- Key Laboratory of Aquacultural Biotechnology Ministry of Education, School of Marine Sciences, Ningbo University, Ningbo, China
Insulin-like peptides (ILPs) are essential to the animal kingdom for regulating growth and development, reproduction, behavior, metabolism, and lifespan. In crustaceans, the most well-known ILP is insulin-like androgenic gland hormone, a key hormone in regulating sex differentiation and reproduction. Identification of other ILPs and their functions are still limited. In this study, an insulin-like peptide gene of the swimming crab Portunus trituberculatus was cloned and characterized. Its transcripts were mainly found in nerve tissues and expression could be induced by glucose, implying a putative role in glucoregulation. After depletion of endogenous ILP, injection of ILP dsRNA (dsILP) significantly elevated blood glucose levels and recombinant ILP (rILP) decreased hemolymph glucose levels, further clarifying the involvement of acquired ILP in hemolymph glucose regulation. Injection of dsILP decreased PtAkt, PtGS, PtPFK and increased PtGSK and PtPEPCK gene expression. The opposite profile was observed after glucose and rILP injection, indicating that PtILP might negatively regulate hemolymph glucose levels via the IIS (insulin/IGF-1 signaling) pathway by inhibiting gluconeogenesis and promoting glycogen synthesis and glycolysis. This study has refined the mechanism of glucose regulation in crustaceans and laid the foundation for further studies on ILP function.
1 Introduction
Glucose is the primary energy source for metabolism, and balanced hemolymph glucose levels are essential to maintain a stable physiological state and to help cope with the metabolic demands imposed by processes such as growth and reproduction (Nelson and Sheridan, 2006). In mammals, insulin and glucagon play a central role in maintaining glucose homeostasis (Miyazaki et al., 2020). In insects, adipokinetic hormone (AKH) and insulin-like peptides (ILPs) constitute an energy regulatory system similar to the mammalian insulin-glucagon system (de Brito Sanchez et al., 2021). In crustaceans, elevation of hemolymph glucose levels is believed to be directly induced by crustacean hyperglycaemic hormone (CHH) (Chen et al., 2020), but to date, the mechanisms involved in lowering hemolymph glucose levels are not fully understood. Recently, several ILPs have been shown to be capable of fulfilling this function, suggesting that the crustacean mechanism may be similar to insects.
ILPs belong to the insulin superfamily, which is found in a wide range of invertebrates, including mollusks (Cherif–Feildel et al., 2019) nematodes (Yohei and Tsuyoshi, 2018) and arthropods (Chowański et al., 2021). Many previous studies have shown that ILPs play a crucial role in regulating development, reproduction, behavior, metabolism, and lifespan (Semaniuk et al., 2021). Injection of ILPs into Bombyx mori (Nagasawa et al., 1984) and Aedes aegypti (Brown et al., 2008) reduced their levels of hemolymph trehalose, the main blood sugar in various insect species. In Drosophila melanogaster, mutated DILP2 can increase blood sugar levels significantly (Broughton et al., 2008). Moreover, in other species, RNA interference shows that ILPs affect the levels of trehalose in the hemolymph of Spodoptera exigua (Al Baki et al., 2019), Maruca vitrata (Kim and Hong, 2015) and Rhodnius prolixus (Defferrari et al., 2016).
In crustaceans, ILP may also be involved in the regulation of hemolymph glucose homeostasis. In early reports, it was noted that there were substances with insulin activity in the methanol extract of the lobster Homarus americanus (Sanders, 1983) and the in shore crabs Carcinus maenas (Gallardo et al., 2003), which could stimulate the synthesis of glycogen and glucose oxidation to carbon dioxide in mouse and rat tissues. An ILP has been reported for Callinectes sapidus, where after RNA interference, hemolymph glucose levels were higher than the control group (Chung, 2014). Over the last few years, an increasing number of crustacean ILPs have been reported to be involved in the regulation of hemolymph glucose. When recombinant ILP (rILP) was injected into Eriocheir sinensis (Wang et al., 2020) and Litopenaeus vannamei (Su et al., 2022), hemolymph glucose levels in these two species decreased significantly. Interestingly, unlike the observation with C.sapidus (Chung, 2014), ILP RNAi did not increase hemolymph glucose levels in Macrobrachium rosenbergii (Jiang et al., 2020) and L.vannamei (Su et al., 2022). Therefore, there is still insufficient understanding of the mechanism used by ILPs to regulate hemolymph glucose levels and further research on this matter is imperative.
The swimming crab (Portunus trituberculatus) is major commercial crab species and has been extensively artificially propagated and cultivated. It is still unclear whether and how ILP is involved in regulating P. trituberculatus hemolymph glucose levels. In the present study, an ILP (PtILP) was identified from P. trituberculatus. We measured changes in the level of hemolymph glucose after injection of exogenous glucose, RNA interference and injection of recombinant PtILP protein. To investigate the function of PtILP in regulating hemolymph glucose, changes in expression levels of genes related to the IIS pathway were also examined.
2 Materials and methods
2.1 Experimental animals and tissue sampling
Healthy swimming crabs were purchased from the local aquatic market in Guoju District, Ningbo City, Zhejiang Province, China. Before the experiments were initiated, the crabs were placed in aerated fresh seawater at 24-26°C. All crabs were fed twice daily with Manila clams (Ruditapes philippinarum) purchased from local aquatic market and subjected to natural photoperiod conditioning for three days. Mature male and female crabs (body weight, 200-250 g) were used for cDNA cloning and tissue distribution analysis. In addition, mature male crabs (body weight, 120-150 g) were used for injection experiments. All tissues were sampled on ice and stored in RNA preservation fluid (Cwbiotech, China) at −20 °C for further analysis.
2.2 Total RNA extraction and cDNA synthesis
Total RNA was isolated from different samples using the TRIzol Reagent (Cwbiotech, China) according to the manufacturer’s instructions, genomic DNA contamination within the total RNA was removed by DNaseI (Takara, Japan). RNA concentrations and quality were determined using a NanoDrop 2000 UV Spectrophotometer (Thermo Fisher, USA). The first strand of cDNA was prepared from 1 μg RNA using the Perfect Real Time PrimeScript® reagent Kit (Takara, Japan) and stored at −80 °C until further use.
2.3 Molecular cloning and characterization
The gene sequence of PtILP was obtained from available P. trituberculatus transcriptome sequencing data (SRR13870346). To confirm the gene sequence of PtILP, specific primers (Supplementary Table 1) were designed based on the transcriptome sequence. The PCR products were separated on a 1.5% agarose gel (Vazyme, China) by electrophoresis, and the bands corresponding to the expected size were excised, purified, and the amplicon was ligated into the pMD19-T vector (Takara, Japan), Finally, the ligated product was transformed into competent Escherichia coli DH5α cells and five positive clones were selected for sequencing. ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to determine the open reading frame (ORF) in the PtILP cDNA. The BLAST program (http://www.ncbi.nlm.nih.gov/blast) was used to analyze sequence similarities and the signal peptide was predicted using the SignalP 5.0 server (https://www.cbs.dtu.dk/services). C-peptide potential cleavage sites were predicted using the CBS prediction server (http://www.cbs.dtu.dk/services). For the automatic prediction of disulfide bridges, we used the tool DISULFIND (https://predictprotein.org/). The 3D structure of the PtILP protein was predicted by the Alphafold2 (Jumper et al., 2021; Varadi et al., 2021) web server (https://alphafold.ebi.ac.uk/) and edited by PyMOL Software (https://pymol.org/2/). The phylogenetic tree was constructed using the Neighbor-Joining (NJ) method by MEGA7.0 (https://www.megasoftware.net/).
2.4 Preparation of recombinant protein
The plasmid pGEX-4t-2 (Miaoling Biology, China) was digested with restriction endonucleases SmaI and XhoI to generate a linearized vector. Primers PtILP-pGEX-F/R were designed to amplify the nucleotide sequence. All primers were synthesized with 21-bp extensions, which were homologous to the linearized vector 5’ and 3’ ends to generate the amplification products. The gene-specific primers are listed in Supplementary Table 1. The purified PCR products was inserted into the corresponding linearized vector by setting up an in-fusion cloning reaction using the ClonExpress II One Step Cloning Kit (Vazyme, China). The recombinant plasmids were transformed into E. coli DH5α competent cells (Vazyme, China) and sequenced to verify the cloned gene. The confirmed clones were selected and cultured for plasmid extraction. Plasmids were extracted using the E.Z.N.A. Plasmid Mini Kit I (Omega Bio-Tek, USA). The recombinant plasmid clone was transformed into E. coli Rosetta gami2(DE3) cells (Weidi, China) to express the GST-PtILP protein (rGST-PtILP). The vector pGEX-4t-2 was also transformed for expression of the GST tag protein (rGST). The protein was expressed by 0.8mM IPTG induction (16°C,16 h). All recombinant proteins were purified using the GST-tag Purification Resin (Beyotime, China) and verified by SDS-PAGE.
