- 1Key Laboratory of Freshwater Fish Reproduction and Development (Ministry of Education), College of Fisheries, Southwest University, Chongqing, China
- 2Key Laboratory of Aquatic Science of Chongqing, College of Fisheries, Southwest University, Chongqing, China
- 3Guangxi Key Laboratory of Aquatic Genetic Breeding and Healthy Aquaculture, Guangxi Academy of Fisheries Science, Nanning, China
The rice flower carp (Cyprinus carpio) is an important fish in integrated rice-fishery farming. Here, we performed the first genome-wide association study (GWAS) for seven growth traits (including body mass, total length, body length, body height, body width, caudal-peduncle depth, and eye spacing) in 200 rice flower carp samples using 369,688 high-quality SNPs and 42,225 indels obtained by double-digest genotyping-by-sequencing (ddGBS). The morphometrics of these traits were highly correlated (Pearson’s correlation coefficients = 0.74–0.99, p < 0.001). GWAS detected 15, 5, 4, 26, 7, 16, and 17 loci significant associated (-log10P ≥ 5) with body mass, total length, body length, body width, body height, caudal-peduncle depth, and eye spacing, respectively. Subsequently, within the 50 kb upstream and downstream regions surrounding these significant loci, 38, 19, 18, 20, 52, 27, and 37 candidate genes for the seven growth traits were detected, respectively. Importantly, B6_4352672 and A8_4978825 were significantly associated with more than five growth traits. These results showed loci significantly associated with more than five growth traits will be helpful for future marker-assisted selection (MAS). Interestingly, chromosomes A8 and B25 had many loci significantly associated with growth traits, most of which were shared among multiple growth-related traits. These results indicated that chromosome A8 and B25 may be closely related to growth traits. Our findings not only help understand the genetic architecture of growth traits in fish but facilitate the identification of candidate genes for marker-assisted selection towards breeding faster-growing rice flower carp in the future.
1 Introduction
The rice flower carp (Cyprinus carpio), belonging to the family Cyprinidae, is an essential economic fish species in the integrated rice-fish culture in China, which has been cultured in paddy fields for over 1000 years, and the Quanzhou rice flower carp of Guangxi Zhuang Autonomous Region in China was also listed as an imperial tribute in the Qing Dynasty (Jiang and Yan, 2009). Through long-term cultivation and artificial selection in the rice field environment, the rice flower carp gradually formed unique morphological characteristics of body color and shape, which distinguishes it from other cultured common carp (Cyprinus carpio) groups (Wu et al., 2021). Because the rice flower carp eat the rice flower, they have many desired qualities, such as high protein content, unique flavor with no muddy smell, and soft and edible intermuscular spines (Yang and Jiang, 2009). These qualities promote the development of integrated rice-fish culture and increase the demand and commercial value. However, the rapid expansion of rice flower carp aquaculture has led to tremendous challenges, including slow growth rates, large individual differences, and uneven listing specifications, which have seriously limited the sustainable development of this species’ industry (Li et al., 2022). Therefore, with increasing market demand, selection for fast growth would be one of the most important breeding goals for the rice flower carp.
Molecular marker-assisted breeding can increase the speed of breeding by nearly 11% compared with traditional methods (Gomez and Klemetsdal, 1999). In addition, the heritability of body mass, body length, and body width of rice flower carp were 0.45, 0.42, and 0.36, respectively (Xu et al., 2022), suggesting that molecular marker-assisted breeding can be used for the selection of superior strains with a rapid growth rate of rice flower carp because of the high heritability of growth traits. Single nucleotide polymorphisms (SNPs), the most reliable third-generation molecular marker, have been widely used in candidate-marker/gene association studies of important traits, and molecular marker-assisted breeding (Vignal et al., 2002). Identification of SNP markers associated with target traits is key for molecular marker-assisted breeding.
Currently, genome-wide association studies (GWAS) based on a large number of SNP markers have been widely used for identifying markers/genetic variants that are significantly associated with various economic traits in fish, such as sex, stress tolerance, and growth (Berghof et al., 2018; Ji et al., 2019; Gao and Yang, 2021; Luo et al., 2022; Mou et al., 2022). In recent years, SNPs have mainly been detected and genotyped at the genome-wide level using gene chips, simplified genome sequencing, and whole genome resequencing. In reduced-representation genome sequencing (RRGS), high-density molecular markers have been obtained at the level of the whole genome (Miller et al., 2007). Genotyping-by-sequencing (GBS) is a simplified genome sequencing method that is inexpensive and reliable (Davey et al., 2011; Bian and Wang, 2017), and is widely used in population genetics (Nunez et al., 2015; Drury et al., 2016) and genome-wide association studies (Wu et al., 2019; Habimana et al., 2021). With the development of GBS technology, double-digest genotyping-by-sequencing (ddGBS) can effectively reduce genome complexity and make genome-wide genetic analysis detection more convenient and reliable (Elshire et al., 2011). In recent years, ddGBS genotyping has been applied for SNP marker development (Jia et al., 2020; Liu et al., 2022), genetic linkage map construction(Li et al., 2018), and GWAS (Gileta et al., 2022). However, the study of growth-associated marker of rice flower carp using GWAS is lacking so far.
In this study, 200 rice flower carp individuals of a population were sequenced by ddGBS, and GWAS was first conducted based on mixed linear model for identify seven growth-traits (including body mass, total length, body length, body height, body width, caudal-peduncle depth and eye spacing) associated markers and candidate genes. The findings of this study will be helpful for future marker-assisted selection (MAS) and speculating on the genetic mechanisms underlying the growth regulation of rice flower carp.
2 Materials and methods
2.1 Ethics statement
All experiments in this study were conducted in strict accordance with the ethical guidelines and regulations of the Animal Care and Use Committee of Southwest University and were approved by the local ethics committee of the Animal Welfare and Ethical Committee of Southwest University.
2.2 Sample preparation
The experimental fish parents were obtained from Quanzhou County, Guilin City, Guangxi Zhuang Autonomous Region, and 120 pairs of rice flower carp (1:1 male to female ratio) were introduced from the Guangxi Agricultural Fine Variety Hainan South Propagation and Breeding Base, San Ya, China. The experimental fish were bred after artificial fertilization. Two hundred 20-month-old fish were randomly collected for measuring seven body size traits, including body mass (BM), total length (TL), body length (BL), body height (BH), body width (BW), caudal-peduncle depth (CPD), and eye spacing (ES). The tail fins of each individual were collected and preserved in 100% ethanol at 4°C for DNA extraction. Genomic DNA of each sample was extracted using DNAsecure Plant Kit (TIANGEN) according to the manufacturer’s protocols. The quantity and quality were assessed using 2.0 Flurometer (Life Technologies, CA, USA) and 1.5% agarose gel electrophoresis, respectively.
2.3 GBS library construction and sequencing
Genomic DNA from each sample was used to prepare the GBS library with the NEB Next® Ultra DNA Library Prep Kit for Illumina® (NEB, USA). In brief, 100 ng genomic DNA from each fish was digested with EcoRI and NlaIII at 37°C for 1 h, and then added End Prep Enzyme Mix at 20°C for 30 s, 65°C 30 s for terminal repair, phosphorylation and addition of A-tail. The double DNA fragments were mixed with EcoRI adapter, NlaIII adapter and T4 DNA ligase at 20°C for 1 h, then added 3 μL USER enzyme at 37°C for 15 min. The DNA fragments with 400-600 bp filtered by AMPure XP beads were used to construct PCR library using universal PCR primers and index primers. After purifying and quality inspection, sequencing was carried out on an Illumina HiSeq 4000 platform.
