
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 06 April 2023
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1158446
This article is part of the Research Topic Environment-Microbe-Host Interactions in Aquaculture Ecosystems View all 5 articles
 David J. Bradshaw II1*
David J. Bradshaw II1* Carlie S. Perricone1
Carlie S. Perricone1 Laura E. King1
Laura E. King1 Elizabeth B. Allmon2
Elizabeth B. Allmon2 Maria Sepúlveda2,3
Maria Sepúlveda2,3 Marty Riche1
Marty Riche1 Paul S. Wills1
Paul S. Wills1 Nicole Kirchhoff4
Nicole Kirchhoff4 Sahar Mejri1
Sahar Mejri1Introduction: Salinity presents economic and technical challenges in land-based recirculating aquaculture systems (RAS) in the U.S. warm water marine finfish aquaculture industry. Many studies have shown euryhaline fish reared at salinities closer to their iso-osmotic salinity can yield enhanced production performance as well as potential reduced costs to farms. However, there is potential for osmotic stress in fish larvae to negatively impact larvae microbiome and innate immune system. Florida pompano (Trachinotus carolinus) is a popular sportfish has been targeted for land-based RAS due to its impressive market value and euryhaline capacity. This study investigated the impacts of rearing Florida pompano larvae at salinities closer to their iso-osmotic salinity.
Materials and methods: Larvae were cultured at 10, 20, and 30 ppt in triplicates, and larvae samples were collected for histopathology, microbiome, and whole transcriptomics analysis every three days from hatching until the time of weaning (24 days post hatch [DPH]). Water samples were also taken for microbiome analysis on every other larval sampling day.
Discussion: These changes were driven more by metamorphosis, causing an increase in expression of antioxidant genes (cat, gss, gsto1, and scara3) than by the presence of potentially pathogenic genera, which failed to induce an immune response (low or unchanged expression of downstream elements of the NOD1 or TLR5 pathways). These findings provide baseline information on Florida pompano low salinity tolerance in larviculture during early developmental stages. In addition, we have shown minimal effects on the immune system at salinities as low as 10 ppt. This work has important implications for larval health management and can be used to refine and direct future research regarding improving commercial production of warm water marine species
Aquaculture producers are attempting to expand the number of marine fish species which can be successfully and efficiently raised via land-based recirculating aquaculture systems. However, due to the high real estate costs associated with a farm being located near the ocean, cost of supplies needed to make saltwater, and the issues related to saltwater discharge when located inland, it would be advantageous for an aquaculture farm to be able to utilize low salinity to economically enhance grow-out operations. Culture at a lower salinity than 32 ppt might increase performance in aquaculture, where growth at intermediate salinities has been found to increase in several species such as Atlantic cod (Lambert et al., 2011). Variation in growth due to salinity is related to the metabolic cost of osmoregulation, estimated to be 2-4% of the resting metabolic rate and is known to be 20-30% higher in true fresh or seawater compared to an iso-osmotic environment, i.e. where the external concentration of ions equals the internal concentration, therefore the osmoregulatory cost greatly reduced (Withers, 1992). These potential performance improvements may also carry over into overall larval health.
Thus, it is important to identify species who can withstand low salinities without causing undo stress to help mitigate some of the costs associated with inland recirculating aquaculture systems. One potential candidate is the euryhaline teleost the Florida pompano (Trachinotus carolinus). It inhabits marine and estuarine waters along the U.S. Atlantic Coast and Gulf of Mexico, as well as some areas of the West Indies and Brazil (Zhang et al., 2022). This species is also one of the most expensive marine warm water food fish available at market (Weirich et al., 2021; Zhang et al., 2022). Its economic value, wide salinity range, and fast growth make Florida pompano a desirable candidate for commercialized aquaculture. The optimal grow out salinity of Florida pompano is not well-defined, with limited available research on its tolerance to different salinities in an aquaculture farm setting. Weirich et al. (2009) revealed that juveniles and adults could tolerate salinities as low as 5 ppt while Riche et al. (2012) showed that osmoregulatory stress could be relieved when salinity was raised to 12 ppt. However, there are no published studies to date on the histopathological, microbiological, or immunological effects of low salinities in Florida pompano larvae in a commercial production setting.
Suboptimal rearing salinities cause osmoregulatory stress in teleost fish, which can lead to abnormal gill development (Abou Anni et al., 2016). The external location of the gills and immediate contact with the water contribute to gill structure vulnerability (Reis et al., 2009). Morphological alterations can be easily recognized in gills (Fanta et al., 2003) and are signs of stress-mediated damage (Laurén and Wails, 2018). Many studies have reported that deleterious morphological alterations in fish gills often coincide with unfavorable environmental factors and can indicate a generalized stress response (Heath, 1995; Humphrey, 2006). Since the gills perform vital functions including respiration, osmoregulation, and waste excretion, histopathological changes in the structure of these organs usually co-occur with respiratory and osmoregulatory stress (Smith and Piper, 1975; Cerqueira and Fernandes, 2002).
Osmoregulatory stress can also have impacts on immunological mechanisms and functions at the molecular level in fish. RNA sequencing is a powerful tool that provides whole transcriptomics information and has been widely used to characterize changes in gene regulation and expression related to the immune system in fish (Zhang et al., 2022). This high-throughput sequencing technology can help elucidate the molecular mechanisms in specific tissues and cells in response to environmental stressors such as suboptimal salinity. Previous transcriptomics studies have demonstrated that salinity fluctuations can lead to changes in expression of immune-related genes in other euryhaline species (Boutet et al., 2006; Li et al., 2012; Nguyen et al., 2016; Cao et al., 2020). Changes in expression and regulation of genes linked to immunity can affect the overall health of the fish, in addition to community structure of the fish microbiome (Pérez-Pascual et al., 2020).
The taxonomic composition of the fish microbiome can be identified using 16S DNA sequencing, which is a promising tool for regular health surveillance in marine warm water aquaculture. Fish microbiomes are directly involved in their host’s metabolism, growth, development, and immunity, while the fish supports the nutrition and colonization of its internal and external microbiota (Rawls et al., 2004; Wang et al., 2018; Sehnal et al., 2021). Maintaining a microbiome of beneficial microbes is important as it suppresses disease through competitive exclusion or toxic secondary metabolites (Llewellyn et al., 2014). An imbalance or disruption of the microbiome is referred to as dysbiosis and is associated with impaired immunocompetence (Llewellyn et al., 2014). Since microbiota found in epidermal mucosa (skin and gills) serve as the first line of defense against pathogens, stress-related dysbiosis in the mucosa can increase susceptibility to disease (Llewellyn et al., 2014). Sequencing based studies of fish mucosal and gut microbiota have revealed that bacterial pathogens can exist as a constituent of a healthy microbiome but can become virulent under stressful conditions (Llewellyn et al., 2014; Sehnal et al., 2021). Ubiquitous bacteria, that are often opportunistically pathogenic in Trachinotus species in aquaculture systems, include Vibrio, Pseudomonas, Flavobacterium, Photobacterium, and Mycobacterium (Llewellyn et al., 2014; Sehnal et al., 2021).
There is a complex interplay among the structure of fish microbiomes, the local environment, and their diet. Microbiomes in the surrounding water are shaped by water quality parameters, since different bacterial species have specific growth requirements for salinity, pH, dissolved oxygen, and available nutrients (Webster et al., 2018). Any fluctuations in these physical and chemical properties can change the taxonomy of the fish microbiome, since the water serves as a reservoir for bacteria that colonize fish (Legrand et al., 2020). However, the fish microbiome also shifts across life stages, becoming more colonized by bacteria found in diet over time rather than those in the environment (Burgos et al., 2018).
The goal of this study is not to identify the low salinity tolerance of Florida pompano, but instead to investigate potential impacts of moderate, i.e. near iso-osmotic, salinity on larval rearing. This research work contains data that was collected during the same study as data reported in a previous manuscript, that assessed the effects of salinity on biometrics, fatty acids, and genes linked to osmoregulation and fatty acids in Florida pompano larviculture (Bradshaw et al., 2023). In this study, we have provided a multidisciplinary view of how Florida pompano larvae respond to low salinities using histological techniques, 16S DNA sequencing, and whole transcriptomics. We report important changes in gill histopathology, water and larval microbiome structure, and immunological gene expression in Florida pompano larvae reared in three different salinities (10, 20, and 30 ppt) for up to 24 days post hatch (DPH). The osmotic salinity of most euryhaline fish species ranges between 11-13 ppt, therefore this study chose salinities bridging this range (10 and 20 ppt) while using 30 ppt as a representation of the normal larviculture salinity. This study is further representation of the benefits of designing studies around the needs of aquaculture producers and provided information regarding the viability of rearing larvae at lower salinities.
This study was approved by the Institutional Animal Care and Use Committee (IACUC-A20-29). All experimental procedures involving animals were conducted following IACUC guidelines and approved protocols. At the end of the trial, all larvae were euthanized.
Eggs, provided by a partnering hatchery (Proaquatix, Vero Beach, FL), were disinfected and stocked (stocking density of 30 eggs L-1) into two upwelling 100-gal egg cones supplied with filtered natural seawater at ~32 ppt at a partnering farm (Live Advantage Bait, Fort Pierce, FL). After hatching (hatching success of ~60%), larvae at 1 DPH were moved into nine 416 L (110 gal) flow-through tanks, filled to 330 L (stocking density of 20 larvae L-1) with three salinities (10, 20, and 30 ppt) in triplicate (total tanks, n=9). Tank water was made using natural seawater and city water that was filtered to remove chloramines and chlorines utilizing the Stealth-RO™ and smallBoy® ChloraShield® carbon filter (HydroLogic Purification Systems, Santa Cruz, CA, USA), smallBoy® Dechlorinator and Sediment System with upgraded KDF85/Catalytic Carbon Filter, 60 GPH (HydroLogic). Prior to utilizing seawater for mixing, removal of chlorines and chloramines was checked using Insta-Test Free & Total Chlorine & Chloramine Strips (LaMotte Company, Chestertown, MD, USA). Florida pompano are normally reared in ~30 ppt, thus 20 and 10 ppt represented the lower treatment salinities. To maintain salinity, tanks were operated as individual flow through units with salinity validated daily and at each water change using a HACH HQ14d Portable Meter with CDC401 Conductivity Cell (Hach Company, Ames, IA, USA).
Eggs and whole larvae were sampled at 1 DPH from the hatching cone and culture tanks were sampled every three days after hatching (3, 6, 9, 12, 15, 18, 20, and 24 DPH) for water and larvae microbiome, larvae whole transcriptomics, and gill histopathology analysis (Table 1). Whole larvae that were sampled for microbiome and transcriptomics analysis were placed into one sterile, DNase- and RNase-free tube per tank containing RNAlater (Invitrogen, Waltham, MA, USA). These samples were first stored at 4°C for 24 h and then stored at -20°C either at the farm or lab until nucleic acid extraction per RNAlater manufacturer instructions. Samples were transferred on ice between the farm and lab. Whole larvae histological samples were preserved in 10% neutral buffered formalin and stored at room temperature until transferred into ethanol within a week. The larval feeding schedule at Florida Atlantic University at Harbor Branch Oceanographic Aquaculture Park was used as our presumptive model for data interpretation (the feeding schedule at partnering farms is proprietary and could not be shared). The schedule is as follows: green water (Nannochloropsis algae paste) from 0-12 DPH, enriched rotifers from 1-10 DPH, Artemia nauplii from 9-15 DPH, enriched Artemia from 12-21 DPH, and micro feeds from 13-24 DPH (Weirich et al., 2021).
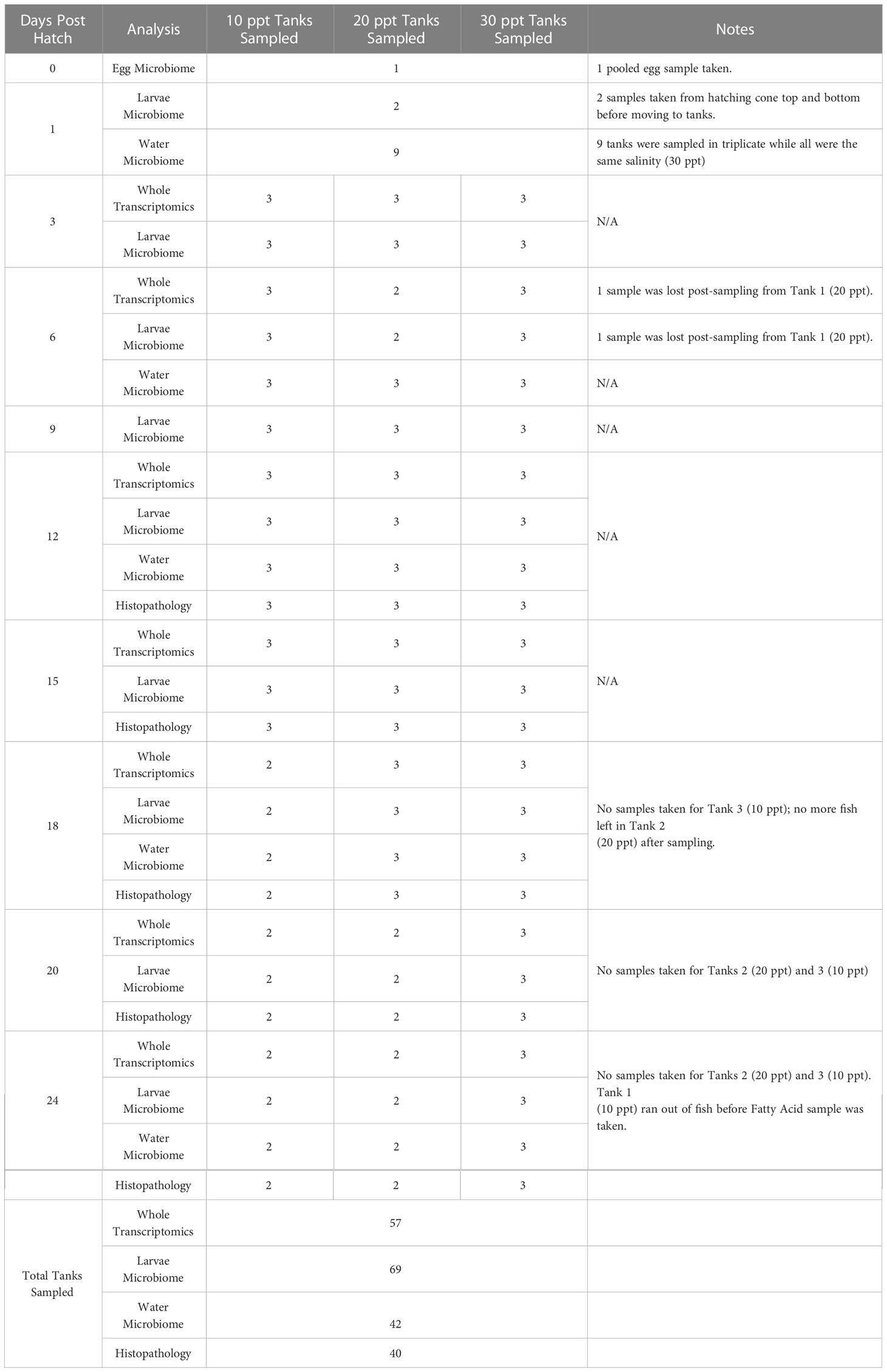
Table 1 Summary of tanks sampled for whole transcriptomics, larvae microbiome, water microbiome, and histopathology in Florida pompano (Trachinotus carolinus) larvae sampled from 0 to 24 days post hatch at three salinities (10, 20, and 30 ppt).
A total of 68 larvae were sampled for histological analysis throughout the duration of the study (10 ppt, n = 22; 20 ppt, n = 20; 30 ppt, n = 26) (Supplementary Table 1). Although samples were taken for histopathology every three days post hatching, only larvae from 12, 15, 18, 20, and 24 dph were used due to the size of fish from 3, 6, and 9 dph not being large enough for analysis. Larvae samples were dehydrated in a graded series of ethanol solutions and embedded in paraffin using a Thermo HistoStar Embedding Workstation, cut in serial transverse and sagittal cuts (4-5 μm) with a Thermo Microm HM 355S-2, mounted on glass slides, and allowed to air dry (Thermo Fisher Scientific, Waltham, MA). Finally, they were stained with hematoxylin and eosin using a Leica Autostainer XL for general visualization of the affected tissues and organs (Leica, Buffalo Grove, IL). The morphology and the pathological changes of larval secondary gills were observed using an optic microscope Olympus BX51, and any pathological alterations recorded. Interpretation of the histopathology results followed previous studies as a reference (Bernet et al., 1999).
Water and larvae were collected at the same time at 5 sampling points (1, 6, 12, 18, and 24 DPH), while larvae were additionally sampled during 0, 3, 9, 15, and 20 DPH. Up to 500 mL of water was filtered in triplicate from each tank through sterile 55 µM banjo filters to prevent particulates and food particles from being collected and then 0.45 µM inline membrane filters (Smith-Root, Vancouver, WA, USA) to concentrate microbial cells. These filters were kept at room temperature until time of extraction. Smith-Root conducted a study showing that DNA did not degrade within 6 months at room temperature (Thomas et al., 2019) and these filters have also been used in other studies (Loeza-Quintana et al., 2020; Skinner et al., 2020; Banks et al., 2021; Nolan et al., 2022). These self-preserving eDNA filters are made of hydrophilic plastic which absorbs any remaining moisture into the package and preserves the DNA and prokaryotes via desiccation which has been shown as an effective preservation technique in other studies (García, 2011; Manzanera, 2021).
