- 1CSIRO Environomics Future Science Platform, Indian Oceans Marine Research Centre, Commonwealth Scientific and Industrial Research Organisation Commonwealth Scientific and Industrial Research Organisation (CSIRO), Crawley, WA, Australia
- 2Cayman Turtle Conservation and Education Centre, Grand Cayman, Cayman Islands
- 3Centre d’Etude et de Découverte des Tortues Marines (CEDTM), Piton Saint-Leu, La Réunion, France
- 4Kélonia l’observatoire des tortues marines, Saint-Leu, La Réunion, France
- 5Commonwealth Scientific and Industrial Research Organisation Commonwealth Scientific and Industrial Research Organisation (CSIRO), Crawley, WA, Australia
- 6Marine Science Program, Department of Biodiversity, Conservation and Attractions, Kensington, WA, Australia
In marine turtles, the sex of hatchlings is determined by their egg incubation temperature. Global warming may increase the extinction risk by skewing hatchling sex ratios. Assessment of this risk at the population level requires the identification of sex in hatchlings and juveniles. However, available methods are typically lethal, highly invasive, or difficult to conduct at a large scale. Changes in DNA methylation, an epigenetic modification, have been characterized as part of sex differentiation pathways in some species with environmentally determined sex, but so far not in marine turtles. Neither have epigenetic biomarkers for sex been developed into rapid assays suited to research on wildlife. In this study, we aimed to develop a rapid, minimally invasive, and inexpensive method to identify the sex of marine turtles. We used reduced representation bisulfite sequencing DNA methylation data from adult green sea turtle (Chelonia mydas) skin biopsies to identify 16 genomic regions exhibiting differential methylation between males and females (adjusted p-value < 0.01). We designed methylation sensitive qPCR assays for these regions and tested their capacity to identify the sex of turtles ranging in age between 3-34 years. The qPCR assay identified the correct sex in turtles > 17 years. However, the sex of younger turtles could not be accurately identified. This suggests the sex differences distinguishable by the assay were adult specific, reflecting the training data on which the sex-specific regions were identified, and likely linked to late-stage ontogenetic changes associated with sexual maturity. Epigenetic biomarkers are a promising tool for wildlife research because they can be minimally invasive and high throughput. Future research into sex-specific differentially methylated regions in hatchlings and juveniles should be based on genome-wide DNA methylation data from a wider age range, which includes hatchlings.
Introduction
Global warming is a risk to all species. One potential group of concern is reptiles with temperature dependent sex determination (Janzen, 1994; Mitchell and Janzen, 2010; Blechschmidt et al., 2020). These animals do not have sex chromosomes and instead, sex is determined by the temperature during egg incubation. For example, in green sea turtles (Chelonia mydas), an egg incubation below 29.2°C results in only male hatchlings and only females above 30°C (Yntema and Mrosovsky, 1980). A similar trend is observed in other marine turtle species, where higher egg incubation temperatures have a bias towards female hatchlings and lower temperatures towards males (Yntema and Mrosovsky, 1980; Miller, 1997). Global warming is predicted to lead to a biased sex ratio and over time result in a population decline (Berec et al., 2001). This effect is likely to be region and population specific and depend on local conditions that dictate the nest temperatures (Maurer et al., 2021). Therefore, there is a need to monitor marine turtle hatchling sex ratios to identify populations at risk of sex bias and decline.
