- 1Guangdong Research Center on Reproductive Control and Breeding Technology of Indigenous Valuable Fish Species, Guangdong Provincial Key Laboratory of Aquatic Animal Disease Control and Healthy Culture, Fisheries College, Guangdong Ocean University, Zhanjiang, Guangdong, China
- 2State Key Laboratory of Biocontrol, School of Ecology, Sun Yat-sen University, Shenzhen, Guangzhou, China
- 3Development and Research Center for Biological Marine Resources, Southern Marine Science and Engineering Guangdong Laboratory (Zhanjiang), Zhanjiang, Guangdong, China
- 4Guangdong Provincial Key Laboratory of Marine Resources and Coastal Engineering, Guangzhou, Guangdong, China
Greater amberjack (Seriola dumerili) is an important commercial fish for its high growth rate and excellent flesh quality. However, its sensitivity to variations of water salinity poses challenges to the cage culture. In this study, the greater amberjack were reared in the optimum salinity (30 ppt, CK) and undesired regimes (10 and 40 ppt) for 72 hours. The molecular adaptive mechanisms to salinity stress were revealed by the comparative transcriptome analysis for the gills and kidneys. In gills, a total of 445 and 423 differentially expressed genes (DEGs) were identified in 10 and 40 ppt salinity stress groups, respectively. Those DEGs were involved in cartilage and skeletal development, ions transport, and immune response. The major ion secretion and osmoregulation transport proteins gene slc12a2/nkcc1 and cftr expression levels were significantly down-regulated at 10 ppt, but slightly activated at 40 ppt, compared with the control group. The expression changes in response to the Na+, K+ movement, and Cl- ion secretion reduced under the hypo-osmotic exposure and ion excretion boost upon hyper-salinity stress. Meanwhile, the cartilage and skeletal development were enhanced in the gills by hypo- or hyper-salinity stimuli, which is critical for maintaining gill structures and improving respiration and osmoregulation under salinity stress. In kidneys, 600 and 539 DEGs were identified in 10 and 40 ppt groups, respectively. Those DEGs were enriched in oxygen transport, pronephros development, regulation of growth, blood coagulation, ion transmembrane transport, and immune response. While the known renal Na+/Cl– co-transporter gene slc12a3/ncc expression level was significantly down-regulated at 10 ppt, the organic cation transporter 2 gene slc22a2, ammonium transmembrane transport gene rhd and rhag expression levels were overexpressed under the hyper-salinity condition at 40 ppt, contributing to the salts secretion and ammonium transport regulation, to combat the osmotic influx of salts following the drink of seawater and elevated ammonia production upon high salinity stress. These findings advance our knowledge of adaptative mechanisms to the salinity stress and provide theoretical guidance for the optimal breeding mode for the aquaculture of greater amberjack.
1 Introduction
Fish is an important resource of protein and nutrient supply to humans. Environmental factors, such as temperature, salinity, and food availability, affect fish growth and survival, especially under artificial cultural conditions. Salinity is recognized as critical and substantial affect factor (Gonzalez, 2012). Teleost fishes grown in freshwater maintain a higher blood osmolality than the surrounding environment. They enhance ion uptake in gills, increase ion reabsorption, and produce diluted urine in the kidneys to compensate for ion loss (Chen et al., 2009; Kültz, 2012). In contrast, marine fish have a lower body salt concentration than seawater. To combat the challenges of hyperosmolarity, fish may excrete more salt through gills and reduce the glomerular filtration rates in the kidneys to maintain the fluid and ion balance (Evans, 2008; Laverty and Skadhauge, 2012; Gui et al., 2016).
Saline-alkali fluctuations beyond the osmoregulation capacity of fishes may pose adverse impacts on their growth and survival rates, metabolism, immunity, and reproduction (Lehtonen et al., 2016; Guo et al., 2020; Galkanda-Arachchige et al., 2021; Lu et al., 2022), and affect fish performance, flesh quality and production in aquaculture. Freshwater fish species, such as Nile tilapia (Oreochromis niloticus) (Pepe et al., 2022), grass carp (Ctenopharyngodon idella) (Liu et al., 2023), African catfish (Clarias gariepinus) (Zidan et al., 2022), and rohu (Labeo rohita) (Sarma et al., 2020) exhibit a higher growth rate and better physiological performance when reared under low salinity environment than that of high salinity, which is because of the resource allocation trade-off between the energy spent on osmoregulation for growth and development under the desirable ambient salinity. In contrast, the growth of marine fish species, including Brazilian flounder (Paralichthys orbignyanus) (Sampaio and Bianchini, 2002), black seabream (Acanthopagrus schlegelii) (Li et al., 2022), and yellowtail kingfish (Seriola lalandi) (Morgenroth et al., 2022) were suppressed when switched from seawater to freshwater. Deviating from ionic and osmotic homeostasis under unfavorable salinity conditions can interfere with fish energy supply and hormone production (Tsuzuki et al., 2007).
Furthermore, salinity may directly impact the fish’s immune systems (Gu et al., 2018). In pipefish (Syngnathus typhle), the elevated saline-alkali stress impaired the immune resistance upon infection and increased the risk of susceptibility (Birrer et al., 2012). The innate immunity of striped catfish (Pangasianodon hypophthalmus) has been demonstrated to be substantially affected by the changes in osmotic environments (Hieu et al., 2021). The activities of lysozyme and alternative complement pathway (ACP) were substantially elevated in mandarin fish (Siniperca chuatsi) (Ouyang et al., 2023), Asian seabass (Lates calcarifer) (Azodi et al., 2021) and Mozambique tilapia (Oreochromis mossambicus) (Jiang et al., 2008) when exposed to high salinity environments. In recent years, with the assistance of high-throughput sequencing technologies, many studies have unveiled the molecular mechanisms underlying the adaptive responses to salinity changes in teleost fish, such as spotted sea bass (Lateolabrax maculatus) (Zhang et al., 2017), half-smooth tongue sole (Cynoglossus semilaevis) (Si et al., 2018), silver pomfret (Pampus argenteus) (Li et al., 2020), Mozambique tilapia (Kammerer et al., 2009) and Atlantic salmon (Salmo salar) (Gjessing et al., 2020). Two novel nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs) were identified in grass carp, and NLRs can enhance their resistance to bacterial infection upon hyper-salinity stress (Fang et al., 2022). Although many studies have been performed in the common fish species, more efforts are still required in other fishes to broaden our understanding.
Greater amberjack (Seriola dumerili), the largest species in Seriola, is widely distributed across the Indo-Pacific Ocean, Mediterranean Sea, and Atlantic Ocean (Zupa et al., 2017; Araki et al., 2018). Due to its rapid growth and excellent flesh quality, greater amberjack exhibits a high commercial value, and more attention is dedicated to its marine cage culture (Mazzola et al., 2000). Greater amberjack inhabits the upper and middle layers of warm seas, and its reproduction and survival are dependent on the suitable water temperature and salinity (Fernández-Montero et al., 2018). The oscillation in water salinity can alter the energy metabolism, ion transport, and electrophysiological processes in greater amberjack (Barany et al., 2021; Peng et al., 2022). However, previous studies only focused on the responses in gills or livers. The kidneys are another important osmoregulatory organ in teleost fishes. In freshwater, fish kidneys up-regulates salt reabsorption to compensate for salt loss while enhancing the excretion of excess divalent ions and water conservation in seawater to maintain the proper blood osmolarity (Chen et al., 2009). A comprehensive study is required to understand the molecular mechanisms that respond to salinity fluctuations in the kidneys of greater amberjack, which brings new insights into the aquacultural strategy.
In this study, the transcriptomic dynamics in the gill and kidney tissues of greater amberjack juveniles reared under different salinity regimes were investigated. Our findings unravelled the adaptive mechanism involved in coping with the impacts of salinity stress on greater amberjack, which provide theoretical guidance for the optimal breeding mode for greater amberjack.
2 Materials and methods
2.1 Ethics statement
All the experiments were implemented according to the guidelines and regulations of the Animal Research and Ethics Committee of Guangdong Ocean University (NIH Pub. No. 85-23, revised 1996) and the laws and regulations of China on biological research. No endangered or protected species were involved in this study.
2.2 Experimental fish and salinity stress
Greater amberjack juveniles (body weight: 6.94 ± 0.45 g; body length: 8.40 ± 0.54 cm) were reared in tanks at the temperature of 22 ± 1.0°C in Donghai Island, Guangdong, China. All fish were fed on a commercial diet (Guangdong Yuequn Biotechnology Co., Ltd, Jieyang, China) twice daily. They were randomly divided into 12 cylindrical 1000 L tanks (10 individuals per tank) at different salinities: 40, 30, and 10 parts per thousand (ppt), where all the saline was prepared with commercial seawater salty crystal and aerated tap water. The fish reared in the water with a median salinity of 30 ppt, the natural seawater condition, were set as the control group (CK). The water was changed once a day (every 24 h). All animal experiment protocols were authorized by the Institutional Animal Care and Use Committee of Guangdong Ocean University (Zhanjiang, China). At 72 h treatment, six fish (biological replicates) were randomly selected from each group. All fish were anesthetized using 100 mg/L tricaine methane sulfonate (MS 222; Sigma-Aldrich, St. Louis, MO, USA) and then dissected. The mixed partial gill filament from each gill arch and whole kidney tissues were taken from each individual of each group, collected in centrifuge tubes containing 1 mL RNA stabilization reagent (Accurate Biology, Changsha, China) overnight, and stored at -80°C for further experiments.
2.3 Total RNA extraction, library construction, and sequencing
For each individual, total RNA was extracted from the gill and kidney tissues of each collected individual using a Trizol reagent (Invitrogen, Carlsbad, CA, USA), following the manufacturer’s instructions. The cleavage of tissue samples, RNA extraction, RNA purity, degradation, and contamination examinations were performed according to the previous study (Shi et al., 2022). The RNA integrity was examined using an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Total RNA with an RNA integrity number (RIN) score >7 was used for sequencing.
According to the manufacturer’s instructions, a total of 1 µg RNA was used for sequencing library construction using NEBNext UltraTM RNA Library Prep Kit for Illumina® (NEB, Ipswich, MA, USA). The messenger RNA (mRNA) was purified using poly-T oligo-attached magnetic beads (Illumina, San Diego, CA, USA) after treating the RNA samples with DNase I (NEB, Ipswich, MA, USA). A fragmentation buffer reagent (New England Biolabs (NEB), Ipswich, MA, USA) was used to fragment mRNA into short fragments. The library fragments were purified using the AMPure XP Reagent (Beckman Coulter, Beverly, USA). USER Enzyme (NEB, Ipswich, MA, USA) was applied to make the adaptor-ligated complementary DNA (cDNA) for the downstream PCR. Then, polymerase chain reaction (PCR) was carried out using Phusion High-Fidelity DNA polymerase (NEB, Ipswich, MA, USA), with universal PCR primers and index (X) primer. PCR products were purified using the AMPure XP system (NEB, Ipswich, MA, USA). The library quantity and quality were assessed on the Agilent Bioanalyzer 2100 system (Agilent Technologies, Palo Alto, CA, USA) and StepOnePlus Real-Time PCR System (Thermo Fisher Scientific, Santa Clara, CA, USA), respectively. The index-coded samples were clustered using the cBot Cluster Generation System with the TruSeq PE Cluster Kit v4-cBot-HS (Illumina, San Diego, CA, USA). All cDNA libraries were sequenced on the Illumina NovaSeq 6000 platform (Illumina, San Diego, CA, USA).
