
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 01 March 2023
Sec. Marine Molecular Biology and Ecology
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1113278
This article is part of the Research Topic Marine Omics in a Changing Ocean: Modelling Molecular Pathways and Networks to Understand Species Acclimation and Adaptation View all 10 articles
Mangroves are an important component of coastal wetland ecosystems, and low temperature is the main factor that limits their extension to higher latitudes. Kandelia obovata as one of the most cold-tolerant species in mangrove ecosystems can provide basis for the northward migration of mangrove ecosystems. We took K. obovata seedlings from Zhoushan (Zhoushan, Zhejiang, China) as the research object in this study. Transcriptome sequencing based on the Illumina HiSeqTM 2500 platform was performed to compare the transcriptome changes of roots, stems, and leaves before and after freezing and to reveal the molecular mechanisms of frost resistance. A total of 1560, 370, and 416 genes were differentially expressed in the roots, stems, and leaves before and after cold snaps, respectively. Among these differentially expressed genes, 13 positive and negative regulators were attributed to the CBL-CIPK signaling network and MAPK cascade, which might be related to the frost resistance mechanism of K. obovata Transcription factors such as AP2/EREBP and bHLH were involved in regulating the synthesis pathways of ethylene, cytokinin, growth hormone, and flavonoids. Results provide new insights into the frost resistance mechanism of K. obovata seedlings.
Mangrove forests are woody plant communities that are naturally distributed throughout the tropics and subtropics and are a unique ecosystem in the sea-land transition zone, in which marine organisms, such as birds, fish, shrimps, and shellfish thrive. Moreover, mangroves can promote siltation, protect beaches and purify the environment (Wang et al., 2011). Mangrove ecosystems play an irreplaceable role in protecting coastal wetland ecosystems and biodiversity and maintaining coastal ecological balance. They are also important ecological security systems in the coastal region of southeastern China (Wang et al., 2011; Su et al., 2019). Among mangrove plants, Kandelia obovata has the highest natural distribution latitude in China and is one of the most cold-resistant mangrove species (Wang et al., 2011). In the 1950s, this species was successfully transplanted from the northernmost part of its natural distribution in Fujian Province (27° 17′ N) to further up north in Yueqing in the Zhejiang Province (28° 20′ N) (Su et al., 2019; Fei et al., 2021b). However, the persistent phenomenon of climate change has led to a tendency for mangrove species to expand to higher latitudes. The introduction of mangrove K. obovata has been recorded in the Zhoushan of Zhejiang Province (29°30′N) (Osland et al., 2013; Zheng C. et al., 2016). During their northward journey, mangroves are susceptible to extreme weather conditions.
In the context of global climate change, the frequency and intensity of extreme weather, particularly in the tropical-temperate transition zone, limits plant productivity and affects species distribution, ecosystem structure, and ecosystem function (Chen et al., 2017). The tendency of North Atlantic and East Asian mangroves to spread northward is significantly influenced by the frequency and severity of cold snaps (Cavanaugh et al., 2014; Cavanaugh et al., 2015; Osland et al., 2017). Cold stress is divided into chilling stress (0 °C-15 °C) or freezing stress (<0 °C). Chilling stress may impede the growth and development of plants, and extended exposure to temperatures below 0 °C can cause ice crystals to accumulate in the cytoplasm of plant cells. This phenomenon can dehydrate cells or rupture cell membranes or walls, causing irreparable harm to the plant (Shi et al., 2018). The various physiological and metabolic processes that occur in plants at low temperatures are very intricate regulatory processes (Guy, 1990). Numerous investigations into the physiological and molecular processes underlying the hypothermic response of K. obovata have been conducted and yielded insightful results (Peng et al., 2015; Liu et al., 2019; Wang et al., 2019; Fei et al., 2021a). For example, the KoWRKY40 gene in WRKY transcription factors plays key roles in responses to biotic and abiotic stresses (Liu et al., 2019; Du Z. K. et al., 2022). Among the C-repeat binding factor (CBF), KoCBF1 and KoCBF3 may have functions in plant growth and cold resistance and are involved in responses to salinity and heavy metal stress (Peng et al., 2020). Ko-Aquaporins (KoAQPs) aid in the transport of water and solutes for adaptation to coastal intertidal habitats and have complex responses to environmental factors, such as salt and temperature (Guo et al., 2022). KoOsmotin, a class of proteins with osmotic functions, is highly induced in K. obovata leaves during cold stress (Fei et al., 2021a). KoHsp70, stress-inducible heat shock protein gene, plays a role in protective response to cold stress in K. obovata (Fei et al., 2015). Moreover, the photosynthetic system, enzymes, and non-enzymatic antioxidants of K. obovata are extensively involved in stress response at low temperatures to increase the plant’s potential for cold adaptation (Peng et al., 2015; Chen et al., 2017). Gene expression regulation tends to respond rapidly to external environmental changes by regulating protein abundance. The six levels of plant gene expression control include mRNA degradation control, protein activity control, RNA processing control, RNA transport, localization control, transcriptional control, and translational control. As transcriptional regulation is a multi-faceted, deep-level process, further research is needed in this field (Alberts, 2015). Although the gene expression profiles of many tissues have been disclosed by sequencing of critical genes for cold adaptation in adult K. obovata, information on the two important cold stress nodes of the B-like protein-interacting protein kinase (CBL-CIPK) network and mitogen-activated protein kinase (MAPK) cascade has not been completely reported (Su et al., 2019).
The CBL-CIPK network plays a key role in cold stress response, where CBL proteins interact with CBL-interacting protein kinases (CIPK) or SOS Ras/Rho Guanine Nucleotide Exchange Factor 2 (SOS2) protein kinase to form different signaling cascades. A complex CBL-CIPK network can regulate Ca2+ transport in response to low temperatures and freezing injury to plant cells (Batistic and Kudla, 2004; Kolukisaoglu et al., 2004; Buchwal et al., 2020). Ca2+ affects the cold tolerance of plant cells by improving the stability of the cell wall and promoting the secretion of peroxidase (Batistic and Kudla, 2004; Kolukisaoglu et al., 2004). The typical MAPK cascade signaling network is composed of three specific protein kinases, namely, MAPK kinase kinases (MKKKs), MAPK kinases (MKKs), and MAPKs. These protein kinases are activated by sequential phosphorylation at conserved activation sites, and the interaction between SaMKK2 and SaMAPK4/7 increases the plant’s resistance to cold temperatures (Teige et al., 2004; Chen et al., 2022). Currently, the CBL-CIPK network and the MARK cascade have been isolated from many plants, and their regulatory pathways have been well studied. However, the regulated transcriptional signaling pathways of K. obovata are still poorly understood, especially during its seedling growth phase.
Due to large-scale ecological restoration operations along the Chinese coast, K. obovata has been introduced as a non-native species in Zhoushan beyond the limits of their natural range. However, the area is more susceptible to extreme cold snaps. Extreme cold events have a significant effect on the abundance and productivity of mangrove plants. However, as plants grow and mature, adults have stronger resistance and recovery from cold events than seedlings. Given that the survival and development of seedlings often determine the rate at which the population grows, additional research is required on mechanisms by which K. obovata withstand cold events during northern migration (Coldren and Proffitt, 2017). In this context, field-based experiments will be more helpful in understanding how K. obovata seedlings react to cold events or freezing damage because extreme cold snap events are relatively rare. It is difficult to replicate a natural environment similar to freezing stress on K. obovata seedlings in an artificial environment. (Osland et al., 2015). In this study, transcriptome changes were examined in the root, stem, and leaf tissues of K. obovata seedlings transplanted in the Zhoushan intertidal zone before and after a cold snap. Given the relevance of CBL-CIPK network and MAPK signaling pathways on the cold tolerance of plants, we then identified the genes involved in these pathways in K. obovata and compared in detail the gene expression of these two pathways before and after the cold snap to provide insights into the freezing resistance mechanisms of K. obovata seedlings under extremely cold events.
