- School of Marine Sciences, Ningbo University, Key Laboratory of Marine Biotechnology of Zhejiang Province, Ningbo, Zhejiang, China
The economically important seaweed Gracilariopsis lemaneiformis (Gp. lemaneiformis) is the main resource for agar production and abalone bait. Few studies have focused on the functions of glycosyltransferases (GTs) and glycoside hydrolases (GHs) involved in saccharometabolism, including that of agar. To investigate the possible functions of glycosyltransferase family 7 (GT7) and glycoside hydrolase family 16 (GH16) members in agar metabolism, analyses of these gene families were conducted using bioinformatics-based and physiological methods. In total, five GlGT7 genes and four GlGH16 genes from the Gp. lemaneiformis genome were identified and analyzed. These GT7 and GH16 members are absent in higher plants, and the majority of GlGT7s and GlGH16s have no introns. Catalytic pocket residue analysis revealed that GlGT7s may function as β-1,4-galactosyltransferases and that GlGH16s play roles as agarases. Promoter prediction and qRT-PCR experiments verified that these genes can be regulated by light and phytohormones. GlGT7s were predicted to interact with carbohydrate sulfotransferases (STs), whereas yeast two-hybrid (Y2H) assays revealed no interactions between these proteins. Under heat stress, no significant difference was found in agar content; however, the expression of GlGT7s fluctuated and that of GlGH16 increased. Low nitrogen stress significantly increased the agar content, and the expression of GlGT7s increased, whereas that of GlGH16s decreased. In three cultivars of Gp. lemaneiformis, the expression of GlGT7 genes in cultivars 981 and Lulong No. 1 was upregulated relative to that in the wild type, whereas GlGH16 expression levels were significantly decreased. Fluorescence microscopy further showed that β-1,4-galactose accumulation was consistent with increases in agar content and GlGT7 expression. Pearson correlation analysis confirmed that the expression levels of GlGT7s and GlGH16s were positively and negatively correlated with agar accumulation, respectively. Taken together, these results demonstrated that GlGT7 and GlGH16 are intimately correlated with agar metabolism; in particular, GlGT7-2, GlGT7-5, and GlGH16-4 could act as molecular markers to indicate agar yield. This study will provide a valuable basis for breeding new cultivars with high agar content.
Introduction
Agar, a sulfated polysaccharide that is abundant in the cell walls of agarophytes, is industrially produced from various kinds of red seaweeds, such as members of the families Gelidiaceae, Gracilariaceae, Gelidiellaceae, and Pterocladiaceae. Because of its excellent rheological properties, bioactive functions, and good compatibility with other bioactive compounds, agar has been widely applied in the fields of food, pharmaceutical, and chemical industries (Lee et al., 2017a; Kim et al., 2021). In addition, as the main component of cell walls in red seaweed, agar can be synthesized and remodeled to protect against extreme environments occurring in the intertidal zone, enabling seaweeds to remain flexible enough to withstand the physical challenges imposed by waves and tides. The biosynthesis of agar from red seaweeds represents an important strategy to address environmental variability (Ficko-Blean et al., 2015). Despite the great importance of agar in applications and algal physical processes, this material cannot be synthesized artificially and the complete synthesis pathways and regulatory mechanisms of agar are still unknown. The basic structure of agar, a type of conglomerate of linear glucan chains, involves the cross-linking of D-galactose and L-galactose through α-1,3- and β-1,4-glycosidic bonds, which are catalyzed by galactosyltransferases (GalTs) and hydrolyzed by glycoside hydrolases (GHs), respectively (Lee et al., 2017a; Hu et al., 2018). However, studies related to the effects of candidates of GalTs and GHs on agar metabolism are still rare.
Glycosyltransferases (GTs) constitute a large multigene family that consists of more than 110 subfamilies whose members are categorized according to the similarity of their amino acid sequences, substrate specificity, reaction mechanism, and three-dimensional structure. GTs are responsible for catalyzing the synthesis of various glycans, which play important roles in plant growth, reproduction, and stress resistance. Among GTs, members of the glycosyltransferase 7 family (GT7) are purportedly involved in the biosynthesis of sulfated polysaccharides such as chondroitin and heparin (Cantarel et al., 2008). In humans, GT7 sequences were annotated as N-acetylgalactosaminyltransferases, which are used for chondroitin synthesis (Brawley et al., 2017). In Florideophytes and Bangiophytes of red algae, N-acetylgalactosaminyltransferases are sulfate chondroitin synthases and can transfer acetyl galactosamine to core tetrasaccharides to extend chondroitin sulfate chains (Sato et al., 2011). GT7 sequences have been annotated in the genomes of Chondrus crispus and Porphyra umbilicalis (Collén et al., 2013; Ho, 2020). Although the knowledge of genomes of red macroalgae addresses the possibility of GT7 being involved in sulfated galactan synthesis, few studies have discussed the connection between agar and GT7.
GHs, which are responsible for hydrolyzing the glycosidic bond between a sugar molecule and other moieties, are likely to be involved in cell wall remodeling and recycling (Cantarel et al., 2008). In higher plants, xyloglucan endotransglucosylase/hydrolase (XTH), which belongs to the glycoside hydrolase family 16 (GH16), is responsible for cleaving cellulose and hemicellulose cross-linkages and is necessary for cell wall loosening and cell expansion (Ezquer et al., 2020). In marine environments, macroalgae, especially red and brown seaweeds, are important carbon sources for microbial hydrolysis and utilization (Rhein-Knudsen and Meyer, 2021). Agar can be cleaved into agar oligosaccharides by agarases from marine bacteria for industrial use and carbon recycling (Jiang et al., 2020). To date, large numbers of microorganisms have been identified and isolated for cleaving agar or agarose for commercial production (Kim et al., 2010; Seo et al., 2014; Gao et al., 2019). The red algae C. crispus and P. umbilicalis harbor genes that appear to encode GH16 enzymes such as carrageenase and porphyranase, and these GH16s are believed to participate in cell wall expansion and recycling (Lipinska et al., 2020). However, no studies have revealed the connection between agar and GH16 in vivo.
The agarophyte Gracilariopsis lemaneiformis (Gp. lemaneiformis) is mainly used for agar production and for abalone feed and food industries. Wild thalli of Gp. lemaneiformis have gradually become scarce because of human disturbance and difficulty surviving heat stress (Zhou et al., 2016). To meet the increasing demand of the agar industry, the cultivars Gp. lemaneiformis 981 and Lulong No. 1, which have excellent high temperature tolerance, are highly productive and produce high agar yields, have been bred. The cultivation of Gp. lemaneiformis has become the second largest algal industry in China (Wang et al., 2016). However, problems such as germplasm degradation, unstable agar production, and changing environments still hinder the development of the Gp. lemaneiformis industry. To identify molecular markers indicating agar content, we characterized the GT7 and GH16 members in Gp. lemaneiformis and analyzed their sequence features, promoter elements, phylogenetic relationships, interacting proteins, morphology, and changes in transcription profiles under heat and low nitrogen stresses. In this study, the candidates for agar polymerization and modification were identified from Gp. lemaneiformis, which provides clues for the clarification of the functions of agar metabolism-related enzymes and their metabolic pathways. Taken together, these findings will provide a valuable basis for breeding new cultivars with high agar content.
Materials and methods
Seaweed collection and culture
Wild-type Gp. lemaneiformis was collected from Zhanshan Bay in Qingdao, Shandong Province, China (36° 03′ N, 120° 22′ E), in May 2020. Gp. lemaneiformis cultivars 981 and Lulong No. 1 were collected from the coasts of Xiapu (26° 42′ N, 119° 59′ E) and Lianjiang (26° 39′ N, 119° 72′ E), Fujian Province, China, respectively. In the laboratory, the thalli were cultivated in transparent aquaria at an irradiance of 40 µmol photons m-2 s-1, under a 12h: 12h light: dark photoperiod, and at 23°C for 1 week of adaptation. The tank containing 5 l of natural seawater (salinity of 25‰) was filled with filtered air, and the seawater was changed every day. The algae used for treatments were mainly those of cultivar 981.
Heat and low nitrogen treatments
The cultivar 981 was exposed to heat and low nitrogen stresses for agar and related gene correlation analysis in the laboratory. Approximately 10 g of fresh thalli of each biological replicate was placed into 8 l of seawater, and each treatment was replicated three times. During the 11 days of heat-stress treatment, the thalli were cultured with normal Provasoli medium at 23°C (the control treatment, CK) or at 28°C (high temperature treatment, HT). During the 21 days of the nitrogen treatment, the thalli were cultured with normal Provasoli media (CK) and Provasoli media without ammonium nitrate (low nitrogen treatment, LN). After the experiments, samples were collected for agar and gene expression analyses. Apart from these factors, the other culture conditions were consistent with the adaptive culture conditions.
Light and hormone treatments
To explore the responses of different treatments according to promoter predictions, the thalli were cultivated under 40 μmol photons m-2 s-1 (CK) and 180 μmol photons m-2 s-1 light intensity (high light treatment, HL). For the hormone treatments, the thalli were cultivated in media without hormone addition (CK), with 50 μmol·l-1 abscisic acid treatment (ABA), and with 50 μmol·l-1 methyl jasmonate treatment (MeJA) (Hou et al., 2018; Sun et al., 2022). Apart from the light and hormone factors, the other culture conditions were the same as the adaptation culture conditions. After 6 h of treatment, samples were collected for relative expression analyses of GlGT7s and GlGH16s.
Agar extraction
Approximately 5 g of fresh thalli was used to assay the agar content according to the alkali treatment method (Chen Q. L. et al., 2022). The agar content was calculated using the following formula: agar content (%) = (dry weight of agar/dry weight of thalli) × 100.
Observation of thalli after treatments
The Ricinus communis agglutinin I (RCA120)-fluorescein staining method was used but was modified according to the methods of Lin et al. (2020). The thalli were cut into approximately 0.1 mm slices and then washed twice with phosphate-buffered saline (PBS) solution. The slices were resuspended in staining buffer and incubated for 1 h in the dark, with inverting occurring every 20 min. After incubation, the stained slices were collected and washed three times with PBS solution to remove the free RCA120-fluorescein. The stained slices were ultimately observed under a Nikon NI-U Normal Fluorescence Microscope (Japan).
Identification of GlGT7 and GlGH16 members in Gp. lemaneiformis
Based on the carbohydrate-active enzymes (CAZy) database (http://www.cazy.org/), the protein sequences of GT7 and GH16 were downloaded from UniProt (https://www.uniprot.org/) and used to generate the GT7- and GH16-BLAST databases, respectively, as referenced by Ulvskov et al. (2013). To identify the predicted GT7 and GH16 sequences in Gp. lemaneiformis (GlGT7s and GlGH16s), the genomic sequence of this alga reported by Sun et al. (2018) was used as the query sequence to search against the GT7- and GH16-BLAST databases using the BLASTP tool with an e-value ≤ e-10. After obtaining the predicted GlGT7s and GlGH16s, the Pfam database (http://pfam.xfam.org/) and SMART (http://smart.embl-heidelberg.de/) were then used to confirm the conserved domains, and those sequences lacking the conserved domains were excluded.
Sequence analysis and structural characterization
The open reading frames (ORFs) of the GlGT7 and GlGH16 genes were predicted using ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/). The theoretical molecular weight (MW) and isoelectric point (pI) of GT7 and GH16 proteins were calculated by ProtParam (https://web.expasy.org/protparam/). The MEME program (http://alternate.meme-suite.org/tools/meme) was used to identify the conserved motifs, with the following parameters: any number of repetitions, maximum of 10 misfits and an optimum motif width of 6–200 amino acid residues. The exon–intron structures were determined via Gene Structure Display Server (GSDS, http://gsds.cbi.-pku.edu.cn/).
