
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 07 March 2023
Sec. Aquatic Physiology
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1138074
This article is part of the Research Topic Integration of Development, Physiology and Responses to Environmental Change in Aquatic Invertebrates View all 13 articles
 Cong Zhou1,2,3,4,5,6
Cong Zhou1,2,3,4,5,6 Zhi-shu Lin7
Zhi-shu Lin7 Ying Shi7
Ying Shi7 Jie Feng8
Jie Feng8 Zhi Hu1,2,3,4,5,6
Zhi Hu1,2,3,4,5,6 Mei-jie Yang1,2,3,4,5,6
Mei-jie Yang1,2,3,4,5,6 Pu Shi1,2,3,4,5,6
Pu Shi1,2,3,4,5,6 Yong-ren Li9
Yong-ren Li9 Yong-jun Guo9
Yong-jun Guo9 Tao Zhang1,2,3,4,6*
Tao Zhang1,2,3,4,6* Hao Song1,2,3,4,6*
Hao Song1,2,3,4,6*Aquaporins (AQPs) are highly-selective transmembrane water transporters that are involved in the adaptation to environmental challenges. However, the structure, function, and evolution of AQPs in bivalves remain largely unknown. In this study, AQP gene family members were identified in nine bivalve species, and their abundance rangs from 7 to 15. Nine AQPs (MmAQPs) were identified in the genome of hard clam (Mercenaria mercenaria), which is a euryhaline bivalve that has evolved sophisticated osmoregulatory mechanisms and salinity adaptation. Structurally, all MmAQPs contain 6 or 12 transmembrane α-helices, a major intrinsic protein (MIP) domain, and 2 asparagine-proline-alanine (NPA) motifs. MmAQPs were classified into three subfamilies based on phylogenetic analysis: AQP1-like, AQP3-like, and AQP8-like. No AQP11-like subfamily member was identified in the genome of hard clam. Tandem duplication resulted in a lineage-specific expansion in AQP8-like subfamily in hard clams. MmAQP8 genes showed different expression sensitivity to different environmental stressors. The gene expression patterns of three MmAQP8 were similar under heat, hypoxia, and air exposure stress, but differed greatly under salinity stress, indicating that tandem duplication events may accelerate the functional divergence of AQP8 genes in hard clams. AQP3-like members may have undergone gene loss during evolution, resulting in weakened glycerol and urea penetration in hard clams. Three orthologs of MmAQPs were detected in the genomes of Cyclina sinensis and Archivesica marissinica through synteny analyses. Tissue expression profiles showed that MmAQP genes were highly expressed in the foot and hepatopancreas. Under environmental stress, the expression levels of most of the MmAQP genes changed significantly to maintain metabolic homeostasis. Several MmAQP genes were downregulated to reduce water permeability under salinity and air exposure stress. Several MmAQP genes were significantly upregulated to promote the transmembrane transport of ammonia and reactive oxygen species and activate anti-apoptotic responses to resist stress. This study provides a comprehensive understanding of the AQP gene family in hard clams, and lays a foundation for further studies to explore the functions of AQPs in bivalves.
The water permeability of cell membranes is highly dependent on the presence of water channel proteins, also known as aquaporins (AQPs). In 1988, the first AQP was isolated from human erythrocytes and changed the concept of transmembrane water transport from involving simple diffusion across the lipid bilayer to more strictly-regulated processes (Denker et al., 1988; Preston et al., 1992). Structurally, AQPs are tetrameric membrane proteins composed of four identical 30-kDa monomers (Benga, 2012). Each monomers harbor a central water-transporting pore surrounded by six transmembrane α-helices (Gonen and Walz, 2006). The pore structure determines the substrate selectivity of AQPs and is characterised by two highly-conserved asparagine-proline-alanine (NPA) motifs (Törnroth-Horsefield et al., 2006; Ishibashi et al., 2011). The pore structure allows different small molecules, including water, glycerol, ammonia, carbon dioxide, urea, and hydrogen peroxide, to specifically penetrate the biological membranes (Krane and Goldstein, 2007; Groszmann et al., 2017). Based on the distinct sequence characteristics and substrate permeability, AQPs can be classified into four subfamilies: AQP1, AQP3, AQP8, and AQP11-like. AQP1-like subfamily consists of AQP0, 1, 2, 4, 5 and 6, which can only permeate water. AQP3-like subfamily members are AQP3, 7, 9, and 10, which can permeate glycerol, urea, and ammonia. AQP8-like subfamily member (AQP8) shows poor ability to transport water across membranes. AQP11 and 12 are the members of AQP11-like subfamily, which are also known as unorthodox AQPs and are characterized by highly-variable NPA motifs (Soto et al., 2012; FinnCerda, 2015).
AQPs are present in almost all the kingdoms of life (Heymann and Engel, 1999), among which mammalian AQPs have been the most extensively studied. To date, 13 AQPs (AQP0–AQP12) have been identified in humans (Ishibashi et al., 2009). Increasing evidence has suggested that human AQPs are involved in osmoregulation, lipid metabolism, cell adhesion and migration, and celluar stress responses (Hara-ChikumaVerkman, 2006; Maeda et al., 2008; Ishibashi et al., 2011; Tamma et al., 2018). With the increasing availability of genome and transcriptome sequencing projects, numerous homologues of human AQPs have been identified in aquatic animals (Cao and Shi, 2019). For instance, 37 AQPs have been identified in the fish Cyprinus carpio (Dong et al., 2016). The transcriptional regulation of AQPs has been found to be closely associated with environmental salinity changes in the crab Portunus trituberculatus and shrimp Litopenaeus vannamei (Wang et al., 2015). These studies imply that AQPs are important elements for osmoregulation and salinity adaptation in aquatic animals.
Bivalves are the second class of mollusca and one of the oldest and most evolutionarily-successful groups of invertebrates (Wang et al., 2013). Bivalves are osmoconformers whose hemolymph osmolality changes rapidly with environmental salinity fluctuations (Koprivnikar and Poulin, 2009; Sokolov and Sokolova, 2019). The osmotic gradient between hemolymph and tissues causes cellular swelling or shrinkage and disrupts normal cellular functions. Bivalves can maintain osmotic equilibrium via regulation of water transport and intracellular osmolytes, such as free amino acids, glycerol, and various inorganic ions (Berger and Kharazova, 1997). Hence, bivalves provide suitable models to investigate the evolutionary diversification and roles of AQPs in mollusca due to their characteristic requirement for sustained water homeostasis. To date, AQPs in mollusca have been studied mainly in gastropods. The first AQPs in gastropods was reported more than two decades later after the discovery of human AQPs (Pieńkowska et al., 2014). Genome-wide identification of AQPs and their structural and functional diversity have been reported in abalone Haliotis discus hannai (Jia et al., 2022). In bivalves, knowledge of AQPs remains largely limited, with only a few studies suggesting that their gene expression is related to salinity stress. For instance, a transcriptomic study reported that 7 days of hyposalinity stress induced a dwonregulation in AQP gene expression in Crassostrea gigas (Meng et al., 2013). Three cDNAs encoding AQPs (AQP1, AQP8, and AQP11) were cloned from Sinonovacula constricta, and their expression levels were significantly upregulated under salinity stress (Ruan et al., 2022). With the completion of the assembly of the reference genomes in various bivalve species, it is necessary to study the structure, function and evolution of the AQP gene family in bivalves, especially the euryhaline species that have evolved sophisticated osmoregulatory mechanisms and salinity adaptation.
The hard clam (Mercenaria mercenaria) is a euryhaline bivalve species that naturally lives along the eastern coasts of the United States and Canada. Hard clams have emerged as an important pond-farmed bivalves since they have been imported into China in 1997. Salinity, temperatures and dissolved oxygen are major abiotic factors that affect the physiological status of pond-farmed hard clams. Hard clam is well-known for its “hardiness” and shows strong adaptations to salinity and other environmental stressors (Song et al., 2021). Hard clam may represent an attractive model for studying the osmoregulation and stress resistance mechanisms in bivalves. Previous studies have reported that the powerful anti-apoptotic system (Song et al., 2021), massive expansion of the heat shock protein 70 gene family (Hu et al., 2022b), and transmembrane transport of inorganic ions and free amino acids (Zhou et al., 2022) are vital stress resistance mechanisms in hard clams. However, AQP gene family and AQP-related osmoregulation and stress responses remain largely unknown in this hardy species. In this study, genome-wide identification and phylogenetic analysis of AQP gene family were performed in hard clam and eight other bivalve species. The AQPs in hard clams (MmAQPs) were selected to investigate the chromosomal locations, gene and protein structures, and evolutionary characteristics. To investigate the potential involvement of MmAQPs in stress resistance, the expression patterns of MmAQPs under salinity, air exposure, heat and hypoxia stress were examined using transcriptome data. This study provides a comprehensive understanding of AQP gene family in hard clams, and lays a foundation for further research to expolre the functions of AQPs in bivalves.
The whole-genome protein sequences of M. mercenaria were downloaded from Figshare (https://Figshare.com/s/a8378910b437fc843a46). The whole-genome protein sequences of C. gigas, C. virginica, Pinctada fucata, Argopecten purpuratus, Chlamys farreri, Patinopecten yessoensis, Modiolus philippinarum, and Sinonovacula constricta were downloaded from the MolluscDB database (http://mgbase.qnlm.ac/home). The known full-length AQPs sequences in Homo sapiens (13), Danio rerio (10), H. discus hannai (18), C. gigas (3), C. hongkongensis (2), C. virginica (1), Ostrea edulis (3), and Sinonovacula constricta (3) were downloaded from the UniProt database (https://www.uniprot.org/). These sequences were used as a query database to conduct a BLASTP search against the M. mercenaria genome, with a cut-off E-value of 1E-10 and identity of 30%. The hidden Markov model (HMM) profile of the conserved major intrinsic protein (MIP) domain (PF00230) was downloaded from the UniProt database. The putative M. mercenaria AQPs (MmAQPs) were identified using HMMER software (v3.3), with a cut-off E-value of 1E-5. The protein sequences that showed high similarities with the query sequences and conserved MIP domain were identified as candidate MmAQPs. The NCBI CD-Search tool (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) was used to confirm the reliability of candidate MmAQPs (Marchler-Bauer et al., 2015). Given the molecular weights (MWs) of the water channel monomers (26–34 kDa), the MmAQPs that comprised more than 200 amino acids were selected for further analysis (Heymann and Engel, 1999). To further study the evolutionary relationship of AQPs in bivalves, AQP gene family members were also identified from the genomes of C. gigas, C. virginica, P. fucata, A. purpuratus, C. farreri, P. yessoensis, M. philippinarum, and S. constricta using the same methods described above.
