- 1CAS Key Laboratory of Marine Ecology and Environmental Sciences, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 2Laboratory for Marine Science and Technology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao, China
- 4CAS Engineering Laboratory for Marine Ranching, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 5Shandong Province Key Laboratory of Experimental Marine Biology, Qingdao, China
- 6Qingdao Marine Management Support Center, Qingdao, China
- 7University of Chinese Academy of Sciences, Beijing, China
- 8Research and Development Center for Efficient Utilization of Coastal Bioresources, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai, China
As a vital developmental event, metamorphosis controls the population dynamics of most marine invertebrates and affects the breeding of economic shellfish. Rapana venosa is an economically important species in China, but artificial aquaculture has hampered its metamorphosis process. Previous studies have found that juvenile oysters can effectively induce the metamorphosis of R. venosa, but the specific induction mechanism is not clear. Here, we investigated the mechanism underlying the response of R. venosa to juvenile oysters through the RNA-seq analysis. In this study, the gene set responses to metamorphosis cues (juvenile oysters) in R. venosa were identified, and GO and KEGG enrichment analyses were further performed on these gene sets. The results showed that the expression of the prototype of the class of immediate early genes, the transcription factor AP-1, was rapidly and significantly increased, and the molecular chaperone of NOS, HSP90, exhibited lower expression in the M12 group than in the control group. In contrast, the expression of inhibitors of apoptosis (IAPs) was significantly increased upon exposure to juvenile oysters. Additionally, the Wnt signaling pathway and MAPK signaling pathway were enriched in the trend analysis. These pathways may also play critical regulatory roles in the response to juvenile oysters. Taken together, the results show that competent larvae rapidly respond to the inducing effects of oysters via some immediate early genes, such as the transcription factor AP-1, which may further regulate downstream pathways such as the MAPK signaling pathway to cause subsequent changes, including a decrease in HSP90 and an increase in IAPs. These changes together may regulate the metamorphosis of R. venosa. This study provides further evidence that juvenile oysters are the metamorphosis cues of R. venosa, which may enhance our understanding of the metamorphosis mechanism in this marine invertebrate.
1. Introduction
Rapana venosa is an economically important gastropod in China. Recently, with the increasing market demand, fishing of R. venosa has intensified, which has severely damaged wild resources. These situations have attracted widespread attention from domestic researchers, and the artificial culture industry of R. venosa will continue to develop in the future (Yu et al., 2020). However, R. venosa is a successful worldwide invader, including in the Black Sea, the Mediterranean Sea, the Adriatic and Aegean seas, the coasts of France and the Netherlands, the Chesapeake Bay on the Atlantic coast of the United States, and the Río de la Plata between Uruguay and Argentina, and it causes great damage to the wild resources of oysters and other bivalves in the above areas (Xue et al., 2018). This problem has also attracted widespread concern. Therefore, the invasion mechanism urgently needs to be elucidated.
Given the special ecological status of this species, in recent years, there have been a number of studies on the behavior, reproduction and development, environmental suitability, ecological effects, and technology of artificial breeding and culturing of R. venosa (Song et al., 2016a; Song et al., 2016b; Song et al., 2016c; Zhang et al., 2017; Xu et al., 2019; Yu, 2019; Yang et al., 2020a; Yang et al., 2020b; Zhang et al., 2020; He et al., 2021; Shi et al., 2022; Yang et al., 2022). However, we have found that the metamorphosis rate of R. venosa has become extremely low in its early development stage with the development of the artificial breeding and culturing industry, resulting in a low breeding survival rate (Yu et al., 2020). This low survival rate severely restricts the development of this industry.
Metamorphosis is critical for most marine invertebrates due to the sensitivity and vulnerability of organisms during this time, which affect the population dynamics and resource recruitment of marine invertebrates (Song et al., 2021). Therefore, it is very important to reveal the regulatory mechanisms of metamorphosis for the development of the artificial culture industry and the illumination of the invasion mechanism. In previous studies, we have found that the metamorphosis rate of R. venosa increases by more than 60% when juvenile oysters (Crassostrea gigas, with a shell length less than 3 cm) are present, although the metamorphosis process cannot be completed spontaneously (Cavalcanti et al., 2020). Meanwhile, Xu et al. (2019) indicated that oyster reefs can significantly promote the recovery of R. venosa resources. This may partly explain why these mollusks are destroying oyster resources. However, the specific mechanism by which oysters induce metamorphosis of R. venosa larvae is not clear.
Considering the importance of metamorphosis, its regulatory mechanism has been widely researched in species including sponges, tubeworm, coral and mollusks, especially in bivalves and herbivorous gastropods (Cavalcanti et al., 2020). Most of these species use cues from bacteria. For example, Song et al. (2021) showed that in the marine sponge Amphimedon queenslandica, bacterial symbionts can play a critical role in animal development by providing their host with the arginine needed for larval settlement and metamorphosis. Shikuma et al. (2016) found that larvae of the tubeworm Hydroides elegans metamorphose in response to surface-bound Pseudoalteromonas luteoviolacea bacteria, further inducing the regulation of mitogen-activated protein kinase (MAPK) signaling. Conspecific adults and food are also important cues of metamorphosis in some species. The metamorphosis of Mytilopsis sallei and Balanus amphitrite is stimulated by conspecific adults (Matsumura et al., 1998; He et al., 2021). Metamorphosis is usually food-stimulated in some gastropods, including the herbivorous gastropods Haliotis rufescens and Crepidula fornicata (Morse et al., 1979; Taris et al., 2010) and the carnivorous gastropod Onchidoris bilamellata, whose metamorphosis cues are juvenile barnacles (Rodriguez et al., 1993).
As R. venosa is a typical carnivorous gastropod, its metamorphosis cue is also its food, juvenile oysters (Yu et al., 2020), consistent with that in O. bilamellata. We have also performed many studies on the metamorphosis of R. venosa, including studies on its morphology, behavior, enzyme kinetics, symbiotic microbiota, metabolome, proteome, transcriptome and critical genes (Song et al., 2016a; Song et al., 2016b; Song et al., 2016c; Zhang et al., 2017; Yu et al., 2020; Yang et al., 2020a; Yang et al., 2020b; Shi et al., 2022; Yang et al., 2022). However, a comprehensive analysis of the metamorphosis regulation mechanism in R. venosa has not been conducted.
Transcriptomics has been widely applied to investigate the mechanisms underlying metamorphosis in invertebrates, revealing the critical pathways and genes that regulate metamorphosis and providing novel insights (Shikuma et al., 2016; He et al., 2021). Herein, to further investigate the mechanism by which juvenile oysters induce the metamorphosis of R. venosa larvae, we obtained the mRNA expression profiles of competent larvae exposed to juvenile oysters for 2 hours (M2) and 12 hours (M12) and before exposure to juvenile oysters (control, Con) via sequencing on an Illumina NovaSeq 6000 platform. Furthermore, we identified the differentially expressed mRNAs (differentially expressed genes, DEGs) and performed trend analysis of gene expression based on their transcripts per million (TPM) values to further screen out the genes that were consistently regulated during exposure to juvenile oysters. Then, these genes were subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses. The findings of this study will enhance understanding of the metamorphosis mechanism of R. venosa, which may not only aid in the development of artificial breeding and culturing but also reveal the invasion mechanism and support the protection of wild populations of oysters and other bivalves.
2. Materials and methods
2.1. Larval culture and sample collection
Culturing of R. venosa larvae and sample collection were performed according to Yang et al. (2022), and the competent larvae (shell height > 1,200 μm) were used to conduct the assay. Three cement pools (3.5 × 5.2 × 1.5 m) were used to culture these larvae, and the assay was performed with a larval density of 0.1 ind/mL. First, we randomly collected 100 larvae from each pool as the Con group. Then, the juvenile oysters (Crassostrea gigas) were placed evenly in pools (50 oysters on each scallop, added about 25000 scallop shells to each pool), and 100 larvae were collected at 2 and 12 hours thereafter from each pool as the oyster-exposed groups (M2 and M12, respectively). During the whole process of experiment and sampling, the three pools were closed with no water in or out, and maintain a stable experimental conditions (23 ± 1°C, 30‰ salinity, 7mg/L dissolved oxygen). A total of nine samples were obtained from the Con, M2 and M12 groups, which were washed with PBS and stored at -80°C for RNA extraction.
