
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Mar. Sci., 03 August 2023
Sec. Marine Biology
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1176006
This article is part of the Research TopicInteractions between the Adaptability of Marine Organisms and Environmental FactorsView all 5 articles
Cysteine sulfinate decarboxylase (CSAD) is a rate-limiting enzyme in taurine biosynthesis. In this study, the structure and expression characteristics of the RpCSAD gene in Ruditapes philippinarum were analyzed. The pyridoxal phosphate (PLP)-dependent amino acid carboxylase structural domains and catalytic active sites of six copies of RpCSAD were highly conserved. Tissue expression analysis demonstrated that RpCSAD1 and RpCSAD3-6 was primarily expressed in the mantle tissue, and RpCSAD2 in the water tube and gonad tissue. After exposure to hyposalinity stress, the expression levels of RpCSADs were found to be upregulated in all three shell-colored groups of clams. Remarkably, there was a pronounced increase in the expression of RpCSAD1-6 observed specifically in the zebra shell-color groups. These findings strongly suggest the involvement of taurine metabolism in the response of zebra shell-color clams to low salinity stress, thereby emphasizing its critical role in the adaptation mechanisms. These findings could contribute to a better understanding the mechanisms of osmotic stress tolerance in shellfish.
As tides, seasonal rainfall, evaporation, and terrestrial freshwater runoff can all lead to periodic changes in the salinity of the intertidal environment, marine organisms inhabiting the coastal intertidal zone must be able to adapt their physiological activities to changes in salinity. Invertebrates such as shellfish living in the intertidal zone can adapt to fluctuations in seawater salinity by altering the osmotic pressure of their internal environment (Yancey et al., 1982; Gilles, 1987). Free amino acids, including alanine, glycine, proline, taurine, and glutamate, are involved in maintaining the osmotic pressure balance of the cells (Allen and Garrett, 1972; Zurburg and De Zwaan, 1981). Taurine is the most abundant free amino acid in bivalve shellfish (Neufeld and Wright, 1995; Na et al., 2015; Zhu et al., 2018), and plays an important role in processes such as resistance, growth, and development. In oysters, taurine can increase the activity of the mitochondrial electron transport system and increase the amount of ATP synthesis under hyperosmotic stress, providing more energy to cope with osmotic stress (Sokolov and Sokolova, 2019). It was found that the mussel Mytilus galloprovincialis juveniles with a larger gill surface area had higher filter feeding efficiency and grew much faster than individuals with a smaller gill surface area (Prieto et al., 2020). There exists a significant negative correlation between the surface area of gill tissue and taurine content in the mussel M. galloprovincialis (Babarro et al., 2011). Transcriptome analysis of the pearl mussel Pinctada fucata also revealed significant differences in the expression of cysteine sulfite decarboxylase between different sizes of pearl mussel (Shi and He, 2014), Therefore, the synthesis of taurine in shellfish may affect the efficiency of filter feeding by influencing the surface area of the gills, and thus their growth process. Haliotis rufescens needs to maintain a constant level of taurine in its body from embryo to larvae before metamorphosis to ensure normal development of the embryo (Jaeckle and Manahan, 1989).The level of taurine in the planktonic larval stage of the oyster Crassostrea gigas assists it in adapting to dramatic changes in environmental osmolarity during development (Welborn and Manahan, 1995), confirming that taurine also plays an important role in shellfish development similar to that of vertebrates. Therefore, a better understanding of taurine metabolism in shellfish will help to elucidate the mechanisms of growth, development, and stress resistance in shellfish.
Taurine is mainly synthesized in shellfish from cysteine (Allen and Garrett, 1972). There are two synthetic pathways involved in this process. The first pathway converts cysteamine to taurine via cysteamine dioxygenase ADO. The second pathway involves the oxidation of cysteine to cysteine sulfite via cysteine dioxygenase CDO. In this pathway, cysteine is oxidized to cysteine sulfinic acid (CSA) by cysteine dioxygenase CDO, followed by decarboxylation to taurine by cysteine sulfinic acid decarboxylase (CSAD), which is the rate-limiting enzyme for taurine biosynthesis (Peck and Awapara, 1967; Jacobsen and Smith, 1968). Taurine transport protein (TAUT) is primarily involved in the cellular transport of taurine (Tappaz, 2004). However, relatively little research has been conducted on taurine synthase genes in shellfish, and only cysteamine dioxygenase has been cloned in mussels (Nagasaki et al., 2015). Recently, cysteamine dioxygenase CDO and cysteine sulfinic acid decarboxylase (CSAD) have been cloned from the deep-sea clam Phreagena okutanii (Kuroda et al., 2021). Additionally, in the mussel Bathymodiolus septentrionalis, cysteine dioxygenase CDO and cysteine sulfinic acid decarboxylase (CSAD) have been cloned (Kuroda et al., 2021). The taurine transporter protein (TAUT) found in the mussel B. septemdierum, the oyster C. gigas, the hard-shell clam Meretrix lusoria, and the razor clam Sinonovacula constricta has been shown to be involved in adaptation to changes in environmental osmolality (Hosoi et al., 2007; Lin et al., 2016; Nakamura-Kusakabe et al., 2016; Huang et al., 2021).
