- 1College of Oceanography, Hohai University, Nanjing, Jiangsu, China
- 2Jiangsu Province Engineering Research Center for Marine Bio-resources Sustainable Utilization, Hohai University, Nanjing, China
- 3West China School of Public Health and West China Fourth Hospital, Sichuan University, Chengdu, China
- 4School of Chemical Engineering and Technology, Sun Yat-sen University, Zhuhai, Guangdong, China
Chiton teeth in the radula are one of the hardest biomaterials in nature. Chiton uses radula to scrape algae on hard surfaces. The ultrastructure and composition of teeth are well known while how they move is less clear. This study used an array of material characterizations including soft-tissue micro-computed tomography (micro-CT), histology, scanning electron microscopy, and proteomics to investigate the tissue that may control the movement of the radula of Acanthopleura loochooana. Surprisingly, unusually complex muscle fibers were found around the radula. 54 muscle fibers with diameters of around 130 µm were anchored to the second and third shell plates. These muscle fibers are in close contact with the radula and cartilage beneath the radula. Proteomics using a recently published chiton genome as a reference confirmed the proteins related to energy metabolism, calcium metabolism, as well as a cartilage oligomeric matrix protein in the muscle. qPCR found that the above proteins were highly expressed in the radula muscle compared to the foot. Taken together, this study provides insights into the complex tissue structures that control the movement of the radula, which may inspire robotics design relating to hard-soft tissue interfaces.
Introduction
Chitons are important Mollusca living in diverse marine environments from the intertidal regions to the deep sea. They are ancient Mollusca with eight shell plates dating back to the Cambrian (Smith et al., 2011; Wanninger and Wollesen, 2019). They use radulae to graze algae, diatoms, and foraminiferans on rocks (Lobo-da-Cunha et al., 2022). The radula contains hundreds of mineralized teeth that are made of mostly magnetite and apatite, which have been discovered since the 1960s (Lowenstam, 1967). The teeth are reported to exhibit one of the largest hardness and stiffness among biomaterials on Earth and attracted the great attention of materials scientists (Brooker et al., 2003; Weaver et al., 2010; Stegbauer et al., 2021; Krings et al., 2022; Wang et al., 2022). State-of-the-art modern materials characterization such as electron microscopy and spectroscopy were used to probe the ultrastructure, composition, and formation of radular teeth from Cryptochiton stelleri, Acanthopleura echinate, and Lepidochitona cinereal (Brooker et al., 2003; Weaver et al., 2010; Stegbauer et al., 2021; Krings et al., 2022; Wang et al., 2022). Additionally, molecular biology and omics methods were used to reveal the proteins in the teeth of chiton C. stelleri (Nemoto et al., 2012; Nemoto et al., 2019).
Despite such great efforts, the feeding process is still unclear because teeth are only a part of the feeding apparatus for chitons. To fulfill the function of feeding, chitons require multiple tissues to work together besides the teeth. During grazing, chitons pushed the radula quickly out of the mouth and then retracted back into the mouth, bringing food into the esophagus within seconds (Scheel et al., 2020). By performing the process repeatedly, food is processed. The radula is supported by certain connective tissues such as cartilage and muscles. In gastropods (e.g. limpets or snails), the teeth slide over on a cartilaginous base (the odontophore) (Guralnick and Smith, 1999; Golding et al., 2009). The odontophore protractor muscle, the radula protractor, and the radula retractor muscle are responsible to push and retract radulae into and out of the mouth, which had been intensively studied in gastropods (Graham, 1973; Kehl et al., 2019). However, recent studies showed that the moving pattern of the radula in chitons is complex containing various motions like rotating and twisting (Pohl et al., 2020; Scheel et al., 2020). It is unclear what tissues are in play in chitons. Hence, this study examined the feeding apparatus of chiton Acanthopleura loochooana, a typical species living in the southeast coastal areas of China, in detail by various and multiscale approaches including histology, soft-tissue micro-computed tomography (micro-CT), proteomics, and molecular biology. Interestingly, for the first time, complex muscle fibers were found in the chiton that may be critical in controlling the movement of the radula.
Results
The complex movement of radulae in chitons
Chiton A. loochooana has eight overlapping shell plates (Liu et al., 2022a) (Figure 1A). They had radula composed of iron-based minerals similar to others. The mineralization degree was intensified as the growth continues (Figure 1B). Interestingly, in the mature region of the radula, a narrowed band was observed (Figure 1B, red arrows), which was deduced to be generated from unknown forces exerted on the radula membrane. During feeding, the whole radula of chiton C. stelleri was pushed back and forward of the mouth repeatedly, and sometimes, the rolling of the radula membrane can be observed to scrape algae on solid surfaces. The whole process happened within several seconds (Figure 1C), similar to the feeding process of A. loochooana (lab observation). The teeth movement is obviously under the exquisite control of the organism. A question, therefore, arouses: what organ or tissue controls the movement of the radula, and thus, the feeding of chitons?
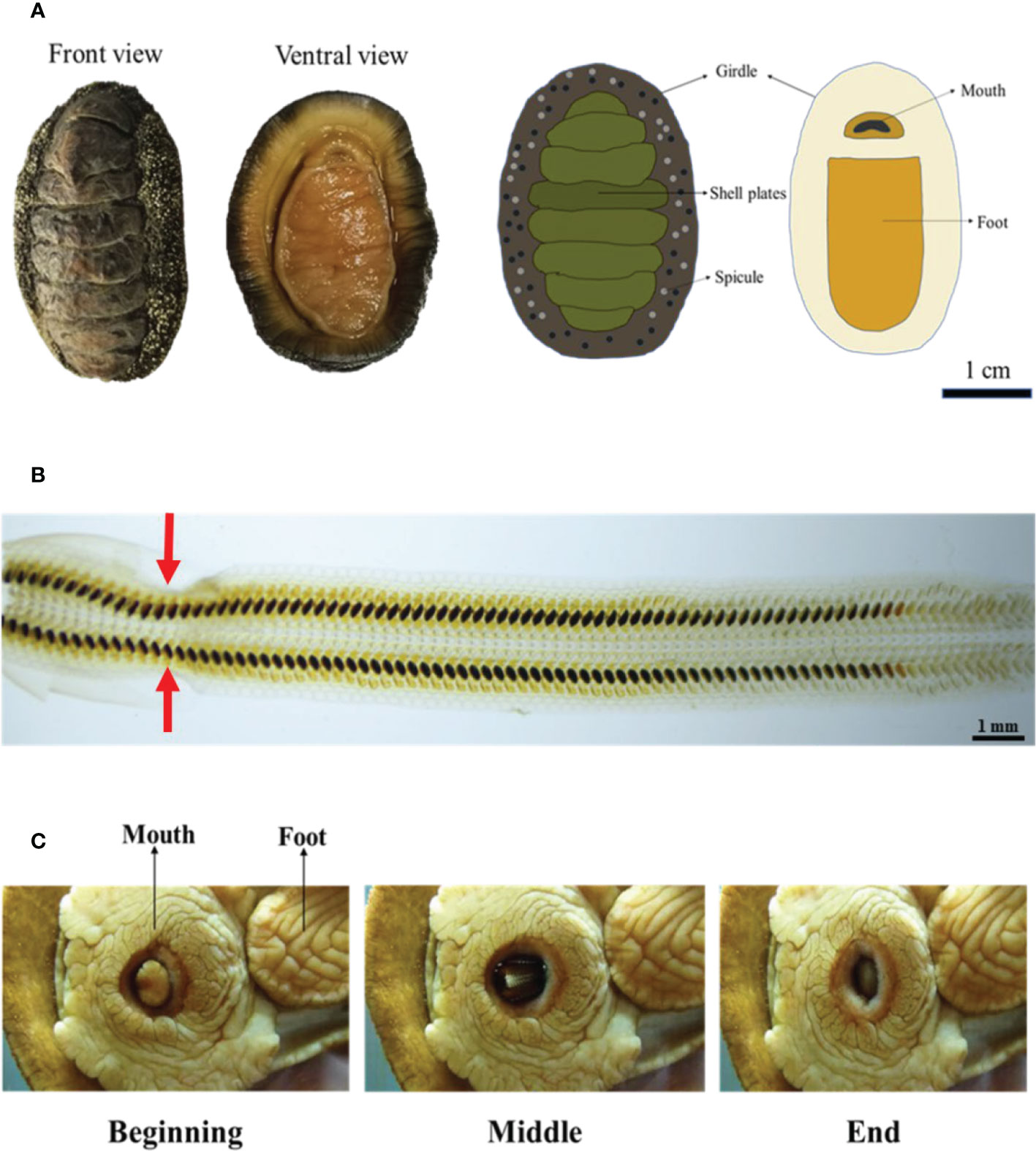
Figure 1 The radula and its motion in chitons. (A) Optical image and the basic anatomy of chiton Acanthopleura loochooana. (B) The radula of the chiton A. loochooana. The red arrows indicated the narrow band (C) The feeding process from chiton Cryptochiton stelleri. Images were made based on the video from https://www.youtube.com/watch?v=8JqgCXsq1OM. Image credit: VicHigh Marine/David Young.
