- 1Development and Research Center for Biological Marine Resources, Southern Marine Science and Engineering Guangdong Laboratory, Zhanjiang, China
- 2Fisheries College, Guangdong Ocean University, Zhanjiang, China
- 3Guangdong Provincial Engineering Laboratory for Mariculture Organism Breeding, Guangdong Ocean University, Zhanjiang, China
- 4Guangdong Research Center on Reproductive Control and Breeding Technology of Indigenous Valuable Fish Species, Guangdong Ocean University, Zhanjiang, China
Circular RNAs (circRNAs) play key roles in several biological processes in animals and their regulatory mechanism in body color formation or pigmentation in fish remains unclear. Here, circRNAs from black and red individuals of Plectropomus leopardus were identified to clarify the mechanism of circRNAs and the competing endogenous RNA (ceRNA) network (circRNA-microRNA (miRNA)-messenger RNA (mRNA)) in body color formation. We detected a total of 1,424 novel circRNAs. Expression analysis of circRNAs in black vs. red P. leopardus revealed 24 differentially expressed circRNAs (DECs), and 11 and 13 of these DECs were up-regulated and down-regulated in red individuals relative to black individuals, respectively (P<0.05 and |log2 Fold Change (FC)|>1). We identified a total of 19 significant miRNA-circRNA-mRNA ceRNA networks through the analysis of DECs, differentially expressed miRNAs (DEMs) and differentially expressed genes (DEGs). Pathway enrichment analyses of the DEGs involved in the ceRNA network revealed that they were mainly involved in melanin metabolism and immune response. Our findings showed the possibility of the regulatory functions of circRNAs and the corresponding ceRNA network in the body color formation process and will aid the breeding selection process of P. leopardus.
1 Introduction
Fisheries and aquaculture provide important sources of food and income, and their importance continues to grow as wild fish stocks decline (Jennings et al., 2016; Belton et al., 2018). Body color is an essential phenotypic trait that has implications for predator avoidance and thermoregulation in animals (Leclercq et al., 2010). In fish, body color can be an indicator of the quality and economic value of fish, as fish with brighter, more vibrant colors are generally preferred by consumers (Vissio et al., 2021). Several genes, miRNAs, and metabolites that affect body color have been identified in previous studies (Curran et al., 2009; Zhu et al., 2021; Hao et al., 2022a). For example, Forkhead box D3 (Foxd3) controls melanophore specification in the zebrafish neural crest by regulation of microphthalmia transcription factor (Mitf) which upstream of multiple genes necessary for melanin production (Curran et al., 2009). Zhu et al. (2021) reported that low-density lipoproteins receptor adapter 1, guanosine triphosphatase, and regulator of G-protein signaling genes and metabolites including tyrosine, lecithin, prostacyclin may contribute to skin color differences in P. leopardus. A total of 158 differentially expressed miRNAs were identified in the skin color differentiation of red tilapia and among them, miR-138-5p and miR-722 were predicted to play important roles in regulating the pigmentation process (Wang et al., 2018a). However, the molecular regulatory mechanisms of these genes have not yet been clarified (Bertolini et al., 2020).
Circular RNAs (circRNAs) are a large class of non-coding RNAs with covalent bonds that link the 3´ and 5´ ends by back splicing; they are involved in the regulation of various biological processes in eukaryotic cells (Zhang et al., 2014). CircRNAs are highly stable and resistant to degradation by ribonucleases because they lack free 3′ or 5´ ends (Lasda and Parker, 2016). CircRNAs perform their functions in various biological processes by binding to their source genes via the formation of an R-loop, which results in the cessation of transcription and even changes in phenotype (Xu et al., 2020). CircRNAs might also function as microRNA (miRNA) sponges and hinder miRNA-mediated gene inhibition or silencing through competing endogenous RNA (ceRNA) networks (Salmena et al., 2011; Memczak et al., 2013). Ribonucleoprotein complexes can be formed by the binding of circRNAs to transcription factors and RNA-binding proteins (Aktas et al., 2017; Quan et al., 2021). Several studies have reported that circRNAs function in muscle development (Kotb et al., 2015), transcriptional regulation (Zhang et al., 2013), as well as cellular communication and signal transduction (Li et al., 2015). Studies of circRNAs in fish have mainly examined the roles of circRNAs in the pathogenesis of various diseases in grass carp (Ctenopharyngodon idellus) (He et al., 2017; Liu et al., 2019), flounder (Paralichthys olivaceus) (Xiu et al., 2019), and Nile tilapia (Oreochromis niloticus) (Fan et al., 2019). However, few studies have examined the function of circRNAs in body color formation.
Several miRNAs involved in body color formation have been documented in fish (Wang et al., 2018a; Dong et al., 2012; Kennell et al., 2012; Dong et al., 2020). Multiple miRNAs in red tilapia and common carp have been reported to regulate the expression of body color formation related genes (Wang et al., 2018a; Yan et al., 2013; Luo et al., 2018). Recent studies have shown that messenger RNAs (mRNAs) are not the only molecules regulated by miRNAs (Poliseno et al., 2010; Quinn and Chang, 2016). CircRNAs can mediate decreases in the expression of miRNAs via ceRNA networks by acting as miRNA sponges, and this can have downstream regulatory effects on the expression of target mRNAs (Jiang et al., 2020).
P. leopardus is an important tropical and subtropical fish that varies in body color (Zhu et al., 2021). Several body color-related genes of P. leopardus have been identified in previous transcriptome analyses (Zhu et al., 2021; Hao et al., 2022b; Yang et al., 2020a). However, the specific regulatory mechanism of these genes has not yet been clarified. Here, we identified several circRNAs in black and red P. leopardus to clarify the molecular mechanism by which circRNAs, via their corresponding ceRNAs, regulate the expression of mRNAs involved in body color formation in P. leopardus.
2 Materials and methods
2.1 Experimental animals
Black and red P. leopardus fish (four months old) were obtained and fed with Dongwan grouper feed (Guangdong Yuequn Biotechnology Co., Ltd., China) twice per day (10 a.m. and 4 p.m.). Six red-colored and six black-colored individuals were utilized for this experiment. Both the red-colored and black-colored groups were obtained from base stock established with the breeders collected from Hainan and Taiwan provinces of China, Australia and Philippines. They were reared in the same conditions with studies of Zhu et al. (2021) and Hao et al. (2022a). After temporary rearing for one week, eugenol was used to anesthetize fish; fish were then sacrificed, and skin tissue samples were taken. All animal experiments were conducted in accordance with the guidelines and approval of the respective Animal Care and Use Committee of Guangdong Ocean University, China.
2.2 Sequencing of circRNAs and bioinformatics analysis
Total RNA was extracted from skin tissue samples, and linear RNAs were removed via treatment with RNase R. A strand-specific library of the present study was constructed and sequenced using an Illumina NovaSeq_6000 platform with the purified RNAs. The sequenced data were then filtered by fastp (version 0.18.0) (Chen et al., 2018). TopHat2 (version 2. 1.1) (Kim et al., 2013) was used to map these filtered reads to the reference genome (Zhou et al., 2020). The software find_circ (version 1) (Memczak et al., 2013) was used to identify circRNAs. Types of circRNAs, their distribution on chromosomes, and their length distribution were analyzed; circBase (Glažar et al., 2014) annotations of the circRNAs were obtained via BLAST searches. Novel circRNAs were circRNAs that could not be annotated.
2.3 Quantification of circRNA abundance
Reads per million mapped reads of circRNAs were calculated using edgeR package (version 3.12.1) (http://www.r-project.org/) to quantify the abundance of circRNAs. DECs were identified (|log2FC|>1 and P < 0.05). The genes from which circRNAs were derived were referred to as source genes. Gene ontology (GO) and Kyoto Encylopaedia of Genes and Genomes (KEGG) enrichment analyses of DECs source genes were conducted to clarify the role of identified circRNAs.