Manila clam, R. philippinarum, is one of the most extensively cultivated bivalves along the Chinese coast. Our previous study on the R. philippinarum genome identified several genes involved in taurine synthesis and metabolism (Jian-feng et al., 2013; Dong-min et al., 2017). In this study, we aim to identify the key enzyme gene for taurine synthesis, RpCSAD, which is annotated in the genome sequence, and elucidate the role of the RpCSAD gene in the low-salt adaptation process of clams and investigate its contribution to the development of stress tolerance in clams with different shell colors.
A total of 300 clams with intact and vigorous shells, measuring (3.0 ± 0.5) cm in length, were temporarily reared for one week in a flat-bottomed plastic tank containing 50 L of water maintained at a temperature of (18.0 ± 0.5) °C and a salinity of 30 ppt. The water was changed once daily, and Spirulina powder was fed to the clams once in the morning and once in the evening.
The identification of the RpCSAD gene was primarily based on the genome data of the R. philippinarum (NCBI accession number PRJNA479743) (Yan et al., 2019). The CSAD sequences obtained from the clam genome annotation results were reconfirmed by the online conserved region analysis tool CDD and the sequence alignment tool BlastX on the NCBI website (https://www.ncbi.nlm.nih.gov). Amino acid alignment of the putative proteins was conducted using the Clustalx1.83 software. Phylogenetic trees were constructed based on the neighbor-joining method using MEGA 11.0 software. Amino acid secondary structures were mapped using the online tool ESPript 3.0 (https://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi).
Four clams, each of the three shell-color group, were dissected on ice trays. Tissues including the foot, mantle, gill, water tube, digestive glands, gonads, and labial palps were carefully extracted and snap-frozen in liquid nitrogen. These tissue samples were subsequently stored at -80°C in a refrigerator for future tissue expression analysis.
Sixty individuals from each shell-color group of clams, namely orange, white, and zebra-shell color groups, were selected and divided into two parallel groups, consisting of 30 individuals each. The experimental control group was maintained at a salinity of 30 ppt, while the low-salt treatment group was exposed to a salinity of 14 ppt, which was determined to be the threshold value that the clams could tolerate. Four individuals were randomly selected from each treatment group at 0, 8, 16, 24, 48, 72, and 96 hours after treatment, and their water tube tissues were collected, frozen in liquid nitrogen, and stored at -80°C for subsequent gene expression analysis.
The total RNA extraction was performed using an RNA extraction kit according to the manufacturer’s protocol(Tiangen, Bejing, China). To assess the RNA integrity, 1.5% agarose gel electrophoresis was performed. The quality of RNA was further evaluated by measuring the A260/A280 ratio using a nucleic acid analyzer(Bio-Rad, Hercules, CA, USA). Reverse transcription was conducted using a reverse transcription kit(Takara, Dalian, China).
The gene expression assay was performed via RT-qPCR using the Tubulin gene as an internal reference for analyzing the expression of the RpCSAD gene. Primers were designed using Primer 5.0 software (Premier Biosoft International, Palo Alto, CA), and the specific primer sequences are presented in Table 1. To calculate the relative gene expression, the 2-δδCt method was utilized (Livak and Schmittgen, 2001).
Gene expression data were analyzed using SPSS 20.0 software. Tissue expression analysis was conducted with water tube tissue expression as a reference, and salinity stress expression analysis was performed using the expression of the 0h control as a reference. The experimental results were expressed as mean ± standard deviation (Means ± S.D.) and analyzed using one-way ANOVA with Duncan’s method for multiple comparisons. The significance level was set at 0.05.
Amino acid comparisons revealed that the putative RpCSAD proteins exhibited 50.2% identity with other vertebrate and invertebrate CSAD. Additionally, the identified RpCSAD proteins all contained structural domains containing pyridoxal phosphate (PLP)-dependent amino acid carboxylase and catalytic active sites (Figure 1).
