- 1Laboratory of Viral Ecology, Research and Production Center for Microbiology and Virology, Almaty, Kazakhstan
- 2Institute of Hydrobiology and Ecology, Almaty, Kazakhstan
- 3Leeds Institute of Medical Research at St. James’s, St. James’s University Hospital, Leeds, United Kingdom
- 4School of Biology, Faculty of Biological Sciences, University of Leeds, Leeds, United Kingdom
Disease surveillance of marine mammal populations is essential to understand the causes of strandings, identify potential threats to animal health, and to support development of conservation strategies. Here we report the first large multi-pathogen screening of prevalence for viruses, bacteria and parasites in a sample of 177 live, healthy, wild Caspian seals (Pusa caspica), captured and released during satellite telemetry studies 2007-2017. Employing molecular and serological assays we assess prevalence of pathogens known to be of significance for marine mammal health worldwide, and evaluate the results in relation to Caspian seal health and conservation. RT-PCR, and PCR assays find evidence for infection by Canine Distemper Virus (CDV), Phocine herpes virus, phocine adenovirus and Influenza A at prevalences of 5%, 6.4%, 21.7%, and 4% respectively. The genomes of CDV isolates collected in 2008 showed 99.59% identity with the 2000 Caspian seal CDV epizootic strain. A partial coding sequence for the Us2 gene from the Caspian seal herpes virus was identical to PhHV-1 isolate PB84, previously reported from a harbor seal (Phoca vitulina), while amplicon sequences for the adenovirus polymerase gene indicated a novel strain. ELISA assays detected exposure to Influenza A (55% of tested samples), adenovirus (25%), coronavirus (6%), CDV (8%), herpes virus (94%), Toxoplasma gondii (2.6%) and heartworm (1%). Hemagglutination inhibition (HI) tests detected exposure to Influenza B at a prevalence of 20%, and Leptospira microscopic agglutination tests detected suspected exposure to Leptospira serovars in 9% of tested samples. Overall, the risks, profile and prevalence of pathogens in Caspian seals appear comparable to other wild phocid seal populations. Our results suggest Caspian seals have exposure pathways to pathogens with epizootic potential or ability to cause significant morbidity, and that disease impacts could reduce the resilience of the population to other conservation threats. Caspian seals are listed as Endangered by the International Union for Conservation of Nature (IUCN), and we recommend that resources are invested to support further surveillance programs and to understand how anthropogenic pressures may influence future disease risks. A translated version of this abstract is available in Russian and Kazakh in the Supplementary Material (Presentation 1 and Presentation 2)
Introduction
Monitoring the health status of marine mammal populations helps with understanding the causes of stranding events, identification of potential threats to human and animal health, and supports development of conservation and management strategies (Greig et al., 2014; Rosales and Vega Thurber, 2015). Marine mammal pathogens can cause mass mortality events involving thousands of individuals or underlie chronic morbidity, influencing the demography and resilience of populations (Kennedy, 1998; Dobson and Foufopoulos, 2001; Di Guardo et al., 2005). They can also act as selective agents on genetic variation (Stephens et al., 2014; Batley et al., 2021), influencing the evolution of populations. Baseline information on the prevalence of commensal, conditionally pathogenic, and pathogenic microflora is important for evaluating their role in epizootics, and individual health (Bogomolni et al., 2008). In this study, we report the first large multi-pathogen screening of prevalence for viruses, bacteria and parasites for live, healthy, wild Caspian seals (Pusa caspica), and evaluate their significance in relation to Caspian seal health and conservation.
Viral, bacterial, and parasitic infections can all cause significant mortality and morbidity in pinniped populations. In particular, influenza and morbilliviruses have frequently been associated with mass mortality events in phocid seals. Epizootics due to influenza A subtypes of avian origin, including highly pathogenic strains (H5N8) (Shin et al., 2019), have been documented among harbor seals (Phoca vitulina) on the Atlantic coast of North America (Hinshaw et al., 1984; Callan et al., 1995; Anthony et al., 2012), and in Europe (Zohari et al., 2014), while “swine” influenza A (H1N1) has been detected in Northern elephant seals (Mirounga angustirostris) in California (Goldstein et al., 2013). This suggests continuous influenza spill over risks for seal populations along bird migration routes, and as a result of human activities.
Morbilliviruses have caused several high profile epizootic events in phocid seals involving tens of thousands of individuals. Phocine Distemper Virus (PDV) was first identified following a mass mortality of harbor seals in the North Sea in 1988 (Markussen and Have, 1992; Kennedy et al., 2000), followed by a further European mass mortality in 2002 (Jensen et al., 2002; Duignan et al., 2014). PDV is now known to circulate widely in pinnipeds across the North Atlantic and North Eastern Pacific (VanWormer et al., 2019; Puryear et al., 2021). Additionally, Canine Distemper Virus (CDV), a morbillivirus of terrestrial carnivores, has caused die-offs in land-locked Baikal (Pusa sibirica) and Caspian seals (Grachev et al., 1989).
Herpesviruses are another common source of viral infections for phocid seals. Infections in harbor and grey seals (Halichoerus grypus) are characterized by respiratory symptoms with interstitial pneumonia and coagulative necrosis of the adrenal tissue and liver (Osterhaus et al., 1985; Gulland et al., 1997). At present, seven species of herpesviruses (PhHV-1 - PhHV-7), belonging to the genera alpha and gamma herpesviruses (Harder et al., 1996; Maness et al., 2011; Bodewes et al., 2015b), have been isolated from seals and sea lions (Gulland et al., 2020).
Bacteria are frequently a cause of primary disease, or found as secondary infections on a background of viral diseases. Commonly reported infections include leptospirosis, brucellosis, and pasteurellosis (Tryland et al., 2018). In a systematic review of marine mammal disease incidence from North America 1972-2012, bacterial disease represented approximately 20% of the total number of cases, with 60% of bacterial cases associated with clinical symptoms (Simeone et al., 2015).
Parasitic protozoan and helminth infections can have both direct and indirect effects on pinniped health status and population dynamics (Measures, 2018). Pinniped endo- and ectoparasites can have their origins in both terrestrial and marine environments (Leidenberger et al., 2007). The most abundant and pathogenic parasites for seals are lung and gastrointestinal nematodes (Measures, 2001; Ulrich et al., 2016). Lungworm infections in seals often exacerbate secondary, opportunistic bacterial infections and bronchopneumonia (Lehnert et al., 2007). For protozoan parasites, Toxoplasma gondii has caused fatal infections in harbor seals (Sonne et al., 2020).
The first documented mass mortality of Caspian seals occurred on the Tulenii islands (Dagestan, Russia) in 1931, where up to six carcasses per 100 meters were recorded as stranded due to an unidentified cause (Timoshenko and Yu, 1969). Another mass death was noted in 1955-56 when about 30 thousand animals died, with a diplococci infection implicated as the cause of the mortality on the basis of basic bacteriological analyses (Vylegzhanin, 1965; Vylegzhanin, 1967). Such infections are now recognized as secondary to viral epizootics, but at the time there was no precedent for viral involvement, and no virological investigation was carried out.
The first virological studies of Caspian seals focused on influenza A virus circulation, with Yamnikova et al. reporting negative results for 152 samples collected between 1976 and 1999, after culturing them in embryonated chicken eggs (Yamnikova et al., 2001). However, Ohishi et al. (2002) found a high prevalence of antibodies to influenza A and B viruses in serum samples of Caspian seals collected between 1993-2000. Influenza A (H4N6) virus was isolated from Caspian seals in Russia in 2012 (Gulyaeva et al., 2018), with the isolate being closely related to avian-derived influenza viruses of the classical Eurasian lineage.
High profile mass mortality events in Caspian seals occurred between 1997 and 2001, with the stranding of 1000s of carcasses widely reported in international media. CDV was identified as the etiological agent in 1997, in carcasses from Azerbaijan, on the basis of positive PCR results, with serological surveys suggesting the virus was circulating in the seal population before the spring of 1997 (Ohashi et al., 2001). A further CDV epizootic occurred in the spring of 2000, resulting in an estimated mortality of 10,000 animals, with infections characterized by broncho-interstitial pneumonia, lymphocytic necrosis and depletion in lymphoid organs. CDV infection was confirmed by RT-PCR, immunohistochemically, and phylogenetic analysis (Kennedy et al., 2000; Kuiken et al., 2006; Jo et al., 2019).
Concurrent bacterial infection of Bordetella bronchiseptica, Streptococcus phocae, Salmonella dublin, and S. choleraesuis were identified in seals that died in CDV epizootics in 2000 (Kuiken et al., 2006). Anti-Brucella antibodies have been detected in the blood serum of Caspian seals sampled in 1993-2002, with up to 4% seroprevalence (Durymanova et al., 2004; Ohishi et al., 2018). Parasitic diseases such as pseudo-amphistomosis (caused by the trematode Pseudamphistomum truncatum), anisakidosis – (nematode Anisakis schupakovi), eustrongylidosis (nematode Eustrongylides excisus), and corynosomosis (Acanthocephalan parasite - Corinosoma strumosum) have also been described in Caspian seals (Popov et al., 1989; Kuiken et al., 2006; Amin et al., 2011; Volodina and Grushko, 2012; Heckmann et al., 2014), and antibodies to Leptospira and Toxoplasma gondii have been reported for seals undergoing rehabilitation in Iran after release from fishing nets (Namroodi et al., 2018).
Caspian seals are endemic to the Caspian Sea in Central Asia, and are the only marine mammal present in the world’s largest landlocked water body. The species has been listed as Endangered by the International Union for Conservation of Nature (IUCN) since 2008 (Goodman and Dmitrieva, 2016), and has been included in the Red Books of all the Caspian littoral states since 2020. The current primary threats are considered to be high rates of mortality due to entanglement in gear set for sturgeon poaching and habitat degradation (Dmitrieva et al., 2013; Goodman and Dmitrieva, 2016). Disease is assessed as a potential threat, but more information is needed to fully evaluate the consequences of disease for Caspian seal population dynamics (Goodman and Dmitrieva, 2016).
The aim of the study is to assess baseline prevalence of pathogens of Caspian seals using molecular and serological analyses of samples collected from wild, live individuals caught and released during deployments of satellite telemetry tags in Kazakhstan. We screen for the key pathogens previously involved in mass mortality events, or with potential implications for seal and human health, known from pinniped studies elsewhere in the world. We evaluate the results in relation to the implications for Caspian seal health and conservation management.
Methods
Sampling locations and dates
Samples were collected at seal haul out aggregations at three locations in the Kazakh part of the Caspian Sea, annually between 2007 and 2017 (Supplementary Table 1). Molting seals (n=46) were sampled on the Durnev Islands (45°30′ N, 52°37′ E) in Komsomolets Bay in spring 2011. Post-molting sampling (n=4) was carried out on the Kendirli islets in Kazakh Bay (42°44′ N, 52°32′ E) in late April and May of 2014-2015. Additional post-molting samples (n=23) were obtained from islets in the vicinity of Prorva (45°53′ N 52°73′ E) in May of 2016. Samples were also taken during the autumn migration of seals at Rybachi sandbank (n=21) to the south of the Kulaly island group (44°76′ N, 50°37′ E) in November of 2007-2008, as well as at Kendirli (n=79) in October and November of 2009-2010, 2012-2013, and 2016-2017.
Seal capture, handling, ethical review, and sample collection
Live sampling of seals was carried out during satellite tagging studies (Dmitrieva et al., 2016). Seal capture was conducted by deploying seine nets from inflatable boats around haul out sites to entangle seals, or by ‘rush and grab’ of seals on sandbanks with hoop nets. Animals were restrained and handled without the use of chemical sedation, and were released immediately after the completion of sampling and tagging procedures. Animal sampling was regulated by the Resolution of the Government of the Republic of Kazakhstan (Government of the Republic of Kazakhstan, 2004) and the Order of Minister of Agriculture of the Republic of Kazakhstan (Government of the Republic of Kazakhstan, 2004), according to the legislation “Rules for conducting biomedical experiments, preclinical (non-clinical) and clinical studies (№697, 12 November 2007, Republic of Kazakhstan)”, and were approved by the Research and Production Center for Microbiology and Virology (RPCMV) Local Ethics Committee (Approval number: 2015-04-№1-ICPI). All animal handling and sampling procedures were also reviewed by the University of Leeds Animal Welfare Ethical Review Committee.
Body length (from nose tip to tail tip), girth and weight, and sex was recorded for each individual. Seals were categorised as juveniles (<1 year; body length 70–90 cm), sub-adults (older than one year of age; length >91–109 cm), adults (mature, length >110–140 cm). Complete external body examination was made for skin ulcers, parasites, trauma lesions and any visible alterations. Swab samples (Supplementary Table 2) were collected from body orifices (conjunctival, nasal, buccal, rectal, and urogenital) using sterile cotton swabs, which were placed into cryovials with a viral transport medium 199 containing antibiotics (penicillin, 2000 U/ml streptomycin, 2 mg/ml, gentamicin 50 µg/ml nystatin 50 U/ml) and bovine serum albumin at a final concentration of 0.5%. Samples were stored in liquid nitrogen (-196°C) until analyzed. Duplicates of specimens for molecular analyses were preserved in RNAlater.