K. obovata embryos were collected from Lei Zou of Guangdong in March 2020 and temporarily nurtured in an outdoor nursery in the Zhoushan area for 6 months before being moved to the intertidal zone of the Zhoushan main island in September 2020 (Figure S1). We transplanted 300 seedlings on about 600 m2 of tidal flat. We determined the sampling time range according to Zhoushan’s local historical meteorological data. To predict the best sampling times, we followed the weather forecast to know when a cold snap would occur. Three tissues including the root, stem, and leaf were freshly sampled from different healthy K. obovata seedlings on December 29 and December 31 before and after the cold snaps (Figure S2). The lowest temperature registered during the cold snap was -2.10°C, measured with an automatic temperature recorder (HOBO MX2201) placed on the sampling sites for the period (2020.12.16-2021.01.13). After collection, samples were washed with water, dried with paper, and stored in liquid nitrogen in an ultra-low temperature refrigerator at -80°C. At least three biological replicates were collected from the field at each sampling time, with each plant representing an independent biological replicate. In total, we analyzed 18 samples (3 biological replicates × 2 sampling times × 3 tissues).
Extraction was performed according to the manufacturer’s instructions using the RNeasy Plant RNA Extraction Kit (Qiagen, Dusseldorf, Germany) to extract RNA from each of the six unique tissues mentioned above. RNA concentration was measured using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). NanoPhotometer® spectrophotometer (IMPLEN, CA, USA) was used to check RNA purity.
We constructed cDNA library and conducted RNA sequences at Biomarker Biotechnology Corporation in Beijing, China. About 3 μg of RNA per sample was used for RNA preparation. A poly-T oligo-attached magnetic bead method was used to purify mRNA from total RNA. Dividing the mRNA with divalent cations was conducted at elevated temperatures using NEBNext First-Strand Synthesis Reaction Buffer (5×). The first-strand cDNA was synthesized using random hexamer primers and M-MuLV reverse transcriptase (RNase H-). DNA polymerase I and RNase H were used to synthesize cDNA on the second strand while exonucleases and polymerases were used to turn the remaining overhangs into blunt ends. To prepare samples for hybridization, the 3′ ends of the DNA fragments were adenylated and ligated to NEBNext adaptors with hairpin loop structures. In the following step, 150–200 bp-long cDNA fragments were selected from the library and purified using a Beckman Coulter Agencourt AMPure XP system (Brea, California, USA). The size-selected, adaptor-ligated cDNA was added with 3 μL of USER enzyme (New England Biolabs, Ipswich, MA, USA) at 37 °C for 15 min followed by 5 min at 95 °C. PCR analysis was performed using Phusion High-Fidelity DNA polymerase (Thermo Fisher, Waltham, MA, USA), universal PCR primers, and index (X) primer. A Bioanalyzer 2100 was used to evaluate the quality of the library constructed from the PCR products purified with the AMPure XP system. Clustering of index-coded samples was performed using a cBot Cluster Generation System and a TruSeq PE Cluster Kit v3-cBot-HS (Illumina, San Diego, CA, USA). The manufacturer’s instructions were followed for all experimental procedures. Using the Illumina Hiseq 2000 platform, paired-end reads were generated from the library preparations.
Raw data in fastq format were processed using in-house Perl scripts. Clean data were obtained by removing low-quality reads and those containing the adapters or poly-N sequences. We checked the quality of the unassembled read dataset by examining Q20, Q30, GC-content, and sequence duplication. All the downstream analyses were performed using high-quality clean data.
Genes from 18 transcriptomes were previously annotated in seven public databases including NR (NCBI, Non-Redundant Protein Database), Swiss-Prot, GO (Gene Ontology), Pfam database, KOG database, COG (Clusters of Orthologous Groups), and KEGG (Kyoto Encyclopedia of Genes and Genomes). Based on the annotation results of 18 transcriptomes, the unigene annotation information of the K. obovata seedling transcriptome was screened and analyzed. FPKM method was used to calculate gene expression based on the annotation results. The relative measure of transcript abundance was fragmented per kilobase of transcript per million mapped reads (FPKM) (Florea et al., 2013). Correlation and principal component (PCA) analyses were also performed for the 18 transcriptomes of genes by using the latic, cluster package.
Differentially expressed genes (DEGs) were analyzed using the DESeq 2 package to visualize the results, with FDR < 0.01 and |log2FoldChange| ≥ 2.00 as the screening criteria to obtain the set of DEGs among samples. DEGs were classified as up- or down-regulated genes based on significant positive or negative log changes in their values. The screened DEGs were subjected to GO and KEGG pathway enrichment analyses, where q-value < 0.05 indicated the significant enrichment of the DEGs.
The TF family of K. obovata seedlings was predicted in different tissue samples by using the Plant Transcription Factor Database (version 3.0). The DEGs were submitted to iTAK software for prediction and further classification. The dataset of the TFs of K. obovata seedlings was used as reference, and the default values of the parameters “substitution matrix” and “E-value” were used (Zheng Y. et al., 2016).
The NGDC website (https://ngdc.cncb.ac.cn) contains information on the sequences of K. obovata genes. The CBL-CIPK network and MAPK cascade genes of K. obovata can be identified by comparing the protein sequences generated by the genes of K. obovata seedlings in the database with those in the experiment. When the similarity of gene sequences was higher than 80% compared with the database, it was identified as the corresponding genes with the close homology relationship in the k. obovata.
The expression of 13 key genes in the CBL-CIPK network and MAPK cascade in K. obovata seedling was studied under control and cold stress conditions. The fpkm values of gene expression were normalized by z-score, and the expression was plotted using the stats package for heat mapping. We performed Pearson correlation analysis to obtain correlation matrices between gene pairs. The network relationship graph between 13 genes was determined using the igraph package in R language.
We generated 18 cDNA libraries in 18 samples of various tissues of K. obovata seedlings at different temperatures by using RNA-seq technique. A total of 117.37 Gb of raw data were obtained to gain insights into the response network of genes in the root, stem, and leaf of K. obovata seedlings before and after cold stress. After removing low-quality sequences and adapter sequences from the raw data obtained by sequencing, the GC rates of the 18 samples were 45.58%–46.5%, and all Q30s were greater than 90%, indicating the sufficient accuracy and quality of the sequencing data for further analysis. The general sequencing statistics are shown in Table S1. On the PCA of the gene expression profiles of the 18 samples, three biological replicates of each tissue clustered together (Figure S3), indicating the reliability of all mRNA sequence library replicates. The Pearson correlation analysis of the dataset revealed gene expression patterns and significant tissue specificity, with identical tissues clustered together (Figure S4), consistent with the phenomenon in Figure S3. To gain insight into single genes, GO was classified using WEGO, resulting in 53 GO terms in a three-level ontology (Figure 1A) (Ye et al., 2018). The category of biological process was dominated by “cellular process” (GO:0009987, 6249 unigenes), “metabolic process” (GO:0008152, 5723 unigenes), “single-organism process” (GO:0044699, 4353 unigenes) dominated. In the category of cellular component, “cell” (GO:0005623, 5901 unigenes), “cellular part” (GO:0044464, 5901 unigenes), “membrane” (GO:0016020, 5088 unigenes), “organelle” (GO:0043226, 4583 unigenes), and “membrane part” (GO: 0044425, 4445 unigenes) had important roles. In the GO molecular function, a large number of single genes were annotated as “binding” (GO:0005488, 8107 unigenes) and “catalytic activity” (GO:0 003824, 6915 unigenes).