Complex muscle fibers of chiton A. loochooana in vitro and ex vivo
To answer this question, we first performed anatomical dissection on the animal under stereo microscopy due to the small size of the animal (2-5 cm). Interestingly, we found two nearly symmetric bundles of dark red muscle-like tissue wrapped around the teeth and attached to the shell plate in the distal end (Figure 2A). In the magnified image, dozens of muscle-like fibrous tissue were clearly observed (Figure 2B). The isolated single fiber was around 100 and 50 µm under optical and scanning electron microscopy (SEM) respectively (Figure 2C). The smaller size of fibers under SEM may be due to the loss of water during drying. The cross-section of the fiber indicated that it was made of thinner small fibers, resembling the muscle of vertebrates (Figure 2D).
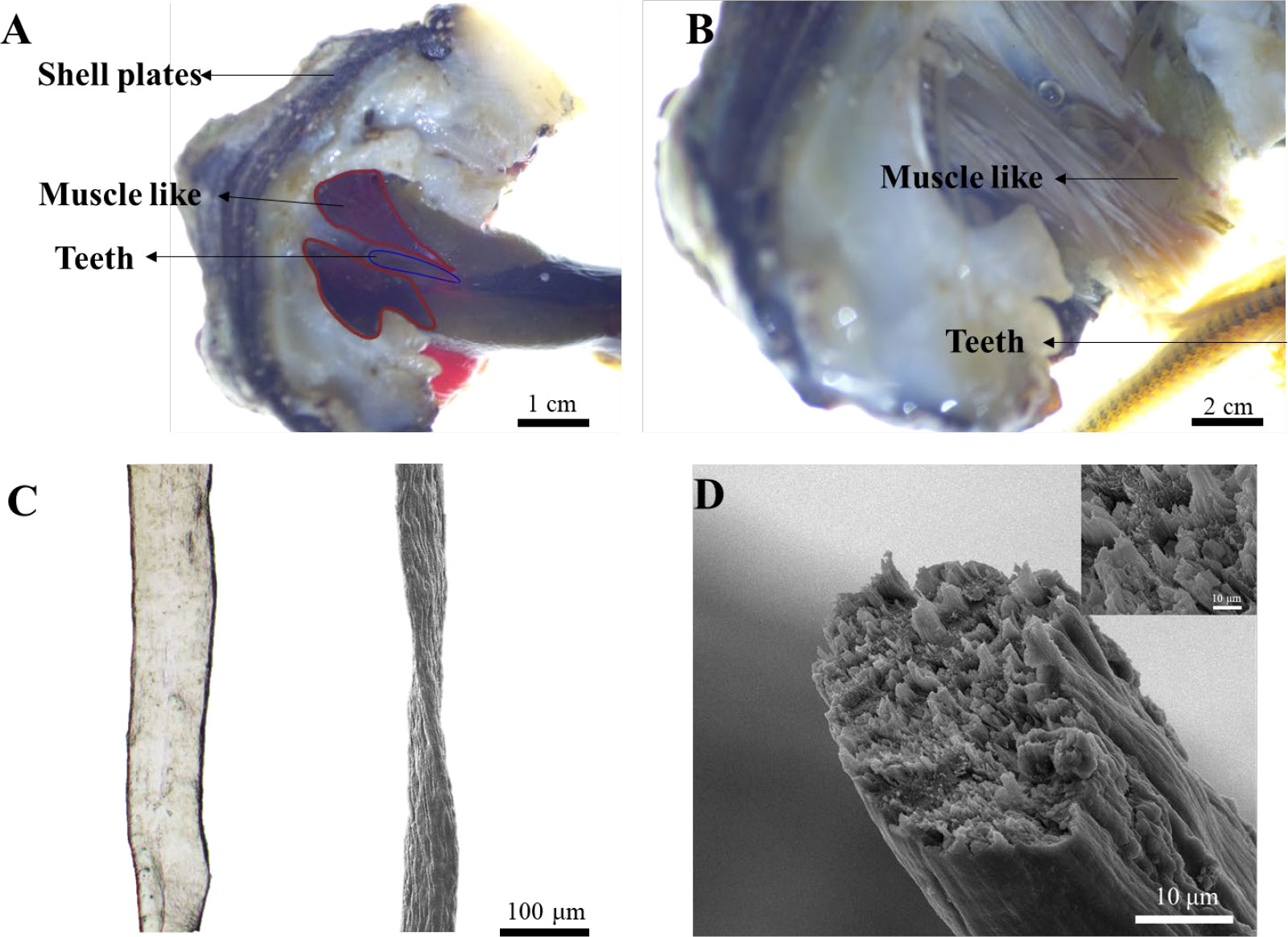
Figure 2 Isolation, dissection, and visualization of radular muscle-like tissues. (A) Dissection of muscle-like tissue around the radula. (B) The magnified image of muscle-like tissue and the teeth under a stereo microscope. (C) The optical (left) and SEM image (right) of a single radular muscle-like fiber (D) The cross-sectional SEM image of a single fiber. The inset is the magnified image.
The above in vitro anatomic results revealed the existence of muscle-like tissues around the teeth, however, how these tissues interacted with the teeth in vivo or ex vivo is unclear. The anatomical dissection is destructive in nature and could not reveal the interaction of tissues in vivo. Micro-CT is a valuable approach for digital anatomy and soft-tissue micro-CT by staining can be useful to reveal the interaction of soft and hard tissues. Reanalyzing and segmenting a previous dataset of soft-tissue micro-CT data from a closely related chiton species Acanthochitona crinite (Ziegler et al., 2018), the relationship of the teeth and muscle-like tissue was revealed. Based on a recent paper (Liu et al., 2022b), A. loochooana and A. crinita are in the same order Chitonida but in different suborders. A. loochooana and A. crinita belong to Chitonina and Acanthochitonina respectively. Based on the bottom and side view of 3D rendering images, it can be seen that the muscle-like fibers (orange color) were wrapped around the teeth (green color) at the position close to the narrow band area seen in the optical image of the radula (Figures 3A–E). From the coronal view, it can be seen that muscle-like tissues were in close contact with the teeth (Figure 3B). Additionally, muscle-like tissues anchored to second and third shell plates were seen from the sagittal view (Figure 3C). Two clusters of muscles with different angles with respect to the teeth were identified, 23 fibers with around 77° and 21 fibers with around 50° (Figures 3D, E). The diameters of fibers were 70-190 μm with an average of approximately 130 μm (Figure 3F), in agreement with SEM and optical images considering the loss of water and shrinking of fibers in vitro. The length of the fibers was around 2.5 mm (Figure S1).