The sex of adult turtles can be identified by visual inspection, as the males have a large and more muscular tail compared to females (Eckert et al., 1999). However, turtles are long lived, and to estimate future population trends it is necessary to monitor hatchling and juvenile sex ratios. Yet, morphological differences between sexes in turtles may not become obvious for many years after hatching (Berry and Shine, 1980). Several approaches exist to identify hatchling sex. For example, histology on the gonads can be used, but is lethal (Mrosovsky et al., 1984; Mrosovsky and Provancha, 1992) and particularly undesirable for endangered species. Laparoscopic examination of the gonads is non-lethal and accurate on juveniles, but is highly invasive, impractical on a large scale, and cannot be carried out on small individuals (Wyneken et al., 2007). Less invasive methods include blood hormone assays. In a recent example, a Western blot assay of anti-Mullerian Hormone was demonstrated to be an accurate marker of sex in 1-2 day old loggerhead hatchlings (Tezak et al., 2020). However, the accuracy dropped to 90% in 83–177 day old individuals and has not been confirmed to work in other species. The cost of this hormone assay was approximately $18.00 US per samples, excluding labor and equipment. This method may be ideal where appropriate laboratory facilities are not available but is unlikely to be suited to large scale monitoring. Finally, because marine turtles do not exhibit genotypic sex determination (GSD), sex identification tests based on differences in DNA sequences between sexes, which are commonly applied in organisms such as birds and mammals (Hrovatin and Kunej, 2018; Purwaningrum et al., 2019), are not appropriate.
Another approach to characterizing population sex ratios is to predict them with developmental models calibrated from sexes identified via one of the above methods and incorporate nest temperatures and incubation durations (Godfrey et al., 1996; Marcovaldi et al., 1997; Godley et al., 2001a; Godley et al., 2001b). If calibrations are transferable between populations, these models can predict hatchling sex ratios from beach temperatures alone. However, this does not take into account other factors such as hatchling success (Schwanz et al., 2010), and may not reflect final clutch sex ratios. These model also crucially depend on accurate known sex calibration data from hatchlings, and as described, none of the available methods meet the most desirable characteristics by being minimally invasive, accurate, high-throughput, and low cost.
In other vertebrate species, changes in an epigenetic modification known as DNA methylation are known to occur throughout the genome (Waters et al., 2018). The most common form of DNA methylation is the addition of a methyl group to cytosine residues within cytosine phosphate guanine (CpG) sites (Bird, 1993). DNA methylation is well known for its role in regulating gene expression but has been used as a biomarker for life history traits including sex. The methylation state of specific genes is a characteristic of the sex determination cascade in several species exhibiting temperature dependent sex determination (Garcia-Moreno et al., 2018). An example is the European sea bass (Dicentrarchus labrax), where higher temperatures during development induce increased DNA methylation within the promoter of the aromatase gene, known for its role in sexual development of females (Ramsey and Crews, 2009; Navarro-Martín et al., 2011). As far as we are aware, however, this class of biomarkers has not been explored for sex identification purposes in wildlife species exhibiting TSD, including marine turtles. This is likely owing to the lack of appropriate whole genome methylation data, and the relative novelty of methylation analysis in non-model organisms (Bock et al., 2022).
To explore the potential of this novel class of biomarkers, we identified DNA methylation markers of sex in green sea turtles (Chelonia mydas) sampled from skin biopsies. Then, seeking to produce a fast, low cost and high throughput assay suitable for population monitoring we developed a quantitative PCR assay and evaluated its accuracy when applied to known sex individuals at multiple life stages of green sea turtles.
Methods
DNA methylation dataset
To identify differentially methylated regions between sex of green sea turtles we used reduced representation bisulfite sequencing data from a previous study (Mayne et al., 2022). The previous study contained sequencing data for 63 green sea turtles skin biopsies ranging between 1-43.8 years of age and with 22 unknown sex, 31 females, and 10 males. Samples with known sex (15.8-43.8 years) were only used in this study (Supplementary Table 1). The sequencing data was prepared using the restriction enzyme MspI digestion which cleaves DNA at a C^CGG sequence as previously described (Smallwood et al., 2011). The library was constructed using the Ovation Methyl-Seq system from NuGEN.
