
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 11 April 2023
Sec. Marine Molecular Biology and Ecology
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1156831
This article is part of the Research Topic Spiralian Genomics in a Changing World View all 5 articles
Since the Industrial Revolution, the concentration of atmospheric carbon dioxide (CO2) due to anthropogenic activities has increased at unprecedented rates. One-third of the atmospheric anthropogenic CO2 emissions are dissolved in the oceans affecting the chemical equilibrium of seawater, which in turn leads to a decrease in pH and carbonate ion (CO32-) concentration, a phenomenon known as ocean acidification (OA). This chemical disequilibrium can be detrimental to marine organisms (e.g., mollusks) that fabricate mineralized structures based on calcium carbonate (CaCO3). Most studies on the effect of reduced pH in seawater have been conducted on the early developmental stages of shell-building invertebrates, given less attention to how adult individuals face OA stress. Here, we evaluate histological, secretory, and transcriptional changes in the mantle of adult oysters (Crassostrea gigas) exposure to ambient (8.0 ± 0.2) and reduced (7.6 ± 0.2) pH during 20 days. Most histological observations did not show differences in terms of mantle cell morphology. However, Alcian Blue/PAS staining revealed significant differences in the number of Alcian Blue positive cells in the mantle edge, suggesting a decrease in the secretory activity in this morphogenetic zone. Transcriptomic analysis revealed 172 differentially expressed genes (DEGs) between mantle tissues from adult oysters kept in normal and reduced pH conditions. Almost 18% of the DEGs encode secreted proteins that are likely to be contributing to shell fabrication and patterning. 17 of 31 DEGs encoding secreted proteins correspond to oyster-specific genes, highlighting the fact that molluscan shell formation is underpinned by a rapidly evolving secretome. The GO analysis of DEGs encoding secreted proteins showed that they are involved in the cellular response to stimulus, response to stress, protein binding, and ion binding, suggesting these biological processes and molecular functions are altered by OA. This study demonstrates that histology and gene expression profiling can advance our understanding of the cellular and molecular mechanisms underlying adult oyster tolerance to low pH conditions.
Since the beginning of the Industrial Revolution, carbon dioxide (CO2) levels in the atmosphere have been increasing from 280- to 400-ppmv due to human activities (Falkowski et al., 2000). Most global CO2 emitted from anthropogenic activities is eventually absorbed by the oceans, producing a progressive increase in ocean inorganic carbon concentrations, which in turn leads to changes in the carbonate chemistry through a process called ocean acidification (OA) (Caldeira and Wickett, 2003; Doney et al., 2009). Consequently, the acid-base chemistry of seawater tends towards more acidic conditions, reducing the pH and saturation state of carbonate minerals (Doney et al., 2009; Hurd et al., 2019). The subsequent increase in carbonic acid reduces the availability of carbonate ions (CO32-) in oceans, thus affecting marine organisms that use calcium carbonate (CaCO3) as building blocks to fabricate their shells (Orr et al., 2005; Fabry et al., 2008; Guinotte and Fabry, 2008; Gazeau et al., 2013; Hurd et al., 2019). Ocean acidification has been reported to affect ocean biota, but the effects of reduced pH depend greatly on the marine organisms studied (Ries et al., 2009). For instance, some marine organisms are resilient and can acclimate to OA (Kroeker et al., 2010; Munday et al., 2011; McCulloch et al., 2012), while most shell-building invertebrates, including corals, echinoderms, and mollusk face trouble fabricating their shells in the presence of elevated CO2 levels and reduced pH in seawater (Gazeau et al., 2007; Kurihara et al., 2007; Miller et al., 2009; Talmage and Gobler, 2009; Todgham and Hofmann, 2009; Albright et al., 2010; Brennand Sheppard et al., 2010; Gazeau et al., 2010; Albright and Langdon, 2011; Parker et al., 2011; Gazeau et al., 2013; Parker et al., 2013; Bhadury, 2015; Doney et al., 2020). Accumulating evidence suggests that mollusks find it more difficult to deposit their CaCO3 shells and suffer a range of negative impacts including smaller-sized embryos and larvae, decreased shell thickness, increased larval developmental time, shell abnormalities, and reduced survival and metamorphosis rates (Beniash et al., 2010; Parker et al., 2013; Ko et al., 2014). This has been used to argue that the early developmental stages of mollusks are more sensitive to ocean acidification than the late (juvenile/adult) stages (Fabry et al., 2008; Ross et al., 2011; Gazeau et al., 2013; Parker et al., 2013; Przeslawski et al., 2015; Bhadury, 2015).
The adult mollusk shell is a remarkably stable organo-mineral biocomposite, in which the mineral phase (CaCO3) makes up 95-99% by weight and the remainder is composed of organic macromolecules such as proteins, polysaccharides, and lipids (Marin et al., 2012). Molluscan shells are assembled extracellularly and are the result of the secretory activity of the outer epithelium of the mantle (Kocot et al., 2016; McDougall and Degnan, 2018), which consists of highly specialized cell types (Sud et al., 2002; McDougall et al., 2011; Budd et al., 2014) and is divided into distinct morphogenetic zones (Jolly et al., 2004; Takeuchi and Endo, 2005; Jackson et al., 2006; Jackson et al., 2007); each responsible for the secretion of shell matrix proteins (SMPs) that influences the formation of specific shell layers (Takeuchi and Endo, 2005; Gardner et al., 2011; Marie et al., 2012; Herlitze et al., 2018). Therefore, the control of specific shell layers is provided by the coordinated expression of a suite of genes encoding secreted proteins, the so-called “mantle secretome” (Jackson et al., 2006; Kocot et al., 2016; Aguilera et al., 2017). Previous studies that analyzed the effect of OA on late developmental stages (juvenile/adult) found a reduction in the calcification rate, growth, strength, and increased dissolution of molluscan shells (Parker et al., 2013; Avignon et al., 2020). However, it remains unclear how OA affects the cellular morphology, secretory activity, and gene expression profiles of the outer epithelium of the mantle tissue in adult mollusks.
Given the different physiological, behavioral, and habitat requirements that exist between early (embryonic/larval) and late (juvenile/adult) stages in mollusks, the effects of OA are likely to differ depending on their stage of development (Parker et al., 2013; Ramos-Silva et al., 2022). Therefore, a thorough understanding of the mechanisms that control cellular and molecular response to low pH, at different developmental stages, is needed to uncover the sensitivity of marine organisms exposed to OA. Here, we evaluate histological, secretory, and transcriptional changes in the mantle tissues of adult oysters (Crassostrea gigas) exposed to seawater at a pH of 8.0 ± 0.2 and 7.6 ± 0.2 for 20 days to determine the underpinning molecular mechanisms and physiological responses of oysters coping with environmental pH variations, and thus to provide an integrated view of the cellular response and molecular pathways affected by OA.
Ethical review and approval were not required for the animal study because for research on oysters is not required under Chilean research legislation law. Adult oysters (Crassostrea gigas) were purchased from a culture farm located in Coliumo Bay (36.5431°S, 72.9589°W), Concepcion, Chile. Oceanographic parameters of Coliumo Bay (i.e., natural pH, salinity, and temperature) were measured in situ using a waterproof 4-in-1 pH/EC/TSD/Temperature tester (MW804, Milwaukee). Natural pH, salinity, and temperature values were 8.1, 35‰, and 16°C, respectively. The same parameters were used for the acclimation of adult oysters in the laboratory for subsequent experiments. Adult oysters were acclimated in 30L tanks with artificial seawater (Instant Ocean Salt) adjusted to 35‰ and constant aeration for two weeks before starting exposure experiments (Figure 1A). After acclimation, adult oysters were randomly divided into ten individual 2L flasks with artificial seawater at 35‰ (Figure 1A). Two pH treatment levels (pH 7.6 ± 0.2 and pH 8.0 ± 0.2) were targeted with five replicate tanks per treatment (Figure 1A). The normal pH seawater was produced by bubbling individual tanks with atmospheric air through air diffusers, whereas low pH seawater was made with a system based on the controlled injection of CO2 to produce CO2-enriched air until the desired pH value (7.6 ± 0.2). These pH levels were chosen because encompass potential mean oceanic pH levels estimated for current oceans (pH 8.0) and reflect atmospheric CO2 for future oceans (pH 7.6) predicted by the Intergovernmental Panel on Climate Change IPCC for the year 2100 (Caldeira and Wickett, 2005).
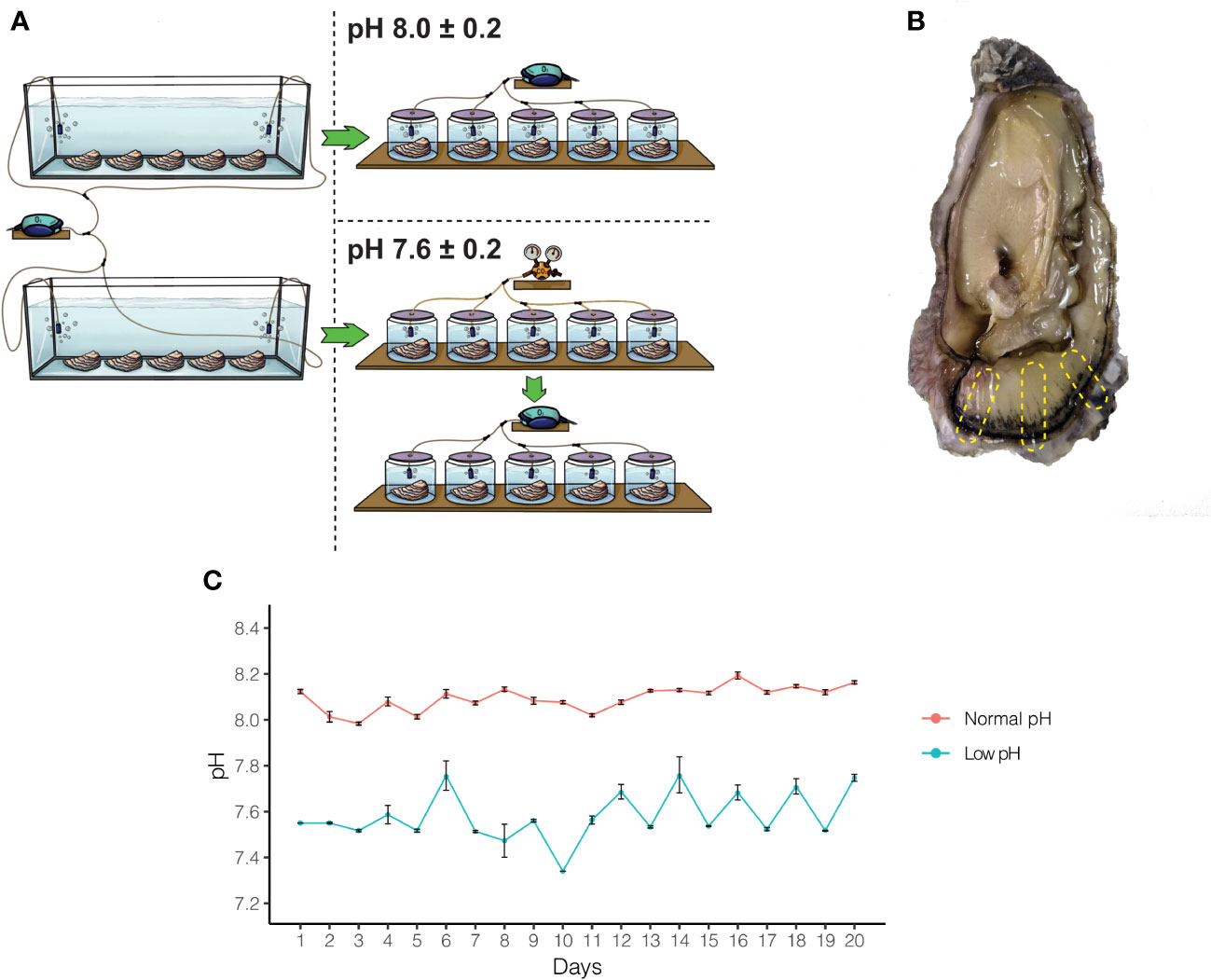
Figure 1 Experimental design and pH measurement to study the effect of OA on adult oysters. (A) A total of ten oysters were acclimated for two weeks and then split into two experimental treatments. After acclimation, five oysters were kept under normal pH (8.0 ± 0.2) and low pH (7.6 ± 0.2) conditions for 20 days. (B) After the experimental period, oysters were sacrificed, and different regions of the mantle tissue were removed to conduct histological and transcriptome analyses. (C) pH on each experimental flask was measured three times daily for 20 days.
