
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 12 September 2023
Sec. Marine Molecular Biology and Ecology
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1190475
This article is part of the Research Topic Understanding the Effects of Climate Change on Marine Biodiversity through Genomics and Transcriptomics View all 6 articles
Introduction: The coastal ecosystem is a hub of both marine organisms and human activities. It plays a crucial role in human food production and affects facilities through biofouling. Long-read amplicon sequencing provides more accurate species identification and demonstrates numerous advantages in community diversity studies, making it an effective tool for ecological monitoring.
Methods: To investigate the zooplankton community characteristics in the oyster aquaculture area, the composition and temporal dynamics of zooplankton near Longwan Bay, Qingdao, China, were determined by the 18S rRNA gene long reads sequencing technique.
Results: A total of 89 zooplankton species were identified, among which copepods were the most abundant. Zooplankton composition and proportional abundances showed significant seasonal variations. The species richness in summer was the highest, while the species abundance in spring was the highest. Oyster farming showed weak influence on the zooplankton community variation. Paracalanus parvus abundance was higher in most assayed months. Chthamalus stellatus showed a strong temporal preference, with the highest percentage in May. Sea water temperature and species-species interactions were revealed to be the main contributors to the shifts in the community composition.
Discussion: The ubiquitous positive correlation between zooplankton suggests that species interactions are important in adaptation to the changing environment. The results reveal the seasonal occurrence of several major biofouling organisms and help improve biofouling management efficiency.
Zooplankton functions as a bridge between producers and consumers. The changes in the zooplankton abundance affect primary and higher-order productivity in the oceans (Hernández-MIranda et al., 2022). The study of zooplankton composition is of great value to support research on ecosystem dynamics (Shi et al., 2020) and biofouling management. Many zooplankton species have short life cycles, and their richness and distribution are greatly affected by environmental factors. Community structure can serve as an indicator of local ecological conditions and environmental health and provide basic data for ecosystem detection (Pearson, 1978; Zhao et al., 2022). Oysters are the predominantly cultured species in China, potentially changing the composition of zooplanktonic and biofouling organisms around the oyster farm (Das et al., 2012). However, the structure and dynamic patterns of the zooplankton community in the oyster farm are poorly studied.
DNA barcoding technology has become increasingly important in ecological research (Valentini et al., 2009; Mugnai et al., 2021). Compared with traditional methods, it has the advantages of fast identification and high accuracy (Savolainen et al., 2005). The 18S rRNA gene includes conserved and highly variable regions, which can reflect genetic relationships among species and show the differences between species. It has been successfully applied in many studies: the discovery of new species (Kakui and Munakata, 2022; Kumar et al., 2022), the assessment of marine biological diversity (Liu et al., 2020), community characteristics and environmental correlation analysis (Liu et al., 2022). This study analyzed the species richness, diversity, and temporal dynamics of zooplankton in an oyster farm area using the third-generation sequencing-based DNA barcoding technique. The correlation analysis indicated significant interactions between zooplankton species and provided a basis for in-depth research on the interactions between human activities and marine ecosystems.
The study was carried out from May 2020 to May 2021 in the Pacific oyster farm located in Longwan Bay, the northern part of the Yellow Sea (Figure 1A). A total of 33 monthly zooplankton samples were collected from three sites by a shallow water type- II plankton net (∼50 μm mesh size) at a depth of ~1 m where the Pacific oysters were cultured. Samples were fixed in 70% ethanol at 4°C for further processing. Water temperature, salinity, pH, and dissolved oxygen (DO) were measured in situ with a multi-parameter portable meter (Multi 3630 IDS, Germany).
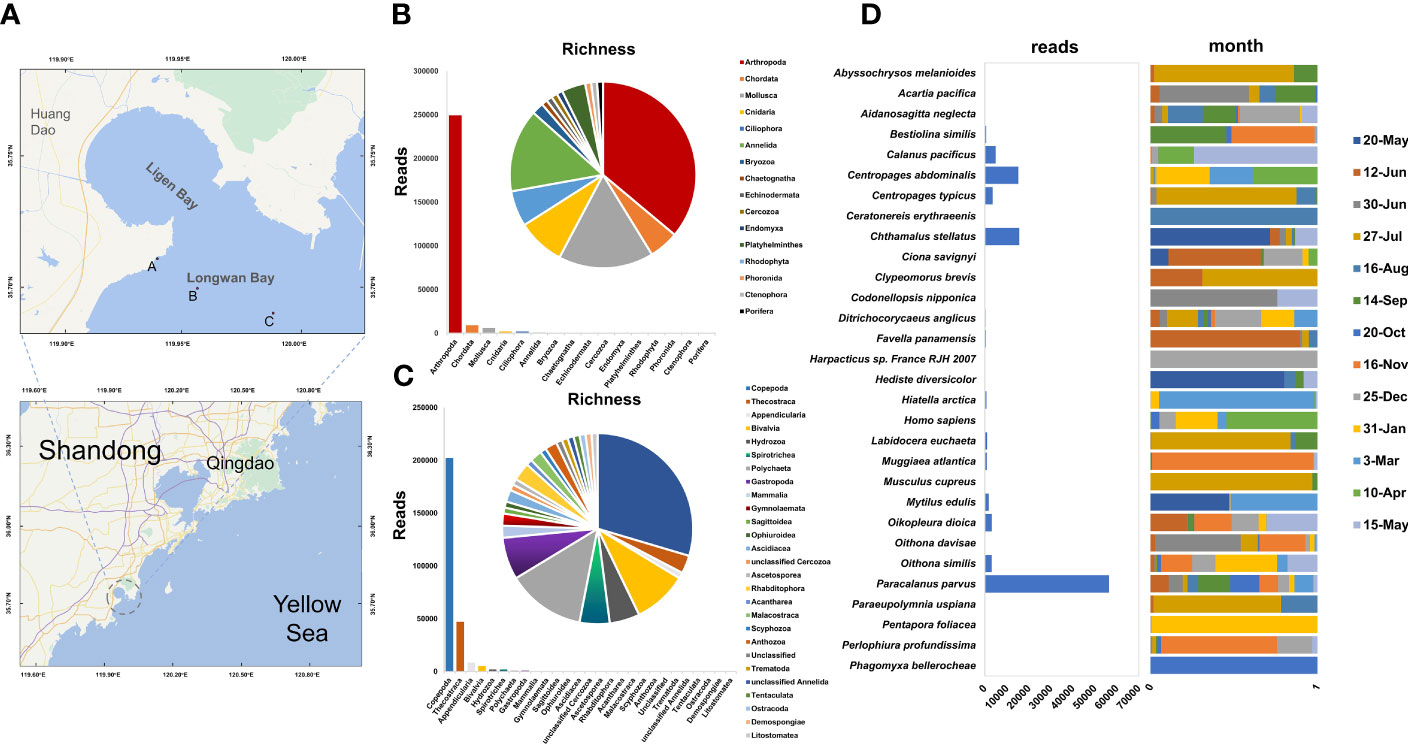
Figure 1 Zooplankton richness and relative abundances. (A) Locations of 3 sampling sites (point A, point B and C) in the aquaculture area. (B, C) Operational taxonomic units (OTUs) richness and reads numbers of zooplankton phyla and classes. The pie chart represents richness. The bar chart represents abundance (the number of reads). (D) Reads number and the percentage distribution of reads per month changes for Top30 species (ranking based on reads number).