Handling & processing of blood samples
Blood samples (Supplementary Table 2) were collected from the epidural venous sinuses of the spinal canal (Geraci and Smith, 1975), using the Vacutаiner blood collection system. The venipuncture site was disinfected with 70% ethanol. Depending on the nutritional condition of animals, spinal needles of 18G × 75 mm or 18G × 90 mm size were used for blood drawing. After coagulation, the blood samples were centrifuged (15 min at 3000 rpm), the resulting plasma was transferred into cryogenic polypropylene vials and transported in liquid nitrogen (-196°C).
List of target pathogens
Swab samples were screened for infection by influenza A and B viruses, morbilliviruses, seal herpesvirus [Phocine herpesviruses] 1 (PhHV-1), and canine coronavirus, parvovirus, adenoviruses, caliciviruses, and Brucella canis at the Laboratory of Viral Ecology of the authors’ institute (RPCMV, Almaty). Animals were visually examined for pox and calicivirus specific skin lesions (vesicles, nodules), seal lice (Echinophthirius horridus) and other ectoparasites. External physical inspection was also conducted for ophthalmic diseases, trauma, clinical signs of infections, and occurrence of aborted fetuses on haul out sites. Presence of IgG antibodies in seal sera was tested for influenza A and B viruses, canine distemper virus (CDV), canine herpes-, corona-, parvo-, adenoviruses, Brucella canis, Leptospira spp. Toxoplasma gondii, and heartworm Acanthocheilonema spirocauda.
Isolation of total nuclei acids from biological samples for PCR research
Isolation of RNA from blood or swab samples was carried out using a QIAamp Viral RNA Mini Kit (Qiagen GmbH, Hilden), in accordance with the manufacturer’s recommendations (Qiagen Ltd, 2005). DNA was extracted using a Qiagen DNeasy blood and tissue kit.
PCR protocols & Sanger sequencing
Reverse transcriptase polymerase chain reaction (RT-PCR) assays were performed on the basis of one-step protocols (Payungporn et al., 2004) using appropriate RT-PCR kits (Access RT-PCR System, Promega) according to the manufacturers’ instructions. Previously published oligonucleotide primers (Supplementary Table 3) were used for amplification of the targeted genes of viruses with a concentration of 0.5 μM each primer, and 5× AMV/Tfl buffer, 5 U inhibitor of RNase, 5 U AMV reverse transcriptase, 5 U Tn DNA polymerase, 1.0 mM MgSO4 and RNase-free water in a final volume of 25 μl. The reactions were performed in an Eppendorf Gradient thermocycler under the following conditions: reverse transcription at 48°C for 45 min, initial denaturation at 95°C for 2 min and amplification over 40 cycles, including denaturation, primer annealing and elongation as specified in Supplementary Table 4, followed by final elongation at 72°C for 10 min. PCR for DNA viruses was performed with the Q5® Hot Start High-Fidelity 2X Master Mix (NEB #M0492S) according to the manufacturer’s protocol with an initial denaturation at 98°C 30 seconds and other cycle details given in Supplementary Table 4.
Live infectious bronchitis virus (IBV) vaccine (Lohman Animal Health, Germany); PhHV-1 (from the European Virus Archive); local isolates of influenza A and B virus; live vaccine containing attenuated canine distemper virus (CDV; Vanguard Plus® (Pfizer, New York, NY, USA)); canine adenovirus type 1; canine adenovirus type 2; canine parainfluenza virus; canine coronavirus; canine parvovirus; and Leptospira spp. were used as a positive control for PCR screening of viral and bacterial pathogens, and sterile ultrapure water samples were applied as negative controls.
Horizontal gel electrophoresis was performed in a 2% agarose gel (Sigma, USA) stained with ethidium bromide in tris-acetate buffer at a voltage of 88V (8 volts/cm) on a Biostep apparatus (UK). The gel electrophoresis was visualized and documented in the GelMax® 125 Imager (Upland, CA, USA). The PCR products were sequenced using the fluorescent dideoxy-terminator method on ABI 3500 Genetic Analyzer (Applied Biosystems).
Genomic sequencing of viruses
The quality and quantity of viral RNA samples was determined using a Bioanalyzer (Agilent) with 100 ng used to make individual TruSeq Stranded Total RNA with Ribo-Zero Gold rRNA depletion (Illumina, USA) libraries according to manufacturer’s recommendations. Each library was sequenced on a HiSeq 3000, 150 bp paired end lane producing approximately 30 million read pairs per sample, at the University of Leeds Sequencing Facility. The genomes were then assembled with the CLC Genomics Workbench software (Qiagen) using a CDV reference sequence (AF014953) as a guide.
For herpes virus, DNA extracted from swabs was quantified using a Tapestation and NEBNext Ultra whole genome libraries were generated according to the manufacture’s protocol. Libraries were sequenced on a MiSeq, 150bp paired end lane, at RPCMV, Almaty, and contigs assembled with the CLC Genomics Workbench software (Qiagen).
Analysis of sequence data
Consensus sequences for PCR amplicons were assembled in MEGA X (Kumar et al., 2018). Sequence identity was confirmed using the BLAST tool suite at https://blast.ncbi.nlm.nih.gov/. For phylogenetic analyses relevant reference sequences were imported into MEGA X, and sequence alignments were generated using the MUSCLE application within MEGA X. The most suitable sequence evolution model was then evaluated with ModelTest within MEGA X. Maximum Likelihood phylogenies were generated for each data set using the indicated best-fit sequence evolution model, with 1000 bootstrap replicates. Trees were visualized and annotated with MEGA X’s Tree Explorer application.
Serological protocols
Hemagglutination inhibition test
Serum samples were tested for the presence of antibodies (antihemagglutins) against the hemagglutinin (HA) of influenza A virus HA subtypes: H1-H7, H9-H11, H13, H16 using the hemagglutination inhibition (HI) test. HI assays were performed on microtiter plates using a 0.5% suspension of chicken red blood cells and 4 HA units of the viral antigens: A/black-headed gull/Atyrau/2010 (Н1N2); А/duck/Germany/1215/73 (Н2NЗ); А/duck/California/72 (НЗN8); А/duck/Czech/56 (Н4N6); A/flamingo/Aktau/6570/15 (Н5N1) А/turkey/Mass./3740/65 (Н6N2); А/FPV/Rostok/34 (H7N7); А/pintail/North KZ/6401/14 (Н10N7); A/black-headed gull/Atyrau/07 (Н11N2); А/Pallas’s gull/Atyrau/2913/08 (Н13N6); А/Gull/Atyrau/07 (Н16NЗ). For influenza B viruses, strains B/Yamagata/16/88 and B/Florida/04/06 were used as antigens. The reaction was accompanied by controls for the antigen, red blood cells and test sera. An HI titer of 1:20 or higher was considered positive.
Enzyme-linked immunosorbent assay
Sixty-four sera collected between 2007 and 2013 were tested using the ID Screen® Influenza A Antibody Competition Multi-species ELISA kit (De Benedictis et al., 2010) at the Friedrich Loeffler Institute, Insel Riems, Germany. These 64 samples and 12 sera collected during 2014-2016 were tested at RPCMV (Almaty, Kazakhstan) again with another ELISA test kit according to Bodewes et al. (2015c). ELISA assays were carried out with manufacturers’ kits following the specified protocols: IDEXX Influenza A Ab Test Cat.# 99-53101; ID Screen® Influenza A Antibody Competition Multi-species ELISA kit; CAV (Canine Adenovirus) IgG Ab ELISA Cat.# DE2480; CHV (Canine Herpes Virus) IgG Ab ELISA Cat.# DE2481; CCV (Canine Corona Virus) IgG Ab ELISA Cat.# DE2482; CDV (Distemper Virus) IgG antibodies ELISA Cat.# DE2478; Rapid CDV Ab Test Kit 2.0 (RB2152DD); Rapid Canine Brucella Ab Test Kit (RB2103DD); Carnivore Brucella IgG ELISA (Sibitek LLC, Russia); Carnivore Leptospira (serogroup Pomona, Tarassovi, Hebdomadis, Icterohaemorrhagiae, Grippotyphosa, Sejroe and canicola) IgG ELISA (Sibitek LLC, Russia); Carnivore Toxoplasma IgG ELISA (Sibitek LLC, Russia).
Leptospira microscopic agglutination tests (LMAT) were carried out at the Laboratory of Anti-Bacteriosis Biotechnology, Faculty of Veterinary Medicine of the Kazakh National Agrarian University with eight Leptospira serogroups, using 10-day old cultures with at least 100 leptospira in the field of view at 40×15 magnification. Leptospira in sodium chloride physiological solution were used as negative control, and positive controls consisted of homologous rabbit serum from a set of known agglutinating leptospirose sera.
Results
Numbers of seals captured and sampled
A total of 177 seals were live captured, sampled, and released 2007-2017 (Supplementary Table 1). In total, we obtained 678 swab and 97 serum samples (Supplementary Table 2).
Virus detections from RT-PCR and PCR screening
A summary of virus detections via RT-PCR and PCR is given in Table 1. RT-PCR assays for the CDV P gene detected the expected 429 bp product in five of 13 seals sampled in 2008. All the positive samples were derived from animals under two years old. No PCR positive specimens were detected in any other samples for the subsequent 2009-2017 survey period. Further testing of samples returning negative results with the Pan-paramyxovirus primers specific for the L-gene of all Paramyxoviridae family members also gave negative results.
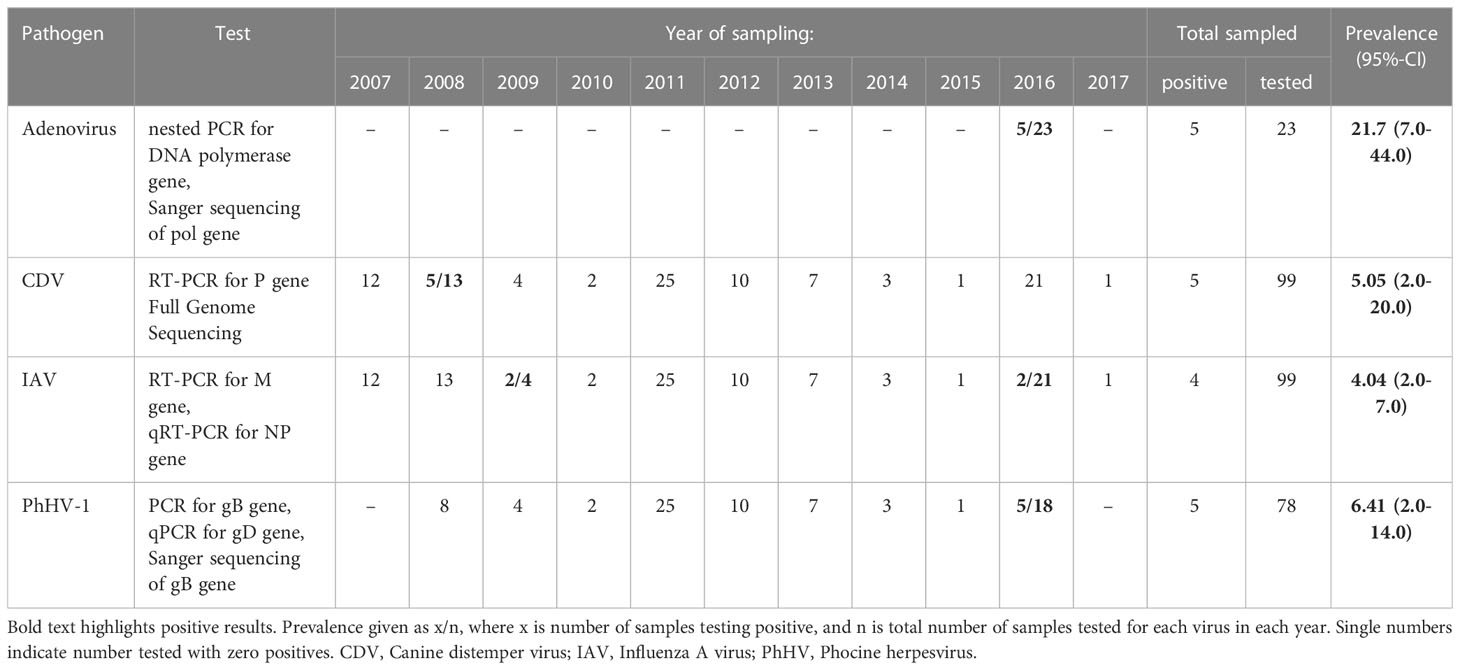
Table 1 Prevalence of CDV, Influenza A Virus, PhHV-1 virus, and Adenovirus detected via RT-PCR and PCR in Caspian seals.
