- 1Department of Biological Sciences, California State University, Chico, CA, United States
- 2Department of Chemistry and Biochemistry, California State University, Chico, CA, United States
- 3Department of Biology, University of San Diego, San Diego, CA, United States
We examined the response of microbial communities in the model sea anemone Exaiptasia diaphana (Aiptasia) to short-term thermal elevation. Through 16S rRNA gene sequencing, we characterized the microbiomes of symbiotic (with algal symbionts) and aposymbiotic (bleached) anemones under ambient (27°C) and heat-stressed (34°C) conditions for 8-10 days, using both replicated endpoint and non-replicated time-course approaches. Consistent with prior studies, we observed a stable abundance of bacteria from the families Alteromonadaceae and Rhodobacteraceae, though with wide variation among individual anemones. We observed that symbiotic state conferred a larger impact on the microbiome than heat stress, implying the microbiome may play a metabolic role in the maintenance of cnidarian-dinoflagellate symbiosis. In particular, Pelobacter, an anaerobic sulfate reducer that is also a potential nitrogen fixer, was present only in symbiotic anemones, and its abundance decreased with initial exposure to 34°C, but recovered after 7 days. In aposymbiotic anemones, the added heat stress appeared to result in a large increase of rare bacterial taxa, which included potential pathogens such as Vibrio following bleaching. We also observed several archaea, the first reported for this model, but only in the seawater surrounding aposymbiotic Aiptasia, where abundance increased dramatically following heat stress. We further explored the diazotrophic (nitrogen fixation) potential of diverse bacteria associated with symbiotic and aposymbiotic Aiptasia, under both ambient and heat-stressed conditions, using nifH-PCR and qPCR and the acetylene reduction assay (ARA). In contrast to some stony corals, nifH was barely expressed in both anemone types, and under ambient conditions, diazotrophic activity was not detectable via ARA. Thus, although this research contributes to the growing knowledge of the bacterial community associated with a prominent model used in coral-symbiosis research, our results also suggest using caution when making direct comparisons between Aiptasia and different coral species in microbiome studies.
Introduction
An “oceanic rainforest,” tropical-reef ecosystems are one of the most diverse and productive ecosystems in the world, harbouring >25% of marine species while covering <1% of the ocean floor (Knowlton, 2001; Rosenberg et al., 2007; Veron et al., 2009; Knowlton et al., 2010; Burke et al., 2011; Santos et al., 2014; Pogoreutz, 2016). Rich in biodiversity and productivity, coral reefs are found in oligotrophic waters in the tropics that are extremely scarce in nutrients (Fiore et al., 2010; Ceh et al., 2013; Rädecker et al., 2015; Pogoreutz, 2016). This strange phenomenon of being highly productive in nutrient wastelands represents the “Darwin Paradox” (Rädecker et al., 2015; Pogoreutz, 2016), largely successful due to the symbiotic interactions occurring within the coral holobiont (Wang and Douglas, 1998; Knowlton, 2001; Muller-Parker and Davy, 2001; Baumgarten et al., 2015; Rädecker et al., 2015; Röthig et al., 2016; Rädecker and Pogoreutz, 2019; Tilstra et al., 2019; Liang et al., 2020).
The coral holobiont is a symbiotic network in which the coral host, photosynthetic dinoflagellates from the family Symbiodiniaceae (hereinafter ‘algal symbionts’), and other related microorganisms (i.e., bacteria, archaea, viruses, fungi, and protists) benefit from and interact with each other. (Rohwer et al., 2002; Rosenberg et al., 2007; Davy et al., 2012; Lema et al., 2012; Rädecker et al., 2015; Peixoto et al., 2017). The coral host offers inorganic nutrients from their metabolic waste to their microbial symbionts for their growth, along with protection from environmental predation (Weis, 2008; Weis et al., 2008; Lema et al., 2012; Santos et al., 2014; Rädecker et al., 2015; Röthig et al., 2016; Pogoreutz et al., 2017; Cui et al., 2019; Tilstra et al., 2019). In exchange, the algal symbionts provide the coral host with essential nutrients in the form of amino acids and photosynthates (i.e., photosynthetically fixed carbon) (Rädecker et al., 2015; Pogoreutz et al., 2017; Rädecker et al., 2018; Tilstra et al., 2019). Prokaryotes, such as bacteria and archaea, offer the coral host and their algal symbionts fundamental micronutrients and antimicrobial properties to fend off diseases and infections (Rohwer et al., 2002; Croft et al., 2005; Grant et al., 2014). The multiparty interactions between the coral host and its microbial symbionts are crucial towards sustaining a healthy holobiont in nutrient-poor environments.
Studies on marine-bacterial communities are becoming increasingly important in coral-reef research (Ainsworth and Hoegh-Guldberg, 2009; Bourne et al., 2009; Vega Thurber et al., 2009; Littman et al., 2011; Garren et al., 2014; Rädecker et al., 2015; Pogoreutz, 2016; Ziegler et al., 2017; Tran, 2022). Corals exhibit an altered microbiome in response to varying thermal conditions and disruptions to integral nutrient cycles (Ainsworth and Hoegh-Guldberg, 2009; Raina et al., 2010; Littman et al., 2011; Ziegler et al., 2017). In the face of climate change, both pathogens and potentially beneficial bacteria are important to study in the context of holobiont health. For example, dimethylsulfoniopropionate (DMSP) present in the coral mucus induces a chemotactic response in the coral pathogen Vibrio coralliilyticus, providing a chemical cue that progresses infection. Corals undergoing heat stress can have up to a five-fold increase in DMSP in their mucus, causing a corresponding increase in pathogen response (Garren et al., 2014). To complement this study, Rosado et al. (2019) have identified strong candidates of the coral microbiome that can increase the thermotolerance of the host, reducing the severity of bleaching by multiple means, one of which being the degradation of DMSP. By doing so, the infochemical that V. coralliilyticus uses to target its host is metabolized, preventing the progression of infection (Rosado et al., 2019). While pathogen prevalence is shown to be linked to holobiont health, potentially beneficial bacteria involved in the nutrient cycle should not be overlooked.
Diazotrophic (nitrogen-fixing) bacteria form site- and species-specific associations with various corals (Lesser et al., 2004; Lesser et al., 2007, Olson et al., 2009; Lema et al., 2012; Olson and Lesser, 2013; Lema et al., 2014; Santos et al., 2014; Benavides et al., 2017; Lesser et al., 2018). These supply the coral host and algal symbionts with an additional source of nitrogen when availability of dissolved inorganic nitrogen is scarce (Wang and Douglas, 1998; Reshef et al., 2006; Rädecker et al., 2015; Benavides et al., 2016; McDevitt-Irwin et al., 2017; Pupier et al., 2019). For the coral host, access to fixed forms of nitrogen is vital for the production of cellular and molecular components. (Pupier et al., 2021). For the algal symbionts, bioavailable nitrogen is required for growth, maintenance of chlorophyll-a content, thermal tolerance, and translocation of fixed carbon to the coral (Rädecker et al., 2015; Morris et al., 2019; Matthews et al., 2020; Xiang et al., 2020). Interestingly, fixed nitrogen supplied by the diazotrophs is assimilated at higher rates by algal symbionts than the coral host (Pernice et al., 2012; Benavides et al., 2016). However, nitrogen-fixation rates appear to vary: Although coral-associated diazotrophs contribute up to 18% of the overall nitrogen pool even in nitrogen-replete reefs, scleractinian (stony) corals appear to have much higher nitrogen-fixation rates than soft corals (Pupier et al., 2019). Diazotrophs are specifically associated with the scleractinian coral endolithic layer (Moynihan et al., 2021) and mucus, and much lower nitrogen-fixation rates in soft corals have been hypothesized as possibly due to differing nutritional strategies (Pupier et al., 2021). Even then, nutritional strategies for obtaining nitrogen are highly variable from species to species within both hard and soft corals (Pupier et al., 2021). Given this wide variation in the diazotroph abundance and nitrogen-fixation activity across different types of corals, a central laboratory model to elucidate the role of diazotrophs within the cnidarian holobiont seems necessary.
The sea anemone Exaiptasia diaphana (hereafter referred to as ‘Aiptasia’) has been accepted as an informative laboratory model for coral-symbiosis research because it shares similar characteristics with corals (Weis et al., 2008). Aiptasia, a cnidarian like corals, consists of the same key members of the holobiont (cnidarian host, algal symbionts, and other associated microbes) that contribute essential resources for the stability of nutrient cycling (Voolstra, 2013; Baumgarten et al., 2015; Rädecker et al., 2015; Rädecker et al., 2018; Rädecker et al., 2019). Aiptasia is fast-growing under minimal conditions, making it suitable for diverse laboratory experiments (Voolstra, 2013). In addition, Aiptasia can be maintained in symbiotic (with algal symbionts) and aposymbiotic (without algal symbionts) states for long periods (Weis et al., 2008; Voolstra, 2013; Rädecker et al., 2018). Like corals, Aiptasia also offers some degree of algal-symbiont flexibility, in which it can establish symbiosis with similar strains of Symbiodiniaceae associated with corals (Thornhill et al., 2013). Finally, Aiptasia lacks a calcium-carbonate skeleton, making it easier to process tissue for cellular and molecular work in the laboratory (Weis et al., 2008; Voolstra, 2013; Baumgarten et al., 2015; Rädecker et al., 2018). As a result, Aiptasia is a powerful system for studying symbiosis and metabolic interactions between animal and microbial partners.
Although microbiome research pertaining to corals has long been explored, similar studies applied to the Aiptasia model system are only recently gaining traction. Röthig et al. (2016) reported a ‘core’ microbiome, but also observed notable differences in the bacterial community associated with Aiptasia in aposymbiotic and symbiotic states, suggesting algal presence may play a role in influencing the bacterial community’s composition. In symbiotic anemones, Herrera et al. (2017) showed distinct bacterial communities associated with the clonal lines H2 (Hawaii) and CC7 (Florida), demonstrating a lack of uniformity in bacterial community composition across laboratory clonal lines. Subsequently, Hartman et al. (2020a) compared these microbiomes with those of wild Aiptasia sourced from the Great Barrier Reef. They found a comparable composition of dominant taxa, albeit with lower evenness and perhaps lower bacterial diversity in laboratory-cultured strains of Aiptasia. Characterization of the microbiomes associated with defensive tissues (acontia) observed similarity across wild Aiptasia sourced from the Great Barrier Reef (Maire et al., 2021).
Aiptasia also appears to be a suitable model for coral diseases. Bacterial pathogens such as the genus Vibrio infected Aiptasia in comparable ways (Zaragoza et al., 2014), with disease progressing along the same necrotizing pattern, and comparable glycosidase activity. These complementary studies highlight how the bacterial community influences, both directly and indirectly, the maintenance of cnidarian symbioses and their role in the prevention of dysbiosis (whereby the partnership between the symbiotic dinoflagellates and their cnidarian host collapses, leading to rapid death of coral ecosystems). Current interest in aiding endangered coral-reef systems using microbiome transplants as ‘probiotics’ has motivated manipulation studies to produce gnotobiotic strains (Dungan et al., 2021b; Hartman et al., 2022), to perform microbiome transplants from coral taxa (Costa et al., 2021), or to try and mitigate oxidative stress (Dungan et al., 2021a; Dungan et al., 2022).
Temperature stress, currently occurring globally as a result of anthropogenic climate change, causes dysbiosis. This occurs by one or more cellular mechanisms: (i) in situ degradation of the algal cell, (ii) exocytic release of algae from the gastroderm, (iii) detachment of host cells containing algae, or (iv) death of the host cell (Bieri et al., 2016). Recent work has shown changes in nutrient cycling prior to overt bleaching (Rädecker et al., 2021). The impact of temperature stress on the coral holobiont and the role of the microbiome in mediating sensitivity to temperature stress (Morrow et al., 2018) have motivated studies with Aiptasia. Hartman et al. (2020b) observed a decline in microbiome diversity in Aiptasia from the Great Barrier Reef in response to a 7°C elevation over 2 weeks, along with bleaching. In contrast, Ahmed et al. (2019) reared Aiptasia strains from North America at ambient (25°C) and elevated (32°C) temperatures for >2 years and observed higher richness (number of taxa) in heat-stressed polyps. These studies reflect likely differences in short- and long-term acclimations, but also possibly changes due to different Aiptasia lines.
In the present study, we examined the bacterial communities associated with symbiotic and aposymbiotic Aiptasia of the CC7 clonal line (Sunagawa et al., 2009) during exposure to ambient (27°C) and elevated (34°C) temperatures over 8-10 days. We implemented 16S and 18S rRNA gene sequencing to characterize the abundance of various prokaryotic and eukaryotic taxa respectively. We predicted differential responses of the Aiptasia microbiome to temperature changes and symbiotic state, with an impact on bacterial taxa that have major functions in nutrient cycling within the holobiont.
With little known about the nitrogen-fixation potential of bacteria associated with Aiptasia, we also screened symbiotic and aposymbiotic anemones specifically for the abundance of the nifH gene and its expression via PCR and quantitative PCR (qPCR). nifH encodes for the iron protein of nitrogenase, the enzyme that converts nitrogen to ammonium, and is conserved across all diazotrophs (Zehr et al., 2003; Gaby and Buckley, 2012), thereby making it an effective biomarker for their detection. We also assessed nitrogen-fixation activity in symbiotic and aposymbiotic Aiptasia under ambient (27°C) and elevated (34°C) temperatures, expecting differences across symbiotic state and temperature.
Materials and methods
Animal-rearing conditions
Symbiotic and aposymbiotic Aiptasia belonging to the CC7 clonal line (Sunagawa et al., 2009) were reared in 1-L polystyrene tanks filled with 700 mL of artificial seawater (ASW) (Red Sea Coral Pro Salt) at 27°C while being fed Artemia nauplii and supplied with fresh ASW twice per week. Symbiotic Aiptasia were raised in an incubator on a 12 h light:12 h dark cycle subjected to 25 μmol photons m-² s-¹ of white LED light, while aposymbiotic Aiptasia, previously derived from 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) treatment according to established methods (Xiang et al., 2013), remained in the dark. Ten days prior to the beginning of each experiment, anemones allocated for the experiment were deprived of food to ensure Artemia were not present in the anemones’ gastric cavities.
