- 1Guangdong Provincial Key Laboratory of Marine Biotechnology, Shantou University, Shantou, China
- 2STU-UMT Joint Shellfish Research Laboratory, Shantou University, Shantou, China
- 3Laboratory of Arthropod Behavior and Ecology, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Mengla, Yunnan, China
- 4Higher Institute Centre of Excellence (HICoE), Institute of Tropical Aquaculture and Fisheries, Universiti Malaysia Terengganu, Kuala Nerus, Terengganu, Malaysia
Introduction: Crustacean predators exhibit diverse trophic interactions across various habitats in aquatic ecosystems. To assess their ecological roles, it is essential to understand their complete dietary spectrum. Recently, DNA-based techniques such as DNA metabarcoding and high-throughput sequencing have provided insights into trophic networks and their response to environmental drivers.
Methods: In this study, we used these methods to investigate the impact of salinity gradients on the trophic networks of mud crabs (Scylla paramamosain) in marine ecosystems.
Results: Our results revealed that mud crabs prey on a wide range of taxa, including crabs (other than the host species), fish, shrimps, and other prey factions. Salinity gradients had a significant influence on the specific prey items, such as high predation on Metapenaeus, and Trigonopterus species in high salinity sites. Notably, crabs exhibited a greater infestation of Portunion parasites in environments with high salinity levels, in contrast to environments with lower salinity levels. Ordination analysis showed that mud crabs adapt their diet according to environmental constraints.
Discussion: Furthermore, the results indicate that mud crabs showed opportunistic predation behavior as they were observed preying on other Portunidae species sharing the same trophic guild. Overall, these findings demonstrate the dietary plasticity and associated parasite infestation of an important crustacean predator, and provide evidence that environmental changes may affect incidences of the associated parasites as well as the specific predator-prey interactions indirectly through fluctuations in marine ecosystems.
1 Introduction
Comprehensive knowledge regarding the complex interactions between predators and prey and their position in the trophic networks is a crucial area of research in ecology (Symondson, 2002; Michalko and Pekár, 2016). The insights derived from these trophic network studies provide a deeper understanding of the functioning of ecosystems, community interactions, and their response to environmental changes. Previously, predator-prey interaction networks were assumed to have a static structure, but recent ecological studies have revealed the dynamic nature of trophic interactions across environmental gradients (Pellissier et al., 2018). The dynamic nature of these interactions is important in determining the abundance, distribution, and behavior of both predator and prey species in a trophic network and help to comprehend the distribution of species in predator-prey networks (Saqib et al., 2022). The strength and characteristics of the relationships between species in predator-prey networks are essential and play a fundamental role in delivering ecosystem services. Such as for omnivores and generalist predators, the preferences and availability of prey species are the critical factors that determine their predator-prey range (Saqib et al., 2021). The availability and accessibility of specific prey species can also play a significant role in determining the prey consumption of generalist predators at any given time (Friman et al., 2008; Michalko and Pekár, 2016). Measuring and understanding the contribution of each consumer taxon or trophic guild is critical (Saqib et al., 2021), and traditional methods of prey fragment identification in predator guts are logistically challenging and time-consuming (Williams et al., 2012). This significance becomes particularly pronounced for opportunistic predators in dynamic marine ecosystems, especially where both prey and predators exhibit a wide range of sizes, life cycles, and trophic roles.
The dynamics and strength of relationships between predators and prey in a trophic network is likely affected by environmental variability (Poisot et al., 2012). This impact is particularly notable in aquatic ecosystems, where various environmental drivers can impede the ability of both predators and prey to detect each other, and also determine the nature of the relationships (Abrahams and Kattenfeld, 1997). For example, intraguild predation in a trophic network occurs when species compete for the same resources, and the prey availability and diversity is limited (Arim and Marquet, 2004). Both empirical evidence and theoretical models have shown that intraguild predation can significantly influence population abundances and shape animal communities in an ecosystem (Holt and Polis, 1997). The intensity and outcome of intraguild predator interactions may also change over time due to variations in environmental conditions. Therefore, studying intraguild predation by crustaceans in diverse marine ecosystems with high biodiversity and fluctuating environmental factors is essential for exploring the broad range of trophic interactions.
Trophic interactions, which are interactions between species in an ecosystem based on their feeding relationships, often require the use of precise techniques and tools to analyze the entire dietary spectrum of generalist predators. DNA-based approaches have become widely used in dietary analysis research in recent years and have significantly improved the assessment of trophic interactions (King et al., 2008; Pompanon et al., 2012). Conventional approaches based on prey-specific primers are no longer useful for dietary analysis of generalist predators or in environments with poorly described prey ranges (Harper et al., 2005; De Barba et al., 2014). DNA metabarcoding, a method that combines amplification and sequencing of small DNA sections from samples, has emerged as a powerful tool for detecting a wide range of trophic interactions in complex food webs when combined with high-throughput sequencing (Brown et al., 2012; Littlefair and Clare, 2016). This combined approach improves the detection of prey species in the diet of predators over conventional morphological identification of consumed taxa, or simple PCR-based detection of prey species which may fail to perceive the full range of prey (Valentini et al., 2009). By removing many of the drawbacks of conventional methodology, and by improving the detection of prey species in diet of predators, DNA metabarcoding in combination with high-throughput sequencing technologies has altered our understanding of the dynamics in predator-prey relationships and can be applied to expand our understanding of trophic interactions in highly complex predator-prey networks, such as inter- and intra-guild interaction.