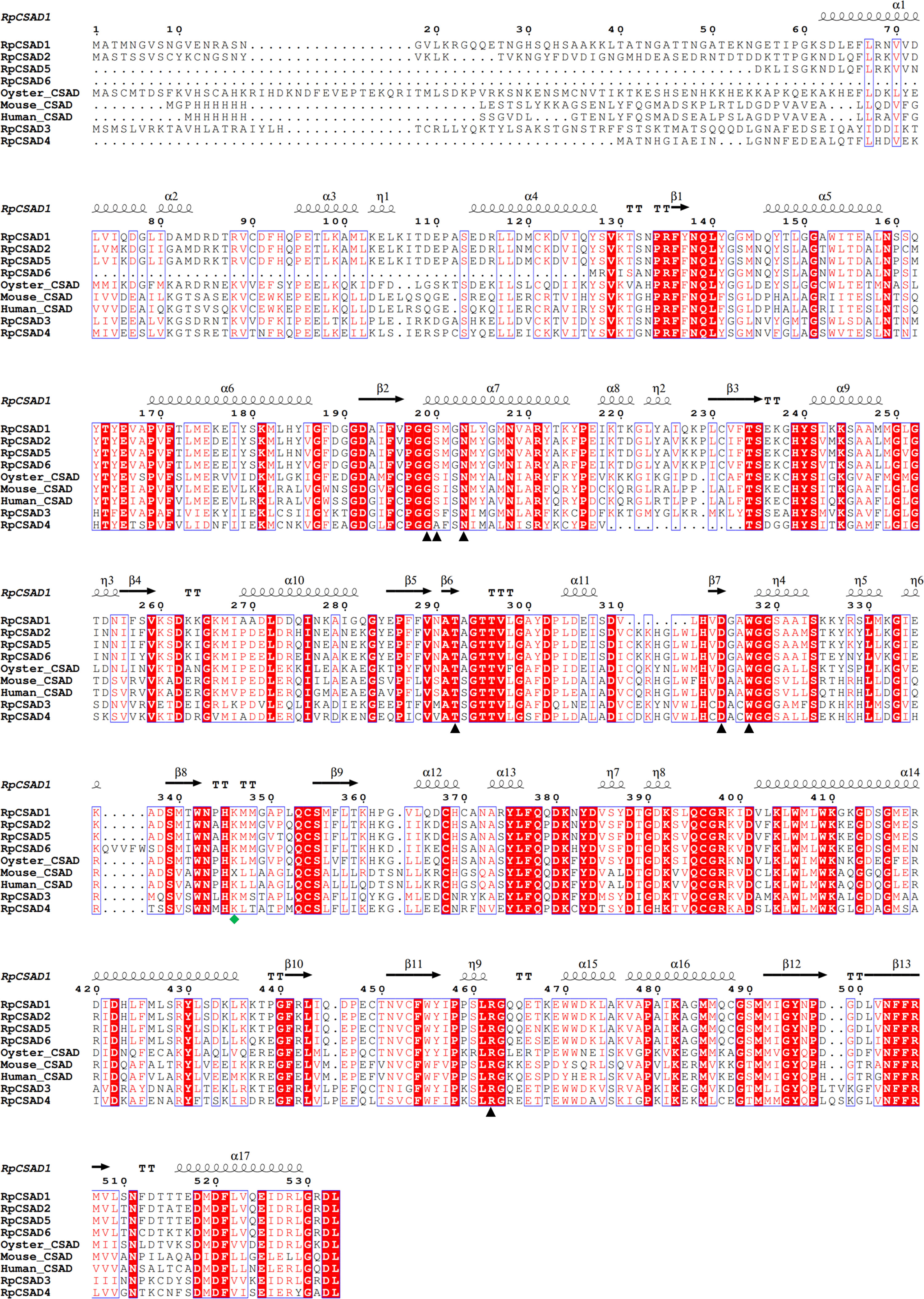
Figure 1 Multiple Comparison of RpCSAD Amino Acids in R. philippinarum Pyridoxal 5’-phosphate binding sites are marked with black triangles, Lysine residues in the catalytically active site are marked with green quadrilateral symbols.
The phylogenetic tree constructed using the CSAD gene sequences of 18 species including mammals and mollusks (Figure 2) showed that the CSAD of vertebrates and invertebrates were clustered independently, and the CSAD of crustaceans and shellfish were also clustered independently. The CSAD of the shellfish was divided into two branches, and the RpCSAD1, RpCSAD2, RpCSAD5 and RpCSAD6 of the Manila clam clustered together, while RpCSAD3 and RpCSAD4 clustered in the other branch of the mollusks.
The expression of RpCSAD1 in different tissues was significantly different (P < 0.05) with the highest expression in mantle tissue, the lowest expression in gonad, labial palps and gill (Figure 3). The expression of RpCSAD2 was significantly higher (P < 0.05) in water tube and gonad tissue than in labial palps tissue, while the expression of RpCSAD3, RpCSAD4, RpCSAD5 and RpCSAD6 was highest (P<0.05) in mantle tissue, followed by water tube, foot, gonad, labial palps, gill and digestive gland. In particular, the expression of RpCSAD5 was highest (P < 0.05) in water tube tissue, while the expression of RpCSAD6 was highest (P < 0.05) in mantle tissue.
