
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 24 February 2023
Sec. Marine Pollution
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1086166
Introduction: In marine ecosystems, microbial communities are important drivers of material circulation and energy flow. The complex interactions between phytoplankton and bacterial communities constitute one of the most crucial ecological relationships in the marine environment. Inorganic nitrogen can affect the type of relationship between algae and bacteria. However, the quantitative relationship between the bacterial communities, inorganic nitrogen, and phytoplankton remains unclear.
Methods: Under laboratory conditions, we altered the forms (nitrate and ammonium) and amounts of nitrogen sources to study the dynamics of bacterial biomass, diversity, and community structure in the phycosphere of the marine model species Phaeodactylum tricornutum. The bacterial community structure during P. tricornutum growth was analyzed using Illumina HiSeq sequencing of 16S rDNA amplicons.
Results: The results indicated that inorganic nitrogen concentration was the main factor promoting P. tricornutum biomass growth. The change in the algal biomass would significantly increase the phycosphere bacterial biomass. The bacterial biomass in the algal-bacteria co-culture system was 1.5 ~ 5 times that of the conditional control groups without microalgae under the same culture conditions. The variation of P. tricornutum biomass also affected the bacterial communities in the phycosphere. When P. tricornutum was in the exponential phase (96 ~ 192 h), the bacterial community structure differed between the high- and low-concentration groups. The difference in the bacterial communities over time in the high-concentration groups was more prominent than in the low-concentration groups. Under high-concentration groups (HA and HN), the relative abundance of Marivita and Marinobacter, engaged in the transformation of aquatic inorganic nitrogen, gradually decreased with time. However, the relative abundance of Oceanicaulis, closely related to algal growth, gradually increased with time.
Discussion: The above phenomena might be related to the change in P. tricornutum biomass. Our results explain when and how the phycosphere bacterial communities responded to algal biomass variations. The study provides a foundation for the quantitative relationship among nutrients, microalgae, and bacteria in this system.
Microorganisms are an important part of marine ecosystems. They are major participants in biogeochemical cycles and energy flows. Bacteria complex community structure, functions, interactions, and dynamic changes are crucial to the maintenance of ecological function (Grossart et al., 2020; Frazão et al., 2021). In marine habitats, environmental factors can affect the construction and function of microbial communities through processes like selection, drift, dispersal, and diversification (Grossart et al., 2020). The interaction between phytoplankton and bacteria also affected the community assembly. The phycosphere is a diffusion-limited layer that surrounds a phytoplankton cell. It is enriched in organic molecules secreted by phytoplankton and increases the possibility of interactions between phytoplankton and bacteria (Seymour et al., 2017). Bacteria that colonize the phycosphere are known as phycosphere bacteria. These bacteria include attached and free-living bacteria (Seymour et al., 2017). Park et al. (2020) discovered that the significant increase in phytoplankton abundance might be a key cause of the increase in bacterial abundance and changes in the communities along the southern Korean coast. Phytoplankton can secrete metabolites to provide nutrients for the phycosphere bacteria. Phytoplankton can also contribute to the establishment of bacterial communities that support microalgal growth (Fu et al., 2020). This nutrient-based bacterial community construction mechanism may be the foundation for the evolution and maintenance of microbial communities. The relationship between phytoplankton and bacteria is also based on the supply of resources (Grossart et al., 2020). Specifically, bacteria in the phycosphere can secrete bioactive substances like polyunsaturated fatty acids, vitamins, phytohormones, antibiotics, and quorum-sensing signaling molecules. These substances can promote algal growth or strengthen their defense mechanisms. They can also release antialgal compounds to lyse algal cells and cause resource competition. At the same time, phytoplankton can also secrete organic carbon compounds and other metabolites that affect the growth, physiological processes, and community components of bacteria in the phycosphere (Grossart et al., 2020; Variem and Kizhakkedath, 2021). The complex interactions between microalgae and bacteria constitute one of the most crucial ecological relationships in the aquatic environment (Grossart et al., 2020). They can regulate the primary productivity of the marine system and the stability of the food web through cascading bottom-up influences on ecosystem-scale processes (Seymour et al., 2017). Inter-organism association is crucial to shaping aquatic ecosystem function (Variem and Kizhakkedath, 2021).
When nutrient concentrations are relatively stable in the environment, bacterial growth, physiological states, and ecological strategies change over time. The time required for the bacterial metabolism process changes and algal cell division is long (from hours to days, Brumley et al., 2020). Consequently, it is vital to research the long-term changes in phytoplankton-bacteria relationships, and short-term kinetic investigations are also necessary to reveal these interactions. Steichen et al. (2020) used outdoor reactors to study the changes in the bacterial composition of the Chlorella sorokiniana phycosphere during different growing seasons. They discovered that the diversity of phycosphere bacteria increased with time during the culture cycle of different reactors. The community composition of phycosphere bacteria was closely related to the efficiency of microalgal biomass production and microalgal health (Steichen et al., 2020). To some extent, algae can explain shifts in the phycosphere bacterial communities better than environmental factors. Bacterial communities shift and eventually stabilize over time. The effect of algae on bacteria lasts for dozens of generations and enhances their tolerance to algae. Meanwhile, some phycosphere bacteria can modify algal physiology to maximize bacterial growth (Jackrel et al., 2021). Aylward et al. (2015) reported that phytoplankton and phycosphere bacteria exhibit similar transcriptional patterns in coastal and pelagic ecosystems, and these patterns have clear diel cycles. Linz et al. (2020) also observed a similar pattern in lakes with different trophic statuses. Namely, genes involved in photosynthesis, sugar transport, and carbon fixation have prominent diel cycles, and all three metabolic processes occur in a relationship between phytoplankton and heterotrophic bacteria. Bacterioplankton also showed significant temporal heterogeneity in different stages of harmful algal blooms. The succession of microbial communities is determined by the interactions between environmental parameters and phytoplankton-bacteria relationships (Zhou et al., 2020).
Nutrient distribution in marine ecosystems is highly structured, varying with depth and time, across the globe (Brumley et al., 2020). Nitrogen is the principal limiting resource in marine ecosystems compared to other nutrients (Rogato et al., 2015). Investigating the impact of nitrogen on the relationship between algae and bacteria is of great significance for maintaining and restoring marine ecological balance. Phytoplankton can uptake nitrogen in its mineral and organic forms, and bacteria can also use both inorganic and organic nitrogen. There is a complex relationship between algae and bacteria for nitrogen that can shift from a competitive to a mutualistic relationship, driven by mineral nitrogen availability (Le Chevanton et al., 2016). Amin et al. (2015) showed that Sulfitobacter (SA11) could uptake nitrate and release ammonium for microalgal growth in co-culture with Pseudo-nitzschia multiseries under nitrate supersaturation. P. multiseries preferentially utilizes bacterial-derived ammonium for growth rather than exogenous nitrate. However, bacterial-derived ammonium was not the main factor in promoting the growth of P. multiseries (Amin et al., 2015). Compared to nitrate, algae use less energy to absorb ammonium, and ammonium is easier to utilize (Markou et al., 2014). Therefore, we hypothesized that microalgae might preferentially utilize ammonium in the aquatic environment. If ammonium becomes scarce, phycosphere bacteria may convert nitrate to ammonium for algal growth. However, in previous studies, we found that nitrogen limitation (i.e., nitrogen being relatively insufficient compared to other elements in solution) can inhibit microalgal growth. Algal biomass may be the primary factor influencing bacterial communities at 168 h (this time point corresponded to the exponential phase of microalgal growth in N-sufficient conditions and the stationary phase in N-deficient conditions). The content and shape of inorganic nitrogen have no apparent influence on the bacterial community composition, and Proteobacteria are the dominating flora in phycosphere bacteria (Shi et al., 2018). Proteobacteria (primarily classes Alphaproteobacteria and Gammaproteobacteria) are one of the most widely distributed and abundant microbial groups in marine ecosystems. They are also an essential part of phycosphere bacteria and include phototrophs, autotrophs, and heterotrophs. Proteobacteria are involved in sulfur, methane, and hydrogen oxidation, sulfate reduction, and denitrification (Zhou et al., 2020). Interactions between specific taxa (e.g., Marivita, Oceanicaulis, and Marinobacter) and algae vary with the microalgal growth cycle (Bolch et al., 2017). Sun et al. (2021) found a strong correlation between the relative abundance of Marivita and the inorganic nitrogen content. This result might be a critical factor influencing nitrogen transformation in water. The research (Guidi et al., 2018; Deschaseaux et al., 2019) revealed that the relative abundance of Oceanicaulis varied with different growth periods of phytoplankton, which may be associated with changes in the microalgal physiological status. Marinobacter biomass is linked to the amount of inorganic nitrogen in aqueous environments (Zhang et al., 2016), and the gene abundance of its assimilating nitrate reductase is proportional to the nitrate concentration in water (Allen et al., 2005). Marinobacter has growth-promoting potential toward their algal host (Yang et al., 2021). When Ostreococcus tauri was in the latency and decline phases, Marinobacter abundance was higher. Bacterial diversity (mostly Marinobacter) was also higher in the exponential, stationary, and decline phases of the microalgae. The type of algal-bacterial interaction might vary with environmental circumstances (Lupette et al., 2016).
