- 1School of Marine Sciences, Nanjing University of Information Science & Technology, Nanjing, Jiangsu, China
- 2Jiangsu Province Engineering Research Center for Aquatic Animals Breeding and Green Efficient Aquacultural Technology, College of Marine Science and Engineering, Nanjing Normal University, Nanjing, Jiangsu, China
- 3Freshwater Fisheries Research Institute of Jiangsu Province, Nanjing, Jiangsu, China
Hepatopancreatic necrosis disease (HPND) is a serious threat to the stable and sustainable development of the Chinese mitten crab farming industry. Studies have found that the hepatopancreas is the main lesion organ in HPND crabs, capable of causing metabolic abnormalities and non-feeding (starvation state). Here, four amylases (named as EsAmy1-4) and four cellulases (endo-beta-1, 4-glucanase, named as EsGlu1-4) that relate to carbohydrate metabolism were cloned from Eriocheir sinensis. Our studies found that the expressions of EsAmy1-4 and EsGlu1-4 were significantly decreased in the hepatopancreas of crabs with HPND. Moreover, the expressions of two important regulatory factors of carbohydrate metabolism, hormone receptor 38 (EsHR38) and ecdysone receptor (EsEcR), were remarkably decreased in the hepatopancreas of crabs with HPND. Further studies found that knockdown of EsHR38 and EsEcR could down-regulated the expressions of EsAmy1-4 and EsGlu1-4. In addition, downregulation of EsEcR could decrease the expression of EsHR38. Hence, HPND causes down-regulation of EsEcR expression, inhibits the transcription of EsHR38, further inhibits the expression of EsAmy1-4 and EsGlu1-4, and then leads to the abnormal carbohydrate metabolism. The abnormalities of carbohydrate metabolism may result in the abnormal food intake in crabs. This study provides new evidence for further understanding the mechanism of HPND.
1 Introduction
Chinese mitten crab, Eriocheir sinensis, is an important economic breeding species with high nutritional value, delicious taste, and rapid growth (Cheng et al., 2008). In the middle and lower reaches of the Yangtze River, especially in Jiangsu of China, Chinese mitten crabs are widely farmed (Dai et al., 2021). However, the expansion of crab farming scale and the increase of farming density has caused the outbreak of various diseases, such as tremor disease (TD) (Huang et al., 2019), edema disease, black gill disease (Wang, 2011), hepatopancreatic necrosis disease (HPND), and so on. HPND, commonly known as “shui biezi”, has received a lot of attention since its large-scale outbreak in Xinghua, Jiangsu Province in 2015 because of the huge economic losses it causes (Yang et al., 2016). There are different opinions on the possible causes of HPND, including the microsporidia fungus (Ding et al., 2016), abnormal fatty acid metabolism, high concentration of alanine (Gao et al., 2018), imbalance of hepatopancreatic flora (Shen et al., 2021), abiotic factors related to cyanobacteria bloom (Chi et al., 2021), environment and nutrition (Huang et al., 2021). Until now, there is no unanimous conclusion about the cause of HPND. Therefore, the cause and mechanism of HPND need to be urgently explored to ensure healthy crab production.
The main target and lesion organ of HPND is the hepatopancreas, which is an important digestive and immune organ of crustaceans with high digestive enzyme activity (Yang et al., 2018; Huang et al., 2021). Hepatopancreatic lesions are characterized by degeneration and atrophy, and the color changes from gold to light yellow and white (Pan et al., 2017). Studies have also found that there was abnormal nutrient metabolism in hepatopancreas and signs of non-eating and starvation in crabs with HPND (Ding et al., 2018; Yang et al., 2020). However, how HPND affects carbohydrate metabolism is not clear. Amylase and cellulase are two common digestive enzymes in carbohydrate metabolism, in which α-amylase is responsible for the hydrolysis of α-1,4-glucosidic bonds in starch and related α-glucans (Janecek et al., 2014), and endo-β-1, 4-glucanase is responsible for the hydrolysis of cellulose like β-1,4-glucans (Watanabe and Tokuda, 2001). In the current study, we explored the effects of HPND on the expression of these two kinds of digestive enzymes for explaining the influences of HPND on hepatopancreas carbohydrate metabolism.
Studies have found that the carbohydrate metabolism can be regulated by the 20-hydroxyecdysone (20E), which is an insect steroid hormone that involves in the regulation of molting (Colombani et al., 2005; Kh and Keshan, 2021). 20E is able to induce hunger by slowly reducing food consumption (Liu et al., 2010), and can inhibit the transcription of key glycolytic enzymes (Tian et al., 2010). 20E-regulated gene expression is accomplished by ecdysone receptor (EcR), where EcR is a homolog of the vertebrate FXR/LXR receptor (Riddiford et al., 2000; King-Jones and Thummel, 2005). EcR belongs to the nuclear receptor superfamily and has been proved to be a key regulator of carbohydrate metabolism in mosquitoes (Dong et al., 2018). Therefore, whether the expressions of amylase and cellulase genes are regulated by EsEcR in E. sinensis is studied to reveal the possible roles of EcR in hepatopancreas carbohydrate metabolism.
The NR4A subfamily is the orphan nuclear receptor and belongs to the larger nuclear receptor (NRs) superfamily of eukaryotic transcription factors (Ranhotra, 2015). The NR4A subfamily includes three members, namely Nur77 (NR4A1), Nurr1 (NR4A2) and Nor1 (NR4A3) (Kurakula et al., 2014), which have been shown to be transcriptional regulators of hepatic glucose metabolism (Pei et al., 2006). In arthropods, hormone receptor 38 (HR38) is the only homolog of mammalian NR4As (Nakagawa and Henrich, 2009). Studies have shown that HR38 plays a key role in carbohydrate metabolism and is evolutionarily conserved (Ruaud et al., 2011). We studied the effects of HR38 on the expression of amylase and cellulase for clarifying the roles of HR38 in carbohydrate metabolism. In addition, HR38 has been suggested to mediate the 20E/EcR/USP signaling cascade that regulates carbohydrate metabolism in response to 20E (Dong et al., 2018). Hence, we further explored the possible role of EcR in the expression of HR38.
In this study, four amylases (EsAmy1-4), four cellulases (endo-beta-1, 4-glucanase, EsGlu1-4), one EcR gene (EsEcR), and one HR38 gene (EsHR38) from E. sinensis were identified. Quantitative Real-Time PCR (RT-qPCR) was used to analyze the tissue distribution of EsAmy1-4 and EsGlu1-4. Moreover, the expression levels of EsAmy1-4, EsGlu1-4, EsEcR, and EsHR38 in the hepatopancreas of crabs with HPND were detected by RT-qPCR. RNA interference (RNAi) was utilized to study the effects of EsEcR and EsHR38 knockdown on the expressions of EsAmy1-4 and EsGlu1-4 in the hepatopancreas. Furthermore, we explored whether EsEcR has a regulatory effect on the transcription of EsHR38 by using RNAi. This study will reveal the effects of HPND on the carbohydrate metabolism, and provide new knowledge of HPND.
2 Materials and methods
2.1 Experimental animals and tissues collection
Crabs with HPND (10 ± 2 g each) were obtained from crab farming base in Xinghua, Jiangsu Province, China. The hepatopancreas of sick or healthy crabs were collected for subsequent RNA extraction. Healthy crabs (12 ± 2 g each) were provided by Freshwater Fisheries Research Institute of Jiangsu Province, Nanjing, China, and temporary farming in an aerated water tank filled with freshwater at 25 °C-26 °C for 7 days before processing. Six kinds of tissues, including hemocytes, heart, hepatopancreas, gills, stomach, and intestine, were collected from three random healthy crabs. For hemocytes collection, the hemolymph was extracted from three crabs and placed in an equal volume of precooled anticoagulant solution (glucose, 1.47 g; citric acid, 0.48 g; trisodium citrate, 1.32 g; prepared in ddH2O and added to 100 mL, pH 7.3). The mixture was centrifuged at 4 °C, 2000 rpm for 10 min to precipitate the hemocytes. All the tissue samples were stored briefly at –80 °C before RNA extraction.
2.2 Total RNA extraction and cDNA synthesis
Total RNA was extracted from each collected sample using the High Purity Total RNA Rapid Extraction Kit (BioTeke, Beijing, China) in accordance with the manufacturer’s instructions. The quality of RNA was evaluated by 1% agarose gel electrophoresis. The concentration of RNA was obtained by measuring the absorbance at a wavelength of 260:280 nm (OD260/OD280 = 1.8-2.0) using Nanodrop 2000 (Thermo Fisher Scientific, USA). About 1 μg of total RNA was used to synthesize the first-strand cDNA using TransScript® All-In-one First Strand cDNA Synthesis SuperMix (One-step gDNA removal) (Transgen Biotech, Beijing, China) in accordance with the manufacturer’s instructions. Extracted RNA and synthesized cDNA were stored at -80 °C and -20 °C, respectively.
