- 1College of Ocean and Earth Sciences, Xiamen University, Xiamen, China
- 2Key Laboratory of Marine Chemistry Theory and Technology, Ministry of Education, and Institute for Advanced Ocean Study, Ocean University of China, Qingdao, China
- 3College of Chemistry and Chemical Engineering, Ocean University of China, Qingdao, China
- 4College of Marine Life Sciences, Ocean University of China, Qingdao, China
Green tides Ulva prolifera have broken out in the Yellow Sea for more than 10 years, becoming a periodic ecological disaster. The largest-ever green tide that occurred in 2021 promoted innovation in treatment methods. Different from the traditional harvest-disposal method, a microbial complex formulation was firstly sprayed on the harvest U. prolifera that promotes rapid degradation, and then fermented and disposed into the sea. At present, little was known about the ecological effects of those different treatment methods. In order to examine this hypothesis, we run an in-lab incubation of 60 days to simulate the two methods to degrade U. prolifera, with focuses on the degradation ensued impacts on water quality. The degradation process of fresh U. prolifera over two months was dominated by the continuous and slow release of DOM, and the concentration of DOM in the water column was hardly observed to decrease within two months. The pre-discomposed-disposal method also significantly altered microbial community structure. The pre-decomposing treatment with microbial complex formulations destroyed U. prolifera cell tissues and changed its physical state in seawater from floating to fast depositing, and increased the degradation rate by about 14 times. The rapid decomposition of the released bioactive organic matter consumed a substantial amount of dissolved oxygen in local seawater, which has the potential risk of causing local hypoxia and acidification in a short-term. The pre-decomposition treatment of U. prolifera could be a practical and efficient countermeasures to U. prolifera blooming. After the complete degradation of the pre-decomposed U. prolifera, the resulting dissolved organic matter could increase TA to resist acidification. Overall, compared with traditional harvest-packing-disposal method, the pre-decomposing-disposal treatment is an efficient and environmental-friendly disposal method to deal with the U. prolifera “green tide”, but it should be used cautiously.
1 Introduction
The bloom of Ulva prolifera (abbreviated U. prolifera) is one of the most famous “green tides”. Large-scale and frequent outbreaks of “green tides” in the coastal waters of many countries have made it a global marine ecological problem (Smetacek and Zingone, 2013). The increase in Ulva blooms on European and American beaches began in the 1970s, and the frequencies and scales of green tides have increased rapidly since the 1990s (Schramm, 1996; Charlier et al., 2008). In China, the “green tide” has erupted annually in the Yellow Sea since 2007, causing severe ecological and economic damage to the southern coastal region of the Shandong Peninsula (He et al., 2019). The “green tide” consists of different species of Ulva, such as Ulva prolifera, Ulva linza, Ulva intestinalis, and Ulva compressa (Wu et al., 2018). Based on previous research, U. prolifera is the predominant species of the “green tide” in the Yellow Sea (Gao et al., 2016; Gao et al., 2017). It has become the largest “green tide” disaster in the world (Hu et al., 2017; Zhang et al., 2017). According to the Bulletin of China Marine Disaster (2021), the distribution of U. prolifera reached 61,898 km2 during the “green tide” of 2021, the highest values recorded in history (https://www.mnr.gov.cn/sj/sjfw/hy/gbgg/zghyzhgb/). “Green tides” of U. prolifera have a variety of ecological impacts on natural landscapes and marine life, resulting in imbalances in marine ecosystems (Ye et al., 2011; Le Luherne et al., 2016; Qi et al., 2016; Glibert, 2017; Miao et al., 2018; Chen et al., 2020; Liu et al., 2021). At the end of a “green tide” (late July to early August), a large amount of U. prolifera settles on the seafloor and gradually decomposes. This process consumes oxygen and releases carbon dioxide, leading to hypoxia and acidification of coastal waters (Zhang et al., 2019; Li et al., 2021).
The coastal waters of Qingdao (China) are one of the areas most affected by the “green tides”. The Qingdao Municipal Government has taken aggressive measures to reduce the harmful effects of U. prolifera (Sun et al., 2022). The most common measure is harvesting, packaging, and disposal in nearshore waters (Li et al., 2022b). Some of the U. prolifera recovered from the sea or collected from the beach was processed into algal powder, algal polysaccharide, or biofertilizer (Zhuang et al., 2012; Yuan et al., 2018; Chen et al., 2022). In 2021, a new treatment method was applied. A microbial complex formula was sprayed on the harvested U. prolifera. After being fermented in fermentation tanks, the U. prolifera was transported back and disposed of into the sea, where it naturally degraded.
The “green tides” have long-lasting effects on the environment. Massive U. prolifera fronds die and are decomposed gradually by microorganisms at the end of “green tides” (mid-July to early August). The natural degradation of dead U. prolifera will continue to release dissolved organic matter and consume dissolved oxygen, which may lead to hypoxia and ocean acidification (Zhang et al., 2019; Liang et al., 2021). Therefore, we propose that pre-decomposition treatment could be an effective and useful measure to control the “green tide” of U. prolifera by accelerating the cycling of materials and shortening the time during which the green tide of U. prolifera continues to negatively impact the marine environment. But does this treatment also pose potential risks? It is still unclear how the degradation of pre-decomposed U. prolifera differs from that of naturally degraded U. prolifera. Whether the effects of the two treatments on the marine environment are significantly different? In this study, we conducted a long-term (60 days) degradation experiment to investigate the two different treatment methods and the resulting ecological impacts. Based on the results, we propose reasonable suggestions for treatment strategies to deal with the future bloom of U. prolifera.
2 Materials and methods
2.1 Experimental materials
Ten kilograms of fresh U. prolifera were collected on July 29, 2021from the Yellow Sea near Qingdao (120.37°E, 36.06°N) when the “green tide” broke. The salinity of the seawater was measured at 31.0 with a conductivity salinity meter, and the pH of the seawater was measured at 7.98 with a pH meter in the lab. U. prolifera with emerald green color and no “white coloration” were selected, washed with seawater on site to remove impurities, and pre-cultured in the laboratory (20°C) for 24 hours. The temperature of the seawater was measured at 20.2°C with a thermometer on site. These ten kilos of fresh U.prolifera was used for the naturally degraded experiment.
To obtain the pre-decomposing U. prolifera samples, the harvest U. prolifera were collected in the fermentation unit, where a microbial complex formulation was sprayed, consisting of various microorganisms such as Bacillus and Yeast. After complete mixing, the U. prolifera was fermented in fermentation tanks at a temperature of about 30°C for more than 7 days. Samples whose color and physical condition were relatively uniform were selected for the experiment. The samples were stored in the laboratory at a constant temperature (20°C) for 24 hours without any special treatment before the experiment began.
About 200 liters of seawater was collected off the coast of Qingdao (120.44°E, 36.06°N), far from the sewage discharge and other human-influenced areas. It was poured into a large HDPE bucket and left for 12 hours to remove floating objects and settle the sediment. The seawater was then filtered through a sieve with a pore size of 20 µm to remove particles and marine plankton and retain most microorganisms in situ. The filtered seawater was kept at a constant temperature overnight to reach the incubation temperature (20°C).
2.2 Experimental design
2.2.1 Incubation conditions and samples collection
A one-time incubation method was applied using a 30-liter suspension culture system (SI Figure SI2). A 250 g sample of fresh and pre-decomposed U. prolifera was separately added to 25 L of filtered seawater to simulate the natural conditions for U. prolifera degradation (Chen et al., 2020). Three parallel groups were set up for fresh and pre-decomposed U. prolifera, separately. Incubation was performed in the dark at a temperature of 20°C.
Both water and microbial samples were collected at 0 h, 12 h, 24 h (1 day), 36 h, 3 day, 5 d, 8 d, 12 d, 20 d, 35 d, and 60 d. Samples for alkalinity were collected in 100 mL glass bottles and analyzed as soon as possible. One liter of water was collected from each parallel experimental group at each time interval and analyzed for DOC, DON, NO3-, NO2-, and NH4+. Water samples were first filtered with GF/F filters (Whatman, pre-combustion at 450°C) with a pore size of 0.7 µm to remove particulate matter, then with PC filters (Millipore) with a pore size of 0.2 µm, and finally with PDVF filters (Millipore) with a pore size of 0.1 µm. Water samples were stored in a pre-cleaned 1 liter high-density polyethylene (HDPE) bottle and frozen at -20°C. The filtered PC filter membrane with a diameter of 0.2 µm was folded and placed in a cryopreservation tube, later soaked in RNA (Thermo Co.) and quickly stored in the refrigerator at -80°C for 16S rDNA sequencing.