Total DNA was extracted from the filters using the Qiagen DNeasy PowerWater Kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions and then stored at -20°C. Up to 0.25 g of whole larvae was homogenized from each tank using the Qiagen TissueRuptor II. Multiple larvae were used to get to a weight of at least 0.02g if needed. Total DNA was extracted from the homogenized larval samples using the Qiagen DNeasy Blood and Tissue Kit following an optimized protocol, and then kept at -20°C. All nucleic acid extracts were checked for concentration (ng. µL-1) using a Qubit Fluorometer 3 (ThermoFisher Scientific, Waltham, MA, USA) and quality (A260/A280) using a ThermoFisher NanoDrop Spectrophotometer 2000.
Purified DNA samples (69 larvae and 126 water samples) were sent to Azenta (South Plainfield, NJ, USA) for high-throughput sequencing. The 16S V3V4 region (341f-805r) was amplified using proprietary primers and sequenced on the Illumina HiSeq 2500 platform (San Diego, CA, USA). Demultiplexed raw sequences were analyzed using a snakemake (Koster and Rahmann, 2012) (v. 6.0.2) workflow (Bradshaw et al., 2020) in a conda (Anaconda Inc., 2021) (v. 4.9.2) environment with biopython (Cock et al., 2009) (v. 1.78), trim-galore (Krueger, 2020) (v. 0.6.6), fastx_toolkit (Gordon and Hannon, 2010) (v. 0.0.14), and mamba (v. 0.7.14) installed within the original QIIME2 (Bolyen et al., 2019) 2021.2 environment. Trim Galore was used to trim primers and sequences based on Phred quality scores (>20) and length (>20 base pairs) from paired reads, while FastQC (Andrews, 2010) was used to check various quality metrics. Once imported into QIIME2, the trimmed sequences were joined and denoised by Dada2 (Callahan et al., 2016) with default parameters, except the truncating length for forward and reverse reads was set to 225 base pairs. Dada2 summarizes the sequences into amplicon sequence variants (ASVs), which represent 100% unique sequences. ASVs were annotated using the Scikit-learn system (Pedregosa et al., 2011) and a 16S V3V4 region (341f-805r) (Herlemann et al., 2011) classifier trained on the SILVA 138.1 database (Quast et al., 2012; Yilmaz et al., 2014) using the RESCRIPt (Robeson et al., 2021) plugin pipeline. Mitochondria, chloroplasts, and unassigned sequences were filtered from the annotated table.
Outputs from QIIME2 were imported into RStudio (version 1.4.1106, R version 4.0.4) for data manipulation, visualization, and statistical testing. The main libraries loaded into the session included phyloseq (1.34.0), vegan (2.5-7), ggplot2 (3.3.3), plyr (1.8.6), tidyverse (1.3.0), FSA (0.8.32), reshape (0.8.8), DESeq2 (1.30.1), metagenomeSeq (1.32.0), ggvenn (0.1.8), readxl (1.3.1), and rcompanion (2.4.0) ((Wickham, 2007; Wickham, 2011; McMurdie and Holmes, 2013; Love et al., 2014; Wickham, 2016; Oksanen et al., 2019; Wickham and Bryan, 2019; Mangiafico, 2021; Ogle et al., 2021; Yan, 2021). To remove very rare ASVs, any ASV that did not occur across greater than 10 samples were removed (Ziegler et al., 2016). The filtered, untransformed ASV counts were used to calculate alpha diversity and relative percentages for comparison across metadata categories. Shannon diversity was statistically correlated with Fisher, Simpson, and observed ASVs based on the Spearman Rank method (p values < 2.2e-16), thus Shannon diversity was used moving forward to represent alpha diversity. We will refer to the following nine genera as potentially pathogenic genera (PPG) throughout this manuscript since they have been shown in the literature to be pathogenic for Trachinotus species: Nocardia (seriolae), Mycobacterium (marinum), Pseudomonas (alcaligenes, maltophilia), Vibrio (anguillarum, harveyi, ponticus, vulnificus), Streptococcus (agalactiae), Aeromonas (hydrophilia), Flexibacter, Flavobacterium, and Photobacterium (damsela) (Aronson, 1926; Hawke, 1976; Giavenni et al., 1980; Zhou et al., 2002; Wang et al., 2009; Yanong et al., 2010; Xia et al., 2015; Chong et al., 2017; Zhu et al., 2018; Food and Agriculture Organization of the United Nations, 2023).
RNA extraction and transcriptomics analysis occurred as previously reported in another article published on this study (Bradshaw et al., 2023). Briefly, total RNA was extracted from up to 0.25 g whole homogenized larvae with the RNeasy Mini Kit (Qiagen, Hilden, Germany). Multiple larvae were used to get to a weight of at least 0.02g if needed. The purified RNA was assessed for quality and quantity with a Qubit Fluorometer 3 (ThermoFisher Scientific, Waltham, MA, USA) and ThermoFisher NanoDrop Spectrophotometer 2000, respectively, and then stored at -20°C. A total of 57 samples were sent to Azenta (South Plainfield, NJ, USA) for high-throughput paired-end RNA sequencing on an Illumina HiSeq after library preparation with a NEBNext Ultra II RNA Library Prep Kit (NEB, Ipswich, MA, USA). Raw sequences were imported into CLC Genomics Workbench (Qiagen, Hilden, Germany) for quality control and trimming of low-quality reads and adapter sequences. Filtered reads were mapped against the Seriola dumerili reference genome (NCBI Genome: 12614) (Araki et al., 2018). Pairwise log-fold changes between salinities were generated when calculating differentially expressed genes with CLC Genomics Workbench’s algorithm. Once imported into RStudio, the transcript-level counts and transcripts per million abundances were converted to gene-level counts, which were variance-stabilizing transformed in DESeq2 (see GitHub page for R session information). Log-fold changes were separately imported into RStudio and used in Gene Set Enrichment Analysis. General whole transcriptomics results and interpretation of the DEGs, Gene Set Enrichment Analysis, and fatty acid biosynthesis genes can be found in our previous article (Bradshaw et al., 2023). In this article we further analyzed the trends for 12 target genes that were chosen due to past studies demonstrating that their expression was significantly affected by salinity changes or pathogenic infection (Zhou et al., 2015; Maekawa et al., 2017; Si et al., 2018; Gong et al., 2020; Lin et al., 2020; Peng et al., 2022). The genes affected by salinity changes includes: tlr2 (toll-like receptor 2), lyz (lysozyme), cat (catalase), gsto1 (glutathione S-transferase omega 1), gss (glutathione synthetase), scara3 (scavenger receptor class A member 3), il6l (interleukin-6-like [LOC111223243 in Seriola dumerili genome], il8l (interleukin-8-like [LOC111231462]), and tlr5 (toll-like receptor 5). While the genes affected by pathological infections includes: nod1 (nucleotide-binding oligomerization domain containing 1), ripk2 (receptor interacting serine/threonine kinase 2), il6l, il8l, tlr5, and tab1 (TGF-β activated kinase 1/MAP3K7 binding protein 1).
A Fisher’s exact test was performed using the statistical package SPSS 28.0 to calculate the effect of different salinity levels on the frequency of gill pathologies. An online epidemiological software WinEpi (www.winepi.net) was used to calculate the Odds Ratio (chi-squared test), i.e., the potential role of salinity as a risk factor for the presence of gill abnormalities. The Odds Ratio test was calculated between 10 ppt and 20/30 ppt samples.
Statistical significance among metadata categories for Shannon diversity and relative abundance values were determined using Kruskal-Wallis testing across all samples and Dunn pairwise testing between groups. The Benjamini-Hochberg (BH) method was used to adjust for multiple testing and all p values less than 0.05 were considered statistically significant (Benjamini and Hochberg, 1995; Elliott and Hynan, 2011). Abundance counts were variance-stabilized, transformed using DESeq2 before beta diversity analysis (McMurdie and Holmes, 2013; Love et al., 2014). Bray-Curtis was used to transform these values into a dissimilarity matrix which was then visualized using principal coordinates of analysis. PRIMER7/PERMANOVA+ was used to conduct permutational analysis of variance (PERMANOVA) tests across all samples and pairwise with 9999 permutations, the unrestricted method, the Type III Sum of Squares, and the p-values adjusted with the Monte Carlo method (Anderson et al., 2008; Clarke and Gorley, 2015a; Clarke and Gorley, 2015b). A repeated measures design was used for PERMANOVA analysis with Salinity and DPH as fixed variables and Tanks nested in Salinity as a random variable. Kruskal-Wallis and Dunn, analysis was performed by testing among salinities without separating by DPH, among DPH without separating by salinity, and among salinities at each DPH.
Gene trends were tested using Kruskal-Wallis to determine the statistical significance across all samples, Dunn to execute pairwise analysis between groups, and Benjamini Hochberg as the multiple testing p-value adjustments.
The R scripts and metadata for our previous article and this present article can be found on GitHub (https://github.com/aqua-omics/stakeholder_salinity_trial_physiology and https://github.com/aqua-omics/stakeholder_salinity_trial_immunology). The transcriptomics sequences were uploaded into the NCBI Sequence Read Archive using the Gene Expression Omnibus (GSE190968). The 16S amplicon sequences have been uploaded into the NCBI Sequence Read Archive (PRJNA776842).
Pathological alterations in the secondary gill lamellae of the fish were observed and considered as (i) progressive changes (epithelial hyperplasia and mucous cell hyperplasia); (ii) regressive changes (epithelial lifting and atrophy/necrosis); and (iii) circulatory disturbances (hyperemia/hemorrhage) (see Supplementary Figure 1 for examples of select pathologies). Three of these histological changes were remarkably more abundant in our results: hyperemia/hemorrhage (blood congestion and leaking), mucous cells hyperplasia (excessive presence of mucus) and, especially, epithelial lifting (detachment of the epithelial cells of the secondary lamellae) (Supplementary Table 1, Table 2). The number of gills with epithelial lifting was significantly different across salinities (Fisher’s exact test p-value =0.01466) with larvae reared at 10 ppt having a higher percentage (45.45% of total larvae) compared to larvae reared at 20 and 30 ppt (15.00 and 15.22% of total larvae, respectively) (Table 2). No other histological change was significantly different across salinities (Fisher’s exact test p-values > 0.05). The fish reared at 10 ppt were almost 5 times (Odds Ratio = 4.6429, p-value = 0.007) more likely to suffer from morphological alterations such as gill epithelial lifting than fish reared at medium to high salinities (20 or 30 ppt).
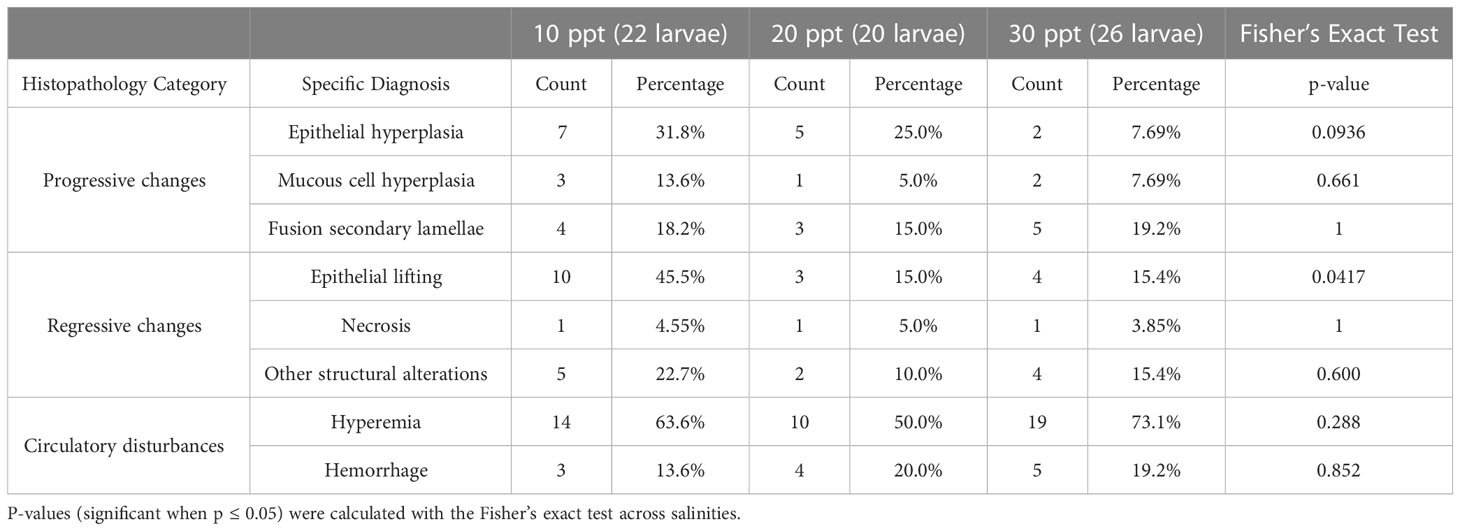
Table 2 Counts and percentages of histopathological diagnoses associated with progressive and regressive changes, and circulatory disturbances, observed on Florida pompano (Trachinotus carolinus) larvae secondary gill lamellae for each of the tested salinities (10, 20 and 30 ppt).
A total of 195 tank water and whole-body larval fish samples were taken over the course of the experiment (Table 1) for 16S amplicon sequencing. One water sample was removed due to contamination (LSE1DP2a). Across these 194 remaining samples, there were 18,794,113 sequences representing 15,642 ASVs after removal of ASVs that did not occur more than 10 times across all samples. After being separated by sample type, there were 8,369 ASVs representing 583 genera across all 125 water samples and 10,386 ASVs representing 946 genera across all 69 larvae samples (Supplementary Table 2). Out of these total ASVs, 3,113 appeared in both datasets with 481 common genera. This means that 30% of larvae ASVs and 37% of water ASVs, in addition to 51% of larvae genera and 83% of water genera, were found in both datasets.
The Shannon diversity was statistically higher in the larvae samples compared to the water samples according to Kruskal-Wallis testing (chi2 = 60.839, BH adjusted p-value = 9.72E-14) (Supplementary Table 3). Visualization of sample differences using a principal coordinates of analysis showed that there was a distinctive separation of samples between the water and larvae samples as well as a separation of the 1 DPH water samples away from the other samples (Supplementary Figure 2). PERMANOVA analysis on the Bray-Curtis dissimilarity matrix with the egg and 1 dph samples removed since they were taken before salinity changes revealed that there was a significant difference between sample types (Pseudo-F = 11.44, permutational P-value = 0.0001, unique permutations = 9552, P(MC) [Monte Carlo] = 0.0001) (Figure 1). The relative percentage of the Vibrio genera was 24 times higher in larvae than in water samples and it should be noted that no other genera occurred greater than two times more in larvae versus water samples. Seven genera occurred two times or more higher in water samples than in larvae samples, with the top three including Idiomarina (8.8x), NS3a marine group (5.6x), and Winogradskyella (5.8x). The most common genera in larvae samples included Vibrio (35%), NS3a marine group from the Flavobacteriaceae family (3.4%), and Pseudomonas (2.8%). For water samples, the top three genera were the NS3a marine group (20%), the NS9 marine group from the Flavobacteriales order (9.4%), and Pseudomonas (7.2%) (Figure 1).
