- 1Department of BioSciences, University of Melbourne, Parkville, VIC, Australia
- 2College of Built Environments, University of Washington, Seattle, WA, United States
- 3Department of Biology, University of North Florida, Jacksonville, FL, United States
- 4Oregon Institute of Marine Biology, University of Oregon, Charleston, OR, United States
- 5Bigelow Laboratory for Ocean Sciences, East Boothbay, ME, United States
- 6Scottish Association for Marine Science, Scottish Marine Institute, Oban, United Kingdom
- 7Department of Marine Sciences, University of Gothenburg, Tjärnö, Sweden
- 8Department of Ecology and Evolutionary Biology, Cornell University, Ithaca, NY, United States
- 9School of Life and Environmental Sciences, Centre for Integrative Ecology, Deakin University, Burwood, VIC, Australia
Labyrinthula are unicellular protists occupying diverse spatial and functional niches, including various roles in host and ecological function, fatty acid production, pandemic marine disease and saprobic decomposition. Labyrinthula species span tropical and temperate climates and have been isolated from each marine coastal ecosystem tested. Our understanding of primary cellular and molecular functions of Labyrinthula has substantially progressed through a combination of increased global investments, research interest and technological advances. Recent advances in molecular techniques provide a toolkit for advancing ecological questions in marine infectious disease in seagrass meadows around the world. Here we provide a comprehensive review of relevant ecological and molecular techniques used in long-term research and the progression of Labyrinthula scholarship. Our aims in preparing this review are to: 1) share, compare and advance global Labyrinthula protocols, 2) increase accessibility to robust methodology to encourage the uptake of Labyrinthula-based questions into marine studies of molecular and ecological qualities of Labyrinthula and 3) encourage uptake of robust Labyrinthula-based questions into coastal marine studies, while also encouraging international collaborative networks across multiple fields. Lastly, we discuss gaps in the over 100 years of Labyrinthula research and opportunities for expanding research on this model marine organism.
1 Introduction to Labyrinthula sp.
Labyrinthulea are a class of heterotrophic protists containing two orders of Bigyra (Labyrinthulida and Thraustochytrida) known for their unique ability to produce exogenous sagenetosomal networks (also known as ‘slime nets’ or extracellular network) and biflagellated zoospores. Labyrinthulida are classified as Chromista and are common saprobic decomposers, parasites and pathogens of other organisms (Cavalier-Smith and Chao, 2006). They can be found in terrestrial and marine habitats, where they have been sampled from every ocean on the world, including temperate, tropical and polar regions (Raghukumar and Damare, 2011). Labyrinthulida comprises two families, Labyrinthulaceae and Aplanochytridiaceae, with the former containing the sole genus Labyrinthula (Anderson and Cavalier-Smith, 2012; Beakes et al., 2014; Pan et al., 2017).
Labyrinthula is a genus in Labyrinthulaceae described by unique ‘spindle-shaped’, ‘fungal-like’ qualities and production of distinctive colonies of transparent net-plasmodium (Pokorny, 1967; Porter, 1969). Labyrinthula spp. demonstrate considerable tolerance across a wide range of ecological conditions, such as variable temperature, salinity, light and nutritional resources (Vishniac, 1955; Sykes and Porter, 1973; Vergeer et al., 1995). Labyrinthula are also able to swiftly and efficiently transfer between a variety of biological hosts and sediment substrates (Muehlstein, 1992) and their potential roles in global disease phenomena are well documented, including observations of epidemic events in seagrass and turfgrass systems (Sullivan et al., 2013; Sullivan et al., 2018).
Labyrinthula was originally described from a marine alga (Cienkowski, 1867), and subsequently has been found in association with all three major algal groups, including one freshwater alga (Zopf, 1892), and even blue-green algae. In many of these observations Labyrinthula was considered to be pathogenic (Raghukumar 1987; Raghukumar, 1986) yet, to our knowledge Labyrinthula are not etiological agents of mass die-offs in marine algae. Rapid Blight (RB) however afflicts a variety of turfgrasses and is a lesser-known and reviewed pathosystem than Labyrinthula-driven Seagrass Wasting Disease (SWD). It was first noted in 1995, in the U.S. (Entwistle et al., 2014), with Labyrinthula terrestris later confirmed as the disease agent (Olsen et al., 2003; Bigelow et al., 2005). It is predominately associated with elevated salinity attributed to irrigation water (Camberato et al., 2006; Kerrigan et al., 2012) and considered an emergent disease driven by human activity (Douhan et al., 2009; Entwistle et al., 2014). Given the genetic distinctness observed between European and U.S. Labyrinthula terrestris, and the genetic diversity likely still hidden within the genus (Douhan et al., 2009; Chitrampalam et al., 2015; Martin et al., 2016; Lohan et al., 2020), exploring the wild host-origin(s) of the species affecting turfgrasses would be valuable (Douhan et al., 2009).
Tolerance to wide ranging ecological conditions and discoveries in advanced pathology support recognition of Labyrinthula as a model organism for studying infectious disease in halophytes (broadly defined here as salt-tolerant plants) across marine and terrestrial systems (Cienkowski, 1867; Watson and Raper, 1957; Pokorny, 1967; Dick, 2001; Olsen et al., 2003; Martin et al., 2016; Popova et al., 2020). Labyrinthula also plays an important and lesser-known role as a saprobic decomposer and remineralizer in coastal marine ecosystems, where researchers have discovered potential ecological value derived from its position in the food web, where it decomposes organic carbon and produces the nutritious fatty acid, DHA (Kumon et al., 2003; Armenta and Valentine, 2013; Yoshioka et al., 2019).
Difficulties in detecting and culturing Labyrinthula and a paucity of global monitoring protocols for SWD creates a steep learning curve for examination of emerging seagrass disease events when they do occur, and the lack of local skills may hinder timely detection of seagrass die-offs following regional and localized outbreaks of SWD. Increasing anthropogenic pressure on marine coastal ecosystems, including global warming, has created an increased risk of introduction or migration by non-native biota, such as pathogenic microbes (Harvell et al., 1999; Lafferty et al., 2004; Lafferty, 2009). It is prudent to inventory, review and share current methodologies in Labyrinthula and SWD research to highlight key research gaps that may bottleneck progress of evaluations and timely response to increasing instance of localized epidemics and loss events. Our review and syntheses provide a globally relevant summary of methodological research in support of advancement in examinations of Labyrinthula impacting conservation biology and ecology.
2 Ecological investigations
Despite their ecological value and ubiquity in the environment, co-infections and phenotypic variability can make Labyrinthula species challenging to identify and evaluate in the field. As genetic signatures and ecological datasets grow (Collado-Mercado et al., 2010; Martin et al., 2016; Lohan et al., 2020) combined molecular and ecological approaches will enhance our understanding of both the biology and ecology of cultured, as-yet uncultured, or unculturable, genotypes of Labyrinthula, whose hidden diversity has been predicted to be significant (Adl et al., 2012). Much remains to be discovered regarding the broader ecological characteristics of ‘elusive’ protists like Labyrinthulea, including their role in the food web, connections to marine disease ecology, pathogen transmission, distribution patterns and diversity in seagrass and other marine ecosystems. Identification of pathways for maintaining resilient seagrass and macroalgal ecosystems are critically needed to protect and restore global habitats, especially those recognized and conserved as high-value resources in marine areas.
2.1 Microbiome
The role of Labyrinthula in the seagrass and macroalgae microbiome and biofilm production is gaining attention (Hurtado-McCormick et al., 2020; Popova et al., 2020; Trevathan-Tackett et al., 2020; Wang et al., 2020; Beatty et al., 2022). The larger role of Labyrinthula in microbial ecology, including pathology of other organisms (O’Kelly, 2005) and trophic food webs is largely unknown. However, microbial drivers of seagrass health have been discovered to increase host resilience (Martin et al., 2019), suggesting that, similar to marine corals (Rosado et al., 2019), development of a pro-biotic treatment may provide some support for seagrass or other populations facing exacerbating disease. Relatedly, host microbiome changes were associated with prevalence of SWD in eelgrass, with the potential of some enriched taxa (e.g. Cellvibrionaceae, Colwelliaceae, and Granulosicoccaceae) to exacerbate disease (Beatty et al., 2022).
2.2 Signs of disease
SWD appears in infected seagrass plants as small dark patches of black or brown spots or streaks, typically in leaf tissues. Within days, the streaks expand and coalesce, causing rapid loss of photosynthetic ability and buoyancy in leaves. The ‘stains’ symptomatic of SWD may eventually take over the entire leaf, and in the process, cells may be transferred in the water column or by leaf-to-leaf contact. RB in turfgrasses progresses in a very similar fashion to SWD, with water-logged grass tissues becoming discolored (often chlorotic to near transparent) and eventually dying (Kopec et al., 2004). These patches may eventually join to form larger areas of collapsed turf (Olsen et al., 2003). Extended periods of time between large-scale disease events has hindered long-term data collection.
2.3 Pathosystems
Interactions among hosts, pathogens and their environment are known to regulate disease in plants, including seagrasses (Sullivan et al., 2013). SWD is sometimes described as a syndrome, rather than a specific ‘disease’ as there are often co-factors implicated in disease progression (Porter, 1986; Brakel et al., 2019; Duffin et al., 2020). The pathosystems model recognizes the complex and intersectional roles of multiple environmental parameters and thresholds in determining the health of both pathogen and host, including their co-interactions. The core relationships between age, salinity, temperature, light and associated host-pathogen (Seagrass species–Labyrinthula species) relationships have well-demonstrated impacts on occurrences and severity of disease (Groner et al., 2014; Groner et al., 2016; Dawkins et al., 2018).
2.4 Transmission
Labyrinthula are ubiquitous in plant, water and sediment samples from a variety of saline terrestrial and marine environments, including halophytes in salt lakes (Amon, 1978), golf courses (Olsen et al., 2003), diatoms, algae (Jepps, 1931) and seagrass around the world (Vergeer and den Hartog, 1994). Inhabiting a range of halophytes, and even amoeba (Dyková et al., 2008), Labyrinthula have been identified from all nearshore marine environments of the world it has been sampled (Vergeer and den Hartog, 1994; Raghukumar and Damare, 2011). Evidence of both leaf-to-leaf and water borne disease transmission have been demonstrated (Muehlstein et al., 1988; Martin et al., 2016). Research initially focused on leaf-to-leaf transfer as a primary mode of transmission, thus pathogenicity studies were initiated using replicate inoculated leaf tissue vectors aimed at mimicking natural direct contact conditions (Muehlstein et al., 1988). Newly developed metabarcoding and molecular investigation techniques targeting Labyrinthula spp. indicate alternate transmission pathways in life-cycle and various microhabitats, including sediment and water columns (Lohan et al., 2020), and ballast water samples (Galil and Hülsmann, 1997; Lohan et al., 2016). Variability in transmission mode and potential vectors indicate unknown modes of transmission might affect Labyrinthula pathosystems, especially in the wild. Pathways for transmissions are an active area of research in Labyrinthula.
