
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 02 June 2023
Sec. Aquatic Physiology
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1195016
This article is part of the Research Topic Impact of Heavy Metal on Aquatic Life and Human Health View all 9 articles
 Hassan Ahmed1*
Hassan Ahmed1* Karima A. Bakry2
Karima A. Bakry2 Ahmed Abdeen3*
Ahmed Abdeen3* Halla E. K. El bahgy4
Halla E. K. El bahgy4 Mohamed Abdo5,6
Mohamed Abdo5,6 Florin Imbrea7*
Florin Imbrea7* Liana Fericean8
Liana Fericean8 Mona A. Elshemy9
Mona A. Elshemy9 Samah F. Ibrahim10
Samah F. Ibrahim10 Mustafa Shukry11
Mustafa Shukry11 Samar H. Baloza12
Samar H. Baloza12 Walaa F. A. Emeish2
Walaa F. A. Emeish2Salinity stress is one of the marked influencing factors on the ecophysiology of aquaculture and is considered an important reason for the retreat of the fish industry. The current study is an endeavor to elucidate the molecular mechanisms that underlie the response to salinity stress in common carp. Fish (Average weight 5 ± 2 g) were randomly distributed into two groups; the 1st is a control was exposed to tap water (0.2 ppt salinity) and the 2nd is a treated was exposed to hypersalinity (10 ppt salinity) for five days. Serum biochemical indicators including total protein, albumin, globulins, A/G ratio, blood glucose, cortisone, Na+, K+, and Cl- levels were evaluated. Besides, Tumor necrosis factor-α, interleukin-1β, corticotropin-releasing hormone, and catalase enzyme mRNA expression levels were assessed in lymphoid and immunocompetent organs (liver and spleen) and osmoregulatory organs (kidney and gills) by using Real-time qPCR. Hypersalinity adversely affected the biochemical markers; total protein, albumin, and globulins decreased significantly; however, blood glucose, serum cortisol, and sodium markedly increased in fish exposed to hypersalinity compared with the control. In addition, from the molecular point of view, all the evaluated genes were upregulated at a high expression rate in the liver compared with other studied organs after the salinity challenge. On the contrary, hypersalinity modulated the expression of immune-related genes (Tumor necrosis factor-α and interleukin-1β) in the kidney and spleen and upregulated corticotropin-releasing hormone mRNA in all studied organs except gills. In conclusion, the obtained data clarified the molecular and biochemical mechanisms of salinity stress on the liver, kidney, spleen, and gills. Furthermore, it strongly suggests the implication of neural, endocrine, and immune systems in the responsive mechanisms to the salinity stress in carp.
Aquaculture is one of the world’s fastest-growing industries as 50% of fish production yield is achieved by fish farms (Fitzsimmons et al., 2015). However, the fish farming industry is facing a considerable challenge, in particular, water salinity and the scarcity of freshwater (FAO, 2003). This is owing to water shortage and climate changes including temperature elevation, which directly impacts water evaporation and, subsequently, higher salinity concentrations. Besides, the freshwater ecosystem is polluted by anthropogenic salts derived from industrial and agricultural waste (Velasco et al., 2019). In addition, salinity has a marked impact on the ecophysiology of fish and is considered an essential ecological factor in the retreat of the fish industry. Moreover, salinity change is the main cause of deleterious effects on physiological hormones and enzyme activities, reflecting fish bioactivity and survival (Cao et al., 2022). Furthermore, hypersalinity stress is considered the harshest aquatic environment disturbing the physiological functions of growth performance, metabolic rate, food intake, and hormonal stimulation in aquaculture (Gonzalez, 2012).
Common carp (Cyprinus Carpio) is a stenohaline freshwater fish with a low endurance ability to salinity. Salinity is one of the common causes of ecosystem disturbance and physiological homeostasis imbalance, ending in high mortalities of carp due to osmotic shock (Evans and Kültz, 2020). Research has shown that when salinity is elevated, homeostasis is achieved by increasing the concentrations of Na+ and K+ in the blood, and there is enhanced activity of gills Na+/K+ ATPase (Salati et al., 2011). Moreover, Alkhshali and Al-Hilalli (2019) reported increases in RBCs, HB, PCV, and WBCs in common carp in response to steady elevations in the salt concentrations (from 5 to 10 to 15 g/L) as a physiological response to environmental salinity perturbations (Al-Khshali and Al Hilali, 2019). In addition, the chronic salinity change contributes to the disturbance of liver function in common carp (Alkhshali and Al-Hilalli, 2019). The adverse effect of salt stress was extended to general performance in common carp as it reduced food intake and growth rates with unfavorable effects on survivability after that, with depletion of both muscle and liver glycogen ending with energy expenditure (Sharma et al., 2020). Furthermore, drastic salinity change has been associated with oxidative stress and the production of reactive oxygen species (ROS) in fish accompanied by suppressed immune functions and antioxidant defense abilities (Kim and Kang, 2015).
Exposure to salinity stress is associated with genetic response and the modulation of gene expression in hypersalinity tolerance. For instance, the function of Na+/K+ ATPase (NKA) has been studied in the gill and kidney of multiple species of fish including silver catfish (Marx et al., 2022), Nile tilapia (Oreochromis niloticus) (Mohamed et al., 2021), Japanese eel (Tang et al., 2012), Senegalese sole (Armesto et al., 2015), and pufferfish (Tetraodon nigroviridis) (Wang et al., 2017). Since NKA is in the gills, increased expression of NKA in gills after 14 days of exposure to 10 ppt saline is probably the result of the excretion of ions that flow into the body across the thin epithelia of the gills. Also, aquaporins are water channels that play a key role in water transportation in hypersalinity exposure. The gene expression of aquaporin 3 is upregulated in gills 6 days after transfer from 5 ppt to 30 ppt saline in pufferfish. Moreover, the change in environmental salinity (10 and 25%) enhanced immune response by the elevation of mRNA expression profiles of cytokine genes IL-1β, IL-6, IL-10, and TNF-α in coastal fish Scatophagus argus (Lu et al., 2022).
As an adaptive and hemostasis response from the fish against salinity challenge, antioxidant enzymes, immunostimulants (pro-inflammatory cytokines), stress hormones, and non-specific immunity are elevated (Tapia-Paniagua et al., 2011). Therefore, evaluating these parameters in fish is an appropriate marker for measuring the potential influence of higher salinity exposure on innate immunity and antioxidant tolerance (Saurabh and Sahoo, 2008).
The hypothesis of the current study is to explore the molecular and biochemical mechanism by which the common carp respond to the hypersalinity and change in ecosystem. Furthermore, it is expected that the fish being studied brings about the change in the disturbed environment through homeostasis by modulating the gene expression of proinflammatory cytokines (TNF and IL-1β), stress hormones (CRH) and antioxidants (CAT) in the osmoregulatory and immune organs, mainly the liver, kidney, spleen, and gills. In addition, the molecular mechanism that explains the adaptation of common carp to hypersalinity is elusive; therefore, the present study is designed to bridge the gap between hypersalinity and homeostasis through molecular and biochemical mechanisms.
Although many studies have manipulated the effect of hypersalinity on common carp, molecular analysis is still elusive. The current study is conducted to figure out the ability of common carp – depending on their economic importance – to acclimate to hypersalinity, especially in the harshest change in ecosystem conditions such as freshwater salinization, pollution, and water scarcity problems
Subsequently, the current study endeavors to investigate the physiological mechanisms depending on the molecular and biochemical aspects in the rescue of the disturbed ecosystem due to the change in environmental salinity. The biochemical markers including total protein, albumin, globulins, A/G ratio, blood glucose, cortisone, Na+, K+, and Cl- levels were identified. Furthermore, the mRNA expression of pro-inflammatory cytokines (TNF and IL-1β), corticotropin-releasing hormone (CRH; a stress hormone), and catalase enzyme (CAT; an antioxidant marker) were assessed in the gills, liver, kidney, and spleen.
Common carp (Cyprinus carpio) were obtained from a commercial fish farm with an average weight of (5 ± 2 g). The fish were acclimated for 3 weeks under laboratory conditions in the Department of Fish Diseases and Aquatic Laboratory of the Faculty of Veterinary Medicine, South Valley University, Qena, Egypt, in accordance with the guidelines on the care and use of experimental animals for scientific purposes set by South Valley University. During the acclimation period, the water temperature was kept at 22°C, the dissolved oxygen at 6.1 ± 0.5 mg/L, and the pH at 7.1. The fish were fed a commercial diet (Table 1) twice daily at 3% of their body weight, maintained on a 12 h:12 h light/dark cycle, and reared in a porcelain tank re-circulation system (260 × 65 × 70 cm) provided with pump aeration and filled with fresh dechlorinated water (salinity = 0.2 ppt).
The fish were randomly distributed into two experimental groups (18 fish for each) and stocked in six fiberglass aquaria (60 × 30 × 40 cm) containing 120 L (six fish per tank in triplicates) of dechlorinated tap water.