2.5 Glucose injection
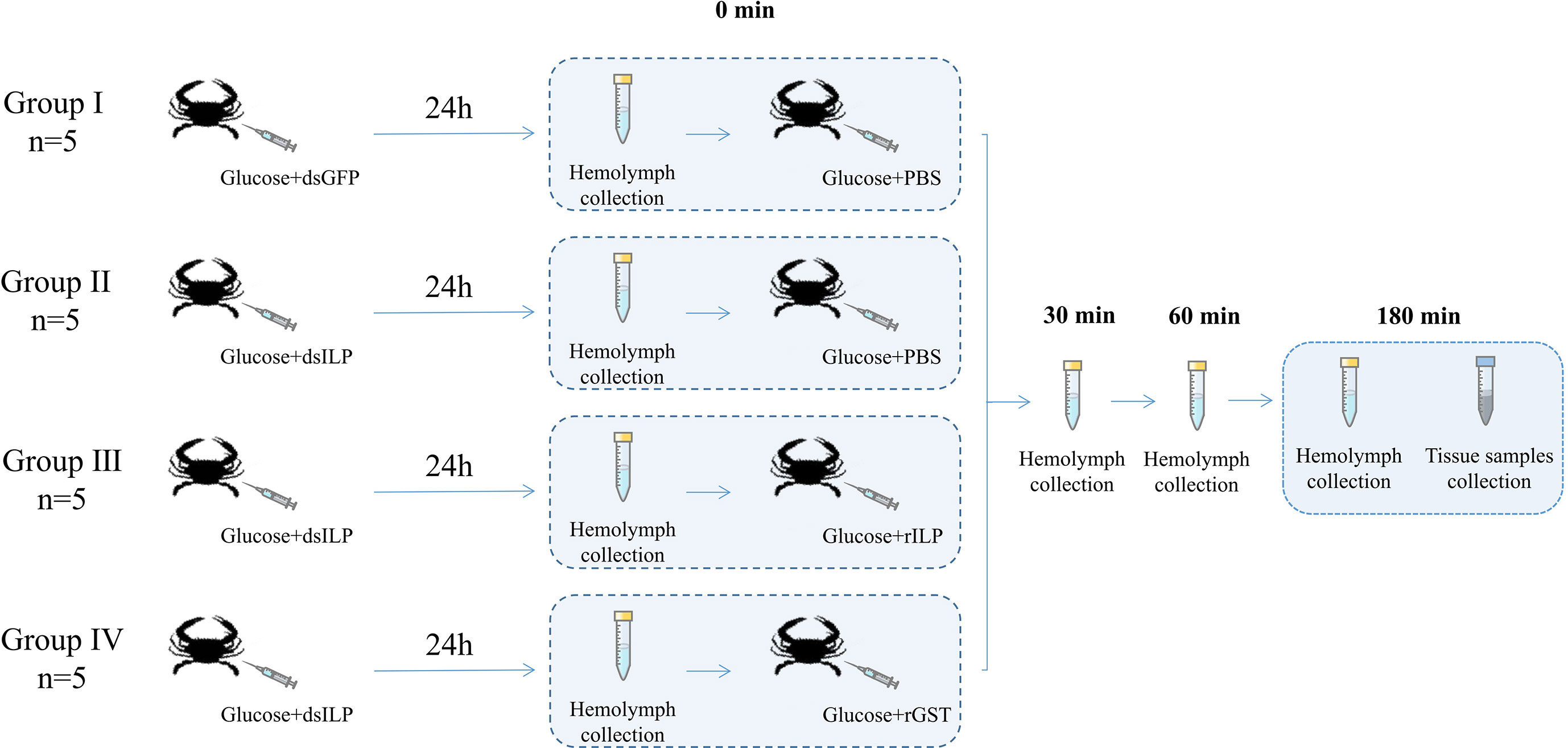
Twenty crabs were randomly divided into four groups: Group I and Group II were injected with 100 µl crab saline (CS), Group III and Group IV were injected with 18 mg glucose/100 µl crab saline (Chung, 2014). Prior to the injection, hemolymph samples were taken at 0 min (t0), and 100 µL hemolymph was taken from crabs in Group I and Group III at 5 (t1), 15 (t2), 30 (t3) and 60 (t4) min after the injection while group II and Group IV were only sampled for the time points up to t3. All crabs were anesthetized on ice and dissected after obtaining the last hemolymph sample. The eyestalk ganglia, hepatopancreas, and muscle were used for total RNA extraction and cDNA synthesis. Seven genes were selected which were PtILP, PtAkt (threonine-serine protein kinase), glycogen synthesis-related genes PtGS (glycogen synthase) and PtGSK3 (glycogen synthase kinase 3), PtGLUT4 (glucose transporter-related gene glucose transporter 4) glycolysis and gluconeogenesis-related genes PtPFK (phosphofructokinase) and PtPEPCK (phosphoenolpyruvate carboxy kinase).
2.6 RNA interference
The dsRNA synthesis of PtILP (dsILP) and green fluorescent protein (dsGFP) was performed as described previously (Xie et al., 2016). For dsRNA injection, the domesticated crabs were randomly divided into the PtILP dsRNA injection group and the GFP dsRNA injection control group. Each crab was injected with 2 μg/g body weight of dsRNA at the base of the last swimming leg. Five crabs from each injection group were sacrificed at 24 h after injection. 100 μL of hemolymph were extracted for the blood glucose assay. Gene expression levels in the eyestalk ganglion, hepatopancreas and muscle were analyzed as above. In addition, 100 mg of muscle and hepatopancreas were used to determine glycogen content.
2.7 In vivo recombinant protein injection
Twenty crabs were randomly divided into four groups, group I: dsGFP injection, followed by PBS after 12 h; Group II: dsPtILP injection, followed by PBS after 12 h; Group III: dsPtILP injection, followed by rGST-ILP after 12 h; Group IV: dsPtILP injection, followed by rGST after 12 h. All crabs were also simultaneously injected with glucose at each time of injection (Figure 1). dsRNA was injected at 2 μg/g body weight, and recombinant protein was injected at 1μmol dissolved in 200 μl of PBS. Hemolymph was collected at 30, 60, and 180 minutes after PtILP recombinant protein injection. Tissue samples were dissected and sampled after the last hemolymph collection.
2.8 Glucose and glycogen assays
The hemolymph samples were measured using the Glucose Assay Kit purchased from Nanjing Jiancheng Institute of Biological Engineering (Nanjing, China). To inhibit coagulation, the hemolymph samples were immediately diluted with saline containing 0.01 M EDTA at a 1:1 ratio. The mixtures were centrifuged (2000 g, 20 min, 4 °C), and the resultant supernatants were collected and immediately used for glucose analysis according to the manufacturer’s instructions. Glycogen levels in muscle and hepatopancreas tissues were measured according to the manufacturer’s instructions using the Liver/muscle Glycogen Analysis Kit produced by Nanjing Jiancheng Institute of Biological Engineering (Nanjing, China).
2.9 Quantitative real-time PCR
This study’s relative gene expression level was detected using quantitative real-time PCR (qPCR). PCR was carried out using the SYBR®Premix Ex TaqTM II kit (Takara, Japan) according to the manufacturer’s instructions. PCR conditions were as follows: 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s, and 58 °C for 20 s. The β-actin (Supplementary Table 1) was carried out as a reference gene. The mRNA expression levels of gene data were analyzed by the comparative Ct (2−ΔΔCt) method.
2.10 Statistical analysis
All data were presented as mean ± standard deviation (SD). The statistical differences were analyzed using independent-samples t-test between two groups or one-way ANOVA followed by Duncan test among multiple groups. Prior to all statistical tests. The data were analyzed for homogeneity of variance (Levene’s test). Significant differences were accepted at P < 0.05. The statistical significance of the data was analyzed using the SPSS 20.0 software.