2.4 Transcriptome assembly and functional gene annotation
All the generated raw sequencing data were deposited and submitted to Gene Expression Omnibus (GEO) of the National Center for Biotechnology Information (NCBI) with an accession number GSE220485 (GSM6805719-GSM6805730). The raw reads were pre-processed for each library, including removing adapter contamination and filtering out the reads containing ploy-N and the low-quality bases through in-house perl scripts. The Q20, Q30, GC-content, and sequence duplication levels of the clean data were assessed to illustrate the data quality after quality control. Then, the clean reads from each sample were mapped to the reference genome of S. dumerili using HISAT2 (Kim et al., 2019).
Gene functions were annotated against the NCBI’s non-redundant (NR; ftp://ftp.ncbi.nih.gov/blast/db/), Swiss-Prot (http://www.uniprot.org/), Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) and Gene Ontology (GO; http://www.geneontology.org/) databases, using the BLASTx (v. 2.2.26; https://blast.ncbi.nlm.nih.gov/) with an E-value cutoff of 1 × e-5.
2.5 Data structure and differential expression analysis
The gene expression level was inferred by the read abundance computed using featureCounts (Liao et al., 2014). The data structure was then assessed by principal component analysis (PCA) using the DESeq2 package (v. 1.38.3) (Love et al., 2014). The expression data were first log2 transformed and the genes were sorted based on their variance across samples. The top 500 genes with the largest variance were selected for PCA using the prcomp function. DESeq2 was used to identify differentially expressed genes (DEGs) under each of salinity stress conditions (10 or 40 ppt) against CK (30 ppt), where Wald test was employed to compute the p-value for each tested gene. Benjamini and Hochberg (BH) method was used to adjust the p-value to correct for multiple testing. Genes with adjusted p-value < 0.05 and |log2 fold change| > 1.0 were significantly differentially expressed. DEGs showing higher or lower expression levels than CK for each stress condition were denoted as “up-regulated” or “down-regulated” genes, respectively. GO and KEGG pathway enrichment analyses were performed on up- and down-regulated DEGs using clusterProfiler (Yu et al., 2012). The statistical significance (p-value) was measured based on hypergeometric distribution, and the GO terms and KEGG pathways with p-value < 0.05 were significantly over-represented.
2.6 Trend analysis across different salinity regimes
Trend analysis was carried out for the gills and kidneys across the salinity conditions (10, 30, and 40 ppt) using the Mfuzz package (Futschik and Carlisle, 2005; Kumar and Futschik, 2007). The gene expression level was normalized within each sample as transcripts per million (TPM). Genes of low variation among conditions were filtered out from the subsequent steps. The retained genes were clustered into six groups of different expression trends across different salinities using the Mfuzz function for each tissue (Supplementary Figure S1, S2). Fisher’s exact test was used to assess the statistical significance of enrichment of each cluster following the strategy as described previously (Qian et al., 2020). Our null hypothesis is that there is no difference in gene numbers across all the trends. To test the hypothesis, all the tested genes were first randomly assigned into six clusters 10,000 times, and the median of the number of genes in each cluster was set as the background. For each cluster, the significance level of the over-representation was tested using one-tail Fisher’s exact test. The clusters with p-value < 0.05 were considered to have a significantly higher proportion of genes than the random assignment.
2.7 Real-time quantitative polymerase chain reaction (qRT-PCR) validation
To validate the DEGs from RNA sequencing (RNA-seq) data, a total of 10 DEGs were randomly selected (five for the gills and five for the kidneys), and their expression levels were assessed by qRT-PCR. The procedure of qRT-PCR is as follows: initial denaturation at 94°C for 5 min, followed by 35 cycles of 20 s denaturation at 94°C, 20 s annealing at 55°C and 20 s extension at 72°C. The primers of all selected genes were designed by Primer Premier software v5.0 (Premier Biosoft International, Palo Alto, CA, USA) and listed in Supplementary Table S1. A light CyclerTM96 (Roche, Indianapolis, IN, USA) was employed for qRT-PCR using SYBR Green Real-time PCR Master Mix (TaKaRa Biotechnology, Dalian, China). All samples were validated in triplicates. β-actin was used as the reference gene, which was identified to be stably expressed in greater amberjack (Djellata et al., 2021). The relative expression levels of DEGs were estimated using the 2-ΔΔCt method. Data were expressed as means ± standard error (SE) (n = 3). The statistical analysis was performed in the SPSS v. 16.0 (SPSS Inc., Chicago, IL, USA).
3 Results
3.1 Illumina sequencing and annotation
A total of 12 cDNA libraries (G10, G30, G40, K10, K30, and K40; Table 1) were constructed to assess the transcriptional responses to salinity stress in the gill and kidney tissues of greater amberjack. After quality control, 134.02 and 136.21 million clean reads were retained for the downstream transcriptomic analyses of the gills and kidneys, respectively. The Q30 values of the clean reads were greater than 91% in all the samples, indicating good data quality. Summary statistics for all the 12 cDNA libraries, including the number of clean reads and bases, Q20 and Q30 values, and GC content, are listed in Table 1.
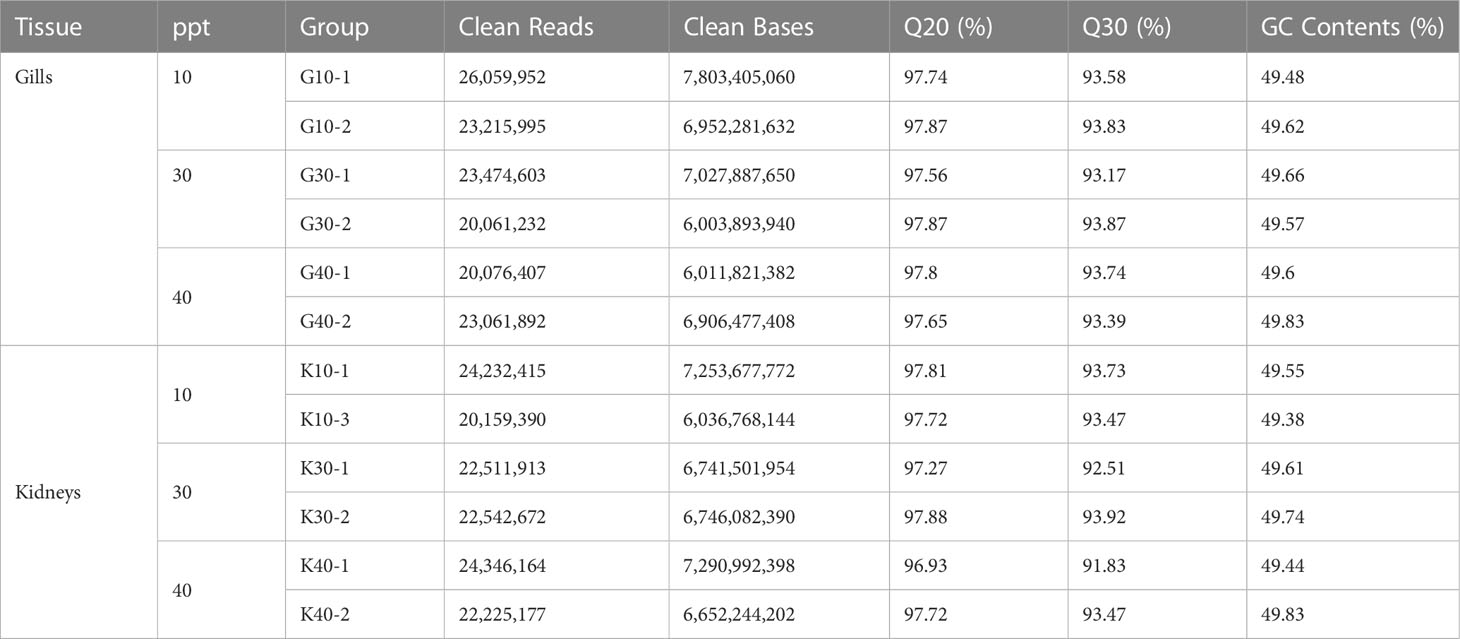
Table 1 Summary statistics for the RNA sequencing (RNA-seq) data of the gills and kidneys in greater amberjack.
3.2 Transcriptional dynamics in response to salinity alterations in gills of greater amberjack
PCA was conducted to explore the consistency in gene expression profiles between samples of each treatment. All the biological replicates were aggregated, and different treatments were substantially distinct from each other (Figure 1A), where 35% and 23% of the variations were explained by the first and second principal components (PCs), respectively. In the gills, 445 DEGs were identified under 10 ppt salinity stress, including 309 up-regulated and 136 down-regulated genes, compared to the CK (Figure 1B; Supplementary Figure S3A). In the 40 ppt saline treatment group, a total of 320 up-regulated and 103 down-regulated genes were detected (Figure 1B; Supplementary Figure S3B). GO enrichment analysis showed that the down-regulated DEGs were enriched in the lipoprotein metabolic process, inorganic anion transport, chloride, and ammonium transmembrane transport (Figure 1C). The up-regulated DEGs under 10 ppt saline were overexpressed in the biological processes involved in cartilage development, oxygen transport, canonical Wnt signaling pathway, transport and microtubule-based process (Figure 1D). Under the salinity stress of 40 ppt, the DEGs, involved in the immune response processes, regulation of anion transmembrane transport, and stress response, were substantially inhibited (Figure 1E). The up-regulated DEGs were found to participate in extracellular matrix organization, skeletal system development, inorganic anion transport, cartilage development, and oxygen transport (Figure 1F).