One control group was kept in dechlorinated tap water (salinity = 0.2 ppt) and another salinity-exposed group was reared at 10 ppt for 5 days. The salinity is obtained by mixing an adequate amount of commercial-grade NaCl with dechlorinated tap water. Water salinity levels were gradually raised in the experimental tanks by adding NaCl to water at 3 g/l daily until reaching the proposed levels of salinity and the concentration was selected after the preliminary test run using a salinometer to determine salinity concentrations (Thermo Electron corporation model Orion 150 A+).
The water quality indices (temperature, pH, dissolved oxygen, and nitrite) were checked daily during the study using Martini Instruments Model 201/digital to detect the temperature, dissolved oxygen, salinity, and pH. The total ammonia level was detected calorimetrically using a HI-83303–02 Aquaculture Multi-parameter Photometer (Woonsocket, RI, USA). The aquarium was kept at 20 ± 1°C under natural light cycle conditions. The fish were fed twice a day with commercial floating powder feed containing 45% protein at a feeding rate of 3% of their body weight. During the experimental period, fish were checked twice a day for uncommon clinical symptoms and mortality rates (Emeish, 2019).
Fish were euthanized using 0.033 mL/L eugenol (Hamáčková et al., 2006) (clove oil) on the 5th day after exposure to salinity stress. Blood samples were collected in Eppendorf tubes from the caudal vein by severing the tail and centrifuging at 3000 rpm for 15 min. The collected sera were stored at -20°C until they were used in biochemical assays. The liver, kidney, spleen, and gills (30 mg/organ) were collected from control and salinity-exposed groups and stored in RNA later (Omega BIO-TEK, GA, USA) at −80°C until quantitative qPCR gene expression analysis.
All biochemical parameters were monitored spectrophotometrically according to standard methods using commercial laboratory kits. Serum total proteins, albumin, sodium, and potassium chloride kits (Spectrum, Egyptian Company for Biotechnology, Obour City, Egypt) were used. Globulin level (g/dl) was calculated mathematically by subtracting albumin from the total protein value. In addition, blood glucose levels in whole blood samples were checked using the Gluco Dr. auto blood glucose monitoring system and gold-plated test strips (All Medicus Co., Ltd., Germany) following the manufacturer’s description. All biochemical parameters were carried out by using T80 UV/VIS spectrophotometer (PG Instruments, UK). Serum cortisol levels were measured using enzyme-linked immunosorbent assay (ELISA) kits according to the manufacturer’s instructions (#CAN-C-270; Diagnostics Biochem Canada Inc., Canada). A cortisol assay was done by a microplate reader (Infinite 50, Männedorf, Switzerland) at a wavelength of 450 nm according to the manufacturer’s instructions.
Total RNA was extracted from the liver, kidney, spleen, and gill tissues using RNeasy® Mini kit (QIAGEN, Germany) following the manufacturer’s instructions, and the quantity of the RNA was estimated by using Nanophotometer (NanoDrop Technologies, Wilmington, USA). For cDNA synthesis, 1μg of each RNA sample was reverse transcribed to cDNA in the Vetiti™ 96 Well Thermal Cycler, according to the instructions using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, MA, USA).
RNA expressions of TNF-α, IL-1β, CRH, and CAT mRNA were measured using a real-time qRT-PCR. The cDNA products were amplified using the HERAPLUS SYBR® Green qPCR Kit (Willowfort, England), utilizing a Stratagene Mx3005p® real-time qPCR detection system (Agilent Technologies, MA, USA). Each 20 μL reaction mixture contained 10 μL Sybr Green Master Mix, 1.5 μL cDNA, the gene-specific primers (0.5 μM each), and 1 μL of each primer (10 μM of forward and reverse) of the proposed genes (Table 2), and the volume was completed up to 20 μL with sterile nuclease-free PCR grade water, and no template PCR control was included. The cycling conditions were the initial enzyme activation step at 95°C/3 min followed by 40 cycles of 95°C for 10 s and 60°C for 1 min. To confirm the amplification of specific products, cycles were continued to check the melting curve.
Each sample was normalized to the expression level of the housekeeping genes Beta-Actin (β-actin) and Elongation Factor 1-α (EF1α) to correct for differences in template input and reverse transcriptase efficiencies. All reactions were assayed in duplicate on triplicate samples, and the threshold cycle (Ct) value was obtained from the automatic setting on the Mx3005p® real-time qPCR detection system and calculated. The fold change of target gene expression in a target sample relative to a reference sample, normalized to a reference gene. The relative gene expression was calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
All data were analyzed with GraphPad Prism software (GraphPad 8.0.1 Software, San Diego, CA, USA) using the student’s t-test. Data were expressed as the mean ± standard error, with significant differences defined as p < 0.05. RStudio (R version 4.0.2) was used to generate the correlation heatmap between variables, clustering heatmap among different treatments, and the variable important projection. The current data were validated for normality and homogeneity of variance using the Shapiro-Wilk normality test and the Kolmogorov-Smirnov normality test.
The mortality rate was recorded in the 0.2 and 10 ppm salinity-exposed fish during the experimental period. Fish exposed to 0.2 ppm groups survived during the experimental period with a mortality rate of 0%; however, those challenged with 10 ppm disclosed different mortality rates; for instance, the death percentage on the 3rd day was 11.11%, was 16.66% on the 4th day, and was 33.33% on the 5th day, as shown in Figure 1. These data suggest the existence of a negative impact of hypersalinity medium on fish survival rate.
The hypersalinity-exposed fish denoted significant (P < 0.05) decreases in the serum levels of total protein, albumin, and globulins when compared with the corresponding control, as depicted in Figures 2A–C, respectively. Conversely, the A/G ratio elevated non-significantly after salinity exposure compared with control fish (Figure 2D). Furthermore, blood glucose and serum cortisol levels markedly (P < 0.05) increased after exposure to salinity stress when compared with control (Figures 2E, F, respectively). As a response to the change in the salinity environment, circulating Na+, K+, and Cl- exhibited remarkable (P < 0.05) increases compared with the corresponding controls (Figures 2G-I, respectively). These findings proposed the effect of salinity exposure on the biochemical parameters measured in this study.
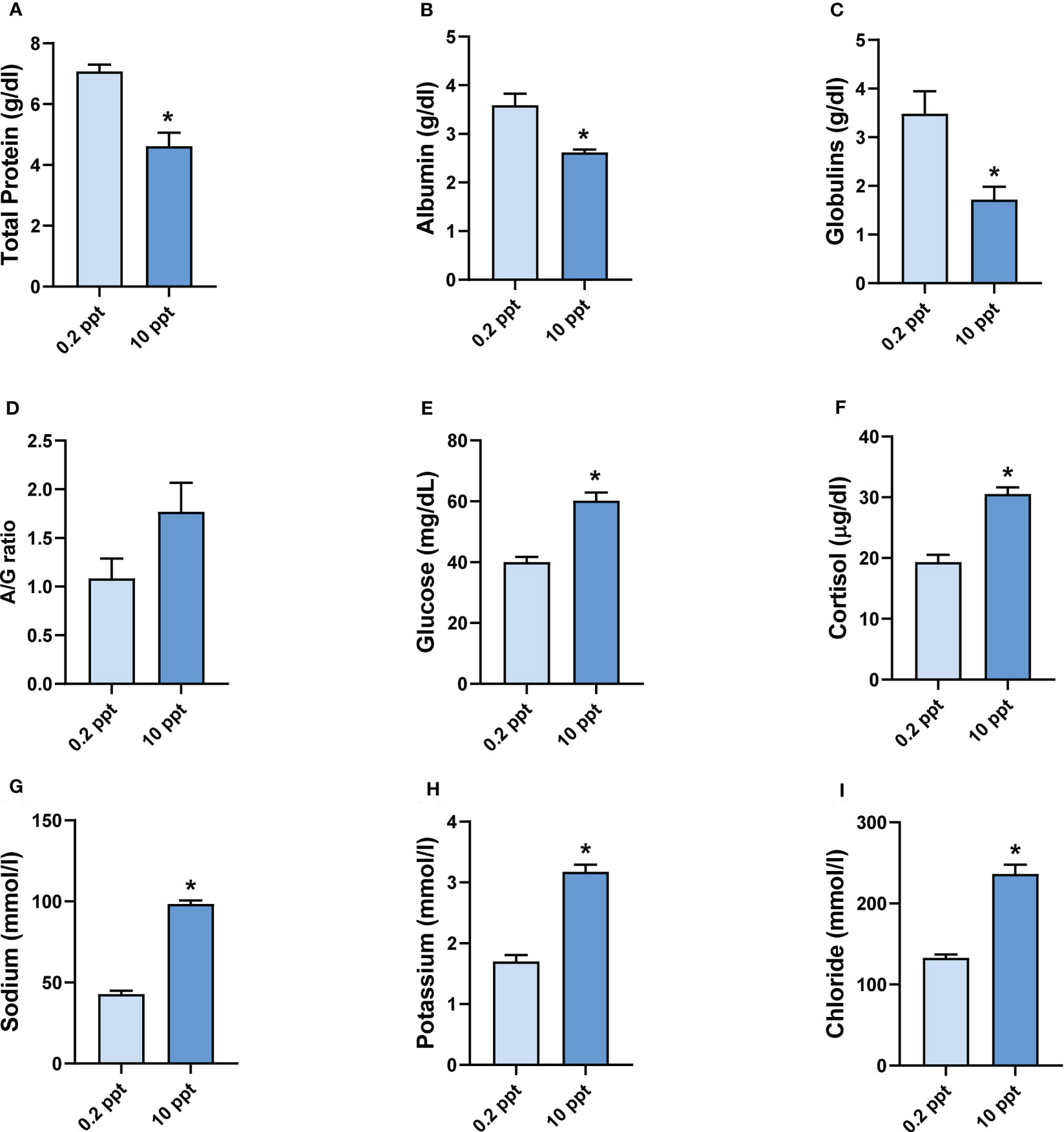
Figure 2 Biochemical analysis in common carp after exposure to 0.2 and 10 ppt salinity. (A) Serum total protein level (g/dl). (B) Serum albumin level (g/dl). (C) Serum level of globulins (g/dl). (D) A/G ratio (%). (E) Blood glucose level (mg/dl). (F) Serum cortisol level (µg/dl). (G) Serum level of sodium (mmol/l). (H) Serum level of potassium (mmol/l). (I) Serum level of chloride (mmol/l). Data are expressed as mean ± SEM. *P < 0.05 vs. control group.