2.2.2 Analytical methods
Water samples were thawed and brought to room temperature and then shaken well. The sum of the concentrations of nitrate and nitrate was measured by a chemiluminescence method using a NOx analyzer (TELEDYNE, API-200E) with an accuracy of 3% (Yan et al., 2021). Nitrite concentration was determined by colorimetry. Ammonium concentration was determined by the sodium hypobromite oxidation method (Grasshoff et al., 2009). Dissolved inorganic nitrogen concentration (DIN) was the sum of the concentrations of nitrate, nitrite, and ammonium. Total dissolved nitrogen (TDN) was determined using the alkaline potassium persulfate oxidation method (Grasshoff et al., 2009). Dissolved organic nitrogen (DON) was the difference between TDN and DIN. The DOC was measured using a Total Organic Carbon Analyzer (Shimadzu, TOC-L) with CRM (Hansel Lab) as an internal standard to correct the reported DOC data. The TA was determined by gran titration using an automatic potentiometric titrator with an accuracy of 0.1%.
Echo Laboratories RVL-100-G Microscope with the built-in camera was used to observe the U. prolifera cells. The organic alkalinity was determined by back titration (Cai et al., 1998), using automatic point titrator (ZDJ-5B, Shangai INESA Scientific Instrument Co., Ltd).
The fluorescence excitation emission matrix (EEM) of the water samples was measured in a 1-cm quartz cuvette using a spectrofluorometer (Hitachi, F-4600) with 5-nm steps, where the excitation wavelength was between 200 and 450 nm and the emission wavelength was between 250 and 580 nm. The scan speed was 1200 nm/min. For each sample, the EEM was subtracted from Milli-Q water. UV absorbance was measured using a UV spectrophotometer (Shimadzu, UV-1900). Modeling by parallel factor decomposition (PARAFAC) was then performed in R (Mathworks) using the staRdom toolbox (Pucher et al., 2019).
For bacterial community structure analysis, the 16S rDNA gene was amplified with primers targeting the hypervariable regions V3-V4 341F (CCTAYGGGRBGCASCAG) and 806R (GGACTACVSGGGTATCTAAT). Whole genome DNA from the samples was extracted by Shenzhen Weikemeng Technology Group Co, Ltd, using the CTAB method. The Mixture PCR products was purified with Qiagen Gel Extraction Kit (Qiagen, Germany), and amplicons were sequenced on an Illumina MiSeq platform.
2.3 Data processing and analysis
SPSS (statistics 26) was used to perform statistical analysis on the data. ANOVA was applied to examine the statistical differences in the parameters of the two processing methods. The bioinformatics analysis was conducted by following the “Atacama soil microbiome tutorial” of Qiime2docs along with customized program scripts (https://docs.qiime2.org/2019.1/).
3 Results
3.1 Morphological differences in the fresh and pre-decomposed Ulva prolifera
The color of fresh U. prolifera was emerald green with a unique smell of seaweed. Fresh U. prolifera was fibrous, hollow, and floated in seawater. The decomposed U. prolifera was yellow-brown and had a strong sour odor, but a relatively complete shape of the algal body could still be seen. After being put into sea water, the decomposed U. prolifera settled to the bottom (Support Information, Figure SI1).
Under the microscope, the cell structure of fresh U. prolifera was intact, while that of decomposed U. prolifera had been damaged. After 35 days of degradation in the laboratory, the structure of fresh U. prolifera was partially decomposed, but some fibers were still visible, and the color turned yellow and white. The microscope examined that the cell structure was damaged and the algae had died. However, after 35 days of degradation, there was no fiber structure in the decomposed U. prolifera. Most algae had decomposed into dissolved organic matter and debris (Figure 1). After 60 days of degradation, most of the fresh and decomposed U. prolifera were completely decomposed and no tissues were observed visually (SI, Figure SI2).
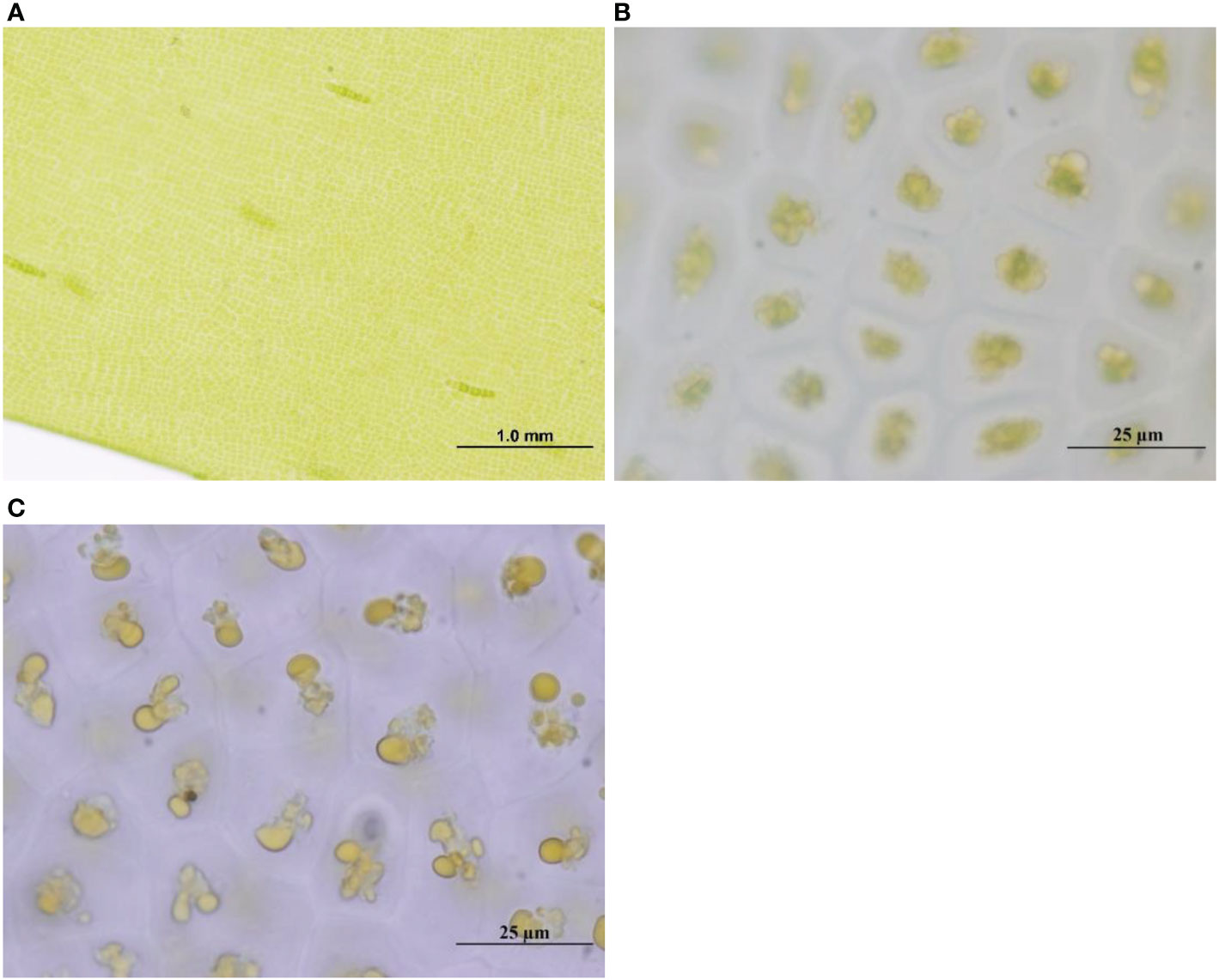
Figure 1 The cell structure of fresh U. prolifera at the initial stage of degradation (A) and 35 days (B) under the microscope, and that of pre-decomposed U. prolifera at the initial stage (C).
3.2 Variations of nitrogen nutrients during the degradation
For the degradation process of fresh U. prolifera, the nitrite was always lower than 0.10 µmol/L over 60 days. The concentration of ammonium increased slightly at 12 h and 5 d (0.34 and 0.84 µmol/L, respectively), and remained lower than 0.10 µmol/L over the rest of the time. The nitrate increased slightly from the 5th to the 12th day, but the maximum concentration was only 1.64 µmol/L (Figure 2). Over the 60 days of degradation, the concentration of DON was 13−955 times that of DIN. The mineralization of organic matter to form inorganic nutrients was not significant.