Conventional RT-PCR testing for the influenza A virus M gene revealed positive ocular and buccal swabs for two animals sampled in 2009. qRT-PCR assays for the Influenza A virus NP gene resulted in specific fluorescent amplification signals (Ct values 23-34) in two swabs collected in October 2016. However, no haemagglutination agents could be isolated following inoculation and passage of material from the 2009 and 2016 samples PCR-positive for the presence of influenza A virus nucleic acids in 10-11 day-old chick embryos.
The expected 290 bp PCR product for PhHV-1 gD gene was detected in oral swabs from five animals sampled in May (three) and October (two) 2016. Positives were verified via real time - PCR with primers to the gD gene of PhHV-1 and Sanger sequencing.
Adenovirus DNA was amplified by consensus nested PCR (Wellehan et al., 2004) targeting the pol gene from pooled nasal and buccal swabs of five Caspian seals sampled in May 2016.
PCR screening for influenza B viruses, coronaviruses were negative in all tested samples.
Sequence identity and phylogeny of DNA sequences
CDV (Canine Morbillivirus)
Complete CDV genome sequences were recovered for 2 of the 5 positive 2008 samples, CDV/Pusa caspica/2008-1AP and CDV/Pusa caspica/2008-1BN, consisting of 15,697 nucleotides. The sequences closely matched the genomes of the 2000 Caspian CDV strain reported by Jo et al. (2019), with 99.59% identity, Figure 1.
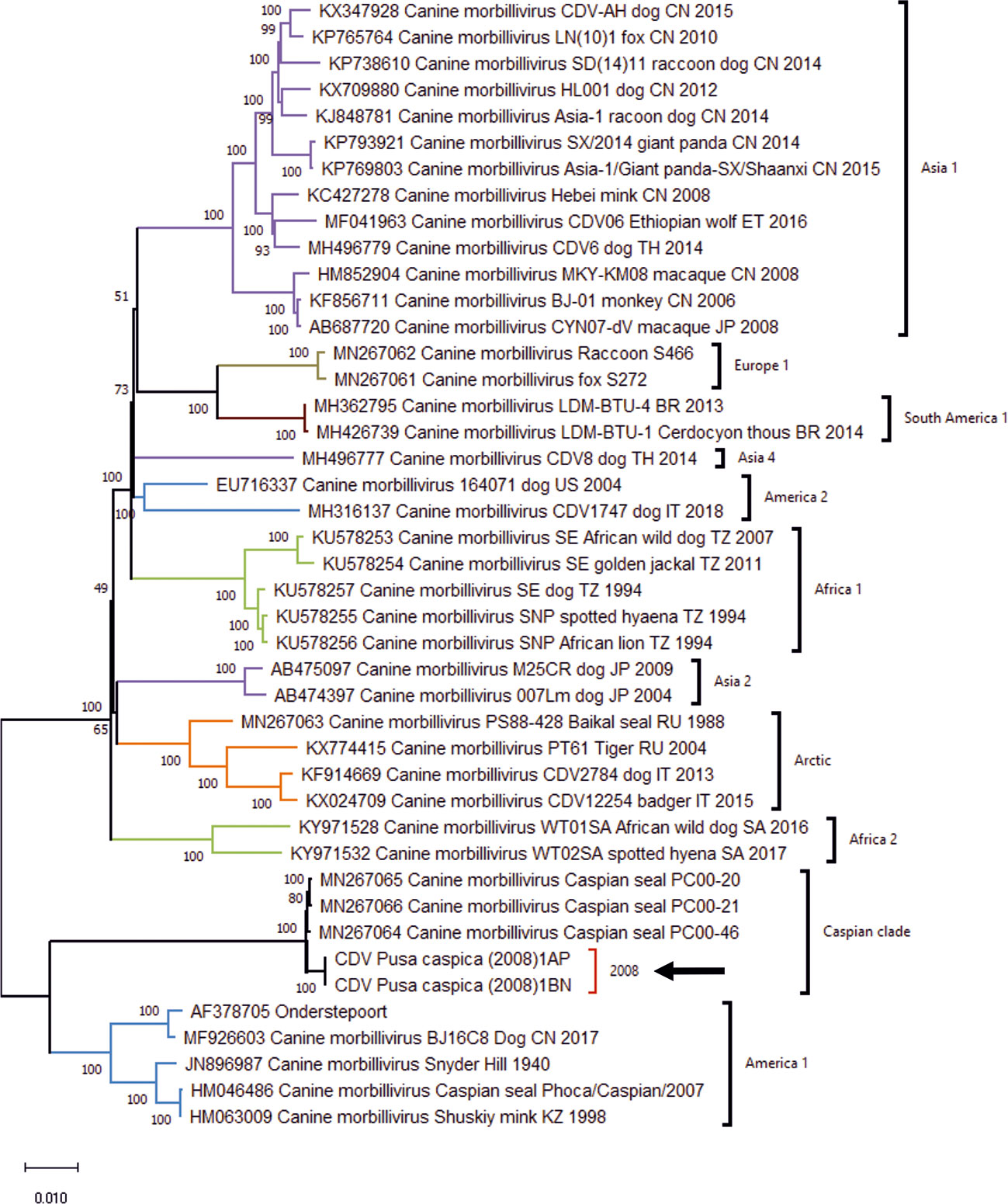
Figure 1 Maximum Likelihood phylogenetic tree for Canine Distemper Virus whole genome sequences, generated using the General Time Reversible model with invariant sites and gamma distribution for evolutionary rates (GTR+I+G), with 1000 bootstrap replicates. Position of the Caspian clade, and genomes for the 2008 isolates is highlighted by arrow. Regional annotation of clades follows Jo et al. (2019).
The nucleotide sequences of the P gene from the CDV/Pusa caspica/2008 genomes, and the 2008 RT-PCR P gene amplicons show a close relationship with the epizootic strain CDV/Caspian seal/Baku/1997 (Forsyth et al., 1998; Kennedy et al., 2000) and 2000 complete genomes (Jo et al., 2019), with sequence identity ranging from 99.44% to 100%, average 99.73% (Supplementary Table 5). Together these results suggest a highly similar virus circulated in seals from Azerbaijan and Kazakhstan, and across an 11 year interval.
PhHV-1
BLAST analysis of a 770 bp contig recovered from the DNA sequencing library returned 100% sequence identity to the Us2 gene of phocid alphaherpesvirus type-1 PhHV-1 isolate PB84 (Accession: NC_043410.1) derived from a harbor seal, 99.7% similarity with Us2 of 2 other phocid alphaherpesvirus type-1 isolates, and 78.7% similarity with next closet matching strains, which are all Canid alphaherpesviruses (Supplementary Figure 1). No other pinniped Us2 alphaherpesvirus type-1 sequences are available in Genbank at the time of writing.
Adenovirus
The PCR positive samples from 2016 yielded a 261bp amplicon. BLAST analysis of the pol gene amplicon sequence using the blastn option, returned top 10 hits with percentage identities ranging from 77.1% to 73%.3% (coverage 94%-76%) including Lutrine adenovirus type 1, Phocine adenovirus 2, Bat mastadenovirus isolates, and Otarine adenovirus 2. Phylogenetic analysis suggested the closest related sequences were derived from Lutrine adenovirus type 1, bovine adenovirus, bat mastadenoviruses, Felis catus adenovirus, and bottlenose dolphin adenovirus 1, although relationships were poorly resolved (Supplementary Figure 2). In combination, this indicates the presence of a novel adenovirus in Caspian seals, which appears to be distinct from currently known members of the Adenoviridae family.
Incidence of seal lice, pox lesions, cataracts, and injuries
Pox-like lesions of approximately 2-3cm diameter were observed in three animals sampled in 2010 and 2011, with one having bald skin patches on the head and neck. Other vesicle-like skin lesions were seen in one animal in 2015. Ophthalmic pathologies representing keratopathy or cataracts were noted in four seals sampled in 2007, 2010, 2016, and 2017 (Supplementary Figure 2A). No visible ectoparasite infestations or their characteristic dermatological lesions were observed among any captured seals. Potential predator (wolf/jackal) bites were observed on single individuals sampled in 2011 and 2016 (Supplementary Figure 2B). Monofilament net entanglement injuries and scars were observed on individuals during most sampling expeditions.
Serological evidence for infections
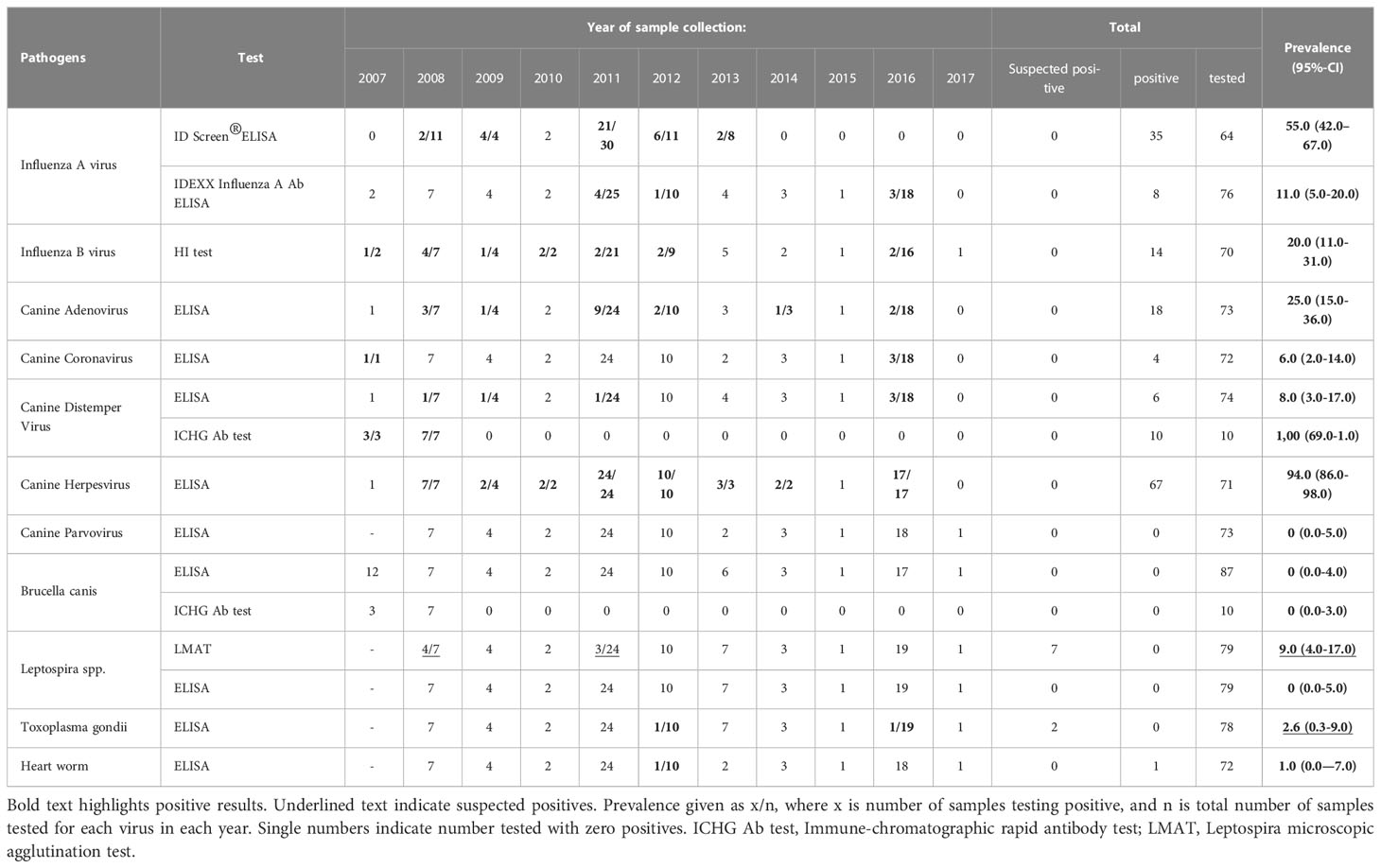
A summary of IgG Antibody prevalence from tested samples is given in Table 2.
Influenza A
Initially, antibodies against the influenza A virus NP protein were detected in 35 (55%) out of 64 Caspian seal serum samples collected from 2007 to 2013. Prevalence for NP-specific IgG antibodies in years with positive detections were 2008 - 2/11 (18%); 2009 – 4/4 (100%); 2011 – 21/30 (70%); 2012 – 6/11 (5.5%); 2013 2/8 (25%).
IDEXX Influenza A Ab ELISA kit Flu A/NP-protein specific IgG antibodies were detected in eight out of 76 serum samples tested (10.5%). The prevalence in years with positive detections were 2011 - 4/27 (18.5%); 2012 1/10 (10%); 2016 – 3/18 (17%).
