
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 01 May 2023
Sec. Marine Molecular Biology and Ecology
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1154968
Mollusk shells are characterized by hierarchical aggregation of calcium carbonate and organic matrix, and matrix protein is considered as a key active ingredient to understand shell biomineralization. In this study, a total of 21 proteins, including a novel matrix protein Hcprismatin-14 were identified in the EDTA-soluble matrix of the prismatic layer of the mussel Hyriopsis cumingii by liquid chromatography tandem mass spectrometry (LC-MS/MS). The full length of Hcprismatin-14 cDNA was cloned from the mantle of H. cumingii. Hcprismatin-14 contains a high proportion of Gly, Tyr, Arg and Asp residues, their concentrated distribution forms three structurally characteristic regions: a Gly/Tyr-rich region, a WDD-repeat region and a C-terminal basic tail. Hcprismatin-14 expression was high in mantle edge tissue in a tissue-specific analysis, and during disordered crystal deposition in a saibo transplantation assay. Knocking down Hcprismatin-14 expression with double-stranded RNA induced subgrains deposition inhibition and lost contact with chitinous scaffold. In addition, the WDD-repeat region polypeptide was involved in morphological regulation of calcite and had dose-dependent inhibitory activity against aragonite deposition in vitro. Based on these results, Hcprismatin-14 appears to be a dual-function prismatic-layer matrix protein, responsible for both framework recognition and crystal deposition. These findings contribute to understanding the relationship between the modular structure of matrix protein and their regulation mechanism during shell biomineralization in mollusks.
Biomineralization is an elaborate process involved in the orderly deposition of inorganic minerals under the control of biomacromolecule. Hence, biominerals of living organisms are usually characterized by complex and hierarchical aggregation of crystals and bio-macromolecule, and are widely involved in biological functions including support, protection and gravity sensing. The unique biomineral shell of mollusks lies external to the mantle cavity. The inert periostracum provides an enclosed space to induce calcium carbonate deposition and also protect inner calcified layers from corrosion. The prismatic and nacreous layers comprise an orderly deposition of crystals, resulting in a structure that is strong, resistant to erosion and long-lasting. Thus, the mechanisms involved in shell formation mechanism have become a focus in material and life science research (Weiner et al., 1984).
The prismatic layer is a transitional structure, which initially nucleates and grows vertically on the periostracum, forming a base for nacre tablet deposition. It was originally thought that the mature prismatic layer comprised competitively growing prisms, with prisms growing together but being separated by an organic sheath (Liu and Li, 2015). Follow-up experiments showed that a single prism could be further subdivided into nano-sized subgrains after enzymatic hydrolysis (Dauphin, 2003). The organic matrix accounts for only ~5% of the prismatic layer, but numerous matrix proteins have been isolated from shell prismatic layer. Thus, the organic matrix is now considered vital for prismatic layer biomineralization. MSI31 was the first prismatic-layer matrix protein isolated from Pinctada fucata. The poly-Gly in the N-terminus adopts a beta-sheet conformation and is involved in framework formation (Sudo, 1997). Similarly, prismalin-14, prisilkin-39, KRMR and shematrin family in P. fucata are glycine and tyrosine rich, mainly involved in framework formation (Suzuki et al., 2004; Yano et al., 2006; Zhang et al., 2006; Suzuki and Nagasawa, 2007; Kong et al., 2009). In addition, some unusual acidic matrix proteins with high proportion of Asp have been isolated from the prismatic layer, and their Asp-rich region were considered to be involved in crystal nucleation and to be essential for calcite formation in marine shellfish (Sarashina and Endo, 2001; Tsukamoto et al., 2004; Gotliv et al., 2005; Hasegawa and Uchiyama, 2010).
The mussel H. cumingii is used extensively for freshwater pearl cultivation in China, with its pearls characterized by a lustrous, colorful nacreous layer. The aquaculture of this species supplies over 95% of the total output of pearls worldwide. Over the past decade, research has increasingly focused on the shell organic matrix of H. cumingii to explore the nacre formation mechanism to further improve the quality of freshwater pearls (Luo et al., 2022a). Ma et al. compared the organic matrix of aragonite and vaterite pearls and further explored its involvement in crystal nucleation and growth (Ma et al., 2012; Ma et al., 2013). Bai et al. selected a subset of genes differentially expressed in white and purple nacre (Bai et al., 2013). Moreover, numerous nacreous layer matrix proteins have been isolated and shown to have an essential role in constructing the matrix framework, regulating nucleation and controlling crystal morphology (Jin et al., 2019b; Jin et al., 2020; Luo et al., 2022b). By contrast, few prismatic layer matrix proteins have been isolated given that the brown prismatic layer is almost covered by a lustrous nacreous layer. Silkmapin is a silk-like matrix protein that is mainly involved in the organic framework of the prismatic layer and in the crystal nucleation of the nacreous layer (Liu et al., 2015a). Hic31 is specificity expressed in the mantle edge and acts as a framework protein during prismatic layer formation (Liu et al., 2015b). Previous studies showed the prismatic layer of freshwater mollusks comprised aragonite crystal, which was distinct from the calcite prismatic layer of seawater shellfish (Liu and Li, 2015). Prismatic layer matrix proteins are also involved in pearl formation, and their expression is increased significantly before regular nacre deposition and is usually negatively correlated with pearl quality (Inoue et al., 2011; Liu et al., 2015a; Liu et al., 2015b; Blay et al., 2018; Jin et al., 2019a; Le Luyer et al., 2019). Therefore, there is a need to identify and determine the function of prismatic layer matrix proteins to determine the specific mechanism of prismatic layer formation, such proteins could also be used as a molecular marker of pearl quality. In this study, a novel prismatic layer matrix protein Hcprismatin-14 was identified in prismatic layer proteomics and the full-length nucleotide sequence of Hcprismatin-14 cDNA was isolated from the mantle of H. cumingii, and its putative functions in prismatic layer formation were elucidated.
Forty artificially cultured H. cumingii (four-year old, shell length 13-15 cm) were purchased from Shanxiahu Town, Zhuji City, Zhejiang Province, China. The shells were washed with distilled water to remove the surface attached silt and preserved in 5% sodium hydroxide (Figure 1A). The periostracum was removed by abrasion on Day 2, and the prismatic layer was peeled off mechanically on Day 5 (Figure 1B, C). The dried prismatic layer was ground to a fine powder using a pestle and mortar and sieved through a 200-mesh screen. Powder (20 g) was decalcified with 0.5M EDTA (200 ml) at 4 °C for 48 h with continuous stirring. The supernatant containing proteins was collected after centrifugation at 13,000 rpm for 30 min, and dialyzed against 5L of ultrapure water at 4 °C in a 3.5 kDa molecular weight cut-off dialysis tubing and then concentrated and freeze-dried to obtain EDTA-soluble matrix protein of prismatic layer.
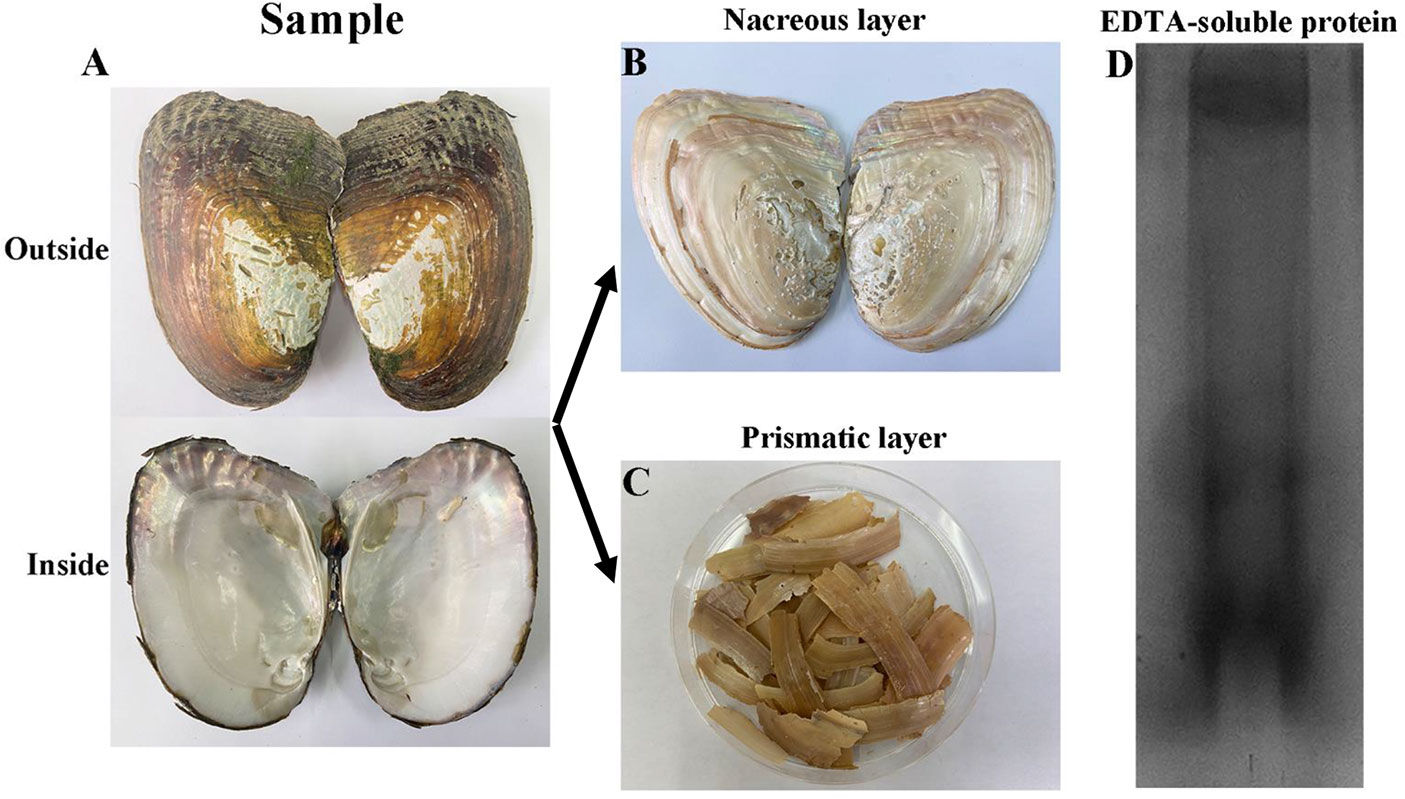
Figure 1 Extraction of EDTA-soluble matrix proteins of prismatic layer. (A) The inner and outer surfaces of H. cumingii; (B) The nacreous layer of shell; (C), The prismatic layer of shell; (D) EDTA-soluble extracts from prismatic layer of H. cumingii were fractionated by SDS-PAGE.
