- 1Ocean Science Department, University of California at Santa Cruz, Santa Cruz, CA, United States
- 2Flathead Lake Biological Station, University of Montana, Polson, MT, United States
Dinitrogen (N2) fixation is carried out by specialized microbes, called diazotrophs, and is a major source of nitrogen supporting primary production in oligotrophic oceans. One of the best-characterized diazotroph habitats is the North Pacific Subtropical Gyre (NPSG), where warm, chronically N-limited surface waters promote year-round N2 fixation. At Station ALOHA (A Long-Term Oligotrophic Habitat Assessment) in the NPSG, N2 fixation is typically ascribed to conspicuous, filamentous cyanobacterial diazotrophs (Trichodesmium and Richelia), unicellular free-living Crocosphaera, and the UCYN-A/haptophyte symbiosis, based on using microscopy and quantitative PCR (qPCR). However, the diazotroph community in this ecosystem is diverse and includes non-cyanobacterial diazotrophs (NCDs). We investigated the diversity, depth distributions, and seasonality of diazotroph communities at Stn. ALOHA using high throughput sequencing (HTS) of nifH gene fragments from samples collected throughout the euphotic zone (0-175 m) at near-monthly intervals from June 2013 to July 2016. The UCYN-A symbioses and Trichodesmium sp. consistently had the highest relative abundances and seasonal patterns that corroborated qPCR-based analyses. Other prevalent community members included a new Crocosphaera-like species, and several NCDs affiliated with γ- and δ-proteobacteria. Notably, some of the NCDs appear to be stable components of the community at Stn. ALOHA, having also been reported in prior studies. Depth and temporal patterns in microdiversity within two major diazotroph groups (Trichodesmium and UCYN-A) suggested that sub-populations are adapted to time- and depth-dependent environmental variation. A network analysis of the upper euphotic (0-75 m) HTS data identified two modules that reflect a diazotroph community structure with seasonal turnover between UCYN-A/Gamma A, and Trichodesmium/Crocosphaera. It also reveals the seasonality of several important cyanobacteria and NCDs about which little is known, including a putative δ-proteobacterial phylotype originally discovered at Stn. ALOHA. Collectively, these results underscore the importance of coupling nifH gene HTS with other molecular techniques to obtain a comprehensive view of diazotroph community composition in the marine environment and reveal several understudied diazotroph groups that may contribute to N2 fixation in the NPSG.
1 Introduction
Biological dinitrogen (N2) fixation, the reduction of N2 gas into ammonia, supplies biologically available nitrogen (N) to N-limited ecosystems (Falkowski, 1997; Galloway et al., 2004). This process is carried out by a specialized group of Bacteria and Archaea called diazotrophs. In the oceans, one of the best-characterized diazotroph habitats is the North Pacific Subtropical Gyre (NPSG), where warm, chronically N-limited surface waters promote year-round N2 fixation supporting up to half of new production (Karl et al., 1997; Böttjer et al., 2017). N2 fixation in the NPSG had historically been ascribed to filamentous diazotrophs visible using microscopy, which includes colony-forming Trichodesmium spp. and heterocyst-forming symbionts of diatoms (diatom diazotroph associations, DDAs) (Mague et al., 1974; Venrick, 1974; Villareal et al., 1993; Letelier & Karl, 1996). However, it is now recognized that unicellular taxa (both cyanobacteria and non-cyanobacteria) often dominate diazotroph communities in the NPSG, and that small diazotrophs also likely contribute substantially to bulk water N2 fixation (Church et al., 2009).
Knowledge of diazotroph community structure in marine systems has improved through applications of molecular approaches targeting the nifH gene, which encodes the two identical subunits of the iron protein of the nitrogenase enzyme that catalyzes N2 fixation (Zehr and Capone, 2021). Phylogeny constructed from nifH gene sequences agrees fairly well with 16S rRNA gene-based phylogeny making nifH a useful phylogenetic marker gene (Zehr et al., 2003). Sequencing nifH genes from pelagic ocean samples has shown that marine diazotrophs include unicellular cyanobacteria and non-cyanobacteria in addition to the conspicuous filamentous cyanobacterial groups (Zehr et al., 1998). Unicellular cyanobacterial diazotrophs include three major groups: UCYN-A (obligate symbionts of haptophytes), UCYN-B (Crocosphaera spp.; free-living, colonial, or symbionts of Climacodium), and UCYN-C (Cyanothece-like organisms, free-living and symbionts of diatoms) (Zehr et al., 2001; Foster et al., 2007; Zehr and Capone, 2020, Schvarcz et al., in press). Filamentous cyanobacteria include Trichodesmium and DDAs, which are classified into three groups based on phenotypic and phylogenomic criteria: Richelia intracellularis associated with Rhizosolenia (Het-1), Richelia euintracellularis associated with Hemiaulus (Het-2), and Richelia rhizosoleniae associated with Chaetoceros (Het-3) (Foster and Zehr, 2006; Foster et al., 2022). Non-cyanobacterial diazotrophs (NCDs) include representatives from diverse taxa, including the α-, β-, δ-, and γ-proteobacteria, firmicutes, and planctomycetes (Zehr et al., 1998; Delmont et al., 2018), although it is still uncertain whether these organisms contribute substantively to N2 fixation in pelagic open ocean ecosystems (Moisander et al., 2017). Marine diazotroph taxa differ not only in taxonomy, but also in size, lifestyle, and ecology, making it important to understand their different distributions and environmental drivers.
One important application of the nifH gene has been the development of quantitative PCR (qPCR) and digital droplet PCR (ddPCR) assays to target and quantify specific taxa. At Stn. ALOHA (A Long-term Oligotrophic Habitat Assessment, 22°45’N, 158°00’W), a field site within the NPSG, cyanobacterial diazotrophs have been quantified via nifH qPCR on near-monthly Hawaii Ocean Time-series (HOT) cruises since 2006 (Church et al., 2009; dataset doi: 10.5281/zenodo.4477269). The six groups targeted (UCYN-A1, Crocosphaera, Trichodesmium, Het-1, Het-2, and Het-3) are detectable year-round, but display different seasonality and depth-distributions. For instance, abundances of UCYN-A1, a small sub-lineage within the UCYN-A group, peak in the spring, while Crocosphaera, Trichodesmium, and DDAs generally reach peak abundances in summer or fall (Church et al., 2009; Böttjer et al., 2014), when bulk N2 fixation rates are the highest (Böttjer et al., 2017). The mechanisms driving these patterns are complex but appear at least partially related to seasonal changes in sea surface temperature and nutrient availability, as well as episodic changes attributable to mesoscale physical features (Church et al., 2009).
Despite the utility of qPCR, there are disadvantages to using this method as the only determinant of diazotroph community structure. Namely, qPCR assays are designed to amplify targets with low nucleotide diversity in primer/probe binding sites, and thus only target specific, known nifH gene sequence types. A complementary approach is sequencing the nifH gene using degenerate, nearly universal primers, which provides qualitative information on diazotroph diversity and the relative abundances of taxa. HTS technology now generates orders of magnitude more data than original clone library sequencing approaches, enabling the recovery of sequences from rare taxa (Pedros-Alio, 2012). Applications of nifH gene HTS in the NPSG have revealed diverse diazotroph communities (Farnelid et al., 2011; Gradoville et al., 2017b; Cornejo-Castillo et al., 2019; Farnelid et al., 2019; Gradoville et al., 2020), including many NCD taxa for which qPCR primer/probe sets are currently unavailable. HTS analysis of the nifH gene also show fine-scale nifH gene diversity, such as different clades within the Trichodesmium genus (Hynes et al., 2012; Gradoville et al., 2017a) that can have different distributions and environmental drivers (Hutchins et al., 2013; Rouco et al., 2014) but are not distinguished from one another by existing Trichodesmium nifH-based qPCR assays.
While previous nifH gene HTS studies have revealed diverse diazotroph communities at and near Stn. ALOHA (Farnelid et al., 2011; Gradoville et al., 2017b; Farnelid et al., 2019; Gradoville et al., 2020), these studies reflect snapshots in time and do not address seasonality or environmental factors driving variability in diazotroph community structure. We present a three-year time series of nifH gene diversity at Stn. ALOHA. We performed nifH gene HTS using DNA samples collected at nearly-monthly intervals from HOT cruises from June 2013 to July 2016 and analyzed results in the context of hydrographic and biogeochemical data available from the HOT dataset. Our results reveal new co-occurrence patterns within the diazotroph community at Stn. ALOHA, identify understudied taxa (both cyanobacterial and non-cyanobacterial) with strong depth- and seasonality-patterns, and reveal microdiversity within two major diazotroph taxa (Trichodesmium and UCYN-A) suggesting that distinct lineages are adapted to different niches within this oligotrophic habitat.
2 Materials and methods
2.1 Diazotroph community composition using nifH gene high throughput sequencing
The seasonal and temporal diversity of diazotrophs at Stn. ALOHA were characterized using nifH gene HTS as previously described (Turk-Kubo et al., 2015; Cabello et al., 2020). For DNA samples, seawater (~2 L) was collected from 5, 25, 45, 75, 100, 125, 150, and 175 m depths using Niskin® bottles attached to the CTD rosette and sampled into acid-washed polycarbonate bottles. Water was immediately filtered through 25 mm diameter 0.2 µm pore size polyethersulfone filters using gentle peristaltic pumping. Filters were placed in lysis buffer AP1 (DNeasy Plant Kit, Qiagen, Germantown, MD, USA), flash frozen, and stored at -80°C until processing. DNA was extracted using the DNeasy Plant Kit (Qiagen) following the protocol detailed in Paerl et al. (2008), which includes repeated freeze-thaw cycles, agitation with a bead beater, and a proteinase K digestion to improve lysis of cyanobacterial cell walls.
Partial nifH gene sequences were amplified using a universal nested nifH PCR assay (Zehr and McReynolds, 1989; Zani et al., 2000). The first round of reactions contained 2 µL of template DNA, Platinum™ Taq DNA polymerase (6 units; Invitrogen, Carlsbad, CA), 1X PCR Buffer (-MgCl2), 4 mM MgCl2, 400 µM dNTP mix, and 0.5 µM of primers nifH3 and nifH4 in a reaction brought up to 20 µL with RT-PCR grade water (Applied Biosystems, Waltham, MA, USA). Second round reactions contained 2 µL of the first-round reaction, 10 units of DNA polymerase, 1X PCR Buffer, 4 mM MgCl2, 200 µM dNTP mix, and 1 µM of nifH1 and nifH2 primers in a 15 µL reaction volume. To minimize contamination, all PCR reactions were carried out in an amplicon-free UV-hood as described by Turk-Kubo et al. (2014), and no-template-controls were processed with each set of PCR reactions. Products were amplified using the following thermocycling parameters: 95°C for 3 min, followed by 25 (round 1) or 30 cycles (round 2) of 95°C for 30s, annealing at 55°C (round 1) or 57°C (round 2), and 72°C for 45s. All samples were amplified in duplicate, screened for the correct products using gel electrophoresis, and pooled prior to sequencing. The no-template-controls did not yield any amplification. A targeted amplicon sequencing approach was used to create barcoded libraries as described in Green et al. (2015), using 5’ common sequence linkers (Moonsamy et al., 2013) on second round primers, nifH1 and nifH2. Sequence libraries were prepared at the DNA Service Facility at the University of Illinois at Chicago, and multiplexed amplicons were bidirectionally sequenced (2 x 300 bp) using the Illumina MiSeq platform at the W.M. Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana-Champaign. Samples were multiplexed to achieve ca. 40,000 high quality merged reads per sample. Demultiplexed raw sequences are available under BioProject PRJNA913939 in the Sequence Read Archive at NCBI.
De-multiplexed raw paired end reads were merged, quality filtered (trimming reads after two consecutive bases with quality scores <20), and size selected (300-400 bp) using Paired-End reAd mergeR (Zhang et al., 2014). Primers were removed using CLC Genomics workbench (Qiagen). Chimera removal and determination of operational taxonomic units (OTUs; 97% nucleotide identity) were conducted in QIIME 1 (Caporaso et al., 2010) using UCHIME and USEARCH v6.1 (Edgar, 2010; Edgar et al., 2011). Representative sequences from OTUs with greater than 100 sequences were imported into ARB (Ludwig et al., 2004), translated into amino acid sequences, and non-nifH sequences and sequences with frameshifts were removed. After these quality control steps, a total of 4,758,763 nifH sequences remained, ranging from 5,019-33,963 sequences per sample (averaging 18,662). Samples were then rarefied to 5019 sequences using the QIIME script multiple_rarefactions_even_depth.py. OTUs were screened for known reagent contaminants (>92% nucleotide identity using the QIIME script exclude_seqs_by_blast.py) but none were found. Taxonomy was assigned via blastx of representative OTU sequences against a curated nifH database containing full length nifH sequences (www.jzehrlab.com/nifh) where nifH cluster designations have been assigned based on the convention outlined in Zehr et al. (2003). Phylogenetic trees based on partial nifH nucleotide sequences were calculated in ARB.