Artificial seawater was renewed every day with pre-bubbled seawater that had been equilibrated previously, according to the experimental trials (pH 8.0 ± 0.2 or pH 7.6 ± 0.2). The pH was measured during each seawater renewal with a pH meter (MW804, Milwaukee). During the acclimation and experimental period, adult oysters were fed daily with a commercial algal mix (PhytoBlast, Continuum), containing a mixture of Nannochloropsis sp., Tetraselmis sp., Pavlova sp., and Thalassiosira sp. microalgae. After 20 days of cultivation, 10 individuals (6 for histology and secretory assays, and 4 for transcriptomic analysis) were sacrificed. Before the dissection of the mantle tissues, animals were relaxed in 1M MgCl2 in filtered artificial seawater.
Samples from different regions of the mantle tissue (Figure 1B) were dissected and fixed in 4% paraformaldehyde in PBS for general histology. Tissue samples were embedded in paraffin and sectioned at 3-5µm using a microtome (Microm HM325, Thermo Scientific). Before histological staining, tissue sections were treated with xylene to remove paraffin, rehydrated through a graded ethanol series, and running tap water. To characterize mantle histology, tissue sections were stained using the hematoxylin and eosin (H&E) staining kit (ab245880, abcam) following the manufacturer’s instructions. To characterize the cellular nucleus, tissue sections were stained with toluidine blue as described by Sridharan and Shankar (2012). Briefly, dehydrated tissue sections were treated with 0.1% toluidine blue for 30 minutes, and staining excess was removed through three washes with distilled water. Finally, tissue sections were dehydrated at 60°C to remove distilled water before mounting.
To characterize connective and muscular tissues in the mantle, we used the protocol described by Masson (1929). Briefly, tissue sections were incubated in Bouin solution at 60°C, and Bouin excess was removed using 70% ammoniac-alcohol solution and washed using running tap water. After washing, tissue sections were stained with Weigert’s Hematoxylin for 30 minutes and differentiation was performed using a 70% acid-alcohol solution. Tissue sections were then plunged in Biebrich Scarlet for 3 minutes and treated with phosphotungstic and phosphomolybdic acids for 3 minutes for allowing the binding of aniline blue. Tissue sections stained with aniline blue were differentiated using 1% acetic acid and dehydrated at 60°C before mounting.
After staining procedures, tissue sections were dehydrated using xylene and mounted using DPX mounting media (06522, Sigma). Tissue sections were observed under a microscope Axiolab 5 (Zeiss) and images were processed using ImageJ v1.53 (Schneider et al., 2012) for adjusting contrast and lightness.
To identify possible secretory cells in the mantle tissue, the histochemical procedure based on the periodic acid-Schiff stain (PAS) and Alcian Blue (AB) was used for neutral and acid mucopolysaccharide (mucin) staining, respectively. We used the AB/PAS staining kit (ab245876, abcam) following the manufacturer’s instructions, with some modifications. Briefly, tissue sections were plunged into 3% acetic acid for 2 minutes; the acetic acid excess was rinsed, and tissue sections were incubated in AB (pH 2.5) for 30 minutes. Staining excess was removed using running tap water and distilled water; then, tissue sections were plunged in the periodic acid solution for 5 minutes, washed in distilled water, and incubated in Schiff’s solution for 20 minutes. Finally, tissue sections were stained using Mayer’s Hematoxylin for 2 minutes, rinsed with running tap water, dehydrated using a graded ethanol series followed by xylene, and mounted with DPX mounting media (06522, Sigma). Digital images were captured using a microscope Axiolab 5 (Zeiss) equipped with an Axiocam 208 color camera (Zeiss). Images were processed using ImageJ v1.53 (Schneider et al., 2012) for adjusting contrast and lightness.
To calculate the number of secretory cells in mantle tissue exposed to normal and low pH, we used light microscopy at 40X magnification and went through the outer mantle epithelium to identify AB-positive cells. These cells were counted and based on their location along the outer mantle epithelium were classified as secretory cells belonging to the mantle edge or mantle pallial zone. This procedure was performed using six individuals, and for analysis, three sections from the mantle were selected as shown in Figure 1B. The number of AB-positive cells was compared between morphogenetic zones from the six individuals that were kept under normal and low pH conditions. A t-test was used to evaluate the difference between groups with a significance of p<0.05. Statistical analysis was performed using R v4.1.3 (R Development Core Team, 2022) and plots were made using ggplot (Wickham, 2016).
After relaxation, whole mantle tissues, encompassing the mantle edge and mantle pallial regions, were dissected, and stored immediately at -80°C until further RNA extraction. Total RNA was extracted from mantle tissues of adult oysters kept under normal and low pH conditions with TriReagent (T9424, Sigma) following a modified protocol described by Gao et al. (2001) to remove inhibitory pigments. RNA was quantified using a NanoDrop™ Lite Spectrophotometer (Thermo Scientific) and shipped to Macrogen (Korea) to perform cDNA library preparation and sequencing in an Illumina NovaSeq6000 paired-end mode.
Sequencing quality was performed using FastQC v0.11.9 (Andrews, 2010), and reads were trimmed with Trim Galore! v0.67 (Krueger, 2012). High-quality reads were mapped to the C. gigas genome v9 (accessed at http://gigadb.org/dataset/100030) (Zhang et al., 2012) using STAR v2.6.1a_08-27 (Dobin et al., 2013), and those reads that mapped to transcripts were counted with featureCounts v1.6.3 (Liao et al., 2014). The differences between each pair of replicates were quantified by Pearson’s correlation coefficient, calculated across all genes with non-zero count in at least one replicate. Counted reads were also used for identifying differentially expressed genes (DEGs) using edgeR v3.36.0 (Robinson et al., 2010) and an FDR < 0.05 and an absolute value of log2-fold change ≥ 2. DEGs were visualized as volcano plot using the VolcaNoseR web app (Goedhart and Luijsterburg, 2020). Gene Ontology (GO) annotation of C. gigas gene models was performed following the Trinotate pipeline (Haas et al., 2013). Those GO terms associated with DEGs were analyzed using WEGO v2.0 (Ye et al., 2018). In addition, we assigned the KO (KEGG Orthology) number for DEGs through BlastKOALA web service and the family eukaryotes KEGG GENES database (Kanehisa et al., 2016).
Proteins encoded by those genes identified as differentially expressed were classified as cytosolic, secreted, or transmembrane following the procedure described by Aguilera et al. (2017). Briefly, C. gigas proteins were searched for signal peptides using a local installation of SignalP v4.1 (Petersen et al., 2011), with the following parameters: -s best -t euk -u 0.45 -U 0.50. Proteins predicted as non-secretory were classified as cytosolic proteins. Signal peptide-positive proteins were additionally screened for transmembrane domains using a local installation of THMHH v2.0c (Krogh et al., 2001), or for mitochondrial, Golgi, or lysosomal targeting using the TargetP v2.0 web server (Emanuelsson et al., 2007). Positive proteins were classified as transmembrane or organelle targeting, respectively. Transcript counts (normalized using TMM transformation in edgeR) from DEGs encoding secreted proteins were used to generate a heatmap for visualization of differentially expressed genes with the ClustVis web tool (Metsalu and Vilo, 2015). GO terms associated with DEGs that encode secreted proteins were analyzed using WEGO v2.0 (Ye et al., 2018).
DEGs encoding secreted proteins were further investigated whether they (i) were likely to encode shell matrix proteins (SMPs) based upon similarity to proteins that had previously been identified from C. gigas shells, or they had transcriptional regulatory or signaling activity roles. Similarity to SMPs was performed by BLASTP searchers (Camacho et al., 2009) against an in-house database of published proteins that had previously been identified from C. gigas larval and adult shells (Marie et al., 2011; Zhang et al., 2012; Zhao et al., 2018), using an e-value of 1e-100. As many SMPs contain repetitive low-complexity domains (RLCDs) (Jackson et al., 2010; McDougall et al., 2013; Aguilera et al., 2017), BLASTP searches were conducted without filtering for low-complexity regions and without compositional adjustment. Potential transcription factor or signaling activity was ascertained by searching GO term annotations for GO:000370 (DNA-binding transcription factor activity), or the word “signal” in the GO description.
Ocean acidification is a concerning and global-scale phenomenon with negative impacts on diverse animal groups that form calcified structures based on CaCO3, being mollusks one of the groups most affected (Gazeau et al., 2007; Gazeau et al., 2013). Many studies have shown these negative impacts in the early-life stages of mollusk (Parker et al., 2013); however, the impact of ocean acidification on adult individuals has not been well explored yet. To achieve a better understanding of the effects of OA in adult mollusks, we maintained adult oysters under normal (8.0 ± 0.2) and low (7.6 ± 0.2) pH for 20 days (Figure 1C). To evaluate the effects of low pH, we employed an integrative approach, combining histological and transcriptomic analyses, to investigate the cellular and molecular responses of adult mollusks exposure to OA condition.
The mantle is composed of epithelia, loose stroma, muscular, adipose, and connective tissues (Figure 2A). Histological observations show that epithelial cells are closely aligned to form the epidermis of the mantle that extends into the connective tissue (Figures 2B, C). H&E and toluidine blue staining reveal numerous invaginations along the outer mantle epithelium covering the morphogenetic zones known as mantle edge and mantle pallial (Figure 2A and Supplementary Figure 1). The inner fold is similar in terms of length and shape compared to the other mantle folds (Figure 2A and Supplementary Figure 1). The outer fold is composed of a columnar epithelium (Figures 2B, D), whereas the middle fold is characterized by a pseudostratified columnar epithelium, with nuclei at different heights (Figure 2D). This epithelial transition in the periostracal groove may be relevant because this mantle region is responsible for the secretion of the proteinaceous periostracum layer (McDougall et al., 2011; Marin et al., 2012; Kocot et al., 2016).