The EDTA-soluble matrix proteins of prismatic layer were redissolve in ultrapure water and subjected to SDS-PAGE analysis. The protein strips were vacuum freeze-dried after decolorization and dehydration in a 1.5 ml centrifuge tube, and then digested in digestion buffer (0.02μg/μL trypsin, 25mM NH4HCO3) at 37 °C for 16h. The peptide desalting was performed with the Pierce C18 spin tips (ThermoFisher Scientific, Waltham, MA, USA). The nano-HPLC liquid phase system UltiMate 3000 RSLCnano was used for separation. The samples were loaded into a trap column and separated on a RP-C18 analytical column (75 μm×150mm) at a flow rate of 300nL/min and then analyzed by Q-Exactive plus mass spectrometry (Thermo Scientific). The mass spectrometer was operated in data dependent acquisition (DDA) mode. MS1 scans were acquired at the m/z range of 300-1500, a resolution of 70000, an AGC target of 3e6 and a maximum injection time of 100 ms. Following each full scan, 20 strongest fragment profiles (MS2 Scan) were acquired at a resolution of 17500, an AGC target of 1e5 and a maximum injection time of 50 ms.
MS/MS spectra were searched using the MaxQuant against the local proteomic database deduced from transcriptome database of H. cumingi mantle and pearl sac (Bai et al., 2013). The search database specific parameters are set as follows: Fixed modifications: Carbamidomethyl (C); Variable modification: Oxidation (M) and Acetyl (Protein N-term); digestion: trypsin; First search peptide tolerance: 20ppm; Main search peptide tolerance: 4.5 ppm; Max Missed: 2. The protein identification was performed using BLASTp tool of National Center for Biotechnology Information (NCBI, http://blast.ncbi.nlm.nih.gov/blast.cgi).
Three hundred artificially cultured H. cumingii (1-year old, shell length 7-8 cm) were purchased from Shanxiahu Town, Zhuji City, Zhejiang Province, China. Nine mussels were harvested and tissues, including mantle edge (ME), mantle pallium (MP), mantle central (MC), foot (F), adductor muscle (AM), intestines (I), blood (BL), and gonads (G) were stored in RNAStore (TianGen Biotech, Beijing, China) and preserved in an ultra-low temperature freezer until RNA extraction. Tissues were ground by grinding rods and RNA extraction was performed using Trizol reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The concentration and quality of sample RNA was detected by absorbance at A260/280 and A260/230 using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific).
The qualified RNA of multiple tissues were prepared and 3′- and 5′-Ready cDNA syntheses were performed using a SMARTer RACE 5′/3′ Kit according to the manufacturer’s instructions (Clontech Bio. Inc., Dalian, China). The specific primers for 3′- and 5′-RACE (Table 1) were designed based on the partial segment in the H. cumingii mantle transcriptome library (Bai et al., 2013). The touchdown PCR program was as follows: initial denaturation for 5 min at 94°C, 5 cycles at 94°C for 30 s and 72°C for 3 min, 5 cycles at 94°C for 30 s, 70°C for 30 s and 72°C for 3 min, 27 cycles at 94°C for 30 s, 68°C for 30 s and 72°C for 3 min, and a final elongation at 72°C for 10 min. The target band was recovered and purified using a DNA Gel/PCR Purification Kit (Novoprotein Scientific, Jiangsu, China) and directly ligated using a pLB cloning vector. The products were transformed into BL21 (DE3) competent cells (Novoprotein Scientific) on coated plates; monoclonal colonies were selected by PCR and sequenced by Sangon Biotech (Shanghai). Full-length Hcprismatin-14 was obtained by splicing the 3′/5′-end sequences with the known partial segment from the H. cumingii mantle transcriptome library. The accuracy of the splicing sequence was further detected by a pair of specific primers (Table 1), the amplification product of which contained the whole open reading frame (ORF).
The ORF and its potential protein-encoding segments were predicted by ORF finder (www.ncbi.nlm.nih.gov/orffinder/). The presence of signal peptides and the location of their cleavage sites in putative proteins were predicted by using the SignalP - 5.0 Server (https://services.healthtech.dtu.dk/service.php?SignalP-5.0). The theoretical isoelectric point (pI), molecular weight (MW), and amino acid composition of mature protein were predicted by using the ProtParam tool (https://web.expasy.org/protparam/). The hydrophilic and hydrophobic regions of mature protein were predicted by using ProtScale (https://web.expasy.org/protscale/). The potential domain and repeat motifs were predicted by using a Simple Modular Architecture Research Tool (SMART) (http://smart.embl-heidelberg.de/). Protein similarity and homology were determined using Protein BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and the protein secondary and tertiary structures were predicted by using Protein Homology/analogy Recognition Engine V 2.0 (Phyre2) (www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index). Multiple sequence alignments of GY rich region (residues 56–111) of Hcprismatin-14 were conducted by BioEdit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html).
The RNA extracted from various tissues as described in Section 2.4 were reverse-transcribed to cDNA for tissue-specific gene expression analysis using the PrimeScript™ RT reagent kit (TaKaRa, Dalian, China). The expression profile of Hcprismatin-14 in multiple tissues was detected by reverse transcription (RT)-PCR and quantitative real-time (qRT)-PCR. A pair of specific primers generating a 199 bp product was designed based on the full-length Hcprismatin-14, and the expression of house-keeping gene EF1a among tissues was used as a control for normalization (Table 1). The PCR reaction system comprised: 10 μL 2× Taq master mix (for RT-PCR) or SYBR qPCR super mix (for qRT-PCR) (Novoprotein Scientific), 1 μL upstream and downstream primers, 2 μL tissues cDNA (100 ng/μL), and 6 μL RNase-free water. The amplification procedure was initial denaturation for 3 min at 94°C, 40 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. After a final elongation at 72°C for 10 min, the amplification products of RT-PCR were detected by agarose gel electrophoresis. The dissociation curve and cycle threshold (Ct) values of qRT-PCR were detected in an additional step from 65°C to 95°C. The relative expression of Hcprismatin-14 in different tissues was analyzed by equation 2−ΔΔCt, and expressed as the means ± standard error (SE) (Livak and Schmittgen, 2001). Significant tests were conducted with one-way ANOVA with a p<0.05 considered significant.
Two hundred adult H. cumingii were purchased from a pearl farm in Shanxiahu Town and the saibo transplantation was performed as described by Jin et al. (Jin et al., 2019a). The posterior segment of the mantle pallium of donor mussels was collected and the epithelial cells were made into saibos. Six saibos from four recipient mussels were sampled on Day 0. Twenty saibos from one donor mussels were implanted into the mantle center of each recipient mussel. The operation described above was performed by one expert. Four recipient mussels were harvested on Day 3, 6, 9, 12, 15, 18, 24, 30, and 38 after implantation. Six pearl sacs from each mussel were collected and total RNA was extracted for expression pattern analysis. EF1a served as the internal reference gene, whereas the prismatic layer matrix protein gene Hic31 and nacreous layer matrix protein gene Hc-upsalin were used as negative controls. The qRT-PCR reaction system, amplification procedure, and significant tests were conducted as described in Section 2.7.
Based on the full-length cDNA of Hcprismatin-14, a pair of specific primers was synthesized with an additional T7 promoter sequence in the 5′-end of the upstream and downstream primers. The purified PCR products were transcribed in vitro, and the following double-stranded (ds) RNA synthesis and purification were performed using a T7 high-efficiency transcription kit according to manufacturer’s instructions (TransGen Biotech, Beijing, China). The Hcprismatin-14-dsRNA concentration was diluted to 40 μg/60 μL and 80 μg/60 μL with phosphate-buffered saline (PBS). Four H. cumingii in the experimental group were injected with Hcprismatin-14-dsRNA, and mussels in blank and negative control groups were injected with PBS and green fluorescent protein (GFP)-dsRNA. On Day 7, the mantle of each mussel was collected for RNA extraction and qRT-PCR detection and the shell was harvested for scanning electron microscopy (SEM) observation of the microstructure of calcified layer.
Based on the bioinformatics analysis of Hcprismatin-14, a 21-amino acid polypeptide including a WDD-repeat region (residues 112–132) was synthesized by GL Biochem (Shanghai, China). Cleavage from resin and high performance liquid chromatography (HPLC) purification were performed to obtain the purified polypeptide (purity = 95.17%), and mass spectrometric detection was performed to ensure the correct amino acid sequence of polypeptide. The polypeptide was dissolved in an acetonitrile/water mixture (1:3) with a final concentration 1 mg/mL and stored at -80°C.
