
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 21 July 2023
Sec. Marine Conservation and Sustainability
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1185111
Chelonia mydas (green turtles) are being threatened worldwide by fibropapillomatosis (FP), which has seriously affected their survival. The presence of FP on the body surface and visceral organs of green turtles found dead was confirmed, causing obstruction of the gastrointestinal tract, changes in foraging behavior, and reduction of visceral functions. The etiology of FP has not yet been elucidated, and previous research generally considers that the occurrence of FP is related to the chelonid alphaherpesvirus 5 (ChHV5), associated with low animal immunity, and also with marine environmental factors, such as poor water quality and eutrophication. However, there is no evaluation on the induction of FP pathogenesis associated with the green turtle. In this study, we evaluated blood samples from green turtles with and without FP using de novo transcriptome assembly. Results indicated that 3,090 differentially expressed genes (DEGs) (p < 0.05) were identified, including 1,357 upregulated genes and 1,733 downregulated genes in turtles with or without FP. We observed that DEGs, which are significantly upregulated, are found in cancer development, namely, MAPK1IP1L and APAF1. Furthermore, the infected green turtle indicated that the greater number of DEGs was contributed by the NOD-like receptor signaling pathway, which can be activated through an endocytosis of the viral particle by the immune system cells, and the Wnt signaling pathway, which is believed to have played a role in FP tumorigenesis. We validated the more upregulated/downregulated DEGs in cancer development and immunization, and DEGs such as LEF1, BTRC, and FOSL1 participating in the NOD-like receptor signaling pathway, as well as ERBIN, TRAF6, and NFKB1 in the Wnt signaling pathway, using real-time quantitative polymerase chain reaction (RT-qPCR). Altogether, this study provided some genes as potential markers during FP infection and a further evidence of FP in endangered green turtles in Taiwan.
There are a total of seven species of sea turtles in the world, five of which have been found in the sea off the coast of Taiwan, namely, green turtles (Chelonia mydas), hawksbill turtles (Eretmochelys imbricate), olive ridley turtles (Lepidochelys olivacea), loggerhead turtles (Caretta caretta), and leatherback turtles (Dermochelys coriacea) (Cheng and Chen, 1997; Kuo et al., 2017). The green turtle is the most common species and the only one that nests in Taiwan (Cheng, 2000; Cheng et al., 2018). In fact, the green turtle has been listed as a globally endangered species and is listed in the Red List of the IUCN (International Union for Conservation of Nature; IUCN, 2016). Furthermore, anthropogenic impacts, including artificial coastal development, fisheries by-catch, global environmental change, illegal egg harvesting, marine debris, environmental pollution, and diseases, pose a major threat to green turtle populations all over the world (Aguirre and Lutz, 2004; Hamann et al., 2010; Shigenaka, 2010; Jones et al., 2016; Li et al., 2017; Perrault et al., 2017; Godoy and Stockin, 2018; Ng et al., 2018; Li and Chang, 2020). Among the different threats to turtles, as a consequence of the aforementioned circumstances, the recent study focused on threats from the diseases of marine turtles, which is a priority research area for sea turtle’s conservation (Hamann et al., 2010; Rees et al., 2016). Fibropapillomatosis (FP) is a tumor-forming disease that globally affects marine turtles (Herbst, 1994; Alfaro-Nunez et al., 2014). However, the reports of this disease in Asia are still very limited (Hargrove et al., 2015; Li et al., 2017; Li and Chang, 2020; Dujon et al., 2021; Li et al., 2022). This disease was first discovered in Florida in 1938 (Smith and Coates, 1938); there are still many cases until the present day and the geographic distribution of the disease was moving northward from the equatorial region (Duffy et al., 2018). Together with multiple anthropogenic pressures impacting sea turtles such as ingestion of marine litter and interaction with fishing gear (Dimitriadis et al., 2022; Jo et al., 2022; Rodríguez et al., 2022; Mghili et al., 2023), FP is one of the main causes of stranding in sea turtles (Foley et al., 2005; Chaloupka et al., 2008; Da Silva-Júnior et al., 2019). FP could occur in all these seven species of sea turtles, but it is more common in green turtles (Jones et al., 2016; Dujon et al., 2021). FP cases were more likely observed in immature turtles and less common in adults (Jones et al., 2016; Lawrance et al., 2018). In sea turtles with FP, tumors can appear on the body surface (such as the eyes, oral cavity, skin, and carapaces) or in internal organs such as lungs and kidney (Herbst, 1994; Aguirre and Lutz, 2004; Rossi et al., 2021), preventing foraging, reducing sea turtle mobility and visceral function of sea turtles, and causing death from infirmity (Work et al., 2001).
A higher prevalence of FP has been reported in highly contaminated marine environments or environments with poor water quality (Formia et al., 2007; Van Houtan et al., 2010; Guimarães et al., 2013; Van Houtan et al., 2014; Baines et al., 2021; Manes et al., 2022). Furthermore, a recent global meta-analysis study further indicated that a high prevalence of sea turtle FP is thought to be associated with exposure to toxic phytoplankton bloom events (Dujon et al., 2021). In contrast, a relationship between sea turtle FP prevalence and water quality indices rankings at each site along the Queensland coastline could not be quantified (Jones et al., 2022). A study conducted in Taiwan on FP in green turtles described that all green turtles with FP were discovered in eastern Taiwan (Li and Chang, 2020). However, the study by Liu et al. (2015) indicated that the highly problematic region of coastal eutrophication is located in southwestern Taiwan. Hence, the exact etiology of FP is not well understood. Thus, Li et al. (2017) also concluded that for the conservation of endangered sea turtles, more studies of this nature are recommended for Asian marine turtle populations to improve our understanding of the infection. Marine environmental factors or infectious agents, among which Chelonid herpesvirus 5 (ChHV5), have been hypothesized to be associated with sea turtle FP (Jones et al., 2016; Duffy et al., 2018). A strong association between sea turtle FP and the presence of ChHV5 have been reported. For example, ChHV5 DNA has been distinguished molecularly from diseased tissues of sea turtles with FP (Lackovich et al., 1999; Page-Karjian et al., 2012; Alfaro- Nunez and Gilbert, 2014; Li et al., 2017). However, sea turtles have been diagnosed with the occurrence of FP, and ChHV5 nucleic acid can also be detected in tumor-free (asymptomatic) sea turtles (Page-Karjian et al., 2020a; Loganathan et al., 2021; Li et al., 2022). Nevertheless, the mode of transmission route of ChHV5 among wild sea turtles is still unknown (Dujon et al., 2021). Ozobranchus spp. (marine leeches) were commonly found in sea turtles with FP, and the large amount of ChHV5 DNA was detected in Ozobranchus leech parasite in green turtles that died from FP (Greenblatt et al., 2004) while leeches removed from healthy sea turtles did not test positive for ChHV5 (Farrell et al., 2021). Therefore, the Ozobranchus spp. was suggested to be a possible vector of ChHV5 transmitting between sea turtles (Greenblatt et al., 2004). Nowadays, several studies suggested that FP in sea turtles is mainly caused by the interaction of ChHV5 and environmental factors such as water quality and autoimmunity (Aguirre et al., 1995; Alfaro-Nunez et al., 2016; Bruno et al., 2021; James et al., 2021; Sposato et al., 2021; Nash & Ryan, 2023). Sea turtles with FP were shown to have immunosuppression and reduced activity of natural killer cells (Work et al., 2001; Li and Chang, 2020; Perrault et al., 2021). Hence, some concern further exists regarding FP in sea turtle rehabilitation facilities (Page-Karjian et al., 2014; Li and Chang, 2020; Page-Karjian et al., 2020b). Jones et al. (2016) also concluded that molecular investigations on ChHV5 from different areas are critical to improve our understanding of the epidemiology and pathogenesis of this virus. As a consequence of the aforementioned circumstances, it is extremely important and timely to clarify the pathogenesis of this viral disease.
Previous studies have shown that the prevalence of FP is higher in juvenile green turtles than in other size/age groups (Jones et al., 2016; Perrault et al., 2021; Manes et al., 2023). So far, there is evidence that green turtles develop FP only after recruitment to nearshore habitats (Jones et al., 2016). This is supported by the fact that juvenile green turtles, the most vulnerable individuals to FP, are coastal foragers living in coastal areas (Work et al., 2005; Hazel et al., 2009) that have undergone significant anthropogenic change in recent decades (Dujon et al., 2021; Jones et al., 2022; Manes et al., 2023). In fact, in the same feeding ground, there were more than one sea turtle at the same time, and ChHV5 could be transmitted horizontally through seawater (Farrell et al., 2021) to other sea turtles in the same feeding area. Hence, the migratory habit of sea turtles may also play an important role in distributing ChHV5 worldwide (Jones et al., 2016).
