- 1Key Laboratory of Healthy Mariculture for the East China Sea, Ministry of Agriculture and Rural Affairs, Ornamental Aquarium Engineering Research Centre in University of Fujian Province, Fisheries College, Jimei University, Xiamen, Fujian, China
- 2Key Laboratory of Estuarine Ecological Security and Environmental Health, Tan Kah Kee College, Xiamen University, Zhangzhou, Fujian, China
- 3College of Marine Science, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China
Tumor necrosis factor receptor-associated factors (TRAFs) play vital roles in tumor necrosis factor receptor (TNF-R) and interleukin-1 receptor/Toll-like receptor (IL-1R/TLR) mediated signaling pathway. However, the role that TRAF7 plays in the host immune responses is largely unknown in comparison to the extensive and in-depth research that has been conducted on other members of the TRAF family. Notably, Lc-TRAF7, a cloned TRAF7 ortholog, was discovered in the large yellow croaker (Larimichthys crocea) in the current study, which has an open reading frame (ORF) of 1,962 base pairs and encodes a protein of 653 amino acids (aa). Lc-TRAF7 is consisted of a RING finger domain, a coiled-coil domain, and seven WD40 domains, with the genomic organization consisted of 20 exons and 19 introns. According to the expression analysis, Lc-TRAF7 was presented in a wide range of detected organs and tissues of the healthy fish, and was able to significantly induced by stimulations of poly I:C, LPS, PGN, and Pseudomonas plecoglossicida infection. Subcellular distribution analysis revealed that Lc-TRAF7 was a cytoplasmic protein, with the RING finger and coiled-coil domain function importantly in its subcellular localization. Luciferase assays demonstrated that Lc-TRAF7 overexpression significantly induced the activation of NF-κB, IRF3, IRF7, and IFN1 promoters. In addition, the WD40 domains play a pivotal role in the NF-κB promoter activation, whereas the RING finger and coiled-coil domain were essential in the IRF3, IRF7, and IFN1 promoter activation. Notably, Lc-TRAF7 overexpression could suppress SVCV proliferation in EPC cells, and the expression levels of IRF3, IRF7, ISG15, ISG56, RSAD2, and TNF-α were up-regulated under Lc-TRAF7 overexpression in LYCMS cells. These findings collectively implied that Lc-TRAF7 may function as an important regulator in the host antiviral responses via the NF-κB as well as IRF3/7 involved signaling pathways.
1 Introduction
As the first line of defense against pathogen invasion and tissue damage, the innate immune system plays vital roles in the host immune responses (Langevin et al., 2013; Kordon et al., 2018; Stosik et al., 2021), which is depended on the recognition of various pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs) through the pattern recognition receptors (PRRs) (Kumar et al., 2011; Brubaker et al., 2015; Stosik et al., 2021). Tumor necrosis factor receptor-associated factor (TRAF) proteins are evolutionarily conserved intracellular regulators that mediate a variety of signaling transduction, especially in those PRRs, such as Toll-like receptor (TLR), retinoic acid inducible gene I (RIG-I)-like receptor (RLR), and nucleotide-biding oligomerization domain (NOD)-like receptor (NLR) mediated singling pathways (Inoue et al., 2000; Dhillon et al., 2019; Arkee and Bishop, 2020). The TRAF proteins are widely involved in the host immune defense, inflammatory response, autophagy, and apoptosis (Lalani et al., 2018; Shi and Sun, 2018; Dhillon et al., 2019). Till now, seven members have been identified in mammalian TRAF family, reported as TRAF1-7, respectively (Zotti et al., 2012; Park, 2018; Park, 2021).
TRAF7, the unique noncanonical TRAF family member, was discovered by searching for a new protein containing the RING finger-like structure of the TRAF family, which was firstly recognized as an E3 ubiquitin ligase (Xu et al., 2004; Zotti et al., 2017). It has been demonstrated that TRAF7 interacts with mitogen activated protein kinase kinase kinase (MEKK3) to enhance MEKK3-induced activation of AP-1 and C/EBP homologous protein (CHOP) in mammals (Bouwmeester et al., 2004; Xu et al., 2004; Zotti et al., 2011). Notably, most of the other members of the TRAF family interact with a variety of signaling molecules and even protein kinases through their TRAF domain, whereas TRAF7 interacts with MEKK3 through the WD40 domains (Bouwmeester et al., 2004; Zotti et al., 2017). Overexpression of TRAF7 could induce caspase-dependent apoptosis, and the deletion of the RING finger domain significantly abolish the induction, suggesting the crucial role of the RING finger domain in TRAF7-mediated apoptosis (Xu et al., 2004). In addition, TRAF7 was found to associate with RelA/p65 (p65) members of the NF-κB transcription factor family and IκB kinase (IKK)/NF-κB essential modulator (NEMO), which could promote Lys-29-linked polyubiquitylation of NEMO and p65, leading to the degradation of these two molecules, and also affecting the nuclear localization of p65 (Zotti et al., 2011). In TLR2-mediated signaling cascade, the interaction of TRAF6 and TRAF7 initiates the activation of NF-κB and MAPK kinase (MKK) 3/6-p38 pathways (Zotti et al., 2012). Interestingly, cylindromatosis (CYLD) could inhibit the activation of TRAF6 and TRAF7 through a deubiquitination-dependent mechanism to negatively regulate TLR2-mediated signaling pathway (Yoshida et al., 2005).
Although the function of mammalian TRAF7 has been widely studied, few orthologs of TRAF7 have been characterized in teleosts. One investigation in half-smooth tongue sole (Cynoglossus semilaevis) showed that TRAF7 was widely expressed in various tissues of the healthy half-smooth tongue sole, and the expression profile of TRAF7 was significantly increased under the infection of Vibrio harveyi, suggesting the possible immune-related function of TRAF7 in teleosts (Li et al., 2020). Nevertheless, more research is needed to determine the precise mechanism by which TRAF7 mediated the signaling pathway.
Large yellow croaker (Larimichthys crocea) is a popularly cultivated species of commercially important marine fish in China (Zhang et al., 2019; Li et al., 2020). With the fast advancement of large yellow croaker aquaculture industry, diseases caused by viruses (such as iridovirus), bacteria (such as Pseudomonas plecoglossicida), and parasites (such as Cryptocaryon irritans) occurred frequently, which seriously affected the aquaculture industry and caused huge economic losses (Chen et al., 2003; Zheng et al., 2020; Zou et al., 2022). Therefore, in order to benefit the further control and prevention of such fish diseases, it is essential to comprehend the host fundamental immune defense against the invading pathogens. In this study, one ortholog named Lc-TRAF7 was characterized in large yellow croaker. The protein sequence feature of Lc-TRAF7 was compared and the phylogenetic relationship was analyzed among those of other vertebrate TRAF7 proteins. The genome organization as well as the subcellular localization of Lc-TRAF7 was determined, and the expression profiles of Lc-TRAF7 in a variety of organs and tissues as well as under various PAMPs stimulations were also examined. Additionally, the present study revealed the activation of Lc-TRAF7 in the NF-κB, IRF3, IRF7, and type I IFN promoters. The antiviral effect of Lc-TRAF7, and the expression patterns of downstream immune-related molecules under Lc-TRAF7 overexpression were also determined. It is thus possible to provide a new and essential perspective on the roles of TRAF7 in teleost immune-related signaling.
2 Materials and methods
2.1 Experimental fish, cell lines, virus, and transfection
Large yellow croakers used in this study were purchased from Ningde Fufa Fishing Co., Ltd, Ningde, Fujian Province, with the length measured as 18.0 ± 1.5 cm, weight as 60.0 ± 15.0 g. The fish were kept in the recirculated seawater system with 3,000-L tanks which maintained at 24-26°C and feeded with commercial diet. After temporary rearing for about two weeks, the healthy fish were used in the subsequent experiments as previously mentioned (Zou et al., 2019; Zou et al., 2020).
Human embryonic kidney 293T (HEK 293T) cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Invitrogen-Gibco) and 100 U/mL penicillin (P) and streptomycin (S) as described previously. The cells were then kept in an incubator at 37°C and 5% CO2 as our previous reports (Zou et al., 2019; Zou et al., 2020). Large yellow croaker muscle (LYCMS) cells were maintained in L15 Medium (Boster, USA) containing 10% FBS and 100 U/mL PS at 27°C as our previous report (Chen et al., 2022). Epithelioma papulosum cyprinid (EPC) cells were cultured at 25°C in M199 medium (Procell, Wuhan, China) containing 10% FBS and 100 U/mL PS. Spring viraemia of carp virus (SVCV), which is infective in EPC cells, was propagated in EPC cells at 25°C. Followed the manufacturer’s instructions, Lipofectamine®3000 (Invitrogen, Carlsbad, CA) was used to transfect the plasmids into the above-mentioned cells.
2.2 Gene cloning and plasmids constructions
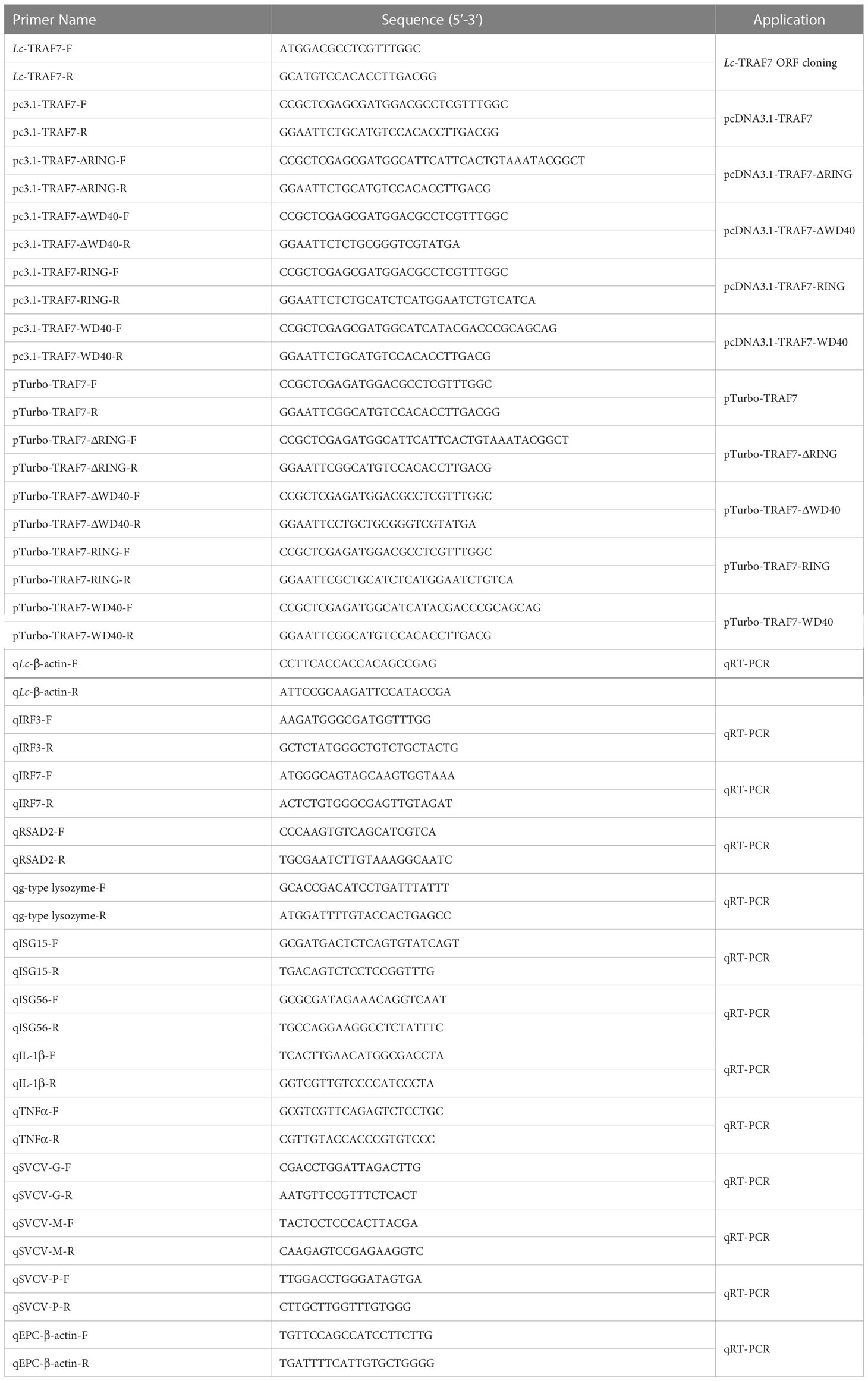
Based on the transcriptome information for the large yellow croaker on the National Center for Biotechnology Information website (NCBI, GenBank accession No. XM_027288056.1), specific primers were used to amplify the full-length ORF of large yellow croaker TRAF7. For eukaryotic expression and subcellular localization analysis, the full-length ORF of TRAF7 and its truncated forms were cloned and inserted into the pcDNA3.1/myc-His (-) A vector that contains a C-terminal c-Myc tag (Invitrogen, Carlsbad, CA, USA) and pTurboGFP-N vector that contains a C-terminal TurboGFP tag (Evrogen, Moscow, Russia), respectively. All plasmid constructs were confirmed by sequencing analysis in Sangon Biotech Co., Ltd. (Shanghai, China) and Western blotting analysis. Primers used in the present study are listed in Table 1.