A saturated calcium bicarbonate solution was prepared as follows: 1 g calcium carbonate power was added to 80 mL Milli-Q water, followed by magnetic stirring and introducing carbon dioxide overnight (8 h). The muddy calcium bicarbonate solution was filtered by syringe filter (Aquo-system, 0.22 μm) to remove redundant calcium bicarbonate, and the filter liquor was exposed to carbon dioxide for another 2 h to prepare a saturated calcium bicarbonate solution. In vitro crystallization assays were performed in calcite and aragonite systems. In the calcite crystallization system, WDD-repeat region polypeptide was mixed with saturated calcium bicarbonate solution to various final concentrations in a 20 μL system. In the aragonite crystallization system, 1 μL magnesium chloride (1 M) was added to the saturated calcium bicarbonate solution to induce aragonite formation. The crystallization systems were dripped on borosilicate cover glass slips and stored in a glass desiccator for crystallization. After 48 h, the crystal was cleaned with Milli-Q water and prepared for polymorph and morphology detection by Raman spectrum and SEM, respectively.
The calcium carbonate crystallization rate of the WDD-repeat region polypeptide was determined using the method of Suzuki et al. (Suzuki and Nagasawa, 2007). A 10 μL polypeptide solution and 100 μL of 70 mM NaHCO3 were mixed on an ELISA plate, and 100 μL of 70 mM CaCl2 was then added. The acetonitrile/water mixtures (1:3) and bovine serum albumin (BSA) was used as blank and negative groups, respectively. The turbidity of the solution, as an indicator of the crystallization rate, was detected by absorbance at 570 nm/min in a spectrophotometer (for a total of 5 min).
The prismatic layer was collected by soaking the shells in 5% sodium hydroxide for 5 days and then decalcified with 0.5M EDTA (Figure 1A-C). SDS-PAGE of the EDTA soluble organic matrix of prismatic layer showed there are three major bands (Figure 1D). The MS/MS data were searched against the local proteomic database deduced from transcriptome database of H. cumingii mantle and pearl sac. As a result, there were total 21 matrix proteins that were identified in current study (Table 2). According to the biological functions of proteins, the identified proteins were classified into two broad categories. The first category included silkmapin, silkmaxin, nasilin 1 and nasilin 2 that participate in framework formation in a parallel beta-sheet conformation during shell biomineralization (Liu et al., 2015a; Xia et al., 2019). The second class contained cytoplasmic and cytoskeletal proteins. In addition, 10 predictive proteins showed no homology with any known protein in publically available databases. Among them, contig7882_4 was rich in Gly and Tyr with repeated sequence of GY consistent with the general characteristics of Gly/Tyr-rich framework matrix proteins. Therefore, we conducted follow-up research only on the contig7882_4.
A 197-base pair (bp) and a 457-bp fragment were generated by 5′-RACE and 3′-RACE, respectively. The spliced full-length cDNA was 710 bp and included a 37-bp 5′-untranslated region (UTR), a 423-bp ORF and a 226-bp 3′-UTR (including a putative polyadenylation signal AATAAA and a 26-bp poly A tail). The cDNA sequence of Hcprismatin-14 was submitted to GenBank (accession no. OK300051). Further bioinformatics analysis indicated that the protein was a secreted protein with a signal peptide of 24 amino acid residues located in the N terminus (Figure 2A). The mature protein comprised 124 amino acids with a theoretical molecular weight of 14.1 kDa and an isoelectric point of 8.77 (Figure 2B). Analysis of the amino acid composition showed a high content of Gly (28.2%), Tyr (14.5%), Arg (10.5%), and Asp (9.7%), the concentrative distribution of which resulted in a modular structure for Hcprismatin-14, including a 24-amino acid signal peptide in the N terminus (residues 1–24), a Gly/Tyr-rich region (residues 56–111), a WDD-repeat region (residues 112–132), and a basic region in the C terminus (residues 133–148) (Figure 2C). ProtScale analysis revealed that the hydrophobic part of mature protein was in the Gly/Tyr-rich region and the hydrophilic region comprised the WDD-repeat region and the basic tail (Figure 2D). Protein structure prediction showed that the Gly/Tyr-rich region and the basic region in the C terminus comprised beta strands, and WDD-repeat region comprised α-helices (Figure 2E). Protein BLAST analysis showed that Hcprismatin-14 had no domain or significant similarity with any other known protein. Multiple sequence alignment revealed that the GY-rich region of Hcprismatin-14 displays a high degree of similarity with GY region of Prismalin-14 and GYS-region I of Prisilkin-39 (Figure 2F).
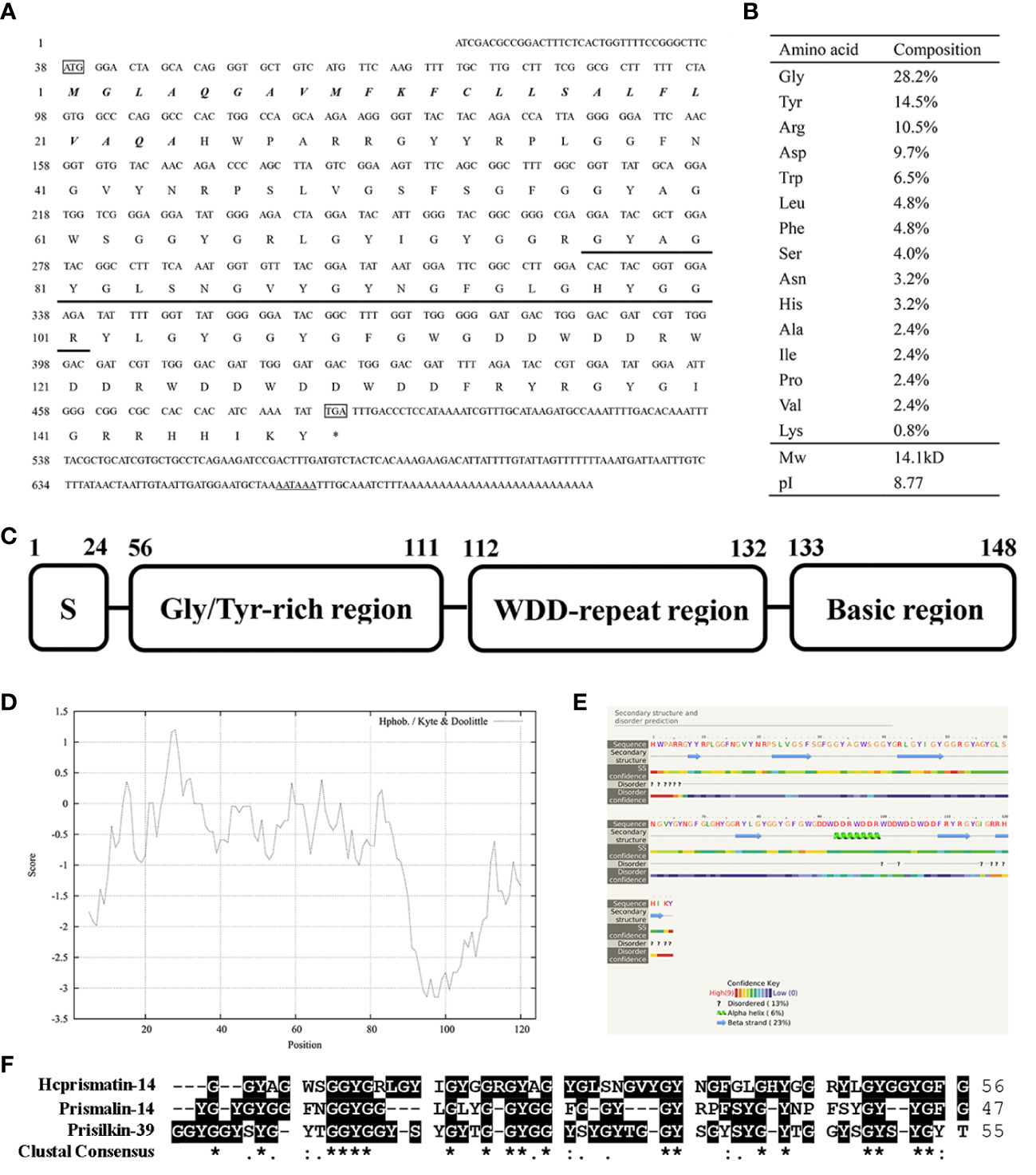
Figure 2 Sequence analysis of Hcprismatin-14. (A) The full-length cDNA and putative amino acid sequence of Hcprismatin-14. The initiation codon ATG and termination codon TGA are boxed. The signal peptide is bold italic. The amino acid sequence detected by liquid chromatography tandem mass spectrometry is underlined. The putative polyadenylation signal AATAAA is underlined by a double solid line; (B) Amino acid composition of Hcprismatin-14; (C) The modular structure of Hcprismatin-14. S, signal peptide. (D) The hydrophobicity analysis by ProtScale; (E) The secondary structure prediction performed by Phyre2. The regions adopting putative α-helix and β-sheet conformations are represented as green spiral and blue arrow, respectively. (F) Alignment of GY rich region of Hcprismatin-14 with repeats 3-7 of Prisilkin-39 (residues 83-138) and GY rich region of Prismalin-14 (residues 67-113).
The relative mRNA expression of Hcprismatin-14 in various tissues was first detected by RT-PCR. Reference gene EF1a was consistent expressed in various tissues, and a single band of Hcprismatin-14 was obtained on agarose gel electrophoresis only from the mantle edge, no expression was detected in any other tissues (Figure 3A). The qRT-PCR results were consistent with those from RT-PCR, thus, Hcprismatin-14 was expressed only in the mantle edge (Figure 3B).