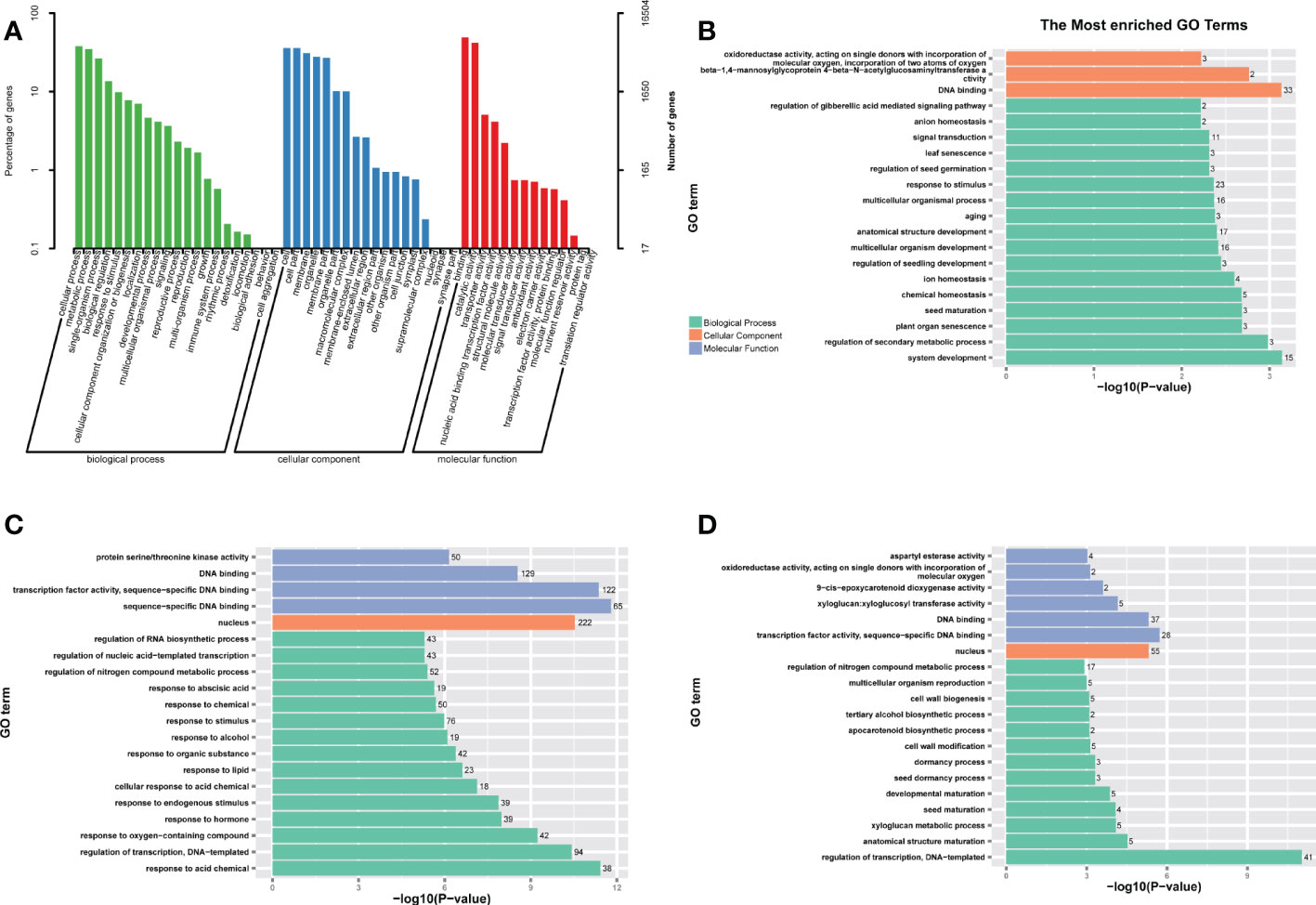
Figure 1 (A) Gene ontology (GO) belonging to the functional classification of all unigenes of the K. obovata seedlings transcriptome, the right y-axis indicates the number of genes in each category and the left y-axis indicates the percentage of genes. (B-D) Gene ontology (GO) enrichment analysis of differentially expressed genes (DEG) in three tissues under cold stress conditions. (B) Representative enrichment (p-value < 0.05) GO terms in (B) leaf (C) root (D) stem DEGs, three colors indicate three different categories and horizontal coordinates indicate p-values.
The KEGG pathway annotations were used to identify biochemical pathways in the K. obovata seedling transcriptome. A total of 6,902 single genes were matched to 136 pathways. The major pathways included “plant hormone signal transduction” (524 single genes), “plant–pathogen interaction” (494 single genes), “MAPK signaling pathway–plant” (318 single genes), and “carbon metabolism” (311 single genes) pathways (Table S2).
RNAseq was used to identify DEGs in different tissues of K. obovata seedlings before and after the cold snaps. According to the RNA-seq results, gene expression in the seedlings changed significantly before and after the cold snap, with a total of 1560, 370, and 416 DEGs in the root, stem, and leaf respectively. The Volcano map and Differential genetic Wayne plots showed the aggregation of DEGs in the roots, stems, and leaves, respectively (Figures 2 and S5). The DEGs included 346 up-regulated genes and 1214 down-regulated genes in the roots, 183 up-regulated genes and 187 down-regulated genes in the stems, and 281 up-regulated genes and 135 down-regulated genes in the leaves (Figure 2).

Figure 2 Volcano plot showing the numbers of DEGs among the three K.obovata seedlings organizations. The x-axis represents the fold-change (FC) of gene expression among samples. The more different genes are at either end. The y-axis represents the significance of the shift in gene expression, which is indicated by -log10. The y is the P-value (t-test) negative log, as adjusted by the false discovery rate (FDR). Genes that were significantly differentially expressed are represented by red dots (up-regulated) and green dots (down-regulated), while insignificantly differentially expressed genes are represented by black dots. Leaf: 281 up-regulated genes and 135 down-regulated genes. Root: 346 up-regulated genes and 1214 down-regulated genes. Stem: 183 up-regulated genes and 187 down-regulated genes.
The DEGs of each tissue were subjected to GO and KEGG pathway enrichment analyses to determine the possible functions of the cold response genes. In GO enrichment, the leaf response annotations were mainly focused on biological processes, with “system development” and “regulation of secondary metabolic processes” as the most abundant. The most abundant GO terms in molecular functions were related to “DNA binding.” The root and stem response annotations tended to be consistent, and the molecular function of “transcription factor activity” dominated. The cellular component was mainly annotated as “nuclear,” and the “management of transcription factors” was highly represented in biological processes. The terms “sequence-specific DNA binding” and “response to acidic chemicals” were unique to the root (Figures 2B-D). Among the KEGG pathway enrichments, circadian-vegetative (ko04712), taurine, and hypotaurine metabolism (ko00430) were significantly enriched in the leaves. Plant–pathogen interactions (ko04626), MAPK signaling pathway (ko04016), biosynthesis of scopolamine, piperidine, and pyridine alkaloids (ko00960), beta-Alanine metabolism (ko00410), and amino and nucleotide sugar metabolism (ko00520) were significantly enriched in the roots. Starch and sucrose metabolism (ko00500), phenylpropanoid biosynthesis (ko00940), and carotenoid biosynthesis (ko00906) were enriched in the stems. Phytohormone signaling (ko04075) was enriched in all tissues (Figures 3A-C).