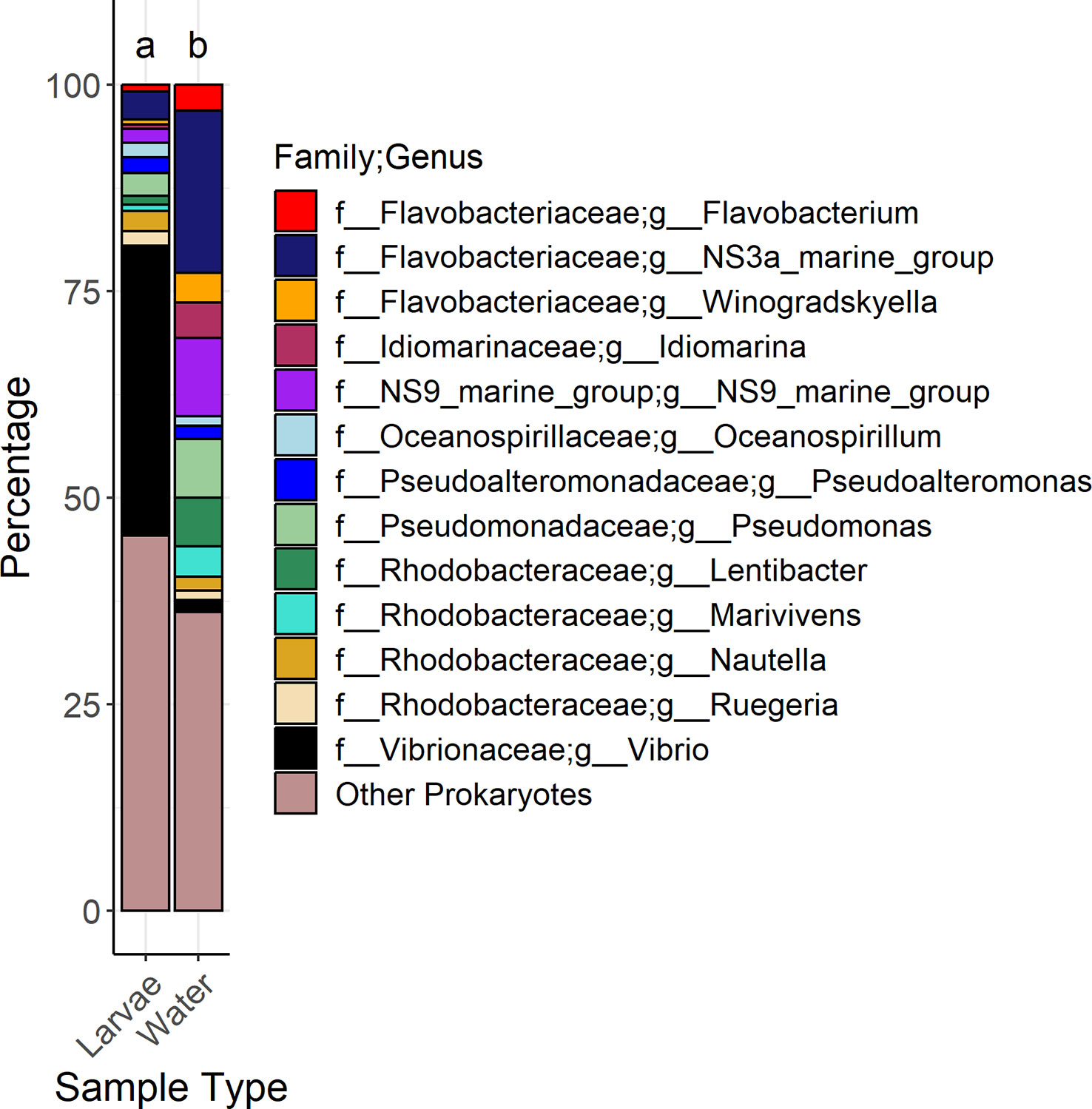
Figure 1 Stacked bar graphs showing the genera that had a mean relative abundance greater than 1% across all samples associated with water and larvae samples. Genera and associated families are represented by color. Letters above the bar plots indicate results of permutational analysis of variance testing. Different letters between bar plots means that the sample types were statistically different (Pseudo-F value < 0.05).
In water samples, there were 225 genera shared among all salinities: 64% (10 ppt), 62% (20 ppt), and 68% (30 ppt) of the genera (Supplementary Figure 3). Samples from 10, 20, and 30 ppt had 20%, 12%, and 14% of unique genera, respectively. Kruskal-Wallis tests on Shannon diversity revealed that there were no significant differences among salinities across all DPH (BH p-value = 0.13) and among salinities at 6 DPH (0.090) (Supplementary Table 3). However, there were significant differences among salinities at 12 (0.001), 18 (0.00019), and 24 (0.00097) DPH. Pairwise Dunn testing of pooled samples showed that 10 ppt samples had a significantly lower Shannon diversity than samples at 20 and 30 ppt. Although samples at 10 and 30 ppt were statistically different from one another during each set of DPH samples, there was no consistent pattern as to which one had a larger Shannon diversity. There was no other notable pattern in the Dunn test results since the statistical pattern among salinities was different during each set of DPH samples. In terms of samples separated by DPH, the alpha diversity at 18 and 24 DPH were similar to one another yet statistically different from 6 or 12 DPH (Figure 2). The latter was statistically lower than the former (Supplementary Table 3).
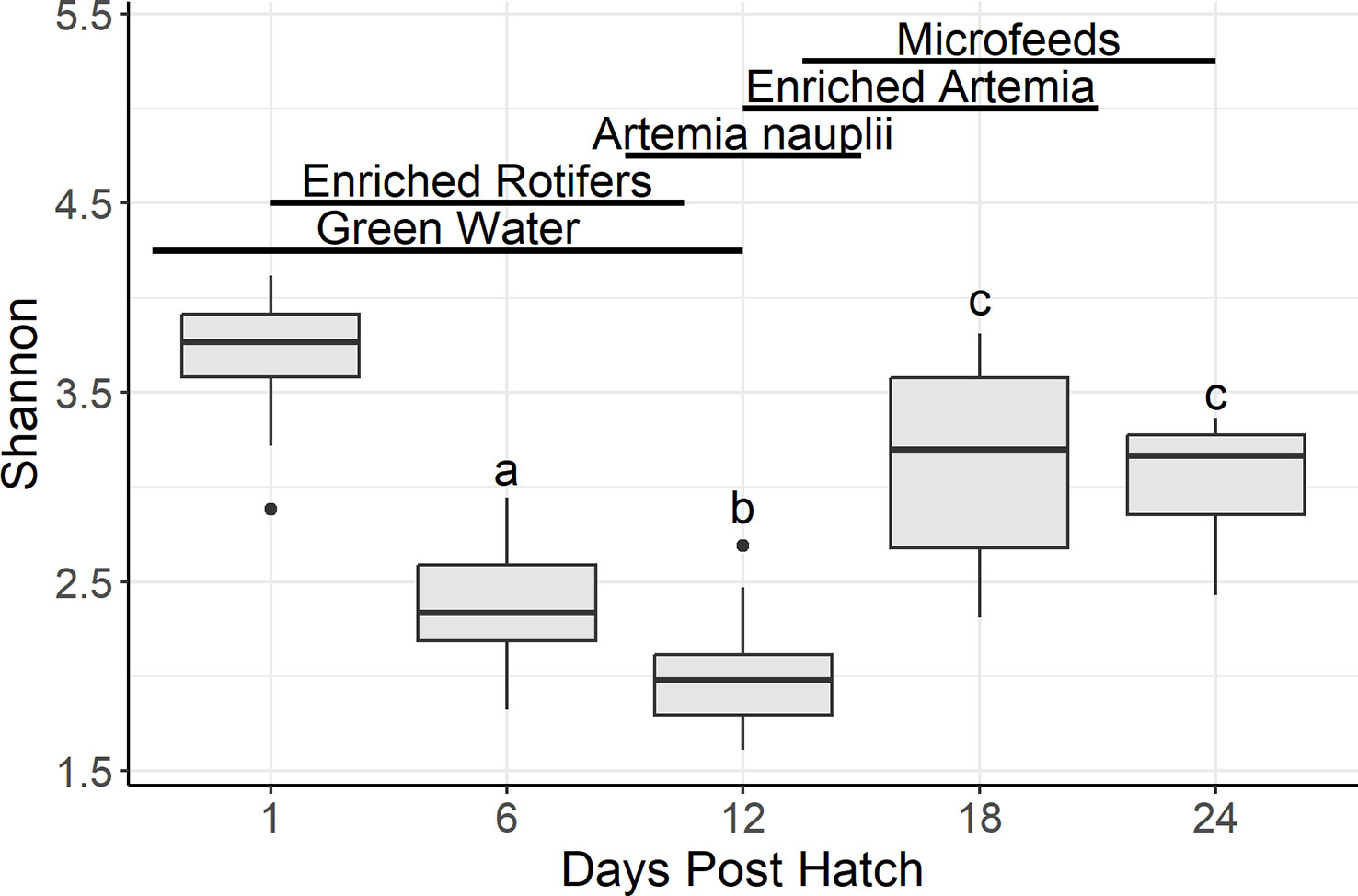
Figure 2 Boxplots summarizing water Shannon diversity by days post hatch. Bars denote largest and smallest values within 1.5 times the interquartile range with dots representing values outside that range, middle line is the medium, and ends of boxes are the first and third quartiles. Letters above the boxplots indicate results of pairwise Dunn testing with Benjamini-Hochberg adjusted p-values. If there is a shared letter between boxplots, that means that they were statistically similar (Benjamini-Hochberg adj. p-value > 0.05). Lines and labels above the boxplots indicate the presumptive feeding schedule of the larvae through 24 days post hatch. Due to the actual feeding regimen being proprietary, this feeding regimen is based off the Florida Atlantic University at Harbor Branch Oceanographic Aquaculture Department’s hatchery routine for Florida pompano larvae.
The principal coordinates of analysis shows a distinct visual separation of samples based on DPH as well as salinity (Figure 3). As confirmed by repeated measures PERMANOVA analysis, the samples from the four different DPH (6, 12, 18, and 24) and the three salinities (with just the 6, 12, 18, and 24 DPH samples) were all statistically different from one another across all samples (DPH Pseudo-F =4.8988, P(MC) = 0.0001; salinity Pseudo-F = 3.5806, P(MC) = 0.0001) and pairwise (all PP(MC) < 0.05) (Supplementary Tables 4, 5). When just comparing samples taken on the same DPH, salinities were significantly different (P(MC) < 0.05) from each pairwise between all salinities at 6 DPH and 12 DPH while at 18 DPH only 20 ppt was significantly different from 30 ppt (P(MC) = 0.033) and at 24 DPH only 10 ppt was significantly different from 30 ppt (P(MC) = 0.0483) (Supplementary Table 5). Samples were statistically different across tanks across all samples (Pseudo-F = 2.4177, P(MC) = 0.0001), but not all pairwise comparisons were significantly different. All the tanks from treatments at 10 ppt (P3, P5, P6), 20 ppt (P1, P2, P7), and 30 ppt (P4, P8, P9) were significantly similar to one another and different from the other six tanks.
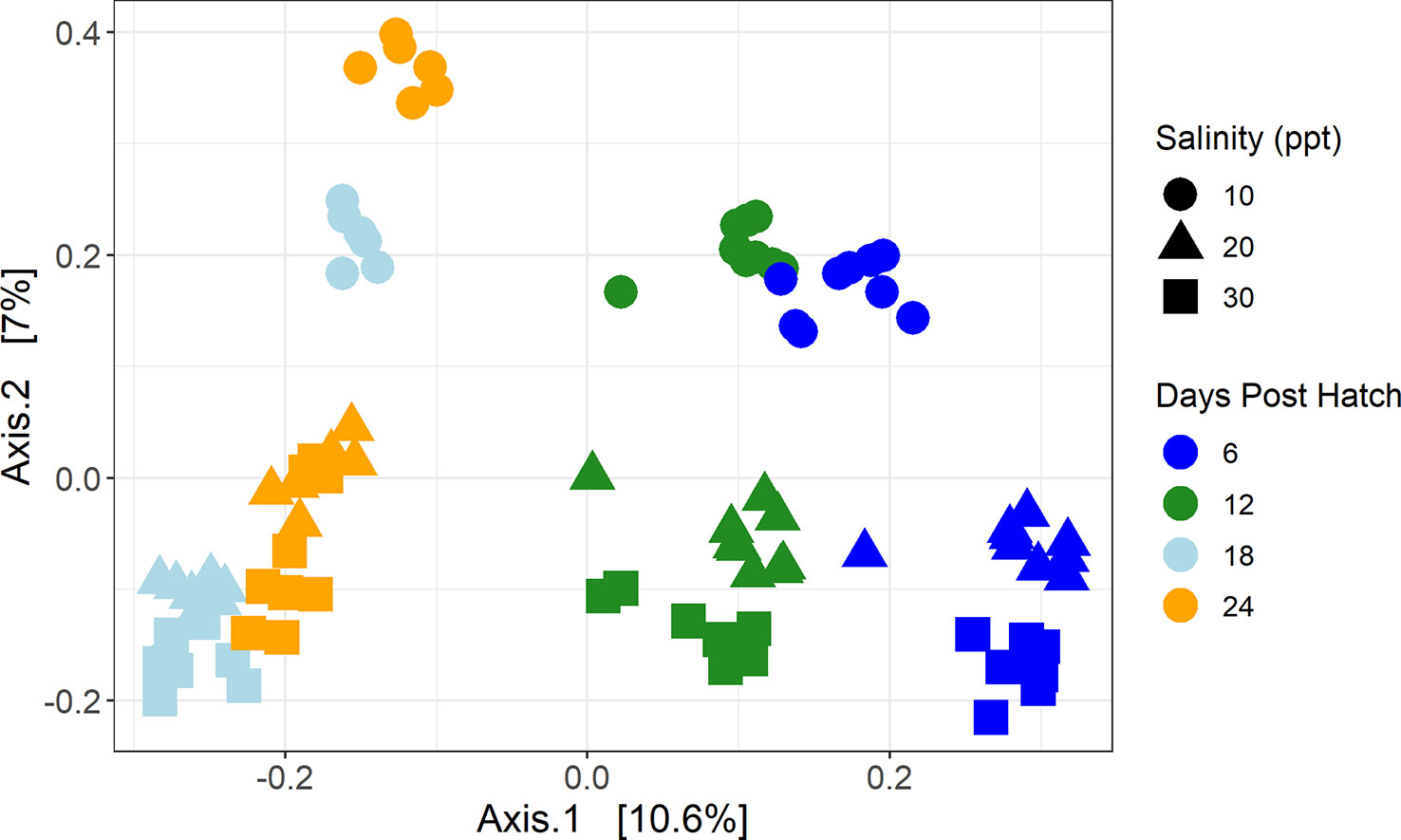
Figure 3 Principal coordinates of analysis of water samples using a Bray-Curtis dissimilarity matrix. Color represents days post hatch with dark blue representing samples taken 6 days post hatch, green 12 days post hatch, light blue 18 days post hatch, and orange 24 days post hatch. Shape represents salinity with circles representing samples that were at 10 ppt, triangles 20 ppt, and squares 30 ppt.
The NS3a marine group was among the most common genera in all three salinities (10 ppt= 14%, 20 ppt= 24%, 30 ppt= 20%) (Supplementary Figure 4). The top three were rounded out by NS9 marine group (14%) and Lentibacter (5.4%) for samples at 30 ppt, by Lentibacter (11%) and NS9 marine group (8.8%) for samples at 20 ppt, and Pseudomonas (19%) and Idiomarina (14%) for samples at 30 ppt. Four genera (>1%) were greater than two times higher in 10 ppt samples than 20 and 30 ppt samples. These included Flavobacterium (10 ppt samples were 702x 20 ppt samples; 10 ppt samples were 557x 30 ppt samples), Idiomarina (275x, 285x), Pseudomonas (5.1x, 52x), and Marivivens (3.4x, 2.1x). Both 20 and 30 ppt samples had greater percentages of Marivita (20 ppt 79x 10 ppt; 30 ppt 8.7x 10 ppt) and Lentibacter (31x, 15x) than 10 ppt samples, although 20 ppt samples had 2.1x more Lentibacter than 30 ppt samples. The 20 ppt samples had a greater percentage of Winogradskyella (20 ppt 13x 10 ppt, 20 ppt 2.2x 30 ppt), and SAR92 clade (3.1x, 2.2x) than 10 and 30 ppt samples. The 30 ppt samples had more Oceanospirillum than 10 ppt (5.2x) and 20 ppt (2.7x) samples.
The percentage of PPG in water across salinities was statistically higher in samples at 10 ppt than 20 and 30 ppt per Dunn testing (Figure 4A, Supplementary Table 3) as well as samples at 6 DPH compared to all other DPH (Figure 4C). There were spikes of Pseudomonas spp. in 6 DPH samples in 10 and 20 ppt; there were also spikes of Flavobacterium spp. at 12 and 24 DPH in 20 ppt samples (Figures 4B, D, Supplementary Figure 5).
