
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 16 June 2023
Sec. Microbial Symbioses
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1099478
Ulva is a commercially important marine macroalga. It hosts both epiphytes and endophytes. The latter are assumed to protect Ulva through secondary metabolites. Previously, we demonstrated bioactive endophytes from macroalgae with great potential to control diseases of aquaculture. In this study, we introduced a bioactive bacterial endophyte back into its original host (Ulva sp.) and demonstrated its survival over time in fresh and freeze-dried Ulva sp. We visualized the endophyte’s location and survival in the seaweed using a Green Fluorescent Protein (GFP) reporter gene. The isolate colonized the intercellular space and survived for at least 5 months in fresh, and 12 months in freeze-dried algae, while maintaining its bioactivity against the aquaculture pathogen Streptococcus iniae. We studied the influence of the endophyte on the bacterial community in the Ulva sp. We found that once introduced, the endophyte significantly changed algal microbiota diversity and abundance. Two of Ulva’s associated bacterial species were quantified over time, suggesting different trends in absolute abundance of these bacteria between treatments. To the best of our knowledge, this is the first report of the successful introduction of an endophytic microorganism into macroalgal tissue. These findings may be useful in applied research for the potential management of aquaculture diseases.
Endophytes are microorganisms that colonize the intercellular tissue of plants with no deleterious effects on their host. They are found in all plants, including algae. Many endophytes secrete bioactive compounds, termed secondary metabolites (Bacon and White, 2016; Gouda et al., 2016), which may inhibit the growth of other microorganisms in their habitat (the endosphere) (Kharwar and Strobel, 2010; Vallet et al., 2018; Trivedi et al., 2020). Research on endophytes has typically focused on those in terrestrial plants (Liarzi and Ezra, 2014), with less attention to aquatic plants, and algae in particular, despite their having been shown to harbor endophytes. The study of algal endophytes may open new prospects for future use of endophytes in various biotechnological applications (Sarasan et al., 2020). The use of algal endophytes as biological control agents and as a source for secondary metabolites could provide potential benefits in the aquaculture industry. Our previous study on algal endophytes collected from the Israeli coastline revealed bioactive endophytes from an Ulva sp. (Deutsch et al., 2021). Ulva is a green seaweed of the phylum Chlorophyta that has great importance in macroalgal growth applications and has more recently been developed into a suitable model system for studying the interactions between bacteria and macroalgae (Wichard, 2015; Kessler et al., 2018). Ulva spp. and their associated endophytes are considered a rich source for bioactive compounds (Dhanya et al., 2016; Habbu et al., 2016). Like other macroalgae, Ulva lacks obvious structural defense mechanisms and utilizes chemical defenses, produced by itself or by its associated microorganisms, to survive in varying habitats (Jensen and Fenical, 1994; Dominguez and Loret, 2019). Ulva’s microbiota typically shows common associated bacterial taxa, including Maribacter sp. and Roseobacter sp. (Marshall et al., 2006; Burke et al., 2010; Liu et al., 2011; Fu et al., 2018). These taxa secrete morphogenesis-causing compounds that impact Ulva’s morphology (Wichard, 2015; Ramanan et al., 2016; Kessler et al., 2018; Califano et al., 2020; Ren et al., 2022). Associate bacteria maintain a synergistic effect responsible for a proper thallus development. In the absence of those, the Ulva fails to form its typical morphology and proliferates as an undifferentiated clump of callus cells (Wichard et al., 2015).
As already noted, endophytes may serve as biocontrol agents against aquaculture pathogens that impact fish health and can devastate aquaculture yields. The rationale for using these microorganisms lies mainly in the fact that they evolved mechanisms aiding them in competing with other microorganisms (Gouda et al., 2016) including fish pathogens that may be present in their environment (i.e., in the water, on and in algae, but not within the animal host)(Klerks et al., 2007). Thus, supplementing these active endophytes in fish feed might be beneficial for the fish and other aquatic organisms of interest. Feed additives have been used in aquaculture to induce disease resistance since the early 1990s. One very promising genus of bacterial endophyte which we isolated in our previous study is Bacillus, consisting of Gram-positive bacteria of the phylum Firmicutes. They are rod-shaped, obligate aerobes or facultative anaerobes. Bacillus spp. can be found in many living niches, including fresh and marine water, soil, plants, and animal intestines. They are known for their bioactive metabolites and are frequently used in agriculture for the biological control of pathogens and pests (Jacobsen et al., 2004; Ongena and Jacques, 2008). In recent years, spore-forming Bacillus spp. have become important biocontrol agents and candidate probiotics in aquaculture (Tarnecki et al., 2019; Thurlow et al., 2019; Olmos et al., 2020; Santos et al., 2021). To simplify the study of these microbes in algae, Bacillus isolates can be visualized by transducing a Green Fluorescent Protein (GFP) marker gene alongside an antibiotic resistance gene. A phage is used to transform genes from a donor Bacillus to an acceptor Bacillus. The phage attacks the donor and transforms selected genes to the acceptor, allowing the transformed Bacillus to grow on selective media amended with the antibiotic chloramphenicol (Chu et al., 2006).
Terrestrial plants with introduced endophytes for the biocontrol of pathogens have been widely used since the early 1980s. Endophytes are used for the delivery of bioactive compounds and genes to enhance biotic and abiotic stress resistance, and improve seedling emergence, plant growth and nutritional qualities, resulting in higher yields (Myers and Strobel, 1983; Hall et al., 1986; Kuldau and Bacon, 2008; Ding et al., 2011; Li et al., 2012). However, to the best of our knowledge, the successful artificial introduction of target endophytes into macroalgae has never been reported. An attempt to reveal reintroduced endophytes in macroalgal (Ascophyllum) tissue was made (Thorén-Tolling, 1978), but it was unsuccessful. Co-inoculation of macroalgae with an endophytic fungus and a pathogen for the purpose of biological control was successful in mitigating disease (Vallet et al., 2018). However, this co-inoculation did not represent an actual introduction into algal tissue.
Algae serve as hosts for many microorganisms, both endophytes and epiphytes. Various factors can influence the algal microbiota. Abiotic factors (e.g., temperature, nutrients, geographical and temporal gradients, seasonality) and biotic factors (e.g., symbionts, competitors, pathogens, parasites, grazers) can differ between samples of the same algal species. Microorganisms—pathogenic or beneficial—play a role in influencing the composition of the algal microbiota (Flewelling et al., 2013; Califano et al., 2020). Biologically active metabolite-secreting endophytes can affect the distribution of other microorganisms in their host. Mitter et al. (2017) demonstrated modification of the microbiota of various plant seeds by introducing beneficial endophytes into parent plants at flowering. They concluded that this approach to changing the seed microbiota can improve the traits of seeds of agriculturally important plants and can overcome today’s limitations in crop production. Following this approach, we hypotheses that the introduction of an active endophyte into the alga will affect its microbiota and may benefit the host.
In this study, we demonstrate that an endophytic bacterium (Bacillus subtilis) isolated from an Ulva sp. can be reintroduced into its original host and impact the algal microbiome. We also hypothesized that the active endophyte may survive in the introduced algae for long periods of time, under the right conditions, while retaining its bioactivity against fish pathogens. We suggest that this is a prerequisite for the use of macroalgal endophytes as probiotic feed additives for disease management in aquaculture.
The B. subtilis isolate PU10 was originally obtained from an Ulva sp. sampled off the Mediterranean coast of Israel (Deutsch et al., 2021). The isolation procedure, and bioactivity assays against major aquaculture pathogens were performed as described previously (Deutsch et al., 2021). Isolates were identified using polymerase chain reaction (PCR) amplification and multiple sequencing analysis (Deutsch et al., 2021).
The Bacillus isolate, PU10 was transformed with a chloramphenicol-resistance gene and GFP marker (Pspank-gfp), using the transduction method and termed PU10GFP. The transduction procedure for the GFP construct (Duanis-Assaf et al., 2016) was performed according to Shemesh and Chaia (2013). Transformation was validated by visualizing GFP activity in the bacteria using a fluorescence microscope (BX63F, Olympus, Tokyo, Japan) equipped with a DP80 Olympus camera, and growth on antibiotic-amended culture media: nutrient agar (NA) or Luria–Bertani (LB) agar (Acumedia, Lansing, MI, USA) with 10 µg/mL chloramphenicol (Thermo Fisher Scientific, Geel, Belgium).
A single colony of PU10GFP was incubated for 48 h at 25°C, 150 rpm in LB broth medium. After 48 h, the bacterial cells were washed twice with artificial seawater (ASW) (3.7% NaCl) as follows: centrifugation at 3944 g for 10 min, discard supernatant, centrifuge again at 3944 g for 10 min after rinsing with ASW, discard supernatant and re-suspend the bacterial pellet in ASW at half the volume of the initial growth media.
