
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 27 April 2023
Sec. Deep-Sea Environments and Ecology
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1170238
This article is part of the Research Topic Bioremediation for a Greener Planet: Removal of Excess Nutrients, Heavy Metals, Hydrocarbons, and Other Pollutants by Aquatic Microorganisms View all 4 articles
 Tong Hao1
Tong Hao1 Zhisong Cui2,3*
Zhisong Cui2,3* Xiao Luan4
Xiao Luan4 Guangzhu Zhou1
Guangzhu Zhou1 Yingchao Li2
Yingchao Li2 Jinyan Liu2
Jinyan Liu2 Junhui Chen2,3
Junhui Chen2,3 Zongling Wang2,3*
Zongling Wang2,3*Hydrocarbons are ubiquitous in marine environments and might fuel hydrocarbon-metabolizing microbes in the ocean. Numerous studies have documented microbial hydrocarbon degradation in water columns and deep-sea surface sediment. However, the degradation potential and biogeochemical cycling of hydrocarbons in subsurface sediments remain largely unknown. In this study, we used two different hydrocarbons, n-hexadecane (HEX) and methylcyclohexane (MCH), to investigate the distribution and diversity of hydrocarbon-consuming bacteria in a core sediment sample from the Central Indian Ridge (CIR), which is adjacent to mid-ridge hydrothermal vents in the Indian Ocean. We observed different vertical profiles of HEX- and MCH-degrading bacteria in the core sediments. Specifically, HEX-degrading bacteria were universally distributed, while MCH-degrading bacteria were found only in the intermediate layers of the core sediments. Changing factors including dissolved oxygen might affect the natural distribution of different hydrocarbon consumers. We found that a novel species of the genus C1-B045 might play a pivotal role in metabolizing MCH in the CIR deep biosphere. Through amino acid identity comparison with published sequences, we determined that C1-B045 harbors two novel classes of cyclohexanone monooxygenases involved in MCH metabolism. This study sheds light on the structure and function of hydrocarbon-consuming microbes in deep biospheres.
Hydrocarbons are constantly introduced into the marine environment via both natural oil seeps and anthropogenic oil spills and are therefore universally distributed in the ocean. A diverse assemblage of hydrocarbon-containing crude oils regularly escapes into the marine environment via natural seepage from underground reservoirs (Atlas and Hazen, 2011). In addition, the hydrocarbon biosynthesis by cyanobacteria also contributes alkanes and alkenes (C13–C17) to the ocean (Schirmer et al., 2010). Love et al. (2021) estimated that the global flux of pentadecane (C15) produced by cyanobacteria exceeds the total oil input in the ocean by at least 100-fold (Love et al., 2021). Alkaline hydrothermal systems also contribute to the abiotic synthesis of hydrocarbons (even long-chain alkanes up to C24) and other organic compounds under hydrothermal conditions (Konn et al., 2009; Colman et al., 2017; He et al., 2021). Therefore, the environmental impact, biogeochemical cycling, and fate of hydrocarbons in the ocean have drawn a considerable amount of attention. Microorganisms play a pivotal role in driving the ocean hydrocarbon cycle (Lea-Smith et al., 2015; LaRowe et al., 2020; Zhou et al., 2020). To date, numerous studies have documented microbial hydrocarbon degradation in the marine environment, including in the water column and surface sediment (Yuan et al., 2015; Zhang et al., 2018). However, insufficient attention has been paid to studies on the biodegradation of hydrocarbons in subsurface sediment.
Previous studies have described the abiotic source of hydrocarbons in subsurface environments. McCollom and Seewald (2001) reported that the reduction of CO2 during serpentinization produces methane and other hydrocarbons abiotically (McCollom and Seewald, 2001). Moreover, Foustoukos and Seyfried (2004) suggested that chromium-bearing minerals catalyzed a Fischer-Tropsch type of synthesis of hydrocarbons (methane, ethane, and propane) through water–rock interactions in mid-ocean ridge hydrothermal systems. The abiotically produced hydrocarbons maybe sustain the growth of microbes in the subsurface environment at hydrothermal zones (Foustoukos and Seyfried, 2004). Bains et al. (2015) also indicated that hydrocarbons maybe fuel microorganisms in the deep biosphere to a depth of 10 km, except when the temperature was higher than a hypothetical life-maximum of 150°C (Bains et al., 2015).
The oceanographic context of this work was the Central Indian Ridge (CIR), which is proximate to the Edmond and Kairei hydrothermal vents; the distance from our sampling site to these two hydrothermal vents is within 200 km. Therefore, we speculated that the geochemistry of the CIR subsurface sediments might be impacted by matter (e.g., hydrocarbons and metals) released from nearby hydrothermal systems (Resing et al., 2015). Of note, Peng et al. (2011) analyzed the aliphatic hydrocarbons in the Kairei sediments and found that C16 and C17 alkanes exhibited the highest concentrations in the CIR hydrothermal field (Peng et al., 2011). Therefore, the CIR subsurface sediment might house distinct microorganisms fueled by abiotic hydrocarbons related to hydrothermal activity. This study could provide new insight into the geochemical cycle and fate of hydrocarbon in deep biospheres.
The purpose of this study was to 1) investigate the biodiversity, vertical distribution, and degradation potential of hydrocarbon consumers in a 4.8 m long gravity core from the CIR by adopting two different substrates as sole carbon sources (n-hexadecane as labile hydrocarbon and methylcyclohexane as recalcitrant hydrocarbon); 2) identify the key uncultured cycloalkane consumers in deep-sea subsurface sediment through metagenomics; and 3) explore the key functional genes and metabolic pathways in cycloalkane-consuming bacteria derived from deep subsurface habitats.
A sediment sample (CIR-GC01) was collected from the CIR in 2019 using a gravity column sampler on board the R/V Da-Yang-Yi-Hao. The sampling site (24°37′S, 68°46′E, 4034 m depth) was proximate to the Edmond (23°53′S, 69°36′E, 3300 m depth) and Kairei (25°19′S, 70°02′E, 2415–2460 m depth) hydrothermal vents. The depth of this core sediment was approximately 4.8 m below the seafloor (mbsf). Core sediments were subsampled at various depths, including at 0 m, 1.0 mbsf, 2.0 mbsf, 3.0 mbsf, and 4.8 mbsf. Then, these subsamples were divided into three groups for the enrichment experiment and downstream data analysis, namely the shallow layer (0 m and 1.0 mbsf), the intermediate layer (2.0 mbsf and 3.0 mbsf), and the deep layer (4.8 mbsf). The samples were kept at −20°C on board prior to enriching hydrocarbon-consuming bacteria.
We measured the hydrocarbons in the core sediment as described by Zheng et al. (2022). More specifically, sediment samples were prepared by freeze-drying for 24 h. Next, 5.0 g (dw) of each sample was placed into a 50 mL quartz colorimetric tube (with stopper) and 10 mL of hexane (≥ 97%)/dichloromethane (≥ 99.5%) solvent mixture (1:1, v/v) was added for ultrasonic extraction. Then, the supernatant was transferred into a 60 mL flask. The extraction procedures were repeated and the supernatant was combined. After that, the extract was concentrated to 2 mL at 40°C using rotary evaporation. Finally, the eluent was purged to 1 mL using N2 at a flow rate of 30–50 mL·min−1.