2.4 SNP discovery and genotyping
The short-read data obtained from the Illumina HSseq 4000 platform were filtered using FASTP (v 0.18.0) (Chen et al., 2018) according to the following steps: (a) removing reads linked with the adapter sequences; (b) discarding paired reads with more than 10% ambiguous bases; (c) removing low quality reads with ≥ 50% bases harboring Phred scores of ≤ 20. Subsequently, the clean reads were aligned to the common carp genome (GCF_018340385.1) (Li et al., 2021) using BWA software (v 0.7.12) (Li and Durbin, 2009) with parameters “-k 32 -M”, and then the results were marked by Picard (v 1.129) (Picard: Http://sourceforge.net/projects/picard/). SNPs were called using GATK (v 3.4-46) (Der-Auwera et al., 2013), and SNPs with the number of alleles > 2, minor allele frequency (MAF) < 0.05, missing rate > 0.9, and heterozygote ratio > 0.8 were filtered using PLINK 2.0 (Chang et al., 2015) to gain high-quality SNPs. Finally, the retained SNPs were annotated by ANNOVAR program (Wang et al., 2010).
2.5 Analysis of the population structure
Neighbor-joining (NJ) phylogeny construction was performed by MEGA-X (Kumar et al., 2018) with model of p-distance, bootstrap runs: 500. Population structure of the 200 fish was calculated by Admixture (v 1.3) (Alexander et al., 2009) with maximum likelihood approach. The putative number of subgroup (K-value) was set from one to nine, and the optimal K-value was determined by the lowest cross-validation error. The genetic composition of each sample in subgroup was mapped using Pophelper (v 2.2.7) (Francis, 2017). Principal component analysis (PCA) and kinship analysis of the population were performed using GCTA (v1.93.2) with the default settings (Yang et al., 2011).
2.6 Genome-wide association study
Pearson correlation coefficients between seven traits and principal component analysis (PCA) were calculated using R (v 3.5.0) (https://www.R-project.org/) and SPSS20.0, respectively. Principal components were extracted according to the principle of an 85% cumulative contribution rate. Seven growth- associated traits from 200 individuals were selected for GWAS analysis using GEMMA (v 0.98.1) based on mixed linear model (MLM) (Zhou and Stephens, 2012). The p value of genome-wide significance threshold was calculated by Bonferroni correction based on the formula p value = 0.05/n, n represents the number of markers used for GWAS. Manhattan plot representing the -log10(p) for threshold of extremely significant association and significant association were set as 6.9 and 5, respectively. Quantile-quantile (Q-Q) plot expressing the observed and expected -log10(p) values were produced using R software.
2.7 Identification of candidate genes
The candidate genes associated with growth were obtained by mapping significant markers onto the common carp genome (GCF_018340385.1) (Li et al., 2021) with scanning up- and down-stream 50 kb regions from the significant markers. Then, obtained candidate genomic regions were mapped to the contig by local sequence similarity searches and annotated with an in-house script.
3 Results
3.1 Statistics of growth traits
In this study, seven morphological traits for growth traits of 200 rice flower carp were measured, as shown in Table 1. The mean values of BM, TL, and BL traits (± SD) were 638.38 ± 302.33 g, 32.72 ± 4.37 cm, and 24.60 ± 3.52 cm, respectively. Pearson’s correlation coefficients between all pairs of the seven morphological traits ranged from 0.74 to 0.99 (Figure 1). These seven traits were strongly and positively correlated with each other.

Table 1 Summary of morphometric statistics for seven growth traits in 200 rice flower carp for GWAS.
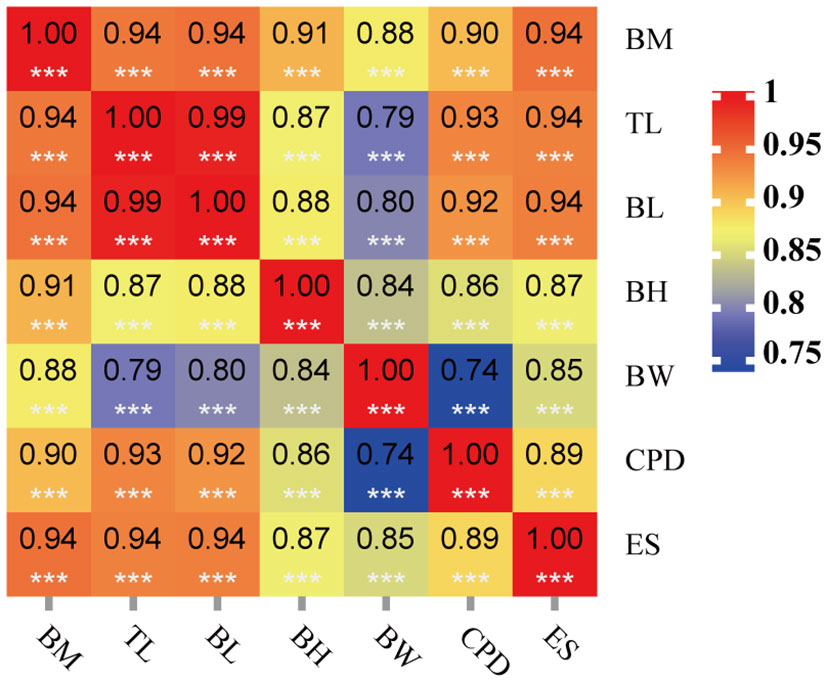
Figure 1 Correlation coefficients between seven traits of 200 rice flower carp. *** means significantly different at the p < 0.001 level.
3.2 Sequencing data and population structure
Using the Illumina sequencing platform, a total of 1,919,365,466 clean reads and 270.12 Gb clean data with 43.15% GC content were obtained from 200 rice flower carp. The average Q20 and Q30 were 97.32% and 95.02%, respectively (Table S1). After mapping all clean reads on to the common carp genome (GCF_018340385.1) (Li et al., 2021) and filtering low-quality SNPs, a total of 369,688 high-quality SNPs and 42,225 indels were used to analyze the population structure and GWAS.
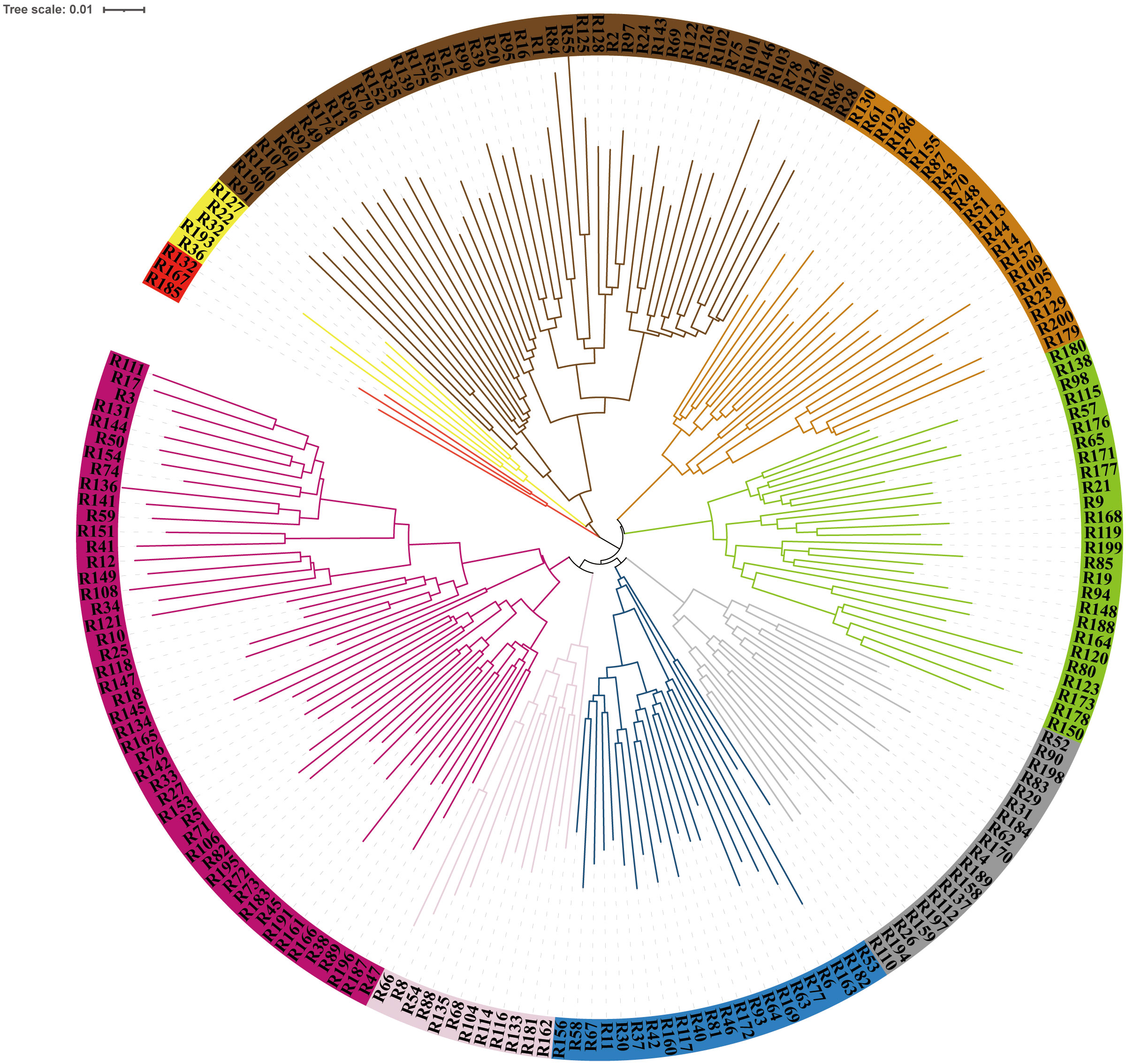
To understand the genetic relationships among these experimental individuals, we conducted phylogenetic tree, population structure, PCA, and kinship analyses. Phylogenetic tree analysis indicated that these 200 rice flower carp were mainly divided into nine branches (Figure 2). In the population structure analysis, the optimal K-value (K = 9) was determined using the lowest cross-validation error (Figure S1). The results of the population genetic structure and PCA are shown in Figure S2 and Figure S3, respectively. Apparently, the two analysis methods yielded consistent results. The genetic relationship coefficient ranged from -0.24 to 0.69 (Figure S4).