For the Trichodesmium and UCYN-A microdiversity analyses, OTUs representing >0.1% of the total nifH sequence dataset were included. Trichodesmium OTUs were resolved to the species level if the OTU cluster representative sequence shared >98% nucleotide (nt) identity over the full length of the nifH gene fragment to nifH from a Trichodesmium isolate.
2.2 Diazotroph community composition using nifH-based quantitative PCR techniques
UCYN-A1, Crocosphaera spp. (UCYN-B), Trichodesmium spp., and the DDA groups Het-1, Het-2, and Het-3 were enumerated as part of a prior study using qPCR targeting nifH (dataset doi: 10.5281/zenodo.4477269). Details on qPCR assays are summarized in Church et al. (2009), with commonly used primers/probes (Church et al., 2005a; Church et al., 2005b). Two additional NCD groups were enumerated using ddPCR as part of this study based on previously developed qPCR assays, one targeting a cluster III sequence type (Church et al., 2005a) and the other targeting γ-24774A11 (or gamma A; Moisander et al., 2008). All aspects of the ddPCR protocol, including reaction conditions, droplet generation, thermocycling parameters, thresholding, and detection limits, are detailed in Gradoville et al. (2021).
The nifH gene HTS dataset was generated from the same DNA extracts used for qPCR and ddPCR allowing for useful comparisons of the nifH gene HTS- and nifH qPCR-based approaches. For comparisons between nifH gene HTS- and qPCR-based abundances, the dominant OTU for a given target group was selected, e.g. UCYN-A1_otu0 relative abundances vs. UCYN-A1 qPCR-based abundances, after verifying the OTU had no significant mismatches to the qPCR primers/probe (Table S1).
2.3 Environmental data from Station ALOHA
Environmental data from Stn. ALOHA (Table S2) were obtained from the Hawaii Ocean Time-series Data Organization & Graphical System (HOT-DOGS; hahana.soest.hawaii.edu/hot/hot-dogs/interface.html), except for sea level anomaly (SLA) data, which are described below. Environmental data included in this analysis are from Niskin bottle collections (hahana.soest.hawaii.edu/FTP/hot/water) and included the following parameters: temperature (°C); salinity; dissolved oxygen (µmol kg-1); pH; depth (m); particulate carbon (PC, µmol kg-1); particulate nitrogen (PN, µmol kg-1), particulate phosphorus (PP, nmol kg-1); low-level nitrate+nitrite (N+N, µmol kg-1), low-level soluble reactive phosphorus (SRP, µmol kg-1), dissolved organic N (DON, µmol kg-1); dissolved organic C (DOC, µmol kg-1); total dissolved N (TDN, µmol kg-1); heterotrophic bacteria, Prochlorococcus, Synechococcus and photosynthetic picoeukaryote abundances (x 105 mL-1); and chlorophyll a concentrations (chl a, ng L-1). Missing data were imputed by depth via the R package bcv (https://arxiv.org/abs/0908.2062) using multivariate singular value decomposition (Table S2). The mixed layer depth (MLD) for each cruise was defined by a potential density offset of 0.03 kg m-3 relative to 10 m (de Boyer Montégut et al., 2004). For each HOT cruise, the mean potential densities at each pressure level over all CTD casts were used.
Measurements of photosynthetically active radiation (PAR) at the sea surface were conducted using a shipboard LI-COR I-1500 Light Sensor Logger (https://hahana.soest.hawaii.edu/FTP/hot/light/licor/). The resulting measurements were used to derive the mean daily-integrated PAR fluxes for each cruise. The coefficient describing the vertical attenuation of PAR (KPAR) was computed based on measurements of downwelling PAR through the upper ~150 m using a profiling HyperPro (hp, Satlantic, Halifax, Novia Scotia) (hahana.soest.hawaii.edu/hot/hot-dogs/interface.html). Downwelling PAR was converted to scalar PAR by multiplying by 1.2 (Wozniak et al., 1992). For each cruise, we calculated the average daily integrated PAR at discrete depths (PARdepth, mol quanta m-2 d-1) as the product of daily integrated incident PAR (PARsurf, mol quanta m-2 d-1) and the derived PAR isopleths (% PAR), similar to Letelier et al. (2004).
Multimission satellite altimetric observations of sea level anomalies were obtained from the Copernicus Marine and Environment Monitoring Service (http://www.marine.copernicus.eu). Modifications were applied as published in Barone et al. (2019) that corrected for both the long-term linear trend and seasonal cycle; consistent with that study, the resulting SLA product is referred to here as SLAcorr.
2.4 Weighted gene correlation network analyses and correlations to environment
Weighted gene correlation network analysis (WGCNA) was used to identify OTUs with highly correlated relative abundances over the time series, using the approach of Langfelder and Horvath (2008) as implemented in the WGCNA R package (v1.70.3). Of the 2916 total OTUs, the analysis included 131 non-rare OTUs (>10 reads in ≥10 samples) which accounted for 87.5% of all the reads in the rarefied dataset. Due to the observed bias between relative abundances and qPCR absolute abundances in lower euphotic zone samples (when comparisons can be made; Figure S1), network analysis focused on data from the 128 upper euphotic zone samples (≤ 75 m), which contained 128 non-rare OTUs (44.3% of all rarefied reads). On the log2 abundances (+1) for the 128 OTUs, we used blockwiseConsensusModules() to create a signed network with minModuleSize = 20 and power = 4, selected based an evaluation of different soft thresholds using pickSoftThreshold(). Two modules were identified which had 43 OTUs (M1) and 37 OTUs (M2). Pearson correlations between these OTUs and environmental variables were considered significant for p-values < 0.05, calculated using corPvalueStudent() with nSamples=128. These correlations were hierarchically clustered for OTUs as well as environmental variables with the pvclust R package v2.2 (correlation-based distances, average linkage clustering, 2000 bootstraps, cluster significance alpha = 0.95; Suzuki and Shimodaira, 2006).
3 Results
3.1 Biological, chemical, and physical conditions at Stn. ALOHA
Over the course of the study (Jun 2013 - Jul 2016), biological, chemical, and physical conditions were largely consistent with previously reported climatological records at Stn. ALOHA (Karl and Church, 2017; Table S2). The upper euphotic zone (≤75 m) was characterized by high-light and low-nutrient concentrations, while the lower euphotic zone (100-175 m) was characterized by lower-light and higher-nutrient concentrations. Variation in mixed layer depths (MLD) were typical for Stn. ALOHA, with deeper mixing in the winter and spring (mean of 60 m) and shoaling in the summer and fall (mean of 25 m). Over the study period, uncharacteristically shallow mixing for the winter season was observed in 2014 (Table S2). SLAcorr ranged from -16 to 14 cm, with 75% of measured values falling between -4 to 6 cm.
In the upper euphotic zone (≤75 m), temperatures ranged from 22.4-27.5°C (Table S2), with the highest temperatures occurring in summer-early fall (Aug-Oct). Mean daily integrated PAR at the sea surface (PARsurf) was highest (>50 mol quanta m-2 d-1) in May-August, which is consistent with Stn. ALOHA historical climatology (Letelier et al., 2004). Cooler temperatures in the upper euphotic zone occurred during winter months, when PARsurf was at a minimum (Jan, 26.3 mol quanta m-2 d-1). In the lower euphotic zone (100-175 m), temperatures ranged from 17.7-24.7°C.
Nutrient and chlorophyll a concentrations also followed a seasonal cycle, most pronounced in the lower euphotic zone (Table S2). In the upper euphotic zone, N+N remained low year-round, ranging from 0.001-0.013 µmol kg-3. In the lower euphotic zone, N+N ranged from 0.001-4.6 µmol kg–1 and generally peaked in late fall to early winter, consistent with patterns described by Venrick (1988) and Letelier et al. (2004). In the lower euphotic zone, N+N concentrations were inversely correlated to SLAcorr (Pearson correlation, r(122) = -0.348, p<0.001). SRP concentrations ranged from 0.02-0.15 µmol kg–1 in the upper euphotic zone and from 0.03-0.37 µmol kg–1 in the lower euphotic zone. Like N+N, SRP concentrations were inversely correlated to SLAcorr (Pearson correlation, r(127) = -0.232, p<0.01) in the lower euphotic zone. The resulting N+N:SRP values were well below Redfield stoichiometry in both the upper and lower euphotic zone, averaging 0.05 and 0.65, respectively. Chl a ranged from 43-415 ng L-1 in the upper euphotic zone and from 8-535 ng L-1 in the lower euphotic zone, with concentrations of chl a demonstrating a consistent maximum near 100 m (Table S2).
3.2 Major constituents of the diazotroph community at Stn. ALOHA
The most highly recovered nifH sequence types at Stn. ALOHA included both cyanobacteria (nifH cluster 1B) and NCDs (nifH clusters 1G and 3) (Figure 1A). In the rarefied dataset, 12 OTUs accounted for 79.1% of the total nifH sequences; 7 of these were cyanobacteria (59.5% of total), and 5 were NCDs (19.5%) (Table S3). Five of the major OTUs have been enumerated using qPCR/ddPCR for this time series, enabling direct comparisons of patterns observed using both approaches.

Figure 1 Overview of the diversity, depth distributions, and relative abundances of major diazotrophs at Stn. ALOHA. (A) Maximum likelihood (RAxML; Stamatakis, 2014)) tree of partial nifH gene nucleotide sequences (clustered at 97% nt identity) including representative sequences corresponding to the OTUs with high relative abundances, along with reference sequences from closely related organisms. The phylogenetic tree was built based on 341 nifH nucleotide positions and the GTR GAMMA model of nucleotide rate heterogeneity (Yang, 1994) was used to determine branch lengths. The percentage of calculated trees in which sequences clustered together in the bootstrap test (100 replicates) is shown next to the node when greater than 50%. Genbank accession numbers of reference sequences are in parenthesis. OTUs with existing qPCR assays are indicated with grey diamonds and white diamonds indicate OTUs with no known qPCR assays. Cluster designations follow the convention established in Zehr et al. (2003). (B) Depth patterns in the euphotic zone of the selected OTUs are plotted as solid lines representing mean values of relative abundances across the time series with shading representing +/- 1 standard deviation. (C) Relative abundances of select OTUs summed across depths for each cruise. Rows represent diazotroph OTUs, and the size of the bubble scales to relative abundances for each group (columns).
The most highly recovered diazotroph was UCYN-A1_otu0 (31.9% total rarefied dataset), which had 100% nucleotide identity with the haptophyte symbiont UCYN-A1 (Candidatus Atelocyanobacterium thalassa, CP001842.1) originally discovered at Stn. ALOHA (Zehr et al., 1998). This sequence type is targeted with the UCYN-A (grpA) qPCR assay described originally in Church et al. (2005a) and used to enumerate this group at Stn. ALOHA. UCYN-A1_otu0 had highest relative abundances in the upper euphotic zone, with a subsurface maximum (Figure 1B) that was most prevalent in the spring and summer (Mar-Jul; Figure S2). These depth and seasonal patterns are broadly consistent with those determined using qPCR (Figures S1, S3).
UCYN-A3 (UCYN-A3_otu5) and UCYN-A2 (UCYN-A2_otu11) sublineages were also observed, but at much lower relative abundances (2.9% and 1.0% total sequences, respectively; Table S3). These groups cannot currently be differentiated using qPCR that targets nifH gene fragments (Farnelid et al., 2016) thus no comparisons can be made. UCYN-A3_otu5 had peak relative abundances between 0-75 m (Figure 1B) but persisted in the lower euphotic zone during the fall and spring (Figure S2). Relative abundances of UCYN-A3_otu5 were generally low throughout the year (Figure 1C) but were slightly higher in the euphotic zone between Feb-May (Figure S2). Depth distribution patterns of UCYN-A2_otu11 were distinct from the other two UCYN-A sequence types, with generally low relative abundances through the upper euphotic zone and peak abundances in the lower euphotic zone (Figure 1B), although this is driven largely by high relative abundances at 150 m in April and October (Figure S2).
Trichodesmium sp. (Tricho_otu1) was the next most abundant of all sequence-types (13.0% total sequences) and identical to T. thiebautii H9-4 within Clade I of Trichodesmium. Tricho_otu1 is enumerated by the qPCR assay described in (Church et al., 2005a; Table S1). The seasonality of Tricho_otu1 was distinct from that of UCYN-A1_otu0, with peaks in relative abundances beginning in the late summer through early winter (Jul-Dec, Figure 1C), consistent with qPCR patterns (Figure S3). Relative abundances of Tricho_otu1 were highest in the upper 45 m of the water column and lowest at depths ≥ 125 m (Figure 1B).
