- 1Research Unit Marine Symbioses, GEOMAR Helmholtz Centre for Ocean Research Kiel, Kiel, Germany
- 2Institut de Ciències del Mar–Spanish National Research Council (CSIC), Marine Biology and Oceanography, Marine Biogeochemistry, Atmosphere and Climate, Barcelona, Spain
- 3Christian-Albrechts-Universität Kiel, Kiel, Germany
Introduction: Sponges harbor diverse, specific, and stable microbial communities, but at the same time, they efficiently feed on microbes from the surrounding water column. This filter-feeding lifestyle poses the need to distinguish between three categories of bacteria: food to digest, symbionts to incorporate, and pathogens to eliminate. How sponges discriminate between these categories is still largely unknown. Phagocytosis is conceivable as the cellular mechanism taking part in such discrimination, but experimental evidence is missing. We developed a quantitative in-vivo phagocytosis assay using an emerging experimental model, the sponge Halichondria panicea.
Methods: We incubated whole sponge individuals with different particles, recovered the sponge (host) cells, and tracked the incorporation of these particles into the sponge cells. Fluorescence-activated cell sorting (FACS) and fluorescent microscopy were used to quantify and verify phagocytic activity, defined here as the population of sponge cells with incorporated particles. Sponges were incubated with a green microalgae to test if particle concentration in the seawater affects the percentage of phagocytic activity, and to determine the timing where the maximum of phagocytic cells are captured in a pulse-chase experiment. Lastly, we investigated the application of our phagocytic assay with other particle types (i.e., fluorescently-labelled bacteria and fluorescent beads).
Results and discussion: The percentage of sponge cells that had incorporated algae, bacteria, and beads ranged between 5 to 24%. These phagocytic sponge cells exhibited different morphologies and sizes depending on the type of particle presented to the sponge. Particle incorporation into sponge cells was positively related to algal concentration in the seawater, suggesting that sponge cells adjust their phagocytic activity depending on the number of particles they encounter. Our results further revealed that sponge phagocytosis initiates within minutes after exposure to the particles. Fluorescent and TEM microscopy rectified algal internalization and potential digestion in sponge cells. To our knowledge, this is the first quantitative in-vivo phagocytosis assay established in sponges that could be used to further explore phagocytosis as a cellular mechanism for sponges to differentiate between different microorganisms.
Introduction
Early branching metazoans provide an opportunity to investigate the evolution of host-microbe interactions. Sponges (phylum Porifera) are benthic suspension feeders that actively filter large volumes of water. Due to this filter-feeding lifestyle, they are constantly exposed to high numbers of particles, which poses the question of how sponges distinguish and process between the different types of microbes they encounter. Sponges exhibit three well-defined cell layers: the external pinacoderm composed of T-shaped or flattened cells (i.e., pinacocytes) which covers the outside of the animal; the internal choanoderm containing the flagellated cells (i.e., choanocytes); and the mesohyl, a matrix within which other sponge cells, skeletal components, and most symbiotic microbes reside (Simpson, 1984; Leys and Hill, 2012). These filter feeders evolved a complex branched water canal system (i.e., the aquiferous system) comprised of several choanocytes arranged into hollow chambers which generate water flow by the beating of their flagella. Water enters the sponge through small pores, or ostia, which spread along the animals’ outer surface, circulates through the incurrent canals into the choanocyte chambers, and exits through excurrent canals to larger outflow openings, or oscula. The particles (e.g., bacterio- and phytoplankton) that are filtered by the choanocytes are translocated to amoebocyte-like cells (i.e., archaeocytes) which are regarded as potential nutrient transporters within the sponge (Imsiecke, 1993; Leys and Eerkes-Medrano, 2006; Steinmetz, 2019). The participation of both choanocytes and archaeocytes in intracellular digestion is supported by single-cell transcriptomic data which shows enrichment in genes related to e.g., engulfment and motility, lysosomal enzymes, and phagocytic vesicles (Sebé-Pedrós et al., 2018; Musser et al., 2021). Despite this filter-feeding lifestyle, sponges harbor a highly diverse community of associated microbes in their mesohyl that is remarkably different from the bacterioplankton in the surrounding seawater (Hentschel et al., 2006; Thomas et al., 2016; Moitinho-Silva et al., 2017). It remains enigmatic if, and how, sponges distinguish between food to digest, symbionts to acquire, and pathogens to eliminate.
Selectivity in particle uptake continues to be a controversial topic in sponge physiology. Some studies regard particle size as the main parameter driving the selection of microbes during the filtering process (Reiswig, 1971; Turon et al., 1997; Ribes et al., 1999; Pile and Young, 2006), whereas others propose a size-independent discrimination between particles involving individual particle recognition, sorting, and transport through the sponge tissue during the digestion process (Reiswig, 1971; Leys and Eerkes-Medrano, 2006; Yahel et al., 2006; Maldonado et al., 2010; McMurray et al., 2016). Furthermore, two feeding experiments with culturable bacteria provide first evidence that sponges take up seawater bacteria but not sponge-associated bacteria (Wilkinson et al., 1984; Wehrl et al., 2007). The latter studies hypothesized that the lower uptake rates of sponge-associated bacteria isolates result from slime capsules or secondary metabolites that protected symbionts from being recognized and ingested by the sponge. How the sponge host exactly discriminates microbes at the cellular level is still unclear, yet we posit that this cellular host-microbe recognition mechanism must be essential for establishing and maintaining symbiotic interactions and a stable microbiome in sponges.
We thus hypothesize that phagocytosis will play a major role in sponge-microbe interactions. Hijacking of phagocytosis promotes the colonization and maintenance of microbes during symbiotic interactions (Nyholm and Graf, 2012). Phagocytosis includes the recognition and ingestion of particles larger than 0.5 µm within a plasma-membrane envelope (i.e., phagosome) (Rosales and Uribe-Querol, 2017). It is a highly conserved cellular process from unicellular to multicellular organisms, involved in nutrition, defense, homeostasis and symbiosis (Nyholm and Graf, 2012; Lim et al., 2017; Hartenstein and Martinez, 2019). Symbionts from diverse hosts such as amoeba, leech and squid are capable of escaping different stages of the phagocytic process, avoiding either incorporation or digestion by host cells (e.g., Silver et al., 2007; Nyholm et al., 2009; Nguyen et al., 2014), in their way to colonize and persist in the animal host.
In sponges, transcriptomic studies have revealed that upon exposure to microbes or microbial elicitors the host activates phagocytic-related genes (Yuen, 2016; Pita et al., 2018; Geraghty et al., 2021; Schmittmann et al., 2021). Furthermore, sponge-associated bacteria are enriched in ankyrin proteins compared to non-associated sponge bacteria (Silver et al., 2007; Thomas et al., 2010; Siegl et al., 2011; Díez-Vives et al., 2017). These proteins modulate host phagocytosis in humans assisting the infection and intracellular survival of symbionts and pathogens (Al-Khodor et al., 2008; Habyarimana et al., 2008; Pan et al., 2008). Functional evidence on how ankyrins from sponge symbionts modulate host phagocytosis is only probed in non-sponge systems, in particular in free-living amoeba and murine macrophages (Nguyen et al., 2014; Jahn et al., 2019), due to the lack of experimental assays in sponges. Interestingly, bacteria expressing ankyrin-repeat proteins were phagocytized, but not digested by the amoeba Acanthamoeba castellanii (Nguyen et al., 2014), whereas bacteria decorated with phage encoded ankyrins evaded phagocytosis in murine macrophages (Jahn et al., 2019). These pioneering studies serve as evidence that the host phagocytic machinery is targeted by sponge-associated microbes and highlights the importance to develop an assay that allows to test these functions testing these functions in the sponge host.
Feeding experiments shed some light on the filtration and particle internalization by sponges, a process usually referred to in a generic way as “phagocytosis” in sponge literature (Imsiecke, 1993; Leys and Eerkes-Medrano, 2006; Maldonado et al., 2010). However, the laborious and time-consuming microscopic observations in combination with the lack of well-established methodologies to study cellular processes in sponges hampers our current understanding of the implications of phagocytosis on sponge-microbe interactions. In other eukaryote phyla such as Amoebozoa and Cnidaria, advances in genetic manipulation, isolation of symbiont strains and identification of cellular markers have started to pave the way to study phagocytosis as the underlying mechanism for microbe discrimination (e.g., Dictyostelium sp., Aiptasia sp., Pocillopora damicornis, and Nematostella vectensis) (Sattler et al., 2013; Bucher et al., 2016; Rosental et al., 2017; Jacobovitz et al., 2021; Jauslin et al., 2021; Snyder et al., 2021). In hexacorallians, fluorescence-activated cell sorting (FACS) and microscopy allowed further identification and mechanistic characterization of specialized phagocytic cells capable of lysosomal degradation and production of reactive oxygen species (ROS) upon microbe engulfment (Snyder et al., 2021). The development of methods and establishment of model systems are thus fundamental for studying the function of host phagocytosis in microbe selectivity.
In the present study, we describe the development of an in-vivo phagocytosis assay based on combining live sponge incubation experiments with fluorescent particles followed by sponge cell dissociation and flow cytometry to quantify the incorporation of different particles into sponge cells (from now on termed phagocytosis). We used the Baltic Sea sponge Halichondria panicea. This shallow water sponge is widely distributed along the North Atlantic and its ecology and physiology have been studied both in natural and aquaria conditions (e.g., feeding experiments with microalgae and bacteria (Barthel, 1988; Riisgård et al., 2016; Lüskow et al., 2019)). H. panicea is a low microbial abundance (LMA) sponge with a dominant and stable bacterial symbiont (i.e., Candidatus Halichondribacter symbioticus, relative abundance approx. 25%–80% based on 16S rRNA gene amplicon data) (Knobloch et al., 2019), and presumably also harbors intracellular green algae (Vethaak et al., 1982; Goldstein and Funch, 2022). The sponge microbiome can be experimentally manipulated (Schmittmann et al., 2022) and the host transcriptomic response has been characterized using microbial elicitors (Schmittmann et al., 2021). These features make H. panicea a promising model for studying sponge symbiosis (Pita et al., 2016). We quantified the incorporation of inert (beads) and natural particles (microalgae and bacteria) into H. panicea cells. We chose these particles because of their size (1-3 µm in diameter), clear fluorescence signal, and ecological relevance [i.e., H. panicea naturally feeds on phytoplankton and bacteria (Riisgård et al., 2016; Lüskow et al., 2019)]. Fluorescence-activated cell sorting (FACS) served to quantify, in a high-throughput way, particle incorporation into sponge cells, which was also confirmed by fluorescence and transmission electron microscopy. To our knowledge, this is the first quantitative in-vivo phagocytosis assay established in sponges which could be used to further explore the mechanistic underpinnings of recognition and differentiation between sponges and diverse microorganisms (food, friend, or foe).
Material and methods
Sponge collection
Individuals of the breadcrumb sponge Halichondria panicea (Pallas, 1766) were collected for establishing the assay between Jun 2020 to Jun 2022. The sponge individuals for which data is presented were collected in Aug 2022 at the coast of Schilksee (54.424278 N, 10.175794 E; Kiel, Germany). Sponges were collected at water depths of between 1-3 m and were carefully removed from the crevices of a breakwater structure using a metal spatula. The individuals were directly transferred to the KIMMOCC climate chamber facilities at GEOMAR Helmholtz Centre for Ocean Research (Kiel, Germany), and maintained in 10 L tanks in a semi-flow through aquaria system supplied with sand-filtered seawater pumped from the Kiel fjord at 6 m depth. The sponges (n = 10 individuals) were cut with a scalpel into approximately equal-sized fragments (3.9 ± 1.0 g wet weight [average ± S.D.]), cleared of epibionts, and randomly placed back in the 10 L tanks supplied with the natural seawater from the flow-through system at KIMOCC facilities (3-4 sponge fragments per tank). The water flow in the tanks was 0.5 L min-1 and a mini-pump with a maximum flow rate of 300 L h-1 (Dupla TurboMini) enhanced further water movement in each tank. Sponges were left on top of clay tiles (6.3 x 1 cm - Ø x H) to heal and attach for 8 days at 10°C room temperature, 17°C water temperature, and a salinity of 16 PSU.