Crabs are itinerant indicator and keystone species with a generalist predation behavior, possessing a broad diet preferences that can cause wide-ranging trophic cascades and exerting substantial top-down control over benthic organisms on a large scale (Kotta et al., 2018; Cuthbert and Briski, 2022). One such commercially important crustacean is the mud crab (Scylla paramamosain), which is widely distributed, rapidly developed, and cultured in Indo-Pacific countries and along the southeast coastline of China (Deng et al., 2019). This euryhaline species can tolerate a wide range of salinities and is economically significant as well as a popular seafood in Southeast Asian countries. It is found in diverse environments such as mangrove systems, inner bays, shallow seas, and estuaries, where it encounters frequent changes in salinity (Le Vay et al., 2007; Li et al., 2018). Moreover, mud crabs species are known to migrate in response to environmental cues influenced by factors such as reproductive needs, food availability, environmental conditions and seasonal changes. These migrations can involve movements between different habitats, such as from estuaries and mangroves to offshore areas or vice versa, where individuals can move 500–1,000 m over a 12h period (Le Vay et al., 2007). Given the migratory behavior of mud crabs, the direct observation of predation events in such varied marine environments with differing salinity conditions is challenging. Therefore, a precise analytical strategy, such as DNA metabarcoding for whole dietary analysis, is necessary to better understand the trophic guilds of mud crabs and emphasize their trophic interactions. Hence, this study aims to explore trophic guilds (inter- and intra-guild predation) of S. paramamosain in marine ecosystems across varying salinity gradients.
2 Methods
2.1 Study system and samples collection
Field sites were located on the eastern coast of Guangdong, China, and samples were collected May 2022. To reduce the impact of spatial autocorrelation, the 10 fields were marked out using GPS (GPSMAP 60CSx-Garmin) and were at least 1 kilometer apart. Moreover, spatial autocorrelation was tested using the gearymoran function from the ade4 package, and no spatial autocorrelation was detected between sites. Sites were classified into two groups based on salinity; five sites were in a high salinity region (21-23‰) and five sites were in a low salinity region (1-2‰) (see Figure S1 for more sampling site details). Water samples were taken from each sampling point three times over the course of a day for quantification of salinity utilizing a handheld refractometer (ATAGO MASTER-series).
A total of 150 S. paramamosain wild crabs were collected, all of which exhibited no sign of disease. A total of 75 crabs (15 crabs/site) were selected to represent each group (high and low salinity). The crabs were quickly cryoanesthesized upon arrival at the laboratory, followed by proper surface disinfection with 75% ethanol for two minutes and cleaning with sterile water. The crabs were dissected using sterile lancets, and the gut and stomach were collected in a new sterile 5 mL microcentrifuge tubes. All the dissecting tools were sterilized after performing each dissection with an alcohol flame. The dissected samples were stored at −80°C until further analysis. The animal experiments were conducted in full compliance with the College of Science and approved by the ethics committee at Shantou University.
2.2 DNA extraction, PCR and library preparation
Of the 15 S. paramamosain collected at each site, all 15 crabs were dissected at every site and their stomach contents pooled across groups of three individuals for DNA extraction (n = 5 genetic samples/site) (Sunnucks and Hales, 1996). To detect barcoded prey DNA a pair of previously developed primers (F-GCATTYCCACGAATAAATAAYATAAG and R-TAAACTTCAGGGTGACCAAARAAYCA) designed to amplify a fragment in subunit I (COI) of cytochrome c oxidase. The PCR amplification was performed with a final volume of 10 µL, containing 1 µL template DNA (>50 ng µL−1), 5 µL KOD FX Neo buffer, 2 µL (2 mM) dNTPs, 0.3 µL (10 µM) each primer, 0.2 µL KOD FX Neo DNA Polymerase, and 1.2 µL ddH2O. The PCR was performed by initial denaturation for 300 seconds at 95°C, 25 cycles of denaturation for 30 seconds at 95°C, annealing for 30 seconds at 50°C, and extension for 40 seconds at 72°C, with a final extension for 300 seconds at 72°C. To purify the successfully amplified PCR products, VAHTSTM Magnetic DNA Beads were used to remove primers, dimers, salts, and deoxynucleoside triphosphates (dNTPs). Equally molar DNA libraries were prepared for paired-end sequencing using the Illumina Novaseq 6000 platform. For each batch of DNA extraction and PCR amplification, a negative control was used to detect possible contaminants, all of which were negative.
2.3 Bioinformatics
The raw sequence data was filtered based on the quality single nucleotide using Trimmomatic (version 0.33) (Bolger et al., 2014). Primer sequence identification and deletion were performed using Cutadapt (version 1.9.1) (Martin, 2011). Demultiplexing and combining paired-end reads were done using USEARCH (version 10) (Edgar, 2010), and chimeras were removed using UCHIME (version 8.1) (Edgar et al., 2011), followed by identification of high-quality readings. For analysis of prey read in each sample, clean tags with 97% sequence similarity were pooled using the USEARCH function of QIIME. The NCBI database was used for classification and annotation of the assigned operational taxonomic units (OTUs).
2.4 Data analyses
The study aimed to compare the prey community composition of crabs and examine the effect of salinity on prey assemblage. All of the statistical analyses were conducted using the R software (version 3.6.3). The relative abundance of consumed prey between high and low salinity was compared. The β-dissimilarities in prey assemblages were assessed using NMDS (non-metric multidimensional scaling) with the ‘vegan’ package based on Bray-Curtis differences and species sample count data. The NMDS scores were used to plot the values of all variables against each axis to determine their correlation with explanatory variables. The ‘DESeq2’ package was used to identify differentially abundant prey taxa between high and low salinity groups at the genus, family, and order level. The results were plotted based on the adjusted p-values (using the Benjamini-Hochberg method) and log-fold change values obtained from differential abundance analysis.