Samples were pretreated by centrifugation at 5000 ×g, and the pellets were washed twice with PBS. The total genomic DNA was then extracted using the Tissue DNA Kit (Omega, USA) according to the manufacturer’s protocol. DNA quality and quantity were examined by 1% agarose gel electrophoresis and NanoDrop 2000 (Thermo Fisher Scientific, USA). The 18S rRNA genes were amplified using universal primers: the forward primer 5’-AACCTGGTTGATCCTGCCAGT-3’, and the reverse primer 5 ‘-GATCCTTCTGCAGGTTCACCTAC-3’ (Bradley et al., 2016). The specific steps of PCR are as below: one cycle of denaturation at 95°C for 4 min, 32 cycles of amplification (95°C for 1 min, 65°C for 2 min, at 72°C for 2 min), and one cycle of extension at 72°C for 10 min. The products were confirmed by 1% agarose gel electrophoresis and purified by the Qiagen Gel Extraction Kit (Qiagen, Germany). The high-throughput sequencing of 18S rRNA gene was conducted on the PacBio Sequel platform by the BioMarker Technologies Company (Beijing, China).
Raw sequences were firstly trimmed to remove the adapters and primers using Cutadapt v2.7 (Martin, 2011) and filtered by LImA v1.7.0 (https://lima.how/) and UCHIME v4.2 (Edgar, 2016) to eliminate most of the low-quality data. All low-quality sequences or unassembled sequences, including chimeric sequences, fuzzy sequences, short sequences (<1200 bp), long sequences (> 2000 bp), and non-specific amplifications, were eliminated for further analysis (Chen et al., 2022). The sequences were clustered into Operational Taxonomic Units (OTUs) based on 97% similarity by using the USEARCH (version 10.0) (Edgar, 2010; Edgar, 2013). Species annotation was mainly conducted in two steps: The feature sequences of OTUs gained by Blast in the QIIME2 software package (Bolyen et al., 2019) were aligned to the SILVA_138 reference database, and then supplementary annotations were obtained with Feature-classifier Classify-sklearn at the confidence threshold of 70% in the QIIME2 software package.
The richness and abundance of species were calculated to reflect the composition characterization of zooplankton communities. Spearman correlations between zooplankton at the phylum or class levels and environmental factors were analyzed by the R package Corrplot (Wei et al., 2021). Correlations between zooplankton OTUs and environmental factors were analyzed and visualized by the R packages Psych (Revelle, 2017) and ggplot2 (Wickham, 2016) respectively. Correlations between zooplankton from phyla to OTU levels were analyzed by R Package Psych and visualized using R Package Circlize (Gu et al., 2014) and ggplot2.
A total of 369,875 circular consensus sequences (CCSs) were obtained through PacBio Sequel sequencing, of which 344,712 were retained after quality filtering. After cluster analysis by Usearch, 179 OTUs were obtained by filtering with a threshold of 0.005%. Twelve OTUs were annotated at the kingdom level, while others were annotated to the phylum level or below the phylum levels: Arthropoda (35 OTUs, 92.21%), Chordata (5 OTUs, 3.34%), Mollusca (16 OTUs, 2.22%), Cnidaria (8 OTUs, 0.77%), Ciliophora (6 OTUs, 0.72%), Annelida (14 OTUs, 0.31%), Bryozoa (2 OTUs, 0.12%), Chaetognatha (1 OTU, 0.093%), Echinodermata (1 OTU, 0.079%), Cercozoa (1 OTU, 0.067%), Endomyxa (1 OTU, 0.036%), Platyhelminthes (4 OTUs, 0.032%), Phoronida (1 OTU, 0.007%), Ctenophora (1 OTU, 0.003%), and Porifera (1 OTU, 0.002%) (Figure 1B). The top five zooplankton classes included Copepoda (29 OTUs, 74.76%), Thecostraca (3 OTUs, 17.41%), Appendicularia (1 OTU, 3.14%), Bivalvia (9 OTUs, 1.92%), and Hydrozoa (5 OTUs, 0.74%) (Figure 1C).
A total of 93 OTUs were annotated at the genera and species level, including 86 genera and 89 species. Most genera contain only one species, except for Centropages (3 species), Oithona (2 species) and Ciona (2 species). The Paracalanus parvus, Chthamalus stellatus, and Centropages abdominalis were the top 3 species with extremely high abundance (Figure 1D).
The species richness (species-level OTU numbers) and relative abundance (total reads of species-level OTUs) analysis of zooplankton samples from May 2020 to May 2021 revealed obvious temporal dynamics (Figure 2). The ANOSIM and NMDS analysis results indicated that samples from the same season were clustered together, and samples in autumn, winter, and spring had overlap, summer samples were independent of other seasons (Figure 2A). The highest richness value appeared in September, while the relative abundance showed the highest in March. Species richness in summer and autumn is much higher than that in spring and winter. Many species exhibit strong temporal dynamics with a seasonal preference, while others are evenly distributed throughout each month. For example, the arthropod C. stellatus mainly presented in May, while Paracalanus parvus was detected in each month with little difference in proportion (Figure 2B).