Tracer preparation for the in-vivo phagocytosis assay
The particles included the microalgae Nannochloropsis sp., the Vibrio sp. isolate PP-XX7 (16S rRNA gene sequence similarity of 98.6% with Vibrio sp. NBRC 101805 as the next related strain in the NCBI database based on blastn), and 1 µm polystyrene-based latex beads. The Nannochloropsis sp. live culture was purchased from BlueBio Tech (Germany). The stock algal culture concentration was 12 x 109 algae cells mL-1, and was kept at 4°C, protected from light until the experiments were performed, as recommended by the manufacturer. The Vibrio strain was initially isolated from the top 1 cm sediment sampled in our sponge collection site. The bacteria culture was prepared by inoculating 100 mL of liquid marine broth (Zobell 2216) with a culture that grew for 24h on an agar plate. The liquid culture was incubated on a shaker at 120 rpm, at 25°C, for 48 hours until the culture reached the mid to late exponential phase. After this incubation time, the culture OD600 was measured (OD600 = 1.45) to estimate the aimed bacteria concentration of approx. 105-106 bacteria mL-1. The culture was centrifuged at 5000 x g for 5 min to recover the bacteria pellet, resuspended in 0.22 μm-filtered artificial seawater (FASW), and stained the same day of the experiment with the fluorescent dye 5 -TAMRA/SE™ (Thermo Fisher Scientific, C1171) at a final concentration of 1 μM (as in (Wehrl et al., 2007)). The bacteria suspension was incubated with the dye for 90 min in the dark at room temperature, the excess dye was washed off by centrifuging at 5000 x g for 5 min, and the bacteria pellet was resuspended in FASW. The bacteria suspension was kept at 4°C in the dark until the experiment was performed. The bead stock solution (Fluoresbrite YG microsphere, Cat. 17154-10, Polyscience) with a concentration of 4.55 x 1010 particles mL-1, was sonicated for 5 min and vortexed immediately before the experiment.
In-vivo sponge phagocytosis assay
The phagocytosis assay consisted of incubating individual fragments of H. panicea with a specific particle for 30 min and tracking its incorporation into the sponge (host) cells. The sponges attached to the ceramic tiles were placed in individual 1 L straight-sided polypropylene wide-mouth Nalgene bottles (ThermoFisher; cat. no. 2118-0032) filled with unfiltered natural seawater from the aquaria system inlet. The tiles were laid on a PVC support under which a magnetic stirring bar was positioned, and the bottles were placed on top of a stirring plate to ensure constant water flow and uniform mixing of the particles during the incubation. Incubations were performed in a climate chamber in which temperature was kept at 10°C. In all cases, sponges incubated in the absence of tracer particles and natural seawater incubations served as controls (n = 4 biological replicates per control and sponges).
First, we tested whether initial particle concentration in seawater had an effect on the percentage of sponges incorporating particles. We incubated the sponge with three different Nannochloropsis sp. concentrations, as this was the natural particle given the clearest readout. We added 10 µL, 100 µL, or 1000 µL of the live algal stock culture to each incubation chamber to get a final concentration of approx. 105, 106 and 107 algae mL-1, respectively. Secondly, we performed a pulse-chase experiment using Nannochloropsis sp. to identify the time when we could capture the highest percentage of cells with incorporated particles. Sponges were incubated separately with algae (106 algae cells mL-1 final concentration) over a 30 min pulse phase, and then transferred to the flow-through aquaria with clean natural seawater for a 30 min and 150 min chase phase. Sponges were sampled at t = 0 min (immediately after the 30 min pulse phase was over), at t = 30 min, and at t = 150 during the chase phase (n = 4 biological replicates per time point). Lastly, we aimed to extend the application of our phagocytic assay to other particles. We incubated for 30 min H. panicea individuals with TAMRA-stained bacteria and fluorescent bead solution, separately (n = 4 biological replicates per tracer). We aimed at a particle concentration similar to the bacteria concentration naturally encountered by the sponge (Lüskow et al., 2019) (i.e., final start concentration of approx. 105 bacteria cells mL-1 and 106 beads mL-1). The sponge cell dissociation was performed immediately after the incubation time (i.e., no pulse-chase design was applied in this case).
Particle uptake estimation
Water samples (1.8 mL) were taken along the incubation period at 0 min, 2 min, 5 min, 7 min, 14 min, 22 min and 30 min to assess whether sponges were filtering the particles from the water column (i.e., particle uptake) during each in-vivo assay. The sampled water was immediately fixed with a mixture of paraformaldehyde (PFA) and glutaraldehyde in 1x PBS (final concentration 1% and 0.05%, respectively), kept in the dark for 30 min, and stored at -80°C until flow cytometry analysis was performed (following (Gasol and Del Giorgio, 2000)). Algal, bacterial, and bead concentrations from the water samples were estimated using the Accuri C6 Plus flow cytometer (BD Biosciences). Each sample was run for 1 min at a flow rate of 14 µL min-1. The cell populations of interest were identified based on the fluorescence of each tracer particle and the side scatter (SSC) using the BD Accuri C6 Plus Software. Differences in particle uptake between sponge and control incubations were analyzed using an exponential regression approach (as in (Scheffers et al., 2004; De Goeij and Van Duyl, 2007; Yahel et al., 2007)). In each case, the concentrations were corrected based on the average initial concentration of all incubations. The standardized data was fitted to an exponential model (as proposed by (De Goeij et al., 2008)) and this was used to estimate the concentration of particles in the seawater at the start (C0) and end (C30) of the incubation. The total number of particles that were taken up by the sponge was calculated as (C0 – C30) x volume of incubation (i.e., 1000 mL), and with this the percentage of particles reduced by the end of the incubation was estimated. The regression fits and values used for estimating particle uptake can be found in https://doi.org/10.1594/PANGAEA.956281.
Sponge cell dissociation, preparation, and staining
Immediately after each in-vivo assay, H. panicea fragments were used to extract the sponge cell fraction by tissue dissociation and centrifugation methods [adapted from (Fieseler et al., 2004; Wehrl et al., 2007)]. All buffers and solutions used during the dissociation were adjusted to the salinity (16 PSU) and pH (8.1) of the aquaria at the moment of running the experiment to prevent the cells from undergoing an osmotic or pH shock. Calcium- and magnesium-free artificial seawater (CMFASW; as in (Rottmann et al., 1987)) was used through the whole dissociation process as it prevents sponge cells from reaggregating and minimize unspecific cell-cell interactions. The entire sponge fragments were rinsed in sterile, ice-cold CMFASW, and subsequently cut with a disposable scalpel into small pieces while removing any leftover epibionts or dirt. The tissue fragments were transferred into 50 mL sterile Falcon tubes prefilled with 25 mL of sterile fresh ice-cold CMFASW containing EDTA (25mM) and incubated on ice while gently shaking the tubes horizontally for 15 min. Samples were filtered through 40 μm cell strainers (Corning Inc.) into 50 mL sterile Falcon tubes by gently squeezing the tissue against the walls of the sieve with sterile forceps to remove dissociated tissue fragments and spicules. Ice-cold CMFASW was added to the resulting cell suspension until reaching a total volume of 25 mL and the samples were centrifuged for 5 minutes at 500 x g at 4°C in a swinging rotor. The supernatant was discarded, and the sponge cell pellet was resuspended in 4 mL of fresh sterile ice-cold CMFASW resulting in a total sponge cell suspension of approx. 5 mL, which was divided into 1 mL aliquots in sterile Eppendorf tubes. The aliquots were fixed by adding PFA to the cell suspension (final concentration 4% in CMFASW) and stored at 4°C in the dark overnight. The fixative was washed off the cells by centrifugation for 5 minutes at 500 x g at 4°C. Finally, the pellet was resuspended in 1 mL of ice-cold CMFASW. The concentration of sponge cells for each sample was estimated by an automated cell counter (Fluidlab R-300, Anvajo) and adjusted to approx. 5 x 107 cells mL-1 by diluting the cells with ice-cold CMFASW. The fixed cell suspensions were stored at 4°C in the dark for later FACS quantification. For each sample, 200 µL of cell suspension was diluted into 4 mL of ice-cold CMFASW and filtered through a 40 μm-cell strainer into a 5 mL round-bottom Falcon tube. From the filtered cell suspension, an 2 mL aliquot was used for staining sponge cell nuclei with 14 µL of 100 ng mL-1 DAPI in a 2 mL cell suspension (final concentration 0.7 ng uL-1). The cells were gently mixed by pipetting and incubated for 30 min at 4°C in the dark. The remaining cell suspension aliquot served as control for the DAPI staining (i.e., non-stained aliquot).
FACS quantification of phagocytic active cells
We define phagocytic active cells as those that had incorporated the tracer particle presented to the sponge during the incubation. Sponge cells were analyzed on a MoFlo Astrios EQ® cell sorter (Beckman Coulter) fitted with a 70 µm nozzle and 355nm, 488nm, 561nm and 640 nm wavelength lasers to identify and quantify phagocytic cells. Each sample was analyzed three times (i.e., technical replicates) with FACS for 1 min, and the voltage and pressure were adjusted to ensure to record a similar number of events (i.e., approx. 5k) per second for all samples. The gating was performed with the Summit software (V6.3.1) based on the emission of DAPI and the fluorescence of the respective particle used during the incubation. The strategy we developed for detecting and quantifying the phagocytic active sponge cells of interest followed the subsequent steps. First, to differentiate debris from the “bulk” sponge cells we compared the DAPI emission of non-stained cell aliquots with the DAPI stained aliquots by using the 355 nm UV laser and a filter with a band pass of 448/59 nm (Figure 1A). A scatter plot was created by selecting the DAPI filter channel on the x-axis and the forward scatter (FCS) on the y-axis, and the DAPI positive cell population was gated. Second, to identify the relative percent of “bulk” sponge cells that had incorporated Nannochloropsis sp. cells, the DAPI gate was used to create a new scatter using the laser (561 nm) and filter settings (692/75 nm) in the y-axis to detected algae fluorescence (Figure 1B). This approach allowed us to distinguish between sponge cells that had incorporated algae from the rest of the sponge cells (i.e., phagocytic from non-phagocytic sponge cells, respectively). We confirmed this gating by comparing the fluorescence of the control sponges (i.e., without algae) with the sponges provided with algae (Figure 1B). To identify sponge cells that had incorporated TAMRA-labelled Vibrio sp. bacteria from the other sponge cells a scatter plot was created using the side scatter (SSC) on the x-axis and the bacteria fluorescence (laser 561 nm and filter 692/75 nm) in the y-axis (Figure 1C). On the other hand, sponge cells phagocytizing beads were detected by plotting the particle size [forward scatter (FCS): x-axis] against the bead fluorescence (laser 488 nm and filter 526/52 nm: y-axis) (Figure 1D). In all cases, the phagocytic and non-phagocytic sponge cell populations were gated and sorted directly onto microscopy slides (2k-3k cells) for fluorescent microscopy inspection of each tracer (Figure 4). After this verification, the number of events of each gate was used to estimate the relative (%) phagocytic and non-phagocytic cell fraction in relation to the total number of events from these two gates (the complete data set can be found in https://doi.org/10.1594/PANGAEA.956281).
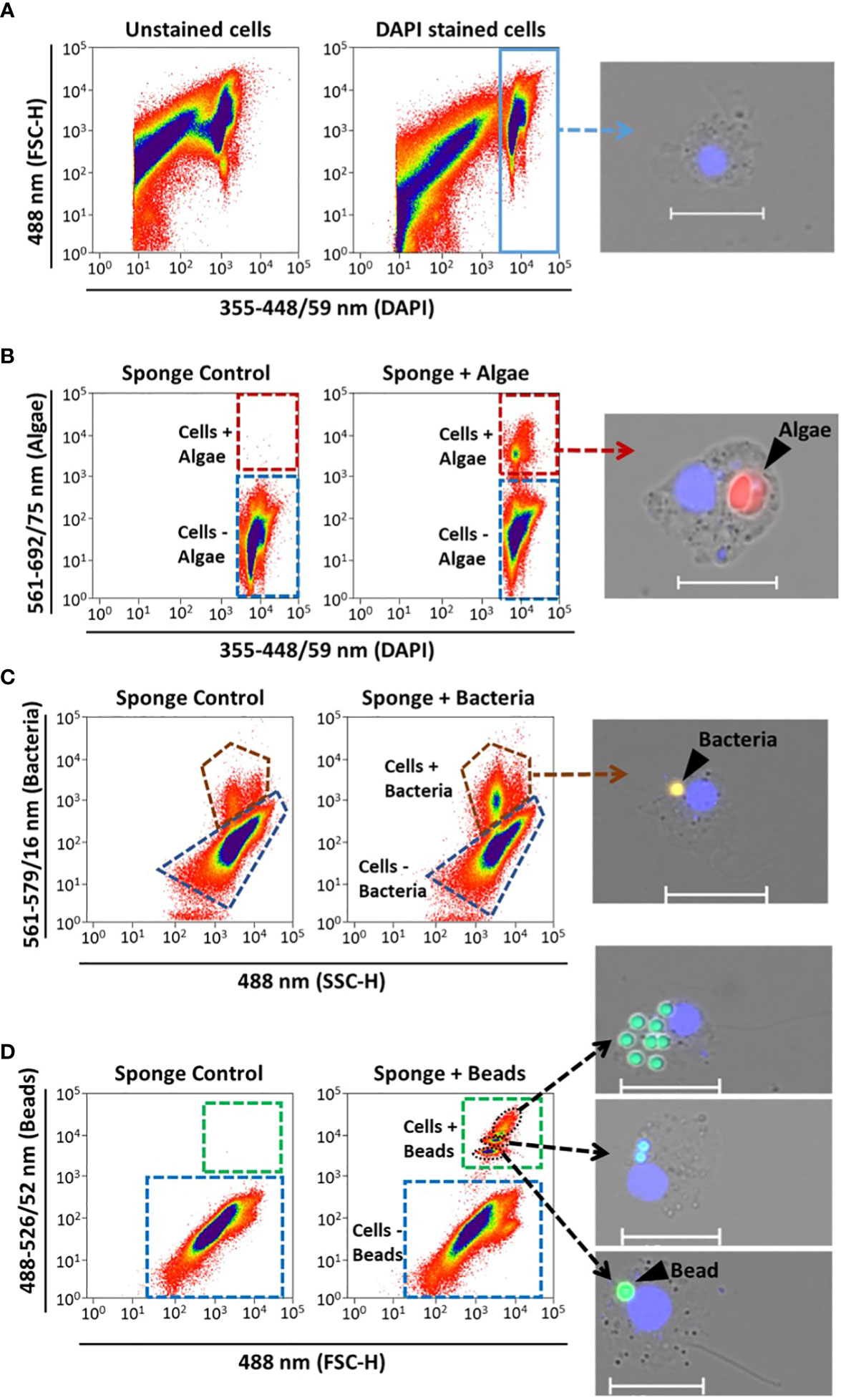
Figure 1 Sponge cell gating strategy for identification and quantification of phagocytic active sponge cells using FACS. Representative cytograms show (A) identification of the “bulk sponge cells” population after DAPI staining (blue rectangle). Gates of phagocytic cells with (+) incorporation of (B) algae, (C) bacteria, and (D) beads (red, brown, and green dashed outlines, respectively). Fluorescent microscopy pictures of sorted cells to verify the different gates. Sponge cell nuclei (blue) stained with DAPI. Scale bars: 10 µm. Gates for cells without (–) particle incorporation are shown in each case (blue dashed outlines). Sponges incubated without particles served as controls.