3 Results
3.1 Community composition of prey
A total of 7,695,292 high-quality sequencing reads were obtained, representing 59 unique operational taxonomic units (OTUs). Of 59 detected OTUs, were found to contain DNA of four different prey phyla representing 12 classes, 25 orders, 34 families, 44 genera and 53 species (excluding the host species and unclassified taxa) (see Supplementary Table 1 for more details). Among these reads, 58.488% belonged to the predators (host) DNA, 39.096% belonged to prey contents, and 2.416% belonged to associated parasites. Arthropoda and Chordata were the most abundant phyla, representing 60.857% and 38.709% of the reads, respectively. At the class level, Malacostraca and Actinopteri were the most dominant prey classes, accounting for 56.075% and 34.340% of the reads, respectively. The most common prey orders were Decapoda (53.631%) and Cypriniformes (34.311%), and the most frequent prey families were Portunidae (52.017%) and Xenocyprididae (33.979%). Scylla and Hypophthalmichthys were the most abundant prey genera, representing 47.769% and 33.438% of the reads, respectively, followed by Charybdis (4.248%) and Mus (4.138%) (Figure 1). While a crab endoparasite genera, Portunion, was found to comprise 2.416% of the reads (Figure 1).
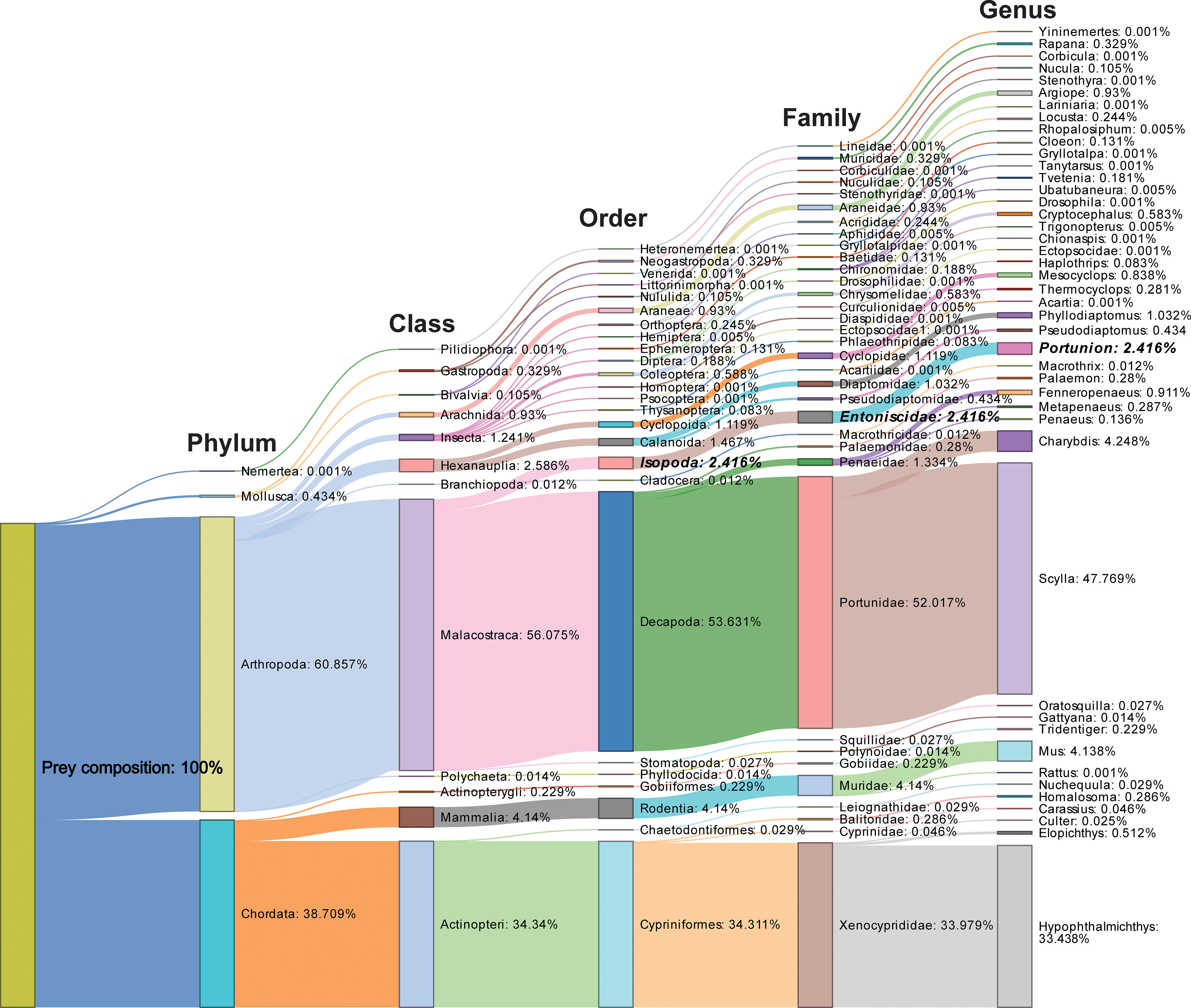
Figure 1 Sankey diagram illustrating the percentage of dominant prey taxa (including phyla, classes, orders, families, and genera) detected in the diet of mud crabs captured from both salinity gradients. The taxa names in bold and italic represent crab endoparasites.
3.2 Comparison of prey composition in higher and lower salinities
The trophic patterns of mud crabs indicate that the most important prey items quantitatively were other crabs, fish, rodents, copepods, and shrimps (Figure 2A). Our results indicated that mud crabs exhibited the highest levels of predation on Scylla and Hypophthalmichthys species, which belong to the Decapoda and Cypriniformes orders, respectively, regardless of variations in the salinity. Furthermore, mud crabs showed relatively higher infestation by the Entoniscidae family of Isopods (characterized as an endoparasite) in high salinity environments, in contrast to low salinity environments (Figures 2B–E). Non-metric multidimensional scaling (NMDS) revealed differences in beta dissimilarities among the prey species communities in different salinities (Figure 3). In ordination space, predation by crabs in different salinities formed different clusters of prey items in terms of their assemblages. For prey families, Portunidae, Chrysomelidae and Araneidae were distinctly clustered away from the other top prey families (Muridae, Muricidae, Penaeidae, Diaptomidae, Xenocyprididae and Pseudodiaptomidae) across different salinities (Figure 3).