2.2. RNA extraction, library preparation and sequencing
Total RNA was extracted from the larval samples using TRIzol® Reagent (Invitrogen, USA) according the manufacturer’s recommendations, and 1% agarose gels were used to monitor the degradation and contamination of RNA. The integrity and quantification of RNA samples were evaluated using a 2100 Bioanalyzer (Agilent Technologies, USA), and RNA samples with integrity numbers (RINs) > 8.0 were used for construction of an mRNA library. The library was prepared using a TruSeq™ RNA Sample Preparation Kit (Illumina, USA) and sequenced on an Illumina NovaSeq 6000 platform (Majorbio. Shanghai, China), and 125-bp paired-end reads were generated and deposited in the Sequence Read Archive (ncbi.nlm.nih.gov/sra) under BioProject PRJNA855327.
2.3. Read mapping and quantification of gene expression levels
The raw reads were trimmed and quality-controlled via SeqPrep and Sickle with default parameters, and the clean reads were separately aligned to the reference genome of R. venosa (unpublished data) with orientation mode using HISAT2 software (Kim et al., 2015). The mapped reads of each sample were assembled by StringTie via a reference-based approach (Pertea et al., 2015), and then the paired-end clean reads were aligned to the reference genome of R. venosa (unpublished data) using TopHat v2.0.12. HTSeq v0.6.1 was used to count the reads mapped to each gene. Known genes and novel transcripts were identified by Reference Annotation Based Transcript (RABT) assembly method, the Cufflinks v2.1.1. Subsequently, the TPM of each gene was calculated based on the length of the gene and the number of reads mapped to the gene.
2.4. Differential expression analysis and trend analysis of genes
DEG analysis was conducted among the three groups, and the TPM algorithm was used to normalize the expression level of each transcript. RSEM was used to quantify gene abundances. Briefly, DEG analysis was performed using the DESeq2 R package with the significance thresholds set to a P-adjusted value < 0.05 and a |log2FC| ≥ 1. Additionally, after filtering out the genes with TPM = 0 and normalizing the TPM by log2 transformation, the trend analysis of R. venosa transcriptome data was performed on the Majorbio Cloud Platform by Short Time-series Expression Miner (STEM) software.
2.5. Functional enrichment analysis
To further explore the biological functions of the DEGs and these genes with significant trends, GO and KEGG enrichment analyses were performed. GO enrichment analysis was implemented using the GOseq package in R, and KOBAS was used to analyze the statistical enrichment of genes in KEGG pathways. GO terms and KEGG pathways with the significance enrichment threshold set to a P-adjusted value < 0.05 corrected by the Bonferroni method.
2.6. Verification by quantitative real-time PCR (qRT−PCR)
To validate the accuracy of the RNA-Seq profiling results, critical genes were selected for qRT−PCR analysis. The cDNA for qRT−PCR was synthesized using the Prime Script™ RT Reagent Kit with gDNA Eraser (TaKaRa, Japan). The primers used in the mRNA qRT−PCR assay were designed using Primer 5, and 60S ribosomal protein L28 (RL28) was selected as a housekeeping gene to normalize the data (Song et al., 2017). A SYBR PrimeScript RT−PCR Kit II (TaKaRa, Japan) was used to quantify the expression levels. The relative expression levels of genes were estimated by the 2−ΔΔCT method. All data are presented as the means ± SEs (N = 3). Statistical significance was analyzed using SPSS v.19, with a P value < 0.05 considered to indicate significance.
3. Results
3.1. Overview of RNA-seq and quantitative RT-PCR validation
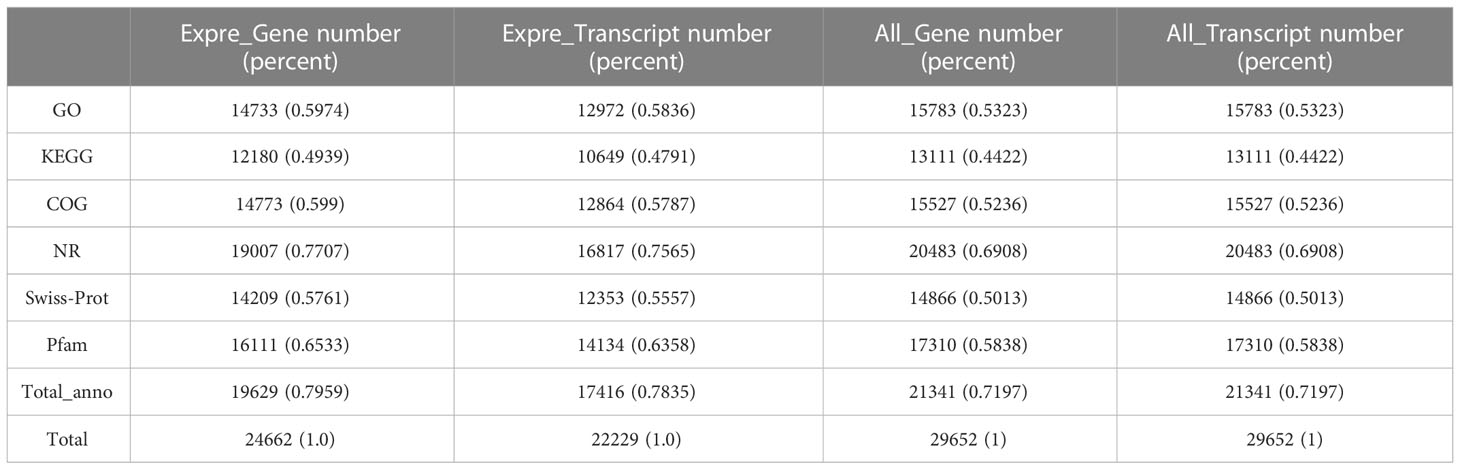
Nine libraries of R. venosa larvae were constructed from the Con group and oyster-exposed groups (M2 and M12), from which 63.84 Gb of clean reads were generated. Each cDNA library was greater than 6.09 Gb in size (Q30 > 93.36%), and the rate of successful mapping to the reference genome ranged from 75.32 to 76.71% (Table 1). A total of 37,493 genes were obtained, including 24,662 known genes and 12,831 novel genes. Gene function annotations showed that 19,629 genes (79.59% of known genes) had significant matches in the COG, GO, KEGG, KOG, Pfam, SwissProt, eggNOG, or NR databases (Table 2). Nine random genes were then chosen to verify the accuracy of the transcriptome sequence data by qRT-PCR. The results were shown in Figure 1. Altogether, the transcriptome data were consistent with the qRT-PCR results, which further reinforced our findings.
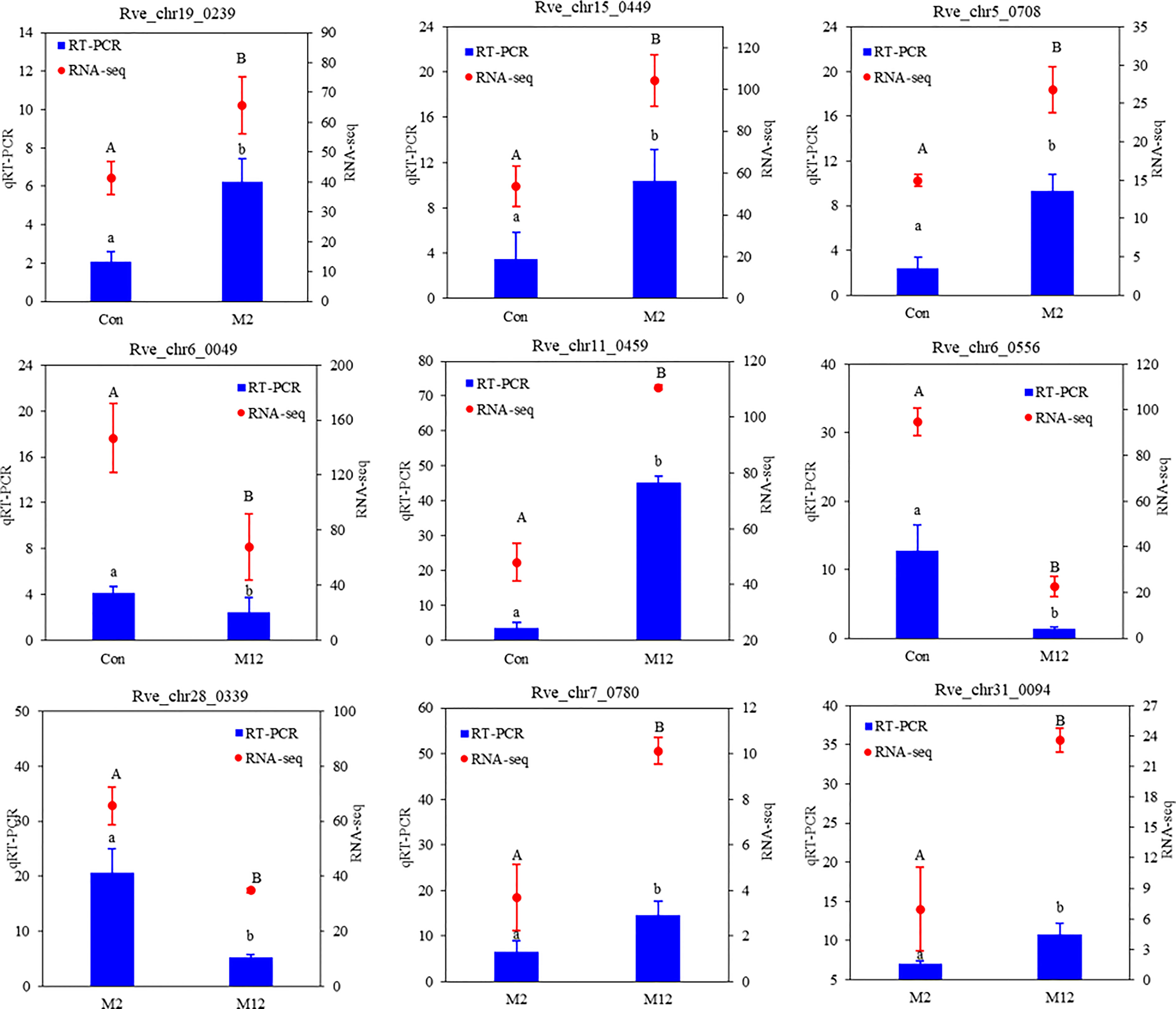
Figure 1 Validation of the 9 random DEGs involved in different pairwise comparisons by qRT-PCR. RL28 was selected to normalize the gene expression levels. The data are shown as means ( ± SE) of three replicates. Different superscripted letters indicate significant differences (P < 0.05).