Fluorescence microscopy
A sub-sample of the sponge cell suspension was inspected by fluorescent microscopy to gain insights into the types of cells involved in the phagocytic process. The cell suspension was mounted on microscopy slides using ROTI mount FluorCare DAPI and 20 x 20 mm cover slides. The slides were air-dried for 30 minutes and examined under an inverted fluorescence microscope equipped with a camera (Axio Observer Z1 with Axiocam 506 and HXP-120 light; Zeiss), at a total magnification of 100x. The following filters and excitations were used: 49 DAPI (335-383 nm for sponge nuclei), 00 Propidium Iodide (530-585 nm for Nannochloropsis sp.), 43 HE DsRed (538-562 nm for TAMRA-stained Vibrio sp.), and 38 HE GFP (450-490 nm for fluorescent beads). Pictures and z-stack series were acquired using the ZEN Blue Edition software (Zeiss).
Transmission electron microscopy
In order to locate the particles and verify algal incorporation into the sponge cells, a small portion of tissue from two individuals per treatment was collected and processed [following (Busch et al., 2020)]. The tissue was fixed for transmission electron microscopy (TEM; in 2.5% glutaraldehyde in 1x PBS) overnight at 4°C. Samples were washed three times with 1x PBS for 15 min and then postfixed with 2% osmium tetroxide for 2 h at room temperature, gently shaking the tissue. The sponge tissue was rinsed with 1x PBS three times on ice for 15 min and partial dehydration was performed with an ascending ethanol series (2x 30%, 2x 50%, and 2x 70%). Sponge pieces were treated with 5% hydrofluoric acid (in 70% ethanol) overnight at room temperature to remove any silica spicules from their skeleton. Subsequently, samples were rinsed with ethanol (6x 70% for 15 min), further dehydrated through an increasing series of ethanol (1x 90% and 2x 100% for 30 min), and after embedded in Epon epoxy resin. Ultrathin sections were obtained from region of interest using a Ultracut UC7 (Leica Microsystems) and TEM analysis was performed using a J1010 (Jeol) at the TEM-SEM Electron Microscopy Unit from Scientific and Technological Centers (CCiTUB), Universitat de Barcelona.
Statistical analyses
To test the effect of algal concentration, chase-time and particle type on the percentage of sponge phagocytic cells, One-way ANCOVA analyses were performed. Particle uptake was used as covariate as we expected that this would influence the estimation of phagocytic sponge cells. ANCOVA assumptions were checked, and significance was determined at the α = 0.05 level. Statistical analysis was performed in R-studio (V4.2.1; Rstudio Team 2022) by fitting an analysis of variance model (aov () function).
Results
We estimated the sponge’s phagocytic activity as the percentage of sponge cells with incorporated fluorescent particles over the total of sponge cells recovered by coupling incubation experiments with whole H. panicea individuals with sponge cell dissociation and FACS. Sponges were incubated with three types of fluorescent particles: Nannochloropsis sp., TAMRA-stained Vibrio sp., and fluorescent latex beads. Our optimized cell dissociation protocol for H. panicea host cells yielded an average recovery of 1.4 x 107 ± 5.5 x 106 cells g-1 (sponge wet weight) (Figure S1). The recovered cell suspension was analyzed by FACS (Figure 1) to quantify the relative number (%) of phagocytic active (+fluorescent signal) to the total of DAPI+ cells. Fluorescent microscopy z-stack images of the sorted phagocytic active cells supported the incorporation of the particles into the sponge cells (Figure 2).
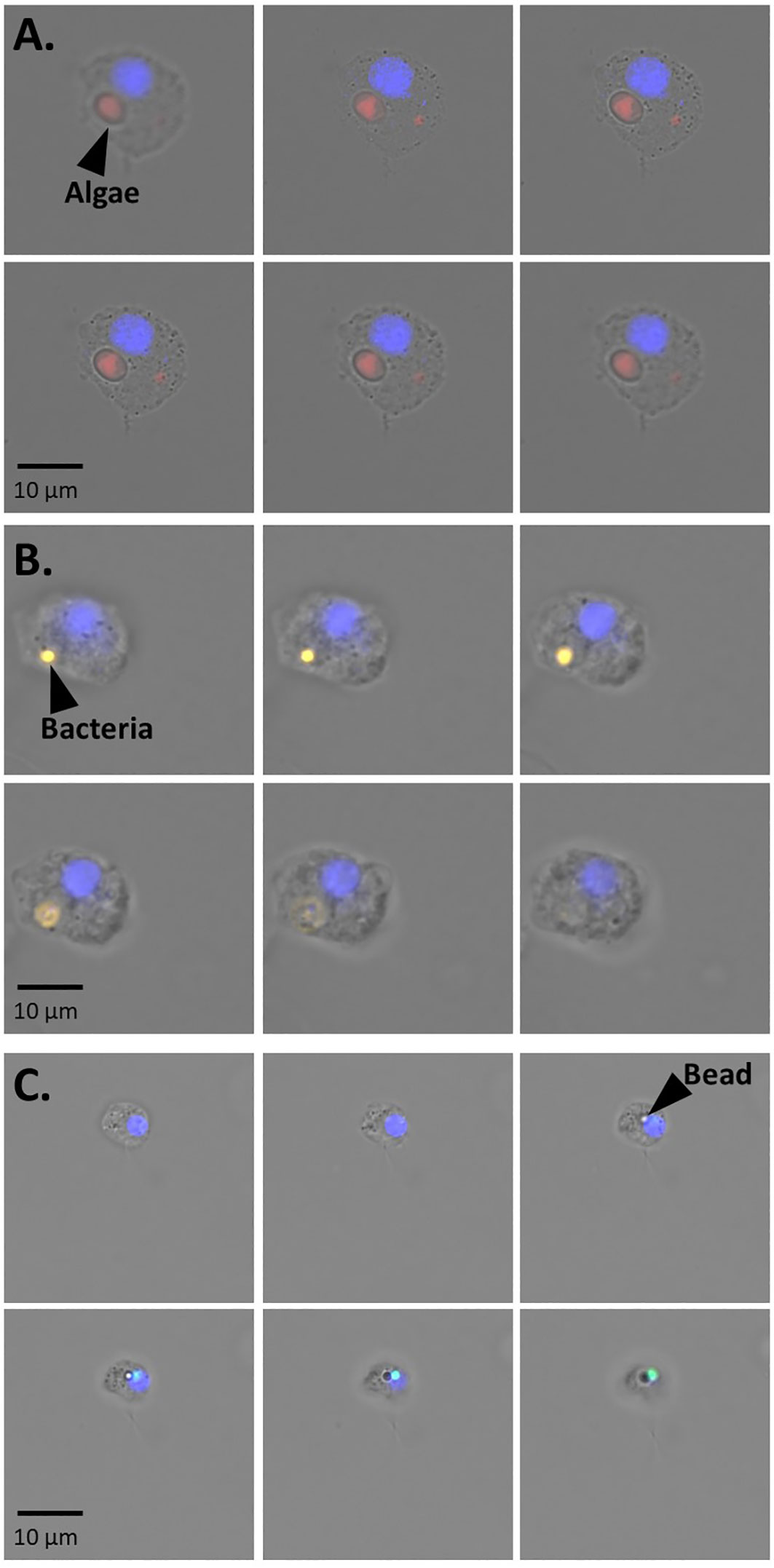
Figure 2 Representative fluorescent microscopy pictures of z-stacks series showing the internalization of (A) algae (B) bacteria and (C) beads into H. panicea cells. The series consists of photos taken every 4 to 5 µm. Sponge cell nuclei (blue).
Testing tracer concentration for the phagocytic assay
We tested the effect of different tracer concentration on phagocytosis and particle uptake. The total amount of Nannochloropsis sp. cells (mean ± SD throughout the text, unless stated otherwise) taken up by H. panicea was 4.9 x 104 ± 2.5 x 104 cells and 2.4 x 105 ± 3.5 x 105 cells (Figure 3A) during the incubation with low (105 cells mL-1) and medium (106 cells mL-1) algae concentration, respectively. This uptake corresponds to a reduction in algal concentration in seawater of 19.6 ± 9.4% and 12.2% ± 14.6%, respectively. The algal uptake for the highest (107 cells mL-1) Nannochloropsis sp. concentration used during the phagocytic assay was inconclusive (Figure 3A) presumably because algal cells might have clumped together and the sponge became oversaturated during the incubation.
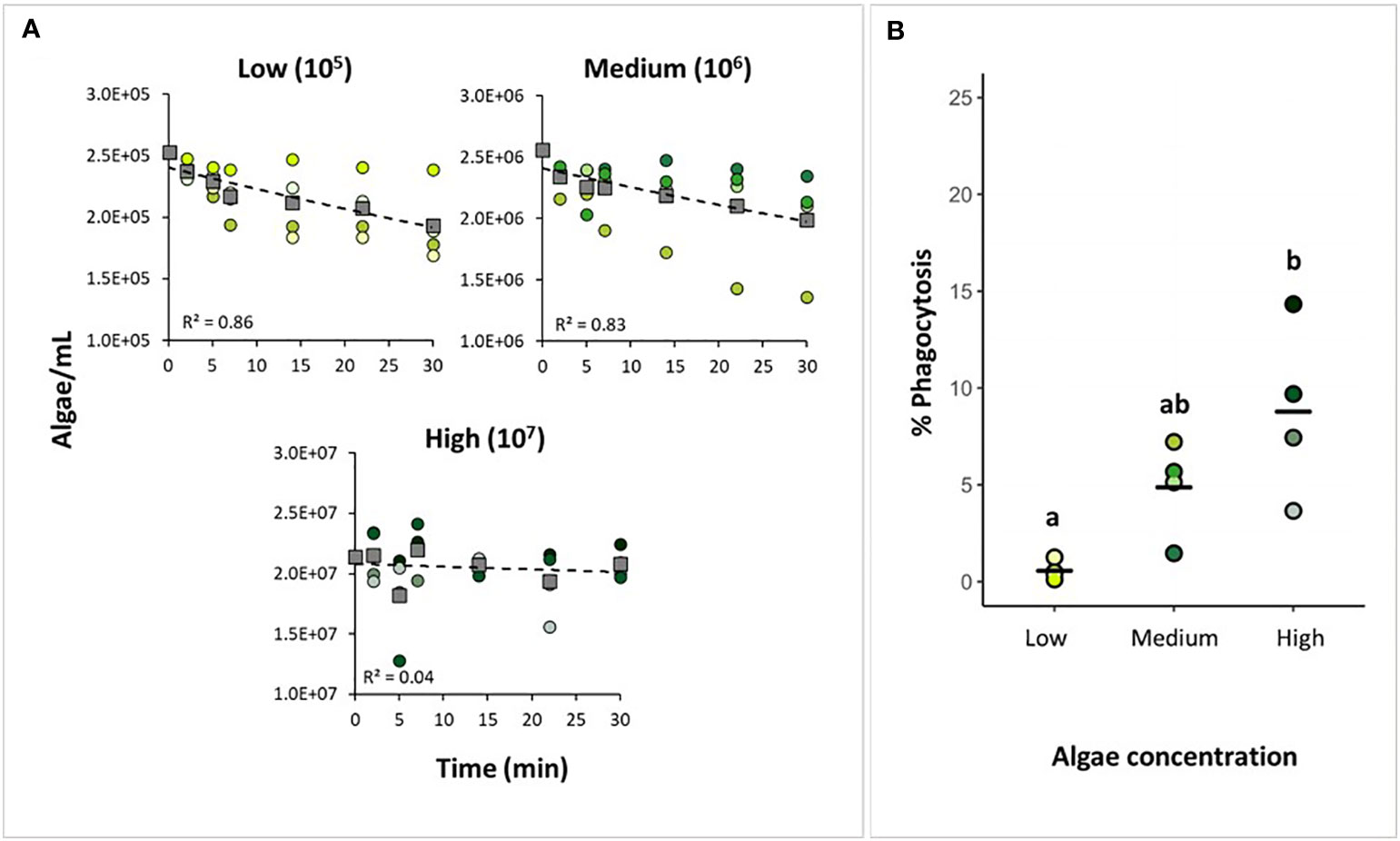
Figure 3 Testing tracer concentration for the phagocytic assay. (A) Algal uptake by H. panicea individuals incubated at three algal concentrations (105, 106, and 107 algae mL-1) based on water sample analyses. Dots of the same color: biological replicates (n = 4 per treatment). Squares and dash lines: averaged data fitted to an exponential model. (B) Estimates of phagocytic active sponge cells based on FACS analyses. Bold line: average for the 4 biological replicates. Treatments marked with different letters are significantly different at α=0.05.
