- 1Key Laboratory of Green Mariculture (Co-construction by Ministry and Province), Ministry of Agriculture and Rural, Ningbo University, Ningbo, Zhejiang, China
- 2Key Laboratory of Aquacultural Biotechnology Ministry of Education, Ningbo University, Ningbo, Zhejiang, China
Portunus trituberculatus, an economically important marine crab that was widely distributed along the coast of China, has been in short supply of high-quality crab seedlings in recent years. The larval development of P. trituberculatus is regulated by numerous biological processes at different biological levels, particularly regarding gene expression during the larval development process. Understanding the changes in these critical genes can help further regulate these biological processes to increase crab seedlings’ survival and improve crab quality. In this study, the molecular mechanisms of morphological and physiological changes in larval stages of P. trituberculatus were investigated using DNA microarrays. A total of 109,533 unigenes were obtained, of which 16 genes were differentially expressed during the whole development stages with evident timeliness. The enrichment analysis and comparison of differentially expressed genes (DEGs) showed three types of physiological activities, chitin development, energy metabolism, and neuroendocrine development. The physiological activities were evident in the early development of P. trituberculatus, especially at the Z4-M stage, which is crucial for the growth and development of crab larvae. These DEGs, cuticle proteins, retinol dehydrogenase (RDHs), Juvenile Hormone Epoxide Hydrolase (JHEH), and other vital genes were differentially expressed at the Z4-M stage. Moreover, the insulin-like androgenic gland (IAG), which was reported to be associated with sex differentiation, was highly expressed in the megalopa stage, indicating that the gender differentiation divide may occur at the beginning of this stage. Overall, this research provides a theoretical basis for the development of the swimming crab larvae, which could contribute to the artificial growing seedlings technology of P. trituberculatus.
1 Introduction
Portunus trituberculatus (Crustacea: Decapoda: Portunidae) is a giant offshore crab widely distributed in the Bohai Sea, Yellow Sea, East China Sea, and South China Sea (Xu et al., 2009). Due to its rapid growth, delicious taste, and high breeding profits, it is a commercially important species artificially cultured along East China’s coastal regions (Wang et al, 2010). Its culture yield in 2022 reached 105,283 tons in China, marking a 4.35% increase from the previous year (Fishery and Fisheries Administrative Bureau, 2023). To meet the gradual increase in market demand, the cultivation and breeding of this species became popularized in China since the 1990s (Wang, 2010). However, with the expansion of the artificial crab breeding scale and the intensification of stocking in recent years, the accompanying demand for quality larvae of P. trituberculatus has exceeded the supply (Wu et al., 2010). The larval development of P. trituberculatus goes through four zoeal stages (Z1-4) to one megalopa stage (M) before they metamorphose to the first juvenile (C) (Hamasaki, 1996; Anger et al., 2015). It is a fast successional process that takes 16-22 days (Lu et al., 2022). Several critical biological processes accompany the larval development, such as molting, organ formation, food transformation, and immune system formation. The morphological structure of P. trituberculatus is slowly being perfected and the habits are adapted during the process (Dan et al., 2013; Dan et al., 2016). However, mass mortality is frequent during seed production of P. trituberculatus, and the average survival rate of larvae is only 11% (Hamasaki et al., 2011). It has been reported that morphological abnormalities, deficiency of essential nutrients, and bacterial and fungal diseases are possible causes of the mass mortality events that occur during swimming crab seed production (Arai et al., 2007; Hamasaki et al., 2011; Dan et al., 2013). In addition to the survival rate, the phenotypic characteristics (neat size, intact appendages) and vitality of swimming crab larvae are essential criteria for judging the quality of the larvae (Huang et al., 2015).
Therefore, it is necessary to improve our understanding of the molecular processes occurring during the development of the larvae of this marine species, to improve the survival rate and quality of P. trituberculatus larvae. At present, domestic and foreign research on swimming crab larvae is mainly focused on external factors, such as larval rearing (Dan et al., 2017), diseases (Muroga et al., 1989), morphology (Hamasaki et al., 2011; Dan et al., 2013), and environmental factors (Ren et al., 2017; Gao and Wang, 2021; Zhu et al., 2022). In recent years, genomics has been widely applied in studying crustaceans to elucidate the molecular mechanisms of different physiological processes (Li et al., 2014; Huerlimann et al., 2018; Zhao et al., 2021; Zhang et al., 2022). Compared to other sequencing methods, gene microarray technology can rapidly and efficiently obtain expression results for many genes at the transcriptional level, enabling genome-wide gene expression analysis (Bellin et al., 2009). The identification and analysis of the genes involved in the ontogeny of P. trituberculatus will contribute to improving the survival of larvae yielding a commercial benefit.
Therefore, in this research, the cDNA libraries were constructed by using specimens from each larval stage of the swimming crab P. trituberculatus (Z1-C). DNA Microarrays technology was utilized to compare and analyze the transcriptome during the larval developmental stages of P. trituberculatus. Various differentially expressed genes between these stages were screened, validated, and functionally predicted using GO and KEGG pathway localization. These findings may help elucidate the molecular mechanisms involved in the larval development of P. trituberculatus. Overall, this study provides a panoramic insight into the gene expression of the complex changes that occurred during the larval development.
2 Materials and methods
2.1 Animals and samples preparation
The larvae culture of P. trituberculatus was carried out at Jing-Ye Nursery Farm (Ningbo, China). Several artificial-cultivated spawning crabs were cultured in a cement tank with a volume of 30 m3. Water salinity and temperature were maintained at 24 ± 1 and 27 ± 2°C, respectively. The spawning crabs were fed daily with fresh mussels and chilled fish at 5-8% of the total weight of the crab. One spawning crab, which larvae were going to hatch, was transferred to a plastic basket with an aeration device in the cement tank, and it was not fed before hatching. After hatching, the larvae were freely scattered into a cement pool. The larval-rearing method refers to the process described by Wu et al. (2013). Briefly, rotifers (Branchionus plicatilis) 30-40 ind./mL and microalga (Platymonas subcordiformis) (5-10) ×104/mL were fed every 2 h during Z1-2, while Artemia nauplii 2-3 ind./mL was provided to Z3-C. The seawater in the pond was not replaced during the experiment. The larvae from the first zoea (Z1) to the first juvenile crab (C) were sampled repeatedly at each stage. Samples were collected when 70-80% of the population molted to the next stage using Sun’s methods (Sun et al., 1984). The samples were immediately rinsed with sterilized seawater, snap-frozen in liquid nitrogen, and then stored at -80°C. Three biological replicates of six stages were used for the following analysis.
2.2 Microarray hybridization and statistical analysis
The total RNA was extracted by using the TRIzol reagent. The RNA integrity was checked by Agilent Bioanalyzer 2100 (Agilent Technologies). After assessing the RNA quality, the sample labeling, chip hybridization and elution follow the standard chip procedure. Firstly, the total RNA is reverse-transcribed into double-stranded cDNA, and then the cRNA labeled with Cyanine-3-CTP (Cy3) is further synthesized. The probes were designed based on the unigenes of the P. trituberculatus cDNA library and NCBI. The cRNA was purified by RNeasy® Kit (QIAGEN) and subjected to quality control before the Cy3-labeled cDNAs were used as microarray targets. The labeled cRNA hybridizes to the chip, and after elution, the original image is obtained by scanning with an Agilent Scanner G2505C (Agilent Technologies).
Feature Extraction software (version 10.7.1.1, Agilent Technologies) was used to process the original image to extract the original data. Quantile standardization and subsequent processing were performed through Genespring software (version 12.5; Agilent Technologies). The normalized data were filtered, and at least one set of 75% probes labeled “Detected” in each collection of samples for comparison was stored for subsequent analysis. Microarray data of the present study were submitted to GEO and are accessible through GEO series accession number GSE226561.
In this study, differentially expressed genes (DEGs) were obtained by comparing the gene expression levels of the latter growth stage with the former growth stage of swimming crab larvae (five comparative groups: Z2 vs Z1; Z3 vs Z2; Z4 vs Z3; M vs Z4; C vs M). The DEGs screening was performed using the p-value and Fold Change of the t-test. The screening criteria were |Fold Change (FC)| > 2.0 and p-value < 0.05. The DEGs with FC > 2 / FC < -2 were assigned as up-regulated/down-regulated differential genes.
2.3 Cluster analysis, GO, and KEGG enrichment
The STEM Clustering Method performed a clustering analysis of DEGs trends (Ernst and Bar-Joseph, 2006). Trend analysis is the assignment of DEGs to pre-defined representative trends. If the expression trends of a particular class of DEGs are similar during the development of swimming crab larvae, they will be concentrated in a specific trend. It will result in the number of DEGs in this trend being more significant than the expected value of the random distribution. After completing the enrichment analysis, STEM software will determine the trend of significant enrichment according to the set significance threshold (p = 0.05). Statistically significant clustering (p < 0.05, significant temporal characteristics) was highlighted with a colored background. Next, Fisher’s exact test was used to calculate the significance of each pathway enriched for DEGs in the clusters to obtain the enriched pathway bubble chart. GO enrichment and KEGG pathway enrichment analysis of DEGs was performed using R based on the hypergeometric distribution.
2.4 qRT-PCR confirmation of differentially expressed genes
In order to test the reliability of Microarray, 5 DEGs were selected to confirm the accuracy of different expression patterns by qRT-PCR. Specific primers were designed according to the sequences in the cDNA library of P. trituberculatus by using Primer Premier 5.0 (Table S1). cDNA was generated by reverse transcription of RNA using HiFiScript cDNA Synthesis Kit. The qPCR program was under the following conditions: 95°C for 5 min; 95°C for 15 s, 60°C for 15s, and 72°C for 15s (40 cycles). The ribosomal protein L18 (RPL-18) was used as the reference gene for expression analysis (Xu and Liu, 2011), which is stably expressed during the development of swimming crab larvae (Table S2). The qRT-PCR for each gene was performed in triplicate on Light Cycler® 480. Real-time quantitative PCR results were analyzed by 2−ΔΔCT method (Livak and Schmittgen, 2002). Statistical analysis was performed with SPSS13.0.
3 Results
3.1 Global gene expression analysis
A total of 109,533 unigenes were obtained from the chip. Using two public databases (NR and SwissProt), 28,020 (25.58%) genes in the chip were annotated to some extent, of which 14,975 were mapped to at least one GO term and 10,953 genes were annotated to KEGG managed genes.