2.3 Bioinformatics analysis
After searching the transcriptome data of crab hepatopancreas, EsAmy1-4, EsGlu1-4, EsHR38 and EsEcR were found. The homologs of EsAmy1-4 and EsGlu1-4 were acquired by the basic local alignment search tool (BLAST) (http://www.ncbi.nlm.nih.gov/BLAST/). The amino acids composition of EsAmy1, 2, 4 and EsGlu1-4 were obtained using an Expert Protein Analysis System (https://web.expasy.org/translate/). The protein structures of EsAmy1, 2, 4 and EsGlu1-4 were predicted using a Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de/). Phylogenetic tree was built by the neighbor-joining (NJ) algorithm using MEGA 7.0 software.
2.4 Tissue distribution analysis of EsAmy1-4 and EsGlu1-4
The expression levels of EsAmy1-4 and EsGlu1-4 in the hemocytes, heart, hepatopancreas, gills, stomach, and intestine of healthy crabs were analyzed. Specific EsAmy1-4-qF/qR and EsGlu1-4-qF/qR primers (Table 1) were designed for tissue distribution analysis by performing RT-qPCR using TransStart®Top Green qPCR SuperMix kit (TransGen Biotech, China). The reaction system included 5 μL of 2×TransStart Top Green qPCR SuperMix, 0.4 μL of (10 mM) each qF and qR primers, 1 μL of cDNA template, and 3.2 μL of PCR-grade Water. PCR procedure was performed as follows: 95 °C for 60 s, 1 cycle; 95 °C for 15 s, 60 °C for 30 s, 40 cycles. Melting curve analysis was performed from 60°C to 95°C. The β-actin gene in E. sinensis was amplified as internal reference using Esβ-actin-qF and Esβ-actin-qR primers (Table 1).
2.5 Expression levels of EsAmy1-4, EsGlu1-4, EsHR38, and EsEcR in the crabs with or without HPND
The expression levels of EsAmy1-4, EsGlu1-4, EsHR38, and EsEcR in the hepatopancreas of carbs with or without HPND were detected by RT-qPCR. Samples from healthy crabs were set as control. Esβ-actin gene was amplified as an internal reference. Specific RT-qPCR primers, including EsAmy1-4-qF/qR, EsGlu1-4-qF/qR, EsHR38-qF/qR, EsEcR-qF/qR, and Esβ-actin-qF/qR were listed in Table 1. All experiments were repeated three times, and the data were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Student’s t-test was conducted for statistical analysis, and p < 0.05 was considered statistically significant.
2.6 RNAi of EsEcR or EsHR38 and detection of EsAmy1-4 and EsGlu1-4
Gene-specific EsHR38-iF/iR and EsEcR-iF/iR primers were designed to amplify the fragment of EsHR38 and EsEcR, respectively. The purified fragments were used as templates to synthesize the EsHR38-dsRNA and EsEcR-dsRNA by using HiScribe T7 Quick High-yield RNA Synthesis Kit (BioLabs, USA) in accordance with the instruction manual. The dsRNA of green fluorescent protein (GFP) was synthesized as control by using GFP-iF and GFP-iR primers (Table 1). The quality and concentration of dsRNA were separately assessed by electrophoresis on 1% agarose gel and the absorbance from Nanodrop 2000 (Thermo Fisher Scientific, USA) at a wave-length of 260:280 nm (OD260/OD280 = 1.8-2.0). The RNAi experiments were divided into three groups, including the EsHR38-dsRNA-injection, EsEcR-dsRNA-injection, and GFP-dsRNA-injection groups. 20 μg of EsHR38-dsRNA, EsEcR-dsRNA or GFP-dsRNA was injected into the healthy crabs. After 24 h, 20 μg of EsHR38-dsRNA, EsEcR-dsRNA or GFP-dsRNA was injected into the same crab. At 24 h after the second dsRNA injection, the hepatopancreas from three random crabs in each group were collected for RNA extraction and cDNA synthesis. The expression level of EsHR38 in the hepatopancreas of EsHR38-dsRNA-injected and GFP-dsRNA-injected crabs were detected by RT-qPCR. The expression level of EsEcR or EsHR38 in the hepatopancreas of EsEcR-dsRNA-injected and GFP-dsRNA-injected crabs were detected by RT-qPCR. The expression levels of EsAmy1-4 and EsGlu1-4 in the hepatopancreas after knockdown of EsHR38 or EsEcR were detected by RT-qPCR.
3 Results
3.1 Sequence characters and evolutionary analysis of amylases in crabs
The full-length cDNA of EsAmy1 was 1909 bp with a 1554 bp open reading frame (ORF) that encodes a protein containing 517 amino acids (Figure S1A). The full-length cDNA of EsAmy2 was 2541 bp with a 2169 bp ORF that encodes a protein containing 722 amino acids (Figure S1B). The partial cDNA of EsAmy3 was 1637 bp (Figure S1C). The full-length cDNA of EsAmy4 was 2560 bp with a 2112 bp ORF that encodes a protein containing 703 amino acids (Figure S1D). Phylogenetic analysis showed that four amylases form two groups. Among which, EsAmy2-4 and alpha-amylases from arthropods formed one group, whereas EsAmy1 and alpha-amylases from arthropods formed another group (Figure 1A). SMART prediction showed that EsAmy1, EsAmy2, and EsAmy4 all contain signal peptide, Aamy domain, and Aamy_C domain. Besides, EsAmy2 and EsAmy4 contained the low complexity regions (Figure 1B).
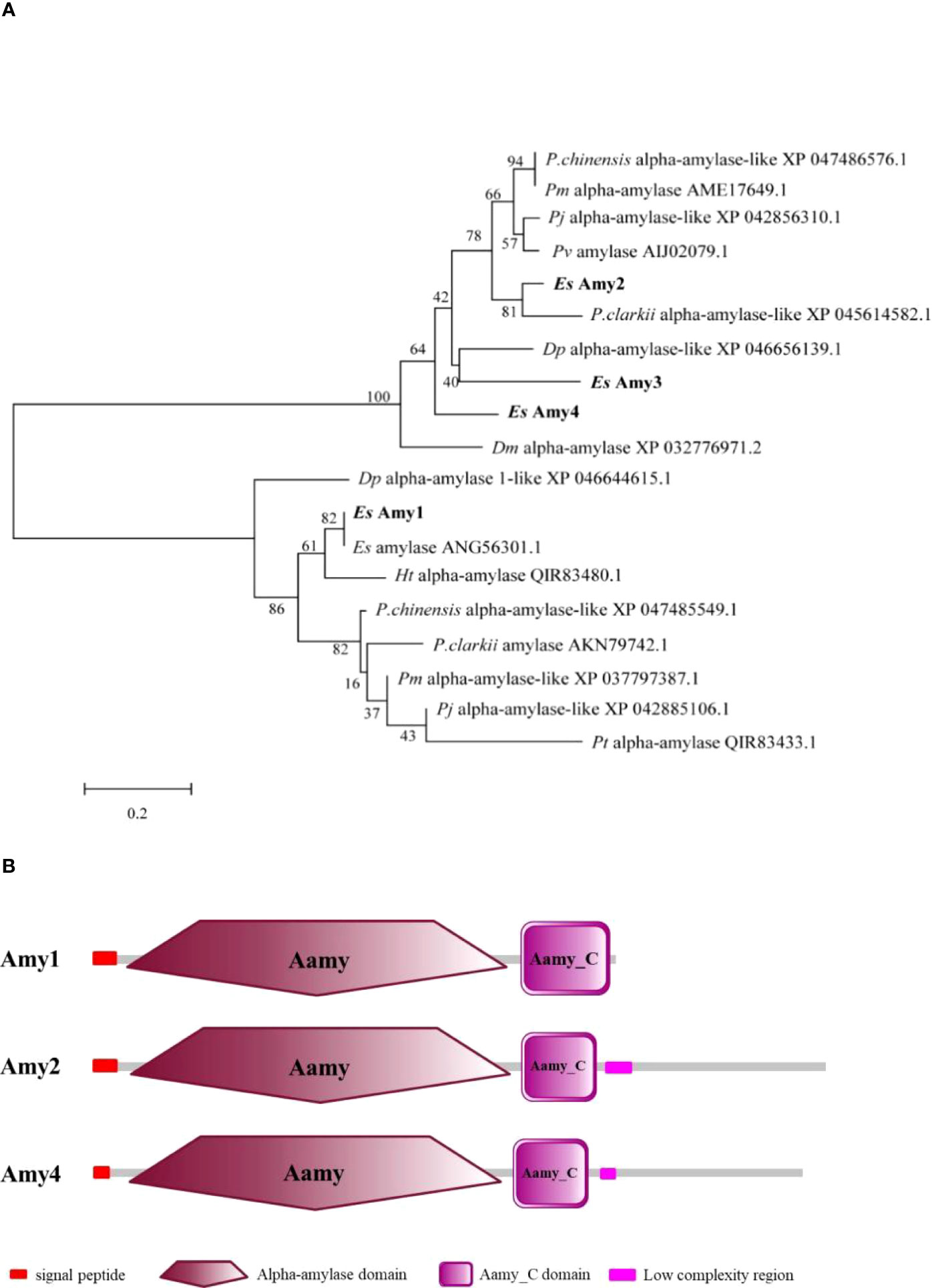
Figure 1 Evolutionary analysis and protein domains prediction of amylases. (A) Evolutionary analysis of EsAmy1-4 and their homologs was performed by MEGA 7.0. The EsAmy1-4 are shown in bold. Abbreviation of species, protein name, and GenBank accession number were provided in the evolutionary tree. The full name of each species name is listed as follows: P. chinensis (Penaeus chinensis), Pm (Penaeus monodon), Pj (Penaeus japonicas), Pv (Penaeus vannamei), P.clarkii (Procambarus clarkia), Dm (Daphnia magna), Dp (Daphnia pulicaria), Ht (Helice tientsinensis), Pt (Portunus trituberculatus), and Es (E. sinensis). (B) Protein structure of EsAmy1, EsAmy2 and EsAmy4. Signal peptide, Alpha-amylase domain (Aamy), Aamy_C domain, and Low complexity region are respectively marked in red box, dark red polygon, purple box, and pink box.