2.3 Bioinformatics analysis
For ORF determination and protein sequence prediction of the obtained large yellow croaker TRAF7, the Translate tool at the ExPASy Bioinformatics Resource Portal (https://web.expasy:translate) was used (Duvaud et al., 2021), and the conserved domain structures were detected according to the Simple Modular Architecture Research Toll (SMART) (http://smart.embl.de) and the Conserved Domain Database (CDD) on the NCBI (http://www.ncbi.nlm.nih.gov/cdd) (Lu et al., 2020). The Basic Local Alignment Search Tool (BLAST) analysis tool on the NCBI (http://www.ncbi.nlm.nih.gov/blast) was utilized for the search of vertebrate TRAF7 orthologs. The Clustal X program was used to conduct amino acid alignments of vertebrate TRAF7, with the results edited using the GeneDoc software (Larkin et al., 2007). The phylogenetic tree of vertebrate TRAF7 was constructed using the MEGA7 software with the neighbor-joining method (Kumar et al., 2016). The vertebrate TRAF7 gene sequences were analyzed online using the Splign tool (https://www.ncbi.nlm.nih.gov/sutils/splign/splign.cgi) following the gene search on the NCBI genome database (https://www.ncbi.nlm.nih.gov/genome).
2.4 Constitutive and inductive expression analysis
To understand the constitutive expression profiles of TRAF7 in large yellow croaker, various organs and tissues, such as the head kidney, trunk kidney, liver, gill, spleen, heart, muscle, blood, intestine, skin, and brain were dissected from randomly selected six healthy fish, then immediately frozen in liquid nitrogen for RNA extraction and qPCR analysis.
In order to determine the expression profiles of TRAF7 in large yellow croakers that subjected to a variety of immune stimuli, the fish were divided into five groups, each with 50 fish, and each fish of the group was separately taken an injection with different PAMPs (Zou et al., 2021a; Zou et al., 2021b). In brief, 100 μL of phosphate buffered saline (PBS) was injected intraperitoneally into each fish of the control group. Simultaneously, each fish in the challenge groups was taken an injection of the same amount of lipopolysaccharides (LPS) (L3024, Sigma, 0.5 mg/mL), polyinosinic-polycytidylic acid potassium salt (poly I:C) (P9582, Sigma, 1 mg/mL), peptidoglycan (PGN) (69554, Sigma, 1 mg/mL), or P. plecoglossicida suspension in PBS (5 × 105 CFU/mL), respectively (Zou et al., 2021a; Zou et al., 2021b). After that, six fish from each group were randomly selected and anaesthetized in 0.01% eugenol at 6, 12, and 24 h post injection (hpi), the intestine, gill, blood, spleen, and head kidney were collected for the subsequent experiments as described above.
To further reveal the role of large yellow croaker TRAF7 in the host immune responses, especially on the expression profiles of antiviral and inflammatory-related genes including g-type lysozyme, IL-1β, TNF-α, ISG15, ISG56, IRF3, IRF7, and RSAD2, LYCMS cells seeded on the 6-well plates (5 × 105 cells per well) overnight were transiently transfected with 5 μg of pcDNA3.1-TRAF7 or pcDNA3.1 empty vector (control), respectively. After that, the cells were collected at 48 h post-transfection (hpt), with the expression patterns of TRAF7 and the antiviral and inflammatory-related genes mentioned above detected by qRT-PCR analysis. All primers used are listed in Table 1 and could also refer to our previous reports (Chen et al., 2022; Luo et al., 2022).
2.5 RNA extraction and qRT-PCR analysis
Total RNA was isolated from the different organ/tissue and the LYCMS cell samples described above by using the Eastep™ Super Total RNA Extraction Kit (Promega, Beijing, China), then the first-stand cDNA synthesis kit (RevertAid First Stand cDNA Synthesis Kit, #K1622, Thermo Scientific™) was used to reverse transcribed the total RNA into cDNA according to the manufacturer’s protocol. The synthetic cDNA was then kept at -80°C and used as a template for full-length ORF cloning of target genes or qRT-PCR analysis.
qRT-PCR was performed on a Roche LightCycler® 480 II quantitative real-time detection system (Roche, Switzerland) using Go Taq® qPCR Master Mix (Promega, Madison, USA) with the following procedure: 5 min at 95°C, followed by 40 cycles of 20 s at 95°C, 15 s at 60°C, and 15 s at 72°C, with a final extension at 72°C for 10 min. Then, the melting curve was conducted and analyzed as the reaction temperature was raised from 72°C to 95°C at a rate of 0.5°C per second. All reactions were performed in a 384-well plate, with the mean value recorded. The comparative Ct method (2-ΔΔCt) was used to analyze the relative expression of target gene, which was normalized to the expression of β-actin (Livak and Schmittgen, 2001; Zou et al., 2022).
2.6 Confocal microscopy
To investigate the subcellular distribution of large yellow croaker TRAF7 and its truncated forms, HEK 293T cells seeded on cell slides in 6-well plates (2 × 105 cells per well) overnight were separately transfected with 5 μg constructed plasmid of pTurbo-TRAF7-GFP, pTurbo-TRAF7-ΔRING-GFP, pTurbo-TRAF7-WD40-GFP, pTurbo-TRAF7-ΔWD40-GFP, pTurbo-TRAF7-RING-GFP, or pTurboGFP-N (control) using Lipofectamine 3000 in accordance with the manufacturer’s protocols. At 24 hpt, the cells on the coverslips were washed with 1 mL PBS, fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100 for 10 min at room temperature, respectively. After that, the cell slides were washed with PBS, followed by overlaying with one drop of mounting media (VECTASHIELDR Hard Set™ Mounting media with DAPI, Vector Laboratories, CA). Finally, a confocal microscope (Leica TCS SP8, Germany) was used for examining and photographing the cells.
2.7 Luciferase activity assays
To evaluate the roles of large yellow croaker TRAF7 on the induction of IRF3, IRF7, NF-κB, and IFN1 promoter activation, HEK 293T cells cultured in 24-well plates (1 × 105 cells per well) for one night, and then the cells were transiently transfected with 100 ng of pGL4-IRF3-pro (Chinese invention patent number: ZL201710457836.8), pGL4-IRF7-pro (Chinese invention patent number: ZL201710457820.7), pNF-κB-luc (Clontech, Palo Alto, CA), or pGL4-IFN1-pro (Chinese invention patent application number: 201710456729.3), 10 ng of pRL-TK (Promega, Madison, WI) and together with pcDNA3.1-TRAF7 or pcDNA3.1 empty vector (control) at increasing concentrations (10, 50, 100, and 200 ng) using Lipofectamine 3000. At 24 hpt, the cells were lysed for subsequent luciferase activity assays using Dual-Luciferase Reporter System (Promega), with luciferase activity measured on a Promega GloMax® 20/20 luminometer (Promega). The results were normalized to the Renilla luciferase activities and expressed as the fold in comparison to the control group.
To further determine the roles of various domains of large yellow croaker TRAF7 on IRF3, IRF7, NF-κB, and IFN1 promoter activation, HEK 293T cells were transiently transfected with 100 ng of pGL4-IRF3-pro, pGL4-IRF7-pro, pNF-κB-luc, or pGL4-IFN1-pro, 10 ng of pRL-TK together with 100 ng of pcDNA3.1-TRAF7, pcDNA3.1-TRAF7-ΔRING, pcDNA3.1-TRAF7-WD40, pcDNA3.1-TRAF7-ΔWD40, pcDNA3.1-TRAF7-RING, or pcDNA3.1 empty vector (control), respectively. At 24 hpt, the cells were collected for luciferase activity assays as described above.
2.8 Antiviral activity assays
For antiviral activity assays, EPC cells seeded in 6-well plates (2 × 106 cells per well) overnight were transfected with 2.5 μg pcDNA3.1-TRAF7 or pcDNA3.1 empty vector (control). At 24 hpt, the transfected cells were washed and infected with SVCV at an MOI of 1, with the cells collected for total RNA extraction at 24 h post-infection, followed by subsequent mRNA expression pattern evaluation of SVCV Glycoprotein (SVCV-G), Matrix protein (SVCV-M), and Phosphoprotein (SVCV-P) by qRT-PCR analysis. The expression data was calculated using the comparative Ct method (2−ΔΔCt) as described above, with the viral genes normalized to the β-actin of EPC. The primers used for qRT-PCR analysis are shown in Table 1.
2.9 Western blotting analysis
To confirm the expression of the constructed plasmids described above, Western blotting analysis was performed as per our previous reports (Zou et al., 2019; Zou et al., 2020). In brief, the cells transfected with the constructed plasmids were collected and lysed using the RIPA buffer (Beyotime, Shanghai, China), with the proteins separated by 10% SDS-PAGE gels and then transferred to polyvinylidene difluoride (PVDF) membrane with pore size of 0.45 μm (Millipore Corporation). The transferred membrane was blocked with 5% non-fat dry milk in PBS for 1 h at room temperature. After one wash with PBS containing 0.1% Tween-20 (PBST) and two washes with PBS, each for 5 min, the membrane was incubated with rabbit polyclonal Anti-TurboGFP antibody (Evrogen, Moscow, Russia) or mouse monoclonal Anti-c-Myc antibody (Beyotime, Shanghai, China) that diluted with antibody diluent at 1:5000 at 4°C overnight, respectively. After that, the membrane was washed three times as described above, and then incubated with the secondary antibody (diluted with antibody diluent at 1:2000) for 1 h at room temperature, followed by washing for three times. Subsequently, the bands were visualized using WesternBright™ ECL HRP substrate (Advansta, San Jose, USA) and ECL Western blotting system (LAS-4000mini, Fujifilm, Tokyo, Japan) according to the manufacturers’ instructions.
2.10 Statistical analysis
All data acquired from the qRT-PCR as well as dual-luciferase reporter assays were recorded as mean of three repeated experiments, with the bars above the histogram representing the standard error (SE). One-way analysis of variance (ANOVA) was used for statistical analysis, followed by Duncan’s multiple range test on the SPSS version 25. The different superscripts above the bars indicate statistically different (P < 0.05), * P < 0.05, and ** P < 0.01 are considered statistically significant and remarkably significant, respectively.
3 Results
3.1 Identification and sequence analysis of large yellow croaker TRAF7
The full-length ORF of TRAF7 gene was cloned and named as Lc-TRAF7 (GenBank Accession No. ON357640), which was constituted of 1,962 bp and encoded a 653 aa protein. It was revealed that Lc-TRAF7 contained a RING finger domain (101-134 aa), a coiled-coil domain (292-356 aa), and seven WD40 domains (368-407, 411-448, 451-487, 492-528, 531-568, 571-612, and 615-652 aa) (Supplementary Figure 1).