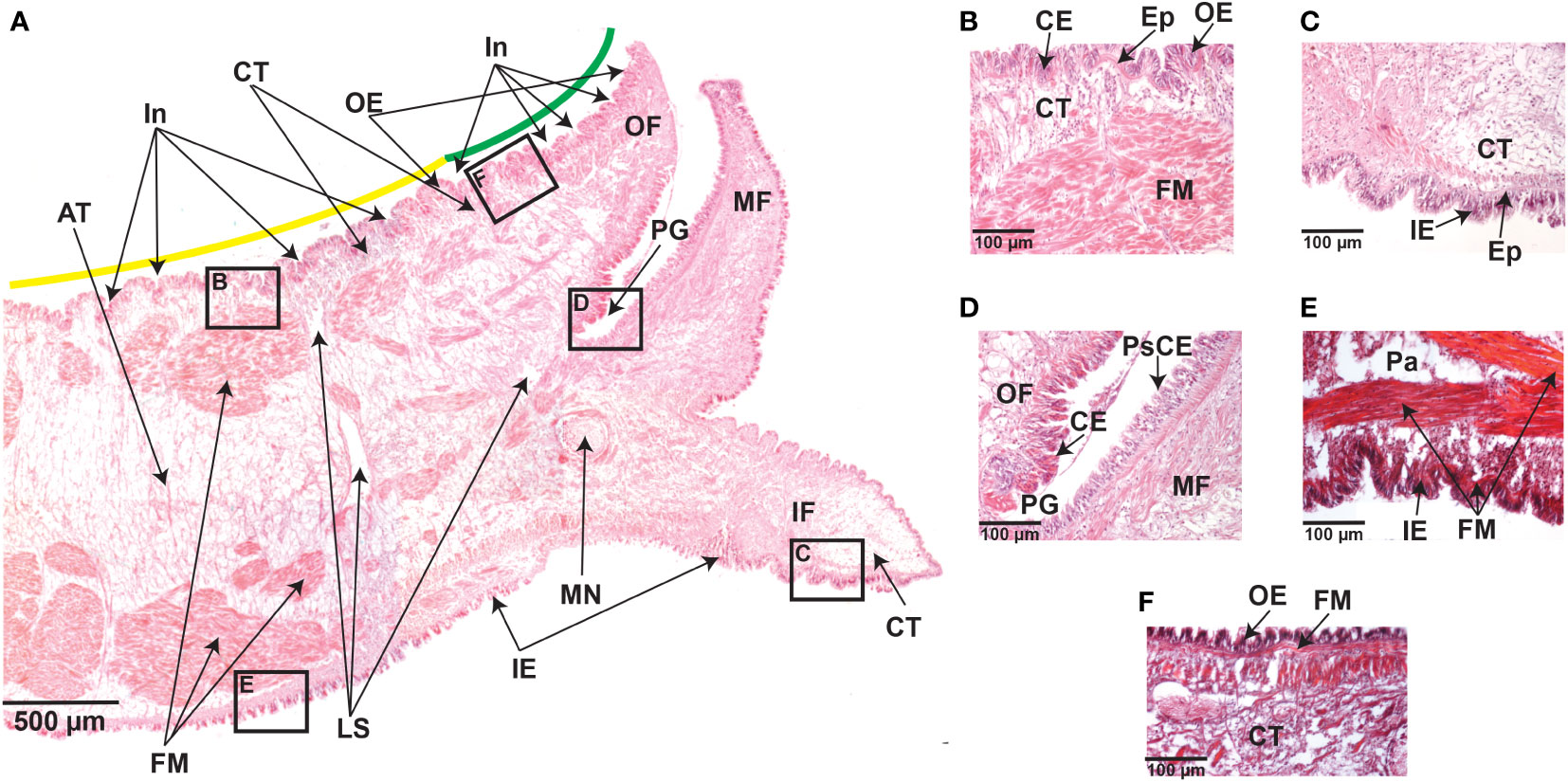
Figure 2 Transverse sections of the mantle tissue of C. gigas adult oyster were kept under normal pH (8.0 ± 0.2) conditions. (A) Hematoxylin and Eosin-stained section through the mantle depicting various structures, including the outer epithelium (OE), outer fold (OF), periostracal groove (PG), middle fold (MF), inner fold (IF), the inner epithelium (IE), mantle nerve (MN), loose stroma (LS), fibers muscle (FM), adipose tissue (AT), and connective tissue (CT). Along the outer epithelium (OE), there are several invaginations covering the mantle zones known as mantle edge (green line) and mantle pallial (yellow line). Scale bar = 500µm. (B) Higer magnification of the boxed area in A, showing the outer epithelium (OE) and the presence of aligned columnar epithelial (CE) cells forming the epidermis, which extends into the connective tissue (CT). Hematoxylin and Eosin staining. Scale bar = 100µm. (C) Higher magnification of the boxed area in A, showing aligned columnar epithelial (CE) cells that form the epidermis (Ep) extending into the connective tissue (CT) in the inner epithelium (IE). Hematoxylin and Eosin staining. Scale bar = 100µm. (D) Higher magnification of the boxed area in A, showing the base of the periostracal groove (PE) depicting the presence of columnar epithelium (CE) on the outer fold (OF) side, but not on the middle fold (MF) side that is characterized by a pseudostratified columnar epithelium (PsCE) with nuclei at different heights. Hematoxylin and Eosin staining. Scale bar = 100µm. (E) Higher magnification of the boxed area in A, depicting bundles of fibers muscle (FM) that underlie the inner epithelium (IE) and near the parenchyma (Pa). Trichome-Masson staining. Scale bar = 100µm. (F) Higher magnification of the boxed area in A, showing the presence of fibers muscle (FM) underlying the outer epithelium (OE). Trichome-Masson staining. Scale bar = 100µm.
The mantle musculature consists of bundles of fibers muscle located in the parenchyma and underneath the outer and inner epithelia (Figures 2A, E,F). Within the parenchyma, numerous long muscle fibers are densely organized and longitudinally distributed, with apparently no striated (Figure 2E). The outer epithelium adjoins a layer of longitudinal muscles, without apparent striated, embedded in connective tissue (Figure 2F). These histological observations were similar in the mantle tissues from adult oysters kept in low pH conditions (Supplementary Figures 2 and Supplementary Figure 3), suggesting that ocean acidification does not affect mantle tissue anatomy and cell morphology, at least for 20 days of exposure to reduced (7.6 ± 0.2) pH.
To identify between secretory and non-secretory cells in the mantle tissue, we used AB-PAS staining technique, in the same fashion as previous studies in mollusks (Lee et al., 2014; Wang et al., 2022a; Wang et al., 2022b). It is worth mentioning that our approach does not discriminate among difference types of secretory cells. We can only ensure that AB-PAS positive cells correspond to cells containing mucins, which are glycosylated proteins involved in controlling mineralization (Midura and Hascall, 1996; Marin et al., 2000; Boskey, 2003; Marin et al., 2007; Petrou and Crozier, 2018), including shell formation (Marin et al., 2000; Marin et al., 2007). Staining with AB at pH 2.5 show that acidic mucins (stained in blue) produced by mucin(s)-secreting cells are distributed at the base of the epithelium of the three folds of the mantle (Figures 3A, B). These secretions are equally evident in outer and inner epithelia compared to the epithelium lining the middle fold (Figures 3A, B). Mucin(s)-secreting cells are observed in the mantle edge and mantle pallial zones of adult oysters maintained at normal (8.0 ± 0.2) pH, but with a high concentration of those cells lining the outer epithelium of the mantle and the near connective tissue (Figure 3A). In contrast, there is a low number of mucin(s)-secreting cells in the mantle tissues of adult oysters kept at a low (7.6 ± 0.2) pH condition (Figure 3B). We did not observe any cells containing AB-positive signal in the connective tissue of adult oysters kept at a reduced (7.6 ± 0.2) pH condition (Figure 3B). Remarkably, the difference in terms of the number of secretory cells is related to a particular morphogenetic zone of the mantle (n = 54; p 2.2e-16) (Figure 3C), suggesting a low capacity to secrete glycosylated proteins in the mantle edge region in adult oysters kept under reduced (7.6 ± 0.2) pH condition for 20 days. Given that different secretory repertoire control the formation of prismatic and nacreous layers (Takeuchi and Endo, 2005; Gardner et al., 2011; Marie et al., 2012), our results indicate that an ocean acidification condition of 7.6 ± 0.2 pH for 20 days has negative effects in the secretory activity of the mantle edge, affecting potentially its ability to secrete macromolecules that might be part of the organic matrix of the foliated layer in C. gigas.
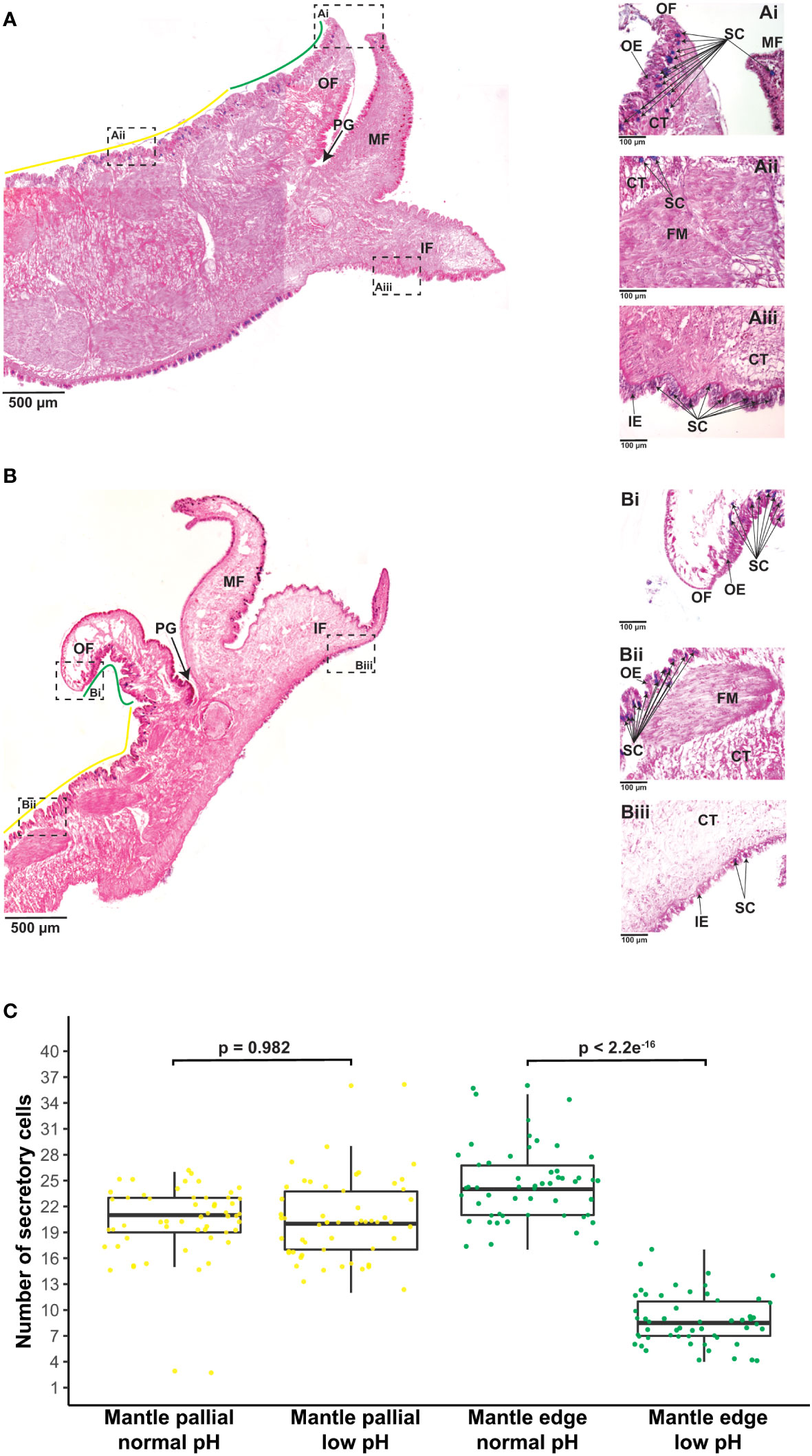
Figure 3 Effect of ocean acidification on the secretory activity of mantle cells. (A) Example of Alcian Blue-stained section through the mantle tissue of one adult individual of C. gigas kept under normal pH (8.0 ± 0.2) condition showing the mantle edge (green line) and mantle pallial (yellow line) morphogenetic zones. Scale bar = 500µm. In addition, three mantle regions are shown to indicate: (Ai) A region of the outer fold belonging to the mantle edge zone, highlighting secretory cells in blue. Scale bar = 100µm. (Aii) A region of the outer epithelium belonging to the mantle pallial zone, showing secretory cells in blue. Scale bar = 100µm. (Aiii) A region of the inner fold that shows secretory cells in blue. Scale bar = 100µm. (B) Example of Alcian Blue-stained section through the mantle tissue of one adult individual of C. gigas kept under low pH (7.6 ± 0.2) condition showing the mantle edge (green line) and mantle pallial (yellow line) morphogenetic zones. Scale bar = 500µm. Three mantle regions are also shown to indicate: (Bi) A region of the outer fold belonging to the mantle edge zone, highlighting secretory cells in blue. Scale bar = 100µm. (Bii) A region of the outer epithelium belonging to the mantle pallial zone, showing secretory cells in blue. Scale bar = 100µm. (Biii) A region of the inner fold that shows secretory cells in blue. Scale bar = 100µm. OF, outer fold; PG, periostracal groove; MF, middle fold; IF, inner fold; OE, outer epithelium; CT, connective tissue; FM, fibers muscle; IE, inner epithelium; SC, secretory cells. (C) Number of secretory cells along the outer epithelium covering the mantle edge and mantle pallial zones in adult oysters kept under normal and low pH conditions. Yellow dots indicate secretory cells found in the mantle pallial zone, whereas green dots correspond to secretory cells in the mantle edge zone.