3.2 Identification and bioinformatics analysis of cellulases in crabs
The full-length cDNA of EsGlu1 was 2014 bp with a 1377 bp ORF that encodes a protein containing 458 amino acids (Figure S2A). The partial cDNA of EsGlu2 was 2202 bp (Figure S2B). The full-length cDNA of EsGlu3 was 1840 bp with a 1677 bp ORF that encodes a protein containing 558 amino acids (Figure S2C). The full-length cDNA of EsGlu4 was 2269 bp with a 1722 bp ORF that encodes a protein containing 573 amino acids (Figure S2D). As shown in the Figure 2A, there are two branches in the phylogenetic tree. Among which, endo-beta-1, 4-glucanases from termites form one branch, whereas EsGlu1-4 and endoglucanases from crabs form another branch. The protein structures of four cellulases all contain a Glyco_hydro_9 domain, which has hydrolase activity (Figure 2B). In addition, EsGlu1, EsGlu3, and EsGlu4 contain a signal peptide, whereas EsGlu2 lacks a signal peptide because of incomplete sequences.
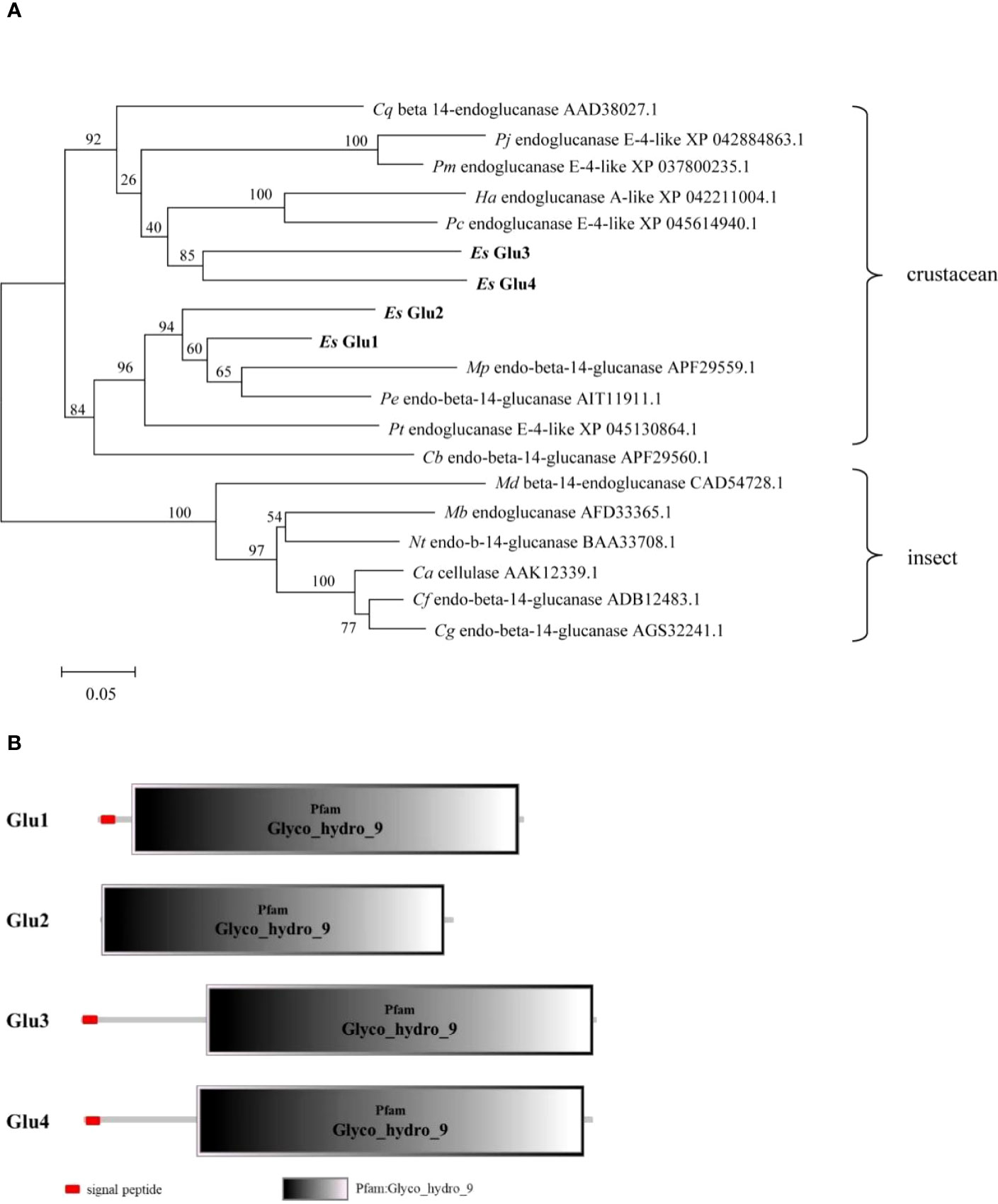
Figure 2 Evolutionary analysis and protein domains prediction of cellulase. (A) Evolutionary analysis of EsGlu1-4 and their homologs was performed by MEGA 7.0. The EsAmy1-4 are shown in bold. Abbreviation of species, protein name, and GenBank accession number were provided in the evolutionary tree. The full name of each species name is listed as follows: Cq (Cherax quadricarinatus), Pj (P. japonicas), Pm (P. monodon), Pc (P. chinensis), Ha (Homarus americanus), Es (E. sinensis), Mp (Mictyris platycheles), Pe (Parasesarma erythrodactyla), Pt (Portunus trituberculatus), Cb (Coenobita brevimanus), Md (Mastotermes darwiniensis), Mb (Macrotermes barneyi), Nt (Nasutitermes takasagoensis), Ca (Coptotermes acinaciformis), Cf (Coptotermes formosanus), and Cg (Coptotermes gestroi). (B) The protein structures of EsGlu1-4. Signal peptide and Glyco_hydro_9 domain are marked in red box and gray box.
3.3 The expressions of amylases, cellulases, HR38, and EcR were inhibited in the crabs with HPND
As shown in Figures 3A–H, four amylases (EsAmy1-4) and four cellulases (EsGlu1-4) were mainly expressed in the hepatopancreas and intestine, and nearly not expressed in the hemocytes, heart, gills, and stomach. The highest expression level in hepatopancreas of EsAmy1-4 and EsGlu1-4 suggested that the hepatopancreas is the major organ for producing amylases and cellulases. RT-qPCR was used to explore the effects of HPND on the expressions of amylases, cellulases, HR38, and EcR. The results showed that the expression levels of four amylases (EsAmy1-4) (Figures 4A–D), four cellulases (EsGlu1-4) (Figures 4E–H), EsHR38 (Figure 4I) and EsEcR (Figures 4E–J) in the hepatopancreas of crabs with HPND were significantly decreased than that in the normal crabs (as control). These findings indicate that the expressions of amylases, cellulases, HR38, and EcR in the hepatopancreas were inhibited in the crabs with HPND.