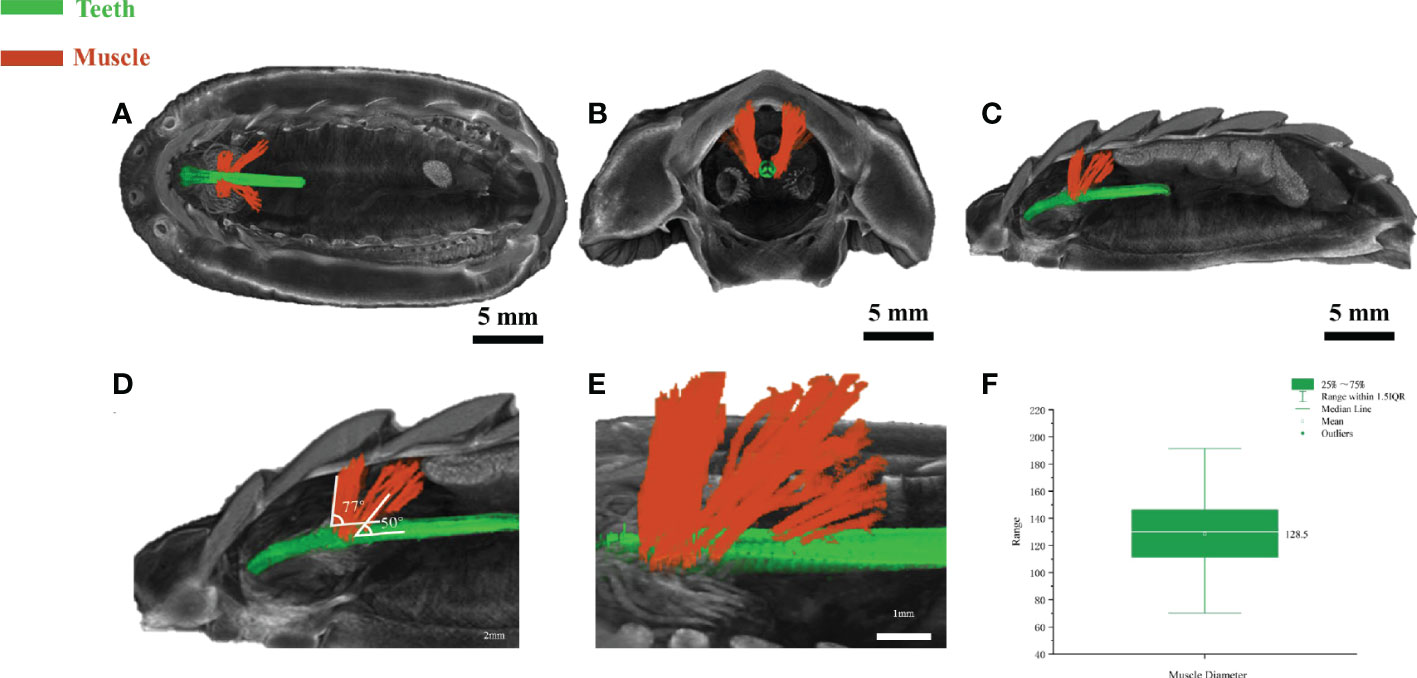
Figure 3 Soft-tissue micro-CT of radular muscles ex vivo. The bottom view (A), coronal (B), and the sagittal view (C) of the chiton. The magnified image (D, E) of sagittal view highlighting the radula and muscle angles. (F) Statistical analysis of muscle diameter in the chiton (n=54). Green=radular teeth, orange=muscle. Soft-tissue micro-CT data of chiton Acanthochitona crinita was directly retrieved from the Morphobank database (https://morphobank.org/) with an accession number of 3107 (Ziegler et al., 2018).
Next, to determine the nature of these muscle-like fibers, histology and Masson staining were performed on fixed tissues in both longitudinal and cross-sections (Figure 4, Figure S2). Masson stained muscle-like tissues to be red while some nearby tissues to be blue (Figure 4, Figure S2). The red indicated the tissue to be muscle while the blue indicated the presence of collagen. Hematoxylin-eosin (H&E) stain showed the multiple nuclei in the cells (black arrows). The cross-section showed the near-circle shape of fibers with diameters of around 100 μm, in agreement with the above micro-CT and SEM results. The staining also indicated the absence of any tissues between the muscle fibers, different from vertebrate muscles. From the histology, it was also seen that the muscle was in close contact with potential cartilage (blue color), similar to the structure found in the gastropod radular apparatus (Guralnick and Smith, 1999; Katsuno and Sasaki, 2008). They may provide physical support for the radula, which needs to be studied in the future. Additionally, at the interface of cartilage-like tissue and muscle tissue, they were intertwined, indicating a gradient composition.
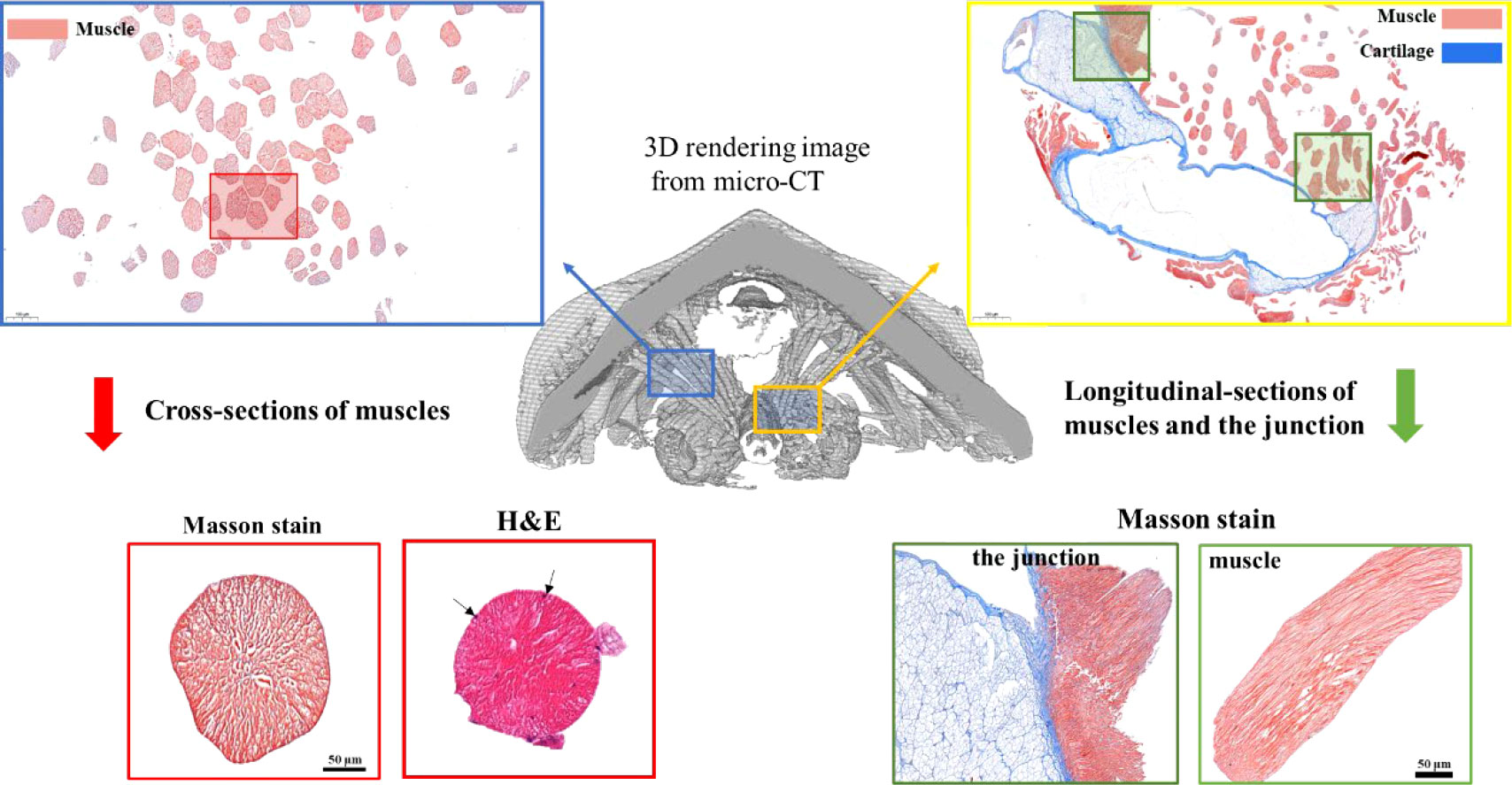
Figure 4 Masson staining of radular muscles and their surrounding tissues. The middle is the 3D rendering micro-CT image. The upper left and the lower left is the cross-section of muscle fibers and one of the magnifications. The upper right and the lower right are the longitudinal sections of muscle fibers and the junction regions between the muscle and the supporting cartilage. For the cross-sectional sections of muscles, both Masson staining and H&E staining were shown. Black arrows indicated multiple cell nuclei.
Molecules underlying complex muscle fibers of chiton A. loochooana
After establishing that the complex muscle tissue is potentially involved in the control of radular movement, we were wondering about the molecular mechanism underlying such function. To solve this question, proteomics (Figure 5, Tables 1, 2) was used to examine the abundant molecules at protein levels while qPCR (Figure 6) was used to investigate the molecules at transcriptional levels in the muscle and the differential expressed genes compared to the foot, a less mobile tissue.