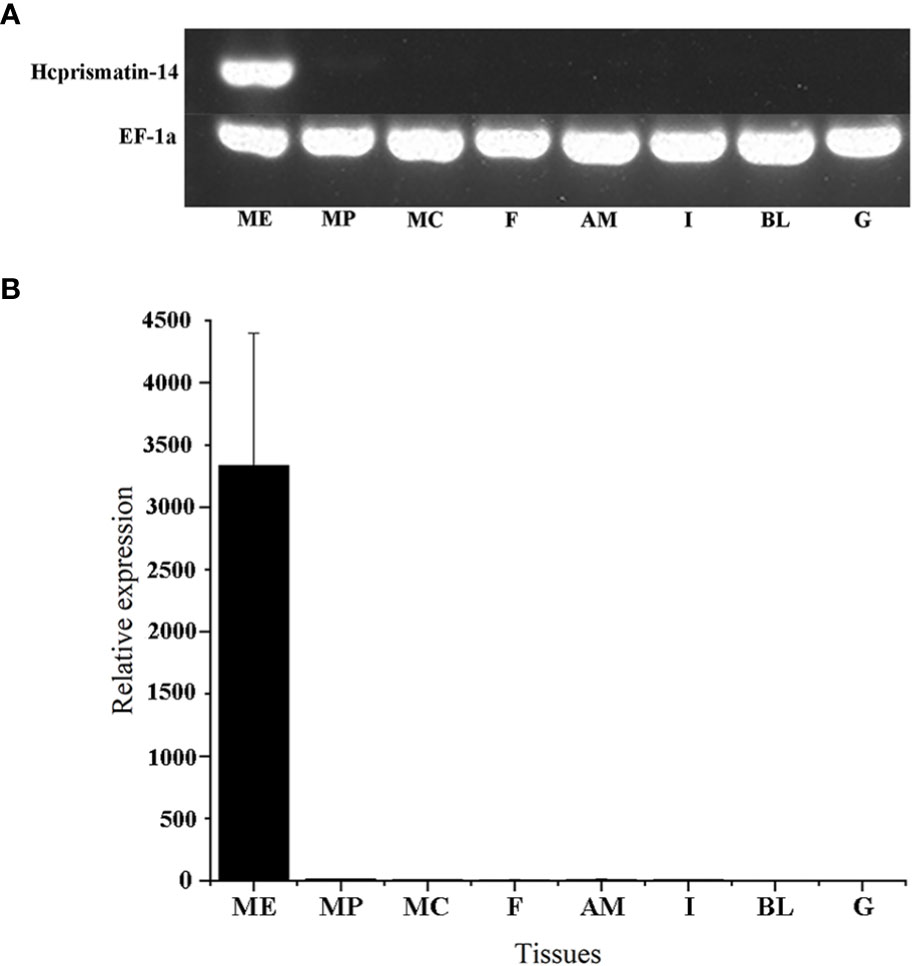
Figure 3 RT-PCR and qRT-PCR of Hcprismatin-14 expression in H. cumingii. ME, mantle edge; MP, mantle pallial; MC, mantle center; F, foot; AM, adductor muscle; I, intestines; BL, blood; G, gonad. The housekeeping gene EF-1α was used as a positive control.
The saibos of donor mussels were transplanted into the mantle of recipients (Figure 4A). The pearl sacs developed during pearl formation were collected for total RNA extraction, and further used to detect the expression pattern of Hcprismatin-14 by qRT-PCR. The prismatic layer matrix protein gene Hic31 and nacreous layer matrix protein gene Hc-upsalin were used as controls, and EF1a was used as a housekeeping gene (Liu et al., 2015b; Liu et al., 2019). The results showed that Hcprismatin-14 transcripts were low from Day 0 to Day 6, gradual increased between Day 9 and Day 15, peaked on Day 18, and gradually declined between Days 24 and 38. Expression of Hic31 was low from Day 0 to Day 6, peaked on Day 9, then gradually declined between Day 12 and 30, reaching a minimum by Day 38. The expression of Hc-upsalin was decreased on Day 3, and remained low from Day 6 to Day 15, then gradually increased from Day 18 to Day 38 (Figure 4B).
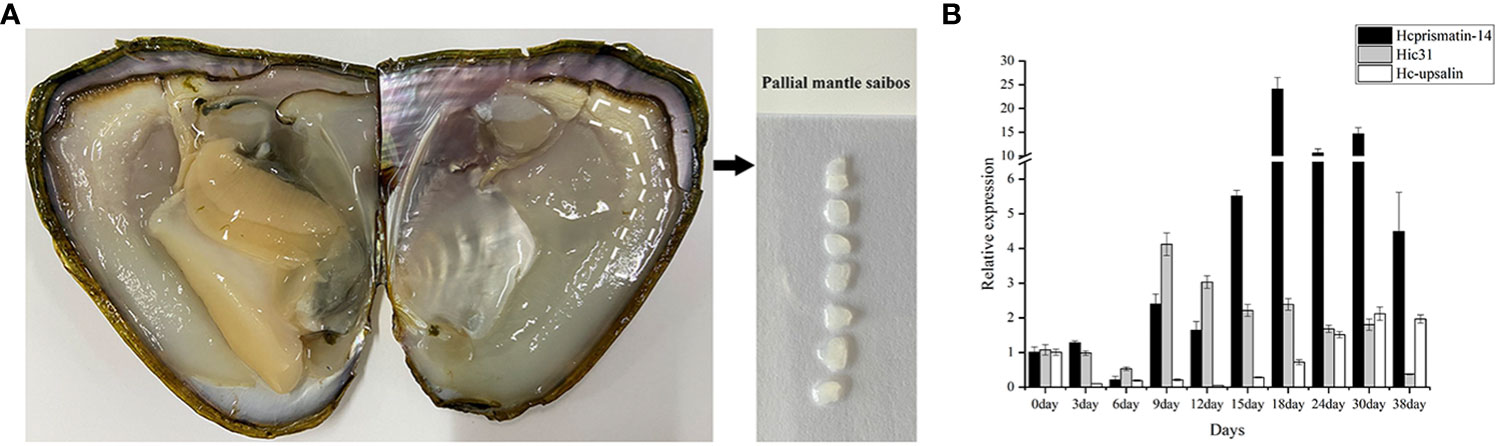
Figure 4 qRT-PCR analysis of Hcprismatin-14 gene expression in pearl culture assay. (A) The schematic diagram of saibos; (B) Expression analysis of Hcprismatin-14 in pearl sac during pearl formation.
The role of Hcprismatin-14 in shell biomineralization in vivo was verified by an RNAi assay. After the injection of 40 μg/60 μL Hcprismatin-14-dsRNA into the adductor muscle of the mussels, the relative expression of Hcprismatin-14 in mantle decreased to ~25% of the control group. When the dose of Hcprismatin-14-dsRNA was increased to 80 μg/60 μL dsRNA, the expression level of Hcprismatin-14 further decreased to ~14% of the control group (Figure 5A).
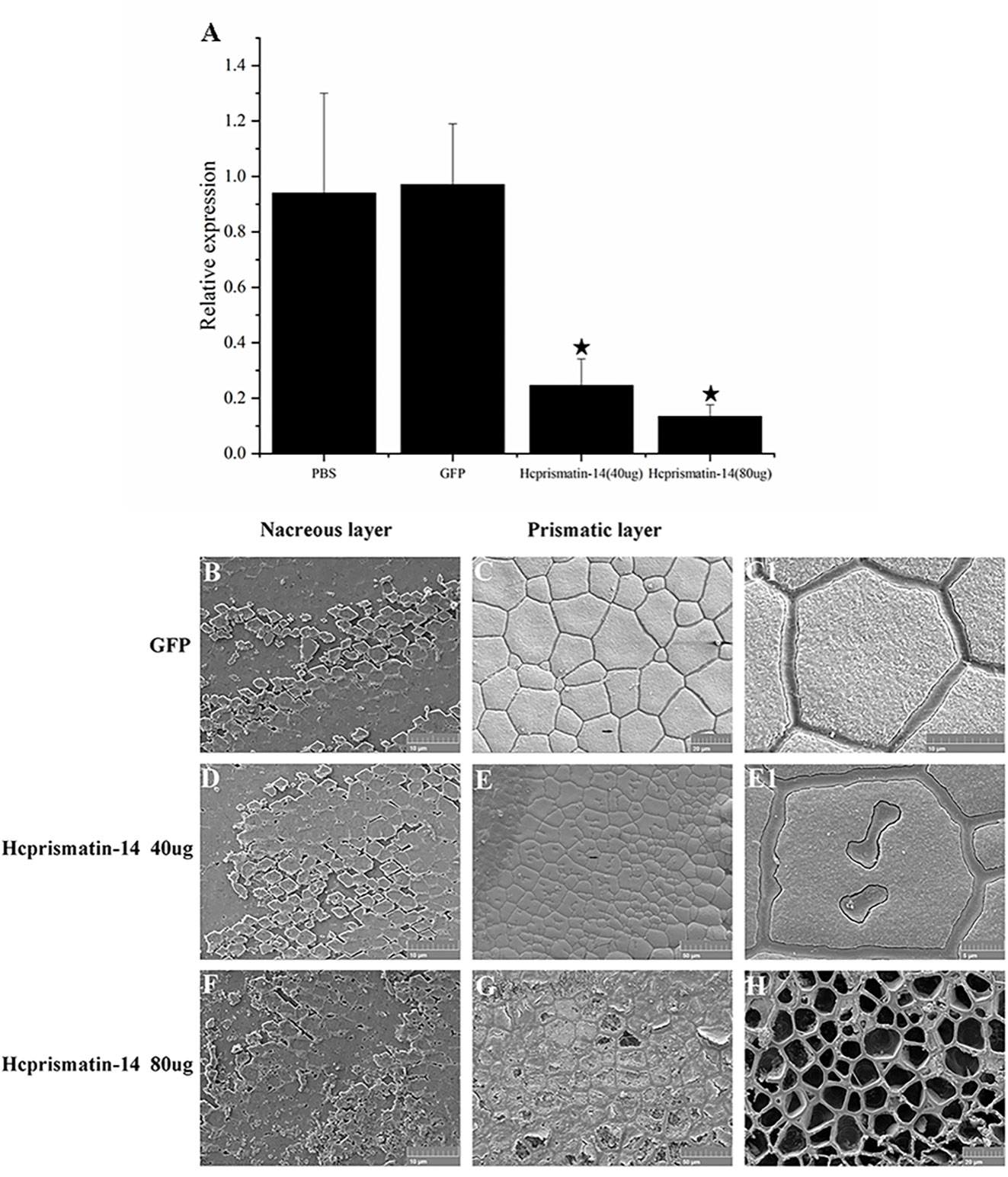
Figure 5 Knockdown of Hcprismatin-14 in RNAi assay. (A), The relative expression of Hcprismatin-14 in mantle 7 days after injection, and the expression level in the PBS-injected group were attributed a relative value of 1.0; B–C1, Microstructure of shell inner surface in control groups. Scanning electron microscopy (SEM) observation of: (B) the nacreous layer in the control group characterized by a brick-wall structure comprising smooth aragonite and (C) prismatic layer in the control group characterized by the competitive growth of prisms with a smooth surface; (C1) enlargement of the arrow shown in (C). D–E1, Microstructure of the shell inner surface in 40 μg/60 μL Hcprismatin-14-dsRNA injection groups. SEM observation of: (D) nacreous layer comprising smooth aragonite and (E) prismatic layer, in which irregular sinuous lacunae appear prominently in the center of mature prisms. (E1) Enlargement of the arrow shown in (E); F–H, Microstructure of shell inner surface in the 80 μg/60 μL Hcprismatin-14-dsRNA injection groups. SEM observation of: (F) nacreous layer comprising smooth aragonite and (G) prismatic layer, intra-column organic framework was destroyed, and the nano-sized subgrains became sparse and disordered in the center of prisms. (H) crystals have been either completely or nearly completely disappeared, leaving only thick polygonal organic sheaths.