3.3 GWAS of growth traits
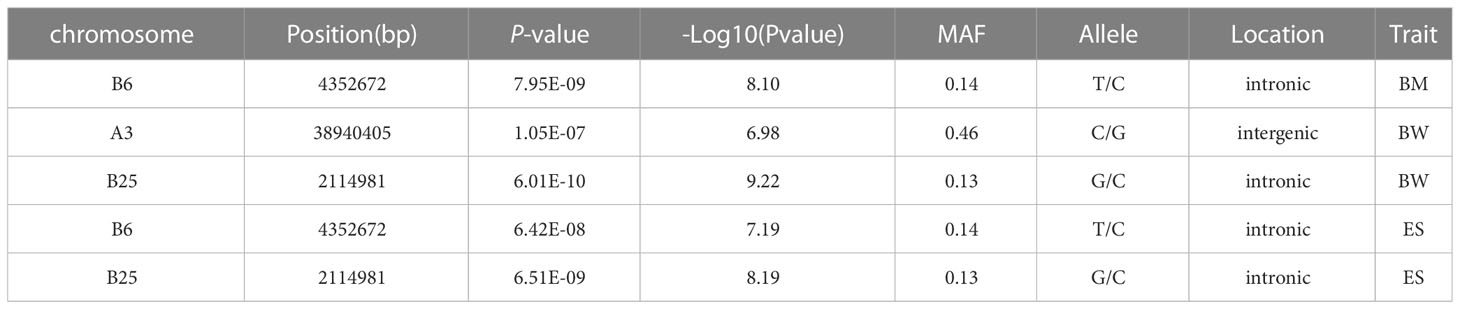
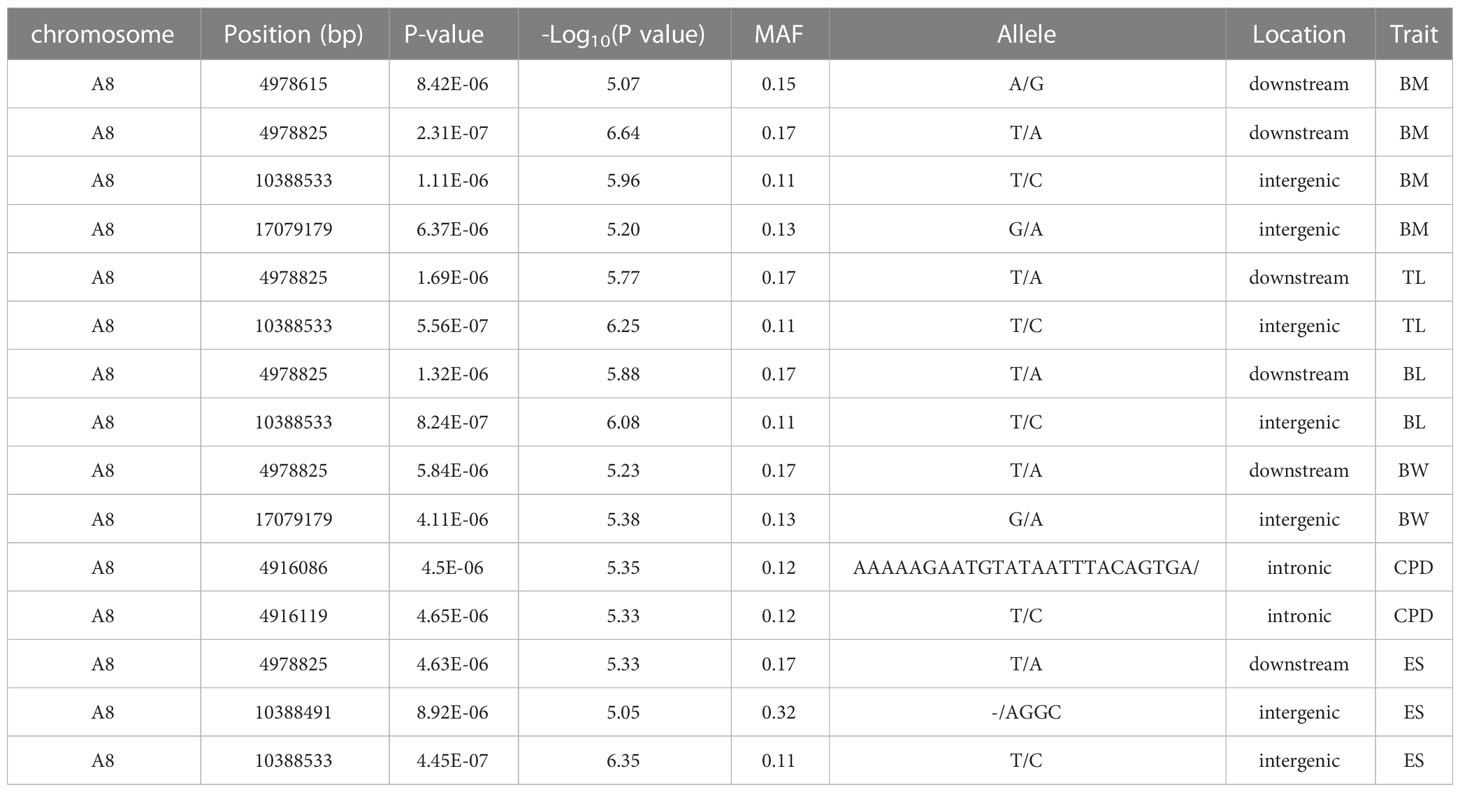
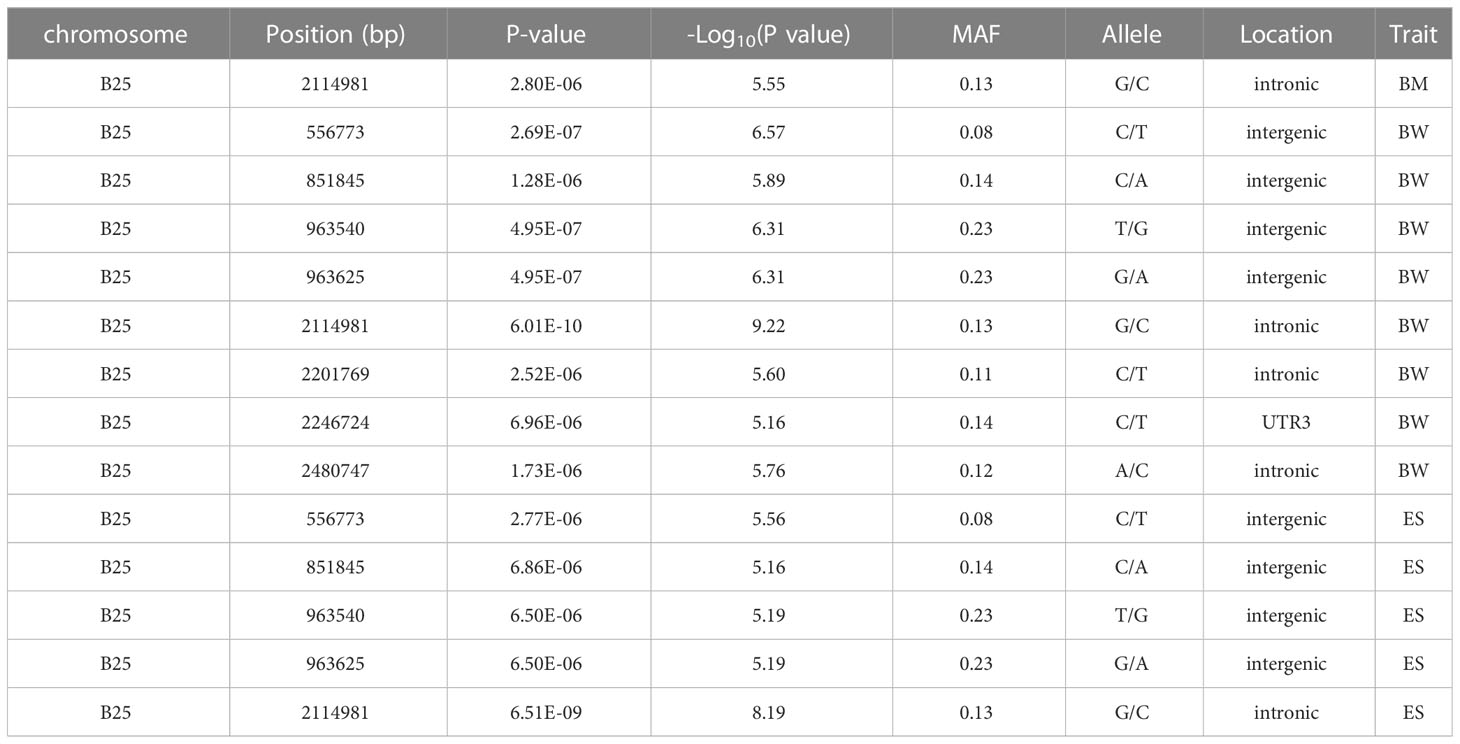
Based on 369,688 high-quality SNPs and 42,225 indels, GWAS of seven growth-associated traits from 200 individuals was carried out using GEMMA (v 0.98.1) based on a mixed linear model (MLM). The results showed that five extremely significant associated (-log10P ≥ 6.9) (Table 2) and 85 significant associated (-log10P ≥ 5.0) loci with growth-associated traits (Table S2, Figure 3). Among these loci, one locus located on chromosome B6, two loci located on chromosome B25 and A3, and two loci located on chromosome B25 and B6 were extremely significantly associated with BM, BW, and ES, respectively (Table 2). Additionally, 15, 5, 4, 26, 7, 16, and 17 loci were significant associated with BM, TL, BL, BW, BH, CPD, and ES, respectively. BW had the greatest number of extremely significant and significant loci, followed by ES and BM. Interestingly, three chromosomes of A3, B6, and B25 had significant loci associated with growth traits, in which chromosome B25 has more significant loci than other chromosomes, except for chromosome A8. Fifteen loci were located on chromosome A8, and locus A8_10388533 was associated with BM, TL, BL, and ES (Table 3). Among the loci located on chromosome B25, B25_2114981 was shared among three traits including BM, BW, and ES. B25_556773, B25_851845, B25_963540, and B25_963625 were related to BW and ES traits (Table 4).
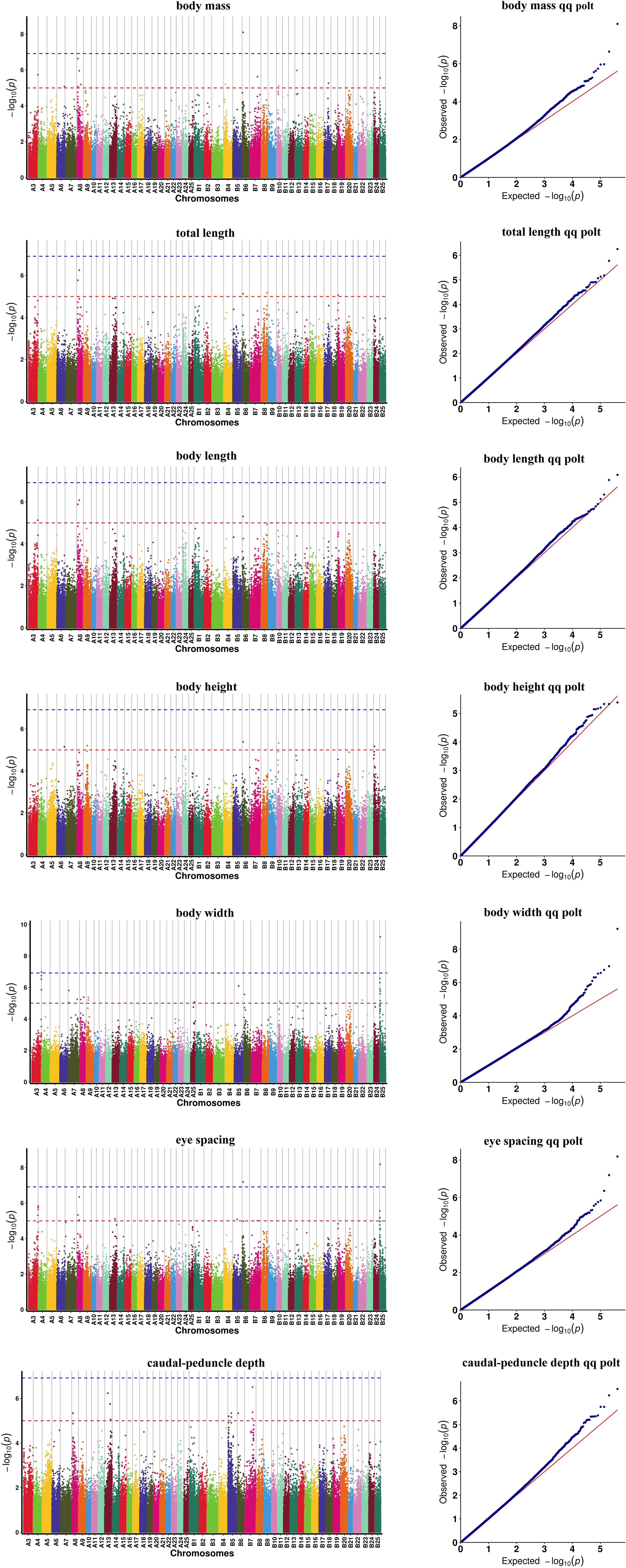