Hydrocarbon concentrations in core sediments were analysed using an Agilent 7890A gas chromatograph (GC) coupled with a micro-electron capture detector. The GC is equipped with an HP-5MS fused silica capillary column. The temperature of the injector and detector was adjusted to 300°C. The oven temperature of the GC was set to start at 70°C, rise to 150°C at 20°C·min−1, then to 300°C at 7°C·min−1, and finally hold at 300°C for 7 min. High-purity helium was set at a flow rate of 1 mL·min−1. The concentrations of standard solutions for HEX were 0, 1, 2, 5, and 10 mg·L−1. The concentrations of standard solutions for MCH were 200, 400, 500, 800, and 1000 μg·L−1. Each standard was measured in triplicates. The correlation coefficients of the calibration regression curves were 0.9965 for HEX and 0.9942 for MCH. The hydrocarbons in the abovementioned samples were identified according to the retention time of the peak area. Finally, the data were processed to determine the concentrations of hydrocarbons in the core sediment using Chemstation software (F.01.03.2357).
The different hydrocarbon-degrading bacteria from the core sediment sampled at the CIR were enriched in accordance with the approach described by Cui et al.(Cui et al., 2022). Two species of hydrocarbons, namely n-hexadecane (HEX; Chemic, purity > 99%) and methylcyclohexane (MCH; Aladdin, purity 99%), were used as the sole sources of carbon and energy, respectively. 0.5 g of sediment was inoculated into the serum bottle containing 150 mL of ONR7a medium (Wang et al., 2008) amended with HEX/MCH (final concentration 0.23 mM/0.52 mM). Two duplicates were prepared for each treatment. The dissolved oxygen (DO) in the closed serum bottles was monitored daily via a highly sensitive fiber-optic oxygen meter (Pyroscience, FireStingO2). The enrichment cultures were incubated in the dark at 10°C without shaking until the microbial hydrocarbon respiration drew the DO concentration down to 0 μM in the closed serum bottles (bloomed hydrocarbon-consuming cultures). After incubation, microbial biomass was collected by filtration of the enrichment cultures using a 0.22-μm polycarbonate membrane (Whatman) and kept these samples at −80°C prior to DNA extraction.
The bloomed enrichment cultures were inoculated into the abovementioned media at an inoculum size of 7% (v/v). Then, the subcultures were incubated under the abovementioned conditions until they bloomed. The microbial biomass of the hydrocarbon-consuming subcultures was collected and stored as described above.
Microbial DNA was extracted from the core sediment subsamples described in 2.1 (two duplicates for the treatment of original bacterial communities) and enrichment cultures/subcultures described in 2.3 (two duplicates for each hydrocarbon-spiked treatment) using the OMEGA soil DNA kit (Omega Bio-Tek, Norcross, GA, USA) according to the manufacturer’s protocols. The concentration of extracted DNA was measured using a NanoDrop NC2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) and the DNA integrity was assessed using electrophoresis on a 0.8% (w/v) agarose gel.
Next, the V3-V4 region of the 16S rRNA gene was amplified using PCR with primer pair 338F (5`-ACTCCTACGGGAGGCAGCA-3`) and 806R (5`-GGACTACHVGGGTWTCTAAT-3`). Specifically, PCR reactions were performed in triplicate using 25 μL of the mixture containing 0.25 μL of Q5 high-fidelity DNA polymerase, 5 μL of 5× reaction buffer, 5 μL of 5× high GC buffer, 2 μL of dNTPs (10 mM), 1 μL of each primer (10 μM), and 2 μL of DNA template. The PCR reagents were purchased from NEB and Invitrogen, USA. Thermal cycling consisted of initial denaturation at 98°C for 5 min, followed by 25 cycles consisting of denaturation at 98°C for 30 s, annealing at 53°C for 30 s, and extension at 72°C for 45 s, with a final extension of 5 min at 72°C, then hold at 12°C.
The amplicons were extracted and purified from 2% (w/v) agarose gels using the Axygen DNA gel recovery kit (Axygen Biosciences, Union City, CA, USA) according to the manufacturer’s protocols. Then, purified amplicons were sequenced with paired-end (PE) 250 strategy on an Illumina NovaSeq platform using the NovaSeq 6000 SP reagent kit according to the manufacturer’s protocols at Personal Biotechnology Co., Ltd. (Shanghai, China). The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database under accession number PRJNA876038.
Microbial DNA of the MCH-degrading bacterial communities was extracted from the membrane samples (eight samples derived from the MCH-spiked enrichment cultures and subcultures) described in 2.3 using the OMEGA Mag-Bind soil DNA kit (Omega Bio-Tek, Norcross, GA, USA) according to the manufacturer’s protocols. The extracted DNA was measured (optical concentration > 2.5 ng·μL−1 and total content > 200 ng) using a NanoDrop NC2000 spectrophotometer and agarose gel electrophoresis. Next, libraries with insert sizes of 400 bp were constructed for metagenome shotgun sequencing using the Illumina TruSeq Nano DNA LT library preparation kit. Each library was sequenced with the PE150 strategy on an Illumina NovaSeq platform at Personal Biotechnology Co., Ltd. The raw reads of metagenomics were deposited in the NCBI SRA database under accession number PRJNA876059.
Non-target fragments were removed from the raw sequences using Cutadapt (v1.17) and FASTP (v0.20.0), and the clean reads were obtained using SortMeRNA and bmtagger. Next, we assembled each sample, removed the unmatched sequences, and obtained the contigs set without redundancy using MEGAHIT (Li et al., 2015). MetaWRAP was used to performed the metagenomic binning (https://github.com/bxlab/metaWRAP).The alleles were sorted to obtain the original bins using MaxBin2, metaBAT2, and CONCOCT with default settings. An improved bin set was produced using the bin_refine module, which combines two or three bin sets obtained by different software (or the same software with different parameters). Then, CheckM was used to assess the completeness and incompleteness of the bins. The genomic features (protein-coding genes and RNA) and their functions were annotated with rapid annotation using the subsystem technology (RAST) annotation engine. The phylogenetic information of the bins were annotated using the classify_wf model in GTDB-tk, which classifies genomes using the Genome Classification Database(Chaumeil et al., 2020). In addition, phylogenetic trees were constructed directly from the genomes using the de_novo_wf model in GTDB-tk. All parameters in de_novo_wf were set to the default values. The assembled bins of metagenomic datasets were deposited into the NCBI GenBank database (accession number PRJNA892034).
The average nucleotide identity (ANI) was analysed to illustrate the phylogenetic relationship between the bins using ANI web-based software provided by Kostas lab (http://enve-omics.ce.gatech.edu/ani/index). The software calculator estimated the ANI between two bins using both best hits (one-way ANI) and reciprocal best hits (two-way ANI). In addition, the amino acid identity (AAI) was analysed between the key genes involved in MCH metabolism using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/).
Statistical analyses were performed using R (v3.6), QIIME2 (2021.8), and Vegan package (v2.5-6). Specifically, stacked histograms of species composition was plotted using QIIME2. The alpha diversity index with a normalized amplicon sequence variant table was calculated using QIIME2. Non-metric multidimensional scaling (NMDS) analysis was performed to present beta diversity using the Vegan package. The statistical significance was determined using the Dunn test and the Kruskal– Wallis test. Finally, pheatmap package was used for visualization.
The core sediment collected from the CIR was pale yellow with a water content of 46–58% (shallow layer to deep layer). We detected the presence of HEX and MCH at various depths of the core sediment. Specifically, the concentration of HEX in the core sediment ranged from 2.33 mg·kg−1 to 3.05 mg·kg−1. The concentration of MCH in the core sediment ranged from 111.53 μg·kg−1 to 591.22 μg·kg−1 (Figure S1). The average concentration of HEX was 6.65 times higher than that of MCH. The source of these hydrocarbons is unknown. However, we speculated that they might come from the process of long-term sedimentation affected by proximate hydrothermal activities.