Using HI assays, eight sera positive for Flu A/NP-protein specific IgG were subtyped with a set of influenza A virus antigens (Table 3). Antihemagglutinins were detected at a low level (1:40) against influenza A virus subtype Н10N7 in two seal sera sampled in 2011, and notably high titers ranging from 1:80 to 1:1280 were observed for three serum samples of 2016. Those three seropositive seals of 2016 also had antihemagglutinins against Н1N2 subtype at a threshold level (1:20), and two additionally possessed antibodies against Н4N6 at titers of 1:40-80. Three seal sera collected in 2011, 2014, and 2016 did not react with any of the tested HA antigen subtypes. Antibodies to influenza A virus HA subtypes H1, H2, H3, H5-H9, H11, H13, H16 were not detected in any tested sera (Table 2). Viral antigens of the H8, H12, H14 and H15 subtypes were not available for HI assays.
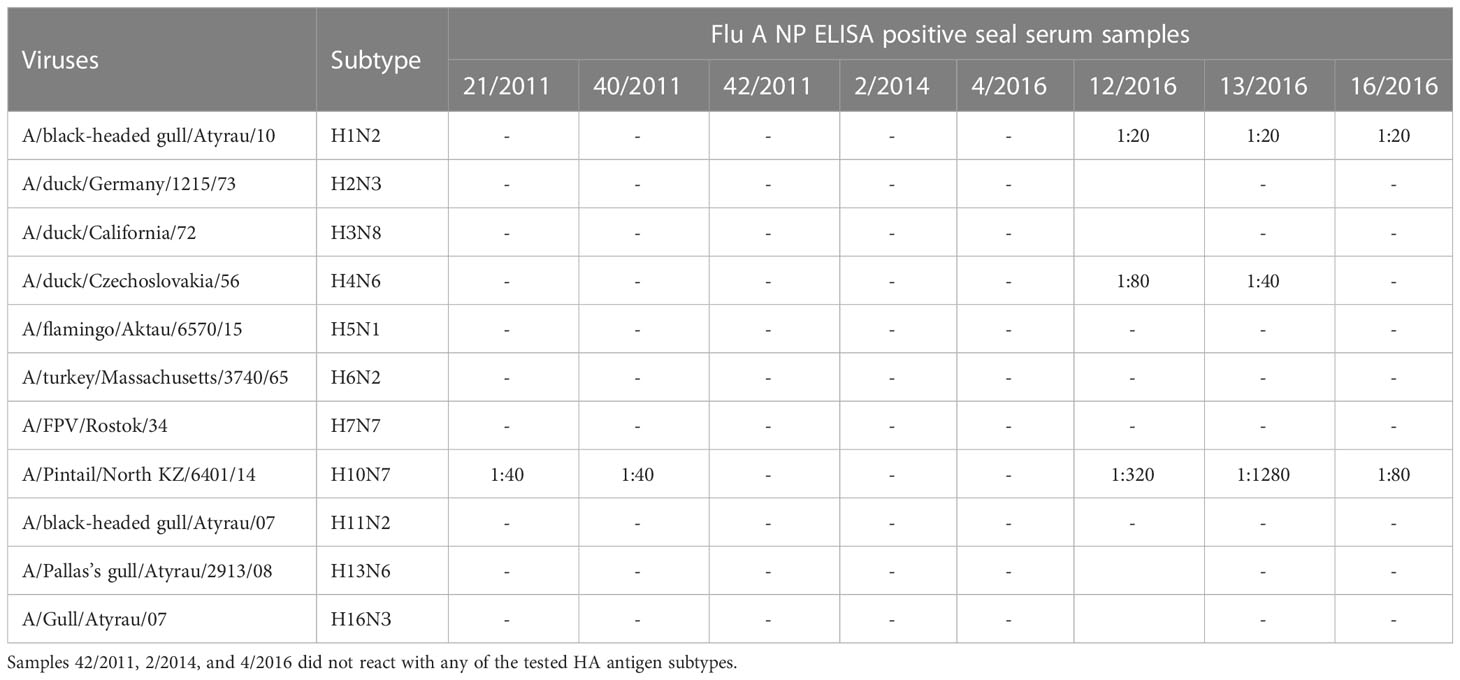
Table 3 Hemagglutination inhibition (HI) titers of Influenza A (NP) for 8 ELISA positive Caspian seal serum samples.
Influenza B
Influenza B specific hemagglutinin antibodies were detected in 14 seal sera (19,7%, confidence interval (CI): 12.0% – 24.7%) from 71 investigated samples (Table 2). These anti-influenza B antibodies were detected in serum samples collected each year in the period 2007-2012 and once in 2016 for the period 2013-2017 (Table 2, Supplementary Figure 3), where the proportion of seropositivity varied from 9.5% up to 100% depending on the sample size per year. Overall the prevalence 2007-2012 was 12/45 (26%) compared to 2/25 (8%) for 2013-2017, but this does not present a significant difference in frequency (Fisher’s exact test P>0.05).
Canine distemper Virus
Antibodies to CDV were detected in ELISA tests for six sera out of 74 (8.1%) seals sampled from 2007 to 2017, with 1/7 (14%) in 2008; 1/4 (25%) in 2008; 1/24 (4%) in 2011 and 3/18 (17%) in 2016. All three positives from 2016 belonged to adult seals (Supplementary Table 6).
Phocine herpesvirus
Nearly all tested samples reacted positively for the presence of antibodies against canine herpesvirus in ELISA tests. Canine herpesvirus has antigenic similarity to PhHV-1 and is highly cross-reactive in serological tests (Osterhaus et al., 1985; Lebich et al., 1994). Specific IgG antibodies to canine herpes virus were detected in 67 samples from 71 sera (94.4%, CI: 61.5% – 99.9%), indicating persistent exposure to PhHV-1 in the Caspian seal population.
Other pathogens
Antibodies to canine adenovirus were detected in 18 seal sera representing 25% (CI: 2.09% – 3.39%) of investigated samples. The highest number (9) of positives (37.5%) was detected in the samples of adult animals (24) collected in spring of 2011 (Table 2, Supplementary Table 6), while the lowest seroprevalence was found in the samples of 2016 (2/18, 11.1%).
Canine Coronavirus-specific IgG were found in one seal serum sample from 2007 and three samples from 2016.
No antibodies against canine parvovirus were detected in any of the investigated sera with ELISA tests. Similarly, none of the Caspian seal serum samples were positive for anti-Brucella canis antibodies with either ELISA or immune-chromatographic rapid Ab tests.
No positives were detected for Leptospira spp. in ELISA screening. In contrast, Leptospira MAT analysis revealed seven suspected positive cases (considered positive at a dilution of 1:50 and 1: 100, i.e. less than 25% of leptospira agglutination) for serogroups Pomona, Hebdomadis (Kabura), Grippotyphosa (Moskva V), Tarassovi (Perepellicin), Icterohaemorrhagiae (M-20). Four sera were suspected positive for L. hebdomadis (Kabura), one simultaneously reacting with L. icterohaemorrhagiae (M-20), and another in combination with L. tarassovi. One sample appeared positive for both L. pomona and L. tarassovi, the last sample separately demonstrated 25% microagglutination of L. grippotyphosa.
Toxoplasma gondii – specific antibodies were detected in two seal sera sampled in October of 2012 and in May of 2016. The optical density of the two sera was close to the threshold for a positive result and therefore were considered as suspected rather than definitive detections.
Antibodies related to heartworm Acanthocheilonema spirocauda were detected in one seal serum sample from October of 2012.
Comparison of pathogen prevalence among seasonal sampling periods
The main periods in which high density of aggregations of Caspian seal occur and provide opportunities for pathogen transmission are during the spring molt, and autumn pre-breeding season period. We aggregated spring and autumn samples across years and compared prevalence of each tested pathogen using Fisher’s exact tests (Supplementary Tables 7 and 8). Only the detections of influenza B virus in HI tests varied significantly (P=0.03), with a higher prevalence in autumn samples (33%) compared to spring (10%). However, these comparisons should be treated cautiously due to small sample sizes in some cases and opportunistic sampling. Similarly, we choose not to formally compare prevalence differences between sex and age classes (Supplementary Table 6) due to the biased nature of sampling during satellite telemetry deployments.
Discussion
As a transboundary, top trophic level predator species, the Caspian seal is a key indicator of ecosystem status (Ivanov et al., 2000; Goodman and Dmitrieva, 2016), but is exposed to anthropogenic stressors, including high levels of mortality from bycatch associated with illegal fisheries (Dmitrieva et al., 2013), and habitat degradation arising from historical and contemporary industrial development (Wilson et al., 2014; Wilson et al., 2017b). The CDV mass mortalities between 1997 and 2001 raised concerns about pathogen impacts on the population, but the current health status of the population and the relative importance of disease as a conservation issue remains uncertain. The present study expands knowledge on baseline prevalence of pathogens known to be important for pinniped health worldwide, and provides a basis for evaluating conservation risks associated with infectious disease.
Until 2000 knowledge of the health status of Caspian seals was limited primarily to opportunistic parasitological (Shchupakov, 1936; Kurochkin and Badamshin, 1968; Kurochkin, 1975) and bacteriological assessments during the historical commercial seal harvest and seal strandings. Historically, regional veterinary laboratories lacked diagnostic capacity for wildlife and viral diseases, being focused on confirming or excluding common bacterial pathogens of livestock in the event of wildlife disease outbreaks. Consequently, opportunistic bacterial pathogens were frequently declared as causes of seal mortalities (The Economist, 2000). The first virological surveys in Caspian seals were carried out for Influenza A virus monitoring between 1976-1999 (Yamnikova et al., 2001). The CDV die-offs in 1997-2001 led to the first comprehensive investigations of viral diseases of Caspian seals (Kennedy et al., 2000; Kuiken et al., 2006), but focused on explaining the epizootic mortality, rather quantifying baseline pathogen prevalence in the wider population.
CDV infection
Morbilliviruses of the of Paramyxoviridae family are one of the most frequent causes of marine mammal mass mortality events (Jo et al., 2018). Canine Distemper Virus (CDV; otherwise referred to as canine morbillivirus), has caused confirmed mass mortalities of the Baikal seal (Pusa sibirica) in 1987-1988 in Siberia (Grachev et al., 1989), and was the mostly likely agent responsible for a mortality of Antarctic Crabeater seals (Lobodon carcinophagus) in 1955 (Laws and Taylor, 2009). Antibodies to CDV have subsequently been found in recent serological surveys of Arctic and Antarctic pinnipeds (Kennedy et al., 2019). Canine morbillivirus infection in Caspian seals has been reported since 1997 and caused mass mortalities estimated at thousands of seals in 2000 and 2001 (Forsyth et al., 1998; Kennedy et al., 2000; Kuiken et al., 2006). Recent phylogenetic studies indicated that the CDV isolate recovered from the epizootic in the Caspian Sea in 2000 belongs to a previously undetected novel clade designated as “Caspian” (Jo et al., 2019), which is basal to other known CDV strains.
In our investigation, CDV was found by RT-PCR in five individuals out of 11 seals sampled in 2008. All positive specimens came from individuals under two years old, suggesting an age-dependent susceptibility of animals to the virus, consistent with exposure of naïve juveniles, and resistance in older age classes. No further RT-PCR positive individuals were detected in samples from subsequent years (2009-2017). The lack of CDV-positives in RT-PCR-screening after 2008 may be explained by preferential sampling of adult individuals for satellite tag deployments rather than juveniles.
The RT-PCR positive individuals sampled in 2008 were asymptomatic, and appeared otherwise healthy. However, phylogenetic analysis of complete genomes for the 2008 viruses showed they were derived (average 99.73% identity) from the same epizootic causing “Caspian” clade from 2000, suggesting possible adaptation to reduce virulence. Further characterization of the 2008 strain sequence data is required to evaluate this possibility further. The 2000 “Caspian” strain is not closely related to known CDV strains from terrestrial carnivores (Jo et al., 2019), but there has been limited assessment of CDV strains circulating among wild and domestic carnivores within the Caspian region. Available sequence data for small surveys of CDV strains circulating among dogs in northern Iran in the vicinity of the Caspian shore, indicate they are derived from Arctic and European clades (Namroodi et al., 2015). At this time it is not possible to conclusively determine if the 2000 “Caspian” CDV clade derives from a recent spill over from a terrestrial source, or if it is an endemic strain with long term circulation in the seal population.
Our serological findings indicated maintenance of CDV in the Caspian seal population; antibodies to CDV were detected in six out of 74 seal serum samples (8.1%) between 2007 to 2017, with the most recent positives detected in three adult seals out of 18 sampled in 2016. These findings were obtained with a commercially available ELISA test kit based on monoclonal antibodies against a common epitope of CDV, although a microplate virus neutralization test is considered as “gold standard” in serological surveys of morbillivirus antibodies due to their high sensitivity and specificity. Monoclonal antibody-mediated ELISA tests are also recommended as a useful tool for epidemiological surveillance of morbillivirus infections in marine mammal populations for their comparable sensitivity and specificity to virus neutralization tests, while being simpler to perform and less time consuming (Saliki and Lehenbauer, 2001). Additionally, Saliki and Lehenbauer (2001) demonstrated that Pinniped samples achieve similar levels of performance as Canid sera in ELISA. Hence, we assume the frequency of CDV antibody detection in the Caspian seal serum samples accurately reflects the age class profile for CDV seen in the PCR screening.
