- 1Department of Health Management, Atlantic Veterinary College, University of Prince Edward Island, Charlottetown, PE, Canada
- 2Department of Animal Science and Aquaculture, Faculty of Agriculture, Dalhousie University, Bible Hill, NS, Canada
Introduction: The native Atlantic rock crab (Cancer irrotatus) and the invasive European green crab (Carcinus maenas) are commercially and ecologically important crustacean species in Atlantic Canada. The importance of microbiomes for host health and ecology has been recognized in many species, although very few studies have focused on crustaceans or their external shell microbiome. This is the first-ever study to characterize and analyze the microbial communities associated with the external carapace of C. irrotatus and C. maenas.
Methods: Microbiome samples were collected from three locations in Atlantic Canada, processed using standard 16S Illumina MiSeq PE250 sequencing and analyzed with the open-access QIIME2 software.
Results: Taxonomic classification of the microbial compositions, as well as alpha- and beta diversities, reveal that the shell microbiome differs by host species between C. irrotatus and C. maenas sampled from the same location and between C. irrotatus sampled from different locations. Interestingly, the differences are greater between species at the same location than between locations for the same species.
Discussion: These are the first-ever results showing that the crustacean shell microbiome not only depends on geographical factors but also on intrinsic factors specific to the host species. This implies that crustaceans exert some impact on their shell microbiome, potentially selecting beneficial taxa. These are important findings that could elucidate contributing factors of crustacean shell diseases that are still poorly understood.
1 Introduction
The native Atlantic rock crab (Cancer irrotatus) and the invasive European green crab (Carcinus maenas) are common crustacean species in Atlantic Canada that share similar near-shore habitats (Bélair and Miron, 2009). Rock crabs are an important food source for the commercially valuable American lobster Homarus americanus (Gendron et al., 2001; Fisheries and Oceans, 2022). The green crab was first reported in eastern Canadian waters in the 1950s in southern Nova Scotia and has since then spread throughout Atlantic Canada. Due to its tolerance for extremes of salinity, temperature and emersion and its aggressive nature, this species can disrupt marine ecosystems by predation on the local benthic fauna or by destroying nursery ground for juvenile fishes (Fisheries and Oceans, 2011; Matheson et al., 2016). This and their competition with native crustacean species (e.g. rock crab and lobster) for food or habitat makes green crabs an ecologically and economically high-risk species. For example, green crabs have been shown to prey on newly settled American lobster and adult lobsters are less likely to enter traps if green crabs are present (Sigurdsson and Rochette, 2013; Rayner and McGaw, 2019).
It is widely recognized that host-microbiome interactions play significant roles in host health and physiology. Interactions between a host and its microbial communities are intricate and range from beneficial to adverse (Wahl et al., 2012; Egan and Gardiner, 2016). Research in humans has suggested an association between microbiome changes and several chronic diseases such as obesity, diabetes or Crohn’s disease (Clemente et al., 2012; Benahmed et al., 2021; Kwan et al., 2022). Unlike in human research, microbiomes of marine animals remain largely underexplored. On the other hand, marine diseases are increasingly being reported indicating they are becoming more frequent and prevalent (Harvell et al., 2002; Lafferty et al., 2004; Egan and Gardiner, 2016; Groner et al., 2016). Knowledge gaps in aquatic microbial communities associated with marine animals inhibit our ability to understand, manage and limit associated disease outbreaks; especially in the context of climate change (Groner et al., 2016; Groner et al., 2018).
A baseline understanding of the microbial composition in healthy hosts is necessary to detect changes in the microbiome and their potential significance for host ecology. Previous microbiome research in crustaceans has primarily focused on the gut, hemolymph and shell (Feinmann et al., 2017; Holt et al., 2020a; Jung et al., 2021).The microbial community associated with healthy lobster (H. americanus) carapaces mainly consists of Rhodobacteraceae (Alphaproteobacteria), Thiotrichaceae (Gammaproteobacteria) and Bacteroidetes (Chistoserdov et al., 2012; Feinmann et al., 2017) whereas, in healthy brown crab Cancer pagurus Acidobacteria, Actinobacteria, Bacteroidetes, Proteobacteria and Cyanobacteria make up most of the shell microbiome (Kraemer et al., 2020). It has also been shown that the microbial community and composition change according to the developmental stages of the host, environmental conditions or disease in shrimp and lobster. (Meres et al., 2012; Cheung et al., 2015; Ooi et al., 2019; Holt et al., 2020b). The shell microbiome has gotten increased attention in the past few decades due to the rise in shell disease outbreaks in American lobsters on the U.S./Canadian east coast (Smolowitz et al., 2005; Quinn et al., 2013). Dysbiosis is a negative shift in the e.g. shell microbiome from a diverse and abundant bacterial community to a less diverse community dominated by few potentially pathogenic species (Egan and Gardiner, 2016: Mousa et al., 2022). This has been hypothesized as a key factor for disease proliferation including shell disease (Meres et al., 2012; Meres, 2016; Wynne et al., 2020). A healthy and functioning microbiome is generally considered beneficial to its hosts due to its resilience against invading pathogens and its molecular functional potential to maintain a stable host-associated ecology (Lloyd-Price et al., 2016; Rajeev et al., 2021). Accordingly, insight into the shell microbiome and factors that influence its diversity and composition is crucial for ecologically important species. So far, there have been no microbiome studies for either rock or green crabs; constituting a significant knowledge gap, especially regarding microbiome-associated diseases in species that share their habitat.
Recent advances in affordable, fast and accurate culture-independent analyses of bacterial communities associated with terrestrial and marine organisms (Lederberg and McCray, 2001; Knight et al., 2018; Petersen and Osvatic, 2018) allowed us to describe the shell microbial community of apparently healthy Atlantic rock crab and European green crab for the first time in Atlantic Canada. Our goal is to generate baseline microbiome data in the region for these species and to determine the impact that species and location have on the carapace microbiome of these crabs.
2 Materials and methods
2.1 Sample collection
Sampling was conducted in the summer and fall of 2018. Atlantic rock crabs (C. irrotatus) were sampled from traps in three locations: Charlo (New Brunswick; latitude: 47.9922, longitude: -66.2658) in the western Gulf of St. Lawrence, Malagash (Nova Scotia; 45.7747, -63.2956) in the central Northumberland Straight and Tracadie (Prince Edward Island; 46.4033, -63.0287) in the southern Gulf of St. Lawrence, whereas the European green crabs (C. maenas) were only present in traps at the Malagash site (Nova Scotia) which enabled a single sampling of this species there. Rock and green crabs in Malagash were sampled during the same sampling event. All the locations were sampled during one sampling event. All crabs were sampled in <4m water depth, but no data on sea bottom temperature was collected during the survey. The carapace of apparently healthy animals (no visible signs of shell disease) was rinsed with 5 mL of molecular biology-grade water filtered through a 0.22 µm filter. A sterile cotton swab was then dipped in buffer A (20 mM Tris-HCl, 2 mM sodium EDTA, 1.2% Triton X-100, pH 8.0) (Birer et al., 2017) and used to swab the rinsed area of the carapace. Seawater control samples were collected by submerging a sterile cotton swab in seawater at the respective sampling locations. Swabs were placed in 100% ethanol immediately and stored in -20°C until further processing.
2.2 DNA extraction, 16S-amplification and sequencing
Bacterial DNA from the collected swab samples was extracted using a 96-well plate extraction protocol modified from Ivanova et al. (2006). Briefly, 50 µL of 1 mg/mL lysozyme in a lysozyme reaction mix (50 mM NaCl, 4 mM EDTA, 30 mM Tris-HCl, pH 8.0) was added to each swab tip in a sterile well of a 96-well plate, sealed with sealing film and incubated at 37°C for 30 min, then the plates were unsealed so 50 µL of lysis mix (1.4 M GuSCN, 60 mM EDTA, 60 mM Tris-HCl, 1% Triton X-100, 10% Tween 20, pH 8.0) containing 1.8 mg/mL proteinase K, resealed and incubated overnight at 56°C. Plates were centrifuged at 1500 x g for two min and unsealed and 150 µL of a binding mix (6 M GuSCN, 20 mM EDTA, 10 mM Tris-HCl, 4% Triton X-100, pH 6.4) was added to the swab and solution. The lysate was removed and placed in a corresponding well of a glass fibre plate (AcroPrep™ 96 filter plate; 3.0 µm GF/0.2 µm BioInert, NTRL), the plate was sealed and centrifuged at 4500 x g for 7 min. The eluate was discarded and 180 µL of protein wash buffer (1.56 M GuSCN, 5.2 mM EDTA, 2.6 mM Tris-HCl, 1% Triton X-100, 67.2% EtOH) was added, the plate sealed and was centrifuged again at 4500 x g for 5 min. The eluate was discarded, and the well was washed twice more with 750 µL wash buffer (60% EtOH, 50 mM NaCl 10 mM Tris-HCl, 0.5 mM EDTA, pH 7.4) at 4500 x g for 5 min. After the eluate was discarded, the unsealed plate was placed in a 37°C incubator and covered with a sterile pipet box lid for 10 min to evaporate residual ethanol. DNA was eluted using 50 µL of pre-heated molecular biology grade water (56°C) in each well incubating at room temperature and centrifuging at 4500 x g for 5 min and collecting the eluate in a sterile 96 well collection plate. Samples were stored by adding 5.5 µL of 10X TE buffer (100 mM Tris-HCl, 1 mM EDTA, pH 8.0) to each sample, sealing the plate and storing at -20°C until use.