Heat-stress microbiome studies
Two different temperature studies were conducted, each with an ambient treatment at 27°C, and a heat-stress treatment at 34°C, which prompts bleaching (Bieri et al., 2016; Esherick et al. in revision). A preliminary 10-day endpoint study with triplicate replication was performed to assess variance. Following this experiment, a subsequent 8-day time-series experiment was conducted without replication, but with much greater numbers of polyps per time point and associated observation of bleaching. This experiment, which added sequencing of eukaryotic 18S rRNA, was designed to give distinct but complementary results that could be intercompared with the endpoint experiment.
Endpoint experiment. Thirty symbiotic and 30 aposymbiotic polyps were split into twelve separate 700-mL tanks, each containing 5 individuals. Half of the tanks were placed in a 34°C incubator while the other half were kept at 27°C for the entire duration of the 10-day trial. On the final day, triplicate samples, each consisting of a mixture of 5 individual polyps, were collected for DNA extraction (Figure 1A). Triplicate samples of seawater were also filtered for DNA extraction for seawater bacterial communities, although only one was sequenced due to cost limitations.
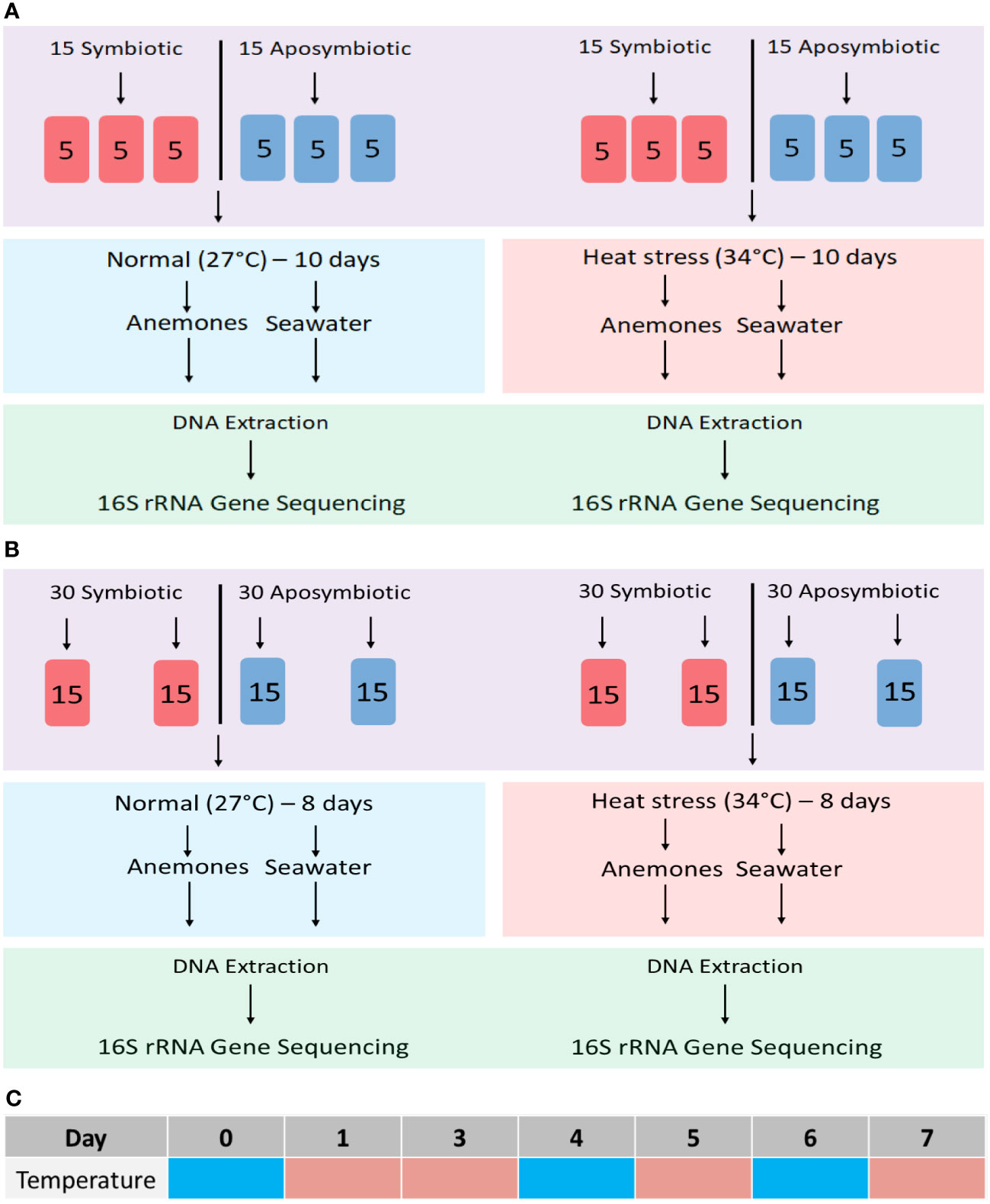
Figure 1 Experimental workflows for microbiome community profiling in the sea anemone Aiptasia and its surrounding seawater. (A) Initial 10-day endpoint experiment; (B) 8-day time-series experiment. (C) Time-series sampling from tanks at 27°C on days 0, 4, and 6 (cyan) and from tanks at 34°C on days 1, 3, 5 and 7 (peach).
Time-series experiment. Sixty aposymbiotic and 60 symbiotic anemones were incubated at an ambient temperature of 27°C, with four tanks from each symbiotic state harbouring 15 individuals each (Figure 1B). On day 0, half of the tanks were moved to a heat-stress temperature of 34°C for the remainder of the 8-day trial. Throughout the 8-day time series, algal fluorescence was used as a qualitative proxy for algal abundance as anemones bleached during thermal stress. Five images per anemone were taken at each time point using a Leica MC190 HD camera equipped to Leica M165FC fluorescence stereo microscope with a GFP longpass filter. All images were standardized using 125 ms exposure, 3.4X magnification, and 10X gain.
DNA samples were collected from Aiptasia and their surrounding seawater throughout the 8-day period. In ambient-temperature treatments, samples were collected on day 0, 4 and 6 (Figure 1C). In the heat-stress treatment, samples were collected on days 1, 3, 5, and 7 to observe community responses of both prokaryotes and eukaryotes before and after peak bleaching (Figure 1C).
Microbiome community profiling
DNA extraction from Aiptasia and seawater
For DNA extractions, polyps were placed in a 1.5-mL microcentrifuge tube and washed two times with 500 µL of sterilized seawater (SSW). After washing, 200 µL of SSW was added to each tube, processed with an OMNI Tissue Master homogenizer, then needle-aspirated (25G) further to ensure the sample was properly homogenized. Next, 300 µL of lysis buffer (Buffer AP1, DNeasy Plant Mini Kit, Qiagen) were added and the remainder of the DNA extraction was performed according to the manufacturer’s instructions.
Seawater was sampled to isolate bacterial DNA representative of the surrounding environment of the anemones. Each tank’s surface was thoroughly agitated with a micropipette tip to release any bacterial cells that had adhered to the tanks prior to sampling. In the endpoint experiment (described above), 1.4 L of seawater from two separate tanks were filtered through a single 0.4-µm Durapore PVFD filter on day 10 of the trial. Filters were cut into strips using a sterile razor blade and placed in a 1.5-mL microcentrifuge tube with 500 µL of lysis buffer (DNeasy Plant Mini Kit, Qiagen). For the time-series experiment (described above), the same process was repeated instead with 120 mL of water filtered on each sampling day of the 8-day trial from the same tank, showing changes in the bacterial community within a single tank for the duration of the trial. The remainder of the DNA extraction for both experiments was performed according to the manufacturer’s instructions.
DNA quality control and gene sequencing
Quality of the DNA extracted was carefully assessed for each experiment, in particular for the time-series experiment. Multiple DNA extractions were conducted from the same samples and analyzed via gel electrophoresis and NanoDrop (Thermo Fisher Scientific) to screen for low recoveries. Samples with adequate recoveries were pooled.
All DNA samples from both experiments were also verified with initial PCR tests using rRNA primers for all three domains, then stored at -20°C until submission for gene sequencing. Community rRNA gene sequencing was conducted at MR DNA (www.mrdnalab.com) on an Ion Torrent PGM following the manufacturer’s guidelines. The prokaryote 16S rRNA gene V4 region was amplified using primers 515F/806R (Caporaso et al., 2011), while for eukaryotes the 18S rRNA gene V4 region was amplified using primers V4F/V4R (Stoeck et al., 2010). Eukaryotic gene sequencing was performed only on symbiotic samples.
Data analysis
Reads from 16S and 18S rRNA gene sequencing were processed with a proprietary analysis pipeline (MR DNA). Sequences with less than 150 bp after barcode and primer removal were discarded, as well as sequences containing ambiguous base identification and homopolymer runs greater than 6 bp. Operational taxonomic units (OTUs) were generated with the parameter of 97% sequence homology after conducting BLASTn searches in an aggregate database consisting of GreenGenes, RDPII, and NCBI. From these taxonomic assignments, raw OTU read counts were organized at different phylogenetic levels. For comparison with prior studies, we focused on OTUs classified at the family level for this study (90-99% sequence identity).
Statistical assessment of bacterial OTU variance in the endpoint experiment was done with PERMANOVA using the R Vegan package. Bacterial family-level composition was examined both untransformed and square-root-transformed, with 9999 permutations; both analyses gave similar results.
Detection of nitrogen-fixation potential by Aiptasia microbiomes
Detection of nifH genes and expression
Development of nifH methods was conducted after the microbiome studies and involved separate experiments with polyps of the same clonal line (CC7) that were reared under the same laboratory conditions as described above. Genomic DNA (gDNA) from Aiptasia was used to quantify nifH abundance, while complementary DNA (cDNA) was used to quantify nifH expression. Total DNA and RNA were extracted from homogenized symbiotic and aposymbiotic anemones and fava-bean root nodules (as a positive control), following the manufacturer’s instructions using a ZymoBIOMICS DNA miniprep kit (Zymo Research) and RNAqueous-4PCR Total Isolation kit (Thermo Fisher Scientific) respectively. DNA and RNA concentrations from all samples were verified via a NanoDrop (Thermo Fisher Scientific). To synthesize cDNA, ~500 ng of total RNA of each sample type was processed with the Superscript III First-Strand Synthesis kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. Before qPCR analysis, gDNA and cDNA were diluted to 5 ng µL-1 for all samples.
Initial PCR tests were conducted to choose the best nifH primer set (Table S1 and Figure S1), Ueda19F/R6 (Angel et al., 2018). This primer set was then used for real-time PCR analysis (Eppendorf MasterCycler). All qPCR reactions used 5 µl of PowerTrack SYBR Green Master Mix (Invitrogen), 0.5 µL of each primer at a concentration of 8 µM, 2 µL of template, and 2 µL of nuclease-free water. A 4X 1:5 dilution series of cDNA was used to generate standard calibration curves for testing primer efficiency. qPCR was conducted with the following thermal-cycler conditions: 95°C for 2 min, followed by 50 cycles of 95°C for 30 sec, 54°C for 90 sec, and 72°C for 1 min. The 2-∆∆Ct method (Livak and Schmittgen, 2001) was used to determine potential differences in relative fold-change of nifH, normalized to the 16S-V3 rRNA gene copy numbers from aposymbiotic anemones and root nodules.
Detection of nitrogenase activity
The acetylene reduction assay (ARA) was applied to Aiptasia by modifying the parameters previously used on coral colonies or fragments (Shashar et al., 1994; Wilson et al., 2012; Rädecker et al., 2014; Pogoreutz et al., 2017; Tilstra et al., 2019) to accommodate the smaller size of the anemones (Figure 2). Three replicate 60-mL serum vials, each containing 48 mL ASW, of which 10% (4.8 mL) had been previously saturated with acetylene, were prepared for Aiptasia of the CC7 clonal line. Symbiotic and aposymbiotic anemones (average pooled weights 1.11 g and 0.87 g, respectively) were added to vials and allowed to acclimate for 10-15 min. Control vials were prepared with 48 mL of artificial seawater, of which 10% was previously saturated with acetylene alone (no Aiptasia, negative control), or with freshly collected fava-bean root nodules in ambient air as a positive control (triplicate; average pooled weight 4.87 g), as previous studies have used them to assess nitrogen-fixation rates (Hudd et al., 1980; Herdina and Silsbury, 1990).