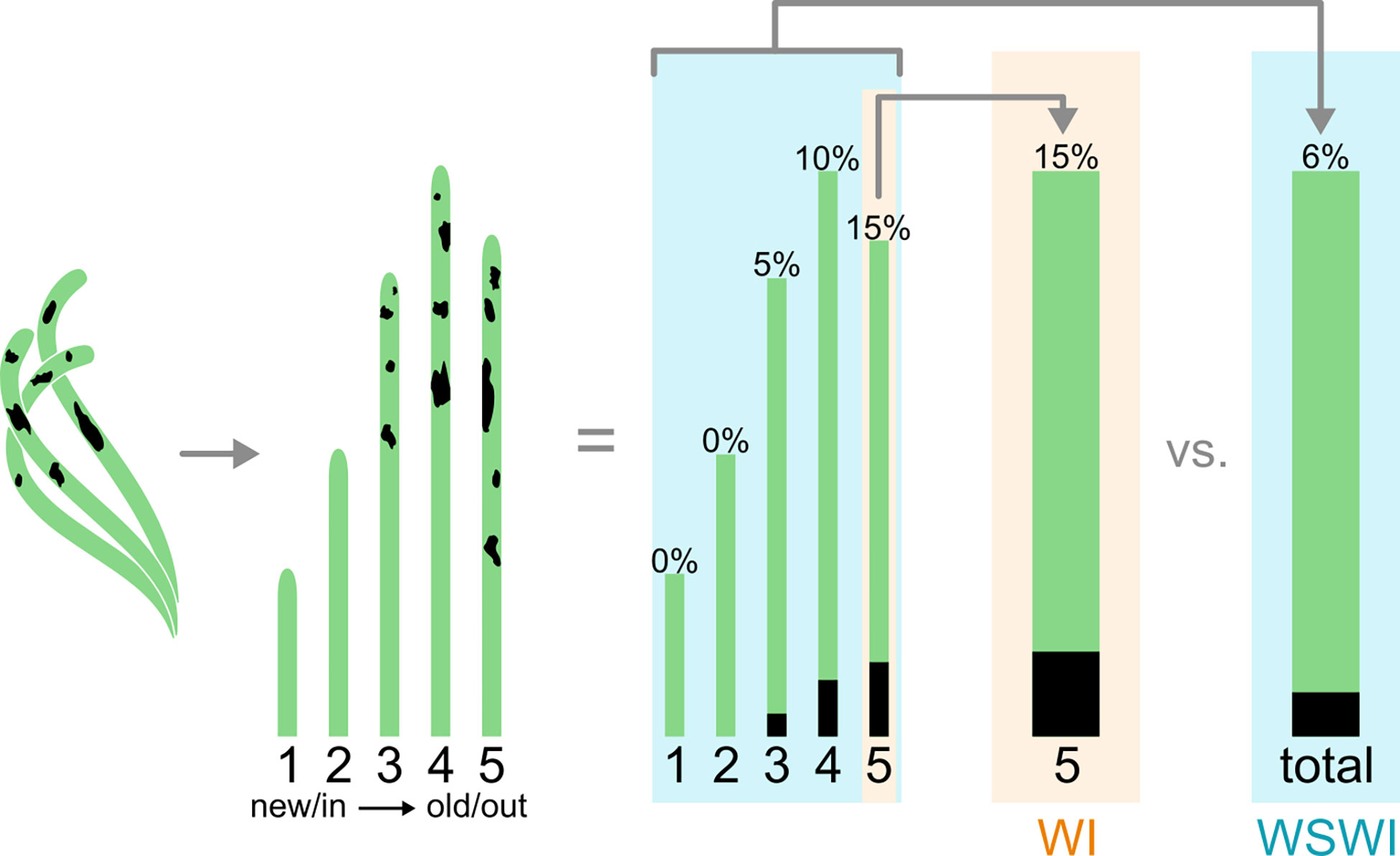
2.5 Wasting disease index
Leaf-based analyses of lesion size and disease severity metrics can be visually estimated using the Wasting Disease Index (WI) method (Burdick et al., 1993). The WI reveals characteristics about the most severe infections of host tissue and may be valuable at a molecular or physiological level. This metric may be used to model a worst-case epidemic infection. Still, a high WI does not say anything about how the infection is affecting the rest of the shoot, since most other leaves can be healthy even if the WI is high on a given leaf. On the other hand, the Whole Shoot Wasting Index (WSWI) determines to what extent the infection affects the whole plant, which can be averaged over a sample population to understand how the infection is affecting population dynamics and to understand resilience (Figure 1). Burdick et al. (1993) argue that WI is a better measurement of infection than WSWI since the range of disease values is larger, making it easier to monitor variation in disease over time. A major limitation with the WI more broadly is that it only measures the visual outcome of the infection. It does not reveal anything about the abundance of Labyrinthula cells or physiological responses to infection on the leaf, shoots, or below-ground energy reserves. More recently, the WI has been coupled with histology (Groner et al., 2014) and quantitative PCR methods (Bergmann et al., 2011; Duffin et al., 2020) to provide greater resolution for measures of density and abundance of Labyrinthula associated with a specific leaf or plant. For example, researchers found Labyrinthula load and areal lesion extent in turtlegrass exhibit a logistic relationship, suggesting pathogen load does not scale linearly across lesion sizes (Duffin et al., 2020). Investigations of cell counts reveal threshold levels of infection may exist before a leaf shows any symptoms and that cell counts may experience a dramatic decline in abundance when they are associated with larger lesion areas (> ~1/3 of leaf area). The relative presence of Labyrinthula may be further moderated through host immunity, which can be spatially and temporally variable (Bockelmann et al., 2013; Duffin et al., 2020). An additional constraint with WI is that black lesions can be a result of necrosis by other reasons besides Labyrinthula infection. Depending on the plant species and health of the plant it can be difficult to distinguish between lesions, heat stress and hydrogen sulfide toxicity. It is therefore difficult to be 100% sure lesion measurements give a fair picture of the infection severity. By including a second measuring technique, e.g., qPCR, plants with necrosis can be examined for the presence and abundance of the pathogen, hence corroborating the WI findings. Thus, it is important for researchers to distinguish between the different indexing methods and in either case use a quantitative cell measurement when investigating Labyrinthula infection. Improved measurement techniques have included taking photographs of each leaf used in WI and WSWI and assessing visual characteristics with image software capable of analyses, such as ImageJ (Trevathan-Tackett et al., 2018a) and emerging ‘artificial intelligence’ tools, such as EeLISA (Rappazzo et al., 2021; Aoki et al., 2022).
2.6 Collection guidance
Place-based, or ‘field’ sampling has often been the first step in initiating Labyrinthula collections and investigations worldwide. Responsible scientific investigations (Minteer and Collins, 2008) in Labyrinthula, wasting disease and other related research may include obtaining legal permits and conducting sampling practices that encourage the recognition and conservation of natural processes, and local communities. This is especially true on long-term monitoring sites; while minimizing harm to vulnerable plant roots and sediments, we can honor and respect local traditional ecological knowledge, people and society (Pierotti and Wildcat, 2000). Practically, when sampling eelgrass for Labyrinthula, take care to avoid sampling the outer-most leaves as they are older and could be senescing or turning colors for any number of reasons. Older leaves are further likely to have higher densities of fungi and other microbial contaminants as they naturally acquire epiphytes and fouling organisms with age (Opsahl and Benner, 1993). Specific recommendations for collecting methods are provided in the Supplemental Manual.
2.7 Considerations for sampling design
Field collections targeting Labyrinthula in monocultural halophytes (such as seagrass and turf grass) generally involves cluster sampling or modified cluster sampling (Madden and Hughes, 1999; Garcias-Bonet et al., 2011). Sampling individuals and populations across variable spatial and temporal conditions can be performed through stratified or randomized grid, transect or other data point locations. The total number of samples required to test hypotheses may be quite variable based on the type of samples being collected, Labyrinthula lifecycle, or nuances of the specific questions being asked.
3 Laboratory protocols
Many techniques have been utilized to study the biology of Labyrinthula in the laboratory, including culture-based isolation, molecular and morphological characterization, pathogenicity assays, transmission dynamics, host immunity responses, fatty acid analyses, microscopy, and virulence. More recently, emerging research on host immunity (Duffin et al., 2020), discovery of Labyrinthula’s role in primary DHA production (Kumon et al., 2002; Yoshioka et al., 2019) and the recent sequencing of the first Labyrinthula genome (Tan et al., 2021) have highlighted the importance and potential for advancement of discoveries related to this unique protist.
3.1 Species identification and classification
Labyrinthula spp. (Cienkowski) were discovered through early field sampling and laboratory culture of marine microbes, and are described in the literature as early as 1867 (Cienkowski, 1867). The family only consists of one genus and has been variably classified between fungi and slime molds since that time, though more recent evaluations have established their position among the Stramenopiles, adjacent to the oomycetes (Porter, 1990; Leander and Porter, 2001; Tsui et al., 2009; Leano and Damare, 2012; Beakes et al., 2014).
3.1.1 Morphology
Labyrinthula can be identified in culture through observation of distinctive fusiform cells embedded within hyaline ectoplasmic networks, or ‘slimeways’ (Porter, 1969). Individual Labyrinthula cells are colorless, though dense aggregations may have some yellowish/orangish coloring (Figure 2), measuring between 5 µm – 30 µm long by 3-8 µm wide (Pokorny, 1967; Muehlstein et al., 1988; Leano and Damare, 2012). Each Labyrinthula cell contains a single large central nucleus, flanked by 2 large vacuoles where stores of lipid bodies are easily visible (Jepps, 1931; Young, 1943; Porter, 1969). Labyrinthula cells also contain specialized posteriorly oriented organelles, including dictyosomes and an endoplasmic reticulum (Porter, 1969).
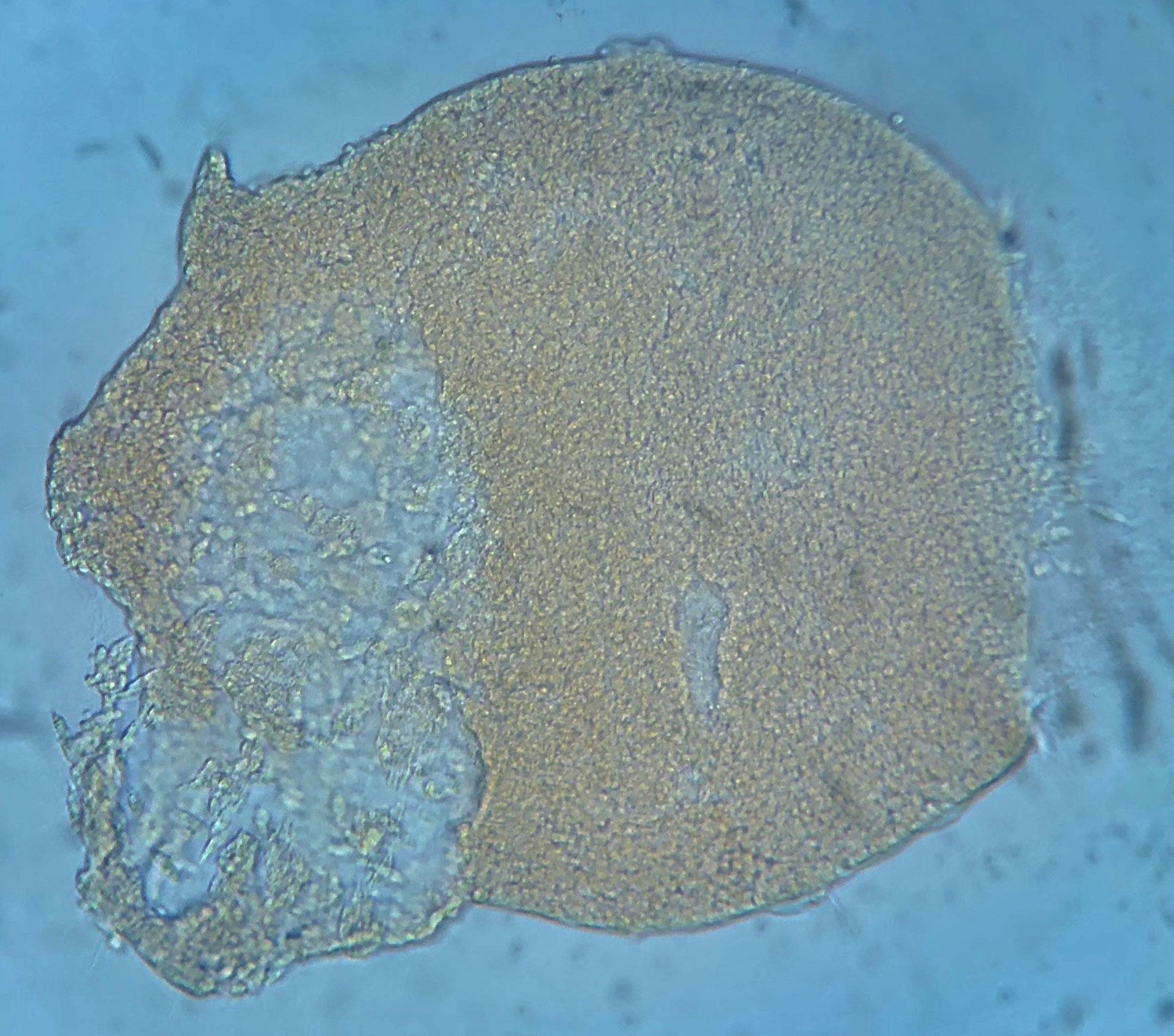
Figure 2 Image of large aggregate/sorus-like mounding of Labyrinthula sp. cells from liquid culture, isolated from diseased tissue of Thalassia testudinum. Yellowish or orangish color is common in such aggregates.
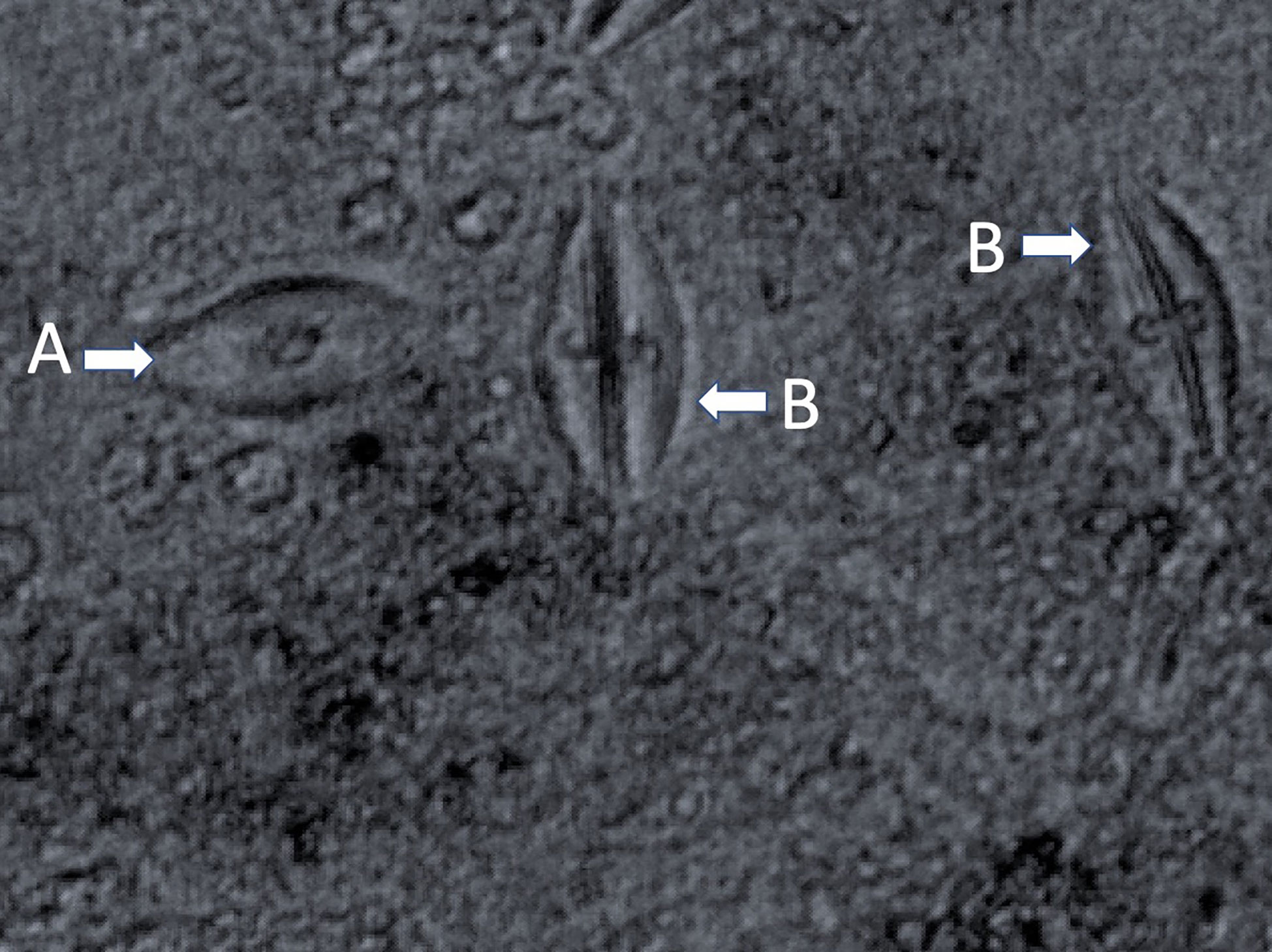
Early in isolation, Labyrinthula cells appear enigmatic, as actin-myosin reactions in the ectoplasm generate distinctive gliding motion of cells within the colony. Active cells may travel up to 50 µm per minute (Dietz and Schnetter, 1999; Preston and King, 2005). The plasmodium generating organelle allows the cells to develop sometimes extensive networks, which are used by the cells for both motility and osmotic feeding (Porter, 1969) by secreting exoenzymes for digestion during nutrient uptake (Tsui et al., 2009). The unique ectoplasmodic networks generated by Labyrinthula grow in root-like, or ‘lacey’ patterns (Vergeer and den Hartog, 1994). They are most often observed following a period of intensive axenic culturing and growth in a laboratory setting. Cell colonies are readily identifiable through microscopy where they can be observed forming distinctive and robust fractal-like vegetative cell communities in culture (Figure 3) (Jepps, 1931; Young, 1943; Trevathan-Tackett et al., 2018a). In the literature, Labyrinthula colonies have been variably described by morphological qualities, however there is agreement in their tendency to branch out from single-point infections. Growth characteristics of the colony may be characterized by cellular alignments, density, agar penetration and cellular sizes (Sullivan et al., 2013; Sullivan et al., 2017; Trevathan-Tackett et al., 2018a). Finally, care should be taken to avoid confusing Labyrinthula with a closely-related and commonly co-occurring taxa, such as aplanochytrids, especially for smaller-bodied Labyrinthula spp. (Figure 2, Leander et al., 2004). With experience and under similar culture conditions, aplanochytrids can appear to have a more distinct nucleus, be more rounded, and have a more ‘dotted line’ like presentation when observing continuity among cells in the leading edge/arm of a colony on agar (Leander et al., 2004).