To obtain a comprehensive snapshot of the molecular changes influenced by OA, we conducted transcriptome sequencing of adult mantle tissues from oysters exposed to normal (8.0 ± 0.2) and low (7.6 ± 0.2) pH. Raw transcriptome data are publicly available under the following NCBI BioProject PRJNA904561 and read mapping statistics to the C. gigas genome (Zhang et al., 2012) are shown in Supplementary Table 1. In general, RNA-Seq replicates show high Pearson correlation coefficients (Supplementary Figure 4). Genes significantly differentially expressed in mantle tissues under different pH conditions were identified using edgeR. We found 172 genes with differential expression profiles, of which 78 genes showed a down-regulation under low pH conditions, while 94 genes are over-expressed by ocean acidification (Figure 4A and Supplementary Figure 5). Given the low number of differentially expressed genes (172 in total), we were unable to find any enriched GO term. Nevertheless, most GO terms associated with up-regulated genes (>50%) belong to binding (GO:0005488), biological regulation (GO:0065007), regulation of biological processes (GO:0050789), response to stimulus (GO:0050896), and cellular process (GO:0009987) (Supplementary Figure 6). While most down-regulated genes (>50%) are related to the catalytic activity (GO:0003824), binding (GO:0005488), cellular process (GO:0009987), and metabolic process (GO:0008152) (Supplementary Figure 7). In addition, we conducted KEGG annotation to provide a deeper functional understanding of DEGs. KEGG pathway analysis indicates that 25 out of 94 up-regulated genes were assigned to eight categories (Supplementary Figure 8), highlighting the presence of genes involved in carbohydrate metabolism and environmental information processing (Supplementary Figure 8 and Supplementary Table 2); whereas down-regulated genes are mainly assigned to protein families: genetic information processes (Supplementary Figure 8).
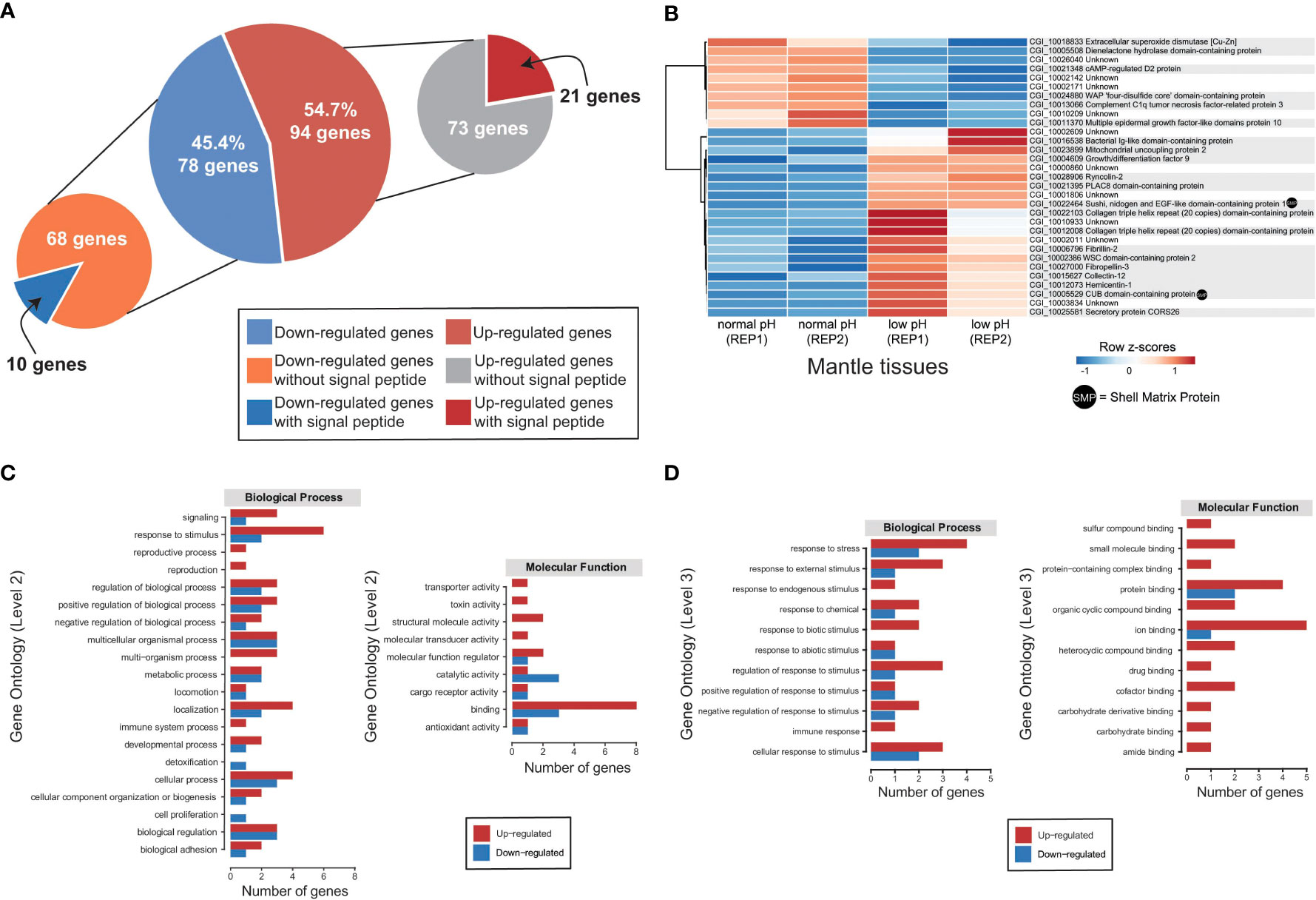
Figure 4 Transcriptomic response of mantle cells of oysters exposed to ocean acidification condition. (A) A total of 172 genes were identified as differentially expressed in adult oysters reared under normal (8.0 ± 0.2) and low (7.6 ± 0.2) pH conditions and placed into two categories according to the presence or absence of a signal sequence. (B) Heatmap of significantly differentially expressed genes encoding secreted proteins. At the bottom of each column is depicted the mantle tissue and the corresponding pH condition used to generate the RNA-Seq data. Genes are displayed as horizontal rows clustered by similar expression profiles, represented by the dendrogram to the left of the heatmap. Red indicates higher expression, while blue illustrates lower expression. Gene annotations are indicated to the right of the heatmap, and gene products (proteins) that have been previously identified as part of the organic matrix of larval/adult shells are indicated by a black dot. (C) Distribution of differentially expressed genes encoding secreted proteins with annotated biological process GO terms (Level 2) and molecular function GO terms (Level 2). See Supplementary Table 4 for gene name information. (D) Distribution of differentially expressed genes encoding secreted proteins with annotated biological process GO terms (Level 3) and molecular function GO terms (Level 3). See Supplementary Table 5 for gene name information.
Based on the presence of conserved signal sequences, we estimate that 31 of 172 genes show differential expression between normal and decreased pH conditions (Figure 4A), of which 21 have associated Swissprot or PFAM annotations (Figure 4B and Supplementary Table 3). No specific GO term (biological process or molecular function) is over-represented in this dataset, most likely due to the low number of annotated genes. Furthermore, two DEGs encoding secreted proteins have previously been reported as SMPs in C. gigas (Figure 4B and Supplementary Table 3).
Ten differentially expressed genes encoding secreted proteins are downregulated in the mantle tissues of oysters reared at low pH (Figure 4B). Six differential expressed genes with low expression at low pH condition have Swissprot or PFAM annotations (Supplementary Table 3). Down-regulated genes include extracellular superoxide dismutase [Cu-Zn], complement C1q tumor necrosis factor-related protein 3, cAMP-regulated D2 protein, and multiple epidermal growth factor-like domains protein 10 (Figure 4B and Supplementary Table 3). By contrast, 21 DEGs encoding secreted proteins are upregulated in the mantle tissues of oyster exposure to low pH (Figure 4A), and two (Sushi, nidogen and EGF-like domain-containing protein 1 and CUB domain-containing protein) are like known SMPs (Figure 4B and Supplementary Table 3). Aside from SMPs, 13 genes have Swissprot or PFAM annotations (Figure 4B and Supplementary Table 3). Up-regulated genes include ryncolin-2, fibrillin-2, mitochondrial uncoupling protein 2, collectin-12, fibropellin-3, growth/differentiation factor 9, hemicentin-1, secretory protein CORS26, and WSC domain-containing protein 2 (Figure 4B and Supplementary Table 3).
The analysis of those DEGs encoding secreted proteins reveals several functional domains associated with cys-rich proteins (PLAC8 motif-binding domain), extracellular structural proteins (collagen triple helix repeat, CUB domain, F5/8 type C domain, fibrinogen beta and gamma chains C-terminal globular domain, VWA domain containing CoxE-like, von Willebrand factor type A domain, nidogen-like, AMOP domain, sushi repeat), immune proteins (bacterial Ig-like domain, C1q domain, IgGFc domain), carbohydrate-binding proteins (WSC domain, lectin C-type domain), calcium-binding proteins (complement Clr-like EGF-like, calcium-binding EGF domain, EGF domain, human growth factor-like EGF, EGF-like domain), substrate carrier proteins (mitochondrial carrier domain), chitin-binding proteins (chitin binding Peritrophin-A domain), and enzymes (dienelactone hydrolase, WAP ‘four-disulfide core’, carboxylesterase, BD-FAE, abhydrolase 3 alpha/beta hydrolase fold, copper/zinc superoxide dismutase) (Supplementary Table 3).
The GO analysis revealed that most DEGs (>50%) encoding secreted proteins are involved in binding (GO:0005488), response to stimulus (GO:0050896), localization (GO:0051179), cellular process (GO:0009987), and biological regulation (GO:0065007) (Figure 4C and Supplementary Table 4). In the binding category, six genes are likely to have protein binding function (GO:0005515) and the same number of genes seem to have ion binding (GO:0043167) function (Figure 4D and Supplementary Table 5). Whereas in the response to stimulus category, six genes are associated with response to stress (GO:0006950) and five genes with cellular response to stimulus (GO:0051716) (Figure 4D and Supplementary Table 5). Most genes related to protein binding, ion binding, response to stress, and cellular response to stimulus are upregulated in mantle cells of oysters kept at 7.6 pH (Figures 4C, D), suggesting expression shifts in these genes could be responsible for regulating pH in response to OA. Notably, three genes are found to be downregulated in mantle cells exposed to OA that exhibit catalytic functions (Figure 4C and Supplementary Table 4).
Four genes (fibrillin-2, secretory protein CORS26, collectin-12, and complement C1q tumor necrosis factor-related protein 3) with potential signaling functions and encoding secreted proteins are differentially expressed (Figure 4B and Supplementary Table 3). Three of them fibrillin-2, secretory protein CORS26, and collectin-12) are upregulated in the mantle cells of oysters reared at low pH, while complement C1q tumor necrosis factor-related protein 3 is downregulated. This result suggests a putative role of these signaling genes in the genetic regulatory network that affects OA; even though their functions are still unstudied in mollusks. No specific transcriptional regulatory genes encoding secreted proteins were found as differentially expressed (Supplementary Table 3).
We conducted an integrative analysis of histological and transcriptomic data in the mantle tissue of Pacific oyster (Crassostrea gigas) exposure to normal (8.0) and low (7.6) pH conditions to elucidate how oysters cope with OA and examine the cellular and molecular underpinnings of this response. At the cellular level, oysters maintained in low pH conditions do not show significant differences in terms of mantle tissue anatomy, although the secretory activity of the mantle cells is diminished by OA. At the molecular level, C. gigas exhibited a subtle transcriptomic response to OA, and surprisingly few shifts occur in terms of gene expression.
Molluscan mantle epithelia have long been studied to understand the biological control of shell formation, as reviewed by Suzuki and Nagasawa (2013) and Kocot et al. (2016). Mantle tissue encloses all internal organs and is organized into different regions namely mantle edge and mantle pallial, with the mantle edge in direct contact with the prismatic shell layer and the mantle pallial with the nacreous layer (Kocot et al., 2016; Clark, 2020). Within the mantle edge, there is a series of folds with the inner and middle folds involved in water inflow and sensory functions, respectively, whilst the outer fold is involved in shell secretion (Clark, 2020; Clark et al., 2020). Most bivalves have three mantle folds (Suzuki and Nagasawa, 2013; Clark et al., 2020), although there are exceptions where the outer fold is duplicated (Waller, 1980) or the inner fold is fused (Sleight et al., 2016). In bivalves, the shell varies greatly in terms of shape, color, size, and thickness, which might be related to the different morphologies of the mantle tissue (Parvizi et al., 2017). The mantle of C. gigas possesses many similarities to that of pearl oysters (Pinctada spp.), including a single periostracal groove, three folds in the mantle edge, and an outer fold epithelium encompasses mainly columnar epithelial cells (Dix, 1972; Jabbour-Zahab et al., 1992; Parvizi et al., 2017; Zhu et al., 2021; Han et al., 2022). However, the structure of the periostracal groove is somehow different in that two cell types are distinguishable in C. gigas, columnar cells to the base and planar cells to the tip of the periostracal groove, which are not observed in pearl oysters (Jabbour-Zahab et al., 1992; Parvizi et al., 2017). Given the morphological similarities in the mantle tissue between C. gigas and pearl oysters, it is likely that the differences observed, in terms of the shape and microstructure of their shells (Farre et al., 2011; Lee et al., 2011; Marie et al., 2011; Marie et al., 2012; Mouchi et al., 2016; Checa et al., 2018; Deng et al., 2022), are the product of different shell matrix proteins secreted by the mantle in a species-specific manner (Aguilera et al., 2017), rather than as a product of mantle morphology. Inside the mantle tissue, there are contractile muscle fibers possibly involved in mantle retraction and adipose tissue containing high glycogen content (Berthelin et al., 2000; Álvarez Nogal and Molist García, 2015; Parvizi et al., 2017). It is well known that shell formation is a high-demand energy process (Palmer, 1992; Waldbusser et al., 2013; Clark et al., 2020); therefore, the use of glycogen as an energy resource is no doubt an economic method employed by mollusks to construct their shells.
