- 1School of Marine Sciences, Ningbo University, Ningbo, China
- 2Department of Agriculture and Rural Affairs of Henan Province, Henan Fishery Technical Extension Station, Zhengzhou, China
- 3Key Laboratory of Aquacultural Biotechnology Ministry of Education, Ningbo University, Ningbo, China
- 4Key Laboratory of Green Mariculture (Co-construction by Ministry and Province), Ministry of Agriculture and Rural, Ningbo, China
- 5Aquatic Technology Promotion Station, Yanjin Rural Bureau, Xinxiang, China
Introduction: The intestinal microbiota acts as an additional “organ” that performs a variety of fu\nctions for the host’s health. However, the composition and role of the intestinal microbiota in Scylla paramamosain cultivated in inland low salinity saline-alkaline water are unknown.
Methods: Accordingly, from the perspective of practical production, we explored the intestinal microbiota communities and the critical bacteria of S. paramamosain in normal salinity seawater (NS), coastal low salinity seawater (CS), acute low salinity seawater (AS) and inland low salinity saline-alkaline water (IS).
Results: Results showed that there were significant differences in the diversity composition of intestinal microbiota and the relative abundance of dominant taxa in each group of cultured crabs. Firmicutes, Proteobacteria, Bacteroidota and Campilobacterota were shown to be the major phyla shared by the four groups, with Bacteroidota having the highest relative abundance (27.10%) in the inland low salinity saline-alkaline water group (IS). Fusobacteriota had the highest proportion in IS group compared with other low salinity groups. A total of 284 indicator bacteria were identified, belonging to eight phyla, and their relative abundances were varied significantly (P < 0.05). Genus Carboxylicivirga, as the indicator bacterium of the IS group, may play a critical role in the adaptation of crab to saline-alkaline water environment. Moreover, salinity may exert considerable selective pressure on the entire microbial community.
Discussion: These findings revealed the features of the intestinal microbiome in S. paramamosain in multiple low salinity patterns, and provided candidate probiotics and basic information for crab farming in saline-alkaline water, which was conducive to the development and perfection of mud crab culturing technology in inland low salinity saline-alkaline water.
1 Introduction
Over the past few decades, the contemporary aquaculture industry has grown tremendously, becoming one of the fastest growing sectors in world food production, increasing from 25.7% in 2000 to 46.8% in 2018 (FAO, 2020). Blue foods make a great contribution to global food security and nutrition (Tigchelaar et al., 2022). The importance of aquaculture in the food supply is increasing due to the significant decline in wild capture fisheries. The mud crab, Scylla paramamosain has become a vital species in aquaculture along the southeast coast of China because of its rapid growth, delicious meat, strong adaptability, elevated nutritional and economic value (Meng et al., 2017). In 2020, the output of S. paramamosain increased to 159,433 tons, accounting for more than half of the total marine crab farming industry in China (China fishery statistical yearbook, 2021). However, the yield cannot meet the ever-increasing market demand, becoming the bottleneck of the development of the crab industry (Wang et al., 2018a). With the current saturation of aquaculture space along the coast, it is an effective way to break through the bottleneck to expand inland saline-alkaline areas and other low salinity waters (Liang et al., 2022; Niu et al., 2022). However, only metabolic (Niu et al., 2022) and transcriptomic (Liang et al., 2022) analysis have been conducted on the adaptation mechanism of the crab to saline-alkaline water, while studies on intestinal microbiota and its role have not been reported.
The growth, immunity, metabolism, nutrition and development of the host are all affected by the gut microbiota (Fabbiano et al., 2013; Chevalier et al., 2015; Singh et al., 2015; Chen et al., 2017). Studies indicated that changes in environmental parameters such as salinity, temperature, type of diet, pH, and water chemistry lead to rapid and profound changes in the gut microbiota of organisms (Chen et al., 2017; Fan et al., 2018; Martínez-Mota et al., 2019; Sun et al., 2020). Host habitat is the major determinant of fish intestinal microbes (Kim et al., 2021). And salinity is one of the key environmental variables influencing bacterial community composition (Rudi et al., 2017), various microorganisms may grow better at different salinities. More importantly, the host shows a close and complex interaction with the microorganisms in its gastrointestinal tract (McFall-Ngai et al., 2013). The gut microbiota further participates in and maintains host physiological activities by generating a range of different small-molecule metabolites obtained from diet, medications, and host-generated components (Krautkramer et al., 2021). These metabolites, in turn, serve an essential role in maintaining microbial homeostasis by engaging in host energy metabolism and regulation of immune function (Canfora et al., 2019; Garrett, 2019), thereby helping the host to adapt to different habitats. Recent research has revealed that the gut microbiome plays a vital role in local adaptation to environments such as pollution (Henry et al., 2019), drought (Ribeiro et al., 2019) and salinity (Fietz et al., 2018; Rennison et al., 2019). However, relative to vertebrates, the intestinal microbiota function of crustaceans, especially mud crabs, has been poorly studied. It is known that the intestinal microbiota operates as an additional “organ” that performs a variety of functions for the host’s health, such as obtaining nutrients and preventing pathogen invasion (Dai et al., 2018; Xiong et al., 2019b; Huang et al., 2020). Under these circumstances, it is crucial to systematically characterize the gut microbiome composition of mud crabs in inland low salinity saline-alkaline water (ILSW).
In this study, we comparatively characterized the gut bacterial communities of crabs in multiple low salinity aquaculture patterns using Illumina sequencing technology, which further revealed the features of the intestinal microbiome of S. paramamosain in saline-alkaline water environment and provided candidate probiotics and basic information for the crab farming in ILSW. As far as we know, this is the first attempt to systematically characterize the gut bacterial communities of S. paramamosain under multiple low-salinity patterns, revealing the specialization of intestinal microbiota to ILSW.
2 Materials and methods
2.1 Experimental animals and design
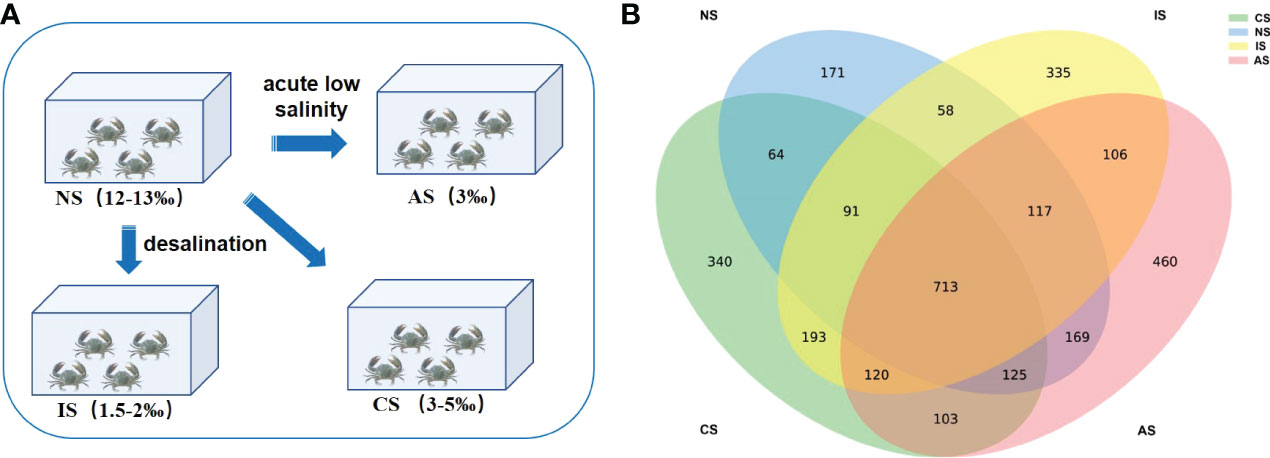
The random selection of 600 mud crabs from Sanmen Bay with 30 ± 5 g bodyweight was carried out. Then the crabs were separated into the following three groups: NS, CS and IS groups, with 150 crabs in each group, and placed into the pools (6 m × 3 m × 1.5 m) under the same rearing conditions (three replicates per group). The NS (normal salinity) group was located on Sanmen Bay, the east coast of Zhejiang Province, which is one of the major producing areas of mud crabs in China. The salinity of sea water is around 12‰, which is the optimum salinity for mud crab growth. The CS (coastal low salinity) group was located on Hangzhou Bay, the estuary and bay of Qiantang River in the northeast of Zhejiang Province. The salinity of seawater is low and unstable (3-5‰) because of river infiltration and precipitation, and crabs can grow naturally here. The IS (inland low salinity) group was located on the north bank of the Yellow River, Henan Province, where there are large areas of saline-alkaline land. The groundwater is saline-alkaline water with a salinity of 1.5-2‰, crabs from seawater could grow normally here after gradient desalination. The desalination method was as follows: The juvenile crabs from Sanmen Bay were acclimated to the original salinity for 12 h, the water salinity was decreased to 12, then to 8 at a rate of 2/6 h, and finally to 1 at a rate of 1/6 h. The desalination process was completed after 6 h of stability. After that, place them in the foam box (45 cm × 45 cm × 10 cm) with a density of 150 per box. The foam box has air vents all around it, and a layer of moist sponge cushion was placed at the bottom. The humidity was kept above 90%, and the foam box was flown to the saline-alkaline water pond via air (Figure 1A; Table S1).