We determined the microbial respiration of hydrocarbons in the enrichment cultures by monitoring the remaining DO level throughout incubation. The initial DO concentration was 223 ± 14 μM (n = 10) for the enrichment cultures with HEX as the sole carbon source (Figure S2A). All the HEX-spiked enrichment cultures completely consumed the oxygen in the medium within 25 days of incubation (bloomed cultures; 0 μM, n = 10), indicating microbial growth and respiration on alkane as the sole carbon source. Therefore, HEX-degrading bacteria derived from various depths of the CIR core sediment proliferated in the enrichment cultures after incubation.
The initial DO concentration was 223 ± 12 μM (n = 10) for the MCH-spiked enrichment cultures (Figure S2B). The DO in the MCH-spiked enrichment cultures derived from two shallow sediment layers (0 m and 1.0 mbsf) and one deep sediment layer (4.8 mbsf) did not fall below 92 ± 24 μM (n = 6) after incubation (> 70 days). These enrichment cultures failed to achieve substantial respiration of the hydrocarbons. In contrast, the MCH-spiked enrichment cultures derived from two intermediate sediment layers (2.0 mbsf and 3.0 mbsf) bloomed within 50 days of incubation (0 μM, n = 4). Therefore, MCH-degrading bacteria derived from the intermediate sediment layer proliferated preferentially in the enrichment cultures after incubation. In addition, we observed that the average incubation time for bloomed cultures of the MCH treatment was two times longer than that of the HEX treatment. In other words, MCH is more recalcitrant than HEX for microbes.
The initial DO concentration for the HEX/MCH-spiked subcultures was comparable to that of the enrichment cultures (Figures S2C, D). After incubation, all the HEX-spiked subcultures bloomed within 12 days of incubation (0 μM, n = 10). Similarly, the MCH-spiked subcultures derived from two intermediate sediment layers bloomed within 15 days of incubation (0 μM, n = 4). In addition, we observed that the incubation time for the bloomed MCH-spiked subcultures was at least 30 days less than that of the corresponding enrichment cultures.
The alpha diversity of the hydrocarbon-degrading bacterial communities derived from the CIR core sediment was significantly lower (Kruskal– Wallis test, P < 0.01) than that of the blank treatment of the original bacterial communities in the subsurface sediment (Table S1). The supplement of HEX/MCH as the sole source of carbon and energy probably fuelled the growth of specific microbes derived from the subsurface sediment and decreased the alpha diversity of hydrocarbon-spiked enrichment cultures and subcultures compared with the original bacterial communities. Furthermore, the result of the NMDS analysis showed the divergence of bacterial community structure between hydrocarbon-spiked treatments and blank treatment (Figure 1, stress value < 0.2). Specifically, both bacterial community structures of the hydrocarbon-spiked cultures exhibited a noticeable shift compared with the original bacterial communities and formed two separate clusters on the plot. In addition, the MCH-spiked communities exhibited different bacterial community structure from that of the HEX-spiked communities suggested by the result of ANOSIM (P < 0.01, Table S2), indicating that distinct bacterial taxa were involved in the consumption of labile/refractory hydrocarbons in the subsurface sediment.
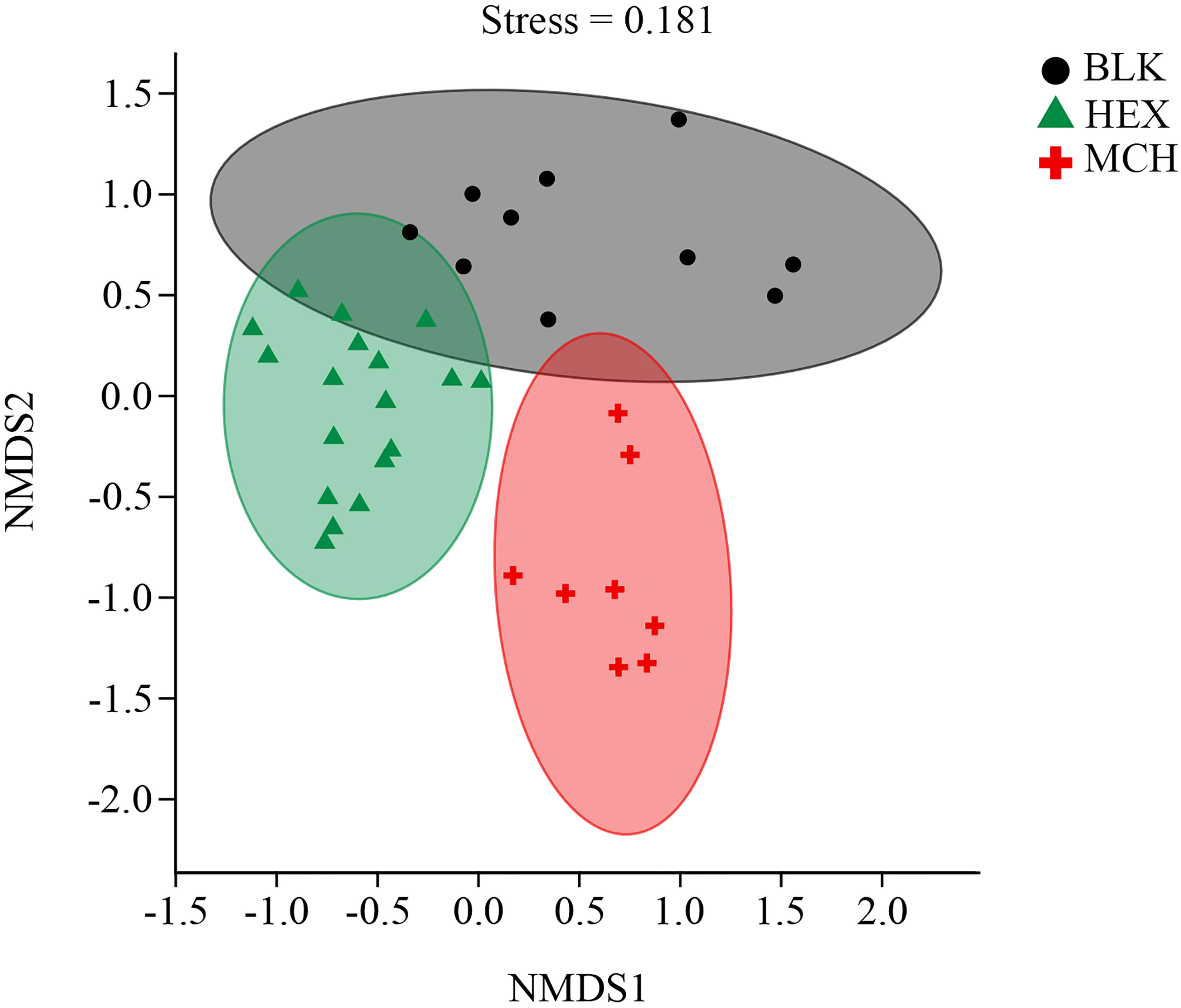
Figure 1 Divergence of different hydrocarbon-degrading bacterial communities derived from the CIR core sediment. The colored solid symbols denote the bacterial community structures of different treatments. MCH, methylcyclohexane-spiked enrichment cultures, and subcultures; HEX, n-hexadecane-spiked enrichment cultures, and subcultures; BLK, the original bacterial communities of the CIR core sediment without the addition of any carbon source.