Extracted DNA samples were sequenced at Gloucester Marine Genomics Institute (GMGI) where they were amplified prior to sequencing with a two-step PCR protocol using the primers 515F (5’– GTGYCAGCMGCCGCGGTAA–3’) and 806R-B (5’–GGACTACNVGGGTWTCTAAT–3’) specific to the 16S rRNA V4 region (Apprill et al., 2015; Parada et al., 2015). The amplicon size was confirmed on a Fragment Analyzer and paired-end 250 base-pair sequencing was performed on an Illumina MiSeq, version 2 kit.
2.3 Bioinformatic analysis
All bioinformatics (trimming, denoising, alignment) were performed using QIIME2 on the open-source Galaxy interface run through Docker on Windows (https://github.com/qiime2/q2galaxy) (Afgan et al., 2018; Bolyen et al., 2019). The demultiplexed paired-end forward and reverse sequences were truncated (forward reads at 250 bp, reverse reads at 187 bp) based on the quality profiles and the 515F/806R primer sequences were trimmed off (forward reads at 52 bp, reverse reads 49 bp). Reads were then filtered, denoised (error correction) and merged using the DADA2 pipeline implemented in QIIME2, which also removed chimeric sequences. The taxonomy of the resulting amplicon sequence variants (ASVs) was assigned with the 16S Greengenes v.138.99 trained classifier (McDonald et al., 2011).
2.4 Statistical analysis
To visualize the most important microbial taxa in the microbial dataset, the relative abundance was calculated (Table S1). The 20 most frequent ASVs and bacterial classes were displayed using bar plots. The dataset was rarefied to a read depth of 10,000 with the rarefy tool in QIIME2, which retained 18% of features in all samples and rarefaction curves were used to verify that the number of observed ASVs level out at that read depth in all samples (Figure S1). For each sample, three alpha diversity indices were estimated in QIIME2: estimated richness (Chao1), the Pielou evenness and the Shannon diversity. For each of these numerical diversity indices, linear regression models were fitted with crab sex, size and either location (when comparing rock crabs from Charlo NB, Malagash NS and Tracadie NS) or species (when comparing green and rock crabs from Malagash NS) as fixed effects explanatory variables. These models were fit using Stata (v. 17; StataCorp, 2021: https://stata.com) and the significance level for all analyses was set at 0.05. Beta diversity was calculated and visualized using emperor plots in QIIME2. Bray-Curtis, weighted and unweighted UniFrac distances between the samples were calculated based on the dissimilarity of their microbial composition (Lozupone et al., 2007). Principal Coordinate Analyses (PCoA) were applied to multivariate distance matrices to extract the dimensions that account for the maximum distances enabling a map-based visualization in the QIIME2 emperor plot tool. To test for significant differences in the microbial composition between location- (rock crabs) and species (rock and green crab) PERMANOVAs (permutational multivariate analysis of variance) with 999 permutations were performed in QIIME2 (Anderson, 2017).
3 Results
3.1 Descriptives
Table 1 summarizes the number, mean size and sex ratio of sampled rock and green crabs for each sampled location. Summary statistics on the number of raw, quality filtered, merged and chimeric reads are presented in Supplementary Table S1. The quality filtering removed 12% of paired reads; on average, 81% of reads were successfully merged. The DADA2 algorithm also removed chimeric reads which were 20% of all reads. In total, 20,489 ASVs (amplicon sequence variants) were created, of which 18,846 were taxonomically assigned to at least the phylum level, 16,997 to class level, 14,162, to order level, 9,577 to family level, 3,420 to genus level and 673 down to species level.
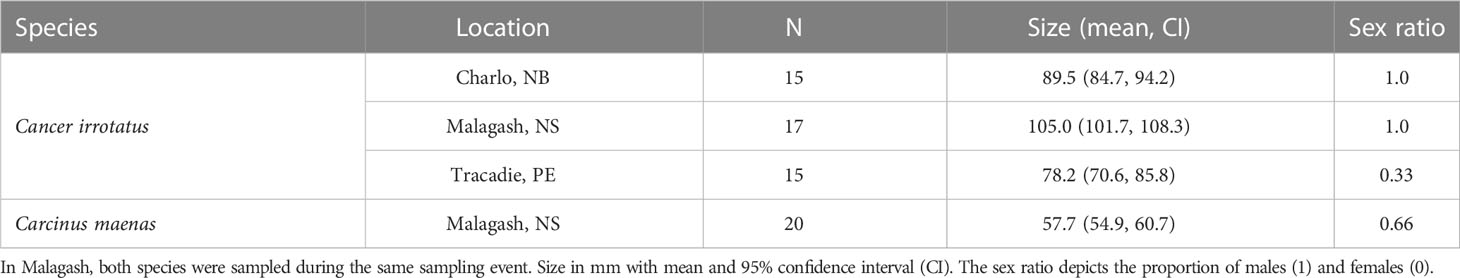
Table 1 Sampling details and biological parameters of sampled rock crab (C. irrotatus) and green crab (C. maenas).
3.2 Microbial community
The three most abundant bacterial classes across all samples were Gammaproteobacteria, Alphaproteobacteria and Flavobacteria. Together they made up around 60% of the observed microbiome. Their proportions were similar across sampling locations and host species. As only the 20 most common bacterial classes and taxa were presented in the taxonomic bar graphs (Figures 1, 2), their relative abundance did not sum up to 100% and the less common taxa were represented by white space. There were some differences in the less frequent bacterial classes between the three sampling locations for rock crabs, and between the two host species in Malagash NS (Figure 1). The shell microbiome of rock crabs from Malagash had a higher relative abundance of Verrucomicrobiae (4.9%) compared to animals from Charlo (2.1%) and Tracadie (1.4%). In Tracadie, the rock crab shell microbiome had a high relative abundance of Deltaproteobacteria (8.8%) and Epsiolonproteobacteria (2.6%), while these classes were not as common in Charlo (2.9%, 0.2%) and Malagash (1.8%, 0.2%). As for inter-species differences in the shell microbial composition, green crabs from Malagash hosted more Epsilonproteobacteria (5.9%), but fewer Verrucomicrobiae (1.9%) and Oscillatoriophycidaea (0.9%) than rock crabs from the same location (0.2%, 4.9%, 4.0%) (Table S1). Figure 2 displays the relative abundances of the 20 most abundant microbial taxa of which six were assigned to genus level (Aquimarina, Cocleimonas, Loktanella, Maribacter, Psychrobacter, Winogradskyella). Aquimarina and Loktanella had the highest relative abundance in C. irrotatus from Charlo (1.5%, 5.7%) and Malagash (2.0%, 4.2%) compared to Tracadie (0.5%, 1.5%), whereas Cocleimonas had a much higher relative abundance in Tracadie (8.0%) compared to the other two sampling locations. When comparing the most common genera between C. irrotatus and C. maenas from Malagash, C. maenas hosted a higher abundance of Cocleimonas (9.7% vs. 1.7%) but a lower abundance of Loktanella (0.2% vs. 4.2%), Maribacter (0.3% vs. 1.8%) and Winogradskyella (1.0% vs. 2.1%).