2.4 CeRNA network analysis
The ceRNA network was constructed via analysis of DEGs (|log2FC|>1 and false discovery rate (FDR) < 0.05), DECs (|log2FC|>1 and P < 0.05), and DEMs (|log2FC|>1 and P < 0.05) between black and red fish. miRNAs target genes were predicted using Mireap, Miranda (version 3.3a), and TargetScan (version 7.0). The ceRNA network was constructed using the following rules: regulator–target relationships must exist between miRNAs and ceRNAs, and their expression must be negatively correlated; there are only positive correlations in the expression of ceRNAs; and there is competition among ceRNAs for binding to the same miRNA. Significant correlations between mRNAs and miRNAs, between circRNAs and miRNAs, and between circRNAs and mRNAs were identified (Spearman Rank correlation coefficient (SCC) < -0.7 and Pearson correlation coefficient > 0.9). The significance of the common miRNA sponges between two genes was evaluated using a hypergeometric cumulative distribution function test (P < 0.05). DEGs from the ceRNAs were analyzed via GO and KEGG pathway enrichment analyses.
3 Results
3.1 Identification of circRNAs
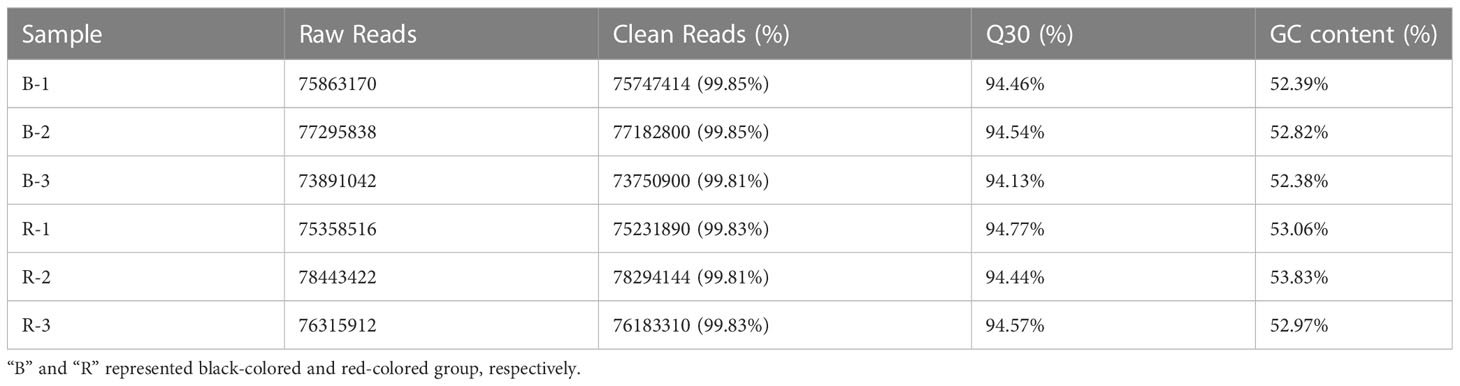
The skin tissues of black and red P. leopardus were used to construct circRNA libraries. A total of 75,683,350 and 76,705,950 reads were obtained from skin tissues of black and red fish, and the number of clean reads for black and red fish samples was 75,560,371 and 76,569,781, respectively (Table 1). The raw data presented in the study are deposited in the Sequence Read Archive of the NCBI (SRR22522288, SRR22522289, SRR22522290, SRR22522291, SRR22522292, SRR22522293). The accession number is PRJNA908242. The Q30 (the accuracy of each base is 99.9%) values for the black and red fish were 94.4% and 94.6%, respectively, which suggests that the data were sufficiently robust for subsequent analysis. We identified a total of 1,424 circRNAs, including 1,086 and 1,084 in the black and red group, respectively. Analysis of the spliced lengths of the candidate circRNAs (which ranged from 89 nucleotides (nt) to 72,726 nt) revealed that approximately 90.9% and 49.0% of circRNAs had spliced lengths less than 2,000 nt and between 201 and 500 nt, respectively (Figure 1A). Analysis of the genes from which circRNAs were derived revealed that most circRNAs (55.76%, 794 circRNAs) were spliced from annot_exons, and only 1.69% of circRNAs (24 circRNAs) were intronic circRNAs (Figure 1B). Analysis of the chromosomal distribution of circRNAs revealed that they were present on every chromosome (Figure 1C).
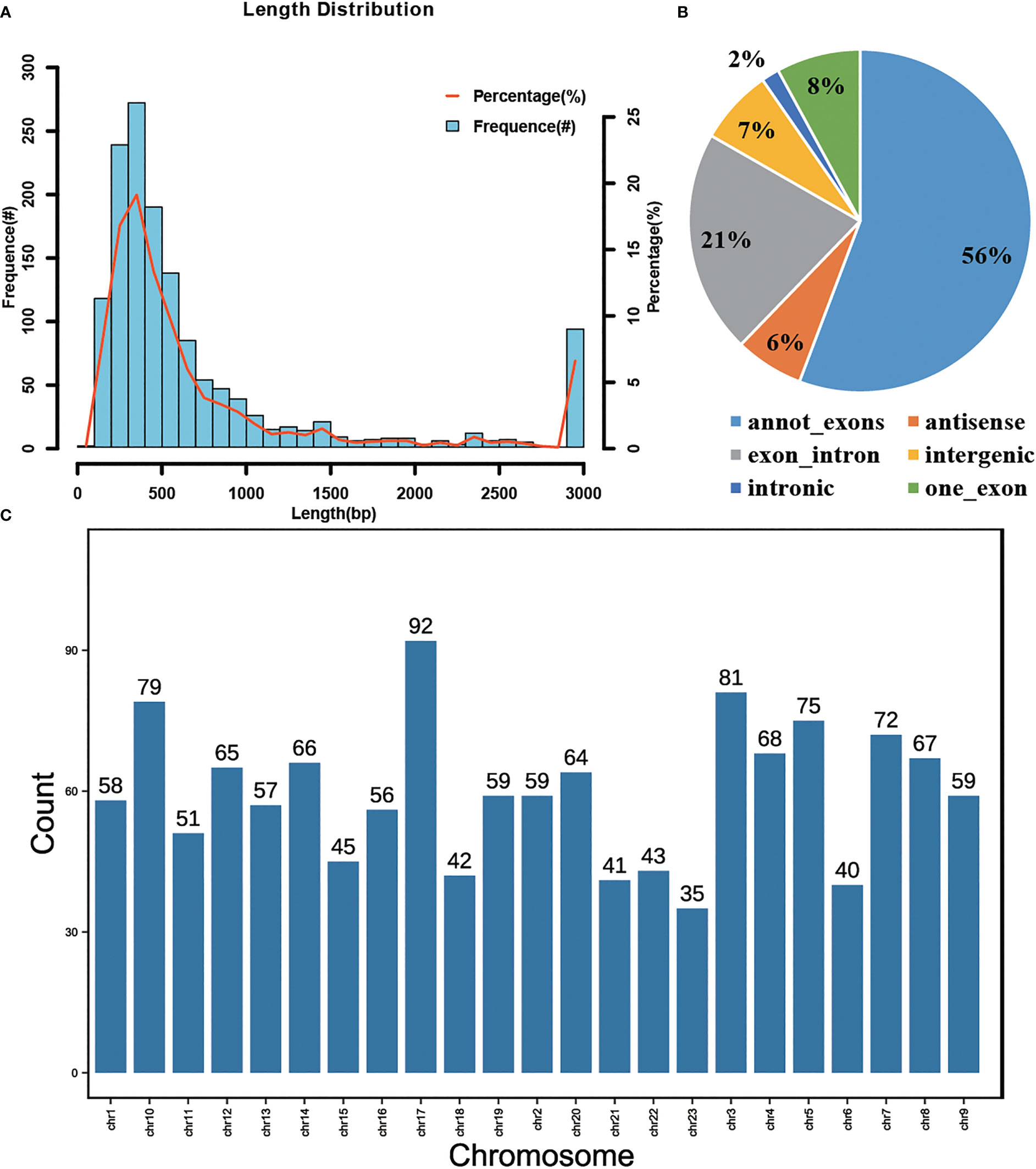
Figure 1 Analysis of the circRNAs identified in P. leopardus. (A) Length distribution of circRNAs. The column represents the frequency of length (left longitudinal axis), and the curve represents the percentage of length (right longitudinal axis) (B) Categories of circRNAs including annot_exons, antisense, exon_intron, intergenic, intronic and one_exon. (C) The chromosomal locations of circRNAs which showed that cirRNAs were present on every chromosome.