Figure 3 Manhattan plot for seven growth traits of rice flower carp. Dots with different colors denote loci on different chromosomes. The blue horizontal line represents the significance threshold (5.0), and the red line represents the extremely significance threshold (6.9). The red horizontal line represents the significance threshold (5.0), and the blue line represents the extremely significance threshold (6.9).
3.4 Candidate genes related to growth traits
Genes potentially related to the seven growth-associated traits in rice flower carp were explored after annotating the upstream and downstream 50 kb genome sequences based on significant loci. The number of candidate genes associated with the seven growth traits was 38, 19, 18, 20, 52, 27, and 37 for BM, TL, BL, BH, BW, CPD, and ES, respectively (Table S3). Based on the analysis above, we found that 14 and 18 genes harboring significant loci were associated with growth traits in chromosomes A8 and B25, respectively (Table 5). A total of 40 candidate genes were associated with at least two growth traits, of which five candidate genes, including tripartite motif containing 25 (trim25l), trafficking kinesin protein 2 (trak2), STE20 related adaptor beta (stradb), neurobeachin like 1 (nbeal1), and RNA binding motif protein 26 (rbm26), were shared among the seven traits. Ubiquitin carboxyl-terminal hydrolase 11 (usp11), ER membrane protein complex subunit 3 (emc3), UXT (uxt), ETS domain-containing protein Elk-1 (elk1), leucine-rich repeat neuronal protein 1 (lrrn1), and OTU domain-containing protein 5-A (otud5a) were shared among the five traits (BM, TL, BL, BW, and ES).