Additional cyanobacterial sequence types that were among the most highly recovered included three unicellular cyanobacterial groups: Crocosphaera (Croco_otu9, 2.0% total sequences), a Crocosphaera-like group (Croco_otu3, 5.9% total sequences), and the newly described spheroid body of the marine diatom Epithemia pelagica strain UMH3203 (EpSB_otu8, 2.8% total sequences; Schvarcz et al., 2022). Croco_otu3 has >99% nucleotide identity to nifH sequences reported at Stn. ALOHA (DQ088688; J. Zehr, unpublished) as well as the South Pacific (HQ229012; Moisander et al., 2014) and the Arabian Sea (JX064489; Bird & Wyman, 2012). Both Croco_otu3 and EpSB_otu8, neither of which were targeted by qPCR assays routinely used at Stn. ALOHA previously, were distinct in having higher relative abundances in the lower euphotic zone (125-175 m; Figure 1B). This is consistent across all seasons at Stn. ALOHA; relative abundances for both groups were low in euphotic zone waters year-round, with maxima in the lower euphotic zone in the late winter, spring, and early summer (Feb-Jul; Figure S2). Croco_otu9, which clusters with Crocosphaera watsonii (Figure 1A) and is targeted by the UCYN-B qPCR assay (Church et al., 2005a), generally co-occurred with Tricho_otu1 (Figure 1C) consistent with qPCR patterns (Figures S1, S3). However, Croco_otu9 was underrepresented in the nifH HTS dataset when compared to the qPCR dataset, where UCYN-B (grpB) nifH-based abundances were of similar magnitude to UCYN-A (grpA) and Trichodesmium (Figure S1), suggesting the nifH gene HTS PCR primers used in this study do not amplify Crocosphaera nifH as well as other diazotroph groups.
All three major groups of DDAs (Het-1, Het-2, Het-3) were detected but had comparatively low relative abundances. Het-1 had the highest relative abundances of the DDAs (0.6% total sequences, Het-1_otu12), Het-3 were present at low relative abundances (0.1% total sequences, Het-3_otu27), and Het-2 sequences were recovered but not retained in the rarefied dataset (data not shown).
Although a majority of the sequences were from cyanobacterial diazotrophs, NCDs were also present. Four NCD groups affiliated with nifH cluster 1G (primarily γ-proteobacteria) had high relative abundances (Figure 1). The putative γ-proteobacterium “gamma A” was recovered from nearly every sample, and a single OTU accounted for 8.5% of the total sequences (gammaA_otu2; Table S3). GammaA_otu2 clusters within the Marine 1 group of γ-proteobacteria (defined in Langlois et al., 2015) that also contains AO15 (Zehr et al., 1998), proteo_1 (Fong et al., 2008), γ-proteo_1 (Church et al., 2008), γ-24774A11 (Moisander et al., 2008), as well as gamma B and gamma C sequence types (Langlois et al., 2015) (Figure 2A). This group is targeted by the γ-24774A11 qPCR assay (Moisander et al., 2008). GammaA_otu2 was a persistent member of the diazotroph community, with high relative abundances found throughout the year (Figure 1C) predominantly in the upper euphotic zone (Figure 1B). Gamma A depth and seasonal patterns from the nifH HTS are largely consistent with qPCR-based patterns, particularly in upper euphotic zone waters (Figures S1, S4).
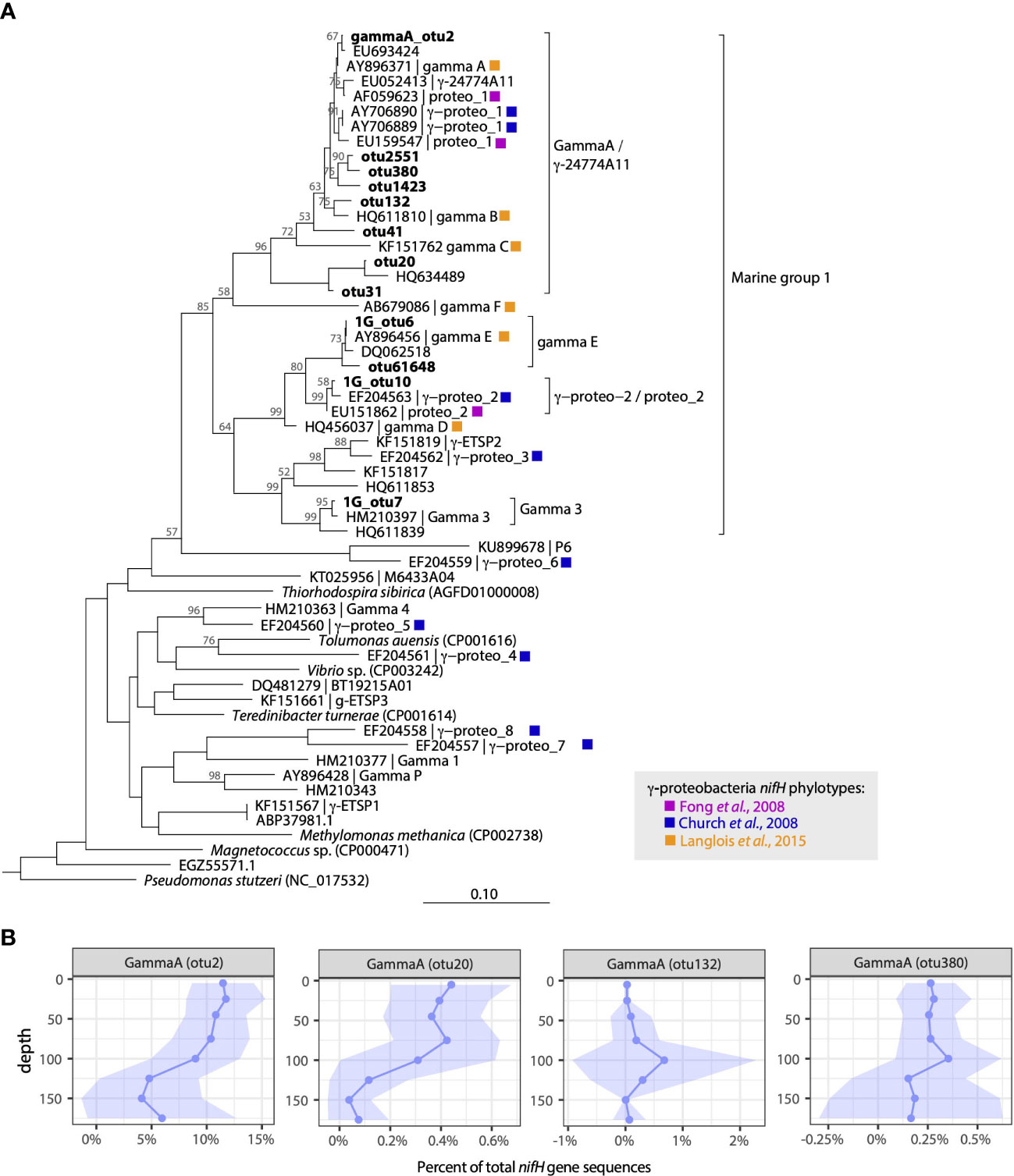
Figure 2 Diverse γ-proteobacterial diazotrophs inhabit waters at Stn. ALOHA. (A) Neighbor-joining nucleotide tree of γ-proteobacterial OTUs along with representative sequences from prior studies in the North Pacific. Cluster designations follow those defined in Langlois et al. (2015) or reflect naming conventions from Fong et al. (2008) and Church et al. (2008). (B) Depth profiles of selected gamma A OTUs plotted as a percent of total nifH gene sequences in the rarefied dataset. Solid lines represent mean values, shading represents ± 1 standard deviation.
Three additional γ-proteobacterial NCDs had high relative abundances across the dataset. 1G_otu7, which accounted for 1.8% of the total sequences, is identical to sequences recovered in the Eastern Tropical North Pacific (ETNP) Oxygen Minimum Zone (OMZ) actively transcribing nifH (e.g. KY968013.1; Jayakumar et al., 2017) and shares 99% nucleotide identity with a previously described γ-proteobacterial group Gamma 3 (Halm et al., 2012). 1G_otu6 accounted for 1.7% of the total sequences and is identical to sequences recovered from the NPSG (DQ062533.1; Zehr et al., 2007) and clusters with gamma E (Langlois et al., 2015). Finally, 1G_otu10 (0.9% total sequences) is identical to sequences reported from the South China Sea (HQ456044.1; Kong et al., 2011) and clusters with sequence types originally reported in the vicinity of Stn. ALOHA, proteo_2 (Fong et al., 2008) and γ-proteo-2 (Church et al., 2008) (Figure 2A). These 1G phylotypes were found throughout the water column and, except for 1G_otu7, did not have strong depth (Figure 1B) or seasonal patterns (Figure S2).
An additional NCD affiliated with cluster III (which contains mainly anaerobes including δ-proteobacteria), cIII_otu4, was also among the most highly abundant sequence types. Accounting for 6.5% of the total nifH sequences (Table S3), cIII_otu4 had 99.7% nucleotide identity to a cluster III sequence originally described and targeted with qPCR by Church et al. (2005a). In the nifH HTS dataset, this group had a distinct depth profile, with peak relative abundances in the lower euphotic zone (Figure 1B), but ddPCR-based abundances were generally low and did not exhibit distinct depth patterns (Figures S1, S3). However, both nifH HTS and ddPCR datasets captured a strong seasonal pattern, with highest abundances in the fall (Sept-Dec; Figures 1C, S4).
3.3 Microdiversity of major diazotroph groups at Station ALOHA
This dataset provided a unique opportunity to characterize the nifH-based microdiversity of Trichodesmium and UCYN-A in the NPSG (Figure 3). Tricho_otu1 (T. thiebautii) was the dominant Trichodesmium OTU (representing 13% of total sequences); however, >100 Trichodesmium OTUs were present in the dataset (Table S3). Among these, three additional OTUs represented >0.1% of the total nifH gene sequences: Tricho_otu46 (undefined species, 0.6% of total sequences), Tricho_otu42 (T. erythraeum, 0.5% of total sequences), and Tricho_otu16 (undefined species, 0.3% of total sequences) (Figure 3A). These four Trichodesmium OTUs had different seasonal and depth-distribution patterns (Figure 4). Tricho_otu16 was significantly, positively correlated to Tricho_otu1 (T. thiebautii) (Figure S5). Relative abundances of Tricho_otu16 were highest in the fall and lowest in the spring and decreased with depth throughout the year (Figure 4). In contrast, T. erythraeum (Tricho_otu42) was negatively correlated to T. thiebautii (Figure S5), with highest relative abundances in the spring and summer, and often a subsurface maximum in relative abundance (Figure 4). Tricho_otu1 and Tricho_otu16 sequences have no mismatches to the Trichodesmium qPCR primer/probe set routinely used at Stn. ALOHA (Church et al., 2005a). Tricho_otu46 and Tricho_otu42 have one and two mismatches to the forward primer, respectively, and both have one mismatch to the probe (Table S1).
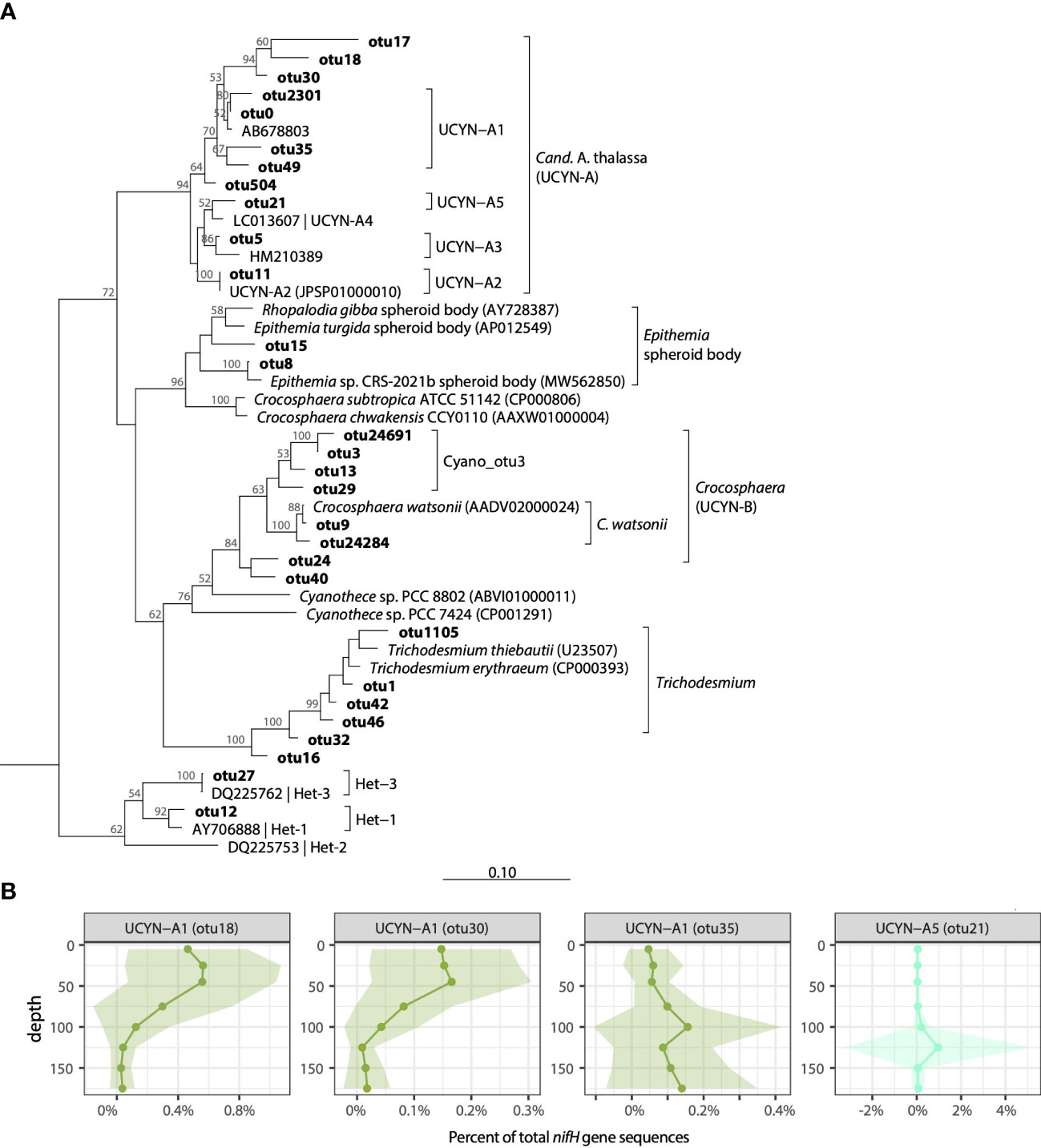
Figure 3 Cyanobacterial diazotroph diversity at Stn. ALOHA. (A) Neighbor-joining nucleotide tree of cyanobacterial OTUs along with cultivated representatives. (B) Depth profiles of selected UCYN-A OTUs plotted as a percent of total nifH gene sequences in the rarefied dataset. Solid lines represent mean values, shading represents ± 1 standard deviation.