Cornwell et al. (1992) reported the persistence of PDV antibodies in harbor and grey seals at moderate to high titers in recovered animals for at least 6.5 months. After administration of an inactivated CDV vaccine to harbor seals, CDV antibody levels declined rapidly to very low levels within the same period, while titres in grey seals persisted at detectable levels for at least 12 months (Cornwell et al., 1992).
Namroodi et al. (2018) also recently reported CDV antibody prevalence from virus neutralization tests, with serum from 12/36 (33%) individuals, sampled 2015-2017 after release from accidental entanglement in fishing gear, testing positive, and concluded this provided evidence for persistence of CDV infections in the population. In principle, the current estimated population size for Caspian seals at approximately 168,000 individuals (Goodman and Dmitrieva, 2016), is above the threshold necessary for the virus to persist as endemic infection (Kuiken et al., 2006), but further systematic serological and genetic surveillance is necessary to fully understand CDV dynamics in the Caspian seal population. Comparisons of CDV antibody prevalence between our studies should be treated with caution due to differences in detection methods, sampling locations and periods, and sex-age class composition of the animals tested.
Influenza A virus infection
Avian influenza A viruses frequently jump from birds into various mammalian species, including pinnipeds (Repérant et al., 2009; Anthony et al., 2012; Bodewes et al., 2015a), with spill over events being associated with close contact with avian reservoirs. In the case of pinnipeds, this may occur through sharing of haul out sites with infectious birds shedding virus in aerosols and feces.
Four Caspian seals yielded suspected positives for influenza A virus on RT-PCR. Virus cultivation attempts from samples with Ct values <35 for further analysis failed. Similar results with non-cultivable influenza A viruses were described for qRT-PCR positive samples in live-captured North Atlantic gray seals (Puryear et al., 2016), broadly reflecting difficulties growing and sequencing influenza viruses from wild reservoirs.
Antibody screening using ELISA test kits from two independent manufacturers suggested regular exposure of Caspian seals to influenza A infection. Our results may be downwardly biased as there are no approved ELISA test-kits for monitoring of anti-influenza A antibody levels of marine mammals, although equivalent kits to those used here have previously been used successfully in other pinniped studies (Bodewes et al., 2015c; Puryear et al., 2016).
The results of the commercially available ID Screen® Influenza A Antibody Competition Multi-species ELISA kit were verified by an IDEXX Influenza A Ab ELISA kit. Differences in antibody prevalence in test kit results may be partially accounted by deterioration of specific IgG after freeze-thawing cycles and the duration of storage. Our HI assay results revealed a narrow range of antibodies against influenza A virus hemagglutinin subtypes including H1, H4 and H10 in ELISA positive sera of Caspian seals. Elevated titers to influenza A (H10N7) were detected in samples collected in 2016. The absence of antibodies to this strain in serum samples from the preceding years may indicate recent exposure of the population, although no reports of elevated morbidity and mortality were recorded among Caspian seals at the time. Increased seropositivity in seal rehabilitation center patients was also reported in the Netherlands in 2015 (Bodewes et al., 2015c) after an influenza A (H10N7) virus outbreak among harbor seals in the North Sea in the spring/summer of 2014 (Bodewes et al., 2015a).
Three influenza A/NP-positive serum samples did not react with any hemagglutinin subtype antigens in HI assays, suggesting that these sera came from Caspian seals which encountered other antigenically different influenza A viruses. Previously a possible transmission of human influenza A and B viruses to Caspian seals was serologically characterized (Ohishi et al., 2002). Serological evidence for influenza viruses infection with avian-associated hemagglutinin subtypes H1, H2, H3, H4, H6, H7, H8, H9, H10, H11, H12, H13, and H16 has been obtained in marine mammals in a number of studies (De Boer et al., 1990; Steuen et al., 1994; Danner and McGregor, 1998; Nielsen et al., 2001; Puryear et al., 2016). The North Caspian region is covered by intersection of the Black Sea-Mediterranean and East African-West Asian migratory flyways, meaning Caspian seals could consistently interface with avian reservoirs of influenza viruses on their haul-out sites. Thus the detection of antibodies against avian-associated influenza A hemagglutinin subtypes, and potential for regular bird-seal interactions in their environment, strongly supports transmission of influenza viruses from avian reservoirs to Caspian seals rather than being derived from mammalian sources.
Influenza B virus infection
In contrast to influenza A viruses, which have been isolated from many different species, influenza B viruses are human pathogens which had no or unknown reservoirs in nature until 1999, when it was reported that harbor and grey seals can become infected with this virus (Osterhaus et al., 2000), with Influenza virus B isolated from a juvenile harbor seal with symptoms of respiratory disease. Since the acknowledgment of marine mammals as new hosts, there have been several reports on the detection of antibodies to the influenza B virus in phocid and otariid species (Ohishi et al., 2002; Blanc et al., 2009), with evidence for infection of harbor seals in the Netherlands on a regular basis (Bodewes et al., 2013a).
Our serology results provide evidence for influenza B virus infection in Caspian seals. The highest antibody titers were found against influenza B/Almaty/8/18 strain belonging to B/Victoria-linage viruses, whereas lower antibody titers were detected for B/Florida/4/2006 strain of B/Yamagata-lineage viruses. Antibodies at high titers were found in serum samples from juvenile seals suggesting recent influenza B infection in those animals. The proportion of influenza B positive seal sera ranged from 9.5% up to 100% (in 2010) per sampling time point, indicating regular exposure of Caspian seals. The negative results of influenza B virus PCR screening in Caspian seals probably relates to low viral load, dependence on sampling time, and if infection has a seasonal character. Transmission is most likely to occur when seals are hauled out in high densities. Haul out densities are highest during the spring molt, and to a lesser extent during the autumn. Winter transmission could occur during breeding, but normally there is some separation between females hauled out on the ice sheet during lactation (Wilson et al., 2017a). Resolving whether influenza B is endemic within Caspian seals, or if there is recurrent infection either from a human or other reservoir requires phylogenomic analysis of influenza B viruses from seals and additional serology analysis with a broad panel of epidemic strains.
Adenovirus infection
Adenovirus infection is one of the most common diseases in carnivores. There are two types of this disease - infectious hepatitis and infectious laryngotracheitis, which are caused by adenovirus type I and II, respectively. Adenoviral infections with clinical signs of hepatitis and enteritis have been noted in sea lions and fur seals (Goldstein et al., 2011; Inoshima et al., 2013). Phocine adenoviruses PhAdV-1 and PhAdV-2 were isolated from ocular swabs of northern elephant seals and Pacific harbor seals with ophthalmic disorders (Wright et al., 2015).
Antibodies to canine adenovirus type I (CAV-1) were detected in 25% of Caspian seal sera predominantly in adult animals. The anti-adenovirus antibody detection frequency in Caspian seals is comparable with a 29% prevalence reported in Otariid serological surveys for CAV-1 (Burek et al., 2005). The antigenic cross-reactivity of CAV and CAV-related adenovirus of pinnipeds is discussed by Inoshima et al. (2013). Results of viral surveys in Northern elephant seals and Pacific harbor seals (Wright et al., 2015) demonstrate the existence species-specific adenoviruses, Our preliminary sequencing of PCR amplicons from the Caspian seal swab samples of 2016 suggests the existence of a novel adenovirus specific to Caspian seals, related to Lutrine adenovirus type 1, bovine adenovirus 10 and bat mastadenovirus, but requires further full-genome characterization to fully understand its relationship to other adenoviruses.
Coronavirus infection
Coronaviruses are common pathogens of the respiratory system, gastrointestinal tract and cause systemic infections of domestic and wild animals (Buonavoglia et al., 2006). Harbor seal coronavirus (HSCoV) is manifested by hemorrhagic pneumonia and causes epizootics with high mortality (Nollens et al., 2010). Canine respiratory coronavirus is a new coronavirus of dogs, which is widespread in North America, Japan, and several European countries, and is genetically and antigenically distinct from enteric canine coronavirus (Erles and Brownlie, 2008). Modified pan-coronavirus PCR-screening of the Caspian seal swab and post-mortem samples were negative for the poly L gene of this virus. However, Canine Coronavirus (CCV)-specific IgG were found in one seal serum from 2007 and three samples collected in 2016. The CCV ELISA test kit is designed to detect antibodies against CCV proteins (mostly glycoproteins). CCV is a highly infectious but usually self-limiting disease characterized by diarrhea, intestinal disease, weakness and loss of appetite in dogs. Taking account of the cross-species spill over risks associated with coronaviruses, our findings demonstrate a potential exposure risk of Caspian seals to coronaviruses, which should be closely monitored in future surveillance activities.
PhHV-1 infection
Antibodies reacting to PhHV-1 were detected in Caspian seal sera from 68 of 72 samples (94.7%) tested, representing all age classes. This suggests persistent, stable circulation of PhHV-1 in the Caspian seal population, with potential recurrent exposure of individuals. Previous studies have demonstrated significant homology of alphaherpesviruses of terrestrial carnivores and phocid-herpesvirus at the antigenic and immunogenic levels (Osterhaus et al., 1985; Lebich et al., 1994; Martina et al., 2003), and found strong correlation between ELISA and serum neutralisation tests for PhHV-1 (Roth et al., 2013). Therefore, we suggest that the canine herpesvirus ELISA employed here can serve as a useful proxy for PhHV-1. Presence of PhHV-1 was confirmed by molecular tests. Analysis of a partial sequence of the Us2 gene suggests the Caspian seal strain is highly similar to Phocine alphaherpesvirus PhHV-1 isolate PB84 derived from a harbor seal, but sequencing of the full genome is necessary to fully characterise the virus and its relationship to other phocine herpes viruses.
Herpesvirus PhHV-1 was first isolated from newborn habor seals with clinical signs of acute pneumonia and hepatitis at a rehabilitation center in the Netherlands in 1985 (Osterhaus et al., 1985). In the Pacific harbor seal (Phoca vitulina richardsi), this virus is associated with focal caogulative necrosis of the adrenal cortex and liver. In phylogenetic studies of the DNA polymerase (gD) gene of PhHV-1, their relationship with herpes viruses of dogs and cats was established (Martina et al., 2003). Genital herpesvirus otarine herpesvirus 1 is strongly associated as trigger of cervical tumors in California sea lions (Zalophus californianus) (Deming et al., 2021).
Serological studies have shown widespread infection of PhHV-1 or other closely related α-herpes viruses in seal populations in the North Sea, as well as in the waters of the Antarctic and the North Pacific (Harder et al., 1991; Have et al., 1991; Zarnke et al., 1997). The serological prevalence of 94% in our study is similar to PhHV-1 antibody prevalence in harbor seals from Svalbard (Norway) where detection frequency ranged from 72 to 100% in each study year (Roth et al., 2013). The annual variation in seroprevalence in harbor seals was concluded as reflecting different frequencies of reactivation of the latent virus, influenced by factors impacting on stress and immune-competence of the animals (Roth et al., 2013). PhHV-1 infection in Caspian seals likely represents a consistent viral burden on health which may be exacerbated by triggering factors. The pathological implications, of PhHV-1 virus infection in Caspian seals are likely to be similar as for other phocid species, but further research is needed for these to be established.
Other pathogens
Parvovirus
Parvoviruses are known as a major pathogen of several terrestrial carnivore species (Hoelzer and Parrish, 2010), but little is known of their impact on marine mammals. Recently a novel parvovirus belonging to the same genus as human parvovirus B19, was detected in the brain of a young harbor seal with chronic meningo-encephalitis that was rehabilitated at the Netherlands (Bodewes et al., 2013b; Bodewes et al., 2014). Parvovirus sequences have also been detected in sub-Antarctic and South American fur seals. Members of the same genus within the Parvoviridae should share at least 30% amino acid identity in the predicted NS1 sequence (Kluge et al., 2016). No antibodies against canine parvovirus in the ELISA test were detected in any Caspian seal sera. Virus screens in PCR were also negative for parvoviruses in all swab samples. The negative results for PCR screening in our study indicate a deep metagenomics survey of Caspian seals may be required to detect any distantly related parvovirus.
Brucella canis
None of the Caspian seal serum samples were positive for anti-Brucella canis antibodies with either ELISA or immune-chromatographic rapid Ab tests. Although up to 4% seroprevalence in Caspian seals for Brucella antibodies have been reported in previous studies (Durymanova et al., 2004; Ohishi et al., 2018).
Leptospirosis
Leptospirosis has frequently been reported in otariid species, with the Pomona strain considered enzootic in the California sea lion population (Lloyd-Smith et al., 2007; Tryland et al., 2018). Among phocid seals, Pomona, Grippotyphosa, Bratislava, and Icterohaemorrhagiae serovars have been isolated (Cilia et al., 2021).