Few studies from European and American regions (Duffy et al., 2018; Blackburn et al., 2021; Yetsko et al., 2021) used C. mydas skin samples to perform transcriptome profiling and discovered that there were similar signaling pathways between green sea turtles FP and human basal cell carcinoma (BCC). Currently, there is no research on transcriptome analysis of FP in C. mydas in the Asian context; the results of this study may provide additional information of this disease. In the present study, blood samples of FP-afflicted and clinically healthy C. mydas were used to evaluate immune-related gene expression and tumor-related gene expression using transcriptome analysis. This study aims to evaluate and understand the possible pathogenesis of the formation of FP with the gene expression results of transcriptome analysis. Detection of risk genes may help early identification of FP in green turtles, improve the management of infected sea turtles, and further develop effective treatment method against green turtle FP in the future.
C. mydas, including eight green sea turtles with FP tumors (n = 1 with tumor score 1; n = 1 with tumor score 2; n = 6 with tumor score 3) was used in this study (Work and Balazs, 1999). FP tumors were detected through visual inspection of external masses by veterinarians (Figure 1). The green turtles analyzed showed a size range of 29.4–86.5 cm of curved carapace length (Figure 1).
All individuals in this study were discovered and rescued through the official reporting system of the Marine Animal Rescue Network (established by the Ocean Conservation Administration) and transported to the National Museum of Marine Biology and Aquarium (NMMBA) for further rehabilitation/medical care. Therefore, blood samples were necessarily collected under humane procedures from an external jugular vein (Dutton, 1996; Day et al., 2010; Li et al., 2015) by our licensed veterinarians (Dr. Tsung-Hsien Li or Dr. I-Chun Chen) for routine medical care. Hence, no individuals of green turtles were specifically captured from the wild for the purpose of this study. As a consequence of the aforementioned circumstances, the excess of blood was obtained and utilized. The scientific permit numbers for sample use were obtained from the Ocean Conservation Administration (Permit nos. 1090009887 and 1100012644).
Total DNA was extracted from whole blood samples of 24 green turtles using the DNeasy Blood & Tissue kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. The chelonid alphaherpesvirus 5 (ChHV5) infection status of all 24 green turtles was determined by polymerase chain reaction (PCR) using three genes associated with ChHV5. Three independent primers sets were designed and targeted on UL18 (capsid protein gene), UL22 (glycoprotein H gene), and UL27 (glycoprotein B gene) regions of the ChHV5 (Alfaro-Nunez and Gilbert, 2014). PCR amplifications were prepared in a total reaction volume of 50 μl. The reactions included 1 μl of the template DNA, 1 μl of each primer (10 μM), 22 μl of distilled water (DDW), and 25 μl of enzyme of AmpliTaq Gold® 360 Master Mix (Life Technologies, Valencia, CA, USA). The reaction conditions were as follows: Initial denaturing at 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 62°C for 30 s, and 72°C for 60 s, and a final extension step of 72°C for 7 min. Amplified PCR products were then visualized by gel electrophoresis (2% agarose) with SYBR Safe DNA Gel Stain (Invitrogen, Carlsbad, CA, USA).
Total RNA was extracted using TRIzol® LS Reagent according to the manufacturer’s protocol. RNA samples were subjected to removal of DNA contamination using DNase I (BioLab, London, UK) and 5 μg of total RNA was used for cDNA synthesis using the iScript® cDNA Synthesis Kit (Bio-Rad, California, USA) according to the manufacturer’s instructions.
Total RNA samples were sent to sequencing company Genomics Bioscience Technology Co Ltd. (Taipei, Taiwan). Total RNA was used for library construction using the TrueSeq library construction kit. The library was sequenced on the Illumina HiSeq™4000 (Illumina, Inc., San Diego, USA) platform. After that, “Trimmomatic” was used for read-quality filtering, and low-quality bases and adaptors were removed from Illumina sequences. Trinity (Illumina, Inc) was used to assemble the RNA sequence data, and de novo assembly was later performed (Haas et al., 2013; Bolger et al., 2014). The data after de novo transcriptome assembly was used for annotation (Supek et al., 2011). Owing to the lack of detailed genetic data on green sea turtles, the human genetic database was chosen for alignment. Aligning human genetic sequence to assembly result evaluated the gene expression according to the number of sequences aligned. Differential expression calculated by the software “edgeR v3.5” and functional annotation of Gene Ontology (GO) term (Kanehisa and Goto, 2000; Kanehisa, 2019) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment (Mi et al., 2018) were also conducted. Differentially expressed genes (DEGs) were filtered with the parameter false discovery rate (FDR), in which the p-adjusted value (padj) should be smaller than 0.05.
DEGs with a large difference in expression between tumor and healthy green sea turtles were selected. Upregulated genes related to cancer-forming and other DEGs participate in the Wnt signaling pathway, the NOD-like receptor signaling pathway was selected, and β-actin was used as control. Primer sequences were designed using Pimer3 Input (Reference or weblink) and the NCBI primer designing tool. The details of the primers tested, including MPAK1IP1L, TBRG4, NFIA, APAF1, ZBTB41, and LATS2, are mentioned in Table 1. Real-time quantitative polymerase chain reaction (RT- qPCR) was performed using iQ™ SYBR® Green Supermix (Bio-Rad, California, USA), and each sample was performed in quadruplicate. Cycling parameters are as follows: 95°C for 2 min, 45 cycles of 95°C for 10 s, 60°C for 20 s, 72°C for 20 s, and a melting curve (Duffy et al., 2018). The fold expressions of target genes were calculated using the 2ΔΔCt (Livak and Schmittgen, 2001).
From the PCR result for the detection of ChHV5 DNA in blood samples (Table 2), eight green sea turtles involved in this study afflicted with FP were all positive for ChHV5 (viremia); the positive rate was 100.0% (8/8). For those who were clinically healthy (without FP), the positive rate of viral infection was 0.0% (0/16).
Four blood samples, two from green turtles with FP and two from green turtles without FP, were included in transcriptome analysis. In total, 3,090 DEGs (DEGs were calculated using edgeR v3.22.5.; p < 0.05) (Robinson et al., 2010; Britt et al., 2020), 1,357 upregulated genes and 1,733 downregulated genes, were identified (Figure 2). In reference to human orthologs, comparing gene expression between two sample groups, the 10 most expressed differential genes of up- and downregulation were shown (Table 3). Upregulated genes related to tumor formation and immunity were selected and quantified using RT-qPCR.