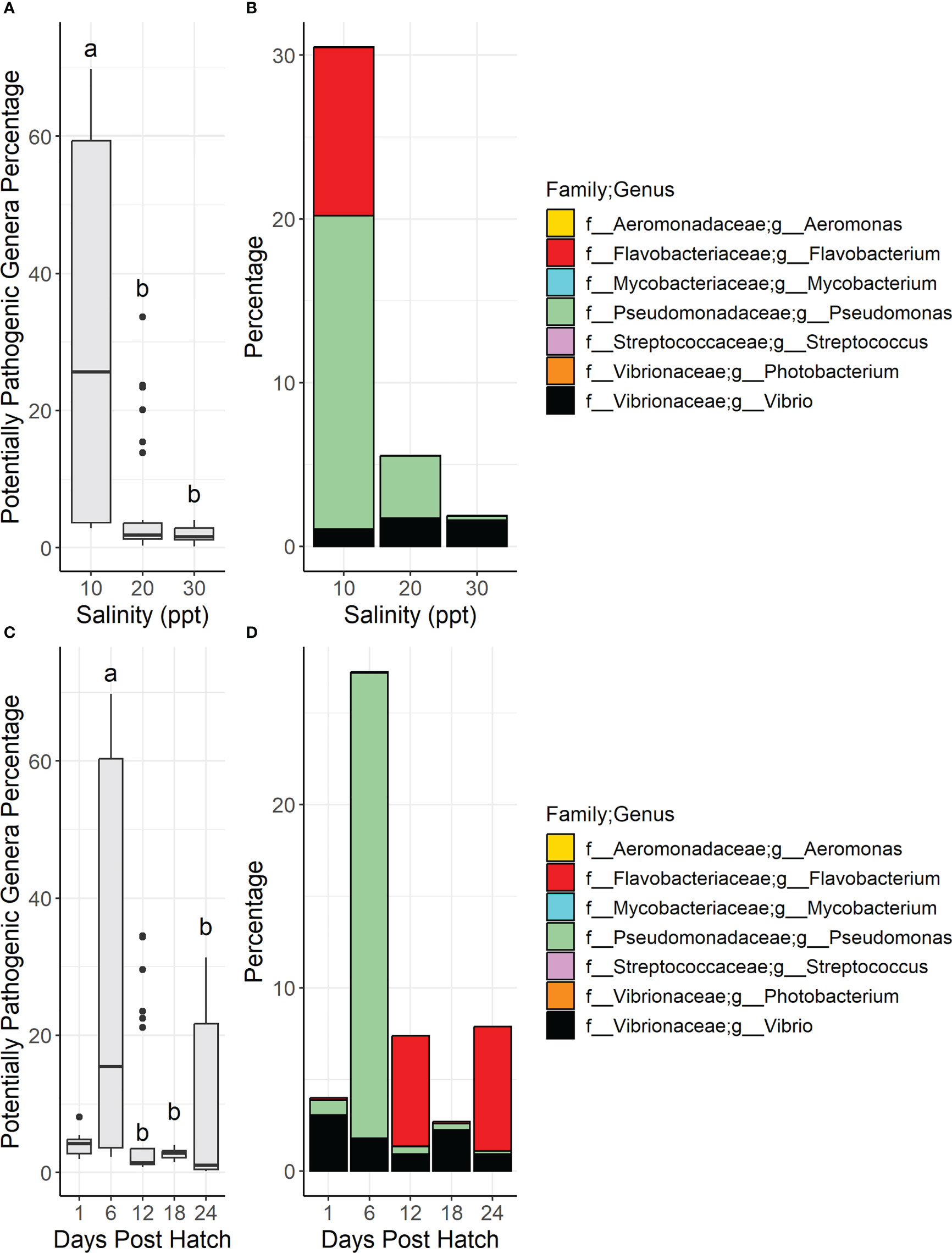
Figure 4 Boxplots show the total relative diversity of the nine potentially pathogenic genera in water samples summarized by salinity (A) and days post hatch (C). In the boxplot figures (A, C) bars denote largest and smallest values within 1.5 times the interquartile range with dots representing values outside that range, middle line is the medium, and ends of boxes are the first and third quartiles. Letters above the boxplots indicate results of pairwise Dunn testing with Benjamini-Hochberg adjusted p-values. If there is a shared letter between boxplots, it means that they were statistically similar (Benjamini-Hochberg adj. p-value > 0.05). Stacked bar graphs of the total relative diversity of the potentially pathogenic genera in larvae samples summarized by salinity (B) and days post hatch (D). Genera and their associated families are represented by color.
Across all larvae samples, there were 413 shared genera among all salinities representing 62% (10 ppt), 68% (20 ppt), and 61% (30 ppt) of genera (Supplementary Figure 6). At 10 ppt, 21% of genera were unique, 14% for 20 ppt samples, and 19% for 30 ppt samples. Kruskal-Wallis and Dunn tests indicated that Shannon diversity had no statistical differences among salinities across all samples and pairwise in all larvae samples as well as when focused on samples in each individual DPH dataset (Figure 5, Supplementary Table 3). The samples from 3, 6 and 9 DPH were all statistically similar to one another and the Shannon diversity was statistically higher than the samples from 15, 18, and 20 DPH. Both 12 and 24 DPH were statistically similar to each other as well as at least one sample in each of the other two groups.
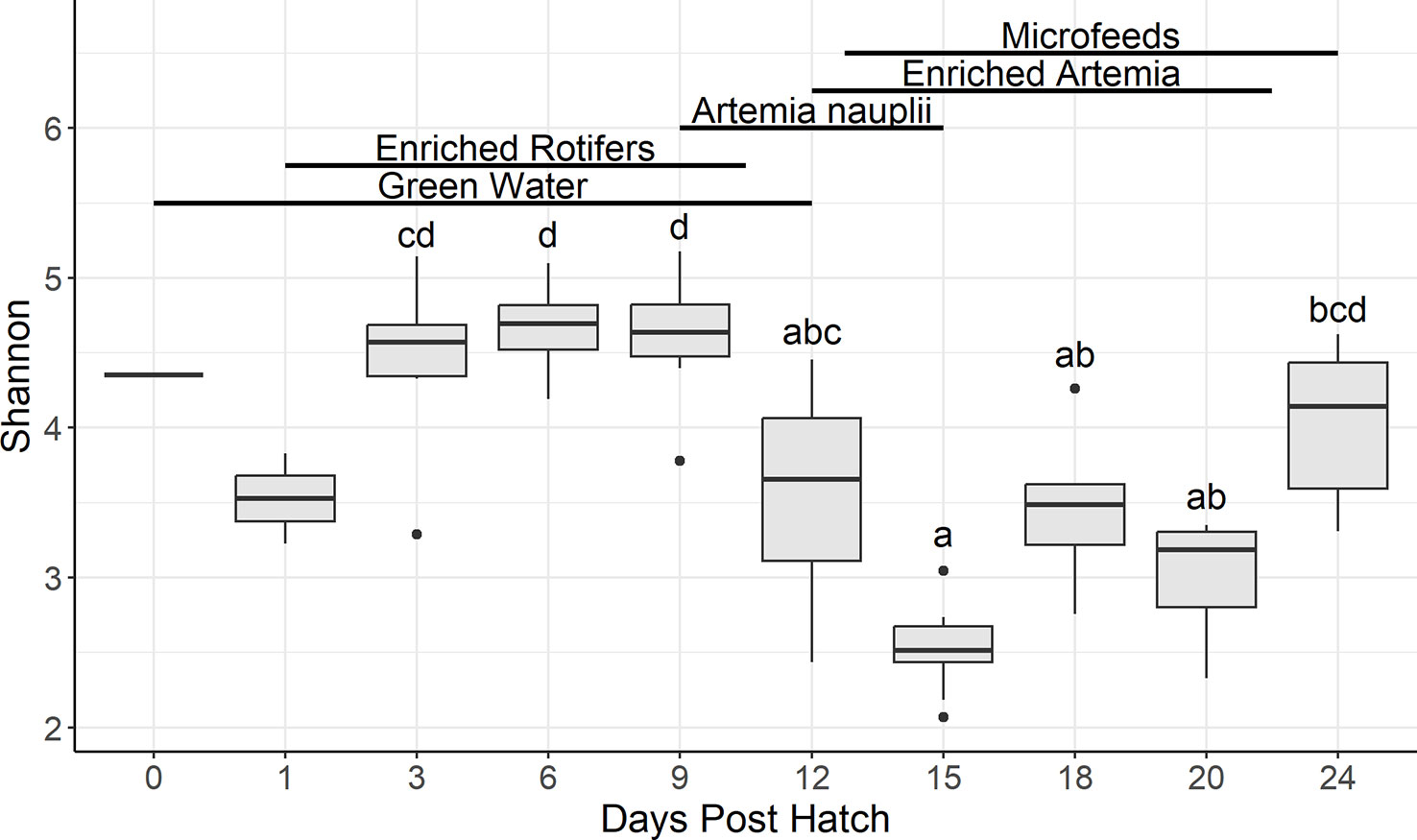
Figure 5 Boxplots summarizing Shannon diversity in larvae samples by days post hatch. Bars denote largest and smallest values within 1.5 times the interquartile range with dots representing values outside that range, middle line is the medium, and ends of boxes are the first and third quartiles. Letters above the boxplots indicate results of pairwise Dunn testing with Benjamini-Hochberg adjusted p-values. If there is a shared letter between boxplots, it means that they were statistically similar (Benjamini-Hochberg adj. p-value > 0.05). Lines and labels above the boxplots indicate the presumptive feeding schedule of the larvae through 24 days post hatch. Due to the actual feeding regimen being proprietary, this feeding regimen is based off the Florida Atlantic University at Harbor Branch Oceanographic Aquaculture Department’s hatchery routine for Florida pompano larvae.
The principal coordinates of analysis shows a clustering of 15, 18, and 20 DPH samples away from the other samples (Figure 6). The larvae samples from all DPH (3, 6, 9, 12, 15, 18, 20, and 24) and the three salinities (10, 20, and 30 ppt) were all statistically different from one another proven by PERMANOVA tests across all samples (DPH Pseudo-F =2.1457, P(MC) = 0.0001; salinity Pseudo-F = 1.6249, P(MC) = 0.0026) (Supplementary Table 4). Pairwise analysis was not as straightforward as with the water samples since the 20 and 30 ppt samples were statistically similar (t=1.1172, P(MC) = 0.2053), while 10 ppt samples were significantly different from 20 (1.2763, 0.0484) and 30 (1.408, 0.0066) ppt samples. In terms of pairwise DPH tests, samples from D15 and D18 were significantly different from D3 and D9 while both sets of DPH were significantly similar to one another. D12, D20, D24, and D6 were significantly similar to each other and the other four DPH (Supplementary Table 5). When comparing samples taken on the same DPH, salinities were significantly similar to each other across all samples and pairwise (P(MC) < 0.05) (Supplementary Table 5).
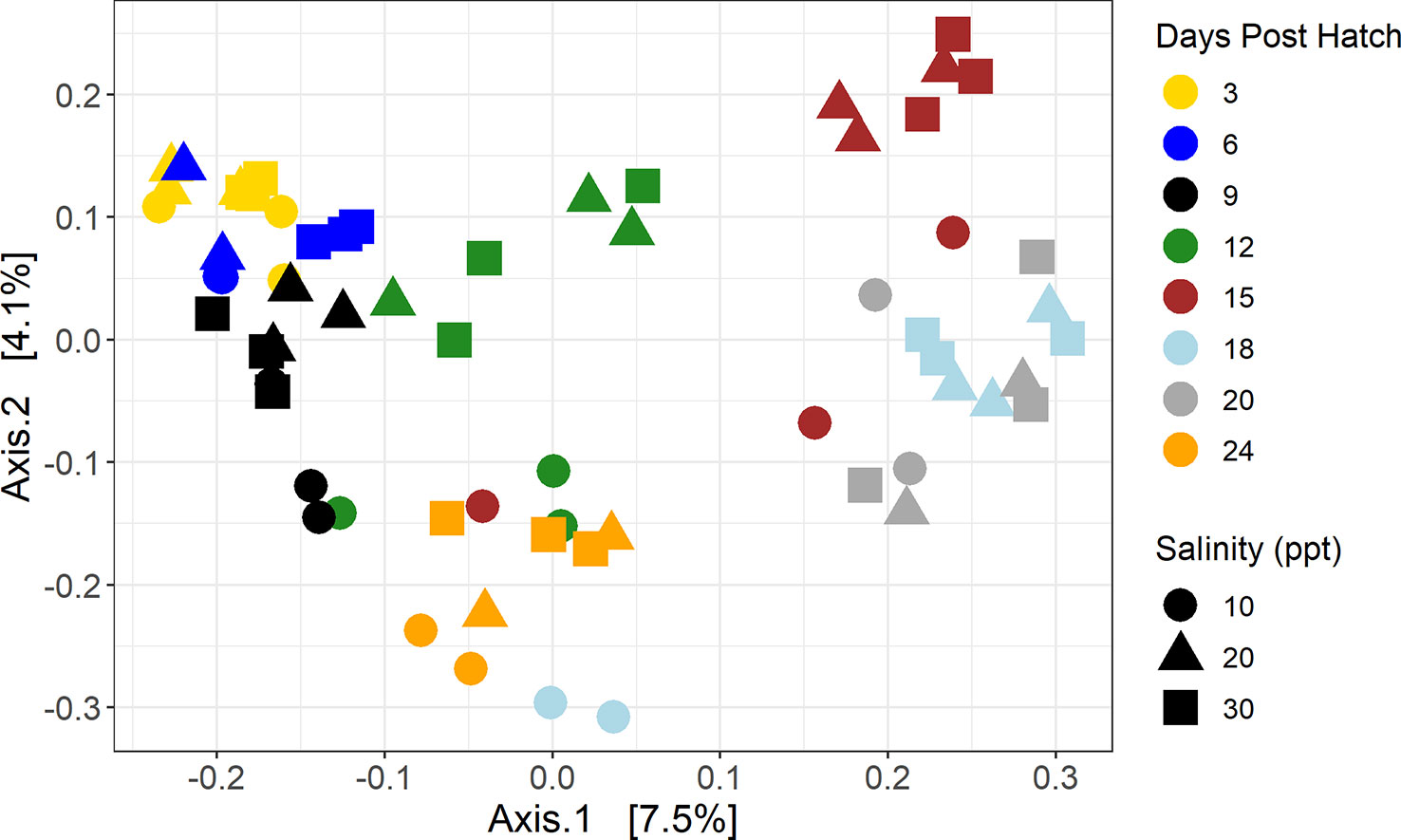
Figure 6 Principal coordinates of analysis of larvae samples using a Bray-Curtis dissimilarity matrix. Color represents days post hatch with yellow representing samples taken 3 days post hatch, dark blue 6 days post hatch, black 9 days post hatch, green 12 days post hatch, brown 15 days post hatch, light blue 18 days post hatch, gray 20 days post hatch, and orange 24 days post hatch. Shape represents salinity with circles representing samples that were at 10 ppt, triangles 20 ppt, and squares 30 ppt.
Vibrio was the genus with the highest relative percentage in all three salinities: 29% in 10 ppt, 41% in 20 ppt, and 35% in 30 ppt samples (Supplementary Figure 7). The top three genera were rounded out by Ruegeria (4.5%) and Pseudoalteromonas (4.4%) in 10 ppt samples, Nautella (4.8%) and Pseudomonas (3.3%) in 20 ppt samples, and NS9 marine group (3.7%) and NS3 marine group (3.1%) in 30 ppt samples. The 10 ppt samples had more Ruegeria (10 ppt 6.9x 20 ppt; 10 ppt 32x 30 ppt), Pseudoalteromonas (6.9x, 5.3x), and Flavobacterium (29x, 93x) than 20 and 30 ppt samples, but less Lentibacter (20 ppt 28x 10 ppt; 30 ppt 44x 10 ppt) and Nautella (9.1x, 4.0x). Pseudomonas was found less in 30 ppt samples than 10 (3.7x) and 20 ppt (3.0x) samples, while Oceanospirillum was more abundant in 30 ppt samples than 10 (3.2x) and 20 ppt (3.5x) samples.
PPG percentages were statistically similar both across all samples (chi2 = 0.70, BH adj. p value = 0.70) and pairwise (BH adj. p value > 0.05) across salinities (Figure 7A, Supplementary Table 3). Of the nine PPG, Vibrio, Pseudomonas, and Flavobacterium were the most common genera across salinities (Figure 7B). There were statistically significant differences across all samples (chi2 = 47, BH adj. p value = 1.0e-7) and pairwise differences between the different DPH samples with the PPG percentages being statistically lower in the 3, 6, and 9 DPH samples than the 15, 18, and 20 DPH samples (Figure 7C, Supplementary Table 3). Across all salinities, Pseudomonas was generally higher in the early DPH (from 3 to 12 DPH) while Vibrio was higher in the later DPH (12 to 20 DPH) (Figure 7D, Supplementary Figure 8). There were spikes in Flavobacterium in the 12 and 24 DPH samples, specifically in the 10 ppt samples. The only bacterial genus with members tested and proven as probiotics in Trachinotus sp. was Bacillus (Zhang et al., 2014; Hauville et al., 2016; Liu et al., 2020). Bacillus was not found in any water samples and was only found in 29 out of 68 larval samples (43% of samples) with a max relative percentage of 0.74% and an average across all samples of 0.060%. Kruskal Wallis testing revealed that there were no significant differences in the relative percentages of Bacillus among salinities (chi-square = 0.48, p-value = 0.79) or DPH (chi-square = 7.2, p-value = 0.41).