The experiment was conducted during the natural photoperiod at room temperature. And water quality parameters were measured every day. The parameters including: pH 7.8~8.6, water temperature 25°C~30°C, dissolved oxygen > 7.0 mg·L-1, nitrite concentration < 0.15 mg·L-1, ammonia nitrogen concentration < 0.8 mg·L-1 (Table S1). The crabs were fed once a day at 5:00 p.m. Depending on local conditions, low-value and fresh shellfish were selected as food sources, such as clams and river snails, which are commonly used as feed in mud crab farming due to their high nutritional value and palatability. After 90 days, five active, healthy adult crabs with uniform body weight (165 g-297 g) were randomly selected from NS, CS and IS group in October 2020, respectively. Then 30 crabs from NS group were randomly selected and fed in seawater diluted to 3‰ for one week (three replicates), and defined as AS (acute low salinity) group, under the same feeding conditions as above. At the end of the culture, crabs were killed after being anesthetized in ice water, and then the intestine and its contents were quickly taken out and stored in liquid nitrogen for 16S rRNA gene sequencing (n=5).
2.2 DNA extraction, PCR amplification and Illumina MiSeq sequencing
The bacterial genomic DNA of the gut samples was extracted by DNeasy PowerLyzer PowerSoil Kit (QIAGEN) in accordance with the manufacturer’s protocol. The quality of extracted DNA was assessed with a NanoDrop 2000 instrument (Thermo Fisher Scientific, Wilmington, MA, USA) based on absorption ratios at 260/280 nm and 260/230 nm (Lin et al., 2022). The 16S rRNA genes were partially amplified with the bacterial universal V3-V4 primer 343F (5′-TACGGRAGGCAGCAG-3′) and 798R (5′- AGGGTATCTAATCCT-3′). A 30 μL polymerase chain reaction (PCR) system, including 11 µL ddH2O, 1.0 μL of each primer (10 mmol/L), 15 μL 2 × SYBR Prime-Script™ Master Mix (TaKaRa Biotech, Japan), and 2 μL of template DNA. The PCR reaction procedure was performed as described by Wei et al. (Wei et al., 2019). Agencourt Ampure XP beads (Beckman Coulter Co., USA) were used to purify PCR products. Following assessment of fragment size and quantification, and then these amplicons were sequenced in equimolar and paired-end (2 × 250) using the Illumina MiSeq platform (Illumina, San Diego, CA, USA).
2.3 Processing of Illumina sequencing data
PE readings were preprocessed using Trimmomatic software (Bolger et al., 2014). After trimming, FLASH software was used to compile paired-end reads (Reyon et al., 2012). The Vsearch program was used to eliminate the primer sequences and cluster sequences into generated operational taxa taxonomic units (OTUs) with a 97% similarity cut-off value (Rognes et al., 2016). The QIIME software suite was used to pick representative contigs for each OTU. The Silva database (version 138) was annotated and parsed using the RDP classifier (70% confidence threshold) (Wang, 2007).
2.4 Statistical analyses
Mothur software (version 1.32.1) was used to analyze Alpha diversity indexes, including Chao 1, observed species, Simpson, Shannon diversity and Good’s coverage estimate using an OTU specified at the 97% identity level (Schloss et al., 2009). Based on Bray-Curtis distance metrics, principal co-ordinates analysis (PCoA) and analysis of variance (ANOVA) were performed to examine the overall differences in the bacterial community of the crab guts (Clarke, 1993). The linear discriminant analysis (LDA) coupled with effect size measurements (LEfSe) were used to distinguish the differences of dominant bacterial communities. The indicator values (IndVal) of OTU in each group were calculated, and then the indicators were screened based on the relative abundance of OTU in any group exceeded than 1% and IndVal was significant between groups (P < 0.05) (Dufrêne and Legendre, 1997). Additionally, heat map analysis was applied to analyze the differences in microorganisms in four groups. To explore the variations in intestinal bacteria composition among the three low salinity groups, the ternary plot was generated using package ggtern in R. In order to explore the correlation between bacterial abundance and salinity, logistic regression curve analysis was performed in R.
2.5 Prediction of functional potentials
The Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt2) (Douglas et al., 2020) was performed to predict the functional composition of known microbial genes based on 16S sequencing data annotated in the Greengenes database (Desantis et al., 2006), to count the functional differences between different groups. The KEGG pathways were retrieved according to the KEGG database (http://www.kegg.jp/). Multiple comparisons were performed using the Kruskal-Wallis test and ggplots2 in R was used to construct the plots.
3 Results
3.1 Data summary
We analyzed the intestinal microbiota community composition of 20 crabs inhabiting in four different salinity environments (Figure 1A). A total of 1,463,579 raw 16S rRNA gene sequence tags were generated from the 20 samples, and obtained 1,340,133 high-quality sequence tags, with 59,675-71,365 sequences per sample after the removal of low-quality and chimeric sequences. High-quality sequences were clustered into 3165 OTUs, applying a threshold of 97% sequence identity (Supplementary Table S2). These OTUs were classified into 33 phyla and 682 genera. The characterization of bacterial community richness, diversity and sequencing depth were compared between four groups (Supplementary Figure S1, Table S3). According to the Shannon, Simpson, Chao1 and Observed species index, there was no significant difference in the diversity of the crabs’ intestinal microbial in four groups, suggesting that geographical location and salinity had no influence on the diversity and richness of the intestinal microbiota in S. paramamosain. The Goods coverage index > 99% in all samples, demonstrating that the sequencing was efficient (Supplementary Table S3). There were 712 OTUs shared among the four groups, with 120 OTUs shared by the three low salinity groups, and just 58 shared by the IS and NS groups. Each group has 171 (NS), 340 (CS), 335 (IS) and 460 (AS) unique OTUs, respectively (Figure 1B).
3.2 Composition of the microbial communities
At phylum level, the four groups of crab gut microbiomes were similar in composition, but had significant differences in relative abundance. The predominant phyla (relative abundance >5% at least in one group) shared by the four groups of crabs’ intestinal bacteria were Firmicutes, Proteobacteria, Bacteroidota and Campilobacterota (Figures 2A, S2). In the NS and IS groups, Fusobacteriota also accounted for a large proportion, accounting for 11.35% and 7.25%, respectively, whereas only 0.74% and 0.47% in the CS and AS groups. Bacteroidota had the highest percentage in the IS group, accounting for 27.10% (Figures 2A, S2A, C). Dependentiae was found exclusively in the CS group, and in extremely low abundance. Interestingly, the relative abundances of most phyla in the AS group were higher than that of the other three groups (Figure S3A). There were 682 genera identified in all, but only 20 genera had an abundance of more than 1%. At genus level, the four groups showed distinct bacterial community structure (Figure 2B). The relative abundance of Candidatus_Hepatoplasma was higher in NS (31.11%), CS (12.89%) and AS (18.02%), but very low in IS group (0.16%). The results indicated that Candidatus_Hepatoplasma may be an important gut bacterium in mariculture crabs. Photobacterium was abundant in the NS and CS groups, occupying approximately 17.39% and 15.96%, respectively, but lower in the IS and AS groups, accounting for just 0.04% and 0.47%. Carboxylicivirga (21.12%) and Candidatus Hepatoplasma (31.22%) were the most abundant in the IS and NS groups, respectively. In particular, Psychrilyobacter, accounting for the second-largest proportion in the NS group, which was essentially non-existent in the other groups (Figure 2B).