The principal components analysis (PCA) showed the spatial distribution of the microarray data and revealed the presence of six clear clusters, which correspond to each development stage respectively (Figure 1). The results showed that there were significant differences in gene expression patterns at different developmental stages. The zoeal larval stages (Z1-Z4) were distinguished from the megalopa stage (M) and the first juvenile crab stage (C) by the PC1 component. In contrast, the Z4 was distinguished from the other zoeal larval stages (Z1-Z3), M, and C, by the PC2 component. Notably, there is a clear distinction between zoeal and subsequent development stages, indicating that significant changes may experience in gene expression at development stages, consistent with the morphological, physiological, and biochemical changes of the actual larval development.
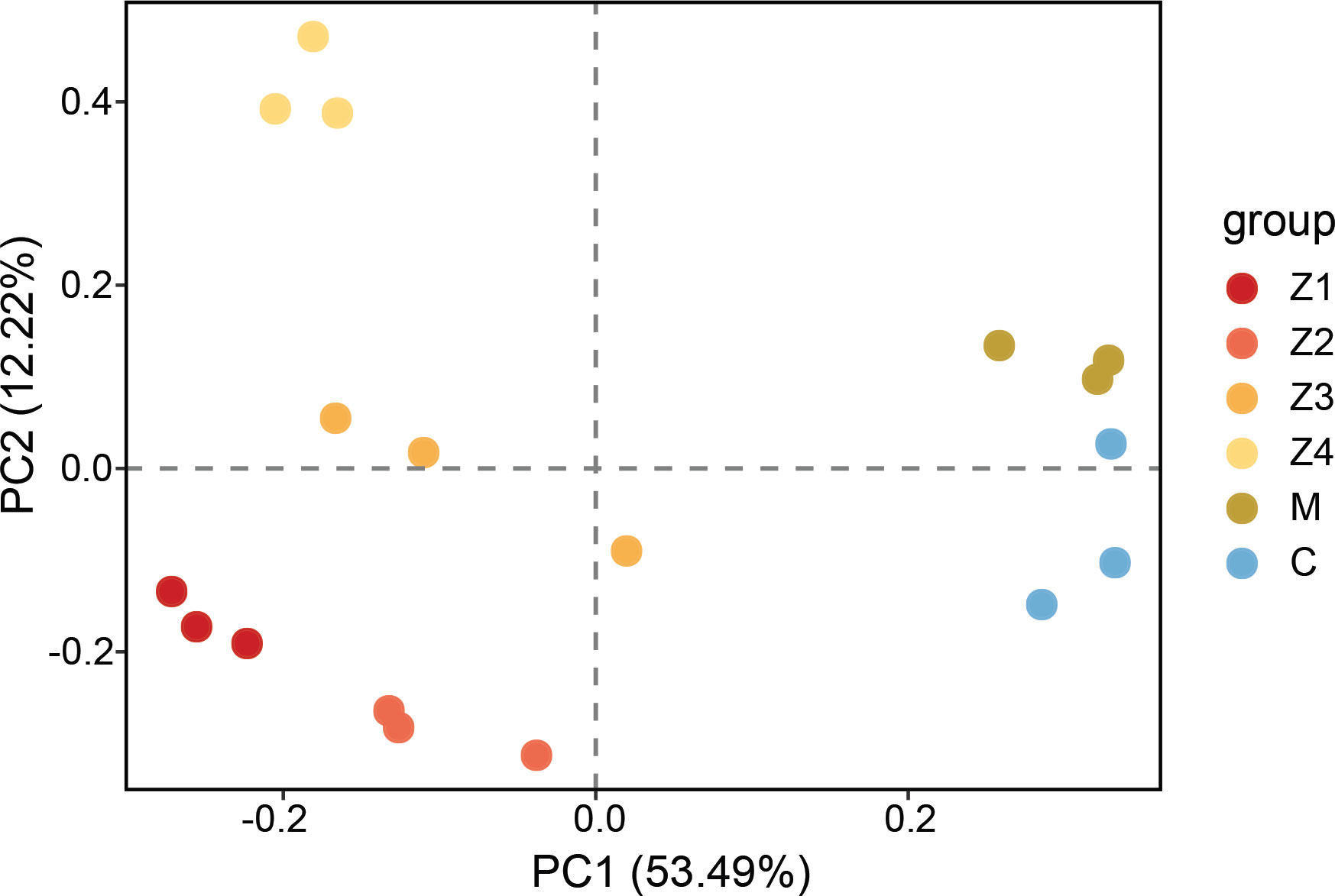
Figure 1 The principal component analysis (PCA) of the microarray data. Group indicates the development stages of P. trituberculatus larvae (Z1-C).
A total of 25,629 DEGs were identified across the five comparative groups (Z2 vs Z1; Z3 vs Z2; Z4 vs Z3; M vs Z4; C vs M) (Figure 2A). The highest number of DEGs was found in M vs Z4, with 18,036 DEGs (10,796 up-regulated and 7,240 down-regulated). In Z2 vs Z1, Z4 vs Z3, and C vs M, a less but considerable number of DEGs was observed (n = 7,057, 4,050, 3,330, respectively). The fewest number of differentially expressed genes was observed in Z3 vs Z2, with 1,367 DEGs (871 up-regulated and 496 down-regulated). It indicates that the amount of differential gene expression was highest in the Z4-M period and least in the Z2-Z3 period during the development of swimming crab larvae.
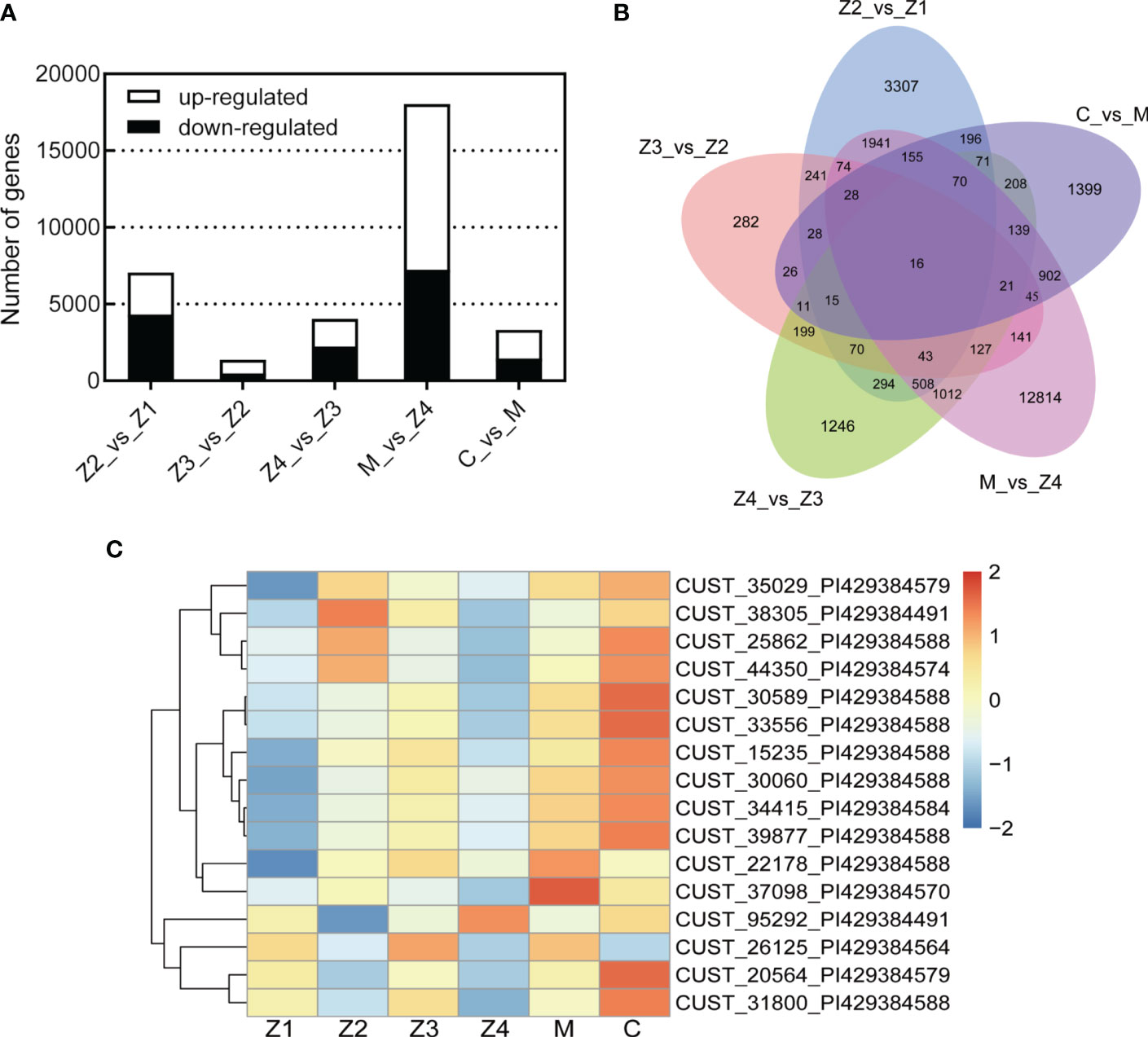
Figure 2 Differentially expressed genes (DEGs) statistics. (A) Bar Chart. (B) Venn Graph. (C) Heat map of common DEGs. |Fold Change(FC)| > 2.0 and adjusted p-value < 0.05.
Only 16 genes were differentially expressed in all five comparative groups by Venn diagram analysis (Figure 2B). This result indicates that the 16 DEGs were always differentially expressed throughout the development of swimming crab larvae. Further heat map clustering analysis of these DEGs revealed that the expression of the DEGs was temporally sequential (Figure 2C). Most of the DEGs were highly expressed in the C stage (11/16) and lowly expressed in Z1 and Z4 stages (12/16; 13/16, respectively). Sequence alignment analysis of the DEGs revealed that the databases annotated the most (13/16). Five of these DEGs have functions related to immune response (solute carrier family 15), olfactory development (UDP-glucosyltransferase 2), and chitin development (protein obstructor-E) (Table S3), and the remaining genes (3/16) have yet to be studied.