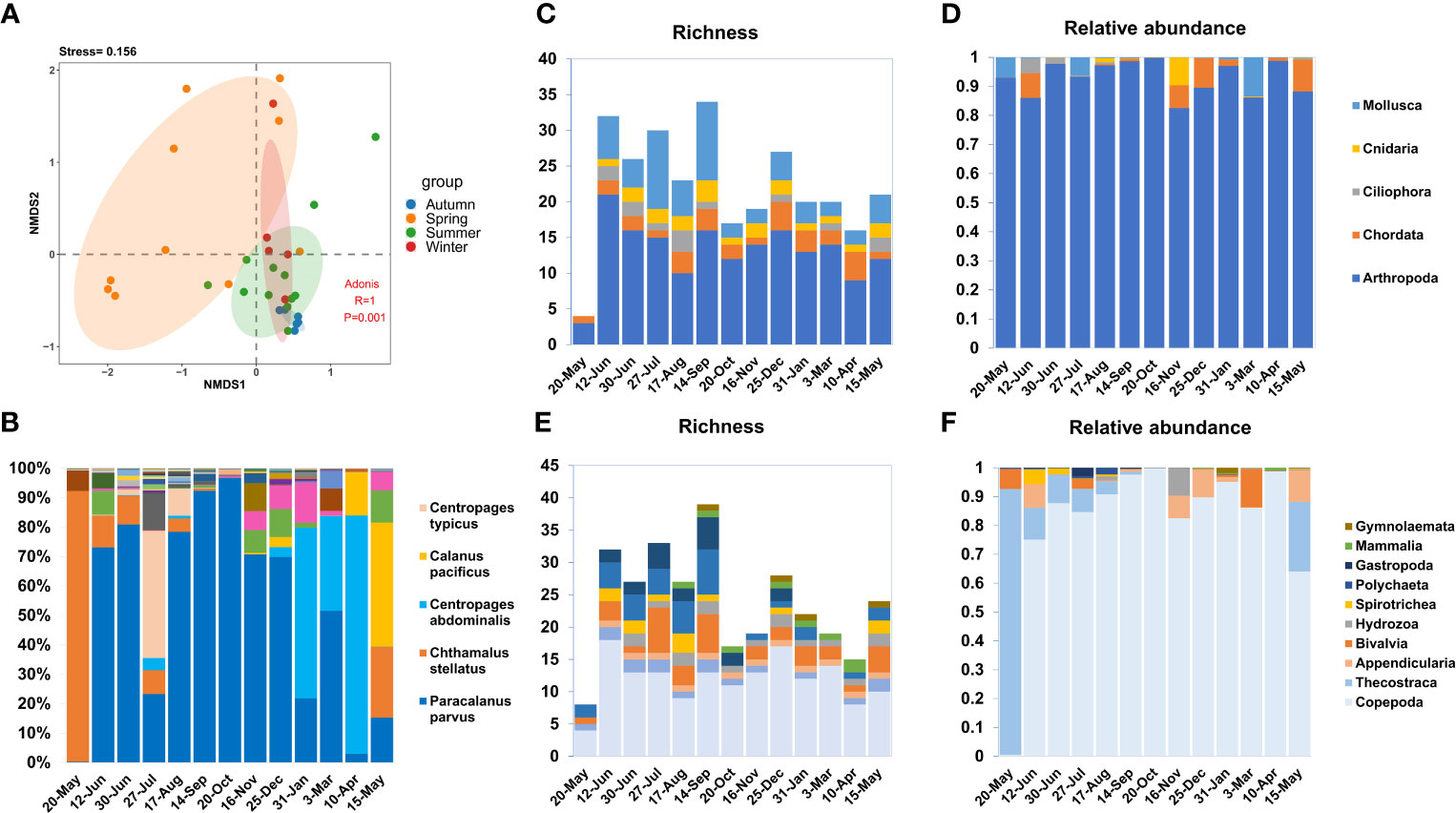
Figure 2 Temporal dynamics for zooplankton community. (A) NMDS and ADONIS analysis for the zooplankton community, demonstrated the differences in zooplankton composition in different seasons. (B) The number of reads in each month is used as a whole to show the proportion of each species in each month, the legend only shows the species with the largest proportion in each month. (C, D) Relative abundance and richness of top 5 phyla, two charts use the same legend. (E, F) Relative abundance and richness of top 10 class, two charts use the same legend.
With copepods being the dominant species, the species richness of Arthropoda is consistently the highest throughout the year. However, molluscan richness was mainly dominated by Bivalvia and Gastropoda, and showed significant seasonal patterns (high abundance in summer) (Figure 2C). Changes in the abundance of zooplankton communities also showed temporal variations. Among all detected OTUs, the relative abundance of Arthropoda was the highest throughout the year (>83%) (Figure 2D), with Copepoda being the most dominant Arthropoda taxon.
The annual changes in the seawater temperatures showed an opposite pattern with the DO (Figure 3A), while the salinity (29~31.2) and pH (7.9~8.4) were relatively stable (Figure 3B). The highest temperature was recorded in August, with an average value of 28.2˚C. An abnormally high DO value was recorded during the August sampling, while a low salinity was also recorded. It was suspected to be caused by river flow or windy weather. The canonical correlation analysis (Kordas et al.) indicated that temperature, DO, and salinity significantly influenced the composition of zooplankton communities, in which, the temperature exerts a dominant influence (Figure 3C). Further correlation analysis with the spearman method on the class-level taxa and environmental factors indicated that the abundance of 48 taxa had significant correlations with environmental factors (50 negatives and 43 positives) (Figure 3D). The abundances of different taxa showed different relationships with the environmental parameters. For example, the Acantharea showed an extremely significant (p<0.01) correlation between salinity and temperature, while the Gastropoda abundance was significantly affected by dissolved oxygen and temperature (p<0.01). Gymnolaemata also showed an extremely significant correlation with salinity (p<0.01) besides a significant correlation with temperature (p<0.05) (Figure 3E).
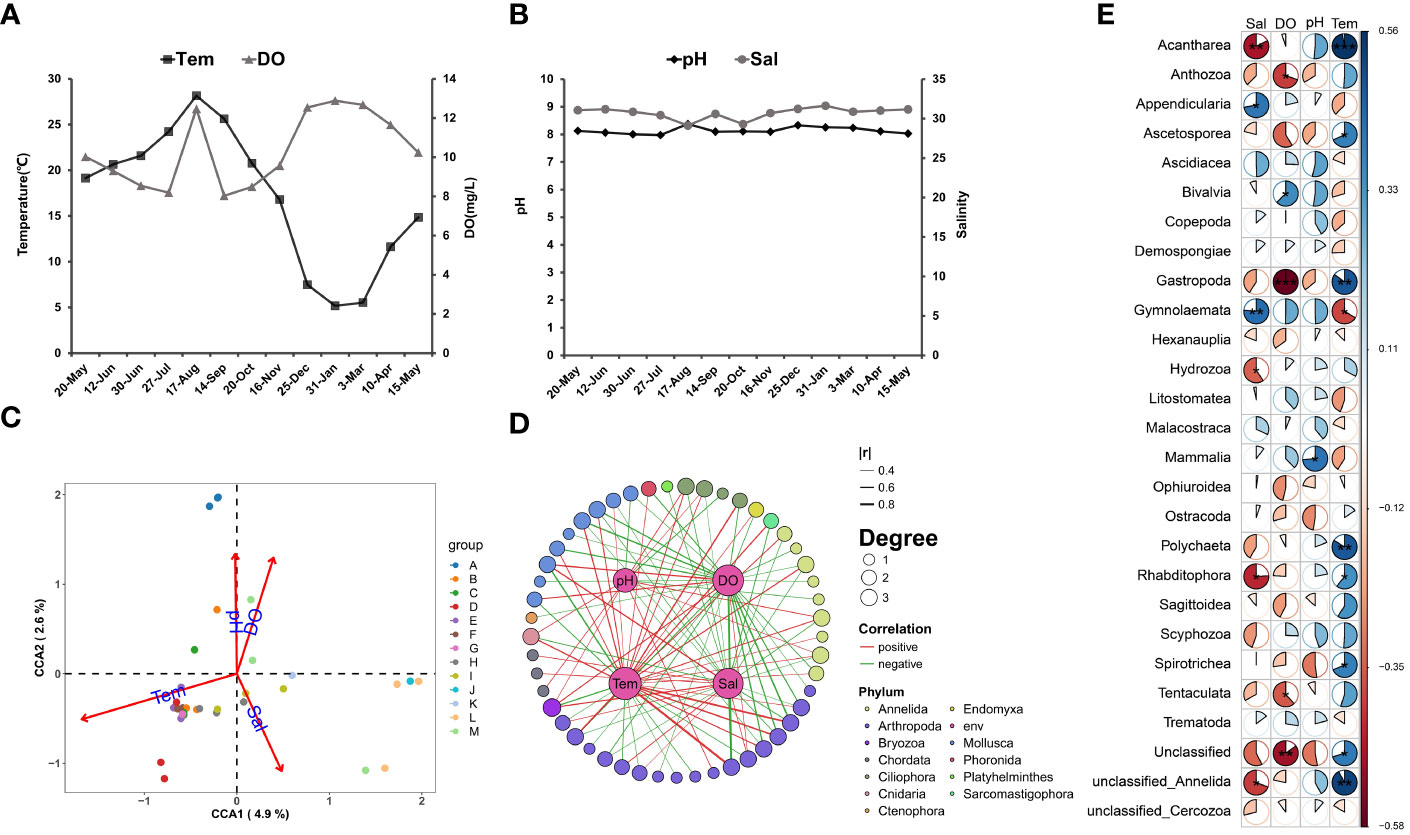
Figure 3 Annual changes in environmental factors and correlation analysis with zooplankton. (A, B) Variation of the environmental factors (Tem, DO, pH and Sal) in the 13 months, take the numerical average of the three sample points. (C) Based on the scatterplot analyzed by CCA, A-M represents the sample collection of different months, showing the similarity between samples in each month and the influence of environmental factors. (D) The Edge Bundling plot based on spearman’s method shows the correlation between each environmental factor and phylum, which only shows a significant correlation (p<0.05), |r| indicates the strength of the correlation, represented by the thickness of the line segment, and Degree represents the number of environmental factors affecting OTU, represented by the size of the point. (E) Based on the sector chart of Spearman’s method, whether the correlation between different classes and environmental factors is significant, red represents negative correlation, blue represents positive correlation, ** represents p<0.01, * represents p<0.05.