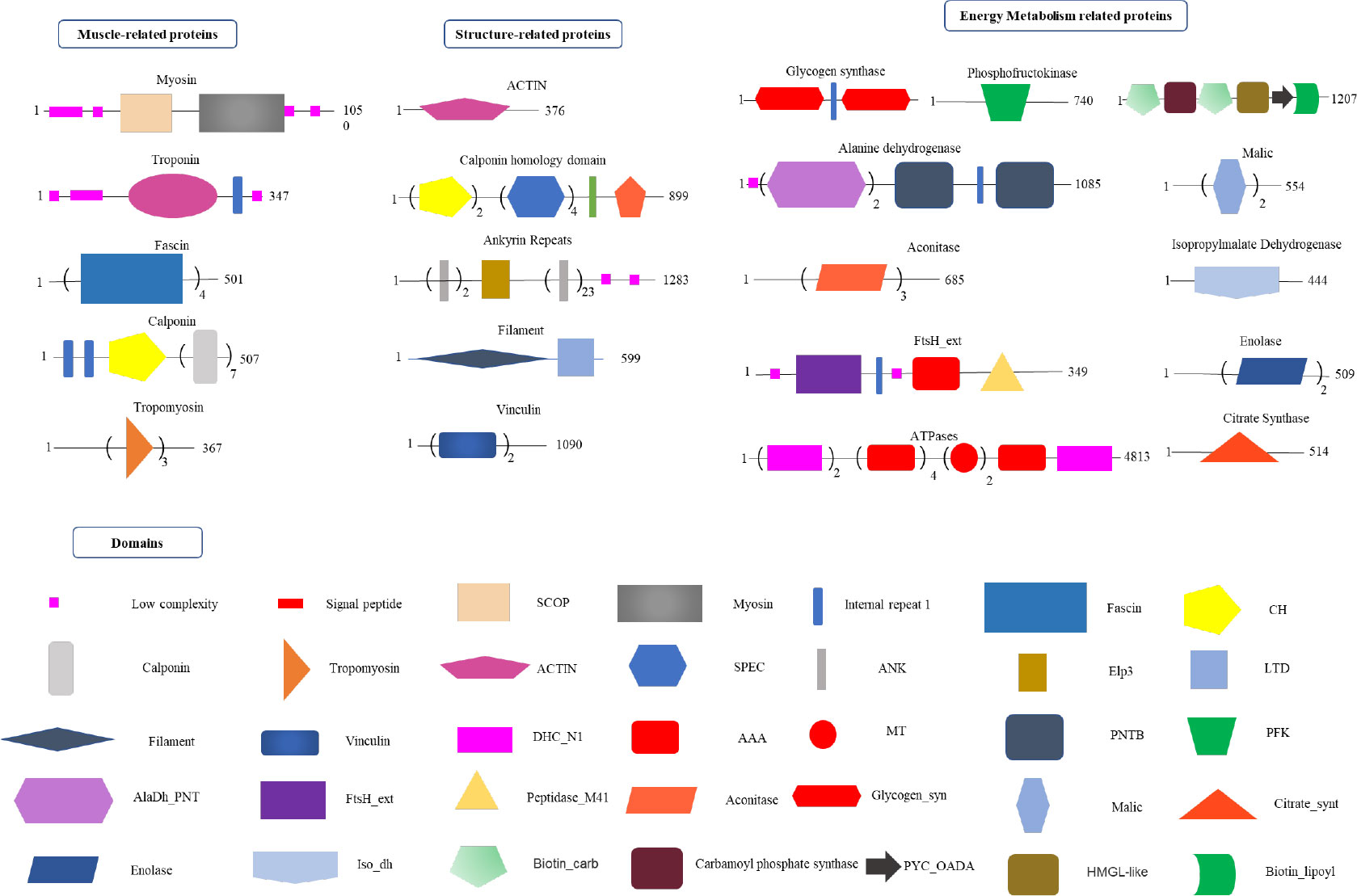
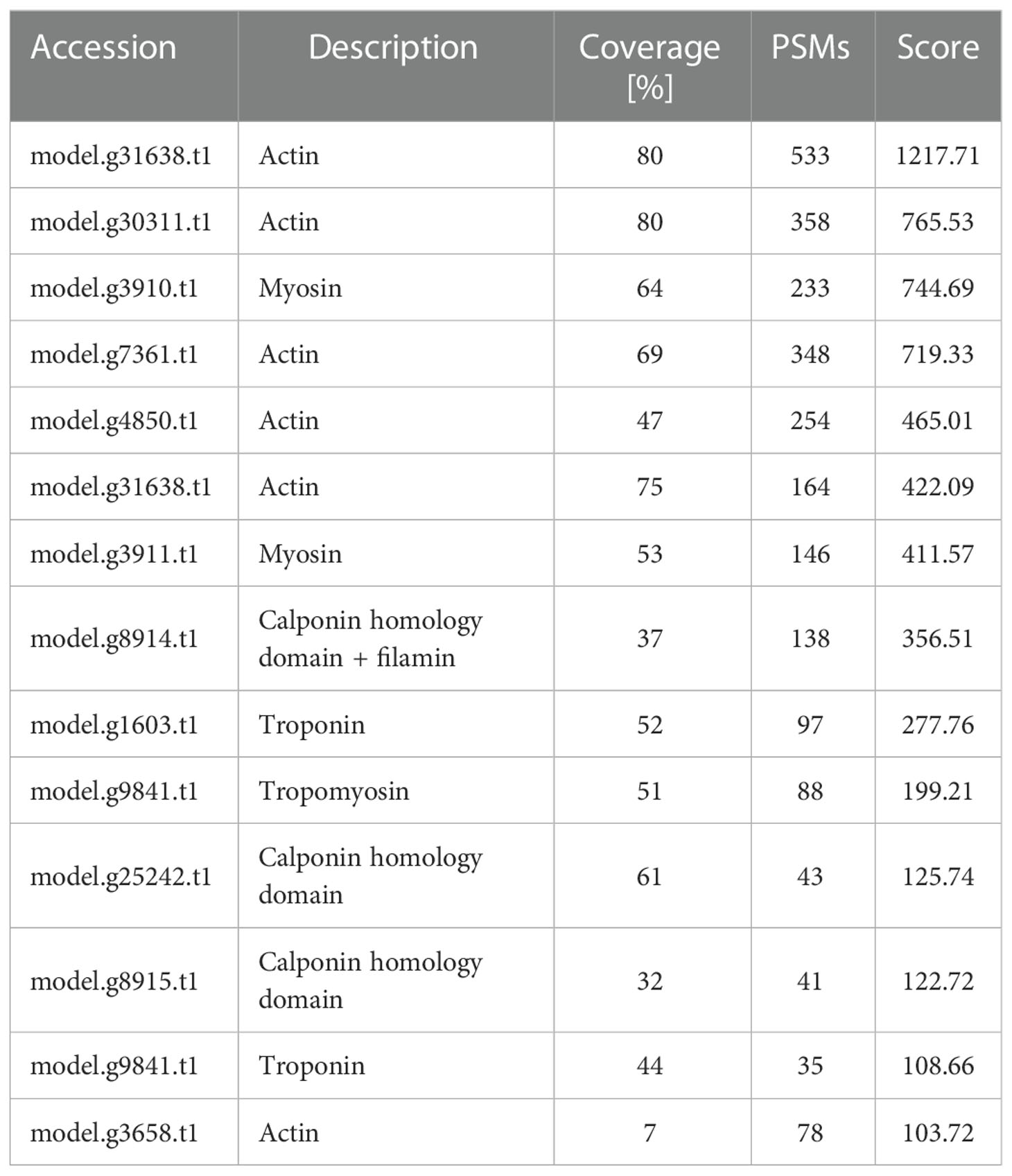
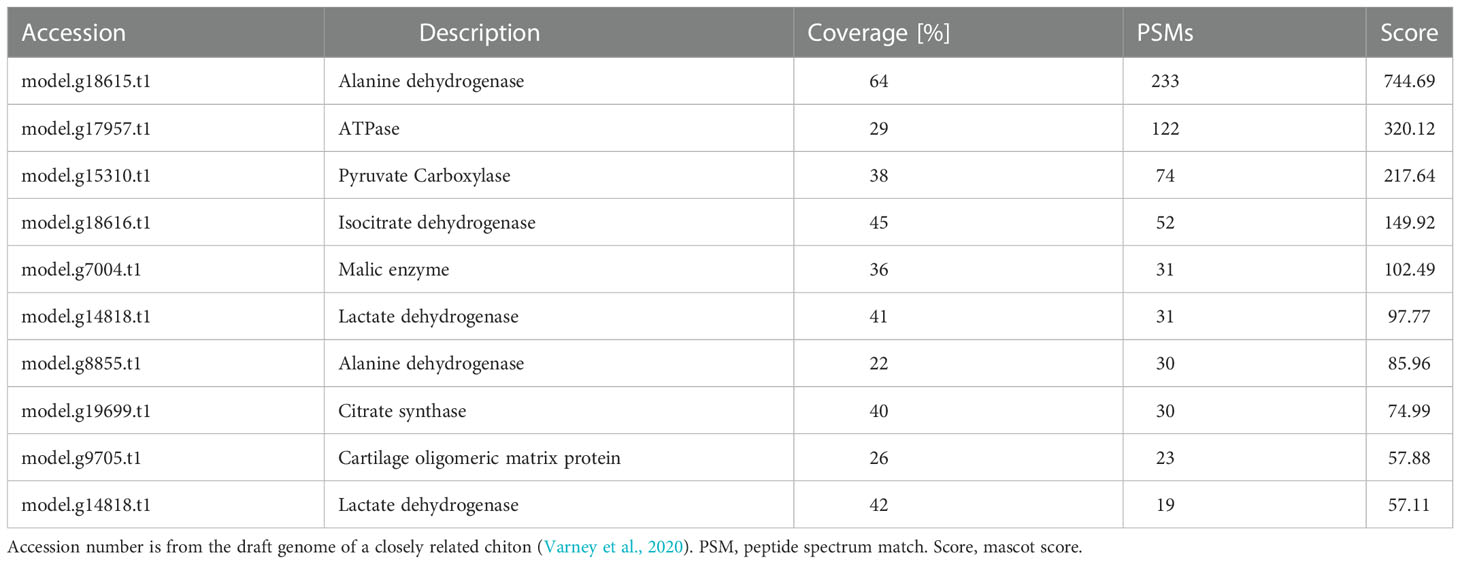
Figure 5 Proteomics analysis of radular muscles. Three types of proteins that are abundant in muscles were manually divided including muscle-related, structure-related, and energy metabolism-related proteins. Domains were predicted by the SMART website. AAA, ATPases associated with a variety of cellular activities; AlaDh_PNT, Alanine dehydrogenase/PNT, N-terminal domain; ANK, ankyrin repeats; Biotin_carb, Biotin carboxylase; CH, Calponin homology domain; DHC_N1, Dynein heavy chains; Elp3,Elongator protein 3; FtsH_ext, FtsH Extracellular; HMGL-like, Pyruvate carboxylase; Iso_dh, Isocitrate/isopropylmalate dehydrogenase; LTD, C-terminal lamin-tail domain; MT, Microtubule-binding stalk of dynein motor; PNTB, NAD(P) transhydrogenase beta subunit; PFK, Phosphofructokinase; PYC_OADA, Conserved carboxylase domain; SPEC, Spectrin repeats;.