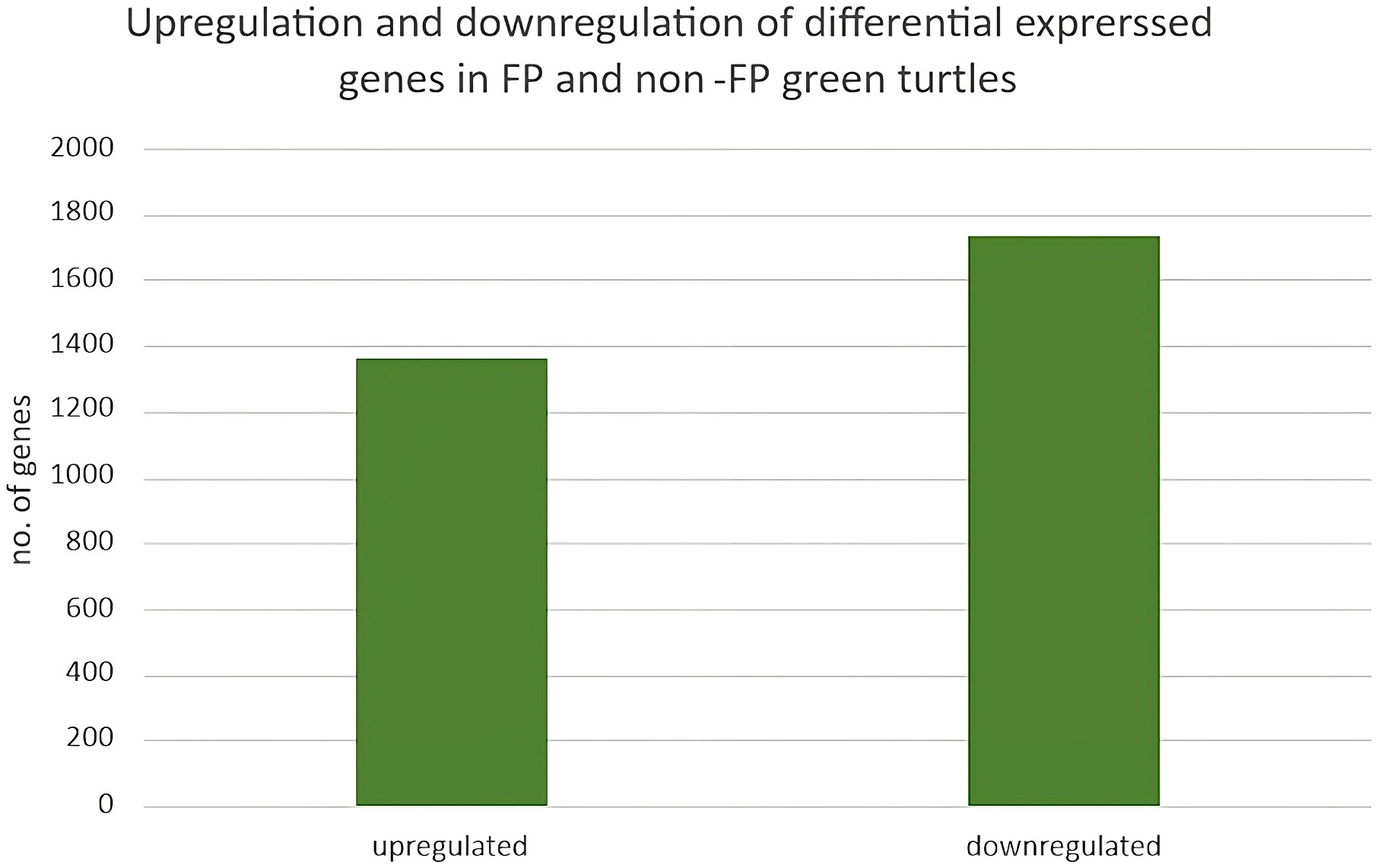
Figure 2 Upregulation and downregulation of differentially expressed genes in FP and non-FP green turtles.
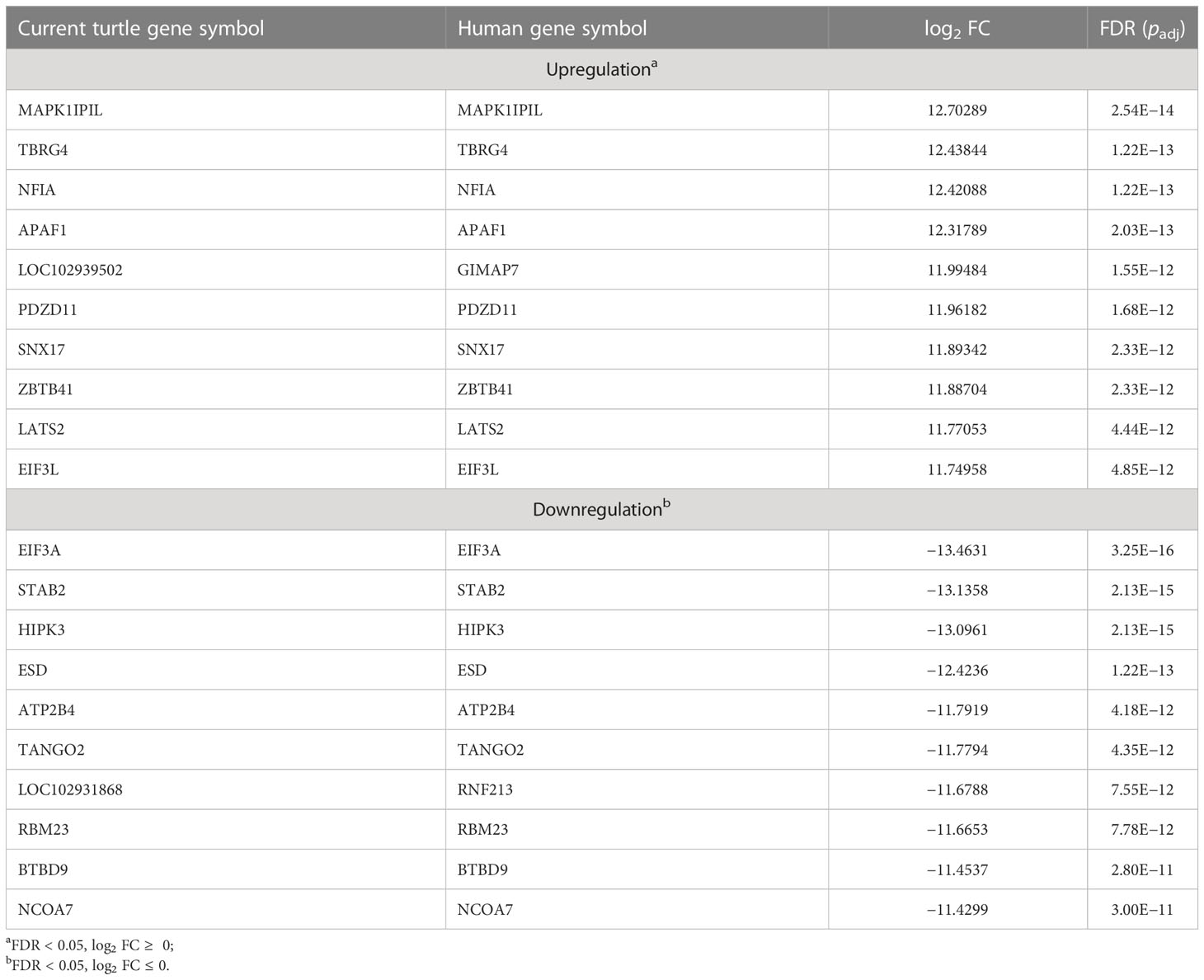
Table 3 Top 10 most up- and downregulated differentially expressed genes of clinically healthy green sea turtles and those with tumors.
From the annotation result, functions of DEGs mainly included cancer formation and immunity. Among the top 10 upregulated genes, with higher performance in blood samples of green sea turtles with FP, more than half of the DEGs took part in tumorigenesis or associated with some other type of cancer, namely, MAPK1IP1L, NFIA, APAF1, PDZD11, ZBTB41, and LATS2. Other than promoting tumor growth, PDZD11 has also a correlation with immune cell infiltrating levels. For downregulated genes, with a higher performance in blood samples of clinically healthy green sea turtles, the top three have a connection with the development of cancer, namely, EIF3A, STAB2, and HIPK3.
Dot plots of significant GO ontology enriched among DEGs in three domains—cellular component (Figure 3A), molecular function (Figure 3B), and biological process (Figure 3C)—were conducted.
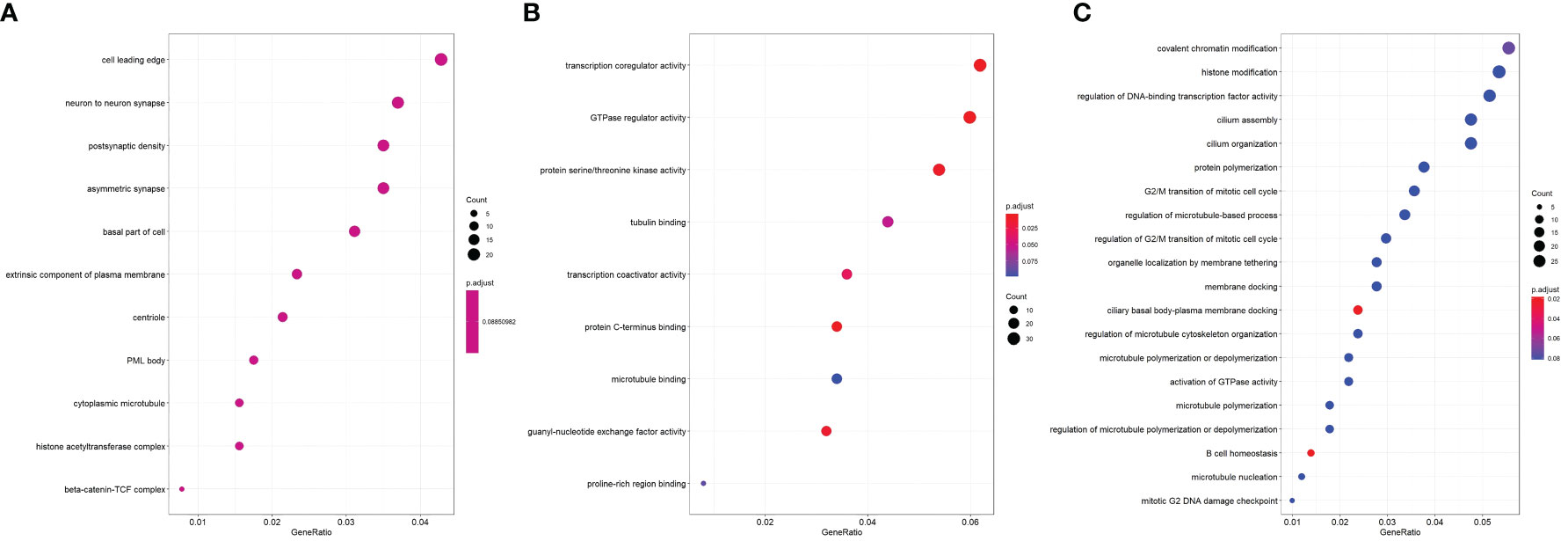
Figure 3 Dot plot of significant GO ontology enriched among DEGs. (A) Cellular component; (B) molecular function; (C) biological process.