3.2. Sample clustering and heatmap analysis
Hierarchical clustering analysis (HCA) showed that the groups exposed to oysters for 12 hours (M12) were clustered as one branch, while the group exposed to oysters for 2 hours (M2) was clustered with the Con group (Figure 2A). Additionally, principal component analysis (PCA) showed that all three groups were significantly separated from each other (Figure 2B). These results may mean that exposure to oysters has a significant effect on the mRNA profile of R. venosa larvae and that the effect increases with time.
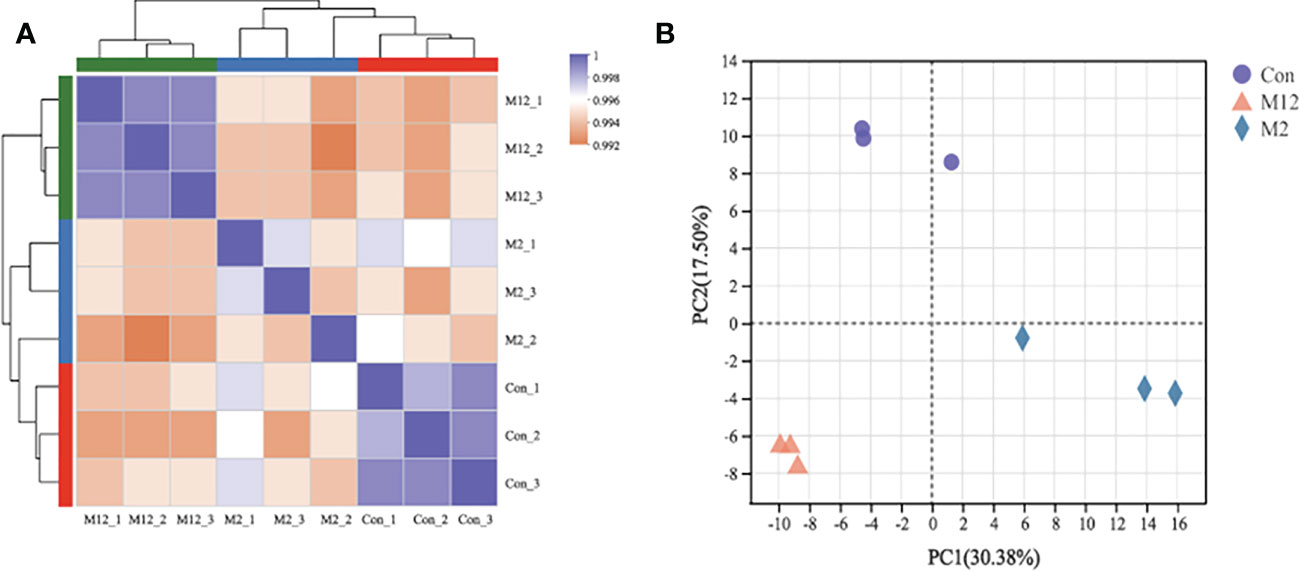
Figure 2 Relationships analysis between samples: Hierarchical clustering analysis (A) and principal component analysis (B).
3.3. Differential expression analysis and functional enrichment analysis
Differential expression analysis of genes among the three groups was also performed based on the TPM value. The DEGs in the Con vs. M2, Con vs. M12 and M2 vs. M12 comparisons were regarded as those induced by juvenile oysters but participating in different time periods of the induced-regulation process. In total, there were 189 DEGs in Con vs. M2, 597 DEGs in Con vs. M12 and 744 DEGs in M2 vs. M12 (Figure 3A). Additionally, a Venn diagram showed that 42 DEGs were shared between the Con vs. M2 and Con vs. M12 comparisons, 55 DEGs were shared between the Con vs. M2 and M2 vs. M12 comparisons, and 292 DEGs were shared between the Con vs. M12 and M2 vs. M12 comparisons (Figure 3B). In contrast, 92 DEGs were unique to Con vs. M2 comparisons, 263 DEGs were unique to Con vs. M12 comparisons, and 397 DEGs were unique to M2 vs. M12 comparisons.
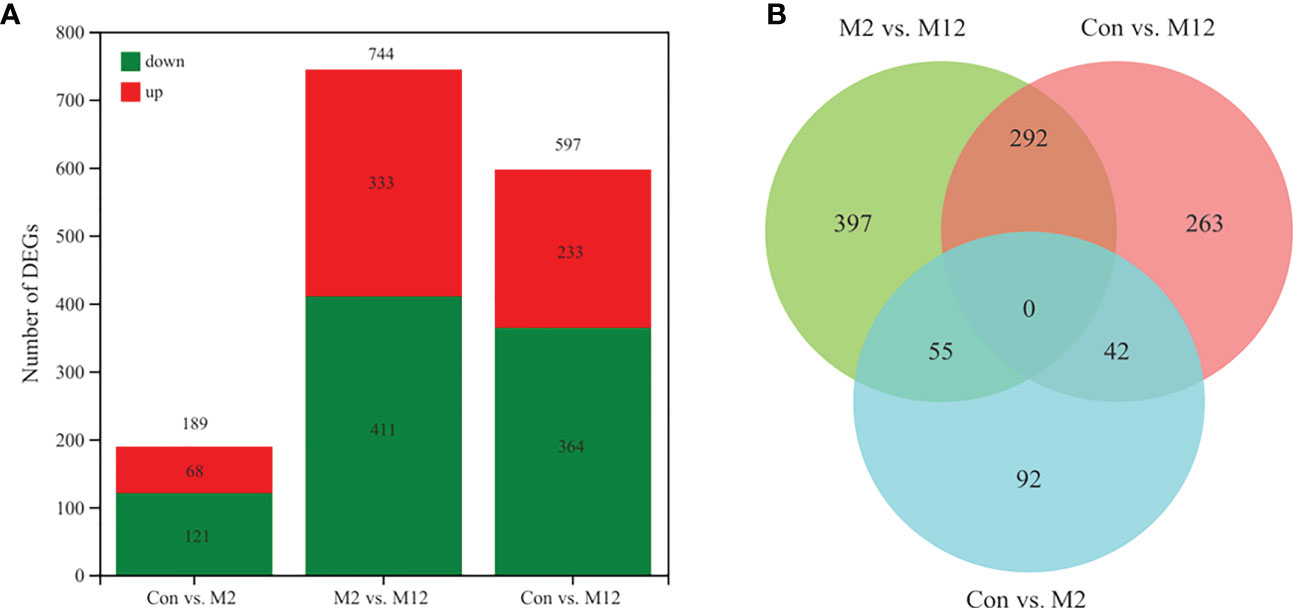
Figure 3 Statistics of differentially expressed genes (A) and Venn diagram of differentially expressed genes (B).