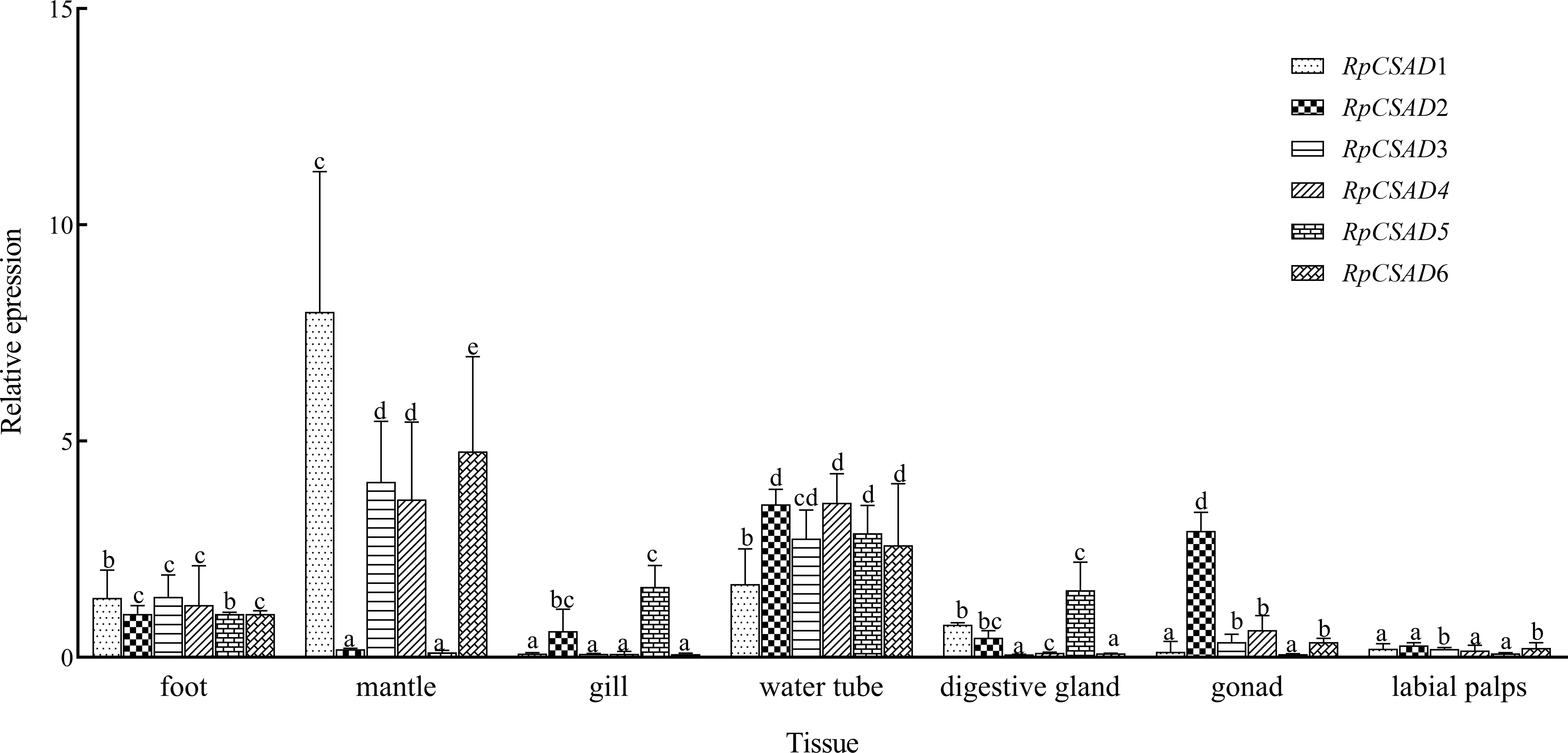
Figure 3 Tissue Expression of RpCSAD in R. philippinarum Different lowercase letters indicate that the expression of the same gene is significantly different in different tissues (P<0.05).
Under low salt stress, the RpCSAD1 gene expression increased significantly in the orange and zebra shell-color groups at 72h and 8h, respectively (P < 0.05), and decreased in the white shell-color group with the lowest value observed at 48h (P < 0.05). The RpCSAD2 gene expression was significantly higher in the orange shell-color group at 24h and 48h (P < 0.05) with the highest value observed at 72h (P < 0.05). The white and zebra shell-color groups showed the highest expression at 96h (P<0.05). The RpCSAD3 gene expression was significantly higher in the white shell-color group at 8h, 72h, and 96h (P < 0.05), while the orange and zebra shell-color groups showed the highest expression at 24h and 16h, respectively (P < 0.05). The RpCSAD4 gene expression was significantly higher in the orange and white shell-color groups at 48h and 8h, respectively (P < 0.05), and in the zebra shell-color group at 96h (P<0.05). The RpCSAD5 gene expression was significantly higher in the orange shell-color group at 8h (P < 0.05) and in the orange and zebra shell-color groups at 24h and 16h, respectively (P < 0.05). The RpCSAD6 gene expression was highest at 24h in both orange and zebra shell-color groups of clams (P < 0.05), and significantly higher in the white shell-color group at 8h (P < 0.05) (Figure 4).