In the cellular component, the most enriched ontology was the cell leading edge. In molecular function, the most enriched ontology was transcription coregulator activity. Lastly, in the domain of biological process, covalent chromatin modification and histone modification were the first and second enriched ontology in green sea turtles with tumor (Figure 3C).
KEGG pathway was conducted, and as shown in Figure 4, the top 20 KEGG pathway permutations according to their gene ratio took part in the specific pathways in green sea turtles with tumors. The NOD-like receptor signaling pathway was the most enriched pathway. The second is Epstein–Barr infection and endocytosis in the third.
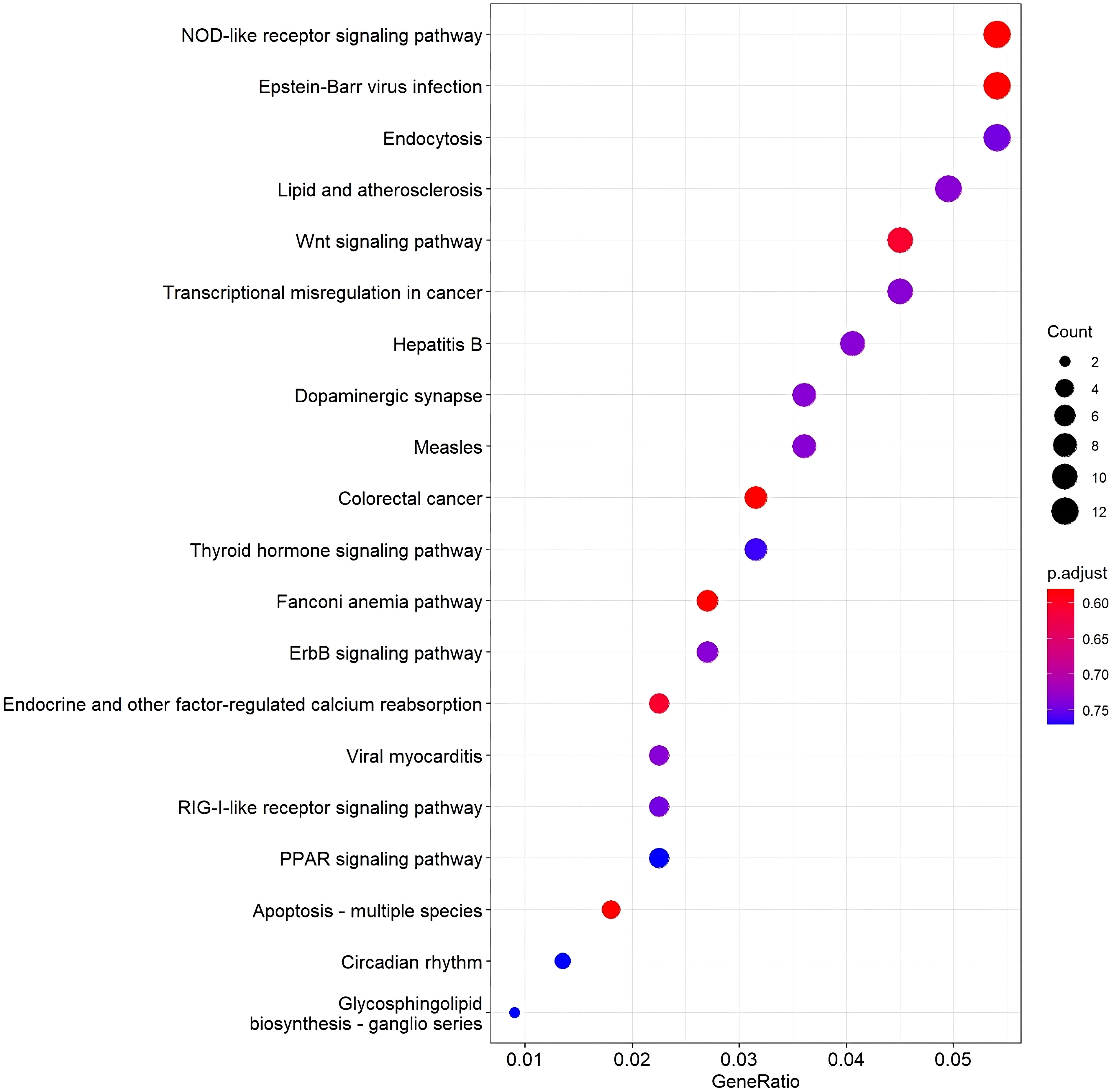
Figure 4 A dot plot of top 20 KEGG pathway permutations according to gene ratio. Dot size shows the number of DEG, and the color shows the adjusted p-value of the gene set enrichment.
In the NOD-like receptor signaling pathway, genes NFKB1 and TRAF6 have the highest upregulation whereas the gene ERBIN has the highest expression in downregulation (Figure 5). In the Wnt signaling pathway, genes BTRC and FOSL1 have the highest expression in upregulation, whereas LEF1 has the highest expression in downregulation (Figure 6).
Expressions of genes selected from upregulated DEGs, NOD-like receptor signaling pathway, and Wnt signaling pathway were validated through RT-qPCR. Because of the small sample size, non-parametric statistical tests (Mann–Whitney U test) (Mansfield et al., 2017; Tanabe et al., 2022; Fumero-Hernández et al., 2023) were used in this study. There were no major significant findings (using Mann–Whitney U test) for the comparison of gene expression profiles between green turtles with and without FP. As shown in Figure 7, the quantification of ERBIN, MAPK1P1IL, APAF1, and LATS2 in green turtles with FP was much higher than in green turtles without FP, though not showing statistical significance. The quantification of ZBTB41 and NFKB1 was slightly higher in green turtles with FP when compared with green turtles without FP. The results of the other two genes, BTRC and NFIA, were similar between turtles with and without FP.