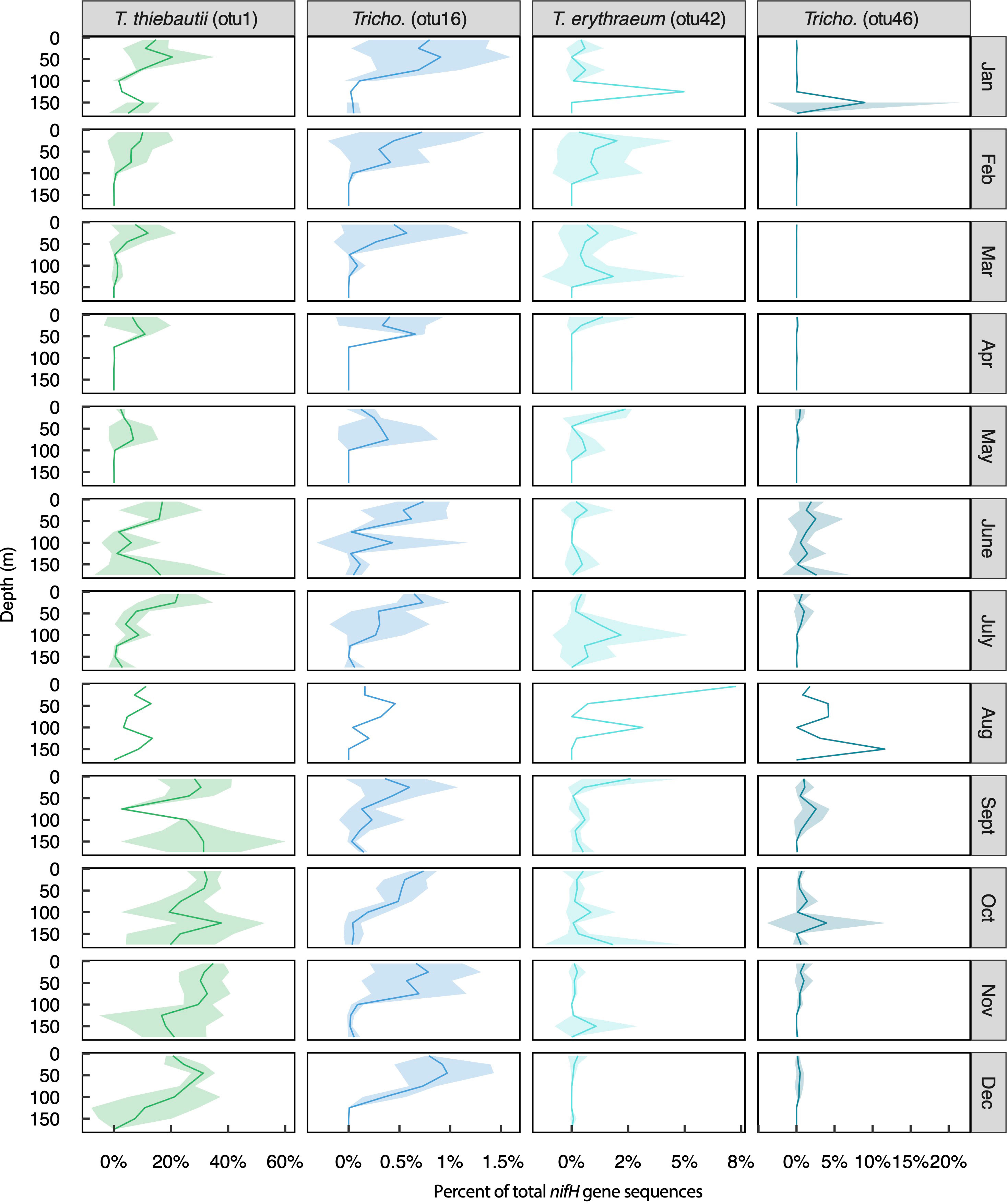
Figure 4 Relative abundances of the four most abundant Trichodesmium OTUs across months and depths. Solid lines represent mean values, shading represents +/- 1 standard deviation. Note that this timeseries includes only one observation during the month of August.
As with Trichodesmium, the dataset contained many UCYN-A OTUs (>100), however microdiversity analyses focused on the 11 UCYN-A OTUs each representing >0.1% of the total nifH gene sequences (Figure 3A). These included 5 OTUs with >96% nt similarity to UCYN-A1 (UCYN-A1_otu0, UCYN-A1_otu2301, UCYN-A1_otu35, UCYN-A1_otu49, and UCYN-A1_otu30) which together comprised 32.2% of total sequences, single OTUs that clustered with UCYN-A2 (UCYN-A2_otu11; 1.0% of total sequences), UCYN-A3 (UCYN-A3_otu5; 2.9% of total UCYN-A), and UCYN-A5 (UCYN-A5_otu21; 0.2% of total sequences), as well as 2 OTUs from unknown UCYN-A sublineages (UCYN-A_otu17, UCYN-A_otu18; both 0.3% of total sequences) (Figure 3A; Table S3).
Distinct seasonal and depth distribution patterns were seen within UCYN-A1 sublineages. In contrast to UCYN-A1_otu0, which had high relative abundances in euphotic zone waters in the late winter-early summer (Jan-Jul; Figure S2), UCYN-A1_otu30 was found to have highest relative abundances beginning in the summer and continuing through the fall (June-Dec), and a shallower maximum in the upper euphotic zone (≥50 m; Figure S6). These differences are reflected in correlations between relative abundances and temperature: UCYN-A1_otu30 had a positive correlation with temperature, whereas UCYN-A1_otu0 was negatively correlated (Figure S7). Additionally, several UCYN-A1 OTUs had highest relative abundances in the lower euphotic zone, including UCYN-A1_otu35 and UCYN-A1_otu49 (Figures 3B, S6). Notably, qPCR primers targeting UCYN-A1 enumerate all the UCYN-A1 OTUs (as well as UCYN-A_otu17 and UCYN-A_otu18; Table S1).
3.4 Environmental determinants for diazotroph community composition
WGCNA and correlation analyses were conducted for the upper euphotic zone samples (≤75 m) to gain insight into shifts in diazotroph community composition in response to changing environmental conditions. Network analysis produced 2 modules of nifH gene OTUs based on highly correlated abundances over the time series (Table S4; samples 5-75 m; 128 non-rare OTUs comprising 88% of all reads). Module 1 (M1; 43 OTUs) and was comprised of Trichodesmium (Tricho_otu1, Tricho_otu16, Tricho_otu46), Crocosphaera (Croco_otu9, Croco_otu40), in addition to several UCYN-A OTUs (UCYN-A1_otu18, UCYN-A1_otu30) and NCDs (cIII_otu4, 1G_otu7). Module 2 (M2; 37 OTUs) contained mostly UCYN-A (UCYN-A1_otu0, UCYN-A3_otu5, UCYN-A2_otu11, UCYN-A_otu17, UCYN-A1_otu21, UCYN-A1_otu49, UCYN-A1_otu504, UCYN-A1_otu2301) and γ-proteobacteria affiliated with gamma A (gammaA_otu2, gammaA_otu20, gammaA_otu31) OTUs, but also notably EpSB (EpSB_otu8). The remaining 48 OTUs had abundance patterns that correlated poorly to OTUs in M1 or M2 and were assigned by WGCNA to the catch-all module M0 (Table S4). Some OTUs in M0 were rare in the upper euphotic zone but had high relative abundances in the lower euphotic zone (depths which were not included in these analyses), including Croco_otu3 and 1G_otu6.
In general, OTUs in M1 and M2 modules had opposite correlations to environmental variables (Figure 5), reflecting different seasonal patterns which appear to be driven by the cyanobacterial OTUs (Figure S8). Many of the OTUs in M1 had significant positive correlations to temperature, particulate matter concentrations (i.e., PC, PN, PP), and heterotrophic bacteria abundances. M1 OTUs also had significant negative correlations to SLAcorr, as well as to photosynthetic picoeukaryote (peuk) and Synechococcus abundances. M2 OTUs had largely opposite correlation patterns with significant positive correlations to SLAcorr, photosynthetic picoeukaryote and Synechococcus abundances, along with significant negative correlations to temperature and concentrations of chl a, PN, PP, and PC, as well as Prochlorococcus and heterotrophic bacteria abundances. Beyond these general trends, this analysis revealed other correlations between specific OTUs and environmental conditions. For example, in M1, both cIII_otu4 and Het-1_otu12 had significant positive correlations to chl a, reflecting the high relative abundances of these organisms in the lower euphotic zone. Likewise, in M2, UCYN-A2_otu11 correlation patterns were quite different than other M2 OTUs, with significant positive correlations to PARdepth, KPAR, and SRP, and inverse correlations with depth suggesting that UCYN-A2 lives in the upper sunlit layers.
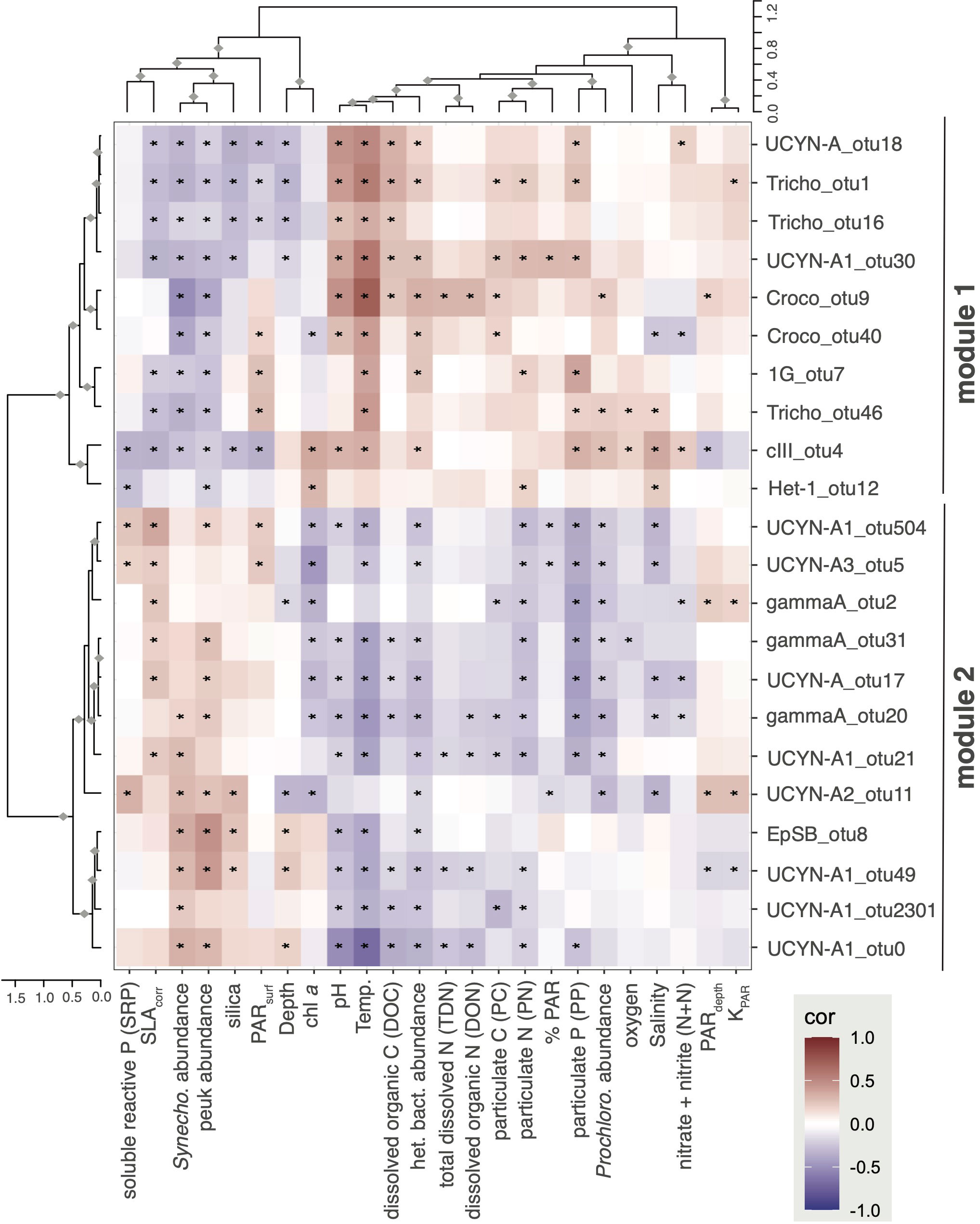
Figure 5 Weighted gene co-expression network analysis identify co-occurring diazotroph taxa and OTU-level correlations to environmental parameters. The network analysis partitioned diazotrophs into two main modules based on abundance pattern similarity over the time series (with abundances summed from 5-75 m on each date). Dominant Trichodesimum and Crocosphaera OTUs were in module 1 (M1) while most UCYN-A OTUs were in module 2 (M2). Generally, diazotrophs in M1 and M2 had opposite correlations to environmental variables. ♦Row or column cluster is significant (bootstrapping, alpha ≥ 0.95); *Pearson correlation is significant (p < 0.05).