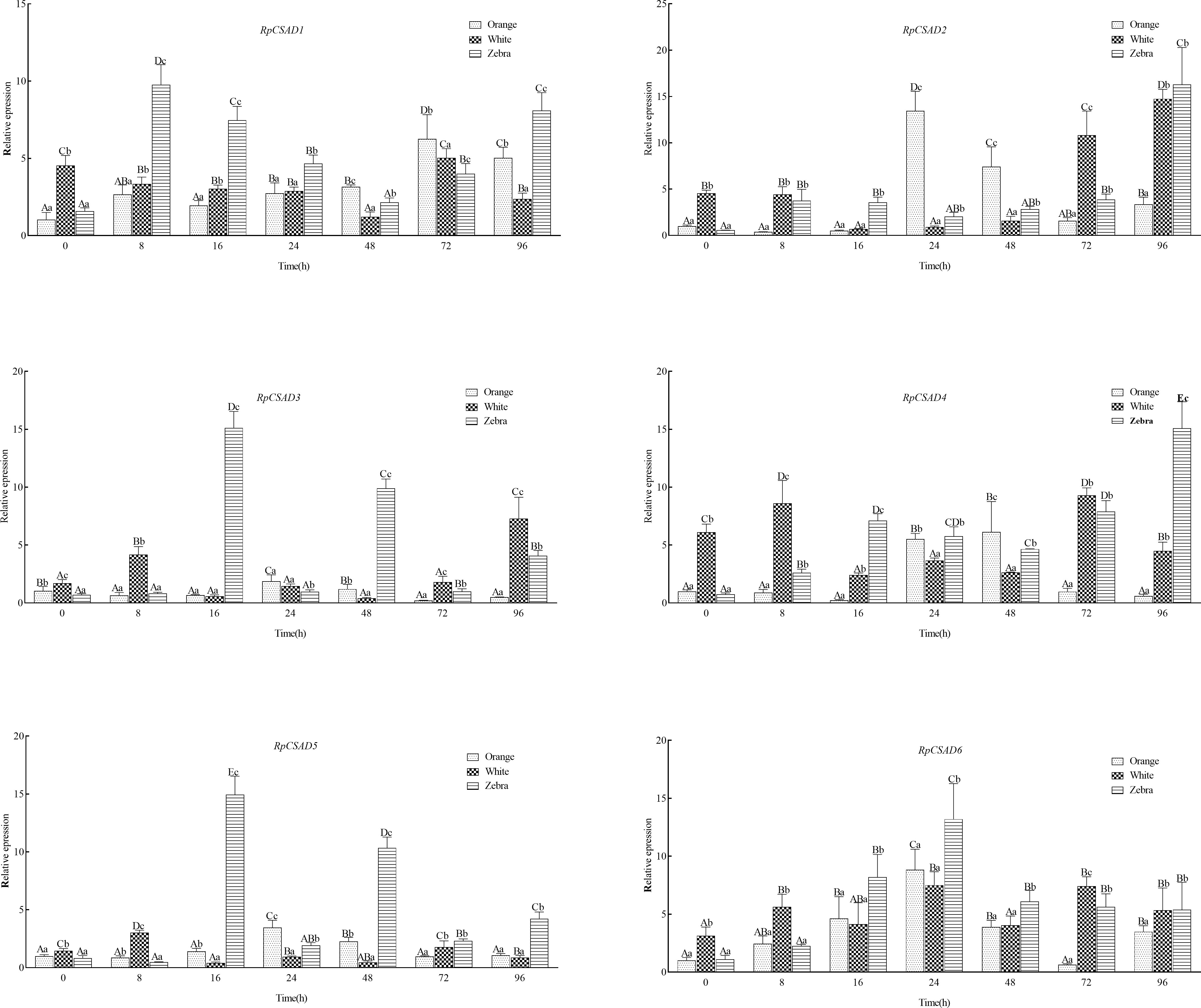
Figure 4 Relative expression of RpCSAD gene in three shell colors groups of R. philippinarum under low-salt stress Different lowercase letters indicate significant differences between different shell colors at the same time; Different capital letters indicate significant differences between different time points of the same shell color clam (P<0.05).
In our study, a genome-wide analysis was conducted to identify six RpCSAD genes in the clam genome. CSAD belongs to the type II PLP-dependent amino acid decarboxylase carboxylase family (Sandmeier et al., 1994). Structural domains, conserved motifs, and amino acid structures of RpCSAD were highly conserved and similar to CSAD proteins in human, rat, and oyster species. Evolutionary analysis also supports this hypothesis by demonstrating that RpCSAD in R. philippinarum has a closer genetic distance to bivalves.