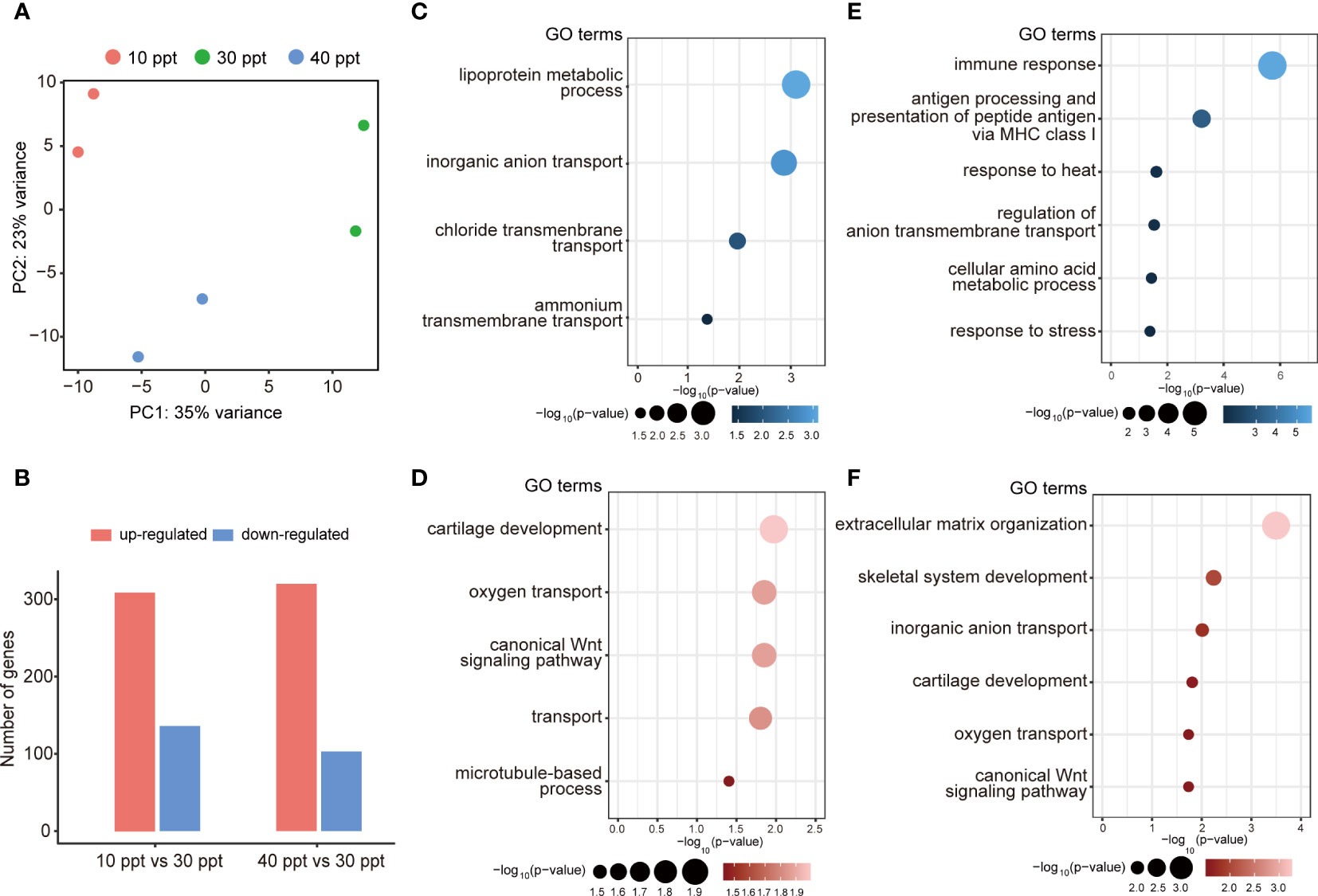
Figure 1 Summary of differentially expressed genes (DEGs) in the gills under different salinity stress. (A) Principal component analysis (PCA). (B) The number of up- and down-regulated DEGs for each comparison in gills. (C, D) Featured GO terms enriched for the down-regulated DEGs (C) and up-regulated DEGs (D) under 10 ppt condition compared to 30 ppt. (E, F) Featured GO terms enriched for the down-regulated DEGs (E) and up-regulated DEGs (F) under 40 ppt condition compared to 30 ppt.
Compared to the CK, the expression levels of 185 DEGs were enhanced in both 10 and 40 ppt salinity groups, which indicates that these genes participate in the responses to both hypo- and hyper-salinity stress (Figure 2A). Of these, the expression levels of 135 and 124 DEGs were specifically induced in 40 and 10 ppt treatment groups, respectively. The significant candidate genes in response to salinity stress in gills are listed in the heatmap, including slc9a3, loxa, mmp14b, col4a5, and so on (Figure 2B). To investigate the transcriptional dynamics across different salinities, we grouped the genes into six clusters according to the similarities in their expression profiles. Of these, three clusters (1, 3, and 6) were statistically significant (p-value = 8.55e-06, 4.12e-07, and 6.21e-14; Figures 2C–E). In cluster 1, gene expression was increased when the salinity varied from 10 to 30 ppt but reduced as the salinity increased from 30 to 40 ppt. GO enrichment analysis suggested that genes in cluster 1 were involved in lipid metabolism and transport, Golgi organization, and response to hypoxia (Figure 2C). In contrast, the expression levels of genes in cluster 3 were enhanced when the salinity altered from 30 ppt to either 10 or 40 ppt. These genes were significantly enriched in the immune response, cartilage development, extracellular matrix organization, skeletal system development, and actin cytoskeleton organization (Figure 2D). Genes in cluster 6 were substantially down-regulated following the salinity change from 10 to 30 ppt and retained a similar level between 30 and 40 ppt salinity. These genes participated in the mitogen-activated protein kinase (MAPK) cascade, response to stimulus, retrograde transport, endosome to Golgi, cellular iron ion homeostasis, and cytoskeleton organization (Figure 2E).
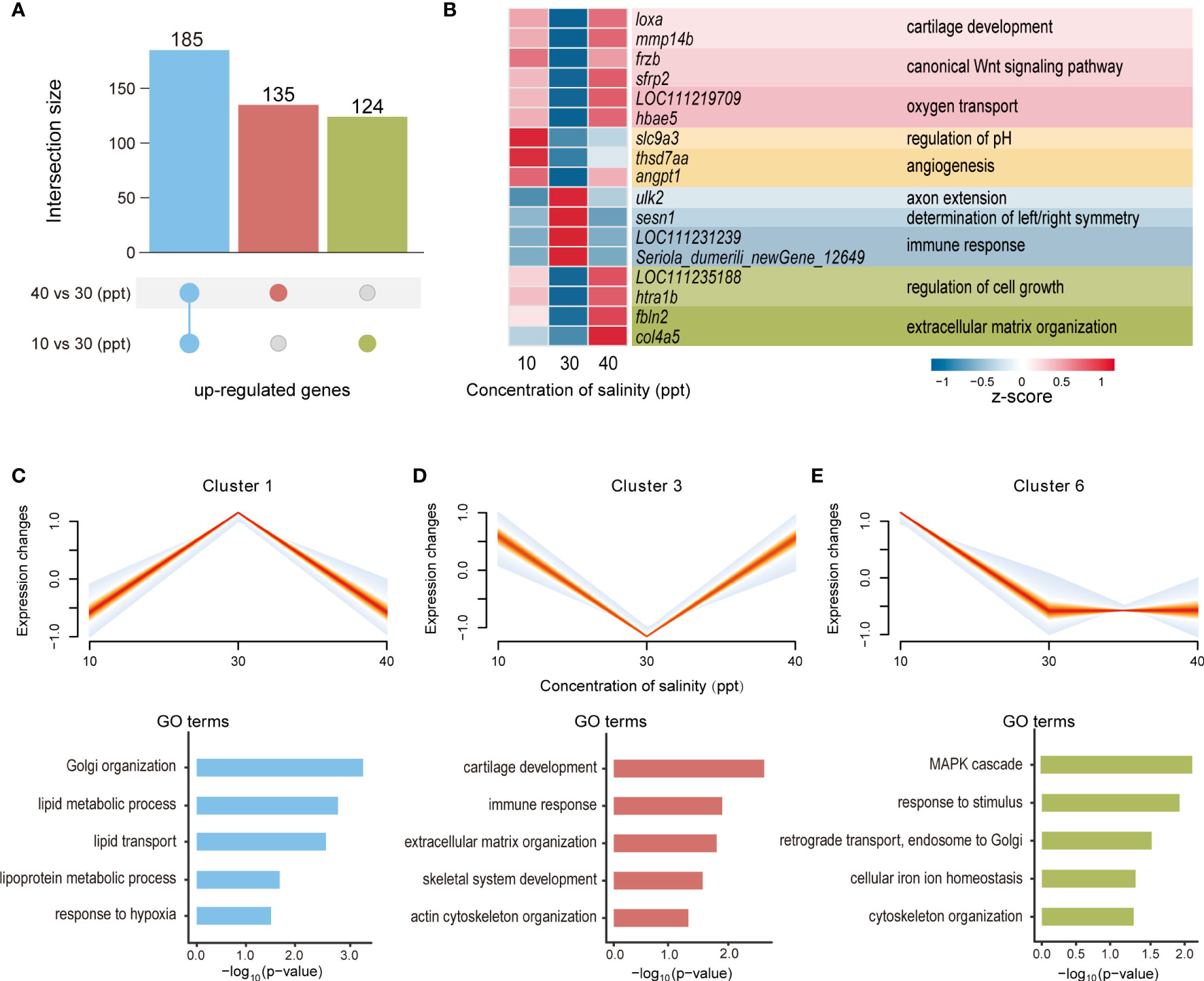
Figure 2 Transcriptome dynamics across different salinity in the gills. (A) Overlap of differentially expressed genes (DEGs) up-regulated under 10 ppt treatment and 40 ppt treatment. (B) Heatmap showing gene expression profiles of DEGs under three salinity concentrations (10, 30, and 40 ppt). (C–E) Significantly enriched clusters of genes with similar expression profiles across three salinity concentrations (10, 30, and 40 ppt). For each cluster, the expression trend of each gene was represented by one line, and the density of lines was highlighted by the intensity of colors (the denser, the more red). Featured GO terms enriched for the genes of each cluster are shown below.
We further compared the gene expression levels between 10 and 40 ppt salinity groups. A total of 118 DEGs were significantly overexpressed in the gills under 40 ppt treatment compared to that of the 10 ppt group (Supplementary Figure S4A). Functional enrichment analysis showed that the up-regulated DEGs were enriched in fatty acid metabolism, inorganic anion transport, and lipoprotein metabolic process (Supplementary Figure S4B). The down-regulated DEGs were associated with extracellular matri (ECM)-receptor interaction, chloride transmembrane transport, and nucleoside metabolic process (Supplementary Figure S4C).
3.3 Transcriptional dynamics induced by salinity changes in the kidneys of greater amberjack
In the kidneys of greater amberjack, a total of 600 (206 up- and 394 down-regulated) and 539 (312 up-regulated and 227 down-regulated) DEGs were detected under the salinity stress of 10 and 40 ppt, respectively (Figure 3A; Supplementary Figures S3C, D). The representative DEGs involved in salinity stress resistance in the kidneys are highlighted in the heatmap, such as hbae5, c5, c4b, itih2, and so on (Figure 3B). GO enrichment analysis identified 23 significantly enriched GO terms in 10 ppt saline, where the up-regulated DEGs were involved in oxygen transport and transmembrane transport, while the down-regulated DEGs were highly represented in immune response, negative regulation of endopeptidase activity and lipid transport (Figure 3C). Under the salinity stress of 40 ppt, the up-regulated genes were enriched in 16 GO terms, including those related to the regulation of cell growth, microtubule cytoskeleton organization, and ammonium transmembrane transport. In contrast, blood coagulation, lipid metabolic process, and cellular iron ion homeostasis were enriched for the down-regulated DEGs (Figure 3D).