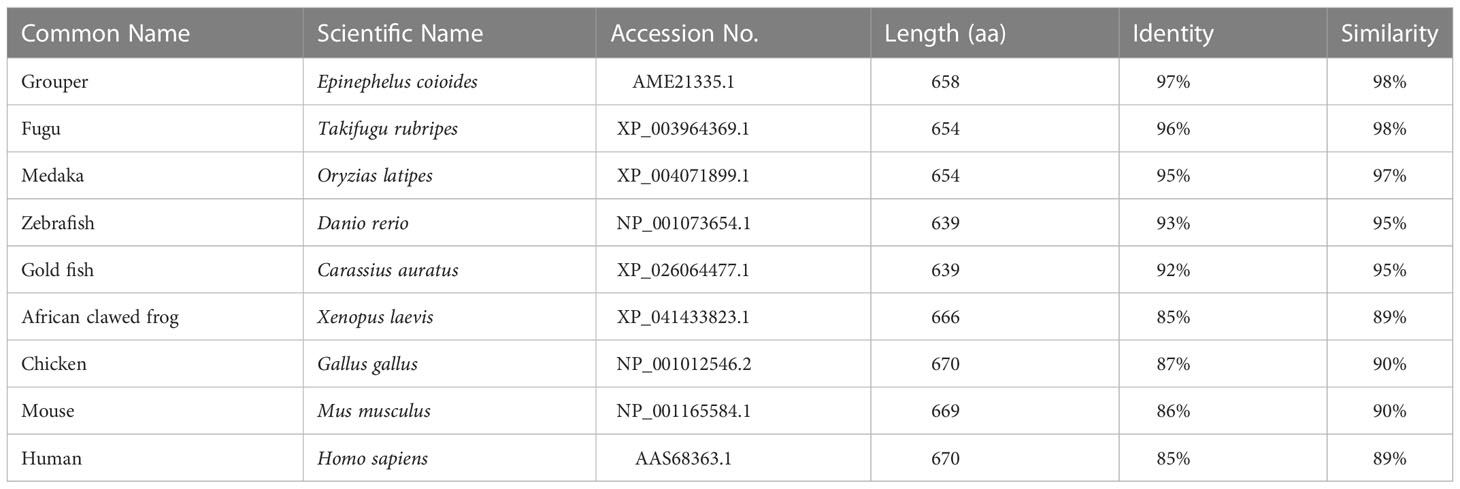
The amino acid sequence of Lc-TRAF7 was compared with TRAF7 of other vertebrates, including grouper (Epinephelus coioides), medaka (Oryzias latipes), zebrafish (Danio rerio), fugu (Takifugu rubripes), gold fish (Carassius auratus), African clawed frog (Xenopus laevis), chicken (Gallus gallus), mouse (Mus musculus), and human (Homo sapiens) (Supplementary Figure 1), which revealed a relatively conservative RING finger domain, coiled-coil domain, and WD40 domains. Additionally, the amino acid sequence of Lc-TRAF7 had an identity of 97% with the TRAF7 of grouper, 96% with fugu, 95% with medaka, 93% with zebrafish, 92% with gold fish, 87% with chicken, 86% with mouse, and 85% with human and African clawed frog (Table 2).
To comprehend the phylogenic relationship of TRAF7 in vertebrates, a phylogenetic tree of TRAF7 was constructed using the amino acid sequences of Lc-TRAF7 and the TRAF7 orthologs from other vertebrates, such as teleosts, amphibians, birds, and mammals. It was shown that TRAF7 orthologs could be isolated into two gatherings, with various fishes bunching in one clade, while amphibians, birds, and mammals gathering into another clade (Figure 1).
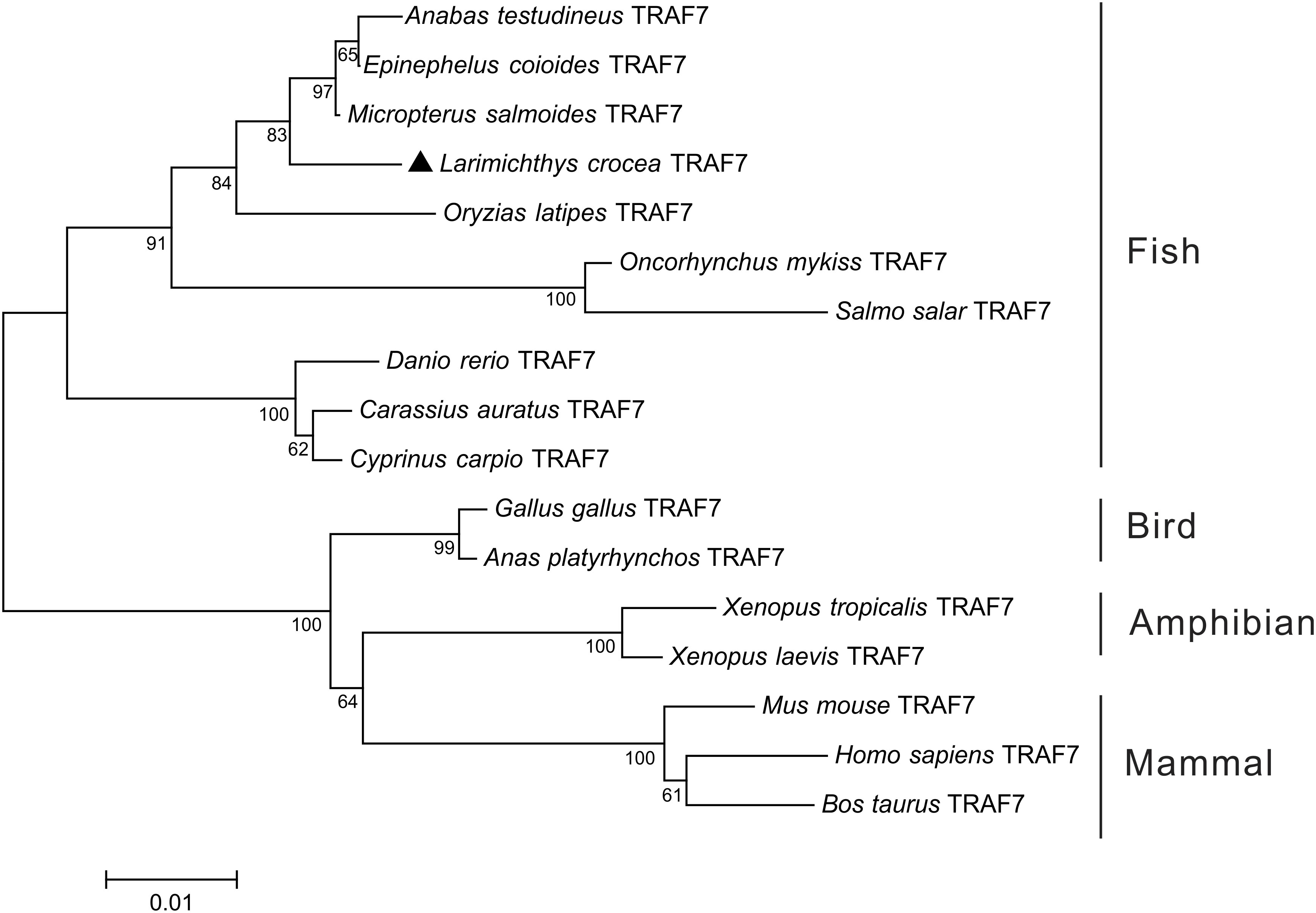
Figure 1 Phylogenetic analysis of vertebrate TRAF7 orthologs. The phylogenetic tree was constructed based on the alignment of amino acid sequences of Lc-TRAF7 with other vertebrate TRAF7 orthologs using the neighbor-joining method of MEGA 7.0 software with 10,000 replications of the bootstrap test. Lc-TRAF7 is marked with a triangle, and the numbers on the branch represent the bootstrap values. The GenBank association numbers for TRAF7 orthologs used are as follows: L. crocea, ON357640; E. coioides, AME21335.1; Anabas testudineus, XP_026211477.1; Micropterus salmoides, XP_038548977.1; O. latipes, XP_004071899.1; Oncorhynchus mykiss, XP_036829477.1; Salmo salar, XP_045571926.1; D. rerio, NP_001073654.1; Cyprinus carpio, XP_042578745.1; C. auratus, XP_026064477.1; Anas platyrhynchos, XP_027324633.1; G. gallus, NP_001012546.2; Xenopus tropicalis, XP_012825628.1; X. laevis, XP_041433823.1; Bos taurus, XP_024840564.1; M. musculus, NP_001165584.1; H. sapiens, AAS68363.1.
3.2 Genomic organization of TRAF7 genes in vertebrates
The TRAF7 genomic structures of the large yellow croaker, medaka, grouper, African clawed frog, chicken, mouse, and human were identified by comparing the vertebrate TRAF7 genomic sequences. It was demonstrated that the genomic sequences of TRAF7 ranged from 9,378 bp to 30,654 bp in length, with the longest sequence found in the African clawed frog and the shortest in the grouper (Figure 2). The large yellow croaker TRAF7 gene was consisted of 20 exons and 19 introns, which was the same from that found in medaka, grouper, African clawed frog, chicken, mouse, and human (Figure 2). In addition, the sizes of the exons of TRAF7 gene in large yellow croaker, grouper, and medaka were nearly the same, only the sixth exon showed some different. Meanwhile, the lengths of the exons of TRAF7 gene in African clawed frog, chicken, mouse, and human were also conserved, except for the first exon, the lengths of other exons were completely the same. However, the lengths of the introns were varied greatly from different species (Figure 2).