Micro-structure observations of shell inner surface were detected by SEM. In the GFP-dsRNA injection group, the nacreous layer had a brick-wall structure comprising smooth aragonite (Figure 5B). The prismatic layer was characterized by competitive growth of prisms encased in thick polygonal organic sheaths (Figures 5C and C1). In Hcprismatin-14-dsRNA injection groups, the nacreous layer also comprised nearly hexagonal aragonite tablets (Figures 5D, F). By contrast, significant changes were observed in the micro-structure of mature prisms. In the 40 μg/60 μL Hcprismatin-14-dsRNA injection group, the prismatic layer comprised squashed prisms, whereas irregular sinuous lacunae were appeared prominently in the center of mature prisms, rather than the edge of mature prisms, and several organic pits appeared around the prism centers (Figures 5E and E1). In the 80 μg/60 μL Hcprismatin-14-dsRNA injection group, the nano-sized subgrains became sparse and disordered in the middle of prisms (Figure 5G), even in some shell edge areas that has been either completely or nearly completely disappeared, leaving only thick polygonal organic sheaths (Figure 5H).
The role of the WDD-repeat region polypeptide in controlling crystal polymorphy and morphology was detected by mixing the polypeptide with a saturated calcium carbonate solution for in vitro crystallization. In the calcite crystallization system, the Raman spectra of crystals in the control and experimental groups contained characteristic calcite peak at 156, 283, 713, and 1087 cm-1 (Figures 6I, J). BSA was added in the negative group and SEM observation showed the crystals were typical rhombohedra with smooth surfaces (Figures 6A and A1). In experiment group, remarkable changes in the crystal morphology were observed as the WDD-repeat region polypeptide concentration increased. In the 5 μg/ml polypeptide group, the crystals were typical rhombohedra (Figures 6B and B1). In 10 μg/ml polypeptide group, the crystals were displayed an irregular shape characterized by the disordered superposition of several cylinder-like crystals (Figures 6C and C1). In 20 μg/ml polypeptide group, the crystals were cylinder-shaped with additional scaly crystals deposition on the surface (Figures 6D–D2). In 40 μg/ml polypeptide group, the crystals were spherical-shaped and their surface were covered with a denser scaly crystals (Figures 6E–E2). These results indicated that the WDD-repeat region is not involved in polymorph transformation, but is involved in morphology fine-tuning as the polypeptide concentration increased. In aragonite crystallization system, SEM observations showed that the crystals of the control group (BSA addition) were spheres and had a characteristic aragonite peak at 154, 210, 705, and 1086 cm-1 by Raman detection (Figures 6F, K). In the experiment group, the shape and size of crystals were the same as in the control group (Figures 6G, H). However, the number of crystals decreased gradually as the concentration of polypeptide increased from 5μg/mL to 10μg/mL, and there was no aragonite deposition in the 20μg/mL polypeptide group (data not shown). These results indicated that the WDD-repeat region had no effect on polymorph and morphology regulation, but could inhibit aragonite nucleation.
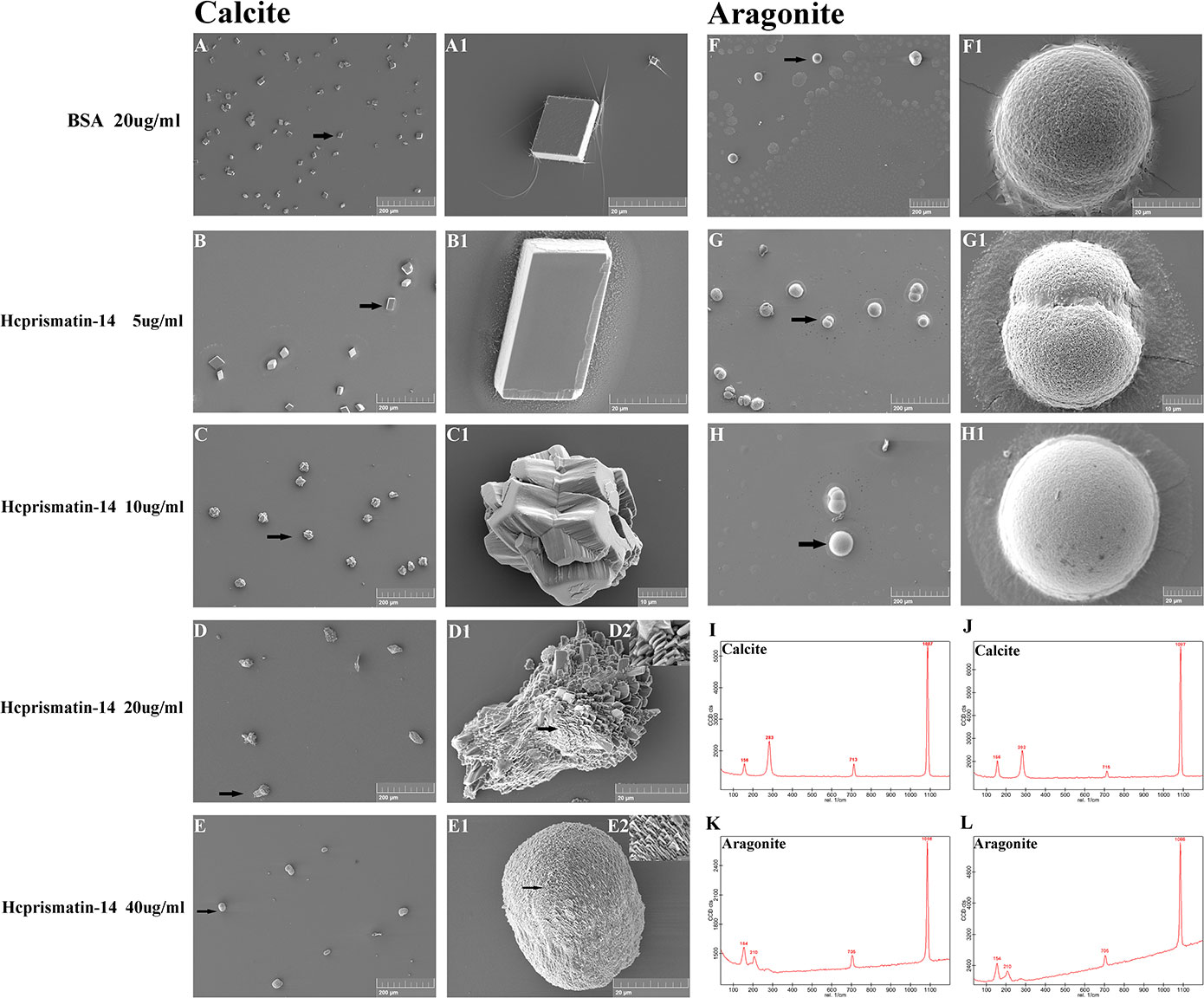
Figure 6 Scanning electron microscopy (SEM) observation and Raman spectra detection of in vitro crystallization assay in the present of WDD-repeat region polypeptide. (A–E) SEM observation of the calcite system. Crystallization with: (A, B) 20 μg/mL BSA as a negative control and 5 μg/mL WDD-repeat region polypeptide; crystals were typical rhombohedra with a smooth surface; (C) 10 μg/mL WDD-repeat region polypeptide, crystals were irregular shape; (D) 20 μg/mL WDD-repeat region polypeptide, the crystals were cylinder-shaped; and (E) 40 μg/mL WDD-repeat region polypeptide, the crystals were spherical-shaped. (A1–E2) Enlargement of the arrows shown in A–E: (A1-B1) the surface of rhombohedra was smooth; (C1) the crystals were consisted of several cylinder-like crystals; (D1-D2) the cylinder-shaped crystals were consisted of sparse scaly plates; (E1-E2) the spherical-shaped crystals were consisted of denser and finer scaly plates. (F–H) SEM observation of the aragonite system. Crystallization with 20 μg/mL BSA in (F), 5 μg/mL WDD-repeat region polypeptide in (G), 10 μg/mL WDD-repeat region polypeptide in (H). (F1–H1) Enlargement of the arrows shown in F–H. The number of hemispherical crystals decreased gradually as the concentration of polypeptide increased; (I, J) Raman spectra of the crystals formed in calcite systems. (K, L) Raman spectra of the crystals formed in aragonite systems.