3.2 Expression analysis of circRNAs
Expression of circRNAs in skin tissues of black and red P. leopardus were analyzed. Principal component analysis (PCA) revealed clear differences in circRNA expression between black and red fish (Figure 2A). The numbers and distribution of circRNAs are shown in a volcano plot (Figure 2B). We identified a total of 24 circRNAs (P < 0.05 and |log2 FC| > 1) (Supplemental Table 1; Figure 2C). The expression of 11 and 13 DECs was up-regulated and down-regulated in red fish compared with black fish, respectively. A heatmap was made to visualize the expression profiles of these DECs (Figure 2D). The heatmap revealed that the expression profiles of DECs differed in black and red fish.
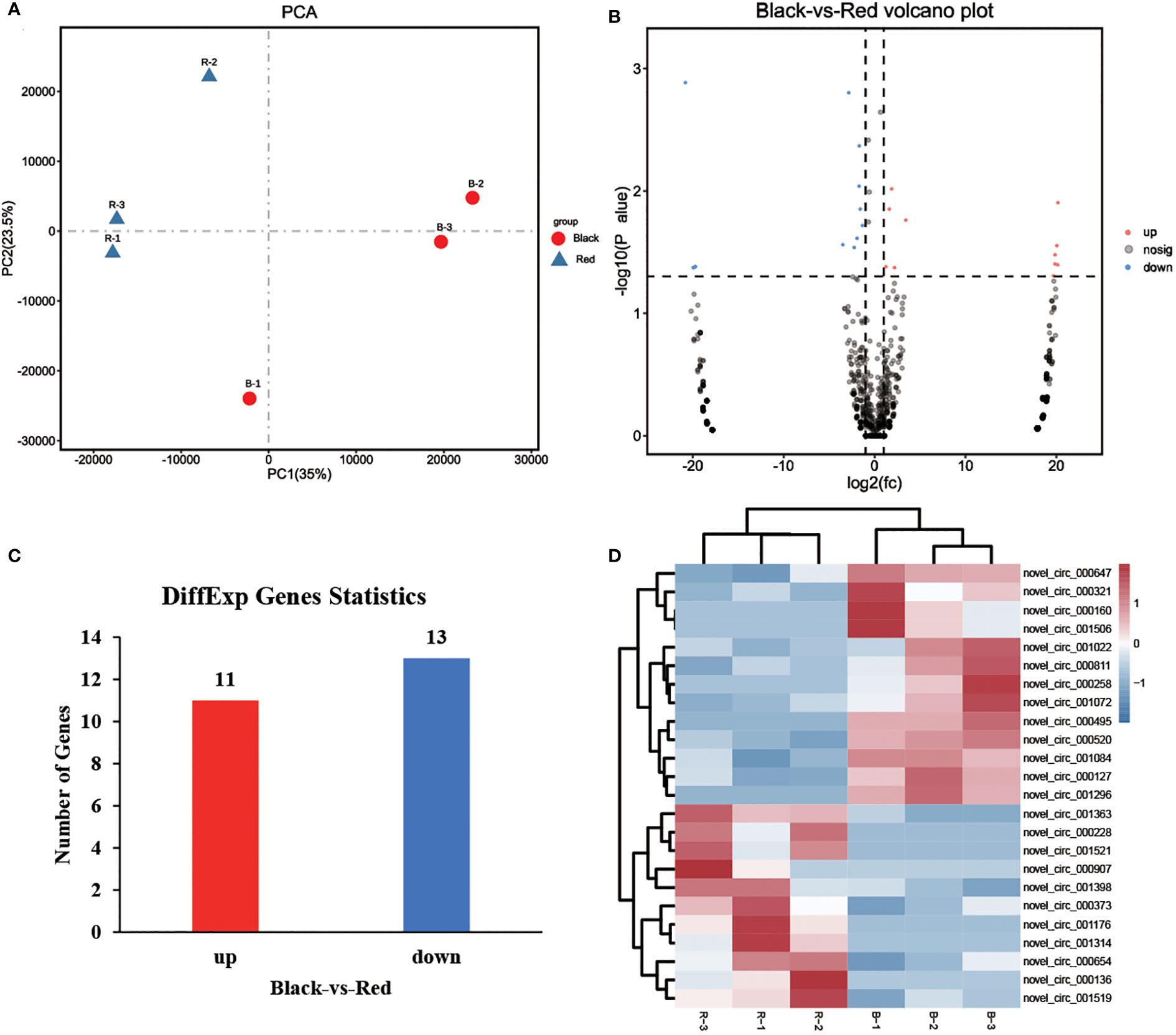
Figure 2 Analysis of the expression of circRNAs. (A) Results of a PCA of the expression levels of circRNAs in black and red P. leopardus. (B) Volcano plot of circRNAs. Red and blue dot represented up-regulated and down-regulated genes in the red-colored group compared with black-colored group, respectively. (C) Analysis of DEGs in black and red P. leopardus. Red and blue columns represented the number of up-regulated and down-regulated genes in the red-colored group compared with black-colored group, respectively. (D) Heatmap of the expression profiles of circRNAs. Red and blue represented high expression and low expression, respectively. “B” and “R” represented black-colored and red-colored group, respectively.
3.3 Pathway analysis of DEC source genes
A total of four DECs of the 24 DECs identified were circular intronic RNAs with no source gene; the remaining 20 DECs were circular exonic circRNAs and exon-intron circRNAs derived from 20 source genes. GO analysis showed that DEC source genes were mainly involved in cellular component, molecular function, and biological process (P < 0.5) and enriched in Golgi related pathway including Golgi apparatus part, Golgi membrane, and Golgi apparatus (Figure 3A; Supplemental Table 2). KEGG pathway enrichment of DEC source genes indicated that they were enriched in several pathways, including bacterial invasion of epithelial cells, chemokine signaling pathway, cellular senescence, and ubiquitin-mediated proteolysis (Figure 3B).
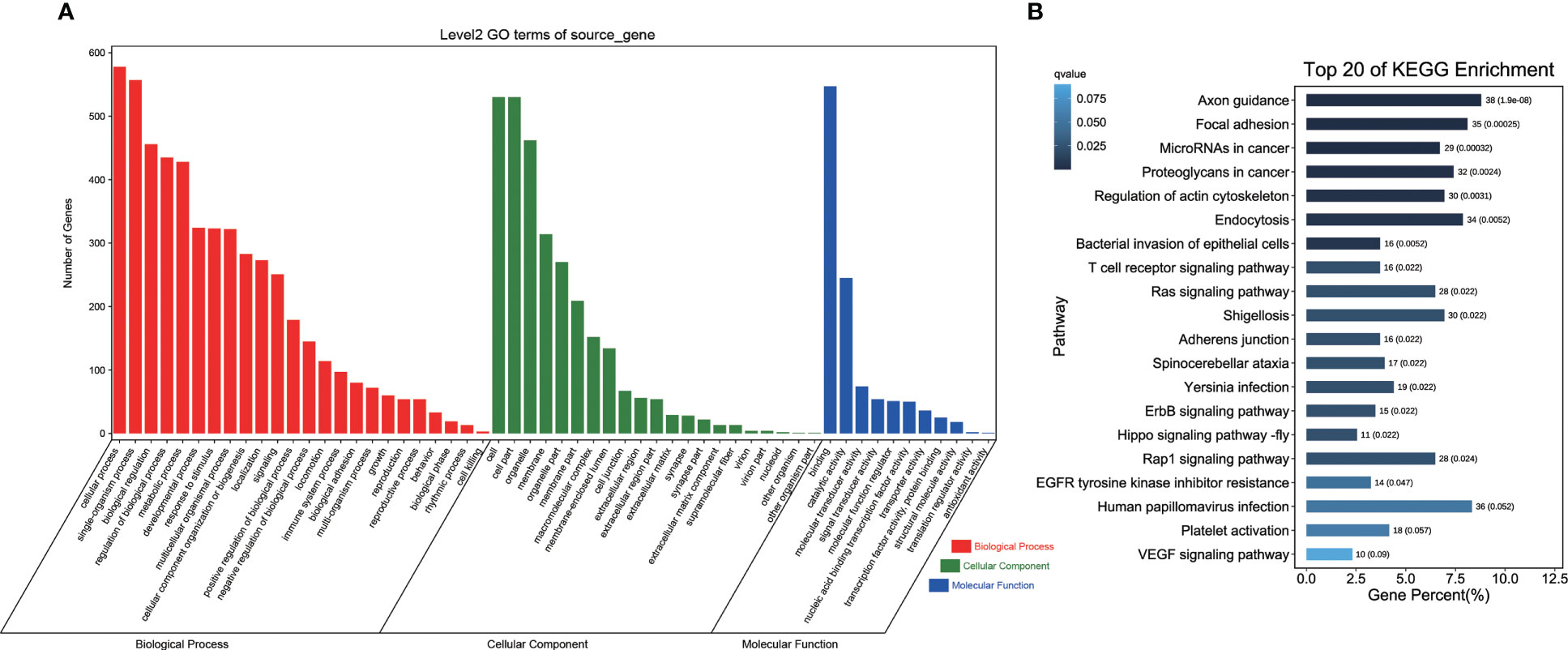
Figure 3 Enrichment analysis of circRNA source genes. (A) GO terms analysis of circRNA source genes. Red, green and blue represented the biological process, cellular component, and molecular function terms, respectively. (B) KEGG pathway enrichment analysis of circRNA source genes. The length of column showed the number and percent of pathway, and the depth of color showed q-value of pathway.