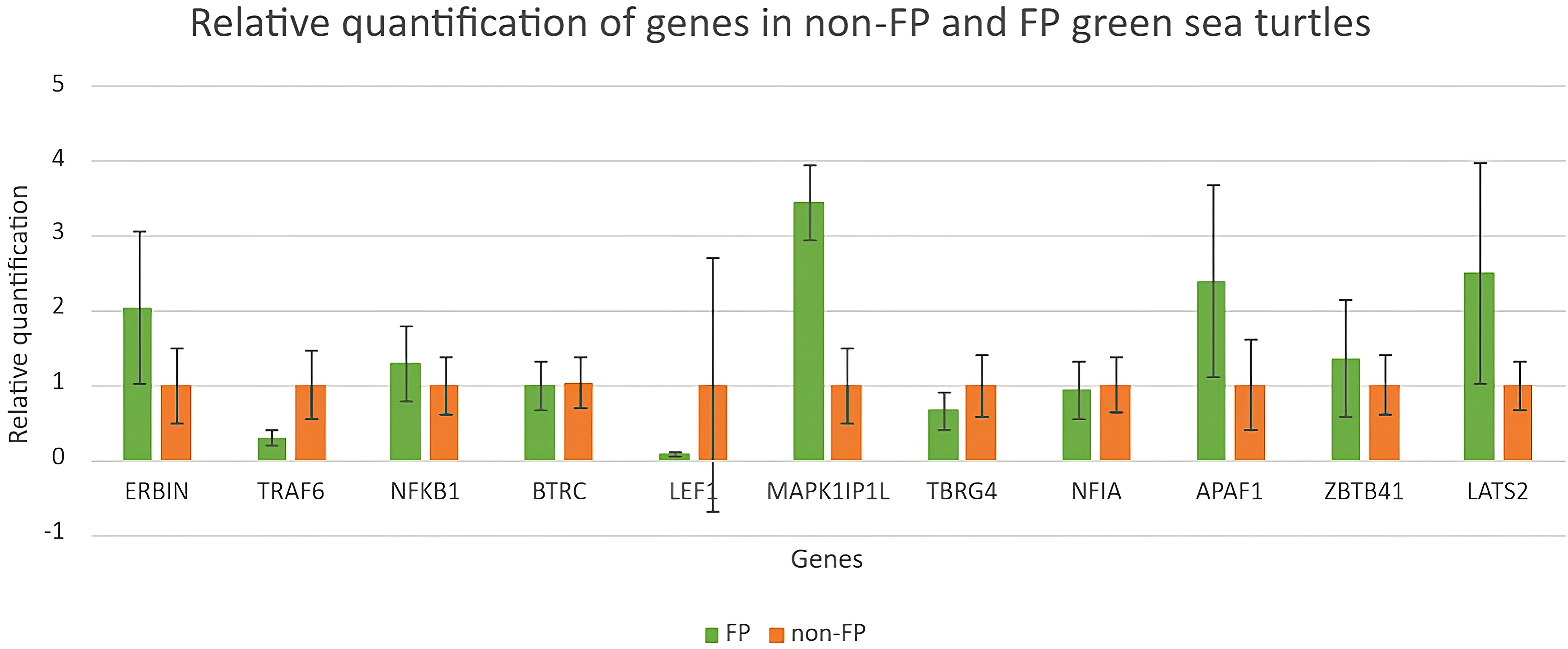
Figure 7 Relative quantification of genes in green turtles with and without FP. Data indicate mean ± SD from n = 8 (FP) and n = 16 (non-FP).
The relative quantifications of DEGs in the NOD-like receptor signaling pathway as well as the Wnt signaling pathway are shown in Figure 7. Relative quantification of TRAF6 and LEF1 was much higher in clinically healthy green turtles. The quantification of TBRG4 was slightly higher in green turtles without those afflicted with FP.
Green turtles involved in this study were detected with ChHV5 using PCR (Table 2); the positive rate of green turtles afflicted with FP and green turtles without FP was 100.0% (8/8) and 0.0% (0/16), respectively. Several studies have previously reported the low ChHV5 detection rate in green turtles without external FP, including wild, rehabilitating, and nesting green turtles (Page-Karjian et al., 2020a; Page-Karjian et al., 2020b; James, 2020; Perrault et al., 2021). Among 24 green sea turtles, 8 developed FP: 12.5% (1/8) was lightly afflicted, 12.5% (1/8) was moderately afflicted, and 75% (6/8) was heavily afflicted. Sixteen of them were not afflicted with FP.
In this study, blood samples of green turtles with and without FP were evaluated using de novo transcriptome assembly. From the annotation result, functions of DEGs mainly included cancer formation and immunity. The most enriched KEGG pathway was the NOD-like receptor signaling pathway. The Wnt signaling pathway, which ranked fifth, indicates a potential role in C. mydas while regulating FP (Duffy et al., 2018; Blackburn et al., 2021; Yetsko et al., 2021). However, there were no statistically significant results in KEGG enrichment. The DEGs in the NOD-like receptor signaling pathway and Wnt signaling pathway are shown in Figures 5 and 6; genes NFKB1 and TRAF6 have the highest upregulation whereas gene ERBIN has the highest expression in downregulation in the NOD-like receptor signaling pathway. Genes BTRC and FOSL1 have the highest expression in upregulation whereas LEF1 has the highest expression in downregulation in the Wnt signaling pathway. The ability of tumorigenesis or expression as a tumor suppressor gene which may have, the presence of LEF1 indicating clinical value to become a biomarker of FP (Santiago et al., 2017).
In the DEGs’ annotation result, over half of the upregulated DEGs took part in tumorigenesis or were associated with some other type of cancer. The first three most downregulated genes have a connection or are associated with the development of cancer. MAPK1IP1L, encoded for mitogen-activated protein kinase 1 interacting protein 1 like the most expressed genes in green sea turtles with FP, has the ability to develop as an auxiliary diagnostic tool for lung cancer detection in human as it significantly overexpressed in the lung cancer group (Zhang et al., 2018). In our qPCR result, this gene has a significant increase in green turtles with FP, which we believe is worthy of further investigation. APAF1 encoded apoptotic peptidase activating factor 1, which appeared to be a tumor-suppressor gene, and research reported that the expression of APAF1 has an inverse correlation with the progression of tumor (Zlobec et al., 2007); however, we have a quite different result in our qPCR quantification: the expression of APAF1 was higher in green sea turtles with FP. LATS2 encoded for large tumor suppressor kinase 2, similar to APAF1, and it played a role in tumor suppression by inhibiting cell proliferation (Li et al., 2003); at the same time, it has the same quantification result as APAF1. We believed that it has a connection with the health status or tumor progression of green sea turtles and further research was needed.
Relative quantification of DEGs annotated in the NOD-like receptor signaling pathway and Wnt signaling pathway was evaluated using RT-qPCR. Expression of ERBIN was higher in green sea turtles with FP; this result did not agree with the annotation result in which ERBIN has a higher expression in clinically healthy green sea turtles. ERBIN was the most downregulated DEG in the NOD-like signaling pathway; loss of ERBIN expression increases cell migration and invasion, which is an early event in tumorigenesis (Stevens et al., 2018). TRAF6 has a higher expression, and this result did not agree with the annotation result in which TRAF6 has a higher expression in green sea turtles with FP. Overexpression of TRAF6 may result in NF-κB activation in human lung cancer and promoted proliferation of colon cancer cells (Starczynowski et al., 2011; Sun et al., 2014). NF-κB was a pathway that regulated cancer cell proliferation, survival, and angiogenesis, and it also regulated immune responses and inflammation (Dolcet et al., 2005; Tilborghs et al., 2017; Soleimani et al., 2020); the expression of NF-κB reflected the potential risk of developing malignant tumors (Xia et al., 2018). Another activated NF-κB gene is TRAF6, which is also involved in angiogenesis and one of the key elements of tumorigenesis. TRAF6 was suggested to serve as a therapeutic target suppressing cancer formation for cancer patients (Sun et al., 2014).
Relative quantification of LEF1 in the Wnt signaling pathway was the highest in green sea turtles without FP. This agreed with the annotation where LEF1 is expressed in downregulated DEGs, meaning it was more significantly expressed in clinically healthy green turtles. Santiago et al. (2017) stated that the presence of LEF1 may have clinical value to be a potential biomarker of FP.
Understanding the cause of FP in endangered sea turtles is crucial for sea turtle conservation works. Sea turtles at rehabilitation facilities provide us with an opportunity to advance our knowledge and understanding of FP from wild green turtles. Moreover, with the rise in FP in recent years within the Taiwan waters, and based on the findings, green turtles with FP can be identified and thus can be rehabilitated before releasing them back to the wild. In this study, we have listed some of the risk genes that may participate in the occurrence and established possible pathogenesis of FP in green turtles according to the transcriptome analysis result. In conclusion, most of the DEGs annotated participated in the NOD-like receptor and Wnt signaling pathway. Genes expressed in these two pathways may cause either tumor formation (tumorigenesis) or suppression. Among these genes, the presence of LEF1 may have a clinical value as a biomarker of FP (Santiago et al., 2017). Additionally, studies of FP are recommended for marine turtle populations in Asia to improve our understanding of the infection.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethical review and approval was not required for the animal study because All individuals in this study were discovered and rescued through the official reporting system of the Marine Animal Rescue Network (established by the Ocean Conservation Administration) and transported to the National Museum of Marine Biology and Aquarium (NMMBA) for further rehabilitation/medical care. Therefore, blood samples were necessarily collected by our licensed veterinarians (Dr. Tsung-Hsien Li or Dr. I-Chun Chen) for routine medical care. Hence, no individuals of green turtles were specifically captured from wild for the purpose of this study. As a consequence of the aforementioned circumstances, the excess of the blood was obtained and utilized from blood samples taken for routine medical care by our veterinarians. The scientific permit numbers for sample use were obtained from the Ocean Conservation Administration (Permit no.1090009887 and 1100012644).