In the Pacific oyster C. gigas, CSAD was found to be highly expressed in gonad, hemolymph, and adductor muscle tissues, while lower expression was observed in gill tissues (Zhao et al., 2017). Conversely, in the deep-sea mussel Bathymodiolus septemdierum, CSAD gene expression was higher in the gill, followed by the gonad, foot, and mantle (Haga et al., 2015). In our study, the expression of the clam RpCSAD gene was found to be higher in the mantle and water tube tissues, which are in direct contact with the external environment where osmotic pressure varies. This suggests that RpCSAD plays an important role in regulating osmotic pressure in clams.
Cysteine sulfonic acid decarboxylase (CSAD) plays a crucial role in the hypersaline adaptation of certain shellfish. In juvenile S. constricta, one copy was significantly up-regulated only under acute high salt stress, while the remaining copies were significantly up-regulated under both acute low and high salt stress (Ma et al., 2019). In the oyster C. gigas, one copy of the CSAD gene was significantly reduced under low salt stress and significantly increased under high salt stress, while the other copy was significantly increased under low salt stress conditions (Meng et al., 2013). In our study, six copies of the RpCSAD gene also showed different patterns of expression changes following low salt stress. This divergence in expression is a possible sub-functionalization of genes following replication, as confirmed in oyster genomic studies where stress response genes are more likely to retain similar replication than all genes on average and expression differences exist in homologues with highly similar sequences (Zhang et al., 2012).
Intracellular taurine exhibits antioxidant properties by diminishing superoxide production in the electron transport chain and augmenting the efficacy of antioxidant enzymes (Shimada et al., 2015). Shellfish frequently respond to sudden alterations in salinity by closing their shells, resulting in transient insufficiency of oxygen uptake and subsequent accumulation of electrons, which promotes the formation of reactive oxygen species (ROS) (Storey, 1996). Studies have preliminarily substantiated this process, revealing that a reduction in salinity from 19.6‰ to 8‰ led to heightened ROS levels in Anadara kagoshimensis hemolymphocytes, resulting in a 3.5-fold increase in ROS production (Kladchenko et al., 2021). Similarly, low salt stress can induce increased ROS production in abalone (Martello et al., 2000; Boamah et al., 2022). Furthermore, in R. philippinarum, genes encoding antioxidant enzymes initially demonstrate heightened expression followed by recovery after exposure to hypoxic stress, signifying clams’ capacity to elevate ROS production under hypoxic conditions (Dong-min et al., 2017). Therefore, the observed upregulation of RpCSAD expression in clams during the brief period following hyposalinity stress in this study may be ascribed to the compensatory effect of taurine depletion induced by heightened ROS levels due to hypoxia. Prior studies have reported varying levels of stress resistance among different shell-colored R. philippinarum individuals under low salt stress conditions (Jian-feng et al., 2013; Dong-min et al., 2017). Additionally, significant differences in taurine content have been observed among oysters and noble scallops with varying shell colors (Zhu et al., 2018; Tan et al., 2019). Consequently, the observed variability in RpCSAD gene expression among the three shell-colored groups of clams during low salt stress could contribute to the differences in stress resistance observed in R. philippinarum with varying shell colors. Nevertheless, further experimental data are required to validate this hypothesis.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by Dalian Ocean University Animal Ethics Committee.
ZZ and JW designed and conducted the experiments. ZH and HN performed data analysis and visualizations. XY refined the papers and supervised the data analysis. JD wrote and edited the paper. All authors contributed to the article and approved the submitted version.
This work was supported by the Natural Science Foundation of Liaoning Province (2020-MS-279), the Chinese Ministry of Science and Technology through the National Key Research and Development Program of China (2018YFD0901400), and the Dalian High-level Talent Innovation Programs (2021RJ09).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Allen J. A., Garrett M. R. (1972). Studies on taurine in the euryhaline bivalve Mya arenaria. Comp. Biochem. Physiol. – Part A: Physiol. 41, 307–317. doi: 10.1016/0300-9629(72)90062-X
Babarro J. M. F., FernÁNdez Reiriz M. J., Labarta U., Garrido J. L. (2011). Variability of the total free amino acid (TFAA) pool in Mytilus galloprovincialis cultured on a raft system. Effect of body size. Aquaculture Nutr. 17, e448–e458. doi: 10.1111/j.1365-2095.2010.00781.x
Boamah G. A., Huang Z., Shen Y., Lu Y., Wang Z., Su Y., et al. (2022). Transcriptome analysis reveals fluid shear stress (FSS) and atherosclerosis pathway as a candidate molecular mechanism of short-term low salinity stress tolerance in abalone. BMC Genomics 23, 1–22. doi: 10.1186/s12864-022-08611-8
Dong-min Y., Yan-li Z., Jian-feng D., Feng Y., Zhong-ming H., Hong-tao N., et al. (2017). Synergistic effects of high temperature and low salinity on immunity of Manila clam Ruditapes philippinarum. J. Dalian Ocean Univ. 32, 302–309. (In Chinese).