Promoter response elements
The template DNA from Gp. lemaneiformis cultivar 981 was isolated via a RaPure Plant DNA Mini Kit (Magen, Guangzhou, China) according to the manufacturer’s protocol. The predicted promoter region of each sequence was limited to 2,000 bp upstream from the transcription start site of each sequence. The actual sequence used for validating the promoter region was 2,015–2,135 bp, including the beginning 15–135 bp of the ORFs, so that we could ensure that the promoter region of each sequence was sufficient. The primers used for each sequence are listed in Supplementary Table S1. Promoter elements were predicted using the online software PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Phylogenetic analysis
GT7 sequences from the genomes of Ochrophyta and Rhodophyta were retrieved from the NCBI (https://www.ncbi.nlm.nih.gov/), JGI (https://genome.jgi.doe.gov/portal/), and Ensembl (https://plants.ensembl.org/index.html) databases. The other protein sequences annotated as belonging to the GT7 family were retrieved from CAZy and downloaded from UniProt. GH16 sequences, considered to encode β-agarases from marine bacteria, were retrieved from the NCBI database. The other GH16 sequences were retrieved from the genomes of Ochrophyta, Rhodophyta, and terrestrial plants in the NCBI, JGI, and Ensembl databases. These sequences are presented in Supplementary Tables S2, S3. Multiple-sequence alignment involving different species was performed using Clustal X (Dublin, Republic of Ireland). A phylogenetic tree was subsequently constructed using the maximum likelihood method with 1,000 bootstrap replicates by MEGA 7 (Auckland, New Zealand).
Interaction network analysis and yeast two-hybrid assays
Interaction networks comprising chondroitin synthase and other proteins and comprising β-agarase and other proteins were constructed using the STRING database v11.0 (https://string-db.org/). To verify the interactions between GlGT7 proteins and carbohydrate sulfotransferase (GlST) proteins, putative GlST sequences were retrieved by searching the genome of Gp. lemaneiformis using the hidden Markov model (HMM) of sulfotransferase (ST) (PF03567), with an e-value ≤ e-10. The full-length sequences of GlGT7s and GlSTs were obtained by PCR with gene-specific primers (Supplementary Table S4) and then cloned and ligated into pGBKT7 and pGADT7 vectors (Clontech), respectively. Autoactivation and toxicity tests were performed following the protocols of the Matchmaker Gold Yeast Two-Hybrid System (Clontech, Mountain View, CA, USA). Then, one-to-one verification assays were performed. The recombinant vectors pGADT7-GlST-1, pGADT7-GlST-2, and pGADT7-GlST-3 were co-transformed with pGBKT7-GlGT7-1, pGBKT7-GlGT7-2, pGBKT7-GlGT7-3, pGBKT7-GlGT7-4, and pGBKT7-GlGT7-5 into the yeast two-hybrid (Y2H) yeast. The resulting yeast cells were subsequently incubated on SD/-Trp/-Leu/-His/-Ade/x-α-gal/AbA media to validate their interactions.
Total RNA extraction and expression analysis of the GlGT7 and GlGH16 genes
After the different treatments of high light, hormone, heat, and low nitrogen, the total RNA was extracted using a Plant RNA kit (OMEGA, Georgia, USA). The extracted RNA was then quantified by a NanoDrop 2000 Spectrophotometer (Thermo Scientific, MA, USA). First-strand cDNA was synthesized using HiScript II Q RT SuperMix for qPCR with a gDNA wiper (Vazyme, Nanjing, China) and stored at -20°C for subsequent analysis. All the procedures were performed following the manufacturers’ instructions.
Primer Premier 5 was used to design primers specific to the full-length sequences of the GlGT7 and GlGH16 genes (Supplementary Table S5), and the specificity of the primers was determined by NCBI Primer-BLAST. qRT-PCR was carried out using PerfectStart Green qPCR SuperMix (TransGen, Beijing, China) on a Mastercycler RealPlex2 real-time PCR system (Eppendorf, Germany) with the following program: 95°C for 30 s followed by 40 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 10 s. For each treatment, three biological replicates, each with two technical replicates (n = 6), were evaluated to acquire reliable results. The housekeeping gene β-actin was used as an internal reference gene (Ding et al., 2014). The relative expression levels of the GlGT7s and GlGH16s were calculated by the 2-ΔΔCT method (Livak and Schmittgen, 2001).
Statistical analysis
The correlations between the expression profiles of GlGT7s or GlGH16s and agar contents under heat and low nitrogen treatments were determined by Pearson correlation analysis (n = 6 of each treatment, SPSS 16.0, USA). The normal distribution of all the data under each treatment and the homogeneity of variance were confirmed by a Shapiro–Wilk test (p > 0.05) and the Levene test (p > 0.05), respectively. The effects of the treatments were assessed by independent t tests and one-way analysis of variance (ANOVA). A Tukey post-hoc test (Tukey HSD) was performed to determine the differences between the treatments. The significant and very significant levels were set at p < 0.05 and p < 0.01, respectively.
Results
Identification and characterization of GlGT7 and GlGH16 genes
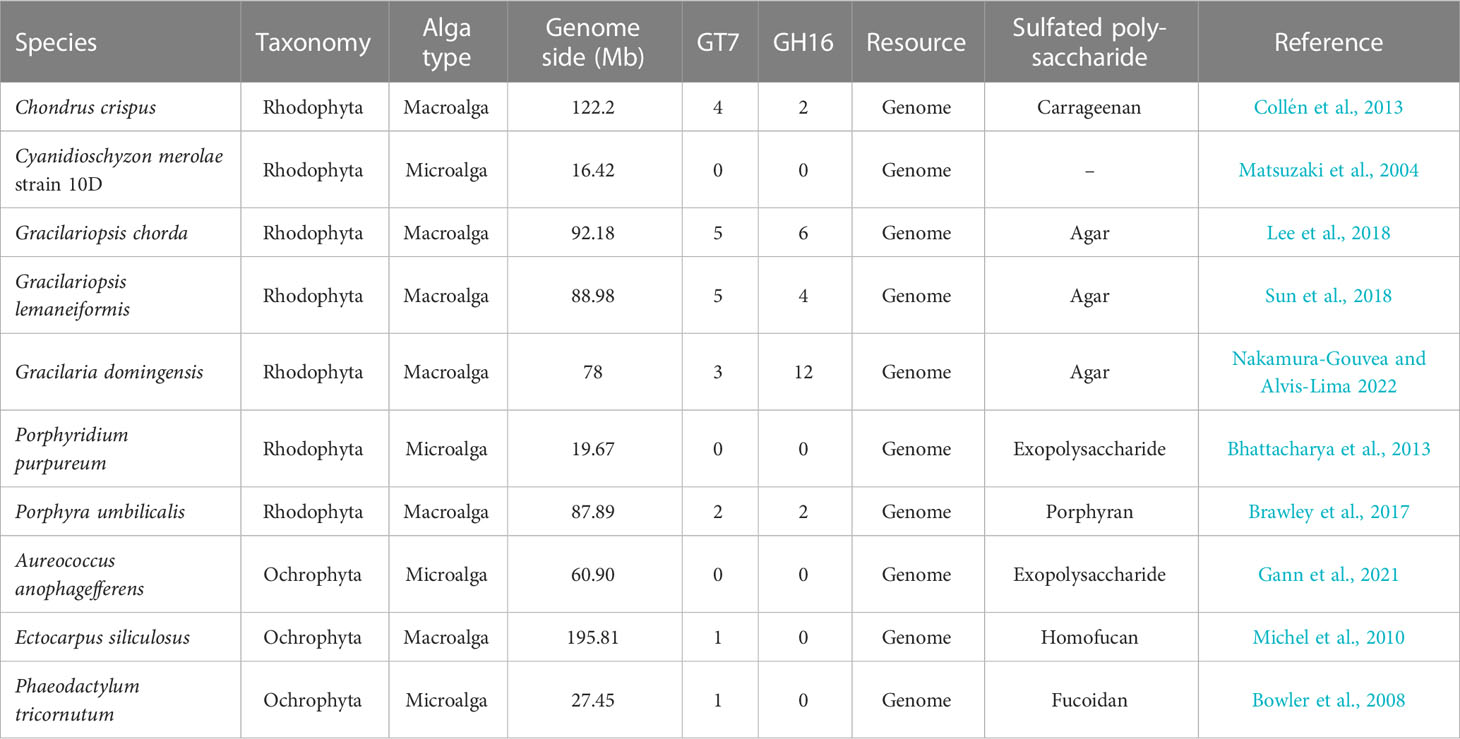
The available GT7 and GH16 genes in red algae, brown algae, and diatoms were identified (Table 1). In Rhodophyta, C. crispus, Gp. chorda, Gracilaria domingensis, and P. umbilicalis have 2-5 GT7 genes. However, only one GT7 gene was found in Ectocarpus siliculosus and Phaeodactylum tricornutum. For GH16, the red macroalga G. domingensis has the most genes (12), followed by Gp. chorda (6) and Gp. lemaneiformis (4), whereas no GH16 gene was found in Aureococcus anophagefferens, E. siliculosus, or P. tricornutum. However, in the genomes of red microalgae, such as Cyanidioschyzon merolae strain 10D and Porphyridium purpureum, neither GT7 nor GH16 genes were found.
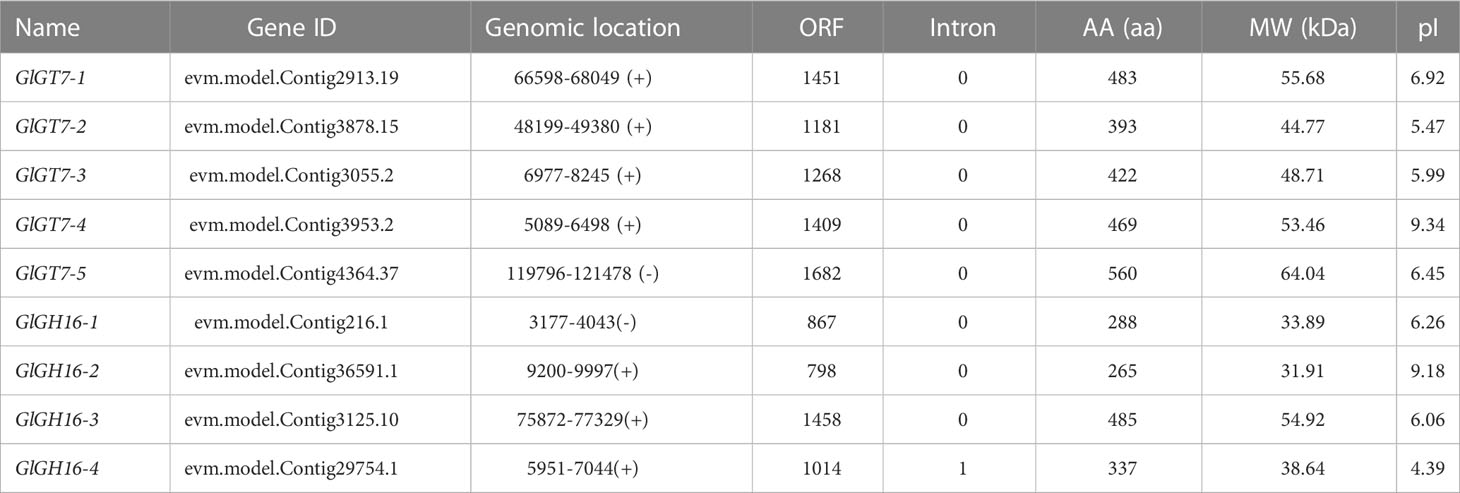
In Gp. lemaneiformis, a total of five GlGT7 members were annotated in the genome (Table 2). The ORF lengths of GlGT7 genes varied from 1,181 to 1,682 bp, the MWs of GlGT7s were between 44.77 and 64.04 kDa, and the predicted pI values of the GlGT7s were between 5.47 and 9.34. Four GlGH16 sequences identified in Gp. lemaneiformis encoded 265 amino acids (aa) to 485 aa, with MWs of 31.91 to 54.92 kDa and pIs between 4.39 and 9.18. Given that red algae typically have a small genome, except for GlGH16-4, which has one intron, no other GlGT7 and GlGH16 genes have introns.