Most studies evaluating the effect of OA in adult individuals have reported negative outcomes, including reduced calcification rates as well as decreased shell growth and strength, but also increased shell dissolution, reviewed by Parker et al. (2013). However, the impact of OA in terms of mantle morphology is scarce (Wei et al., 2015). We here found that the effect of reduced pH in the mantle tissue of C. gigas for 20 days was undetectable with no apparent detrimental effects on cell morphology between hypercapnic and normocapnic oysters, which is in concordance with a previous study exposing adult oysters to 28 days (Wei et al., 2015). This might indicate that hypercapnic adult individuals develop physiological strategies to allocate extra energy for pH homeostasis under a decreased pH external environment. However, this mechanism remains speculative and requires further investigation.
Ocean acidification (OA) triggered by anthropogenic activities is accompanied by an increase of CO2, which when dissolved in water becomes carbonic acid (H2CO3) (Falkowski et al., 2000; Caldeira and Wickett, 2003; Doney et al., 2009). This carbonic acid dissociates into bicarbonate (HCO3-) and a hydrogen ion (H+), the latter of which makes the water more acidic (Doney et al., 2009; Hurd et al., 2019). The fabrication of the mollusk shell involves the selective uptake of calcium (Ca2+) and bicarbonate (HCO3-) ions from the environment that are eventually transported through the outer epithelium of the mantle into the mineralization front (Jodrey, 1953; Sillanpää et al., 2016; Sillanpää et al., 2018; Sillanpää et al., 2020). The HCO3- is used as the carbon source by mollusks to produce the shell through a process during which another hydrogen ion (H+) is released, and thus consequently alters the acid-base homeostasis of the mantle cells (Ramesh et al., 2020). Therefore, it is essential to understand the cellular response and secretory activity employed by mantle cells to cope with OA.
Most mollusks possess a multi-layered shell that is formed by specialized secretion of distinct morphogenetic zones of the mantle (Kocot et al., 2016). The outermost layer, known as the periostracum, comprises proteins secreted from the periostracal groove (Zhang et al., 2006; Checa and Harper, 2010; Nakayama et al., 2013). The middle prismatic layer is thought to be controlled by genes expressed in the mantle edge region, whereas the inner nacreous layer is likely controlled by genes expressed in the mantle pallial (Takeuchi and Endo, 2005; Gardner et al., 2011; Marie et al., 2012; Herlitze et al., 2018). This spatial dichotomy in expression is corroborated by ultrastructural investigations of the mantle epithelium, indicating cellular differentiation of prism- and nacre-secreting mantle cells (Fang et al., 2008). In pearl oysters, two major secretory cells have been described – basophilic and acidophilic – in the mantle, where the former type is characterized by sulfated acidic mucosubstances (Parvizi et al., 2017). We found that most of the cells in the mantle stained blue with the combined technique of AB-PAS showing that they contain acidic mucosubstances. These AB-PAS positive cells are distributed along the outer mantle epithelium, covering the mantle edge and mantle pallial zones. These acidic mucosubstances (probably mucins) might presumably have an active role in the mineralization of the shell by aiding with the absorption or binding of calcium ions (Breedham, 1958; Kapur and Gibson, 1967; Marin et al., 2020; Álvarez Nogal and Molist García, 2015).
The mineralogical composition of adult oyster shell is mostly calcite and a small portion of aragonite in the myostracum layer, which is located at the shell inner surface in intimate connection with the adductor muscle (Lee et al., 2011; Marie et al., 2011; Mouchi et al., 2016; Checa et al., 2018). In contrast to most bivalves, C. gigas adult shell textures correspond to a shell-like foliae layer and lenses of chalk (Marie et al., 2011; Checa et al., 2018), a highly porous and poorly organized microstructure that is only found in Ostreidae (Mouchi et al., 2016; Checa et al., 2018). Recent structural and crystallographic observations have depicted that growth lines are continuous between the foliae and chalk, suggesting both layers are made simultaneously by different zones of the mantle tissue, where the foliated layer is made by the mantle edge and the chalky layer by the mantle pallial (Banker and Sumner, 2020). In oysters kept in low pH conditions, the outer epithelium of the mantle edge shows fewer AB-PAS positive cells compared with the mantle pallial zone, suggesting a decrease in the secretory activity in mantle cells underlying the foliated layer that possibly affects its formation.
Transcriptional profiling has been performed to provide a more detailed picture of the molecular events underpinning the physiological response of marine animals to OA (Strader et al., 2020). Here, we found a relatively low number of genes (172) that were significantly differentially expressed under OA, suggesting a subtle transcriptomic response to OA in C. gigas mantle cells after 20 days of low-pH exposure.
Gene ontology classification of these DEGs showed that low pH (7.6) triggers the regulation of cellular and biological processes but prominently suppresses some metabolic processes and catalytic activities. These results were consistent with previous studies on C. gigas larvae at 14-16 hours post-fertilization exposed to similar levels of low-pH stress (de Wit et al., 2018). In addition, we found that response to stimulus and immune-related genes are up-regulated in mantle tissues of oysters in the low pH group. Immune-related genes have been found down-regulated in the hemocytes of adult C. gigas, as well as in C. gigas larvae, exposed to OA (Wang et al., 2016; Wang et al., 2020; Dineshram et al., 2021), but it has also been seen that immune-related genes are up-regulated in other oysters (Goncalves et al., 2016; Goncalves et al., 2017). We also found that catalytic and oxidoreductase genes were downregulated in oysters kept in low pH conditions, which agrees with what happens before larval shell formation in C. gigas (Liu et al., 2020); but it is inconsistent with what has been observed in the adult mantle tissue of C. virginica (Downey-Wall et al., 2020). In contrast to previous studies that showed higher expression of genes involved in metabolism and oxidative stress under OA in either C. gigas larvae or hemolymph from adult oysters (Wang et al., 2016; Cao et al., 2018; Dineshram et al., 2021), we did not find such changes in mantle cells of C. gigas after exposure to low pH during 20 days. Notably, we found that genes involved in environmental information processing and carbohydrate metabolism are up-regulated by low pH, revealing the importance of polysaccharides components in molluscan shells (Marin et al., 2000; Marin et al., 2007). Together, these data, and previous studies in other marine metazoans (Strader et al., 2020), suggest that marine organisms vary dramatically in their transcriptomic responses to low pH, with differences among closely related animal groups and even between life-history stages.
To examine the effects of OA on the mantle secretory activity at the molecular level, we focused on DEGs encoding secreted proteins – the so-called mantle secretome (Jackson et al., 2006; Kocot et al., 2016; Aguilera et al., 2017) – because those proteins are likely to be embedded within the shell. Among genes encoding secreted proteins that were significantly differentially expressed, we found the presence of domains of diverse nature that cover a large set of functions. Some are collagen-like, von Willebrand factor type A, and sushi-like domains that are associated with extracellular matrix binding molecules (Marin, 2020). Von Willebrand type A domains are usually found in most - if not all - molluscan organic matrices and exhibit an adhesion function through protein-protein interactions, playing a role in 3D framework structuring (Whittaker and Hynes, 2002; Arivalagan et al., 2017; Marin, 2020). Saccharide domains comprise the WSC and lectin C-type domains. Many molluscan shell-associated proteins have lectins that are calcium-dependent but do not have a structural role as framework constituents; indeed, they may exert a function in immunity and protection (Marin, 2020). Calcium-binding domains include several kinds of EGF domains. These domains are involved in a diverse array of functions, including intracellular signaling mediated by calcium (Wouters et al., 2005). Notably, the EGF-like domain has been reported in tandem repeats in Pinctada and Crassostrea shells (Marie et al., 2011; Marie et al., 2012), and this repeating arrangement is favored during evolution due to its stability against enzyme attack through the formation of compact dimeric or tetrameric pseudo-symmetrical structures (Arivalagan et al., 2017). Domains related to immunity and protective mechanisms comprise C1q and WAP ‘four-disulfide core’ domains. The C1q domain that belongs to the C1q protein family (Ghebrehiwet et al., 2012) has been previously identified in the C. gigas shell (Arivalagan et al., 2017). WAPs are highly conserved proteinase inhibitor domains found in vertebrate and invertebrate species (Smith, 2011). WAP domain-containing proteins play major roles in gastropod shell formation (Shen et al., 1997; Treccani et al., 2006; Marie et al., 2010), inhibiting calcium carbonate crystal growth in vitro (Treccani et al., 2006). However, the function of WAP domain-containing proteins in Crassostrea is unknown.
Most secreted genes annotated as downregulated do not possess similarity with previously described Swissprot sequences, even though some of them have functional domains that give some glimpse into their putative role. Aside from unknown genes, other down-regulated genes exhibited similarity to complement C1q tumor necrosis factor-related proteins; multiple growth factor-like domains protein 10, and metabolic proteins such as cAMP-regulated D2 gene and extracellular superoxide dismutase [Cu-Zn]. Regarding annotated upregulated genes encoding secreted proteins, these include ryncolin-2, fibrillin-2, mitochondrial uncoupling protein 2, collectin-12, fibropellin-3, growth/differentiation factor 9, hemicentin-1, secretory protein CORS26, and WSC domain-containing protein 2. Although the function of most - if not all - of these genes is unknown in mollusks, there are some upregulated DEGs encoding secreted proteins exhibiting similarity with genes that have been implicated in biomineralization in other species, for example, secretory protein CORS26 (Maeda et al., 2001), fibrillin-2 (Arteaga-Solis et al., 2011; Mead et al., 2022), and hemicentin-1 (Ramos-Silva et al., 2013; Germer et al., 2015; Jackson et al., 2015). Indeed, hemicentin-1, a multimodular extracellular matrix protein that interacts with basement membrane proteins to confer tissue linkage (Zhang et al., 2022), has been reported as downregulated in C. virginica larvae exposed to low pH (Barbosa et al., 2022), but also as upregulated in C. gigas larvae and adults exposed to low pH (Timmins-Schiffman et al., 2014; Dineshram et al., 2021), as in this study as well.
Furthermore, secreted proteins involved in protein binding, ion binding, response to stress, and cellular response to stimulus were upregulated in mantle tissues of oysters kept at 7.6 pH. However, a previous study on C. gigas larvae has shown a suppression of proteins involved in response to stimulus at a 7.4 pH (Dineshram et al., 2016). Our findings are not in line with those of Dineshram et al. (2016) and strongly suggest that the effects of OA vary considerably between different life-story stages in the same species. Within ion binding GO terms, calcium ion binding genes (fibrillin-2, hemicentin-1, and sushi, nidogen and EGF-like domain-containing protein 1) were positively regulated by OA. The stimulation of calcium-related genes was shown in C. gigas larvae to survive and develop under OA but at a slower rate (de Wit et al., 2018). In a previous study in adult C. gigas but focusing on the hemocytes response, a series of calcium-binding genes and calcium signaling pathways were also upregulated in response to OA indicating the regulation of calcium homeostasis under low pH (Wang et al., 2020). By contrast, calcium-dependent signaling pathways were suppressed in the pearl oyster Pinctada fucata under OA (Li et al., 2016) but whereas in the case of C. virginica, no significant difference in terms of calcium-related gene expression was observed upon exposure to low pH (Downey-Wall et al., 2020). Thus, mantle cells likely activate different cellular stress responses and modulate genes involved in calcium metabolism and pathways to cope with low pH. Some secreted genes encoding shell matrix proteins embedded in the calcified adult shell of C. gigas (Zhang et al., 2012) were also found to be significantly differentially expressed between hypercapnic and normocapnic oysters, suggesting that physical or mechanical properties of the shell might be affected to some degree by OA.