3 Results
3.1 Molecular characteristics and tissue distribution of PtILP
The PtILP cDNA contains a 609 bp open reading frame encoding a 202 amino acid-containing protein with a predicted molecular weight of 22.64 kDa (Figure 2A). A 20 aa N-terminal signal peptide was predicted by signalP-5.0, and SMART analysis showed that PtILP contained an IIGF (insulin/IGF) structural domain. PtILP pre-propeptide contains four segments from the N-terminal to the C-terminal in the order of signal peptide, A-chain, C-peptide, and B-chain (Figure 2B). The signal peptide and C-peptide are excised in the mature PtILP, while six cysteine residues form two inter-chain disulfide bonds and one intra-chain disulfide bond (Figures 2C, D). Phylogenetic analysis based on amino acid sequences showed that PtILP was more closely related to the three crustacean ILPs, M. rosenbergii ILP, Cherax quadricarinatus ILP1, and Sagmariasus verreauxi ILP2 (Figure 3B).
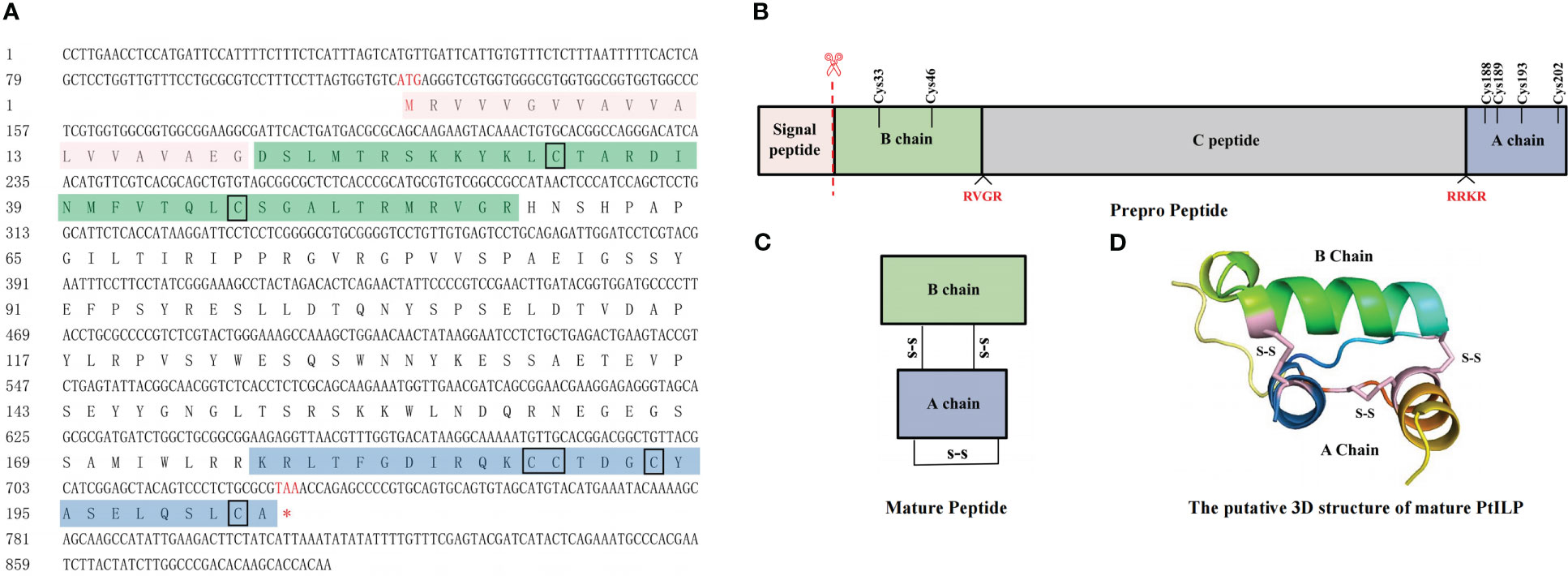
Figure 2 Identification and characterization of PtILP. (A) Nucleotide and amino acid sequences of PtILP. Marked in red are the nucleotide and amino acid sequences encoding the start codon (ATG) and the stop codon (TAA). The predicted signal peptide sequences are shown in red boxes. The predicted B and A chains are shown in green and blue boxes, respectively, with the C peptide between the B and A chains and the cysteine residues marked by black boxes. (B) Structure of prepro-PtILP. The predicted cleavage sites RVGR and RRKR are marked in red. (C) Structure of PtILP mature peptide. The signal peptide and the C chain are excised in the mature peptide, and the six cysteine residues of the B and A chains form two interchain disulfide bonds and one intrachain disulfide bond. (D) The putative 3D structure of mature PtILP. The three disulfide keys are shown as solid pink lines.
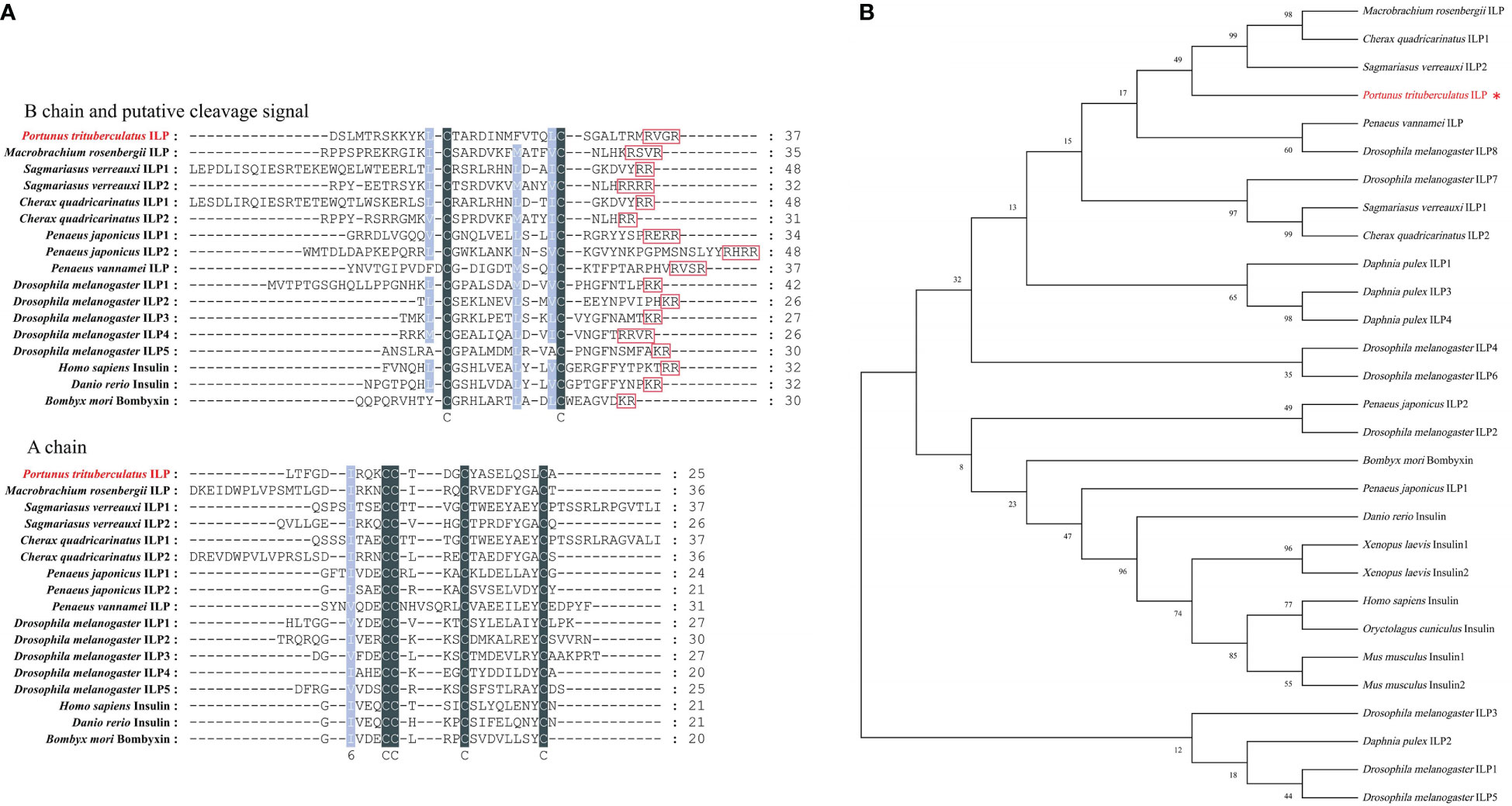
Figure 3 PtILP multiple sequence alignment and phylogenetic analysis. (A) Multiple amino acid sequence alignment of insulin family peptides. Amino acid sequences of putative B and A chains have been shown. Putative cleavage sites have been boxed. Identical sequences are shown in gray and similar sequences in light blue. (B) Neighbor-joining phylogenetic tree of insulin family peptides based on amino acid sequences. The PtILP proteins was marked with red. The sequences used in the phylogenetic tree are summarized in Supplementary Table 2.