The bacterial community dynamics (phylum level) results showed that Proteobacteria and Firmicutes were predominant in the hydrocarbon-degrading enrichment cultures and subcultures derived from the CIR core sediment (Table S3). Furthermore, the bacterial community dynamics (genus level) results showed that each group of hydrocarbon-spiked bacterial communities derived from the CIR core sediment harbored distinct dominant species (Figure 2A; e.g., top three most abundant genera). More specifically, the dominant genera in the treatments of HEX-degrading communities were Alcanivorax, Marinobacter, and Pseudomonas, the total relative abundance of which accounted for 77.61% in enrichment cultures and 85.60% in subcultures, respectively. In contrast, the dominant genera in the treatment of MCH-degrading communities were the novel genus C1-B045, Halomonas, and Paenibacillus, the total relative abundance of which accounted for 55.41% in enrichment cultures and 83.34% in subcultures, respectively. Therefore, the degradation of labile/recalcitrant hydrocarbons was ascribed to distinct hydrocarbon consumers derived from the same core sediment. It is worth noting that the relative abundance of C1-B045 increased dramatically from extremely low relative abundance (< 0.001%) in the original bacterial community to 71.99% in the MCH-degrading subcultures, implying it plays a key role in cycloalkane decomposition.
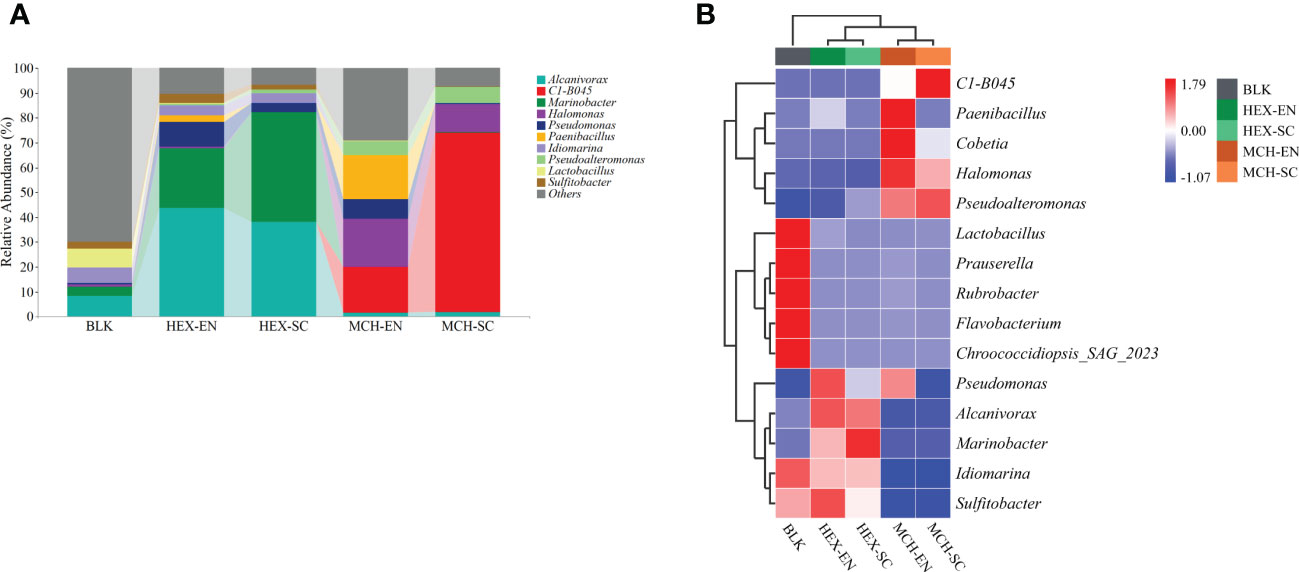
Figure 2 Profile of different hydrocarbon-degrading bacterial communities derived from the CIR core sediment. (A) The bacterial community structures of the HEX/MCH-degrading enrichment cultures and subcultures. The top 10 most dominant genera are illustrated in the plot. MCH denotes methylcyclohexane-spiked cultures; HEX denotes n-hexadecane-spiked cultures; BLK denotes the original bacterial communities of the CIR core sediment without the addition of any carbon source; EN denotes enrichment cultures; SC denotes subcultures. (B) Differentiated genera in the HEX/MCH-spiked treatments demonstrated by a standardized heatmap. The tree on the left of the plot is generated based on clustering the relative abundance of the top 15 most dominant genera in each treatment. Red color denotes higher abundance, while blue color denotes lower abundance.
The differentiated genera in the hydrocarbon-spiked treatments were illustrated using a standardized heatmap (Figure 2B). Specifically, strains of Alcanivorax, Marinobacter, and Pseudomonas in the HEX-degrading enrichment cultures and subcultures exhibited higher relative abundance than the rest of the treatments, which reinforced that they were cosmopolitan alkane consumers in the marine environment. Remarkably, strains of C1-B045, Cobetia, Halomonas, Paenibacillus, and Pseudoalteromonas in the MCH-degrading enrichment cultures and subcultures exhibited higher relative abundance than in the rest of the treatments, suggesting the pivotal role of these bacteria to metabolize MCH in the core sediment.
We reconstructed 24 high-quality metagenome-assembled genomes (MAGs) (completeness > 51.72% and contamination < 8.62%) by binning from seven metagenomic datasets derived from the MCH-consuming enrichment cultures and subcultures (Figure 3; Table S4). One out of the 24 bins was classified using GTDB-Tk as novel genus C1-B045 (completeness > 55.17% and contamination < 1.72%), which has been previously reported as one of the pivotal players in MCH decomposition (Cui et al., 2022). Moreover, five out of the 24 bins were classified as Pseudomonas (completeness > 53.44% and contamination < 4.83%) and Alcanivorax (completeness > 51.72% and contamination < 8.62%), respectively. In addition, another three bins were classified as genus Marinobacter (completeness > 60.34% and contamination < 1.30%). All these bacteria have been previously reported as hydrocarbon consumers, though their capacity to degrade cycloalkanes has not yet been characterized.