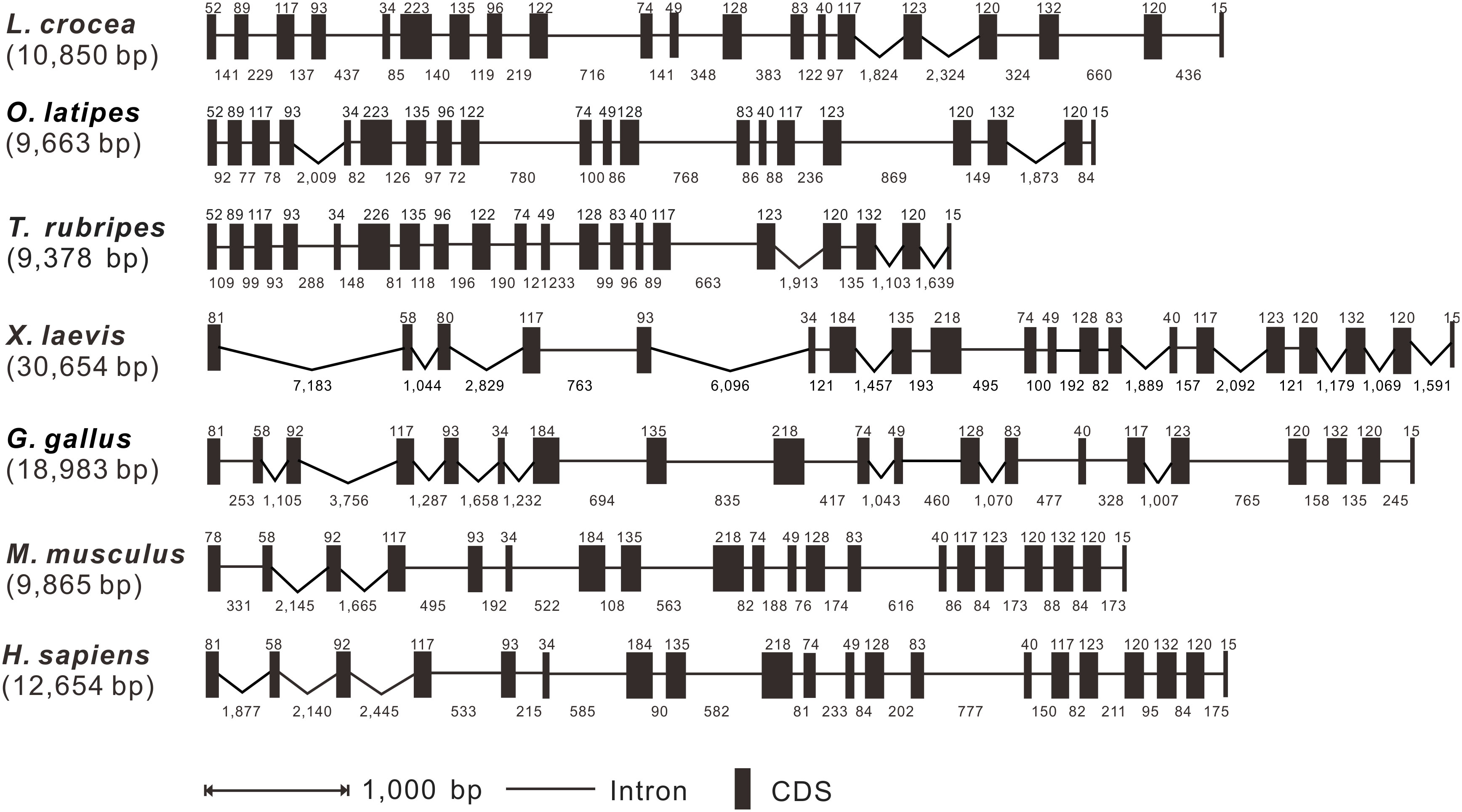
Figure 2 Genomic organization comparison of Lc-TRAF7 with other vertebrates. Comparison of exons and introns of TRAF7 gene in L. crocea, O. latipes, T. rubripes, X. laevis, G. gallus, M. musculus, and H. sapiens. Black boxes indicate the exons with the length in base pairs (bp) shown above the black boxes. The lines represent the introns with the lengths in bp shown below the lines. Gene sequences information and their GenBank association numbers are shown as follows: L. crocea, NC_040022.1 (12369484-12384469); O. latipes, NC_019866.2 (23006181-23019805); T. rubripes, NC_042289.1 (2970858-2983898); X. laevis, NC_054388.1 (102661726-102704069); G. gallus, NC_052545.1 (6421549-6450335); M. musculus, NC_000083.7 (24727824-24746912); H. sapiens, NC_000016.10 (2155778-2178129).
3.3 Expression profiles of TRAF7 in organs/tissues and challenged with PAMPs
The mRNA expression levels of TRAF7 in various organs/tissues were detected by qRT-PCR analysis. The findings demonstrated that Lc-TRAF7 was widely expressed in all the detected eleven organs/tissues of the healthy fish. In addition, the expression level of Lc-TRAF7 in the liver was significantly higher than that found in other organs/tissues, whereas the lowest expression level was observed in the brain (Figure 3).
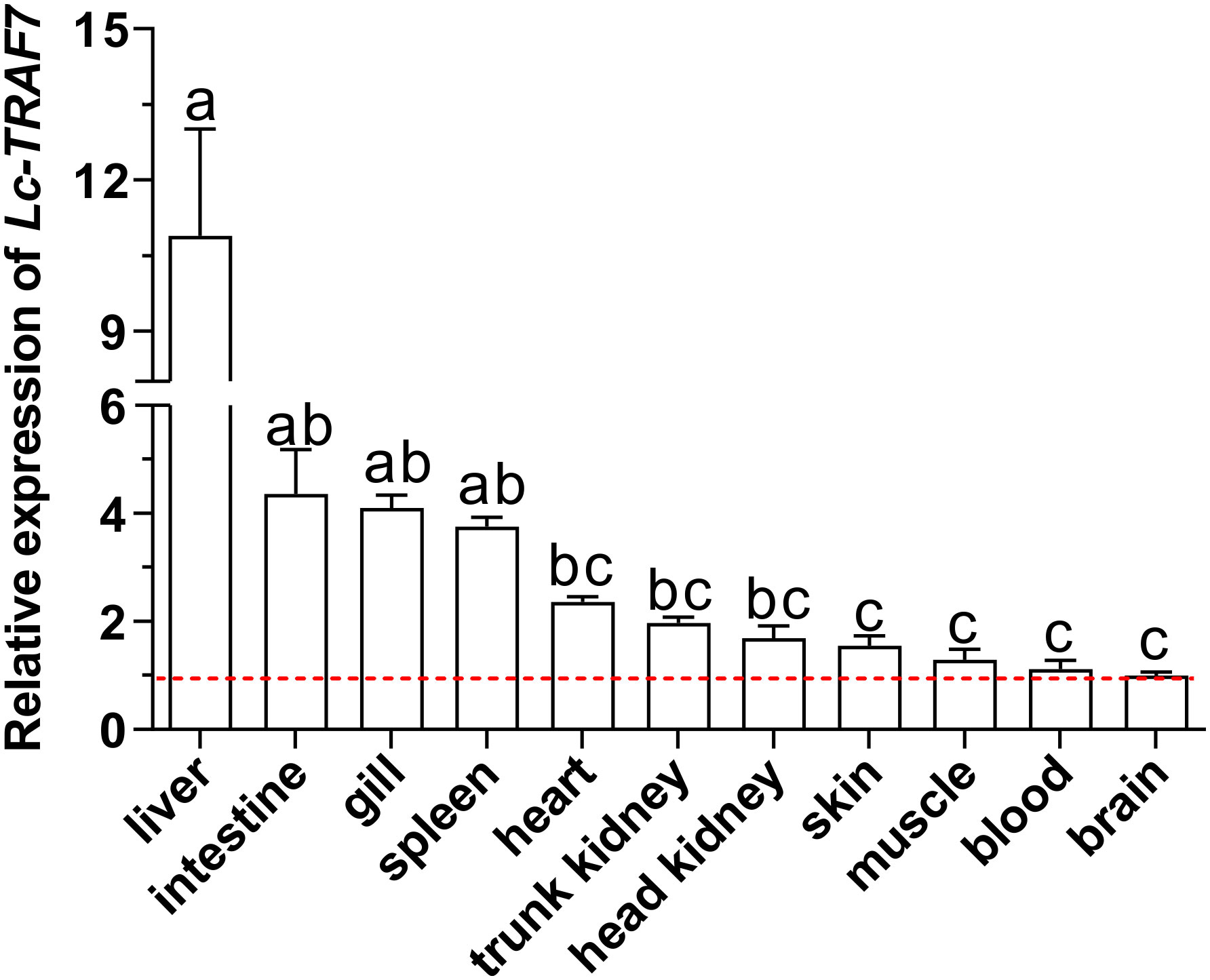
Figure 3 Organs/tissues distribution analysis of Lc-TRAF7 mRNA in large yellow croaker. The mRNA expression levels of Lc-TRAF7 were detected in eleven various organs/tissues of the healthy fish by qRT-PCR, with normalizing to the expression of L. crocea β-actin. The mRNA expression level of Lc-TRAF7 in the brain was set as 1-fold, and the mRNA expression levels of Lc-TRAF7 in other organs/tissues were recorded as multiples of the expression level in the brain, and the baseline of 1-fold was marked with the red dotted line. All data were expressed as mean of three repeated experiments, with the bars representing the SE (N = 6). Different superscripts indicate statistical differences between the groups (P < 0.05), whereas the same superscript indicates no statistical difference.
To further determine the role of Lc-TRAF7 in the host immune responses, qRT-PCR was used to detect the expression of Lc-TRAF7 in the gill, intestine, spleen, head kidney, and blood under different PAMPs stimulation or bacterial infection. The findings demonstrated that poly I:C stimulation significantly increased the mRNA expression level of Lc-TRAF7 in the gill, intestine, spleen, head kidney, and peripheral blood, with a 2.0-, 2.2-, and 3.0-fold increase at 6, 12, and 24 hpi in the gill (Figure 4A), a 5.0-fold increase at 12 hpi in the intestine (Figure 4B), a 2.4- and 2.7-fold increase at 12 and 24 hpi in the spleen (Figure 4C), a 4.1- and 2.4-fold increase at 6 and 12 hpi in the head kidney (Figure 4D), and a 2.8- and 3.7-fold increase at 6 and 12 hpi in the peripheral blood (Figure 4E), respectively. Upon LPS challenge, the expression levels of Lc-TRAF7 were also significantly increased, which was a 4.5-, 4.0-, and 2.8-fold increase at 6, 12, and 24 hpi in the gill (Figure 4A), a 2.9- and 3.6-fold increase at 6 and 12 hpi in the intestine (Figure 4B), a 4.5- and 3.6-fold increase at 6 hpi in the spleen and peripheral blood (Figures 4C, E), a 3.4- and 2.5-fold increase at 6 and 12 hpi in the head kidney (Figure 4D), respectively. In addition, Lc-TRAF7 was significantly up-regulated in response to PGN stimulation, with a 7.8-fold at 6 hpi in the gill (Figure 4A), a 2.1- and 3.1-fold increase at 6 and 12 hpi in the spleen (Figure 4C), a 2.1- and 1.9-fold increase at 12 and 24 hpi in the head kidney (Figure 4D), a 3.1- and 5.4-fold increase at 12 hpi in the intestine and peripheral blood (Figures 4B, E), respectively. Additionally, Lc-TRAF7 was also significantly induced upon P. plecoglossicida infection, with a 2.3- and 1.9-fold at 6 and 12 hpi in the gill (Figure 4A), a 2.9- and 2.1-fold at 6 and 12 hpi in the intestine (Figure 4B), a 1.9-fold at 6 hpi in the spleen (Figure 4C), a 2.0- and 1.9-fold at 12 hpi in the head kidney and peripheral blood (Figures 4C, E), respectively.