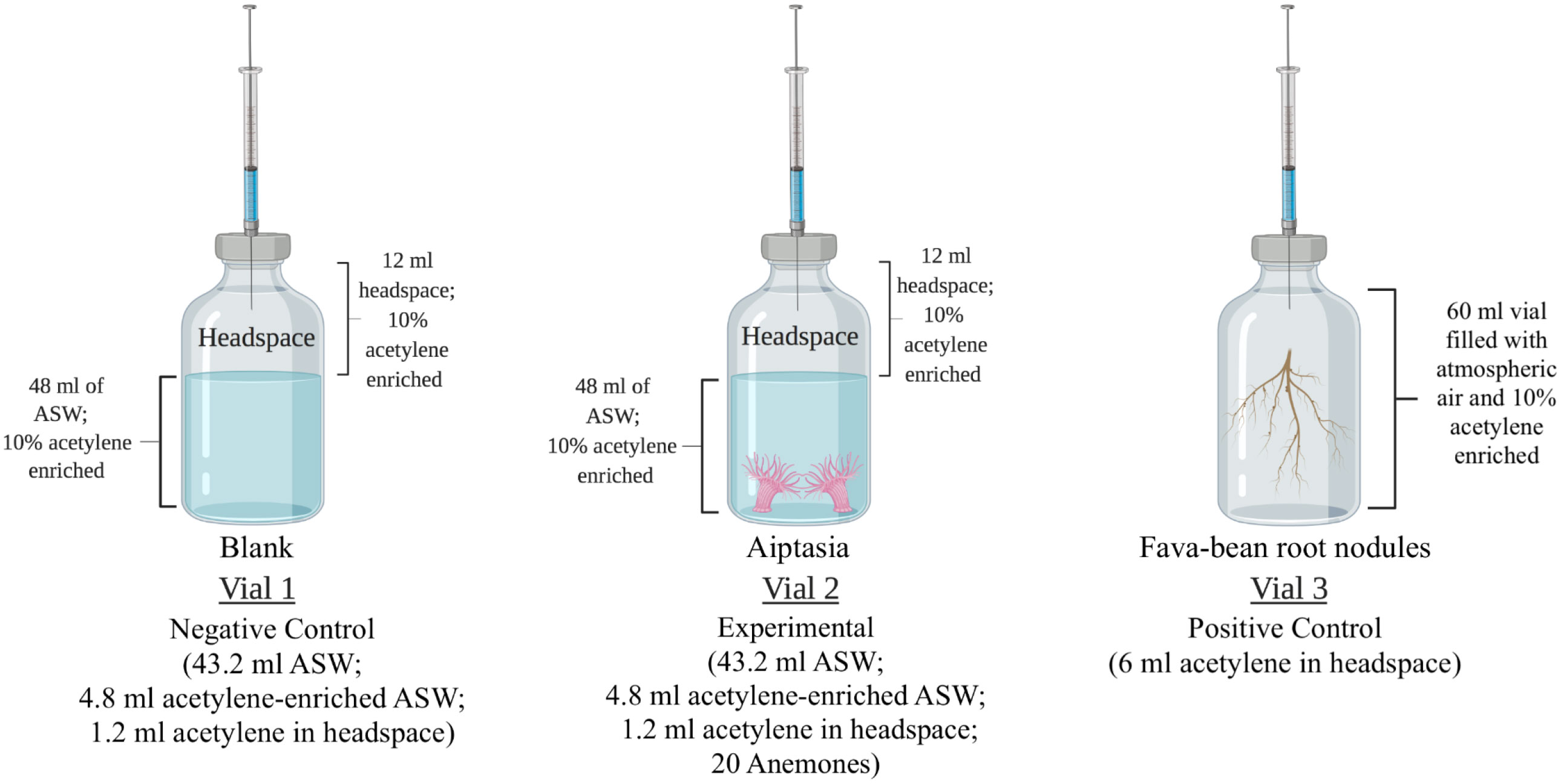
Figure 2 Experimental design for measuring nitrogenase activity using the acetylene reduction assay (ARA). Vial 1: negative control for the detection of acetylene gas in the headspace. Vial 2: experimental vials in which twenty anemones (either symbiotic or aposymbiotic) were added. Vial 3: positive control containing fava-bean root nodules. Gas samples were collected at the start and end of a 24-hour incubation period for the detection of acetylene and ethylene using gas chromatography. ASW, artificial seawater. Created with BioRender.com.
Vials were crimp-sealed with rubber septa, and 1.2 mL of the headspace was removed with a gas-tight syringe and replaced with acetylene. This created a headspace environment with 10% acetylene. From each vial, three replicate 100-µL gas samples were immediately collected via the rubber septa and analyzed for acetylene and ethylene by gas chromatography (HP 6890 equipped with a GS-CarbonPLOT column and a flame ionization detector). All gas samples were analyzed under the following optimized parameters for a 6-minute run time: inlet temperature of 250°C, split mode with a 50:1 split ratio, helium as a carrier gas, a flow rate of 2.1 ml min-1, initial oven temperature of 45°C with a hold time of 3.80 minutes, a ramp-up time of 25°C min-1, a final oven temperature of 100°C, and a detector temperature of 250°C (Table S2).
Subsequently, vials containing aposymbiotic anemones were covered with aluminum foil to prevent light exposure. All vials were then incubated at 27°C, shaking at 130 rpm, and exposed to a 12 h light:12 h dark cycle with white LED lights (25 μmol photons m-2 s-1). After 24 h, triplicate gas samples were again collected and analyzed by gas chromatography. A separate ARA experiment was conducted at an incubation temperature of 34°C with only one replicate each on symbiotic and aposymbiotic Aiptasia (pooled weights 1.17 g and 1.25 g, respectively) and fava-bean root nodules (pooled weight 5.32 g).
Results
Predominant members of two bacterial families make up Aiptasia’s microbiome
The initial 10-day endpoint experiment, which assessed Aiptasia bacterial microbiomes, yielded 2,508 bacterial OTUs. However, even though each replicate was a mixture of 5 anemones, they showed huge variance, with a minority of OTUs present in all replicates. Alpha diversity, as measured by the Shannon-Weaver index, revealed a slightly significant change due to symbiotic state only in the 34°C treatment (Figure 3, p=0.02). A PERMANOVA test showed no significant effect of heat stress on microbiome composition, but a modestly significant effect of symbiotic state (Table 1, p=0.01).
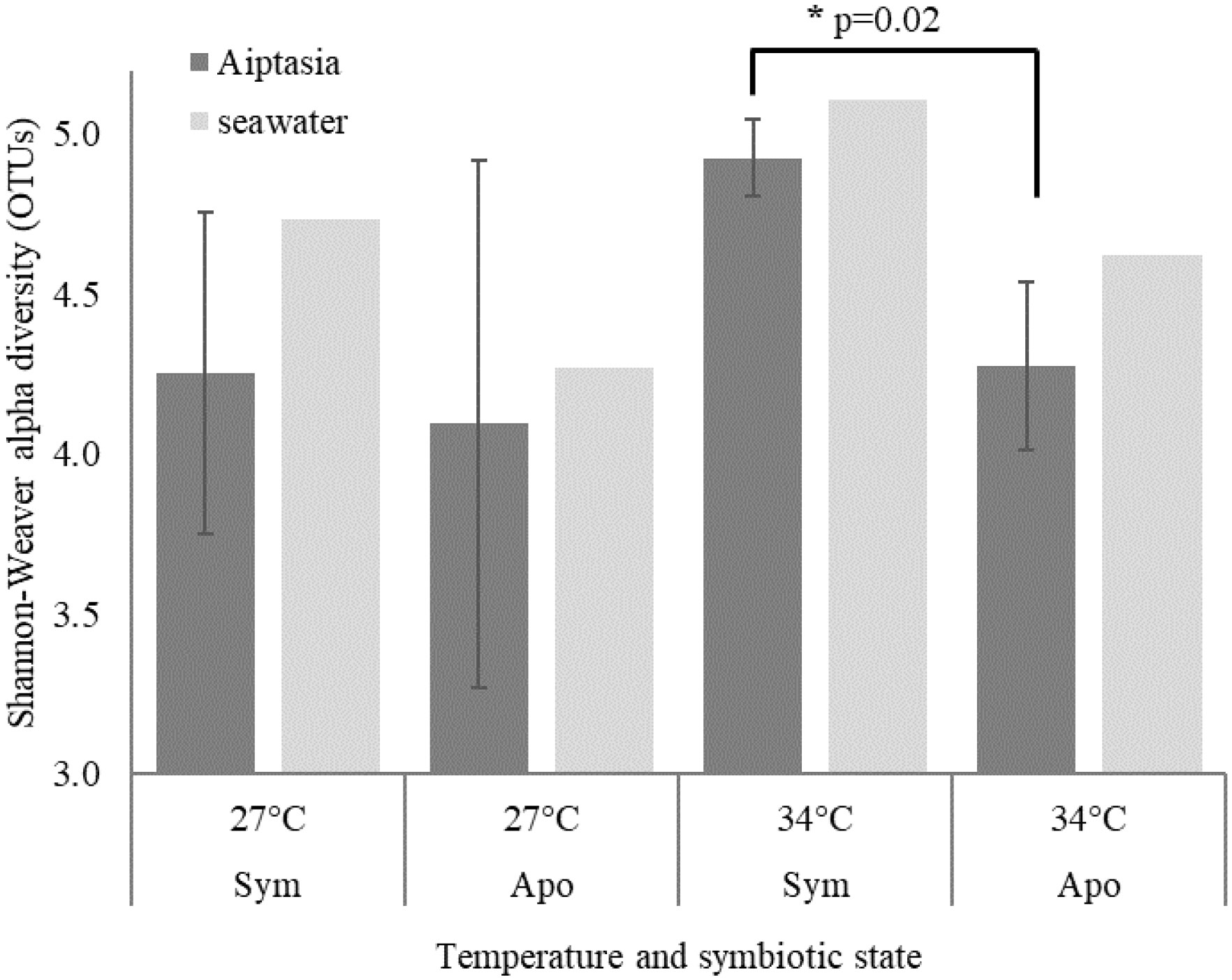
Figure 3 Shannon-Weaver alpha diversity based on bacterial operational taxonomic units (OTUs) for the 10-day endpoint experiment. Means ± 1 standard deviation are shown for 3 Aiptasia replicates, each containing 5 polyps. Although triplicate seawater samples were collected, only one was sequenced due to cost. Sym, symbiotic anemones; apo, aposymbiotic anemones. * denotes p=0.02.
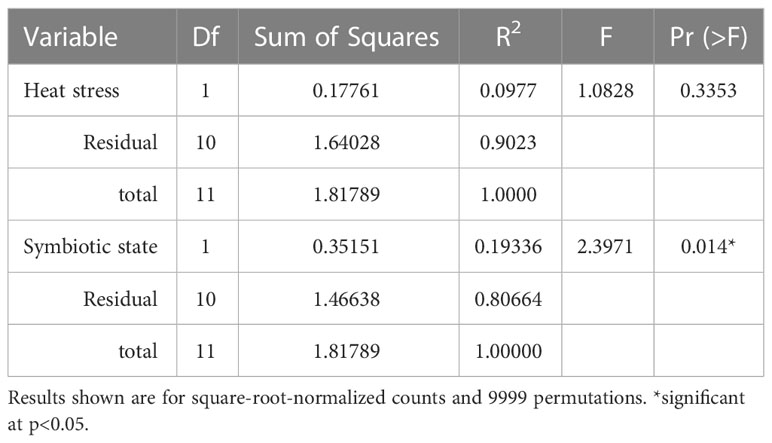
Table 1 PERMANOVA test results depicting impact of heat stress and symbiotic state on the Aiptasia bacterial microbiome (at the family level) in the 10-day endpoint experiment with triplicates.
The subsequent 8-d time-series experiment took a complementary approach. We used twice as many polyps and merged 15 anemones per time point to observe changes during and after bleaching. We also added sequencing of eukaryotes and archaea. This experiment yielded considerably more bacterial OTUs (11,787), likely due to increased quantity of Aiptasia DNA extracted. Although it lacked replication, this experiment showed consistent patterns across time points and in response to bleaching. Rhodobacteraceae and Alteromonadaceae dominated Aiptasia bacterial microbiomes in both experiments, but the first experiment showed a higher fraction of low-abundance families (40-60%) due to variance, as well as possible contamination, so we will focus on the time-series results here, as they contain more detailed information.
Across algal-symbiotic states and temperature treatments in both Aiptasia and seawater, 24-40% of all OTUs belong to bacterial families Alteromonadaceae and Rhodobacteracaeae (Figure 4). At the start of the experiment prior to temperature shift (day 0), we observed that symbiotic state determined bacterial microbiome composition in Aiptasia (Figure 4A) but not the surrounding seawater (Figure 4B). Symbiotic Aiptasia were dominated by members of Pelobacteraceae (30%) and Rhodobacteraceae (17%), while aposymbiotic Aiptasia were dominated by members of Alteromonadaceae (22%), with smaller fractions of Rhodobacteraceae (10%) and Kordiimonadaceae (9%), a group that was <1% of the microbiome in symbiotic anemones (Figure 4A). In contrast, bacterial communities of the seawater surrounding symbiotic and aposymbiotic animals were less distinct (Figure 4B), and were dominated by members of Alteromonadaceae and Rhodobacteraceae, but with significant contributions from members of Sphingomonadaceae and Chitinophagaceae, two sugar-consuming families.
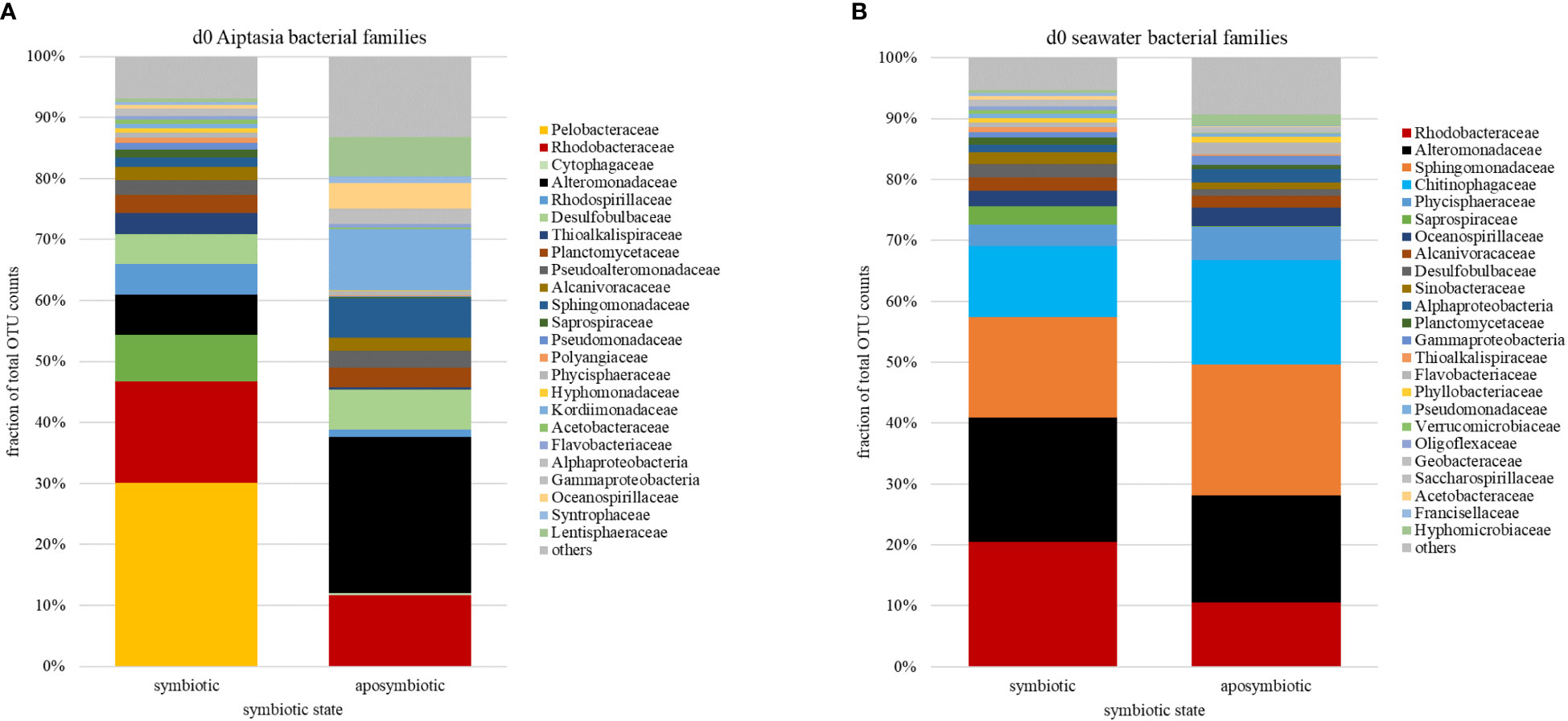
Figure 4 Relative abundances of bacterial families (based on counts of operational taxonomic units, OTU) associated with the sea anemone Aiptasia (A) and surrounding seawater (B) at the start (day 0) of the 8-day time-series experiment, prior to temperature stress. Families are ranked by abundance in symbiotic Aiptasia or their seawater. d, day.