To classify MmAQPs, a multiple sequence alignment of the MmAQPs and other known AQPs in H. sapiens, D. rerio and Caenorhabditis elegans was carried out using ClustalW algorithm. An unrooted phylogenetic tree was constructed using maximum-likelihood (ML) algorithm in MEGA 11.0 software (Tamura et al., 2021). The MmAQPs were named based on the phylogenetic tree branching, BLASTP search against Swiss-Prot database, and gene positions on chromosomes. The subfamily classification and protein nomenclature of the AQPs in eight other bivalve species were performed using the same methods. A multiple sequence alignment of all bivalve AQPs was performed using ClustalW, and a phylogenetic tree was constructed using neighbor-joining (NJ) algorithm in MEGA 11.0. We further constructed an NJ tree of MmAQPs used for further analysis. To assess the reliability, the bootstrap value of each phylogenetic tree was set at 1,000 replications. The phylogenetic trees were imported into the iTOL website for embellishment (https://itol.embl.de/itol.cgi).
The MWs, isoelectric points (pI), and grand averages of hydropathicity (GRAVY) of the MmAQPs were predicted using ExPASy-ProtParam tool (https://web.expasy.org/protparam/). The transmembrane structures of the MmAQPs were predicted and analyzed using DeepTMHMM software (https://dtu.biolib.com/DeepTMHMM). The subcellular localization of the MmAQPs was predicted using WoLF PSORT online software (https://wolfpsort.hgc.jp/).
The genome annotation file (GFF3 format) of M. mercenaria was downloaded from Figshare (https://Figshare.com/s/a8378910b437fc843a46). Based on the genome annotation information, the chromosomal localization of MmAQPs was visualized using the gene location visualize from GTF/GFF tool in TBtools software (v1.1043) (Chen et al., 2020). The whole-genome protein sequences of hard clams were compared in pairs using DIAMOND software with the cut-off max-target-seqs of 5 and E-value of 1E-5 (Buchfink et al., 2015). MCScanX software was used to search for tandemly-duplicated and collinear gene pairs (Wang et al., 2012). The collinear gene pairs were visualized using the advanced circos tool in TBtools.
Gene structures, including untranslated regions (UTR), coding sequences (CDS), and introns, were displayed using GSDS 2.0 (http://gsds.gao-lab.org/). To better understand the structural diversity of MmAQPs, a conserved motif analysis of MmAQPs was conducted using MEME (v5.5.0) with a maximum number of motifs of 10 (https://meme-suite.org/meme/tools/meme) (Bailey et al., 2015). The results were visualized using the simple MEME wrapper tool in TBtools.
To detect the conserved NPA motifs in MmAQPs, a multiple sequence alignment was performed using MUSCLE algorithm in MEGA 11.0. The secondary structures of MmAQPs were predicted using SOPMA web tool (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html) with a cut-off similarity threshold of 8. The three-dimensional (3D) tertiary structures of MmAQPs were predicted using SWISS-MODEL (https://swissmodel.expasy.org/), which is a fully-automated protein structure homology-modelling online tool (Arnold et al., 2006). The 3D structures of MmAQPs were visually edited and mapped using PyMOL molecular graphics system (v2.2.0).
Synteny analyses were performed to detect the orthologs of MmAQPs in four chromosome-level bivalve genomes. The genome sequence and annotation file of P. yessoensis were downloaded from the MolluscDB database. The genome sequence and annotation file of C. gigas were downloaded from the NCBI genome database (https://www.ncbi.nlm.nih.gov/genome/?term=Crassostrea+gigas). The genome sequence of Cyclina sinensis genome was downloaded from NCBI (project PRJNA612143), and the genome annotation file was obtained from the DRYAD website (https://doi.org/10.5061/dryad.44j0zpcb5). The genome sequence and annotation file of Archivesica marissinica were downloaded from Figshare (https://figshare.com/articles/dataset/Host-Endosymbiont_Genome_Integration_in_a_Deep-Sea_Chemosymbiotic_Clam/12198987). Diamond software was used to perform a two-way BLASTP analysis between M. mercenaria and P. yessoensis, M. mercenaria and C. gigas, M. mercenaria and C. sinensis, and M. mercenaria and A. marissinica, with a cut-off max-target-seqs of 5 and E-value of 1E-5. Then MCScanX software was used to screened out the collinear large fragments consisting of multiple neighboring collinear gene pairs in two genomes. According to the gene ID of MmAQPs, the collinear AQP gene pairs in two genomes were identified and visualized using the systeny plot for MCScanX tool in TBtools.
The gills, mantles, feet, adductor muscles, hemolymph, liver, stomach, intestines, testes, and ovaries were sampled from the healthy hard clams under suitable living conditions (salinity: 30 ± 0.5 psu, temperature: 15 ± 1 °, dissolved oxygen: 8.0 ± 1.0 mg/L). The Illumina paired-end transcriptome libraries of these tissues were constructed in our previous study (Song et al., 2021). Raw data were obtained from the NCBI SRA database (accession number: PRJNA596049). The raw data were pre-processed to remove adapters, poly-N, and low-quality reads using Cutadapt (v3.3). After performing a quality control check using FastQC (v0.11.8), the high-quality reads were mapped to the M. mercenaria reference genome using TopHat (v2.0.12) with default parameters. HTSeq (v0.6.1) was used to count the number of reads that were mapped to each gene. FPKM value represented the gene expression levels and was calculated based on the length of the gene and its read count. The expression levels of MmAQP genes in the different tissues were standardized by Z-scores and visualized using pheatmap package in R (v4.2.2).
To clarify the role of MmAQPs in osmoregulation and stress responses, hard clams were exposed to various environmental stressors, including hyposalinity, hypersalinity, air exposure, heat, and hypoxia. Gills were selected as the target tissue because they can exchange substances with the external environment and are vital for stress resistance in bivalves (Hosoi et al., 2007). Moderate and severe hyposalinity were set at 15 practical salinity units (psu) and 5 psu, respectively. The hypersalinity was set at 40 psu (Song et al., 2021). Four groups were established in this experiment: 30 psu (control group, S30), 5 psu (S5), 15 psu (S15), and 40 psu (S40). After 10 days of salinity stress, gills were aseptically dissected from the clams and immediately stored at –80°C. To examine the effects of air exposure stress on MmAQPs, hard clams were exposed to air in a thermostatic incubator at 15°C and 50% humidity. Gills were dissected from the clams on days 0 (control group, PreAE), day 8 (PostAE8), and day 16 (PostAE16). Additional experimental details are available in Song et al. (2021). In heat and hypoxia stress experiments, the temperature was set to 35°, and the dissolved oxygen concentration was set to 0.2 mg/L (Hu et al., 2022a). Four groups were established in this experiment: 20°, 6 mg/L (control group, C_6); 35°, 6 mg/L (heat group, H_6); 20°, 0.2 mg/L (hypoxia group, C_02); and 35°, 0.2 mg/L (combined heat and hypoxia group, H_02). After three days of stress treatments, gills were dissected from the clams and stored at –80°. Further experimental details can be found in Hu et al. (2022a). Each experimental group had three biological replicates. The frozen gill samples were used for RNA extraction and transcriptome sequencing. Raw data were obtained from the NCBI SRA database (accession number: PRJNA596049, PRJNA764366, and PRJNA764372). The quality control for the reads, genome mapping, reads counts, and gene expression level calculations were performed using the same methods described in section 2.8. The expression levels of MmAQP genes under various environmental stressors were standardized and visualized using pheatmap package in R.
Statistical analyses were performed using IBM SPSS Statistics 25 (IBM Corp., US). To evaluate statistical differences, the expression levels of MmAQP genes were tested using one-way analysis of variance with Tukey’s test. P values less than 0.05 were considered to be statistically significant.
Nineteen potential AQPs were screened out from the genome of hard clam through BLASTP analysis. Sixteen of the nineteen potential AQPs comprised typical MIP domains. Moreover, seven AQPs were filtered out because their MWs were much smaller than those of other known AQPs. Therefore, nine AQPs were identified in the genome of hard clam (MmAQPs). Additionally, we identified 13 AQPs in C. gigas, 15 AQPs in C. virginica, 7 AQPs in P. fucata, 10 AQPs in A. purpuratus, 9 AQPs in C. farreri, 11 AQPs in P. yessoensis, 9 AQPs in M. philippinarum, and 12 AQPs in S. constricta.
To classify MmAQPs, an unrooted phylogenetic tree was constructed based on the AQPs from the hard clam and three model organisms, including H. sapiens, D. rerio, and C. elegans (Figure 1A). The tree consisted of four distinct clades, representing four subfamilies. MmAQPs and their homologues in model organisms were clustered on the same branch. Hence, MmAQPs could be classified into three subfamilies: AQP1-like (MmAQP1_1, MmAQP1_2, MmAQP4, MmAQP5_1, and MmAQP5_2), AQP3-like (MmAQP3), and AQP8-like (MmAQP8_1, MmAQP8_2, and MmAQP8_3). A lineage-specific expansion was observed in AQP8-like subfamily in hard clams. However, AQP11-like subfamily members were not identified in the genome of hard clams.