The transcript levels of PtILP were measured in different tissues obtained from adult crabs (Figure 4). The results showed that PtILP transcripts were detected in all the tissues tested, including the eyestalk ganglia, hepatopancreas, brain, muscle, heart, thoracic ganglion, testis, androgenic glands and ovary. Among them, PtILP mRNA expression was significantly higher in the eyestalk ganglion than in other tissues while relatively high expression levels were also noted in brain and muscle.
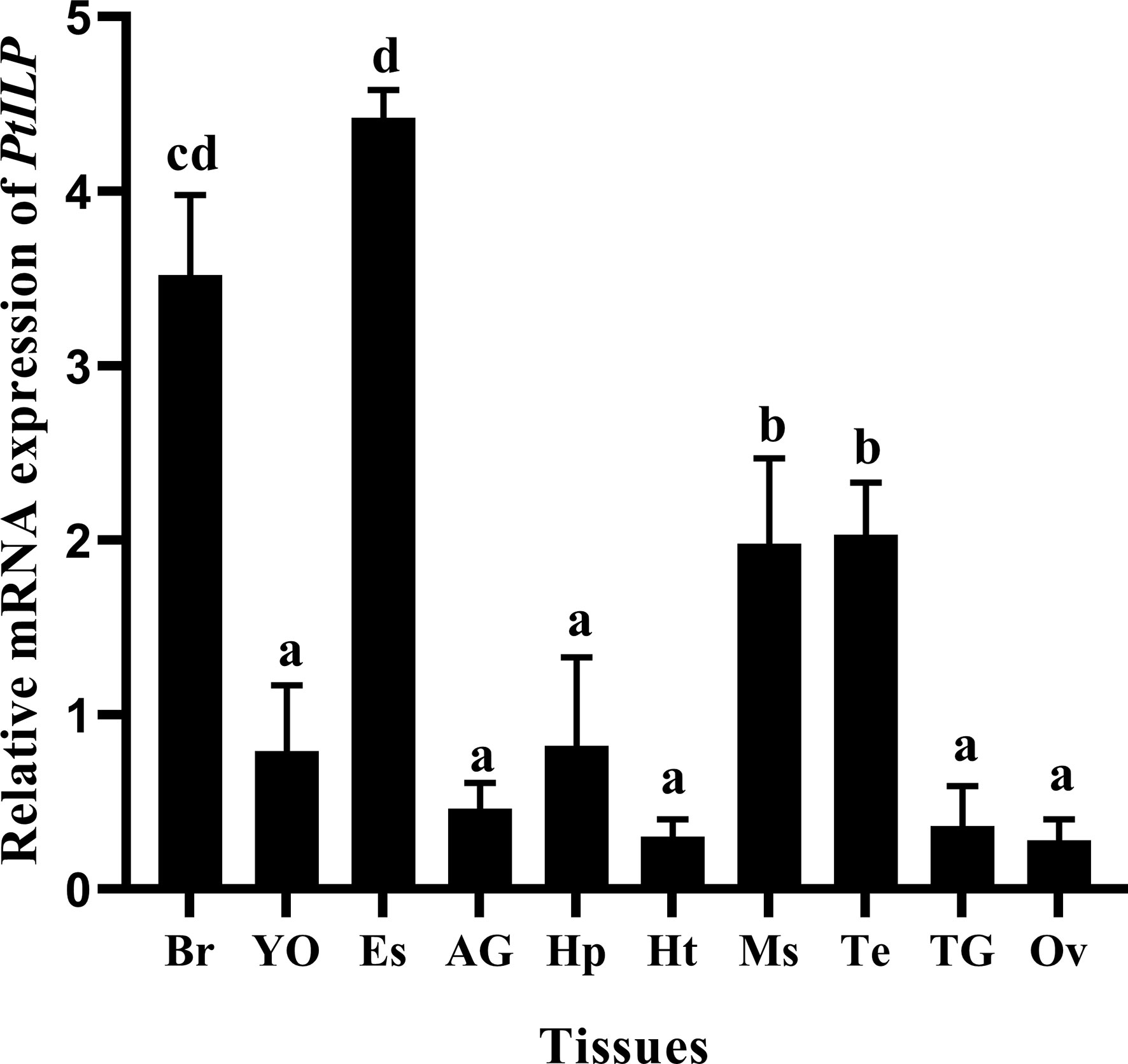
Figure 4 Tissue distributions of PtILP transcripts in male and female P. trituberculatus. Tissue abbreviations are as follows: Br, brain; YO, Y-organ; Es, eyestalk ganglia; AG, androgenic gland; Hp, hepatopancreas, Ht, heart; Ms, muscle; Te, testis; TG, thoracic ganglion; Ov, ovary. Vertical bars represent means ± SD (n=4). Data were analyzed by one-way ANOVA and post hoc Duncan’s multiple group analysis, with significant differences indicated by different letter labels (P < 0.05).
3.2 Effect of exogenous glucose on hemolymph glucose levels and PtILP expression
After injection of exogenous glucose, the hemolymph glucose levels at 5 min increased about 6.8 times than the saline injection group (Figure 5A). However, it seemed that the hemolymph glucose failed to return to the basal level in the subsequent 60 min, as the hemolymph glucose levels in glucose injection group at different time points were all significantly higher than in the control group, and there was no significant difference between each time point. We speculated that this may be due to the stress caused by repeated hemolymph collection in a short period of time, since the net change in Figure 5B showed the glucose levels fell from 5.41 mmol/L (5 min) to 3.49 mmol/L (15 min), 3.42 mmol/L (30 min), 3.82 mmol/L (60 min).
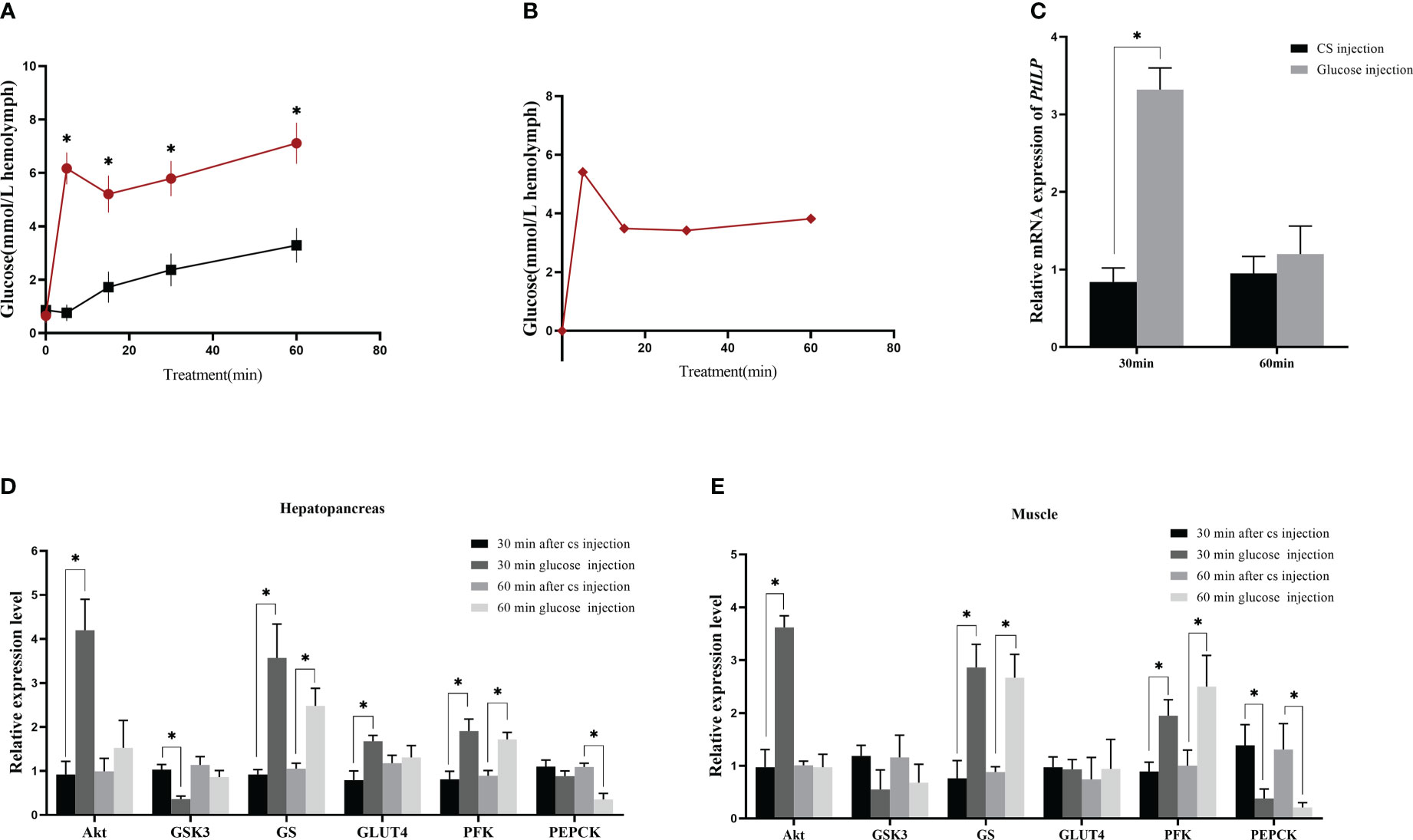
Figure 5 Effect of glucose injection in P. trituberculatus on hemolymph glucose and related gene expression. (A) Changes in hemolymph glucose levels after glucose injection. The “*” symbol mean difference between the glucose group and the CS group at the same time (B) Glucose levels in the hemolymph after glucose injection exclude crab saline (CS) injection changes. (C) Changes in PtILP expression in the eyestalk ganglia at 30 min and 60 min after glucose injection. (D) Changes in expression of genes related to glucose metabolism in hepatopancreas 30 min and 60 min after glucose injection. (E) Changes in expression of genes related to glucose metabolism in muscle at 30 min and 60 min after glucose injection. Data were analyzed by independent samples t-tests and significant differences was indicated by “*” (P < 0.05).