Bleaching event following temperature shift is supported by algal fluorescence but not 18S OTU abundance shifts
We observed a decrease in chlorophyll fluorescence in heat-stressed Aiptasia, occurring most dramatically between days 3 and 5, most notably in the tentacles (Figure 5). In contrast, fluorescence remained constant in anemones that remained at ambient temperature (Figure 5). 18S sequencing of Aiptasia and seawater revealed consistent shifts in the eukaryotic community over this event. Aiptasia samples were dominated, as expected, by anemone and Symbiodiniaceae DNA, with only traces of other eukaryotes. In seawater, we found little Symbiodiniaceae DNA at day 0 at 27°C, as expected. Diatoms (Nitschzia sp.) were detected on day 0, but they rapidly declined by day 3 (Figure 6A), likely due to grazing by a gastrotrich (Chaetonotus), which increased on day 4 (Figure 6B). Exclusive to the 34°C treatments, we observed an increase of Symbiodiniaceae (normalized to Aiptasia OTU counts) in seawater from days 3-5, corresponding to Aiptasia bleaching (Figure 6C). This declined by days 6-7, following a second bloom of the gastrotrich. Curiously, though, counts of Symbiodiniaceae OTUs in Aiptasia (normalized to Aiptasia counts) did not decrease in the 34°C treatments (Figure 6D).
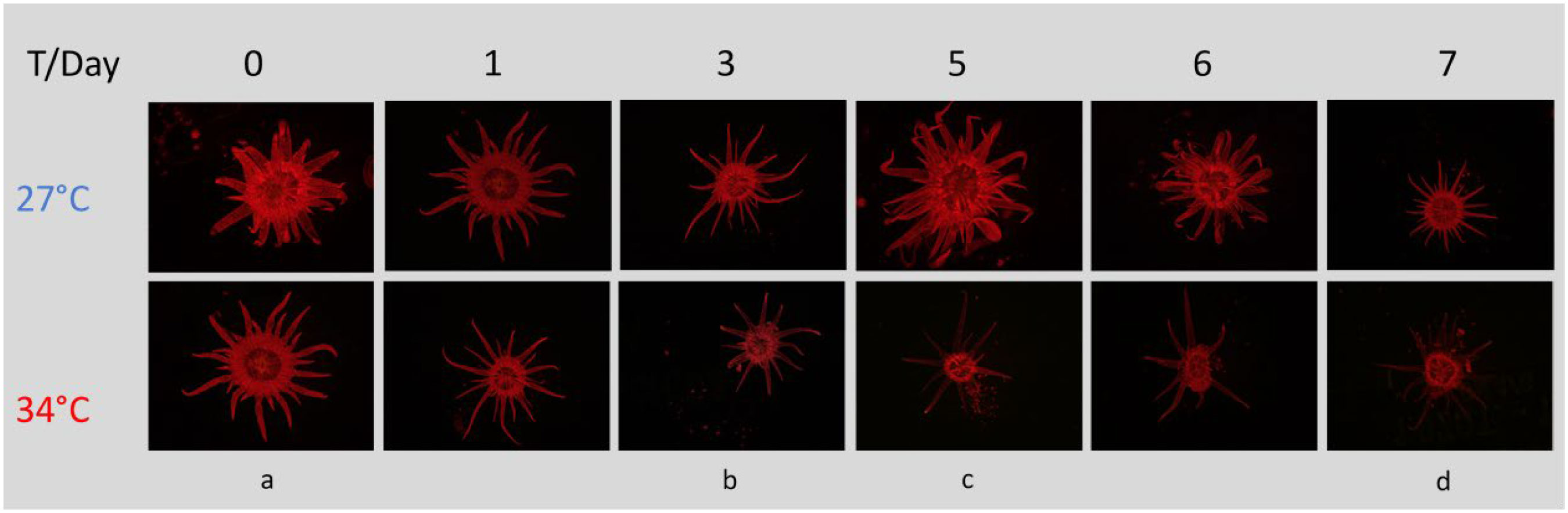
Figure 5 Chlorophyll fluorescence in the sea anemone Aiptasia decreases over time while undergoing heat stress, most notably in the tentacles. Experimental stages denoted by letters: (a) just before onset of heat stress; (b, c) before and after peak expulsion of algae; (d) end of experiment. All images were taken with the same settings (125 ms exposure, 3.4X magnification, 10X gain) using a Leica MC190 HD camera equipped to a Leica M165FC fluorescence stereo microscope with GFP longpass filter. T, temperature.
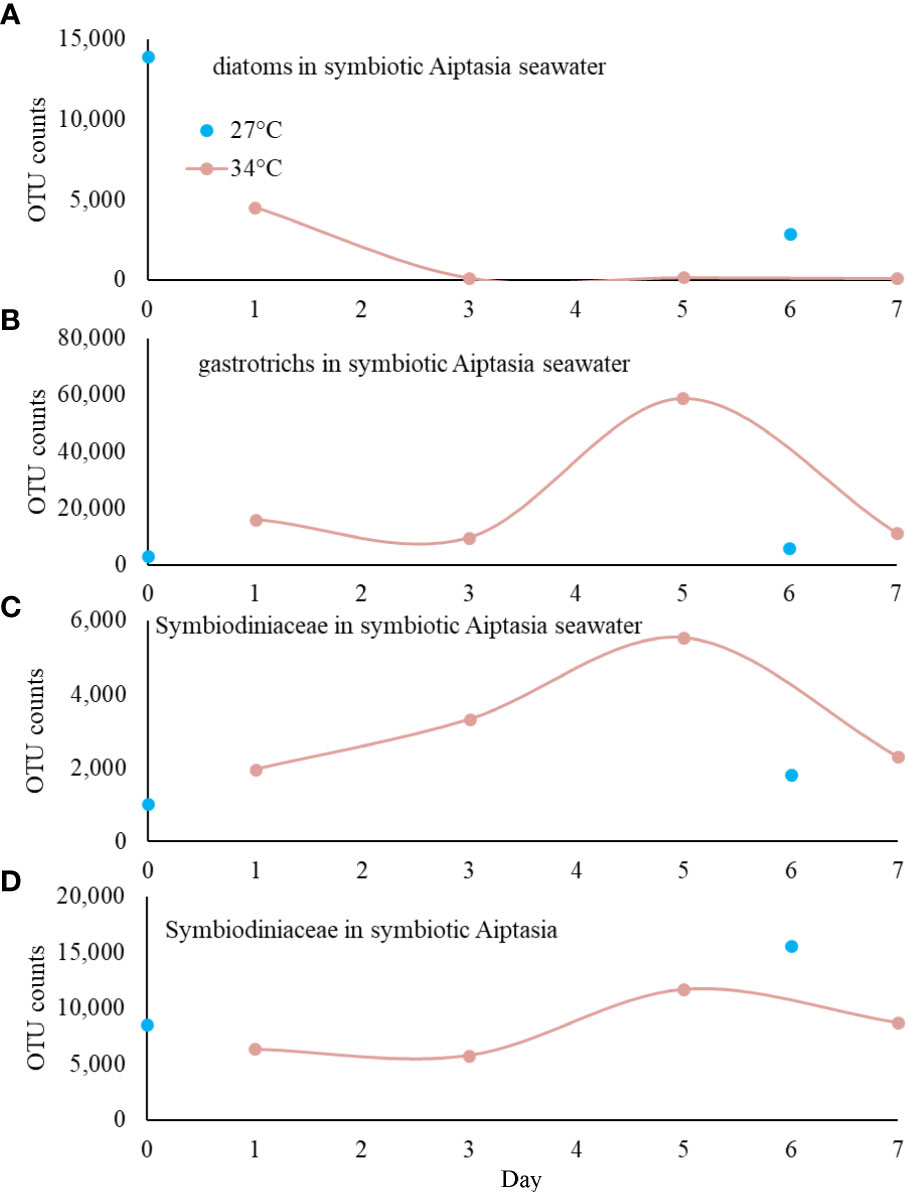
Figure 6 18S V4 profiling of eukaryotes associated with symbiotic Aiptasia over 8 days (based on counts of operational taxonomic units, OTU); heat stress was applied immediately after sampling at day 0. (A) Diatoms were observed at the start of the experiment, but only in symbiotic cultures. They decreased rapidly, likely from grazing by the gastrotrich Chaetonotus (B). Symbiodiniaceae found in seawater (C) increased during bleaching (days 3-5) in heat-stress treatments only, but there was no corresponding decrease of Symbiodiniaceae in Aiptasia anemones (D).
Bacterial and archaeal microbiome shifts follow heat stress and bleaching
In 27°C samples over 8 days, we observed variability, but no consistent shifts in bacterial microbiomes for Aiptasia, which retained the symbiotic signatures of day 0 (Figure S2A). In contrast, seawater samples showed clear shifts. In seawater surrounding symbiotic anemones, the fraction of Rhodobacteraceae and Saprospiraceae members increased, while those of Alteromonadaceae, Sphingomonadaceae and Chitinophagaceae decreased (Figure S2B). Even more dramatic was a greater representation of Alteromonadaceae in seawater surrounding aposymbiotic anemones on days 4 and 6, which dominated the community (Figure S2B). Reflecting this, alpha diversity indices remained fairly consistent throughout the experiment, aside from the seawater associated with aposymbiotic Aiptasia (Table 2), which decreased markedly.
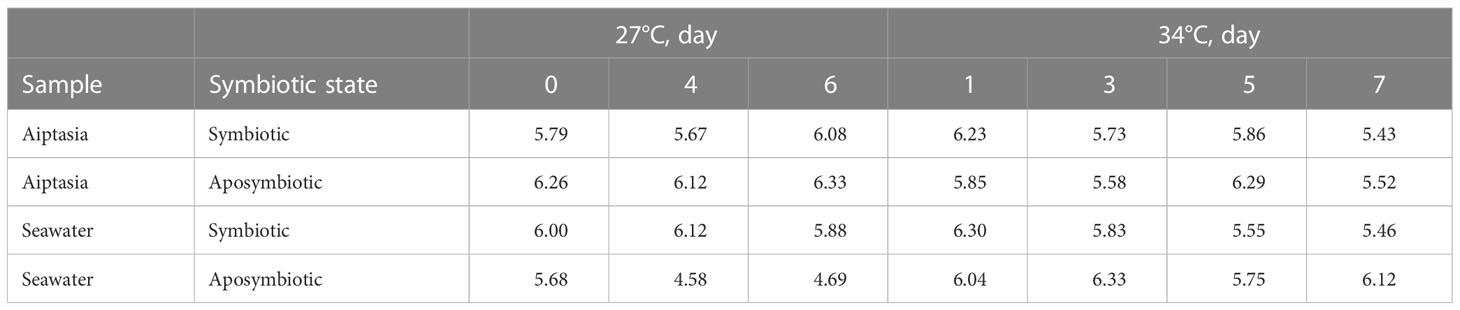
Table 2 Bacterial community Shannon-Weaver alpha-diversity indices for the 8-day time-point experiment, calculated based on operational taxonomic units (OTUs), and stratified by sample type, symbiotic state, and temperature.
Under heat stress (34°C), we observed microbiome changes in both symbiotic states of Aiptasia. Just one day following heat stress, symbiotic anemones lost almost all their Pelobacteraceae. This loss extended into bleaching, but recovered after day 5 (Figure 7A), along with more Saprospiraceae members present. Seawater from symbiotic anemones also showed a slightly greater abundance of Saprospiraceae following heat stress (Figure 7B), particularly around days 3-5 when bleaching was observed, with a corresponding decline of Alteromonadaceae members.