Promoter response elements
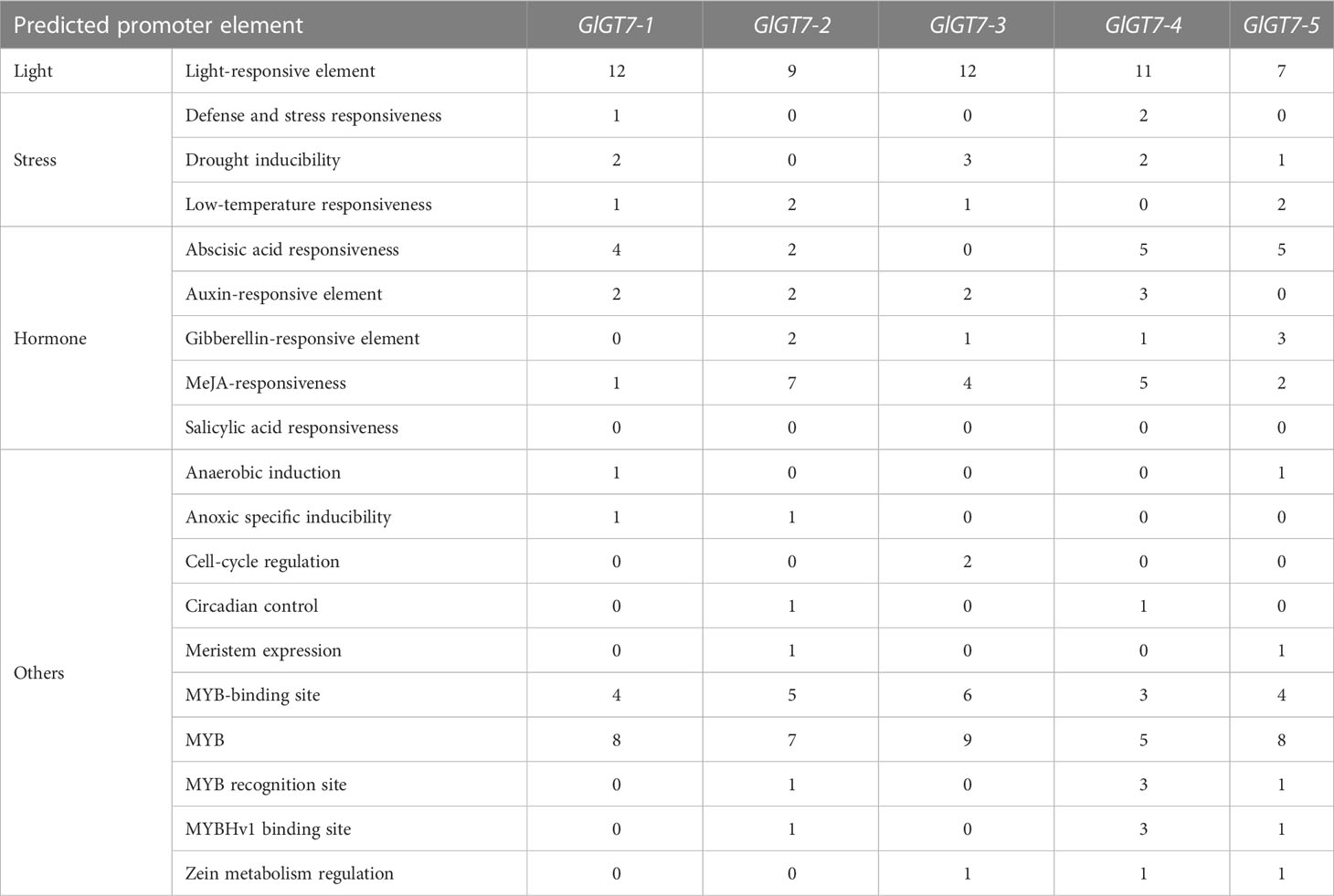
To explore the expression regulatory patterns of the GlGT7 and GlGH16 genes, a total of 19 kinds of promoter elements, including light-, stress-, and hormone-responsive elements, were predicted (Tables 3-1, 3-2). Multiple light-responsive elements were found in the promoter regions of the five GlGT7 and four GlGH16 genes, indicating that the expression of GlGT7 and GlGH16 may be regulated by light. Moreover, all the genes contained at least one stress-responsive element, such as those that respond to low temperature, drought, defense, or stress. Among the hormone-responsive elements, elements that respond to ABA and MeJA were the most frequently predicted ones. Regarding the other elements, MYB and MYB-related sites were the only elements related to transcription factors predicted in the GlGT7s and GlGH16s.
Light and hormone signaling responses of GlGT7 and GlGH16
To further determine what factors regulate the expression of the GlGT7 and GlGH16 genes, we selected the most frequently predicted factors, such as light and hormone elements. Under high light conditions (Figure 1A), the expression levels of five GlGT7 genes were decreased, indicating that the expression of GlGT7 members could be inhibited by high light, which was consistent with the results that the agar contents significantly decreased (Supplementary Figure S1A). However, diverse expression modes were observed for GlGH16s, namely, GlGH16-2 was downregulated by high light, whereas GlGH16-4 expression was enhanced 2.00-fold, and the expression of the other two GlGH16 genes was not significantly altered by high light.
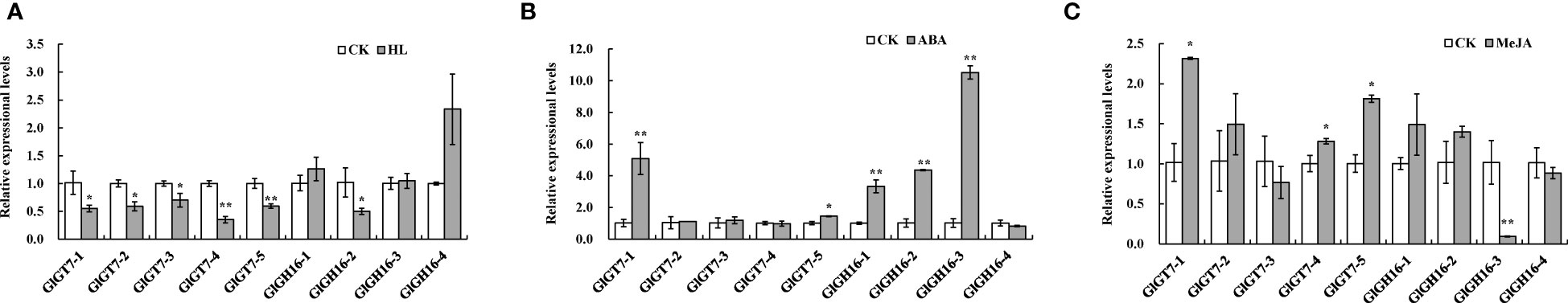
Figure 1 Expressions of the GlGT7 and GlGH16 genes under high light, ABA, and MeJA treatments. (A), high light treatment (HL); (B), ABA treatment (ABA); (C), MeJA treatment (MeJA); CK, the control treatment. * and ** represent significant (p < 0.05) and very significant (p < 0.01) differences between the treatments.
After ABA treatment, the expression levels of GlGT7-1 and GlGT7-5 were significantly increased, especially for GlGT7-1, with a 3.17-fold expression increase. In addition, GlGH16-1, GlGH16-2, and GlGH16-3 were upregulated at least threefold by ABA treatment relative to that of the CK (Figure 1B).
In the presence of MeJA, the expression of GlGT7-1, GlGT7-4, and GlGT7-5 was enhanced and GlGT7-1 displayed a maximal expression fold change of 2.00, whereas the other two GlGT7 genes demonstrated no significant changes in expression (Figure 1C). The majority of the GlGH16 genes did not significantly change after the addition of MeJA, except that GlGH16-3 expression was significantly downregulated.
Evolutionary relationships among GT7 and GH16 members
To determine the evolutionary relationships among the GT7 members from different species, an unrooted phylogenetic tree was constructed. A total of 63 GT7 sequences from red and brown seaweeds, marine bacteria, Homo sapiens, Drosophila melanogaster, Gallus gallus, and Mus musculus were analyzed in the phylogenetic tree (Figure 2). According to the sequence homology data, these GT7 sequences could be divided into two main clusters: clades I and II. Marine organisms such as Styela clava, the photosynthetic Ochrophyta A. anophagefferens, and red seaweeds clustered in clade I, and the majority of GT7 proteins of H. sapiens and M. musculus clustered in clade II. Interestingly, E. siliculosus, P. tricornutum, and D. melanogaster clustered in clade II. All the GT7 sequences contained the highly conserved motif 1, which encodes the catalytic pocket residue WGGEDDD (Figure 3).
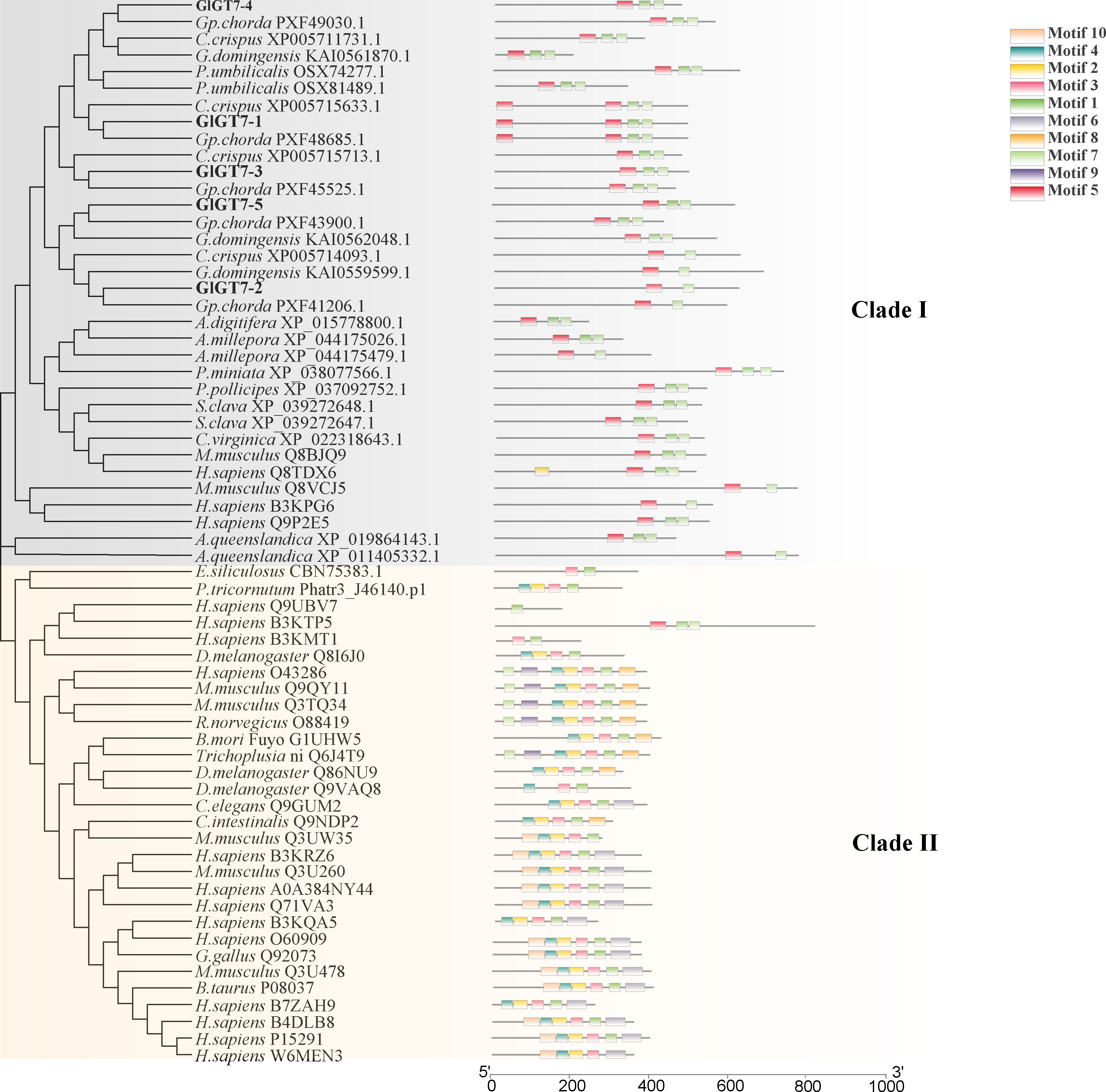
Figure 2 Phylogenetic tree and conserved motifs of GT7 members constructed based on the protein sequences using the maximum likelihood method. The bootstrap analysis was carried out with 1,000 replicates.