Altogether, we show that the transcriptome response of mantle cells under OA condition for 20 days is subtle, but there are significant changes in metabolic genes, shell-building genes, and calcium-dependent pathways.
We evaluated the effects of low pH in terms of cell morphology, secretory activity, and gene expression profiles in the mantle cells of adult oysters exposed to OA conditions for 20 days. During this time, we found that the morphology of mantle cells is not affected by exposure to OA. Nevertheless, mantle cell secretory activity is influenced by OA but in a mantle zone specific manner. It appears likely that mantle edge cells are more susceptible to OA. The modular nature of the mantle tissue (Herlitze et al., 2018), along with the difference in secretory activity in mantle edge cells, suggests that the foliated shell layer might be the first compromised due to OA.
Furthermore, our study demonstrated a subtle transcriptomic response of adult mantle cells with few shifts in gene expression profiles. However, we identified several OA-responsive genes related to cellular and biological processes involved in the metabolic process, catalytic activity, and ion binding. Once we focused on DEGs encoding secreted proteins, mantle cells differentially expressed genes involved in metabolic and calcium-dependent pathways, and shell biomineralization. Since most OA studies have been conducted on whole embryos or non-calcifying tissues (Leung et al., 2022), this study provides valuable morphological, secretory, and transcriptomic information on the mantle tissue for understanding the capacity of C. gigas to respond to OA.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethical review and approval was not required for the animal study because approval for research on oysters is not required under Chilean research legislation law.
FA conceived the study, conducted bioinformatics and data analysis, and wrote the manuscript with contributions from all other authors. NZ-S performed fertilization experiments under normal and low pH conditions with assistance from FA and conducted the histological analysis. IP-B performed histological analysis. CQ performed bioinformatic analysis. All authors contributed to the article and approved the submitted version.
This research was funded by the following Agencia Nacional de Investigación (ANID) grants: Fondecyt Iniciación 11180084, Fondecyt Regular 1220708, PAI79170033, and Fondequip EQM200056 to FA. ANID Beca Doctorado Nacional 21211666 to NZ-S and ANID Beca Doctorado Nacional 21221268 to IP-B.
We acknowledge all members from the Group of Developmental Processes (GDeP) at the University of Concepcion for fruitful discussions on this project. We would also like to thank the two reviewers for their insightful comments that improved our manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1156831/full#supplementary-material
Aguilera F., McDougall C., Degnan B. M. (2017). Co-Option and de novo gene evolution underlie molluscan shell diversity. Mol. Biol. Evol. 34, 779–792. doi: 10.1093/molbev/msw294
Albright R., Langdon C. (2011). Ocean acidification impacts multiple early life history processes of the Caribbean coral Porites astreoides. Global Change Biol. 17, 2478–2487. doi: 10.1111/j.1365-2486.2011.02404.x
Albright R., Mason B., Miller M., Langdon C. (2010). Ocean acidification comprimises recruitment sucess of the threatened Caribbean coral Acropora palmata. Proc. Natl. Acad. Sci. U.S.A. 107, 20400–20404. doi: 10.1073/pnas.1007273107
Álvarez Nogal R., Molist García P. (2015). The outer mantle epithelium of Haliotis tuberculata (Gastropoda: Haliotidae): An ultrastructural and histochemical study using lectins. Acta Zool. 96, 452–459. doi: 10.1111/azo.12090
Andrews S. (2010) FastQC: A quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Arivalagan J., Yarra T., Marie B., Sleight V. A., Duvernois-Berthet E., Clark M. S., et al. (2017). Insights from the shell proteome: Biomineralization to adaptation. Mol. Biol. Evol. 34, 66–77. doi: 10.1093/molbev/msw219
Arteaga-Solis E., Sui-Arteaga L., Kim M., Schaffler M. B., Jepsen K. J., Pleshko N., et al. (2011). Material and mechanical properties of bones deficient for fibrillin-1 or fibrillin-2 microfibrils. Matrix Biol. 30, 188–194. doi: 10.1016/j.matbio.2011.03.004
Avignon S., Auzoux-Bordenave S., Martin S., Dubois P., Badou A., Coheleach M., et al. (2020). An integrated investigation of the effects of ocean acidification on adult abalone (Haliotis tuberculata). ICES J. Mar. Sci. 77, 757–772. doi: 10.1093/icesjms/fsz257
Banker R. M. W., Sumner D. Y. (2020). Structure and distribution of chalky deposits in the pacific oyster using x-ray computed tomography (CT). Sci. Rep. 10, 12118. doi: 10.1038/s41598-020-68726-4
Barbosa M., Schwaner C., Pales Espinosa E., Allam B. (2022). A transcriptomic analysis of phenotypic plasticity in Crassostrea virginica larvae under experimental acidification. Genes 13, 1529. doi: 10.3390/genes13091529
Beniash E., Ivanina A., Lieb N. S., Kurochkin I., Sokolova I. M. (2010). Elevated level of carbon dioxide affects metabolism and shell formation in oysters Crassostrea virginica. Mar. Ecol. Progs. Ser. 419, 95–108. doi: 10.3354/meps08841
Berthelin C., Kellner K., Mathieu M. (2000). Histological characterization and glucose incorporation into glycogen of the pacific oyster Crassostrea gigas storage cells. Mar. Biotechnol. 2, 136–145. doi: 10.1007/s101269900017
Bhadury P. (2015). Effects of ocean acidification on marine invertebrates - a review. Indian J. Geo-Mar Sci. 44, 454–464.
Boskey A. L. (2003). Biomineralization: An overview. Connect Tissue Res. 44, 5–9. doi: 10.1080/03008200390152007
Breedham G. E. (1958). Observations on the mantle of the Lamellibranchia. Q J. Microsc Sci. 99, 181–197. doi: 10.1242/jcs.s3-99.46.181
Brennand Sheppard H., Soars N., Dworjanyn S. A., Davis A. R., Byrne M. (2010). Impact of ocean warming and ocean acidification on larval development and calcification in the sea urchin Tripneustes gratilla. PloS One 5, e11372. doi: 10.1371/journal.pone.0011372
Budd A., Mcdougall C., Green K., Degnan B. M. (2014). Control of shell pigmentation by secretory tubules in the abalone mantle. Front. Zool. 11, 62. doi: 10.1186/s12983-014-0062-0
Caldeira K., Wickett M. E. (2003). Anthropogenic carbon and ocean pH. Nature 425, 365. doi: 10.1038/425365a
Caldeira K., Wickett M. E. (2005). Ocean model predictions of chemistry changes from carbon dioxide emissions to the atmosphere and ocean. J. Geophys. Res. 110, 2671. doi: 10.1029/2004JC002671
Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST+: architecture and applications. BMC Bioinf. 10, 421. doi: 10.1186/1471-2105-10-421
Cao R., Liu Y., Wang Q., Zhang Q., Yang D., Liu H., et al. (2018). The impact of ocean acidification and cadmium on the immune responses of pacific oyster, Crassostrea gigas. Fish Shellfish Immunol. 81, 456–462. doi: 10.1016/j.fsi.2018.07.055
Checa A. G., Harper E. M. (2010). Spikey bivalves: Intra-periostracal crystal growth in anomalodesmatans. Biol. Bull. 219, 231–248. doi: 10.1086/BBLv219n3p231
Checa A. G., Harper E. M., González-Segura A. (2018). Structure and crystallography of foliated and chalk shell microstructures of the oyster Magallana: The same materials grown under different conditions. Sci. Rep. 8, 7507. doi: 10.1038/s41598-018-25923-6
Clark M. S. (2020). Molecular mechanisms of biomineralization in marine invertebrates. J. Exp. Biol. 223, jeb206961. doi: 10.1242/jeb.206961
Clark M. S., Peck L. S., Arivalagan J., Backeljau T., Berland S., Cardoso J. C. R., et al. (2020). Deciphering mollusc shell production: The roles of genetic mechanisms through to ecology, aquaculture and biomimetics. Biol. Rev. 95, 1812–1837. doi: 10.1111/brv.12640
Deng Z., Sun J., Wei H., Zhao W., Chen M., Li Y., et al. (2022). Shell colors and microstructures of four pearl oyster species in the south China sea. Aquac. Rep. 25, 101214. doi: 10.1016/j.aqrep.2022.101214
de Wit P., Durland E., Ventura A., Langdon C. J. (2018). Gene expression correlated with delay in shell formation in larval pacific oysters (Crassostrea gigas) exposed to experimental ocean acidification provides insights into shell formation mechanisms. BMC Genomics 19, 160. doi: 10.1186/s12864-018-4519-y
Dineshram R., Chandramouli K., Kuen Ko G. W., Zhang H., Qian P.-Y., Ravasi T., et al. (2016). Quantitative analysis of oyster larval proteome provides new insights into the effects of multiple climate change stressors. Global Change Biol. 22, 2054–2968. doi: 10.1111/gcb.13249
Dineshram R., Xiao S., Kuen Ko G. W., Li J., Smrithi K., Thiyagarajan V., et al. (2021). Ocean acidification triggers cell signaling, supress immune and calcification in the pacific oyster larvae. Front. Mar. Sci. 8, 782583. doi: 10.3389/fmars.2021.782583
Dix T. G. (1972). Histochemistry of mantle and pearl sac secretory cells in Pinctada maxima (Lamellibranchia). Aust. J. Zool. 20, 359–368. doi: 10.1071/ZO9720359
Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Doney S. C., Busch D. S., Cooley S. R., Kroeker K. J. (2020). The impacts of ocean acidification on marine ecosystems and reliant human communities. Annu. Rev. Env. Resour. 45, 83–112. doi: 10.1146/annurev-environ-012320-083019
Doney S. C., Fabry V. J., Feely R. A., Kleypas J. A. (2009). Ocean acidification: The other CO2 problem. Annu. Rev. Mar. Sci. 1, 169–192. doi: 10.1146/annurev.marine.010908.163834
Downey-Wall A. M., Cameron L. P., Ford B. M., Mcnally E. M., Venkataraman Y. R., Roberts S. B., et al. (2020). Ocean acidification induces subtle shifts in gene expression and DNA methylation in mantle tissue of the Eastern oyster (Crassostrea virginica). Front. Mar. Sci. 7, 566419. doi: 10.3389/fmars.2020.566419
Emanuelsson O., Brunak S., Von Heijne G., Nielsen H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971. doi: 10.1038/nprot.2007.131
Fabry V. J., Seibel B. A., Feely R. A., Orr J. C. (2008). Impacts of ocean acidification on marine fauna and ecosystem processes. ICES J. Mar. Sci. 65, 414–432. doi: 10.1093/icesjms/fsn048
Falkowski P., Scholes R. J., Boyle E., Canadell J., Canfield D., Elser J., et al. (2000). The global carbon cycle: A test of our knowledge of earth as a system. Science 290, 291–296. doi: 10.1126/science.290.5490.291