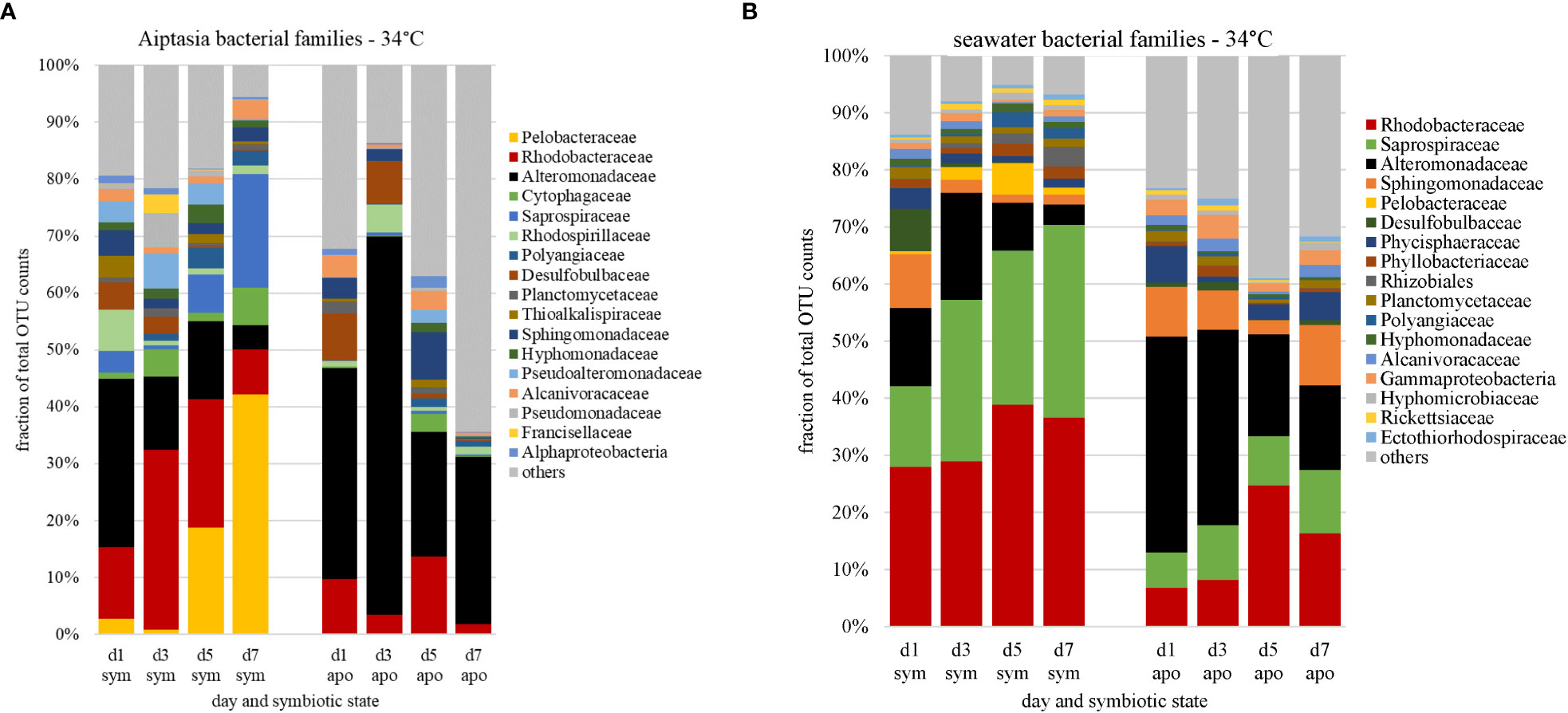
Figure 7 Changes in relative abundances of bacterial families (based on counts of operational taxonomic units, OTU) associated with Aiptasia (A) and surrounding seawater (B) in response to temperature stress, stratified by symbiotic state. Families are ranked by abundance from day 0 at 27°C (Figure 4); major families shown constitute 87% of all families observed; the remaining 13% are grouped as ‘other’ (grey). d, day; sym, symbiotic anemones; apo, aposymbiotic anemones.
The aposymbiotic anemones, which started with a different microbiome as explained above, also showed strong changes, with an initial larger then smaller fraction of Alteromonadaceae (Figure 7A). Most striking was the larger proportion of community members that were barely present at the start of the experiment (‘others’), increasing to over 60% of the total microbiome by day 7 (Figure 7A). As with symbiotic anemones, the microbial community in the surrounding seawater reflected these changes, with the proportion of Alteromonadaceae diminishing over time (Figure 7B) and a greater representation of ‘others’ to about 40% of the total by days 5-7. Although day 4 of the aposymbiotic anemones at 27°C also showed a large burden of ‘others’ (Figure S2A), there was no consistent trend, and ‘others’ in most samples were <15% of the communities.
We also detected several archaeal families, but only in seawater, with one possible exception (Figure 8, day 4 of the aposymbiotic Aiptasia at 27°C, but likely a contaminant). The most abundant OTUs resembled Aciduliprofundum, a thermophilic euryarchaeote associated with deep-sea hydrothermal vents, but only at about 85% similarity, with another group mapping to Methanobacteriaceae. Although present at both temperatures and in both symbiotic states, we observed a dramatic increase of both groups in seawater of aposymbiotic anemones following heat stress, which remained high for the duration of the experiment (Figure 8). To our knowledge, this is the first observation of archaea associated with Aiptasia, albeit only in surrounding seawater.
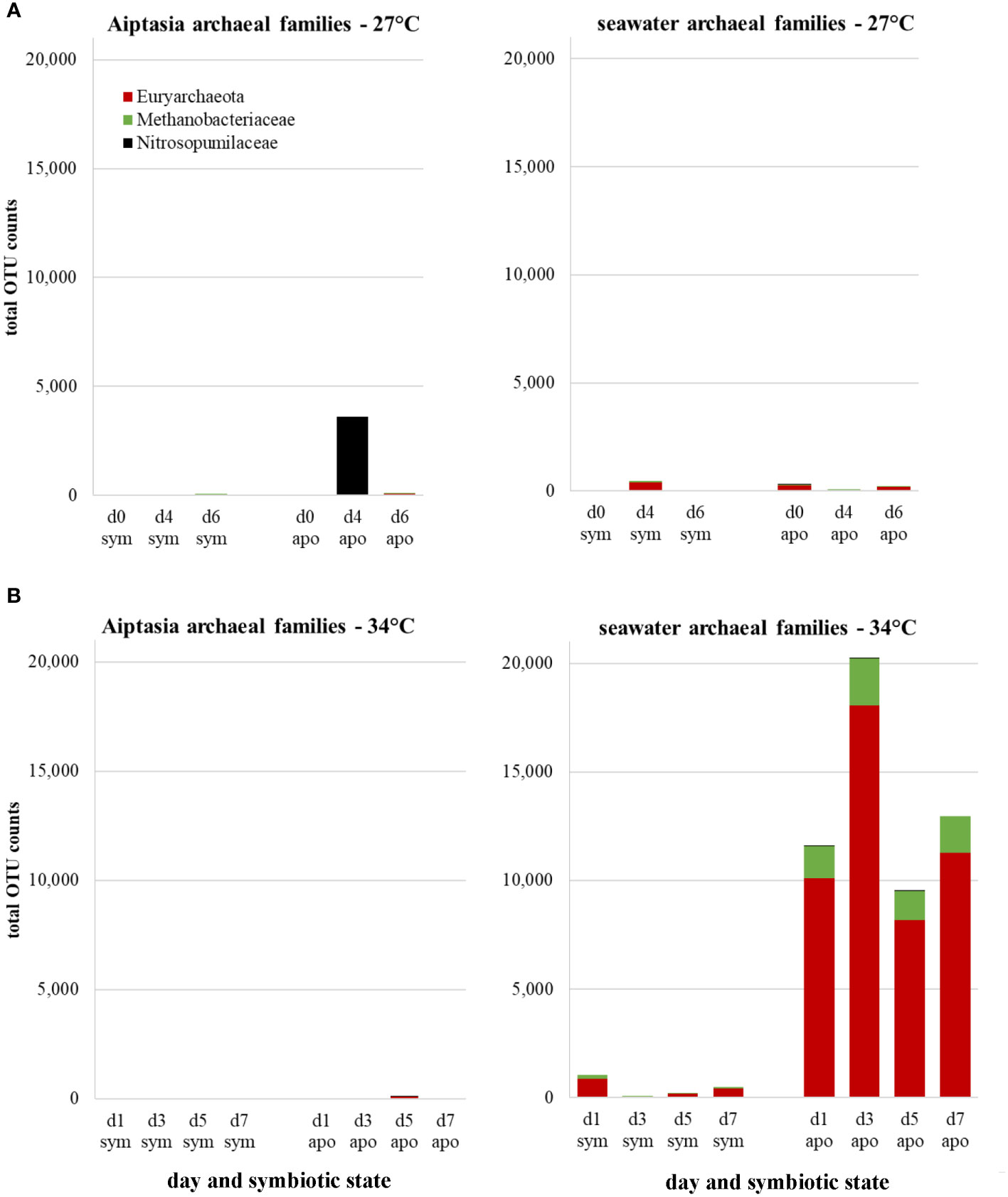
Figure 8 Changes in archaeal families expressed as counts of total operational taxonomic unit (OTU) represented in Aiptasia and surrounding seawater at 27°C (A) or in response to a temperature stress of 34°C (B), stratified by symbiotic state. d, day; sym, symbiotic anemones; apo, aposymbiotic anemones.
nifH genes are detected, but not expressed in Aiptasia microbiomes
Given that Pelobacteraceae, a family containing potential nitrogen fixers, was present only in symbiotic anemones and responded dramatically to heat stress, we further examined the nitrogen-fixation potential of diverse bacteria associated with the alga-symbiotic state of Aiptasia. To determine the presence of the nifH genes, we tested four degenerate primer sets universal for known nifH genes (Table S1), selecting Ueda19F/R6 as the optimal primer set with an amplicon size of ~454 bp (Figure S1, lanes 14-16). These primers amplify with known Pelobacter nifH genes (Figure S3). Sequencing of nifH from root-nodule samples confirmed the diazotroph, Rhizobium leguminosarum, to be the bacterial symbiont of fava-bean plants, as expected. Furthermore, sequencing confirmed the amplification of nifH sequences in symbiotic and aposymbiotic Aiptasia, and multiple diazotroph species were apparent in these samples (Phillips et al., unpublished). nifH expression, assessed via qPCR using cDNA from Aiptasia, showed no difference between symbiotic and aposymbiotic anemones, as nifH was barely expressed in either (Figure 9A). To detect whether nifH genes were even present, we attempted qPCR of gDNA from Aiptasia, but the primer efficiencies were out of range when using gDNA from all samples. However, gel analysis of nifH-PCR products using a dilution series from anemone gDNA suggested detectable nifH in Aiptasia microbiomes, with more possibly in aposymbiotic anemones (Figure S4, lane 9).
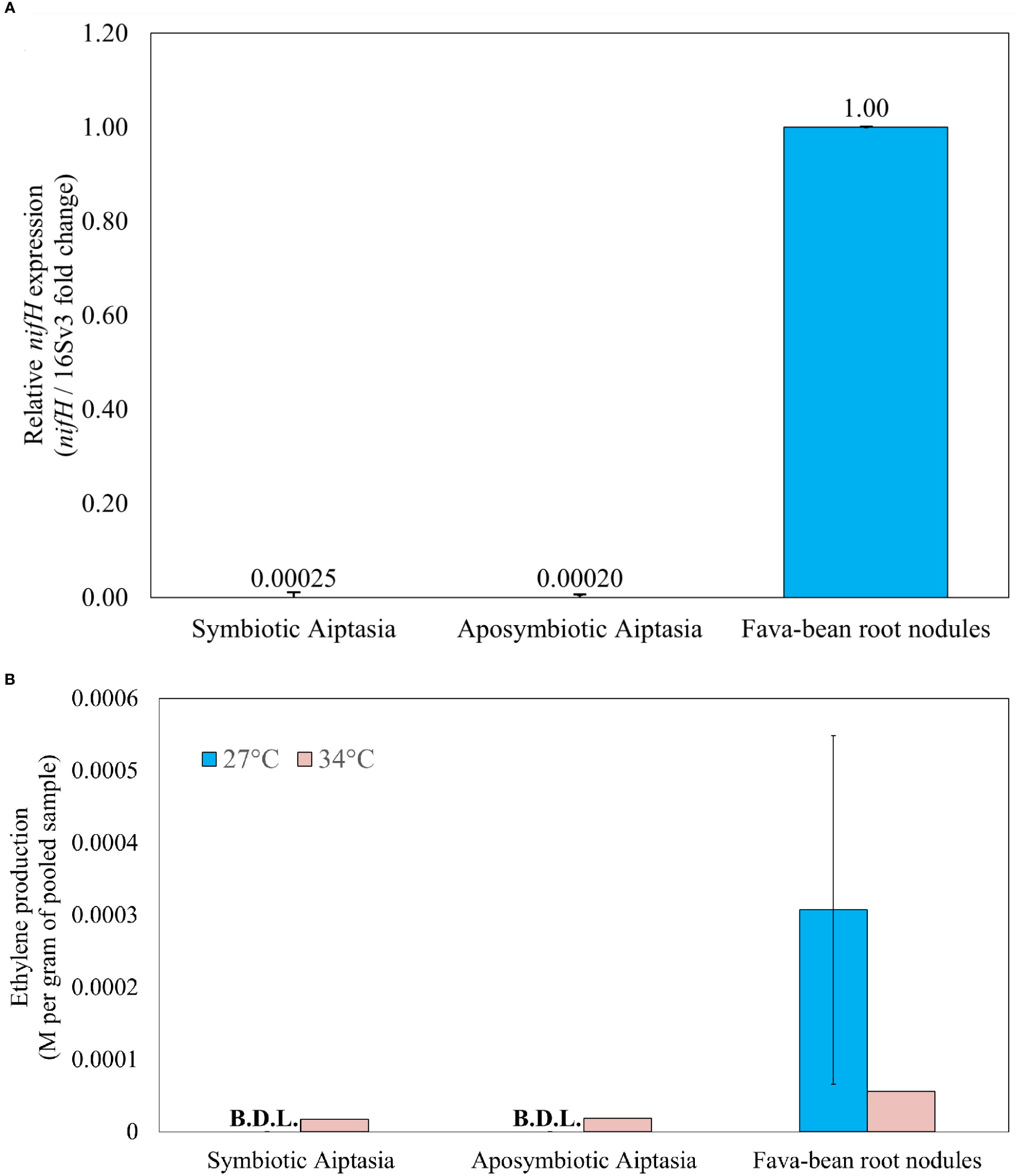
Figure 9 Nitrogenase gene expression (A) and activity (B) in Aiptasia, relative to fava-bean controls. (A) qPCR products for nifH using 5 ng µL-1 cDNA. Relative nifH expression for all samples was referenced against the V3 region of the 16S rRNA gene and normalized to aposymbiotic-anemone and fava-bean samples accordingly. (B) Nitrogenase activity, as inferred from the acetylene reduction assay, in 24-hour incubations at 27°C or 34°C. Ethylene production was normalized to the average pooled weight for each sample type. The error bar represents one standard deviation from the mean of three gas replicates taken from 3 vials of Aiptasia or 6 vials of root nodules. Only one experimental replicate was done under temperature elevation across all sample types. BDL, below detection limit.
Nitrogenase activity is barely detectable in Aiptasia, though possibly present in aposymbiotic anemones under temperature elevation
The ARA was applied to determine if differences in nitrogenase activity were present in symbiotic versus aposymbiotic Aiptasia at ambient temperature (27°C). Over 24 h, conversion to ethylene was easily detected in fava-bean root nodules. Although acetylene was observed to decrease over 24 h in all Aiptasia samples, conversion to ethylene was not detected at 27°C (Figure 9B). The ARA was then repeated at 34°C. Root nodules showed a decrease in nitrogenase activity at elevated temperature, but was still easily detectable (Figure 9B). Both symbiotic and aposymbiotic Aiptasia produced similar but barely detectable amounts of ethylene (17.4 and 18.8 µM ethylene per gram anemone respectively, Figure 9B). However, not all anemones remained alive after 24 h at 34°C.