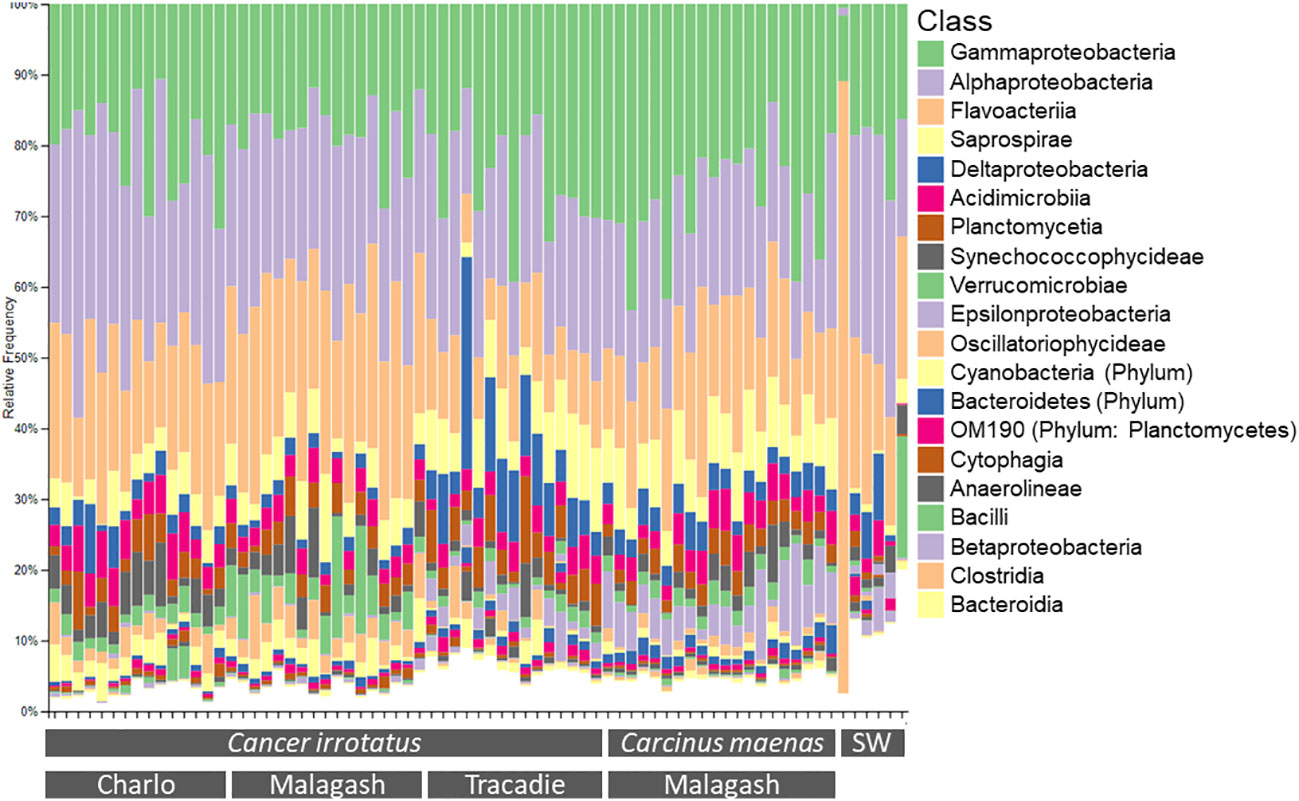
Figure 1 Relative abundance of the twenty most abundant bacterial classes of the 16S community on the shell of each of the sampled rock (C. irrotatus) and green crabs (C. maenas) and in the seawater controls (SW) from the three sampling locations.
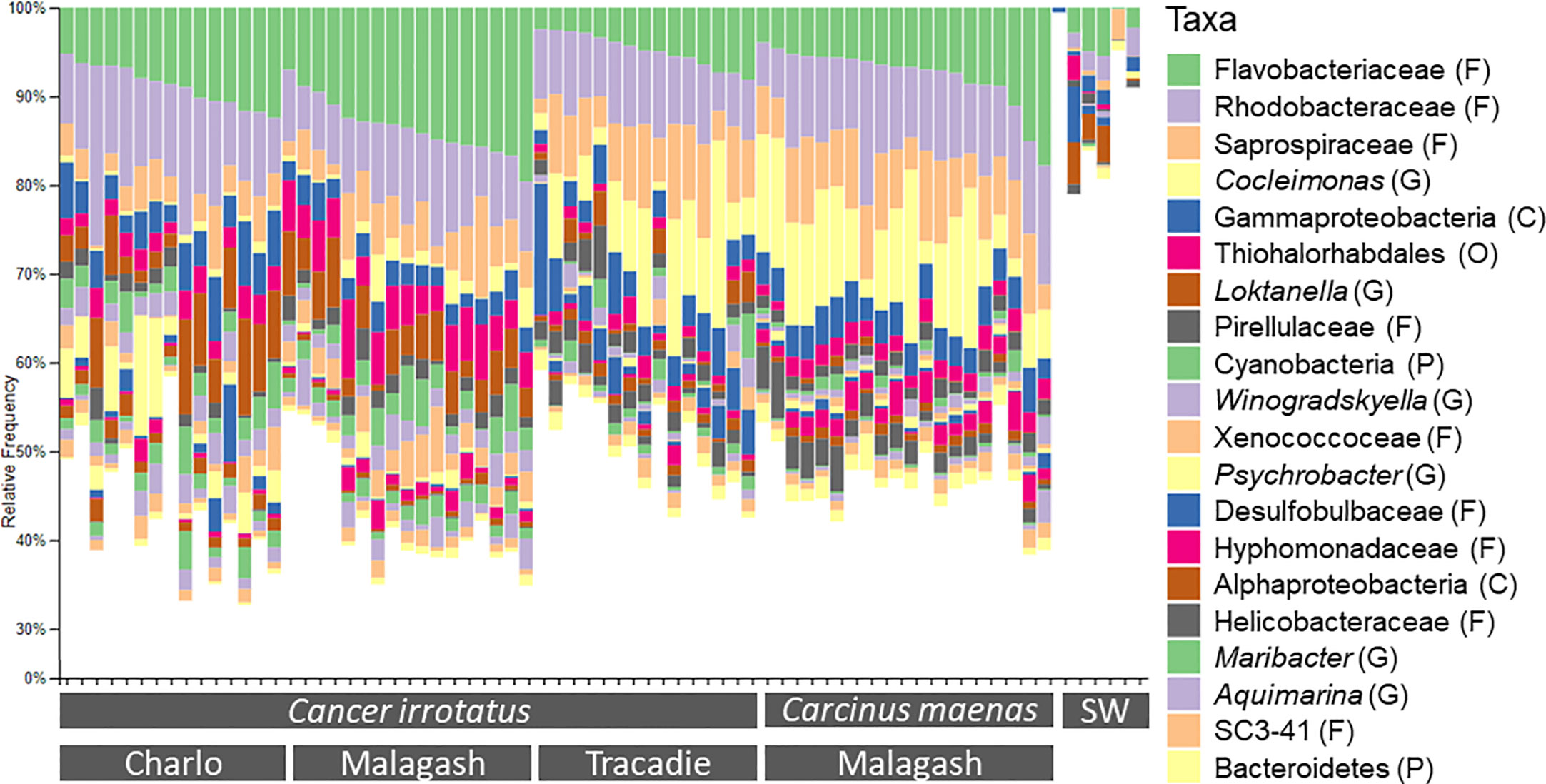
Figure 2 Relative abundance of the twenty most abundant bacterial taxa of the 16S community on the shell rock (C. irrotatus) and green crabs (C. maenas) and seawater controls (SW) from the three sampling locations. The presented taxa were assigned to the lowest possible taxonomic level and taxonomic ranks were indicated as: P, phylum; C, class; O, order; F, family; G, genus.
On class level, the microbial community in seawater controls consisted mainly of the same bacterial classes as the shell samples, for example Alpha- and Gammaprotebacteria and Flavobacteriia but with different relative abundances. On the lowest taxonomic level, the microbial composition of the sea water does not resemble that of the shell samples as only a small proportion of seawater microbial community (<20%) belongs to the twenty overall most common bacterial taxa of the shell samples (Figure 2).
3.3 Alpha diversity
Differences in microbiome richness, evenness and diversity between location and host species were assessed by linear regression models. The estimated richness and Shannon diversity, were significantly different between the sampling locations for rock crabs (p = 0.001, p = 0.000) and between rock and green crabs in Malagash (p = 0.019, p = 0.003). For the Pielou evenness, there was a significant difference between sampling locations (p = 0.005), but not between host species (p = 0.066). The model parameters are summarized in Tables 2, 3.