Gilles R. (1987). “Volume regulation in cells of euryhaline invertebrates,” in Current topics in membranes and transport (Academic Press) 30, 205–247.
Haga Y., Kondo H., Kumagai A., Satoh N., Hirono I., Satoh S. (2015). Isolation, molecular characterization of cysteine sulfinic acid decarboxylase (CSD) of red sea bream Pagrus major and yellowtail Seriola quinqueradiata and expression analysis of CSD from several marine fish species. Aquaculture 449, 8–17. doi: 10.1016/j.aquaculture.2015.04.004
Hosoi M., Shinzato C., Takagi M., Hosoi-Tanabe S., Sawada H., Terasawa E., et al. (2007). Taurine transporter from the giant Pacific oyster Crassostrea gigas: function and expression in response to hyper-and hypo-osmotic stress. Fisheries Sci. 73, 385–394. doi: 10.1111/j.1444-2906.2007.01346.x
Huang S., Jiang H., Zhang L., Gu Q., Wang W., Wen Y., et al. (2021). Integrated proteomic and transcriptomic analysis reveals that polymorphic shell colors vary with melanin synthesis in Bellamya purificata snail. J. Proteomics 230, 103950. doi: 10.1016/j.jprot.2020.103950
Jacobsen J. G., Smith L. H. (1968). Biochemistry and physiology of taurine and taurine derivatives. Physiol. Rev. 48, 424–511. doi: 10.1152/physrev.1968.48.2.424
Jaeckle W. B., Manahan D. T. (1989). Feeding by a “nonfeeding” larva: uptake of dissolved amino acids from seawater by lecithotrophic larvae of the gastropod Haliotis rufescens. Mar. Biol. 103, 87–94. doi: 10.1007/BF00391067
Jian-feng D., Rui W., Xi-wu Y., Li-qiang Z., Feng Y., Lian-shun W. (2013). Comparative tolerance to low salinity stress in Manila clam Ruditapes philippinarum with three shell colors. J. Dalian Ocean Univ. 28, 264–268. (In Chinese).
Kladchenko E. S., Andreyeva A. Y., Kukhareva T. A., Rychkova V. N., Soldatov A. A., Mindukshev I. V. (2021). Impact of low salinity on hemocytes morphology and functional aspects in alien clam Anadara kagoshimensis (Tokunaga, 1906). Russian J. Biol. Invasions 12, 203–212. doi: 10.1134/S2075111721020089
Kuroda M., Nagasaki T., Koito T., Hongo Y., Yoshida T., Maruyama T., et al. (2021). Possible roles of hypotaurine and thiotaurine in the vesicomyid clam phreagena okutanii. Biol. Bull. 240, 34–40. doi: 10.1086/712396
Lin C. H., Yeh P. L., Lee T. H. (2016). Ionic and amino acid regulation in hard clam (Meretrix lusoria) in response to salinity challenges. Front. Physiol. 7, 368. doi: 10.3389/fphys.2016.00368
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma B., Ran Z., Xu X., Xu J., Liao K., Cao J., et al. (2019). Comparative transcriptome analyses provide insights into the adaptation mechanisms to acute salt stresses in juvenile Sinonovacula constricta. Genes Genomics 41, 599–612. doi: 10.1007/s13258-019-00805-x
Martello L. B., Friedman C. S., Tjeerdema R. S. (2000). Combined effects of pentachlorophenol and salinity stress on phagocytic and chemotactic function in two species of abalone. Aquat. Toxicol. 49, 213–225. doi: 10.1016/S0166-445X(99)00075-2
Meng J., Zhu Q., Zhang L., Li C., Li L., She Z., et al. (2013). Genome and transcriptome analyses provide insight into the euryhaline adaptation mechanism of Crassostrea gigas. PloS One 8, e58563. doi: 10.1371/journal.pone.0058563
Li N., Wang X. Z., Wu H., Yi R. Z., Liu R., Wang L. C., et al. (2015). Study on the dynamic accumulation of taurine in four shellfish flesh of Jiangsu east costal and the appropriate harvest times. Sci. Technol. Food Industry 36, 54–59.