M-AT and T-HL conceived the ideas and designed the methodology. T-HL and I-CC collected the data. I-IL, OB, and M-AT analyzed the data. M-AT, T-HL, and I-IL led the writing of the manuscript. All authors contributed critically to the drafts and gave final approval for publication.
The rehabilitation work for stranding marine turtles that were rescued in Taiwan by NMMBA was authorized and granted by the Ocean Conservation Administration, Taiwan, through funding support. This study was supported by the research grants (MOST1092313B291001) from the Ministry of Science and Technology, Executive Yuan, Taiwan, and by intramural funding from the NMMBA.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aguirre A. A., Balazs G. H., Spraker T. R., Gross T. S. (1995). Adrenal and hematological responses to stress in juvenile green turtles (Chelonia mydas) with and without fibropapillomas. Physiol. Zool 68 (5), 831–854. doi: 10.1086/physzool.68.5.30163934
Aguirre A. A., Lutz P. L. (2004). Marine turtles as sentinels of ecosystem health: is fibropapillomatosis an indicator? EcoHealth 1 (3), 275–283. doi: 10.1007/s10393-004-0097-3
Alfaro-Nunez A., Bojesen A. M., Bertelsen M. F., Wales N., Balazs G. H., Gilbert M. T. P. (2016). Further evidence of chelonid herpesvirus 5 (ChHV5) latency: high levels of ChHV5 DNA detected in clinically healthy marine turtles. PeerJ 4, e2274. doi: 10.7717/peerj.2274
Alfaro-Nunez A., Frost Bertelsen M., Bojesen A. M., Rasmussen I., Zepeda-Mendoza L., Tange Olsen M., et al. (2014). Global distribution of chelonid fibropapillomaassociated herpesvirus among clinically healthy sea turtles. BMC Evol. Biol. 14 (1), 206. doi: 10.1186/s12862-014-0206-z
Alfaro-Nunez A., Gilbert M. T. P. (2014). Validation of a sensitive PCR assay for the detection of chelonid fibropapilloma-associated herpesvirus in latent turtle infections. J. Virol. Methods 206, 38–41. doi: 10.1016/j.jviromet.2014.05.019
Baines C., Lerebours A., Thomas F., Fort J., Kreitsberg R., Gentes S., et al. (2021). Linking pollution and cancer in aquatic environments: a review. Environ. Int. 149, 106391. doi: 10.1016/j.envint.2021.106391
Blackburn N. B., Leandro A. C., Nahvi N., Devlin M. A., Leandro M., Martinez Escobedo I., et al. (2021). Transcriptomic profiling of fibropapillomatosis in green sea turtles (Chelonia mydas) from south Texas. Front. Immunol. 12, 410. doi: 10.3389/fimmu.2021.630988
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Britt J. L., Noorai R. E., Duckett S. K. (2020). Differentially expressed genes in cotyledon of ewes fed mycotoxins. BMC Genomics 21 (1), 680. doi: 10.1186/s12864-020-07074-z
Bruno D., Willmer I. Q., Pereira L., Rocha R. C. C., Saint’Pierre T. D., Baldassin P., et al. (2021). Metal and metalloid contamination in green Sea turtles (Chelonia mydas) found stranded in southeastern Brazil. Front. Mar. Sci. 8, 608253. doi: 10.3389/fmars.2021.608253
Chaloupka M., Work T. M., Balazs G. H., Murakawa S. K. K., Morris R. (2008). Cause-specific temporal and spatial trends in green sea turtle strandings in the Hawaiian archipelago, (1982-2003). Mar. Biol. 154 (5), 887–898. doi: 10.1007/s00227-008-0981-4
Cheng I. J. (2000). Post-nesting migrations of green turtles (Chelonia mydas) at wanan island, penghu archipelago, Taiwan. Mar. Biol. 137 (4), 747–754. doi: 10.1007/s002270000375
Cheng I. J., Chen T. H. (1997). The incidental capture of five species of sea turtles by coastal setnet fisheries in the eastern waters of Taiwan. Biol. Conserv. 82 (2), 235–239. doi: 10.1016/S0006-3207(97)00027-X
Cheng I. J., Cheng W. H., Chan Y. T. (2018). Geographically closed, yet so different: contrasting long-term trends at two adjacent sea turtle nesting populations in Taiwan due to different anthropogenic effects. PloS One 13 (7), e0200063. doi: 10.1371/journal.pone.0200063
Da Silva-Júnior E. S., De Farias D. S. D., Bomfim A. D. C., Freire A. C. D. B., Revorêdo R.Â., Rossi S., et al. (2019). Stranded marine turtles in northeastern Brazil: incidence and spatial–temporal distribution of fibropapillomatosis. Chelonian Conserv. Biol. 18 (2), 249–258. doi: 10.2744/CCB-1359.1
Day R. D., Keller J. M., Harms C. A., Segars A. L., Cluse W. M., Godfrey M. H., et al. (2010). Comparison of mercury burdens in chronically debilitated and healthy loggerhead sea turtles (Caretta caretta). J. Wildl. Dis. 46 (1), 111–117. doi: 10.7589/0090-3558-46.1.111
Dimitriadis C., Mazaris A. D., Katsanevakis S., Iosifakis A., Spinos E., Kalli E., et al. (2022). Stranding records and cumulative pressures for sea turtles as tools to delineate risk hot spots across different marine habitats. Ocean Coast. Manage. 217, 106017. doi: 10.1016/j.ocecoaman.2021.106017
Dolcet X., Llobet D., Pallares J., Matias-Guiu X. (2005). NF-kB in development and progression of human cancer. Virchows Archiv. 446 (5), 475–482. doi: 10.1007/s00428-005-1264-9
Duffy D. J., Schnizler C., Karpinski L., Thomas R., Whilde J., Eastman C., et al. (2018). Sea Turtle fibropapilloma tumours share genomic drivers and therapeutic vulnerabilities with human cancers. Commun. Biol. 1, 63. doi: 10.1038/s42003-018-0059-x
Dujon A. M., Schofield G., Venegas R. M., Thomas F., Ujvari B. (2021). Turtles in the cancer risk landscape: a global meta-analysis of fibropapillomatosis prevalence and associated risk factors. Sea Pathogens 10 (10), 1295. doi: 10.3390/pathogens10101295
Dutton P. H. (1996). “Methods for collection and preservation of samples for sea turtle genetic studies,” in Proceedings of the international symposium on Sea turtle genetics. Eds. Bowen B. W., W. N. (Miami, Florida: NOAA Technical Memorandum), 17–24.
Farrell J. A., Yetsko K., Whitmore L., Whilde J., Eastman C. B., Ramia D. R., et al. (2021). Environmental DNA monitoring of oncogenic viral shedding and genomic profiling of sea turtle fibropapillomatosis reveals unusual viral dynamics. Commun. Biol. 4 (1), 1–17. doi: 10.1038/s42003-021-02085-2
Foley A. M., Schroeder B. A., Redlow A. E., Fick-Child K. J., Teas W. G. (2005). Fibropapillomatosis in stranded green turtles (Chelonia mydas) from the eastern united state-98): trends and associations with environmental factors. J. Wildl Dis. 41 (1), 29–41.
Formia A., Deem S., Billes A., Ngouessono S., Parnell R., Collins T., et al. (2007). Fibropapillomatosis confirmed in Chelonia mydas in the gulf of Guinea, West Africa. Mar. Turtle Newslett. 116, 20.
Fumero-Hernández M., Encinoso Quintana M., Ramírez A. S., Morales Fariña I., Calabuig P., Jaber J. R. (2023). Morphometric study of the eyeball of the loggerhead turtle (Caretta caretta) using computed tomography (CT). Animals 13, 1016. doi: 10.3390/ani13061016
Godoy D. A., Stockin K. A. (2018). Anthropogenic impacts on green turtles Chelonia mydas in new zealand. endanger. Species Res. 37, 1–9. doi: 10.3354/esr00908
Greenblatt R. J., Work T. M., Balazs G. H., Sutton C. A., Casey R. N., Casey J. W. (2004). The Ozobranchus leech is a candidate mechanical vector for the fibropapilloma-associated turtle herpesvirus found latently infecting skin tumours on Hawaiian green turtles (Chelonia mydas). Virology 321 (1), 101–110. doi: 10.1016/j.virol.2003.12.026
Guimarães S. M., Gitirana H. M., Wanderley A. V., Monteiro-Neto C., Lobo-Hajdu G. (2013). Evidence of regression of fibropapillomas in juvenile green turtles Chelonia mydas caught in niterói, southeast Brazil. Dis. Aquat. Org. 102 (3), 243–247. doi: 10.3354/dao02542
Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat. Protoc. 8 (8), 1494–1512. doi: 10.1038/nprot.2013.084
Hamann M., Godfrey M. H., Seminoff J. A., Arthur K., Barata P. C. R., Bjorndal K. A., et al. (2010). Global research priorities for sea turtles: informing management and conservation in the 21st century. Endanger. Species Res. 11 (3), 245e269. doi: 10.3354/esr00279
Hargrove S., Work T. M., Brunson S., Foley A. M., Balazs G. H. (2015). Proceedings of the 2015 international summit on fibropapillomatosis: global status, trends, and population impacts (NOAA Technical Memorandum NMFS-PIFSC-054).