Differential methylation
Sequencing data was trimmed using trimmomatic v0.38 with a sliding window of 4-15bp and a minimum read length of 36bp (Bolger et al., 2014). The trimmed reads were aligned to the green sea turtle genome (rCheMyd1.pri.v2, NCBI) using Bismark v0.23.0 with default parameters and bowtie2 v2.4.4 as an aligner (Krueger and Andrews, 2011; Langmead and Salzberg, 2012). Methylation calling was carried out using Bismark’s bismark_methylation_extractor function and returns methylation values as a percentage. Methylation calling output was read into R v4.2.0 using BiSeq (Hebestreit et al., 2013). Differentially methylated regions were also identified using BiSeq using default parameters in the recommended user’s guide. Regions were considered significant with a false discover rate < 0.05. Global methylation differences were tested by calculating the mean DNA methylation average for each CpG site and using a student unpaired t-test between groups. All analyses were carried out in R v4.2.0.
Gene ontology
The closest genes to the differentially methylated regions were identified using the closest function in bedtools v2.29.1. To group the closest genes into functional pathways, gene ontology enrichment was carried out with EnrichR using 2018 terms (Kuleshov et al., 2016). Terms were considered significant if p < 0.01.
Sample collection and animal ethics
Skin biopsies used for qPCR were from either Kélonia (Réunion; captive individuals) or Ningaloo (Western Australia; wild individuals). Réunion samples were conducted under permits from Kélonia, Réunion Island Prefecture and Department of the Environment (09-1405/SG/DRCTCV; DA 2017-01). Western Australian samples were collected during mark-recapture investigations under permits from the Department of Biodiversity, Conservation and Attractions (U10/2020-2022) and the CSIRO Large Animal Ethics Committee (AEC: 14-07).
Methylation sensitive qPCR
A methylation sensitive qPCR assay was chosen as it is a cost-effective and high-throughput assay. DNA from samples where sex is known was extracted using the DNeasy Blood and Tissue Kit (Qiagen) as instructed in the manufacturer’s protocol. DNA (150ng) was bisulfite treated using an in-house protocol (Supplementary Information).
Primer design was carried out using MethPrimer v2.0 (Li and Dahiya, 2002). Differentially methylated regions with a difference in magnitude of 10% were targeted for primer design. This was done as changes less than 10% have been found not to be detectable in methylation sensitive qPCR (Kurdyukov and Bullock, 2016). MethPrimer produces a primer pair for when the DNA is methylated and unmethylated (Supplementary Table 2). Primers were tested with an annealing gradient (55-60°C) and qPCR was carried out using Bio-Rad SsoAdvanced™ Universal SYBR Green Supermix (Cat# 172-5271). The mastermix and cycling conditions were carried out as instructed by the manufacturer’s protocol (Supplementary Tables 3, 4). qPCR for the methylated and unmethylated alleles were performed in separate wells. To determine the DNA methylation state of the amplicon, ΔCq between primer pairs were used as indicators for methylation. The performance of the assay was assessed using a receiver operating characteristic (ROC) curve. A general logistic regression model was also applied between the ΔCq and age to determine if there was significant difference in methylation differences with age. All analyses were carried out in R version 4.2.0.
Results
Differentially methylated regions
As described previously, the sequencing data aligned to the green sea turtle reference genome with an alignment rate of 88.6% (Mayne et al., 2022). No statistical difference was found between sex for global methylation (unpaired t-test, p-value = 0.14, Supplementary Figure 1). Differential methylation analysis identified 16 regions to be significantly different between sex (adjusted p-value < 0.05, Supplementary Table 5).
Gene enrichment analysis
Gene enrichment analysis found seven of the 16 differentially methylated regions to overlap with genes, five to be within 10,000bp of the gene and the remaining four to be further than 10,000bp (Supplementary Table 5). Gene ontology analysis resulted in a total of 22 significant terms with 32% being related to an immune response and a further 32% relating to epigenetic regulation (Supplementary Table 6). For example, genes such as PTPN2, a known negative regulator of T cell signaling and an association with inflammation was exhibited a hypermethylated region in males (Spalinger et al., 2021; James et al., 2022). The promoter of DNMT1 was hypermethylated in females. DNMT1 is a DNA methyltransferase, involved in the addition of methyl groups to CpG sites (Svedružić, 2011; Ito et al., 2022) and was enriched for terms relating to histone methylation (GO:0031061) and chromatin silencing (GO:0005677).