In recent years, more studies have been conducted on the link between nutrients, phytoplankton, and bacteria. Several scholars employed axenic microalgae and artificially added single bacterial species as study subjects. Although it is a better approach to conduct research with a controlled microalgal-bacteria consortium, skepticism about the experimental results is unavoidable compared to the actual microalgal-bacteria state. Microalgal cells that were damaged during separation may also have affected the experimental outcomes (Vu et al., 2018). In addition, lab-cultured algal-bacterial symbiotic consortia have been acknowledged and widely employed by researchers in this field due to their stable, rich species, nutrient sufficiency, and lack of interference from exogenous organisms (Amin et al., 2015; Shi et al., 2018; Shibl et al., 2020). However, it is well known that the bacterial communities in the phycosphere in a realistic marine environment are more complex than those in a laboratory. Kublanovskaya et al. (2020) discovered Acidobacteria, Actinobacteria, Bacteroidetes, Chlamydiae, Chloroflexi, Proteobacteria, and others in the Haematococcus lacustris phycosphere of ambient samples. In the laboratory, however, H. lacustris cultures included exclusively Actinobacteria and Proteobacteria.
Taken together, phycosphere bacteria and phytoplankton have different sensitivities to changes in environmental factors, resulting in different responses. This difference may affect the relationship between algae and bacteria, resulting in changes in the phycosphere bacterial communities over time. Prior research has demonstrated that the changes in algal biomass caused by nitrogen concentration may lead to significant changes in the phycosphere bacterial communities at a certain time point (Shi et al., 2018). However, the temporal dynamics of inorganic nitrogen, microalgae, and associated phycosphere bacteria are poorly understood. Lab-cultured algal-bacterial symbiotic consortia can reflect the relationship between microalgae and bacteria in the marine environment to a certain extent. As a result, we investigated the changes of lab-cultured phycosphere bacterial communities over time under different forms and concentrations of inorganic nitrogen. The aim of this study was to understand when and to what extent the phycosphere bacterial communities respond to changes in algal biomass, as well as the changing trends and interrelationship of microalgae and bacterial communities over time. This study gives us a basis for learning more about how nutrients, phytoplankton, and bacteria in this system interact quantitatively.
The marine model organism Phaeodactylum tricornutum was selected as the experimental algal species. It was obtained from the Marine Algae Stock Culture Collection Center (MASCCC, Qingdao, China), and its strain number was MASCC17. The algae were isolated from the Yellow Sea in China without any sterile treatment. The bacteria were the original phycosphere bacteria of P. tricornutum, and no bacteria were added or specially manipulated in this experiment.
The culture media for all experimental groups were sterilized artificial seawater (Guillard, 1975) and f/2 modified medium (Guillard and Ryther, 1962). The form and concentration of the nitrogen sources are shown in Table 1. To simulate the total amount of inorganic nitrogen in offshore waters, the concentrations of low-nitrogen experimental groups were set to 40 μmol/L (Ji, 2011). According to the results of previous studies, we found that 500 μmol/L was the optimum content for P. tricornutum growth in our laboratory conditions. The bacterial communities did not change significantly under the same concentration and different form conditions (Shi et al., 2018). On the other hand, there were differences in the utilization of different forms of inorganic nitrogen by algae, and their effects on bacteria might also vary. As a result, this experiment established two methods of adding nitrogen sources: ammonium alone and nitrate alone, neither of which set the cultivation mode for mixed nitrogen sources. The microalgal-bacteria consortium was domesticated for 30 days under the same conditions as the experimental settings to ensure they adapted to our experimental conditions. During the acclimation period, the freshly sterile media replacement for three-quarters of the mother liquor was performed every 10 days. To avoid external contamination, the domestication and all experiments were conducted under strict sterile conditions, following the biological test criteria in the marine monitoring specification (AQSIQ and SAC, 2008).
Furthermore, two control groups were set up in this experiment. The blank control groups evaluated the influence of inorganic nitrogen on the abundance of algae and bacteria. There were similar bacterial communities between the blank control groups and the low-nitrogen experimental groups (Shi et al., 2018). Thus, this paper no longer analyzes the bacterial community composition of the blank control groups. In the condition control groups, ultrasonic vibration and filtration were used to get rid of the algae in the consortium of domesticated microalgae and bacteria.
Following acclimation, the media were immediately used as mother liquor for the experiment. Each trial (including experimental groups and control groups) consisted of three replicates and was cultured at an 800 mL volume in glass conical flasks (1 L). These flasks were placed in an artificial climate box (BIC-400, Shanghai Boxun) at (22 ± 1) °C on a 12 h (light): 12 h (dark) cycle. The light intensity was 80 μmol/(m2·s). The initial algal density was about 4 × 104 cells/mL. From the day of inoculation, the biomass of algae and bacteria in the experimental and two control groups was measured every 2 days until the nitrogen source was exhausted, across 12 days total.
Algal cells were counted by microscopy (ECLIPSE Ci-S; Nikon, Japan). Samples of 200 μL were fixed with Lugol and counted in a hemocytometer. Each sample must have at least 400 cells counted to assure accuracy.
We counted the phycosphere bacteria using 4, 6-diamidino-2-phenylindole (DAPI) fluorescence microscopy (Eclipse 80i; Nikon, Japan). The samples were fixed with 5% (v/v) formalin. At least 10-20 randomly chosen fields were counted, and bacterial biomass was calculated according to Shi et al. (2018).
According to earlier studies (Shi et al., 2018), the lag phase, exponential phase, end exponential phase, and stationary phase of P. tricornutum occurred at 24, 96, 192, and 288 h, hence the experimental groups were sampled at these four-time points. The conditional control groups were sampled according to the lag, exponential, and stationary phases of algal growth, namely 24, 96, and 288 h. Samples were filtered on a sterilized 0.22 μm polycarbonate membrane (Merkmillipore Inc., USA) to analyze the phycosphere bacterial communities using 16S rDNA amplicon paired-end sequencing according to the method described by Shi et al. (2018). Total bacterial genome DNA was extracted from filters with a FastDNA SPIN Kit for 160 Soil (MP Biomedicals, USA) and purified with a TaKaRa MiniBEST Agarose Gel DNA Extraction Kit (Takara Bio Inc., Japan). The sequencing platform was the Illumina HiSeq2500 platform (Novogene Co., Ltd., China). The sequencing region is the 16S rDNA V4 region.
We relied on the size of the effect estimates and 95% confidence intervals when reporting the differential analysis (Gardner and Altman, 1986; Laslett et al., 2022). They were calculated by IBM SPSS 22. The correlation analysis of algal and bacterial biomass was performed by Spearman correlation analysis using IBM SPSS 22. The data analysis for the phycosphere bacterial communities was performed as described previously (Shi et al., 2018).
We used the Shannon-Weaver index to evaluate the alpha diversity of the bacterial communities. To investigate the beta diversity of bacterial communities, we ran the unweighted pair-group method with arithmetic means (UPGMA), with the weighted Unifrac distance. To explore species with statistical differences among different samples, we used the linear discriminant analysis (LDA) effect size (LefSe). All gene sequences obtained in this study have been deposited with the National Center for Biological Information (accession number: PRJNA803332).
In the algal-bacterial co-culture system, P. tricornutum grew fast from 0 h under low concentration (LA and LN) and reached a stationary phase at 144 h (LA) and 96 h (LN), respectively (Figure 1A). Maximum algal density was 1.47×106 cells/mL (LA) and 1.70×106 cells/mL (LN). The bacterial biomass of the LA groups also rose and reached a stabilization growth phase at 144 h, with a maximum bacterial density of 4.51 × 106 cells/mL. Meanwhile, the bacterial biomass in the LN groups kept growing, with a maximum bacterial density of 4.52 × 106 cells/mL (Figure 1B). Compared with the blank control groups, the density of algae and bacteria in the low-concentration experimental groups increased significantly (the effect was outside zero, Supplementary Figure 1). The algae density of the low-concentration experimental groups was 3–13 times (LA) and 6–15 times (LN) of the control groups, whereas the bacterial density was 1–2 times that of the control groups (LA and LN).