Discussion
In cnidarian research, Aiptasia is growing as a tractable model organism for corals, primarily due to its shared symbiotic relationship with Symbiodiniaceae and ease of culturing in a laboratory setting (Weis et al., 2008; Davy et al., 2012). More recently, research into its microbiome has contributed to the development of the model by highlighting similarities of the bacterial communities between Aiptasia and corals (Röthig et al., 2016; Herrera et al., 2017; Ahmed et al., 2019; Hartman et al., 2020b). Cnidarian-associated bacterial communities may confer heat tolerance to the cnidarian host (Ziegler et al., 2017) and likely respond rapidly to changes in the environment. In corals, multiple stressors have been known to cause the associated bacterial community to change (Ainsworth and Hoegh-Guldberg, 2009; Raina et al., 2010; Littman et al., 2011; Ziegler et al., 2017). These changes to the bacterial community can disrupt the health of the coral holobiont (Thompson et al., 2015; Bourne et al., 2016; Peixoto et al., 2017); however, the mechanisms as to how this happens are still unknown.
In this study, we aimed to describe the impact of temperature on Aiptasia’s microbiome by characterizing the microbial communities of symbiotic and aposymbiotic Aiptasia under short-term heat stress at 34°C. We also examined in detail one potential microbiome benefit, nitrogen fixation, that is thought to be important for coral symbioses (Moynihan et al., 2021; Pupier et al., 2021).
Removal of algal symbionts and elevated temperature are potentially synergistic stressors that alter Aiptasia’s microbiome and pave the way for pathogens
Our study confirms prior reports that Aiptasia maintains core members of two key families, Alteromonadaceae and Rhodobacteraceae, as the most abundant across both symbiotic and aposymbiotic anemones, and at ambient and elevated temperatures. This is consistent with findings from Röthig et al. (2016) and suggests that these core members may not only be stable across multiple rearing conditions and research laboratories, but also might be specific to this strain of Aiptasia, as both studies use the CC7 clonal line. Host specificity, as opposed to environmental conditions, was once suggested not to be a major factor in bacterial community structure (Brown et al., 2017), but distinct microbiomes between the CC7 and H2 clonal lines of Aiptasia indicate otherwise (Herrera et al., 2017). Our study helps clarify some of these previously contrasting interpretations, affirming that stable members of two predominant bacterial families exist in Aiptasia and perhaps even within a specific clonal line.
However, the bacterial communities also varied dramatically across individual anemones, suggesting either tank effects, as observed previously (Hartman et al., 2020a; Xiang et al., 2022), or a stochastic element in microbiome acquisition and development, as supported by recent transplant studies (Damjanovic et al., 2017; Ziegler et al., 2017). Because of this variability, we focus primarily on the time-series experiment, which merged 15 anemones for each time point sample. Although lacking replication due to limitations of funds at the time of this experiment, multiple quality-control assessments of the DNA extractions were conducted to confidently pool anemones and sample multiple time points (see Materials and Methods). The strength of this experiment is the added capacity to observe trends in both prokaryotic (bacterial and archaeal) and eukaryotic microbial communities over time following temperature stress, with a much greater yield of bacterial OTUs and more consistent results (i.e., less variability between individual anemones or tanks).
Beyond Alteromonadaceae and Rhodobacteraceae, bacterial community structure appears rather flexible. Other bacterial families show considerable variation: Whereas Röthig et al. (2016) identified 14 other families across all their samples, we found 20 other families in ours. Although most families occurred in both studies, albeit at different levels of abundance, our control Aiptasia (symbiotic, 27°C) have microbiomes dominated by Pelobacteraceae (Figure 4A), represented in the community by the singular genus Pelobacter, a group not detected by Röthig et al. (2016). This family of Deltaproteobacteria, belonging to the order Desulfuromonadales, are fermenting anaerobes, and also have the ability to fix nitrogen (Schink, 1992; Akob et al., 2017). They were absent from aposymbiotic Aiptasia, suggesting that nitrogen fixation is more likely to occur when symbiosis with the algae is intact. This implication is in line with the findings of Xiang et al. (2022) in which diazotrophs and denitrifying bacteria were abundantly associated with symbiotic Aiptasia. In contrast, the family Kordiimonadaceae, represented by the singular genus Kordiimonas, showed the opposite pattern, being the second-most abundant family in aposymbiotic samples while absent in symbiotic anemones (Figure 4A). These bacteria are aerobic polycyclic aromatic hydrocarbon (PAH)-degrading bacteria found in a variety of shallow-water environments (Kim and Kwon, 2010; Math et al., 2012). The strong associations of Pelobacter and Kordiimonas with different symbiotic states are not described in any prior research on Aiptasia’s microbiome. Hence, caution must be taken in microbiome studies as we consider variability in the bacterial communities of various Aiptasia cultures maintained long-term in different laboratories conducting these studies, as well as the impacts of producing aposymbiotic states (e.g., use of the photosystem-II inhibitor DCMU). Interestingly, these dramatic microbiome shifts due to symbiotic state did not extend to surrounding seawater communities, which were quite similar (Figure 4B), but with large contributions from families not found in the anemones (e.g., Sphingomonadaceae, Chitinophagaceae) or in prior studies, though data are sparse.
Despite the different microbiomes present in symbiotic versus aposymbiotic animals, we observed clear trends in bacterial-community responses to heat stress corresponding to the period of bleaching in symbiotic anemones (Figures 5, 7). The dominant Pelobacteraceae in symbiotic Aiptasia dropped dramatically following heat stress, recovering only after bleaching on days 5-7 (Figure 7A), with several other families also being more (e.g., Saprospiraceae) or less (e.g., Alteromonadaceae) represented. Previous research shows coral- and Aiptasia-associated microbiomes exhibit higher diversity while undergoing bleaching caused by heat stress (Zaneveld et al., 2017; Ahmed et al., 2019). This change in diversity has been attributed to the Anna Karenina Principle (AKP), in which animals undergoing dysbiosis possess higher variation within microbial community composition compared to healthy animals (Zaneveld et al., 2017), attributed to the environmental stress limiting the ability of the host or its microbiome to regulate community composition. The outcome of a multi-year heat-stress experiment in Aiptasia (Ahmed et al., 2019) was in accordance with the AKP, but in our short-term heat-stress study, alpha-diversity indices during bleaching of symbiotic anemones were largely unchanged under heat stress (Table 2).
However, we did observe increased abundance of rare taxa during the combined treatments of (i) lack of algal symbionts and (ii) animals and seawater under elevated temperatures (Figures 7A, B). Therefore, if we interpret aposymbiotic Aiptasia to be analogous to bleached corals and potentially experiencing some stress in this state, our results suggest that the AKP may explain the combined effects of multiple short-term stressors (this study) or a sustained single stressor (Ahmed et al., 2019). In aposymbiotic Aiptasia, the most notable change was a larger proportion of rare community members, which came to dominate the heat-stressed anemones by days 5-7 (Figure 7B). Among these were potential pathogens including Vibrio species, which exhibited a steady increase in aposymbiotic animals undergoing heat stress, as well as their surrounding seawater (Figure 10A), but not in symbiotic Aiptasia or any non-heat-stressed animals (Figure 10B). Dominant Vibrio species identified included V. harveyi, V. parahemolyticus, and V. alginolyticus. In tropical corals, V. harveyi is a known causative agent of white band disease, resulting in a general loss of tissue and algal pigment (Luna et al., 2010). Similar necrotizing pathogens V. coralliialyticus and V. shiloi, originally isolated from tropical corals and inoculated into Aiptasia, also cause disease (Zaragoza et al., 2014). As we identified V. harveyi exclusively in aposymbiotic samples, this suggests that the dual stressors (i.e., lack of algae and elevated temperature) are synergistic. Microbiome-transplant studies have shown that aposymbiotic Aiptasia have a ~10X lower carrying capacity than symbiotic Aiptasia (Costa et al., 2021), and perhaps the combination of thermal stress and lower microbiome occupancy can enhance the success of pathogens in the holobiont. Follow-up studies using a growth-inhibition assay with cultured V. harveyi in the presence of Symbiodiniaceae could isolate a potentially interesting algal-bacterial interaction within the holobiont.
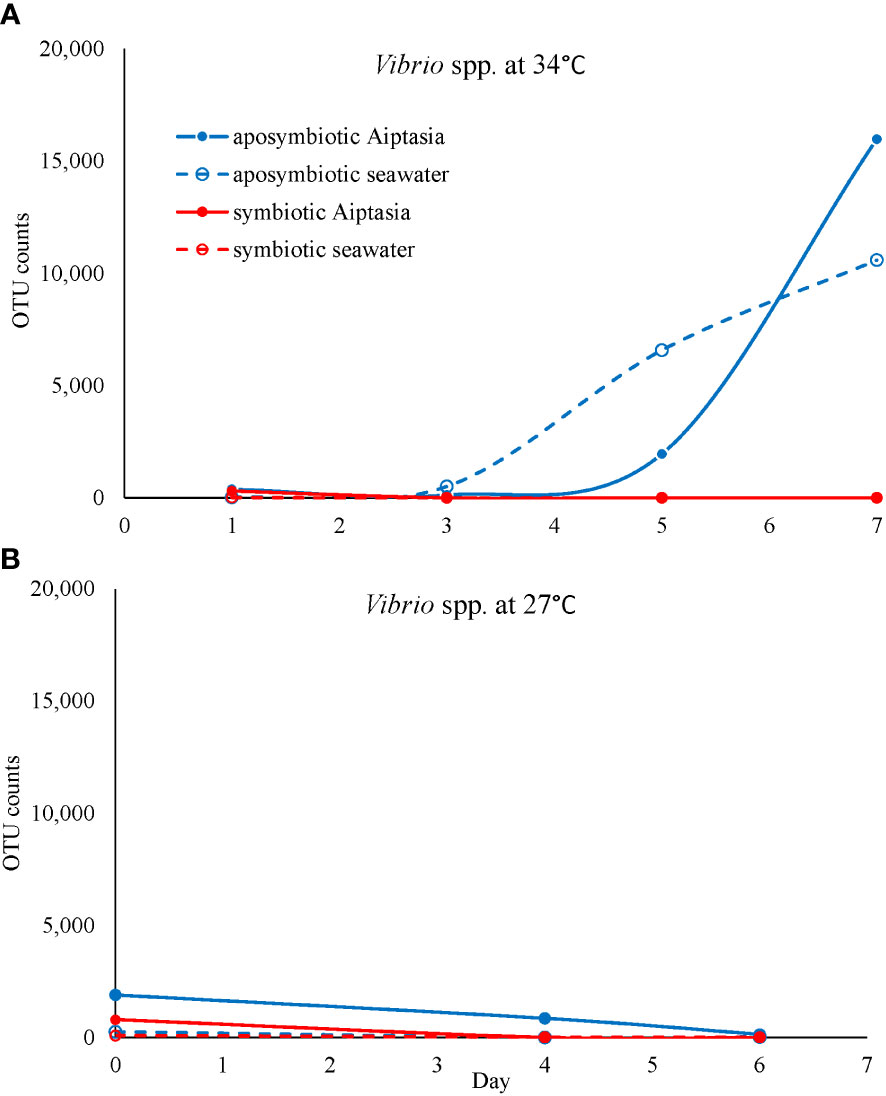
Figure 10 The potential pathogen Vibrio increases in abundance (based on counts of operational taxonomic units, OTU) exclusively in heat-stressed aposymbiotic Aiptasia and their seawater (A) but not in symbiotic Aiptasia or their seawater, or any non-heat-stressed treatments (B). Dominant Vibrio species identified included V. harveyi, V. parahemolyticus, and V. alginolyticus.
Archaea have seldom been described in the coral holobiont, and never in Aiptasia (Puntin et al., 2022). Crenarchaeota and euryarchaeota have been found in close association with the tropical coral Porites asteroides, accounting for upwards of 50% of their prokaryotic community (Wegley et al., 2004). Archaea may have a role in methanol oxidation and ammonia oxidation (Siboni et al., 2008). In this study, we observed euryarchaeota present in the seawater, but not polyps, of heat-stressed aposymbiotic Aiptasia, although low sequence homology (87%) precluded identification. The presence of archaea exclusively in the environment but not in anemones themselves aligns with observations of archaea’s tendency to reside exclusively in coral mucus but not corals themselves (Frade et al., 2016). Further studies distinguishing anemone mucus from the seawater environment are warranted to understand the role of these novel archaea associated with the Aiptasia holobiont.
Little evidence of nitrogen fixation in Aiptasia suggests a more heterotrophic lifestyle and its use as a model for only some (not all) coral types
Since Pelobacter, a potential nitrogen fixer, was specifically associated with symbiotic anemones, and more present following heat stress at 34°C, we decided to further investigate the role of nitrogen fixation in Aiptasia. But despite using PCR primers that should easily amplify Pelobacter nifH (Figure S3), we found, if anything, more evidence for genomic nifH in aposymbiotic Aiptasia (Figure S4), but little evidence of actual nifH expression (Figure 9A) or nitrogen fixation, as measured by the ARA (Figure 9B), for both symbiotic states. Our results are vastly different from what has been observed in autotrophic coral species that display both nifH expression and nitrogenase activity (Pogoreutz et al., 2017), but this difference may also be attributed to the different nifH primers used between the two studies. Interestingly, our findings are more consistent with the highly heterotrophic coral species that Pogoreutz et al. (2017) examined, in which nifH abundance was low and nitrogen-fixation activity was also below the detectable limit. Thus, it would appear that cultured laboratory strains of Aiptasia may rely on heterotrophy when nitrogen is limiting. Furthermore, our observations of possible increased nitrogen fixation in heat-stressed Aiptasia (Figure 9B) align with observations of increased nitrogen-fixation activity under heat stress in certain corals (Rädecker et al., 2015; Cardini et al., 2016; Bednarz et al., 2019; Rädecker et al., 2021). But the lack of replication and obvious death of anemones during the 24-h heat stress in the ARA makes this observation very tenuous.
As Aiptasia laboratory cultures have been maintained for more than 30 years (Weis et al., 2008), it is possible that lack of diazotrophy is an adaptation to a more heterotrophic lifestyle, and its reputation as a notorious fast-growing contaminant of marine aquaria also suggests a different trophic strategy. Wild strains should also be examined for nitrogen-fixing microbiomes, but our results emphasize using caution when extrapolating results from laboratory strains of Aiptasia to coral-reef ecosystems.