The percentage of phagocytic cells was 0.6 ± 0.4% for the low concentration (105 cells mL-1), 4.9 ± 2.1% for the medium concentration (106 cells mL-1), and 8.8 ± 3.9 for the high-concentration (107 cells mL-1) (Figure 3B). There was a significant effect of algal concentration in seawater on the percentage of phagocytic cells in H. panicea (ANCOVA, F = 11.03, p < 0.01; df = 2). The high algal concentration yielded on average an approx. 15-fold increase in the percentage of phagocytic cells compared to the lowest concentration. At the medium concentration, the percentage of phagocytic cells appeared to yield on average an approx. 8-fold and 2-fold increase compared to the low and high concentration, respectively. However, the medium treatment was neither statistically different from the low (p = 0.09) nor the high (p = 0.10) algal treatment. (Figure 3B). For the high concentration treatment, it is worth noting that even though the algal uptake estimates based on flow cytometry measurements were inconclusive (Figure 3A), we did detect an increase in phagocytic activity, and observed incorporation of algae into the sponge cells (Figure 3B).
Assessing timing of algal phagocytosis
We performed a pulse-chase experiment using Nannochloropsis sp. to assess at what time point after presenting the algae to H. panicea yield the highest percentage of phagocytic sponge cells. The total number of algal cells taken up by the sponges after the 30 min pulse period was 3.3 ± 2.5 x 105 cells, which translates to a 13.7 ± 10.6% algal reduction (Figure 4A). The percentage of phagocytic cells was 4.9 ± 2.1% after 0 min, 2.2 ± 0.78% after 30 min, and 2.0 ± 0.4% after 150 min chase-period (Figure 4B). The percentage of phagocytic cells was positively related with algal uptake (ANCOVA, F = 6.53, p = 0.03; df = 1). After removing the effect of particle uptake on the response variable, phagocytic activity significantly decreased during the chase period (i.e., phagocytosis decreased with time after initial particle exposure) (ANCOVA, F = 9.80, p < 0.01; df = 2). The phagocytic activity peaked at 0 min chase, then significantly decreased by approx. 50% at 30 min chase, but there was no further significant decline in the percentage of phagocytic cells after 150 min chase.
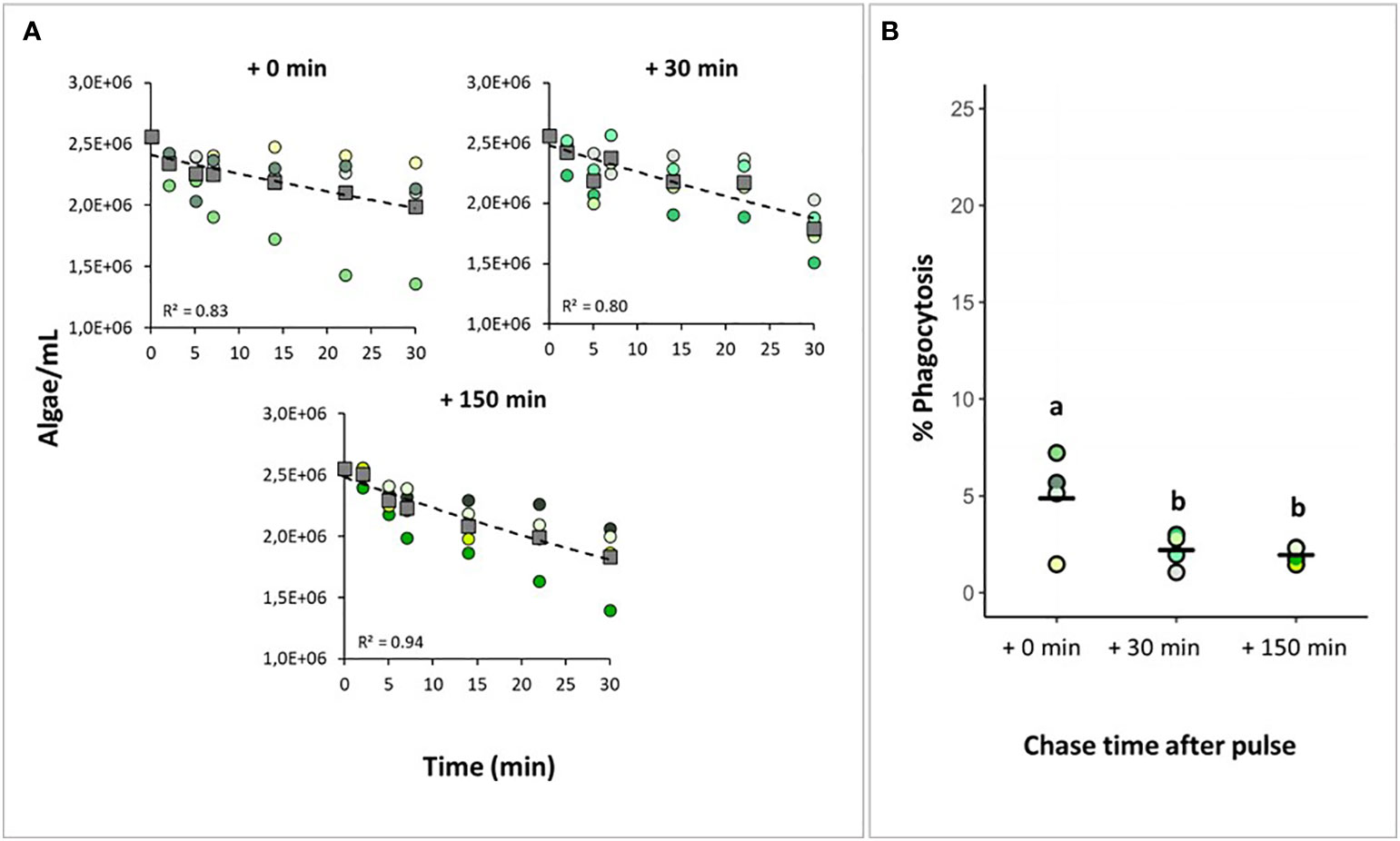
Figure 4 Assessing the starting time of algal phagocytosis. H. panicea individuals were incubated for 30 min (pulse period) with an initial algal concentration of 106 algae mL-1 and sampled after three chase periods (+ 0 min, + 30 min, and + 150 min). (A) Algal uptake by sponges during the pulse period based on flow cytometry analysis of water samples taken at time intervals. Dots of the same color: biological replicates (n = 4 per treatment). Squares and dash lines: averaged data fitted to an exponential model. (B) Estimates of phagocytic active sponge cells based on FACS analyses. Bold line: average for the 4 biological replicates. Treatments marked with different letters are significantly different at α=0.05.
Bacteria and latex beads as tracers of phagocytosis
When bacteria and beads were provided as tracers, the total number of particles taken up by the sponge was 2.7 x 104 ± 7.9 x 103 bacteria and 2.4 x 105 ± 2.3 x 105 beads, corresponding to a 31.0 ± 8.6% and 14.3 ± 16.0% reduction of particle concentration in seawater after each assay, respectively (Figure 5A). Particle uptake varied among H. panicea individuals. Some sponge individuals only reduced the particle concentration by 1 to 4%, while other individuals decreased the concentration by 22 to 37%. The differences in particle uptake between individuals could derived from variations in the filtration activity of the sponges. It is known that filtration rates of H. panicea can considerably change over time due to osculum contractions [e.g (Riisgård et al., 2016; Goldstein et al., 2020)], and we cannot discard this to be the cause of the observed variations. Despite this, there was overall no significant difference in particle uptake between bacteria, beads, and algae (ANOVA, F = 0.57, p = 0.59; df = 2). In the bacteria experiment, sponge control samples (i.e., individuals incubated without bacteria) showed some events in the gate used for quantifying cells with incorporated fluorescent signals (Figure 1C). Based on our microscopy observations using the filter set 43 HE DsRed (538-562 nm), the signal may correspond to natural auto-fluorescent granules present in the H. panicea cells. After subtracting the events estimated in the control samples, the percentage of cells phagocytizing bacteria in the treated sponges was 8.0 ± 2.3% (Figure 5B). In the assay with the beads, we identified a distinct population of phagocytic sponge cells consisting of at least three subpopulations (Figure 1D). Fluorescent microscopy of the sorted cells within these three subpopulations revealed differences in the number of beads phagocytized per sponge cell. Most cells from the lower subpopulation in the y-axis (i.e., green fluorescence) showed incorporation of one bead, while cells from the medium and highest subpopulation contained 2-3 and > 3 beads per cell, respectively (Figure 1D). Overall, the percentage of sponge cells phagocytizing beads was 11.6 ± 7.6% (Figure 5B). When comparing the phagocytic activity of H. panicea individuals exposed to bacteria, beads, and algae, we detected no significant difference between tracers after controlling for particle uptake (ANCOVA, F = 1.52, p = 0.28; df = 2).