The 189 DEGs in the Con vs. M2 comparison were regarded as the initial-response genes (I-DEGs) to oyster presence, which were significantly enriched with 2 GO terms: DNA-binding transcription factor activity and transcription regular activity (Figure 4A). There were no significantly enriched pathways (P adjust < 0.05). We further analyzed the expression of these genes enriched with the 2 GO terms. As shown in Figure 4B, 12 genes were upregulated in the M2 group compared with the Con group, including some nuclear receptor genes (nuclear receptor subfamily 1 and nuclear hormone receptor E75), CREBRF, the transcription factor Maf/MafK/AP-1, dendritic arbor reduction protein 1, cAMP-responsive element-binding protein, CCAAT/enhancer-binding protein beta and transforming protein v-Fos/v-Fox. Five other genes were downregulated in the M2 group, including the transcription factor MafAa, forkhead box protein B1, vitamin D3 receptor, deformed epidermal autoregulatory factor 1 and nuclear factor interleukin-3-regulated protein.
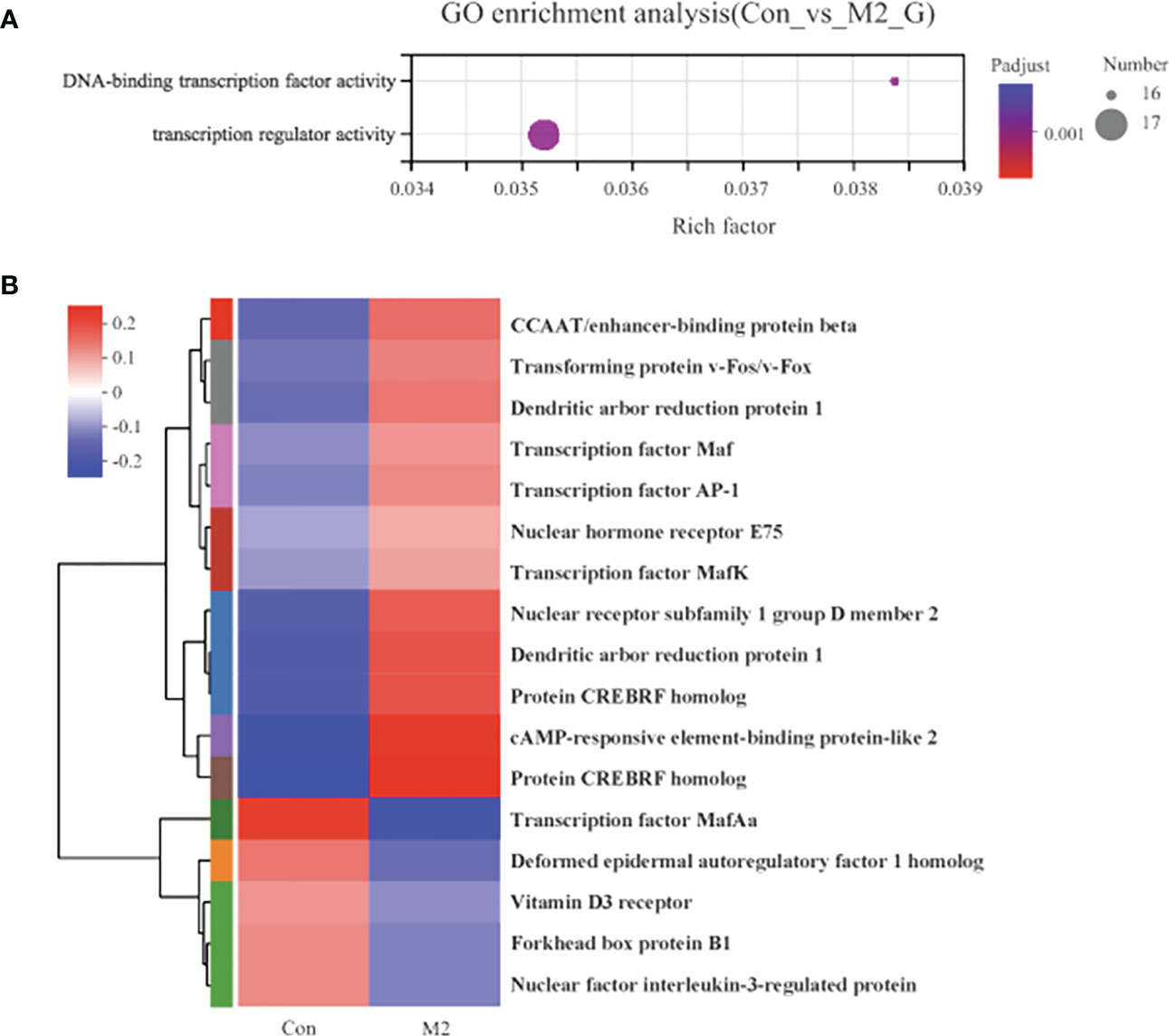
Figure 4 The functional enrichment and expression analysis of initial-response genes to oyster induction. (A) GO enrichment results of 92 DEGs in Con vs. M2 comparison. (P adjust < 0.05). (B) Heat map of the expression analysis of 17 DEGs in GO:0003700 (DNA-binding transcription factor activity) and GO:0140110 (transcription regulator activity).
The 597 DEGs in Con vs. M12 were regarded as the final-response genes (F-DEGs) to oyster presence, which were significantly enriched in 3 pathways, including ribosome biogenesis in eukaryotes, antigen processing and presentation, and RNA transport (P adjusted < 0.05) (Figure 5A). There were 36 DEGs in the 3 pathways, and all of them were downregulated in the M12 group compared with the Con group, which suggested that these 3 pathways were suppressed in the presence of oysters or were inhibited by oyster exposure. There were 36 DEGs in the 3 pathways, including many heat shock proteins (HSPs), such as HSP70 proteins (heat shock protein 68, heat shock 70 kDa protein A1, heat shock cognate 71 kDa protein and so on) and HSP90 proteins (97 kDa heat shock protein and heat shock protein HSP 90-beta) (Figure 5C). These DEGs were significantly enriched with 52 GO terms (top 20 were shown), and the small-subunit processome term was the most enriched (Figure 5B).
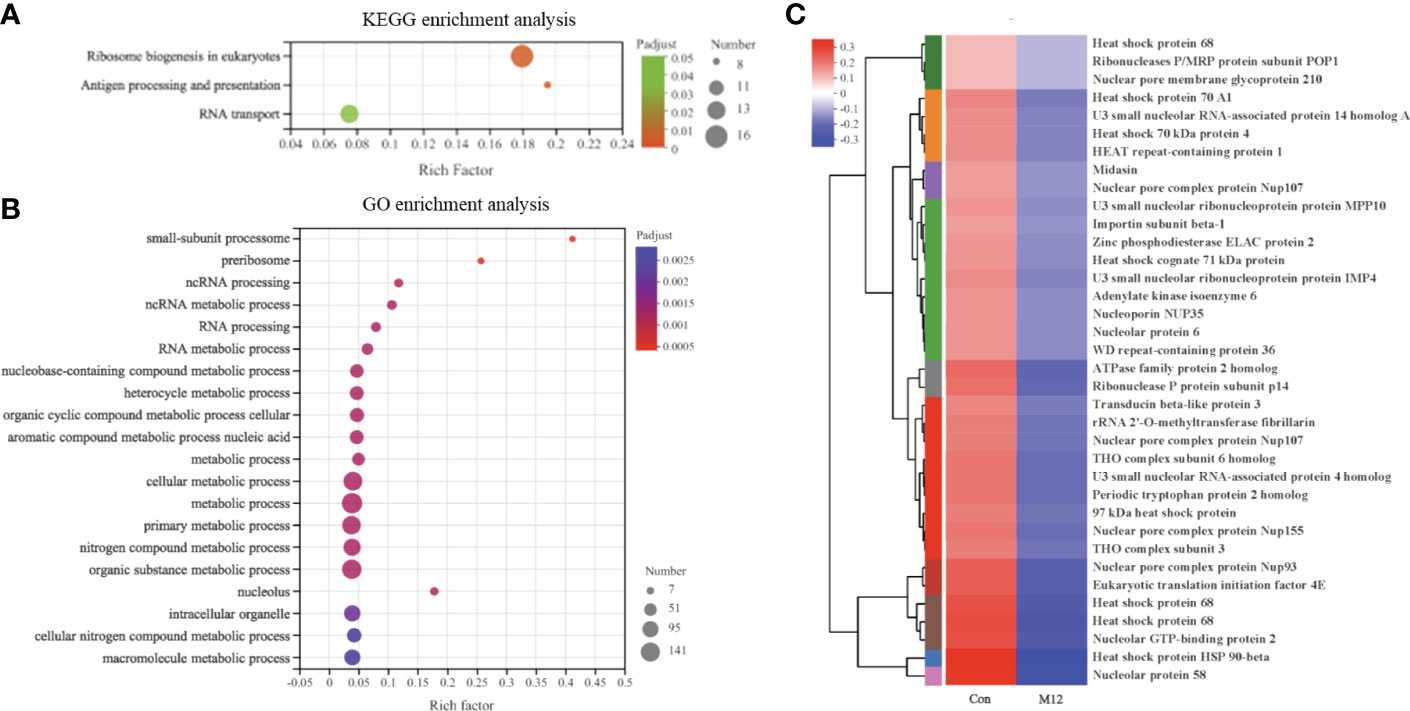
Figure 5 The functional enrichment and expression analysis of final-response genes to oyster induction. (A) KEGG enrichment results of 597 DEGs in Con vs. M12 comparison. (P adjust < 0.05). (B) GO enrichment results of 597 DEGs in Con vs. M12 comparison. (P adjust < 0.05). (C) Heat map of the expression analysis of 36 DEGs in the enriched pathways: “Ribosome biogenesis in eukaryotes”, “Antigen processing and presentation” and “RNA transport”.