Fang Z., Feng Q., Chi Y., Xie L., Zhang R. (2008). Investigation of cell proliferation and differentiation in the mantle of Pinctada fucata (Bivalve, Mollusca). Mar. Biol. 153, 745–754. doi: 10.1007/s00227-007-0851-5
Farre B., Brunelle A., Laprévote O., Cuif J.-P., Williams C. T., Dauphin Y. (2011). Shell layers of the black-lip pearl oyster Pinctada margaritifera: Matching microstructure and composition. Comp. Biochem. Phys. B 159, 131–139. doi: 10.1016/j.cbpb.2011.03.001
Gao J., Liu J., Li Z. (2001). Isolation and purification of functional total RNA from blue-grained wheat endosperm tissues containing high levels of starches and flavonoids. Plant Mol. Biol. Rep. 19, 185a–185i. doi: 10.1007/BF02772163
Gardner L. D., Mills D., Wiegand A., Leavesley D., Elizur A. (2011). Spatial analysis of biomineralization associated gene expression from the mantle organ of the pearl oyter Pinctada maxima. BMC Genomics 12, 455. doi: 10.1186/1471-2164-12-455
Gazeau F., Gattuso J.-P., Dawber C., Pronker A. E., Peene F., Peene J., et al. (2010). Effect of ocean acidification on the early life stages of the blue mussel Mytilus edulis. Biogeosciences 7, 2051–2060. doi: 10.5194/bg-7-2051-2010
Gazeau F., Parker L. M., Comeau S., Gattuso J.-P., O’connor W. A., Martin S., et al. (2013). Impacts of ocean acidification on marine shelled molluscs. Mar. Biol. 160, 2207–2245. doi: 10.1007/s00227-013-2219-3
Gazeau F., Quiblier C., Jansen J. M., Gattuso J.-P., Middelburg J. J., Heip C. H. R. (2007). Impact of elevated CO2 on shellfish calcification. Geophy Res. Lett. 34, e028554. doi: 10.1029/2006GL028554
Germer J., Mann K., Wörheide G., Jackson D. J. (2015). The skeleton forming proteome of an early branching metazoan: A molecular survey of the biomineralization components employed by the coralline sponge Vaceletia sp. PloS One 10, e0140100. doi: 10.1371/journal.pone.0140100
Ghebrehiwet B., Hosszu K. K., Valentino A., Peerschke E. I. B. (2012). The C1q family of proteins: insights into the emerging non-traditional functions. Front. Immunol. 3, 52. doi: 10.3389/fimmu.2012.00052
Goedhart J., Luijsterburg M. (2020). VolcaNoseR is a web app for creating, exploring, labeling, and sharing volcano plots. Sci. Rep. 10, 20560. doi: 10.1038/s41598-020-76603-3
Goncalves P., Anderson K., Thompson E. L., Melwani A., Parker L. M., Ross P. M., et al. (2016). Rapid transcriptional acclimation following transgenerational exposure of oysters to ocean acidification. Mol. Ecol. 25, 4836–4849. doi: 10.1111/mec.13808
Goncalves P., Jones D. B., Thompson E. L., Parker L. M., Ross P. M., Raftos D. A. (2017). Transcriptomic profiling of adaptive responses to ocean acidification. Mol. Ecol. 26, 5974–5988. doi: 10.1111/mec.14333
Guinotte J. M., Fabry V. J. (2008). Ocean acidification and its potential effects on marine ecosystems. Ann. N Y Acad. Sci. 1134, 320–342. doi: 10.1196/annals.1439.013
Haas B. J., Papanicolaou A., Yassour M., Grabherr M. G., Blood P. D., Bowden J., et al. (2013). De novo transcript sequences reconstruction from RNA-seq using trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Han Y., Xie C., Fan N., Song H., Wang X., Zheng Y., et al. (2022). Identification of melanin in the mantle of the pacific oyster Crassostrea gigas. Front. Mar. Sci. 9, 880337. doi: 10.3389/fmars.2022.880337
Herlitze I., Marie B., Marin F., Jackson D. J. (2018). Molecular modularity and asymmetry of the molluscan mantle revealed by a gene expression atlas. GigaScience 7, 1–15. doi: 10.1093/gigascience/giy056
Hurd C. L., Beardall J., Comeau S., Cornwall C. E., Havenhand J. N., Munday P. L., et al. (2019). Ocean acidification as a multiple driver: How interactions between changing seawater carbonate parameters affect marine life. Mar. Freshw. Res. 71, 263–274. doi: 10.1071/MF19267
Jabbour-Zahab R., Chagot D., Blanc F., Grizel H. (1992). Mantle histology, histochemistry and ultrastructure of the pearl oyster Pinctada margaritifera (L.). Aquat Living Resour. 5, 287–298. doi: 10.1051/alr:1992027
Jackson D. J., Mann K., Häussermann V., Schihabel M. B., Lüter C., Griesshaber E., et al. (2015). The Magellania venosa biomineralizing proteome: A window into brachiopod shell evolution. Genome Biol. Evol. 7, 1349–1362. doi: 10.1093/gbe/evv074
Jackson D. J., McDougall C., Green K., Simpson F., Worheide G., Degnan B. M. (2006). A rapidly evolving secretome builds and patterns a sea shell. BMC Biol. 4, 40. doi: 10.1186/1741-7007-4-40
Jackson D. J., McDougall C., Woodcroft B., Moase P., Rose R. A., Kube M., et al. (2010). Parallel evolution of nacre building gene sets in molluscs. Mol. Biol. Evol. 27, 591–608. doi: 10.1093/molbev/msp278
Jackson D. J., Worheide G., Degnan B. M. (2007). Dynamic expression of ancient and novel molluscan shell genes during ecological transitions. BMC Evol. Biol. 7, 160. doi: 10.1186/1471-2148-7-160
Jodrey L. H. (1953). Studies on shell formation. III. measurement of calcium deposition in shell and calcium turnover in mantle tissue using the mantle-shell preparation Ca45. Biol. Bull. 104, 398–407. doi: 10.2307/1538493
Jolly C., Berland S., Milet C., Borzeix S., Lopez E., Doumenc D. (2004). Zonal localization of shell matrix proteins in mantle of Haliotis tuberculata (Mollusca, Gastropoda). Mar. Biotechnol. 6, 541–551. doi: 10.1007/s10126-004-3129-7
Kanehisa M., Sato Y., Morishima K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Bio 428, 726–731. doi: 10.1016/j.jmb.2015.11.006
Kapur S. P., Gibson M. A. (1967). A histological study of the development of the mantle-edge and shell in the freshwater gastropod, Helisoma duryi eudiscus (Pilsbry). Can. J. Zool. 45, 1169–1181. doi: 10.1139/z67-126
Ko G. W. K., Dineshram R., Campanati C., Chan V. B. S., Havenhand J., Thiyagarajan V. (2014). Interactive effects of ocean acidification, elevated temperatura, and reduced salinity on early-life stages of the pacific oyster. Environ. Sci. Technol. 48, 10079–10088. doi: 10.1021/es501611u
Kocot K. M., Aguilera F., Mcdougall C., Jackson D. J., Degnan B. M. (2016). Sea Shell diversity and rapidly evolving secretomes: Insights into the evolution of biomineralization. Front. Zool. 13, 23. doi: 10.1186/s12983-016-0155-z
Kroeker K. J., Kordas R. L., Crim R. N., Singh G. G. (2010). Meta-analysis reveals negative yet variable effects of ocean acidification on marine organisms. Ecol. Lett. 13, 1419–1434. doi: 10.1111/j.1461-0248.2010.01518.x
Krogh A., Larsson B., Von Heijne G., Sonnhammer E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Krueger F. (2012) Trim galore!. Available at: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/.
Kurihara H., Kato S., Ishimatsu A. (2007). Effects of increased seawater pCO2 on early development of the oyster Crassostrea gigas. Aquat Biol. 1, 91–98. doi: 10.3354/ab00009
Lee S.-W., Jang Y.-N., Kim J.-C. (2011). Characteristics of the aragonitic layer in adult oyster shells, Crassostrea gigas: Structural study of myostracum including the adductor muscle scar. Evid Based Complement Alternat Med. 2011, 742963. doi: 10.1155/2011/742963
Lee D. G., Park M. W., Kim B. H., Jeon M. A. (2014). Microanatomy and ultrastructure of outer mantle epidermis of the cuttlefish, Sepia esculenta (Cephalopoda: Sepiidae). Micron 58, 38–46. doi: 10.1016/j.micron.2013.11.004
Leung J. Y. S., Zhang S., Connell S. D. (2022). Is ocean acidification really a threat to marine calcifiers? a systematic review and meta-analysis of 980+ studies spanning two decades. Small 35, e2107407. doi: 10.1002/smll.202107407
Li S., Liu C., Huang J., Liu Y., Zhang S., Zheng G., et al. (2016). Transcriptome and biomineralization responses of the pearl oyster Pinctada fucata to elevated CO2 and temperature. Sci. Rep. 6, 18943. doi: 10.1038/srep18943
Liao Y., Smyth G. K., Shi W. (2014). featureCounts: An efficient general purpose program for assigning sequences reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liu Z., Zhang Y., Zhou Z., Zong Y., Zheng Y., Liu C., et al. (2020). Metabolomic and transcriptomic profiling reveals the alteration of energy metabolism in oyster larvae during initial shell formation and under experimental ocean acidification. Sci. Rep. 10, 6111. doi: 10.1038/s41598-020-62963-3
Maeda T., Abe M., Kurisu K., Jikko A., Furukawa S. (2001). Molecular cloning and characterization of a novel gene, CORS26, encoding a putative secretory protein and its possible involvement in skeletal development. J. Biol. Chem. 276, 3628–3634. doi: 10.1074/jbc.M007898200
Marie B., Joubert C., Tayalé A., Zanella-Cléon I., Belliard C., Piquemal D., et al. (2012). Different secretory repertories control the biomineralization processes of prism and nacre deposition of the pearl oyster shell. Proc. Natl. Acad. Sci. U.S.A. 109, 20986–20991. doi: 10.1073/pnas.1210552109
Marie B., Marie A., Jackson D. J., Dubost L., Degnan B. M., Milet C., et al. (2010). Proteomic analysis of the organic matrix of the abalone Haliotis asinina calcified shell. Proteome Sci. 8, 54. doi: 10.1186/1477-5956-8-54
Marie B., Zanella-Cleon I., Guichard N., Becchi M., Marin F. (2011). Novel proteins from the calcifying shell matrix of the pacific oyster Crassostrea gigas. Mar. Biotechnol. 13, 1159–1168. doi: 10.1007/s10126-011-9379-2
Marin F. (2020). Mollusc shellomes: Past, present and future. J. Struct. Biol. 212, 107583. doi: 10.1016/j.jsb.2020.107583
Marin F., Corstjens P., de Gaulejac B., de Vrind-De Jong E., Westbroek P. (2000). Mucins and molluscan calcification: Molecular characterization of mucoperlin, a novel mucin-like protein from the nacreous shell layer of the fan mussel, Pinna nobilis (Bivalvia, pteriomorphia). J. Biol. Chem. 275, 20667–20675. doi: 10.1074/jbc.M003006200
Marin F., Le Roy N., Marie B. (2012). The formation and mineralization of mollusk shell. Front. Biosci. 4, 1099–1125. doi: 10.2741/s321
Marin F., Luquet G., Marie B., Medakovic D. (2007). Molluscan shell proteins: Primary structure, origin, and evolution. Curr. Top. Dev. Biol. 80, 209–276. doi: 10.1016/S0070-2153(07)80006-8
Masson P. (1929). Some histological methods: Trichrome stainings and their preliminary technique. J. Tech. Methods 12, 75–90.