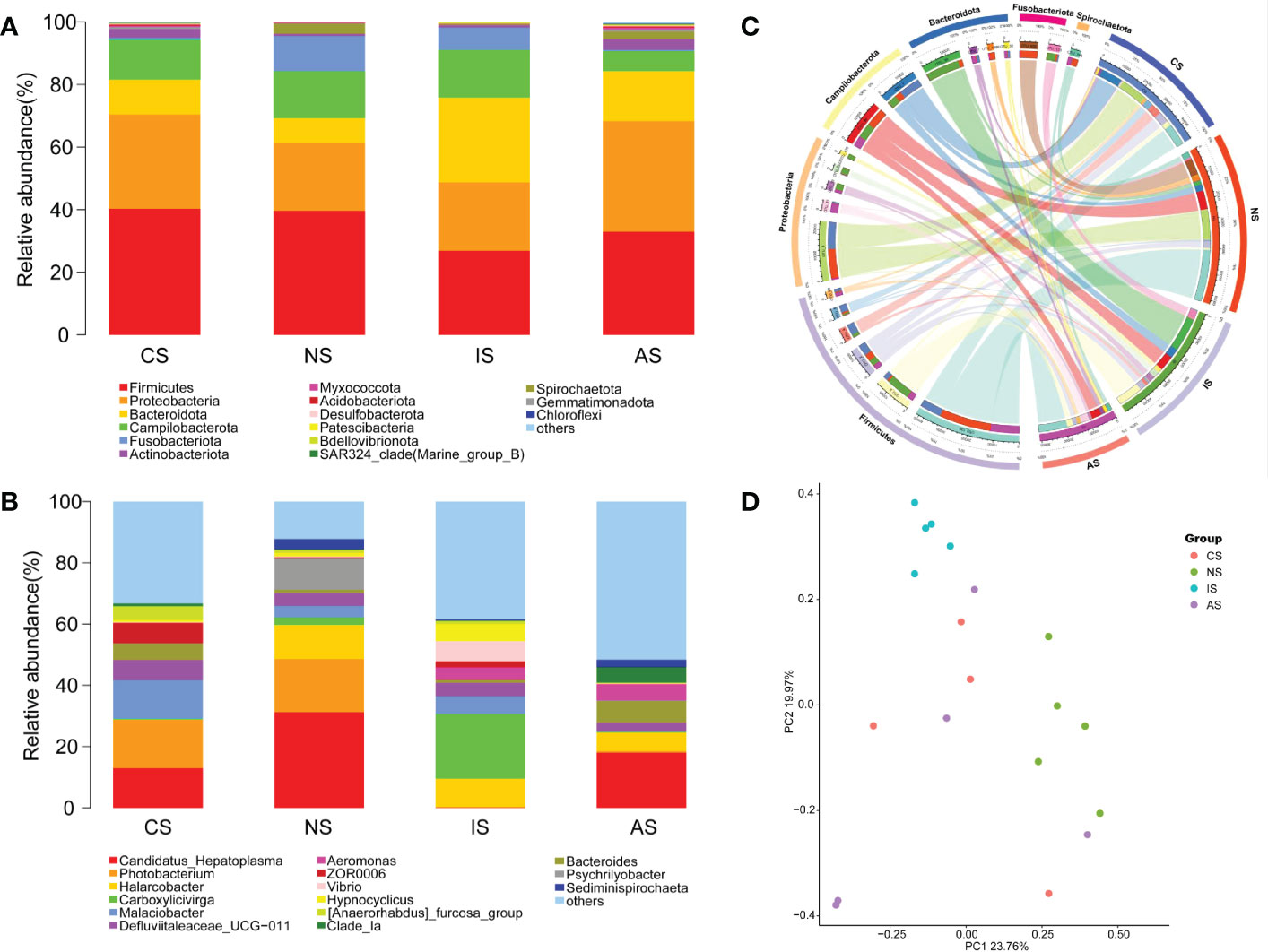
Figure 2 Composition and abundance of the intestinal microbiota. (A) Compositions and abundances of the gut microbiome communities at the top15 phylum level; (B) Compositions and abundances of the microbiome communities at the top 15 genus level; (C) Circos plot of the proportion of gut microbiome in the four groups at phylum level; (D) Principal coordinates analysis (PCoA) of the bacterial communities based on the Bray-Curtis distances.
3.3 Differential intestinal microbiota of S. paramamosain in response to multiple low salinity modes
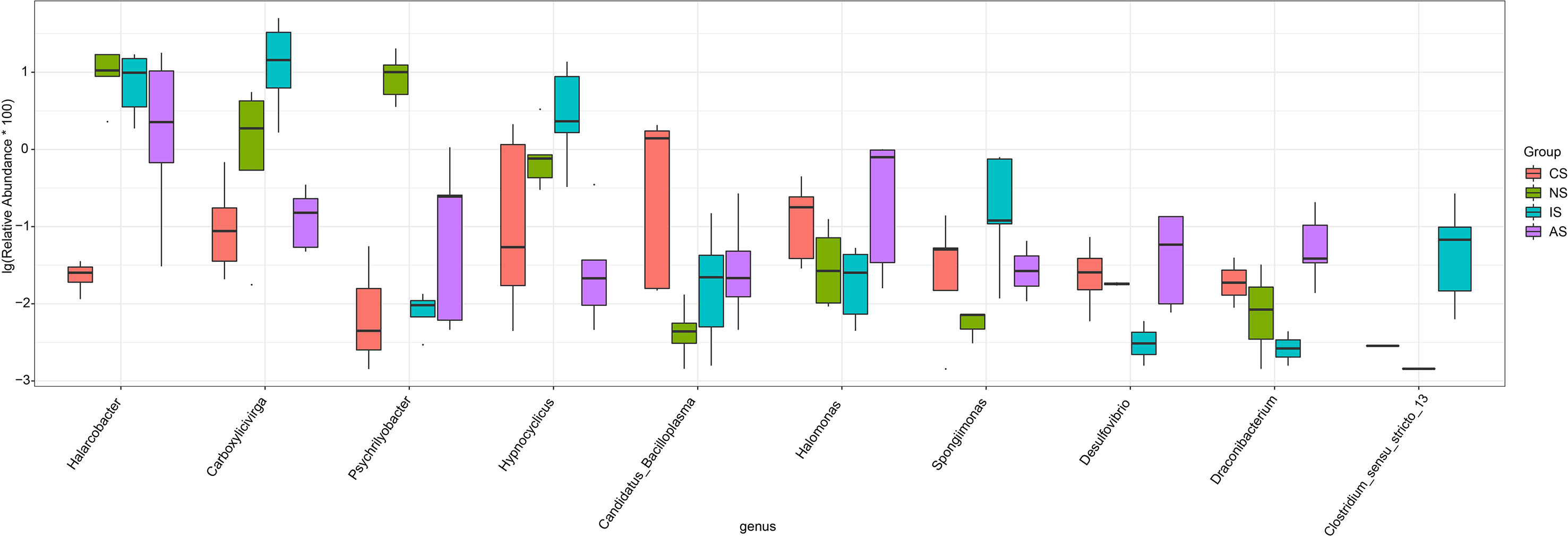
We found significant differences in gut bacterial community composition between IS and other groups (Figure 2C). In the PCoA, principal coordinate 1(PCo1) explained 23.76% of the variance (Figure 2D), indicating that the bacterial communities clustered according to the salinity, aside from CS and AS groups, and the IS group was separated clearly from the other groups. These finding suggested that saline-alkaline water may be the most important driver of variance in crab gut microbiota. ANOVA analysis was used to examine species that differed significantly across groups. There were 129 differential OTUs, belonging to 37 differential genera (Figure S4). Of the top 10 differential genera, Carboxylicivirga, Hypnocyclicus, Spongiimona and Clostridium_sensu_stricto_13 were significantly elevated in the IS group, while Desulfovibrio and Draconibacterium were significantly less abundant. In the NS group, the relative abundance of Psychrilyobacter was significantly increased. When compared to the other three groups, the relative abundance of Halarcobacter was significantly lower in the CS group, while the relative abundance of Candidatus Bacilloplasma was greatest in the CS group. In the AS group, the relative abundance of Draconibaterium was significantly increased (Figure 3).
