- Avram and Stella Goldstein–Goren Department of Biotechnology Engineering, Ben-Gurion University of the Negev, Beer-Sheva, Israel
Gene silencing by dsRNA is well documented in crustaceans, but RNA interference (RNAi) in developing oocytes is yet to be achieved. The main obstacle to RNAi in the oocytes of oviparous animals derives from their protective layers, including the cytosolic membrane, the vitelline envelope, and a layer of follicular cells. These layers form a barrier preventing the entry of large nonspecific molecules, such as double-stranded RNA (dsRNA). This article describes a sophisticated tool – designated OSDel [oocyte-specific delivery] – for the delivery of dsRNA for gene silencing in the developing oocyte. The methodology exploits the process of receptor-mediated endocytosis (RME) taking place in the crustacean female for internalizing vitellogenin (Vg) (the precursor of the major yolk protein, vitellin) into oocytes. In this process, the extracellular domain of the Vg receptor (VgR) interacts with a distinct amino acid sequence of Vg and internalizes it to form yolk droplets. Here, we tested the premise that a distinct 24 amino-acid peptide derived from Macrobrachium rosenbergii Vg (designated VgP) would interact with VgR to form particles of a size suitable for piggy-backing dsRNA into oocytes via RME. We found that fluorescently labeled VgP had a micromolar affinity for the ligand-binding domain (LBD) of the VgR and could indeed be delivered to and internalized in oocytes. As model system to illustrate the applicability of the OSDel, we injected vitellogenic females with dsRNA encoding the eye development gene PAX6 piggybacked on VgP. Proof that the dsRNA had been successfully internalized into the developing oocytes and had silenced the PAX6 gene was derived from impaired eye development in 87% of the embryos. The ability to manipulate embryos by simple injection into vitellogenic crustacean females may prove to be a powerful high throughput tool for functional genomics investigation in crustacean embryos and for silencing genes relevant to crustacean aquaculture and biotechnology.
Introduction
Gene silencing by double-stranded RNA (dsRNA) is well documented in arthropods (Aronstein et al., 2011). In particular, in crustaceans, effective silencing of specific genes has been achieved by injecting dsRNA into juveniles and adults (Sagi et al., 2013). Despite the success of early work in crustaceans, gene silencing through RNA interference (RNAi) in embryos has been achieved only by manual dsRNA injection into newly laid fertilized eggs at early developmental stages, and gene silencing in developing oocytes remains elusive. The lack of progress in gene silencing in oocytes is, to some extent, a function of their specific physiology and morphology. Specifically, the oocytes possess copious amounts of yolk to provide nutrients for their proper development into embryos, which are held in a pouch outside the female body until they hatch (Lodé, 2012). In addition, each oocyte is enclosed inside a protective cover, termed the vitelline envelope, which is an acellular layer that surrounds the chorion; together with the cell membrane and a layer of follicle cells, the vitelline envelope prevents the penetration of pathogens and other molecules into the oocytes (Mazzini et al., 1984). The protective role of the vitelline envelope thus complicates genetic and biochemical manipulations of the embryo at the oocyte stage.
In oviparous animals, such as crustaceans, oocyte maturation is accompanied by vitellogenesis, a process in which large amounts of vitellogenin (Vg) – the major circulating yolk lipoprotein – are accumulated rapidly and stored in the oocyte as vitellin. After fertilization, vitellin serves as the main nutrient for embryonic development (Subramoniam, 2011). In decapod crustaceans, Vg is typically synthesized in the hepatopancreas and circulates in the hemolymph as lipoprotein particles made up of several protein subunits (Okuno et al., 2002; Subramoniam, 2011). A specific vitellogenin receptor (VgR) recognizes and internalizes Vg into yolk droplets in the oocytes (Okuno et al., 2002) in a process termed receptor-mediated endocytosis (RME) (Subramoniam, 2011). This process begins with the interaction between the ligand (protein) and its specific receptor, and the ligand-receptor complex is then internalized into the cell, in the form of clathrin-coated vesicles (Goldstein et al., 1979), and delivered to cytoplasmic endosomes (Brown and Goldstein, 1976) (Goldstein et al., 1985). Small endosomes then merge to form larger endosomes that store vitellin as yolk droplets. The pH in the endosome is typically lower than the physiological pH, leading to conformational changes in the receptor, which, in turn, facilitate the release of the ligand from the receptor-ligand complex. After detachment from the ligands, receptors are recirculated to the membrane to continue the cycle of receptor-ligand interactions and Vg internalization (Warrier and Subramoniam, 2002).
VgR belongs to the low-density lipoprotein receptor (LDLR) superfamily and is synthesized exclusively in the ovary and targeted to the oocyte surface (Roth and Khalaila, 2012). In contrast to the homologous vertebrate LDLRs and insect lipophorin receptors that possess a single ligand-binding domain (LBD), insect and crustacean VgRs contain two LBDs (Mekuchi et al., 2008; Tiu et al., 2008; Tufail and Takeda, 2009). Despite differences in the LBD compositions of VgRs in vertebrates and invertebrates, the mode of VgR-Vg interactions and sequence homology in lipoproteins and LDLRs of vertebrates and invertebrates appears to be conserved (Li et al., 2003); for example, Xenopus laevis Vg interacts with chicken VgR and vice versa, and crab Scylla serrata VgR cross-reacts with mammalian apo-B and apo-E lipoproteins (Stifani et al., 1990; Warrier and Subramoniam, 2002).
A break-through study on the Vg-VgR interaction was conducted on the blue tilapia Oreochromis aureus; in that study, it was shown that VgR binds an 84-amino-acid domain in the Vg N-terminal region (Li et al., 2003). It was also shown that a point mutation at Vg K185A, i.e., a change from a positively charged to a neutral amino acid, attenuated the Vg-VgR interaction, indicating the electrostatic nature of the interaction between the ligand and its receptor. In that study, it was suggested that the 84-amino-acid sequence is a conserved domain throughout the animal kingdom (Li et al., 2003). Importantly, a later study on the Vg of the freshwater prawn Macrobrachium rosenbergii (BAB69831.1) revealed that it is a 24-amino-acid encompassing amino acids 237-260 peptide (designated VgP) of the C-terminal of the conserved 84-amino-acid stretch that interacts with the VgR (Roth et al., 2013).
In the current study, we sought to leverage the above-described body of knowledge to develop the means to deliver dsRNA into M. rosenbergii oocytes. As mentioned above, the vitelline envelope constitutes a significant barrier to nucleic acid delivery into the eggs of oviparous animals. Hence, the delivery of proteins and nucleic acids into the eggs is currently performed mainly by microinjection (Reid and O'brochta, 2016; Xu et al., 2020), a procedure with significant disadvantages: it is time intensive and requires experienced personnel, and survival rates are low (Cheers and Ettensohn, 2004; Xu et al., 2020). An alternative with exciting potential would be delivery via RME.
We thus aimed to investigate the possibility of exploiting the RME for delivery of oligonucleotide-based molecules into M. rosenbergii oocytes. We posited that the 24-amino-acid M. rosenbergii VgP, described above, would be able to penetrate M. rosenbergii oocytes—both alone and piggybacking different cargos. As proof of concept, this study focused on dsRNA as a candidate delivery cargo. We report here the successful and specific delivery of dsRNA into oocytes (Khalaila et al., 2020), where it effectively silenced PAX6, a gene that belongs to the family of developmental transcription factors and is responsible for eye development in early embryonic developmental stages (Cvekl and Callaerts, 2017). We found that injection of vitellogenic females with PAX6-dsRNA (dsPAX6) piggybacked on VgP led to defective eye development of the embryos (87% exhibited impaired eye development). The results obtained from this model system support the notion that an RME-based delivery tool can be used for silencing aquaculture-relevant genes, such as growth genes, sex-determining or sterility genes, and different pathogens. A single dsRNA-treated crustacean female could produce thousands of embryos with a particular desired trait. Therefore, a dsRNA delivery tool for short-term silencing directly in the oocytes could make a massive impact on sustainable crustacean aquaculture.
Materials and methods
Animals
For the in vitro and in vivo experiments, 100 M. rosenbergii females were collected from the Aquaculture Station of the Ministry of Agriculture (Dor, Israel) (Levy et al., 2017) and then transferred to a holding facility at Ben-Gurion University of the Negev (Beer-Sheva, Israel) and acclimated for two weeks. During the acclimation and throughout the experiment, Females were maintained in tanks at 27°C, 12 h daylight, and fed ad libitum with Polychaeta and protein-rich pellets. Imitation black weeds and PVC tubes were included in the tanks as hiding substrates to minimize cannibalism.
Peptide synthesis
The fluorophores labeled (TAMRA or FITC) VgP and scrambled VgP (Table 1), and KH9-VgP were purchased from Peptron (Daejeon 34054, South Korea).