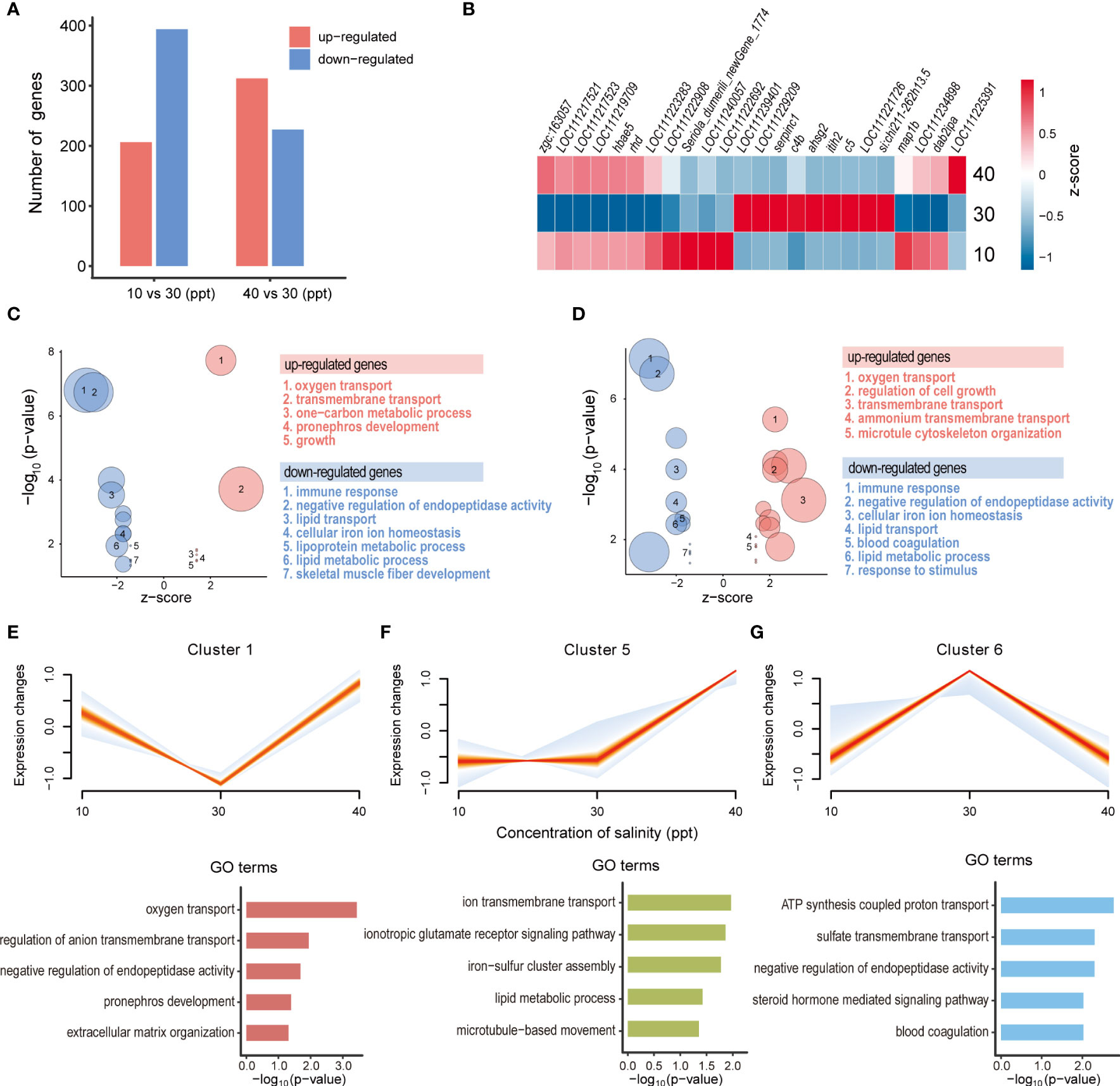
Figure 3 Transcriptomic alterations in the kidneys under different salinity stress. (A) The number of up- and down-regulated differentially expressed genes (DEGs) for each comparison in the kidneys. (B) Heatmap showing gene expression profiles of DEGs under three salinity concentrations (10, 30, and 40 ppt). (C, D) Featured GO terms enriched for the up- and down-regulated DEGs under 10 ppt (C) and 40 ppt conditions (D) compared to 30 ppt. Bubble plot showing the featured GO terms enriched for the DEGs induced by 10 (C) and 40 ppt (D) salinity treatments. (E-G) Significantly enriched clusters of genes with similar expression profiles across three salinity concentrations (10, 30, and 40 ppt). For each cluster, the expression trend of each gene was represented by one line, and the density of lines was highlighted by the intensity of colors (the denser, the more red). Featured GO terms enriched for the genes of each cluster are shown below.
In the kidneys, genes exhibited three major trends of expression profiles across different salinities (p-value < 0.05; Figures 3E-G). In cluster 1 (p-value = 0.016), gene expression was increased when switched from 30 ppt to 10 or 40 ppt salinity, and these genes participate in oxygen transport, pronephros development, and extracellular matrix organization (Figure 3E). For cluster 5 (p-value = 4.56e-07), the expression levels of genes showed no apparent difference between 10 and 30 ppt salinities, while they were induced by the increased salinity from 30 to 40 ppt. These genes were significantly enriched in ion transmembrane transport, ionotropic glutamate receptor signaling pathway, lipid metabolic process, and microtubule-based movement (Figure 3F). Genes in cluster 6 (p-value = 5.20e-09) exhibited the up-regulated gene expression levels from 10 to 30 ppt salinity, while these were down-regulated from 30 to 40 ppt. GO enrichment analysis displayed that these genes were involved in adenosine triphosphate (ATP) synthesis coupled proton transport, negative regulation of endopeptidase activity, steroid hormone-mediated signaling pathway, and blood coagulation (Figure 3G).
In the kidneys, 196 DEGs were detected under hyper-salinity condition (40 ppt) over hypo-salinity scenario (10 ppt) (Supplementary Figure S4D). Functional enrichment analysis revealed that the up-regulated DEGs were significantly enriched in the MAPK signaling pathway, Herpes simplex infection, cell growth regulation, and intracellular receptor signaling pathway (Supplementary Figure S4E). On the other hand, the down-regulated DEGs were enriched in the cysteine and methionine metabolism, PPAR (peroxisome proliferators-activated receptors) signaling pathway, glycine, serine and threonine metabolism, and lipoprotein metabolic process (Supplementary Figure S4F).
3.4 Comparative transcriptome analysis between gills and kidneys of greater amberjack under the hypo- and hyper-salinity
To explore the similarity and specificity in stress-response and resistance mechanisms between the different tissues, gills and kidneys, a comparative transcriptome analysis was carried out. When exposed to 10 ppt saline, 24 up-regulated DEGs were overexpressed in both gills and kidneys, which were significantly enriched in oxygen transport, blood vessel development and growth regulatory process (Supplementary Figure S5A). A total of 285 and 182 DEGs were up-regulated in gills or kidneys, respectively, mirroring the substantial difference between different tissues. The gill-specific DEGs were enriched in cartilage development, focal adhesion and canonical Wnt signaling pathway, while those specifically up-regulated in kidneys were over-represented in pronephros development, transmembrane transport and nervous system development (Supplementary Figure S5A). Similarly, 363 and 105 down-regulated DEGs were specific to gills or kidneys, and only 31 DEGs were overexpressed in both of tissues (Supplementary Figure S5B). Functional enrichment analyses illustrated that the DEGs specific to gills were associated with chloride transmembrane transport, ammonium transmembrane transport, and inorganic anion transport, whereas those in the kidneys were enriched in amino acid transmembrane transport, response to stress, and cellular iron ion homeostasis. Those co-expressed DEGs were involved in immune response, cell chemotaxis, and metal ion transport (Supplementary Figure S5B).
Under 40 ppt salinity stress, 80 up- and 14 down-regulated DEGs were overlapped in gills and kidneys, while 21 - 73% of the DEGs were identified in the two tissues (Supplementary Figure S5C, D). The commonly overexpressed DEGs were enriched in oxygen transport, ECM-receptor interaction, and transport (Supplementary Figure S5C), while those of down-regulation were over-represented in immune response, antigen processing, and presentation of peptide antigen via MHC class I and response to stimulus (Supplementary Figure S5D). The gill-specific up-regulated DEGs were involved in skeletal system development, inorganic anion transport, and hydrogen ion transmembrane transport, whereas those specific to the kidneys were highly represented in microtubule cytoskeleton organization, microtubule-based process, and AGE-RAGE (advanced glycation end product (AGE)-receptor for AGE (RAGE) signaling) signaling pathway in diabetic complications (Supplementary Figure S5C). The gill-specific down-regulated DEGs were enriched in blood coagulation, cellular iron ion homeostasis, and primary bile acid biosynthesis, while those DEGs in kidneys were involved in the regulation of anion transmembrane transport, response to stress and toll-like receptor (TLR) signaling pathway (Supplementary Figure S5D).
3.5 Validation by qRT-PCR
The expression levels of five randomly selected DEGs were validated in the gills (Figure 4A) and kidneys (Figure 4B), respectively, by qRT-PCR. In both tissues, the fold changes of the 10 DEGs under the stress of hypo- (10 ppt) and hyper-salinity (40 ppt) were of high consistency between RNA-seq and qRT-PCR (Figure 4), which suggests the reliability of our RNA-seq results.
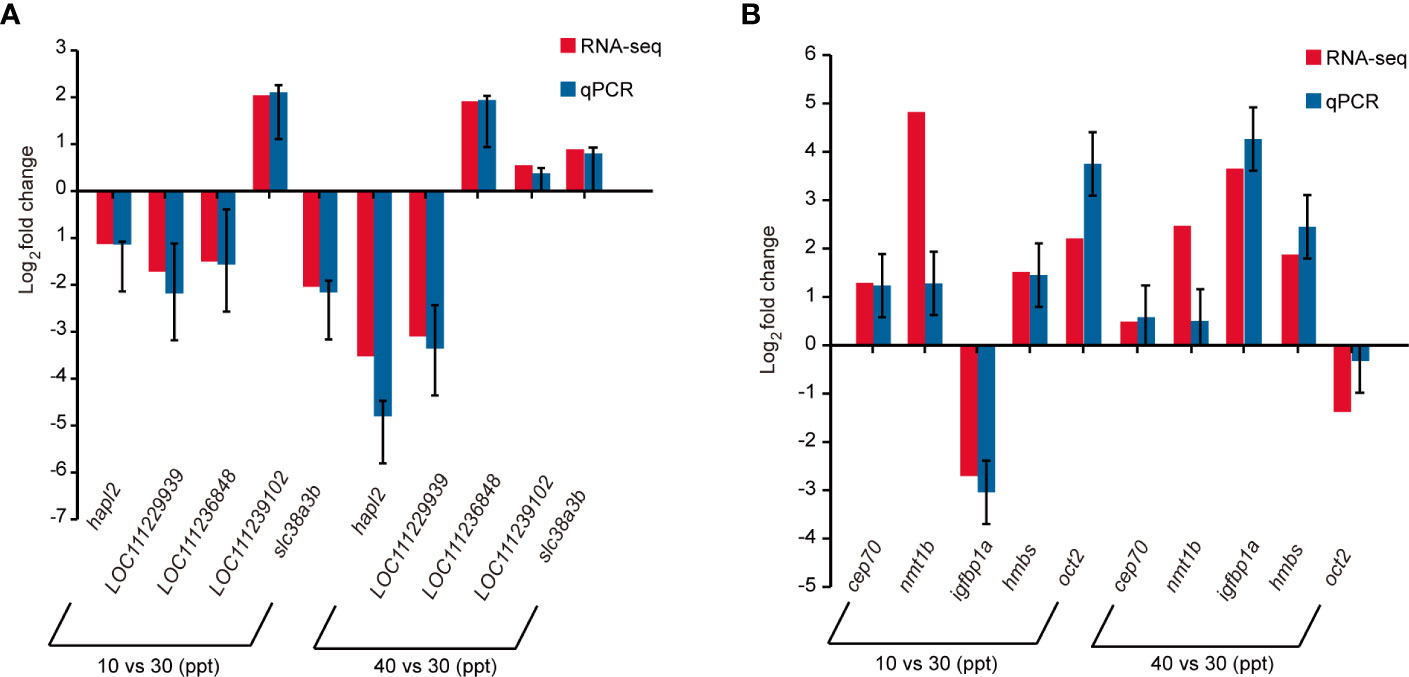
Figure 4 Comparison of gene expression alterations under salinity stress between RNA sequencing (RNA-seq) and quantitative polymerase chain reaction (qRT-PCR) in the gills (A) and kidneys (B), respectively. The relative expression level of messenger RNA (mRNA) transcripts was detected using qRT-PCR via the 2−ΔΔCt method. Data are expressed as means ± standard error (SE) (n = 3). β-actin was used as the reference gene.