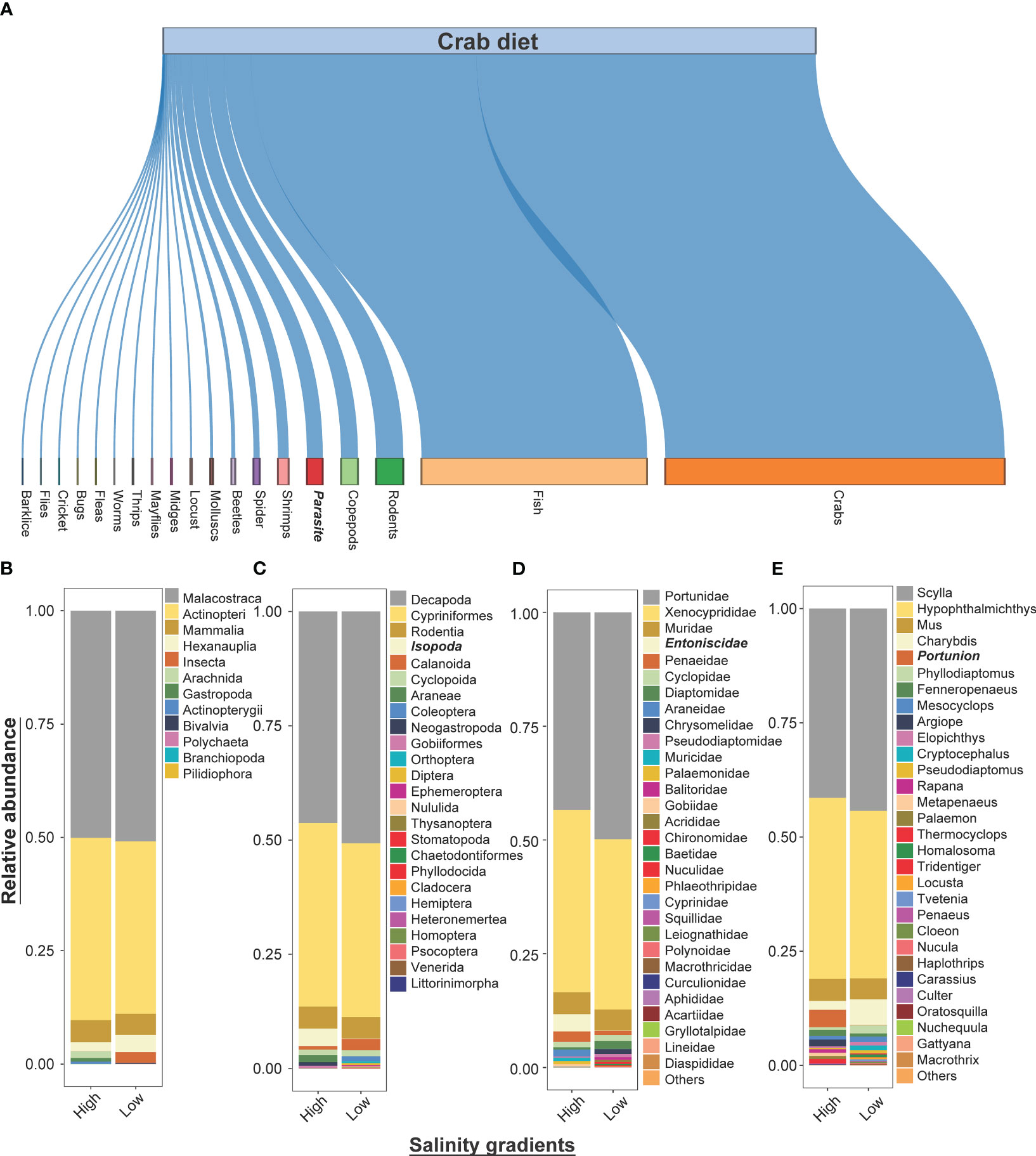
Figure 2 The figure shows the (A) dietary compositions of mud crabs captured from both salinity gradients. It also shows the relative abundance (%) of prey (B) classes, (C) orders, top 30 (D) families and (E) genera detected in diet of mud crabs. The mud crabs were captured from marine environment with varying salinities, with some having high-salinity and other low-salinity levels. The taxa names in bold and italic represent crab endoparasites.