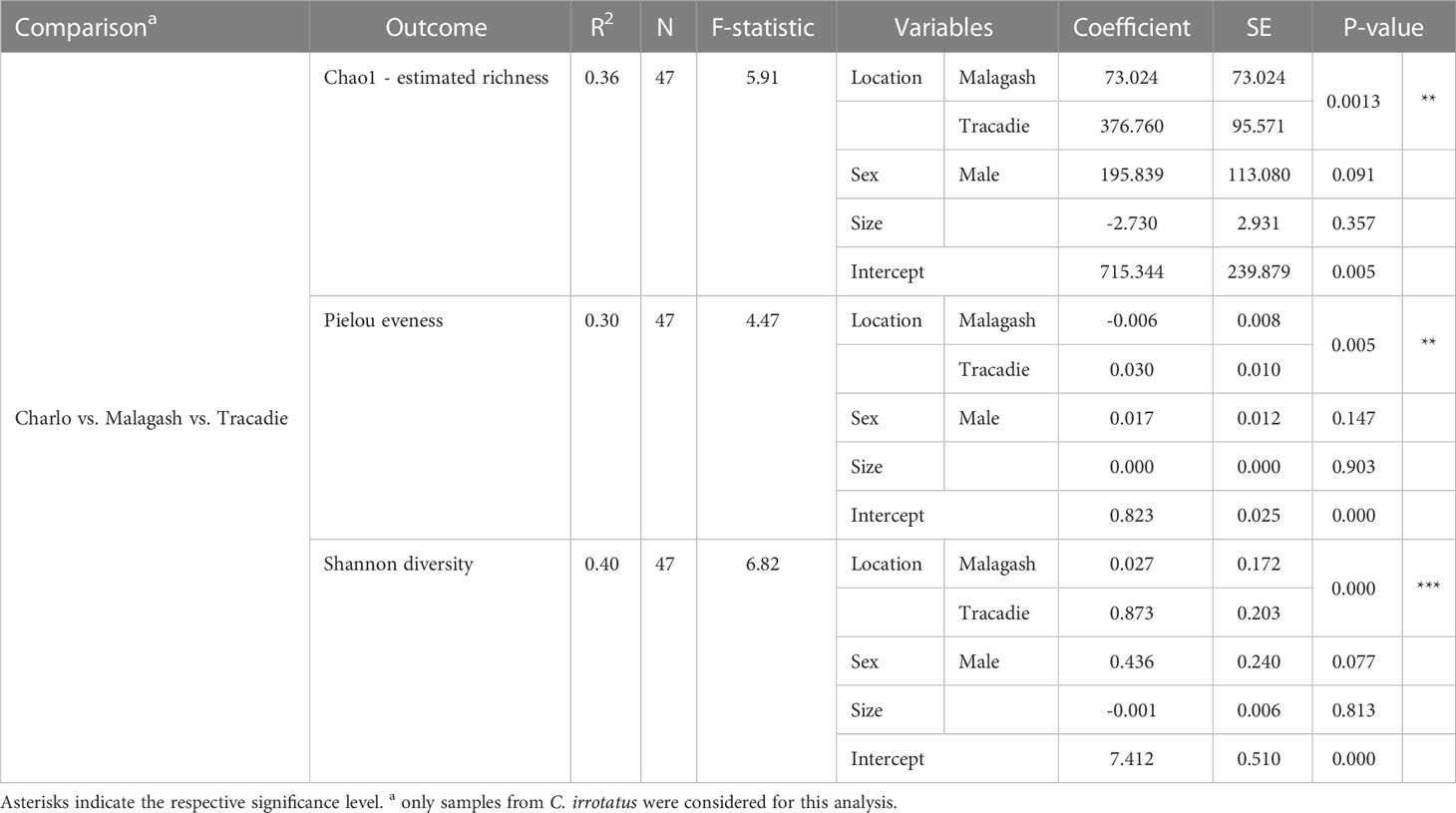
Table 2 The parameters and outputs from linear multivariable models evaluating the location effect on three diversity measures for the shell microbiome of rock crabs (C. irrotatus).
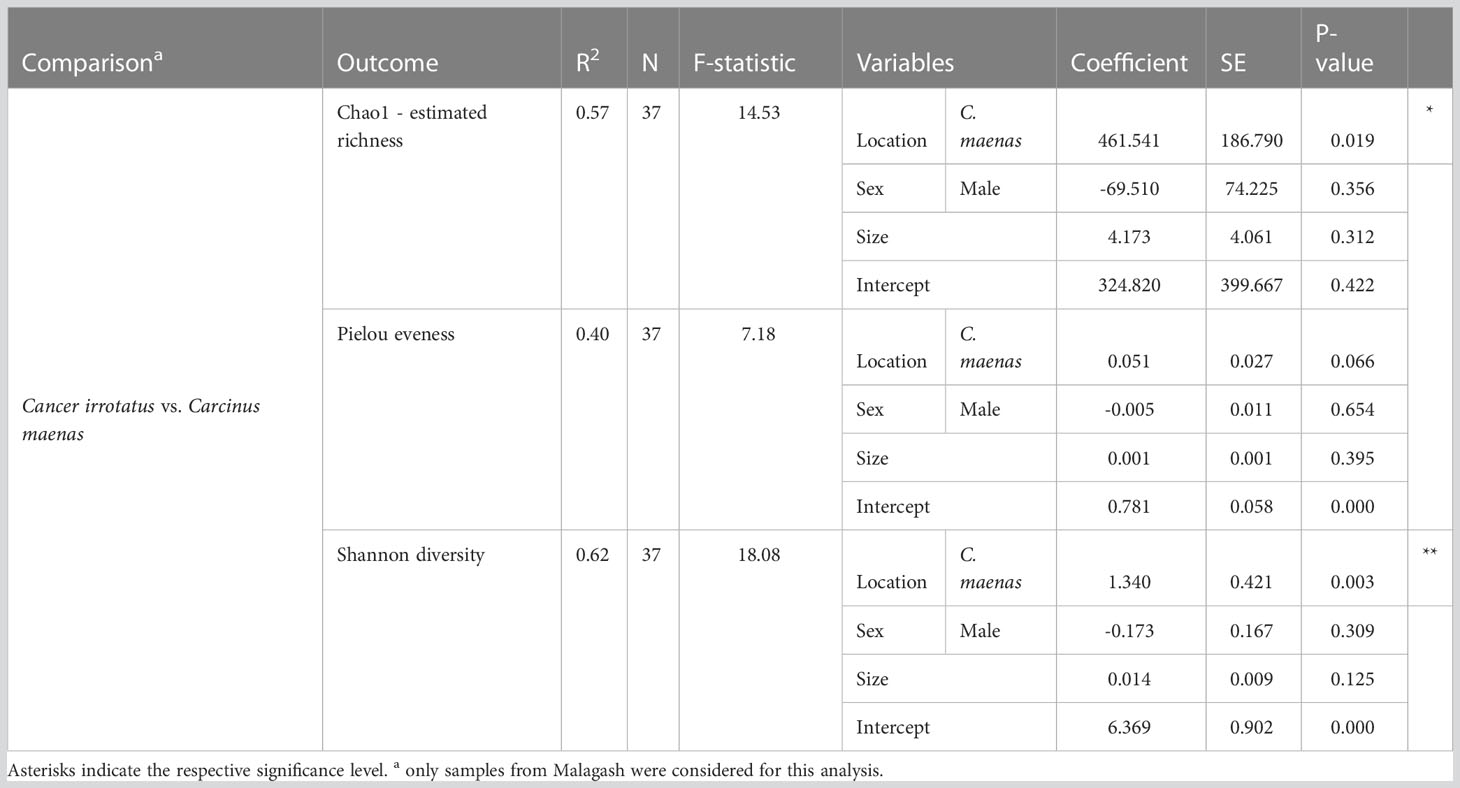
Table 3 The parameters and outputs from the multivariable linear models evaluating the host effect on three diversity measures in the shell microbiome of rock crabs (C. irrotatus) and green crabs (C. maenas) sampled in Malagash.
When looking at the shell microbiome of rock crabs, Figure 3 shows that the estimated species richness, Pielou evenness as well as Shannon diversity, were higher in Tracadie than in Charlo and Malagash. For the comparison between green and rock crabs sampled in Malagash, the shell microbiome of green crab was richer, more diverse, and more evenly distributed than the one from the native rock crab (Figure 4). The microbial diversity and richness of the seawater controls were significantly lower than of samples taken from crab shells but there was no significant difference observed when it came to the evenness of the microbial community.