3.1.2 Reproduction
Labyrinthula cells reproduce both sexually and asexually (Figure 4). Vegetative cells multiply through binary fission or mitotic division of vegetative cells. This is considered to be the most commonly used strategy for replication and propagation of Labyrinthula cells in naturally occurring colonies (Pokorny, 1967). A cyst-like resting phase, possibly asexual, is also reported (Pokorny, 1967; Amon, 1978). Sexual modes of reproduction have been described through accounts of aggregating plasmodia (Pokorny, 1967), sporulating cells, and zoospore recovery (Amon and Perkins, 1968; Perkins and Amon, 1969). Several accounts of cells restructuring in culture to form dense aggregations of cells with nucleated walls are described as ‘sori’ (Pokorny, 1967). The ‘spores’ aggregating inside the ‘sori’ are extremely difficult to visualize and often usual staining methods are not effective (Jepps, 1931). However, bi-flagellated zoospores were isolated from a sorus and photographed using a scanning electron microscope (Amon and Perkins, 1968). Imaging and assessment of chemical signaling and quorum-sensing behaviors has been described for other organisms, such as bacteria (De Kievit and Iglewski, 2000), but is generally lacking for stramenopiles (Hassani et al., 2018). The role of chemotaxis in Labyrinthula life cycle and behavior are unknown.
3.1.3 Systematics
The first attempt to morphologically and taxonomically describe separate Labyrinthula (Cienkowski) species evaluated and summarized cell structure, colony morphology, physiology, ecology and pathogenic considerations, for 10 species (L. macrocystis, L. minuta, L. vitellina var. vitellina, L. vitellina var. pacifica, L. cienkowski, L. zopfii, L. valkonovii, L. chattonii, L. algeriensis, L. roscoffensis, L. coenosystis) of known isolates (Pokorny, 1967). A review of taxonomy reveals early nomenclature was abandoned when two epithets emerged in the literature as pathogenic phenomenon were discovered in Labyrinthula, including L. zosterae (Porter, 1990) affecting seagrasses and L. terrestris (Bigelow et al., 2005) affecting turf grasses. A third species, Labyrinthula diatomea, is more recently described and is associated with marine sediments where it primarily, if not entirely, consumes diatoms (Popova et al., 2020). There may be redundancy in the descriptions of cultured specimens based on morphological features, however, molecular sequence-based approaches help clarify, replicate and underpin resolution efforts in this arena (e.g. Martin et al., 2016).
Metabarcoding studies commonly target the V9 section of the 18S rDNA gene to describe protist communities (Amaral-Zettler et al., 2009). Huge data collection surveys like the Tara Ocean Expedition or the Earth Microbiome Project accumulate an immense database of 18S rDNA sequences and associated environmental data. These efforts drive advances in protistan phylogeny and accelerate our understanding of ecological needs for otherwise cryptic taxa, including Labyrinthulomycetes (Pan et al., 2017). Global Labyrinthula surveys, including two nuclear DNA sequencing methods (18S and ITS amplicons) report there are at least 16-21 Labyrinthula species covering both terrestrial and marine habitats (Bigelow et al., 2005; Martin et al., 2016). ITS sequences are more commonly used as a barcode for fungi phylogeny (Schoch et al., 2012). Therefore, both currently used molecular markers have high quality reference data and are a good tool for discriminating taxonomic units and to describe diversity within the genus Labyrinthula. Newer whole genome approaches will eventually resolve phylogenetic uncertainties.
Given the plastic and somewhat featureless nature of Labyrinthula cells at the level of gross morphology, genetic signatures are a critical pathway for resolution. In the last decade, molecular techniques have been more commonly used to identify Labyrinthula spp. cultured from field samples (see Section 3 for details on molecular markers). Progress has been made in classifying Labyrinthula in the context of other genera and families in this order (Pan et al., 2016; Pan et al., 2017). The most recent key developed to describe distinguishable morphological characteristics of species in this genus was prepared over 20 years ago (Dick, 2001). Genetic evidence for Labyrinthulomycetes species diversity has recently leaned more on barcoding-type approaches, but also ecological context, to establish speciation and develop a stable taxonomic key (Martin et al., 2016). Given the robust record of publishing species descriptions, recent advances in genome sequencing (Tan et al., 2021) and success in developing prototypic genetic-level species delineations (Martin et al., 2016), there are new opportunities to resolve historic and ongoing discoveries into speciation and evolution of Labyrinthula.
3.2 From leaf to culture
To isolate and identify Labyrinthula from field collections, targeted sections of leaf tissue may be extricated from leaf samples and transferred to sterile agar media for growth and isolation (Young, 1943; Muehlstein, 1992). The use of high-quality clear agar results in the best colony growth visualizations. Isolated cultures can be used for cellular and molecular investigations, including diagnostics, growth assays, fatty acid analyses, visualizations and long-term sample storage.
3.2.1 Tissue preparation and growth media
Prior to sampling collected plant material for transfer onto agar media, it is advisable to pre-treat tissues to reduce contaminants (See Supplemental Manual). Labyrinthulids are able to utilize a range of microbial or plant substrates (See Supplemental Manual) for survival (Sykes and Porter, 1973). This is consistent with findings about their diverse biogeographic range, hosts, and trophism. Variabilities in nutrients and other additives in media may have a substantial impact on the cell and colony characteristics of isolates. Accordingly, care should be taken with the intent of using any culture medium, as the consequences of different media on growth results remains an open question, especially with regards to affecting strain virulence. A summary of standard agar and liquid culture methods successfully used to culture Labyrinthula can be found in the Supplementary Manual.
3.3 Axenic culture
After Labyrinthula is identified growing away from the host tissues and onto selected sterile culture media, it can be transferred repeatedly to new petri-dishes. Active and relatively rapid identification and transfer of Labyrinthula is required in initial phases of axenic cell and colony isolation. A common source of contamination in cultures are fungi, bacteria and other protists from the original seagrass segments. The use of antibiotics and germanium dioxide in the sterile seawater agar media helps to avoid and minimize bacterial and diatom contaminations (See Supplementary Manual).
3.4 Quantifying cell and colony growth
When examining one or more factors suspected of impacting Labyrinthula cell and colony growth, assays may be required. Assays provide a tool for experimentation of environmental and laboratory conditions that may favor or hinder the growth or pathogeneses of Labyrinthula.
3.4.1 Growth assay
Since Labyrinthula does not grow fully homogeneous in liquid culture or on agar plates, it is difficult to control Labyrinthula cell densities at the start of any assay. For this reason, it is important to advance protocol to standardize precise conditions and controls to account for the overall variance in Labyrinthula growth between replicates. The length of the assay will depend on how fast isolates are growing. All assays should be stopped before the colony reaches the edge of the tank, petri dish or well-plate. Those types exhibiting more agar-surface growth tend to grow faster and often reach the edge within days, while predominantly in-agar types are usually slower and may never reach the edge. Given the phenotypic plasticity observed in Labyrinthula, care should be taken to make sure the Labyrinthula isolate used for all replicates of the assay exhibit similar cell size and shape, and that they are growing at roughly the same rate. Thus, colonies used in assays should be the same age, and maintained by transferring to new agar plates at the same time (ideally, not more than 1 week or so old). Finally, as noted by Martin et al. (2009), abiotic conditions (and likely biotic, as well) can alter both colony extent and cell densities within colonies/ectoplasmic networks, with experimental designs and questions dictating whether both measures are necessary. Three key parameters (dish size, borer size and incubation time) can be altered to fit the need of the assay. For example, if the assay is used to investigate Labyrinthula growth on seagrass extracts it can be beneficial to use smaller petri dishes or well-plates to minimize the amount of extracts used for each replicate (Trevathan-Tackett et al., 2015; Jakobsson-Thor et al., 2018). If long term data are required, a larger dish and longer incubation time may be needed.
3.5 Cell quantification by hemocytometer
Cell counts using hemocytometers is a common, easy and inexpensive (though sometimes time-consuming) method for quantifying cell numbers. Due to the adhesive nature of Labyrinthula cells and colonies to surfaces and each other, accurate quantification of Labyrinthula in suspension interferes with both direct counting and absorbance-based cell quantifications. Through informal tests of counting cells via hemocytometer in suspension, 25-second vortex durations of Labyrinthula with 100-µm glass or zirconia/silica beads emerged as a useful method of separating cells to facilitate counting (Dawkins et al., 2018).
3.6 Visualization
Given Labyrinthula’s generally hyaline appearance, histological and microscopic investigations have been critical to species discoveries, including the creation of detailed drawings of Labyrinthula cellular and colony morphology (Jepps, 1931; Young, 1943). It can be very difficult to detect Labyrinthula colonies with the naked eye, though it is possible with experience. Methods to track live Labyrinthula cells and visualize their distinct qualities and behaviors will improve our knowledge of cellular and colony functional traits.
3.6.1 Histology
Staining of plant tissues has been effective in revealing important cellular characteristics of Labyrinthula, including descriptions of key structures and functions (Jepps, 1931; Young, 1938; Watson and Raper, 1957; Armstrong et al., 2000; Preston and King, 2005). Early visualizations were drawn by Jepps (1931) observed from stained and fixed Labyrinthula cells, as viewed through a microscope. More recently, color images of histological investigations of seagrass demonstrate the presence of Labyrinthula cells within the leaf tissue of its seagrass host (Groner et al., 2014; Groner et al., 2016). For example, in our experience and that of Harrington and Hageage (2003) when utilizing a fluorescing stain, it is strongly advised to carefully evaluate stain performance under particular conditions, as it may vary by organism, pH, production batch, and many other factors. Notably for seagrasses, background fluorescence can vary significantly by species (e.g., Figure 5A, B), leaf age (possibly related to cuticle thickness and/or composition), degree of fouling and damage to the epidermis (Figures 5A–C), and microscope filters used.