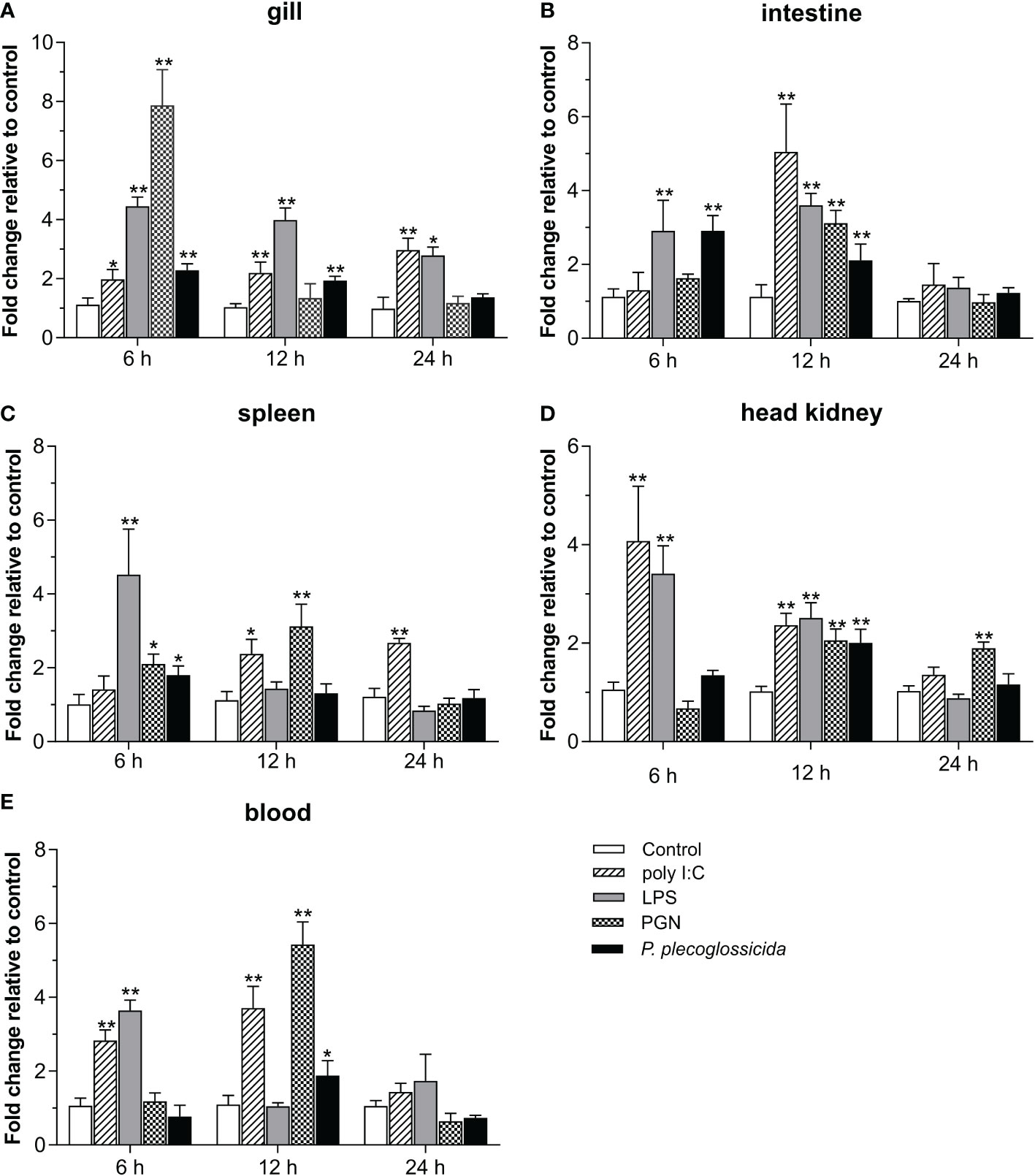
Figure 4 Expression analysis of Lc-TRAF7 under the stimulations of poly I:C, LPS, PGN, and P. plecoglossicida. Each fish of the challenge group was intraperitoneally injected with 100 μL of poly I:C (1 mg/mL), LPS (0.5 mg/mL), PGN (1 mg/mL), or P. plecoglossicida (5 × 105 CFU/mL), respectively. The control group was injected with the same volume of PBS solution. The expression levels of Lc-TRAF7 in the gill (A), intestine (B), spleen (C), head kidney (D), and blood (E) of the fish were detected by qRT-PCR at 6, 12, and 24 hpi. The expression level changes were calculated by normalization to the expression of L. crocea β-actin and then recorded as fold induction compared to the PBS injection group (control group) at the same time point. All data were shown as mean of three repeated experiments, with bars representing the SE. *P < 0.05, **P < 0.01.
3.4 Function of Lc-TRAF7 and its domains in NF-κB, IRF3, IRF7, and type I IFN promoter activation
The results of dual-luciferase reporter assays revealed that Lc-TRAF7 overexpression could significantly induce the NF-κB, IRF3, IRF7, and IFN1 promoter activation, and such induction was presented as a plasmid dose-dependent manner (Figures 5A–D). To further ascertain the roles that various domains of Lc-TRAF7 play in the activation of NF-κB, IRF3, IRF7, or IFN1 promoter, four truncated forms of expression vectors were established, including TRAF7-ΔRING (with RING finger domain deletion), TRAF7-WD40 (only containing seven WD40 domains), TRAF7-ΔWD40 (deletion of seven WD40 domains), and TRAF7-RING (only containing RING finger domain) (Figure 6A). Along with the full-length TRAF7, it was found that the truncated forms of TRAF7-ΔRING and TRAF7-WD40 could significantly trigger the activation of NF-κB promoter, whereas TRAF7-ΔWD40 and TRAF7-RING could not induce the NF-κB promoter activation (Figure 6B). Moreover, the truncated form of TRAF7-ΔWD40 could effectively initiate IRF3, IRF7, and IFN1 promoter activation, whereas other truncated forms including TRAF7-ΔRING, TRAF7-WD40, and TRAF7-RING had no significant activity to induce such promoter activation (Figures 6C–E). Additionally, the expressions of target proteins including the full-length Lc-TRAF7 as well as its truncated forms were further detected by Western blotting analysis using Anti-c-Myc antibody, which confirmed the successful transfection and expression of the constructed plasmids (Figure 6F).
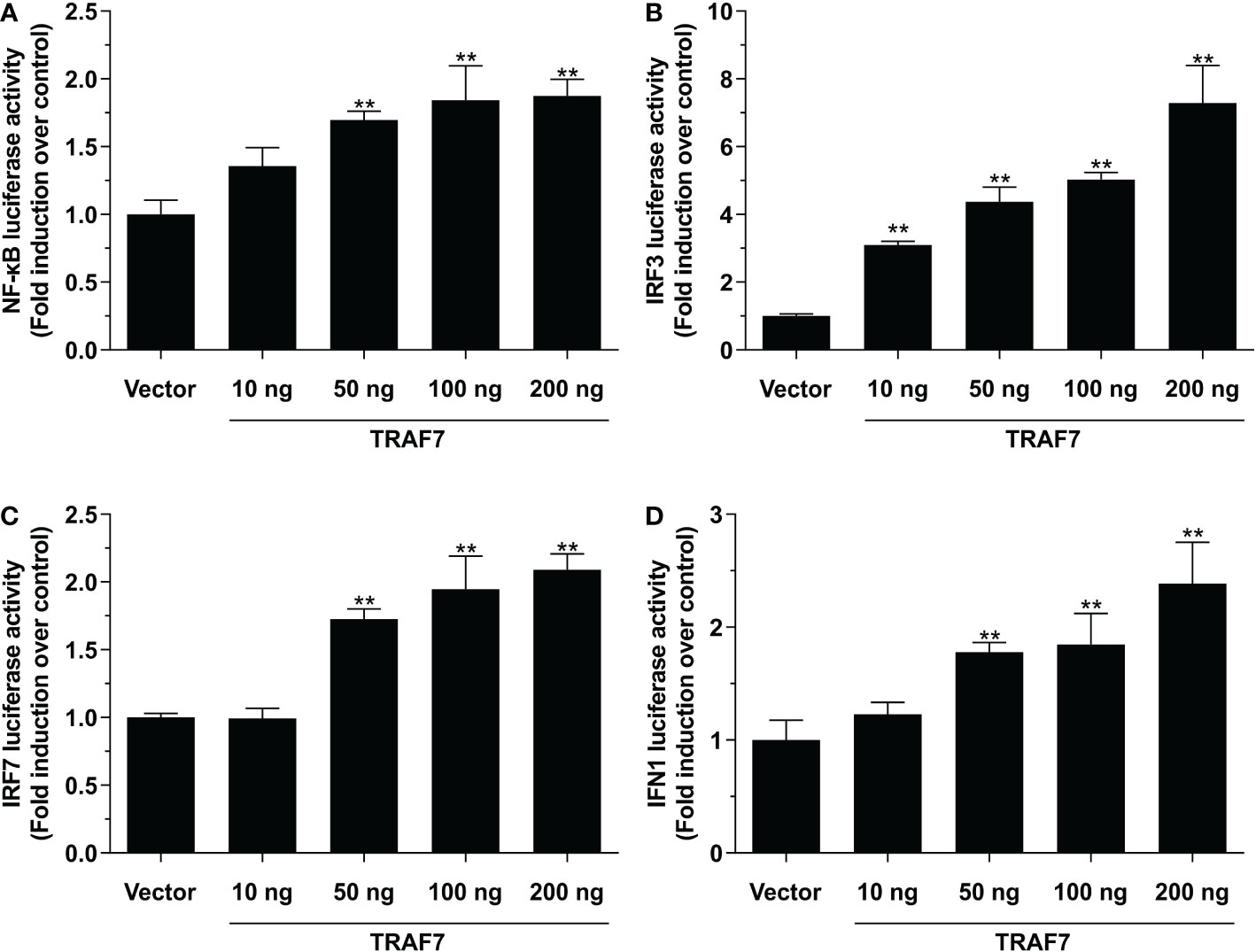
Figure 5 Activation of NF-κB (A), IRF3 (B), IRF7 (C), and IFN1 (D) promoters by Lc-TRAF7. HEK 293T cells seeded in 24-well plates were transiently transfected with 100 ng of pNF-κB-luc, pGL4-IRF3-pro, pGL4-IRF7-pro, or pGL4-IFN1-pro, 10 ng of pRL-TK together with the increasing amounts of pcDNA3.1-TRAF7 or pcDNA3.1 (vector control). At 24 hpt, the cells were harvested for detecting luciferase activity, with the results normalized to the Renilla luciferase activities and exhibited as the fold induction relative to the control group transfected with the pcDNA3.1 empty vector. All data were shown as mean of three independent experiments, with bars representing the SE. **P < 0.01.
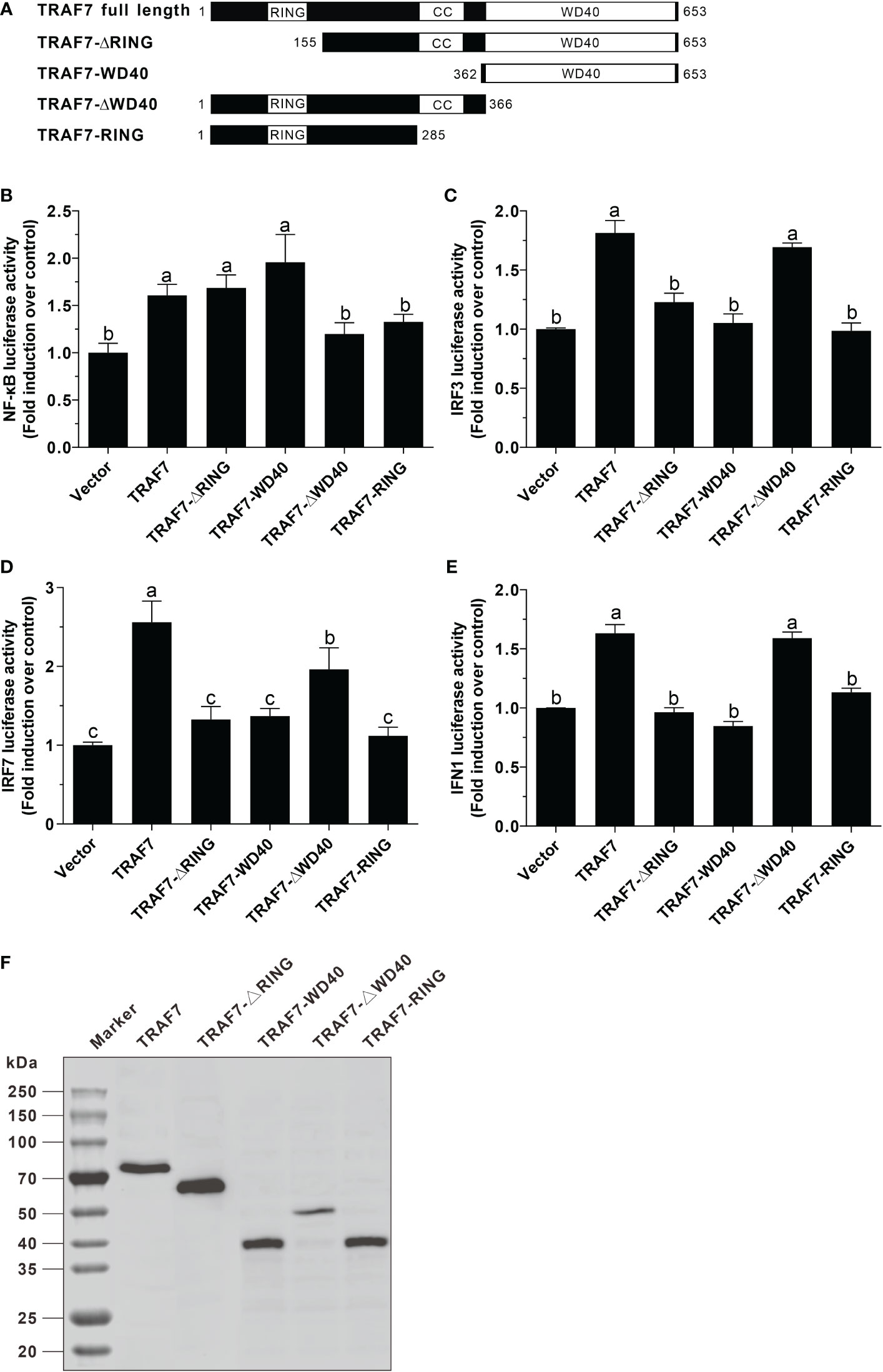
Figure 6 Function of Lc-TRAF7 and its domains in NF-κB, IRF3, IRF7, and IFN1 promoter activation. (A) Schematic representation of large yellow croaker TRAF7 full length and the truncated forms used in this study. Induction of NF-κB (B), IRF3 (C), IRF7 (D), and IFN1 (E) promoter activity by Lc-TRAF7 and its truncated forms. HEK 293T cells seeded in 24-well plates were co-transfected with 100 ng of pNF-κB-luc, pGL4-IRF3-pro, pGL4-IRF7-pro, or pGL4-IFN1-pro, 10 ng of pRL-TK together with 100 ng of the constructed plasmids including pcDNA3.1-TRAF7, pcDNA3.1-TRAF7-ΔRING, pcDNA3.1-TRAF7-WD40, pcDNA3.1-TRAF7-ΔWD40, or pcDNA3.1-TRAF7-RING. At 24 hpt, the cells were collected for detecting luciferase activity, with the results normalized to the Renilla luciferase activities and presented as the fold induction relative to the control group transfected with the pcDNA3.1 empty vector. (F) Lysates of the cells transfected with pcDNA3.1-TRAF7, pcDNA3.1-TRAF7-ΔRING, pcDNA3.1-TRAF7-WD40, pcDNA3.1-TRAF7-ΔWD40, or pcDNA3.1-TRAF7-RING plasmids were analyzed for the expression of the full-length Lc-TRAF7 or its truncated forms by Western blotting analysis using a mouse monoclonal Anti-c-Myc antibody. All data were shown as mean of three independent experiments, with bars representing the SE. Different letters above the bars indicate statistically significant differences between the groups (P < 0.05).