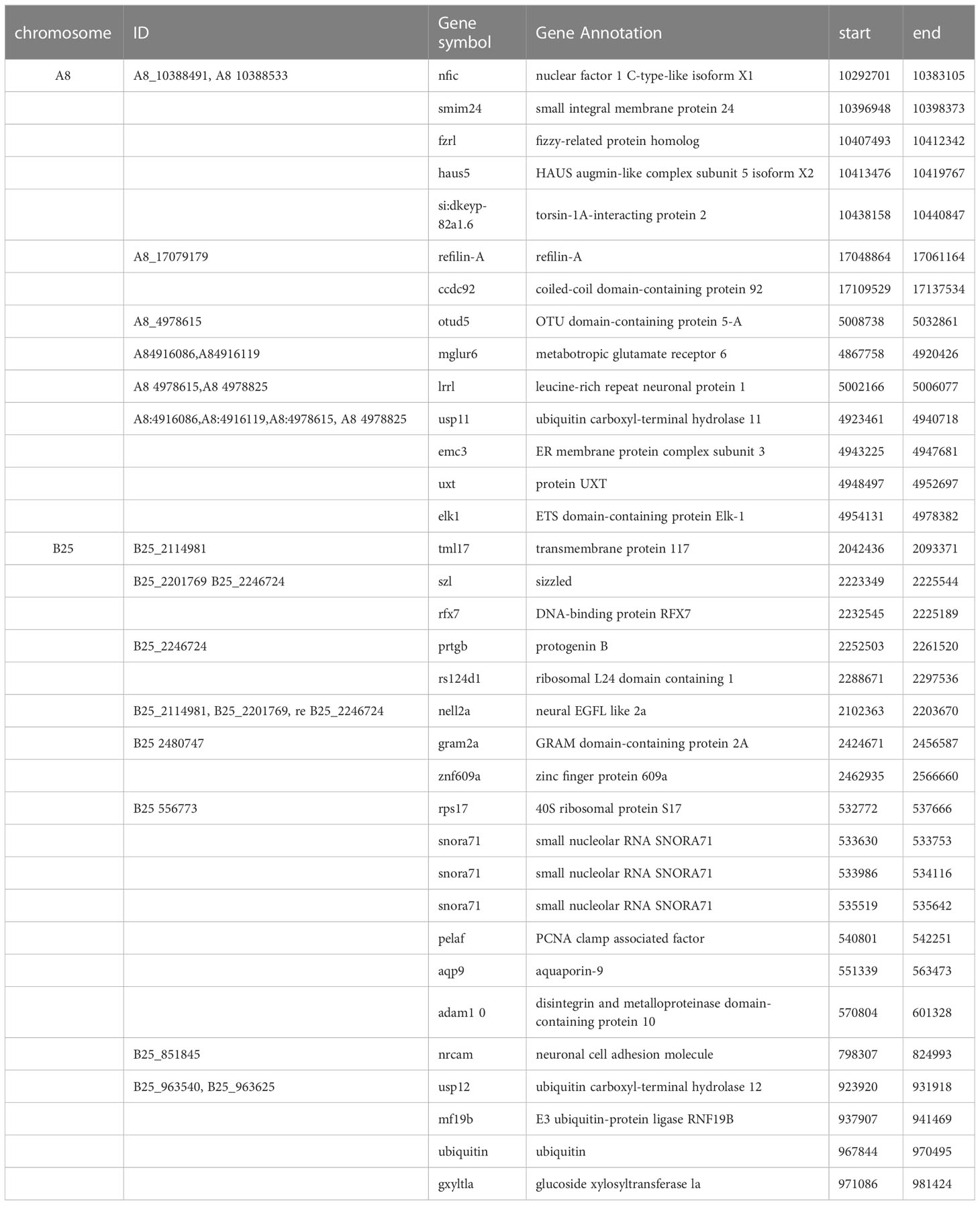
Table 5 The annotation information of candidate genes associated with growth traits on chromosome A8 and B25.
4 Discussion
The growth properties of the rice flower carp, an economically important fish species in integrated rice-fishing culture in China, affects fish production and economic value. In the present study, the first GWAS for growth traits in rice flower carp was conducted to identify markers and genes related to growth traits. A total of 90 loci that were significantly related to growth traits were detected in multiple chromosomes (Table S3), indicating that growth traits were complex quantitative traits controlled by multiple loci and multiple genes (Löhr et al., 2018; Yin et al., 2020). Similar results have been reported for many aquatic animals. For example, in Takifugu rubripes, 394 SNPs associated with growth traits were distributed among all 16 chromosomes (Wang, 2022); and in Epinephelus lanceolatus, a total of 36 growth-related SNPs spread over 14 chromosomes were detected (Wu et al., 2019).
The location of loci showed 31 loci associated with growth-related traits were concentrated on chromosomes A8 and B25, which indicated that chromosomes A8 and B25 may be closely related to growth traits of rice flower carp. Similar results have been reported in many fish species. In common carp, a total of 10 SNP loci distributed on LG8 for body mass were identified based on an ultra-high density linkage map and QTL mapping (Peng et al., 2016). In Larimichthys longirostris, the most obvious relationship to growth traits were located on chromosome 16 (Mou et al., 2022). Most growth-associated loci were clustered on chromosomes A8 and B25, suggesting that these two chromosomes may be central to identify growth-related genes for rice flower carp. In this study, 14 and 18 candidate genes were identified surrounding these loci in chromosomes A8 and B25, respectively. Among them, some candidate genes were involved in the regulation of the cell cycle and cells activity. Fizzy-related protein homolog (fzr1), a critical cell cycle regulator, plays a role in regulating the cell cycle by participating in the ubiquitin proteasome pathway (Manchado et al., 2010; Lu, 2013). Usp11 regulates BRCA2 expression levels by deubiquitinating it and participates in the cellular response to DNA damage (Schoenfeld et al., 2004). Uxt is a component of centrosome and is essential for cell viability (Zhao et al., 2005). Lrrn1 could suppresses GC cell apoptosis by downregulating AP‐1 (Liu et al., 2019). And some candidate genes were associated with visual function, innate immunity, osteogenic differentiation, and metabolic activity. Mutation of the zebrafish emc3 homolog, partial optokinetic response b (pob), causes the degeneration of photoreceptor cells (Taylor et al., 2005). Metabotropic glutamate receptor 6-like (mglur6) mediates excitatory chemo-synaptic transmission, and is a critical site for regulating information transmission in the optic rod pathway (Xu et al., 2000). Otud5 negatively regulates natural immunity by inhibiting the production of type I interferons (Kayagaki et al., 2007). Elk1 has been reported to be involved in osteoblast differentiation (Kousteni et al., 2003; Wiren et al., 2004; Kim et al., 2014). Nuclear factor 1 C-type-like isoform X1 (nfic) is associated with organismal osteogenesis (Zhang, 2017). Coiled-coil domain-containing protein 92 (ccdc92) is a protein-coding gene, and GWAS of several dyslipidemia and coronary heart disease in humans showed that the ccdc92 gene is localized in or adjacent to a susceptible region, including triglycerides and high-density lipoprotein cholesterol (Teslovich et al., 2010; Willer et al., 2013; Gupta et al., 2015; Spracklen et al., 2018).