Methylation sensitive qPCR
A total of 10 regions exhibiting highly differentiated methylation between the sexes in the reduced representation bisulfite sequencing data were targeted for primer design (see methods), however primers with desirable amplification properties could only be designed for one region (Supplementary Table 2). The ΔCq between the methylated and unmethylated primers was used to infer a methylation level over the targeted level. A positive ΔCq was suggestive of a female and negative value is a male. In total, skin biopsies from 35 females and 13 males ranging between 3-34 years of age were tested. The assay correctly identified sex in 43 of the 48 samples (Supplementary Table 7). The oldest incorrectly identified sample was 16 years. The accuracy of the assay with samples less than 17 years of age was 44%, whereas, for 17 and older the accuracy of the assay was 100% (Figure 1).
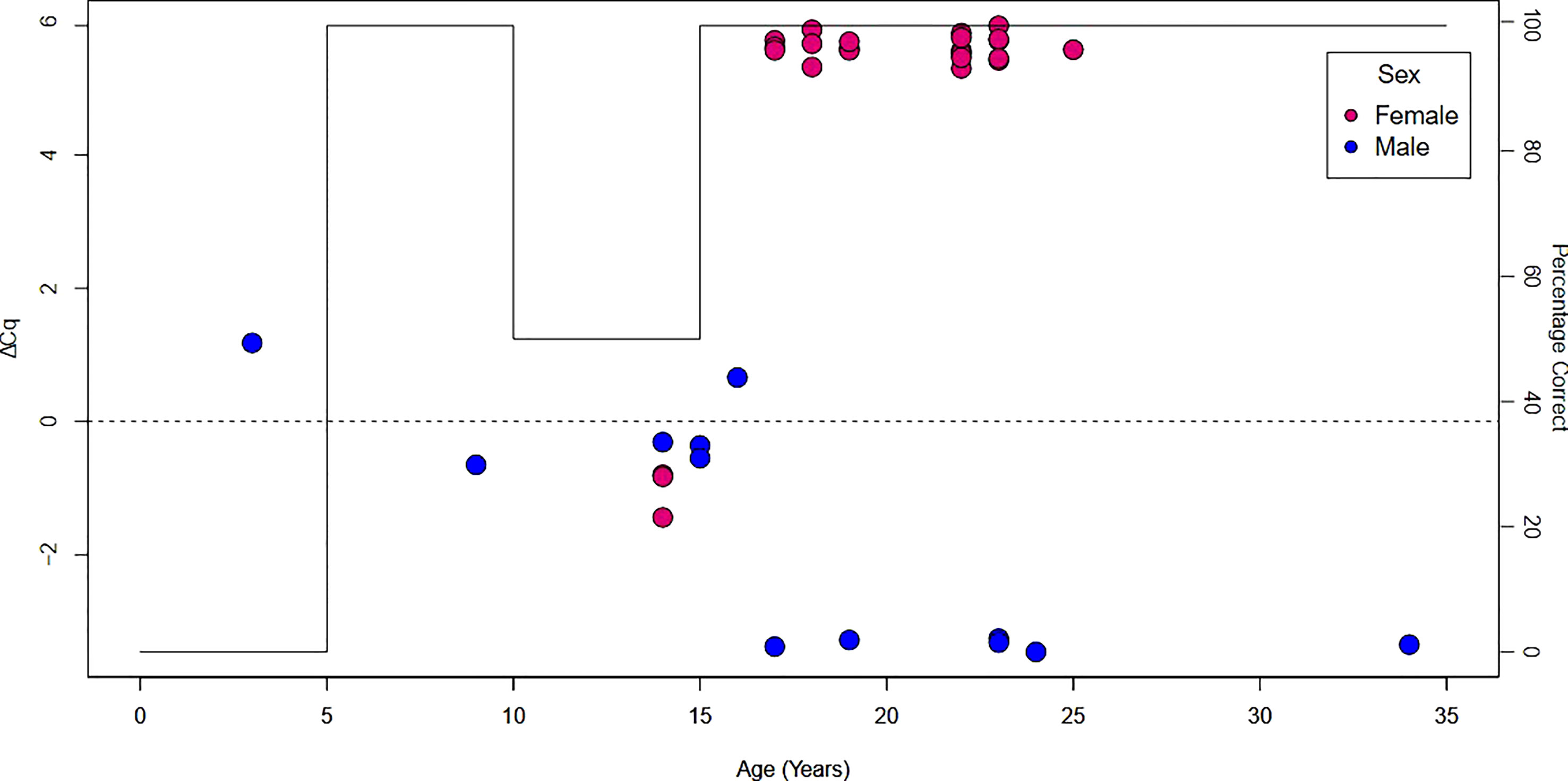
Figure 1 Performance of the methylation sensitive qPCR to identify sex in green sea turtle skin biopsies. Each dot represents an individual turtle and is colored by the known sex. The horizontal axis is the known age of the turtle. The left vertical axis details the ΔCq between the methylated and unmethylated primers. The right vertical axis shows the percentage of correctly predicted sex and is the black continuous line through the plot.