4 Discussion
4.1 Characterization of diazotroph community composition benefits from complementary approaches
Diazotroph community composition at Stn. ALOHA has been previously explored using two main approaches that target known organisms. Large, conspicuous diazotrophs, such as Trichodesmium and DDAs, have been enumerated using microscopy (Letelier and Karl, 1996; White et al., 2018). Additionally, diazotroph community composition has commonly been assessed using qPCR targeting the nifH gene from six cyanobacterial diazotrophs (Church et al., 2009; dataset doi: 10.5281/zenodo.4477269) based on their presence in several early clone libraries constructed from nifH gene transcripts (suggesting active synthesis of nitrogenase) from a small number of samples (Church et al., 2005a; Church et al., 2005b). The present study shows that using a newer approach, nifH gene HTS, enables analysis of the seasonality, depth distributions, and environmental drivers of potentially important diazotrophs that are not (or cannot be) targeted by microscopy or qPCR assays.
Abundance patterns derived from nifH gene HTS agree with those from qPCR/ddPCR-based abundance trends for UCYN-A1, Trichodesmium, gamma A, and cIII (Figures S1, S3, S4), but not for other important diazotroph groups. DDAs (Het-1, Het-2, Het-3), which have long been established as quantitatively significant N2-fixers at Stn. ALOHA (White et al., 2007; Karl et al., 2012) have low amplification efficiency using the universal nifH primers used in this study (discussed in Caputo et al., 2018). As such, seasonal and depth patterns for Het-1 and Het-3 are not consistent with qPCR-based patterns (Figure S1), indicating this nifH HTS approach is not suitable for DDAs-specific inquiries. Furthermore, Crocosphaera is unexpectedly underrepresented in the nifH gene HTS, despite being detectable in the same DNA extracts using qPCR, which suggests a methodological artifact such as low PCR amplification efficiency.
The nifH gene HTS dataset also has a skew in depth distributions, that is most evident for some taxa, e.g. cIII_otu4 in samples from >100 m depths (Figure S1). This likely arises from applying end-point PCR to samples having low starting nifH gene copy numbers, as might be expected in the lower euphotic zone. Although relative abundance data need to be interpreted with caution, this approach does provide qualitative information on how the overall community changes with depth and which organisms persist into the lower euphotic zone or have a preferred niche at these depths.
Furthermore, the nifH gene HTS dataset also highlights the extensive nifH diversity in this region, revealing several potentially important diazotroph taxa and potential environmental controls (discussed below). Of particular interest is the new Crocosphaera-like group (Croco_otu3) that were highly abundant in this dataset yet are not captured by current UCYN-B qPCR assays. Furthermore, this amplicon-based study allows characterization of diazotroph microdiversity of major taxa at Stn. ALOHA, including the UCYN-A/haptophyte symbiosis sublineages and Trichodesmium (discussed below). Collectively, this work illustrates the complementary role that HTS analyses can and should play in studying microbial diversity. Furthermore, coupling qPCR/ddPCR with nifH gene HTS offers insights into absolute gene abundances of specific, often relatively abundant, diazotrophs, while also providing a detailed look at diversity (including microdiversity) and relative abundances of the “entire” diazotroph community.
4.2 Stn. ALOHA diazotroph community includes poorly characterized cyanobacteria and non-cyanobacteria
It is well-established that cyanobacterial diazotrophs are important in the NPSG, and Stn. ALOHA is one of the best characterized N2-fixing ecosystems. While the nifH gene HTS dataset supports the prevalence of previously identified cyanobacterial diazotrophs (discussed above), this study provides insight on the seasonality, depth distributions, and environmental factors influencing the distribution of several diazotroph taxa that are poorly-characterized in the North Pacific.
4.2.1 UCYN-A/haptophyte symbioses include coastal and oligotrophic sublineages
The UCYN-A1 haptophyte symbiosis is well established as an important N2-fixing organism in the NPSG (Church et al., 2009; Shiozaki et al., 2017; Gradoville et al., 2020), and is routinely monitored at Stn. ALOHA using qPCR. Seasonal and depth-distributions patterns for UCYN-A1_otu0 in this time series are consistent with previously reported observations (Church et al., 2005a; Church et al., 2009). However, little is known about other UCYN-A sublineages because UCYN-A2 and UCYN-A3 are not differentiated by existing nifH gene qPCR assays, thus nifH gene HTS is needed to gain insights into environmental conditions favorable to the UCYN-A sublineages at Stn. ALOHA. UCYN-A1_otu0 and UCYN-A3_otu5 have similar temporal patterns (M2; Figure 5) consistent with prior observations of co-occurrence in open ocean ecosystems (Turk-Kubo et al., 2017). However, UCYN-A1_otu0 and UCYN-A3_otu5 fall into different subclusters within M2, distinguished by UCYN-A3_otu5 correlations with SLAcorr (positive) and chl a (negative). This reflects broad seasonal patterns that are consistent between the two sublineages, but suggests UCYN-A3_otu5 may be favored in anticyclonic eddies.
Previous work suggested that the UCYN-A2 symbiosis was rare at Stn. ALOHA (Turk-Kubo et al., 2017; Cornejo-Castillo et al., 2019). Although UCYN-A2 was not among the most highly abundant sequence types, it was detected in more than half of the samples and occasionally reached high relative abundances in the lower euphotic zone (e.g. HOT255; up to 34% at 150 m, Table S3). Thus, it is a regular member of the diazotroph community, present at low abundances in the upper euphotic zone and persisting into the lower euphotic zone. Given that the UCYN-A2 sublineage is commonly found in temperate, coastal, and even neritic environments (e.g. Hagino et al., 2013; Shiozaki et al., 2018; Cabello et al., 2020; Selden et al., 2021; Turk-Kubo et al., 2021), its year-round presence at Stn. ALOHA is somewhat surprising. Network analyses indicate it co-occurs with the dominant UCYN-A sublineages (M2); however, distinct correlation patterns are observed for UCYN-A2_otu11, and it differs from most other M2 OTUs with correlations to depth (negative), SRP (positive), PARdepth (positive), and KPAR (positive) and no strong correlations with either temperature or particulate nutrients. Collectively, these patterns suggest that UCYN-A2 inhabits shallow sunlit waters during the winter-spring season, and its relative abundances increase with the concentration of SRP. It is important to note that significant gaps remain in our understanding of the diversity of UCYN-A hosts, so we cannot rule out the possibility that there are host lineages with distinct symbionts that simply cannot be differentiated using partial nifH gene fragments, i.e. the UCYN-A2_otu11 has a genetically distinct host from those found in coastal ecosystems. More work is needed to validate whether the haptophyte hosting UCYN-A2_otu11 is identical to coastal lineages.
4.2.2 Additional unicellular cyanobacterial taxa may be important in oligotrophic ecosystems
This study is the first to report the prevalence of a Crocosphaera-like organism, Croco_otu3, at Stn. ALOHA. Although sequences closely related to this organism have been reported in the North and South Pacific and Mediterranean Sea, this organism is not targeted by existing UCYN-B or UCYN-C qPCR assays nor have they previously been emphasized as important marine cyanobacterial diazotrophs. Peak relative abundances of Croco_otu3 are at depths > 100 m, suggesting that it occupies a different niche than Croco_otu9 (C. watsonii), which is most abundant in the upper euphotic zone based on both nifH NGS and UCYN-B qPCR data (Figures 1B, S1). Targeted approaches are needed to confirm the seasonality of Croco_otu9 but relative abundances in the lower euphotic zone (Figures 1B, S2) and summed across depths for each cruise (Figure 1C) suggest it may prefer warmer summer-fall conditions at Stn. ALOHA. Given the light-dependency of cyanobacterial diazotrophs, the presence of Croco_otu3 deeper in the water column may indicate adaptation to low light, which would confer an ecological advantage in the lower euphotic zone, as has been hypothesized for UCYN-A1 (Gradoville et al., 2021). Alternatively, it may simply persist longer than other cyanobacteria taxa on particles sinking out of the euphotic zone, a process demonstrated to transport diazotrophs into the mesopelagic (Farnelid et al., 2019; Poff et al., 2021), or as self-aggregates, which are formed by some Crocosphaera strains (Bench et al., 2016). It is also possible that absolute abundances have a different pattern, but relative abundances appear to peak in the lower euphotic zone as abundances of other groups decrease. Quantitative methods (e.g., qPCR/ddPCR) are needed for estimating abundances and clarifying these patterns.
The dataset also expands our knowledge of the seasonal and depth distributions of EpSB_otu8, a symbiont (or “spheroid body”) of a recently isolated marine lineage of Epithemia pelagica (strain UMH3203) from Station ALOHA (Schvarcz et al., 2022). Like Croco_otu3, EpSB_otu8 had markedly different depth and temporal patterns compared to other cyanobacterial taxa, likely reflecting the preferred ecological niche of the host diatom. Although generally present at low relative abundances in the upper euphotic zone, network analysis indicated it co-occurs with the UCYN-A symbiosis and gamma A, reflecting presence in the spring (Mar–Jun), and preference for cooler waters (significant negative correlation with temperature). E. pelagica has been demonstrated to be globally distributed and the symbiont, EpSB_otu8, has been shown to actively fix N2 in culture (Schvarcz et al., 2022) thus is likely to be an important contributor to marine N2 fixation. However, further work is needed to determine under what environmental conditions this Epithemia-spheroid body symbiosis actively fixes N2, and to characterize its relative contribution to bulk N2 fixation and the export of PN in subtropical oceans.
It appears that several cyanobacterial diazotophs have preference for or persist in lower euphotic zone waters. Although the vast majority of N2 fixation occurs in the upper 75 m at Stn. ALOHA, low rates of N2 fixation are routinely measured deeper in the water column (Böttjer et al., 2017), where the UCYN-A symbiosis has been recently demonstrated to fix N2 (Gradoville et al., 2021). However, the extent and importance of contributions from other diazotrophs like UCYN-A2, Croco_otu3 and EpSB_otu8 in the lower euphotic zone will need to be determined in future studies.
4.2.3 NCDs are prevalent and stable parts of the diazotroph community at Stn. ALOHA
NCDs are known to inhabit well-oxygenated photic waters in all major oceanic regions (e.g. Bird et al., 2005; Man-Aharonovich et al., 2007; Church et al., 2008; Fong et al., 2008; Farnelid et al., 2011; Blais et al., 2012; Halm et al., 2012; Delmont et al., 2018; Chen et al., 2019; Delmont et al., 2022) yet very little is known about the abundances, activities and metabolic diversity of this group. Although commonly assumed to be chemoheterotrophs, thus requiring exogenous organic C to fuel their metabolism and N2 fixation activity, there are also cultivated NCDs that are capable of utilizing alternative energy sources (e.g. such as photoheterotrophs that use light energy, or chemolithotrophs that gain energy through the oxidation of inorganic molecules). The enzymatic activity of nitrogenase is irreversibly inactivated by O2, thus many NCDs are thought to be limited by high O2 concentrations in addition to the requirement for exogenous organic C or other energy sources needed to meet the energetic demands of N2 fixation. NCDs are hypothesized to meet these challenges in surface waters by finding low O2, high organic matter niches, such as suspended particles (Riemann et al., 2010; Riemann et al., 2022), or by living in symbiosis with another organism (e.g. Farnelid et al., 2010).