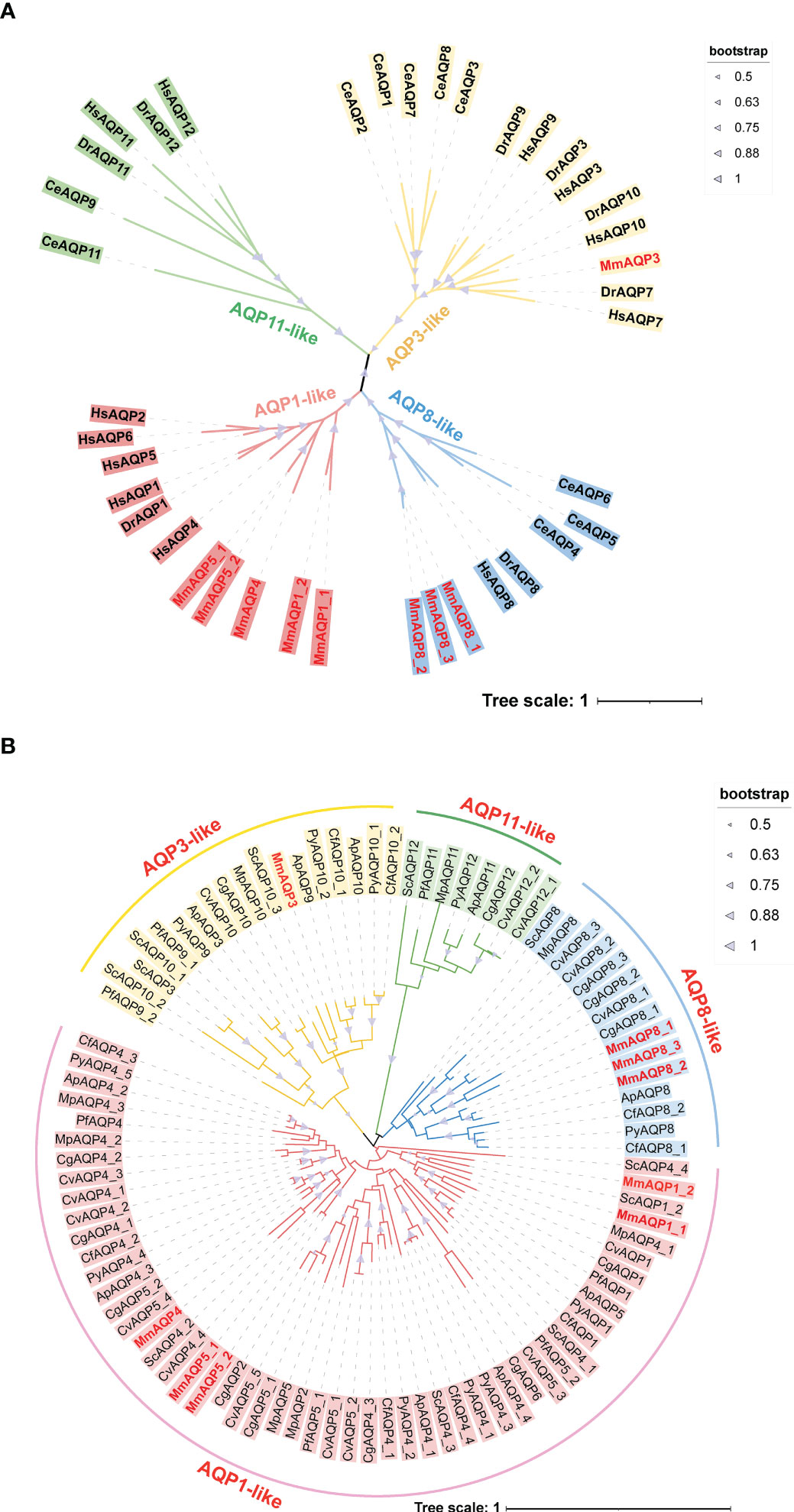
Figure 1 Phylogenetic analysis of MmAQPs and other known AQPs in three model organisms (A), and the AQPs identified in nine bivalves (B). The species names are abbreviated as follows, H. sapiens, Hs; D. rerio, Dr; C. elegans, Ce; C. gigas, Cg; C. virginica, Cv; P. fucata, Pf; A. purpuratus, Ap; C. farreri, Cf; P. yessoensis, Py; M. philippinarum, Mp; and S. constricta, Sc. AQP3-like, AQP1-like, AQP8-like, and AQP11-like subfamily members are marked in yellow, pink, blue, and green, respectively. The MmAQPs are highlighted in red. The bootstrap values greater than 50 are marked with purple triangles.
Another phylogenetic tree was constructed based on 95 AQPs that were identified in nine bivalves to investigate their evolutionary relationships (Figure 1B). The AQPs belonging to different subfamilies identified from different bivalve species clustered together to form four major branches on the phylogenetic tree. The AQP1-like subfamily members accounted for the largest proportion of all bivalve AQPs (55.79%), whereas AQP-11 like subfamily members accounted for the smallest proportion (8.42%). Each bivalve species had only one AQP-11 like member, except for C. virginica (2), C. farreri (0), and hard clam (0). MmAQP1_1 and MmAQP1_2 clustered most closely with ScAQP1_2, which was consistent with the evolutionary relationship between the two species. Similarly, MmAQP3 clustered closely with ScAQP10_3. MmAQP4, MmAQP5_1 and MmAQP5_2 clustered together with most of the bivalve AQP4. As opposed to that in the other bivalves, the AQP8-like subfamily has undergone expansion in C. gigas and C. virginica. The AQP4 had more than three copies in bivalves, except for P. fucata and hard clam. The AQP3-like subfamily had the largest number of members in S. constricta, and AQP5 was the most abundant (five copies) in C. virginica compared to that in other bivalves.
As shown in Table 1, the predicted MWs of MmAQPs ranged from 24.79 to 52.59 kDa, encoding a maximum of 234 amino acids and minimum of 505 amino acids. The pI of MmAQPs ranged from 7.7 to 7.7. The GRAVY values were between 0.349 and 0.806, suggesting that MmAQPs are hydrophobic proteins. All MmAQPs were located on the cell membrane and contained 6 transmembrane α-helices structure, except MmAQP8_2, which contained 12 transmembrane α-helices. In general, the physicochemical properties of MmAQPs were found to be consistent with those of other known AQPs.
Nine MmAQP genes were unevenly distributed on four chromosomes in hard clam (Figure 2A). The MmAQPs clustered on the same branch of the phylogenetic tree were located adjacent on the same chromosome. MmAQP4 and two MmAQP5 genes were located on chr4. Three MmAQP8 genes were located on chr9. MmAQP3 was distributed on chr10, and two MmAQP1 genes were observed on chr18. Through a gene duplication analysis using MCScanX software, we identified 3,962 pairs of tandemly-duplicated genes in M. mercenaria genome, including two MmAQP5 genes and three MmAQP8 genes distributed on chr4 and chr9, respectively. Additionally, we detected 135 pairs of collinear genes in M. mercenaria genome, among which no MmAQPs were found (Figure 2B).
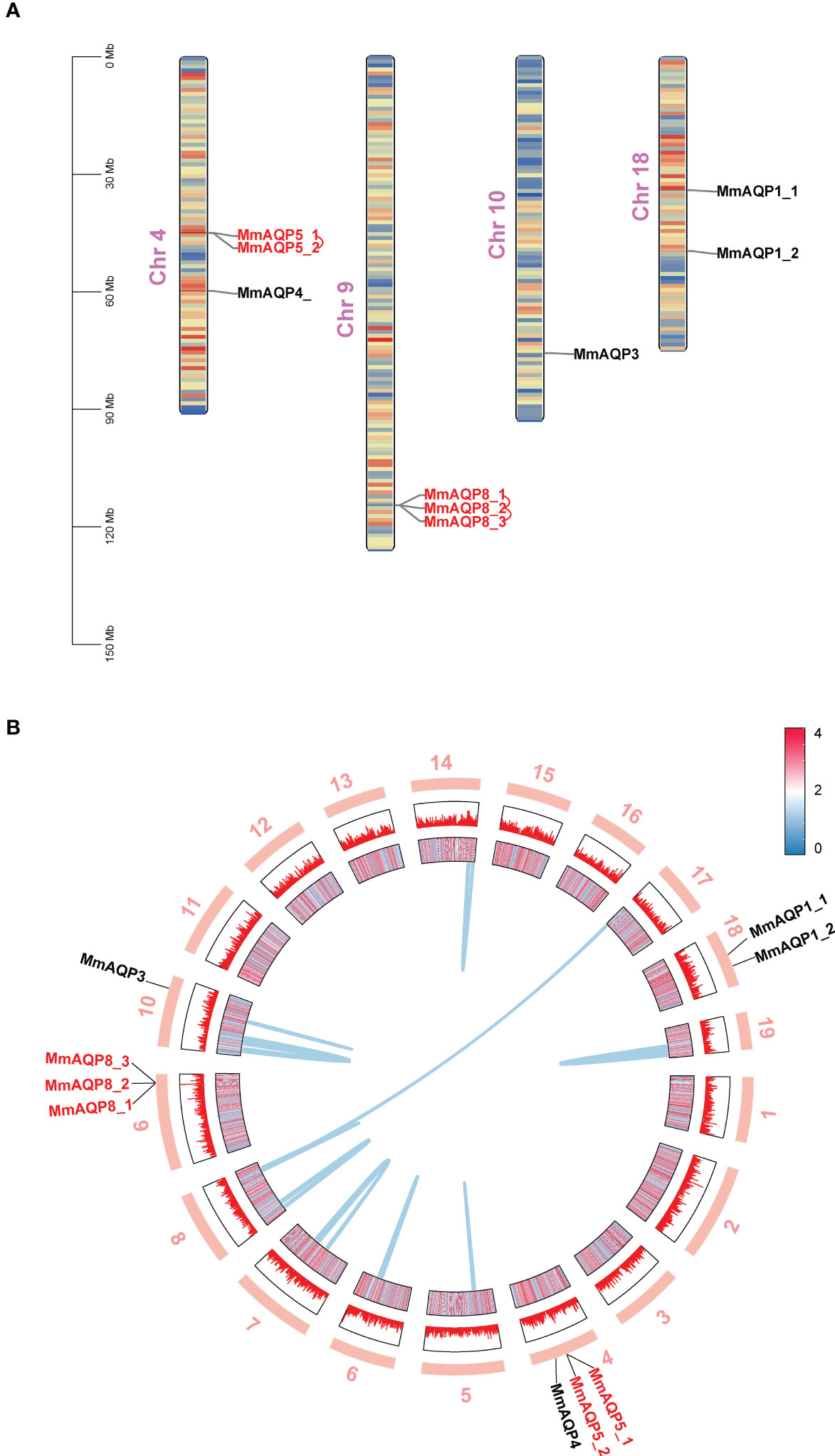
Figure 2 Chromosomal distribution, gene duplication and collinearity of MmAQPs. (A) Chromosome location of MmAQP genes. Tandemly-duplicated MmAQP genes are marked in red. (B) Collinear gene pairs on different chromosomes. The outer circle represents 19 chromosomes in hard clams. The inner circle represents the gene density of each chromosome. Red represents high, and blue represents low. The collinear gene pairs are linked by blue lines.