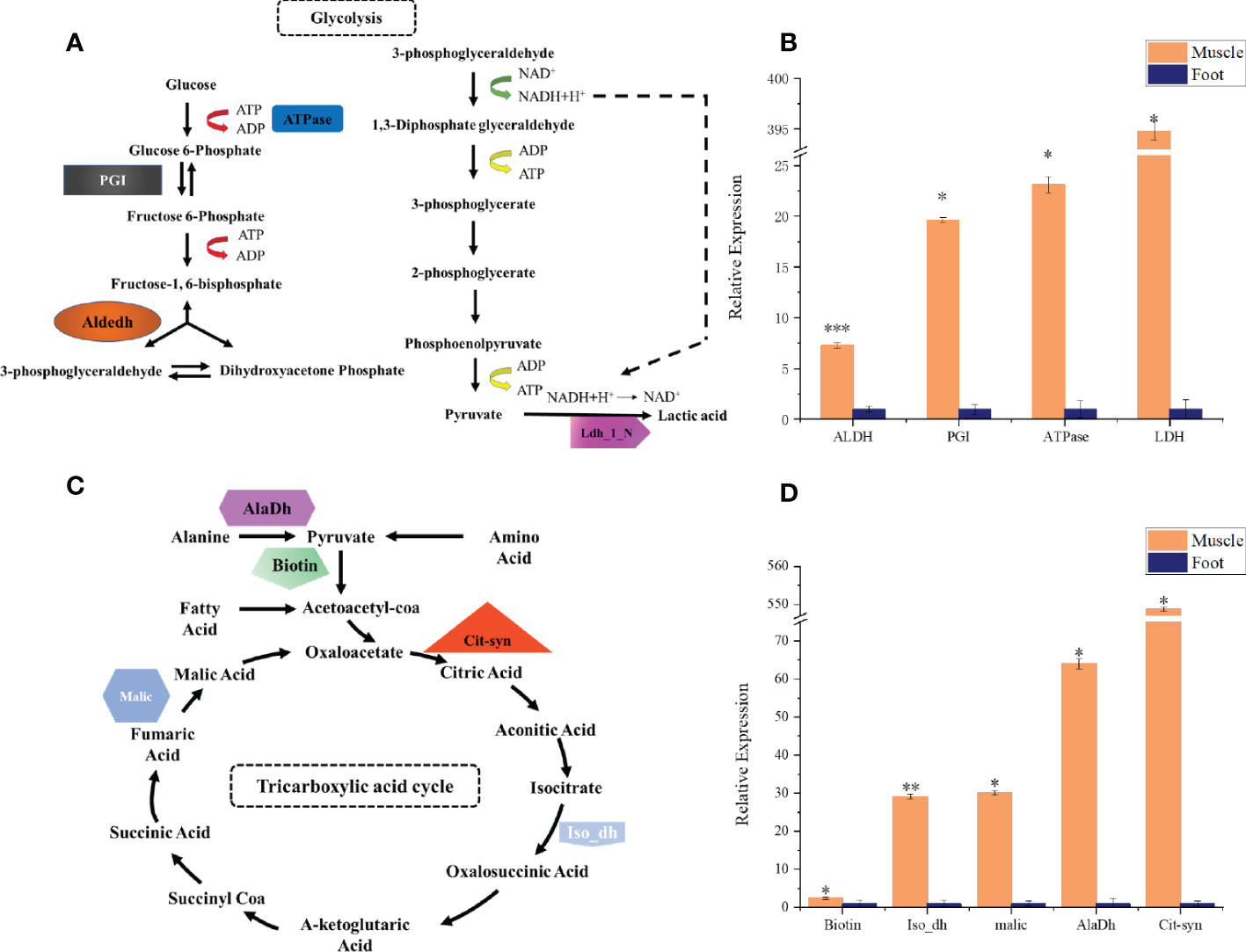
Figure 6 qPCR analysis of key energy metabolism-related genes in radular muscles and foot. (A) the glycolysis pathway and selected genes (B) qPCR analysis of aldehyde dehydrogenases (Aldedh), phosphoglucose isomerase (PGI), ATPase, and lactate dehydrogenase (Ldh_1_N) (C) the TCA cycle and selected genes (D) qPCR analysis of Biotin, isocitrate dehydrogenase (Iso_dH), Malic enzyme, alanine dehydrogenase (AlaDh), and citrate synthase (Cit-syn). All gene expressions are relative to the foot tissues. *P<0.05; **P<0.01; ***P<0.001, t-test.
By using the recently published chiton genome of a closely related species Acanthopleura granulata (Varney et al., 2020), more proteins were detected. In total, 1195 proteins were identified (score above 5), among them, 163 proteins were manually annotated by domain analysis (score above 50). In general, highly abundant proteins can be divided into three types: muscle, structure, and energy metabolism-related proteins (Figure 5, Tables 1, 2). Myosin, actin, and other cytoskeletal proteins were found in large quantities with high mascot scores and peptide spectrum matches (Table 1). These proteins are mainly involved in muscle synthesis and movement in the organism. For example, troponin is integral to muscle contraction in skeletal muscle and cardiac muscle (Gomes et al., 2002) while calponin is a smooth muscle-specific protein (Gimona et al., 1990), confirming the muscle nature of the tissue. Additionally, ATPase, tricarboxylic acid (TCA) cycle-related enzymes, and glycolysis enzymes were also identified (Figure 5). Since muscle movement may require mountable energy, qPCR was used to quantify the mRNA expression levels of some key enzymes in the energy production process (Figure 6). The first step in the energy production using sugar is glycolysis. Compared to the foot, ATPase [catalyzing adenosine triphosphate (ATP) to form adenosine diphosphate (ADP)], phosphoglucose isomerase (PGI, catalyzing an aldose-ketose isomerization), aldehyde dehydrogenases (ALDH, catalyzing the oxidation of aldehydes), and lactate dehydrogenase (LdH, catalyzing the conversion of lactate to pyruvate and back) were highly expressed with 25 ± 2, 20 ± 1, 7 ± 1, and 395 ± 3 times fold than that in the foot respectively (Figures 6A, B) (Achari et al., 1981; Smith and Doolittle, 1992; Axelsen and Palmgren, 1998). In the oxidative energy production by the TCA cycle, alanine dehydrogenase (AlaDH, catalyzing a reversible conversion of L-alanine to pyruvate), Biotin carboxylase (Biotin, oxidizing acetyl-CoA to pyruvate), citrate synthase (Cit-syn, catalyzing the irreversible condensation of acetyl-CoA with oxaloacetate to form citrate), isocitrate dehydrogenase (Iso_dH, catalyzing the oxidative decarboxylation of isocitrate, producing alpha-ketoglutarate and CO2), and malic enzyme (catalyzing the conversion of malate to pyruvate) were highly expressed with 65 ± 2, 2 ± 0.5, 560 ± 1, 30 ± 1, and 30 ± 1 times fold in the muscle compared to the foot respectively (Figures 6C, D) (Hurley et al., 1989; Long et al., 1994; Tran et al., 2015). The above results suggested that the energy production process in the muscle is extremely dynamic, which may provide a high energy supply for muscles.