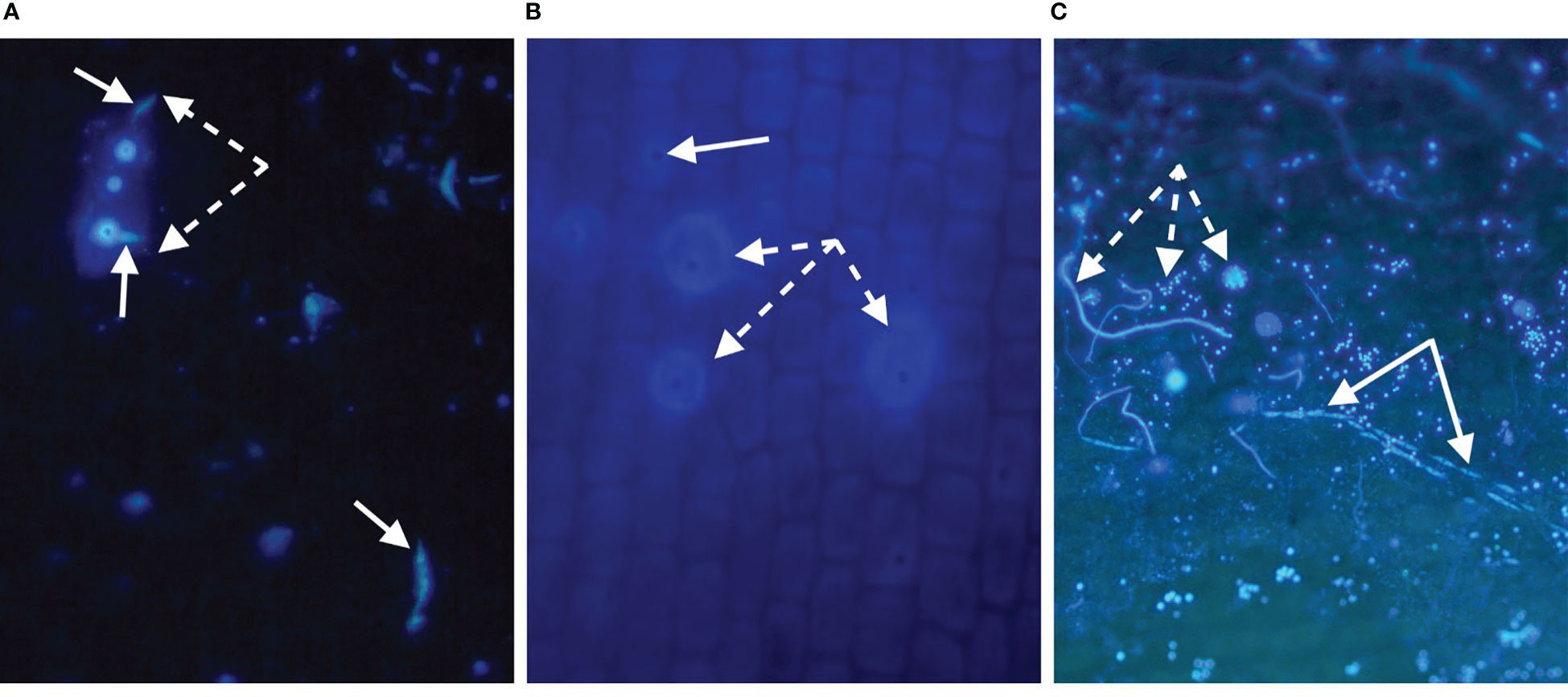
Figure 5 (A-C). Evidence of Labyrinthula sp. infection using u/v light microscopy with the fluorescing stain, Calcofluor White (CW). CW binds cellulose of host cell walls (less so for waxy cuticle regions), and the chitin within Labyrinthula sp. and fungal cell wall structures. (A) Thalassia testudinum surface showing rectangular epidermal cell (dashed arrows), infected with Labyrinthula sp. in vitro, fluorescing from the inside where CW has gained access to the inner walls, and thus is brighter than neighboring cells. Individual Labyrinthula sp. cells (solid arrows), some of which appear to be entering (or exiting) epidermal holes (bright spots, for which fluorescing edges have bound stain, and dark center/hole with no stain). (B) Field-collected Zostera marina epidermal surface from diseased (bleached/lesioned) tissue, where apparent Labyrinthula sp. hole-like damage (solid arrow) has allowed stain to bind inner structures of some cells (dashed arrows); focused slightly below the surface wall, hence rounder appearance. (C) Field-collected Thalassia testudinum epidermal surface of green tissue located 3 mm from diseased (brown/lesioned) tissue edge; stained with CW and showing trail of Labyrinthula sp. cells (solid arrows) co-occurring with apparent fungal structures (dashed arrows: hyphae, spores [white spots without dark centers], appressoria), emphasizing the potential for interactions among the host and its microbiome.
3.6.2 Microscopy
An inverted microscope with phase contrast allows for the best visualization of cell and colony formations and measurements needed to verify the presence of Labyrinthula organisms. Scanning electron microscopy (SEM) has been successfully used to view zoospores (Amon and Perkins, 1968; Perkins and Amon, 1969) and ultrastructure (Perkins, 1972) in Labyrinthulomycota. To improve microscopic images of cultured colonies, staining and light manipulations may be required. Exploration and discovery of these and other visualization tools may aid in further discoveries of temporal and spatial qualities of host-pathogen relationships and trophic transfers.
3.7 Culture maintenance and storage
As isolates are acquired, it may be desirable to slow Labyrinthula colony growth while maintaining genotypes or healthy populations in the laboratory. There are storage options for researchers who may no longer require rapid and intensive growth in their collection. Short-term (day-to-day) storage of sealed isolates is possible for rapid growth of cultured colonies. Temperature thresholds can be strain specific and thus, the optimum temperature for culturing isolates will be different depending on climatic adaptation. Generally, cultures can be grown at approximately 20 °C. When longer time periods between transfers are needed, this can be achieved by storing growth plates under chilled conditions (~4 °C). Samples may also be stored for longer periods, (month-month) in controlled light and temperature chambers. Cryopreservation allows for samples to be stored at -80 °C for up to six months (Trevathan-Tackett et al., 2018b). However, regular thawing, culturing and re-cryopreservation may keep viable isolates vital for longer timescales. For important isolates, duplicating efforts to maintain genotypes, especially through collaborative storage agreements with other institutions and facilities has been important in long-term studies of Labyrinthula, as cultures can die off for no obvious reasons and historical knowledge and testing of the same isolates has a powerful effect on background needed for effective experimental design and interpretation of statistical results.
3.8 Laboratory experiments
3.8.1 Pathogenicity
Research on Labyrinthula disease and ecology have primarily focused on laboratory pathogenicity assays. Investigations into seagrass pathogenicity testing of Labyrinthula spp. was pioneered by Charles Renn (Renn, 1936, Watson and Ordal 1957, Muehlstein, 1992; Martin et al., 2009; Sullivan et al., 2013) using Labyrinthula to test Koch’s postulate. Direct observation of infection and pathogenicity are most common (Figure 6, See Supplemental Manual). Laboratory and field assays are both used to investigate a) pathogenicity and virulence of newly isolated cultures (Vergeer and den Hartog, 1994; Martin et al., 2016; Trevathan-Tackett et al., 2018a), b) how infection develops naturally on infected plants under various environmental conditions (Buchsbaum et al., 1990; Vergeer et al., 1995; Beets et al., 2014; Dawkins et al., 2018), and c) variability in infectiousness of healthy or asymptomatic plants by inoculating under different environmental conditions (McKone and Tanner, 2009; Trevathan et al., 2011). Pathogenicity in seagrass has also been researched indirectly through correlation of environmental parameters, and spatial distribution and coverage (Cottam and Munro, 1954; Vergeer and den Hartog, 1994; Lathrop et al., 2001).
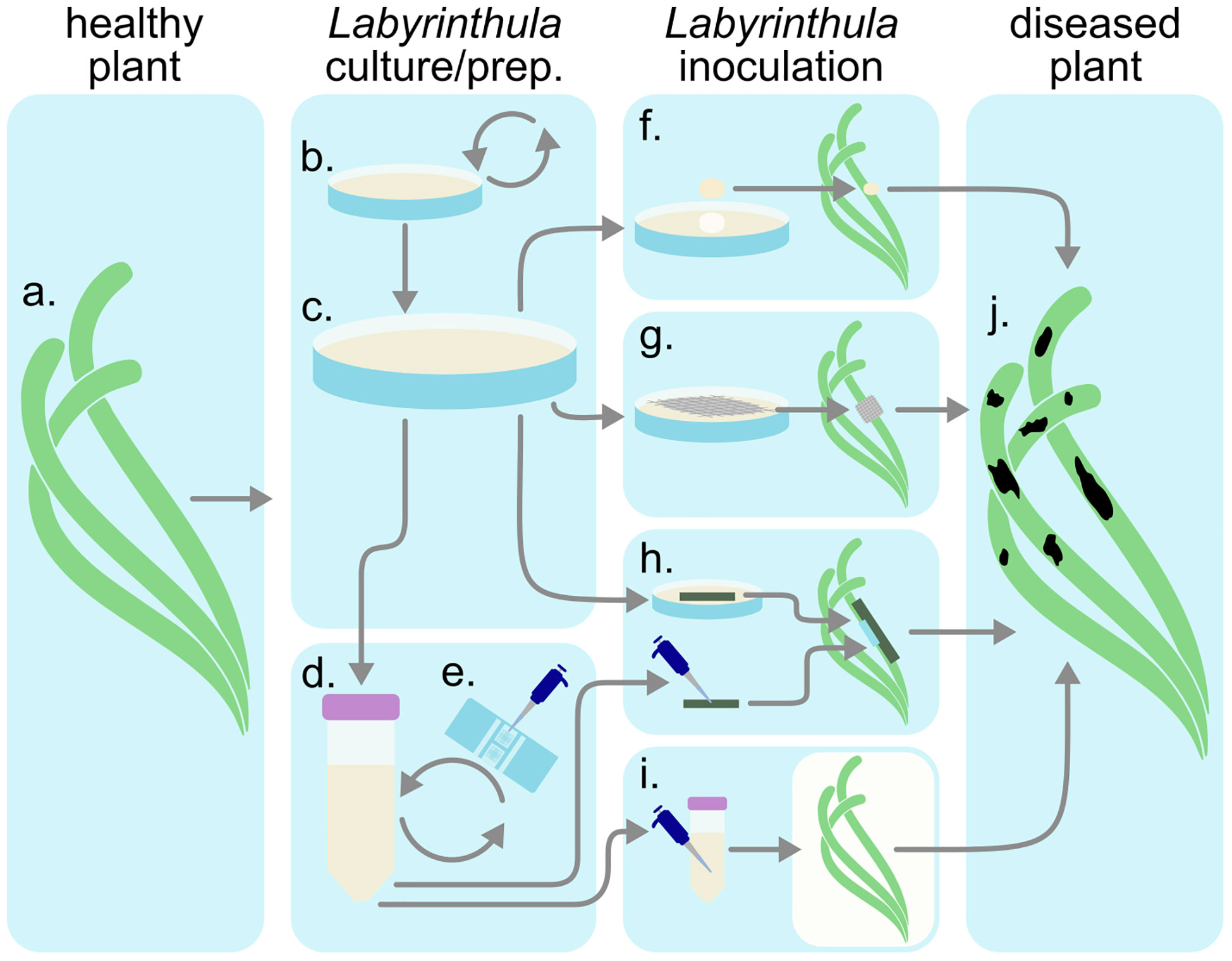
Figure 6 Inoculation procedure diagram. (A) Collect healthy plant tissues. (B) Utilize Labyrinthula isolate(s). Curate media (C) plates or (D) liquids. (E) Inocula concentration may be quantified via hemacytometer before introducing infected materials (F–I) to the healthy plant. After observing infection (J), sample diseased tissue and reisolate Labyrinthula to fulfill Koch’s postulates.
3.8.2 Experimental vectors
Different kinds of L. zosterae vectors, including inoculated leaf tissues, agar plugs (Jakobsson-Thor et al., 2019), gauze and water-baths (Groner et al., 2016) have been used to perform infection experiments on healthy seagrass shoots. It is important to standardize experimental vector infection protocol for pathogenicity assays. While several techniques have been used to infect plants with Labyrinthula (Martin et al., 2016), the majority of pathogenicity assay techniques use surface-to-surface transfer. A limitation with the method above is that it is difficult to control for L. zosterae quantities added during inoculation. It is important to be able to control pathogen loads when studying disease severity and other density-dependent effects. Only the in vitro water-borne method by Groner et al. (2016) makes it possible to evaluate and control the concentration of the Labyrinthula cells when performing infection experiments prior to treatment. In this method, a hemacytometer is used to determine the Labyrinthula cell dosage in either a concentrated inoculum applied directly to the plant or as an inoculum bath in which the ramet is immersed. Similar assays have been performed on fresh leaf sections thereby providing a relative measure of potential virulence or severity (Groner et al., 2014; Dawkins et al., 2018). This method can be used for transferring cells as mentioned above, but concentrations can also be sampled and assessed with this method at any time to understand pathogen loading and variability due to concentration.
3.8.3 Mesocosms
Infection experiments have so far mainly been used to investigate direct infection effects on a given seagrass host (Short et al., 1993; Steele et al., 2005; McKone and Tanner, 2009; Garcias-Bonet et al., 2011; Trevathan et al., 2011). Recent studies on the effect of physical and chemical environmental stressors, e.g. temperature, salinity, light, pH and interactions of these factors, have shed some light on how a changing environment will affect L. zosterae infection (Bishop et al., 2017; Dawkins et al., 2018; Brakel et al., 2019; Jakobsson-Thor et al., 2020).
4 Emerging molecular and -omic toolbox
To better understand ecological and evolutionary characteristics of an organism, knowledge about multi-omics is essential. Molecular approaches are rapidly advancing research into Labyrinthula genomic, transcriptomic, and metabolomic analyses. Advances in capabilities and reduced costs for molecular tools are leading to rapid advancement of discoveries related to speciation, classification, quantification, and phylogeny of Labyrinthula (Lohan et al., 2020). In addition, there is increasing recognition for the importance of Labyrinthula in host-pathogen relationships, trophic food webs, DHA production, and microbial symbioses in peer reviewed literature. Further advancement and refinement of key genetic markers and regions will continue to help researchers better understand Labyrinthula biology and allow for better assessments of the roles Labyrinthula play in marine and terrestrial ecology.
4.1 Extraction
High quality RNA and DNA sequences are important for successful genetic investigations. This can prove challenging because Labyrinthula are regularly found growing in close association with other microbes, such as other stramenopiles, bacteria, yeasts, and other contaminants (See Supplementary Manual). As such, Labyrinthula DNA needs to be lysed from axenic, liquid cultures (See Section 2.3.2 Growth Media) (Martin et al., 2009; Martin et al., 2016) or from over-growth agar cultures, taking care to scrape the surface to avoid unnecessary agar collection (Trevathan-Tackett personal observation). Labyrinthula DNA has been extracted with repeated success using standard or modified cetyl trimethylammonium bromide (CTAB) in sufficient amounts and required quality for analyses (Honda et al., 1999; Leander and Porter, 2001). DNA extraction kits have also been repeatedly successful for isolating Labyrinthula genetic profiles (Bockelmann et al., 2013; Lohan et al., 2020).