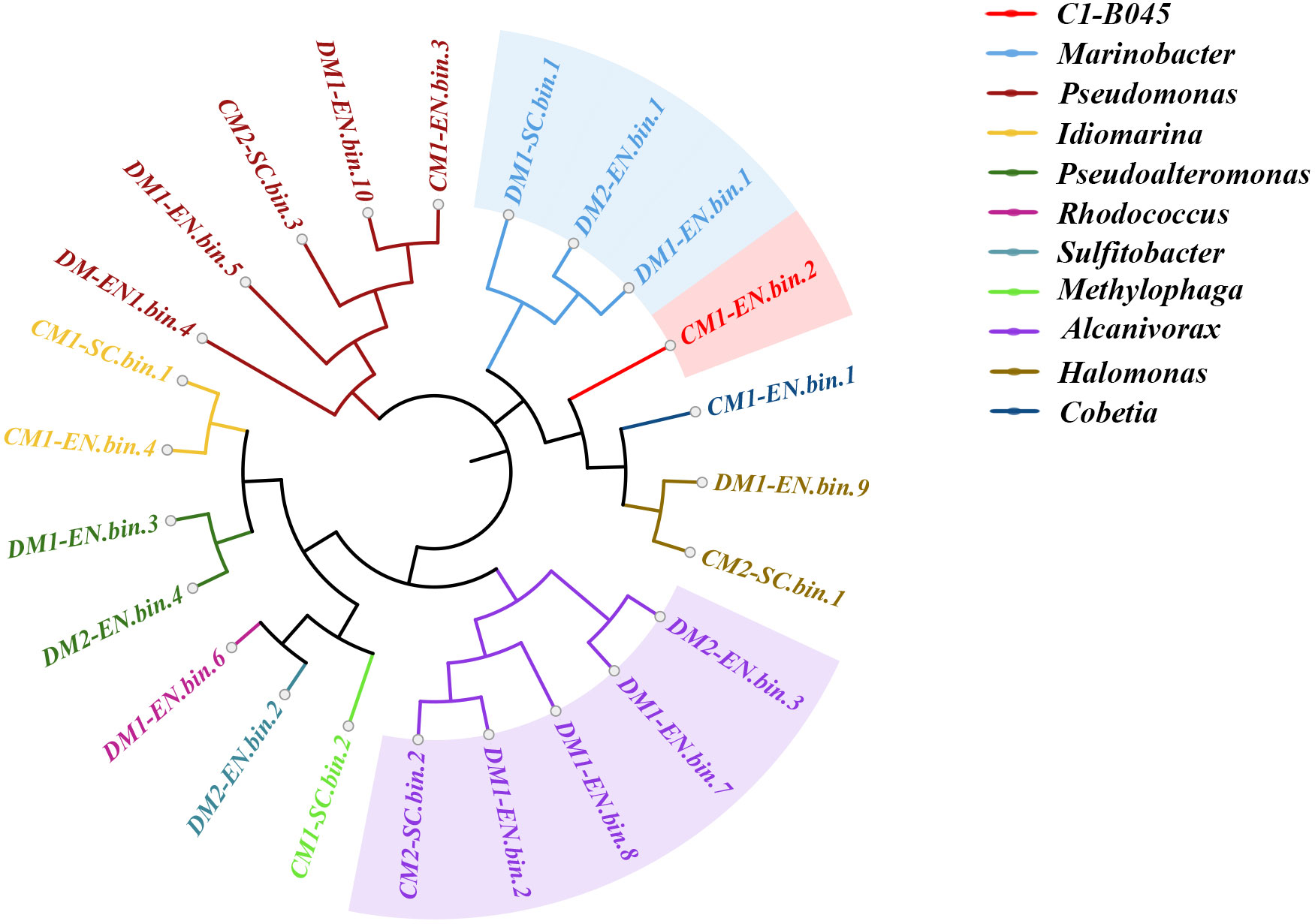
Figure 3 Maximum likelihood phylogenomic tree of the 24 bins reconstructed from seven metagenomic datasets derived from the MCH-consuming cultures. The bins of the genus Alcanivorax are highlighted in light purple; the bins of the genus Marinobacter are highlighted in light blue; the bin of genus C1-B045 is highlighted in light red.
The genus C1-B045 exhibited the highest abundance in MCH-consuming subcultures, implying that it might be a key MCH consumer in the intermediate layer of the CIR core sediment (Figure 2A). The genome size of the C1-B045 MAG was 3,294,068 bp, and the GC content was 50.2%. A total of 3,216 coding sequences were predicted in this MAG based on the RASTtk annotation scheme (Figure 4A). Specifically, we found two copies of alkane monooxygenase-like genes that activate the initial oxidation of MCH in the MAG of C1-B045. Moreover, we annotated eight copies of genes encoding cyclohexanone monooxygenase and one copy of the gene encoding 6-hexanolactone hydrolase in the C1-B045 genome. We proposed an MCH degradation pathway based on the key enzymes annotated in the MAG of C1-B045 and the intermediates experimentally detected in MCH metabolism, including 4-methyl-2-oxepanone, 4-methylcyclohexanone, and 3-methyladipic acid (Donoghue and Trudgill, 1975; Koma et al., 2004) (Figure 4B).
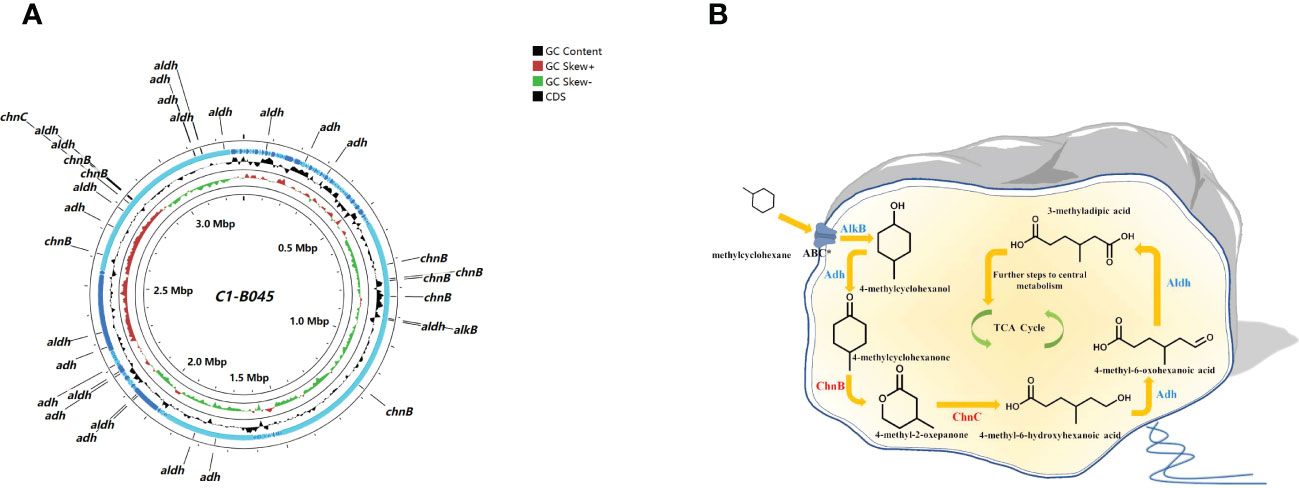
Figure 4 Key genes encoding degradation pathway for MCH in the MAG of C1-B045 derived from the CIR core sediment. (A) circular representation of the MAG of C1-B045. CGView map represents the coding sequence (annotated by RAST), contigs, GC skew, GC content, and genome backbone from outside to inside, respectively. alkB, alkane 1-monooxygenase; chnB, cyclohexanone monooxygenase; chnC, 6-hexanolactone hydrolase; adh, alcohol dehydrogenase; aldh, aldehyde dehydrogenase. (B) putative MCH degradation pathway with key enzymes predicted in the MAG of C1-B045. * indicates the transporter ABC is not annotated in this MAG.
The source of HEX and MCH measured in the core sediment in this study might derive from either biogenetic hydrocarbons or abiogenetic hydrocarbons. Given the proximate hydrothermal activities from the Kairei and Edmond hydrothermal zones, we speculate that they probably originated from abiogenesis of hydrocarbon. In addition, Yamashita et al. (2023) also proposed that hydrothermal-derived black carbon was a source of recalcitrant dissolved organic carbon in the ocean, and could migrate long distances (4,440km) in the deep ocean with hydrothermal plumes. However, source analysis is needed to further validate the origin of these hydrocarbons in the CIR core sediment in future studies.