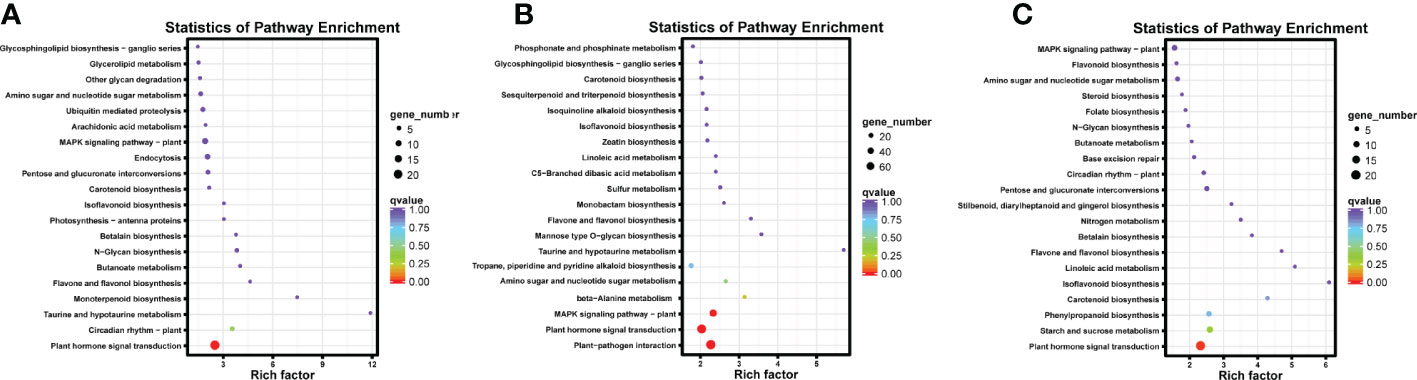
Figure 3 Results of gene and genome enrichment analysis of DEGs in leaves (A) and roots (B) and stems (C) of K. obovata seedlings before and after the cold snaps and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. The horizontal coordinates represent the degree of enrichment, the different colors represent the size of the p-value, and the size of the dots represents the number of genes.
Transcription factors regulate gene expression and are thus regulators of cellular processes and environmental responses. Transcription factors can control the expression of many target genes by specifically binding to their various promoters (Nakashima et al., 2009). To clarify the transcriptional regulation of cold stress-responsive genes, we identified DEGs encoding TFs under cold stress. Among transcription factors, AP2/ERF, WRKY, C2H2, and NAC were concentrated at the root. And the enrichment of these transcription factors in the root was much higher than in the stem and leaves. The major transcription factors in the stems were the same as in the roots, except for the increase in HD-ZIP transcription factors. WRKY did not appear in the stems. The transcription factors NAC, AP2/ERF, bHLH, and GARP were mainly present in the leaves (Figures 4A-C).
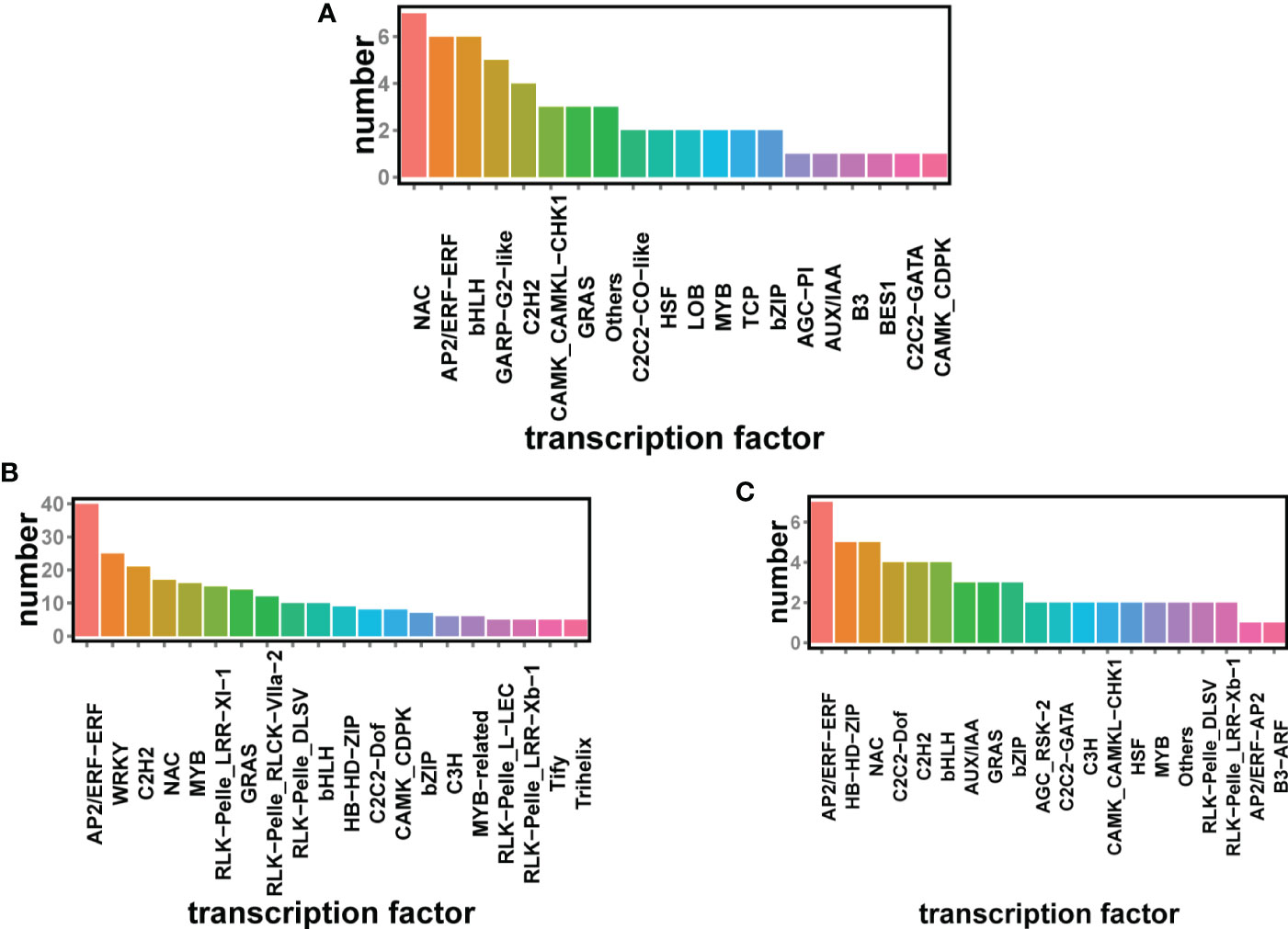
Figure 4 Significant transcription factor (TF) classification in three tissues of K. obovata seedlings, leaves (A) and roots (B) and stems (C), before and after chilling, with vertical coordinates representing the number of transcription factors (P < 0.05 and |LOG2FC| > 2).