Nagasaki T., Hongo Y., Koito T., Nakamura-Kusakabe I., Shimamura S., Takaki Y., et al. (2015). Cysteine dioxygenase and cysteine sulfinate decarboxylase genes of the deep-sea mussel Bathymodiolus septemdierum: possible involvement in hypotaurine synthesis and adaptation to hydrogen sulfide. Amino Acids 47, 571–578. doi: 10.1007/s00726-014-1891-z
Nakamura-Kusakabe I., Nagasaki T., Kinjo A., Sassa M., Koito T., Okamura K., et al. (2016). Effect of sulfide, osmotic, and thermal stresses on taurine transporter mRNA levels in the gills of the hydrothermal vent-specific mussel Bathymodiolus septemdierum. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 191, 74–79. doi: 10.1016/j.cbpa.2015.09.013
Neufeld D. S., Wright S. H. (1995). Basolateral transport of taurine in epithelial cells of isolated, perfused mytilus californianus gills. J. Exp. Biol. 198, 465–473. doi: 10.1242/jeb.198.2.465
Peck E. J. Jr., Awapara J. (1967). Formation of taurine and isethionic acid in rat brain. Biochim. Biophys. Acta (BBA)-General Subj. 141, 499–506. doi: 10.1016/0304-4165(67)90178-X
Prieto D., Tamayo D., Urrutxurtu I., Navarro E., Ibarrola I., Urrutia M. B. (2020). Nature more than nurture affects the growth rate of mussels. Sci. Rep. 10, 1–13. doi: 10.1038/s41598-020-60312-y
Sandmeier E., Hale T. I., Christen P. (1994). Multiple evolutionary origin of pyridoxal-5’-phosphate-dependent amino acid decarboxylases. Eur. J. Biochem. 221, 997–1002. doi: 10.1111/j.1432-1033.1994.tb18816.x
Shi Y., He M. (2014). Differential gene expression identified by RNA-Seq and qPCR in two sizes of pearl oyster (Pinctada fucata). Gene 538, 313–322. doi: 10.1016/j.gene.2014.01.031
Shimada K., Jong C. J., Takahashi K., Schaffer S. W. (2015). Role of ROS production and turnover in the antioxidant activity of taurine. Adv. Exp. Med. Biol. 803, 581–596. doi: 10.1007/978-3-319-15126-7_47
Sokolov E. P., Sokolova I. M. (2019). Compatible osmolytes modulate mitochondrial function in a marine osmoconformer Crassostrea gigas (Thunberg, 1793). Mitochondrion 45, 29–37. doi: 10.1016/j.mito.2018.02.002
Storey K. B. (1996). Oxidative stress: animal adaptations in nature. Braz. J. Med. Biol. Res. 29, 1715–1733.
Tan K. S., Leng X., Zhao Y., Hongxing L., Cheng D., Ma H., et al. (2019). Amino acid variations in polymorphic noble scallops, Chlamys nobilis. J. Food Process. Preservation 43, e14262. doi: 10.1111/jfpp.14262
Tappaz M. L. (2004). Taurine biosynthetic enzymes and taurine transporter: molecular identification and regulations. Neurochemical Res. 29, 83–96. doi: 10.1023/B:NERE.0000010436.44223.f8
Welborn J., Manahan D. (1995). Taurine metabolism in larvae of marine invertebrate molluscs (Bilvalvia, Gastropoda). J. Exp. Biol. 198, 1791–1799. doi: 10.1242/jeb.198.8.1791
Yan X., Nie H., Huo Z., Ding J., Li Z., Yan L., et al. (2019). Clam genome sequence clarifies the molecular basis of its benthic adaptation and extraordinary shell color diversity. iScience 19, 1225–1237. doi: 10.1016/j.isci.2019.08.049
Yancey P. H., Clark M. E., Hand S. C., Bowlus R. D., Somero G. N. (1982). Living with water stress: evolution of osmolyte systems. Science 217, 1214–1222. doi: 10.1126/science.7112124
Zhang G., Fang X., Guo X., Li L., Luo R., Xu F., et al. (2012). The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54. doi: 10.1038/nature11413
Zhao X., Li Q., Meng Q., Yue C., Xu C. (2017). Identification and expression of cysteine sulfinate decarboxylase, possible regulation of taurine biosynthesis in Crassostrea gigas in response to low salinity. Sci. Rep. 7, 5505. doi: 10.1038/s41598-017-05852-6
Zhu Y., Li Q., Yu H., Kong L. (2018). Biochemical composition and nutritional value of different shell color strains of pacific oyster crassostrea gigas. J. Ocean Univ. China 17, 897–904. doi: 10.1007/s11802-018-3550-6
Keywords: Ruditapes philippinarum, taurine, cysteine sulfinate decarboxylase, hyposalinity stress, expression profile
Citation: Zhang Z, Wang J, Huo Z, Nie H, Yan X and Ding J (2023) Identification and expression of cysteine sulfite decarboxylase genes and their response to hyposalinity stress in Ruditapes philippinarum. Front. Mar. Sci. 10:1176006. doi: 10.3389/fmars.2023.1176006
Received: 28 February 2023; Accepted: 17 July 2023;
Published: 03 August 2023.
Edited by:
Pengzhi Qi, Zhejiang Ocean University, ChinaCopyright © 2023 Zhang, Wang, Huo, Nie, Yan and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianfeng Ding, amZkaW5nQGRsb3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.