The gene lengths and exon-intron structures varied greatly among different MmAQP genes (Figure 3). Apart from MmAQP8_2, each MmAQP gene had 3–5 exons. Although the gene lengths of MmAQP5_1 and MmAQP5_2 were different, they exhibited similar gene structures (consisting of three exons and three introns). The CD-Search analysis revealed that each MmAQP had 1–2 conserved MIP domains. Motif analysis showed that motifs 3, 10, 4, and 1, and motifs 5, 6, and 2 were highly conserved among different MmAQPs. Motif 8 was only present in MmAQ8. The similar motifs distribution was observed among the MmAQPs belonging to the same subfamily (Figure 3).
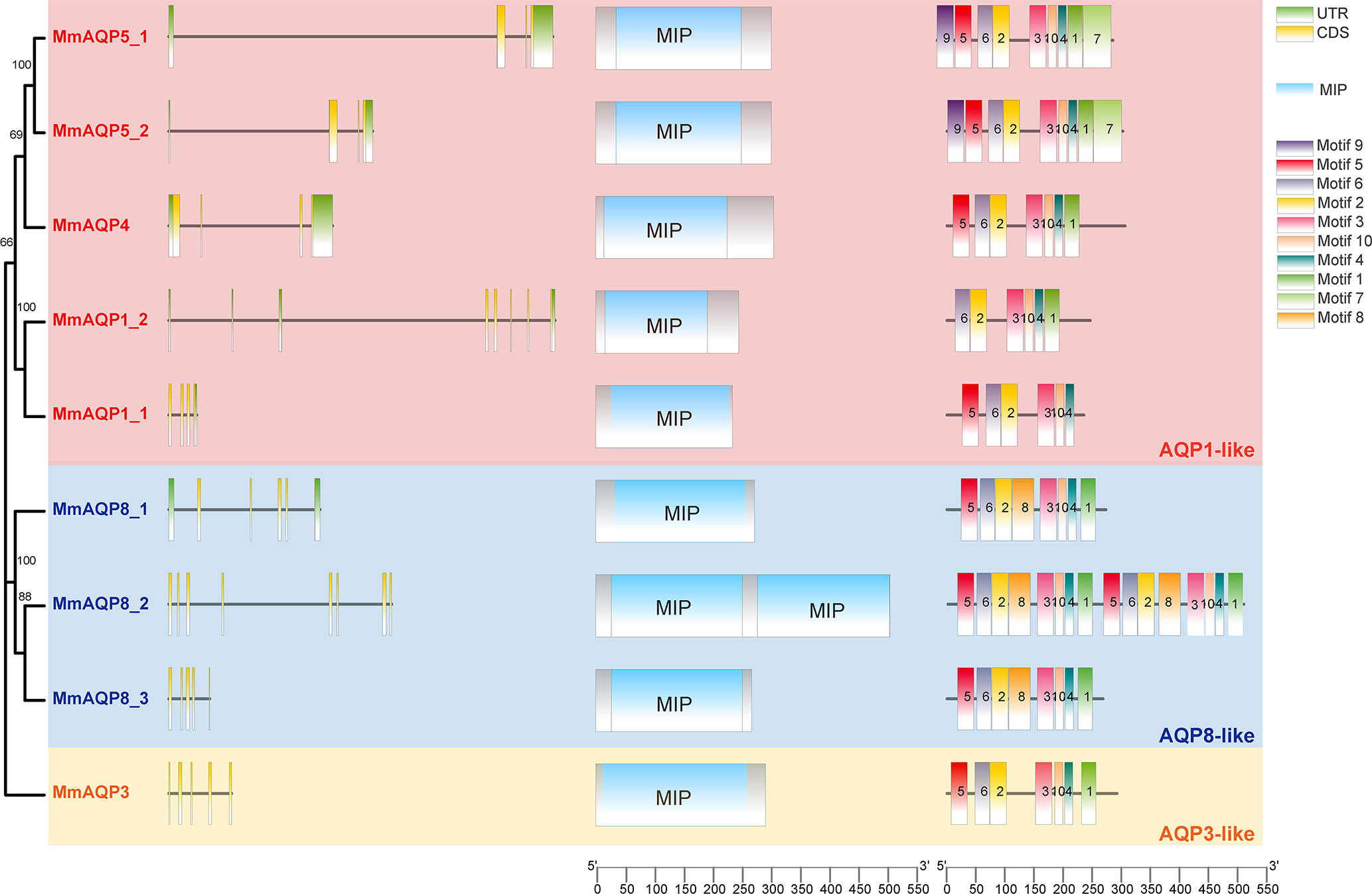
Figure 3 Phylogenetic tree, exon-intron structures, conserved domains and motifs of MmAQPs. On the left side of the diagram, green boxes, black lines, and yellow boxes represent untranslated regions (UTRs), introns, and exons, respectively. In the middle, blue boxes represent the MIP domains. On the right, 10 boxes of different colors represent different motifs. The AQP1-like, AQP8-like, and AQP3-like subfamilies are marked with pink, blue, and yellow, respectively.
As shown in Figure 4A, two highly-conserved NPA motifs were observed in each MmAQP. The composition of the secondary structural unit of each MmAQP exhibited high similarity (Figure S1). The α-helices and random coil structures were the most abundant (32.65–45.51% and 30.89–41.63%, respectively) secondary structural units in MmAQPs, while β-turns accounted for the smallest proportion (2.33–4.75%). Notably, each secondary structural unit accounted for almost the same proportion of the tandemly-duplicated MmAQP5_1 and MmAQP5_2. The 3D homology modeling of each MmAQP was performed using SWISS-MODEL. The crystal structure of the Bos taurus AQP1 (PDBID: 1J4N) was found to be the best structural template for MmAQP1_1 and MmAQP1_2. The H. sapiens AQP10 (PDBID: 6F7H) was the most suitable template for MmAQP3. The Rattus norvegicus AQP4 (PDBID: 2ZZ9) was selected as the template for MmAQP4. The H. sapiens AQP5 (PDBID: 5C5X), and Anabas testudineus AQP (PDBID: 7W7S) were the two best templates for the two MmAQP5, and three MmAQP8, respectively. Different MmAQPs that were modeled homologically with the same template showed almost identical 3D structures (Figure 4B). All MmAQPs displayed six typical transmembrane domains and a water-transporting pore.
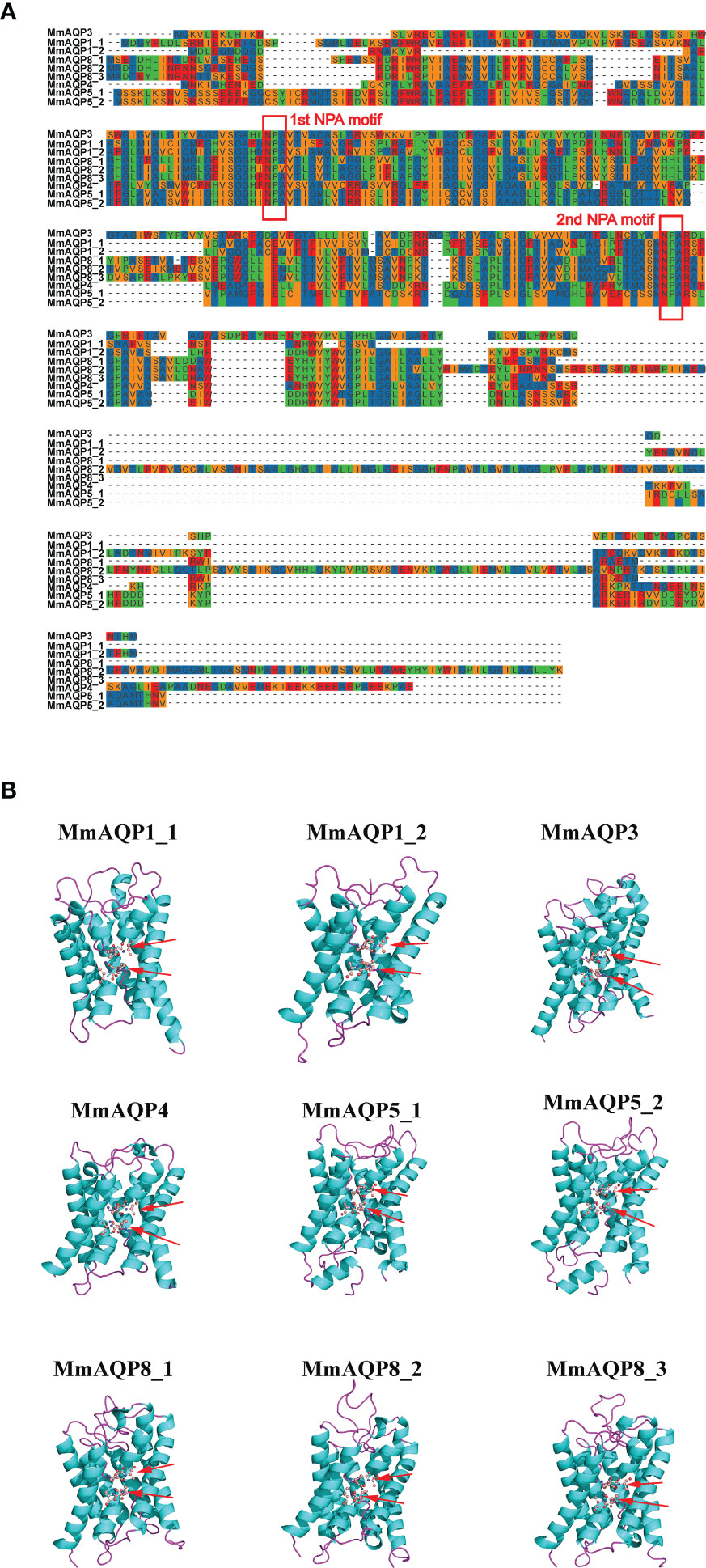
Figure 4 Amino acid composition and tertiary structures of MmAQPs. (A) Multiple sequence alignment of MmAQPs to detect the conserved NPA motifs. The two NPA motifs in each MmAQP are marked with red rectangles. (B) Homology modelling of the 3D structures of MmAQPs. Six transmembrane α-helices in each MmAQP are labeled with cyan, and the linker loops are labeled with purple. The NPA motifs are marked with red arrows.