Discussion
We aimed to develop a qPCR assay to identify the sex of green sea turtles from patterns of DNA methylation in skin biopsies. This would offer a rapid, inexpensive, scalable, and minimally invasive way to characterize sex ratios, which is potentially at risk from global climate change. Our results indicate that the sex of turtles >17 years could be identified with high accuracy, which is encouraging as a proof of concept for this unique class of epigenetic diagnostic tests applied to skin biopsies. Yet, adults exhibit morphological differences between sexes, diminishing the need for diagnostic tests.
In contrast, hatchling and juvenile green turtles lack sex-specific morphological features, and therefore are the age classes where diagnostic sex tests would be most valuable. This suggests that the differentially methylated region characterized by our qPCR assay reflects a late ontogenetic stage in sexual differentiation and maturation. Gene ontology analysis suggests that on a DNA methylation level, the main differences in skin biopsies between sex are associated with genes relating to the immune response and epigenetic regulation. Since only skin biopsies were used it is unknown if this result would be similar in other tissues. Since only one tissue type was investigated it is difficult to draw any conclusions on what genes may be involved on a mechanistic level of why some parts of the genome may have methylation differences. However, it does provide a potential future approach to investigate genes relating to the immune response and epigenetic regulation as potential biomarkers of sex. Potentially, expression differences could exist too, within these classes of genes as DNA methylation is regulator of gene expression. RNA sequencing or targeted qPCR for genes could also be used as an alternative to DNA methylation to develop a molecular based assay for sex identification. Whether these differences are maintained throughout all life stages is unknown as genomic and expression across all life stages of marine turtles are not readily available.
If a rapid epigenetic test can be developed for the identification of hatchling turtles, we envisage that it would have two significant modes of application. The first is to permit on-ground testing of large numbers of hatchlings to produce accurate empirical data on current sex ratios and variation between populations. Minimal laboratory equipment is typically required to undertake qPCR assays. Therefore, they could be conducted in the field if there were benefits in receiving fast results. The second is to provide additional calibration data for developmental models (Stubbs and Mitchell, 2018), so that not only can estimates of turtle sex ratio be extrapolated over large geographic regions, but that forecasts into the future can also be made with greater certainty than is currently possible. Methylation sensitive PCR has commonly been used in biomedical diagnostics from detecting genetic abnormalities to cancer diagnosis (Zhang et al., 2012; Mehta et al., 2014; Massen et al., 2022). However, it has not been taken up as a commonly used molecular tool in an environmental or wildlife management setting.
Future research should focus on developing more genomic data on marine turtles for hatchlings and juveniles. Such data can then be combined with the data presented in this study to identify sex specific markers. By not having genome-wide bisulfite sequencing data of younger turtles is a limitation of this study and is most likely the reason why the assay was biased to adults. Future research should also focus on using tissues such as blood as sex-specific methylation may potentially be more distinct than in skin biopsies. Future research should also focus on using a qPCR-based assays as it is relatively inexpensive compared to DNA sequencing and to ensure it is non-lethal.