In vitro and in vivo assays for peptide endocytosis into M. rosenbergii oocytes
For the in-vitro assay, slices of M. rosenbergii ovaries, 1-3 mm thick, containing oocytes of diameter ~150 µm [from early vitellogenic ovaries stage II, (Huang et al., 2010)], were incubated at room temperature for 24 h in a biological hood in a sterile 48-well plate having a 1-cm² growth area in each well (Biofil®). Each slice was incubated with 250 µL of Schneider’s Drosophila Medium (Biological Industries), of a fixed osmolality of 420 mOsm/L (Huong et al., 2010), supplemented with 10% fetal bovine serum (FBS; Biological Industries), PSA (final concentration: penicillin, 100 units/mL; streptomycin, 0.1 mg/mL; amphotericin B, 0.25 µg/mL; Biological Industries), and bovine insulin (Morris and Spradling, 2011), final concentration 0.2 mg/mL (Sigma-Aldrich). In addition, to verify the role of the amino acid sequence of VgP in its internalization into the oocytes, the VgP 24-amino-acid sequence was synthesized in scrambled order (scVgP) to be used as a control. The ovary slices were incubated with 6 µM each VgP-TAMRA or scVgP-TAMRA, or 6 µM VgP-TAMRA + 6 µM scVgP-FITC (Table 1).
For the in-vivo assay, a solution (2 µl/g body weight) containing VgP-TAMRA + scVgP-FITC (10 µg/g body weight for each peptide, dissolved in DMSO and mixed with crustacean saline 420 mOsm/L) was injected with a manual insulin syringe (BD®) into the hemolymph sinus at the base of the fifth walking leg of M. rosenbergii reproductive females [each weighing approximately 15 g and bearing opaque white to yellowish ovaries stage II, (Huang et al., 2010)]. After injection, the animals were kept at 28°C for 24 h in the culture facility for M. rosenbergii of Ben-Gurion University of the Negev. Thereafter, the ovaries and gills were dissected out for confocal imaging.
Confocal imaging of M. rosenbergii oocytes
Ovary pieces from the in-vitro and in-vivo experiments that were incubated with the fluorescing molecules were each transferred to a slide and incubated with Hoechst staining solution (5 µg/mL) for 7 min. Thereafter, the pieces were washed with saline buffer for 1 min ×3. The slides were inspected, and images were obtained with a confocal microscope FV1000 (Olympus) at ×60 magnification. The excitation laser wavelengths were 405 for Hoechst dye, 561 nm for the TAMRA-labeled peptides, and 488 nm for the FITC-labeled control peptide, and emission was obtained at 460-490 nm, 540-640 nm, and 490-530 nm, respectively.
Binding affinity test by micro-scale thermophoresis
MST was used to evaluate the affinity of VgP and scVgP to VgR-LBD-I and VgR-LBD-II. VgR-LBD-I and II were expressed and purified as described in supplementary methods. The peptides were diluted in Tris-buffered saline containing Tween and Ca2+ (TBS, Tris 20 mM pH 7.4, NaCl 150 mM, TWEEN 0.1%, CaCl2 5 mM) to 500 to 1000 fluorescence units. A serial dilution for each LBD protein was prepared in the same buffer. Ten microliters of each dilution of the peptide of interest were added to each LBD-containing tube. The experiments were performed with MST Monolith NT.115, NanoTemper.
dsRNA synthesis
The open reading frame (ORF) of M. rosenbergii PAX6-like was sequenced (GenBank OP292287). Two dsRNA: dsPAX6A (230bp) and dsPAXB (198bp), distinct parts of the ORF sequence were then synthesized in our laboratory (Supplementary Figure 1). The templates for the transcription were amplified by adding the following primer pairs to the PCR: the sense strand was synthesized using a gene-specific forward primer containing the T7 promoter sequence vs. a gene-specific template reverse primer. The reverse primer containing the T7 promoter sequence was used vs. the gene-specific template forward primer to synthesize the antisense strand. Primers and T7 promoter sequences for dsRNA synthesis are given in Table 2. PCR amplicons were separated on a 1.3% agarose gel. DNA bands were visualized with SYBR Safe DNA Gel Stain (Invitrogen). Amplicons were purified with a PCR purification kit (NucleoSpin Gel and PCR Clean-up, Machery-Nagel). A TranscriptAid T7 High Yield Transcription Kit (Thermo Scientific) was used (according to the manufacturer’s instructions) to generate single-stranded RNA. Synthesized RNA molecules were purified by phenol:chloroform (1:1) and sodium acetate extraction. After purification, RNA was precipitated with ethanol. Reconstituted sense and antisense RNA strands were hybridized by incubation at 70°C for 15 min, 65°C for 15 min, and room temperature for 30 min. dsRNA quality was assessed on an agarose gel. dsRNA was then diluted to 5 mg/ml and stored at -80°C until use. FITC-labeled dsPAX6A (dsPAX6A-FITC) was synthesized as described above by incorporating UTP-FITC (Sigma-Aldrich) into the synthesized dsRNA. FITC-labeled UTP was used in an equal amount to free UTP in the dsRNA synthesis reaction. RNA molecules were purified by the RNeasy MinElute Cleanup Kit (QIAGEN), followed by hybridization and quality assessment, as described above.
Peptide-dsRNA characterization
In this work, we chose to bind the VgP to dsRNA by leveraging an electrostatic interaction between VgP and the negatively charged phosphoryl groups of dsRNA. To facilitate such an electrostatic interaction, a poly cationic tail composed of 9 KH repeats was synthesized on the N-terminal side of VgP ((KH)9VgP).
Agarose gel electrophoresis was used to evaluate the migration characteristics of the peptide-dsRNA particles. A constant amount of dsPAX6 (25 pmole) solution was mixed with increasing amounts of peptide (dsRNA/peptide ratio: 1:1, 1:5, 1:10, 1:20 and 1:50) in Eppendorf vials. For each vial, the total reaction volume was made up to 15 µL with doubly distilled water, and the mixtures were incubated at room temperature for 20 min. An equal volume of loading buffer (50% glycerol and 0.5 M EDTA) was added to each mixture, and the total volume of each mixture was separated on a 1.3% agarose gel in TAE buffer for 30 min at 90 V. Fluorescence emissions of the compounds in the gels were scanned by a Typhoon™ FLA 9500 gel system (GE Healthcare Life Sciences). Thereafter, the gels were stained with SYBR Safe DNA Gel Stain (Invitrogen) and visualized with UV light.
The morphology and particle size of the (KH)9VgP-dsRNA compounds were investigated by TEM (FEI Talos F200C). Samples with dsRNA:(KH)9VgP molar ratios of 1:5 and 1:10 were prepared, and 2.5-µL of each were taken for TEM sample preparation on a 300-mesh copper grid. Excess liquid was blotted with filter paper after 1 min, and the grid was dried in air for 1 min. Contrast enhancement was achieved by negative staining with 5 µL of 2% uranyl acetate. Thereafter, the grid was blotted again to remove excess uranyl acetate. Finally, the grid was air dried before insertion into the microscope. The images were obtained with an FEI Ceta 16M CMOS camera.
In-vitro and in-vivo endocytosis of dsRNA piggybacked on VgP
The ability of VgP to piggyback dsRNA into oocytes by endocytosis in vitro was investigated by incubating ovary slices with dsPAX6-FITC linked to (KH)9VgP-TAMRA (molar ratio 1:10) at room temperature for 5 h, and confocal images of oocytes were obtained. As the control, ovary slices were incubated with free dsPAX6-FITC.
For the in-vivo experiment, the silencing of PAX6 piggybacked on VgP was evaluated in developing embryos. In this experiment, 20 previtellogenic females (14.96± 0.41 g) were injected with ds-vitellogenesis-inhibiting hormone (dsVIH) to induce ovary maturation (Cohen et al., 2021). Each day, the females were inspected visually in the daylight to determine ovary maturation. As soon as the ovaries acquired a yellowish appearance, the females (n=10) were injected with a solution containing (KH)9-VgP linked to dsPAX6A and dsPAX6B (10 µg dsPAX6/g body weight, 5 µg of each dsPAX6A and dsPAX6B, dsPAX6-peptide molar ratio 1:10). The control group (n=10) was injected with free dsRNA. Chloroquine (2 mM final concentration) was incorporated into the injection mixtures as an endosomal escape reagent. The female animals were monitored daily, and upon appearance of the reproductive molt, each female was released into a tank with a male for mating and fertilization. The development of the embryos on the pleopods of each female was monitored under a stereoscope (Figure 1). The effect of PAX6 silencing on embryo eye development was followed by sampling 10 embryos from the berried females on the day after eye appearance and examining them under a stereoscope. Embryos were counted and categorized according to eye development, i.e., normally developing, retarded, remnant eyes, or no eye development. Additionally, the dimensions (length and width) of the pigmented area of 10 eyes from 10 different embryos of each female were measured under a microscope, and an eye length/width index was calculated.