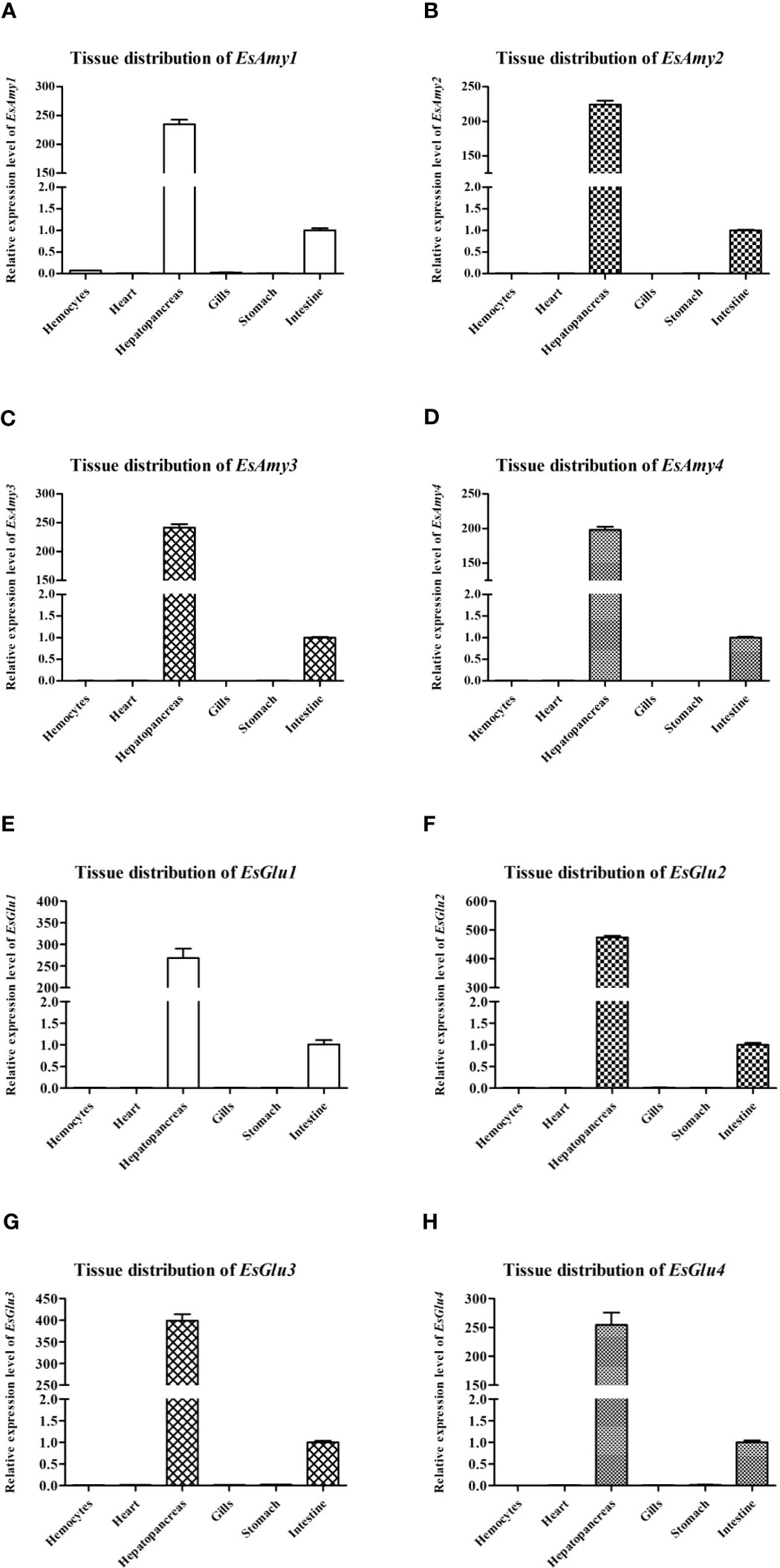
Figure 3 The expression levels of (A-D) amylases (EsAmy1-4) and (E-H) cellulases (EsGlu1-4) in multiple tissues of healthy crabs. The tested tissues included the hemocytes, heart, hepatopancreas, gills, stomach, and intestine. Esβ-Actin gene was amplified as an internal reference.
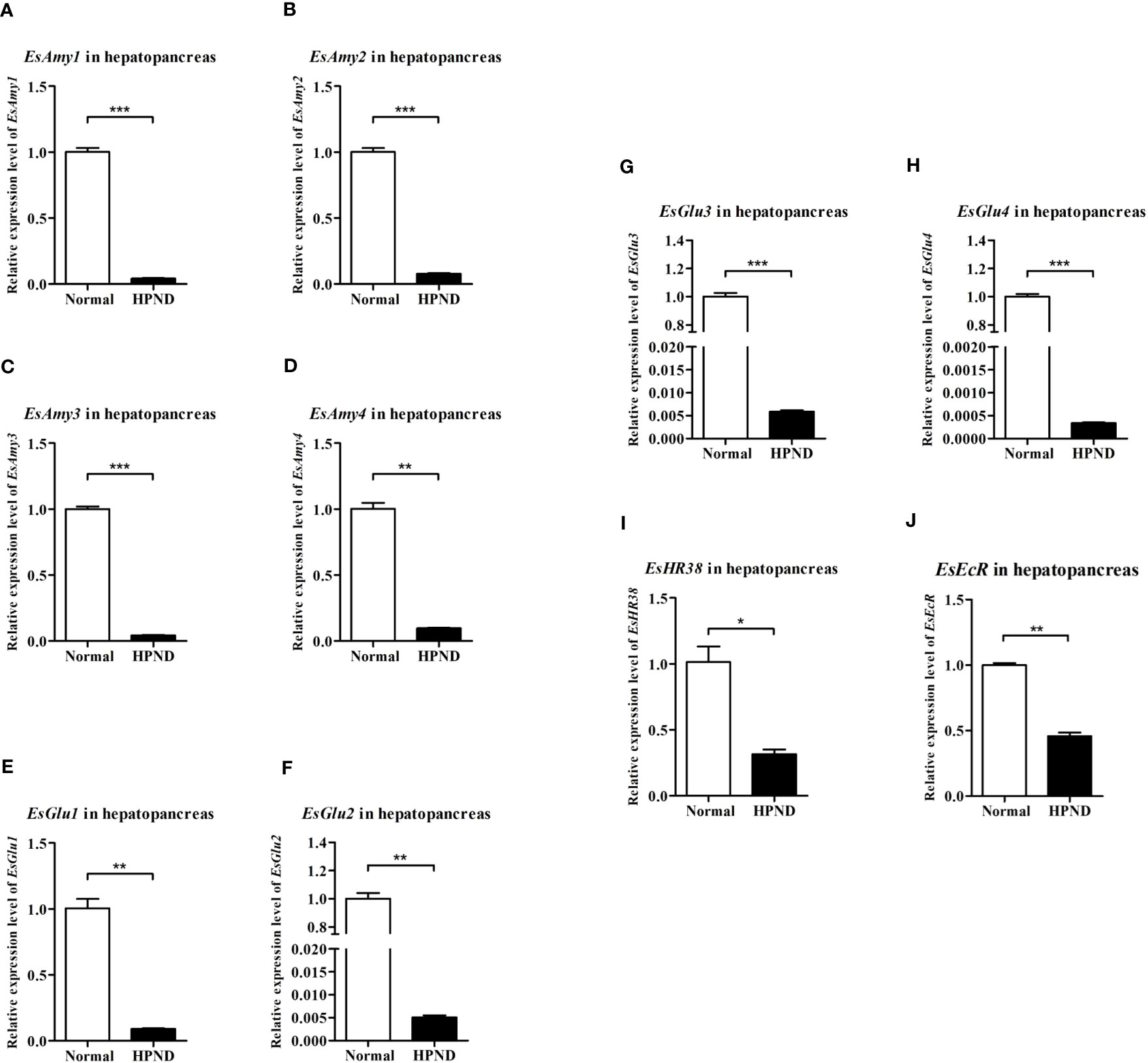
Figure 4 The expression levels of (A-D) amylases (EsAmy1-4), (E-H) cellulases (EsGlu1-4), (I) EsHR38, and (J) EsEcR in the hepatopancreas of crabs with or without HPND. Sample from healthy crabs (Normal group) was set as control. Asterisks indicate significant differences compared with the values of the control (*p < 0.05, **p < 0.01, and ***p<0.001). The data represents the mean ± SD of three biological repeats.