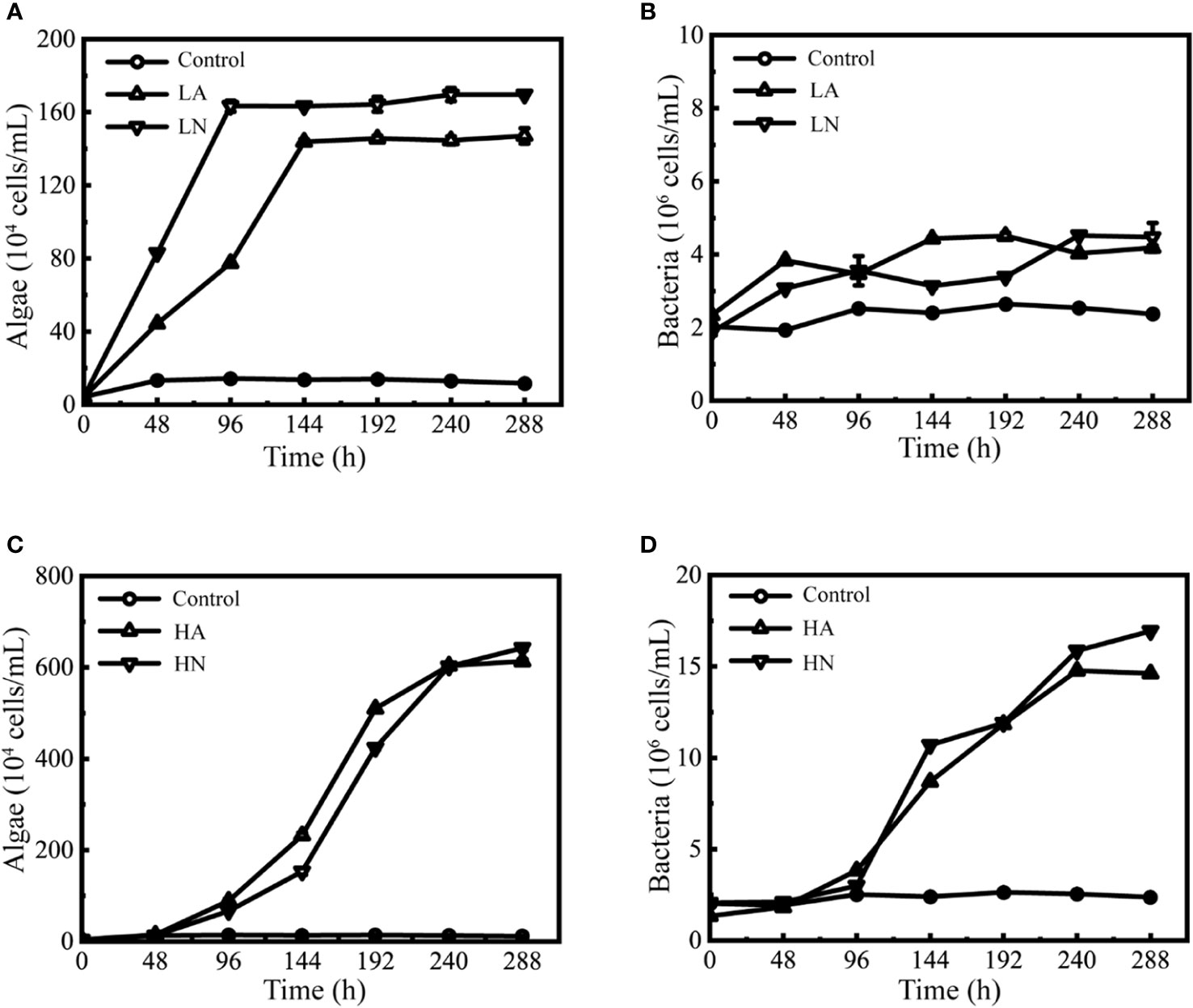
Figure 1 Growth curves of Phaeodactylum tricornutum and associated phycosphere bacteria co-cultured in different conditions. Biomass of P. tricornutum under different conditions (A, C). Biomass of the phycosphere bacteria under different conditions (B, D). The meaning of each group abbreviation is shown in Table 1. Error bars indicate the standard deviation (SD) of three independent cultures.
Under high concentration (HA and HN) conditions, P. tricornutum grew fast from 48 h until it entered a stationary phase at 240 h (Figure 1C). The maximum algal density was 6.13×106 cells/mL (HA) and 6.42×106 cells/mL (HN), respectively. The bacteria also grew rapidly in this state, and their biomass quickly increased from 96 h to 240 h (Figure 1D). The maximum bacterial density was 1.48×107 cells/mL (HA) and 1.69×107 cells/mL (HN), respectively. Compared with the blank control groups, the algal-bacterial density in the high-concentration experimental groups increased significantly (the effect was outside zero, Supplementary Figure 1). The algae density of the high-concentration experimental groups was 6 ~ 53 times (HA) and 5 ~ 55 times (HN) that of the control groups, and the bacterial density was 1 ~ 6 times (HA) and 1 ~ 7 times (HN) that of the control groups.
The statistical results revealed that there were significant differences in the algal-bacterial density under different nitrogen concentrations (the effect was outside zero, Supplementary Figure 2) but not under various forms (the effect was near zero, Supplementary Figure 2). The Spearman correlation analysis (Table 2) showed that P. tricornutum growth was significantly correlated with bacterial growth in all treatment groups and blank control groups. Under experimental conditions, algal-bacterial correlation coefficients exceeded 0.6, indicating a significant positive connection.

Table 2 Results of a correlation analysis between Phaeodactylum tricornutum abundance and the phycosphere bacterial abundance in different culture systems.
In this work, a total of 3,801,259 high-quality sequences from the microalgal-bacteria consortium were acquired, with an average length of 253 bp (base pairs) (Supplementary Table 1). We obtained a total of 1,468 OTUs at 97% sequence similarity. Rarefaction curves showed that the sequencing could represent most of the microbial diversity in each sample (Supplementary Figure 3).
Based on the species annotation analysis of the SILVA database, the obtained 1468 OTUs were classified into 27 phyla, 53 classes, 115 orders, 175 families, 315 genera, and 119 identifiable species (the rest were unidentifiable species). The results demonstrated that the proportion of Proteobacteria in all treatment groups was the highest, accounting for 79.41% of all sequences, followed by Bacteroidetes and Firmicutes, which accounted for 10.05% and 8.09%, respectively. In Proteobacteria, Alphaproteobacteria and Gammaproteobacteria accounted for 63.08% and 16.11% of the total sequences, respectively, as shown in Figure 2.
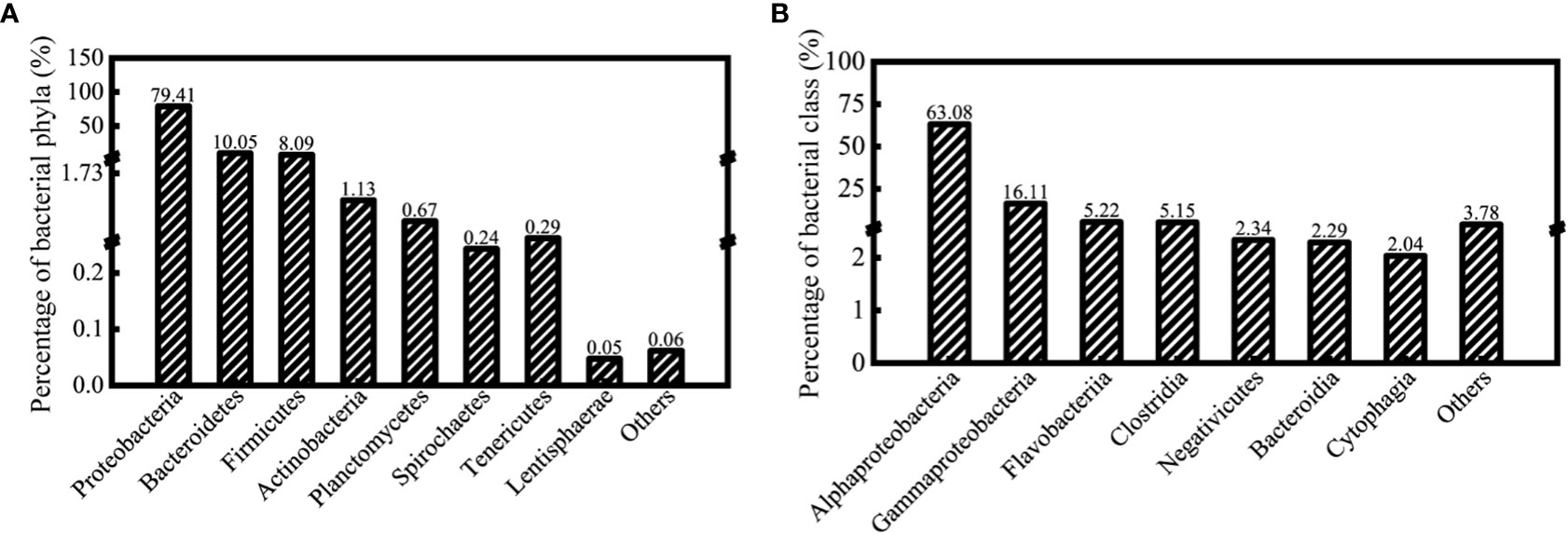
Figure 2 The operational taxonomic units (OTUs) identified under the algal-bacteria co-culture system that was assigned to major bacterial phyla (A) and classes (B) (97% similarity cutoff).