In this study, a HEX/MCH-spiked enrichment experiment was used to investigate the vertical distribution of different hydrocarbon-degrading bacteria in a CIR core sediment. We found that the labile hydrocarbon (HEX) consumers were widely distributed in the core sediment (0–4.8 mbsf), whereas the refractory hydrocarbon (MCH) consumers were stratified only in the intermediate layer of the core sediment (2.0 and 3.0 mbsf) (Figure S2). The vertical heterogeneity of the different hydrocarbon consumers might be ascribed to the gradient of DO along the vertical core sediment. Durbin and Teske (2011) reported that the concentration of DO in subsurface sediments decreased with increasing depth. They found that the DO dropped from 115 μM at 5 cm below the sediment (cmbsf) to 2–10 μM (an apparent detection limit) at 100–150 cmbsf (Durbin and Teske, 2011).This suggests that the absence of oxygen in the deep layers of the CIR core sediment (4.8 mbsf) was unfavorable to the aerobic degradation of the refractory hydrocarbons and the growth of refractory hydrocarbon consumers. MCH-consuming bacteria could not be enriched in the shallow layer of the core sediment. Therefore, we speculated that the labile hydrocarbon consumers outcompeted their counterparts of refractory hydrocarbon consumers in the shallow layer of the original core sediment where the DO was sufficient. In contrast, decreased DO in the intermediate layer of the original core sediment might have limited the growth of fast-growing labile hydrocarbon consumers (Massmig et al., 2019). Therefore, the slow-growing refractory hydrocarbon consumers co-existed with their counterparts of labile hydrocarbon consumers in the intermediate layer of the original core sediment. Among these refractory hydrocarbon consumers, we found the occurrence of cycloalkane-degrading bacteria C1-B045 in the deep-sea subsurface environment for the first time (Figure 2A). Importantly, the abundance of C1-B045 surged to 71.99% in MCH-consuming subcultures compared to < 0.001% in the original bacterial community, implying that it might be a potential key cycloalkane consumer in the deep biosphere of the CIR.
To the best of our knowledge, the genus C1-B045 has not yet been cultured or characterized in detail since the first report of their near-complete 16S rRNA gene sequences in the clone library derived from the Guaymas Basin sediment (Teske et al., 2002). Several of the most recent studies have demonstrated that it might also be involved in the microbial degradation of light crude oil (Uribe-Flores et al., 2019), polycyclic aromatic hydrocarbons (Peng et al., 2020), and short-chain n-alkylcyclohexanes (Arrington et al., 2022; Cui et al., 2022). Due to the poor culturability of genus C1-B045, we obtained only MAGs rather than pure cultures by binning from metagenomic datasets in this study. The ANI value between the MAG of C1-B045 derived from the CIR deep biosphere and its counterpart derived from Bohai Sea (BS) (unpublished data) was 85.84%, implying that they represent two different species of genus C1-B045 (Figure S3; cutoff value of ANI for species delineation < 95%)(Parks et al., 2020). Therefore, strains of the genus C1-B045 are phylogenetically diverse and might represent cosmopolitan cycloalkane consumers in different marine habitats. In addition to C1-B045, almost all the other differentiated bacteria in MCH-spiked cultures (Figure 2B) have been characterized to be capable of metabolizing hydrocarbons, implying they might take part in MCH metabolism. For instance, Govarthanan et al. (2020) reported that strains of Halomonas were capable of utilizing both low and high molecular weight polycyclic aromatic hydrocarbons for growth. Hedlund and Staley (2006) found that members of Pseudoalteromonas contain dioxygenase genes and can effectively decompose naphthalene.
Hitherto, two MCH degradation pathways have been proposed in previous studies, including lactone formation and aromatization. The lactone formation pathway, characterized by Koma et al. (2004) in Rhodococcus sp.NDKK48, might represent a primary mechanism for cycloalkane degradation. Moreover, the aromatization pathway was found in Rhodococcus sp. EC1, which might be relatively rare in aerobic bacteria (Yi et al., 2011). In our study, we proposed a lactone formation pathway in the MAG of C1-B045 (Figure 4B), which is similar to that of Rhodococcus sp.NDKK48 (Koma et al., 2004). More recently, we discovered two nearly complete MCH degradation pathways, lactone formation and aromatization, concurrently in the MAG of Roseovarius derived from Antarctic surface water (unpublished data). Therefore, future research should be devoted to the metabolic diversity of cycloalkanes by marine microorganisms.
Remarkably, we annotated abundant copies of genes encoding cyclohexanone monooxygenase (CHMO/ChnB) in the MAGs of C1-B045 derived from the CIR and BS. CHMOs belong to Baeyer-Villiger monooxygenases (BVMOs) and can catalyze a variety of oxidation reactions, including Baeyer-Villiger oxidations. This reaction involves the conversion of cyclic ketones into lactones or ketones into esters by introducing an oxygen atom adjacent to the carbonyl group (Yachnin et al., 2012). In this study, we discovered the presence of two classes of CHMOs based on phylogenetic analysis with the available amino acid sequences of CHMOs (Figure 5; Table S5). Specifically, class I CHMOs derived from the MAGs of C1-B045 exhibited low AAI values with that of Rhodococcus sp. HI-31(29.04–41.71%). Similarly, class II CHMOs derived from the MAGs of C1-B045 exhibited low AAI values with that of Acinetobacter calcoaceticus CA16 (28.60–31.87%). This indicates that CHMOs derived from genus C1-B045 represent two novel classes of enzymes (Kim et al., 2013; Walock et al., 2014), which exhibited different primary and advanced protein structures from the published reference CHMOs. In addition, eight pairwise CHMOs (one from the CIR and the other from the BS) were seen in the phylogenetic tree and exhibited high similarity to one another (90.53–99.36%).