Seven samples of Caspian seal sera were suspected for the presence of antibodies against Leptospira interrogans serogroups Pomona, Hebdomadis (Kabura), Grippotyphosa, Tarassovi, Icterohaemorrhagiae. Four samples of the probable positives reacted with two different strains simultaneously (cross-reaction). Here the masked or low level of leptospira antibodies is possibly related to limitations of the leptospira microscopic agglutination test (LMAT), serum deterioration or antibodies lacking in the acute phase of infection. Caspian seal sera studied in Iran by LMAT revealed exposure to the serovars L. Pomona and L. Canicola (Namroodi et al., 2018). Our results for seals sampled in the northern part of the Caspian Sea suggest that Caspian seals are susceptible to more than two serovars of Leptospira interrogans depending on sampling site and season. Possible sources of environmental exposure to Leptospira spp. in Caspian seals are discussed by Namroodi et al. (2018).
Toxoplasmosis
Exposure to Toxoplasma gondii or Toxoplasma gondii–like protozoan parasites has been documented for a wide species range of Pinnipeds (Martins et al., 2021). Parasite-associated disease among phocid seals has been reported for harbor, Hawaiian monk (Monachus schauinslandi) seals and northern elephant seals (Miller et al., 2018). In our studies, the presence of Toxoplasma gondii antibodies was marginal in two serum samples. Studies in Iran demonstrated high antibody frequency in Caspian seals housed in rehabilitation, despite the absence of clinical signs of toxoplasmosis (Namroodi et al., 2018). Significant variability of toxoplasma antibody frequency among animals sampled in opposite parts of the Caspian Sea requires further analysis, paying attention to potential disease association.
Overall disease status of Caspian seals and implications for conservation management
Overall, the risks, profile and prevalence of pathogens in healthy, live, Caspian seals appears comparable to other wild phocid seal populations (Simeone et al., 2015). Where sequence data are available (e.g. for CDV, herpes, and adenoviruses) they suggest that most strains circulating are unique to Caspian seals. There are limited surveys of domestic and wild carnivore populations around the Caspian Sea, and so it is presently not possible to definitively say whether most pathogens in Caspian seals are endemic or spill over from terrestrial sources. However, the current Caspian seal population size is likely to be sufficient to sustain endemic infections for at least some viral pathogens (Kuiken et al., 2006). For influenza A, Caspian seals likely have frequent exposure to migratory birds at haul out sites, providing opportunities for spill over. This presents a future epizootic risk, should a strain highly pathogenic for seals arise.
Caspian seals came to global attention due to the mass mortalities from CDV 1997-2001 (Kuiken et al., 2006), but there have been no confirmed further CDV outbreaks since then despite evidence for its continuing circulation from this and other recent studies (Namroodi et al., 2018). Our observation of apparently asymptomatic infection in juveniles from 2008 suggest potential development of reduced virulence, which could partially account for the absence of recurrent large outbreaks. However, more research is needed to further characterise currently circulating CDV strains and understand future risks.
Mass strandings of Caspian seals, involving hundreds or thousands of individuals are a frequent occurrence (Wilson et al., 2014). For example, in December 2020 an estimated 300 to 2000 carcasses stranded along the coast of Dagestan, Russia News18.com, 2020. Such strandings are rarely investigated in depth due to difficulties in accessing remote locations, carcasses being in advanced states of decomposition, lack of resources, and there being few researchers with expertise in marine mammal pathology in the region (Authors’ observations). Where investigation of carcasses has taken place, the role of disease has often proven inconclusive, but evidence of fisheries related mortality is sometimes recorded (Dmitrieva et al., 2013). Stranding rates increase in the spring and autumn, which could be accounted for by natural mortality due to post-breeding/molting stress, increased disease transmission on haul out sites, and are also coincident peaks in fishing activity (Wilson et al., 2014).
Future research priorities should focus on determining the potential for the pathogens identified here to cause mortality and morbidity in the Caspian seal population, and how this might influence resilience to other conservation threats. The sources of future epizootic risks should be better quantified, and how these relate to anthropogenic environmental pressures such as human population density, pollution (sewage, agricultural and industrial effluent, heavy metals, hydrocarbons and persistent organic pollutants), habitat loss and degradation, and other potential sources of stress arising from human activities. Climate change and decline in the Caspian Sea sea level have potential to increase disease exposure or increase stress and reduce resilience to disease.
Building on this work, it is important to expand routine disease surveillance for Caspian seals, to facilitate both pre-emptive and reactive management of disease risks. At present there are no well-funded, routine, coordinated stranding investigation programs in any Caspian countries. It will be important to provide resources to support stranding investigations, and build capacity for veterinary pathology. This has potential to strengthen overall monitoring and understanding of ecosystem health for the Caspian Sea, and identify and respond to emerging conservation issues.
Data availability statement
The novel Canine Distemper Virus isolate Illumina short read data is deposited in the NCBI short read archive (https://www.ncbi.nlm.nih.gov/sra), with accession number PRJNA933140. Other sequence data is deposited in the NCBI Genbank nucleotide database (https://www.ncbi.nlm.nih.gov/nucleotide/) with accession numbers: Canine Distemper Virus assembled genomes, OQ442787, OQ442788; Phocine herpes virus (PhHV-1) Us2 gene contig, OQ511816; Caspian seal Adenovirus pol gene partial CDS PCR amplicon, OQ511815.
Ethics statement
The animal study was reviewed and approved by Institute of Microbiology and Virology Local Ethics Committee (Approval number: 2015-04-№1-ICPI).
Author contributions
Conceptualization: SG, AK. Data curation: AK, YK, KK, SG, MK, IC. Investigation: AK, KK, YK, SS, MK, SM, IC. Methodology: KK, AK, IC, SG. Resources: AK, KK, YK, SS, MB. Statistical analysis: MK, SM, AK, SG. Software: MK, SM. Writing original draft: AK, SG. Writing review & editing: SG. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Science Committee of the Ministry of Education and Science of the Republic of Kazakhstan (Grant No. AP08856073), and the University of Leeds. A grant from the UK government’s Darwin Initiative to SG (ref: 15-024), held at the University of Leeds provide support for fieldwork 2007-2008, and Agip KCO, NCOC and Tengizchevroil supported fieldwork 2008-2017.
Acknowledgments
We thank Timur Baimukanov, Lilia Dmitrieva, Mikhail Verevkin, Mart Jüssi, and Ivar Jüssi for help with capture and sampling of seals during fieldwork.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1087997/full#supplementary-material
References
Amin O. M., Heckmann R. A., Halajian A., El-Naggar A. M. (2011). The morphology of an unique population of corynosoma strumosum (Acanthocephala, polymorphidae) from the Caspian seal, pusa caspica, in the land-locked Caspian Sea using SEM, with special notes on histopathology. Acta Parasit 56, 438. doi: 10.2478/s11686-011-0070-6
Anthony S. J., St Leger J. A., Pugliares K., Ip H. S., Chan J. M., Carpenter Z. W., et al. (2012). Emergence of fatal avian influenza in new England harbor seals. mBio 3 (4), e00166-12. doi: 10.1128/mBio.00166-12
Batley K. C., Sandoval-Castillo J., Kemper C. M., Zanardo N., Tomo I., Beheregaray L. B., et al. (2021). Whole genomes reveal multiple candidate genes and pathways involved in the immune response of dolphins to a highly infectious virus. Mol. Ecol. 30 (23), 6434–6448. doi: 10.1111/mec.15873
Blanc A., Ruchansky D., Clara M., Achaval F., Le Bas A., Arbiza J. (2009). Serologic evidence of influenza a and В viruses in south American fur seals (Arctocephalus australis). J. Wildl Dis. 45, 519–521. doi: 10.7589/0090-3558-45.2.519
Bodewes R., Bestebroer T. M., van der Vries E., Verhagen J. H., Herfst S., Koopmans M. P., et al. (2015a). Avian influenza A(H10N7) virus-associated mass deaths among harbor seals. Emerg. Infect. Dis. 21 (4), 720–722. doi: 10.3201/eid2104.141675
Bodewes R., Contreras G. J., Garcia A. R., Hapsari R., van de Bildt M. W., Kuiken T., et al. (2015b). Identification of DNA sequences that imply a novel gammaherpesvirus in seals. J. Gen. Virology. 96, 1109–1114. doi: 10.1099/vir.0.000029
Bodewes R., Hapsari R., Rubio García A., Sánchez Contreras G. J., van de Bildt M. W., de Graaf M., et al. (2014). Molecular epidemiology of seal parvovirus, 1988–2014. PloS One 9 (11), e112129. doi: 10.1371/journal.pone.0112129
Bodewes R., Morick D., de Mutsert G., Osinga N., Bestebroer T., van der Vliet S., et al. (2013a). Recurring influenza В virus infections in seals. Emerging Infect. Dis. 19(3):511–512. doi: 10.3201/eid1903.120965
Bodewes R., Rubio García A., Brasseur S. M., Sanchez Conteras G. J., van de Bildt M. W. G., Koopmans M. P. G., et al. (2015c). Seroprevalence of antibodies against seal influenza A(H10N7) virus in harbor seals and Gray seals from the Netherlands. PloS One 10 (12), e0144899. doi: 10.1371/journal
Bodewes R., Rubio García A., Wiersma L. C. M., Getu S., Beukers M., Schapendonk C. M., et al. (2013b). Novel B19-like parvovirus in the brain of a harbor seal. PLoSONE 8 (11), e79259. doi: 10.1371/journal.pone.0079259
Bogomolni A. L., Gast R. J., Ellis J. C., Dennett M., Pugliares K. R., Lentell B. J., et al. (2008). Victims or vectors: a survey of marine vertebrate zoonoses from coastal waters of the Northwest Atlantic. Dis. Aquat Org 81, 13–38. doi: 10.3354/dao01936
Buonavoglia C., Decaro N., Martella V., Elia G., Campolo M., Desario C., et al. (2006). Canine coronavirus highly pathogenic for dogs. Emerging Infect. Dis. 12 (3), 492–494. doi: 10.3201/eid1203.050839
Burek K. A., Gulland F. M. D., Sheffield G., Beckmen K. B., Keyes E., Spraker T. R., et al. (2005). Infectious disease and the decline of steller sea lions (Eumetopias jubatus) in Alaska, USA: insights from serologic data. J. Wildlife Dis. 41, 512–524. doi: 10.7589/0090-3558-41.3.512
Callan R. G., Early C., Rida H., Hinshaw V. S. (1995). The appearance of H3 influenza viruses in seals. J. Gen. Virol. 76 (Part 1), 199–203. doi: 10.1099/0022-1317-76-1-199
Cilia G., Bertelloni F., Albini S., Fratini F. (2021). Insight into the epidemiology of leptospirosis: A review of leptospira isolations from “Unconventional” hosts. Animals. 11 (1), 191. doi: 10.3390/ani11010191
Cornwell H. J., Anderson S. S., Thompson P. M., Mayer S. J., Ross H. M., Pomeroy P. P., et al. (1992). The serological response of the common seal (Phoca vitulina) and the grey seal (Halichoerus grypus) to phocine distemper virus as measured by a canine distemper virus neutralisation test. Sci. Total Environ. 115 (1-2), 99–116. doi: 10.1016/0048-9697(92)90036-r
Danner G. R., McGregor M. (1998). Serologic evidence of influenza virus infection in a ringed seal (Phoca hispida) from Alaska. Mar. Mammal Sci. 14, 380–384. doi: 10.1111/j.1748-7692.1998.tb00730.x
De Benedictis P., Anderson T. C., Perez A., Viale E., Veggiato C., Tiozzo Caenazzo S., et al. (2010). A diagnostic algorithm for detection of antibodies to influenza a viruses in dogs in Italy (2006-2008). J. Vet. Diagn. Invest. 22, 914–920. doi: 10.1177/104063871002200610
De Boer G. F., Back W., Osterhaus A. D. M. E. (1990). An ELISA for detection of antibodies against influenza a nucleoprotein in humans and various animal species. Arch. Virol. 115, 47–61. doi: 10.1007/BF01310622
Deming A. C., Wellehan J. F. X., Colegrove K. M., Hall A., Luff J., Lowenstine L., et al. (2021). Unlocking the role of a genital herpesvirus, otarine herpesvirus 1, in California Sea lion cervical cancer. Animals. 11 (2), 491. doi: 10.3390/ani11020491
Di Guardo G., Marruchella G., Agrimi U., Kennedy S. (2005). Morbillivirus infections in aquatic mammals: a brief overview. J. Vet. Med. A Physiol. Pathol. Clin. Med. 52 (2), 88–93. doi: 10.1111/j.1439-0442.2005.00693.x
Dmitrieva L., Jüssi M., Jüssi I., Kasymbekov Y., Verevkin M., Baimukanov M., et al. (2016). Individual variation in seasonal movements and foraging strategies of a land-locked, ice-breeding pinniped. Mar. Ecol. Prog. Ser. 554, 241–256. doi: 10.3354/meps11804
Dmitrieva L., Kondakov A. A., Oleynikov E., Kydyrmanov A., Karamendin K., Kasimbekov Y., et al. (2013). Assessment of Caspian seal by-catch in an illegal fishery using an interview-based approach. PloS One 8 (6), e67074. doi: 10.1371/journal.pone.0067074
Dobson A., Foufopoulos J. (2001). Emerging infectious pathogens of wildlife. Phil. Trans. R. Soc Lond. B 356, 1001–1012. doi: 10.1098/rstb.2001.0900
Duignan P. J., Van Bressem M.-F., Baker J. D., Barbieri M., Colegrove K. M., De Guise S., et al. (2014). Phocine distemper virus: Current knowledge and future directions. Viruses. 6 (12), 5093–5134. doi: 10.3390/v6125093
Durymanova A. A., Dimov S. K., Kuznetsov V. N., Khuras'kin L. N., Zolotykh S. I., Shestopalov A. M. (2004). Antibodies to brucella in Caspian seals. Zh Mikrobiol Epidemiol. Immunobiol. 6), 99–101.