To investigate the differences in bacterial community structure across various samples, we used UPGMA with the weighted Unifrac distance, namely beta diversity analysis. The results are presented in Figure 3.
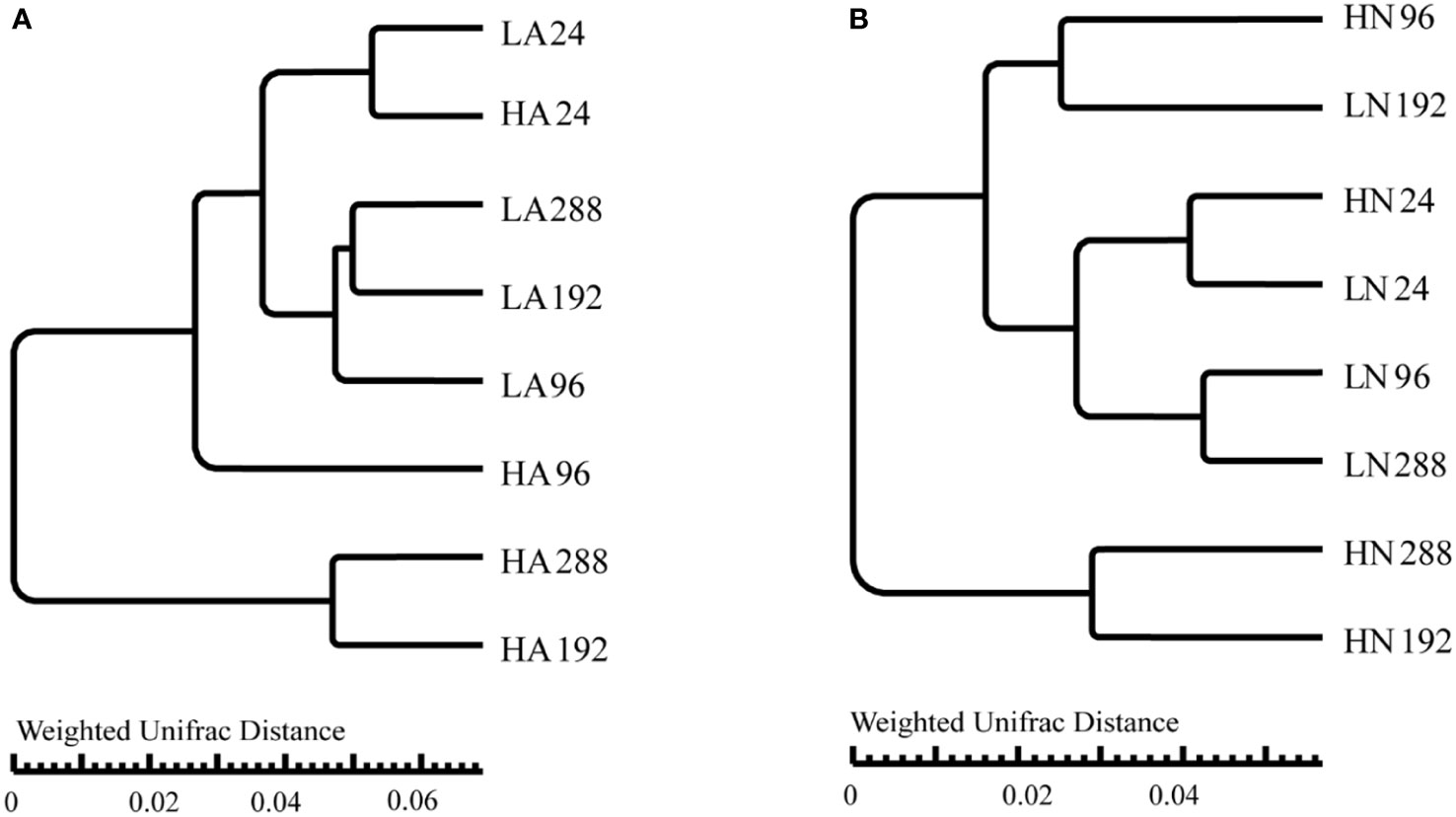
Figure 3 UPGMA analysis of different treatment groups under the algal-bacteria co-culture system, using the weighted Unifrac distance (each leaf represents three independent experiments). (A) UPGMA analysis of the comparison between the low concentration (LA) and the high concentration (HA) groups with ammonium. (B) UPGMA analysis of the comparison between the low concentration (LN) and the high concentration (HN) groups with nitrate.
Under the algal-bacteria co-culture system, we compared samples (LA, LN, HA, and HN) between different time points in each group. All the time points in the LA groups belonged to different subclasses except those at 192 and 288 h. Except for the 96 and 288 h, all of the remaining time points in the LN groups belonged to distinct subclasses. In the HA and HN experimental groups, all time points except 192 and 288 h were divided into various subclasses. The HA and HN groups had farther-apart samples at each time point than the LA and LN groups.
When comparing treatment groups with the same nitrogen form and varied concentrations (LA vs. HA and LN vs. HN), bacterial community structures at all times except 24 h belonged to various subclasses (Figure 3). Additionally, the comparison of forms at the same concentration (Supplementary Figure 4) revealed that, except for the 24 h phycosphere bacteria, which belonged to a category, all others belonged to different subcategories (LA vs. LN). HA vs. HN revealed that the bacterial communities at 24 and 96 h were clustered into one category, respectively, but at 192 and 288 h, they were clustered into different subcategories.
In conclusion, in the algal-bacteria consortium of this experiment, time and nitrogen concentration are the main factors that cause differences in phycosphere bacterial communities among treatment groups. In contrast, the nitrogen form has less of an effect. Under low nitrogen conditions (LA and LN), the bacterial community structure was different at times. Still, the distance between the bacterial community structures was closer than in the high nitrogen groups, showing that the bacterial community composition was more similar. Under high nitrogen conditions (HA and HN), the bacterial communities changed from 24 to 192 h, but they were similar after 192 h.
The Shannon-Weaver index is commonly used to quantify bacterial alpha diversity, representing the richness and variety of bacterial communities. The Shannon-Weaver diversity index of all treatment groups ranged from 3.20 to 4.39 (Figure 4). The Shannon diversity index of phycosphere bacteria increased gradually over time under the same growth conditions. Under low nitrogen circumstances, the diversity index grew by 11% (LA) and 18% (LN), and the difference was not obvious. Under high nitrogen conditions, the diversity index increased by 24% (HA) and 29% (HN), and the difference was more obvious.
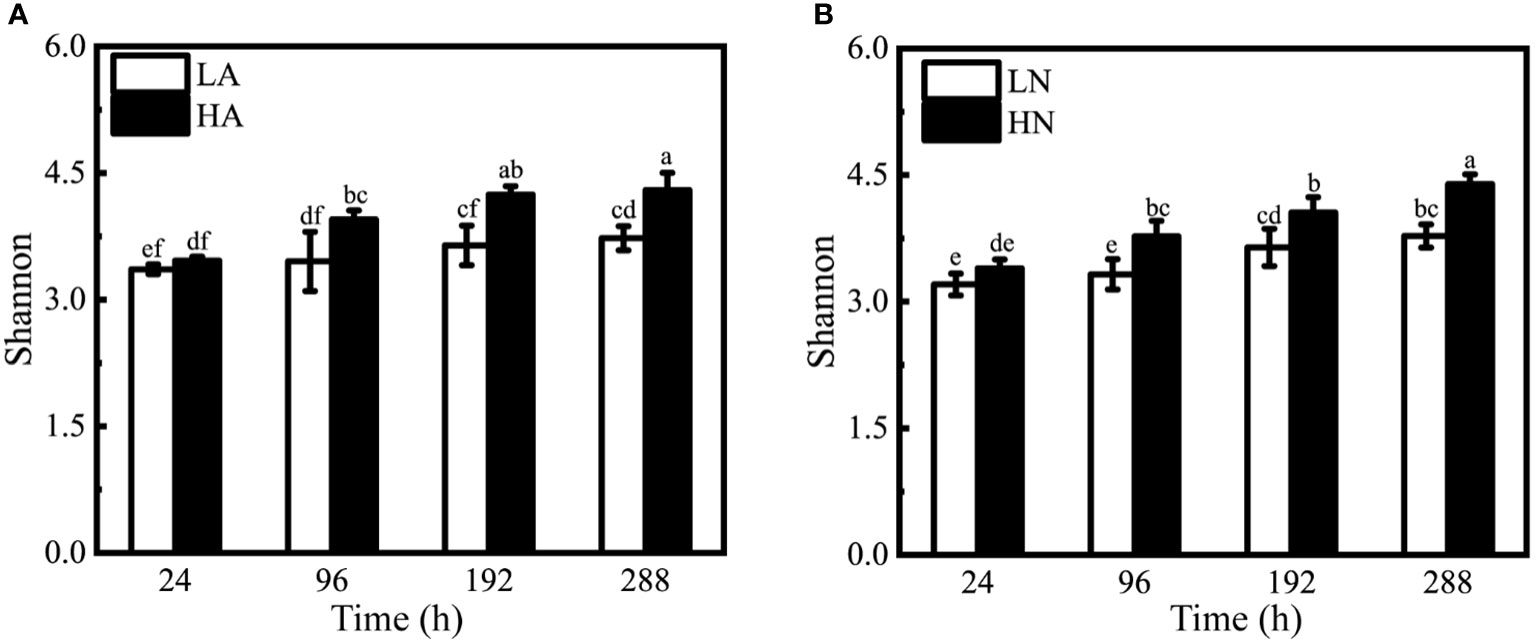
Figure 4 Shannon-Weaver indices of different treatment groups under the algal-bacteria co-culture system. (A) The comparison between the low concentration (LA) and the high concentration (HA) groups with ammonium. (B) The comparison between the low concentration (LN) and the high concentration (HN) groups with nitrate. Different lowercase letters indicate statistically significant differences.