3.4 RNAi of EsHR38 or EsEcR inhibited the expression of amylases and cellulases
DsRNA-mediated RNAi was utilized to study the relationship between EsHR38 or EsEcR and the production of amylases and cellulases. As shown in Figure 5A, specific EsHR38-dsRNA injection could remarkably reduce the transcription of EsHR38 gene in the hepatopancreas of healthy crabs. Further RT-qPCR detection found that knockdown of EsHR38 significantly downregulate the expressions of EsAmy1-4 (Figure 5B) and EsGlu1-4 (Figure 5C) in the hepatopancreas of healthy crabs. In addition, knockdown of EsEcR (Figure 6A) could also evidently decrease the expressions of EsAmy1-4 (Figure 6B) and EsGlu1-4 (Figure 6C) in the hepatopancreas of healthy crabs. These results suggest that EsHR38 and EsEcR play positive regulatory roles in the synthesis of amylases and cellulases in the hepatopancreas.
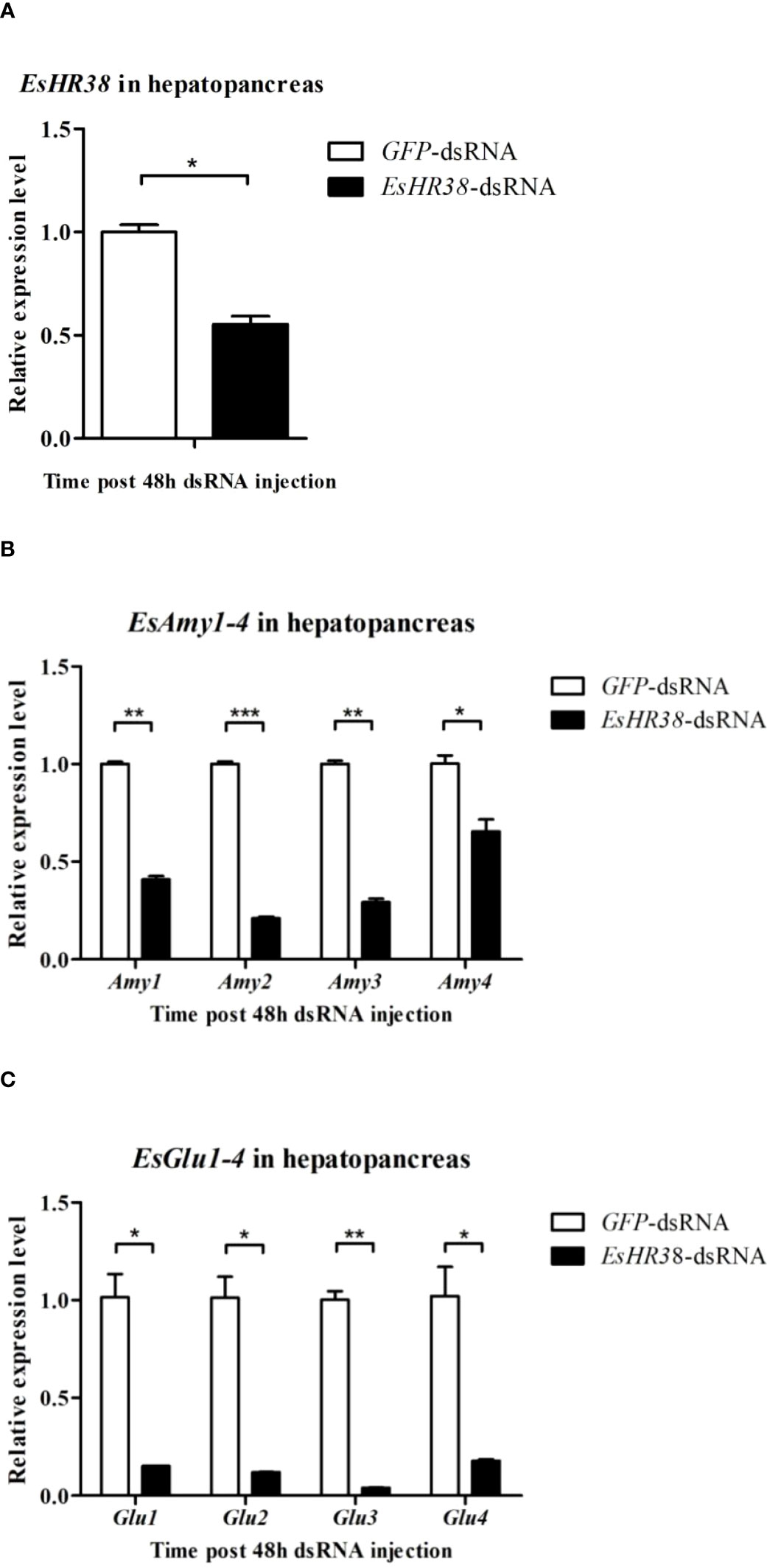
Figure 5 Knockdown of EsHR38 gene inhibited the expressions of amylases and cellulases in the hepatopancreas of healthy crabs. (A) The expression level of EsHR38 in the hepatopancreas of EsHR38-dsRNA-injected and GFP-dsRNA-injected crabs. (B) The expression levels of EsAmy1-4 in the hepatopancreas of EsHR38-dsRNA-injected and GFP-dsRNA-injected crabs. (C) The transcriptional levels of EsGlu1-4 in the hepatopancreas of EsHR38-dsRNA-injected and GFP-dsRNA-injected crabs. Injection of GFP-dsRNA was set as control group. Three random crabs were selected to eliminate individual differences. Asterisks indicate the significant differences (*p < 0.05, **p < 0.01, and ***p<0.001) compared with the values of the control. The data represents the mean ± SD of three biological repeats.
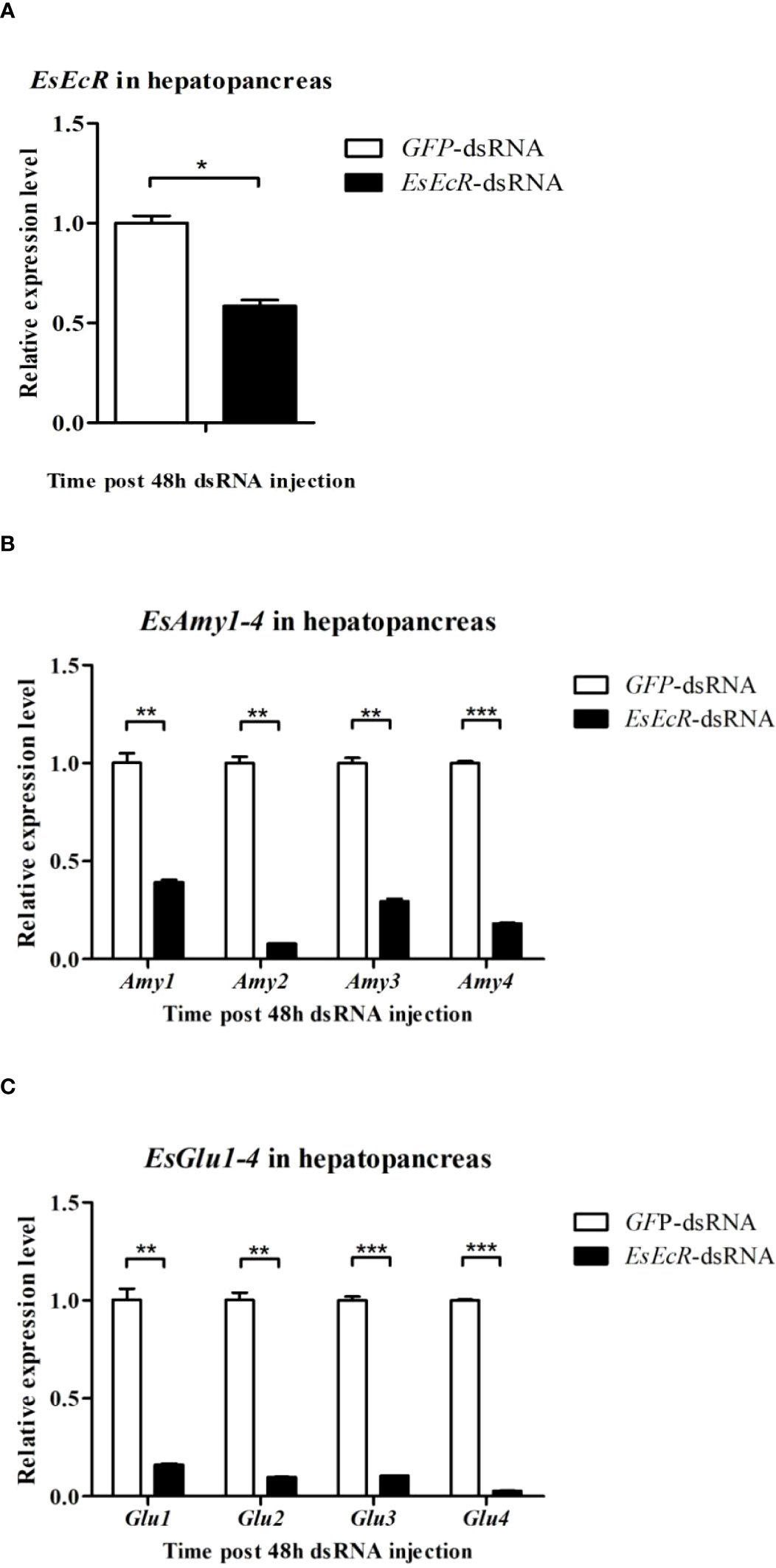
Figure 6 Knockdown of EsEcR gene inhibited the expressions of amylases and cellulases in the hepatopancreas of healthy crabs. (A) The expression level of EsEcR in the hepatopancreas of EsEcR-dsRNA-injected and GFP-dsRNA-injected crabs. (B) The expression levels of EsAmy1-4 in the hepatopancreas of EsEcR-dsRNA-injected and GFP-dsRNA-injected crabs. (C) The expression levels of EsGlu1-4 in the hepatopancreas of EsEcR-dsRNA-injected and GFP-dsRNA-injected crabs. Injection of GFP-dsRNA was set as control group. Three random crabs were selected to eliminate individual differences. Asterisks indicate the significant differences (*p < 0.05, **p < 0.01, and ***p<0.001) compared with the values of the control. The data represents the mean ± SD of three biological repeats.