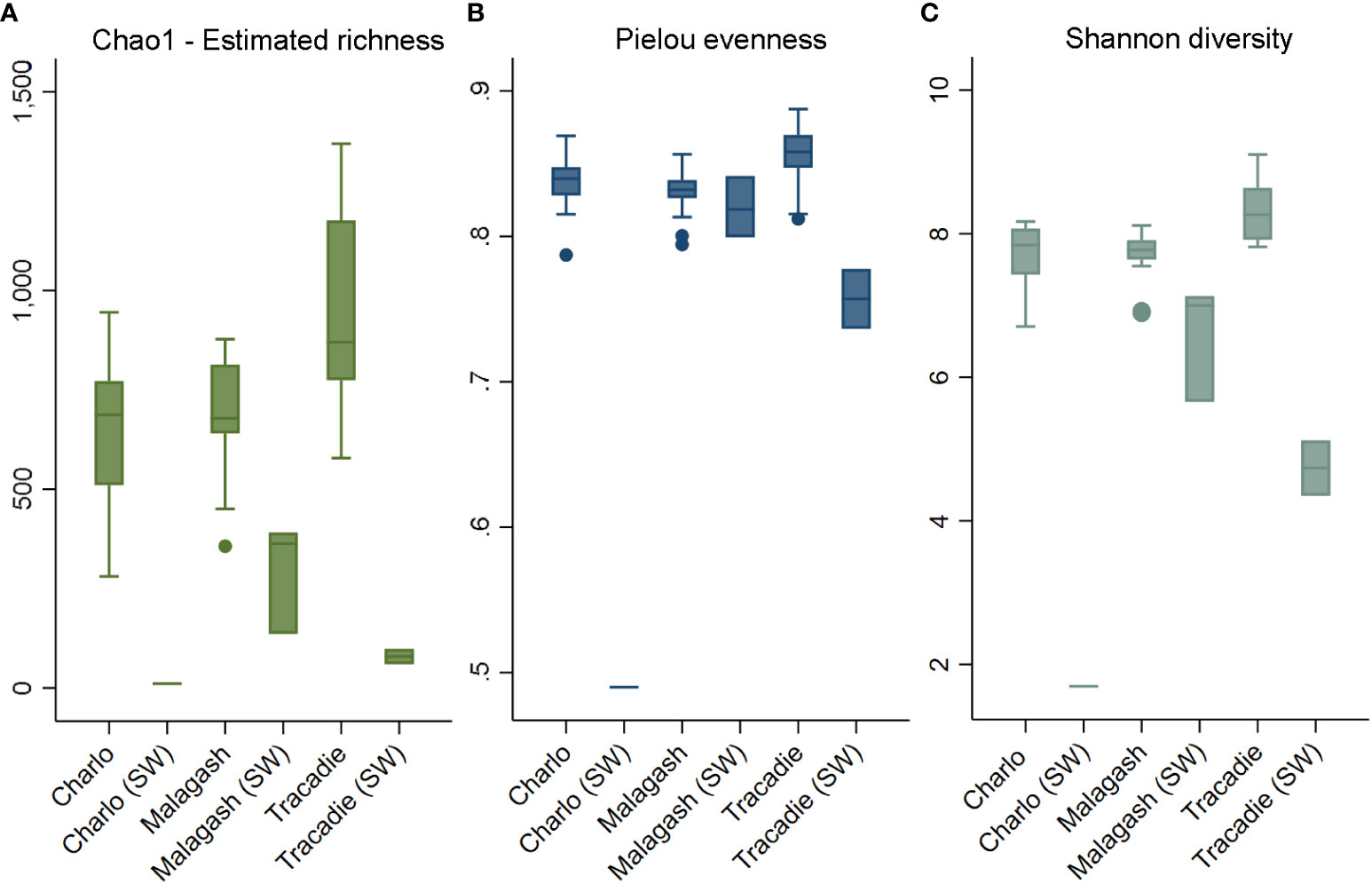
Figure 3 Alpha diversity indices of shell microbiome from rock crab (C. irrotatus) sampled in three locations in Atlantic Canada next to the respective seawater control (SW) from that location. Chao1 estimated species richness (A), Pielou’s index of species evenness (B), and Shannon’s index of diversity (C).
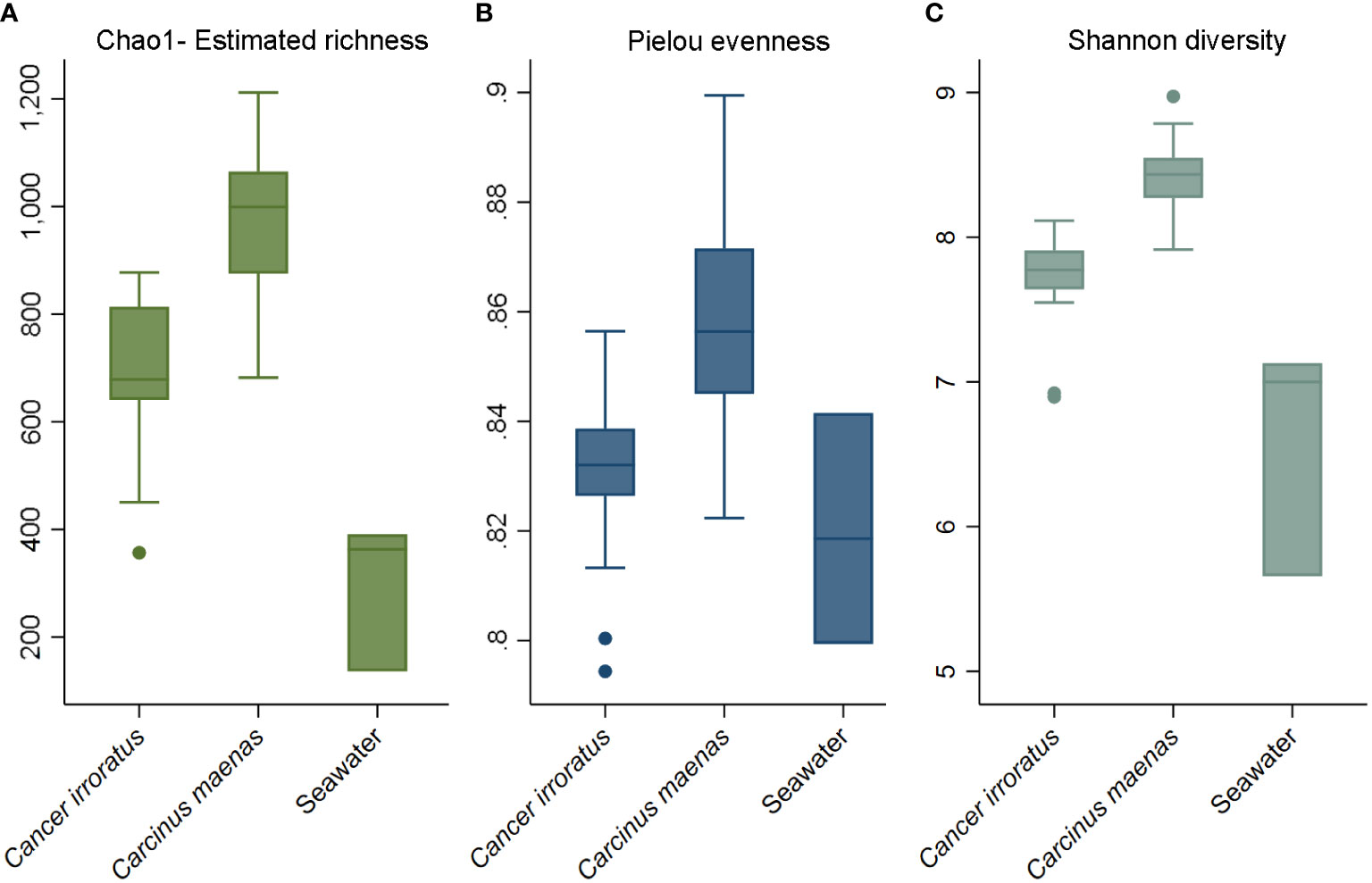
Figure 4 Alpha diversity indices of shell microbiome from rock crab (C. irrotatus), green crab (C. maenas) and seawater control (SW) sampled in Malagash, NS. Chao1 estimated species richness (A), Pielou’s index of species evenness (B), and Shannon’s index of diversity (C).
3.4 Beta diversity
The Bray-Curtis, weighted and unweighted UniFrac distances are presented in three respective ordination plots (Figure 5). Bray-Curtis ordination shows that the microbiome samples formed clusters both by sampling location (shapes) and by host species (color), which is concordant to the weighted and unweighted UniFrac metrics. The shell microbial composition of green crab from Malagash clustered distinctly from the shell microbiome of rock crab from the same location. But, as seen in the previous analyses, distinct clustering was also present when looking at the shell microbial composition in different locations. The PERMANOVA analysis for all distances showed significant group differences by location (rock crab only) and host species (Malagash only) (Figure 5).