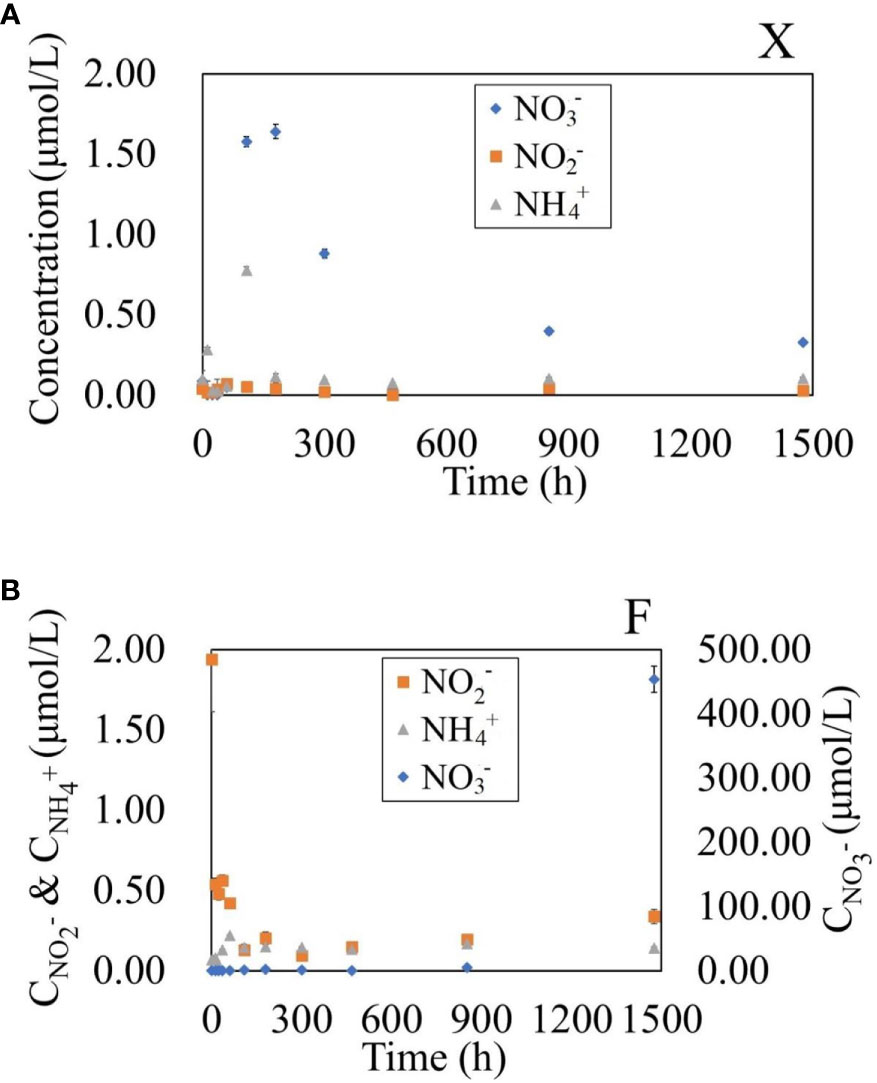
Figure 2 Variation of NO3-, NO2- and NH4+ concentrations in water during the degradation of fresh U. prolifera (A) and pre-decomposed U. prolifera (B).
For the degradation process of decomposed U. prolifera, the ammonium was always lower than 0.20 µmol/L over 60 days. Nitrite showed a rapid decreasing trend similar to that of DON in the first five days, from 1.65 to 0.10 µmol/L. The nitrate was always lower than 2.5 µmol/L within the first 20 days. It rose to 5.44 µmol/L on the 35th day, and continued rising rapidly to 478.53 µmol/L on the 60th day. DON decreased rapidly from the 35th to the 60th day. From 0 to 35 days, the concentration of DON in water was 46−903 times that of DIN, but the concentration of DIN was nine times that of DON on the 60th day. The degradation process of decomposed U. prolifera was dominated by the rapid decomposition of organic matter, but the inorganic nitrogen nutrients had not accumulated significantly within the first 30-day. In the second 30-day, with the complete disappearance of algae, nitrate increased rapidly. There were significant differences in nitrate and nitrite, but not in ammonium, between the two treatments at the 0.05 level (One-Way ANOVA).
3.3 Variations of DOC and DON during the degradation
As fresh U. prolifera degraded, the DOC and DON showed an overall increasing trend. However, as the pre-decomposed U. prolifera degraded, the DOC and DON showed a general decreasing trend (Figure 3). The DOC and DON concentrations of the fresh U. prolifera fluctuated during the first five days. The U. prolifera might still be alive. From the 5th day to the 35th day, the concentration of DOC and DON continued to increase rapidly. On the 35th day of degradation, the concentration of DOC reached its highest value, while the increase in DON slowed down. After 60 days of degradation, the concentrations of DOC and DON were 10 and 12 times higher than the initial seawater concentrations, respectively.
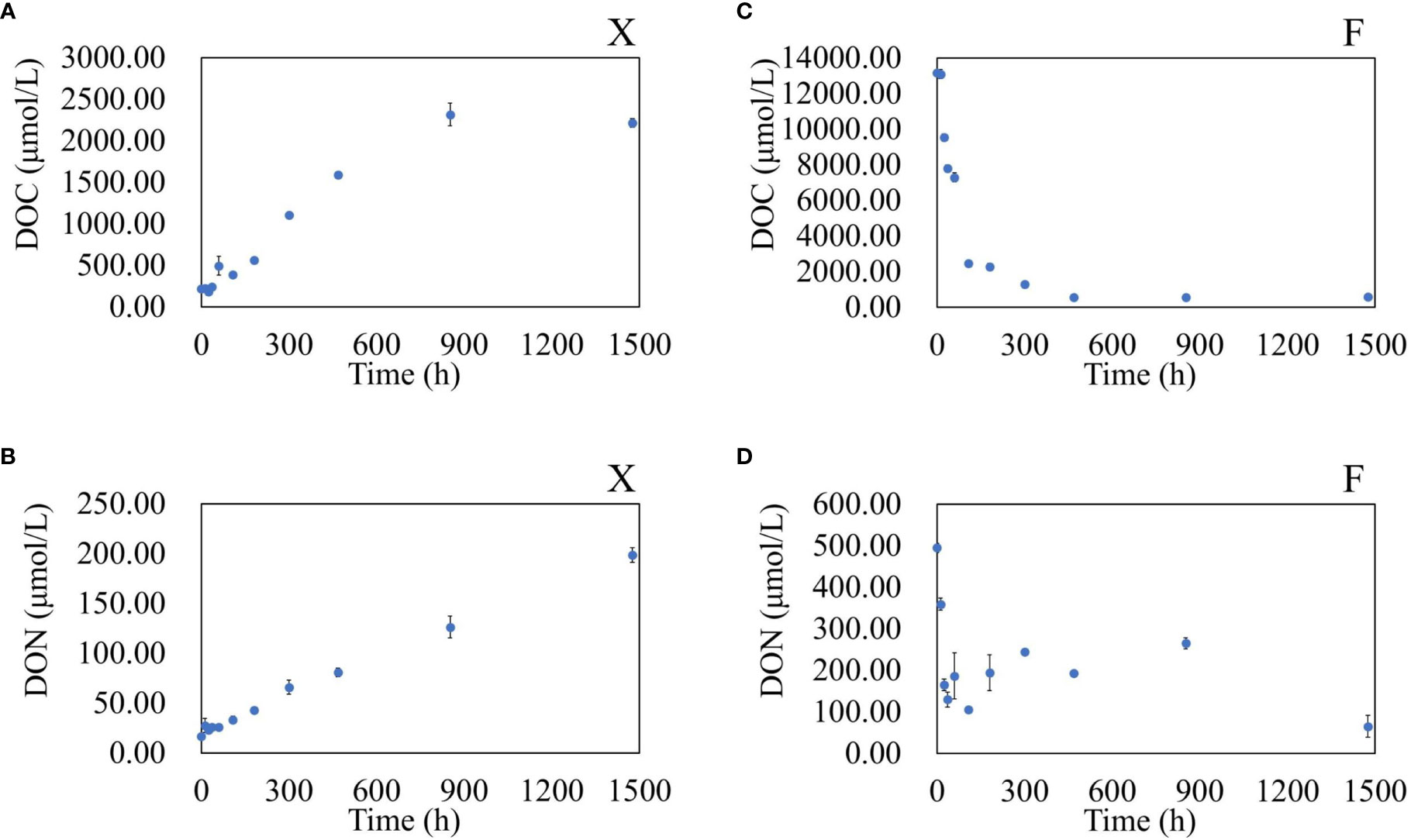
Figure 3 Variation in DOC and DON concentrations in water during the degradation of fresh U. prolifera (A, B) and pre-decomposed U. prolifera (C, D).
The DOC and DON reached the maximal instantaneously. At the beginning, the DOC was 13086 ± 53 µmol/L, and the DON was 491.96 ± 9.75 µmol/L. The concentration of DOC and DON reduced by 80% within 5 days. The concentration of DOC tended to be stable after 20 days. The concentration of DON in water fluctuated slightly between 5−35 days, and decreased significantly again between 35−60 days. There were significant differences in DOC and DON between the two treatments, with a significant difference at the 0.05 level (One-Way ANOVA).
3.4 Variations of FDOM during the degradation
Using EEM-PARAFAC, four kinds of fluorescent active components were found in the degradation products of U. prolifera. Among them were C1, a tyrosine-like protein component and C3, a tryptophan-like protein component (Yamashita et al., 2008). These two components represented organic matter with high activity. C2 and C4, two humic components representing refractory organic matter (Stedmon and Markager, 2005; Yamashita et al., 2008), were also present (SI, Figure SI3).
During the degradation of fresh U. prolifera, the FDOM in the water showed an increasing trend for the first 12 days (Figure 4) and the proportion of refractory components (C2+C4) was stable at about 14%, indicating that the U. prolifera released dissolved organic matter into water continuously. After 12 days, the amount of FDOM decreased and the proportion of refractory components increased (38−52%).