Research has concentrated on the CBL-CIPK network and MAPK cascade of Arabidopsis thaliana and some model plants. No study has been conducted on thorough identification and functional analysis of the expression and regulation of the CBL-CIPK network and MAPK cascade-related genes in K. obovata seedlings under cold stress. In the present work, we found that K. obovata seedlings contain 13 CBL-CIPK networks and MAPK cascade-related genes (Figure 5A). The heat map shows that CBL4, CBL3, and CIPK2 are grouped together, but they are separated from CBF1, meanwhile, CBL2, CBL3, and MAPK6 are grouped together. The gene functions of the CBL-CIPK network and MAPK cascade are connected in the K. obovata seedlings. The co-expression regulatory network of the 13 genes in K. obovata was established using this dataset, and the mutual regulatory relationships between among the genes were visualized. The gray line represents the negative regulatory relationship between genes, the blue line represents the positive regulatory relationship between genes, and the increase in line thickness represents a stronger prediction relationship (Figure 5B). Among the co-expression regulatory networks, genes in the CBL-CIPK network and MAPK cascade, for instance CRLK1 with MEKK1 and MEKK1 with MAPK3/6, had strong correlations. The co-expression relationship among the 13 genes could provide insights into the correlation between the CBL-CIPK network and MAPK cascade genes in K. obovata seedlings under cold stress.
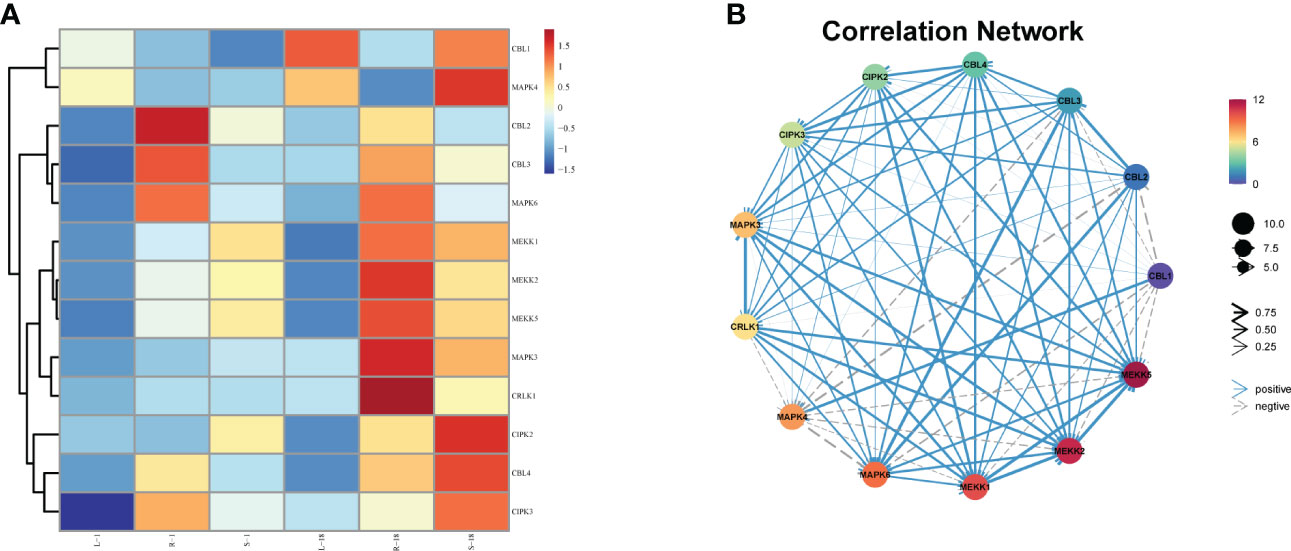
Figure 5 Identification of CBL-CIPK network and MAPK cascade gene genes in different tissues of K. obovata seedlings and visualization of their co-expression networks. (A) Hierarchical clustering analysis of 13 CBL-CIPK networks and MAPK cascade genes. The values in the heat map indicate the z-normalized values of FPKM in different samples. Red and blue colors indicate high and low expression levels, respectively. (B) Thirteen ICE- CBL-CIPK network and MAPK cascade gene co-expression networks.
Under cold stress, plants can trigger the expression of genes involved in multiple signal transduction pathways and further activate downstream regulatory processes related to physiological adaptation. Cold stress generates downstream responses in plants by modifying membrane flow in plant cells (Orvar et al., 2000). Important processes, such as nutrient and ion transport, solute uptake, and osmotic pressure, are affected by the properties of cell membranes, particularly their structure and composition, proper membrane fluidity is necessary to maintain the structure and function of the membrane (Valerica Raicu, 2008). According to our studies, there were 463 DEGs related to membrane composition altered in various tissues (101, 367, and 64 in the leaves, roots, and stems, respectively), among which 69 were up-regulated in the leaves, 46 in the roots, and 27 in the stems. This finding suggests that different tissues of K. obovata seedlings respond differently to cold stress. The membrane fluidity increases in the leaves but decreases in the roots and stems. Changes in membrane fatty acid composition and content can affect membrane fluidity, thereby influencing the adaptation of K. obovata seedlings to cold stress (Zhao et al., 2012). Membrane fluidity initiates cold signaling by affecting the transport of ions and metabolites (Jeon and Kim, 2013). Ion transport mainly affects Ca2+ ions released from apoplastic, endomembrane systems, and organelles, and CIPK acts as a Ca2+ ions sensor in response to elevated cellular solute Ca2+ levels (Tähtiharju et al., 1997). Under cold stress, four genes associated with Ca2+ signaling were found in K. obovata seedlings, consistent with the expected role of Ca2+ in early cold signaling. The results are in line with Su’s findings in his study of adult hardiness (Su et al., 2019). Ca2+ induces intracellular biochemical responses through sensing and transduction of downstream CDPKs, thereby regulating plant responses to various abiotic stress signals (Boudsocq and Sheen, 2013). CBL is a specific calcium sensor with non-classical EF-hand domains that can capture calcium signals in different plant species, including Arabidopsis (Ma X. et al., 2022). These CBLs/SCaBPs do not have enzymatic activity of their own, but CBLs are triggered by changes in Ca2+ characteristics and interact precisely with CIPK (CBL-interacting protein kinase) or SOS2 protein kinase (Ma X. et al., 2022). Increasing lines of evidence indicate that the CBL-CIPK network plays a key role in cold stress response. In the present study, one CDPK, twoCMLs, three CBLs, and three CIPKswere found to be up-regulated, indicating the critical function of Ca2+-mediated signaling pathways in the response of K. obovata seedlings to cold stress.
Plants produce metabolites to adapt to a stressful, changing growing environment, which may involve interactions through signaling processes and pathways (Edreva et al., 2008). In the present study, the term “regulation of secondary metabolic processes” was a rich category in GO classification. The results of the KEGG pathway enrichment analysis showed that starch and sucrose metabolism, amino saccharide, and nucleotide sugar metabolism were enriched in different tissues. Plant responses to cold stress include sucrose accumulation due to the induction of sucrose phosphate synthase and sucrose catabolic enzymes (convertases and sucrose synthases) (Coleman et al., 2009). Moreover, nucleotide sugars are key intermediates in the starch and sucrose biosynthesis pathways (Coleman et al., 2009; Figueroa et al., 2021). At low temperatures, starch hydrolyzes into soluble sugars (sucrose), improving the osmotic pressure in cells and preventing protoplasm from dehydration and solidification. Dissolution of sucrose increases the concentration of the cell fluid, reducing the freezing point, avoiding the possibility of protein precipitation and solidification and salting, and simultaneously increasing the cell’s structural stability (Sun et al., 2015). We found the enrichment of metabolic pathways for starch and sucrose metabolism as well as amino and nucleotide sugars in the stems and roots. These metabolic pathways are significantly enriched in cold-domesticated K. obovata and play an important role in its resistance to cold stress.