3.4 Integrated analysis of DEMs with DEGs and DECs
The DEG, DEM, and DEC datasets for red and black P. leopardus were used to conduct an integrated analysis of DEMs with DECs and DEGs. A total of 489 DEGs (Supplemental Table 3, |log2FC|>1 and FDR < 0.05), 60 DEMs (Hao et al., 2022a, |log2FC|>1 and P < 0.05), and 24 DECs (Supplemental Table 1, |log2FC|>1 and P < 0.05) were analyzed. A total of 2,173 mRNA–miRNA pairs, including 379 DEGs and 59 DEMs, and 104 circRNA-miRNA pairs, including 21 DECs and 53 DEMs, were identified in red and black P. leopardus. SCC was less than -0.7 for 605 miRNA-mRNA pairs and 31 miRNA-circRNA pairs, and network analysis was conducted using these miRNA-mRNA and miRNA-circRNA pairs (Supplemental Tables 4, 5).
3.5 Analysis of the regulatory ceRNA network (DECs-DEMs-DEGs)
The network analysis revealed 357 circRNA-mRNA pairs, containing eight circRNAs and 272 mRNAs with an SCC greater than 0.9. The final ceRNA network containing 19 miRNA-circRNA and miRNA-mRNA pairs (six circRNAs, 16 miRNAs, and 18 mRNAs) was generated by conducting hypergeometric cumulative distribution function tests (P < 0.05, Supplemental Table 6). Connectivity analysis was conducted on the hub genes of the net-work with important functions. In the ceRNA network, APC membrane recruitment 2 (Dxb_GLEAN_10011350) was regulated by five miRNAs (miR-192-z, miR-466-x, miR-625-x, novel-m0120-5p, and novel-m0121-3p) and one circRNA (novel_circ_000127) (Figure 4). The tyrosine family member genes tyrosinase related protein 1 (TYRP1) (Dxb_GLEAN_10020133; Dxb_GLEAN_10003424) and tyrosinase related protein 2 (TYRP2) (Dxb_GLEAN_10014015) were regulated by one circRNA (novel_circ_000495) and one miRNA (novel-m0095-3p).
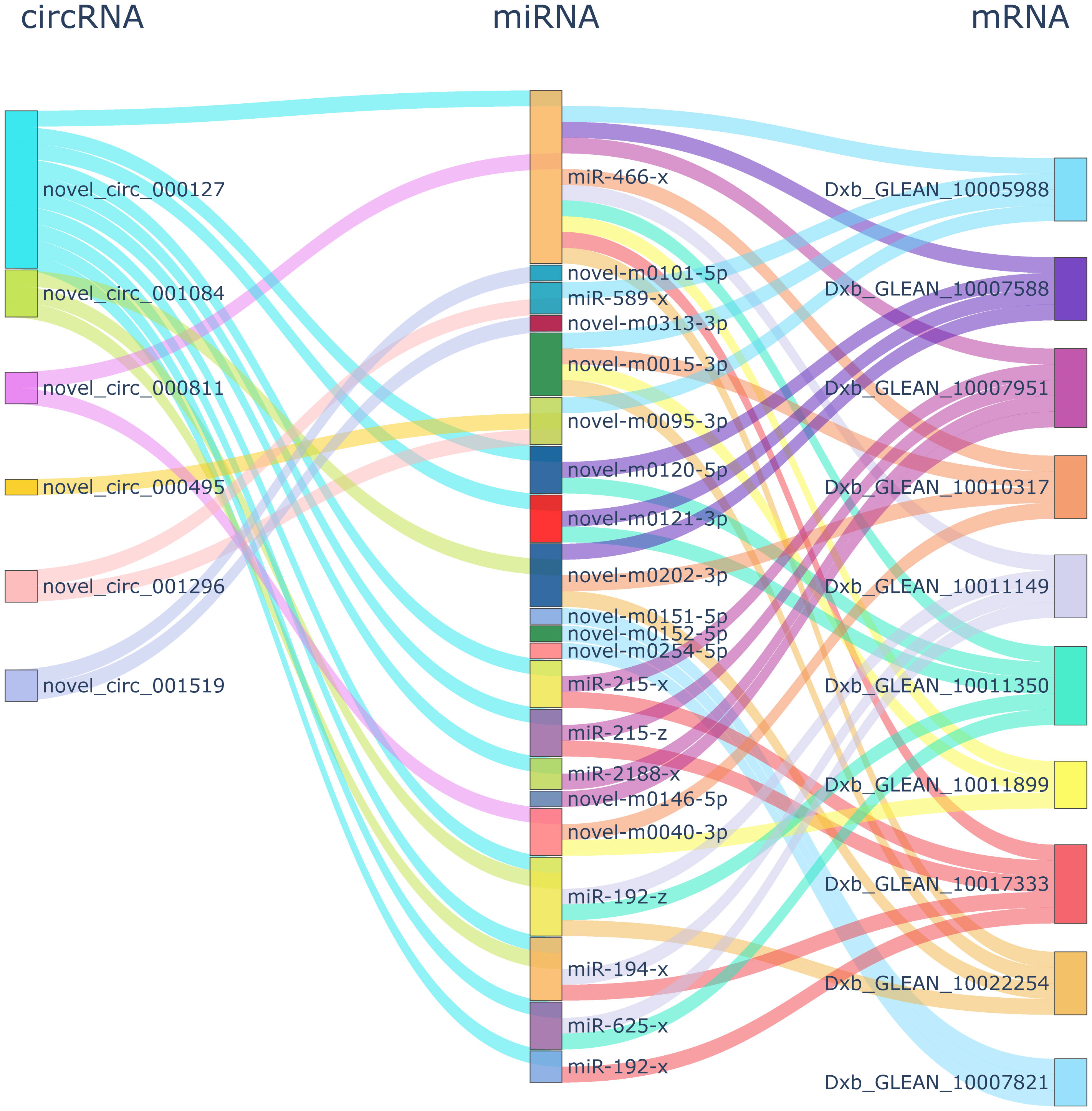
Figure 4 ceRNA network connectivity analysis. The first, second, and third column showed the DECs, DEMs, and DEGs, respectively. In the first and third column, the same color showed the same DECs and DEGs network, respectively.
3.6 GO analysis of ceRNA network DEGs
GO analysis revealed that DEGs of the ceRNA network were significantly enriched in 71 GO terms (P < 0.05; Figure 5). These 71 GO terms included pigment-related terms (melanin metabolic process, tyrosine metabolic process, pigment metabolic process, pigment granule, pigmentation, pigment granule organization, cellular pigmentation, pigment cell differentiation, and developmental pigmentation), as well as immune-related terms or stress response terms containing cellular response to glucocorticoid stimulus, response to ammoniumion, cellular response to corticosteroid stimulus, and response to glucagon.