4.2 Genetic markers for phylogeny and population genetics
Phylogenetic analyses within the genus Labyrinthula have been performed based on several molecular markers, including 1) small subunit (SSU) ribosomal genes in 18S rDNA (Douhan et al., 2009; Bockelmann et al., 2012; Chitrampalam et al., 2015; Martin et al., 2016; Sullivan et al., 2017), 2) internal transcribed spacer (ITS) regions (Martin et al., 2016) and SL1, NS4/CITS5, actin, tubulin and Ef-alpha sequences (Tsui et al., 2009). As of November 2022, the search terminus “Labyrinthula” applied to NCBI GenBank nucleotide database retrieves 572 sequences of linear DNA/RNA with around 1700+ base pairs, 1 linear DNA/RNA submitted with nearly 4,758 base pairs and one complete genome containing 26,822 base pairs (https://www.ncbi.nlm.nih.gov/nuccore/?term=Labyrinthula). These entries clearly reflect the dominance of ITS and 18S markers in research. Though generally regarded as a viable marker for population level genetic analyses, the ITS regions for Labyrinthula zosterae are surprisingly similar between isolates procured from different oceans (Bockelmann et al., 2013; Martin et al., 2016). Still, most of the published studies on Labyrinthula specific diversity (Douhan et al., 2009; Bockelmann, 2012; Chitrampalam et al., 2015; Martin et al., 2016; Sullivan et al., 2017) have been tested on Labyrinthula cultured prior to sequencing. Thus, uncultivable taxa were undiscovered prior to sequencing efforts. While ITS and 18S are the most common regions used to evaluate Labyrinthula thus far (Lohan et al., 2016; Lohan et al., 2020), these regions are not necessarily the “best” markers to understand speciation. Areas of the genome used to assess specific diversity in the future will depend on the questions being asked (e.g., taxonomic level), and will likely include massive multi-gene analyses from whole-genome efforts.
4.3 Cell quantification
Quantitative PCR (qPCR) is commonly used to detect, identify and quantify presence of oomycetes in host tissue and other environmental samples (See Supplemental Manual). Unidentified Labyrinthula presented in metagenomic data sets are growing rapidly in terms of both quality and quantity (Karst et al., 2018; Lohan et al., 2020), demonstrating how much remains to be discovered with regards to the biological and ecological characteristics of this elusive group of microbes. There are important applications for qPCR in plant pathology and microbial ecology (Lievens et al., 2006), including Labyrinthula (Bockelmann et al., 2013; Chitrampalam et al., 2015; Martin et al., 2016; Duffin et al., 2020; Duffin et al., 2021). The great advantage of this technique is the fast and sensitive detection of Labyrinthula cells in host tissue without time intensive staining or culturing procedures. Further, detection of Labyrinthula can be achieved independent from the detection of host symptoms, which may be ambiguous. Although it would be logistically impossible to verify every strain and pathosystem, qPCR primers are likely to provide valuable first steps for various seagrass pathosystems, especially those harboring more than one pathogenic phylotype (e.g., turtlegrass; Martin et al., 2016). At least one set of primers has been designed to partly address this specific issue (Duffin et al., 2020; Duffin et al., 2021; Duffin et al 2022). Gene expression has also been explored with qPCR in an attempt to detect immunity and stress markers in halophytes (Brakel et al., 2014).
Still, qPCR for Labyrinthula studies have some challenges. First, results can depend on the DNA extraction method used (Klein, 2002), making cross-study comparisons difficult. Further, false negatives can occur when the DNA sequences vary at primer binding sites (Klein, 2002). Thus, good validation with isolated Labyrinthula strains is necessary before developing effective qPCR assays (Duffin et al., 2020; Duffin et al., 2021; Duffin et al., 2022). Standard cell solutions need to be developed with locally occurring Labyrinthula isolates, as variation in copy numbers of ITS genes have been detected in other organisms (Rogers and Bendich, 1987). However, such efforts may be balanced against other assumptions, for instance, relative loading may suffice for some study questions, but provide misleading perspective for others when considering wild populations that may include more and less virulent strains (Dawkins et al., 2018; Trevathan-Tackett et al., 2018a).
4.4 Genome sequencing
The first whole genome for Labyrinthula has been assembled and published, sourced from axenic culture from southeast Australia (Tan et al., 2021) and previously identified within the pathogenic clade and confirmed via pathogen testing (Martin et al., 2016; Trevathan-Tackett et al., 2018a). The first genome and transcriptome provide novel insight into Labyrinthula phylogeny and an opportunity to explore the genes available and used by Labyrinthula. In the future, emerging DNA-based techniques and assessments will further improve our evolutionary, biological and ecological understanding of Labyrinthula, especially with regards to reproduction, growth, speciation, immunity, and virulence.
4.5 Biochemical analyses and metabolomics
4.5.1 Use
Compounds produced by Labyrinthula spp. and their hosts may provide valuable information on their interactions. Measurement of particular compounds or classes thereof (e.g. phenolics) may inform targeted questions, while overall metabolic signatures (usually of a certain class of compounds) of a given sample may provide broader or more exploratory information. Metabolomics has been useful in the examination of phylogeny and halophyte defense compounds. For instance, Huang et al. (2003) used fatty acid (FA) signatures to cluster unknown isolates of thraustochytrids against a phylogeny constructed with the 18S rRNA gene. For such methods to be applicable for Labyrinthula phylogeny, a precise understanding of how FA are synthesized and how production may change through disease progression is required. Also, though the presence of defense compounds can be detected if there is a change in concentration between different treatments, such as between an infected and uninfected plant (Brakel et al., 2014), it is difficult to detect these defense compounds if the compounds are at the same levels and always active (constitutive defense).
4.5.2 Host response mechanisms
It has been hypothesized that seagrasses defend against pathogenic Labyrinthula species by the production of chemical defense compounds. Overall, secondary metabolites, including chemical defenses against Labyrinthula are poorly understood (Zidorn, 2016). Despite research, no direct links between defense compounds and resilience have been identified thus far.
4.5.3 Phenolic acids
Several studies have investigated the effect of phenolic acids on Labyrinthula, a well-known group of defense compounds in terrestrial plants. While some have found a positive correlation between Labyrinthula infection and phenolic acid production in seagrasses (Vergeer and Develi, 1997; McKone and Tanner, 2009), others suggest phenolics accumulation in diseased leaves is ‘pseudo-induced’ due to lesions disrupting the allocation and storage of leaf resources (Steele et al., 2005). These methods have made it possible to compare phenolic acids in seagrass shoots collected from the field, or from shoots used in different kinds of infection experiments (Groner et al., 2016; Groner et al., 2018). Still, the role of phenolics in moderating Labyrinthula is not certain.
4.5.4 Additional defense compounds
Additional compounds with inhibitory effects on Labyrinthula growth have been isolated from T. testudinum, but the structures have not been confirmed (Trevathan-Tackett et al., 2015). The isolation was done using bioassay-guided fractionation, a common method for isolation of bioactive compounds. This method allows detection and isolation of inhibitory compounds from complex samples without targeting a specific group of compounds. However, the extraction technique used by Trevathan-Tackett et al. (2015) did not target phenolic acids. It would be beneficial for future studies to apply a broader extraction technique that also includes phenolic acids. This would better resolve whether phenolic acids are actively inhibiting Labyrinthula growth in seagrasses or if other compounds are responsible for this interaction. Immune-related enzymes represent another group of compounds that may act on and in response to Labyrinthula, at least for turtlegrass, which is thought to impact virulence. Duffin et al. (2020) developed assays for four enzymes (peroxidase, exochitinase, polyphenol oxidase, and lysozyme activity), two of which correlate with Labyrinthula load. Further, all are expressed constitutively in turtlegrass, and appear useful in terms of elucidating Labyrinthula defense strategies such as tolerance (Duffin et al., 2020), or even defense ‘syndromes’ (Agrawal and Fishbein, 2006; Defossez et al., 2018), in wild seagrass populations. Duffin et al. (2021) subsequently combined all four markers when evaluating host immune activity relative to Labyrinthula load in the field.
4.5.5 Non-structural carbohydrates
Generally, the production of secondary metabolites is considered a cost to a plant, with multiple functions as a potential means to offset its toll (Erb and Kliebenstein, 2020). Still, the production of defensive compounds in plants afflicted with SWD, along with reduced host photosynthetic ability (Ralph and Short, 2002), may in turn reduce host energetic resources such as non-structural carbohydrates. As valuable measures of seagrass fitness broadly, assessments of non-structural carbohydrates (i.e. starch and/or sugar content) have also been applied to SWD systems. Brakel et al. (2019) found limited effects of Labyrinthula inoculation on fitness measures, including rhizome starch, leaf starch, and leaf sucrose concentrations, in eelgrass relative to other stressors (temperature, light, and salinity) in a factorial, multi-stressor experiment. In contrast, Graham et al. (2021) found that rhizome sugar content decreased with the severity of SWD in field-collected eelgrass plants.
4.5.6 DHA production
Long-chain polyunsaturated FA (LCPUFA), especially the omega-3 FA docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA), are valuable dietary constituents for human and aquatic life (Parrish, 2009). As effective synthesizers of LCPUFA, thraustochytrids have received considerable research and industry attention (Marchan et al., 2018). Labyrinthulids are also known to produce LCPUFA, notably DHA, with much research focused on potential industrial applications of high-LCPUFA producing strains (Sakata et al., 2000; Kumon et al., 2002; Kumon et al., 2003; Kumon et al., 2005; Kumon et al., 2006; Wang et al., 2019). However, most FA studies on Labyrinthulids use single-taxon microbial (bacteria or yeasts) or derived (serum, yeast, and/or peptone-based) substrates for culture (Sakata et al., 2000; Kumon et al., 2002; Kumon et al., 2003; Kumon et al., 2005; Kumon et al., 2006; Wang et al., 2019; Yoshioka et al., 2019). Consequently, little is known about Labyrinthula LCPUFA production in situ.
4.5.7 DHA and pathogenic Labyrinthula
Pathogenic Labyrinthula can produce substantial amounts of DHA (Yoshioka et al., 2019). Grown on a modified serum seawater agar and an eelgrass-based agar (see Growth Media), Labyrinthula isolates from diseased Z. marina produced DHA as their dominant FA. In both laboratory-inoculated and field-collected eelgrass tissues, DHA content was greater in diseased tissue than healthy tissue, suggesting that pathogenic Labyrinthula produces DHA in situ (Yoshioka et al., 2019). For Yoshioka et al. (2019), plant DHA content associated with disease was detectable but modest. Fatty acid changes in diseased plants are likely subject to other variables that may affect the plant-pathogen relationships. Lastly, surface metabolites, including FA of Z. marina, can inhibit or modulate microbes that in turn may affect disease (Papazian et al., 2019), suggesting more work is needed to understand production and the role of microbes in modulating FA levels in plants.
5 Review, synthesis and future directions
Molecular work describing microbe-microbe and microbe-plant interactions in Labyrinthula has led to ongoing technological advances, shedding light on important topics related to Labyrinthula, such as speciation (Martin et al., 2016; Popova et al., 2020), gene expression (Brakel et al., 2014; Jakobsson-Thor et al., 2020), global pathogenicity (Sullivan et al., 2017; Trevathan-Tackett et al., 2018a) and host immunity (Duffin et al., 2020; Duffin et al., 2021). Thus, research attentive to Rapid Blight, Seagrass Wasting Disease, and broader ecological implications of Labyrinthula in critical ecosystem functions, such as disease, decomposition, host immunity and primary production and transfer of fatty acids are ongoing.
A significant gap remaining in biological research of Labyrinthula, requiring both cellular and molecular advancement, are life-cycle studies. Heterokont flagella are a defining feature of the stramenopiles, yet there is a distinct lack of information about zoospore production and identification in Labyrinthula, and it is entirely lacking in relation to those taxa thought to be pathogenic. Sporulation has been reported in Labyrinthula, but recovery and positive identification of motile zoospores has not been made or imaged with SEM in a laboratory since initial reports (Amon and Perkins, 1968). Subsequent attempts to observe and replicate sporulation and visualize the results have been unsuccessful. Understanding life phases of Labyrinthula is critical for advancing our understanding of complex marine systems. Further, testing the possible roles zoospores play in progressive phases of disease, or seagrass and marine ecology more broadly (including saprobic decay, remineralization and trophic transfer), may also include fixed factors such as seasonality and phases of growth. Thus, confirmation of a flagellated dispersive stage and a consistent laboratory method for obtaining zoospores is needed.
Additionally, more work is needed to understand the connections between Labyrinthula and DHA production, including potential roles for pathogenic strains in trophic transfer. If fatty acid (FA) production is increased in pathogenic Labyrinthula, questions remain about its biological and ecological relevance. For instance, Jain et al. (2005) suggest that lipids, including DHA, are important as energy reserves and for building the ectoplasmic network in thraustochytrids. The ectoplasmic network enzymatically breaks down plant tissues through Labyrinthula infection (Muehlstein, 1992), so FA production may be related to pathogenesis in seagrass, or other organisms. If FA production is indeed pathology-related, then Labyrinthula may have additional ecological consequences or advantages for nutritional resources it provides to its consumers. Saprotrophic Labyrinthulomycetes and similar microbes are known to be an important link between detrital resources and higher consumers (Raghukumar, 2002). Whether pathogenic Labyrinthula provide substantial FA in situ and how production relates to infection should be verified. Future work with FA analyses should aim to understand how tissue- and plant-level changes to FA correspond with disease, and whether such changes may have individual, community and ecosystem-level effects. Clarity at both scales of Labyrinthula FA production will be informative for understanding infection and developing new tools (possibly in concert with genomic analyses) while providing a more holistic view of Labyrinthula ecology.
Coupled research in seagrass immunity, pathogen virulence, transmission (Dawkins et al., 2018; Duffin et al., 2020), stressor identification and environmental threshold assessments (Brakel et al., 2019) are of great importance to seagrass conservation and restoration efforts. Host susceptibility and immunity remain important areas for ongoing research. As more halophyte pathosystems are investigated, it will be interesting to assess variability in tolerance and defense mechanisms between different families of seagrass and other hosts to determine how interactive pathosystems work in favor or against Labyrinthula and other disease organisms. For instance, how do environmental conditions mediate gene-expression? What affect may Labyrinthula have on seagrass co-infections with other pathogenic organisms? Studies such as these may reveal important or different immune responses in seagrasses as well. Since seagrasses are not monophyletic, discoveries of variability in host immune defenses and vulnerabilities between individuals and populations will provide useful insight into observed variabilities in transmission, symptoms and virulence for this organism (Trevathan-Tackett et al., 2018a) and those impacts on community ecology. Further, exploration into how disease events impact ecosystem biodiversity and host coevolution long-term might be shaped by Labyrinthula associations (Duffin et al., 2020) and also warrants attention.