Under the same form and different concentrations of nitrogen culture conditions, the bacterial diversity index at the same time point was significantly different (the effect was near zero, Supplementary Figure 5), except for the initial time (24 h). In conditions with ammonium added alone, the high-concentration groups had 1.14, 1.16, and 1.15 times the bacterial diversity index of the low-concentration groups at 96, 192, and 288 h (Figure 4A). In conditions with nitrate added alone, the high-concentration groups had 1.13, 1.11, and 1.16 times the low concentration’s bacterial diversity index at 96, 192, and 288 h (Figure 4B). Under the same concentration and various forms, the bacterial diversity index did not vary at the same time point (the effect was outside zero, Supplementary Figures 6, 7).
According to the LefSe analysis, the bacterial taxa that changed significantly (species with an average relative abundance greater than 1%) appeared primarily in high-concentration conditions, and the main bacterial taxa that changed over time belonged to Proteobacteria. Two of them belonged to the class Alphaproteobacteria (Marivita and Oceanicaulis; Figure 5A, B), and the other belonged to the class Gammaproteobacteria (Marinobacter, Figure 5C).
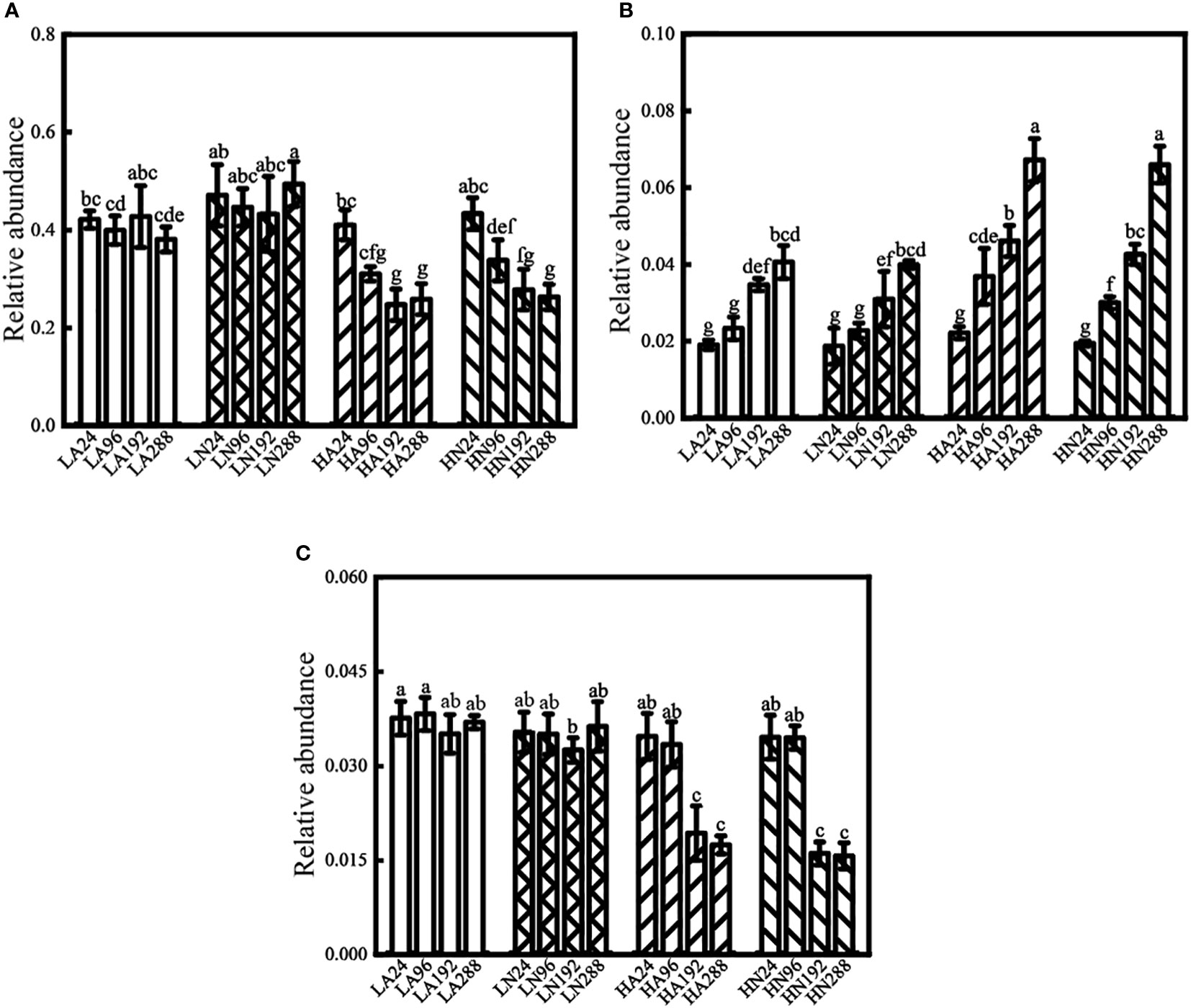
Figure 5 Relative abundance of Marivita (A), Oceanicaulis (B), and Marinobacter (C) under different conditions in the algal-bacteria co-culture system. Error bars represent the standard error of the mean of three independent experiments. Different lowercase letters indicate statistically significant differences.
Under low nitrogen conditions (LA and LN), the relative abundances of Marivita and Marinobacter did not change significantly with time (the effect was near zero; Supplementary Figures 8A, B, Supplementary Figures 9A, B). In contrast, the relative abundance of Oceanicaulis increased (most of the effect was outside zero, Supplementary Figures 10A, B) by 1.13 times (LA) and 1.14 times (LN). In the HA groups, the relative abundances of Marivita and Marinobacter declined by 37% and 50%, respectively (part of the effect was outside zero, Supplementary Figures 8C, 9C), whereas the relative abundance of Oceanicaulis increased by 2.03 times (the effect was outside zero, Supplementary Figure 10C). In the HN groups, the relative abundances of Marivita and Marinobacter decreased by 39% and 55%, respectively (most of the effect was outside zero, Supplementary Figures 8D, 9D), while the relative abundance of Oceanicaulis bacteria increased by 2.40 times (the effect was outside zero, Supplementary Figure 10D).
Under all conditions (including high nitrogen, low nitrogen, and blank control groups), the phycosphere bacterial growth was significantly related to algal growth (Figure 1, Table 2). Moreover, as the initial inorganic nitrogen concentration rose, the biomass of P. tricornutum and associated phycosphere bacteria in the high-concentration groups (HA and HN) was 1–4 times that of the low-concentration groups. In contrast to the condition control groups, the bacterial abundance of the algal-bacteria co-culture system was significantly increased (the effect was outside zero, Supplementary Figure 11) in the same culture conditions (Supplementary Figure 12). Under different conditions without algae, bacterial biomass did not differ with time (most of the effect was near zero, Supplementary Figure 11). It followed that the concentration of inorganic nitrogen was the critical factor for promoting P. tricornutum growth. The phycosphere bacterial growth in this system might be regulated by the extracellular polymeric substances (EPS) secreted by P. tricornutum. The f/2 medium is presently widely used by marine algae (Hasan et al., 2022). However, it lacked the carbon source required for bacterial development. Studies have shown that EPS released by algae is the primary carbon source for bacterial development in the phycosphere (Bohórquez et al., 2017; Behringer et al., 2018; Ye et al., 2020). Kim et al. (2022) found that Marinobacter co-cultured with algae was more numerous than without, indicating that the bacteria also utilized dissolved organic carbon from P. tricornutum for growth. This result is consistent with our research.