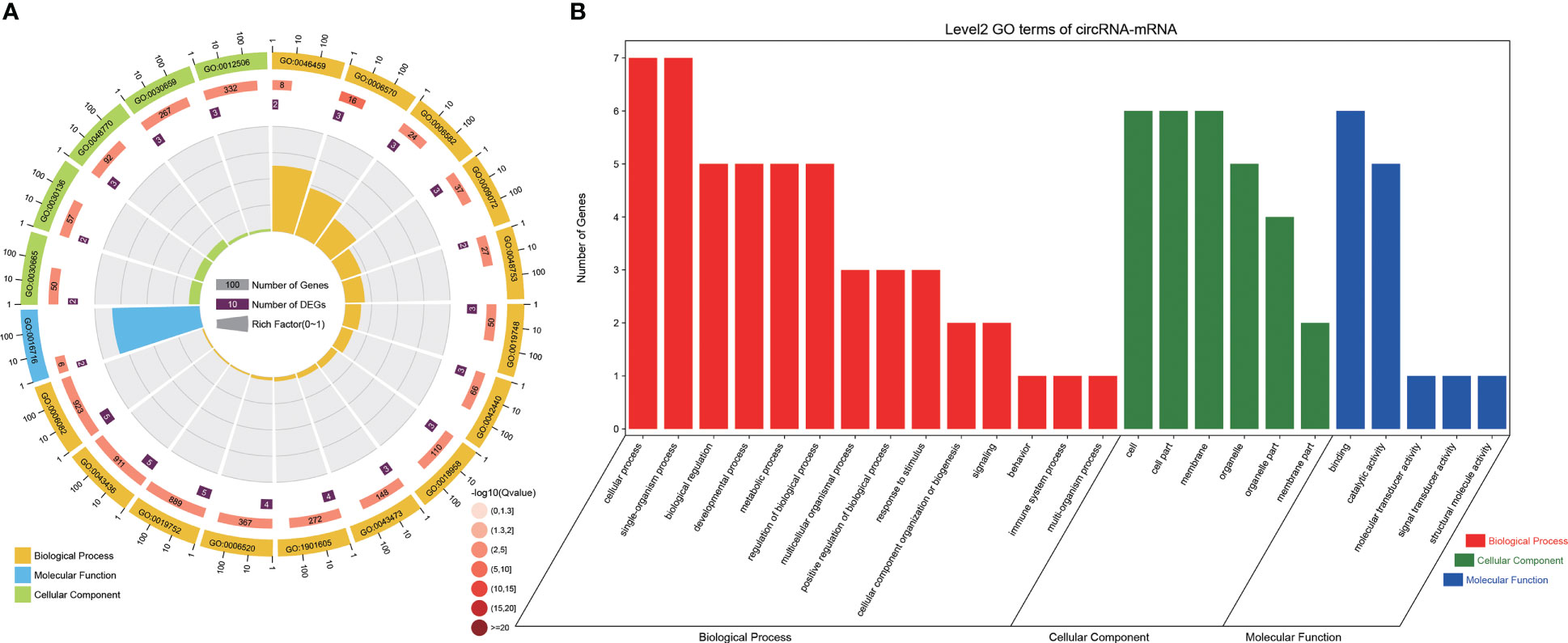
Figure 5 GO analysis of DEGs from the ceRNA network. (A) Analysis of the GO terms enrichment of DEGs from the ceRNA network. The first circle: the top 20 GO terms in the enrichment analysis, and the outside circle is the coordinate scale for the number of DEGs. Different colors represent different Ontology; Circle 2: The DEGs number and Q value of the GO term in background. The more the number of DEGs, the longer the bars, and the smaller the Q value, the redder the color; Circle 3: The number of DEGs in the GO term; Circle 4: RichFactor value of each GO term. (B) GO terms analysis of DEGs from the ceRNA network. Red, green and blue represented the biological process, cellular component, and molecular function terms, respectively.
3.7 KEGG pathway analysis of ceRNA network DEGs
A total of 24 enriched pathways, including pigmentation-related pathways (tyrosine metabolism and melanogenesis) and immune system pathways (C-type lectin receptor signaling pathway, nucleotide-binding, oligomerization domain (NOD)-like receptor signaling pathway, and chemokine signaling pathway) (Figure 6) were detected in the KEGG pathway analysis of ceRNA network DEGs. Infectious disease-related pathways such as pertussis, Yersinia infection, and Salmonella infection were also enriched.
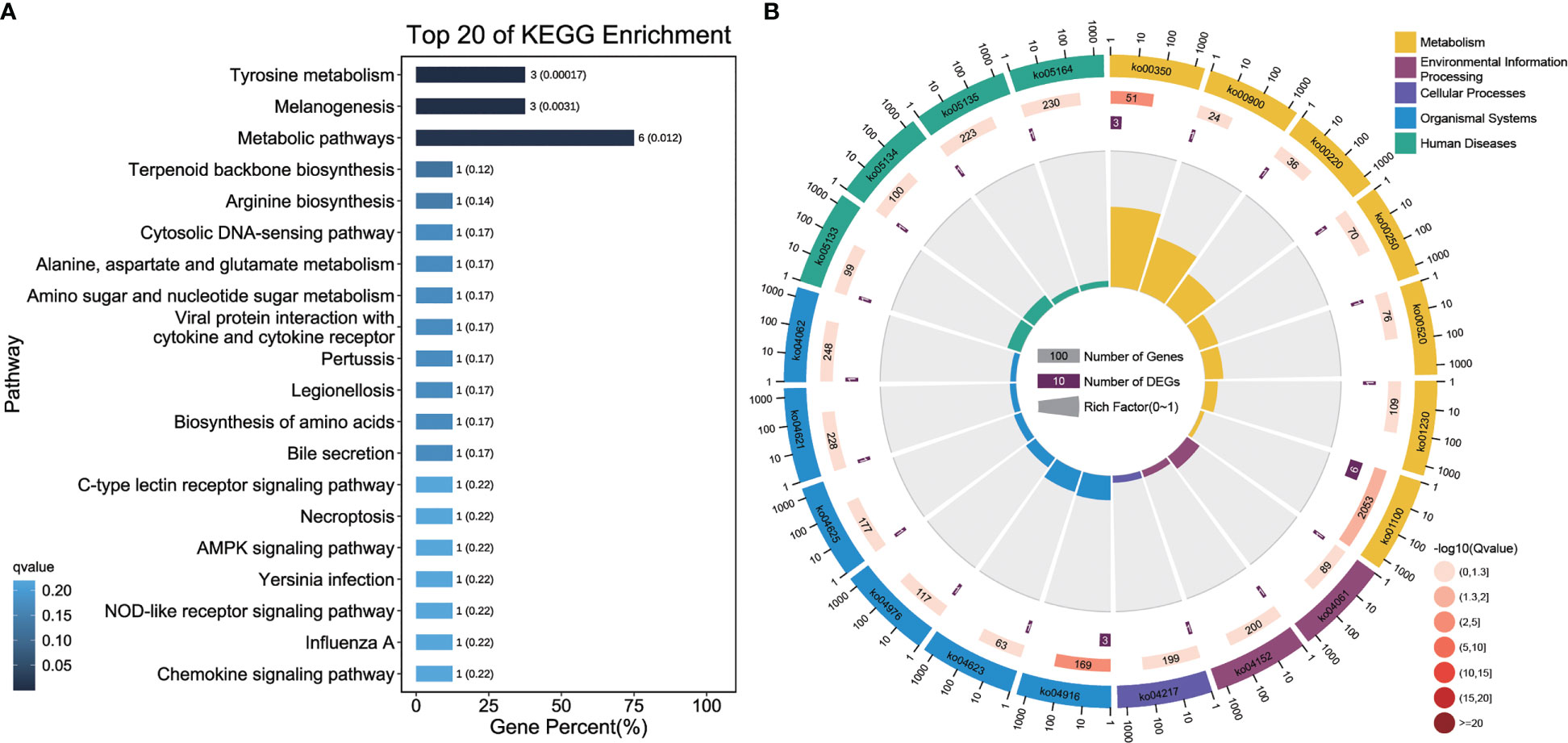
Figure 6 KEGG pathway analysis of DEGs from the ceRNA network. (A) KEGG pathway enrichment analysis of DEGs from the ceRNA network. The length of column showed the number and percent of pathway, and the depth of color showed q-value of pathway. (B) Analysis of the KO enrichment of DEGs from the ceRNA network. The first circle: the top 20 pathway in the enrichment analysis, and the outside circle is the coordinate scale for the number of DEGs. Different colors represent different class; Circle 2: The DEGs number and Q value of the pathway in background. The more the number of DEGs, the longer the bars, and the smaller the Q value, the redder the color; Circle 3: The number of DEGs in the pathway; Circle 4: RichFactor value of each pathway.