The ternary plot demonstrated that the dominating species at phylum level were distinct between three low salinity groups (Figure S5). Fusobacteriota had the largest proportion and relative abundance in the IS group, while Spirochaetota have the highest proportion in the AS group, indicating that Fusobacteriota may play an important role in IS group. Firmicutes, Proteobacteria, Bacteroidota and Campilobacterota were the dominant phyla shared by the three groups, which was the same as the above results. Meanwhile, the proportion of nine phyla were higher in the AS group, but the relative abundance was lower, indicating that the intestinal microbial of the AS group was disordered. LEfSe analyses was performed to identify differentially represented OTUs across four groups (Figure 4). The analysis revealed that there were 3, 15, 13, and 6 biomarkers (LDA score >3) in the CS, NS, IS and AS groups, respectively. The family Marinilabiliaceae and genus Carboxylicivirga made a significant contribution to the IS group. The top featured microbes in the NS and AS groups were genus Photobacterium and class Alphaproteobacteria, respectively. Logistic regression curve analysis indicated that the phylum Fusobacteriota and genus Carboxylicivirga were shown to be significantly positively correlated with salinity (P < 0.05, Figure S6).
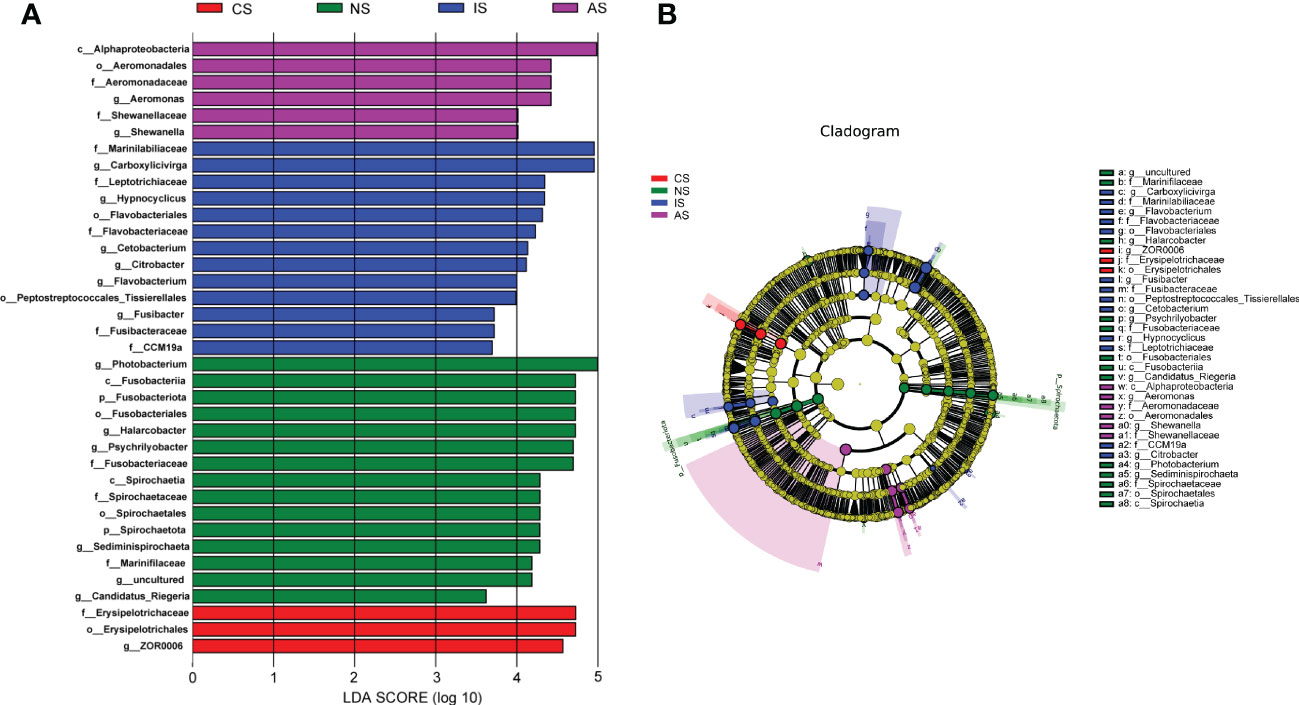
Figure 4 Differential bacterial composition of the four groups based on LEfSe Analysis. (A) Histogram of LDA analysis. Different colors represent different groups, different colored bars indicate species with relatively high abundance in different groups; (B) The cladograms of differential bacterial. Different colors represent different groups, color dots corresponding to groups indicate species with relatively high abundance and significant differences in that group, the yellow dots represent species that were not significantly different between the four groups, node diameter size is proportional to relative abundance size.
Indicator mainly refer to species, genera, or communities that can have a great impact on the growing environment. We analyzed indicator bacteria that significantly contributed to the differentiation of intestinal samples from multiple low salinity modes at the OTU level using FDR > 0.50. A total of 284 indicator bacteria were identified, belonging to eight phyla, and their relative abundances were varied significantly (P < 0.05). The relative abundance of 64 indicator bacteria detected in the IS group samples was higher than that of the NS, CS and AS groups, with Carboxylicivirga (OTU_38) being the highest. However, the relative abundance of 15 indicator bacteria detected in the NS group was higher than that of the IS, CS and AS groups, with Psychrilyobacte (OTU_809) being the most abundant (Figure 5). Meanwhile, Carboxylicivirga (OTU_38) and Psychrilyobacte (OTU_809) showed high indicator values, indicating that the two OTUs may be the representative bacterium of IS and NS groups, respectively.
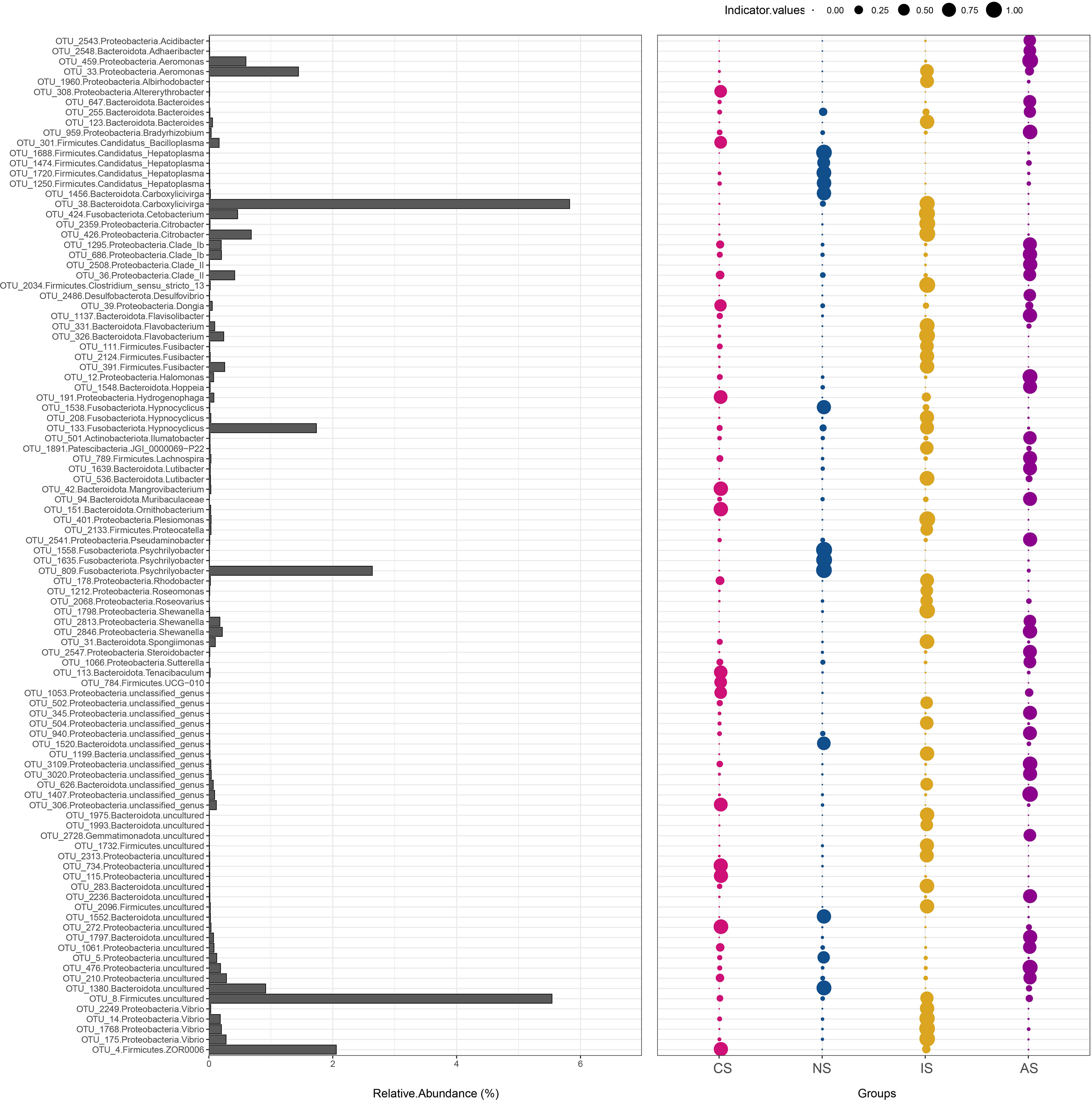
Figure 5 Gut bacterial indicators in response to multiple low salinity modes. The bar graph represents the relative abundance of each OTU, and the bubble size represents the indicator value of each species in the sample group, that is, the indicative size of the species in the group.