In addition to the initial-response and final-response genes that were altered in response to oysters, 397 DEGs existed only in the M2 vs. M12 comparison. These genes may respond to oyster presence slowly and ultimately not significantly change but still have an important regulatory effect; therefore, they should not be ignored. We regarded them as late-response genes (L-DEGs) to oyster exposure. The 397 DEGs were significantly enriched with 3 GO terms, including rRNA metabolic process, ncRNA metabolic process and rRNA processing. Furthermore, we analyzed the expression of 14 genes enriched with 3 GO terms (Figure 6A). All 14 genes were downregulated in the M12 group compared with the M2 group, which may mean that the metabolic processes related to rRNA and ncRNA were slowed down in the later stage in the presence of oysters (Figure 6B).
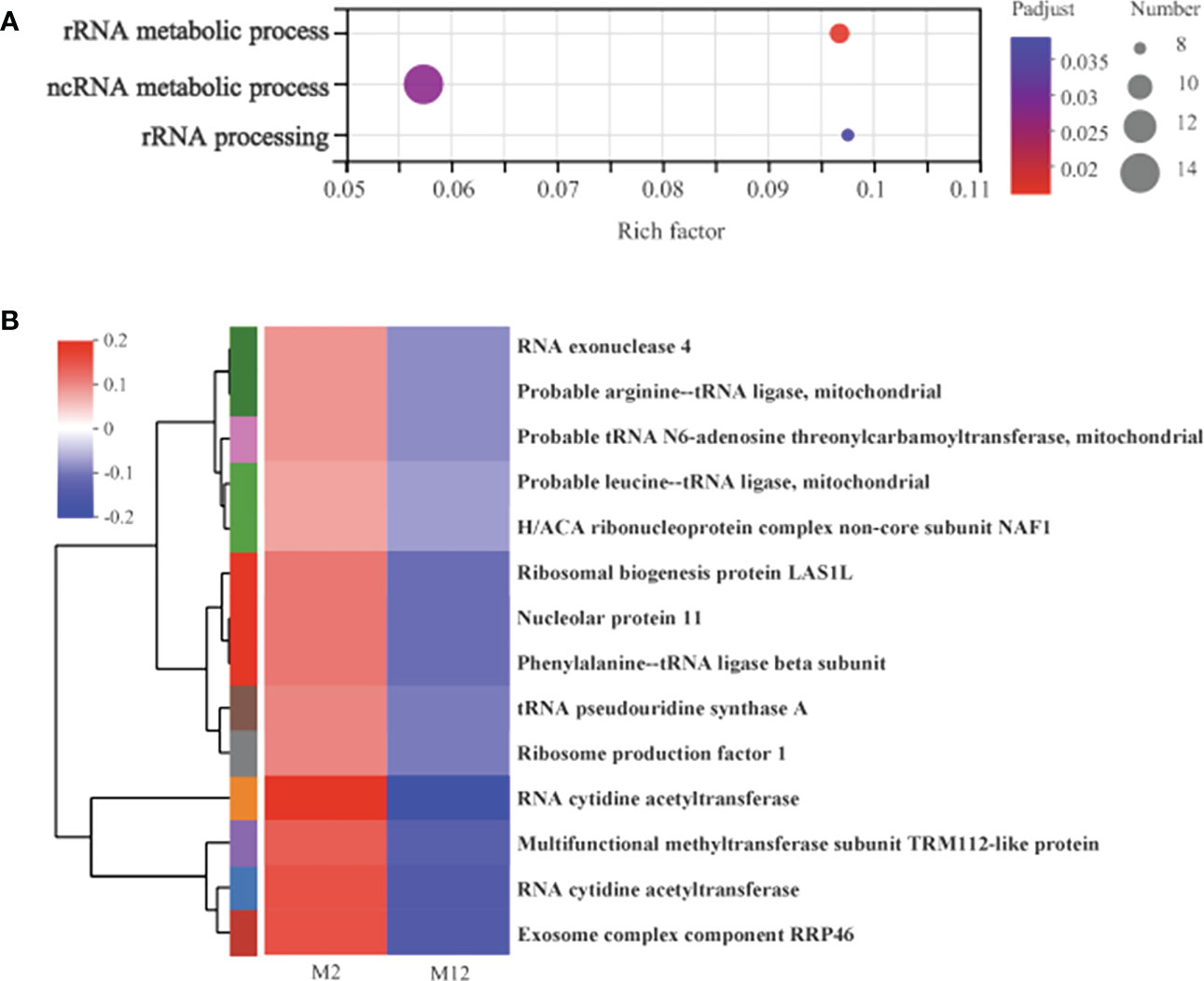
Figure 6 The functional enrichment and expression analysis of late-response genes to oyster induction. (A) GO enrichment results of 397 DEGs in M2 vs. M12 comparison. (P adjust < 0.05). (B) Heat map of the expression analysis of 14 DEGs in GO:0016072 (rRNA metabolic process) and GO:0034660 (ncRNA metabolic process), GO:0006364 (rRNA processing).
3.4. Analysis of gene expression trends and functional enrichment analysis
A total of 22,662 genes were used to perform trend analysis, and 8 model profiles were used to summarize the expression patterns. The genes in the same profile presented similar expression patterns. As shown in Figure 7A, 3 profiles labeled with different colors were statistically significant, including profile 2 (3563 genes), profile 4 (1308 genes), and profile 1 (751 genes). These genes with different expression trends may have different functions in the metamorphosis induced by oysters. To further understand the functions of these genes in the 3 profiles, GO and KEGG enrichment analyses were performed. The top 20 significantly enriched GO terms and pathways are illustrated. For the genes in profile 2, 10 of the significantly enriched pathways were in the “human disease” category, 5 were in the “organismal systems” category, and only one was in the “cellular process” category. Four were in the “Environmental Information Processing” category, including the Ras signaling pathway (62 genes with adjusted P < 0.05), the Wnt signaling pathway (46 genes with adjusted P < 0.05), the MAPK signaling pathway (62 genes with adjusted P < 0.05), and the Rap1 signaling pathway (64 genes with adjusted P < 0.05) (Figure 7B). In addition, the genes in profile 2 were significantly enriched in 10 GO terms belonging to the “biological process” category, 1 belonging to the “cellular component” category, and 9 belonging to the “molecular function” category, which also contained the Wnt signaling pathway (22 genes with adjusted P < 0.05) (Figure 7C). For the genes in profile 4, only one pathway, autophagy - animal (18 genes with adjusted P < 0.05), was significantly enriched, and no significantly enriched GO term was identified. For the genes in profile 1, there were also no significantly enriched GO terms or pathways. Furthermore, we analyzed the expression of the genes in the Wnt signaling pathway and animal autophagy.