4 Discussion
Salinity stress poses various consequences on fish, including growth, metabolism, and nutrition (Paital and Chainy, 2010; Kültz, 2015). In the current study, we described the global transcriptional responses in greater amberjack to the hypo-and hyper-salinity stress and provided new insights into the regulatory mechanisms of the gill and kidney tissues in cell growth and development, ion transport and osmoregulation, and immunity.
4.1 Cell growth and development
Insulin-like growth factors (IGFs) signal functions in regulating the cell cycle and cellular repair is essential for surviving salinity stress (Evans and Kültz, 2020). It also plays a key role in cortisol-mediated cell differentiation into mitochondria-rich cells (MRCs) (Gonzalez, 2012). In half-smooth tongue sole, gill igfbp5 (insulin-like growth factors binding protein 5) and igfbp2 were significantly up-regulated under the high and low salinity, responsible for the up-regulation of igf1 expression and regulation of cell growth and proliferation (Zhao et al., 2022). Meanwhile, MRCs were enriched in gills under the high salinity stress. The boosting MRCs would make an important contribution to the osmotic adjustment in teleost fishes upon the increased ambient salinity (Varsamos et al., 2005) by multiple ion channels anchored on the surface. In goby fish (Gillichthys mirabilis), gill igfbp1 mRNA expression is increased in response to the hyper-osmotically (Evans and Somero, 2008). In this study, when exposed to a higher salinity condition (40 ppt), the mRNA abundance of igfbp2 and ifgbp5 showed no change in gills, while ifgbp5 and igfbp6 expression levels were substantially up-regulated in the kidneys. The enhanced expression of Igfbps facilitates cell growth, proliferation, and migration, and increases cellular tolerance to rising ambient salinity (Tipsmark et al., 2007; Seale et al., 2020). In this study, the growth-related processes, cartilage development, skeletal system development and extracellular matrix organization in greater amberjack gills were activated in response to salinity alterations. The cartilage and skeletal development were induced in the gills of greater amberjack by hypo- or hyper-salinity stimuli. The enhanced development of the gill skeletal system may play a key role in maintaining gill structures and improving respiration and osmoregulation under salinity stress, similar to the associations between gill development and the tolerance capacity to hyper-salinity in Persian sturgeon (Acipenser persicus) (Shirangi et al., 2016).
In the kidneys, pronephros development was induced by lower salinity (10 ppt) compared to the optimum growth condition (30 ppt). Pronephros, which is also known as the primordial kidneys, mainly functions in ultrafiltration and urine production before being taken over by the definitive kidneys, and plays a crucial role in osmoregulation throughout the early development stages of fishes (Varsamos et al., 2005). In addition, the expression level of cep70 (centrosomal protein 70) mRNA was significantly increased in response to salinity reduction. Cep70 is involved in cilia assembly and essential for kidney development and homeostasis (Shi et al., 2011; Marra et al., 2016). The expression of ccn2 (cellular communication network factor 2) mRNA was significantly up-regulated in response to the increased salinity. It also known as the connective tissue growth factor that mediates the proliferation and adhesion of vascular endothelial cells. Thus, its enhancement can encourage the growth of blood vessels and reduce tissue damage caused by salinity (Pan et al., 2019). The expression level of mfap2 (microfibril-associated protein 2), a gene associated with blood vessel development, was overexpressed in both gills and kidneys under hypo-salinity stress, indicative of a similar regulatory route for the growth of blood vessels in both tissues. These results indicate that the undesired salinity impacts on the normal kidneys development for adaption to the salinity stress.
4.2 Ion transport and osmoregulation
Ion transport is important for osmoregulation, which plays a critical role in the adaptive response to changes in external salinity (Deane and Woo, 2006; Tomy et al., 2009; Knowles et al., 2014; Vij et al., 2020). Gills are the main osmotic organs, which face directly to the external aqueous environment, responsible for maintaining the ion-osmotic balance of fish (Alkan and Oğuz, 2021; Zhang et al., 2021). In teleosts, Nkcc1 (Na-K-Cl cotransporter-1, encoded by the gene slc12a2), Nka (Na+/K+-ATPase), and Cftr (cystic fibrosis transmembrane conductance regulator) are the major ion transport proteins participated in ion secretion and osmoregulation in gills. Nkcc1 is crucial for inward movements of Na+, K+, and transcellular secretion of Cl− in teleosts (Takvam et al., 2021). Nka is used for the active uptake of K+ and extrusion of Na+ in cells through protein conformational changes fueled by ATP (Mobasheri et al., 2000; Pierre and Blanco, 2021). Cftr contributes to the across-membrane conduction of Cl- efflux from the epithelial cell (McCormick et al., 2013). In our study, gill slc12a2/nkcc1 and cftr mRNA expression levels were significantly down-regulated under the hypo-salinity stress at 10 ppt, while these levels were substantially activated in response to the elevated salinity from 30 to 40 ppt at 72 hours in great amberjack, except for the nka gene. The above results suggested that the lower gill permeability to reduce the Na+, K+ movement and Cl- ion secretion was induced by hypo-osmotic exposure, while ion excretion upon hyper-salinity stress was boosted and a comparatively steady blood osmolality was maintained (Cao et al., 2022), through Nkcc1 and Cftr ion transport proteins in great amberjack, coincided with Mozambique tilapia gill ion regulation mechanism (Hiroi et al., 2008; Hiroi and McCormick, 2012). In half-smooth tongue sole, gill cftr expression was significantly down-regulated under low salinity but up-regulated under high salinity for 10 days. Gill nkcc and nkain (Na+/K+ transporting ATPase interacting) mRNA expression showed no significant changes. The results indicate that Cftr is the major ion transporter protein for regulating Cl−, Na+, and K+ ions transport and osmolality balance under the hypo- and hyper-salinity stress (Zhao et al., 2022). Similarly, in freshwater climbing perch (Anabas testudineus), the expression of gill cftr mRNA was up-regulated after exposure to seawater for 6 days (Chen et al., 2018). In European seabass (Dicentrarchus labrax), gill cftr and nkcc1 mRNA expression levels were decreased, while nka mRNA expression level was increased within 4 days post-acute transfer from seawater to freshwater (Maugars et al., 2018), suggesting that Cftr and Nkcc1 were mainly responsible for preventing the ions loses and osmolality out-of-balance under the low-salinity condition. Taken together, gills play a dominant role in osmotic and ionic regulation, but the molecular mechanism was divergent to some degree in different fish species.
Fish kidneys function as the main osmoregulatory organ in integrating ion and water transport to maintain internal homeostasis in osmotic environments. The renal transporters include Nka enzymes, monovalent ion transporters, water transport proteins, and divalent ion transporters. Na+/Cl– co-transporter (Ncc, encoded by the gene slc12a3), is a monovalent ion-absorptive co-transporter, expressed in the kidneys, especially in the mammalian distal convoluted tubule and teleost fish distal nephrons (Gamba, 2005; Hiroi et al., 2008; Hsu et al., 2014). Ncc is essential in producing hypotonic urine in teleost fishes when grown in a hypoosmotic external environment (Takvam et al., 2021). In bull shark (Carcharhinus leucas), kidney ncc mRNA expression in freshwater sharks is higher than in seawater sharks, contributing to NaCl reabsorption from the urine and tolerance to the freshwater environment (Imaseki et al., 2019). In pufferfish (Tetradon nigroviridis) and European eel (Anguilla anguilla), ncc mRNA expression levels and apical location of the Ncc protein in the kidneys were significantly decreased following acclimation to seawater, which demonstrated the essential roles of Ncc in production of hypotonic urine and reabsorbed salts, responded to the freshwater environment (Cutler and Cramb, 2008; Kato et al., 2011). However, in this study, the gene expression level of slc12a3/ncc was significantly down-regulated in the kidneys under the hypo-salinity condition at 10 ppt in great amberjack. We speculated whether some other factors in kidneys were compensable for regulating the water and salt balance under the decreased salinity. Oct2 (organic cation transporter 2, encoded by the gene slc22a2) mediates in the initiation of the renal secretion of organic cations. In great amberjack, slc22a2 was overexpressed at 40 ppt, suggesting an induced secretion of organic cations might be response to the hyper-osmoregulation, to combat the osmotic influx of salts following the drink of seawater upon high salinity stress. When grown in 40 ppt saline, the increased external salinity largely induced the ammonium transmembrane transport gene rhd (Rh blood group, D antigen) and rhag (Rhesus-associated glycoprotein) expression in the kidneys. The up-regulated ammonium transport may mirror an elevated amount of produced ammonia (Wood and Nawata, 2011; Bucking, 2017). It is further supported by the overexpression of the genes encoding solute transport proteins (slc4a3, slc4a5, slc24a2, and slc22a2) under the treatment of 40 ppt saline, which may promote the transmembrane transport of amino acids and other substances. The metabolism of amino acids is specifically enhanced in the kidneys to provide more energy for coping with hyper-salinity stress and function as osmolytes for cell volume regulation (Aragão et al., 2010). Previous studies have demonstrated that amino acid levels are positively correlated with salinity (Aragão et al., 2010; Cheng et al., 2022). Together, our findings offer a molecular insight into the osmoregulatory mechanisms of greater amberjack kidneys against the adverse impacts of ambient salinity alterations.
Major osmotic organs, including gills and kidneys, play critical roles in regulating ion and water transport to maintain internal homeostasis and adapt to the external environment salinity changes. The molecular mechanisms in the gills and kidneys were interconnecting and distinguishing from each other, demonstrated by the expression pattern of the same transporters in different tissues or the different functional transporters, response to the hyper- and hypo-salinity stress.