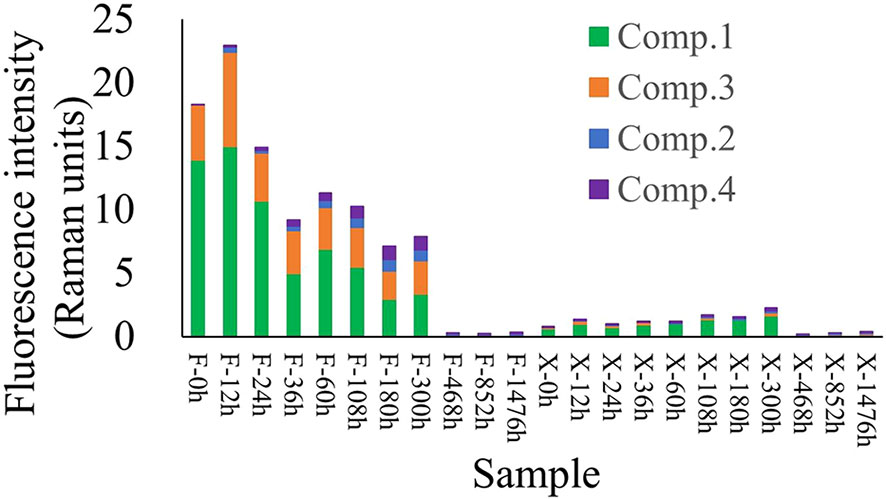
Figure 4 Variation in fluorescent active components over time during the degradation of U. prolifera. The abscissa represents the samples at different times (h).
During the degradation of pre-decomposed U. prolifera, the total FDOM showed a decreasing trend, while the proportion of refractory components continued to increase (Figure 4). The proportion of protein-like components gradually decreased from nearly 100% at the beginning to about 20%, while the proportion of humic-like components gradually increased from 0% to ~80% (SI, Figure SI4).
3.5 Variations of TA during the degradation
In the initial stage (the first five days), the degradation of both fresh and decomposed U. prolifera caused a downward fluctuation of TA (Figure 5). From day five until the end of the degradation experiment, the degradation of fresh U. prolifera did not cause significant changes in the total alkalinity, which was basically the same as the initial state of incubation culture. However, the continuous degradation of the pre-decomposed U. prolifera resulted in an increase of 13% in the total alkalinity.
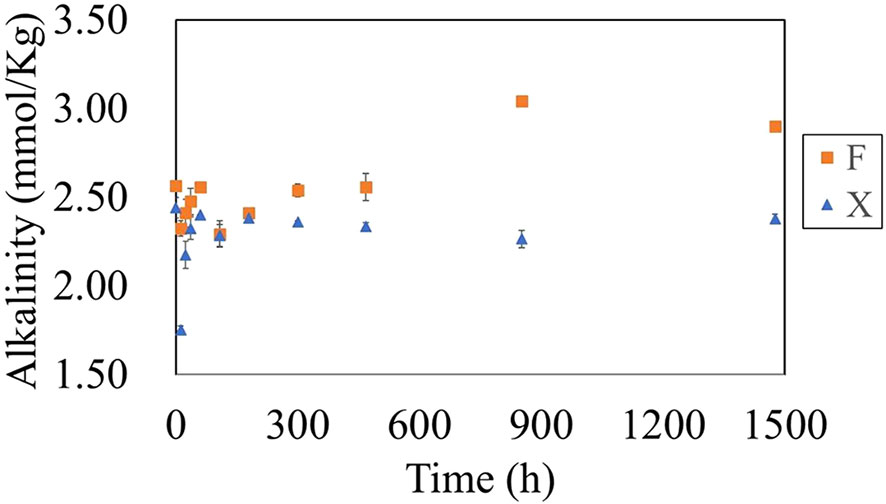
Figure 5 Variation in alkalinityin water during the degradation of fresh U. prolifera (X) and pre-decomposed U. prolifera (F).
3.6 Variations in microbial community structure during degradation
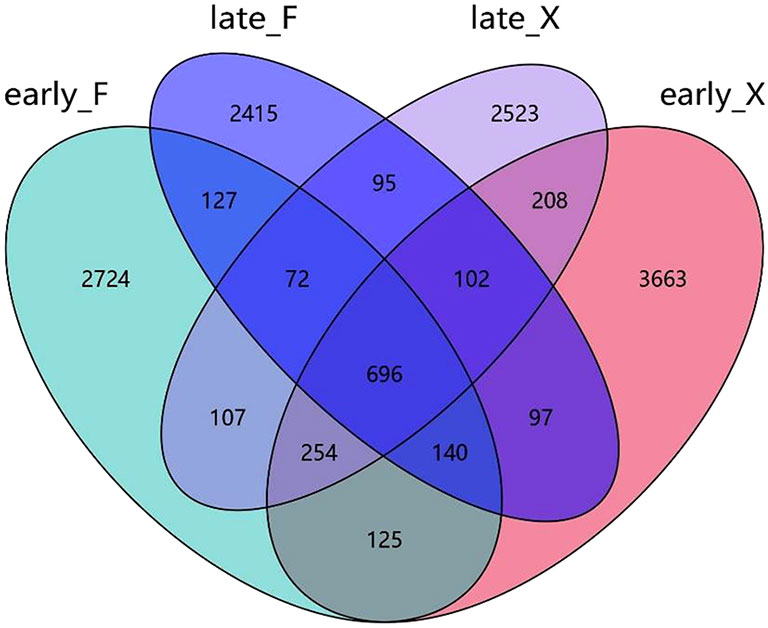
Based on the variations in concentrations of DOC, DON and DIN during the degradation process, the degradation experiment can be divided into two stages up to day 20, namely the early stage (early_X for fresh U. prolifera, and early_F for pre-decomposed U. prolifera) and the late stage (late_X for fresh U. prolifera, and late_F for pre-decomposed U. prolifera).
Using the sequencing results, differences in bacterial species between the two groups of fresh and pre-decomposed U. prolifera in the early and late stages of degradation were counted. There were a large number of endemic species in different stages. In the early and late stages of the degradation of fresh U. prolifera and in the early and late stages of the degradation of pre-decomposed U. prolifera, the number of specific bacterial species were 3663 and 2523 and 2415 and 2714, respectively (Figure 6). The number of endemic species was far more than the common species between any two groups.
Based on the relative abundance of various groups of microorganisms at the phylum level (Figure 7), it can be seen that the bacterial structure at different stages of degradation was significantly different. Except for the common Proteobacteria (more than 57% for each group) in the seawater, the flora typically with high abundance in the early stage of the degradation of pre-decomposed U. prolifera (early_F) was Bacteroidetes (9.4%) and Bacillus (GN02, 4.0%). At the late stage of the degradation of decomposed U. prolifera (late_F), the abundance of Bacteroidetes and Planctomycetes increased significantly (9.4% to 20.2% and 0.4 to 5.2, respectively). At the early stage of degradation of fresh U. prolifera (early_X), Actinobacteria (7.6%) and Bacteroidetes (5.3%) had relatively high abundance. However, in the late stage of degradation of fresh U. prolifera (late_X), the abundance of these two groups decreased, and the abundance of Parcubacia (OD1, 19.2%) increased significantly.