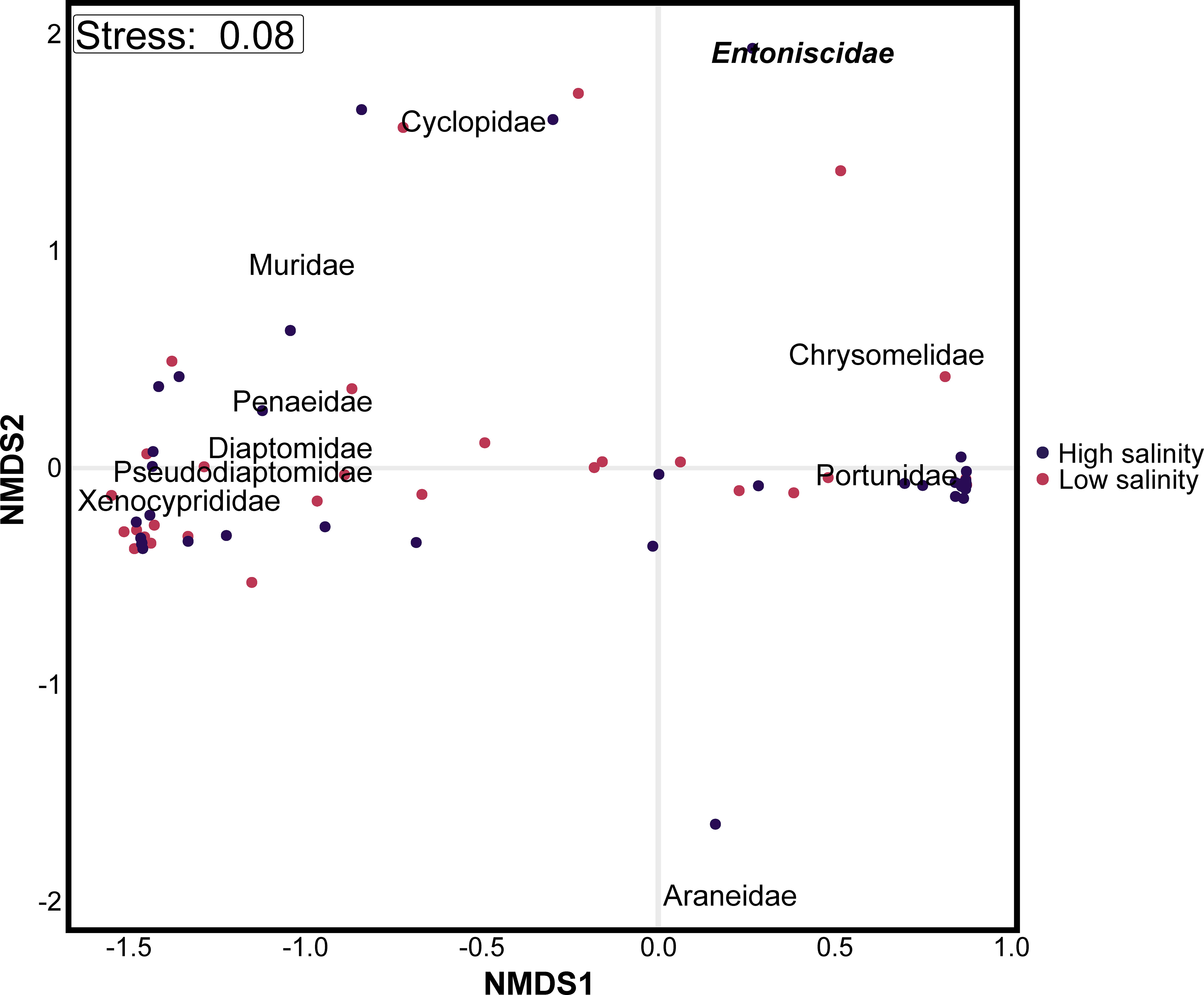
Figure 3 Non-metric dimensional scaling (NMDS) ordination plot depicting the Bray-cutis dissimilarities of prey taxa detected in the diet of mud crabs, captured from two distinct salinity levels (high and low salinity). The taxa names in bold and italic represent crab endoparasites.
3.3 Differential abundances of prey taxa in higher and lower salinities
When analyzing the differential abundances of prey taxa in the diet of crabs at various taxonomic levels (such as species, genus, and family), the trophic preferences (i.e., over- and under-representations) were apparent among mud crabs captured from different salinities (Figure 4). The mud carbs collected from high salinity locations had significantly higher abundance of Penaeidae (prey), and Entoniscidae (endoparasite) than the crabs captured from low salinity locations (Figure 4A). Likewise, the predation on Metapenaeus and Trigonopterus by mud crabs and infestation of Portunion were significantly higher in high salinity as compared to the low salinity (Figure 4B). Similarly, highest infestations of Portunion sp. were detected in mud crabs captured from high salinities (Figure 4C). Overall, these results clearly highlighted the generalist predation patterns and the diet preferences as well as the varying incidences of an endoparasite of mud crabs captured from high and low salinities.
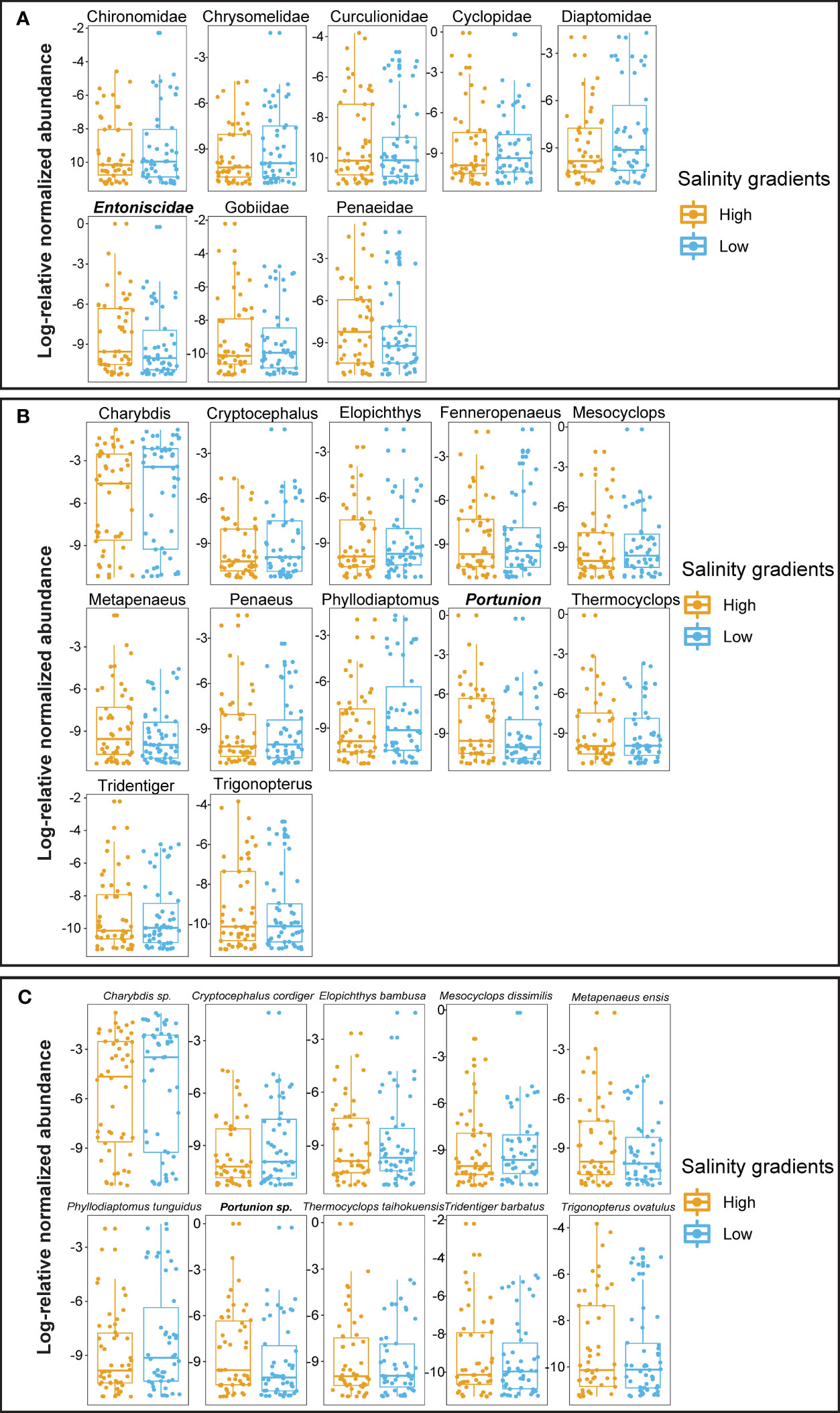
Figure 4 Differential relative abundances of highly significant (padj < 0.001) prey (A) families, (B) genera and (C) species detected in the diet of mud crabs collected from high and low salinity. The Pearson correlation test was conducted using DESeq. padj corresponds to the p-value adjusted for multiple correlation testing using the Benjamini–Hochberg method with a significance level of <0.001. The taxa names in bold and italic represent crab endoparasites.