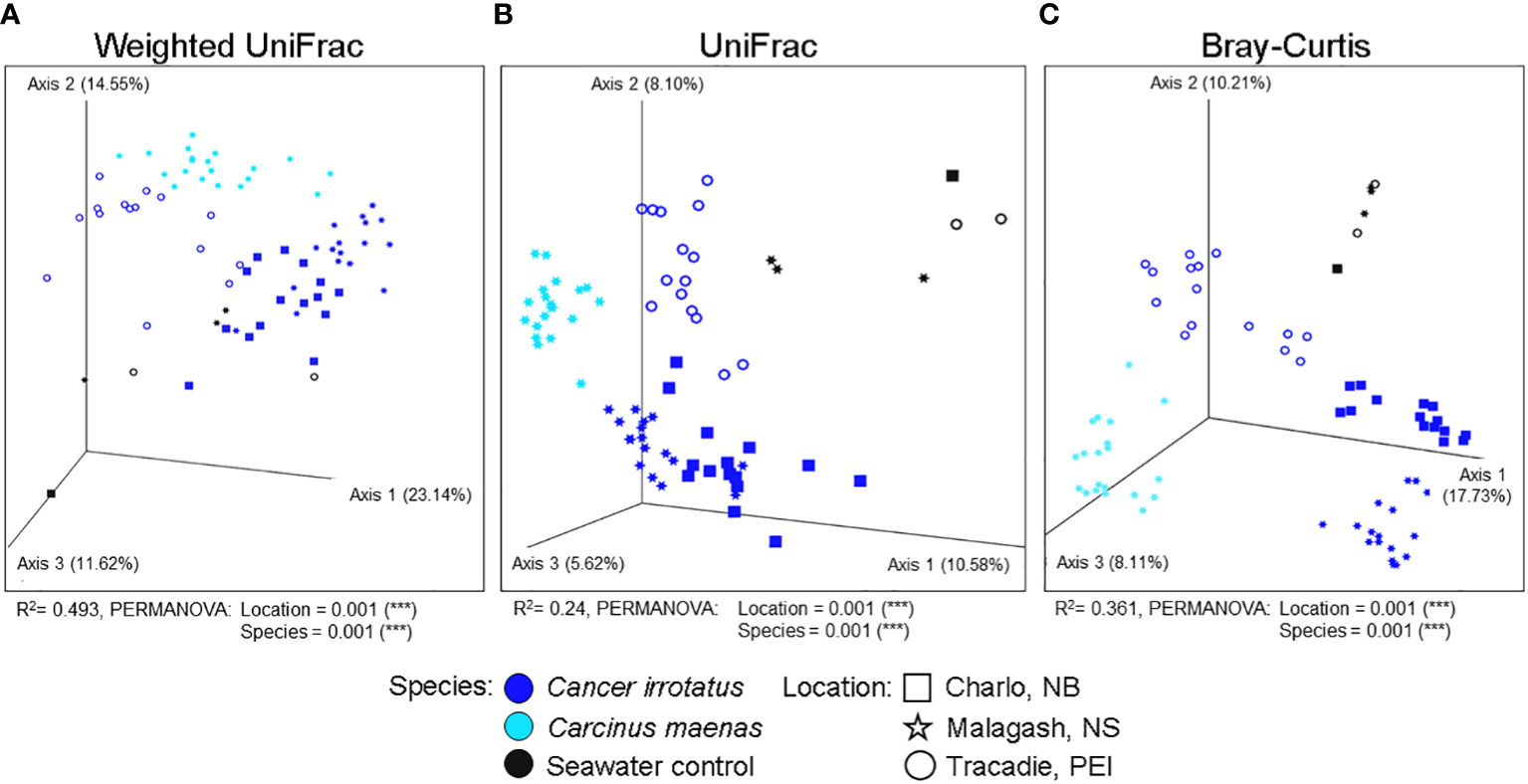
Figure 5 Visualization of shell microbial beta diversity in three-dimensional non-metric dimensional scaling (NMDS) plots by species (color) and sampling location (shape) including seawater controls based on (A) a weighted UniFrac, (B) an unweighted UniFrac and (C) a Bray-Curtis dissimilarity. Values in brackets indicate the proportion of explained variation by the respective axis.
4 Discussion
We present the first characterization of the shell microbiome of apparently healthy rock (C. irrotatus) and green (C. maenas) crabs in Canadian waters. Our results show that of the 20 most common bacterial taxa making up the crab shell microbiome, 15 have not yet been classified at the genus level by previous studies, hence demonstrating the substantial gap in our understanding and recording of marine microbes. This study provides important novel insights into the microbial populations on brachyuran carapaces and demonstrates that the microbial composition differs by both the sampling location and host species, expanding from the traditional view that surface microbiomes are governed mainly by their environment.
Overall, the microbiome of the seawater controls featured similar bacterial taxa as were observed in the carapace swab samples, but had a lower relative abundance of these taxa, as well as a lower microbial diversity, richness, and evenness. Taken together with the ordination plots, where seawater microbiome samples clustered apart from the shell microbiome samples, it is sensible to assume that the surface microbiome of the carapace distinctly differs from what is present in the surrounding seawater in terms of species composition. Some bacterial species may lack the ability to live in the water column and need a surface to colonize, or vice versa, which leads to differences in the microbiome between swabs and control samples.
So far, lobsters (H. americanus) have been the main species of focus regarding shell microbial research in the northwestern Atlantic (Chistoserdov et al., 2012; Meres et al., 2012; Quinn et al., 2013). The bacterial classes that were found, with high frequencies of Alpha- and Gammaproteobacteria, on rock and green crabs were also identified on wild U.S. and Canadian lobster carapaces in culture-independent high throughput (Meres et al., 2012) and medium throughput (Chistoserdov et al., 2012; Quinn et al., 2013) approaches. Here, two of the most common bacterial genera on rock and green crab were Aquimarina and Maribacter (Flavobacteriia) which have been associated with shell disease in lobsters from shell disease-affected regions in southern New England (Chistoserdov et al., 2012; Meres et al., 2012). Isolating these taxa from other apparently healthy crustacean species in Atlantic Canada highlights the importance of microbial monitoring to determine risk factors for shell disease occurrence in local crustacean populations. In both humans and crustaceans, certain bacteria in the gut and hemolymph microbiome have been linked to higher immunity, survival and resistance against pathogens (Daniels et al., 2010; Lawley and Walker, 2012; Wang and Wang, 2015). Although evidence is still scarce, the composition of the shell microbiome likely influences the susceptibility and proliferation of dermal/shell disease in crustaceans (Chistoserdov et al., 2012; Quinn et al., 2012).
Our sampling design enabled a spatial comparison of the shell microbiome between the sampling locations of rock crabs, and an interspecies comparison between rock and green crabs in one location, Malagash. The regional comparison of rock crabs from Charlo, Malagash and Tracadie, shows that alpha diversity indices and microbial composition significantly differed between sampling locations. As the shell microbiome interacts closely with the surrounding seawater, its composition should be influenced by regional differences in environmental conditions. Sampling environments can shape the composition of the gut microbiome of wild and farmed shrimp (Chen et al., 2017; Zhao et al., 2018). A previous study on catfish found that the microbial structure of the skin differed by sampling location (Chiarello et al., 2019). It is suggested that biotic and abiotic factors, which likely differ regionally, affect the population of microbes present in the environment, in turn changing the bacterial taxa and population structure of host microbiomes (Chen et al., 2017).
Arguably of equal importance, our analyses also revealed that even in the same sampling location from the same sampling event, the shell microbiome of rock and green crabs differed significantly from each other. In the ordination plots, microbiome samples from the rock crabs form one large cluster with the three locations as subclusters that overlap to some extent, while samples from green crabs are somewhat separate. This indicates that the rock crab microbiome from Malagash is more similar to rock crab microbiomes from other locations than it is to the green crab microbiome from the same location. This is the first inter-species comparison of crustacean shell microbiomes, though a previous study has found differences in surface microbiota between two ant species (Birer et al., 2017), and that it can be shaped by host genotypes in fish (Boutin et al., 2014). It has also been shown that crustaceans are able to regulate the microbial assembly in their gut and the hemolymph (Cuellar-Gempeler and Leibold, 2018; Zhang et al., 2018; Cuellar-Gempeler and Leibold, 2019; Cannicci et al., 2020). It is hypothesized that harboring commensal bacteria in the microbiome protects against disease agents (Scarborough et al., 2005; Wang and Wang, 2015).
Taken together with our results of certain host-specificity of microbial taxa and given that the crustacean shell is the most important protection against invading pathogens, it seems plausible that rock and green crabs can select microbial communities that are beneficial for host health and immunity. In the crustacean hemolymph, as well as the human skin, the regulation of bacterial growth or settlement is linked to the production of antimicrobial factors (Handfield et al., 2018; Zhang et al., 2018). While it is not yet clear if or how antimicrobials or specific taxa-promoting factors are transported onto the shell, the pore canals in the crustacean shell are a plausible source. Crustacean shell pore canals which likely transport materials during moult (Cheng et al., 2008) may also be involved in moving selective bacterial factors that promote the colonization of certain bacterial taxa, to the shell.