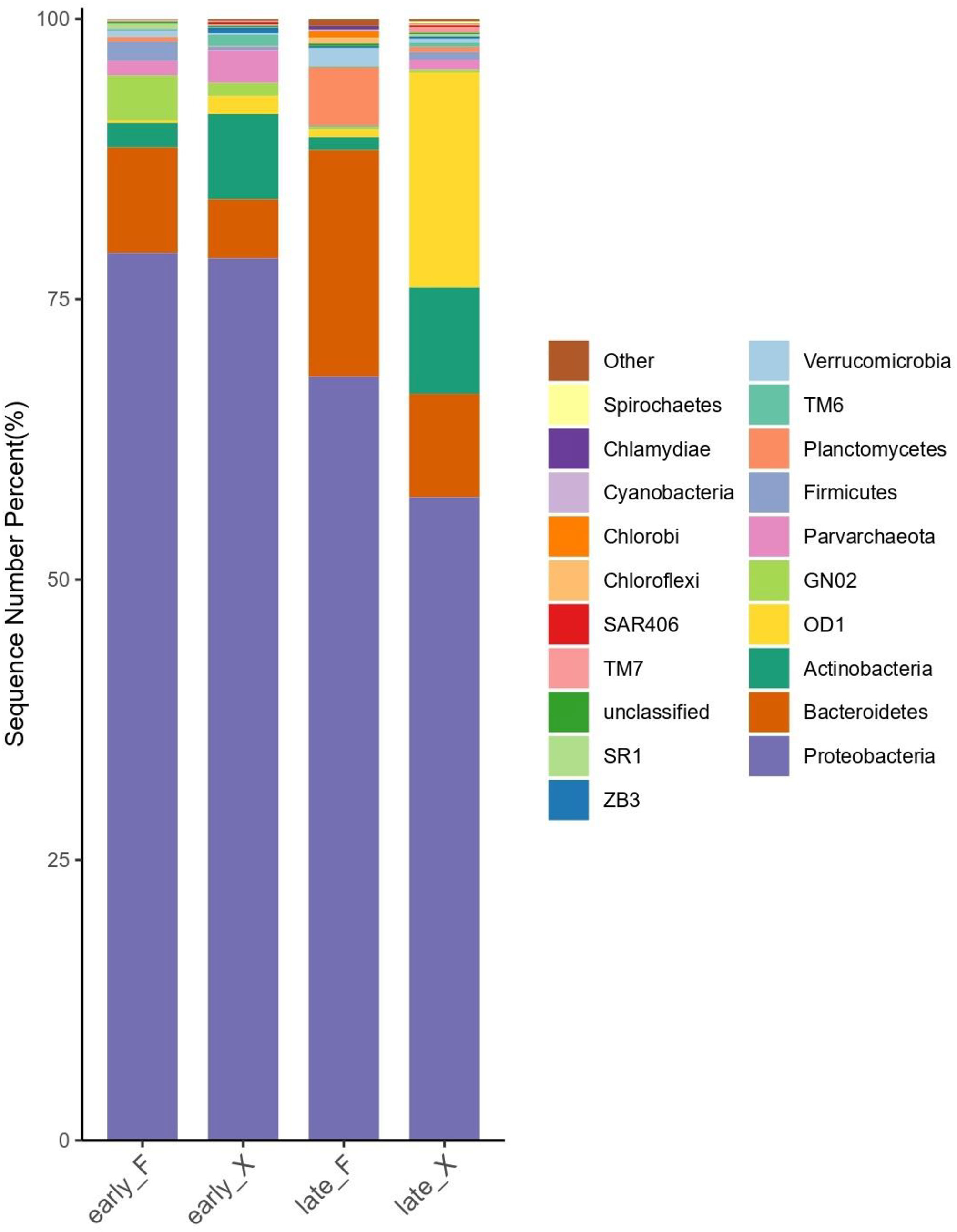
Figure 7 Column chart of relative distribution of microbes in each grouping at the phylum level (Top 20 Species in Relative Abundance).
4 Discussions
4.1 Differences in the DOM dynamics of the two treatment methods
During the degradation of fresh U. prolifera, the concentration of DOC in water increased continuously for the first 20 days. The average net release rate of DOC was 58.7 μmol/(L·d). The concentration of DON increase continuously for all 60 days, with an average DON net release rate of 3.9 µmol/(L·d). The released dissolved organic matter (DOM) experienced an incomplete microbial degradation (Zhang et al., 2021) in the early 20 days, with the proportion of the refractory components in FDOM increasing to 45%. Although the algal fibers visually disappeared after 60 days, the degradation of debris and particles continued. It took about two months for the fresh U. prolifera to experience natural death and decompose until the algal structure visually disappeared. According to Chen et al. (2020), a natural complete degradation of fresh U. prolifera takes about 7.5 months. The results of our experiment suggest a different time frame. The density of fresh U. prolifera in the Chen et al. experiment was the same as in our experiment, so the main reasons for the discrepancy may be the different physiological condition of the U. prolifera used and the difference in experimental temperature (20°C in ours vs 25°C in Chen et al., 2020). U. prolifera died faster at 25°C (Han et al., 2021).
Different from the fresh U. prolifera degradation, large amounts of organic matter was released immediately from the decomposed U. prolifera upon the start of degradation incubation. When pre-decomposed U. prolifera was put into water, the DOC concentration reached highest immediately and then declined rapidly over the next five days, with an average degradation rate of 2708.6 μmol/(L·d). Then the average degradation rate of DOC decreased to120.2 μmol/(L·d) from days 8–20. After 20 days, the DOC remained basically stable (Figure 3), approximately 4% of the initial concentration. Meanwhile, more than 80% of the FDOM components were identified as refractory components, indicating that the degradation process of pre-decomposed U. prolifera was close to completeness. Similar to the DOC dynamics, DON was rapidly degraded with an average degradation rate of 139.2 μmol/(L·d). After five days, the degradation rate approached nearly zero.
The degradation process of fresh U. prolifera over two months was dominated by the continuous and slow release of DOM, and the concentration of DOM in the water column was hardly observed to decrease within two months. For pre-decomposed U. prolifera, the special DOC and DON dynamics was due to the fact that the pre-treatment via microbial complex formula had destroyed the cellular tissue of U. prolifera, large amounts of DOM, including DOC and DON, was rapidly released. More than 80% of the released DOM was degraded within 5 days, and the degradation was basically completed within 20 days. Since the DOC concentration of fresh U. prolifera increased within 2 months in this experiment, the degradation rate could not be estimated accurately. Previous research results also showed that the natural degradation of fresh U. prolifera was a long-term process (Zhang et al., 2019; Chen et al., 2020), which took about 7.5 months, and the average degradation rate of DOC within 4 weeks of rapid degradation was about 194.5 μmol/L·d (Chen et al., 2020). The average degradation rate of DOC in the first 5 days (2708.6 μmol/L·d) of pre-decomposed U. prolifera far exceeded the previous research result on the degradation process of fresh U. prolifera, indicating that the pre-decomposing treatment speeds up the degradation rate of U. prolifera, which was about 14 times faster than the traditional method.
4.2 Potential risks of hypoxia by the pre-decomposing treated U. prolifera
The maximum concentration of DOC (13086.16 µmol/L) produced by pre-decomposed U. prolifera was much higher than that of fresh U. prolifera degradation (2234.48 µmol/L). When the pre-decomposed U. prolifera is disposed into the sea, the rapid oxygen consuming and CO2 producing undoubtedly puts huge ecological pressure on the marine environment. We assume that the degradation of DOM follows the principle that 138 molar of O2 is consumed for 1 molar DOM decomposition (Redfield, 1958):
(CH2)106(NH3)16(H3PO4) + 138O2 = 106CO2 + 16HNO3 + H3PO4 + 122H2O
According to the average degradation rate of DOC and the density of decomposed U. prolifera put in the sea, the estimated oxygen consumption rate per unit mass of decomposed U. prolifera averaged about 1196 mg/(g·d) during the first five days and about 53 mg/(g·d) from the 8th to 20th days.
In summer, the dissolved oxygen (DO) in the bottom water outside Qingdao was about 6.2 mg/L (Wei et al., 2019). In the first five days after pre-decomposed U. prolifera is disposed into the sea, each kilogram would consume dissolved oxygen in 193 m3 seawater every day. During the 8th to 20th days, this consumption could be 8.5 m3 every day. Therefore, we have to carefully choose where U. prolifera is disposed, because concentrated disposal of the pre-decomposed U. prolifera in a constrained area most likely cause regional short-term hypoxia. Therefore, the pre-decomposed U. prolifera should not be disposed in a narrow location with poor hydrological dynamics. Instead, a wide coastal shelf with good water exchange is a recommended disposal location.
4.3 Changes in the bioactivity of organic matter
Based on the content of FDOM and the relative proportion of proteins and humic substances (Figure 4; SI, Figure SI4), it can be seen that the degradation process of fresh U. prolifera first releases bioactive organic matter and then gradually degrades. With the degradation of the pre-decomposed U. prolifera, the released DOM gradually changed from the highly concentrated and reactive matter to the low concentrated and refractory matter. During the degradation of pre-decomposed U. prolifera, the proportion of protein-like components gradually decreased from nearly 100% at the beginning to about 20%, while the proportion of humic-like components gradually increased from 0% to ~80% (SI, Figure SI4).
In the first 12 days, FDOM continued to increase during the degradation process of fresh U. prolifera (Figure 4), and the relative proportion of protein-like components remained at about 85% (SI, Figure SI4). After 12 days, the proportion of protein-like components decreased gradually, and the proportion of humic-like components began to increase. This was consistent with previous research results that FDOM increased to a maximum within 3 weeks and then gradually decreased (Chen et al., 2020). The degradation process of pre-decomposed U. prolifera was completely different. The FDOM value at the beginning of the degradation experiment of pre-decomposed U. prolifera was more than 10 times higher than the highest value of FDOM of fresh U. prolifera. The FDOM of pre-decomposed U. prolifera reached the highest value within 12 hours, and then gradually decreased until 60 days later to slightly lower than that of fresh U. prolifera. The reason for this difference lay in the difference in organic matter release process and microbial activity. Fresh U. prolifera was complete in its cell structure at the initial stage, and it slowly and continuously released DOM into the surrounding water. However, the cell structure of pre-decomposed U. prolifera has been destroyed during the pretreatment process, and once it was thrown into water, almost all organic matter was released rapidly. Different concentrations and compositions of organic matter also affected microbial community succession (Liang et al., 2021; Li et al., 2022a), leading to different trends in FDOM components.