Data availability statement
The datasets presented in this study are available in online repositories. The 16S and 18S rRNA data sets from the time-series experiment are considered a Targeted Locus Study project that has been deposited at DDBJ/EMBL/GenBank under the accession KHVH00000000, associated with BioProject PRJNA951783 and BioSample SAMN34059703. Additionally, the 16S rRNA data set from the endpoint experiment is available under accession numbers OQ822830 - OQ824899.
Author contributions
JS, JL, LO, GW, and CT conceived the ideas and designed the experiments. JS, JL, LO, and GW performed the experiments and analyzed the data. JS, JL, GW, and CT wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Student Awards in Research and Creativity to JS and JL, and a CSU Program for Education and Research in Biotechnology grant to CT. Publication cost was supported by the University of San Diego and an award from the Faculty Research Publications Fund to CT.
Acknowledgments
The authors would like to acknowledge the students of the 2018 BIOL 402 (Microbial Ecology) class for helping with DNA extractions, as well as Ryan Phillips and Francisco Benjie Cabrera for their laboratory assistance to JL. Nicholas Lytal, CSU Chico Dept. Mathematics & Statistics, provided help with the R Vegan package for PERMANOVA testing. We would also like to thank Emily Fleming, Larry Hanne, Amanda Banet, and members of the Tran Laboratory research group for helpful discussions about this work. Some content of this manuscript previously appeared online in a different form as part of JS’s Master’s thesis (Sydnor, 2020) and JL’s Master’s thesis (Lopez, 2022).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1130964/full#supplementary-material
References
Ahmed H. I., Herrera M., Liew Y. J., Aranda M. (2019). Long-term temperature stress in the coral model aiptasia supports the “Anna karenina principle” for bacterial microbiomes. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00975
Ainsworth T. D., Hoegh-Guldberg O. (2009). Bacterial communities closely associated with coral tissues vary under experimental and natural reef conditions and thermal stress. Aquat. Biol. 4, 289–296. doi: 10.3354/ab00102
Akob D. M., Baesman S. M., Sutton J. M., Fierst J. L., Mumford A. C., Shrestha Y., et al. (2017). Detection of diazotrophy in the acetylene-fermenting anaerobe pelobacter sp. strain SFB93. Appl. Environ. Microbiol. 83, e01198–e01117. doi: 10.1128/AEM.01198-17
Angel R., Nepel M., Panhölzl C., Schmidt H., Herbold C. W., Eichorst S. A., et al. (2018). Evaluation of primers targeting the diazotroph functional gene and development of NifMAP – a bioinformatics pipeline for analyzing nifH amplicon data. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00703
Baumgarten S., Simakov O., Esherick L. Y., Liew Y. J., Lehnert E. M., Michell C. T., et al. (2015). The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc. Natl. Acad. Sci. 112, 11893–11898. doi: 10.1073/pnas.1513318112
Bednarz V. N., van de Water J. A. J. M., Rabouille S., Maguer J.-F., Grover R., Ferrier-Pagès C. (2019). Diazotrophic community and associated dinitrogen fixation within the temperate coral Oculina patagonica: N 2 fixation in Oculina patagonica. Environ. Microbiol. 21, 480–495. doi: 10.1111/1462-2920.14480
Benavides M., Bednarz V. N., Ferrier-Pagès C. (2017). Diazotrophs: Overlooked key players within the coral symbiosis and tropical reef ecosystems? Front. Mar. Sci. 4. doi: 10.3389/fmars.2017.00010
Benavides M., Houlbrèque F., Camps M., Lorrain A., Grosso O., Bonnet S. (2016). Diazotrophs: A non-negligible source of nitrogen for the tropical coral Stylophora pistillata. J. Exp. Biol 219 (17), 2608–2612. doi: 10.1242/jeb.139451
Bieri T., Onishi M., Xiang T., Grossman A. R., Pringle J. R. (2016). Relative contributions of various cellular mechanisms to loss of algae during cnidarian bleaching. PloS One 11, e0152693. doi: 10.1371/journal.pone.0152693
Bourne D. G., Garren M., Work T. M., Rosenberg E., Smith G. W., Harvell C. D. (2009). Microbial disease and the coral holobiont. Trends Microbiol. 17, 554–562. doi: 10.1016/j.tim.2009.09.004
Bourne D. G., Morrow K. M., Webster N. S. (2016). Insights into the coral microbiome: Underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Brown T., Otero C., Grajales A., Rodriguez E., Rodriguez-Lanetty M. (2017). Worldwide exploration of the microbiome harbored by the cnidarian model, exaiptasia pallida (Agassiz in verrill 1864) indicates a lack of bacterial association specificity at a lower taxonomic rank. PeerJ 5, e3235. doi: 10.7717/peerj.3235
Burke L., Reytar K., Spalding M., Perry A. (2011) Reefs at risk revisited (World Resources Institute). Available at: https://digitalarchive.worldfishcenter.org/handle/20.500.12348/1107 (Accessed 2, 2022).
Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Cardini U., van Hoytema N., Bednarz V. N., Rix L., Foster R. A., Al-Rshaidat M. M. D., et al. (2016). Microbial dinitrogen fixation in coral holobionts exposed to thermal stress and bleaching: Microbial dinitrogen fixation in stressed corals. Environ. Microbiol. 18, 2620–2633. doi: 10.1111/1462-2920.13385
Ceh J., Kilburn M. R., Cliff J. B., Raina J.-B., van Keulen M., Bourne D. (2013). Nutrient cycling in early coral life stages: Pocillopora damicornis larvae provide their algal symbiont (Symbiodinium ) with nitrogen acquired from bacterial associates. Ecol. Evol. 3, 2393–2400. doi: 10.1002/ece3.642
Costa R. M., Cárdenas A., Loussert-Fonta C., Toullec G., Meibom A., Voolstra C. R. (2021). Surface topography, bacterial carrying capacity, and the prospect of microbiome manipulation in the Sea anemone coral model aiptasia. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.637834
Croft M. T., Lawrence A. D., Raux-Deery E., Warren M. J., Smith A. G. (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438, 90–93. doi: 10.1038/nature04056
Cui G., Liew Y. J., Li Y., Kharbatia N., Zahran N. I., Emwas A.-H., et al. (2019). Host-dependent nitrogen recycling as a mechanism of symbiont control in aiptasia. PloS Genet. 15, e1008189. doi: 10.1371/journal.pgen.1008189
Damjanovic K., Blackall L. L., Webster N. S., van Oppen M. J. H. (2017). The contribution of microbial biotechnology to mitigating coral reef degradation. Microb. Biotechnol. 10, 1236–1243. doi: 10.1111/1751-7915.12769
Davy S. K., Allemand D., Weis V. M. (2012). Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol. Mol. Biol. Rev. 76, 229–261. doi: 10.1128/MMBR.05014-11
Dungan A. M., Bulach D., Lin H., van Oppen M. J. H., Blackall L. L. (2021a). Development of a free radical scavenging bacterial consortium to mitigate oxidative stress in cnidarians. Microb. Biotechnol. 14, 2025–2040. doi: 10.1111/1751-7915.13877
Dungan A. M., Hartman L. M., Blackall L. L., van Oppen M. J. H. (2022). Exploring microbiome engineering as a strategy for improved thermal tolerance in exaiptasia diaphana. J. Appl. Microbiol. 132, 2940–2956. doi: 10.1111/jam.15465
Dungan A. M., van Oppen M. J. H., Blackall L. L. (2021b). Short-term exposure to sterile seawater reduces bacterial community diversity in the Sea anemone, exaiptasia diaphana. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.599314
Esherick L. Y., DeNofrio J. C., Krediet C. J., Xiang T., Tolleter D., Grossman A. G., et al. The role of host and symbiont genotype in the thermal tolerance of a model cnidarian-dinoflagellate symbiosis.
Fiore C. L., Jarett J. K., Olson N. D., Lesser M. P. (2010). Nitrogen fixation and nitrogen transformations in marine symbioses. Trends Microbiol. 18, 455–463. doi: 10.1016/j.tim.2010.07.001
Frade P. R., Roll K., Bergauer K., Herndl G. J. (2016). Archaeal and bacterial communities associated with the surface mucus of Caribbean corals differ in their degree of host specificity and community turnover over reefs. PloS One 11, e0144702. doi: 10.1371/journal.pone.0144702
Gaby J. C., Buckley D. H. (2012). A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenase. PloS One 7, e42149. doi: 10.1371/journal.pone.0042149
Garren M., Son K., Raina J.-B., Rusconi R., Menolascina F., Shapiro O. H., et al. (2014). A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J. 8, 999–1007. doi: 10.1038/ismej.2013.210
Grant M. A., Kazamia E., Cicuta P., Smith A. G. (2014). Direct exchange of vitamin B12 is demonstrated by modelling the growth dynamics of algal–bacterial cocultures. ISME J. 8, 1418–1427. doi: 10.1038/ismej.2014.9
Hartman L. M., Blackall L. L., van Oppen M. J. H. (2022). Antibiotics reduce bacterial load in exaiptasia diaphana, but biofilms hinder its development as a gnotobiotic coral model. Access Microbiol. 4. doi: 10.1099/acmi.0.000314
Hartman L. M., van Oppen M. J. H., Blackall L. L. (2020a). Microbiota characterization of exaiptasia diaphana from the great barrier reef. Anim. Microbiome 2, 10. doi: 10.1186/s42523-020-00029-5
Hartman L. M., van Oppen M. J. H., Blackall L. L. (2020b). The effect of thermal stress on the bacterial microbiome of exaiptasia diaphana. Microorganisms 8, 20. doi: 10.3390/microorganisms8010020
Herdina, Silsbury J. H. (1990). Estimating nitrogenase activity of faba bean (Vicia faba) by acetylene reduction (Ar) assay. Funct. Plant Biol. 17, 489–502. doi: 10.1071/pp9900489
Herrera M., Ziegler M., Voolstra C. R., Aranda M. (2017). Laboratory-cultured strains of the Sea anemone exaiptasia reveal distinct bacterial communities. Front. Mar. Sci. 4. doi: 10.3389/fmars.2017.00115
Hudd G. A., Lloyd-Jones C. P., Hill-Cottingham D. G. (1980). Comparison of acetylene-reduction and nitrogen-15 techniques for the determination of nitrogen fixation by field bean (Vicia faba) nodules. Physiol. Plant 48, 111–115. doi: 10.1111/j.1399-3054.1980.tb03227.x
Kim S.-J., Kwon K. K. (2010). “Marine, hydrocarbon-degrading alphaproteobacteria,” in Handbook of hydrocarbon and lipid microbiology. Ed. Timmis K. N. (Berlin, Heidelberg: Springer Berlin Heidelberg), 1707–1714. doi: 10.1007/978-3-540-77587-4_120
Knowlton N. (2001). The future of coral reefs. Proc. Natl. Acad. Sci. U.S.A. 98, 5419–5425. doi: 10.1073/pnas.091092998
Knowlton N., Brainard R. E., Fisher R., Moews M., Plaisance L., Caley M. J. (2010). “Coral reef biodiversity,” in Life in the world’s oceans (Hoboken, New Jersey, U.S.: John Wiley & Sons, Ltd), 65–78. doi: 10.1002/9781444325508.ch4
Lema K. A., Willis B. L., Bourne D. G. (2012). Corals form characteristic associations with symbiotic nitrogen-fixing bacteria. Appl. Environ. Microbiol. 78, 3136–3144. doi: 10.1128/AEM.07800-11
Lema K. A., Willis B. L., Bourne D. G. (2014). Amplicon pyrosequencing reveals spatial and temporal consistency in diazotroph assemblages of the acropora millepora microbiome. Environ. Microbiol. 16, 3345–3359. doi: 10.1111/1462-2920.12366
Lesser M., Falcón L., Rodríguez-Román A., Enríquez S., Hoegh-Guldberg O., Iglesias-Prieto R. (2007). Nitrogen fixation by symbiotic cyanobacteria provides a source of nitrogen for the scleractinian coral montastraea cavernosa. Mar. Ecol. Prog. Ser. 346, 143–152. doi: 10.3354/meps07008
Lesser M. P., Mazel C. H., Gorbunov M. Y., Falkowski P. G. (2004). Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science 305, 997–1000. doi: 10.1126/science.1099128
Lesser M. P., Morrow K. M., Pankey S. M., Noonan S. H. C. (2018). Diazotroph diversity and nitrogen fixation in the coral stylophora pistillata from the great barrier reef. ISME J. 12, 813–824. doi: 10.1038/s41396-017-0008-6
Liang J., Yu K., Wang Y., Huang X., Huang W., Qin Z., et al. (2020). Diazotroph diversity associated with scleractinian corals and its relationships with environmental variables in the south China Sea. Front. Physiol. 11. doi: 10.3389/fphys.2020.00615
Littman R., Willis B. L., Bourne D. G. (2011). Metagenomic analysis of the coral holobiont during a natural bleaching event on the great barrier reef. Environ. Microbiol. Rep. 3, 651–660. doi: 10.1111/j.1758-2229.2010.00234.x
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lopez J. (2022). Nitrogen-fixation potential in bacteria associated with the sea anemone, exaiptasia pallida (Chico (CA: California State University, Chico). Available at: scholarworks.calstate.edu.