Figure 3 Conserved motifs of the GT7 and GH16 families corresponding to the proteins. The red asterisks indicate the catalytic sites.
A total of 50 sequences were retrieved for constructing the phylogenetic tree of the GH16 family members (Figure 4). In clade II, Zea mays harbors conserved motifs 4, 5, and 8, which are completely different from those of other species, representing a separate evolutionary direction. A close relationship between red seaweeds and marine bacteria was found (clade I). In clade I, the majority of GH16 members contained the common sequence motif 6-EIDVFE (Figure 3).
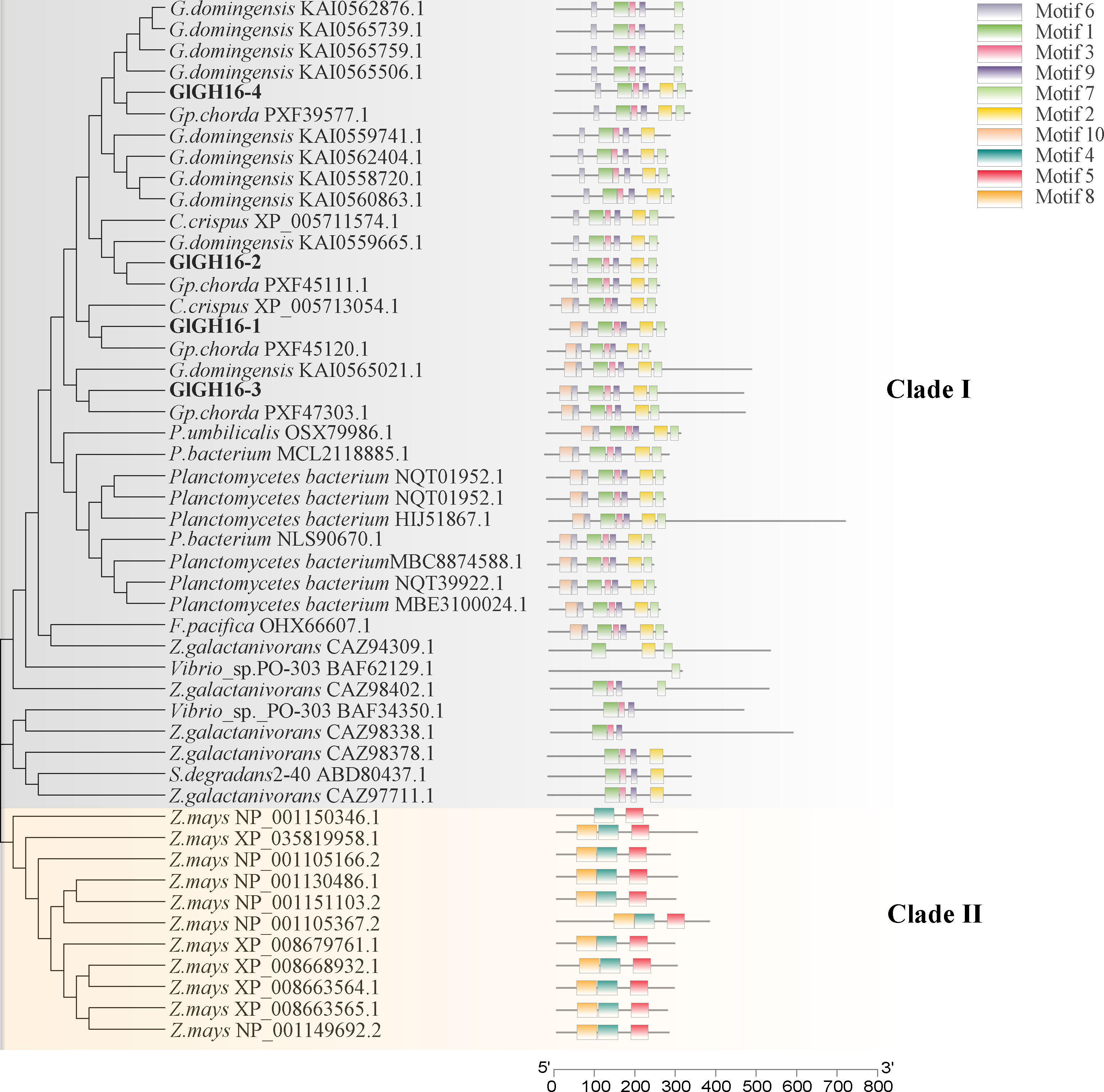
Figure 4 Phylogenetic tree and conserved motifs of GH16 members constructed based on the protein sequences using the maximum likelihood method. The bootstrap analysis was carried out with 1,000 replicates.
Interaction network of the GT7 and GH16 families
No genomic information of Gp. lemaneiformis is present in the STRING database; however, Gp. lemaneiformis and Gp. chorda are closely evolutionarily related. In this study, to reveal the functional role of the GT7 and GH16 families, protein–protein interactions (PPIs) were predicted using the STRING database information about Gp. chorda. The GT7 (annotated as chondroitin sulfate synthase) interaction network contained three nodes, and the interactions among chondroitin synthase and its functional partners had a confidence score of 0.501 (no significance). Three carbohydrate STs (A0A2V3J539, A0A2V3IMF1, and A0A2V3J7C1) were the only proteins predicted to be incorporated into the interaction network (Figure 5A).
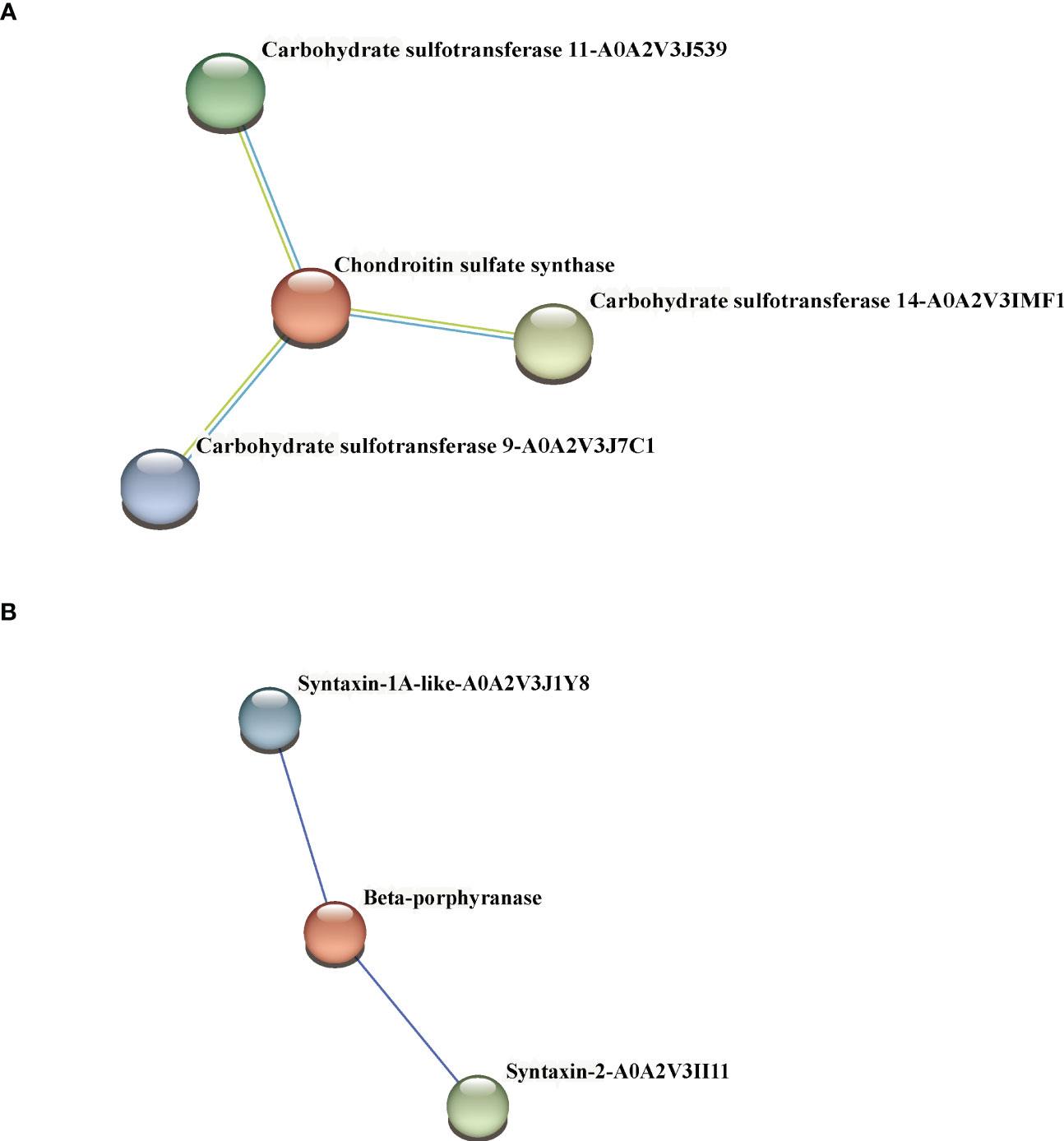
Figure 5 Protein–protein interactions among the GT7 (A) and GH16 (B) members with other proteins in the STRING database. The nodes with various colors represent the proteins that interact with the GT7 and GH16 members. The thickness of the lines represents the confidence scores of the protein–protein interactions.
GH16 members (annotated as β-porphyranases) in Gp. chorda were chosen to construct the protein interaction network (Figure 5B). The interaction network contained two nodes and interactions among β-porphyranases and syntaxin family proteins with a confidence score of 0.407 (no significance).
Validation of GlGT7-interacting proteins
The full-length sequence of GlGT7 was cloned into a pGBKT7 bait vector, and the sequence of GlST was cloned into a pGADT7 prey vector. Before validation, the toxicity and autoactivation of these recombinants were tested in Y2H yeast. The clones displayed no autoactivation on SD/-Leu/-Trp/-His/-Ade media (Supplementary Figure S2). After cotransformation of pGBKT7-GlGT7s and pGADT7-GlSTs, no clone was found to be positive on SD/-Leu/-Trp/-His/-Ade/x-α-gal/-AbA media, indicating that there are no interactions between GlGT7s and GlSTs (Supplementary Figure S3).