4 Discussion
Body color is an essential phenotypic trait that affects the adaptation of P. leopardus to its environment (Chen et al., 2019). An increasing number of studies have examined the regulatory roles of circRNAs in body color formation (Zhu et al., 2020). However, the special mechanism by which circRNAs regulate body color formation in marine animals has not been extensively studied. We identified circRNAs involved in body color formation and constructed circRNA-miRNA-mRNA networks in P. leopardus. Pathway enrichment analyses were clarified to show the functions of circRNAs in body color formation; the results of these analyses provide data that could be used in future studies of the regulatory mechanism by which circRNAs participate in marine animal body color formation. The potential for miRNAs and mRNAs to be used to alter the body color of fish is low given that their half-lives in cells and tissues are short. The potential for circRNAs to be used for this purpose through targeted miRNA silencing is higher than that of linear RNAs given that circRNAs are more chemically stable than linear RNAs (Holdt et al., 2018; Rbbani et al., 2021).
We identified circRNAs in individuals of P. leopardus varying in body color; these circRNAs were classified as exonic circRNAs (one_exon, annot_exon, and exon_intron), intronic circRNAs, and intergenic circRNAs. Exonic circRNAs were the most common, and these findings are consistent with the results of previous studies of tilapia and mice (Jeck et al., 2013; Fan et al., 2019). In tilapia, multiple circRNAs can be derived from a single gene (Fan et al., 2019). In our study, one source gene could generate multiple circRNA isoforms in P. leopardus, and this was associated with alternative back splicing.
CircRNAs are synthesized by back splicing, which differs from the traditional splicing mechanism of linear mRNAs; study of the function of the source mRNAs can provide insights into the functions of circRNAs. A total of 24 DECs were identified via analysis of the expression of DECs between black and red P. leopardus. CircRNAs can be co-expressed with their source genes in the same locus (Huang et al., 2017), and they can play a role in regulating the expression of their source genes by mediating decreases in the quantity of pre-mRNA for canonical splicing (Salzman et al., 2012). In addition, the linear transcripts of the source gene might regulate the expression of circRNAs. The novel_circ_001296 DEC was transcribed from the foxp1b (Dxb_GLEAN_10021390) gene. Previous studies have shown that miR-429 silencing can lead to an increase in Foxd3 expression in vivo and inhibit MITF expression, which leads to decreases in the expression of downstream genes, including the melanin synthesis-related genes tyrosinase (TYR), TYRP1, and TYRP2 (Yan et al., 2013). The novel_circ_000160 DEC was transcribed from the SLC9A7 (Dxb_GLEAN_10021160) gene. Solute carriers (SLCs) play a role in skin pigmentation or body color formation, and many SLC genes are differentially expressed in chicken, tilapia, and leopard grouper individuals varying in color (Hao et al., 2022b; Hoglund et al., 2011; Zhu et al., 2016; Chen et al., 2020). The source genes of the DECs are also involved in protein ubiquitination. E3 ubiquitin-protein ligase CBL (Dxb_GLEAN_10018739) is involved in the process of ubiquitin-mediated protein degradation (Hochrainer et al., 2005), and the up-regulation of the gene encoding this protein in black fish relative to red fish suggests that protein degradation has occurred. However, additional studies are needed to clarify the relationships between source genes and circRNAs.
The activity and expression of miRNAs can be inhibited by circRNAs, and this can alter the expression of the target genes of miRNAs (Hansen et al., 2011; Hansen et al., 2013). Genetics plays a role in trait inheritances and the SNP detection of trait-related genes or regulatory molecules to obtain potential markers for phenotype in breeding has been widely used in aquaculture (Fan et al., 2017; Lei et al., 2019; Yang et al., 2020b). In the present study, differential expression of circRNAs and genes in the two phenotypes is evident, which may suggest a direction for the SNPs detection which were associated with body color formation. Color phenotype is determined by the combination of genetics and environmental factors, and changes in environmental factors can also trigger variations in hormone and neuron secretion, resulting in different skin color responses (Luo et al., 2021). For example, background color signals are transmitted to the brain via vision, and are processed by the central nervous system to act on skin pigment cells to adjust the body color to the background color (Zimmermann et al., 2018). Dietary Cys and Tyr could affect the melanin synthesis pathway in the red tilapia and participate in the skin color differentiation (Wang et al., 2018b). Therefore, it is reasonable to hypothesize that body color changes triggered by environmental factor is partially related to the expression level changes in molecules including circRNAs, miRNAs and mRNAs through participating in or regulating pigmentation related pathway.
GO analysis of DEC source genes revealed that genes related to body color variation were involved in Golgi apparatus part, Golgi membrane, and Golgi apparatus. Tyrosinase family proteins involved in melanin biosynthesis might be trafficked through the Golgi apparatus and then to type II melanosomes, where they promote melanin deposition (Dooley et al., 2013). Therefore, novel_circ_001363 might regulate the expression of its source gene to modify the transport of TYR and TYRP1 through the Golgi apparatus (Hu et al., 2022). Black body color is highly associated with melanin synthesis (Henning et al., 2013); in our study, DEGs in the ceRNA network were enriched in melanin synthesis-related pathways, including melanogenesis and tyrosine metabolism (Kelsh, 2004).
RNAs with the same miRNA-binding site may compete with each other for miRNA binding and function as ceRNAs. Previous studies have shown that circRNAs play a regulatory role in determining the color of the hair on the skin of mice via the circRNA-miRNA-mRNA ceRNA network (Zhu et al., 2018; Zhu et al., 2020). In our study, the ceRNA network was constructed, and related DEGs were enriched in several immune pathways, indicating that circRNAs might regulate the immunity of fish varying in body color. Higher levels of melanin promote immune defense and wound healing in Amphiprion percula (Smith et al., 2018). Darker eumelanic individuals show higher immune activity than lighter individuals (McGraw, 2005; Ducrest et al., 2008). However, additional studies are necessary to determine whether these genes are targeted by circRNAs or whether they affect the body color of P. leopardus.
5 Conclusions
A total of 1,424 novel circRNAs were identified in black and red P. leopardus. A total of 24 circRNAs were detected as DECs. Integrated analysis of DECs, DEMs, and DEGs yielded a ceRNA network with 19 significant miRNA-circRNA and miRNA-mRNA pairs. Pathway enrichment of ceRNA network DEGs revealed that they may participate in the regulation of the immune response and melanin-related metabolism. The results indicated the potential functions of circRNAs and the ceRNA network in body color formation and will aid future efforts to breed P. leopardus individuals with colors.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was reviewed and approved by the respective Animal Care and Use Committee of Guangdong Ocean University.