The inhibitory activity of the WDD-repeat region polypeptide on the crystallization rate of calcium carbonate was reflected by a test absorbance at 570 nm. The addition of acetonitrile/water mixtures (1:3) to a control group resulted in the absorbance slowly increasing from ~0.048 at 0 min to ~0.134 at 5 min. The addition of BSA (20 μg/mL) in the negative group induced the slow increase in absorbance from ~0.0463 at 0 min to ~0.125 at 5 min. There was is no significant difference in precipitation rate between the two control groups. In the experimental groups, the absorbance was ~0.0687 at 5 min in the 5 μg/mL WDD-repeat region polypeptide group. There was a similar trend in absorbance in the 10 μg/mL WDD-repeat region polypeptide group and the 20 μg/mL WDD-repeat region polypeptide group, the absorbance of which was ~0.0490 at 5 min (Figure 7). These results indicated that the WDD-repeat region of Hcprismatin-14 had inhibitory activity on calcium carbonate precipitation.
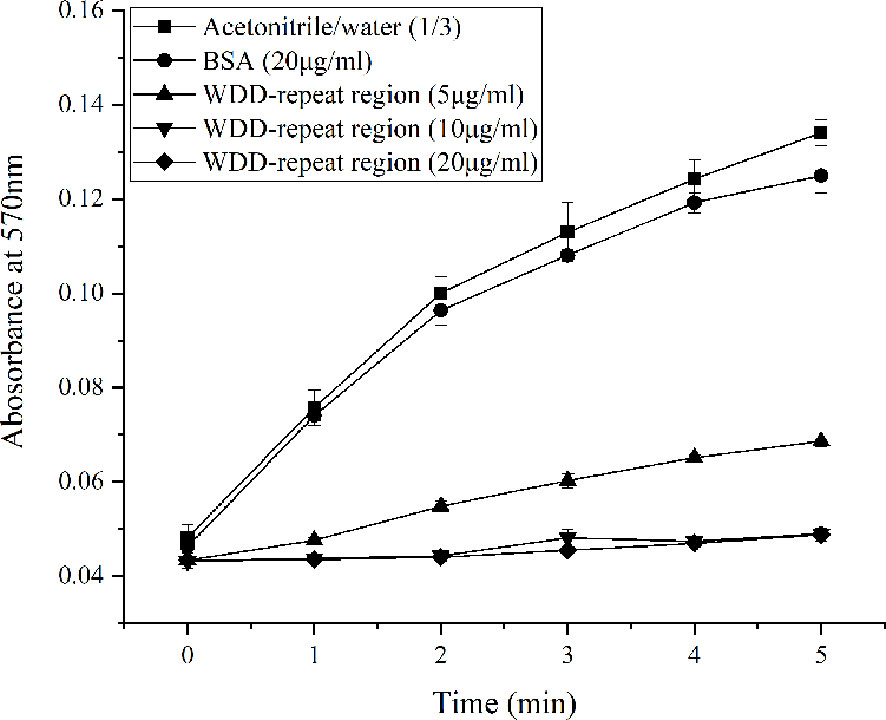
Figure 7 Inhibitory activity of WDD-repeat region polypeptide on the crystallization rate of calcium carbonate., Acetonitrile/water (1/3) mixture;, BSA (20μg/mL);, WDD-repeat region (5μg/mL);, WDD-repeat region (10μg/mL);, WDD-repeat region (20μg/mL).
The central role of matrix protein in shell biomineralization has been widely acknowledged as a result of isolation and functional studies of hundreds of novel matrix proteins. Previous researches mainly focused on acidic matrix proteins of the nacreous layer and their direct interaction with calcium ions, revealing them to be involved in crystal nucleation, growth and morphology, ultimately affecting the value of the resulting pearl. So far, numerous models for matrix proteins regulated aragonite tablets deposition tried to recreate the process of nacreous layer formation (Levi-Kalisman et al., 2001). At the same time, a number of matrix proteins of the prismatic layer have been identified from different marine shellfish (Suzuki et al., 2004; Takagi and Miyashita, 2010). In the current study, a total of 21 proteins were identified in the EDTA-soluble matrix of the prismatic layer. Interestingly, it contained 4 known silk-like matrix proteins such as silkmapin, silkmaxin, nasilin 1 and nasilin 2 (Liu et al., 2015a; Xia et al., 2019). The hydrated silk-like protein is an important component in chitin-silk fibroin gel proteins–acidic macromolecules model, which acts as a space filler between chitinous framework and prevents uncontrolled crystallization during initial nacreous layer formation (Addadi et al., 2006). This result revealed that the prismatic layer biomineralization pattern is somewhat comparable to the nacreous layer due to the same organic matrix.
In order to further study the role of silk-like protein in prismatic layer biomineralization, a novel matrix protein, Hcprismatin-14, was identified in the EDTA-soluble matrix of the prismatic layer and the full length of Hcprismatin-14 cDNA was cloned from the mantle of H. cumingii. Tissue-specific expression analysis showed that it is specifically expressed in the mantle edge, which was consistent with previous studies suggesting that the mantle is a complex organ responsible for shell biomineralization and the matrix proteins secreted by the mantle edge are usually involved in prismatic layer formation (Marie et al., 2012). Therefore, the current results indicated that Hcprismatin-14 is synthesized in mantle edge, secreted into the extrapallial fluid and incorporated into the prismatic layer.
Pearl is a nacre biomineral controlled strictly by the matrix proteins secreted by the pearl sac. As well as superficial nacre tablet deposition, a thin calcified layer comprising prisms or irregular deposited block-like crystals occur around the pearl core. Previous studies showed that this irregular deposition of block-like crystals occurred before Day 18 after saibo transplantation, which corresponded to the high expression of prismatic layer matrix proteins. Nacre tablets then regularly deposited under the control of nacreous layer matrix proteins (Liu et al., 2012; Lin et al., 2013; Jin et al., 2019a). In the current study, expression of Hcprismatin-14 gradually increased after calcium carbonate deposition, peaking on Day 18, and gradually decreased with nacre deposition. Similarly, the expression of prismatic layer matrix protein gene Hic31 peaked on Day 9, and gradually decreased as nacre deposition increased. By contrast, the nacreous layer matrix protein gene Hc-upsalin showed low expression during irregular crystal deposition and gradually increased during nacre deposition. Hence, its expression pattern during early pearl formation also indicated that Hcprismatin-14 is a prismatic layer matrix protein involved in irregular crystal deposition during pearl formation.
Hcprismatin-14 had a modular structure with high proportions of Gly (28.2%) and Tyr (14.5%). Previous studies showed that known matrix proteins contain several specific amino acids to promote their biological functions during shell formation. The poly-Gly blocks in matrix proteins are considered to be involved in framework construction in a parallel beta-sheet conformation (Sudo, 1997). In the amino acid sequence of Hcprismatin-14, Tyr was always behind Gly, leading to some repeats of Glym(X)Yn (m=1,2; n=1,2; X=Val/His/Arg) with a beta-sheet conformation in Gly/Tyr-rich region. The brown prismatic layer lay between the periostracum and nacreous layer, the microstructure of which was characterized by the competitive growth of prisms. Each prism was separated by intercolumn organic matrix and its subgrains were also enveloped in an intra-column organic matrix (Okumura et al., 2012). Chitin acting as a structural polysaccharide has been suggested as the main ingredient necessary for chitinous scaffold construction of prismatic layers during initial biomineralization, with other matrix proteins then binding to chitin to control the crystal nucleation and growth that follows, which is consistent with the features of nacreous layer (Suzuki et al., 2007; Kintsu et al., 2017). Among the known nacreous layer matrix proteins, the chitin binding domain (ChtBD2) was considered to be a site for recognizing and binding chitinous scaffold (Suzuki et al., 2009; Jin et al., 2019b; Fan et al., 2020). In addition, the prismatic layer matrix proteins prismalin-14 and prisilkin-39 of P. fucata could also have the ability to bind chitin via their Gly/Tyr-rich regions (Suzuki and Nagasawa, 2007; Kong et al., 2009). In another study, the Gly/Tyr-rich region in the KRMP family proteins in P. fucata was originally thought to be involved in protein crosslinks, and later shown to be involved in chitin binding (Zhang et al., 2006; Liang et al., 2015). The heterogeneous distribution of matrix proteins in the prismatic layer showed that basic matrix proteins rich in Gly and Try, such as Shematrin, KRMP family proteins, and prismalin-14, were distributed in the inter-column organic matrix (Huang et al., 2021). Moreover, the expression of Hic31, the tertiary structure of which is similar to collagen I and which is mainly involved in prismatic layer framework construction, peaked on Day 9 of pearl formation, whereas the expression level of Hcprismatin-14 gradually increased and peaked on Day 18, indicating that the latter might interact sequentially with the organic framework of the prismatic layer. In addition, intra-column subgrains lost contact with chitinous scaffold as high-dose Hcprismatin-14 inhibition in the RNAi assay. Therefore, it is reasonable to assume that the Gly/Tyr-rich region of Hcprismatin-14 is a potential motif for chitin binding in the prismatic layer.