3.4 Predicted function of intestinal microbiota community
The function was predicted by PICRUSt2 to compare the functional potentials of crabs’ intestinal microbiota community under multiple low-salinity environments (Figure 6). A total of 41 functional pathways were predicted, and most of the pathways enriched in the AS group were significantly increased than those in other groups, especially amino acid metabolism, lipid metabolism, energy metabolism and environmental adaptation, while CS group was the next (Figure 6). The KEGG pathways in the IS group were significantly enriched in comparison to the NS group. For instance, the KEGG pathways involved in immune system, transport and catabolism, metabolism of terpenoids and polyketides, amino acid metabolism, and biosynthesis of other secondary metabolites increased in IS group compared with NS group, while the pathways involved in signaling molecules and interaction, and immune system diseases exhibited an opposite trend (Figure S7).
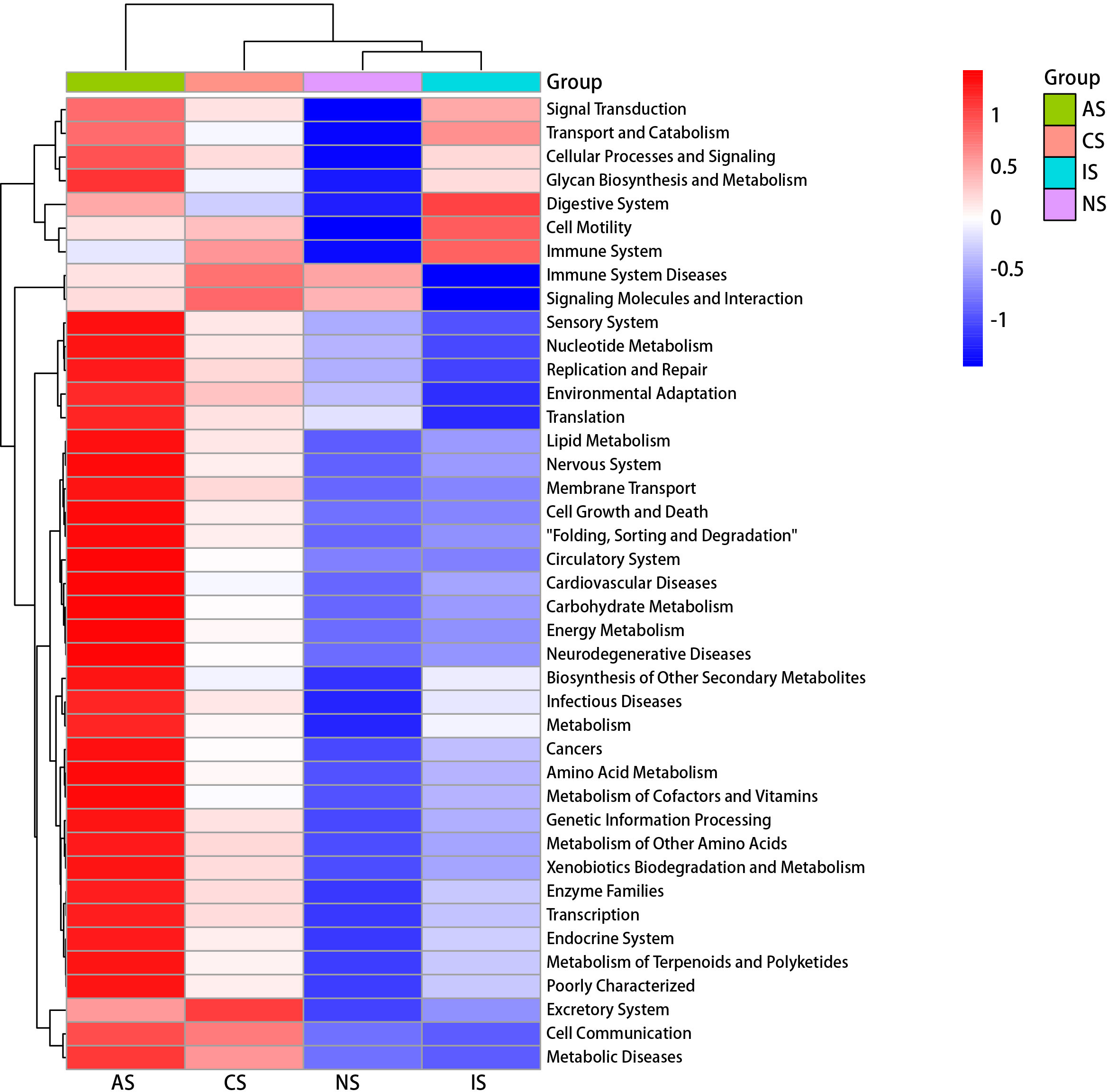
Figure 6 Heat map of KEGG function prediction of the four groups. Red indicates a higher relative abundance of species and blue indicates a lower relative abundance.
4 Discussion
Given the significant role of the microbiota in aquatic crustacean health, a thorough understanding of the intestinal microbiota is the first step in regulating bacteria for use in crab farming (Anita et al., 2016; Deng et al., 2019). Despite recent progress, we do not yet have a fundamental understanding of the role of the invertebrate gut microbiota in environmental adaptation. Additionally, few studies have been done on the composition and function of the gut microbial of mud crabs in ILSW. In this study, the bacterial community in the gut of S. paramamosain was explored for the first time under variable low salinity patterns.
A large number of studies have demonstrated that changes in habitat can alter the gut microbiota rapidly and profoundly, which in turn affects the growth and development of the host (Alberdi et al., 2016; Rudi et al., 2017; Rothman et al., 2019). The gut microbiota inhabits the intestinal tract of the host, forming a relatively stable gut ecological environment to adapt to the diversified environment (Smith et al., 2015). In this study, there were some similarities in intestinal microbes of crabs from four different environments at the phylum level. Firmicutes, Proteobacteria, Bacteroidota and Campilobacterota were the dominant phyla shared by the four groups, and the first three phyla are common dominant phyla in crustaceans (Chen et al., 2022), Campilobacterota is also a common phylum of intestinal bacteria (Hassan et al., 2021; Qi et al., 2022; Xiao et al., 2022). Previous studies have indicated that the gut microbes of Litopenaeus vannamei were high consistency despite distinct species pool (Xiong et al., 2019), and S. paramamosain in different coastal regions of southern China shared the same core gut microbiota (Wei et al., 2019). Meanwhile, the relative abundances of most phyla and functional pathways enriched in the AS group were higher than that of other groups, the reason is that under the condition of acute low salinity stress condition, the intestinal microbial of crabs in AS group were in a dynamic change process to adapt to the environment, while crabs in NS, CS and IS groups have adapted to the environment respectively, with more stable intestinal microbiota. Research showed that the balance of the gut ecosystem can be disrupted under the disturbance of external factors, from steady state to transient non-steady state, and finally either returns to the original state or enters another steady state (Fassarella et al., 2021). Amino acid and energy metabolism are the main factors for S. paramamosain to adapt to acute salinity decline, and lipid metabolites play an auxiliary role (Yao et al., 2021; Niu et al., 2022).