Relations of agar and GlGT7/GlGH16 expression levels among three cultivars of Gp. lemaneiformis
Under the experimental conditions used in this study, the agar contents of Gp. lemaneiformis 981 and Lulong No. 1 were 113% and 37% higher, respectively, than that of the wild-type seaweed (Figure 6A). Agar contents were positively correlated with the expression variations of GlGT7s and negatively related to those of GlGH16s (Table 4). The relative expression levels of four GlGT7 genes in cultivar 981 were all significantly increased compared with those in the other two cultivars; this was especially true for GlGT7-4, whose expression increased 6.02-fold relative to that of the wild-type thalli. In the cultivar Lulong No. 1, only GlGT7-2 and GlGT7-4 displayed expression levels higher than those of the wild-type thalli. In contrast to the GlGT7s, the GlGH16 genes showed the response opposite to those of the agar; the GlGH16 expression levels in cultivar 981 and Lulong No. 1 were all decreased compared with those in the wild-type thalli (Figure 6B).
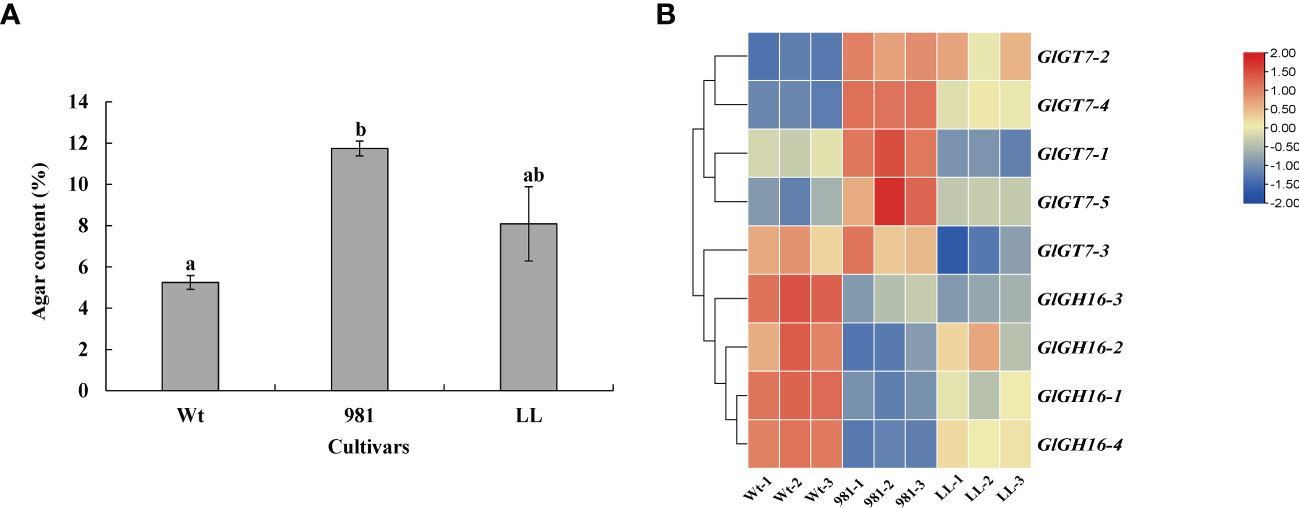
Figure 6 Agar contents (A) and expression levels of GlGT7s and GlGH16s (B) in the three cultivars of Gracilariopsis lemaneiformis. Wt, wild-type; 981, cultivar 981; LL, cultivar Lulong No. 1. The different lowercase letters represent significant differences.
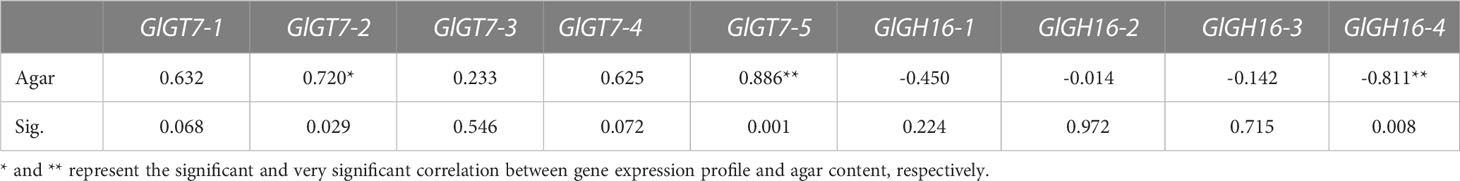
Table 4 Pearson correlations between expression profiles of GlGT7 and GlGH16 genes and agar contents in different treatments.
Relations of agar and GlGT7/GlGH16 transcription under abiotic stresses
To explore the relations between agar and the expression profiles of the GlGT7 and GlGH16 genes, agar accumulation and gene expression levels were assessed under heat and low nitrogen stresses. In this study, the agar contents did not significantly change, whereas the expression levels of several GlGT7 genes fluctuated under moderate heat stress (Figure 7). GlGT7-1, GlGT7-2, and GlGT7-5 were downregulated in response to heat stress compared with the CK, whereas GlGT7-3 and GlGT7-4 were upregulated—notably the expression of GlGT7-4 increased 8.70-fold. Four GlGH16 genes were all upregulated in response to heat stress; this was especially true for GlGH16-2 and GlGH16-3, whose expression significantly increased 4.14- and 2.64-fold, respectively.
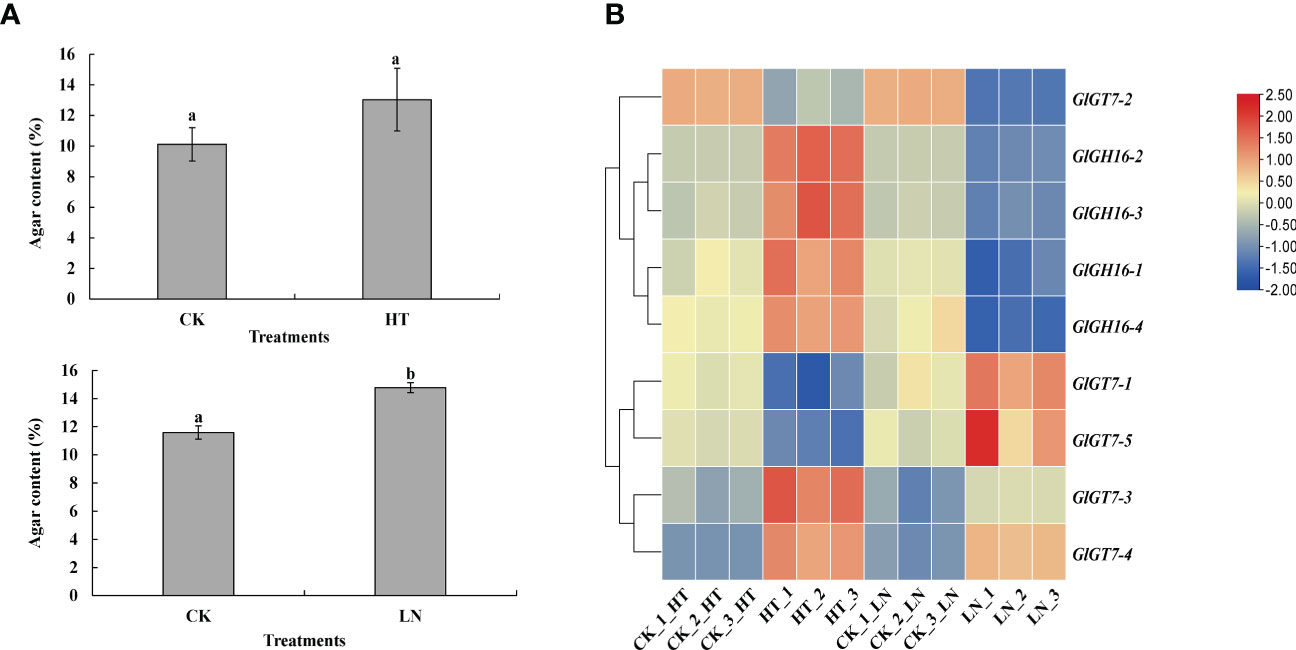
Figure 7 Agar content (A) and expression levels of GlGT7s and GlGH16s (B) under heat and nitrogen stresses. CK, the control treatment; HT, high-temperature treatment; LN, low-nitrogen treatment. The different lowercase letters represent significant differences.
Compared with the unchanged agar content under heat stress, the agar content increased 115% under low nitrogen conditions (Figure 7A). The expression changes of the GlGT7 and GlGH16 genes showed distinct expression change patterns under low nitrogen stress (Figure 7B). Low nitrogen stress stimulated the expression of four out of five GlGT7 genes, especially GlGT7-4, whose expression increased 5.90-fold, whereas all four GlGH16 genes were significantly downregulated by low nitrogen stress.
Pearson correlation analysis was performed on the agar variation data and expression profile data of GlGT7s and GlGH16s under different treatments (abiotic stresses and for the different cultivars of Gp. lemaneiformis). The expression profiles of GlGT7s were positively correlated with agar contents; this was especially true for GlGT7-2 and GlGT7-5, whose correlation coefficients were 0.720 and 0.903, respectively. GlGH16s were negatively correlated with agar content, and this was especially the case for GlGH16-4, which had a correlation coefficient of -0.811.
Cell morphology and fluorescence observations under abiotic stresses
After cultivation under heat and low nitrogen stresses, the thalli were observed with RCA120-fluorescein, which can label β-1,4-galactosidic bonds (Figure 8). The thalli are cylindrical, and the algal cells are mainly concentrated around the circle. Stress treatments had no significant effect on the morphology of the thalli. However, β-1,4-galactan, the main structural component of agar chains, accumulated to significant levels after the thalli were exposed to heat or low nitrogen stresses.