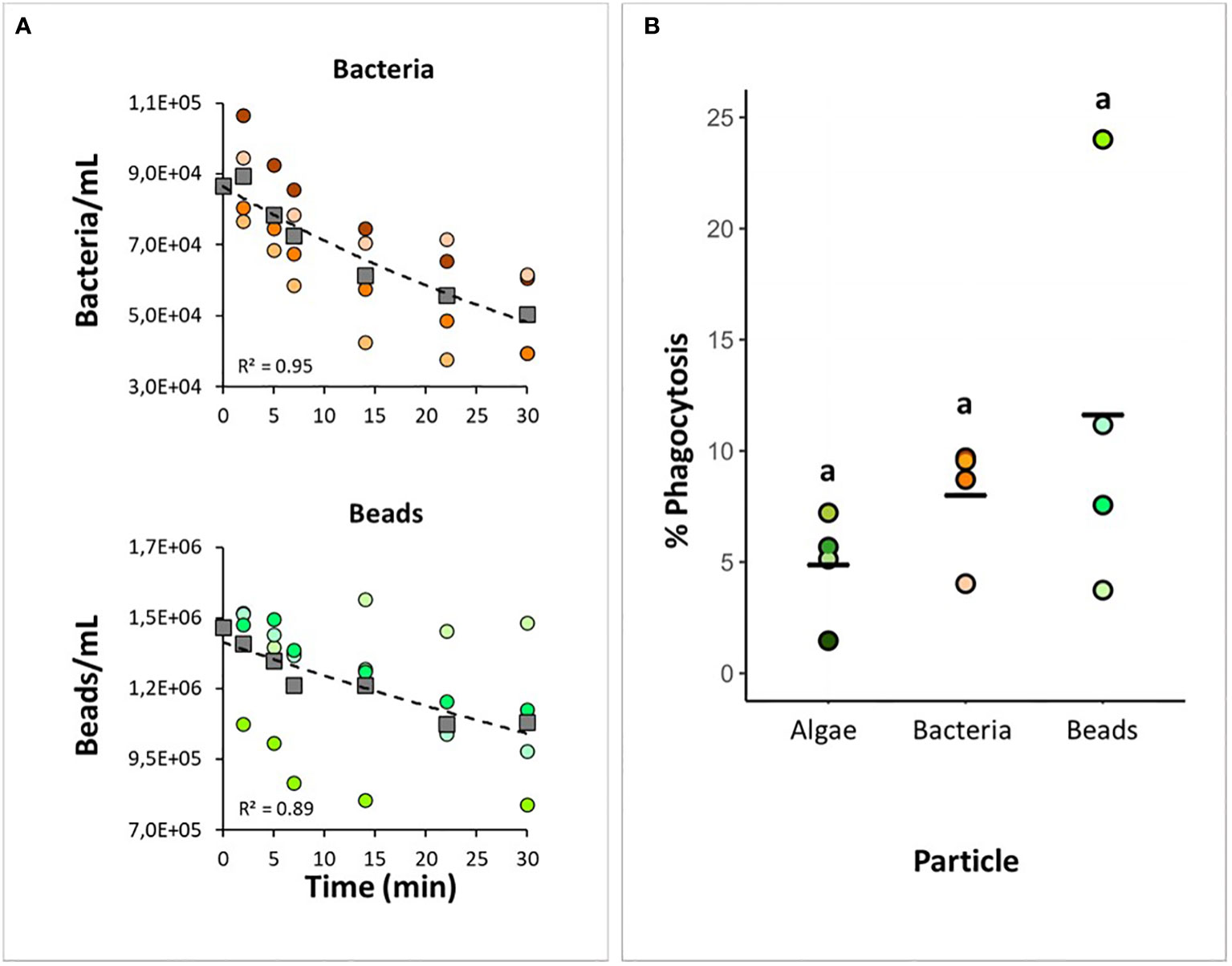
Figure 5 Testing bacteria and beads as tracers for the phagocytic assay. H panicea individuals were incubated for 30 min with TAMRA-stained bacteria (Vibrio sp.) and fluorescent latex beads (1 µm) at an initial concentration of around 105 to 106 particles mL-1. (A) Bacterial and bead uptake by sponges during the incubation period based on flow cytometry analysis of water samples taken at time intervals. Dots of the same color: biological replicates (n = 4 per treatment). Squares and dash lines correspond to the averaged data fitted to an exponential model. (B) Estimates of phagocytic active sponge cells in comparison to the algal assay. Bold line: average for the 4 biological replicates. There was no significant difference between samples.
Cellular insights into the phagocytic process
Fluorescent microscopy of H. panicea dissociated cells revealed differences in terms of morphology and size of the sponge cells engaged in phagocytosis (Figures 6A–C). In general, we observed relatively small cells (approx. 5 µm) with a clear nucleus of around 2 µm and a flagellum of various length, which we presume are choanocytes. Medium to big cells (approx. 6 to 10 µm, and 10 to 12 µm, respectively) with a nucleus of around 5 µm, and no flagella that resemble archaeocyte-like cells were also visible. For all particle types (i.e., algae, bacteria, and beads), 39 to 50% of the cells performing phagocytosis were choanocyte-like cells (Table S1). However, medium to big phagocytic cells (10 to 12 µm) engaged more often in algal phagocytosis (29 and 18% of total phagocytic cells, respectively) compared to bacteria (13 and 4%, respectively) and beads (10 and 3%, respectively) (Table S1).
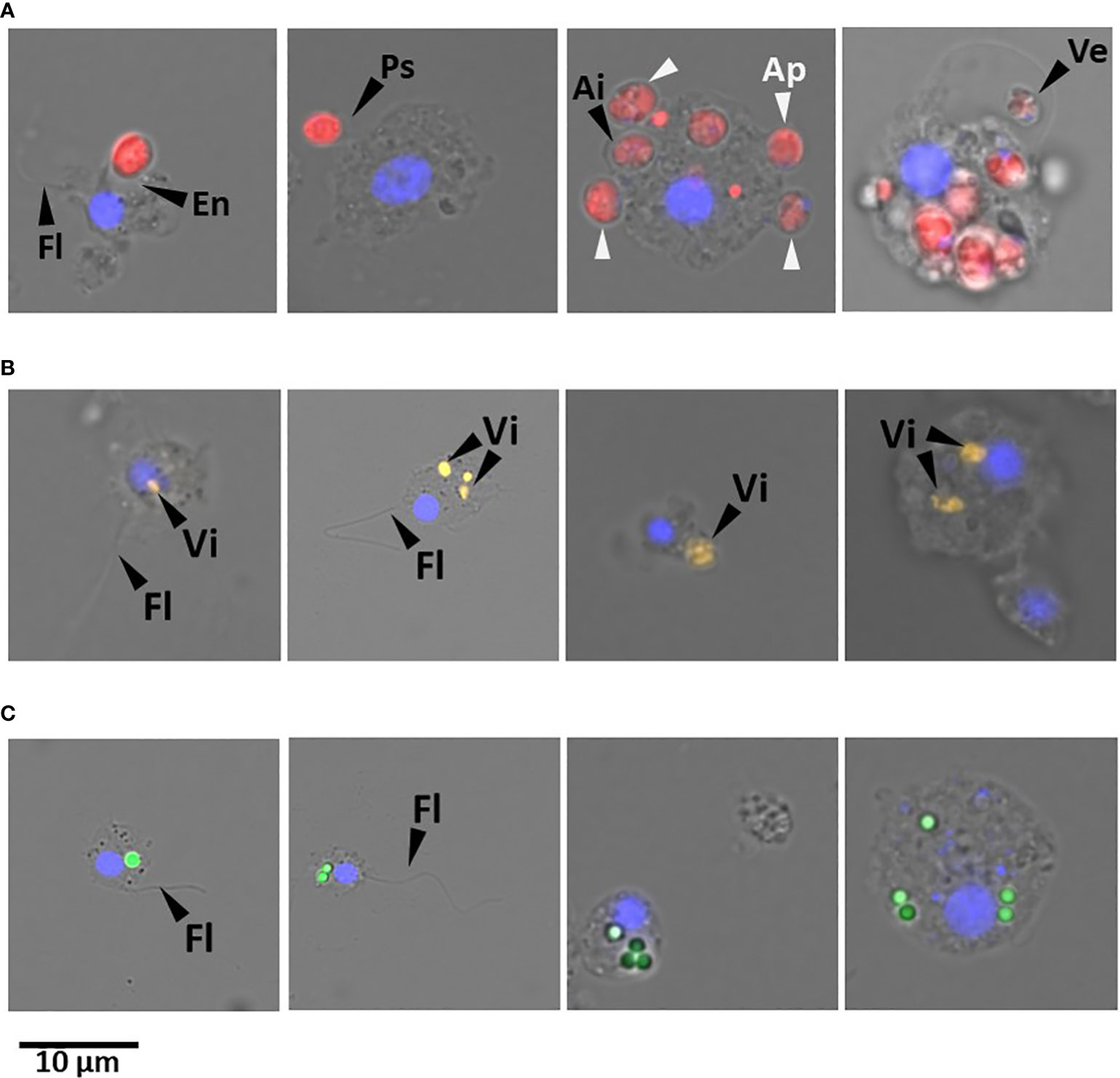
Figure 6 Representative fluorescent microscopy pictures of cells dissociated from H. panicea tissue after phagocytic assays using different tracers. Sponge cells phagocytizing (A) microalgae (Nannochloropsis sp.; red). Flagellum (Fl); evagination (Ev) and pseudopodium (Ps) of sponge cell membrane for incorporating algae; algal cells in the periphery (Ap) and internalized (Ai) in the sponge cell; and vesicle (Ve) with algal cell. (B) TAMRA-stained Vibrio sp. (Vi; yellow) and (C) fluorescent latex beads (1 µm; green) internalized in different cell types. Sponge cell nuclei (blue) stained with DAPI. Scale bar: 10 µm in all cases.
In the pulse-chase experiment with alga incubations, we identified potential early stages of algal phagocytosis at + 0 min chase, in which the sponge cell membrane evaginates, protrudes into pseudopodia-like structures, or extends vesicles to incorporate Nannochloropsis sp. cells (Figure 6A). Furthermore, the percentage of phagocytic choanocyte-like cells showed a 3 to 7-fold significant reduction (Table S1) one and three hours after the assay started (i.e., + 30 min and + 150 min time point, respectively). In contrast, the proportion of archaeocyte-like cells continued to increase significantly by up to 30% at + 150 min (Table S1).
TEM observations on H. panicea tissue samples from the algal assays provided additional evidence of Nannochloropsis sp. incorporation and processing into the sponge cells. Intact algal cells were observed in the extracellular matrix of the sponge (i.e., the mesohyl), exhibiting characteristic structures like the cell wall, thylakoids, and vacuole (Figure 7A). Whole Nannochloropsis sp. cells internalized in sponge cells were detected in the inspected samples (Figures 7B, C). We also observed phagosome-like structures with potential remnants of cell wall and thylakoids, which we presume, is the result of algal digestion (Figures 7D–F).
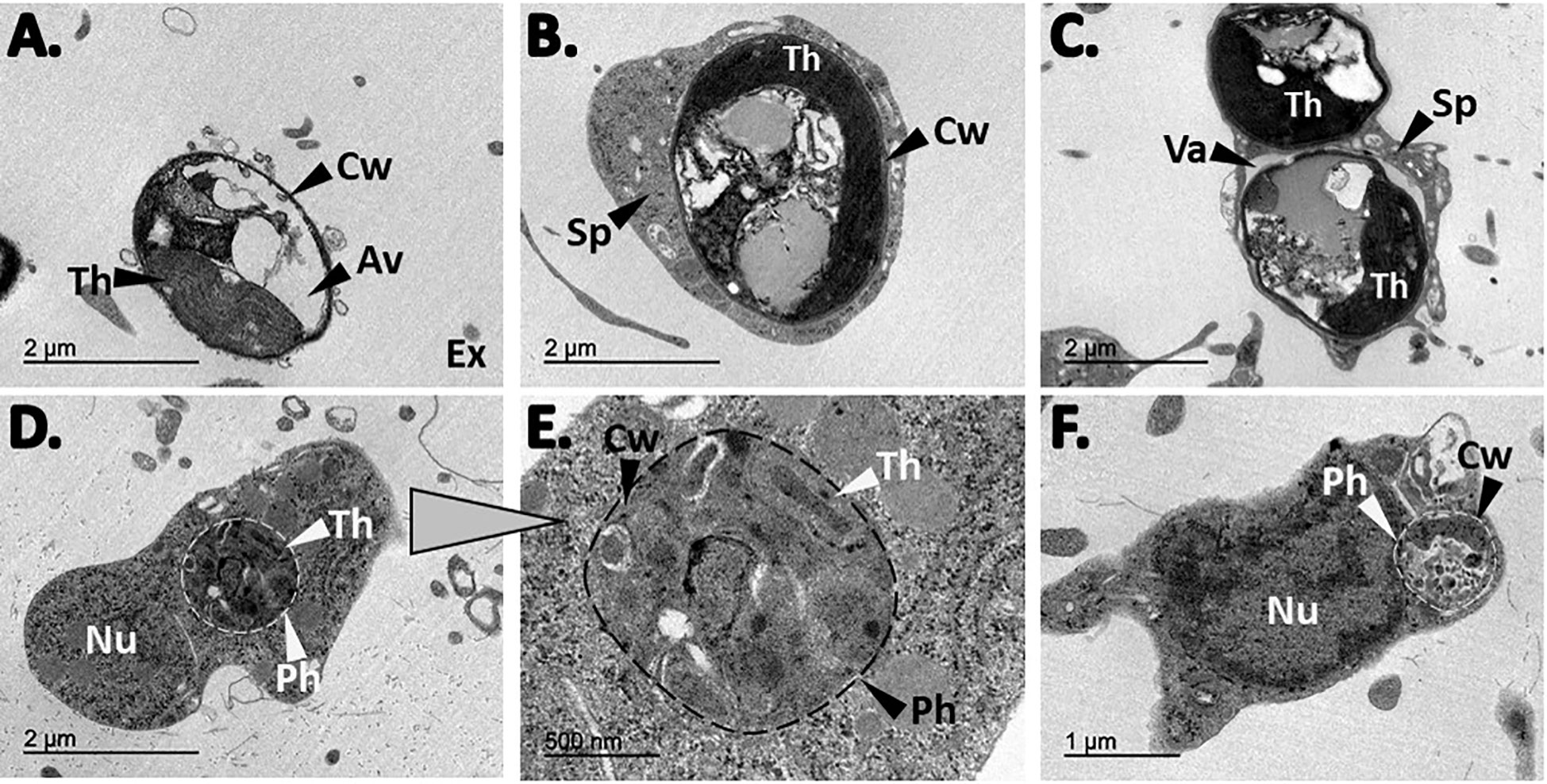
Figure 7 TEM observations from H. panicea tissue after algal phagocytosis. (A) Free, intact Nannochloropsis sp. cell in the sponge extracellular (Ex) matrix. The cell wall (Cw), thylakoids (Th), and the algae vacuole is visible. (B, C) sponge cells (Sp) with one and two internalized algal cells, respectively. One of the algae is inside a vacuole (Va). (D–F) Potential algal digestion in which possible remnants of algal cells are observed in phagosome-like structures (Ph). Nucleus (Nu).