Figure 3 shows the expression of the studied genes in liver tissue after exposure to 0.2 (Control) and 10 ppt for 5 days. The liver in salinity-exposed fish presented a marked (P < 0.05) upregulation of TNF-α, IL-1β, CRH, and CAT mRNA expression levels when compared with the control. Furthermore, the kidney and spleen of common carp exposed to salinity 10 ppt revealed dramatic (P < 0.05) upregulations of TNF-α, IL-1β, and CRH mRNA expression in comparison to controls. However, both organs revealed slightly increased mRNA expressions of antioxidant markers (CAT) compared to controls.
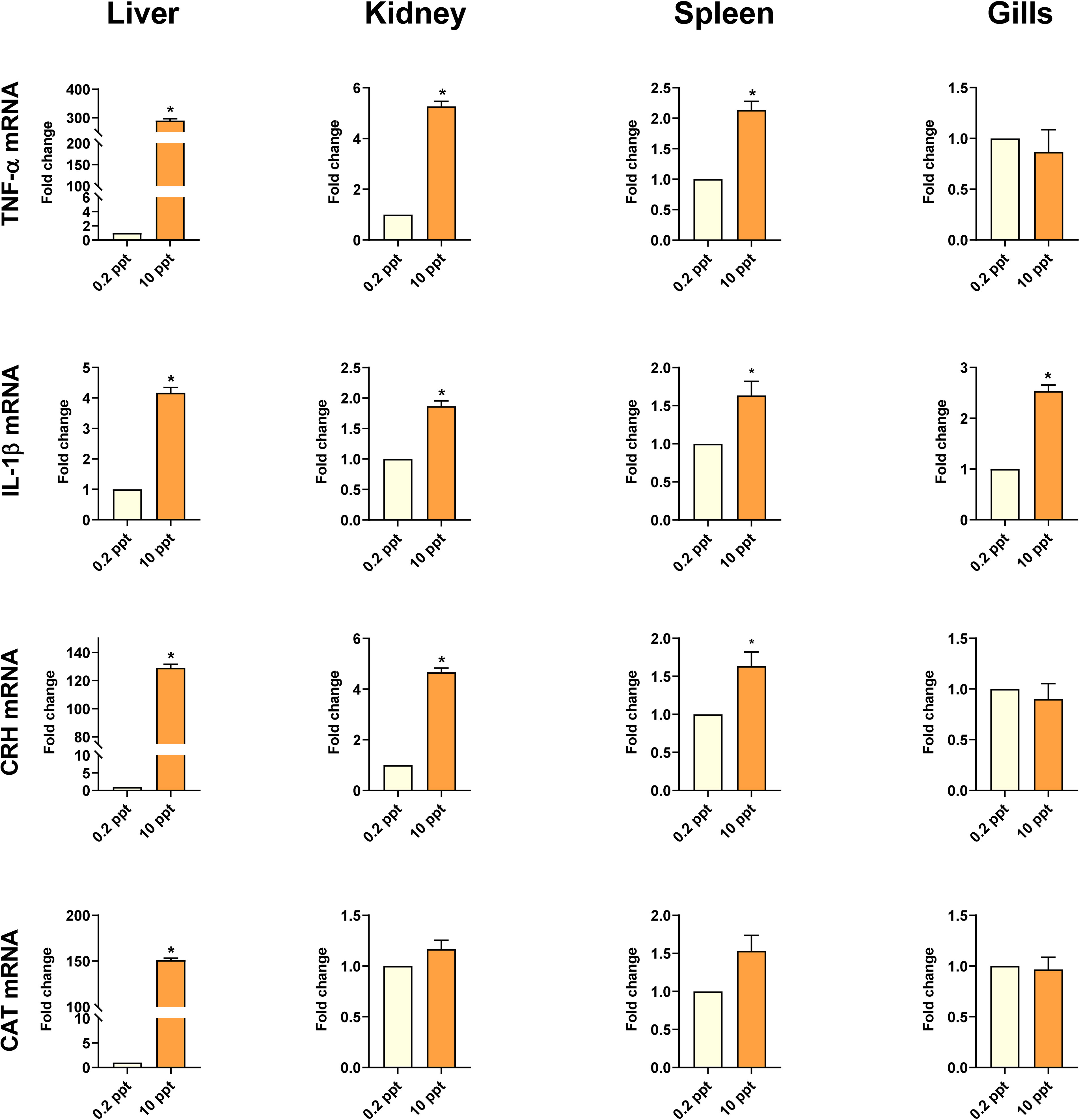
Figure 3 Cytokines (TNF-α and IL-1β), CRH, and CAT gene expression (relative to β-actin and EF1α) in common carp’s liver, kidney, spleen, and gills exposed to 0.2 and 10 ppt salinity. Data are expressed as mean ± SEM. *P < 0.05 vs. control group.
In gills tissue, the mRNA expression of IL-1β revealed a significant (P < 0.05) enhancement in the high salinity-exposed fish in comparison with the control group. In contrast, the mRNA expression of TNF-α, CRH, and CAT showed a non-significant change (Figure 3).
The results in Figure 4 summarize the expression of proposed genes in the studied organs. TNF-α mRNA was markedly upregulated (P < 0.05) in the liver compared with the kidney, spleen, and gills. At the same time, the expression of the same gene revealed no significant elevation in the kidney compared with splenic and gill tissues, and the spleen compared with gills. Concerning IL-1β gene expression, the highest expression was exemplified in the liver, which showed a non-significant upregulation compared with other proposed organs. Furthermore, IL-1β gene expression slightly increased in the gills compared with the liver and spleen. Similar to TNF-α, CRH gene expression recorded a significant (P < 0.05) upregulation in the liver compared with the expression of the other three organs.
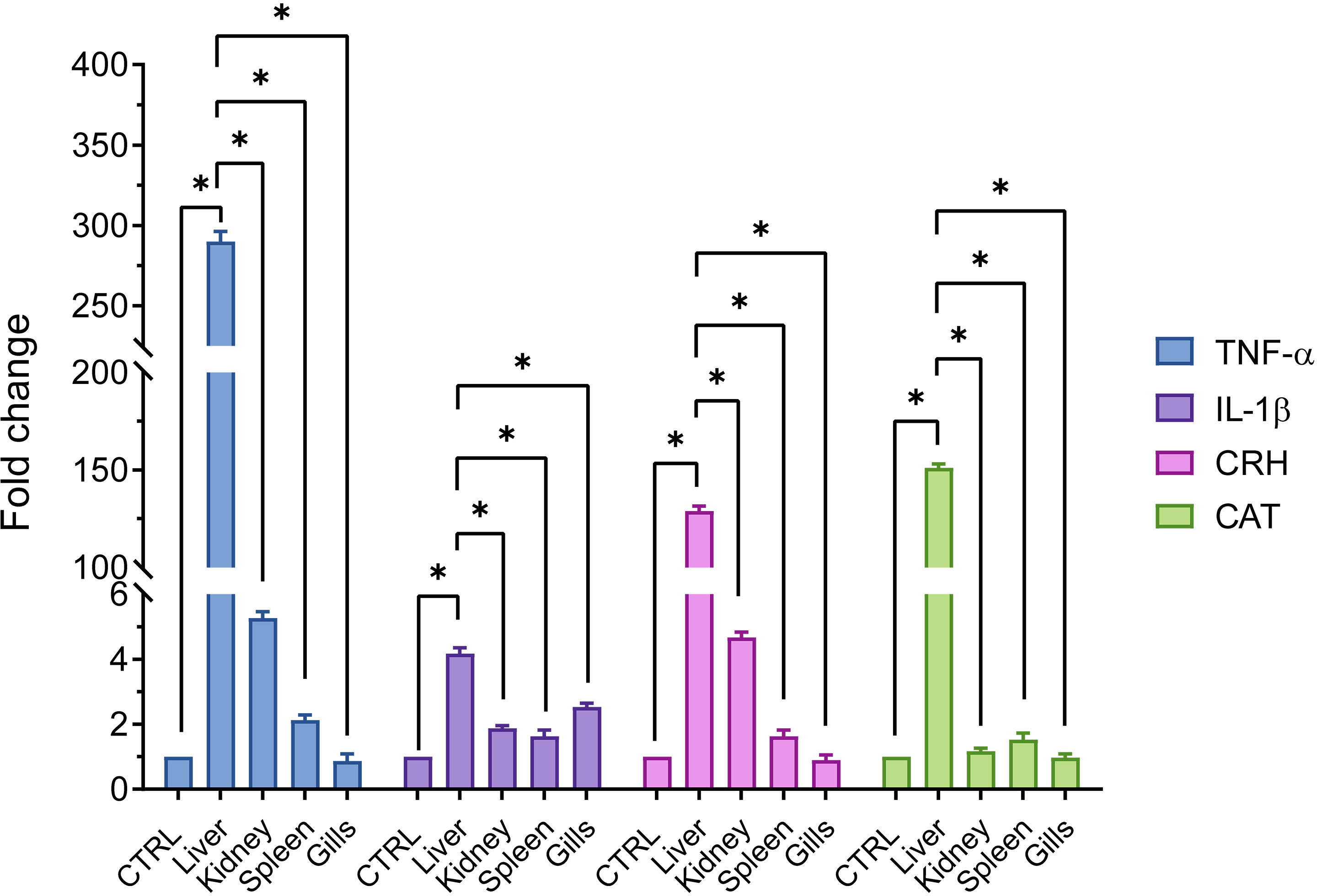
Figure 4 Comparison of gene expressions (relative to β-actin and EF1α) in common carp’s different organs (liver, kidney, spleen, and gills) exposed to 0.2 and 10 ppt salinity. Data are expressed as mean ± SEM. *P < 0.05 vs. control group.