SUVA254 can be used to estimate the percentage of aromatic carbon content of humic acid and it was also an indicator of aromaticity and chemical reactivity for dissolved organic matters (Comstock et al., 2010; He et al., 2011; Abd et al., 2020). (Pifer and Fairey, 2012). SUVA254 of DOM from fresh U. prolifera first decreased and then increased slightly on the 60th day of the degradation experiment (Figure 8), indicating that the time window of bioactive organic matter releasing was much longer. The SUVA254 of pre-decomposed U. prolifera increased with time during the degradation process, indicating that the aromaticity of dissolved organics in water increased and the reactivity decreased during the degradation process. At the same time, the SUVA254 through the degradation process of the two treatment methods was all lower than 3 L/mg·m, indicating that the FDOM components are closely related to the microbial contribution (Boyer et al., 2008).
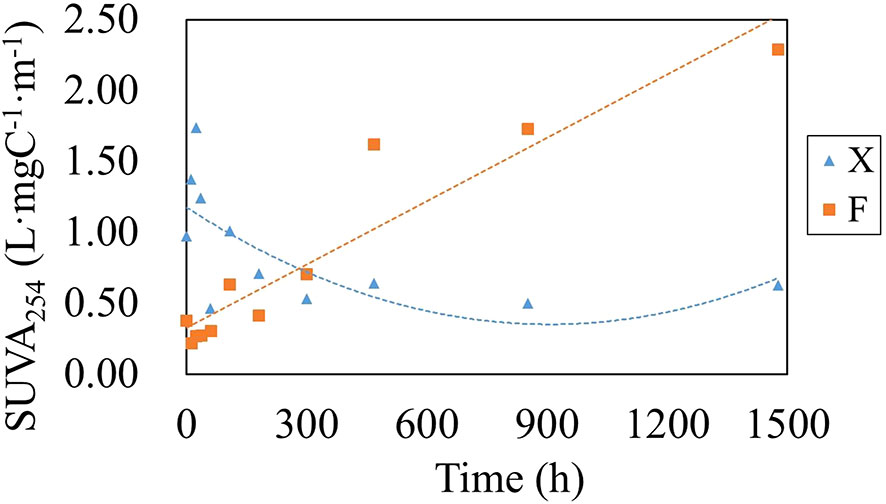
Figure 8 Variation in SUVA254 in water during the degradation of fresh U. prolifera (X) and pre-decomposed U. prolifera (F).
4.4 Distinctive impacts on microbial community structure
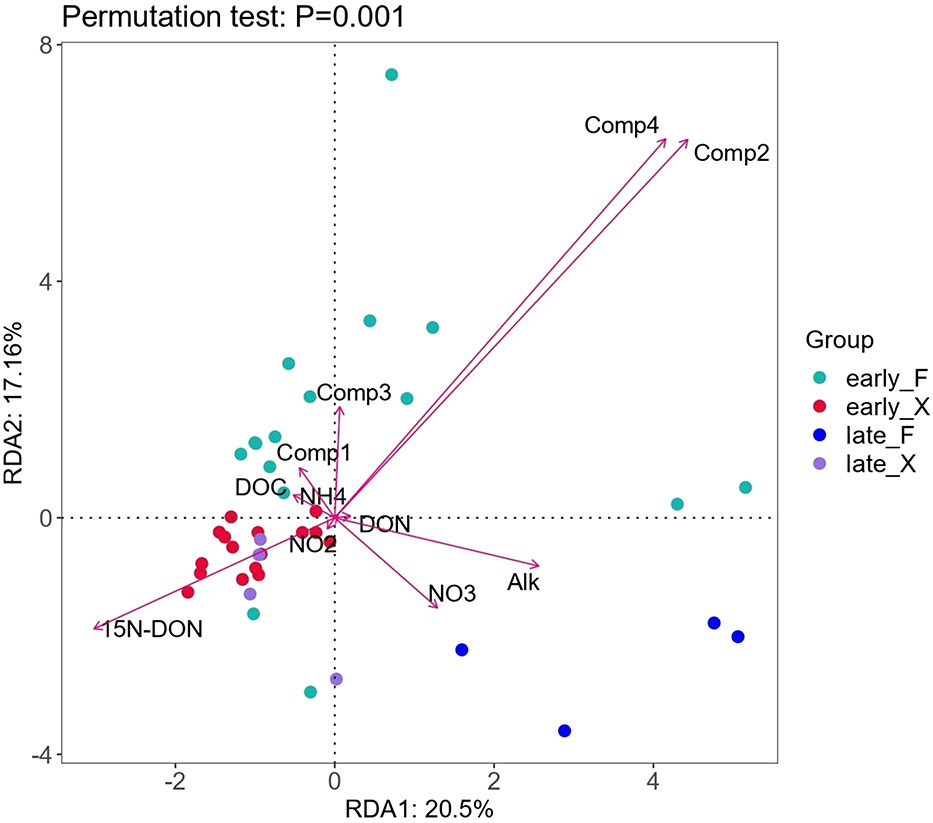
To investigate the relationship between microbial communities and degradation progress, we applied Redundancy Analysis (R language, VEGAN package) to DOC, DON, NOX, NH4+, alkalinity and fluorescent active components (Comp 1 ~ Comp 4) of fresh and pre-decomposed U. prolifera. Comp2 and Comp4 had the longest arrow and DOC and DON had very short arrow (Figure 9), which suggested that DOM compositions, rather than the DOM concentration, had strong relationship with microbial communities. The microbial community structure of stage early_F was closely related to all the four components of FDOM. Due to the drastical changing in FDOM compositions, early_F samples was scattered in the first to third quadrants. The late_F samples was distributed in the fourth quadrant, and the microbial community changed drastically compared with the early stage of degradation. The nitrate concentration were closely related to the microbial community of late_F, and were positively correlated with Comp2 and Comp4 (Figure 9), indicating that the increase of nitrate in the later stage was caused by the degradation of nitrogen containing organic matter (Comp2 and Comp4) under the action of microorganisms. The Bacteroidetes with relatively high abundance in late_F (Figure 7) was a type of bacteria that plays an important role in nitrification (Burut-Archanai et al., 2021; Lei et al., 2021).
During 2-month degradation of the fresh U. prolifera, the variances in the DOM composition characteristics was smaller than that of pre-decomposed U. prolifera, so the microbial communities of early_X and late_X were closely distributed in the second quadrant (Figure 9). After 60 days, the relative abundance of OD1 in seawater degraded with fresh U. prolifera increased significantly (Figure 7). In a broad range of anoxic environments, OD1 bacteria (also known as Parcubacteria) had been identified through community survey analysis (Harris et al., 2004; Rinke et al., 2013; Brown, et al., 2015; Nelson and Stegen, 2015), so this was a kind of anaerobic bacteria (Elshahed et al., 2005). After 60 days of degradation, aerobic nitrifying bacteria in the environment of pre-decomposed U. prolifera increased. On the contrary, anaerobic bacteria increased significantly during the degradation of fresh U. prolifera. This indicated that most of the organic matter from the pre-decomposed U. prolifera had rapidly degraded, and the pressure on the ecological environment had reduced. However, the organic matter from fresh U. prolifera still degraded continuously and consumed oxygen in water.
The dynamic changes in DOM composition was the driving force for the bacterial community (Liang et al., 2021; Li et al., 2022a). Proteobacteria, which widely existed in the environment, were always in relative high abundance in the degradation process of fresh and the pre-decomposed U. prolifera. The difference was that the rapid degradation of pre-decomposed U. prolifera promoted the increase of the relative abundance of ammonia-oxidizing microorganisms and accelerated the cycle of organic matter such as DON; while the degradation of fresh U. prolifera made the relative abundance of anaerobic microorganisms higher for a long period of time. The results of microbial community changes seemed to indicate that the degradation of fresh U. prolifera might lead to a longer period of hypoxia, while the dissolved oxygen concentration and aerobic microorganisms could recover faster in degradation of pre-decomposed U. prolifera.
4.5 U. prolifera degradation affects seawater alkalinity
The alkalinity in the seawater where the fresh U. prolifera and the pre-decomposed U. prolifera were both decreased in the first five days, due to the rapid release of CO2, but then recovered (Figure 5). It was worthy of noticing that with the complete degradation of the pre-decomposed U. prolifera, alkalinity increased about 13% (Figure 5). As shown in Figure 9, alkalinity was positively correlated with Comp 2, Comp 4 and DON, indicating that the increase of alkalinity was related to nitrogen-containing organic matters. It has been notified that DOM molecules may cause an increase in seawater alkalinity (Cai et al., 1998; Ko et al., 2016). Martín Hernández-Ayon et al. (2007) reported that alkalinity increased by 400 and 800 μmol/kg in microalgae culture experiments. Lozovik (2005) pointed out that the contribution of organic acid anions to alkalinity in natural water was close to 10% of its total concentration. The organic alkalinity was determined by back titration (Cai et al., 1998). Our results showed that the organic alkalinity increased by 60 μmol/kg (Figure 10) during the degradation process of the pre-decomposed U. prolifera. Organic alkalinity explained some of the elevated alkalinity, but not all. In any case, the complete degradation of the decomposed U. prolifera would increase the organic alkalinity and alkalinity of seawater, which had a certain significance for anti-acidification.