To resist and adapt to cold stress, plants form complex and efficient regulatory networks, with transcriptional regulation playing a key role. Transcription factors regulate gene expression by binding to cis-acting elements in the promoter region and have an important function in the plant abiotic stress response network (Wang et al., 2020). Furthermore, TFs highlight the role of multiple transcriptional mechanisms in stress signaling pathways and are linked to the regulation of various abiotic stress signals and gene transcription (Shanker and Venkateswarlu, 2011). Many TF families, including AP2/EREBP, bHLH, WRKY, MYB, NAC, and MYC, coordinate the signals transduced by plants in response to different environmental stresses (Liu et al., 2014). According to our results, the largest TF families induced by cold stress are AP2-ERF and bHLH, which consist of 20 members, respectively. AP2-ERF and bHLH-TFs are involved in a variety of physiological processes, and overexpression of AP2/ERF in red pine and PtrbHLH in lemon and tobacco can enhance the cold tolerance of plants at freezing temperatures (Huang et al., 2013; Wang et al., 2020). According to Hajar’s research results, the expression level of plant AP2/ERF transcription factors in roots was higher than that in stems and leaves under cold stress, which is similar to our research results (Alberts, 2015). WRKY is the largest TF family in plants, and it controls plant growth and development and plays a critical part in many stress responses. NAC is a class of transcription factors that are unique to plants.It is involved in various cold stress regulatory pathways, particularly ABA-dependent signaling pathways, which interact with the gibberellin and Auxin signaling pathways and control lateral root development (Nakashima et al., 2012; Wang et al., 2018). Since the samples we collected were seedlings, the over-expression of WRKY and NAC transcription factors in the roots might improve the plant’s cold tolerance and promote root development (Du Z. et al., 2022; Fei et al., 2022; Sun et al., 2022). Cold stress in plants first damages the structure of the cell membrane, thus affecting its function of the cell membrane. C2H2-ZF transcription factors can enhance the ability of plants to resist cold stress by maintaining the stability of cell membranes (Ma L. et al., 2022). At the same time, C2H2-ZF transcription factors contain cis-elements related to plant hormones or abiotic stress, which are involved in tissue and organ development, especially root and flower (Liu et al., 2015). Therefore, C2H2-ZF transcription factors were highly enriched in roots. Other TFs, including DEGs of the GRAS, bZIP, MYB, and WHSF families, also respond to cold stress in K. obovata seedlings. Hence, these TFs may be essential regulators that drive downstream gene expression cascades.
The MAPK cascade is a crucial plant response to abiotic stress. The MAPK1-MKK4/5-MAPK3/6 module of Arabidopsis thaliana was the first MAPK signaling module identified in a plant. It mediates the transcriptional up-regulation of genes encoding transcription factors WRKY22 and WRKY29 in response to fungal and bacterial pathogen assault (Asai et al., 2002; Galletti et al., 2011). The MKK4/5-MAPK3/6, MKK1-MAPK3/6, and MKK6-MAPK4 modules are key players in plant response to cold stress (Wang et al., 2021). We identified 13 genes in the CBL-CIPK network and MAPK cascade signaling module of K. obovata seedlings and investigated their relevance under cold stress. When subjected to cold stress conditions, MEKK1-MKK2-MPK4 confers cold sensitivity and freezing tolerance in K. obovata seedlings (Zhao et al., 2017). Low temperatures change the fluidity of the cell membrane, which Ca2+ channels and proteins can sense on the membrane. The calcium response protein and MAPK cascade genes are then activated, leading to downstream modulation of genes that regulate the low-temperature response (Zhao et al., 2017). MAPK contains a group of tertiary phosphorylation-dependent kinases: MEKK or MKKK, MEK or Mkk, and MPK. The MAPK cascade activates and mediates signals from the cell membrane to the nucleus, regulating physiological activity (Furuya et al., 2013). When K. obovata seedlings are exposed to cold stress conditions, ca2+/cams regulate the activity of receptor-like kinases (CRLK1 and CRLK2) and phosphorylates MEKK1, thereby activating the MeKK1-MKK2-MPK4 cascade. Activated MEKK1-MKK2-MPK4 inhibits the protein abundance and kinase activity of Mpk3 and MPK6, actively modulating the hypothermia response, inhibiting cold-mediated activation, and enhancing the cold resistance of plants. (Furuya et al., 2013; Zhao et al., 2017).
Mangroves are an important component of the wetland ecosystem. Kandelia. obovata, as one of the cold-resistant mangrove species, will experience more severe extreme low temperatures as it moves north. We compared gene expression differences in three tissues of K. obovata seedlings before and after cold snaps and found 1,560, 370, and 416 DEGs in the roots, stems, and leaves, respectively. The gene expression strategies of K. obovata seedlings in response to winter cold snaps are summarized as follows: (1)In regulating transcription factors in various tissues, AP2-ERF, NAC, and C2H2 are the most involved transcription factor families in the cold stress response. In addition, bHLH, WRKY, GRAS, bZIP, MYB, and WHSF also participate in the cold stress response. (2)The fluidity of cell membranes in different seedling tissues varies in response to cold pressure, being faster in leaves and slower in roots and stems. Changes in the membrane composition affect Ca2+ ion transport and, therefore, the CBL-CIPK network. Moreover, (3) the upregulation of secondary metabolism-related genes resulted in the accumulation of starch and sucrose in a seedling’s root and stem tissues. In K. obovata seedlings, 13 cold stress-responsive CBL-CIPK network and MAPK cascade genes were identified and used to construct a paired co-expression network model. The findings provide insights into molecular regulatory mechanisms governing the cold stress response of K. obovata seedlings. This study can be used as an empirical reference for further research into the CBL-CIPK network and MAPK cascade in plants.
The data presented in the study are deposited in the NCBI repository, accession number PRJNA927281. https://www.ncbi.nlm.nih.gov/sra/PRJNA927281.
KT, and QL: conceptualization, investigation, validation, data analysis, methodology, and visualization, writing-original draft. XZ, PC, SX, HG, and YW: conceptualization, experiment design, reviewing and revising the manuscript. WL: collection of samples, funding acquisition. All authors contributed to the article and approved the submitted version.
This research was supported by Zhejiang Provincial Natural Science Foundation of China under Grant NO. LGN18D060001, LGN20C190002.