Four bivalve species with different taxonomic statuses, including P. yessoensis (Autobranchia, Pectinidae), C. gigas (Autobranchia, Ostreida), C. sinensis (Autobranchia, Venerida, Veneridae, Cyclina), and A. marissinica (Autobranchia, Venerida, Vesicomyidae, Archivesica) were selected to perform synteny analyses with M. mercenaria (Autobranchia, Venerida, Veneridae, Mercenaria). A total of 130 and 123 collinear gene pairs were detected between M. mercenaria and P. yessoensis and M. mercenaria and C. gigas, respectively; however, no ortholog of MmAQPs was identified in the genomes of P. yessoensis and C. gigas (Figure 5A). The syntenic conservation was observed between M. mercenaria and C. sinensis as well as M. mercenaria and A. marissinica. Three orthologs of MmAQPs were detected in the genome of C. sinensis, which were collinear with MmAQP4, MmAQP8_3, and MmAQP1_1. Similarly, the orthologs of MmAQP5_1, MmAQP4, and MmAQP8_1 was identified in the genome of A. marissinica (Figure 5B).
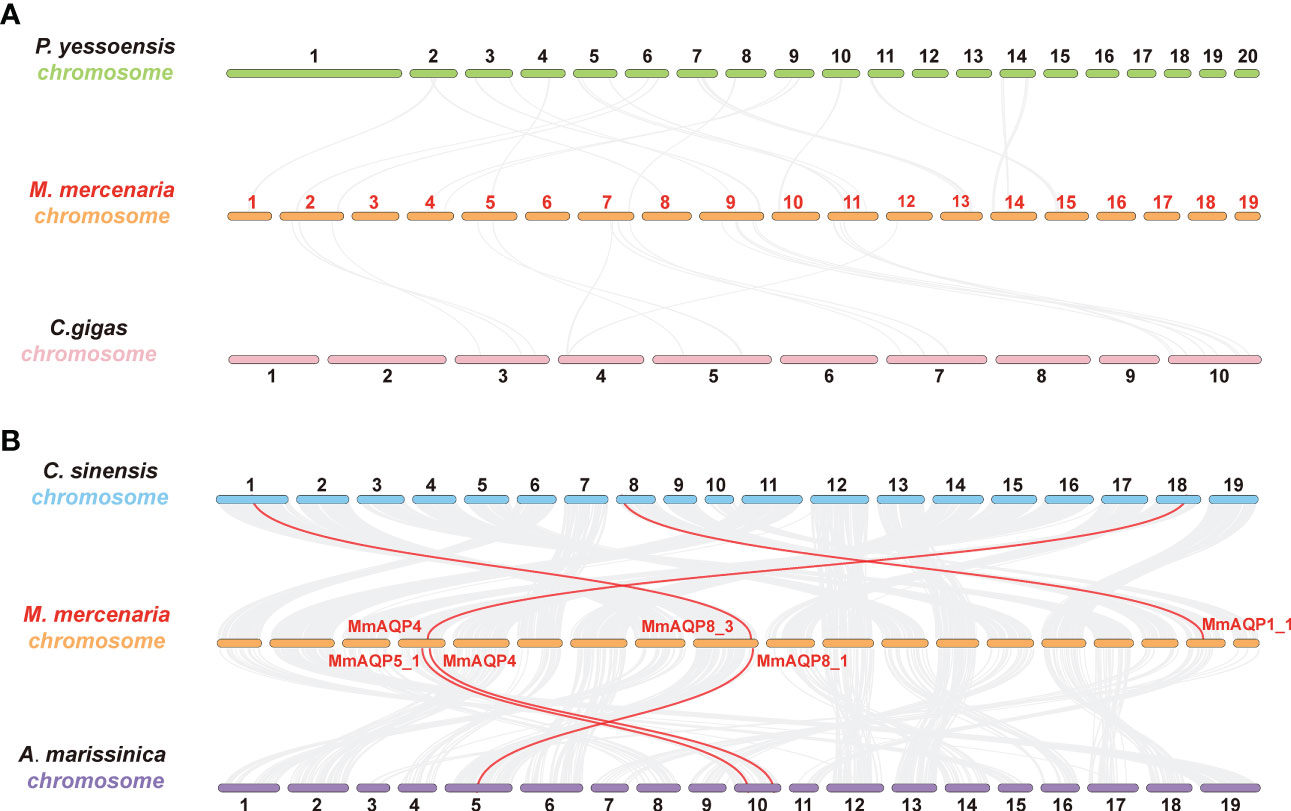
Figure 5 Synteny analysis to detect the orthologs of MmAQPs in the geome of P. yessoensis and C. gigas (A), as well as C. sinensis and A. marissinica (B). Grey lines in the background represent the collinear gene pairs between the two genomes. The orthologous AQP gene pairs are linked by red lines.
The transcriptional profiles of MmAQP genes were characterized in nine tissues to determine their spatial expression patterns. The AQP genes showed tissue-specific expression in hard clams. Most of the MmAQP genes were expressed in the foot and hepatopancreas tissues. MmAQP1_1 was the only gene that was highly expressed in the gills, and MmAQP1_2 was specifically expressed in the foot. MmAQP4 gene expression was detected in the foot and adductor muscles.The tandemly-duplicated MmAQP8_2 and MmAQP8_3 genes showed almost identical expression levels in different tissues (Figure 6).
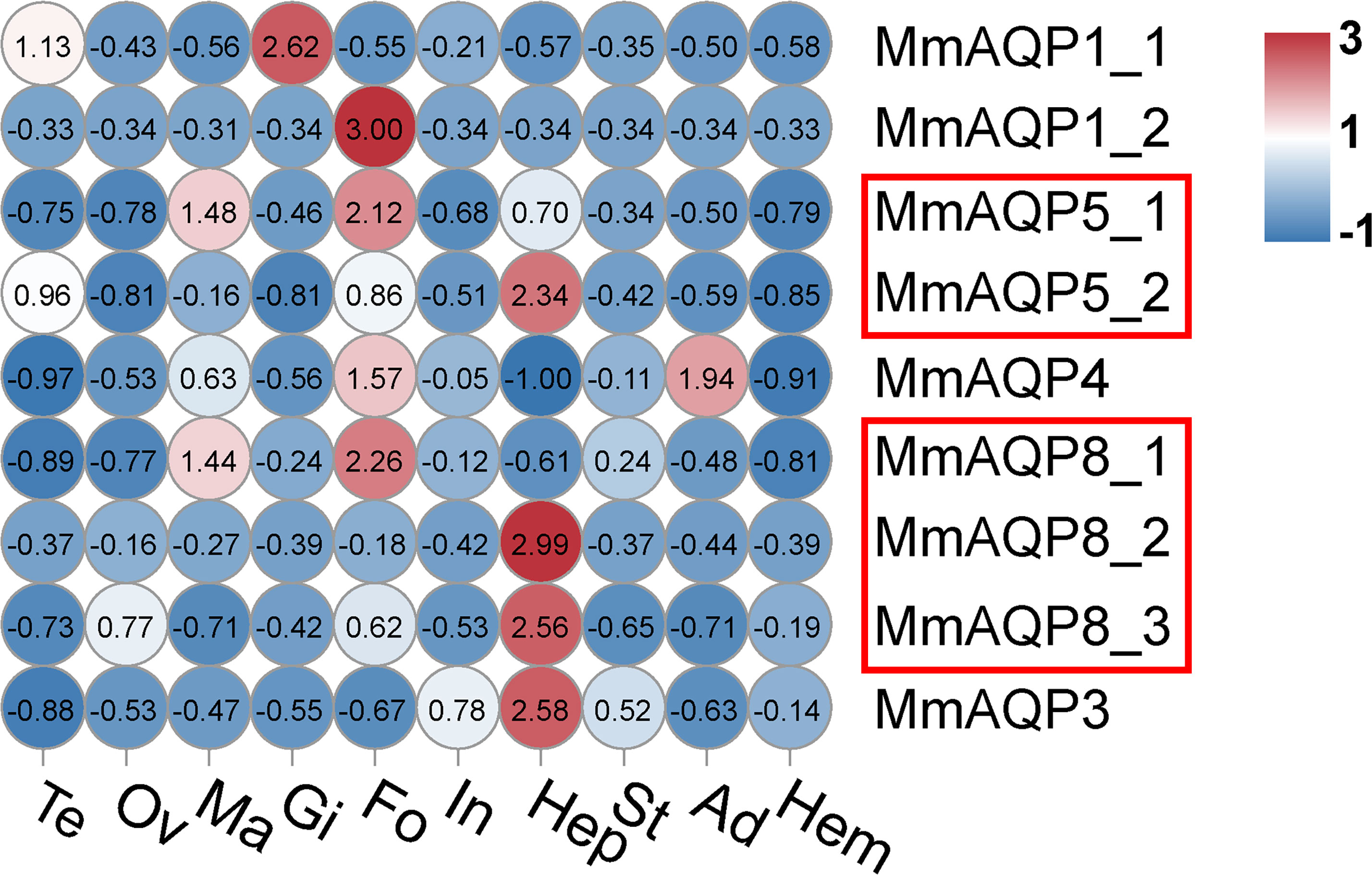
Figure 6 Heatmap analysis of MmAQP genes in different tissues. The name of each tissue is abbreviated as follows: Te, testis; Ov, ovary; Ma, mantle; Gi, gill; Fo, foot; In, intestine; Hep, hepatopancreas; St, stomach; Ad, adductor muscle; and Hem, hemolymph. Red indicates a high level of gene expression, and blue indicates low expression. The tandemly-duplicated genes are marked with red rectangles.