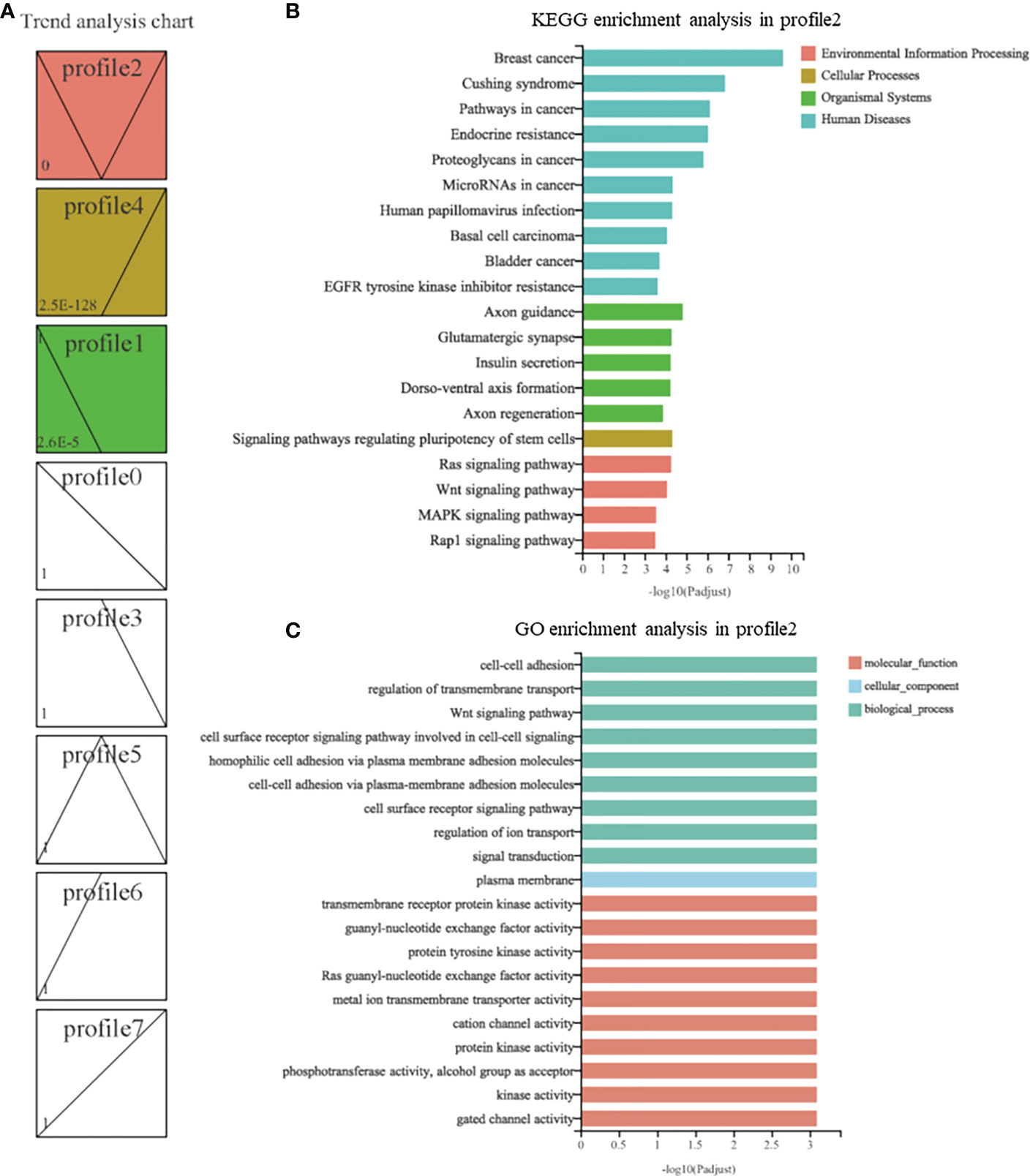
Figure 7 Trend analysis of genes in R. venosa transcriptome. (A) Different expression patterns of all genes. Each box represents a model profile. The upper number in the box represents the serial number of profiles and the lower one is the P value. Boxes with P values <0.05 are colored. GO (B) and KEGG (C) enrichment analysis of genes in profile 2.
4. Discussion
The food or prey of some animals not only provides them with the necessary nutrients for growth and development but also plays an important role in their development at a critical time. One of the most dramatic examples is when the food or prey induces animal metamorphosis (Morse et al., 1979; Rodriguez et al., 1993; Taris et al., 2010). Although representative species from diverse taxa undergo food-induced metamorphosis, little is known about the processes that occur within the animals as they detect the cues produced by their food. To further understand the mechanism by which juvenile oysters induce the metamorphosis of R. venosa larvae, we performed RNA-seq and analyzed changes in gene expression at different time points upon exposure to metamorphosis cues, the juvenile oysters. Then, we conducted functional enrichment for these genes and identified the critical signaling pathways in the process. The present results may provide new insights into the regulatory mechanisms of metamorphosis in marine invertebrates.
In the present study, GO enrichment analysis revealed that 189 DEGs that responded early to the presence of juvenile oysters were significantly enriched with two GO terms: DNA-binding transcription factor activity and transcription regulator. These genes included some critical transcription factor genes, such as Maf, AP-1, MafK, MafAa, and transforming protein v-Fos/v-Fox, and most of the genes were upregulated. A previous study has also indicated that these genes are Jun or Fos genes (transcription factor AP-1 gene family), which are prototypical immediate early genes. Immediate early genes are characterized by a rapid and transient activation of transcription in response to changes of environmental conditions, such as growth factors, cytokines, tumor promoters, carcinogens, and expression of certain oncogenes (Angel and Hess, 2010). The transcription factor AP-1 gene family participates in many pathways, such as the MAPK signaling pathway, Wnt signaling pathway and apoptosis, which have been proven to be important in the metamorphoses of marine invertebrate animals (Angel and Hess, 2010). Therefore, we speculate that the transcription factor AP-1 may be a critical initiator in competent larvae of R. venosa in response to the metamorphosis cues of juvenile oysters that regulates other subsequent processes.
A previous study has indicated that the most critical members of the class of protein kinases regulating the activity of AP-1 in response to extracellular stimuli are MAPKs (Angel and Hess, 2010). In addition, researchers have shown that the MAPK signaling pathway is induced by the metamorphosis cue in tubeworm Hydroides elegans and regulates the downstream process of metamorphosis (Shikuma et al., 2016; Wang and Qian, 2010), which also occurs in other marine invertebrate animals, such as corals, ascidians and barnacles (Chambon et al., 2007; He et al., 2012; Siboni et al., 2014). Interestingly, we also found that the MAPK signaling pathway was significantly enriched in the gene set of profile 2 in the trend analysis, which may mean that in competent larvae of R. venosa, MAPK signaling participates in the response to metamorphosis cues.
Chambon et al. (2007) found that the MAPK signaling pathway regulates gene networks that stimulate metamorphosis and apoptosis in tail tissues of ascidian tadpoles, and apoptosis occurs during metamorphosis in almost all invertebrates, including R. venosa (Song et al., 2016a). In the present study, we found that the expression of IAPs was significantly increased upon exposure to juvenile oysters. IAP expression is also upregulated after metamorphosis in juveniles of R. venosa (Shi, 2022). IAPs are important regulators that participate in the sophisticated molecular machineries in cells that guard against inappropriate or premature apoptosis to counter various stimuli capable of triggering death (Song et al., 2021). IAPs are also highly upregulated in response to various environmental stresses, including heat, air exposure, and low-salinity stress, in some marine invertebrate animals (Hu et al., 2022). The presence of oysters can also be considered an environmental factor for competent R. venosa larvae, and the upregulation of IAPs in the present results indicates that apoptosis is tightly regulated during the metamorphosis of R. venosa. The observed upregulation also suggests that apoptosis is induced by the presence of oysters during the metamorphosis of R. venosa. Furthermore, we found that the transcription factor AP-1 affected the expression of some proapoptotic genes, including p53, Fas, Fas-L, Bim and HRK, which supports the findings of a previous study showing that AP-1 target genes are involved in the regulation of apoptosis (Angel and Hess, 2010).
Past studies have revealed that, as a molecular chaperone of nitric oxide synthase (NOS), HSP90 is also a negative regulator (repressor) of metamorphosis in a diverse range of marine invertebrates (Bishop and Brandhorst, 2001; Bishop et al., 2001; Hens et al., 2006; Taris et al., 2009). However, Ueda and Degnan (2014) had a different discovery: that inhibition of NOS reduces rates of metamorphosis, which means that NO facilitates, rather than represses, induction of metamorphosis in Haliotis asinina. Additionally, Ueda and Degnan (2014) tested the hypothesis of NOS-HSP90 interaction at the initiation of settlement and metamorphosis because they found that the spatial and temporal expression trends of NOS and HSP90 were not consistent. In the present study, we found that HSP90 genes were significantly downregulated after exposure to oysters for 12 hours (Figure 4C), which might be considered to be a signal of metamorphosis induced by the oysters, but the specific function of HSP90 needs further investigation.