Ulva sp. was grown in the Department of Plant Pathology and Weed Research, Agricultural Research Organization, Volcani Center, Israel, under controlled conditions (23 ± 2°C, daylight, and constant air bubbling). Ulva thalli were cut into approximately 4 cm x 4 cm pieces. Cut thalli were either scratched using a thin needle or introduced into a rotating container with ASW and 10 cm x 10 cm rocks, for 30 min, simulating the natural damage that can occur in the ambient habitat. Thalli were placed in an empty petri plate and covered with PU10GFP solution. ASW alone was used in the control treatment. The thalli were incubated for 4 days at room temperature. To initiate the experiment, thalli were washed four times with ASW and placed in a glass beaker with air-bubbled ASW and a concentration of 0.1 mL/L “Shaphir brand” Nitrate Solution fertilizer (Gat Fertilizers, Kiryat Gat, Israel). Endophytic colonization of Ulva sp. by PU10GFP was validated 24 h postwashing, by microscopic imaging of the Ulva thalli (Figure 1). Images were acquired with a Leica SP8 laser-scanning microscope (Leica, Wetzlar, Germany) equipped with a solid-state laser, HC PL APO CS2 40x/1.10-water objective (Leica) and Leica Application Suite X software. The GFP signal was imaged by excitation with 488 nm laser light and the emission was detected at 500-525 nm. Chloroplast autofluorescence was detected at 650–700 nm. GFP intensity was measured using ImageJ software (NIH.gov).
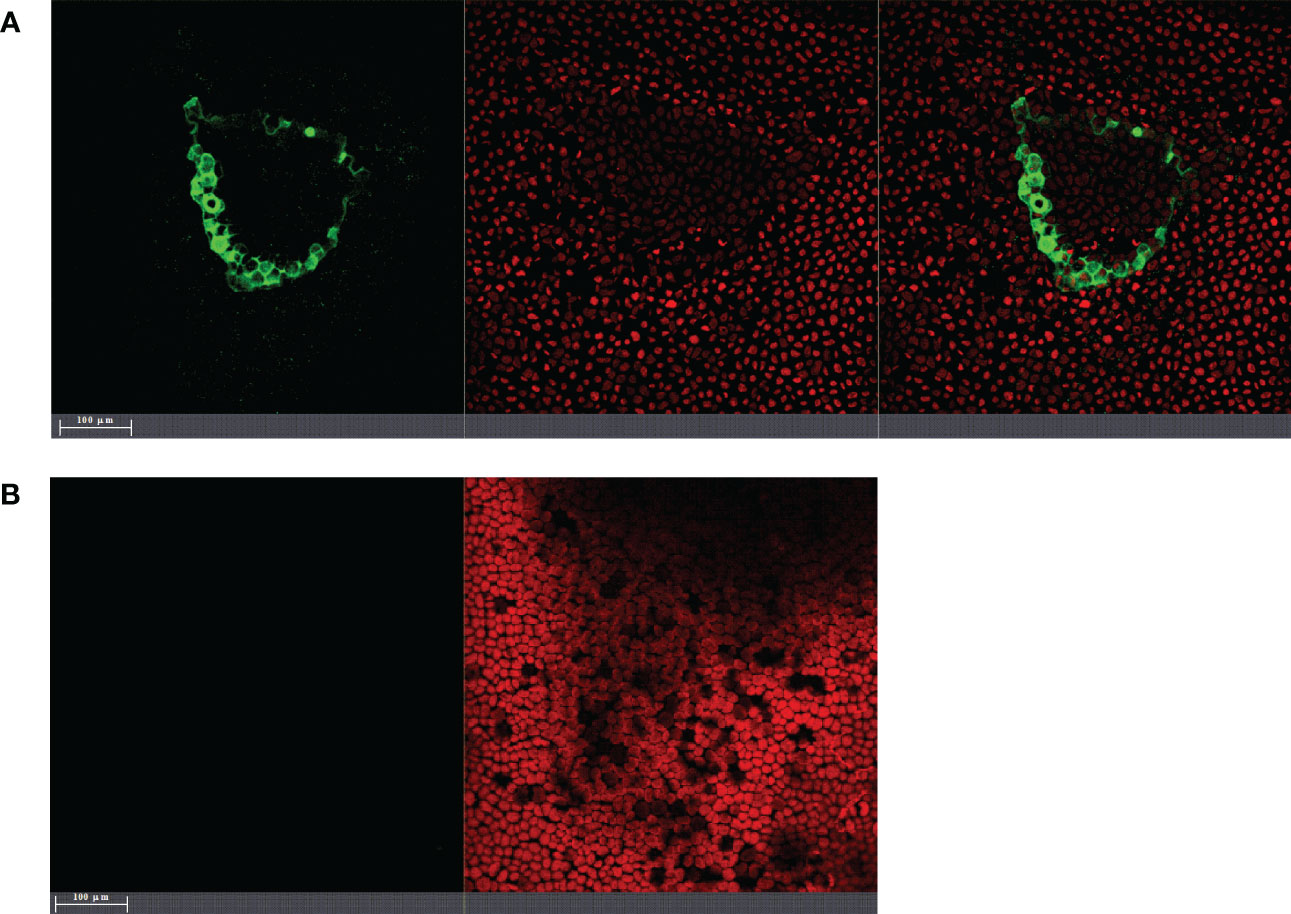
Figure 1 Laser-scanning fluorescence microscopy images of Ulva sp. inoculated with gfp-transformed endophytic PU10 (PU10GFP) 24 h postwashing of inoculation solution. (A) Ulva tissue after introducing PU10GFP isolate. GFP emission at 500–525 nm visualizing the colonization of PU10GFP (left image), emission range of 650–700 nm visualizing autofluorescence of Ulva’s chloroplasts (middle image) and an overlay of both (right image). (B) Control treatment, non-inoculated Ulva tissue (only artificial seawater). Emission range of 500–525 nm showing no GFP (left image) and Ulva chloroplast autofluorescence in emission range of 650–700 nm (right image).
The fluorescence intensity of treated and untreated Ulva thalli was measured every 2-3 days for 2 weeks by laser-scanning microscopy (Figure 2A). In addition to monitoring the fluorescent signal, bacterial colony-forming units (CFU) per sample of treated Ulva thalli was evaluated (and compared to CFU absence in the untreated samples) using antibiotic resistance (chloramphenicol) of the transformed bacteria (Figure 2B). Ulva thalli were sampled on days 0, 4, 7, 14, and then every 15 days for over 5 months. At each sampling time, ~130 mm2 Ulva sp. samples (in triplicate) were crushed with 500 µL sterile saline (0.9% NaCl) in a small 4 cm x 4 cm double-layer nylon sleeve. The crushed Ulva sp. was cultured on LB agar plates amended with chloramphenicol (10 µg/mL) and incubated at 25°C for 72 h, followed by the evaluation of CFU. Fluorescence intensity and CFU/sample experiments were repeated three times each.
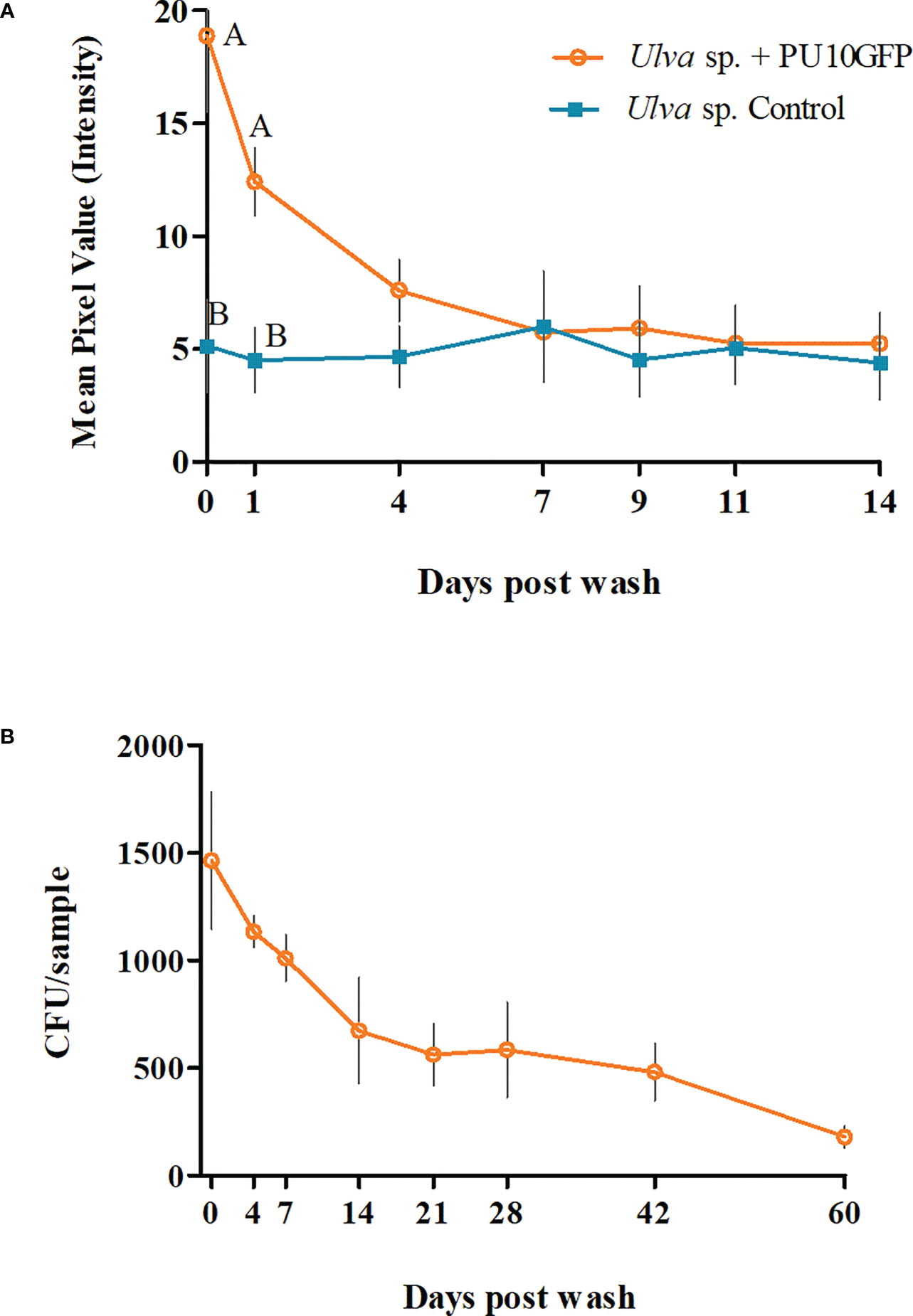
Figure 2 Survival dynamics of endophyte PU10GFP introduced in live fresh Ulva sp. (A) Fluorescence intensity measured as mean pixel value (MPV) of treated (orange) and non-treated (light blue) Ulva sp. during the 14 days of the experiment. Light blue line represents the auto fluorescence received from the algae tissue. Significant differences in fluorescence intensity between Ulva inoculated with PU10GFP and control Ulva on the day of the wash and 1 day postwashing of inoculation solution are demonstrated (P = 0.0254 and P = 0.0193, respectively). (B) Presence of PU10GFP in Ulva sp. tissue, measured as colony forming units (CFU) per Ulva sp. sample (approx. 130 mm2) isolated on antibiotic-amended selective medium over time. Points on curves of both A and B graphs represent average counts and standard error of three experiments.