4 Discussion
In this study, we investigated the inter- and intra-guild predation by S. paramamosain in marine ecosystems across a salinity gradient in the South China Sea. The DNA metabarcoding approach was used to understand trophic networks of crabs in different marine environments with varying salinity levels. DNA metabarcoding approaches used in this study allow us to comprehensively examine the trophic networks (i.e., inter and intra guild predation) of generalist predators, especially mud crabs, and provide a detailed picture of consumed prey in highly dynamic marine environments. In trophic structure and environment relationship studies, choosing the right effect variables (varying environmental conditions and community structure of prey species) may influence the trophic structure of mud crabs. In our study, the effects of salinity gradients on the inter- and intra-guild predation as well as the detection of associated endoparasites of mud crabs were evident across our two sample regions. Although there are a number of other variables besides salinity (i.e., temperature and habitat composition) that may have influenced crab diet across this spatial grouping, they are unlikely to be responsible for the observed trend because of the relatively close proximity of the two sampling regions to each other. However, approaches used and results highlighted in this smaller scale study could provide a solid foundation to investigate the feeding adaptability of mud carbs at large spatial scales across other unexamined variables (i.e., competition, seasonal changes, predation pressure, prey population density, prey preferences and others).
The current study aimed to map the trophic niche breadth in a highly dynamic marine environment, and the results demonstrated evidence of inter- and intra-guild predation. This study detected high predation (52.017%) on crabs (excluding the host species) while other consumed prey taxa comprised 45.567% of the diet. Among all the detected prey taxa, Arthropoda and Chordata were the major contributors to prey contents. A wide range of prey detected in the diet of mud crabs indicated that mud crabs may use different predatory strategies, such as actively hunting near prey species and frequently moving around their enclosures. Similarly, Cordone et al. (2022) found using DNA metabarcoding that green crabs in the Atlantic Patagonian region exhibit a highly diverse dietary spectrum with distinct dietary preferences between males and females. These results, coupled with ours, illustrate how crabs can be active hunters with a wide dietary spectrum, highlighting their role as major food web nodes in various marine environments.
The occurrence of crab parasites is a complex ecological phenomenon influenced by various environmental factors, with salinity being a significant variable. In this study different salinity levels play a pivotal role in distribution of crab parasites. This may be because parasite species can exhibit specific salinity tolerances (Ford and Haskin, 1988; Gauthier et al., 1990; Moussa, 2018). Some parasites may thrive in environments with higher salinity levels, while others may be more adapted to lower salinities. One likely explanation for why our mud crabs exhibited substantially higher Portunion sp. (isopod) infestation in the high salinity sites is that the parasite is physiologically unable to tolerate the reduced salinity levels at the low salinity sites. Likewise, Ford and Haskin (1988) also observed that oyster parasites have low occurrences in intermediate salinity in contrast to the extreme salinity environment. In summary, the relationship between salinity and extent of aquatic parasite infestation is multifaceted. Further investigation on how salinity affects crab parasite occurrence is essential for understanding the broader ecological dynamics, and for designing effective management strategies to minimize the impact of parasitic infestation on crabs.
The impact of intraguild predators on ecosystems can be significant, with intraguild predation having both direct and indirect effects on prey populations (Silliman and Bertness, 2002; Silliman et al., 2004; Kurle et al., 2008). In the current study, we found that variations in predation were linked to the consumption of specific prey types, with mud crabs consuming a range of prey including crabs, fish, and copepods. Non-metric multidimensional scaling ordinations confirmed that predation by mud crabs in different salinities was variable. The intra-guild predation might be as a result of the predators sharing similar hunting strategies or lacking choices in their prey (Lima, 2002). Studies also suggest that the diverse nature of prey across trophic levels is important for controlling the structure of food webs (Wissinger and McGrady, 1993; Peacor and Werner, 2001; Preisser et al., 2005). The strong intraguild and cross-trophic level predation observed in our survey emphasizes that further dietary studies examining prey characteristics (e.g., examining both inter- and intra-guild prey simultaneously) would be helpful in delineating the role of mud crabs in aquatic ecosystems.
Our study highlighted the prey preferences of mud crabs at different salinities and found distinct abundances of various prey taxa at different taxonomic levels. Our findings suggest that the opportunistic predation nature of mud crabs may serve as a key factor in determining the size, abundance, and composition of coexisting prey populations. Overall, the genetic approaches used and the predator-prey interactions observed in our study could help further investigate how the trophic networks can drive the coexistence of different prey species and predators, and how inter- and intra-guild predation benefits each group in a highly dynamic marine ecosystem. Such future investigations will contribute to a better understanding of the functioning of marine ecosystems and the role of predator-prey dynamics within them.