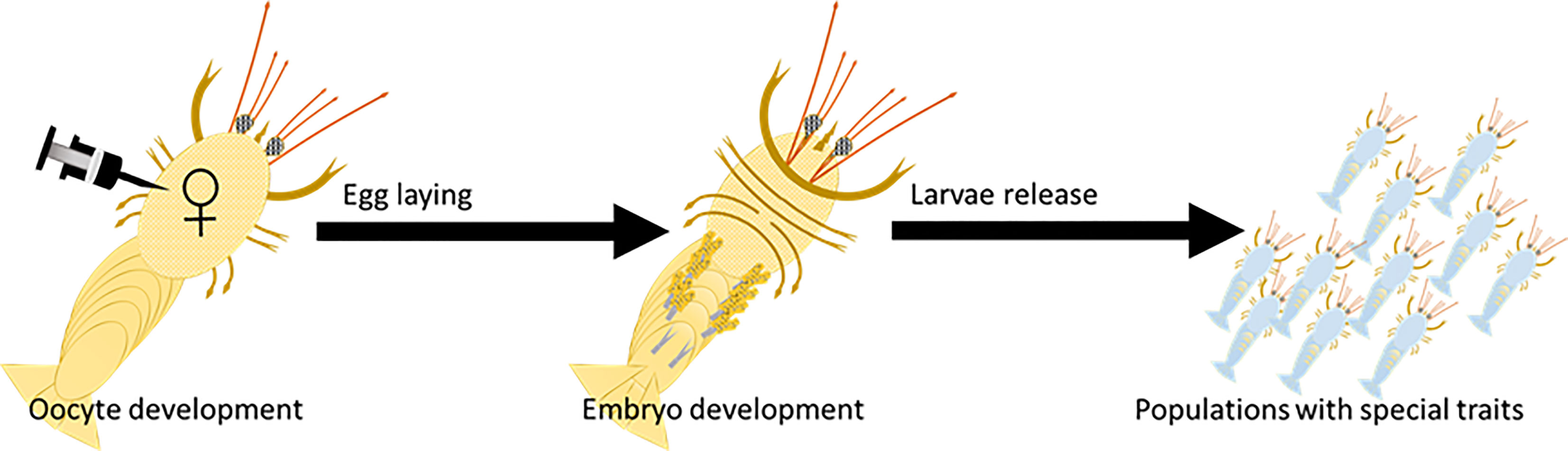
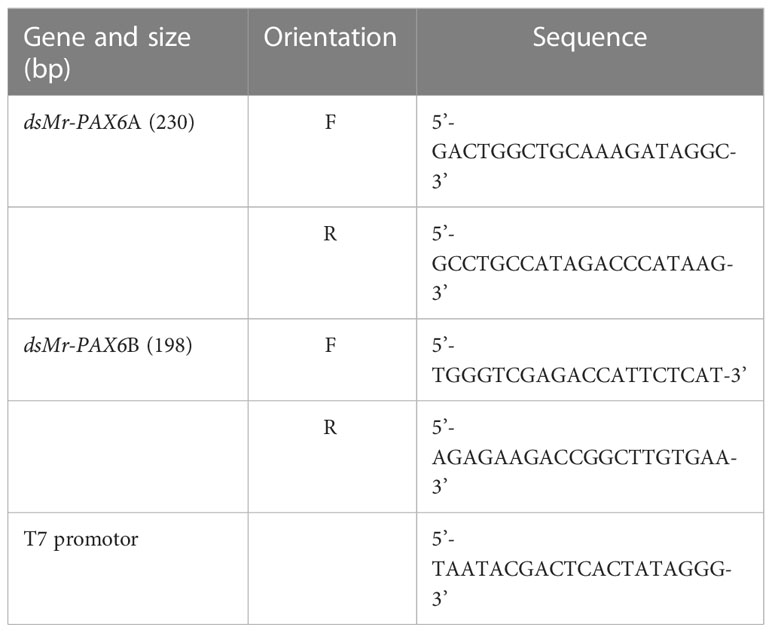
Figure 1 Oocyte-specific delivery tool (designated OSDel) for gene silencing and regulation. Figure shows OSDel injection into the hemolymph of M. rosenbergii vitellogenic females. The regulated trait can be monitored by inspecting the developing embryos, larvae, or post-larvae.
SEM sample preparation
Larvae from the VgP-dsPAX6 injected females and from the control group were fixed and dehydrated by immersion in increasing ethanol concentrations (50%, 70%, 80%, 90%, 95% and twice in 100%; 15 min each). The samples were dried in a critical point dryer, placed on stubs, and coated with gold. The specimens were examined with a scanning electron microscope (SEM; JEOL model JSM-7400F).
Statistical analysis
Graphical and statistical analyses were performed using OriginPro and SPSS. For multiple comparisons, statistical differences were determined by one-way ANOVA. Differences were considered statistically significant at P < 0.05. The most important evaluation metric for checking any classification model’s performance is the Receiver Operating Characteristics (ROC). ROC is a probability curve, and the Area Under the Curve (AUC) represents the degree or measure of separability. It tells how much the model is capable of distinguishing between classes. The higher the AUC, the better the model predicts impaired eye development. The classes are the (KH)9VgP-dsPAX6 injected versus the control ds injected females. The classifier is the length/width index of the eye pigment. ROC analysis was performed to define the percentage of embryos possessing impaired eye development and therefore, the efficiency of the treatment.
Results
The Vg-derived peptide VgP is capable of endocytosing into oocytes in vitro and in vivo
To enable evaluation of the endocytosis ability of VgP into early vitellogenic oocytes, both VgP and the control scrambled peptide, scVgP, were labeled with the TAMRA fluorophore. When early vitellogenic oocytes were incubated with the peptides and inspected under a confocal microscope, fluorescence emission at 564 nm was detected. Both VgP and scVgP were visible in the vicinity of the oocyte membrane (Figures 2A–D). VgP was distributed mainly on the cytosolic side of the oocyte membrane where yolk droplets were forming (Figures 2A, C, black arrow). Figure 2C shows that VgP accumulated mainly in the peripheral area of the oocyte, near the membrane (white arrow), being scattered on the external surface of newly formed droplets. In contrast, scVgP was dispersed mainly on the outer surface of the oocyte (Figures 2B, D).
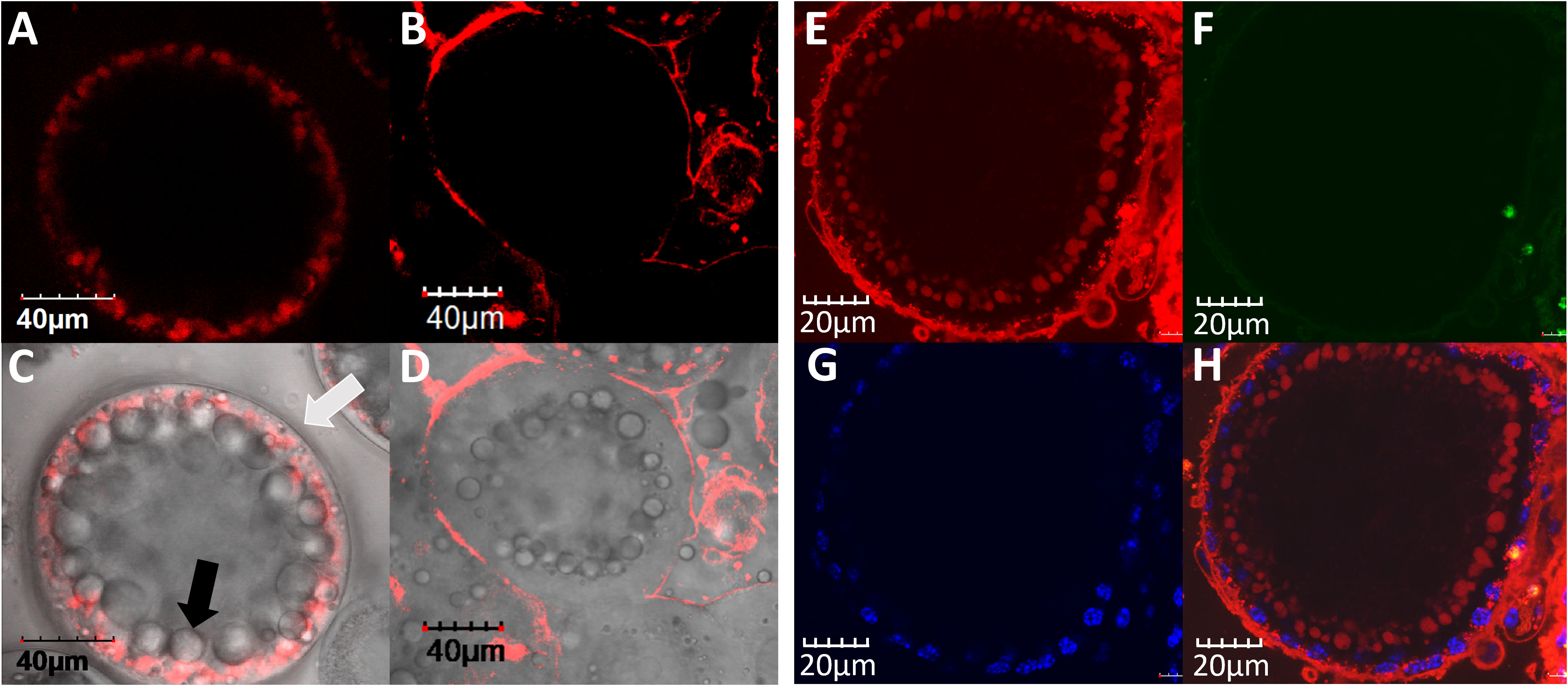
Figure 2 VgP endocytoses into M. rosenbergii early vitellogenic oocytes. In-vitro incubation of ovary pieces in the presence of VgP-TAMRA (A, C), ScVgP-TAMRA (B, D), or VgP-TAMRA + ScVgP-FITC (E–H). A and B are TAMRA fluorescence emission images. (C, D) show the overlay images of (A, B), respectively, with the bright field image of the same oocyte. The white arrow in (C) points to the oocyte membrane, and the black arrow points to Vg accumulation inside the oocyte. (E, F) are confocal images of the VgP-TAMRA (red) and scVgP-FITC (green) emissions, respectively. (G) shows the nuclear DNA of follicular cells with Hoechst staining (blue). (H) shows the overlay image of (E, G).