3.2 Enrichment and cluster analyses of the DEGs
All 25,629 DEGs were clustered by their variation in abundance using the STEM software program to obtain gene dynamic expression patterns during the larval development (Figure 3). Forty-nine profiles were produced according to their normalized signal value, in which significant expression profiles (Profile 22, Profile 38, Profile 8, Profile 15, Profile 1, Profile 36, Profile 26 and Profile 46, p < 0.05) were identified and marked with different background colors. The results showed that the clusterings of DEGs of these eight trend expression profiles were significantly enriched in the larval crab development time series. Among these profiles, the most significantly enriched trend expression, Profile 22, clustered to 4,573 DEGs. Secondly, Profile 8, Profile 15, Profile 1, Profile 36, Profile 26, and Profile 46 were enriched with 2,758, 2,210, 1,888, 1,781, 1,283, and 725 DEGs, respectively (Figure 3).
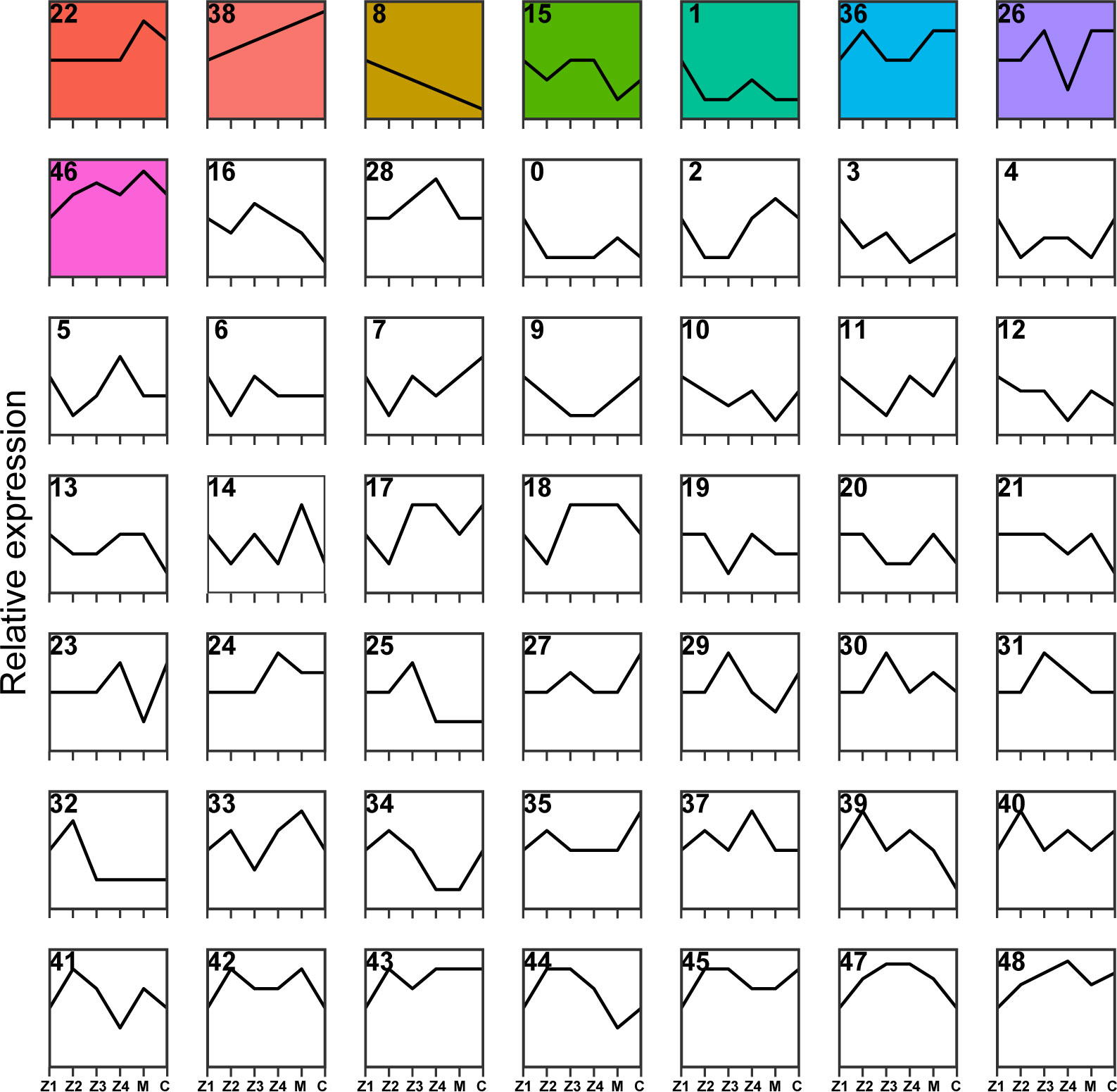
Figure 3 Cluster analysis of DEGs during P. trituberculatus larval development. Relative expression of genes is depicted on the vertical axis, and development stages of P. trituberculatus larvae are depicted on the horizontal axis. Colored backgrounds represent significant enrichment of differential genes in this trend and are arranged in ascending order of p-value.
Consistent with the trend of larval morphology changes in P. trituberculatus, the expression levels of DEGs of Profile 22 showed no noticeable difference from Z1 to Z4 stage. However, a marked increase and a decline in expression levels were noted during the transition from Z4 to C stage (Figure 3). In Profile 22, 28 pathways were significantly enriched (p < 0.05), with 21 pathways related to metabolism. Lysosome (ko04142) and pentose and glucuronate interconversions (ko00040) were the most significantly enriched pathways. In conjunction with the trend changes described above, this finding suggests that the Z4 stage may prepare for significant morphological changes that occur at the M stage (Figure 4A). The expression trend of Profile 26 is at a critical point in the Z4 stage. The expressions of DEGs in Profile 26 reached the minimum expression at the Z4 stage and then recovered at the M stage (Figure 3). In Profile 26 (Figure 4B), Cyanoamino acid metabolism (ko00460) and AMPK signaling pathway (ko04152) related to energy metabolism were significantly enriched.
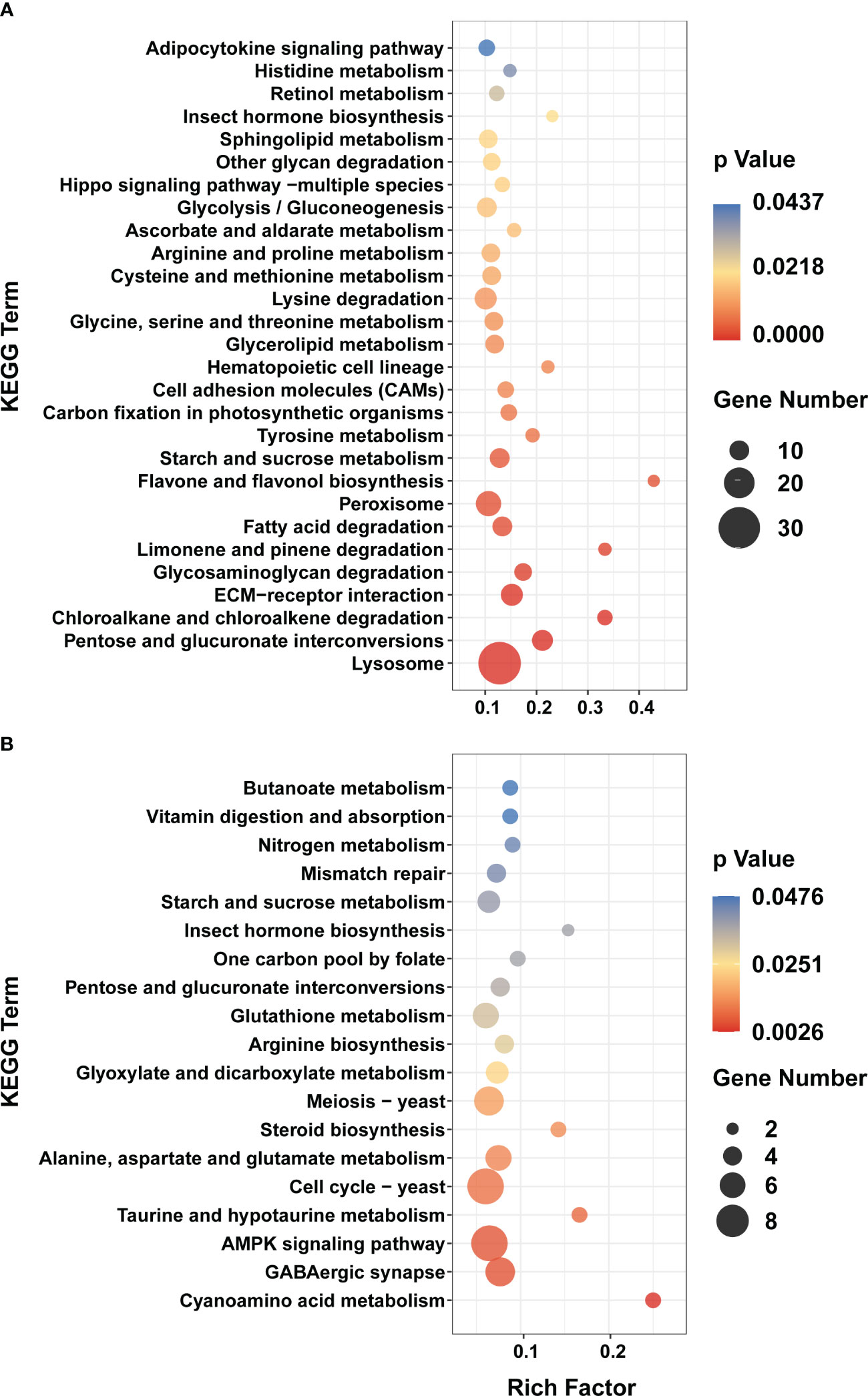
Figure 4 KEGG enrichment analyses of DEGs in Profile 22 and 26. (A) Profile 22. (B) Profile 26. Y axis represents pathway name. The color indicates the p-value, the lower p-value indicates the more significant enrichment. Point size indicates DEG number (The larger dots refer to a larger amount).
3.3 Functional enrichment analysis for DEGs
To acquire complete functional information and pathway-based analysis of DEGs, GO and KEGG databases were annotated for gene classification and pathway analysis.