We further investigated the variations in gene expression of MmAQPs in the gills of hard clams under various environmental stressors. Upon exposure to severe hyposalinity (5 psu) for 10 days, the expression of MmAQP1_1 and MmAQP5_2 was significantly upregulated, and that of MmAQP3 was significantly downregulated (P <0.05). The expression of these three MmAQP genes was not significantly different between the S15 group and the control groups. The expression levels of MmAQP5_1 and MmAQP8_2 reached the highest after 10 days of moderate hyposalinity stress. Moreover, the expression of MmAQP4 and MmAQP3 were significantly upregulated, and MmAQP8_1 was significantly downregulated in the S40 group (Figure 7).
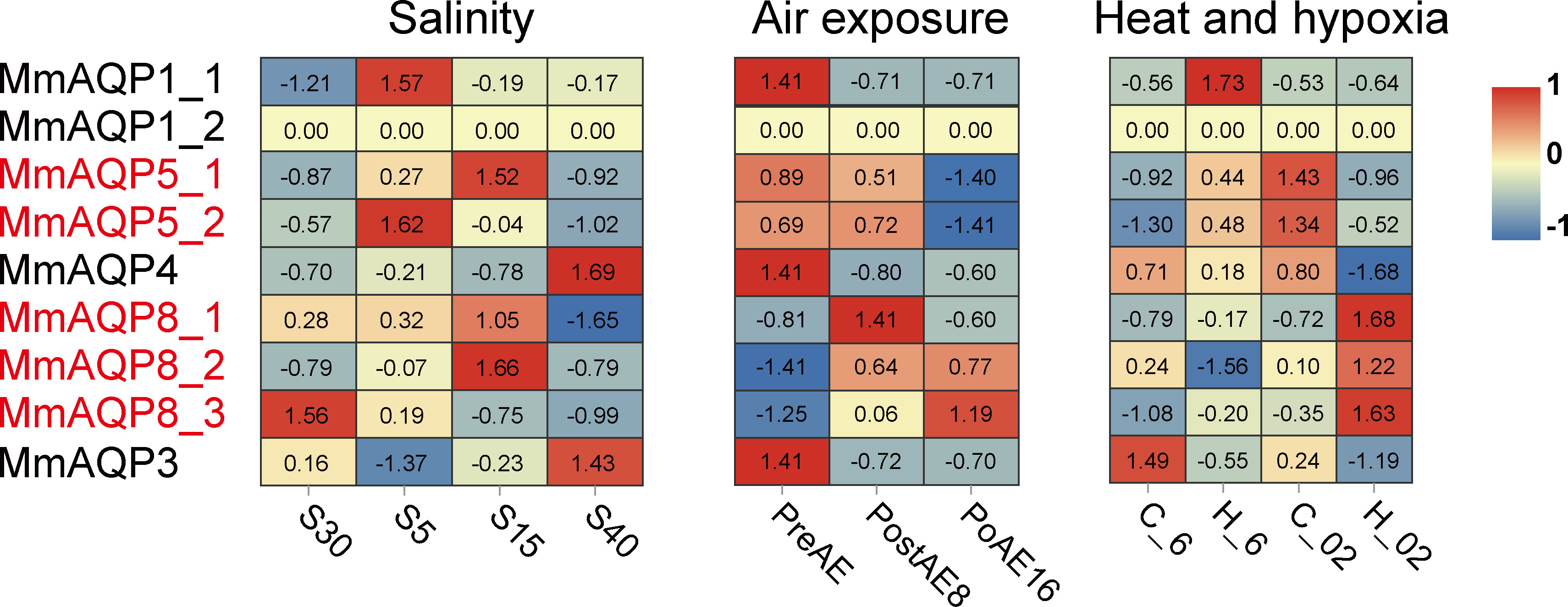
Figure 7 Heatmap analysis of MmAQP genes in response to salinity, air exposure, and heat and hypoxia stress. Red indicates a high level of gene expression, and blue indicates low expression. The tandemly-duplicated genes are marked in red.
During air exposure stress, the expression levels of MmAQP1_1, MmAQP4, and MmAQP_3 were significantly downregulated on the 8th day and remained at low levels until the 16th day. The expression patterns of tandemly-duplicated MmAQP5_1 and MmAQP5_2 was almost identical in each group; both were downregulated to the lowest level on the 16th day. The expression levels of tandemly-duplicated MmAQP8_2 and MmAQP8_3 was continuously upregulated during air exposure stress (Figure 7).
MmAQP1_1 expression was significantly induced under heat stress (P <0.05), but remained at the control level under hypoxia and combined heat and hypoxia stress. The highest expression levels of tandemly-duplicated MmAQP5_1 and MmAQP5_2 was observed in the C_02 group. MmAQP4 expression was significantly downregulated, whereas three tandemly-duplicated MmAQP8 genes were significantly upregulated under combined heat and hypoxia stress (P <0.05). Unlike that of the other MmAQP genes, MmAQP3 expression was inhibited under heat, hypoxia, and combined stress (Figure 7). Additionally, we did not detect the MmAQP1_2 expression in any group, likely because it was specifically expressed in the feet of hard clams (Figure 6).
AQPs are members of the major intrinsic protein superfamily that mediate the specific permeation of water and other small solutes across the cell membrane (Hara-Chikuma and Verkman, 2006). The structures and functions of AQPs have been thoroughly studied in humans and other animal models, such as mice and zebra fish, uncovering their important roles in osmoregulation and metabolic homeostasis maintenance (Saparov et al., 2007). Euryhaline bivalves showed extraordinary osmoregulation ability and have evolved a wide range of salinity adaptation. However, knowledge of AQP gene family remains limited in bivalves. To fill this gap, we investigated the genome-wide identification, gene and protein structures, evolutionary relationships, and expression profiles of AQP gene family members in hard clams. This study provides a comprehensive understanding of AQPs in bivalves and highlights their roles in osmoregulation and stress responses.
The number of AQP genes is highly diverse among different groups of animals (Agre, 2004). The number of bivalve AQPs was similar to that in mammals but much less than that in fish (Cao and Shi, 2019). Oysters (C. gigas and C. virginica) had the highest number of AQPs (13 and 15, respectively), whereas clams (M. philippinarum and M. mercenaria) had the lowest (9), suggesting that the number of AQPs is more consistent among bivalve species with close phylogenetic relationships. A previous study identified more than 20 AQPs from the genome of oyster using BLASTP method (Jia and Liu, 2022). Our study combined BLASTP and hmmsearch, and filtered out the short AQPs to strictly identify the AQP gene family members in bivalves. Structurally, although the overall primary sequences of the AQPs were not well conserved in different animals (approximately 30% identity), all MmAQPs exhibited the conserved structural characteristics, including six or twelve transmembrane α-helices, one MIP domain, and two NPA motifs. Based on phylogenetic analysis, MmAQPs could be classified into three subfamilies: AQP1-like, AQP3-like, and AQP8-like. The same classification of AQPs was observed in C. gigas and C. virginica. Only one AQP3-like AQP was identified in hard clams, which was far less than that in other animal models. This indicates that AQP7, AQP9, and AQP10 may have undergone gene loss during evolution, resulting in a weakened function of glycerol and urea penetration in hard clams. Similarly, AQP10 was found to be lost or turned into a non-functional pseudogene in mice (Morinaga et al., 2002), and chickens (Isokpehi et al., 2009). The presence of AQP11-like subfamily members in molluscs remains debatable (Colgan and Santos, 2018). In this study, we did not identify any AQP11-like member in the genome of hard clam, possibly because AQP11 and AQP12 were distinguished on the basis of their NPA structures and showed low sequence identities with other AQPs.
Compared with that in other bivalves, AQP8-like subfamily has undergone a considerable lineage-specific expansion in hard clams. The expansion of the AQP8-like subfamily was also observed in H. discus hannai (Jia et al., 2022), and C. gigas (Jia and Liu, 2022). The expansion of the AQP8-like subfamily in hard clam may have been related to the evolutionary process of adaptation to dynamic environments. In this study, the expression patterns of three tandemly-duplicated MmAQP8 genes showed high similarity under heat, hypoxia, and air exposure stress, reflexing their functional conservation in resistance to these stressors. Tandem duplication is a powerful driving force for evolutionary novelty (Chen et al., 2013). Tandemly-duplicated genes may obtain new functions through neofunctionalization and sub-functionalization (Sémon and Wolfe, 2007). In hard clam, we observed that three tandemly-duplicated MmAQP8 showed significant differences in gene expressions under salinity stress. MmAQP8_3 expression was specifically downregulated upon exposure to severe hyposalinity stress, whereas the expression levels of MmAQP8_1 and MmAQP8_2 remained at the control level. When the hard clams were exposed to moderate hyposalinity stress, the expression of MmAQP8_1 and MmAQP8_2 was specifically upregulated, whereas MmAQP8_3 was downregulated. Moreover, the expression of MmAQP8_1 and MmAQP8_3 was significantly inhibited under hypersalinity stress, whereas MmAQP8_2 did not change. These results indicate that MmAQP8 genes are more sensitive to salinity stress compared to heat, hypoxia, and air exposure stress. Tandem duplication events may acclerate the functional divergence of AQP8 genes in hard clams. Additionally, synteny analyses were performed to detect the orthologs of MmAQPs in different bivalves to further understand the origin and evolution of MmAQPs. No orthologous AQP gene pairs were detected among the different chromosomes in hard clams. This may have been because only a few whole-genome duplication events occurred in molluscs during evolution (Liu et al., 2021). Additionally, low collinearity in the gene arrangements and no orthologous AQP gene pairs were observed between P. yessoensis and hard clam as well as C. gigas and hard clam. This indicates that the evolution and diversification of AQP genes occurred independently in Autobranchia, the common ancestor of P. yessoensis, C. gigas, and hard clam. The orthologs of MmAQP4, MmAQP8_3, and MmAQP1_1 were detected in the genome of C. sinensis, which belongs to the same family (Veneridae) with hard clams. Moreover, the orthologs of MmAQP5_1, MmAQP4, and MmAQP8_1 were detected in the genome of A. marissinica, which belongs to same order (Venerida) with hard clams but different families (Vesicomyidae). These results indicates that the AQP4 and AQP8 genes in bivalves are highly orthologous at the order level.