Discussion
The aim of this study was to establish a high-throughput method for quantifying particle incorporation into the cells of whole sponge individuals by coupling live sponge incubations using fluorescent tracers, with sponge cell (host) dissociation, FACS analysis, and microscopy inspections. We adopted an in-vivo rather than an in-vitro cellular assay approach to replicate and accurately predict, as much as possible, the natural behavior of cells in H. panicea. In-vitro work in sponges is challenging mainly because their cells reaggregate fast, and the use of chemicals to induce disaggregation can reduce cell viability and inhibit essential cellular processes (Pomponi, 2006). With the established in-vivo phagocytic assay, we were able to successfully quantify the incorporation of algae, bacteria, and beads into H. panicea cells, and identify different sponge cell types involved in the process of particle incorporation.
Even though the incorporation of the different tracers into the sponge cells was evident via z-stack images (Figure 2), we cannot discard that some of the particles were not internalized but rather attached to the cell’s surface. Yet, our cell dissociation protocol applies CMFASW which, besides preventing sponge cells from aggregating, should also minimize unspecific cell-cell interactions. Thus, we speculate that the cells identified as phagocytic were indeed in the process of internalizing particles (e.g., Figure 6A). Besides phagocytosis, there are other endocytic pathways (e.g., micropinocytosis and macropinocytosis) by which particles can be internalized into cells. Based on the size of the particles we used during our assays (1µm to 3µm in diameter), internalization must happen either via phagocytosis or macropinocytosis. We acknowledge that evidence regarding the evolvement of ligand-receptors, which is the main difference between these two endocytic processes, is not provided in the manuscript. In the sponge research field, particle internalization into the cells has been, to our knowledge, extensively termed and described as phagocytosis (e.g., Imsiecke, 1993; Leys and Eerkes-Medrano, 2006; Maldonado et al., 2010; Leys et al., 2018) based on microscopy observations. Transcriptomic data in H. panicea and in other sponge species have already pointed to the activation of phagocytic-related genes upon sponge encounter with microbes or microbial elicitors (Yuen, 2016; Pita et al., 2018; Geraghty et al., 2021; Schmittmann et al., 2021). Indeed, it is very plausible that phagocytosis is the mechanism by which particles enter the sponge cells. Unlike other invertebrates (e.g., amoeba, corals, mussels, insects, etc.), detailed studies on the phagocytic process have not been conducted in sponges yet. We are convinced that the developed assay in H. panicea has enormous potential to be used as a tool to investigate how sponge phagocytosis works by combining, for example, dyes for lysosomal activity, actin filaments, and oxidative stress, among others, which could even be coupled with FACS analysis to resolve subpopulations of phagocytic sponge cells [as done e.g., in cnidarians (Snyder et al., 2021)].
Tracer concentration and phagocytic activity
In each experiment, we quantified clearance of particles as indicator for sponge filtering activity (as in (Leys et al., 2011; Mueller et al., 2014; Riisgård et al., 2016; Lüskow et al., 2019)). We observed that an increase in tracer concentration resulted in a linear increase in algal removal by H. panicea. This is consistent with previous findings of concentration-dependent particle removal rates in sponges (e.g (Yahel et al., 2006; Mueller et al., 2014)). In our algal assays, most of the sponge individuals were actively filtering the tracer during the 30 min incubations with low and medium Nannochloropsis sp. concentrations (105 and 106 cells mL-1, respectively), as the algal abundance declined exponentially over time. The algal uptake for the highest (107 cells mL-1) Nannochloropsis sp. concentration used during the phagocytic assay was inconclusive. Algal cells might have clumped hindering the detection of changes in algal concentration during the assay and thus, we recommend working with particle concentrations below 107 cells mL-1. The overall (mean ± SD) algal clearance rate of our sponges (1.5 ± 0.2 mL water mL sponge-1 min-1) is also in line with previous reports for H. panicea (1.5 to 1.9 ± 0.8 to 1.1 mL water mL sponge-1 min-1 (Scheffers et al., 2004)). Filtration rates of H. panicea can considerably vary in laboratory conditions (Riisgård et al., 2016), and even though we observed variation in the filtration activity of certain individuals, our incubation setup for the phagocytic assay proved overall to ensure the sponge filtration activity, which is an important factor to control.
We further hypothesized initial tracer concentration to positively affect the percentage of phagocytic active cells since this relation has been observed in other phagocytic assays (e.g (Lindner et al., 2020)). Indeed, we found a positive relation between algal concentration in the seawater and the percentage of sponge cells that incorporated particles. Studies in cnidaria (Bucher et al., 2016), fish (Li et al., 2006), and mice (Tartaro et al., 2015) also reported phagocytosis to increase with particle concentration (i.e., when using fluorescent beads, bacteria and algae). In our assay, increasing the algal concentration from 105 to 107 algae mL-1 (i.e., low to high concentration treatment) resulted in a significant 15-fold increase in the sponge phagocytic activity. While the phagocytic activity seemed to also increase when increasing the concentration from low to medium (8-fold), and from medium to low (2-fold), this trend was not significant. This indicates that sponge cells can adjust their phagocytic activity depending on the number of algal cells they encounter. These results suggest that an algal concentration of approx. 106 cells mL-1 is optimal for performing algal phagocytosis assays in H. panicea.
Onset of phagocytosis
The observations from our pulse-chase experiment revealed incorporation of algae into the sponge cells at + 0 min chase, then after 30 min chase phagocytic activity significantly decreased 2-fold, but then remained constant (i.e., + 150 min chase) (Figure 7B). After 30 min exposure to the algae, during the chase phase, sponges were transferred to particle-free water, yet our FACS results showed that approx. 75% of the algae that were taken up by the sponge were found as “intact algae” (i.e., present in the sponge fraction but not incorporated in sponge cells) at + 0 min and + 30 min chase (Figures 7A, S2). Our dissociation protocol was designed to be gentle enough to keep sponge cells intact and this was evident since we observed delicate structures like flagella (Figure 6). We suggest that intact algae are either algae that were loosely attached to sponge cells (e.g., Figure 6A) and got detached during the sponge cell dissociation process or during the FACS. Interestingly, at + 150 min chase the percentage of intact algae decreased to 56% (Figure S2), supporting that intact Nannochloropsis sp. cells were taken up by the sponge during the pulse chase and were in the process of incorporation during the chase phase.
We propose that the decrease in phagocytic activity between + 0 min chase and + 30 min chase is the consequence of algal digestion (e.g., Figures 7D–F), and during this process, internalization of intact algae might decrease. Once digestion is completed the process of internalization could be resumed and hence, the maintenance of phagocytic activity between + 30 min and + 150 min chase. In freshwater sponges, algae internalization and translocation between sponge cells occurs within minutes after feeding, whereas digestion takes a couple of hours. For example, in young individuals of Spongilla lacustris hatched from gemmules, extensive digestion of algal cells by phagocytes was evident 5 h after feeding (Imsiecke, 1993). Likewise, in algal-free (i.e., “aposymbiotic”) Ephydatia muelleri intracellular algal symbionts were observed to be internalized in archaeocytes 4 h post-infection (Geraghty et al., 2021). Our findings together with the above observations reveal that algal phagocytosis in sponges initiates within minutes after particle exposure and takes a couple of hours to be completed. We further suggest that the fact that some Nannochloropsis sp. cells were taken up by H. panicea but not internalized, due to potential digestion events of other algal cells, indicates a decoupling in time between algal uptake, internalization, and digestion by sponge cells.
Algae, bacteria, and beads as tracers for the phagocytic assay
The percentage of phagocytic cells estimated with our assay for H. panicea using algae, bacteria, and beads as tracer particles ranged overall between 5 to 24%. It is difficult to compare our estimates to other studies since, to our knowledge, similar quantifications of phagocytic sponge cells have not been done yet. Thus, we have compared our results to well-established models for phagocytosis. For example, amoebal phagocytic activity of Dictyostelium discoideum upon exposure to GFP-tagged Legionella pneumophila was estimated to be < 2% based on FACS counts (Hägele et al., 2000), whereas in Acanthamoeba castellanii microscopy counts showed 15-35% of amoeba cells phagocytizing E. coli (Nguyen et al., 2014). In-vitro phagocytosis experiments in cnidarians estimated with FACS percentages of coral and sea anemone phagocytic cells between 2.2 to 7.8% and 9 to 18% after feeding cells with 1 µm latex beads and fluorescently labeled Escherichia coli, respectively (Rosental et al., 2017; Snyder et al., 2021). Although the aforementioned assays diverge from ours in the sense that they were performed in cell suspensions, under different conditions, and not in whole sponge individuals, the reported phagocytic activity in those studies is comparable to our estimates in H. panicea.
Our data suggest that phagocytosis differs depending on the sponge filtration activity (i.e., particle uptake) and on the type of particle used during the assay. The percentage of phagocytic cells tends to increase with increased particle uptake in all tracer types, but this relationship seems to be tracer-specific (Figure S3). The increase in phagocytic cells appeared to be faster for beads, followed by bacteria, and at a lower degree for algae (Figure S3). The beads tend to accumulate in the cells (Figures 1D, 6C) as they cannot be digested by the sponge. For H. panicea, we speculate that once the sponge cells are saturated with beads (≥ 5 beads per cell) new cells need to engage in the phagocytic process, which could explain the steeper increase in bead incorporation (Figure S3). In E. muelleri high accumulation of beads in the choanocytes occurs within 13 min of exposure to the particle, and after 15 min bead incorporation extends to the archaeocytes (Funayama et al., 2005). Thus, the fast uptake and high number of sponge cells with incorporated beads may be the result of the participation of more choanocytes within the first 30 min of exposure to the particles.
In the case of bacteria, the number of Vibrio cells incorporated per sponge cell was difficult to resolve because the resolution acquired with the fluorescent microscope was not high enough, and it was difficult to accurately distinguish our fluorescent bacteria from natural fluorescence occurring in the sponge cells. However, FACS allowed us to subtract the natural fluorescence based on the signal in control samples and we estimate that an approx. 50% increase in bacterial uptake would yield a 50% increase in the percentage of phagocytic cells (Figure S3). NanoSIMS experiments in the marine encrusting sponge Halisarca caerulea using isotopically labeled bacteria indicate that bacteria are rapidly phagocytized by choanocytes (Hudspith et al., 2021). Within 15 min after feeding the sponge with labeled bacteria, 90 to 100% of the choanocyte cells incorporated bacteria and individual bacteria cells were visible in intracellular vesicles. Moreover, bacterial digestion and assimilation processes can vary depending on the bacteria encountered by the sponge. For example, Hymeniacidon perlevis can rapidly process E. coli, whereas assimilation of Vibrio anguillarum is more laborious. Vibrio cells are semi-digested by choanocytes 4 h after feeding and digestion is further completed by amoeba-like cells (Maldonado et al., 2010).
In the case of the algae, the percentage of sponge cells engaged in phagocytosis tended to be lower compared to the other tracers, however this trend was not significant. Algal phagocytosis only increased by 1.5% despite an approx. 30% increase in algal uptake (Figure S3). It is plausible to suggest that the bigger size and cell wall structure of the Nannochloropsis cells requires more time for the sponge to digest the algae (see previous section for details). Overall, our results indicate that the phagocytic response of H. panicea depends on the nature of the particle the sponge encounters. Indigestible particles (i.e., latex beads) trigger a faster incorporation into the sponge cells (Figure S3) and likely involved different cell types (Figure 6C). Whereas for microorganisms that can be digested (i.e., bacteria and algae), phagocytosis seems to vary with particle size. Big algal cells are incorporated at a slower rate than smaller bacterial cells (Figure S3), and a higher rate of translocation between sponge cell types is evident (Table S1).
Prospects of the phagocytosis assay to investigate microbial discrimination by sponges
The established phagocytosis assay is a high-throughput method for quantifying particle incorporation in cells of whole sponge individuals, which can be applied to a variety of sponge species, and even other marine invertebrates. Other established methodologies like NanoSIMS can provide microscopic details into cellular mechanisms, but only a small portion of the organisms can be analyzed, and the methodology is expensive and not easily accessible. Although our assay may require the FACS for initial validations and certain applications, estimates of phagocytic cells could be done in a normal flow cytometer, an instrument that is common in most marine institutes and which can even be used in research vessels.