Ulva sp. with introduced PU10GFP was cultured for 3 days (postwashing), sampled, freeze-dried (Alpha 1-4, Martin Christ, Osterode am Harz, Germany) and kept at 4°C. The Ulva sp. was sampled before and after freeze-drying (3 days fresh and dry, respectively) and 30, 60, 90, 120 and 180 days postwashing. Samples were crushed and the algal powder was plated on LB agar amended with chloramphenicol (10 µg/mL) and incubated at 25°C for 3 days, and then CFU were counted. The experiment was repeated three times.
A single colony of PU10GFP (verified by GFP emission), isolated from both fresh and freeze-dried PU10GFP-augmented Ulva sp. samples, was grown on LB agar as a single line in the middle of the plate (Figures 3B, C, respectively) and incubated at 25°C for 1 week. As a positive control, a single colony of PU10GFP stock from the freezer (pre-introduction) was grown on LB agar for 1 week as well (Figure 3A). The fish pathogen Streptococcus iniae was inoculated on both sides of the plates about 1 cm from the endophyte colony. As a negative control, S. iniae was inoculated on LB agar in the absence of PU10GFP under the same conditions (Figure 3D). The cultures were incubated for an additional week (25°C) and examined for pathogen growth during that time.
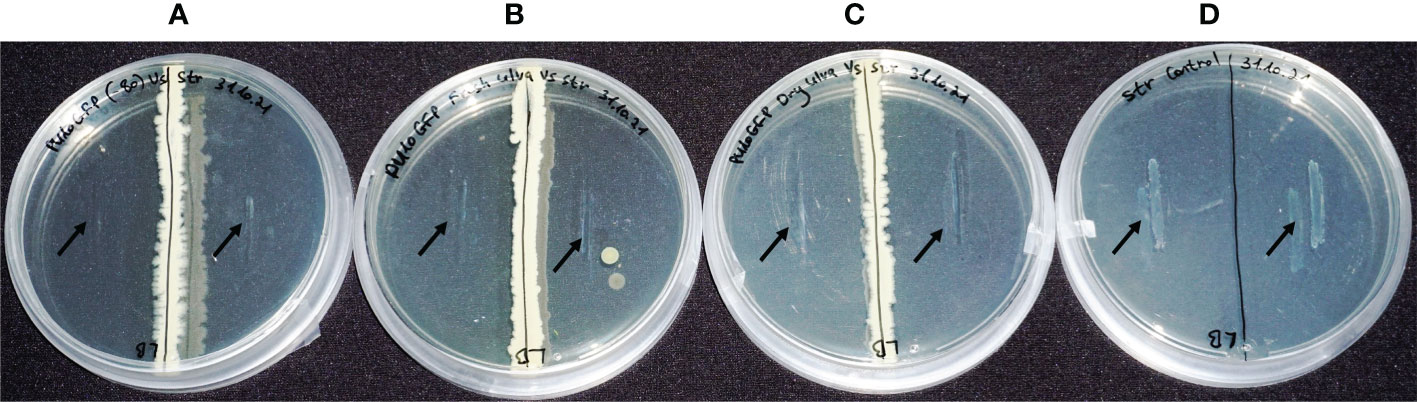
Figure 3 In-vitro bioactivity assays of PU10GFP against the fish pathogen Streptococcus iniae under different storage conditions. (A) S. iniae vs. PU10GFP isolated from Ulva sp. at Palmachim shore and kept at -80°C. (B) S. iniae vs. PU10GFP reisolated from inoculated Ulva sp., 14 days postwashing of inoculated solution. (C) S. iniae vs. PU10GFP reisolated from inoculated, freeze-dried Ulva sp. 3 months postinoculation. (D) S. iniae control, growing in the absence of PU10GFP. Black arrows mark the location of pathogen streaking on the growth medium.
For bacterial microbiota analysis, PU10GFP was introduced into Ulva sp. as described above. Inoculated and non-inoculated Ulva sp. were sampled on days 0 (after washing the excess endophytes from the alga), 4 and 14 postwash. Samples were stored at -80°C until processing.
DNA was extracted from the samples using the GenElute™ Plant Genomic DNA Miniprep kit (Sigma-Aldrich, St. Louis, MO, USA). Genomic DNA was used to amplify by PCR- the V4 region of microbial small subunit ribosomal RNA (rRNA) genes. The amplicons were generated using a two-stage PCR amplification protocol as described in Naqib et al. (2018)]using primers CS1_515FB and CS2_806RB [modified from the primer set employed by the Earth Microbiome Project (EMP; ACACTGACGACATGGTTCTACAGTGYCAGCMGCCGCGG- TAA and TACGGTAGCAGAGACTTGGTCTGGACTACNVGGGTWTCTAAT); for the first stage of the microbiome amplification, as described previously (Moonsamy et al., 2013). with the following conditions: 95°C for 5 min, and 28 cycles of 95°C for 45 s, 50°C for 60 s and 72°C for 90 s Deutsch et al. (2021).
The second PCR amplification, library construction, pooling and sequencing were performed at the Genome Research Core of the Research Resources Center, University of Illinois at Chicago. The second PCR amplification was performed in 10-µL reactions in 96-well plates. A mastermix for the entire plate was made using MyTaq HS 2X Mix. Each well received a separate primer pair with a unique 10-base barcode, obtained from the Access Array Barcode Library for Illumina (Fluidigm, San Francisco, CA, USA; Item #100-4876). These Access Array primers contained the CS1 and CS2 linkers at the 3’ ends of the oligonucleotides. Cycling conditions were: 95°C for 5 min, followed by 8 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 30 s. Samples were then pooled in equal volumes using an EpMotion5075 liquid handling robot (Eppendorf, Hamburg, Germany). The pooled library was purified using the AMPure XP cleanup protocol (0.6X, v/v; Agencourt, Beckman Coulter, Brea, CA, USA) to remove fragments smaller than 300 bp. The pooled libraries, with a 20% phiX spike-in, were loaded into an Illumina MiniSeq mid-output flow cell (2 x 153 bp paired-end reads). Based on the distribution of reads per barcode, the amplicons (before purification) were repooled to generate a more balanced distribution of reads. The repooled library was purified using AMPure XP cleanup, as described above. The repooled libraries, with a 20% phiX spike-in, were loaded into a Miniseq flow cell, and sequenced (2 x 153 bp paired-end reads). Fluidigm sequencing primers, targeting the CS1 and CS2 linker regions, were used to initiate sequencing. De-multiplexing of reads was performed on-instrument. Sequence data were delivered as pairs of fastq-formatted files.
The Dada2 pipeline (Callahan et al., 2016) (dada2 package version 1.14.0) was used for sequence data processing. Sequences were filtered and trimmed for quality using the ‘filterAndTrim’ command with the parameters maxN set to zero, maxEE set to 2, trimLeft set to 15 bp. The truncLen was set to 150 bases for both forward and reverse reads. A sequence error-estimation model was calculated using the ‘learnErrors’ option with default parameters. Then, the dada2 algorithm for error correction was applied with the ‘dada’ command using default parameters. Sequences were merged using the ‘mergePairs’ command with minimum overlap set at 8 bp. Suspected chimera were then detected and removed using the command ‘removeBimeraDenovo’. A count table including each amplicon sequence variant (ASV) in each sample was then produced. Taxonomic annotation of ASVs was done using ‘assignTaxonomy’ against the Silva non-redundant rRNA database (v132) with minimum bootstrap value set at 80.