A dose-dependent test using VgP-TAMRA and scVgP-TMARA was performed to determine the optimal peptide concentration for in-vitro incubation of the peptide with the ovary slices (Supplementary Figure 2). Unlike the distribution patterns for scVgP, the distribution patterns of VgP in the oocyte remained similar, irrespective of the peptide concentrations (Supplementary Figure 2, upper row). For VgP, a clear fluorescence signal was detected at concentrations of 12, 6, and 1.5 µM, while the fluorescence signal for scVgP was weak at high concentrations (12 µM and 6 µM), and no signal was detected at 1.5 µM. No signal was obtained at 0.75 µM for both peptides (Supplementary Figure 2).
To evaluate the binding affinity of VgP and scVgP to the two LBDs (LBDI and LBDII) of VgR, the kinetic dissociation constant (Kd) was calculated from kinetic curves obtained by microscale thermophoresis (MST) (Table 3 and Supplementary Figure 3). For VgP, Kd values of 3.42 ± 2.16 and 50.89 ± 26.47 μM for binding to LBDII and LBDI, respectively, were obtained (Table 3). In contrast, there was no apparent interaction of scVgP with the receptor LBDs.
VgP and its control counterpart (scVgP) were labeled with different fluorophores, TAMRA and FITC, respectively, to enable differentiation of the specific localization of each in the oocyte. In accordance with the results obtained from incubating ovary slices with the peptides labeled with the same TAMRA fluorophore, the fluorophore experiment showed the distributions of the two fluorescently labeled peptides in the same oocyte. Endocytosis of VgP-TAMRA into the oocytes was evident, with the labeled peptide being distributed along the inner side of the membranes (Figures 2E, H). In contrast, scVgP-FITC was visible only on the membrane’s outer surface, overlapping with the outer surface of the follicular cell nuclei stained with Hoechst stain (Figures 2F–H).
In in-vivo experiments designed to determine whether VgP-TAMRA was internalized specifically into oocytes, a mixture of the VgP-TAMRA and the control-FITC peptides was injected into the circulatory system of a female prawn. The control-FITC peptide was absent from oocytes dissected 24 h after injection (Figure 3B). For VgP-TAMRA, fluorescence emission was evident inside the oocytes (Figures 3A, C), mostly inside yolk droplets. To verify that the two peptides did indeed circulate through the female prawn body, the prawn gills were dissected out and examined by confocal microscopy. Clear dual (TAMRA and FITC) fluorescence emission was evident, indicating that most of the injected peptide had indeed been washed out through the osmoregulatory system (namely, the gills; Figures 3D–F).
Figure 3 VgP circulates through the hemolymph, reaches the ovary and is specifically internalized into the oocytes. In vivo injection of both VgP-TAMRA and ScVgP-FITC peptides shows directed delivery of VgP to the oocytes. Fluorescence images of oocytes and gills were taken from an early vitellogenic female injected simultaneously with VgP-TAMRA and ScVgP-FITC. Confocal images of the accessory cells surrounding the oocytes (Hoechst, blue), (A) VgP-TAMRA (red dots), (B) ScVgP-FITC. (C) the overlay of images (A, B) Both VgP-TAMRA and ScVgP-FITC were detected in the gills (D, E), respectively). (F) overlaid images of (D, E).
A dsRNA-(KH)9VgP complex forms particles suitable for receptor-mediated delivery
VgP-TAMRA and scVgP-FITC were synthesized with cationic poly (KH)9 tails to facilitate electrostatic interactions between the peptides and the negatively charged dsRNA. dsRNA-peptide complexes were prepared at different molar ratios (dsRNA/peptide ratio: 1, 1:5, 1:10, 1:20 and 1:50) and characterized by two different means, separation on agarose gel and TEM. In the agarose gel assay, gradual retardation of the dsRNA, indicative of increasing particle size or decreasing negative charge, was distinguished as the interaction ratios increased (Figures 4A, B). The fluorescence gel image in Figure 4A shows the migration of the cationic VgP-TAMRA toward the cathode. At a 1:50 dsRNA:peptide ratio, a sharp fluorescence band may be seen on the loading well, which probably indicates the formation of large dsRNA-peptide complex aggregates or charge-neutralized molecules (Figure 4A). However, at ratios of 1:5 and 1:10 a clear shift was evident in comparison to dsRNA alone (Figure 4B). The (KH)9Vg-dsRNA complex was further characterized by TEM for evaluating the morphology and particle size of the dsRNA-peptide complex at molar ratios of 1:5 and 1:10. Free dsRNA appeared as long thin filaments (Figure 4C). (KH)9VgP appeared as helical 50- to 450-nm filaments (oval shapes, Figure 4D). Incubation of the peptide with dsRNA at both indicated ratios led to the formation of almost spherical particles of 100 nm diameter, surrounded by filaments (Figures 4E, F).
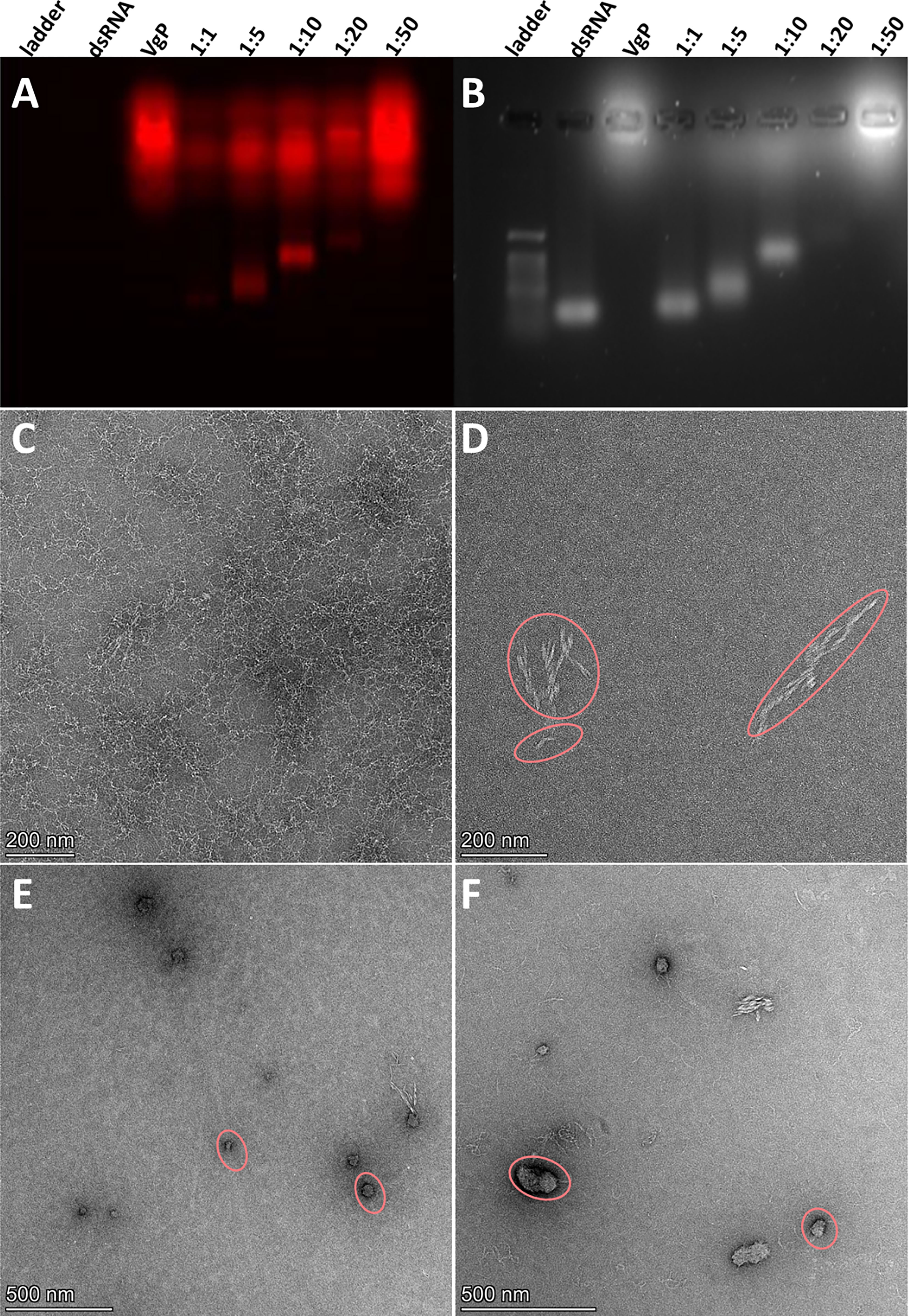
Figure 4 VgP-dsRNA complex validation and characterization. (A) Fluorescence emission (564 nm) image and (B) UV image of an agarose gel loaded with (KH)9VgP-TAMRA mixed with dsRNA. A constant amount (25 pmole) of dsRNA was mixed with VgP in increasing molar ratios of dsRNA : VgP (1:1, 1:5, 1:10, 1:20 and 1:50 as indicated on each lane). (C–F) TEM images of (C) dsRNA, (D) (KH)9VgP, (E) dsRNA complexed with (KH)9VgP in a 1:5 molar ratio, and (F) dsRNA complexed with (KH)9VgP in a 1:10 molar ratio.