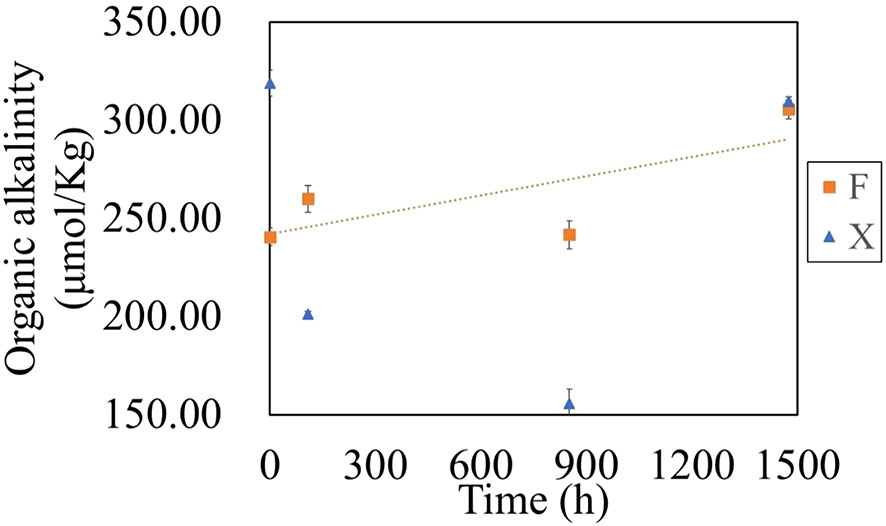
Figure 10 Variation of organic alkalinity during the degradation of fresh U. prolifera (X) and decomposed U. prolifera (F).
5 Conclusion
This paper compared the ecological effects of two different treatment methods to deal with U. prolifera “green tide” in the marine environment. The degradation process of fresh U. prolifera over two months was dominated by the continuous and slow release of DOM, and the concentration of DOM in the water column was hardly observed to decrease within two months. The pre-decomposing treatment with microbial complex formulations destroyed U. prolifera cell tissues and changed its physical state in seawater from floating to fast depositing, and increased the degradation rate by about 14 times. The rapid decomposition of the released bioactive organic matter consumed a substantial amount of dissolved oxygen in local seawater, which has the potential risk of causing local hypoxia and acidification in a short-term. This pre-discomposed-disposal method also significantly altered microbial community structure.
The pre-decomposition treatment of U. prolifera could be a practical and efficient countermeasures to U. prolifera blooming. After the complete degradation of the pre-decomposed U. prolifera, the resulting dissolved organic matter could increase TA to resist acidification. Future studies should focus on the molecular compositions of the organic matter that produces organic alkalinity. Overall, compared with traditional harvest-packing-disposal method, the pre-decomposing-disposal treatment is an efficient and environmental-friendly disposal method to deal with the U. prolifera “green tide”, but it should be used cautiously. Locations with dynamical water changing are recommended as suitable disposal sites, based on the results of this study.
Data availability statement
The 16S rDNA sequencing data presented in the study are deposited in the Figshare repository, accession number 21747266 (https://figshare.com/articles/dataset/16S_RawData_zip/21747266). And other data presented in the study are deposited in the Figshare repository, accession number 22046492 (https://figshare.com/articles/dataset/Data_of_Ulva_prolifera_degradation_zip/22046492).
Author contributions
SD: experiments conducting, data analysis, Writing – original draft. YX: Conceptualization, Methodology, Writing – review and editing. CL: Methodology. YQX: Samples determination. XF: Samples determination. TL: Conceptualization, Resources, Methodology, Project administration, Writing – review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the fund of Zhejiang Provincial “Lingyan” R&D Program (No.2022C03041), NSFC general projects (No. 92051115), the Focus on research and development plan in Shandong province (No. 2019GHZ026) and Asian Cooperation Fund Program (Contract No. 2021245).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1084519/full#supplementary-material
References
Abd Manan T. S. B., Khan T., Mohtar W. H. M. W., Beddu S., Kamal N. L. M., Yavari S., et al. (2020). Dataset on specific UV absorbances (SUVA254) at stretch components of perak river basin. Data Brief 30, 105518. doi: 10.1016/j.dib.2020.105518
Boyer T. H., Singer P. C., Aiken G. R. (2008). Removal of dissolved organic matter by anion exchange: effect of dissolved organic matter properties. Environ. Sci. Technol. 42 (19), 7431–7437. doi: 10.1021/es800714d
Brown C. T., Hug L. A., Thomas B. C., Sharon I., Castelle C. J., Singh A., et al. (2015). Unusual biology across a group comprising more than 15% of domain bacteria. Nature 523 (7559), 208–211. doi: 10.1038/nature14486
Burut-Archanai S., Ubertino D., Chumtong P., Mhuantong W., Powtongsook S., Piyapattanakorn S. (2021). Dynamics of microbial community during nitrification biofilter acclimation with low and high ammonia. Mar. Biotechnol. 23 (4), 671–681. doi: 10.1007/s10126-021-10056-1
Cai W. J., Wang Y., Hodson R. E. (1998). Acid-base properties of dissolved organic matter in the estuarine waters of Georgia, USA. Geochimica Cosmochimica Acta 62 (3), 473–483. doi: 10.1016/S0016-7037(97)00363-3
Charlier R. H., Morand P., Finkl C. W. (2008). How Brittany and Florida coasts cope with “green tides”. Int. J. Environ. Stud. 65 (2), 191–208. doi: 10.1080/00207230701791448
Chen J., Li H., Zhang Z., He C., Shi Q., Jiao N., et al. (2020). DOC dynamics and bacterial community succession during long-term degradation of ulva prolifera and their implications for the legacy effect of “green tides” on refractory DOC pool in seawater. Water Res. 185, 116268. doi: 10.1016/j.watres.2020.116268
Chen Z., Liu M., Yang Y., Bi M., Li M., Liu W. (2022). Environmental and economic impacts of different disposal options for Ulva prolifera green tide in the yellow Sea, China. ACS Sustain. Chem. Eng. 10 (35), 11483–11492. doi: 10.1021/acssuschemeng.2c02638
Comstock S. E., Boyer T. H., Graf K. C., Townsend T. G. (2010). Effect of landfill characteristics on leachate organic matter properties and coagulation treatability. Chemosphere 81 (7), 976–983. doi: 10.1016/j.chemosphere.2010.07.030
Elshahed M. S., Najar F. Z., Aycock M., Qu C., Roe B. A., Krumholz L. R. (2005). Metagenomic analysis of the microbial community at zodletone spring (Oklahoma): Insights into the genome of a member of the novel candidate division OD1. Appl. Environ. Microbiol. 71 (11), 7598–7602. doi: 10.1128/AEM.71.11.7598-7602.2005
Gao G., Liu Y., Li X., Feng Z., Xu Z., Wu H., et al. (2017). Expected CO2-induced ocean acidification modulates copper toxicity in the green tide alga ulva prolifera. Environ. Exp. Bot. 135, 63–72. doi: 10.1016/j.envexpbot.2016.12.007
Gao G., Zhong Z., Zhou X., Xu J. (2016). Changes in morphological plasticity of ulva prolifera under different environmental conditions: a laboratory experiment. Harmful algae 59, 51–58. doi: 10.1016/j.hal.2016.09.004
Glibert P. M. (2017). Eutrophication, harmful algae and biodiversity–challenging paradigms in a world of complex nutrient changes. Mar. pollut. Bull. 124 (2), 591–606. doi: 10.1016/j.marpolbul.2017.04.027
Grasshoff K., Kremling K., Ehrhardt M. (Eds.) (2009). Methods of seawater analysis. (John Wiley & Sons), 198–207.