To normalize the microbiome’s relative abundance, a series of qPCR analyses were performed using Rotor-Gene Q (Qiagen, Sao Paulo, Brazil). The qPCR results were calculated for absolute quantification based on standard curves for each DNA region (as described by Karlsson et al., 2012) and calculation was performed using the following formula:
For total bacterial DNA in the samples, qPCRs were performed with HDA primers (ACTCCTACGGGAGGCAGC and GTATTACCGCGGCTGCTGGCA) targeting the V2-V3 region of the 16S (rDNA) gene (Walter et al., 2000). Ulva’s chloroplast DNA was deducted from the total bacterial DNA. The primers used for chloroplast qPCR (UChc_F: GAATGATTGGGCGTAAAGCG and UChp_R: TCGCCATTGGTGTTCTTACTGA) were designed with NCBI GenBank using the Primer-BLAST tool. The amount of bacterial DNA (without chloroplast DNA) was divided by the qPCR result of Ulva DNA in the sample, obtained using the Primer-BLAST tool on the U. rigida 5.8S rRNA gene (Ur5.8S_F: ACTCTTCGGAGGAGACCACA and Ur5.8S_R: AGCTACCTACCTAGTCGGGG). The qPCR conditions were as follows: 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 30 s and a melt curve of 60°C to 99°C in 1°C increments every 10 s. The above formula was inserted as a factor for each sample of relative abundance results from the microbiota sequencing.
To analyze the differences between the microbiota of PU10GFP-introduced and non-introduced Ulva sp., we used alpha diversity parameters. Three alpha diversity indices—Shannon H’ (Figure 4A left), Simpson (Figure 4A middle) and the number of observed amplicon sequence variants (ASVs) (Figure 4A right) were calculated. Count data for microbiota composition were normalized to abundance using a sample-specific size factor determined by qPCR quantification of total bacteria. For calculation of alpha diversity parameters, normalized counts were first subsampled to 25,000 using command ‘rrarefy’ in R package ‘vegan’ [version 2.5-7 (Oksanen et al., 2019)]. The Shannon H’ index and the Simpson index were calculated with command ‘diversity’ and the number of observed species was calculated with command ‘specnumber’ in R package ‘vegan’. We then applied aligned rank transform (ART) ANOVA (Wobbrock et al., 2011) using R package ‘ARTool’ (version 0.11.1) to test the effect of PU10GFP inoculation and time (0, 4 and 14 days after washing of inoculation solution) and their interaction on these alpha diversity parameters (Table S1).
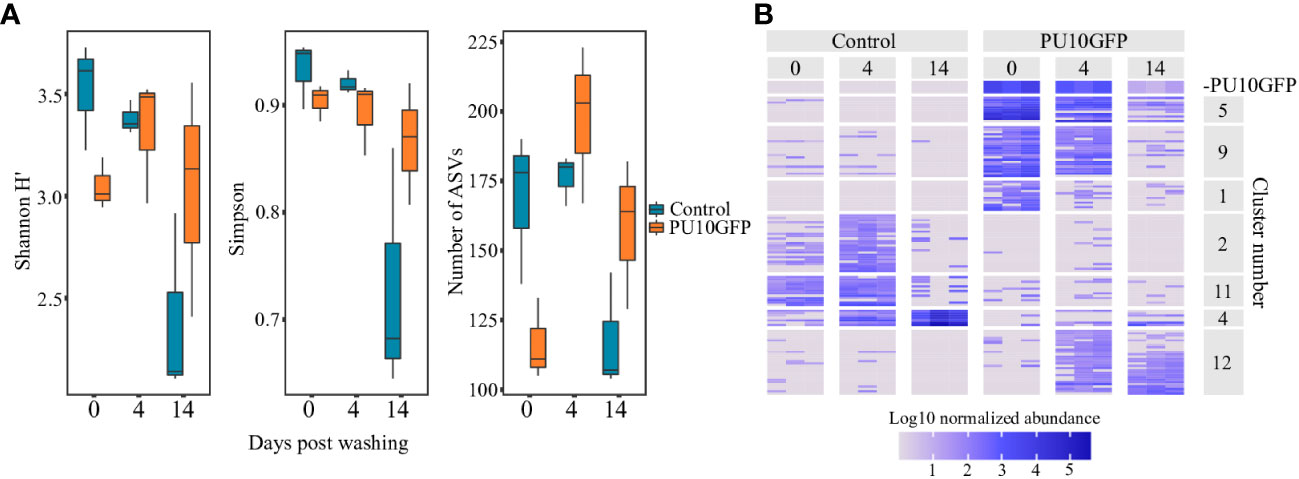
Figure 4 Changes in Ulva sp. microbiota as result of PU10GFP inoculation. Ulva thalli were sampled 0, 4 and 14 days postwashing of inoculation solution. Microbiota composition was determined by sequencing 16S rRNA gene amplicons (V4 region). Read counts were normalized by total counts of 16S rRNA quantified by qPCR and normalized to U. rigida 5.8S rRNA gene. (A) Alpha diversity parameters Shannon H’, Simpson and number of observed ASVs were calculated and compared by ART-ANOVA. Significant effect of days postwashing was found for all three indices; Shannon H’ index (F = 5.83, P< 0.05), Simpson index (F = 5.39, P< 0.05) and number of ASVs observed (F = 7.06, P< 0.01). Significance of Day x Treatment was found for both the Simpson index (F = 5.16, P< 0.05) and number of ASVs observed (F = 8.76, P< 0.01). (B) Heat map displaying changes in normalized abundance of differentially abundant ASVs (ART-ANOVA Benjamini–Hchberg adjusted P< 0.05). Top panel displays population dynamics of the introduced strain PU10GFP. Displayed clusters include 121 ASVs out of 204 ASVs found to be affected by treatment, days postwashing or their interaction.
To assess the effect of treatment (i.e., inoculation with PU10GFP) and time on each of the identified bacterial populations (ASVs), we applied ART-ANOVA with the formula qPCR-normalized abundance ~ Treatment x Day postwash. P values were adjusted for false discovery by the Benjamini–Hochberg method. Effects were considered significant for Benjamini–Hochberg adjusted P value<0.05. In total, a significant effect for any of the terms tested was found for 204 ASVs (Table S2). To demonstrate different trends of population-abundance divergence, log-transformed normalized abundances of differentially abundant ASVs were clustered into 12 clusters using the K-means method of the R package ‘superheat’. The 12 clusters represent 2 treatments x 3 time points x 2 change-direction possibilities. A heat map was then drawn using log-transformed abundances that included ASVs in clusters showing a marked treatment effect.
Maribacter sp. and Roseobacter sp. were used as markers to study PU10GFP’s influence on the abundance of other bacteria in the seaweed. These two bacteria are known to be associated with Ulva spp. We followed their abundance in both PU10GFP-introduced and non-introduced Ulva sp. over time. Inoculated and non-inoculated Ulva sp. were sampled on days 0, 4, and 14 postwash. Samples were stored at -80°C until further use. At the end of each experiment, DNA was extracted from the samples and 2 µL of the pure DNA was subjected to qPCR analysis using the Ulva rigida 5.8S rRNA gene under the qPCR conditions described in the supporting information methods. To track Maribacter sp., Roseobacter sp. and PU10GFP in the samples, primers were designed with NCBI GenBank using the Primer-BLAST tool. Another 2 µL of pure DNA was subjected to qPCR analysis with the same conditions by using MariF: GGCGGGCGATTAAGTCAGAG and MariR: CTAATCCTGTTCGCTCCCCA primers. The same was done for RoseoF: GGGGTTAG- CGTTGTTCGGA, RoseoR: CAGCGTCAGTATCGAGCCAG primers and for PU10GFPF: ACTGGGGAACTTGAGTGCAG and PU10GFPR: GCTCCTCAGCGTCAGTTACA primers. Absolute abundance of Maribacter sp., Roseobacter sp. and PU10GFP in the samples was normalized for sample size using the qPCR results of U. rigida 5.8S rRNA.
GFP intensity values were analyzed with the JMP 10 software package (SAS Inc., Cary, NC, USA). Mean numbers of Ulva sp. pixel values (MPV) with the addition or in the absence of PU10GFP endophyte were subjected to one-way analysis of variance (ANOVA), followed by Tukey-Kramer multiple comparison test, with significance set at P< 0.05.
A highly active isolate of a bacterial endophyte (B. subtilis) was successfully transformed with a gfp marker gene and antibiotic (chloramphenicol) resistance gene using a transduction method (Shemesh and Chaia, 2013).
Introduction of PU10GFP into Ulva sp. is demonstrated in Figure 1A displaying the colonization of a PU10GFP isolate next to a random wound. GFP expression can be observed in the seaweed’s intercellular spaces, whereas no GFP expression was detected in the control Ulva sp. (Figure 1B).