The present study showed that the change in inorganic nitrogen concentration had a significant effect on the phycosphere bacterial diversity. In contrast, the nitrogen form had a negligible effect. This finding was also reported by Shi et al. (2018). In the microalgal-bacteria consortium, the diversity index of phycosphere bacteria at the same time point showed a significant difference from 96 h among the treatment groups with the same form and different concentrations (Figure 4). At the same time, P. tricornutum entered the exponential growth phase under different conditions (Figure 1). Therefore, microalgae might be responsible for the significant difference in bacterial diversity index with time at high and low concentrations. The decomposition and mineralization of dissolved organic materials by heterotrophic bacteria are essential to their survival and proliferation (Falkowski et al., 2008). Due to the limited available nutrients in the culture medium, the phycosphere bacteria primarily interacted with algae in the indoor culture environment, and the usage of organic substances generated by algae was the primary survival method of the bacteria (Daly et al., 2021; Kim et al., 2022). In this experiment, as P. tricornutum entered the exponential growth phase, a large number of metabolites secreted by algae might have increased the bacterial diversity index. The biomass of P. tricornutum differed significantly among treatment groups with different concentrations and the same form. That is, compared with the high-concentration groups, P. tricornutum released relatively fewer organic substrates due to the limited algal growth in the low-concentration groups. The limitation of nutrients resulted in differences in bacterial diversity among treatment groups with different concentrations and the same form. Kim et al. (2022) also discovered that the bacterial community structure was indeed affected by algal photosynthetic activity and exudate production; specifically, the bacterial diversity was shown to be greater in cocultures with algae compared to without. This result is similar to that of our research.
Proteobacteria dominated the microbiomes of P. tricornutum in seawater, followed by Bacteroidetes (Kimbrel et al., 2019). Proteobacteria dominated the culture system without P. tricornutum, followed by Bacteroides and Firmicutes (Supplementary Figure 13), which was consistent with the bacterial communities of the algal-bacteria co-culture system. In the condition control groups, different growth conditions and time had no significant influence on the bacterial community composition (AMOVA, P > 0.05).
In the co-culture system of algae and bacteria, P. tricornutum grew rapidly from 48h to 240h and reached the plateau phase under high-concentration conditions (HA and HN, Figure 1C). But under the same condition, the phycosphere bacterial community structure varied with time, and their communities were similar after 192 h (Figure 3). This might be owing to the progressive rise in metabolites during algal growth and reproduction. On the other hand, the metabolism and natural death of microalgae could generate organic matter. These organic substances offered a good nutritional foundation for phycosphere bacterial growth and reproduction. In this system, there are some bacteria that have a symbiotic relationship with algae. This bacterial biomass, which relied primarily on these substances, had risen over this period. In contrast, bacterial biomass that might compete with algae for nutrition declined due to a competitive disadvantage. These changes in bacterial biomass altered the overall bacterial community structure. Thiele et al. (2022) discovered that carbon sources released by diatoms could enhance the number of Gammaproteobacteria and Bacteroidia. By contrast, the abundance of certain Betaproteobacteria decreased due to competition for ammonium with phytoplankton (Christman et al., 2011). As a result, changes in phytoplankton abundance can influence variations in the phycosphere bacterial communities (Park et al., 2020), which is consistent with the findings of our study. Under nitrogen limitation, algae reached stability earlier (96h and 144h, Figure 1A). P. tricornutum might have low biomass and activity in this condition. Because of this, microalgae might provide limited nutrients to the phycosphere bacteria. Consequently, the microbial community structure did not change significantly.
From the above, algae growth was primarily responsible for the shift in the phycosphere bacterial communities. Adequate nitrogen sources could promote algal development. The change in the biomass and activity of algae might affect the diversity and composition of the phycosphere bacterial community. The change in the biomass of algae and bacteria also supported this conclusion. These results showed that, under the circumstances of the experimental culture system, bacteria tended to rely on algae for survival due to long-term adaptation to the culture environment and natural selection.
Rhodobacteraceae exist in different phycospheres (Schäfer et al., 2002; Patzelt et al., 2013; Amin et al., 2015; Limardo and Worden, 2015) and can secrete N-acyl-homoserine lactones (AHLs) signaling molecules to mediate quorum sensing systems to regulate their physiological traits and engage in signal transduction between algae and bacteria (Doberva et al., 2017). The research showed that (Maki et al., 2016) higher nitrate levels in seawater can stimulate the growth of microalgae cells, leading to a rise in Rhodobacteraceae abundance. Marivita is the most abundant genus in Rhodobacteraceae (the proportion of Marivita in different samples was 62%–91% of Rhodobacteraceae). Previous research has discovered that Rhodobacteraceae are ubiquitous in all samples and are one of the most dominant taxa (Shi et al., 2018). This also accords with our observations, which showed that Rhodobacteraceae accounted for 21%–64% of all sample. Marivita, the most important genus of Rhodobacteraceae, is widely distributed in different phycospheres. Marivita can participate in the absorption and metabolism of inorganic and organic compounds containing carbon, nitrogen, or phosphorus in the ocean. When Marivita enters the late-exponential growth phase, it may compete with phytoplankton for essential nutrients like inorganic nitrogen (Zheng et al., 2019). Previous studies using LEfSe revealed that Marinobacter was one of the species with significant differences among the groups (Shi et al., 2018). Marinobacter can dissimilarly reduce nitrate to ammonium (DNRA) for microalgae growth (Poddar et al., 2018).
In this study, the biomass of Marivita and Marinobacter decreased progressively over time in high-concentration groups. However, the change in biomass over time was not apparent in low-concentration groups (Figure 5A, C). This phenomenon might be due to the nutritional competition between the two types of bacteria and microalgae. P. tricornutum might form massive blooms in the presence of enough nitrogen, disrupting the original equilibrium between microalgae and bacteria. During the competition, Marivita and Marinobacter were gradually disadvantaged, so their relative abundance was significantly reduced. The competition between algae and bacteria for limiting nutrients is widespread in the phycosphere. Sun et al. (2020) discovered that diatoms compete strongly with dominant bacteria such as Marivita and Marinobacter in red tides with low inorganic nitrogen concentrations, and their abundance is negatively correlated. Therefore, the decline in the relative abundance of Marivita and Marinobacter over time might be driven by algal growth. These results stayed in line with the analysis results of the phycosphere bacterial communities. Furthermore, in the previous study (Shi et al., 2018), when Marinobacter were at a single sampling point for 168 h, their relative abundance under low-concentration conditions (LA, LN, LAN) was significantly higher than under high-concentration conditions (HA, HN, HAN). In this study, low-concentration groups also had a far higher relative abundance than high-concentration groups (the effect was outside zero, Supplementary Figures 9A, B) at similar periods (LA192 vs HA192, LN192 vs HN192, Figure 5C). Changes in algal biomass may explain why the relative abundance of Marinobacter shifts in different concentration conditions. Marinobacter strains were disadvantaged when competing for resources with microalgae in high-concentration groups, so their relative abundance gradually decreased with time. The relative abundance of Marinobacter had not changed significantly because algal growth was limited by nitrogen sources, algal biomass was low, and its competition was weak in the low-concentration groups. Therefore, at 168 h (or 192 h), the relative abundance of Marinobacter showed significant differences between different concentration groups. Xu et al. (2022) showed that Marinobacter was associated with Phaeocystis globose growth. Moreover, Marinobacter also inhibited P. globose growth and colony formation at a particular density. These findings might be owing to the competitive relationship between the two. The research results are similar to the findings of our study.
The changing trend of Oceanicaulis over time was utterly different from the previous two strains. The relative abundance of Oceanicaulis gradually increased with time under different conditions. In contrast to low-concentration groups, the increasing trend was more evident in high-concentration groups (Figure 5B). That is, the increase in the relative abundance of high-concentration groups was almost twice as much as that of low-concentration groups. It is well known that different phycosphere bacteria can form various associations with microalgae. Competition, antagonism, mutualism, and commensalism were among them (Amin et al., 2012). Contrary to the prior two strains, Oceanicaulis might prefer the metabolites secreted by algae to survive in this experimental system. That is, there existed a mutualism between Oceanicaulis and P. tricornutum. Therefore, their relative abundance increased with algal growth under different conditions. Jasti et al. (2005) revealed that many long-term lab-cultured phycosphere bacteria relied on algae-released organic substances for growth and reproduction. They could form a sustained symbiotic relationship with microalgae. The relative abundance of Oceanicaulis was also strongly associated with Ostreopsis cf. ovata over the algal growth (Guidi et al., 2018). The research findings are consistent with our findings.
From the above, different inorganic nitrogen concentrations made a significant difference in the biomass of P. tricornutum, and changes in microalgal biomass affected the structure of the phycosphere bacterial community. The composition of the phycosphere bacterial community differed between the high- and low-concentration groups as P. tricornutum entered the exponential growth phase. The variation of the bacterial community in the high-concentration groups over time was greater than that in the low-concentration groups. The relative abundance of Marivita and Marinobacter gradually decreased with time under the high-concentration inorganic nitrogen culture, whereas the relative abundance of Oceanicaulis rose. These results might be related to changes in algal biomass. This might also be due to competition and symbiosis between algae and bacteria.