Furthermore, the former gene increased non-significantly in the renal tissue by three and five folds compared to the spleen and gill expression. Finally, CAT gene expression revealed prominent and significant (P < 0.05) upregulations in the hepatic tissue compared with the renal, splenic, and gill tissues, which expressed almost the same values of the CAT gene. The abovementioned gene expression data indicated that the recorded gene alterations were organ dependent.
The clustering heatmap depicted in Figure 5 provides an intuitive visualization of all datasets which summarizes the correlation between all measured biochemical variables and mRNA expression of target genes in response to 0.2 and 10 ppt salt concentrations for 5 days. The contribution is explicated by a colored scale ranging from the highest (red indicates positive correlation) to the lowest (blue indicates negative correlation). The degree of correlation is shown by the color’s intensity. The various squares are colored in accordance with Pearson’s correlation coefficient between all measured variables. These data exhibit strong positive correlations between stress, proinflammatory cytokines, and antioxidants-expressed genes in all targeted tissues and the exitance of negative correlations between those genes and the total protein, albumin, and globulin concentrations in blood. In addition, Figure 6A summarizes the variable averages in response to different salinity concentrations in the current study. These data suggest the occurrence of physiopathological alterations in salinity-stressed fish.
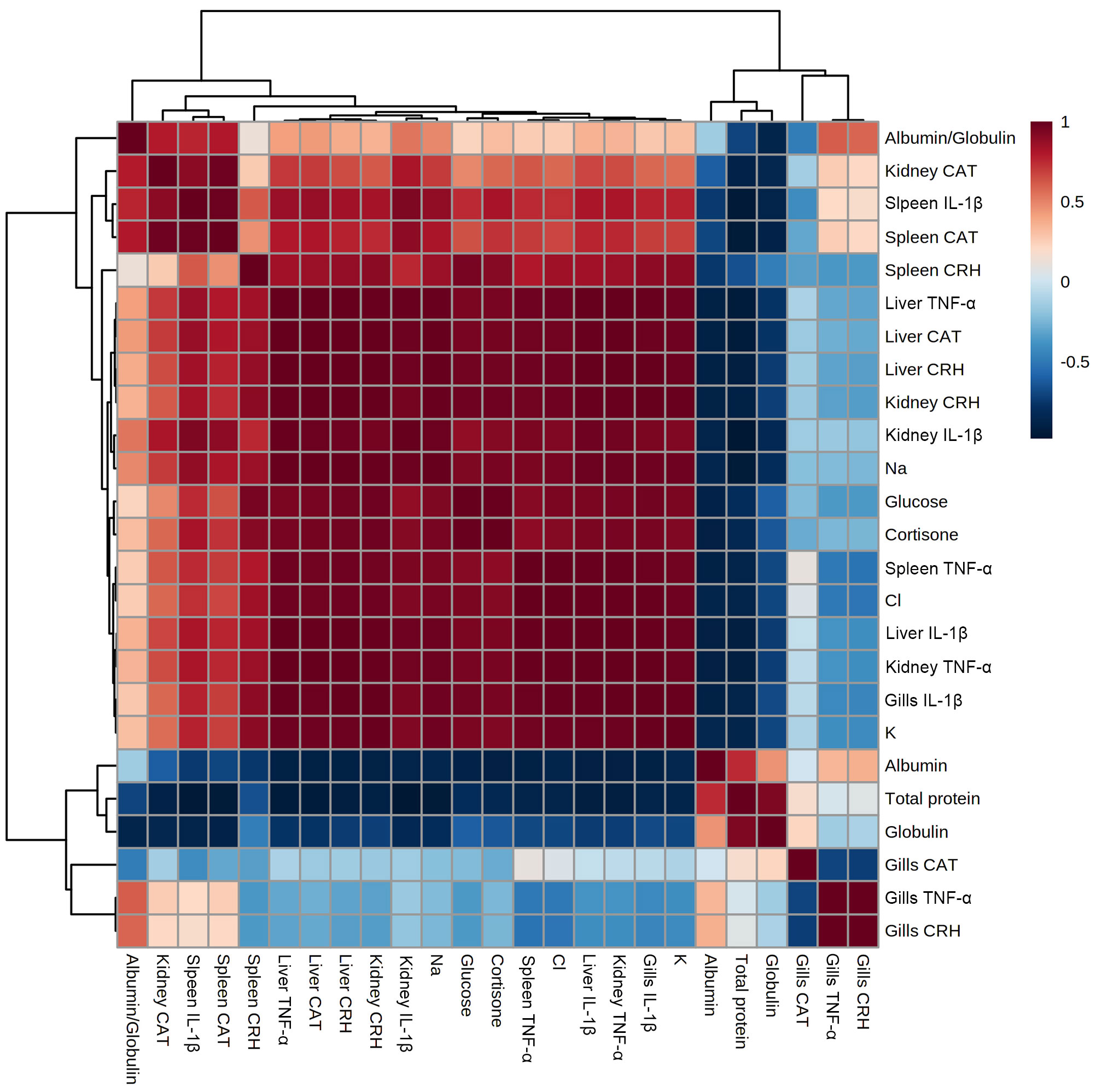
Figure 5 Correlation heatmap of all variables. The clustering heatmap provides an intuitive visualization of all data sets which summarizes the correlation between all measured biochemical variables and mRNA expression of target genes in response to 0.2 and 10 ppt salt concentrations for 5 days in each sample. The contribution is explicated by a colored scale ranging from the highest (red indicates positive correlation) to the lowest (blue indicates negative correlation). The degree of correlation is shown by the color’s intensity. The various squares are colored in accordance with Pearson’s correlation coefficient between all measured variables.