4.3 Immune response
In the current study, we also found that changes in external salinity would down-regulated the immune function in both the gills and kidneys of greater amberjack juveniles, which is consistent with the previous observations in other marine fishes, spotted scat (Scatophagus argus) (Zhong et al., 2021) and striped eel catfish (Plotosus lineatus) (Hieu et al., 2021). In particular, in the kidneys, the genes involved in immunological responses and blood coagulation were substantially suppressed by hypo- and hyper-salinity stress. In particular, the expression of mhc1 (the major histocompatibility complex (MHC) class I) mRNA, which mainly mediates antigen presentation, was suppressed in the kidneys under 10 or 40 ppt condition at 72 h, suggesting the down-regulated recognition of intracellular pathogen infection and activation of the downstream immune cascade under hypo- and hyper-salinity stress (Jiang et al., 2015). It was different from the observation of Nile tilapia (El-Leithy et al., 2019). High immune-related genes expression levels have indicated the fish is vulnerable to infect agents owing to the immune suppression under hyper-salinity stress (Bowden, 2008). Moreover, the expression levels of genes within or around the complement system, such as those encoding complement component 3 (c3) and vitronectin b (vtnb), were also significantly inhibited in greater amberjack by either decreased or increased ambient salinity at 72 h, indicating the repression of the inflammatory response, impede pathogen clearance, and disrupt cellular homeostasis (Liu et al., 2019; Wang et al., 2019). In puffer (Takifugu fasciatus), the expression of c3 in the gills and kidneys is negatively correlated with salinity (Wang et al., 2019). In this study, the mRNA expression of several genes participating in blood coagulation, such as coagulation factor II (f2) and hemopexin (hpx), were depressed in the greater amberjack when exposed to hyper-salinity stress, which is consistent with the observation in Nile tilapia (Xu et al., 2018). Because of the direct crosstalk between the coagulation and the immune system, the activation of blood coagulation positively contributes to pathogen clearance during infections (Antoniak, 2018). The gene expression of zap70, encoding an enzyme belonging to the protein tyrosine kinase family, was down-regulated in the kidneys of greater amberjack under the hypo-salinity situation. Similarly, in marbled eel (Anguilla marmorata), gill zap70 expression was decreased following the salinity (Chan et al., 1992; Cao et al., 2021). The repression of Zap60 perhaps leads to a disruption in T cell development and lymphocyte activation (Chan et al., 1992; Cao et al., 2021). It is worth noting that the gene fabp4 expression (fatty acid-binding protein-4) was induced in the gills of greater amberjack when exposed to 10 or 40 ppt saline. In fish, Fabp4 can bind to the marine environmental toxins perfluoroalkyl acids (PFAAs), trigger the undesirable uptake and accumulation (Lei et al., 2022), which has been demonstrated to suppress lysozyme activity in marine organisms, then interrupt their resistance to bacterial infection (Betts, 2007). Furthermore, PFAA can easily enter human bodies via the food chain and disturb human hormonal signaling, which poses potential risks to food safety and public health (Betts, 2007; McComb et al., 2020), which highlights the importance of a well-controlled rearing condition during the aquaculture to ensure the food safety of greater amberjack. The immune related genes in teleost gills or/and kidneys were influenced by the environmental salinity, which may impact the immune function, increase the risk of pathogen infection, and reduce the fresh security and yield of greater amberjack. These results help understand the mechanisms of the adaptive immunology response to the environmental salinity stress.
5 Conclusion
In this study, we performed comparative transcriptome analysis for gills and kidneys in greater amberjack between the optimum salinity (30 ppt) and undesired regimes (10 and 40 ppt). In gills, The identified DEGs were involved in cartilage and skeletal development, ions transport, and immune response. The down-regulation of slc12a2/nkcc1 and cftr expression levels in response the reduced Na+, K+ movement, and Cl- ion secretion under the hypo-osmotic exposure and ion excretion boost upon hyper-salinity stress. Meanwhile, the cartilage and skeletal development were enhanced in the gills by hypo- or hyper-salinity stimuli, which is critical for maintaining gill structures and improving respiration and osmoregulation under salinity stress. In kidneys, the DEGs were enriched in oxygen transport, pronephros development, regulation of growth, blood coagulation, ion transmembrane transport, and immune response. The expression levels of slc22a2, rhd and rhag mRNA was overexpressed under the hyper-salinity condition at 40 ppt, mainly contributing to the salt secretion and ammonium transport regulation, to combat the osmotic influx of salts following the drink of seawater and elevated ammonia production upon high salinity stress. These novel findings advance our knowledge of adaptative mechanisms to salinity stress and provide theoretical guidance for the optimal breeding mode for the aquaculture of greater amberjack.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/search/all/?term=GSE220485.
Ethics statement
The animal study was reviewed and approved by the Animal Research and Ethics Committee of Guangdong Ocean University (NIH Pub. No. 85-23, revised 1996).
Author contributions
Conceptualization: YL, HS and GL. Methodology: YP, RQ and YL. Data processing and figure preparation: YL, YP, HS and YY. Funding acquisition: GL. Experimental fish culture: YH and CZ. Writing-original draft: YL, HS, DJ and YY. Writing-review and editing: YL, HS, YY and DJ. All authors contributed to the article and approved the submitted version.
Funding
This research was financed by grants from the National Natural Science Foundation of China (32002367 and 32201420), the Natural Science Foundation of Guangdong Province (2023A1515010890), the Fund of Southern Marine Science and Engineering Guangdong Laboratory (Zhanjiang) (ZJW-2019-06), Key Project of ‘Blue Granary Science and Technology Innovation” of the Ministry of Science and Technology (2018YFD0901203). The Provincial College Students Innovation and Entrepreneurship Training Program (S202110566008).
Acknowledgments
We thank the Guangdong Research Center on Reproductive Control and Breeding Technology of Indigenous Valuable Fish Species, Fisheries College, Guangdong Ocean University, State Key Laboratory of Biocontrol, School of Ecology, Sun Yat-sen University for the technical support. We also thank Southern Marine Science and Engineering Guangdong Laboratory-Zhanjiang, for the experimental animals breeding.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1185015/full#supplementary-material
References
Alkan Z., Oğuz A. R. (2021). Investigation of gill mucus cells of lake van fish (Alburnus tarichi) during reproductive migration. Fish Physiol. Biochem. 47 (2), 409–419. doi: 10.1007/s10695-020-00921-6
Antoniak S. (2018). The coagulation system in host defense. Res. Pract. Thromb. Haemostasis 2 (3), 549–557. doi: 10.1002/rth2.12109
Aragão C., Costas B., Vargas-Chacoff L., Ruiz-Jarabo I., Dinis M. T., Mancera J. M., et al. (2010). Changes in plasma amino acid levels in a euryhaline fish exposed to different environmental salinities. Amino Acids 38, 311–317. doi: 10.1007/s00726-009-0252-9
Araki K., Aokic J.-y., Kawase J., Hamada K., Ozaki A., Fujimoto H., et al. (2018). Whole genome sequencing of greater amberjack (Seriola dumerili) for SNP identification on aligned scaffolds and genome structural variation analysis using parallel resequencing. Int. J. Genomics 2018, 7984292. doi: 10.1155/2018/7984292
Azodi M., Bahabadi M. N., Ghasemi A., Morshedi V., Mozanzadeh M. T., Shahraki R., et al. (2021). Effects of salinity on gills’ chloride cells, stress indices, and gene expression of Asian seabass (Lates calcarifer, Bloch 1790). Fish Physiol. Biochem. 47 (6), 2027–2039. doi: 10.21203/rs.3.rs-534301/v1
Barany A., Gilannejad N., Alameda-López M., Rodríguez-Velásquez L., Astola A., Martínez-Rodríguez G., et al. (2021). Osmoregulatory plasticity of juvenile greater amberjack (Seriola dumerili) to environmental salinity. Animals 11 (9), 2607. doi: 10.3390/ani11092607
Betts K. S. (2007). Perfluoroalkyl acids: what is the evidence telling us? Natl. Instit. Environ. Health Sci. 115 (7), A344. doi: 10.1289/ehp.115-a250
Birrer S. C., Reusch T. B., Roth O. (2012). Salinity change impairs pipefish immune defence. Fish Shell. Immunol. 33 (6), 1238–1248. doi: 10.1016/j.fsi.2012.08.028
Bowden T. J. (2008). Modulation of the immune system of fish by their environment. Fish Shell. Immunol. 25 (4), 373–383. doi: 10.1016/j.fsi.2008.03.017
Bucking C. (2017). A broader look at ammonia production, excretion, and transport in fish: a review of impacts of feeding and the environment. J. Comp. Physiol. B 187 (1), 1–18. doi: 10.1007/s00360-016-1026-9
Cao W., Bi S., Chi C., Dong Y., Xia S., Liu Z., et al. (2022). Effects of high salinity stress on the survival, gill tissue, enzyme activity and free amino acid content in razor clam Sinonovacula constricta. Front. Mar. Sci. 131. doi: 10.3389/fmars.2022.839614
Cao Q., Wang H., Fan C., Sun Y., Li J., Cheng J., et al. (2021). Environmental salinity influences the branchial expression of TCR pathway related genes based on transcriptome of a catadromous fish. Comp. Biochem. Physiol. D-Genomics Proteomics. 38, 100815. doi: 10.1016/j.cbd.2021.100815
Chan A. C., Iwashima M., Turck C. W., Weiss A. (1992). ZAP-70: a 70 kd protein-tyrosine kinase that associates with the TCR ζ chain. Cell 71 (4), 649–662. doi: 10.1016/0092-8674(92)90598-7
Chen X. L., Lui E. Y., Ip Y. K., Lam S. H. (2018). RNA Sequencing, de novo assembly and differential analysis of the gill transcriptome of freshwater climbing perch Anabas testudineus after 6 days of seawater exposure. J. Fish Biol. 93 (2), 215–228. doi: 10.1111/jfb.13653