Erles K., Brownlie J. (2008). Canine respiratory coronavirus: an emerging pathogen in the canine infectious respiratory disease complex. Vet. Clin. North Am. Small Anim. Pract. 38 (4), 815–25, viii. doi: 10.1016/j.cvsm.2008.02.008
Forsyth M. A., Kennedy S., Wilson S., Eybatov T., Barrett T. (1998). Canine distemper virus in a Caspian seal. Vet. Rec. 143 (24), 662–664. doi: 10.1136/vr.143.24.662
Geraci J. R., Smith T. G. (1975). Functional hematology of ringed seals in the Canadian Arctic. J. Fish. Res.Board Can. 32, 2559–2564. doi: 10.1139/f75-302
Government of the Republic of Kazakhstan. (2004). Order of the minister of agriculture of the republic of Kazakhstan no. 652 of November 10, 2004–on approval of the rules for using animals, except rare and endangered species, for scientific, cultural-educational, up- bringing, and esthetic purposes, as well as for creation of zoological collections. Available at: http://adilet.zan.kz/rus/origins/V14F0010168.
Government of the Republic of Kazakhstan. (2004). Resolution of the government of the republic of Kazakhstan no. 1469 of December 31, 2004–on establishment of the rules for granting permission to use animals. Available at: https://online.zakon.kz/document/?doc_id=1052607#pos=0;0.
Goldstein T., Colegrove K. M., Hanson M., Gulland F. M. (2011). Isolation of a novel adenovirus from California sea lions zalophus californianus. Dis.Aquat.Organ. 94, 243–248. doi: 10.3354/dao02321
Goldstein T., Mena I., Anthony S. J., Medina R., Robinson P. W., Greig D. J., et al. (2013). Pandemic H1N1 influenza isolated from free-ranging northern elephant seals in 2010 off the central California coast. PloS One 8 (5), e62259. doi: 10.1371/journal.pone.0062259
Goodman S., Dmitrieva L. (2016). Pusa caspica. IUCN Red List Threatened Species 2016, e.T41669A45230700. doi: 10.2305/IUCN.UK.2016-1.RLTS.T41669A45230700.en
Grachev M. A., Kumarev V. P., Mamaev L. V., Zoryn V. L., Baranova L., Denikina N. N., et al. (1989) Distemper virus in baikal seals. Nature, 338, 209. doi: 10.1038/338209a0
Greig D. J., Gulland F. M., Smith W. A., Conrad P. A., Field C. L., Fleetwood M., et al. (2014). Surveillance for zoonotic and selected pathogens in harbor seals phoca vitulina from central California. Dis. Aquat Organ. 111 (2), 93–106. doi: 10.3354/dao02762
Gulland F. M. D., Hall A. J., Ylitalo G. M., Colegrove K. M., Norris T., Duignan P. J., et al. (2020). Persistent contaminants and herpesvirus OtHV1 are positively associated with cancer in wild California Sea lions (Zalophus californianus). Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.602565
Gulland F. M. D., Lowenstine L. J., Lapointe J. M., Spraker T., King D. P. (1997). Herpesvirus infection in stranded pacific harbor seals of coastal California. J. Wildlife Dis. 33 (3), 450–458. doi: 10.7589/0090-3558-33.3.450
Gulyaeva M., Sobolev I., Sharshov K., Kurskaya O., Alekseev A., Shestopalova L., et al. (2018). Characterization of avian-like influenza a (H4N6) virus isolated from Caspian seal in 2012. Virol. Sin. 33 (5), 449–452. doi: 10.1007/s12250-018-0053-y
Harder T. C., Harder M., Vos H., Kulonen K., Kennedy-Stoskopf S., Liess B., et al. (1996). Characterization of phocid herpesvirus-1 and-2 as putative alpha- and gammaherpesviruses of north American and European pinnipeds. J. Gen.Virol. 77, 27–35. doi: 10.1099/0022-1317-77-1-27
Harder T. C., Plötz P., Liess B. (1991). Detection of antibodies against European seal herpesviruses in sera of Antarctic seals. Polar Biol. 34, 43–47. doi: 10.1007/BF00233087
Have P., Nielsen J., Botner A. (1991). The seal death in Danish waters 1988. II. virological studies. Acta veterinaria Scandinavica 32, 211–219. doi: 10.1186/BF03546983
Heckmann R., Halajian A., El-Naggar A., Luus-Powell W. (2014). A histopathology study of Caspian seal (Pusa caspica) (Phocidae, Mammalia) liver infected with trematode, pseudamphistomum truncatum (Rudolphi, 1819) (Opisthorchidae, trematoda). Iran J. Parasitol. 9 (2), 266–275.
Hinshaw V. S., Bean W. J., Webster R. G., Rehg J. E., Fiorelli P., Early G., et al. (1984). Are seals frequently infected with avian influenza viruses? J. Virol. 51, 863–865. doi: 10.1128/jvi.51.3.863-865.1984
Hoelzer K., Parrish C. R. (2010). The emergence of parvoviruses of carnivores. Vet. Res. 41, 39. doi: 10.1051/vetres/2010011
Inoshima Y., Murakami T., Ishiguro N., Hasegawa K., Kasamatsu M. (2013). An outbreak of lethal adenovirus infection among different otariid species. Vet. Microb. 165, 455–459. doi: 10.1016/j.vetmic.2013.04.013
Ivanov V., Kamakin A., Ushivstsev V., Shiganova T., Zhukova O., Aladin N., et al. (2000). Invasion of the Caspian Sea by the comb jellyfish mnemiopsisleidyi (Ctenophora). Biol. Invasions 2, 255–258. doi: 10.1023/A:1010098624728
Jensen T., van de Bildt M., Dietz H. H., Andersen T. H., Hammer A. S., Kuiken T., et al. (2002). Another phocine distemper outbreak in Europe. Science. 297 (5579), 209. doi: 10.1126/science.1075343
Jo W. K., Osterhaus A. D., Ludlow M. (2018). Transmission of morbilliviruses within and among marine mammal species. Curr. Opin. Virol. 28, 133–141. doi: 10.1016/j.coviro.2017.12.005
Jo W. K., Peters M., Kydyrmanov A., van de Bildt M. W. G., Kuiken T., Osterhaus A., et al. (2019). The canine morbillivirus strain associated with an epizootic in Caspian seals provides new insights into the evolutionary history of this virus. Viruses 11 (10), 894. doi: 10.3390/v11100894
Kennedy S. (1998). Morbillivirus infections in marine mammals. J. Comp. Pathol. 119, 201–225. doi: 10.1016/S0021-9975(98)80045-5
Kennedy J. M., Earle J., Omar S., Abdullah H., Nielsen O., Roelke-Parker M. E., et al. (2019). Canine and phocine distemper viruses: Global spread and genetic basis of jumping species barriers. Viruses 11 (10), 944. doi: 10.3390/v11100944
Kennedy S., Kuiken T., Jepson P., Deaville R., Forsyth M., Barrett T., et al. (2000). Mass die-off of Caspian seals caused by canine distemper virus. Emerg. Infect. Dis. 6 (6), 637–639. doi: 10.3201/eid0606.000613
Kluge M., Campos F. S., Tavares M., de Amorim D. B., Valdez F. P., Giongo A., et al. (2016). Metagenomic survey of viral diversity obtained from feces of subantarctic and south American fur seals. PloS One 11 (3), e0151921. doi: 10.1371/journal.pone.0151921
Kuiken T., Kennedy S., Barrett T., van de Bildt M. W. G., Borgsteede F. H., Brew S. D., et al. (2006). The 2000 canine distemper epidemic in Caspian seals (Phoca caspica): Pathology and analysis of contributory factors. Vet. Pathol. 43, 321–338. doi: 10.1354/vp.43-3-321
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi: 10.1093/molbev/msy096
Kurochkin Y. V. (1975). Parasites of the Caspian seal pusa caspica. rapports et proces-verbaux des réunions. Conseil Int. pour l’éxploration la Mer 169, 363–365.
Kurochkin Y. V., Badamshin B. I. (1968). Finding of the seal lice echinophthirius horridus olfers. 1816 on the Caspian seal and the problem of origin of its parasite fauna. Tr Astrakh Zapovod 11, 199–209.
Laws R. M., Taylor R. J. F. (2009). A mass dying of crabeater seals, lobodon carcinophagus (Gray). J. Zool 129, 315–324. doi: 10.1111/j.1096-3642.1957.tb00296.x
Lebich M., Harder T. C., Frey H.-R., Visser I. K. G., Osterhaus A. D. M. E., Liess B. (1994). Comparative immunological characterization of type-specific and conserved b-cell epitopes of pinniped, felid and canid herpesviruses. Arch. Virol. 136, 335–347. doi: 10.1007/BF01321062
Lehnert K., Raga J. A., Siebert U. (2007). Parasites in harbour seals (Phoca vitulina) from the German wadden Sea between two phocine distemper virus epidemics. Helgol Mar. Res. 61, 239–245. doi: 10.1007/s10152-007-0072-9
Leidenberger S., Harding K., Härkönen T. (2007). Phocid seals, seal lice and heartworms: a terrestrial host-parasite system conveyed to the marine environment. Dis. Aquat Organ. 77 (3), 235–253. doi: 10.3354/dao01823
Lloyd-Smith J. O., Greig D. J., Hietala S., Ghneim G. S., Palmer L., St Leger J., et al. (2007). Cyclical changes in seroprevalence of leptospirosis in California sea lions: endemic and epidemic disease in one host species? BMC Infect. Dis. 7, 125. doi: 10.1186/1471-2334-7-125
Maness H. T. D., Nollens H. H., Jensen E. D., Goldstein T., LaMere S., Childress A., et al. (2011). Phylogenetic analysis of marine mammal herpesviruses. Veterinary Microbiol. 149, 23–29. doi: 10.1016/j.vetmic.2010.09.035
Markussen N. H., Have P. (1992). Phocine distemper virus infection in harp seals (Phoca groenlandica). Mar. Mamm. Sci. 8, 19–26. doi: 10.1111/j.1748-7692.1992.tb00121x
Martina B. E. E., Harder T. C., Osterhaus A. D. M. E. (2003). Genetic characterization of the unique short segment of phocid herpesvirus type 1 reveals close relationships among alphaherpesviruses of hosts of the order Carnivora. J. Gen. Virol. 84 (Pt 6), 1427–1430. doi: 10.1099/vir.0.19005-0
Martins M., Urbani N., Flanagan C., Siebert U., Gross S., Dubey J. P., et al. (2021). Seroprevalence of toxoplasma gondii in pinnipeds under human care and in wild pinnipeds. Pathogens. 10 (11), 1415. doi: 10.3390/pathogens10111415
Measures L. N. (2001). “Lungworms of marine mammals,” in Parasitic diseases of wild mammals. Eds. Samuel W. M., Pybus M. J., Kocan A. A. (Ames, Iowa, USA: Iowa State University Press), 279–300. doi: 10.1002/9780470377000.ch10
Measures L. N. (2018). “Helminths and parasitic arthropods,” in CRC Handbook of marine mammal medicine, 3d ed (Boca Raton: Taylor & Francis, CRC Press), 471–497.
Miller M., Shapiro K., Murray M. J., Haulena M., Raverty S. (2018). “Protozoan parasites of marine mammals,” in CRC Handbook of marine mammal medicine, 3d ed (Boca Raton: Taylor & Francis, CRC Press), 425–469. doi: 10.1201/9781315144931
Namroodi S., Rostami A., Majidzadeh-Ardebili K., Ghalyanchi Langroudi A., Morowati A. (2015). Detection of Arctic and European cluster of canine distemper virus in north and center of Iran. Veterinary Res. Forum. 6 (3), 199–204.