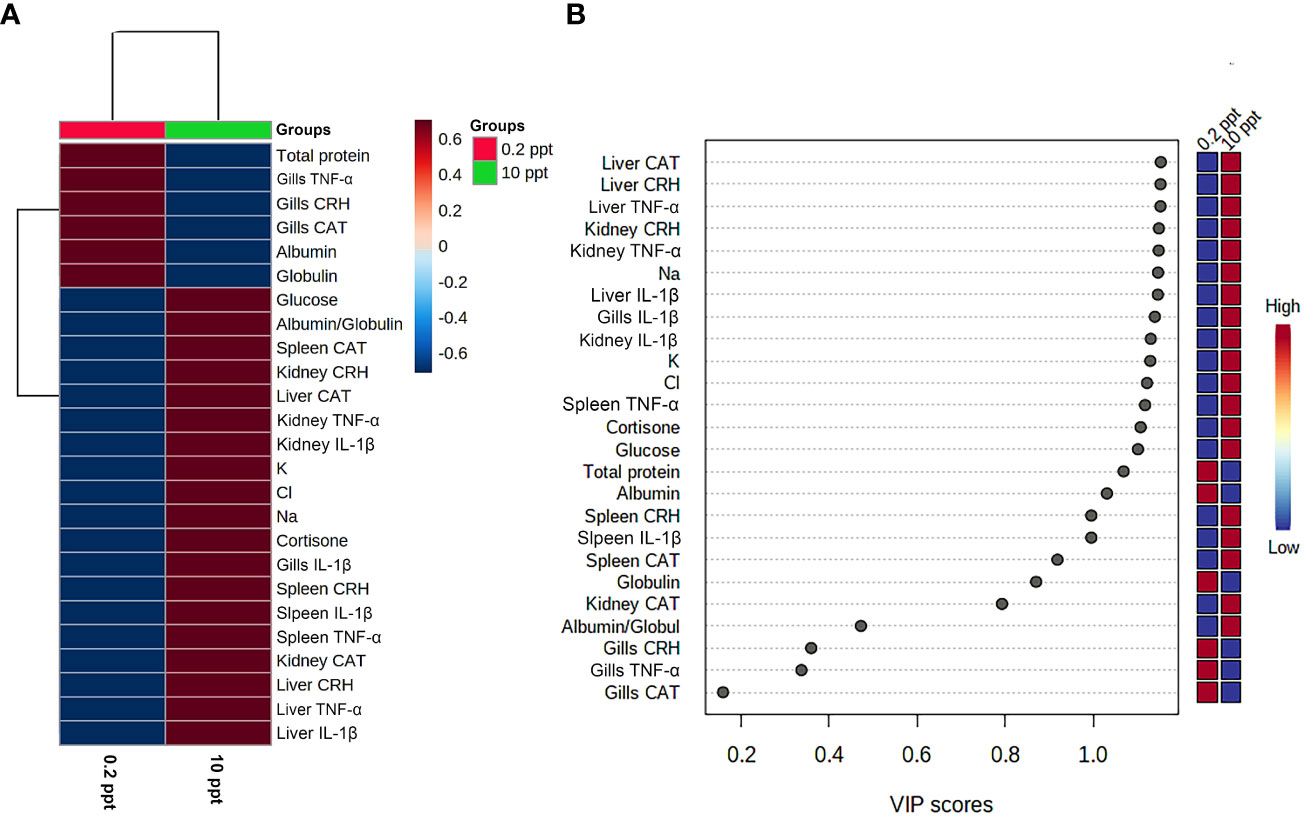
Figure 6 Clustering heatmap and variable importance in projection (VIP) scores. (A) Hierarchical clustering heatmap enables intuitive visualization of all data sets. Each colored cell on the map denotes the concentration values, with variable averages in rows and different treatment sets in columns. Dark red is the highest value on the gradation scale, and blue represents the lowest value. (B) Variable importance in projection (VIP) scores: the colored boxes on the right display the relative concentrations of the relevant measured parameters in each study group, while the contribution intensity is indicated by a colored scale spanning from the highest (red) to the lowest (blue).
Next, in order to reveal the most influencing variables in response to salinity exposure, the VIP score was calculated. As presented in Figure 6B, TNF-α, IL-1β, CRH, and CAT mRNA in liver, kidney, and spleen tissue along with blood Na+, K+, Cl-, and cortisone strongly contributed to the mechanism involved in the salinity-induced physiopathological alterations in common carp.
Salinity change is a major eco-stress factor affecting marine and estuarine organisms. However, many species can tolerate salinity fluctuation depending on how they cope. The current study is an in-depth understanding of the molecular mechanism response to salinity stress for 5 days by evaluating pro-inflammatory cytokines (TNF-α and IL-1β), the stress hormone (CRH), and antioxidant capacity (CAT) in the liver, kidney, spleen, and gills. Salinity challenge disrupts most physiological functions and biological processes, including hepatic, renal, and gill functions, blood dissolved gases, blood parameters, and hormonal regulation (Mohamed et al., 2021).
In addition, the tolerance to salinity stress is varied according to the aquatic species. In this study, mortalities started on the 3rd day and increased gradually until reaching 33.33% on the 5th day, indicating the limited ability of common carp to salinity adaptation. Therefore, the current study is necessary to shed light on the physiological mechanism depending on molecular and biochemical aspects in the rescue of the disturbed ecosystem and the change in environmental salinity. The current results are matching with that recorded by Akther et al., 2013 who found that acute exposure of Barbodes gonionotus to 15 ppt salinity showed a mortality rate of 20% after 24 h and 100% after 4 days.
The liver is the most affected organ by salinity stress, which is linked with a noticeable decrease in serum total protein, albumin, and globulins (Alkhshali and Al-Hilalli, 2019). The alteration of hepatic function is caused by oxidative stress and reactive oxygen species production (Kim and Kang, 2015), which is confirmed by the upregulation of CAT gene expression in the liver. Moreover, the change in circulating total protein, albumin, and globulin levels in the abovementioned results could be proof of water and electrolyte (dehydration) perturbation resulting from hypersalinity stress. In addition, the elevation of stress factors (CRH gene, cortisol, and blood glucose) in response to salinity challenges that are linked with the hypothalamic-pituitary-interrenal (HPI) axis often results in tissue modulation. Consequently, elevated corticosteroids reduce protein synthesis, enhance catabolism, and accelerate energy expenditure (Sopinka et al., 2016).
In addition, there was also a dynamic elevation in blood glucose levels. A stress reaction is expressed as increased blood glucose levels associated with high CRH gene expression and high circulating cortisol level, which is assumed to be a result of the hypothalamic-pituitary-interrenal (HPI) axis activity as evidenced by Wang et al., 2020. Interestingly, the elevation of blood glucose levels after salinity exposure is accompanied by catecholamine secretion, which is part of the mechanism to counteract abrupt changes in environmental salinity. Furthermore, catecholamine release is followed by increased gill perfusion facilitating ion and gas exchange between fish bodies and the external environment (Demska-Zakęś et al., 2021).
The most influenced aspect after the salinity challenge is the ionic basis as common carp showed significant elevations of Na+, K+, and Cl-. The primary organs for ion transport and water exchange in aquaculture are the gill epithelium, kidney, and intestinal epithelium (Evans and Claiborne, 2009). In this study, the elevation of Na+ and Cl- is a preparatory stage for ionic equilibrium and water exchange by activating Na+/K+ ATPase and upregulating aquaporin3, especially in gills tissue (Jumah, 2020). The preceding studies denoted the elevation of Na+, K+, and Cl- in pikeperch exposed to high salinity, matching the current findings (Demska-Zakęś et al., 2021).
The liver is a fundamental organ that plays a crucial role in vital functions in living organisms, including metabolism, immunity, digestion, and detoxification (Atrees and Rabia, 2021). A stressful environment for aquatic organisms contributes to the deterioration of physiological cellular function by developing oxidative stress (Zhou et al., 2020). As a response to salinity stress, the present study demonstrates an immune response represented by the upregulation of TNF-α and IL-1β gene expression in hepatic tissue. The upregulation of these genes is a potent inflammation and pro-inflammatory response to regulate immunity and cope with stressors (Yahfoufi et al., 2018).
Furthermore, the results emphasized the upregulation of CRH gene expression in hepatic tissue. Oxidative stress induced by hypersalinity is associated with the activation of antioxidant enzymes involving CAT gene expression trying to overcome the adverse effect of ROS on DNA hepatocytes (Baldissera et al., 2019; Xavier et al., 2020).
The kidney is the second osmoregulatory organ after the gills, representing the crucial line (Buddington and Krogdahl, 2004). The osmoregulatory role of the kidney in stenohaline is prominent in environmental salinity stress. This role is represented in the excretion of a large amount of water through high glomerular filtration and activity reabsorption of the most ions to maintain fluid and electrolytes hemostasis (Takvam et al., 2021). TNF-α, IL-1β, CRH, and CAT monitored the molecular mechanism of renal tissue.
Like other organs, the kidney is affected adversely by hypersalinity, starting with the activation of pro-inflammatory cytokines (Brocker et al., 2012), supporting the current results by the elevation of TNF-α and IL-1β gene expression. In fish, the hypothalamic-pituitary-interrenal axis is a primary neuroendocrine modulatory system of immunity to tolerate stress (Gallo and Civinini, 2003). Moreover, it is stimulated by variable stressors via IL-1β, subsequently enhancing CRH release from the hypothalamic preoptic nucleus and cortisol hormone synthesis and secretion from inter-renal cells of the head kidney (Metz et al., 2006).
On the other hand, salinity stress has no effect on CAT gene expression in the kidney. According to Wang et al. (2016), antioxidant enzyme activity, including SOD gene expression, is downregulated in juvenile marbled eels after a high salinity challenge but upregulated after a low salinity challenge. The fact that the relationship between enzymatic activity and mRNA expression is not linear (Chambers and Matrisian, 1997; Wang et al., 2016) explains the inexplicable changes in CAT gene expression.
The spleen is the main lymphoid and immunocompetent organ (Zapata et al., 2006). So, it is more sensitive and is rapidly affected by salinity stress, as shown in the present study; TNF-α and IL-1β gene expression is upregulated, agreeing with Xu et al. (2018) and Xu et al. (2020), who reported the upregulation of both genes in the spleen of Tilapia nilotica after exposure to 16% salinity. Similarly, CRH gene expression was upregulated in the spleen as a response to salinity stress. Harlé et al. (2018) confirmed that CRH receptors-2 are expressed in splenic B-cells, supporting our results. Likewise, other organs and the spleen suffer from oxidative stress after salinity stress, which is observed by enhancing antioxidant capacity through the upregulation of CAT.