3.3 Changes in the expression of P. trituberculatus PtILP and glucose metabolism-related genes induced by exogenous glucose injection
The results of exogenous glucose injection experiment showed that the expression of PtILP in the experimental group was significantly higher than that in the control group 30 min post-glucose injection. However, there was no significant difference after 60 min (Figure 5C). We examined the changes in expression for genes related to glycogen synthesis and glucose uptake in both muscle and hepatopancreas. PtGSK3 was significantly decreased at 30 min, PtGS was significantly increased at both 30 min and 60 min, and PtGLUT4 was significantly increased at 30 min in hepatopancreas, while PtGSK3 and PtGLUT4 showed no significant difference in muscle. Furthermore, in both muscle and hepatopancreas, the glycolysis-related gene PtPFK increased significantly at 30 min, the gluconeogenesis-related gene PtPEPCK decreased significantly at 60 min, and the PtAkt gene in the insulin pathway also showed a significant increase at 30 min (Figures 5D, E).
3.4 Effect of PtILP silencing on hemolymph glucose and related genes
Compared with the control dsGFP, PtILP mRNA expression levels significantly decreased after 24 h of dsILP injection, and 64% silencing efficiency was reached (Figure 6A). After a brief knock-down of PtILP, expression levels of PtAkt, PtPFK and PtGS decreased sharply in the hepatopancreas, as did PtAkt and PtPFK in muscle, while the expression of PtGSK3 and PtPEPCK increased significantly in hepatopancreas, and PtPEPCK as well as PtGS was also up-regulated in muscle. Expression levels of PtGLUT4 were not significantly changed in muscle and hepatopancreas and neither were PtGSK3 transcript levels in muscle (Figures 5E, 6D). Hepatopancreas and muscle glycogen were dramatically reduced in the experimental group (Figure 6C). However, there was no notable change in hemolymph glucose levels (Figure 6B).
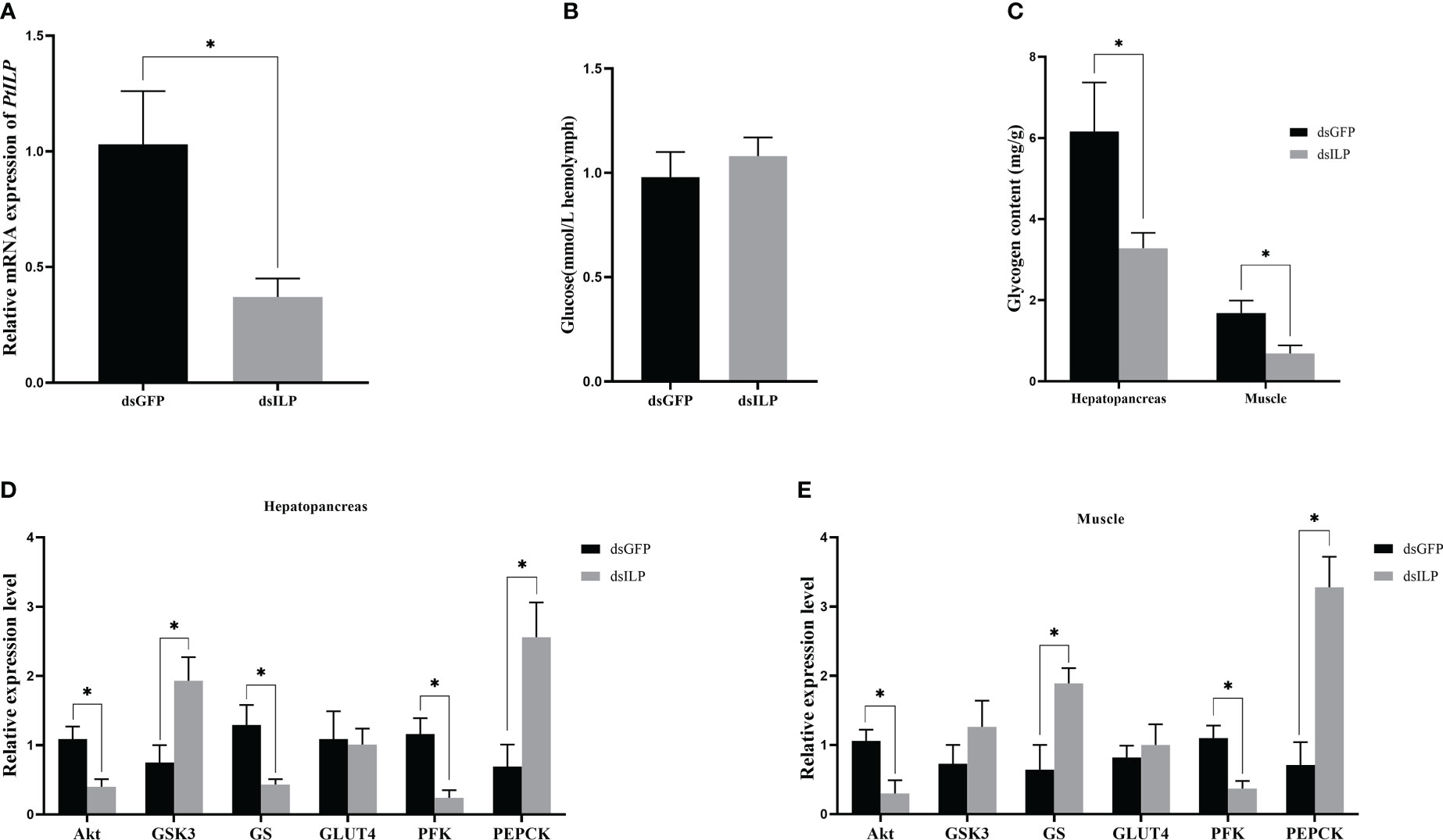
Figure 6 Effect of PtILP interference after 24h on hemolymph glucose, glycogen and related gene expression in P. trituberculatus. (A) Relative expression levels of PtILP in the eyestalk ganglia after RNA interference 24h. (B) Hemolymph glucose levels after 24h of PtILP interference. (C) Hepatopancreas and muscle glycogen content after 24h of PtILP interference. (D) Expression of genes related to glucose metabolism in the hepatopancreas after 24h of PtILP interference. (E) Expression of genes related to glucose metabolism in hepatopancreas and muscle after 24h of PtILP interference. Data were analyzed by independent samples t-tests and significant differences was indicated by “*” (P < 0.05).
3.5 Recombinant PtILP protein preparation and injection
The SDS-PAGE profile showed that the rGST and rGST-PtILP proteins were present after induction with IPTG compared to the non-induced strains. The molecular weights of the proteins were consistent with the expected molecular sizes (Figure 7A).