The NCD with highest relative abundances at Stn. ALOHA, gammaA_otu2, is widely distributed and actively transcribing nifH in warm subtropical surface waters (Moisander et al., 2014; Langlois et al., 2015; Cornejo-Castillo and Zehr, 2021). Gamma A is speculated to associate with small suspended particles (Cornejo-Castillo and Zehr, 2021) or photosynthetic eukaryotes (Benavides et al., 2016). The predominant gamma A OTU at Stn. ALOHA, gammaA_otu2, shows strong seasonal and depth patterns in the nifH HTS dataset. It co-occurs with UCYN-A1 symbioses with highest relative abundances in the winter-spring, but is one of only two M2 members with a significant negative correlation with depth, reflecting high relative abundances in well-lit surface waters (0-75 m; Figure 1B). These depth patterns are confirmed by nifH-based abundance of gamma A (via ddPCR; Figure S1), however ddPCR analyses also indicate relatively consistent abundances in the upper euphotic zone throughout the year (ca. 104 nifH copies L-1; Figure S4). Thus, seasonal relative abundance patterns reflect a greater contribution to the diazotroph community in the winter-spring, not peak abundances.
Additional gamma proteobacteria from Marine group 1 (Langlois et al., 2015) are potentially important in the North Pacific, including 1G_otu6 (gamma E), 1G_otu7 (gamma 3), and 1G_otu10 (γ-proteo_2/proteo_2). Of these, only gamma 3 (1G_otu7) showed strong seasonality in the upper euphotic zone, clustering in M1 along with Trichodesmium and Crocosphaera OTUs. Most members of this cluster have strong positive correlations to temperature, but 1G_otu7 is among only a few OTUs that have a significant and positive correlation to PARsurf, suggesting that 1G_otu7 is adapted for higher light and temperature conditions than other N2-fixing γ-proteobacteria. In contrast, 1G_otu6 and 1G_otu10 lacked the distinct seasonal trends present for other diazotroph groups, at least in the upper euphotic zone, and thus did not cluster with either M1 or M2. They both had peak relative abundances in lower euphotic zone waters (Figures 1B, S2), which suggests that like Croco_otu3 and EpSB_otu8, these γ-proteobacteria either are adapted to low-light niches or persist into the deep euphotic zone attached to sinking particles.
Very little is known about diazotrophs from cluster III in the surface ocean, but the ecology of cIII_otu4 is arguably the best characterized. It was originally described at Stn. ALOHA by Church et al. (2005a) and was detected in both the 0.2-10 µm and >10 µm size classes, at moderate abundances (ca. 104 nifH gene L-1) that peaked in the mixed layer but persisted in deeper waters (125-200 m). Both nifH HTS and ddPCR datasets indicate that cIII_otu4 exhibits strong seasonality, with peak abundances in the fall (ca. 103 nifH copies L-1; Figure S4), and co-occurs with Trichodesmium and Crocosphaera OTUs in M1 (Figure 5). In the North Atlantic, this phylotype has been associated with the spring bloom at 40°N, in the presence of nitrate (Langlois et al., 2008). Thus, increasing relative abundances observed with depth (Figure 1B) and a positive correlation with concentrations of chl a (Figure 5) are consistent with prior observations. Furthermore, nifH genes of this phylotype have been detected in Trichodesmium colonies from Stn. ALOHA (OTU 1527; Gradoville et al., 2017a), thus co-occurrence of cIII_otu4 with Trichodesmium in this study provides further support for potential metabolic interdependencies. Whether cIII_otu4 is actively fixing N2 is yet to be determined, but active nifH transcription has been measured in the South Pacific Gyre (Halm et al., 2012).
The detection of several NCD taxa present at high relative abundances at Stn. ALOHA underscores their potential ecological relevance in well-oxygenated photic waters. Moreover, the recovery of closely related or identical sequences reported as far back as 2005 in the North Pacific, e.g. gamma_otu2, 1G_otu10, and cIII_otu4 indicates they are a stable component of the microbial community. Importantly, we do not yet know whether these organisms actively fix N2. NCD nifH gene sequences have been reported in oceanic regions where no N2 fixation was detected, including the California Current System and high latitude seas (Gradoville et al., 2017b; Shiozaki et al., 2017), suggesting some NCDs may rely on other less-energetically expensive N-sources (Inomura et al., 2018). Indeed, a recent analysis of N-acquisition strategies among NCD metagenome assembled genomes constructed from Tara Oceans metagenomes, suggests that many of these taxa may be facultative N2 fixers (Turk-Kubo et al., 2022). Direct demonstrations of NCD N2 fixation activity, including temporal and spatial variability, are needed to inform interpretations about the importance of NCDs in the marine N-cycle.
4.3 Trichodesmium and UCYN-A microdiversity at Station ALOHA
There were diverging abundance patterns of diazotrophs at the species- or clade-level. For instance, Trichodesmium are typically enumerated at the genus level (Letelier and Karl, 1996; Church et al., 2005a; White et al., 2018), but this group includes species from multiple clades that differ in their physiology and ecology (Hynes et al., 2012; Hutchins et al., 2013) and cannot be reliably classified morphologically (Hynes et al., 2012). In the present time-series, the dominant Trichodesmium OTU (Tricho_otu1) was closely related to T. thiebautii (Clade I), agreeing with previous nifH sequencing-based studies near Stn. ALOHA (Gradoville et al., 2014; Gradoville et al., 2017b). The especially high relative abundance of Tricho_otu1 sequences from Sept-Dec 2015 corresponds to a period of high Trichodesmium cell abundances and an abnormally large number of colonies (as opposed to free filaments) reported in a microscopy-based study that used samples from the same HOT cruises (White et al., 2018), suggesting that this OTU may be linked to colony formation. Many other Trichodesmium OTUs were present, and the most abundant representatives appear to differ in their seasonality and depth distributions (Figure 4). Relative abundances of Tricho_otu1 were negatively correlated with relative abundances of Tricho_otu42 (T. erythraeum) (Figure S5), agreeing with previous suggestions of niche partitioning of these two species (Rouco et al., 2014). The mechanisms driving the different abundance patterns of these two species remain unknown. The vast majority of culture studies have used laboratory strains of Clade III (e.g. T. erythraeum IMS101); however, a study by Hutchins et al. (2013) showed that T. thiebautii and T. erythraeum isolates respond differently to pCO2, suggesting that Trichodesmium clades/species respond differently to variations in the environment. More studies on the environmental drivers of Trichodesmium Clade I are needed, since T. thiebautii appears to be the dominant species in open-ocean environments (Chappell et al., 2012; Rouco et al., 2014) and perhaps globally (Marumo and Nagasawa, 1976; Carpenter and Price, 1977).
Amplicon- or metagenomic/metatranscriptomic- based studies are currently required to characterize abundance patterns that incorporate the known microdiversity of the UCYN-A/haptophyte symbiosis sublineages, which are composed of numerous sequence types that display niche and biogeographical partitioning (Turk-Kubo et al., 2017; Turk-Kubo et al., 2021). Similar to other open ocean regions, the UCYN-A symbiosis assemblage at Stn. ALOHA is primarily composed of major (globally abundant) and minor (rare) OTUs that cluster with UCYN-A1 sublineages (Figure 3). The UCYN-A1 symbiosis is characteristically found in cooler waters than other diazotrophs (Moisander et al., 2010), with temperature optima <25°C. However, nifH gene HTS revealed two OTUs, UCYN-A_otu18 and UCYN-A1_otu30, that have opposite patterns, clustering with M1 (Figure 5) and with highest relative abundances in waters >25°C (Figure S7). The contrasting seasonal and depth distribution patterns for these minor UCYN-A1 OTUs suggest they occupy different niches and have different temperature optima. Given that the UCYN-A cyanobacterium is hypothesized to be an obligate symbiont, these patterns are likely to be driven by the ecophysiology of the prymnesiophyte hosts, which are not well understood. A recently obtained isolate of the B. bigelowii genotype III, which hosts the UCYN-A2 sublineage, is leading to important insights into B. bigelowii biology including validating its insensitivity to the presence of nitrate described in Mills et al. (2020) (Suzuki et al., 2021). However there are multiple B. bigelowii strains that have been morphologically and genetically characterized by Hagino et al. (2009) that have not yet been linked to nifH-based sublineages, limiting interpretations based solely on nifH or microscopic observations of B. bigelowii.
5 Conclusions
Leveraging nifH gene HTS and qPCR provides an unprecedented view of the temporal and spatial (depth) dynamics associated with the diazotroph community at Stn. ALOHA and provides important insights into distribution patterns among poorly characterized diazotrophs. These include the co-occurrence of most UCYN-A and gamma A OTUs in the winter-early summer, and the identification of several taxa, including a Crocosphera-like (Croco_otu3) taxa and the Epithemia pelagica spheroid body (EtSB_otu8), that may preferentially inhabit the lower euphotic zone or associate with particles sinking out of the euphotic zone. Furthermore, analyzing this time series using nifH gene HTS reveals that within Trichodesmium and UCYN-A, changes in populations (at the OTU level) can be observed that appear consistent with adaption to time- and depth-dependent environmental variation.
Furthermore, analysis of nifH-based diazotroph community composition at Stn. ALOHA reveals that diazotroph diversity extends beyond commonly studied groups (e.g., Trichodesmium, Crocosphaera, UCYN-A, etc.), and identifies several potentially important diazotroph groups for future monitoring. Importantly, this nifH gene HTS dataset validates that early efforts to characterize diazotroph diversity in the North Pacific successfully identified many of the dominant phylotypes. The suite of qPCR assays commonly used to characterize this region target the top two most abundant OTUs in our time series (UCYN-A1, Trichodesmium) and important groups not well-captured by nifH gene HTS (Crocosphaera, Het-1, Het-2, Het-3). However, several other highly abundant OTUs would be targeted by existing qPCR assays and should be considered for inclusion for future studies in the North Pacific. Specifically, we recommend incorporating the following diazotroph qPCR/ddPCR assays in future studies: gamma A and cIII (used in this study); UCYN-A2 (Thompson et al., 2014) which targets UCYN-A2/A3/A4 (Farnelid et al., 2016); UCYN-C (Foster et al., 2007) to target EpSB_otu8; and Gamma 3 to target 1G_otu7. Further work is needed to determine whether the newly identified cyanobacterial groups and NCDs actively fix N2 at Stn. ALOHA. Findings from this study underscore that we currently lack a complete understanding of diazotrophy in this ecosystem.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
KT-K, MC and JZ conceptualized the study. KT-K and BH processed the samples and KT-K and MG analyzed the amplicon data. JM and BH performed statistical analysis. KT-K, BH, and MG wrote the first draft of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Science Foundation (OCE-2023498 to KT-K) and the Simons Foundation (#72440 and #824082 to JZ and #721221 to MC).