5 Conclusion
In summary, the complementary genetic approaches used in this study enable us to better comprehend the trophic networks of the mud crabs across different salinity gradients. This study revealed that mud crabs have a broad dietary range in marine ecosystems, highlighting previously understudied differences in feeding patterns of crabs between high and low salinities. In future investigations, combining the trophic networks of predators with taxonomic and functional compositions of prey populations and the hunting behavior of predators will help to better predict their ecological role and responses to disturbances such as salinity and temperature variations, habitat destruction, and species invasions.
Data availability statement
The data presented in the study are deposited in the NCBI repository, accession numbers SAMN37670375-SAMN37670474.
Ethics statement
The animal study was reviewed and approved by the Animal Ethics and Use Committee of Shantou University (Approval Number: 202304001).
Author contributions
HS, YY and HM conceived and designed the experiments. HS, YY, SK and ZX conducted field sampling and lab experiments. HS performed the data analyses, prepared figure and tables. HS, SK and HM interpreted the data and wrote the manuscript. MI and MT gave inputs in revising the manuscript. All authors revised the final version and gave their approval for submission.
Funding
This work was supported by the Special Projects in Key Fields of Colleges and Universities in Guangdong Province [2020ZDZX1001], the Program of Agricultural and Rural Department of Guangdong Province (Yuenongnonghan-2022-1144, 2022-SPY-00-014), the Science and Technology Project of Guangdong Province [STKJ202209029, KTP20210376], and the Leading Talent Project of Special Support Plan of Guangdong Province [2019TX05N067] and STU Scientific Research Initiation Grant (NTF21050).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1206004/full#supplementary-material
References
Abrahams M., Kattenfeld M. (1997). The role of turbidity as a constraint on predator-prey interactions in aquatic environments. Behav. Ecol. Sociobiol. 40, 169–174. doi: 10.1007/s002650050330
Arim M., Marquet P. A. (2004). Intraguild predation: a widespread interaction related to species biology. Ecol. Lett. 7, 557–564. doi: 10.1111/j.1461-0248.2004.00613.x
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Brown S. D., Jarman S. N., Symondson W. O. C. (2012). Pyrosequencing of prey DNA in reptile faeces: analysis of earthworm consumption by slow worms. Mol. Ecol. Resour. 12, 259–266. doi: 10.1111/j.1755-0998.2011.03098.x
Cordone G., Lozada M., Vilacoba E., Thalinger B., Bigatti G., Lijtmaer D. A., et al. (2022). Metabarcoding, direct stomach observation and stable isotope analysis reveal a highly diverse diet for the invasive green crab in Atlantic Patagonia. Biol. Invasions 24, 505–526. doi: 10.1007/s10530-021-02659-5
Cuthbert R. N., Briski E. (2022). Functional responses of an invasive mud crab across a salinity gradient. Sci. Total Environ. 818, 151684. doi: 10.1016/j.scitotenv.2021.151684
De Barba M., Miquel C., Boyer F., Mercier C., Rioux D., Coissac E., et al. (2014). DNA metabarcoding multiplexing and validation of data accuracy for diet assessment: application to omnivorous diet. Mol. Ecol. Resour. 14, 306–323. doi: 10.1111/1755-0998.12188
Deng Y., Cheng C., Xie J., Liu S., Ma H., Feng J., et al. (2019). Coupled changes of bacterial community and function in the gut of mud crab (Scylla Paramamosain) in response to Baimang disease. AMB Express 9, 18. doi: 10.1186/s13568-019-0745-1
Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Ford S. E., Haskin H. H. (1988). Comparison of in vitro salinity tolerance of the oyster parasite, Haplosporidium nelsoni (msx) and hemocytes from the host, Crassostrea virginica. Comp. Biochem. Physiol. Part A Physiol. 90, 183–187. doi: 10.1016/0300-9629(88)91025-0
Friman V. P., Hiltunen T., Laakso J., Kaitala V. (2008). Availability of prey resources drives evolution of predator-prey interaction. Proc. R. Soc B. Biol. Sci. 275, 1625–1633. doi: 10.1098/rspb.2008.0174
Gauthier J. D., Soniat T. M., Rogers J. S. (1990). A parasitological survey of oysters along salinity gradients in Coastal Louisiana. J. World Aquac. Soc 21, 105–115. doi: 10.1111/j.1749-7345.1990.tb00530.x
Harper G. L., King R. A., Dodd C. S., Harwood J. D., Glen D. M., Bruford M. W., et al. (2005). Rapid screening of invertebrate predators for multiple prey DNA targets. Mol. Ecol. 14, 819–827. doi: 10.1111/j.1365-294X.2005.02442.x
Holt R. D., Polis G. A. (1997). A theoretical framework for intraguild predation. Am. Nat. 149, 745–764. doi: 10.1086/286018
King R. A., Read D. S., Traugott M., Symondson W. O. C. (2008). INVITED REVIEW: Molecular analysis of predation: a review of best practice for DNA-based approaches. Mol. Ecol. 17, 947–963. doi: 10.1111/j.1365-294X.2007.03613.x
Kotta J., Wernberg T., Jänes H., Kotta I., Nurkse K., Pärnoja M., et al. (2018). Novel crab predator causes marine ecosystem regime shift. Sci. Rep. 8, 4956. doi: 10.1038/s41598-018-23282-w