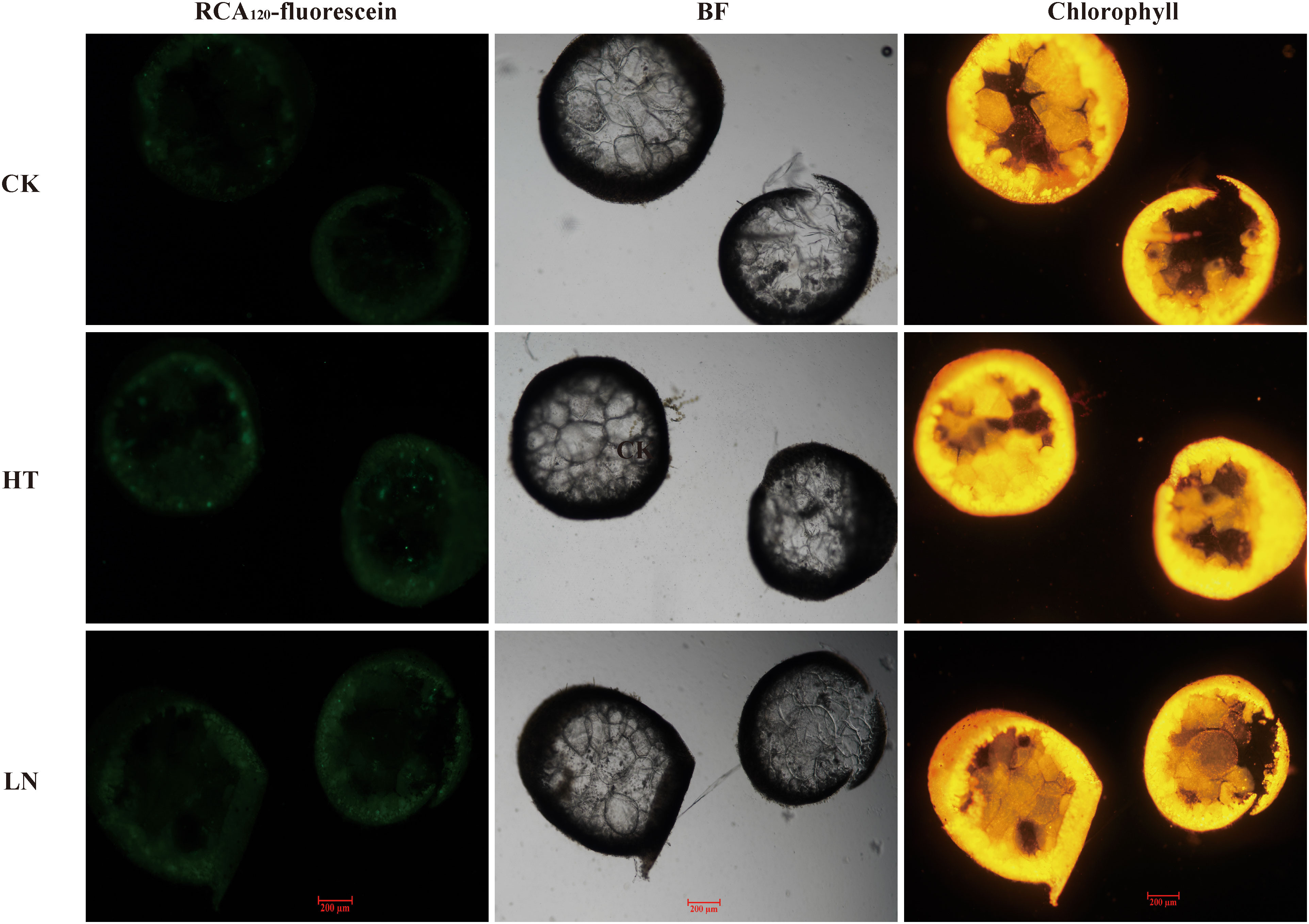
Figure 8 Cell morphology and fluorescence observations under abiotic stresses. RCA120-fluorescein and chlorophyll were excited at 488 and 561 nm, and their corresponding emitted fluorescence was detected at 530/30 and 695/40 nm, respectively. BF, bright field; CK, the control treatment; HT, high-temperature treatment; LN, low-nitrogen treatment.
Discussion
Evolution and function of GT7 members
The majority of agar is extracted from agarophytes, in which Gracilaria/Gracilariopsis contribute up to 60%–80% of the agar produced owing to the abundant agar yields and excellent agar properties. Some studies have explored the correlations between agar yield and the expression of genes, such as those that encode galactose-1-phosphate uridylyltransferase (Li et al., 2010), UDP-glucose pyrophosphorylase (Chang et al., 2014), phosphoglucomutase and phosphomannomutase (Tang et al., 2015), mannose-6-phosphate isomerase and GDP-mannose-3′,5′-epimerase (Hu et al., 2018), and UDP-galactose-4-epimerase (Lim et al., 2019). However, limited research has focused on the influence of GTs on sulfated polysaccharide synthesis. Ficko-Blean et al. (2015) suggested that at least two classes of GTs were required for the formation of α-1,3- and β-1,4-galactopyranosyl linkages in agar polysaccharides. In the present study, we identified five GT7 genes in the Gp. lemaneiformis genome that may be involved in agar biosynthesis. Compact genes are typical for this group, as there is no intron in any of the GlGT7 genes (Table 2), which is consistent with previous observations in other red algae (Brawley et al., 2017; Chen H. M. et al., 2022). Collén et al. (2013) proposed a possible evolutionary model of red algae: the ancestor of red algae was presumed to have a genome that was rich in introns, and genome reduction may occur in extreme environments, followed by genome expansion through the activity of transposable elements.
In our study, no GT7 sequences were found in red microalgae (Table 1), suggesting that the acquisition of these genes could have occurred after divergence from the red algae Cyanidiales (Lee and Ho, 2022). According to the phylogenetic tree (Figure 2), GlGT7 proteins clustered with GT7 proteins from other red algae, followed by those of marine organisms, such as S. clava and sea urchins, which are considered to have entered the eukaryotic lineage through secondary endosymbiosis (Keeling, 2010). Interestingly, the GT7 sequences from brown algae, diatoms, and animals clustered together as candidate genes involved in fucoidan biosynthesis; these genes are present in mammals, plants, and most bacteria. This may indicate the ancestral pathway of fucoidans (Han et al., 2019).
The putative chondroitin sulfate synthase was speculated to participate in carrageenan biosynthesis, as this enzyme has β-1,4-GalT activity, which is necessary for the biosynthesis of carrageenan and agar (Ho, 2020). Here, the catalytic pocket residue WGGEDDD was present in all GT7 proteins (Figure 3). This domain is conserved across most β-1,4-GalT-T7 proteins (Ramakrishnan and Qasba, 2010), demonstrating that the GT7 gene products in red seaweeds have the capacity for β-1,4-galactan transfer. To elucidate the possible regulatory mechanism of agar biosynthesis, using chondroitin sulfate synthase of Gp. chorda as the target protein, we conducted online protein–protein interaction predictions (Figure 5A). Here, ST was predicted to interact with GT7 proteins. The enzymes such as ST, galactosyltransferase, and sulfurylases are the main classes responsible for sulfated polysaccharide formation (Garcia-Jimenez et al., 2020). ST can transfer the sulfate group from adenosine-3′-phosphate-5′-phosphate sulfate (PAPS) to an agar chain (Lee et al., 2017a). The physicochemical properties and biological activities of sulfated polysaccharides are affected by the pattern and number of sulfate moieties (Ho, 2015). Different life stages and living niches can lead to differential carrageenan qualities, among which ST plays an essential role (Helbert, 2017). The interaction between chondroitin sulfate synthase and ST provides a possible biosynthetic mechanism underlying agar chain production.
Due to the importance of sulfation in agar composition, to validate the possible biosynthetic mechanism of the agar chain, we conducted Y2H assays to validate the interactions between GlGT7s and GlSTs. Although GalTs are in the Golgi apparatus, at a location close to STs, they facilitate fast assembly of these nucleotide sugars into polysaccharide chains (Collén et al., 2013), and the STRING database revealed interactions between GT7s and STs in Macaca mulatta, Latimeria chalumnae, and Ornithorhynchus anatinus. In this study, one-to-one verifications revealed no interactions between these proteins (Supplementary Figure S3), which was consistent with that carbohydrate STs may independently transfer sulfate groups from PAPS to growing agar chains (Lipinska et al., 2020). However, due to the uncertainty of the assembled genome and the lack of experimental research on the agar synthesis mechanism, whether ST and GT function synergistically in the synthesis of agar chains remains to be further explored.
Evolution and function of GH16 members
Sulfated polysaccharides in red seaweeds provide important food and industrial resources. Furthermore, many marine bacteria produce specific GHs to break down these bioactive compounds for carbon use (Hehemann et al., 2012). As agar is the main component of the cell wall in agarophyte seaweeds, its remodeling is vital to regulate agar yield and quality, resulting in the changes in the growth and physiology of seaweeds.
To explore the function of GH16s in algae, GH16 genes from the genomes of different species were retrieved (Table 1). Sequences of the proteins annotated as β-agarase, β-porphyranase, and carrageenase in CAZy were downloaded. In higher plants, no agarase-related sequences were retrieved from the Arabidopsis thaliana genome; however, sequences of 18 genes belonging to the GH16 family were identified at The Arabidopsis Information Resource (TAIR, (https://www.arabidopsis.org/browse/genefamily/index.jsp). In Z. mays, 11 GH16 sequences were found, but no similar conserved motifs that matched the sequences of red seaweeds and marine bacteria were detected, which proved the absence of agarase in higher plants. In brown algae, no GH16 sequences related to β-agarase, β-porphyranase, and carrageenase were found; in addition, XTH in E. siliculosus is absent, implying that brown algae have evolved different molecular mechanisms for cell wall stress relaxation and expansion (Michel et al., 2010). In red algae, four GH16 genes in Gp. lemaneiformis were identified, and their structures are compact, with either no introns or only one intron within GlGH16 genes.
Red seaweeds and marine bacteria have similar conserved motif compositions, indicating their possibility of origin and horizontal gene transfer (HGT). Furthermore, the classic catalytic site EIDVFE is conserved in all red seaweeds, and this module is the classic site in AgaC-type proteins in some marine bacteria (Figure 3), suggesting that the GH16 sequences behave as does β-agarase acting on agar (Naretto et al., 2019). This type of GH is now widely present in marine bacteria and effectively breaks down the structure of G-L6S, the typical backbone of agar in agarophyte (Jiang et al., 2020; Rhein-Knudsen and Meyer, 2021). Because of the essential function of these agarases from bacteria, these GH16 sequences from agarophytes may play a role in cleaving and remodeling agar. Additionally, the protein interaction prediction results demonstrated that the majority of proteins interacting with GH16s are syntaxin proteins (Figure 5B); however, the interaction network between GH16 sequences and predicted proteins needs to be further verified.
Responses of GT7/GH16 to light and phytohormone signals
Promoters are crucial elements that regulate gene expression, and their regulatory effect is the result of multilevel and multifactor interactions. Here, notwithstanding that the most predicted promoter elements responded to light, high light suppressed the expression of GT7s, which was consistent with the decrease in agar content (Supplementary Figure S1A). However, the reports about the effect of high light on agar accumulation were few. Several reports can be found about the influence of low light to agar production, e.g., low light conditions decreased the agar content (Lee et al., 2017b); optimum illumination decreased the agar content, whereas low light resulted in agar accumulation in Gelidium coulteri (Macler, 1986). Here, the expression levels of four GlGH16 genes varied in response to high light, and the correlation of GlGH16s with the cell wall or agar production was unclear, although GH16s, just like XTH (family GH16), are putatively involved in wall remodeling and expansion (Henrissat et al., 2001; Cosgrove, 2005).
Plant hormones such as auxins, cytokinins, and ethylene are broadly defined in terms of their roles in growth and development, and some are considered to be “stress” hormones, such as brassinosteroids (BRs), jasmonic acid (JA), salicylic acid (SA), and ABA (Santner et al., 2009). Hormone signaling pathways play vital roles in cell wall formation to help resist stress (Wang et al., 2019). Gibberellin levels or signaling can alter cellulose and lignin accumulation in plant secondary cell walls (Wang et al., 2017). The downregulation of genes involved in BR synthesis leads to a reduction in cellulose and lignin contents (Hossain et al., 2012). Cellulose production in A. thaliana is regulated via BR signaling, with cellulose synthases (CESAs) modulated at both the transcriptional and posttranscriptional levels. The transcription factor BES1 is activated through BR signaling and can directly bind to CANNTG-E motifs in the promoters of CESA genes (Yennawar et al., 2006). Overexpression of XTH1 can be regulated by hormones including ABA and auxins, resulting in a high cell wall density and thus enhancing tolerance to salt stress (Han et al., 2017). In this study, ABA and MeJA fluctuated the transcription of GlGT7s and GlGH16s (Figures 1B, C), and yet the agar production almost unchanged after the hormone treatments (Supplementary Figures S1B, C). This result might be partially caused by the treatment time or other reasons, and the potential role of GlGT7s and GlGH16s in agar metabolism after hormone application still needs further exploration.