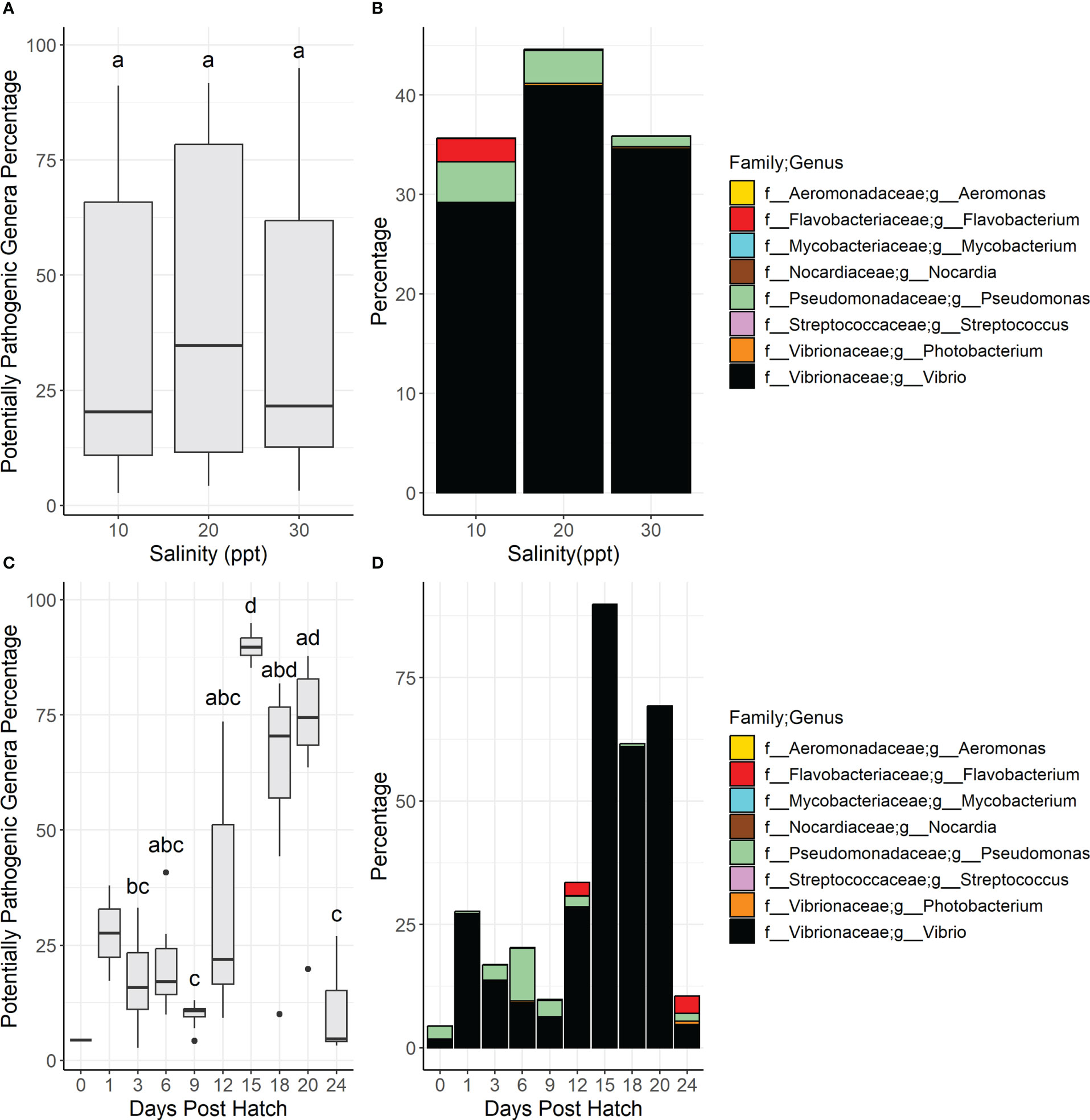
Figure 7 Boxplots show the total relative diversity of the nine potentially pathogenic genera in larvae samples summarized by salinity (A) and days post hatch (C). In the boxplot figures (A, C) bars denote largest and smallest values within 1.5 times the interquartile range with dots representing values outside that range, middle line is the medium, and ends of boxes are the first and third quartiles. Letters above the boxplots indicate results of pairwise Dunn testing with Benjamini-Hochberg adjusted p-values. If there is a shared letter between boxplots, it means that they were statistically similar (Benjamini-Hochberg adj. p-value > 0.05). Stacked bar graphs of the total relative diversity of the potentially pathogenic genera in larvae samples summarized by salinity (B) and days post hatch (D). Genera and their associated families are represented by color.
The only immune-related gene tested that was significantly different across all samples and pairwise across salinities was nod1, which had a significantly lower expression in 10 versus 30 ppt samples (Supplementary Table 6, Supplementary Figure 9). All immune related genes tested were significantly different across DPH besides il6l and tlr2 (Supplementary Table 6, Figures 8C, 9A). Eight of the twelve genes (nod1, il8l, tlr5, lyz, cat, gsto1, gss, and scara3) had expression levels that generally increased from 12 DPH to a peak at 18 or 20 or 24 DPH (Figures 8A, D, E, 9B–F). Although only nod1, cat, gsto1, and gss were statistically different between 12 DPH and the peak value. Two genes (ripk2 and tab1) had the opposite trend with expression levels that generally decreased over time (Figures 8B, F).
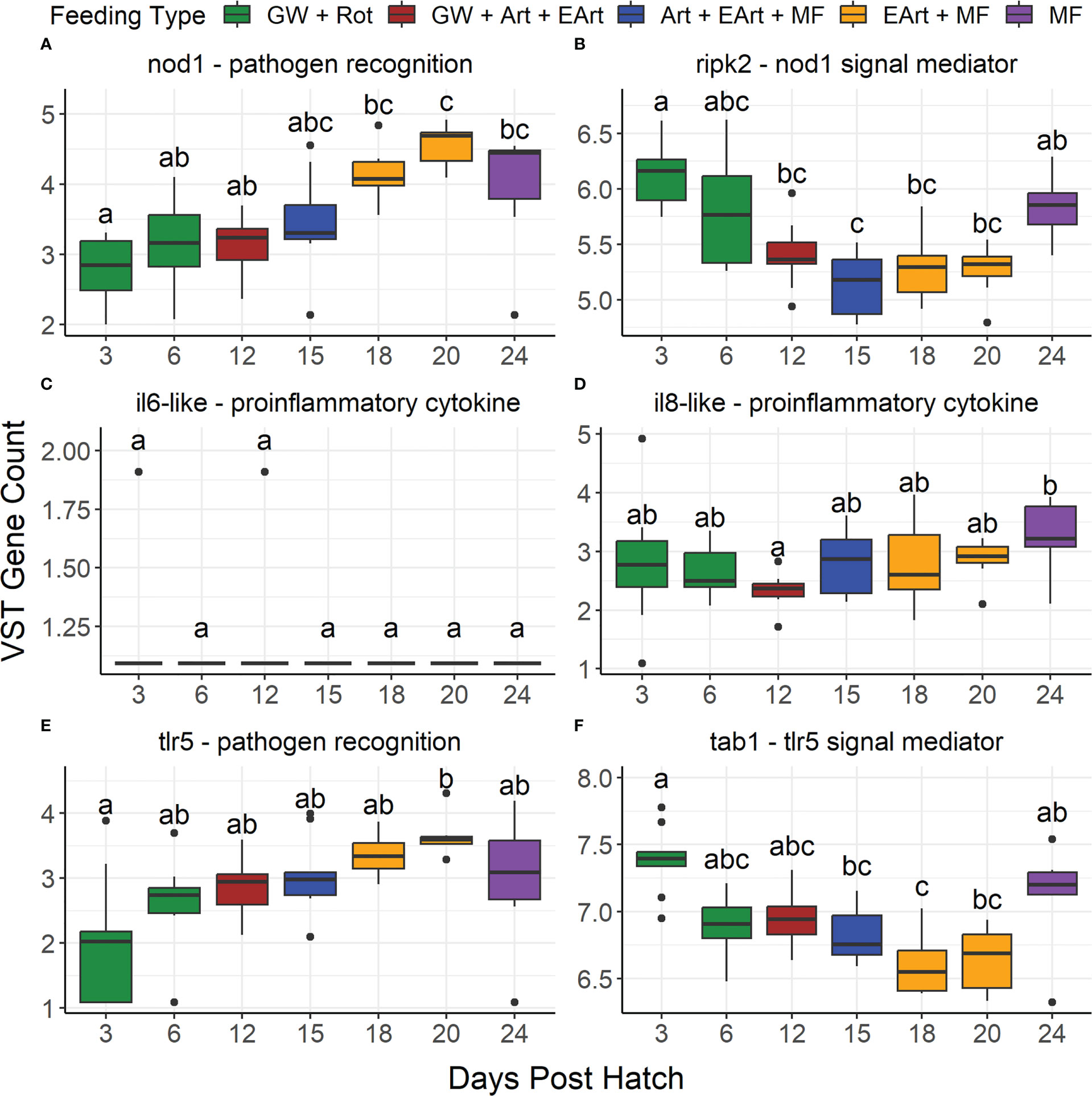
Figure 8 Boxplots show the variance stabilizing transformed gene counts of six immune-related genes found in the literature to be differentially expressed in infected fish grouped by days post hatch. These genes include: (A) nucleotide binding oligomerization domain containing 1 (nod1), (B) receptor interacting serine/threonine kinase 2 (ripk2), (C) interleukin 6-like (il6-like), (D) interleukin 8-like (il8-like), (E) toll-like receptor 5 (tlr5), and (F) TGF-beta activated kinase 1/MAP3K7 binding protein 1 (tab1). Color represents what the larvae were likely being fed on those days: green water/algae paste (GW), rotifers (Rot), Artemia (Art), enriched Artemia (EArt), or microfeeds (MF). Due to the actual feeding regimen being proprietary, this feeding regimen is based off the Florida Atlantic University at Harbor Branch Oceanographic Aquaculture Department’s hatchery routine for Florida pompano larvae. Bars denote largest and smallest values within 1.5 times the interquartile range with dots representing values outside that range, middle line is the medium, and ends of boxes are the first and third quartiles. Letters above the boxplots indicate results of pairwise Dunn testing with Benjamini-Hochberg (BH) adjusted p-values. If there is a shared letter between boxplots, it means that they were statistically similar (BH adj. p-value > 0.05).
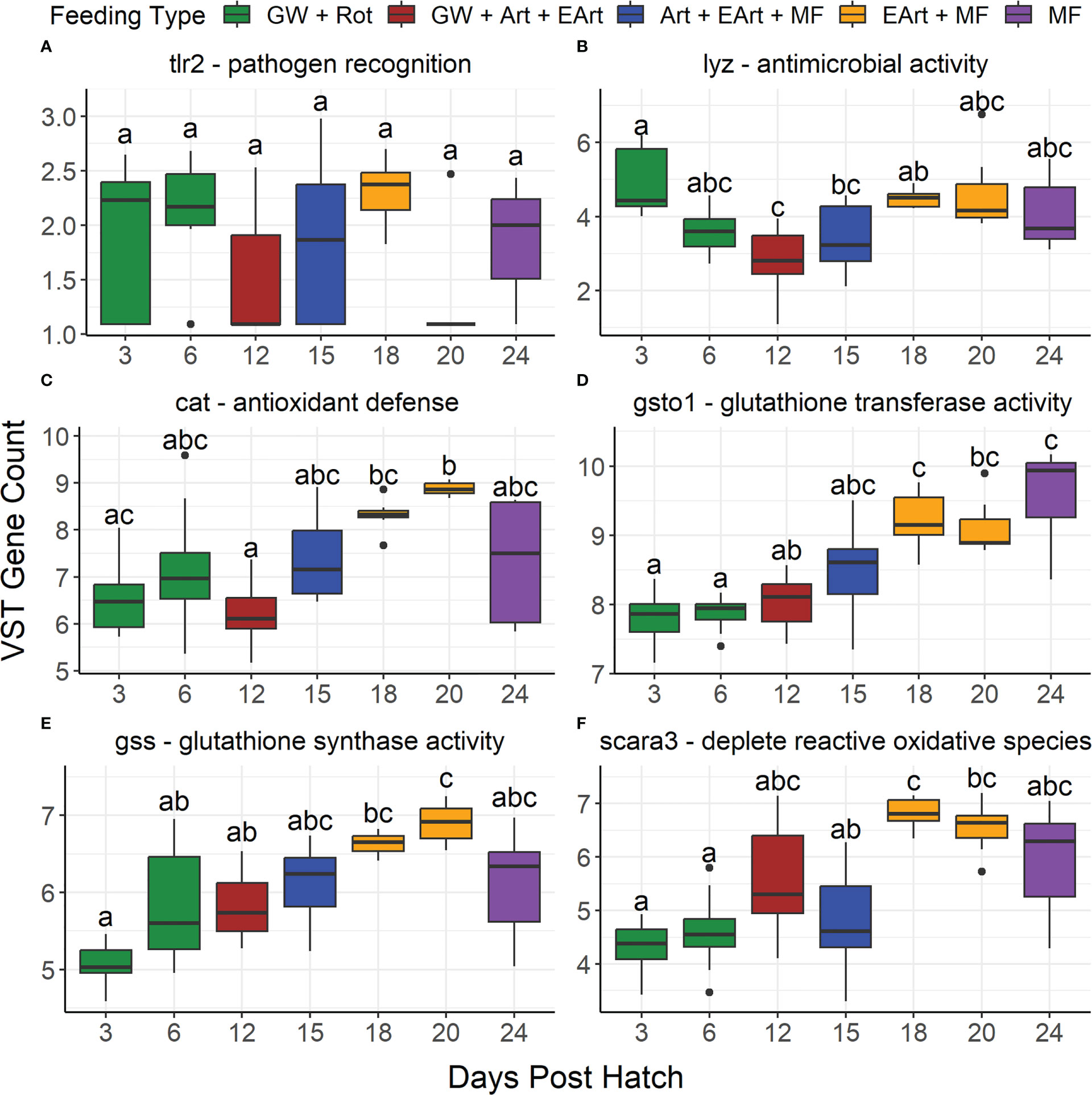
Figure 9 Boxplots show the variance stabilizing transformed gene counts of six immune-related genes found in the literature to be differentially expressed based upon salinity differences grouped by days post hatch. These genes include: (A) toll-like receptor 2 (tlr2), (B) lysozyme (lyz), (C) catalase (cat), (D) glutathione S-transferase omega 1 (gsto1), (E) glutathione synthetase (gss), and (F) scavenger receptor class A member 3 (scara3). Color represents what the larvae were likely being fed on those days: green water/algae paste (GW), rotifers (Rot), Artemia (Art), enriched Artemia (EArt), or microfeeds (MF). Due to the actual feeding regimen being proprietary, this feeding regimen is based off the Florida Atlantic University at Harbor Branch Oceanographic Aquaculture Department’s hatchery routine for Florida pompano larvae. Bars denote largest and smallest values within 1.5 times the interquartile range with dots representing values outside that range, middle line isthe medium, and ends of boxes are the first and third quartiles. Letters above the boxplots indicate results of pairwise Dunn testing with Benjamini-Hochberg (BH) adjusted p-values. If there is a shared letter between boxplots, it means that they were statistically similar (BH adj. p-value > 0.05).
This study offered considerable evidence that Florida pompano larval rearing can be conducted at near iso-osmotic salinity of 10ppt without significant impacts on histology, microbiome, or immunology. While some changes were noted, none significantly impacted fish production through larval rearing. This paper also offered one of the first descriptions of changes in microbiome and immunological gene expression over the ontogeny of Florida pompano. The significance of these findings as well as impact on broader since will be discussed.
This study was run in collaboration with aquaculture producers at their farm, thus we worked within their standard operating procedures and honored their proprietary information. We acknowledge our interpretations based upon our presumptive feeding schedule instead of their proprietary feeding schedule may not be accurate, although they have confirmed that it generally agrees with theirs. Also, in relation to feeding, sampling requirements for this study required that sampling took anywhere from 1-3 hours to complete larvae sampling which did overlap with feeding times. We did not want to change feeding times to accommodate sampling due to wanting to keep the fish on an appropriate and consistent feeding schedule. We acknowledge that sampling during different stages of feeding could have influenced the larvae microbiome results, but by focusing analysis on pooling by salinity or days post hatch, that influence will be lessened by replication.
Larviculture is associated with differences in survival and hatching densities among tanks, thus we ran our experiment in triplicate to address this. Although due to loss of tanks throughout the study, we acknowledge that our study has less statistical power than originally designed. A large amount of sampling in addition to inferior egg quality could have contributed to the loss of tanks and overall survival. The 60% hatch rate (average for pompano should instead exceed 80%), also meant tanks were stocked with less fish than intended and may also indicate potential reduced larval survival to first feeding (~4dph), further reducing fish densities. Survival results were also not recorded due to a large amount of sampling, causing any measure of survival to be instead a measure of pseudo-survival. Despite this, we did not observe any major mortalities when fish were transferred from the hatching cone to the test salinities. Future studies should incorporate additional tanks that are not sampled to be used as an assessment of true survival.
Our results showed that rearing Florida pompano under 10 ppt salinity conditions may impact the structural integrity of gills during early development. This has been frequently described in previous studies as associated with respiratory and osmoregulatory distress and a higher susceptibility to pathogen infections, resulting in health impairment and even mortality (Hugh, 2006; Harper and Wolf, 2009). In our study, alterations in gill membrane permeability presented as swelling of lamellar epithelial cells or oedema of the subepithelial space, particularly in the 10 ppt samples. This gill alteration has been traditionally defined as ‘epithelial lifting’ (Roberts, 2012). Our findings agree with other studies that observed epithelial lifting in pompano larvae, suggesting a generalized stress response to salinity (Hugh, 2006; Harper and Wolf, 2009; Takishita et al., 2015). However, salinity is likely not the sole contributing factor of these stress-induced gill alterations as we also identified the presence of potentially pathogenic genera, that could also cause epithelial lifting (Yandi et al., 2017; Singh et al., 2021). However, epithelial lifting is a common artifact for samples fixed and stored in formalin for extended periods of time (Wolf et al., 2015). Due to picking up samples once a week from the farm, some samples were in formalin for up to a week. This could have led to artifacts, although that would have affected all salinities equally. While we had sufficient replication to make conclusions based upon histological changes at the overall salinity level, we did not have enough replication to draw significant conclusions nor statistically test differences between salinities at each days post hatch level. Thus we could not assess if histopathological changes worsened over time at a particular salinity versus another. Future studies will put more emphasis on gathering enough histological samples to provide enough replication to test salinity at a dph level and to run the study for longer than 24 dph to assess long term cultivation.
This is the first study to explore the recirculating aquaculture system water and whole-body microbiomes of Florida pompano larvae. It should be noted that the use of whole larvae restricted our interpretation of the results due to the mixture of internal (gut) and external (skin and gills) microbial communities. Our comprehensive analysis revealed that the water microbiome was significantly dissimilar from the whole-larval microbiome with lower microbial diversity in water than larvae. A previous study on cultured gibel carp (Carassius auratus gibelio) and bluntnose black bream (Megalobrama amblycephala) established that bacteria in the rearing medium exhibited greater diversity than those on gibel carp skin, but the opposite pattern was observed on bream skin (Wang et al., 2010). While disparities in taxonomic composition and diversity exist between environment and host, water microbiomes are involved in determining the fish microbiomes. This concept has been substantiated through comparisons between water and larvae communities associated with wild-caught and farm-raised fish, as well as between those of freshwater and marine fish (Webster et al., 2018; Sehnal et al., 2021).