The fluorescence intensity measured in Ulva sp. inoculated with endophyte PU10GFP on the day of washing (day 0) was nearly four times higher than in the control; 18.87 and 5.16 MPV, respectively (P = 0.0254). The next day (day 1 postwash), the fluorescence intensity value of PU10GFP in Ulva decreased to 12.42 MPV, which was still ~3-fold greater than that measured for the auto fluorescence in the control (Ulva sp. with no introduced endophytes; 4.52 MPV) (P = 0.0193). Seven days postwash, the fluorescence intensity had declined and equaled the auto fluorescence measured for the control samples (Figure 2A). Along with the GFP gene, PU10GFP acquired an antibiotic (chloramphenicol) resistance gene. This resistance enabled PU10GFP growth on selective media during the 60 days of the experiment, in which we tried to isolate the bacteria from the introduced algae. The presence of the endophytes was measured as CFU/Ulva sp. sample (approx. 130 mm2) (Figure 2B). On day 0 (day of wash from inoculation), there were 1500 CFU/sample. One-week postwash, 1000 CFU of PU10GFP were isolated from each sample on the selective media. By day 21 postwash, the number of colonies isolated from the introduced algae had declined to 550 CFU/sample and by day 60, it was 130 CFU/sample (Figure 2B). We isolated PU10GFP from Ulva sp. for up to 150 days postwash, at which point we obtained 30 CFU/sample (data not shown). There was no growth from the non-inoculated Ulva sp. samples on the selective media plates.
Bacterial numbers (CFU/mg) isolated from the freeze-dried tissue, were recorded for 180 days (Figure 5). The isolated bacteria’s bioactivity was evaluated by their ability to inhibit and kill S. iniae at each time point. We found that the number of PU10GFP was stable and equal to the numbers recorded for both fresh and dry seaweed 3 days postwash (Figure 5). One experiment was continued for more than a year with the same results (data not shown). This supports the notion that, if kept under controlled conditions, the freeze-dried Ulva may be stored for at least a year without losing PU10 as an active endophyte.
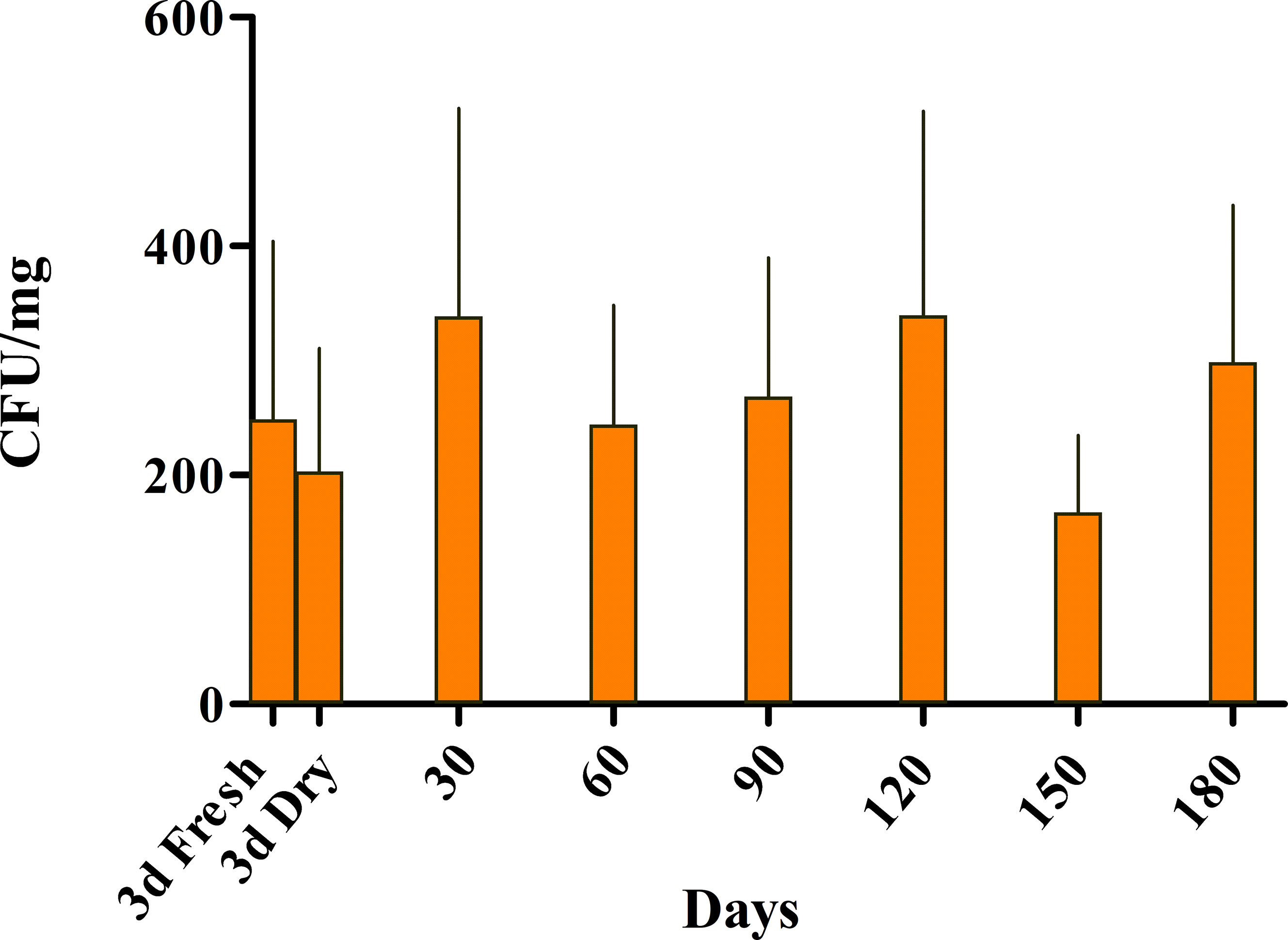
Figure 5 Dynamics of survival of the introduced endophyte PU10GFP from freeze-dried Ulva sp. The presence of PU10GFP in freeze-dried Ulva sp. was evaluated as colony forming units (CFU) per milligram of dry Ulva. Ulva thalli were freeze-dried 3 days postwashing of inoculation solution. Before freeze drying, a few pieces were used for PU10GFP reisolation on selective growth medium, and CFU were counted (3d Fresh). Immediately after drying (3d Dry), and every 30 days for 180 days, a few dry pieces were taken out of the 4°C fridge for PU10GFP reisolation on selective growth medium, and CFU were counted. Bars represent average counts and standard error of three experiments.
The endophytes from both the fresh (Figure 3B) and freeze-dried (Figure 3C) seaweed were as active against S. iniae as a colony of PU10GFP that was not introduced into Ulva sp. but grown fresh from a stock at -80°C (Figure 3A). Figure 3D presents the normal growth of S. iniae on LB agar after 1 week. The pathogen did not grow on the plates and was not viable postexposure to the endophyte under any of the treatments (data not shown and Deutsch et al., 2021).
The highest diversity was observed for the control treatments at the start of the experiment (day 0), with lower diversity calculated for the PU10GFP-treated samples. On day 4, diversity seemed to have high values in both control and treated samples (Figure 4A left and middle). Fourteen days after the wash, each treatment (PU10GFP-introduced and non-introduced control) acted differently for all indices; the control samples showed very low diversity values due to domination of one particular genus in Ulva tissues, whereas the PU10GFP-treated samples demonstrated medium-to-high diversity values (Figure 4A). A significant effect of time (days postwashing) was found for all three indices; Shannon H’ index (F = 5.83, P< 0.05), Simpson index (F = 5.39, P< 0.05) and number of ASVs observed (F = 7.06, P< 0.01). Significance of the interaction Day x Treatment was found for both the Simpson index (F = 5.16, P< 0.05) and number of ASVs observed (F = 8.76, P< 0.01).
Raw sequence data has been deposited to the NCBI SRA database under Bioproject accession:PRJNA913475.
The heat map presents the log10-normalized abundance values of each ASV for each sample on days 0, 4 and 14 postwash. Hierarchical clustering generated 12 clusters with distinct population dynamics. We focused on those best exemplifying certain dynamics (Figure 4B): (a) ASVs showing similar dynamics as the inoculated PU10GFP (cluster 5); (b) ASVs strongly associated with PU10GFP and abundant on day 0, but declining toward day 14 (clusters 1 and 9); (c) ASVs abundant in controls on days 0 and 4 but missing or nearly missing in the PU10GFP inoculation treatment (clusters 2 and 11); (d) ASVs dominating controls on day 14 but not dominant in PU10GFP treatment (cluster 4); (e) ASVs dominating PU10GFP on days 4 and 14 but missing or nearly missing in controls (cluster 12).
Data from the microbiome analysis (Figure 6A) and from qPCR analysis of two introduction trials (Figure 6B) were used to determine the absolute abundance of Maribacter sp. and Roseobacter sp., as well as that of the isolate PU10GFP in the samples. Results of data from the microbiome analysis (Figure 6A) showed that when PU10GFP was introduced into the alga, Maribacter sp. and Roseobacter sp. abundance declined with time. This response contrasted with that in non-treated control samples where Maribacter sp. and Roseobacter sp. thrived with time (1 × 104 and 1 × 106 on day 14, respectively) (Figure 6A). This was validated by the two qPCR experiments (Figure 6B). The non-treated samples in Figure 6B showed constant abundance of the two bacterial populations throughout the experimental period. The treated samples displayed increasing abundance of the two bacteria from days 0 to 4, followed by a decrease from day 4 to 14. In the two qPCR trials, pretreatment Ulva sp. samples were analyzed to determine the initial non-influenced presence of Maribacter sp. and Roseobacter sp. (marked as day -4, Figure 6B). At that time point, we found at least tenfold lower abundance compared to day 0 (Figure 6B). Note that the abundance of PU10GFP decreased with time in both the microbiome experiment and the qPCR experiments (Figures 6A, B); this observation is supported by the results obtained in afore described GFP-intensity and CFU-isolation assays. Furthermore, the PU10GFP strain could not be detected in the non-treated samples in any of the three trials (data not shown).