Correlation analysis among 12 phyla indicated significant taxa interactions. The correlation between Mollusca and Annelida was extremely significant (p<0.01), while Ctenophora, Endomyxa and Phoronida had a significant positive correlation (p<0.05) (Figure 4A). Significant correlations were also identified among 14 classes. Except for the negative correlation between Spirotrichea and Copepoda, the other pairs were positively correlated (Figure 4B). Similarly, among the 898 OTU-level correlations, 868 were positively correlated, while only 30 were negatively correlated.
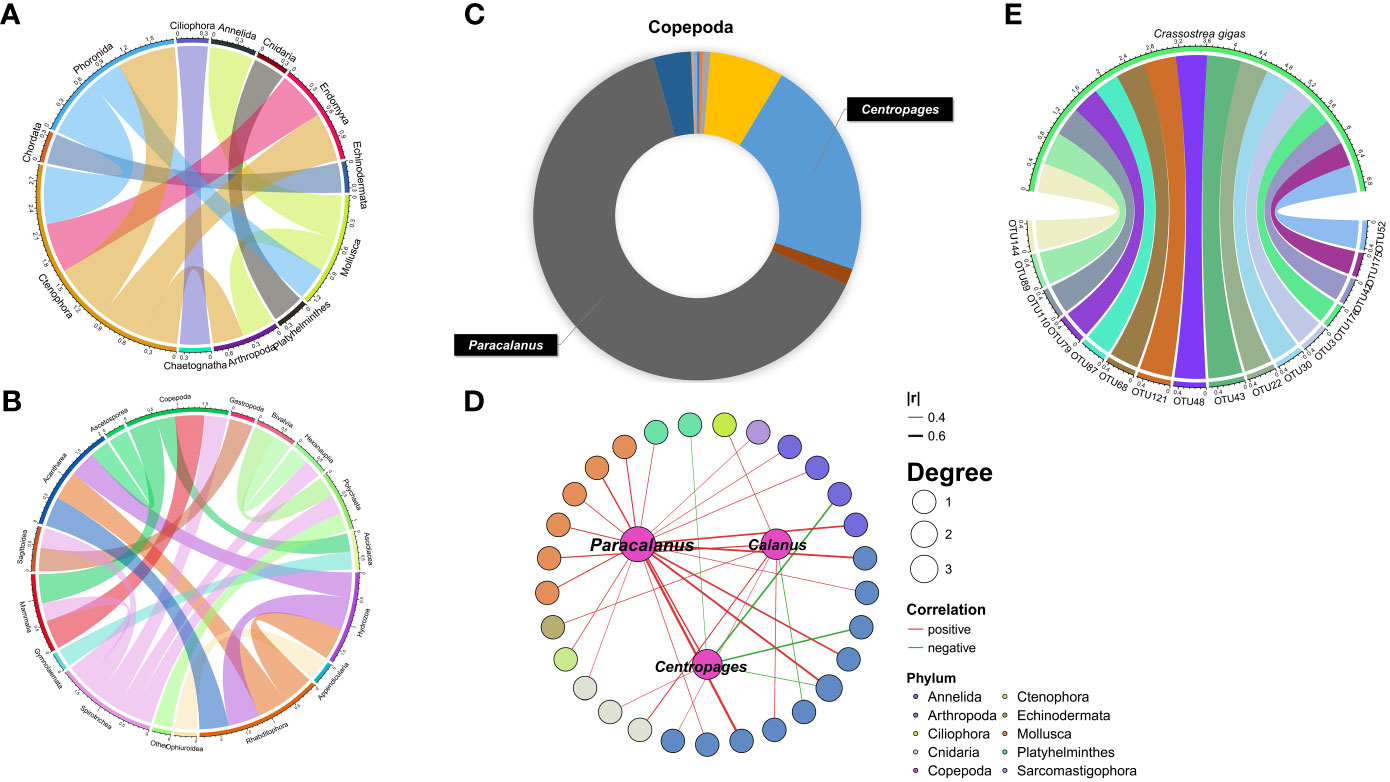
Figure 4 Interaction networks between zooplankton OTUs. (A, B) The Spearman method was used to analyze the correlation between zooplankton inside (phylum and class). (C) Pie chart based on the reads number showing the quantitative composition of each species in the dominant class. (D, E) Analysis of inter-species correlation based on OTU data shows the relationship between C. gigas and OTU, the most dominant species and OTU.