Previous research has shown that a multi-trait comprehensive breeding strategy should be adopted for the breeding of rice flower carp (Xu et al., 2022). In this study, loci B6_4352672 was shared among seven growth-related traits, and A8_4978825 was shared among five growth-related traits, suggesting that B6_4352672 and A8_4978825 may be suitable for future molecular marker-assisted selection (MAS). Five candidate genes were identified surrounding B6_4352672, including trim25l, trak2, stradb, nbeal1, and RNA-binding motif protein 26 (rbm26). Trim25 is an E3 ubiquitin ligase involved in regulating the innate immune response against viruses (Martín-Vicente et al., 2017). In aquatic animals, Trim25 has been reported to play an important role in antiviral innate immunity in common carp (Palaiokostas et al., 2018), Epinephelinae (Yang et al., 2016), and Danio rerio (Jin et al., 2019). Nbeal1 is a Golgi-associated protein required for the regulation of cholesterol metabolism (Bindesbøll et al., 2020).
In addition to the candidate genes mentioned above, there are some candidate genes that are involved in fat uptake, myogenesis, growth, and immunity. Parkin RBR E3 ubiquitin protein ligase (prkn) encodes the E3-ubiquitin ligase PARKIN, which regulates systemic fat uptake via ubiquitin ligase-dependent effects (Kim et al., 2011) and changes ubiquitin labeling of cell cycle proteins (Wahabi et al., 2018). Adiponectin receptor protein 2-like (adipor2), an important adipocytokine, is associated with feeding, reproduction, immunity, and glycolipid metabolism (Kubota et al., 2007; Dobrzyn et al., 2018; Peng et al., 2018; Jian et al., 2019). Toll-like receptor 13 (tlr13) has been reported to recognize bacterial RNA, and Epinephelus coioides tlr13 is involved in recognizing the RNA of Vibrio parahaemolyticus and can induce the production of inflammatory cytokines (Zhou, 2017). Ubiquitin carboxyl-terminal hydrolase 49-like (usp49) negatively regulates cellular antiviral responses by deconjugating K63-linked ubiquitination of MITA (Ye et al., 2019). Sox-9-B-like (sox9b) was associated with skeletal development (Dalcq et al., 2012), pancreatic duct development (Manfroid et al., 2012), gonadal development (Meyer et al., 1997; Chiang et al., 2001; Chiang et al., 2001) and retinal development (Yokoi et al., 2009). Interestingly, many ubiquitin proteasome pathway-associated genes were detected, including ubiquitin carboxyl-terminal hydrolase FAF-X (usp9x), usp11, trim25l, ubiquitin-conjugating enzyme E2 D4 (ube2d4), prkn, usp49, ubiquitin carboxyl-terminal hydrolase 12-like (usp12), E3 ubiquitin-protein ligase RNF19B-like (rnf19b), and ubiquitin, which is similar to the previous study on the muscle transcriptome of rice flower carp with different sizes (Li et al., 2022). This suggests that the ubiquitin-proteasome pathway is involved in the growth regulation of rice flower carp. In this study, the function of most candidate genes may play important roles in the growth of rice flower carp should be determined in the future studies.
5 Conclusion
Rice flower carp is an important commercial fish species used in integrated rice-fishery farming. Improving the growth ratio using growth-trait associated markers could increase supply to match increasing market demand. In this study, 411,913 loci were obtained from the reduced-representation genome of rice flower carp and used to conduct a genome-wide association analysis to identify growth-trait associated markers. A total of 90 loci were significantly associated with growth traits in the rice flower carp. A series of candidate genes involved in metabolism, immunity, osteogenic differentiation, and visual function were extracted near the significant loci, including ccdc92, nbeal1, prkn, adipor2, trim25, tlr13, usp49, otud5, elk1, and nfic. This study provides important insights into the genetic architecture of growth traits in this fish species, as well as facilitate marker-assisted selection for growth improvement in rice flower carp.
Data availability statement
The data presented in the study are deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation, Chinese Academy of Sciences repository, accession number PRJCA014203.
Ethics statement
All experiments in this study were conducted in strict accordance with the ethical guidelines and regulations of the Animal Care and Use Committee of Southwest University and were approved by the local ethics committee of the Animal Welfare and Ethical Committee of Southwest University.
Author contributions
HL, XD, HY, DW, and YL designed the experiments. HL, YH, ZL, XD, HY, XP, KZ, LW, JQ, and WH performed the experiments and/or analyzed data. HL, YH, XP, ZL, and YL wrote the paper. HY, XD, DW, and YL revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (31960730), Natural Science Foundation of Chongqing (cstc2021jcyj-msxmX0837), and Chongqing Alliance for Aquatic Science and Technology Innovation, Guangxi characteristic freshwater fish industry innovation team (nycytxgxcxtd-2021-08).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1130667/full#supplementary-material
Supplementary Figure 1 | The change of lowest cross validation error at different K-value.
Supplementary Figure 2 | The population genetic structure of 200 rice flower carp, when K = 2-9.
Supplementary Figure 3 | Principal component analysis 3D plot of 200 rice flower carp.
Supplementary Figure 4 | The sequencing information of 200 rice flower carp.
Supplementary Table 2 | Information of loci significantly associated with growth traits.
Supplementary Table 3 | Candidate genes information near the loci associated with growth-related traits.
References
Alexander D. H., Novembre J., Lange K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. doi: 10.1101/gr.094052.109
Berghof T. V. L., Visker M. H. P. W., Arts J. A. J., Parmentier H. K., van der Poel J. J., Vereijken A. L. J., et al. (2018). Genomic region containing toll-like receptor genes has a major impact on total IgM antibodies including KLH-binding IgM natural antibodies in chickens. Front. Immunol. 8. doi: 10.3389/fimmu.2017.01879
Bian L., Wang J. (2017). The reduced-representation sequencing technique and its applications in marine organisms. J. Xiamen Univ. Nat. Sci. 56 (1), 3–12. doi: 10.6043/j.issn.0438-0479.201607015
Bindesbøll C., Aas A., Ogmundsdottir M. H., Pankiv S., Reine T., Zoncu R., et al. (2020). NBEAL1 controls SREBP2 processing and cholesterol metabolism and is a susceptibility locus for coronary artery disease. Sci. Rep. 10, 4528. doi: 10.1038/s41598-020-61352-0
Chang C. C., Chow C. C., Tellier L. C. A. M., Vattikuti S., Purcell S. M., Lee J. J. (2015). Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, 7. doi: 10.1186/s13742-015-0047-8
Chen S., Zhou Y., Chen Y., Gu J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, 884–890. doi: 10.1093/bioinformatics/bty560
Chiang E. F.-L., Pai C.-I., Wyatt M., Yan Y.-L., Postlethwait J., Chung B. (2001). Two Sox9 genes on duplicated zebrafish chromosomes: Expression of similar transcription activators in distinct sites. Dev. Biol. 231, 149–163. doi: 10.1006/dbio.2000.0129
Dalcq J., Pasque V., Ghaye A., Larbuisson A., Motte P., Martial J. A., et al. (2012). RUNX3, EGR1 and SOX9B form a regulatory cascade required to modulate BMP-signaling during cranial cartilage development in zebrafish. PloS One 7, e50140. doi: 10.1371/journal.pone.0050140
Davey J. W., Hohenlohe P. A., Etter P. D., Boone J. Q., Catchen J. M., Blaxter M. L. (2011). Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 12, 499–510. doi: 10.1038/nrg3012
Der-Auwera G. A. V., Carneiro M. O., Hartl C., Poplin R., Thibault J. (2013). From FastQ data to high confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinforma. 43, 11.10.1. doi: 10.1002/0471250953.bi1110s43
Dobrzyn K., Smolinska N., Kiezun M., Szeszko K., Rytelewska E., Kisielewska K., et al. (2018). Adiponectin: A new regulator of female reproductive system. Int. J. Endocrinol. 2018, 7965071. doi: 10.1155/2018/7965071
Drury C., Dale K. E., Panlilio J. M., Miller S. V., Lirman D., Larson E. A., et al. (2016). Genomic variation among populations of threatened coral: Acropora cervicornis. BMC Genomics 17, 286. doi: 10.1186/s12864-016-2583-8
Elshire R. J., Glaubitz J. C., Sun Q., Poland J. A., Kawamoto K., Buckler E. S., et al. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PloS One 6, e19379. doi: 10.1371/journal.pone.0019379
Francis R. M. (2017). POPHELPER: An r package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 17, 27–32. doi: 10.1111/1755-0998.12509
Gao J., Yang R. (2021). Joint genome-wide association study of body mass and morphological traits in turbot (Scophthalmus maximus). Prog. Fish. Sci. 42, 63–70. doi: 10.19663/j.issn2095-9869.20200202001
Gileta A. F., Fitzpatrick C. J., Chitre A. S., St. Pierre C. L., Joyce E. V., Maguire R. J., et al. (2022). Genetic characterization of outbred sprague dawley rats and utility for genome-wide association studies. PloS Genet. 18, e1010234. doi: 10.1371/journal.pgen.1010234
Gomez R. L., Klemetsdal G. (1999). Two-stage selection strategies utilizing marker-quantitative trait locus information and individual performance. J. Anim. Sci. 77, 2008. doi: 10.2527/1999.7782008x
Gupta G. D., Coyaud É., Gonçalves J., Mojarad B. A., Liu Y., Wu Q., et al. (2015). A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell 163, 1484–1499. doi: 10.1016/j.cell.2015.10.065
Habimana R., Ngeno K., Okeno T. O., Hirwa C. D. A., Keambou Tiambo C., Yao N. K. (2021). Genome-wide association study of growth performance and immune response to newcastle disease virus of indigenous chicken in rwanda. Front. Genet. 12. doi: 10.3389/fgene.2021.723980
Ji J., Luo C. L., Zou X., Lv X. H., Xu Y. B., Shu D. M., et al. (2019). Association of host genetics with intestinal microbial relevant to body weight in a chicken F2 resource population. Poult. Sci. 98, 4084–4093. doi: 10.3382/ps/pez199
Jia C., Zhu X., Wang L., Zhou C., Weng Q., Gan S., et al. (2020). SNPs discovery and phylogenetic analysis in eucalypts using ddGBS. J. Cent. South Univ. For. Technol. 40, 101–108. doi: 10.14067/j.cnki.1673-923x.2020.10.012
Jian M., Kwan J. S.-C., Bunting M., Ng R. C.-L., Chan K. H. (2019). Adiponectin suppresses amyloid-β oligomer (AβO)-induced inflammatory response of microglia via AdipoR1-AMPK-NF-κB signaling pathway. J. Neuroinflamm. 16, 110. doi: 10.1186/s12974-019-1492-6
Jiang Y., Yan X. (2009). Talking about the history, current situation and industrialization of farming rice flower carp in rice fields in quanzhou county. Fish. Guide Be Rich, (16), 20–21.