3.5 Knockdown of EsEcR inhibited the expression of EsHR38
There are studies showing that HR38 and EcR are linked (Baker et al., 2003; Dong et al., 2018). However, the relationship between them is unclear. In this study, we studied the effect of knockdown of EsEcR on the expression of EsHR38 in the hepatopancreas of healthy crabs. The result showed that the expression level of EsHR38 in the hepatopancreas of EsEcR-dsRNA-injected crabs was significantly decreased compared with that of GFP-dsRNA-injected crabs (Figure 7), indicating that EsEcR positively regulates the expression of EsHR38 in the hepatopancreas.
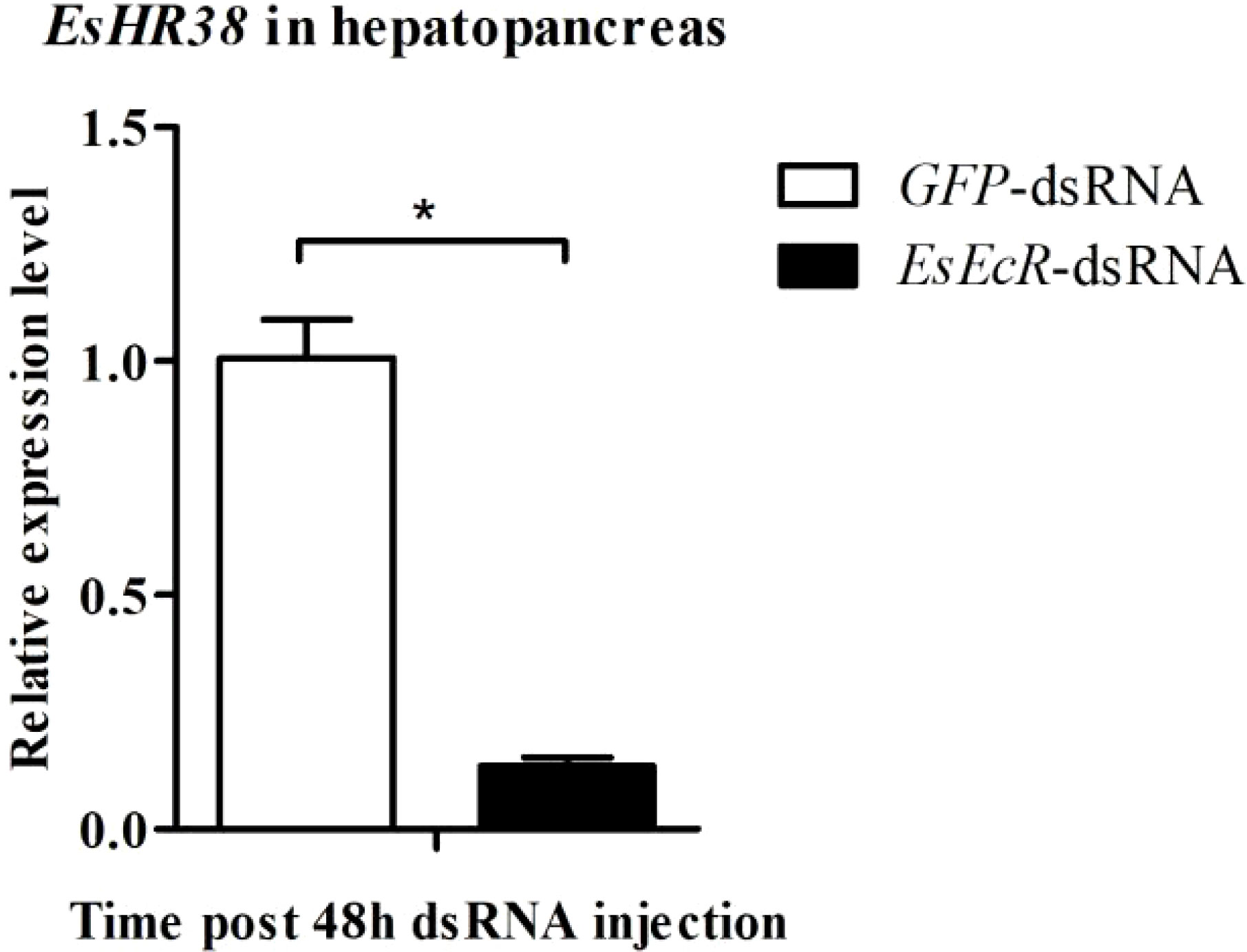
Figure 7 RNAi of EsEcR inhibited the expression of EsHR38 in the hepatopancreas of healthy crabs. Samples from the GFP-dsRNA injection group were used as control. Three random crabs were selected to eliminate individual differences. Asterisks indicate the significant differences (*p < 0.05) compared with the values of the control. The data represents the mean ± SD of three biological repeats.
4 Discussion
In decapods, glucose homeostasis is necessary to maintain normal organ function and response to external stimuli (Asaro et al., 2018). The hepatopancreas is an important organ for glucose homeostasis and the main site of digestion (Asaro et al., 2018; Wang et al., 2020). The hepatopancreatic F-cells are the synthesis site of digestive enzymes including amylases and cellulases (Vogt, 2019; Vogt, 2021). Amylase and cellulase are present in the hepatopancreas of many decapod species. And, some types of digestive enzymes in the hepatopancreas are more active than those in the gastric juice (Toullec et al., 1992; Linton et al., 2006). In this study, four amylases (named as EsAmy1-4) and four cellulases (named as EsGlu1-4) were identified from E. sinensis. Tissue distribution analysis showed that EsAmy1-4 and EsGlu1-4 were highly expressed in the hepatopancreas. HPND threatens the healthy farming of crabs, and the main diseased organ of crabs with HPND is the hepatopancreas (Huang et al., 2021). Our studies found that the expression levels of EsAmy1-4 and EsGlu1-4 in the hepatopancreas of crabs with HPND were significantly decreased than that of healthy crabs, indicating that the occurrence of HPND inhibits the synthesis of amylases and cellulases. Since amylase and cellulase are closely related to carbohydrate metabolism, HPND may lead to imbalance of carbohydrate metabolism by inhibiting the synthesis of amylases and cellulases.
HR38 is a homologue of NR4As, whereas NR4As is a subfamily of orphan nuclear receptors, which contain the conserved zinc finger DNA binding domain (DBD) and C-terminal ligand binding domain (LBD) (Ruaud et al., 2011). Moreover, the family of NR4As plays a key role in carbohydrate metabolism, especially in the glucose transport and glucose utilization (Fu et al., 2007; Zhang et al., 2020). In mice, NR4A receptor has been identified as a downstream mediator of cyclic adenosine monophosphate (cAMP) in gluconeogenic control (Pei et al., 2006), in which NR4A1(Nur77) can affect the glucose metabolism in skeletal muscle and hepatopancreas (Chao et al., 2007; Chao et al., 2009). In Drosophila, NR4A homologue Dhr38 plays a key role in carbohydrate metabolism, such as glycogen synthesis (Nakagawa and Henrich, 2009; Ruaud et al., 2011). The research found that the expression of Dhr38 was up-regulated in the fed larvae and down-regulated in starvation (Ruaud et al., 2011). In our study, the expression level of EsHR38 was significantly down-regulated in the crabs with HPND. In mosquitoes, HR38 was shown to affect the expression of key carbohydrate metabolizing enzyme genes (Dong et al., 2018). Similarly, our study also found that EsHR38 could positively regulate the expressions of amylases and cellulases in E. sinensis. These findings suggest that HR38 may influence the carbohydrate metabolism by regulating the synthesis of amylases and cellulases in crabs. The decreased expression of EsHR38 in the crabs with HPND may be one of the causes of carbohydrate metabolic abnormalities.