Copepods were the year-round dominant species in this study except for May, when the barnacle C. stellatus boomed (Figure 2B). A total of 21 Crustacea species from 18 genera were identified, with the most abundant species from copepods Paracalanus and Centropages (Figure 4C). The maximum abundance of Centropages typicus was at the end of July, with the maximum richness of Centropages abdominalis was observed in January and April. Paracalanus parvus was the most abundant species in other months (Figure 2B). Spearman correlation analysis indicated that the abundances of Labidocera euchaeta, C. typicus, Canuella perplexa, and Acartia pacifica were significantly correlated with seawater temperature (p<0.01). The dominant genera Paracalanus, Centropages, Calanus showed significant correlations with 29 OTUs (Figure 4D). The abundance of C. gigas peaked in July, and showed a significantly positive correlation with 16 OTUs (Figure 4E)
A total of 89 zooplankton species were identified with the highly reliable 18S rRNA gene long reads sequencing, including the dominant species P. parvus (OTU1 + 172), Oithona similis (OTU7), C. abdominalis (OTU3), Ditrichocorycaeus anglicus (OTU20), and C. typicus (OTU124 + 136). The distribution of these species was consistent with their temperature preference (Hsieh and Chiu, 2004). The species number identified in this study was higher than some other similar studies on aquaculture areas (Guo et al., 2015), which may be a result of different local hydrological conditions (Domínguez et al., 2017) or species identification techniques (Gao et al., 2020). Compared to the traditional morphological identification, the metabarcoding technique has a great advantage in the case of zooplankton larvae or rare species, which leads to the discovery of a larger number of species and facilitates subsequent differential studies (Ayala et al., 2016; Yin et al., 2022). For example, Centropages hamatus and Phyllodoce groenlandica were rarely reported in the studied sea area by traditional morphological identification. However, Acartia bifilosa and Calanus sinicus, the dominant species in the Yellow Sea (Huo et al., 2012; Shi et al., 2020), were not identified in this study. This should be the consequence of the incomplete database which did not deposit the reference sequences of corresponding species (Djurhuus et al., 2018). With the improvement of the molecular marker database, the sequencing result can be re-annotated, and more comprehensive survey results are possibly revealed.
Copepoda is widespread (Cornils and Held, 2014) and one of the most abundant marine organisms in many marine ecosystems (Escribano and Hidalgo, 2000; Hwang et al., 2006). Copepods are thus often used as indicator species for water monitoring (Hsieh et al., 2005; Tseng et al., 2008; Shin et al., 2022). While early studies of copepods are mainly on large copepods (Shimode et al., 2012; Gong et al., 2013), the roles of small and medium-sized zooplankton in marine ecosystems were gradually realized (Middelbo et al., 2019; Tarrant, 2020). Many studies have been conducted on tiny zooplankton species composition, community characteristics, and life habits (Tiselius et al., 2013; Van and Park, 2016; Uttieri et al., 2021). Paracalanus (4 OTUs identified in this study) and Centropages (2 OTUs) are the most common small and medium-sized copepods and are the main contributors to the richness of copepods. They are dominant species in many ocean areas, such as Helgoland, Germany and Maryland Coastal Bays, USA (Oghenekaro et al., 2018; Hirche et al., 2019; Prusova and Galagovets, 2022), as well as the Bohai, Yellow Sea, and East China Sea (Wang et al., 2001; Wang et al., 2002). Previous studies have shown that copepods’ abundance change dramatically impacts the density and diversity of marine zooplankton (Michel and Herring, 1984; Zakaria et al., 2016). Centropages are omnivorous animals, usually preying on eggs and other copepods (Turner et al., 1985; Slater and Hopcroft, 2005). They have been widely identified in multiple sea areas (Dagg and Grill, 1980; Liang et al., 1994). The aquaculture area provides extraordinary biomass and may provide appropriate conditions for the growth and development of Centropages.
Tiny marine organisms are highly susceptible to environmental factors and show significant seasonal variation (Zhang et al., 2019). The abundance of P. parvus exhibited seasonal variations, consistent with the observations from other areas of the China Seas (Ke et al., 2001; Rong et al., 2002; Zheng et al., 2022). However, the maximum abundance season of P. parvus varied among different reports. The optimum temperature range for the growth of P. parvus is 13-24°C (Zhang et al., 2006), which should be responsible for the different peak occurrences for different sea areas. The abundance of C. typicus in the Longwan Bay reached the highest in summer, differing from the April boom in the southern Mediterranean Sea (Halsband-Lenk et al., 2001). It should also be caused by the local temperature of the corresponding seasons. At the same time, the growth of C. abdominalis starts at 5°C, and the optimal temperature is between 10-15°C (Slater and Hopcroft, 2005). It is also consistent with the seasonal distribution of its abundance observed in this study.
Barnacles (Thecostraca) are one of the most common marine biofouling crustaceans, with planktonic larvae and sessile adults (Pérez-Losada et al., 2012). The abundance of Thecostraca was mainly contributed by C. stellatus in this study. The interaction between oysters and barnacles may impact their abundance and distribution. They are all habited in the intertidal zone, forming partial competition on the limited substrates, which will then lead to a decrease in the population. However, barnacles and oysters showed significant niche differences in this area. The barnacle larvae occurrence (May) was earlier than oyster larvae (July).
Furthermore, barnacles adapt to longer dry exposure environments and mainly distribute around high tide lines. The high abundance of C. stellatus larvae in Longwan Bay indicated that oyster farming did not adversely affect its living. The relative abundance of other Mollusa peaks in March, May, and July, mostly were Mytilus edulis, Musculus cupreus, and Hiatella arctica. The molluscan abundance peaks in March and May were mainly contributed by the larvae of mussel M. edulis, which is consistent with its common spawning season (Nordsieck, 2006; Tyler-Walters, 2008). At the same time, the July molluscan occurrence peak was mainly contributed by M. cupreus larvae. The study results indicated a relatively low proportion of oyster larvae in the collected zooplankton samples, even within the oyster farm region. Oyster culture in Longwan Bay should have a limited impact on the marine zooplankton community. It should also benefit from the well-controlling aquaculture density in Longwan Bay.
The diversity of species distribution in marine ecosystems is affected by physicochemical and biological factors (Tittensor et al., 2010), where temperature is the main controlling factor (Gillooly, 2000; Lewandowska et al., 2014; Yasuhara and Danovaro, 2016). Temperature determines the general dissolved oxygen level and affects the composition and species interactions of the zooplankton community (Portner and Farrell, 2008; Kordas et al., 2011). Temperature is also a fundamental determinant of organisms’ life cycle. Too high or too low temperatures can be associated with a high mortality rate (Heinle, 1969). Studies have suggested that increasing temperature accelerates zooplankton metabolism and sexual maturation and reduces zooplankton lifespan (Huang, 1985).
Adult survival and larval occurrence were also influenced by changing ocean temperatures. The Pacific oyster C. gigas is one of the main mariculture species in Longwan Bay, whose optimal water temperature for growth is 15-25°C. According to our field investigation, this bay is very close (~100 km) to the southern distribution limit (around Lianyungang, Jiangsu Province) of C. gigas. Severe summer mortality occurred in recent years, possibly because of environmental changes caused by global warming. The high temperature observed in August (~28°C) has exceeded the optimal temperature range of the Pacific oyster and could cause heat stress. Correspondingly, the development and maturity of oyster gonads may also be influenced by environmental change. The swimming larvae of C. gigas were mainly sampled at the end of July, indicating a dominant mass spawning during July. The newborn oyster spats will then face heat stress during August. If global warming persists, the wild Pacific oyster population and distribution limits may change in this area.
The composition of tiny zooplankton communities is also driven by their biological activities, which are sensitive to temperature and dissolved oxygen (Bērziņš and Pejler, 1989; Zhao et al., 2020). In marine ecosystems, many animals (e.g. ctenophores) feed on zooplankton and have a wide range of adaptations to DO. However, the movement frequency of copepods is reduced in the relatively low DO environment, which improves the predation rate and thus affects the composition of the community (Mary Beth et al., 2004). A previous study indicated that both the survival rate and the reproduction of the copepod Acartia tona and Oithona colcarva were inhibited at a low DO state (Roman et al., 1993).