Acknowledgments
We gratefully acknowledge all the HOT program participants, including the many captains, ship’s crew, and chief scientists that have enabled this long-term monitoring program. We also thank Benedetto Barone (University of Hawai’i at Mānoa) for consultations about SLA data. Environmental data was obtained via the Hawaii Ocean Time-series HOT-DOGS portal hosted at the University of Hawai’i at Mānoa (National Science Foundation Award # 1756517).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1130158/full#supplementary-material
References
Barone B., Coenen A. R., Beckett S. J., McGillicuddy D. J., Weitz J. S., Karl D. M. (2019). The ecological and biogeochemical state of the North Pacific Subtropical Gyre is linked to sea surface height. J. Mar. Res. 77, 215–245. doi: 10.1357/002224019828474241
Benavides M., Moisander P. H., Daley M. C., Bode A., Arístegui J. (2016). Longitudinal variability of diazotroph abundances in the subtropical North Atlantic Ocean. J. Plankton Res. 38, 662–672. doi: 10.1093/plankt/fbv121
Bench S. R., Frank I., Robidart J., Zehr J. P.. (2016). Two subpopulations of Crocosphaera watsonii have distinct distributions in the North and South Pacific. Environ. Microbiol. 18, 514–524. doi: 10.1111/1462-2920.13180
Bird C., Martinez J., O'Donnell A. G., Wyman M.. (2005). Spatial distribution and transcriptional activity of an uncultured clade of planktonic diazotroph γ-proteobacteria in the Arabian Sea. Appl. Environ. Microbiol. 71, 2079–2085. doi: 10.1128/AEM.71.4.2079-2085.2005
Bird C., Wyman M. (2012). Transcriptionally active heterotrophic diazotrophs are widespread in the upper water column of the Arabian Sea. FEMS Microbiol. Ecol. 84, 189–200. doi: 10.1111/1574-6941.12049
Blais M., Tremblay JÉ, Jungblut A. D., Gagnon J., Martin J., Thaler M., et al. (2012). Nitrogen fixation and identification of potential diazotrophs in the Canadian Arctic. Global Biogeochem. Cy 26, GB3022. doi: 10.1029/2011GB004096
Böttjer D., Dore J. E., Karl D. M., Letelier R. M., Mahaffey C., Wilson S. T., et al. (2017). Temporal variability of nitrogen fixation and particulate nitrogen export at station ALOHA. Limnol. Oceanogr. 62, 200–216. doi: 10.1002/lno.10386
Böttjer D., Karl D. M., Letelier R. M., Viviani D. A., Church M. J. (2014). Experimental assessment of diazotroph responses to elevated seawater pCO2 in the North Pacific Subtropical Gyre. Global Biogeochem. Cy 28, 601–616. doi: 10.1002/2013GB004690
Cabello A. M., Turk-Kubo K. A., Hayashi K., Jacobs L., Kudela R. M., Zehr J. P. (2020). Unexpected presence of the nitrogen-fixing symbiotic cyanobacterium UCYN-A in Monterey bay, California. J. Phycol. 56, 1521–1533. doi: 10.1111/jpy.13045
Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 1548–7091. doi: 10.1038/nmeth.f.303
Caputo A., Stenegren M., Pernice M. C., Foster R. A. (2018). A short comparison of two marine planktonic diazotroph symbioses highlights an un-quantified disparity. Front. Mar. Sci. 5, 1–8. doi: 10.3389/fmars.2018.00002
Carpenter E. J., Price C. C. IV (1977). Nitrogen fixation, distribution, and production of Oscillatoria (Trichodesmium) spp. in the western Sargasso and Caribbean seas. Limnol. Oceanogr. 22, 60–72. doi: 10.4319/lo.1977.22.1.0060
Chappell P. D., Moffett J. W., Hynes A. M., Webb E. A. (2012). Molecular evidence of iron limitation and availability in the global diazotroph Trichodesmium. ISME J. 6, 1728–1739. doi: 10.1038/ismej.2012.13
Chen T.-Y., Y-lL C., Sheu D.-S., Chen H.-Y., Lin Y.-H., Shiozaki T. (2019). Community and abundance of heterotrophic diazotrophs in the Northern South China Sea: revealing the potential importance of a new alphaproteobacterium in N2 fixation. Deep Sea Res. Part I.: Oceanographic Res. Papers 143, 104–114. doi: 10.1016/j.dsr.2018.11.006
Church M. J., Bjorkman K. M., Karl D. M., Saito M. A., Zehr J. P. (2008). Regional distributions of nitrogen-fixing bacteria in the Pacific Ocean. Limnol. Oceanogr. 53, 63–77. doi: 10.4319/lo.2008.53.1.0063
Church M. J., Jenkins B. D., Karl D. M., Zehr J. P. (2005a). Vertical distributions of nitrogen-fixing phylotypes at Stn ALOHA in the oligotrophic North Pacific Ocean. Aquat. Microb. Ecol. 38, 3–14. doi: 10.3354/ame038003
Church M. J., Mahaffey C., Letelier R. M., Lukas R., Zehr J. P., Karl D. M. (2009). Physical forcing of nitrogen fixation and diazotroph community structure in the North Pacific Subtropical Gyre. Global Biogeochem. Cycles 23, GB2020. doi: 10.1029/2008GB003418
Church M. J., Short C. M., Jenkins B. D., Karl D. M., Zehr J. P. (2005b). Temporal patterns of nitrogenase gene (nifH) expression in the oligotrophic North Pacific Ocean. Appl. Environ. Microbiol. 71, 5362–5370. doi: 10.1128/AEM.71.9.5362-5370.2005
Cornejo-Castillo F. M., Munoz-Marin M. D. C., Turk-Kubo K. A., Royo-Llonch M., Farnelid H., Acinas S. G., et al. (2019). UCYN-A3, a newly characterized open Ocean sublineage of the symbiotic N2-fixing cyanobacterium Candidatus Atelocyanobacterium thalassa. Environ. Microbiol. 21, 111–124. doi: 10.1111/1462-2920.14429
Cornejo-Castillo F. M., Zehr J. P. (2021). Intriguing size distribution of the uncultured and globally widespread marine non-cyanobacterial diazotroph gamma-a. ISME J. 15, 124–128. doi: 10.1038/s41396-020-00765-1
de Boyer Montégut C., Madec G., Fischer A. S., Lazar A., Ludicone D.. (2004). Mixed layer depth over the global Ocean: an examination of profile data and a profile-based climatology. J. Geophysical Res.: Oceans 109, C12003. doi: 10.1029/2004JC002378
Delmont T. O., Karlusich J. J. P., Veseli I., Fuessel J., Eren A. M., Foster R. A., et al. (2022). Heterotrophic bacterial diazotrophs are more abundant than their cyanobacterial counterparts in metagenomes covering most of the sunlit Ocean. ISME J. 16, 927–936. doi: 10.1038/s41396-021-01135-1
Delmont T. O., Quince C., Shaiber A., Esen Ö. C., Lee S. T. M., Rappé M. S., et al. (2018). Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface Ocean metagenomes. Nat. Microbiol. 3, 804–813. doi: 10.1038/s41564-018-0176-9
Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 1367–4803. doi: 10.1093/bioinformatics/btq461
Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Falkowski P. (1997). Evolution of the nitrogen cycle and its influence on the biological sequestration of CO2 in the Ocean. Nature 387, 272–275. doi: 10.1038/387272a0
Farnelid H., Andersson A. F., Bertilsson S., Al-Soud W. A., Hansen L. H., Sorensen S., et al. (2011). Nitrogenase gene amplicons from global marine surface waters are dominated by genes of non-cyanobacteria. PloS One 6, e19223. doi: 10.1371/journal.pone.0019223
Farnelid H., Tarangkoon W., Hansen G., Hansen P. J., Riemann L. (2010). Putative N2-fixing heterotrophic bacteria associated with dinoflagellate–cyanobacteria consortia in the low-nitrogen Indian Ocean. Aquat. Microb. Ecol. 61, 105–117. doi: 10.3354/ame01440
Farnelid H., Turk-Kubo K., Munoz-Marin M. D., Zehr J. P. (2016). New insights into the ecology of the globally significant uncultured nitrogen-fixing symbiont UCYN-A. Aquat. Microb. Ecol. 77, 125–138. doi: 10.3354/ame01794
Farnelid H., Turk-Kubo K., Ploug H., Ossolinski J. E., Collins J. R., Van Mooy B. A. S., et al. (2019). Diverse diazotrophs are present on sinking particles in the North Pacific Subtropical Gyre. ISME J. 13, 170–182. doi: 10.1038/s41396-018-0259-x
Fong A. A., Karl D., Lukas R., Letelier R. M., Zehr J. P., Church M. J. (2008). Nitrogen fixation in an anticyclonic eddy in the oligotrophic North Pacific Ocean. ISME J. 2, 663–676. doi: 10.1038/ismej.2008.22
Foster R. A., Subramaniam A., Mahaffey C., Carpenter E. J., Capone D. G., Zehr J. P. (2007). Influence of the Amazon river plume on distributions of free-living and symbiotic cyanobacteria in the western tropical North Atlantic Ocean. Limnol. Oceanogr. 52, 517–532. doi: 10.4319/lo.2007.52.2.0517
Foster R. A., Villareal T. A., Lundin D., Waterbury J. B., Webb E. A., Zehr J. P. (2022). “Richelia,” in Bergey's manual of systematics of archaea and bacteria. Eds. Trujillo M. E., Dedysh S., DeVos P., Hedlund B., Kämpfer P., Rainey FA & Whitman W. B. (John Wiley & Sons, Inc., in association with Bergey's Manual Trust).
Foster R. A., Zehr J. P. (2006). Characterization of diatom-cyanobacteria symbioses on the basis of nifH, hetR, and 16S rRNA sequences. Environ. Microbiol. 8, 1913–1925. doi: 10.1111/j.1462-2920.2006.01068.x
Galloway J. N., Dentener F. J., Capone D. G., Boyer E. W., Howarth R. W., Seitzinger S. P., et al. (2004). Nitrogen cycles: past, present and future. Biogeochem. 70, 153–226. doi: 10.1007/s10533-004-0370-0
Gradoville M. R., Bombar D., Crump B. C., Letelier R. M., Zehr J. P., White A. E. (2017b). Diversity and activity of nitrogen-fixing communities across Ocean basins. Limnol. Oceanogr. 62, 1895–1909. doi: 10.1002/lno.10542
Gradoville M. R., Cabello A. M., Wilson S. T., Turk-Kubo K. A., Karl D. M., Zehr J. P. (2021). Light and depth dependency of nitrogen fixation by the non-photosynthetic, symbiotic cyanobacterium UCYN-A. Environ. Microbiol. 23, 4518–4531. doi: 10.1111/1462-2920.15645
Gradoville M. R., Crump B. C., Letelier R. M., Church M. J., White A. E. (2017a). Microbiome of Trichodesmium colonies from the North Pacific Subtropical Gyre. Front. Microbiol. 8, 1–16. doi: 10.3389/fmicb.2017.01122
Gradoville M. R., Farnelid H., White A. E., Turk‐Kubo K. A., Stewart B., Ribalet F., et al. (2020). Latitudinal constraints on the abundance and activity of the cyanobacterium UCYN-A and other marine diazotrophs in the North Pacific. Limnol. Oceanogr. 65, 1858–1875. doi: 10.1002/lno.11423
Gradoville M. R., White A. E., Böttjer D., Church M. J., Letelier R. M. (2014). Diversity trumps acidification: Lack of evidence for carbon dioxide enhancement of Trichodesmium community nitrogen or carbon fixation at Station ALOHA. Limnol. Oceanogr. 59, 645–659. doi: 10.4319/lo.2014.59.3.0645
Green S. J., Venkatramanan R., Naqib A. (2015). Deconstructing the polymerase chain reaction: understanding and correcting bias associated with primer degeneracies and primer-template mismatches. PloS One 10, e0128122. doi: 10.1371/journal.pone.0128122
Hagino K., Onuma R., Kawachi M., Horiguchi T. (2013). Discovery of an endosymbiotic nitrogen-fixing cyanobacterium UCYN-A in Braarudosphaera bigelowii (Prymnesiophyceae). PloS One 8, e81749. doi: 10.1371/journal.pone.0081749
Hagino K., Takano Y., Horiguchi T. (2009). Pseudo-cryptic speciation in Braarudosphaera bigelowii (Gran and braarud) deflandre. Mar. Micropaleontol. 72, 210–221. doi: 10.1016/j.marmicro.2009.06.001
Halm H., Lam P., Ferdelman T. G., Lavik G., Dittmar T., LaRoche J., et al. (2012). Heterotrophic organisms dominate nitrogen fixation in the South Pacific gyre. ISME J. 6, 1238–1249. doi: 10.1038/ismej.2011.182
Hutchins D. A., Fu F. X., Webb E. A., Walworth N., Tagliabue A. (2013). Taxon-specific response of marine nitrogen fixers to elevated carbon dioxide concentrations. Nat. Geosci. 6, 790–795. doi: 10.1038/ngeo1858
Hynes A. M., Webb E. A., Doney S. C., Waterbury J. B. (2012). Comparison of cultured Trichodesmium (Cyanophyceae) with species characterized from the field. J. Phycol. 48, 196–210. doi: 10.1111/j.1529-8817.2011.01096.x
Inomura K., Bragg J., Riemann L., Follows M. J. (2018). A quantitative model of nitrogen fixation in the presence of ammonium. PloS One 13, e0208282. doi: 10.1371/journal.pone.0208282
Jayakumar A., Chang B. X., Widner B., Bernhardt P., Mulholland M. R., Ward B. B. (2017). Biological nitrogen fixation in the oxygen-minimum region of the eastern tropical North Pacific Ocean. ISME J. 11, 2356–2367. doi: 10.1038/ismej.2017.97
Karl D. M., Church M. J. (2017). Ecosystem structure and dynamics in the North Pacific Subtropical Gyre: new views of an old Ocean. Ecosyst. 20, 433–457. doi: 10.1007/s10021-017-0117-0
Karl D. M., Church M. J., Dore J. E., Letelier R. M., Mahaffey C. (2012). Predictable and efficient carbon sequestration in the North Pacific Ocean supported by symbiotic nitrogen fixation. Proc. Natl. Acad. Sci. 109, 1842–1849. doi: 10.1073/pnas.1120312109
Karl D., Letelier R., Tupas L., Dore J., Christian J., Hebel D. (1997). The role of nitrogen fixation in biogeochemical cycling in the subtropical North Pacific Ocean. Nature 388, 533–538. doi: 10.1038/41474
Kong L., Jing H., Kataoka T., Sun J., Liu H. (2011). Phylogenetic diversity and spatio-temporal distribution of nitrogenase genes (nifH) in the Northern South China Sea. Aquat. Microbial. Ecol. 65, 15–27. doi: 10.3354/ame01531
Langfelder P., Horvath S. (2008). WGCNA: an r package for weighted correlation network analysis. BMC Bioinf. 9, 1–13. doi: 10.1186/1471-2105-9-559
Langlois R., Grosskopf T., Mills M., Takeda S., LaRoche J. (2015). Widespread distribution and expression of gamma a (UMB), an uncultured, diazotroph, gamma-proteobacterial nifH phylotype. PloS One 10, e0128912. doi: 10.1371/journal.pone.0128912
Langlois R. J., Hummer D., LaRoche J. (2008). Abundances and distributions of the dominant nifH phylotypes in the Northern Atlantic Ocean. Appl. Environ. Microbiol. 74, 1922–1931. doi: 10.1128/AEM.01720-07
Letelier R. M., Karl D. M. (1996). Role of Trichodesmium spp. in the productivity of the subtropical North Pacific Ocean. Mar. Ecol. Prog. Ser. 133, 263–273. doi: 10.3354/meps133263
Letelier R. M., Karl D. M., Abbott M. R., Bidigare R. R. (2004). Light driven seasonal patterns of chlorophyll and nitrate in the lower euphotic zone of the North Pacific Subtropical Gyre. Limnol. Oceanogr. 49, 508–519. doi: 10.4319/lo.2004.49.2.0508
Ludwig W., Strunk O., Westram R., Richter L., Meier H., Yadhukumar , et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371. doi: 10.1093/nar/gkh293
Mague T. H., Weare M. M., Holm-Hansen O. (1974). Nitrogen fixation in the North Pacific Ocean. Mar. Biol. 24, 109–119. doi: 10.1007/BF00389344
Man-Aharonovich D., Kress N., Bar Zeev E., Berman-Frank I., Beja O. (2007). Molecular ecology of nifH genes and transcripts in the eastern Mediterranean Sea. Environ. Microbiol. 9, 2354–2363. doi: 10.1111/j.1462-2920.2007.01353.x
Marumo R., Nagasawa S. (1976). Seasonal variation of the standing crop of a pelagic blue-green alga, Trichodesmium in the Kuroshio water. Bull. Plankton Soc. Japan, 23, 19–25.