Jin Y., Jia K., Zhang W., Xiang Y., Jia P., Liu W., et al. (2019). Zebrafish TRIM25 promotes innate immune response to RGNNV infection by targeting 2CARD and RD regions of RIG-I for K63-linked ubiquitination. Front. Immunol. 10. doi: 10.3389/fimmu.2019.02805
Kayagaki N., Phung Q., Chan S., Chaudhari R., Quan C., O’Rourke K. M., et al. (2007). DUBA: A deubiquitinase that regulates type I interferon production. Science 318, 1628–1632. doi: 10.1126/science.1145918
Kim H. K., Kim M.-G., Leem K.-H. (2014). Collagen hydrolysates increased osteogenic gene expressions via a MAPK signaling pathway in MG-63 human osteoblasts. Food Funct. 5, 573–578. doi: 10.1039/c3fo60509d
Kim K.-Y., Stevens M. V., Akter M. H., Rusk S. E., Huang R. J., Cohen A., et al. (2011). Parkin is a lipid-responsive regulator of fat uptake in mice and mutant human cells. J. Clin. Invest. 121, 3701–3712. doi: 10.1172/JCI44736
Kousteni S., Han L., Chen J.-R., Almeida M., Plotkin L. I., Bellido T., et al. (2003). Kinase-mediated regulation of common transcription factors accounts for the bone-protective effects of sex steroids. J. Clin. Invest. 111, 1651–1664. doi: 10.1172/JCI17261
Kubota N., Yano W., Kubota T., Yamauchi T., Itoh S., Kumagai H., et al. (2007). Adiponectin stimulates AMP-activated protein kinase in the hypothalamus and increases food intake. Cell Metab. 6, 55–68. doi: 10.1016/j.cmet.2007.06.003
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li Z., Du X., Wen L., Li Y., Qin J., Chen Z., et al. (2022). Transcriptome analysis reveals the involvement of ubiquitin-proteasome pathway in the regulation of muscle growth of rice flower carp. Comp. Biochem. Physiol. Part D Genomics Proteomics 41, 100948. doi: 10.1016/j.cbd.2021.100948
Li H., Durbin R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li C., Wang J., Song K., Meng J., Xu F., Li L., et al. (2018). Construction of a high-density genetic map and fine QTL mapping for growth and nutritional traits of crassostrea gigas. BMC Genomics 19, 626. doi: 10.1186/s12864-018-4996-z
Li J.-T., Wang Q., Yang M.-D. H., Li Q.-S., Cui M.-S., Dong Z.-J., et al. (2021). Parallel subgenome structure and divergent expression evolution of allo-tetraploid common carp and goldfish. Nat. Genet. 53, 1493–149+. doi: 10.1038/s41588-021-00933-9
Liu F., Jiang X., Weng W., Gong X., Xu G. (2022). Development of SNP markers in section aigeiros poplar based on ddGBS technology. J. Cent. South Univ. For. Technol. 42, 178–185. doi: 10.14067/j.cnki.1673-923x.2022.07.019
Liu B., Zhang Y., Fan Y., Wang S., Li Z., Deng M., et al. (2019). Leucine-rich repeat neuronal protein-1 suppresses apoptosis of gastric cancer cells through regulation of Fas/FasL. Cancer Sci. 110, 2145–2155. doi: 10.1111/cas.14042
Löhr H., Hess S., Pereira M. M. A., Reinoß P., Leibold S., Schenkel C., et al. (2018). Diet-induced growth is regulated via acquired leptin resistance and engages a pomc-Somatostatin-Growth hormone circuit. Cell Rep. 23, 1728–1741. doi: 10.1016/j.celrep.2018.04.018
Lu L. (2013). Novel function of hect-type ubiquitin ligase nedl2 in cell cycle regulation (Beijing: Beijing university of technology).