The primary osmoregulatory organs are the gills (Hwang et al., 2018). They control ion transport via the Na+/K+ ATPase pump and water exchange via aquaporin channels (Li et al., 2006). Furthermore, TNF-α is the main pro-inflammatory cytokine associated with apoptosis and autoimmune conditions (Gao et al., 2015). The present results are consistent with El-Leithy et al. (2019), who confirmed the elevation of IL-1β gene expression in Tilapia nilotica after high salinity exposure. Moreover, salinity stress has a dual effect on immune response as it down-regulates or activates some immune mediators according to salinity dose and exposure time (Tort, 2011; Schmitz et al., 2017). In addition, some stress conditions induce salinity that can suppress or stimulate a specific pathway of cytokine production (Dhabhar, 2008). Furthermore, as reported by Castillo et al. (2008), cortisol secreted in response to stress has an influential inhibitory role in some cytokines, specifically TNF-α (Saeij et al., 2003).
Salinity stress is considered the hypothalamic-pituitary-interrenal axis activator represented by the neuroendocrine response that involves instant secretion of CRH by the preoptic nucleus of the hypothalamus, ACTH from the pituitary gland, and, finally, cortisol hormone from the head kidney (Tort, 2011). CRH allocation has been found in the common carp spleen and kidney, indicating that CRH receptors are highly expressed in these organs. Huising et al., 2004 explain that no change in CRH gene expression in gills tissue is due to a negative feedback mechanism. Hence, enhancing antioxidant enzyme activity can cause a disturbance in the balance between ROS production and removal, resulting in oxidative stress (Chowdhury and Saikia, 2020). Unexpectedly, the current study showed no change in gills CAT gene expression after exposure to salinity challenge due to no change in TNF-α. Pro-inflammatory cytokines (mainly TNF-α) trigger oxidative stress and stimulate the regulation and adaptation of the immune system to tolerate environmental stressors (Yahfoufi et al., 2018).
The activity of antioxidant enzymes including CAT after salinity challenge depending on the salinity dose and time of exposure to hypersalinity; antioxidant capacity has been improved in Tilapia nilotica after exposure to high salinity for a short time (Baldissera et al., 2019; Xavier et al., 2020). The current results agree with Xu et al. (2018) who denoted that hypersalinity for a long time has no effect on CAT activity in Tilapia nilotica. In this study, the powerful upregulation of CAT gene expression in the liver may oppose its expression in the kidney, spleen, and gills via a negative feedback mechanism.
Hypersalinity induced an osmoregulatory modification in the gills including the activation of the Na+/K+ ATPase pump which is considered a compensatory response to the increased absorption of ions, so the elevation of gill’s NKA activity is an integral part of a successful acclimation to increased salinity (Sangiao-Alvarellos et al., 2006) besides the high density of mitochondria in the gills and the elongation of the cristae and acquiring tubular profile. Subsequently, there is an increase in the ability to produce energy by ATPase synthase, which is necessary for the generation of ATP. Thus, high energy production is the key activator of the Na+/K+ ATPase pump which regulates ion exchange during hypersalinity ending with tolerance (Paumard et al., 2002). Moreover, the elevation of environmental salinity changes the ultrastructure of chloride cells which perform an integral role in acid-base regulation during exposure to high salinity by the regulation of Cl-/HCO3- exchange (Perry, 1997). Previous studies have proved the increase in the number and activity of chloride cells after salinity exposure in the gills of common carp (Al-Hilalli and Alkhshali, 2019), Oreochromis mossambicus (Lee et al., 2000), and juvenile Australian snapper (Fielder et al., 2007).
Exposure to high salinity requires a marked change in the osmoregulatory mechanism to maintain osmotic homeostasis by excessive water intake and ion excretion. For the active and passive transport of ions and water, all the epithelia in the intestine, gills, and kidney play an essential role in the exchange between water and ions, but the intestinal epithelia are predominant in the regulation of fluid intake among the previous organs (Evans, 2008). The intestine tight junction consists of several physiologically regulated proteins forming the circumferential seals around the adjacent epithelial cells called claudins. Claudins gene expression was upregulated during hypersalinity as they regulated the permeability of charged and uncharged molecules (Van Itallie et al., 2008).
The multivariate analyses represented by heatmaps and VIP scores affirm the abovementioned data. The clustering heatmap cleared observable differences between the concentration values of all variables in response to different changes in salinity % exposure. These data strongly confirm the responsive adaptive mechanisms of common carp to changes in salinity concentration. Therefore, the VIP scores revealed that the TNF-α, IL-1β, CRH, and CAT mRNA in the liver, kidney, and spleen tissue along with blood Na+, K+, Cl-, and cortisone were the most contributing factors in the adaptive mechanism of common carp to salinity alterations.
The current experiment is considered a novel study that explains the molecular and biochemical mechanisms for the acclimation to hypersalinity stress in common carp. Total serum protein, albumin, and globulins decreased significantly, while blood glucose, circulating cortisol, Na+, K+, and Cl- increased, by the adverse effect of hypersalinity. In addition, hypersalinity is considered an immune and stress stimulant by upregulating gene expression of pro-inflammatory cytokines, TNF-α, and IL-1β, as well as CRH genes in the liver, kidney, and spleen. Moreover, the acclimation of common carp to hypersalinity is represented by a marked upregulation of CAT gene expression in the liver without changes in the kidney and spleen. It is worth mentioning that the salinity challenge had a minimum effect on the gill tissue by regulating the IL-1β gene only without changes in TNF-α, CRH, and CAT gene expression. Furthermore, future study is required, using advanced techniques such as electron microscopy for an in-depth clarification of the activity and number of chloride cells inside the gill’s microstructure as the main osmoregulatory organ in hypersalinity conditions. In addition, further molecular study is necessary for Na+/K+ ATPase and Aquaporin gene expression which are the main contributors of homeostasis to hypersalinity.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
The animal study was reviewed and approved by the Ethical Research Committee of Faculty of Veterinary Medicine in South Valley University, Egypt under approval number (No. 48/12.9.2022).
HA, KB, and WE: Conceptualization, Formal analysis, Methodology, Investigation, Writing—original draft, and Writing – review and editing. AA, MA, and MS: Conceptualization, Formal analysis, Methodology, Data curation, Formal analysis, Writing—original draft, and Writing – review and editing. HE, ME, and SB: Conceptualization, Methodology, Investigation, Data curation, Formal analysis, Validation, and Writing—original draft. LF and SI: Resources, Software, Data curation, Validation, Visualization, Writing – original draft, Writing – review and editing, and Funding acquisition. FI: Software, Data curation, Validation, Writing – review and editing, and Funding acquisition. All authors contributed to the article and approved the submitted version.
This research was funded by the Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2023R127), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia; and the project 6PFE of the University of Life Sciences "King Mihai I" from Timisoara and Research Institute for Biosecurity and Bioengineering from Timisoara.
The authors are grateful for the resources provided by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2023R127), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. This paper is published by the project 6PFE of the University of Life Sciences “King Mihai I” from Timisoara and the Research Institute for Biosecurity and Bioengineering from Timisoara. Moreover, the authors would like to thank Prof. Ahmad Abd Elhady Elkamel, Professor and head of the Aquatic Animal Medicine and Management Department, Faculty of Veterinary Medicine, Assiut University, for his technical assistance with RT-qPCR.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Akther M., Mollah A., Kadir M. (2013). Laboratory investigation on salinity tolerance to Barbodes gonionotus (Bleeker). Progressive Agric. 20, 193–200. doi: 10.3329/pa.v20i1-2.16871
Al-Hilalli H., Alkhshali M. (2019). Influence of transfer to high salinity on chloride cells, oxygen and energy consumption in common carp Cyprinus carpio. J. Anim. Sci. products 2, 1–12.