Namroodi S., Shirazi A. S., Khaleghi S. R., N. Mills J., Kheirabady V. (2018). Frequency of exposure of endangered Caspian seals to canine distemper virus, leptospira interrogans, and toxoplasma gondii. PloS One 13 (4), e0196070. doi: 10.1371/journal.pone.0196070
News18.com. Almost 300 endangered seals found dead along Caspian Sea coast, Russia begins probe. (2020) Available at: https://www.news18.com/news/buzz/almost-300-endangered-seals-found-dead-along-caspian-sea-coast-russia-begins-probe-3170486.html.
Nielsen O., Clavijo A., Boughen J. A. (2001). Serologic evidence of influenza a infection in marine mammals of Arctic Canada. J. Wildl Dis. 37 (4), 820–825. doi: 10.7589/0090-3558-37.4.820
Nollens H. H., Wellehan J. F., Archer L., Lowenstine L. J., Gulland F. M. (2010). Detection of a respiratory coronavirus from tissues archived during a pneumonia epizootic in free-ranging pacific harbor seals phoca vitulina richardsii. Dis. Aquat Organ. 90 (2), 113–120. doi: 10.3354/dao02190
Ohashi K., Miyazaki N., Tanabe S., Nakata H., Miura R., Fujita K., et al. (2001). Seroepidemiological survey of distemper virus infection in the Caspian Sea and in lake baikal. Vet. Microbiol. 82 (3), 203–210. doi: 10.1016/S0378-1135(01)00371-6
Ohishi K., Abe E., Amano M., Miyazaki N., Boltunov A., Katsumata E., et al. (2018). Detection of serum antibodies to brucella in Russian aquatic mammals. J. Vet. Med. Sci. 80 (11), 1696–1701. doi: 10.1292/jvms.18-0330
Ohishi K., Ninomiya A., Kida H., Park C. H., Maruyama T., Arai T., et al. (2002). Serological evidence of transmission of human influenza a and b viruses to Caspian seals (Phoca caspica). Microbiol. Immunol. 46 (9), 639–644. doi: 10.1111/j.1348-0421.2002.tb02746.x
Osterhaus A. D. M. E., Rimmelzwaan G. F., Martina B. E. E., Bestebroer T. M., Fouchier R. A. (2000). Influenza b virus in seals. Science 288, 1051–1053. doi: 10.1126/science.288.5468.1051
Osterhaus A. D., Yang H., Spijkers H. E., Groen J., Teppema J. S., van Steenis G. (1985). The isolation and partial characterization of a highly pathogenic herpesvirus from the harbor seal (Phoca vitulina). Arch. Virol. 86, 239–251. doi: 10.1007/BF01309828
Payungporn S., Phakdeewirot P., Chutinimitkul S., Theamboonlers A., Keawcharoen J., Oraveerakul K., et al. (2004). Single-step multiplex reverse transcription–polymerase chain reaction (RT-PCR) for influenza a virus subtype H5N1 detection. Viral Immunol. 17, 588–593. doi: 10.1089/vim.2004.17.588
Popov V. N., Korolev V. A., Nesterov E. N., Skorokhod L. A., Kupsha E. I. (1989). Anisakiasis in the Caspian seal. Parazitologiia. 23 (2), 178–181.
Puryear W. B., Keogh M., Hill N., Moxley J., Josephson E., Davis K. R., et al. (2016). Prevalence of influenza a virus in live-captured north Atlantic gray seals: a possible wild reservoir. Emerg. Microbes Infect. 5 (8), e81. doi: 10.1038/emi.2016.77
Puryear W., Sawatzki K., Bogomolni A., Hill N., Foss A., Stokholm I., et al. (2021). Longitudinal analysis of pinnipeds in the northwest Atlantic provides insights on endemic circulation of phocine distemper virus. Proc. Biol. Sci. 288 (1962), 20211841. doi: 10.1098/rspb.2021.1841
Qiagen Ltd. QIAamp viral RNA vini handbook. (2005). Available at: https://www.qiagen.com.
Repérant L. A., Rminielzwaan G. F., Kuiken T. (2009). Avian influenza viruses in mammals. Rev. Sei Tech. 28, 137–159. doi: 10.20506/rst.28.1.1876
Rosales S. M., Vega Thurber R. (2015). Brain meta-transcriptomics from harbor seals to infer the role of the microbiome and virome in a stranding event. PloS One 10 (12), e0143944. doi: 10.1371/journal.pone.0143944
Roth S. J., Tischer B. K., Kovacs K. M., Lydersen C., Osterrieder N., Tryland M. (2013). Phocine herpesvirus 1 (PhHV-1) in harbor seals from Svalbard, Norway. Vet. Microbiol. 164 (3-4), 286–292. doi: 10.1016/j.vetmic.2013.03.008
Saliki J. T., Lehenbauer T. W. (2001). Monoclonal antibody-based competitive enzyme-linked immunosorbent assay for detection of morbillivirus antibody in marine mammal sera. J. Clin. Microbiol. 39 (5), 1877–1881. doi: 10.1128/JCM.39.5.1877-1881.2001
Shin D. L., Siebert U., Lakemeyer J., Grilo M., Pawliczka I., Wu N. H., et al. (2019). Highly pathogenic avian influenza A(H5N8) virus in Gray seals, Baltic Sea. Emerg. Infect. Dis. 25 (12), 2295–2298. doi: 10.3201/eid2512.181472
Simeone C. A., Gulland F. M. D., Norris T., Rowles T. K. (2015). A systematic review of changes in marine mammal health in north America, 1972-2012: The need for a novel integrated approach. PloS One 10 (11), e0142105. doi: 10.1371/journal.pone.0142105
Sonne C., Lakemeyer J., Desforges J. P., Eulaers I., Persson S., Stokholm I., et al. (2020). A review of pathogens in selected Baltic Sea indicator species. Environ. Int. 137, 105565. doi: 10.1016/j.envint.2020.105565
Stephens N., Duignan P. J., Wang J., Bingham J., Finn H., Bejder L., et al. (2014). Cetacean morbillivirus in coastal indo-pacific bottlenose dolphins, Western Australia. Emerging Infect. Dis. 20 (4), 666–670. doi: 10.3201/eid2004.131714
Steuen S., Have - P., Osterhaus A. D. M. E., Arnemo J. M., Moustgaard A. (1994). Serological investigation of virus infections in harp seals (Phoca groenlandica) and hooded seals (Cystophora cristata). Veterinary Rec. 134, 502–503. doi: 10.1136/vr.134.19.502
The Economist. (2000). Oil and the seals of Kazakhstan. Available at: https://www.economist.com/asia/2000/06/01/oil-and-the-seals-of-kazakhstan.
Timoshenko Y. K. (1969). “Questions of rational use of stocks of the Caspian seal. Marine mammals,” in Materials of the 3rd All-Union Conference. (Vladivostok, Moscow: Morskiye mlekopitayushchiye). pp. 255–260.
Tryland M., Larsen A. K., Nymo I. H. (2018). “Bacterial infections and diseases,” in CRC Handbook of marine mammal medicine, 3d ed (Boca Raton: Taylor & Francis, CRC Press), 367–388. doi: 10.1201/9781315144931
Ulrich S. A., Lehnert K., Rubio-Garcia A., Sanchez-Contreras G. J., Strube C., Siebert U. (2016). Lungworm seroprevalence in free-ranging harbour seals and molecular characterisation of marine mammal MSP. Int. J. Parasitology: Parasites Wildlife 5 (1), 48–55. doi: 10.1016/j.ijppaw.2016.02.001
VanWormer E., Mazet J. A. K., Hall A., Gill V. A., Boveng P. L., London J. M., et al. (2019). Viral emergence in marine mammals in the north pacific may be linked to Arctic sea ice reduction. Sci. Rep. 9 (1), 15569. doi: 10.1038/s41598-019-51699-4
Volodina V. V., Grushko M. P. (2012). Invasion diseases of Caspian seal. Actual Questions Veterinary Biol. 2 (14), 25–29.
Vylegzhanin A. F. (1965). Diplococcal disease of Caspian seals. (Moscow: Rybnoe hozyajstvo) 41, 31–33.
Vylegzhanin A. F. (1967). Infectious diseases of fish and seals in the Caspian Sea (ulcerative and dermatological, fish disease, diplococcal and vibrionic disease of seals) Thesis for a degree doctor of veterinary sciences. (Kharkov: Kharkiv Zooveterinary Institute).
Wellehan J. F., Johnson A. J., Harrach B., Benkö M., Pessier A. P., Johnson C. M., et al. (2004). Detection and analysis of six lizard adenoviruses by consensus primer PCR provides further evidence of a reptilian origin for the atadenoviruses. J. Virol. 78 (23), 13366–13369. doi: 10.1128/JVI.78.23.13366-13369.2004
Wilson S. C., Dolgova E., Trukhanova I., Dmitrieva L., Crawford I., Baimukanov M., et al. (2017a). Breeding behavior and pup development of the Caspian seal, pusa caspica. J. Mammalogy 98 (1), 143–153. doi: 10.1093/jmammal/gyw176
Wilson S. C., Eybatov T. M., Amano M., Jepson P. D., Goodman S. J. (2014). The role of canine distemper virus and persistent organic pollutants in mortality patterns of Caspian seals (Pusa caspica). PloS One 9 (7), e99265. doi: 10.1371/journal.pone.0099265
Wilson S. C., Trukhanova I., Dmitrieva L., Dolgova E., Crawford I., Baimukanov M., et al. (2017b). Assessment of impacts and potential mitigation for icebreaking vessels transiting pupping areas of an ice-breeding seal. Biol. Conserv. 214, 213–222. doi: 10.1016/j.biocon.2017.05.028
Wright E. P., Waugh L. F., Goldstein T., Freeman K. S., Kelly T. R., Wheeler E. A., et al. (2015). Evaluation of viruses and their association with ocular lesions in pinnipeds in rehabilitation. Veterinary Ophthalmol. 18 Suppl 1, 148–159. doi: 10.1111/vop.12235
Yamnikova S. S., Gambaryan A. S., Fedyakina I. T., Shilov A. A., Ye.S. P., Lvov D. K. (2001). Monitoring of influenza a virus circulation in a population of wild birds in northern Caspian region. Vopr. virusol 4, 39–43.
Zarnke R. L., Harder T. C., Vos H. W., Ver Hoef J. M., Osterhaus A. D. (1997). Serologic survey for phocid herpesvirus-1 and -2 in marine mammals from Alaska and Russia. J. Wildlife Dis. 33, 459–465. doi: 10.7589/0090-3558-33.3.459
Keywords: Caspian seal (Pusa caspica), marine mammals, one health, pinniped, disease ecology, disease surveillance, wildlife disease, wildlife health
Citation: Kydyrmanov A, Karamendin K, Kassymbekov Y, Kumar M, Mazkirat S, Suleimenova S, Baimukanov M, Carr IM and Goodman SJ (2023) Exposure of wild Caspian seals (Pusa caspica) to parasites, bacterial and viral pathogens, evaluated via molecular and serological assays. Front. Mar. Sci. 10:1087997. doi: 10.3389/fmars.2023.1087997
Received: 02 November 2022; Accepted: 07 March 2023;
Published: 17 April 2023.
Edited by:
Jose Carlos Báez, Spanish Institute of Oceanography (IEO), SpainReviewed by:
Nicole L. Gottdenker, University of Georgia, United StatesWendy Blay Puryear, Tufts University, United States
Copyright © 2023 Kydyrmanov, Karamendin, Kassymbekov, Kumar, Mazkirat, Suleimenova, Baimukanov, Carr and Goodman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aidyn Kydyrmanov, a3lkeXJtYW5vdkB5YW5kZXgua3o=; Simon J. Goodman, cy5qLmdvb2RtYW5AbGVlZHMuYWMudWs=
†ORCID: Aidyn Kydyrmanov, orcid.org/0000-0002-8374-6128
Kobey Karamendin, orcid.org/0000-0003-0829-3330
Yermukhammet Kassymbekov, orcid.org/0000-0002-8773-4984
Marat Kumar, orcid.org/0000-0001-6148-8204
Shynar Mazkirat, orcid.org/0000-0003-1768-3779
Symbat Suleimenova, orcid.org/0000-0003-4107-0681
Mirgaliy Baimukanov, orcid.org/0000-0002-5039-3901
Ian M. Carr, orcid.org/0000-0001-9544-1068
Simon J. Goodman, orcid.org/0000-0003-4118-8575
 Aidyn Kydyrmanov
Aidyn Kydyrmanov Kobey Karamendin
Kobey Karamendin Yermukhammet Kassymbekov
Yermukhammet Kassymbekov Marat Kumar
Marat Kumar Shynar Mazkirat
Shynar Mazkirat Symbat Suleimenova
Symbat Suleimenova Mirgaliy Baimukanov
Mirgaliy Baimukanov Ian M. Carr
Ian M. Carr Simon J. Goodman
Simon J. Goodman