We thank Dr. Zhiqiang Han for his writing guidance on this manuscript. Dr. Zhiqiang Han and multiple reviewers were especially helpful in shaping this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1113278/full#supplementary-material
Alberts B. (2015). Molecular biology of the cell, sixth edition (W.W. Norton & Company), 131–135. doi: 10.1201/9781315735368/molecular-biology-cell-bruce-alberts
Asai T., Tena G., Plotnikova J., Willmann M. R., Chiu W. L., Gomez-Gomez L., et al. (2002). MAP kinase signalling cascade in arabidopsis innate immunity. Nature 415, 977–983. doi: 10.1038/415977a
Batistic O., Kudla J. (2004). Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta 219, 915–924. doi: 10.1007/s00425-004-1333-3
Boudsocq M., Sheen J. (2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18, 30–40. doi: 10.1016/j.tplants.2012.08.008
Buchwal A., Sullivan P. F., Macias-Fauria M., Post E., Myers-Smith I. H., Stroeve J. C., et al (2020). Divergence of Arctic shrub growth associated with sea ice decline. Proc. Natl. Acad. Sci. U. S. A. 117, 33334–33344. doi: 10.1073/pnas.2013311117
Cavanaugh K. C., Kellner J. R., Forde A. J., Gruner D. S., Parker J. D., Rodriguez W., et al (2014). Poleward expansion of mangroves is a threshold response to decreased frequency of extreme cold events. Proc. Natl. Acad. Sci. U. S. A. 111, 723–727. doi: 10.1073/pnas.1315800111
Cavanaugh K. C., Parker J. D., Cook-Patton S. C., Feller I. C., Williams A. P., Kellner J. R. (2015). Integrating physiological threshold experiments with climate modeling to project mangrove species' range expansion. Global Change Biol. 21, 1928–1938. doi: 10.1111/gcb.12843
Chen Y., Chen L., Sun X. M., Kou S., Liu T. T., Dong J. K., et al. (2022). The mitogen-activated protein kinase kinase MKK2 positively regulates constitutive cold resistance in the potato. Environ. Exp. Botany 194, 10. doi: 10.1002/ecs2.1865
Chen L. Z., Wang W. Q., LI Q. S. Q., Zhang Y. H., Yang S. C., Osland M. J., et al. (2017). Mangrove species' responses to winter air temperature extremes in China. Ecosphere 8, 14. doi: 10.1002/ecs2.1865
Coldren G. A., Proffitt C. E. (2017). Mangrove seedling freeze tolerance depends on salt marsh presence, species, salinity, and age. Hydrobiologia 803, 159–171. doi: 10.1007/s10750-017-3175-6
Coleman H. D., Yan J., Mansfield S. D. (2009). Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc. Natl. Acad. Sci. U. S. A. 106, 13118–13123. doi: 10.1073/pnas.0900188106
Du Z., You S., Yang D., Tao Y., Zhu Y., Sun W., et al (2022). Comprehensive analysis of the NAC transcription factor gene family in Kandelia obovata reveals potential members related to chilling tolerance. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1048822
Du Z. K., You S. X., Zhao X., Xiong L. H., Li J. M. (2022). Genome-wide identification of WRKY genes and their responses to chilling stress in Kandelia obovata. Front. Genet. 13. doi: 10.3389/fgene.2022.875316
Edreva A., Velikova V., Tsonev T., Dagnon S., Gurel A., Aktas L., et al. (2008). Stress protective role of secondary metabolite: Diversity of functions and mechanisms. Gen. Appl. Plant Physiol. Special issue, 34 (1-2), 67–78. Available at: https://www.researchgate.net/publication/251847574.
Fei J., Wang Y. S., Cheng H., Su Y. B., Zhong Y. J., Zheng L. (2021a). Cloning and characterization of KoOsmotin from mangrove plant Kandelia obovata under cold stress. BMC Plant Biol. 21, 12. doi: 10.1186/s12870-020-02746-0
Fei J., Wang Y. S., ChenG H., Su Y. B., Zhong Y. J., Zheng L. (2022). The Kandelia obovata transcription factor KoWRKY40 enhances cold tolerance in transgenic arabidopsis. BMC Plant Biol. 22, 15. doi: 10.1186/s12870-022-03661-2
Fei J., Wang Y. S., Cheng H., Sun F. L., Sun C. C. (2021b). Comparative physiological and proteomic analyses of mangrove plant Kandelia obovata under cold stress. Ecotoxicology 30, 1826–1840. doi: 10.1007/s10646-021-02483-6
Fei J., Wang Y. S., Zhou Q., Gu J. D. (2015). Cloning and expression analysis of HSP70 gene from mangrove plant Kandelia obovata under cold stress. Ecotoxicology 24, 1677–1685. doi: 10.1007/s10646-015-1484-y
Figueroa C. M., Lunn J. E., Iglesias A. A. (2021). Nucleotide-sugar metabolism in plants: the legacy of Luis f. leloir. J. Exp. Botany 72, 4053–4067. doi: 10.1093/jxb/erab109
Florea L., Song L., Salzberg S. L. (2013). Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Research 2, 188. doi: 10.12688/f1000research.2-188.v2
Furuya T., Matsuoka D., Nanmori T. (2013). Phosphorylation of Arabidopsis thaliana MEKK1 via Ca2+ signaling as a part of the cold stress response. J. Plant Res. 126, 833–840. doi: 10.1007/s10265-013-0576-0
Galletti R., Ferrari S., De Lorenzo G. (2011). Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against botrytis cinerea. Plant Physiol. 157, 804–814. doi: 10.1104/pp.111.174003
Guo Z. J., Ma D. N., LI J., Wei M. Y., Zhang L. D., Zhou L. C., et al. (2022). Genome-wide identification and characterization of aquaporins in mangrove plant Kandelia obovata and its role in response to the intertidal environment. Plant Cell Environ. 45, 1698–1718. doi: 10.1111/pce.14286
Guy C. L. (1990). Cold-acclimation and freezing stress tolerance - role of protein-metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41, 187–223. doi: 10.1146/annurev.pp.41.060190.001155
Huang X.-S., Wang W., Zhang Q., Liu J.-H. (2013). A basic helix-Loop-Helix transcription factor, ptrbhlh, of poncirus trifoliata confers cold tolerance and modulates peroxidase-mediated scavenging of hydrogen peroxide. Plant Physiol. 162, 1178–1194. doi: 10.1104/pp.112.210740
Jeon J., Kim J. (2013). Cold stress signaling networks in arabidopsis. J. Plant Biol. 56, 69–76. doi: 10.1007/s12374-013-0903-y
Kolukisaoglu U., Weinl S., Blazevic D., Batistic O., Kudla J. (2004). Calcium sensors and their interacting protein kinases: Genomics of the arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 134, 43–58. doi: 10.1104/pp.103.033068
Liu J. H., Peng T., Dai W. S. (2014). Critical cis-acting elements and interacting transcription factors: Key players associated with abiotic stress responses in plants. Plant Mol. Biol. Reporter 32, 303–317. doi: 10.1007/s11105-013-0667-z
Liu Q., Wang Z., Xu X., Zhang H., LI C. (2015). Genome-wide analysis of C2h2 zinc-finger family transcription factors and their responses to abiotic stresses in poplar (Populus trichocarpa). PloS One 10, e0134753. doi: 10.1371/journal.pone.0134753
Liu W. C., Zheng C. F., Chen J. N., Qiu J. B., Huang Z. X., Wang Q., et al. (2019). Cold acclimation improves photosynthesis by regulating the ascorbate-glutathione cycle in chloroplasts of Kandelia obovata. J. Forest. Res. 30, 755–765. doi: 10.1007/s11676-018-0791-6