The top 5 most significantly enriched GO terms sorted by p-value were shown in Table 1, belonging to three categories. The most significant GO terms were related to signal transduction in comparing Z2 vs Z1. In addition, the top 5 most significant GO biological process terms involved with energy metabolism and others in comparing C vs M.
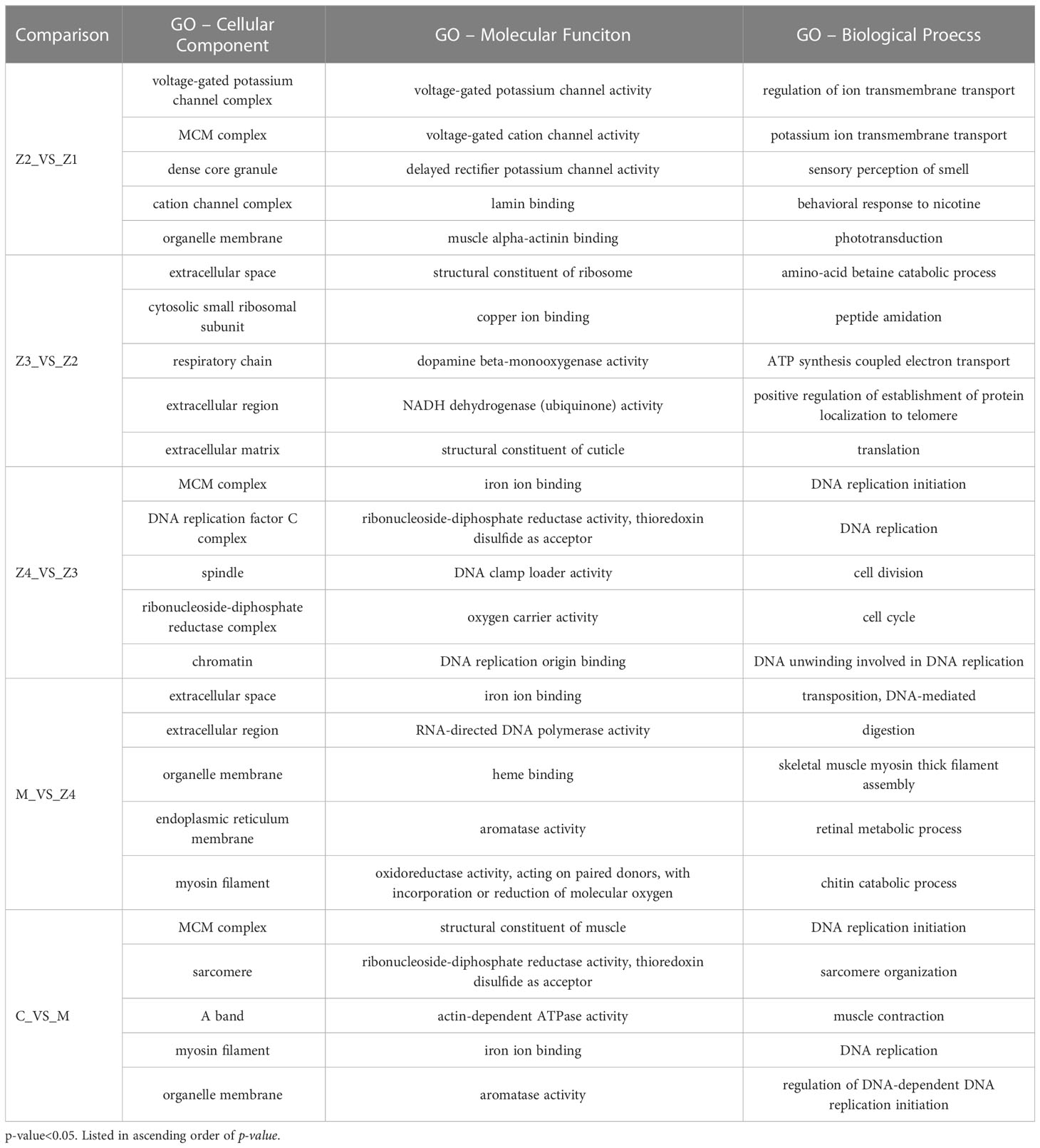
Table 1 The top 5 significantly enriched gene ontology (GO) terms of different functional classes at P. trituberculatus larval development stages.
For Z4 vs Z3 comparison, the top 5 most significant GO-BP terms are associated with DNA replication, cell division and cell cycle. For M vs Z4 comparison, the most significant GO-BP terms were “transposition, DNA-mediated,” followed by “digestion,” and “skeletal muscle myosin thick filament assembly.” “Retinal metabolic process” and “chitin catabolic process,” related to larval perception and morphological development, was also significant during this stage. In addition, the GO-BP term “phototransduction” is rich in Z2 vs Z1.
Interestingly, there is a GO-MF term (GO: 0008061, chitin binding) that is enriched in every comparison (Table 2). 12, 4, 6, 18, and 6 genes were enriched in the comparison of Z2 vs Z1, Z3 vs Z2, Z4 vs Z3, M vs Z4, and C vs M, respectively. The enriched genes mainly include chitinase, obstructor, cuticle protein, chitin-binding protein, peritrophin and filaggrin-1-like. Among them, the most up-regulated is the chitinase 3 gene (FC = 1986) in M vs Z4 comparison, while the most down-regulated is the obstructor A gene (FC = −14.5) in Z4 vs Z3 comparison. In the Z4 vs Z3 comparison, only one (chitin-binding protein) of the six genes was up-regulated. In contrast, in the other comparisons, only one gene was down-regulated in the Z2 vs Z1 comparison (chitinase 3) and the M vs Z4 comparison (acidic mammalian chitinase-like).
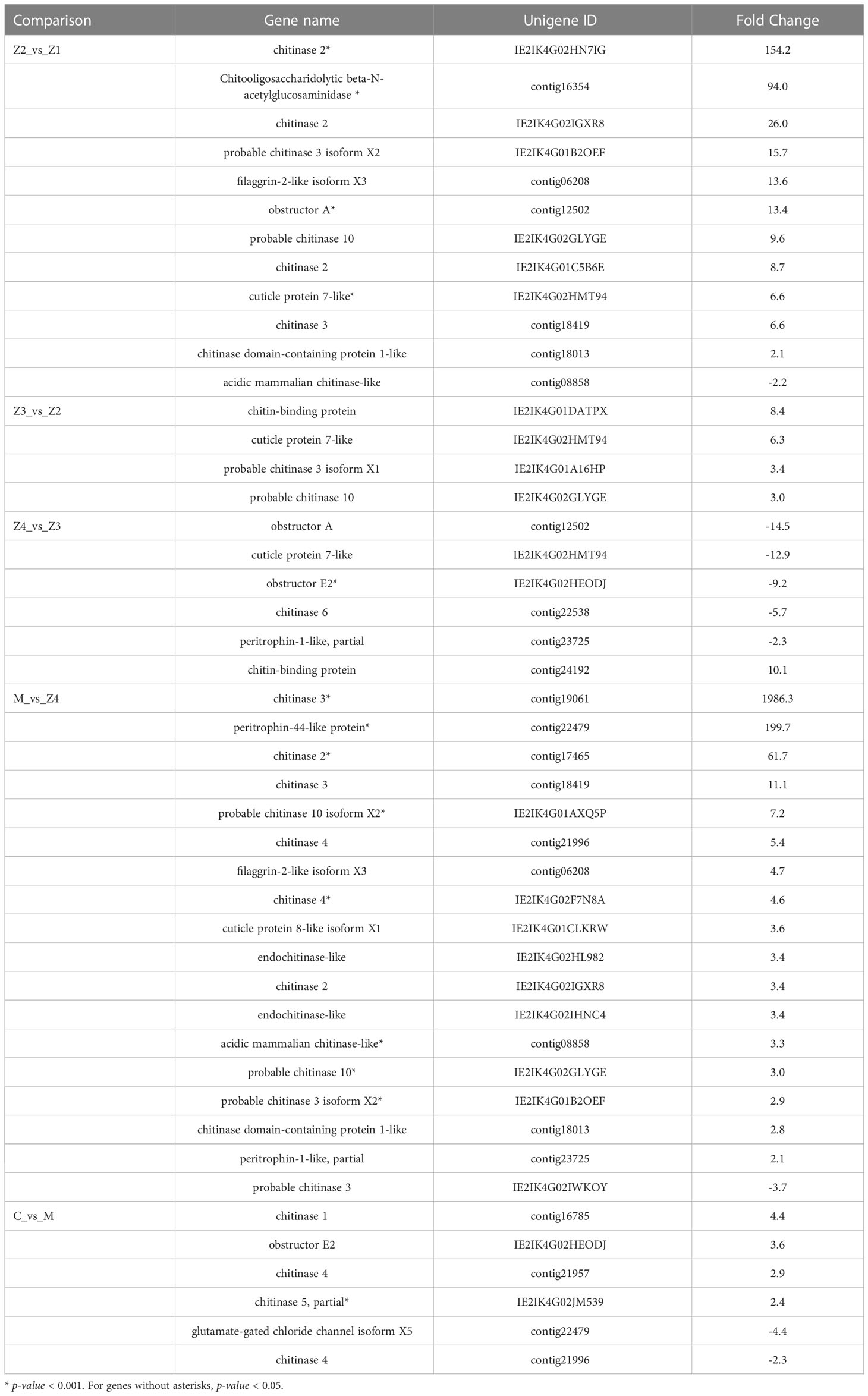
Table 2 Differentially expressed genes in “chitin binding” term among the adjacent larval stages of P. trituberculatus.
The KEGG pathway enrichment analysis was conducted to evaluate the pathways associated with DEGs. Pathway analysis can further understand genes’ biological functions (Kanehisa and Goto, 2000). The top ten enriched pathways of P. trituberculatus larval development stages sorted by p-value are listed in Table 3. Among the top ten pathways in each group, some occur multiple times, such as “DNA replication,” “cell cycle,” and related metabolic pathways. However, some, such as “retinol metabolism,” “insect hormone biosynthesis,” and “hematopoietic cell lineage” occurred only in the M vs Z4 comparison. More significant KEGG pathways were associated with DNA replication and meiosis in the Z4 vs Z3 and C vs M. The most important pathways were mainly related to signal transduction in the Z3 vs Z2 comparison.