Besides the high proportion of Gly and Tyr, Hcprismatin-14 is also rich in Asp (9.7%). Asp together with Trp formed an unusually hydrophilic acidic WDD-repeat region (residues 112–132) behind the Gly/Tyr-rich region. Although Hcprismatin-14 contains Asp, it does not contain Glu, consistent with the finding that known matrix proteins of the prismatic layer always contain more Asp in shellfish. The Asp-rich regions in matrix proteins are known as regulatory elements in shell formation because of their calcium-binding capacity. On the one hand, aspartic acid has a strong calcium-concentrating effect, which combined with sulfated polysaccharides, create the supersaturation necessary for nucleation (Addadi et al., 1987). Previous studies that showed aspartic acids are a dominant component of the intra-column organic matrix of the prismatic layer, and that soluble intra-column proteins are oriented almost perpendicular to the prism axes to promote the interaction between the carboxylate groups of aspartic acid and calcium ions (Berman et al., 1993; Huang et al., 2021), and in situ chemical speciation showed the heterogeneous distribution of sulfate in prisms, with high amounts in inter-column organic matrices and a center core in intra-column organic matrices (Dauphin et al., 2003). In the current study, several irregular sinuous lacunae were appeared in the center of mature prisms, and subgrains became sparse and even has been either completely or nearly completely disappeared as Hcprismatin-14 inhibition increases in RNAi assay, reflecting the inability of crystal deposition on organic membranes. In subsequent experiments, the WDD-repeat region polypeptide significantly reduced the calcium carbonate precipitation rate. These results indicated that the WDD-repeat region of Hcprismatin-14 is the calcium-binding site and is crucial for crystal deposition. Moreover, previous studies showed that the Asp-rich family of Atrina rigida regulates calcite growth via the joint effects of magnesium ions (Kim et al., 2008; Kim et al., 2012). Aspein of P. fucata enhances the conversion of amorphous calcium carbonate to calcite by regulating the Mg2+/Ca2+ ratio in extrapallial fluid (Tsukamoto et al., 2004; Takeuchi et al., 2008). Thus, on the other hand, the Asp-rich region in prismatic layer matrix proteins of marine shellfish are essential for calcite formation because of a reduction in the Mg2+/Ca2+ ratio as a result of selective calcium binding, which is consistent with our in vitro crystallization results showing that the WDD-repeat region polypeptide participated in morphological regulation and crystal growth of calcite on a nanometer scale. While, the concentration of Mg2+ in freshwater is significantly higher than in seawater, making the prismatic layer of H. cumingii comprised aragonite crystal (Liu and Li, 2015). The high concentration of Mg2+ in extrapallial fluid might lead to aragonite deposition by the WDD-repeat region rather than controlling calcite morphology in H. cumingii. Moreover, Hcprismatin-14 is also rich in Arg (10.5%), which with other basic amino acids (His and Lys) mainly formed a basic tail located in the C terminus. A similar outcome was reported for Shematrin from P. fucata and Lustrin A from Haliotis rufescens, which assist the interaction between bicarbonate ions and other acidic proteins to induce calcium carbonate deposition (Shen et al., 1997; Yano et al., 2006). Therefore, the hydrophilic WDD-repeat region and the basic region in the C terminus would be the specific parts involved in aragonite nucleation and interaction with other matrix proteins, respectively.
In conclusion, the silk-like protein seems to be an important 7ingredient among the prismatic layer organic matrix. Hcprismatin-14 is a multifunctional matrix protein of prismatic layer with three structurally characteristic regions. A Gly/Tyr-rich region with a beta-sheet conformation serves as a structure for recognition and interaction with the chitinous framework. A WDD-repeat region acts as a Ca2+ carrier and is crucial for crystal deposition and growth, whereas the basic region in the C terminus assists the interaction with other acidic proteins for further regulation of crystal growth. Our findings will contribute to a deeper understanding of the molecular mechanism of shell biomineralization and promote the development of pearl industry.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
CJ and WL designed the experiment. YZ and KC performed the experiments. RJ, SJ and YS analyzed the data. CJ wrote the original manuscript. WL and GR revised and approved the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by The National Natural Science Foundation of China (32202921); The Sci-tech Special Commissioners Project of Zhejiang Province-Pearl Quality Improvement and Industrialization (2020-2024).
We thank the Shanghai Luming Biological Technology Co., Ltd (Shanghai, China) for providing proteomics services. We also thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1154968/full#supplementary-material
Addadi L., Joester D., Nudelman F., Weiner S. (2006). Mollusk shell formation: a source of new concepts for understanding biomineralization processes. Chemistry 12 (4), 980–987. doi: 10.1002/chem.200500980
Addadi L., Moradian J., Shay E., Maroudas N. G., Weiner S. (1987). A chemical model for the cooperation of sulfates and carboxylates in calcite crystal nucleation: relevance to biomineralization. P. Natl. Acad. Sci. U.S.A. 84 (9), 2732–2736. doi: 10.2307/29257
Bai Z. Y., Zheng H. F., Lin J. Y., Wang G. L., Li J. L. (2013). Comparative analysis of the transcriptome in tissues secreting purple and white nacre in the pearl mussel Hyriopsis cumingii. PloS One 8 (1), e53617. doi: 10.1371/journal.pone.0053617
Berman A., Hanson J., Leiserowitz L., Koetzle T. F., Weiner S., Addadi L. (1993). Biological control of crystal texture: a widespread strategy for adapting crystal properties to function. Science 259 (5096), 776–779. doi: 10.2307/2880829
Blay C., Planes S., Ky C. L. (2018). Cultured pearl surface quality profiling by the shell matrix protein gene expression in the biomineralised pearl sac tissue of Pinctada margaritifera. Mar. Biotechnol. 20 (4), 490–501. doi: 10.1007/s10126-018-9811-y
Dauphin Y. (2003). Soluble organic matrices of the calcitic prismatic shell layers of two pteriomorphid bivalves Pinna nobilis and Pinctada margaritifera. J. Biol. Chem. 278 (17), 15168–15177. doi: 10.1074/jbc.m204375200
Dauphin Y., Cuif J.-P., Doucet J., Salomé M., Susini J., Terry Willams C. (2003). In situ Chemical speciation of sulfur in calcitic biominerals and the simple prism concept. J. Struct. Biol. 142 (2), 272–280. doi: 10.1016/s1047-8477(03)00054-6
Fan S. S., Zheng Z., Hao R. J., Du X. D., Jiao Y., Huang R. L. (2020). PmCBP, a novel poly (chitin-binding domain) gene, participates in nacreous layer formation of Pinctada fucata martensii. Comp. Biochem. Phys. B., 240, 110374. doi: 10.1016/j.cbpb.2019.110374
Gotliv B. A., Kessler N., Sumerel J. L., Morse D. E., Weiner S. (2005). Asprich: a novel aspartic acid-rich protein family from the prismatic shell matrix of the bivalve Atrina rigida. ChemBioChem 6 (2), 304–314. doi: 10.1002/cbic.200400221
Hasegawa Y., Uchiyama K. (2010). cDNA clonings of shell matrix proteins from scallop shell. Fisheries Sci. 71 (5), 1174–1178. doi: 10.1111/j.1444-2906.2005.01078.x
Huang J. L., Liu Y. J., Liu C., Xie L. P., Zhang R. Q. (2021). Heterogeneous distribution of shell matrix proteins in the pearl oyster prismatic layer. Int. J. Biol. Macromol. 189, 641–648. doi: 10.1016/j.ijbiomac.2021.08.075
Inoue N., Ishibashi R., Ishikawa T., Atsumi T., Aoki H., Komaru A. (2011). Can the quality of pearls from the Japanese pearl oyster (Pinctada fucata) be explained by the gene expression patterns of the major shell matrix proteins in the pearl sac? Mar. Biotechnol. 13 (1), 48–55. doi: 10.1007/s10126-010-9267-1
Jin C., Li J. L., Liu X. J. (2020). Teosin, a novel basic shell matrix protein from Hyriopsis cumingii induces calcium carbonate polycrystal formation. Int. J. Biol. Macromol. 150, 1229–1237. doi: 10.1016/j.ijbiomac.2019.10.134
Jin C., Zhao J. Y., Liu X. J., Li J. L. (2019a). Expressions of shell matrix protein genes in the pearl sac and its correlation with pearl weight in the first 6months of pearl formation in Hyriopsis cumingii. Mar. Biotechnol. 21, 240–249. doi: 10.1007/s10126-019-09876-z