Chen J., Wu H. Q., Shi Y. H., Li C. H., Li M. Y. (2009). The effects of environmental salinity on trunk kidney proteome of juvenile ayu (Plecoglossus altivelis). Comp. Biochem. Physiol. Part D: Genomics Proteomics 4 (4), 263–267. doi: 10.1016/j.cbd.2009.06.003
Cheng Y., Zhao J., Ayisi C. L., Cao X. (2022). Effects of salinity and alkalinity on fatty acids, free amino acids and related substance anabolic metabolism of Nile tilapia. Aquacult. Fish. 7 (4), 389–395. doi: 10.1016/j.aaf.2020.06.005
Cutler C. P., Cramb G. (2008). Differential expression of absorptive cation-chloride-cotransporters in the intestinal and renal tissues of the European eel (Anguilla anguilla). Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 149 (1), 63–73. doi: 10.1016/j.cbpb.2007.08.007
Deane E. E., Woo N. (2006). Tissue distribution, effects of salinity acclimation, and ontogeny of aquaporin 3 in the marine teleost, silver sea bream (Sparus sarba). Mar. Biotechnol. 8 (6), 663–671. doi: 10.1007/s10126-006-6001-0
Djellata A., Sarih S., Hernández-Cruz C. M., Martínez-Rodríguez G., Gilannejad N., Roo J. (2021). The effect of different co-feeding protocols on greater amberjack (Seriola dumerili, risso 1810) larvae. Aquacult. Nutr. 27 (5), 1761–1776. doi: 10.1111/anu.13313
El-Leithy A. A. A., Hemeda S. A., El Naby W., El Nahas A. F., Hassan S. A. H., Awad S. T., et al. (2019). Optimum salinity for Nile tilapia (Oreochromis niloticus) growth and mRNA transcripts of ion-regulation, inflammatory, stress- and immune-related genes. Fish Physiol. Biochem. 45 (4), 1217–1232. doi: 10.1007/s10695-019-00640-7
Evans D. H. (2008). Teleost fish osmoregulation: what have we learned since august krogh, homer smith, and ancel keys. Am. J. Physiology-Regulatory Integr. Comp. Physiol. 295 (2), R704–R713. doi: 10.1152/ajpregu.90337.2008
Evans T. G., Kültz D. (2020). The cellular stress response in fish exposed to salinity fluctuations. J. Exp. Zool. A. Ecol. Integr. Physiol. 333 (6), 421–435. doi: 10.1002/jez.2350
Evans T. G., Somero G. N. (2008). A microarray-based transcriptomic time-course of hyper- and hypo-osmotic stress signaling events in the euryhaline fish Gillichthys mirabilis: osmosensors to effectors. J. Exp. Biol. 211 (Pt 22), 3636–3649. doi: 10.1242/jeb.022160
Fang H., Yang Y. Y., Wu X. M., Zheng S. Y., Song Y. J., Zhang J., et al. (2022). Effects and molecular regulation mechanisms of salinity stress on the health and disease resistance of grass carp. Front. Immunol. 13. doi: 10.3389/fimmu.2022.917497
Fernández-Montero A., Caballero M. J., Torrecillas S., Tuset V. M., Lombarte A., Ginés R. R., et al. (2018). Effect of temperature on growth performance of greater amberjack (Seriola dumerili risso 1810) juveniles. Aquacult. Res. 49 (2), 908–918. doi: 10.1111/are.13537
Futschik M. E., Carlisle B. (2005). Noise-robust soft clustering of gene expression time-course data. J. Bioinf. Comput. Biol. 3 (04), 965–988. doi: 10.1142/s0219720005001375
Galkanda-Arachchige H. S. C., Davis R. P., Nazeer S., Ibarra-Castro L., Davis D. A. (2021). Effect of salinity on growth, survival, and serum osmolality of red snapper, Lutjanus campechanus. Fish Physiol. Biochem. 47 (5), 1687–1696. doi: 10.1007/s10695-021-01009-5
Gamba G. (2005). Molecular physiology and pathophysiology of electroneutral cation-chloride cotransporters. Physiol. Rev. 85 (2), 423–493. doi: 10.1152/physrev.00011.2004
Gjessing M. C., Krasnov A., Timmerhaus G., Brun S., Afanasyev S., Dale O. B., et al. (2020). The Atlantic salmon gill transcriptome response in a natural outbreak of salmon gill pox virus infection reveals new biomarkers of gill pathology and suppression of mucosal defense. Front. Immunol. 11. doi: 10.3389/fimmu.2020.02154
Gonzalez R. (2012). The physiology of hyper-salinity tolerance in teleost fish: a review. J. Comp. Physiol. B 182 (3), 321–329. doi: 10.1007/s00360-011-0624-9
Gu J., Dai S., Liu H., Cao Q., Yin S., Lai K. P., et al. (2018). Identification of immune-related genes in gill cells of Japanese eels (Anguilla japonica) in adaptation to water salinity changes. Fish Shell. Immunol. 73, 288–296. doi: 10.1016/j.fsi.2017.12.026
Gui L., Zhang P., Liang X., Su M., Wu D., Zhang J. (2016). Adaptive responses to osmotic stress in kidney-derived cell lines from Scatophagus argus, a euryhaline fish. Gene 583 (2), 134–140. doi: 10.1016/j.gene.2016.02.026
Guo T., Yang Y., Meng F., Wang S., Xia S., Qian Y., et al. (2020). Effects of low salinity on gill and liver glycogen metabolism of great blue-spotted mudskippers (Boleophthalmus pectinirostris). Comp. Biochem. Physiol. Part C: Toxicol. Pharmacol. 230, 108709. doi: 10.1016/j.cbpc.2020.108709
Hieu D. Q., Hang B. T. B., Huong D. T. T., Kertaoui N. E., Farnir F., Phuong N. T., et al. (2021). Salinity affects growth performance, physiology, immune responses and temperature resistance in striped catfish (Pangasianodon hypophthalmus) during its early life stages. Fish Physiol. Biochem. 47 (6), 1995–2013. doi: 10.1007/s10695-021-01021-9
Hiroi J., McCormick S. D. (2012). New insights into gill ionocyte and ion transporter function in euryhaline and diadromous fish. Respir. Physiol. Neurobiol. 184 (3), 257–268. doi: 10.1016/j.resp.2012.07.019
Hiroi J., Yasumasu S., McCormick S. D., Hwang P. P., Kaneko T. (2008). Evidence for an apical Na-cl cotransporter involved in ion uptake in a teleost fish. J. Exp. Biol. 211 (Pt 16), 2584–2599. doi: 10.1242/jeb.018663
Hsu H. H., Lin L. Y., Tseng Y. C., Horng J. L., Hwang P. P. (2014). A new model for fish ion regulation: identification of ionocytes in freshwater- and seawater-acclimated medaka (Oryzias latipes). Cell Tissue Res. 357 (1), 225–243. doi: 10.1007/s00441-014-1883-z
Imaseki I., Wakabayashi M., Hara Y., Watanabe T., Takabe S., Kakumura K., et al. (2019). Comprehensive analysis of genes contributing to euryhalinity in the bull shark, Carcharhinus leucas; na(+)-cl(-) co-transporter is one of the key renal factors upregulated in acclimation to low-salinity environment. J. Exp. Biol. 222 (Pt 12), jeb201780. doi: 10.1242/jeb.201780
Jiang F., Kumar V. B., Lee D.-N., Weng C.-F. (2008). Acute osmotic stress affects tilapia (Oreochromis mossambicus) innate immune responses. Fish Shell. Immunol. 25 (6), 841–846. doi: 10.1016/j.fsi.2008.09.006
Jiang C., Zhang J., Yao J., Liu S., Li Y., Song L., et al. (2015). Complement regulatory protein genes in channel catfish and their involvement in disease defense response. Dev. Comp. Immunol. 53 (1), 33–41. doi: 10.1016/j.dci.2015.06.002
Kammerer B. D., Sardella B. A., Kültz D. (2009). Salinity stress results in rapid cell cycle changes of tilapia (Oreochromis mossambicus) gill epithelial cells. J. Exp. Zool. Part A: Ecol. Genet. Physiol. 311 (2), 80–90. doi: 10.1002/jez.498
Kato A., Muro T., Kimura Y., Li S., Islam Z., Ogoshi M., et al. (2011). Differential expression of na+-cl- cotransporter and na+-K+-Cl- cotransporter 2 in the distal nephrons of euryhaline and seawater pufferfishes. Am. J. Physiol. Regul. Integr. Comp. Physiol. 300 (2), R284–R297. doi: 10.1152/ajpregu.00725.2009
Kim D., Paggi J. M., Park C., Bennett C., Salzberg S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37 (8), 907–915. doi: 10.1038/s41587-019-0201-4
Knowles G., Handlinger J., Jones B., Moltschaniwskyj N. (2014). Hemolymph chemistry and histopathological changes in pacific oysters (Crassostrea gigas) in response to low salinity stress. J. Invertebrate Pathol. 121, 78–84. doi: 10.1016/j.jip.2014.06.013
Kültz D. (2012). The combinatorial nature of osmosensing in fishes. Physiology 27 (4), 259–275. doi: 10.1152/physiol.00014.2012
Kültz D. (2015). Physiological mechanisms used by fish to cope with salinity stress. J. Exp. Biol. 218 (12), 1907–1914. doi: 10.1242/jeb.118695
Kumar L., Futschik M. E. (2007). Mfuzz: a software package for soft clustering of microarray data. Bioinformation 2(1), 5–7. doi: 10.6026/97320630002005
Laverty G., Skadhauge E. (2012). Adaptation of teleosts to very high salinity. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 163 (1), 1–6. doi: 10.1016/j.cbpa.2012.05.203
Lehtonen T. K., Wong B. B. M., Kvarnemo C. (2016). Effects of salinity on nest-building behaviour in a marine fish. BMC Ecol. 16, 7. doi: 10.1186/s12898-016-0067-y
Lei C.-x., Xie Y.-j., Li S.-j., Jiang P., Du J.-x., Tian J.-j. (2022). Fabp4 contributes toward regulating inflammatory gene expression and oxidative stress in Ctenopharyngodon idella. Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 259, 110715. doi: 10.1016/j.cbpb.2022.110715
Li X., Shen Y., Bao Y., Wu Z., Yang B., Jiao L., et al. (2022). Physiological responses and adaptive strategies to acute low-salinity environmental stress of the euryhaline marine fish black seabream (Acanthopagrus schlegelii). Aquaculture 554, 738117. doi: 10.1016/j.aquaculture.2022.738117
Li J., Xue L., Cao M., Zhang Y., Wang Y., Xu S., et al. (2020). Gill transcriptomes reveal expression changes of genes related with immune and ion transport under salinity stress in silvery pomfret (Pampus argenteus). Fish Physiol. Biochem. 46 (4), 1255–1277. doi: 10.1007/s10695-020-00786-9
Liao Y., Smyth G. K., Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30 (7), 923–930s. doi: 10.1093/bioinformatics/btt656
Liu Y., Song Q., Li D., Zou R., Zhang Y., Hao S., et al. (2019). A novel complement C3 like gene (Lv-C3L) from Litopenaeus vannamei with bacteriolytic and hemolytic activities and its role in antiviral immune response. Fish Shell. Immunol. 91, 376–387. doi: 10.1016/j.fsi.2019.05.045
Liu D., Zhang Z., Song Y., Yang J., Lu Y., Lai W., et al. (2023). Effects of salinity on growth, physiology, biochemistry and gut microbiota of juvenile grass carp (Ctenopharyngodon idella). Aquat. Toxicol. 258, 106482. doi: 10.1016/j.aquatox.2023.106482
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 1–21. doi: 10.1186/s13059-014-0550-8
Lu M., Su M., Liu N., Zhang J. (2022). Effects of environmental salinity on the immune response of the coastal fish Scatophagus argus during bacterial infection. Fish Shell. Immunol. 124, 401–410. doi: 10.1016/j.fsi.2022.04.029