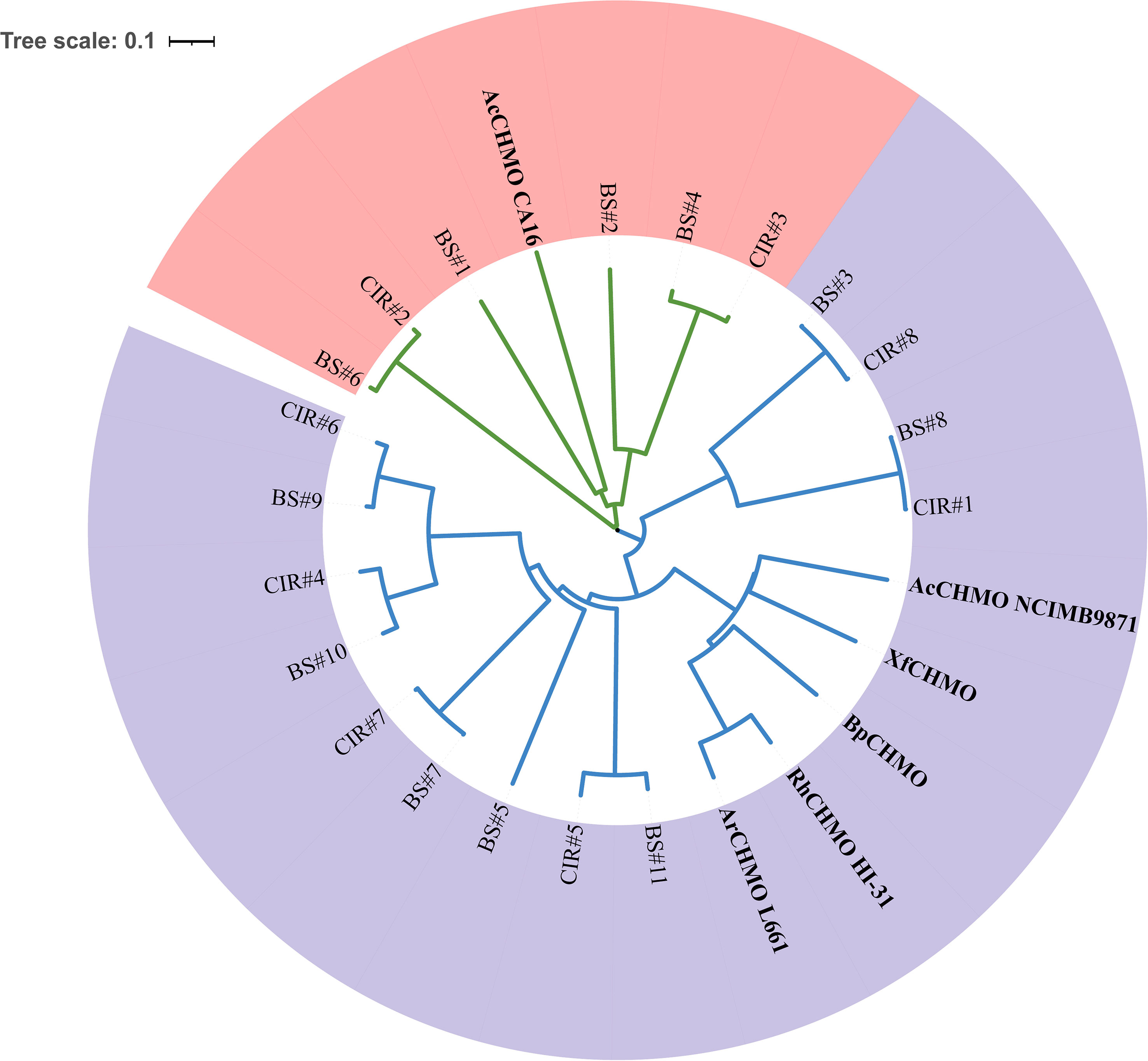
Figure 5 Phylogeny of CHMOs in MCH-consuming bacteria. CIR, CHMOs annotated in genus C1-B045 derived from the Central Indian Ridge; BS, CHMOs annotated in genus C1-B045 derived from the Bohai Sea; AcCHMO CA16, CHMO of Acinetobacter calcoaceticus CA16; RhCHMO HI-31, CHMO of Rhodococcus sp. HI-31; AcCHMO NCIMB9871, CHMO of Acinetobacter calcoaceticus NCIMB9871; XfCHMO, CHMO of Xanthobacter flavus; BpCHMO, CHMO of Brachymonas petroleovorans; ArCHMO L661, CHMO of Arthrobacter sp. L661. Class I of CHMOs is highlighted in light purple; The class II of CHMOs is highlighted in light red.
In this study, we found that HEX-consuming bacteria were ubiquitous in the CIR core sediment. The vertical profile of the MCH-consuming bacteria was sandwich-like in the CIR core sediment. Specifically, the MCH-consuming bacteria were stratified only in the intermediate layer of the core sediment. We also discovered that a novel species of genus C1-B045 might be involved in the degradation of cycloalkane in the deep biosphere of the CIR. The bacteria of genus C1-B045 harbored two novel classes of CHMOs to break down the metabolites of MCH.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
TH and ZC contributed to conception and design of the study. ZC and ZW led funding acquisition and project management. JC managed the resources, and guaranteed their availability to perform the experiments and analysis. TH and XL performed the experiments and analyzed the data. TH, ZC and XL wrote the first draft of the manuscript. TH, ZC and GZ are responsible for the review and editing of article. ZC and ZW supervised the writing of the entire manuscript. All authors contributed to the article and approved the submitted version.
This work was financially supported by the China Ocean Mineral Resources Research and Development Association (DY135-E2−1-03) and the National Natural Science Foundation of China (42076165). This study used core sediment samples provided by COMRA. This cruise was conducted onboard R/V Da-Yang-Yi-Hao.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1170238/full#supplementary-material
Arrington E., Tarn J., Kittner H., Kivenson V., Liu R., Valentine D. (2022). Methylated cycloalkanes fuel a novel genera in the porticoccaceae family and inform substrate affinity for a unique copper membrane monooxygenase. bioRxiv. doi: 10.1101/2022.11.07.515388
Atlas R. M., Hazen T. C. (2011). Oil biodegradation and bioremediation: a tale of the two worst spills in us history. Environ. Sci. Technol. 45, 6709–6715. doi: 10.1021/es2013227
Bains W., Xiao Y., Yu C. (2015). Prediction of the maximum temperature for life based on the stability of metabolites to decomposition in water. Life (Basel Switzerland) 5, 1054–1100. doi: 10.3390/life5021054
Chaumeil P., Mussig A. J., Hugenholtz P., Parks D. H. (2020). Gtdb-tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36, 1925–1927. doi: 10.1093/bioinformatics/btz848
Colman D. R., Poudel S., Stamps B. W., Boyd E. S., Spear J. R. (2017). The deep, hot biosphere: twenty-five years of retrospection. Proc. Natl. Acad. Sci. U S 114, 6895–6903. doi: 10.1073/pnas.1701266114
Cui Z., Luan X., Li S., Li Y., Bian X., Li G., et al. (2022). Occurrence and distribution of cyclic-alkane-consuming psychrophilic bacteria in the yellow sea and east china sea. J. Hazard Mater 427, 128129. doi: 10.1016/j.jhazmat.2021.128129
Donoghue N. A., Trudgill P. W. (1975). The metabolism of cyclohexanol by acinetobacter ncib 9871. Eur. J. Biochem. 60, 1–7. doi: 10.1111/j.1432-1033.1975.tb20968.x
Durbin A. M., Teske A. (2011). Microbial diversity and stratification of south pacific abyssal marine sediments. Environ. Microbiol. 13, 3219–3234. doi: 10.1111/j.1462-2920.2011.02544.x
Foustoukos D. I., Seyfried W. E. J. (2004). Hydrocarbons in hydrothermal vent fluids: the role of chromium-bearing catalysts. Sci. (New York N.Y.) 304, 1002–1005. doi: 10.1126/science.1096033
Govarthanan M., Khalifa A. Y., Kamala-Kannan S., Srinivasan P., Selvankumar T., Selvam K., et al. (2020). Significance of allochthonous brackish water halomonas sp. on biodegradation of low and high molecular weight polycyclic aromatic hydrocarbons. Chemosphere 243, 125389. doi: 10.1016/j.chemosphere.2019.125389
He D., Wang X., Yang Y., He R., Zhong H., Wang Y., et al. (2021). Hydrothermal synthesis of long-chain hydrocarbons up to c-24 with nahco3-assisted stabilizing cobalt. Proc. Natl. Acad. Sci. U S 118, 51. doi: 10.1073/pnas.2115059118
Hedlund B. P., Staley J. T. (2006). Isolation and characterization of pseudoalteromonas strains with divergent polycyclic aromatic hydrocarbon catabolic properties. Environ. Microbiol. 8, 178–182. doi: 10.1111/j.1462-2920.2005.00871.x
Kim D. H., Patnaik B. B., Seo G. W., Kang S. M., Lee Y. S., Lee B. L., et al. (2013). Identification and expression analysis of a novel r-type lectin from the coleopteran beetle, tenebrio molitor. J. Invertebr Pathol. 114, 226–229. doi: 10.1016/j.jip.2013.08.002
Koma D., Sakashita Y., Kubota K., Fujii Y., Hasumi F., Chung S. Y., et al. (2004). Degradation pathways of cyclic alkanes in rhodococcus sp. Ndkk48. Appl. Microbiol. Biotechnol. 66, 92–99. doi: 10.1007/s00253-004-1623-5
Konn C., Charlou J. L., Donval J. P., Holm N. G., Dehairs F., Bouillon S. (2009). Hydrocarbons and oxidized organic compounds in hydrothermal fluids from rainbow and lost city ultramafic-hosted vents. Chem. Geol 258, 299–314. doi: 10.1016/j.chemgeo.2008.10.034
LaRowe D. E., Arndt S., Bradley J. A., Estes E. R., Hoarfrost A., Lang S. Q., et al. (2020). The fate of organic carbon in marine sediments - new insights from recent data and analysis. Earth Sci. Rev. 204. 103146 doi: 10.1016/j.earscirev.2020.103146