An assessment of the gene expression patterns provided insights into the possible physiological roles of MmAQPs. The AQP genes showed tissue-specific expression in hard clams. Almost no MmAQP gene was expressed in the testes, ovaries, mantles, intestines, stomachs, adductor muscles, or hemolymph under suitable living conditions. Consistent with this result, inactive AQPs gene expression was also observed in most of the tissues of S. constricta (Ruan et al., 2022), and nile tilapia (Ni et al., 2022). MmAQP5_2, MmAQP8_2, MmAQP8_3, and MmAQP3 were highly expressed in the hepatopancreas, and the expression levels of MmAQP1_2, MmAQP5_1, and MmAQP8_1 were highest in the foot, suggesting that these MmAQPs play an important role in maintaining the basic physiological processes in the hepatopancreas and foot. AQPs are closely related to environmental adaptation in broader phyla (Finn et al., 2014). We further investigated the variations in the gene expression of MmAQP under various environmental stressors. MmAQP genes belonging to the same subfamily showed diverse expression patterns under salinity stress. Six MmAQP genes specifically responded to hyposalinity stress, among which four were significantly upregulated and two were downregulated. MmAQP4 and MmAQP3 showed the highest expression levels under hypersalinity, whereas MmAQP8_1 and MmAQP8_3 showed the lowest. Under salinity stress, the downregulation of MmAQP genes expressions resulted in a widespread reduction in water permeability. This regulation was vital to maintain and rebuilt the osmotic gradient between the gill cells and hemolymph in hard clams. Consistent with this result, the expression levels of three AQP genes in C. gigas decreased to protect against water currents and cell swelling under hyposalinity stress (Meng et al., 2013). Additionally, our previous study reported that ammonia was significantly accumulated in the gills of hard clams after 5 days of hypersalinity exposure (Zhou et al., 2023). Therefore, the significant upregulation of MmAQP3 expression under hypersalinity stress may be closely related to its specific function in ammonia transport.
The roles of AQPs in drought resistance have mainly been reported in plants (Zhang et al., 2021). Except for three MmAQP8 genes, the gene expressions of the remaining MmAQPs were significantly inhibited after air exposure, implying that MmAQPs also function to maintain the intracellular water contents during drought stress. Decreased expression of AQPs can enhance anti-apoptotic responses and affect the cell apoptosis rate in mammals (Jablonski et al., 2004; Jablonski et al., 2007). The stress resistance role of inhibitor of apoptosis has been highlighted in hard clams (Song et al., 2021). Hence, we speculate that the AQP-related anti-apoptotic responses may be activated in hard clams during air exposure stress. Additionally, hypoxia has been reported to alter the gene expressions of AQPs in the rat astrocytes (Yamamoto et al., 2001). In hard clams, two MmAQP5 genes were highly expressed under hypoxia stress and three MmAQP8 genes were significantly upregulated under the combined stress of heat and hypoxia. Oxidative stress and the accumulation of reactive oxygen species (ROS) in the gills are common physiological changes in bivalves upon exposure to heat and hypoxia stress (Chen et al., 2007). The enhancement of antioxidant enzyme activity and gene expression is vital for bivalves to resist environmental stress (Freitas et al., 2017). Human AQP8 has been reported to regulate the diffusion of H2O2 across cell membranes (Bienert et al., 2007). Results of this study imply that the overexpression of the three MmAQP8 genes under air exposure and the combined stress of heat and hypoxia were important responses to eliminate excess intracellular ROS in hard clams. Our previous metabolomics studies have reported that environmental stress can cause alterations in various metabolic processes in hard clams, including osmoregulation, lipid metabolism, ROS generation, ammonia transport, and membranes stabilization (Zhou et al., 2022; Zhou et al., 2023). These metabolic alterations were highly related to the roles of human AQPs (Hara-Chikuma and Verkman, 2006; Bienert et al., 2007; Maeda et al., 2008; Ishibashi et al., 2011; Tamma et al., 2018). Therefore, the diverse MmAQPs may play critical and non-redundant roles in maintaining the metabolic homeostasis in hard clams under environmental stress. Functional experiments, such as the gene knockdown or overexpression of MmAQPs, will further be conducted to investigate the osmoregulation and stress resistance roles of MmAQPs.
In the present study, nine MmAQPs were identified in the genome of M. mercenaria, which were classified into AQP1-like, AQP3-like, and AQP8-like subfamilies. Structurally, MmAQPs showed typical characteristics of AQPs, including transmembrane α-helices, MIP domains, and NPA motifs. A lineage-specific expansion of AQP8-like subfamily and gene loss of AQP3-like subfamily was observed in hard clams. Three orthologs of MmAQPs were detected in the genome of C. sinensis and A. marissinica, suggesting that AQP4 and AQP8 genes were highly orthologous in the order Venerida. Tissue expression profiles showed that MmAQP genes were highly expressed in the foot and hepatopancreas. Three tandemly-duplicated MmAQP8 genes showed different expression sensitivity to different environmental stressors. The gene expression patterns of three MmAQP8 were similar under heat, hypoxia, and air exposure stress, but differed greatly under salinity stress. Tandem duplication events may acclerate the functional divergence of MmAQP8 genes. Under salinity and air exposure stress, several MmAQP genes in gills were significantly downregulated to reduce the water permeability and maintain the osmotic equilibrium. Other MmAQP genes were significantly upregulated to promote the transport of ammonia and ROS, and activate the anti-apoptotic responses to resist hypersalinity, heat, hypoxia, and air exposure stress. This study provides a comprehensive understanding of AQP gene family in hard clams and is of great significance for further studies to explore the AQPs-related stress resistance mechanisms in bivalves.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
CZ: Investigation, data curation, methodology, writing – original draft. Z-SL: Data curation, writing – review & editing. YS: Methodology, investigation. JF: Methodology, investigation. ZH: Methodology. M-JY: Methodology. PS: Investigation, Data curation. Y-RL: Investigation. Y-JG: Investigation, methodology. TZ, HS: Conceptualization, supervision, funding acquisition, writing – review & editing. All authors contributed to the article and approved the submitted version.
This research was supported by Tianjin science and technology commission project (20YFZCSN00240), Tianjin Agricultural Committee Project (202103010), the earmarked fund for CARS (CARS-49), Shandong Province Agricultural Major Applied Technology Innovation Project (Grant No. SF1405303301), the Key Research and Development Program of Guangxi Province (Grant No. 2021AB34014), Taishan Industrial Leading Talents Project (Grant No. LJNY201704, Recipient: TZ), the ‘Double Hundred’ Blue Industry Leader Team of Yantai (Recipient: TZ), the Creative Team Project of the Laboratory for Marine Ecology and Environmental Science, Qingdao Pilot National Laboratory for Marine Science and Technology (Grant No. LMEES-CTSP-2018-1), the Young Elite Scientists Sponsorship Program by CAST (2021QNRC001), and the Youth Innovation Promotion Association CAS. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors acknowledge the Oceanographic Data Center, IOCAS for providing assistance of bioinformatics analyses.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1138074/full#supplementary-material
Supplementary Figure 1 | The composition of the secondary structural unit of MmAQPs.
Agre P. (2004). Aquaporin water channels (Nobel lecture). Angew. Chem. Int. Ed. Engl. 43, 4278–4290. doi: 10.1002/anie.200460804
Arnold K., Bordoli L., Kopp J., Schwede T. (2006). The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201. doi: 10.1093/bioinformatics/bti770
Bailey T. L., Johnson J., Grant C. E., Noble W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Benga G. (2012). The first discovered water channel protein, later called aquaporin 1: molecular characteristics, functions and medical implications. Mol. Aspects Med. 33, 518–534. doi: 10.1016/j.mam.2012.06.001
Berger V. J., Kharazova A. (1997). “Mechanisms of salinity adaptations in marine molluscs,” in Interactions and adaptation strategies of marine organisms (Springer), 115–126.