Marra A. N., Li Y., Wingert R. A. (2016). Antennas of organ morphogenesis: the roles of cilia in vertebrate kidney development. Genesis 54 (9), 457–469. doi: 10.1002/dvg.22957
Maugars G., Manirafasha M. C., Grousset E., Boulo V., Lignot J. H. (2018). The effects of acute transfer to freshwater on ion transporters of the pharyngeal cavity in European seabass (Dicentrarchus labrax). Fish Physiol. Biochem. 44 (5), 1393–1408. doi: 10.1007/s10695-018-0529-6
Mazzola A., Favaloro E., Sarà G. (2000). Cultivation of the Mediterranean amberjack, Seriola dumerili (Risso 1810), in submerged cages in the Western Mediterranean Sea. Aquaculture 181 (3-4), 257–268. doi: 10.1016/s0044-8486(99)00243-4
McComb J., Mills I., Berntsen H. F., Ropstad E., Verhaegen S., Connolly L. (2020). Human-based exposure levels of perfluoroalkyl acids may induce harmful effects to health by disrupting major components of androgen receptor signalling in vitro. Exposure Health 12 (3), 527–538. doi: 10.1007/s12403-019-00318-8
McCormick S. D., Regish A. M., Christensen A. K., Björnsson B. T. (2013). Differential regulation of sodium–potassium pump isoforms during smolt development and seawater exposure of Atlantic salmon. J. Exp. Biol. 216 (7), 1142–1151. doi: 10.1242/jeb.080440
Mobasheri A., Avila J., Cózar-Castellano I., Brownleader M. D., Trevan M., Francis M. J., et al. (2000). Na+, k+-ATPase isozyme diversity; comparative biochemistry and physiological implications of novel functional interactions. Biosci Rep. 20 (2), 51–91. doi: 10.1023/a:1005580332144
Morgenroth D., McArley T., Danielo Q., Harford A., Hickey A. J., Khan J., et al. (2022). Kingfish (Seriola lalandi) adjust to low salinity with only subtle effects to cardiorespiratory and growth performance. Aquaculture 556, 738268. doi: 10.1016/j.aquaculture.2022.738268
Ouyang H., Deng N., Xu J., Huang J., Han C., Liu D., et al. (2023). Effects of hyperosmotic stress on the intestinal microbiota, transcriptome, and immune function of mandarin fish (Siniperca chuatsi). Aquaculture 563, 738901. doi: 10.1016/j.aquaculture.2022.738901
Paital B., Chainy G. (2010). Antioxidant defenses and oxidative stress parameters in tissues of mud crab (Scylla serrata) with reference to changing salinity. Comp. Biochem. Physiol. Part C: Toxicol. Pharmacol. 151 (1), 142–151. doi: 10.1016/j.cbpc.2009.09.007
Pan W.-Q., Wang J.-P., Tu Z.-H., Gan T., Hu J., Wei J., et al. (2019). Cloning, molecular characterization, and tissue differential expression of connective tissue growth factor (ctgf) of grass carp. Fish Physiol. Biochem. 45 (4), 1431–1443. doi: 10.1007/s10695-019-00653-2
Peng Y., Shi H., Liu Y., Huang Y., Zheng R., Jiang D., et al. (2022). RNA Sequencing analysis reveals divergent adaptive response to hypo-and hyper-salinity in greater amberjack (Seriola dumerili) juveniles. Animals 12 (3), 327. doi: 10.3390/ani12030327
Pepe N., Canli E. G., Canli M. (2022). Salinity and/or nanoparticles (Al2O3, TiO2) affect metal accumulation and ATPase activity in freshwater fish (Oreochromis niloticus). Environ. Toxicol. Pharmacol. 94, 103931. doi: 10.1016/j.etap.2022.103931
Pierre S. V., Blanco G. (2021). Na/K-ATPase ion transport and receptor-mediated signaling pathways. J. Membr. Biol. 254 (5-6), 443–446. doi: 10.1007/s00232-021-00207-9
Qian J., Zhang X., Yan Y., Wang N., Ge W., Zhou Q., et al. (2020). Unravelling the molecular mechanisms of abscisic acid-mediated drought-stress alleviation in pomegranate (Punica granatum l.). Plant Physiol. Biochem. 157, 211–218. doi: 10.1016/j.plaphy.2020.10.020
Sampaio L. A., Bianchini A. (2002). Salinity effects on osmoregulation and growth of the euryhaline flounder Paralichthys orbignyanus. J. Exp. Mar. Biol. Ecol. 269 (2), 187–196. doi: 10.1016/s0022-0981(01)00395-1
Sarma K., Dey A., Kumar S., Chaudhary B., Mohanty S., Kumar T., et al. (2020). Effect of salinity on growth, survival and biochemical alterations in the freshwater fish Labeo rohita (Hamilton 1822). Indian J. Fish 67 (2), 41–47. doi: 10.21077/ijf.2019.67.2.86894-06
Seale A. P., Pavlosky K. K., Celino-Brady F. T., Lerner D. T. (2020). Sex, salinity and sampling period dependent patterns of growth hormone mRNA expression in mozambique tilapia. Aquaculture 519, 734766. doi: 10.1016/j.aquaculture.2019.7347
Shi H., Ru X., Pan S., Jiang D., Huang Y., Zhu C., et al. (2022). Transcriptomic analysis of pituitary in female and male spotted scat (Scatophagus argus) after 17β-estradiol injection. Comp. Biochem. Physiol. Part D: Genomics Proteomics 41, 100949. doi: 10.1016/j.cbd.2021.100949
Shi X., Sun X., Liu M., Li D., Aneja R., Zhou J. (2011). CEP70 protein interacts with γ-tubulin to localize at the centrosome and is critical for mitotic spindle assembly. J. Biol. Chem. 286 (38), 33401–33408. doi: 10.1074/jbc.m111.252262
Shirangi S. A., Kalbassi M. R., Khodabandeh S., Jafarian H., Lorin-Nebel C., Farcy E., et al. (2016). Salinity effects on osmoregulation and gill morphology in juvenile persian sturgeon (Acipenser persicus). Fish Physiol. Biochem. 42 (6), 1741–1754. doi: 10.1007/s10695-016-0254-y
Si Y., Wen H., Li Y., He F., Li J., Li S., et al. (2018). Liver transcriptome analysis reveals extensive transcriptional plasticity during acclimation to low salinity in Cynoglossus semilaevis. BMC Genomics 19 (1), 1–14. doi: 10.1186/s12864-018-4825-4
Takvam M., Wood C. M., Kryvi H., Nilsen T. O. (2021). Ion transporters and osmoregulation in the kidney of teleost fishes as a function of salinity. Front. Physiol. 12. doi: 10.3389/fphys.2021.664588
Tipsmark C. K., Luckenbach J. A., Madsen S. S., Borski R. J. (2007). IGF-I and branchial IGF receptor expression and localization during salinity acclimation in striped bass. Am. J. Physiology-Regulatory Integr. Comp. Physiol. 292 (1), R535–R543. doi: 10.1152/ajpregu.00915.2005
Tomy S., Chang Y.-M., Chen Y.-H., Cao J.-C., Wang T.-P., Chang C.-F. (2009). Salinity effects on the expression of osmoregulatory genes in the euryhaline black porgy Acanthopagrus schlegeli. Gen. Comp. Endocrinol. 161 (1), 123–132. doi: 10.1016/j.ygcen.2008.12.003
Tsuzuki M. Y., Sugai J. K., Maciel J. C., Francisco C. J., Cerqueira V. R. (2007). Survival, growth and digestive enzyme activity of juveniles of the fat snook (Centropomus parallelus) reared at different salinities. Aquaculture 271 (1-4), 319–325. doi: 10.1016/j.aquaculture.2007.05.002
Varsamos S., Nebel C., Charmantier G. (2005). Ontogeny of osmoregulation in postembryonic fish: a review. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 141 (4), 401–429. doi: 10.1016/j.cbpb.2005.01.013
Vij S., Purushothaman K., Sridatta P. S. R., Jerry D. R. (2020). Transcriptomic analysis of gill and kidney from Asian seabass (Lates calcarifer) acclimated to different salinities reveals pathways involved with euryhalinity. Genes 11 (7), 733. doi: 10.3390/genes11070733
Wang D., Cao Q., Zhu W., Hu Y., Zhang X., Yin S., et al. (2019). Individual and combined effects of salinity and lipopolysaccharides on the immune response of juvenile Takifugu fasciatus. Fish Physiol. Biochem. 45 (3), 965–976. doi: 10.1007/s10695-018-0607-9
Wood C. M., Nawata C. M. (2011). A nose-to-nose comparison of the physiological and molecular responses of rainbow trout to high environmental ammonia in seawater versus freshwater. J. Exp. Biol. 214 (21), 3557–3569. doi: 10.1242/jeb.057802
Xu C., Li E., Suo Y., Su Y., Lu M., Zhao Q., et al. (2018). Histological and transcriptomic responses of two immune organs, the spleen and head kidney, in Nile tilapia (Oreochromis niloticus) to long-term hypersaline stress. Fish Shell. Immunol. 76, 48–57. doi: 10.1016/j.fsi.2018.02.041
Yu G., Wang L.-G., Han Y., He Q.-Y. (2012). clusterProfiler: an r package for comparing biological themes among gene clusters. Omics: J. Integr. Biol. 16 (5), 284–287. doi: 10.1089/omi.2011.0118
Zhang X., Wen H., Wang H., Ren Y., Zhao J., Li Y. (2017). RNA-Seq analysis of salinity stress–responsive transcriptome in the liver of spotted sea bass (Lateolabrax maculatus). PloS One 12 (3), e0173238. doi: 10.1371/journal.pone.0173238
Zhang T., Yao J., Xu D., Ma X., Jin W., Lv G., et al. (2021). Gill physiological and transcriptomic response of the threatened freshwater mussel Solenaia oleivora to salinity shift. Comp. Biochem. Physiol. Part D: Genomics Proteomics. 40, 100913. doi: 10.1016/j.cbd.2021.100913
Zhao H., Wang Q., Zhao H., Chen C. (2022). Transcriptome profiles revealed high- and low-salinity water altered gill homeostasis in half-smooth tongue sole (Cynoglossus semilaevis). Comp. Biochem. Physiol. Part D: Genomics Proteomics. 42, 100989. doi: 10.1016/j.cbd.2022.100989
Zhong Y., Duan Z., Su M., Lin Y., Zhang J. (2021). Inflammatory responses associated with hyposaline stress in gill epithelial cells of the spotted scat Scatophagus argus. Fish Shell. Immunol. 114, 142–151. doi: 10.1016/j.fsi.2021.04.023
Zidan E. M., Goma A. A., Tohamy H. G., Soliman M. M., Shukry M. (2022). Insight study on the impact of different salinity levels on behavioural responses, biochemical stress parameters and growth performance of African catfish (Clarias gariepinus). Aquacult. Res. 53 (7), 2750–2759. doi: 10.1111/are.15790
Keywords: salinity stress, osmoregulation, greater amberjack, gills, kidneys
Citation: Liu Y, Yang Y, Qin R, Peng Y, Huang Y, Zhu C, Li G, Jiang D and Shi H (2023) Divergent molecular responses of greater amberjack (Seriola dumerili) to acute salinity stress revealed by comparative transcriptome analysis. Front. Mar. Sci. 10:1185015. doi: 10.3389/fmars.2023.1185015
Received: 13 March 2023; Accepted: 20 June 2023;
Published: 25 July 2023.
Edited by:
Melissa Diogo Faria, Leitat Technological Center, SpainReviewed by:
Shaowu Yin, Nanjing Normal University, ChinaChunyan Zhao, Qingdao Agricultural University, China
Copyright © 2023 Liu, Yang, Qin, Peng, Huang, Zhu, Li, Jiang and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuchen Yang, eWFuZ3ljaDY4QG1haWwuc3lzdS5lZHUuY24=; Hongjuan Shi, c2hpaGpAZ2RvdS5lZHUuY24=
 Yuqi Liu1,2,4
Yuqi Liu1,2,4 Yuchen Yang
Yuchen Yang Chunhua Zhu
Chunhua Zhu Guangli Li
Guangli Li Hongjuan Shi
Hongjuan Shi