Author contributions
CZ designed and supervised the research. RH conducted the research, analyzed the data and wrote the manuscript. XZ, CT, YH nd GL contributed to the final writing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Nature Science Foundation of China (Grant No. 32102766); Southern Marine Science and Engineering Guangdong Laboratory (Zhanjiang) (Grant No. ZJW-2019-06); Jieyang City Science and Technology Plan Project (Grant No. 2019066).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1201726/full#supplementary-material
References
Aktas T., Avsar I., Maticzka D., Bhardwaj V., Pessoa R., Mittler G., et al. (2017). DHX9 suppresses RNA processing defects originating from the alu invasion of the human genome. Nature 544, 115–119. doi: 10.1038/nature21715
Belton B., Bush S. R., Little D. C. (2018). Not just for the wealthy: rethinking farmed fish consumption in the global south. glob. Food Secur. 16, 85–92. doi: 10.1016/j.gfs.2017.10.005
Bertolini F., Ribani A., Capoccioni F., Buttazzoni L., Utzeri V. J., Bovo S., et al. (2020). Identification of a major locus determining a pigmentation defect in cultivated gilthead seabream (Sparus aurata). Anim. Genet. 51, 319–323. doi: 10.1111/age.12890
Chen Y., Gong Q., Lai J., Song M., Liu Y., Wu Y., et al. (2020). Transcriptome analysis identifies candidate genes associated with skin color variation in Triplophysa siluroides. Comp. Biochem. Phys. D 35, 100682. doi: 10.1016/j.cbd.2020.100682
Chen S., Xu W., Liu Y. (2019). Fish genomic research: decade review and prospect. J. Fish China 43, 1–14. doi: 10.11964/jfc.20181211599
Chen S., Zhou Y., Chen Y., Gu J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Curran K., Raible D. W., Lister J. A. (2009). Foxd3 controls melanophore specification in the zebrafish neural crest by regulation of mitf. Dev. Biol. 332, 408–417. doi: 10.1016/j.ydbio.2009.06.010
Dong Z., Luo M., Wang L., Yin H., Zhu W., Fu J. (2020). MicroRNA-206 regulation of skin pigmentation in koi carp (Cyprinus carpio l.). Front. Genet. 11. doi: 10.3389/fgene.2020.00047
Dong C., Wang H., Xue L., Dong Y., Yang L., Fan R., et al. (2012). Coat color determination by miR-137 mediated down-regulation of microphthalmia-associated transcription factor in a mouse model. RNA 18, 1679–1686. doi: 10.1261/rna.033977.112
Dooley C., Schwarz H., Mueller K., Mongera A., Konantz M., Neuhauss S., et al. (2013). Slc45a2 and V-ATPase are regulators of melanosomal pH homeostasis in zebrafish, providing a mechanism for human pigment evolution and disease. Pigment Cell Res. 26, 205–217. doi: 10.1111/pcmr.12053
Ducrest A. L., Keller L., Roulin A. (2008). Pleiotropy in the melanocortin system, coloration and behavioural syndromes. Trends Ecol. Evol. 23, 502–510. doi: 10.1016/j.tree.2008.06.001
Fan B., Chen F., Li Y., Wang Z., Wang Z., Lu Y., et al. (2019). A comprehensive profile of the tilapia (Oreochromis niloticus) circular RNA and circRNA-miRNA network in the pathogenesis of meningoencephalitis of teleosts. Mol. Omics 15, 233–246. doi: 10.1039/c9mo00025a
Fan S., Xu Y., Liu B., He W., Zhang B., Su J., et al. (2017). Molecular characterization and expression analysis of the myostatin gene and its association with growth traits in noble scallop (Chlamys nobilis). Comp. Biochem. Physiol. B 212, 24–31. doi: 10.1016/j.cbpb.2017.07.004
Glažar P., Papavasileiou P., Rajewsky N. (2014). circBase: a database for circular RNAs. RNA 20, 1666–1670. doi: 10.1261/rna.043687.113
Hansen T. B., Jensen T. I., Clausen B. H., Bramsen J. B., Finsen B., Damgaard C. K. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. doi: 10.1038/nature11993
Hansen T. B., Wiklund E. D., Bramsen J. B., Villadsen S. B., Statham A. L., Clark S. J., et al. (2011). MiRNA-dependent gene silencing involving ago2-mediated cleavage of a circular antisense RNA. EMBO J. 30, 4414–4422. doi: 10.1038/emboj.2011.359
Hao R., Zhu X., Tian C., Jiang M., Huang Y., Zhu C. (2022a). Integrated analysis of the role of miRNA-mRNA in determining different body colors of leopard coral grouper (Plectropomus leopardus). Aquaculture 548, 737575. doi: 10.1016/j.aquaculture.2021.737575
Hao R., Zhu X., Tian C., Zhu C., Li G. (2022b). Analysis of body color formation of leopard coral grouper Plectropomus leopardus. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.964774
He L., Zhang A., Xiong L., Li Y., Huang R., Liao L., et al. (2017). Deep circular RNA sequencing provides insights into the mechanism underlying grass carp reovirus infection. Int. J. Mol. Sci. 18, 1977. doi: 10.3390/ijms18091977
Henning F., Jones J. C., Franchini P., Meyer A. (2013). Transcriptomics of morphological color change in polychromatic Midas cichlids. BMC Genomics 14, 171. doi: 10.1186/1471-2164-14-171
Hochrainer K., Mayer H., Baranyi U., Binder B., Lipp J., Kroismayr R. (2005). The human HERC family of ubiquitin ligases: novel members, genomic organization, expression profiling, and evolutionary aspects. Genomics 85, 153–164. doi: 10.1016/j.ygeno.2004.10.006
Hoglund P. J., Nordstrom K. J., Schioth H. B., Fredriksson R. (2011). The solute carrier families have a remarkably long evolutionary history with the majority of the human families present before divergence of bilaterian species. Mol. Biol. Evol. 28, 1531–1541. doi: 10.1093/molbev/msq350
Holdt L. M., Kohlmaier A., Teupser D. (2018). Circular RNAs as therapeutic agents and targets. Front. Physiol. 9. doi: 10.3389/fphys.2018.01262
Hu Y., Li Y., Cheng P., Chen S. (2022). Comprehensive analysis of circular RNAs to decipher the potential roles in blind-side hypermelanosis in Chinese tongue sole (Cynoglossus semilaevis). Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.868987
Huang S., Yang B., Chen B., Bliim N., Ueberham U., Arendt T., et al. (2017). The emerging role of circular RNAs in transcriptome regulation. Genomics 109, 401–407. doi: 10.1016/j.ygeno.2017.06.005
Jeck W. R., Sorrentino J. A., Wang K., Slevin M. K., Burd. C. E., Liu. J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. doi: 10.1261/rna.035667.112
Jennings S., Stentiford G. D., Leocadio A. M., Jeffery K. R., Metcalfe J. D., Katsiadaki I., et al. (2016). Aquatic food security: insights into challenges and solutions from an analysis of interactions between fisheries, aquaculture, food safety, human health, fish and human welfare, economy and environment. Fish Fish. 17, 893–938. doi: 10.1111/faf.12152
Jiang L., Huang J., Hu Y., Lei L., Ouyang Y., Long Y., et al. (2020). Identification of the ceRNA networks in a-MSH-Induced melanogenesis of melanocytes. Aging 13, 2700–2726. doi: 10.18632/aging.202320
Kelsh R. N. (2004). Genetics and evolution of pigment patterns in fish. Pigment Cell Res. 1, 326–336. doi: 10.1111/j.1600-0749.2004.00174.x
Kennell J. A., Cadigan K. M., Shakhmantsir I., Waldron E. J. (2012). The MicroRNA miR-8 is a positive regulator of pigmentation and eclosion in drosophila. Dev. Dyn. 241, 161–168. doi: 10.1002/dvdy.23705
Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. doi: 10.1186/gb-2013-14-4-r36
Kotb A., Panda A. C., Supriyo D., Ioannis G., Jiyoung K., Jun D., et al. (2015). Circular RNAs in monkey muscle: age-dependent changes. Aging 7, 903–910. doi: 10.18632/aging.100834
Lasda E., Parker R. (2016). Circular RNAs co-precipitate with extracellular vesicles: a possible mechanism for circRNA clearance. PloS One 11, e0148407. doi: 10.1371/journal.pone.0148407