Kurle C. M., Croll D. A., Tershy B. R. (2008). Introduced rats indirectly change marine rocky intertidal communities from algae- to invertebrate-dominated. Proc. Natl. Acad. Sci. 105, 3800–3804. doi: 10.1073/pnas.0800570105
Le Vay L., Ut V. N., Walton M. (2007). Population ecology of the mud crab Scylla paramamosain (Estampador) in an estuarine mangrove system; a mark-recapture study. Mar. Biol. 151, 1127–1135. doi: 10.1007/s00227-006-0553-4
Li Y., Ai C., Liu L. (2018). “Mud crab, scylla paramamosain China’s leading maricultured crab,” in Aquaculture in China. eBook, Wiley. 226–233. doi: 10.1002/9781119120759.ch3_4
Lima S. L. (2002). Putting predators back into behavioral predator–prey interactions. Trends Ecol. Evol. 17, 70–75. doi: 10.1016/S0169-5347(01)02393-X
Littlefair J. E., Clare E. L. (2016). Barcoding the food chain: from Sanger to high-throughput sequencing. Genome 59, 946–958. doi: 10.1139/gen-2016-0028
Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17 (1), 10–12. doi: 10.14806/ej.17.1.200
Michalko R., Pekár S. (2016). Different hunting strategies of generalist predators result in functional differences. Oecologia 181, 1187–1197. doi: 10.1007/s00442-016-3631-4
Moussa R. M. (2018). The potential impacts of low and high salinities on salinity tolerance and condition index of the adult pearl oyster Pinctada imbricata radiata (Leach 1814). J. Basic Appl. Zool. 79, 12. doi: 10.1186/s41936-018-0021-y
Peacor S. D., Werner E. E. (2001). The contribution of trait-mediated indirect effects to the net effects of a predator. Proc. Natl. Acad. Sci. 98, 3904–3908. doi: 10.1073/pnas.071061998
Pellissier L., Albouy C., Bascompte J., Farwig N., Graham C., Loreau M., et al. (2018). Comparing species interaction networks along environmental gradients. Biol. Rev. 93, 785–800. doi: 10.1111/brv.12366
Poisot T., Canard E., Mouillot D., Mouquet N., Gravel D. (2012). The dissimilarity of species interaction networks. Ecol. Lett. 15, 1353–1361. doi: 10.1111/ele.12002
Pompanon F., Deagle B. E., Symondson W. O. C., Brown D. S., Jarman S. N., Taberlet P. (2012). Who is eating what: diet assessment using next generation sequencing. Mol. Ecol. 21, 1931–1950. doi: 10.1111/j.1365-294X.2011.05403.x
Preisser E. L., Bolnick D. I., Benard M. F. (2005). Scared to death? The effects of intimidation and consumption in predator–prey interactions. Ecology 86, 501–509. doi: 10.1890/04-0719
Saqib H. S. A., Liang P., You M., Gurr G. M. (2021). Molecular gut content analysis indicates the inter- and intra-guild predation patterns of spiders in conventionally managed vegetable fields. Ecol. Evol. 11, 9543–9552. doi: 10.1002/ece3.7772
Saqib H. S. A., Sun L., Pozsgai G., Liang P., You M., Gurr G. M., et al. (2022). DNA metabarcoding of gut contents reveals key habitat and seasonal drivers of trophic networks involving generalist predators in agricultural landscapes. Pest Manage. Sci. 78, 5390–5401. doi: 10.1002/PS.7161
Silliman B. R., Bertness M. D. (2002). A trophic cascade regulates salt marsh primary production. Proc. Natl. Acad. Sci. 99, 10500–10505. doi: 10.1073/pnas.162366599
Silliman B. R., Layman C. A., Geyer K., Zieman J. C. (2004). Predation by the black-clawed mud crab, Panopeus herbstii, in Mid-Atlantic salt marshes: Further evidence for top-down control of marsh grass production. ESTUARIES 27, 188–196. doi: 10.1007/BF02803375
Sunnucks P., Hales D. F. (1996). Numerous transposed sequences of mitochondrial cytochrome oxidase I-II in aphids of the genus Sitobion (Hemiptera: Aphididae). Mol. Biol. Evol. 13, 510–524. doi: 10.1093/oxfordjournals.molbev.a025612
Symondson W. O. C. (2002). Molecular identification of prey in predator diets. Mol. Ecol. 11, 627–641. doi: 10.1046/j.1365-294X.2002.01471.x
Valentini A., Pompanon F., Taberlet P. (2009). DNA barcoding for ecologists. Trends Ecol. Evol. 24, 110–117. doi: 10.1016/j.tree.2008.09.011
Williams R. L., Goodenough A. E., Stafford R. (2012). Statistical precision of diet diversity from scat and pellet analysis. Ecol. Inform. 7, 30–34. doi: 10.1016/j.ecoinf.2011.08.004
Keywords: high-throughput sequencing, trophic interactions, marine ecosystem, mud crab, food web
Citation: Saqib HSA, Yuan Y, Kazmi SSUH, Tayyab M, Xiang Z, Ikhwanuddin M and Ma H (2023) DNA metabarcoding reveals evidence of inter- and intra-guild predation by Scylla paramamosain in a marine ecosystem. Front. Mar. Sci. 10:1206004. doi: 10.3389/fmars.2023.1206004
Received: 14 April 2023; Accepted: 26 September 2023;
Published: 12 October 2023.
Edited by:
Yiqin Deng, Chinese Academy of Fishery Sciences (CAFS), ChinaReviewed by:
Yushun Tian, Dalian Ocean University, ChinaBenjamin Belgrad, Dauphin Island Sea Lab, United States
Copyright © 2023 Saqib, Yuan, Kazmi, Tayyab, Xiang, Ikhwanuddin and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongyu Ma, bWFoeUBzdHUuZWR1LmNu
†These authors have contributed equally to this work
 Hafiz Sohaib Ahmed Saqib
Hafiz Sohaib Ahmed Saqib Ye Yuan
Ye Yuan Syed Shabi Ul Hassan Kazmi1,2
Syed Shabi Ul Hassan Kazmi1,2 Muhammad Tayyab
Muhammad Tayyab Mhd Ikhwanuddin
Mhd Ikhwanuddin Hongyu Ma
Hongyu Ma