Although, this method was only able to correctly predict sex in adults for which external morphology already allows visual sex determination, with additional training data with known sex in hatchlings and juveniles, better biomarkers for sex identification could potentially be identified. This would enable researchers to readily identify populations that are at risk of becoming sex-biased so that management can be more targeted.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author. The raw demultiplex sequencing data was from a publicly available database and can be accessed from: https://data.csiro.au/collections/collection/CIcsiro:49784v1
Ethics statement
The animal study was reviewed and approved by Kélonia, 110 Réunion Island Prefecture and Department of the Environment.
Author contributions
BM designed the study, performed the bioinformatic analysis and laboratory work, and wrote the manuscript. WM, VB, FC, KB, MB, MV, and AT were involved in sample collection, study design and provided discussion into the manuscript. OB was involved in study design and provided discussion into the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was funded by the CSIRO Environomics Future Science Platform.
Acknowledgments
The authors would also like to thank all individuals who were involved in the collection of the samples used in this study, especially Stéphane Ciccione from Kélonia, La Réunion. We would also like to thank Chloe Anderson for assistance with laboratory work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1169808/full#supplementary-material
References
Berec L., Boukal D. S., Berec M. (2001). Linking the allee effect, sexual reproduction, and temperature-dependent sex determination via spatial dynamics. Am. Nat. 157, 217–230. doi: 10.1086/318626
Berry J. F., Shine R. (1980). Sexual size dimorphism and sexual selection in turtles (order testudines). Oecologia 44, 185–191. doi: 10.1007/BF00572678
Bird A. P. (1993). Functions for DNA methylation in vertebrates. Cold Spring Harb. Symp. Quant Biol. 58, 281–285. doi: 10.1101/SQB.1993.058.01.033
Blechschmidt J., Wittmann M. J., Blüml C. (2020). Climate change and green Sea turtle sex ratio-preventing possible extinction. Genes (Basel) 11. doi: 10.3390/genes11050588
Bock S. L., Smaga C. R., McCoy J. A., Parrott B. B. (2022). Genome-wide DNA methylation patterns harbour signatures of hatchling sex and past incubation temperature in a species with environmental sex determination. Mol Ecol. 31, 5487–5505. doi: 10.1111/mec.16670
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Eckert K. L., Bjorndal K. A., Abreu-Grobois F. A., Donnelly M. (1999). Research and management techniques for the conservation of sea turtles. Marine Turtle Specialist Group. Vol 1, pg 1–248.
Garcia-Moreno S. A., Plebanek M. P., Capel B. (2018). Endocrinology c: epigenetic regulation of male fate commitment from an initially bipotential system. Mol Cell Endocrinol 468, 19–30. doi: 10.1016/j.mce.2018.01.009
Godfrey M. H., Mrosovsky N., Barreto R. (1996). Estimating past and present sex ratios of sea turtles in Suriname. Can J Zool. 74, 267–277. doi: 10.1139/z96-033
Godley B., Broderick A., Downie J., Glen F., Houghton J., Kirkwood I., et al. (2001a). Ecology: thermal conditions in nests of loggerhead turtles: further evidence suggesting female skewed sex ratios of hatchling production in the Mediterranean. J Exp Mar Biol Ecol. 263, 45–63. doi: 10.1016/S0022-0981(01)00269-6
Godley B., Broderick A., Mrosovsky N. (2001b). Estimating hatchling sex ratios of loggerhead turtles in Cyprus from incubation durations. Marine Ecology Progress Series 210, 195–201. doi: 10.3354/meps210195
Hebestreit K., Dugas M., Klein H. U. (2013). Detection of significantly differentially methylated regions in targeted bisulfite sequencing data. Bioinformatics 29, 1647–1653. doi: 10.1093/bioinformatics/btt263