The next step was to evaluate the ability of VgP to piggyback dsRNA into oocytes. For this purpose, dsRNA-FITC, which exhibits green fluorescence, was synthesized. dsRNA-FITC was then linked to (KH)9VgP-TAMRA and incubated in vitro with ovary pieces for 5 h. The green fluorescence of dsRNA-FITC can be seen within the oocyte boundary (Figure 5C, arrows), and the green puncta appear to overlap with the red fluorescence of (KH)9VgP-TAMRA, with the overlap indicating a stable complex and internalization of the two molecules (Figure 5D, arrows). However, no green fluorescence puncta could be seen in the oocytes from ovary pieces incubated with dsRNA alone (Figure 5A).
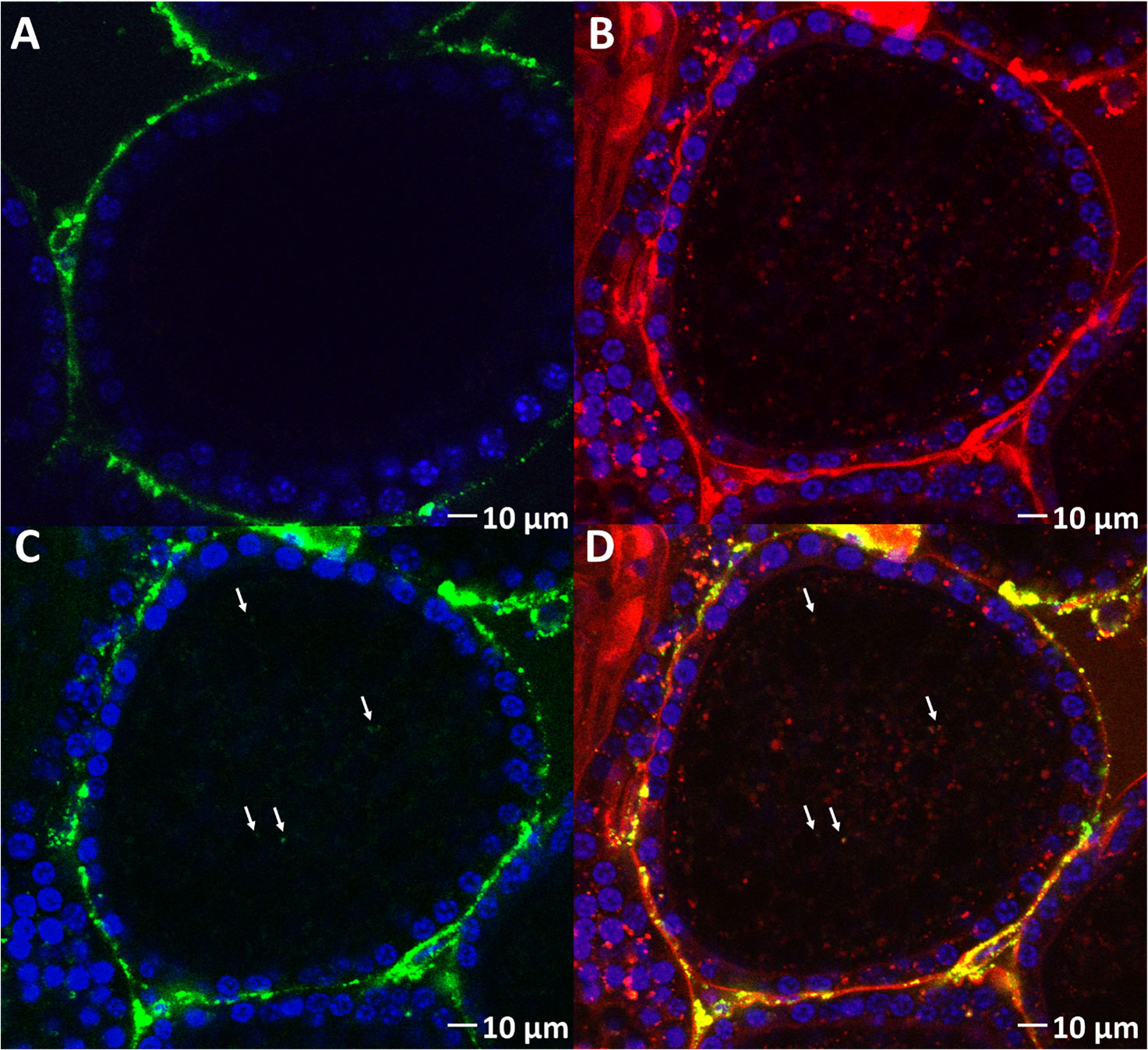
Figure 5 (KH)9VgP-TAMRA can piggyback dsRNA-FITC into oocytes in vitro. (A) Fluorescence images of oocytes from ovary pieces incubated with dsRNA-FITC. (B) Red fluorescence image of an oocyte from ovary pieces incubated with (KH)9VgP-TAMRA mixed with dsRNA-FITC. (C) Green fluorescence image of the same preparation as in B; white arrows directed toward green puncta indicate the endocytosis of dsRNA. (D) Overlay of (B, C) images; white arrows directed toward orange puncta indicate the endocytosis of dsRNA and peptide together.
dsPAX6 piggybacked into the oocyte on VgP retards eye development
To confirm that VgP can piggyback dsRNA into oocytes, which would lead to gene silencing, PAX6, which controls eye development in embryos, was selected as a candidate gene for illustrating gene silencing (Supplementary Figure 1). In M. rosenbergii embryos, PAX6 gene expression begins on day 5 (Supplementary Figure 4), and eyes appear on day 9. In our study, vitellogenic females were injected with (KH)9VgP-dsPAX6 or free dsPAX6, and the development of the embryos (on the pleopods of the injected females) was monitored. Embryos were examined one day after eye appearance, and eye development was documented (Figure 6). The embryos from (KH)9VgP-dsPAX6-injected females showed either impaired eye development or a lack of eye development. A thin eye pigment was abundant in those embryos (Figures 6A, B, black arrowhead), and maldeveloped eyes (Figures 6A–C, red arrowhead) or a complete lack of eyes were also documented (Figures 6A–C, white arrowhead). To identify embryos with retarded eye development, an eye pigment length/width index was calculated. The average ratio for embryos taken from a (KH)9VgP-dsPAX6-injected female was significantly higher (4.55 ± 0.19, p<0.0001) than that for the embryos taken from a dsPAX6-injected female (3.48 ± 0.05, Figure 6G). ROC analysis revealed that the AUC reached 0.93, which means there is a 93% chance that the model will be able to distinguish between impaired versus normally developed eyes. Indeed 87% of embryos taken from the (KH)9VgP-dsPAX6-injected female possessed length/width eye pigment value above the AUC threshold (0.93), indicating impaired eye development (Supplementary Figure 5). The eye phenotype was also examined at advanced developmental stages. A SEM investigation of the eye surface of larvae at stage 11 (11 days after hatching) from (KH)9VgP-dsPAX6-injected females revealed irregular, elongated ommatidia, with continuous or elevated hinges between ommatidia (Figures 6I, J). In contrast, the control larvae demonstrated well-shaped hexagonal ommatidia (Figure 6H).