Al-Khshali M. S., Al-Hilali H. A. (2019). Some physiological changes (ALP, AST AND ALT) of common carp (Cyprinus carpio) caused by high salinity. Biochem. Cell. Arch. 2, 4605–4610. doi: 10.35124/bca.2019.19.2.4605
Armesto P., Cousin X., Salas-Leiton E., Asensio E., Manchado M., Infante C. (2015). Molecular characterization and transcriptional regulation of the renin–angiotensin system genes in Senegalese sole (Solea senegalensis Kaup 1858): differential gene regulation by salinity. Comp. Biochem. Physiol. Part A.: Mol. Integr. Physiol. 184, 6–19. doi: 10.1016/j.cbpa.2015.01.021
Atrees S. S., Rabia H. M. (2021). Herbs and supplements for liver toxicity: a review on mode of action of herbs and supplements on liver toxicity. Med. J. Cairo Univ. 89, 2179–2183. doi: 10.21608/mjcu.2021.203687
Baldissera M. D., Souza C. F., Descovi S. N., Petrolli T. G., Da Silva A. S., Baldisserotto B. (2019). A caffeine-supplemented diet modulates oxidative stress markers and prevents oxidative damage in the livers of Nile tilapia (Oreochromis niloticus) exposed to hypoxia. Fish Physiol. Biochem. 45, 1041–1049. doi: 10.1007/s10695-019-00616-7
Bernier N. J., Gorissen M., Flik G. (2012). Differential effects of chronic hypoxia and feed restriction on the expression of leptin and its receptor, food intake regulation and the endocrine stress response in common carp. J. Exp. Biol. 215, 2273–2282. doi: 10.1242/jeb.066183
Brocker C., Thompson D. C., Vasiliou V. (2012). The role of hyperosmotic stress in inflammation and disease. BioMol. Concepts 3, 345–364. doi: 10.1515/bmc-2012-0001
Buddington R. K., Krogdahl Å. (2004). Hormonal regulation of the fish gastrointestinal tract. Comp. Biochem. Physiol. Part A.: Mol. Integr. Physiol. 139, 261–271. doi: 10.1016/j.cbpb.2004.09.007
Cao W., Bi S., Chi C., Dong Y., Xia S., Liu Z., et al. (2022). Effects of high salinity stress on the survival, gill tissue, enzyme activity and free amino acid content in razor clam Sinonovacula constricta. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.839614
Castillo J., Castellana B., Acerete L., Planas J. V., Goetz F. W., Mackenzie S., et al. (2008). Stress-induced regulation of steroidogenic acute regulatory protein expression in head kidney of gilthead seabream (Sparus aurata). J. Endocrinol. 196, 313–322. doi: 10.1677/JOE-07-0440
Chambers A. F., Matrisian L. M. (1997). Changing views of the role of matrix metalloproteinases in metastasis. J. Natl. Cancer Inst. 89, 1260–1270. doi: 10.1093/jnci/89.17.1260
Chowdhury S., Saikia S. K. (2020). Oxidative stress in fish: a review. J. Sci. Res. 12, 145–160. doi: 10.3329/jsr.v12i1.41716
Demska-Zakęś K., Gomułka P., Rożyński M., Zakęś Z. (2021). Effect of a short-term sodium chloride bath on juvenile pikeperch (Sander lucioperca) welfare. Aquac. Rep. 19, 100569. doi: 10.1016/j.aqrep.2020.100569
Dhabhar F. S. (2008). Enhancing versus suppressive effects of stress on immune function: implications for immunoprotection versus immunopathology. Allergy Asthma Clin. Immunol. 4, 2–11. doi: 10.1186/1710-1492-4-1-2
El-Leithy A. A. A., Hemeda S. A., El Naby W. S. H. A., El Nahas A. F., Hassan S. A. H., Awad S. T., et al. (2019). Optimum salinity for Nile tilapia (Oreochromis niloticus) growth and mRNA transcripts of ion-regulation, inflammatory, stress- and immune-related genes. Fish Physiol. Biochem. 45, 1217–1232. doi: 10.1007/s10695-019-00640-7
Emeish W. (2019). ADAPTATION OF COMMON CARP TO SALINITY. Assiut. Vet. Med. J. 65, 101–110. doi: 10.21608/avmj.2019.168957
Evans D. H. (2008). Teleost fish osmoregulation: what have we learned since august krogh, homer smith, and ancel keys. Am. J. Physiol.-Regulatory Integr. Comp. Physiol. 295, R704–R713. doi: 10.1152/ajpregu.90337.2008
Evans D., Claiborne J. (2009). “Osmotic and ionic regulation infishes,” in Osmotic and ionic regulation:cells and animals. Ed. Evans D. E. (Boca Raton: CRC Press), 295–366.
Evans T. G., Kültz D. (2020). The cellular stress response in fish exposed to salinity fluctuations. J. Exp. Zool. Part A.: Ecol. Integr. Physiol. 333, 421–435. doi: 10.1002/jez.2350
FAO (2003). Unlocking the water potential of agriculture (Italy: Food and Agriculture Organization United Nations), 62.
Fielder D. S., Allan G. L., Pepperall D., Pankhurst P. M. (2007). The effects of changes in salinity on osmoregulation and chloride cell morphology of juvenile Australian snapper, pagrus auratus. Aquaculture 272, 656–666. doi: 10.1016/j.aquaculture.2007.08.043
Fitzsimmons K., Janjua R. S. N., Ashraf M. (2015). “Aquaculture handbook, fish farming and nutrition in Pakistan,” in SoyPak (Pakistan: ASA/WISHH), 441.
Gallo V. P., Civinini A. (2003). Survey of the adrenal homolog in teleosts. Int. Rev. Cytol. 230, 89–187. doi: 10.1016/S0074-7696(03)30003-8
Gao J., Wang D., Liu D., Liu M., Ge Y., Jiang M., et al. (2015). Tumor necrosis factor-related apoptosis-inducing ligand induces the expression of proinflammatory cytokines in macrophages and re-educates tumor-associated macrophages to an antitumor phenotype. Mol. Biol. Cell 26, 3178–3189. doi: 10.1091/mbc.e15-04-0209
Ghelichpour M., Mirghaed A. T., Hoseinifar S. H., Khalili M., Yousefi M., Van Doan H., et al. (2019). Expression of immune, antioxidant and stress related genes in different organs of common carp exposed to indoxacarb. Aquat. Toxicol. 208, 208–216. doi: 10.1016/j.aquatox.2019.01.011
Gonzalez R. J. (2012). The physiology of hyper-salinity tolerance in teleost fish: a review. J. Comp. Physiol. B. 182, 321–329. doi: 10.1007/s00360-011-0624-9
Hamáčková J., Kouril J., Kozák P., Stupka Z. (2006). Clove oil as an anaesthetic for different freshwater fish species. Bulgarian J. Agric. Sci. 12, 185–194. doi: 10.1007/s10695-012-9682-5
Harlé G., Kaminski S., Dubayle D., Frippiat J.-P., Ropars A. (2018). Murine splenic b cells express corticotropin-releasing hormone receptor 2 that affect their viability during a stress response. Sci. Rep. 8, 143. doi: 10.1038/s41598-017-18401-y
Huising M. O., Metz J. R., Van Schooten C., Taverne-Thiele A. J., Hermsen T., Verburg-Van Kemenade B. M., et al. (2004). Structural characterisation of a cyprinid (Cyprinus carpio l.) CRH, CRH-BP and CRH-R1, and the role of these proteins in the acute stress response. J. Mol. Endocrinol. 32, 627–648. doi: 10.1677/jme.0.0320627
Hwang J., Kim S., Seo Y., Lee K., Park C., Choi Y., et al. (2018). Mechanisms of salinity control in Sea bass. Biotechnol. Bioprocess Eng. 23, 271–277. doi: 10.1007/s12257-018-0049-3
Jumah Y. (2020). A review on the activity of Na+/K+-ATPase in branchial ionocytes and its role in salinity adaptation among diadromous species. World J. Advanced Res. Rev. 6, 201–211. doi: 10.30574/wjarr.2020.6.2.0158
Kim J. H., Kang J. C. (2015). Oxidative stress, neurotoxicity, and non-specific immune responses in juvenile red sea bream, Pagrus major, exposed to different waterborne selenium concentrations. Chemosphere 135, 46–52. doi: 10.1016/j.chemosphere.2015.03.062
Lee T. H., Hwang P. P., Shieh Y. E., Lin C. H. (2000). The relationship between `deep-hole’ mitochondria-rich cells and salinity adaptation in the euryhaline teleost, Oreochromis mossambicus. Fish Physiol. Biochem. 23, 133–140. doi: 10.1023/A:1007818631917
Li D. Q., Luo L., Chen Z., Kim H. S., Song X. J., Pflugfelder S. C. (2006). JNK and ERK MAP kinases mediate induction of IL-1beta, TNF-alpha and IL-8 following hyperosmolar stress in human limbal epithelial cells. Exp. Eye Res. 82, 588–596. doi: 10.1016/j.exer.2005.08.019
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu M., Su M., Liu N., Zhang J. (2022). Effects of environmental salinity on the immune response of the coastal fish Scatophagus argus during bacterial infection. Fish Shellfish Immunol. 124, 401–410. doi: 10.1016/j.fsi.2022.04.029
Marx M. T. S., Souza C. D. F., Almeida A. P. G., Descovi S. N., Bianchini A. E., Martos-Sitcha J. A., et al. (2022). Expression of ion transporters and Na+/K+-ATPase and H+-ATPase activities in the gills and kidney of silver catfish (Rhamdia quelen) exposed to different pHs. Fishes 7, 261. doi: 10.3390/fishes7050261
Metz J. R., Huising M. O., Leon K., Verburg-Van Kemenade B. M., Flik G. (2006). Central and peripheral interleukin-1beta and interleukin-1 receptor I expression and their role in the acute stress response of common carp, Cyprinus carpio l. J. Endocrinol. 191, 25–35. doi: 10.1677/joe.1.06640
Mohamed N. A., Saad M. F., Shukry M., El-Keredy A. M. S., Nasif O., Van Doan H. (2021). Physiological and ion changes of Nile tilapia (Oreochromis niloticus) under the effect of salinity stress. Aquac. Rep. 19, 100567. doi: 10.1016/j.aqrep.2020.100567
Mráz J. (2012). Lipids in common carp (Cyprinus carpio) and effects on human health. Doctoral Thesis (Uppsala:Swedish University of Agricultural Sciences) 36.