Agar regulatory mechanisms of the three cultivars in response to abiotic stress
Agarophytes with higher agar yield and superior gelling properties are desired due to the growing demand for agar in the global market. The synthesis of sulfated polysaccharides (mainly agar, carrageenan, and alginate) can be altered by environmental changes and geographical variations (Lee et al., 2016; Véliz et al., 2019). Seasons and life cycle stages of red macroalgae differ from the contents and chemical structures of agar and carrageenan to adapt to the environment (Imbs et al., 2009; Ciancia et al., 2021). Agar can accumulate to resist stress conditions, including acidification (Lee et al., 2019), osmotic stress (Hu et al., 2018), high or low light (Silva-Brito et al., 2021), nutrient deficiency (Rocha et al., 2019), and temperature extremes (Tang et al., 2015). To understand the regulation of agar yield, we investigated the expression profiles of putative GT7 agar synthesis-related genes and GH16 cell wall-remodeling genes under high-temperature and low nitrogen stresses. Temperature is one of the most important factors affecting the growth and physiology of Gp. lemaneiformis. Agar yield increased with increasing water temperature (Figure 7A). Martin et al. (2013) found that the highest agar yield of G. gracilis occurred in summer. Friedlander (1991) reported that both the agar yield and the gel strength of G. conferta cultured in tanks were positively correlated with water temperature. However, in the present study, the agar content did not differ compared with that of the CK, which may be due to the differences in culture conditions.
Nitrogen can influence agar yield (Chen Q. L. et al., 2022). The agar contents of Gracilaria sordida was shown to be negatively correlated with the nitrogen level of the environment (Pickering et al., 1990). In our study, the agar content of Gp. lemaneiformis increased in response to low nitrogen stress (Figure 7A). Combining our expression profile data, we found that the GlGT7 and GlGH16 genes co-participated in moderating agar production. Among these GlGT7 genes, GlGT7-4 is highly correlated with agar variation. In the three cultivars of Gp. lemaneiformis, the agar content of cultivar 981 was higher than that of wild-type thalli, and this phenomenon is in accordance with the increasing expression of GlGT7s and the decreasing expression of GlGH16s (Figure 7B).
Agar chains are mainly composed of β-1,4- and α-1,3-galactose, which account for approximately 20%–30% of the algal dry biomass. RCA120 lectin recognizes carbohydrate chains with non-reducing terminal β-1,4-galactose specific to sulfated galactose (Wang et al., 2011). In this study, the fluorescence intensity of RCA120-fluorescein was enhanced under heat or nitrogen stresses, the phenomenon of which is in accordance with the results of agar content and GlGT7 expression (Figure 8). This result further verified the correlations between GlGT7s and agar accumulation.
Conclusion
This study first reports the identification of the most promising candidates for agar polymerization and degradation, and systematic bioinformatic analysis of GT7 and GH16 in Gp. lemaneiformis. Altogether, five GlGT7 genes and four GlGH16 genes were identified through whole-genome searching. The compact features, conserved catalytic pocket residues, and evolutionary relationships of the GT7 and GH16 sequences suggested that they function as β-1,4-galactosyltransferases and agarases, respectively. The protein interaction network revealed that GlGT7 can function cooperatively with ST in agar biosynthesis, although the Y2H assays did not support this. GlGT7 and GlGH16 gene expressions were sensitive to heat and low nitrogen stress. The fluorescence staining results indicated that β-1,4-galactose accumulated under heat and nitrogen stresses, which was consistent with the expressional variation of the GlGT7s. The positive relations of GlGT7s and the negative relations of GlGH16s with agar yield were confirmed by Pearson correlation analysis. Especially, GlGT7-2, GlGT7-5, and GlGH16-4 could be used as molecular markers for indicating agar yield. This study therefore opens up more opportunities for others to confirm their involvement in the agar metabolism.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
Conceptualization, XS and QC; data curation, QC and XY; formal analysis, QC; funding acquisition, NX and XS; investigation, CH and QC; methodology, SL and QC; project administration, NX software, QC, XY and SL; writing—original draft, QC; writing—review and editing, SL, CH, NX, and XS. All authors have reviewed and agreed to the published version of the manuscript.
Funding
This research was funded by Key Program of Science and Technology Innovation in Ningbo (2019B10009) and Major Project of Science and Technology in Ningbo (2021Z114, 2022Z172).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1156490/full#supplementary-material
References
Bhattacharya D., Price D. C., Chan C. X., Qiu H., Rose N., Ball S., et al. (2013). Genome of the red alga porphyridium purpureum. Nature 4, 1941. doi: 10.1038/ncomms2931
Bowler C., Allen A. E., Badger J. H., Grimwood H., Jabbari K., Kuo A., et al. (2008). The phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. doi: 10.1038/nature07410
Brawley S. H., Blouin N. A., Ficko-Blean E., Wheeler G. L., Lohr M., Goodson H. V., et al. (2017). Insights into the red algae and eukaryotic evolution from the genome of Porphyra umbilicalis (Bangiophyceae, Rhodophyta). PNAS 114, E6361–E6370. doi: 10.1073/pnas.1703088114
Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2008). The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D2338. doi: 10.1093/nar/gkn663
Chang L. P., Sui Z. H., Fu F., Zhou W., Wang J. G., Kang K. H., et al. (2014). Relationship between gene expression of UDP-glucose pyrophosphorylase and agar yield in Gracilariopsis lemaneiformis (Rhodophyta). J. Appl. Phycol. 26, 2435–2441. doi: 10.1007/s10811-014-0277-7
Chen H. M., Chu J. S. C., Chen J. J., Luo. Q. J., Wang H., Lu R., et al. (2022). Insights into the ancient adaptation to intertidal environments by red algae based on a genomic and multiomics investigation of Neoporphyra haitanensis. Mol. Biol. Evol. 39, msab315. doi: 10.1093/molbev/msab315
Chen Q. L., Yu X. L., Liu S. X., Luo S. Y., Chen X. J., Xu N. J., et al. (2022). Identification, characteristic and function of phosphogluco-mutase (PGM) in the agar biosynthesis and carbon flux in the agarophyte Gracilariopsis lemaneiformis (Rhodophyta). Mar. Drugs 20, 442. doi: 10.3390/md20070442
Ciancia M., Matulewicz M. C., Tuvikene R. (2021). Structural diversity in galactans from red seaweeds and its influence on rheological properties. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.559986
Collén J., Porcel B., Carré W., Ball S. G., Chaparro C., Tonon T., et al. (2013). Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the archaeplastida. PNAS 110, 5247–5252. doi: 10.1073/pnas.1221259110
Cosgrove D. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Bio 6, 850–861. doi: 10.1038/nrm1746
Ding Y., Sun H., Zhang R., Yang Q., Liu Y. T., Zang X. N., et al. (2014). Selection of reference gene from Gracilaria lemaneiformis under temperature stress. J. Appl. Phycol. 27, 1365–1372. doi: 10.1007/s10811-014-0423-2
Ezquer I., Salame E., Colombo L., Colombo P. (2020). Plant cell walls tackling climate change: insights into plant cell wall remodeling, its regulation, and biotechnological strategies to improve crop adaptations and photosynthesis in response to global warming. Plants 9, 212. doi: 10.3390/plants9020212
Ficko-Blean E., Hervé C., Michel G. (2015). Sweet and sour sugars from the sea: the biosynthesis and remodeling of sulfated cell wall polysaccharides from marine macroalgae. Perspect. Phycol. 2, 51–64. doi: 10.1127/pip/2015/0028
Friedlander M. (1991). Growth rate, epiphyte biomass and agar yield of Gracilaria conferta in an annual outdoor experiment. 1. irradiance and nitrogen. Biores. Technol. 38, 203–208. doi: 10.1016/0960-8524(91)90155-D
Gann E. R., Truchon A. R., Papoulis S. E., Dyhrman S. T., Gobler C. J., Wihelm S. W. (2021). Aureococcus anophagefferens (Pelagophyceae) genomes improve evaluation of nutrient acquisition strategies involved in brown tide dynamics. J. Phycol. 58, 146–160. doi: 10.1111/jpy.13221
Gao B. L., Li L., Wu H., Zhu D., Jin M., Qu W., et al. (2019). A novel strategy for efficient agaro-oligosaccharide production based on the enzymatic degradation of crude agarose in Flammeovirga pacifica WPAGA1. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01231
Garcia-Jimenez P., Mantesa S. R., Robaina R. R. (2020). Expression of genes related to carrageenan synthesis during Carposporogenesis of the red seaweed Grateloupia imbricata. Marine drugs 18, 432. doi: 10.3390/md18090432
Han W. T., Fan X., Teng L. H., Kaczurowski M. J. S., Zhang X. W., Xu D., et al. (2019). Identification, classification, and evolution of putative xylosyltransferases from algae. Protoplasma 256, 1119–1132. doi: 10.1007/s00709-019-01358-2
Han Y., Han S., Ban Q., He Y., Jin M., Rao J. (2017). Overexpression of persimmon DkXTH1 enhanced tolerance to abiotic stress and delayed fruit softening in transgenic plants. Plant Cell Rep. 36, 583–596. doi: 10.1007/s00299-017-2105-4
Hehemann J. H., Correc G., Thomas F., Barbeyron T., Jam M., Helbert W., et al. (2012). Biochemical and structural characterization of the complex agarolytic enzyme system from the marine bacterium Zobellia galactanivorans. J. Biol. Chem. 287, P30571–P30584. doi: 10.1074/jbc.M112.377184
Helbert W. (2017). Marine polysaccharide sulfatases. Front. Mar. Sci. 4. doi: 10.3389/fmars.2017.00006
Henrissat B., Coutinho P., Davies G. (2001). A census of carbohydrate-active enzymes in the genome of Arabidopsis thaliana. Plant Mol. Biol. 47, 55–72. doi: 10.1023/A:1010667012056
Ho C. L. (2015). Phylogeny of algal sequences encoding carbohydrate sulfotransferases, formylglycine-dependent sulfatases, and putative sulfatase modifying factors. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.01057
Ho C. L. (2020). Comparative genomics reveals differences in algal galactan biosynthesis and related pathways in early and late diverging red algae. Genomics 112, 1536–1544. doi: 10.1016/j.ygeno.2019.09.002
Hossain Z., McGarvey B., Amyot L., Gruber M., Jung J., Hannoufa A. (2012). DIMINUTO 1 affects the lignin profile and secondary cell wall formation in Arabidopsis. Planta 235, 485–498. doi: 10.1007/s00425-011-1519-4
Hou S. N., Lin L. C., Lv Y., Xu N. J., Sun X. (2018). Responses of lipoxygenase, jasmonic acid and salicylic acid to temperature and exogenous phytohormone treatments in Gracilariopsis lemaneiformis (Rhodophyta). J. Appl. Phycol. 30, 3387–3394. doi: 10.1007/s10811-018-1514-2