3.5 Subcellular localization of Lc-TRAF7
The full-length ORF of Lc-TRAF7 was subcloned into the pTurboGFP-N vector and then transfected into HEK 293T cells to determine the subcellular localization. The results showed that, except for the nucleus, the pTubo-TRAF7-GFP fusion protein was distributed throughout the cytoplasm, with a lot of bright green spots found around the nucleus (Figure 7A). To further ascertain the potential impact that various domains might have on the subcellular distribution of Lc-TRAF7, a panel of TRAF7 truncated forms including TRAF7-ΔRING, TRAF7-WD40, TRAF7-ΔWD40, and TRAF7-RING were also inserted into the pTurboGFP-N vector, followed by transiently transfected into HEK 293T cells. It was found that the mutants of TRAF7-ΔRING, TRAF7-ΔWD40, and TRAF7-RING had a similar subcellular localization as that found in the full-length Lc-TRAF7, which were all found in the cytoplasm with some aggregations around the nucleus. However, TRAF7-WD40 was not only distributed in the cytoplasm, but also located in the nucleus. Conversely, the control cells transfected with the pTurboGFP-N vector (control) had an entire GFP distribution throughout the whole cell (Figure 7A). In addition, the presence of the GFP fusion proteins, including the pTurboGFP (control), the full-length of Lc-TRAF7, TRAF7-ΔRING, TRAF7-WD40, TRAF7-ΔWD40, and TRAF7-RING was also confirmed by an Anti-TurboGFP polyclonal antibody using Western blotting analysis, which revealed the successful expression of target proteins (Figure 7B).
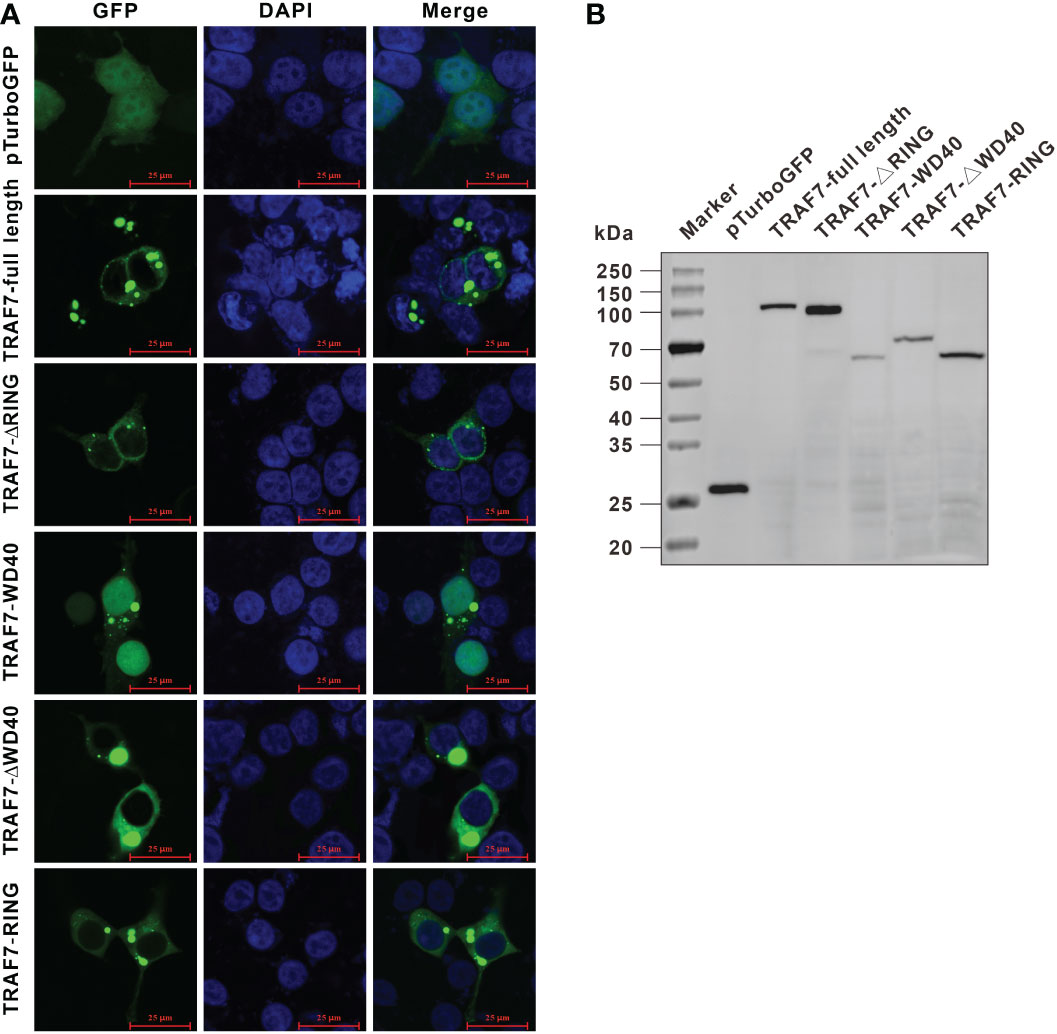
Figure 7 Subcellular localization of Lc-TRAF7 and its truncated forms. (A) HEK 293T cells were transfected with pTurboGFP-N (vector control), pTurbo-TRAF7-GFP, pTurbo-TRAF7-ΔRING-GFP, pTurbo-TRAF7-WD40-GFP, pTurbo-TRAF7-ΔWD40-GFP, and pTurbo-TRAF7-RING-GFP, respectively. At 24 hpt, the cells were stained with DAPI, followed by detecting under a confocal microscope and photographed. (B) The confirmation of the expression of GFP fusion proteins including the pTurboGFP, the full-length TRAF7, TRAF7-ΔRING, TRAF7-WD40, TRAF7-ΔWD40, and TRAF7-RING was conducted by Western blotting analysis using the Anti-TurboGFP antibody.
3.6 Lc-TRAF7 functions in the host antiviral signaling
To determine the role of Lc-TRAF7 in the host antiviral response, EPC cells were transiently transfected with pcDNA3.1-TRAF7 or pcDNA3.1 empty vector (control) before being infected by SVCV. It was revealed that the EPC cells overexpression of Lc-TRAF7 could significantly decreased the mRNA expression levels of SVCV-G, SVCV-M, and SVCV-P compared to the control cells transfected with the empty pcDNA3.1 vector at 24 h post-infection, which was 0.16-, 0.28-, and 0.09-fold relative to the control cells, respectively (Figure 8).
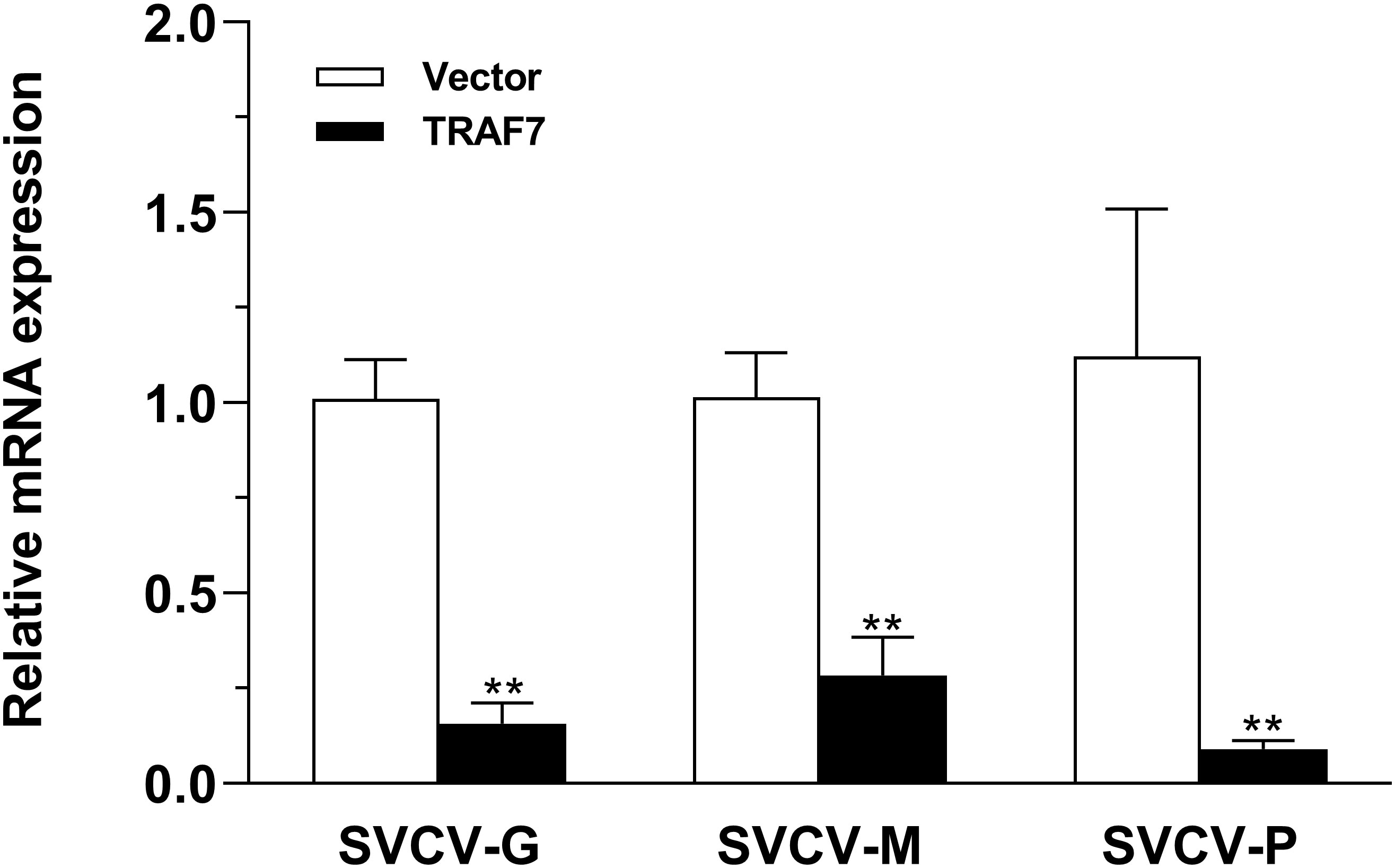
Figure 8 Antiviral assay of Lc-TRAF7 in fish cells. EPC cells in 6-well plates (2 × 106 cells/well) were transfected with 2.5 μg of pcDNA3.1-TRAF7 or pcDNA3.1 empty vector (control). At 24 hpt, the cells were infected with SVCV at an MOI of 1, the cells were then collected at 24 h post-infection for total RNA extraction, and the mRNA expression levels of SVCV-G, SVCV-M, and SVCV-P were detected by qRT-PCR analysis, with normalizing to the expression of EPC β-actin using the 2–ΔΔCt method. All data are obtained from three individual experiments, with bars representing the SE. **P < 0.01.