Interestingly, a cartilage oligomeric matrix protein (COMP) was also found in the proteomic analysis of the muscle (Table 2). Sequence analysis showed that it was predicted to have a signal peptide, a COMP domain, five epidermal growth factor (EGF) domains, and a type 3 thrombospondin domain (Figure 7A). qPCR showed that it was highly expressed in the muscle, 45 times higher compared to the foot (Figure 7B). COMP, also known as thrombospondin-5, was an extracellular matrix (ECM) protein primarily present in cartilage, ligament, and tendon (Posey et al., 2018). The phylogenetic tree showed that the COMP of chitons was closer to that in the cephalopod, and was found to be expanded in gastropods and reduced in bivalves (Figure 7C), consistent with the expansion and loss of radulae in gastropods and bivalves respectively. Additionally, COMP was also found in Crustacean and Annelida (Figure 7C). This suggested an ancestral origin of COMP, which may play roles in cartilage formation at the interface of cartilage and muscles.
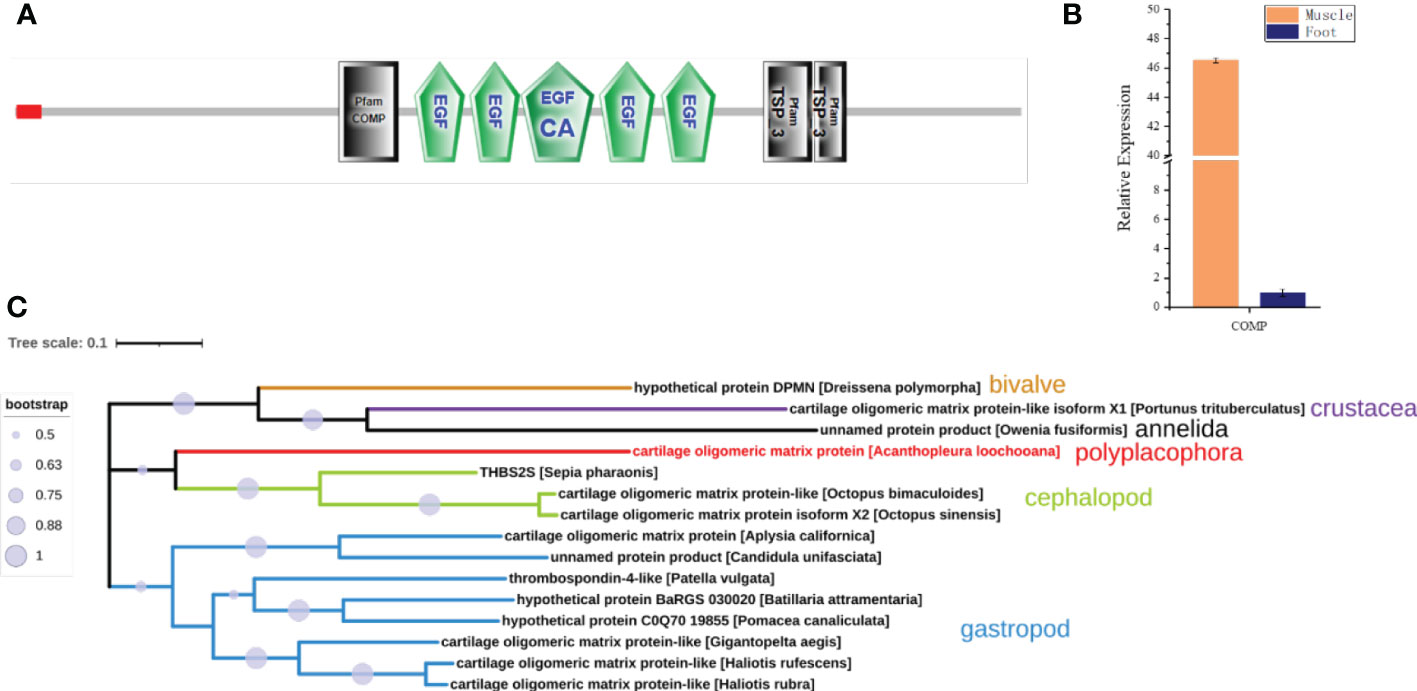
Figure 7 Cartilage oligomeric matrix protein (COMP) analysis. (A) The molecular architecture of COMP. EGF, Epidermal growth factor-like domain; EGF_CA, Calcium-binding EGF-like domain; TSP_3, type 3 thrombospondin. (B) The relative expression of COMP in the muscle compared to the foot by qPCR. (C) The phylogenetic tree of COMP in Mollusca, Crustacean, and Annelida.
Discussion
This study revealed an unknown detail of the feeding apparatus from the chiton from a multiscale perspective. Previous studies focused on iron-based teeth in chitons (Lowenstam, 1967; Weaver et al., 2010) and cartilage tissues in gastropods (Graham, 1973; Kehl et al., 2019). This limited our understanding of the whole feeding process. The moving pattern of the radula in chitons is found to be complex containing various motions such as bending, rotating, and twisting (Pohl et al., 2020; Scheel et al., 2020). The complex muscle fibers found may control the rotating and/or twisting of the radula membrane in chitons. 54 muscle fibers with diameters of ca. 130 μm had two different groups of angles relative to the long axis of the radula (Figure 8A). All these muscles were anchored on the second and third shell plates, which is relatively hard and stiff (Figure 8A). It may provide a fixed point like a swing to contract the muscles, and thus, the radula.
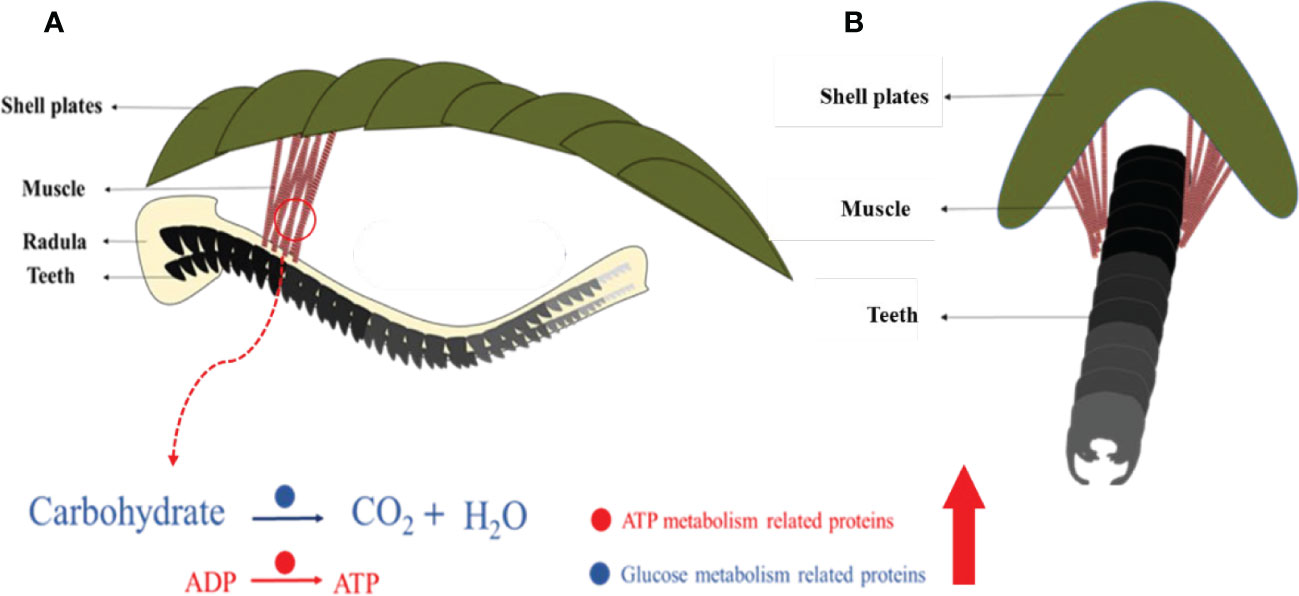
Figure 8 Schematic images of complex muscles and their relationship with the radula. Coronal (A) and sagittal (B) view. Molecules that are abundant in muscles are related to glucose metabolism and ATP generation.