Indirect and direct species interactions are widely present in marine communities and play an important role in the formation and maintenance of biodiversity (Sun, 2011). Zooplankton is crucial for the marine ecosystem as a hub in species interaction networks (Greve, 1977), where Spirotrichea is significantly associated with Copepoda. Spirotrichea is a microplankton which feeds on photosynthetic algae and bacteria and then as the primary food for copepods and young fish (Calbet and Saiz, 2005; Lima-Mendez et al., 2015; Santoferrara et al., 2017). OTUs in Ascetosporea, the parasitic protozoans infecting aquatic invertebrates (Bass et al., 2019), were significantly correlated with OTUs in Tentaculata. The species interactions revealed by this study indicated that food chain and parasite-host relationships also play important roles in stabilizing plankton communities.
The special and complex environment of the ocean is an important driving force for the formation and evolution of marine biodiversity. Human activities have created new challenges for marine zooplanktons, which were driven to constantly evolve many adaptive traits: morphological, behavioural, physiological traits and reproductive patterns (Shen and Shi, 2002). Species living in the same environmental pressure also exert selective pressure on each other to form a relatively stable biome. The metabarcoding analyses of the zooplankton community in this study not only lay the foundation for the spatiotemporal dynamic mechanism of zooplankton in the aquaculture area but also provide supporting evidence for further understanding of the impact of human activities and global climate change on biome composition. This study provides highly reliable 18s full-length sequence data and reveals major planktonic organisms’ structure and seasonal dynamics in Longwan Bay. The results will be helpful for further monitoring the planktonic community change and improving biofouling management efficiency.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: BioProject, PRJNA947076.
FX conceived and designed the study. XM and BQ collected and processed samples. BQ conducted data analysis. BQ, XM and FX wrote the paper. All authors contributed to the article and approved the submitted version.
This research was supported by the Key Research and Development Program of Shandong (2022LZGC015), the Key Deployment Project of Centre for Ocean Mega-Research of Science, Chinese Academy of Science (No. COMS2019Q11), and the Taishan Scholars Program.
We acknowledge Chonglin Li, Liangheng Li and Shaoxi Deng for their assistance with the sample collection. Most of the calculations were supported by the Oceanographic Data Center, IOCAS.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ayala D., Riemann L., Munk P. (2016). Species composition and diversity of fish larvae in the Subtropical Convergence Zone of the Sargasso Sea from morphology and DNA barcoding. Fish. Oceanogr. 25, 85–104. doi: 10.1111/fog.12136
Bass D., Ward G. M., Burki F. (2019). Ascetosporea. Curr. Biol. 29, R7–R8. doi: 10.1016/j.cub.2018.11.025
Bērziņš B., Pejler B. (1989). Rotifer occurrence in relation to oxygen content. Hydrobiologia 183, 165–172. doi: 10.1007/BF00018721
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bradley I. M., Pinto A. J., Guest J. S. (2016). Design and evaluation of Illumina MiSeq-compatible, 18S rRNA gene-specific primers for improved characterization of mixed phototrophic communities. Appl. Environ. Microbiol. 82, 5878–5891. doi: 10.1128/AEM.01630-16
Calbet A., Saiz E. (2005). The ciliate-copepod link in marine ecosystems. Aquat. Microb. Ecol. 38, 157–167. doi: 10.3354/ame038157
Chen Y., Xu C., Li Q. (2022). Genetic diversity in a genetically improved line of the Pacific oyster Crassostrea gigas with orange shell based on microsatellites and mtDNA data. Aquaculture 549, 737791. doi: 10.1016/j.aquaculture.2021.737791
Cornils A., Held C. (2014). Evidence of cryptic and pseudocryptic speciation in the Paracalanus parvus species complex (Crustacea, Copepoda, Calanoida). Front. Zool. 11, 1–17. doi: 10.1186/1742-9994-11-19
Dagg M., Grill D. (1980). Natural feeding rates of Centropages typicus females in the New York Bight 1. Limnol. Oceanogr. 25, 597–609. doi: 10.4319/lo.1980.25.4.0597
Das P., Mandal S. C., Bhagabati S., Akhtar M., Singh S. (2012). Important live food organisms and their role in aquaculture. In: Sundaray JK, Sukham M, Mohanty RK, Otta SK, editors Front in Aquacult. (New Delhi, India: Narendra Publishing House). p69–86.
Djurhuus A., Pitz K., Sawaya N. A., Rojas-Márquez J., Michaud B., Montes E., et al. (2018). Evaluation of marine zooplankton community structure through environmental DNA metabarcoding. Limnol. Oceanogr.: Methods 16, 209–221. doi: 10.1002/lom3.10237
Domínguez R., Garrido S., Santos A. M. P., Dos Santos A. (2017). Spatial patterns of mesozooplankton communities in the Northwestern Iberian shelf during autumn shaped by key environmental factors. Estuarine Coast. Shelf Sci. 198, 257–268. doi: 10.1016/j.ecss.2017.09.008
Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar R. C. (2016). UCHIME2: improved chimera prediction for amplicon sequencing. BioRxiv 074252. doi: 10.1101/074252
Escribano R., Hidalgo P. (2000). Spatial distribution of copepods in the north of the Humboldt Current region off Chile during coastal upwelling. J. Mar. Biol. Assoc. United Kingdom 80, 283–290. doi: 10.1017/S002531549900185X
Gao Y., Li H., Wang X., Sun Y., Zhan A., Aileen T. S.-H., et al. (2020). Research on zooplankton diversity using DNA-based metabarcoding technique: a case study in the Yalvjiang Estuary. Acta Ecol. Sin. 40 (11), 3822–3832. (in Chinese). doi: 10.5846/stxb201903100451
Gillooly J. F. (2000). Effect of body size and temperature on generation time in zooplankton. J. Plankton Res. 22, 241–251. doi: 10.1093/plankt/22.2.241
Gong C., Wang A., Chen H., Jiang W., Tan L., Hu Z. (2013). Effects of UV-C radiation on the survival of copepods and oocysts: implications for drinking water processing facilities. Hydrobiologia 714, 1–11. doi: 10.1007/s10750-013-1495-8
Greve W. (1977). Interspecific interaction: the analysis of complex structures in carnivorous zooplankton populations. Helgolander Wiss. Meeresunters 30, 83–91. doi: 10.1007/BF02207827
Gu Z., Gu L., Eils R., Schlesner M., Brors B. (2014). Circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812. doi: 10.1093/bioinformatics/btu393
Guo Y., Li J., Luo Z., Jie X., Chen S., Zhu C., et al. (2015). Characteristics of zooplankton community in aquaculture areas of Liusha bay. Prog. In Fish. Sci. 36, 8–18. (in Chinese). doi: 10.11758/yykxjz.20150502
Halsband-Lenk C., Nival S., Carlotti F., Hirche H.-J. (2001). Seasonal Cycles of Egg Production of Two Planktonic Copepods, Centropages typicus and Temora stylifera, in the North-western Mediterranean Sea. J. Plankton Res. 23, 597–609. doi: 10.1093/plankt/23.6.597
Hernández-MIranda E., Betancourt I., Sobarzo M., Vergara O., Iturra C., Quiñones R. A. (2022). Spatial dissimilarity of zooplankton and hydrodynamic conditions in a Patagonian channel used intensely by aquaculture: the influence of a geomorphological constriction. Rev. Chil. Hist. Natural 95, 8. doi: 10.1186/s40693-022-00111-z
Hirche H.-J., Boersma M., Wiltshire K. H. (2019). Partial decoupling from the temperature size rule by North Sea copepods. Mar. Biol. 166, 1–13. doi: 10.1007/s00227-019-3503-7
Hsieh C.-H., Chen C.-S., Chiu T.-S. (2005). Composition and abundance of copepods and ichthyoplankton in Taiwan Strait (western North Pacific) are influenced by seasonal monsoons. Mar. Freshw. Res. 56, 153–161. doi: 10.1071/MF04058
Hsieh C.-H., Chiu T.-S. (2004). Summer spatial distribution of copepods and fish larvae in relation to hydrography in the northern Taiwan Strait. Zool. Stud. 41, 85–98.