Bienert G. P., Møller A. L., Kristiansen K. A., Schulz A., Møller I. M., Schjoerring J. K., et al. (2007). Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J. Biol. Chem. 282, 1183–1192. doi: 10.1074/jbc.M603761200
Buchfink B., Xie C., Huson D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Cao J., Shi F. (2019). Comparative analysis of the aquaporin gene family in 12 fish species. Animals 9, 233. doi: 10.3390/ani9050233
Chen C., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen S., Krinsky B. H., Long M. (2013). New genes as drivers of phenotypic evolution. Nat. Rev. Genet. 14, 645–660. doi: 10.1038/nrg3521
Chen M., Yang H., Delaporte M., Zhao S. (2007). Immune condition of Chlamys farreri in response to acute temperature challenge. Aquaculture 271, 479–487. doi: 10.1016/j.aquaculture.2007.04.051
Colgan D., Santos R. (2018). A phylogenetic classification of gastropod aquaporins. Mar. Genomics 38, 59–65. doi: 10.1016/j.margen.2017.12.002
Denker B. M., Smith B. L., Kuhajda F. P., Agre P. (1988). Identification, purification, and partial characterization of a novel Mr 28,000 integral membrane protein from erythrocytes and renal tubules. J. Biol. Chem. 263, 15634–15642. doi: 10.1016/S0021-9258(19)37635-5
Dong C., Chen L., Feng J., Xu J., Mahboob S., Al-Ghanim K., et al. (2016). Genome wide identification, phylogeny, and expression of aquaporin genes in common carp (Cyprinus carpio). PloS One 11, e0166160. doi: 10.1371/journal.pone.0166160
Finn R. N., Cerda J. (2015). Evolution and functional diversity of aquaporins. Biol. Bull. 229, 6–23. doi: 10.1086/BBLv229n1p6
Finn R. N., Chauvigné F., Hlidberg J. B., Cutler C. P., Cerdà J. (2014). The lineage-specific evolution of aquaporin gene clusters facilitated tetrapod terrestrial adaptation. PloS One 9, e113686. doi: 10.1371/journal.pone.0113686
Freitas R., De Marchi L., Bastos M., Moreira A., Velez C., Chiesa S., et al. (2017). Effects of seawater acidification and salinity alterations on metabolic, osmoregulation and oxidative stress markers in Mytilus galloprovincialis. Ecol. Indic. 79, 54–62. doi: 10.1016/j.ecolind.2017.04.003
Gonen T., Walz T. (2006). The structure of aquaporins. Q. Rev. Biophys. 39, 361–396. doi: 10.1017/S0033583506004458
Groszmann M., Osborn H. L., Evans J. R. (2017). Carbon dioxide and water transport through plant aquaporins. Plant Cell Environ. 40, 938–961. doi: 10.1111/pce.12844
Hara-Chikuma M., Verkman A. (2006). Physiological roles of glycerol-transporting aquaporins: the aquaglyceroporins. Cell Mol. Life Sci. 63, 1386–1392. doi: 10.1007/s00018-006-6028-4
Heymann J. B., Engel A. (1999). Aquaporins: phylogeny, structure, and physiology of water channels. News Physiol. Sci. 14, 187–193. doi: 10.1152/physiologyonline.1999.14.5.187
Hosoi M., Shinzato C., Takagi M., Hosoi-Tanabe S., Sawada H., Terasawa E., et al. (2007). Taurine transporter from the giant pacific oyster Crassostrea gigas: function and expression in response to hyper-and hypo-osmotic stress. Fisheries Sci. 73, 385–394. doi: 10.1111/j.1444-2906.2007.01346.x
Hu Z., Feng J., Song H., Zhou C., Yu Z., Yang M., et al. (2022a). Mechanisms of heat and hypoxia defense in hard clam: Insights from transcriptome analysis. Aquaculture 549, 737792. doi: 10.1016/j.aquaculture.2021.737792
Hu Z., Song H., Feng J., Zhou C., Yang M.-J., Shi P., et al. (2022b). Massive heat shock protein 70 genes expansion and transcriptional signatures uncover hard clam adaptations to heat and hypoxia. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.898669
Ishibashi K., Hara S., Kondo S. (2009). Aquaporin water channels in mammals. Clin. Exp. Nephrol. 13, 107–117. doi: 10.1007/s10157-008-0118-6
Ishibashi K., Kondo S., Hara S., Morishita Y. (2011). The evolutionary aspects of aquaporin family. Am. J. Physiol. Regul. Integr. Comp. Physiol. 300, R566–R576. doi: 10.1152/ajpregu.90464.2008
Isokpehi R. D., Rajnarayanan R. V., Jeffries C. D., Oyeleye T. O., Cohly H. H. (2009). Integrative sequence and tissue expression profiling of chicken and mammalian aquaporins. BMC Genomics 10, 1–12. doi: 10.1186/1471-2164-10-S2-S7
Jablonski E. M., Mattocks M. A., Sokolov E., Koniaris L. G., Hughes F. M. Jr, Fausto N., et al. (2007). Decreased aquaporin expression leads to increased resistance to apoptosis in hepatocellular carcinoma. Cancer Lett. 250, 36–46. doi: 10.1016/j.canlet.2006.09.013
Jablonski E. M., Webb A. N., McConnell N. A., Riley M. C., Hughes F. M. Jr. (2004). Plasma membrane aquaporin activity can affect the rate of apoptosis but is inhibited after apoptotic volume decrease. Am. J. Physiol. Cell Physiol. 286, C975–C985. doi: 10.1152/ajpcell.00180.2003
Jia Y., Liu X. (2022). Diversification of the aquaporin family in geographical isolated oyster species promote the adaptability to dynamic environments. BMC Genomics 23, 1–20. doi: 10.1186/s12864-022-08445-4
Jia Y., Xu F., Liu X. (2022). Duplication and subsequent functional diversification of aquaporin family in pacific abalone Haliotis discus hannai. Mol. Phylogenet. Evol. 168, 107392. doi: 10.1016/j.ympev.2022.107392
Koprivnikar J., Poulin R. (2009). Effects of temperature, salinity, and water level on the emergence of marine cercariae. J. Parasitol. Res. 105, 957–965. doi: 10.1007/s00436-009-1477-y
Krane C. M., Goldstein D. L. (2007). Comparative functional analysis of aquaporins/glyceroporins in mammals and anurans. Mamm. Genome. 18, 452–462. doi: 10.1007/s00335-007-9041-5
Liu C., Ren Y., Li Z., Hu Q., Yin L., Wang H., et al. (2021). Giant African snail genomes provide insights into molluscan whole-genome duplication and aquatic–terrestrial transition. Mol. Ecol. Resour. 21, 478–494. doi: 10.1111/1755-0998.13261
Maeda N., Funahashi T., Shimomura I. (2008). Metabolic impact of adipose and hepatic glycerol channels aquaporin 7 and aquaporin 9. Nat. Clin. Pract. Endoc. 4, 627–634. doi: 10.1038/ncpendmet0980
Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., Geer L. Y., et al. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, D222–D226. doi: 10.1093/nar/gku1221
Meng J., Zhu Q., Zhang L., Li C., Li L., She Z., et al. (2013). Genome and transcriptome analyses provide insight into the euryhaline adaptation mechanism of Crassostrea gigas. PloS One 8, e58563. doi: 10.1371/journal.pone.0058563
Morinaga T., Nakakoshi M., Hirao A., Imai M., Ishibashi K. (2002). Mouse aquaporin 10 gene (AQP10) is a pseudogene. Biochem. Biophys. Res. Commun. 294, 630–634. doi: 10.1016/S0006-291X(02)00536-3
Ni P., Zhao X., Liang Y. (2022). Genome-wide identification and expression analysis of the aquaporin gene family reveals the role in the salinity adaptability in Niletilapia (Oreochromis niloticus). Genes Genom. 4(12), 1457–1469. doi: 10.1007/s13258-022-01324-y
Pieńkowska J. R., Kosicka E., Wojtkowska M., Kmita H., Lesicki A. (2014). Molecular identification of first putative aquaporins in snails. J. Membr. Biol. 247, 239–252. doi: 10.1007/s00232-014-9629-0
Preston G. M., Carroll T. P., Guggino W. B., Agre P. (1992). Appearance of water channels in xenopus oocytes expressing red cell CHIP28 protein. Science 256, 385–387. doi: 10.1126/science.256.5055.385
Ruan W., Dong Y., Lin Z., He L. (2022). Molecular characterization of aquaporins genes from the razor clam Sinonovacula constricta and their potential role in salinity tolerance. Fishes 7, 69. doi: 10.3390/fishes7020069
Saparov S. M., Liu K., Agre P., Pohl P. (2007). Fast and selective ammonia transport by aquaporin-8. J. Biol. Chem. 282, 5296–5301. doi: 10.1074/jbc.M609343200
Sémon M., Wolfe K. H. (2007). Consequences of genome duplication. Curr. Opin. Genet. Dev. 17, 505–512. doi: 10.1016/j.gde.2007.09.007
Sokolov E. P., Sokolova I. M. (2019). Compatible osmolytes modulate mitochondrial function in a marine osmoconformer Crassostrea gigas (Thunber). Mitochondrion 45, 29–37. doi: 10.1016/j.mito.2018.02.002
Song H., Guo X., Sun L., Wang Q., Han F., Wang H., et al. (2021). The hard clam genome reveals massive expansion and diversification of inhibitors of apoptosis in bivalvia. BMC Biol. 19, 1–20. doi: 10.1186/s12915-020-00943-9
Soto G., Alleva K., Amodeo G., Muschietti J., Ayub N. D. (2012). New insight into the evolution of aquaporins from flowering plants and vertebrates: orthologous identification and functional transfer is possible. Gene 503, 165–176. doi: 10.1016/j.gene.2012.04.021
Tamma G., Valenti G., Grossini E., Donnini S., Marino A., Marinelli R. A., et al. (2018). Aquaporin membrane channels in oxidative stress, cell signaling, and aging: recent advances and research trends. Oxid. Med. Cell. Longev. doi: 10.1155/2018/1501847
Tamura K., Stecher G., Kumar S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Törnroth-Horsefield S., Wang Y., Hedfalk K., Johanson U., Karlsson M., Tajkhorshid E., et al. (2006). Structural mechanism of plant aquaporin gating. Nature 439, 688–694. doi: 10.1038/nature04316
Wang S., Hou R., Bao Z., Du H., He Y., Su H., et al. (2013). Transcriptome sequencing of zhikong scallop (Chlamys farreri) and comparative transcriptomic analysis with yesso scallop (Patinopecten yessoensis). PloS One 8, e63927. doi: 10.1371/journal.pone.0063927
Wang Y., Tang H., DeBarry J. D., Tan X., Li J., Wang X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49–e49. doi: 10.1093/nar/gkr1293
Wang X., Wang S., Li C., Chen K., Qin J. G., Chen L., et al. (2015). Molecular pathway and gene responses of the pacific white shrimp Litopenaeus vannamei to acute low salinity stress. J. Shellfish Res. 34, 1037–1048. doi: 10.2983/035.034.0330
Yamamoto N., Yoneda K., Asai K., Sobue K., Tada T., Fujita Y., et al. (2001). Alterations in the expression of the AQP family in cultured rat astrocytes during hypoxia and reoxygenation. Mol. Brain Rec. 90, 26–38. doi: 10.1016/S0169-328X(01)00064-X
Zhang G., Yu Z., da Silva J. A. T., Wen D. (2021). Identification of aquaporin members in Acacia auriculiformis and functional characterization of AaPIP1-2 involved in drought stress. Environ. Exp. Bot. 185, 104425. doi: 10.1016/j.envexpbot.2021.104425
Zhou C., Song H., Feng J., Hu Z., Yang M., Shi P., et al. (2022). Metabolomics and biochemical assays reveal the metabolic responses to hypo-salinity stress and osmoregulatory role of cAMP-PKA pathway in Mercenaria mercenaria. Comput. Struct. Biotechnol. J. 20, 4110–4121. doi: 10.1016/j.csbj.2022.08.004
Keywords: aquaporin, evolutionary relationship, tandem duplication, environmental stress, expression pattern
Citation: Zhou C, Lin Z-s, Shi Y, Feng J, Hu Z, Yang M-j, Shi P, Li Y-r, Guo Y-j, Zhang T and Song H (2023) Genome-wide identification, structural and evolutionary characteristics, and expression analysis of aquaporin gene family members in Mercenaria mercenaria. Front. Mar. Sci. 10:1138074. doi: 10.3389/fmars.2023.1138074
Received: 05 January 2023; Accepted: 22 February 2023;
Published: 07 March 2023.
Edited by:
Ming Li, Ningbo University, ChinaCopyright © 2023 Zhou, Lin, Shi, Feng, Hu, Yang, Shi, Li, Guo, Zhang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Zhang, dHpoYW5nQHFkaW8uYWMuY24=; Hao Song, aGFvc29uZ0BxZGlvLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.