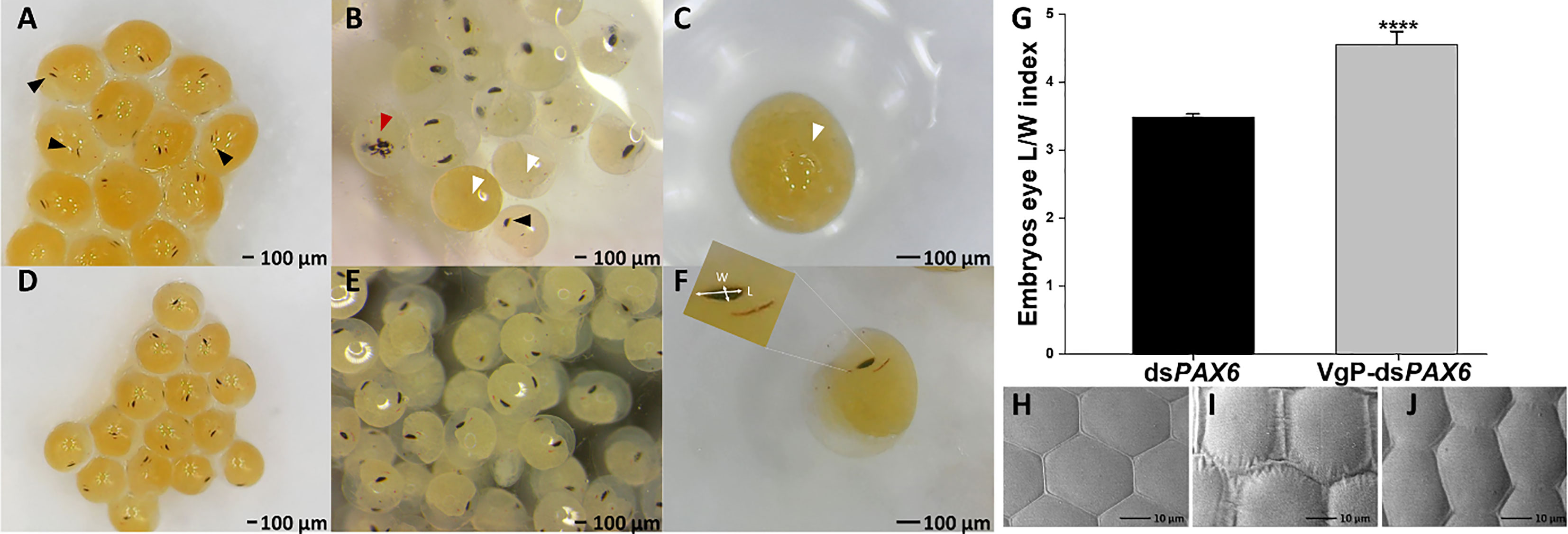
Figure 6 Vg-dsRNA complex injected into reproductive females leads to retarded eye development in the embryos and affects the shape of the ommatidium in advanced larva stages. Embryos detached from a mother injected with (KH)9VgP-dsPAX6 (A–C) or dsPAX6 (D–F). Embryos possessing remnant or no eye development (white arrowheads), partially developed eyes (black arrowheads), and embryo with a cyclops eye (red arrowhead). (G) Average length/width index of eye pigmented area of embryos (n=10 from each female) from dsPAX6- (black column, n=3) or (KH)9VgP-dsPAX6- (gray column, n=4) treated females. Asterisks indicate a significant difference between (KH)9VgP-dsPAX6 and dsPAX6 groups (P< 0.0001) four asterisks indicate high significant difference (P< 0.0001). (H–J) SEM images of the normally developed eye (H, control) and irregularly developed eye in larvae of treated females (I, J).
Discussion
Yolk accumulation inside the eggs of oviparous animals is an intensive process that last for several days spanning the onset of vitellogenesis to egg laying. The process involves receptor-mediated recognition of the yolk protein Vg and endocytosis of the ligand-receptor complex into the developing oocytes. The main aim of the current research was to exploit the intensive RME-mediated accumulation of Vg in the oocytes for piggybacking dsRNA into developing oocytes to facilitate gene silencing. Our starting point for this study was our previous work in which we characterized an 84-amino-acid stretch on the N-terminus of M. rosenbergii Vg (Roth et al., 2013). This 84-amino-acid sequence was found to be highly homologous with specific amino acid stretches of Vg sequences of other oviparous species (Roth et al., 2013). Further work involving an M. rosenbergii Vg-VgR ligand-blot interaction study revealed that a 24-amino-acid peptide, designated VgP, located at the C-terminal of the Vg-retrieved 84 amino acid stretch (Roth et al., 2013). Following this discovery, we instituted a research program directed at utilizing this specific sequence for internalizing various molecules into M. rosenbergii oocytes. In the current study, we first synthesized VgP and labeled it with the TAMRA fluorophore. The same 24-amino-acid composition was synthesized in scrambled order (scVgP) and labeled with the FITC fluorophore to enable us to differentiate between the localization of the original 24-amino-acid sequence, labeled with TAMRA (red fluorescence), and the control scrambled sequence, labeled with FITC (green fluorescence), in the same oocyte. When the two peptides were incubated in vitro with ovary slices or injected in vivo into the hemolymph of vitellogenic females, only VgP was visible as microscopic puncta inside the M. rosenbergii oocytes (Figures 2A, C, E–H, and 3A–C), supporting the notion that the peptide sequence is essential for RME into the oocytes. The microscopic puncta and their distribution resemble the shape and distribution of the yolk droplets accumulating in the oocytes during vitellogenesis. Yolk droplets generally have a small diameter upon formation early in the RME. Later, several droplets merge to form larger and denser mature droplets (Raikhel and Dhadialla, 1992). The fact that two different fluorescing molecules, FITC and TAMRA, were used could lead to the question of whether internalization is affected by the different spatial structures of these fluorophores. We note, however, that scVgP did not appear inside the oocytes, whether labeled with TAMRA (Figures 2B, D) or FITC (Figures 2F, H). These results support the premise that internalization is related mainly to the amino acid sequence and not to the labeling molecule. Therefore, it may indeed be concluded that the amino acid sequence is central to the interaction with the receptor.
In addition, MST affinity tests were performed to reveal the affinity of VgP and scVgP for the LBDs of the VgR. The scrambled peptide did not interact with either of the VgR LBDs, which corroborates the scVgP location obtained in the in-vitro assay, namely, outside the oocyte boundaries. The higher affinity of VgP for VgR-LBD-II (Kd 3.42 ± 2.16 µM) than for VgR-LBD-I (Kd 50.89 ± 26.47 µM) indicates that VgR-LBD-II plays a significant role in the ligand-receptor interaction between the N-terminal peptide of Vg and VgR. The Kd values for the Vg-VgR interaction in other species (A. aegypti, S. serrata and X. laevis) ranged between 0.18 and 1.3 µM (Opresko and Wiley, 1987; Dhadialla and Raikhel, 1991; Warrier and Subramoniam, 2002), namely, values similar to the micromolar affinity of VgP to VgR-LBDII obtained in this work.
In invertebrates, specifically in M. rosenbergii, gene silencing by dsRNA in vivo has been used to elucidate gene function and for silencing of genes relevant to biotechnology (Sagi et al., 2013). Nevertheless, direct silencing of oocyte genes in the ovary in M. rosenbergii and, indeed in crustaceans in general, has not been documented. The obstacles to gene silencing in oocytes may be understood by examining the morphology and developmental physiology of oocytes. These complex, specialized cells, which selectively store yolk proteins and other maternal factors, are surrounded, in the ovary, by accessory cells that protect and nourish them (Eckelbarger and Hodgson, 2021). The accessory cells form a barrier that regulates the internalization into the oocytes of large molecules, such as proteins and dsRNA (Conine and Rando, 2022). Upon spawning, an eggshell is formed to protect the embryo from physical and chemical insults (Mazzini et al., 1984; Tommasini and Sabelli, 1989). Therefore, the standard method currently used to expose the developing embryo to different molecules is injection directly into the fertilized egg or electroporation; both techniques have been applied successfully in different invertebrate species (Etkin et al., 1984; Houdebine and Chourrout, 1991; Xu et al., 2020). However, the major disadvantage of these methods in crustacean embryos is the low survival rates (Yazawa et al., 2005; Kato et al., 2012). Moreover, injection into individual embryos is a cumbersome and labor-intensive process and is inefficient for large populations. Therefore, for large populations, development of a specific tool facilitating delivery to tens of thousands of oocytes could be an easy way to introduce macromolecules. To develop such a tool for gene silencing, we sought to exploit the 24-amino-acid VgP that specifically interacts with VgR, by piggybacking dsRNA onto VgP for delivery into oocytes and hence into developing embryos. In the current study, we leveraged the electrostatic bonding between the negatively charged phosphate groups of dsRNA and the positively charged lysine and histidine residues (at pI 9.74 and 7.59, respectively) in the KH tail added to the VgP to form dsRNA-VgP nanoparticles that could be used as a delivery tool. Upon mixing of the KH-tailed peptide with dsRNA, retardation of dsRNA in the agarose gel was observed (Figures 4A, B), with the retardation being positively correlated with increasing amounts of the interacting KH-peptide; this retardation could be attributed to the formation of larger nanoparticles, in line with previously published studies (Unnamalai et al., 2004; Mo et al., 2012; De Schutter et al., 2022).