Jin C., Zhao J. Y., Pu J. W., Liu X. J., Li J. L. (2019b). Hichin, a chitin binding protein is essential for the self-assembly of organic frameworks and calcium carbonate during shell formation. Int. J. Biol. Macromol. 135, 745–751. doi: 10.1016/j.ijbiomac.2019.05.205
Kim I. W., Collino S., Evans J. S. (2012). Cooperative modulation of mineral growth by prismatic-associated asprich sequences and Mg(II). Int. J. Mol. Sci. 13 (3), 3949–3958. doi: 10.3390/ijms13033949
Kim I. W., Giocondi J. L., Orme C., Collino S., Evans J. S. (2008). Morphological and kinetic transformation of calcite crystal growth by prismatic-associated asprich sequences. Cryst. Growth Des. 8 (4), 1154–1160. doi: 10.1021/cg070099m
Kintsu H., Okumura T., Negishi L., Ifuku S., Kogure T., Sakuda S., et al. (2017). Crystal defects induced by chitin and chitinolytic enzymes in the prismatic layer of Pinctada fucata. biochem. Bioph. Res. Co. 489 (2), 89–95. doi: 10.1016/j.bbrc.2017.05.088
Kong Y. W., Jing G., Yan Z. G., Li C. Z., Gong N. P., Zhu F. J., et al. (2009). Cloning and characterization of prisilkin-39, a novel matrix protein serving a dual role in the prismatic layer formation from the oyster Pinctada fucata. J. Biol. Chem. 284 (16), 10841–10854. doi: 10.1074/jbc.M808357200
Le Luyer J., Auffret P., Quillien V., Leclerc N., Reisser C., Vidal-Dupiol J., et al. (2019). Whole transcriptome sequencing and biomineralization gene architecture associated with cultured pearl quality traits in the pearl oyster, Pinctada margaritifera. BMC Genomics 20 (1). doi: 10.1186/s12864-019-5443-5
Levi-Kalisman Y., Falini G., Addadi L., Weiner S. (2001). Structure of the nacreous organic matrix of a bivalve mollusk shell examined in the hydrated state using cryo-TEM. J. Struct. Biol. 135 (1), 8–17. doi: 10.1006/jsbi.2001.4372
Liang J., Xu G. R., Xie J., Lee I., Xiang L., Wang H. Z., et al. (2015). Dual roles of the lysine-rich matrix protein (KRMP)-3 in shell formation of pearl oyster, Pinctada fucata. . PloS One 10 (7), e0131868. doi: 10.1371/journal.pone.0131868
Lin J. Y., Ma K. Y., Bai Z. Y., Li J. L. (2013). Molecular cloning and characterization of perlucin from the freshwater pearl mussel, Hyriopsis cumingii. Gene 526 (2), 210–216. doi: 10.1016/j.gene.2013.05.029
Liu X. J., Dong S. J., Jin C., Bai Z. Y., Wang G. L., Li J. L. (2015a). Silkmapin of Hyriopsis cumingii, a novel silk-like shell matrix protein involved in nacre formation. Gene 555 (2), 217–222. doi: 10.1016/j.gene.2014.11.006
Liu X. J., Jin C., Guo W., Li J. L. (2019). Identification and molecular characterization of hc-upsalin, a novel matrix protein involved in nacreous-layer biomineralization in Hyriopsis cumingii. Thalassas 35, 143–150. doi: 10.1007/s41208-018-0102-1
Liu X. J., Li J. L. (2015). Formation of the prismatic layer in the freshwater bivalve Hyriopsis curninqii: the feedback of crystal growth on organic matrix. Acta Zool. 96, 30–36. doi: 10.1111/azo.12048
Liu X. J., Li J. L., Xiang L., Sun J., Zheng G. L., Zhang G. Y., et al. (2012). The role of matrix proteins in the control of nacreous layer deposition during pearl formation. P. R. Soc B-Biol. Sci. 279 (1730), 1000–1007. doi: 10.1098/rspb.2011.1661
Liu X. J., Zeng S. M., Dong S. J., Jin C., Li J. L. (2015b). A novel matrix protein hic31 from the prismatic layer of Hyriopsis cumingii displays a collagen-like structure. PloS One 10 (8), e0135123. doi: 10.1371/journal.pone.0135123
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Luo W., Chen Y., Chen C., Ren G. (2022a). The proteomics of the freshwater pearl powder: insights from biomineralization to biomedical application. J. Proteomics 265. doi: 10.1016/j.jprot.2022.104665
Luo W., Jiang R., Ren G., Jin C. (2022b). Hic12, a novel acidic matrix protein promotes the transformation of calcite into vaterite in Hyriopsis cumingii. Comp. Biochem. Phys. B. 261. doi: 10.1016/j.cbpb.2022.110755
Ma Y. F., Berland S., Andrieu J. P., Feng Q. L., Bedouet L. (2013). What is the difference in organic matrix of aragonite vs. vaterite polymorph in natural shell and pearl? study of the pearl-forming freshwater bivalve mollusc Hyriopsis cumingii. Mat. Sci. Eng. C-Mater. 33 (3), 1521–1529. doi: 10.1016/j.msec.2012.12.057
Ma Y. F., Qiao L., Feng Q. L. (2012). In-vitro Study on calcium carbonate crystal growth mediated by organic matrix extracted from fresh water pearls. Mat. Sci. Eng. C-Mater. 32 (7), 1963–1970. doi: 10.1016/j.msec.2012.05.030
Marie B., Joubert C., Tayale A., Zanella-Cleon I., Belliard C., Piquemal D., et al. (2012). Different secretory repertoires control the biomineralization processes of prism and nacre deposition of the pearl oyster shell. P. Natl. Acad. Sci. U.S.A. 109 (51), 20986–20991. doi: 10.1073/pnas.1210552109
Okumura T., Suzuki M., Nagasawa H., Kogure T. (2012). Microstructural variation of biogenic calcite with intracrystalline organic macromolecules. Cryst. Growth Des. 12 (1), 224–230. doi: 10.1021/cg200947c
Sarashina I., Endo K. (2001). The complete primary structure of molluscan shell protein 1 (MSP-1), an acidic glycoprotein in the shell matrix of the scallop Patinopecten yessoensis. Mar. Biotechnol. 3 (4), 362–369. doi: 10.1007/s10126-001-0013-6
Shen X., Belcher A. M., Hansma P. K., Stucky G. D., Morse D. E. (1997). Molecular cloning and characterization of lustrin a, a matrix protein from shell and pearl nacre of haliotis rufescens. J. Biol. Chem. 272 (51), 32472–32481. doi: 10.1074/jbc.272.51.32472
Sudo S. (1997). Structures of mollusc shell framework proteins. Nature 387 (6633), 563–564. doi: 10.1038/42391
Suzuki M., Murayama E., Inoue H., Ozaki N., Tohse H., Kogure T., et al. (2004). Characterization of prismalin-14, a novel matrix protein from the prismatic layer of the Japanese pearl oyster (Pinctada fucata). Biochem. J. 382 (1), 205–213. doi: 10.1042/bj20040319
Suzuki M., Nagasawa H. (2007). The structure–function relationship analysis of prismalin-14 from the prismatic layer of the Japanese pearl oyster, Pinctada fucata. FEBS J. 274 (19), 5158–5166. doi: 10.1111/j.1742-4658.2007.06036.x
Suzuki M., Sakuda S., Nagasawa H. (2007). Identification of chitin in the prismatic layer of the shell and a chitin synthase gene from the Japanese pearl oyster, Pinctada fucata. Biosci. Biotech. Bioch. 71 (7), 1735–1744. doi: 10.1271/bbb.70140
Suzuki M., Saruwatari K., Kogure T., Yamamoto Y., Nishimura T., Kato T., et al. (2009). An acidic matrix protein, pif, is a key macromolecule for nacre formation. Science 325, 1388–1390. doi: 10.1126/science.1173793
Takagi R., Miyashita T. (2010). Prismin: a new matrix protein family in the Japanese pearl oyster (Pinctada fucata) involved in prismatic layer formation. Zool Sci. 27 (5), 416–426. doi: 10.2108/zsj.27.416
Takeuchi T., Sarashina I., Iijima M., Endo K. (2008). In vitro Regulation of CaCO3 crystal polymorphism by the highly acidic molluscan shell protein aspein. FEBS Lett. 582 (5), 591–596. doi: 10.1016/j.febslet.2008.01.026
Tsukamoto D., Sarashina I., Endo K. (2004). Structure and expression of an unusually acidic matrix protein of pearl oyster shells. Biochem. Bioph. Res. Co. 320 (4), 1175–1180. doi: 10.1016/j.bbrc.2004.06.072
Weiner S., Traub W., Parker S. B. (1984). Macromolecules in mollusc shells and their functions in biomineralization [and discussion]. Philos. T. R. Soc B. 304(1121), 425–434. doi: 10.2307/2396111
Xia Z., Liu X., Li J. (2019). The role of Hyriopsis cumingii shell silk-like matrix protein gene silkmaxin in the shell and pearl biomineralization. J. Fish. China. 43 (12), 2449–2458. doi: 10.11964/jfc.20181111542
Yano M., Nagai K., Morimoto K., Miyamoto H. (2006). Shematrin: a family of glycine-rich structural proteins in the shell of the pearl oyster Pinctada fucata. Comp. Biochem. Phys. B. 144 (2), 254–262. doi: 10.1016/j.cbpb.2006.03.004
Keywords: matrix protein, prismatic layer, framework, silk-like, Hcprismatin-14, Hyriopsis cumingii
Citation: Jin C, Zhang Y, Cheng K, Jiang R, Jiang S, Shi Y, Ren G and Luo W (2023) Hcprismatin-14 of Hyriopsis cumingii, a novel matrix protein is crucial for framework recognition and crystal deposition during prismatic layer formation. Front. Mar. Sci. 10:1154968. doi: 10.3389/fmars.2023.1154968
Received: 31 January 2023; Accepted: 10 April 2023;
Published: 01 May 2023.
Edited by:
Andrew Stanley Mount, Clemson University, United StatesReviewed by:
Eva Martins, Universidade Católica Portuguesa, PortugalCopyright © 2023 Jin, Zhang, Cheng, Jiang, Jiang, Shi, Ren and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen Luo, bHVvd2VuQHVzeC5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.