Ecdysone receptor (EcR) is a member of nuclear receptor superfamily, which is composed of N-terminal A/B domain, the DNA binding C domain (DBD), the D domain (hinge region), ligand binding E domain (LBD), and C-terminal F domain (Mangelsdorf et al., 1995). As a ligand-controlled transcription factor (Panguluri et al., 2006), EcR forms heterodimers with ultraspiracle (USP) (Palli et al., 2005) and participates in key biological processes such as development and reproduction (Seliga et al., 2013). Study found that EcR is involved in the regulation of carbohydrate metabolism, and the silence of EcR can lead to the reduction of transcription level of genes involved in carbohydrate metabolism (Hou et al., 2015). In Drosophila, EcR can bind to estrogen-related receptor (ERR) and participate in the regulation of carbohydrate metabolism (Kovalenko et al., 2019). In the current study, RT-qPCR analysis showed that the expression of EsEcR was significantly decreased in the crabs with HPND, indicating that the occurrence of HPND inhibits the expression of EsEcR in crabs. Further studies found that knockdown of EsEcR remarkably down-regulated the expressions of amylases and cellulases in healthy crabs, suggesting that EsEcR plays a positive role in the synthesis of amylases and cellulases. These findings suggest that HPND may lead to carbohydrate metabolic abnormalities by inhibiting the expression of EcR. Finally, the production of amylases and cellulases in crabs was inhibited in HPND crabs.
Some studies have shown that HR38 is a target gene of EcR (Dong et al., 2018), and there is a strong interaction between HR38 and USP (Sutherland et al., 1995). In Drosophila, it has been confirmed that HR38 can form a complex with USP and its DBD can interact with EcR (Pakula et al., 2012). Furthermore, the hinge region of EcR can mediate the formation of specific complexes between EcR and HR38 (Zoglowek et al., 2012). Therefore, HR38 may modulate its activity by forming complexes with two components of the USP-ECR heterodimer (Zoglowek et al., 2012). In our study, knockdown of EsEcR could significantly decrease the expression of EsHR38, indicating that EcR paly a positive regulatory role in the expression of HR38 in E. sinensis. So, we can conclude that HPND causes the down-regulated expression of EsEcR. Down-regulation of EsEcR inhibits the transcription of EsHR38, and further leads to the reduced production of amylases (EsAmy1-4) and cellulases (EsGlu1-4) in crabs. The abnormal expressions of amylases and cellulases in crabs with HPND may destroy the normal carbohydrate metabolism and result in the abnormal feeding of crabs, which may be the reason why HPND crabs do not eat and are in a hungry state.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
YZ: Conceptualization, Methodology, Formal analysis, Investigation, Data Curation, and Writing—Original Draft. XM: Validation, Formal analysis, Investigation, and Data Curation. QR: Conceptualization, Supervision, Writing—review & editing. XH: Conceptualization, Supervision, Resources, and Writing—review & editing. JG: Conceptualization, Supervision, Resources, and Writing—review & editing. All authors contributed to the article and approved the submitted version.
Funding
The current study was supported by the Jiangsu Provincial Department of Science and Technology (BE2019393), the National Natural Science Foundation of China (Grant Nos. 31902397), the Ecological Environment Scientific Research Project of Jiangsu Province (No. JSZC-G2021-291), the Interdisciplinary Research Project of Nanjing Normal University (No. 164320H1847), the Natural Science Foundation of Jiangsu Province (BK20190698), and the Start-up Fund of Research of Nanjing Normal University (184080H202B220), the Startup Foundation for Introducing Talent of NUIST.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1195099/full#supplementary-material
References
Asaro A., Paggi R. A., Del Valle J. C., Lopez Mananes A. A. (2018). Glucose homeostasis in the euryhaline crab cytograpsus angulatus: effects of the salinity in the amylase, maltase and sucrase activities in the hepatopancreas and in the carbohydrate reserves in different tissues. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 216, 39–47. doi: 10.1016/j.cbpb.2017.11.012
Baker K. D., Shewchuk L. M., Kozlova T., Makishima M., Hassell A., Wisely B., et al. (2003). The drosophila orphan nuclear receptor DHR38 mediates an atypical ecdysteroid signaling pathway. Cell. 113, 731–742. doi: 10.1016/S0092-8674(03)00420-3
Chao L. C., Wroblewski K., Zhang Z., Pei L., Vergnes L., Ilkayeva O. R., et al. (2009). Insulin resistance and altered systemic glucose metabolism in mice lacking Nur77. Diabetes. 58, 2788–2796. doi: 10.2337/db09-0763
Chao L. C., Zhang Z., Pei L., Saito T., Tontonoz P., Pilch P. F. (2007). Nur77 coordinately regulates expression of genes linked to glucose metabolism in skeletal muscle. Mol. Endocrinol. 21, 2152–2163. doi: 10.1210/me.2007-0169
Cheng Y., Wu X., Yang X., Hines A. H. (2008). Current trends in hatchery techniques and stock enhancement for Chinese mitten crab, eriocheir japonica sinensis. Rev. Fish. Sci. 16, 377–384. doi: 10.1080/10641260701681698
Chi C., Yu X. W., Zhang C. Y., Liu J. D., Ye M. W., Zhang D. D., et al. (2021). Acute exposure to microcystin-LR induces hepatopancreas toxicity in the Chinese mitten crab (Eriocheir sinensis). Arch. Toxicol. 95, 2551–2570. doi: 10.1007/s00204-021-03061-9
Colombani J., Bianchini L., Layalle S., Pondeville E., Dauphin-Villemant C., Antoniewski C., et al. (2005). Antagonistic actions of ecdysone and insulins determine final size in drosophila. Science. 310, 667–670. doi: 10.1126/science.1119432
Dai H., Sun Y., Ren N., Lu X. (2021). Investigation of Chinese hairy crab industry and analysis of development strategies. Jiangsu Agric. Sci. 49, 248–252.
Ding Z., Meng Q., Liu H., Yuan S., Zhang F., Sun M., et al. (2016). First case of hepatopancreatic necrosis disease in pond-reared Chinese mitten crab, eriocheir sinensis, associated with microsporidian. J. Fish Dis. 39, 1043–1051. doi: 10.1111/jfd.12437
Ding Z., Pan J., Huang H., Jiang G., Chen J., Zhu X., et al. (2018). An integrated metabolic consequence of hepatospora eriocheir infection in the Chinese mitten crab eriocheir sinensis. Fish Shellfish Immunol. 72, 443–451. doi: 10.1016/j.fsi.2017.11.028
Dong D., Zhang Y., Smykal V., Ling L., Raikhel A. S. (2018). HR38, an ortholog of NR4A family nuclear receptors, mediates 20-hydroxyecdysone regulation of carbohydrate metabolism during mosquito reproduction. Insect Biochem. Mol. Biol. 96, 19–26. doi: 10.1016/j.ibmb.2018.02.003
Fu Y., Luo L., Luo N., Zhu X., Garvey W. T. (2007). NR4A orphan nuclear receptors modulate insulin action and the glucose transport system: potential role in insulin resistance. J. Biol. Chem. 282, 31525–31533. doi: 10.1074/jbc.M701132200
Gao T. H., Xu Y., Wang K., Deng Y. F., Yang Y. J., Lu Q. P., et al. (2018). Comparative LC-MS based non-targeted metabolite profiling of the Chinese mitten crab eriocheir sinensis suffering from hepatopancreatic necrosis disease (HPND). Aquaculture. 491, 338–345. doi: 10.1016/j.aquaculture.2018.03.030
Hou Y., Wang X. L., Saha T. T., Roy S., Zhao B., Raikhel A. S., et al. (2015). Temporal coordination of carbohydrate metabolism during mosquito reproduction. PloS Genet. 11, e1005309. doi: 10.1371/journal.pgen.1005309
Huang X., Feng Y., Duan H., Zhao L., Yang C., Geng Y., et al. (2021). Evaluation of pathology and environmental variables contributing to hepatopancreatic necrosis syndrome of Chinese mitten crab, eriocheir sinensis. Ecotoxicol. Environ. Safety. 215, 112157. doi: 10.1016/j.ecoenv.2021.112157