In addition to HSP90 genes, several HSP70 genes were also significantly downregulated in oysters (Figure 4C). Similar trends of downregulation were observed in a study on gene expression during the metamorphosis of R. venosa (Shi et al., 2022). HSP70 first received attention for its role in the response to heat shock (Welch, 1993) and has also been found to be critical for the folding and assembly of other cellular proteins (Ritossa, 1962). HSP70 genes also participate in the regulation of the MAPK signaling pathway (Patel, 2009), which has been widely reported to participate in the regulation of metamorphosis in several marine invertebrate animals mentioned above. Porto et al. (2018) reported that HSP70 can act as a signaling molecule, modulating MAPK downstream signaling during memory consolidation in the hippocampus. Patel (2009) also indicated that HSP70 plays a role in the induction of p38MAPK in animals to respond differently to high osmotic stress. Therefore, we speculate that HSP70 may also participate in the MAPK signaling pathway and be related to the metamorphosis of R. venosa; however, the specific effect of HSP70 on metamorphosis needs further investigation, especially since no previous studies have reported the relationship between HSP70 and metamorphosis in marine invertebrates.
The Wnt signaling pathway, which has been widely studied, regulates cell proliferation, migration, differentiation and survival during animal development as well as the maintenance of homeostasis and regeneration (Liu et al., 2008; De Robertis, 2010; Clevers and Nusse, 2012). In the trend analysis, we found that the Wnt signaling pathway was significantly enriched in profile 2, a gene set that was decreased at the early stage during exposure to juvenile oysters. This finding may suggest that the Wnt signaling pathway participates in the metamorphosis of R. venosa. Chandramouli et al. (2013) also found Wnt signaling pathway enrichment during the larval metamorphosis of the polychaete worm Pseudopolydora vexillosa in transcriptome studies. Furthermore, Yue et al. (2012) have reported that Wnt signaling pathways may be involved in the metamorphosis of the bryozoan Bugula neritina. However, the specific function of the Wnt signaling pathway in the metamorphosis of marine invertebrates needs further investigation.
5. Conclusion
In this study, the gene set responses to metamorphosis cues (juvenile oysters) in R. venosa (I-DEGs and F-DEGs) were identified, and GO and KEGG enrichment analyses were further performed on these gene sets. We found that DNA-binding transcription factor activity and transcription regulator activity were enriched for the I-DEGs, such as the transcription factor AP-1, which is a prototypical immediate early gene that rapidly and transiently activates transcription in response to changes in environmental conditions. The molecular chaperone of NOS, HSP90, exhibited lower expression in the M12 group than in the Con group, while the expression of IAPs was significantly increased upon exposure to juvenile oysters. Additionally, the Wnt signaling pathway and MAPK signaling pathway were enriched in the trend analysis. These pathways may also play critical regulatory roles in the response to juvenile oysters. Taken together, the results show that competent larvae rapidly respond to the presence of oysters by expressing some immediate early genes, such as the transcription factor AP-1. These genes may further regulate downstream pathways such as the MAPK signaling pathway and result in subsequent changes, including a decrease in HSP90 and an increase in IAPs. These changes together may regulate the metamorphosis of the larvae.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving animals were reviewed and approved by the Animal Welfare Committee of the Institute of Oceanology, Chinese Academy of Sciences.
Author contributions
TZ and HS conceived and designed the experiments. M-JY and HS conducted the experiments. M-JY analyzed the data. YS, Z-SL, PS, ZH, CZ, P-PH and Z-LY contributed reagents, materials, and analytical tools. M-JY wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (Grant No. 42206086 & 31972814 & 32002409 & 32002374), the China Postdoctoral Science Foundation (Grant No. 2021M703248), the China Agriculture Research System of MOF and MARA, the Young Elite Scientists Sponsorship Program by CAST (Grant No. 2021QNRC001), the ‘Double Hundred’ Blue Industry Leader Team of Yantai (Recipient: Tao Zhang), and the Creative Team Project of the Laboratory for Marine Ecology and Environmental Science, Qingdao National for Marine Science and Technology (no. LMEESCTSP-2018). The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1122668/full#supplementary-material
References
Angel P., Hess J. (2010). The multi-gene family of transcription factor AP-1. Handb. Cell Signaling (Second Edition) 152 (1), 2059–2068. doi: 10.1016/B978-0-12-374145-5.00251-5
Bishop C. D., Bates W. R., Brandhorst B. P. (2001). A. regulation of metamorphosis in ascidians involves NO/cGMP signaling and HSP90. J. Exp. Zool. 289 (6), 374–384. doi: 10.1002/jez.1019
Bishop C. D., Brandhorst B. P. (2001b). NO/cGMP signaling and HSP90 activity represses metamorphosis in the sea urchin lytechinus pictus. Biol. Bull. 201 (3), 394–404. doi: 10.2307/1543617
Cavalcanti G. S., Alker A. T., Delherbe N., Malter K. E., Shikuma N. J. (2020). The influence of bacteria on animal metamorphosis. Annu. Rev. Microbiol. 74, 137–158. doi: 10.1146/annurev-micro-011320-012753
Chambon J. P., Nakayama A., Takamura K., McDougall A., Satoh N. (2007). ERK- and JNK signalling regulate gene networks that stimulate metamorphosis and apoptosis in tail tissues of ascidian tadpoles. Development 134 (6), 1203–1219. doi: 10.1242/dev.002220
Chandramouli K. H., Sun J., Mok. F. S. Y., Liu L. L., Qiu J. W., Ravasi T., et al. (2013). Transcriptome and quantitative proteome analysis reveals molecular processes associated with larval metamorphosis in the polychaete pseudopolydora vexillosa. J. Proteome Res. 12 (3), 1344–1358. doi: 10.1021/pr3010088
Clevers H., Nusse R. (2012). Wnt/β-catenin signaling and disease. Cell 149, 1192–1205. doi: 10.1016/j.cell.2012.05.012
De Robertis E. M. (2010). Wnt signaling in axial patterning and regeneration: lessons from planaria. Sci. Signal. 3 (127), pe21. doi: 10.1126/scisignal.3127pe21
He L. S., Xu Y., Matsumura K., Zhang Y., Zhang G., Qi S. H., et al. (2012). Evidence for the involvement of p38 MAPK activation in barnacle larval settlement. PloS One 7 (10), e47195. doi: 10.1371/journal.pone.0047195
He J., Wu Z. W., Chen L. Y., Dai Q., Hao H. H., Su P., et al. (2021). Adenosine triggers larval settlement and metamorphosis in the mussel Mytilopsis sallei through the ADK-AMPK-FoxO pathway. ACS Chem. Biol. 16 (8), 1390–1400. doi: 10.1021/acschembio.1c00175
Hens M. D., Fowler K. A., Leise E. M. (2006). Induction of metamorphosis decreases nitric oxide synthase gene expression in larvae of the marine mollusc Ilyanassa obsoleta (Say). Biol. Bull. 211, 208–211. doi: 10.2307/4134543
Hu Z., Feng J., Song H., Zhou C., Yu Z. L., Yang M. J., et al (2022). Mechanisms of heat and hypoxia defense in hard clam: Insights from transcriptome analysis. Aquaculture 549, 737792.
Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Meth. 12, 357–360. doi: 10.1038/nmeth.3317
Liu Y., Wang X., Lu C. C., Kerman R., Steward O., Xu X. M., et al. (2008). Repulsive wnt signaling inhibits axon regeneration after CNS injury. J. Neurosci. 28 (33), 8376–8382. doi: 10.1523/JNEUROSCI.1939-08.2008
Matsumura K., Nagano M., Fusetani N. (1998). Purification of a larval settlement-inducing protein complex (SIPC) of the barnacle, Balanus amphitrite. J. Exp. Zool. 281, 12–20. doi: 10.1002/(SICI)1097-010X(19980501)281:1<12::AID-JEZ3>3.0.CO;2-F
Morse D. E., Hooker N., Duncan H., Jensen J. (1979). γ-aminobutyric acid, a neurotransmitter, induces planktonic abalone larvae to settle and begin metamorphosis. Science 204, 407–410. doi: 10.1126/science.204.4391.407
Patel P. L. (2009). Role of p38MAPK, heat shock proteins, HSP27 and HSP70 in osmotic stress in renal vs. blood cells: A comparative study (University of the Sciences in Philadelphia).