Ma X., Gai W. X., Li Y., Yu Y. N., Ali M., Gong Z. H. (2022). The CBL-interacting protein kinase CaCIPK13 positively regulates defence mechanisms against cold stress in pepper. J. Exp. Botany 73, 1655–1667. doi: 10.1093/jxb/erab505
Ma L., Xu J., Tao X., Wu J., Wang W., Pu Y., et al. (2022). Genome-wide identification of C2H2 ZFPs and functional analysis of BRZAT12 under low-temperature stress in winter rapeseed (Brassica rapa). Int. J. Mol. Sci. 23 (20), 12218. doi: 10.3390/ijms232012218
Nakashima K., ITO Y., Yamaguchi-Shinozaki K. (2009). Transcriptional regulatory networks in response to abiotic stresses in arabidopsis and grasses. Plant Physiol. 149, 88–95. doi: 10.1104/pp.108.129791
Nakashima K., Takasaki H., Mizoi J., Shinozaki K., Yamaguchi-SHinozaki K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Orvar B. L., Sangwan V., Omann F., Dhindsa R. S. (2000). Early steps in cold sensing by plant cells: the role of actin cytoskeleton and membrane fluidity. Plant J. 23, 785–794. doi: 10.1046/j.1365-313x.2000.00845.x
Osland M. J., Day R. H., From A. S., Mccoy M. L., Mcleod J. L., Kelleway J. J. (2015). Life stage influences the resistance and resilience of black mangrove forests to winter climate extremes. Ecosphere 6, 15. doi: 10.1890/ES15-00042.1
Osland M. J., Day R. H., Hall C. T., Brumfield M. D., Dugas J. L., Jones W. R. (2017). Mangrove expansion and contraction at a poleward range limit: climate extremes and land-ocean temperature gradients. Ecology 98, 125–137. doi: 10.1002/ecy.1625
Osland M. J., Enwright N., Day R. H., Doyle T. W. (2013). Winter climate change and coastal wetland foundation species: salt marshes vs. mangrove forests in the southeastern united states. Global Change Biol. 19, 1482–1494. doi: 10.1111/gcb.12126
Peng Y. L., Wang Y. S., Fei J., Sun C. C. (2020). Isolation and expression analysis of two novel c-repeat binding factor (CBF) genes involved in plant growth and abiotic stress response in mangrove Kandelia obovata. Ecotoxicology 29, 718–725. doi: 10.1007/s10646-020-02219-y
Peng Y. L., Wang Y. S., Fei J., Sun C. C., Cheng H. (2015). Ecophysiological differences between three mangrove seedlings (Kandelia obovata, aegiceras corniculatum, and avicennia marina) exposed to chilling stress. Ecotoxicology 24, 1722–1732. doi: 10.1007/s10646-015-1488-7
Shanker A. K., Venkateswarlu B. (2011). Abiotic stress response in plants-physiological, biochemical and genetic perspectives. London, UK: IntechOpen. Abiotic Biotic Stress Response Crosstalk Plants 1, 3–26. doi: 10.5772/1762
Shi Y. T., Ding Y. L., Yang S. H. (2018). Molecular reculation of CBF sicnalinc in colc acclimation. Trends Plant Sci. 23, 623–637. doi: 10.1016/j.tplants.2018.04.002
Su W. Y., Ye C. T., Zhang Y. H., Hao S. Q., Li Q. S. Q. (2019). Identification of putative key genes for coastal environments and cold adaptation in mangrove Kandelia obovata through transcriptome analysis. Sci. Total Environ. 681, 191–201. doi: 10.1016/j.scitotenv.2019.05.127
Sun Y. M., Liu L. J., Feng M. F., Wang J. H., Cang ,. J., Li S., et al. (2015). Research progress of sugar metabolism of plants under cold stress. J. Northeast Agric. Univ. 46 (7), 95–102. doi: 10.3969/j.issn.1005-9369.2015.07.015
Sun M. M., Liu X., Huang X. J., Yang J. J., Qin P. T., Zhou H., et al. (2022). Genome-wide identification and expression analysis of the NAC gene family in Kandelia obovata , a typical mangrove plant. Curr. Issues Mol. Biol. 11, 5622–5637. doi: 10.3390/cimb44110381
Tähtiharju S., Sangwan V., Monroy A. F., Dhindsa R. S., Borg M. (1997). The induction of kin genes in cold-acclimating Arabidopsis thaliana. evidence of a role for calcium. Planta 203, 442–447. doi: 10.1007/s004250050212
Teige M., Scheikl E., Eulgem T., Doczi F., Ichimura K., Shinozaki K., et al. (2004). The MKK2 pathway mediates cold and salt stress signaling in arabidopsis. Mol. Cell. 15, 141–152. doi: 10.1016/j.molcel.2004.06.023
Valerica Raicu A. P. (2008). “Cell membrane: Structure and physical properties,” in Integrated molecular and cellular biophysics (Dordrecht: Springer). doi: 10.1007/978-1-4020-8268-9_3
Wang F., Chen S., Liang D. Y., Qu G. Z., Chen S., Zhao X. Y. (2020). Transcriptomic analyses of Pinus koraiensis under different cold stresses. BMC Genomics 21, 14. doi: 10.1186/s12864-019-6401-y
Wang Z., Wan Y. Y., Meng X. J., Zhang X. L., Yao M. N., Miu W. J., et al. (2021). Genome-wide identification and analysis of MKK and MAPK gene families in brassica species and response to stress in brassica napus. Int. J. Mol. Sci. 22, 21. doi: 10.3390/ijms22020544
Wang W. Q., You S. Y., Wang Y. B., Huang L., Wang M. (2011). Influence of frost on nutrient resorption during leaf senescence in a mangrove at its latitudinal limit of distribution. Plant Soil 342, 105–115. doi: 10.1007/s11104-010-0672-z
Wang Z., Yu D., Zheng C., Wang Y., Cai L., Guo J., et al. (2019). Ecophysiological analysis of mangrove seedlings Kandelia obovata exposed to natural low temperature at near 30°N. J. Mar. Sci. Engineering 7, 292. doi: 10.3390/jmse7090292
Wang P., Zhao Y., Li Z., Hsu C. C., Liu X., Fu L., et al. (2018). Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell. 69, 100–112.e6. doi: 10.1016/j.molcel.2017.12.002
Ye J., Zhang Y., Cui H., Liu J., Wu Y., Cheng Y., et al. (2018). WEGO 2.0: a web tool for analyzing and plotting GO annotations 2018 update. Nucleic Acids Res. 46, W71–W75. doi: 10.1093/nar/gky400
Zhao Z. G., Tan L. L., Dang C. Y., Zhang H., Wu Q. B., An L. Z. (2012). Deep-sequencing transcriptome analysis of chilling tolerance mechanisms of a subnival alpine plant, Chorispora bungeana. BMC Plant Biol. 12, 17. doi: 10.1186/1471-2229-12-222
Zhao C. Z., Wang P. C., Si T., Hsu C. C., Wang L., Zayed O., et al. (2017). MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev. Cell. 43, 618–629. doi: 10.1016/j.devcel.2017.09.024
Zheng Y., Jiao C., Sun H. H., Rosli H. G., Pombo M. A., Zhang P. F., et al. (2016). iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 9, 1667–1670. doi: 10.1016/j.molp.2016.09.014
Keywords: Kandelia obovata seedlings, transcriptome, cold stress, CBL-CIPK network, MAPK cascade
Citation: Tian K, Li Q, Zhang X, Guo H, Wang Y, Cao P, Xu S and Li W (2023) Analysis of the expression and function of the CBL-CIPK network and MAPK cascade genes in Kandelia obovata seedlings under cold stress. Front. Mar. Sci. 10:1113278. doi: 10.3389/fmars.2023.1113278
Received: 01 December 2022; Accepted: 08 February 2023;
Published: 01 March 2023.
Edited by:
Diana Sofia Madeira, University of Aveiro, PortugalReviewed by:
Kai Xu, Jimei University, ChinaCopyright © 2023 Tian, Li, Zhang, Guo, Wang, Cao, Xu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiye Li, bGl3ZWl5ZTE5OTlAc2luYS5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.