Mills M. M., Turk-Kubo K. A., Wilson S. T., Arrigo K. R., Zehr J. P. (2020). Unusual marine cyanobacteria/haptophyte symbiosis relies on N2 fixation even in N-rich environments. ISME J 14, 2395–2406. doi: 10.1038/s41396-020-0691-6
Moisander P. H., Beinart R. A., Hewson I., White A. E., Johnson K. S., Carlson C. A., et al. (2010). Unicellular cyanobacterial distributions broaden the Oceanic N2 fixation domain. Science 327, 1512–1514. doi: 10.1126/science.1185468
Moisander P. H., Beinart R. A., Voss M., Zehr J. P. (2008). Diversity and abundance of diazotroph microorganisms in the South China Sea during intermonsoon. ISME J. 2, 954–967. doi: 10.1038/ismej.2008.51
Moisander P. H., Benavides M., Bonnet S., Berman-Frank I., White A. E., Riemann L. (2017). Chasing after non-cyanobacterial nitrogen fixation in marine pelagic environments. Front. Microbiol. 8, 1736. doi: 10.3389/fmicb.2017.01736
Moisander P. H., Serros T., Paerl R. W., Beinart R. A., Zehr J. P.. (2014). Gammaproteobacterial diazotrophs and nifH gene expression in surface waters of the South Pacific Ocean. ISME J. 8, 1962–1973. doi: 10.1038/ismej.2014.49
Moonsamy P. V., Williams T., Bonella P., Holcomb C. L., Hoglund B. N., Hillman G., et al. (2013). High throughput HLA genotyping using 454 sequencing and the fluidigm access array system for simplified amplicon library preparation. Tissue Antigens 81, 141–149. doi: 10.1111/tan.12071
Paerl R. W., Foster R. A., Jenkins B. D., Montoya J. P., Zehr J. P. (2008). Phylogenetic diversity of cyanobacterial narB genes from various marine habitats. Environ. Microbiol. 10, 3377–3387. doi: 10.1111/j.1462-2920.2008.01741.x
Pedros-Alio C. (2012). The rare bacterial biosphere. Ann. Rev. Mar. Sci. 4, 449–466. doi: 10.1146/annurev-marine-120710-100948
Poff K. E., Leu A. O., Eppley J. M., Karl D. M., DeLong E. F. (2021). Microbial dynamics of elevated carbon flux in the open Ocean's abyss. Proc. Natl. Acad. Sci. U.S.A. 118, e2018269118. doi: 10.1073/pnas.2018269118
Riemann L., Farnelid H., Steward G. F. (2010). Nitrogenase genes in non-cyanobacterial plankton: prevalence, diversity and regulation in marine waters. Aquat. Microb. Ecol. 61, 225–237. doi: 10.3354/ame01431
Riemann L., Rahav E., Passow U., Grossart H. P., de Beer D., Klawonn I., et al. (2022). Planktonic aggregates as hotspots for heterotrophic diazotrophy: the plot thickens. Front. Microbiol. 13, 1–9. doi: 10.3389/fmicb.2022.875050
Rouco M., Warren H. J., McGillicuddy D. J., Waterbury J. B., Dyhrman S. T. (2014). Trichodesmium Sp. clade distributions in the western North Atlantic Ocean. Limnol. Oceanogr. 59, 1899–1909. doi: 10.4319/lo.2014.59.6.1899
Schvarcz C. R., Wilson S. T., Caffin M., Stancheva R., Li Q., Turk-Kubo K. A., et al. (2022). Overlooked and widespread pennate diatom-diazotroph symbioses in the sea. Nat. Commun. 13, 1–9. doi: 10.1038/s41467-022-28065-6
Selden C. R., Chappell P. D., Clayton S., Macías‐Tapia A., Bernhardt P. W., Mulholland M. R., et al. (2021). A coastal N2 fixation hotspot at the Cape Hatteras front: Elucidating spatial heterogeneity in diazotroph activity via supervised machine learning. Limnol. Oceanogr. 66, 1832–1849. doi: 10.1002/lno.11727
Shiozaki T., Bombar D., Riemann L., Hashihama F., Takeda S., Yamaguchi T., et al. (2017). Basin scale variability of active diazotrophs and nitrogen fixation in the North Pacific, from the tropics to the subarctic Bering Sea. Global Biogeochem. Cy. 31, 996–1009. doi: 10.1002/2017GB005681
Shiozaki T., Fujiwara A., Ijichi M., Harada N., Nishino S., Nishi S., et al. (2018). Diazotroph community structure and the role of nitrogen fixation in the nitrogen cycle in the Chukchi Sea (western Arctic Ocean). Limnol. Oceanogr. 63, 2191–2205. doi: 10.1002/lno.10933
Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Suzuki S., Kawachi M., Tsukakoshi C., et al. (2021). Unstable relationship between Braarudosphaera bigelowii (= Chrysochromulina parkeae) and its nitrogen-fixing endosymbiont. Front. Plant Sci. 12, 749895. doi: 10.3389/fpls.2021.749895
Suzuki R., Shimodaira H. (2006). Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542. doi: 10.1093/bioinformatics/btl117
Thompson A., Carter B. J., Turk-Kubo K., Malfatti F., Azam F., Zehr J. P., et al. (2014). Genetic diversity of the unicellular nitrogen-fixing cyanobacteria UCYN-A and its prymnesiophyte host. Environ. Microbiol. 16, 3238–3249. doi: 10.1111/1462-2920.12490
Turk-Kubo K. A., Farnelid H. M., Shilova I. N., Henke B., Zehr J. P.. (2017). Distinct ecological niches of marine symbiotic N2-fixing cyanobacterium Candidatus Atelocyanobacterium thalassa sublineages. J. Phycol. 53, 451–461. doi: 10.1111/jpy.12505
Turk-Kubo K. A., Frank I. E., Hogan M. E., Desnues A., Bonnet S., Zehr J. P. (2015). Diazotroph community succession during the VAHINE mesocosm experiment (New Caledonia lagoon). Biogeosciences 12, 7435–7452. doi: 10.5194/bg-12-7435-2015
Turk-Kubo K. A., Gradoville M. R., Cheung S., Cornejo-Castillo F., Harding K. J., Morando M., et al. (2022). Non-cyanobacterial diazotrophs: Global diversity, distribution, ecophysiology, and activity in marine waters. FEMS Microbiol. Rev. fuac046. doi: 10.1093/femsre/fuac046
Turk-Kubo K. A., Karamchandani M., Capone D. G., Zehr J. P. (2014). The paradox of marine heterotrophic nitrogen fixation: abundances of heterotrophic diazotrophs do not account for nitrogen fixation rates in the Eastern Tropical South Pacific. Environ. Microbiol. 16, 3095–3114. doi: 10.1111/1462-2920.12346
Turk-Kubo K. A., Mills M. M., Arrigo K. R., van Dijken G., Henke B. A., Stewart B., et al. (2021). UCYN-A/haptophyte symbioses dominate N2 fixation in the Southern California current system. ISME Commun. 1, 1–13. doi: 10.1038/s43705-021-00039-7
Venrick E. L. (1974). The distribution and significance of Richelia intracellularis Schmidt in the North Pacific central gyre. Limnol. Oceanogr. 19, 437–445. doi: 10.4319/lo.1974.19.3.0437
Venrick E. L. (1988). The vertical distributions of chlorophyll and phytoplankton species in the North Pacific central environment. J. Plankton Res. 10, 987–998. doi: 10.1093/plankt/10.5.987
Villareal T. A., Altabet M. A., Culver-Rymzsa K. (1993). Nitrogen transport by vertically migrating diatom mats in the North Pacific Ocean. Nature 363, 709–712. doi: 10.1038/363709a0
White A. E., Spitz Y. H., Letelier R. M. (2007). What factors are driving summer phytoplankton blooms in the North Pacific Subtropical Gyre? J. Geophysical Res.-Oceans 112, C12006. doi: 10.1029/2007JC004129
White A. E., Watkins-Brandt K. S., Church M. J. (2018). Temporal variability of Trichodesmium spp. and diatom-diazotroph assemblages in the North Pacific Subtropical Gyre. Front. Mar. Sci. 5, 27.
Wozniak B., Dera J., Koblentz-Mishke O. J. (1992). Bio-optical relationships for estimating primary production in the Ocean. Oceanologia 33, 5–38.
Yang Z. (1994). Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J. Mol. Evol. 39, 306–314. doi: 10.1007/BF00160154
Zani S., Mellon M. T., Collier J. L., Zehr J. P. (2000). Expression of nifH genes in natural microbial assemblages in Lake George, NY detected with RT-PCR. Appl. Environ. Microbiol. 66, 3119–3124. doi: 10.1128/AEM.66.7.3119-3124.2000
Zehr J. P., Capone D. G. (2020). Changing perspectives in marine nitrogen fixation. Science 368, eaay9514. doi: 10.1126/science.aay9514
Zehr J., Jenkins B., Short S., Steward G. F. (2003). Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 5, 539–554. doi: 10.1046/j.1462-2920.2003.00451.x
Zehr J., McReynolds L. (1989). Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl. Environ. Microbiol. 55, 2522–2526. doi: 10.1128/aem.55.10.2522-2526.1989
Zehr J., Mellon M., Zani S. (1998). New nitrogen-fixing microorganisms detected in oligotrophic Oceans by amplification of nitrogenase (nifH) genes. Appl. Environ. Microbiol. 64, 3444–3450. doi: 10.1128/AEM.64.9.3444-3450.1998
Zehr J. P., Montoya J. P., Jenkins B. D., Hewson I., Mondragon E., Short C. M., et al. (2007). Experiments linking nitrogenase gene expression to nitrogen fixation in the North Pacific Subtropical Gyre. Limnol. Oceanogr. 52, 169–183. doi: 10.4319/lo.2007.52.1.0169
Zehr J., Waterbury J., Turner P., Montoya J. P., Omoregie E., Steward G. F., et al. (2001). Unicellular cyanobacteria fix N2 in the subtropical North Pacific Ocean. Nature 412, 635–638. doi: 10.1038/35088063
Keywords: nitrogen fixation, diazotroph community structure, nifH diversity, Trichodesium, UCYN-A, UCYN-A sublineages
Citation: Turk-Kubo KA, Henke BA, Gradoville MR, Magasin JD, Church MJ and Zehr JP (2023) Seasonal and spatial patterns in diazotroph community composition at Station ALOHA. Front. Mar. Sci. 10:1130158. doi: 10.3389/fmars.2023.1130158
Received: 22 December 2022; Accepted: 12 April 2023;
Published: 01 May 2023.
Edited by:
Alexander Eiler, University of Oslo, NorwayReviewed by:
Lauren Frances Messer, University of Stirling, United KingdomConnie Lovejoy, Laval University, Canada
Yanying Zhang, Yantai University, China
Irina N. Shilova, Second Genome, United States
Copyright © 2023 Turk-Kubo, Henke, Gradoville, Magasin, Church and Zehr. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kendra A. Turk-Kubo, a3R1cmtAdWNzYy5lZHU=
†Present address: Mary R. Gradoville, Columbia River Inter-Tribal Fish Commission, Portland, OR, United States
 Kendra A. Turk-Kubo
Kendra A. Turk-Kubo Britt A. Henke
Britt A. Henke Mary R. Gradoville1†
Mary R. Gradoville1† Jonathan D. Magasin
Jonathan D. Magasin Matthew J. Church
Matthew J. Church Jonathan P. Zehr
Jonathan P. Zehr