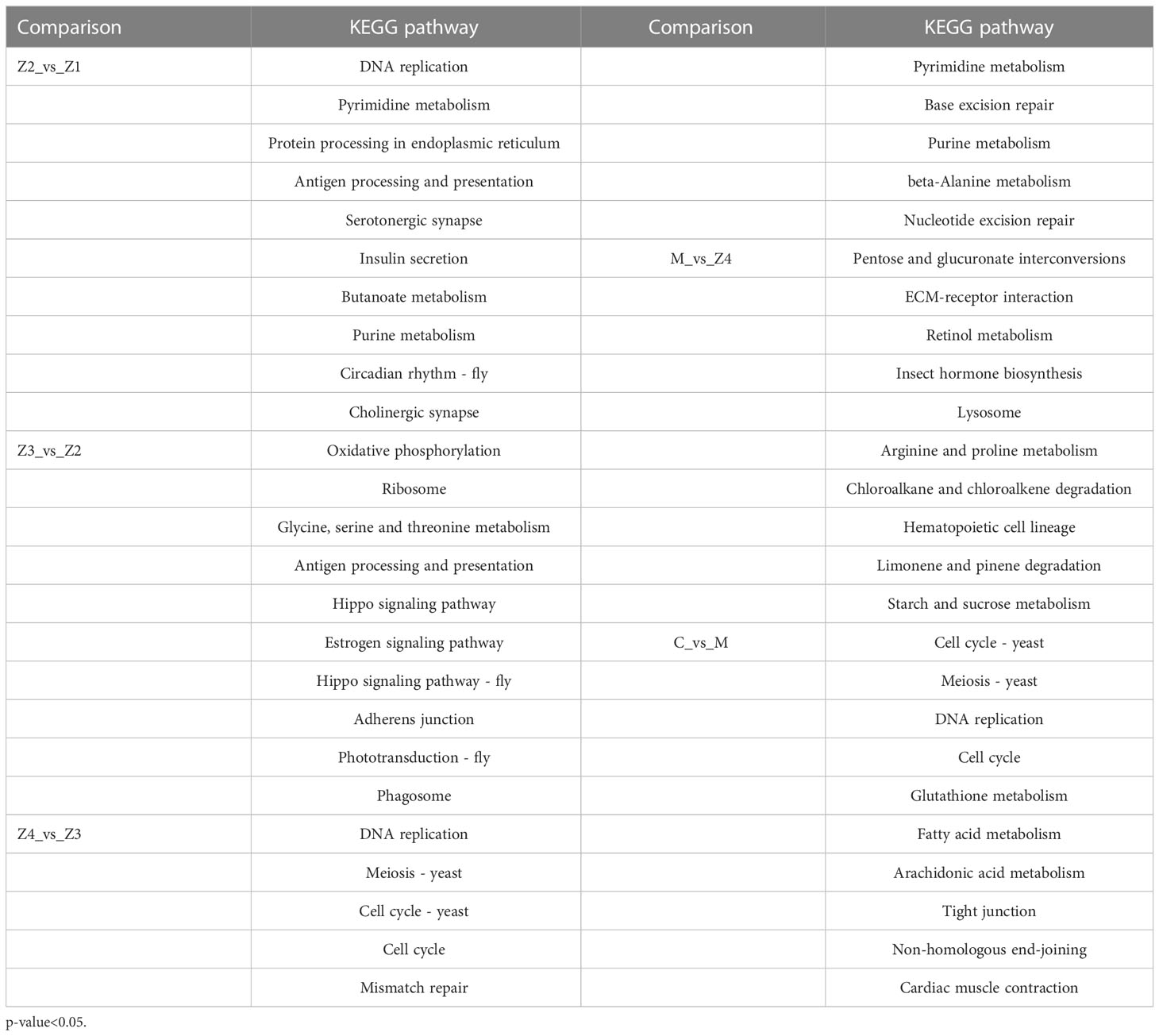
Table 3 The top 10 significantly enriched KEGG pathways at P. trituberculatus larval development stages.
3.4 qRT-PCR validation
To verify the accuracy of the sequencing data, five genes, chitooligosaccharidolytic beta-N-acetylglucosaminidase, peroxinectin, prostaglandin D synthase, heat shock protein 90-1 and pyruvate kinase, that were differentially expressed at most developmental stages, were selected for qRT-PCR. The gene expression trend (Figure 5) showed that the results were considered reliable for research.
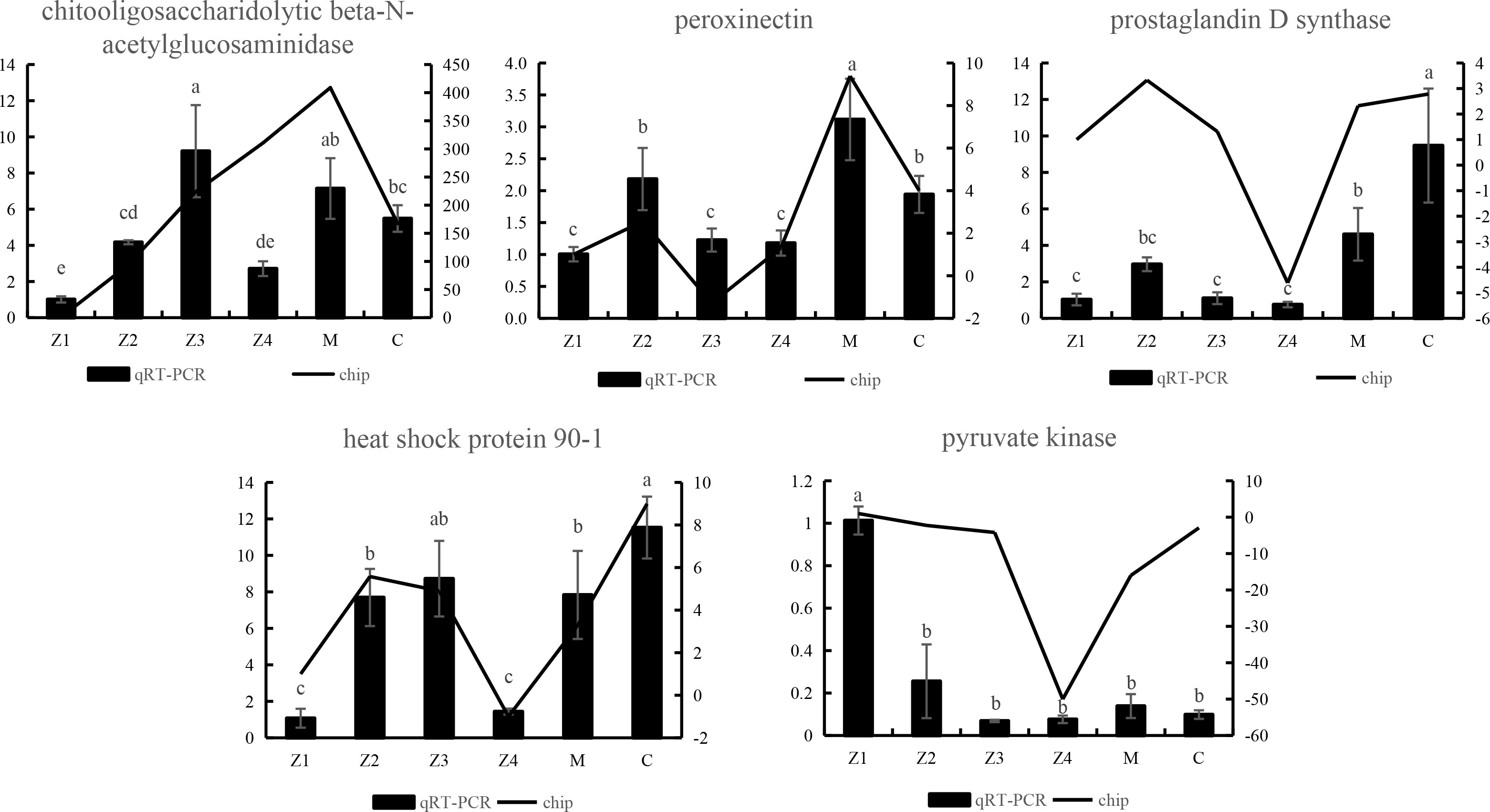
Figure 5 qRT-PCR verification of differentially expressed unigenes of P. trituberculatus at larvae development stages. Different letters indicate significant differences between qRT-PCR groups (p < 0.05).
4 Discussion
The larval development stage of P. trituberculatus is the most vulnerable stage in its life history, in which body structure and physiological traits change dramatically through metamorphosis (Lim and Hirayama, 1991). Larval development is a complex process involving multiple genes and is influenced by external environmental factors. In this study, the transcriptome of six developmental stages (Z1-C) of P. trituberculatus juveniles was sequenced to understand better the molec ular mechanism driving the change from one larval stage to the next in the early life cycle. The gene function enrichment and short time-series clustering analyses of different development stages of swimming crab larvae enabled us to explore the growth mechanism of larval development from the perspective of multi-gene interaction.
Like other invertebrates, swimming crab lack an adaptive immune system and rely on various innate immune responses to combat invading pathogens (Ren et al., 2016), especially during the larval development. In this study, we found that solute carrier family 15 (SLC15), the gene related to immune response, was always differentially expressed in all five comparative groups (Table S3). The solute carrier family (SLCs) are important membrane transport proteins transporting diverse solutes across biological membranes (Bai et al., 2017). The study of Litopenaeus vannamei suggests that SLC15A4 is involved in L. vannamei AMP regulation and anti-microbial response (Chen et al., 2016). In this study, SLC15 was significantly down-regulated at the Z4 stage of swimming crab larvae (Figure 2C). It suggests that most of the energy may be used for morphological changes during the metamorphic development (Dan et al., 2013), which leads to reduced immunity in Z4 stage.
4.1 Chitin development
Structural molting is a complex but essential process for crustaceans, mainly including the degradation of the old epidermis and the formation of new epidermis. The epidermis is a dynamic organism component, mainly composed of chitin, protein and calcium carbonate. In this research, the GO term chitin binding was enriched throughout the larval development. Proteins involved in this GO term mainly include cuticle protein, chitin-binding protein, chitinase, obstructor, peritrophin-1\44-like protein, and filaggrin-2-like.