To further delineate the function of Lc-TRAF7 on the induction of downstream immune-related molecules, qRT-PCR analysis was performed to detect the expression profiles of antiviral and inflammatory-related genes including ISG15, ISG56, IRF3, IRF7, RSAD2, g-type lysozyme, IL-1β, and TNF-α under the overexpression of Lc-TRAF7. By transfecting the pcDNA3.1-TRAF7 plasmid into LYCMS cells, it was discovered that Lc-TRAF7 was successfully overexpressed (Figure 9A). Interestingly, the overexpression of Lc-TRAF7 significantly increased the mRNA expression level of the six detected immune-related molecules, such as IRF3, IRF7, ISG15, ISG56, RSAD2, and TNF-α, whereas the expression patterns of g-type lysozyme and IL-1β were not significantly affected (Figure 9B).
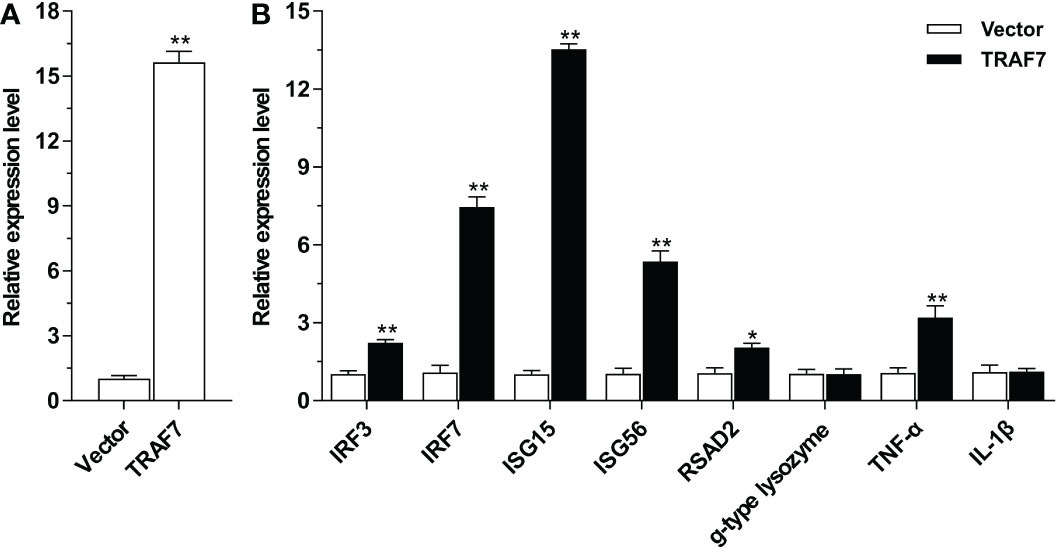
Figure 9 Expression patterns of antiviral and inflammation-related molecules under overexpression of Lc-TRAF7. (A) The confirmation of the overexpression of Lc-TRAF7. (B) The relative expression patterns of antiviral and inflammation-related molecules under Lc-TRAF7 overexpression. The LYCMS cells were transfected with pcDNA3.1-TRAF7 or pcDNA3.1 empty vector (control), respectively. At 48 hpt, the cells were then collected for the extraction of total RNA, follow by reverse transcription into cDNA and qRT-PCR analysis. The mRNA expression levels of large yellow croaker TRAF7, IRF3, IRF7, ISG15, ISG56, RSAD2, g-type lysozyme, TNF-α, and IL-1β were normalized to the expression of L. crocea β-actin, and then recorded as the fold relative to the group transfected with the pcDNA3.1 empty vector. All data are shown as mean of three repeated experiments, with bars representing the SE. *P < 0.05, **P < 0.01.
4 Discussion
As the latest discovered TRAF family member, TRAF7 appears to be involved in diverse biological processes. However, as the animal models genetically modified the encoding of TRAF7 are still limited, there are still many pieces missing to the picture that describes the function of TRAF7 even in mammals (Zotti et al., 2012). In this study, the ortholog of TRAF7, namely Lc-TRAF7 was characterized in large yellow croaker. The expression patterns, subcellular distribution, and the role of Lc-TRAF7 in NF-κB, IRF3, IRF7, and IFN1 promoter activation was investigated, the antiviral effect of Lc-TRAF7, and the induction of immune-related genes under Lc-TRAF7 overexpression was also determined, which providing a reference for the in-depth functional characterization of TRAF7 in the host immune responses in teleosts.
As that has been shown in our present results, Lc-TRAF7 was constitutively expressed in various organs and tissues of the healthy large yellow croakers, with the lowest expression level found in the brain, whereas the highest expression level detected in the liver. Other report in half-smooth tongue sole showed that TRAF7 was expressed highest in the gill (Li et al., 2020), indicating that the expression pattern of TRAF7 in organs/tissues may differ between species in teleosts. Moreover, the Lc-TRAF7 expression profiles in the gill, spleen, head kidney, intestine, and blood were significantly up-regulated under poly I:C, LPS, PGN, and P. plecoglossicida stimulations. Similarly, studies in half-smooth tongue sole also showed that Vibrio harveyi infection could significantly enhance the expression level of TRAF7 (Li et al., 2020). These results together suggesting the potential roles of teleost TRAF7 in the host immune responses, including antibacterial and antiviral responses.
In mammals, TRAF7 was found to be distributed in the cytoplasm as well as the nucleus (Zotti et al., 2011). However, our present results showed that Lc-TRAF7 had a cytoplasmic distribution in HEK 293T cells, with aggregation around the nucleus, such phenomenon was also detected in the truncated forms including the TRAF7-ΔRING, TRAF7-ΔWD40, and TRAF7-RING, indicating the deletion of RING finger or the coiled-coil domain did not affect the subcellular location of TRAF7, which was different from that in mammals (Zotti et al., 2011). Nevertheless, the truncated form of TRAF7-WD40, which has the simultaneous loss of RING finger and the coiled-coil domain, presented a global subcellular distribution including the nucleus. It is thus hypothesized that the RING finger and coiled-coil domain function importantly in the subcellular distribution of Lc-TRAF7.
Mammalian TRAF7 was defined as a protein which down-regulated the transcriptional activity of NF-κB by promoting the Lys-29-linked ubiquitination of NEMO and p65 (Zotti et al., 2011). In contrast, our present results indicated that Lc-TRAF7 overexpression could significantly induce NF-κB promoter activation. In addition, the deletion of the RING finger domain or the coiled-coil domain did not affect the induction activity, whereas the mutants with the WD40 domains deletion significantly abolished the role in NF-κB promoter activation. It is thus speculated that Lc-TRAF7 function distinctively in NF-κB activation from that in mammals, and the WD40 domains is essential in such signaling. Meanwhile, our results for the first time revealed that teleost TRAF7 could activate the IRF3, IRF7, and IFN1 promoters, with the RING finger and coiled-coil domain function essentially in the process. Nevertheless, the mechanism that involved in such singling activation remains more study.
Previous studies have demonstrated that IRF3 and IRF7 are transcription factors in regulating type I IFNs and IFN-stimulated genes (ISGs) production, which play important roles in the host antiviral immunity (Sun et al., 2010; Perng and Lenschow, 2018). In particular, ISG15 has been implicated as an crucial player in the host antiviral responses (Perng and Lenschow, 2018), RSAD2 exhibited antiviral activity against a wide range of viruses (Honarmand Ebrahimi, 2018), whereas ISG56 was determined to act as a negative-feedback regulator of virus-triggered induction of type I IFNs and antiviral responses (Li et al., 2009). Meanwhile, g-type lysozyme is a ubiquitous enzyme with hydrolytic activity of bacterial peptidoglycan, which plays an important role in protecting organisms against bacterial pathogens (Yin et al., 2003; Zhao et al., 2007; Wei et al., 2014), and TNF-α and IL-1β are important inflammatory-related factors and play key roles in the host inflammatory responses (Wu and Zhou, 2010; Lopez-Castejon and Brough, 2011). Those molecules mentioned above are vital regulators of the host immune responses, which are widely used in the detection of immune-related signaling. Notably, our present data revealed that overexpression of Lc-TRAF7 could significantly suppress the proliferation of SVCV in EPC cells, and the mRNA expression levels of antiviral molecules like IRF3, IRF7, ISG15, ISG56, RSAD2, and TNF-α were dramatically up-regulated under Lc-TRAF7 overexpression, indicating that Lc-TRAF7 may play an important role in the host antiviral immunity. Together with the findings that Lc-TRAF7 overexpression could induce NF-κB, IRF3, IRF7, and IFN1 promoter activation, it is thus assumed that Lc-TRAF7 may via NF-κB and/or IRF3/7-related singling to initiate the host antiviral signaling.
In summary, the present study cloned and identified TRAF7 gene from large yellow croaker and conducted a preliminary study on its function. Lc-TRAF7 was widely distributed in all the detected organs and tissues of large yellow croaker, and the expression of Lc-TRAF7 were significantly up-regulated under poly I:C, LPS, PGN, and P. plecogssida stimulations. Defined as a cytoplasmic protein, Lc-TRAF7 can induce the activation of NF-κB, IRF3, IRF7, and IFN1 promoters. Interestingly, Lc-TRAF7 overexpression can significantly restrain SVCV proliferation, and up-regulate the mRNA expression profiles of IRF3, IRF7, ISG15, ISG56, RSAD2, and TNF-α. These results preliminarily determine the role of Lc-TRAF7 in the host antiviral responses. Nevertheless, the signaling pathway that mediated and the molecular mechanism that involved still need further investigation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was reviewed and approved by Animal Administration and Ethics Committee of Jimei University (Permit No. 2021-4).