Bacteroidota showed the highest relative abundance in IS group, indicating its high richness in the crab intestine, as well as its notable adaptation potential to saline-alkaline water. Meanwhile, members of Bacteroidota are significant heterotrophic bacteria involved in the cycling of organic carbons and protein (Heike et al., 2010; Regueiro et al., 2012). For instance, in the human colon, Bacteroides can manufacture various capsular polysaccharides and vary their surface antigenicity (Krinos et al., 2001). Additionally, it can regulate the environment by interacting with the immune system of the host to restrict the proliferation of other bacteria (Wexler, 2007). Species of Bacteroides contain vitamins, glycans, and coenzymes that are crucial to digestion, as well as rich in enzymes involved in carbohydrate transport and protein metabolism (Karlsson et al., 2011). Thus, the enrichment of this phylum can facilitate digestion and absorption for the host, and indicated the adaptation of crabs to the saline-alkaline water environment. Fusobacteriota can produce a large amount of butyrate, which can regulate transepithelial transport, strengthen mucosal barrier, improving intestinal mucosal oxidation and inflammatory state (Sossai, 2012). Thereby their enrichment may enhance the disease resistance of crab in low salinity saline-alkaline water environment. As a symbiotic bacterium, Candidatus_Hepatoplasma is a relatively stable intestinal member in marine invertebrates, such as isopods (Cheng et al., 2019), shrimp (Parlapani et al., 2020) and sea urchins (Li et al., 2018). It is crucial in promoting the growth and survival rate of the host under poor nutrition conditions (Fraune and Zimmer, 2010; Horváthová et al., 2015). However, in this study, its abundance was lower in IS group, the reason may be that it needs some substance in seawater to grow, or that the saline-alkaline water environment cannot provide the conditions for its symbiosis.
Indicator refers to species that are used as ecological indicators of community or habitat type, environmental conditions, or environmental changes (Cáceres et al., 2010). Indicator analysis showed a strong association between the IS and NS group and putative bacteria. In the current study, the abundance of Carboxylicivirga significantly increased in the IS group, which contributed the most to this group, and was also the indicator species for IS group. Hence, Carboxylicivirga may play a critical role in the adaptation of crabs to saline-alkaline water environment. The genus Carboxylicivirga, a member of the family Marinilabiliaceae in the phylum Bacteroidota, which was first proposed by Yang et al. (Yang et al., 2014), includes, at the time of writing, five species with validly published names. Interestingly, these five species were all isolated from marine environments. Cells are strictly aerobic or facultatively anaerobic, NaCl is required for active growth (optimum 2.0-3.0%) (Yang et al., 2014; Wang et al., 2016). And Carboxylicivirga probably consumed carbohydrate, protein and organic acids in sludge for growth (Cai et al., 2021). The role of Carboxylicivirga in the intestine has not been reported so far. Thus, the mechanism of its function in the adaptation of crabs to low salinity saline-alkaline water environment needs further study.
Adaptation is a prerequisite for survival when a species moves to a different environment. Studies have shown that the habitat environment has a significant impact on the intestinal microbial community of Eriocheir sinensis (Chen et al., 2021). The gut microbiota of the four groups showed significantly different diversity, thus reflecting their adaptation to the environment. Multiple of the biotic and abiotic variables of the four habitat types considered in this study may potentially affect intestinal microflora. Significant differences in abiotic variables are, salinity, ionic components in water, and the biotic features was type of diet. Instead of temperature, pH, or other physical and chemical parameters, research has shown that salinity is the key environmental determinant of microbial community composition (Lozupone and Knight, 2007). Consistently, differences in salt concentration may affect the energy costs and metabolic pathways of microorganisms, potentially affecting the distribution and evolution of taxa (Oren, 2001). The change of salinity will lead to the increase of oxygen consumption rate, energy demand, metabolism and physiological function disorder of crustaceans (Tu et al., 2017; Wang et al., 2018b). Although S. paramamosain is a euryhaline species, acute low salinity often leads to its rapid death, resulting in economic loss (Romano and Zeng, 2012; Wang et al., 2019). Collectively, these studies suggest that salinity may exert considerable selective pressure on the entire microbial community.
Diversity and stability of the gut microbiome, as well as microbial interactions and host-microbe symbiotic relationships, appear to be key factors in restoring health. From a production perspective, being able to predict the resilience of the crab gut microbiome before stress, or in a stressed environment, would have significant benefits. The next step is to further understand the microbial interactions and the function of the microbiome, as well as the mechanisms associated with health and disease. These results offer hope for new strategies for culturing crabs in ILSW, that is, the characteristics of individual gut microbiome and personalized treatments, whether targeting key bacteria or involving specific combinations of prebiotics and probiotics, to restore the stability and functionality of intestinal ecosystem.
5 Conclusion
In summary, the present study systematically characterized the intestinal microbiota communities of S. paramamosain under multiple low salinity modes, which revealed the significant differences in the gut microflora structure among four groups. Firmicutes, Proteobacteria, Bacteroidota and Campilobacterota were the dominant phyla shared by the four groups, and Bacteroidota was the highest in the IS group. Additionally, Fusobacteriota had the highest proportion in IS group compared with other low salinity groups. Genus Carboxylicivirga, as an indicator bacterium for the IS group, may play a critical role in the adaptation of crabs to saline-alkaline water environment, and more attention should be paid for screening candidate probiotics from Carboxylicivirga species.
Collectively, this study revealed the features of the intestinal microbiome of S. paramamosain under multiple low salinity modes, and provided candidate probiotics and basic information for the crab farming in saline-alkaline water, which is conducive to the development and perfection of mud crab breeding technology in ILSW.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical review and approval was not required for the animal study because the committee on the Ethics of Animal Experiments of Ningbo University does not require approval for invertebrates. All animal work was conducted according to the relevant national and international guidelines.
Author contributions
HW and RZ designed the research. MN, XL, YC, KQ, and GL performed the experiments. MN and HW analyzed the data and wrote the manuscript. CW and CM reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (42276106), the Key Scientific and Technological Grant of Zhejiang for Breeding New Agricultural Varieties (2021C02069-6), the Major Science and Technology Innovation Tackling Project of Wenzhou (ZF2022008), the earmarked fund for CARS (CARS-48), and K. C. Wong Magna Fund in Ningbo University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1153326/full#supplementary-material
References
Alberdi A., Aizpurua O., Bohmann K., Zepeda-Mendoza M. L., Gilbert M. (2016). Do vertebrate gut metagenomes confer rapid ecological adaptation? Trends Ecol. Evol. 31 (9), 689–699. doi: 10.1007/s10499-016-0070-5
Anita T., Kwong K. O., Md A. C., Wan M. W. N., Khairun Y. (2016). The beneficial effects of multispecies Bacillus as probiotics in enhancing culture performance for mud crab Scylla paramamosain larval culture. Aquacult. Int. 25 (2), 1–18. doi: 10.1007/s10499-016-0070-5
Bolger A. M., Marc L., Bjoern U. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Cáceres M. D., Legendre P., Moretti M. (2010). Improving indicator species analysis by combining groups of sites. OIKOS 119(10), 1674–1684. doi: 10.1111/j.1600-0706.2010.18334.x
Cai G., Ebrahimi M., Zheng G., Kaksonen A. H., Zhang Z. (2021). Effect of ferrous iron loading on dewaterability, heavy metal removal and bacterial community of digested sludge by Acidithiobacillus ferrooxidans. J. Environ. Manage. 295, 113114. doi: 10.1016/j.jenvman.2021.113114
Canfora E. E., Meex R., Venema K., Blaak E. E. (2019). Gut microbial metabolites in obesity, NAFLD and T2DM. Nat. Rev. Endocrinol 119, 261–273. doi: 10.1038/s41574-019-0156-z
Chen X., Chen H., Liu Q., Ni K., Wang C. (2021). High plasticity of the gut microbiome and muscle metabolome of chinese mitten crab (Eriocheir sinensis) in diverse environments. J. Microbiol. Biotechnol. 31 (2), 240–249. doi: 10.4014/jmb.2011.11018
Chen C. Y., Chen P. C., Weng C. H., Tzun-Wen S. G., Wang D., Jadhao S. B. (2017). Habitat and indigenous gut microbes contribute to the plasticity of gut microbiome in oriental river prawn during rapid environmental change. PLoS One 12 (7), e0181427. doi: 10.1371/journal.pone.0181427
Chen C. Z., Li P., Liu L., Li Z. H. (2022). Exploring the interactions between the gut microbiome and the shifting surrounding aquatic environment in fisheries and aquaculture: A review. Environ. Res. 214 (Pt 4), 114202. doi: 10.1016/j.envres.2022.114202
Cheng X. Y., Wang Y., Li J. Y., Yan G. Y., He L. S. (2019). Comparative analysis of the gut microbial communities between two dominant amphipods from the challenger deep, Mariana trench. Deep Sea Res. Part I: Oceanographic Res. Papers 151, 103081. doi: 10.1016/j.dsr.2019.103081
Chevalier C., Stojanović O., Colin D. J., Suarez-Zamorano N., Trajkovski M. (2015). Gut microbiota orchestrates energy homeostasis during cold. Cell 163 (6), 1360–1374. doi: 10.1016/j.cell.2015.11.004
China fishery statistical yearbook (2021). China Fishery statistical yearbook (Beijing: China: Agriculture Press). Available at: http://www.stats.gov.cn/tjsj/ndsj/2021/indexeh.htm.