This study has shown that the inorganic nitrogen concentration was the main factor promoting the increase of P. tricornutum biomass. The biomass of P. tricornutum in the high-concentration groups (HA and HN) was 1 ~ 4 times that of the low-concentration groups (LA and LN). The significant increase in P. tricornutum biomass could affect the phycosphere bacterial biomass and community structure. As P. tricornutum reached the exponential growth phase, the bacterial community composition differed between the high- and low-concentration groups from 96 h. The variation in bacterial community composition over time was greater in the high-concentration groups than in the low-concentration groups. The phycosphere bacterial diversity index in the same form and different concentration groups also showed significant differences from 96 h, with high-concentration groups having 1.11 ~ 1.16 times the bacterial diversity index of low-concentration groups.
Under the high-concentration groups (HA and HN), the relative abundance of Marivita and Marinobacter decreased gradually with time, while Oceanicaulis increased. That is, the relative abundances of Marivita and Marinobacter declined by 37% ~ 55%, whereas the relative abundance of Oceanicaulis increased by 2.03~2.40 times. There were three possible explanations for the above phenomenon. First of all, the change in P. tricornutum biomass. Second, the substances that algae secrete. Third, algae and bacteria interacted in different ways. The study contributes to our understanding of when and to what extent the phycosphere bacterial communities respond to algal biomass variations in this system. The present study provides a quantitative reference for the relationship among nutrients, microalgae, and bacteria in this experimental system.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA803332.
LZ and JF conceived the research. XW and FS designed and performed the experiments. XW drafted the manuscript. FS analyzed the statistics. ZC uploaded the original data to the National Center for Biological Information. LZ, JF, FS, and XW contributed to revising the final manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by funds from the Tianjin Key Research and Development Program (No. 21YFSNSN00230), the National Key Research and Development Program of China (No. 2019YFE0122300, 2019YFC1406403) and the National Natural Science Foundation of China (No. 31470536).
The authors would like to thank everyone who contributed to phasic outcomes in the first stages of this study, especially FS. The authors deeply thank Scott L. Collins for the time-lag analysis of bacterial communities. The authors are also grateful to Novogene Co., Ltd. for providing the sequencing platform and bioinformatic analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1086166/full#supplementary-material
Allen A. E., Booth M. G., Verity P. G., Frischer M. E. (2005). Influence of nitrate availability on the distribution and abundance of heterotrophic bacterial nitrate assimilation genes in the barents Sea during summer. Aquat. Microb. Ecol. 39 (3), 247–255. doi: 10.3354/ame039247
Amin S. A., Hmelo L. R., Van Tol H. M., Durham B. P., Carlson L. T., Heal K. R., et al. (2015). Interaction and signaling between a cosmopolitan phytoplankton and associated bacteria. Nature 522, 98–101. doi: 10.1038/nature14488
Amin S. A., Parker M. S., Armbrust E. V. (2012). Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 76 (3), 667–684. doi: 10.1128/MMBR.00007-12
AQSIQ (General Administration of Quality Supervision, Inspection and Quarantine of the People’s Republic of China), SAC (Standardization Administration of the People’s Republic of China) (2008). The specification for marine monitoring: Part 7. GB, 17378.7–2007.
Aylward F. O., Eppley J. M., Smith J. M., Chavez F. P., Scholin C. A., DeLong E. F. (2015). Microbial community transcriptional networks are conserved in three domains at ocean basin scales. Proc. Natl. Acad. Sci. U. S. A. 112 (17), 5443–5448. doi: 10.1073/pnas.1502883112
Behringer G., Ochsenkühn M. A., Fei C., Fanning J., Koester J. A., Amin S. A. (2018). Bacterial communities of diatoms display strong conservation across strains and time. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00659
Bohórquez J., McGenity T. J., Papaspyrou S., García-Robledo E., Corzo A., Underwood G. J. C. (2017). Different types of diatom-derived extracellular polymeric substances drive changes in heterotrophic bacterial communities from intertidal sediments. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00245
Bolch C. J. S., Bejoy T. A., Green D. H. (2017). Bacterial associates modify growth dynamics of the dinoflagellate Gymnodinium catenatum. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00670
Brumley D. R., Carrara F., Hein A. M., Hagstrom G. I., Levin S. A., Stocker R. (2020). Cutting through the noise: bacterial chemotaxis in marine microenvironments. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00527
Christman G. D., Cottrell M. T., Popp B. N., Gier E., Kirchman D. L. (2011). Abundance, diversity, and activity of ammonia-oxidizing prokaryotes in the coastal Arctic ocean in summer and winter. Appl. Environ. Microbiol. 77 (6), 2026–2034. doi: 10.1128/AEM.01907-10
Daly G., Perrin E., Viti C., Fondi M., Adessi A. (2021). Scaling down the microbial loop: data-driven modelling of growth interactions in a diatom–bacterium co-culture. Environ. Microbiol. Rep. 13 (6), 945–954. doi: 10.1111/1758-2229.13010
Deschaseaux E., O’Brien J., Siboni N., Petrou K., Seymour J. R. (2019). Shifts in dimethylated sulfur concentrations and microbiome composition in the red-tide causing dinoflagellate Alexandrium minutum during a simulated marine heatwave. Biogeosciences 16 (22), 4377–4391. doi: 10.5194/bg-16-4377-2019
Doberva M., Stien D., Sorres J., Hue N., Sanchez-Ferandin S., Eparvier V., et al. (2017). Large Diversity and original structures of acyl-homoserine lactones in strain MOLA 401, a marine Rhodobacteraceae bacterium. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01152
Falkowski P. G., Fenchel T., Delong E. F. (2008). The microbial engines that drive earth’s biogeochemical cycles. Sci. 320 (5879), 1034–1039. doi: 10.1126/science.1153213
Frazão L. R., Penninck S. B., Michelazzo L. S., Moreno G., Guimarães C., Lopes R. M., et al. (2021). Microbial ecology of the south Atlantic subtropical gyre: a state-of-the-art review of an understudied ocean region. Ocean Coast. Res. 69, 21027. doi: 10.1590/2675-2824069.20026lrf
Fu H., Uchimiya M., Gore J., Moran M. A. (2020). Ecological drivers of bacterial community assembly in synthetic phycospheres. Proc. Natl. Acad. Sci. United States America 117 (7), 3656–3662. doi: 10.1073/pnas.1917265117
Gardner M. J., Altman D. G. (1986). Confidence intervals rather than p values: estimation rather than hypothesis testing. Br. Med. J. 292, 746–750. doi: 10.1136/bmj.292.6522.746
Grossart H. P., Massana R., McMahon K. D., Walsh D. A. (2020). Linking metagenomics to aquatic microbial ecology and biogeochemical cycles. Limnol. Oceanogr. 65, S2–S20. doi: 10.1002/lno.11382
Guidi F., Pezzolesi L., Vanucci S. (2018). Microbial dynamics during harmful dinoflagellate Ostreopsis cf. ovata growth: Bacterial succession and viral abundance pattern. MicrobiologyOpen 7, 584. doi: 10.1002/mbo3.584
Guillard R. R. L. (1975). Culture of phytoplankton for feeding marine invertebrates (United States of America: Springer US Press).
Guillard R. R. L., Ryther J. H. (1962). Studies of marine planktonic diatoms. i. Cyclotella nana hustedt and Detonula confervacea cleve. Can. J. Microbiol. 8 (2), 229–239. doi: 10.1139/m62-029
Hasan J., Islam Z., Rabby A. F., Sonia S. S., Aktaruzzaman M., Rahman T., et al. (2022). Dataset describing the growth pattern, amino acid and fatty acid profile of five indigenous marine microalgae species of Bangladesh. Data Brief 45, 108643. doi: 10.1016/j.dib.2022.108643
Jackrel S. L., Yang J. W., Schmidt K. C., Denef V. J. (2021). Host specificity of microbiome assembly and its fitness effects in phytoplankton. ISME J. 15, 774–788. doi: 10.1038/s41396-020-00812-x
Jasti S., Sieracki M. E., Poulton N. J., Giewat M. W., Rooney-Varga J. N. (2005). Phylogenetic diversity and specificity of bacteria closely associated with Alexandrium spp. and other phytoplankton. Appl. Environ. Microbiol. 71 (7), 3483–3494. doi: 10.1128/AEM.71.7.3483-3494.2005
Ji W. D. (2011). Research on the current status and background value of china’s offshore marine environmental quality (Beijing: Ocean Press).