There were several sampling events throughout our study in which patterns between water and larvae were comparable with one another. A significant shift was seen in the microbial community among salinity treatments, as the water and larval samples derived from 10 ppt were clearly distinct from 30 ppt samples in terms of beta diversity. This indicates that the microbial communities of Florida pompano larvae and those found in their rearing medium interact with each other and respond accordingly to different salinities. These microbial composition patterns are informative in aquaculture because consistent microbial responses to salinity have been regarded as phylogenetically conserved (Sehnal et al., 2021). Water and larval samples were significantly different across all days post hatch (DPH), which suggests that developmental life stages in tandem with an evolving diet influenced the microbiome as described in other studies (Nikouli et al., 2019; Zhao et al., 2020). Further, microbial alpha diversity dropped in the later DPH for larvae, while it rose in the corresponding water samples.
Our study showed that both the water and larvae microbiomes changed over time, presumably in response to the feeding schedule, providing information that can be used to design future experiments focused on diet and nutrition requirements. Microbiome shifts can modify feed efficiency, metabolism, immunity, disease susceptibility, and gut function in teleost fish (Sehnal et al., 2021). We had anticipated significant microbiome changes among salinities and DPH, since the captive-state, life-stage, salinity, and diet of the fish all have a strong impact on its microbial community diversity and structure (Egerton et al., 2018; Legrand et al., 2020). Similar to our study, Zheng et al. (2019) demonstrated that the age of fish had a more noticeable impact on the gut microbiome of farmed Chinook salmon (Oncorhynchus tshawytscha) than the salinity, although this pattern was captured over a one-year sampling period, and in older fish.
Pooling samples into salinities across all DPH allowed us to make general predictions about microbial taxonomic differences among salinities. However, we acknowledge that our findings may be affected by significantly higher abundances in certain samples from one DPH compared to others, which was demonstrated by the wide range in Vibrio percentages across DPH (i.e., 90% versus 4.7% at 15 and 24 DPH, respectively). The greater abundance of Ruegeria in the lower salinity larvae samples matched the pattern seen in the water samples, suggesting a synergistic influence between the microbiomes. This genus is normally halophilic and requires salt to grow, so our results deviated from what was expected (Arahal et al., 2018). The typical pattern of higher Ruegeria in 30 ppt was observed in another study focused on the changes in the Oryzias melastigma gut microbiome (Lai et al., 2020). Like our study, the abundance decreased from seawater to freshwater in the Nautella genus. whose type species Nautella italica has an optimal growth salinity between 20-30 ppt (Vandecandelaere et al., 2009). The genus Lentibacter, made up of a single species (L. algarum) that grows at higher salinities, also increased in the 20 and 30 ppt larvae and water samples but represented a smaller percentage of the total larvae microbiome than in the water samples. This bacterium has been isolated from marine and coastal waters as well as from the gut microbiome of various invertebrates (Li et al., 2012). Identifying the core microbiome of farmed fish is important to establish a baseline for analysis, determine the presence of beneficial and potentially pathogenic community members, and understand how the microbial ecology changes in response to fish stress.
Following stress-mediated dysbiosis, fish may become more susceptible to infection by some groups of bacteria that comprise the fish mucosa including PPG such as Vibrio, Flavobacterium, and Pseudomonas (Roberts, 2012). Nevertheless, Vibrio and Pseudomonas species are among the most common microbes of healthy marine fish (Vine et al., 2006). Vibrio dominated the composition of all three salinities with much higher percentages in the later DPH (15, 18, and 20 DPH) in larval samples. In contrast to the Vibrio trends in our study, a whole-body study on Mexican mollies (Poecilia sphenops) described a turnover of Aeromonas and Cetobacterium to Vibrio and an unknown Enterobacteriaceae genus from lower (0 and 5 ppt) to higher (18 to 30 ppt) salinities (Schmidt et al., 2015). Vibrio has also been identified as a dominant genus in multiple fish gut microbiome studies (Egerton et al., 2018) and in general, most species require or are stimulated by the presence of salt (Garrity, 2007). The rise in Vibrio contributed to the significant loss of alpha diversity in larvae samples and to the clustering of the later DPH samples away from the earlier DPH samples. Vibrio drove the changes in the percentage of PPG in larvae samples as well. Shifts in Vibrio as a constituent in the skin or gut microbiome in response to fluctuating salinities and temperatures can lead to pathogenesis in fish (Sehnal et al., 2021). For example, V. anguillarum causes classic vibriosis in farmed marine fish such as turbot (Scophtalmus maximus) under suboptimal rearing conditions (Frans et al., 2011). A source of the Vibrio spike could have been Artemia nauplii, which was likely fed to the larvae during the later DPH (i.e., 15-20 DPH), as Vibrio is a normal constituent of the Artemia gut microbiome (Tkavc et al., 2011). Since live feeds like Artemia can be vectors for Vibrio and other PPG, this introduction of bacteria from feed to fish could be investigated using shotgun metagenomics to understand Vibrio species and virulence (Sehnal et al., 2021).
Our analysis demonstrated that PPG containing species known to infect Trachinotus spp. had higher relative percentages at lower salinities in water and larvae samples. These opportunistic pathogens can proliferate and cause damage to the epithelial surfaces (Handlinger et al., 1997; Austin et al., 2007). Flavobacterium and Pseudomonas were found in higher abundances in both water and larvae samples at 10 ppt compared to the 30 ppt salinity samples. There were higher abundances of Flavobacterium in the 10 ppt whole-larval and water samples taken at 12 and 24 DPH, which also coincided with increased epithelial lifting in Florida pompano larval gills. Bacterial gill disease, caused by F. branchiophilum and F. succinicans, is a typical problem in farmed fish worldwide including recirculating aquaculture systems cultured rainbow trout (Oncorhynchus mykiss) (Good et al., 2015). Steinum et al. (2009) found that cultured Atlantic salmon (Salmo salar) with degenerative gill inflammation had different gill microbiomes in comparison to the healthy fish, with a lower abundance of Gammaproteobacteria and a higher abundance of PPG including Flavobacterium and Tenacibaculum. In addition, species of Flavobacterium, like F. psychrophilum and F. columnare, are etiological agents of freshwater fish diseases, including infections of the gills, that lead to massive mortality in fish stock and economic loss at commercial farms, while other species have shown antagonistic activity against pathogens and are part of the core microbiome of fish skin (Kunttu et al., 2012; Takeuchi et al., 2021).
Certain Pseudomonas species are disease-causing bacteria in freshwater, brackish, and marine farmed fish including trout species and Atlantic salmon (Wiklund and Wiklund, 2016; Kuebutornye et al., 2020). Some studies have focused on the use of probiotics to prevent Pseudomonas infections in the aquaculture industry (Kuebutornye et al., 2020). This PPG showed an opposite temporal trend from Vibrio, with higher abundances in the earlier DPH water and larvae samples (3 through 12 DPH) and in larvae samples from 10 and 20 ppt versus 30 ppt. The decrease in abundance of Pseudomonas in later DPH samples could be explained by a change in feed types such as the switch of live feeds from rotifers to Artemia, or the introduction of a pelleted diet. As a result, these growth conditions were either disadvantageous to genera like Pseudomonas or optimal for those like Vibrio. With a wide salinity tolerance, Pseudomonas has been a dominant genus in the gut and skin microbiome of a diverse array of fish species, and studies have indicated that Pseudomonas growth is variable in response to salinity (Kuebutornye et al., 2020). Pseudomonas exhibited no growth differences among salinities in the gut microbiome of barramundi (Lates calcarifer), while it was found in a higher abundance in the gut microbiome of Chinook salmon when cultured in saltwater compared to freshwater (Zheng et al., 2019; Zhao et al., 2020). Understanding the ecology of these PPG and how they manifest disease following dysbiosis could help inform aquaculture on how to prevent stressful conditions and how to monitor for signs of disease using histopathological markers. Overall, our findings offer sufficient evidence that the Florida pompano larval microbiome is impacted by different salinities and diet, with increased growth of PPG and potential gill alterations at low salinities such as 10 ppt. Although, the actual impact of these changes on the larvae can be assessed using whole transcriptomics.
Several studies have shown that salinity influences the regulation of genes associated with immunity in euryhaline species (Boutet et al., 2006; Nguyen et al., 2016; Cao et al., 2020). An upregulation in immune-related genes due to osmotic stress was revealed in both cobia, (Rachycentron canadum) (Cao et al., 2020), and silvery pomfret (Pampus argenteus) (Li et al., 2020). In contrast, we have observed a downregulation of immune response gene sets in the 10 ppt larvae (Bradshaw et al., 2023). This downregulation in immune response genes may indicate that Florida pompano has an isosmotic point close to 10 ppt. One study found the isosmotic point of another Trachinotus species, T. marginarus, to be approximately 13 ppt (Abou Anni et al., 2016). Rearing the larvae at a salinity around its isosmotic point may reduce stress responses, which would explain the downregulation of gene sets related to immunity.
In previous studies, the genes tlr2, lyz, cat, gsto1, gss, scara3, il6l, il8l, and tlr5 were found to be differentially expressed due to changes in salinity (Si et al., 2018; Lin et al., 2020; Peng et al., 2022). However, in our study, the expression of these genes did not differ statistically among different salinity treatments. The expression of nod1 was the only immune-related gene tested that was significantly different among salinities; it was lower in larvae reared at 10 ppt versus 30 ppt. Nod-like receptor signaling pathways, including nod1, are part of the innate immune defense against bacterial and protozoan parasitic infections. NOD1 specifically has been shown to recognize the G-d-glutamyl-meso-diaminopimelic acid moieties that are components of peptidoglycan as well as lipopolysaccharides (Bi et al., 2018). Peptidoglycan is found on both gram-negative and gram-positive bacteria, whereas lipopolysaccharide is found on the outer membrane of only gram-negative bacteria, such as some of our PPG: Vibrio, Flavobacterium, and Pseudomonas. NOD1 activates RIPK2, which in turn activates the NF-κB signaling pathway. This leads to the expression of pro-inflammatory cytokines such as TNFα, IL-1β, IL-6, and IL-8 (Bi et al., 2018). In a study on silvery pomfret (Pampus argenteus), members of the NOD-like receptor signaling pathway (card8, hsp90a, and il1b) were enriched in the gill tissue from individuals undergoing high salinity stress (Li et al., 2020). None of these genes were found in our transcriptome, but other genes involved in the NOD1 pathway, including ripk2, il6l, and il8l, were. Their expression levels were not significantly different among salinities. Although there was a higher percentage of gram-negative PPG along with more apparent epithelial lifting at lower salinities, the downregulation of immune response related genes sets (Bradshaw et al., 2023) and limited upregulation of NOD1-related genes at lower salinities suggests that either these bacteria were nonvirulent, or the rearing conditions were not optimal for infection. On the other hand, RNA-Seq as performed on whole larvae, thus if there was stress in a particular body part, like the gills, its responses may be masked by the rest of the whole body gene expression generally not being in a stressed state.
In our study, there was a significant change in the expression of most of the immune-related genes we tested in the larvae samples across DPH. This was potentially significant since there was also a rise in Vibrio abundance during 15-20 DPH as well as spikes of Flavobacterium and Pseudomonas during certain DPH. Several studies have shown upregulation of immune-related genes in response to pathogenic members of these PPGs. One study saw the expression of genes associated with the NOD-like receptor and toll-like receptor pathways were upregulated in orange-spotted grouper (Epinephelus coioides) after the introduction of V. harveyi (Maekawa et al., 2017). Toll-like signaling pathways such as tlr5, which reacts to bacterial flagellin, and tlr2, which responds to peptidoglycans and lipoteichoic acids, are important in the activation of innate immunity (Sahoo, 2020). There was also an upregulation of several cytokines (IL-1β, IL-6, IL-8, IL-10, and TNFα) or their related genes in zebrafish (Danio rerio) in response to Vibrio alginolyticus (Gong et al., 2020) as well as in Mandarin fish (Siniperca chuatsi) (IL-1β, IL-8) in response to infection by Flavobacterium columnare (Zhou et al., 2015). Although nod1 and one of its downstream genes, il8l, increased over time, the expression of other nod1 downstream genes differed. There was a decrease in ripk2 over time, while il6l had low expression that was not significantly different across DPH. This suggests that the NOD1 pathway was not activated despite the higher percentage of potentially pathogenic gram-negative bacteria (Sahoo, 2020). While tlr2 did not statistically change across days post hatch, tlr5 steadily increased over time from 3 to 20 DPH. The gene associated with its main downstream mediator, myd88, was not found in our transcriptome, but one of its further downstream signal mediators, tab1, was present. The expression of tab1 decreased over time, which, similar to NOD1, suggests that the TLR5 pathway was not activated in response to bacteria.
In other studies, some of the genes we explored showed differential expression due to changes in salinity, however our study showed changes in their expression over time. The genes cat, gss, gsto1, and scara3 are all related to antioxidant defense, and all increased over time from 12-20 DPH. In their source studies, gst, gss, and cat were upregulated due to low salinity stress, while scara3 was downregulated (Si et al., 2018; Lin et al., 2020). Catalase (cat) decomposes hydrogen peroxide and SCARA3 will scavenge reactive oxygen species and other products of oxidation (Si et al., 2018; Devi et al., 2019). Glutathione, which is synthesized in part by glutathione synthase (gss), is oxidized by glutathione peroxidases to neutralize hydrogen peroxide (Devi et al., 2019; Hoseinifar et al., 2020). Glutathione S-transferases (gsto1) will assist with regeneration of reduced glutathione by catalyzing its conjugation to several substrates to make it more soluble allowing it to be acted upon by glutathione reductase (Birnie-Gauvin et al., 2017; Parolini et al., 2019). (Kalaimani et al., 2008) demonstrated that antioxidant enzymatic activity increased during metamorphosis and Artemia feeding in Asian seabass (Lates calcarifer). Increases in antioxidant enzyme activities during metamorphosis has also been noted in Atlantic cod (Gadus morhua) and Senegal sole (Solea senegalensis) (Fernández-Díaz et al., 2006; Hamre et al., 2014). Studies have suggested that the strong metabolic changes that occur during metamorphosis due to high energy demand and oxygen uptake lead to an increase in antioxidant defense (Mourente et al., 1999; Solé et al., 2005; Fernández-Díaz et al., 2006). The increased expression of antioxidant genes tested in our study generally corresponds with the metamorphosis period of Florida pompano which occurs approximately during 18-25 DPH (Weirich et al., 2021). This increase in antioxidant gene expression during metamorphosis suggests that further enrichment of pelleted feeds with antioxidants, such as polyunsaturated fatty acids, may help with stress management during larval rearing (Kalaimani et al., 2008). Probiotics, prebiotics, and synbiotics have also been shown to potentially increase the activities of enzymes associated with antioxidant defense (Hoseinifar et al., 2020). However, it is unknown currently if the detected increased expression in antioxidant genes is biologically significant in terms of production performance and survival. Future studies can explore this further.
The antimicrobial activity of lysozyme (lyz) involves the hydrolysis of the beta-(1,4) linked glycoside bonds of the peptidoglycans in bacterial cell walls (Magnadottir et al., 2005). Its activity was also increased during 12-20 DPH, suggesting that the innate immune system of the larvae was developing during this time. Similarly, in a transcriptomic study on zebrafish larvae, immune related genes were significantly enriched from hatching through 7 DPH (Yang et al., 2013). This suggests that immune system development after the larvae have hatched can lead to changes in gene expression (Zapata et al., 2006).
On-farm research trials directly with commercial producers offer science many benefits not limited to study results being directly applicable to commercial production. In addition, the expertise of the farmers in larval rearing enable project to overcome hurtles and make contributions to study design and interpretation. However, having the trial completed on the farm instead of in the lab offered many logistical limitations to the study. Primarily logistical, researchers were not present at the time the samples were taken often meaning on-site decisions needed to be made by farmers regarding exact sample collection order and procedures. It is important both researchers and industry partners agree upon the sampling plan, logistics (both on farm production as well as sampling), hierarchy of sampling, and have open communication before, during and after the trial to ensure the best most accurate results and interpretation. Many lessons were learned in the performance of this study, which will be used in future studies with stakeholders. For example, sampling for water microbiome was time consuming and intensive on the farm, taking up to 4 hours total to accomplish. This meant that a farm employee was dedicated to this task, and that sampling water from all tanks could not be completed before feeding. Although this method meant that water did not need to be transferred from the farm to the lab, future studies decreased the time limit of filtering from 15 to 3 minutes. We may also explore using the more classical vacuum filtration method in the future to standardize the timing of sampling (i.e. before feeding, after feeding, etc.) for all tanks within the study to limit these potential variable impacts. The use of banjo filters helped mitigate the possible effects of food particulates on the water microbiome and statistics showed that tanks of the same salinity were still statistically similar to each other but different from other salinities in terms of beta diversity.