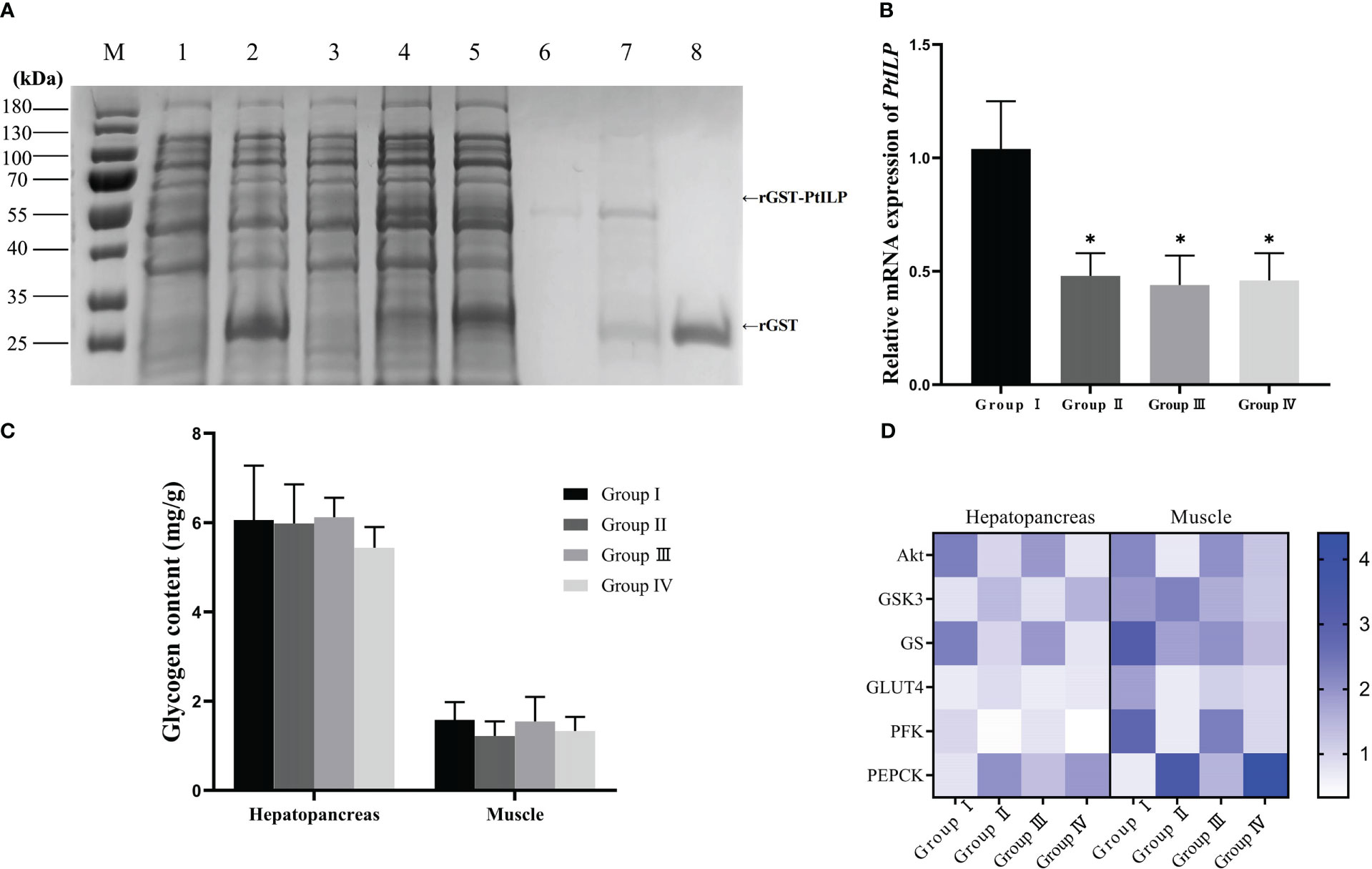
Figure 7 Effect of dsILP and recombinant PtILP injection on hemolymph glucose, glycogen and related gene expression in P. trituberculatus. (A) SDS-PAGE analysis of recombinant GST-PtILP. Protein marker (lane M); proteins of Rosetta gami2 (DE3) containing control vector pGEX-4t-2, non-induced (lane 1) and induced by 0.8mM IPTG (lane 2); proteins of Rosetta gami2 (DE3) containing recombinant pGEX-4P-2-PtILP, non-induced (lane 3) and induced by 0.8mM IPTG (lane 4); Supernatant of expressed recombinant bacterial extracts after ultrasonic fragmentation (lane 5) and precipitation (lane 6); purified rGST-PtILP protein (lane 7); purified GST (lane 8). (B) Relative expression levels of PtILP in the eyestalk ganglia after RNA interference 12h. Data were analyzed by one-way ANOVA and post hoc Duncan’s multiple group analysis, with significant differences indicated by “*” (P < 0.05). (C) Hepatopancreas and muscle glycogen content after dsILP and recombinant PtILP protein injections. (D) Hepatopancreas and muscle glycogen content after dsILP and recombinant PtILP protein injections.
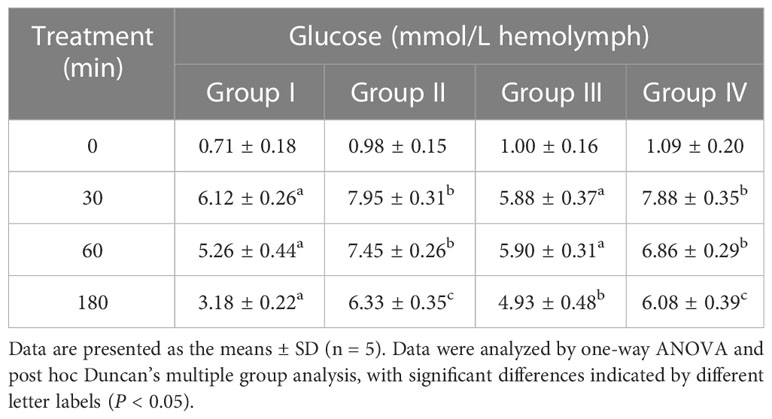
Glucose and rILP were injected 12 hours after the injection of glucose and dsPtILP (Figure 7B), and the changes in hemolymph glucose levels were detected. A significant difference could be seen at 30 min between the experimental group injected with rILP and the two control groups injected with rGST and PBS. At the same time, no statistically significant difference was noted when compared with Group I, which suggests that rILP may contribute to the reduction of glucose in the hemolymph. However, after 180 min treatment, hemolymph glucose levels in Group I was 3.18 mmol/L, and that of the group in which ILP was silenced followed by treatment with rILP, was 4.93 mmol/L, which indicated that endogenous ILP may be more efficient in hemolymph glucose removal compared to rILP (Table 1). There was no significant difference in glycogen content (Figure 7C)
The gene expression results showed that for hepatopancreas, PtAkt, PtGS and PtPFK expression in the rILP injected group was significantly higher than in the control group, while PtGSK3 was significantly lower than that in the control group. Furthermore, there was no obvious difference in PtGLUT4 as well as PtPEPCK expression between the two groups. In muscle, PtAkt and PtPFK expression levels were significantly higher than that in the control group, while PtPEPCK expression was significantly lower than that in the control group. No significant difference in expression was observed for the other genes (Figure 7D).
Discussion
In the present study, an insulin-like peptide gene was cloned from P. trituberculatus (PtILP) and amino acid sequence analysis showed that the encoded protein contained typical structural features of the insulin superfamily peptides which were signal peptide, B chain, C peptide and A chain (Wong et al., 2016). Furthermore, PtILP also has six conserved cysteine sites (Figure 3A), which form three disulfide bonds, of which two were inter-chain with one intra-chain (Andoh, 2016). Based on phylogenetic analysis, it was shown that PtILP primarily clustered with other crustaceans, and secondarily clustered with other members of the insulin superfamily of invertebrates (Figure 3B).
PtILP was mainly expressed at higher levels in neural tissues, including eyestalk ganglia and brain (Figure 4). In crustaceans, MrILP from M. rosenbergii (Jiang et al., 2020) and SvILP2 from S.verreauxi (Chandler et al., 2017) were also expressed mainly in the neural tissues, while in E.sinensis (Wang et al., 2020) and C.sapidus (Chung, 2014), ILPs expressed mainly in the hepatopancreas. In D. melanogaster, four DILPs (1, 2, 3, and 5) expressed in clusters of median neurosecretory cells (mNSCs) have blood sugar-regulating functions (Zhang et al., 2009). Other ILPs (DILP 6, 7) are mainly expressed in the fat body or midgut and are thought to function similarly to insulin-like growth factor (IGF) and relaxin (Brogiolo et al., 2001). This implies that PtILP, which is mainly expressed in neural tissues, has a potential insulin-like function. However, the ILPs of E. sinensis (Wang et al., 2020) and C. sapidus (Chung, 2014),which were expressed in the hepatopancreas, have also been reported to be involved in blood glucose regulation. This suggests that the mechanisms involved in the regulation of hemolymph glucose by ILPs may vary in different crustacean species and deserve further investigation.