Histology and proteomics confirmed the muscle to be more similar to skeletal muscles with cylindrical shapes, multi-nucleated cells, and abundant muscle-specific proteins such as troponin. However, it did not have the typical striated pattern in skeletal muscles of vertebrates and contained calponin, molecules that are smooth muscle-specific. These mixed features may indicate a transit muscle fiber between smooth and skeletal muscles in vertebrates.
The genes related to energy production (both anaerobic and aerobic respiration) were highly expressed in the muscle, compared to the foot, a tissue that is less mobile (Figure 8B). Notably, lactate dehydrogenase, responsible for the conversion of pyruvate, the end product of glycolysis, into lactic acid, was ~400 times expressed in the muscle compared to the foot. In aerobic conditions, citrate synthase is a central enzyme in this process of sugar oxidation. In the muscle, it was 560 times expressed in the muscle compared to the foot. This indicated an extremely adapted molecular mechanism to high energy demand under various conditions.
Moreover, we have identified COMP in the muscle at both transcript and protein levels. COMP is commonly found in the extracellular matrix of cartilage, tendon, and ligament (Posey et al., 2018). By Masson staining, we also saw that some chondrocytes were present at the junction of the radula and the muscle. It is deduced that COMP produced in the muscle may promote the formation of the radula.
Future functional studies should be carried out to investigate how these complex muscles control the movement of the radula membrane, in what direction, and its consequence on the function of radulae. Additionally, the supporting tissue (potentially cartilage) that is in close contact with the muscle and the teeth, should be scrutinized as well. The nervous system controlling the muscle, cartilage, and radula would also be interesting to be known. Thus, the nerves-muscle-cartilage-mineralization system is analogous to that in vertebrate skeletal systems. The information from the chiton radula system will not only be helpful for us to understand how the radula is moved but also inspire novel and efficient soft-hard tissue interface design in cut devices or robotics.
Conclusions
In summary, through multiscale analyses from the macroscale to the molecular levels, complex muscle fibers were found to be in close contact with the radula of the chiton. These muscle fibers with diameters of around 130 μm may be responsible for the bending movement of the radula, given their relative spatial relationship with the radula and their highly adapted molecular traits for energy production.
Materials and methods
Animals
Chiton Acanthopleura loochooana was collected from the coastal area of Zhangzhou, Fujian Province, China. They were cultured in seawater for future use.
Radula and muscle extraction
A scalpel was used to dissect the connecting tissue between the shell plates along the second shell plate. the tooth and the surrounding muscle tissue were isolated under a stereo microscopy (Shangguang Suzhou FTL-7063A). For the protein extraction, they were stored at -20°C for further use. For histology, they were fixed in 4% paraformaldehyde immediately after dissection.
Optical image
Fresh muscles were observed under an optical microscopy (Nexcope NE610) with a magnification of 400X.
Scanning electron microscopy
Air-dried fixed muscles were spurred with gold nanoparticles for 30 s before imaging using Hitachi S4800 at 5 kV.
Histology
Tissues were fixed for 24 h and were then dehydrated gradually with ethanol (75%, 80%, 90%, 95%) followed by paraffin embedding. The block was then cut in both cross and longitudinal-sectional directions with a thickness of 5 μm. Standard Masson staining protocol was used to stain the tissue. Deparaffinized and rehydrated tissue sections were used.
Micro-CT data acquisition and analysis
Soft-tissue micro-CT data of chiton Acanthochitona crinita was directly retrieved from the Morphobank database (https://morphobank.org/) with an accession number of 3107 (Ziegler et al., 2018). Formalin-fixed, ethanol-preserved specimen was placed in 50 ml plastic tubes and stained using an 0.3% phosphotungstic acid (PTA) solution in 70% ethanol. Specimens were stained for four weeks and placed inside plastic tubes filled with clean 70% ethanol directly before scanning. It was originally scanned with SkyScan 1272, 100 kV, with a resolution of 12 μm. The 3D renderings were created and analyzed using the commercial software Dragonfly Pro.
Protein extraction and proteomics
Protein extraction
The muscle tissue was cut into small pieces with a length of less than 1 cm. Appropriate amounts of cell lysate (200 μL per 20 mg tissue) and 4 mM proteinase inhibitor (AEBSF, Shanghai Sangon) were added. The muscle tissue was homogenized with a glass homogenizer until fully cracked. After adequate lysis, the solution was centrifuged at 10,000 g for 5 minutes. The supernatant was collected and approximately 20 μg proteins were subjected to SDS-PAGE gel separation.
Proteomics analysis
The process was according to a previously established protocol in chitons (Liu et al., 2022a). Briefly, proteins were stained with Coomassie Blue and the entire protein lane (Figure S3) was excised in four pieces, which were completely destained by washing with 50 μL of 50 mM NH4HCO3/acetonitrile (50/50) mixture for 30 min at 37 °C. Reduction was conducted with 50 μL of 10 mM DTT in 50 mM NH4HCO3 for 1 h at 57 °C, followed by alkylation with 50 μL of 100 mM iodoacetamide for 30 min at room temperature in the dark. The cut gels pieces were dried in acetonitrile and then treated with 0.4 μg trypsin (Proteomics Grade, Sigma) in 10 μL of 50 mM NH4HCO3 for 12 h at 37 °C. The suspension was treated with 50 μL of 1% formic acid at 30 °C for 30 min under agitation. The digests were then lyophilized and suspended in 30 μL of 0.1% trifluoroacetic acid and 4% acetonitrile for LC-MS/MS analysis. Five μL of the samples were injected into the LTQ Orbitrap Velos mass spectrometer with Dionex U-3000 Rapid Separation nano-LC system (Thermo Scientific) for analysis. MS data were acquired automatically using Analyst QS 1.1 software (Applied Biosystems) following an MS survey scan over m/z 350-1500 at a resolution of 60,000 for full scan and 2, 000 for MS/MS measurements.
The LC-MS/MS data were searched against the translated proteome from the genome of chitons (Varney et al., 2020) using a Mascot 2.1 search engine with carbamidomethylated cysteine as a fixed modification and oxidized methionine and tryptophan as variable modifications. No other post-translation modification has been performed. The peptide MS and MS/MS tolerances were set to 0.5 Da. Finally, sequences with mascot scores of at least 5.0 and with at least two matched peptide fragments were considered valid. The search results can be downloaded from the website of ProteomeXchange.
Bioinformatic analysis
Identification of proteins from above was performed using Blastp and tBlastn searches against the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Conserved domains were predicted using SMART (http://smart.embl-heidelberg.de/) (Letunic and Bork, 2017; Letunic et al., 2020). Phylogenetic trees were conducted using MEGA-X software (https://megasoftware.net/) (Tamura et al., 2021). Maximum likelihood method was used with bootstrap replicates of 1000.
Quantitative real-time PCR (qPCR)
Total RNA was extracted from the muscle and foot tissues of freshly sacrificed chiton using Trizol reagent (Invitrogen) following the manufacturers’ protocol. The cDNA template was synthesized using AMV First Strand cDNA Synthesis Kit (Sangon Biotech). qPCR was performed with three independent replicates by using SYBR Green qPCR mix (Transgen) on a LightCycler 480 II Real-time PCR System (Roche). The program used for the experiment: 30 s at 94°C and 40 cycles (each cycle was for 30 s at 94°C, 15 s at 60°C, and 10 s at 72°C). Gene expression level was calculated by the 2−ΔΔCT method with β-actin as the reference gene. Gene expression is relative to the foot sample. All primers used in this study are listed in Table S1.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/pride/archive/, PXD037323.