Luna G. M., Bongiorni L., Gili C., Biavasco F., Danovaro R. (2010). Vibrio harveyi as a causative agent of the white syndrome in tropical stony corals. Environ. Microbiol. Rep. 2, 120–127. doi: 10.1111/j.1758-2229.2009.00114.x
Maire J., Blackall L. L., van Oppen M. J. H. (2021). Microbiome characterization of defensive tissues in the model anemone exaiptasia diaphana. BMC Microbiol. 21, 152. doi: 10.1186/s12866-021-02211-4
Math R. K., Jeong S. H., Jin H. M., Park M. S., Kim J. M., Jeon C. O. (2012). Kordiimonas aestuarii sp. nov., a marine bacterium isolated from a tidal flat. Int. J. Syst. Evol. Microbiol. 62, 3049–3054. doi: 10.1099/ijs.0.038943-0
Matthews J. L., Raina J., Kahlke T., Seymour J. R., Oppen M. J. H., Suggett D. J. (2020). Symbiodiniaceae-bacteria interactions: Rethinking metabolite exchange in reef-building corals as multi-partner metabolic networks. Environ. Microbiol. 22, 1675–1687. doi: 10.1111/1462-2920.14918
McDevitt-Irwin J. M., Baum J. K., Garren M., Vega Thurber R. L. (2017). Responses of coral-associated bacterial communities to local and global stressors. Front. Mar. Sci. 4. doi: 10.3389/fmars.2017.00262
Morris L. A., Voolstra C. R., Quigley K. M., Bourne D. G., Bay L. K. (2019). Nutrient availability and metabolism affect the stability of coral–symbiodiniaceae symbioses. Trends Microbiol. 27, 678–689. doi: 10.1016/j.tim.2019.03.004
Morrow K. M., Muller E., Lesser M. P. (2018). “How does the coral microbiome cause, respond to, or modulate the bleaching process?,” in Coral bleaching: Patterns, processes, causes and consequences ecological studies. Eds. van Oppen M. J. H., Lough J. M. (Cham: Springer International Publishing), 153–188. doi: 10.1007/978-3-319-75393-5_7
Moynihan M. A., Goodkin N. F., Morgan K. M., Kho P. Y. Y., Lopes dos Santos A., Lauro F. M., et al. (2021). Coral-associated nitrogen fixation rates and diazotrophic diversity on a nutrient-replete equatorial reef. ISME J. 16, 233–246, 1–14. doi: 10.1038/s41396-021-01054-1
Muller-Parker G., Davy S. K. (2001). Temperate and tropical algal-sea anemone symbioses. Invertebr. Biol. 120, 104–123. doi: 10.1111/j.1744-7410.2001.tb00115.x
Olson N. D., Ainsworth T. D., Gates R. D., Takabayashi M. (2009). Diazotrophic bacteria associated with Hawaiian montipora corals: Diversity and abundance in correlation with symbiotic dinoflagellates. J. Exp. Mar. Biol. Ecol. 371, 140–146. doi: 10.1016/j.jembe.2009.01.012
Olson N. D., Lesser M. P. (2013). Diazotrophic diversity in the Caribbean coral, montastraea cavernosa. Arch. Microbiol. 195, 853–859. doi: 10.1007/s00203-013-0937-z
Peixoto R. S., Rosado P. M., Leite D. C., de A., Rosado A. S., Bourne D. G. (2017). Beneficial microorganisms for corals (BMC): Proposed mechanisms for coral health and resilience. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00341
Pernice M., Meibom A., Van Den Heuvel A., Kopp C., Domart-Coulon I., Hoegh-Guldberg O., et al. (2012). A single-cell view of ammonium assimilation in coral–dinoflagellate symbiosis. ISME J. 6, 1314–1324. doi: 10.1038/ismej.2011.196
Pogoreutz C. (2016) Coral holobiont functioning under global environmental change. Available at: https://media.suub.uni-bremen.de/handle/elib/1130 (Accessed 6, 2022).
Pogoreutz C., Rädecker N., Cárdenas A., Gärdes A., Wild C., Voolstra C. R. (2017). Nitrogen fixation aligns with nifH abundance and expression in two coral trophic functional groups. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01187
Puntin G., Sweet M., Fraune S., Medina M., Sharp K., Weis V. M., et al. (2022). Harnessing the power of model organisms to unravel microbial functions in the coral holobiont. Microbiol. Mol. Biol. Rev. 86, e00053–e00022. doi: 10.1128/mmbr.00053-22
Pupier C. A., Bednarz V. N., Grover R., Fine M., Maguer J.-F., Ferrier-Pagès C. (2019). Divergent capacity of scleractinian and soft corals to assimilate and transfer diazotrophically derived nitrogen to the reef environment. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01860
Pupier C. A., Grover R., Fine M., Rottier C., van de Water J. A. J. M., Ferrier-Pagès C. (2021). Dissolved nitrogen acquisition in the symbioses of soft and hard corals with symbiodiniaceae: A key to understanding their different nutritional strategies? Front. Microbiol. 12. doi: 10.3389/fmicb.2021.657759
Rädecker N., Chen J. E., Pogoreutz C., Herrera M., Aranda M., Voolstra C. R. (2019). Nutrient stress arrests tentacle growth in the coral model aiptasia. Symbiosis 78, 61–64. doi: 10.1007/s13199-019-00603-9
Rädecker N., Meyer F., Bednarz V., Cardini U., Wild C. (2014). Ocean acidification rapidly reduces dinitrogen fixation associated with the hermatypic coral seriatopora hystrix. Mar. Ecol. Prog. Ser. 511, 297–302. doi: 10.3354/meps10912
Rädecker N., Pogoreutz C. (2019). Why are coral reefs hotspots of life in the ocean? Front. Young Minds 7. doi: 10.3389/frym.2019.00143
Rädecker N., Pogoreutz C., Gegner H. M., Cárdenas A., Roth F., Bougoure J., et al. (2021). Heat stress destabilizes symbiotic nutrient cycling in corals. Proc. Natl. Acad. Sci. 118, e2022653118. doi: 10.1073/pnas.2022653118
Rädecker N., Pogoreutz C., Voolstra C. R., Wiedenmann J., Wild C. (2015). Nitrogen cycling in corals: The key to understanding holobiont functioning? Trends Microbiol. 23, 490–497. doi: 10.1016/j.tim.2015.03.008
Rädecker N., Raina J.-B., Pernice M., Perna G., Guagliardo P., Kilburn M. R., et al. (2018). Using aiptasia as a model to study metabolic interactions in cnidarian-symbiodinium symbioses. Front. Physiol. 9. doi: 10.3389/fphys.2018.00214
Raina J.-B., Dinsdale E. A., Willis B. L., Bourne D. G. (2010). Do the organic sulfur compounds DMSP and DMS drive coral microbial associations? Trends Microbiol. 18, 101–108. doi: 10.1016/j.tim.2009.12.002
Reshef L., Koren O., Loya Y., Zilber-Rosenberg I., Rosenberg E. (2006). The coral probiotic hypothesis. Environ. Microbiol. 8, 2068–2073. doi: 10.1111/j.1462-2920.2006.01148.x
Rohwer F., Seguritan V., Azam F., Knowlton N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10. doi: 10.3354/meps243001
Rosado P. M., Leite D. C. A., Duarte G. A. S., Chaloub R. M., Jospin G., Nunes da Rocha U., et al. (2019). Marine probiotics: Increasing coral resistance to bleaching through microbiome manipulation. ISME J. 13, 921–936. doi: 10.1038/s41396-018-0323-6
Rosenberg E., Koren O., Reshef L., Efrony R., Zilber-Rosenberg I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. doi: 10.1038/nrmicro1635
Röthig T., Costa R. M., Simona F., Baumgarten S., Torres A. F., Radhakrishnan A., et al. (2016). Distinct bacterial communities associated with the coral model aiptasia in aposymbiotic and symbiotic states with symbiodinium. Front. Mar. Sci. 3. doi: 10.3389/fmars.2016.00234
Santos H. F., Carmo F. L., Duarte G., Dini-Andreote F., Castro C. B., Rosado A. S., et al. (2014). Climate change affects key nitrogen-fixing bacterial populations on coral reefs. ISME J. 8, 2272–2279. doi: 10.1038/ismej.2014.70
Schink B. (1992). “The genus pelobacter,” in The prokaryotes. Eds. Balows A., Trüper H. G., Dworkin M., Harder W., Schleifer K.-H. (New York, NY: Springer New York), 3393–3399. doi: 10.1007/978-1-4757-2191-1_24
Shashar N., Feldstein T., Cohen Y., Loya Y. (1994). Nitrogen fixation (acetylene reduction) on a coral reef. Coral Reefs 13, 171–174. doi: 10.1007/BF00301195
Siboni N., Ben-Dov E., Sivan A., Kushmaro A. (2008). Global distribution and diversity of coral-associated archaea and their possible role in the coral holobiont nitrogen cycle. Environ. Microbiol. 10, 2979–2990. doi: 10.1111/j.1462-2920.2008.01718.x
Stoeck T., Bass D., Nebel M., Christen R., Jones M. D., Breiner H. W., et al. (2010). Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 19, 21–31. doi: 10.1111/j.1365-294X.2009.04480.x
Sunagawa S., Wilson E. C., Thaler M., Smith M. L., Caruso C., Pringle J. R., et al. (2009). Generation and analysis of transcriptomic resources for a model system on the rise: The sea anemone aiptasia pallida and its dinoflagellate endosymbiont. BMC Genomics 10, 258. doi: 10.1186/1471-2164-10-258
Sydnor J. R. (2020). The bacterial community associated with the model sea anemone exaiptasia pallida: Response to rising ocean temperature (Chico (CA: California State University, Chico). Available at: scholarworks.calstate.edu.
Thompson J. R., Rivera H. E., Closek C. J., Medina M. (2015). Microbes in the coral holobiont: partners through evolution, development, and ecological interactions. Front. Cell. Infect. Microbiol. 4. doi: 10.3389/fcimb.2014.00176
Thornhill D. J., Xiang Y., Pettay D. T., Zhong M., Santos S. R. (2013). Population genetic data of a model symbiotic cnidarian system reveal remarkable symbiotic specificity and vectored introductions across ocean basins. Mol. Ecol. 22, 4499–4515. doi: 10.1111/mec.12416
Tilstra A., El-Khaled Y. C., Roth F., Rädecker N., Pogoreutz C., Voolstra C. R., et al. (2019). Denitrification aligns with N2 fixation in red Sea corals. Sci. Rep. 9, 19460. doi: 10.1038/s41598-019-55408-z
Tran C. (2022). Coral–microbe interactions: Their importance to reef function and survival. Emerg. Top. Life Sci. 6, 33–44. doi: 10.1042/ETLS20210229
Vega Thurber R., Willner-Hall D., Rodriguez-Mueller B., Desnues C., Edwards R. A., Angly F., et al. (2009). Metagenomic analysis of stressed coral holobionts. Environ. Microbiol. 11, 2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x
Veron J. E. N., Hoegh-Guldberg O., Lenton T. M., Lough J. M., Obura D. O., Pearce-Kelly P., et al. (2009). The coral reef crisis: The critical importance of<350ppm CO2. Mar. pollut. Bull. 58, 1428–1436. doi: 10.1016/j.marpolbul.2009.09.009
Voolstra C. R. (2013). A journey into the wild of the cnidarian model system aiptasia and its symbionts. Mol. Ecol. 22, 4366–4368. doi: 10.1111/mec.12464
Wang J., Douglas A. (1998). Nitrogen recycling or nitrogen conservation in an alga-invertebrate symbiosis? J. Exp. Biol. 201, 2445–2453. doi: 10.1242/jeb.201.16.2445
Wegley L., Yu Y., Breitbart M., Casas V., Kline D. I., Rohwer F. (2004). Coral-associated archaea. Mar. Ecol. Prog. Ser. 273, 89–96. doi: 10.3354/meps273089
Weis V. M. (2008). Cellular mechanisms of cnidarian bleaching: stress causes the collapse of symbiosis. J. Exp. Biol. 211, 3059–3066. doi: 10.1242/jeb.009597
Weis V. M., Davy S. K., Hoegh-Guldberg O., Rodriguez-Lanetty M., Pringle J. R. (2008). Cell biology in model systems as the key to understanding corals. Trends Ecol. Evol. 23, 369–376. doi: 10.1016/j.tree.2008.03.004
Wilson S. T., Böttjer D., Church M. J., Karl D. M. (2012). Comparative assessment of nitrogen fixation methodologies, conducted in the oligotrophic north pacific ocean. Appl. Environ. Microbiol. 78, 6516–6523. doi: 10.1128/AEM.01146-12
Xiang T., Hambleton E. A., DeNofrio J. C., Pringle J. R., Grossman A. R. (2013). Isolation of clonal axenic strains of the symbiotic dinoflagellate symbiodinium and their growth and host specificity1. J. Phycol. 49, 447–458. doi: 10.1111/jpy.12055
Xiang T., Lehnert E., Jinkerson R. E., Clowez S., Kim R. G., DeNofrio J. C., et al. (2020). Symbiont population control by host-symbiont metabolic interaction in symbiodiniaceae-cnidarian associations. Nat. Commun. 11, 108. doi: 10.1038/s41467-019-13963-z
Xiang N., Rädecker N., Pogoreutz C., Cárdenas A., Meibom A., Wild C., et al. (2022). Presence of algal symbionts affects denitrifying bacterial communities in the sea anemone aiptasia coral model. ISME Commun. 2, 105. doi: 10.1038/s43705-022-00190-9
Zaneveld J. R., McMinds R., Vega Thurber R. (2017). Stress and stability: Applying the Anna karenina principle to animal microbiomes. Nat. Microbiol. 2, 17121. doi: 10.1038/nmicrobiol.2017.121
Zaragoza W. J., Krediet C. J., Meyer J. L., Canas G., Ritchie K. B., Teplitski M. (2014). Outcomes of infections of Sea anemone aiptasia pallida with vibrio spp. pathogenic to corals. Microb. Ecol. 68, 388–396. doi: 10.1007/s00248-014-0397-2
Zehr J. P., Jenkins B. D., Short S. M., Steward G. F. (2003). Nitrogenase gene diversity and microbial community structure: A cross-system comparison. Environ. Microbiol. 5, 539–554. doi: 10.1046/j.1462-2920.2003.00451.x
Keywords: corals, cnidarian, microbiome, diazotroph, nitrogen fixation, symbiosis, holobiont, climate change
Citation: Sydnor JR, Lopez J, Wolfe GV, Ott L and Tran C (2023) Changes in the microbiome of the sea anemone Exaiptasia diaphana during bleaching from short-term thermal elevation. Front. Mar. Sci. 10:1130964. doi: 10.3389/fmars.2023.1130964
Received: 24 December 2022; Accepted: 01 March 2023;
Published: 05 May 2023.
Edited by:
Stephane Roberty, University of Liège, BelgiumReviewed by:
Nan Xiang, The Chinese University of Hong Kong, ChinaTrent Haydon, New York University Abu Dhabi, United Arab Emirates
Copyright © 2023 Sydnor, Lopez, Wolfe, Ott and Tran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cawa Tran, Y2F3YXRyYW5Ac2FuZGllZ28uZWR1
†These authors have contributed equally to this work and share first authorship
 Jamison R. Sydnor
Jamison R. Sydnor Jaime Lopez1†
Jaime Lopez1† Gordon V. Wolfe
Gordon V. Wolfe Lisa Ott
Lisa Ott Cawa Tran
Cawa Tran