As mentioned above, upon interaction with the receptor, Vg is internalized into the cell via RME vesicles. Different studies indicate that RME vesicles are ≤200 nm in diameter (Rejman et al., 2004; Jafari et al., 2012), e.g., 150 nm in chicken oocytes (Pearse, 1980). The current work, aimed to exploit RME for delivery, is the first in crustaceans; thus, the diameter size of microspheres that could be internalized via Vg-VgR endocytosis in crustaceans was hitherto unknown. Theoretically, internalization via a RME vesicle would be possible as long as the nanoparticle diameter size was less than 200 nm. Thus, a size limit of ≤200 nm for dsRNA-peptide nanoparticles guided our experimental work. TEM analysis revealed spherical nanoparticles with a maximal size of 100 nm for 1:5 and 1:10 dsRNA:peptide molar ratios (Figures 4C–F). The results obtained in this study are similar to previously reported data on peptide-dsRNA interactions (Numata et al., 2014; Margus et al., 2016). However, as mentioned above, particle size would be a limitation in any attempts to exploit the RME; the results presented here indicate that dsRNA-VgP nanoparticles are indeed of a suitable size for internalization into oocytes (Figure 5) and that the internalization is most likely facilitated via the VgR. These results constitute the first report the successful internalization via RME of large dsRNA molecules (230 bp) into oocytes in general and crustacean oocytes in particular. We note that earlier works on mammalian and plant cells described the internalization of siRNA molecules (21 bp) via cell-penetrating peptides (Endoh and Ohtsuki, 2009; Presente and Dowdy, 2013). The idea of utilizing of VgR-interacting peptides to internalize proteins has been applied in earlier studies; for example, the Cas9 enzyme was recombinantly expressed with a Vg-derived peptide, and its internalization and capability to edit genes in mosquito oocytes were demonstrated (Chaverra-Rodriguez et al., 2018).
Silencing ovary-expressed genes in crustaceans has not been reported to date. Our results are the first to demonstrate the ability of the Vg-derived peptides to internalize dsRNA and silence genes in oocytes of crustaceans. As a model system, we monitored eye development following administration of dsPAX6 to oocytes via the Vg-derived delivery tool OSDel. We showed successful delivery of functional dsPAX6 and consequent gene silencing, as exemplified by the appearance of developmentally retarded eyes in the embryos of treated females (Figure 6). When vitellogenic females received the dsPAX6-OSDel, impaired eye development appeared in 87% of the embryos of the treated females (Supplementary Figure 5).
It is our belief that the novel delivery tool and the proof of concept of silencing the PAX6 gene will be useful for silencing other aquaculture-relevant genes. In the future, manipulating thousands of embryos by treating the mother animal with the OSDel during vitellogenesis will prove to be a powerful means for gene manipulation and population intervention in aquaculture.
Data availability statement
The data presented in the study are deposited in the https://www.ncbi.nlm.nih.gov/genbank/, repository, accession number OP292287.
Author contributions
SC was responsible for project administration, data curation, formal analysis, investigation, methodology, validation, visualization, and initial draft preparation. MH carried out the cloning and expression of LBD in yeast and did the MST data curation, analysis, and visualization. NF designed the time and dose-dependent methodology, validation, and visualization. IK is responsible for idea conceptualization, funding acquisition, supervision and writing review and editing. All authors contributed to the article and approved the submitted version.
Funding
This research was supported in part by the Israel innovation authority (Grant no. 70021) and by the National Institute for Biotechnology in the Negev (NIBN).
Acknowledgments
We would like to thank Uri Abdu for his advice in selecting the genes for silencing, Yair Noyman for the statistical analysis advice and evaluation, Ms. Inez Mureinik for styling the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1128524/full#supplementary-material
Abbreviations
dsPAX6, PAX6 dsRNA; dsRNA, double-stranded RNA; LBD, ligand-binding domain; LDLR, low-density lipoprotein receptor; MST, microscale thermophoresis; OSDel, oocyte-specific delivery; RME, receptor-mediated endocytosis; Vg, vitellogenin; VgP, 24 amino-acid sequence in M. rosenbergii Vg; VgR, vitellogenin receptor.
References
Aronstein K., Oppert B., Lorenzen M. D. (2011). “RNAi in agriculturally-important arthropods,” in RNA Processing. Ed. PAULA G. (Rijeka: IntechOpen).
Brown M. S., Goldstein J. L. (1976). Receptor-mediated control of cholesterol metabolism: Study of human mutants has disclosed how cells regulate a substance that is both vital and lethal. Science 191, 150–154. doi: 10.1126/science.174194
Chaverra-Rodriguez D., Macias V. M., Hughes G. L., Pujhari S., Suzuki Y., Peterson D. R., et al. (2018). Targeted delivery of CRISPR-Cas9 ribonucleoprotein into arthropod ovaries for heritable germline gene editing. Nat. Commun. 9, 3008. doi: 10.1038/s41467-018-05425-9
Cheers M. S., Ettensohn C. A. (2004). Rapid microinjection of fertilized eggs. Methods Cell Biol. 74, 287–310. doi: 10.1016/S0091-679X(04)74013-3
Cohen S., Ilouz O., Manor R., Sagi A., Khalaila I. (2021). Transcriptional silencing of vitellogenesis-inhibiting and molt-inhibiting hormones in the giant freshwater prawn, macrobrachium rosenbergii, and evaluation of the associated effects on ovarian development. Aquaculture 538, 736540. doi: 10.1016/j.aquaculture.2021.736540
Conine C. C., Rando O. J. (2022). Soma-to-germline RNA communication. Nat. Rev. Genet. 23, 73–88. doi: 10.1038/s41576-021-00412-1
Cvekl A., Callaerts P. (2017). PAX6: 25th anniversary and more to learn. Exp. Eye Res. 156, 10–21. doi: 10.1016/j.exer.2016.04.017
De Schutter K., Verbeke I., Kontogiannatos D., Dubruel P., Swevers L., Van Damme E. J. M., et al. (2022). Use of cell cultures in vitro to assess the uptake of long dsRNA in plant cells. In Vitro Cell. Dev. Biology-Plant 58, .511–520. doi: 10.1007/s11627-022-10260-1
Dhadialla T. S., Raikhel A. S. (1991). Binding of vitellogenin to membranes isolated from mosquito ovaries. Arch. Insect Biochem. Physiol. 18, 55–70. doi: 10.1002/arch.940180106
Eckelbarger K. J., Hodgson A. N. (2021). Invertebrate oogenesis–a review and synthesis: comparative ovarian morphology, accessory cell function and the origins of yolk precursors. Invertebrate Reprod. Dev. 65, 71–140. doi: 10.1080/07924259.2021.1927861
Endoh T., Ohtsuki T. (2009). Cellular siRNA delivery using cell-penetrating peptides modified for endosomal escape. Advanced Drug delivery Rev. 61, 704–709. doi: 10.1016/j.addr.2009.04.005
Etkin L. D., Pearman B., Roberts M., Bektesh S. L. (1984). Replication, integration and expression of exogenous DNA injected into fertilized eggs of xenopus laevis. Differentiation 26, 194–202. doi: 10.1111/j.1432-0436.1984.tb01395.x
Goldstein J. L., Anderson R. G., Brown M. S. (1979). Coated pits, coated vesicles, and receptor-mediated endocytosis. Nature 279, 679–685. doi: 10.1038/279679a0
Goldstein J. L., Brown M. S., Anderson R. G., Russell D. W., Schneider W. J. (1985). Receptor-mediated endocytosis: concepts emerging from the LDL receptor system. Annu. Rev. Cell Biol. 1, 1–39. doi: 10.1146/annurev.cb.01.110185.000245
Houdebine L., Chourrout D. (1991). Transgenesis in fish. Experientia 47, 891–897. doi: 10.1007/BF01929879
Huang K.-H., Wu J.-P., Wang S.-Y., Huang D.-J., Chen H.-C. (2010). Ovarian development in the freshwater prawn macrobrachium asperulum (Decapoda: Palaemonidae). J. Crustacean Biol. 30, 615–623. doi: 10.1651/09-3246.1
Huong D. T. T., Wang T., Bayley M., Phuong N. T. (2010). Osmoregulation, growth and moulting cycles of the giant freshwater prawn (Macrobrachium rosenbergii) at different salinities. Aquaculture Res. 41, e135–e143. doi: 10.1111/j.1365-2109.2010.02486.x
Jafari M., Xu W., Naahidi S., Chen B., Chen P. (2012). A new amphipathic, amino-acid-pairing (AAP) peptide as siRNA delivery carrier: physicochemical characterization and in vitro uptake. J. Phys. Chem. B 116, 13183–13191. doi: 10.1021/jp3072553
Kato Y., Matsuura T., Watanabe H. (2012). Genomic integration and germline transmission of plasmid injected into crustacean daphnia magna eggs. PLoS One 7, e45318. doi: 10.1371/journal.pone.0045318
Khalaila I., Sagi A., Cohen S. (2020). Delivery peptides and methods of using the same. PCT/IL2020/050897.