It is notable in this age of heightened awareness and risks of global pandemics that little research has looked at the role and relationships of Labyrinthula colonies to surrounding zoological communities, including its role in the microbiome, such as roles for moderating or exacerbating agents of human diseases in marine environments (Lamb et al., 2017), or the creation of ‘biofilms’ that serve as antimicrobial barriers or primary colonizers of marine environments. Thus, research is urgently needed to advance of our understanding about the role that Labyrinthula and other marine pathogens play in basic ecology of coastal ecosystems, especially when assessing issues of human health and threats of extinction to rarer halophytes, especially in monocultures. Though research has demonstrated that seagrass meadows reduce the presence of harmful microbes (Lamb et al., 2017), the mechanisms for this reduction, including the role of other seagrass-associated microbes in that capture, are unknown.
The potential for Labyrinthula to cause widespread loss of seagrasses worldwide has been established, yet even in critically valuable meadows, disease monitoring and inclusion in management objectives are often scant or missing. The presence of Labyrinthula has been confirmed in all non-polar major coastal systems, including mangroves, marshes, algae and seagrasses and with various pathogenicity (Martin et al., 2016). What we are still unclear about is whether severe infections with pathogenic Labyrinthula are a result of emerging pathogenic species described with clear genetic and morphological characteristics in the literature, or if environmental anomalies are driving virulence or mutation of these (more typically) saprobic organisms. Such a perspective may be slightly clearer for the Rapid Blight caused by Labyrinthula, as it is considered to be an emergent disease and an advancing threat to higher latitude cool-season turfgrasses in both the U.S. and Europe, and probably globally (Entwistle et al., 2014). A remaining knowledge gap is identifying the host(s) these terrestrial types may be derived from. The phylogenetic work presented in Martin et al. (2016), hints at saltmarsh plants as a possible starting point for such an investigation. Finally, advances are needed to understand deeper phylogenetic relationships and evolutionary history relative to marine Labyrinthula, especially in regards to the freshwater origins of the only fully marine angiosperms – seagrasses.
6 Summary
Researchers have repeatedly found Labyrinthulomycetes are biologically and ecologically important microorganisms (Gleason et al., 2013; Sullivan et al., 2013; Gleason et al., 2014; Scholz et al., 2016). Improved techniques for sampling, isolation, culture, measurement, visualizations, and long-term storage of Labyrinthula and other co-occurring pathogens are necessary to advance our capacity for answering lingering questions about Labyrinthula’s lifecycle, trophic roles, pathogenicity and virulence, microbiome, and other ecological relationships. Advances in available methods and tools for cellular and molecular investigations of Labyrinthula have progressed our understanding of this microbe’s importance and the need for continued research. We reviewed complex ecological roles for Labyrinthula in saprobic decomposition, creation of biofilms, primary trophic pathways for DHA in marine systems and incidence of pandemic diseases in halophytes, and presence of emerging diseases afflicting saline, brackish and terrestrial systems. Labyrinthula mediate structural ecosystems and are also mediated by conditions in the ecosystems themselves. For these reasons Labyrinthula has become a model marine microbe for investigating questions at a variety of functional scales with regards to halophytic environments, as it is not wholly host-specific and plays several key roles in the environment, including remineralization, trophic transfer and infectious disease.
A lack of capacity related to genome sequencing has previously stunted studies needed to advance research into Labyrinthula phylogeny, cellular function, virulence evolution, epidemiology, host-immunity and gaps in our understanding of Labyrinthula life cycle and gene expression. Now that the genome of Labyrinthula has been mapped (Tan et al., 2021), we expect even more break-throughs are possible. Advancements in species level discussions about the genetic basis and specific relevance of observed plasticity in phenotypic and trophic characteristics, especially related to environmental or biological stressors/factors are forthcoming as work on gene expression continues. In addition, continued advancements in ecological research of Labyrinthula will require effective mobilization and automation of these biological investigation protocols and techniques. Basic biological investigations are still required to describe and study Labyrinthula speciation, life cycles and pathosystems before critical ecological questions, such as relative importance in coastal carbon cycling, trophic pathways, and critical host-pathogen thresholds can be answered.
Through this review we have increased international capacity, collaboration and knowledge for the roles for Labyrinthula in complex global systems. It is critically important for researchers to collaborate at the international level and to share methodological resources needed to answer globally relevant questions about Labyrinthula biology and ecology as we balance an awareness of risk for catastrophic losses alongside obvious benefits from potential trophic contributions of DHA, enzyme production and important nutrient and mineral cycling.
Author contributions
BS conceptualized, developed the outline and managed authors for this paper; BS, ST-T, RY, JB, SJ-T, ME and DM contributed to writing and editing the manuscript; RY, BS and DM created figures; BS, RY, DM and STT contributed funds for publication and ST-T wrote the Statement of Contributions. All authors contributed to the article and approved the submitted version.
Funding
This work was not supported by specific funding. S.T-T. was supported by the Australian Research Council DECRA Fellowship (DE210101029). R.Y. was supported by the National Science Foundation Graduate Research Fellowship Program (Grant No. 1309047).
Acknowledgments
The authors wish to dedicate this manuscript to the memory of our compassionate and revolutionary scholar and mentor Dr. Frank Gleason. We also wish to thank Professor Carlos Duarte and Professor Emeritus Fred T. Short who were present when five international PhD students from all over the world originally came together to develop this review and continued to support and guide us as we prepared this collaborative and extensive manuscript for publication. ST-T. was supported by the Australian Research Council DECRA Fellowship (DE210101029). RY was supported by the National Science Foundation Graduate Research Fellowship Program (Grant No. 1309047).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1092587/full#supplementary-material
References
Adl S. M., Simpson A. G., Lane C. E., Lukeš J., Bass D., Bowser S. S., et al. (2012). The revised classification of eukaryotes. J. Eukaryot. Microbiol. 59, 429–514. doi: 10.1111/j.1550-7408.2012.00644.x
Agrawal A. A., Fishbein M. (2006). Plant defense syndromes. Ecology 87, S132–S149. doi: 10.1890/0012-9658(2006)87[132:PDS]2.0.CO;2
Amaral-Zettler L. A., McCliment E. A., Ducklow H. W., Huse S. M. (2009). A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PloS One 4, e6372. doi: 10.1371/annotation/50c43133-0df5-4b8b-8975-8cc37d4f2f26
Amon J. P. (1978). Thraustochytrids and labyrinthulids of terrestrial, aquatic and hypersaline environments of the great salt lake, USA. Mycologia 70, 1299–1301. doi: 10.1080/00275514.1978.12020358
Amon J. P., Perkins F. O. (1968). Structure of Labyrinthula sp. zoospores. J. protozool. 15, 543–546. doi: 10.1111/j.1550-7408.1968.tb02172.x
Anderson O. R., Cavalier-Smith T. (2012). Ultrastructure of Diplophrys parva, a new small freshwater species, and a revised analysis of Labyrinthulea (Heterokonta). Acta Protozool. 51, 291–304. doi: 10.4467/16890027AP.12.023.0783
Aoki L. R., Rappazzo B., Beatty D. S., Domke L. K., Eckert G. L., Eisenlord M. E., et al. (2022). Disease surveillance by artificial intelligence links eelgrass wasting disease to ocean warming across latitudes. Limnol. Oceanogr., 1577–1589 doi: 10.1002/lno.12152
Armenta R. E., Valentine M. C. (2013). Single-cell oils as a source of omega-3 fatty acids: an overview of recent advances. J. Am. Oil Chem. Soc. 90, 167–182. doi: 10.1007/s11746-012-2154-3
Armstrong E., Rogerson A., Leftley J. W. (2000). Utilisation of seaweed carbon by three surface-associated heterotrophic protists, Stereomyxa ramosa, Nitzschia alba and Labyrinthula sp. Aquat. Microbial. Ecol. 21, 49–57. doi: 10.3354/ame021049
Beakes G. W., Honda D., Thines M. (2014). “3 systematics of the Straminipila: Labyrinthulomycota, Hyphochytriomycota, and Oomycota,” in Systematics and evolution (Springer: Berlin, Heidelberg), 39–97.
Beatty D. S., Aoki L. R., Rappazzo B., Bergman C., Domke L. K., Duffy J. E., et al. (2022). Predictable changes in eelgrass microbiomes with increasing wasting disease prevalence across 23° latitude in the northeastern pacific. Msystems 7, e00224–e00222. doi: 10.1128/msystems.00224-22
Beets J., Johnson E., Dixon J., Wyllie-Echeverria S. (2014). Effects of increased nitrate concentrations on Zostera marina health and prevalence of Labyrinthula zosterae Friday Harbor Laboratories, 20.
Bergmann N., Fricke B., Schmidt M. C., Tams V., Beining K., Schwitte H., et al. (2011). A quantitative real-time polymerase chain reaction assay for the seagrass pathogen Labyrinthula zosterae. Mol. Ecol. Resour. 11, 1076–1081. doi: 10.1111/j.1755-0998.2011.03051.x
Bigelow D., Olsen M., Gilbertson R. (2005). Labyrinthula terrestris sp. nov., a new pathogen of turf grass. Mycologia 97, 185–190. doi: 10.1080/15572536.2006.11832852
Bishop N., Martin D. L., Ross C. (2017). Effects of multi-stress exposure on the infection dynamics of a Labyrinthula sp.-turtle grass pathosystem. Mar. Ecol. Prog. Ser. 581, 119–133. doi: 10.3354/meps12318
Bockelmann A.-C. (2012). Widespread occurrence of endophytic Labyrinthula spp. in northern European eelgrass Zostera marina beds. Eds. Beining K., Reusch T. B. H.
Bockelmann A. C., Beining K., Reusch T. B. (2012). Widespread occurrence of endophytic Labyrinthula spp. in northern European eelgrass Zostera marina beds. Mar. Ecol. Prog. Ser. 445, 109–116. doi: 10.3354/meps09398
Bockelmann A.-C., Tams V., Ploog J., Schubert P. R., Reusch T. B. (2013). Quantitative PCR reveals strong spatial and temporal variation of the wasting disease pathogen, Labyrinthula zosterae in northern European eelgrass (Zostera marina) beds. PloS One 8, e62169. doi: 10.1371/journal.pone.0062169
Brakel J., Jakobsson-Thor S., Bockelmann A.-C., Reusch T. B. (2019). Modulation of the eelgrass–Labyrinthula zosterae interaction under predicted ocean warming, salinity change and light limitation. Front. Mar. Sci. 6, 268. doi: 10.3389/fmars.2019.00268
Brakel J., Werner F. J., Tams V., Reusch T. B., Bockelmann A.-C. (2014). Current European Labyrinthula zosterae are not virulent and modulate seagrass (Zostera marina) defense gene expression. PloS One 9, e92448. doi: 10.1371/journal.pone.0092448
Buchsbaum R. N., Short F. T., Cheney D. P. (1990). Phenolic-nitrogen interactions in eelgrass, Zostera marina l.: possible implications for disease resistance. Aquat. Bot. 37, 291–297. doi: 10.1016/0304-3770(90)90075-V
Burdick D. M., Short F. T., Wolf J. (1993). An index to assess and monitor the progression of wasting disease in eelgrass Zostera marina. Mar. Ecology-Progress Ser. 94, 83–83. doi: 10.3354/meps094083
Camberato J. J., Peterson P. D., Bruce Martin S. (2006). Salinity and salinity tolerance alter rapid blight in Kentucky bluegrass, perennial ryegrass, and slender creeping red fescue. Appl. Turfgrass Sci. 3, 1–14. doi: 10.1094/ATS-2006-0213-01-RS
Cavalier-Smith T., Chao E. E. (2006). Phylogeny and megasystematics of phagotrophic heterokonts (kingdom Chromista). J. Mol. Evol. 62, 388–420. doi: 10.1007/s00239-004-0353-8
Chitrampalam P., Goldberg N., Olsen M. W. (2015). Labyrinthula species associated with turfgrasses in Arizona and new Mexico. Eur. J. Plant Pathol. 143, 485–493. doi: 10.1007/s10658-015-0701-0
Cienkowski L. v. (1867). Ueber den bau und die entwickelung der labyrinthuleen. archiv für mikroskopische anatomie Archiv für mikroskopische Anatomie 3, 274–310. doi: 10.1007/BF02960460
Collado-Mercado E., Radway J. C., Collier J. L. (2010). Novel uncultivated Labyrinthulomycetes revealed by 18S rDNA sequences from seawater and sediment samples. Aquat. Microbial. Ecol. 58, 215–228. doi: 10.3354/ame01361
Cottam C., Munro D. A. (1954). Eelgrass status and environmental relations. J. Wildl. Manage. 18, 449–460. doi: 10.2307/3797080
Dawkins P., Eisenlord M., Yoshioka R., Fiorenza E., Fruchter S., Giammona F., et al. (2018). Environment, dosage, and pathogen isolate moderate virulence in eelgrass wasting disease. Dis. Aquat. Org. 130, 51–63. doi: 10.3354/dao03263
Defossez E., Pellissier L., Rasmann S. (2018). The unfolding of plant growth form-defence syndromes along elevation gradients. Ecol. Lett. 21, 609–618. doi: 10.1111/ele.12926
De Kievit T. R., Iglewski B. H. (2000). Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68, 4839–4849. doi: 10.1128/IAI.68.9.4839-4849.2000
Dietz C., Schnetter R. (1999). Interaction of two myosins with microfilaments causes locomotion in Labyrinthula sp. Protoplasma 206, 97–104. doi: 10.1007/BF01279256
Douhan G. W., Olsen M. W., Herrell A., Winder C., Wong F., Entwistle K. (2009). Genetic diversity of Labyrinthula terrestris, a newly emergent plant pathogen, and the discovery of new labyrinthulid organisms. Mycol. Res. 113, 1192–1199. doi: 10.1016/j.mycres.2009.08.002
Duffin P., Martin D. L., Furman B. T., Ross C. (2021). Spatial patterns of Thalassia testudinum immune status and Labyrinthula spp. load implicate environmental quality and history as modulators of defense strategies and wasting disease in Florida bay, united states. Front. Plant Sci. 12, 100. doi: 10.3389/fpls.2021.612947
Duffin P., Martin D. L., Furman B. T., Ross C. (2022). Corrigendum: Spatial patterns of Thalassia testudinum immune status and Labyrinthula spp. load implicate environmental quality and history as modulators of defense strategies and wasting disease in Florida bay, united states. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.877673
Duffin P., Martin D. L., Pagenkopp Lohan K. M., Ross C. (2020). Integrating host immune status, Labyrinthula spp. load and environmental stress in a seagrass pathosystem: Assessing immune markers and scope of a new qPCR primer set. PloS One 15, e0230108. doi: 10.1371/journal.pone.0230108
Dyková I., Fiala I., Dvořáková H., Pecková H. (2008). Living together: The marine amoeba Thecamoeba hilla Schaeffer 1926 and its endosymbiont Labyrinthula sp. Eur. J. Protistol. 44, 308–316. doi: 10.1016/j.ejop.2008.04.001
Entwistle K., Fleming T., Kerr R., Maule A., Martin T., Hainon-McDowell M., et al. (2014). Biosecurity and emerging plant health problems in turf production and maintenance. Eur. J. Hortic. Sci. 79, S108–S115.