Huang X. (1985). Studies of the biology of three freshwater clades. Oceanol. Et Limnol. Sin. 16, 188–195. (in Chinese).
Huo Y., Sun S., Zhang F., Wang M., Li C., Yang B. (2012). Biomass and estimated production properties of size-fractionated zooplankton in the Yellow Sea, China. J. Mar. Syst. 94, 1–8. doi: 10.1016/j.jmarsys.2011.09.013
Hwang J.-S., Souissi S., Tseng L.-C., Seuront L., Schmitt F. G., Fang L.-S., et al. (2006). A 5-year study of the influence of the northeast and southwest monsoons on copepod assemblages in the boundary coastal waters between the East China Sea and the Taiwan Strait. J. Plankton Res. 28, 943–958. doi: 10.1093/plankt/fbl031
Kakui K., Munakata M. (2022). A new Sphaeronella species (Copepoda: Siphonostomatoida: Nicothoidae) parasitic on Euphilomedes sp.(Ostracoda: Myodocopa: Philomedidae) from Hokkaido, Japan, with an 18S molecular phylogeny. System. Parasitol. 100, 1–11. doi: 10.1007/s11230-022-10075-z
Kordas R. L., Harley C. D. G., O’connor M. I. (2011). Community ecology in a warming world: The influence of temperature on interspecific interactions in marine systems. J. Exp. Mar. Biol. Ecol. 400, 218–226. doi: 10.1016/j.jembe.2011.02.029
Kumar B., Maharana B. R., Thakre B., Brahmbhatt N. N., Joseph J. P. (2022). 18S rRNA gene-based piroplasmid PCR: an assay for rapid and precise molecular screening of Theileria and Babesia species in animals. Acta Parasitol. 67, 1697–1707. doi: 10.1007/s11686-022-00625-2
Lewandowska A. M., Hillebrand H., Lengfellner K., Sommer U. (2014). Temperature effects on phytoplankton diversity—The zooplankton link. J. Sea Res. 85, 359–364. doi: 10.1016/j.seares.2013.07.003
Liang D., Uye S., Onbé T. (1994). Production and loss of eggs in the calanoid copepod Centropages abdominalis Sato in Fukuyama Harbor, the Inland Sea of Japan. Bull. Plankton Soc. Japan (Japan) 41 (2), 131–142.
Lima-Mendez G., Faust K., Henry N., Decelle J., Colin S., Carcillo F., et al. (2015). Determinants of community structure in the global plankton interactome. Science 348, 1262073. doi: 10.1126/science.1262073
Liu S., Cui Z., Zhao Y., Chen N. (2022). Composition and spatial-temporal dynamics of phytoplankton community shaped by environmental selection and interactions in the Jiaozhou Bay. Water Res. 218, 118488. doi: 10.1016/j.watres.2022.118488
Liu S., Gibson K., Cui Z., Chen Y., Sun X., Chen N. (2020). Metabarcoding analysis of harmful algal species in Jiaozhou Bay. Harmful Algae 92, 101772. doi: 10.1016/j.hal.2020.101772
Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Mary Beth D., Denise L. B., Jennifer E. P. (2004). Effects of low dissolved oxygen on zooplankton predation by the ctenophore Mnemiopsis leidyi. Mar. Ecol. Prog. Ser. 280, 163–172. doi: 10.3354/meps280163
Michel H. B., Herring D. C. (1984). Diversity and abundance of Copepoda in the northwestern Arabian Gulf. Crustaceana. Supplement, 326–335.
Middelbo A. B., Møller E. F., Arendt K. E., Thyrring J., Sejr M. K. (2019). Spatial, seasonal and inter-annual variation in abundance and carbon turnover of small copepods in young sound, Northeast Greenland. Polar Biol. 42, 179–193. doi: 10.1007/s00300-018-2416-0
Mugnai F., Meglécz E., Abbiati M., Bavestrello G., Bertasi F., Bo M., et al. (2021). Are well-studied marine biodiversity hotspots still blackspots for animal barcoding? Global Ecol. Conserv. 32, e01909. doi: 10.1016/j.gecco.2021.e01909
Nordsieck R. (2006). The Living World of Molluscs. Available at: https://animaldiversity.org/accounts/Mytilus_edulis/.
Oghenekaro E. U., Chigbu P., Oseji O. F., Tang K. W. (2018). Seasonal factors influencing copepod abundance in the Maryland coastal bays. Estuaries Coasts 41, 495–506. doi: 10.1007/s12237-017-0285-3
Pearson T. H. (1978). Macrobenthic succession in relation to organic enrichment and pollution of the marine environment. Oceanogr. Mar. Biol. Ann. Rev. 16, 229–311.
Pérez-Losada M., Høeg J. T., Crandall K. A. (2012). Deep phylogeny and character evolution in Thecostraca (Crustacea: Maxillopoda). Integr. Comp. Biol. 52, 430–442. doi: 10.1093/icb/ics051
Portner H. O., Farrell A. P. (2008). Physiology and climate change. Science 322, 690–692. doi: 10.1126/science.1163156
Prusova I. Y., Galagovets E. A. (2022). Sex ratios of calanoid copepods in the northern Black Sea. Regional Stud. Mar. Sci. 55, 102576. doi: 10.1016/j.rsma.2022.102576
Revelle W. (2017). psych: Procedures for Personality and Psychological Research. Available at: http://CRAN.R-project.org/package=psych.