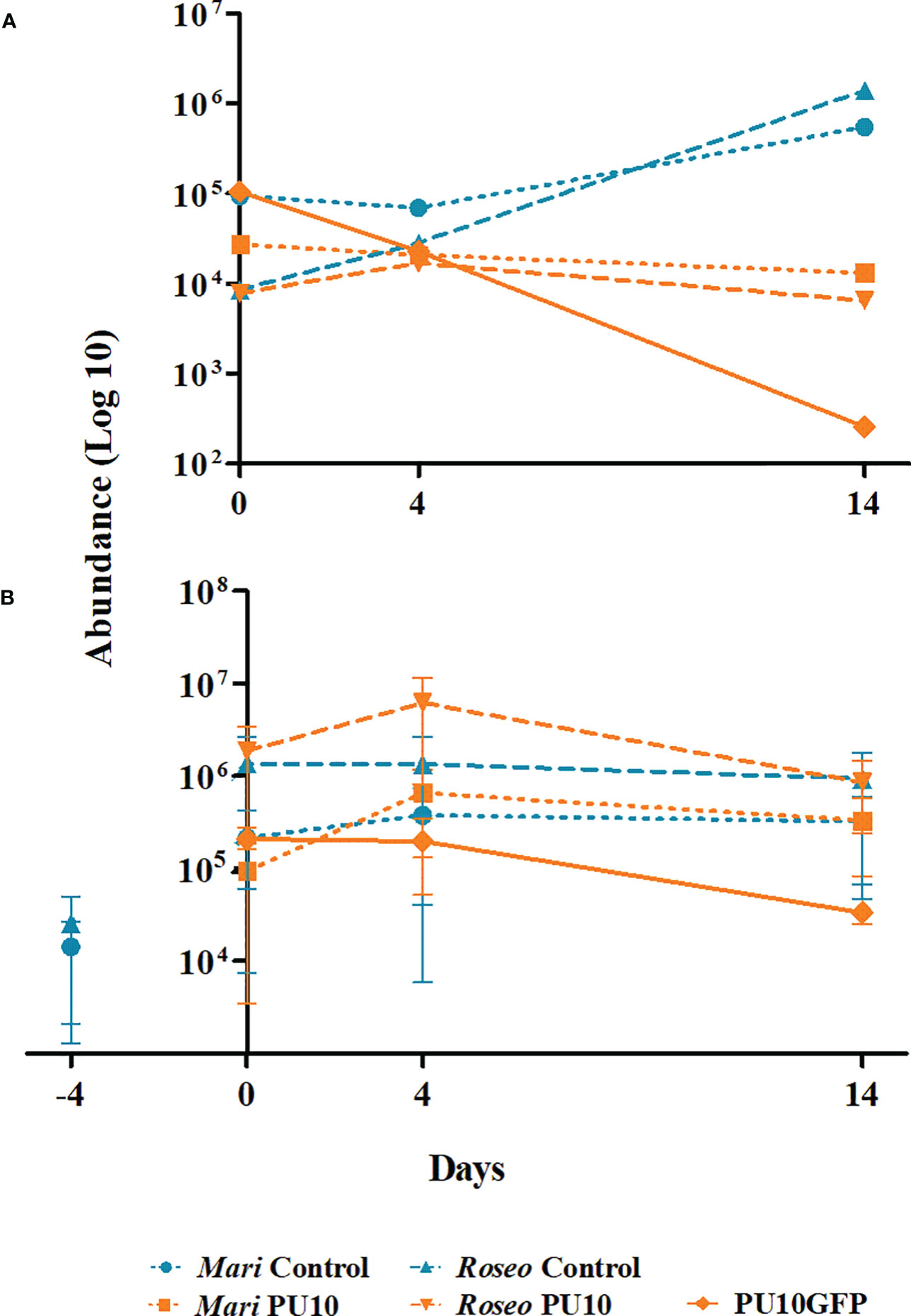
Figure 6 Abundance of PU10GFP, Maribacter sp. and Roseobacter sp. in Ulva sp. as result of PU10GFP inoculation. Control samples (light blue) represent abundance of native Maribacter sp. (Mari control) and Roseobacter sp. (Roseo control) from Ulva thalli in the absence of PU10GFP. Treatment samples (orange) represent abundance of Maribacter sp. (Mari PU10), Roseobacter sp. (Roseo PU10) and PU10GFP from Ulva thalli that were inoculated with PU10GFP. (A) Absolute abundance was determined by using microbiome analysis data (as described in Figure 4) followed by a series of qPCR analyses for the normalized results. (B) Ulva thalli were sampled before inoculation (-4) and then at 0, 4 and 14 days postwash. Absolute abundance was obtained by a series of qPCR analyses for normalized results. Data are presented in log10 scale.
This work was a continuation of our previous study describing macroalgal endophytes (Deutsch et al., 2021). PU10, a Bacillus sp., transformed with antibiotic resistance and gfp reporter genes was introduced into its native (original) seaweed (Ulva sp.). The endophyte had been found colonizing the algal tissue mainly in wounds, scratches, cuts and exposed tissues, probably presenting easier paths for bacterial penetration of the tissue and subsequent colonization of the intercellular space. The localization of the bacteria inside the algae tissue and not on its surface was demonstrated by using Laser-scanning fluorescence microscopy. Another support to PU10GFP location inside the tissue as an endophyte, is by the fact that the GFP signal was located only in between the cells and not equally spread in the algae sample. We repeated the introduction of PU10GFP into Ulva more than twenty independently times. With successful results when we used the protocol we developed for this purpose. It needs to be emphasized that the successful introduction of algae with endophytes was not demonstrated before, unlike with the case of terrestrial plants where introduction of endophytes has become a common practice for many (Chen et al., 1995; Ding et al., 2011; Mitter et al., 2017). Although each introduction resulted in different concentration of the endophyte in the tissue (Figures 2B, 5, error bars), the overall survival trends of the endophyte were kept over time in every specific introduction. We noticed that the endophyte’s presence in the inoculated tissue was rather high (~1500 CFU per sample of 130 mm2) at the end of the inoculation (when the bacterial supernatant was washed off of the algal thalli), and then declined over time (1000 and 675 CFU per sample after 1 and 2 weeks, respectively). This decline in endophyte concentration in the tissue may be controlled or initiated by the seaweed, as previously demonstrated in other seaweeds (Hollants et al., 2011; Wichard, 2015). Alternatively, lack of resources in the seaweed’s intercellular space may limit the potential growth of the endophytic bacteria, because the endophyte biomass (CFU/sample) declined continuously during the 60 days of the experiment (Figure 2B). Yet, even after 150 days, viable CFU of the endophyte were still found in the algal tissue (data not shown). Importantly, the bioactivity of the endophyte was not reduced after reintroduction into the seaweed, and it did not decline after freeze-drying, when the endophytes were re-cultured on new media. The endophytes that survived freeze drying of the Ulva sp. grew viable colonies when plated on fresh media. Some bacteria (Bacillus in our case) have the ability to form endospores to survive drought and other stress conditions. Viable bacterial endospores have been isolated from specimens such as dried plant samples (reviewed Mckenney et al., 2013). We assume that the ability of the introduced endophytes to grow on selective media upon which the dried inoculated alga was plated was due to the formation of endospores, which survive the freeze-drying process. We were able to grow active endophytes from dried seaweed stored at 4°C for more than a year. To obtain a potential feed additive consisting of Ulva powder containing a high concentration of endophytes, we intentionally freeze-dried the Ulva with introduced PU10GFP 3 days after washing off the inoculation suspension, when the concentration of the endophytes in the algal tissue was highest.
Another aspect to be considered is the possibility that the endophyte will change the seaweed’s microbial composition. This change may be beneficial for the seaweed as the endophyte may protect it against unwanted pathogens, or destructive to the seaweed if the change is major and negatively affects growth and survival of the seaweed. To test the effect of active endophytes on the original host (Ulva sp.) microbiome, PU10GFP was introduced into Ulva sp., and the Ulva sp. microbiomes in the presence and absence of the isolate were compared. We found significant differences in diversity between control (host only) and endophyte-introduced seaweed (treatment) at different time points. The treated samples had a certain group of microbes (Bacillus spp., among others) dominating the seaweed’s tissues on day 0 (clusters 5, 9, 1, Figure 4B). With time, this group decreased (in correlation with the PU10GFP isolate) and other microbes filled their place. A community shift occurred in the control samples as well, but involved different species of microbes. On day 14 of the experiment, one main group of microbes dominated the control samples, and the presence of almost all other genera decreased. This might be the reason for the low diversity in the control treatment on day 14. The predominant genus (highest ASVs) in the control sample on day 14 was Aquimarina sp., a bacterium known to have algicidal activity (Chen et al., 2012). Interestingly, the control samples on day 14 revealed wounds and dead tissue, whereas the treated samples did not. The same trend was observed in the qPCR results, where a high concentration of bacterial DNA and low concentration of Ulva DNA were detected compared to the treated samples at the same time point. It may be that when the Aquimarina sp. attacks the alga, it weakens the latter’s defense system and allows other opportunistic microbes to thrive on day 14 (cluster 4, control samples, Figure 4B). In the treated samples, we barely detected the Aquimarina sp.