Clarke K. R. (1993). Non-parametric multivariate analyses of changes in community structure. Austral Ecol. 18 (1), 117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x
Dai W. F., Zhang J. J., Qiu Q. F., Chen J., Yang W. (2018). Starvation stress affects the interplay among shrimp gut microbiota, digestion and immune activities. Fish Shellfish Immunol. 80, 191–199. doi: 10.1016/j.fsi.2018.05.040
Deng Y., Cheng C. H., Xie J. W., Liu S. L., Ma H. L., Feng J., et al. (2019). Coupled changes of bacterial community and function in the gut of mud crab (Scylla paramamosain) in response to baimang disease. AMB Express 9 (1), 18. doi: 10.1186/s13568-019-0745-1
Desantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., et al. (2006). Greengenes, a chimera-checked 16s rrna gene database and workbench compatible with ARB. Am. Soc. Microbiol. 72 (7), 5069–5072. doi: 10.1128/AEM.03006-05
Douglas G. M., Maffei V. J., Zaneveld J. R., Yurgel S. N., Brown J. R., Taylor C. M., et al. (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688. doi: 10.1038/s41587-020-0548-6
Dufrêne M., Legendre P. (1997). Species assemblages and indicator species: The need for a flexible asymmetrical approach. Ecol. Monogr. 67 (3), 345–366. doi: 10.2307/2963459
Fabbiano S., Tarallo V., Hagemann S., Chevalier C., Stojanovic O., Montet X., et al. (2013). Gut microbiota orchestrates energy homeostasis during cold. Cell 163 (6), 1360–1374. doi: 10.1016/j.cell.2015.11.004
Fan Y., Yae Z., Jidong W., Yufan L., Ying Z., Yalun S., et al. (2018). Comparison of microbial diversity and composition in jejunum and colon of the alcohol-dependent rats. J. Microbiol. Biotechnol. 8 (11), 1883–1895. doi: 10.4014/jmb.1806.06050
FAO (2020) The state of world fisheries and aquaculture 2020. Available at: https://www.fao.org/publications/sofia/2020/en/.
Fassarella M., Blaak E. E., Penders J., Nauta A., Zoetendal E. G. (2021). Gut microbiome stability and resilience: elucidating the response to perturbations in order to modulate gut health. Gut 70 (3), 595–605. doi: 10.1136/gutjnl-2020-321747
Fietz K., Rye Hintze C. O., Skovrind M., Kjærgaard Nielsen T., Limborg M. T., Krag M. A., et al. (2018). Mind the gut: genomic insights to population divergence and gut microbial composition of two marine keystone species. Microbiome 6 (1), 82. doi: 10.1186/s40168-018-0467-7
Fraune S., Zimmer M. (2010). Host-specificity of environmentally transmitted mycoplasma-like isopod symbionts. Environ. Microbiol. 10 (10), 2497–2504. doi: 10.1111/j.1462-2920.2008.01672.x
Garrett W. S. (2019). Immune recognition of microbial metabolites. Nat. Rev. Immunol. 20 (2), 91–92. doi: 10.1038/s41577-019-0252-2
Hassan F. U., Guo Y., Li M., Tang Z., Yang C. (2021). Effect of methionine supplementation on rumen microbiota, fermentation, and amino acid metabolism in in vitro cultures containing nitrate. Microorganisms 9 (8), 1717. doi: 10.3390/microorganisms9081717
Heike S., Matthias S., Meinhard S., Thorsten B. (2010). Phylogeny of Proteobacteria and Bacteroidetes from oxic habitats of a tidal flat ecosystem. FEMS Microbiol. Ecol. 54 (3), 351–365. doi: 10.1016/j.femsec.2005.04.008
Henry L., Bruijning M., Forsberg S., Ayroles J. (2019). Can the microbiome influence host evolutionary trajectories? bioRxiv, 700237. doi: 10.1101/700237
Horváthová T., Kozłowski J., Bauchinger U. (2015). Growth rate and survival of terrestrial isopods is related to possibility to acquire symbionts. Eur. J. Soil Biol. 69, 52–56. doi: 10.1016/j.ejsobi.2015.05.003
Huang Z., Zeng S., Xiong J., Hou D., He J. (2020). Microecological koch's postulates reveal that intestinal microbiota dysbiosis contributes to shrimp white feces syndrome. Microbiome 8 (1), 32. doi: 10.1186/s40168-020-00802-3
Karlsson F. H., Ussery D. W., Nielsen J., Nookaew I. (2011). A closer look at bacteroides: Phylogenetic relationship and genomic implications of a life in the human gut. Microbial. Ecol. 61 (3), 473–485. doi: 10.1007/s00248-010-9796-1
Kim P. S., Shin N. R., Lee J. B., Kim M. S., Bae J. W. (2021). Host habitat is the major determinant of the gut microbiome of fish. Microbiome 9 (1), 166. doi: 10.1186/s40168-021-01113-x
Krautkramer K. A., Fan J., Backhed F. (2021). Gut microbial metabolites as multi-kingdom intermediates. Nat. Rev. Microbiol. 19 (2), 77–94. doi: 10.1038/s41579-020-0438-4
Krinos C. M., Coyne M. J., Weinacht K. G., Tzianabos A. O., Kasper D. L., Comstock L. E. (2001). Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414, 555–558. doi: 10.1038/35107092
Li G., Shishun L. I., Min L. I., Deliang L., Ding J., Chang Y., et al. (2018). Effects of dietary arachidonic acid on survival and growth performance and the intestinal bacterial profile of adult sea urchin (Strongylocentrotus intermedius). Chin. Fishery Qual. Standards.