Author contributions
PZ conceived and designed the research. PZ, PL, YL, YC, JZ, ZL, YZ, and JJ performed the experiments and analyzed the data. PZ, YC, and YL wrote the manuscript. PZ, YL, ZZ, YJ, and YW revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a grant (31772878) from the National Natural Science Foundation of China, grants (2021J02046, 2021J01824) from the Natural Science Foundation of Fujian Province of China, a grant (3502Z20206017) from the Youth Innovation Foundation of Xiamen City of China, a grant (2021FJSCZY01) from the Seed Industry Innovation and Industrialization Project of Fujian Province, and a grant (2022ESHML08) from the Key Laboratory of Healthy Mariculture for the East China Sea, Ministry of Agriculture and Rural Affairs, China.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1092732/full#supplementary-material
References
Arkee T., Bishop G. A. (2020). TRAF family molecules in T cells: Multiple receptors and functions. J. Leukoc. Biol. 107, 907–915. doi: 10.1002/JLB.2MR1119-397R
Bouwmeester T., Bauch A., Ruffner H., Angrand P. O., Bergamini G., Croughton K., et al. (2004). A physical and functional map of the human TNF-alpha/NF-kappa b signal transduction pathway. Nat. Cell Biol. 6, 97–105. doi: 10.1038/ncb1086
Brubaker S. W., Bonham K. S., Zanoni I., Kagan J. C. (2015). Innate immune pattern recognition: A cell biological perspective. Annu. Rev. Immunol. 33, 257–290. doi: 10.1146/annurev-immunol-032414-112240
Chen Y., Li Y., Li P. T., Luo Z. H., Zhang Z. P., Wang Y. L., et al. (2022). Novel findings in teleost TRAF4, a protein acts as an enhancer in TRIF and TRAF6 mediated antiviral and inflammatory signaling. Front. Immunol. 13. doi: 10.3389/fimmu.2022.944528
Chen X. H., Lin K B., Wang X. W. (2003). Outbreaks of an iridovirus disease in maricultured large yellow croaker, Larimichthys crocea (Richardson), in China. J. Fish. Dis. 26, 615–619. doi: 10.1046/j.1365-2761.2003.00494.x
Dhillon B., Aleithan F., Abdul-Sater Z., Abdul-Sater A. A. (2019). The evolving role of TRAFs in mediating inflammatory responses. Front. Immunol. 10. doi: 10.3389/fimmu.2019.00104
Duvaud S., Gabella C., Lisacek F., Stockinger H., Ioannidis V., Durinx C. (2021). Expasy, the Swiss bioinformatics resource portal, as designed by its users. Nucleic Acids Res. 49, W216–W227. doi: 10.1093/nar/gkab225
Honarmand Ebrahimi K. (2018). A unifying view of the broad-spectrum antiviral activity of RSAD2 (viperin) based on its radical-SAM chemistry. Metallomics 10, 539–552. doi: 10.1039/C7MT00341B
Inoue J., Ishida T., Tsukamoto N., Kobayashi N., Naito A., Azuma S., et al. (2000). Tumor necrosis factor receptor-associated factor (TRAF) family: Adapter proteins that mediate cytokine signaling. Exp. Cell Res. 254, 14–24. doi: 10.1002/JLB.2MR1119-397R
Kordon A. O., Karsi A., Pinchuk L. (2018). Innate immune responses in fish: Antigen presenting cells and professional phagocytes. Turk. J. Fish. Aquat. Sci. 18, 1123–1139. doi: 10.4194/1303-2712-v18_9_11
Kumar H., Kawai T., Akira S. (2011). Pathogen recognition by the innate immune system. Int. Rev. Immunol. 30, 16–34. doi: 10.3109/08830185.2010.529976
Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lalani A. I., Zhu S., Gokhale S., Jin J., Xie P. (2018). TRAF molecules in inflammation and inflammatory diseases. Curr. Pharmacol. Rep. 4, 64–90. doi: 10.1007/s40495-017-0117-y
Langevin C., Aleksejeva E., Passoni G., Palha N., Levraud J. P., Boudinot P. (2013). The antiviral innate immune response in fish: Evolution and conservation of the IFN system. J. Mol. Biol. 425, 4904–4920. doi: 10.1016/j.jmb.2013.09.033
Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Li K. M., Li M., Wang N., Chen Y. D., Xu X. W., Xu W. T., et al. (2020). Genome-wide identification, characterization, and expression analysis of the TRAF gene family in Chinese tongue sole (Cynoglossus semilaevis). Fish. Shellfish Immunol. 96, 13–25. doi: 10.1016/j.fsi.2019.11.029
Li Y., Li C., Xue P., Zhong B., Mao A. P., Ran Y., et al. (2009). ISG56 is a negative-feedback regulator of virus-triggered signaling and cellular antiviral response. Proc. Natl. Acad. Sci. U.S.A. 106, 7945–7950. doi: 10.1073/pnas.0900818106
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Li C., Wang S., Ren Q., He T., Chen X. (2020). An outbreak of visceral white nodules disease caused by Pseudomonas plecoglossicida at a water temperature of 12°C in cultured large yellow croaker (Larimichthys crocea) in China. J. Fish. Dis. 43, 1353–1361. doi: 10.1111/jfd.13206
Lopez-Castejon G., Brough D. (2011). Understanding the mechanism of IL-1β secretion. Cytokine Growth Factor Rev. 22, 189–195. doi: 10.1016/j.cytogfr.2011.10.001
Luo Z. H., Li Y., Wang Y. L., Zhang Z. P., Zou P. F. (2022). Molecular cloning and functional characterization of HMGB1 and HMGB2 in large yellow croaker. Larimichthys crocea. Fish. Shellfish Immunol. 127, 855–865. doi: 10.1016/j.fsi.2022.07.018
Lu S., Wang J., Chitsaz F., Derbyshire M. K., Geer R. C., Gonzales N. R., et al. (2020). CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268. doi: 10.1093/nar/gkz991
Park H. H. (2018). Structure of TRAF family: Current understanding of receptor recognition. Front. Immunol. 9. doi: 10.3389/fimmu.2018.01999
Park H. H. (2021). Structural feature of TRAFs, their related human diseases and therapeutic intervention. Arch. Pharm. Res. 44, 475–486. doi: 10.1007/s12272-021-01330-w
Perng Y. C., Lenschow D. J. (2018). ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 16, 423–439. doi: 10.1038/s41579-018-0020-5
Shi J. H., Sun S. C. (2018). Tumor necrosis factor receptor-associated factor regulation of nuclear factor κB and mitogen-activated protein kinase pathways. Front. Immunol. 9. doi: 10.3389/fimmu.2018.01849
Stosik M., Tokarz-Deptuła B., Deptuła W. (2021). Immunological memory in teleost fish. Fish. Shellfish Immunol. 115, 95–103. doi: 10.1016/j.fsi.2021.05.022
Sun F., Zhang Y. B., Liu T. K., Gan L., Yu F. F., Liu Y., et al. (2010). Characterization of fish IRF3 as an IFN-inducible protein reveals evolving regulation of IFN response in vertebrates. J. Immunol. 185, 7573–7582. doi: 10.4049/jimmunol.1002401
Wei S., Huang Y., Huang X., Cai J., Wei J., Li P., et al. (2014). Molecular cloning and characterization of a new G-type lysozyme gene (Ec-lysG) in orange-spotted grouper. Epinephelus coioides. Dev. Comp. Immunol. 46, 401–412. doi: 10.1016/j.dci.2014.05.006
Wu Y., Zhou B. P. (2010). TNF-alpha/NF-kappaB/Snail pathway in cancer cell migration and invasion. Br. J. Cancer 102, 639–644. doi: 10.1038/sj.bjc.6605530
Xu L. G., Li L. Y., Shu H. B. (2004). TRAF7 potentiates MEKK3-induced AP1 and CHOP activation and induces apoptosis. J. Biol. Chem. 279, 17278–17282. doi: 10.1074/jbc.C400063200
Yin Z. X., He J. G., Deng W. X., Chan S. M. (2003). Molecular cloning, expression of orange-spotted grouper goose-type lysozyme cDNA, and lytic activity of its recombinant protein. Dis. Aquat. Organ 55, 117–123. doi: 10.3354/dao055117
Yoshida H., Jono H., Kai H., Li J. D. (2005). The tumor suppressor cylindromatosis (CYLD) acts as a negative regulator for toll-like receptor 2 signaling Via negative cross-talk with TRAF6 and TRAF7. J. Biol. Chem. 280, 41111–41121. doi: 10.1074/jbc.M509526200
Zhang X., Li W., Liu M., Zhang X., Yin X., Xu Z., et al. (2019). Transcriptomic analysis of the cold-pretreated Larimichthys crocea showing enhanced growth fitness in cold water. Mar. Biotechnol. (NY) 21, 791–805. doi: 10.1007/s10126-019-09924-8
Zhao J., Song L., Li C., Zou H., Ni D., Wang W., et al. (2007). Molecular cloning of an invertebrate goose-type lysozyme gene from Chlamys farreri, and lytic activity of the recombinant protein. Mol. Immunol. 44, 1198–1208. doi: 10.1016/j.molimm.2006.06.008
Zheng L. B., Hong Y. Q., Sun K. H., Wang J., Hong Y. J. (2020). Characteristics delineation of piscidin 5 like from Larimichthys crocea with evidence for the potent antiparasitic activity. Dev. Comp. Immunol. 113, 103778. doi: 10.1016/j.dci.2020.103778
Zotti T., Scudiero I., Vito P., Stilo R. (2017). The emerging role of TRAF7 in tumor development. J. Cell Physiol. 232, 1233–1238. doi: 10.1002/jcp.25676
Zotti T., Uva A., Ferravante A., Vessichelli M., Scudiero I., Ceccarelli M., et al. (2011). TRAF7 protein promotes lys-29-linked polyubiquitination of IkappaB kinase (IKKgamma)/NF-kappaB essential modulator (NEMO) and p65/RelA protein and represses NF-kappaB activation. J. Biol. Chem. 286 (26), 22924–22933. doi: 10.1074/jbc.M110.215426
Zotti T., Vito P., Stilo R. (2012). The seventh ring: Exploring TRAF7 functions. J. Cell Physiol. 227, 1280–1284. doi: 10.1002/jcp.24011
Zou P. F., Li K. Q., Li Y., Shen Y. J., Zhang Z. P., Wang Y. L. (2021a). RIP3 associates with RIP1, TRIF, MAVS, and also IRF3/7 in host innate immune signaling in large yellow croaker Larimichthys crocea. Antibiotics 10, 1199. doi: 10.3390/antibiotics10101199
Zou P. F., Li K. Q., Li Y., Shen Y. J., Zhang Z. P., Wang Y. L. (2022). Molecular cloning and functional characterization of RIP1 in large yellow croaker Larimichthys crocea. Fish. Shellfish Immunol. 122, 386–398. doi: 10.1016/j.fsi.2022.02.024
Zou P. F., Shen J. J., Li Y., Yan Q., Zou Z. H., Zhang Z. P., et al. (2019). Molecular cloning and functional characterization of TRIF in large yellow croaker Larimichthys crocea. Fish. Shellfish Immunol. 91, 108–121. doi: 10.1016/j.fsi.2019.05.011
Zou P. F., Shen J. J., Li Y., Zhang Z. P., Wang Y. L. (2020). TRAF3 enhances TRIF-mediated signaling Via NF-κB and IRF3 activation in large yellow croaker Larimichthys crocea. Fish. Shellfish Immunol. 97, 114–124. doi: 10.1016/j.fsi.2019.12.024
Keywords: TRAF7, NF-κB, IRF3, IRF7, antiviral responses, large yellow croaker
Citation: Li PT, Li Y, Chen Y, Zhang JX, Luo ZH, Zhang YF, Jiang J, Wang YL, Zhang ZP, Jiang YH and Zou PF (2023) Teleost TRAF7, a protein functions in the host antiviral responses via NF-κB and IRF3/7 mediated signaling. Front. Mar. Sci. 10:1092732. doi: 10.3389/fmars.2023.1092732
Received: 08 November 2022; Accepted: 23 January 2023;
Published: 06 February 2023.
Edited by:
Yafei Duan, South China Sea Fisheries Research Institute, ChinaCopyright © 2023 Li, Li, Chen, Zhang, Luo, Zhang, Jiang, Wang, Zhang, Jiang and Zou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Li, eWluZ2xpQHh1amMuY29t; Peng Fei Zou, cGVuZ2ZlaXpvdUBqbXUuZWR1LmNu
†These authors have contributed equally to this work
 Peng Tian Li
Peng Tian Li Ying Li
Ying Li Ying Chen
Ying Chen Jia Xi Zhang1
Jia Xi Zhang1 Zi Hao Luo
Zi Hao Luo Yi Lei Wang
Yi Lei Wang Yong Hua Jiang
Yong Hua Jiang Peng Fei Zou
Peng Fei Zou