In our study, after the injection of exogenous glucose (Figure 5B), P. trituberculatus hemolymph glucose levels decreased after a sharp climb which was similar to that previously reported for C. sapidus, L. vannamei, and M. rosenbergii (Chung, 2014; Jiang et al., 2020; Su et al., 2022), suggesting that a mechanism for regulating blood glucose may also exist in P. trituberculatus. In addition, PtILP mRNA levels increased rapidly at 30 min after glucose injection but decreased at 60 min (Figure 5C). This implies that P. trituberculatus may also be able to decrease its hemolymph glucose via PtILP.
Notably, there were no changes in hemolymph glucose levels after 24 h of dsILP injection (Figure 6B), and the observations were also reported for L. vannamei and M. rosenbergii (Jiang et al., 2020; Su et al., 2022). One possible reason for this is that the ILPs usually have a pattern for storage and secretion, and in the RNAi group, the stored ILP may be released into the hemolymph to downregulate glucose levels. Therefore, we pre-injected exogenous glucose in order to deplete the stored ILP in parallel with the RNAi treatment (Table 1). We observed that hemolymph glucose levels were significantly higher in the RNAi group (group II) than in the control GFP group (group I). The recovery test via rPtILP injection (group III) showed a significant decrease in hemolymph glucose levels when compared to the GFP group (group II) and GST injection group (group IV). The combined results confirmed the importance of PtILP in hemolymph glucose clearance for P. trituberculatus.
The IIS signaling pathway (insulin/insulin-like growth factor signaling pathway) is an evolutionarily conserved pathway that regulates various critical physiological functions (Semaniuk et al., 2018). It contains two major intracellular pathways: PI3K/Akt (phosphatidylinositol-3kinase/protein kinase B/Fork head box transcription factor) and Ras/MAPK (mitogen-activated protein kinase) pathways where the PI3K/Akt pathway regulates metabolism, while the MAPK pathway mainly controls development and reproduction (Sang et al., 2016). In mammals, insulin enhances glucose uptake, promotes glycogen synthesis, inhibits gluconeogenesis and facilitates the glycolytic process through the PI3K/Akt signaling pathway. Collectively, an organism’s energy homeostasis is regulated by insulin (Roach, 2002; Ferrer et al., 2003).
To investigate how PtILP clears hemolymph glucose, the expression levels of related genes in the IIS pathway were examined after glucose injection, RNA interference and injection of recombinant protein. For the glycogen synthesis-related genes, glycogen synthase kinase 3 (GSK3) is a ubiquitous serine/threonine kinase and glycogen synthase (GS) is an important rate-limiting enzyme, and together, they play an important role in glycogen metabolism (Wang et al., 2017). In this study, GSK3 expression was negatively correlated with PtILP levels and GS expression was positively correlated with PtILP levels in hepatopancreas. While somewhat different results were observed for muscle (Figure 6E), this was probably due to the fact that the regulatory mechanism of glycogen synthase is different in liver and muscle (Adeva-Andany et al., 2016). Meanwhile, the results from the glycogen assay after RNA interference showed that the hepatopancreas glycogen content in the inhibited group was significantly lower than the control group, and similar results were reported in M. rosenbergii (Jiang et al., 2020). Interestingly, a significant decrease in glycogen content in muscle was also observed after the interference (Figure 6C), implying that ILP also affects myoglycogen synthesis, but by a different mechanism than that in the hepatopancreas. These results suggest that PtILP may regulate hemolymph glucose by promoting glycogen synthesis.
In addition to the glycogen synthesis pathway, gluconeogenesis and glycolysis are two metabolic mechanisms responsible for maintaining glucose homeostasis. In all organisms, glycolysis is the only pathway for glucose catabolism, and phosphofructokinase (PFK) is a central rate-limiting enzyme in the process of glycolysis (Wegener and Krause, 2002). Gluconeogenesis is the reverse pathway of glycolysis that allows cells to produce endogenous glucose from pyruvate and gluconeogenic amino acids, and phosphoenolpyruvate carboxykinase (PEPCK) is an important gluconeogenesis rate-limiting enzyme (Yabaluri and Bashyam, 2010). In this study, PtPEPCK expression was negatively correlated with PtILP levels, while PtPFK expression was positively correlated with PtILP levels. This study has shown that PtILP is involved in the gluconeogenesis and glycolytic pathways and may reduce hemolymph glucose levels by inhibiting gluconeogenesis and promoting glycolysis, similar to that reported for M. rosenbergii, L. vannamei, and Carassius gibelio (Jin et al., 2018; Jiang et al., 2020; Su et al., 2022). In terms of glucose transport, the PI3K/Akt signaling pathway enhances GLUT4 transport to the cell surface to stimulate glucose uptake. However, changes in GLUT4 expression levels in our study were only noted in the hepatopancreas 30 min after glucose injection (Figure 5C), with no significant changes in other treatments. As GLUT4 secretion and storage in vesicles is primarily for glucose transport by via exocytosis, the gene may not be regulated at the mRNA level (Li et al., 2017).
In conclusion, the P. trituberculatus ILP was identified, cloned and expressed in the recombinant form. The function of PtILP in reducing glucose levels in P. trituberculatus hemolymph was confirmed by experiments involving glucose injection, RNA interference and recombinant protein injection into the swimming crabs. Based on the expression profile of genes related to the IIS pathway, we inferred that PtILP might negatively regulate hemolymph glucose levels via the IIS pathway by inhibiting gluconeogenesis and promoting glycogen synthesis and glycolysis. These findings provide vital clues to the function of crustacean ILP in crustacean glucose metabolism.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics statement
The animal study was reviewed and approved by Committee on the Ethics of Animal Experiments of the Ningbo University.
Author contributions
RX: conceptualization, sample collection, experiment, data analysis and writing-original draft. ST and MW: experiment. XX and DZ: editing and supervision. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National natural Science Foundation of China (Nos. 41776165 and 31802265), Natural Science Foundation of Zhejiang province (LY20C190004), and the K. C. Wong Magna Fund in Ningbo University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1144781/full#supplementary-material
References
Adeva-Andany M. M., González-Lucán M., Donapetry-García C., Fernández-Fernández C., Ameneiros-Rodríguez E. (2016). Glycogen metabolism in humans. BBA Clin. 5, 85–100. doi: 10.1016/j.bbacli.2016.02.001
Al Baki M. A., Lee D.-W., Jung J. K., Kim Y. (2019). Insulin-like peptides of the legume pod borer, Maruca vitrata, and their mediation effects on hemolymph trehalose level, larval development, and adult reproduction. Arch. Insect Biochem. Physiol. 100 (2), e21524. doi: 10.1002/arch.21524
Andoh T. (2016). “Chapter 19 - insulin family,” in Handbook of hormones. Eds. Takei Y., Ando H., Tsutsui K. (San Diego: Academic Press), 155–e119-151.