Liang G., Qin K., Chen Y., Niu M., Wang H., Wang C., et al. (2022). Transcriptomic analysis of adaptive mechanisms in response to inland saline-alkaline water in the mud crab, Scylla paramamosain. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.974501
Lin W. C., He. Y. M., Shi C., Mu C. K., Wang C. L., Li R. H., et al. (2022). ATP catabolism and bacterial succession in postmortem tissues of mud crab (Scylla paramamosain) and their roles in freshness. Food Res. Int. 155, 110992. doi: 10.1016/j.foodres.2022.110992
Lozupone C. A., Knight R. (2007). Global patterns in bacterial diversity. PNAS 104 (27), 11436–11440. doi: 10.1073/pnas.0611525104
Martínez-Mota R., Kohl K. D., Orr T. J., Dearing M. (2019). Natural diets promote retention of the native gut microbiota in captive rodents. ISME J. 14 (5), 67–68. doi: 10.1038/s41396-019-0497-6
McFall-Ngai M., Hadfield M. G., Bosch T. C. G., Carey H. V., Domazet-Lošo T., Douglas A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. 110 (9), 3229–3236. doi: 10.1073/pnas.1218525110
Meng F., Gao H., Tang X., Wang A., Yao X., Liu C., et al. (2017). Biochemical composition of pond-cultured vs. wild gravid female mud crab Scylla paramamosain in hainan, china: evaluating the nutritional value of cultured mud crab. J. @ Shellfish Res. 36 (2), 445–452. doi: 10.2983/035.036.0216
Niu M., Gao G., Qin K., Chen Y., Wang H., Li X., et al. (2022). Multiple low salinity stress modes provided novel insight into the metabolic response of Scylla paramamosain adapting to inland saline-alkaline water. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.977599
Oren A. (2001). The bioenergetic basis for the decrease in metabolic diversity at increasing salt concentrations: implications for the functioning of salt lake ecosystems. Hydrobiologia 466, 61–72. doi: 10.1023/A:1014557116838
Parlapani F. F., Ferrocino I., Michailidou S., Argiriou A., Boziaris I. S. (2020). Microbiota and volatilome profile of fresh and chill-stored deepwater rose shrimp (Parapenaeus longirostris). Food Res. Int. 132, 109057. doi: 10.1016/j.foodres.2020.109057
Qi L., Lian C. A., Zhu F. C., Shi M., He L. S. (2022). Comparative analysis of intestinal microflora between two developmental stages of rimicaris kairei, a hydrothermal shrimp from the central indian ridge. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.802888
Regueiro L., Veiga P., Figueroa M., Alonso-Gutierrez J., Stams A., Lema J. M., et al. (2012). Relationship between microbial activity and microbial community structure in six full-scale anaerobic digesters. Microbiol. Res. 167 (10), 581–589. doi: 10.1016/j.micres.2012.06.002
Rennison D. J., Rudman S. M., Dolph S. (2019). Parallel changes in gut microbiome composition and function during colonization, local adaptation and ecological speciation. Proc. Biol. Sci. 286 (1916), 20191911. doi: 10.1098/rspb.2019.1911
Reyon D., Tsai S. Q., Khayter C., Foden J. A., Sander J. D., Joung J. K. (2012). FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 30 (5), 460–465. doi: 10.1038/nbt.2170
Ribeiro N. M., Puetz L., Pattinson N. B., Dálen L., Gilbert M. (2019). 31° south: The physiology of adaptation to arid conditions in a passerine bird. Mol. Ecol. 28 (16), 3709–3721. doi: 10.1111/mec.15176
Rognes T., Flouri T., Nichols B., Quince C., Mahé F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4 (10), e2584. doi: 10.7717/peerj.2584
Romano N., Zeng C. (2012). Osmoregulation in decapod crustaceans: implications to aquaculture productivity, methods for potential improvement and interactions with elevated ammonia exposure. Aquaculture 334-337 (7), 12–23. doi: 10.1016/j.aquaculture.2011.12.035
Rothman J. A., Leger L., Kirkwood J. S., Mcfrederick Q. S. (2019). Cadmium and selenate exposure affects the honey bee microbiome and metabolome, and bee-associated bacteria show potential for bioaccumulation. Appl. Environ. Microbiol. 85 (21), e01411–e01419. doi: 10.1128/AEM.01411-19
Rudi K., Angell I. L., Pope P. B., Vik J. O., Sandve S. R., Snipen L. G. (2017). Stable core gut microbiota across the freshwater-to-saltwater transition for farmed atlantic salmon. Appl. Environ. Microbiol. 84 (2), e01974–e01917. doi: 10.1128/AEM.01974-17
Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75 (23), 7537–7541. doi: 10.1128/AEM.01541-09
Singh P., Teal T. K., Marsh T. L., Tiedje J. M., Mosci R., Jernigan K., et al. (2015). Intestinal microbial communities associated with acute enteric infections and disease recovery. Microbiome 3 (1), 45. doi: 10.1186/s40168-015-0109-2
Smith C. C., Snowberg L. K., Caporaso J. G., Knight R., Bolnick D. I. (2015). Dietary input of microbes and host genetic variation shape among-population differences in stickleback gut microbiota. ISME J. 9, 2515–2526. doi: 10.1038/ismej.2015.64
Sossai P. (2012). Butyric acid: what is the future for this old substance? Swiss Med. Weekly 142, w13596. doi: 10.4414/smw.2012.13596
Sun Y., Han W., Liu J., Liu F., Cheng Y. (2020). Microbiota comparison in the intestine of juvenile Chinese mitten crab Eriocheir sinensis fed different diets. Aquaculture 515, 734518–734518. doi: 10.1016/j.aquaculture.2019.734518
Tigchelaar M., Leape J., Micheli F., Allison E. H., Basurto X., Bennett A., et al. (2022). The vital roles of blue foods in the global food system. Global Food Secur. 33, 100637. doi: 10.1016/j.gfs.2022.100637
Tu D.-d., Xu B.-p., Yan M.-c., Shu M.-a., Shao Q.-j. (2017). Molecular characterization of a cDNA encoding Na+/K+/2Cl(-) cotransporter in the gill of mud crab (Scylla paramamosain) during the molt cycle: Implication of its function in osmoregulation. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 203, 115–125. doi: 10.1016/j.cbpa.2016.08.019
Wang Q. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73 (16), 5261–5267. doi: 10.1128/AEM.00062-07
Wang H., Chen W., Qi C., Dong W., Hu X. (2016). Carboxylicivirga flava sp. nov., a novel bacterium isolated from surface sediment of the bohai Sea, China. Int. J. Systematic Evolutionary Microbiol. 66 (12), 5412–5416. doi: 10.1099/ijsem.0.001533
Wang H., Lei T., Hongling W., Junkai L., Changkao M., Chunlin W. (2018b). Transcriptomic analysis of adaptive mechanisms in response to sudden salinity drop in the mud crab, Scylla paramamosain. BMC Genomics 19 (1), 421. doi: 10.1186/s12864-018-4803-x
Wang H., Wei H. L., Tang L., Lu J. K., Mu C. K., Wang C. L. (2018a). A proteomics of gills approach to understanding salinity adaptation of Scylla paramamosain. Gene 677, 119–131. doi: 10.1016/j.gene.2018.07.059
Wang H., Wei H., Tang L., Lu J., Mu C., Wang C. (2019). Gene identification and characterization of correlations for deps_degs same trend responding to salinity adaptation in Scylla paramamosain. Int. J. Genomics 2019, 7940405. doi: 10.1155/2019/7940405
Wei H., Wang H., Tang L., Mu C., Ye C., Chen L., et al. (2019). High-throughput sequencing reveals the core gut microbiota of the mud crab (Scylla paramamosain) in different coastal regions of southern China. BMC Genomics 20, 829. doi: 10.1186/s12864-019-6219-7
Wexler H. M. (2007). Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20 (4), 593–621. doi: 10.1128/CMR.00008-07
Xiao L., Zheng H., Li J., Zeng M., He D., Liang J., et al. (2022). Targeting NLRP3 inflammasome modulates gut microbiota, attenuates corticospinal tract injury and ameliorates neurobehavioral deficits after intracerebral hemorrhage in mice. Biomed. Pharmacother. 149, 112797. doi: 10.1016/j.biopha.2022.112797
Xiong J. B., Nie L., Chen J. (2019b). Current understanding on the roles of gut microbiota in fish disease and immunity. Zoological Res. 40 (2), 70–76. doi: 10.24272/j.issn.2095-8137.2018.069
Xiong J., Xuan L., Yu W., Zhu J., Qiu Q., Chen J. (2019). Spatiotemporal successions of shrimp gut microbial colonization: high consistency despite distinct species pool. Environ. Microbiol. 21 (4), 1383–1394. doi: 10.1111/1462-2920.14578
Yang S. H., Seo H. S., Woo J. H., Oh H. M., Jang H., Lee J. H., et al. (2014). Carboxylicivirga gen. nov. in the family Marinilabiliaceae with two novel species, carboxylicivirga mesophila sp. nov. and Carboxylicivirga taeanensis sp. nov., and reclassification of Cytophaga fermentans as Saccharicrinis fermentans gen. nov., comb. nov. Int. J. Systematic Evolutionary Microbiol. 64 (Pt 4), 1351–1358. doi: 10.1099/ijs.0.053462-0
Keywords: Scylla paramamosain, intestinal microbiota, low salinity, saline-alkaline water, probiotic
Citation: Niu M, Li X, Chen Y, Qin K, Liang G, Hu Y, Jiang X, Wang H, Zhu R, Wang C and Mu C (2023) Response of intestinal microbiota to saline-alkaline water in mud crab (Scylla paramamosain) based on multiple low salinity culture modes. Front. Mar. Sci. 10:1153326. doi: 10.3389/fmars.2023.1153326
Received: 29 January 2023; Accepted: 13 March 2023;
Published: 22 March 2023.
Edited by:
Hongyu Ma, Shantou University, ChinaReviewed by:
Yafei Duan, South China Sea Fisheries Research Institute, ChinaNgoc Tuan Tran, Shantou University, China
Copyright © 2023 Niu, Li, Chen, Qin, Liang, Hu, Jiang, Wang, Zhu, Wang and Mu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huan Wang, d2FuZ2h1YW4xQG5idS5lZHUuY24=; Ritong Zhu, eWFuamluc2h1aWNoYW5AMTYzLmNvbQ==
 Mingming Niu1
Mingming Niu1 Huan Wang
Huan Wang Chunlin Wang
Chunlin Wang Changkao Mu
Changkao Mu