Levy T., Rosen O., Eilam B., Azulay D., Zohar I., Aflalo E. D., et al. (2017). All-female monosex culture in the freshwater prawn macrobrachium rosenbergii–a comparative large-scale field study. Aquaculture 479, 857–862. doi: 10.1016/j.aquaculture.2017.07.039
Li A., Sadasivam M., Ding J. L. (2003). Receptor-ligand interaction between vitellogenin receptor (VtgR) and vitellogenin (Vtg), implications on low density lipoprotein receptor and apolipoprotein B/E: the first three ligand-binding repeats of VtgR interact with the amino-terminal region of vtg. J. Biol. Chem. 278, 2799–2806. doi: 10.1074/jbc.M205067200
Lodé T. (2012). Oviparity or viviparity? that is the question…. Reprod. Biol. 12, 259–264. doi: 10.1016/j.repbio.2012.09.001
Margus H., Arukuusk P., Langel U., Pooga M. (2016). Characteristics of cell-penetrating peptide/nucleic acid nanoparticles. Mol. Pharmaceutics 13, 172–179. doi: 10.1021/acs.molpharmaceut.5b00598
Mazzini M., Callaini G., Mencarelli C. (1984). A comparative analysis of the evolution of the egg envelopes and the origin of the yolk. Ital. J. Zoology 51, 35–101. doi: 10.1080/11250008409439457
Mekuchi M., Ohira T., Kawazoe I., Jasmani S., Suitoh K., Kim Y. K., et al. (2008). Characterization and expression of the putative ovarian lipoprotein receptor in the kuruma prawn, marsupenaeus japonicus. Zoological Sci. 25, 428–437. doi: 10.2108/zsj.25.428
Mo R. H., Zaro J. L., Shen W. C. (2012). Comparison of cationic and amphipathic cell penetrating peptides for siRNA delivery and efficacy. Mol. Pharm. 9, 299–309. doi: 10.1021/mp200481g
Morris L. X., Spradling A. C. (2011). Long-term live imaging provides new insight into stem cell regulation and germline-soma coordination in the drosophila ovary. Development 138, 2207–2215. doi: 10.1242/dev.065508
Numata K., Ohtani M., Yoshizumi T., Demura T., Kodama Y. (2014). Local gene silencing in plants via synthetic dsRNA and carrier peptide. Plant Biotechnol. J. 12, 1027–1034. doi: 10.1111/pbi.12208
Okuno A., Yang W. J., Jayasankar V., Saido-Sakanaka H., Huong D. T. T., Jasmani S., et al. (2002). Deduced primary structure of vitellogenin in the giant freshwater prawn, macrobrachium rosenbergii, and yolk processing during ovarian maturation. J. Exp. Zoology 292, 417–429. doi: 10.1002/jez.10083
Opresko L. K., Wiley H. S. (1987). Receptor-mediated endocytosis in xenopus oocytes. i. characterization of the vitellogenin receptor system. J. Biol. Chem. 262, 4109–4115. doi: 10.1016/S0021-9258(18)61318-3
Pearse B. (1980). Coated vesicles. Trends Biochem. Sci. 5, 131–134. doi: 10.1016/0968-0004(80)90055-9
Presente A., Dowdy S. F. (2013). PTD/CPP peptide-mediated delivery of siRNAs. Curr. Pharm. design 19, 2943–2947. doi: 10.2174/1381612811319160008
Raikhel A. S., Dhadialla T. S. (1992). Accumulation of yolk proteins in insect oocytes. Annu. Rev. Entomol 37, 217–251. doi: 10.1146/annurev.en.37.010192.001245
Reid W., O'brochta D. A. (2016). Applications of genome editing in insects. Curr. Opin. Insect Sci. 13, 43–54. doi: 10.1016/j.cois.2015.11.001
Rejman J., Oberle V., Zuhorn I. S., Hoekstra D. (2004). Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem. J. 377, 159–169. doi: 10.1042/bj20031253
Roth Z., Khalaila I. (2012). Identification and characterization of the vitellogenin receptor in macrobrachium rosenbergii and its expression during vitellogenesis. Mol. Reprod. Dev. 79, 478–487. doi: 10.1002/mrd.22055
Roth Z., Weil S., Aflalo E. D., Manor R., Sagi A., Khalaila I. (2013). Identification of receptor-interacting regions of vitellogenin within evolutionarily conserved β-sheet structures by using a peptide array. Chembiochem 14, 1116–1122. doi: 10.1002/cbic.201300152
Sagi A., Manor R., Ventura T. (2013). Gene silencing in crustaceans: from basic research to biotechnologies. Genes (Basel) 4, 620–645. doi: 10.3390/genes4040620
Stifani S., Nimpf J., Schneider W. (1990). Vitellogenesis in xenopus laevis and chicken: cognate ligands and oocyte receptors. the binding site for vitellogenin is located on lipovitellin I. J. Biol. Chem. 265, 882–888. doi: 10.1016/S0021-9258(19)40132-4
Subramoniam T. (2011). Mechanisms and control of vitellogenesis in crustaceans. Fisheries Sci. 77, 1–21. doi: 10.1007/s12562-010-0301-z
Tiu S. H. K., Benzie J., Chan S.-M. (2008). From hepatopancreas to ovary: molecular characterization of a shrimp vitellogenin receptor involved in the processing of vitellogenin. Biol. Reprod. 79, 66–74. doi: 10.1095/biolreprod.107.066258
Tommasini S., Sabelli F. (1989). Eggshell origin and structure in two species of conchostraca (Crustacea, phyllopoda). Zoomorphology 109, 33–37. doi: 10.1007/BF00312181
Tufail M., Takeda M. (2009). Insect vitellogenin/lipophorin receptors: molecular structures, role in oogenesis, and regulatory mechanisms. J. Insect Physiol. 55, 88–104. doi: 10.1016/j.jinsphys.2008.11.007
Unnamalai N., Kang B. G., Lee W. S. (2004). Cationic oligopeptide-mediated delivery of dsRNA for post-transcriptional gene silencing in plant cells. FEBS Lett. 566, 307–310. doi: 10.1016/j.febslet.2004.04.018
Warrier S., Subramoniam T. (2002). Receptor mediated yolk protein uptake in the crab Scylla serrata: crustacean vitellogenin receptor recognizes related mammalian serum lipoproteins. Mol. Reprod. Dev. 61, 536–548. doi: 10.1002/mrd.10106
Xu S., Pham T., Neupane S. (2020). Delivery methods for CRISPR/Cas9 gene editing in crustaceans. Mar. Life Sci. Technol. 2, 1–5. doi: 10.1007/s42995-019-00011-4
Keywords: RNAi, dsRNA, gene silencing, receptor, endocytosis, vitellogenin, targeted-delivery
Citation: Cohen S, Hasan M, Frishman N and Khalaila I (2023) A crustacean vitellogenin-derived peptide as an oocyte-specific delivery vehicle for gene silencing. Front. Mar. Sci. 10:1128524. doi: 10.3389/fmars.2023.1128524
Received: 20 December 2022; Accepted: 06 March 2023;
Published: 16 March 2023.
Edited by:
Tomer Ventura, University of the Sunshine Coast, AustraliaReviewed by:
J. Sook Chung, University of Maryland, College Park, United StatesRaul Llera-Herrera, Mazatlan Academic Unit, National Autonomous University of Mexico, Mexico
Copyright © 2023 Cohen, Hasan, Frishman and Khalaila. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Isam Khalaila, aXNhbUBiZ3UuYWMuaWw=
 Shany Cohen
Shany Cohen Isam Khalaila
Isam Khalaila