While we accounted for unbalanced sampling and used accomplished bioinformatics tools, it is important to acknowledge the limitations of this study. It needs to be noted that the rock crabs were not sampled at the same time. Rock crabs were sampled in August from Tracadie (PEI), in September from Charlo (NB) and in October from Malagash (NS). This potentially results not only in spatial effects on their shell microbiome but also temporal effects due to temperature fluctuations or other seasonal factors. That being said, the analysis of location effects was intended to assess the influence of the environment on the shell microbiome. This would include all environmental factors that are present at a geographical location, such as water temperature, sampling depth and ocean currents, which were not captured in this study but likely influence the shell microbiome of crustaceans. As with all marker gene analyses, sequencing of the 16S bacterial gene is associated with an inherent degree of primer bias, as primers have unequal affinities for different sequences that lead to certain bacterial genes being amplified more than others (Knight et al., 2018). To minimize primer bias and enable comparability of our results to other microbiome studies using similar methods, we used well-validated primers, standardized sample processing methods and sequencing protocols as well as advanced bioinformatic tools.
5 Conclusion
Using next-generation sequencing and open-source bioinformatic tools, we provide evidence that the shell microbial community of two crustacean species is affected by geographical factors and appears to be host species-specific. While the seawater control samples share a similar microbial community on class level with shell swab samples (Alpha- and Gammaproteobacteria, Flavobacteriia), not many taxa were shared on the lower taxonomic levels the microbial diversity of seawater samples was markedly lower. This means that the shell microbiome is determined by a combination of microorganisms present in the surrounding seawater, and inherent factors controlled by the host. Our results contribute to the understanding of the complex relationships that influence marine microbiomes and the role of host regulation on the composition of their associated surface microbial communities.
Data availability statement
The data presented in the study are deposited in the NCBI Sequence Read Archive, accession number PRJNA986134.
Author contributions
KFC and SKe collected the data; KFC, SKo, SKe, and KT designed the research; SKo and SKe analyzed the data and SKo wrote the manuscript. All authors revised and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project received funding from the Natural Sciences and Engineering Research Council of Canada (RGPIN-2018-05894).
Acknowledgments
We would like to thank all Clark Lab members at the Dalhousie University Faculty of Agriculture for their help in collecting and processing the samples as well as the Gloucester Marine Genomics Institute (GMGI) for their support in sequencing. We would also like to acknowledge UseGalaxy for their free and intuitive bioinformatics webtool.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1152544/full#supplementary-material
References
Afgan E., Baker D., Batut B., Van Den Beek M., Bouvier D., Ech M., et al. (2018). The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544. doi: 10.1093/NAR/GKY379
Anderson M. J. (2017). “Permutational multivariate analysis of variance (PERMANOVA),” in Wiley Stats ref: statistics reference online. Eds. Balakrishnan N., Colton T., Everitt B., Piegorsch W., Ruggeri F., Teugels J. L., (Auckland, New Zealand: John Wiley & Sons, Ltd) 1–15. doi: 10.1002/9781118445112.stat07841
Apprill A., Mcnally S., Parsons R., Weber L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microbial Ecol. 75, 129–137. doi: 10.3354/AME01753
Bélair M. C., Miron G. (2009). Time budget of Cancer irroratus (Say) and Carcinus maenas (L.) under various temperature and prey density conditions during conspecific and heterospecific challenges. J. Shellfish Res. 28, 923–930. doi: 10.2983/035.028.0423
Benahmed A. G., Gasmi A., Doşa A., Chirumbolo S., Mujawdiya P. K., Aaseth J., et al. (2021). Association between the gut and oral microbiome with obesity. Anaerobe 70, 102248. doi: 10.1016/j.anaerobe.2020.102248
Birer C., Tysklind N., Zinger L., Duplais C. (2017). Comparative analysis of DNA extraction methods to study the body surface microbiota of insects: a case study with ant cuticular bacteria. Mol. Ecol. Resour 17, e34–e45. doi: 10.1111/1755-0998.12688
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Boutin S., Sauvage C., Bernatchez L., Audet C., Derome N. (2014). Inter individual variations of the fish skin microbiota: host genetics basis of mutualism? PloS One 9, e102649. doi: 10.1371/journal.pone.0102649
Cannicci S., Fratini S., Meriggi N., Bacci G., Iannucci A., Mengoni A., et al. (2020). To the land and beyond: crab microbiomes as a paradigm for the evolution of terrestrialization. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.575372
Chen C. Y., Chen P. C., Weng F. C. H., Shaw G. T. W., Wang D. (2017). Habitat and indigenous gut microbes contribute to the plasticity of gut microbiome in oriental river prawn during rapid environmental change. PloS One 12, e0181427. doi: 10.1371/JOURNAL.PONE.0181427
Cheng L., Wang L., Karlsson A. M. (2008). Image analyses of two crustacean exoskeletons and implications of the exoskeletal microstructure on the mechanical behavior. J. Material Res. 23, 2854–2872. doi: 10.1557/JMR.2008.0375
Cheung M. K., Yip H. Y., Nong W., Law P. T. W., Chu K. H., Kwan H. S., et al. (2015). Rapid change of microbiota diversity in the gut but not the hepatopancreas during gonadal development of the new shrimp model Neocaridina denticulata. Mar. Biotechnol. 17, 811–819. doi: 10.1007/S10126-015-9662-8
Chiarello M., Paz-Vinas I., Veyssière C., Santoul F., Loot G., Ferriol J., et al. (2019). Environmental conditions and neutral processes shape the skin microbiome of European catfish (Silurus glanis) populations of southwestern France. Environ. Microbiol. Rep. 11, 605–614. doi: 10.1111/1758-2229.12774
Chistoserdov A., Quinn R., Gubbala S., Smolowitz R. (2012). Bacterial communities associated with lesions of shell disease in the American lobster, Homarus americanus. J. Shellfish Res. 31, 449–462. doi: 10.2983/035.031.0205
Clemente J. C., Ursell L. K., Parfrey L. W., Knight R. (2012). The impact of the gut microbiota on human health: an integrative view. Cell 148, 1258–1270. doi: 10.1016/J.CELL.2012.01.035
Cuellar-Gempeler C., Leibold M. A. (2018). Multiple colonist pools shape fiddler crab-associated bacterial communities. ISME J. 3 (12), 825–837. doi: 10.1038/s41396-017-0014-8
Cuellar-Gempeler C., Leibold M. A. (2019). Key colonist pools and habitat filters mediate the composition of fiddler crab–associated bacterial communities. Ecology 100, e02628. doi: 10.1002/ECY.2628
Daniels C. L., Merrifield D. L., Boothroyd D. P., Davies S. J., Factor J. R., Arnold K. E. (2010). Effect of dietary Bacillus spp. and mannan oligosaccharides (MOS) on European lobster (Homarus gammarus l.) larvae growth performance, gut morphology and gut microbiota. Aquaculture 304, 49–57. doi: 10.1016/j.aquaculture.2010.03.018
Egan S., Gardiner M. (2016). Microbial dysbiosis: rethinking disease in marine ecosystems. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00991
Feinmann S., Martínez A., Bowen J., Tlusty M. (2017). Fine-scale transition to lower bacterial diversity and altered community composition precedes shell disease in laboratory-reared juvenile American lobster. Dis. Aquat Organ 124, 41–54. doi: 10.3354/dao03111
Fisheries and Oceans (2011). Ecological assessment of the invasive European green crab (Carcinus maenas) in Newfoundland 2007–2009. Canadian science advisory secretariat rep. 2010 (Ottawa: Canadian Science Advisory Secretariat).