Huang P., Zhang X., Ame K. H., Shui Y., Xu Z., Serwadda A., et al. (2019). Genomic and phylogenetic characterization of a bunya-like virus from the freshwater Chinese mitten crab eriocheir sinensis. Acta Virologica. 63, 433–438. doi: 10.4149/av_2019_410
Janecek S., Svensson B., MacGregor E. A. (2014). α-amylase: an enzyme specificity found in various families of glycoside hydrolases. Cell. And Mol. Life Sci. CMLS. 71, 1149–1170. doi: 10.1007/s00018-013-1388-z
Kh S. D., Keshan B. (2021). The feeding status regulates the action of insulin and 20-hydroxyecdysone on haemolymph trehalose level in bombyx larvae. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 255, 110579. doi: 10.1016/j.cbpb.2021.110579
King-Jones K., Thummel C. S. (2005). Developmental biology. less steroids make bigger flies. Science. 310, 630–631. doi: 10.1126/science.1120410
Kovalenko E. V., Mazina M. Y., Krasnov A. N., Vorobyeva N. E. (2019). The drosophila nuclear receptors EcR and ERR jointly regulate the expression of genes involved in carbohydrate metabolism. Insect Biochem. Mol. Biol. 112, 103184. doi: 10.1016/j.ibmb.2019.103184
Kurakula K., Koenis D. S., van Tiel C. M., de Vries C. J. (2014). NR4A nuclear receptors are orphans but not lonesome. Biochim. Biophys. Acta 1843, 2543–2555. doi: 10.1016/j.bbamcr.2014.06.010
Linton S. M., Greenaway P., Towle D. W. (2006). Endogenous production of endo-beta-1,4-glucanase by decapod crustaceans. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 176, 339–348. doi: 10.1007/s00360-005-0056-5
Liu Y., Zhou S., Ma L., Tian L., Wang S., Sheng Z., et al. (2010). Transcriptional regulation of the insulin signaling pathway genes by starvation and 20-hydroxyecdysone in the bombyx fat body. J. Insect Physiol. 56, 1436–1444. doi: 10.1016/j.jinsphys.2010.02.011
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-delta delta c) method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schutz G., Umesono K., et al. (1995). The nuclear receptor superfamily: the second decade. Cell. 83, 835–839. doi: 10.1016/0092-8674(95)90199-X
Nakagawa Y., Henrich V. C. (2009). Arthropod nuclear receptors and their role in molting. FEBS J. 276, 6128–6157. doi: 10.1111/j.1742-4658.2009.07347.x
Pakula S., Orlowski M., Rymarczyk G., Krusinski T., Jakob M., Zoglowek A., et al. (2012). Conformational changes in the DNA-binding domains of the ecdysteroid receptor during the formation of a complex with the hsp27 response element. J. Biomol. Struct. Dyn. 30, 379–393. doi: 10.1080/07391102.2012.682215
Palli S. R., Hormann R. E., Schlattner U., Lezzi M. (2005). Ecdysteroid receptors and their applications in agriculture and medicine. Insect Hormones 73, 59. doi: 10.1016/S0083-6729(05)73003-X
Pan Z., Song X., Hu X., Xue R., Cao G., Sahib Zar M., et al. (2017). Pathological changes and risk factors of hepatopancreas necrosis disease of mitten crab, Eriocheir sinensis. Fish. Aquac. J. 8, 220. doi: 10.4172/2150-3508.1000220
Panguluri S. K., Kumar P., Palli S. R. (2006). Functional characterization of ecdysone receptor gene switches in mammalian cells. FEBS J. 273, 5550–5563. doi: 10.1111/j.1742-4658.2006.05545.x
Pei L., Waki H., Vaitheesvaran B., Wilpitz D. C., Kurland I. J., Tontonoz P. (2006). NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nat. Med. 12, 1048–1055. doi: 10.1038/nm1471
Ranhotra H. S. (2015). The NR4A orphan nuclear receptors: mediators in metabolism and diseases. J. Recept. Signal Transduct. Res. 35, 184–188. doi: 10.3109/10799893.2014.948555
Riddiford L. M., Cherbas P., Truman J. W. (2000). Ecdysone receptors and their biological actions. Vitamins Hormones. 60, 1–73. doi: 10.1016/S0083-6729(00)60016-X
Ruaud A. F., Lam G., Thummel C. S. (2011). The drosophila NR4A nuclear receptor DHR38 regulates carbohydrate metabolism and glycogen storage. Mol. Endocrinol. 25, 83–91. doi: 10.1210/me.2010-0337
Seliga J., Bielska K., Wieczorek E., Orlowski M., Niedenthal R., Ozyhar A. (2013). Multidomain sumoylation of the ecdysone receptor (EcR) from drosophila melanogaster. J. Steroid Biochem. Mol. Biol. 138, 162–173. doi: 10.1016/j.jsbmb.2013.05.007
Shen Z., Kumar D., Liu X., Yan B., Fang P., Gu Y., et al. (2021). Metatranscriptomic analysis reveals an imbalance of hepatopancreatic flora of Chinese mitten crab eriocheir sinensis with hepatopancreatic necrosis disease. Biology 10, 462. doi: 10.3390/biology10060462
Sutherland J. D., Kozlova T., Tzertzinis G., Kafatos F. C. (1995). Drosophila hormone receptor 38: a second partner for drosophila USP suggests an unexpected role for nuclear receptors of the nerve growth factor-induced protein b type. Proc. Natl. Acad. Sci. U.S.A. 92, 7966–7970. doi: 10.1073/pnas.92.17.7966
Tian L., Guo E., Wang S., Liu S., Jiang R. J., Cao Y., et al. (2010). Developmental regulation of glycolysis by 20-hydroxyecdysone and juvenile hormone in fat body tissues of the silkworm, bombyx mori. J. Mol. Cell Biol. 2, 255–263. doi: 10.1093/jmcb/mjq020
Toullec J. Y., Chikhi M., van Wormhoudt A. (1992). In vitro protein synthesis and alpha amylase activity in f cells from hepatopancreas of palaemon serratus (Crustacea; decapoda). Experientia. 48, 272–277. doi: 10.1007/BF01930474
Vogt G. (2019). Functional cytology of the hepatopancreas of decapod crustaceans. J. Morphol. 280, 1405–1444. doi: 10.1002/jmor.21040
Vogt G. (2021). Synthesis of digestive enzymes, food processing, and nutrient absorption in decapod crustaceans: a comparison to the mammalian model of digestion. Zoology. 147, 125945. doi: 10.1016/j.zool.2021.125945
Wang W. (2011). Bacterial diseases of crabs: a review. J. Invertebrate Pathol. 106, 18–26. doi: 10.1016/j.jip.2010.09.018
Wang Z. F., Tang D., Guo H. Y., Shen C. C., Wu L., Luo Y. Q. (2020). Evolution of digestive enzyme genes associated with dietary diversity of crabs. Genetica. 148, 87–99. doi: 10.1007/s10709-020-00090-7
Watanabe H., Tokuda G. (2001). Animal cellulases. Cell. Mol. Life Sci. CMLS. 58, 1167–1178. doi: 10.1007/PL00000931
Yang Z., Hu K., Hou Y., Wang Y., Yao Y., Lei X., et al. (2020). Transcriptome analysis of hepatopancreas of eriocheir sinensis with hepatopancreatic necrosis disease (HPND). PloS One 15, e0228623. doi: 10.1371/journal.pone.0228623
Yang Z., Zhang Y., Hu K., Liu L., Cai H., Zhang F., et al. (2018). Etiological and histopathological study on hepatopancreatic necrosis syndrome in Eriocheir sinensis. Acta Hydrobiologica Sin. 42, 17–25. doi: 10.7541/2018.003
Yang Z., Zhang Y., Liu L., Zhang F., Cai H., Yang X. (2016). The incidence of river crab HPND in 2015-2016. Sci. Fish Farming 32, 54.
Zhang C., Zhang B., Zhang X., Sun G., Sun X. (2020). Targeting orphan nuclear receptors NR4As for energy homeostasis and diabetes. Front. Pharmacol. 11, 587457. doi: 10.3389/fphar.2020.587457
Keywords: hepatopancreatic necrosis disease, amylase, cellulase, hormone receptor 38, ecdysone receptor, Eriocheir sinensis
Citation: Ren Q, Zhao Y, Ma X, Ge J and Huang X (2023) Inhibited expression of amylases and cellulases in crabs with hepatopancreatic necrosis disease and their regulation by EcR and HR38. Front. Mar. Sci. 10:1195099. doi: 10.3389/fmars.2023.1195099
Received: 28 March 2023; Accepted: 20 June 2023;
Published: 04 July 2023.
Edited by:
Yafei Duan, South China Sea Fisheries Research Institute, ChinaCopyright © 2023 Ren, Zhao, Ma, Ge and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Ren, cmVucWlhbjA0MDJAMTI2LmNvbQ==; Xin Huang, aHVhbmd4aW44MTQxMDJAMTI2LmNvbQ==; Jiachun Ge, Z2pjMDlAaG90bWFpbC5jb20=
†These authors have contributed equally to this work
 Qian Ren
Qian Ren Yuqi Zhao2†
Yuqi Zhao2†