McCulloch M., Falter J., Trotter J., Montagna P. (2012). Coral resilience to ocean acidification and global warming through pH up-regulation. Nat. Clim. Change 2, 623–627. doi: 10.1038/nclimate1473
McDougall C., Aguilera F., Degnan B. M. (2013). Rapid evolution of pearl oyster shell matrix proteins with repetitive, low-complexity domains. J. R Soc. Interface 10, 20130041. doi: 10.1098/rsif.2013.0041
McDougall C., Degnan B. M. (2018). The evolution of mollusc shells. WIREs Dev. Biol. 7, e313. doi: 10.1002/wdev.313
McDougall C., Green K., Jackson D. J., Degnan B. M. (2011). Ultrastructure of the mantle of the gastropod Haliotis asinina and mechanisms of shell regionalization. Cells Tissues Organs 194, 103–107. doi: 10.1159/000324213
Mead T. J., Martin D. R., Wang L. W., Cain S. A., Gulec C., Cahill E., et al. (2022). Proteolysis of fibrillin-2 microfibrils is essential for normal skeletal development. eLife 11, e71142. doi: 10.7554/eLife.71142.sa2
Metsalu T., Vilo J. (2015). ClustVis: A web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 43, W566–W570. doi: 10.1093/nar/gkv468
Midura S., Hascall V. C. (1996). Bone sialoprotein - a mucin in disguise? Glycobiology 6, 677–681. doi: 10.1093/glycob/6.7.677
Miller A. W., Reynolds A. C., Sobrino C., Riedel G. F. (2009). Shellfish face uncertain future in high CO2 world: Influence of acidification on oyster larvae calcification and growth in estuaries. PloS One 4, e5661. doi: 10.1371/journal.pone.0005661
Mouchi V., Lartaud F., Guichard N., Immel F., De Rafélis M., Broussard C., et al. (2016). Chalky versus foliated: A discriminant immunogold labelling of shell microstructures in the edible oyster Crassostrea gigas. Mar. Biol. 163, 256. doi: 10.1007/s00227-016-3040-6
Munday P. L., Gagliano M., Donelson J. M., Dixson D., Thorrold S. R. (2011). Ocean acidification does not affect the early life history development of a tropical marine fish. Mar. Ecol. Prog. Ser. 423, 211–221. doi: 10.3354/meps08990
Nakayama S., Suzuki M., Endo H., Iimura K., Kinoshita S., Watabe S., et al. (2013). Identification and characterization of a matrix protein (PPP-10) in the periostracum of the pearl oyster, Pinctada fucata. FEBS Open Bio 3, 421–427. doi: 10.1016/j.fob.2013.10.001
Orr J. C., Fabry V. J., Aumont O., Bopp L., Doney S. C., Feely R. A., et al. (2005). Anthropogenic ocean acidification over the twenty-first century and its impact on calcifying organisms. Nature 437, 681–686. doi: 10.1038/nature04095
Palmer A. R. (1992). Calcification in marine molluscs: How costly is it? Proc. Natl. Acad. Sci. U.S.A. 89, 1379–1382. doi: 10.1073/pnas.89.4.1379
Parker L. M., Ross P. M., O´Connor W. A., Borysko L., Raftos D. A., Pörtner H.-O. (2011). Adult exposure influences offspring response to ocean acidification in oysters. Global Change Biol. 18, 82–92. doi: 10.1111/j.1365-2486.2011.02520.x
Parker L. M., Ross P. M., O´Connor W. A., Pörtner H. O., Scanes E., Wright J. M. (2013). Predicting the response of molluscs to the impact of ocean acidification. Biology 2, 651–692. doi: 10.3390/biology2020651
Parvizi F., Monsefi M., Noori A., Ranjbar M. S. (2017). Mantle histology and histochemistry of three pearl oysters: Pinctada persica, pinctada radiata and Pteria penguin. Molluscan Res. 38, 11–20. doi: 10.1080/13235818.2017.1387039
Petersen T. N., Brunak S., Von Heijne G., Nielsen H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Petrou G., Crozier T. (2018). Mucins as multifunctional building blocks of biomaterials. Biomater Sci-UK 6, 2282. doi: 10.1039/C8BM00471D
Przeslawski R., Byrne M., Mellin C. (2015). A review and meta-analysis of the effects of multiple abiotic stressors on marine embryos and larvae. Global Change Biol. 21, 2122–2140. doi: 10.1111/gcb.12833
Ramesh K., Hu M. Y., Melzner F., Bleich M., Himmerkus N. (2020). Intracellular pH regulation in mantle epithelial cells of the pacific oyster, Crassostrea gigas. J. Comp. Physiol. B 190, 691–700. doi: 10.1007/s00360-020-01303-3
Ramos-Silva P., Kaandorp J., Huisman L., Marie B., Zanella-Cleon I., Guichard N., et al. (2013). The skeletal proteome of the coral acropora millepora: the evolution of calcification by co-option and domain shuffling. Mol. Biol. Evol. 30, 2099–2112. doi: 10.1093/molbev/mst109
Ramos-Silva P., Odendaal M.-L., Wall-Palmer D., Mekkes L., Peijnenburg K. T. C. A. (2022). Transcriptomic responses of adult versus juvenile atlantids to ocean acidification. Front. Mar. Sci. 9, 801458. doi: 10.3389/fmars.2022.801458
R Development Core Team (2022). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
Ries J. B., Cohen A. L., Mccorkle D. C. (2009). Marine calcifiers exhibit mixed responses to CO2-incuded ocean acidification. Geology 37, 1131–1134. doi: 10.1130/G30210A.1
Robinson M. D., Mccarthy D. J., Smyth G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Ross P. M., Parker L., O´Connor W. A., Bailey E. A. (2011). The impact of ocean acidification on reproduction, early development and settlement of marine organisms. Water 3, 1005–1030. doi: 10.3390/w3041005
Schneider C. A., Rasband W. S., Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Shen X., Belcher A. M., Hansma P. K., Stucky G. D., Morse D. E. (1997). Molecular cloning and characterization of lustrin a, a matrix protein from shell and pearl nacre of Haliotis rufescens. J. Biol. Chem. 272, 32472–32481. doi: 10.1074/jbc.272.51.32472
Sillanpää J. K., Dos Reis Cardoso J. C., Catelo Félix R., Anjos L., Mary Power D., Sundell K. (2020). Dilution of seawater affects the Ca2+ transport in the outer mantle epithelium of Crassostrea gigas. Front. Physiol. 11, 2020.00001. doi: 10.3389/fphys.2020.00001
Sillanpää J. K., Ramesh K., Melzner F., Sundh H., Sundell K. (2016). Calcium mobilisation following shell damage in the pacific oyster, Crassostrea gigas. Mar. Genom. 27, 75–83. doi: 10.1016/j.margen.2016.03.001
Sillanpää J. K., Sundh H., Sundell K. (2018). Calcium transfer across the outer mantle epithelium in the pacific oyster, Crassostrea gigas. Proc. Biol. Sci. B 285, 20181676. doi: 10.1098/rspb.2018.1676
Sleight V. A., Marie B., Jackson D. J., Dyrynda E. A., Marie A., Clark M. S. (2016). An Antarctic molluscan biomineralisation tool-kit. Sci. Rep. 6, 36978. doi: 10.1038/srep36978
Smith V. J. (2011). Phylogeny of whey acidic protein (WAP) four-disulfide core proteins and their role in lower vertebrates and invertebrates. Biochem. Soc. T 39, 1403–1408. doi: 10.1042/BST0391403
Sridharan G., Shankar A. A. (2012). Toluidine blue: A review of its chemistry and clinical utility. J. Oral. Maxil. Pathol. 16, 251–255. doi: 10.4103/0973-029X.99081
Strader M. E., Wong J. M., Hofmann G. E. (2020). Ocean acidification promotes broad transcriptomic responses in marine metazoans: A literature survey. Front. Zool. 17, 7. doi: 10.1186/s12983-020-0350-9
Sud D., Poncet J.-M., Saïhi A., Lebel J.-M., Doumenc D., Boucaud-Camou E. (2002). A cytological study of the mantle edge of Haliotis tuberculata l. (Mollusca, Gastropoda) in relation to shell structure. J. Shellfish Res. 21, 201–210.
Suzuki M., Nagasawa H. (2013). Mollusk shell structures and their formation mechanism. Can. J. Zool. 91, 349–366. doi: 10.1139/cjz-2012-0333
Takeuchi T., Endo K. (2005). Biphasic and dually coordinated expression of the genes encoding major shell matrix proteins in the pearl oyster Pinctada fucata. Mar. Biotechnol. 8, 52–61. doi: 10.1007/s10126-005-5037-x
Talmage S. C., Gobler C. J. (2009). The effects of elevated carbon dioxide concentrations on the metamorphosis, size, and survival of larval hard clams (Mercenaria mercenaria), bay scallops (Argopecten irradians), and Eastern oysters (Crassostrea virginica). Limnol. Oceanogr. 54, 2072–2080. doi: 10.4319/lo.2009.54.6.2072
Timmins-Schiffman E., Coffey W. D., Hua W., Nunn B. L., Dickinson G. H., Roberts S. B. (2014). Shotgun proteomics reveals physiological response to ocean acidification in Crassostrea gigas. BMC Genomics 15, 951. doi: 10.1186/1471-2164-15-951
Todgham A. E., Hofmann G. E. (2009). Transcriptomic response of sea urchin larvae Strongylocentrotus purpuratus to CO2-driven seawater acidification. J. Exp. Biol. 212, 2579–2594. doi: 10.1242/jeb.032540
Treccani L., Mann K., Heinemann F., Fritz M. (2006). Perlwapin, an abalone nacre protein with three four-disulfide core (whey acidic protein) domains, inhibits the growth of calbium carbonate crystals. Biophys. J. 91, 2601–2608. doi: 10.1529/biophysj.106.086108
Waldbusser G. G., Brunner E. L., Haley B. A., Hales B., Langdon C. J., Prahl F. G. (2013). A developmental and energetic basis linking larval oyster shell formation to acidification sensitivity. Geophys. Res. Lett. 40, 2171–2176. doi: 10.1002/grl.50449
Waller T. R. (1980). Scanning electron microscopy of shell and mantle in the order arcoida (Mollusca: Bivalvia). Sm C Zool. 313, 1–58. doi: 10.5479/si.00810282.313
Wang Q., Cao R., Ning X., You L., Mu C., Wang C., et al. (2016). Effects of ocean acidification on immune responses of the pacific oyster Crassostrea gigas. Fish Shellfish Immun. 49, 24–33. doi: 10.1016/j.fsi.2015.12.025
Wang X., Li P., Cao X., Liu B., He S., Cao Z., et al. (2022a). Effects of ocean acidification and tralopyril on bivalve biomineralization and carbon cycling: A study of the pacific oyster (Crassostrea gigas). Environ. pollut. 313, 120161. doi: 10.1016/j.envpol.2022.120161
Wang X., Li P., He S., Zing S., Cao Z., Cao X., et al. (2022b). Effects of tralopyril on histological, biochemical and molecular impacts in pacific oyster, Crassostrea gigas. Chemosphere 289, 133157. doi: 10.1016/j.chemosphere.2021.133157
Wang X., Wang M., Wang W., Liu Z., Xu J., Jia Z., et al. (2020). Transcriptional changes of pacific oyster Crassostrea gigas reveal essential role of calcium signal pathway in response to CO2-driven acidification. Sci. Total Environ. 741, 140177. doi: 10.1016/j.scitotenv.2020.140177
Wei L., Wang Q., Ning X., Mu C., Wang C., Cao R., et al. (2015). Combined metabolome and proteome analysis of the mantle tissue from pacific oyster Crassostrea gigas exposed to elevated pCO2. Comp. Biochem. Physiol. D 13, 16–23. doi: 10.1016/j.cbd.2014.12.001
Whittaker C. A., Hynes R. O. (2002). Distribution and evolution of von Willebrand/Integrin a domains: Widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell 13, 3369–3387. doi: 10.1091/mbc.e02-05-0259
Wickham H. (2016). ggplot2: Elegant graphics for data analysis, (New York:Springer-Verlag). Available at: https://ggplot24272.tidyverse.org.
Wouters M. A., Rigoutsos I., Chu C. K., Feng L. L., Sparrow D. B., Dunwoodie S. L. (2005). Evolution of distinct EGF domains with specific functions. Protein Sci. 14, 1091–1103. doi: 10.1110/ps.041207005
Ye J., Zhang Y., Cui H., Liu J., Wu Y., Cheng Y., et al. (2018). WEGO 2.0: A web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 46, W71–W75. doi: 10.1093/nar/gky400
Zhang G., Fang X., Guo X., Li L., Luo R., Xu F., et al. (2012). The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54. doi: 10.1038/nature11413
Zhang J.-L., Richetti S., Ramezani T., Welcker D., Lütke S., Pogoda H.-M., et al. (2022). Vertebrate extracellular matrix protein hemicentin-1 interacts physically and genetically with basement membrane protein nidogen-2. Matrix Biol. 112, 132–154. doi: 10.1016/j.matbio.2022.08.009
Zhang C., Xie L., Huang J., Chen L., Zhang R. (2006). A novel putative tyrosinase involved in periostracum formation from the pearl oyster (Pinctada fucata). Biochem. Biophys. Res. Commun. 342, 632–639. doi: 10.1016/j.bbrc.2006.01.182
Zhao R., Takeuchi T., Luo Y.-J., Ishikawa A., Kobayashi T., Koyanagi R., et al. (2018). Dual gene repertoires for larval and adult shells reveal molecules essential for molluscan shell formation. Mol. Biol. Evol. 35, 2751–2761. doi: 10.1093/molbev/msy172
Keywords: Crassostrea gigas, ocean acidification, adult oyster, secretory cells, transcriptome, mantle secretome, shell formation
Citation: Zúñiga-Soto N, Pinto-Borguero I, Quevedo C and Aguilera F (2023) Secretory and transcriptomic responses of mantle cells to low pH in the Pacific oyster (Crassostrea gigas). Front. Mar. Sci. 10:1156831. doi: 10.3389/fmars.2023.1156831
Received: 01 February 2023; Accepted: 27 March 2023;
Published: 11 April 2023.
Edited by:
Andrew Stanley Mount, Clemson University, United StatesReviewed by:
Zhi Liao, Zhejiang Ocean University, ChinaCopyright © 2023 Zúñiga-Soto, Pinto-Borguero, Quevedo and Aguilera. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Felipe Aguilera, ZmFndWlsZXJhQHVkZWMuY2w=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.