Author contributions
CL conceived the project. DS performed the experiments. CL, ZW, JH analyzed the data. CL and DS wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
CL gratefully acknowledges the support of the National Natural Science Fund of China 42206081 and Natural Science Foundation of Jiangsu Province BK20210363. JH gratefully acknowledges the support of the National Natural Science Fund of China 42106091.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1107714/full#supplementary-material
References
Achari A., Marshall S., Muirhead H., Palmieri R., Noltmann E. (1981). Glucose-6-phosphate isomerase. Phil. Trans. R. Soc London. B 293, 145–157. doi: 10.1098/rstb.1981.0068
Axelsen K. B., Palmgren M. G. (1998). Evolution of substrate specificities in the p-type ATPase superfamily. J. Mol. Evol. 46, 84–101. doi: 10.1007/PL00006286
Brooker L., Lee A., Macey D., Van Bronswijk W., Webb J. (2003). Multiple-front iron-mineralisation in chiton teeth (Acanthopleura echinata: Mollusca: Polyplacophora). Mar. Biol. 142, 447–454. doi: 10.1007/s00227-002-0957-8
Gimona M., Herzog M., Vandekerckhove J., Small J. V. (1990). Smooth muscle specific expression of calponin. FEBS Lett. 274, 159–162. doi: 10.1016/0014-5793(90)81353-P
Golding R. E., Ponder W. F., Byrne M. (2009). Three-dimensional reconstruction of the odontophoral cartilages of caenogastropoda (Mollusca: Gastropoda) using micro-CT: Morphology and phylogenetic significance. J. Morphology 270, 558–587. doi: 10.1002/jmor.10699
Gomes A. V., Potter J. D., Szczesna-Cordary D. (2002). The role of troponins in muscle contraction. IUBMB Life 54, 323–333. doi: 10.1080/15216540216037
Graham A. (1973). The anatomical basis of function in the buccal mass of prosobranch and amphineuran molluscs. J. Zoology 169, 317–348. doi: 10.1111/j.1469-7998.1973.tb04560.x
Guralnick R., Smith K. (1999). Historical and biomechanical analysis of integration and dissociation in molluscan feeding, with special emphasis on the true limpets (Patellogastropoda: Gastropoda). J. Morphology 241, 175–195. doi: 10.1002/(SICI)1097-4687(199908)241:2<175
Hurley J. H., Thorsness P. E., Ramalingam V., Helmers N. H., Koshland D. Jr., Stroud R. M. (1989). Structure of a bacterial enzyme regulated by phosphorylation, isocitrate dehydrogenase. Proc. Natl. Acad. Sci. U.S.A. 86, 8635–8639. doi: 10.1073/pnas.86.22.8635
Katsuno S., Sasaki T. (2008). Comparative histology of radula-supporting structures in Gastropoda. Malacologia 50, 13–56. doi: 10.4002/0076-2997-50.1.13
Kehl C. E., Wu J., Lu S., Neustadter D. M., Drushel R. F., Smoldt R. K., et al. (2019). Soft-surface grasping: radular opening in aplysia californica. J. Exp. Biol. 222, 1–14. doi: 10.1242/jeb.191254
Krings W., Brütt J.-O., Gorb S. N. (2022). Ontogeny of the elemental composition and the biomechanics of radular teeth in the chiton lepidochitona cinerea. Front. Zool. 19, 19. doi: 10.1186/s12983-022-00465-w
Letunic I., Bork P. (2017). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi: 10.1093/nar/gkx922
Letunic I., Khedkar S., Bork P. (2020). SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Liu C., Liu H., Huang J., Ji X. (2022a). Optimized sensory units integrated in the chiton shell. Mar. Biotechnol. 24, 380–392. doi: 10.1007/s10126-022-10114-2
Liu X., Sigwart J. D., Sun J. (2022b). Phylogenomic analyses shed light on the relationships of chiton superfamilies and shell-eye evolution. bioRxiv. doi: 10.1101/2022.12.12.520088
Lobo-da-Cunha A., Alves A., Oliveira E., Calado G. (2022). Functional histology and ultrastructure of the digestive tract in two species of chitons (Mollusca, polyplacophora). J.Mar. Sci. Eng. 10, 160. doi: 10.1016/S0021-9258(17)42017-5
Long J. J., Wang J.-L., Berry J. O. (1994). Cloning and analysis of the C4 photosynthetic NAD-dependent malic enzyme of amaranth mitochondria. J. Biol. Chem. 269 (4), 2827–2833. doi: 10.1016/S0021-9258(17)42017-5
Lowenstam H. A. (1967). Lepidocrocite, an apatite mineral, and magnetite in teeth of chitons (Polyplacophora). Science 156, 1373–1375. doi: 10.1126/science.156.3780.1373
Nemoto M., Ren D., Herrera S., Pan S., Tamura T., Inagaki K., et al. (2019). Integrated transcriptomic and proteomic analyses of a molecular mechanism of radular teeth biomineralization in cryptochiton stelleri. Sci. Rep. 9, 856. doi: 10.1038/s41598-018-37839-2
Nemoto M., Wang Q., Li D., Pan S., Matsunaga T., Kisailus D. (2012). Proteomic analysis from the mineralized radular teeth of the giant pacific chiton, cryptochiton stelleri (Mollusca). Proteomics 12, 2890–2894. doi: 10.1002/pmic.201100473
Pohl A., Herrera S. A., Restrepo D., Negishi R., Jung J.-Y., Salinas C., et al. (2020). Radular stylus of cryptochiton stelleri: A multifunctional lightweight and flexible fiber-reinforced composite. J. Mech. Behav. Biomed. Mater. 111, 103991. doi: 10.1016/j.jmbbm.2020.103991
Posey K. L., Coustry F., Hecht J. T. (2018). Cartilage oligomeric matrix protein: COMPopathies and beyond. Matrix Biol. 71-72, 161–173. doi: 10.1016/j.matbio.2018.02.023
Scheel C., Gorb S. N., Glaubrecht M., Krings W. (2020). Not just scratching the surface: distinct radular motion patterns in Mollusca. Biol. Open 9, 1–11. doi: 10.1242/bio.055699
Smith M. W., Doolittle R. F. (1992). Anomalous phylogeny involving the enzyme glucose-6-phosphate isomerase. J. Mol. Evol. 34 (6), 544–545. doi: 10.1007/BF00160467
Smith S. A., Wilson N. G., Goetz F. E., Feehery C., Andrade S. C., Rouse G. W., et al. (2011). Resolving the evolutionary relationships of molluscs with phylogenomic tools. Nature 480, 364–367. doi: 10.1038/nature10526
Stegbauer L., Smeets P. J. M., Free R., Wallace S. G., Hersam M. C., Alp E. E., et al. (2021). Persistent polyamorphism in the chiton tooth: From a new biomineral to inks for additive manufacturing. Proc. Natl. Acad. Sci. U.S.A. 118, e2020160118. doi: 10.1073/pnas.2020160118
Tamura K., Stecher G., Kumar S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tran T. H., Hsiao Y.-S., Jo J., Chou C.-Y., Dietrich L. E., Walz T., et al. (2015). Structure and function of a single-chain, multi-domain long-chain acyl-CoA carboxylase. Nature 518, 120–124. doi: 10.1038/nature13912
Varney R. M., Speiser D. I., Mcdougall C., Degnan B. M., Kocot K. M. (2020). The iron-responsive genome of the chiton acanthopleura granulata. Genome Biol. Evol. 13, evaa263. doi: 10.1093/gbe/evaa263
Wang T., Huang W., Pham C. H., Murata S., Herrera S., Kirchhofer N. D., et al. (2022). Mesocrystalline ordering and phase transformation of iron oxide biominerals in the ultrahard teeth of cryptochiton stelleri. Small Struct. 3, 2100202. doi: 10.1002/sstr.202100202
Wanninger A., Wollesen T. (2019). The evolution of molluscs. Biol. Rev. 94, 102–115. doi: 10.1111/brv.12439
Weaver J. C., Wang Q., Miserez A., Tantuccio A., Stromberg R., Bozhilov K. N., et al. (2010). Analysis of an ultra hard magnetic biomineral in chiton radular teeth. Mater. Today 13, 42–52. doi: 10.1016/S1369-7021(10)70016-X
Keywords: radula, muscle, chiton, micro-CT, hard-soft tissue interfaces
Citation: Sun D, Liu C, Wang Z and Huang J (2023) Multiscale analysis of the unusually complex muscle fibers for the chiton radulae. Front. Mar. Sci. 10:1107714. doi: 10.3389/fmars.2023.1107714
Received: 25 November 2022; Accepted: 09 January 2023;
Published: 23 January 2023.
Edited by:
Ciro Rico, Institute of Marine Sciences of Andalusia, Spanish National Research Council (CSIC), SpainReviewed by:
Guoyong Yan, Hong Kong University of Science and Technology, Hong Kong SAR, ChinaMarina Pozzolini, University of Genoa, Italy
Copyright © 2023 Sun, Liu, Wang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuang Liu, Y2h1YW5nbGl1MjAyMEBoaHUuZWR1LmNu
 Dawei Sun1,2
Dawei Sun1,2 Chuang Liu
Chuang Liu Zhenglu Wang
Zhenglu Wang Jingliang Huang
Jingliang Huang