We demonstrated that the whole-larval and water microbiome of Florida pompano changed in response to rearing salinity and diet in an aquaculture farm environment. Part of the microbial changes among salinities was driven by the higher abundances of PPGs such as Flavobacterium and Pseudomonas at the lowest salinity (10 ppt) in both sample types. Although there was an increase in stress-induced gill alterations as well as these PPGs at lower salinities, the expression of immune-related genes suggest that there was little pathogenic activity, since only one of the twelve genes tested was significantly different across salinities.
Changes due to diet was particularly evident in the patterns of PPGs, such as Vibrio, which spiked in larval microbiomes during 12-20 DPH when Artemia were likely being fed, as well as the increase in Pseudomonas in earlier DPH in both sample types. Despite this, as well as the increased expression of pathogen recognition receptors during later DPH, the lower or unchanged expression of the downstream elements of pathogen recognition pathways also suggested little pathogenic activity. This shows that the PPGs present were part of the normal flora and that the larvae were not stressed enough to induce disease either due to diet or salinity.
The gene expression results do indicate that metamorphosis was likely responsible for the increase in antioxidant-related genes during later DPH. Future larval studies encompassing metamorphosis should consider measuring indicators of oxidative stress such as lipid peroxidation/malondialdehyde and glutathione as well as the activity levels of antioxidant enzymes. Our results show that there was more stress associated with metamorphosis than the salinities tested or the presence of PPGs, suggesting that farmers could save time and resources by growing Florida pompano larvae at salinities as low as 10 ppt. Although further studies are needed to assess the long-term effects of histopathological changes. The larvae’s tolerance of lower salinities influences husbandry management decisions such as now having the ability to use the reduction of salinity to potentially treat a halophilic pathogen or knowing possible alternative salinities should a farm’s seawater source is compromised.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by Florida Atlantic University Institutional Animal Care and Use Committee.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication. All authors contributed to the article and approved the submitted version.
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer. This work was supported by the U.S. Department of Agriculture, Agricultural Research Service by cooperative agreement number 59-6034-9-007 with Florida Atlantic University’s Harbor Branch Oceanographic Institute.”
The authors would like to thank all the personnel from Live Advantage Bait, Stuart, FL, and Proaquatix, Vero Beach, FL farms. We are grateful to Avery Davis, Brent Rupnik, Eric Wagner, Dr. Dan Woltering, and Lindsey Romano for their extensive help with raising larvae and collecting samples. We also want to thank Florida Atlantic University at Harbor Branch Oceanographic personnel including Brandon McHenry for processing samples and optimizing our extraction methods, Caitlyn Courtemanche for processing samples for histology, Victoria Uribe for processing of histology samples and initial edits, and Gonzalo Illán for processing of histology samples, statistical analysis, and initial edits.
Dr. Nicole Kirchhoff is the founder, owner, and CEO of Live Advantage Bait, LLC. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1158446/full#supplementary-material
DPH, days post hatch; BH, Benjamini-Hochberg; PERMANOVA, permutational analysis of variance; MC, Monte Carlo; PPG, potentially pathogenic genera.
Abou Anni I. S., Bianchini A., Barcarolli I. F., Varela A. S., Robaldo R. B., Tesser M. B., et al. (2016). Salinity influence on growth, osmoregulation and energy turnover in juvenile pompano Trachinotus marginatus cuvier 1832. Aquaculture 455, 63–72. doi: 10.1016/J.AQUACULTURE.2016.01.010
Anaconda Inc. (2021). Anaconda software distribution. Available at: https://www.anaconda.com.
Anderson M. J., Gorley R. N., Clarke K. R. (2008). PERMANOVA+ for PRIMER: Guide to software and statistical methods. Plymouth: PRIMER-E. doi: 10.13564/j.cnki.issn.1672-9382.2013.01.010
Andrews S. (2010). FastQC. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
Arahal D. R., Lucena T., Rodrigo-Torres L., Pujalte M. J. (2018). Ruegeria denitrificans sp. nov., a marine bacterium in the family rhodobacteraceae with the potential ability for cyanophycin synthesis. Int. J. Syst. Evol. Microbiol. 68, 2515–2522. doi: 10.1099/IJSEM.0.002867/CITE/REFWORKS
Araki K., Aokic J. Y., Kawase J., Hamada K., Ozaki A., Fujimoto H., et al. (2018). Whole genome sequencing of greater amberjack (Seriola dumerili) for SNP identification on aligned scaffolds and genome structural variation analysis using parallel resequencing. Int. J. Genomics 2018. doi: 10.1155/2018/7984292
Aronson J. D. (1926). Spontaneous tuberculosis in salt water fish. J. Infect. Dis. 39, 315–320. doi: 10.1093/INFDIS/39.4.315
Austin B., Austin D., Munn C. (2007). Bacterial fish pathogens: disease of farmed and wild fish (Chichester, UK: Springer).
Banks J. C., Kelly L. T., Falleiros R., Rojahn J., Gabrielsson R., Clapcott J. (2021). Detecting the pest fish, Gambusia affinis from environmental DNA in new Zealand: a comparison of methods. N. Z. J. Zool. 48, 202–216. doi: 10.1080/03014223.2020.1858880/SUPPL_FILE/TNZZ_A_1858880_SM6928.DOCX
Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc Ser. B Stat. Methodol. 57, 289–300. doi: 10.1111/J.2517-6161.1995.TB02031.X
Bernet D., Schmidt H., Meier W., Burkhardt-Holm P., Wahli T. (1999). Histopathology in fish: proposal for a protocol to assess aquatic pollution. J. Fish. Dis. 22, 25–34. doi: 10.1046/J.1365-2761.1999.00134.X
Bi D., Wang Y., Gao Y., Li X., Chu Q., Cui J., et al. (2018). Recognition of lipopolysaccharide and activation of NF-κB by cytosolic sensor NOD1 in teleost fish. Front. Immunol. 9. doi: 10.3389/FIMMU.2018.01413/BIBTEX
Birnie-Gauvin K., Costantini D., Cooke S. J., Willmore W. G. (2017). A comparative and evolutionary approach to oxidative stress in fish: A review. Fish Fish. 18, 928–942. doi: 10.1111/FAF.12215
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/S41587-019-0209-9
Boutet I., Long Ky C. L., Bonhomme F. (2006). A transcriptomic approach of salinity response in the euryhaline teleost, dicentrarchus labrax. Gene 379, 40–50. doi: 10.1016/J.GENE.2006.04.011
Bradshaw D. J., Dickens N. J., Trefry J. H., McCarthy P. J. (2020). Defining the sediment prokaryotic communities of the Indian river lagoon, FL, USA, an estuary of national significance. PloS One 15, e0236305. doi: 10.1371/journal.pone.0236305
Bradshaw D. J., Uribe V., King L. E., Perricone C. S., Illán G., Allmon E., et al. (2023). Effects of low salinities on growth, fatty acid composition, and transcriptome in Florida pompano (Trachinotus carolinus) at early developmental stages. Aquaculture 563, 738964. doi: 10.1016/j.aquaculture.2022.738964
Burgos F. A., Ray C. L., Arias C. R. (2018). Bacterial diversity and community structure of the intestinal microbiome of channel catfish (Ictalurus punctatus) during ontogenesis. Syst. Appl. Microbiol. 41, 494–505. doi: 10.1016/j.syapm.2018.04.006
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cao D., Li J., Huang B., Zhang J., Pan C., Huang J., et al. (2020). RNA-Seq analysis reveals divergent adaptive response to hyper- and hypo-salinity in cobia, Rachycentron canadum. Fish Physiol. Biochem. 46, 1713–1727. doi: 10.1007/s10695-020-00823-7
Cerqueira C. C. C., Fernandes M. N. (2002). Gill tissue recovery after copper exposure and blood parameter responses in the tropical fish Prochilodus scrofa. Ecotoxicol. Environ. Saf. 52, 83–91. doi: 10.1006/EESA.2002.2164
Chong S. M., Wong W. K., Lee W. Y., Tan Z. B., Tay Y. H., Teo X. H., et al. (2017). Streptococcus agalactiae outbreaks in cultured golden pomfret, Trachinotus blochii (Lacépède), in Singapore. J. Fish. Dis. 40, 971–974. doi: 10.1111/JFD.12570
Cock P. J. A., Antao T., Chang J. T., Chapman B. A., Cox C. J., Dalke A., et al. (2009). Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423. doi: 10.1093/bioinformatics/btp163
Devi G., Harikrishnan R., Paray B. A., Al-Sadoon M. K., Hoseinifar S. H., Balasundaram C. (2019). Effect of symbiotic supplemented diet on innate-adaptive immune response, cytokine gene regulation and antioxidant property in Labeo rohita against Aeromonas hydrophila. Fish Shellfish. Immunol. 89, 687–700. doi: 10.1016/J.FSI.2019.04.036
Egerton S., Culloty S., Whooley J., Stanton C., Ross R. P. (2018). The gut microbiota of marine fish. Front. Microbiol. 9. doi: 10.3389/FMICB.2018.00873/BIBTEX
Elliott A. C., Hynan L. S. (2011). A SAS® macro implementation of a multiple comparison post hoc test for a kruskal–Wallis analysis. Comput. Methods Programs Biomed. 102, 75–80. doi: 10.1016/j.cmpb.2010.11.002
Fanta E., Rios F. S., Romão S., Vianna A. C. C., Freiberger S. (2003). Histopathology of the fish Corydoras paleatus contaminated with sublethal levels of organophosphorus in water and food. Ecotoxicol Environ. Saf. 54, 119–130. doi: 10.1016/S0147-6513(02)00044-1
Fernández-Díaz C., Kopecka J., Cañavate J. P., Sarasquete C., Solé M. (2006). Variations on development and stress defences in Solea senegalensis larvae fed on live and microencapsulated diets. Aquaculture 251, 573–584. doi: 10.1016/J.AQUACULTURE.2005.06.014
Food and Agriculture Organization of the United Nations (2023) Cultured aquatic species information programme - trachinotus blochii (Lacépède 1801). Available at: https://www.fao.org/fishery/en/culturedspecies/trachinotus_spp/en.
Frans I., Michiels C. W., Bossier P., Willems K. A., Lievens B., Rediers H. (2011). Vibrio anguillarum as a fish pathogen: virulence factors, diagnosis and prevention. J. Fish Dis. 34, 643–661. doi: 10.1111/J.1365-2761.2011.01279.X
García A. H. (2011). Anhydrobiosis in bacteria: From physiology to applications. J. Biosci. 36, 939–950. doi: 10.1007/S12038-011-9107-0/TABLES/1
Garrity G. (2007). Bergey’s manual® of systematic bacteriology volume 2: The proteobacteria, part b: The gammaproteobacteria (New York, NY, USA: Springer Science & Business Media).
Giavenni R., Finazzi M., Poli G., Grimaldi E. (1980). Tuberculosis in marine tropical fishes in an aquarium. J. Wildl. Dis. 16, 161–168. doi: 10.7589/0090-3558-16.2.161
Gong Q.y., Yang M.j., Yang L.f., Chen Z.g., Jiang M., Peng B. (2020). Metabolic modulation of redox state confounds fish survival against Vibrio alginolyticus infection. Microb. Biotechnol. 13, 796–812. doi: 10.1111/1751-7915.13553
Good C., Davidson J., Wiens G. D., Welch T. J., Summerfelt S. (2015). Flavobacterium branchiophilum and F. succinicans associated with bacterial gill disease in rainbow trout Oncorhynchus mykiss (Walbaum) in water recirculation aquaculture systems. J. Fish Dis. 38, 409. doi: 10.1111/JFD.12249
Gordon A., Hannon G. J. (2010) Fastx-toolkit. FASTQ/A short-reads pre-processing tools. Available at: http://hannonlab.cshl.edu/fastx_toolkit.
Hamre K., Penglase S. J., Rasinger J. D., Skjærven K. H., Olsvik P. A. (2014). Ontogeny of redox regulation in Atlantic cod (Gadus morhua) larvae. Free Radic. Biol. Med. 73, 337–348. doi: 10.1016/J.FREERADBIOMED.2014.05.017
Handlinger J., Soltani M., Percival S. (1997). The pathology of Flexibacter maritimus in aquaculture species in Tasmania, Australia. J. Fish Dis. 20, 159–168. doi: 10.1046/J.1365-2761.1997.00288.X
Harper C., Wolf J. C. (2009). Morphologic effects of the stress response in fish. ILAR J. 50, 387–396. doi: 10.1093/ILAR.50.4.387
Hauville M. R., Zambonino-Infante J. L., Gordon Bell J., Migaud H., Main K. L. (2016). Effects of a mix of Bacillus sp. as a potential probiotic for Florida pompano, common snook and red drum larvae performances and digestive enzyme activities. Aquac. Nutr. 22, 51–60. doi: 10.1111/ANU.12226
Hawke J. P. (1976). “A survey of the diseases of striped bass, Morone saxatilis and pompano, Trachinotus carolinus cultured in earthen ponds,” in Proceedings of the annual meeting – World Mariculture Society. Vol. 7. 495–509. doi: 10.1111/j.1749-7345.1976.tb00084.x
Heath A. G. (1995). Water pollution and fish physiology. 2nd ed (Boca Raton, FL, USA: CRC Press). doi: 10.1201/9780203718896/WATER-POLLUTION-FISH-PHYSIOLOGY-ALAN-HEATH
Herlemann D. P. R., Labrenz M., Jürgens K., Bertilsson S., Waniek J. J., Andersson A. F. (2011). Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5, 1571–1579. doi: 10.1038/ismej.2011.41
Hoseinifar S. H., Yousefi S., van Doan H., Ashouri G., Gioacchini G., Maradonna F., et al. (2020). Oxidative stress and antioxidant defense in fish: the implications of probiotic, prebiotic, and synbiotics. Rev. Fish. Sci. Aquac. 29, 198–217. doi: 10.1080/23308249.2020.1795616
Humphrey J. D. (2007). Systemic pathology of fish: a text and atlas of normal tissues in teleosts and their response in disease. J. Fish. Dis. 30, 381–382. doi: 10.1111/j.1365-2761.2007.00825.x
Kalaimani N., Chakravarthy N., Shanmugham R., Thirunavukkarasu A. R., Alavandi S. v., Santiago T. C. (2008). Anti-oxidant status in embryonic, post-hatch and larval stages of Asian seabass (Lates calcarifer). Fish Physiol. Biochem. 34, 151–158. doi: 10.1007/S10695-007-9155-4/FIGURES/5
Koster J., Rahmann S. (2012). Snakemake–a scalable bioinformatics workflow engine. Bioinformatics 28, 2520–2522. doi: 10.1093/bioinformatics/bts480
Krueger F. (2020). Trim galore. Available at: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/.
Kuebutornye F. K. A., Abarike E. D., Lu Y., Hlordzi V., Sakyi M. E., Afriyie G., et al. (2020). Mechanisms and the role of probiotic Bacillus in mitigating fish pathogens in aquaculture. Fish Physiol. Biochem. 46, 819–841. doi: 10.1007/S10695-019-00754-Y
Kunttu H. M. T., Sundberg L. R., Pulkkinen K., Valtonen E. T. (2012). Environment may be the source of Flavobacterium columnare outbreaks at fish farms. Environ. Microbiol. Rep. 4, 398–402. doi: 10.1111/J.1758-2229.2012.00342.X
Lai K. P., Lin X., Tam N., Ho J. C. H., Wong M. K., Gu J., et al. (2020). Osmotic stress induces gut microbiota community shift in fish. Environ. Microbiol. 22, 3784–3802. doi: 10.1111/1462-2920.15150
Lambert Y., Dutil J. D., Munro J. (2011). Effects of intermediate and low salinity conditions on growth rate and food conversion of Atlantic cod (Gadus morhua). Can. J. Fish. Aquat. Sci. 51, 1569–1576. doi: 10.1139/F94-155
Laurén D. J., Wails D. (2018). “Liver structural alterations accompanying chronic toxicity in fishes: potential biomarkers of exposure,” in Biomarkers of environmental contamination (Boca Raton, FL, USA: CRC Press), 17–57.
Legrand T. P. R. A., Wynne J. W., Weyrich L. S., Oxley A. P. A. (2020). A microbial sea of possibilities: current knowledge and prospects for an improved understanding of the fish microbiome. Rev. Aquac. 12, 1101–1134. doi: 10.1111/raq.12375
Li Z., Qu Z., Zhang X., Zhang X. H. (2012). Lentibacter algarum gen. nov., sp. nov., isolated from coastal water during a massive green algae bloom. Int. J. Syst. Evol. Microbiol. 62, 1042–1047. doi: 10.1099/IJS.0.029868-0
Li J., Xue L., Cao M., Zhang Y., Wang Y., Xu S., et al. (2020). Gill transcriptomes reveal expression changes of genes related with immune and ion transport under salinity stress in silvery pomfret (Pampus argenteus). Fish Physiol. Biochem. 46, 1255–1277. doi: 10.1007/S10695-020-00786-9/FIGURES/17
Lin G., Zheng M., Li S., Xie J., Fang W., Gao D., et al. (2020). Response of gut microbiota and immune function to hypoosmotic stress in the yellowfin seabream (Acanthopagrus latus). Sci. Total Environ. 745, 140976. doi: 10.1016/J.SCITOTENV.2020.140976
Liu S., Wang S., Cai Y., Li E., Ren Z., Wu Y., et al. (2020). Beneficial effects of a host gut-derived probiotic, Bacillus pumilus, on the growth, non-specific immune response and disease resistance of juvenile golden pompano, Trachinotus ovatus. Aquaculture 514, 734446. doi: 10.1016/J.AQUACULTURE.2019.734446
Llewellyn M. S., Boutin S., Hoseinifar S. H., Derome N. (2014). Teleost microbiomes: the state of the art in their characterization, manipulation and importance in aquaculture and fisheries. Front. Microbiol. 5. doi: 10.3389/fmicb.2014.00207
Loeza-Quintana T., Abbott C. L., Heath D. D., Bernatchez L., Hanner R. H. (2020). Pathway to increase standards and competency of eDNA surveys (PISCeS)–advancing collaboration and standardization efforts in the field of eDNA. Environ. DNA 2, 255–260. doi: 10.1002/EDN3.112
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Maekawa S., Byadgi O., Chen Y. C., Aoki T., Takeyama H., Yoshida T., et al. (2017). Transcriptome analysis of immune response against Vibrio harveyi infection in orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol. 70, 628–637. doi: 10.1016/J.FSI.2017.09.052
Magnadottir B., Lange S., Gudmundsdottir S., Bøgwald J., Dalmo R. A. (2005). Ontogeny of humoral immune parameters in fish. Fish Shellfish Immunol. 19, 429–439. doi: 10.1016/J.FSI.2005.03.010
Mangiafico S. (2021) Rcompanion: functions to support extension education program evaluation. Available at: https://CRAN.R-project.org/package=rcompanion.
