAdGlvLm9yZy5jbg==; Zongze Shao, c2hhb3p6QDE2My5jb20=
 Jun Wang
Jun Wang Qiang Zheng
Qiang Zheng Shasha Wang
Shasha Wang Jialing Zeng
Jialing Zeng Qing Yuan1,3,4
Qing Yuan1,3,4 Lijing Jiang
Lijing Jiang Zongze Shao
Zongze Shao