Pertea M., Pertea G. M., Antonescu C. M., Chang T. C., Mendell J. T., Salzberg S. L., et al. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Porto R. R., Dutra F. D., Crestani A. P., Holsinger R. M. D., Quillfeldt J. A., Bittencourt P. D., et al. (2018). HSP70 facilitates memory consolidation of fear conditioning through mapk pathway in the hippocampus. Neuroscience 375, 108–118. doi: 10.1016/j.neuroscience.2018.01.028
Ritossa F. (1962). A new puffing pattern induced by temperature shock and DNP in drosophila. Experientia 18 (12), 571–573. doi: 10.1007/BF02172188
Rodriguez S. R., Ojeda F. P., Inestrosa N. C. (1993). Settlement of benthic marine invertebrates. Mar. Ecol. Prog. Ser. 97, 193–207. doi: 10.3354/meps097193
Shi P. (2022). Effects of autophagy and apoptosis on the regulation of metamorphosis and seedling of Rapana venosa. Institute Oceanol. Chin. Acad. Sci. doi: 10.1111/1749-4877.12675
Shi P., Song H., Jie F., Yang M. J., Yu Z. L., Hu Z., et al. (2022). Molecular response and developmental speculations in metamorphosis of the veined rapa whelk, Rapana venosa (Valenciennes 1846). Integr. Zool. doi: 10.1111/1749-4877.12675
Shikuma N. J., Antoshechkin I., Medeiros J. M., Pilhofer M., Newman D. K. (2016). Stepwise metamorphosis of the tubeworm hydroides elegans is mediated by a bacterial inducer and MAPK signaling. PNAS 113 (36), 10097–10102. doi: 10.1073/pnas.1603142113
Siboni N., Abrego D., Motti C. A., Tebben J., Harder T. (2014). Gene expression patterns during the early stages of chemically induced larval metamorphosis and settlement of the coral Acropora millepora. PloS One 9 (3), e91082. doi: 10.1371/journal.pone.0091082
Song H., Dang X., He Y. Q., Zhang T., Wang H. Y. (2017). Selection of housekeeping genes as internal controls for quantitative rt-pcr analysis of the veined rapa whelk (rapana venosa). PeerJ 5 (7), e3398. doi: 10.7717/peerj.3398
Song H., Hewitt O. H., Degnan S. M. (2021). Arginine biosynthesis by a bacterial symbiont enables nitric oxide production and facilitates larval settlement in the marine-sponge host. Curr. Biol. 31, 1–5. doi: 10.1016/j.cub.2020.10.051
Song H., Wang H. Y., Zhang T. (2016b). Comprehensive and quantitative proteomic analysis of metamorphosis-related proteins in the veined rapa whelk, rapana venosa. Int. J. Mol. Sci. 17, 924. doi: 10.3390/ijms17060924
Song H., Yu Z. L., Sun L. N., Xue D. X., Zhang T., Wang H. Y., et al. (2016c). Transcriptomic analysis of differentially expressed genes during larval development of rapana venosa by digital gene expression profiling. G3-Gene. Genom. Genet. 6, 2181–2193. doi: 10.1534/g3.116.029314
Song H., Zhang T., Hadfield M. G. (2021). Metamorphosis in warming oceans: a microbe-larva perspective. Trends Ecol. Evol. 36, 976–977. doi: 10.1016/j.tree.2021.07.010
Song H., Zhang T., Yu Z. L., Sun L. N., Xue D. X., Zhang T., et al. (2016a). Metabolomic analysis of competent larvae and juvenile veined rapa whelks (rapana venosa). Mar. Biol. 163, 145. doi: 10.1007/s00227-016-2919-6
Taris N., Comtet T., Stolba R., Lasbleiz R., Pechenik J. A., Viard F., et al. (2010). Experimental induction of larval metamorphosis by a naturally-produced halogenated compound (dibromomethane) in the invasive mollusc Crepidula fornicata (L.). J. Exp. Mar. Biol. Ecol. 393, 71–77. doi: 10.1016/j.jembe.2010.07.001
Taris N., Comtet T., Viard F. (2009). Inhibitory function of nitric oxide on the onset of metamorphosis in competent larvae of Crepidula fornicata: a transcriptional perspective. Mar. Genom. 2, 161–167. doi: 10.1016/j.margen.2009.08.002
Ueda N., Degnan S. M. (2014). Nitric oxide is not a negative regulator of metamorphic induction in the abalone haliotis asinine. Front. Mar. Sci. 1, 21. doi: 10.3389/fmars.2014.00021
Wang H., Qian P. Y. (2010). Involvement of a novel p38 mitogen-activated protein kinase in larval metamorphosis of the polychaete Hydroides elegans (Haswell). J. Exp. Zool. Part B. 314 (5), 390–402. doi: 10.1002/jez.b.21344
Welch W. J. (1993). How cells respond to stress. Sci. Am. 268 (5), 56–56. doi: 10.1038/scientificamerican0593-56
Xu M., Qi L., Zhang L. B., Zhang T, Yang H. S., Zhang Y. L. (2019). Ecosystem attributes of trophic models before and after construction of artificial oyster reefs using ecopath. Aquacult. Env. Interac. 11, 111–127. doi: 10.3354/aei00284
Xue D. X., Graves J., Carranza A., Sylantyev S., Snigirov S., Zhang T., et al. (2018). Successful worldwide invasion of the veined rapa whelk, Rapana venosa, despite a dramatic genetic bottleneck. Biol. Invasions 20, 3297–3314. doi: 10.1007/s10530-018-1774-4
Yang M. J., Song H., Feng J., Yu Z. L., Shi P., Liang J., et al. (2022). Symbiotic microbiome and metabolism profiles reveal the effects of induction by oysters on the metamorphosis of the carnivorous gastropod Rapana venosa. Comput. Struct. Biotec. 20, 1–14. doi: 10.1016/j.csbj.2021.11.041
Yang M. J., Song H., Yu Z. L., Bai Y. C., Hu Z., Hu N., et al. (2020a). Expression and activity of critical digestive enzymes during early larval development of the veined rapa whelk, Rapana venosa (Valenciennes 1846). Aquaculture 519, 734722. doi: 10.1016/j.aquaculture.2019.734722
Yang M. J., Song H., Yu Z. L., Hu Z., Zhou C., Wang X. L., et al. (2020b). Changes in symbiotic microbiota and immune responses in early development stages of Rapana venosa (Valenciennes 1846) provide insights into immune system development in gastropods. Front. Microbiol. 11, 1265. doi: 10.3389/fmicb.2020.01265
Yu Z. L. (2019). A study on the behavior of the early developmental stage of Rapana venosa. Doctoral dissertation Institute of Oceanology, Chinese Academy of Science.
Yu Z. L., Yang M. J., Song H., Hu Z., Zhou C., Wang X. L., et al. (2020). Settlement and metamorphosis of Rapana venosa (Gastropoda: Muricidae) with implications for artificial culture. J. Oceanol. Limnol. 38, 249–259. doi: 10.1007/s00343-019-9107-8
Yue H. W., Wang H., Ravasi T., Qian P. Y. (2012). Involvement of wnt signaling pathways in the metamorphosis of the bryozoan Bugula neritina. PLoS One 7 (3), e33323. doi: 10.1371/journal.pone.0033323
Zhang T., Song H., Bai Y. C., Sun J. C., Zhang X. F., Ban S. J., et al. (2017). Effects of temperature, salinity, diet and stocking density on development of the veined rapa whelk, Rapana venosa (Valenciennes 1846) larvae. Aquacult. Int. 25, 1577–1590. doi: 10.1007/s10499-017-0140-3
Keywords: rapana venosa, metamorphic cue, RNA-Seq, juvenile oyster, trend analysis
Citation: Yang M-J, Shi Y, Lin Z-S, Shi P, Hu Z, Zhou C, Hu P-P, Yu Z-L, Zhang T and Song H (2023) RNA-seq analysis reveals the effect of the metamorphic cue (juvenile oysters) on the Rapana venosa larvae. Front. Mar. Sci. 10:1122668. doi: 10.3389/fmars.2023.1122668
Received: 13 December 2022; Accepted: 27 January 2023;
Published: 13 February 2023.
Edited by:
Pedro Morais, Florida International University, United StatesCopyright © 2023 Yang, Shi, Lin, Shi, Hu, Zhou, Hu, Yu, Zhang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Zhang, emhhbmd0YW9jYXNAMTI2LmNvbQ==; Hao Song, aGFvc29uZ0BxZGlvLmFjLmNu
 Mei-Jie Yang
Mei-Jie Yang Ying Shi6
Ying Shi6 Pu Shi
Pu Shi Zhi Hu
Zhi Hu Zheng-Lin Yu
Zheng-Lin Yu Tao Zhang
Tao Zhang Hao Song
Hao Song