Luo H., Li Y., Zheng S., Zhou J., Zou X., Li C., et al. (2022). Identification of male sex-specific markers using genome re-sequencing in the Chinese longsnout catfish leiocassis longirostris. Aquaculture 558, 738392. doi: 10.1016/j.aquaculture.2022.738392
Manchado E., Eguren M., Malumbres M. (2010). The anaphase-promoting complex/cyclosome (APC/C): Cell-cycle-dependent and -independent functions. Biochem. Soc Trans. 38, 65–71. doi: 10.1042/BST0380065
Manfroid I., Ghaye A., Naye F., Detry N., Palm S., Pan L., et al. (2012). Zebrafish sox9b is crucial for hepatopancreatic duct development and pancreatic endocrine cell regeneration. Dev. Biol. 366, 268–278. doi: 10.1016/j.ydbio.2012.04.002
Martín-Vicente M., Medrano L. M., Resino S., García-Sastre A., Martínez I. (2017). TRIM25 in the regulation of the antiviral innate immunity. Front. Immunol. 8. doi: 10.3389/fimmu.2017.01187
Meyer J., Südbeck P., Held M., Wagner T., Schmitz M. L., Bricarelli F. D., et al. (1997). Mutational analysis of the SOX9 gene in campomelic dysplasia and autosomal sex reversal: Lack of genotype/phenotype correlations. Hum. Mol. Genet. 6, 91–98. doi: 10.1093/hmg/6.1.91
Miller M. R., Dunham J. P., Amores A., Cresko W. A., Johnson E. A. (2007). Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res. 17, 240–248. doi: 10.1101/gr.5681207
Mou C.-Y., Li Y., Zhou J., Li Q., Zhou B., Wei Z., et al. (2022). Genome-wide association study reveals growth-related markers and candidate genes for selection in Chinese longsnout catfish (Leiocassis longirostris). Aquaculture 560, 738513. doi: 10.1016/j.aquaculture.2022.738513
Nunez J. C. B., Seale T. P., Fraser M. A., Burton T. L., Fortson T. N., Hoover D., et al. (2015). Population genomics of the euryhaline teleost poecilia latipinna. PloS One 10, e0137077. doi: 10.1371/journal.pone.0137077
Palaiokostas C., Robledo D., Vesely T., Prchal M., Pokorova D., Piackova V., et al. (2018). Mapping and sequencing of a significant quantitative trait locus affecting resistance to koi herpesvirus in common carp. G3 Bethesda Md 8, 3507–3513. doi: 10.1534/g3.118.200593
Peng Y.-J., Shen T.-L., Chen Y.-S., Mersmann H. J., Liu B.-H., Ding S.-T. (2018). Adiponectin and adiponectin receptor 1 overexpression enhance inflammatory bowel disease. J. Biomed. Sci. 25, 24. doi: 10.1186/s12929-018-0419-3
Peng W., Xu J., Zhang Y., Feng J., Dong C., Jiang L., et al. (2016). An ultra-high density linkage map and QTL mapping for sex and growth-related traits of common carp (Cyprinus carpio). Sci. Rep. 6, 26693. doi: 10.1038/srep26693
Schoenfeld A. R., Apgar S., Dolios G., Wang R., Aaronson S. A. (2004). BRCA2 is ubiquitinated in vivo and interacts with USP11, a deubiquitinating enzyme that exhibits prosurvival function in the cellular response to DNA damage. Mol. Cell. Biol. 24, 7444–7455. doi: 10.1128/MCB.24.17.7444-7455.2004
Spracklen C. N., Chen P., Kim Y. J., Wang X., Cai H., Li S., et al. (2018). Association analyses of East Asian individuals and trans-ancestry analyses with European individuals reveal new loci associated with cholesterol and triglyceride levels. Hum. Mol. Genet. 27, 1122. doi: 10.1093/hmg/ddx439
Taylor M. R., Kikkawa S., Diez-Juan A., Ramamurthy V., Kawakami K., Carmeliet P., et al. (2005). The zebrafish pob gene encodes a novel protein required for survival of red cone photoreceptor cells. Genetics 170, 263–273. doi: 10.1534/genetics.104.036434
Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M., et al. (2010). Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713. doi: 10.1038/nature09270
Vignal A., Milan D., SanCristobal M., Eggen A. (2002). A review on SNP and other types of molecular markers and their use in animal genetics. Genet. Sel. Evol. GSE 34, 275–305. doi: 10.1186/1297-9686-34-3-275
Wahabi K., Perwez A., Rizvi M. A. (2018). Parkin in parkinson’s disease and cancer: a double-edged sword. Mol. Neurobiol. 55, 6788–6800. doi: 10.1007/s12035-018-0879-1
Wang Z. (2022). Genome-wide association study of growth traits of takifugu rubripes (Dalian Ocean University).
Wang K., Li M., Hakonarson H. (2010). ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164. doi: 10.1093/nar/gkq603
Willer C. J., Schmidt E. M., Sengupta S., Peloso G. M., Gustafsson S., Kanoni S., et al. (2013). Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45, 1274–1283. doi: 10.1038/ng.2797
Wiren K. M., Toombs A. R., Zhang X.-W. (2004). Androgen inhibition of MAP kinase pathway and elk-1 activation in proliferating osteoblasts. J. Mol. Endocrinol. 32, 209–226. doi: 10.1677/jme.0.0320209
Wu X., Xv Y., Wen L., Huang Y., Qin J., Zhou K., et al. (2021). Observation on the composition, distribution and pigment content of chromatophores in rice flower carp in quanzhou. J. Guangxi Norm. Univ. Sci. Ed. 40 (6), 222–229 doi: 10.16088/j.issn.1001-6600.2021081001
Wu L., Yang Y., Li B., Huang W., Wang X., Liu X., et al. (2019). First genome-wide association analysis for growth traits in the largest coral reef-dwelling bony fishes, the giant grouper (Epinephelus lanceolatus). Mar. Biotechnol. N. Y. N 21, 707–717. doi: 10.1007/s10126-019-09916-8
Xu Y., Chen Z., Qin J., Wen L., Pan X., Zhou K., et al. (2022). Genetic parameters estimation for main growth traits of (Cyprinuscarpio var. quanzhouensis). Guangxi Sci. 29, 801–808. doi: 10.13656/j.cnki.gxkx.20220919.023
Xu H., Mi M., Lang H., Yu X., Wei N., Huang G. (2000). Effects of taurine and micronutrient complex on metabotropic glutamate receptor mglur6 mRNA in rat retina. Acta Nutr. Sin., 22 (4), 303–307. doi: 10.13325/j.cnki.acta.nutr.sin.2000.04.004
Yang Y., Huang Y., Yu Y., Yang M., Zhou S., Qin Q., et al. (2016). RING domain is essential for the antiviral activity of TRIM25 from orange spotted grouper. Fish Shellfish Immunol. 55, 304–314. doi: 10.1016/j.fsi.2016.06.005
Yang S., Jiang A. (2009). Analysis on the flesh rate and the muscle nutrition component in procypris merus. J. Hydroecol. 2 (2), 154–157. doi: 10.15928/j.1674-3075.2009.02.026
Yang J., Lee S. H., Goddard M. E., Visscher P. M. (2011). GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82. doi: 10.1016/j.ajhg.2010.11.011
Ye L., Zhang Q., Liuyu T., Xu Z., Zhang M.-X., Luo M.-H., et al. (2019). USP49 negatively regulates cellular antiviral responses via deconjugating K63-linked ubiquitination of MITA. PloS Pathog. 15, e1007680. doi: 10.1371/journal.ppat.1007680
Yin Y., Jiang W., Pan X., Yang J. (2020). Recent progress in growth trait of aquaculture fish. J. Fish. Sci. China 27, 463–484. doi: 10.3724/SP.J.1118.2020.19150
Yokoi H., Yan Y.-L., Miller M. R., BreMiller R. A., Catchen J. M., Johnson E. A., et al. (2009). Expression profiling of zebrafish sox9 mutants reveals that Sox9 is required for retinal differentiation. Dev. Biol. 329, 1–15. doi: 10.1016/j.ydbio.2009.01.002
Zhang F. (2017). The effect of aging on the expression of osteogenic and adipogenic factors and nfic in rats (Beijing: Chinese People’s Liberation Army (PLA) Medical School).
Zhao H., Wang Q., Zhang H., Liu Q., Du X., Richter M., et al. (2005). UXT is a novel centrosomal protein essential for cell viability. Mol. Biol. Cell 16, 5857–5865. doi: 10.1091/mbc.E05-08-0705
Zhou Y. (2017). Recognition of them vibrio parahaemolyticus RNA via TLR13 in orange-spotted grouper (Epinephelus coioid) (Haikou: Hainan university).
Keywords: rice flower carp, Cyprinus carpio, GWAS, growth traits, SNP, candidate genes
Citation: Luo H, Pan X, Huang Y, Li Z, Ye H, Zhou K, Wen L, Qin J, He W, Du X, Wang D and Lin Y (2023) Genome-wide association study reveals markers and candidate genes associated with growth in the rice flower carp, an economic fish species of integrated rice-fish culture in China. Front. Mar. Sci. 10:1130667. doi: 10.3389/fmars.2023.1130667
Received: 23 December 2022; Accepted: 06 February 2023;
Published: 21 February 2023.
Edited by:
Qian Lu, Jiangsu University of Science and Technology, ChinaReviewed by:
Zhiyong Wang, Jimei University, ChinaYu Xiao, Chinese Academy of Sciences, China
Tao Zhou, Xiamen University, China
Lin Chen, Xiamen University, China
Copyright © 2023 Luo, Pan, Huang, Li, Ye, Zhou, Wen, Qin, He, Du, Wang and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuesong Du, NjI3MzgwNzU2QHFxLmNvbQ==; Dapeng Wang, b3Vjd2RwQDE2My5jb20=; Yong Lin, bGlubm4yMDA1QDEyNi5jb20=
†These authors have contributed equally to this work
 Hui Luo
Hui Luo Xianhui Pan3†
Xianhui Pan3† Zhe Li
Zhe Li Hua Ye
Hua Ye Yong Lin
Yong Lin