Roman M. R., Gauzens A. L., Rhinehart W. K., White J. R. (1993). Effects of low oxygen waters on Chesapeake Bay zooplankton. Limnol. Oceanogr. 38, 1603–1614. doi: 10.4319/lo.1993.38.8.1603
Rong W., Hong-Yan Z., Ke W., Tao Z. (2002). Distribution and population dynamics of Paracalanus parvus, Paracalanus crassirostris, and Acartia bifilosa (copepoda, calanoida) in the Bohai Sea. Chin. J. Oceanol. Limnol. 20, 348–357. doi: 10.1007/BF02847926
Santoferrara L. F., Alder V. V., Mcmanus G. B. (2017). Phylogeny, classification and diversity of Choreotrichia and Oligotrichia (Ciliophora, Spirotrichea). Mol. Phylogenet. Evol. 112, 12–22. doi: 10.1016/j.ympev.2017.03.010
Savolainen V., Cowan R. S., Vogler A. P., Roderick G. K., Lane R. (2005). Towards writing the encyclopedia of life: an introduction to DNA barcoding. Philos. Trans. R Soc. Lond. B Biol. Sci. 360, 1805–1811. doi: 10.1098/rstb.2005.1730
Shi Y., Wang J., Zuo T., Shan X., Jin X., Sun J., et al. (2020). Seasonal changes in zooplankton community structure and distribution pattern in the Yellow Sea, China. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00391
Shimode S., Takahashi K., Shimizu Y., Nonomura T., Tsuda A. (2012). Distribution and life history of two planktonic copepods, Rhincalanus nasutus and Rhincalanus rostrifrons, in the northwestern Pacific Ocean. Deep Sea Res. Part I: Oceanogr. Res. Papers 65, 133–145. doi: 10.1016/j.dsr.2012.03.008
Shin S. S., Choi S. Y., Seo M. H., Lee S. J., Soh H. Y., Youn S. H. (2022). Spatiotemporal distribution characteristics of copepods in the water masses of the Northeastern East China sea. J. Mar. Sci. Eng. 10, 754. doi: 10.3390/jmse10060754
Slater L. M., Hopcroft R. R. (2005). Development, growth and egg production of Centropages abdominalis in the eastern subarctic Pacific. J. Plankton Res. 27, 71–78. doi: 10.1093/plankt/fbh152
Sun J. (2011). Marine biodiversity: Why is there a high level of diversity? Biodivers. Sci. 19, 611. doi: 10.1111/mec.14988
Tarrant A. M. (2020). Small copepods could play a big role in the marine carbon cycle. BioEssays 42, 2000267. doi: 10.1002/bies.202000267
Tiselius P., Saiz E., Kiørboe T. (2013). Sensory capabilities and food capture of two small copepods, Paracalanus parvus and Pseudocalanus sp. Limnol. Oceanogr. 58, 1657–1666. doi: 10.4319/lo.2013.58.5.1657
Tittensor D. P., Mora C., Jetz W., Lotze H. K., Ricard D., Berghe E. V., et al. (2010). Global patterns and predictors of marine biodiversity across taxa. Nature 466, 1098–1101. doi: 10.1038/nature09329
Tseng L.-C., Souissi S., Dahms H.-U., Chen Q.-C., Hwang J.-S. (2008). Copepod communities related to water masses in the southwest East China Sea. Helgoland Mar. Res. 62, 153–165. doi: 10.1007/s10152-007-0101-8
Turner J. T., Tester P. A., Hettler W. F. (1985). Zooplankton feeding ecology. Mar. Biol. 90, 1–8. doi: 10.1007/BF00428208
Tyler-Walters H. (2008). Mytilus edulis Common mussel. Available at: https://www.marlin.ac.uk/species/detail/1421.
Uttieri M., Hinow P., Pastore R., Bianco G., Ribera D’alcalá M., Mazzocchi M. G. (2021). Homeostatic swimming of zooplankton upon crowding: the case of the copepod Centropages typicus. J. R. Soc. Interface 18, 20210270. doi: 10.1098/rsif.2021.0270
Valentini A., Pompanon F., Taberlet P. (2009). DNA barcoding for ecologists. Trends Ecol. Evol. 24, 110–117. doi: 10.1016/j.tree.2008.09.011
Van S.-H., Park W. (2016). Monthly population changes of Paracalanus parvus near Oryuk islets off Busan, South Korea. Korean Soc. Fish. Aquat. Sci., 134–134.
Wang K., Wang R., Gao S. (2001). Preliminary study on the diurnal vertical migration of zooplankton in the East China Sea. Oceanol. Limnol. Sin. 32, 534–540.
Wei T., Simko V., Levy M. (2021). package “corrplot”: Visualization of a Correlation Matrix. 2017. Version 0.92. Available at: https://github.com/taiyun/corrplot.
Wickham H. (2016). “Data analysis,” in ggplot2: Elegant Graphics for Data Analysis. Ed. Wickham H. (Cham: Springer International Publishing), 189–201.
Yasuhara M., Danovaro R. (2016). Temperature impacts on deep-sea biodiversity. Biol. Rev. 91, 275–287. doi: 10.1111/brv.12169
Yin T., Wang Q., Yang Y., Cen J. (2022). Comparative study on zooplankton community structure in Pearl River Estuary based on morphological and DNA identification. J. Trop. Oceanogr. 41, 172–185. (in Chinese). doi: 10.11978/2021151
Zakaria H. Y., Hassan A.-K. M., Abo-Senna F. M., El-Naggar H. A. (2016). Abundance, distribution, diversity and zoogeography of epipelagic copepods off the Egyptian Coast (Mediterranean Sea). Egyptian J. Aquat. Res. 42, 459–473. doi: 10.1016/j.ejar.2016.11.001
Zhang F., Sun S., Yang B., Ji P. (2006). Seasonal changes in abundance of small copepod Paracalanus parvus in the Yellow sea. Oceanol. Limnol. Sin. 37 (4), 322–329. doi: 10.3321/j.issn:0029-814X.2006.04.006
Zhang K., Jiang F., Chen H., Dibar D. T., Wu Q., Zhou Z. (2019). Temporal and spatial variations in zooplankton communities in relation to environmental factors in four floodplain lakes located in the middle reach of the Yangtze River, China. Environ. pollut. 251, 277–284. doi: 10.1016/j.envpol.2019.04.139
Zhao W., Dai L., Chen X., Wu Y., Sun Y., Zhu L. (2022). Characteristics of zooplankton community structure and its relationship with environmental factors in the South Yellow Sea. Mar. pollut. Bull. 176, 113471. doi: 10.1016/j.marpolbul.2022.113471
Zhao Q., Liu S., Niu X. (2020). Effect of water temperature on the dynamic behavior of phytoplankton–zooplankton model. Appl. Mathematics Comput. 378, 125211. doi: 10.1016/j.amc.2020.125211
Keywords: mariculture, Crassostrea gigas, copepod, metabarcoding, temporal dynamic, biofouling organism
Citation: Qian B, Miao X and Xu F (2023) Temporal dynamics of zooplankton community in an oyster farming area of the Yellow Sea in China via metabarcoding. Front. Mar. Sci. 10:1190475. doi: 10.3389/fmars.2023.1190475
Received: 20 March 2023; Accepted: 16 August 2023;
Published: 12 September 2023.
Edited by:
Andrew Stanley Mount, Clemson University, United StatesCopyright © 2023 Qian, Miao and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Xu, eHVmZWlAcWRpby5hYy5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.