Erb M., Kliebenstein D. J. (2020). Plant secondary metabolites as defenses, regulators, and primary metabolites: the blurred functional trichotomy. Plant Physiol. 184, 39–52. doi: 10.1104/pp.20.00433
Galil B. S., Hülsmann N. (1997). Protist transport via ballast water–biological classification of ballast tanks by food web interactions. Eur. J. Protistol. 33, 244–253. doi: 10.1016/S0932-4739(97)80002-8
Garcias-Bonet N., Sherman T. D., Duarte C. M., Marbà N. (2011). Distribution and pathogenicity of the protist Labyrinthula sp. in western Mediterranean seagrass meadows. Estuaries Coasts 34, 1161–1168. doi: 10.1007/s12237-011-9416-4
Gleason F. H., Chambouvet A., Sullivan B. K., Lilje O., Rowley J. J. L. (2014). Multiple zoosporic parasites pose a significant threat to amphibian populations. Fungal Ecol. 11, 181–192. doi: 10.1016/j.funeco.2014.04.001
Gleason F. H., Van Ogtrop F., Lilje O., Larkum A. W. (2013). Ecological roles of zoosporic parasites in blue carbon ecosystems. Fungal Ecol. 6, 319–327. doi: 10.1016/j.funeco.2013.06.002
Graham O. J., Aoki L. R., Stephens T., Stokes J., Dayal S., Rappazzo B., et al. (2021). Effects of seagrass wasting disease on eelgrass growth and belowground sugar in natural meadows. Front. Mar. Sci. 8, 768668. doi: 10.3389/fmars.2021.768668
Groner M. L., Burge C. A., Couch C. S., Kim C. J., Siegmund G.-F., Singhal S., et al. (2014). Host demography influences the prevalence and severity of eelgrass wasting disease. Dis. Aquat. Org. 108, 165–175. doi: 10.3354/dao02709
Groner M. L., Burge C. A., Cox R., Rivlin N. D., Turner M., Van Alstyne K. L., et al. (2018). Oysters and eelgrass: potential partners in a high pCO 2 ocean. Ecology 99, 1802–1814. doi: 10.1002/ecy.2393
Groner M. L., Burge C. A., Kim C. J., Rees E., Van Alstyne K. L., Yang S., et al. (2016). Plant characteristics associated with widespread variation in eelgrass wasting disease. Dis. Aquat. Org. 118, 159–168. doi: 10.3354/dao02962
Harrington B. J., Hageage G. J. Jr. (2003). Calcofluor white: a review of its uses and applications in clinical mycology and parasitology. Lab. Med. 34, 361–367. doi: 10.1309/EPH2TDT8335GH0R3
Harvell C., Kim K., Burkholder J., Colwell R., Epstein P. R., Grimes D., et al. (1999). Emerging marine diseases–climate links and anthropogenic factors. Science 285, 1505–1510. doi: 10.1126/science.285.5433.1505
Hassani M. A., Durán P., Hacquard S. (2018). Microbial interactions within the plant holobiont. Microbiome 6, 1–17. doi: 10.1186/s40168-018-0445-0
Honda D., Yokochi T., Nakahara T., Raghukumar S., Nakagiri A., Schaumann K., et al. (1999). Molecular phylogeny of labyrinthulids and thraustochytrids based on the sequencing of 18S ribosomal RNA gene. J. eukaryot. Microbiol. 46, 637–647. doi: 10.1111/j.1550-7408.1999.tb05141.x
Huang J., Aki T., Yokochi T., Nakahara T., Honda D., Kawamoto S., et al. (2003). Grouping newly isolated docosahexaenoic acid-producing thraustochytrids based on their polyunsaturated fatty acid profiles and comparative analysis of 18S rRNA genes. Mar. Biotechnol. 5, 450–457. doi: 10.1007/s10126-002-0110-1
Hurtado-McCormick V., Kahlke T., Krix D., Larkum A., Ralph P., Seymour J. (2020). Seagrass leaf reddening alters the microbiome of Zostera muelleri. Mar. Ecol. Prog. Ser. 646, 29–44. doi: 10.3354/meps13409
Jain R., Raghukumar S., Tharanathan R., Bhosle N. B. (2005). Extracellular polysaccharide production by thraustochytrid protists. Mar. Biotechnol. 7, 184–192. doi: 10.1007/s10126-004-4025-x
Jakobsson-Thor S., Brakel J., Toth G. B., Pavia H. (2020). Complex interactions of temperature, light and tissue damage on seagrass wasting disease in Zostera marina. Front. Mar. Sci. 7, 865. doi: 10.3389/fmars.2020.575183
Jakobsson-Thor S., Toth G. B., Brakel J., Bockelmann A.-C., Pavia H. (2018). Seagrass wasting disease varies with salinity and depth in natural Zostera marina populations. Mar. Ecol. Prog. Ser. 587, 105–115. doi: 10.3354/meps12406
Jakobsson-Thor S., Toth G. B., Pavia H. (2019). Seagrass wasting disease along a naturally occurring salinity gradient. Mar. Ecol. Prog. Ser. 614, 67–77. doi: 10.3354/meps12911
Jepps M. W. (1931). Note on a marine Labyrinthula. J. Mar. Biol. Assoc. United Kingdom (New Series) 17, 833–838. doi: 10.1017/S0025315400052000
Karst S. M., Dueholm M. S., McIlroy S. J., Kirkegaard R. H., Nielsen P. H., Albertsen M. (2018). Retrieval of a million high-quality, full-length microbial 16S and 18S rRNA gene sequences without primer bias. Nat. Biotechnol. 36, 190. doi: 10.1038/nbt.4045
Kerrigan J. L., Olsen M. W., Martin S. B. (2012). Rapid blight of turfgrass. The Plant Health Instructor. doi: 10.1094/PHI-I-2012-0621-01
Klein D. (2002). Quantification using real-time PCR technology: applications and limitations. Trends Mol. Med. 8, 257–260. doi: 10.1016/S1471-4914(02)02355-9
Kopec D., Olsen M. W., Gilbert J. J., Bigelow D. M., Kohout M. J. (2004). Cool-season grass response to rapid blight disease. Golf Course Manage. 72, 78–81.
Kumon Y., Yokochi T., Nakahara T. (2005). High yield of long-chain polyunsaturated fatty acids by labyrinthulids on soybean lecithin-dispersed agar medium. Appl. Microbiol. Biotechnol. 69, 253–258. doi: 10.1007/s00253-005-1978-2
Kumon Y., Yokochi T., Nakahara T., Yamaoka M., Mito K. (2002). Production of long-chain polyunsaturated fatty acids by monoxenic growth of labyrinthulids on oil-dispersed agar medium. Appl. Microbiol. Biotechnol. 60, 275–280. doi: 10.1007/s00253-002-1086-5
Kumon Y., Yokoyama R., Haque Z., Yokochi T., Honda D., Nakahara T. (2006). A new labyrinthulid isolate that produces only docosahexaenoic acid. Mar. Biotechnol. 8, 170–177. doi: 10.1007/s10126-005-5098-x
Kumon Y., Yokoyama R., Yokochi T., Honda D., Nakahara T. (2003). A new labyrinthulid isolate, which solely produces n-6 docosapentaenoic acid. Appl. Microbiol. Biotechnol. 63, 22–28. doi: 10.1007/s00253-003-1333-4
Lafferty K. D. (2009). The ecology of climate change and infectious diseases. Ecology 90, 888–900. doi: 10.1890/08-0079.1
Lafferty K. D., Porter J. W., Ford S. E. (2004). Are diseases increasing in the ocean? Annu. Rev. Ecol. Evol. Syst, 31–54. Available at: https://www.jstor.org/stable/30034109.
Lamb J. B., van de Water J. A., Bourne D. G., Altier C., Hein M. Y., Fiorenza E. A., et al. (2017). Seagrass ecosystems reduce exposure to bacterial pathogens of humans, fishes, and invertebrates. Science 355, 731–733. doi: 10.1126/science.aal1956
Lathrop R. G., Styles R. M., Seitzinger S. P., Bognar J. A. (2001). Use of GIS mapping and modeling approaches to examine the spatial distribution of seagrasses in barnegat bay, new Jersey. Estuaries Coasts 24, 904–916. doi: 10.2307/1353181
Leander C. A., Porter D. (2001). The Labyrinthulomycota is comprised of three distinct lineages. Mycologia 93, 459–464. doi: 10.1080/00275514.2001.12063179
Leander C. A., Porter D., Leander B. S. (2004). Comparative morphology and molecular phylogeny of aplanochytrids (Labyrinthulomycota). Eur. J. Protistol. 40, 317–328. doi: 10.1016/j.ejop.2004.07.003
Leano E. M., Damare V. (2012). Labyrinthulomycota. In: (Jones E. B. G., Pang K. L., eds.) Marine fungi and fungal-like organisms. Walter de Gruyter, Berlin. pp. 215–244. doi: 10.1515/9783110264067.215
Lievens B., Brouwer M., Vanachter A. C. R. C., Cammue B. P. A., Thomma B. P. H. J. (2006). Real-time PCR for detection and quantification of fungal and oomycete tomato pathogens in plant and soil samples. Plant Sci. 171, 155–165. doi: 10.1016/j.plantsci.2006.03.009
Lohan K. M. P., DiMaria R., Martin D. L., Ross C., Ruiz G. M. (2020). Diversity and microhabitat associations of Labyrinthula spp. in the Indian river lagoon system. Dis. Aquat. Org. 137, 145–157. doi: 10.3354/dao03431
Lohan K. P., Fleischer R., Carney K., Holzer K., Ruiz G. (2016). Amplicon-based pyrosequencing reveals high diversity of protistan parasites in ships’ ballast water: implications for biogeography and infectious diseases. Microbial. Ecol. 71, 530–542. doi: 10.1007/s00248-015-0684-6
Madden L., Hughes G. (1999). Sampling for plant disease incidence. Phytopathology 89, 1088–1103. doi: 10.1094/PHYTO.1999.89.11.1088
Marchan L. F., Chang K. J. L., Nichols P. D., Mitchell W. J., Polglase J. L., Gutierrez T. (2018). Taxonomy, ecology and biotechnological applications of thraustochytrids: A review. Biotechnol. Adv. 36, 26–46. doi: 10.1016/j.biotechadv.2017.09.003
Martin D. L., Boone E., Caldwell M. M., Major K. M., Boettcher A. A. (2009). Liquid culture and growth quantification of the seagrass pathogen, Labyrinthula spp. Mycologia 101, 632–635. doi: 10.3852/08-171
Martin B. C., Bougoure J., Ryan M. H., Bennett W. W., Colmer T. D., Joyce N. K., et al. (2019). Oxygen loss from seagrass roots coincides with colonisation of sulphide-oxidising cable bacteria and reduces sulphide stress. ISME J. 13, 707–719. doi: 10.1038/s41396-018-0308-5
Martin D. L., Chiari Y., Boone E., Sherman T. D., Ross C., Wyllie-Echeverria S., et al. (2016). Functional, phylogenetic and host-geographic signatures of Labyrinthula spp. provide for putative species delimitation and a global-scale view of seagrass wasting disease. Estuaries Coasts 39, 1403–1421. doi: 10.1007/s12237-016-0087-z
McKone K. L., Tanner C. E. (2009). Role of salinity in the susceptibility of eelgrass Zostera marina to the wasting disease pathogen Labyrinthula zosterae. Mar. Ecol. Prog. Ser. 377, 123–130. doi: 10.3354/meps07860
Minteer B. A., Collins J. P. (2008). From environmental to ecological ethics: toward a practical ethics for ecologists and conservationists. Sci. Eng. ethics 14, 483–501. doi: 10.1007/s11948-008-9087-0
Muehlstein L. K. (1992). The host–pathogen interaction in the wasting disease of eelgrass, Zostera marina. Can. J. Bot. 70, 2081–2088. doi: 10.1139/b92-258
Muehlstein L., Porter D., Short F. (1988). Labyrinthula sp., a marine slime mold producing the symptoms of wasting disease in eelgrass, Zostera marina. Mar. Biol. 99, 465–472. doi: 10.1007/BF00392553
O’Kelly C. J. (2005). “The lobster back biofilm: possible role of the total microbial community in lobster shell disease,” in Lobster shell disease workshop (Boston, MA: University of Massachusetts), 22–24.