Paumard P., Vaillier J., Coulary B., Schaeffer J., Soubannier V., Mueller D. M., et al. (2002). The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 21, 221–230. doi: 10.1093/emboj/21.3.221
Perry S. F. (1997). The chloride cell: structure and function in the gills of freshwater fishes. Annu. Rev. Physiol. 59, 325–347. doi: 10.1146/annurev.physiol.59.1.325
Saeij J. P. J., Van Muiswinkel W. B., Van De Meent M., Amaral C., Wiegertjes G. F. (2003). Different capacities of carp leukocytes to encounter nitric oxide-mediated stress: a role for the intracellular reduced glutathione pool. Dev. Comp. Immunol. 27, 555–568. doi: 10.1016/S0145-305X(02)00158-1
Salati A. P., Baghbanzadeh A., Soltani M., Peyghan R., Riazi G. (2011). Effect of different levels of salinity on gill and kidney function in common carp Cyprinus carpio (Pisces: cyprinidae). Ital. J. Zool. 78, 298–303. doi: 10.1080/11250003.2011.567400
Sangiao-Alvarellos S., Arjona F. J., Míguez J. M., Del Río M. P. M., Soengas J. L., Mancera J. M. (2006). Growth hormone and prolactin actions on osmoregulation and energy metabolism of gilthead sea bream (Sparus auratus). Comp. Biochem. Physiol. Part A.: Mol. Integr. Physiol. 144, 491–500. doi: 10.1016/j.cbpa.2006.04.015
Saurabh S., Sahoo P. K. (2008). Lysozyme: an important defence molecule of fish innate immune system. Aquac. Res. 39, 223–239. doi: 10.1111/j.1365-2109.2007.01883.x
Schmitz M., Ziv T., Admon A., Baekelandt S., Mandiki S. N., L’hoir M., et al. (2017). Salinity stress, enhancing basal and induced immune responses in striped catfish pangasianodon hypophthalmus (Sauvage). J. Proteomics 167, 12–24. doi: 10.1016/j.jprot.2017.08.005
Schyth B. D., Lorenzen N., Pedersen F. S. (2006). Antiviral activity of small interfering RNAs: specificity testing using heterologous virus reveals interferon-related effects overlooked by conventional mismatch controls. Virology 349, 134–141. doi: 10.1016/j.virol.2006.01.009
Sharma K., Dey A., Kumar S., Choudhary B., Mohanty S., Kumar T., et al. (2020). Effect of salinity on growth, survival and biochemical alterations in the freshwater fish Labeo rohita (Hamilton 1822). Indian J. Fish. 67 (2), 41–47. doi: 10.21077/ijf.2019.67.2.86894-06
Sopinka N. M., Donaldson M. R., O’connor C. M., Suski C. D., Cooke S. J. (2016). “Stress indicators in fish,” in Fish physiology (United Kingdom: Elsevier), 405–462.
Sunarto A., Mccoll K. A. (2015). Expression of immune-related genes of common carp during cyprinid herpesvirus 3 infection. Dis. Aquat. organisms 113, 127–135. doi: 10.3354/dao02824
Takvam M., Wood C. M., Kryvi H., Nilsen T. O. (2021). Ion transporters and osmoregulation in the kidney of teleost fishes as a function of salinity. Front. Physiol. 12, 664588. doi: 10.3389/fphys.2021.664588
Tang C.-H., Lai D.-Y., Lee T.-H. (2012). Effects of salinity acclimation on Na+/K+-ATPase responses and FXYD11 expression in the gills and kidneys of the Japanese eel (Anguilla japonica). Comp. Biochem. Physiol. Part A. Mol. Integr. Physiol. 163, 302–310. doi: 10.1016/j.cbpa.2012.07.017
Tapia-Paniagua S. T., Reyes-Becerril M., Ascencio-Valle F. D. J., Esteban M. Á., Clavijo E., Balebona S. C., et al. (2011). Modulation of the intestinal microbiota and immune system of farmed sparus aurata by the administration of the yeast debaryomyces hansenii L2 in conjunction with inulin. J. Aquac. Res. Dev. 2011, 1–12. doi: 10.4172/2155-9546.S1-012
Tort L. (2011). Stress and immune modulation in fish. Dev. Comp. Immunol. 35, 1366–1375. doi: 10.1016/j.dci.2011.07.002
Van Itallie C. M., Holmes J., Bridges A., Gookin J. L., Coccaro M. R., Proctor W., et al. (2008). The density of small tight junction pores varies among cell types and is increased by expression of claudin-2. J. Cell Sci. 121, 298–305. doi: 10.1242/jcs.021485
Velasco J., Gutiérrez-Cánovas T., Botella-Cruz M., Sánchez-Fernández D., Arribas P., Carbonell J., et al. (2019). Effects of salinity changes on aquatic organisms in a multiple stressor context. Philos. Trans. R. Soc. B. Biol. Sci. 374, 20180011. doi: 10.1098/rstb.2018.0011
Wang L., Lin W., Zha Q., Guo H., Zhang D., Yang L., et al. (2020). Persistent exposure to environmental levels of microcystin-LR disturbs cortisol production via hypothalamic-Pituitary-Interrenal (HPI) axis and subsequently liver glucose metabolism in adult Male zebrafish (Danio rerio). Toxins (Basel) 12 (5), 282. doi: 10.3390/toxins12050282
Wang X., Wang L., Ren Q., Yin S., Liang F., Jia Y. (2016). Two superoxide dismutases (SODs) respond to bacterial challenge identified in the marbled eel Anguilla marmorata. Aquaculture 451, 316–325. doi: 10.1016/j.aquaculture.2015.09.025
Wang P. J., Yang W. K., Lin C. H., Hwang H. H., Lee T. H. (2017). FXYD8, a novel regulator of renal Na(+)/K(+)-ATPase in the euryhaline teleost, Tetraodon nigroviridis. Front. Physiol. 8, 576. doi: 10.3389/fphys.2017.00576
Xavier W. D. S., Leclercq E., Carvalho P. L. P. F., Vicente I. S. T., Guimarães M. G., Rodrigues E. J. D., et al. (2020). The putative effect of a SOD-rich melon pulp-concentrate on growth performance and antioxidant status of Nile tilapia (Oreochromis niloticus) under heat/dissolved oxygen-induced stress. Aquaculture 529, 735669. doi: 10.1016/j.aquaculture.2020.735669
Xu C., Li E., Suo Y., Su Y., Lu M., Zhao Q., et al. (2018). Histological and transcriptomic responses of two immune organs, the spleen and head kidney, in Nile tilapia (Oreochromis niloticus) to long-term hypersaline stress. Fish Shellfish Immunol. 76, 48–57. doi: 10.1016/j.fsi.2018.02.041
Xu C., Suo Y., Wang X., Qin J. G., Chen L., Li E. (2020). Recovery from hypersaline-Stress-Induced immunity damage and intestinal-microbiota changes through dietary β-glucan supplementation in Nile tilapia (Oreochromis niloticus). Animals: an Open Access J. MDPI 10, 2243. doi: 10.3390/ani10122243
Yahfoufi N., Alsadi N., Jambi M., Matar C. (2018). The immunomodulatory and anti-inflammatory role of polyphenols. Nutrients 10, 1618. doi: 10.3390/nu10111618
Zapata A., Diez B., Cejalvo T., Gutiérrez-De Frías C., Cortés A. (2006). Ontogeny of the immune system of fish. Fish Shellfish Immunol. 20, 126–136. doi: 10.1016/j.fsi.2004.09.005
Keywords: salinity stress, oxidative damage, TNF-α, IL-1β, CRH, catalase
Citation: Ahmed H, Bakry KA, Abdeen A, El bahgy HEK, Abdo M, Imbrea F, Fericean L, Elshemy MA, Ibrahim SF, Shukry M, Baloza SH and Emeish WFA (2023) The involvement of antioxidant, stress, and immune-related genes in the responsive mechanisms of common carp (Cyprinus carpio) to hypersalinity exposure. Front. Mar. Sci. 10:1195016. doi: 10.3389/fmars.2023.1195016
Received: 27 March 2023; Accepted: 11 May 2023;
Published: 02 June 2023.
Edited by:
Ahmed H. El-Sappah, Zagazig University, EgyptReviewed by:
Qudratullah Kalwar, Chinese Academy of Agricultural Sciences, ChinaCopyright © 2023 Ahmed, Bakry, Abdeen, El bahgy, Abdo, Imbrea, Fericean, Elshemy, Ibrahim, Shukry, Baloza and Emeish. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hassan Ahmed, aGFzc2FuLXlvdW5lc0B2ZXQuc3Z1LmVkdS5lZw==; Ahmed Abdeen, YWhtZWQuYWJkZWVuQGZ2dG0uYnUuZWR1LmVn; Florin Imbrea, ZmxvcmluX2ltYnJlYUB1c2FiLXRtLnJv
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.