The substantial differences observed between the microbial communities of the inoculated vs. non-inoculated Ulva sp. suggest that the introduced endophytes colonizing the intercellular space of the seaweed in high concentrations could have an effect on the algal microbiota, as reported for terrestrial plants (Mitter et al., 2017). We found dramatically increased abundance of both Maribacter sp. and Roseobacter sp. with time in the non-treated control samples, whereas in the samples inoculated with PU10GFP, these two genera decreased in abundance over time. These findings support our hypothesis that the introduction of an active endophyte into the seaweed will affect its microbiota.
To further verify these results, we conducted two more experiments using qPCR with specific primers to assess Maribacter sp. and Roseobacter sp., as well as PU10GFP abundance, in new inoculated and non-inoculated Ulva samples. In these two experiments, Maribacter sp. and Roseobacter sp. abundance did not increase in the control samples over time. One explanation for this difference between the microbiome experiment and the qPCR experiments may be the different conditions in these trials. In the control samples of the microbiome trial, Aquimarina sp. can become pathogenic to the Ulva sp., enabling Maribacter sp. and Roseobacter sp., among other bacteria, to multiply and flourish; the conditions of the qPCR trials did not allow this. This explanation is supported by the overall appearance of the control seaweed: in the microbiome experiment they seemed morphologically degraded, whereas in the qPCR trials they appeared morphologically intact. Note that the samples of endophyte-introduced seaweed utilized for the microbiome experiment did not display degradation or morphological wounding, presumably due to the presence of the endophyte. We therefore concluded that when the Ulva sp. is in good shape, i.e., not degraded by any pathogen or pest (the qPCR experiments, Figure 6B), the introduction of active PU10GFP has a minimal influence on the two reference bacteria. However, when the Ulva sp. is degraded or stressed (by the algicidal bacterium Aquimarina sp. in this case, microbiome experiment, Figure 6A), the introduction of active PU10GFP plays a substantial role in suppressing the disease-causing agent and as a result, minimizes the proliferation of other bacteria (Maribacter sp. and Roseobacter sp. in this case) in the algal tissue.
We demonstrated that bacterial endophytes introduced into their original seaweed hosts can colonize the intercellular space at high concentrations and retain their activity. Whereas there is a decline in endophyte concentration in the first 2 weeks and up to 2 months postinoculation, a stable but low concentration can be maintained for at least 150 days. We further demonstrated that the initial high concentration of endophytes in the macroalgal tissue can be preserved for at least 1 year if the inoculated algae are freeze-dried within the first few days postwash. The endophytes maintained their bioactivity under all of our treatments: freeze-dried algal tissue, live algal tissue, or storage at -80°C. The endophytes’ ability to remain bioactive through different manipulations may enable their use as biocontrol agents against aquaculture pathogens. Our results also demonstrate that the introduced endophytes may further impact the host microbiome and affect the biodiversity of the associated microbiota. These interactions may potentially help protect the seaweed from pathogens and other opportunistic microbes, as presented in this study, and could lead to novel research in macroalgal culture and maintenance. The rapidly evolving “food from the sea” and aquaculture industries will need more sustainable, healthier and environmentally friendly solutions in the coming years. This research might lay the foundation for achieving these goals.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: BioProject, PRJNA913475.
DE conceived the re-introduction and the use of selective media. MO-L conceived the microbiome analysis and performed all bioinformatic analyses. MB Built and maintained the macroalgal culture facilities. IBF conceived the Maribacter sp. and Roseobacter sp. abundance in Ulva experiments. YD conceived the freeze-drying of the Ulva and performed all other procedures described in the manuscript. YD wrote the first draft of the manuscript with contributions from MO-L, DE, and IB-F. All authors contributed to the article and approved the submitted version.
This work was financed by the Chief Scientist of the Israeli Ministry of Agriculture and Rural Development (MOARD), grant number 20-02-0122, and Copia Agro Israel.
We would like to thank Rita Smirnov and Tamir Ofek from The Central Laboratory for Fish Health, Nir David, Israel for providing the fish pathogen Streptococcus iniae; Doron Ashkenazi from the Israel Oceanographic and Limnological Research (IOLR) institution and Seakura – Sea of Life Ltd. for providing us with Ulva sp. starters; Dr. Ievgeniia Ostrov from Dr. Moshe Shemesh’s laboratory at the Soil, Water and Environmental Sciences Institute, Agricultural Research Organization, Volcani Institute, Rishon LeZion, Israel for providing us with the transduction protocol, the YC161 donor Bacillus and the SPP1 phage; and Dr. Stefan Green from Rush University Medical Center, Chicago, Illinois, USA for the next-generation sequencing for the microbiome analysis. This work is part of COST Action CA18238 (Ocean4Biotech), supported by COST (European Cooperation in Science and Technology) program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1099478/full#supplementary-material
Bacon C.W., White J.F. Functions, mechanisms and regulation of endophytic and epiphytic microbial communities of plants. Symbiosis 68, 87–98 (2016). doi: 10.1007/s13199-015-0350-2
Burke C., Thomas T., Lewis M., Steinberg P., Kjelleberg S. (2010). Composition, uniqueness and variability of the epiphytic bacterial community of the green alga ulva australis. ISME J. 5, 590–600. doi: 10.1038/ismej.2010.164
Califano G., Kwantes M., Abreu M. H., Costa R., Wichard T. (2020). Cultivating the macroalgal Holobiont : effects of integrated multi-trophic aquaculture on the microbiome of ulva rigida (chlorophyta). Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00052
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: high-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chen C., Bauske E. M., Musson G., Rodríguez Kábana R., Kloepper J. W. (1995). Biological control of fusarium wilt on cotton by use of endophytic bacteria. Biol. Control 5, 83–91. doi: 10.1006/BCON.1995.1009
Chen W. M., Sheu F. S., Sheu S. Y. (2012). Aquimarina salinaria sp. nov., a novel algicidal bacterium isolated from a saltpan. Arch. Microbiol. 194, 103–112. doi: 10.1007/s00203-011-0730-9
Chu F., Kearns D. B., Branda S. S., Kolter R., Losick R. (2006). Targets of the master regulator of biofilm formation in bacillus subtilis. Mol. Microbiol. 59, 1216–1228. doi: 10.1111/J.1365-2958.2005.05019.X
Deutsch Y., Gur L., Berman Frank I., Ezra D. (2021). Endophytes from algae, a potential source for new biologically active metabolites for disease management in aquaculture. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.636636
Dhanya K. I., Swati V. I., Swaroop V. K., O. W. J. (2016). Antimicrobial activity of ulva reticulata and its endophytes. J. Ocean Univ. China (Oceanic Coast. Sea Res. 15, 363–369. doi: 10.1007/s11802-016-2803-7
Ding S., Huang C.-L., Sheng H.-M., Song C.-L., Li Y.-B., An L.-Z. (2011). Effect of inoculation with the endophyte clavibacter sp. strain Enf12 on chilling tolerance in chorispora bungeana. Physiol. Plant 141, 141–151. doi: 10.1111/J.1399-3054.2010.01428.X
Dominguez H., Loret E. P. (2019). Ulva lactuca, a source of troubles and potential riches. Mar. Drugs 17, 1–20. doi: 10.3390/md17060357
Duanis-Assaf D., Steinberg D., Chai Y., Shemesh M. (2016). The LuxS based quorum sensing governs lactose induced biofilm formation by bacillus subtilis. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.01517
Flewelling A. J., Johnson J. A., Gray C. A. (2013). Isolation and bioassay screening of fungal endophytes from north Atlantic marine macroalgae. Bot. Mar. 56, 287–297. doi: 10.1515/bot-2012-0224
Fu H., Jiang P., Zhao J., Wu C. (2018). Comparative genomics of pseudomonas sp. strain SI-3 associated with macroalga ulva prolifera, the causative species for green tide in the yellow Sea. Front. Microbiol. 9 1458. doi: 10.3389/FMICB.2018.01458/BIBTEX
Gouda S., Das G., Sen S. K., Shin H. S., Patra J. K. (2016). Endophytes: a treasure house of bioactive compounds of medicinal importance. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01538
Habbu P., Warad V., Shastri R., Savant C., Madagundi S., Kekare P. (2016). In vitro and in vivo antimicrobial activity of ulva lactuca linn. (greer algae) associated endophytic bacterial strains. J. Appl. Pharm. Sci. 6, 138–146. doi: 10.7324/JAPS.2016.601019
Hall T. J., Schreiber L. R., Leben C. (1986). Effects of xylem-colonizing bacillus spp. on verticillium wilt in maples. Plant Dis. 70, 521–524. doi: 10.1094/pd-70-521
Hollants J., Decleyre H., Leliaert F., De Clerck O., Willems A. (2011). Life without a cell membrane: challenging the specificity of bacterial endophytes within bryopsis (Bryopsidales, chlorophyta). BMC Microbiol. 11, 1–10. doi: 10.1186/1471-2180-11-255
Jacobsen B. J., Zidack N. K., Larson B. J. (2004). “The role of bacillus-based biological control agents in integrated pest management systems: plant diseases,” in Phytopathology (American phytopathological society) 94(11), 1272–1275. doi: 10.1094/PHYTO.2004.94.11.12721
Jensen P. R., Fenical W. (1994). Strategies for the discovery of secondary metabolites from marine bacteria: ecological perspectives. Annu. Rev. Microbiol. 48, 559–584. doi: 10.1146/annurev.mi.48.100194.003015
Karlsson C. L. J., Önnerfält J., Xu J., Molin G., Ahrné S., Thorngren-Jerneck K. (2012). The microbiota of the gut in preschool children with normal and excessive body weight. Obesity 20, 2257–2261. doi: 10.1038/OBY.2012.110
Kessler R. W., Weiss A., Kuegler S., Hermes C., Wichard T. (2018). Macroalgal–bacterial interactions: role of dimethylsulfoniopropionate in microbial gardening by ulva (Chlorophyta). Mol. Ecol. 27, 1808–1819. doi: 10.1111/mec.14472
Kharwar R. N., Strobel G. (2010). “Fungal endophytes: an alternative source of bioactive compounds for plant protection,” in Natural products in plant pest management. Ed. Dubey N. K. (Wallingford, UK: CABI), 218–241.