Brogiolo W., Stocker H., Ikeya T., Rintelen F., Fernandez R., Hafen E. (2001). An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 11 (4), 213–221. doi: 10.1016/S0960-9822(01)00068-9
Broughton S., Alic N., Slack C., Bass T., Ikeya T., Vinti G., et al. (2008). Reduction of DILP2 in Drosophila triages a metabolic phenotype from lifespan revealing redundancy and compensation among DILPs. PLoS One 3 (11), e3721. doi: 10.1371/journal.pone.0003721
Brown M. R., Clark K. D., Gulia M., Zhao Z., Garczynski S. F., Crim J. W., et al. (2008). An insulin-like peptide regulates egg maturation and metabolism in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. 105 (15), 5716–5721. doi: 10.1073/pnas.0800478105
Chandler J. C., Gandhi N. S., Mancera R. L., Smith G., Elizur A., Ventura T. (2017). Understanding insulin endocrinology in decapod Crustacea: Molecular modelling characterization of an insulin-binding protein and insulin-like peptides in the Eastern spiny lobster, Sagmariasus verreauxi. Int. J. Mol. Sci. 18 (9), 1832. doi: 10.3390/ijms18091832
Chen H.-Y., Toullec J.-Y., Lee C.-Y. (2020). The crustacean hyperglycemic hormone superfamily: Progress made in the past decade. Front. Endocrinol. 11. doi: 10.3389/fendo.2020.578958
Cherif–Feildel M., Heude Berthelin C., Adeline B., Rivière G., Favrel P., Kellner K. (2019). Molecular evolution and functional characterisation of insulin related peptides in molluscs: Contributions of Crassostrea gigas genomic and transcriptomic-wide screening. Gen. Comp. Endocrinol. 271, 15–29. doi: 10.1016/j.ygcen.2018.10.019
Chowański S., Walkowiak-Nowicka K., Winkiel M., Marciniak P., Urbański A., Pacholska-Bogalska J. (2021). Insulin-like peptides and cross-talk with other factors in the regulation of insect metabolism. Front. Physiol. 12. doi: 10.3389/fphys.2021.701203
Chung J. S. (2014). An insulin-like growth factor found in hepatopancreas implicates carbohydrate metabolism of the blue crab Callinectes sapidus. Gen. Comp. Endocrinol. 199, 56–64. doi: 10.1016/j.ygcen.2014.01.012
de Brito Sanchez G., Expósito Muñoz A., Chen L., Huang W., Su S., Giurfa M. (2021). Adipokinetic hormone (AKH), energy budget and their effect on feeding and gustatory processes of foraging honey bees. Sci. Rep. 11 (1), 18311. doi: 10.1038/s41598-021-97851-x
Defferrari M. S., Orchard I., Lange A. B. (2016). An insulin-like growth factor in Rhodnius prolixus is involved in post-feeding nutrient balance and growth. Front. Neurosci. 10. doi: 10.3389/fnins.2016.00566
Ferrer J. C., Favre C., Gomis R. R., Fernández-Novell J. M., García-Rocha M., de la Iglesia N., et al. (2003). Control of glycogen deposition. FEBS Lett. 546 (1), 127–132. doi: 10.1016/s0014-5793(03)00565-9
Gallardo N., Carrillo O., Molto E., Deas M., Gonzalez-Suarez R., Carrascosa J. M., et al. (2003). Isolation and biological characterization of a 6-kDa protein from hepatopancreas of lobster panulirus argus with insulin-like effects. Gen. Comp. Endocrinol. 131 (3), 284–290. doi: 10.1016/S0016-6480(03)00014-5
Jiang Q., Jiang Z., Gu S., Qian L., Li X., Gao X., et al. (2020). Insights into carbohydrate metabolism from an insulin-like peptide in Macrobrachium rosenbergii. Gen. Comp. Endocrinol. 293, 113478. doi: 10.1016/j.ygcen.2020.113478
Jin J., Yang Y., Zhu X., Han D., Liu H., Xie S. (2018). Effects of glucose administration on glucose and lipid metabolism in two strains of gibel carp (Carassius gibelio). Gen. Comp. Endocrinol. 267, 18–28. doi: 10.1016/j.ygcen.2018.05.023
Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596 (7873), 583–589. doi: 10.1038/s41586-021-03819-2
Kim Y., Hong Y. (2015). Regulation of hemolymph trehalose level by an insulin-like peptide through diel feeding rhythm of the beet armyworm, Spodoptera exigua. PEPTIDES 68, 91–98. doi: 10.1016/j.peptides.2015.02.003
Li R., Tian J.-Z., Wang M.-R., Zhu L.-N., Sun J.-S. (2017). EsGLUT4 and CHHBP are involved in the regulation of glucose homeostasis in the crustacean Eriocheir sinensis. Biol. Open 6 (9), 1279–1289. doi: 10.1242/bio.027532
Miyazaki M., Hayata M., Samukawa N., Iwanaga K., Nagai J. (2020). Pharmacokinetic–pharmacodynamic modelling of the hypoglycaemic effect of pulsatile administration of human insulin in rats. Sci. Rep. 10 (1), 18876. doi: 10.1038/s41598-020-76007-3
Nagasawa H., Kataoka H., Isogai A., Tamura S., Suzuki A., Ishizaki H., et al. (1984). Amino-terminal amino acid sequence of the silkworm prothoracicotropic hormone: homology with insulin. Sci. (New York N.Y.) 226 (4680), 1344–1345. doi: 10.1126/science.226.4680.1344
Nelson L. E., Sheridan M. A. (2006). Gastroenteropancreatic hormones and metabolism in fish. Gen. Comp. Endocrinol. 148 (2), 116–124. doi: 10.1016/j.ygcen.2006.01.011
Roach P. J. (2002). Glycogen and its metabolism. Curr. Mol. Med. 2 (2), 101–120. doi: 10.2174/1566524024605761
Sanders B. (1983). Insulin-like peptides in the lobster Homarus americanus. III. no glucostatic role. Gen. Comp. Endocrinol. 50 (3), 378–382. doi: 10.1016/0016-6480(83)90259-9
Sang M., Li C., Wu W., Li B. (2016). Identification and evolution of two insulin receptor genes involved in Tribolium castaneum development and reproduction. Gene 585 (2), 196–204. doi: 10.1016/j.gene.2016.02.034
Semaniuk U. V., Gospodaryov D. V., Feden'ko K. M., Yurkevych I. S., Vaiserman A. M., Storey K. B., et al. (2018). Insulin-like peptides regulate feeding preference and metabolism in Drosophila. Front. Physiol. 9. doi: 10.3389/fphys.2018.01083
Semaniuk U., Piskovatska V., Strilbytska O., Strutynska T., Burdyliuk N., Vaiserman A., et al. (2021). Drosophila insulin-like peptides: from expression to functions - a review. Entomologia Experimentalis Applicata 169 (2), 195–208. doi: 10.1111/eea.12981
Su M., Zhang X., Yuan J., Zhang X., Li F. (2022). The role of insulin-like peptide in maintaining hemolymph glucose homeostasis in the pacific white shrimp Litopenaeus vannamei. Int. J. Mol. Sci. 23 (6), 3268. doi: 10.3390/ijms23063268
Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., et al. (2021). AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50 (D1), D439–D444. doi: 10.1093/nar/gkab1061
Wang L., Chen H., Wang L., Song L. (2020). An insulin-like peptide serves as a regulator of glucose metabolism in the immune response of Chinese mitten crab Eriocheir sinensis. Dev. Comp. Immunol. 108, 103686. doi: 10.1016/j.dci.2020.103686
Wang Y., Wang Y., Zhong T., Guo J., Li L., Zhang H., et al. (2017). Transcriptional regulation of pig GYS1 gene by glycogen synthase kinase 3β (GSK3β). Mol. Cell. Biochem. 424 (1), 203–208. doi: 10.1007/s11010-016-2856-1
Wegener G., Krause U. (2002). Different modes of activating phosphofructokinase, a key regulatory enzyme of glycolysis, in working vertebrate muscle. Biochem. Soc. Trans. 30 (2), 264–270. doi: 10.1042/bst0300264
Wong Y. H., Yu L., Zhang G., He L. S., Qian P. Y. (2016). In silico prediction of Neuropeptides/Peptide hormone transcripts in the cheilostome bryozoan Bugula neritina. PLoS One 11 (8), e0160271. doi: 10.1371/journal.pone.0160271
Xie X., Liu Z., Liu M., Tao T., Shen X., Zhu D. (2016). Role of Halloween genes in ecdysteroids biosynthesis of the swimming crab (Portunus trituberculatus): Implications from RNA interference and eyestalk ablation. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 199, 105–110. doi: 10.1016/j.cbpa.2016.06.001
Yabaluri N., Bashyam M. D. (2010). Hormonal regulation of gluconeogenic gene transcription in the liver. J. Biosci. 35 (3), 473–484. doi: 10.1007/s12038-010-0052-0
Yohei M., Tsuyoshi K. (2018). The C. elegans insulin-like peptides (ILPs). AIMS Biophysics 5 (4), 217–230. doi: 10.3934/biophy.2018.4.217
Keywords: Portunus trituberculatus, insulin-like peptide, glucoregulation, IIS pathway, RNAi
Citation: Xu R, Wang M-e, Tu S, Xie X and Zhu D (2023) Molecular identification of an insulin-like peptide from the swimming crab Portunus trituberculatus and evidence for its glucoregulation function. Front. Mar. Sci. 10:1144781. doi: 10.3389/fmars.2023.1144781
Received: 15 January 2023; Accepted: 24 February 2023;
Published: 08 March 2023.
Edited by:
Rui Jia, Freshwater Fisheries Research Center (CAFS), ChinaReviewed by:
Jie Gong, Nantong University, ChinaJorge Felipe Argenta Model, Federal University of Rio Grande do Sul, Brazil
Copyright © 2023 Xu, Wang, Tu, Xie and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xi Xie, eGlleGlAbmJ1LmVkdS5jbg==; Dongfa Zhu, emh1ZG9uZ2ZhQG5idS5lZHUuY24=
 Rui Xu
Rui Xu Meng-en Wang
Meng-en Wang Shisheng Tu
Shisheng Tu Xi Xie
Xi Xie Dongfa Zhu
Dongfa Zhu