Furthermore, our assay could be used to investigate the mechanistic bases of phagocytosis on microbe recognition and differentiation by sponges. For example, the in-vivo assay can be used to compare sponge phagocytosis of symbiont vs. non-symbiont bacteria and would aid to unveil if sponges discriminate “friend” from “foe” through this conserved cellular mechanism. It is known that sponge-associated bacteria express ankyrin-repeat proteins which can interfere with phagocytosis (e.g., (Nguyen et al., 2014; Jahn et al., 2019)). Utilizing the established assay to query whether bacteria expressing ankyrins evade phagocytosis would provide further evidence on the role of ankyrin proteins as mediators of symbiont recognition by sponges. Moreover, combining the developed in-vivo assay with cell markers would aid to resolve which types of sponge cells activate a phagocytic response upon bacterial encounter, and whether the population of phagocytic cells change when the sponge is presented with different types of microbes.
Conclusion and perspectives
Here we present a novel, in-vivo assay to quantify phagocytic active sponge cells in whole H. panicea individuals. Coupling sponge incubations with sponge cell (host) dissociation and FACS analysis proved to be a suitable approach to track fluorescent tracers (i.e., algae, bacteria, and beads) from the surrounding seawater into the sponge cells, and to quantify the relative proportion of H. panicea cells incorporating those tracers. The percentage of cells incorporating particles ranged between 5 to 24% and was not significantly different between algae, bacteria and beads. The number of particles filtered by H. panicea and the degree of digestion by the sponge cells seemed to be tracer-dependent. Furthermore, our method gave us hints on the process of particle incorporation itself. Particle incorporation by sponges is fast, initiating within minutes and concluding within < 60 min of exposure to the tracers, and involves different cell types (presumably choanocyte- and archaeocyte-like cells) and possible translocation between cells based on our microscopy observations.
Sponges as ecologically relevant basal metazoans are ideal models to investigate the evolution of host-microbe interactions. We envision our high-throughput assay as a tool to query whether sponges process different microbes (e.g., food, symbiont, and pathogens) distinctively through the conserved cellular mechanism of phagocytosis. Based on our observations, we speculate that particle discrimination might occur upon internalization and the difference might rely on how each particle type is processed (e.g., digested or expelled). Hence, we hypothesize that symbionts are incorporated and/or digested to a lesser extent by sponge cells compared to non-symbionts, and that certain microbial structures (e.g., ankyrins) mediate phagocytosis evasion. Using the developed assay to compare phagocytosis of symbiont vs. non-symbiont (e.g., bacterioplankton) bacteria, as well as bacteria encoding ankyrin proteins, would expand on previous works to better understand the role of phagocytosis in sponge-microbe interactions. Furthermore, to fully characterize the diversity of sponge cells capable of engaging in the phagocytic process developing of cellular marker for marine sponges is needed (e.g., for detecting lysosomal activity and reactive oxygen species (ROS) production (Snyder et al., 2021)). Fluorescent in-situ hybridization probes (e.g (Musser et al., 2021)) together with our experimental approach and FACS analysis could aid to identify sponge cell types responsible for different steps of the phagocytosis process, and to investigate if their activity changes after encountering different microbes or environmental stressors (e.g., elevated temperatures, ocean acidification, sedimentation) impair this process. As phagocytosis is an essential component of the innate immune system responsible to recognize and protect against foreign particles and damage cells (Uribe-Querol and Rosales, 2020), we expect sponge phagocytic activity to increase upon exposure to foreign bacteria and under environmental conditions inducing cellular damage. Lastly, coupling single-cell RNA sequencing of populations of phagocytic cells could shed light on the molecular machinery behind sponge phagocytosis. Adapting our assay to other early evolutionary metazoans (e.g., the freshwater sponge .E. muelleri., the sea anomes Nematostella sp. and Aiptasia sp., and the Mytilid mussel Bathymodiolus sp.) and adopting comparative analyses could further help to better understand this conserved cellular mechanism and may therefore have the potential to unravel the role of phagocytosis in basal animal-microbe interactions.
Data availability statement
Data is publicly available in Pangea at https://doi.org/10.1594/PANGAEA.956281.
Author contributions
AM-G, LP and UH conceived the idea. AM-G planned and conducted the experiments. KB and AM-G performed the FACS analysis and fluorescent microscopy. LP performed the TEM inspections. The initial draft of the manuscript was written by AM-G and UH. All authors contributed to improving the article and approved the submitted version.
Funding
UH was supported by the DFG (“Origin and Function of Metaorganisms”, CRC1182-TP B01) and the Gordon and Betty Moore Foundation (“Symbiosis in Aquatic Systems Initiative”, GBMF9352). LP received supported by “la Caixa” Foundation (ID 10010434), co-financed by the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement No 847648), fellowship code is 104855. Additional funding support to LP was provided by the “Severo-Ochoa Centre of Excellence” accreditation (CEX2019-000928-S). This is a contribution from the Marine Biogeochemistry and Global Change research group (Grant 2021SGR00430, Generalitat de Catalunya).
Acknowledgments
We are grateful to Dr. Lara Schmittmann for the field collection support, and Dr. Ben Mueller for helpful discussions. We further acknowledge Sabrina Jung for technical assistance in the lab, Andrea Hethke for support with the cell dissociations, Fabian Wendt for technical support with the algae culture and experimental setup logistics, Janis Müller for his experimental support. We thank the TEM-SEM Electron Microscopy Unit from Scientific and Technological Centers (CCiTUB), Universitat de Barcelona, for their support and advice on TEM technique, and the International Max Planck Research School for Evolutionary Biology for supervision effort of AM-G.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1176145/full#supplementary-material
References
Al-Khodor S., Price C. T., Habyarimana F., Kalia A., Abu Kwaik Y. (2008). A Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Mol. Microbiol. 70 (4), 908–923. doi: 10.1111/j.1365-2958.2008.06453.x
Barthel D. (1988). On the ecophysiology of the sponge Halichondria panicea in Kiel Bight. II. Biomass, production, energy budget and integration in environmental processes. Mar. Ecol. Prog. Ser. 43, 87–93. doi: 10.3354/meps043087
Bucher M., Wolfowicz I., Voss P. A., Hambleton E. A., Guse A. (2016). Development and symbiosis establishment in the cnidarian endosymbiosis model Aiptasia sp. Sci. Rep. 6, 1–11. doi: 10.1038/srep19867
Busch K., Wurz E., Rapp H. T., Bayer K., Franke A., Hentschel U. (2020). Chloroflexi dominate the deep-sea golf ball sponges craniella zetlandica and craniella infrequens throughout different life stages. Front. Mar. Sci. 7, 1–12. doi: 10.3389/fmars.2020.00674
De Goeij J. M., Van Den Berg H., Van Oostveen M. M., Epping E. H. G., Van Duyl F. C. (2008). Major bulk dissolved organic carbon (DOC) removal by encrusting coral reef cavity sponges. Mar. Ecol. Prog. Ser. 357, 139–151. doi: 10.3354/meps07403
De Goeij J. M., Van Duyl F. C. (2007). Coral cavities are sinks of dissolved organic carbon (DOC). Limnol. Oceanogr. 52 (6), 2608–2617. doi: 10.4319/lo.2007.52.6.2608
Díez-Vives C., Moitinho-Silva L., Nielsen S., Reynolds D., Thomas T. (2017). Expression of eukaryotic-like protein in the microbiome of sponges. Mol. Ecol. 26 (5), 1432–1451. doi: 10.1111/mec.14003
Fieseler L., Horn M., Wagner M., Hentschel U. (2004). Discovery of the novel candidate phylum “Poribacteria”in marine sponges. Appl. Environ. Microbiol. 70 (6), 3724–3732. doi: 10.1128/AEM.70.6.3724-3732.2004
Funayama N., Nakatsukasa M., Hayashi T., Agata K. (2005). Isolation of the choanocyte in the fresh water sponge, Ephydatia fluviatilis and its lineage marker, Ef annexin. Dev. Growth Differ. 47 (4), 243–253. doi: 10.1111/j.1440-169X.2005.00800.x
Gasol J. M., Del Giorgio P. A. (2000). Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci. Mar. 64 (2), 197–224. doi: 10.3989/scimar.2000.64n2197
Geraghty S., Koutsouveli V., Hall C., Chang L., Sacristan-Soriano O., Hill M., et al. (2021). Establishment of host-algal endosymbioses: genetic response to symbiont versus prey in a sponge host. Genome Biol. Evol. 13 (11), 1–16. doi: 10.1093/gbe/evab252
Goldstein J., Bisbo N., Funch P., Riisgård H. U. (2020). Contraction-expansion and the effects on the aquiferous system in the demosponge Halichondria panicea. Front. Mar. Sci. 7, 1–12. doi: 10.3389/fmars.2020.00113
Goldstein J., Funch P. (2022). A review on Genus halichondria (Demospongiae, Porifera). J. Mar. Sci. Eng. 10 (9), 1312.
Habyarimana F., Al-Khodor S., Kalia A., Graham J. E., Price C. T., Garcia M. T., et al. (2008). Role for the Ankyrin eukaryotic-like genes of Legionella pneumophila in parasitism of protozoan hosts and human macrophages. Environ. Microbiol. 10 (6), 1460–1474. doi: 10.1111/j.1462-2920.2007.01560.x
Hägele S., Köhler R., Merkert H., Schleicher M., Hacker J., Steinert M. (2000). Dictyostelium discoideum: A new host model system for intracellular pathogens of the genus Legionella. Cell Microbiol. 2 (2), 165–171. doi: 10.1046/j.1462-5822.2000.00044.x
Hartenstein V., Martinez P. (2019). Phagocytosis in cellular defense and nutrition: a food-centered approach to the evolution of macrophages. Cell Tissue Res. 377 (3), 527–547. doi: 10.1007/s00441-019-03096-6
Hentschel U., Usher K. M., Taylor M. W. (2006). Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 55 (2), 167–177. doi: 10.1111/j.1574-6941.2005.00046.x
Hudspith M., Rix L., Achlatis M., Bougoure J., Guagliardo P., Clode P. L., et al. (2021). Subcellular view of host–microbiome nutrient exchange in sponges: insights into the ecological success of an early metazoan–microbe symbiosis. Microbiome 9 (1), 1–15. doi: 10.1186/s40168-020-00984-w
Imsiecke G. (1993). Ingestion, digestion, and egestion in Spongilla lacustris (Porifera, Spongillidae) after pulse feeding with Chlamydomonas reinhardtii (Volvocales). Zoomorphology 113 (4), 233–244. doi: 10.1007/BF00403314
Jacobovitz M. R., Rupp S., Voss P. A., Maegele I., Gornik S. G., Guse A. (2021). Dinoflagellate symbionts escape vomocytosis by host cell immune suppression. Nat. Microbiol. [Internet]. 6 (6), 769–782. doi: 10.1038/s41564-021-00897-w
Jahn M. T., Arkhipova K., Markert S. M., Stigloher C., Lachnit T., Pita L., et al. (2019). A phage protein aids bacterial symbionts in eukaryote immune evasion. Cell Host Microbe 26 (4), 542–550.e5. doi: 10.1016/j.chom.2019.08.019
Jauslin T., Lamrabet O., Crespo-Yañez X., Marchetti A., Ayadi I., Ifrid E., et al. (2021). How phagocytic cells kill different bacteria: A quantitative analysis using Dictyostelium discoideum. MBio 12 (1), 1–13. doi: 10.1128/mBio.03169-20
Knobloch S., Jóhannsson R., Marteinsson V. (2019). Genome analysis of sponge symbiont “Candidatus Halichondribacter symbioticus” shows genomic adaptation to a host-dependent lifestyle. Environ. Microbiol. [Internet]. 22, 1462–2920.14869. doi: 10.1111/1462-2920.14869
Leys S. P., Eerkes-Medrano D. I. (2006). Feeding in a calcareous sponge: Particle uptake by pseudopodia. Biol. Bull. 211 (2), 157–171. doi: 10.2307/4134590
Leys S. P., Hill A. (2012). “The physiology and molecular biology of sponge tissues,” in Advances in Marine Biology, 1st ed (Elsevier Ltd), 1–56. doi: 10.1016/B978-0-12-394283-8.00001-1
Leys S. P., Yahel G., Reidenbach M. A., Tunnicliffe V., Shavit U., Reiswig H. M. (2011). The sponge pump: The role of current induced flow in the design of the sponge body plan. PloS One 6 (12). doi: 10.1371/journal.pone.0027787
Leys S. P., Kahn A. S., Fang J. K. H., Kutti T., Bannister R. J. (2018). Phagocytosis of microbial symbionts balances the carbon and nitrogen budget for the deep‐water boreal sponge Geodia barretti. Limnol.Oceanogr. 63 (1), 187–202.