Hrovatin K., Kunej T. (2018). Genetic sex determination assays in 53 mammalian species: literature analysis and guidelines for reporting standardization. Ecol. Evol. 8, 1009–1018. doi: 10.1002/ece3.3707
Ito T., Kubiura-Ichimaru M., Murakami Y., Bogutz A. B., Lefebvre L., Suetake I., et al. (2022). DNMT1 regulates the timing of DNA methylation by DNMT3 in an enzymatic activity-dependent manner in mouse embryonic stem cells. PloS One 17, e0262277. doi: 10.1371/journal.pone.0262277
James J., Chen Y., Hernandez C. M., Forster F., Dagnell M., Cheng Q., et al. (2022). Redox regulation of PTPN22 affects the severity of T-cell-dependent autoimmune inflammation. eLife 11, e74549. doi: 10.7554/eLife.74549.sa2
Janzen F. J. (1994). Climate change and temperature-dependent sex determination in reptiles. Proc. Natl. Acad. Sci. U.S.A. 91, 7487–7490. doi: 10.1073/pnas.91.16.7487
Krueger F., Andrews S. R. (2011). Bismark: A flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics 27, 1571–1572. doi: 10.1093/bioinformatics/btr167
Kuleshov M. V., Jones M. R., Rouillard A. D., Fernandez N. F., Duan Q., Wang Z., et al. (2016). Et al: enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97. doi: 10.1093/nar/gkw377
Kurdyukov S., Bullock M. (2016). DNA Methylation analysis: choosing the right method. Biol. (Basel) 5. doi: 10.3390/biology5010003
Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li L. C., Dahiya R. (2002). MethPrimer: designing primers for methylation PCRs. Bioinformatics 18, 1427–1431. doi: 10.1093/bioinformatics/18.11.1427
Marcovaldi M. Â., Godfrey M. H., Mrosovsky N. (1997). Estimating sex ratios of loggerhead turtles in Brazil from pivotal incubation durations. Can J Zool 75, 755–770. doi: 10.1139/z97-097
Massen M., Lommen K., Wouters K. A. D., Vandersmissen J., van Criekinge W., Herman J. G., et al. (2022). Technical considerations in PCR-based assay design for diagnostic DNA methylation cancer biomarkers. Clin. Epigenet. 14, 56. doi: 10.1186/s13148-022-01273-z
Maurer A. S., Seminoff J. A., Layman C. A., Stapleton S. P., Godfrey M. H., Reiskind M. O. B. (2021). Population viability of Sea turtles in the context of global warming. BioScience 71, 790–804. doi: 10.1093/biosci/biab028
Mayne B., Mustin W., Baboolal V., Casella F., Ballorain K., Barret M., et al. (2022). Age prediction of green turtles with an epigenetic clock. Mol. Ecol. Resour. 00, 1–10. doi: 10.1111/1755-0998.13621
Mehta A., Mielnik A., Schlegel P. N., Paduch D. A. (2014). Novel methylation specific real-time PCR test for the diagnosis of klinefelter syndrome. Asian J. Androl. 16, 684–688. doi: 10.4103/1008-682X.125914
Miller J. (1997). “Reproduction in sea turtles,” in The biology of sea turtles. Eds. Lutz P. L., Musick J. A. (Boca-Raton, Florida: CRC Press).
Mitchell N. J., Janzen F. J. (2010). Temperature-dependent sex determination and contemporary climate change. Sex. Dev. 4, 129–140. doi: 10.1159/000282494
Mrosovsky N., Hopkins-Murphy S. R., Richardson J. I. (1984). Sex ratio of sea turtles: seasonal changes. Science 225, 739–741. doi: 10.1126/science.225.4663.739
Mrosovsky N., Provancha J. (1992). Sex ratio of hatchling loggerhead sea turtles: data and estimates from a 5-year study. Can J Zool 70, 530–538. doi: 10.1139/z92-080
Navarro-Martín L., Viñas J., Ribas L., Díaz N., Gutiérrez A., Di Croce L., et al. (2011). DNA Methylation of the gonadal aromatase (cyp19a) promoter is involved in temperature-dependent sex ratio shifts in the European sea bass. PLoS Genet 7. doi: 10.1371/journal.pgen.1002447
Purwaningrum M., Nugroho H. A., Asvan M., Karyanti K., Alviyanto B., Kusuma R., et al. (2019). Molecular techniques for sex identification of captive birds. Vet. World 12, 1506–1513. doi: 10.14202/vetworld.2019.1506-1513
Ramsey M., Crews D. (2009). “Steroid signaling and temperature-dependent sex determination–reviewing the evidence for early action of estrogen during ovarian determination in turtles,” in Seminars in cell & developmental biology (United States of America: Elsevier), 283–292.