Klerks M. M., Franz E., van Gent-Pelzer M., Zijlstra C., Van Bruggen A. H. (2007). Differential interaction of salmonella enterica serovars with lettuce cultivars and plant-microbe factors influencing the colonization efficiency. ISME J. 1, 620–631. doi: 10.1038/ismej.2007.82
Kuldau G., Bacon C. (2008). Clavicipitaceous endophytes: their ability to enhance resistance of grasses to multiple stresses. Biol. Control 46, 57–71. doi: 10.1016/J.BIOCONTROL.2008.01.023
Li X., Bu N., Li Y., Ma L., Xin S., Zhang L. (2012). Growth, photosynthesis and antioxidant responses of endophyte infected and non-infected rice under lead stress conditions. J. Hazard. Mater. 213–214, 55–61. doi: 10.1016/J.JHAZMAT.2012.01.052
Liarzi O., Ezra D. (2014). “Endophyte-mediated biocontrol of herbaceous and non-herbaceous plants,” in Advances in endophytic research. Eds. Verma V., Vijay C., Gange Alan C. (New Delhi: Springer), 335–369.
Liu M., Dong Y., Zhao Y., Zhang G., Zhang W., Xiao T. (2011). Structures of bacterial communities on the surface of ulva prolifera and in seawaters in an ulva blooming region in jiaozhou bay, China. World J. Microbiol. Biotechnol. 27, 1703–1712. doi: 10.1007/S11274-010-0627-9/FIGURES/3
Marshall K., Joint I., Callow M. E., Callow J. A. (2006). Effect of marine bacterial isolates on the growth and morphology of axenic plantlets of the green alga ulva linza. Microb. Ecol. 52, 302–310. doi: 10.1007/S00248-006-9060-X/FIGURES/3
Mckenney P. T., Driks A., Eichenberger P. (2013). The bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat. Rev. Microbiol. 11, 33–44. doi: 10.1038/nrmicro2921
Mitter B., Pfaffenbichler N., Flavell R., Compant S., Antonielli L., Petric A., et al. (2017). A new approach to modify plant microbiomes and traits by introducing beneficial bacteria at flowering into progeny seeds. Front. Microbiol. 0. doi: 10.3389/FMICB.2017.00011
Moonsamy P. V., Williams T., Bonella P., Holcomb C. L., Höglund B. N., Hillman G., et al. (2013). High throughput HLA genotyping using 454 sequencing and the fluidigm access ArrayTM system for simplified amplicon library preparation. Tissue Antigens 81, 141–149. doi: 10.1111/tan.12071
Myers D. F., Strobel G. A. (1983). Pseudomonas syringae as a microbial antagonist of ceratocystis ulmi in the apoplast of American elm. Trans. Br. Mycol. Soc 80, 389–394. doi: 10.1016/S0007-1536(83)80034-5
Naqib A., Poggi S., Wang W., Hyde M., Kunstman K., Green S. J. (2018). Making and sequencing heavily multiplexed. Gene Expr. Anal. Methods Protoc. 1783, 149–169.
Oksanen A. J., Blanchet F. G., Friendly M., Kindt R., Legendre P., Mcglinn D., et al. (2019). Vegan. Encycl. Food Agric. Ethics, 2395–2396. doi: 10.1007/978-94-024-1179-9_301576
Olmos J., Acosta M., Mendoza G., Pitones V. (2020). Bacillus subtilis, an ideal probiotic bacterium to shrimp and fish aquaculture that increase feed digestibility, prevent microbial diseases, and avoid water pollution. Arch. Microbiol. 202, 427–435. doi: 10.1007/s00203-019-01757-2
Ongena M., Jacques P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. doi: 10.1016/j.tim.2007.12.009
Ramanan R., Kim B. H., Cho D. H., Oh H. M., Kim H. S. (2016). Algae-bacteria interactions: evolution, ecology and emerging applications. Biotechnol. Adv. 34, 14–29. doi: 10.1016/j.biotechadv.2015.12.003
Ren C.-G., Liu Z.-Y., Wang X.-L., Qin S. (2022). The seaweed holobiont: from microecology to biotechnological applications. Microb. Biotechnol. 15, 738–754. doi: 10.1111/1751-7915.14014
Santos R. A., Oliva-Teles A., Pousão-Ferreira P., Jerusik R., Saavedra M. J., Enes P., et al. (2021). Isolation and characterization of fish-gut bacillus spp. as source of natural antimicrobial compounds to fight aquaculture bacterial diseases. Mar. Biotechnol. 23, 276–293. doi: 10.1007/S10126-021-10022-X
Sarasan M., Job N., Puthumana J., R R., P. M. P., Thomas L. C., et al. (2020). Exploration and profiling of hidden endophytic mycota of marine macroalgae with potential drug leads. FEMS Microbiol. Lett. 367, 1–9. doi: 10.1093/femsle/fnaa078
Shemesh M., Chaia Y. (2013). A combination of glycerol and manganese promotes biofilm formation in bacillus subtilis via histidine kinase KinD signaling. J. Bacteriol. 195, 2747–2754. doi: 10.1128/JB.00028-13
Tarnecki A. M., Wafapoor M., Phillips R. N., Rhody N. R. (2019). Benefits of a bacillus probiotic to larval fish survival and transport stress resistance. Sci. Rep. 2019 91 9, 1–11. doi: 10.1038/s41598-019-39316-w
Thorén-Tolling K. (1978). Identity of the fungal endophyte of ascophyllum with mycosphaerella ascophylli established by means of fluorescent antibody technique. Bot. Mar. 21, 409–412. doi: 10.1515/botm.1978.21.7.409
Thurlow C. M., Williams M. A., Carrias A., Ran C., Newman M., Tweedie J., et al. (2019). Bacillus velezensis AP193 exerts probiotic effects in channel catfish (Ictalurus punctatus) and reduces aquaculture pond eutrophication. Aquaculture 503, 347–356. doi: 10.1016/j.aquaculture.2018.11.051
Trivedi P., Leach J. E., Tringe S. G., Sa T., Singh B. K. (2020). Plant–microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621. doi: 10.1038/s41579-020-0412-1
Vallet M., Strittmatter M., Murúa P., Lacoste S., Dupont J., Hubas C., et al. (2018). Chemically-mediated interactions between macroalgae, their fungal endophytes, and protistan pathogens. Front. Microbiol. 9. doi: 10.3389/FMICB.2018.03161
Walter J., Tannock G. W., Tilsala-Timisjarvi A., Rodtong S., Loach D. M., Munro K., et al. (2000). Detection and identification of gastrointestinal lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66, 297–303. doi: 10.1128/AEM.66.1.297-303.2000
Wichard T. (2015). Exploring bacteria-induced growth and morphogenesis in the green macroalga order ulvales (Chlorophyta). Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00086
Wichard T., Charrier B., Mineur F., Bothwell J. H., De Clerck O., Coates J. C. (2015). The green seaweed ulva: a model system to study morphogenesis. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00072
Keywords: endophyte, seaweed, macroalga, Ulva, microbiome, aquaculture, Bacillus, disease
Citation: Deutsch Y, Ofek-Lalzar M, Borenstein M, Berman-Frank I and Ezra D (2023) Re-introduction of a bioactive bacterial endophyte back to its seaweed (Ulva sp.) host, influences the host’s microbiome. Front. Mar. Sci. 10:1099478. doi: 10.3389/fmars.2023.1099478
Received: 15 November 2022; Accepted: 24 May 2023;
Published: 16 June 2023.
Edited by:
Robert Czajkowski, University of Gdansk, PolandReviewed by:
Soizic Prado, Muséum National d’Histoire Naturelle, FranceCopyright © 2023 Deutsch, Ofek-Lalzar, Borenstein, Berman-Frank and Ezra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Ezra, ZGV6cmFAdm9sY2FuaS5hZ3JpLmdvdi5pbA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.