The complex of chitin-binding protein and chitin provides an organic scaffold for calcification in crustaceans (Glazer et al., 2013). Currently, proteins that can bind to chitin mainly include cuticle proteins, chitinase, 30K protein, ATP synthase, and so on (Tang et al., 2010). The cuticle proteins combined with chitin to form a fibrous skeleton control the calcification of the epidermis and play essential roles in the formation of a new epidermis (Faircloth and Shafer, 2007). In the present study, the cuticle protein 7-like gene was only differentially expressed during the zoeal stages, with the highest expression in the Z3 stage, while the cuticle protein 8-like gene only significantly increased from Z4 to M stage. In Anopheles gambiae, some cuticle protein encoding genes were expressed exclusively at one metamorphic stage, while other cuticle proteins were expressed at single or multiple periods associated with molting (Togawa et al., 2008). The study of shrimp found that the diversity of cuticle proteins at the transcriptional level may help to rapidly generate exoskeletons with other physical properties at different growth stages (Wei et al., 2014). In Tribolium castaneum, the cuticle proteins in the elytra and hindwings were different, and their expression profiles were also different (Dittmer et al., 2012). Due to the various combinations of cuticle proteins and their effects on the arrangement of chitin bundles, the cuticles show other physical properties at each growth stage of arthropods (Tajiri et al., 2017). Thus, the results indicate that swimming crab larvae have different physical properties of exoskeleton between the zoeal stages and the megalopa stage.
The Obstructor family is a novel multigene family, representing an evolutionary conserved multigene family in invertebrates that encodes several putatively secreted chitin-binding proteins (Behr and Hoch, 2005). In Drosophila melanogaster, Obstructor-E (Obst-E) localizes to the larval cuticle, regulates the arrangement of chitin in the cuticle, and determines the pupal body shape by conferring oriented contractility/expandability on the cuticle (Tajiri et al., 2017). Moreover, its loss-of-function mutations cause body shape deformation at the level of the cuticle (Behr and Hoch, 2005). In Cherax quadricarinatus, Obstructor-A (Obst-A) proteins play a significant role in cuticle formation, which is related to their predicted ability to bind three chitin strands(Abehsera et al., 2018). Obst-A is required for extracellular matrix dynamics in cuticle-forming organs. Loss of obstructor-A causes severe defects during cuticle molting, wound protection, tube expansion, and larval growth control (Petkau et al., 2012). In the study, Obst-A and Obst-E genes were significantly down-regulated at the Z4 stage of swimming crab larvae (Table 2, Table S3). It indicates that the chitin cuticular of larvae crabs may undergo drastic changes (deformation or shedding) during the Z4 stage in preparation for metamorphosis development. In addition, the Obst-E gene was significantly highly expressed in the C stage (Table 2, Table S3), suggesting that the juvenile crab has a more vital mechanical ability to control their exoskeleton at this stage.
Peritrophin was first isolated from insect peritrophic membrane (PM) and was thought to protect insects from the invasion of microorganisms and stimulate food digestion (Huang et al., 2015). In Eriocheir Sinensis, peritrophin-44 could bind to different microbes, and enhance the clearance of Vibrio parahaemolyticus in vivo, indicating that it is involved in the anti-bacterial innate immunity of crabs (Huang et al., 2015). The peritrophin-44 protein was up-regulated 199.7-fold and 3.2-fold in the M vs Z4 and C vs M comparison, respectively. The gene expression is significantly increased from the Z4 to the C stage, indicating that these stages may induce a robust immune response from external stimuli. From the Z4 stage to the juvenile crab stage, it is already at the late stage of the nursery, and the water quality is relatively poorer than in the early stage.
Moreover, the larvae have undergone two significant morphological changes, consuming more energy and materials. Therefore, the high expression of peritrophin-44 may be a reaction of larvae crabs to these situations during the Z4-C stages. It is known that the hydrolysis of chitin is performed by chitinase, of which 14 molecular types have been identified in crustaceans (Huang et al., 2010). Multiple chitinase genes are usually present in a single individual, and some are responsible for chitin decomposition in the cuticle. In contrast, others are related to immunity and digestion (Huang et al., 2010). Throughout the development process, seven kinds of chitinases are differentially expressed at different stages. Both Cht-1 (chitinase 1) and Cht-3 (chitinase 3) are mainly involved in the digestion of chitin in food and intestinal peritrophic membrane in Penaeus japonicus, and Cht-1 were also found in the defense against the virus in P. trituberculatus (Watanabe et al., 1996; Watanabe et al., 1998; Song et al., 2020). Cht-2 (chitinase 2) played an essential role in the decomposition and reabsorption of chitin in the epidermis during the molting of Penaeus monodon (Proespraiwong et al., 2010). In addition, Cht-4 was expressed in the site of synthesis for digestive enzymes in Penaeus monodon (Huang et al., 2010). These chitinase genes are more differentially expressed and have higher FC during the later stages of larval development (M and C stages) of P. trituberculatus, which might be due to the different shell compositions of swimming crab larvae at different growth stages. They may also be related to the typical diet at the different stages. At the later stages of larval breeding, Artemia nauplii were used as the diet, which chitinases should decompose. Moreover, the water environment became more complex in the later stages of larval breeding. These genes also play an immune role, consistent with the enrichment of immune-related pathways in swimming crab larvae.
In addition, the analysis revealed that the expression of the beta-N-acetylglucosaminidase (NAG) gene was significantly up-regulated in the Z2 stage compared to the Z1 stage (Table 2). This result was also verified in the qPCR analysis (Figure 5). NAG genes are the critical enzyme for chitin degradation, mainly involved in the functions such as molting and food digestion, just like chitinases (Zhang et al., 2021). The expression of NAG is known to fluctuate during the crustacean molting cycle (Meng and Zou, 2009). The expression of NAG is significantly up-regulated in the pre-molt stage of the Uca pugilator, E. sinensis, and Scylla serrata (Salaenoi et al., 2006; Meng and Zou, 2009; Li et al., 2015), which is also the stage when various physiological functions and epidermal structures undergo significant changes. Significantly high expression of the NAG gene during the pre-molt stage ensures effective degradation of the old exoskeleton. Therefore, the results suggest that swimming crab larvae may undergo more thorough exoskeletal remodeling during Z2 than the Z1 stage.
4.2 Energy metabolism
The early development of P. trituberculatus gets through zoeal stages and megalopa stage. The unique early developmental mode implies dramatic changes and indicates the enormous energy required to support it. In the study, enrichment analysis of DEGs showed that plenty of the differentially expressed genes were involved in energy metabolic processes.
In this research, the genes activating “AMPK signaling pathway” (ko04152) (TGF-β-activated kinase 1, G6PC, and PCK, etc.) (Xie et al., 2006; Ju et al., 2012) clustered in Profile 26, and were significantly up-regulated between M vs Z4 stages (Table S4). AMPK is a “metabolic master switch” that mediates the cellular adaptation to nutritional and environmental variations depleting intracellular ATP levels, including heat shock, hypoxia, starvation, or prolonged exercise. Regardless, AMPK activation results in the inhibition of energy-consuming biosynthetic pathways and the activation of ATP-producing catabolic pathways. AMPK can also modulate the transcription of specific genes involved in energy metabolism, thereby exerting long-term metabolic control (Viollet et al., 2009). The result indicates that Z4-M is a stage of high energy demand, which may be related to the drastic morphological changes of larvae crabs.
4.3 Neuroendocrine development
As P. trituberculatus larvae developed, the gene expression of pathways related to the endocrine and nervous systems showed significant differences between adjacent stages.
The gene related to olfactory function (UDP-glucosyltransferase 2) was differentially expressed in all five comparative groups (Table S3). In particular, UDP-glucosyltransferase 2 (UGT2) was highly expressed in stage C but lowly in stage Z4 (Figure 2C). UDP-glucosyltransferases (UGTs) are the group of enzymes linked to odorant and xenobiotic degradation (Ahn et al., 2012). The role of UGTs in vertebrate olfaction is well established (Heydel et al., 2010). In the study of Drosophila melanogaster, several UGT genes were preferentially expressed in the olfactory organ (antenna), suggesting that UGTs may be involved in deactivation of pheromones (Wang et al., 1999; Younus et al., 2014). Thus, the expression of UGT2 during the larval development indicates that the olfactory function of larvae crab may not be broadly developed until the C stage.
Meanwhile, during larval metamorphosis development (Z4-M), several genes involved in retinol metabolism (ko00830) were differentially significantly up-regulated (retinol dehydrogenase11, 12, 13 and 14 and β-carotene 9’,10’-oxygenase) (Table S5). It is worth noting that RDHs mediate key retinoid transformations in vision and critical functions in eye development by regulating retinol metabolism during embryonic development (Haeseleer et al., 2002; Belyaeva et al., 2008). In addition, as a potential retinoic acid-producing enzyme during embryonic development, BCO2 is also crucial for the visual cycle and eye development (Spiegler et al., 2013). Thus, this finding may indicate that the M stage is critical for developing vision in larvae crabs.
Crustaceans and insects both belong to the phylum Arthropoda. There are similarities in their physiological processes, such as growth and molting. It is worth noting that the DEGs of insect hormone biosynthesis (ko00981) were differentially expressed at M-Z4 stages (Table S6). Cytochrome P450 (CYP15) and Juvenile Hormone Epoxide Hydrolase (JHEH) were enriched in this pathway, and their expressions were significantly higher in M stage than in Z4 stage. It has been found that CYP15 and JHEH might be involved in the degradation of the endocrine hormone Methyl farnesoate (MF) in crustaceans (Qiu et al., 2015; Tu et al., 2022). Like the pleiotropic juvenile hormone (JH) in insects, MF can promote the occurrence of molting, control morphogenesis, and gonadal development, and regulate the stress regulation to the external environment of crustaceans (Qiu et al., 2015; Tu et al., 2022). This result indicated that these crucial physiological processes might be enhanced in the Z4 stage of P. trituberculatus, which made adequate preparations for transforming into megalopa larvae.
The insulin-like androgenic gland (IAG) is a key regulator of crustacean reproductive processes such as spermatogenesis, sexual differentiation, and sexual transition (Huang et al., 2014). The IAG gene was differentially expressed at the Z4-M stage (Table S6). Studies have found that the transcripts of IAG in Scylla paramamosain and E. sinensis are widely distributed in various tissues of male and female individuals (Huang et al., 2014; Liu et al., 2015). Moreover, the study found that the knockdown of IAG can lead to sex reversal in Macrobrachium rosenbergii (Lezer et al., 2015). This study indicates that the megalopa stage might represent a critical phase for the development and high expression of secondary sex characteristics in swimming crabs. These findings establish a groundwork for investigating the mechanisms of sex determination in crustaceans.