Kim H., Kimbrel J. A., Vaiana C. A., Wollard J. R., Mayali X., Buie C. R. (2022). Bacterial response to spatial gradients of algal-derived nutrients in a porous microplate. ISME J. 16, 1036–1045. doi: 10.1038/s41396-021-01147-x
Kimbrel J. A., Samo T. J., Ward C., Nilson D., Thelen M. P., Siccardi A., et al. (2019). Host selection and stochastic effects influence bacterial community assembly on the microalgal phycosphere. Algal. Res. 40, 101489. doi: 10.1016/j.algal.2019.101489
Kublanovskaya A., Solovchenko A., Fedorenko T., Chekanov K., Lobakova E. (2020). Natural communities of carotenogenic chlorophyte Haematococcus lacustris and bacteria from the white Sea coastal rock ponds. Microb. Ecol. 79, 785–800. doi: 10.1007/s00248-019-01437-0
Laslett A. M., Kuntsche S., Wilson I. M., Taft A., Fulu E., Jewkes R., et al. (2022). The relationship between fathers’ heavy episodic drinking and fathering involvement in five Asia-pacific countries: An individual participant data meta-analysis. Alcoholism Clin. Exp. Res. 46, 2137–2148. doi: 10.1111/acer.14955
Le Chevanton M., Garnier M., Lukomska E., Schreiber N., Cadoret J.-P., Saint-Jean B., et al. (2016). Effects of nitrogen limitation on Dunaliella sp.–Alteromonas sp. interactions: From mutualistic to competitive relationships. Front. Mar. Sci. 3. doi: 10.3389/fmars.2016.00123
Limardo A. J., Worden A. Z. (2015). Exclusive networks in the sea. Nature 522, 36–37. doi: 10.1038/nature14530
Linz A. M., Aylward F. O., Bertilsson S., McMahon K. D. (2020). Time-series metatranscriptomes reveal conserved patterns between phototrophic and heterotrophic microbes in diverse freshwater systems. Limnol. Oceanogr. 65, S101–S112. doi: 10.1002/lno.11306
Lupette J., Lami R., Krasovec M., Grimsley N., Moreau H., Piganeau G., et al. (2016). Marinobacter dominates the bacterial community of the Ostreococcus tauri phycosphere in culture. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01414
Maki T., Ishikawa A., Mastunaga T., Pointing S. B., Saito Y., Kasai T., et al. (2016). Atmospheric aerosol deposition influences marine microbial communities in oligotrophic surface waters of the western pacific ocean. Deep Sea Res. Part I: Oceanogr. Res. Pap. 118, 37–45. doi: 10.1016/j.dsr.2016.10.002
Markou G., Vandamme D., Muylaert K. (2014). Microalgal and cyanobacterial cultivation: The supply of nutrients. Water Res. 65, 186–202. doi: 10.1016/j.watres.2014.07.025
Park B. S., Lee M., Shin K., Baek S. H. (2020). Response of the bacterioplankton composition to inorganic nutrient loading and phytoplankton in southern Korean coastal waters: A mesocosm study. Mar. Ecol. 41, 12591. doi: 10.1111/maec.12591
Patzelt D., Wang H., Buchholz I., Rohde M., Gröbe L., Pradella S., et al. (2013). You are what you talk: quorum sensing induces individual morphologies and cell division modes in Dinoroseobacter shibae. ISME J. 7, 2274–2286. doi: 10.1038/ismej.2013.107
Poddar N., Sen R., Martin G. J. O. (2018). Glycerol and nitrate utilisation by marine microalgae Nannochloropsis salina and Chlorella sp.and associated bacteria during mixotrophic and heterotrophic growth. Algal. Res. 33, 298–309. doi: 10.1016/j.algal.2018.06.002
Rogato A., Amato A., Iudicone D., Chiurazzi M., Ferrante M. I., d’Alcalà M. R. (2015). The diatom molecular toolkit to handle nitrogen uptake. Mar. Genomics 24 (1), 95–108. doi: 10.1016/j.margen.2015.05.018
Schäfer H., Abbas B., Witte H., Muyzer G. (2002). Genetic diversity of ‘satellite’ bacteria present in cultures of marine diatoms. FEMS Microbiol. Ecol. 42 (1), 25–35. doi: 10.1111/j.1574-6941.2002.tb00992.x
Seymour J. R., Amin S. A., Raina J. B., Stocker R. (2017). Zooming in on the phycosphere: the ecological interface for phytoplankton–bacteria relationships. Nat. Microbiol. 2, 17065. doi: 10.1038/nmicrobiol.2017.65
Shi F., Wei X. X., Feng J. F., Sun Y. X., Zhu L. (2018). Variation of bacterial community associated with Phaeodactylum tricornutum in response to different inorganic nitrogen concentrations. Acta Oceanol. Sin. 37 (12), 118–128. doi: 10.1007/s13131-018-1272-7
Shibl A. A., Isaac A., Ochsenkühn M. A., Cárdenas A., Fei C., Behringer G., et al. (2020). Diatom modulation of select bacteria through use of two unique secondary metabolites. Proc. Natl. Acad. Sci. U. S. A. 117 (44), 27445–27455. doi: 10.1073/pnas.2012088117
Steichen S. A., Gao S., Waller P., Brown J. K. (2020). Association between algal productivity and phycosphere composition in an outdoor Chlorella sorokiniana reactor based on multiple longitudinal analyses. Microb. Biotechnol. 13 (5), 1546–1561. doi: 10.1111/1751-7915.13591
Sun F. L., Wang C. Z., Wang Y. S., Tu K., Zheng Z. P., Lin X. F. (2020). Diatom red tide significantly drive the changes of microbiome in mariculture ecosystem. Aquaculture 520, 734742. doi: 10.1016/j.aquaculture.2019.734742
Sun F. L., Wang C. Z., Yang H. Q. (2021). Physicochemical factors drive bacterial communities in an aquaculture environment. Front. Environ. Sci. 9. doi: 10.3389/fenvs.2021.709541
Thiele S., Storesund J. E., Fernández-Méndez M., Assmy P., Øvreås L. (2022). A winter-to-Summer transition of bacterial and archaeal in Arctic sea ice. Microorganisms 10 (8), 1618. doi: 10.3390/microorganisms10081618
Variem S. S., Kizhakkedath V. K. (2021). Phycosphere associated bacteria; a prospective source of bioactive compounds. Biologia 76 (3), 1095–1098. doi: 10.2478/s11756-020-00640-6
Vu C. H. T., Lee H. G., Chang Y. K., Oh H. M. (2018). Axenic cultures for microalgal biotechnology: Establishment, assessment, maintenance, and applications. Biotechnol. Adv. 36 (2), 380–396. doi: 10.1016/j.biotechadv.2017.12.018
Xu S. S., Wang X. D., Liu J., Zhou F. L., Guo K. L., Chen S. Z., et al. (2022). Bacteria associated with Phaeocystis globosa and their influence on colony formation. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.826602
Yang Q., Feng Q., Zhang B. P., Gao J. J., Sheng Z., Xue Q. P., et al. (2021). Marinobacter alexandrii sp. nov., a novel yellow-pigmented and algae growth-promoting bacterium isolated from marine phycosphere microbiota. Antonie van Leeuwenhoek 114 (6), 709–718. doi: 10.1007/s10482-021-01551-5
Ye T. R., Zhao Z., Bai L. L., Song N., Jiang H. L. (2020). Characteristics and bacterial community dynamics during extracellular polymeric substance (EPS) degradation of cyanobacterial blooms. Sci. Total Environ. 748, 142309. doi: 10.1016/j.scitotenv.2020.142309
Zhang X. X., Fang J., Bach W. G., Edwards K. J., Orcutt B. N., Wang F. P.. (2016). Nitrogen stimulates the growth of subsurface basalt-associated microorganisms at the western flank of the mid-Atlantic ridge. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00633
Zheng Q., Lu J. Y., Wang Y., Jiao N. Z. (2019). Genomic reconstructions and potential metabolic strategies of generalist and specialist heterotrophic bacteria associated with an estuary Synechococcus culture. FEMS Microbiol. Ecol. 95 (3), 1–13. doi: 10.1093/femsec/fiz017
Zhou J., Lao Y. M., Song J. T., Jin H., Zhu J. M., Cai Z. H. (2020). Temporal heterogeneity of microbial communities and metabolic activities during a natural algal bloom. Water Res. 183, 116020. doi: 10.1016/j.watres.2020.116020
Keywords: bacterial community structure, ammonium, nitrate, Phaeodactylum tricornutum, marine
Citation: Wei X, Shi F, Chen Z, Feng J and Zhu L (2023) Response of bacterial communities (Marivita, Marinobacter, and Oceanicaulis) in the phycosphere to the growth of Phaeodactylum tricornutum in different inorganic nitrogen sources. Front. Mar. Sci. 10:1086166. doi: 10.3389/fmars.2023.1086166
Received: 01 November 2022; Accepted: 06 February 2023;
Published: 24 February 2023.
Edited by:
Hernando Bacosa, Mindanao State University-Iligan Institute of Technology (MSU-IIT), PhilippinesCopyright © 2023 Wei, Shi, Chen, Feng and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Zhu, emh1bGluQG5hbmthaS5lZHUuY24=; Jianfeng Feng, ZmVuZ2pmQG5hbmthaS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.