Manzanera M. (2021). Dealing with water stress and microbial preservation. Environ. Microbiol. 23, 3351–3359. doi: 10.1111/1462-2920.15096
McMurdie P. J., Holmes S. (2013). Phyloseq: an r package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8, e61217. doi: 10.1371/journal.pone.0061217
Mourente G., Tocher D. R., Diaz E., Grau A., Pastor E. (1999). Relationships between antioxidants, antioxidant enzyme activities and lipid peroxidation products during early development in Dentex eggs and larvae. Aquaculture 179, 309–324. doi: 10.1016/S0044-8486(99)00167-2
Nguyen T. V., Jung H., Nguyen T. M., Hurwood D., Mather P. (2016). Evaluation of potential candidate genes involved in salinity tolerance in striped catfish (Pangasianodon hypophthalmus) using an RNA-seq approach. Mar. Genomics 25, 75–88. doi: 10.1016/J.MARGEN.2015.11.010
Nikouli E., Meziti A., Antonopoulou E., Mente E., Kormas K. A. (2019). Host-associated bacterial succession during the early embryonic stages and first feeding in farmed gilthead sea bream (Sparus aurata). Genes 10, 483. doi: 10.3390/GENES10070483
Nolan K. P., Loeza-Quintana T., Little H. A., McLeod J., Ranger B., Borque D. A., et al. (2022). Detection of brook trout in spatiotemporally separate locations using validated eDNA technology. J. Environ. Stud. Sci. 13, 66–82. doi: 10.1007/S13412-022-00800-X/FIGURES/9
Ogle D., Doll J., Wheeler P., Dinno A. (2021) FSA: simple fisheries stock assessment methods. Available at: https://github.com/droglenc/FSA.
Oksanen J., Blanchet F. G., Kindt R., Legendre P., McGlin D., Minchin P. R., et al. (2019). Vegan: Community ecology package. Available at: https://CRAN.R-project.org/package=vegan.
Parolini M., Iacobuzio R., de Felice B., Bassano B., Pennati R., Saino N. (2019). Age- and sex-dependent variation in the activity of antioxidant enzymes in the brown trout (Salmo trutta). Fish Physiol. Biochem. 45, 145–154. doi: 10.1007/S10695-018-0545-6/TABLES/2
Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., et al. (2011). Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830. doi: 10.48550/arXiv.1201.0490
Peng Y., Shi H., Liu Y., Huang Y., Zheng R., Jiang D., et al. (2022). RNA Sequencing analysis reveals divergent adaptive response to hypo-and hyper-salinity in greater amberjack (Seriola dumerili) juveniles. Animals 12, 327. doi: 10.3390/ani12030327
Pérez-Pascual D., Estellé J., Dutto G., Rodde C., Bernardet J.-F., Marchand Y., et al. (2020). Growth performance and adaptability of European sea bass (Dicentrarchus labrax) gut microbiota to alternative diets free of fish products. Microorganisms 8, 1346. doi: 10.3390/MICROORGANISMS8091346
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
R Core Team. (2021). R: A language and environment for statistical computing. Available at: URL https://www.R-project.org/.
Rawls J. F., Samuel B. S., Gordon J. I. (2004). Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. PNAS 101, 4596–4601. doi: 10.1073/pnas.0400706101
Reis A. B., Sant’Ana D. M. G., Azevedo J. F., Merlini L. S., Araújo E. J. A. (2009). The influence of the aquatic environment in tanks sequentially interconnected with PVC pipes on the gill epithelium and lamellas of tilapia (Oreochromis niloticus). Braz. J. Vet. Res. 29, 303–311. doi: 10.1590/S0100-736X2009000400005
Riche M. A., Pfeiffer T. J., Wills P. S., Amber J. J., Sepúlveda M. S. (2012). Inland marine fish culture in low salinity recirculating aquaculture systems. Bull. Fish. Res. Agen. 35, 65–75.
Robeson M. S., O’Rourke D. R., Kaehler B. D., Ziemski M., Dillon M. R., Foster J. T., et al. (2021). RESCRIPt: Reproducible sequence taxonomy reference database management. PloS Comput. Biol. 17, e1009581. doi: 10.1371/JOURNAL.PCBI.1009581
Sahoo B. R. (2020). Structure of fish toll-like receptors (TLR) and NOD-like receptors (NLR). Int. J. Biol. Macromol. 161, 1602–1617. doi: 10.1016/J.IJBIOMAC.2020.07.293
Schmidt V. T., Smith K. F., Melvin D. W., Amaral-Zettler L. A. (2015). Community assembly of a euryhaline fish microbiome during salinity acclimation. Mol. Ecol. 24, 2537–2550. doi: 10.1111/mec.13177
Sehnal L., Brammer-Robbins E., Wormington A. M., Blaha L., Bisesi J., Larkin I., et al. (2021). Microbiome composition and function in aquatic vertebrates: Small organisms making big impacts on aquatic animal health. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.567408
Si Y., Wen H., Li Y., He F., Li J., Li S., et al. (2018). Liver transcriptome analysis reveals extensive transcriptional plasticity during acclimation to low salinity in Cynoglossus semilaevis. BMC Genomics 19, 1–14. doi: 10.1186/S12864-018-4825-4/FIGURES/5
Singh S., Mallik S. K., Kala K., Shahi N., Pathak R., Giri A. K., et al. (2021). Characterization of flavobacterium columnare from farmed infected rainbow trout, oncorhynchus mykiss (Walbaum, 1792) of central Indian Himalayan region, India. Aquaculture 544, 737118. doi: 10.1016/J.AQUACULTURE.2021.737118
Skinner M., Murdoch M., Loeza-Quintana T., Crookes S., Hanner R. (2020). A mesocosm comparison of laboratory-based and on-site eDNA solutions for detection and quantification of striped bass (Morone saxatilis) in marine ecosystems. Environ. DNA 2, 298–308. doi: 10.1002/EDN3.61
Smith C. E., Piper R. G. (1975). “Lesions associated with chronic exposure to ammonia,” in The pathology of fishes. Eds. Ribelin W. E., Migaki G. (Madison, WI, USA: Madison: University of Wisconsin Press), 497–514.
Solé M., Potrykus J., Fernández-Díaz C., Blasco J. (2005). Variations on stress defences and metallothionein levels in the Senegal sole, Solea senegalensis, during early larval stages. Fish Physiol. Biochem. 30, 57–66. doi: 10.1007/S10695-004-6786-6
Steinum T., Sjåstad K., Falk K., Kvellestad A., Colquhoun D. J. (2009). An RT PCR-DGGE survey of gill-associated bacteria in Norwegian seawater-reared Atlantic salmon suffering proliferative gill inflammation. Aquaculture 293, 172–179. doi: 10.1016/j.aquaculture.2009.05.006
Takeuchi M., Fujiwara-Nagata E., Katayama T., Suetake H. (2021). Skin bacteria of rainbow trout antagonistic to the fish pathogen Flavobacterium psychrophilum. Sci. Rep. 11, 1–11. doi: 10.1038/s41598-021-87167-1
Takishita S. S., de Oliveira K. F., Flores-Lopes F., Tavares-Braga L. G., Teixeira-Lanna E. A., de Azevedo R. V. (2015). Responses of Nile tilapia to different levels of water salinity. Lat. Am. J. Aquat. Res. 43, 828–835. doi: 10.3856/vol43-issue5-fulltext-3
Thomas A. C., Nguyen P. L., Howard J., Goldberg C. S. (2019). A self-preserving, partially biodegradable eDNA filter. Methods Ecol. Evol. 10, 1136–1141. doi: 10.1111/2041-210X.13212
Tkavc R., Ausec L., Oren A., Gunde-Cimerman N. (2011). Bacteria associated with Artemia spp. along the salinity gradient of the solar salterns at eilat (Israel). FEMS Microbiol. Ecol. 77, 310–321. doi: 10.1111/J.1574-6941.2011.01112.X
Vandecandelaere I., Nercessian O., Segaert E., Achouak W., Mollica A., Faimali M., et al. (2009). Nautella italica gen. nov., sp. nov., isolated from a marine electroactive biofilm. Int. J. Syst. Evol. Microbiol. 59, 811–817. doi: 10.1099/IJS.0.002683-0/CITE/REFWORKS
Vine N. G., Leukes W. D., Kaiser H. (2006). Probiotics in marine larviculture. FEMS Microbiol. Rev. 30, 404–427. doi: 10.1111/J.1574-6976.2006.00017.X
Wang R. X., Guo Z. X., Feng J., Lin H. Z., Li Z. J. (2009). Screening of the effective traditional Chinese herbal medicines on the pathogenic bacteria of seawater-cultured animals. South China Fisheries Sci. 6, 19–24.
Wang A. R., Ran C., Ringø E., Zhou Z. G. (2018). Progress in fish gastrointestinal microbiota research. Rev. Aquac. 10, 626–640. doi: 10.1111/raq.12191
Wang W., Zhou Z., He S., Liu Y., Cao Y., Shi P., et al. (2010). Identification of the adherent microbiota on the gills and skin of poly-cultured gibel carp (Carassius auratus gibelio) and bluntnose black bream (Megalobrama amblycephala yih). Aquac. Res. 41, e72–e83. doi: 10.1111/j.1365-2109.2009.02459.x
Webster T. M. U., Consuegra S., Hitchings M., de Leaniz C. G. (2018). Interpopulation variation in the Atlantic salmon microbiome reflects environmental and genetic diversity. Appl. Environ. Microbiol. 84, e00691–e00618. doi: 10.1128/AEM
Weirich C. R., Riley K. L., Riche M., Main K. L., Wills P. S., Illán G., et al. (2021). The status of Florida pompano, Trachinotus carolinus, as a commercially ready species for U.S. marine aquaculture. J. World Aquac. Soc 52, 731–763. doi: 10.1111/JWAS.12809
Weirich C. R., Wills P. S., Baptiste R. M., Woodward P. N., Riche M. A. (2009). Production characteristics and body composition of Florida pompano reared to market size at two different densities in low-salinity recirculating aquaculture systems. N. Am. J. Aquac. 71, 165–173. doi: 10.1577/A08-016.1
Wickham H. (2007). Reshaping data with the reshape package. J. Stat. Software 21, 1–20. doi: 10.18637/JSS.V021.I12
Wickham H. (2011). The split-apply-combine strategy for data analysis. J. Stat. Software 40, 1–29. doi: 10.18637/jss.v040.i01
Wickham H., Bryan J. (2019). Readxl: Read excel files. Available at: https://CRAN.R-project.org/package=readxl.
Wiklund T., Wiklund T. (2016). Pseudomonas anguilliseptica infection as a threat to wild and farmed fish in the Baltic Sea. Microbiol. Aust. 37, 135–136. doi: 10.1071/MA16046
Wolf J. C., Baumgartner W. A., Blazer V. S., Camus A. C., Engelhardt J. A., Fournie J. W., et al. (2015). Nonlesions, misdiagnoses, missed diagnoses, and other interpretive challenges in fish histopathology studies: A guide for investigators, authors, reviewers, and readers. Toxicol. Pathol. 43, 297–325. doi: 10.1177/0192623314540229/ASSET/IMAGES/LARGE/10.1177_0192623314540229-FIG2.JPEG
Xia L., Cai J., Wang B., Huang Y., Jian J., Lu Y. (2015). Draft genome sequence of Nocardia seriolae ZJ0503, a fish pathogen isolated from Trachinotus ovatus in China. Genome Announc. 3, e01223–e01214. doi: 10.1128/GENOMEA.01223-14
Yan L. (2021). Ggvenn: Draw Venn diagram by ggplot2. Available at: https://CRAN.R-project.org/package=ggvenn.
Yandi I., Kayis S., Er A. (2017). Histopathological effects of etiological and non-etiological agents in some fish gills. Fresenius Environ. Bull. 26, 995–1000.
Yang H., Zhou Y., Gu J., Xie S., Xu Y., Zhu G., et al. (2013). Deep mRNA sequencing analysis to capture the transcriptome landscape of zebrafish embryos and larvae. PloS One 8, e64058. doi: 10.1371/JOURNAL.PONE.0064058
Yanong R. P., Pouder D. B., Falkinham J. O. III (2010). Association of mycobacteria in recirculating aquaculture systems and mycobacterial disease in fish. J. Aquat. Anim. Health 22, 219–223. doi: 10.1577/H10-009.1
Yilmaz P., Parfrey L. W., Yarza P., Gerken J., Pruesse E., Quast C., et al. (2014). The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Zapata A., Diez B., Cejalvo T., Gutiérrez-De Frías C., Cortés A. (2006). Ontogeny of the immune system of fish. Fish Shellfish Immunol. 20, 126–136. doi: 10.1016/J.FSI.2004.09.005
Zhang D., Mohammed H., Ye Z., Rhodes M. A., Thongda W., Zhao H., et al. (2022). Transcriptomic profiles of Florida pompano (Trachinotus carolinus) gill following infection by the ectoparasite Amyloodinium ocellatum. Fish Shellfish Immunol. 125, 171–179. doi: 10.1016/J.FSI.2022.05.017
Zhang Q., Yu H., Tong T., Tong W., Dong L., Xu M., et al. (2014). Dietary supplementation of Bacillus subtilis and fructooligosaccharide enhance the growth, non-specific immunity of juvenile ovate pompano, Trachinotus ovatus and its disease resistance against Vibrio vulnificus. Fish Shellfish Immunol. 38, 7–14. doi: 10.1016/j.fsi.2014.02.008
Zhao R., Symonds J. E., Walker S. P., Steiner K., Carter C. G., Bowman J. P., et al. (2020). Salinity and fish age affect the gut microbiota of farmed Chinook salmon (Oncorhynchus tshawytscha). Aquaculture 528, 735539. doi: 10.1016/J.AQUACULTURE.2020.735539
Zheng X., Yang R., Hu J., Lin S., Gu Z., Ma Z. (2019). The gut microbiota community and antioxidant enzymes activity of barramundi reared at seawater and freshwater. Fish Shellfish Immunol. 89, 127–131. doi: 10.1016/J.FSI.2019.03.054
Zhou Y., Zhang B., Chen X., Qian J. (2002)Preparation of Pseudomonas maltophilia lipopolysaccharidae and immunological analysis of the lipopolysaccharide against Trachinotus ovatus. (Accessed November 20, 2021).
Zhou W., Zhang Y., Wen Y., Ji W., Zhou Y., Ji Y., et al. (2015). Analysis of the transcriptomic profilings of mandarin fish (Siniperca chuatsi) infected with Flavobacterium columnare with an emphasis on immune responses. Fish Shellfish Immunol. 43, 111–119. doi: 10.1016/J.FSI.2014.12.006
Zhu K., Yu W., Guo H., Zhang N., Jiang S., Zhang D. (2018). The polymorphisms of MHCIIß gene of Trachinotus ovatus and their association with resistance/susceptibility to Photobacterium damsela. Aquaculture 485, 160–165. doi: 10.1016/J.AQUACULTURE.2017.11.045
Keywords: Trachinotus carolinus, salinity, microbiome, whole transcriptome, gill histopathology, Vibrio, Flavobacterium, immunology
Citation: Bradshaw DJ II, Perricone CS, King LE, Allmon EB, Sepúlveda M, Riche M, Wills PS, Kirchhoff N and Mejri S (2023) Commercial production of Florida pompano (Trachinotus carolinus) larvae at low salinity induces variable changes in whole-larvae microbial diversity, gene expression, and gill histopathology. Front. Mar. Sci. 10:1158446. doi: 10.3389/fmars.2023.1158446
Received: 03 February 2023; Accepted: 24 March 2023;
Published: 06 April 2023.
Edited by:
Yale Deng, University of Minnesota Twin Cities, United StatesReviewed by:
Zhenhua Ma, South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences (CAFS), ChinaCopyright © 2023 Bradshaw, Perricone, King, Allmon, Sepúlveda, Riche, Wills, Kirchhoff and Mejri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David J. Bradshaw II, ZGJyYWRzaGF3MjAxNUBmYXUuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.