Hazel J., Lawler I. R., Hamann M. (2009). Diving at the shallow end: green turtle behaviour in near-shore foraging habitat. J. Exp. Mar. Bio. Ecol. 371, 84–92. doi: 10.1016/j.jembe.2009.01.007
Herbst L. H. (1994). Fibropapillomatosis of marine turtles. Annu. Rev. Fish Dis. 4, 389–425. doi: 10.1016/0959-8030(94)90037-X
IUCN (2016) The IUCN red list of threatened species. Available at: http://www.iucnredlist.org/ (Accessed 7 Feb. 2023).
James A. (2020). Prevalence of chelonid alphaherpesvirus 5 and associated disease in Sea turtles in Grenada, West indies (True Blue, Grenada: St. George’s University).
James A., Page-Karjian A., Charles K. E., Edwards J., Gregory C. R., Cheetham S., et al. (2021). Chelonid alphaherpesvirus 5 prevalence and first confirmed case of Sea turtle fibropapillomatosis in Grenada, West indies. Animals 11 (6), 1490. doi: 10.3390/ani11061490
Jo K., Im J., Park B. Y., Cho B., Joo S., Kim B. Y., et al. (2022). Possible link between derelict fishing gear and sea turtle strandings in coastal areas. Mar. pollut. Bull. 185, 114240. doi: 10.1016/j.marpolbul.2022.114240
Jones K., Ariel E., Burgess G., Read M. (2016). A review of fibropapillomatosis in green turtles (Chelonia mydas). J. Vet. Med. 212, 48–57. doi: 10.1016/j.tvjl.2015.10.041
Jones K., Limpus C. J., Brodie J., Jones R., Read M., Shum E., et al. (2022). Spatial distribution of fibropapillomatosis in green turtles along the Queensland coast and an investigation into the influence of water quality on prevalence. Conserv. Sci. Pract. 4 (8), e12755. doi: 10.1111/csp2.12755
Kanehisa M. (2019). Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28 (11), 1947–1951. doi: 10.1002/pro.3715
Kanehisa M., Goto S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28 (1), 27–30. doi: 10.1093/nar/28.1.27
Kuo F. W., Fan T. Y., Ng C. K. Y., Cai Y., Balazs G. H., Li T. H. (2017). Tale of the unlucky tags: the story of a rescued, rehabilitated, and released green sea turtle (Chelonia mydas) in southern Taiwan. Bull. Mar. Sci. 93 (3), 689–690. doi: 10.5343/bms.2016.1108
Lackovich J. K., Brown D. R., Homer B. L., Garber R. L., Mader D. R., Moretti R. H., et al. (1999). Association of herpesvirus with fibropapillomatosis of the green turtle Chelonia mydas and the loggerhead turtle Caretta in Florida. Dis. Aquat. Org. 37 (2), 89–97. doi: 10.3354/dao037089
Lawrance M. F., Mansfield K. L., Sutton E., Savage A. E. (2018). Molecular evolution of fibropapilloma-associated herpesviruses infecting juvenile green and loggerhead sea turtles. Virology 521, 190–197. doi: 10.1016/j.virol.2018.06.012
Li T. H., Chang C. C. (2020). The impact of fibropapillomatosis on clinical characteristics, blood gas, plasma biochemistry, and hematological profiles in juvenile green turtles (Chelonia mydas). Bull. Mar. Sci. 96 (4), 723–734. doi: 10.5343/bms.2019.0120
Li T. H., Chang C. C., Cheng I. J., Lin S. C. (2015). Development of a summarized health index (SHI) for use in predicting survival in sea turtles. PloS One 10 (3), e0120796. doi: 10.1371/journal.pone.0120796
Li T. H., Hsu W. L., Chen C. Y., Chen Y. C., Wang Y. C., Tsai M. A., et al. (2022). Preparation of recombinant glycoprotein b (gB) of chelonid herpesvirus 5 (ChHV5) for antibody production and its application for infection detection in sea turtles. Sci. Rep. 12 (1), 1–11. doi: 10.1038/s41598-022-15281-9
Li T. H., Hsu W. L., Lan Y. C., Balazs G. H., Work T. M., Tseng C. T., et al. (2017). Identification of chelonid herpesvirus 5 (ChHV5) in endangered green turtles (Chelonia mydas) with fibropapillomatosis in Asia. Bull. Mar. Sci. 93 (4), 1011–1022. doi: 10.5343/bms.2017.1018
Li Y., Pei J., Xia H., Ke H., Wang H., Tao W. (2003). Lats2, a putative tumour suppressor, inhibits G1/S transition. Oncogene. 22 (28), 4398–4405. doi: 10.1038/sj.onc.1206603
Liu T. K., Chen P., Chen H. Y. (2015). Comprehensive assessment of coastal eutrophication in Taiwan and its implications for management strategy. Bull. Mar. Sci. 97, 440–450. doi: 10.1016/j.marpolbul.2015.05.055
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408.
Loganathan A. L., Palaniappan P., Subbiah V. K. (2021). First evidence of chelonid herpesvirus 5 (ChHV5) infection in green turtles (Chelonia mydas) from sabah, Borneo. Pathogens. 10 (11), 1404. doi: 10.3390/pathogens10111404
Manes C., Carthy R. R., Hull V. (2023). A coupled human and natural systems framework to characterize emerging infectious diseases–the case of fibropapillomatosis in marine turtles. Animals. 13 (9), 1441. doi: 10.3390/ani13091441
Manes C., Pinton D., Canestrelli A., Capua I. (2022). Occurrence of fibropapillomatosis in green turtles (Chelonia mydas) in relation to environmental changes in coastal ecosystems in Texas and Florida: a retrospective study. Animals. 12 (10), 1236. doi: 10.3390/ani12101236
Mansfield K. L., Mendilaharsu M. L., Putman N. F., dei Marcovaldi M. A. G., Sacco A. E., Lopez G., et al. (2017). First satellite tracks of south Atlantic sea turtle ‘lost years’: seasonal variation in trans-equatorial movement. Proc. R. Soc B 284, 20171730. doi: 10.1098/rspb.2017.1730
Mghili B., Benhardouze W., Aksissou M., Tiwari M. (2023). Sea Turtle strandings along the northwestern Moroccan coast: spatio-temporal distribution and main threats. Ocean Coast. Manage. 237, 106539. doi: 10.1016/j.ocecoaman.2023.106539
Mi H., Muruganujan A., Ebert D., Huang X., Thomas P. D. (2018). Panther version 14: more genomes, a new panther go-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47 (D1), D419–D426.. doi: 10.1093/nar/gky1038
Nash A., Ryan E. J. (2023). Immunity in sea turtles: review of a host-pathogen arms race millions of years in the running. Animals. 13 (4), 556. doi: 10.3390/ani13040556
Ng C. K. Y., Lam J. C. W., Zhang X. H., Gu H. X., Li T. H., Ye M. B., et al. (2018). Levels of trace elements, methylmercury and polybrominated diphenyl ethers in foraging green turtles in the south China region and their conservation implications. Environ. pollut. 234, 735–742. doi: 10.1016/j.envpol.2017.11.100
Page-Karjian A., Chabot R., Stacy N. I., Morgan A. S., Valverde R. A., Stewart S., et al. (2020a). Comprehensive health assessment of green turtles Chelonia mydas nesting in southeastern Florida, USA. Endang. J. Species Res. 42, 21–35. doi: 10.3354/esr01036
Page-Karjian A., Norton T. M., Krimer P., Groner M., Nelson S. E., Gottdenker N. L. (2014). Factors influencing survivorship in rehabilitating green sea turtles (Chelonia mydas) with fibropapillomatosis. J. Zoo Wildl. Med. 45, 507–519. doi: 10.1638/2013-0132R1.1
Page-Karjian A., Serrano M. E., Cartzendafner J., Morgan A., Ritchie B. W., Gregory C. R., et al. (2020b). Molecular assessment of chelonid alphaherpesvirus 5 infection in tumour-free green (Chelonia mydas) and loggerhead (Caretta caretta) sea turtles in north Carolina, USA 2015–2019. Animals. 10 (11), 1964. doi: 10.3390/ani10111964
Page-Karjian A., Torres F., Zhang J., Rivera S., Diez C., Moore P. A., et al. (2012). Presence of chelonid fibropapilloma-associated herpesvirus in tumoured and non-tumoured green turtles, as detected by polymerase chain reaction, in endemic and non-endemic aggregations (Puerto Rico: SpringerPlus), 1–8.