Han L., Yang G. P., Liu C. Y., Jin Y. M., Liu T. (2021). Emissions of biogenic sulfur compounds and their regulation by nutrients during an ulva prolifera bloom in the yellow Sea. Mar. pollut. Bull. 162, 111885. doi: 10.1016/j.marpolbul.2020.111885
Harris J. K., Kelley S. T., Pace N. R. (2004). New perspective on uncultured bacterial phylogenetic division OP11. Appl. Environ. Microbiol. 70 (2), 845–849. doi: 10.1128/AEM.70.2.845-849.2004
He Y., Ao Y., Yin Y., Yuan A., Che T., Li L., et al. (2019). Comparative transcriptome analysis between floating and attached Ulva prolifera in studying green tides in the yellow Sea. Algal Res. 44, 101712. doi: 10.1016/j.algal.2019.101712
He X. S., Xi B. D., Wei Z. M., Jiang Y. H., Yang Y., An D., et al. (2011). Fluorescence excitation–emission matrix spectroscopy with regional integration analysis for characterizing composition and transformation of dissolved organic matter in landfill leachates. J. hazardous materials 190 (1-3), 293–299. doi: 10.1016/j.jhazmat.2011.03.047
Hu L., Hu C., Ming-Xia H. E. (2017). Remote estimation of biomass of ulva prolifera macroalgae in the yellow Sea. Remote Sens. Environ. 192, 217–227. doi: 10.1016/j.rse.2017.01.037
Ko Y. H., Lee K., Eom K. H., Han I. S. (2016). Organic alkalinity produced by phytoplankton and its effect on the computation of ocean carbon parameters. Limnology Oceanography 61 (4), 1462–1471. doi: 10.1002/lno.10309
Lei L., Gu J., Wang X., Song Z., Yu J., Wang J., et al. (2021). Effects of phosphogypsum and medical stone on nitrogen transformation, nitrogen functional genes, and bacterial community during aerobic composting. Sci. Total Environ. 753, 141746. doi: 10.1016/j.scitotenv.2020.141746
Le Luherne E., Réveillac E., Ponsero A., Sturbois A., Ballu S., Perdriau M., et al. (2016). Fish community responses to green tides in shallow estuarine and coastal areas. Estuarine Coast. Shelf Sci. 175, 79–92. doi: 10.1016/j.ecss.2016.03.031
Li H., Feng X., Xiong T., He C., Wu W., Shi Q., et al. (2022a). Green tides significantly alter the molecular composition and properties of coastal DOC and perform dissolved carbon sequestration. Environ. Sci. Technol 57 (1), 770–779. doi: 10.1021/acs.est.2c05684
Li B. H., Liu C. Y., Deng X., Wang K. K., Han L., Huang Y. H., et al. (2021). Responses of the marine carbonate system to a green tide: A case study of an ulva prolifera bloom in qingdao coastal waters. Harmful Algae 110, 102133. doi: 10.1016/j.hal.2021.102133
Li S., Xu Z., Wang C. (2022b). Public's preference for the treatment of ulva prolifera blooms: A choice experiment study in China. Algal Res. 66, 102776. doi: 10.1016/j.algal.2022.102776
Liang J., Liu J., Zhan Y., Zhou S., Xue C. X., Sun C., et al. (2021). Succession of marine bacteria in response to ulva prolifera-derived dissolved organic matter. Environ. Int. 155, 106687. doi: 10.1016/j.envint.2021.106687
Liu J., Xia J., Zhuang M., Zhang J., Yu K., Zhao S., et al. (2021). Controlling the source of green tides in the yellow Sea: NaClO treatment of ulva attached on pyropia aquaculture rafts. Aquaculture 535, 736378. doi: 10.1016/j.aquaculture.2021.736378
Lozovik P. A. (2005). Contribution of organic acid anions to the alkalinity of natural humic water. J. Analytical Chem. 60 (11), 1000–1004. doi: 10.1007/s10809-005-0226-3
Martín Hernández-Ayon J., Zirino A., Dickson A. G., Camiro-Vargas T., Valenzuela-Espinoza E. (2007). Estimating the contribution of organic bases from microalgae to the titration alkalinity in coastal seawaters. Limnology Oceanography: Methods 5 (7), 225–232. doi: 10.4319/lom.2007.5.225
Miao X., Xiao J., Pang M., Zhang X., Wang Z., Miao J., et al. (2018). Effect of the large-scale green tide on the species succession of green macroalgal micro-propagules in the coastal waters of qingdao, China. Mar. pollut. Bull. 126, 549–556. doi: 10.1016/j.marpolbul.2017.09.060
Nelson W. C., Stegen J. C. (2015). The reduced genomes of parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front. Microbiol. 6, 713. doi: 10.3389/fmicb.2015.00713
Pifer A. D., Fairey J. L. (2012). Improving on SUVA254 using fluorescence-PARAFAC analysis and asymmetric flow-field flow fractionation for assessing disinfection byproduct formation and control. Water Res. 46 (9), 2927–2936. doi: 10.1016/j.watres.2012.03.002
Pucher M., Wünsch U., Weigelhofer G., Murphy K., Hein T., Graeber D. (2019). staRdom: Versatile software for analyzing spectroscopic data of dissolved organic matter in r. Water 11 (11), 2366. doi: 10.3390/w11112366
Qi L., Hu C., Xing Q., Shang S. (2016). Long-term trend of ulva prolifera blooms in the western yellow Sea. Harmful Algae 58, 35–44. doi: 10.1016/j.hal.2016.07.004
Redfield A. C. (1958). The biological control of chemical factors in the environment. Am. scientist 46 (3), 230A–2221.
Rinke C., Schwientek P., Sczyrba A., Ivanova N. N., Anderson I. J., Cheng J. F., et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499 (7459), 431–437. doi: 10.1038/nature12352
Schramm W. (1996). Marine benthic vegetation: Recent changes and the effects of eutrophication. (Springer Science & Business Media), 127, 7–43.
Smetacek V., Zingone A. (2013). Green and golden seaweed tides on the rise. Nature 504 (7478), 84–88. doi: 10.1038/nature12860
Stedmon C. A., Markager S. (2005). Resolving the variability in dissolved organic matter fluorescence in a temperate estuary and its catchment using PARAFAC analysis. Limnology oceanography 50 (2), 686–697. doi: 10.4319/lo.2005.50.2.0686
Sun Y., Yao L., Liu J., Tong Y., Xia J., Zhao X., et al. (2022). Prevention strategies for green tides at source in the southern yellow Sea. Mar. pollut. Bull. 178, 113646. doi: 10.1016/j.marpolbul.2022.113646
Wei Q., Wang B., Yao Q., Yu Z., Fu M., Sun J., et al. (2019). Physical-biogeochemical interactions and potential effects on phytoplankton and ulva prolifera in the coastal waters off qingdao (Yellow Sea, China). Acta Oceanologica Sin. 38 (2), 11–23. doi: 10.1007/s13131-019-1344-3
Wu H., Gao G., Zhong Z., Li X., Xu J. (2018). Physiological acclimation of the green tidal alga ulva prolifera to a fast-changing environment. Mar. Environ. Res. 137, 1–7. doi: 10.1016/j.marenvres.2018.02.018
Yamashita Y., Jaffé R., Maie N., Tanoue E. (2008). Assessing the dynamics of dissolved organic matter (DOM) in coastal environments by excitation emission matrix fluorescence and parallel factor analysis (EEM-PARAFAC). Limnology oceanography 53 (5), 1900–1908. doi: 10.4319/lo.2008.53.5.1900
Yan Z., Yang N., Liang Z., Yan M., Zhong X., Zhang Y., et al. (2021). Active dissolved organic nitrogen cycling hidden in large river and environmental implications. Sci. Total Environ. 795, 148882. doi: 10.1016/j.scitotenv.2021.148882
Ye N. H., Zhang X. W., Mao Y. Z., Liang C. W., Xu D., Zou J., et al. (2011). ‘Green tides’ are overwhelming the coastline of our blue planet: Taking the world’s largest example. Ecol. Res. 26 (3), 477–485. doi: 10.1007/s11284-011-0821-8
Yuan Y., Xu X., Jing C., Zou P., Zhang C., Li Y. (2018). Microwave assisted hydrothermal extraction of polysaccharides from Ulva prolifera: Functional properties and bioactivities. Carbohydr. polymers 181, 902–910. doi: 10.1016/j.carbpol.2017.11.061
Zhang Y., He P., Li H., Li G., Liu J., Jiao F., et al. (2019). Ulva prolifera green-tide outbreaks and their environmental impact in the yellow Sea, China. Natl. Sci. Rev 6 (4), 825–838. doi: 10.1093/nsr/nwz026
Zhang P., Xin Y., Zhong X., Yan Z., Jin Y., Yan M., et al. (2021). Integrated effects of ulva prolifera bloom and decay on nutrients inventory and cycling in marginal sea of China. Chemosphere 264, 128389. doi: 10.1016/j.chemosphere.2020.128389
Zhang J., Zhao P., Huo Y., Yu K., He P. (2017). The fast expansion of pyropia aquaculture in “Sansha” regions should be mainly responsible for the ulva blooms in yellow Sea. Estuarine Coast. Shelf Sci. 189, 58–65. doi: 10.1016/j.ecss.2017.03.011
Keywords: Ulva prolifera, green tide, pre-decomposing treatment, hypoxia, acidification
Citation: Dong S, Xin Y, Liu C, Xiao Y, Feng X and Liu T (2023) Two treatment methods on Ulva prolifera bloom result in distinctively different ecological effects in coastal environment. Front. Mar. Sci. 10:1084519. doi: 10.3389/fmars.2023.1084519
Received: 09 December 2022; Accepted: 02 February 2023;
Published: 09 March 2023.
Edited by:
Christophe Brunet, Anton Dohrn Zoological Station Naples, ItalyReviewed by:
Yunyan Deng, Institute of Oceanology (CAS), ChinaMeilin Wu, South China Sea Institute of Oceanology (CAS), China
Andrzej Stanisław Rybak, Adam Mickiewicz University, Poland
Copyright © 2023 Dong, Xin, Liu, Xiao, Feng and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Xin, eGlueXUzMTJAb3VjLmVkdS5jbg==; Tao Liu, bGl1dGFvMjAyMUB4bXUuZWR1LmNu
 Shuhang Dong1
Shuhang Dong1 Yu Xin
Yu Xin Chunying Liu
Chunying Liu