Leclercq E., Taylor J. F., Migaud H. (2010). Morphological skin colour changes in teleosts. Fish Fish. 11, 159–193. doi: 10.1111/j.1467-2979.2009.00346.x
Lei C., Li J. H., Zheng Z., Du X. D., Deng Y. W. (2019). Molecular cloning, expression pattern of β-carotene 15, 15-dioxygenase gene and association analysis with total carotenoid content in pearl oyster Pinctada fucata martensii. comp. Biochem. Physiol. B 229, 34–41. doi: 10.1016/j.cbpb.2018.11.006
Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., et al. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22, 256–264. doi: 10.1038/nsmb.2959
Liu B., Yuan R., Liang Z., Zhang T., Zhu M., Zhang X., et al. (2019). Comprehensive analysis of circRNA expression pattern and circRNA-mRNA-miRNA network in Ctenopharyngodon idellus kidney (CIK) cells after grass carp reovirus (GCRV) infection. Aquaculture 512, 734349. doi: 10.1016/j.aquaculture.2019.734349
Luo M., Lu G., Yin H., Wang L., Atuganile M., Dong Z. (2021). Fish pigmentation and coloration: molecular mechanisms and aquaculture perspectives. Rev. Aquacult. 13, 2395–2412. doi: 10.1111/raq.12583
Luo M. K., Wang L. M., Zhu W. B., Fu J. J., Song F. B., Fang M., et al. (2018). Identification and characterization of skin color microRNAs in koi carp (Cyprinus carpio l.) by illumina sequencing. BMC Genomics 19, 779. doi: 10.1186/s12864-018-5189-5
McGraw K. J. (2005). The antioxidant function of many animal pigments: are there consistent health benefits of sexually selected colourants? Anim. Behav. 69, 757–764. doi: 10.1016/j.anbehav.2004.06.022
Memczak S., Jens M., Elefsinioti A., Tprto F., Krueger J., Rybak A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. doi: 10.1038/nature11928
Poliseno L., Salmena L., Zhang J., Carver B., Haveman W. J., Pandolfi P. P. (2010). A coding independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038. doi: 10.1038/nature09144
Quan J., Kang Y., Luo Z., Zhao G., Li L., Liu Z. (2021). Integrated analysis of the responses of a circRNA-miRNA-mRNA ceRNA network to heat stress in rainbow trout (Oncorhynchus mykiss) liver. BMC Genomics 22, 48. doi: 10.1186/s12864-020-07335-x
Quinn J., Chang H. (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62. doi: 10.1038/nrg.2015.10
Rbbani G., Nedoluzhko A., Galindo-Villegas J., Fernandes J. M. O. (2021). Function of circular RNAs in fish and their potential application as biomarkers. Int. J. Mol. Sci. 22, 7119. doi: 10.3390/ijms22137119
Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P. P. (2011). A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell 146, 353–358. doi: 10.1016/j.cell.2011.07.014
Salzman J., Gawad C., Wang P. L., Lacayo. N., Brown P. O. (2012). Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One 7, e30733. doi: 10.1371/journal.pone.0030733
Smith G. E., Maytin A. K., Davies S. W., Mullen S. P., Buston P. M. (2018). De novo transcriptome assembly of the clown anemonefish (Amphiprion percula): a new resource to study the evolution of fish color. Front. Mar. Sci. 5. doi: 10.3389/fmars.2018.00284
Vissio P. G., Darias M. J., Di Yorio M. P., Perez Sirkin D. I., Delgadin T. H. (2021). Fish skin pigmentation in aquaculture: the influence of rearing conditions and its neuroendocrine regulation. Gen. Comp. Endocrinol. 301, 113662. doi: 10.1016/j.ygcen.2020.113662
Wang L., Zhu W., Dong Z., Song F., Dong J., Fu J. (2018a). Comparative microRNA-seq analysis depicts candidate miRNAs involved in skin color differentiation in red tilapia. Int. J. Mol. Sci. 19, 1209. doi: 10.3390/ijms19041209
Wang L., Zhu W., Yang J., Miao L., Dong J., Song J., et al. (2018b). Effects of dietary cystine and tyrosine on melanogenesis pathways involved in skin color differentiation of Malaysian red tilapia. Aquaculture 490, 149–155. doi: 10.1016/j.aquaculture.2018.02.023
Xiu Y., Jiang G., Zhou S., Diao J., Liu H., Su B., et al. (2019). Identification of potential immune-related circR-NA-miRNA-mRNA regulatory network in intestine of Paralichthys olivaceus during Edwardsiella tarda infection. Front. Genet. 10. doi: 10.3389/fgene.2019.00731
Xu X., Zhang J., Tian Y., Gao Y., Dong X., Chen. W., et al. (2020). CircRNA inhibits DNA damage repair by interacting with host gene. Mol. Cancer 19, 128. doi: 10.1186/s12943-020-01246-x
Yan B., Liu B., Zhu C. D., Li K. L., Yue L. J., Zhao J. L., et al. (2013). microRNA regulation of skin pigmentation in fish. J. Cell Sci. 126, 3401–3408. doi: 10.1242/jcs.125831
Yang Y., Wu L., Chen J., Wu X., Xia J., Meng Z., et al. (2020a). Whole-genome sequencing of leopard coral grouper (Plectropomus leopardus) and exploration of regulation mechanism of skin color and adaptive evolution. Zool. Res. 41, 328–340. doi: 10.24272/j.issn.2095-8137.2020.038
Yang C., Yang J., Hao R., Du X., Deng Y. (2020b). Molecular characterization of OSR1 in Pinctada fucata martensii and association of allelic variants with growth traits. Aquaculture 516, 734617. doi: 10.1016/j.aquaculture.2019.734617
Zhang X., Wang H., Zhang Y., Lu X., Chen L., Yang L. (2014). Complementary sequence-mediated exon circularization. Cell 159, 134–147. doi: 10.1016/j.cell.2014.09.001
Zhang Y., Zhang X., Chen T., Xiang J., Yin Q., Xing Y., et al. (2013). Circular intronic long noncoding RNAs. mol. Cell 51, 792–806. doi: 10.1016/j.molcel.2013.08.017
Zhou Q., Guo X. Y., Huang Y., Gao H. Y., Xu H., Liu S. S., et al. (2020). De novo sequencing and chromosomal-scale genome assembly of leopard coral grouper, Plectropomus leopardus. Mol. Ecol. Resour. 20, 1403–1413. doi: 10.1111/1755-0998.13207
Zhu X., Hao R., Tian C., Zhang J., Zhu C., Li G. (2021). Integrated transcriptomics and metabolomics analysis of body color formation in the leopard coral grouper (Plectropomus leopardus). Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.726102
Zhu Z., Li Y., Liu W., He J., Zhang L., Li H., et al. (2018). Comprehensive circRNA expression profile and construction of circRNA-associated ceRNA network in fur skin. Exp. Dermatol. 27, 251–257. doi: 10.1111/exd.13502
Zhu Z., Ma Y., Li Y., Li P., Cheng Z., Li H., et al. (2020). The comprehensive detection of miRNA, lncRNA, and circRNA in regulation of mouse melanocyte and skin development. Biol. Res. 53, 4. doi: 10.1186/s40659-020-0272-1
Zhu W., Wang L., Dong Z., Chen X., Song F., Liu N., et al. (2016). Comparative transcriptome analysis identifies candidate genes related to skin color differentiation in red tilapia. Sci. Rep. 6, 31347. doi: 10.1038/srep31347
Keywords: Plectropomus leopardus, circRNA, ceRNA, body color, pigmentation
Citation: Hao R, Zhu X, Tian C, Huang Y, Li G and Zhu C (2023) Identification and characterization of circRNAs from different body color leopard coral grouper (Plectropomus leopardus). Front. Mar. Sci. 10:1201726. doi: 10.3389/fmars.2023.1201726
Received: 11 April 2023; Accepted: 10 May 2023;
Published: 25 May 2023.
Edited by:
Adriana Muhlia, National Council of Science and Technology (CONACYT), MexicoReviewed by:
Yong Zhang, Sun Yat-sen University, ChinaCarlos Alfonso Alvarez-González, Universidad Juárez Autónoma de Tabasco, Mexico
Copyright © 2023 Hao, Zhu, Tian, Huang, Li and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunhua Zhu, Y2h6NDE2QDE2My5jb20=
 Ruijuan Hao
Ruijuan Hao Xiaowen Zhu
Xiaowen Zhu Changxu Tian
Changxu Tian Yang Huang1,2
Yang Huang1,2 Guangli Li
Guangli Li Chunhua Zhu
Chunhua Zhu