Perrault J. R., Levin M., Mott C. R., Bovery C. M., Bresette M. J., Chabot R. M., et al. (2021). Insights on immune function in free-ranging green sea turtles (Chelonia mydas) with and without fibropapillomatosis. Animals. 11 (3), 861. doi: 10.3390/ani11030861
Perrault J. R., Stacy N. I., Lehner A. F., Mott C. R., Hirsch S., Gorham J. C., et al. (2017). Potential effects of brevetoxins and toxic elements on various health variables in kemp’s ridley (Lepidochelys kempii) and green (Chelonia mydas) sea turtles after a red tide bloom event. Sci. Total Environ. 605, 967–979. doi: 10.1016/j.scitotenv.2017.06.149
Rees A. F., Alfaro-Shigueto J., Barata P. C. R., Bjorndal K. A., Bolten A. B., Bourjea J., et al. (2016). Are we working towards global research priorities for management and conservation of sea turtles? Endanger. Species Res. 31, 337–382. doi: 10.3354/esr00801
Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26 (1), 139–140. doi: 10.1093/bioinformatics/btp616
Rodríguez Y., Vandeperre F., Santos M. R., Herrera L., Parra H., Deshpande A., et al. (2022). Litter ingestion and entanglement in green turtles: an analysis of two decades of stranding events in the NE Atlantic. Environ. pollut. 298, 118796. doi: 10.1016/j.envpol.2022.118796
Rossi S., Zamana R. R., Santos P. P. D. A., Bomfim A. D. C., Farias D. S. D. D., Freire A. C. D. B., et al. (2021). Visceral neoplasms and chelonid alphaherpesvirus 5 in green turtles with fibropapillomatosis. Arch. Vet. Sci. 26 (1), 63–79.
Santiago L., Daniels G., Wang D., Deng F. M., Lee P. (2017). Wnt signaling pathway protein LEF1 in cancer, as a biomarker for prognosis and a target for treatment. Am. J. Cancer Res. 7 (6), 1389.
Shigenaka G. (2010). Oils and sea turtle: biology, planning, and response (Seattle, Washington: Office of Response and Restoration, NOAA Ocean Service).
Smith G. M., Coates C. W. (1938). Fibro-epithelial growths of the skin in large marine turtles, Chelonia mydas (Linnaeus). Zoologica 23 (4), 93–98. doi: 10.5962/p.203654
Soleimani A., Rahmani F., Ferns G. A., Ryzhikov M., Avan A., Hassanian S. M. (2020). Role of the NF-κB signaling pathway in the pathogenesis of colorectal cancer. Gene 726, 144132. doi: 10.1016/j.gene.2019.144132
Sposato P., Keating P., Lutz P. L., Milton S. L. (2021). Evaluation of immune function in two populations of green sea turtles (Chelonia mydas) in a degraded versus a nondegraded habitat. J. Wildl. Dis. 57 (4), 761–772. doi: 10.7589/JWD-D-20-00204
Starczynowski D. T., Lockwood W. W., Deléhouzée S., Chari R., Wegrzyn J., Fuller M., et al. (2011). TRAF6 is an amplified oncogene bridging the RAS and NF-κB pathways in human lung cancer. J. Clin. Investig. 121 (10), 4095–4105. doi: 10.1172/JCI58818
Stevens P. D., Wen Y. A., Xiong X., Zaytseva Y. Y., Li A. T., Wang C., et al. (2018). Erbin suppresses KSR1-mediated RAS/RAF signaling and tumourigenesis in colorectal cancer. Cancer Res. 78 (17), 4839–4852. doi: 10.1158/0008-5472.CAN-17-3629
Sun H., Li X., Fan L., Wu G., Li M., Fang J. (2014). TRAF6 is upregulated in colon cancer and promotes proliferation of colon cancer cells. Int. J. Biochem. Cell Biol. 53, 195–201. doi: 10.1016/j.biocel.2014.04.010
Supek F., Bošnjak M., Škunca N., Šmuc T. (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PloS One 6 (7), e21800. doi: 10.1371/journal.pone.0021800
Tanabe L. K., Scott K., Dasari V., Berumen M. L. (2022). An assessment of heavy metals in green sea turtle (Chelonia mydas) hatchlings from Saudi arabia’s largest rookery, ras baridi. PeerJ 10, e13928. doi: 10.7717/peerj.13928
Tilborghs S., Corthouts J., Verhoeven Y., Arias D., Rolfo C., Trinh X. B., et al. (2017). The role of nuclear factor-kappa b signaling in human cervical cancer. Crit. Rev. Oncol. Hematol. 120, 141–150. doi: 10.1016/j.critrevonc.2017.11.001
Van Houtan K. S., Hargrove S. K., Balazs G. H. (2010). Land use, macroalgae, and a tumour-forming disease in marine turtles. PloS One 5, e12900. doi: 10.1371/journal.pone.0012900
Van Houtan K. S., Smith C. M., Dailer M. L., Kawachi M. (2014). Eutrophication and the dietary promotion of sea turtle tumours. PeerJ 2, e602. doi: 10.7717/peerj.602
Work T. M., Balazs G. H. (1999). Relating tumour score to hematology in green turtles with fibropapillomatosis in Hawaii. J. Wildl. Dis. 35, 804–807. doi: 10.7589/0090-3558-35.4.804
Work T. M., Balazs G. H., Schumacher J. L., Marie A. (2005). Epizootiology of spirorchiid infection in green turtles (Chelonia mydas) in Hawaii. J. Parasitol. 91, 871–876. doi: 10.1645/GE-454R.1
Work T. M., Rameyer R. A., Balazs G. H., Cray C., Chang S. P. (2001). Immune status of free-ranging green turtles with fibropapillomatosis from Hawaii. J. Wildl. Dis. 37 (3), 574–581. doi: 10.7589/0090-3558-37.3.574
Xia L., Tan S., Zhou Y., Lin J., Wang H., Oyang L., et al. (2018). Role of the NFκB-signaling pathway in cancer. OncoTargets Ther. 11, 2063–2073. doi: 10.2147/OTT.S161109
Yetsko K., Farrell J. A., Blackburn N. B., Whitmore L., Stammnitz M. R., Whilde J., et al. (2021). Molecular characterization of a marine turtle tumour epizootic, profiling external, internal and postsurgical regrowth tumours. Commun. Biol. 4, 152. doi: 10.1038/s42003-021-01656-7
Zhang C., Leng W., Sun C., Lu T., Chen Z., Men X., et al. (2018). Urine proteome profiling predicts lung cancer from control cases and other tumours. EBioMedicine 30, 120–128. doi: 10.1016/j.ebiom.2018.03.009
Keywords: marine turtle, fibropapillomatosis, de novo transcriptome assembly, differentially expressed genes, NOD-like receptor signaling pathway, pathogenesis
Citation: Li T-H, Lei I-I, Byadgi OV, Chen I-C and Tsai M-A (2023) Evidence of chelonid herpesvirus 5 infection in green turtle (Chelonia mydas) indicated a possible tumorigenesis activation by transcriptome analysis. Front. Mar. Sci. 10:1185111. doi: 10.3389/fmars.2023.1185111
Received: 13 March 2023; Accepted: 26 June 2023;
Published: 21 July 2023.
Edited by:
Salvatore Siciliano, Fundação Oswaldo Cruz (Fiocruz), BrazilReviewed by:
Hong Meiling, Hainan Normal University, ChinaCopyright © 2023 Li, Lei, Byadgi, Chen and Tsai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming-An Tsai, d2lsbGlhbTg3ODU4OEBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.