5 Conclusion
The present study describes the comprehensive transcriptomic sequence characterization at the larval development stages of P. trituberculatus, in which many differentially expressed growth and development genes were identified. The differentially expressed genes are involved in morphological change, growth metabolism, sensory ability, and neuroendocrine system of larvae crab. Furthermore, the Z4-M stage is the crucial stage for the growth and development of larvae crab based on the comparison of DEGs and functional enrichment analysis. Overall, these findings provided an essential basis for clarifying the growth and development mechanisms of larvae crab, contributing to promoting the upgrading of artificial breeding technology of P. trituberculatus.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Gene Expression Omnibus, GSE226561.
Author contributions
CM conceived and designed the study. JC performed the cultivation and sample collection of experimental animals. JC, XC, YY, RL, WS, CS, LL, HW and CW performed and analyzed all other experiments. JC and CM wrote the manuscript with support from all authors. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (No. 42076087 and No. 42276101), the Modern Technology System of Agricultural Industry (No. CARS- 48), and the K C Wong Magana Fund at Ningbo University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1172214/full#supplementary-material
References
Abehsera S., Zaccai S., Mittelman B., Glazer L., Weil S., Khalaila I., et al. (2018). CPAP3 proteins in the mineralized cuticle of a decapod crustacean. Sci. Rep. 8 (1), 2430. doi: 10.1038/s41598-018-20835-x
Ahn S. J., Vogel H., Heckel D. G. (2012). Comparative analysis of the UDP-glycosyltransferase multigene family in insects. Insect Biochem. Mol. Biol. 42 (2), 133–147. doi: 10.1016/j.ibmb.2011.11.006
Anger K., Queiroga H., Calado R. (2015). “Larval development and behaviour strategies in brachyura,” in Treatise on zoology-anatomy, taxonomy, biology. the Crustacea. (Leiden: Brill), vol. 9, 317–374.
Arai D., Takeuchi T., Sugita D., Hamasaki K., Maruyama K. (2007). Effect of n-3 HUFA-chlorella supplied to rearing tank on morphogenesis of swimming crab zoeas of the Portunus trituberculatus (Brachyura: portunidae). Aquac Sci. 55, 133–134. doi: 10.2508/chikusan.69.875
Bai X., Moraes T. F., Reithmeier R. A. F. (2017). Structural biology of solute carrier (SLC) membrane transport proteins. Mol. Membr Biol. 34 (1-2), 1–32. doi: 10.1080/09687688.2018.1448123
Behr M., Hoch M. (2005). Identification of the novel evolutionary conserved obstructor multigene family in invertebrates. FEBS Lett. 579 (30), 6827–6833. doi: 10.1016/j.febslet.2005.11.021
Bellin D., Ferrarini A., Chimento A., Kaiser O., Levenkova N., Bouffard P., et al. (2009). Combining next-generation pyrosequencing with microarray for large scale expression analysis in non-model species. BMC Genomics 10 (1), 1–9. doi: 10.1186/1471-2164-10-555
Belyaeva O. V., Korkina O. V., Stetsenko A. V., Kedishvili N. Y. (2008). Human retinol dehydrogenase 13 (RDH13) is a mitochondrial short-chain dehydrogenase/reductase with a retinaldehyde reductase activity. FEBS J. 275 (1), 138–147. doi: 10.1111/j.1742-4658.2007.06184.x
Chen Y. G., Yuan K., Zhang Z. Z., Yuan F. H., Weng S. P., Yue H. T., et al. (2016). Identification and functional characterization of a solute carrier family 15, member 4 gene in Litopenaeus vannamei. Dev. Comp. Immunol. 57, 57–66. doi: 10.1016/j.dci.2015.12.005
Dan S., Kaneko T., Takeshima S., Ashidate M., Hamasaki K. (2013). Variations in larval morphology and their relationships to survival during mass seed production by the swimming crab, Portunus trituberculatus (Brachyura, portunidae). Aquaculture 414-415, 109–118. doi: 10.1016/j.aquaculture.2013.07.029
Dan S., Sui Y., Kaneko T., Takeshima S., Ojima D., Miyoshi T., et al. (2016). Abnormal morphology in megalopae of the swimming crab, Portunus trituberculatus during seed production: causes and prevention. Aquaculture Res. 47 (10), 3189–3202. doi: 10.1111/are.12771
Dan S., Yamazaki H., Hamasaki K. (2017). Effects of mother and postnatal food condition on larval performance of swimming crab Portunus trituberculatus. Fisheries Sci. 84 (1), 113–126. doi: 10.1007/s12562-017-1151-8
Dittmer N. T., Hiromasa Y., Tomich J. M., Lu N., Beeman R. W., Kramer K. J., et al. (2012). Proteomic and transcriptomic analyses of rigid and membranous cuticles and epidermis from the elytra and hindwings of the red flour beetle, Tribolium castaneum. J. Proteome Res. 11 (1), 269. doi: 10.1021/pr2009803
Ernst J., Bar-Joseph Z. (2006). STEM: a tool for the analysis of short time series gene expression data. BMC Bioinf. 7 (1), 1–11. doi: 10.1186/1471-2105-7-191
Faircloth L. M., Shafer T. H. (2007). Differential expression of eight transcripts and their roles in the cuticle of the blue crab, Callinectes sapidus. Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 146 (3), 370–383. doi: 10.1016/j.cbpb.2006.11.008
Fishery and Fisheries Administrative Bureau (2023). Ministry of agriculture. China fishery statistics yearbook of 202 (Beijing: Chinese Agricultural Press).
Gao L., Wang Y. (2021). Influences of environmental factors on the spawning stock-recruitment relationship of Portunus trituberculatus in the northern East China Sea. Acta Oceanologica Sin. 40 (8), 145–159. doi: 10.1007/s13131-021-1801-7
Glazer L., Tom M., Weil S., Roth Z., Khalaila I., Mittelman B., et al. (2013). Hemocyanin with phenoloxidase activity in the chitin matrix of the crayfish gastrolith. J. Exp. Biol. 216 (10), 1898–1904. doi: 10.1242/jeb.080945
Haeseleer F., Jang G. F., Imanishi Y., Driessen C. A. G. G., Matsumura M., Nelson P. S., et al. (2002). Dual-substrate specificity short chain retinol dehydrogenases from the vertebrate retina. J. Biol. Chem. 277 (47), 45537–45546. doi: 10.1074/jbc.M208882200
Hamasaki K. (1996). Study on the reproduction and development of the swimming crab, Portunus trituberculatus. Special Sci. Rep. Japan Sea-Farming Assoc. 8, 1–124.
Hamasaki K., Obata Y., Dan S., Kitada S. (2011). A review of seed production and stock enhancement for commercially important portunid crabs in Japan. Aquaculture Int. 19 (2), 217–235. doi: 10.1007/s10499-010-9387-7
Heydel J. M., Holsztynska E. J., Legendre A., Thiebaud N., Artur Y., LeBon A. M. (2010). UDP-Glucuronosyltransferases (UGTs) in neuro-olfactory tissues: expression, regulation, and function. Drug Metab. Rev. 42 (1), 74–97. doi: 10.3109/03602530903208363
Huang Y., Ma F. T., Wang W., Ren Q. (2015). Identification and molecular characterization of a peritrophin-like gene, involved in the antibacterial response in Chinese mitten crab, Eriocheir sinensis. Dev. Comp. Immunol. 50 (2), 129–138. doi: 10.1016/j.dci.2015.01.002
Huang Q. S., Yan J. H., Tang J. Y., Tao Y. M., Xie X. L., Wang Y., et al. (2010). Cloning and tissue expressions of seven chitinase family genes in Litopenaeus vannamei. Fish shellfish Immunol. 29 (1), 75–81. doi: 10.1016/j.fsi.2010.02.014
Huang X., Ye H., Huang H., Yang Y., Gong J. (2014). An insulin-like androgenic gland hormone gene in the mud crab, Scylla paramamosain, extensively expressed and involved in the processes of growth and female reproduction. Gen. Comp. Endocrinol. 204, 229–238. doi: 10.1016/j.ygcen.2014.06.002
Huerlimann R., Wade N. M., Gordon L., Montenegro J. D, Goodall J., McWilliam S., et al. (2018). De novo assembly, characterization, functional annotation and expression patterns of the black tiger shrimp (Penaeus monodon) transcriptome. Sci. Rep. 8 (1), 1–14. doi: 10.1038/s41598-018-31148-4
Ju T. C., Lin Y. S., Chern Y. (2012). Energy dysfunction in huntington’s disease: insights from PGC-1α, AMPK, and CKB. Cell. Mol. Life Sci. 69 (24), 4107–4120. doi: 10.1007/s00018-012-1025-2
Kanehisa M., Goto S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28 (1), 27–30. doi: 10.1093/nar/28.1.27
Lezer Y., Aflalo E. D., Manor R., Sharabi O., Abilevich L. K., Sagi A. (2015). On the safety of RNAi usage in aquaculture: the case of all-male prawn stocks generated through manipulation of the insulin-like androgenic gland hormone. Aquaculture 435, 157–166. doi: 10.1016/j.aquaculture.2014.09.040
Li E., Wang S., Li C., Chen K., Chen L. (2014). Transcriptome sequencing revealed the genes and pathways involved in salinity stress of Chinese mitten crab, Eriocheir sinensis. Physiol. Genomics 46 (5), 177–190. doi: 10.1152/physiolgenomics.00191.2013
Li X., Xu Z., Zhou G., Lin H., Zhou J., Zeng Q., et al. (2015). Molecular characterization and expression analysis of five chitinases associated with molting in the Chinese mitten crab, Eriocheir sinensis. Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 187, 110–120. doi: 10.1016/j.cbpb.2015.05.007
Lim B. K., Hirayama K. (1991). Growth and elemental composition (C, n, p) during larval developmental stages of mass-cultured swimming crab Portunus trituberculatus. Mar. Ecol. Prog. 78 (2), 131–137. doi: 10.3354/meps078131
Liu Y., Hui M., Cui Z., Luo D., Song C., Li Y., et al. (2015). Comparative transcriptome analysis reveals sex-biased gene expression in juvenile Chinese mitten crab Eriocheir sinensis. PloS One 10 (7), e0133068. doi: 10.1145/2818302
Livak K. J., Schmittgen T. D. (2002). Analysis of relative gene expression data using real-time quantitative PCR. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Lu Z. B., Ren Z. M., Lin W. C., Shi C., Mu C. K., Wang C. L., et al. (2022). Succession, sources, and assembly of bacterial community in the developing crab larval microbiome. Aquaculture 548, 737600. doi: 10.1016/j.aquaculture.2021.737600
Meng Y., Zou E. (2009). Impacts of molt-inhibiting organochlorine compounds on epidermal ecdysteroid signaling in the fiddler crab, Uca pugilator, in vitro. Comp. Biochem. Physiol. Part C: Toxicol. Pharmacol. 150 (4), 436–441. doi: 10.1016/j.cbpc.2009.06.009
Muroga K., Suzuki K., Ishibashi N., Nogami K. (1989). A vibriosis occurring in zoeal larvae of swimming crab Portunus trituberculatus. Aquaculture Sci. 37, 133–141. doi: 10.11233/aquaculturesci1953.37.133
Petkau G., Wingen C., Jussen L. C., Radtke T., Behr M. (2012). Obstructor-a is required for epithelial extracellular matrix dynamics, exoskeleton function, and tubulogenesis. J. Biol. Chem. 287 (25), 21396–21405. doi: 10.1074/jbc.M112.359984
Proespraiwong P., Tassanakajon A., Rimphanitchayakit V. (2010). Chitinases from the black tiger shrimp Penaeus monodon: phylogenetics, expression and activities. Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 156 (2), 86–96. doi: 10.1016/j.cbpb.2010.02.007
Qiu X. E., Zhu D. F., Tang J., Cui X. Y., Zhou Y. Q., Liu Z. Y. (2015). Review of the physiological functions and their mechanisms of methyl farnesoate in crustacean. J. Biol. 32 (1), 6.