Schwanz L. E., Spencer R. J., Bowden R. M., Janzen F. J. (2010). Climate and predation dominate juvenile and adult recruitment in a turtle with temperature-dependent sex determination. Ecology 91, 3016–3026. doi: 10.1890/09-1149.1
Smallwood S. A., Tomizawa S., Krueger F., Ruf N., Carli N., Segonds-Pichon A., et al. (2011). Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 43, 811–814. doi: 10.1038/ng.864
Spalinger M. R., Schwarzfischer M., Niechcial A., Atrott K., Laimbacher A., Gottier C., et al. (2021). Loss of PTPN22 promotes intestinal inflammation by compromising granulocyte-mediated antibacterial defence. J. Crohn’s Colitis. 15, 2118–2130. doi: 10.1093/ecco-jcc/jjab098
Stubbs J. L., Mitchell N. J. (2018). The influence of temperature on embryonic respiration, growth, and sex determination in a Western Australian population of green turtles (Chelonia mydas). Physiol Biochem Zool 91, 1102–1114. doi: 10.1086/700433
Tezak B., Sifuentes-Romero I., Milton S., Wyneken J. (2020). Identifying sex of neonate turtles with temperature-dependent sex determination via small blood samples. Sci. Rep. 10, 5012. doi: 10.1038/s41598-020-61984-2
Waters S. A., Capraro A., McIntyre K. L., Marshall Graves J. A., Waters P. D. (2018). The methylome of vertebrate sex chromosomes. Genes (Basel) 9. doi: 10.3390/genes9050230
Wyneken J., Epperly S. P., Crowder L. B., Vaughan J., Blair Esper K. (2007). Determining sex in posthatchling loggerhead sea turtles using multiple gonadal and accessory duct characteristics. J Herpetologica 63, 19–30. doi: 10.1655/0018-0831(2007)63[19:DSIPLS]2.0.CO;2
Yntema C. L., Mrosovsky N. (1980). Sexual differentiation in hatchling loggerheads (Caretta caretta) incubated at different controlled temperatures. Herpetologica 36, 33–36.
Keywords: turtle, DNA methylation, sex, PCR, molecular
Citation: Mayne B, Mustin W, Baboolal V, Casella F, Ballorain K, Barret M, Vanderklift MA, Tucker AD and Berry O (2023) Differential methylation between sex in adult green sea turtle skin biopsies. Front. Mar. Sci. 10:1169808. doi: 10.3389/fmars.2023.1169808
Received: 20 February 2023; Accepted: 26 April 2023;
Published: 09 May 2023.
Edited by:
Michael Paul Jensen, National Oceanic and Atmospheric Administration (NOAA), United StatesReviewed by:
Daniel Garcia-Souto, University of Vigo, SpainMbaye Tine, Gaston Berger University, Senegal
Copyright © 2023 Mayne, Mustin, Baboolal, Casella, Ballorain, Barret, Vanderklift, Tucker and Berry. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Benjamin Mayne, YmVuamFtaW4ubWF5bmVAY3Npcm8uYXU=
 Benjamin Mayne
Benjamin Mayne Walter Mustin
Walter Mustin Vandanaa Baboolal2
Vandanaa Baboolal2 Mathew A. Vanderklift
Mathew A. Vanderklift Anton D. Tucker
Anton D. Tucker Oliver Berry
Oliver Berry