Olsen M., Bigelow D., Gilbertson R., Stowell L., Gelernter W. (2003). First report of a Labyrinthula sp. causing rapid blight disease of rough bluegrass and perennial ryegrass. Plant Dis. 87, 1267–1267. doi: 10.1094/PDIS.2003.87.10.1267B
Opsahl S., Benner R. (1993). Decomposition of senescent blades of the seagrass Halodule wrightii in a subtropical lagoon. Mar. Ecol. Prog. Ser. 94, 191. doi: 10.3354/meps094191
Pan J., Campo J., Keeling P. J. (2016). Reference tree and environmental sequence diversity of Labyrinthulomycetes. J. Eukaryot. Microbiol, 88–96. doi: 10.1111/jeu.12342
Papazian S., Parrot D., Burýšková B., Weinberger F., Tasdemir D. (2019). Surface chemical defence of the eelgrass Zostera marina against microbial foulers. Sci. Rep. 9, 1–12. doi: 10.1038/s41598-019-39212-3
Parrish C. C. (2009). Essential fatty acids in aquatic food webs. In: Kainz M., Brett M., Arts M. (eds) Lipids Aquat. Ecosyst. Springer, New York, NY. doi: 10.1007/978-0-387-89366-2_13
Perkins F. O. (1972). The ultrastructure of holdfasts,”rhizoids”, and “slime tracks”. thraustochytriaceous fungi Labyrinthula spp. Archiv für Mikrobiol. 84, 95–118. doi: 10.1007/BF00412431
Perkins F. O., Amon J. P. (1969). Zoosporulation in Labyrinthula sp.; an electron microscope study. J. protozool. 16, 235–257. doi: 10.1111/j.1550-7408.1969.tb02265.x
Pierotti R., Wildcat D. (2000). Traditional ecological knowledge: the third alternative (commentary). Ecol. Appl. 10, 1333–1340. doi: 10.1890/1051-0761(2000)010[1333:TEKTTA]2.0.CO;2
Pokorny K. S. (1967). Labyrinthula. J. protozool. 14, 697–708. doi: 10.1111/j.1550-7408.1967.tb02065.x
Popova O. V., Belevich T. A., Golyshev S. A., Kireev I. I., Aleoshin V. V. (2020). Labyrinthula diatomea n. sp.–a labyrinthulid associated with marine diatoms. J. Eukaryot. Microbiol. 67, 393–402. doi: 10.1111/jeu.12789
Porter D. (1986). Mycoses of marine organisms: An overview of pathogenic fungi (Cambridge, Great Britain: Cambridge University Press).
Porter D. (1990). Phylum Labyrinthulomycota Margulis L., Corliss J. O., Melkonian M., Chapman D. J. (Eds.), Handbook of Protoctista, Jones and Bartlett Publishers, Boston
Preston T. M., King C. A. (2005). Actin-based motility in the net slime mould Labyrinthula: Evidence for the role of myosin in gliding movement. J. Eukaryot. Microbiol. 52, 461–475. doi: 10.1111/j.1550-7408.2005.00064.x
Raghukumar C. (1986). Fungal parasites of the marine green algae, Cladophora and Rhizoclonium Cladophora and Rhizoclonium 29 (4), 289–298. doi: 10.1515/botm.1986.29.4.289
Raghukumar C. (1987). Fungal parasites of marine algae from Mandapam (South India). Dis. Aquat. Org. 3, 137–145. doi: 10.3354/dao003137
Raghukumar S. (2002). Ecology of the marine protists, the Labyrinthulomycetes (thraustochytrids and labyrinthulids). Eur. J. Protistol. 38, 127–145. doi: 10.1078/0932-4739-00832
Raghukumar S., Damare V. S. (2011). Increasing evidence for the important role of Labyrinthulomycetes in marine ecosystems. Botanica Marina 54, 3–11. doi: 10.1515/bot.2011.008
Ralph P. J., Short F. T. (2002). Impact of the wasting disease pathogen, Labyrinthula zosterae, on the photobiology of eelgrass Zostera marina. Mar. Ecol. Prog. Ser. 226, e271. doi: 10.3354/meps226265
Rappazzo B. H., Eisenlord M. E., Graham O. J., Aoki L. R., Dawkins P. D., Harvell D., et al. (2021). EeLISA: Combating Global Warming Through the Rapid Analysis of Eelgrass Wasting Disease. Proceedings of the AAAI Conference on Artificial Intelligence 35(17), 15156–15165. doi: 10.1609/aaai.v35i17.17779
Renn C. E. (1936). The wasting disease of Zostera marina. i. a phytological investigation of the diseased plant. Biol. Bull. 70, 148–158. doi: 10.2307/1537320
Rogers S. O., Bendich A. J. (1987). Heritability and variability in ribosomal RNA genes of Vicia faba. Genetics 117, 285–295. doi: 10.1093/genetics/117.2.285
Rosado P. M., Leite D. C., Duarte G. A., Chaloub R. M., Jospin G., da Rocha U. N., et al. (2019). Marine probiotics: increasing coral resistance to bleaching through microbiome manipulation. ISME J. 13, 921–936. doi: 10.1038/s41396-018-0323-6
Sakata T., Fujisawa T., Yoshikawa T. (2000). Colony formation and fatty acid composition of marine labyrinthulid isolates grown on agar media. Fish. Sci. 66, 84–90. doi: 10.1046/j.1444-2906.2000.00012.x
Schoch C. L., Seifert K. A., Huhndorf S., Robert V., Spouge J. L., Levesque C. A., et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci. 109, 6241–6246. doi: 10.1073/pnas.1117018109
Scholz B., Guillou L., Marano A. V., Neuhauser S., Sullivan B. K., Karsten U., et al. (2016). Zoosporic parasites infecting marine diatoms–a black box that needs to be opened. Fungal Ecol. 19, 59–76. doi: 10.1016/j.funeco.2015.09.002
Short F., Porter D., Iizumi H., Aioi K. (1993). Occurrence of the eelgrass pathogen Labyrinthula zosterae in Japan. Dis. Aquat. Org. 16, 73–77. doi: 10.3354/dao016073
Steele L. T., Caldwell M., Boettcher A., Arnold T. (2005). Seagrass-pathogen interactions:'pseudo-induction' of turtlegrass phenolics near wasting disease lesions. Mar. Ecol. Prog. Ser. 303, 123–131. doi: 10.3354/meps303123
Sullivan B. K., Robinson K. L., Trevathan-Tackett S. M., Lilje E. S., Gleason F. H., Lilje O. (2017). The first isolation and characterisation of the protist Labyrinthula sp. in southeastern Australia. J. Eukaryot. Microbiol, 504–513. doi: 10.1111/jeu.12387
Sullivan B. K., Sherman T. D., Damare V. S., Lilje O., Gleason F. H. (2013). Potential roles of Labyrinthul aspp. in global seagrass popul. declines. Fungal Ecol. 6, 328–338. doi: 10.1016/j.funeco.2013.06.004
Sullivan B. K., Trevathan-Tackett S. M., Neuhauser S., Govers L. L. (2018). Host-pathogen dynamics of seagrass diseases under future global change. Mar. pollut. Bull. 134, 75–88. doi: 10.1016/j.marpolbul.2017.09.030
Sykes E. E., Porter D. (1973). Nutritional studies of Labyrinthula sp. mycologia Mycologia. 65, 1302–1311. doi: 10.1080/00275514.1973.12019553
Tan M. H., Loke S., Croft L. J., Gleason F. H., Lange L., Pilgaard B., et al. (2021). First genome of Labyrinthula sp., an opportunistic seagrass pathogen, reveals novel insight into marine protist phylogeny, ecology and CAZyme cell-wall degradation. Microbial. Ecol. 82, 498–511. doi: 10.1007/s00248-020-01647-x
Trevathan S. M., Kahn A., Ross C. (2011). Effects of short-term hypersalinity exposure on the susceptibility to wasting disease in the subtropical seagrass Thalassia testudinum. Plant Physiol. Biochem. 49, 1051–1058. doi: 10.1016/j.plaphy.2011.06.006
Trevathan-Tackett S. M., Allnutt T. R., Sherman C. D., Richardson M. F., Crowley T. M., Macreadie P. I. (2020). Spatial variation of bacterial and fungal communities of estuarine seagrass leaf microbiomes. Aquat. Microbial. Ecol. 84, 59–74. doi: 10.3354/ame01926
Trevathan-Tackett S. M., Lane A. L., Bishop N., Ross C. (2015). Metabolites derived from the tropical seagrass Thalassia testudinum are bioactive against pathogenic Labyrinthula sp. Aquat. Bot. 122, 1–8. doi: 10.1016/j.aquabot.2014.12.005
Trevathan-Tackett S. M., Sullivan B. K., Robinson K., Lilje O., Macreadie P. I., Gleason F. H. (2018a). Pathogenic Labyrinthula associated with Australian seagrasses: Considerations for seagrass wasting disease in the southern hemisphere. Microbiol. Res. 206, 74–81. doi: 10.1016/j.micres.2017.10.003
Trevathan-Tackett S. M., Treby S., Gleason F. H., Macreadie P. I., Loke S. (2018b). Cryopreservation methods are effective for long-term storage of Labyrinthula cultures. Dis. Aquat. Org. 130, 65–70. doi: 10.3354/dao03266
Tsui C. K., Marshall W., Yokoyama R., Honda D., Lippmeier J. C., Craven K. D., et al. (2009). Labyrinthulomycetes phylogeny and its implications for the evolutionary loss of chloroplasts and gain of ectoplasmic gliding. Mol. Phylogenet. Evol. 50, 129–140. doi: 10.1016/j.ympev.2008.09.027
Vergeer L. H. T., Aarts T. L., de Groot J. D. (1995). The ‘wasting disease’ and the effect of abiotic factors (light intensity, temperature, salinity) and infection with Labyrinthula zosterae on the phenolic content of Zostera marina shoots. Aquat. Bot. 52, 35–44. doi: 10.1016/0304-3770(95)00480-N
Vergeer L. H. T., den Hartog C. (1994). Omnipresence of Labyrinthulaceae in seagrasses. Aquat. Bot. 48, 1–20. doi: 10.1016/0304-3770(94)90070-1
Vergeer L. H., Develi A. (1997). Phenolic acids in healthy and infected leaves of Zostera marina and their growth-limiting properties towards Labyrinthula zosterae. Aquat. Bot. 58, 65–72. doi: 10.1016/S0304-3770(96)01115-1
Vishniac H. S. (1955). The nutritional requirements of isolates of Labyrinthula spp. J. Gen. Microbiol. 12, 455–463. doi: 10.1099/00221287-12-3-455
Wang L., Tomas F., Mueller R. S. (2020). Nutrient enrichment increases size of Zostera marina shoots and enriches for sulfur and nitrogen cycling bacteria in root-associated microbiomes. FEMS Microbiol. Ecol. 96, fiaa129. doi: 10.1093/femsec/fiaa129
Wang Q., Ye H., Xie Y., He Y., Sen B., Wang G. (2019). Culturable diversity and lipid production profile of labyrinthulomycete protists isolated from coastal mangrove habitats of China. Mar. Drugs 17, 268. doi: 10.3390/md17050268
Watson S. W., Ordal E. J. (1957). Techniques for the isolation of Labyrinthula and Thraustochytrium in pure culture. J. bacteriol. 73, 589. doi: 10.1128/jb.73.4.589-590.1957
Watson S., Raper K. (1957). Labyrinthula minuta sp. nov. Microbiology 17, 368–377. doi: 10.1099/00221287-17-2-368
Yoshioka R., Schram J., Galloway A. (2019). Eelgrass pathogen Labyrinthula zosterae synthesizes essential fatty acids. Dis. Aquat. Org. 135, 89–95. doi: 10.3354/dao03382
Young III, E. L. (1938). Labyrinthula on Pacific coast eel-grass. Can. J. Res. 16, 115–117. doi: 10.1139/cjr38c-008
Young E. L. (1943). Studies on Labyrinthula. the etiologic agent of the wasting disease of eel-grass. Am. J. Bot. 30, 586–593. doi: 10.1002/j.1537-2197.1943.tb10303.x
Zidorn C. (2016). Secondary metabolites of seagrasses (Alismatales and Potamogetonales; Alismatidae): Chemical diversity, bioactivity, and ecological function. Phytochemistry 124, 5–28. doi: 10.1016/j.phytochem.2016.02.004
Keywords: molecular protocols, bioassay, pathogenicity, Labyrinthula sp, seagrass, disease ecology, pathosystems, sequencing
Citation: Sullivan BK, Martin DL, Yoshioka RM, Brakel J, Jakobsson-Thor S, Eisenlord M and Trevathan-Tackett SM (2023) Standard ecological and molecular research methods and techniques for Labyrinthula spp.. Front. Mar. Sci. 10:1092587. doi: 10.3389/fmars.2023.1092587
Received: 08 November 2022; Accepted: 19 January 2023;
Published: 15 February 2023.
Edited by:
Adriana Vallesi, University of Camerino, ItalyReviewed by:
Varada Damare, Goa University, IndiaVladimir V. Aleoshin, Lomonosov Moscow State University, Russia
Copyright © 2023 Sullivan, Martin, Yoshioka, Brakel, Jakobsson-Thor, Eisenlord and Trevathan-Tackett. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brooke K. Sullivan, c3VsbGlAdXcuZWR1
 Brooke K. Sullivan
Brooke K. Sullivan Daniel L. Martin
Daniel L. Martin Reyn M. Yoshioka
Reyn M. Yoshioka Janina Brakel
Janina Brakel Stina Jakobsson-Thor7
Stina Jakobsson-Thor7 Stacey M. Trevathan-Tackett
Stacey M. Trevathan-Tackett