Ren X., Wang Z., Gao B., Liu P., Li J. (2017). Toxic responses of swimming crab (Portunus trituberculatus) larvae exposed to environmentally realistic concentrations of oxytetracycline. Chemosphere 173, 563–571. doi: 10.1016/j.chemosphere.2017.01.078
Ren X., Yu X., Gao B., Liu P. (2016). The immune responses and antioxidant status of Portunus trituberculatus individuals with different body weights. Fish Shellfish Immunol. 51, 337–345. doi: 10.1016/j.fsi.2016.03.004
Salaenoi J., Bootpugdeethum J., Mingmuang M., Thongpan A. (2006). Chitobiase, proteinase, glycogen and some trace elements during molting cycle of mud crab (Scylla serrata forskål 1775). Agric. Natural Resour. 40 (2), 517–528.
Song L., Lv J., Wang L., Sun D., Gao B., Liu P. (2020). Characterization of a chitinase-1 gene (PtCht-1) from a marine crab Portunus trituberculatus and its response to immune stress. Gene 741, 144523. doi: 10.1016/j.gene.2020.144523
Spiegler E., Kim Y., Quadro L. (2013). Does beta-carotene-9’,10’-oxygenase (CMO2) generate retinoic acid during embryonic development? The FASEB Journal. 27 (S1), 32.7–32.7. doi: 10.1096/fasebj.27.1_supplement.32.7
Sun Y. M., Song Z. L., Yan R. S., Gong M. J., Sun J. J. (1984). Preliminary study on the growth of Portunus trituberculatus. ActaEcologicaSinica 4 (1), 57–64.
Tajiri R., Ogawa N., Fujiwara H., Kojima T. (2017). Mechanical control of whole body shape by a single cuticular protein obstructor-e in Drosophila melanogaster. PloS Genet. 13 (1), e1006548. doi: 10.1371/journal.pgen.1006548
Tang L., Liang J., Zhan Z., Xiang Z., He N. (2010). Identification of the chitin-binding proteins from the larval proteins of silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 40 (3), 228–234. doi: 10.1016/j.ibmb.2010.01.010
Togawa T., Dunn W. A., Emmons A. C., Nagao J., Willis J. H. (2008). Developmental expression patterns of cuticular protein genes with the R&R consensus from Anopheles gambiae. Insect Biochem. Mol. Biol. 38 (5), 508–519. doi: 10.1016/j.ibmb.2007.12.008
Tu S. S., Tuo P., Xu D. J., Wang Z., Wang M., Xie X., et al. (2022). Molecular characterization of the cytochrome P450 epoxidase (CYP15) in the swimming crab portunus trituberculatus and its putative roles in methyl farnesoate metabolism. Biol. Bull. 242 (2), 75–86. doi: 10.1086/719047
Viollet B., Lantier L., Devin-Leclerc J., Hebrard S., Amouyal C., Mounier R., et al. (2009). Targeting the AMPK pathway for the treatment of type 2 diabetes. Front. bioscience (Landmark edition) 14, 3380. doi: 10.2741/3460
Wang Q., Hasan G., Pikielny C. W. (1999). Preferential expression of biotransformation enzymes in the olfactory organs of Drosophila melanogaster, the antennae. J. Biol. Chem. 274 (15), 10309–10315. doi: 10.1074/jbc.274.15.10309
Wang C., Jiang L. X., Wang R., Quan L. Y. (2010). Effect of abrupt and gradual changes in salinity on development and feeding in juvenile swimming crab (Portunus trituberculatus). Fisheries Sci. (Dalian) 29 (9), 510–514.
Watanabe T., Kono M., Aida K., Nagasawa H. (1996). Isolation of cDNA encoding a putative chitinase precursor in the kuruma prawn Penaeus japonicus. Mol. Mar. Biol. Biotechnol. 5 (4), 299–303.
Watanabe T., Kono M., Aida K., Nagasawa H. (1998). Purification and molecular cloning of a chitinase expressed in the hepatopancreas of the penaeid prawn Penaeus japonicus. Biochim. Biophys. Acta (BBA)-Protein Structure Mol. Enzymology 1382 (2), 181–185. doi: 10.1016/S0167-4838(97)00184-2
Wei J., Zhang X., Yu Y., Huang H., Li F., Xiang J. (2014). Comparative transcriptomic characterization of the early development in pacific white shrimp Litopenaeus vannamei. PloS One 9 (9), e106201. doi: 10.1371/journal.pone.0106201
Wu X., Cheng Y., Zeng C., Wang C. L., Yang X. Z. (2010). Reproductive performance and offspring quality of wild-caught and pond-reared swimming crab Portunus trituberculatus broodstock. Aquaculture 301 (1-4), 78–84. doi: 10.1016/j.aquaculture.2010.01.016
Wu X., Zeng C. S., Southgate P. C. (2013). Ontogenetic patterns of growth and lipid composition changes of blue swimmer crab larvae: insights into larval biology and lipid nutrition. Mar. Freshw. Res. 65 (3), 228. doi: 10.1071/MF13078
Xie M., Zhang D., Dyck J R. B., Li Y., Zhang H., Morishima M., et al. (2006). A pivotal role for endogenous TGF-β-activated kinase-1 in the LKB1/AMP-activated protein kinase energy-sensor pathway. Proc. Natl. Acad. Sci. 103 (46), 17378–17383. doi: 10.1073/pnas.0604708103
Xu Q., Liu Y. (2011). Gene expression profiles of the swimming crab Portunus trituberculatus exposed to salinity stress. Mar. Biol. 158 (10), 2161–2172. doi: 10.1007/s00227-011-1721-8
Xu Q., Liu R., Liu Y. (2009). Genetic population structure of the swimming crab, Portunus trituberculatus in the East China Sea based on mtDNA 16S rRNA sequences. J. Exp. Mar. Biol. Ecol. 371 (2), 121–129. doi: 10.1016/j.jembe.2009.01.014
Younus F., Chertemps T., Pearce S. L., Pandey G., Bozzolan F., Coppin C. W., et al. (2014). Identification of candidate odorant degrading gene/enzyme systems in the antennal transcriptome of Drosophila melanogaster. Insect Biochem. Mol. Biol. 53 (4), 30–43. doi: 10.1016/j.ibmb.2014.07.003
Zhang W., Lv J., Lan W., Gao B. Q., Liu P. (2022). Discovery of sex-determining genes in Portunus trituberculatus: a comparison of Male and female transcriptomes during early developmental stages. Front. Mar. Sci. 8, 811052. doi: 10.3389/fmars.2021.811052
Zhang X., Yuan J., Li F., Xiang J. (2021). Chitin synthesis and degradation in crustaceans: a genomic view and application. Mar. Drugs 19 (3), 153. doi: 10.3390/md19030153
Zhao J., He Z., Chen X., Huang Y., Xie J., Qin X., et al. (2021). Growth trait gene analysis of kuruma shrimp (Marsupenaeus japonicus) by transcriptome study. Comp. Biochem. Physiol. Part D: Genomics Proteomics 40, 100874. doi: 10.1016/j.cbd.2021.100874
Keywords: Portunus trituberculatus, larval development, transcriptomic analysis, differentially expressed gene, microarray
Citation: Chen J, Chen X, Mu C, Wang C, Ye Y, Li R, Song W, Shi C, Liu L and Wang H (2023) Comparative transcriptome analysis reveals the growth and development in larval stages of the swimming crab Portunus trituberculatus. Front. Mar. Sci. 10:1172214. doi: 10.3389/fmars.2023.1172214
Received: 23 February 2023; Accepted: 02 June 2023;
Published: 23 June 2023.
Edited by:
Adriana Muhlia, National Council of Science and Technology (CONACYT), MexicoReviewed by:
Omar Mendoza-Porras, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaArturo Sanchez, Laboratorio de Virología, Mexico
Copyright © 2023 Chen, Chen, Mu, Wang, Ye, Li, Song, Shi, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changkao Mu, bXVjaGFuZ2thb0BuYnUuZWR1LmNu; Chunlin Wang, d2FuZ2NodW5saW5AbmJ1LmVkdS5jbg==
 Jiameng Chen1
Jiameng Chen1 Changkao Mu
Changkao Mu Chunlin Wang
Chunlin Wang Yangfang Ye
Yangfang Ye Ronghua Li
Ronghua Li Weiwei Song
Weiwei Song Ce Shi
Ce Shi Lei Liu
Lei Liu Huan Wang
Huan Wang