Hu Y. Y., Du Q. W., Mi P., Shang E. L., Sui Z. H. (2018). Gene cloning and expression regulation in the pathway of agar and floridean starch synthesis of Gracilariopsis lemaneiformis (Rhodophyta). J. Appl. Phycol. 31, 1889–1896. doi: 10.1371/journal.pone.0069909
Imbs T. I., Shevchenko N. M., Sukhoverkhov S. V., Semenova T. L., Skriptsova A. V., Zvyagintseva T. N. (2009). Seasonal variations of the compostion and structural characteristics of polysaccharides from the brown alga Costaria costata. Chem. Nat. Compd. 45, 786–791. doi: 10.1007/s10600-010-9507-7
Jiang C. C., Liu Z., Cheng D. Y., Mao X. Z. (2020). Agarose degradation for utilization: enzymes, pathways, metabolic engineering methods and products. Biotechnol. Adv. 45, 107641. doi: 10.1016/j.biotechadv.2020.107641
Keeling P. J. (2010). The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. Lond. Ser. B. Biol. Sci. 365, 729–748. doi: 10.1098/rstb.2009.0103
Kim H. T., Lee S., Lee D., Kim H. S., Bang W. G., Kim K. H., et al. (2010). Overexpression and molecular characterization of Aga50D from Saccharophagus degradans 2-40: an exo-type β-agarase producing neoagarobiose. Appl. Microbiol. Biotechnol. 86, 227–234. doi: 10.1007/s00253-009-2256-5
Kim D. H., Mahomoodally M. F., Sadeer N. B., Seok P. G., Zengin G., Palaniveloo K., et al. (2021). Nutritional and bioactive potential of seagrasses: a review. South Afr. J. Bot. 137, 216–227. doi: 10.1016/j.sajb.2020.10.018
Lee W. K., Ho C. L. (2022). Ecological and evolutionary diversification of sulphated polysaccharides in diverse photosynthetic lineages: a review. Carbohyd. Polym. 277, 118764. doi: 10.1016/j.carbpol.2021.118764
Lee W. K., Lim Y. Y., Ho C. L. (2019). pH affects growth, physiology and agar properties of agarophyte Gracilaria changii (Rhodophyta) under low light intensity from morib, Malaysia. Reg. Stud. Mar. Sci. 30, 100738. doi: 10.1016/j.rsma.2019.100738
Lee W. K., Lim Y. Y., Leow A. T., Namasivayam P., Abdullah J. O., Ho C. L. (2016). Factors affecting yield and gelling properties of agar. J. Appl. Phycol. 29, 1527–1540. doi: 10.1007/s10811-016-1009-y
Lee W. K., Lim Y. Y., Leow A. T., Namasivayam P., Abdullah J. O., Ho C. L. (2017a). Biosynthesis of agar in red seaweeds: a review. Carbohyd. Ploym. 164, 23–30. doi: 10.1016/j.carbpol.2017.01.078
Lee W. K., Lim Y. Y., Leow A. T., Namasivayam P., Abdullah J. O., Ho C. L. (2017b). Factors affecting yield and gelling properties of agar. J. Appl. Phycol. 29, 1527–1540. doi: 10.1007/s10811-016-1009-y
Lee J. M., Yang E. C., Graf L., Yang J. H., Qiu H., Zelzion U., et al. (2018). Analysis of the draft genome of the red seaweed Gracilariopsis chorda provides insights into genome size evolution in rhodophyta. Mol. Biol. Evol. 35, 1869–1886. doi: 10.1093/molbev/msy081
Li M., Sui Z. H., Kang K. H., Zhang X. C., Zhu M., Yan B. L. (2010). Cloning and analysis of the galactose-1-phosphate uridylyltransferase (Galt) gene of Gracilariopsis Lemaneiformis (Rhodophyta) and correlation between gene expression and agar synthesis. J. Appl. Phycol. 22, 157–164. doi: 10.1007/s10811-009-9435-8
Lim Y. Y., Lee W. K., Lim P. E., Phang S. M., Leow A. T.C., Namasivayam P., et al. (2019). Expression analysis of potential transcript and protein markers that are related to agar yield and gel strength in gracilaria changii (Rhodophyta). Algal. Res. 41, 101532. doi: 10.1016/j.algal.2019.101532
Lin J., Chen Y., Yan H. L., Nahidian B., Hu Q., Han D. X. (2020). High-throughput fluorescence-activated cell sorting for cell wall-deficient microalgal mutants screening. Algal. Res. 50, 102011. doi: 10.1016/j.algal.2020.102011
Lipinska A. P., Collén J., Krueger-Hadfield S. A., Mora T., Ficko-Blean E. (2020). To gel or not to gel: differential expression of carrageenan-related genes between the gametophyte and tetrasporophyte life cycle stages of the red alga Chondrus crispus. Sci. Rep. 10, 11498. doi: 10.1038/s41598-020-67728-6
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Macler B. A. (1986). Regulation of carbon flow by nitrogen and light in the red alga, Gelidium coulteri. New Physiol. 82, 136–141. doi: 10.1104/pp.82.1.136
Martin L. A., Rodriguez M. C., Matulewicz M. C., Fissore E. N., Gerschenson L. N., Leonardi P. I. (2013). Seasonal variation in agar composition and properties from Gracilaria gracilis (Gracilariales, Rhodophyta) of the Patagonian coast of Argentina. Phycol. Res. 61, 163–171. doi: 10.1111/pre.12000
Matsuzaki M., Misumi O., Shin-I T., Maruyama S., Takahara M., Miyagishima S., et al. (2004). Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653–657. doi: 10.1038/nature02398
Michel G., Tonon T., Scornet D., Cock M. J., Kloareg B. (2010). The cell wall polysaccharide metabolism of the brown alga Ectocarpus siliculosus. insights into the evolution of extracellular matrix polysaccharides in eukaryotes. New Phytol. 188, 82–97. doi: 10.1111/j.1469-8137.2010.03374.x
Nakamura-Gouvea N., Alves-Lima C. (2022). Insights into agar and secondary metabolite pathways from the genome of the red alga Gracilaria domingensis (Rhodophyta, gracilariales). J. Phycol. 58, 406–423. doi: 10.1111/jpy.13238
Naretto A., Fanuel M., Ropartz D., Rogniaux H., Larocque R., Czjzek M., et al. (2019). The agar-specific hydrolase ZgAgaC from the marine bacterium Zobellia galactanivorans defines a new GH16 protein subfamily. J. Biol. Chem. 294, P6923–P6939. doi: 10.1074/jbc.RA118.006609
Pickering T. D., Gordon M. E., Tong L. J. (1990). Seasonal growth, density, reproductive phenology and agar quality of Gracilaria sordida (Gracilariales, Rhodophyta) at mokomoko inlet, new Zealand. Hydrobiologia 204, 253–262. doi: 10.1007/BF00040242
Ramakrishnan B., Qasba P. K. (2010). Crystal structure of the catalytic domain of Drosophila β-1,4-galactosyltransferase-7. J. Biol. Chem. 285, P15619–P15626. doi: 10.1074/jbc.M109.099564
Rhein-Knudsen N., Meyer A. S. (2021). Chemistry, gelation, and enzymatic modification of seaweed food hydrocolloids. Trends Food Sci. Tech. 109, 608–621. doi: 10.1016/j.tifs.2021.01.052
Rocha C. M., Sousa A. M., Kim J. K., Magalhães J. M., Yarish C., do Pilar Gonçalves M. (2019). Characterization of agar from Gracilaria tikvahiae cultivated for nutrient bioextraction in open water farms. Food Hydrocoll. 89, 260–271. doi: 10.1016/j.foodhyd.2018.10.048
Santner A., Calderon-Villalobos L. I. A., Estelle M. (2009). Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 5, 301–307. doi: 10.1038/nchembio.165
Sato T., Kudo T., Ikehara Y., Ogawa H., Hirano T., Kiyohara K., et al. (2011). Chondroitin sulfate n-acetylgalactosaminyltransferase 1 is necessary for normal endochondral ossification and aggrecan metabolism. J. Biol. Chem. 286, 5803–5812. doi: 10.1074/jbc.M110.159244
Seo Y. B., Lu Y., Chi W. J., Park H. R., Jeong K. J., Hong S. K., et al. (2014). Heterologous expression of a newly screened β-agarase from Alteromonas sp. GNUM1 in Escherichia coli and its application for agarose degradation. Process. Biochem. 49, 430–436. doi: 10.1016/j.procbio.2013.12.014
Silva-Brito F., Pereira S. G., Rocha C. M. R., da Costa E., Domingues M. R., Azevedo A., et al. (2021). Improving agar properties of farmed Gracilaria gracilis by using filtered sunlight. J. Appl. Phycol. 33, 3397–3411. doi: 10.1007/s10811-021-02497-x
Sun P., Chen Q. L., Luo S. Y., Yu X. L., Zhang X. Q., Xu N. J., et al. (2022). The role and mechanism of abscisic acid in mitigating the adverse impacts of high temperature in Gracilariopsis lemaneiformis. J. Appl. Phycol. 34, 1073–1087. doi: 10.1007/s10811-021-02606-w
Sun X., Wu J., Wang G., Kang Y., Ooi H. S., Shen T., et al. (2018). Genomic analyses of unique carbohydrate and phytohormone metabolism in the macroalga Gracilariopsis lemaneiformis (Rhodophyta). BMC Plant Biol. 18, 94. doi: 10.1186/s12870-018-1309-2
Tang X. B., Xu N. J., Sun X., Li Y. H., Zhang B. (2015). Effects of 24-epibrassinolide on the agar synthesis and expression of genes involved in marine alga Gracilariopsis lemaneiformis. J. Fish. China 39, 1788–1798. doi: 10.11964/jfc.20150309742
Ulvskov P., Paiva D. S., Domozych D., Harholt J. (2013). Classification, naming and evolutionary history of glycosyltransferases from sequenced green and red algal genomes. PloS One 8, e76511. doi: 10.1371/journal.pone.0076511
Véliz K., Chandía N., Karsten U., Lara C., Thiel M. (2019). Geographic variation in biochemical and physiological traits of the red seaweeds Chondracanthus chamissoi and Gelidium lingulatum from the south east pacific coast. J. Appl. Phycol. 31, 665–682. doi: 10.1007/s10811-018-1532-0
Wang G. L., Que F., Xu Z. S., Wang F., Xiong A. S. (2017). Exogenous gibberellin enhances secondary xylem development and lignification in carrot taproot. Protoplasma 254, 839–848. doi: 10.1007/s00709-016-0995-6
Wang G. L., Ren X. Q., Liu J. X., Yang F., Wang Y. P., Xiong A. S. (2019). Transcript profiling reveals an important role of cell wall remodeling and hormone signaling under salt stress in garlic. Plant Physiol. Biochem. 135, 87–98. doi: 10.1016/j.plaphy.2018.11.033
Wang J. G., Sui Z. H., Hu Y. Y., Zhou W., Wei H. H., Du Q. W., et al. (2016). Assessment of photosynthetic performance, carboxylase activities, and ATP content during tetrasporic development in Gracilariopsis lemaneiformis (Gracilariaceae, Rhodophyta). Appl. J. Phycol. 28, 2939–2952. doi: 10.1007/s10811-016-0834-3
Wang Y. F., Yu G. L., Han Z. R., Yang B., Hu Y. N., Zhao X., et al. (2011). Specificities of Ricinus communis agglutinin 120 interaction with sulfated galactose. FEBS Lett. 585, 3927–3934. doi: 10.1016/j.febslet.2011.10.035
Yennawar N. H., Li L. C., Dudzinski D. M., Tabuchi A., Cosgrove D. J. (2006). Crystal structure and activities of EXPB1 (Zea m1), a β-expansin and group-1 pollen allergen from maize. PNAS 103, 14664–14671. doi: 10.1073/pnas.0605979103
Keywords: Gracilariopsis lemaneiformis, agar metabolism, glycosyltransferase family 7 (GT7), glycoside hydrolase family 16 (GH16), correlation analysis
Citation: Chen Q, Ying X, Hu C, Liu S, Xu N and Sun X (2023) Genome-wide identification of the GT7 and GH16 genes and their correlations with agar metabolism in the agarophyte Gracilariopsis lemaneiformis. Front. Mar. Sci. 10:1156490. doi: 10.3389/fmars.2023.1156490
Received: 01 February 2023; Accepted: 05 May 2023;
Published: 02 June 2023.
Edited by:
Puja Kumari, University of Aberdeen, United KingdomReviewed by:
Lee Wei-Kang, Codon Genomics, MalaysiaPeng Jiang, Chinese Academy of Sciences (CAS), China
Amit Kumar Chaturvedi, Weizmann Institute of Science, Israel
Copyright © 2023 Chen, Ying, Hu, Liu, Xu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xue Sun, c3VueHVlQG5idS5lZHUuY24=
 Qionglin Chen
Qionglin Chen Xiao Ying
Xiao Ying Shixia Liu
Shixia Liu Xue Sun
Xue Sun