Lea-Smith D. J., Biller S. J., Davey M. P., Cotton C. A. R., Sepulveda B. M. P., Turchyn A. V., et al. (2015). Contribution of cyanobacterial alkane production to the ocean hydrocarbon cycle. Proc. Natl. Acad. Sci. U S 112, 13591–13596. doi: 10.1073/pnas.1507274112
Li D., Liu C., Luo R., Sadakane K., Lam T. (2015). Megahit: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de bruijn graph. Bioinformatics 31, 1674–1676. doi: 10.1093/bioinformatics/btv033
Love C. R., Arrington E. C., Gosselin K. M., Reddy C. M., Van Mooy B. A. S., Nelson R. K., et al. (2021). Microbial production and consumption of hydrocarbons in the global ocean. Nat. Microbiol. 6, 489. doi: 10.1038/s41564-020-00859-8
Massmig M., Piontek J., Le Moigne F. A. C., Cisternas-Novoa C., Enge A. (2019). Potential role of oxygen and inorganic nutrients on microbial carbon turnover in the baltic sea. Aquat Microb. Ecol. 83, 95–108. doi: 10.3354/ame01902
McCollom T. M., Seewald J. S. (2001). A reassessment of the potential for reduction of dissolved co2 to hydrocarbons during serpentinization of olivine. Geochim Cosmochim Acta 65, 3769–3778. doi: 10.1016/S0016-7037(01)00655-X
Parks D. H., Chuvochina M., Chaumeil P. A., Rinke C., Mussig A. J., Hugenholtz P. (2020). A complete domain-to-species taxonomy for bacteria and archaea. Nat. Biotechnol. 38, 1079–1086. doi: 10.1038/s41587-020-0501-8
Peng X., Li J., Zhou H., Wu Z., Li J., Chen S., et al. (2011). Characteristics and source of inorganic and organic compounds in the sediments from two hydrothermal fields of the central indian and mid-atlantic ridges. J. Asian Earth Sci. 41, 355–368. doi: 10.1016/j.jseaes.2011.03.005
Peng C., Tang Y., Yang H., He Y., Liu Y., Liu D., et al. (2020). Time- and compound-dependent microbial community compositions and oil hydrocarbon degrading activities in seawater near the chinese zhoushan archipelago. Mar. pollut. Bull. 152, 110907. doi: 10.1016/j.marpolbul.2020.110907
Resing J. A., Sedwick P. N., German C. R., Jenkins W. J., Moffett J. W., Sohst B. M., et al. (2015). Basin-scale transport of hydrothermal dissolved metals across the south pacific ocean. Nature 523, 140–200. doi: 10.1038/nature14577
Schirmer A., Rude M. A., Li X., Popova E., Del Cardayre S. B. (2010). Microbial biosynthesis of alkanes. Science 329, 559–562. doi: 10.1126/science.1187936
Teske A., Hinrichs K., Edgcomb V., de Vera Gomez A., Kysela D., Sylva S. P., et al. (2002). Microbial diversity of hydrothermal sediments in the guaymas basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68, 1994–2007. doi: 10.1128/AEM.68.4.1994-2007.2002
Uribe-Flores M. M., Cerqueda-Garcia D., Hernandez-Nunez E., Cadena S., Garcia-Cruz N. U., Trejo-Hernandez M. R., et al. (2019). Bacterial succession and co-occurrence patterns of an enriched marine microbial community during light crude oil degradation in a batch reactor. J. Appl. Microbiol. 127, 495–507. doi: 10.1111/jam.14307
Walock C. N., Kittilson J. D., Sheridan M. A. (2014). Characterization of a novel growth hormone receptor-encoding cdna in rainbow trout and regulation of its expression by nutritional state. Gene 533, 286–294. doi: 10.1016/j.gene.2013.09.046
Wang B., Lai Q., Cui Z., Tan T., Shao Z. (2008). A pyrene-degrading consortium from deep-sea sediment of the west pacific and its key member cycloclasticus sp. P1. Environ. Microbiol. 10, 1948–1963. doi: 10.1111/j.1462-2920.2008.01611.x
Yachnin B. J., Sprules T., McEvoy M. B., Lau P. C. K., Berghuis A. M. (2012). The substrate-bound crystal structure of a baeyer-villiger monooxygenase exhibits a criegee-like conformation. J. Am. Chem. Soc. 134, 7788–7795. doi: 10.1021/ja211876p
Yamashita Y., Mori Y., Ogawa H. (2023). Hydrothermal-derived black carbon as a source of recalcitrant dissolved organic carbon in the ocean. Sci. Adv. 9, 3807. doi: 10.1126/sciadv.ade3807
Yi T., Lee E., Ahn Y. G., Hwang G., Cho K. (2011). Novel biodegradation pathways of cyclohexane by rhodococcus sp. Ec1. J. Hazard Mater 191, 393–396. doi: 10.1016/j.jhazmat.2011.04.080
Yuan J., Lai Q., Sun F., Zheng T., Shao Z. (2015). The diversity of pah-degrading bacteria in a deep-sea water column above the southwest indian ridge. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00853
Zhang X., Xu W., Liu Y., Cai M., Luo Z., Li M. (2018). Metagenomics reveals microbial diversity and metabolic potentials of seawater and surface sediment from a hadal biosphere at the yap trench. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02402
Zheng Y., Han B., Xu X., Liu A., Zheng L. (2022). Distribution characteristics, source analysis and risk assessment of organochlorine pesticides in the fildes peninsula and Adelaide island. Mar. pollut. Bull. 181, 114284. doi: 10.1016/j.marpolbul.2022.113862
Keywords: deep biosphere, subsurface sediment, biodegradation, cycloalkane, cyclohexanone monooxygenases, metagenomics
Citation: Hao T, Cui Z, Luan X, Zhou G, Li Y, Liu J, Chen J and Wang Z (2023) Vertical heterogeneity of hydrocarbon-degrading bacteria in a core sediment sample from the Central Indian Ridge. Front. Mar. Sci. 10:1170238. doi: 10.3389/fmars.2023.1170238
Received: 20 February 2023; Accepted: 11 April 2023;
Published: 27 April 2023.
Edited by:
Clara F. Rodrigues, University of Aveiro, PortugalReviewed by:
Willian Garcia Birolli, Barretos Cancer Hospital, BrazilCopyright © 2023 Hao, Cui, Luan, Zhou, Li, Liu, Chen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhisong Cui, Y3pzQGZpby5vcmcuY24=; Zongling Wang, d2FuZ3psQGZpby5vcmcuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.