Fisheries and Oceans (2022). Rock crab (Cancer irrotatus) Newfoundland and Labrador region. Available at: https://www.dfo-mpo.gc.ca/fisheries-peches/ifmp-gmp/rock-crab-commun/2022/index-eng.html
Gendron L., Fradette P., Godbout G. (2001). The importance of rock crab (Cancer irroratus) for growth, condition and ovary development of adult American lobster (Homarus americanus). J. Exp. Mar. Biol. Ecol. 262, 221–241. doi: 10.1016/S0022-0981(01)00297-0
Groner M. L., Maynard J., Breyta R., Carnegie R. B., Dobson A., Friedman C. S., et al. (2016). Managing marine disease emergencies in an era of rapid change. Philos. Trans. R. Soc. B: Biol. Sci. 371, 20150364. doi: 10.1098/RSTB.2015.0364
Groner M. L., Shields J. D., Landers D. F., Swenarton J., Hoenig J. M. (2018). Rising temperatures, molting phenology, and epizootic shell disease in the American lobster. Am. Nat. 192, 163–177. doi: 10.5061/dryad.n102b1s
Handfield C., Kwock J., MacLeod A. S. (2018). Innate antiviral immunity in the skin. Trends Immunol. 39, 328–340. doi: 10.1016/J.IT.2018.02.003
Harvell C. D., Mitchell C. E., Ward J. R., Altizer S., Dobson A. P., Ostfeld R. S., et al. (2002). Climate warming and disease risks for terrestrial and marine biota. Science 296, 2158–2162. doi: 10.1126/science.1063699
Holt C. C., Bass D., Stentiford G. D., van der Giezen M. (2020a). Understanding the role of the shrimp gut microbiome in health and disease. J. Invertebr Pathol. 186, 107387. doi: 10.1016/J.JIP.2020.107387
Holt C. C., van der Giezen M., Daniels C. L., Stentiford G. D., Bass D. (2020b). Spatial and temporal axes impact ecology of the gut microbiome in juvenile European lobster (Homarus gammarus). ISME J. 14, 531–543. doi: 10.1038/s41396-019-0546-1
Ivanova N. V., Dewaard J. R., Hebert P. D. N. (2006). An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol. Ecol. Notes 6, 998–1002. doi: 10.1111/J.1471-8286.2006.01428.X
Jung J., Gillevet P., Sikaroodi M., Andrews J., Song B., Shields J. (2021). Comparative study of the hemolymph microbiome between live and recently dead American lobsters Homarus americanus. Dis. Aquat Organ 143, 147–158. doi: 10.3354/dao03568
Knight R., Vrbanac A., Taylor B. C., Aksenov A., Callewaert C., Debelius J., et al. (2018). Best practices for analysing microbiomes. Nat. Rev. Microbiol. 16, 410–422. doi: 10.1038/s41579-018-0029-9
Kraemer P., Gerlach G., Brinkhoff T. (2020). Diversity and flexibility of the bacterial communities on Cancer pagurus at different temperatures. J. Sea Res. 156, 101827. doi: 10.1016/j.seares.2019.101827
Kwan S.-Y., Sabotta C. M., Joon A., Wei P., Petty L. E., Below J. E., et al. (2022). Gut microbiome alterations associated with diabetes in Mexican americans in south Texas. mSystems 7 (3), e00033–e00022. doi: 10.1128/msystems.00033-22
Lafferty K. D., Porter J. W., Ford S. E. (2004). Are diseases increasing in the ocean? Annu. Rev. Ecol. Evol. Syst. 35, 31–54. doi: 10.1146/annurev.ecolsys.35.021103.105704
Lawley T., Walker A. (2012). Intestinal colonization resistance. Immunology 138, 1–11. doi: 10.1111/j.1365-2567.2012.03616.x
Lloyd-Price J., Abu-Ali G., Huttenhower C. (2016). The healthy human microbiome. Genome Med. 8 (1), 1–11. doi: 10.1186/S13073-016-0307-Y
Lozupone C. A., Hamady M., Kelley S. T., Knight R. (2007). Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73 (5), 1576–1585. doi: 10.1128/AEM.01996-06
Matheson K., McKenzie C. H., Gregory R. S., Robichaud D. A., Bradbury I. R., Snelgrove P. V. R., et al. (2016). Linking eelgrass decline and impacts on associated fish communities to European green crab Carcinus maenas invasion. Mar. Ecol. Prog. Ser. 548, 31–45. doi: 10.3354/MEPS11674
McDonald D., Price M. N., Goodrich J., Nawrocki E. P., Desantis T. Z., Probst A., et al. (2011). An improved greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
Meres N. J. (2016). “Surface biofilm interactions in epizootic shell disease of the American lobster (Homarus americanus),” in Microbial biofilms - importance and applications (London: UK via online publisher InTechOpen Limited), 383–421. doi: 10.5772/63498
Meres N., Ajuzie C., Sikaroodi M., Vemulapalli M., Shields J. D., Gillevet P. (2012). Dysbiosis in epizootic shell disease of the American lobster (Homarus americanus). J. Shellfish Res. 31, 463–472. doi: 10.2983/035.031.0206
Mousa W. K., Chehadeh F., Husband S. (2022). Microbial dysbiosis in the gut drives systemic autoimmune diseases. Front. Immunol. 13. doi: 10.3389/fimmu.2022.906258
Ooi M. C., Goulden E. F., Smith G. G., Bridle A. R. (2019). Haemolymph microbiome of the cultured spiny lobster Panulirus ornatus at different temperatures. Sci. Rep. 9, 1–13. doi: 10.1038/s41598-019-39149-7
Parada A., Needham D., Fuhrman J. (2015). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Petersen J. M., Osvatic J. (2018). Microbiomes in natura: importance of invertebrates in understanding the natural variety of animal-microbe interactions. mSystems 3, e00179–e00117. doi: 10.1128/MSYSTEMS.00179-17
Quinn R., Cawthorn R., Summerfield R., Smolowitz R., Chistoserdov A. (2013). Bacterial communities associated with lesions of two forms of shell disease in the American lobster (Homarus americanus, Milne edwards) from Atlantic Canada. Can. J. Microbiol. 59, 380–390. doi: 10.1139/cjm-2012-0679
Quinn R., Metzler A., Tlusty M., Smolowitz R., Leberg P., Chistoserdov A. (2012). Lesion bacterial communities in American lobsters with diet-induced shell disease. Dis. Aquat. Organisms 98, 221–233. doi: 10.3354/dao02446
Rajeev R., Adithya K. K., Kiran G. S., Selvin J. (2021). Healthy microbiome: a key to successful and sustainable shrimp aquaculture. Rev. Aquac 13, 238–258. doi: 10.1111/RAQ.12471
Rayner G., McGaw I. J. (2019). Effects of the invasive green crab (Carcinus maenas) on American lobster (Homarus americanus): food acquisition and trapping behaviour. J. Sea Res. 144, 95–104. doi: 10.1016/j.seares.2018.10.004
Scarborough C. L., Ferrari J., Godfray H. C. J. (2005). Aphid protected from pathogen by endosymbiont. Science 310, 1781. doi: 10.1126/science.1120180
Sigurdsson G. M., Rochette R. (2013). Predation by green crab and sand shrimp on settling and recently settled American lobster postlarvae. J. Crustacean Biol. 33 (1), 10–14. doi: 10.1163/1937240X-00002107
Smolowitz R., Chistoserdov A. Y., Hsu A. (2005). A description of the pathology of epizootic shell disease in the American lobster (Homarus americanus), h. Milne edwards 1837. J. Shellfish Res. 24 (3). doi: 10.3389/fmicb.2012.00292
Wahl M., Goecke F., Labes A., Dobretsov S., Weinberger F. (2012). The second skin: ecological role of epibiotic biofilms on marine organisms. Frontiers in Microbiology 3, 329. doi: 10.3389/fmicb.2012.00292
Wang X. W., Wang J. X. (2015). Crustacean hemolymph microbiota: endemic, tightly controlled, and utilization expectable. Mol. Immunol. 68, 404–411. doi: 10.1016/j.molimm.2015.06.018
Wynne J. W., Thakur K. K., Slinger J., Samsing F., Milligan B., Powell J. F. F., et al. (2020). Microbiome profiling reveals a microbial dysbiosis during a natural outbreak of tenacibaculosis (yellow mouth) in Atlantic salmon. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.586387
Zhang X., Sun Z., Zhang X., Zhang M., Li S. (2018). Hemolymph microbiomes of three aquatic invertebrates as revealed by a new cell extraction method. Appl. Environ. Microbiol. 84, e02824-17. doi: 10.1128/AEM.02824-17
Keywords: crustacean microbiome, microbial diversity, QIIME2, microbial ecology, Illumina MiSeq
Citation: Koepper S, Kelley S, Thakur KK and Clark KF (2023) Interspecies and spatial differences in the shell microbiome of Atlantic rock crab Cancer irrotatus and European green crab Carcinus maenas from Atlantic Canada. Front. Mar. Sci. 10:1152544. doi: 10.3389/fmars.2023.1152544
Received: 07 March 2023; Accepted: 12 June 2023;
Published: 10 July 2023.
Edited by:
Crisalejandra Rivera-Perez, Centro de Investigación Biológica del Noroeste (CIBNOR), MexicoReviewed by:
José Q. García-Maldonado, Departamento de Recursos del Mar. Cinvestav Unidad Mérida, MexicoWendy Espinosa De Aquino, Universidad Autónoma de Baja California Sur, Mexico
Copyright © 2023 Koepper, Kelley, Thakur and Clark. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Svenja Koepper, c2tvcHBlckB1cGVpLmNh
 Svenja Koepper
Svenja Koepper Shannon Kelley2
Shannon Kelley2 Krishna Kumar Thakur
Krishna Kumar Thakur K. Fraser Clark
K. Fraser Clark