Li J., Barreda D. R., Zhang Y. A., Boshra H., Gelman A. E., LaPatra S., et al. (2006). B lymphocytes from early vertebrates have potent phagocytic and microbicidal abilities. Nat. Immunol. 7 (10), 1116–1124. doi: 10.1038/ni1389
Lim J. J., Grinstein S., Roth Z. (2017). Diversity and versatility of phagocytosis: Roles in innate immunity, tissue remodeling, and homeostasis. Front. Cell Infect. Microbiol. 7, 1–12. doi: 10.3389/fcimb.2017.00191
Lindner B., Burkard T., Schuler M. (2020). Phagocytosis assays with different pH-sensitive fluorescent particles and various readouts. Biotechniques 68 (5), 245–250. doi: 10.2144/btn-2020-0003
Lüskow F., Riisgård H. U., Solovyeva V., Brewer J. R. (2019). Seasonal changes in bacteria and phytoplankton biomass control the condition index of the demosponge Halichondria panicea in temperate Danish waters. Mar. Ecol. Prog. Ser. 608, 119–132. doi: 10.3354/meps12785
Maldonado M., Zhang X., Cao X., Xue L., Cao H., Zhang W. (2010). Selective feeding by sponges on pathogenic microbes: A reassessment of potential for abatement of microbial pollution. Mar. Ecol. Prog. Ser. 403, 75–89. doi: 10.3354/meps08411
McMurray S. E., Johnson Z. I., Hunt D. E., Pawlik J. R., Finelli C. M. (2016). Selective feeding by the giant barrel sponge enhances foraging efficiency. Limnol. Oceanogr. 61 (4), 1271–1286. doi: 10.1002/lno.10287
Moitinho-Silva L., Nielsen S., Amir A., Gonzalez A., Ackermann G. L., Cerrano C., et al. (2017). The sponge microbiome project. Gigascience 6 (10), 1–7. doi: 10.1093/gigascience/gix077
Mueller B., De Goeij J. M., Vermeij M. J. A., Mulders Y., van der Ent E., Ribes M., et al. (2014). Natural diet of coral-excavating sponges consists mainly of dissolved organic carbon (DOC). PloS One 9 (2). doi: 10.1371/journal.pone.0090152
Musser J. M., Schippers K. J., Nickel M., Mizzon G., Kohn A. B., Pape C., et al. (2021). Profiling cellular diversity in sponges informs animal cell type and nervous system evolution. Sci. (80- ). 374 (6568), 717–723. doi: 10.1126/science.abj2949
Nguyen M. T. H. D., Liu M., Thomas T. (2014). Ankyrin-repeat proteins from sponge symbionts modulate amoebal phagocytosis. Mol. Ecol. 23 (6), 1635–1645. doi: 10.1111/mec.12384
Nyholm S. V., Graf J. (2012). Knowing your friends: Invertebrate innate immunity fosters beneficial bacterial symbioses. Nat. Rev. Microbiol. 10 (12), 815–827. doi: 10.1038/nrmicro2894
Nyholm S. V., Stewart J. J., Ruby E. G., McFall-Ngai M. J. (2009). Recognition between symbiotic Vibrio fischeri and the haemocytes of Euprymna scolopes. Environ. Microbiol. 11 (2), 483–493. doi: 10.1111/j.1462-2920.2008.01788.x
Pan X., Lührmann A., Satoh A., Laskowski-Arce M. A., Roy C. R. (2008). Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Sci. (80- ). 320 (5883), 1651–1654. doi: 10.1126/science.1158160
Pile A. J., Young C. M. (2006). The natural diet of a hexactinellid sponge: Benthic-pelagic coupling in a deep-sea microbial food web. Deep Res. Part I Oceanogr. Res. Pap. 53 (7), 1148–1156. doi: 10.1016/j.dsr.2006.03.008
Pita L., Fraune S., Hentschel U. (2016). Emerging sponge models of animal-microbe symbioses. Front. Microbiol. 7, 1–8. doi: 10.3389/fmicb.2016.02102
Pita L., Hoeppner M. P., Ribes M., Hentschel U. (2018). Differential expression of immune receptors in two marine sponges upon exposure to microbial-associated molecular patterns. Sci. Rep. 8 (1), 1–15. doi: 10.1038/s41598-018-34330-w
Pomponi S. A. (2006). Biology of the porifera: cell culture. Can. J. Zool. 84 (2), 167–174. doi: 10.1139/z05-188
Reiswig H. M. (1971). Particle feeding in natural populations of three marine demosponges. Biol. Bull. 141 (3), 568–591. doi: 10.2307/1540270
Ribes M., Coma R., Gili J. M. (1999). Natural diet and grazing rate of the temperate sponge Dysidea avara (Demospongiae, Dendroceratida) throughout an annual cycle. Mar. Ecol. Prog. Ser. 176, 179–190. doi: 10.3354/meps176179
Riisgård H. U., Kumala L., Charitonidou K. (2016). Using the F/R-ratio for an evaluation of the ability of the demosponge Halichondria panicea to nourish solely on phytoplankton versus free-living bacteria in the sea. Mar. Biol. Res. 12 (9), 907–916. doi: 10.1080/17451000.2016.1206941
Rosales C., Uribe-Querol E. (2017). Phagocytosis: A fundamental process in immunity. BioMed. Res. Int. 2017, 1–18. doi: 10.1155/2017/9042851
Rosental B., Kozhekbaeva Z., Fernhoff N., Tsai J. M., Traylor-Knowles N. (2017). Coral cell separation and isolation by fluorescence-activated cell sorting (FACS). BMC Cell Biol. 18 (1), 1–12. doi: 10.1186/s12860-017-0146-8
Rottmann M., Schröder H. C., Gramzow M., Renneisen K., Kurelec B., Dorn A., et al. (1987). Specific phosphorylation of proteins in pore complex-laminae from the sponge Geodia cydonium by the homologous aggregation factor and phorbol ester. Role of protein kinase C in the phosphorylation of DNA topoisomerase II. EMBO J. 6 (13), 3939–3944. doi: 10.1002/j.1460-2075.1987.tb02735.x
Sattler N., Monroy R., Soldati T. (2013). Quantitative analysis of phagocytosis and phagosome maturation. Methods Mol. Biol. 983, 383–402. doi: 10.1007/978-1-62703-302-2_21
Scheffers S. R., Nieuwland G., Bak R. P. M., Van Duyl F. C. (2004). Removal of bacteria and nutrient dynamics within the coral reef framework of Curaçao (Netherlands Antilles). Coral Reefs. 23 (3), 413–422. doi: 10.1007/s00338-004-0400-3
Schmittmann L., Franzenburg S., Pita L. (2021). Individuality in the immune repertoire and induced response of the sponge Halichondria panicea. Front. Immunol. 12, 1–13. doi: 10.3389/fimmu.2021.689051
Schmittmann L., Rahn T., Busch K., Fraune S., Pita L., Hentschel U. (2022). Stability of a dominant sponge-symbiont in spite of antibiotic-induced microbiome disturbance. Environ. Microbiol. 24 (12), 6392–6410. doi: 10.1111/1462-2920.16249
Sebé-Pedrós A., Chomsky E., Pang K., Lara-Astiaso D., Gaiti F., Mukamel Z., et al. (2018). Early metazoan cell type diversity and the evolution of multicellular gene regulation. Nat. Ecol. Evol. 2 (7), 1176–1188. doi: 10.1038/s41559-018-0575-6
Siegl A., Kamke J., Hochmuth T., Piel J., Richter M., Liang C., et al. (2011). Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J. 5 (1), 61–70. doi: 10.1038/ismej.2010.95
Silver A. C., Kikuchi Y., Fadl A. A., Sha J., Chopra A. K., Graf J. (2007). Interaction between innate immune cells and a bacterial type III secretion system in mutualistic and pathogenic associations. Proc. Natl. Acad. Sci. U.S.A. 104 (22), 9481–9486. doi: 10.1073/pnas.0700286104
Simpson T. L. (1984). The cell biology of sponges (New York: Springer Verlag). Available at: http://journal.um-surabaya.ac.id/index.php/JKM/article/view/2203.
Snyder G. A., Eliachar S., Connelly M. T., Talice S., Hadad U., Gershoni-Yahalom O., et al. (2021). Functional characterization of hexacorallia phagocytic cells. Front Immunol 12, 1–13. doi: 10.3389/fimmu.2021.662803
Steinmetz P. R. H. (2019). A non-bilaterian perspective on the development and evolution of animal digestive systems. Cell Tissue Res. 377 (3), 321–339. doi: 10.1007/s00441-019-03075-x
Tartaro K., Vanvolkenburg M., Wilkie D., Coskran T. M., Kreeger J. M., Kawabata T. T., et al. (2015). Development of a fluorescence-based in vivo phagocytosis assay to measure mononuclear phagocyte system function in the rat. J. Immunotoxicol. 12 (3), 239–246. doi: 10.3109/1547691X.2014.934976
Thomas T., Moitinho-Silva L., Lurgi M., Björk J. R., Easson C., Astudillo-García C., et al. (2016). Diversity, structure and convergent evolution of the global sponge microbiome. Nat. Commun. 7, 1–12. doi: 10.1038/ncomms11870
Thomas T., Rusch D., DeMaere M. Z., Yung P. Y., Lewis M., Halpern A., et al. (2010). Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. ISME J. 4 (12), 1557–1567. doi: 10.1038/ismej.2010.74
Turon X., Galera J., Uriz M. J. (1997). Clearance rates and aquiferous systems in two sponges with contrasting life-history strategies. J. Exp. Zool. 278 (1), 22–36. doi: 10.1002/(SICI)1097-010X(19970501)278:1<22::AID-JEZ3>3.0.CO;2-8
Uribe-Querol E., Rosales C. (2020). Phagocytosis: our current understanding of a universal biological process. Front. Immunol. 11, 1–13. doi: 10.3389/fimmu.2020.01066
Vethaak A. D., Cronie R. J. A., van Soest R. W. M. (1982). Ecology and distribution of two sympatric, closely related sponge species, Halichondria panicea (Pallas 1766) and H. bowerbanki burton 1930 (Porifera, demospongiae), with remarks on their speciation. Bijdr tot Dierkd. 52 (2), 82–102. doi: 10.1163/26660644-05202002
Wehrl M., Steinert M., Hentschel U. (2007). Bacterial uptake by the marine sponge Aplysina aerophoba. Microb. Ecol. 53 (2), 355–365. doi: 10.1007/s00248-006-9090-4
Wilkinson C. R., Garrone R., Vacelet J. (1984). Marine sponges discriminate between food bacteria and bacterial symbionts: Electron microscope radioautography and in situ evidence. Proc. R Soc. London - Biol. Sci. 220 (1221), 519–528. doi: 10.1098/rspb.1984.0018
Yahel G., Eerkes-Medrano D. I., Leys S. P. (2006). Size independent selective filtration of ultraplankton by hexactinellid glass sponges. Aquat Microb. Ecol. 45 (2), 181–194. doi: 10.3354/ame045181
Yahel G., Whitney F., Reiswig H. M., Eerkes-Medrano D. I., Leys S. P. (2007). In situ feeding and metabolism of glass sponges (Hexactinellida, Porifera) studied in a deep temperate fjord with a remotely operated submersible. Limnol. Oceanogr. 52 (1), 428–440. doi: 10.4319/lo.2007.52.1.0428
Keywords: sponge-microbe symbiosis, phagocytosis, fluorescence-activated cell sorting (FACS), particle uptake, sponge cells
Citation: Marulanda-Gomez AM, Bayer K, Pita L and Hentschel U (2023) A novel in-vivo phagocytosis assay to gain cellular insights on sponge-microbe interactions. Front. Mar. Sci. 10:1176145. doi: 10.3389/fmars.2023.1176145
Received: 16 March 2023; Accepted: 05 July 2023;
Published: 09 August 2023.
Edited by:
Shirley A. Pomponi, Florida Atlantic University, United StatesReviewed by:
Stéphanie Barnay-Verdier, Sorbonne Université (CNRS), FranceSandie M. Degnan, The University of Queensland, Australia
Copyright © 2023 Marulanda-Gomez, Bayer, Pita and Hentschel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lucia Pita, bHVjaWFwaXRhQGNtaW1hLmNzaWMuZXM=; Ute Hentschel, dWhlbnRzY2hlbEBnZW9tYXIuZGU=
‡ORCID: Angela M. Marulanda-Gomez, orcid.org/0000-0002-9760-0779
Kristina Bayer, orcid.org/0000-0003-2789-9805
Lucia Pita, orcid.org/0000-0003-0163-1587
Ute Hentschel, orcid.org/0000-0003-0596-790X
 Angela M. Marulanda-Gomez1‡
Angela M. Marulanda-Gomez1‡ Kristina Bayer
Kristina Bayer Lucia Pita
Lucia Pita Ute Hentschel
Ute Hentschel